Abstract
The intraflagellar transport (IFT) machinery consists of the anterograde motor kinesin‐II, the retrograde motor IFT dynein, and the IFT‐A and ‐B complexes. However, the interaction among IFT motors and IFT complexes during IFT remains elusive. Here, we show that the IFT‐B protein IFT54 interacts with both kinesin‐II and IFT dynein and regulates anterograde IFT. Deletion of residues 342–356 of Chlamydomonas IFT54 resulted in diminished anterograde traffic of IFT and accumulation of IFT motors and complexes in the proximal region of cilia. IFT54 directly interacted with kinesin‐II and this interaction was strengthened for the IFT54Δ342–356 mutant in vitro and in vivo. The deletion of residues 261–275 of IFT54 reduced ciliary entry and anterograde traffic of IFT dynein with accumulation of IFT complexes near the ciliary tip. IFT54 directly interacted with IFT dynein subunit D1bLIC, and deletion of residues 261–275 reduced this interaction. The interactions between IFT54 and the IFT motors were also observed in mammalian cells. Our data indicate a central role for IFT54 in binding the IFT motors during anterograde IFT.
Keywords: cilia, flagella, IFT dynein, IFT54, intraflagellar transport, kinesin‐II
Subject Categories: Cell Adhesion, Polarity & Cytoskeleton; Membrane & Intracellular Transport
A single subunit of the anterograde intraflagellar transport (IFT) complex IFT‐B bridges both anterograde and retrograde IFT motor proteins to regulate ciliary protein trafficking and ciliogenesis in Chlamydomonas.
Introduction
Cilia are microtubule‐based organelles that function cellular motility and signaling (Roy, 2009; Bangs & Anderson, 2017; Reiter & Leroux, 2017). The assembly, maintenance and disassembly of cilia require intraflagellar transport (IFT), the bi‐directional movement of large protein complexes between the ciliary membrane and the axonemal microtubules (Rosenbaum & Witman, 2002; Scholey, 2003; Liang et al, 2016; Taschner & Lorentzen, 2016). IFT was first observed in Chlamydomonas, a unicellular green alga using video‐enhanced differential contrast microscopy (DIC) (Kozminski et al, 1993). Subsequently, most of the components of the IFT machinery have been identified (Morris & Scholey, 1997; Piperno & Mead, 1997; Cole et al, 1998; Pazour et al, 1999; Porter et al, 1999; Signor et al, 1999; Taschner & Lorentzen, 2016). In Chlamydomonas and mammalian cells, anterograde IFT employs heterotrimeric kinesin‐2 (kinesin‐II) while in Caenorhabditis elegans a homodimeric kinesin‐2 is also involved (Kozminski et al, 1995; Scholey, 2013; Engelke et al, 2019); retrograde IFT is powered by cytoplasmic dynein 2/1b (also termed IFT dynein) (Pazour et al, 1999; Porter et al, 1999; Signor et al, 1999; Lechtreck, 2019). The IFT protein complex that is associated with the motors—the IFT complex consists of 22 proteins, organized into IFT‐A and IFT‐B subcomplexes (Cole et al, 1998; Ou et al, 2005; Behal et al, 2012; Taschner & Lorentzen, 2016). IFT‐B can be further separated into the IFT‐B1 core complex consisting of 10 proteins and the IFT‐B2 peripheral complex encompassing six proteins (Lucker et al, 2010; Taschner et al, 2014; Katoh et al, 2016; Taschner et al, 2016).
During anterograde transport, kinesin‐II carries the IFT complex and inactive dynein to the ciliary tip. During remodeling at the ciliary tip, inactive dynein becomes active and mediates retrograde IFT to return IFT complexes to the cell body while kinesin‐II returns either by diffusion in Chlamydomonas or by association with the IFT trains in worm and mammals (Signor et al, 1999; Pedersen et al, 2006; Engel et al, 2012; Prevo et al, 2015; Chien et al, 2017; Jordan et al, 2018). Because mutations in IFT‐A proteins often induce accumulation of IFT‐A and IFT‐B complexes at the ciliary tip (Piperno et al, 1998; Iomini et al, 2001; Iomini et al, 2009; Liem et al, 2012; Zhu et al, 2017a; Picariello et al, 2019), IFT dynein is thought to bind to IFT‐A during retrograde transport, carrying the IFT‐B complex along as a cargo.
How does the IFT complex interact with the IFT motors during anterograde IFT? Analysis of anterograde IFT trains by cryo‐electron tomography (Cryo‐ET) indicates that kinesin‐II interacts with IFT‐B (Jordan et al, 2018), which is supported by genetic data where mutations in IFT‐B often abolish anterograde IFT (Pedersen & Rosenbaum, 2008) and biochemical studies that show possible associations of kinesin‐II with IFT‐B (Baker et al, 2003; Krock & Perkins, 2008; Liang et al, 2018). Recently, in vitro binding studies showed that kinesin‐II strongly interacts with IFT‐B1 and IFT‐B2 connecting tetramer consisting of IFT52, IFT88, IFT38, and IFT57 and may also bind to other IFT‐B2 peripheral subunits including IFT20, IFT54, IFT80, and IFT172 (Funabashi et al, 2018). However, which subunit of this peripheral IFT‐B2 module directly interacts with kinesin‐II and the physiological consequence for IFT and ciliary assembly entailed by an alteration of the kinesin‐II/IFT‐B2 interaction are unknown.
For the interaction of IFT dynein with IFT complex during anterograde IFT, Cryo‐ET of anterograde IFT trains revealed that IFT‐A and IFT dynein bind independently of each other to IFT‐B (Jordan et al, 2018). The large IFT dynein complex bridges above several IFT‐B complexes indicating that its transport involves heterogenous interactions with the IFT‐B complex (Jordan et al, 2018; Toropova et al, 2019). One such interaction likely involves the IFT‐B subunit IFT172, which co‐immunoprecipitated specifically with IFT dynein (Pedersen et al, 2006). Furthermore, the amount of IFT dynein in cilia was decreased at restrictive temperature in a temperature‐sensitive mutant ift172‐fla11 (Pedersen et al, 2006; Williamson et al, 2012). However, it is unknown whether IFT172 directly interacts with IFT dynein. Thus, how IFT‐B interacts with IFT dynein remains elusive. Also, it remains unknown which parts or subunits of the IFT dynein complex bind to IFT‐B.
In this work, we show that Chlamydomonas IFT54 directly binds to IFT dynein via its D1bLIC subunit; further, IFT54 also directly interacts with kinesin‐II. Targeted disruption of these interactions impairs anterograde IFT and ciliogenesis, indicating a central role of IFT54 in organizing the IFT motors during anterograde IFT.
Results
Identification of two conserved regions of IFT54 that are critical for ciliogenesis
Sequencing of genomic DNA and cDNA showed that Chlamydomonas IFT54 is a protein of 509 residues (Appendix Fig S1). It has two well‐established domains: a tubulin‐binding calponin homology domain (CH domain) at the N terminus and a coiled‐coil domain at the C terminus (Taschner et al, 2016) (Fig 1A). However, deletion of the N‐terminal sequences up to 250 residues does not affect ciliogenesis (Zhu et al, 2017b), indicating that the CH domain is not essential for ciliogenesis. In contrast, the coiled‐coil domain binds and stabilizes IFT20 and is required for ciliogenesis (Kunitomo & Iino, 2008; Taschner et al, 2016; Zhu et al, 2017b). The expression of the IFT54 coiled‐coil domain in an ift54 null mutant rescues the stability of IFT20 but not ciliogenesis (Zhu et al, 2017b), suggesting that additional portions of IFT54 located between the N‐terminal 250 residues and the C‐terminal coiled‐coil domain are critical for ciliogenesis independent of stabilizing IFT20. Within this region, an alignment of IFT54 sequences from various species revealed three highly conserved short sequences corresponding to residues 261–275, 309–330, and 342–356 of Chlamydomonas IFT54 (Fig 1A).
Figure 1. Two previously undefined and conserved regions of IFT54 are required for proper ciliogenesis.
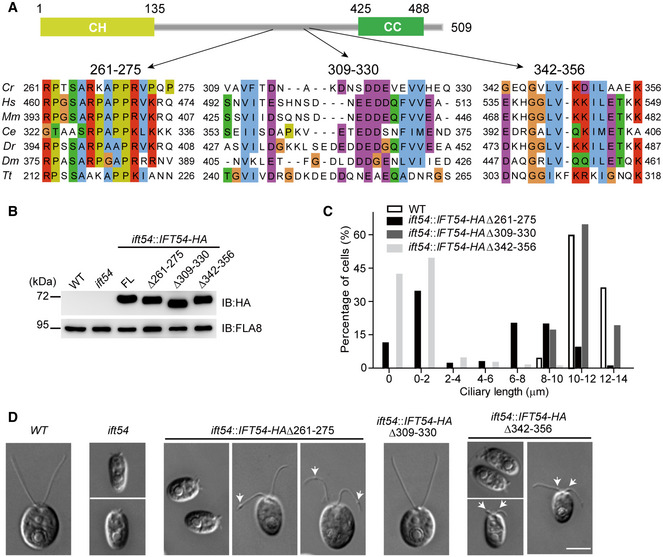
- IFT54 has three previously undefined and conserved regions. Schematic representation of the primary structure of Chlamydomonas IFT54 is shown on the top. Three conserved regions are shown on the bottom. Abbreviations for the species and protein entries: Cr, Chlamydomonas reinhardtii (Cre11.g467739); Hs, Homo sapiens (NP_056465.2); Mm, Mus musculus (NP_082994.1); Ce, Caenorhabditis (NP_508149.1); Dr, Danio rerio (ABW96885); Dm, Drosophila melanogaster (NP_650353); Tt, Tetrahymena thermophila (XP_001026831.1).
