Abstract
Long non-coding RNA (lncRNA) small nucleolar RNA host gene 7 (SNHG7) plays a crucial role in the progression of pancreatic cancer (PC). SNHG7 is upregulated in PC; therefore, the purpose of the present study was to investigate the role and underlying mechanism of SNHG7 on PC progression. In the present study, the mRNA expression levels of SNHG7, microRNA(miR)-146b-5p and roundabout homolog 1 (Robo1) were measured via reverse transcription-quantitative PCR. Moreover, cell viability and apoptosis were assessed by MTT and flow cytometry assays, respectively. The ability of cells to migrate and invade was evaluated by Transwell assays. In addition, dual-luciferase reporter, RNA immunoprecipitation and RNA pull-down assays were conducted to assess the interaction between miR-146b-5p and SNHG7 or Robo1. The protein expression of Robo1 was measured via western blotting. Furthermore, mouse xenograft models were established to further investigate the effect of SNHG7 on PC progression in vivo. The results indicated that SNHG7 was highly expressed in PC tissues and cells. It was also found that SNHG7 was sponged by miR-146b-5p and that Robo1 was a target of miR-146b-5p. Moreover, it was demonstrated that SNHG7 knockdown inhibited cell proliferation, migration and invasion, as well as tumorigenesis and apoptosis of PC cells in vitro and in vivo by regulating miR-146b-5p. The results also suggested that miR-146b-5p overexpression inhibited the progression of PC cells by modulating Robo1. Furthermore, silencing of SNHG7 downregulated Robo1 expression by sponging miR-146b-5p. Collectively, the present results indicate that SNHG7 promotes PC progression by sponging miR-146b-5p and upregulating Robo1.
Keywords: long non-coding RNA small nucleolar RNA host gene 7, microRNA-146b-5p, roundabout homolog 1, pancreatic cancer, cancer progression
Introduction
Pancreatic cancer (PC), a lethal tumor type occurring worldwide, is associated with a poor patient prognosis and a high morbidity rate among elderly patients, with an average age of 71 years at diagnosis (1,2). Currently, resection is the most effective treatment to prolong the survival time of patients with PC (3). However, early-stage diagnosis has not substantially improved in the past decades (4) and ~80% of patients with PC miss the optimal time for surgical resection (5). Therefore, it is important to identify novel biomarkers for early-stage diagnosis of PC.
Long non-coding RNAs (lncRNAs), >200 nucleotides in length, are a type of transcript that lack translation capacity and can affect target genes at the transcriptional or post-transcriptional stages (6). It has been shown that numerous lncRNAs are involved in PC progression. For example, lncRNA differentiation antagonizing non-protein coding RNA (DANCR) promotes cell proliferation and invasion in PC by sponging microRNA (miRNA/miR)-135a (7). In addition, other lncRNAs, such as CCDC26 lncRNA (8), Pvt1 oncogene (PVT1) (9), urothelial cancer associated 1 (UCA1) (10) and ZEB2 antisense RNA 1 (ZEB2-AS1) (11) show similar effects in PC. Small nucleolar RNA host gene 7 (SNHG7), located on chromosome 9q34.3 with 984 bp, has been identified to be aberrantly expressed in several types of tumor, including lung cancer (12), gastric cancer (13), glioblastoma (14) and PC (15). However, to the best of our knowledge, there have been few studies examining the biological mechanism of SNHG7 in PC.
miRNAs, ~22 nucleotides in length, are small non-coding RNAs with no translation ability that constrain gene expression by inhibiting mRNA translation or mediating mRNA degradation (16). Previous studies have shown that several miRNAs, such as miR-135b-5p (17), miR-613(18) and miR-1181(19), are dysregulated and affect PC progression. It has also been revealed that miR-146b-5p is implicated in PC progression (20). Moreover, roundabout homolog 1 (Robo1), located on human chromosome 3p12.3, is related to the processes of tumor progression in PC (21). Therefore, the aims of the present study were to investigate the functions and mechanism of SNHG7 in PC, thus providing a novel therapeutic target for patients with PC.
Materials and methods
Tissue collection
The study was approved by the Ethics Committee of Jingzhou Central Hospital (Jingzhou, Hubei, China) and performed according to the Declaration of Helsinki Principles. The age of the patients ranged from 20-81 years, including 18 females and 32 males. In total, 50 PC tissue samples, including 20 I-II stage PC samples and 30 stage III-IV PC samples, were obtained from Jingzhou Central Hospital between March 2016 and August 2018, as well as 50 corresponding adjacent healthy tissue samples. The staging system used for PC was the American Joint Committee on Cancer tumor node metastasis (TNM)system (22). The collected PC tissue samples were divided into two groups based on the median value of the expression levels of SNHG7 as determined by RT-qPCR assays: i) Patients with PC with high SNHG7 expression (n=25); and ii) Patients with PC with low SNHG7 expression (n=25). All tissues were frozen at 80˚C until further analysis. Before the experiments, written informed consent was provided by all patients with PC.
Cell culture and transfection
In total, four PC cell lines PANC-1, SW1990, BxPC-3 and AsPC-1, as well as the healthy pancreas ductal epithelial cell line HPDE were purchased from Tong Pai Technology. Cells were cultured in DMEM [Wokawi (Beijing) Biotechnology Co., Ltd.] supplemented with 10% FBS (Beijing BioDee Biotechnology Co., Ltd.) in a 5% CO2 incubator at 37˚C.
Short hairpin RNA (shRNA) targeting SNHG7 (sh-SNHG7) and its negative control (sh-control), miR-146b-5p mimics (miR-146b-5p: 5'-UGAGAACUGAAUUCCAUAGGCU-3') and its control (miR-control: 5'-UUCUCCGAACGUGUCACGUTT-3'), miR-146b-5p inhibitor (5'-AGCCUAUGGAAUUCAGUUCUCA-3') and its control (inhibitor-control: 5'-CAGUACUUUUGUGUAGUACAA-3') were obtained from Shanghai GenePharma Co., Ltd. The sequences of SNHG7 (Accession: NR_024543.1) and Robo1 (Accession: GBYX01104758.1) were inserted into pcDNA vectors (Invitrogen; Thermo Fisher Scientific, Inc.) to construct overexpression plasmids, referred to as pcDNA-SNHG7 and Robo1, respectively. SW1990 and AsPC-1 cells were transfected with the constructed vectors or miRs (final concentration 50 µM) by using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions 48 h after transfection, the cells were used for further assays.
Reverse transcription-quantitative PCR (RT-qPCR)
RNA from PC tissue samples or cells was extracted using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.). Specific primers (Takara Biotechnology Co., Ltd) were used targeting RT SNHG7 and Robo1 with the PrimeScript™RT Master Mix kit (Takara Biotechnology Co., Ltd.). Briefly, the transcription was performed in a 10 µl reaction mixture, including polyadenylated RNA (100 ng), 5XPrimeScript Buffer (2 µl), PrimeScript RT Enzyme Mix I (0.5 µl), RT primer mixture (1 µl) and RNase-free water. The total reaction mixture was incubated at 50˚C for 15 min and 85˚C for 5 sec.
While RT for miR-146b-5p was conducted using TaqMan miRNA assays (Applied Biosystems; Thermo Fisher Scientific, Inc.). qPCR was performed using SYBR Premix Ex Taq II (Takara Biotechnology Co., Ltd.). The amplification parameters were as follows: Denaturation at 95˚C for 10 min, followed by 40 cycles of denaturation at 95˚C for 30 sec, annealing at 60˚C for 30 sec and extension at 72˚C for 1 min. The relative expression levels of SNHG7, miR-146b-5p and Robo1 were normalized by GAPDH or small nuclear RNA U6, and then calculated by the 2-ΔΔCq method (23). The primers were synthesized by Songon Biotechj Co., Ltd. and are listed in Table I.
Table I.
