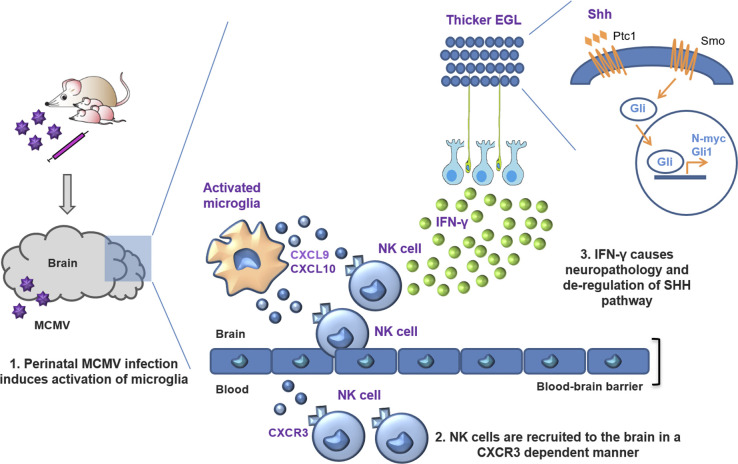
Using a mouse model of congenital cytomegalovirus infection, evidence is provided for a pathogenic role of NK and ILC1 cells in the brain. Although brain-infiltrating innate immune cells fail to control infection, they orchestrate pathological inflammatory responses that lead to altered cerebellar development.
Abstract
Congenital human cytomegalovirus (cHCMV) infection of the brain is associated with a wide range of neurocognitive sequelae. Using infection of newborn mice with mouse cytomegalovirus (MCMV) as a reliable model that recapitulates many aspects of cHCMV infection, including disseminated infection, CNS infection, altered neurodevelopment, and sensorineural hearing loss, we have previously shown that mitigation of inflammation prevented alterations in cerebellar development, suggesting that host inflammatory factors are key drivers of neurodevelopmental defects. Here, we show that MCMV infection causes a dramatic increase in the expression of the microglia-derived chemokines CXCL9/CXCL10, which recruit NK and ILC1 cells into the brain in a CXCR3-dependent manner. Surprisingly, brain-infiltrating innate immune cells not only were unable to control virus infection in the brain but also orchestrated pathological inflammatory responses, which lead to delays in cerebellar morphogenesis. Our results identify NK and ILC1 cells as the major mediators of immunopathology in response to virus infection in the developing CNS, which can be prevented by anti–IFN-γ antibodies.
Graphical Abstract
Introduction
A large amount of data on models of neuroinflammation induced by viruses have been accumulated (Klein et al., 2019; Klein and Hunter, 2017). Most of these models were developed to study acute central nervous system (CNS) infections that follow peripheral infections with neurotropic viruses in adults and were not intended to model infection of the developing nervous system. Human CMV (HCMV) is the most common causative agent of congenital viral infections in humans that may lead to long-term CNS abnormalities such as hearing and vision impairments, epilepsy, and disorders of motor and cognitive functions (Boppana et al., 2013). Despite fundamental advancements in clinical medicine, investigations of CNS infection in infants with congenital HCMV (cHCMV) infection have been limited to observational studies, and very little improvement has been made regarding prevention and treatment. A major obstacle in the study of the pathogenesis of HCMV infection is the strict species specificity exhibited by the virus, which restricts the scope of studies of neuropathogenesis of cHCMV infection to animal models (Reddehase and Lemmermann, 2018). Mouse CMV (MCMV), the most commonly used animal model in studies of HCMV infection, cannot pass through the placenta and thus cannot cause intrauterine infection (Britt et al., 2013). To circumvent this obstacle, we previously developed an experimental model of a cHCMV infection by infecting newborn mice with MCMV via an i.p. route (Koontz et al., 2008). Neurodevelopmentally, the newborn mouse closely parallels that of an early human second-trimester fetus (Clancy et al., 2001), and this model recapitulates several features of CNS infection in second-trimester human fetuses, including the route of viral neuroinvasion and neuropathological findings. Following i.p. inoculation of newborn mice with MCMV, the virus disseminates to the CNS via blood, replicates in the brain parenchyma, and causes altered cerebellar development, most evident in the smaller size of the cerebellum and thicker external granular layer (EGL; Koontz et al., 2008). In addition, MCMV infection in newborn mice induces a strong inflammatory response that deregulates brain homeostasis, leading to the activation of microglia and influx of innate immune cells (Bantug et al., 2008; Brizić et al., 2019; Brizić et al., 2018b). These immune cells can further produce proinflammatory cytokines, such as IFN-γ and TNF-α, which can then exacerbate cerebellar developmental problems (Seleme et al., 2017). This is underscored by the finding that the treatment of infected animals with glucocorticoids or blocking of TNF-α attenuates neuroinflammation and limits deficits in cerebellar development (Kosmac et al., 2013; Seleme et al., 2017), indicating that host inflammatory responses to MCMV infection, rather than the cytopathic effect of virus on infected cells, are important drivers of deficits in cerebellar development. The exact mechanisms and critical components involved are, however, largely unknown.
Group 1 innate lymphoid cells (ILCs) respond to intracellular pathogens such as viruses, intracellular bacteria, and certain parasites. They comprise natural killer (NK) cells and type 1 ILCs (ILC1s; Vivier et al., 2018). NK cells rapidly respond to infections by producing cytokines such as IFN-γ or exhibiting cytotoxicity, whereas ILC1s produce IFN-γ but are generally noncytotoxic. It is generally accepted that ILC1s are tissue-resident cells, whereas NK cells circulate in the bloodstream. Here, we show that activated newborn NK and ILC1 cells mediate immunopathology instead of controlling the infection and limiting tissue damage. Immunopathology is preceded by activation of microglia and production of CXCL9 and CXCL10, chemokines that recruit NK/ILC1 cells to the brain of MCMV-infected newborns in a CXCR3-dependent manner. IFN-γ released by highly activated, brain-infiltrating NK/ILC1 cells is a major contributing factor to the altered cerebellar development. This study is the first to demonstrate the immune-pathogenic action of immature newborn NK/ILC1 cells in the brain. Thus, this paper provides an important contribution to understanding the pathogenesis of cHCMV infection, which can be harnessed to design novel therapeutic targets.
Results and discussion
MCMV infection of newborn mice induces early activation of microglia and causes substantial alterations of their transcriptional landscape
Microglia are the only brain-resident immune cells and are thus the first responders to a virus infection of the CNS (Goldmann et al., 2016; Rock et al., 2004). Activation of microglia and infiltration of CD45hi leukocytes in the brain are considered as markers of neuroinflammation (Prinz and Priller, 2017; Streit et al., 2004). We have previously described that perinatal MCMV infection leads to infiltration of various leukocyte populations into the brain and activation of microglia, as indicated by up-regulation of MHC-II expression (Bantug et al., 2008; Kosmac et al., 2013).
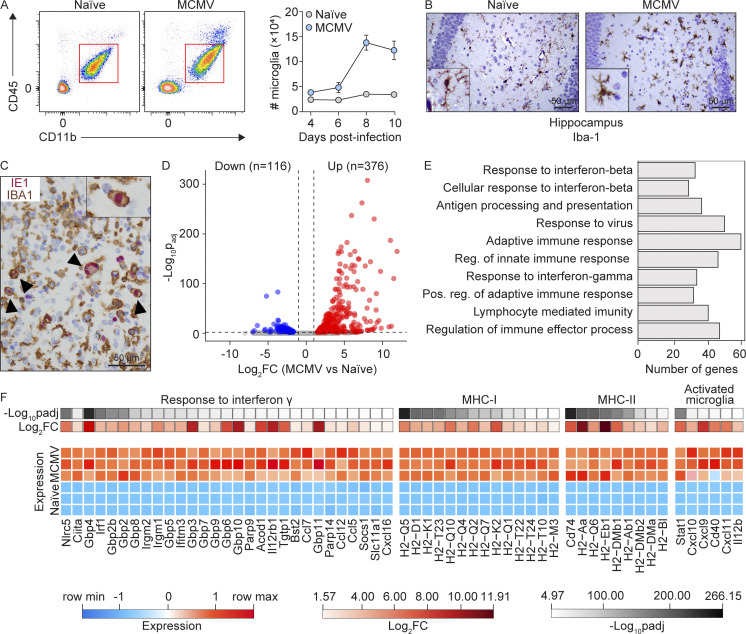
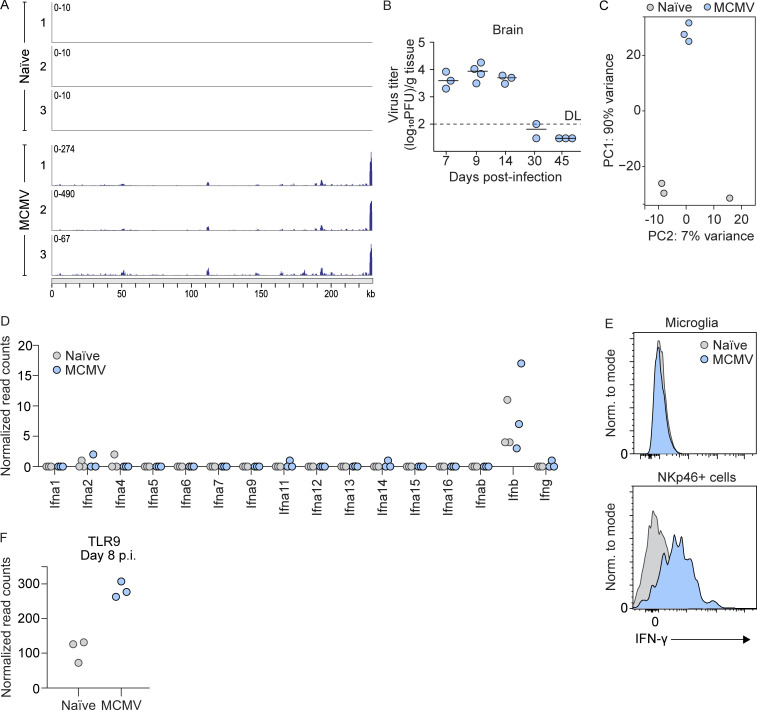
To investigate microglia behavior during MCMV infection, we i.p. infected newborn BALB/c mice and monitored the number of microglia during the infection (Fig. 1 A). In agreement with previous findings, we observed that perinatal MCMV infection leads to increased numbers of microglia starting from day 8 postinfection (p.i.; Fig. 1 A). In addition, brain sections from naive and MCMV-infected animals were stained with mAbs to ionized calcium-binding adaptor protein 1 (IBA-1), an accepted microglia marker (Fig. 1 B). The number of IBA-1+ cells was significantly increased following infection. Using immunohistochemistry, we showed that the proportion of microglia also expresses MCMV IE1 protein, confirming that microglia can be infected in vivo (Fig. 1 C). Moreover, a proportion of RNA sequencing (RNA-seq) reads, obtained from microglia of infected mice, mapped to the MCMV genome with a pattern consistent with the previously published transcriptomic profile of MCMV infection (Juranic Lisnic et al., 2013; Fig. S1 A), confirming that active viral transcription occurs in the microglia of infected mice.
Figure 1.