- Immunoblot analysis of whole cell samples of the HA‐tagged full‐length (FL) IFT54, and its deletion mutants as indicated, each expressed in the ift54 null mutant; wild‐type (WT) and ift54 mutant cells were used as control. Anti‐HA was use to visualize the transgenic proteins, and antibodies to FLA8/KIF3B were used as a loading control.
- Histogram showing ciliary length distribution in populations of ift54 mutant cells transformed with the deletion mutant constructs as indicated. Wild‐type (WT) cells were used as a control.
- Representative DIC images of wild‐type (WT), ift54, and ift54 cells transformed with the deletion constructs as indicated. Arrows indicate cilia bulges. Scale bar, 5 µm.
To determine whether these regions are crucial for the function of IFT54 in ciliogenesis, we expressed the HA‐tagged deletion constructs IFT54Δ261–275, IFT54Δ309–330, and IFT54Δ342–356, respectively, in the ift54 null mutant; HA‐tagged full‐length (FL) IFT54 was used as a control (Fig 1B). Expression of full‐length and the IFT54Δ309–330 deletion construct restored normal ciliogenesis. In contrast, the IFT54Δ261–275 and IFT54Δ342–356, when expressed in ift54 at similar levels as the wild‐type proteins, resulted in cells with shorter than normal cilia or lacking cilia entirely (Fig 1C and D). Closer examination of ciliary morphology of these two mutants by DIC revealed the formation of ciliary bulges along the ciliary length: the IFT54Δ261–275 mutant developed bulges, mostly near the ciliary tips, in 73.4% of the cells analyzed while 86.4% of the cells of the IFT54Δ342–356 mutant showed bulges in the proximal region of the cilium near its base (Fig 1D). Taken together, we identified two conserved regions of IFT54 that are critical for ciliogenesis but likely affect ciliogenesis by distinct mechanisms as evident by the different positions of the ciliary bulges.
Accumulation of IFT motors and IFT complex at ciliary bulges in IFT54Δ261–275 and IFT54Δ342–356 mutants
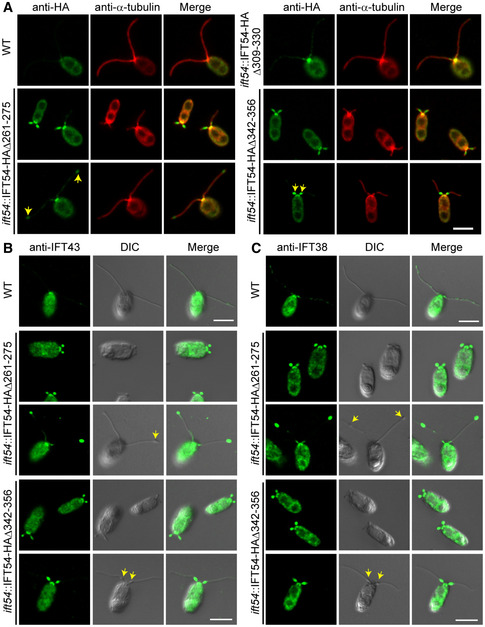
Ciliary bulges often result from an abnormal accumulation of IFT proteins between the ciliary membrane and the axoneme (Piperno et al, 1998; Tam et al, 2007; Iomini et al, 2009; Meng & Pan, 2016; Zhu et al, 2017a; Picariello et al, 2019). We used immunostaining to examine whether the mutated IFT54 proteins were enriched abnormally in cilia. As expected, IFT54Δ309–330‐HA which fully restores ciliogenesis in ift54 (Fig 1C) showed a distribution of IFT proteins typical for wild‐type cells with enrichment at the ciliary base and spotted presence along the cilium (Fig 2A). Both IFT54Δ261–275‐HA and IFT54Δ342–356‐HA, which were present in apparently normal amounts at the ciliary base, were highly enriched in stumpy cilia (Fig 2A). In mutant cells with longer cilia, IFT54Δ261–275‐HA was enriched near the ciliary tip whereas IFT54Δ342–356‐HA was enriched in the proximal part of cilia, indicating that both mutant proteins are present in the ciliary bulges.
Figure 2. IFT‐A and IFT‐B proteins accumulate at ciliary tip in the IFT54Δ261–275 mutant while at proximal end of cilia in IFT54Δ342–356 mutant.
-
AImmunostaining of IFT54 mutated proteins in various deletion mutants as indicated; wild‐type (WT) cells were used as control. Specimens were stained with anti‐HA and anti‐α‐tubulin antibodies followed by imaging using an epifluorescence microscope. Arrows indicate protein accumulation at ciliary tip (left) or proximal end of cilia (right). Scale bars, 5 µm.
-
B, CImmunostaining analysis of IFT43 and IFT38 in the deletion mutants as indicated. Cells were fixed and stained with antibodies against IFT43 (an IFT‐A subunit) (B) or against IFT38 (an IFT‐B subunit) (C) followed by imaging using both DIC and epifluorescence microscopy. IFT43 and IFT38 accumulated at the ciliary tip in IFT54Δ261–275 mutant but at proximal end of cilia in IFT54Δ342–356 mutant. Arrows indicate ciliary bulges. WT, wild‐type cells. Scale bars, 5 µm.
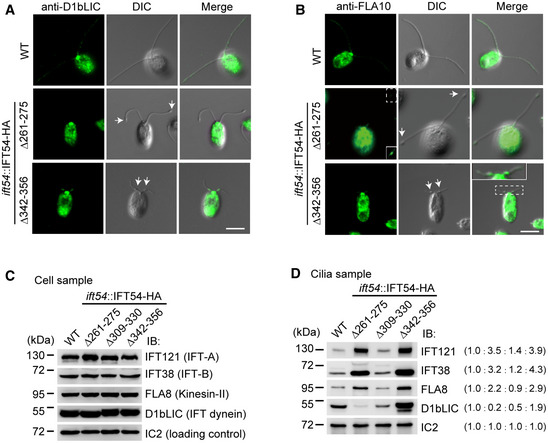
To determine whether other IFT complex proteins were similarly enriched, we examined the ciliary distribution of IFT43, a component of IFT‐A, and IFT38, a component of IFT‐B (Fig 2B and C). In the IFT54Δ261–275 mutant, both IFT43 and IFT38 were enriched in bulges located near the ciliary tips. In contrast, IFT43 and IFT38 were enriched in bulges near the proximal end of cilia in the IFT54Δ342–356 mutant. Next, we examined the distribution of IFT motors in the cilia of the deletion mutants by immunostaining. In the IFT54Δ261–275 mutant, the kinesin‐II subunit FLA10 was slightly enriched in bulges near the ciliary tip while the IFT dynein subunit D1bLIC was not enriched (Fig 3A and B). In the IFT54Δ342–356 mutant, however, both FLA10 and D1bLIC were enriched in the proximal ciliary bulges (Fig 3A and B).
Figure 3. Analysis of ciliary accumulation of IFT motors and IFT complex proteins in IFT54 deletion mutants.
-
AImmunostaining of cells with antibody against IFT dynein subunit D1bLIC. D1bLIC is absent in the cilia bulges at the ciliary tip in IFT54Δ261–275 mutant while enriched at the cilia bulges at proximal end of cilia in IFT54Δ342–356 mutant. WT, wild‐type cells. Arrows indicate ciliary bulges. Scale bar, 5 µm.
-
BImmunostaining of cells with antibody against kinesin‐II subunit FLA10. FLA10 is enriched in cilia bulges at the ciliary tip in IFT54Δ261–275 mutant or at proximal end of cilia in IFT54Δ342–356 mutant. WT, wild‐type cells. The insets show enhanced and/or enlarged images. Arrows indicate ciliary bulges. Scale bar, 5 µm.
-
C, DImmunoblot analysis of whole cells (C) and isolated cilia (D) of wild‐type and the three IFT54 deletion mutants. The membranes were probed with antibodies against IFT complex and motor proteins as indicated. Wild‐type levels of the various IFT components were detected in the whole cell samples of the deletion strains (C). IFT121, IFT38, and FLA8 accumulated in cilia of both IFT54Δ261–275 and IFT54Δ342–356 mutants. D1bLIC was relatively reduced in IFT54Δ261–275 mutant cilia but was substantially increased in IFT54Δ342–356 mutant cilia (D). Ratios of protein amounts from one representative experiment are in parentheses.
Source data are available online for this figure.
We used immunoblotting analyses with antibodies against representative IFT complex proteins and motor proteins to provide a quantitative view of IFT protein presence in the mutants. In whole cell samples, the IFT‐A protein IFT121, the IFT‐B protein IFT38, the kinesin‐II subunit FLA8, and the IFT dynein subunit D1bLIC were present in wild‐type amounts revealing that the changes to IFT54 did not affect the expression or stability of these proteins (Fig 3C). In isolated cilia of the IFT54Δ309–330 mutant, the levels of these IFT proteins were similar to those in the control cilia, as expected considering that ciliogenesis is normal in this strain (Fig 3D). In contrast, the levels of IFT121, IFT38, and FLA8 were relatively increased in the cilia of IFT54Δ261–275 and IFT54Δ342–356 mutants. In contrast, the level of D1bLIC was greatly reduced in the cilia of IFT54Δ261–275 mutant but strongly increased in those of the IFT54Δ342–356 mutant, indicating that the two mutations differentially affect the behavior of IFT dynein.