Oligonucleotide sequences used in the present study.
| Gene | Sequences |
|---|---|
| SNHG7 | F: 5'-AGGCTGGCTGGAATAAAGGT-3' |
| R: 5'-TATGAAAAGGGAGGCGTGGT-3' | |
| miR-146b-5p | F: 5'-GATGAGAAGGTATTTCTGCT-3' |
| R: 5'-GAGAAATTGAAGGTCATAAA-3' | |
| Robo1 | F: 5'-GAAACAGCGACAGCAACCT-3' |
| R: 5'-TGACAAAACGCCCATCCT-3' | |
| GAPDH | F: 5'-TGTTCGTCATGGGTGTGAAC-3' |
| R: 5'-ATGGCATGGACTGTGGTCAT-3' | |
| U6 | F: 5'-CTCGCTTCGGCAGCACA-3' |
| R: 5'-AACGCTTCACGAATTTGCGT-3' |
SNHG7, small nucleolar RNA host gene 7; Robo1, roundabout homolog 1; F, forward; R, reverse.
MTT assay
MTT (Sigma-Aldrich; Thermo Fisher Scientific, Inc.) was used to assess the viability of SW1990 and AsPC-1 cells. Cells (6x103 per well) were seeded in 96-well plates and incubated for 24 h at 37˚C. Following transfection, cells were cultured for another 0, 24, 48 or 72 h at 37˚C. Subsequently, MTT (5 mg/ml) was added into each well and incubated at 37˚C for 4 h. Then, 150 µl DMSO (Sigma-Aldrich; Merck KGaA) was added to solubilize formazan. The absorbance at 490 nm was measured by a Multiscan Spectrum (Beijing Putian New Bridge Technology Co., Ltd.).
Flow cytometry analysis of cell apoptosis
An Annexin V-FITC/propidium iodide (PI) apoptosis detection kit (Beijing Solarbio Science & Technology Co., Ltd.) was used to evaluate the apoptotic rate. Transfected SW1990 and AsPC-1 cells (1x105) were re-suspended in binding buffer (Beijing Solarbio Science & Technology Co., Ltd.), incubated with 5 µl Annexin V-FITC for 10 min and then with 10 µl PI for 5 min in the dark at room temperature. Cell apoptotic rate was assessed using flow cytometry (Agilent 2100 Bioanalyzer; Agilent Technologies, Inc.) and analyzed using CELL Quest 3.0 software (BD Biosciences).
Transwell assay
Transwell chambers (Corning, Inc.) with inserts of 8-µm pore were used to assess the migratory and invasive abilities of SW1990 and AsPC-1 cells. For migratory ability, transfected SW1990 and AsPC-1 cells at a density of 1x105 cells/well were plated into the upper chamber supplemented with non-FBS DMEM, while the lower chamber contained DMEM supplemented with 10% FBS. After 24 h culture, cells were fixed in 4% methanol for 20 min at room temperature, and stained with 0.1% crystal violet for 30 min at room temperature. Cells in 10 random fields were analyzed with a fluorescent microscope (magnification, x100; Olympus Corporation). For cell invasive ability, the upper chamber was coated with Matrigel matrix (BD Biosciences) at 4˚C, followed by incubation for 5 h at 37˚C.
Dual-luciferase reporter assay
The interaction between miR-146b-5p and SNHG7 or Robo1 was predicted by starBase v2.0 (http://starbase.sysu.edu.cn/) and TargetScan (http://www.targetscan.org/vert_72/) online databases. The wild-type (WT) and mutant (MUT) fragments of SNHG7 were amplified and then inserted into pGL3 vector (Promega Corporation) to construct the luciferase reporter, referred to as SNHG7 WT and SNHG7 MUT, respectively. The luciferase reporter (0.1 µg) and miR-146b-5p mimics (40 nM) or miR-control (40 nM) were co-transfected into SW1990 and AsPC-1 cells using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). The construction of the luciferase reporter Robo1 3'untranslated region (UTR) WT (GUUCUCA) or Robo1 3'UTR MUT (ACCAAUG) were similar to that of SNHG7 WT (CAGTTCTC) or SNHG7 MUT (ATCAAGCA) reporter. The luciferase activity was assessed using a dual-luciferase assay kit (Beijing Solarbio Science & Technology Co., Ltd.). Renilla luciferase activities were used as the internal control for the normalization of firefly luciferase activity.
RNA immunoprecipitation (RIP) assay
Following lysis of the transfected SW1990 and AsPC-1 cells, the lysate samples were incubated with magnetic beads labelled with anti-argonaute-2 (Ago2; BIOSS) or negative control anti-IgG antibody (BIOSS) for 4 h at 4˚C, and a 10 µl aliquot of RIP mixture was saved as input. Then, DNAse I (20 U; Sigma-Aldrich; Merck KGaA) for 15 min at 37˚C, and proteinase K (0.5 mg/ml; Sigma-Aldrich; Merck KGaA) for 15 min at 55˚C, were used to treat the immunoprecipitates for 20 min. Input functioned as a positive control. Subsequently, the expression of SNHG7 was detected by RT-qPCR as aforementioned.
RNA pull-down assay
Biotin (Bio)-labelled miR-146b-5p (Bio-miR-146b-5p; forward: 5'-ugagaacuc gccgcgggaccg c-3'; reverse: 5'-gcggucccgcggcgaguucuca-3') and its negative control (Bio-NC; forward: 5'-uucuccgaacguguc acgutt-3'; reverse: 5'-acgugacacguucgg agaatt-3') were obtained from Sangon Biotech Co., Ltd. SW1990 and AsPC-1 cells (3x106) were transfected with Bio-miR-146b-5p or Bio-NCata final concentration of 100 nM using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). Subsequently, cells were lysed in specific lysis buffer (Ambion; Thermo Fisher Scientific, Inc.) for 10 min, and the lysate (the RNA-RNA complex) was conjugated with streptavidin magnetic beads (Dyna beads M-280 Streptavidin; Invitrogen; Thermo Fisher Scientific, Inc.), followed by incubation at 4˚C for 3 h. After elution twice with 500 µl pre-cooled lysis buffer, thrice with low salt buffer and once with high salt buffer in succession, the bound RNA was purified using Trizol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and then the expression of SNHG7 was measured by RT-qPCR.
Western blotting
Protein samples were extracted with RIPA lysis buffer (Beyotime Institute of Biotechnology) and then quantified using the BCA Protein Assay kit (Beyotime Institute of Biotechnology). Next, 50 µg of each protein samples were separated by 10% SDS-PAGE and then transferred onto a PVDF (Absin Bioscience, Inc.) membrane. Subsequently, the membrane was blocked in 5% skimmed milk for 2 h at 37˚C and incubated with primary antibodies against Robo1 (cat. no. ab7279; 1:1,000; Abcam) and GAPDH (cat. no. ab8227; 1:5,000; Abcam), which acted as an internal reference, at 4˚C overnight. Then, the membrane was incubated with secondary antibodies (cat. no. ab205178; 1:10,000; Abcam) for a further 2 h at 37˚C. Band intensity was detected using an eyoECL Plus kit (Beyotime Institute of Biotechnology) and analyzed with Quantity One v4.6.2 software (Bio-Rad Laboratories, Inc.).
Mouse xenograft models
The experiment in nude mice was approved by the Animal Care Committee of Jingzhou Central Hospital and performed in accordance with the guidelines of National Institutes of Health (24). A total of 10 male nude mice (age, 6 weeks, weight 16-22 g; n=5 per group) were purchased from Shanghai Laboratory Animal Center, followed by maintained under specific pathogen-free conditions at a temperature of 25˚C and relative air humidity between 45 and 50%. Subsequently, 100 µl SW1990 cells (5x106 cells/ml) stably transfected with sh-SNHG7 were subcutaneously injected into a single side of the dorsal flank of nude mice. After inoculation, the tumor volumes were calculated every 5 days in accordance with the following formula: Volume (mm3) = width2 x length/2. At 30 days after cell injection, mice were euthanized using 2% methoxyflurane and cervical dislocation. Subsequently, xenograft tumor samples were excised for weight measurement, followed by analysis with RT-qPCR and western blot assays.
