Abstract
Background
Hepatocellular carcinoma (HCC) causes a heavy disease burden worldwide. Cell division cycle 45 (Cdc45) and its encoding gene (CDC45) have been studied for a long time, but their expression patterns and roles in liver carcinogenesis and advanced HCC deterioration are still incompletely understood. This study integrated tissue microarray and bioinformatics analyses to explore the expression and clinical value of CDC45 and Cdc45 in HCC.
Material/Methods
In HCC, the expression and relationships with clinic-pathological parameters of CDC45 and Cdc45 were investigated by integrating the RNA-sequencing data, downloaded from The Cancer Genome Atlas and Oncomine databases, and tissue microarray with immunohistochemistry staining. Co-expressed genes and genetic alterations of CDC45 separately obtained from Oncomine and cBioPortal databases were identified to shed light on the potential mechanisms of CDC45 in HCC.
Results
CDC45 and Cdc45 were both overexpressed in HCC tissues, and the CDC45 level progressively increased from stage I to III. The survival outcomes of the group with high CDC45 expression were significantly worse compared with the group with low expression. Amplification and deep deletion were 2 major significant alteration types in HCC patients, and the outcomes were worse in patients with altered versus unaltered CDC45. NUDT1, E2F1, CCNE2, MCM5, and CENPM were identified as the most significantly co-expressed genes.
Conclusions
CDC45 and Cdc45 were both upregulated in HCC, and increased expression levels and genetic alternations of CDC45 were correlated with worse prognosis in HCC patients. CDC45 may promote HCC by co-expressing with NUDT1, E2F1, CCNE2, MCM5, and CENPM.
Keywords: Carcinoma, Hepatocellular; Immunohistochemistry; Tissue Array Analysis
Background
In the United States, liver cancer has increased in mortality and morbidity more than any other human cancer during the past decade [1]. In China, new cases of hepatocellular carcinoma (HCC) represent about 45% of the total HCC cases reported worldwide annually [2,3]. Liver cancer has long been recognized as a serious threat to human health [4]. HCC is the most common subtype of liver cancer, and it is well known for its high heterogeneity, aggressiveness, fatality rate, and incidence, as well as a lack of acceptable and efficient biomarkers to predict disease progression and guide its treatment. Thus, most HCC cases are diagnosed at an advanced stage, and the 5-year survival rate of HCC patients in both the United States and China is less than 20% [1,3].
Growing research on tumor signal transduction pathways has demonstrated that HCC progression results from the aberrant activation of several molecules in various signaling pathways controlling cell cycle progression or arrest, proliferation, differentiation, cell survival, and apoptosis [5]. Therefore, it is urgent and essential to research cancer genes to better understand their involvement in the underlying molecular mechanisms of tumorigenesis and cancer progression and to develop more effective diagnosis and treatment options for HCC.
Cancer is characterized by uncontrolled cell proliferation and DNA replication catalyzed by DNA helicases, and in-depth research on these characteristic molecular events is needed to probe their underlying causes and design novel therapeutic targets. The gene cell division cycle 45 (CDC45), also known as CDC45L, MGORS7, CDC45L2, and PORC-PI-1, is located at chromosome 22q11.21, contains 21 exons, and encodes the protein Cdc45. Cdc45 combines with helicases MCM2-7 and GINS to form the Cdc45-MCM2-7-GINS (CMG) helicase complex, and it plays a key role in the early stages of DNA replication in eukaryotes [6], especially in regulating the initiation and elongation stages of eukaryotic chromosomal DNA replication [7]. From G1 to G2, Cdc45 exists only in the nucleus. However, it is distributed throughout the cell after the nuclear membrane is broken down during mitosis, which explains the change of Cdc45 distribution in the S-phase, from dispersion to local accumulation. Previous studies have shown that CDC45, Cdc45, or both are upregulated in various human carcinomas, leukemia, and lymphoma [6] and are limited or even undetectable in nonproliferating cells [8]. Cdc45 can be a protein marker for cell proliferation because its expression is positively correlated with cell proliferation [8]. Furthermore, studies have shown that CDC45 is one of the target genes of the myc gene [9] and has a critical effect on myc-dependent DNA replication stress, in addition to limiting the rate of replication origin activation [10,11]. In particular, the overexpression of CDC45 “recapitulates all c-myc-induced replication and damage phenotypes” [12]. For these reasons, CDC45 might play a critical role during tumorigenesis.
However, agreement has not been reached on the expression and functions of CDC45 and Cdc45 in HCC [13,14]. Therefore, this study investigated the expression, prognostic functions, and potential mechanisms of CDC45 with bioinformatics analysis and evaluated the expression of Cdc45 by immunohistochemistry (IHC) in HCC with the goal of exploring the roles of CDC45 and Cdc45 in HCC.
Material and Methods
Clinical Implications of CDC45 mRNA Expression Levels in HCC Tissues
CDC45 mRNA Expression Levels in HCC Tissues Based on RNA-sequencing Data
In this study, FireBrowse [15] (http://firebrowse.org/) and GEPIA (Gene Expression Profiling Interactive) [16] (http://gepia.cancer-pku.cn/) were used to analyze all CDC45 mRNA expression data. Both are based on the RNA-sequencing data from The Cancer Genome Atlas (TCGA), and GEPIA also contains the data from the Genotype-Tissue Expression (GTEx) project. To identify the expression level of CDC45 among different types of cancer, RNA-Seq by Expectation-Maximization (RSEM) (log2) was used, which was shown in FireBrowse. With regard to GEPIA and the data from GTEx, the TPM (transcripts per million) format for the relevant CDC45 mRNA expression levels were calculated. In total, 369 cases of HCC and 160 cases of non-HCC liver samples were examined. The clinicopathological information for the HCC patients was downloaded from TCGA, including their clinical stages, overall survival rates, and disease-free survival numbers.
