Abstract
Autoinducer 2 (AI-2) is a ubiquitous metabolite but, instead of acting as a “universal signal,” relatively few phenotypes have been associated with it, and many scientists believe AI-2 is often a metabolic byproduct rather than a signal. Here, the aim is to present evidence that AI-2 influences both biofilm formation and motility (swarming and chemotaxis), using Escherichia coli as the model system, to establish AI-2 as a true signal with an important physiological role in this bacterium. In addition, AI-2 signaling is compared to the other primary signal of E. coli, indole, and it is shown that they have opposite effects on biofilm formation and virulence.
Keywords: AI-2, chemotaxis, aggregation, biofilm, motility, Escherichia coli
1. Introduction
Quorum sensing (QS) is the process by which bacteria communicate via secreted signals (autoinducers); once the concentration of the autoinducers reaches a threshold, the signal is detected, and gene expression is altered [1]. The roles of QS are diverse and include population density detection, virulence, biofilm formation, and the maintenance of the stress response [2]. Although inhibitors of QS (quorum-quenching compounds) are still promoted as a means to reduce virulence without promoting resistance [3], these compounds will indubitably and unfortunately fail. The main problem is that the inhibition of QS leads to pleiotropic effects that affect growth; hence, lab strains and clinical isolates rapidly evolve resistance to these compounds [4,5,6]. Clearly, it is imperative to have a better understanding of QS in order to be in a position to better control bacteria to prevent diseases, such as stomach cancer and ulcers caused by Helicobacter pylori and Lyme disease by Borrelia burgdorferi [7], and to utilize them for synthetic biology applications. Therefore, in this opinion piece, we probe the physiological role of AI-2 by focusing on the best-studied bacterium, Escherichia coli.
2. Autoinducer-2
Commensal E. coli has several QS pathways, including one system based on indole (Figure 1) [8,9,10], which is produced by TnaA from tryptophan, and another system based on autoinducer 2 (AI-2) (Figure 1) [11], which is produced by LuxS from S-ribosylhomocysteine [12]. It appears AI-2 is used primarily for communication inside the gastrointestinal tract at 37 °C, while indole is used primarily at lower temperatures (30 °C and lower) when the bacterium is outside of its eucaryotic host [9]. Although E. coli can detect homoserine lactones through the autoinducer-1 sensor SdiA (a LuxR homolog), it lacks a homoserine lactone synthase to produce the homoserine lactone signal, so E. coli uses SdiA to eavesdrop on signals of other bacteria [13]. Moreover, there is an interaction between these systems in that SdiA has been shown to be important for indole signaling in E. coli [8].
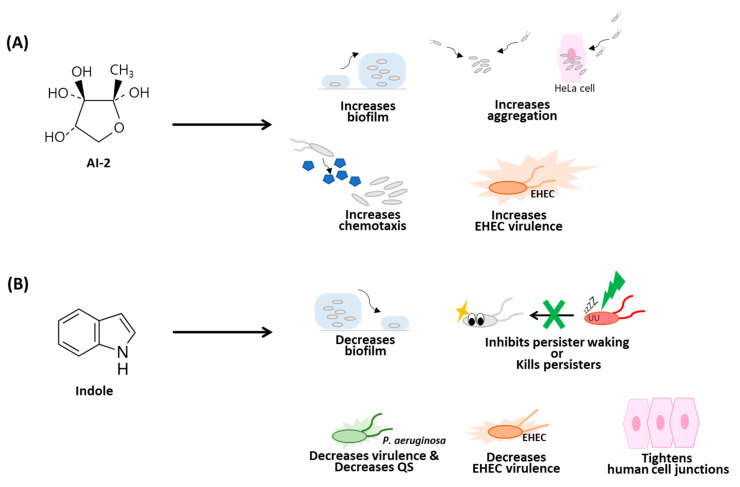
Figure 1.
Comparison of the phenotypes affected by (A) autoinducer 2 (AI-2) and (B) indole. Curved black arrows indicate cell motility/movement, QS is quorum sensing, EHEC is Escherichia coli O157:H7, and flagella are indicated by two lines at one of the cell poles. Human cells are indicated by pink hexagons. Green lightning indicates the application of indole. The R-2-methyl-2,3,3,4-tetrahydroxytetrahydrofuran (R-THMF) form of AI-2 is shown.