Statistical analysis
Data analysis was performed using GraphPad Prism 7 (GraphPad Software, Inc.). All experiments were repeated three times and data are presented as the mean ± SD. χ2 test was applied to analyze the correlations between the SNHG7 expression and clinicopathological characteristics of PC patients. The overall survival curve was plotted using Kaplan-Meier method and assessed with the log-rank test. Pearson correlation analysis was used to analyze the expression association. Comparison between two groups was analyzed via Student's t-test, while comparisons among multiple groups were calculated by one-way ANOVA followed by Tukey's test. P<0.05 was considered to indicate a statistically significant difference.
Results
SNHG7 is significantly upregulated in PC tissues and correlates with the pathological characteristics of tumor size, distant metastasis, lymph node metastasis and TNM stage of patients with PC
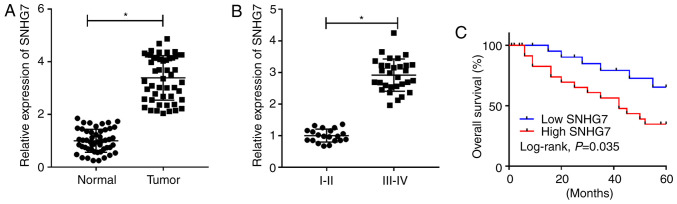
To assess the role of SNHG7 in PC, the expression of SNHG7 was detected in 50 PC tissue samples and corresponding adjacent healthy tissue samples. It was found that the expression of SNHG7 was significantly enhanced in PC tissues compared with that noted in the healthy tissue samples (Fig. 1A). Moreover, SNHG7 was more highly expressed in III-IV stage samples compared with I-II stage samples (P<0.05; Fig. 1B). Kaplan-Meier survival analysis results indicated that patients with PC with high expression of SNHG7 had a low survival rate compared with patients with PC with low expression of SNHG7 (Fig. 1C). In addition, the χ2 test results identified that high expression of SNHG7 was closely associated with tumor size (P=0.005), distant metastasis (P=0.011), lymph node metastasis (P=0.004) and TNM stage (P=0.004), while its expression was not associated with sex and age (Table II). Therefore, the results suggested that SNHG7 was significantly elevated in PC tissues and was associated with the pathological characteristics of tumor size, distant metastasis, lymph node metastasis and TNM stage in patients with PC.
Figure 1.
SNHG7 is significantly upregulated in PC tissues and correlates with the pathological characteristics of tumor size, distant metastasis, lymph node metastasis and TNM stage of patients with PC. Expression of SNHG7 in (A) PC tissues or (B) different tumor stages was measured by reverse transcription-quantitative PCR. (C) Association between overall survival rate and the expression of SNHG7. *P<0.05 vs. the respective normal groups. SNHG7, small nucleolar RNA host gene 7.
Table II.
Correlations between SNHG7 expression and clinicopathological characteristics of patients with PC.
| SNHG7 expression | ||||
|---|---|---|---|---|
| Characteristics | Total (n=50) | Low (n=25) | High (n=25) | P-valuea |
| Sex | 0.556 | |||
| Male | 32 | 15 | 17 | |
| Female | 18 | 10 | 8 | |
| Age | 0.564 | |||
| ≤60 | 20 | 9 | 11 | |
| >60 | 30 | 16 | 14 | |
| Tumor size, cm | 0.005b | |||
| ≤2 | 26 | 18 | 8 | |
| >2 | 24 | 7 | 17 | |
| Distant metastasis | 0.011b | |||
| Absent | 25 | 17 | 8 | |
| Present | 25 | 8 | 17 | |
| Lymph node metastasis | 0.004b | |||
| Negative | 30 | 20 | 10 | |
| Positive | 20 | 5 | 15 | |
| TNM stage | 0.004b | |||
| I-II | 28 | 19 | 9 | |
| III-IV | 22 | 6 | 16 | |
aχ2 test;
bP<0.05, indicative of a statistically significant difference. PC, pancreatic cancer; SNHG7, small nucleolar RNA host gene 7.
SNHG7 knockdown inhibits proliferation, migration and invasion of PC cells, and induces cell apoptosis
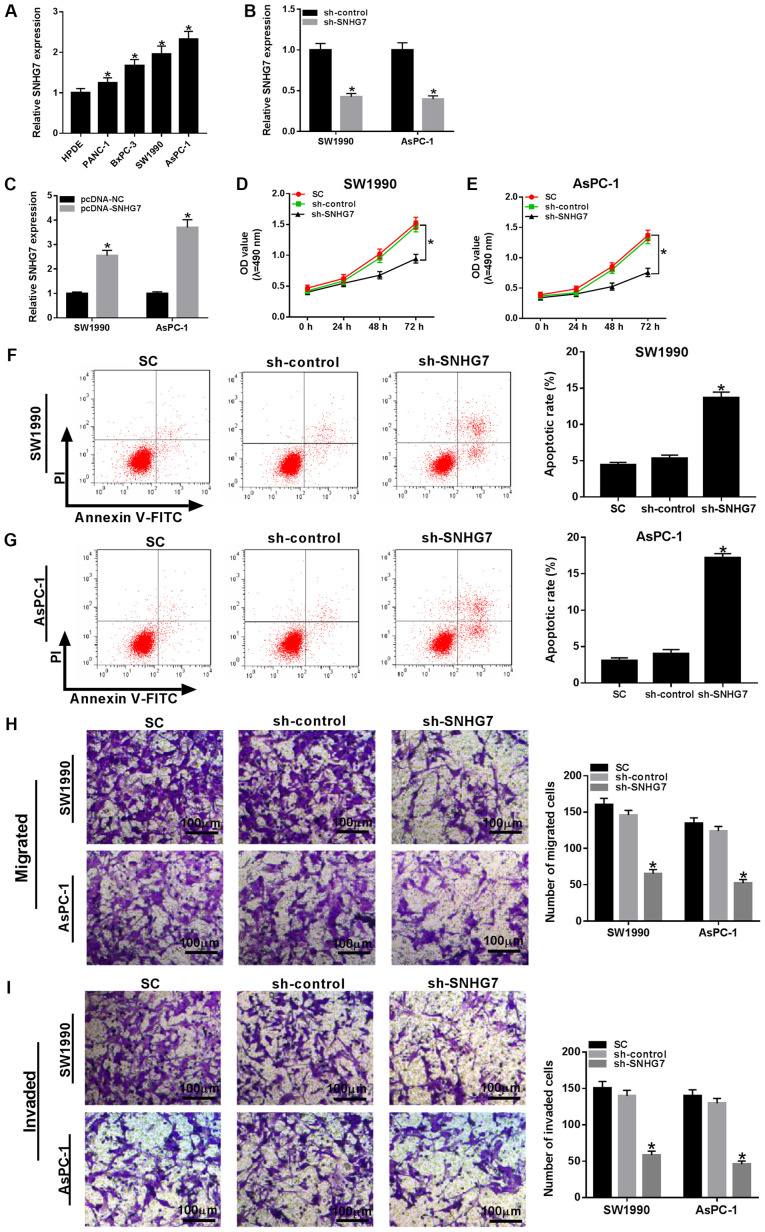
Based on the above results, the present study examined the expression of SNHG7 in PC cells. It was demonstrated that SNHG7 was significantly upregulated in PANC-1, BxPC-3, SW1990 and AsPC-1 cells compared with that noted in the HPDE cells (Fig. 2A), most notably in the SW1990 and AsPC-1 cells. Hence, SW1990 and AsPC-1 cells were chosen for subsequent experiments. Furthermore, the transfection efficiencies of sh-SNHG7 and pc-SNHG7 were examined and are presented in Fig. 2B and C. Subsequently, sh-SNHG7 was transfected into SW1990 and AsPC-1 cells to investigate the function of SNHG7. MTT assay results identified that cell viability was significantly decreased in SW1990 and AsPC-1 cells when SNHG7 was knocked down (Fig. 2D and E). Moreover, it was found that the transfection of sh-SNHG7 resulted in a significant increase in the apoptotic rate of SW1990 and AsPC-1 cells (Fig. 2F and G). In addition, the migratory and invasive abilities were both significantly reduced in SW1990 and AsPC-1 cells compared with the negative controls (Fig. 2H and I). Collectively, it was demonstrated that silencing SNHG7 inhibited proliferation, migration and invasion, and facilitated apoptosis in SW1990 and AsPC-1 cells.
Figure 2.
