Summary
Heterogeneous nuclear ribonucleoproteins (hnRNPs) play critical roles in the nuclear export, splicing, and sensing of RNA. However, the role of heterogeneous nuclear ribonucleoprotein A/B (hnRNPAB) is poorly understood. In this study, we report that hnRNPAB cooperates with nucleoprotein (NP) to restrict viral mRNA nuclear export via inhibiting viral mRNA binding to ALY and NXF1. HnRNPAB restricts mRNA transfer from ALY to NXF1, inhibiting the mRNA nuclear export. Moreover, when cells are invaded by influenza A virus, NP interacts with hnRNPAB and interrupts the ALY-UAP56 interaction, leading to repression of ALY-viral mRNA binding, and then inhibits the viral mRNA binding to NXF1, leading to nuclear stimulation of viral mRNA. Collectively, these observations provide a new role of hnRNPAB to act as an mRNA nuclear retention factor, which is also effective for viral mRNA of influenza A virus, and NP cooperates with hnRNPAB to further restrict the viral mRNA nuclear export.
Subject areas: Molecular Biology, Virology
Graphical Abstract
Highlights
-
•
HnRNPAB inhibits influenza A virus replication by repressing viral mRNA nuclear export
-
•
HnRNPAB interrupts viral mRNA transferring from ALY to NXF1
-
•
NP cooperates with hnRNPAB to further restrict viral mRNA nuclear export
-
•
The ALY-viral mRNA binding is restricted by NP-hnRNPAB complex
Molecular Biology; Virology
Introduction
Influenza A virus (IAV), a member of the Orthomyxoviridae family, poses a serious threat to public health. The genome of IAV consists of eight single-stranded negative-sense RNA segments, which are coated by a nucleoprotein (NP) to form viral ribonucleoprotein complex (vRNP) with viral polymerase (Julia et al., 2014). The vRNP comprises viral RNA (vRNA), NP, and heterotrimeric RNA-dependent RNA polymerase (Fodor, 2013; Resainfante et al., 2011). After IAV infection, the vRNP is transported into the nucleus of the host cell, followed by transcription and viral gene replication in the nucleus. NP is a core component of genomic vRNP and takes part in the organization of RNA packing, nuclear trafficking, and vRNA transcription and replication. RNA binding to NP is critical for the vRNP complex. NP is more than just a structural protein of IAV as it has been reported to interact with multiple cellular factors (Kawaguchi et al., 2011; Luo et al., 2018; Naito et al., 2007; Wang et al., 2009), which influences the replication of IAV. NP contains both a nuclear localization signal (NLS) and nuclear export signals (NES), which are critical for vRNP transporting through the nucleus during the virus life cycle (Li et al., 2015; Pemberton et al., 1998). However, whether NP is involved in modulating the host cell life cycle is still unknown.
Nuclear retention of mRNA is essential for gene expression in cells. Misprocessed RNAs or spurious transcripts from intergenic DNA regions are mostly retained in the nucleus and then degraded. Nuclear retention of mRNA can restrict misprocessed mRNA translation, which might be translated into toxic proteins and be harmful to the cell. Specialized cis-elements in certain RNAs and mistakes in splicing, polyadenylation, and RNA modifications all lead to RNA nuclear retention (Palazzo and Lee, 2018). Except for the misprocessed RNAs, the nuclear export of certain normally spliced and polyadenylated mRNA is also restricted to protect cytoplasmic transcript levels from transcriptional bursts (Halpern et al., 2015). Aberrant mRNA nuclear retention might lead to cell disability or disease. For example, an expansion mutation in polyadenylate RNA-binding protein 1 leads to oculopharyngeal muscular dystrophy (Yotam and Raz, 2014); a mutation in COL1A1 (encoding collagen, type I, alpha 1) affects the splicing of intron 26 and leads to intron retention, which causes type I osteogenesis imperfecta (Carol Johnson et al., 2000). Whether there is a cell factor to restrict viral mRNA nuclear export during infection is still unknown.
The mRNA nuclear export is considered a key step for gene expression, and it requires the mRNA transport receptor NXF1-NXT1 (Bjork and Wieslander, 2014; Walsh et al., 2010). Once the exporting mRNA is delivered to NXF1, the NXF1 binds to FG repeat sequences in the nucleoporins lining the nuclear pore and transports through the nuclear pore (Fribourg et al., 2001). However, the NXF1 displays weak and nonspecific RNA-binding activity (Bjork and Wieslander, 2014; Hautbergue et al., 2008; Zolotukhin et al., 2002). In the nucleus, a set of receptors interact with NXF1 to facilitate its RNA-binding ability and transfer the exporting mRNA to NXF1 (Viphakone et al., 2012; Williams et al., 2018). Transcription-export (TREX) complex is the key adaptor of NXF1, which is mainly composed of AML-1 and LEF-1 (ALY), RNA helicase UAP56 (also known as DExD-box helicase 39B), and THO-sub-complex (including THOC1, 2, 3, 6, 7, and so on) (Bjork and Wieslander, 2014; Hautbergue, 2017; Katja et al., 2002). The UAP56, a core subunit of the TREX complex, binds pre-mRNA during splicing, and on binding to the ALY its bound RNA is transferred to ALY via direct interaction (Chang et al., 2013). NXF1 interaction with ALY and THOC5 is essential to expose NXF1's RNA-binding domain (RBD) to allow RNA transfer from ALY to NXF1 (Viphakone et al., 2012; Wickramasinghe and Laskey, 2015). Increasing evidence indicates that the nuclear export of viral mRNA may be associated with UAP56, ALY, and NXF1 (Read and Digard, 2010; Tunnicliffe et al., 2011). Heterogeneous nuclear ribonucleoprotein AB (hnRNPAB), as a member of the hnRNP family, is an abundant RNA-binding protein first isolated in 1987 from 40S hnRNP particles (Akindahunsi et al., 2005; Dean et al., 2002). HnRNPAB is involved in transcription, alternative splicing, mRNA transport and localization, translation, mRNA instability, and telomere maintenance (Amit et al., 2012; Lampasona and Czaplinski, 2019; Nanaho et al., 2013; Raju et al., 2008; Yang et al., 2019). However, whether and how hnRNPAB is involved in mRNA nuclear export is not clear.
In this study, we demonstrated that hnRNPAB acts as an mRNA nuclear retention factor to regulate gene expression. In IAV-invaded cells hnRNPAB interacts with NP via RNA to restrict viral mRNA nuclear export. Mechanistically, hnRNPAB interacts with the mRNA nuclear export factors, ALY and NXF1, and interrupts the mRNA transfer from ALY to NXF1, leading to mRNA nuclear retention. When cells were infected by the IAV, hnRNPAB interacted with NP of IAV to interrupt the interaction of ALY and UAP56, leading to the repression of ALY-viral mRNA binding, and therefore the viral mRNA could not be transferred to NXF1 for nuclear export successfully.
Results
Cellular hnRNPAB plays an inhibitory role in flu A virus replication
HnRNPs play critical roles in nuclear export, splicing, and sensing of vRNA for IAV replication to trigger innate immune responses (Cao et al., 2019; Mor et al., 2017; Tsai et al., 2013; Wang et al., 2014). The hnRNPAB (AB), a member of hnRNPs family, is abundant but rarely discussed. To explore the role of hnRNPAB during virus replication, we detected the kinetics of hnRNPAB expression in cells infected with different subtypes of IAV using quantitative real-time reverse-transcriptase PCR (qRT-PCR) and western blotting. The results showed that the mRNA level and protein level in H9N2, H7N9, H5N1, H3N2, and PR8 virus-infected cells at 6, 12, and 24 h post-infection both displayed significant increase (Figures 1A and 1B), demonstrating that the expression of hnRNPAB is up-regulated when the cells were infected by IAV. To investigate the function of hnRNPAB during IAV infection, 293T cells transfected with myc-hnRNPAB were infected separately with H9N2, H7N9, H5N1, H3N2, and PR8 viruses, and virus replication was monitored at 24 h post-infection by determining viral protein expression and TCID50 (median tissue culture infectious dose). Data shown in Figures 1C and S1A revealed that the viral titer and protein expression of different subtypes of IAV decreased in hnRNPAB-overexpressing cells when compared with that of wild-type cells. However, in hnRNPAB knockdown cells infected with different subtypes of IAV, the viral titers and protein levels of four IAVs increased significantly (Figures 1D and S1B). Interestingly, in the early stage of infection, the M1 mRNA and vRNA alteration are not significant in both hnRNPAB-overexpressing and knockdown cells; however, in the late stage of infection the mRNA and vRNA levels of M1 have a significant decrease in hnRNPAB-overexpressing cells and a significant increase in hnRNPAB knockdown cells (Figure S1C and S1D). By the way, the cell proliferation assay via CCK-8 proved that expressing or knocking down hnRNPAB showed no significant effect on cell proliferation (Figure 1E). These results demonstrated that hnRNPAB inhibits the replication of different subtypes of IAV.
