Defining the pulmonary cell types infected by SARS‐CoV‐2 and finding ways to prevent subsequent tissue damage are key goals for controlling COVID‐19. Recent work establishing a human lung organoid‐derived air–liquid interface model permissive to SARS‐CoV‐2 infection identifies alveolar type II cells as the primary cell type infected, reports an infection‐induced interferon response and demonstrates the effectiveness of interferon lambda 1 treatment in dampening lung infection.
Subject Categories: Methods & Resources; Microbiology, Virology & Host Pathogen Interaction; Respiratory System
Recent work establishing a human bronchioalveolar culture model that allows in vitro characterization of coronavirus infection and treatment options.

Coronavirus disease‐2019 (COVID‐19) has triggered a global pandemic and is caused by the severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2). This novel coronavirus causes a spectrum of respiratory illnesses ranging from mild upper airway disease to life‐threatening acute respiratory distress syndrome (ARDS), placing the lung as a primary organ to study in the worldwide effort to control this deadly respiratory virus. Within the lung, epithelial cells are the primary cell types that express ACE2 and TMPRSS2, a receptor and a protease essential for viral entry, respectively (Wang et al, 2020). However, how much of their expression level dictates cellular susceptibility to infection remains unclear. At the molecular level, it has been shown that SARS‐CoV‐2 infection induces interferon stimulated gene (ISG) expression. Addressing whether this upregulation occurs in infected cells or bystander cells and defining the cascade of molecular changes that follow is critical for devising prophylactic strategies to prevent severe infection.
Animal models such as transgenic mice expressing human ACE2 and golden hamsters have been used to study COVID‐19, but none was able to recapitulate severe lung infection. This elevated the need to directly use human lung cells, especially non‐immortalized cell lines, to simulate in vivo infection. Several human lung cell culture platforms have been developed. A workhorse system is air–liquid interface (ALI) culture where airway progenitors such as basal cells efficiently differentiate into club, goblet and ciliated cells, recapitulating an in vivo airway organization (Karp et al, 2002). Primary epithelial cells can also be cultured as three‐dimensional (3D) organoids, forming bronchospheres or alveolospheres, depending on the source of the epithelial cells (Barkauskas et al, 2017). Co‐culture with mesenchymal cells further improves the authenticity of the microenvironment and differentiation efficiency. The drawbacks of these primary cell‐based systems are the challenge to obtain human lung tissue and wide donor‐to‐donor variation, in some cases due to underlying disease state. Although 3D organoids more closely recapitulate in vivo signalling, the apical side is oriented towards the centre, thus making it difficult to be used for infection studies. Lung cells derived from pluripotent stem cells (PSCs) including embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs), have allowed investigators to mass produce, store and genetically manipulate human lung cells (Leibel et al, 2020). The primary challenge remains the immaturity of the lung cells generated.
In a recent study, Lamers et al (2021) adapted a 3D lung organoid culture system based on multi‐potent human distal tip cells from fetal lungs (Nikolić et al, 2017). These cells were expanded as submerged organoids and then changed to air–liquid interface to differentiate, further stimulated by niche signals from mesenchymal cells in the bottom compartment. Markers for alveolar type 1 and type 2 cells as well as for airway cells increased, demonstrating formation of mixed bronchioalveolar cell types. The differentiated cells also expressed ACE2 and TMPRSS2, setting the stage for infection (Fig 1).
Figure 1. A bronchioalveolar lung culture platform for the study of SARS‐CoV‐2 lung infection.