SNHG7 knockdown impairs cell proliferation, migration and invasion, but induces apoptosis in SW1990 and AsPC-1 cells. (A) Expression of SNHG7 in human pancreatic cancer cell lines PANC-1, BxPC-3, SW1990 and AsPC-1, and human pancreas normal ductal epithelial cell line HPDE was detected via reverse transcription-quantitative PCR. SNHG7 expression was detected in SW1990 and AsPC-1 cells transfected with (B) sh-control and sh-SNHG7, or (C) pcDNA-NC and pcDNA-SNHG7. SW1990 and AsPC-1 cells were transfected with sh-control, sh-SNHG7 or cultured in normal conditions. Cell viability in (D) SW1990 and (E) AsPC-1 cells was evaluated by MTT assay. Apoptotic rate in (F) SW1990 and (G) AsPC-1 cells was assessed via flow cytometry. (H) Migratory and (I) invasive abilities in transfected cells were measured by Transwell assay. Scale bar, 100 µm. *P<0.05 vs. their respective normal groups. sh, short hairpin RNA; SC, (standard conditions); SNHG7, small nucleolar RNA host gene 7; OD, optical density.
miR-146b-5p sponges SNHG7
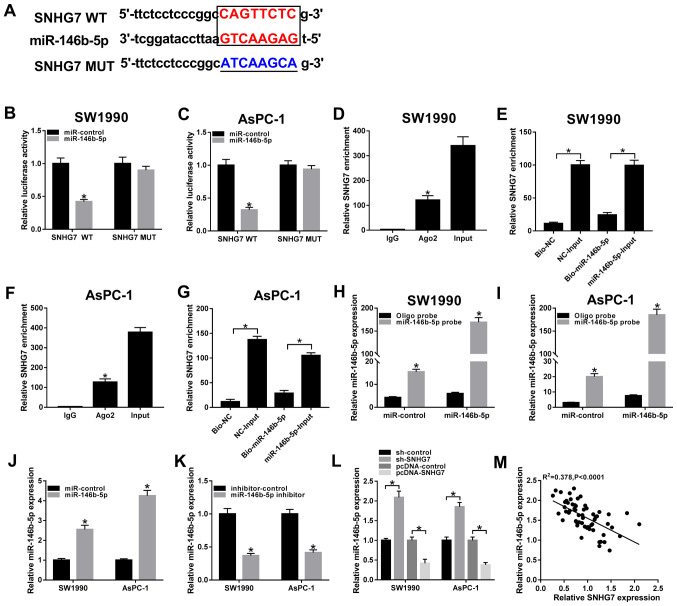
To investigate the biological mechanism of SNHG7 in PC progression starBase v2.0 (http://starbase.sysu.edu.cn) was used to predict the putative targets of SNHG7, and it was identified that miR-146b-5p had complementary sites with SNHG7 (Fig. 3A). Subsequently, dual-luciferase reporter assay results indicated that the transfection of miR-146b-5p significantly downregulated the luciferase activity of the SNHG7 WT reporter in SW1990 and AsPC-1 cells compared with the miR-control group (Fig. 3B and C). However, there were no significant differences in the luciferase activity of the SNHG7 MUT reporter in any group (Fig. 3B and C).
Figure 3.
miR-146b-5p is a direct target of SNHG7. (A) Complementary sequences between miR-146b-5p and SNHG7, and the MUT sequences of SNHG7. Luciferase activity of SNHG7 WT or SNHG7 MUT reporter in (B) SW1990 and (C) AsPC-1 cells transfected with miR-146b-5p or miR-control was assessed via dual-luciferase reporter. (D) Enrichment of SNHG7 in anti-Ago2 or anti-IgG labeled SW1990 cells transfected with miR-146b-5p or miR-control was evaluated by RIP assay. (E) Enrichment of SNHG7 in SW1990 cells transfected with Bio-miR-146b-5p or Bio-NC was analyzed by RNA pull-down assay. (F) Enrichment of SNHG7 in anti-Ago2 or anti-IgG labeled AsPC-1 cells transfected with miR-146b-5p or miR-control was evaluated by RIP assay. (G) Enrichment of SNHG7 in AsPC-1 cells transfected with Bio-miR-146b-5p or Bio-NC was analyzed by RNA pull-down assay. Transfection efficiency of miR-146b-5p probe was examined in (H) SW1990 and (I) AsPC-1 cells transfected with miR-control or miR-146b-5p. miR-146b-5p expression was measured in SW1990 and AsPC-1 cells transfected with (J) miR-control and miR-146b-5p, or (K) inhibitor-control and miR-146b-5p inhibitor. (L) Expression of miR-146b-5p in cells transfected with sh-control, sh-SNHG7, pcDNA-control or pcDNA-SNHG7 were detected via reverse transcription-quantitative PCR. (M) Correlation between SNHG7 and miR-146b-5p was analyzed by Pearson test. *P<0.05 vs. their respective normal groups. WT, wild-type; MUT, mutant; SNHG7, small nucleolar RNA host gene 7; miR, microRNA; sh, short hairpin RNA; NC, negative control; Ago2, argonaute-2.
It was demonstrated that SNHG7 was significantly enriched by the Ago2 antibody in SW1990 and AsPC-1 cells transfected with miR-146b-5p compared with the IgG antibody group (Fig. 3D and F). In addition, the RNA pull-down assay results indicated that Bio-miR-146b-5p had a lower enrichment of SNHG7 in SW1990 and AsPC-1 cells compared with the Bio-miR-146b-5p-Input group, and the Bio-NC group also had a lower enrichment of SNHG7 in PC cells relative to the NC-Input group(Fig. 3E and G). It was also found that the transfection of Bio-miR-146b-5p was successful in SW1990 and AsPC-1 cells (Fig. 3H and I).
Moreover, the transfection efficiency of the miR-146b-5p mimic and inhibitor on the upregulation or downregulation of miR-146b-5p expression, respectively, was examined (Fig. 3J and K). The results indicated the expression of miR-146b-5p was significantly elevated in SW1990 and AsPC-1 cells by silencing of SNHG7, while it was significantly decreased by overexpression of SNHG7 (Fig. 3L). It was also identified that the expression of miR-146b-5p was weakly negatively correlated with SNHG7, based on the interpretation of correlations (25) (Fig. 3M). Therefore, it was speculated that miR-146b-5p may negatively interact with SNHG7.
miR-146b-5p inhibitor relieves the constraint effects on proliferation, migration and invasion, as well as the promotive effect on cell apoptosis in SW1990 and AsPC-1 cells caused by SNHG7 silencing
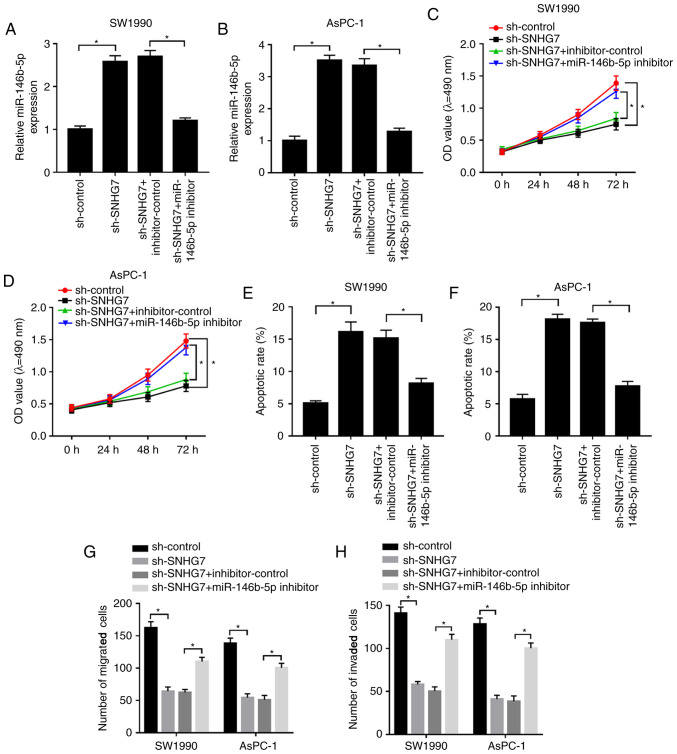
To examine whether the effect of SNHG7 on PC progression was mediated by miR-146b-5p, sh-SNHG7 and the miR-146b-5p inhibitor were co-transfected into SW1990 and AsPC-1 cells. It was found that the expression of miR-146b-5p was significantly increased in the sh-SNHG7-transfected SW1990 and AsPC-1 cells, while it was significantly reduced after miR-146b-5p inhibitor transfection (Fig. 4A and B). Furthermore, the miR-146b-5p inhibitor reduced the suppressive impacts on cell viability and the migratory and invasive abilities of SW1990 and AsPC-1 cells, which were inhibited by SNHG7 knockdown (Fig. 4C and D; G and H). However, transfection of sh-SNHG7 elevated the apoptotic rate of SW1990 and AsPC-1 cells, but this promotion was attenuated by the miR-146b-5p inhibitor (Fig. 4E and F). Collectively, the results suggested that silencing of SNHG7 reduced cell proliferation, migration and invasion, but promoted apoptosis in SW1990 and AsPC-1 cells by regulating miR-146b-5p.
