Abstract
Simple Summary
The potential importance of germline genetic variation for identifying men at increased risk of prostate cancer has become increasingly recognised in recent years. We present an extensive review of the major developments in the identification of genetic loci, genes and individual variants associated with greater risk of prostate cancer, and what is currently known regarding whether these heritable prostate cancer risk factors can also inform likelihood of experiencing clinically significant rather than indolent forms of the disease. We finally discuss how these research discoveries might serve to inform clinical germline genetic testing guidelines and treatment options for prostate cancer in the future.
Abstract
Prostate cancer (PrCa) is a heterogeneous disease, which presents in individual patients across a diverse phenotypic spectrum ranging from indolent to fatal forms. No robust biomarkers are currently available to enable routine screening for PrCa or to distinguish clinically significant forms, therefore late stage identification of advanced disease and overdiagnosis plus overtreatment of insignificant disease both remain areas of concern in healthcare provision. PrCa has a substantial heritable component, and technological advances since the completion of the Human Genome Project have facilitated improved identification of inherited genetic factors influencing susceptibility to development of the disease within families and populations. These genetic markers hold promise to enable improved understanding of the biological mechanisms underpinning PrCa development, facilitate genetically informed PrCa screening programmes and guide appropriate treatment provision. However, insight remains largely lacking regarding many aspects of their manifestation; especially in relation to genes associated with aggressive phenotypes, risk factors in non-European populations and appropriate approaches to enable accurate stratification of higher and lower risk individuals. This review discusses the methodology used in the elucidation of genetic loci, genes and individual causal variants responsible for modulating PrCa susceptibility; the current state of understanding of the allelic spectrum contributing to PrCa risk; and prospective future translational applications of these discoveries in the developing eras of genomics and personalised medicine.
Keywords: prostate cancer, aggressive prostate cancer, prostate cancer susceptibility, prostate cancer genetics, genome-wide association studies, massively parallel sequencing studies
1. Introduction
Prostate cancer (PrCa) is the most frequently diagnosed cancer in males in Europe and North America and second most common worldwide, with over 1.4 million diagnoses recorded in 2020 [1,2]. Although a majority of patients present with indolent, slow developing disease, PrCa remains a substantial cause of mortality with over 375,000 deaths recorded worldwide in 2020. The five-year cause specific survival rate for men diagnosed with localised or regional PrCa is effectively 100%, however this deteriorates to only 30.1% in men with metastatic disease at the point of diagnosis [3,4].
Although several promising molecular and genomic biomarkers for PrCa diagnosis or management have been identified in recent years [5], prostate-specific antigen (PSA) remains the only biomarker to have been widely employed for PrCa detection to date. The ability of PSA to discriminate high-risk disease is however poor and it is therefore no longer widely advocated as a systematic screening tool [6]. Although modest evidence linking smoking [7] and possibly body mass index [8,9] to poorer prognosis in PrCa patients has been presented, few clear, modifiable lifestyle risk factors have been established for either disease development or progression. In the past decade, substantial progress has been made towards identifying heritable PrCa risk factors; however, the ability to discriminate patients or healthy men at greater risk of dying from PrCa remains modest. The identification of genetic risk factors predisposing towards more advanced clinical presentation of PrCa and more rapid disease progression would have the potential to facilitate targeted screening of individuals at higher risk of death from PrCa whilst concurrently reducing overtreatment of men with lower risk disease, or to inform decision making in treatment pathways [10].
No consistent, consensus definition of aggressive PrCa has thus far been adopted within the research or clinical settings, which can hinder comparability of studies to identify risk factors and uniformity of clinical treatment application. Prior to the advent of PSA testing, aggressive PrCa had been considered to encompass only cancers which had advanced beyond the prostate itself [11]. More recently, due to increasing diagnosis of men at younger ages who are presenting at earlier timepoints within their disease course, the definition of aggressive PrCa has routinely been expanded to encompass also men that have localised disease coupled with indicators reflective of higher risks of future progression to lethal phenotype. These updated classifications of aggressive PrCa may consider varying combinations of Gleason score (typically one of Gleason score ≥7, Gleason score ≥8 or Gleason grade group ≥3), T stage (generally either T4 or ≥T3), nodal invasion, metastatic spread, extreme PSA measurements, young age at diagnosis (generally defined as diagnosis at age < or ≤55 years, with ages 60 or 65 also frequently used as cut-offs), and death from PrCa. A recent analysis considering the sensitivity and positive predictive value of different definitions of aggressive disease with respect to discriminating patients that experience death from PrCa within 10 years of diagnosis has proposed the adoption of a standardised definition of aggressive PrCa in etiological research as being any one or more of stage T4 or N1 or M1 or Gleason score ≥8 disease at diagnosis [12].
2. Evidence of a Genetic Basis for Prostate Cancer
The most clearly established risk factors for PrCa are increased age, ethnicity and family history of PrCa and certain other cancers. Familial aggregation of PrCa is one of the strongest risk factors, and provided support for the likely existence of genetic risk factors shared among families. Men with one male first-degree relative (FDR; father or brother) diagnosed with PrCa themselves have an estimated relative risk of approximately 2.5, and risk of diagnosis with PrCa further increases for men with multiple affected FDRs and lower ages at their diagnoses [13,14,15]. Research comparing mono- and dizygotic twin pairs subsequently provided strong evidence for a substantial heritable component in PrCa development [16], believed to be higher than for any other form of common cancer, with the latest estimate of PrCa heritability at 58% and the role of genetic factors consistently high across age groups [17].
Evidence has also been presented for clustering of aggressive clinical presentation of PrCa within families, suggestive for heritability of aggressive PrCa phenotypes. A number of studies from the Swedish population have demonstrated increased risk of high grade PrCa observed among brothers of cases with high grade disease [18], with greater concordance of high risk disease between monozygotic twins [18], familial clustering of fatal PrCa [19], and concordance of good or short PrCa specific survival times between affected fathers and sons [20,21]. A large study that calculated relative risks for lethal PrCa in the United States based on the number of affected FDRs estimated increased relative risks ranging from 2.49 for males with 1 affected FDR to 5.30 for ≥3 affected FDRs, with higher risk also observed for greater numbers of affected second- and third-degree relatives [22]. In the Swedish population, absolute risk of PrCa was calculated to be 12.9% by age 75 and 5.2% for high-risk disease; with these estimates rising with increasing family history of PrCa to 30.3% and 8.9%, respectively, for men with one affected brother, and to 63.6% and 20.5% for men with two affected brothers and an affected father [23].
PrCa also clusters in families with a strong family history of other cancer types, especially breast and ovarian cancer [24,25,26], whilst PrCa is now also widely regarded as among the Lynch syndrome spectrum of cancers [27,28,29,30]. Hereditary breast and ovarian cancer syndrome is frequently associated with germline mutations in the BRCA1 and BRCA2 genes, whilst Lynch syndrome is categorised by germline mutations in DNA mismatch repair genes. A strong family history of PrCa remains a more effective indicator of PrCa risk than family history of other cancer types however, especially with respect to early-onset and lethal disease [31].
PrCa incidence and mortality rates differ substantially across ethnic groups, with the greatest burden of PrCa and highest mortality rates afflicting men of African ancestral origin and lowest observed in men of Asian ancestry [32]. In the United States, PrCa incidence rates are estimated to be approximately 1.76-fold higher in African American men than Caucasians, with relative mortality rates from PrCa elevated 2.20-fold among black men [33]. Whilst these disparities may in part result from socioeconomic, cultural and healthcare availability or treatment preference factors, differences in predisposition towards PrCa development arising through varying allelic frequencies of heritable genetic risk factors between ancestral groups are also likely [34]. Differences in PrCa incidence rates are also observed between men of the same race and ethnicity residing in different countries however, suggesting that these also occur in part due to environmental factors [32].
Age-specific PrCa incidence rates rise steeply after age 50, with a peak incidence rate per 100,000 men in the UK between 2015 and 2017 among the 75–79 age group [35]. The incidence of younger onset PrCa (aged ≤55 years) is however increasing and represents a greater proportion of total new diagnoses [36]. Early-onset PrCa patients generally also demonstrate poorer cause-specific survival than men diagnosed at older age ranges and are likely to represent a cohort enriched for genetic variants that increase susceptibility to PrCa, and which could benefit substantially from well informed screening and genetic testing initiatives [36,37].
3. Initial Approaches for the Identification of PrCa Susceptibility Genes
Substantial advances in the technologies available for application in genomics research have been made during the past quarter of a century, especially since the year 2000. Prior to these developments however, many early efforts to identify PrCa susceptibility genes revolved around genetic association studies examining biologically plausible candidate genes [38]. These approaches demonstrated modest success, although were frequently limited by factors including sample size constraints inherent to the available lower-throughput techniques of the era, population stratification, the genotyping only of specific founder mutations within a cohort rather than screening of full coding sequences of genes, and publication bias against the reporting of negative results. The CAG and GGN trinucleotide repeat polymorphisms within the transactivation domain of the androgen receptor gene are one example of an extensively studied candidate, however evidence for association between repeat length and PrCa risk has been inconsistent and modest [39,40,41,42,43,44]. Several other variants, genes or pathways were also examined under this approach, including the androgen [45] and oestrogen [46] metabolic pathways, and TP53 gene [47]; again with inconsistent evidence of association and magnitude or direction of effect frequently reported between studies. Another widely investigated class of candidate was DNA repair genes, for which associations with overall, aggressive or young onset disease have been reported for a handful of genes including BRCA2 [48,49,50,51,52], BRCA1 [53,54,55], CHEK2 [56], and NBN [57,58].
Techniques such as genetic linkage or admixture analyses also permitted genome level scans and therefore did not require a priori hypotheses of the identity or location of candidate PrCa susceptibility genes. A seminal example of the isolation of a disease gene through linkage methods in a common, complex disease was the mapping in 1990 of a major breast and ovarian cancer predisposition locus to Chr17q21 [59,60], later traced to the BRCA1 gene using positional cloning [61,62]. Linkage studies for non-Mendelian traits in humans are an arduous process, seeking to identify shared stretches of DNA between genetic markers that cosegregate among affected individuals within large family units in which multiple cases of a particular disease cluster [63,64]. Statistically significant linkage intervals may also be long and contain large numbers of candidate genes. Linkage signals within families may also potentially be obscured due to the occurrence of phenocopies (sporadic occurrence of the disease in individuals that did not inherit the susceptibility allele), or by incomplete and age-dependent penetrance of disease among risk allele carriers; therefore these studies are primarily powered for the detection of high penetrance variants. Linkage analyses for PrCa were typically performed using large families that meet criteria for hereditary prostate cancer [65] and from which multiple PrCa patients and unaffected men were willing to contribute DNA. Pedigrees enriched for clustering of clinically significant or young-onset disease were also prioritised, in order to maximise the prospect of identifying genes influencing poorer prognosis PrCa phenotypes and minimise the likelihood of confounding through instances of sporadic, nonhereditary PrCa. In contrast to genetic linkage analyses, admixture mapping is performed using a study cohort that contains admixture from two or more ancestral populations that experience differences in rate of a phenotype or disease, under the hypothesis that casual variants affecting the trait will occur more frequently on segments of DNA inherited from the higher risk ancestral population [66]. The resolution achieved by admixture mapping approaches is substantially greater than that of linkage studies, reducing the likely number of candidate genes at an identified association.
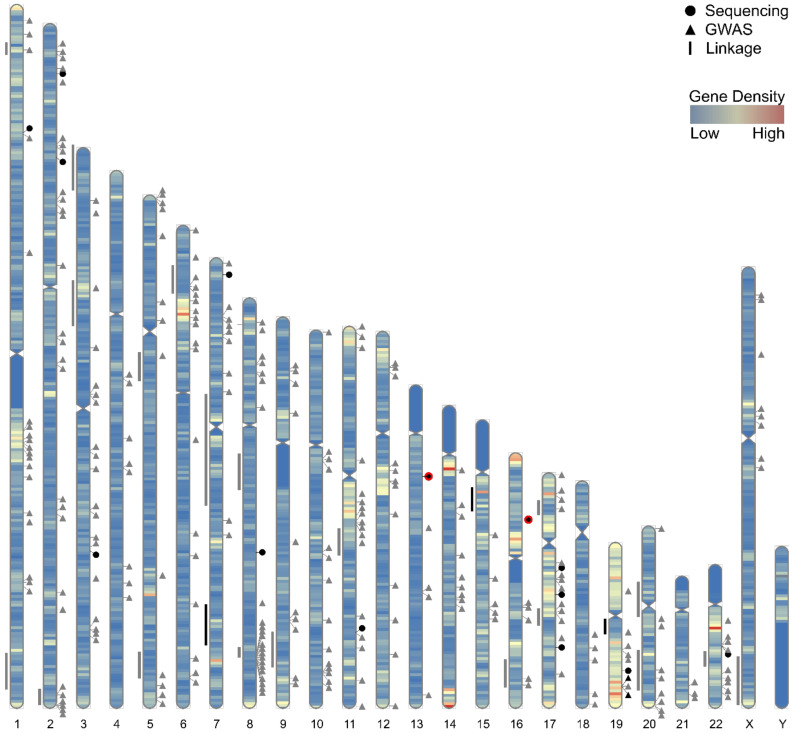
The first putative high risk PrCa locus identified through linkage, located at Chr1q24-25 and denoted HPC1, was reported in 1996 [67]. This signal was observed primarily within families with multiple early-onset cases [68] and subsequently assigned to the RNASEL gene [69]; however, replication of the HPC1 locus in independent cohorts has proven inconsistent [70,71,72,73,74,75,76,77]. Subsequent proposed PrCa susceptibility loci reported in linkage or admixture mapping analyses include Chr1p36 [78,79,80], Chr1q42.2-43 [73,81,82], Chr2q37.3 [83,84,85], Chr3p14 [86], Chr3p25-26 [87,88], Chr5q11-12 [76,89], Chr5q35 [76,89,90], Chr6p22.3 [89,91], Chr7q11-21 [92], Chr7q31-33 [90,93,94,95], Chr8p22-23 [79,96,97,98], Chr8q12-13 [76,89], Chr8q24 [90,99,100], Chr9q34 [101,102], Chr11q14 [88,89,91], Chr15q11-14 [76,103,104,105], Chr16q23 [84,101], Chr17p12 [106,107,108,109], Chr17q21-22 [83,105,110], Chr19q12-13 [93,111,112,113], Chr20p11-q11 [89,91], Chr20q13 [114,115,116], Chr22q12.3 [76,89,117,118,119,120], and ChrXq27-28 [121,122,123,124] (Figure 1). Several of these loci also demonstrated greater evidence for linkage in families with higher numbers of affected relatives or containing multiple early-onset cases, whilst the loci at Chr7q31-33, Chr15q and Chr19q were reported to associate with risk of more aggressive disease. Evidence from validation studies and gene mapping approaches again proved conflicting for many of these loci however [101,125,126]; therefore, although rare higher penetrance risk alleles at these loci may be present within specific families, at population levels the majority of these loci appear unlikely to account for a large proportion genetic susceptibility to PrCa or aggressive disease.
Figure 1.
Karyogram depicting the approximate locations of candidate PrCa risk loci reported through linkage, GWAS and sequencing study approaches. To the right of the chromosome ideogram, triangle symbols indicate the position of 269 reported independent GWAS index variants [127] and circle symbols the locations of DNA repair genes reported as a potential candidate for risk in two or more PrCa sequencing studies (listed within Table 1). To the left of the chromosome ideogram, line symbols show the approximate intervals of linkage peaks according to representative markers for the region or cytogenetic band co-ordinates. Grey coloured symbols indicate no or limited evidence for association with risk of aggressive PrCa, the black colour denotes moderate or conflicting evidence for risk of aggressive disease and black symbols with a red border signify stronger evidence for a contribution towards poorer prognosis phenotypes. The karyogram is overlaid with a heat map depicting gene density across the human genome in 1 Mb windows and was generated using RIdeogram [128] and custom plotting functions.
One notable success for linkage-based analyses in PrCa families, however, was the robust identification of HOXB13 as the first gene accounting for a substantial fraction of familial PrCa. Evidence for linkage on chromosome 17q was first reported based on 175 pedigrees [110], and subsequently refined to a 10cM interval (approximately 15.5Mb) at Chr17q21-22, containing 202 candidate genes [129]. In a seminal report, Ewing et al. performed targeted sequencing of the coding regions of these genes in 94 unrelated men from hereditary PrCa families; identifying a recurrent nonsynonymous HOXB13 variant (rs138213197/G84E) in four of these probands. This variant was observed to cosegregate in all 18 PrCa cases with DNA available within these four families and was also enriched among cases in a replication cohort, particularly among men with either or both young-onset PrCa or a family history of PrCa [130]. HOXB13 G84E was subsequently confirmed as a moderate penetrance PrCa susceptibility variant in European ancestry populations through a number of independent studies, with consistent evidence demonstrated for strong associations with early-onset and familial PrCa, but importantly no evidence for increased risk of poorer prognosis disease in mutation carriers [131,132,133,134,135,136,137,138,139,140]. HOXB13 G84E mutations are estimated to be present in up to 5% of European ancestry families with hereditary PrCa [140], whilst average estimated risks of developing PrCa for heterozygous male carriers are approximately 17% by age 65 rising to 62% by age 85, and further increasing with stronger family history of PrCa [141]. The HOXB13 G84E mutation is found almost exclusively in European ancestry populations [131,132,133,140,142]. Other recurrent HOXB13 mutations have, however, subsequently been observed in Chinese [143] and Japanese [144] men with PrCa, and additional rare nonsynonymous mutations within African ancestry [130] and Portuguese [145] PrCa families, indicating a contribution by germline HOXB13 mutations in susceptibility to PrCa in diverse populations and ethnic groups.
Besides HOXB13 however, linkage approaches were largely unsuccessful at definitively isolating higher penetrance PrCa susceptibility genes attributable to a substantial fraction of disease heritability among populations. These outcomes implied that the allelic spectrum of PrCa risk variants could therefore more closely resemble a ‘Common Disease, Common Variant’ model, under which larger numbers of low penetrance but frequently occurring variants are the main contributors to genetic risk of the disease, rather than a ‘Common Disease, Rare Variant’ hypothesis, in which rare, high penetrance variants are the major initiator. Although soon to be superseded by emerging higher throughput approaches for common variant association analyses, linkage and admixture mapping based techniques also successfully identified the first evidence for common variation conferring susceptibility to PrCa, at the Chr8q24 locus [99,100].
