Abstract
The discovery of high-risk breast cancer susceptibility genes such as BRCA1 DNA Repair Associated (BRCA1) and BRCA2 DNA Repair Associated (BRCA2) has led to accurate identification of individuals for risk management and targeted therapy. The rapid decline in sequencing costs has tremendously increased the number of individuals who are undergoing genetic testing world-wide. However, given the significant differences in population-specific variants, interpreting the results of these tests can be challenging especially for novel genetic variants in understudied populations. Here we report the characterization of novel variants in the Malaysian and Singaporean population that consist of different ethnic groups (Malays, Chinese, Indian and other indigenous groups). We have evaluated the functional significance of 14 BRCA2 VUS by using multiple in silico prediction tools and examined their frequency in a cohort of 7,840 breast cancer cases and 7,928 healthy controls. In addition, we have used a mouse embryonic stem cell (mESC)-based functional assay to assess the impact of these variants on BRCA2 function. We found these variants to be functionally indistinguishable from wild-type BRCA2. These variants could fully rescue the lethality of Brca2-null mESCs and exhibited no sensitivity to six different DNA damaging agents including a PARP inhibitor. Our findings strongly suggest that all 14 evaluated variants are functionally neutral. Our findings should be valuable in risk assessment of individuals carrying these variants.
Keywords: Breast Cancer, BRCA2, Variants of Uncertain Clinical Significance (VUS), Functional evaluation, ES cell-based assay, Molecular dynamic analysis
INTRODUCTION
Breast cancer is one of the leading causes of death in women, with more than 2 million new cases diagnosed each year. Breast cancer incidence varies globally, from 27 per 100,000 in East Asia to 92 per 100,000 in North America (Martin-Sanchez et al., 2018). Notably, while the median age of diagnosis of breast cancer is approximately 60 years in the majority of high income countries of European descent, it is approximately 10 years younger in the majority of low-income countries among women of Asian descent (Yap et al., 2019). Given that prevalence of germline alteration is higher among individuals diagnosed with breast cancer at young ages, the proportion of breast cancers attributable to pathogenic variants in high-risk genes such as BRCA1 DNA Repair Associated (BRCA1) and BRCA2 DNA Repair Associated (BRCA2) is likely to be higher in Asian breast cancer patients compared to patients of European descent (Kemp et al., 2019; Wen et al., 2016).
Unfortunately, in part because of cost and availability of genetic counselling and genetic testing services, there remains a significant gap in provision of genetic services for breast cancer patients in Asia (Nakamura et al., 2016). In addition, because there are relatively few data on the prevalence of variants understudied populations, such as those in Asia, a high proportion of variants are of uncertain significance. This poses a major challenge for physicians and genetic counselors in offering guidance to patients and their family members. Therefore, classification of VUS remains to be of utmost importance. At present, VUS are classified by the ENIGMA (Evidence-based Network for the Interpretation of Germline Mutant Alleles) consortium based on a multifactorial likelihood quantitative analysis model, which takes into account many factors including evolutionary conservation of the amino acid, personal and family history of cancer, prevalence in the population, co-occurrence with a known pathogenic variant and co-segregation of the variant with cancer in the families (Parsons et al., 2019; Spurdle et al., 2012). Databases such as ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) and BRCA Exchange (https://brcaexchange.org/) also provide information on classification of BRCA variants.
In this study, we have functionally characterized 14 BRCA2 VUS that have been found in the different ethnic groups (Malays, Chinese, Indian and other indigenous groups) of the Malaysian population (Lai et al., 2017; Thirthagiri et al., 2008; Toh et al., 2008). We have determined the prevalence of these variants in a cohort of 7,840 breast cancer cases and 7,928 controls in Malaysia and Singapore. We have evaluated the pathogenicity of these variants using three different computational models. Finally, we directly tested the impact of these variants on BRCA2 function using a well-established mouse embryonic stem cell (mESC)-based functional assay (Biswas et al., 2011; Kuznetsov, Chang, & Sharan, 2010; Kuznetsov, Liu, & Sharan, 2008). This functional assay examines the effect of BRCA2 VUS on the viability of mESC and also on the sensitivity of mESC to six different DNA damaging agents..
MATERIALS AND METHODS
Selection of BRCA2 variants
Variants were selected based on a hospital based study where germline DNA from 467 breast cancer patients were analysed using Sanger sequencing to identify germline variants in BRCA1 and BRCA2 in multi-ethic Malaysian population (i.e. Malay, Chinese, Indian and other ethnic groups) (Thirthagiri et al., 2008). As per HGVS nomenclature, nucleotide numbers reflect cDNA numbers with +1 corresponding to the A of the ATG initiation codon in the reference sequence of BRCA2 (GenBank accession number NM_000059.3). The initiation codon is codon 1.
Study subjects in breast cancer case control study
The study participants were women recruited in the Malaysian Breast Cancer Genetic (MyBrCa) study (M. M. Tan et al., 2018) and the Singapore Breast Cancer Cohort (SGBCC) study. Cases were recruited from two hospitals in Malaysia (recruitment started in 2002 in the first hospital and extended to the second hospital in 2012) and six hospitals in Singapore (recruitment started in 2010 in the first hospital and extended to additional five hospitals in 2016). Women diagnosed clinically with breast cancer (invasive and non-invasive) with a mixture of prevalent and incident cases were included as breast cancer cases.
In MyBrCa, controls were healthy women between ages 40 and 74, with no personal history of breast cancer, recruited through a subsidized opportunistic mammographic screening programme that was initiated in the same two hospitals where cases were recruited. The Singaporean controls were from the Singapore Population Health Studies (National University Health System, 2016) and the Singapore Multi-Ethnic Cohort (K. H. X. Tan et al., 2018), and matched by ethnicity and age ±5 years with SGBCC cases.
Participants donated a blood or saliva sample that was processed and stored, completed a questionnaire that included information on lifestyle risk factors for breast cancer, and provided written informed consent. Recruitment and genetic studies have been approved by the Ethics Committees of University Malaya Medical Centre [UM 842.9], Subang Jaya Medical Centre [reference no: 201109.4 and 201208.1], NHG Domain Specific Review Board [NHG DSRB Ref: 2009/00501], SingHealth Centralised Institutional Review Board [CIRB Ref: 2010/632/B], and National University Hospital Singapore [NUS-IRB: 11–075].
Germline analyses
Germline DNA of breast cancer cases and controls were sequenced in two batches, using targeted sequencing panels that target the coding regions and exon-intron boundaries of known and suspected breast cancer susceptibility genes respectively, which included BRCA2 (Dorling, 2020). Target enrichment were performed using the Fluidgm Access Array system (n=5,090) or the Fluidgm Juno system (n=11,342) and sequenced on Illumina HiSeq 2500 or HiSeq 4000. Library preparations were performed according to manufacturer’s protocols as described previously (Dorling, 2020).All pathogenic and variants of uncertain significance were confirmed by Sanger sequencing.
In silico prediction
Potential clinical impact of BRCA2 variants were analyzed using three in silico prediction tools: Alignment-Grantham variation Grantham deviation (Align-GVGD) (Tavtigian et al., 2006), Combined Annotation Dependent Depletion (CADD) (Kircher et al., 2014) and Rare Exome Variant Ensemble Learner (REVEL) (Ioannidis et al., 2016). The GV and GD scores are used to determine the Align-GVGD class (C0, C15, C25, C35, C45, C55, C65). Align-GVGD scores represent impact of amino acid change on protein function (C0 class suggests the change is not predicted to have an impact and C65 suggests a severe impact on protein function). Variants with CADD score ≥ 20 were predicted to be likely deleterious. While the cut-off score for REVEL is set at 0.5 (<0.5 for likely benign and >0.5 for likely pathogenic).