MCMV infection causes substantial alterations in the microglia transcriptome. Newborn BALB/c mice were infected with MCMV. (A) Gating strategy used to define microglia (left). Kinetics of microglia in the brain was analyzed by flow cytometry at indicated time points p.i. (right). Mean values ± SEM are shown (n = 3–6). The data are representative of three independent experiments. (B) IBA-1+ microglia in hippocampus from MCMV-infected and naive mice (n = 5). Scale bars, 50 µm. The data are representative of two independent experiments. (C) Infected microglia were identified with dual staining for IBA-1 (brown) and the intranuclear viral IE1 protein (red). Counterstaining was performed with hematoxylin. Arrowheads point to infected microglia. Scale bar, 50 µm (n = 5). The data are representative of two independent experiments. (D) Volcano plot of DE genes in microglia from MCMV-infected versus microglia from naive mice (n = 3). (E) Gene Ontology overrepresentation analysis showing the top 10 perturbed biological pathways in microglia from infected mice, ranked by padj. padj < 0.01 (n = 3). (F) Heatmap of DE IFN-γ response genes, MHC-I and MHC-II genes, and markers of activated microglia from naive and MCMV-infected mice (n = 3).
Figure S1.
MCMV infection of microglia and immune response in brain. (A) RNA reads were obtained in RNA-seq experiments of microglia sorted from naive or MCMV-infected newborn mice at day 8 p.i. and were mapped to the MCMV genome (n = 3). (B) Newborn BALB/c mice were infected with MCMV. Brains were collected at the indicated time points p.i., and viral titers were determined. Titers in brain of individual mice are shown (circles); horizontal bars indicate the median values; DL, detection limit (n = 3–4). The data are representative of two independent experiments. (C) Principal-component (PC) analysis of RNA-seq data shown in Fig. 1 and Fig. S1. (D) Expression levels of genes encoding type I and type II IFNs in microglia from RNA-seq data (n = 3). (E) Results of intracellular flow cytometric analysis of IFN-γ production by microglia and NKp46+ cells at day 8 p.i. (n = 3 replicates, with four brains pooled per replicate). The data are representative of two independent experiments. (F) Expression level of TLR9 in microglia from RNA-seq data (n = 3).
To gain more insight into the impact of MCMV infection on the transcriptional program of microglia at day 8 p.i., when virus titers reach their peak in the CNS (Fig. S1 B), we isolated microglia from whole brains of naive and MCMV-infected newborn mice and performed RNA-seq. The principal-component analysis showed clear separation of microglia transcriptomes from MCMV-infected versus naive animals (Fig. S1 C), while analysis of differentially expressed (DE) genes revealed that MCMV infection causes significant changes in the expression of 492 genes in microglial cells (log2FC > 1, padj [adjusted P value] < 0.01; Fig. 1 D). Further analysis revealed that among the 10 most significant biological processes affected by MCMV infection were the adaptive and innate immune responses and responses to type I and type II IFNs (Fig. 1 E). Notably, in addition to genes encoding MHC-I and MHC-II and their transcriptional activators, Nlrc5 and Ciita, among the most strongly up-regulated genes, were the markers of activated microglia, including CXCR3-binding chemotactic chemokines Cxcl9, Cxcl10, and Cxcl11 (Fig. 1 F). Indeed, querying the list of DE genes against the Interferome database (Rusinova et al., 2013) suggested that the dysregulation of more than 80% of the genes in microglia during MCMV infection is a consequence of their response to either type I or type II IFNs (Fig. 1 F). Although it was previously reported that microglia produce IFN-γ in response to infection with Toxoplasma gondii (Wang and Suzuki, 2007) or in response to IL-12 and/or IL-18 (Kawanokuchi et al., 2006), we did not detect any significant amounts of either IFN-γ transcripts (Fig. S1 D) or protein products in microglia at day 8 p.i., thus indicating that some other immune cells are the source of IFN-γ (Fig. S1 E). In summary, alterations of the microglial transcriptome in response to MCMV infection are predominantly a consequence of type I and type II IFN signaling.
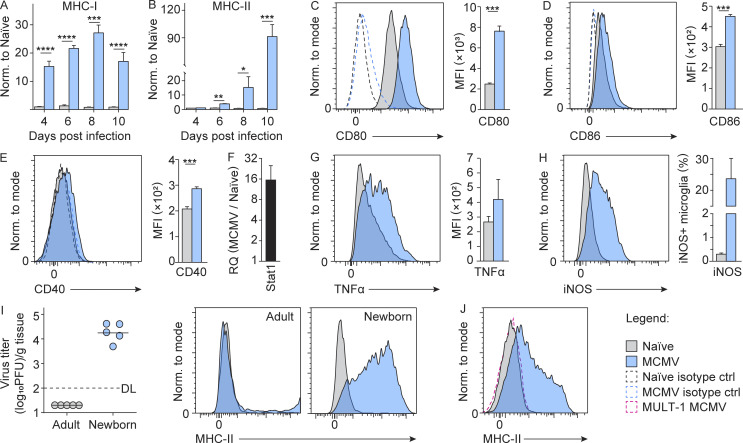
To further validate the impact of MCMV infection on microglia early after infection, we infected newborn BALB/c mice and monitored the activation status of microglia. While surface expression of MHC-I and MHC-II was hardly detectable on the microglia of naive mice, MCMV infection resulted in their significant up-regulation (Fig. 2, A and B). In addition, we observed up-regulated costimulatory molecules CD80, CD86, and CD40 (Fig. 2, C–E) as well as STAT1 expression (Fig. 2 F) following MCMV infection as well as increased production of TNF-α (Fig. 2 G) and inducible nitric oxide synthase (iNOS; Fig. 2 H). Taken together, these data demonstrate that MCMV infection of newborn mice leads to the activation of microglia.
Figure 2.
MCMV infection induces early activation of microglia toward proinflammatory phenotype. (A and B) Newborn BALB/c mice were infected with MCMV. The expression of MHC-I (A) and MHC-II (B) on microglia was analyzed by flow cytometry at indicated time points p.i. Mean values ± SEM are shown (n = 3–6). The data are representative of two or three independent experiments. Unpaired two-tailed Student’s test was used. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P ≤ 0.0001. (C–E) The expression of CD80 (C), CD86 (D), and CD40 (E) on microglia was analyzed by flow cytometry on day 8 p.i. Mean values ± SEM are shown (n = 5). The data are representative of two independent experiments. Unpaired two-tailed Student’s test was used. ***, P < 0.001. Representative histograms showing expression of CD80, CD86, and CD40 are shown (C–E). (F) The expression of Stat1 was determined by qPCR on day 8 p.i. (n = 3). The data are representative of two independent experiments. (G and H) TNF-α (G) and iNOS (H) production on day 8 p.i. was analyzed by flow cytometry (n = 3 replicates, with three brains pooled per replicate). The data are representative of two independent experiments. Unpaired two-tailed Student’s test was used. (I) Newborn and adult BALB/c mice were infected with MCMV. Brains were harvested on day 8 p.i., and the virus titers were determined. Titers in organs of individual mice are shown (circles); horizontal bars indicate the median values; DL, detection limit (left). Representative histograms showing expression of MHC-II on microglia are shown (right; n = 5). The data are representative of two independent experiments. (J) Postnatal day 1 C57BL/6 mice were infected with wild-type MCMV or MULT-1MCMV. Representative histograms showing the expression of MHC-II on microglia are shown (n = 3–5). The data are representative of two independent experiments. MFI, mean fluorescence intensity.
Although the pathogenesis of cHCMV infection has been the subject of intensive studies, very little is known about early events that initiate intrinsic and subsequent innate immune response in the developing brain. We hypothesized that infection of microglia could be the main trigger for initiating the immune response. TLR9 has been shown to be the key sensor of MCMV infection (Brinkmann et al., 2015; Krug et al., 2004). TLR9 induces the proliferation of microglia and the production of TNF-α, IL-6, nitric oxide, and IL-10 (Fiebich et al., 2018). In line with these studies, we also found that microglia derived from MCMV-infected newborn mice shows up-regulation of TLR9 (Fig. S1 F) as well as of IL12p40 transcript (Fig. 1 F). Studies in humans using antivirals have shown that treatment in the postnatal period can reduce virus shedding and improve neurodevelopmental outcomes, suggesting that virus replication is likely required for CNS disease associated with cHCMV infection (Kimberlin et al., 2015; Oliver et al., 2009). To determine whether viral replication in the brain is required for microglial activation, we compared surface levels of MHC-II on microglia from mice infected as newborns with mice infected as adults. In contrast to infection of newborn mice, the virus does not reach the brain in adult mice (Krmpotic et al., 2003), and, consequently, no up-regulation of MHC-II was detected (Fig. 2 I). The same was observed after infection of newborn mice with MULT-1MCMV (Fig. 2 J), an attenuated virus that cannot reach the brain, even in newborns (Brizić et al., 2018b). Altogether, viral replication in the brain is required for the activation of microglia toward a proinflammatory phenotype.
Perinatal MCMV infection causes infiltration of highly activated NK/ILC1 cells in brain
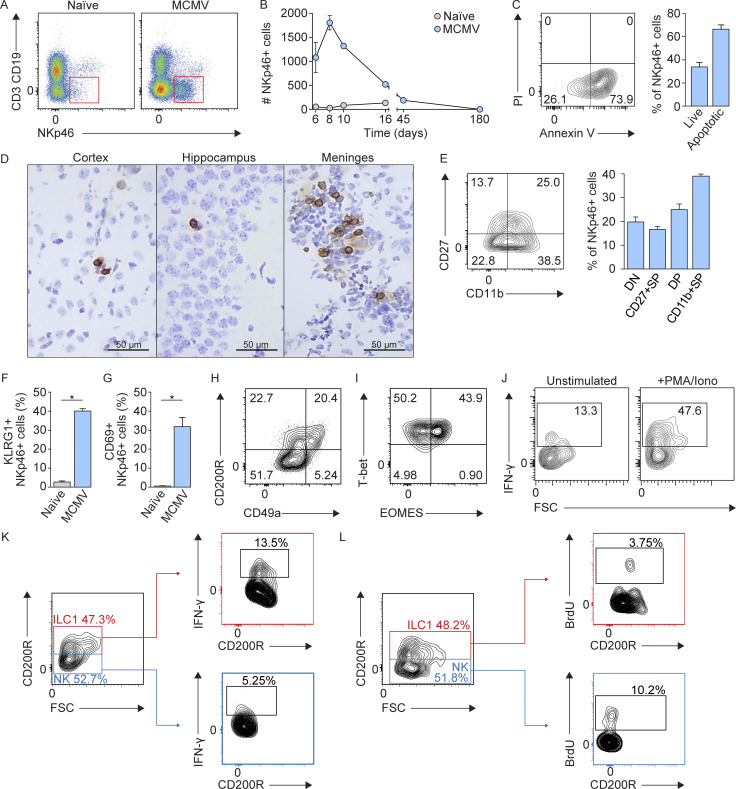
Although NK cells play an important role during MCMV infection in peripheral organs (Bubić et al., 2004; Jonjic et al., 2008; Welsh et al., 1991), their role during perinatal infection in brain has not been demonstrated. This particularly holds true for developing mice, since their NK cells are considered immature (Dorfman and Raulet, 1998). Therefore, the role of NK cells in neuroinflammation after perinatal CMV infection was hardly expected. To investigate the dynamics, phenotype, and distribution of NKp46+ cells (including NK cells and ILC1s) during neuroinflammation, we infected newborn BALB/c mice with MCMV and monitored the number of brain-infiltrating NKp46+ cells during the course of infection. While NKp46+ cells were hardly detectable in the brains of naive mice, MCMV infection resulted in their significant increase (Fig. 3 A). Moreover, kinetics of NKp46+ cell infiltration in the brain coincided with the detection of the virus (Fig. S1 B), peaked at day 8 p.i., and then declined (Fig. 3 B) as a consequence of apoptosis (Fig. 3 C). In addition, immunohistochemical staining of the brain tissue sections showed that NKp46+ cells were located in various regions of the brain at day 8 p.i. (Fig. 3 D), indicating that they leave the blood vessels and enter the brain parenchyma. We next assessed maturation and functionality of brain-infiltrating NKp46+ cells and showed that ∼40% were terminally differentiated NKp46+ cells (CD11bhighCD27low; Fig. 3 E). In addition, we observed that brain-infiltrating NKp46+ cells had up-regulated KLRG1 and CD69 (Fig. 3, F and G). To discriminate between NK cells and ILC1s, we stained CD3−CD19−NKp46+ cells with eomesodermin (EOMES), CD200R, and CD49a, markers previously used to discriminate NK cells from ILC1s in the CNS (Daussy et al., 2014; Gordon et al., 2012; Romero-Suárez et al., 2019). Approximately 40% of NKp46+ cells expressed CD200R (Fig. 3 H). In addition, the expression of EOMES was observed in ∼40% of NKp46+ cells (Fig. 3 I). These data suggest that a considerable fraction of NKp46+ cells present in the MCMV-infected brain represent ILC1s. Thus, in the further text, we refer to brain-infiltrating NKp46+ cells as NK/ILC1 cells. In addition, we observed that NKp46+ cells readily produced IFN-γ after in vitro stimulation (Fig. 3 J) and that both NK cells (CD45hiCD3−CD19−NKp46+CD200R−) and ILC1s (CD45hiCD3−CD19−NKp46+CD200R+) are a source of IFN-γ in the brain following MCMV infection of newborn mice (Fig. 3 K). Both, NK cells and ILC1s proliferate in the brain of newborn infected mice (Fig. 3 L). Even though we have used state-of-the-art markers to differentiate NK and ILC1 cells, it is possible that these markers do not discriminate between NK cells and ILC1s efficiently in newborn mice. To date, no published studies have definitively characterized the expression of NK and ILC1 cell markers in the neonatal period in the setting of inflammation. In adult mice, CD200R was used as a marker to identify ILC1 cells in infection (Weizman et al., 2017) and has thus been used in this study. However, whether CD200R is a reliable marker in the neonatal period remains undetermined. Altogether, these results show the recruitment of immature NK cells and ILC1s to the brain of MCMV-infected newborn mice, and these cells are highly activated and readily produce IFN-γ.