Once produced by LuxS, the AI-2 precursor 4,5-dihydroxy-2,3-pentanedione is converted spontaneously into R-2-methyl-2,3,3,4-tetrahydroxytetrahydrofuran (R-THMF) in E. coli (Figure 1), and R-THMF is the active form of AI-2 [7]. Hydrophilic AI-2 is transported from the cell by the membrane protein TqsA [14]. Once a threshold concentration is reached in the late exponential phase, AI-2 is imported into E. coli through its recognition by the AI-2 receptor LsrB [15]. In addition to LsrB in E. coli, LuxP (e.g., Vibrio harveyi) and the dCACHE-domain proteins PctA/TlpQ (Pseudomonas aeruginosa) are receptors for AI-2 [15], so there are at least three forms of AI-2 receptors in different bacteria. Furthermore, upon import, AI-2 is phosphorylated by LsrK in E. coli, and phosphorylated AI-2 binds and inhibits the repressor LsrR, which leads to changes in gene expression primarily at 37 °C [9].
3. AI-2 and Biofilm Formation
Although indole reduces both pathogenic [16] and non-pathogenic E. coli biofilm formation [17], AI-2 increases E. coli biofilm formation (Figure 1). Initially, QS was linked to biofilm formation using non-E. coli species and based on non-AI-2 signaling, specifically, for homoserine lactone increasing Pseudomonas aeruginosa [18]. Later studies, with Vibrio cholerae [19], Serratia liquefaciens [20], and Streptococcus mutans [21], confirmed the link of QS to biofilm formation.
The first report of AI-2 and biofilm formation was indirect and based on masking AI-2 signaling in E. coli with the QS inhibitor (5Z)-4-bromo-5-(bromomethylene)-3-butyl-2(5H)-furanone (henceforth furanone) from the alga Delisea pulchra; in this report, biofilm formation was reduced by 60 µg/mL furanone [22]. Later reports of AI-2 influencing biofilm formation were based on luxS mutants rather than purified AI-2. For example, a luxS mutation in Streptococcus gordonii influenced mixed-species biofilm formation with Porphyromonas gingivalis [23], a luxS mutation had a small impact on the architecture of Klebsiella pneumoniae (although there was no effect for a luxS mutant for intestinal colonization and colonization on polystyrene) [24], and a luxS mutant increased biofilm formation in Helicobacter pylori [25]. Unfortunately, these early results related to AI-2 via luxS mutations do not provide compelling evidence due to pleiotropic changes resulting from the luxS mutations.
The first direct demonstration that AI-2 was responsible for influencing biofilm formation was the 4- to 24-fold increase in biofilm formation in microtiter plates for three E. coli strains upon the addition of 11 µM of purified AI-2 [11]. Moreover, AI-2 failed to stimulate biofilm formation for an lsrK AI-2 regulation mutant, and AI-2 stimulated biofilm formation five-fold in flow cells [11]. A decade later, the Sourjik group rediscovered that AI-2 increases E. coli biofilm formation and extended the original results to show AI-2 increases aggregation through the adhesin antigen 43 and curli [26]. They [26] also confirmed that the AI-2 Lsr uptake/processing pathway influences E. coli biofilm formation [27].
4. AI-2 and Chemotaxis
The first indication that AI-2 affects E. coli motility was that the QS inhibitor furanone at 13 µg/cm2 inhibited E. coli swarming motility [22]; critically, the furanone also inhibited E. coli AI-2 signaling by 26,600-fold [22]. Next, furanone was shown to repress 44 of the 56 genes induced by AI-2, including those for chemotaxis (e.g., aer, cheABRWYZ, tap, tsr, trg) and motility (e.g., motAB, flgABCDEFGHIJKLMN, fliACDFHIKLMNOPQ) [28]. Therefore, AI-2 induces chemotaxis and motility genes in E. coli, and masking AI-2 signaling with furanone reduces motility and biofilm formation.
