Abstract
Background
Despite the necessity, there is no reliable biomarker to predict disease severity and prognosis of patients with COVID-19. The currently published prediction models are not fully applicable to clinical use.
Objectives
To identify predictive biomarkers of COVID-19 severity and to justify their threshold values for the stratification of the risk of deterioration that would require transferring to the intensive care unit (ICU).
Methods
The study cohort (560 subjects) included all consecutive patients admitted to Dubai Mediclinic Parkview Hospital from February to May 2020 with COVID-19 confirmed by the PCR. The challenge of finding the cut-off thresholds was the unbalanced dataset (eg, the disproportion in the number of 72 patients admitted to ICU vs 488 non-severe cases). Therefore, we customised supervised machine learning (ML) algorithm in terms of threshold value used to predict worsening.
Results
With the default thresholds returned by the ML estimator, the performance of the models was low. It was improved by setting the cut-off level to the 25th percentile for lymphocyte count and the 75th percentile for other features. The study justified the following threshold values of the laboratory tests done on admission: lymphocyte count <2.59×109/L, and the upper levels for total bilirubin 11.9 μmol/L, alanine aminotransferase 43 U/L, aspartate aminotransferase 32 U/L, D-dimer 0.7 mg/L, activated partial thromboplastin time (aPTT) 39.9 s, creatine kinase 247 U/L, C reactive protein (CRP) 14.3 mg/L, lactate dehydrogenase 246 U/L, troponin 0.037 ng/mL, ferritin 498 ng/mL and fibrinogen 446 mg/dL.
Conclusion
The performance of the neural network trained with top valuable tests (aPTT, CRP and fibrinogen) is admissible (area under the curve (AUC) 0.86; 95% CI 0.486 to 0.884; p<0.001) and comparable with the model trained with all the tests (AUC 0.90; 95% CI 0.812 to 0.902; p<0.001). Free online tool at https://med-predict.com illustrates the study results.
Keywords: COVID-19, biotechnology & bioinformatics, infectious diseases, respiratory infections, information technology, biochemistry
Strength and limitations of the study.
The research is based on a unique study cohort that is representative of the entire population because of the national standard that required all patients with confirmed COVID-19 to be admitted to acute care hospitals regardless of their symptoms or illness severity.
To distinguish the patients with the confirmed COVID-19 who may worsen while treated, we justified the threshold values of the laboratory tests done on admission.
The prediction of the future deterioration by the neural network is reliable even with the top three valuable laboratory tests (activated partial thromboplastin time, C reactive protein and fibrinogen) used for training (area under the curve 0.86; 95% CI 0.486 to 0.884; p<0.001).
The limitation of the study was the unbalanced dataset (eg, the disproportion in the number of patients admitted to the intensive care unit vs non-severe cases).
Introduction
Despite the necessity, there is no reliable prognostic biomarker to predict disease severity and prognosis of patients with COVID-19.1 Studies on COVID-19 have built up several types of prediction models. These have been the models designed to indicate the disease risk in the general population, the diagnostic models based on medical imaging and the prognostic models. Unfortunately, these models have had some limitations that have precluded their use in clinical practice.2
Models using laboratory findings as the inputs
Researchers tried to establish the role of laboratory findings in the diagnosis of COVID-19.3 They showed that the severe cases of COVID-19 were associated with D-dimer level over 0.28 µg/L, interleukin (IL)-6 level over 24.3 pg/mL3 and lactate dehydrogenase (LDH) activity with an upper limit cut-off in the range of 240–255 U/L.4 However, the use of these laboratory parameters with the above-mentioned cut-off values was limited for the following reasons. First, these studies were conducted on severe forms of the disease. Limited research was done on patients who were asymptomatic or had mild disease.3 5 Second, the whole spectrum of the regularly used clinical laboratory data is unavailable for non-severe patients. Thus, the published papers add justification on the diagnostic utility of separate laboratory findings, instead of working out reliable diagnostic criteria for a set of them.
Gong et al6 have generated a tool for the early prediction of severe COVID-19 pneumonia out of the following data: age, serum LDH activity, C reactive protein (CRP), the coefficient of variation of red blood cell distribution width, blood urea nitrogen, direct bilirubin, lower albumin. The resulting performance was not high (sensitivity 77.5%, specificity 78.4%).6 Supposedly, this is because the dataset used as the input consists of exceptionally the age and laboratory findings.
In another model, the inputs included basic information, symptoms and the results of laboratory tests. After the feature selection, the number of key features was set to just three laboratory results: LDH, lymphocytes and high-sensitivity CRP. The model was trained with the follow-up studies of the general, severe and critical patients.1 By feeding machine learning (ML) algorithm with the results obtained at the time of admission and in follow-up studies, the authors worked out a decision rule to predict patients at the highest risk. However, physicians are interested in the early prediction of the disease outcomes, and it is highly disputable that the model will not loose its predictive potential if applied exceptionally to the data received on admission.
We believe that a more accurate model can be built based on the simultaneous interpretation of laboratory results, clinical data and physical examination findings (eg, body mass index, body temperature, respiratory rate) at the time of presentation. The analysis using an ML algorithm could provide an accurate prediction of the disease severity.
Data used by clinicians for stratifying risks
Clinicians routinely use physical examination findings and laboratory parameters for risk stratification and hospital resources management. Commonly, each laboratory test kit has the only cut-off value to segregate the normal status from a pathology. We believe that threshold values should be re-adjusted for each disease rather than used as a common cut-off value for all pathologies.
As a standard of care, baseline blood tests and inflammatory markers are obtained on admission to the hospital. The proper approach for the risk assessment should allow physicians to forecast the patient’s future worsening out of the initial findings on admission. This is what we intend to do by applying an ML approach to the predictors routinely used in clinical practice. There are some promising data for the following set of prognostic biomarkers of COVID-19 severity.
Inflammatory markers
There is evidence that IL-6 and tumour necrosis factor (TNF)-α do not indicate the level of COVID-19 progression.7 Some markers of inflammation are elevated in the serum of patients with COVID-19 compared with the healthy people, that is, the serum SARS-CoV-2 viral load (RNAaemia) is closely correlated with drastically elevated IL-6 levels in critically ill patients with COVID-19.8 However, there is no significant difference between severe and mild groups.7 In contrast to this, the indicators are reflective in the progression of the diseases caused by other coronaviruses (eg, Middle East respiratory syndrome (MERS), SARS).9 This may be explained by the huge amino acid differences in viral proteins of distinct coronaviruses. Even with different MERS-CoV strains, common cytokine signalling by TNF and IL-1α results in the differential expression of innate immune genes.10
Ferritin
Ferritin is a marker of iron storage. However, it is also an acute-phase reactant, the level of which elevates in processes of acute inflammation, whether infectious or non-infectious. Marked elevations have been reported in cases of COVID-19 infection.11
D-dimer
A common finding in most patients with COVID-19 is high D-dimer levels (>0.28 mg/L), which are associated with a worse prognosis.3 12 An exceptional interest of physicians in this biomarker comes from the fact that the vast majority of patients who died of COVID-19 fulfilled the criteria for diagnosing the disseminated intravascular coagulation. This is why the incidence of pulmonary embolism in COVID-19 is high. In this condition, the D-dimer concentration will definitely rise up because it is a product of degradation of a blood clot formed out of fibrin protein.13 Thromboembolic complications explain the association of low levels of platelets, increased levels of D-dimer and increasing levels of prothrombin in COVID-19.14 Alternatively, the D-dimer level may go up as a direct consequence of SARS-CoV-2 itself.15
Reasonably, laboratory haemostasis may provide an essential contribution to the COVID-19 prognosis and therapeutic decisions.16 Researchers tried to forecast the severity of COVID-19 with D-dimer as a single predictor. They showed that D-dimer level >0.5 mg/L had a 58% sensitivity, 69% specificity in the forecast of the disease severity.17 In another study, D-dimer level of >2.14 mg/L predicted in-hospital mortality with a sensitivity of 88.2% and specificity of 71.3%.18 Another study highlighted that a D-dimer threshold of >2.66 mg/L detected all patients with a pulmonary embolus on the chest CT.15 So, the high levels of D-dimer are a reliable prognostic biomarker of in-hospital mortality.