CDC45 mRNA Expression Levels in HCC Tissues Based on Other High-throughput Datasets
The Oncomine (https://www.oncomine.org) database contains gene expression measurement from more than 4700 experiments and is a powerful public platform for oncogenomic analyses [17]. Therefore, to validate the CDC45 mRNA expression levels in HCC patients, we searched the Oncomine datasets for the CDC45 expression levels in HCC and non-HCC liver tissues. All microarray platforms and RNA-sequencing data were considered.
Analyzing Cdc45 Expression in HCC With In-house Tissue Microarrays and IHC
IHC was performed according to the manufacturer’s instructions. All patients had given informed consent, and this study was authorized by the Ethics Committee of the First Affiliated Hospital of Guangxi Medical University in Nanning, China (No. 2020 [KY-E-126]). The formalin-fixed paraffin-embedded tissue microarray (TMA) was supplied by Fanpu Biotech, Inc. (LVC1021, LVC1601, Guilin, China) and contained 137 HCC tissues and 62 non-HCC liver tissues. After being deparaffinized and rehydrated, the tissue slides were put into a boiled 0.01 M citrate buffer (pH 6.0) to retrieve the antigen. The endogenous peroxidase activity was blocked with 3% H2O2. The primary antibody against Cdc45 was a rabbit anti-human Cdc45 monoclonal antibody (ab126762) (Abcam, UK, dilution 1: 100), which was incubated overnight at 4°C and then with horseradish peroxidase (HRP)-labeled secondary antibody (ready-to-use, Long Island Antibody, Shanghai, China) at room temperature for 25 min. The last step involved the visualization of the HRP with 3,3′-diaminobenzidine (DAB), followed by dehydration, sealing, and evaluation with a bright-field microscope. Brown-yellow granules in the nucleus and/or cytoplasm indicated positive staining. Negative control sections were incubated with phosphate-buffered saline during the primary antibody incubation.
The 2 stained TMAs were assessed blindly by 2 independent pathologists and were each evaluated at ×400 magnification for the intensity and percentage of positive cells in 10 randomly selected fields. The staining intensity was scored according to a model used in prior studies [18], with the recorded percentage falling into 1 of 4 grades: 0 (<5%), 1 (5–25%), 2 (26–50%), or 3 (>50%). To find the total IHC score, the intensity score was multiplied by the percentage score.
The Potential Molecular Mechanism of CDC45 in HCC
The Co-expressed Genes of CDC45 in HCC Tissues
Since CDC45 may be co-expressed with other genes and mediate a potential mechanism in the pathogenesis and progression of HCC, we investigated the genes co-expressed with CDC45 in 102 cases of HCC from the Chen Liver dataset, 35 cases of HCC from the Wurmbach Liver dataset, and 22 cases of HCC from the Roessler Liver dataset. Heat-maps were drawn to show the most significant co-expressed genes based on their correlation coefficient. All these analyses were based on Oncomine [17].
The Genetic Alterations of CDC45 in HCC Tissues
Genetic alterations also contribute to the function of a gene. Thus, we used multiple datasets to reveal the genetic alterations of CDC45 in HCC. The following datasets from cBioPortal (The cBio Cancer Genomics Portal) [19] (http://cbioportal.org) were included: “Hepatocellular Carcinoma” (MSK, Clin Cancer Res 2018, 127 samples), “Hepatocellular Carcinoma” (INSERM, Nat Genet 2015, 243 samples), “Liver Hepatocellular Adenoma and Carcinomas” (MSK, PLoS One 2018, 19 samples), “Liver Hepatocellular Carcinoma” (AMC, Hepatology 2014, 231 samples), “Liver Hepatocellular Carcinoma” (RIKEN, Nat Genet 2012, 27 samples), and “Liver Hepatocellular Carcinoma” (TCGA, Firehose Legacy, 442 samples). The molecular profiles of the mutations, copy number of the alterations, and mRNA Expression z-Scores (RNA-Seq V2 RSEM) were analyzed [20].
Statistical Analysis
All statistics were analyzed with SPSS Statistics (version 23.0). The results were presented as mean±standard deviation (SD). A t test was used to analyze the differences between the 2 groups, while a 1-way ANOVA was used to analyze ≥3 groups. The association between CDC45 expression levels and the clinicopathological parameters were analyzed by a chi-square test, and the relationships between the different genes were examined using a correlation coefficient test. The survival analysis was performed with the Kaplan-Meier method. The Cox proportional hazards model was used to analyze the effects of multiple factors on the progress and hazard ratio (HR). P<0.05 was considered statistically significant.