The first direct report of AI-2 as a chemoattractant for any species was the 2008 discovery that Escherichia coli O157:H7 (EHEC) is attracted to purified AI-2 [29]. For EHEC, AI-2 also increases both swimming motility and attachment to HeLa cells [29]. For non-pathogenic E. coli, microfluidic devices were used a year later to show AI-2 is an attractant [30]. Later, similar to their studies on biofilm formation, the Sourjik group confirmed that AI-2 attracts E. coli [26]. Furthermore, as with biofilms, indole signaling is opposite that of AI-2 since indole repels enterohemorrhagic EHEC [31], whereas AI-2 attracts EHEC [29] (Figure 1).
The mechanism by which AI-2 is detected in E. coli was determined to be the chemotactic receptor Tsr, which previously was known for its recognition of L-serine [32]; LsrB, the AI-2 receptor, was also shown to be necessary [32]. As with chemotaxis and biofilm formation, chemotaxis through Tsr was corroborated by the Sourjik group [26]. Furthermore, the Manson group also verified that AI-2 increases biofilm formation in E. coli and found that biofilm formation in this strain is enhanced by chemotaxis to AI-2 [33]. Therefore, AI-2 stimulates biofilm formation in E. coli by increasing aggregation and chemotaxis (Figure 1).
5. AI-2 and Virulence
The two main E. coli signals influence pathogens in an opposite manner—indole decreases EHEC chemotaxis, motility, biofilm formation, and adherence to epithelial cells at the physiologically relevant concentration of primarily 0.5 mM [31]; these results that indole decreases EHEC virulence were largely confirmed 12 years later by the Sperandio group [34,35] (Figure 1). Indole from E. coli also reduces the virulence of P. aeruginosa by masking its QS [36], prevents P. aeruginosa from resuscitating [37] from the dormant persister state [38], and tightens the epithelial cell junctions of the human host [39]. Indole and its derivatives also kill persister cells [40,41]. In contrast, AI-2 at 100 µM to 500 µM increases EHEC chemotaxis, motility, and adherence to epithelial cells and induces biofilm-related genes [29]. Moreover, AI-2 induces the expression of 23 genes of the locus of enterocyte effacement of EHEC [29]. Hence, in pathogenic E. coli, indole reduces pathogenicity, while AI-2 increases it.
6. Perspectives
The discovery that the E. coli AI-2 signal secreted by cells attracts other E. coli cells and leads to increased biofilm formation indicates that E. coli cells actively seek other E. coli cells to form communities [42]. Hence, it illustrates how bacteria can seek kin to increase their fitness, i.e., cells seek others to build communities (biofilms) to protect themselves from myriad stresses [43] and to increase their pathogenicity.
The chemoattractant property of AI-2 has also led to several synthetic biology applications. For example, biological nanofactories have been devised that detect and bind cancer cells and then produce AI-2 at the surface of the cancer cells, which attracts E. coli homing cells that internalize the synthesized AI-2 and then produce a biomarker or potentially an anti-cancer compound from an AI-2-induced promoter [44]. In this way, healthy cells could be discriminated from diseased ones. Therefore, the better understanding of the roles AI-2 and indole play in E. coli physiology has had a significant impact, both in our understanding of how communities are formed and in synthetic biology. Hence, AI-2 and indole are true and important signals in E. coli.
Acknowledgments
This work was supported by funds derived from the Biotechnology Endowed Professorship at the Pennsylvania State University.
Funding
This research was funded by a National Research Foundation of Korea (NRF) grant from the Korean government (No. NRF-2020R1F1A1072397).
