Abstract
The major natural auxin in plants, indole-3-acetic acid (IAA), orchestrates a plethora of developmental responses that largely depend on the formation of auxin concentration gradients within plant tissues. Together with inter- and intracellular transport, IAA metabolism—which comprises biosynthesis, conjugation, and degradation—modulates auxin gradients and is therefore critical for plant growth. It is now very well established that IAA is mainly produced from Trp and that the IPyA pathway is a major and universally conserved biosynthetic route in plants, while other redundant pathways operate in parallel. Recent findings have shown that metabolic inactivation of IAA is also redundantly performed by oxidation and conjugation processes. An exquisite spatiotemporal expression of the genes for auxin synthesis and inactivation have been shown to drive several plant developmental processes. Moreover, a group of transcription factors and epigenetic regulators controlling the expression of auxin metabolic genes have been identified in past years, which are illuminating the road to understanding the molecular mechanisms behind the coordinated responses of local auxin metabolism to specific cues. Besides transcriptional regulation, subcellular compartmentalization of the IAA metabolism and posttranslational modifications of the metabolic enzymes are emerging as important contributors to IAA homeostasis. In this review, we summarize the current knowledge on (1) the pathways for IAA biosynthesis and inactivation in plants, (2) the influence of spatiotemporally regulated IAA metabolism on auxin-mediated responses, and (3) the regulatory mechanisms that modulate IAA levels in response to external and internal cues during plant development.
The extensive search for the plant molecule responsible for tropic responses to light and gravity led to the identification of the first auxin molecule, indole-3-acetic acid (IAA), more than 80 years ago (Abel and Theologis 2010). Since then, it has been determined that auxin is not only involved in the control of tropisms, but also regulates numerous plant developmental responses that mainly rely on the spatiotemporal control of cell division, growth, and differentiation (Zhao 2018; Gallei et al. 2020). In addition to IAA, phenylacetic acid (PAA) and 4-chloro-indole-3-acetic acid (4-Cl-IAA) are naturally occurring auxins in plants. Although both are perceived by the auxin signaling machinery (Shimizu-Mitao and Kakimoto 2014; Jayasinghege et al. 2019), 4-Cl-IAA is not widespread (Lam et al. 2015) and PAA has been studied far less than IAA due to generally less potent effects (Cook 2019).
Because plant responses to auxin concentrations are threshold-dependent, IAA levels must be finely regulated during plant growth in response to external and internal cues. Tuning of the IAA concentration within cells and tissues is largely performed by directional transport and localized biosynthesis (Brumos et al. 2018; Robert et al. 2018) as well as inactivation of IAA (Zheng et al. 2016; Di Mambro et al. 2019). A classical model for auxin gradient formation in plants is based on a primary synthesis of IAA in the shoot—mainly in the young leaves and cotyledons—which is then distributed throughout the plant by polar transport with a major rootward component. However, early reports already suggested a role for the root tip in auxin biosynthesis (van Raalte 1936; van Overbeek 1939; Davies and Mitchell 1972). Improvements in the sensitivity of analytical technologies that allowed quantification of IAA from minute amounts of plant tissue, along with the identification of newly synthesized auxin in these tissues using stable isotope labeling (Novák et al. 2012, 2017), have helped researchers establish that auxin biosynthesis occurs locally in different plant organs and, albeit at different rates, in every root cell type (Ljung et al. 2001a, 2005; Petersson et al. 2009). Together with advances in analytical methodologies, the availability of the Arabidopsis genome sequence boosted the identification of genes associated with auxin biosynthesis, conjugation, and degradation. The detailed genetic and biochemical evaluation of these genes and their regulatory networks has revealed the importance of an initially disregarded role of auxin metabolism in plant development.