Figure 1.
HnRNPAB inhibits the replication of different subtypes of influenza A virus
(A) Dynamics of hnRNPAB mRNA in cells infected with influenza A virus. A549 cells were infected with H7N9, H5N1, H9N2, H3N2, or PR8 viruses (MOI = 1.0), respectively. The abundance of viral mRNA at the indicated time was detected using qRT-PCR. Expression was normalized to the mRNA level.
(B) Dynamics of hnRNPAB protein in cells infected with influenza A virus. Cell lysates collected from (A) at the indicated time were analyzed by immunoblotting with rabbit anti-hnRNPAB mAb, mouse anti-NP mAb, and rabbit anti-GAPDH pAbs. Quantitation of the immunoreactive protein bands on the blots was performed using ImageJ software, and the hnRNPAB band was normalized using the GAPDH control.
(C) Inhibition of H7N9 virus replication in hnRNPAB-overexpressing cells. 293T cells were transfected with myc-hnRNPAB. The myc vector was used as a control. At 24 h after transfection, cells were infected with H7N9, H5N1, H9N2, H3N2, or PR8 viruses (MOI = 1.0), respectively, and cultured for 24 h. The TCID50 was measured by IFA (immunofluorescence assay) with the mouse anti-M2 mAb.
(D) Enhancement of H7N9 virus replication in hnRNPAB knockdown cells. Cells were transfected with hnRNPAB small interfering RNA (siRNA) si-AB or control si-NC. At 36 h after transfection cells were infected with the H7N9, H5N1, H9N2, H3N2, or PR8 viruses (MOI = 1.0), respectively and cultured for 24 h. TCID50 was measured as described in (C).
(E) 293T cells transfected with myc vector, myc-tagged hnRNPAB, si-NC, or si-hnRNPAB for 12 h. The cells were countered and added into 96-well plate at 2,000 cells per well. After 12 h CCK-8 was measured at 450 nm. The Error bars: Mean + SD of 3 independent tests. ∗p < 0.05; ∗∗p < 0.01; and ns (nonsignificant), p > 0.05 compared to control.
HnRNPAB binding to viral nucleoprotein enhances the inhibition of IAV replication
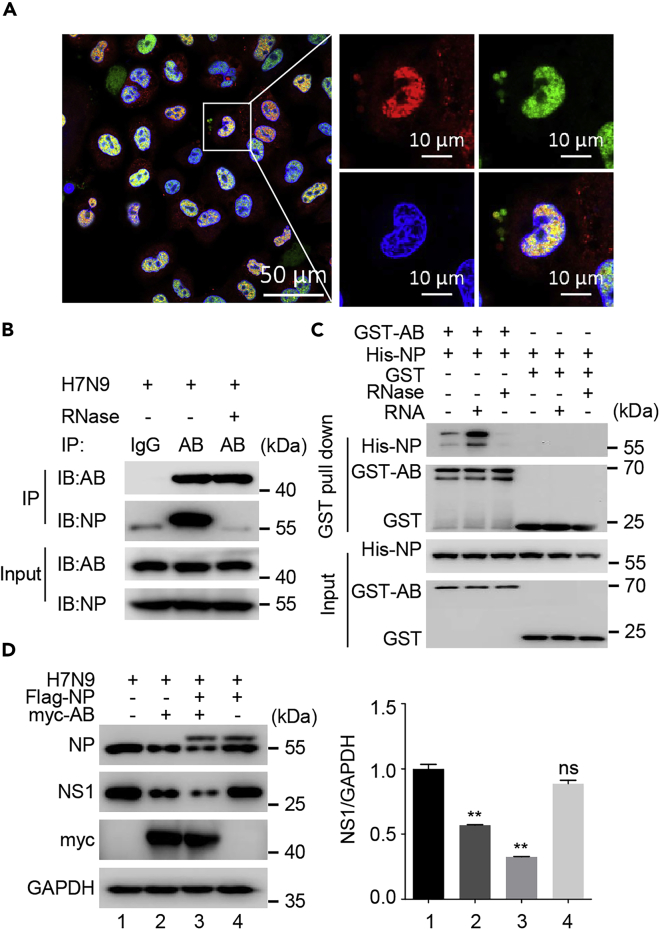
The NP of IAV was reported to be the major component of vRNP complexes and to bind single-stranded RNA without sequence specificity (Klumpp et al., 1997). We wanted to know whether viral NP associates with hnRNPAB-induced inhibition of IAV. For the purpose we investigated the interaction of hnRNPAB and NP during infection of IAV. A549 cells were infected with H7N9 virus as a model virus, and 12 h later the cells were harvested. Confocal microscopy analysis revealed that hnRNPAB overlapped with the NP protein (Figure 2A). Furthermore, co-immunoprecipitation assay showed that the viral protein NP had an interaction with hnRNPAB and the interaction disappeared after RNase A treatment (Figure 2B). RNA immunoprecipitation (RIP) assay further proved that hnRNPAB binds mainly to viral mRNA instead of vRNA or cRNA (Figure S2A). In the GST pull-down experiment, NP was pulled down by GST-hnRNPAB but not GST alone, and the interaction between hnRNPAB and NP disappeared after RNase A treatment and was recovered when RNA was added (Figure 2C). By GST pull-down assay, we further identified the critical region(s) for the interaction between hnRNPAB and NP. The results showed that both of RRMs and Gly domains are critical for hnRNPAB interaction with NP (Figure S2B) and that the interaction between Gly and NP is dependent on RNA (Figure S2C). These data confirmed that RNA mediates the interaction between hnRNPAB and NP.
Figure 2.
hnRNPAB interacts with viral protein NP via RNA
(A) HnRNPAB colocalization with viral protein NP. A549 cells were infected with H7N9 virus for 12 h and were then fixed with 4% paraformaldehyde. Cells were stained with anti-NP and anti-hnRNPAB antibodies. Nuclei were stained with DAPI. Scale bars, 10 µm.
(B) Co-IP assay of viral protein NP and endogenous hnRNPAB in infected cells. 293T cells were infected with H7N9 at MOI = 1.0 for 24 h and immunoprecipitated with anti-mouse IgG and anti-hnRNPAB mAb, followed by western blotting.
(C) Co-IP assay of FLAG-NP and myc-hnRNPAB with RNase treatment. 293T cells were co-transfected with myc-hnRNPAB and FLAG-NP or FLAG for 24 h. Cell lysates were treated with or without RNase for 2 h and immunoprecipitated with the mouse anti-FLAG mAb, followed by western blotting with the anti-FLAG mAb and anti-myc pAbs.
(D) GST pull-down assay of GST-hnRNPAB and His-NP. The purified GST, GST-hnRNPAB, and His-NP were incubated together as indicated. One group was added with the cellular RNA extract, and another group was treated with RNase.
(E) H7N9 virus replication in cells co-expressing hnRNPAB and NP. 293T cells were co-transfected with FLAG-NP and/or myc-hnRNPAB for 24 h and were then infected with H7N9 at MOI = 1.0 for 24 h. Cell lysates were analyzed by western blotting with anti-NP mAb, anti-NS1 mAb, anti-myc, and anti-GAPDH pAbs. The Error bars: Mean + SD of 3 independent tests. ∗p < 0.05; ∗∗p < 0.01; and ns (nonsignificant), p > 0.05 compared to control.
To analyze the impact of hnRNPAB-NP interaction on virus replication, the 293T cells transfected with myc-hnRNPAB and/or FLAG-NP, individually or together, were inoculated with H7N9 virus. The levels of viral protein NS1 were downregulated significantly in hnRNPAB-overexpressing cells, especially in hnRNPAB and NP co-expressing cells (Figure 2D, p < 0.01); however, the alteration of viral protein NS1 was not significant in NP-overexpressing cells. Meanwhile, we tested the viral mRNA and vRNA levels of M1 when hnRNPAB and/or NP were overexpressed. Results showed that in the early stage of infection overexpression of hnRNPAB and/or NP showed no significant effect on the viral mRNA and vRNA levels. However, in the late stage of infection, the viral mRNA and vRNA levels were both inhibited significantly when hnRNPAB was expressed individually or expressed with NP (Figure S2D). Taken together, the results demonstrated that hnRNPAB binding to NP via RNA exacerbates the inhibition of virus replication.