Figure 4.
miR-146b-5p inhibitor relieves the constraint effects on cell proliferation, migration and invasion, as well as reduces the effect on apoptosis in SW1990 and AsPC-1 cells caused by SNHG7 silencing. SW1990 and AsPC-1 cells were transfected with sh-control, sh-SNHG7, sh-SNHG7 + inhibitor-control or sh-SNHG7 + miR-146b-5p inhibitor. Expression of miR-146b-5p was measured in (A) SW1990 and (B) AsPC-1 cells by reverse transcription-quantitative PCR. Cell viability was assessed in (C) SW1990 and (D) AsPC-1 cells via MTT assay. Apoptotic rate was evaluated in (E) SW1990 and (F) AsPC-1 cells via flow cytometry. (G) Migratory and (H) invasive abilities were detected via Transwell assay. *P<0.05 vs. the respective normal groups. SNHG7, small nucleolar RNA host gene 7; miR, microRNA; sh, short hairpin RNA; OD, optical density.
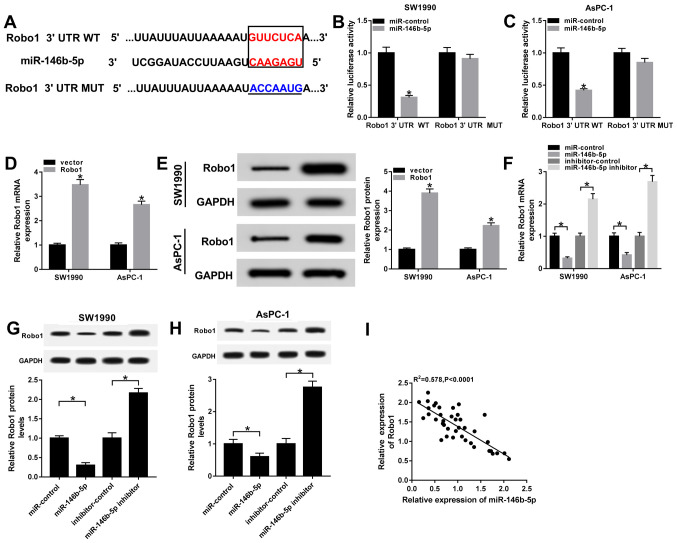
Robo1 negatively interacts with miR-146b-5p
To identify the biological mechanism of miR-146b-5p in PC, the TargetScan online database (http://www.targetscan.org) was used to predict the potential candidate targets of miR-146b-5p. The results suggested that the Robo1 3'UTR had complementary binding sites with miR-146b-5p (Fig. 5A). Moreover, dual-luciferase reporter assay results found that the luciferase activity of Robo1 3'UTR WT reporter was significantly reduced in SW1990 and AsPC-1 cells transfected with miR-146b-5p, while there were no significant changes in the miR-control group. However, the luciferase activity of Robo1 3'UTR MUT reporter had no significant fluctuations in any group (Fig. 5B and C).
Figure 5.
Robo1 negatively interacts with miR-146b-5p. (A) Complementary binding sites between miR-146b-5p and Robo1 3'UTR, and the MUT sequences of Robo1 3'UTR. Luciferase activity of Robo1 3'UTR WT or Robo1 3'UTR MUT reporter in (B) SW1990 and (C) AsPC-1 cells transfected with miR-146b-5p mimics or miR-control was evaluated via dual-luciferase reporter assay. (D) mRNA and (E) protein expression levels of Robo1 were examined in SW1990 and AsPC-1 cells transfected with vector and Robo1. (F) mRNA and protein expression levels of Robo1 in (G) SW1990 and (H) AsPC-1 cells transfected with miR-control, miR-146b-5p, inhibitor-control or miR-146b-5p inhibitor were detected by reverse transcription-quantitative PCR and western blotting, respectively. (I) Correlation between Robo1 and miR-146b-5p was assessed with a Pearson test. *P<0.05 vs. their respective normal groups. 3'UTR, 3'untranslated region; MUT, mutant; WT, wild-type; miR, microRNA; Robo1, roundabout homolog 1; SNHG7, small nucleolar RNA host gene 7.
Furthermore, the transfection efficiency of Robo1 overexpression in SW1990 and AsPC-1 cells was detected by RT-qPCR and western blotting (Fig. 5D and E). It was demonstrated that the mRNA and protein expression levels of Robo1 were significantly decreased in SW1990 and AsPC-1 cells transfected with miR-146b-5p mimics, while both were significant enhanced in the miR-146b-5p inhibitor group (Fig. 5F-H). Moreover, the results indicated that the expression of Robo1 was moderately negatively correlated with miR-146b-5p (Fig. 5I). Thus, it was speculated that Robo1 may be a direct target of miR-146b-5p.
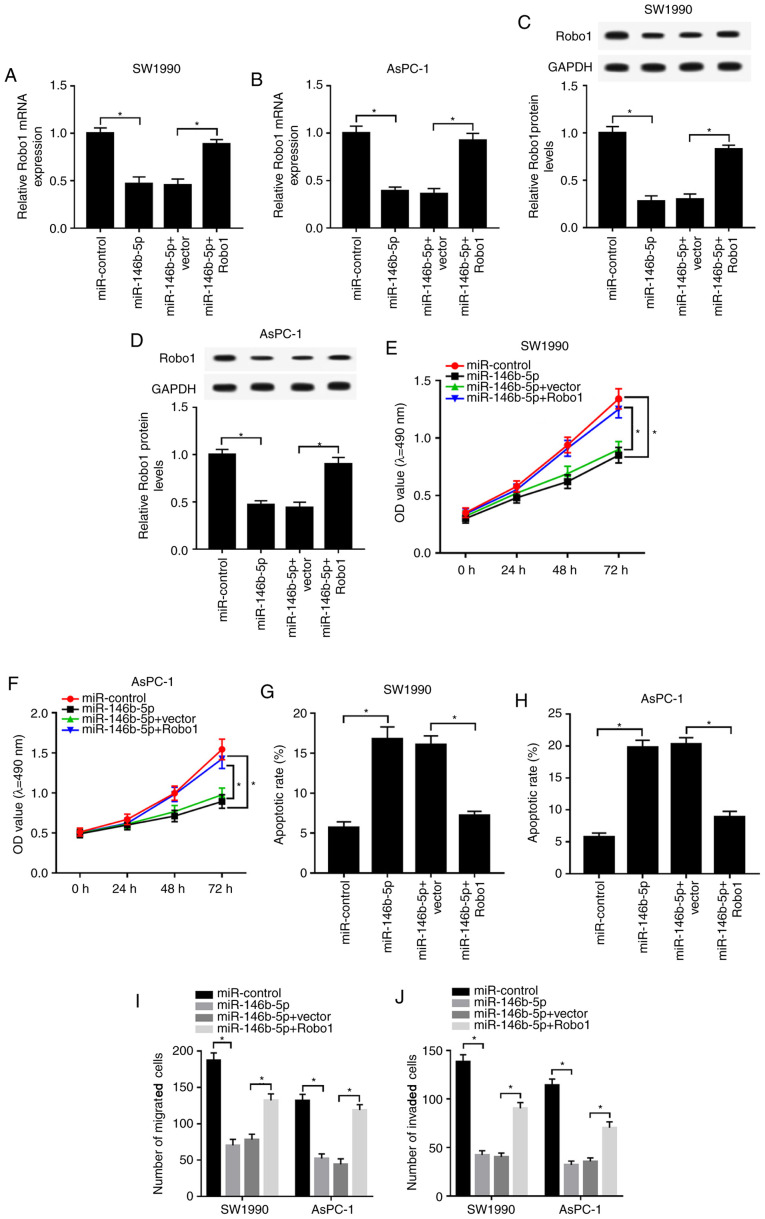
Robo1 overexpression mitigates the inhibitive effect on proliferation, migration and invasion, as well as the enhanced effect on apoptosis in SW1990 and AsPC-1 cells induced by miR-146b-5p mimics
Based on the above results, it was found that Robo1 negatively interacted with miR-146b-5p. Subsequently, the present study detected the functions of miR-146b-5p and Robo1 in PC. It was identified that mRNA and protein expression levels of Robo1 were reduced in miR-146b-5p-transfected SW1990 and AsPC-1 cells, while they were increased by the overexpression of Robo1 (Fig. 6A-D). Furthermore, overexpression of Robo1 reduced the inhibitory effect of miR-146b-5p on viability, migration and invasion in SW1990 and AsPC-1 cells (Fig. 6E and F; I and J). The flow cytometry results also demonstrated that the overexpression of Robo1 reversed the inhibitory effect of miR-146b-5p upregulation on the apoptotic rate in SW1990 and AsPC-1 cells (Fig. 6G and H). Therefore, the results indicated that miR-146b-5p reduced cell proliferation, migration and invasion, but increased apoptosis in PC cells by modulating Robo1.