Fibrinogen
In patients with COVID-19 admitted to ICU for acute respiratory failure, the level of fibrinogen is significantly higher than in healthy controls (517±148 vs 297±78 mg/dL).12 The small vessel thrombi revealed on autopsy in lungs and other organs suggest that disseminated intravascular coagulation in COVID-19 results from severe endothelial dysfunction, driven by the cytokine storm and associated hypoxaemia. As standard-dose deep vein thrombosis prophylaxis cannot prevent the consumptive coagulopathy, monitoring D-dimer and fibrinogen levels are required. This will promote the early diagnostics of hypercoagulability and its treatment with direct factor Xa inhibitors.14 19
Activated partial thromboplastin time
In a study conducted in February 2020, the levels of activated partial thromboplastin time (aPTT) as well as white blood cells (WBC), lymphocytes, aspartate aminotransferase (AST), alanine aminotransferase (ALT) and creatinine, differed negligibly between severe and mild patients.3 At the same time, other researchers showed inconsequential distinction in aPTT in survivors versus non-survivors.20 According to the results of another study published in March 2020, no significant difference in aPTT values were found in the cohort of severe cases versus the non-severe one.6 The results obtained in another study in April in Italy were the same.12 The common limitation of these early studies was a small sample size. Finally, a meta-analysis justified that the elevation of D-dimer, rather than prothrombin time and aPTT, reflects the progression of COVID-19 towards an unfavourable outcome.21
LDH and creatine kinase
Increased levels of the enzymes may reflect the level of the organ damage in a systemic disease.4 22 Reasonably, they may serve as biomarkers for COVID-19 progression.
C reactive protein
In the early stage of COVID-19, CRP levels are positively correlated with the diameter of lung lesions and severe presentation.23
Liver enzymes and total bilirubin
COVID-19 leads to elevated liver biochemistries (eg, the level of AST, ALT, gamma-glutamyl transferase, total bilirubin) in over 50% of patients on admission. AST-dominant aminotransferase elevation reflects the disease severity and true hepatic injury.24 25
Objectives
We decided to identify predictive biomarkers of COVID-19 severity and to justify their threshold values. Hypothetically, the absolute values of the biomarkers on admission to the clinics could provide physicians with an accurate prognosis on the future worsening of the patient that would require transferring the individual to the intensive care unit (ICU). Getting a reliable tool for such a prognosis will support decision making and logistical planning in clinics.
To address the objective, we designed a set of the following tasks:
To study the linear separability of the laboratory findings values in patients with confirmed COVID-19 who were transferred to ICU versus non-severe cases of the disease, and to make the comparative analysis of the ICU department cases (both the deceased and survived cohorts) with other patients with COVID-19.
To identify the risk factors by selecting the most valuable features for predicting the deterioration that would require transferring the patient to ICU.
To work out the threshold criteria for the major clinical data for the early identification of the patients with a high risk of being transferred to ICU.
To identify the accuracy of the prediction of the patient’s deterioration by the ML algorithm and by a set of the newly created threshold values of the laboratory and clinical findings.
Materials and methods
Study design and sample
We did a retrospective analysis of the clinical data obtained as a standard of primary and secondary care. The study sample included all the consecutive patients admitted to Dubai Mediclinic from 24 February to 1 July 2020, who fit the criteria of eligibility (total 560 cases). Using this sample, the intention of the study was met, that is, to allow for the early prognostic stratification.
The inclusion criteria were as follows: age 18 years or older; inpatient admission; SARS-CoV-2-positive real-time reverse transcription PCR from nasopharyngeal swabs only, at our site. Those patients who met the inclusion criteria for our studies were included in the study sample. All the patients were discharged at the time of writing the paper.
The remarkable feature of our study is that at the beginning of the pandemic, all the patients with COVID-19 verified by PCR were hospitalised in the Mediclinic even if they did not present any symptoms. We observed many mild and asymptomatic forms of the disease, with all the required spectrum of analyses being conducted. All patients who were hospitalised stayed in Dubai Mediclinic until they were afebrile for >72 hours and had SpO2 value not <94%.
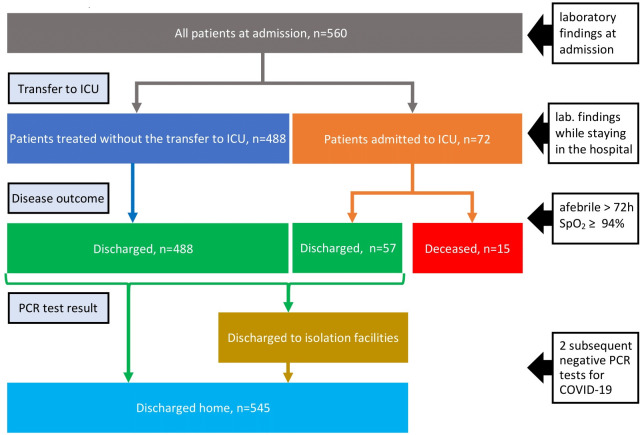
We assessed the duration of viral shedding as the number of days from the disease onset when the diagnosis was confirmed (eg, the first positive PCR test) to the first negative PCR test.26 All the patients hospitalised to the Mediclinics hospital were subject to the regular collection of nasopharyngeal swabs by a standard technique. Furthermore, after the patient stopped presenting disease symptoms, the specimen collection continued on a daily basis until two subsequent negative PCR tests for COVID-19 >24 hours apart. In the case of the mild disease course, patients might be transported to isolation facilities before being discharged home (see the flow chart diagram in figure 1). If the facilities were run by Mediclinic, we had their follow-up PCR results. For those patients who went to other isolation facilities not connected to Mediclinic, we could not study the duration of viral shedding (the data are missing for 27 out of 560 patients).
Figure 1.
The flow of patients with COVID-19 in Dubai Mediclinic. ICU, intensive care unit.
The treatment was administered in full accordance with ‘National Guidelines for Clinical Management and Treatment of COVID-19’. The indications for the supportive oxygen therapy were (a) the oxygen saturation level below 94%, (b) the respiratory rate (RR) above 30 breaths per minute, (c) both of them. In case of suspicion of superimposed bacterial pneumonia, physicians ordered empirical broad-spectrum antibiotics. The administration of the antiviral and antimalarial drugs followed the national guidelines.27
Patient and public involvement
No patient involved. The data were collected retrospectively from the medical record system.
Methods used
To address the first task, we studied the separability of laboratory findings values on admission to Dubai Mediclinic concerning the future transfer of the patient to the ICU department. To carry out the comparative analysis of features with regard to transferring to ICU, we used a set of non-parametric tests. The relationships involving two variables were assessed with the Mann-Whitney U test or Kruskal-Wallis test for the continuous features, and with Fisher’s exact test or χ2 test for the quantitative ones. The data were expressed as IQR, median±SD or number of cases and their percentage. The missing data for the comparative analysis were treated with the complete-case analysis method.