The Gly motif of hnRNPAB is critical to inhibit IAV replication
Whether the hnRNPAB from different species has identical function during virus infection is unknown. We constructed myc-tagged hnRNPAB from mouse (m-AB) and chicken (c-AB). Then, 293T cells were transfected with m-AB or c-AB and infected with the H7N9 virus for 16 h. The immunoblotting results showed that both m-AB and c-AB could restrict the expression of viral proteins NP and NS1 significantly during H7N9 infection (Figures 3A and 3B), indicating that hnRNPABs from different species have a similar function during virus replication. To further characterize the critical domain of hnRNPAB for inhibition of virus replication, we constructed myc-tagged CBFNT fragment (CARG-binding factor N-terminus), RRMs fragment (RNA recognition motif), Gly fragment (Gly) of hnRNPAB, and CBFNT-deficient hnRNPAB (ΔCBFNT), RRMs-deficient hnRNPAB (ΔRRMs), and Gly-deficient hnRNPAB (ΔGly) (Figure 3C). The 293T cells were transfected with these constructs for 24 h and then infected with H7N9 virus (multiplicity of infection [MOI] = 1.0) for 16 h. The results showed that only the Gly domain of hnRNPAB, but not the CBFNT and RRMs, inhibited virus replication. However, in the ΔGly-transfected cells, the expression of viral proteins showed no significant change during infection (Figures 3D and 3E). The TCID50 assay of the cells' supernatant that was transfected with myc-tagged Gly and infected by H7N9 virus showed that Gly inhibited H7N9 virus replication significantly (Figure S2E). These data suggested that the Gly motif was critical for hnRNPAB to inhibit virus replication.
Figure 3.
Identification of the Gly motif of hnRNPAB for the inhibition of flu virus replication
(A and B) Inhibition of H7N9 virus replication in mouse and chicken hnRNPAB-overexpressing cells. 293T cells were transfected with myc-m-AB (A) or myc-c-AB (B) for 24 h and infected with H7N9 virus at MOI = 1.0 for 16 h. Cell lysates was analyzed using western blotting with the indicated antibody.
(C) Schematic representation of the CBFNT, RRM1, RRM2, and Gly domains of hnRNPAB. The numbers indicate amino acid (aa) positions.
(D and E) Gly domain of hnRNPAB is critical to inhibit H7N9 virus replication. 293T cells were transfected with myc-tagged hnRNPAB or the hnRNPAB-truncated mutants CBFNT, RRMs, and Gly (D), or hnRNPAB-deleted mutants ΔCBFNT, ΔRRMs, and ΔGly (E) for 24 h, and these cells were infected with H7N9 virus for 16 h. Cell extracts were analyzed by western blotting using anti-NP mAb, anti-NS1 mAb, and anti-GAPDH pAbs.
(F) The residues 241–263 aa and 312–332 aa are important for Gly function. 293T cells were transfected with the plasmids myc, myc-tagged hnRNPAB, or hnRNPAB-deleted mutants Δ254–254 aa, Δ241–263 aa, and Δ312–332 aa for 24 h and were infected with H7N9 virus at MOI = 1.0 for 16 h. Whole-cell extracts were analyzed by western blotting with anti-NP mAb, anti-NS1 mAb, and anti-GAPDH pAb. The Error bars: Mean + SD of 3 independent tests. ∗p < 0.05; ∗∗p < 0.01; and ns (nonsignificant), p > 0.05 compared to control.
To finely locate the critical sites of the Gly motif, we analyzed the Gly motif of hnRNPAB, including the N-terminal Arg-rich domain (245–254 aa), the N-terminal Arg and Gly-rich domain (241–263 aa), and the C-terminal conservative sequence (312–332 aa). Subsequently, we constructed myc-tagged 245–254 aa-deficient hnRNPAB (Δ245–254 aa), 241–263 aa-deficient hnRNPAB (Δ241–263 aa), and 312–332 aa-deficient hnRNPAB (Δ312–332 aa). 293T cells were transfected with these constructs for 24 h and then infected with H7N9 virus (MOI = 1.0) for 16 h. The results showed that in Δ241–263 aa- and Δ312–332 aa-, rather than Δ245–254 aa-, transfected cells the expression of viral proteins NP and NS1 showed no significant change during infection (Figure 3F). These data suggested that residues 241–263 aa and 312–332 aa within the Gly motif of hnRNPAB are the critical regions to inhibit virus replication.
Nuclear export of virus mRNA is limited by interacting with Gly motif of hnRNPAB
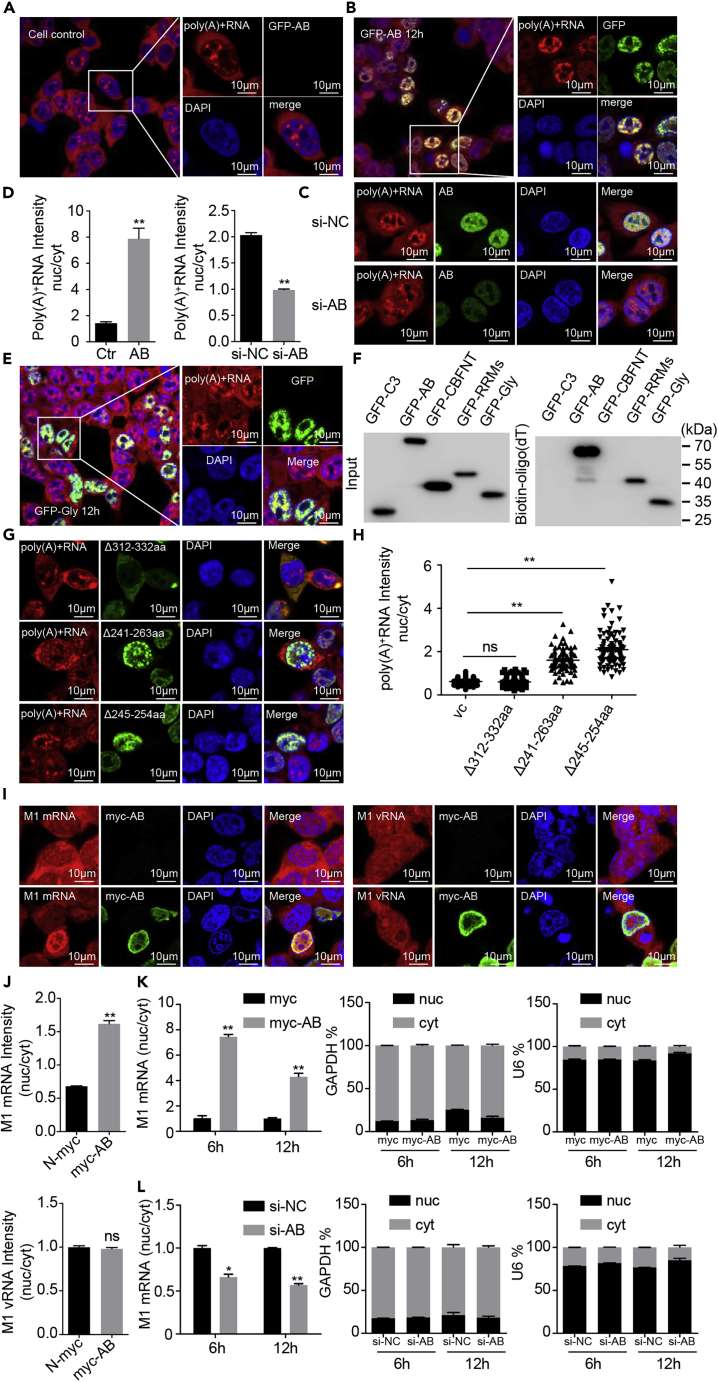
HnRNPAB was reported to bind to poly(A)+RNA (Piergiorgio et al., 2002). To investigate the role of hnRNPAB in the cellular mRNA nuclear export, we checked the distribution of poly(A)+RNA in hnRNPAB-overexpressing and knockdown cells using a cy3-labeled oligo(dT)40 probe. Confocal microscopy observation showed that significant amounts of poly(A)+RNA remained in the nucleus of hnRNPAB-overexpressing cells, which contrasted with poly(A)+RNA being mainly distributed in the cytoplasm of wild-type 293T cells (Figure 4A, 4B, and 4D). However, poly(A)+RNA levels decreased significantly in the nucleus of hnRNPAB knockdown A549 cells (Figures 4C and 4D). The results demonstrated that hnRNPAB limited the nuclear export of poly(A)+RNA. To verify if the intranuclear retention of poly(A)+RNA is involved in the interaction with hnRNPAB, 293T cells were transfected with GFP-tagged CBFNT, RRMs, and Gly of hnRNPAB and the Gly mutants Δ245–254 aa, Δ241–263 aa, and Δ312–332 aa. 293T cells were transfected with these constructs and stained with the cy3-labeled oligo(dT)40 probe. The overlapping analysis showed that the Gly domain overlapped with poly(A)+RNA in the nucleus of the transfected cells; however, CBFNT and RRMS were mainly distributed in the cytoplasm of the transfected cells and were not colocalized with poly(A)+RNA (Figures 4E, S3A, and S3B). However, an RNA pull-down assay demonstrated an interaction between full-length hnRNPAB, Gly, RRMS, and poly(A)+RNA, respectively (Figure 4F), confirming that hnRNPAB interacts with poly(A)+RNA by its Gly and RRM motifs. The colocalization experiment using the Gly mutants revealed that the intranuclear accumulation of poly(A)+RNA decreased significantly in the Δ312–332 aa-transfected cells, but not in Δ245–254 and Δ241–263 aa-transfected cells (Figures 4G and 4H), indicating that the C-terminal residues 312–332 aa within the Gly motif are the NLS involved the intranuclear location of hnRNPAB and the N-terminal residues 245–254 aa and 241–263 aa of the Gly motif are crucial for hnRNPAB binding to poly(A)+RNA.