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Mukherjee S., Bassler B.L. Bacterial quorum sensing in complex and dynamically changing environments. Nat. Rev. Microbiol. 2019;17:371–382. doi: 10.1038/s41579-019-0186-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.García-Contreras R., Nuñez-López L., Jasso-Chávez R., Kwan B.W., Belmont J.A., Rangel-Vega A., Maeda T., Wood T.K. Quorum sensing enhancement of the stress response promotes resistance to quorum quenching and prevents social cheating. ISME J. 2015;9:115–125. doi: 10.1038/ismej.2014.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ellermann M., Sperandio V. Bacterial signaling as an antimicrobial target. Curr. Opin. Microbiol. 2020;57:78–86. doi: 10.1016/j.mib.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.García-Contreras R., Martínez-Vázquez M., Velázquez-Guadarrama N., Villegas Pañeda A.G., Hashimoto T., Maeda T., Quezada H., Wood T.K. Resistance to the quorum-quenching compounds brominated furanone C-30 and 5-fluorouracil in Pseudomonas aeruginosa clinical isolates. Path. Dis. 2013;68:8–11. doi: 10.1111/2049-632X.12039. [DOI] [PubMed] [Google Scholar]
- 5.Maeda T., García-Contreras R., Pu M., Sheng L., Garcia L.R., Tomás M., Wood T.K. Quorum quenching quandary: Resistance to antivirulence compounds. ISME J. 2012;6:493–501. doi: 10.1038/ismej.2011.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kalia V.C., Wood T.K., Kumar P. Evolution of Resistance to Quorum-Sensing Inhibitors. Microb. Ecol. 2014;68:13–23. doi: 10.1007/s00248-013-0316-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pereira C.S., Thompson J.A., Xavier K.B. AI-2-mediated signalling in bacteria. FEMS Microbiol. Rev. 2013;37:156–181. doi: 10.1111/j.1574-6976.2012.00345.x. [DOI] [PubMed] [Google Scholar]
- 8.Lee J., Jayaraman A., Wood T.K. Indole is an inter-species biofilm signal mediated by SdiA. BMC Microbiol. 2007;7:42. doi: 10.1186/1471-2180-7-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee J., Zhang X.-S., Hegde M., Bentley W.E., Jayaraman A., Wood T.K. Indole cell signaling occurs primarily at low temperatures in Escherichia coli. ISME J. 2008;2:1007–1023. doi: 10.1038/ismej.2008.54. [DOI] [PubMed] [Google Scholar]
- 10.Lee J.-H., Wood T.K., Lee J. Roles of indole as an interspecies and interkingdom signaling molecule. Trends Microbiol. 2015;23:707–718. doi: 10.1016/j.tim.2015.08.001. [DOI] [PubMed] [Google Scholar]
- 11.Barrios A.F.G., Zuo R., Hashimoto Y., Yang L., Bentley W.E., Wood T.K. Autoinducer 2 Controls Biofilm Formation in Escherichia coli through a Novel Motility Quorum-Sensing Regulator (MqsR, B3022) J. Bacteriol. 2006;188:305–316. doi: 10.1128/JB.188.1.305-316.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen X., Schauder S., Potier N., Van Dorsselaer A., Pelczer I., Bassler B.L., Hughson F.M. Structural identification of a bacterial quorum-sensing signal containing boron. Nat. Cell Biol. 2002;415:545–549. doi: 10.1038/415545a. [DOI] [PubMed] [Google Scholar]
- 13.Soares J.A., Ahmer B.M.M. Detection of acyl-homoserine lactones by Escherichia and Salmonella. Curr. Opin. Microbiol. 2011;14:188–193. doi: 10.1016/j.mib.2011.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herzberg M., Kaye I.K., Peti W., Wood T.K. YdgG (TqsA) controls biofilm formation in Escherichia coli K-12 by enhancing autoinducer 2 transport. J. Bacteriol. 2006;188:587–598. doi: 10.1128/JB.188.2.587-598.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang L., Li S., Liu X., Wang Z., Jiang M., Wang R., Xie L., Liu Q., Xie X., Shang D., et al. Sensing of autoinducer-2 by functionally distinct receptors in prokaryotes. Nat. Commun. 