Expression of BRCA2 VUS in PL2F7 mES cells
Desired variants were generated in human BRCA2 cloned in a bacterial artificial chromosome clone (CTD-2342K5 with a 127 kb insert) containing full-length BRCA2 in SW102 cells as described previously (Biswas, Das, et al., 2012; Biswas, Stauffer, & Sharan, 2012). Nucleotide changes were introduced by galK selection and counter-selection method (Warming, Costantino, Court, Jenkins, & Copeland, 2005). Oligonucleotide sequences are available upon request. BAC DNA (20μg) carrying various variant alleles of BRCA2 was electroporated into 1.0×107 PL2F7 mES cells, selected in the presence of G418 (Invitrogen) and characterized as described previously (Kuznetsov et al., 2008). BRCA2 expression was confirmed by detecting the presence of BRCA2 transcript using the primers from exon 11 (5′- TGGTTTTGTCAAATTCAAGAATTGG −3′) and exon 14 (5′- CCAATCAAGCAGTAGCTGTAACTTTCAC-3′). RT-PCR was carried out using the Titan One Tube RT-PCR system (Roche) following the manufacturer’s protocol and the amplified 600 bp PCR product was detected by agarose gel electrophoresis.
BRCA2 functional analysis in mES cells
To examine the effect of BRCA2 variants on viability of Brca2ko/ko mES cells, the conditional allele of Brca2 in PL2F7 mES cells expressing each variant was deleted by electroporating 25 μg of Pgk-Cre as described previously (Kuznetsov et al., 2010). Recombinant colonies were selected in HAT media (Gibco) after seeding 1X105 cells in a 100-mm dish. HAT resistant (HATr) colonies were visualized by staining with methylene blue (2% methylene blue (wt/vol) in 70% ethanol) for 15 min followed by washing in 70% ethanol. Loss of conditional allele of Brca2 was confirmed by Southern as described previously (Kuznetsov et al., 2008). The number colonies grown in the presence (HATr recombinant colonies) and absence (number of cells plated) of HAT were counted. Viability percentage was determined using the formula (number of HATr colonies/number of cell plated) X 100. Relative viability compared to WT was calculated by dividing the viability percentage of particular variant with viability percentage of WT used in that batch of experiment. Sensitivity assays to different drugs and ionizing radiation were performed as described previously (Kuznetsov et al., 2008).
Effect of variants on splicing
We examined the effect of potential splice site variants (IVS2–7T>A, IVS7–10insT and IVS22–5DdelAACA) on aberrant splicing by RT-PCR, Total RNA was extracted using RNA-BEE (Tel-Test, Inc.) according to the manufacturer’s protocol. To detect alternatively spliced forms of BRCA2, RT-PCR analysis was performed using the Qiagen one-step RT-PCR kit (Roche) following the manufacturer’s protocol. The sequence of primers used: IVS2–7T>A: Forward exon 1 primer 5´-GCGGTTTTTGTCAGCTTAC-3´; Reverse exon 10 primer 5´-CAGCGTTTGCTTCATGGA-3´, IVS7–10insT: Forward exon 2 primer 5´TAAGACACGCTGCAACAAAGC3-´; Reverse exon 10 primer 5´-CAGCGTTTGCTTCATGGA-3´, IVS22–5DdelAACA: Forward exon 21 primer 5´GCAAGATGGTGCAGAGCTTT3´; Reverse exon 25 primer 5´-GACTTGCCCCTTTCGTCTAT-3´
System Preparation for Molecular Dynamic analysis
The structure of the mouse BRCA2 was retrieved from the structure of the BRCA2/DSS1 complex (PDB id: 1miu). The gaps (ILE2614 - VAL2637; LYS2795 - ARG2809) in the protein were modelled using Modeller (Sali & Blundell, 1993) in UCSF Chimera (Pettersen et al., 2004) using the DOPE-HR (Shen & Sali, 2006) algorithm. The human BRCA2 (UniProt ID: P51587) was modelled using SWISS-MODEL using the mouse BRCA2 as a template (Waterhouse et al., 2018). The Y3035C/F/S substitutions were performed using UCSF chimera (Shapovalov & Dunbrack, 2011). The protein systems for wild-type BRCA2 and the mutants were prepared with the LEaP program in Ambertools18 (Team, 2017). The systems were prepared in a cubic box of TIP4P/2005 water, with a minimum distance of at least 1.2 nm between solute atoms and box edge. Counter ions were added to neutralize each system (Best & Mittal, 2010b).
Molecular Dynamic Simulation
Parameters describing system topology were based on the Amber ff03w force field (Best & Mittal, 2010a). The systems were first relaxed by energy minimization using the Sander module of Amber18 in two stages; in the first stage, the waters were minimized, while in the second stage, both the protein and water were minimized. The systems were then heated from 0 K to 300K incrementally for 5 ns during which positional restraints (20 kcal/mol/Å2) was applied to all protein atoms. The density of the system was equilibrated to 1 g/cm3 for 5ns using canonical ensemble (NVT). Subsequently, NPT equilibrations were run with restraints of the backbone atoms “C, CA, N, O” reduced from 10 to 0 through a series of four 1ns molecular dynamics simulations. Finally, 1μs production was run in PMEMD (Particle Mesh Ewald Molecular Dynamics) module of Amber18 using the NPT ensemble. The distance cut-off for short-range non-bonded interactions was set to 1 nm. The particle mesh Ewald (PME) method was employed to treat long-range electrostatic interactions. The SHAKE algorithm was applied to constrain all bonds involving hydrogen atoms. The temperature was set to 300 K using Langevin thermostat with a collision frequency of 1.0 ps−1. The Pressure was maintained at 1 bar using the Berendsen barostat with a coupling constant of 1.0 ps. Using the hydrogen mass repartitioning (HMR) scheme (Hopkins, Le Grand, Walker, & Roitberg, 2015), the integration time step was set to 4 fs. Dynamics were propagated using the leapfrog integrator. The structure was saved every 40 ps, and the total NPT simulation time was 1μs.
Molecular Dynamic Analysis
The trajectories were analyzed using the Cpptraj module of Ambertools18, and the RMSD graphs were plotted using R software (Roe & Cheatham, 2013; Team, 2017). The Residue interaction network (RIN) analysis was performed on trajectories using UCSF Chimera, StructureViz2, RINalyzer, and the network were visualized using Cytoscape (Doncheva, Klein, Domingues, & Albrecht, 2011; Morris, Huang, Babbitt, & Ferrin, 2007; Shannon et al., 2003). The RIN was created using the “contact” algorithm, for the wild-type and substituted residues with the rest of the protein.
RESULTS AND DISCUSSION
BRCA2 VUS in Malaysian cohort
We selected 14 BRCA2 VUS identified in BRCA1 and BRCA2 testing of 467 breast cancer patients in Malaysian (Lai et al., 2017; Table 1) for further analysis and characterization. The missense variants map to different exons ranging from 10 to 27. Eleven of the variants are missense variants (NM_000059.3:c.1600G>A, (p.Glu534Lys); NM_000059.3:c.3782C>G (p.Ser1261Cys); NM_000059.3:c.6322C>T (p.Arg2108Cys); NM_000059.3:c.6929C>A, (p.Thr2310Asn); NM_000059.3:c.8356G>A (p.Ala2786Thr); NM_000059.3:c.8393C>T (p.Pro2798Leu); NM_000059.3:c.9104A>G (p.Tyr3035Cys); NM_000059.3:c.9106C>G (p.Gln3036Glu); NM_000059.3:c.9344A>G (p.Lys3115Arg); NM_000059.3:c.9538C>T (p.Leu3180Phe) and NM_000059.3:c.9907A>T (p.Ser3303Cys)) and the remaining three are potential splice site variants and map to introns 2, 7 and 22 (NM_000059.3:c.68–7T>A (IVS2–7T>A); NM_000059.3:c.632–10dupT, (IVS7–10insT); NM_000059.3:c.8954–5_8954–2delAACA, (IVS22–5delAACA). Hereafter, for readability, the protein format is used to describe the variants and a single letter amino acid code will be used instead of the three letter code. Similarly, intronic variants will be referred to by their intron number and location within the intron. All variants are listed either as variant of uncertain clinical significance or having conflicting interpretation in ClinVar except for IVS2–7T>A, which is considered benign. IVS7–10insT is not listed in ClinVar (Table 1).