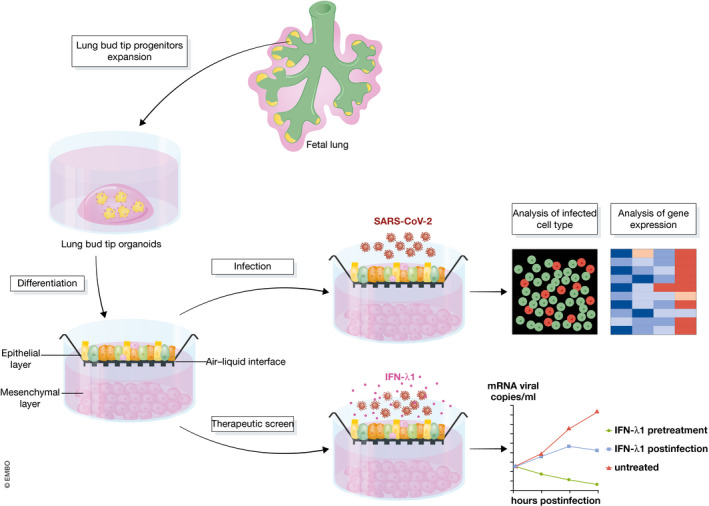
This diagram illustrates the bronchioalveolar culture system established in Lamers et al (2021). Cells in this culture, in particular alveolar type cells, can be efficiently infected by SARS‐CoV‐2. Infection led to an increase in ISG expression. Treatment with IFN‐λ1 either before or after infection abrogated or reduced infection rate, respectively.
Infection by SARS‐CoV‐2 was achieved at a low multiplicity of infection (MOI) of 0.01 and 0.1, suggesting that these cells are readily infected. Most of the infected cells were alveolar type 2 cells, while airway epithelial cells such as pulmonary neuroendocrine cells or lung mesenchymal cells were not infected. The demonstrated susceptibility of alveolar cells to SARS‐CoV‐2 infection is notable, as COVID‐19‐associated ARDS is a pathological condition of the alveolar region, where vital gas exchange occurs.
Bulk RNA‐seq showed an increase of ISG signature following infection. Interestingly, staining for phosphorylated STAT1, a downstream readout of interferon activation, showed an increase of signal in the bystander cells, but not in SARS‐CoV‐2 infected cells. This result suggests that attention should be paid to bystander cells to address downstream interferon response.
To test whether the bronchioalveolar system can be used for SARS‐CoV‐2 drug screens, Lamers et al (2021) investigated the effect of Interferon lambda 1 (IFN‐λ1) in modulating infectability. Treatment of cells 2 h prior to infection completely aborted viral replication. Treatment of cells 24 h after infection reduced infectious virus by ~ 5 logs and virus RNA copies by ~ 3 logs, demonstrating the antiviral activity of IFN‐λ1 and the value of the bronchioalveolar system in drug screens.
Other human lung cell culture platforms have recently been developed to study SARS‐CoV‐2 infection. For example, primary airway cells in air–liquid interface culture were infected with a high viral load and confirmed infection of ciliated and goblet cells (Hao et al, 2020). Cigarette smoke exposure led to goblet cell metaplasia and a higher infection rate (Purkayastha et al, 2020). Primary lung‐derived alveolospheres were infected and mirrored features of COVID‐19 lungs including increased interferon, pro‐inflammatory, and apoptotic pathway genes and decreased surfactants (Katsura et al, 2020; Youk et al, 2020). iPSC‐derived alveolar type 2 cells adapted to air–liquid interface culture were infected and showed an inflammatory phenotype with upregulation of NFkB signalling and loss of mature alveolar program (Huang et al, 2020). The iPSC‐derived alveolar type 2‐like cell cultures also offered an ideal platform for high‐throughput chemical screening (Han et al, 2020).
The human lung cell platforms, including the one established here, open opportunities for in‐depth studies of SARS‐CoV‐2 infection of the lung. For example, having airway luminal cells such as club, ciliated and goblet cells in the same mixed culture with alveolar type 1 and type 2 cells will enable testing of relative infectability of lung epithelial cells, thus allowing assessment of the kinetics of SARS‐CoV‐2 lung infection once the virus is inhaled through the airway. Analysis of bulk RNA‐seq data across multiple time points will chart the time course of molecular impact. Application of single‐cell RNA‐seq to the system will tease out the specific contribution of infected versus bystander cells to inflammatory and antiviral signals. Building on cultures such as the bronchioalveolar system used here, addition of immune cells and endothelial cells will allow testing of how the 3D alveolar and airway units respond to SARS‐CoV‐2 infection. We are hopeful that with vaccinations, COVID‐19 will be better controlled. Once this is achieved, screens for therapeutics will be re‐directed to preventing and reversing lung damage caused by infection. Towards this goal, lung progenitor cultures such as the one established in this study (Lamers et al, 2021) will remain a pivotal tool in the search to repair chronic respiratory dysfunction following infection by not just SARS‐CoV‐2, but also other respiratory pathogens.