To address the second task, we used a set of different methods. First, we trained the neural network (NN) ML model on each variable separately. To come up with laboratory data cut-off levels which may be considered as biomarkers of severe course of the disease we assessed their statistical significance against chance performance. We calculated 95% CI for receiver operating characteristic (ROC) and ROC AUC scores with the bootstrap technique and p values with permutation tests.
Second, we used ML tree-based methods (AdaBoost, Gradient Boosting, Random Forest and Extra Trees) to check if there were unique patterns within the data that could unambiguously identify the event of transferring the patient to ICU from the data obtained on admission. For the list of features used as predictors, see online supplemental appendix 1. To assess the importance of the variables, we ranked all features concerning their impurity-based predictive potential. For ranking, we used a set of classifiers and then averaged all the received scores. Missing data in all ML models were replaced by the mean or median values with regard to the continuous or quantitative feature, respectively using single imputation method.
bmjopen-2020-044500supp009.pdf (1.8MB, pdf)
To tackle the third task, we used two approaches: a threshold moving technique (Youden’s index)28 and a heuristically chosen percentile-based cut-off level. The problem of predicting the transfer to ICU had a severe class imbalance. Therefore, we needed to focus on the performance of the classifier on the minority class (admitted to ICU patients). The sensitivity and specificity of the supervised ML classification model (NN) were used to evaluate the quality of the chosen optimal threshold for each important laboratory finding.
To evaluate the classifier output quality, we trained several ML classification models using a stratified 10-fold cross-validation technique to generalise the models to the true rate error. For each fold, we used 90% of the data to train the model and then tested it with the rest 10%. The decision matrices built on the test dataset for all folds were combined and used to calculate the performance metrics.
Results
Comparison of the ICU versus non-ICU patients
The problem of predicting admission to ICU has a severe class imbalance (488 vs 72). Therefore, we need to focus on the performance of the classifier on the minority class (the patients admitted to ICU).
We look at the linear separability of the groups of numerical data composed from the laboratory findings values with regard to their quartiles. In figure 2, box plots for the laboratory findings data are presented with the red dashed line that marks the 75th percentile for the subjects that were not transferred to ICU. The assumption is to use the third quartile (Q3) start point value as the threshold if there is separability between ICU and non-ICU groups. In diagrams in figure 2, the red line indicates the 75th percentile for not admitted to the ICU group. The exception is the diagram for the lymphocyte count, where it stands for the 25th percentile.
Figure 2.
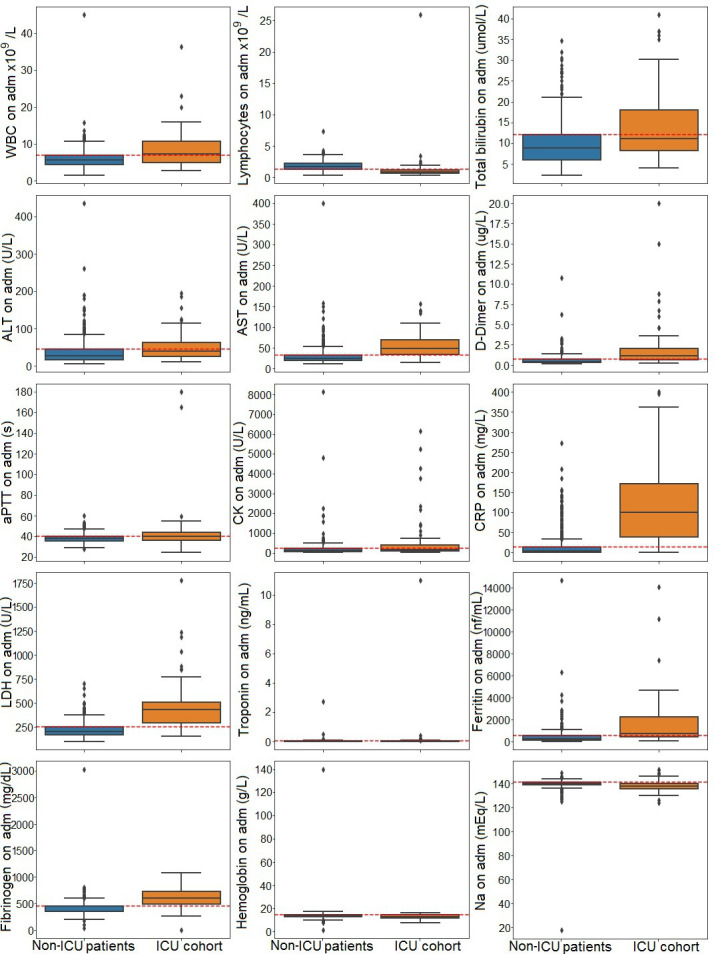
Variation of laboratory findings values in the intensive care unit (ICU) cohort (orange box plot) versus the non-ICU cohort of patients (blue box plot). ALT, alanine aminotransferase; aPTT, activated partial thromboplastin time; AST, aspartate aminotransferase; LDH, lactate dehydrogenase; WBC, white blood cell.
The results of the comparative analysis of features with regard to transferring to ICU and the final outcomes of the disease are presented in table 1. We excluded from further analysis the laboratory findings that did not significantly differ in the distribution of two groups. Therefore, we considered the list of 13 variables: WBC, lymphocyte count, total bilirubin, ALT, AST, D-dimer, aPTT, creatine kinase (CK), CRP, LDH, troponin, ferritin and fibrinogen on admission.
Table 1.
Comparison of the patients hospitalised to ICU concerning the COVID-19 outcomes: comorbidities, the result of physical examination on admission, laboratory findings on admission and deterioration (eg, peak or minimal values), ethnicity and disease course features
| All patients | ICU patients | Missing values, count | |||||||
| Total n1=560 | Not admitted to ICU n2=488 (87.14%) | Admitted to ICU n3=72 (12.86%) | P2-3 | Dead n4=15 (20.83%) | Discharged n5=57 (79.17%) | P4-5 | |||
| Age | 39.0 (33.0–49.0) | 38.0±11.97 | 51.0±13.08 | <0.0001 | 46.0±12.56 | 62.0±11.01 | <0.0018 | ||
| Gender | Female | 189 (33.75%) | 175 (35.86%) | 14 (19.44%) | <0.0072 | 8 (14.04%) | 6 (40.0%) | 0.06 | |
| Male | 371 (66.25%) | 313 (64.14%) | 58 (80.56%) | 49 (85.96%) | 9 (60.0%) | ||||
| Comorbidities | Count | 0.0 (0.0–1.0) | 0.0±1.04 | 1.0±1.22 | <0.0002 | 1.0±1.15 | 0.0±1.45 | 0.4072 | |
| Current smoking | 36 (6.43%) | 34 (6.97%) | 2 (2.78%) | 0.2984 | 2 (3.51%) | ||||
| Chronic cardiac disease | 20 (3.57%) | 15 (3.07%) | 5 (6.94%) | 0.1611 | 4 (7.02%) | 1 (6.67%) | |||
| Hypertension | 115 (20.54%) | 92 (18.85%) | 23 (31.94%) | <0.018 | 18 (31.58%) | 5 (33.33%) | 1 | ||