Results
The Clinical Significance of CDC45 mRNA Expression Levels in HCC
Calculating the RNA-sequencing Data to Identify the CDC45 mRNA Expression Levels in HCC Tissues
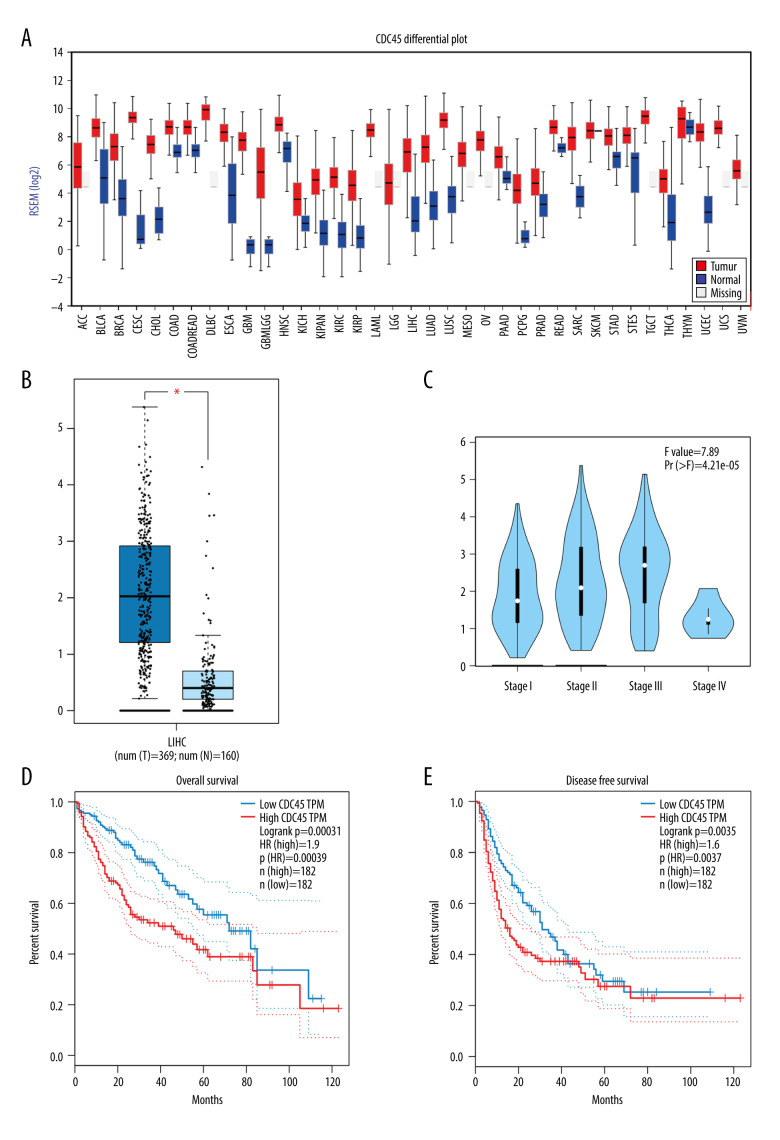
Using the TCGA RNA-Seq data, FireBrowse provided the CDC45 mRNA expression levels for more than 30 types of tumors. In 27 types of tumors with nontumorous controls, the CDC45 mRNA expression levels were clearly upregulated compared with the controls (Figure 1A). To improve the accuracy and validity of the statistical analysis, we increased the number of noncancerous cases used in the comparison by adding normal liver samples from GTEx in GEPIA. The CDC45 mRNA expression levels were predominantly higher in the 369 cases of HCC than in the 160 cases of non-HCC liver controls (Figure 1B). We also found increases in the CDC45 mRNA expression levels from stage I to stage III, indicating that CDC45 might be responsible for tumor progression. However, at stage IV, the CDC45 mRNA expression levels decreased, probably due to the small sample size (Figure 1C). The proposed role of CDC45 mRNA in tumor development was also supported by the prognostic values obtained. The HR was 1.9 (P=0.00039) for the overall survival prediction (Figure 1D) and 1.6 for the disease-free survival prediction (P=0.0037, Figure 1E). The RNA-sequencing data suggested that the upregulation of CDC45 mRNA expression levels might play a part in causing tumorigenesis and poorer prognosis of HCC patients.
Figure 1.
The clinical significance of CDC45 mRNA expression levels in hepatocellular carcinoma (HCC) tissues based on RNA-Seq data. The mRNA expression level of CDC45 was clearly upregulated compared with the controls (A, B). (A) The CDC45 mRNA expression was clearly higher in HCC than in normal tissues. (B) The CDC45 mRNA expression was significantly increased in HCC compared with normal tissues. (C) The CDC45 mRNA expression levels progressively increased from stage I to stage III and decreased in stage IV. A high CDC45 mRNA expression level indicated a negative prognosis (D: for overall survival, HR=1.9, P=0.00039; E: for disease-free survival, HR=1.6, P=0.0037). HR=hazard ratio.
