Abstract
Background
Despite the emerging insights into many snoRNAs (small nucleolar RNAs) which are detectable in body fluids and serve as noninvasive biomarkers, few studies have previously discussed the role of snoRNAs in tumor‐educated platelets (TEPs). Herein, we systematically estimated dysregulation of snoRNAs in non‐small cell lung cancer (NSCLC) and clarified the biomarker potential of SNORD55 in platelets.
Methods
We compared expression of snoRNAs between NSCLC and normal tissues using SNORic datasets. Platelets were isolated from plasma using low‐speed centrifugation and subjected to quantitative polymerase chain reaction (qPCR) for SNORD55 detection.
Results
SNORD55 was significantly decreased in TEPs from NSCLC patients especially in early‐stage patients compared with healthy controls. Importantly, we validated that TEP SNORD55 was capable of acting as a promising biomarker for NSCLC. It exerted diagnostic performance for NSCLC diagnosis, possessing an AUC of 0.803, as well as for early NSCLC diagnosis, possessing an AUC of 0.784. Moreover, the combination of TEP SNORD55 and carcinoembryonic antigen (CEA) improved the diagnostic efficiency of cancer progression. In addition, TEP SNORD55 also potentially acts as a noninvasive early biomarker for lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC) with favorable diagnostic efficiencies.
Conclusions
In summary, TEP SNORD55 could potentially serve as a noninvasive biomarker for NSCLC diagnosis and early diagnosis.
Key points
SNORD55 was significantly decreased in TEPs from NSCLC patients compared to healthy controls and acted as a novel biomarker for early NSCLC.
Keywords: biomarker, diagnosis, SNORD55, snoRNA, TEP
Our findings reveal cancer‐specific changes of SNORD55 in platelets of NSCLC, which paving the way for the exploitation of sensitive and specific NSCLC diagnostic biomarker either alone or in combination, providing certain basis for clinical projects of TEP SNORD55 in early screening of NSCLC.
![]()
INTRODUCTION
Non‐small cell lung cancer (NSCLC) has the highest incidence and ranks first in cancer‐related mortality worldwide. 1 Despite the continuous improvements in standard treatment and low‐dose computed tomography (CT) lung imaging, NSCLC patients are extremely vulnerable to relapse and mortality as most patients have local or distant metastasis at the time of diagnosis 2 due to no obvious symptoms exhibiting at early stage. 3 Therefore, there is an urgent need to discover sensitive, specific, especially noninvasive biomarkers for early detection of NSCLC.
Platelets are small anucleate cells generated from megakaryocytes (MKs), residing in bone marrow or intravascular sites in the lung, 4 traditionally emerging as central players in hemostasis and initiation of wound healing. 5 More recently, platelets have emerged as active players in all steps of tumorigenesis including tumor growth, tumor cell extravasation, and metastasis, thereby sequestering tumor‐specified biomolecules including RNA transcripts and proteins, as well as altering their spliced RNA profiles, called tumor‐educated platelets (TEPs). 6 Emerging data suggests that TEPs may provide abundant biosources for diagnostics and prognostic of multiple kinds of cancer. 6 Differential TEP RNA profiles have been identified in healthy individuals and glioma patients, forming an attractive platform for the companion diagnostics of cancer. 7 In addition, TEP RNA profiles have been reported to precisely pinpoint the primary origin of pan‐cancer, as well as predict the oncogenic status of many tumors including MET or HER2 positivity and the existence of KRAS, EGFR, or PIK3CA mutations. 8 Due to their lifespan of 7–10 days and the structure of platelet membrane, tumor‐specified biosources and biomolecules are enriched in TEPs and protected from circulating RNAs, and thus TEP RNA analysis is capable of reflecting tumor bioactivity up‐to‐date, intensively, and dynamically.