Figure 4.
HnRNPAB blocks nuclear export of viral mRNA
(A and B) FISH analysis of the distribution of poly(A)+ RNA in cells transfected with hnRNPAB. 293T cells were transfected without (A) or with (B) GFP-hnRNPAB for 12 h and then probed with cy3-oligo(dT)40 for poly(A)+RNA. Nuclei were stained with DAPI. Scale bars, 10 µm.
(C) FISH analysis of the distribution of poly(A)+RNA in A549 cells that were transfected with si-hnRNPAB or si-NC as control. Scale bars, 10 µm.
(D) Fluorescence intensity for the nuclear/cytoplasmic ratio of poly(A)+RNA in hnRNPAB-overexpressing and knockdown cells was analyzed by countering 100 cells using ImageJ software.
(E) FISH analysis of poly(A)+RNA distribution in cells transfected with the Gly domain of hnRNPAB. 293T cells were transfected with GFP-Gly of hnRNPAB for 12 h and probed with cy3-oligo(dT)40. Scale bars, 10 µm.
(F) RNA pull-down assay of hnRNPAB. 293T cells were transfected with GFP-tagged hnRNPAB or indicated truncation mutants for 24 h and were then subjected to biotin-oligo(dT) assay and western blotting.
(G) Poly(A)+RNA-distributed FISH analysis in cells transfected with the Gly domain-truncated mutants. 293T cells were transfected with GFP-tagged Gly-truncated mutants Δ241–263 aa, Δ245–254 aa, and Δ312–332 aa and were stained with cy3-oligo(dT)40 and DAPI. Scale bars, 10 µm.
(H) Fluorescence intensity for the nuclear/cytoplasmic ratio of poly(A)+RNA in hnRNPAB mutant-overexpressing cells by ImageJ software.
(I) The viral mRNA-distributed FISH analysis in H7N9 virus-infected cells. 293T cells were transfected with myc vector or myc-tagged hnRNPAB for 24 h and then infected with H7N9 virus for 12 h. Cells were stained with cy3-labeled M1 mRNA probes or cy3-labeled M1 vRNA probes. Scale bars, 10 µm.
(J) Fluorescence intensity for the nuclear/cytoplasmic ratio of M1 mRNA or vRNA in hnRNPAB-overexpressing cells was analyzed by countering 100 cells using ImageJ software.
(K and L) The nuclear-cytoplasmic ratio of viral mRNA. 293T cells were transfected with the indicated plasmids (K) or siRNA (L) for 24 h and were infected with H7N9 at MOI = 1.0 for 6 and 12 h. Cells are subjected to nuclear-cytoplasmic separation and qRT-PCR measurements of the M1 mRNA. The Error bars: Mean + SD of 3 independent tests. ∗p < 0.05; ∗∗p < 0.01; and ns (nonsignificant), p > 0.05 compared to control.
As in the early stage of IAV infection, hnRNPAB showed its significant effect on the viral protein expression instead of viral mRNA and vRNA levels, which proved that hnRNPAB did not affect the virus RNA synthesis, we then determined whether the inhibition of hnRNPAB-induced virus replication is involved in the nuclear export limitation of viral mRNA. HnRNPAB-transfected cells were infected with H7N9 virus and then stained with the cy3-labeled probes specific for the M1, HA, NP, and NS1 mRNA and the cy3-labeled M1 vRNA probes. Confocal microscopy observations showed that after H7N9 virus infection, the level of intranuclear M1, HA, NP, and NS1 mRNA increased markedly in hnRNPAB-overexpressing cells when compared with that in mock-transfected cells, and overexpression of hnRNPAB showed no significant effect on the cellular distribution of vRNA (Figures 4I and 4J and S3C). However, the hnRNPAB knockdown A549 cells with ActD treatment displayed that intranuclear distribution of M1 mRNA decreased significantly (Figure S3D). Subsequently, the nuclear-cytoplasmic separation was carried out in hnRNPAB-overexpressing or knockdown cells with H7N9 virus infection, and total RNA was extracted. The mRNA levels of the H7N9 virus in the cytoplasm and nucleus were analyzed using qRT-PCR with M1 mRNA primers, which showed that when compared with that in mock-transfected cells, the intranuclear levels of the mRNA transcript of M1 increased significantly in hnRNPAB-transfected cells (Figures 4K and S3E, p < 0.01) and decreased significantly in hnRNPAB knockdown cells (Figure 4L and S3F, p < 0.01). These data demonstrated that hnRNPAB blocks the nuclear export of viral mRNA during infection. Taken together, these data allowed us to conclude that nuclear export of viral mRNA is inhibited by interaction with the motif Gly of hnRNPAB.
To further analyze whether hnRNPAB interaction with viral protein NP affects virus replication via viral mRNA nuclear export, we transfected myc-tagged hnRNPAB and FLAG-tagged NP into 293T cells, and the cells were then infected with H7N9 virus. Fluorescence in situ hybridization (FISH) analysis with cy3-labeled M1 mRNA probes indicated that the viral M1 mRNA showed more distribution in the nucleus of cells co-expressing hnRNPAB and NP in contrast to that of the cells expressing hnRNPAB individually (Figure S4A). RNA cytoplasm and nuclear isolation assay proved that expression of NP and hnRNPAB inhibited viral M1 mRNA nuclear export (Figure S4B). The data demonstrated that the NP interaction with hnRNPAB inhibited viral mRNA nuclear export.
HnRNPAB inhibits the interaction of NXF1 with viral mRNA via binding to the NXF1 and TREX complex
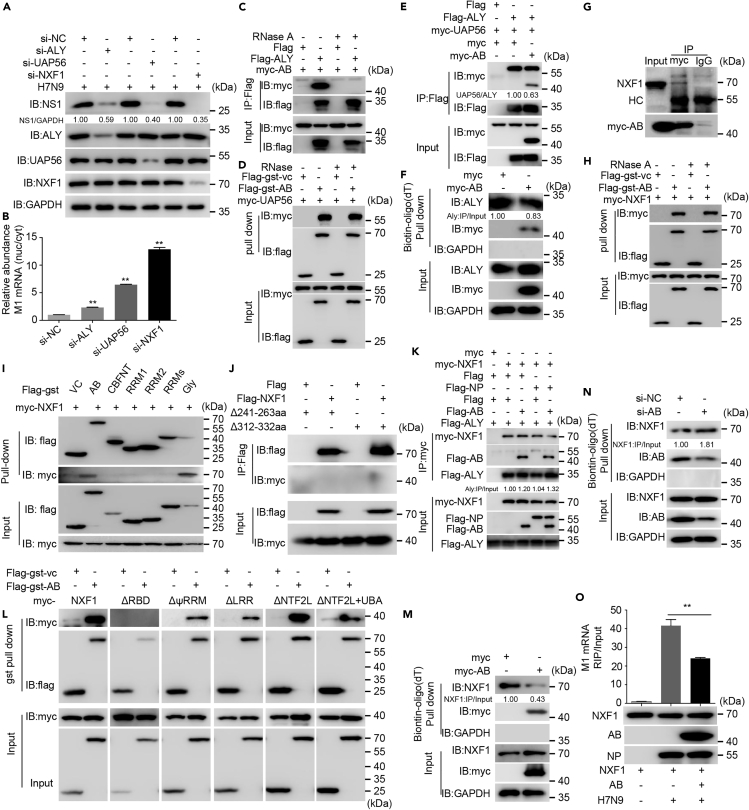
In eukaryotic cells, the nuclear export of mRNA is mediated by the transport receptor NXF1. The TREX complex acts as the key adaptor of NXF1. The TREX complex is mainly composed of ALY, UAP56, and THO complex, and UAP56 is required for the assembly of the complex and recruitment of ALY to mRNA (Luo et al., 2001). To investigate whether the cellular mRNA nuclear export factors are involved in the nuclear export of viral mRNA, the expression of ALY or UAP56 or NXF1 was knocked down, and these cells were then infected with the H7N9 virus. These cells were subjected to immunoblotting and nuclear-cytoplasmic fraction separation for qRT-PCR. In the immunoblotting assay, H7N9 virus protein NS1 levels were significantly reduced in ALY, UAP56, or NXF1 knockdown cells (Figure 5A, p < 0.01). FISH and qRT-PCR showed that the intranuclear concentration of viral mRNA was significantly increased in the ALY, UAP56, or NXF1 knockdown cells in contrast to the control cells (Figure 5B and S5A-B, p < 0.01), especially in the NXF1 knockdown cells. These results showed that the cellular mRNA export pathway participates in regulating the nuclear export of viral mRNA (Read and Digard, 2010). Whether hnRNPAB is related to nuclear export of viral mRNA regulated by NXF1 and TREX complex is unknown. Therefore, we performed immunoprecipitation to detect the interaction of hnRNPAB with ALY or UAP56 in overexpressing cells. In the co-immunoprecipitation (co-IP) assay, hnRNPAB could bind to ALY in ALY and hnRNPAB co-expressing cells without RNase treatment but not with RNase treatment; however, hnRNPAB bound to UAP56 in UAP56 and hnRNPAB co-expressing cells with or without RNase treatment (Figures 5C and 5D), confirming that RNA mediated the interaction of hnRNPAB with ALY, and that hnRNPAB interacts with UAP56 directly. Meanwhile, we also tested hnRNPAB's interaction with both UAP56 and ALY by artificially mixing the myc-hnRNPAB, FLAG-ALY, and myc-UAP56. An anti-FLAG co-IP assay showed that hnRNPAB bound to ALY and UAP56 to form a trimer complex, and that binding of UAP56 to ALY and hnRNPAB in the ALY-UAP56-hnRNPAB mixture decreased when compared with that in the ALY-UAP56 mixture (Figure 5E; p < 0.001), showing that hnRNPAB decreased UAP56 binding to ALY. A biotin-oligo(dT) pull-down assay further showed that the binding of ALY to mRNA decreased in hnRNPAB-overexpressing cells (Figure 5F; p < 0.01), indicating that overexpression of hnRNPAB repressed the binding of ALY to mRNA. Taken together, these results demonstrated that hnRNPAB weakened mRNA transfer from UAP56 to ALY by binding to ALY.