Table 1:
List of BRCA2 VUS and Summary of in silico analysis
| Variant | Protein Change * | Exon/ Intron | Align-GVGD Grade | PRIOR Score | CADD_ Phred | CADD_RAW | REVEL | ClinVAR | Published Functional data (references) |
|---|---|---|---|---|---|---|---|---|---|
| c.1600G>A | p.Glu534Lys(E534K) | Ex 10 | C0 | 0.02 | 13.12 | 0.975605 | 0.207 | VUS | None |
| c.3782C>G | p.Ser1261Cys (S1261C) | Ex 11 | C0 | 0.02 | 14.16 | 1.148972 | 0.161 | VUS | None |
| c.6322C>T | p.Arg2108Cys (R2108C) | Ex 11 | C0 | 0.02 | 11.59 | 0.763875 | 0.240 | Conflicting interpretations | Pathogenic (Balia, Galli, & Caligo, 2011) Benign (Parsons et al., 2019) |
| c.6929C>A | p.Thr2310Asn (T2310N) | Ex 12 | C0 | 0.02 | 24.7 | 3.402632 | 0.423 | VUS | None |
| c.8356G>A | p.Ala2786Thr (A2786T) | Ex 19 | C0 | 0.03 | 23.1 | 2.746416 | 0.705 | VUS | Benign (Parsons et al., 2019) |
| c.8393C>T | p.Pro2798Leu (P2798L) | Ex 19 | C65 | 0.81 | 26.7 | 3.842999 | 0.537 | VUS | None |
| c.9104A>G | p.Tyr3035Cys (Y3035C) | Ex 23 | C55 | 0.66 | 27.4 | 3.931712 | 0.684 | VUS | Likely Deleterious (Karchin, Agarwal, Sali, Couch, & Beattie, 2008) Benign (Parsons et al., 2019) Benign (Guidugli et al., 2018) |
| c.9104A>T | p.Tyr3035Phe (Y3035F) | Ex 23 | C0 | 0.03 | 25.9 | 3.690516 | 0.282 | VUS | None |
| c.9104A>C | p.Tyr3035Ser (Y3035S) | Ex 23 | C55 | 0.66 | 26.9 | 3.87055 | 0.672 | VUS | Likely Deleterious (Karchin et al., 2008) Benign (Parsons et al., 2019) Intermediate (Guidugli et al., 2018) Moderate (Shimelis et al., 2017) Functional class I or Benign (Ikegami et al., 2020) |
| c.9106C>G | p.Gln3036Glu (Q3036E) | Ex 23 | C0 | 0.03 | 26.1 | 3.741717 | 0.201 | Conflicting interpretations | None |
| c.9344A>G | p.Lys3115Arg (K3115R) | Ex 25 | C0 | 0.03 | 22.6 | 2.498113 | 0.456 | VUS | None |
| c.9538C>T | p.Leu3180Phe (L3180F) | Ex 26 | C0 | 0.03 | 26.2 | 3.763685 | NA | VUS | None |
| c.9907A>T | p.Ser3303Cys (S3303C) | Ex 27 | C15 | 0.29 | 23.5 | 2.926882 | 0.095 | VUS | None |
| c.68–7T>A | NA (IVS2–7T>A) | Int 2 | NA | NA | 10.14 | 0.583839 | NA | Benign | No exon skipping (Thery et al., 2011) |
| c.632-10dupT | NA (IVS7–10insT) | Int 7 | NA | NA | NA | NA | NA | NA | None |
| c.8954-5_89542delAACA | NA (IVS22–5delAACA) | Int 22 | NA | NA | NA | NA | NA | Conflicting interpreta tions | None |
Align-GVGD: in silico prediction tool; different C scores have different probabilities of being pathogenic, C0: prior probability of pathogenicity of 0.03; C15: prior probability of pathogenicity of 0.29; C55: prior probability of pathogenicity of 0.66; C65: prior probability of pathogenicity of 0.81. PRIOR Scoresobtained from http://priors.hci.utah.edu/PRIORS/, Probability of pathogenicity scores: 0.02, 0.03 = weak/null, 0.29 = Low, 0.66: Moderate, 0.81 = High probability of pathogenicity; CADD_phred: in silico prediction tool; CADD score≥ 20 = likely to be deleterious; CADD_raw: CADD raw score; Revel: in silico prediction tool; Revel score > 0.5 =pathogenic; NA: No applicable
short variant name used in the text is mentioned within parentheses.
One of the missense variants identified is Y3035C. The 3035 residue is present in a region that contains three oligonucleotide-binding (OB) folds and one helix-turn-helix motif (Suppl. Fig. 1A)(Yang et al., 2002). Two other variants affecting the same residue, Y3035S (NM_000059.3:c.9104A>C; p.Tyr3035Ser) and Y3035F (NM_000059.3:c.9104A>T; p.Tyr3035Phe), are listed in the ClinVar database. Based on the protein likelihood ratios using the bioinformatic information about protein sequence, conservation and structure Y3035C and Y3035S were classified as deleterious, whereas Y3035F was classified as neutral (Karchin, Agarwal, Sali, Couch, & Beattie, 2008) Y3035C was recently reported to have no impact on HR (Guidugli et al., 2018; Shimelis et al., 2017). Y3035S was found to be associated with an increased risk to breast cancer in the European population with an odds ratio 2.52 (p=0.04) and homologous DNA recombination assays showed that it has partial impact on BRCA2 function (Guidugli et al., 2018; Shimelis et al., 2017). However, recent multifactorial likelihood ratio model using functional and clinical data predicted Y3035S to be likely benign (Parsons et al., 2019). This was supported by a functional assay based on sensitivity to multiple PARP inhibitors of BRCA2 (−/−) human DLD1 cells expressing this variant that classified Y3035S as functional class 1 or benign variant (Ikegami et al., 2020). We examined the structural impact by molecular dynamic analysis, which simulates the conformational movements of residues that can change the non-covalent interactions. The Residue Interaction Network (RIN) analysis revealed that Y3035 interacted with hydrophobic residues: I2989, L2996, L2999, L3000, I3008, and A3029 (Suppl. Fig. 1B). Moreover, Y3035 interacted with T3033 and R3007 through hydrogen bonds. Both the hydrophobic substitutions, F3035 and C3035, retained most of the interactions (Suppl Fig.1C and 1D, respectively). However, serine is a polar and hydrophilic amino acid, and the S3035 variant lost all the hydrophobic interactions and maintained only a few contacts (Suppl. Fig. 1E). To examine the functional impact of these variants, we included them in our study for comparison.