Figure 3.
Accumulation of NKp46+ cells in brain following MCMV infection. Newborn BALB/c mice were infected with MCMV. (A) Gating strategy used to define NKp46+ cells in the brain at day 8 p.i. (B) Kinetics of brain-infiltrating NKp46+ cells. Mean values ± SEM are shown (n = 3–5). The data are representative of two or three independent experiments. (C) Early apoptosis was analyzed using annexin V-PI staining by flow cytometry (left) at day 8 p.i. Percentages of live and apoptotic NKp46+ cells are shown (right). The data are shown as means ± SEM; n = 3 replicates, with three brains pooled per replicate. The data are representative of two independent experiments. (D) Immunohistochemical staining of NKp46+ cells in the brain of MCMV-infected mice at day 8 p.i. Scale bars, 50 µm. The data are representative of three independent experiments. (E) Analysis of NKp46+ cell maturation in the brain, as assessed by CD11b in combination with CD27. Mean values ± SEM are shown (n = 3–5). The data are representative of three independent experiments. (F and G) Expression of KLRG1 (F) and CD69 (G). Mean values ± SEM are shown (n = 3–5). The data are representative of three independent experiments. Unpaired two-tailed Student`s test was used. *, P < 0.05. (H and I) Representative contour plots of the expression of CD200R and CD49a (H) and EOMES and T-bet (I) are shown (n = 3–5). The data are representative of two independent experiments. (J) Intracellular flow cytometric analysis of IFN-γ production by brain-infiltrating NKp46+ cells (n = 3 replicates, with four brains pooled per replicate), day 8 p.i. The data are representative of three independent experiments. (K) Intracellular flow cytometric analysis of IFN-γ production by NK cells (CD45hiCD3−CD19−NKp46+CD200R−) and ILC1s (CD45hiCD3−CD19−NKp46+CD200R+; n = 2–3 replicates, with four brains pooled per replicate) at day 8 p.i. The data are representative of two independent experiments. (L) The proliferation of NK cells and ILC1s in the brain following perinatal MCMV infection (n = 5) at day 8 p.i. The data are representative of two independent experiments. FSC, forward scatter; DP, double positive; SP, single positive; Iono, Ionomycin.
NK/ILC1 cells are recruited into MCMV-infected brain in a CXCR3-dependent manner
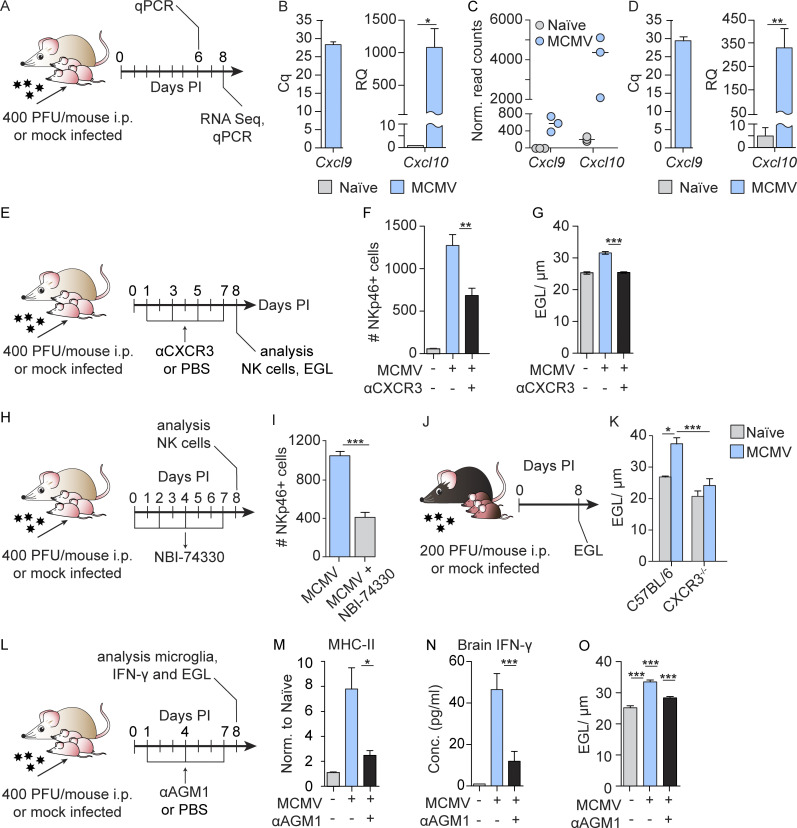
Previous studies have shown that CXCL9 and CXCL10, chemokines that mediate T and NK cell recruitment, are expressed in the brain in response to virus infection (Thapa et al., 2008; Trifilo et al., 2004). Both CXCL9 and CXCL10 selectively bind CXCR3, which is expressed on both NK cells and activated T cells (Thapa et al., 2008). Our transcriptomic data on microglia and quantitative PCR (qPCR) on cerebellar homogenates showed that expression of Cxcl9 and Cxcl10 were up-regulated following perinatal MCMV infection (Fig. 4, A–D), already at day 6 (Fig. 4 B). Thus, the early expression of Cxcl9 and Cxcl10 could explain the recruitment of NK/ILC1 cells into the brain. To address this question, we infected newborn BALB/c mice with MCMV and blocked CXCR3 in vivo by administering anti-CXCR3 antibodies (Fig. 4 E). Blocking of CXCR3 in infected mice resulted in a significantly reduced number of brain-infiltrating NKp46+ cells (Fig. 4 F). In addition, treatment of infected newborn mice with a CXCR3 antagonist (NBI-74330) resulted in a reduced number of brain-infiltrating NKp46+ cells (Fig. 4, H and I). Moreover, we have shown that in newborn mice lacking CXCR3 receptor (CXCR3−/− mice), MCMV infection resulted in reduced EGL thickness (Fig. 4, J and K). Altogether, CXCL9- and CXCL10-mediated signaling through CXCR3 is responsible for recruiting NKp46+ cells to the brain in MCMV-infected newborns.
Figure 4.
NK cells are recruited to the brain in CXCR3-dependent manner and induce early activation of microglia and altered cerebellar development. (A) Experimental scheme for B–D. Newborn BALB/c mice were infected with MCMV. (B) Expression of Cxcl9 and Cxcl10 in the cerebella measured by qPCR at day 6 p.i. Mean values ± SEM are shown (n = 3). The data are representative of two independent experiments. Unpaired two-tailed Student`s test was used. *, P < 0.05. Cq, quantification cycle; RQ, relative quantification. (C) Microglia were isolated from whole brains of naive and MCMV-infected newborn mice, and RNA-seq was performed. Expression of Cxcl9 and Cxcl10 in microglia is shown at day 8 p.i. Horizontal bars indicate the median values (n = 3). (D) Expression of Cxcl9 and Cxcl10 in the cerebella measured by qPCR at day 8 p.i. Mean values ± SEM are shown (n = 4). The data are representative of two independent experiments. Unpaired two-tailed Student’s test was used. **, P < 0.01. (E) Experimental scheme for F and G. Newborn BALB/c mice were infected with MCMV and CXCR3 was blocked in vivo by administration of anti-CXCR3 antibodies. (F and G) Number of brain-infiltrating NKp46+ cells (F) and cerebellar EGL measurements (G) are shown. Mean values ± SEM are shown (n = 3–6). The data are representative of two independent experiments. Unpaired two-tailed Student’s test was used. **, P < 0.01; ***, P < 0.001. (H) Experimental scheme for I. Newborn BALB/c mice were infected with MCMV, and CXCR3 was blocked by the administration of the CXCR3 antagonist (NBI-74330). (I) Number of brain-infiltrating NKp46+ cells. Mean values ± SEM are shown (n = 3). The data are representative of two independent experiments. Unpaired two-tailed Student’s test was used. ***, P < 0.001. (J) Experimental scheme for K. (K) Cerebellar EGL measurements from naive and MCMV-infected CXCR3−/− and C57BL/6 mice are shown (n = 6–9). The results of two pooled experiments are shown. Unpaired two-tailed Student’s test was used. *, P < 0.05; ***, P < 0.001. (L) Experimental scheme for M–O. Newborn BALB/c mice were infected with MCMV, and NK cells were depleted. (M–O) Expression of MHC-II on microglia (M), concentration of IFN-γ in brain homogenates after NK cell depletion (N), and cerebellar EGL measurements (O). Mean values ± SEM are shown (n = 3–5). The data are representative of two (N) or three (M and O) independent experiments. Unpaired two-tailed Student’s test was used. *, P < 0.05; ***, P < 0.001.