Figure 5.
HnRNPAB inhibits viral mRNA binding to NXF1
(A) The impact of NXF1, UAP56, and ALY on H7N9 virus replication. 293T cells were transfected with the indicated siRNAs for 36 h and then infected with H7N9 virus for 12 h. Cell lysates were analyzed with mAbs recognizing ALY, UAP56, NXF1, and NS1 in western blotting.
(B) The nucleo-cytoplasmic ratio of H7N9 viral mRNA in NXF1, UAP56, or ALY knockdown cells. Cells from (A) were subjected to nuclear-cytoplasmic separation. RT-qPCR assays were performed for the H7N9 M1 mRNA analysis.
(C) Co-IP assay of FLAG-ALY and myc-hnRNPAB. 293T cells were co-transfected with myc-hnRNPAB and FLAG-ALY. Cell lysates were added with or without RNase as indicated and then subjected to immunoprecipitation with anti-FLAG mAb and western blotting with anti-FLAG mAb and anti-myc pAb. FLAG was used as control.
(D) Pull-down assay of UAP56 and hnRNPAB. 293T cells were transfected with myc-tagged UAP56 or FLAG-gst-hnRNPAB. Cell lysates were mixed and treated with or without RNase and incubated with glutathione beads. Pull-down and western blotting assays were performed with anti-FLAG mAb and anti-myc pAb. FLAG-gst-vc was used as a control.
(E) Co-IP assay of hnRNPAB's interaction with ALY and UAP56. 293T cells were transfected with FLAG-ALY, myc-UAP56, myc-hnRNPAB, or myc individually for 36 h. Cell lysates were mixed as indicated and then immunoprecipitated.
(F) RNA pull-down analysis of poly(A)+RNA binding to hnRNPAB and ALY. 293T cells were transfected with myc-hnRNPAB or myc for 24 h. Cell lysates were subjected to biotin-oligo (dT) pull-down and western blotting with anti-ALY mAb, anti-myc, and anti-GAPDH pAbs. The quantitation of ALY was analyzed using ImageJ software.
(G) Identification of hnRNPAB interaction with endogenous NXF1. Co-IP assay of myc-hnRNPAB and endogenous NXF1 in 293T cells. 293T cells were transfected with myc-hnRNPAB and subjected to immunoprecipitation and western blotting assays using anti-myc pAbs and anti-NXF1 mAb.
(H) Pull-down assay of NXF1 and hnRNPAB. Pull-down assay of FLAG-gst-hnRNPAB and myc-NXF1. 293T cells were transfected with myc-NXF1 and FLAG-gst-hnRNPAB individually. Pull-down assays were performed as in (D).
(I) Mapping of the NXF1-interacting domain within hnRNPAB. FLAG-gst tagged hnRNPAB and truncated mutants were transfected with myc-NXF1 into 293T cells individually for 24 h. Cell lysates were mixed and were then pulled down, and western blotting were conducted using anti-FLAG mAb and anti-myc pAbs.
(J) The residues 241–263 aa and 312–332 aa of the hnRNPAB Gly domain are critical for the interaction with NXF1. 293T cells were transfected with FLAG-NXF1 and myc-tagged Gly domain of hnRNPAB Δ241–263 aa or Δ312–332 aa, individually or in combination. Cell lysates were subjected to immunoprecipitation and western blotting with anti-FLAG mAb and anti-myc pAbs.
(K) HnRNPAB and NP do not interrupt the interaction of ALY with NXF1. 293T cells were transfected with myc-NXF1, FLAG-hnRNPAB, FLAG-NP, or FLAG-ALY individually for 24 h. Cell lysates were immunoprecipitated using anti-myc mAb and analyzed using western blotting with anti-FLAG mAb and anti-myc pAbs.
(L) Identification of the hnRNPAB-binding domain of NXF1. 293T cells were transfected with FLAG-gst-hnRNPAB, myc-tagged NXF1, and NXF1 deletion mutants ΔRBD, ΔψRRM, ΔLRR, ΔNTF2L, and ΔNTF2L + UBA for 24 h. Cell lysates were subjected to pull-down and western blotting assays using anti-FLAG mAb and anti-myc pAbs.
(M and N) RNA pull-down assay of NXF1 in hnRNPAB-overexpressing or knockdown cells. 293T cells were transfected with myc-hnRNPAB (M) or its siRNA (N). Cell lysates were subjected to RNA pull-down and western blotting assays using anti-NXF1 and anti-hnRNPAB mAbs and anti-myc and anti-GAPDH pAbs.
(O) RIP assay of viral mRNA binding to NXF1. 293T cells were transfected with myc-NXF1 or FLAG-hnRNPAB for 24 h or infected with H7N9 virus for 24 h. Cells were solubilized with RIP lysis buffer and subjected to immunoprecipitation with anti-myc mAb. The NXF1-bound RNAs were analyzed using qRT-PCR.
Next, we investigated if the NXF1's RNA binding is also restricted by hnRNPAB to limit viral mRNA nuclear export. In co-IP and pull-down assays, fine location experiments showed that NXF1 is directly bound to the N-terminal residues 241–263 aa and C-terminal residues 312–332 aa within the Gly motif of hnRNPAB (Figures 5G–5J). NXF1 binds to ALY and exposes its RBD (Viphakone et al., 2012), and viral mRNAs are exported via the NXF1-dependent pathway (Read and Digard, 2010). Therefore, we wanted to know whether NXF1 binding to hnRNPAB blocks the exposing of its RBD and whether the NXF1-viral mRNA binding is restricted. For the purpose, we performed co-IP, GST, and RNA pull-down assays. The co-IP assay showed that hnRNPAB binding to NXF1 did not interrupt NXF1's interaction with ALY when hnRNPAB was incubated with ALY and NXF1 (Figure 5K), suggesting that the hnRNPAB-NXF1 interaction did not interfere with the NXF1-ALY interaction. In the GST pull-down assay, hnRNPAB was only detected to interact with the RBD of NXF1 by incubating separately with NXF1 RBD (ΔRBD), ψRRM (ΔψRRM), LRR (ΔLRR), NTF2L (ΔNTF2L), or NTF2L + UBA(ΔNTF2L + UBA) deletion mutants (Figure 5L). Consistently, in the biotin-oligo(dT) pull-down assay, the RNA-binding activity of NXF1 was upregulated in hnRNPAB knockdown cells, but not in hnRNPAB-overexpressing cells (Figures 5M and 5N). RNA pull-down assay with biotin-labeled M1 mRNA probes further showed that overexpression of hnRNPAB inhibited the NXF1 and ALY binding to M1 mRNA; however, knocking down of hnRNPAB promoted the NXF1 and ALY binding to M1 mRNA (Figures S6A and S6B). Meanwhile, in the RIP assay, we proved that hnRNPAB markedly repressed NXF1 binding to viral mRNA (Figures 5O and S6C), confirming that hnRNPAB binding to the RBD of NXF1 inhibits the NXF1 RNA-binding activity and mRNA transfer from ALY to NXF1. Taken together, the results suggested that hnRNPAB binding to NXF1 and the core components, UAP56 and ALY, of the TREX complex inhibits the transfer of RNA from ALY to NXF1.