Some functional characterization of a few other variants have also been reported (Table 1). Using a functional assay based on the stimulation of homologous DNA recombination activity by transient overexpression of variants in a derivative of HeLa cells (HelaG1), R2108C was classified as likely pathogenic because of stimulation of HR activity comparable to a pathogenic variant although the stimulation was higher than WT control (Balia, Galli, & Caligo, 2011). Recently, based on multifactorial likelihood ratio, this variant as well as A2786T were classified as benign variants (Parsons et al., 2019). One of the splice variants, c.68–7T>A (IVS2–7T>A) was found to increase the level of exon 3 skipping in patient blood samples, but the clinical significance was not clear due to the presence of exon 3 deleted transcript in control samples (Vreeswijk et al., 2009). However, splicing mini-gene assays failed to detect exon 3 skipping, suggesting it to be a neutral variant (Thery et al., 2011).
In silico evaluation of BRCA2 VUS
We used some of the available in silico tools to predict the impact of single amino acid alterations on BRCA2 function (Table 1). Align-GVGD (http://agvgd.hci.utah.edu/), classified all missense variants to be class C0 or C15 and hence likely to be neutral, except for P2798L, Y3035S, and Y3035C that were class C65 or C55 (Tavtigian et al., 2006; Tavtigian, Greenblatt, Lesueur, Byrnes, & Group, 2008). Align-GVGD scores have been found to be relevant mainly in the functionally important DNA binding domain of BRCA2 as variants outside this region are likely to have low prior probability of pathogenicity. This is now taken into consideration in the prior probability scores obtained from the HCI priors database (http://hci-priors.hci.utah.edu/PRIORS/) (Vallee et al., 2016). These scores also predicted P2798L to have high impact and Y3035S and Y3035C to have moderate impact on protein function (Table 1). Most other variants have weak or no impact except S3303C, which is predicted to have low impact.
We examined Combined Annotation-Dependent Depletion (CADD) score (https://cadd.gs.washington.edu/score), which takes into consideration various genomic features that are combined into a single CADD score via a machine learning model (Kircher et al., 2014; Rentzsch, Witten, Cooper, Shendure, & Kircher, 2019). CADD scores are converted into a PHRED-like rank score by utilizing the genome-wide distribution of scores of every possible single nucleotide variant. All variants had a CADD-Phred, score ≥ 20 are predicted to be likely deleterious, except for E534K, S1261C, and R2108C. Finally, we used Rare Exome Variant Ensemble Learner (REVEL), an ensemble method to predict the pathogenicity of variants (https://sites.google.com/site/revelgenomics/). REVEL was reported to have the best overall predictive ability to differentiate between neutral and pathogenic variants as compared to some of the other predictive tools (Ioannidis et al., 2016). Using a REVEL score of greater than 0.5 to classify a variant as pathogenic, A2786T, P2798L, Y3035C and Y3035S were predicted to be pathogenic. Although there was considerable variability in the prediction of different in silico tools, all models predicted P2798L, Y3035C and Y3035S to be likely pathogenic (Table 1).
Epidemiological analysis of BRCA2 VUS
When first reported, no information was available on the frequency of these variants in the Malaysian and Singaporean population and their association with different ethnic groups (Malays, Chinese, Indian and other indigenous groups) (Thirthagiri et al., 2008). To characterize these variants, we have examined the presence of these variants in an ongoing targeted sequencing cohort of 7,840 breast cancer cases (including 22 cases of ductal carcinoma in situ) and 7,928 controls in Malaysia and Singapore. The frequency of 5 variants (R2108C, A2786T, IVS22–5delAACA, T2310N and Y3035C) were similar in cases and controls, suggesting that these are likely to be benign (Table 2). Three rare variants including P2798L, IVS7–10insT and S3303C were found only in the cases. The majority of the variants are present in multiple ethnic groups (Supplementary Information 1). However, some are restricted to specific ethnic groups such as S1261C was found in Malays, T2310N and P2798L in Indians, Y3035C and Q3036E in Chinese. Notably, E534K, R2108C, A2786T, Y3035C, Q3036E, L3180F and IVS22–5delAACA were > 10 times more common in East Asians compared to the Non-Finnish European population (Table 2, Supplementary Information 1).
Table 2:
Prevelance of BRCA2 VUS Identified in Malaysia and Singapore
| Variant | No. of carriers (Case) | No. of carriers (Control) | 1000g_EAS_AF | 1000g_EUR_AF | gnomAD_AF_Overall | gnomAD_ EAS_AF | gnomAD_ SAS_AF | gnomAD_ NFE_AF | sub-pop/general pop | Crude odds ratio | p-value |
|---|---|---|---|---|---|---|---|---|---|---|---|
| c.1600G>A (E534K) | 2 | 1 | 1.00E-03 | NA | 2.05E-05 | 2.92E-04 | NA | NA | 14.23 (EAS) | 2.02 | 0.57 |
| c.3782C>G (S1261C) | 1 | 1 | NA | NA | NA | NA | NA | NA | NA | 1.01 | 0.99 |
| c.6322C>T (R2108C) | 119 | 120 | 3.00E-03 | 2.00E-03 | 5.93E-04 | 6.47E-03 | 6.83E-05 | 2.09E-04 | 10.91 (EAS) | 1.00 | 0.98 |
| c.6929C>A (T2310N) | 4 | 4 | NA | NA | 3.26E-05 | NA | 1.96E-04 | NA | 6.02 (SAS) | 1.01 | 0.99 |
| c.8356G>A (A2786T) | 10 | 11 | NA | NA | 6.09e-05 | 8.12E-04 | 3.27E-05 | NA | 13.32 (EAS) | 0.99 | 0.85 |
| c.8393C>T (P2798L) | 1 | 0 | NA | NA | NA | NA | NA | NA | NA | 3.03 | 0.50 |
| c.9104A>G (Y3035C) | 5 | 4 | NA | NA | 2.05E-05 | 2.34E-04 | NA | 9.07E-06 | 11.43 (EAS) | 1.26 | 0.73 |
| c.9104A>T (Y3035F) | 0 | 0 | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| c.9104A>C (Y3035S) | 0 | 0 | NA | NA | 4.99E-05 | NA | NA | 1.02E-04 | 2.04 (NFE) | NA | NA |
| c.9106C>G (Q3036E) | 4 | 2 | 1.00E-03 | 0 | 3.93E-05 | 5.27E-04 | NA | NA | 13.43 (EAS) | 2.02 | 0.42 |
| c.9344A>G (K3115R) | 1 | 1 | NA | NA | 4.47e-05 | NA | 2.00E-04 | NA | 4.47 (SAS) | 1.01 | 0.99 |
| c.9538C>T (L3180F) | 2 | 2 | 1.00E-03 | NA | 4.06E-05 | 5.22E-04 | NA | NA | 12.85 (EAS) | 1.01 | 0.99 |
| c.9907A>T S3303C) | 1 | 0 | NA | NA | NA | NA | NA | NA | NA | 3.03 | 0.50 |
| c.68–7T>A (IVS2–7T>A) | 5 | 1 | NA | NA | 2.84E-03 | 2.02E-04 | 1.90E-03 | 2.46E-03 | NA* | 5.06 | 0.14 |
| c.632–10dupT (IVS710insT) | 1 | 0 | NA | NA | NA | NA | NA | NA | NA | 3.03 | 0.50 |
| c.8954-5_89542delAACA (IVS225delAACA) | 24 | 26 | NA | NA | 1.06E-04 | 1.51E-03 | NA | NA | 14.14 (EAS) | 0.93 | 0.81 |
1000g_EAS and 1000g_EUR: Allelic frequency in East Asians and Europeans respectively in 1000 genome database; gnomAD_Overall, gnomAD_EAS, gnomAD_SAS, and gnomAD_NFE: Allelic frequency overall, and in East Asians, South Asians and Non-Finnish Europeans respectively in the gnomAD database; Enrichment in Asians: enrichment of variant in either East Asian (EAS) or South Asian (SAS) population compared to the allelic frequency in gnomAD overall
indicates MAF»0.3% in general population; Crude odds ratio were calculated using total breast cases (n=7840) vs controls (n=7928); NA: not applicable.