4. Genome-Wide Association Studies for Common, Low Penetrance PrCa Susceptibility Loci
The completion of the Human Genome Project [146,147] alongside catalogues of common human variation and linkage disequilibrium patterns [148,149] heralded a significant development for research into genetic susceptibility to common diseases. These resources facilitated simultaneous, unbiased and lower cost association testing of large numbers of variants throughout the genome in the form of genome-wide association studies (GWAS) [150]. The construction of SNP-arrays to interrogate large numbers of genetic variants [151], alongside imputation panels and software for the inference of additional nongenotyped variants [152], were accompanied by development of robust methodologies for handling population stratification [153], establishing statistical significance [154,155] and the meta-analysis of data from discrete studies [156]. Although many formative GWAS were underpowered to detect variants with modest effect sizes or minor allele frequencies (MAF) [157], steadily increasing sample sizes have led to the identification of large numbers of phenotype associated loci, the majority of which had not previously been indicated through alternative approaches [158,159] (Figure 1).
The first GWAS conducted for PrCa confirmed the risk locus initially identified through linkage and admixture mapping at Chr8q24, and identified additional independent risk signals within this locus [160,161,162,163,164]. Chr8q24 remains the major locus contributing to PrCa susceptibility [165,166] across ancestral groups [127], with multiple independent risk signals discovered within the locus, including rare or low frequency moderate penetrance susceptibility variants that are population specific or enriched [136,167,168,169]. The Chr8q24 locus is also established as a significant contributor to familial PrCa [167,170,171] and a diverse spectrum of other cancer types [172]. Many of these Chr8q24 risk signals are located a substantial distance from genes, however a number of plausible candidate variants within regulatory elements have been identified [173,174]. These are implicated in regulating the expression of multiple protein coding genes including the MYC proto-oncogene [175,176,177,178], POU5F1B transcriptional activator [165,179] and FAM84B gene [113,180], and long noncoding RNAs [181] including PVT1 [178,182], PCAT1 [178,183] and PRNCR1 [178,184]. This indicates that PrCa risk modulation by the Chr8q24 locus is likely to be influenced through a diverse and complex range of biological mechanisms.
A number of subsequent European ancestry GWAS and meta-analyses of increasingly large sample sizes have reported rapidly expanding numbers of loci associated with PrCa risk outside of the Chr8q24 region, many of which loci also contained multiple independently associated risk signals [185,186,187,188,189,190,191,192,193,194]. As larger sample sizes are employed, the novel loci identified generally exhibit diminishing index variant effect size and/or MAF, however, are identified in greater numbers. Several of these PrCa risk loci contain genes heavily linked to prostate function, or whose expression is enriched in the prostate relative to other tissues [195]; however, the biological mechanisms underpinning the majority of PrCa risk loci identified through GWAS generally remain poorly characterised at present. Functional validation of a small number of candidate causal variants has been performed to date [169,175,178,196,197,198,199,200,201,202,203,204,205], whilst fine-mapping of association signals and in silico annotation procedures have also helped to narrow the pool of likely candidate causal variants and identify prospective target genes and biological mechanisms that may give rise to differential risk [173,206,207,208,209]. A related methodology, transcriptome wide association studies (TWAS), in which GWAS summary statistic datasets and SNP-gene expression data from a reference panel are integrated, for the purpose of imputing gene expression data into phenotyped datasets which lack directly measured expression data, has also been employed in order to identify prospective gene–trait associations and prioritise putative candidate genes at many GWAS loci [210]. For PrCa, individual large TWAS have so far reported between 38 and 217 genes significantly associated with PrCa risk [211,212,213], including a number of the most strongly implicated functional candidates identified through other approaches in addition to candidate genes not previously proposed. PrCa TWAS have also identified a handful of additional novel candidate PrCa predisposition regions and corresponding candidate genes that had not previously been implicated in disease risk through significant GWAS associations [211,212].
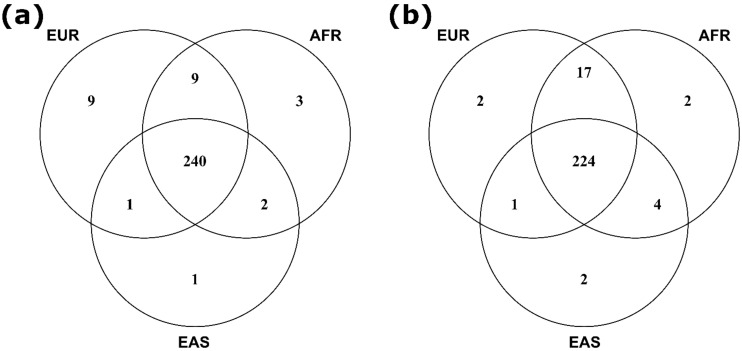
The majority of samples included in published PrCa GWAS to date are of European ancestry; however, a number of studies have also been conducted for men of African [214,215,216,217], Japanese [180,218,219,220], Chinese [220,221], and Latino [222] ancestries. Despite currently unequal power across ancestral populations studied, many susceptibility loci reported as statistically significant in large European ancestry GWAS have also replicated at genome-wide significance in additional populations, whilst further risk signals specific to or that have substantially enriched risk allele frequency within non-European ancestral populations have also been identified. The remaining loci discovered in European ancestry GWAS but which have not yet formally validated across other ancestries have also generally demonstrated consistent directionality of effect in additional populations [223,224,225], indicating a strong likelihood that common functional causal alleles shared across multiple populations at varying allelic frequencies will underlie the majority of GWAS loci reported at this point in time. These observations facilitated the aggregation of samples from multiple ancestries to perform larger multiethnic meta-analyses, which identified additional cross-ancestry risk loci not detectable through existing sample cohort sizes from any individual ancestral population [194]. The largest meta-analysis for PrCa risk so far conducted, comprising a total of 107,247 cases and 127,006 controls from European, African, Asian, and Hispanic ancestry populations, although predominantly European ancestry men, has reported evidence for 269 independent PrCa susceptibility signals (Figure 1), of which 183 had been identified through previous GWAS [127]. These 269 PrCa risk signals are situated within 176 distinct genomic loci when defined as >800kb from any neighbouring independently associated index variant, and the overwhelming majority of index variants are common, with their risk allele frequencies ≥5% in multiple ancestral populations (Figure 2).
Figure 2.
Venn diagrams comparing the proportion of the 269 independent GWAS index variants reported to date that were (a) present at ≥1% risk allele frequency, and (b) present at ≥5% risk allele frequency in European (EUR), African (AFR) and East Asian (EAS) ancestral super-populations.
A recent report estimated the genetic architecture of PrCa to be modulated by approximately 4500 common susceptibility variants, of which a greater proportion of risk variants confer larger effect sizes than observed for other cancer sites with comparably powered GWAS available [226]. This analysis indicates that large numbers of PrCa risk loci potentially remain to be discovered; albeit predominantly with diminishing per variant effect sizes or risk allele frequencies. The same simulation also estimated that approximately 250,000 cases and an equal number of controls would be required for the identification of variants explaining approximately 70% of the GWAS heritability at the standard genome-wide significance threshold, and 500,000 of each cohort to achieve 80% of heritability [226]. According to heritability estimates, this study also estimated a maximum theoretical achievable relative risk due to common variation for a man in the top risk percentile of approximately 5-fold greater than a man at average risk [226]; a relative risk level comparable to carriers of many monogenic susceptibility mutations for diseases, but substantially more prevalent within the general population [227].
Although GWAS have identified a large number of loci associated with risk of developing PrCa and shed light on the biological underpinnings of disease development, the ability of these loci to inform clinical management pathways remains unclear. In particular, whether a specific subset of loci or the cumulative polygenic burden of inheriting greater numbers of risk loci are predictive for risk of developing aggressive disease. A number of studies have reported potential susceptibility loci for aggressive disease based on analyses of cases with aggressive PrCa versus controls [161,228,229,230,231,232,233,234]; however, these comparisons do not definitively demonstrate that a risk locus is associated specifically with the aggressive disease state rather than all cancers. Indeed, case-only analyses comparing PrCa patients with aggressive and low risk phenotypes have so far been unable to provide support for a widespread ability of common risk loci identified in GWAS for overall PrCa to discriminate between patient outcomes [235,236,237].
Case-only GWAS have to date reported at a genome-wide significant threshold two loci associated with Gleason score [238], one with more aggressive phenotype [239] and another with shorter PrCa-specific survival [240]. These loci have not separately associated with risk of overall PrCa through case–control GWAS and could therefore represent disease state specific prognostic markers warranting further investigation. The AOX1 gene locus associated with PrCa survival time is particularly noteworthy, as the index variant also associated with AOX1 expression and AOX1 levels in turn with biochemical recurrence of PrCa [240], whilst the methylation status of this gene had previously been suggested as a candidate biomarker for PrCa outcomes [241,242]. These prospective loci for risk of aggressive disease however remain to be confirmed in independent studies and ancestral populations, and their clinical utility needs to be fully established. A large case-only PrCa GWAS excluding patients with intermediate disease aggressiveness observed evidence for association between aggressive status and variation at the KLK3 locus, however, no other common variants were associated, including those previously reported with respect to overall disease or phenotypic indicators of poorer prognosis [Saunders et al., Manuscript in preparation]. This is consistent with previous reports that the KLK3 GWAS SNP rs2735839 is associated with Gleason score in addition to overall disease risk [243,244,245,246,247]. In all of these studies, the risk allele of rs2735839 for overall PrCa was however overrepresented in patients with nonaggressive disease and thus inversely associated with aggressive status; leading to caution that the associations observed may relate to detection bias of indolent disease due to raised PSA expression in carriers of this genotype [246,247]. In independent study cohorts, the association between KLK3 and less aggressive disease was however not substantially attenuated by adjusting for PSA levels at diagnosis [Saunders et al., Manuscript in preparation] or remained associated with aggressive status among only patients with low PSA levels from two separate ancestral populations [248]. The full role of KLK3 variation in PrCa susceptibility, risk of aggressive disease and serum PSA levels may therefore warrant more extensive investigation, especially in prospective or PSA naïve cohorts, and may have the potential to enable the identification of a subset of individuals at lower risk of poor prognosis disease who could benefit from less interventionist treatment options.
Whilst these initial case-only reports imply that caution is warranted regarding the potential for GWAS loci to be able to accurately stratify PrCa patients more likely to develop clinically significant disease, increasing evidence supports the improving ability of genetic risk scores (GRS; also frequently referred to as polygenic risk scores/PRS) incorporating ever larger numbers of established susceptibility variants to identify a population subset at greater lifetime risk of diagnosis with PrCa of any severity [127,249,250,251,252,253,254,255,256,257]. This suggests the prospect that future PrCa screening programs could be targeted towards only a specific segment of the population at the greatest risk, to facilitate earlier identification of the majority of patients who will progress to develop poorer prognosis disease whilst potentially concurrently reducing levels of overdiagnosis of men with indolent disease [258,259,260]. GRS for complex common diseases comprise the sum of the germline risk alleles for the disease that an individual possesses, weighted by the effect estimates for each risk variant, and are used to estimate the individual’s lifetime risk for developing the disease [261]. GRS have to date primarily been developed using risk variant catalogues and effect estimates compiled from large European ancestry GWAS discovery populations, and perform less optimally when applied to populations with divergent ancestry [262]. The use of trans-ethnic variant discovery and effect size estimation approaches has, however, demonstrated promise for improving cross-population risk prediction performance, with the latest 269 variant PrCa GRS established through a multiethnic meta-analysis framework reporting a mean GRS 2.18-fold higher for men of African ancestry and 0.73-fold lower for men of East Asian ancestry in comparison to men of European ancestry [127].
Although the ability of GRS to predict disease status within cohort studies has been demonstrated for various traits, their potential clinical utility to inform screening decisions for individual members of the population does however largely remain to be established at this point in time. Their potential consideration for implementation as a prospective risk-profiling tool prior to screening for PrCa may, however, soon become warranted. A polygenic hazard score (PHS) has demonstrated initial promise for the detection of clinically significant PrCa at younger age among higher PHS percentiles [263,264], including in men from diverse ancestral groups [265]; however, the PHS associated with PrCa of any severity and was not able to differentiate specifically for the subset of men who develop clinically significant disease. Coupled with advances in prostate imaging techniques, approaches of this nature do, however, hold promise for facilitating personalised, genetically informed screening decisions to enable early detection of cancers in men at higher risk of developing PrCa of any severity, to be followed by the application of appropriate treatment decisions after diagnosis. However, whilst initial studies to assess the feasibility of genetics-informed screening approaches at the population level have been undertaken, further evaluation and refinements are likely to be required before GRS can be widely integrated into public healthcare systems; particularly in respect to equitable applicability across diverse ethnicities and ascertaining appropriate thresholds for benefit–harm trade off and cost effectiveness.
5. Sequencing Studies for Rare, Moderate Penetrance PrCa Susceptibility Genes
Although many common loci have been identified which contribute substantially towards PrCa risk, rare variation is also estimated to play an important role in PrCa heritability [266], especially in men of African ancestry [267]. A number of next-generation sequencing studies reporting rare germline mutation findings in PrCa patients have been conducted in recent years. However, the overall number of samples included in these studies to date remains a fraction of those in GWAS, whilst due to the low allelic frequency of variation primarily examined, large sample sizes are required to achieve sufficient statistical power for moderate penetrance variant detection in complex diseases [268]. To increase power, most sequencing studies, therefore, primarily examine rare protein altering variant frequencies collapsed at the gene or gene-set rather than individual variant level, for which a variety of statistical methodologies have been developed [269]. Many studies also incorporate an extreme phenotype sampling strategy in order to maximise power with limited available sample size [270]. For PrCa, many sequencing studies to date reporting prospective susceptibility genes have taken the form of tumour-sequencing studies that also reported germline findings from matched normal DNA, which were conducted on case cohorts numbering in the hundreds, contained no control cohort and usually specifically examined metastatic castrate resistant (mCRPC) PrCa cases [271,272,273,274,275]. Other similarly sized studies examining germline DNA exclusively have reported mutation frequencies for overall [276,277] and familial [278,279] PrCa, men with PrCa alongside additional primary tumour types [280], or compared rates between aggressive and nonaggressive phenotypes [281,282]. More recently, larger association studies comparing mutation frequencies among cohorts of a few thousand samples have also begun to be completed, predominantly interrogating the coding regions of panels of prospective candidate genes [144,283,284,285]. To date, published PrCa sequencing studies have, however, predominantly either sequenced, analysed or reported only findings primarily related to DNA repair genes, and therefore these remain at present the only class of gene widely scrutinised for rare germline variation in the PrCa setting and for which findings for individual genes may be comparable across a number of separate studies and cohorts.
Sequencing studies of PrCa patients with aggressive disease have consistently reported elevated mutation rates for the BRCA2 gene, confirming earlier observations linking BRCA2 mutation carriers to more aggressive phenotypes through other approaches [286,287,288,289,290,291]. Several additional DNA repair genes have also been implicated in multiple sequencing studies as prospective moderate penetrance PrCa susceptibility genes warranting further investigation in larger sample cohorts, especially ATM, BRCA1 and PALB2 (Table 1, Figure 1). Although these genes are widely included in PrCa sequencing panels in both research and clinical settings, their contribution towards PrCa susceptibility and risk of aggressive disease await definitive confirmation, whilst additional genes associated with risk may remain to be identified through larger study sizes and broader sequencing panels. A recent case-only study comprising 2770 aggressive and 2775 nonaggressive PrCa cases reported statistically significant evidence for substantially increased risk of aggressive disease among germline BRCA2 and PALB2 mutation carriers, with ATM also nominally associated [284], corresponding with observations of high combined germline and somatic mutation frequencies for these genes among mCRPC patients [292]. Pathogenic germline PALB2 mutations were present at a far lower rate than BRCA2 in this study cohort, however, were substantially more enriched among aggressive, metastatic and lethal cases [284]. A number of other studies have also linked ATM mutations to poorer prognosis PrCa phenotypes [281,282,293,294]; although the largest retrospective ATM sequencing study to date comprising 5560 cases and 3353 controls of European ancestry observed only limited support for association with aggressive disease but strong evidence for increased risk of overall PrCa among ATM mutation carriers [295].
Table 1.
DNA repair genes described as candidate PrCa susceptibility and/or poor prognosis genes in more than one next generation sequencing study.
| Gene | Chromosome | Reporting Studies (Reference Number) |
|---|---|---|
| ATM | 11 | [144,271,275,276,277,278,281,282,283,284,285,295,296] |
| ATR | 3 | [271,275,281] |
| BRCA1 | 17 | [271,277,278,282,283] |
| BRCA2 | 13 | [144,271,275,277,278,281,282,283,284,285,296] |
| BRIP1 | 17 | [271,278] |
| CHEK2 | 22 | [271,277,278,283] |
| ERCC2 | 19 | [281,283] |
| GEN1 | 2 | [271,283] |
| MSH2 | 2 | [271,277,283] |
| MUTYH | 1 | [275,277,278] |
| NBN | 8 | [271,281,283,285] |
| PALB2 | 16 | [271,277,278,281,284,285] |
| PMS2 | 7 | [271,277,278,281] |
| RAD51D | 17 | [271,281] |
PrCa sequencing studies reported to date have primarily been performed using men of European ancestry; however, large panel sequencing studies have also been conducted in African ancestry [285] and Japanese populations [144]. These studies have provided supporting evidence for pan-ethnic contributions towards PrCa risk for particular genes, especially ATM and BRCA2; however, ethnic specific differences in mutation carrier rates at individual genes were also observed, indicating that the allelic frequency spectrum of moderate penetrance PrCa risk genes could differ substantially across ancestral groups [297]. A sequencing study of aggressive PrCa cases and disease-free controls also implicated rare variants in the TET2 gene, a locus previously associated with overall PrCa through GWAS [185], as a prospective susceptibility gene for aggressiveness in African American men, with 24.4% of aggressive cases and only 9.6% of controls carrying a rare deleterious TET2 variant [298]. However, the association between deleterious germline variation and aggressive disease was not observed in the European ancestry cohort of the reporting study and remains to be validated in external cohorts.