SnoRNAs, a family of conserved nuclear RNAs (60–300 nt) widely existing in Cajal bodies or nucleolus of eukaryotic cells, 9 are divided into two classes: C/D box snoRNAs (SNORDs) and H/ACA box snoRNAs (SNORAs). SNORD guide−2′‐O‐ribose methylation, and SNORA direct the pseudouridylation of NTs. 10 , 11 More recently, no specific RNA:RNA duplex to ribosomal was observed in numerous SNORDs, suggesting they have more functions other than traditional 2′‐O‐methylation. 12 SNORD27 was reported to regulate the alternative splicing of E2F7 pre‐mRNA through by competing with small nuclear ribonucleoprotein (snRNP) without methylating the RNA 13 ; SNORD115 presented sequence complementarity and bound to splice factors, then influenced alternative splicing of the serotonin receptor 2C (HTR2C) pre‐mRNA. 14 Interestingly, snoRNAs have also been detected within anucleated platelets. 15 The presence of both HTR2C pre‐mRNA and splicing factors in platelets might indicate the snoRNAs in platelets mediate alternative splicing. 16
Small nuclear RNAs (snRNAs) of the spliceosome are the basal factors and present in platelets, as they are required for splicing. 17 In our previous study, we reported snRNAs (U1, U2, U5) in TEPs are downregulated and act as promising biomarkers in lung cancer, indicating their contribution to the specific spliced mRNA signature in TEPs. 18 In the anucleated platelets, snoRNAs might more likely regulate the alternative splicing of pre‐mRNA by competing with snRNP other than direct modification to snRNA. 19 Therefore, we hypothesized that aberration in the expression of snoRNAs might exist in lung cancer, responsible for alteration of TEP mRNA profiles, exerting the potential as a biomarker for lung cancer diagnostics.
In the present study, we processed a series of bioinformatic analysis for snoRNA expression across NSCLC datasets, and identified snoRNA involved in oncogenesis. Furthermore, we demonstrated that SNORD55 in platelets was significantly downregulated in NSCLC patients compared with healthy controls, and confirmed it as a TEP‐based diagnostic biomarker in a large NSCLC cohort including early‐stage patients.
METHODS
Public database
The clinical information of LUAD and LUSC tumor and paired adjacent tissue samples were provided by The Cancer Genome Atlas of the National Cancer Institute (TCGA, http://cancergenome.nih.gov). The downloaded clinical information included TNM stage and so on for 522 LUAD patients and 504 LUSC patients. SNORic (http://bioinfo.life.hust.edu.cn/SNORic) was used to examine the expression of SNORD55 for patients with LUAD (513 tumor tissues and 46 adjacent normal tissues included) and LUSC (476 tumor tissues and 45 adjacent normal tissues included).
Patients and clinical samples
A total of 290 NSCLC patients admitted to Shandong Cancer Hospital and Institute between January 2019 and December 2019, as well as 105 healthy volunteers from the above hospital and 84 from Shandong Provincial Third Hospital excluded from any malignant tumor after examination, were enrolled in current study (Tables S1 and S2). Written informed consent was obtained from all individual participants. For all patients, plasma was collected at diagnosis prior to surgery or any antitumor treatment.
Platelet isolation and RNA extraction
The platelets were isolated using centrifugation as previously described. 20 Plasma samples were collected in EDTAK2‐coated purple‐cap vacutainer tubes (BD) and then centrifuged twice for 10 min at 120 × g to remove cells and debris. The platelet‐rich plasma (PRP) was centrifuged at 360 × g for 20 min to pellet platelets at room temperature. For platelet RNA isolation, total RNA was extracted using TRIzol reagent (Thermo Fisher Scientific, Carlsbad, CA, USA), and stored at −80°C until use.
Liu staining
To assess the quality of RNA, freshly PRP and PBS suspension of platelet samples were subjected to Liu staining and morphological analysis. The sample was added to microscope slides, and the blood smear processed. Briefly, Liu A solution was dropped and covered the whole specimen for 1 min, then Liu B solution was added and fully mixed with A solution for another 3 min, followed by washing stream with water. The smears were observed under an optical microscope.
Reverse transcription and quantification by real‐time PCR
Total RNA was extracted with the TRIzol reagent and reverse‐transcribed into complementary DNA (cDNA) using the Takara PrimeScript RT reagent Kit (TaKaRa Bio) in 20 μl reaction according to the manufacturer's instructions. and then subjected to qPCR using LightCycler 480 qPCR system (Roche Diagnostics). qPCR reactions were carried out using TB‐Green Premix Ex Taq II Reagent (TaKaRa Bio). The SNORD55 primer: 5′‐GACAACTCGGTAATGCTGCATACTC‐3′ (forward) and 5′‐GCTCTCCAAGGTTGGCTTCCC‐3′ (reverse). U6 acted as control, the sequences were listed as follows: 5′‐TGGAACGCTTCACGAATTTGCG‐3′ (forward) and 5′‐GGAACGATACAGAGAAGATTAGC‐3′ (reverse). The relative expression of SNORD55 was evaluated by the comparative cycle threshold (ΔCt) method: (ΔCt = Ct SNORD55–Ct U6) as described previously. 21
Statistical analysis
The statistical analyses were carried out using GraphPad Prism version 6.0 (GraphPad Software, San Diego, CA, USA) and SPSS 22.0 (SPSS Inc., Chicago, IL, US). A Chi‐square test was used to assess the statistical significance between groups, and multigroup analysis was tested by Kruskal‐Wallis test. Receiving operating characteristic (ROC) curve was performed to estimate diagnostic efficiency of the snoRNA signature. All the values were represented as mean ± interquartile range and p‐values < 0.05 were considered statistically significant, and all tests were set as double‐tailed.