Figure 6.
Robo1 overexpression mitigates the restraint effects on cell proliferation, migration and invasion, and reduces the effect on apoptosis in SW1990 and AsPC-1 cells induced by miR-146b-5p mimics. SW1990 and AsPC-1 cells were transfected with miR-control, miR-146b-5p, miR-146b-5p + vector or miR-146b-5p + Robo1. Expression of Robo1 was measured in (A) SW1990 and (B) AsPC-1 cells by reverse transcription-quantitative PCR. Protein expression of Robo1 was detected in (C) SW1990 and (D) AsPC-1 cells via western blotting. Cell viability in (E) SW1990 and (F) AsPC-1 cells was assessed by MTT assay. Apoptotic rate was evaluated in (G) SW1990 and (H) AsPC-1 cells by flow cytometry. (I) Migratory and (J) invasive abilities in transfected SW1990 and AsPC-1 cells were detected via Transwell assay. *P<0.05 vs. the respective normal groups. miR, microRNA; Robo1, roundabout homolog 1; SNHG7, small nucleolar RNA host gene 7; OD, optical density.
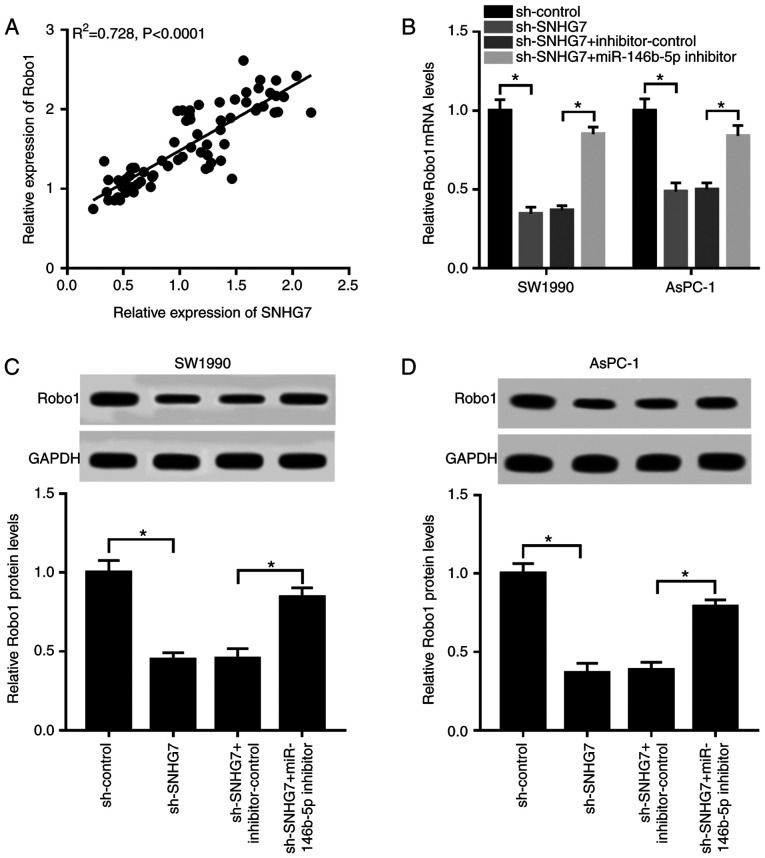
Silencing of SNHG7 downregulates Robo1 expression by sponging miR-146b-5p
The results suggested that the expression of Robo1 was strongly positively correlated with SNHG7 expression (Fig. 7A). To investigate the relationship between SNHG7, miR-146b-5p and Robo1, sh-SNHG7 and miR-146b-5p inhibitor were co-transfected into SW1990 and AsPC-1 cells. It was found that the mRNA and protein expression levels of Robo1 were significantly reduced in the SW1990 and AsPC-1 cells transfected with sh-SNHG7, while they were increased after transfection with the miR-146b-5p inhibitor (Fig. 7B-D). Thus, depletion of SNHG7 reduced Robo1 expression by acting as a miR-146b-5p sponge.
Figure 7.
SNHG7 silencing downregulates Robo1 expression by targeting miR-146b-5p. (A) Correlation between Robo1 and SNHG7 was determined by Pearson test. SW1990 and AsPC-1 cells were transfected with sh-control, sh-SNHG7, sh-SNHG7 + inhibitor or sh-SNHG7 + miR-146b-5p inhibitor. (B) mRNA expression of Robo1 was measured by reverse transcription-quantitative PCR. Protein expression of Robo1 was detected in (C) SW1990 and (D) AsPC-1 cells via western blotting. *P<0.05 vs. the respective normal groups. miR, microRNA; Robo1, roundabout homolog 1; SNHG7, small nucleolar RNA host gene 7; sh, short hairpin RNA.
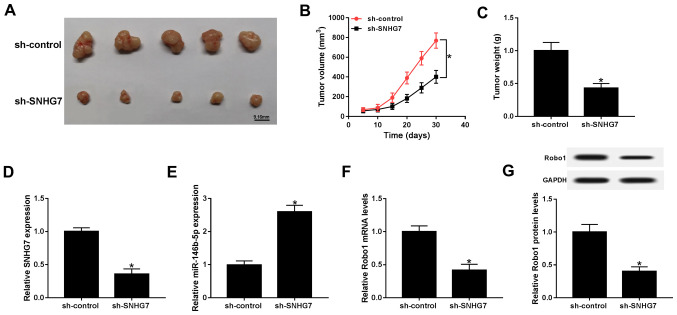
SNHG7 knockdown represses xenograft tumor growth in vivo
To further assess the function of SNHG7 in PC, SW1990 cells were transfected with sh-SNHG7 or sh-control and then injected into nude mice. Following 30 day measurement, the results indicated that tumor volume and weight were both significantly decreased in the sh-SNHG7 group compared with the sh-control group (Fig. 8A-C). Moreover, the maximum diameter and volume of the tumors were 18.32 mm and 768.56 mm3, respectively. Furthermore, the expression of SNHG7 was significantly downregulated, while the expression of miR-146b-5p was significantly elevated in the sh-SNHG7 group (Fig. 8D and E). The results also suggested that the mRNA and protein expression levels of Robo1 were significantly decreased in the sh-SNHG7 group (Fig. 8F and G). Collectively, the results demonstrated that SNHG7 knockdown inhibited xenograft tumor growth in vivo.