Whilst the PrCa sequencing studies conducted to date have achieved some degree of success in identifying a small number of genes linked to substantially increased risks of PrCa onset or poorer prognosis disease phenotypes, and a handful of additional candidates warranting further investigation, the majority of studies were not designed or sufficiently powered to provide accurate risk estimates for individual genes. A recent prospective study of PrCa risk for male BRCA1 and BRCA2 carriers estimated standardised incidence ratios of 2.35 and 4.45, respectively, in addition to a stronger association with higher Gleason score and a standardised mortality rate of 3.85 for BRCA2 carriers [299]. A meta-analysis of mutation data for ATM has also quantified likely pathogenic variation in this gene as a moderate penetrance contributor towards PrCa susceptibility, with an odds ratio of 4.4; however, evidence of association with aggressive or younger onset disease was less distinct [295]. Larger sequencing studies or meta-analyses interrogating broader panels of genes would be required in the future to clarify the definitive set of genes linked to higher risks of developing PrCa and enable accurate quantification of risk experienced by mutation carriers of specific genes. The growth of national Biobanks may in the future provide convenient and cost effective means to validate results from germline sequencing studies, whilst large aggregate sequencing data resources such as The Genome Aggregation Database (gnomAD) [300] also allow efficient comparison of variant frequencies across populations. Caution should however be exercised with regard to the direct inclusion of external cohorts alongside internally sequenced samples in rare variant association studies, especially their application specifically as external control cohorts for case-only sequencing data, due to the potential for artefactual results arising from differences in sequencing depths, QC procedures or population stratification between datasets. A role for linkage analyses for the interrogation of rare variation within PrCa family units may also re-emerge as whole-genome sequencing becomes more widely adopted [301].
6. Translational Potential of Germline PrCa Susceptibility Variation and Conclusions
Evidence exists for a substantial heritable component for overall PrCa risk, and familial clustering of high risk and fatal PrCa phenotypes. A combination of common and rare variants is likely to influence risk of PrCa, with common variants a substantial contributor at the population level and rare variants important within specific families or sub-groups. Current evidence does not support common variants contributing substantially towards risk of aggressive disease individually or cumulatively, however; with this class of variation appearing primarily to be a substantial driver of initiation of tumorigenesis but subsequently having limited influence on progression to aggressive phenotypes. Whilst rare variant studies have to date been underpowered, a handful of DNA repair genes have been implicated as prospective risk factors for predisposition towards development of aggressive phenotypes among PrCa cases who are mutation carriers.
Germline testing for rare moderate penetrance pathogenic variants in specific genes is becoming an increasingly important focus of PrCa management and treatment, with the additional promise of guiding tailored therapeutic interventions appropriate for targeting specific molecular vulnerabilities in individual patients’ tumours. Although few genes have been conclusively identified to date in which rare variants confer higher risk of developing aggressive PrCa, as further larger, cross population sequencing studies and meta-analyses are performed, additional genes for which mutation carriers experience greater risk of aggressive disease and/or more favourable response to particular treatment options are likely to be established [302]. At present, germline testing in relation to PrCa is recommended primarily to inform treatment options or clinical trial eligibility for patients with metastatic or locally advanced disease, screening of PrCa patients or healthy men with a family history suggestive of hereditary PrCa, active surveillance decisions, or for patients with Ashkenazi Jewish ancestry [303,304]. Genes currently widely advocated for definite inclusion or consideration of inclusion in germline PrCa sequencing panels in one or more of these contexts are ATM, BRCA1, BRCA2, DNA mismatch repair genes involved in Lynch syndrome (especially MSH2, however testing of MLH1, MSH6, PMS2 and potentially EPCAM is also generally advised) and HOXB13. The latest National Comprehensive Cancer Network (NCCN) guidelines for germline testing also propose screening CHEK2 and PALB2 as part of their minimum recommended predisposition gene panel [304]. The tumour suppressor gene TP53 plus additional DNA repair genes including BRIP1 and NBN have also been proposed for inclusion on screening panels, but await more definitive evidence to achieve consensus of utility [303]. In patients with metastatic PrCa, among this gene panel for germline screening, ATM, BRCA1, BRCA2 and potentially other DNA repair genes may inform response to PARP inhibitors [296,305,306], BRCA1, BRCA2 and other DNA repair genes sensitivity to platinum chemotherapy [307,308], and DNA mismatch repair genes response to anti-PD-1 immunotherapy [309,310]. Investigation of whether carriers of mutations currently considered actionable specifically in the treatment of metastatic PrCa would also benefit from earlier treatment with targeted therapies prior to progression to incurable metastatic phenotypes may, therefore, also be warranted.
Although family history of the disease is usually the main reason for genetic testing of men without a diagnosis of PrCa, an appreciable proportion of PrCa patients without a family history sufficient to meet NCCN guidelines for germline genetic testing have also been demonstrated to carry rare germline putative PrCa susceptibility variants [277]. Even greater numbers of men towards the upper extremity of common variant GRS distributions may also experience similarly elevated levels of risk to carriers of mutations in moderate penetrance genes [226,227]. Identification of germline variation that modulates PrCa risk therefore holds promise for informing targeted screening programs to facilitate earlier identification of tumours. If coupled with screening for genes specifically linked to higher risks of aggressive disease and appropriate treatment options for early stage disease, such as active surveillance, these insights could simultaneously enable improved survival of patients who would progress towards poorer prognosis phenotypes, alongside reductions in overtreatment of men with clinically insignificant disease. At present, no germline testing guidelines incorporate the use of GRS approaches; however, owing to the common nature of the underlying individual variants of which they are composed, at this point in time this class of variation has actually been more rigorously statistically evaluated for association with phenotypic traits in the research setting than has been the case for many genes in which rare variants are expected to cause non-Mendelian diseases. Given the greater number of men who may experience equivalent levels of risk arising through multiple common, low penetrance variants to those men that are carriers of rare moderate penetrance PrCa susceptibility mutations, and the markedly cheaper cost of genotyping common polymorphisms in comparison to sequencing of gene panels to screen for rare pathogenic mutations, further evaluation of whether GRS may now have developed to sufficiently informative levels to warrant incorporation as a consideration within PrCa germline genetic screening guidelines may soon become appropriate.
The majority of samples included in studies investigating risk factors for PrCa to date have been from populations of European ancestry. However, given differing allelic architecture and frequencies of both rare and common variants between populations, in addition to the higher incidence and poorer prognosis of PrCa among men of African descent, reducing under-representation of additional ethnicities in PrCa research remains an unmet requirement in order to ensure applicability of discoveries across populations and pan-ethnic access to healthcare improvements [311,312,313]. Enabling ubiquitous access to germline genetic testing across national and global healthcare systems would however continue to represent a substantial challenge.
Acknowledgments
We would like to acknowledge NIHR funding to the Biomedical Research Centre at The Royal Marsden and The Institute of Cancer Research.
Author Contributions
Conceptualisation, E.J.S., Z.K.-J. and R.A.E.; writing—original draft preparation, E.J.S.; writing—review and editing, E.J.S., Z.K.-J. and R.A.E.; supervision, Z.K.-J. and R.A.E.; project administration, Z.K.-J. and R.A.E.; funding acquisition, Z.K.-J. and R.A.E. All authors have read and agreed to the published version of the manuscript.
Funding
E.J.S. is funded by CRUK grant CRM077 and PCUK. Z.K.-J. is funded by CRUK grant CRM143.
Conflicts of Interest
R.A.E. has received speaker honoraria from GU-ASCO, The University of Chicago, ESMO (paid by Bayer and Ipsen) and The Royal Marsden NHS Foundation Trust (with support from Janssen). R.A.E. is a member of the AstraZeneca UK Limited Prostate Dx Advisory Panel external expert committee. No organisation had any role in the decision to publish this review or in the writing of the manuscript.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: Globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Global Cancer Observatory. [(accessed on 14 December 2020)]; Available online: https://gco.iarc.fr/today/online-analysis-table.
- 3.Howlader N., Noone A.M., Krapcho M., Miller D., Brest A., Yu M., Ruhl J., Tatalovich Z., Mariotto A., Lewis D.R., et al. Seer Cancer Statistics Review, 1975–2017. National Cancer Institute; Bethesda, MD, USA: [(accessed on 14 December 2020)]. Available online: https://seer.cancer.gov/csr/1975_2017/ [Google Scholar]
- 4.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2020. CA Cancer J. Clin. 2020;70:7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 5.Koo K.M., Mainwaring P.N., Tomlins S.A., Trau M. Merging new-age biomarkers and nanodiagnostics for precision prostate cancer management. Nat. Rev. Urol. 2019;16:302–317. doi: 10.1038/s41585-019-0178-2. [DOI] [PubMed] [Google Scholar]
- 6.Tikkinen K.A.O., Dahm P., Lytvyn L., Heen A.F., Vernooij R.W.M., Siemieniuk R.A.C., Wheeler R., Vaughan B., Fobuzi A.C., Blanker M.H., et al. Prostate cancer screening with prostate-specific antigen (psa) test: A clinical practice guideline. BMJ. 2018;362:k3581. doi: 10.1136/bmj.k3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brookman-May S.D., Campi R., Henriquez J.D.S., Klatte T., Langenhuijsen J.F., Brausi M., Linares-Espinos E., Volpe A., Marszalek M., Akdogan B., et al. Latest evidence on the impact of smoking, sports, and sexual activity as modifiable lifestyle risk factors for prostate cancer incidence, recurrence, and progression: A systematic review of the literature by the european association of urology section of oncological urology (esou) Eur. Urol. Focus. 2019;5:756–787. doi: 10.1016/j.euf.2018.02.007. [DOI] [PubMed] [Google Scholar]
- 8.Harrison S., Tilling K., Turner E.L., Martin R.M., Lennon R., Lane J.A., Donovan J.L., Hamdy F.C., Neal D.E., Bosch J., et al. Systematic review and meta-analysis of the associations between body mass index, prostate cancer, advanced prostate cancer, and prostate-specific antigen. Cancer Causes Control. 2020;31:431–449. doi: 10.1007/s10552-020-01291-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Discacciati A., Orsini N., Wolk A. Body mass index and incidence of localized and advanced prostate cancer--a dose-response meta-analysis of prospective studies. Ann. Oncol. 2012;23:1665–1671. doi: 10.1093/annonc/mdr603. [DOI] [PubMed] [Google Scholar]
- 10.Attard G., Parker C., Eeles R.A., Schroder F., Tomlins S.A., Tannock I., Drake C.G., de Bono J.S. Prostate cancer. Lancet. 2016;387:70–82. doi: 10.1016/S0140-6736(14)61947-4. [DOI] [PubMed] [Google Scholar]
- 11.Moul J.W. The evolving definition of advanced prostate cancer. Rev. Urol. 2004;6(Suppl. 8):S10–S17. [PMC free article] [PubMed] [Google Scholar]
- 12.Hurwitz L.M., Agalliu I., Albanes D., Barry K.H., Berndt S.I., Cai Q., Chen C., Cheng I., Genkinger J.M., Giles G.G., et al. Recommended definitions of aggressive prostate cancer for etiologic epidemiologic research. J. Natl. Cancer Inst. 2020 doi: 10.1093/jnci/djaa154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kicinski M., Vangronsveld J., Nawrot T.S. An epidemiological reappraisal of the familial aggregation of prostate cancer: A meta-analysis. PLoS ONE. 2011;6:e27130. doi: 10.1371/journal.pone.0027130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johns L.E., Houlston R.S. A systematic review and meta-analysis of familial prostate cancer risk. BJU Int. 2003;91:789–794. doi: 10.1046/j.1464-410X.2003.04232.x. [DOI] [PubMed] [Google Scholar]
- 15.Stanford J.L., Ostrander E.A. Familial prostate cancer. Epidemiol. Rev. 2001;23:19–23. doi: 10.1093/oxfordjournals.epirev.a000789. [DOI] [PubMed] [Google Scholar]
- 16.Lichtenstein P., Holm N.V., Verkasalo P.K., Iliadou A., Kaprio J., Koskenvuo M., Pukkala E., Skytthe A., Hemminki K. Environmental and heritable factors in the causation of cancer--analyses of cohorts of twins from sweden, denmark, and finland. N. Engl. J. Med. 2000;343:78–85. doi: 10.1056/NEJM200007133430201. [DOI] [PubMed] [Google Scholar]
- 17.Hjelmborg J.B., Scheike T., Holst K., Skytthe A., Penney K.L., Graff R.E., Pukkala E., Christensen K., Adami H.O., Holm N.V., et al. The heritability of prostate cancer in the nordic twin study of cancer. Cancer Epidemiol. Biom. Prev. 2014;23:2303–2310. doi: 10.1158/1055-9965.EPI-13-0568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jansson K.F., Akre O., Garmo H., Bill-Axelson A., Adolfsson J., Stattin P., Bratt O. Concordance of tumor differentiation among brothers with prostate cancer. Eur. Urol. 2012;62:656–661. doi: 10.1016/j.eururo.2012.02.032. [DOI] [PubMed] [Google Scholar]
- 19.Brandt A., Sundquist J., Hemminki K. Risk for incident and fatal prostate cancer in men with a family history of any incident and fatal cancer. Ann. Oncol. 2012;23:251–256. doi: 10.1093/annonc/mdr056. [DOI] [PubMed] [Google Scholar]
- 20.Hemminki K., Ji J., Forsti A., Sundquist J., Lenner P. Concordance of survival in family members with prostate cancer. J. Clin. Oncol. 2008;26:1705–1709. doi: 10.1200/JCO.2007.13.3355. [DOI] [PubMed] [Google Scholar]
- 21.Lindstrom L.S., Hall P., Hartman M., Wiklund F., Gronberg H., Czene K. Familial concordance in cancer survival: A swedish population-based study. Lancet Oncol. 2007;8:1001–1006. doi: 10.1016/S1470-2045(07)70282-6. [DOI] [PubMed] [Google Scholar]
- 22.Albright F.S., Stephenson R.A., Agarwal N., Cannon-Albright L.A. Relative risks for lethal prostate cancer based on complete family history of prostate cancer death. Prostate. 2017;77:41–48. doi: 10.1002/pros.23247. [DOI] [PubMed] [Google Scholar]
- 23.Bratt O., Drevin L., Akre O., Garmo H., Stattin P. Family history and probability of prostate cancer, differentiated by risk category: A nationwide population-based study. J. Natl. Cancer Inst. 2016 doi: 10.1093/jnci/djw110. [DOI] [PubMed] [Google Scholar]
- 24.Pritchard C.C. New name for breast-cancer syndrome could help to save lives. Nature. 2019;571:27–29. doi: 10.1038/d41586-019-02015-7. [DOI] [PubMed] [Google Scholar]
- 25.Barber L., Gerke T., Markt S.C., Peisch S.F., Wilson K.M., Ahearn T., Giovannucci E., Parmigiani G., Mucci L.A. Family history of breast or prostate cancer and prostate cancer risk. Clin. Cancer Res. 2018;24:5910–5917. doi: 10.1158/1078-0432.CCR-18-0370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cerhan J.R., Parker A.S., Putnam S.D., Chiu B.C., Lynch C.F., Cohen M.B., Torner J.C., Cantor K.P. Family history and prostate cancer risk in a population-based cohort of iowa men. Cancer Epidemiol. Biomed. Prev. 1999;8:53–60. [PubMed] [Google Scholar]
- 27.Dominguez-Valentin M., Sampson J.R., Seppala T.T., Ten Broeke S.W., Plazzer J.P., Nakken S., Engel C., Aretz S., Jenkins M.A., Sunde L., et al. Cancer risks by gene, age, and gender in 6350 carriers of pathogenic mismatch repair variants: Findings from the prospective lynch syndrome database. Genet. Med. 2020;22:15–25. doi: 10.1038/s41436-019-0596-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haraldsdottir S., Hampel H., Wei L., Wu C., Frankel W., Bekaii-Saab T., de la Chapelle A., Goldberg R.M. Prostate cancer incidence in males with lynch syndrome. Genet. Med. 2014;16:553–557. doi: 10.1038/gim.2013.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raymond V.M., Mukherjee B., Wang F., Huang S.C., Stoffel E.M., Kastrinos F., Syngal S., Cooney K.A., Gruber S.B. Elevated risk of prostate cancer among men with lynch syndrome. J. Clin. Oncol. 2013;31:1713–1718. doi: 10.1200/JCO.2012.44.1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bauer C.M., Ray A.M., Halstead-Nussloch B.A., Dekker R.G., Raymond V.M., Gruber S.B., Cooney K.A. Hereditary prostate cancer as a feature of lynch syndrome. Fam. Cancer. 2011;10:37–42. doi: 10.1007/s10689-010-9388-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beebe-Dimmer J.L., Kapron A.L., Fraser A.M., Smith K.R., Cooney K.A. Risk of prostate cancer associated with familial and hereditary cancer syndromes. J. Clin. Oncol. 2020;38:1807–1813. doi: 10.1200/JCO.19.02808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taitt H.E. Global trends and prostate cancer: A review of incidence, detection, and mortality as influenced by race, ethnicity, and geographic location. Am. J. Mens Health. 2018;12:1807–1823. doi: 10.1177/1557988318798279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.DeSantis C.E., Miller K.D., Goding Sauer A., Jemal A., Siegel R.L. Cancer statistics for african americans, 2019. CA Cancer J. Clin. 2019;69:211–233. doi: 10.3322/caac.21555. [DOI] [PubMed] [Google Scholar]
- 34.McGinley K.F., Tay K.J., Moul J.W. Prostate cancer in men of african origin. Nat. Rev. Urol. 2016;13:99–107. doi: 10.1038/nrurol.2015.298. [DOI] [PubMed] [Google Scholar]
- 35.Cancer Research, UK. [(accessed on 14 December 2020)]; Available online: https://www.cancerresearchuk.org/health-professional/cancer-statistics/statistics-by-cancer-type/prostate-cancer/incidence.