The interaction of viral protein NP with hnRNPAB enhances the inhibition of ALY binding to viral mRNA
To analyze how viral protein NP binding to hnRNPAB inhibits H7N9 virus replication, we tested whether the NP-hnRNPAB complex disrupts the mRNA nuclear export pathway. Strikingly, the NP-hnRNPAB complex markedly repressed the interaction between ALY and UAP56 (Figure 6A; p < 0.01). To test whether the NP-hnRNPAB complex could inhibit the RNA-ALY binding significantly, we performed an RNA pull-down assay with biotin-oligo(dT) and biotin-labeled M1 mRNA probes. Results showed that the repression of ALY-mRNA binding was more significant in hnRNPAB-overexpressing cells with H7N9 virus infection (Figure 6B; p < 0.01) and ALY binding to M1 mRNA was also repressed significantly by NP-hnRNPAB complex (Figure S7A). We then tested whether NP affects the interaction between hnRNPAB and ALY. The result showed that the ALY-hnRNPAB interaction is inhibited by viral protein NP (Figure 6C). Meanwhile, the RNA pull-down assays with biotin-oligo(dT) and biotin-labeled M1 mRNA probes revealed that NXF1 binding to viral mRNA decreased more significantly (Figures 6D and S7B). Taken together, the results demonstrated that the NP-hnRNPAB interaction interrupts ALY's interplay with UAP56 and hnRNPAB, which decreases the viral mRNA binding to ALY and the subsequent nuclear export of viral mRNA by repressing viral mRNA binding to NXF1.
Figure 6.
Viral protein NP interaction with HnRNPAB strengthens inhibition of ALY-viral mRNA binding
(A) Detection of viral protein NP interaction with ALY and UAP56. 293T cells were cotransfected with myc-UAP56, FLAG-hnRNPAB, FLAG-NP, or FLAG-ALY for 24 h. Cell lysates were subjected to immunoprecipitation and western blotting with anti-FLAG mAb and anti-myc pAbs.
(B) RNA pull-down assay of ALY. 293T cells were infected with H7N9 virus for 12 h and were then transfected with myc tagged hnRNPAB for 24 h. Cell lysates were subjected to RNA pull-down and western blotting with anti-ALY mAb, anti-GAPDH pAbs, and anti-myc pAbs.
(C) Inhibition of hnRNPAB's interaction with ALY by viral protein NP. 293T cells were transfected with myc-tagged hnRNPAB, FLAG-tagged NP, or ALY for 24 h. Co-IP and western blotting were conducted with anti-FLAG mAb and Anti-myc pAbs.
(D) RNA pull-down assay of NXF1. Cell lysates from (B) were subjected to RNA pull-down and western blotting with anti-NXF1 mAb, anti-GAPDH, and anti-myc pAbs.
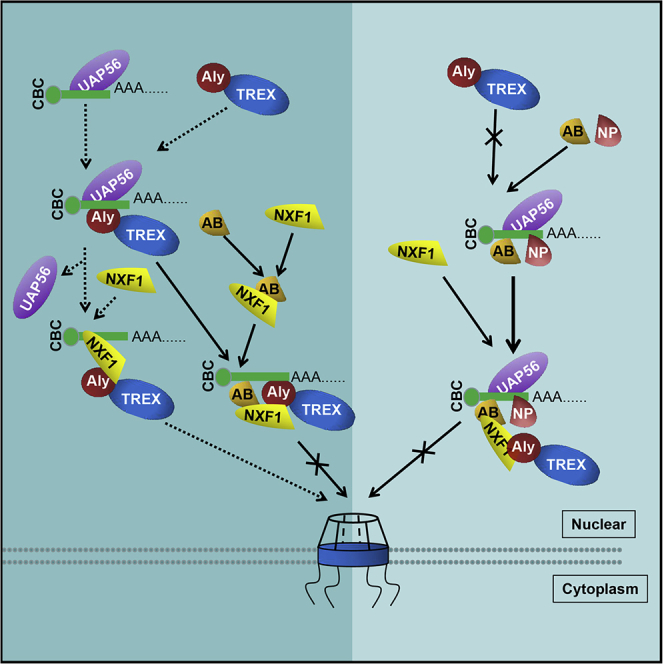
(E) Inhibition model of viral mRNA nuclear export by hnRNPAB interaction with viral protein NP. To limit RNA nuclear export, hnRNPAB weakens the transfer of poly(A)+RNA from UAP56 to ALY via direct interaction with UAP56 and inhibits the NXF1-poly(A)+RNA binding by interacting with the RBD of NXF1. When cells are infected with influenza A virus, the hnRNPAB-recruited viral protein further enhanced the inhibition of ALY and NXF1 binding to viral mRNA to block nuclear export of viral mRNA. The Error bars: Mean + SD of 3 independent tests. ∗p < 0.05; ∗∗p < 0.01; and ns (nonsignificant), p > 0.05 compared to control.
Discussion
HnRNPs A/B is the most abundant sub-family of hnRNPs (He and Smith, 2009) and is involved in regulating the virus life cycle in different ways. HnRNPA1's interaction with nucleocapsid protein of porcine epidemic diarrhea virus impairs viral replication (Li et al., 2018). HnRNPA2B1 inhibits IAV replication by suppressing NS1 mRNA nuclear export (Wang et al., 2014), inhibits DNA virus replication by initiating the innate immune response (Wang et al., 2019), and supports hepatitis E virus replication by interacting with the promoter regions of the hepatitis E virus genome (Pingale et al., 2020). HnRNPAB, as an RNA-binding protein, is considered as a distantly related member of hnRNPs A/B and is involved in the regulation of RNA biological processes. Structurally, hnRNPAB comprises a CARG-binding factor-A domain in the N terminus (CBFNT), two RBDs (RRM1 and RRM2), and a C-terminal glycine-rich region (Gly domain) (Khan et al., 1991). The Gly domain contains a motif characterized by closely spaced clusters of Arg-Gly-Gly tripeptide repeats with interspersed aromatic amino acids and is responsible for RNA binding (Geuens et al., 2016). In the present study, infection with different subtypes of IAV continuously upregulated both mRNA and protein levels of hnRNPAB, and overexpression of hnRNPAB and its Gly domain inhibited IAV replication (Figures 3D and 3E). This led to nuclear accumulation of poly(A)+RNA and enhanced nuclear retention of viral mRNA (Figures 4I–4L). Taken together these results demonstrated that the Gly domain of hnRNPAB is the binding domain of poly(A)+RNA and that hnRNPAB acts as a negative regulator of influenza virus replication by inhibiting mRNA nuclear export.
The TREX complex, composed of ALY, UAP56, and THO complexes, is a key player in mRNA biogenesis and participates in almost all stages of mRNA processing, linking transcription, and export (Katja et al., 2002; Viphakone et al., 2012). During the maturation and nucleo-cytoplasmic transport of poly(A)+mRNA, UAP56 binding to pre-mRNAs is required for the recruitment of ALY to form the TREX complex (Hautbergue et al., 2008), and the RNA bound to UAP56 is transferred to ALY by UAP56 binding to ALY (Klumpp et al., 1997). ALY also interacts with the mRNA export receptor NXF1 and the binding between ALY and NXF1 triggers the transfer of RNA from ALY to NXF1 (Ben-Shem et al., 2011; Ming-Lung et al., 2010). The mRNAs of HA, M1, and NS1 of PR8 virus were shown to be dependent on NXF1 and UAP56 for their nuclear export (Read and Digard, 2010). HIV-1 ANP32A and ANP32B mediate the nuclear export of HIV-1 unspliced or partially spliced mRNA via interactions with Rev and CRM1 (Branche and Falsey, 2016). The limited studies on viral mRNA nuclear export have shown that nuclear export of viral mRNA from different viruses depends on different nuclear export pathways. In the present study, we demonstrated that hnRNPAB markedly repressed NXF1-poly(A)+RNA binding and did not interrupt the interaction of ALY and NXF1 by directly linking to NXF1 and UAP56 (Figures 5K, 5M, and 5N). Meanwhile, hnRNPAB weakens the interaction between ALY and UAP56 by linking to ALY, resulting in poly(A)+RNA not transferring from UAP56 to ALY or from ALY to NXF1. Moreover, we also found that the Gly domain of hnRNPAB directly interacts with the RBD of NXF1 via its N-terminal 241–263 residues and C-terminal 312–332 residues. More importantly, during IAV infection, hnRNPAB binding to viral protein NP enhances the interference with the UAP56-ALY interaction (Figure 6A). Therefore, we proposed that hnRNPAB interaction with NP of IAV strengthens the inhibition of flu virus replication by limiting the nuclear export of viral mRNA.