Taken together, we conclude that IFT54Δ261–275 and the IFT54Δ342–356 deletion mutants interfere with IFT and induce an abnormal accumulation of IFT particle proteins. The accumulation of IFT material at the ciliary tip is reminiscent of retrograde IFT mutants indicating a possible defect in retrograde IFT in the IFT54Δ261–275 strain; this notion is supported by the reduced levels of D1bLIC in the IFT54Δ261–275 cilia. In contrast, an accumulation of IFT proteins and subunits of both motors in the proximal region of cilia, as observed in the IFT54Δ342–356 strain, could result from a failure of IFT to travel toward the ciliary tip.
IFT54 interacts with kinesin‐II to facilitate anterograde IFT
To further understand how IFT54 regulates IFT, we performed live imaging of IFT and analyzed how the mutations affected IFT54’s interactions with other IFT components, starting with the IFT54Δ342–356 mutant. In comparison with an ift43::IFT43‐YFP rescue strain (as a control), both the anterograde velocity and frequency of IFT43‐YFP were relatively decreased in the ift54::IFT54Δ342–356 strain, indicating defects in anterograde IFT (Fig 4A–C, Movie EV1). We also noticed that the velocity and frequency of retrograde IFT were reduced in the IFT54Δ342–356 mutant compared to controls. A defect in retrograde IFT frequency could result from the reduced number of active IFT trains that reach the ciliary tip. The observed decrease in retrograde velocity could suggest a role of IFT54 during retrograde IFT.
Figure 4. IFT54 interacts with kinesin‐II and deletion of residues 342–356 of IFT54 strengthens its interaction with kinesin‐II and impairs anterograde IFT.
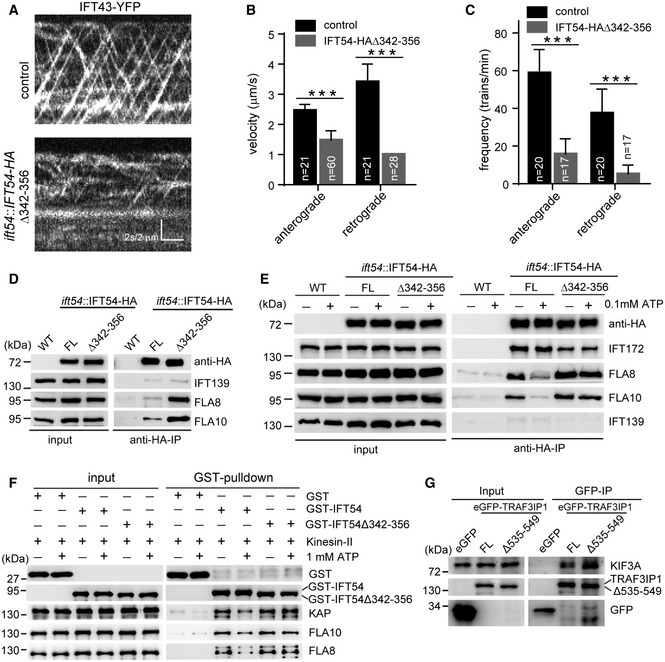
-
AKymograms showing the trajectories of IFT trains inside cilia as visualized with IFT43‐YFP. An ift43 rescue strain expressing IFT43‐YFP (as a control) and the IFT54Δ342–356 mutant expressing IFT43‐YFP were analyzed by live imaging via TIRF microscopy.
-
B, CVelocities (B) and frequencies (C) of anterograde and retrograde IFT of IFT43‐YFP in control and the IFT54Δ342–356 mutant. n represents the number of cilia assayed from three independent experiments. Values show the mean ± SD. Unpaired two‐tailed Student’s t‐test analysis, ***P < 0.0001.
-
DDeletion of residues 342–356 of IFT54 increases its interaction with kinesin‐II. Cell extracts from wild‐type (WT), ift54 expressing HA‐tagged full‐length (FL) IFT54 or IFT54Δ342–356 mutant were subjected to immunoprecipitation with anti‐HA antibody followed by immunoblotting with the indicated antibodies. The normalized ratios of FLA8 and FLA10 in the immunoprecipitates (IFT54 versus the mutant) are 1:3.90 and 1:3.13, respectively.
-
EATP treatment induces dissociation of IFT54 but not IFT54Δ342–356 mutant from kinesin‐II. Immunoprecipitation experiments were performed as shown in (D) in the presence or absence of ATP. The normalized ratios of FLA8 and FLA10 in the immunoprecipitates (IFT54 without ATP/with ATP/mutant without ATP/with ATP) are 1:0.37:1.70:1.38 and 1:0.35:1.47:1.45, respectively.
-
FAnalysis of the interaction of IFT54 and IFT54Δ342–356 mutant with kinesin‐II by GST pull‐down assay. Bacterial expressed GST, GST‐tagged IFT54, or IFT54Δ342–356 mutant was mixed, respectively, with recombinant kinesin‐II purified from insect cells in the presence or absence of ATP followed by GST pull‐down and immunoblotting. The normalized ratios of KAP, FLA10, and FLA8 in the pull‐down samples (GST‐IFT54 without ATP/with ATP/mutant without ATP/with ATP) are 1:0.51:1.31:1.44, 1:0.55:1.29:1.19, and 1:0.50:1.31:1.29, respectively.
-
GInteraction of IFT54 with kinesin‐II is conserved in mammalian cells. Cell extracts from HEK293T cells expressing eGFP, eGFP‐tagged TRAF3IP1, or TRAF3IP1Δ535–549 (corresponding deletion mutant of IFT54Δ342–356) were analyzed by immunoprecipitation with GFP antibody followed by immunoblotting with anti‐GFP and KIF3A antibodies, respectively. Please note that the deletion mutant had an increased interaction with kinesin‐II subunit KIF3A relative to the control. The normalized ratio of KIF3A in the immunoprecipitates (TRAF3IP1 versus mutant) is 1:1.76.
Source data are available online for this figure.
The defects in anterograde IFT in the IFT54Δ342–356 mutant are consistent with the formation of bulges at the proximal end of cilia containing IFT proteins and both IFT motors (Fig 3). As anterograde IFT is powered by kinesin‐II, we wondered if the IFT54Δ342–356 mutation interferes with the movement of kinesin‐II, leading to a frequent cessation of IFT once the trains enter the cilia. We performed immunoprecipitation assays to determine whether the Δ342–356 mutation of IFT54 affects its interaction with kinesin‐II. HA‐tagged full‐length and IFT54Δ342–356, respectively, expressed in ift54 were immunoprecipitated with anti‐HA antibodies and analyzed by immunoblotting with the indicated antibodies; wild‐type cells were used as a control. Interestingly, the Δ342–356 mutation of IFT54 increased rather than reduced its interaction with kinesin‐II motor subunits FLA8 and FLA10 relative to the control (Fig 4D). This result suggests a tighter binding of the mutant with kinesin‐II, which could restrain kinesin‐II movement. To further confirm this result, we included ATP in our immunoprecipitation assay, which is known to facilitate the dissociation of kinesin‐II from IFT complex (Baker et al, 2003; Krock & Perkins, 2008). In agreement with these data, treatment with 0.1 mM ATP induced the dissociation of IFT54 from kinesin‐II motor (Fig 4E). In contrast, this treatment barely affected the interaction of the IFT54Δ342–356 mutant with kinesin‐II, supporting the notion that this mutation strengthens the interaction between kinesin‐II and the IFT‐B complex.
To determine whether IFT54 directly interacted with kinesin‐II, we performed an in vitro pull‐down assay. Bacterial expressed GST‐IFT54, GST‐IFT54Δ342–356 or, as a control, GST alone were mixed, respectively, with purified recombinant kinesin‐II followed by pull‐down with glutathione agarose beads. Both GST‐IFT54 and GST‐IFT54Δ342–356 pulled down kinesin‐II whereas GST alone did not (Fig 4F). Importantly, the addition of ATP decreased the binding of the wild‐type IFT54 to kinesin‐II but not that of the GST‐IFT54Δ342–356 mutant, suggesting that this mutation alters the ATP‐regulated binding of kinesin‐II to IFT54.
Since the region presented by 342–356 residues of Chlamydomonas IFT54 is well conserved across ciliated organisms (Fig 1A), we determined whether this region is functionally preserved in mammalian cells. Full‐length TRAF3IP1, the mammalian homologue of IFT54, and the corresponding deletion mutant TRAF3IP1Δ535–549, each tagged with eGFP, were expressed in HEK293T cells; eGFP alone was expressed as a control. The KIF3A subunit of kinesin‐II co‐immunoprecipitated with anti‐GFP antibody from the eGFP‐TRAF3IP1 cells but not those expressing eGFP alone (Fig 4G). Similar to our observations with the Chlamydomonas proteins, the eGFP‐TRAF3IP1Δ535–549 mutant co‐immunoprecipitated more KIF3A than the full‐length eGFP‐TRAF3IP1. The data suggest that the interaction of IFT54 with kinesin‐II is conserved in mammalian cells.