| Asthma | 38 (6.79%) | 31 (6.35%) | 7 (9.72%) | 0.3121 | 6 (10.53%) | 1 (6.67%) | |||
| Chronic kidney disease | 7 (1.25%) | 5 (1.02%) | 2 (2.78%) | 1 (1.75%) | 1 (6.67%) | ||||
| Diabetes | 98 (17.5%) | 71 (14.55%) | 27 (37.5%) | <0.0001 | 21 (36.84%) | 6 (40.0%) | 1 | ||
| Active malignant cancer | 6 (1.07%) | 4 (0.82%) | 2 (2.78%) | 1 (1.75%) | 1 (6.67%) | ||||
| BMI | adm | 27.0 (23.92–30.44) | 26.84±5.44 | 28.0±4.54 | <0.01 | 27.82±4.7 | 31.14±0.48 | 0.2575 | 278 |
| Body temperature, °C | adm | 37.0 (37.0–37.9) | 37.0±0.63 | 38.0±0.97 | <0.0001 | 38.0±0.97 | 38.0±0.98 | 0.3925 | |
| Heart rate, bmp | adm | 85.0 (78.0–95.0) | 84.5±12.32 | 94.5±19.97 | <0.0001 | 95.0±20.93 | 85.0±15.3 | 0.1589 | |
| SBP | adm | 124.0 (114.0–135.0) | 123.0±16.51 | 126.0±17.31 | 0.2092 | 129.0±16.29 | 120.0±20.58 | 0.2122 | |
| DBP | adm | 78.0 (70.0–84.0) | 78.0±10.92 | 75.0±10.1 | <0.0208 | 75.0±9.46 | 75.0±12.05 | 0.4254 | |
| RR/min | adm | 18.0 (18.0–18.0) | 18.0±1.56 | 25.0±6.74 | <0.0001 | 24.0±6.95 | 28.0±5.62 | 0.1336 | |
| SOFA score | adm | 0.0 (0.0–0.0) | 0.0±0.75 | 3.0±2.85 | <0.0001 | 3.0±2.42 | 4.0±3.69 | <0.0275 | 4 |
| WBC, ×109/L | adm | 5.8 (4.5–7.2) | 5.65±2.68 | 7.35±5.21 | <0.0001 | 7.4±5.34 | 7.0±4.68 | 0.3801 | 3 |
| min | 5.5 (4.1–7.2) | 5.5±7.72 | 7.0±6.68 | <0.0008 | 7.2±6.93 | 5.5±5.38 | 0.0775 | 3 | |
| Platelet, ×109/L | adm | 224.0 (180.25–272.0) | 224.5±78.42 | 222.0±82.13 | 0.4102 | 225.0±86.02 | 196.0±57.76 | 0.0516 | 2 |
| min | 224.0 (178.0–272.0) | 226.0±79.7 | 197.0±123.27 | <0.0049 | 202.0±116.33 | 102.0±84.42 | <0.0001 | 22 | |
| Lymphocyte, ×109/L | adm | 1.56 (1.06–2.1) | 1.66±0.76 | 0.81±2.97 | <0.0001 | 0.83±3.32 | 0.73±0.64 | 0.4806 | 3 |
| min | 1.49 (0.89–2.09) | 1.6±0.8 | 0.49±3.64 | <0.0001 | 0.5±4.07 | 0.38±0.62 | 0.1412 | 3 | |
| Total bilirubin, μmol/L | adm | 9.0 (6.0–12.6) | 8.6±5.24 | 11.0±9.17 | <0.0001 | 11.0±8.6 | 13.0±11.03 | 0.4094 | 11 |
| peak | 9.85 (6.5–14.38) | 9.0±6.55 | 16.3±37.25 | <0.0001 | 16.0±17.77 | 25.0±68.93 | 0.1412 | 10 | |
| ALT, U/L | adm | 28.0 (17.25–47.75) | 27.0±34.84 | 39.0±38.04 | <0.0001 | 39.0±39.5 | 41.0±31.76 | 0.4889 | 10 |
| peak | 32.0 (19.0–67.75) | 28.5±50.05 | 102.5±7266.58 | <0.0001 | 99.0±114.51 | 289.0±15 305.74 | <0.0495 | 10 | |
| AST, U/L | adm | 24.0 (18.0–36.22) | 23.0±24.3 | 47.0±30.9 | <0.0001 | 46.0±30.35 | 63.0±32.56 | 0.3722 | 10 |
| peak | 25.5 (19.0–44.0) | 24.0±29.8 | 82.5±914.01 | <0.0001 | 79.0±69.77 | 200.0±1715.26 | <0.0009 | 10 | |
| D-dimer, mg/L | adm | 0.4 (0.2–0.6) | 0.3±0.72 | 1.15±3.13 | <0.0001 | 1.1±2.96 | 1.4±3.62 | 0.1638 | 86 |
| peak | 0.4 (0.3–0.7) | 0.3±0.73 | 2.6±7.56 | <0.0001 | 1.6±6.37 | 18.0±7.12 | <0.0001 | 86 | |
| aPTT, s | adm | 37.4 (35.0–41.05) | 37.2±4.65 | 40.0±23.0 | <0.0014 | 39.0±19.65 | 41.0±31.76 | 0.14 | 73 73 |
| peak | 38.0 (35.15–42.35) | 37.4±5.14 | 47.0±44.56 | <0.0001 | 45.0±38.41 | 63.0±54.06 | <0.0005 | ||
| Creatinine, μmol/L | adm | 76.1 (67.0–89.0) | 75.4±27.52 | 80.5±54.62 | 0.0767 | 81.0±50.84 | 76.0±66.53 | 0.4448 | 6 6 |
| peak | 78.0 (67.78–91.0) | 76.2±27.74 | 86.5±98.51 | <0.0001 | 83.0±69.12 | 196.0±130.29 | <0.0003 | ||
| CK, U/L | adm | 106.0 (66.0–173.0) | 99.0±529.25 | 173.0±1168.65 | <0.0001 | 174.0±1278.56 | 152.0±561.74 | 0.2269 | 126 |
| peak | 109.5 (66.75–199.75) | 100.0±536.11 | 391.0±10 621.26 | <0.0001 | 391.0±11 963.38 | 370.0±563.66 | 0.4855 | 125 | |
| CRP, mg/L | adm | 5.8 (1.75–27.0) | 4.2±32.27 | 101.0±105.14 | <0.0001 | 102.0±102.19 | 100.0±115.53 | 0.4367 | 5 5 |
| peak | 6.5 (1.9–50.65) | 4.8±45.93 | 157.5±113.35 | <0.0001 | 143.0±108.72 | 219.0±115.19 | <0.0191 | ||
| LDH, U/L | adm | 192.0 (159.0–264.0) | 181.0±80.08 | 445.0±267.95 | <0.0001 | 432.5±284.01 | 480.0±199.68 | 0.2706 | 95 |
| peak | 194.0 (160.0–280.0) | 182.0±83.76 | 538.0±1232.13 | <0.0001 | 490.5±302.93 | 1925.0±2039.83 | <0.0001 | 95 | |
| Troponin, ng/mL | adm | 0.0 (0.0–0.0) | 0.0±0.15 | 0.0±1.31 | <0.0001 | 0.0±0.04 | 0.0±2.73 | 0.0598 | 135 |
| peak | 0.0 (0.0–0.0) | 0.0±0.18 | 0.04±1.85 | <0.0001 | 0.0±0.26 | 0.36±3.66 | <0.0001 | 135 | |
| Ferritin, ng/mL | adm | 216.7 (84.5–475.5) | 181.95±876.92 | 725.0±2282.55 | <0.0001 | 882.0±2480.17 | 612.0±1214.49 | 0.3036 | 53 |
| peak | 230.0 (89.95–595.5) | 196.5±1530.13 | 2258.0±9784.72 | <0.0001 | 2063.5±4781.9 | 4669.0±15 029.77 | <0.0014 | 53 | |
| Fibrinogen, mg/dL | adm | 396.0 (330.0–529.5) | 377.0±187.31 | 610.0±199.71 | <0.0001 | 612.0±204.96 | 567.0±179.01 | 0.3104 | 153 |
| peak | 405.0 (331.25–554.0) | 380.0±130.61 | 700.0±735.07 | <0.0001 | 701.0±816.38 | 692.0±252.63 | 0.1613 | 153 | |
| Clinical severity | asymp/mild | 431 (76.96%) | 431 (88.32%)* | 0 (0.0%)* | <0.0001 | ||||
| severe | 83 (14.82%) | 54 (11.07%)* | 29 (40.28%)* | 29 (50.88%)* | 0 (0.0%)* | <0.0002 | |||
| critical | 46 (8.21%) | 3 (0.61%)* | 43 (59.72%)* | 28 (49.12%)* | 15 (100.0%)* | ||||
| Ethnicity | White | 60 (10.71%) | 53 (10.86%) | 7 (9.72%) | 7 (12.28%) | 0 (0.0%) | |||
| S. Asians | 244 (43.57%) | 206 (42.21%) | 38 (52.78%) | 28 (49.12%) | 10 (66.67%) | ||||
| M. Easterns | 148 (26.43%) | 136 (27.87%)* | 12 (16.67%)* | 0.1102 | 7 (12.28%) | 5 (33.33%) | <0.0219 | ||