Figure 8.
SNHG7 silencing blocks xenograft tumor growth in vivo. Mice were injected with SW1990 cells transfected with sh-SNHG7 or sh-control. (A) Representative images of PC xenograft tissues are presented. (B) Tumor volume in treated mice. (C) Xenograft tumor weight in treated mice. mRNA expression levels of (D) SNHG7, (E) miR-146b-5p and (F) Robo1 in xenograft tumor were detected via reverse transcription-quantitative PCR. (G) Protein expression of Robo1 in xenograft tumor was measured by western blotting. *P<0.05 vs. the respective normal groups. miR, microRNA; Robo1, roundabout homolog 1; SNHG7, small nucleolar RNA host gene 7; sh, short hairpin RNA.
Discussion
PC is a fatal cancer type that occurs worldwide and is often associated with poor prognosis (1,26). Previous studies have shown that lncRNAs are implicated in the progression of various cancers (27-29). For example, Guo et al (30) reported that lncRNA HNF1A-AS1 knockdown suppressed cell migration, invasion and glycolysis through targeting the miR-124/MYO6 in colorectal cancer. Li et al (31), demonstrated that LncRNA FTX played a promoting role in gastric cell proliferation and invasion by regulating the miR144/Axis. Additionally, Feng et al (32) confirmed that lncRNA NEAT1 acted as a carcinogenic factor through facilitating cell proliferation and metastasis in PC development. The present study focused on the function and mechanism of SNHG7 in PC progression, and it was found that lncRNA SNHG7 knockdown inhibited PC progression via a miR-146b-5p/Robo1 axis.
Previous studies revealed that aberrant expression of SNHG1 occurs in various types of cancer, including PC. For instance, a study in melanoma indicated that SNHG7 is significantly elevated in melanoma tissues, and its silencing suppresses cell migration and invasion in vitro (33). Another study in thyroid cancer showed that SNHG7 is upregulated in thyroid cancer tissues; the depletion of SNHG7 inhibits cell proliferation, but induces apoptosis in thyroid cancer cells (34). Xu et al (35), reported that SNHG7 expression is significantly enhanced in human bladder cancer, and SNHG7 knockdown represses cell proliferation and metastasis, but induces apoptosis in SW780, T24, UMUC and 5637 cells. The present results suggested that the expression of SNHG7 was significantly upregulated in PC tissues and cell lines, and was increased in high-grade compared with low-grade tumors. Moreover, the high expression of SNHG7 was found to be associated with the pathological characteristics of PC. Furthermore, functional experiment results identified that SNHG7 silencing reduced cell proliferation, migration and invasion, while it promoted apoptosis in PC cells. It was also demonstrated that the depletion of SNHG7 decreased xenograft tumor growth in vivo. Thus, these results of SNHG7 are in line with a previous study (15) and indicated that SNHG7 may promote PC progression.
lncRNA can function as a competing endogenous RNA to recruit miRNA and further affected target gene expression. For example, a previous study showed that SNHG7 sponges miR-186 to promote breast cancer progression (36). Another study in breast cancer revealed that SNHG7 increases cell proliferation, invasion and epithelial-mesenchymal transition by sponging miR-34a (37). Cheng et al (15), reported that SNHG7 knockdown blocks cell proliferation, migration and invasion via miR-342-3p in PC. In the present study, it was found that miR-146b-5p was sponged to SNHG7. Moreover, the restoration experimental results indicated that the expression of miR-146b-5p was reduced in SW1990 and AsPC-1 cells transfected with sh-SNHG7 induced by the miR-146b-5p inhibitor. It was also found that SNHG7 silencing inhibited PC progression by regulating miR-146b-5p. Therefore, it was speculated that SNHG7 accelerated PC progression by sponging miR-146b-5p.
It has been shown that the dysregulation of Robo1 is associated with cancer progression. For example, a previous study in hepatocellular carcinoma showed that Robo1 is significantly increased in hepatocellular carcinoma tissues and cells, and its overexpression reduces hepatocellular carcinoma progression regulated by miR-490-5p in vitro (38). The present results identified an interaction between miR-146b-5p and Robo1 by dual luciferase reporter assay. Moreover, it was demonstrated that Robo1 overexpression attenuated the restraint impacts on cell viability, migration and invasion, and the promoting action on apoptosis caused by miR-146b-5p mimics. The present results relating to Robo1 in PC were consistent with those of a previous study (21). Furthermore, it was found that SNHG7 increased Robo1 expression by sponging miR-146b-5p in PC cells. Thus, the present results indicated that SNHG7 modulated Robo1 expression to promote cell proliferation, migration and invasion, as well as inhibit apoptosis in PC by sponging miR-146b-5p.
In conclusion, the expression of SNHG7 was significantly increased in PC tissues and cells. Based on the results of the functional and mechanical experiments, it was speculated that SNHG7 positively regulated Robo1 expression by sponging miR-146b-5p, which contributed to the promotion of PC progression. Therefore, this novel regulatory network may provide a new therapeutic target for patients with PC.
Acknowledgements
Not applicable.
Funding Statement
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
Conceptualization of the research design and the execution of the experiment were carried out by YJ. Acquisition of data was conducted by YJ. Writing and review of the manuscript were done by YJ and QF. Study supervision was carried out by YJ. Formal analysis and data curation was carried out by QF. All authors read and approved the manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
The current study was approved by the Ethics Committee of Jingzhou Central Hospital. A written informed consent form was obtained from each patient.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2. Surveillance Epidemiology and End Results (SEER) Fact Sheets: Pancreas. Web site. http://seer.cancer.gov/. Accessed July 15, 2016. [Google Scholar]
- 3.Aarnink A, Richard C, Truntzer C, Vincent J, Bengrine L, Vienot A, Borg C, Ghiringhelli F. Baseline splenic volume as a surrogate marker of FOLFIRINOX efficacy in advanced pancreatic carcinoma. Oncotarget. 2018;9:25617–25629. doi: 10.18632/oncotarget.25424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paulson AS, Tran Cao HS, Tempero MA, Lowy AM. Therapeutic advances in pancreatic cancer. Gastroenterology. 2013;144:1316–1326. doi: 10.1053/j.gastro.2013.01.078. [DOI] [PubMed] [Google Scholar]
- 5.De Luca R, Blasi L, Alù M, Gristina V, Cicero G. Clinical efficacy of nab-paclitaxel in patients with metastatic pancreatic cancer. Drug Des Devel Ther. 2018;12:1769–1775. doi: 10.2147/DDDT.S165851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang G, Lu X, Yuan L. LncRNA: A link between RNA and cancer. Biochim Biophys Acta. 2014;1839:1097–1109. doi: 10.1016/j.bbagrm.2014.08.012. [DOI] [PubMed] [Google Scholar]
- 7.Tang Y, Cao G, Zhao G, Wang C, Qin Q. LncRNA differentiation antagonizing non-protein coding RNA promotes proliferation and invasion through regulating miR-135a/NLRP37 axis in pancreatic cancer. Invest New Drugs. 2019;38:714–721. doi: 10.1007/s10637-019-00798-0. [DOI] [PubMed] [Google Scholar]