Taken together, we have shown that IFT54 directly interacts with kinesin‐II and that the Δ342–356 mutation of Chlamydomonas IFT54 or its corresponding mutation Δ535–549 of mammalian IFT54 results in tighter binding of IFT54 with kinesin‐II. The accumulation of IFT particle and motor proteins near the ciliary base, the reduction in anterograde IFT, and the defects in ciliogenesis in the IFT54Δ342–356 mutant suggest that the increased stability of kinesin‐II/IFT54‐binding interferes with kinesin‐II motor activity.
IFT54 interacts with D1bLIC to mediate anterograde transport of IFT dynein
IFT dynein is required for retrograde transport (Pazour et al, 1999; Porter et al, 1999; Signor et al, 1999; Blisnick et al, 2014). The accumulation of IFT proteins near the tip of IFT54Δ261–275 mutant cilia could result from the decreased ciliary level of IFT dynein (Fig 3B). Since IFT dynein is an inactive cargo of anterograde IFT (Jordan et al, 2018), we hypothesize that deletion of the residues 261–275 of IFT54 could interrupt the binding of IFT dynein to IFT complex during anterograde IFT leading to a decreased entry of IFT dynein into the mutant cilia. To test this hypothesis, we analyzed the transport of IFT dynein as visualized by the expression of D1bLIC‐YFP in wild‐type and ift54::IFT54Δ261–275 mutant cells using TIRF microscopy (Fig 5A, Movie EV2). While the anterograde velocity of D1bLIC‐YFP in the Δ261–275 mutant was similar to that in the control, the frequency of its transport by anterograde IFT was relatively reduced (Fig 5B and C), indicating that this IFT54 deletion mutation indeed affects ciliary entry of IFT dynein. Similarly, the frequency of D1bLIC‐YFP retrograde IFT was reduced, as expected because of the decreased levels of D1bLIC in the mutant cilia (Fig 3B). However, the reduced ciliary entry of dynein could also be caused by the reduction in the numbers of anterograde IFT trains. To exclude this possibility, we measured IFT of IFT43‐YFP in the mutant and control cells (Fig 5D, Movie EV3). Both the anterograde velocity and frequency of IFT43‐YFP were comparable to those in the control cells (Fig 5E and F). Thus, we could reasonably conclude that dynein binding to IFT is disrupted in the IFT54Δ261–275 mutant.
Figure 5. IFT54 interacts with IFT dynein subunit D1bLIC and deletion of residues 261–275 of IFT54 disrupts this interaction and impairs anterograde IFT of D1bLIC.
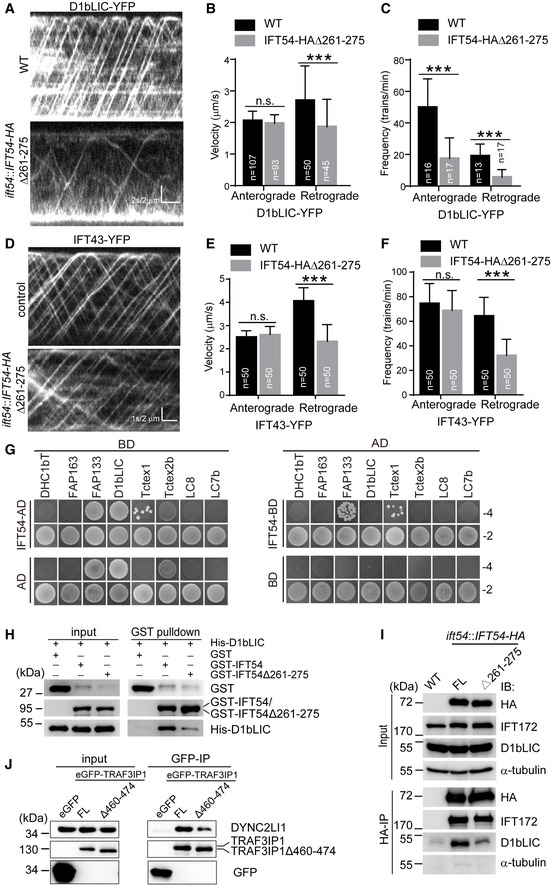
-
AKymograms showing the trajectories of IFT trains inside cilia as visualized with D1bLIC‐YFP. Wild‐type (WT) and IFT54Δ261–275 mutant cells expressing D1bLIC‐YFP were analyzed by live imaging via TIRF microscopy.
-
B, CVelocities (B) and frequencies (C) of anterograde and retrograde IFT of D1bLIC‐YFP. n represents the number of cilia assayed from four independent experiments. Values show the mean ± SD. Unpaired two‐tailed Student’s t‐test analysis, n.s.: no significance; ***P < 0.0001.
-
DKymograms of IFT43‐YFP of WT and mutant cilia.
-
E, FVelocities (E) and frequencies (F) of anterograde and retrograde IFT of IFT43‐YFP. n represents the number of cilia assayed from three independent experiments. Values show the mean ± SD. Unpaired two‐tailed Student’s t‐test analysis, n.s.: no significance; ***P < 0.0001.
-
GYeast two‐hybrid assay for interaction between IFT54 and IFT dynein subunits. Yeast cells that were transformed with each pair of constructs as indicated were grown under selection media lacking leucine, tryptophan, histidine, and adenine (−4) or lacking leucine and tryptophan (−2). DHC1bT, DHC1b tail domain; Empty AD or BD vectors were used as control.
-
HInteraction of IFT54 and IFT54Δ261–275 with D1bLIC by GST pull‐down assay. Bacterial expressed GST, GST‐IFT54, or GST‐IFT54Δ261–275 were mixed, respectively, with His‐D1bLIC followed by GST pull‐down and immunoblotting with anti‐GST and anti‐His antibodies. The normalized ratio of His‐D1bLIC in the pull‐down samples (GST‐IFT54 versus mutant) is 1:0.32.
-
ICo‐immunoprecipitation of IFT54 and IFT54Δ261–275 with D1bLIC. Cell extracts from wild‐type (WT), ift54 expressing HA‐tagged full‐length (FL) IFT54 or IFT54Δ261–275 mutant were subjected to immunoprecipitation with anti‐HA antibody followed by immunoblotting with the indicated antibodies. Please note that IFT54Δ261–275 mutant exhibited weaker interaction with D1bLIC relative to the control. The normalized ratio of D1bLIC in the immunoprecipitates (IFT54‐HA versus mutant) is 1:0.37.
-
JInteraction of IFT54 with D1bLIC is conserved in mammalian cells. Cell extracts from HEK293T cells expressing eGFP, eGFP‐tagged TRAF3IP1, or TRAF3IP1Δ460–474 (corresponding deletion mutant of IFT54Δ261–275) were analyzed by immunoprecipitation with GFP antibody followed by immunoblotting with anti‐GFP and DYNC2LI1, human homologue of D1bLIC, antibodies, respectively. Please note that the deletion mutant weakened the interaction with IFT dynein subunit DYNC2LI1 relative to the control. The normalized ratio of DYNC2LI1 in the immunoprecipitates (TRAF3IP1 versus mutant) is 1:0.49.
Source data are available online for this figure.
How does the Δ261–275 mutation of IFT54 affect ciliary entry of IFT dynein? The mutation could interrupt a direct interaction between IFT54 and IFT dynein, reduce IFT dynein binding by distorting the IFT‐B complex or a combination of the two. To examine whether IFT54 directly interacts with IFT dynein, we used a yeast two‐hybrid assay. The IFT dynein complex encompasses the DHC1b heavy chain, the intermediate chains FAP163 and FAP133, the light intermediate chain D1bLIC, and the light chains Tctex1, Tctex2b, LC8, and LC7b (Asante et al, 2014; Hou & Witman, 2015; Toropova et al, 2019). We analyzed pair‐wise interaction of IFT54 with these subunits except that the tail domain of DHC1b was used rather than the full‐length protein. Based on colony growth, FAP133 and Tctex1 appeared to be the candidates for interacting with IFT54 (Fig 5G); however, GST pull‐down assays did not support this (Appendix Fig S2A–D). The apparent interaction of D1bLIC with IFT54 as observed was likely due to self‐activation of D1bLIC (Fig 5G). However, we reasoned that this did not exclude the possibility that they indeed interact. To test this possibility, GST pull‐down assays were performed. His‐D1bLIC was pulled down with GST‐IFT54 but not GST (Fig 5H). In the reciprocal experiment, His‐IFT54 was also pulled down with GST‐D1bLIC but not GST (Appendix Fig S2E). These data suggest that IFT54 and D1bLIC indeed could interact with each other. To determine whether deletion of residues of 261–275 of IFT54 could impair this interaction, similar pull‐down assays were performed. Indeed, this deletion weakened this interaction (Fig 5H and Appendix Fig S2E). Taken together, the data indicate that IFT54 directly binds D1bLIC and that this interaction is reduced by the deletion of residues 261–275 of IFT54.
To examine whether this interaction also occurred in vivo, HA‐tagged full‐length IFT54 and Δ261–275 mutant were expressed in ift54 null mutant. Immunoprecipitation with anti‐HA was performed with cell extracts of these cells and wild‐type cells as a control. Both IFT172 and D1bLIC were immunoprecipitated in cells expressing HA‐tagged full‐length IFT54 whereas the deletion of residues 261–275 of IFT54‐HA substantially decreased the amount of co‐immunoprecipitated D1bLIC relative to the control cells (Fig 5I). The amount of co‐immunoprecipitated IFT172 was not affected by this deletion arguing against a large‐scale defect in the IFT‐B complex in the IFT54Δ261–275 strain.