NK/ILC1 cells are responsible for the early activation of microglia and altered cerebellar development
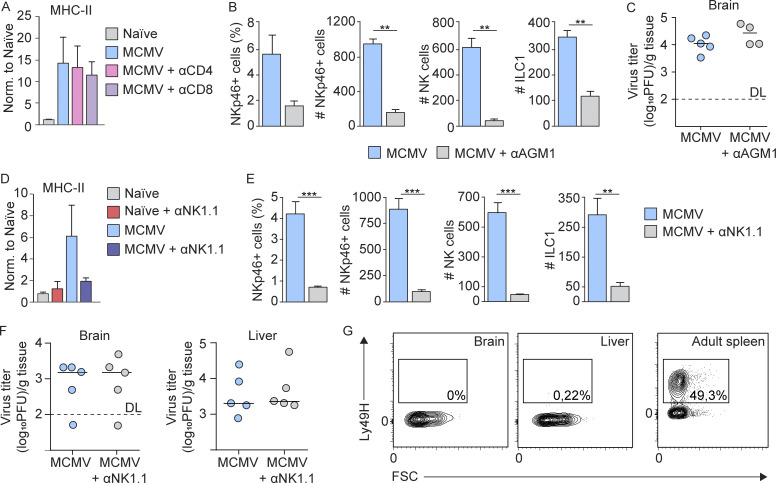
We have previously shown that the MCMV infection of newborn mice leads to impaired cerebellar development (Koontz et al., 2008) and that treatment of infected animals with glucocorticoids or anti–TNF-α antibodies attenuates neuroinflammation and limits deficits in cerebellar development (Kosmac et al., 2013; Seleme et al., 2017). These findings indicate that host inflammatory responses to MCMV infection contribute to deficits in cerebellar development in MCMV-infected mice. Although CD8 and CD4 T cells infiltrate the brain at later time points p.i. (Bantug et al., 2008), when the activation of microglia following MCMV infection is already established, to formally exclude the role of T cells in early brain inflammation, we have depleted CD8 or CD4 T cells in vivo. MCMV induced up-regulation of MHC-II on microglia was not abolished by T cell depletion (Fig. S2 A), indicating that neither CD8 nor CD4 T cells are responsible for early microglial activation. As demonstrated above, the NK/ILC1 cells infiltrated into the brain at day 6 p.i. (Fig. 3 B). To determine whether NKp46+ cells have an early role in neuroinflammation and the pathogenesis of perinatal MCMV infection of the brain, we treated MCMV-infected BALB/c mice with anti-AGM1, which depletes brain-infiltrating NK and ILC1 cells, even though NK cells were depleted more efficiently (Fig. 4 L and Fig. S2 B). Although depletion of NKp46+ cells in mice infected with MCMV did not impact the viral titer in the brain (Fig. S2 C), MCMV-induced up-regulation of MHC-II on microglia was abolished (Fig. 4 M). The same was observed after treatment of MCMV-infected C57BL/6 mice with anti-NK1.1 antibody, which depletes both NK cells and ILC1 (Fig. S2, D and E), demonstrating that this phenomenon is not strain dependent and is not a result of anti-AGM1 depletion of NKT and certain subsets of CD8 T cells (Trambley et al., 1999) or basophils (Nishikado et al., 2011). In addition, NKp46+ cell depletion in MCMV-infected newborn C57BL/6 mice did not have any effect on the virus titer (Fig. S2 F). This is not unexpected, since NK cells do not express the Ly49H receptor at that time (Fig. S2 G). In addition, we observed a decreased concentration of IFN-γ in brain homogenates after anti-AGM1 treatment, which indicates that NKp46+ cells were major producers of IFN-γ (Fig. 4 N). Finally, we demonstrated that cerebellar pathology was significantly reduced by depletion of NK/ILC1 cells (Fig. 4 O). This finding is in agreement with a recent study showing NKp46+ cells in HCMV-infected fetal brains in all severe cases but in only four of seven moderately affected ones, demonstrating a significant association between severe fetal brain damage and high levels of NK cells in the infected brain (Sellier et al., 2020). In addition, blocking of CXCR3 in MCMV-infected newborn mice (Fig. 4 E) resulted not only in a significantly reduced number of NK/ILC1 cells that infiltrate the brain (Fig. 4 F) but also in reduced EGL thickness (Fig. 4 G). Altogether, these results demonstrate that NK/ILC1 cells play a major role in early neuroinflammation and deficits in cerebellar development while being dispensable for virus control in the brain.
Figure S2.
The role of T and NK cells in microglia activation and virus control in brain. (A) CD4 and CD8 T cells are not responsible for early microglia activation. Newborn BALB/c mice were infected with MCMV, and CD4 or CD8 T cells were depleted. Expression of MHC-II on microglia is shown. Mean values ± SEM are shown (n = 4). The data are representative of two independent experiments. Unpaired two-tailed Student’s test was used. (B–G) Depletion of NK cells in mice infected with MCMV did not impact the viral titer in the brain. (B and C) Newborn BALB/c mice were infected with MCMV, and NK cells were depleted. (B) The frequency and number of NKp46+ cells and number of NK cells and ILC1 in the brain on day 8 p.i. are shown. Mean values ± SEM are shown (n = 3). The data are representative of two independent experiments. Unpaired two-tailed Student’s test was used. **, P < 0.01. (C) Brains were collected on day 8 p.i., and viral titers were determined. Titers in brain of individual mice are shown (circles); horizontal bars indicate the median values; DL, detection limit (n = 4–5). The data are representative of two independent experiments. Mann–Whitney (U) test was used. (D–G) Postnatal day 1 C57BL/6 mice were infected with MCMV, and NK cells were depleted (D–F). (D and E) Expression of MHC-II on microglia (D) and the frequency and number of NKp46+ cells and number of NK cells and ILC1s in the brain (E) on day 8 p.i. are shown. Mean values ± SEM are shown (n = 3). The data are representative of two independent experiments. Unpaired two-tailed Student’s test was used. **, P < 0.01; ***, P < 0.001. (F) Brains and liver were collected on day 8 p.i. and viral titers were determined. Titers in brain and liver of individual mice are shown (circles); horizontal bars indicate the median values; DL, detection limit (n = 5). The data are representative of two independent experiments. Mann–Whitney (U) test was used. (G) The flow cytometric analysis of Ly49H expression on NK cells in the brain and liver following perinatal MCMV infection and in the adult spleen are shown (n = 3). The data are representative of two independent experiments.
IFN-γ is a major mediator of early activation of microglia, deregulates the Sonic hedgehog (SHH) pathway, and contributes to altered cerebellar development
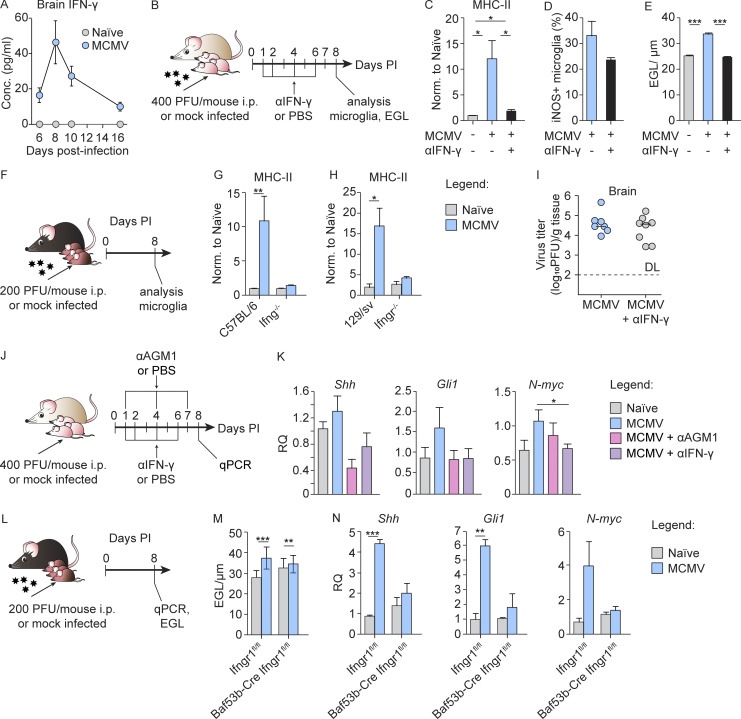
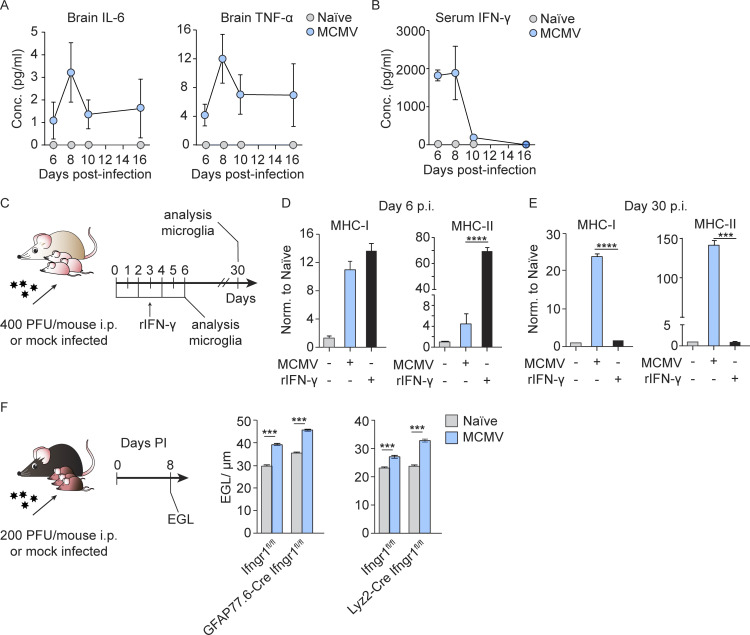
IFN-γ receptor (IFN-γR) is expressed by most CNS cell types, including microglia (Ding et al., 2015). Since high doses of IFN-γ are known to increase MHC-II expression and costimulatory molecules in microglia (Ottum et al., 2015), we measured IFN-γ in brain homogenates at different p.i. time points. The concentration of IFN-γ peaked at day 8 p.i. and then declined (Fig. 5 A). Moreover, the kinetics of IFN-γ in the brain coincided with the kinetics of NK/ILC1 cell infiltration in the brain (Fig. 3 B), and IFN-γ was not detected in the uninfected group, in which hardly any NK/ILC1 cells were detected. Similar kinetics was observed for other proinflammatory cytokines (Fig. S3 A). To determine the role of IFN-γ in the pathogenesis of congenital MCMV infection of the brain, we infected newborn BALB/c mice and neutralized IFN-γ in vivo by administration of anti–IFN-γ mAbs (Fig. 5 B). Neutralization of IFN-γ prevented the up-regulation of MHC-II on microglia observed in MCMV-infected mice (Fig. 5 C). The same effect was observed in Ifng−/− mice (Fig. 5, F and G) and Ifngr−/− mice (Fig. 5, F and H). In addition, neutralization of IFN-γ decreased production of iNOS by microglia (Fig. 5 D). Next, we determined if peripheral IFN-γ could drive microglia activation. We first determined the serum level of this cytokine in infected mice at different time points p.i. (Fig. S3 B). At early days p.i., the level of IFN-γ in the sera was high but significantly decreased after day 8 p.i. It was previously shown that IFN-γ induces proliferation and microglia activation (Ta et al., 2019). Thus, it is likely that peripheral IFN-γ contributes to early microglia activation, perhaps together with other cytokines such as TNF-α released by microglia after MCMV infection. This view is supported by results from an experiment in which naive newborn mice were injected i.p. with recombinant IFN-γ (Fig. S3 C) and high expression of MHC molecules could be detected on microglia on day 6 after treatment (Fig. S3 D). However, the expression of MHC molecules was undetectable at later time points in naive IFN-γ–treated mice, whereas in MCMV-infected mice, this polarization persisted (Fig. S3 E), supporting our conclusion that local infection is essential to maintain microglia activation (Fig. 2, I and J). Similar to depletion of NK/ILC1 cells, neutralization of IFN-γ in MCMV-infected newborn mice resulted in reduced cerebellar pathology, as observed by reduced EGL thickness (Fig. 5 E), but did not impact the viral titer in the brain (Fig. 5 I). These results indicate that IFN-γ released by highly activated NKp46+ cells leads to activation of microglia and altered cerebellar development.