Mining the Microarray Data to Evaluate the CDC45 mRNA Expression Levels in HCC Tissues
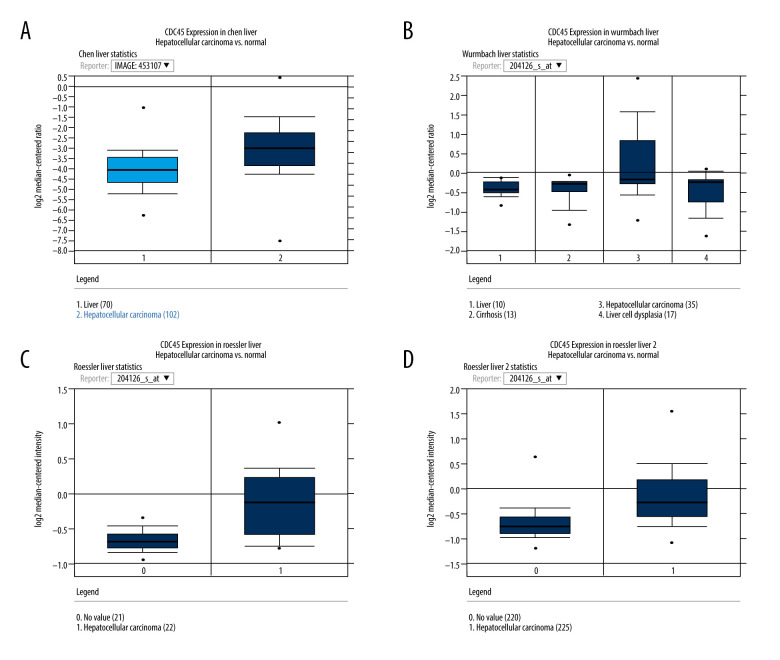
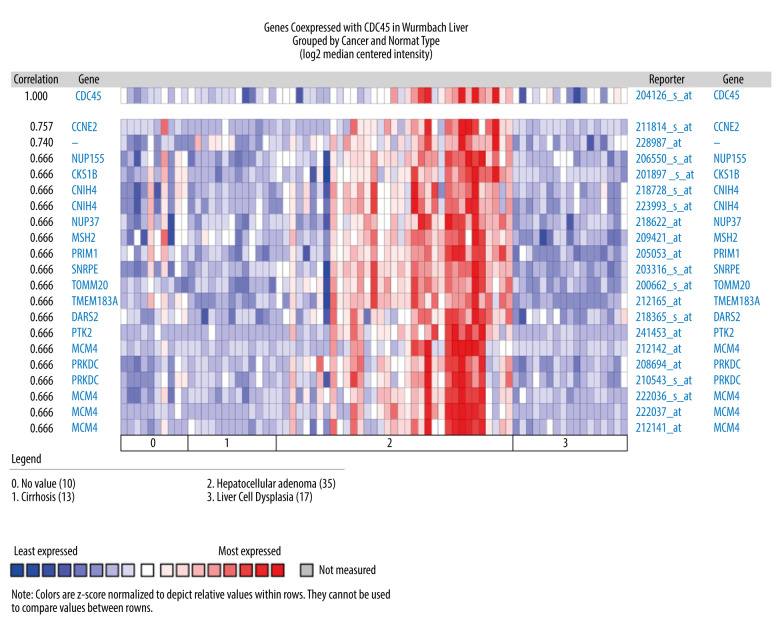
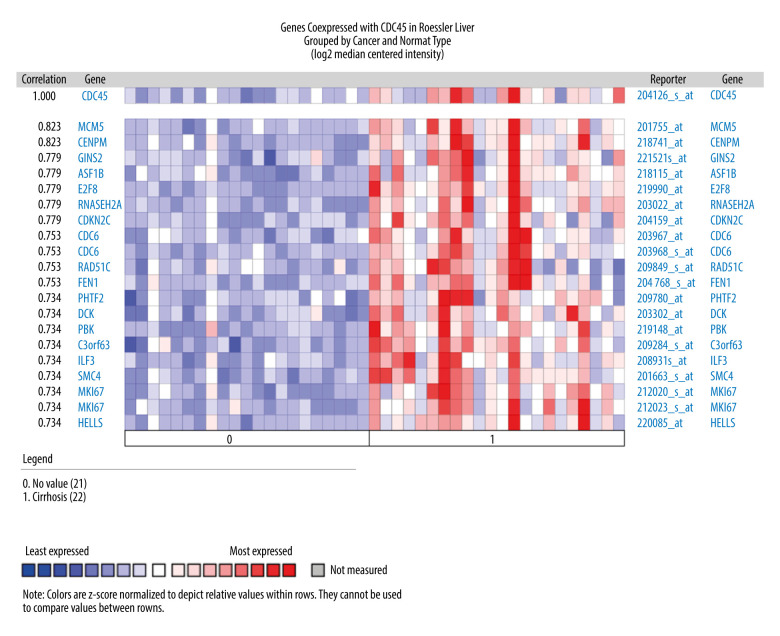
To gather more evidence supporting the proposed oncogenic implications of CDC45 mRNA in HCC, we collected several microarray datasets from Oncomine. Four of these datasets showed that the CDC45 mRNA expression levels were indeed obviously upregulated in the HCC samples compared with the non-HCC controls, including the Chen Liver, Wurmbach Liver, Roessler Liver, and Roessler Liver 2 datasets (Figure 2).
Figure 2.
The CDC45 mRNA expression levels were obviously upregulated in the microarrays from the Oncomine datasets. (A) Chen Liver: fold change=2.135; P=1.16E-10; 197 samples; 10 802 measured genes; platform not predefined in Oncomine. (B) Wurmbach Liver: fold change=1.581; P=4.17E-5; 75 samples; 19 574 measured genes; Human Genome U133 Plus 2.0 Array. (C) Roessler Liver: fold change=1.429; P=2.70E-5; 43 samples; 12 603 measured genes; Human Genome U133A 2.0 Array. (D) Roessler Liver 2: fold change=1.453; P=2.00E-36; 445 samples; 12 624 measured genes; Affymetrix Human Genome HT U133A Array.