To determine the conservation of this interaction, eGFP‐tagged full‐length IFT54 human homologue TRAF3IP1 and the corresponding deletion mutant TRAF3IP1Δ460–474 and, as a negative control, eGFP alone were expressed respectively in HEK293T cells. Using antibodies against GFP, DYNC2LI1/LIC3, the human homologue of D1bLIC, was co‐immunoprecipitated with wild‐type eGFP‐TRAF3IP1; the quantity of precipitated DYNC2LI1/LIC3 was relatively decreased when the eGFP‐TRAF3IP1Δ460–474 deletion mutant was used (Fig 5J). Taken together, we conclude that IFT54 directly binds D1bLIC to link IFT dynein to IFT complex during anterograde IFT and this interaction is also conserved in mammalian cells.
Discussion
The IFT protein complex is the primary cargo of the IFT motors and also functions as a platform to move other proteins in and out of cilia. Thus, many IFT proteins are likely to possess multiple role in the assembly of the IFT complex itself, the interactions with the IFT motors, and the binding of specific cargoes (Lechtreck, 2015; Taschner & Lorentzen, 2016; Wingfield et al, 2018). However, loss of function mutations in most IFT‐B proteins result in a failure of IFT and the complete loss of cilia impedes the analysis of the individual roles of IFT proteins and their domains for IFT and cilia assembly. Here, we continued our analysis of the essential IFT protein IFT54, which possesses a dispensable N‐terminal CH domain and a critical C‐terminal coiled‐coil domain (Kunitomo & Iino, 2008; Taschner et al, 2016; Zhu et al, 2017b). We showed that IFT54 directly interacts with both the IFT dynein subunit D1bLIC and kinesin‐II. We identified two novel conserved regions of IFT54 (corresponding to residues 261–275 and 342–356 of the Chlamydomonas protein) that are critical for proper interaction with IFT dynein and kinesin‐II, respectively. The deletion of residues 261–275 of IFT54 reduced its interaction with D1bLIC and decreased the import of IFT dynein into cilia, indicating that the conserved 261–275 region directly participates in the D1bLIC binding or the deletion affects the conformation of the IFT54 protein and indirectly impaired its binding to D1bLIC. In contrast, deletion of the latter IFT54 region strengthens its interaction with kinesin‐II and interferes with anterograde IFT, as indicated by the accumulation of IFT proteins in the proximal region of the cilia. Similar defects in motor binding were observed for the corresponding mutations in TRAF3IP1 (human IFT54), indicating that the interactions of IFT54 with IFT motors are conserved in mammalian cells. Cryo‐ET data revealed that both IFT dynein and kinesin‐II are in contact with same sub‐region of the IFT‐B complex (Jordan et al, 2018). Our data suggest that this region encompasses IFT54, which would position the protein in the outskirts of the IFT‐B complex (Katoh et al, 2016; Taschner et al, 2016). As depicted in Fig 6, we propose that IFT54 interacts with both kinesin‐II and IFT dynein and is important for anterograde IFT and ciliogenesis.
Figure 6. A model for the function of IFT54 in regulating anterograde IFT.
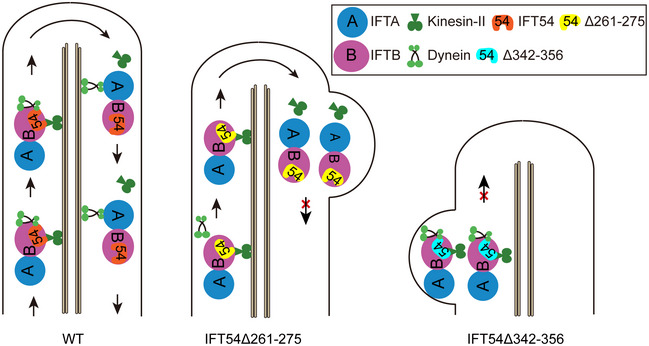
During anterograde IFT, kinesin‐II carries IFT‐A, IFT‐B, and inactive IFT dynein from ciliary base to tip. Inactive dynein is activated at ciliary tip and carries IFT‐A and IFT‐B back for retrograde transport while kinesin‐II diffuses. IFT54 interacts with both kinesin‐II and IFT dynein during anterograde IFT. Deletion of residues 261–275 of IFT54 disrupts its interaction with IFT dynein leading to inhibition of anterograde transport of IFT dynein and accumulation of IFT complexes near the ciliary tip with bulge formation. In contrast, deletion of residues 342–356 of IFT54 strengthens its interaction with kinesin‐II, supposedly inhibiting motility of kinesin‐II via unknown mechanism leading to cessation of IFT shortly after IFT trains enter cilia. IFT complexes and motors thus accumulate at proximal end of cilia with bulge formation. Please note that Δ342–356 mutant shows stronger cilia mutant type relative to Δ261–275 mutant.
During anterograde IFT, kinesin‐II is bound to the IFT‐B complex and several IFT‐B proteins are thought to participate in this interaction (Baker et al, 2003; Krock & Perkins, 2008; Funabashi et al, 2018; Liang et al, 2018). In vitro binding assays showed that a tetramer of IFT52, IFT88, IFT38, and IFT57, which forms the contact module between the IFT‐B1 and IFT‐B2 subcomplexes, also interacts strongly with kinesin‐II. Kinesin‐II may also bind to any of the IFT‐B2 proteins IFT20, IFT54, IFT80 and IFT172 (Funabashi et al, 2018), suggesting that the IFT‐B2 subcomplex is central for IFT–kinesin‐II interactions. In agreement with these studies, we show that IFT54 of Chlamydomonas and mammals binds to kinesin‐II. Interestingly, the deletion of residues 342–356 of IFT54 enhances its interaction with kinesin‐II, leading to accumulation of both IFT motors and IFT trains at the proximal end of cilia. We propose that this increased interaction may inhibit kinesin‐II movement via unknown mechanisms either by interference with kinesin‐II ATPase activity or its conformation change during ATP hydrolysis. Though we showed that IFT54 interacts with kinesin‐II, which motifs or domains of IFT54 mediate this interaction remains a question to address. Intuitively, residues 342–356 is unlikely involved in the direct interaction. Future elucidation of the full structures of IFT54 and kinesin‐II should provide details on how IFT54 and kinesin‐II interact with each other and how deletion of the residues 342–356 of IFT54 interferes with the motor activity of kinesin‐II (Taschner et al, 2016). What is the function of IFT54 in interacting with kinesin‐II? Cargo binding is one of the mechanisms for activation of kinesins (Verhey & Hammond, 2009). In vitro studies show that the non‐motor subunit KAP binding to the two motor subunits of kinesin‐II is sufficient for full‐activation of the motor (Sonar et al, 2020). In this regard, IFT54 binding may not function in activation of kinesin‐II though we could not exclude this possibility in vivo. One possibility is that IFT54, together with the IFT‐B1‐B2 connecting tetramer, stabilize the interaction between IFT complex and kinesin‐II.
Auto‐inhibited IFT dynein is transported as a cargo by anterograde IFT to the ciliary tip, where it is activated to ensure the return of the IFT particles to the cell body (Jordan et al, 2018). Deletion of residues 261–275 of IFT54 interferes with the ciliary entry and anterograde transport of IFT dynein. Because the amount of residual IFT dynein arriving at the ciliary tip is insufficient to ensure complete return of IFT trains to the cell body, accumulation of IFT proteins is provoked near the ciliary tip as previously observed in IFT dynein mutants (Pazour et al, 1998; Pazour et al, 1999; Porter et al, 1999; Signor et al, 1999; Lechtreck, 2019). Recent cryo‐ET data revealed that IFT dynein binds to the IFT‐B complex spanning out over seven to eight IFT‐B repeats (Jordan et al, 2018; Toropova et al, 2019). Thus, binding of IFT dynein likely depends on assembled IFT trains rather than individual IFT‐B complex or proteins. Indeed, during the assembly of IFT trains, IFT dynein is loaded briefly before the trains enter the cilia (Wingfield et al, 2017). The structural analysis of IFT dynein further suggests that each IFT dynein complex makes several distinct interactions with the IFT‐B complex, likely involving different IFT‐B proteins or at least territories on the IFT‐B complex. Here, we show that IFT54 provides one such contact site between IFT‐B and IFT dynein. The severity of the effect of the IFT54Δ261–275 deletion on IFT dynein transport argues against the presence of numerous redundant binding sites. Rather, IFT54 could provide a major contact site for IFT dynein or binding of IFT dynein depends on the cooperative interaction of several binding sites. Residual IFT dynein in the cilia of the IFT54Δ261–275 mutant is likely explained by its interactions with the remaining sites, one of which likely involves the IFT‐B subunit IFT172, which co‐immunoprecipitates with IFT dynein and is required to maintain normal levels of IFT dynein in cilia (Pedersen et al, 2006; Williamson et al, 2012).