RESULTS
Screening differential snoRNA from database
A total of 15 differential snoRNAs were selected based on p < 0.001 and absolute log2 (fold change) >1.5 in LUAD set and LUSC set download from snoRNA dataset (http://bioinfo.life.hust.edu.cn/SNORic), respectively. The heatmaps and volcano plots were constructed to show the expression patterns of these snoRNAs and to filter the differential transcripts for the NSCLC patient group (Figure 1a‐d).
FIGURE 1.
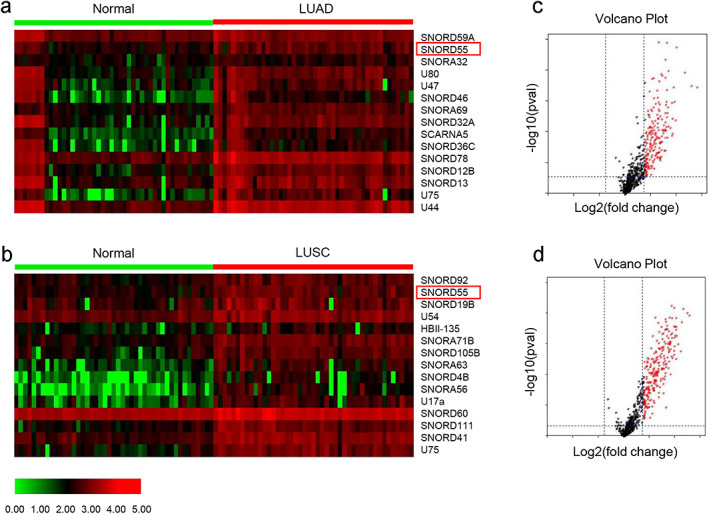
Screening differential small nucleolar RNA (snoRNA) from database. (a, b) Heat maps showed the differential snoRNAs in (a) lung adenocarcinoma (LUAD); and (b) lung squamous cell carcinoma (LUSC), respectively. The red boxes labeled the selected SNORD55. (c, d) Volcano plots compared the expression fold‐change of snoRNAs in LUAD (c) and LUSC (d). The red dots represent the upregulated snoRNAs. LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma
We first examined the expression of SNORD55 for LUAD including 513 tumor tissues and 46 adjacent normal tissues, as well as LUSC patients which including 476 tumor tissues and 45 adjacent normal tissues from SNORic. As shown in Figure S1a–d, we found that SNORD55 was significantly increased expression in LUAD and LUSC tissues as well as with early‐stage tissues than in paracancerous tissues, implying its role in tumorigenesis and development of NSCLC.
TEP SNORD55 downregulated in NSCLC
To value the isolated platelet quality, isolated platelets were stained with Liu staining. As shown in Figure S2, neither red nor white blood cells were observed in PRP and PBS suspension of platelets, consistent with the reports in the literature that a high purity platelet preparation was determined by a ratio of <5 nucleated cells per 10 million platelets. 8 To further determine the differential expression of SNORD55 in TEPs, 189 healthy donors and 290 initial NSCLC patients (those diagnosed with NSCLC before receiving any anticancer treatment) were enrolled, TEPs were then collected and subjected to qPCR analysis. As shown in Figure 2a, the expression of TEP SNORD55 was significantly decreased in NSCLC patients (p < 0.0001, Mann–Whitney test) compared with that in healthy controls. Furthermore, we analyzed the differential expression of TEP SNORD55 between 91 early‐stage NSCLC patients (Tis stage = 9, stage I = 58, stage II = 24) and 189 healthy donors. Consistently, TEP SNORD55 was also dramatically decreased in early‐stage NSCLC patients (p < 0.0001, Mann–Whitney test) compared with those in healthy subjects (Figure 2b). Notably, SNORD55 was upregulated in tumor tissue but downregulated in the TEPs of NSCLC compared with that in the control group, implying its different expression and function pattern.
FIGURE 2.