2020;11:1–13. doi: 10.1038/s41467-020-19243-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee J., Bansal T., Jayaraman A., Bentley W.E., Wood T.K. Enterohemorrhagic Escherichia coli Biofilms Are Inhibited by 7-Hydroxyindole and Stimulated by Isatin. Appl. Environ. Microbiol. 2007;73:4100–4109. doi: 10.1128/AEM.00360-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Domka J., Lee J., Wood T.K. YliH (BssR) and YceP (BssS) Regulate Escherichia coli K-12 Biofilm Formation by Influencing Cell Signaling. Appl. Environ. Microbiol. 2006;72:2449–2459. doi: 10.1128/AEM.72.4.2449-2459.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davies D.G., Parsek M.R., Pearson J.P., Iglewski B.H., Costerton J.W., Greenberg E.P. The Involvement of Cell-to-Cell Signals in the Development of a Bacterial Biofilm. Science. 1998;280:295–298. doi: 10.1126/science.280.5361.295. [DOI] [PubMed] [Google Scholar]
- 19.Hammer B.K., Bassler B.L. Quorum sensing controls biofilm formation in Vibrio cholerae. Mol. Microbiol. 2003;50:101–104. doi: 10.1046/j.1365-2958.2003.03688.x. [DOI] [PubMed] [Google Scholar]
- 20.Labbate M., Queck S.Y., Koh K.S., Rice S.A., Givskov M., Kjelleberg S. Quorum Sensing-Controlled Biofilm Development in Serratia liquefaciens MG1. J. Bacteriol. 2004;186:692–698. doi: 10.1128/JB.186.3.692-698.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Y.-H., Lau P.C.Y., Lee J.H., Ellen R.P., Cvitkovitch D.G. Natural Genetic Transformation of Streptococcus mutans Growing in Biofilms. J. Bacteriol. 2001;183:897–908. doi: 10.1128/JB.183.3.897-908.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ren D., Sims J.J., Wood T.K. Inhibition of biofilm formation and swarming of Escherichia coli by (5Z)-4-bromo-5-(bromomethylene)-3-butyl-2(5H)-furanone. Environ. Microbiol. 2001;3:731–736. doi: 10.1046/j.1462-2920.2001.00249.x. [DOI] [PubMed] [Google Scholar]
- 23.McNab R., Ford S.K., El-Sabaeny A., Barbieri B., Cook G.S., Lamont R.J., Dietrich G., Kurz S., Hübner C., Aepinus C., et al. LuxS-Based Signaling in Streptococcus gordonii: Autoinducer 2 Controls Carbohydrate Metabolism and Biofilm Formation with Porphyromonas gingivalis. J. Bacteriol. 2003;185:274–284. doi: 10.1128/JB.185.1.274-284.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Balestrino D., Haagensen J.A.J., Rich C., Forestier C. Characterization of Type 2 Quorum Sensing in Klebsiella pneumoniae and Relationship with Biofilm Formation. J. Bacteriol. 2005;187:2870–2880. doi: 10.1128/JB.187.8.2870-2880.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cole S.P., Hardwood J., Lee R., She R., Guiney D.G. Characterization of monoespecies biofilm formation by Helicobacter pylori. J. Bacteriol. 2004;186:3124–3132. doi: 10.1128/JB.186.10.3124-3132.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laganenka L., Colin R., Sourjik V. Chemotaxis towards autoinducer 2 mediates autoaggregation in Escherichia coli. Nat. Commun. 2016;7:12984. doi: 10.1038/ncomms12984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li J., Attila C., Wang L., Wood T.K., Valdes J.J., Bentley W.E. Quorum Sensing in Escherichia coli Is Signaled by AI-2/LsrR: Effects on Small RNA and Biofilm Architecture. J. Bacteriol. 2007;189:6011–6020. doi: 10.1128/JB.00014-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ren D., Bedzyk L.A., Ye R.W., Thomas S.M., Wood T.K. Differential gene expression shows natural brominated furanones interfere with the autoinducer-2 bacterial signaling system of Escherichia coli. Biotechnol. Bioeng. 2004;88:630–642. doi: 10.1002/bit.20259. [DOI] [PubMed] [Google Scholar]
- 29.Bansal T., Jesudhasan P., Pillai S., Wood T.K., Jayaraman A. Temporal regulation of enterohemorrhagic Escherichia coli virulence mediated by autoinducer-2. Appl. Microbiol. Biotechnol. 2008;78:811–819. doi: 10.1007/s00253-008-1359-8. [DOI] [PubMed] [Google Scholar]