AUXIN BIOSYNTHESIS IN PLANTS
Several decades of research on auxin metabolism have firmly established the aromatic amino acid l-tryptophan (Trp) as a central precursor for IAA biosynthesis in plants. Trp is produced in chloroplasts via the shikimate pathway, a route through which most living organisms—excluding animals—produce aromatic amino acids (Maeda and Dudareva 2012). Far from being linear, Trp-dependent auxin biosynthesis involves various parallel pathways converging at the production of IAA, being IAOx (indole-3-acetaldoxime), IAM (indole-3-acetamide) and IPyA (indole-3-pyruvic acid) the most common intermediates (Fig. 1). A Trp-independent pathway for auxin synthesis was proposed after finding that maize and Arabidopsis mutants defective in Trp biosynthesis were still producing IAA (Wright et al. 1991; Normanly et al. 1993). It was later suggested that a cytosolic indole synthase (INS) mediates Trp-independent IAA production via the conversion of indole-3-glycerol-phosphate to indole (Zhang et al. 2008; Wang et al. 2015). However, the biochemical pathway for the Trp-independent conversion of indole to IAA remains unclear (Nonhebel 2015). The following section will describe the Trp-dependent pathways for auxin synthesis.
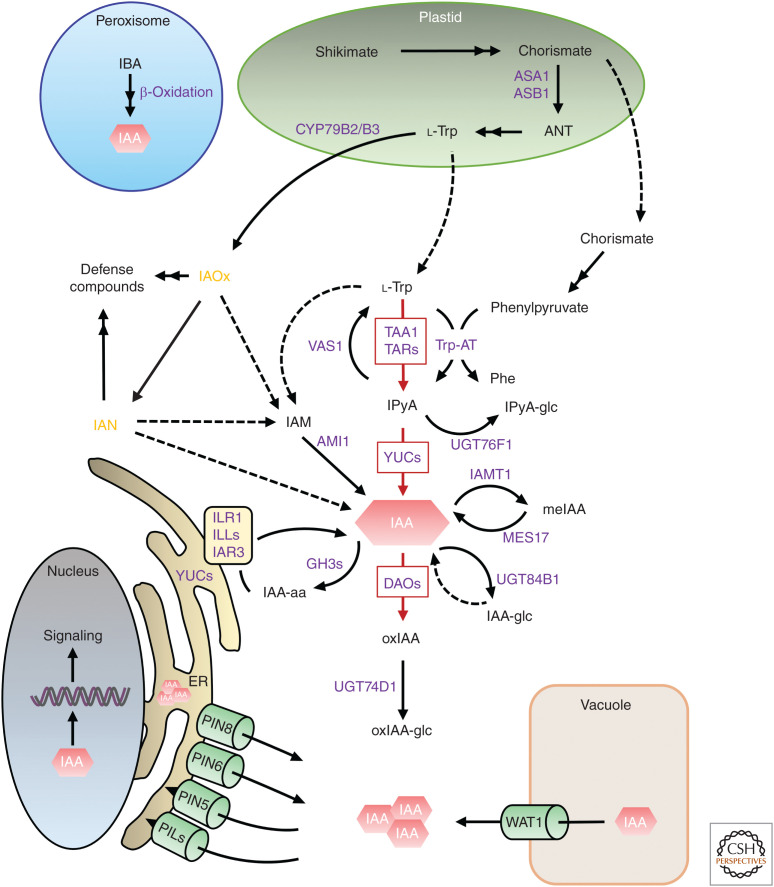
Figure 1.