Also, it remains largely unclear which parts of IFT dynein provide contact with IFT‐B. IFT dynein includes multiple subunits including dynein heavy chains, intermediate chains, light intermediate chains, and light chains (Asante et al, 2014; Hou & Witman, 2015; Toropova et al, 2019). Fitting of the 3D structure of the mammalian IFT dynein complex in the structure of anterograde trains obtained by cryo‐EM from Chlamydomonas suggests that the IFT dynein heavy chains make most of the interactions with the IFT‐B complexes (Jordan et al, 2018; Toropova et al, 2019). The intermediate chains and light chains that are embedded within the two heavy chains participate in controlling IFT dynein asymmetry, oligomerization and activity (Toropova et al, 2019). Thus, the function of the light intermediate chain D1bLIC, which is positioned on the outer surface of the dynein heavy chain stalk domain, is not clear. Here, we show that the D1bLIC subunit binds to IFT54 revealing that at least this non‐motor subunit of IFT dynein participates directly in its interactions with the IFT‐B complex. Similar to the IFT54Δ261–275 mutant, mutants lacking D1bLIC also form bulges near the tip of shortened cilia but the overall amount of dynein is reduced in such cells suggesting an additional role of D1bLIC in stabilization of IFT dynein (Hou et al, 2004; Reck et al, 2016). To summarize, our work identified IFT54 and D1bLIC as the first pair of proteins mediating binding between the IFT‐B complex and IFT dynein.
Materials and Methods
Strains and culture
Chlamydomonas reinhardtii wild‐type strain 21gr (mt+; CC‐1690) is available from the Chlamydomonas Resource Center (University of Minnesota, St. Paul, MN). ift54 and ift54::IFT54‐HA were from a previous study (Zhu et al, 2017b). Cells were cultured on 1.5% agar plates or in liquid M medium at 23°C with aeration under 14/10 h light/dark cycle. For transformation, cells were grown in liquid Tris‐acetate‐phosphate medium. HEK293T cells were cultured in six well plates in DMEM with high glucose supplemented with 10% FBS (Sanyo CO2 Incubator, Japan).
Generation of strains expressing deletion mutants of IFT54
The full‐length IFT54 expression vector was described previously (Zhu et al, 2017b). The deletion constructs of IFT54 were made by using overlapping PCR and conventional molecular methods and were cloned into the original pKH‐IFT54‐HA vector. All constructs were verified by sequencing. The final constructs pKH‐IFT54Δ261–275‐HA, pKH‐IFT54Δ309–330‐HA, and pKH‐IFT54Δ342–356‐HA were linearized with KpnI and transformed into the ift54 mutant cells by electroporation (Liang & Pan, 2013). The hygromycin‐resistant transformants were screened by immunoblotting with the antibody against HA.
Cilia isolation, length measurement, and cell imaging
A previously published protocol was followed for cilia isolation (Craige et al, 2013). Briefly, cells were deflagellated by pH shock, and the detached cilia were further purified by sucrose gradient centrifugation. Isolated cilia were suspended in HMDEK buffer (50 mM HEPES, pH 7.2, 5 mM MgCl2, 0.5 mM EDTA, 1 mM DTT, 25 mM KCl) containing protease inhibitor cocktail (mini‐cOmplete EDTA‐free, Roche) and 20 µM proteasome inhibitor MG‐132 (Selleck, China), flash frozen in liquid nitrogen, and stored at 80°C until use.
For cilia length measurements or phenotype examination, cells were fixed by 1% glutaraldehyde and imaged by DIC microscopy using a 40× or 100× lens on a Zeiss Axio Observer Z1 microscope (Carl Zeiss, Germany) that is equipped with a CCD camera (QuantEM512SC, Photometric, USA). Cilia length was measured using ImageJ (NIH, USA), and the data were plotted using GraphPad Prism 5.01 (GraphPad, USA). Cell images were processes using Photoshop and Illustrator (Adobe, USA).
Antibodies
Details of the primary antibodies used for immunofluorescence and immunoblotting are provided in Table S1. The polyclonal anti‐IFT172 antibody was raised against the IFT172 amino acid residues 1,505–1,729 and affinity purified (ABclonal, China). Monoclonal anti‐IFT139 antibody was kindly provided by Dr Joel Rosenbaum (Yale University, New Haven, CT).
For immunoblotting, the secondary antibodies HRP‐conjugated goat anti‐mouse, goat anti‐rabbit, and goat anti‐rat (1:5,000, EASYBIO, China) were used. And the second antibodies for immunostaining were Texas red–conjugated goat anti‐mouse IgG, Alexa Fluor 488 goat anti‐rat and Alexa Fluor 488–conjugated goat anti‐rabbit IgG (1:200, Life technologies, USA).
SDS–PAGE, immunoblotting, and immunoprecipitation
SDS–PAGE and immunoblotting analysis were performed as described previously (Pan & Snell, 2000). Briefly, cells were lysed in Buffer A (50 mM Tris–HCl pH 7.5, 10 mM MgCl2, 1 mM EDTA, 1 mM DTT) containing protease inhibitor cocktail and 20 µM MG‐132 as described above, boiled in 1×SDS sample buffer for 10 min, separated by SDS–PAGE, transferred to polyvinylidene difluoride membranes, and probed with indicated antibodies.
Immunoprecipitation (IP) using Chlamydomonas was carried out as detailed previously (Zhu et al, 2017b). In brief, 1 × 109 cells were lysed in 1 ml IP buffer (20 mM Hepes, pH 7.2, 5 mM MgCl2, 1 mM DTT, 1 mM EDTA, 10 mM NaF, 0.1 mM Na3VO4, 150 mM NaCl, 20 mM beta‐glycerol phosphate, 5% glycerol, protease inhibitor cocktail and 20 µM MG‐132. The supernatants were pre‐cleared by incubating with 30 µl protein A Sepharose beads (GE, Sweden) for 30 min at 4°C followed by incubating with 30 µl pre‐washed rat anti‐HA Affinity Matrix (Roche) for 3 h at 4°C. After washing four times with IP buffer, the samples were analyzed by SDS–PAGE followed by immunoblotting. For ATP treatment, a final 0.1 mM ATP was added to cell lysates prior to addition of protein A Sepharose beads.
For immunoprecipitation using mammalian cells, human cDNA of TRAF3IP1 was amplified by PCR from a Human Lung cDNA Library (Takara, Japan). Full‐length cDNA and its deletion constructs as indicated were cloned into the EGFP‐C3 vector followed by verification with sequencing. HEK293T cells cultured for 24 h were transfected with the constructs using Lipofectamine 3000 (Invitrogen, USA) and collected after starvation for 48 h. Approximately 1 × 107 cells were lysed in 500 µl lysis buffer (50 mM Tris–HCl pH 8.0, 150 mM KCl, 5 mM MgCl2, 1 mM EDTA, 0.5% NP40) containing protease inhibitor cocktail. After 30 min on ice, the lysates were clarified by centrifugation at 20,817 g for 10 min at 4°C twice. The supernatants were mixed with 30 µl GFP beads (Smart‐life Sciences, China) followed by rotation for 2 h at 4°C. After five times washes each with 1 ml ice‐cold wash buffer (50 mM Tris–HCl, pH 8.0, 150 mM KCl, 5 mM MgCl2, 1 mM EDTA, 0.5% NP40, 0.1% Triton X‐100), the beads were resuspended in 60 µl 1 × SDS sample buffer and boiled for 10 min followed by SDS–PAGE and immunoblotting.
Immunofluorescence microscopy
Immunostaining analysis was performed essentially as described previously (Zhu et al, 2017b). The cells were viewed on a Zeiss LSM780 META Observer Z1 Confocal Laser Microscope (Zeiss, Germany). Images were acquired and processed by ZEN 2012 Light Edition (Zeiss) and Photoshop and assembled in Illustrator (Adobe USA).
Yeast‐based two‐hybrid analysis
The cDNAs of IFT54 and all IFT dynein subunits were amplified from a C. reinhardtii cDNA library (Takara, Japan) and then inserted into yeast expression vector pGBKT7 (BD) and pGADT7 (AD) respectively. For DHC1b, cDNA for the tail domain (1–1,704 aa) was used. All possible pair‐wise combinations of IFT54 with the IFT dynein subunits constructs were co‐transformed into yeast strain AH109, following a standard lithium acetate/PEG‐4000/salmon sperm DNA protocol published previously (Lucker et al, 2005). The transformed yeast cells with the construct pairs as indicated in the text were then cultured on selective media (‐Leu/‐Trp/‐His/‐Ade or –Leu/‐Trp) (Clontech, USA) at 30°C for 2–3 days.
Expression and purification of recombinant proteins from Escherichia coli and insect cells
For bacterial expression, cDNAs of full‐length IFT54, IFT54Δ261–275, IFT54Δ342–356, FAP133, Tctex1, and D1bLIC were cloned into vector pGEX‐6P‐1 harboring N‐terminal GST tag or pET‐28a harboring N‐terminal 6xHis tag. The final constructs were transformed into E. coli Rosetta (DE3) cells (CWBIO, China). The transformants were grown in LB medium overnight at 18°C with addition of 0.5 mM IPTG for induction of expression of the recombinant proteins. The GST‐tagged proteins were purified with Glutathione‐sepharose 4B beads (GE, Sweden) if necessary.