Amino acid sequence analysis revealed that hnRNPAB is conserved among different species and has 91.10% homology between human and mouse and 79.05% homology between human and chicken (data not shown). We also proved that hnRNPAB from different species has the ability to inhibit virus replication (Figures 3A and 3B), indicating that hnRNPAB from different species has a similar function during influenza virus infection. Members of hnRNPs A/B subfamily, such as hnRNPA1 and hnRNPA2/B1, contain a nuclear targeting sequence in their C termini (He and Smith, 2009; Jacques et al., 2013). In the present study, we also found that the C-terminal residues 312–332 in the Gly domain represent the NLS of hnRNPAB and are critical for hnRNPAB to interact with the NXF1 and viral mRNA. Interestingly, C-terminal residues 312–332 aa within the Gly motif are also the binding site for NXF1 (Figure 5J). Combined with the fact that hnRNPAB binds to poly(A)+RNA, our findings suggest that in the nucleus, hnRNPAB, via its NLS, participates in inhibiting IAV replication.
Our data indicated that hnRNPAB acts as a negative regulator of gene expression by inhibiting mRNA nuclear export. In this process, hnRNPAB acts as a valve to calm down the protein expression of both exogenous and endogenous genes when cells are subjected to external stimuli like virus infection for maintaining cell homeostasis. This is consistent with the viewpoint that normal nuclear retention of mRNA enables cells to rapidly respond to stress. Therefore, we inferred that when cells were infected with viruses or invaded by exogenous genes, hnRNPAB regulated negatively the expression of the exogenous genes to gain time for cells to prepare innate immunity or other response and keep the cells in a stable station.
In summary, our study indicated that hnRNPAB could inhibit the replication of IAV via repression of viral mRNA nuclear export. HnRNPAB binding to viral protein NP enhances its repression of viral mRNA nuclear export by interrupting the transfer of poly(A)+RNA from UAP56 to ALY and NXF1 (Figure 6E).
Limitations of the study
First, we demonstrated that hnRNPAB interacts with viral mRNA and inhibits the viral mRNA nuclear export. However, whether the viral mRNA is decayed in the nucleus or is transferred into cytoplasm later is still unclear. Second, we demonstrate that the IAV NP cooperates with hnRNPAB to repress the interaction of UAP56 and ALY, further inhibiting viral mRNA transfer from UAP56 to ALY. However, the mechanism of NP-hnRNPAB complex inhibiting the interaction of UAP56 and ALY is unknown.
Resource availability
Lead contact
Further requests and information should be directed to and will be fulfilled by Min Liao (liaomin4545@zju.edu.cn).
Materials availability
Materials are available upon reasonable request.
Data and code availability
All data are included in the published article and the Supplemental Information, and any additional information will be available from the lead contact upon request.
Methods
All methods can be found in the accompanying Transparent methods supplemental file.
Acknowledgments
This study is supported by grants from the National Key Technology R&D Program of China (grant No.2015BAD12B01) and China Agricultural Research System (Grant No. CARS-40-K13). We thank Ms. Yunqin Li from Bio-ultrastructure Analysis Lab, Center of Agrobiology and Environmental Sciences, Zhejiang University, for technical help with confocal microscopy observations.
Author contributions
X.W., J.Z., and M.L. designed the experiments. X.W., L.L., and Y.Z. equally performed the experiments with the help of M.F., T.Y., Y.Y., and M.L., X.W., and J.Z. wrote the paper.
Declaration of interests
The authors declare no competing interests.
Published: March 19, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.102160.
Contributor Information
Jiyong Zhou, Email: jyzhou@zju.edu.cn.
Min Liao, Email: liaomin4545@zju.edu.cn.
Supplemental information
References
- Akindahunsi A.A., Bandiera A., Manzini G. Vertebrate 2xRBD hnRNP proteins: a comparative analysis of genome, mRNA and protein sequences. Comput. Biol. Chem. 2005;29:13–23. doi: 10.1016/j.compbiolchem.2004.11.002. [DOI] [PubMed] [Google Scholar]
- Amit B., Shahar B., Galit S., Yael G., Geula H., Greenberg D.S., Maya K., Becker A.J., Alon F., Hermona S. Cholinergic-associated loss of hnRNP-A/B in Alzheimer's disease impairs cortical splicing and cognitive function in mice. Embo Mol. Med. 2012;4:730–742. doi: 10.1002/emmm.201100995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shem A., de Loubresse N.G., Melnikov S., Jenner L., Yusupova G., Yusupov M. The structure of the eukaryotic ribosome at 3.0 Å resolution. Science (New York, NY) 2011;334:1524–1529. doi: 10.1126/science.1212642. [DOI] [PubMed] [Google Scholar]
- Bjork P., Wieslander L. Mechanisms of mRNA export. Semin. Cell Dev Biol. 2014;32:47–54. doi: 10.1016/j.semcdb.2014.04.027. [DOI] [PubMed] [Google Scholar]
- Branche A., Falsey A. Parainfluenza virus infection. Semin. Respir. Crit. Care Med. 2016;37:538–554. doi: 10.1055/s-0036-1584798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L., Liu S., Li Y., Yang G., Luo Y., Li S., Du H., Zhao Y., Wang D., Chen J. The nuclear matrix protein SAFA surveils viral RNA and facilitates immunity by activating antiviral enhancers and super-enhancers. Cell Host Microbe. 2019;26:369–384.e8. doi: 10.1016/j.chom.2019.08.010. [DOI] [PubMed] [Google Scholar]
- Carol Johnson D.P., McKinstry M., John McNeil D.R., Lawrence a.J.B. Tracking COL1A1 RNA in osteogenesis imperfecta: splice-defective transcripts initiate transport from the gene but are retained within the SC35 domain. J. Cell Biol. 2000;151:1129. doi: 10.1083/jcb.150.3.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.T., Hautbergue G.M., Walsh M.J., Viphakone N., van Dijk T.B., Philipsen S., Wilson S.A. Chtop is a component of the dynamic TREX mRNA export complex. EMBO J. 2013;32:473–486. doi: 10.1038/emboj.2012.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean J.L.E., Gareth S., Robin W., Lesley R., Clark A.R., Jeremy S. Identification of a novel AU-rich-element-binding protein which is related to AUF1. Biochem. J. 2002;366:709. doi: 10.1042/BJ20020402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fodor E. The RNA polymerase of influenza A virus: mechanisms of viral transcription and replication. Acta Virol. 2013;57:113–122. doi: 10.4149/av_2013_02_113. [DOI] [PubMed] [Google Scholar]
- Fribourg S., Braun I.C., Izaurralde E., Conti E. Structural basis for the recognition of a nucleoporin FG repeat by the NTF2-like domain of the TAP/p15 mRNA nuclear export factor. Mol. Cell. 2001;8:645–656. doi: 10.1016/s1097-2765(01)00348-3. [DOI] [PubMed] [Google Scholar]
- Geuens T., Bouhy D., Timmerman V. The hnRNP family: insights into their role in health and disease. Hum. Genet. 2016;135:851–867. doi: 10.1007/s00439-016-1683-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halpern K.B., Caspi I., Lemze D., Levy M., Landen S., Elinav E., Ulitsky I., Itzkovitz S. Nuclear retention of mRNA in mammalian tissues. Cell Rep. 2015;13:2653–2662. doi: 10.1016/j.celrep.2015.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hautbergue G.M. RNA nuclear export: from neurological disorders to cancer. Adv. Exp. Med. Biol. 2017;1007:89–109. doi: 10.1007/978-3-319-60733-7_6. [DOI] [PubMed] [Google Scholar]
- Hautbergue G.M., Hung M.-L., Golovanov A.P., Lian L.-Y., Wilson S.A. Mutually exclusive interactions drive handover of mRNA from export adaptors to TAP. Proc. Natl. Acad. Sci. U S A. 2008;105:5154–5159. doi: 10.1073/pnas.0709167105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Smith R. Nuclear functions of heterogeneous nuclear ribonucleoproteins A/B. Cell Mol. Life Sci. 2009;66:1239–1256. doi: 10.1007/s00018-008-8532-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacques J.P., Sean P., Massimo C. hnRNP A1: the Swiss army knife of gene expression. Int. J. Mol. Sci. 2013;14:1C8999–19024. doi: 10.3390/ijms140918999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julia D., Olivier T., Manuel R.C. Influenza viruses and mRNA splicing: doing more with less. mBio. 2014;5 doi: 10.1128/mBio.00070-14. e00070-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katja S.S., Seiji M., Paul M., Jens P., Marisa O., Susana R.N., Rondón A.G., Andres A., Kevin S., Robin R. TREX is a conserved complex coupling transcription with messenger RNA export. Nature. 2002;417:304–308. doi: 10.1038/nature746. [DOI] [PubMed] [Google Scholar]