Main pathways for indole-3-acetic acid (IAA) metabolism in plants. Biosynthesis of the IAA precursor l-tryptophan (Trp) takes place in the plastid (green oval). Subsequent IAA biosynthesis, catabolism, and conjugation reactions commonly operate in the cytoplasm. IAA molecules are indicated as red hexagons and IAA-related metabolites are shown in black. Enzymes catalyzing the metabolic reactions are indicated in purple. Major components for IAA biosynthesis and inactivation are indicated with red arrows. Solid arrows indicate pathways in which the enzymes, genes, or intermediates are known, while dashed arrows indicate pathways that are not yet well defined. Brassica-specific metabolites are indicated in orange. Green cylinders indicate intracellular IAA transporters and the attached arrows indicate the IAA movement direction. The blue circle, the gray oval, and the pale orange square indicate peroxisome, nucleus, and vacuole, respectively. Endoplasmic reticulum (ER) is indicated by the dark yellow structure attached to the nucleus. Organelles are not drawn to scale. (AMI1) AMIDASE-LIKE PROTEIN 1, (ANT) anthranilate, (ASA1) ANTHRANILATE SYNTHASE α SUBUNIT 1, (ASB1) ANTHRANILATE SYNTHASE β SUBUNIT 1, (CYP79B2/B3) CYTOCHROME P450, family 79, subfamily B, polypeptides 2 and 3, (DAO) DIOXYGENASE FOR AUXIN OXIDATION, (GH3) GRETCHEN HAGEN3, (IAA-glc) IAA-glucose, (IAM) indole-3-acetamide, (IAMT1) indole-3-acetate O-methyltransferase 1, (IAN) indole-3-acetonitrile, (IAOx) indole-3-acetaldoxime, (IBA) indole-3-butyric acid, (IAR3) IAA-ALANINE RESISTANT3, (ILLs) ILR1-LIKE, (ILR1) IAA-LEUCINE RESISTANT1, (IPyA) indole-3-pyruvic acid, (meIAA) methylindole-3-acetic acid, (MES17) METHYLESTERASE 17, (oxIAA) 2-oxindole-3-acetic acid, (Phe) phenylalanine, (PILS) PIN-likes, (PIN) PIN-FORMED, (TAA1) TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS 1, (TAR) TRYPTOPHAN AMINOTRANSFERASE-RELATED PROTEIN, (Trp-AT) TRYPTOPHAN AMINOTRANSFERASE, (UGTs) URIDINE-DIPHOSPHATE GLYCOSYLTRANSFERASE, (VAS1) REVERSAL OF SAV3 PHENOTYPE 1, (WAT1) WALLS ARE THIN1, (YUC) YUCCA flavin-containing monooxygenases.
The Main Pathway for IAA Biosynthesis
Whereas auxin production from IAOx and IAM is not yet fully understood, the IPyA pathway has been established as the prevailing route for IAA synthesis in plants. It consists of a two-step reaction in which Trp is first deaminated to IPyA by TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS 1 (TAA1) and TAA1-RELATED proteins (TARs) (Stepanova et al. 2008; Tao et al. 2008; Yamada et al. 2009). IPyA is then decarboxylated to IAA in a rate-limiting and irreversible reaction catalyzed by flavin-containing monooxygenases from the YUCCA (YUC) family (Mashiguchi et al. 2011; Stepanova et al. 2011; Won et al. 2011). TAA1 and YUC homologs are found across the genomes of vascular and nonvascular plants (Yue et al. 2014; Eklund et al. 2015; Poulet and Kriechbaumer 2017; Matthes et al. 2019), and functional conservation of TAA1 and YUC homologs has been shown in maize (Gallavotti et al. 2008; Phillips et al. 2011; Bernardi et al. 2012), rice (Yamamoto et al. 2007; Yoshikawa et al. 2014), and Marchantia polymorpha (Eklund et al. 2015). Taken together, this suggests that the IPyA pathway is a universal route for IAA synthesis in land plants. Compared to single mutants in TAA1/TARs and YUCCA genes, which show only subtle developmental phenotypes, higher-order taa1/tar and yuc mutants that bypass functional redundancy result in marked reductions in IAA levels and severe developmental defects in Arabidopsis, including abnormal embryo patterning, reduced stature, aberrant vasculature, defective root growth and gravitropic response, abnormal apical hook formation, and altered leaf and floral patterning (Cheng et al. 2006, 2007; Stepanova et al. 2008; Chen et al. 2014). Loss-of-function of the maize vt2 gene, a TAA1 ortholog, resulted in reduced plant growth and dramatic effects in inflorescence development, which entails a severe sterility of the mutant plants (Phillips et al. 2011). Similar reproductive defects were observed in the maize spi1 mutants, impaired in the function of a monocot-specific YUC-like protein (Gallavotti et al. 2008). In the liverwort Marchantia, the knock-out of its single TAA gene resulted in severe growth and developmental defects caused by a loss of cell and tissue differentiation (Eklund et al. 2015). The severity of the above-mentioned phenotypes, along with the seedling lethality observed in the Arabidopsis taa1 tar1 tar2 triple mutant (Stepanova et al. 2008), points to the IPyA pathway as a major and essential route for IAA biosynthesis in plants.