- 36.Salinas C.A., Tsodikov A., Ishak-Howard M., Cooney K.A. Prostate cancer in young men: An important clinical entity. Nat. Rev. Urol. 2014;11:317–323. doi: 10.1038/nrurol.2014.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lindstrom S., Schumacher F.R., Cox D., Travis R.C., Albanes D., Allen N.E., Andriole G., Berndt S.I., Boeing H., Bueno-de-Mesquita H.B., et al. Common genetic variants in prostate cancer risk prediction--results from the nci breast and prostate cancer cohort consortium (bpc3) Cancer Epidemiol. Biomed. Prev. 2012;21:437–444. doi: 10.1158/1055-9965.EPI-11-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hirschhorn J.N., Lohmueller K., Byrne E., Hirschhorn K. A comprehensive review of genetic association studies. Genet. Med. 2002;4:45–61. doi: 10.1097/00125817-200203000-00002. [DOI] [PubMed] [Google Scholar]
- 39.Weng H., Li S., Huang J.Y., He Z.Q., Meng X.Y., Cao Y., Fang C., Zeng X.T. Androgen receptor gene polymorphisms and risk of prostate cancer: A meta-analysis. Sci. Rep. 2017;7:40554. doi: 10.1038/srep40554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sissung T.M., Price D.K., Del Re M., Ley A.M., Giovannetti E., Figg W.D., Danesi R. Genetic variation: Effect on prostate cancer. Biochim. Biophys. Acta. 2014;1846:446–456. doi: 10.1016/j.bbcan.2014.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schleutker J. Polymorphisms in androgen signaling pathway predisposing to prostate cancer. Mol. Cell Endocrinol. 2012;360:25–37. doi: 10.1016/j.mce.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 42.Price D.K., Chau C.H., Till C., Goodman P.J., Baum C.E., Ockers S.B., English B.C., Minasian L., Parnes H.L., Hsing A.W., et al. Androgen receptor cag repeat length and association with prostate cancer risk: Results from the prostate cancer prevention trial. J. Urol. 2010;184:2297–2302. doi: 10.1016/j.juro.2010.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zeegers M.P., Kiemeney L.A., Nieder A.M., Ostrer H. How strong is the association between cag and ggn repeat length polymorphisms in the androgen receptor gene and prostate cancer risk? Cancer Epidemiol. Biomed. Prev. 2004;13:1765–1771. [PubMed] [Google Scholar]
- 44.Freedman M.L., Pearce C.L., Penney K.L., Hirschhorn J.N., Kolonel L.N., Henderson B.E., Altshuler D. Systematic evaluation of genetic variation at the androgen receptor locus and risk of prostate cancer in a multiethnic cohort study. Am. J. Hum. Genet. 2005;76:82–90. doi: 10.1086/427224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mononen N., Schleutker J. Polymorphisms in genes involved in androgen pathways as risk factors for prostate cancer. J. Urol. 2009;181:1541–1549. doi: 10.1016/j.juro.2008.11.076. [DOI] [PubMed] [Google Scholar]
- 46.Liu X., Huang J., Lin H., Xiong L., Ma Y., Lao H. Esr1 pvuii (rs2234693 t>c) polymorphism and cancer susceptibility: Evidence from 80 studies. J. Cancer. 2018;9:2963–2972. doi: 10.7150/jca.25638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Han P.Z., Cao D.H., Zhang X.L., Ren Z.J., Wei Q. Association between tp53 gene codon72 polymorphism and prostate cancer risk: A systematic review and meta-analysis. Medicine (Baltim.) 2019;98:e16135. doi: 10.1097/MD.0000000000016135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kote-Jarai Z., Leongamornlert D., Saunders E., Tymrakiewicz M., Castro E., Mahmud N., Guy M., Edwards S., O’Brien L., Sawyer E., et al. Brca2 is a moderate penetrance gene contributing to young-onset prostate cancer: Implications for genetic testing in prostate cancer patients. Br. J. Cancer. 2011;105:1230–1234. doi: 10.1038/bjc.2011.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Edwards S.M., Kote-Jarai Z., Meitz J., Hamoudi R., Hope Q., Osin P., Jackson R., Southgate C., Singh R., Falconer A., et al. Two percent of men with early-onset prostate cancer harbor germline mutations in the brca2 gene. Am. J. Hum. Genet. 2003;72:1–12. doi: 10.1086/345310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thompson D., Easton D., Breast Cancer Linkage C. Variation in cancer risks, by mutation position, in brca2 mutation carriers. Am. J. Hum. Genet. 2001;68:410–419. doi: 10.1086/318181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sigurdsson S., Thorlacius S., Tomasson J., Tryggvadottir L., Benediktsdottir K., Eyfjord J.E., Jonsson E. Brca2 mutation in icelandic prostate cancer patients. J. Mol. Med. (Berl.) 1997;75:758–761. doi: 10.1007/s001090050162. [DOI] [PubMed] [Google Scholar]
- 52.Agalliu I., Karlins E., Kwon E.M., Iwasaki L.M., Diamond A., Ostrander E.A., Stanford J.L. Rare germline mutations in the brca2 gene are associated with early-onset prostate cancer. Br. J. Cancer. 2007;97:826–831. doi: 10.1038/sj.bjc.6603929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Leongamornlert D., Mahmud N., Tymrakiewicz M., Saunders E., Dadaev T., Castro E., Goh C., Govindasami K., Guy M., O’Brien L., et al. Germline brca1 mutations increase prostate cancer risk. Br. J. Cancer. 2012;106:1697–1701. doi: 10.1038/bjc.2012.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thompson D., Easton D.F., Breast Cancer Linkage C. Cancer incidence in brca1 mutation carriers. J. Natl. Cancer Inst. 2002;94:1358–1365. doi: 10.1093/jnci/94.18.1358. [DOI] [PubMed] [Google Scholar]
- 55.Ford D., Easton D.F., Bishop D.T., Narod S.A., Goldgar D.E. Risks of cancer in brca1-mutation carriers. Breast cancer linkage consortium. Lancet. 1994;343:692–695. doi: 10.1016/S0140-6736(94)91578-4. [DOI] [PubMed] [Google Scholar]
- 56.Hale V., Weischer M., Park J.Y. Chek2 ( *) 1100delc mutation and risk of prostate cancer. Prost. Cancer. 2014;2014:294575. doi: 10.1155/2014/294575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cybulski C., Wokolorczyk D., Kluzniak W., Jakubowska A., Gorski B., Gronwald J., Huzarski T., Kashyap A., Byrski T., Debniak T., et al. An inherited nbn mutation is associated with poor prognosis prostate cancer. Br. J. Cancer. 2013;108:461–468. doi: 10.1038/bjc.2012.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cybulski C., Gorski B., Debniak T., Gliniewicz B., Mierzejewski M., Masojc B., Jakubowska A., Matyjasik J., Zlowocka E., Sikorski A., et al. Nbs1 is a prostate cancer susceptibility gene. Cancer Res. 2004;64:1215–1219. doi: 10.1158/0008-5472.CAN-03-2502. [DOI] [PubMed] [Google Scholar]
- 59.Narod S.A., Feunteun J., Lynch H.T., Watson P., Conway T., Lynch J., Lenoir G.M. Familial breast-ovarian cancer locus on chromosome 17q12–q23. Lancet. 1991;338:82–83. doi: 10.1097/00006254-199203000-00017. [DOI] [PubMed] [Google Scholar]
- 60.Hall J.M., Lee M.K., Newman B., Morrow J.E., Anderson L.A., Huey B., King M.C. Linkage of early-onset familial breast cancer to chromosome 17q21. Science. 1990;250:1684–1689. doi: 10.1126/science.2270482. [DOI] [PubMed] [Google Scholar]
- 61.Friedman L.S., Ostermeyer E.A., Szabo C.I., Dowd P., Lynch E.D., Rowell S.E., King M.C. Confirmation of brca1 by analysis of germline mutations linked to breast and ovarian cancer in ten families. Nat. Genet. 1994;8:399–404. doi: 10.1038/ng1294-399. [DOI] [PubMed] [Google Scholar]
- 62.Miki Y., Swensen J., Shattuck-Eidens D., Futreal P.A., Harshman K., Tavtigian S., Liu Q., Cochran C., Bennett L.M., Ding W., et al. A strong candidate for the breast and ovarian cancer susceptibility gene brca1. Science. 1994;266:66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- 63.Altshuler D., Daly M.J., Lander E.S. Genetic mapping in human disease. Science. 2008;322:881–888. doi: 10.1126/science.1156409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lander E.S., Schork N.J. Genetic dissection of complex traits. Science. 1994;265:2037–2048. doi: 10.1126/science.8091226. [DOI] [PubMed] [Google Scholar]
- 65.Potter S.R., Partin A.W. Hereditary and familial prostate cancer: Biologic aggressiveness and recurrence. Rev. Urol. 2000;2:35–36. [PMC free article] [PubMed] [Google Scholar]
- 66.Shriner D. Overview of admixture mapping. Curr. Protoc. Hum. Genet. 2013 doi: 10.1002/0471142905.hg0123s76. Chapter 1, Unit 1 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Smith J.R., Freije D., Carpten J.D., Gronberg H., Xu J., Isaacs S.D., Brownstein M.J., Bova G.S., Guo H., Bujnovszky P., et al. Major susceptibility locus for prostate cancer on chromosome 1 suggested by a genome-wide search. Science. 1996;274:1371–1374. doi: 10.1126/science.274.5291.1371. [DOI] [PubMed] [Google Scholar]
- 68.Gronberg H., Smith J., Emanuelsson M., Jonsson B.A., Bergh A., Carpten J., Isaacs W., Xu J., Meyers D., Trent J., et al. In swedish families with hereditary prostate cancer, linkage to the hpc1 locus on chromosome 1q24-25 is restricted to families with early-onset prostate cancer. Am. J. Hum. Genet. 1999;65:134–140. doi: 10.1086/302447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Carpten J., Nupponen N., Isaacs S., Sood R., Robbins C., Xu J., Faruque M., Moses T., Ewing C., Gillanders E., et al. Germline mutations in the ribonuclease l gene in families showing linkage with hpc1. Nat. Genet. 2002;30:181–184. doi: 10.1038/ng823. [DOI] [PubMed] [Google Scholar]
- 70.Eeles R.A., Durocher F., Edwards S., Teare D., Badzioch M., Hamoudi R., Gill S., Biggs P., Dearnaley D., Ardern-Jones A., et al. Linkage analysis of chromosome 1q markers in 136 prostate cancer families. The cancer research campaign/british prostate group u.K. Familial prostate cancer study collaborators. Am. J. Hum. Genet. 1998;62:653–658. doi: 10.1086/301745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.McIndoe R.A., Stanford J.L., Gibbs M., Jarvik G.P., Brandzel S., Neal C.L., Li S., Gammack J.T., Gay A.A., Goode E.L., et al. Linkage analysis of 49 high-risk families does not support a common familial prostate cancer-susceptibility gene at 1q24-25. Am. J. Hum. Genet. 1997;61:347–353. doi: 10.1086/514853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cooney K.A., McCarthy J.D., Lange E., Huang L., Miesfeldt S., Montie J.E., Oesterling J.E., Sandler H.M., Lange K. Prostate cancer susceptibility locus on chromosome 1q: A confirmatory study. J. Natl. Cancer Inst. 1997;89:955–959. doi: 10.1093/jnci/89.13.955. [DOI] [PubMed] [Google Scholar]
- 73.Cancel-Tassin G., Latil A., Valeri A., Mangin P., Fournier G., Berthon P., Cussenot O. Pcap is the major known prostate cancer predisposing locus in families from south and west europe. Eur. J. Hum. Genet. 2001;9:135–142. doi: 10.1038/sj.ejhg.5200592. [DOI] [PubMed] [Google Scholar]
- 74.Xu J. Combined analysis of hereditary prostate cancer linkage to 1q24-25: Results from 772 hereditary prostate cancer families from the international consortium for prostate cancer genetics. Am. J. Hum. Genet. 2000;66:945–957. doi: 10.1086/302807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Neuhausen S.L., Farnham J.M., Kort E., Tavtigian S.V., Skolnick M.H., Cannon-Albright L.A. Prostate cancer susceptibility locus hpc1 in utah high-risk pedigrees. Hum. Mol. Genet. 1999;8:2437–2442. doi: 10.1093/hmg/8.13.2437. [DOI] [PubMed] [Google Scholar]
- 76.Xu J., Dimitrov L., Chang B.L., Adams T.S., Turner A.R., Meyers D.A., Eeles R.A., Easton D.F., Foulkes W.D., Simard J., et al. A combined genomewide linkage scan of 1,233 families for prostate cancer-susceptibility genes conducted by the international consortium for prostate cancer genetics. Am. J. Hum. Genet. 2005;77:219–229. doi: 10.1086/432377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Berry R., Schaid D.J., Smith J.R., French A.J., Schroeder J.J., McDonnell S.K., Peterson B.J., Wang Z.Y., Carpten J.D., Roberts S.G., et al. Linkage analyses at the chromosome 1 loci 1q24-25 (hpc1), 1q42.2-43 (pcap), and 1p36 (capb) in families with hereditary prostate cancer. Am. J. Hum. Genet. 2000;66:539–546. doi: 10.1086/302771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gibbs M., Stanford J.L., McIndoe R.A., Jarvik G.P., Kolb S., Goode E.L., Chakrabarti L., Schuster E.F., Buckley V.A., Miller E.L., et al. Evidence for a rare prostate cancer-susceptibility locus at chromosome 1p36. Am. J. Hum. Genet. 1999;64:776–787. doi: 10.1086/302287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Matsui H., Suzuki K., Ohtake N., Nakata S., Takeuchi T., Yamanaka H., Inoue I. Genomewide linkage analysis of familial prostate cancer in the japanese population. J. Hum. Genet. 2004;49:9–15. doi: 10.1007/s10038-003-0099-y. [DOI] [PubMed] [Google Scholar]
- 80.Badzioch M., Eeles R., Leblanc G., Foulkes W.D., Giles G., Edwards S., Goldgar D., Hopper J.L., Bishop D.T., Moller P., et al. Suggestive evidence for a site specific prostate cancer gene on chromosome 1p36. The crc/bpg uk familial prostate cancer study coordinators and collaborators. The eu biomed collaborators. J. Med. Genet. 2000;37:947–949. doi: 10.1136/jmg.37.12.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gibbs M., Chakrabarti L., Stanford J.L., Goode E.L., Kolb S., Schuster E.F., Buckley V.A., Shook M., Hood L., Jarvik G.P., et al. Analysis of chromosome 1q42.2-43 in 152 families with high risk of prostate cancer. Am. J. Hum. Genet. 1999;64:1087–1095. doi: 10.1086/302342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Berthon P., Valeri A., Cohen-Akenine A., Drelon E., Paiss T., Wohr G., Latil A., Millasseau P., Mellah I., Cohen N., et al. Predisposing gene for early-onset prostate cancer, localized on chromosome 1q42.2-43. Am. J. Hum. Genet. 1998;62:1416–1424. doi: 10.1086/301879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cropp C.D., Simpson C.L., Wahlfors T., Ha N., George A., Jones M.S., Harper U., Ponciano-Jackson D., Green T.A., Tammela T.L., et al. Genome-wide linkage scan for prostate cancer susceptibility in finland: Evidence for a novel locus on 2q37.3 and confirmation of signal on 17q21-q22. Int. J. Cancer. 2011;129:2400–2407. doi: 10.1002/ijc.25906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Suarez B.K., Lin J., Burmester J.K., Broman K.W., Weber J.L., Banerjee T.K., Goddard K.A., Witte J.S., Elston R.C., Catalona W.J. A genome screen of multiplex sibships with prostate cancer. Am. J. Hum. Genet. 2000;66:933–944. doi: 10.1086/302818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pierce B.L., Friedrichsen-Karyadi D.M., McIntosh L., Deutsch K., Hood L., Ostrander E.A., Austin M.A., Stanford J.L. Genomic scan of 12 hereditary prostate cancer families having an occurrence of pancreas cancer. Prostate. 2007;67:410–415. doi: 10.1002/pros.20527. [DOI] [PubMed] [Google Scholar]
- 86.Larson G.P., Ding Y., Cheng L.S., Lundberg C., Gagalang V., Rivas G., Geller L., Weitzel J., MacDonald D., Archambeau J., et al. Genetic linkage of prostate cancer risk to the chromosome 3 region bearing fhit. Cancer Res. 2005;65:805–814. [PubMed] [Google Scholar]
- 87.Rokman A., Baffoe-Bonnie A.B., Gillanders E., Fredriksson H., Autio V., Ikonen T., Gibbs K.D., Jr., Jones M., Gildea D., Freas-Lutz D., et al. Hereditary prostate cancer in finland: Fine-mapping validates 3p26 as a major predisposition locus. Hum. Genet. 2005;116:43–50. doi: 10.1007/s00439-004-1214-7. [DOI] [PubMed] [Google Scholar]
- 88.Schleutker J., Baffoe-Bonnie A.B., Gillanders E., Kainu T., Jones M.P., Freas-Lutz D., Markey C., Gildea D., Riedesel E., Albertus J., et al. Genome-wide scan for linkage in finnish hereditary prostate cancer (hpc) families identifies novel susceptibility loci at 11q14 and 3p25-26. Prostate. 2003;57:280–289. doi: 10.1002/pros.10302. [DOI] [PubMed] [Google Scholar]
- 89.Christensen G.B., Baffoe-Bonnie A.B., George A., Powell I., Bailey-Wilson J.E., Carpten J.D., Giles G.G., Hopper J.L., Severi G., English D.R., et al. Genome-wide linkage analysis of 1,233 prostate cancer pedigrees from the international consortium for prostate cancer genetics using novel sumlink and sumlod analyses. Prostate. 2010;70:735–744. doi: 10.1002/pros.21106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bock C.H., Schwartz A.G., Ruterbusch J.J., Levin A.M., Neslund-Dudas C., Land S.J., Wenzlaff A.S., Reich D., McKeigue P., Chen W., et al. Results from a prostate cancer admixture mapping study in african-american men. Hum. Genet. 2009;126:637–642. doi: 10.1007/s00439-009-0712-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Schaid D.J., McDonnell S.K., Zarfas K.E., Cunningham J.M., Hebbring S., Thibodeau S.N., Eeles R.A., Easton D.F., Foulkes W.D., Simard J., et al. Pooled genome linkage scan of aggressive prostate cancer: Results from the international consortium for prostate cancer genetics. Hum. Genet. 2006;120:471–485. doi: 10.1007/s00439-006-0219-9. [DOI] [PubMed] [Google Scholar]