| E. Asians | 94 (16.79%) | 79 (16.19%) | 15 (20.83%) | 15 (26.32%)* | 0 (0.0%)* | ||||
| Others | 14 (2.5%) | 14 (2.87%) | 0 (0.0%) | ||||||
| Onset to hospitalisation days | 14.0 (8.0–19.0) | 12.0±7.07 | 22.0±16.5 | <0.0001 | 21.0±17.72 | 27.5±10.25 | 0.1336 | 72 | |
| Onset to positive PCR days | 2.0 (1.0–5.0) | 2.0±3.89 | 5.0±4.97 | <0.0001 | 5.0±5.01 | 4.0±4.79 | 0.3425 | 72 | |
| High-risk group patients | 41 (7.32%) | 3 (0.61%) | 38 (52.78%) | <0.0001 | 24 (42.11%) | 14 (93.33%) | <0.0003 | ||
| Discharged alive | 545 (97.32%) | 488 (100.0%) | 57 (79.17%) | <0.0001 | 57 (100.0%) | <0.0001 | |||
| Length of stay in clinics | 7.0 (3.0–12.25) | 6.0±8.25 | 16.0±16.08 | <0.0001 | 16.0±17.34 | 23.0±9.97 | 0.1521 | 94 | |
| Duration of viral shedding, days | 10.0 (6.0–14.0) | 10.5±5.64 | 8.0±9.04 | 0.0714 | 8.0±9.05 | 13.0±8.65 | 0.1304 | 28 | |
| Need for supplementary O2 | 82 (14.64%) | 23 (4.71%) | 59 (81.94%) | <0.0001 | 46 (80.7%) | 13 (86.67%) | 0.7229 | ||
| Any complication | 123 (21.96%) | 53 (10.86%) | 70 (97.22%) | <0.0001 | 55 (96.49%) | 15 (100.0%) | 1 | ||
| ARDS | 76 (13.57%) | 7 (1.43%) | 69 (95.83%) | <0.0001 | 54 (94.74%) | 15 (100.0%) | 1 | ||
| Liver dysfunction | 54 (9.64%) | 23 (4.71%) | 31 (43.06%) | <0.0001 | 23 (40.35%) | 8 (53.33%) | 0.3944 | ||
adm, data on admission; ALT, alanine aminotransferase; aPTT, activated partial thromboplastin time; ARDS, acute respiratory distress syndrome; AST, aspartate aminotransferase; asymp, asymptomatic; BMI, body mass index; bpm, beats per minute; CK, creatine kinase; CRP, C reactive protein; DBP, diastolic blood pressure; ICU, intensive care unit; LDH, lactate dehydrogenase; min, the minimal levels; peak, the peak levels; RR, respiratory rate; SBP, systolic blood pressure; SOFA, sequential organ failure assessment; WBC, white blood cell.
Feature ranking with regard to ML model performance
The features of the dataset listed in online supplemental appendix 1 were ranked with four tree-based ML classifiers (eg, Random Forest, AdaBoost, Gradient Boosting and ExtraTrees). Tree-based models provide measures of feature importances. The classifiers are based on the mean decrease in impurity. The impurity is quantified by the splitting criterion of the decision trees. Averaged values of impurity-based attribute ranks were calculated as the mean of rank values for the algorithms (see online supplemental figure 1). The classification performance is seen in online supplemental figure 2.
bmjopen-2020-044500supp005.pdf (105KB, pdf)
bmjopen-2020-044500supp006.pdf (746.6KB, pdf)
The cut-off levels of the laboratory findings
To come up with laboratory data cut-off levels, which may be considered as biomarkers of the severe course of the disease, we trained the NN ML model on each variable separately and assessed their statistical significance against chance performance. We calculated 95% CI for ROC and AUC scores with the bootstrap technique and p values with permutation tests (see table 2).
Table 2.
Statistical significance of ROC AUC for predicting transfer to ICU out of the laboratory findings on admission
| No | Feature | AUC | 95% CI | P value |
| 1 | AST | 0.4882 | (0.399 to 0.595) | 0.828 |
| 2 | ALT | 0.5057 | (0.482 to 0.538) | 0.331 |
| 3 | Total bilirubin | 0.5573 | (0.443 to 0.557) | 0.077 |
| 4 | LDH | 0.5652 | (0.515 to 0.644) | 0.072 |
| 5 | WBC | 0.5727 | (0.427 to 0.573) | 0.035 |
| 6 | Lymphocyte | 0.5881 | (0.474 to 0.588) | 0.01 |
| 7 | Troponin | 0.6088 | (0.5 to 0.609) | 0.008 |
| 8 | D-dimer | 0.6151 | (0.5 to 0.615) | 0.004 |
| 9 | CK | 0.6918 | (0.6 to 0.725) | <0.001 |
| 10 | Ferritin | 0.6973 | (0.616 to 0.74) | <0.001 |
| 11 | aPTT | 0.7534 | (0.219 to 0.755) | <0.001 |
| 12 | Fibrinogen | 0.7704 | (0.718 to 0.771) | <0.001 |
| 13 | CRP | 0.8194 | (0.798 to 0.822) | <0.001 |
| aPTT+CRP+fibrinogen | 0.8618 | (0.486 to 0.884) | <0.001 | |
| All together | 0.9019 | (0.812 to 0.902) | <0.001 | |
ALT, alanine aminotransferase; aPTT, activated partial thromboplastin time; AST, aspartate aminotransferase; CK, creatine kinase; CRP, C reaction protein; ICU, intensive care unit; LDH, lactate dehydrogenase; WBC, white blood cell.
Table 2 shows that there is a notable difference between the performance of the model in terms of ROC AUC and the performance at chance level. High-performance measures were obtained for aPTT, CRP and fibrinogen values (sensitivity and specificity of 0.9877 and 0.4028, respectively). The values increased to 0.9754 and 0.75, respectively, for all 13 significant tests. So we used the performance of the classification model based on the combination of these 3 and 13 features.
First, we trained the ML classification model based on the data taken from only one lab feature using a stratified 10-fold cross-validation technique. Then, we built ROC for the test data of all 10 folds (see diagrams in online supplemental figure 3).
bmjopen-2020-044500supp007.pdf (229.9KB, pdf)
To improve the model’s efficiency and to choose the cut-off value set for some laboratory findings data, we used a threshold moving technique along with a supervised ML classification model (NN).