Effect of BRCA2 VUS on mESC survival and splicing
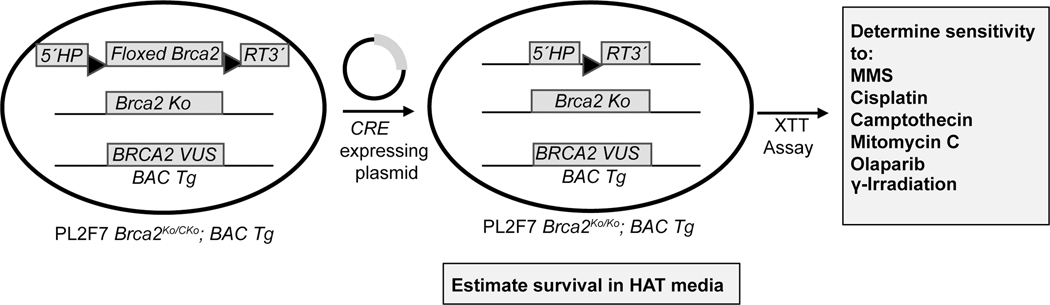
We used a mES cell-based functional assay to examine the functional impact of these variants. All variants were generated in a bacterial artificial chromosome (BAC) containing the full-length human BRCA2 and expressed in PL2F7 mES cells. In these cells, one of the alleles of Brca2 is a functionally null allele (ko) and the other is a conditional allele (cko) flanked by two loxP sites (Fig. 1). We confirmed the expression of BRCA2 transgene by RT-PCR using primers from exons 11 and 14. Three of the potential splice site variants were further examined using primers specific to the exons of the flanking region that may be impacted by aberrant splicing (Supplementary Fig. 2A–C). However, there was no change in the presence or abundance of different alternatively spliced transcript and there was no apparent reduction in the full-length transcript. To examine the ability of the variants to support the viability of Brca2ko/ko mES cells, we deleted the conditional allele in two independently generated mES cell clones expressing each variant. In every case, the number of viable cells obtained was comparable to those obtained from PL2F7 mES cells expressing WT BRCA2 (Supplementary Table 1). The viability of all variants relative to WT control ranged between 0.61 and 2.48 (Table 3). We owe this variability in relative viability to batch to batch experimental variation. However, in every case, at least one clone exhibited relative viability > 0.8. The loss of the conditional allele in viable clones was confirmed by Southern blot analysis (data not shown). We concluded that none of the variants disrupt the ability of BRCA2 to support cell viability and these variants are likely to be hypomorphic or functionally neutral.
Figure 1: Functional analysis of BRCA2 in mouse ES Cells.
Schematic representation of the mouse ES cell-based functional assay. BRCA2 variants are generated in the gene cloned in a BAC. These variants are expressed in PL2F7 mouse ES cell with a functionally null (ko) and a conditional allele (cko) of Brca2. The conditional allele is flanked by loxP sites with a split human HPRT minigene. A functional HPRT is generated when the conditional allele is deleted by Cre-mediated recombination between the loxP sites. Recombinants are selected in HAT media. Viable Brca2ko/ko cells expressing BRCA2 VUS are then tested for sensitivity to six different DNA damaging agents to examine the effect of VUS on BRCA2 function.
Table 3:
Summary of Functional Analysis of Brca2ko/ko mES Cells Expressing BRCA2 VUS
| Variant | Functional Analysis | |||
|---|---|---|---|---|
| Relative Viability | Sensitivity to DNA damaging agents | |||
| Clone 1 | Clone 2 | Clone 1 | Clone 2 | |
| c.1600G>A (E534K) | 0.68 | 0.95 | None | None |
| c.3782C>G (S1261C) | 1.01 | 0.84 | None | None |
| c.6322C>T (R2108C) | 1.26 | 1.75 | None | None |
| c.6929C>A (T2310N) | 0.81 | 0.96 | None | None |
| c.8356G>A (A2786T) | 0.96 | 1.01 | None | None |
| c.8393C>T (P2798L) | 1.00 | 0.99 | None | None |
| c.9104A>G (Y3035C) | 0.95 | 0.87 | None | None |
| c.9104A>T (Y3035F) | 0.90 | 0.92 | None | None |
| c.9104A>C (Y3035S) | 1.35 | 1.87 | None | None |
| c.9106C>G (Q3036E) | 1.15 | 0.91 | None | None |
| c.9344A>G (K3115R) | 0.61 | 0.84 | None | None |
| c.9538C>T (L3180F) | 2.70 | 1.68 | None | None |
| c.9907A>T S3303C) | 1.01 | 0.73 | None | None |
| c.68-7T>A (IVS2-7T>A) | 1.20 | 1.12 | None | None |
| c.632-10dupT (IVS7-10insT) | 1.17 | 0.88 | None | None |
| c.8954-5_89542delAACA (IVS225delAACA) | 2.46 | 2.48 | None | None |
Relative viability was calculated by taking the ratio of percentage of HAT resistant (HATR) cells of respective variant expressing clone over percentage of HATR cells of WT BRCA2 expressing clone after deletion of the conditional allele of endogenous Brca2 by CRE-mediated recombination. Sensitivity of DNA damaging agents refers to sensitivities to camptothecin, mitomycin C, cisplatin, MMS, olaparib and ionizing radiation (IR). For each variant, two independent mES cell clones were tested and referred as Clone 1 and Clone 2.
Effect of BRCA2 VUS in Sensitivity to DNA damaging agents
To test the DNA repair function of the BRCA2 variants, we challenged the viable Brca2ko/ko mES cells expressing each of the variants with poly ADP ribose polymerase (PARP) inhibitor (olaparib), replication stress inducers (cisplatin, camptothecin), DNA inter-strand crosslinking agent (mitomycin C, MMC), alkylating agents (methyl methane-sulfonate, MMS) and DSB inducer (ionizing radiation, IR) (Kuznetsov et al., 2010). Sensitivity of Brca2ko/ko mES cells for each variant was compared with Brca2KO/KO mES cells expressing WT BRCA2. None of the variants exhibited sensitivity to these DNA damaging agents (Supplementary Fig. 3, Table 3). Y3035S, which has been reported to be a moderate-risk variant (OR2.52), also did not exhibit sensitivity to any of the DNA damaging agents including Olaparib (Shimelis et al., 2017). Interestingly, the other two variants affecting the same residue, Y3035C, which was found in the Malaysian population and Y3035F were also not sensitive to any of the DNA damaging agents suggesting that these are all neutral variants (Supplementary Fig 3G and 3I). In spite of the structural differences that were predicted based on molecular dynamic analysis (Supplementary Fig. 1), we did not observe any functional difference between these three variants. If future studies validate Y3035S to be a moderate risk variant, it will reflect a limitation of our assay.