- 92.Friedrichsen D.M., Stanford J.L., Isaacs S.D., Janer M., Chang B.L., Deutsch K., Gillanders E., Kolb S., Wiley K.E., Badzioch M.D., et al. Identification of a prostate cancer susceptibility locus on chromosome 7q11-21 in jewish families. Proc. Natl. Acad. Sci. USA. 2004;101:1939–1944. doi: 10.1073/pnas.0308336100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Witte J.S., Goddard K.A., Conti D.V., Elston R.C., Lin J., Suarez B.K., Broman K.W., Burmester J.K., Weber J.L., Catalona W.J. Genomewide scan for prostate cancer-aggressiveness loci. Am. J. Hum. Genet. 2000;67:92–99. doi: 10.1086/302960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Neville P.J., Conti D.V., Paris P.L., Levin H., Catalona W.J., Suarez B.K., Witte J.S., Casey G. Prostate cancer aggressiveness locus on chromosome 7q32-q33 identified by linkage and allelic imbalance studies. Neoplasia. 2002;4:424–431. doi: 10.1038/sj.neo.7900254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Paiss T., Worner S., Kurtz F., Haeussler J., Hautmann R.E., Gschwend J.E., Herkommer K., Vogel W. Linkage of aggressive prostate cancer to chromosome 7q31-33 in german prostate cancer families. Eur. J. Hum. Genet. 2003;11:17–22. doi: 10.1038/sj.ejhg.5200898. [DOI] [PubMed] [Google Scholar]
- 96.Wiklund F., Jonsson B.A., Goransson I., Bergh A., Gronberg H. Linkage analysis of prostate cancer susceptibility: Confirmation of linkage at 8p22-23. Hum. Genet. 2003;112:414–418. doi: 10.1007/s00439-003-0916-6. [DOI] [PubMed] [Google Scholar]
- 97.Xu J., Zheng S.L., Hawkins G.A., Faith D.A., Kelly B., Isaacs S.D., Wiley K.E., Chang B., Ewing C.M., Bujnovszky P., et al. Linkage and association studies of prostate cancer susceptibility: Evidence for linkage at 8p22-23. Am. J. Hum. Genet. 2001;69:341–350. doi: 10.1086/321967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Xu J., Zheng S.L., Komiya A., Mychaleckyj J.C., Isaacs S.D., Hu J.J., Sterling D., Lange E.M., Hawkins G.A., Turner A., et al. Germline mutations and sequence variants of the macrophage scavenger receptor 1 gene are associated with prostate cancer risk. Nat. Genet. 2002;32:321–325. doi: 10.1038/ng994. [DOI] [PubMed] [Google Scholar]
- 99.Amundadottir L.T., Sulem P., Gudmundsson J., Helgason A., Baker A., Agnarsson B.A., Sigurdsson A., Benediktsdottir K.R., Cazier J.B., Sainz J., et al. A common variant associated with prostate cancer in european and african populations. Nat. Genet. 2006;38:652–658. doi: 10.1038/ng1808. [DOI] [PubMed] [Google Scholar]
- 100.Freedman M.L., Haiman C.A., Patterson N., McDonald G.J., Tandon A., Waliszewska A., Penney K., Steen R.G., Ardlie K., John E.M., et al. Admixture mapping identifies 8q24 as a prostate cancer risk locus in african-american men. Proc. Natl. Acad. Sci. USA. 2006;103:14068–14073. doi: 10.1073/pnas.0605832103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Witte J.S., Suarez B.K., Thiel B., Lin J., Yu A., Banerjee T.K., Burmester J.K., Casey G., Catalona W.J. Genome-wide scan of brothers: Replication and fine mapping of prostate cancer susceptibility and aggressiveness loci. Prostate. 2003;57:298–308. doi: 10.1002/pros.10304. [DOI] [PubMed] [Google Scholar]
- 102.Xu J., Gillanders E.M., Isaacs S.D., Chang B.L., Wiley K.E., Zheng S.L., Jones M., Gildea D., Riedesel E., Albertus J., et al. Genome-wide scan for prostate cancer susceptibility genes in the johns hopkins hereditary prostate cancer families. Prostate. 2003;57:320–325. doi: 10.1002/pros.10306. [DOI] [PubMed] [Google Scholar]
- 103.Fitzgerald L.M., McDonnell S.K., Carlson E.E., Langeberg W., McIntosh L.M., Deutsch K., Ostrander E.A., Schaid D.J., Stanford J.L. Genome-wide linkage analyses of hereditary prostate cancer families with colon cancer provide further evidence for a susceptibility locus on 15q11-q14. Eur. J. Hum. Genet. 2010;18:1141–1147. doi: 10.1038/ejhg.2010.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lange E.M., Ho L.A., Beebe-Dimmer J.L., Wang Y., Gillanders E.M., Trent J.M., Lange L.A., Wood D.P., Cooney K.A. Genome-wide linkage scan for prostate cancer susceptibility genes in men with aggressive disease: Significant evidence for linkage at chromosome 15q12. Hum. Genet. 2006;119:400–407. doi: 10.1007/s00439-006-0149-6. [DOI] [PubMed] [Google Scholar]
- 105.Gillanders E.M., Xu J., Chang B.L., Lange E.M., Wiklund F., Bailey-Wilson J.E., Baffoe-Bonnie A., Jones M., Gildea D., Riedesel E., et al. Combined genome-wide scan for prostate cancer susceptibility genes. J. Natl. Cancer Inst. 2004;96:1240–1247. doi: 10.1093/jnci/djh228. [DOI] [PubMed] [Google Scholar]
- 106.Fujiwara H., Emi M., Nagai H., Nishimura T., Konishi N., Kubota Y., Ichikawa T., Takahashi S., Shuin T., Habuchi T., et al. Association of common missense changes in elac2 ( hpc2) with prostate cancer in a japanese case-control series. J. Hum. Genet. 2002;47:641–648. doi: 10.1007/s100380200099. [DOI] [PubMed] [Google Scholar]
- 107.Camp N.J., Tavtigian S.V. Meta-analysis of associations of the ser217leu and ala541thr variants in elac2 (hpc2) and prostate cancer. Am. J. Hum. Genet. 2002;71:1475–1478. doi: 10.1086/344516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tavtigian S.V., Simard J., Teng D.H., Abtin V., Baumgard M., Beck A., Camp N.J., Carillo A.R., Chen Y., Dayananth P., et al. A candidate prostate cancer susceptibility gene at chromosome 17p. Nat. Genet. 2001;27:172–180. doi: 10.1038/84808. [DOI] [PubMed] [Google Scholar]
- 109.Rebbeck T.R., Walker A.H., Zeigler-Johnson C., Weisburg S., Martin A.M., Nathanson K.L., Wein A.J., Malkowicz S.B. Association of hpc2/elac2 genotypes and prostate cancer. Am. J. Hum. Genet. 2000;67:1014–1019. doi: 10.1086/303096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lange E.M., Gillanders E.M., Davis C.C., Brown W.M., Campbell J.K., Jones M., Gildea D., Riedesel E., Albertus J., Freas-Lutz D., et al. Genome-wide scan for prostate cancer susceptibility genes using families from the university of michigan prostate cancer genetics project finds evidence for linkage on chromosome 17 near brca1. Prostate. 2003;57:326–334. doi: 10.1002/pros.10307. [DOI] [PubMed] [Google Scholar]
- 111.Schaid D.J., Stanford J.L., McDonnell S.K., Suuriniemi M., McIntosh L., Karyadi D.M., Carlson E.E., Deutsch K., Janer M., Hood L., et al. Genome-wide linkage scan of prostate cancer gleason score and confirmation of chromosome 19q. Hum. Genet. 2007;121:729–735. doi: 10.1007/s00439-007-0368-5. [DOI] [PubMed] [Google Scholar]
- 112.Slager S.L., Schaid D.J., Cunningham J.M., McDonnell S.K., Marks A.F., Peterson B.J., Hebbring S.J., Anderson S., French A.J., Thibodeau S.N. Confirmation of linkage of prostate cancer aggressiveness with chromosome 19q. Am. J. Hum. Genet. 2003;72:759–762. doi: 10.1086/368230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Neville P.J., Conti D.V., Krumroy L.M., Catalona W.J., Suarez B.K., Witte J.S., Casey G. Prostate cancer aggressiveness locus on chromosome segment 19q12-q13.1 identified by linkage and allelic imbalance studies. Genes Chrom. Cancer. 2003;36:332–339. doi: 10.1002/gcc.10165. [DOI] [PubMed] [Google Scholar]
- 114.Zheng S.L., Xu J., Isaacs S.D., Wiley K., Chang B., Bleecker E.R., Walsh P.C., Trent J.M., Meyers D.A., Isaacs W.B. Evidence for a prostate cancer linkage to chromosome 20 in 159 hereditary prostate cancer families. Hum. Genet. 2001;108:430–435. doi: 10.1007/s004390100513. [DOI] [PubMed] [Google Scholar]
- 115.Bock C.H., Cunningham J.M., McDonnell S.K., Schaid D.J., Peterson B.J., Pavlic R.J., Schroeder J.J., Klein J., French A.J., Marks A., et al. Analysis of the prostate cancer-susceptibility locus hpc20 in 172 families affected by prostate cancer. Am. J. Hum. Genet. 2001;68:795–801. doi: 10.1086/318797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Berry R., Schroeder J.J., French A.J., McDonnell S.K., Peterson B.J., Cunningham J.M., Thibodeau S.N., Schaid D.J. Evidence for a prostate cancer-susceptibility locus on chromosome 20. Am. J. Hum. Genet. 2000;67:82–91. doi: 10.1086/302994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Johanneson B., McDonnell S.K., Karyadi D.M., Quignon P., McIntosh L., Riska S.M., FitzGerald L.M., Johnson G., Deutsch K., Williams G., et al. Family-based association analysis of 42 hereditary prostate cancer families identifies the apolipoprotein l3 region on chromosome 22q12 as a risk locus. Hum. Mol. Genet. 2010;19:3852–3862. doi: 10.1093/hmg/ddq283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Johanneson B., McDonnell S.K., Karyadi D.M., Hebbring S.J., Wang L., Deutsch K., McIntosh L., Kwon E.M., Suuriniemi M., Stanford J.L., et al. Fine mapping of familial prostate cancer families narrows the interval for a susceptibility locus on chromosome 22q12.3 to 1.36 mb. Hum. Genet. 2008;123:65–75. doi: 10.1007/s00439-007-0451-y. [DOI] [PubMed] [Google Scholar]
- 119.Camp N.J., Cannon-Albright L.A., Farnham J.M., Baffoe-Bonnie A.B., George A., Powell I., Bailey-Wilson J.E., Carpten J.D., Giles G.G., Hopper J.L., et al. Compelling evidence for a prostate cancer gene at 22q12.3 by the international consortium for prostate cancer genetics. Hum. Mol. Genet. 2007;16:1271–1278. doi: 10.1093/hmg/ddm075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Camp N.J., Farnham J.M., Cannon-Albright L.A. Localization of a prostate cancer predisposition gene to an 880-kb region on chromosome 22q12.3 in utah high-risk pedigrees. Cancer. Res. 2006;66:10205–10212. doi: 10.1158/0008-5472.CAN-06-1233. [DOI] [PubMed] [Google Scholar]
- 121.Baffoe-Bonnie A.B., Smith J.R., Stephan D.A., Schleutker J., Carpten J.D., Kainu T., Gillanders E.M., Matikainen M., Teslovich T.M., Tammela T., et al. A major locus for hereditary prostate cancer in finland: Localization by linkage disequilibrium of a haplotype in the hpcx region. Hum. Genet. 2005;117:307–316. doi: 10.1007/s00439-005-1306-z. [DOI] [PubMed] [Google Scholar]
- 122.Farnham J.M., Camp N.J., Swensen J., Tavtigian S.V., Albright L.A. Confirmation of the hpcx prostate cancer predisposition locus in large utah prostate cancer pedigrees. Hum. Genet. 2005;116:179–185. doi: 10.1007/s00439-004-1220-9. [DOI] [PubMed] [Google Scholar]
- 123.Peters M.A., Jarvik G.P., Janer M., Chakrabarti L., Kolb S., Goode E.L., Gibbs M., DuBois C.C., Schuster E.F., Hood L., et al. Genetic linkage analysis of prostate cancer families to xq27-28. Hum. Hered. 2001;51:107–113. doi: 10.1159/000022965. [DOI] [PubMed] [Google Scholar]
- 124.Xu J., Meyers D., Freije D., Isaacs S., Wiley K., Nusskern D., Ewing C., Wilkens E., Bujnovszky P., Bova G.S., et al. Evidence for a prostate cancer susceptibility locus on the x chromosome. Nat. Genet. 1998;20:175–179. doi: 10.1038/2477. [DOI] [PubMed] [Google Scholar]
- 125.Schaid D.J. The complex genetic epidemiology of prostate cancer. Hum. Mol. Genet. 2004;13:R103–R121. doi: 10.1093/hmg/ddh072. [DOI] [PubMed] [Google Scholar]
- 126.Easton D.F., Schaid D.J., Whittemore A.S., Isaacs W.J., International Consortium for Prostate Cancer G. Where are the prostate cancer genes?--a summary of eight genome wide searches. Prostate. 2003;57:261–269. doi: 10.1002/pros.10300. [DOI] [PubMed] [Google Scholar]
- 127.Conti D.V., Darst B.F., Moss L.C., Saunders E.J., Sheng X., Chou A., Schumacher F.R., Olama A.A.A., Benlloch S., Dadaev T., et al. Trans-ancestry genome-wide association meta-analysis of prostate cancer identifies new susceptibility loci and informs genetic risk prediction. Nat. Genet. 2021;53:65–75. doi: 10.1038/s41588-020-00748-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hao Z. Rideogram: Drawing svg graphics to visualize and map genome-wide data on the idiograms. PeerJ. Comput. Sci. 2020 doi: 10.7717/peerj-cs.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lange E.M., Robbins C.M., Gillanders E.M., Zheng S.L., Xu J., Wang Y., White K.A., Chang B.L., Ho L.A., Trent J.M., et al. Fine-mapping the putative chromosome 17q21-22 prostate cancer susceptibility gene to a 10 cm region based on linkage analysis. Hum. Genet. 2007;121:49–55. doi: 10.1007/s00439-006-0274-2. [DOI] [PubMed] [Google Scholar]
- 130.Ewing C.M., Ray A.M., Lange E.M., Zuhlke K.A., Robbins C.M., Tembe W.D., Wiley K.E., Isaacs S.D., Johng D., Wang Y., et al. Germline mutations in hoxb13 and prostate-cancer risk. N. Engl. J. Med. 2012;366:141–149. doi: 10.1056/NEJMoa1110000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Chen Z., Greenwood C., Isaacs W.B., Foulkes W.D., Sun J., Zheng S.L., Condreay L.D., Xu J. The g84e mutation of hoxb13 is associated with increased risk for prostate cancer: Results from the reduce trial. Carcinogenesis. 2013;34:1260–1264. doi: 10.1093/carcin/bgt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Shang Z., Zhu S., Zhang H., Li L., Niu Y. Germline homeobox b13 (hoxb13) g84e mutation and prostate cancer risk in european descendants: A meta-analysis of 24,213 cases and 73, 631 controls. Eur. Urol. 2013;64:173–176. doi: 10.1016/j.eururo.2013.03.007. [DOI] [PubMed] [Google Scholar]
- 133.Witte J.S., Mefford J., Plummer S.J., Liu J., Cheng I., Klein E.A., Rybicki B.A., Casey G. Hoxb13 mutation and prostate cancer: Studies of siblings and aggressive disease. Cancer Epidemiol. Biom. Prev. 2013;22:675–680. doi: 10.1158/1055-9965.EPI-12-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Beebe-Dimmer J.L., Hathcock M., Yee C., Okoth L.A., Ewing C.M., Isaacs W.B., Cooney K.A., Thibodeau S.N. The hoxb13 g84e mutation is associated with an increased risk for prostate cancer and other malignancies. Cancer Epidemiol. Biomed. Prev. 2015;24:1366–1372. doi: 10.1158/1055-9965.EPI-15-0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Huang H., Cai B. G84e mutation in hoxb13 is firmly associated with prostate cancer risk: A meta-analysis. Tumour Biol. 2014;35:1177–1182. doi: 10.1007/s13277-013-1157-5. [DOI] [PubMed] [Google Scholar]
- 136.Gudmundsson J., Sulem P., Gudbjartsson D.F., Masson G., Agnarsson B.A., Benediktsdottir K.R., Sigurdsson A., Magnusson O.T., Gudjonsson S.A., Magnusdottir D.N., et al. A study based on whole-genome sequencing yields a rare variant at 8q24 associated with prostate cancer. Nat. Genet. 2012;44:1326–1329. doi: 10.1038/ng.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kote-Jarai Z., Mikropoulos C., Leongamornlert D.A., Dadaev T., Tymrakiewicz M., Saunders E.J., Jones M., Jugurnauth-Little S., Govindasami K., Guy M., et al. Prevalence of the hoxb13 g84e germline mutation in british men and correlation with prostate cancer risk, tumour characteristics and clinical outcomes. Ann. Oncol. 2015;26:756–761. doi: 10.1093/annonc/mdv004. [DOI] [PubMed] [Google Scholar]
- 138.Laitinen V.H., Wahlfors T., Saaristo L., Rantapero T., Pelttari L.M., Kilpivaara O., Laasanen S.L., Kallioniemi A., Nevanlinna H., Aaltonen L., et al. Hoxb13 g84e mutation in finland: Population-based analysis of prostate, breast, and colorectal cancer risk. Cancer Epidemiol. Biomed. Prev. 2013;22:452–460. doi: 10.1158/1055-9965.EPI-12-1000-T. [DOI] [PubMed] [Google Scholar]
- 139.Stott-Miller M., Karyadi D.M., Smith T., Kwon E.M., Kolb S., Stanford J.L., Ostrander E.A. Hoxb13 mutations in a population-based, case-control study of prostate cancer. Prostate. 2013;73:634–641. doi: 10.1002/pros.22604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Xu J., Lange E.M., Lu L., Zheng S.L., Wang Z., Thibodeau S.N., Cannon-Albright L.A., Teerlink C.C., Camp N.J., Johnson A.M., et al. Hoxb13 is a susceptibility gene for prostate cancer: Results from the international consortium for prostate cancer genetics (icpcg) Hum. Genet. 2013;132:5–14. doi: 10.1007/s00439-012-1229-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Nyberg T., Govindasami K., Leslie G., Dadaev T., Bancroft E., Ni Raghallaigh H., Brook M.N., Hussain N., Keating D., Lee A., et al. Homeobox b13 g84e mutation and prostate cancer risk. Eur. Urol. 2019;75:834–845. doi: 10.1016/j.eururo.2018.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Handorf E., Crumpler N., Gross L., Giri V.N. Prevalence of the hoxb13 g84e mutation among unaffected men with a family history of prostate cancer. J. Genet. Couns. 2014;23:371–376. doi: 10.1007/s10897-013-9672-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Lin X., Qu L., Chen Z., Xu C., Ye D., Shao Q., Wang X., Qi J., Chen Z., Zhou F., et al. A novel germline mutation in hoxb13 is associated with prostate cancer risk in chinese men. Prostate. 2013;73:169–175. doi: 10.1002/pros.22552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Momozawa Y., Iwasaki Y., Hirata M., Liu X., Kamatani Y., Takahashi A., Sugano K., Yoshida T., Murakami Y., Matsuda K., et al. Germline pathogenic variants in 7636 japanese patients with prostate cancer and 12 366 controls. J. Natl. Cancer Inst. 2020;112:369–376. doi: 10.1093/jnci/djz124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Maia S., Cardoso M., Pinto P., Pinheiro M., Santos C., Peixoto A., Bento M.J., Oliveira J., Henrique R., Jeronimo C., et al. Identification of two novel hoxb13 germline mutations in portuguese prostate cancer patients. PLoS ONE. 2015;10:e0132728. doi: 10.1371/journal.pone.0132728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 147.Venter J.C., Adams M.D., Myers E.W., Li P.W., Mural R.J., Sutton G.G., Smith H.O., Yandell M., Evans C.A., Holt R.A., et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 148.Genomes Project C., Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A., et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.International HapMap C. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Hirschhorn J.N., Daly M.J. Genome-wide association studies for common diseases and complex traits. Nat. Rev. Genet. 2005;6:95–108. doi: 10.1038/nrg1521. [DOI] [PubMed] [Google Scholar]