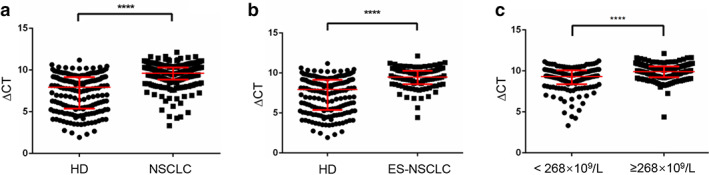
Tumor‐educated platelet (TEP) SNORD55 was downregulated in non‐small cell lung cancer (NSCLC). (a) The expression of TEP SNORD55 was downregulated in NSCLC patients (n = 290) compared with healthy controls (n = 189). (b) The expression of SNORD55 was downregulated in early‐stage NSCLC patients (n = 91) compared with healthy controls (n = 189). (c) TEP SNORD55 expression was downregulated in patients with high platelet counts compared to patients with low platelet counts. ES‐NSCLC, early‐stage non‐small cell lung cancer patients; HD, healthy donors; NSCLC, non‐small cell lung cancer patients. ****p < 0.0001
The relationship between TEP SNORD55 and clinicopathological characteristics of NSCLC patients is shown in Table 1. Thrombocytosis is an important index for the pathological diagnosis and prognosis of NSCLC, 22 thus NSCLC patients were divided into high (n = 146) and low (n = 143) level groups based on their median value 268 × 109/L of platelet counts. As shown in Figure 2c, SNORD55 was dramatically downregulated in the patients with higher platelet counts (p < 0.0001, Mann–Whitney test). Therefore, we selected TEP SNORD55 as a potential biomarker for diagnosis and early diagnosis of NSCLC.
TABLE 1.
Correlation between SNORD55 expression and clinicopathological characteristics of NSCLC patients
| Characteristics | Cases No. (%) | Expression of SNORD55 | |||
|---|---|---|---|---|---|
| Low, No. cases | High, No. cases | p‐value* | |||
| Gender | Male | 184 (63.4) | 85 | 99 | 0.088 |
| Female | 106 (36.6) | 60 | 46 | ||
| Age (year) | ≥62 | 144 (49.7) | 74 | 70 | 0.639 |
| <62 | 146 (50.3) | 71 | 75 | ||
| Smoking | No | 164 (56.6) | 87 | 77 | 0.236 |
| Yes | 126 (43.4) | 58 | 68 | ||
| Drinking | No | 202 (69.7) | 104 | 98 | 0.443 |
| Yes | 88 (30.3) | 41 | 47 | ||
| Histology (NSCLC) | LUAD | 204 (70.3) | 109 | 95 | 0.092 |
| LUSC | 76 (26.2) | 32 | 44 | ||
| Others | 10 (3.4) | 4 | 6 | ||
| Tumor size | <6.292 cm3 | 113 (39.0) | 63 | 50 | 0.206 |
| ≥6.292 cm3 | 114 (39.3) | 54 | 60 | ||
| Unknown | 63 (21.7) | ||||
| Clinical stage | Stage 0 | 9 (3.1) | 6 | 3 | 0.375 |
| Stage I | 58 (20.0) | 30 | 28 | ||
| Stage II | 24 (8.3) | 11 | 13 | ||
| Stage III | 60 (20.7) | 24 | 36 | ||
| Stage IV | 135 (46.6) | 72 | 63 | ||
| Unknown | 4 (1.4) | ||||
| T stage | Tis | 9 (3.1) | 6 | 3 | 0.616 |
| T1 | 79 (27.2) | 44 | 35 | ||
| T2 | 98 (33.8) | 45 | 53 | ||
| T3 | 34 (11.7) | 17 | 17 | ||
| T4 | 57 (19.7) | 28 | 29 | ||
| Unknown | 13 (4.5) | ||||
| Lymph node metastasis | No | 97 (33.4) | 51 | 46 | 0.65 |
| Yes | 183 (63.1) | 91 | 92 | ||
| Unknown | 10 (3.4) | ||||
| Distant metastasis | No | 152 (52.4) | 72 | 80 | 0.313 |
| Yes | 135 (46.6) | 72 | 63 | ||
| Unknown | 3 (1.0) | ||||
Abbreviations: LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; NSCLC, non‐small cell lung cancer.
Chi‐square test was used.
TEP SNORD55 as biomarker for NSCLC diagnosis and early diagnosis
To explore the potential of TEP SNORD55 as a circulating diagnostic marker for NSCLC, a ROC curve was plotted as shown in Figure 3a, and excellent separation between the groups of patients and controls was observed. The AUC of TEP SNORD55 was 0.803 with 79.3% sensitivity and 68.3% specificity. Furthermore, the corresponding ROC curves revealed that the SNORD55 expression was able to discriminate early‐stage NSCLC patients from healthy controls. As shown in Figure 3b, the AUC of SNORD55 was 0.784 with 91.2% sensitivity and 49.7% specificity. Taken together, our data supported that TEP SNORD55 could serve as a promising biomarker for NSCLC diagnosis and early diagnosis.