Parallel Pathways for IAA Biosynthesis
The Trp derivative IAOx is an intermediate in an IAA biosynthetic route that is yet to be fully understood. The conversion of Trp to IAOx is mediated by two isozymes from the cytochrome P450 (CYP) monooxygenase family, CYP79B2 and CYP79B3 (Hull et al. 2000; Mikkelsen et al. 2000; Zhao et al. 2002). Both IAOx and CYP79B2/3 genes have so far only been found in Brassica species (Sugawara et al. 2009), which suggests that this pathway is restricted to the Brassicaceae family (Fig. 1). Moreover, the cyp79b2 cyp79b3 double mutant shows conditional auxin phenotypes when grown at high temperatures (Zhao et al. 2002; Sugawara et al. 2009), further suggesting a role for the IAOx-dependent IAA synthesis specifically during adverse conditions. IAOx is a well-known precursor of indole glucosynolates (IGs) and camalexin, which serve as defense metabolites in plants (Hull et al. 2000; Glawischnig et al. 2004; Nafisi et al. 2007). Nevertheless, an increase in IAOx levels—through either genetic disruption of the IG pathway or overexpression of genes associated with IAOx biosynthesis—results in elevated IAA and high-auxin phenotypes (Boerjan et al. 1995; King et al. 1995; Delarue et al. 1998; Barlier et al. 2000; Zhao et al. 2002; Grubb et al. 2004; Sugawara et al. 2009; Novák et al. 2012; Kong et al. 2015). A study in which isotope-labeled IAOx was fed to Arabidopsis seedlings revealed that IAOx, IAM, and IAN (indole-3-acetonitrile) are intermediates of IAA biosynthesis (Sugawara et al. 2009). IAN-to-IAA conversion by a family of plant nitrilases (NITs) is thought to account for the IAOx-dependent auxin biosynthesis (Lehmann et al. 2017). However, the lack of direct genetic and biochemical evidence for NIT-mediated auxin synthesis in planta, together with the knowledge that nitrilases participate in cyanide and glutathione detoxification (Piotrowski 2008; Niehaus et al. 2019), means that the biochemical route from IAOx to IAA remains unresolved.
IAM is a well-known auxin biosynthesis intermediate in certain plant-associated bacteria (Patten et al. 2013) in which Trp is converted to IAA through the formation of IAM. While it was demonstrated that IAM can be produced from IAOx in Arabidopsis (Sugawara et al. 2009), IAM has also been detected in non-Brassica species that lack IAOx (Pollmann et al. 2002; Sugawara et al. 2009; Novák et al. 2012). IAM application results in classical high-auxin phenotypes (Sugawara et al. 2009; Gao et al. 2020), indicating that IAM-to-IAA conversion operates in planta. However, the disruption of the main IAM hydrolases in Arabidopsis, IAMH1, and IAMH2, did not lead to substantial developmental defects or variations in IAA contents, suggesting that the IAM pathway only plays a minor role in auxin homeostasis under standard growth conditions (Gao et al. 2020).
Indole-3-butyric acid (IBA) is a compound that has been shown to stimulate an auxin response when applied to plants. However, it is very unlikely (1) that IBA itself is perceived by the plant (Strader and Bartel 2011; Uzunova et al. 2016), and (2) that it is transported via polar transport (Liu et al. 2012). Instead, the effects of IBA can be attributed to its conversion to IAA by a group of peroxisomal enzymes (Zolman et al. 2008). IBA-to-IAA conversion has been found to be relevant to plant development (Frick and Strader 2018). How plants synthesize IBA is still unknown and, as such, it is not clear whether IBA is an IAA precursor or storage molecule. Also, whether endogenous IBA is present at physiologically relevant concentrations in plants has been questioned (Novák et al. 2012) and is still under debate (Frick and Strader 2018).