In conclusion, we have examined the functional impact of 14 BRCA2 VUS identified in the Malay, Chinese and Indian population living in Malaysia and Singapore. Most of these were novel variants when first identified in one or two Malaysian families. In the absence of any co-segregation data or co-occurrence with other pathogenic variants, it was not possible to ascertain the clinical significance of these variants. Our epidemiological studies have provided valuable data on the frequency of these variants in various populations, sub-populations and ethnic groups. The classification of the variants based on our findings and following the guidelines of the American College of Medical Genetics and Genomics (ACMG) are summarized in Table 4. As per the guidelines, variants are described using specific standard terminology, “pathogenic”, “likely pathogenic”, “uncertain”, “likely benign” and “benign”. These classifications are based on a number of evidences that are denoted by specific codes (Richards et al., 2015). For example, in several cases, the VUS reported here were observed at a frequency which is greater than expected for breast cancer risk gene, supporting their neutral classification. As per the ACMG guidelines, we have assigned BS1 to these variants (Table 4). Although computational tools can be useful in predicting impact of amino acid changes on the protein structure and function, we did not observe consistency in the in silico prediction of different models, Align-GVGD, prior scores, CADD-Phred and REVEL. Yet, several variants were predicted to be neutral by multiple models and three of the variants, P2798L, Y3035C, and Y3035S were predicted to be pathogenic by all four models (Table 4, BP4, PP3). Our mouse ES cell-based evaluation of the impact of the variants showed that none of the variants affected cell viability or exhibited sensitivity to DNA damaging agents (Table 4, BS3). Taken together, we conclude that the 14 variants identified in the Chinese, Malay and Indian population are likely to be benign and not associated with significant increased disease risk.
Table 4:
Classification of Novel BRCA2 Variants According to the ACMG guidelines
| Variant | Cell viability assay/sensitivity to DNA damaging agents | Classification (evidence) |
|---|---|---|
| c.1600G>A (E534K) | Neutral/Neutral | Likely benign (BS1, BP4, BS3) |
| c.3782C>G (S1261C) | Neutral/Neutral | Likely benign (BP4, BS3) |
| c.6322C>T (R2108C) | Neutral/Neutral | Likely benign (BS1, BP4, BS3) |
| c.6929C>A (T2310N) | Neutral/Neutral | Likely benign (BS1, BS3) |
| c.8356G>A (A2786T) | Neutral/Neutral | Likely benign (BS1, BS3) |
| c.8393C>T (P2798L) | Neutral/Neutral | Likely benign (BP2, PP3, BS3) |
| c.9104A>G (Y3035C) | Neutral/Neutral | Likely benign (BS1, PP3, BS3) |
| c.9104A>T (Y3035F) | Neutral/Neutral | Likely benign (BP4, BS3) |
| c.9104A>C (Y3035S) | Neutral/Neutral | Uncertain (PP3, BS3, PP1) |
| c.9106C>G (Q3036E) | Neutral/Neutral | Likely benign (BS3, BP2) |
| c.9344A>G (K3115R) | Neutral/Neutral | Likely benign (BS1, BS3, BP4) |
| c.9538C>T (L3180F) | Neutral/Neutral | Likely benign (BS1, BS3) |
| c.9907A>T S3303C) | Neutral/Neutral | Likely benign (BP4, BS3) |
| c.68-7T>A (IVS2-7T>A) | Neutral/Neutral | Likely benign (BS1, BS3, BP6) |
| c.632-10dupT (IVS7-10insT) | Neutral/Neutral | VUS (BS3) |
| c.8954-5_8954-2delAACA (IVS22-5delAACA) | Neutral/Neutral | Likely benign (BS1, BS3) |
Description of ACMG codes (Richards et al., 2015):
BS1:frequency is greater than expected for disorder
BS3:Well-established in vitro or in vivo functional studies show no damaging effect on protein function or splicing
BP4:Multiple lines of computational evidence suggest no impact on gene or gene product (conservation, evolutionary, splicing impact, etc.)
BP2:Observed in trans with a pathogenic variant for a fully penetrant dominant gene/disorder or observed in cis with a pathogenic variant in any inheritance pattern
BP6:Reputable source recently reports variant as benign
PP3:Multiple lines of computational evidence support a deleterious effect on the gene or gene product
PP1:Co-segregation with disease in multiple affected family members in a gene definitively known to cause the disease
Supplementary Material
ACKNOWLEDGMENTS
We thank Ms. Laura Brown (NCI-Frederick) for her technical help. SGBCC and MYBRCA thank the participants and all research coordinators for their excellent help with recruitment, data and sample collection. MYBRCA is funded by research grants from the Malaysian Ministry of Higher Education (UM.C/HlR/MOHE/06), the Wellcome Trust [grant no: v203477/Z/16/Z], Yayasan Sime Darby, Yayasan PETRONAS, Estee Lauder Breast Cancer Campaign and Cancer Research Malaysia. MYMAMMO is supported by research grants from Yayasan Sime Darby LPGA Tournament and Malaysian Ministry of Higher Education (RP046B-15HTM). SGBCC was supported by the National Research Foundation Singapore (NRF-NRFF2017–02, awarded to J Li), NUS start-up Grant, National University Cancer Institute Singapore (NCIS) Centre Grant [NMRC/CG/NCIS/2010, NMRC/CG/012/2013, CGAug16M005], Saw Swee Hock School of Public Health Research Programme of Research Seed Funding (Breast Cancer Prevention Program), Asian Breast Cancer Research Fund, and the NMRC Clinician Scientist Award (SI Category) [NMRC/CSA-SI/0015/2017]. Controls from Singapore were recruited by the Singapore Consortium of Cohort Studies-Multi-ethnic cohort (SCCS-MEC), which was funded by the Biomedical Research Council, grant number: 05/1/21/19/425. The sequencing and analysis for this project was funded by the European Union’s Horizon 2020 Research and Innovation Programme (BRIDGES: grant number 634935) and the Wellcome Trust [grant no: v203477/Z/16/Z]. The research was sponsored by the Intramural Research Program, Center for Cancer Research, National Cancer Institute, U.S. National Institutes of Health (SKS).
KEY FUNDING SOURCES: This research was sponsored by the Intramural Research Program of US National Institutes of Health. Malaysian Ministry of Higher Education (UM.C/HlR/MOHE/06), The European Union’s Horizon 2020 Research and Innovation Programme (BRIDGES: grant number 634935) and the Wellcome Trust [grant no: v203477/Z/16/Z]. Malaysian Ministry of Higher Education (RP046B-15HTM). National Research Foundation Singapore (NRF-NRFF2017–02), National University Cancer Institute Singapore (NCIS) Centre Grant [NMRC/CG/NCIS/2010, NMRC/CG/012/2013, CGAug16M005], Saw Swee Hock School of Public Health Research Programme of Research Seed Funding (Breast Cancer Prevention Program), Asian Breast Cancer Research Fund, and the NMRC Clinician Scientist Award (SI Category) [NMRC/CSA-SI/0015/2017].
APPENDIX.