The ML estimator assigns threshold values for interpreting probabilities. The default threshold returned by the estimator to class labels is 0.5. However, when the dataset is unbalanced, tuning this hyperparameter can improve the model’s efficiency by finding the optimal threshold. This is crucial when the importance of predicting the positive class (admitted to ICU) outweighs true negative predictions. Performance metrics calculated for all laboratory features with regard to the optimal threshold value are presented in table 3. The table displays the sensitivity, specificity and AUC values obtained after applying the threshold moving technique. We marked in bold the AUC values which are higher than the ones displayed in online supplemental figure 3A. The optimal cut-off value returned by the technique is shown in the appropriate column.
Table 3.
Justification of the cut-off levels for the admission values of laboratory findings to predict transferring to ICU
| No. | Feature | Normal values | Threshold moving technique | Percentile level | ||||||
| Cut-off | Sensitivity | Specificity | AUC | Cut-off | Sensitivity | Specificity | AUC* | |||
| 1 | WBC (×109/L) | 4.0–11.0 | 45 | 0.6 | 0.5 | 0.5486 | 7 | 0.5278 | 0.75 | 0.6389 |
| 2 | Lymphocytes (×109/L) | 1–4.8 | 0.3 | 0.43 | 0.62 | 0.5267 | 1.24 | 0.7778 | 0.75 | 0.7639 |
| 3 | Total bilirubin (μmol/L) | 3.4–20.5 | 37 | 0.54 | 0.43 | 0.4880 | 11.9 | 0.4861 | 0.7439 | 0.6150 |
| 4 | ALT (U/L) | 0–55 | 435 | 0.29 | 0.68 | 0.4880 | 43 | 0.4583 | 0.7439 | 0.6011 |
| 5 | AST (U/L) | 5–34 | 400 | 0.53 | 0.46 | 0.4944 | 32 | 0.7639 | 0.7418 | 0.7528 |
| 6 | D-dimer (mg/L) | 0.0–0.5 | 15 | 0.35 | 0.7 | 0.5261 | 0.7 | 0.7222 | 0.7234 | 0.7228 |
| 7 | aPTT (s) | 28.0–40.0 | 180 | 0.57 | 0.71 | 0.6413 | 39.9 | 0.5139 | 0.7336 | 0.6237 |
| 8 | CK (U/L) | 30.0–200.0 | 4808 | 0.54 | 0.63 | 0.5864 | 247 | 0.4028 | 0.6619 | 0.5323 |
| 9 | CRP (mg/L) | 0.0–5.0 | 400 | 0.6 | 0.79 | 0.6921 | 14.3 | 0.9306 | 0.75 | 0.8403 |
| 10 | LDH (U/L) | 125–243 | 1778 | 0.21 | 0.88 | 0.5427 | 246 | 0.8889 | 0.6537 | 0.7713 |
| 11 | Troponin (ng/mL) | <0.03 | 11 | 0.33 | 0.75 | 0.5427 | 0.037 | 0.2361 | 0.7172 | 0.4767 |
| 12 | Ferritin (ng/mL) | 21.8–274.6 | 14 025 | 0.35 | 0.82 | 0.5824 | 498 | 0.6667 | 0.75 | 0.7083 |
| 13 | Fibrinogen (mg/dL) | 200–400 | 3030 | 0.33 | 0.89 | 0.6124 | 446 | 0.8611 | 0.4939 | 0.6774 |
*The AUC values marked in bold are higher than the ones displayed in online supplemental figure 3A.
ALT, alanine aminotransferase; aPTT, activated partial thromboplastin time; AST, aspartate aminotransferase; AUC, area under the curve; CK, creatine kinase; CRP, C reactive protein; LDH, lactate dehydrogenase; WBC, white blood cell.
As per the box plots regarding the laboratory findings values in the ICU versus the non-ICU cohort of patients in figure 2, we decided to check whether the performance of the model is good if we applied thresholds in the following manner. For lymphocyte count, we set the cut-off level to the 25th percentile (values lower than or equal to the chosen level were set to 1 or 0 otherwise). For the other features, we set the thresholds to the 75th percentile (values higher or equal to the cut-off limit were set to 1 or 0 otherwise). The performance of the models with regard to the aforementioned cut-off levels is presented in table 3.
Online supplemental figure 4A shows the performance of the logistic regression model built on the binary data by applying the cut-off level for the threshold moving technique. Online supplemental figure 4 illustrates the same information for the percentile’s cut-off levels.
bmjopen-2020-044500supp008.pdf (229.9KB, pdf)
The performance of the classification models
The applied ML algorithms were trained with stratified 10-fold cross-validation technique. The predictors used are listed in online supplemental table 1. The performance of the classification models such as Gradient Boosting, AdaBoost, ExtraTrees, Random Forest, NN, logistic regression with and without L1 regularisation is presented in online supplemental figure 2, online supplemental table 2. It displays all 560 test points concatenated from test (actual and predicted) label values for each fold. Online supplemental tables 3 and 4 show the performance metrics obtained by the NN model with the highest output quality. Online supplemental figure 3 displays ROC curves and AUC for the NN model with different variables, observed on admission, as predictors. Online supplemental figure 4 illustrates the quality of the performance for the binary data obtained by using the threshold moving or percentile-based heuristic approach.
bmjopen-2020-044500supp001.pdf (105KB, pdf)
bmjopen-2020-044500supp002.pdf (746.6KB, pdf)
bmjopen-2020-044500supp003.pdf (746.6KB, pdf)
bmjopen-2020-044500supp004.pdf (746.6KB, pdf)
Discussion
Severity of the disease course in SARS-CoV-2 infection
There are different risk factors for COVID-19 severity. Finding and justifying them are the issues of the ongoing studies because of the persistence of the viral infection. In research on the severe respiratory illness for COVID-19, the authors justified the age above 65 years as a predictor of clinical outcomes of interest.29 The data we received support this fact. In the same study, the authors showed inconsistent results regarding the race of the patient. In the univariate model, the race was a non-significant predictor of the disease severity, however it turned out to be significant in the multivariate prediction. We did not find ethnic differences between ICU and non-ICU cohorts, but observed a notable difference in the outcome of the disease within these groups (eg, discharged vs deceased patients). According to other studies, age is the largest contributor to risk of death for SARS-CoV-2, the impact of the race or ethnicity on the disease course remains not fully understood. The researchers have difficulty adjusting the samples for comorbidities as physicians did not examine all the patients thoroughly before the disease.30 31 Presumably, the same limitations account for disparities between the studies in which the authors try to consider comorbidities (eg, asthma, diabetes, hypertension and chronic kidney disease) as risk factors. To overcome the limitation, we decided to base the prediction on the laboratory findings on admission. They are standardised and unambiguously interpretable.
Biomarkers of the deterioration of the patients
It is common sense that people with unmanaged chronic conditions are more vulnerable to severe outcomes. High-sensitive laboratory findings are a reliable tool for assessing pathologies of these kinds. Reasonably, these findings may serve as predictors of the disease progression.
As it comes from feature selection, LDH activity is the laboratory finding that has maximal informative value for the prediction of worsening of the patient (see online supplemental table 1). This keeps up with the results of a pooled analysis that show an association of elevated LDH values with a sixfold increase in odds of developing severe disease. Notably, the LDH cut-off in the included studies ranged from 240 to 253.2 U/L. The threshold value for the LDH activity in our study is 246 U/L, which is close to the median of the range.4 It is also known to be a predictor of worse outcomes in inpatients.32 In our study, LDH is the top rank predictor of disease severity, CK levels have a medium informativeness. Both of them are non-specific biomarkers of energy deficiency and hypoxia. The levels of CRP have an expectedly high predictive value as they reflect the activity of an inflammatory process.