- 151.LaFramboise T. Single nucleotide polymorphism arrays: A decade of biological, computational and technological advances. Nucl. Acids Res. 2009;37:4181–4193. doi: 10.1093/nar/gkp552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Marchini J., Howie B. Genotype imputation for genome-wide association studies. Nat. Rev. Genet. 2010;11:499–511. doi: 10.1038/nrg2796. [DOI] [PubMed] [Google Scholar]
- 153.Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 154.Fadista J., Manning A.K., Florez J.C., Groop L. The (in)famous gwas p-value threshold revisited and updated for low-frequency variants. Eur. J. Hum. Genet. 2016;24:1202–1205. doi: 10.1038/ejhg.2015.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Pe’er I., Yelensky R., Altshuler D., Daly M.J. Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genet. Epidemiol. 2008;32:381–385. doi: 10.1002/gepi.20303. [DOI] [PubMed] [Google Scholar]
- 156.Evangelou E., Ioannidis J.P. Meta-analysis methods for genome-wide association studies and beyond. Nat. Rev. Genet. 2013;14:379–389. doi: 10.1038/nrg3472. [DOI] [PubMed] [Google Scholar]
- 157.Wang W.Y., Barratt B.J., Clayton D.G., Todd J.A. Genome-wide association studies: Theoretical and practical concerns. Nat. Rev. Genet. 2005;6:109–118. doi: 10.1038/nrg1522. [DOI] [PubMed] [Google Scholar]
- 158.Visscher P.M., Wray N.R., Zhang Q., Sklar P., McCarthy M.I., Brown M.A., Yang J. 10 years of gwas discovery: Biology, function, and translation. Am. J. Hum. Genet. 2017;101:5–22. doi: 10.1016/j.ajhg.2017.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Visscher P.M., Brown M.A., McCarthy M.I., Yang J. Five years of gwas discovery. Am. J. Hum. Genet. 2012;90:7–24. doi: 10.1016/j.ajhg.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Gudmundsson J., Sulem P., Manolescu A., Amundadottir L.T., Gudbjartsson D., Helgason A., Rafnar T., Bergthorsson J.T., Agnarsson B.A., Baker A., et al. Genome-wide association study identifies a second prostate cancer susceptibility variant at 8q24. Nat. Genet. 2007;39:631–637. doi: 10.1038/ng1999. [DOI] [PubMed] [Google Scholar]
- 161.Duggan D., Zheng S.L., Knowlton M., Benitez D., Dimitrov L., Wiklund F., Robbins C., Isaacs S.D., Cheng Y., Li G., et al. Two genome-wide association studies of aggressive prostate cancer implicate putative prostate tumor suppressor gene dab2ip. J. Natl. Cancer Inst. 2007;99:1836–1844. doi: 10.1093/jnci/djm250. [DOI] [PubMed] [Google Scholar]
- 162.Yeager M., Orr N., Hayes R.B., Jacobs K.B., Kraft P., Wacholder S., Minichiello M.J., Fearnhead P., Yu K., Chatterjee N., et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat. Genet. 2007;39:645–649. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- 163.Haiman C.A., Patterson N., Freedman M.L., Myers S.R., Pike M.C., Waliszewska A., Neubauer J., Tandon A., Schirmer C., McDonald G.J., et al. Multiple regions within 8q24 independently affect risk for prostate cancer. Nat. Genet. 2007;39:638–644. doi: 10.1038/ng2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Witte J.S. Multiple prostate cancer risk variants on 8q24. Nat. Genet. 2007;39:579–580. doi: 10.1038/ng0507-579. [DOI] [PubMed] [Google Scholar]
- 165.Matejcic M., Saunders E.J., Dadaev T., Brook M.N., Wang K., Sheng X., Olama A.A.A., Schumacher F.R., Ingles S.A., Govindasami K., et al. Germline variation at 8q24 and prostate cancer risk in men of european ancestry. Nat. Commun. 2018;9:4616. doi: 10.1038/s41467-018-06863-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Al Olama A.A., Kote-Jarai Z., Giles G.G., Guy M., Morrison J., Severi G., Leongamornlert D.A., Tymrakiewicz M., Jhavar S., Saunders E., et al. Multiple loci on 8q24 associated with prostate cancer susceptibility. Nat. Genet. 2009;41:1058–1060. doi: 10.1038/ng.452. [DOI] [PubMed] [Google Scholar]
- 167.Darst B.F., Wan P., Sheng X., Bensen J.T., Ingles S.A., Rybicki B.A., Nemesure B., John E.M., Fowke J.H., Stevens V.L., et al. A germline variant at 8q24 contributes to familial clustering of prostate cancer in men of african ancestry. Eur. Urol. 2020 doi: 10.1016/j.eururo.2020.04.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Han Y., Rand K.A., Hazelett D.J., Ingles S.A., Kittles R.A., Strom S.S., Rybicki B.A., Nemesure B., Isaacs W.B., Stanford J.L., et al. Prostate cancer susceptibility in men of african ancestry at 8q24. J. Natl. Cancer Inst. 2016 doi: 10.1093/jnci/djv431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Hazelett D.J., Coetzee S.G., Coetzee G.A. A rare variant, which destroys a foxa1 site at 8q24, is associated with prostate cancer risk. Cell Cycl. 2013;12:379–380. doi: 10.4161/cc.23201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Dupont W.D., Breyer J.P., Plummer W.D., Chang S.S., Cookson M.S., Smith J.A., Blue E.E., Bamshad M.J., Smith J.R. 8q24 genetic variation and comprehensive haplotypes altering familial risk of prostate cancer. Nat. Commun. 2020;11:1523. doi: 10.1038/s41467-020-15122-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Teerlink C.C., Leongamornlert D., Dadaev T., Thomas A., Farnham J., Stephenson R.A., Riska S., McDonnell S.K., Schaid D.J., Catalona W.J., et al. Genome-wide association of familial prostate cancer cases identifies evidence for a rare segregating haplotype at 8q24.21. Hum. Genet. 2016;135:923–938. doi: 10.1007/s00439-016-1690-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Huppi K., Pitt J.J., Wahlberg B.M., Caplen N.J. The 8q24 gene desert: An oasis of non-coding transcriptional activity. Front. Genet. 2012;3:69. doi: 10.3389/fgene.2012.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Hazelett D.J., Rhie S.K., Gaddis M., Yan C., Lakeland D.L., Coetzee S.G., Ellipse G.-O.N., Practical C., Henderson B.E., Noushmehr H., et al. Comprehensive functional annotation of 77 prostate cancer risk loci. PLoS Genet. 2014;10:e1004102. doi: 10.1371/journal.pgen.1004102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Jia L., Landan G., Pomerantz M., Jaschek R., Herman P., Reich D., Yan C., Khalid O., Kantoff P., Oh W., et al. Functional enhancers at the gene-poor 8q24 cancer-linked locus. PLoS Genet. 2009;5:e1000597. doi: 10.1371/journal.pgen.1000597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Wasserman N.F., Aneas I., Nobrega M.A. An 8q24 gene desert variant associated with prostate cancer risk confers differential in vivo activity to a myc enhancer. Genom. Res. 2010;20:1191–1197. doi: 10.1101/gr.105361.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Sotelo J., Esposito D., Duhagon M.A., Banfield K., Mehalko J., Liao H., Stephens R.M., Harris T.J., Munroe D.J., Wu X. Long-range enhancers on 8q24 regulate c-myc. Proc. Natl. Acad. Sci. USA. 2010;107:3001–3005. doi: 10.1073/pnas.0906067107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Pomerantz M.M., Ahmadiyeh N., Jia L., Herman P., Verzi M.P., Doddapaneni H., Beckwith C.A., Chan J.A., Hills A., Davis M., et al. The 8q24 cancer risk variant rs6983267 shows long-range interaction with myc in colorectal cancer. Nat. Genet. 2009;41:882–884. doi: 10.1038/ng.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Walavalkar K., Saravanan B., Singh A.K., Jayani R.S., Nair A., Farooq U., Islam Z., Soota D., Mann R., Shivaprasad P.V., et al. A rare variant of african ancestry activates 8q24 lncrna hub by modulating cancer associated enhancer. Nat. Commun. 2020;11:3598. doi: 10.1038/s41467-020-17325-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Breyer J.P., Dorset D.C., Clark T.A., Bradley K.M., Wahlfors T.A., McReynolds K.M., Maynard W.H., Chang S.S., Cookson M.S., Smith J.A., et al. An expressed retrogene of the master embryonic stem cell gene pou5f1 is associated with prostate cancer susceptibility. Am. J. Hum. Genet. 2014;94:395–404. doi: 10.1016/j.ajhg.2014.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Takata R., Takahashi A., Fujita M., Momozawa Y., Saunders E.J., Yamada H., Maejima K., Nakano K., Nishida Y., Hishida A., et al. 12 new susceptibility loci for prostate cancer identified by genome-wide association study in japanese population. Nat. Commun. 2019;10:4422. doi: 10.1038/s41467-019-12267-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Xiang J.F., Yang L., Chen L.L. The long noncoding rna regulation at the myc locus. Curr. Opin. Genet. Dev. 2015;33:41–48. doi: 10.1016/j.gde.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 182.Meyer K.B., Maia A.T., O’Reilly M., Ghoussaini M., Prathalingam R., Porter-Gill P., Ambs S., Prokunina-Olsson L., Carroll J., Ponder B.A. A functional variant at a prostate cancer predisposition locus at 8q24 is associated with pvt1 expression. PLoS Genet. 2011;7:e1002165. doi: 10.1371/journal.pgen.1002165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Guo H., Ahmed M., Zhang F., Yao C.Q., Li S., Liang Y., Hua J., Soares F., Sun Y., Langstein J., et al. Modulation of long noncoding rnas by risk snps underlying genetic predispositions to prostate cancer. Nat. Genet. 2016;48:1142–1150. doi: 10.1038/ng.3637. [DOI] [PubMed] [Google Scholar]
- 184.Chung S., Nakagawa H., Uemura M., Piao L., Ashikawa K., Hosono N., Takata R., Akamatsu S., Kawaguchi T., Morizono T., et al. Association of a novel long non-coding rna in 8q24 with prostate cancer susceptibility. Cancer Sci. 2011;102:245–252. doi: 10.1111/j.1349-7006.2010.01737.x. [DOI] [PubMed] [Google Scholar]
- 185.Eeles R.A., Kote-Jarai Z., Al Olama A.A., Giles G.G., Guy M., Severi G., Muir K., Hopper J.L., Henderson B.E., Haiman C.A., et al. Identification of seven new prostate cancer susceptibility loci through a genome-wide association study. Nat. Genet. 2009;41:1116–1121. doi: 10.1038/ng.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Gudmundsson J., Sulem P., Rafnar T., Bergthorsson J.T., Manolescu A., Gudbjartsson D., Agnarsson B.A., Sigurdsson A., Benediktsdottir K.R., Blondal T., et al. Common sequence variants on 2p15 and xp11.22 confer susceptibility to prostate cancer. Nat. Genet. 2008;40:281–283. doi: 10.1038/ng.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Eeles R.A., Kote-Jarai Z., Giles G.G., Olama A.A., Guy M., Jugurnauth S.K., Mulholland S., Leongamornlert D.A., Edwards S.M., Morrison J., et al. Multiple newly identified loci associated with prostate cancer susceptibility. Nat. Genet. 2008;40:316–321. doi: 10.1038/ng.90. [DOI] [PubMed] [Google Scholar]
- 188.Thomas G., Jacobs K.B., Yeager M., Kraft P., Wacholder S., Orr N., Yu K., Chatterjee N., Welch R., Hutchinson A., et al. Multiple loci identified in a genome-wide association study of prostate cancer. Nat. Genet. 2008;40:310–315. doi: 10.1038/ng.91. [DOI] [PubMed] [Google Scholar]
- 189.Kote-Jarai Z., Olama A.A., Giles G.G., Severi G., Schleutker J., Weischer M., Campa D., Riboli E., Key T., Gronberg H., et al. Seven prostate cancer susceptibility loci identified by a multi-stage genome-wide association study. Nat. Genet. 2011;43:785–791. doi: 10.1038/ng.882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Schumacher F.R., Berndt S.I., Siddiq A., Jacobs K.B., Wang Z., Lindstrom S., Stevens V.L., Chen C., Mondul A.M., Travis R.C., et al. Genome-wide association study identifies new prostate cancer susceptibility loci. Hum. Mol. Genet. 2011;20:3867–3875. doi: 10.1093/hmg/ddr295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Gudmundsson J., Sulem P., Gudbjartsson D.F., Blondal T., Gylfason A., Agnarsson B.A., Benediktsdottir K.R., Magnusdottir D.N., Orlygsdottir G., Jakobsdottir M., et al. Genome-wide association and replication studies identify four variants associated with prostate cancer susceptibility. Nat. Genet. 2009;41:1122–1126. doi: 10.1038/ng.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Schumacher F.R., Al Olama A.A., Berndt S.I., Benlloch S., Ahmed M., Saunders E.J., Dadaev T., Leongamornlert D., Anokian E., Cieza-Borrella C., et al. Association analyses of more than 140,000 men identify 63 new prostate cancer susceptibility loci. Nat. Genet. 2018;50:928–936. doi: 10.1038/s41588-018-0142-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Eeles R.A., Olama A.A., Benlloch S., Saunders E.J., Leongamornlert D.A., Tymrakiewicz M., Ghoussaini M., Luccarini C., Dennis J., Jugurnauth-Little S., et al. Identification of 23 new prostate cancer susceptibility loci using the icogs custom genotyping array. Nat. Genet. 2013;45:385–391. doi: 10.1038/ng.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Al Olama A.A., Kote-Jarai Z., Berndt S.I., Conti D.V., Schumacher F., Han Y., Benlloch S., Hazelett D.J., Wang Z., Saunders E., et al. A meta-analysis of 87,040 individuals identifies 23 new susceptibility loci for prostate cancer. Nat. Genet. 2014;46:1103–1109. doi: 10.1038/ng.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.O’Hurley G., Busch C., Fagerberg L., Hallstrom B.M., Stadler C., Tolf A., Lundberg E., Schwenk J.M., Jirstrom K., Bjartell A., et al. Analysis of the human prostate-specific proteome defined by transcriptomics and antibody-based profiling identifies tmem79 and acoxl as two putative, diagnostic markers in prostate cancer. PLoS ONE. 2015;10:e0133449. doi: 10.1371/journal.pone.0133449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Spisak S., Lawrenson K., Fu Y., Csabai I., Cottman R.T., Seo J.H., Haiman C., Han Y., Lenci R., Li Q., et al. Causel: An epigenome- and genome-editing pipeline for establishing function of noncoding gwas variants. Nat. Med. 2015;21:1357–1363. doi: 10.1038/nm.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Huang Q., Whitington T., Gao P., Lindberg J.F., Yang Y., Sun J., Vaisanen M.R., Szulkin R., Annala M., Yan J., et al. A prostate cancer susceptibility allele at 6q22 increases rfx6 expression by modulating hoxb13 chromatin binding. Nat. Genet. 2014;46:126–135. doi: 10.1038/ng.2862. [DOI] [PubMed] [Google Scholar]
- 198.Qian Y., Zhang L., Cai M., Li H., Xu H., Yang H., Zhao Z., Rhie S.K., Farnham P.J., Shi J., et al. The prostate cancer risk variant rs55958994 regulates multiple gene expression through extreme long-range chromatin interaction to control tumor progression. Sci. Adv. 2019;5:eaaw6710. doi: 10.1126/sciadv.aaw6710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Hua J.T., Ahmed M., Guo H., Zhang Y., Chen S., Soares F., Lu J., Zhou S., Wang M., Li H., et al. Risk snp-mediated promoter-enhancer switching drives prostate cancer through lncrna pcat19. Cell. 2018;174:564–575. doi: 10.1016/j.cell.2018.06.014. [DOI] [PubMed] [Google Scholar]
- 200.Gao P., Xia J.H., Sipeky C., Dong X.M., Zhang Q., Yang Y., Zhang P., Cruz S.P., Zhang K., Zhu J., et al. Biology and clinical implications of the 19q13 aggressive prostate cancer susceptibility locus. Cell. 2018;174:576–589. doi: 10.1016/j.cell.2018.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Luo Z., Rhie S.K., Lay F.D., Farnham P.J. A prostate cancer risk element functions as a repressive loop that regulates hoxa13. Cell Rep. 2017;21:1411–1417. doi: 10.1016/j.celrep.2017.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Lou H., Yeager M., Li H., Bosquet J.G., Hayes R.B., Orr N., Yu K., Hutchinson A., Jacobs K.B., Kraft P., et al. Fine mapping and functional analysis of a common variant in msmb on chromosome 10q11.2 associated with prostate cancer susceptibility. Proc. Natl. Acad. Sci. USA. 2009;106:7933–7938. doi: 10.1073/pnas.0902104106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Chang B.L., Cramer S.D., Wiklund F., Isaacs S.D., Stevens V.L., Sun J., Smith S., Pruett K., Romero L.M., Wiley K.E., et al. Fine mapping association study and functional analysis implicate a snp in msmb at 10q11 as a causal variant for prostate cancer risk. Hum. Mol. Genet. 2009;18:1368–1375. doi: 10.1093/hmg/ddp035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Zhang X., Cowper-Sal lari R., Bailey S.D., Moore J.H., Lupien M. Integrative functional genomics identifies an enhancer looping to the sox9 gene disrupted by the 17q24.3 prostate cancer risk locus. Genom. Res. 2012;22:1437–1446. doi: 10.1101/gr.135665.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Kote-Jarai Z., Amin Al Olama A., Leongamornlert D., Tymrakiewicz M., Saunders E., Guy M., Giles G.G., Severi G., Southey M., Hopper J.L., et al. Identification of a novel prostate cancer susceptibility variant in the klk3 gene transcript. Hum. Genet. 2011;129:687–694. doi: 10.1007/s00439-011-0981-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Dadaev T., Saunders E.J., Newcombe P.J., Anokian E., Leongamornlert D.A., Brook M.N., Cieza-Borrella C., Mijuskovic M., Wakerell S., Olama A.A.A., et al. Fine-mapping of prostate cancer susceptibility loci in a large meta-analysis identifies candidate causal variants. Nat. Commun. 2018;9:2256. doi: 10.1038/s41467-018-04109-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Whitington T., Gao P., Song W., Ross-Adams H., Lamb A.D., Yang Y., Svezia I., Klevebring D., Mills I.G., Karlsson R., et al. Gene regulatory mechanisms underpinning prostate cancer susceptibility. Nat. Genet. 2016;48:387–397. doi: 10.1038/ng.3523. [DOI] [PubMed] [Google Scholar]