Expression and purification of kinesin‐II was essentially as described previously (Li et al, 2020). Baculovirus expression system was used for expression of kinesin‐II in Sf‐9 insect cells. cDNAs of full‐length FLA8, FLA10, and KAP were amplified by PCR from a Chlamydomonas cDNA library and subcloned into vectors pOCC25 with RFP tag, pOCC52 with MBP tag, and pOCC8 with GFP‐His tag, respectively (kindly provided by Dr. Xin Liang, Tsinghua University) to generate pOCC25‐FLA8, pOCC52‐FLA10, and pOCC8‐KAP constructs. The resulting constructs were expressed together in Sf‐9 cells using the baculovirus expression system. Kinesin‐II complex was purified first by using Ni‐NTA Agarose (Qiagen, Germany) followed by using Amylose Resin (NEB, USA). The proteins were eluted in 1xBRB80 buffer (80 mM PIPES, 1 mM MgCl2, 1 mM EGTA, pH 6.8) containing 100 mM KCl and 0.1 mM ATP, and were flash frozen in liquid nitrogen and stored at 80°C until use. The protein concentration was determined with Protein Assay Dye Reagent Concentrate (Bio‐Rad, USA).
GST pull‐down assay
A published method was followed with modifications (Taschner et al, 2014). 50 ml E. coli cells expressing recombinant proteins were collected and dissolved in 2 ml lysis buffer (50 mM Tris, pH 8.0, 100 mM NaCl, 1 mM DTT, and 100 mM PMSF). After sonication on ice, the lysates were centrifuged at 20,817 g for 20 min at 4°C (5417R, Eppendorf, Germany). Clear lysates containing GST‐tagged proteins and His‐tagged proteins were mixed, respectively, as indicated in the text followed by incubating with 30 µl Glutathione‐sepharose 4B beads (GE, Sweden) at 4°C for 2 h with rotation. 1 mM ATP was added in some of the experiments as indicated. The protein bound beads were harvested by centrifugation at 3,000 g at 4°C followed by three washes with 1 ml ice‐cold lysis buffer each. The washed beads were processed for SDS–PAGE and immunoblotting. For pull‐down assay involving kinesin‐II, 50–100 µg of GST‐tagged protein and kinesin‐II complex were mixed in a total volume of 500 µl lysis buffer with 30 µl Glutathione‐sepharose 4B beads (GE, Sweden).
TIRF microscopy
For TIRF imaging, previous protocols were followed (Wren et al, 2013; Wingfield et al, 2017). In brief, cells were placed to a home‐made observation chamber using No.1.5 coverslips and then imaged at room temperature on an Eclipse Ti‐U microscope (Nikon, Japan) equipped with a 60× TIRF objective (N.A. 1.49) and a cooled Andor iXon X3 DU897 EMCCD camera. Images were captured at 10 fps and analyzed with NIS‐Elements Advanced Research software (Nikon, Japan). Kymographs were assembled from the obtained data using ImageJ (NIH, USA), and the velocity and frequency of the IFT train were calculated. For the imaging data presented in Fig 5E–G, images were captured at 20 fps on an Olympus microscope (IX83) with a 100× TIRF objective and a cooled Photometrics Prime 95B camera. The strains for analysis were expressing either D1bLIC‐YFP or IFT43‐YFP as indicated. IFT43‐YFP construct and the data for IFT43‐YFP in control cells (Fig 4A–C) were from our previous studies (Wingfield et al, 2017; Zhu et al, 2017a). To generate construct for expressing D1bLIC‐YFP, IFT54 gene in a previous construct was replaced by the genomic DNA of D1bLIC with YFP tag at the 3′ end while the hygromycin‐resistant gene was replaced by bleomycin‐resistant gene (Zhu et al, 2017b).
Quantification and statistical analysis
For all the experiments, two or more independent experiments were performed. Data were presented as mean ± SD. Statistical significance was performed by using unpaired two‐tailed Student’s t‐test analysis. P < 0.01 was considered to be statistically significant. *P < 0.01; **P < 0.001; ***P < 0.0001.
Author contributions
Study design: All authors; Experiments: XZ and JW; Recombinant kinesin‐II: SL; Live imaging of IFT: KL; Resources: JP and KL; Manuscript writing: JP, KL, XZ, and JW.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Review Process File
Appendix
Movie EV1
Movie EV2
Movie EV3
Source Data for Appendix
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 5
Acknowledgements
We are grateful to Drs. Xin Liang (Tsinghua University) for providing plasmids, and Dennis Diener and Joel Rosenbaum for providing antibodies. We also thank Core Facility of Center of Biomedical Analysis (Tsinghua University) for assistance on cell imaging analysis. This work was supported by the National Key R&D Program of China (2017YFA0503500, 2018YFA0902500) and the National Natural Science Foundation of China (31991191, 31671387, 31972888 to JP), and by the National Institutes of Health (GM110413 to KL); the content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The EMBO Journal (2021) 40: e105781.
Data availability
There are no primary datasets or computer codes associated with the study.
References
- Asante D, Stevenson NL, Stephens DJ (2014) Subunit composition of the human cytoplasmic dynein‐2 complex. J Cell Sci 127: 4774–4787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker SA, Freeman K, Luby‐Phelps K, Pazour GJ, Besharse JC (2003) IFT20 links kinesin II with a mammalian intraflagellar transport complex that is conserved in motile flagella and sensory cilia. J Biol Chem 278: 34211–34218 [DOI] [PubMed] [Google Scholar]
- Bangs F, Anderson KV (2017) Primary cilia and mammalian Hedgehog signaling. Cold Spring Harb Perspect Biol 9: a028175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behal RH, Miller MS, Qin H, Lucker BF, Jones A, Cole DG (2012) Subunit interactions and organization of the Chlamydomonas reinhardtii intraflagellar transport complex A proteins. J Biol Chem 287: 11689–11703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blisnick T, Buisson J, Absalon S, Marie A, Cayet N, Bastin P (2014) The intraflagellar transport dynein complex of trypanosomes is made of a heterodimer of dynein heavy chains and of light and intermediate chains of distinct functions. Mol Biol Cell 25: 2620–2633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien A, Shih SM, Bower R, Tritschler D, Porter ME, Yildiz A (2017) Dynamics of the IFT machinery at the ciliary tip. Elife 6: e28606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole DG, Diener DR, Himelblau AL, Beech PL, Fuster JC, Rosenbaum JL (1998) Chlamydomonas kinesin‐II‐dependent intraflagellar transport (IFT): IFT particles contain proteins required for ciliary assembly in Caenorhabditis elegans sensory neurons. J Cell Biol 141: 993–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craige B, Brown JM, Witman GB (2013) Isolation of Chlamydomonas flagella. Curr Protoc Cell Biol Chapter 3: Unit 3 41 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel BD, Ishikawa H, Wemmer KA, Geimer S, Wakabayashi K, Hirono M, Craige B, Pazour GJ, Witman GB, Kamiya R et al (2012) The role of retrograde intraflagellar transport in flagellar assembly, maintenance, and function. J Cell Biol 199: 151–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelke MF, Waas B, Kearns SE, Suber A, Boss A, Allen BL, Verhey KJ (2019) Acute inhibition of heterotrimeric Kinesin‐2 function reveals mechanisms of intraflagellar transport in mammalian cilia. Curr Biol 29: 1137–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funabashi T, Katoh Y, Okazaki M, Sugawa M, Nakayama K (2018) Interaction of heterotrimeric kinesin‐II with IFT‐B‐connecting tetramer is crucial for ciliogenesis. J Cell Biol 217: 2867–2876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y, Pazour GJ, Witman GB (2004) A dynein light intermediate chain, D1bLIC, is required for retrograde intraflagellar transport. Mol Biol Cell 15: 4382–4394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y, Witman GB (2015) Dynein and intraflagellar transport. Exp Cell Res 334: 26–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iomini C, Babaev‐Khaimov V, Sassaroli M, Piperno G (2001) Protein particles in Chlamydomonas flagella undergo a transport cycle consisting of four phases. J Cell Biol 153: 13–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iomini C, Li L, Esparza JM, Dutcher SK (2009) Retrograde intraflagellar transport mutants identify complex A proteins with multiple genetic interactions in Chlamydomonas reinhardtii . Genetics 183: 885–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan MA, Diener DR, Stepanek L, Pigino G (2018) The cryo‐EM structure of intraflagellar transport trains reveals how dynein is inactivated to ensure unidirectional anterograde movement in cilia. Nat Cell Biol 20: 1250–1255 [DOI] [PubMed] [Google Scholar]
- Katoh Y, Terada M, Nishijima Y, Takei R, Nozaki S, Hamada H, Nakayama K (2016) Overall architecture of the intraflagellar transport (IFT)‐B complex containing Cluap1/IFT38 as an essential component of the IFT‐B peripheral subcomplex. J Biol Chem 291: 10962–10975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozminski KG, Johnson KA, Forscher P, Rosenbaum JL (1993) A motility in the eukaryotic flagellum unrelated to flagellar beating. Proc Natl Acad Sci USA 90: 5519–5523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozminski KG, Beech PL, Rosenbaum JL (1995) The Chlamydomonas kinesin‐like protein FLA10 is involved in motility associated with the flagellar membrane. J Cell Biol 131: 1517–1527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krock BL, Perkins BD (2008) The intraflagellar transport protein IFT57 is required for cilia maintenance and regulates IFT‐particle‐kinesin‐II dissociation in vertebrate photoreceptors. J Cell Sci 121: 1907–1915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunitomo H, Iino Y (2008) Caenorhabditis elegans DYF‐11, an orthologue of mammalian Traf3ip1/MIP‐T3, is required for sensory cilia formation. Genes Cells 13: 13–25 [DOI] [PubMed] [Google Scholar]