Figure 5.
IFN-γ induces early activation of microglia and altered cerebellar development. (A) Newborn BALB/c mice were infected with MCMV. Brains were collected at the indicated time points p.i. The concentration of IFN-γ in brain homogenates is shown. Mean values ± SEM are show (n = 3–6). The data are representative of two independent experiments. (B) Experimental scheme for C–E and I. Newborn BALB/c mice were infected with MCMV and IFN-γ was neutralized in vivo by administration of anti–IFN-γ antibody. (C and D) Expression of MHC-II on microglia (C) and iNOS production (D) on day 8 p.i. were analyzed by flow cytometry. Mean values ± SEM are shown (n = 3–5). The data are representative of two (D) or three (C) independent experiments. Unpaired two-tailed Student’s test was used. *, P < 0.05. (E) Cerebellar EGL measurements. Mean values ± SEM are shown (n = 3–5). The data are representative of three independent experiments. Unpaired two-tailed Student’s test was used. ***, P < 0.001. (F) Experimental scheme for G and H. (G and H) Postnatal day 1 C57BL/6 and Ifng−/− (G) or 129/sv and Ifngr−/− mice (H) were infected with MCMV. Expression of MHC-II on microglia from C57BL/6 and Ifng−/− mice (G) and 129/sv and Ifngr−/− mice (H). Mean values ± SEM are shown (n = 3–5). The data are representative of two independent experiments. Unpaired two-tailed Student’s test was used. *, P < 0.05; **, P < 0.01. (I) Brains were collected on day 8 p.i., and viral titers were determined. Titers in brain of individual mice are shown (circles); horizontal bars indicate the median values; DL, detection limit (n = 5–8). The data are representative of three independent experiments. Mann–Whitney (U) test was used. (J) Experimental scheme for K. Newborn BALB/c mice were infected with MCMV. NK cells were depleted in vivo. IFN-γ was neutralized in vivo by administration of anti–IFN-γ antibody. (K) Expression of Shh, N-myc, and Gli1 in the cerebella measured by qPCR. Mean values ± SEM are show (n = 3–5). The data are representative of two independent experiments. Unpaired two-tailed Student’s test was used. *, P < 0.05. RQ (relative quantification). (L) Experimental scheme for M and N. Postnatal day 1 Baf53b-CreIfngr1flox/flox and Ifngr1flox/flox mice were infected with MCMV. (M) Cerebellar EGL measurements (N) expression of Shh, N-myc, and Gli1 in the cerebella measured by qPCR. Mean values ± SEM are shown (n = 3–5). The data are representative of two independent experiments. Unpaired two-tailed Student’s test was used. **, P < 0.01; ***, P < 0.001.
Figure S3.
The level and role of cytokines in brain of MCMV infected newborn mice. Newborn BALB/c mice were infected with MCMV or left uninfected. Brains and sera were collected at the indicated time points p.i. and concentration of proinflammatory cytokines was determined. (A and B) Concentration of proinflammatory cytokines in the brain homogenates (A) and sera (B) was determined by CBA kit. Mean values ± SEM are show (n = 3–6). The data are representative of two independent experiments. (C–E) Peripheral IFN-γ can induce the expression of MHC-I and MHC-II on microglia at early days p.i., but the infection is needed for long-term microglia activation. (C) Experimental scheme for D and E. Newborn BALB/c mice were infected with MCMV or left uninfected. Proportion of naive mice was treated with recombinant IFN-γ for 6 d. (D and E) Brain tissue was harvested, and the expression of MHC I and MHC-II on microglia was analyzed on day 6 p.i. (D) and day 30 p.i. (E). Mean values ± SEM are shown (n = 3–5). The data are representative of two independent experiments. Unpaired two-tailed Student’s test was used. ***, P < 0.001; ****, P ≤ 0.0001. (F) Effect of IFN-γ on GFAP and Lyz2-positive cells is not responsible for delay in EGL maturation. Postnatal day 1 GFAP77.6-CreIfngr1flox/flox and Ifngr1flox/flox or Lyz2-CreIfngr1flox/flox and Ifngr1flox/flox mice were infected with MCMV. Cerebellar EGL measurements are shown. Mean values ± SEM are shown (n = 3–5). The data are representative of two independent experiments. Unpaired two-tailed Student’s test was used. ***, P < 0.001.
Cerebellar development in mice initiates during embryonal development and continues postnatally. At birth, EGL granule neuron progenitor cells (GNPCs) actively proliferate and migrate inward through Bergman glia processes and past Purkinje cell layer to form the inner granular layer (Butts et al., 2014; Sathyanesan et al., 2019). In the developing cerebellum, the SHH is the major mitogen that promotes GNPC proliferation (De Luca et al., 2016; Wechsler-Reya and Scott, 1999). We have previously shown that the SHH pathway is up-regulated in the cerebellum following MCMV infection of newborn mice (Kosmac et al., 2013; Seleme et al., 2017). In addition, Wang et al. (2004) have shown that IFN-γ affects the proliferation of GNPCs in the EGL by activating SHH signaling, which leads to the persistence of the EGL. As shown above, IFN-γ is a major mediator of altered cerebellar development following MCMV infection in newborn mice. Therefore, we tested whether the increased concentration of IFN-γ p.i. affects the proliferation of GNPC via the SHH pathway. To that aim, we performed RT-qPCR analysis from cerebellar tissue lysates and evaluated the expression of Shh, N-myc, and Gli1, three genes in the SHH pathway critical for GNPC proliferation (Wallace, 1999; Fig. 5 J). The expression of Shh, N-myc, and Gli1 was increased by MCMV infection, while neutralization of IFN-γ reduced the transcription of these genes to levels comparable to those in naive mice (Fig. 5 K). Similar to neutralization of IFN-γ, depletion of NK/ILC1 cells in MCMV-infected newborn mice reduced the transcription of Shh, N-myc, and Gli1 to levels comparable to those in naive mice (Fig. 5 K). These results demonstrate that IFN-γ released by highly activated NKp46+ cells deregulates the SHH pathway in the cerebellum.
Next, we assessed on which cell type in cerebellum IFN-γ has a direct effect. By selective removal of IFN-γR1 from neurons (BAF53b-Cre Ifngr1flox/flox mice), astrocytes (GFAP77.6-Cre Ifngr1flox/flox mice), or Lyz2+ cells including microglia (Lyz2-Cre Ifngr1flox/flox mice), we showed that neurons are the main targets of IFN-γ–induced altered EGL development following MCMV infection (Fig. 5, L and M; and Fig. S3 F). Although removal of IFN-γR1 from neurons only moderately corrects EGL thickness (Fig. 5, L and M), this correction was absent after removal of IFN-γR1 from astrocytes or microglia (Fig. S3 F). These results demonstrate that neurons are the main targets of IFN-γ–induced altered cerebellar development following MCMV infection. In addition, we performed RT-qPCR from cerebellar tissue lysates from BAF53b-Cre Ifngr1flox/flox mice and evaluated the expression of Shh, N-myc, and Gli1. Consistent with the results provided after depletion of NK/ILC1 cells or blocking of IFN-γ, neuron-specific absence of IFN-γR in MCMV-infected newborn mice results in a decreased expression of not only Shh but also N-myc and Gli1 (Fig. 5 N). Thus, the data indicate that IFN-γ interferes with the proliferation of GNPCs in the EGL by activating SHH signaling, which leads to the prolonged persistence of the EGL. However, it is important to note that our previous findings in this model have demonstrated that cerebella of infected mice are hypoplastic and that GNPCs in the EGL have decreased proliferation as compared with naive animals (Koontz et al., 2008; Kosmac et al., 2013; Seleme et al., 2017). Thus, the persistence of the EGL in infected animals cannot be linked only to the up-regulation of the SHH pathway. Further studies will be required to precisely define the contribution of dysregulation of this developmental pathway and altered cerebellar development in our model. Lastly, neutralization of IFN-γ did not affect virus titer in the brain but prevented altered cerebellar development, raising the possibility that IFN-γ signaling could be used as novel therapeutic target.
In conclusion, this study provides fundamentally new insight into the role of NK/ILC1 cells in neuroinflammation and pathogenesis of cHCMV infection in the developing brain in a model of perinatal MCMV infection. Apart from its role in microglia polarization, IFN-γ secreted by newborn NK/ILC1 cells could exert a detrimental impact on cerebellar development, which could be abrogated by depletion of IFN-γ. Thus, our study represents an important contribution for a better understanding of the pathogenesis of congenital viral infections.
Materials and methods
Mouse strains
Mice were strictly age matched within experiments and handled in accordance with institutional and national guidelines. All mice were housed and bred under specific pathogen–free condition at the animal facility of the Faculty of Medicine, University of Rijeka. Wild-type C57BL/6J (B6; strain 000664), BALB/c, 129/sv, CXCR3−/− (005796), Ifng−/− (002287), Ifngr−/− (003288), Ifngr1f (IFN-γR floxed; 025394), GFAP-Cre line 77.6 (024098), BAF53b-Cre (027826), and Lyz2-Cre (004781) mice were obtained from The Jackson Laboratory. All animal experiments were approved by the National Committee for Welfare of Animals.
Viruses
For production of virus stocks, the Smith strain of MCMV (ATCC VR-194; American Type Tissue Culture) was propagated in MEFs. Virus stocks for newborn mice were aliquoted, frozen at −80°C, and titrated on MEFs using standard procedures. Viral titers in organs were determined by standard plaque assay (Brizić et al., 2018a). Virus titers are expressed as PFUs and correspond to the amount of infectious virus per gram tissue or per organ. Newborn BALB/c pups (6–18 h postpartum) were inoculated i.p. with 400 PFU of tissue culture–passaged MCMV Smith strain. Postnatal day 1 C57BL/6J, 129/sv, Ifng−/−, Ifngr−/−, Ifngr1f, GFAP-Cre line 77.6 Ifngr1f, BAF53b-Cre Ifngr1f, Lyz2-Cre Ifngr1f, and CXCR3−/− mice were infected i.p. with 200 PFU of MCMV. Adult BALB/c mice were infected i.p. with 2 × 105 PFU of MCMV.
qPCR
RNA was extracted from mouse cerebellum using an RNA extraction kit (RNeasy Mini Kit; Qiagen), and cDNA was generated with a reverse transcription kit (iScript Reverse Transcription Supermix for RT-qPCR; Bio-Rad). The expression of mRNA was examined by qPCR with a 7500 Fast Real Time PCR machine (ABI). TaqMan assays were used to quantify the expression of Cxcl9 (Mm 00434946_m1), Cxcl10 (Mm00445235_m1), Shh (Mm00436528_m1), N-myc (Mm00476449_m1), and Gli1 (Mm00494654_m1). Relative mRNA expression was normalized by quantification of Gusb (Mm01197698_m1) or Gapdh (Mm05724508_g1) RNA in each sample.