- 208.Han Y., Hazelett D.J., Wiklund F., Schumacher F.R., Stram D.O., Berndt S.I., Wang Z., Rand K.A., Hoover R.N., Machiela M.J., et al. Integration of multiethnic fine-mapping and genomic annotation to prioritize candidate functional snps at prostate cancer susceptibility regions. Hum. Mol. Genet. 2015;24:5603–5618. doi: 10.1093/hmg/ddv269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Amin Al Olama A., Dadaev T., Hazelett D.J., Li Q., Leongamornlert D., Saunders E.J., Stephens S., Cieza-Borrella C., Whitmore I., Benlloch Garcia S., et al. Multiple novel prostate cancer susceptibility signals identified by fine-mapping of known risk loci among europeans. Hum. Mol. Genet. 2015;24:5589–5602. doi: 10.1093/hmg/ddv203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Gusev A., Ko A., Shi H., Bhatia G., Chung W., Penninx B.W., Jansen R., de Geus E.J., Boomsma D.I., Wright F.A., et al. Integrative approaches for large-scale transcriptome-wide association studies. Nat. Genet. 2016;48:245–252. doi: 10.1038/ng.3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Wu L., Wang J., Cai Q., Cavazos T.B., Emami N.C., Long J., Shu X.O., Lu Y., Guo X., Bauer J.A., et al. Identification of novel susceptibility loci and genes for prostate cancer risk: A transcriptome-wide association study in over 140,000 european descendants. Cancer Res. 2019;79:3192–3204. doi: 10.1158/0008-5472.CAN-18-3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Mancuso N., Gayther S., Gusev A., Zheng W., Penney K.L., Kote-Jarai Z., Eeles R., Freedman M., Haiman C., Pasaniuc B., et al. Large-scale transcriptome-wide association study identifies new prostate cancer risk regions. Nat. Commun. 2018;9:4079. doi: 10.1038/s41467-018-06302-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Emami N.C., Kachuri L., Meyers T.J., Das R., Hoffman J.D., Hoffmann T.J., Hu D., Shan J., Feng F.Y., Ziv E., et al. Association of imputed prostate cancer transcriptome with disease risk reveals novel mechanisms. Nat. Commun. 2019;10:3107. doi: 10.1038/s41467-019-10808-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Du Z., Lubmawa A., Gundell S., Wan P., Nalukenge C., Muwanga P., Lutalo M., Nansereko D., Ndaruhutse O., Katuku M., et al. Genetic risk of prostate cancer in ugandan men. Prostate. 2018;78:370–376. doi: 10.1002/pros.23481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Conti D.V., Wang K., Sheng X., Bensen J.T., Hazelett D.J., Cook M.B., Ingles S.A., Kittles R.A., Strom S.S., Rybicki B.A., et al. Two novel susceptibility loci for prostate cancer in men of african ancestry. J. Natl. Cancer Inst. 2017;109 doi: 10.1093/jnci/djx084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Cook M.B., Wang Z., Yeboah E.D., Tettey Y., Biritwum R.B., Adjei A.A., Tay E., Truelove A., Niwa S., Chung C.C., et al. A genome-wide association study of prostate cancer in west african men. Hum. Genet. 2014;133:509–521. doi: 10.1007/s00439-013-1387-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Haiman C.A., Chen G.K., Blot W.J., Strom S.S., Berndt S.I., Kittles R.A., Rybicki B.A., Isaacs W.B., Ingles S.A., Stanford J.L., et al. Genome-wide association study of prostate cancer in men of african ancestry identifies a susceptibility locus at 17q21. Nat. Genet. 2011;43:570–573. doi: 10.1038/ng.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Akamatsu S., Takata R., Haiman C.A., Takahashi A., Inoue T., Kubo M., Furihata M., Kamatani N., Inazawa J., Chen G.K., et al. Common variants at 11q12, 10q26 and 3p11.2 are associated with prostate cancer susceptibility in japanese. Nat. Genet. 2012;44:426–429. doi: 10.1038/ng.1104. [DOI] [PubMed] [Google Scholar]
- 219.Takata R., Akamatsu S., Kubo M., Takahashi A., Hosono N., Kawaguchi T., Tsunoda T., Inazawa J., Kamatani N., Ogawa O., et al. Genome-wide association study identifies five new susceptibility loci for prostate cancer in the japanese population. Nat. Genet. 2010;42:751–754. doi: 10.1038/ng.635. [DOI] [PubMed] [Google Scholar]
- 220.Wang M., Takahashi A., Liu F., Ye D., Ding Q., Qin C., Yin C., Zhang Z., Matsuda K., Kubo M., et al. Large-scale association analysis in asians identifies new susceptibility loci for prostate cancer. Nat. Commun. 2015;6:8469. doi: 10.1038/ncomms9469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Xu J., Mo Z., Ye D., Wang M., Liu F., Jin G., Xu C., Wang X., Shao Q., Chen Z., et al. Genome-wide association study in chinese men identifies two new prostate cancer risk loci at 9q31.2 and 19q13.4. Nat. Genet. 2012;44:1231–1235. doi: 10.1038/ng.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Du Z., Hopp H., Ingles S.A., Huff C., Sheng X., Weaver B., Stern M., Hoffmann T.J., John E.M., Van Den Eeden S.K., et al. A genome-wide association study of prostate cancer in latinos. Int. J. Cancer. 2020;146:1819–1826. doi: 10.1002/ijc.32525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Hoffmann T.J., Van Den Eeden S.K., Sakoda L.C., Jorgenson E., Habel L.A., Graff R.E., Passarelli M.N., Cario C.L., Emami N.C., Chao C.R., et al. A large multiethnic genome-wide association study of prostate cancer identifies novel risk variants and substantial ethnic differences. Cancer Discov. 2015;5:878–891. doi: 10.1158/2159-8290.CD-15-0315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Han Y., Signorello L.B., Strom S.S., Kittles R.A., Rybicki B.A., Stanford J.L., Goodman P.J., Berndt S.I., Carpten J., Casey G., et al. Generalizability of established prostate cancer risk variants in men of african ancestry. Int. J. Cancer. 2015;136:1210–1217. doi: 10.1002/ijc.29066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Liu F., Hsing A.W., Wang X., Shao Q., Qi J., Ye Y., Wang Z., Chen H., Gao X., Wang G., et al. Systematic confirmation study of reported prostate cancer risk-associated single nucleotide polymorphisms in chinese men. Cancer Sci. 2011;102:1916–1920. doi: 10.1111/j.1349-7006.2011.02036.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Zhang Y.D., Hurson A.N., Zhang H., Choudhury P.P., Easton D.F., Milne R.L., Simard J., Hall P., Michailidou K., Dennis J., et al. Assessment of polygenic architecture and risk prediction based on common variants across fourteen cancers. Nat. Commun. 2020;11:3353. doi: 10.1038/s41467-020-16483-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Khera A.V., Chaffin M., Aragam K.G., Haas M.E., Roselli C., Choi S.H., Natarajan P., Lander E.S., Lubitz S.A., Ellinor P.T., et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat. Genet. 2018;50:1219–1224. doi: 10.1038/s41588-018-0183-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Pal P., Xi H., Guha S., Sun G., Helfand B.T., Meeks J.J., Suarez B.K., Catalona W.J., Deka R. Common variants in 8q24 are associated with risk for prostate cancer and tumor aggressiveness in men of european ancestry. Prostate. 2009;69:1548–1556. doi: 10.1002/pros.20999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Liu W., Sun J., Li G., Zhu Y., Zhang S., Kim S.T., Sun J., Wiklund F., Wiley K., Isaacs S.D., et al. Association of a germ-line copy number variation at 2p24.3 and risk for aggressive prostate cancer. Cancer Res. 2009;69:2176–2179. doi: 10.1158/0008-5472.CAN-08-3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Sun J., Zheng S.L., Wiklund F., Isaacs S.D., Li G., Wiley K.E., Kim S.T., Zhu Y., Zhang Z., Hsu F.C., et al. Sequence variants at 22q13 are associated with prostate cancer risk. Cancer Res. 2009;69:10–15. doi: 10.1158/0008-5472.CAN-08-3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Amin Al Olama A., Kote-Jarai Z., Schumacher F.R., Wiklund F., Berndt S.I., Benlloch S., Giles G.G., Severi G., Neal D.E., Hamdy F.C., et al. A meta-analysis of genome-wide association studies to identify prostate cancer susceptibility loci associated with aggressive and non-aggressive disease. Hum. Mol. Genet. 2013;22:408–415. doi: 10.1093/hmg/dds425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Nam R.K., Zhang W., Siminovitch K., Shlien A., Kattan M.W., Klotz L.H., Trachtenberg J., Lu Y., Zhang J., Yu C., et al. New variants at 10q26 and 15q21 are associated with aggressive prostate cancer in a genome-wide association study from a prostate biopsy screening cohort. Cancer Biol. Ther. 2011;12:997–1004. doi: 10.4161/cbt.12.11.18366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.FitzGerald L.M., Kwon E.M., Conomos M.P., Kolb S., Holt S.K., Levine D., Feng Z., Ostrander E.A., Stanford J.L. Genome-wide association study identifies a genetic variant associated with risk for more aggressive prostate cancer. Cancer Epidemiol. Biomed. Prev. 2011;20:1196–1203. doi: 10.1158/1055-9965.EPI-10-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Ahn J., Kibel A.S., Park J.Y., Rebbeck T.R., Rennert H., Stanford J.L., Ostrander E.A., Chanock S., Wang M.H., Mittal R.D., et al. Prostate cancer predisposition loci and risk of metastatic disease and prostate cancer recurrence. Clin. Cancer Res. 2011;17:1075–1081. doi: 10.1158/1078-0432.CCR-10-0881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Szulkin R., Karlsson R., Whitington T., Aly M., Gronberg H., Eeles R.A., Easton D.F., Kote-Jarai Z., Al Olama A.A., Benlloch S., et al. Genome-wide association study of prostate cancer-specific survival. Cancer Epidemiol. Biomed. Prev. 2015;24:1796–1800. doi: 10.1158/1055-9965.EPI-15-0543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Penney K.L., Pyne S., Schumacher F.R., Sinnott J.A., Mucci L.A., Kraft P.L., Ma J., Oh W.K., Kurth T., Kantoff P.W., et al. Genome-wide association study of prostate cancer mortality. Cancer Epidemiol. Biomed. Prev. 2010;19:2869–2876. doi: 10.1158/1055-9965.EPI-10-0601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Wiklund F.E., Adami H.O., Zheng S.L., Stattin P., Isaacs W.B., Gronberg H., Xu J. Established prostate cancer susceptibility variants are not associated with disease outcome. Cancer Epidemiol. Biomed. Prev. 2009;18:1659–1662. doi: 10.1158/1055-9965.EPI-08-1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Berndt S.I., Wang Z., Yeager M., Alavanja M.C., Albanes D., Amundadottir L., Andriole G., Beane Freeman L., Campa D., Cancel-Tassin G., et al. Two susceptibility loci identified for prostate cancer aggressiveness. Nat. Commun. 2015;6:6889. doi: 10.1038/ncomms7889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Xu J., Zheng S.L., Isaacs S.D., Wiley K.E., Wiklund F., Sun J., Kader A.K., Li G., Purcell L.D., Kim S.T., et al. Inherited genetic variant predisposes to aggressive but not indolent prostate cancer. Proc. Natl. Acad. Sci. USA. 2010;107:2136–2140. doi: 10.1073/pnas.0914061107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Li W., Middha M., Bicak M., Sjoberg D.D., Vertosick E., Dahlin A., Haggstrom C., Hallmans G., Ronn A.C., Stattin P., et al. Genome-wide scan identifies role for aox1 in prostate cancer survival. Eur. Urol. 2018;74:710–719. doi: 10.1016/j.eururo.2018.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Shui I.M., Wong C.J., Zhao S., Kolb S., Ebot E.M., Geybels M.S., Rubicz R., Wright J.L., Lin D.W., Klotzle B., et al. Prostate tumor DNA methylation is associated with cigarette smoking and adverse prostate cancer outcomes. Cancer. 2016;122:2168–2177. doi: 10.1002/cncr.30045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Haldrup C., Mundbjerg K., Vestergaard E.M., Lamy P., Wild P., Schulz W.A., Arsov C., Visakorpi T., Borre M., Hoyer S., et al. DNA methylation signatures for prediction of biochemical recurrence after radical prostatectomy of clinically localized prostate cancer. J. Clin. Oncol. 2013;31:3250–3258. doi: 10.1200/JCO.2012.47.1847. [DOI] [PubMed] [Google Scholar]
- 243.Li H., Fei X., Shen Y., Wu Z. Association of gene polymorphisms of klk3 and prostate cancer: A meta-analysis. Adv. Clin. Exp. Med. 2020;29:1001–1009. doi: 10.17219/acem/121521. [DOI] [PubMed] [Google Scholar]
- 244.He Y., Gu J., Strom S., Logothetis C.J., Kim J., Wu X. The prostate cancer susceptibility variant rs2735839 near klk3 gene is associated with aggressive prostate cancer and can stratify gleason score 7 patients. Clin. Cancer Res. 2014;20:5133–5139. doi: 10.1158/1078-0432.CCR-14-0661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Pomerantz M.M., Werner L., Xie W., Regan M.M., Lee G.S., Sun T., Evan C., Petrozziello G., Nakabayashi M., Oh W.K., et al. Association of prostate cancer risk loci with disease aggressiveness and prostate cancer-specific mortality. Cancer Prev. Res. (Phila) 2011;4:719–728. doi: 10.1158/1940-6207.CAPR-10-0292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Kader A.K., Sun J., Isaacs S.D., Wiley K.E., Yan G., Kim S.T., Fedor H., DeMarzo A.M., Epstein J.I., Walsh P.C., et al. Individual and cumulative effect of prostate cancer risk-associated variants on clinicopathologic variables in 5895 prostate cancer patients. Prostate. 2009;69:1195–1205. doi: 10.1002/pros.20970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Xu J., Isaacs S.D., Sun J., Li G., Wiley K.E., Zhu Y., Hsu F.C., Wiklund F., Turner A.R., Adams T.S., et al. Association of prostate cancer risk variants with clinicopathologic characteristics of the disease. Clin. Cancer Res. 2008;14:5819–5824. doi: 10.1158/1078-0432.CCR-08-0934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Helfand B.T., Roehl K.A., Cooper P.R., McGuire B.B., Fitzgerald L.M., Cancel-Tassin G., Cornu J.N., Bauer S., Van Blarigan E.L., Chen X., et al. Associations of prostate cancer risk variants with disease aggressiveness: Results of the nci-spore genetics working group analysis of 18,343 cases. Hum. Genet. 2015;134:439–450. doi: 10.1007/s00439-015-1534-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Amin Al Olama A., Benlloch S., Antoniou A.C., Giles G.G., Severi G., Neal D.E., Hamdy F.C., Donovan J.L., Muir K., Schleutker J., et al. Risk analysis of prostate cancer in practical, a multinational consortium, using 25 known prostate cancer susceptibility loci. Cancer Epidemiol. Biomed. Prev. 2015;24:1121–1129. doi: 10.1158/1055-9965.EPI-14-0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Zheng J., Liu F., Lin X., Wang X., Ding Q., Jiang H., Chen H., Lu D., Jin G., Hsing A.W., et al. Predictive performance of prostate cancer risk in chinese men using 33 reported prostate cancer risk-associated snps. Prostate. 2012;72:577–583. doi: 10.1002/pros.21462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Macinnis R.J., Antoniou A.C., Eeles R.A., Severi G., Al Olama A.A., McGuffog L., Kote-Jarai Z., Guy M., O’Brien L.T., Hall A.L., et al. A risk prediction algorithm based on family history and common genetic variants: Application to prostate cancer with potential clinical impact. Genet. Epidemiol. 2011;35:549–556. doi: 10.1002/gepi.20605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Machiela M.J., Chen C.Y., Chen C., Chanock S.J., Hunter D.J., Kraft P. Evaluation of polygenic risk scores for predicting breast and prostate cancer risk. Genet. Epidemiol. 2011;35:506–514. doi: 10.1002/gepi.20600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Lecarpentier J., Silvestri V., Kuchenbaecker K.B., Barrowdale D., Dennis J., McGuffog L., Soucy P., Leslie G., Rizzolo P., Navazio A.S., et al. Prediction of breast and prostate cancer risks in male brca1 and brca2 mutation carriers using polygenic risk scores. J. Clin. Oncol. 2017;35:2240–2250. doi: 10.1200/JCO.2016.69.4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Conran C.A., Na R., Chen H., Jiang D., Lin X., Zheng S.L., Brendler C.B., Xu J. Population-standardized genetic risk score: The snp-based method of choice for inherited risk assessment of prostate cancer. Asian J. Androl. 2016;18:520–524. doi: 10.4103/1008-682X.179527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Szulkin R., Whitington T., Eklund M., Aly M., Eeles R.A., Easton D., Kote-Jarai Z.S., Amin Al Olama A., Benlloch S., Muir K., et al. Prediction of individual genetic risk to prostate cancer using a polygenic score. Prostate. 2015;75:1467–1474. doi: 10.1002/pros.23037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Akamatsu S., Takahashi A., Takata R., Kubo M., Inoue T., Morizono T., Tsunoda T., Kamatani N., Haiman C.A., Wan P., et al. Reproducibility, performance, and clinical utility of a genetic risk prediction model for prostate cancer in japanese. PLoS ONE. 2012;7:e46454. doi: 10.1371/journal.pone.0046454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Sipeky C., Talala K.M., Tammela T.L.J., Taari K., Auvinen A., Schleutker J. Prostate cancer risk prediction using a polygenic risk score. Sci. Rep. 2020;10:17075. doi: 10.1038/s41598-020-74172-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Pashayan N., Duffy S.W., Neal D.E., Hamdy F.C., Donovan J.L., Martin R.M., Harrington P., Benlloch S., Amin Al Olama A., Shah M., et al. Implications of polygenic risk-stratified screening for prostate cancer on overdiagnosis. Genet. Med. 2015;17:789–795. doi: 10.1038/gim.2014.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Callender T., Emberton M., Morris S., Eeles R., Kote-Jarai Z., Pharoah P.D.P., Pashayan N. Polygenic risk-tailored screening for prostate cancer: A benefit-harm and cost-effectiveness modelling study. PLoS Med. 2019;16:e1002998. doi: 10.1371/journal.pmed.1002998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Tasa T., Puustusmaa M., Tonisson N., Kolk B., Padrik P. Precision prostate cancer screening with a polygenic risk score. medRxiv. 2008:20180570. [Google Scholar]