Assessing the Cdc45 Expression Levels in HCC Tissues with In-house Tissue Microarrays and IHC
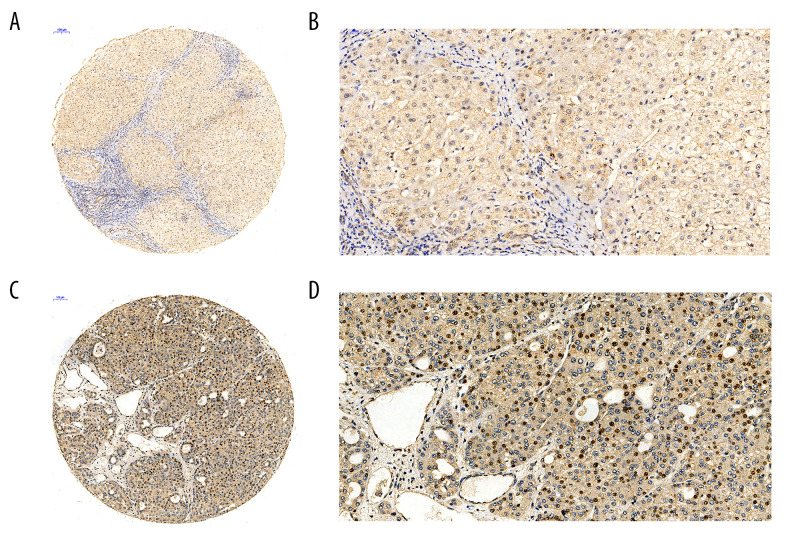
We further validated the CDC45 mRNA expression levels by assessing their protein levels with in-house tissue microarrays and IHC. The positive signaling of the Cdc45 was located in the nucleus and cytoplasm. In line with the mRNA levels, the Cdc45 levels in the HCC tissues were also notably increased in comparison to those in the non-HCC tissues (4.635±2.051 vs 2.565±1.410, respectively; P=4.317E-14; Figure 3).
Figure 3.
The Cdc45 expression levels for the in-house tissue microarrays. Immunohistochemistry staining to evaluate the expression of Cdc45 in hepatocellular carcinoma (HCC) and adjacent tissues. (A, B) The expression of Cdc45 was absent or weak in the non-HCC tissue (2.565±1.410). (C, D) The expression of Cdc45 was obviously high in the HCC tissue (4.635±2.051). Cdc45 was detected in the nucleus and cytoplasm. Magnification, ×20 (A, C) and ×400 (B, D).
The Potential Molecular Mechanism of CDC45 in HCC
The Genes Co-expressed with CDC45 in HCC Tissues
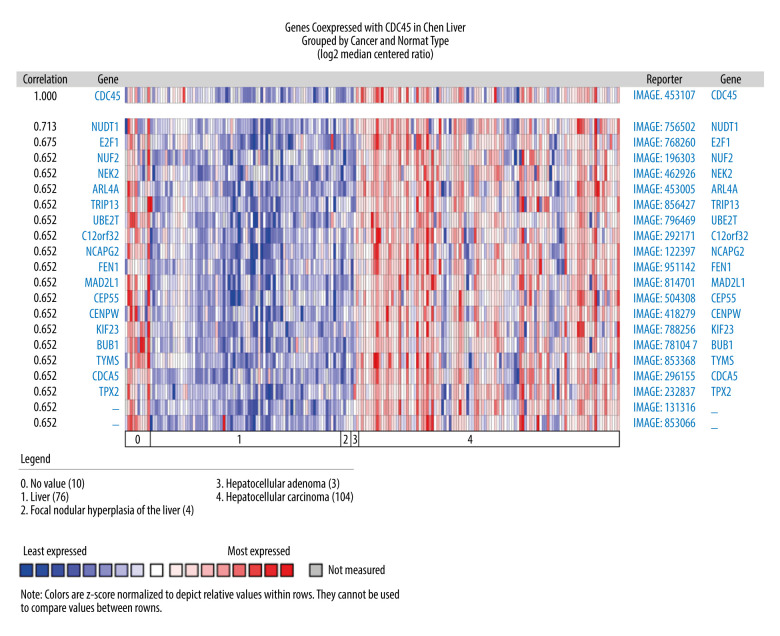
The genes co-expressed with CDC45 could partially reveal the potential functional mechanism of CDC45 in HCC. Two most significant genes co-expressed with CDC45 in the Chen Liver dataset were NUDT1 (r=0.713) and E2F1 (r=0.675). In addition, 16 other genes had the same correlation index (r=0.652), including NUF2, NEK2, and ARL4A (Figure 4). The first co-expressed gene listed in the Wurmbach Liver dataset was CCNE2, followed by 16 genes with the same r value (0.666), including NUP155, CKS1B, and CNIH4 (Figure 5). The first 2 co-expressed genes in the Roessler Liver dataset were MCM5 and CENPM (r=0.823), followed by 5 other genes with the same r value (0.779): GINS2, ASF1B, E2F8, RNASEH2A, and CDKN2C (Figure 6).
Figure 4.
The co-expressed genes based on the Chen Liver microarray. NUDT1 and E2F1 were the top 2 genes on the list of co-expressed genes of CDC45 (r=0.713 and 0.675, respectively). The correlation index of other genes was 0.652.
Figure 5.
The co-expressed genes based on the Wurmbach Liver microarray. All the correlation indexes of co-expressed genes of CDC45 were 0.666, except CCNE2 (r=0.757).
Figure 6.
The co-expressed genes based on the Roessler Liver microarray. MCM5, CENPM, GINS2, ASF1B, E2F8, RNASEH2A, and CDKN2C were the top genes co-expressed with CDC45 in Roessler Liver.