FIGURE 3.
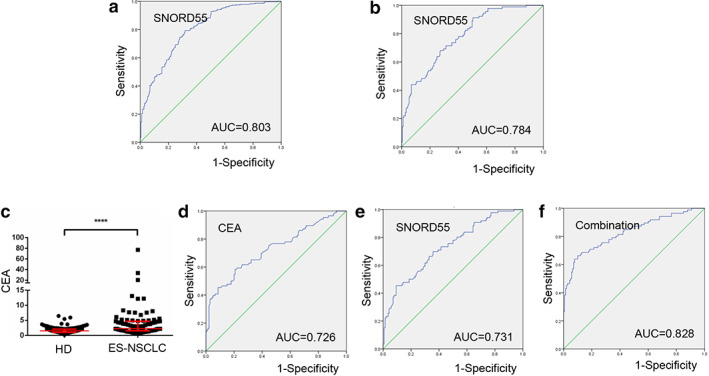
Tumor‐educated platelet (TEP) SNORD55 as a biomarker for non‐small cell lung cancer (NSCLC) diagnosis and early diagnosis. (a) The receiver operating characteristic (ROC) curve analysis of SNORD55 for NSCLC. (b) The ROC curve analysis of NSCLC for early‐stage lung cancer. (c) The levels of carcinoembryonic antigen (CEA) were increased in early‐stage NSCLC patients (n = 86) compared with healthy controls (n = 140). (d–f) The ROC curve analysis of CEA (AUC = 0.726), SNORD55 (AUC = 0.731), and the combination (AUC = 0.828) for early‐stage NSCLC. AUC, area under the curve; CEA, carcinoembryonic antigen; ES‐NSCLC, early‐stage NSCLC patients; HD, healthy donors. ****p < 0.0001
CEA has been shown to be important biomarker in diagnosis prediction of NSCLC, but processes the poor clinical diagnosis efficiency at an early stage of cancer development. 23 We retrospectively included 86 early‐stage NSCLC patients and 140 healthy donors with available CEA levels. As shown in Figure 3c,d, the levels of CEA were highly increased in early‐stage NSCLC patients (p < 0.0001, Mann–Whitney test), and exerted the AUC of 0.726 with 58.1% sensitivity and 79.3% specificity for early NSCLC diagnostics. More importantly, the combination of TEP SNORD55 and CEA improved the diagnostic efficiency of cancer progression, the AUC was 0.828 with 66.3% sensitivity and 90.0% specificity, higher than that for TEP SNORD55 (Figure 3e) and CEA alone (Figure 3f). These data provided compelling evidences that TEP SNORD55 acted as relatively high diagnostic accuracies for early NSCLC.
SNORD55 functions as a diagnostic biomarker in early LUAD and LUSC
LUAD and LUSC are the main subtypes of NSCLC and commonly diagnosed at an advanced stage. 24 Thus, an early diagnosis appears to be a promising measure for improving the prognosis of patients with LUAD and LUSC. First, a Mann–Whitney test was performed to analyze the differential expression between LUAD patients and healthy controls. TEP SNORD55 was significantly decreased in LUAD patients (n = 204, p < 0.0001) (Figure 4a), as well as in 68 early‐stage LUAD patients (Tis stage = 9, stage I = 48, stage II = 11, p < 0.0001) compared with that in healthy controls (n = 189, Figure 4b). As shown in Figure 4c,d, the diagnostic performance of LUAD and early LUAD was also calculated, the AUCs of TEP SNORD55 were 0.791 with 77.9% sensitivity and 68.3% specificity, as well as 0.759 with 89.7% sensitivity and 49.7% specificity, respectively.
FIGURE 4.