- 261.Lewis C.M., Vassos E. Polygenic risk scores: From research tools to clinical instruments. Genom. Med. 2020;12:44. doi: 10.1186/s13073-020-00742-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Martin A.R., Kanai M., Kamatani Y., Okada Y., Neale B.M., Daly M.J. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat. Genet. 2019;51:584–591. doi: 10.1038/s41588-019-0379-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Huynh-Le M.P., Fan C.C., Karunamuni R., Walsh E.I., Turner E.L., Lane J.A., Martin R.M., Neal D.E., Donovan J.L., Hamdy F.C., et al. A genetic risk score to personalize prostate cancer screening, applied to population data. Cancer Epidemiol. Biomed. Prev. 2020;29:1731–1738. doi: 10.1158/1055-9965.EPI-19-1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Seibert T.M., Fan C.C., Wang Y., Zuber V., Karunamuni R., Parsons J.K., Eeles R.A., Easton D.F., Kote-Jarai Z., Al Olama A.A., et al. Polygenic hazard score to guide screening for aggressive prostate cancer: Development and validation in large scale cohorts. BMJ. 2018;360:j5757. doi: 10.1136/bmj.j5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Huynh-Le M.-P., Fan C.C., Karunamuni R., Thompson W.K., Martinez M.E., Eeles R.A., Kote-Jarai Z., Muir K., Schleutker J., Pashayan N., et al. Polygenic hazard score is associated with prostate cancer in multi-ethnic populations. medRxiv. 2020:19012237. doi: 10.1038/s41467-021-21287-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.MacInnis R.J., Antoniou A.C., Eeles R.A., Severi G., Guy M., McGuffog L., Hall A.L., O’Brien L.T., Wilkinson R.A., Dearnaley D.P., et al. Prostate cancer segregation analyses using 4390 families from uk and australian population-based studies. Genet. Epidemiol. 2010;34:42–50. doi: 10.1002/gepi.20433. [DOI] [PubMed] [Google Scholar]
- 267.Mancuso N., Rohland N., Rand K.A., Tandon A., Allen A., Quinque D., Mallick S., Li H., Stram A., Sheng X., et al. The contribution of rare variation to prostate cancer heritability. Nat. Genet. 2016;48:30–35. doi: 10.1038/ng.3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Sham P.C., Purcell S.M. Statistical power and significance testing in large-scale genetic studies. Nat. Rev. Genet. 2014;15:335–346. doi: 10.1038/nrg3706. [DOI] [PubMed] [Google Scholar]
- 269.Lee S., Abecasis G.R., Boehnke M., Lin X. Rare-variant association analysis: Study designs and statistical tests. Am. J. Hum. Genet. 2014;95:5–23. doi: 10.1016/j.ajhg.2014.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Li D., Lewinger J.P., Gauderman W.J., Murcray C.E., Conti D. Using extreme phenotype sampling to identify the rare causal variants of quantitative traits in association studies. Genet. Epidemiol. 2011;35:790–799. doi: 10.1002/gepi.20628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Pritchard C.C., Mateo J., Walsh M.F., De Sarkar N., Abida W., Beltran H., Garofalo A., Gulati R., Carreira S., Eeles R., et al. Inherited DNA-repair gene mutations in men with metastatic prostate cancer. N. Engl. J. Med. 2016;375:443–453. doi: 10.1056/NEJMoa1603144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Schrader K.A., Cheng D.T., Joseph V., Prasad M., Walsh M., Zehir A., Ni A., Thomas T., Benayed R., Ashraf A., et al. Germline variants in targeted tumor sequencing using matched normal DNA. JAMA Oncol. 2016;2:104–111. doi: 10.1001/jamaoncol.2015.5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Robinson D., Van Allen E.M., Wu Y.M., Schultz N., Lonigro R.J., Mosquera J.M., Montgomery B., Taplin M.E., Pritchard C.C., Attard G., et al. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;161:1215–1228. doi: 10.1016/j.cell.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Huang K.L., Mashl R.J., Wu Y., Ritter D.I., Wang J., Oh C., Paczkowska M., Reynolds S., Wyczalkowski M.A., Oak N., et al. Pathogenic germline variants in 10,389 adult cancers. Cell. 2018;173:355–370. doi: 10.1016/j.cell.2018.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Hart S.N., Ellingson M.S., Schahl K., Vedell P.T., Carlson R.E., Sinnwell J.P., Barman P., Sicotte H., Eckel-Passow J.E., Wang L., et al. Determining the frequency of pathogenic germline variants from exome sequencing in patients with castrate-resistant prostate cancer. BMJ Open. 2016;6:e010332. doi: 10.1136/bmjopen-2015-010332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Lu C., Xie M., Wendl M.C., Wang J., McLellan M.D., Leiserson M.D., Huang K.L., Wyczalkowski M.A., Jayasinghe R., Banerjee T., et al. Patterns and functional implications of rare germline variants across 12 cancer types. Nat. Commun. 2015;6:10086. doi: 10.1038/ncomms10086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Nicolosi P., Ledet E., Yang S., Michalski S., Freschi B., O’Leary E., Esplin E.D., Nussbaum R.L., Sartor O. Prevalence of germline variants in prostate cancer and implications for current genetic testing guidelines. JAMA Oncol. 2019;5:523–528. doi: 10.1001/jamaoncol.2018.6760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Leongamornlert D., Saunders E., Dadaev T., Tymrakiewicz M., Goh C., Jugurnauth-Little S., Kozarewa I., Fenwick K., Assiotis I., Barrowdale D., et al. Frequent germline deleterious mutations in DNA repair genes in familial prostate cancer cases are associated with advanced disease. Br. J. Cancer. 2014;110:1663–1672. doi: 10.1038/bjc.2014.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Paulo P., Maia S., Pinto C., Pinto P., Monteiro A., Peixoto A., Teixeira M.R. Targeted next generation sequencing identifies functionally deleterious germline mutations in novel genes in early-onset/familial prostate cancer. PLoS Genet. 2018;14:e1007355. doi: 10.1371/journal.pgen.1007355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Pilie P.G., Johnson A.M., Hanson K.L., Dayno M.E., Kapron A.L., Stoffel E.M., Cooney K.A. Germline genetic variants in men with prostate cancer and one or more additional cancers. Cancer. 2017;123:3925–3932. doi: 10.1002/cncr.30817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Mijuskovic M., Saunders E.J., Leongamornlert D.A., Wakerell S., Whitmore I., Dadaev T., Cieza-Borrella C., Govindasami K., Brook M.N., Haiman C.A., et al. Rare germline variants in DNA repair genes and the angiogenesis pathway predispose prostate cancer patients to develop metastatic disease. Br. J. Cancer. 2018;119:96–104. doi: 10.1038/s41416-018-0141-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Na R., Zheng S.L., Han M., Yu H., Jiang D., Shah S., Ewing C.M., Zhang L., Novakovic K., Petkewicz J., et al. Germline mutations in atm and brca1/2 distinguish risk for lethal and indolent prostate cancer and are associated with early age at death. Eur. Urol. 2017;71:740–747. doi: 10.1016/j.eururo.2016.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Leongamornlert D.A., Saunders E.J., Wakerell S., Whitmore I., Dadaev T., Cieza-Borrella C., Benafif S., Brook M.N., Donovan J.L., Hamdy F.C., et al. Germline DNA repair gene mutations in young-onset prostate cancer cases in the uk: Evidence for a more extensive genetic panel. Eur. Urol. 2019;76:329–337. doi: 10.1016/j.eururo.2019.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Darst B.F., Dadaev T., Saunders E., Sheng X., Wan P., Pooler L., Xia L.Y., Chanock S., Berndt S.I., Gapstur S.M., et al. Germline sequencing DNA repair genes in 5,545 men with aggressive and non-aggressive prostate cancer. J. Natl. Cancer Inst. 2020 doi: 10.1093/jnci/djaa132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Matejcic M., Patel Y., Lilyquist J., Hu C., Lee K.Y., Gnanaolivu R.D., Hart S.N., Polley E.C., Yadav S., Boddicker N.J., et al. Pathogenic variants in cancer predisposition genes and prostate cancer risk in men of african ancestry. JCO Precis. Oncol. 2020;4:32–43. doi: 10.1200/PO.19.00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Castro E., Goh C., Leongamornlert D., Saunders E., Tymrakiewicz M., Dadaev T., Govindasami K., Guy M., Ellis S., Frost D., et al. Effect of brca mutations on metastatic relapse and cause-specific survival after radical treatment for localised prostate cancer. Eur. Urol. 2015;68:186–193. doi: 10.1016/j.eururo.2014.10.022. [DOI] [PubMed] [Google Scholar]
- 287.Castro E., Goh C., Olmos D., Saunders E., Leongamornlert D., Tymrakiewicz M., Mahmud N., Dadaev T., Govindasami K., Guy M., et al. Germline brca mutations are associated with higher risk of nodal involvement, distant metastasis, and poor survival outcomes in prostate cancer. J. Clin. Oncol. 2013;31:1748–1757. doi: 10.1200/JCO.2012.43.1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Taylor R.A., Fraser M., Livingstone J., Espiritu S.M., Thorne H., Huang V., Lo W., Shiah Y.J., Yamaguchi T.N., Sliwinski A., et al. Germline brca2 mutations drive prostate cancers with distinct evolutionary trajectories. Nat. Commun. 2017;8:13671. doi: 10.1038/ncomms13671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Castro E., Jugurnauth-Little S., Karlsson Q., Al-Shahrour F., Pineiro-Yanez E., Van de Poll F., Leongamornlert D., Dadaev T., Govindasami K., Guy M., et al. High burden of copy number alterations and c-myc amplification in prostate cancer from brca2 germline mutation carriers. Ann. Oncol. 2015;26:2293–2300. doi: 10.1093/annonc/mdv356. [DOI] [PubMed] [Google Scholar]
- 290.Akbari M.R., Wallis C.J., Toi A., Trachtenberg J., Sun P., Narod S.A., Nam R.K. The impact of a brca2 mutation on mortality from screen-detected prostate cancer. Br. J. Cancer. 2014;111:1238–1240. doi: 10.1038/bjc.2014.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Thorne H., Willems A.J., Niedermayr E., Hoh I.M., Li J., Clouston D., Mitchell G., Fox S., Hopper J.L., Kathleen Cunningham Consortium for Research in Familial Breast Cancer Consortium et al. Decreased prostate cancer-specific survival of men with brca2 mutations from multiple breast cancer families. Cancer Prev. Res. (Phila) 2011;4:1002–1010. doi: 10.1158/1940-6207.CAPR-10-0397. [DOI] [PubMed] [Google Scholar]
- 292.Lang S.H., Swift S.L., White H., Misso K., Kleijnen J., Quek R.G.W. A systematic review of the prevalence of DNA damage response gene mutations in prostate cancer. Int. J. Oncol. 2019;55:597–616. doi: 10.3892/ijo.2019.4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Wu Y., Yu H., Li S., Wiley K., Zheng S.L., LaDuca H., Gielzak M., Na R., Sarver B.A.J., Helfand B.T., et al. Rare germline pathogenic mutations of DNA repair genes are most strongly associated with grade group 5 prostate cancer. Eur. Urol. Oncol. 2020;3:224–230. doi: 10.1016/j.euo.2019.12.003. [DOI] [PubMed] [Google Scholar]
- 294.Carter H.B., Helfand B., Mamawala M., Wu Y., Landis P., Yu H., Wiley K., Na R., Shi Z., Petkewicz J., et al. Germline mutations in atm and brca1/2 are associated with grade reclassification in men on active surveillance for prostate cancer. Eur. Urol. 2019;75:743–749. doi: 10.1016/j.eururo.2018.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Karlsson Q., Brook M.N., Dadaev T., Wakerell S., Saunders E.J., Muir K., Neal D.E., Giles G.G., MacInnis R.J., Thibodeau S.N., et al. Rare germline variants in atm predispose to prostate cancer: A practical consortium study. Eur. Urol. Oncol. 2021 doi: 10.1016/j.euo.2020.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Mateo J., Carreira S., Sandhu S., Miranda S., Mossop H., Perez-Lopez R., Nava Rodrigues D., Robinson D., Omlin A., Tunariu N., et al. DNA-repair defects and olaparib in metastatic prostate cancer. N. Engl. J. Med. 2015;373:1697–1708. doi: 10.1056/NEJMoa1506859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Oak N., Cherniack A.D., Mashl R.J., Network T.A., Hirsch F.R., Ding L., Beroukhim R., Gumus Z.H., Plon S.E., Huang K.L. Ancestry-specific predisposing germline variants in cancer. Genom. Med. 2020;12:51. doi: 10.1186/s13073-020-00744-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Koboldt D.C., Kanchi K.L., Gui B., Larson D.E., Fulton R.S., Isaacs W.B., Kraja A., Borecki I.B., Jia L., Wilson R.K., et al. Rare variation in tet2 is associated with clinically relevant prostate carcinoma in african americans. Cancer Epidemiol. Biom. Prev. 2016;25:1456–1463. doi: 10.1158/1055-9965.EPI-16-0373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Nyberg T., Frost D., Barrowdale D., Evans D.G., Bancroft E., Adlard J., Ahmed M., Barwell J., Brady A.F., Brewer C., et al. Prostate cancer risks for male brca1 and brca2 mutation carriers: A prospective cohort study. Eur. Urol. 2020;77:24–35. doi: 10.1016/j.eururo.2019.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alfoldi J., Wang Q., Collins R.L., Laricchia K.M., Ganna A., Birnbaum D.P., et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Ott J., Wang J., Leal S.M. Genetic linkage analysis in the age of whole-genome sequencing. Nat. Rev. Genet. 2015;16:275–284. doi: 10.1038/nrg3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Morgans A.K., Szymaniak B.M. Genetically-informed treatment for advanced and metastatic prostate cancer. Can. J. Urol. 2019;26:54–56. [PubMed] [Google Scholar]
- 303.Giri V.N., Knudsen K.E., Kelly W.K., Cheng H.H., Cooney K.A., Cookson M.S., Dahut W., Weissman S., Soule H.R., Petrylak D.P., et al. Implementation of germline testing for prostate cancer: Philadelphia prostate cancer consensus conference 2019. J. Clin. Oncol. 2020 doi: 10.1200/JCO.20.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.National Comprehensive Cancer Network Prostate Cancer (Version 3.2020) [(accessed on 14 December 2020)]; Available online: https://www.nccn.org/professionals/physician_gls/pdf/prostate.pdf.
- 305.de Bono J., Mateo J., Fizazi K., Saad F., Shore N., Sandhu S., Chi K.N., Sartor O., Agarwal N., Olmos D., et al. Olaparib for metastatic castration-resistant prostate cancer. N. Engl. J. Med. 2020;382:2091–2102. doi: 10.1056/NEJMoa1911440. [DOI] [PubMed] [Google Scholar]
- 306.Mateo J., Porta N., McGovern U.B., Elliott T., Jones R.J., Syndikus I., Ralph C., Jain S., Varughese M.A., Parikh O., et al. Toparp-b: A phase ii randomized trial of the poly(adp)-ribose polymerase (parp) inhibitor olaparib for metastatic castration resistant prostate cancers (mcrpc) with DNA damage repair (ddr) alterations. J. Clin. Oncol. 2019;37:5005. doi: 10.1200/JCO.2019.37.15_suppl.5005. [DOI] [Google Scholar]
- 307.Mota J.M., Barnett E., Nauseef J.T., Nguyen B., Stopsack K.H., Wibmer A., Flynn J.R., Heller G., Danila D.C., Rathkopf D., et al. Platinum-based chemotherapy in metastatic prostate cancer with DNA repair gene alterations. JCO Precis. Oncol. 2020;4:355–366. doi: 10.1200/PO.19.00346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Hager S., Ackermann C.J., Joerger M., Gillessen S., Omlin A. Anti-tumour activity of platinum compounds in advanced prostate cancer-a systematic literature review. Ann. Oncol. 2016;27:975–984. doi: 10.1093/annonc/mdw156. [DOI] [PubMed] [Google Scholar]
- 309.Graff J.N., Alumkal J.J., Thompson R.F., Moran A., Thomas G.V., Wood M.A., Drake C.G., Slottke R., Beer T.M. Pembrolizumab (pembro) plus enzalutamide (enz) in metastatic castration resistant prostate cancer (mcrpc): Extended follow up. J. Clin. Oncol. 2018;36:5047. doi: 10.1200/JCO.2018.36.15_suppl.5047. [DOI] [Google Scholar]
- 310.Tucker M.D., Zhu J., Marin D., Gupta R.T., Gupta S., Berry W.R., Ramalingam S., Zhang T., Harrison M., Wu Y., et al. Pembrolizumab in men with heavily treated metastatic castrate-resistant prostate cancer. Cancer Med. 2019;8:4644–4655. doi: 10.1002/cam4.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Tan S.H., Petrovics G., Srivastava S. Prostate cancer genomics: Recent advances and the prevailing underrepresentation from racial and ethnic minorities. Int. J. Mol. Sci. 2018;19:1255. doi: 10.3390/ijms19041255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Bentley A.R., Callier S., Rotimi C.N. Diversity and inclusion in genomic research: Why the uneven progress? J. Commun. Genet. 2017;8:255–266. doi: 10.1007/s12687-017-0316-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Haga S.B. Impact of limited population diversity of genome-wide association studies. Genet. Med. 2010;12:81–84. doi: 10.1097/GIM.0b013e3181ca2bbf. [DOI] [PubMed] [Google Scholar]