- Lechtreck KF (2015) IFT‐cargo interactions and protein transport in cilia. Trends Biochem Sci 40: 765–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechtreck K (2019) Dynein in intraflagellar transport. In Handbook of dynein, Hirose K (ed.), pp 225‐275. Singapore: Pan Stanford Publishing Pte. Ltd. [Google Scholar]
- Li S, Chen W, Wan KW, Tao H, Liang X, Pan J (2020) Functional exploration of heterotrimeric kinesin‐II in IFT and ciliary length control in chlamydomonas. Elife 9: e58868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Pan J (2013) Regulation of flagellar biogenesis by a calcium dependent protein kinase in Chlamydomonas reinhardtii . PLoS One 8: e69902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Meng D, Zhu B, Pan J (2016) Mechanism of ciliary disassembly. Cell Mol Life Sci 73: 1787–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Zhu X, Wu Q, Pan J (2018) Ciliary length sensing regulates IFT entry via changes in FLA8/KIF3B phosphorylation to control ciliary assembly. Curr Biol 28: 2429–2435 [DOI] [PubMed] [Google Scholar]
- Liem KF Jr, Ashe A, He M, Satir P, Moran J, Beier D, Wicking C, Anderson KV (2012) The IFT‐A complex regulates Shh signaling through cilia structure and membrane protein trafficking. J Cell Biol 197: 789–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucker BF, Behal RH, Qin H, Siron LC, Taggart WD, Rosenbaum JL, Cole DG (2005) Characterization of the intraflagellar transport complex B core: direct interaction of the IFT81 and IFT74/72 subunits. J Biol Chem 280: 27688–27696 [DOI] [PubMed] [Google Scholar]
- Lucker BF, Miller MS, Dziedzic SA, Blackmarr PT, Cole DG (2010) Direct interactions of intraflagellar transport complex B proteins IFT88, IFT52, and IFT46. J Biol Chem 285: 21508–21518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng D, Pan J (2016) A NIMA‐related kinase, CNK4, regulates ciliary stability and length. Mol Biol Cell 27: 838–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris RL, Scholey JM (1997) Heterotrimeric kinesin‐II is required for the assembly of motile 9+2 ciliary axonemes on sea urchin embryos. J Cell Biol 138: 1009–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou G, Blacque OE, Snow JJ, Leroux MR, Scholey JM (2005) Functional coordination of intraflagellar transport motors. Nature 436: 583–587 [DOI] [PubMed] [Google Scholar]
- Pan J, Snell WJ (2000) Regulated targeting of a protein kinase into an intact flagellum. An aurora/Ipl1p‐like protein kinase translocates from the cell body into the flagella during gamete activation in chlamydomonas. J Biol Chem 275: 24106–24114 [DOI] [PubMed] [Google Scholar]
- Pazour GJ, Wilkerson CG, Witman GB (1998) A dynein light chain is essential for the retrograde particle movement of intraflagellar transport (IFT). J Cell Biol 141: 979–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour GJ, Dickert BL, Witman GB (1999) The DHC1b (DHC2) isoform of cytoplasmic dynein is required for flagellar assembly. J Cell Biol 144: 473–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen LB, Geimer S, Rosenbaum JL (2006) Dissecting the molecular mechanisms of intraflagellar transport in chlamydomonas. Curr Biol 16: 450–459 [DOI] [PubMed] [Google Scholar]
- Pedersen LB, Rosenbaum JL (2008) Intraflagellar transport (IFT) role in ciliary assembly, resorption and signalling. Curr Top Dev Biol 85: 23–61 [DOI] [PubMed] [Google Scholar]
- Picariello T, Brown JM, Hou Y, Swank G, Cochran DA, King OD, Lechtreck K, Pazour GJ, Witman GB (2019) A global analysis of IFT‐A function reveals specialization for transport of membrane‐associated proteins into cilia. J Cell Sci 132: jcs220749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno G, Mead K (1997) Transport of a novel complex in the cytoplasmic matrix of Chlamydomonas flagella. Proc Natl Acad Sci USA 94: 4457–4462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno G, Siuda E, Henderson S, Segil M, Vaananen H, Sassaroli M (1998) Distinct mutants of retrograde intraflagellar transport (IFT) share similar morphological and molecular defects. J Cell Biol 143: 1591–1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter ME, Bower R, Knott JA, Byrd P, Dentler W (1999) Cytoplasmic dynein heavy chain 1b is required for flagellar assembly in Chlamydomonas . Mol Biol Cell 10: 693–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prevo B, Mangeol P, Oswald F, Scholey JM, Peterman EJ (2015) Functional differentiation of cooperating kinesin‐2 motors orchestrates cargo import and transport in C. elegans cilia. Nat Cell Biol 17: 1536–1545 [DOI] [PubMed] [Google Scholar]
- Reck J, Schauer AM, VanderWaal Mills K, Bower R, Tritschler D, Perrone CA, Porter ME (2016) The role of the dynein light intermediate chain in retrograde IFT and flagellar function in Chlamydomonas . Mol Biol Cell 27: 2404–2422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter JF, Leroux MR (2017) Genes and molecular pathways underpinning ciliopathies. Nat Rev Mol Cell Biol 18: 533–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum JL, Witman GB (2002) Intraflagellar transport. Nat Rev Mol Cell Biol 3: 813–825 [DOI] [PubMed] [Google Scholar]
- Roy S (2009) The motile cilium in development and disease: emerging new insights. BioEssays 31: 694–699 [DOI] [PubMed] [Google Scholar]
- Scholey JM (2003) Intraflagellar transport. Annu Rev Cell Dev Biol 19: 423–443 [DOI] [PubMed] [Google Scholar]
- Scholey JM (2013) Kinesin‐2: a family of heterotrimeric and homodimeric motors with diverse intracellular transport functions. Annu Rev Cell Dev Biol 29: 443–469 [DOI] [PubMed] [Google Scholar]
- Signor D, Wedaman KP, Orozco JT, Dwyer ND, Bargmann CI, Rose LS, Scholey JM (1999) Role of a class DHC1b dynein in retrograde transport of IFT motors and IFT raft particles along cilia, but not dendrites, in chemosensory neurons of living Caenorhabditis elegans . J Cell Biol 147: 519–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonar P, Youyen W, Cleetus A, Wisanpitayakorn P, Mousavi SI, Stepp WL, Hancock WO, Tuzel E, Okten Z (2020) Kinesin‐2 from C. reinhardtii is an atypically fast and auto‐inhibited motor that is activated by heterotrimerization for intraflagellar transport. Curr Biol 30: 1160–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam LW, Wilson NF, Lefebvre PA (2007) A CDK‐related kinase regulates the length and assembly of flagella in Chlamydomonas . J Cell Biol 176: 819–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taschner M, Kotsis F, Braeuer P, Kuehn EW, Lorentzen E (2014) Crystal structures of IFT70/52 and IFT52/46 provide insight into intraflagellar transport B core complex assembly. J Cell Biol 207: 269–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taschner M, Lorentzen E (2016) The intraflagellar transport machinery. Cold Spring Harb Perspect Biol 8: a028092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taschner M, Weber K, Mourao A, Vetter M, Awasthi M, Stiegler M, Bhogaraju S, Lorentzen E (2016) Intraflagellar transport proteins 172, 80, 57, 54, 38, and 20 form a stable tubulin‐binding IFT‐B2 complex. EMBO J 35: 773–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toropova K, Zalyte R, Mukhopadhyay AG, Mladenov M, Carter AP, Roberts AJ (2019) Structure of the dynein‐2 complex and its assembly with intraflagellar transport trains. Nat Struct Mol Biol 26: 823–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhey KJ, Hammond JW (2009) Traffic control: regulation of kinesin motors. Nat Rev Mol Cell Biol 10: 765–777 [DOI] [PubMed] [Google Scholar]
- Williamson SM, Silva DA, Richey E, Qin H (2012) Probing the role of IFT particle complex A and B in flagellar entry and exit of IFT‐dynein in Chlamydomonas . Protoplasma 249: 851–856 [DOI] [PubMed] [Google Scholar]
- Wingfield JL, Mengoni I, Bomberger H, Jiang YY, Walsh JD, Brown JM, Picariello T, Cochran DA, Zhu B, Pan J et al (2017) IFT trains in different stages of assembly queue at the ciliary base for consecutive release into the cilium. Elife 6: e26609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingfield JL, Lechtreck KF, Lorentzen E (2018) Trafficking of ciliary membrane proteins by the intraflagellar transport/BBSome machinery. Essays Biochem 62: 753–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wren KN, Craft JM, Tritschler D, Schauer A, Patel DK, Smith EF, Porter ME, Kner P, Lechtreck KF (2013) A differential cargo‐loading model of ciliary length regulation by IFT. Curr Biol 23: 2463–2471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B, Zhu X, Wang L, Liang Y, Feng Q, Pan J (2017a) Functional exploration of the IFT‐A complex in intraflagellar transport and ciliogenesis. PLoS Genet 13: e1006627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Liang Y, Gao F, Pan J (2017b) IFT54 regulates IFT20 stability but is not essential for tubulin transport during ciliogenesis. Cell Mol Life Sci 74: 3425–3437 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Review Process File
Appendix
Movie EV1
Movie EV2
Movie EV3
Source Data for Appendix
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 5
Data Availability Statement
There are no primary datasets or computer codes associated with the study.