The concentration of D-dimer seems to be a more promising biomarker of COVID-19 severity because of the endothelial dysfunction mechanism which is specific for this viral infection (see ‘Data used by clinicians for stratifying risks’ subsection). For the same reason, aPTT is an interesting predictor for SARS-CoV-2-infected patients. Therefore, recent studies justified the coagulation indicators on admission (eg, D-dimer, aPTT, prothrombin time and fibrinogen) as significant indicators of severe course of COVID-19.33
Online supplemental table 1 shows that fibrinogen values are not predictive of disease severity. The explanation to this discrepancy is many missing values for this indicator in our database. As it is seen from table 1, the total number of 153 cases (27%) were missing. We had to replace them with the mean values to perform the multivariate prediction with the tree-based model. The replacement decreased the real prognostic value, which was expected to be high. In contrast to this, the univariate model based on fibrinogen levels had the best classifying metrics compared with other predictors. Its ROC AUC value is 0.7704 (see table 2).
Threshold criteria for the major clinical data
With the ML approach, we justify the cut-off thresholds for the major laboratory tests regularly done on admission.
The disproportion in the number of patients admitted to ICU versus non-severe cases was challenging. Therefore, we customised the ML algorithms in terms of threshold values used to predict worsening. For each laboratory findings feature, we (1) fit the model to the training dataset using 10-fold cross-validation technique, (2) predicted the probabilities on the test dataset, (3) found the optimal threshold value which maximises the ROC AUC measure.
The optimised threshold values (marked in bold in table 3) can be used to predict the supposed deterioration of the patient from the initial findings at presentation. Some of the thresholds are close to the normal reference values, but not completely. For instance, the cut-off for CRP is 3 times bigger than the top reference value. The cut-offs that we found for WBC and total bilirubin are within the range of normal values for these laboratory findings. That is why it is challenging to interpret them.
The prediction based on CRP with ROC AUC equal to 0.8403 proved to be most accurate. A meta-analysis done by other authors showed the possibility to predict mortality for COVID-19 out of CRP with the same level of accuracy (ROC AUC 0.84).17 Unfortunately, they did not state clearly the time point for collecting the samples.
In our study, the performance of the disease severity prediction based on the coagulation indicators was not so high (eg, D-dimer 0.7228; fibrinogen 0.6774). However, it almost equals the results of ROC analyses for mortality risk by other authors who received AUCs value of 0.742 for D-dimer on admission and 0.643 for aPTT on admission.33 Other authors reached even better performance for the prediction of in-hospital mortality based on D-dimer on admission (AUC 0.85).
Despite the similarities in performance metrics, the studies cannot be compared as they are based on different inclusion criteria, study cohorts and threshold values found. In general, our findings support the idea of other researchers to use laboratory findings on admission for risk stratification. Moreover, they encourage the further studies to implement new biomarkers into prognostic models along with the proven ones.17
The multivariable prediction of the severity of COVID-19
For better prediction, it is recommended that several biomarkers are analysed concomitantly. A combination of 3 and 13 most valuable ones, if fed to the deployed ML algorithm, provides a reliable prognosis. Online supplemental figure 2 clearly shows that there is a separability pattern within all variables used to build the predictive model. When we rank the features in accordance with their importance, most laboratory findings variables are listed at the top (see online supplemental table 1). It also helps to justify the threshold values, presented in this study.
Limitations
There are several limitations in the current study. First, the dataset is unbalanced. Therefore, we customised the supervised ML algorithm in terms of the threshold value used to predict worsening. Second, the severity and mortality of the included patients might not be representative of the community because of the latent course of the mild and asymptomatic cases. Third, the population of Dubai is specific in terms of unequal age distribution and ethnic heterogeneity. However, one may consider the last feature as a strength because we can generalise the results to the world population. Fourth, although other clinical examinations (eg, diagnostic imaging) could provide additional information, we limited the predictors of disease deterioration to laboratory findings. Nonetheless, this was enough to build up an ML algorithm with good performance. The concomitant analysis of the top three valuable biomarkers on admission provided a reliable prognosis without radiological predictors. Another advantage of the choice we made is the high applicability of study results into practice. The justified cut-off thresholds for the laboratory tests are easy to use on admission to the hospital.
Conclusion
By comparing the data for the patients who were transported to ICU with those who did not worsen throughout the hospitalisation, we selected a set of laboratory findings with the significant differences on admission to the clinics. The variables were used as the predictors to build up the classification model. The performance of the models was low, with the default thresholds returned by the ML estimator, we improved it by setting the cut-off level to the 25th percentile for lymphocyte count and the 75th percentile for other features.
To distinguish the patients with confirmed COVID-19 who may worsen while treated, we justified the following threshold values of the laboratory tests done on admission: lymphocyte count <2.59×109/L, and the upper levels for total bilirubin 11.9 μmol/L, ALT 43 U/L, AST 32 U/L, D-dimer 0.7 mg/L, aPTT 39.9 s, CK 247 U/L, CRP 14.3 mg/L, LDH 246 U/L, troponin 0.037 ng/mL, ferritin 498 ng/mL and fibrinogen 446 mg/dL.
The performance of the neural network to predict the future deterioration out of the top three valuable tests (aPTT, CRP and fibrinogen) is admissible (AUC 0.86; 95% CI 0.486 to 0.884; p<0.001). It is comparable with the model trained with all the tests (AUC 0.90; 95% CI 0.812 to 0.902; p<0.001).
Supplementary Material
Acknowledgments
The authors would like to thank the UAE University (Al Ain, UAE) and Mediclinic Parkview Hospital (Dubai, UAE) for providing support and for allowing to use the facilities for conducting this research. The authors would also like to thank the healthcare staff and the patients for their dedication and commitment to this research.
Footnotes
Twitter: @StatsenkoE
Contributors: All authors contributed to the creation of the article as follows: all of them contributed to the conceptual idea of the paper equally; FAZ and YS formulated the objectives; FAZ collected the dataset; YS wrote the manuscript; TH proposed the methodology of the study, and performed the statistical analysis, prepared the figures and tables for data presentation and illustration; FAZ, TH, KN-VG and NZ contributed to the literature review and data analysis. The data were analysed and interpreted by the authors, who also reviewed the manuscript and vouch for the accuracy and completeness of the data and for the adherence of the study to the protocol.
Funding: The authors have not declared a specific grant for this research from any funding agency in the public, commercial or not-for-profit sectors.
Competing interests: None declared.
Patient consent for publication: Not required.