Full list of Singapore Breast Cancer Cohort (SGBCC) Authors
Swee Ho Lim 1,2, Ern Yu Tan 3, Benita Kiat Tee Tan 2,4,5, Su-Ming Tan 6, Veronique Kiak Mien Tan 2,4,5, Ching Wan Chan7, Siau-Wei Tang7, Celene Wei Qi Ng7, Geok Hoon Lim1, Jinnie Siyan Pang1, Jung Ah Lee1, Patrick Mun Yew Chan3, Juliana Chen3, Sarah Qinghui Lu3, Yirong Sim2,4, Wei Sean Yong 2,4,5, Preetha Madhukumar2,4,5, Fuh Yong Wong8, Joanne Yuen Yie Ngeow9,10, Tira Jing Ying Tan9, Wai Peng Lee6, Chi Wei Mok6, Chin Mui Seah6, Linda Tan11, E Shyong Tai11,12, Xueling Sim11, Peh Joo Ho13, Alexis Jiaying Khng13
1 Breast Department, KK Women’s and Children’s Hospital, Singapore
2 SingHealth Duke-NUS Breast Centre, Singapore
3 Department of General Surgery, Tan Tock Seng Hospital, Singapore
4 Division of Surgical Oncology, National Cancer Centre Singapore, Singapore
5 Department of General Surgery, Singapore General Hospital, Singapore
6 Division of Breast Surgery, Department of General Surgery, Changi General Hospital, Singapore
7 Department of Surgery, University Surgical Cluster, National University Hospital and National University Health System, Singapore
8 Division of Radiation Oncology, National Cancer Centre Singapore, Singapore
9 Division of Medical Oncology, National Cancer Centre Singapore, Singapore
10 Cancer Genetics Service, National Cancer Centre Singapore, Singapore
11 Saw Swee Hock School of Public Health, National University of Singapore and National University Health System, Singapore
12 Yong Loo Lin School of Medicine, National University of Singapore and National University Health System, Singapore
13 Genome Institute of Singapore, Human Genetics, Singapore, Singapore
Full list of Malaysian Breast Cancer Study (MyBrCa) Authors
Nur Aishah Taib1,2, Cheng Har Yip3, Sook-Yee Yoon4, Weang Kee Ho5, Pei Sze Ng4, Shivaani Mariapun4, Siti Norhidayu Hassan4, Daphne Lee4, Tiara Hasan4, Meow Keong Thong6, Min Min Tan4, Joanna Lim4, Shao Yan Lao4, Chan Eng Chong4, Eldarina Wijaya4, Nadia Rajaram4, Wei Xiong Wen4, See Mee Hoong1, Suniza Jamaris1, Tania Islam1, Teh Mei Sze1, Teoh Li Ying1, Kartini Rahmat7, Farhana Fadzli7, Caroline J. Westerhout7, Anushya Vijayananthan7, Faizah Harun1, Hanani Che Halim1, Ernie Azwa Yusop1, Zurina Che Rohani1.
1 Department of Surgery, Faculty of Medicine, University Malaya
2 University Malaya Cancer Research Institute, Faculty of Medicine, University Malaya
3 Subang Jaya Medical Centre
4 Cancer Research Malaysia
5 Department of Mathematics, University of Nottingham (Malaysia Campus)
6 Department of Paediatrics, Faculty of Medicine, University Malaya
7 Department of Biomedical Imaging, Faculty of Medicine, University Malaya
Footnotes
A full list of Singapore Breast Cancer Cohort (SGBCC) and Malaysian Breast cancer Genetic study (MYBRCA) investigators is listed in the Acknowledgments section.
CONFLICT OF INTEREST
The authors disclose no conflicts of interest.
DATA AVAILABILITY STATEMENT
The data that supports the findings of this study are available in the supplementary material of this article.
REFERENCES
- Balia C, Galli A, & Caligo MA (2011). Effect of the overexpression of BRCA2 unclassified missense variants on spontaneous homologous recombination in human cells. Breast Cancer Res Treat, 129(3), 1001–1009. doi: 10.1007/s10549-011-1607-y [DOI] [PubMed] [Google Scholar]
- Best RB, & Mittal J. (2010a). Balance between alpha and beta structures in ab initio protein folding. J Phys Chem B, 114(26), 8790–8798. doi: 10.1021/jp102575b [DOI] [PubMed] [Google Scholar]
- Best RB, & Mittal J. (2010b). Protein simulations with an optimized water model: cooperative helix formation and temperature-induced unfolded state collapse. J Phys Chem B, 114(46), 14916–14923. doi: 10.1021/jp108618d [DOI] [PubMed] [Google Scholar]
- Biswas K, Das R, Alter BP, Kuznetsov SG, Stauffer S, North SL, . . . Sharan SK (2011). A comprehensive functional characterization of BRCA2 variants associated with Fanconi anemia using mouse ES cell-based assay. Blood, 118(9), 2430–2442. doi: 10.1182/blood-2010-12-324541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas K, Das R, Eggington JM, Qiao H, North SL, Stauffer S, . . . Sharan SK (2012). Functional evaluation of BRCA2 variants mapping to the PALB2-binding and C-terminal DNA-binding domains using a mouse ES cell-based assay. Hum Mol Genet, 21(18), 3993–4006. doi: 10.1093/hmg/dds222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas K, Stauffer S, & Sharan SK (2012). Using recombineering to generate point mutations:galK-based positive-negative selection method. Methods Mol Biol, 852, 121–131. doi: 10.1007/978-1-61779-564-0_10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doncheva NT, Klein K, Domingues FS, & Albrecht M. (2011). Analyzing and visualizing residue networks of protein structures. Trends Biochem Sci, 36(4), 179–182. doi: 10.1016/j.tibs.2011.01.002 [DOI] [PubMed] [Google Scholar]
- Dorling L. (2020). Breast cancer risk genes: association analysis in more than 113,000 women. New England Journal of Medicine, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidugli L, Shimelis H, Masica DL, Pankratz VS, Lipton GB, Singh N, . . . Couch FJ (2018). Assessment of the Clinical Relevance of BRCA2 Missense Variants by Functional and Computational Approaches. Am J Hum Genet, 102(2), 233–248. doi: 10.1016/j.ajhg.2017.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins CW, Le Grand S, Walker RC, & Roitberg AE (2015). Long-Time-Step Molecular Dynamics through Hydrogen Mass Repartitioning. J Chem Theory Comput, 11(4), 1864–1874. doi: 10.1021/ct5010406 [DOI] [PubMed] [Google Scholar]
- Ikegami M, Kohsaka S, Ueno T, Momozawa Y, Inoue S, Tamura K, . . . Mano H. (2020). High-throughput functional evaluation of BRCA2 variants of unknown significance. Nat Commun, 11(1), 2573. doi: 10.1038/s41467-020-16141-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannidis NM, Rothstein JH, Pejaver V, Middha S, McDonnell SK, Baheti S, . . . Sieh W. (2016). REVEL: An Ensemble Method for Predicting the Pathogenicity of Rare Missense Variants. Am J Hum Genet, 99(4), 877–885. doi: 10.1016/j.ajhg.2016.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karchin R, Agarwal M, Sali A, Couch F, & Beattie MS (2008). Classifying Variants of Undetermined Significance in BRCA2 with protein likelihood ratios. Cancer Inform, 6, 203–216. doi: 10.4137/cin.s618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp Z, Turnbull A, Yost S, Seal S, Mahamdallie S, Poyastro-Pearson E, . . . Rahman N. (2019). Evaluation of Cancer-Based Criteria for Use in Mainstream BRCA1 and BRCA2 Genetic Testing in Patients With Breast Cancer. JAMA Network Open, 2(5), e194428-e194428. doi: 10.1001/jamanetworkopen.2019.4428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, & Shendure J. (2014). A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet, 46(3), 310–315. doi: 10.1038/ng.2892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsov SG, Chang S, & Sharan SK (2010). Functional analysis of human BRCA2 variants using a mouse embryonic stem cell-based assay. Methods Mol Biol, 653, 259–280. doi: 10.1007/978-1-60761-759-4_16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsov SG, Liu P, & Sharan SK (2008). Mouse embryonic stem cell-based functional assay to evaluate mutations in BRCA2. Nature medicine, 14(8), 875–881. doi: 10.1038/nm.1719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai KN, Ho WK, Kang IN, Kang PC, Phuah SY, Mariapun S, . . . Teo SH (2017). Characterization of BRCA1 and BRCA2 variants in multi-ethnic Asian cohort from a Malaysian case-control study. BMC Cancer, 17(1), 149. doi: 10.1186/s12885-017-3099-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Sanchez JC, Lunet N, Gonzalez-Marron A, Lidon-Moyano C, Matilla-Santander N, Cleries R, . . . Martinez-Sanchez JM (2018). Projections in Breast and Lung Cancer Mortality among Women: A Bayesian Analysis of 52 Countries Worldwide. Cancer Res, 78(15), 4436–4442. doi: 10.1158/0008-5472.CAN-18-0187 [DOI] [PubMed] [Google Scholar]