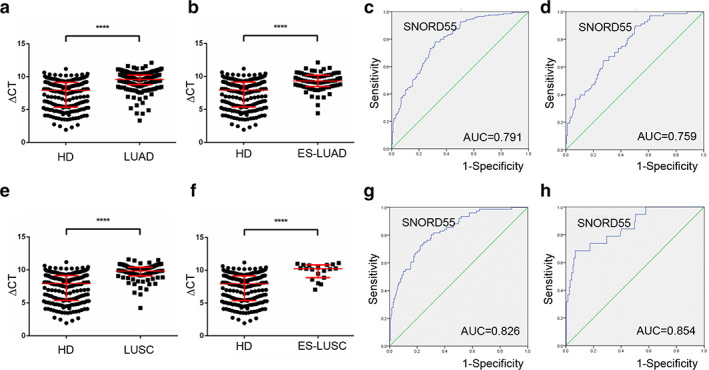
SNORD55 functions as a diagnostic biomarker in early lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC). (a, b) Tumor‐educated platelet (TEP) SNORD55 was downregulated in 204 LUAD patients as well as in 68 early‐stage LUAD patients compared with healthy controls (n = 189). (c) The receiver operating characteristic (ROC) curve analysis of SNORD55 for LUAD. (d) The ROC curve analysis of non‐small cell lung cancer (NSCLC) for early‐stage LUAD. (e,f) TEP SNORD55 was downregulated in 76 LUSC patients as well as in 19 early‐stage LUSC patients compared with healthy controls. (g) The ROC curve analysis of SNORD55 for LUSC. (h) The ROC curve analysis of NSCLC for early‐stage LUSC. AUC, area under the curve; ES‐LUAD, early‐stage lung adenocarcinoma; ES‐LUSC, early‐stage lung squamous cell carcinoma; HD, healthy donors; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma. ****p < 0.0001
Consistently, TEP SNORD55 levels were also significantly decreased in patients with 76 LUSC as well as in 19 early‐stage patients (p < 0.0001, and p < 0.0001, respectively, Mann–Whitney test) (Figure 4e,f). Subsequently, when comparing the patients with LUSC and early stage LUSC to healthy controls, ROC curves demonstrated favorable diagnostic efficiencies of TEP SNORD55, possessing AUCs of 0.826 with 72.4% sensitivity and 77.7% specificity, and 0.854 with 68.4% sensitivity and 93.1% specificity, respectively (Figure 4g,h). Unexpectedly, as shown in Figure S3, its expression exerted no difference between LUAD and LUSC.
DISCUSSION
Over the past decade, although cancer morbidity and mortality has declined annually, 25 lung cancer is still the leading cause of cancer death with approximately 1.8 million new cases and 1.6 million deaths annually worldwide 26 but when found early is highly curable with surgery alone. In the current study, TEP SNORD55 was downregulated in NSCLC and acted as a biomarker for NSCLC diagnosis and early diagnosis.
SNORD55, a member of C/D box SNORNAs family, is in chromosome 1 and also known as U39, U55, RNU39, RNU55 or SNORD39. However, no comprehensive literature has demonstrated the expression and function of this gene. Here, we reported that SNORD55 was downregulated in TEPs from patients with NSCLC, as well as in early‐stage NSCLC compared with that from healthy donors. Notably, it was elevated in LUAD and LUSC tissues as evidenced from the data from the TCGA and SNORic databases. In the process of tumor‐educated platelets, we presumed that several pathways were involved including a direct connection between tumor cells and platelets, extracellular vesicle‐dependent horizontal transmission from tumor cells to platelets, as well as megakaryocytes influenced by tumor cells. In the current study, we observed downregulation of SNORD55 in TEPs and upregulation in tumor tissue. This is probably because TEP SNORD55 might be affected by the tumor in the process of platelet formation from megakaryocytes, and not directly from the tumor.
Importantly, in the current study, we validated that TEP SNORD55 was capable of acting as a promising biomarker for NSCLC. First, TEP SNORD55 exerted diagnostic performance in NSCLC diagnosis, with an AUC of 0.803, sensitivity of 79.3% and specificity of 68.3%, as well as in early NSCLC diagnosis, with an AUC of 0.784, sensitivity of 97.3% and specificity of 52.1%. Moreover, the combination with a standard screening test marker CEA which exerts a poor clinical diagnosis efficiency at an early stage of cancer development, could be an important diagnostic clinical approach. 22 Our data showed the combination of TEP SNORD55 and CEA improved the diagnostic efficiency of cancer progression, with an AUC of 0.828 with 66.3% sensitivity and 90.0% specificity. In addition, TEP SNORD55 also potentially acts as a noninvasive early biomarker for LUAD and LUSC with favorable diagnostic efficiencies.
Despite the emerging insights into many snoRNAs which are detectable in body fluids and serve as noninvasive biomarkers, 27 few studies have previously discussed the role of snoRNAs in platelets. Nevertheless, there are several limitations which should be carefully considered in the present study. Our results included 290 NSCLC patients, and the total sample size was small and lacked statistically vigorous power. In addition, TEP samples enrolled in this study failed to match the paired tissue samples, thus differences between tissues and platelets from the same donor could not be observed directly.