Ethics approval: The study got an ethical review by Dubai Scientific Research Ethics Committee (DSREC), Dubai Health Authority (protocol no. DSREC-05/2020_25) and was approved for the retrospective analysis of the data obtained as a standard of care. No potentially identifiable personal information is presented in the study.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data availability statement: Data are available on reasonable request. The datasets generated for this study are available on request at Data Analytics Group website: https://bi-dac.com. To assess the risk of having complications in a patient with COVID-19, one may use the ML-based free online tool at https://med-predict.com, which illustrates the results of the current study.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
References
- 1.Yan L, Zhang H-T, Goncalves J, et al. An interpretable mortality prediction model for COVID-19 patients. Nat Mach Intell 2020;2:283–8. 10.1038/s42256-020-0180-7 [DOI] [Google Scholar]
- 2.Wynants L, Van Calster B, Collins GS, Bonten MM, et al. Prediction models for diagnosis and prognosis of covid-19 infection: systematic review and critical appraisal. BMJ 2020;369:m1328. 10.1136/bmj.m1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gao Y, Li T, Han M, et al. Diagnostic utility of clinical laboratory data determinations for patients with the severe COVID-19. J Med Virol 2020;92:791–6. 10.1002/jmv.25770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Henry BM, Aggarwal G, Wong J, et al. Lactate dehydrogenase levels predict coronavirus disease 2019 (COVID-19) severity and mortality: a pooled analysis. Am J Emerg Med 2020;38:1722–6. 10.1016/j.ajem.2020.05.073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou R, Li F, Chen F, et al. Viral dynamics in asymptomatic patients with COVID-19. Int J Infect Dis 2020;96:288–90. 10.1016/j.ijid.2020.05.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gong J, Ou J, Qiu X, et al. A tool for early prediction of severe coronavirus disease 2019 (COVID-19): a multicenter study using the risk nomogram in Wuhan and Guangdong, China. Clin Infect Dis 2020;71:833–40. 10.1093/cid/ciaa443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zheng H-Y, Zhang M, Yang C-X, et al. Elevated exhaustion levels and reduced functional diversity of T cells in peripheral blood may predict severe progression in COVID-19 patients. Cell Mol Immunol 2020;17:541–3. 10.1038/s41423-020-0401-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen X, Zhao B, Qu Y, et al. Detectable serum severe acute respiratory syndrome coronavirus 2 viral load (RNAemia) is closely correlated with drastically elevated interleukin 6 level in critically ill patients with coronavirus disease 2019. Clin Infect Dis 2020;71:1937–42. 10.1093/cid/ciaa449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mahallawi WH, Khabour OF, Zhang Q, et al. Mers-Cov infection in humans is associated with a pro-inflammatory Th1 and Th17 cytokine profile. Cytokine 2018;104:8–13. 10.1016/j.cyto.2018.01.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Selinger C, Tisoncik-Go J, Menachery VD, et al. Cytokine systems approach demonstrates differences in innate and pro-inflammatory host responses between genetically distinct MERS-CoV isolates. BMC Genomics 2014;15:1161. 10.1186/1471-2164-15-1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kappert K, Jahić A, Tauber R. Assessment of serum ferritin as a biomarker in COVID-19: bystander or participant? insights by comparison with other infectious and non-infectious diseases. Biomarkers 2020;25:616–25. 10.1080/1354750X.2020.1797880 [DOI] [PubMed] [Google Scholar]
- 12.Spiezia L, Boscolo A, Poletto F, et al. Covid-19-related severe hypercoagulability in patients admitted to intensive care unit for acute respiratory failure. Thromb Haemost 2020;120:998. 10.1055/s-0040-1710018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang N, Li D, Wang X, et al. Abnormal coagulation parameters are associated with poor prognosis in patients with novel coronavirus pneumonia. Journal of Thrombosis and Haemostasis 2020;18:844–7. 10.1111/jth.14768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Griffin DO, Jensen A, Khan M, et al. Pulmonary embolism and increased levels of D-dimer in patients with coronavirus disease. Emerg Infect Dis 2020;26:1941–3. 10.3201/eid2608.201477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Léonard-Lorant I, Delabranche X, Séverac F, et al. Acute pulmonary embolism in patients with COVID-19 at CT angiography and relationship to D-dimer levels. Radiology 2020;296:E189–91. 10.1148/radiol.2020201561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lippi G, Favaloro EJ. D-Dimer is associated with severity of coronavirus disease 2019: a pooled analysis. Thromb Haemost 2020;120:876. 10.1055/s-0040-1709650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang I, Pranata R, Lim MA, et al. C-Reactive protein, procalcitonin, D-dimer, and ferritin in severe coronavirus disease-2019: a meta-analysis. Ther Adv Respir Dis 2020;14:1753466620937175. 10.1177/1753466620937175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yao Y, Cao J, Wang Q, et al. D-Dimer as a biomarker for disease severity and mortality in COVID-19 patients: a case control study. J Intensive Care 2020;8:1–11. 10.1186/s40560-020-00466-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang N, Bai H, Chen X, et al. Anticoagulant treatment is associated with decreased mortality in severe coronavirus disease 2019 patients with coagulopathy. J Thromb Haemost 2020;18:1094–9. 10.1111/jth.14817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang D, Hu B, Hu C, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel Coronavirus–Infected pneumonia in Wuhan, China. JAMA 2020;323:1061–9. 10.1001/jama.2020.1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bashash D, Abolghasemi H, Salari S. Elevation of D-dimer, but not Pt and aPTT, reflects the progression of covid-19 toward an unfavorable outcome: a meta-analysis. Iranian Journal of Blood & Cancer 2020;12:47–53 http://ijbc.ir/article-1-1005-en.html [Google Scholar]
- 22.Quartuccio L, Sonaglia A, McGonagle D, et al. Profiling COVID-19 pneumonia progressing into the cytokine storm syndrome: results from a single Italian centre study on tocilizumab versus standard of care. J Clin Virol 2020;129:104444. 10.1016/j.jcv.2020.104444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang L, Ling W. C-Reactive protein levels in the early stage of COVID-19. Médecine et Maladies Infectieuses 2020;50:332–4. 10.1016/j.medmal.2020.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bloom PP, Meyerowitz EA, Reinus Z, et al. Liver Biochemistries in hospitalized patients with COVID‐19. Hepatology 2020;21. 10.1002/hep.31326 [DOI] [PubMed] [Google Scholar]
- 25.Ali N, Hossain K. Liver injury in severe COVID-19 infection: current insights and challenges. Expert Rev Gastroenterol Hepatol 2020;14:879–84. 10.1080/17474124.2020.1794812 [DOI] [PubMed] [Google Scholar]
- 26.Lee N, Chan PKS, Hui DSC, et al. Viral loads and duration of viral shedding in adult patients hospitalized with influenza. J Infect Dis 2009;200:492–500. 10.1086/600383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.National Emergency Crisis and Disasters Management Authority, . National guidelines for clinical management and treatment of covid-19- version 4.1, 2020. Available: https://www.dha.gov.ae/en/HealthRegulation/Documents/National_Guidelines_of_COVID_19_1st_June_2020.pdf
- 28.Fernández A, García S, Galar M. Herrera learning from imbalanced data sets. New York: Springer, 2018. [Google Scholar]
- 29.Robilotti EV, Babady NE, Mead PA, et al. Determinants of COVID-19 disease severity in patients with cancer. Nat Med 2020;26:1218–23. 10.1038/s41591-020-0979-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ravi K. Ethnic disparities in COVID-19 mortality: are comorbidities to blame? The Lancet 2020;396:22. 10.1016/S0140-6736(20)31423-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baqui P, Bica I, Marra V, et al. Ethnic and regional variations in hospital mortality from COVID-19 in Brazil: a cross-sectional observational study. The Lancet Global Health 2020;8:e1018–26. 10.1016/S2214-109X(20)30285-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Erez A, Shental O, Tchebiner JZ, et al. Diagnostic and prognostic value of very high serum lactate dehydrogenase in admitted medical patients. Isr Med As- soc J 2014;16:439–43. [PubMed] [Google Scholar]
- 33.Long H, Nie L, Xiang X, et al. D-Dimer and prothrombin time are the significant indicators of severe covid-19 and poor prognosis. Biomed Res Int 2020;2020:1–10. 10.1155/2020/6159720 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
bmjopen-2020-044500supp009.pdf (1.8MB, pdf)
bmjopen-2020-044500supp005.pdf (105KB, pdf)
bmjopen-2020-044500supp006.pdf (746.6KB, pdf)
bmjopen-2020-044500supp007.pdf (229.9KB, pdf)
bmjopen-2020-044500supp008.pdf (229.9KB, pdf)
bmjopen-2020-044500supp001.pdf (105KB, pdf)
bmjopen-2020-044500supp002.pdf (746.6KB, pdf)
bmjopen-2020-044500supp003.pdf (746.6KB, pdf)
bmjopen-2020-044500supp004.pdf (746.6KB, pdf)