- 30.Englert D.L., Manson M.D., Jayaraman A. Flow-Based Microfluidic Device for Quantifying Bacterial Chemotaxis in Stable, Competing Gradients. Appl. Environ. Microbiol. 2009;75:4557–4564. doi: 10.1128/AEM.02952-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bansal T., Englert D., Lee J., Hegde M., Wood T.K., Jayaraman A. Differential Effects of Epinephrine, Norepinephrine, and Indole on Escherichia coli O157:H7 Chemotaxis, Colonization, and Gene Expression. Infect. Immun. 2007;75:4597–4607. doi: 10.1128/IAI.00630-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hegde M., Englert D.L., Schrock S., Cohn W.B., Vogt C., Wood T.K., Manson M.D., Jayaraman A. Chemotaxis to the quorum-sensing signal AI-2 requires the Tsr chemo-receptor and the periplasmic LsrB AI-2-binding protein. J. Bacteriol. 2011;193:768–773. doi: 10.1128/JB.01196-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jani S., Seely A.L., Jayaraman A., Manson M.D. Chemotaxis to self-generated AI-2 promotes biofilm formation in Escherichia coli. Microbiology. 2017;163:1778–1790. doi: 10.1099/mic.0.000567. [DOI] [PubMed] [Google Scholar]
- 34.Kumar A., Sperandio V. Indole Signaling at the Host-Microbiota-Pathogen Interface. mBio. 2019;10:e01031-19. doi: 10.1128/mBio.01031-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wood T.K., Lee J. Precedence for the Role of Indole with Pathogens. mBio. 2019;10:e01599-19. doi: 10.1128/mBio.01599-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee J., Attila C., Cirillo S.L.G., Cirillo J.D., Wood T.K. Indole and 7-hydroxyindole diminish Pseudomonas aeruginosa virulence. Microb. Biotechnol. 2008;2:75–90. doi: 10.1111/j.1751-7915.2008.00061.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang W., Yamasaki R., Song S., Wood T.K. Interkingdom signal indole inhibits Pseudomonas aeruginosa persister cell waking. J. Appl. Microbiol. 2019;127:1768–1775. doi: 10.1111/jam.14434. [DOI] [PubMed] [Google Scholar]
- 38.Wood T.K., Song S. Forming and waking dormant cells: The ppGpp ribosome dimerization persister model. Biofilm. 2020;2:100018. doi: 10.1016/j.bioflm.2019.100018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bansal T., Alaniz R.C., Wood T.K., Jayaraman A. The bacterial signal indole increases epithelial-cell tight-junction resistance and attenuates indicators of inflammation. Proc. Natl. Acad. Sci. USA. 2009;107:228–233. doi: 10.1073/pnas.0906112107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Song S., Gong T., Yamasaki R., Kim J., Wood T.K. Identification of a potent indigoid persister antimicrobial by screening dormant cells. Biotechnol. Bioeng. 2019;116:2263–2274. doi: 10.1002/bit.27078. [DOI] [PubMed] [Google Scholar]
- 41.Song S., Wood T.K. Combatting persister cells with substituted indoles. Front. Microbiol. 2020;11:1565. doi: 10.3389/fmicb.2020.01565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Defoirdt T. Can bacteria actively search to join groups? ISME J. 2010;5:569–570. doi: 10.1038/ismej.2010.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang X.-S., Garciía-Contreras R., Wood T.K. YcfR (BhsA) Influences Escherichia coli Biofilm Formation through Stress Response and Surface Hydrophobicity. J. Bacteriol. 2007;189:3051–3062. doi: 10.1128/JB.01832-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu H., Tsao C., Quan D.N., Cheng Y., Servinsky M.D., Carter K.K., Jee K.J., Terrell J.L., Zargar A., Rubloff G.W., et al. Autonomous bacterial localization and gene expression based on nearby cell receptor density. Mol. Syst. Biol. 2013;9:636. doi: 10.1038/msb.2012.71. [DOI] [PMC free article] [PubMed] [Google Scholar]