- Morris JH, Huang CC, Babbitt PC, & Ferrin TE (2007). structureViz: linking Cytoscape and UCSF Chimera. Bioinformatics, 23(17), 2345–2347. doi: 10.1093/bioinformatics/btm329 [DOI] [PubMed] [Google Scholar]
- Nakamura S, Kwong A, Kim SW, Iau P, Patmasiriwat P, Dofitas R, . . . Teo SH (2016). Current Status of the Management of Hereditary Breast and Ovarian Cancer in Asia: First Report by the Asian BRCA Consortium. Public Health Genomics, 19(1), 53–60. doi: 10.1159/000441714 [DOI] [PubMed] [Google Scholar]
- Parsons MT, Tudini E, Li H, Hahnen E, Wappenschmidt B, Feliubadalo L, . . . Spurdle AB (2019). Large scale multifactorial likelihood quantitative analysis of BRCA1 and BRCA2 variants: An ENIGMA resource to support clinical variant classification. Hum Mutat, 40(9), 1557–1578. doi: 10.1002/humu.23818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, & Ferrin TE (2004). UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem, 25(13), 1605–1612. doi: 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- Rentzsch P, Witten D, Cooper GM, Shendure J, & Kircher M. (2019). CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res, 47(D1), D886-D894. doi: 10.1093/nar/gky1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, . . . Committee, A. L. Q. A. (2015). Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med, 17(5), 405–424. doi: 10.1038/gim.2015.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roe DR, & Cheatham TE 3rd. (2013). PTRAJ and CPPTRAJ: Software for Processing and Analysis of Molecular Dynamics Trajectory Data. J Chem Theory Comput, 9(7), 3084–3095. doi: 10.1021/ct400341p [DOI] [PubMed] [Google Scholar]
- Sali A, & Blundell TL (1993). Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol, 234(3), 779–815. doi: 10.1006/jmbi.1993.1626 [DOI] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, . . . Ideker T. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res, 13(11), 2498–2504. doi: 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapovalov MV, & Dunbrack RL Jr. (2011). A smoothed backbone-dependent rotamer library for proteins derived from adaptive kernel density estimates and regressions. Structure, 19(6), 844–858. doi: 10.1016/j.str.2011.03.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen MY, & Sali A. (2006). Statistical potential for assessment and prediction of protein structures. Protein Sci, 15(11), 2507–2524. doi: 10.1110/ps.062416606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimelis H, Mesman RLS, Von Nicolai C, Ehlen A, Guidugli L, Martin C, . . . for, N. C. (2017). BRCA2 Hypomorphic Missense Variants Confer Moderate Risks of Breast Cancer. Cancer Res, 77(11), 2789–2799. doi: 10.1158/0008-5472.CAN-16-2568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spurdle AB, Healey S, Devereau A, Hogervorst FB, Monteiro AN, Nathanson KL, . . . Enigma (2012). ENIGMA--evidence-based network for the interpretation of germline mutant alleles: an international initiative to evaluate risk and clinical significance associated with sequence variation in BRCA1 and BRCA2 genes. Hum Mutat, 33(1), 2–7. doi: 10.1002/humu.21628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan KHX, Tan LWL, Sim X, Tai ES, Lee JJ, Chia KS, & van Dam RM (2018). Cohort Profile: The Singapore Multi-Ethnic Cohort (MEC) study. Int J Epidemiol. doi: 10.1093/ije/dyy014 [DOI] [PubMed] [Google Scholar]
- Tan MM, Ho WK, Yoon SY, Mariapun S, Hasan SN, Lee DS, . . . Teo SH (2018). A case-control study of breast cancer risk factors in 7,663 women in Malaysia. PLoS One, 13(9), e0203469. doi: 10.1371/journal.pone.0203469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavtigian SV, Deffenbaugh AM, Yin L, Judkins T, Scholl T, Samollow PB, . . . Thomas A. (2006). Comprehensive statistical study of 452 BRCA1 missense substitutions with classification of eight recurrent substitutions as neutral. J Med Genet, 43(4), 295–305. doi: 10.1136/jmg.2005.033878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavtigian SV, Greenblatt MS, Lesueur F, Byrnes GB, & Group, I. U. G. V. W. (2008). In silico analysis of missense substitutions using sequence-alignment based methods. Hum Mutat, 29(11), 1327–1336. doi: 10.1002/humu.20892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Team, R. C. (2017). R: A language and environment for statistical computing In R. (Foundation for Statistical Computing). [Google Scholar]
- Thery JC, Krieger S, Gaildrat P, Revillion F, Buisine MP, Killian A, . . . Tosi M. (2011). Contribution of bioinformatics predictions and functional splicing assays to the interpretation of unclassified variants of the BRCA genes. Eur J Hum Genet, 19(10), 1052–1058. doi: 10.1038/ejhg.2011.100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thirthagiri E, Lee SY, Kang P, Lee DS, Toh GT, Selamat S, . . . Teo SH (2008). Evaluation of BRCA1 and BRCA2 mutations and risk-prediction models in a typical Asian country (Malaysia) with a relatively low incidence of breast cancer. Breast Cancer Res, 10(4), R59. doi: 10.1186/bcr2118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh GT, Kang P, Lee SS, Lee DS, Lee SY, Selamat S, . . . Teo SH (2008). BRCA1 and BRCA2 germline mutations in Malaysian women with early-onset breast cancer without a family history. PLoS One, 3(4), e2024. doi: 10.1371/journal.pone.0002024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallee MP, Di Sera TL, Nix DA, Paquette AM, Parsons MT, Bell R, . . . Tavtigian SV (2016). Adding In Silico Assessment of Potential Splice Aberration to the Integrated Evaluation of BRCA Gene Unclassified Variants. Hum Mutat, 37(7), 627–639. doi: 10.1002/humu.22973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vreeswijk MP, Kraan JN, van der Klift HM, Vink GR, Cornelisse CJ, Wijnen JT, . . . Devilee (2009). Intronic variants in BRCA1 and BRCA2 that affect RNA splicing can be reliably selected by splice-site prediction programs. Hum Mutat, 30(1), 107–114. doi: 10.1002/humu.20811 [DOI] [PubMed] [Google Scholar]
- Warming S, Costantino N, Court DL, Jenkins NA, & Copeland NG (2005). Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res, 33(4), e36. doi:33/4/e36 [pii] 10.1093/nar/gni035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, . . . Schwede T. (2018). SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res, 46(W1), W296-W303. doi: 10.1093/nar/gky427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen W, Shu XO, Guo X, Cai Q, Long J, Bolla MK, . . . Zheng W. (2016). Prediction of breast cancer risk based on common genetic variants in women of East Asian ancestry. Breast Cancer Res, 18(1), 124. doi: 10.1186/s13058-016-0786-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Jeffrey PD, Miller J, Kinnucan E, Sun Y, Thoma NH, . . . Pavletich NP (2002). BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA structure. Science, 297(5588), 1837–1848. doi: 10.1126/science.297.5588.1837 [DOI] [PubMed] [Google Scholar]
- Yap YS, Lu YS, Tamura K, Lee JE, Ko EY, Park YH, . . . Lee SC (2019). Insights Into Breast Cancer in the East vs the West: A Review. JAMA Oncol. doi: 10.1001/jamaoncol.2019.0620 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.