Antibodies and flow cytometry
Antibodies for in vivo applications, including anti-CD4 (YTS191.1.2), anti-CD8 (YTS169.4.2), anti-NK1.1 (PK136), anti–IFN-γ (XMG1.2), and anti-CXCR3 (CD183), were produced by our in-house facility or purchased from BioXcell. For flow cytometry, single-cell suspensions of brain were prepared according to standard protocols. Briefly, mice were perfused with cold PBS, and each brain was collected in RPMI 1640 with 3% FCS and mechanically dissociated. A 30% Percoll/brain homogenate suspension was underlaid with 70% Percoll in PBS and then centrifuged at 1,800 rpm for 25 min. Cells in the interphase were collected for further analysis. Flow cytometric analysis were performed using anti-mouse CD45 (30-F11), CD11b (M1/70), MHC-II (M5/114.15.2), MHC-I (34–5-8S), CD3 (145-2C11), CD19 (eBio1D3 (1D3)), NKp46 (29A1.4), NK1.1 (PK136), KLRG1 (2F1), CD69 (H1.2F3), CD27 (O323), CD80 (16-10A1), CD86 (GL1), CD40 (1C10), TNF-α (MP6-XT22), IFN-γ (XMG1.2), iNOS (CXNFT), CD200R (OX110), CD49a (Hma1), T-bet (4B10), EOMES (Dan11mag), BrdU kit, propidium iodide (PI), and Annexin V from eBioscience, preceded by blocking of Fc receptors using 2.4G2 antibodies (in-house generated). To measure cytokine production by NK/ILC1 cells, cells were stimulated with IL-2 for 4 h in the presence of Brefeldin A (10 mg/ml; eBioscience). To measure cytokine and iNOS production by microglia, cells were cultured for 4 h in the presence of Brefeldin A (10 mg/ml; eBioscience). For intracellular staining, permeabilization and fixation of cells was done with the Fix/Perm kit (eBioscience). For detection of apoptotic cells, single-cell suspensions were stained with Annexin V FITC and PI (eBioscience) according to the manufacturer’s instructions. All data were acquired using a FACSAria (BD Biosciences) and analyzed using FlowJo software (Tree Star).
Cytokine measurements
To quantitatively measure protein levels of IL-6, IFN-γ, and TNF-α in brain homogenates and protein levels of IFN-γ in sera by flow cytometry, we used CBA Mouse Inflammation Kit (BD Biosciences) according to the manufacturer’s protocol. Brains were lysed in DMEM with cOmplete protease inhibitor (Roche) with a bead homogenizer.
Methods performed in vivo
NK cells in BALB/c mice were depleted in vivo by administration of 5 µl anti-AGM1 (Wako Chemicals) to MCMV-infected animals on days 1 and 4 p.i. and 10 µl anti-AGM1 to MCMV-infected animals on day 7 p.i. NK cells in C57BL/6 mice were depleted in vivo by administration of 50 µg anti-NK1.1 antibody (PK136) to MCMV-infected animals on days 1, 4, and 7 p.i. CD8 T cells were depleted in vivo by administration of 50 µg anti-CD8 antibody (YTS169.4.2) to MCMV-infected animals on days 4 and 7 p.i. CD4 T cells were depleted in vivo by administration of 50 µg anti-CD4 antibody (YTS191.1.2) to MCMV-infected animals on days 4 and 7 p.i. For neutralization of IFN-γ, 50 µg anti–IFN-γ antibody (XMG1.2) was injected to MCMV-infected animals on days 1, 2, 4, and 6 p.i. CXCR3 was blocked in vivo by administration of anti-CXCR3 antibodies (CD183) to MCMV-infected animals on days 1, 3, 5, and 7 p.i. To assess the proliferation of NK/ILC1 cells, mice were treated i.p. with 50 µg/g BrdU for 3 h before sacrifice. The CXCR3-specific antagonist NBI-74330 was used to block CXCR3-mediated signaling. Briefly, mice received 100 mg/kg/day NBI-74330, beginning from day 1 p.i., for 7 d. Newborn mice were daily injected with 104 IU of mouse recombinant IFN-γ (rIFN-γ), beginning from the day of birth, for 6 d. All listed antibodies, NBI-74330, BrdU, and rIFN-γ were dissolved in PBS and injected i.p.
Histology
Brains were fixed in 4% neutral buffered formalin and embedded in paraffin. For morphometric analyzes of cerebellum, serial sagittal sections (3 µm thick) were stained with cresyl violet. Images of cresyl violet–stained cerebellar sections were acquired by DP 71 camera mounted on an BX 51 microscope (Olympus), and morphometrical measurements were performed with CellSens Dimension software (Olympus). Thickness of the cerebellar EGL was measured at eight points along the primary cerebellar fissure. Immunohistochemical staining for microglia was performed on paraffin sections using anti-mouse Iba-1 antibody (FUJIFILM; Wako Chemicals) and anti-IE1 antibody (clone IE1.01; Center for Proteomics, Faculty of Medicine, University of Rijeka, Rijeka, Croatia). Antibody binding was visualized with peroxidase-conjugated goat anti-rabbit IgG antibody (Abcam), followed with 3,3′-diaminobenzidine as a chromogen (Dako). Slides were counterstained with Shandon hematoxylin. Immunohistochemical staining for NK/ILC1 cells was performed on frozen sections using anti-NCR1 (clone mNCR1.20) antibody (Center for Proteomics, Faculty of Medicine, University of Rijeka, Rijeka, Croatia). To minimize/eliminate the possibility that the cells examined are NK cells trafficking in the brain vasculature, mice were exhaustively perfused with cold PBS. As secondary antibody, peroxidase-conjugated goat anti-mouse (Jackson ImmunoResearch Laboratory) antibody, was used and then visualized with 3,3′-diaminobenzidine and counterstained with Shandon instant hematoxylin.
Sample preparation, quality control of isolated RNA, and RNA-seq
Mononuclear cells were isolated from whole brains of naive or MCMV-infected mice at day 8 p.i. as described previously (Cekinovic et al., 2014). Following isolation, mononuclear cells were labeled with anti-CD45 and anti-CD11b antibodies, and microglia, defined as CD45intCD11bhigh cells, were separated from the mixture using fluorescence-activated cell sorting on Aria IIu at 20 pounds/square inch using a 100-µm nozzle. Sort purity was determined by sorting an aliquot of cells into 10% RPMI and then immediately reanalyzing the sorted aliquot by flow cytometry. In general, we achieved sort purities of over 98%. Microglia for RNA isolation were sorted directly into RLT lysis buffer (Qiagen) and their total RNA isolated using RNeasy Micro Kit (Qiagen), according to the manufacturer’s recommendations. The quality and quantity of isolated total RNA were then estimated on Agilent Bioanalyzer 2100 using the Agilent RNA 6000 Nano Kit, and only samples with RNA integrity number values higher than 9.0 were used. Prior to library generation, RNA was subjected to DNase I digestion (Thermo Fisher Scientific) followed by RNeasy MinElute column clean up (Qiagen). RNA-seq libraries were generated using the SMART-Seq v4 Ultra Low Input RNA Kit (Clontech Laboratories) as per the manufacturer’s recommendations. From cDNA, final libraries were generated using the Nextera XT DNA Library Preparation Kit (Illumina). Concentrations of the final libraries were measured with a Qubit 2.0 Fluorometer (Thermo Fisher Scientific), and fragment length distribution was analyzed with the DNA High Sensitivity Chip on an Agilent 2100 Bioanalyzer (Agilent Technologies). All samples were normalized to 2 nM and pooled at equimolar concentrations. The library pool was sequenced on the NextSeq500 (Illumina) in a single 1 × 76–bp run, producing 16.0–24.9 M reads per sample from a total of six mRNA-seq libraries. Prior to downstream processing, adapter sequences were hard-clipped from raw sequencing reads as part of the bcl2fastq pipeline (version 2.20.0.422). Overall quality of the trimmed sequences was assessed by FastQC v0.11.8 (Andrews, 2010). Where applicable, quality data from individual analyses were aggregated using MultiQC v1.6 (Ewels et al., 2016).
Premapping quality control
Initial quality control of the sequencing reads, performed using FastQC v0.11.8 (Andrews, 2010), indicated that the filtering and/or additional trimming of the reads before mapping is not required. We next performed the screen of the sequencing libraries for contaminating sequences, originating from biological or technical sources, using FastQ Screen v0.14.0 (Wingett and Andrews, 2018) and Bowtie 2 v2.3.5.1 (Langmead and Salzberg, 2012). A total of 448,721 partial or complete nucleotide sequences of the mouse genome and potential technical or biological contaminants in mouse RNA-seq experiments (Bayona-Bafaluy et al., 2003; Frankish et al., 2019; Grozdanov et al., 2003; Mahmood and Ali, 2017; Petrov et al., 2017; Pritchett-Corning et al., 2009; Rawlinson et al., 1996; Rigden and Fernández, 2020) were divided into 14 different categories within the in-house–built FastQ Screen database. Almost all sequences used for the construction of the FastQ Screen database were downloaded from the NCBI website (Sayers et al., 2020) using ncbi-genome-download v0.2.11 (Blin, 2016), except for the genomic sequences of the Paecilomyces sp., which were downloaded from the MycoCosm portal (Grigoriev et al., 2014). Following download, sequences in each category were concatenated into a single multi-FASTA file before the generation of Bowtie 2 indexes and screening. The composition of all sequencing libraries correlated highly with the experimental design and excluded the possibility of significant sample contamination with undesired or wrong nucleic acid molecules.
Read mapping, postmapping quality control, and read counting
Sequencing reads were successfully mapped to concatenated sequence of the MCMV genome (GenBank accession no. NC_004065.1) and mouse GRCm38.p6 (release M23) primary reference genome assembly (Frankish et al., 2019) using STAR v2.7.3a (Dobin et al., 2013; Dobin and Gingeras, 2015; Dobin and Gingeras, 2016) with the following parameters: --runThreadN 32; --readFilesCommand zcat; --genomeLoad LoadAndKeep; --outSAMtype BAM SortedByCoordinate; --limitBamsortRAM 20000000000; --outBAMsortingThreadN 32. Obtained alignment files were indexed using samtools v1.10 (Li et al., 2009), while postmapping quality control of the RNA-seq dataset, performed using QoRTs v1.3.6 (Hartley and Mullikin, 2015), indicated an absence of artifacts, significant systematic biases, or other issues that might negatively affect differential expression analysis. Reads mapping to individual genes were then summarized using featureCounts (Liao et al., 2014), and obtained gene counts, excluding multimapping reads, were used for further analysis.
Differential expression and Gene Ontology overrepresentation analysis
Principal-component analysis of the rlog-transformed raw count data and identification of DE genes, applying padj < 0.01 and log2FC > 1 as cutoffs for statistical significance, were performed using the DESeq2 package (Love et al., 2014). Gene Ontology overrepresentation analysis of the list of DE genes was performed using clusterProfiler v3.14.1 (Yu et al., 2012), a volcano plot was drawn using EnhancedVolcano v1.4.0 (Blighe et al., 2020), and heatmaps were generated with Morpheus (https://software.broadinstitute.org/morpheus).
Data availability
Raw sequencing reads used in this study have been submitted to the European Nucleotide Archive and are available under the accession no. PRJEB38849.
Statistical analyses
Data are presented as mean ± SEM. Statistical significance was determined by either two-tailed unpaired Student’s t test or Mann–Whitney U test using GraphPad Prism 5. A value of P > 0.05 was deemed not statistically significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P ≤ 0.0001.