The Genetic Alterations of CDC45 in HCC Tissues
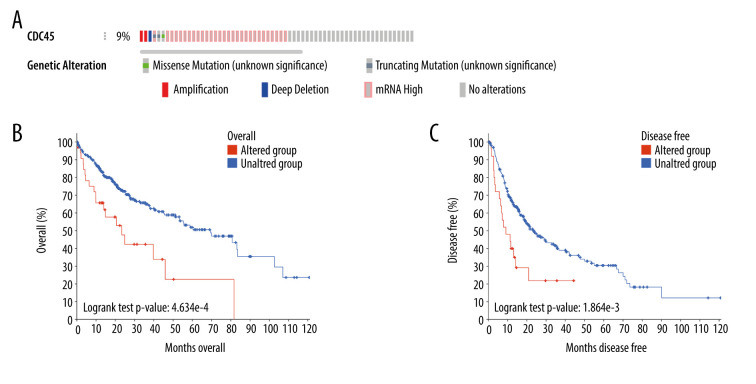
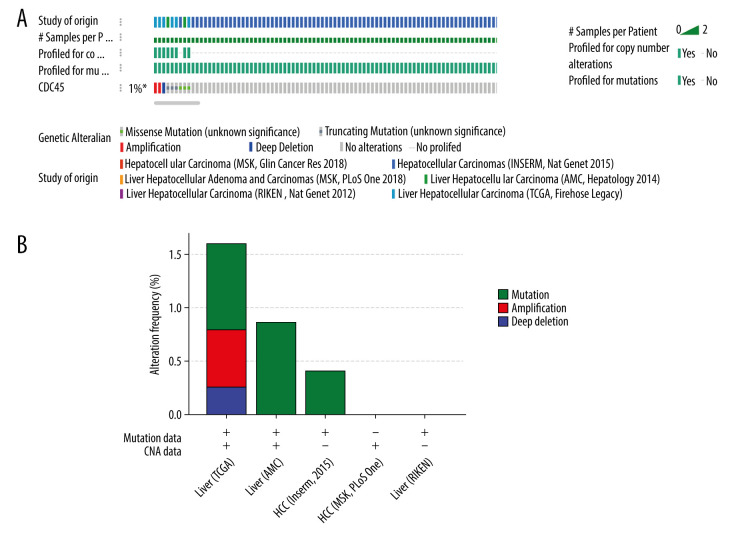
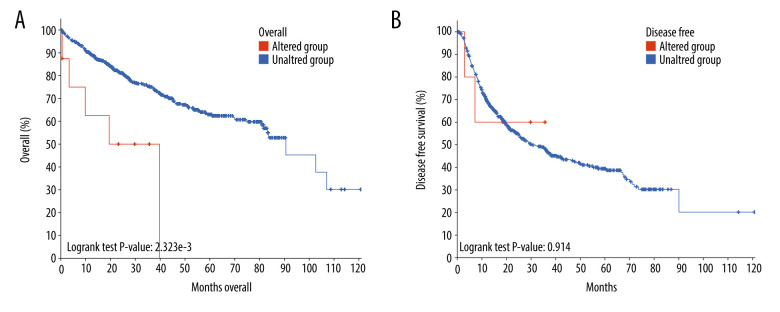
Genetic alterations, such as mutations and amplifications, could contribute to the potential molecular mechanism of CDC45 in HCC. To investigate this idea, we examined the genetic alterations of CDC45 in multiple datasets. CDC45 was found to be altered in 34 of 360 patients/complete samples (9%) included in 442 HCC cases from the TCGA dataset (Figure 7A). The patterns of CDC45 genetic alterations covered missense mutations, truncating mutations, amplification, deep deletion, and higher mRNA expression. Of note, the cases involving CDC45 genetic alterations tended to have more unfavorable overall survival (Figure 7B) and disease-free survival (Figure 7C). In addition to the TCGA datasets, we used 5 other studies to obtain CDC45 genetic alteration information (Figure 8A). CDC45 mutations could also be noted in liver (AMC) and HCC (Inserm, 2015) studies (Figure 8B). Across these datasets of CDC45 genetic alterations, a poorer overall survival rate was observed in the cases involving CDC45 alterations than in the unaltered group (Figure 9A), although a poorer disease-free survival rate was not observed (Figure 9B).
Figure 7.
The clinical implications of CDC45 genetic alterations. (A) The oncoprint of CDC45 in 360 patients/complete samples from 442 cases of hepatocellular carcinoma (HCC). CDC45 was altered in 34 (9%) cases. Patients with the CDC45 genetic alteration were more likely to have poor lifespan (B, for overall survival; C, for disease-free survival).
Figure 8.
The CDC45 genetic alterations in multiple HCC datasets. (A) The oncoprint of CDC45 in 1089 cases of hepatocellular carcinoma (HCC) from multiple datasets involved The Cancer Genome Atlas (TCGA), AMC, Inserm, MSK, and RIKEN. CDC45 was altered in 9 (1%) of the patients. (B) The alteration frequency of CDC45 in HCC and mutation was the most common alternate forms, especially in liver (AMC) and HCC (Inserm, 2015) studies.
Figure 9.
The prognostic values of CDC45 genetic alterations in hepatocellular carcinoma (HCC). The survival time of altered group was less than the unaltered group, both for overall survival (A) and disease-free survival (B).
Discussion
Overexpressed CDC45 has been observed in many kinds of tumors. Previous studies have confirmed that Cdc45 is significantly upregulated in colorectal cancer tissues, papillary thyroid carcinoma tissues, and tongue squamous cell carcinoma tissues compared with noncancerous tissues [21–23]. It has also been reported that CDC45 expression is significantly higher in malignant squamous cell carcinomas of the tongue than in mild precancerous epithelial dysplasia and that the expression levels generally increase with increasing grades of dysplasia [23]. However, the prognoses for patients with different CDC45 expression levels in various tumors can be diverse. For example, the prognosis for colorectal cancer patients was found to be worse in a low than in a high CDC45 expression group [24], while CDC45 was significantly upregulated in prostate cancer metastatic samples [25] and was correlated with late-stage papillary thyroid carcinomas [22].