- Kawaguchi A., Momose F., Nagata K. Replication-coupled and host factor-mediated encapsidation of the influenza virus genome by viral nucleoprotein. J. Virol. 2011;85:6197–6204. doi: 10.1128/JVI.00277-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan F.A., Jaiswal A.K., Szer W. Cloning and sequence analysis of a human type A/B hnRNP protein. FEBS Lett. 1991;290:159–161. doi: 10.1016/0014-5793(91)81249-8. [DOI] [PubMed] [Google Scholar]
- Klumpp K.,., Ruigrok R.W., Baudin F. Roles of the influenza virus polymerase and nucleoprotein in forming a functional RNP structure. Embo J. 1997;16:1248–1257. doi: 10.1093/emboj/16.6.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampasona A.A., Czaplinski K. Hnrnpab regulates neural cell motility through transcription of Eps8. RNA (New York, NY) 2019;25:45–59. doi: 10.1261/rna.067413.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Yu M., Zheng W., Liu W.J.V. Nucleocytoplasmic shuttling of influenza A virus proteins. Viruses. 2015;7:2668–2682. doi: 10.3390/v7052668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Zeng W., Ye S., Lv J., Nie A., Zhang B., Sun Y., Han H., He Q. Cellular hnRNP A1 interacts with nucleocapsid protein of porcine epidemic diarrhea virus and impairs viral replication. Viruses. 2018;10:127. doi: 10.3390/v10030127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M.L., Zhou Z.,., Magni K.,., Christoforides C.,., Rappsilber J.,., Mann M., Reed R. Pre-mRNA splicing and mRNA export linked by direct interactions between UAP56 and Aly. Nature. 2001;413:644–647. doi: 10.1038/35098106. [DOI] [PubMed] [Google Scholar]
- Luo W., Zhang J., Liang L., Wang G., Li Q., Zhu P., Zhou Y., Li J., Zhao Y., Sun N. Phospholipid scramblase 1 interacts with influenza A virus NP, impairing its nuclear import and thereby suppressing virus replication. PLoS Pathog. 2018;14:e1006851. doi: 10.1371/journal.ppat.1006851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming-Lung H., Hautbergue G.M., Snijders A.P.L., Dickman M.J., Wilson S.A. Arginine methylation of REF/ALY promotes efficient handover of mRNA to TAP/NXF1. Nucleic Acids Res. 2010;38:3351–3361. doi: 10.1093/nar/gkq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mor A., White A., Zhang K., Thompson M., Esparza M., Muñozmoreno R., Koide K., Lynch K.W., Garcíasastre A., Fontoura B.M.A. Influenza virus mRNA trafficking through host nuclear speckles. Nat. Microbiol. 2017;1:16069. doi: 10.1038/nmicrobiol.2016.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito T., Kiyasu Y., Sugiyama K., Kimura A., Nakano R., Matsukage A., Nagata K. An influenza virus replicon system in yeast identified Tat-SF1 as a stimulatory host factor for viral RNA synthesis. Proc. Natl. Acad. Sci. U S A. 2007;104:18235–18240. doi: 10.1073/pnas.0705856104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanaho F., Tomoyuki F., John S., Abrahan H.H., Manizheh I., Raju C.S., Kevin C., Piergiorgio P. The transacting factor CBF-A/Hnrnpab binds to the A2RE/RTS element of protamine 2 mRNA and contributes to its translational regulation during mouse spermatogenesis. Plos Genet. 2013;9:e1003858. doi: 10.1371/journal.pgen.1003858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palazzo A.F., Lee E.S.J. Sequence determinants for nuclear retention and cytoplasmic export of mRNAs and lncRNAs. Front. Genet. 2018;9:440. doi: 10.3389/fgene.2018.00440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pemberton L.F., Blobel G., Rosenblum J.S. Transport routes through the nuclear pore complex. Curr. Opin. Cell Biol. 1998;10:392–399. doi: 10.1016/s0955-0674(98)80016-1. [DOI] [PubMed] [Google Scholar]
- Piergiorgio P., Andreas J., Dmitri N., Christina K., Tomas B., Apostolia G., Bertil D.J.N.A.R. Nuclear actin is associated with a specific subset of hnRNP A/B-type proteins. Nucleic Acids Res. 2002;30:1725–1734. doi: 10.1093/nar/30.8.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pingale K.D., Kanade G.D., Karpe Y.A. Heterogeneous nuclear ribonucleoproteins participate in Hepatitis E virus (HEV) replication. J. Mol. Biol. 2020:30202–30203. doi: 10.1016/j.jmb.2020.02.025. [DOI] [PubMed] [Google Scholar]
- Raju C.S., Göritz C., Nord Y., Hermanson O., López-Iglesias C., Visa N., Castelo-Branco G., Percipalle P. In cultured oligodendrocytes the A/B-type hnRNP CBF-A accompanies MBP mRNA bound to mRNA trafficking sequences. Mol. Biol. Cell. 2008;19:3008–3019. doi: 10.1091/mbc.E07-10-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read E.K.C., Digard P. Individual influenza A virus mRNAs show differential dependence on cellular NXF1/TAP for their nuclear export. J. Gen. Virol. 2010;91:1290–1301. doi: 10.1099/vir.0.018564-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resainfante P., Jorba N., Coloma R., Ortín J. The influenza virus RNA synthesis machine: advances in its structure and function. RNA Biol. 2011;8:207–215. doi: 10.4161/rna.8.2.14513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai P.-L., Chiou N.-T., Kuss S., García-Sastre A., Lynch K.W., Fontoura B.M. Cellular RNA binding proteins NS1-BP and hnRNP K regulate influenza A virus RNA splicing. PLoS Pathog. 2013;9:e1003460. doi: 10.1371/journal.ppat.1003460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tunnicliffe R.B., Hautbergue G.M., Kalra P., Jackson B.R., Whitehouse A., Wilson S.A., Golovanov A.P. Structural basis for the recognition of cellular mRNA export factor REF by herpes viral proteins HSV-1 ICP27 and HVS ORF57. PLoS Pathog. 2011;7:e1001244. doi: 10.1371/journal.ppat.1001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viphakone N., Hautbergue G.M., Walsh M., Chang C.T., Holland A., Folco E.G., Reed R., Wilson S.A. TREX exposes the RNA-binding domain of Nxf1 to enable mRNA export. Nat. Commun. 2012;3:1006. doi: 10.1038/ncomms2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh M.J., Hautbergue G.M., Wilson S.A. Structure and function of mRNA export adaptors. Biochem. Soc. Trans. 2010;38:232–236. doi: 10.1042/BST0380232. [DOI] [PubMed] [Google Scholar]
- Wang L., Wen M., Cao X. Nuclear hnRNPA2B1 initiates and amplifies the innate immune response to DNA viruses. Science (New York, NY) 2019;365:eaav0758. doi: 10.1126/science.aav0758. [DOI] [PubMed] [Google Scholar]
- Wang P., Song W., Mok B., Zhao P., Qin K., Lai A., Smith G., Zhang J., Lin T., Guan Y. Nuclear factor 90 negatively regulates influenza virus replication by interacting with viral nucleoprotein. J. Virol. 2009;83:7850–7861. doi: 10.1128/JVI.00735-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zhou J., Du Y. hnRNP A2/B1 interacts with influenza A viral protein NS1 and inhibits virus replication potentially through suppressing NS1 RNA/protein levels and NS1 mRNA nuclear export. Virology. 2014;449:53–61. doi: 10.1016/j.virol.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickramasinghe V.O., Laskey R.A. Control of mammalian gene expression by selective mRNA export. Nat. Rev. Mol. Cell Biol. 2015;16:431–442. doi: 10.1038/nrm4010. [DOI] [PubMed] [Google Scholar]
- Williams T., Ngo L.H., Wickramasinghe V.O. Nuclear export of RNA: different sizes, shapes and functions. Semin. Cell Dev Biol. 2018;75:70–77. doi: 10.1016/j.semcdb.2017.08.054. [DOI] [PubMed] [Google Scholar]
- Yang Y., Chen Q., Piao H.Y., Wang B., Zhu G.Q., Chen E.B., Xiao K., Zhou Z.J., Shi G.M., Shi Y.H. HNRNPAB-regulated lncRNA-ELF209 inhibits the malignancy of hepatocellular carcinoma. Int. J. Cancer. 2019;146:169–180. doi: 10.1002/ijc.32409. [DOI] [PubMed] [Google Scholar]
- Yotam R., Raz V. Oculopharyngeal muscular dystrophy as a paradigm for muscle aging. Front. Aging Neurosci. 2014;6:317. doi: 10.3389/fnagi.2014.00317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolotukhin A.S., Tan W., Bear J., Smulevitch S., Felber B.K. U2AF participates in the binding of TAP (NXF1) to mRNA. J. Biol. Chem. 2002;277:3935–3942. doi: 10.1074/jbc.M107598200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are included in the published article and the Supplemental Information, and any additional information will be available from the lead contact upon request.