Online supplemental material
Fig. S1 shows that microglia can be infected with MCMV and are not an early source of IFN-γ in the brain after MCMV infection. Fig. S2 shows that CD4 and CD8 T cells are not responsible for early microglia activation and that depletion of NK cells in mice infected with MCMV does not impact the viral titer in the brain. Fig. S3 shows that recombinant IFN-γ can induce the expression of MHC-I and MHC-II on microglia at early days p.i., but the infection is needed for long-term microglia activation. In addition, this figure shows that the effect of IFN-γ on GFAP and Lyz2-positive cells is not responsible for delay in EGL maturation.
Acknowledgments
This work was supported by the National Institutes of Health (grant “Inflammation and Hearing Loss Following Congenital CMV Infection” [1 R01 DC015980-01A1] to S. Jonjić and W.J. Britt and grant “Strengthening the capacity of CerVirVac for research in virus immunology and vaccinology” [KK.01.1.1.01.0006] to the Scientific Centre of Excellence for Virus Immunology and Vaccines) and cofinanced by the European Regional Development Fund (S. Jonjić), the Croatian Science Foundation (under project IP-2018-01-4435 to I. Brizić and project IP-2018-01-9086 to A. Krmpotić), and the University of Rijeka (under project uniri-biomed-18-234 to B. Lisnić).
Author contributions: Conceptualization, S. Jonjić, A. Krmpotić, D. Kveštak, and W.J. Britt; investigation, validation, and formal analysis: D. Kveštak, V. Juranić Lisnić, B. Lisnić, I. Brizić, J. Tomac, M. Golemac, D. Indenbirken, M. Cokarić Brdovčak, F. Krstanović, C. Rožmanić, and A. Grundhoff; methodology, D. Kveštak, D. Indenbirken, A. Grundhoff, B. Lisnić, V. Juranić Lisnić, I. Brizić, and M. Golemac; resources, G. Bernardini; writing, D. Kveštak, V. Juranić Lisnić, B. Lisnić, I. Brizić, W.J. Britt, A. Krmpotić, and S. Jonjić; data curation, B. Lisnić and D. Indenbirken; supervision, V. Juranić Lisnić, A. Krmpotić, B. Lisnić, I. Brizić, and S. Jonjić; funding acquisition and project administration, S. Jonjić, A. Krmpotić, I. Brizić, B. Lisnić, and W.J. Britt.
References
- Andrews, S. 2010. FastQC: a quality control tool for high throughput sequence data. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed October 5, 2018).
- Bantug, G.R., Cekinovic D., Bradford R., Koontz T., Jonjic S., and Britt W.J.. 2008. CD8+ T lymphocytes control murine cytomegalovirus replication in the central nervous system of newborn animals. J. Immunol. 181:2111–2123. 10.4049/jimmunol.181.3.2111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayona-Bafaluy, M.P., Acín-Pérez R., Mullikin J.C., Park J.S., Moreno-Loshuertos R., Hu P., Pérez-Martos A., Fernández-Silva P., Bai Y., and Enríquez J.A.. 2003. Revisiting the mouse mitochondrial DNA sequence. Nucleic Acids Res. 31:5349–5355. 10.1093/nar/gkg739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blighe, K., Rana S., and Lewis M.. 2020. EnhancedVolcano: Publication-ready volcano plots with enhanced colouring and labeling. R package version 1.6.0. https://github.com/kevinblighe/EnhancedVolcano (accessed November 1, 2019).
- Blin, K. 2016. NCBI Genome Downloading Scripts. GitHub Repository. https://github.com/kblin/ncbi-genome-download (accessed September 4, 2019).
- Boppana, S.B., Ross S.A., and Fowler K.B.. 2013. Congenital cytomegalovirus infection: clinical outcome. Clin. Infect. Dis. 57(Suppl 4):S178–S181. 10.1093/cid/cit629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkmann, M.M., Dağ F., Hengel H., Messerle M., Kalinke U., and Čičin-Šain L.. 2015. Cytomegalovirus immune evasion of myeloid lineage cells. Med. Microbiol. Immunol. (Berl.). 204:367–382. 10.1007/s00430-015-0403-4 [DOI] [PubMed] [Google Scholar]
- Britt, W.J., Cekinovic D., and Jonjic S.. 2013. Murine Model of Neonatal Cytomegalovirus Infection. In Cytomegaloviruses: From Molecular Pathogenesis to Intervention. Reddehase M.J., editor. Caister Academic Press, UK. pp. 119–141. [Google Scholar]
- Brizić, I., Lisnić B., Brune W., Hengel H., and Jonjić S.. 2018a. Cytomegalovirus Infection: Mouse Model. Curr. Protoc. Immunol. 122:e51. 10.1002/cpim.51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brizić, I., Šušak B., Arapović M., Huszthy P.C., Hiršl L., Kveštak D., Juranić Lisnić V., Golemac M., Pernjak Pugel E., Tomac J., et al. 2018b. Brain-resident memory CD8+ T cells induced by congenital CMV infection prevent brain pathology and virus reactivation. Eur. J. Immunol. 48:950–964. 10.1002/eji.201847526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brizić, I., Hiršl L., Šustić M., Golemac M., Britt W.J., Krmpotić A., and Jonjić S.. 2019. CD4 T cells are required for maintenance of CD8 TRM cells and virus control in the brain of MCMV-infected newborn mice. Med. Microbiol. Immunol. (Berl.). 208:487–494. 10.1007/s00430-019-00601-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubić, I., Wagner M., Krmpotić A., Saulig T., Kim S., Yokoyama W.M., Jonjić S., and Koszinowski U.H.. 2004. Gain of virulence caused by loss of a gene in murine cytomegalovirus. J. Virol. 78:7536–7544. 10.1128/JVI.78.14.7536-7544.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butts, T., Green M.J., and Wingate R.J.. 2014. Development of the cerebellum: simple steps to make a ‘little brain’. Development. 141:4031–4041. 10.1242/dev.106559 [DOI] [PubMed] [Google Scholar]
- Cekinovic, D., Lisnic V.J., and Jonjic S.. 2014. Rodent models of congenital cytomegalovirus infection. Methods Mol. Biol. 1119:289–310. 10.1007/978-1-62703-788-4_16 [DOI] [PubMed] [Google Scholar]
- Clancy, B., Darlington R.B., and Finlay B.L.. 2001. Translating developmental time across mammalian species. Neuroscience. 105:7–17. 10.1016/S0306-4522(01)00171-3 [DOI] [PubMed] [Google Scholar]
- Daussy, C., Faure F., Mayol K., Viel S., Gasteiger G., Charrier E., Bienvenu J., Henry T., Debien E., Hasan U.A., et al. 2014. T-bet and Eomes instruct the development of two distinct natural killer cell lineages in the liver and in the bone marrow. J. Exp. Med. 211:563–577. 10.1084/jem.20131560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Luca, A., Cerrato V., Fucà E., Parmigiani E., Buffo A., and Leto K.. 2016. Sonic hedgehog patterning during cerebellar development. Cell. Mol. Life Sci. 73:291–303. 10.1007/s00018-015-2065-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, X., Yan Y., Li X., Li K., Ciric B., Yang J., Zhang Y., Wu S., Xu H., Chen W., et al. 2015. Silencing IFN-γ binding/signaling in astrocytes versus microglia leads to opposite effects on central nervous system autoimmunity. J. Immunol. 194:4251–4264. 10.4049/jimmunol.1303321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin, A., and Gingeras T.R.. 2015. Mapping RNA-seq Reads with STAR. Curr. Protoc. Bioinformatics. 51:14.1–: 19.. 10.1002/0471250953.bi1114s51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin, A., and Gingeras T.R.. 2016. Optimizing RNA-Seq Mapping with STAR. Methods Mol. Biol. 1415:245–262. 10.1007/978-1-4939-3572-7_13 [DOI] [PubMed] [Google Scholar]
- Dobin, A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., and Gingeras T.R.. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 29:15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorfman, J.R., and Raulet D.H.. 1998. Acquisition of Ly49 receptor expression by developing natural killer cells. J. Exp. Med. 187:609–618. 10.1084/jem.187.4.609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewels, P., Magnusson M., Lundin S., and Käller M.. 2016. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 32:3047–3048. 10.1093/bioinformatics/btw354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiebich, B.L., Batista C.R.A., Saliba S.W., Yousif N.M., and de Oliveira A.C.P.. 2018. Role of Microglia TLRs in Neurodegeneration. Front. Cell. Neurosci. 12:329. 10.3389/fncel.2018.00329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankish, A., Diekhans M., Ferreira A.M., Johnson R., Jungreis I., Loveland J., Mudge J.M., Sisu C., Wright J., Armstrong J., et al. 2019. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 47(D1):D766–D773. 10.1093/nar/gky955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldmann, T., Wieghofer P., Jordão M.J., Prutek F., Hagemeyer N., Frenzel K., Amann L., Staszewski O., Kierdorf K., Krueger M., et al. 2016. Origin, fate and dynamics of macrophages at central nervous system interfaces. Nat. Immunol. 17:797–805. 10.1038/ni.3423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon, S.M., Chaix J., Rupp L.J., Wu J., Madera S., Sun J.C., Lindsten T., and Reiner S.L.. 2012. The transcription factors T-bet and Eomes control key checkpoints of natural killer cell maturation. Immunity. 36:55–67. 10.1016/j.immuni.2011.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoriev, I.V., Nikitin R., Haridas S., Kuo A., Ohm R., Otillar R., Riley R., Salamov A., Zhao X., Korzeniewski F., et al. 2014. MycoCosm portal: gearing up for 1000 fungal genomes. Nucleic Acids Res. 42(Database issue, D1):D699–D704. 10.1093/nar/gkt1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grozdanov, P., Georgiev O., and Karagyozov L.. 2003. Complete sequence of the 45-kb mouse ribosomal DNA repeat: analysis of the intergenic spacer. Genomics. 82:637–643. 10.1016/S0888-7543(03)00199-X [DOI] [PubMed] [Google Scholar]
- Hartley, S.W., and Mullikin J.C.. 2015. QoRTs: a comprehensive toolset for quality control and data processing of RNA-Seq experiments. BMC Bioinformatics. 16:224. 10.1186/s12859-015-0670-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonjic, S., Krmpotic A., Arapovic J., and Koszinowski U.H.. 2008. Dissection of the antiviral NK cell response by MCMV mutants. Methods Mol. Biol. 415:127–149. 10.1007/978-1-59745-570-1_8 [DOI] [PubMed] [Google Scholar]
- Juranic Lisnic, V., Babic Cac M., Lisnic B., Trsan T., Mefferd A., Das Mukhopadhyay C., Cook C.H., Jonjic S., and Trgovcich J.. 2013. Dual analysis of the murine cytomegalovirus and host cell transcriptomes reveal new aspects of the virus-host cell interface. PLoS Pathog. 9:e1003611. 10.1371/journal.ppat.1003611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawanokuchi, J., Mizuno T., Takeuchi H., Kato H., Wang J., Mitsuma N., and Suzumura A.. 2006. Production of interferon-gamma by microglia. Mult. Scler. 12:558–564. 10.1177/1352458506070763 [DOI] [PubMed] [Google Scholar]