The role of CDC45 in HCC remains unclear. Xiang et al [13] found that CDC45 was one of the risk factors in HCC patients and was markedly upregulated in HCC tissues. However, Chen et al [14] found that CDC45 expression was increased while Cdc45 was downregulated in HCC tissues. By integrating data from tissue microarray, TCGA, and Oncomine, the current study provided evidence that both Cdc45 and CDC45 levels increased in HCC tissues compared with non-HCC tissues. Moreover, concerning the expression of CDC45/Cdc45 and clinicopathological features of HCC patients, Xiang et al [13] confirmed that senescence-associated genes including CDC45 can predict the 1-, 3-, and 5-year overall survival of HCC patients more accurately than serum alpha-fetoprotein. Both Chen et al [14] and the current study suggested that higher expression levels of CDC45 indicated more advanced clinicopathological features and shorter survival. Nevertheless, in terms of the expression of Cdc45, this study showed that Cdc45 was clearly higher in HCC than in noncancerous tissues and located in the nucleus and cytoplasm, whereas Chen et al [14] found that Cdc45 was downregulated in HCC tissues and mainly located in the cytoplasm. All in all, these studies indicated that CDC45 was overexpressed in HCC tissues, while expression of Cdc45 varied. The difference in Cdc45 expression patterns might be caused by different primary antibodies, regional disparity, and sample size. In addition, our study suggested that the cases with CDC45 genetic alterations, including missense mutations, truncating mutations, amplification, deep deletion, and higher mRNA expression, tended to have a shorter overall survival. These results suggest that CDC45 might be a cancer-promoting factor in HCC and could help to predict the prognosis of HCC patients. However, the mechanism of the translation of CDC45 to Cdc45 remains unknown.
The mechanism of CDC45 in HCC is unclear. The current study integrated data on gene co-expression and genetic alterations of CDC45 to explore the potential molecular mechanism of CDC45 in HCC tissues. It was found that 5 genes, including NUDT1, E2F1, CCNE2, MCM5, and CENPM, had significant correlation indices with CDC45 and were upregulated in HCC tissues. Numerous studies agree that the increased expression of these 5 genes is associated with a poor prognosis and more advanced tumor stages in HCC patients [13,26–30]. Moreover, research showed that E2F1 regulates DNAJA1 to promote cell proliferation and metastasis by stabilizing CDC45 in colorectal cancer cell lines [21] and that silencing CCNE2 or CENPM could inhibit HCC cell growth [30,31]. Therefore, CDC45 may have a synergistic relationship with NUDT1, E2F1, CCNE2, MCM5, and CENPM that promotes the oncogenesis and progression of HCC.
There were 2 main limitations to this study. First, the roles of CDC45 in vivo and in vitro require further investigation. Second, future experiments need to uncover and validate the exact relationship between the 5 co-expressed genes and CDC45.
Conclusions
The present study confirmed the upregulated expression pattern of CDC45 and Cdc45 in HCC and revealed that increased expression levels and genetic alternations of CDC45 can be associated with a worse prognosis in HCC patients. In addition, CDC45 may promote the growth and progression of HCC through co-expression with NUDT1, E2F1, CCNE2, MCM5, and CENPM. These findings indicate that CDC45 may be a novel prognostic biomarker in HCC patients.
Acknowledgments
The authors thank the people and organizations who maintain the public databases for the relevant data.
Footnotes
Conflict of Interest
None.
Source of support: This study was funded by grants from the Promoting Project of Basic Capacity for Young and Middle aged University Teachers in Guangxi (2018KY0123), Innovation Project of Guangxi Graduate Education (YCBZ2020045), Natural Science Foundation of Guangxi, China (2018GXNSFAA294025), Guangxi Zhuang Autonomous Region Health and Family Planning Commission Self-Financed Scientific Research Project (Z20201233, Z20190594, Z20190523), Guangxi Degree and Postgraduate Education Reform and Development Research Projects, China (JGY2019050), Guangxi Higher Education Undergraduate Teaching Reform Project (2020JGA146), Guangxi Medical University Education and Teaching Reform Project (2019XJGZ04), Guangxi Medical University 2020 Undergraduate Innovation and Entrepreneurship Training Program (202010598002), and the Future Academic Star of Guangxi Medical University (WLXSZX20088)
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. Cancer J Clin. 2020;70(1):7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. Cancer J Clin. 2016;66(2):115–32. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Li Z, Wang Y, Duan S, et al. Expression of TBX3 in hepatocellular carcinoma and its clinical implication. Med Sci Monit. 2018;24:9324–33. doi: 10.12659/MSM.909378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Endeshaw M, Hallowell BD, Razzaghi H, et al. Trends in liver cancer mortality in the United States: Dual burden among foreign- and US-born persons. Cancer. 2019;125(5):726–34. doi: 10.1002/cncr.31869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen C, Wang G. Mechanisms of hepatocellular carcinoma and challenges and opportunities for molecular targeted therapy. World J Hepatol. 2015;7(15):1964–70. doi: 10.4254/wjh.v7.i15.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seo YS, Kang YH. The human replicative helicase, the CMG complex, as a target for anti-cancer therapy. Front Mol Biosci. 2018;5:26. doi: 10.3389/fmolb.2018.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Broderick R, Nasheuer HP. Regulation of Cdc45 in the cell cycle and after DNA damage. Biochem Soc Trans. 2009;37(Pt 4):926–30. doi: 10.1042/BST0370926. [DOI] [PubMed] [Google Scholar]