A previous study has already demonstrated that a significant difference was found to exist in TEP‐mRNA expression profile between tumor patients and healthy volunteers. It could not only be used for tumor diagnosis, but also accurately locate the main origin of pan‐cancer, as well as reveal the status of tumor cell gene variation in real time. 8 In anucleated platelets, snoRNAs are more likely to regulate pre‐mRNA alternative splicing, one of the main reasons for the change of mRNA expression profile. Our results demonstrated that TEP SNORD55 was downregulated in NSCLC patients compared to healthy donors, and valuable in the diagnosis of NSCLC, especially early diagnostics.
In conclusion, collectively, our findings reveal cancer‐specific changes of SNORD55 in the platelets of NSCLC patients, which pave the way for the exploitation of sensitive and specific NSCLC diagnostic biomarkers, either alone or in combination, providing a certain basis for clinical projects of TEP SNORD55 in the early screening of NSCLC.
Supporting information
Figure S1. The differentially expressed SNORD55 in SNORic data between normal tissues and tumor tissues of LUAD (a), LUSC (b), early stage LUAD (c) and early stage LUSC (d).
Figure S2. Platelet purity was detected by Liu staining under a microscope. PRP: platelet rich plasma.
Figure S3. TEP SNORD55 expression exerted no difference between LUAD and LUSC.
Table S1. Clinical and pathological characteristics of NSCLC patients.
Table S2. Clinical and pathological characteristics of healthy controls.
ACKNOWLEDGMENTS
This work was supported by National Natural Science Foundation of China (81972014, 81672104), the Shandong Provincial Natural Science Foundation (ZR2019MH004 and ZR2019LZL016), and National Key Research and Development Program of China (2016YFC0106002).
Dong X, Song X, Ding S, et al. Tumor‐educated platelet SNORD55 as a potential biomarker for the early diagnosis of non‐small cell lung cancer. Thoracic Cancer. 2021;12:659–666. 10.1111/1759-7714.13823
Funding information National Natural Science Foundation of China, Grant/Award Numbers: 81972014, 81672104; Shandong Provincial Natural Science Foundation, Grant/Award Numbers: ZR2019LZL016, ZR2019MH004
REFERENCES
- 1. Adjei AA. Lung cancer worldwide. J Thorac Oncol. 2019;14(6):956. [DOI] [PubMed] [Google Scholar]
- 2. Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Cancer statistics 2006. CA Cancer J Clin. 2006;56(2):106–30. [DOI] [PubMed] [Google Scholar]
- 3. Villalobos P, Wistuba II. Lung cancer biomarkers. Hematol Oncol Clin North Am. 2017;31(1):13–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lefrancais E, Ortiz‐Muñoz G, Caudrillier A, Mallavia B, Liu F, Sayah DM, et al. The lung is a site of platelet biogenesis and a reservoir for haematopoietic progenitors. Nature. 2017;544(7648):105–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Leslie M. Cell biology. Beyond clotting: the powers of platelets. Science. 2010;328(5978):562–4. [DOI] [PubMed] [Google Scholar]
- 6. Best MG, Wesseling P, Wurdinger T. Tumor‐educated platelets as a noninvasive biomarker source for cancer detection and progression monitoring. Cancer Res. 2018;78(13):3407–12. [DOI] [PubMed] [Google Scholar]
- 7. Nilsson RJ, Balaj L, Hulleman E, Van Rijn S, Pegtel DM, Walraven M, et al. Blood platelets contain tumor‐derived RNA biomarkers. Blood. 2011;118(13):3680–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Best MG, Sol N, Kooi I, Tannous J, Westerman BA, Rustenburg F, et al. RNA‐Seq of tumor‐educated platelets enables blood‐based pan‐cancer, multiclass, and molecular pathway cancer diagnostics. Cancer Cell. 2015;28(5):666–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lafontaine DL, Tollervey D. Birth of the snoRNPs: the evolution of the modification‐guide snoRNAs. Trends Biochem Sci. 1998;23(10):383–8. [DOI] [PubMed] [Google Scholar]
- 10. Cavaille J, Nicoloso M, Bachellerie JP. Targeted ribose methylation of RNA in vivo directed by tailored antisense RNA guides. Nature. 1996;383(6602):732–5. [DOI] [PubMed] [Google Scholar]