- 8.Peng W, Jiang A. Long noncoding RNA CCDC26 as a potential predictor biomarker contributes to tumorigenesis in pancreatic cancer. Biomed Pharmacother. 2016;83:712–717. doi: 10.1016/j.biopha.2016.06.059. [DOI] [PubMed] [Google Scholar]
- 9.Zhang X, Feng W, Zhang J, Ge L, Zhang Y, Jiang X, Peng W, Wang D, Gong A, Xu M. Long non-coding RNA PVT1 promotes epithelial-mesenchymal transition via the TGF-β/Smad pathway in pancreatic cancer cells. Oncol Rep. 2018;40:1093–1102. doi: 10.3892/or.2018.6462. [DOI] [PubMed] [Google Scholar]
- 10.Zhang M, Zhao Y, Zhang Y, Wang D, Gu S, Feng W, Peng W, Gong A, Xu M. LncRNA UCA1 promotes migration and invasion in pancreatic cancer cells via the hippo pathway. Biochim Biophys Acta Mol Basis Dis. 2018;1864:1770–1782. doi: 10.1016/j.bbadis.2018.03.005. [DOI] [PubMed] [Google Scholar]
- 11.Gao H, Gong N, Ma Z, Miao X, Chen J, Cao Y, Zhang G. LncRNA ZEB2-AS1 promotes pancreatic cancer cell growth and invasion through regulating the miR-204/HMGB1 axis. Int J Biol Macromol. 2018;116:545–551. doi: 10.1016/j.ijbiomac.2018.05.044. [DOI] [PubMed] [Google Scholar]
- 12.She K, Huang J, Zhou H, Huang T, Chen G, He J. lncRNA-SNHG7 promotes the proliferation, migration and invasion and inhibits apoptosis of lung cancer cells by enhancing the FAIM2 expression. Oncol Rep. 2016;36:2673–2680. doi: 10.3892/or.2016.5105. [DOI] [PubMed] [Google Scholar]
- 13.Wang MW, Liu J, Liu Q, Xu QH, Li TF, Jin S, Xia TS. LncRNA SNHG7 promotes the proliferation and inhibits apoptosis of gastric cancer cells by repressing the P15 and P16 expression. Eur Rev Med Pharmacol Sci. 2017;21:4613–4622. [PubMed] [Google Scholar]
- 14.Ren J, Yang Y, Xue J, Xi Z, Hu L, Pan SJ, Sun Q. Long noncoding RNA SNHG7 promotes the progression and growth of glioblastoma via inhibition of miR-5095. Biochem Biophys Res Commun. 2018;496:712–718. doi: 10.1016/j.bbrc.2018.01.109. [DOI] [PubMed] [Google Scholar]
- 15.Cheng D, Fan J, Ma Y, Zhou Y, Qin K, Shi M, Yang J. LncRNA SNHG7 promotes pancreatic cancer proliferation through ID4 by sponging miR-342-3p. Cell Biosci. 2019;9(28) doi: 10.1186/s13578-019-0290-2. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Z, Che X, Yang N, Bai Z, Wu Y, Zhao L, Pei H. MiR-135b-5p Promotes migration, invasion and EMT of pancreatic cancer cells by targeting NR3C2. Biomed Pharmacother. 2017;96:1341–1348. doi: 10.1016/j.biopha.2017.11.074. [DOI] [PubMed] [Google Scholar]
- 18.Cai H, Yao J, An Y, Chen X, Chen W, Wu D, Luo B, Yang Y, Jiang Y, Sun D, He X. LncRNA HOTAIR acts a competing endogenous RNA to control the expression of notch3 via sponging miR-613 in pancreatic cancer. Oncotarget. 2017;8:32905–32917. doi: 10.18632/oncotarget.16462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang J, Guo XJ, Ding YM, Jiang JX. MiR-1181 inhibits invasion and proliferation via STAT3 in pancreatic cancer. World J Gastroenterol. 2017;23:1594–1601. doi: 10.3748/wjg.v23.i9.1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin F, Wang X, Jie Z, Hong X, Li X, Wang M, Yu Y. Inhibitory effects of miR-146b-5p on cell migration and invasion of pancreatic cancer by targeting MMP16. J Huazhong Univ Sci Technolog Med Sci. 2011;31(509) doi: 10.1007/s11596-011-0481-5. [DOI] [PubMed] [Google Scholar]
- 21.He H, Hao SJ, Yao L, Yang F, Di Y, Li J, Jiang YJ, Jin C, Fu DL. MicroRNA-218 inhibits cell invasion and migration of pancreatic cancer via regulating ROBO1. Cancer Biol Ther. 2014;15:1333–1339. doi: 10.4161/cbt.29706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cuccurullo V, Mansi L. AJCC cancer staging handbook: From the AJCC cancer staging manual (7th edition) Eur J Nucl Med Mol Imaging. 2011;38(408) [Google Scholar]
- 23.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 24. Harden VA and Hannaway C: National Institutes of Health (NIH). In: Encyclopedia of Life Sciences. New York, John Wiley & Sons, Ltd, 2001. [Google Scholar]
- 25.Schober P, Boer C, Schwarte LA. Correlation coefficients: Appropriate use and interpretation. Anesth Analg. 2018;126:1763–1768. doi: 10.1213/ANE.0000000000002864. [DOI] [PubMed] [Google Scholar]
- 26.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 27.Xu W, Chang J, Du X, Hou J. Long non-coding RNA PCAT-1 contributes to tumorigenesis by regulating FSCN1 via miR-145-5p in prostate cancer. Biomed Pharmacother. 2017;95:1112–1118. doi: 10.1016/j.biopha.2017.09.019. [DOI] [PubMed] [Google Scholar]
- 28.Huo X, Wang H, Huo B, Wang L, Yang K, Wang J, Wang L, Wang H. FTX contributes to cell proliferation and migration in lung adenocarcinoma via targeting miR-335-5p/NUCB2 axis. Cancer Cell Int. 2020;20(89) doi: 10.1186/s12935-020-1130-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun J, Zhang P, Yin T, Zhang F, Wang W. Upregulation of LncRNA PVT1 facilitates pancreatic ductal adenocarcinoma cell progression and glycolysis by regulating MiR-519d-3p and HIF-1A. J Cancer. 2020;11:2572–2579. doi: 10.7150/jca.37959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo X, Zhang Y, Liu L, Yang W, Zhang Q. HNF1A-AS1 regulates cell migration, invasion and glycolysis via modulating miR-124/MYO6 in colorectal cancer cells. Onco Targets Ther. 2020;13:1507–1518. doi: 10.2147/OTT.S231249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li H, Yao G, Zhai J, Hu D, Fan Y. LncRNA FTX promotes proliferation and invasion of gastric cancer via miR-144/ZFXAxis. Onco Targets Ther. 2019;12:11701–11713. doi: 10.2147/OTT.S220998. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 32.Feng Y, Gao L, Cui G, Cao Y. LncRNA NEAT1 facilitates pancreatic cancer growth and metastasis through stabilizing ELF3 mRNA. Am J Cancer Res. 2020;10:237–248. [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang C, Zhu B, Li XB, Cao YQ, Yang JC, Li X, Liu YX, Wang YB. Long non-coding RNA SNHG7 promotes migration and invasion of melanoma via upregulating SOX4. Eur Rev Med Pharmacol Sci. 2019;23:4828–4834. doi: 10.26355/eurrev_201906_18069. [DOI] [PubMed] [Google Scholar]
- 34.Wang YH, Huo BL, Li C, Ma G, Cao W. Knockdown of long noncoding RNA SNHG7 inhibits the proliferation and promotes apoptosis of thyroid cancer cells by downregulating BDNF. Eur Rev Med Pharmacol Sci. 2019;23:4815–4821. doi: 10.26355/eurrev_201906_18067. [DOI] [PubMed] [Google Scholar]
- 35.Xu C, Zhou J, Wang Y, Wang A, Su L, Liu S, Kang X. Inhibition of malignant human bladder cancer phenotypes through the down-regulation of the long non-coding RNA SNHG7. J Cancer. 2019;10:539–546. doi: 10.7150/jca.25507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luo X, Song Y, Tang L, Sun DH, Ji DG. LncRNA SNHG7 promotes development of breast cancer by regulating microRNA-186. Eur Rev Med Pharmacol Sci. 2018;22:7788–7797. doi: 10.26355/eurrev_201811_16403. [DOI] [PubMed] [Google Scholar]
- 37.Sun X, Huang T, Liu Z, Sun M, Luo S. LncRNA SNHG7 contributes to tumorigenesis and progression in breast cancer by interacting with miR-34a through EMT initiation and the notch-1 pathway. Eur J Pharmacol. 2019;856(172407) doi: 10.1016/j.ejphar.2019.172407. [DOI] [PubMed] [Google Scholar]
- 38.Chen W, Ye L, Wen D, Chen F. MiR-490-5p inhibits hepatocellular carcinoma cell proliferation, migration and invasion by directly regulating ROBO1. Pathol Oncol Res. 2019;25:1–9. doi: 10.1007/s12253-017-0305-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.