- Kimberlin, D.W., Jester P.M., Sánchez P.J., Ahmed A., Arav-Boger R., Michaels M.G., Ashouri N., Englund J.A., Estrada B., Jacobs R.F., et al. National Institute of Allergy and Infectious Diseases Collaborative Antiviral Study Group . 2015. Valganciclovir for symptomatic congenital cytomegalovirus disease. N. Engl. J. Med. 372:933–943. 10.1056/NEJMoa1404599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein, R.S., and Hunter C.A.. 2017. Protective and Pathological Immunity during Central Nervous System Infections. Immunity. 46:891–909. 10.1016/j.immuni.2017.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein, R.S., Garber C., Funk K.E., Salimi H., Soung A., Kanmogne M., Manivasagam S., Agner S., and Cain M.. 2019. Neuroinflammation During RNA Viral Infections. Annu. Rev. Immunol. 37:73–95. 10.1146/annurev-immunol-042718-041417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koontz, T., Bralic M., Tomac J., Pernjak-Pugel E., Bantug G., Jonjic S., and Britt W.J.. 2008. Altered development of the brain after focal herpesvirus infection of the central nervous system. J. Exp. Med. 205:423–435. 10.1084/jem.20071489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosmac, K., Bantug G.R., Pugel E.P., Cekinovic D., Jonjic S., and Britt W.J.. 2013. Glucocorticoid treatment of MCMV infected newborn mice attenuates CNS inflammation and limits deficits in cerebellar development. PLoS Pathog. 9:e1003200. 10.1371/journal.ppat.1003200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krmpotic, A., Bubic I., Polic B., Lucin P., and Jonjic S.. 2003. Pathogenesis of murine cytomegalovirus infection. Microbes Infect. 5:1263–1277. 10.1016/j.micinf.2003.09.007 [DOI] [PubMed] [Google Scholar]
- Krug, A., French A.R., Barchet W., Fischer J.A., Dzionek A., Pingel J.T., Orihuela M.M., Akira S., Yokoyama W.M., and Colonna M.. 2004. TLR9-dependent recognition of MCMV by IPC and DC generates coordinated cytokine responses that activate antiviral NK cell function. Immunity. 21:107–119. 10.1016/j.immuni.2004.06.007 [DOI] [PubMed] [Google Scholar]
- Langmead, B., and Salzberg S.L.. 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 9:357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., and Durbin R.. 1000 Genome Project Data Processing Subgroup . 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 25:2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao, Y., Smyth G.K., and Shi W.. 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 30:923–930. 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- Love, M.I., Huber W., and Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmood, A., and Ali S.. 2017. Microbial and viral contamination of animal and stem cell cultures: common contaminants, detection and elimination. J. Stem Cell Res. Ther. 2:149–155. [Google Scholar]
- Nishikado, H., Mukai K., Kawano Y., Minegishi Y., and Karasuyama H.. 2011. NK cell-depleting anti-asialo GM1 antibody exhibits a lethal off-target effect on basophils in vivo. J. Immunol. 186:5766–5771. 10.4049/jimmunol.1100370 [DOI] [PubMed] [Google Scholar]
- Oliver, S.E., Cloud G.A., Sánchez P.J., Demmler G.J., Dankner W., Shelton M., Jacobs R.F., Vaudry W., Pass R.F., Soong S.J., et al. National Institute of Allergy, Infectious Diseases Collaborative Antiviral Study Group . 2009. Neurodevelopmental outcomes following ganciclovir therapy in symptomatic congenital cytomegalovirus infections involving the central nervous system. J. Clin. Virol. 46(Suppl 4):S22–S26. 10.1016/j.jcv.2009.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottum, P.A., Arellano G., Reyes L.I., Iruretagoyena M., and Naves R.. 2015. Opposing Roles of Interferon-Gamma on Cells of the Central Nervous System in Autoimmune Neuroinflammation. Front. Immunol. 6:539. 10.3389/fimmu.2015.00539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov, A.I., Kay S.J.E., Kalvari I., Howe K.L., Gray K.A., Bruford E.A., Kersey P.J., Cochrane G., Finn R.D., Bateman A., et al. The RNAcentral Consortium . 2017. RNAcentral: a comprehensive database of non-coding RNA sequences. Nucleic Acids Res. 45(D1):D128–D134. 10.1093/nar/gkw1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinz, M., and Priller J.. 2017. The role of peripheral immune cells in the CNS in steady state and disease. Nat. Neurosci. 20:136–144. 10.1038/nn.4475 [DOI] [PubMed] [Google Scholar]
- Pritchett-Corning, K.R., Cosentino J., and Clifford C.B.. 2009. Contemporary prevalence of infectious agents in laboratory mice and rats. Lab. Anim. 43:165–173. 10.1258/la.2008.008009 [DOI] [PubMed] [Google Scholar]
- Rawlinson, W.D., Farrell H.E., and Barrell B.G.. 1996. Analysis of the complete DNA sequence of murine cytomegalovirus. J. Virol. 70:8833–8849. 10.1128/JVI.70.12.8833-8849.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddehase, M.J., and Lemmermann N.A.W.. 2018. Mouse Model of Cytomegalovirus Disease and Immunotherapy in the Immunocompromised Host: Predictions for Medical Translation that Survived the “Test of Time”. Viruses. 10:693. 10.3390/v10120693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigden, D.J., and Fernández X.M.. 2020. The 27th annual Nucleic Acids Research database issue and molecular biology database collection. Nucleic Acids Res. 48(D1):D1–D8. 10.1093/nar/gkz1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock, R.B., Gekker G., Hu S., Sheng W.S., Cheeran M., Lokensgard J.R., and Peterson P.K.. 2004. Role of microglia in central nervous system infections. Clin. Microbiol. Rev. 17:942–964. 10.1128/CMR.17.4.942-964.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Suárez, S., Del Rio Serrato A., Bueno R.J., Brunotte-Strecker D., Stehle C., Figueiredo C.A., Hertwig L., Dunay I.R., Romagnani C., and Infante-Duarte C.. 2019. The Central Nervous System Contains ILC1s That Differ From NK Cells in the Response to Inflammation. Front. Immunol. 10:2337. 10.3389/fimmu.2019.02337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusinova, I., Forster S., Yu S., Kannan A., Masse M., Cumming H., Chapman R., and Hertzog P.J.. 2013. Interferome v2.0: an updated database of annotated interferon-regulated genes. Nucleic Acids Res. 41(Database issue, D1):D1040–D1046. 10.1093/nar/gks1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathyanesan, A., Zhou J., Scafidi J., Heck D.H., Sillitoe R.V., and Gallo V.. 2019. Emerging connections between cerebellar development, behaviour and complex brain disorders. Nat. Rev. Neurosci. 20:298–313. 10.1038/s41583-019-0152-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayers, E.W., Beck J., Brister J.R., Bolton E.E., Canese K., Comeau D.C., Funk K., Ketter A., Kim S., Kimchi A., et al. 2020. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 48(D1):D9–D16. 10.1093/nar/gkz899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seleme, M.C., Kosmac K., Jonjic S., and Britt W.J.. 2017. Tumor Necrosis Factor Alpha-Induced Recruitment of Inflammatory Mononuclear Cells Leads to Inflammation and Altered Brain Development in Murine Cytomegalovirus-Infected Newborn Mice. J. Virol. 91:e01983-16. 10.1128/JVI.01983-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellier, Y., Marliot F., Bessières B., Stirnemann J., Encha-Razavi F., Guilleminot T., Haicheur N., Pages F., Ville Y., and Leruez-Ville M.. 2020. Adaptive and Innate Immune Cells in Fetal Human Cytomegalovirus-Infected Brains. Microorganisms. 8:176. 10.3390/microorganisms8020176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streit, W.J., Mrak R.E., and Griffin W.S.. 2004. Microglia and neuroinflammation: a pathological perspective. J. Neuroinflammation. 1:14. 10.1186/1742-2094-1-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ta, T.T., Dikmen H.O., Schilling S., Chausse B., Lewen A., Hollnagel J.O., and Kann O.. 2019. Priming of microglia with IFN-γ slows neuronal gamma oscillations in situ. Proc. Natl. Acad. Sci. USA. 116:4637–4642. 10.1073/pnas.1813562116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thapa, M., Welner R.S., Pelayo R., and Carr D.J.. 2008. CXCL9 and CXCL10 expression are critical for control of genital herpes simplex virus type 2 infection through mobilization of HSV-specific CTL and NK cells to the nervous system. J. Immunol. 180:1098–1106. 10.4049/jimmunol.180.2.1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trambley, J., Bingaman A.W., Lin A., Elwood E.T., Waitze S.Y., Ha J., Durham M.M., Corbascio M., Cowan S.R., Pearson T.C., and Larsen C.P.. 1999. Asialo GM1(+) CD8(+) T cells play a critical role in costimulation blockade-resistant allograft rejection. J. Clin. Invest. 104:1715–1722. 10.1172/JCI8082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trifilo, M.J., Montalto-Morrison C., Stiles L.N., Hurst K.R., Hardison J.L., Manning J.E., Masters P.S., and Lane T.E.. 2004. CXC chemokine ligand 10 controls viral infection in the central nervous system: evidence for a role in innate immune response through recruitment and activation of natural killer cells. J. Virol. 78:585–594. 10.1128/JVI.78.2.585-594.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivier, E., Artis D., Colonna M., Diefenbach A., Di Santo J.P., Eberl G., Koyasu S., Locksley R.M., McKenzie A.N.J., Mebius R.E., et al. 2018. Innate Lymphoid Cells: 10 Years On. Cell. 174:1054–1066. 10.1016/j.cell.2018.07.017 [DOI] [PubMed] [Google Scholar]
- Wallace, V.A. 1999. Purkinje-cell-derived Sonic hedgehog regulates granule neuron precursor cell proliferation in the developing mouse cerebellum. Curr. Biol. 9:445–448. 10.1016/S0960-9822(99)80195-X [DOI] [PubMed] [Google Scholar]
- Wang, X., and Suzuki Y.. 2007. Microglia produce IFN-gamma independently from T cells during acute toxoplasmosis in the brain. J. Interferon Cytokine Res. 27:599–605. 10.1089/jir.2006.0157 [DOI] [PubMed] [Google Scholar]
- Wang, J., Lin W., Popko B., and Campbell I.L.. 2004. Inducible production of interferon-gamma in the developing brain causes cerebellar dysplasia with activation of the Sonic hedgehog pathway. Mol. Cell. Neurosci. 27:489–496. 10.1016/j.mcn.2004.08.004 [DOI] [PubMed] [Google Scholar]
- Wechsler-Reya, R.J., and Scott M.P.. 1999. Control of neuronal precursor proliferation in the cerebellum by Sonic Hedgehog. Neuron. 22:103–114. 10.1016/S0896-6273(00)80682-0 [DOI] [PubMed] [Google Scholar]
- Weizman, O.E., Adams N.M., Schuster I.S., Krishna C., Pritykin Y., Lau C., Degli-Esposti M.A., Leslie C.S., Sun J.C., and O’Sullivan T.E.. 2017. ILC1 Confer Early Host Protection at Initial Sites of Viral Infection. Cell. 171:795–808.e12. 10.1016/j.cell.2017.09.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh, R.M., Brubaker J.O., Vargas-Cortes M., and O’Donnell C.L.. 1991. Natural killer (NK) cell response to virus infections in mice with severe combined immunodeficiency. The stimulation of NK cells and the NK cell-dependent control of virus infections occur independently of T and B cell function. J. Exp. Med. 173:1053–1063. 10.1084/jem.173.5.1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingett, S.W., and Andrews S.. 2018. FastQ Screen: A tool for multi-genome mapping and quality control. F1000 Res. 7:1338. 10.12688/f1000research.15931.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, G., Wang L.G., Han Y., and He Q.Y.. 2012. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 16:284–287. 10.1089/omi.2011.0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Raw sequencing reads used in this study have been submitted to the European Nucleotide Archive and are available under the accession no. PRJEB38849.