- 11. Ganot P, Bortolin ML, Kiss T. Site‐specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell. 1997;89(5):799–809. [DOI] [PubMed] [Google Scholar]
- 12. Filipowicz W, Pogacic V. Biogenesis of small nucleolar ribonucleoproteins. Curr Opin Cell Biol. 2002;14(3):319–27. [DOI] [PubMed] [Google Scholar]
- 13. Falaleeva M, Pages A, Matuszek Z, Hidmi S, Agranat‐Tamir L, Korotkov K, et al. Dual function of C/D box small nucleolar RNAs in rRNA modification and alternative pre‐mRNA splicing. Proc Natl Acad Sci U S A. 2016;113(12):E1625–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kishore S, Khanna A, Zhang Z, Hui J, Balwierz PJ, Stefan M, et al. The snoRNA MBII‐52 (SNORD 115) is processed into smaller RNAs and regulates alternative splicing. Hum Mol Genet. 2010;19(7):1153–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bray PF, McKenzie SE, Edelstein LC, Nagalla S, Delgrosso K, Ertel A, et al. The complex transcriptional landscape of the anucleate human platelet. BMC Genomics. 2013;14:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Best MG, Vancura A, Wurdinger T. Platelet RNA as a circulating biomarker trove for cancer diagnostics. J Thromb Haemost. 2017;15(7):1295–306. [DOI] [PubMed] [Google Scholar]
- 17. Hastings ML, Krainer AR. Pre‐mRNA splicing in the new millennium. Curr Opin Cell Biol. 2001;13(3):302–9. [DOI] [PubMed] [Google Scholar]
- 18. Dong X, Ding S, Yu M, Niu L, Xue L, Zhao Y, et al. Small nuclear RNAs (U1, U2, U5) in tumor‐educated platelets are downregulated and act as promising biomarkers in lung cancer. Front Oncol. 2020;10:1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kehr S, Bartschat S, Tafer H, Stadler PF, Hertel J. Matching of Soulmates: coevolution of snoRNAs and their targets. Mol Biol Evol. 2014;31(2):455–67. [DOI] [PubMed] [Google Scholar]
- 20. Best MG, Sol N, In't Veld SGJG, Vancura A, Muller M, ALN N, et al. Swarm intelligence‐enhanced detection of non‐small‐cell lung cancer using tumor‐educated platelets. Cancer Cell. 2017;32(2):238–252 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ko HH, Lee JJ, Chen HM, Kok SH, Kuo MY, Cheng SJ, et al. Upregulation of vascular endothelial growth factor mRNA level is significantly related to progression and prognosis of oral squamous cell carcinomas. J Formos Med Assoc. 2015;114(7):605–11. [DOI] [PubMed] [Google Scholar]
- 22. Pedersen LM, Milman N. Prognostic significance of thrombocytosis in patients with primary lung cancer. Eur Respir J. 1996;9(9):1826–30. [DOI] [PubMed] [Google Scholar]
- 23. Molina R, Filella X, Auge JM, Fuentes R, Bover I, Rifa J, et al. Tumor markers (CEA, CA 125, CYFRA 21‐1, SCC and NSE) in patients with non‐small cell lung cancer as an aid in histological diagnosis and prognosis. Comparison with the main clinical and pathological prognostic factors. Tumour Biol. 2003;24(4):209–18. [DOI] [PubMed] [Google Scholar]
- 24. Lv S, Xue J, Wu C, Wang L, Wu J, Xu S, et al. Identification of a panel of serum microRNAs as biomarkers for early detection of lung adenocarcinoma. J Cancer. 2017;8(1):48–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. [DOI] [PubMed] [Google Scholar]
- 26. Torre LA, Siegel RL, Jemal A. Lung Cancer Statistics. Adv Exp Med Biol. 2016;893:1–19. [DOI] [PubMed] [Google Scholar]
- 27. Egidi MG, Cochetti G, Guelfi G, Zampini D, Diverio S, Poli G, et al. Stability assessment of candidate reference genes in urine sediment of prostate cancer patients for miRNA applications. Dis Markers. 2015;2015:973597. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. The differentially expressed SNORD55 in SNORic data between normal tissues and tumor tissues of LUAD (a), LUSC (b), early stage LUAD (c) and early stage LUSC (d).
Figure S2. Platelet purity was detected by Liu staining under a microscope. PRP: platelet rich plasma.
Figure S3. TEP SNORD55 expression exerted no difference between LUAD and LUSC.
Table S1. Clinical and pathological characteristics of NSCLC patients.
Table S2. Clinical and pathological characteristics of healthy controls.