- 8.Pollok S, Bauerschmidt C, Sanger J, et al. Human Cdc45 is a proliferation-associated antigen. FEBS J. 2007;274(14):3669–84. doi: 10.1111/j.1742-4658.2007.05900.x. [DOI] [PubMed] [Google Scholar]
- 9.Nepon-Sixt BS, Bryant VL, Alexandrow MG. Myc-driven chromatin accessibility regulates Cdc45 assembly into CMG helicases. Commun Biol. 2019;2:110. doi: 10.1038/s42003-019-0353-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Srinivasan SV, Dominguez-Sola D, Wang LC, et al. Cdc45 is a critical effector of myc-dependent DNA replication stress. Cell Rep. 2013;3(5):1629–39. doi: 10.1016/j.celrep.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tanaka S, Nakato R, Katou Y, et al. Origin association of Sld3, Sld7, and Cdc45 proteins is a key step for determination of origin-firing timing. Curr Biol. 2011;21(24):2055–63. doi: 10.1016/j.cub.2011.11.038. [DOI] [PubMed] [Google Scholar]
- 12.Wang Z, Yang B, Zhang M, et al. lncRNA epigenetic landscape analysis identifies EPIC1 as an oncogenic lncRNA that interacts with MYC and promotes cell-cycle progression in cancer. Cancer Cell. 2018;33(4):706–20e9. doi: 10.1016/j.ccell.2018.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiang XH, Yang L, Zhang X, et al. Seven-senescence-associated gene signature predicts overall survival for Asian patients with hepatocellular carcinoma. World J Gastroenterol. 2019;25(14):1715–28. doi: 10.3748/wjg.v25.i14.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang C, Xie S, Wu Y, et al. Prognostic implications of CDC45 expression in hepatocellular carcinoma. Research Square; 2020 https://www.researchsquare.com/article/rs-31516/v1. [Google Scholar]
- 15.Deng M, Bragelmann J, Kryukov I, et al. FirebrowseR: An R client to the Broad Institute’s Firehose Pipeline. Database (Oxford) 2017;2017:baw160. doi: 10.1093/database/baw160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang Z, Li C, Kang B, et al. GEPIA. A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rhodes DR, Kalyana-Sundaram S, Mahavisno V, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9(2):166–80. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo B, Huang L, Gu Y, et al. Expression of exportin-1 in diffuse large B-cell lymphoma: Immunohistochemistry and TCGA analyses. Int J Clin Exp Pathol. 2018;11(12):5547–60. [PMC free article] [PubMed] [Google Scholar]
- 19.Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–4. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang S, Ren X, Liang Y, et al. KNK437 restricts the growth and metastasis of colorectal cancer via targeting DNAJA1/CDC45 axis. Oncogene. 2020;39(2):249–61. doi: 10.1038/s41388-019-0978-0. [DOI] [PubMed] [Google Scholar]
- 22.Sun J, Shi R, Zhao S, et al. Cell division cycle 45 promotes papillary thyroid cancer progression via regulating cell cycle. Tumour Biol. 2017;39(5) doi: 10.1177/1010428317705342. 1010428317705342. [DOI] [PubMed] [Google Scholar]
- 23.Li JN, Feng CJ, Lu YJ, et al. mRNA expression of the DNA replication-initiation proteins in epithelial dysplasia and squamous cell carcinoma of the tongue. BMC Cancer. 2008;8:395. doi: 10.1186/1471-2407-8-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu Y, Wang L, Li Z, et al. Potential prognostic and diagnostic values of CDC6, CDC45, ORC6 and SNHG7 in colorectal cancer. Onco Targets Ther. 2019;12:11609–21. doi: 10.2147/OTT.S231941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li HY, Jin N, Han YP, Jin XF. Pathway crosstalk analysis in prostate cancer based on protein-protein network data. Neoplasma. 2017;64(1):22–31. doi: 10.4149/neo_2017_103. [DOI] [PubMed] [Google Scholar]
- 26.Gu Y, Wang X, Liu H, et al. SET7/9 promotes hepatocellular carcinoma progression through regulation of E2F1. Oncol Rep. 2018;40(4):1863–74. doi: 10.3892/or.2018.6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ou Q, Ma N, Yu Z, et al. Nudix hydrolase 1 is a prognostic biomarker in hepatocellular carcinoma. Aging (Albany NY) 2020;12(8):7363–79. doi: 10.18632/aging.103083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu Z, Wang R, Chen F, et al. Five novel oncogenic signatures could be utilized as AFP-related diagnostic biomarkers for hepatocellular carcinoma based on next-generation sequencing. Dig Dis Sci. 2018;63(4):945–57. doi: 10.1007/s10620-018-4961-3. [DOI] [PubMed] [Google Scholar]
- 29.Sonntag R, Giebeler N, Nevzorova YA, et al. Cyclin E1 and cyclin-dependent kinase 2 are critical for initiation, but not for progression of hepatocellular carcinoma. Proc Natl Acad Sci USA. 2018;115(37):9282–87. doi: 10.1073/pnas.1807155115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiao Y, Najeeb RM, Ma D, et al. Upregulation of CENPM promotes hepatocarcinogenesis through mutiple mechanisms. J Exp Clin Cancer Res. 2019;38(1):458. doi: 10.1186/s13046-019-1444-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun Q, Yu R, Wang C, et al. Circular RNA circ-CSPP1 regulates CCNE2 to facilitate hepatocellular carcinoma cell growth via sponging miR-577. Cancer Cell Int. 2020;20:202. doi: 10.1186/s12935-020-01287-8. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]