The EMBO Journal (2021) 40: e107651.
See also: MM Lamers et al (March 2021)
References
- Barkauskas CE, Chung MI, Fioret B, Gao X, Katsura H, Hogan BL (2017) Lung organoids: current uses and future promise. Development 144: 986–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Duan X, Yang L, Nilsson‐Payant BE, Wang P, Duan F, Tang X, Yaron TM, Zhang T, Uhl S et al (2020) Identification of SARS‐CoV‐2 inhibitors using lung and colonic organoids. Nature 589: 270–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao S, Ning K, Kuz CA, Vorhies K, Yan Z, Qiu J (2020) Long‐term modeling of SARS‐CoV‐2 infection of in vitro cultured polarized human airway epithelium. MBio 11: e02852‐20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Hume AJ, Abo KM, Werder RB, Villacorta‐Martin C, Alysandratos KD, Beermann ML, Simone‐Roach C, Lindstrom‐Vautrin J, Olejnik J et al (2020) SARS‐CoV‐2 infection of pluripotent stem cell‐derived human lung alveolar type 2 cells elicits a rapid epithelial‐intrinsic inflammatory response. Cell Stem Cell 27: 962–973.e967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp PH, Moninger TO, Weber SP, Nesselhauf TS, Launspach JL, Zabner J, Welsh MJ (2002) An in vitro model of differentiated human airway epithelia. Methods for establishing primary cultures. Methods Mol Biol 188: 115–137 [DOI] [PubMed] [Google Scholar]
- Katsura H, Sontake V, Tata A, Kobayashi Y, Edwards CE, Heaton BE, Konkimalla A, Asakura T, Mikami Y, Fritch EJ et al (2020) Human lung stem cell‐based alveolospheres provide insights into SARS‐CoV‐2‐mediated interferon responses and pneumocyte dysfunction. Cell Stem Cell 27: 890–904.e898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamers MM, van der Vaart J, Knoops K, Riesebosch S, Breugem TI, Mykytyn AZ, Beumer J, Schipper D, Bezstarosti K, Koopman CD et al (2021) An organoid‐derived bronchioalveolar model for SARS‐CoV‐2 infection of human alveolar‐type II‐like cells. EMBO J 40: e105912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibel SL, McVicar RN, Winquist AM, Niles WD, Snyder EY (2020) Generation of complete multi‐cell type lung organoids from human embryonic and patient‐specific induced pluripotent stem cells for infectious disease modeling and therapeutics validation. Curr Protoc Stem Cell Biol 54: e118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolić MZ, Caritg O, Jeng Q, Johnson JA, Sun D, Howell KJ, Brady JL, Laresgoiti U, Allen G, Butler R et al (2017) Human embryonic lung epithelial tips are multipotent progenitors that can be expanded in vitro as long‐term self‐renewing organoids. Elife 6: e26575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purkayastha A, Sen C, Garcia G, Langerman J, Shia DW, Meneses LK, Vijayaraj P, Durra A, Koloff CR, Freund DR et al (2020) Direct exposure to SARS‐CoV‐2 and cigarette smoke increases infection severity and alters the stem cell‐derived airway repair response. Cell Stem Cell 27: 869–875.e864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A, Chiou J, Poirion OB, Buchanan J, Valdez MJ, Verheyden JM, Hou X, Kudtarkar P, Narendra S, Newsome JM et al (2020) Single cell multiomic profiling of human lung reveals cell type‐specific and age‐dynamic control of SARS‐CoV2 host genes. Elife 9: e62522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youk J, Kim T, Evans KV, Jeong YI, Hur Y, Hong SP, Kim JH, Yi K, Kim SY, Na KJ et al (2020) Three‐dimensional human alveolar stem cell culture models reveal infection response to SARS‐CoV‐2. Cell Stem Cell 27: 905–919.e910 [DOI] [PMC free article] [PubMed] [Google Scholar]