AUXIN METABOLIC INACTIVATION
Together with directional transport and local biosynthesis, metabolic inactivation of IAA also modulates auxin concentrations across plant cells and tissues. Indeed, research has shown that the majority of plant IAA exists as (1) inactive conjugates and methyl ester forms that can be reversibly converted to IAA (auxin storage forms), to rapidly fine-tune auxin levels without the need for de novo synthesis; and (2) as irreversible inactive IAA (auxin catabolites), which is the result of the removal of excess auxin or a regulated response to create auxin minima. The most extensively studied inactive forms of auxin will be summarized in the following section.
IAA Storage Forms
Many different auxin storage forms have been identified in plants (Korasick et al. 2013). These forms fall into three main groups: ester-linked IAA conjugates, amide-linked IAA conjugates, and methyl IAA (meIAA). The most prevalent and abundant ester-linked auxin in plants is IAA-glucose (IAA-glc), which is present at higher levels than any other directly measured conjugate (Pěnčík et al. 2009, 2018; Brunoni et al. 2020). IAA-glc has been detected in seedling extracts from different plants (Kai et al. 2007), and is the predominant IAA metabolite throughout Arabidopsis tissues (Porco et al. 2016). IAA-glc and its metabolic derivative IAA-myo-inositol (IAA-Ins) are particularly abundant in plant seeds (Hall 1980; Cohen and Bandurski 1982), and are thought to be the main auxin source during early seedling establishment in vascular plants (Bartel et al. 2001; Ljung et al. 2001b). High-molecular-weight IAA-glycan and -glycoprotein conjugates have also been found in plants (Korasick et al. 2013), although their specific roles in auxin homeostasis are not yet understood. Hydrolases that release free IAA from IAA-glc and IAA-Ins were identified in maize kernels (Jakubowska and Kowalczyk 2005), and rice (TGW6; Ishimaru et al. 2013). Specific UDP-glycosyltransferases (UGTs) that produce IAA-glc have been identified in plants (Szerszen et al. 1994; Jackson et al. 2002; Ludwig-Müller et al. 2005; Liu et al. 2019). Overexpression of these UGTs results in increased levels of IAA-glc and reduced levels of amide-linked auxins (Jackson et al. 2002; Ludwig-Müller et al. 2005), indicating that IAA homeostasis was disturbed. The availability of knockout mutants for the known auxin UGTs, together with the discovery of additional IAA glycosylases and hydrolases, will help clarify the roles of ester-linked auxins throughout plant development.
Amide-linked auxins encompass a group of compounds in which IAA is conjugated to amino acids, small peptides and proteins, among which IAA-amino acid (IAA-aa) conjugates are the best characterized. The formation of the IAA-amide bond is catalyzed by a group of IAA acyl acid amido synthetases from the GRETCHEN HAGEN3 (GH3) family (Staswick et al. 2005; Ludwig-Müller et al. 2009). GH3 genes (Terol et al. 2006; Okrent and Wildermuth 2011), along with different IAA-aa conjugates (Korasick et al. 2013; Záveská Drábková et al. 2015), are found all across land plants. GH3 co-orthologs have been functionally characterized in vascular and nonvascular plants (Staswick et al. 2005; Ludwig-Müller et al. 2009; Brunoni et al. 2020). The application of various IAA-aa conjugates results in plant phenotypes that are similar to what can be observed upon the addition of exogenous IAA, which provides strong evidence that IAA-aa conjugates can serve as IAA storage forms (LeClere et al. 2002; Rampey et al. 2004). Several IAA-aa amidohydrolases, including IAA-LEUCINE RESISTANT1 (ILR1), ILR1-LIKE proteins (ILLs), and IAA-ALANINE RESISTANT3 (IAR3), were identified in screens for mutants that are insensitive to IAA-aa conjugates (Bartel and Fink 1995; Davies et al. 1999; LeClere et al. 2002). These amidohydrolases appear to be functionally conserved across plant species. IAR3 was found to be required for root architectural changes under osmotic stress in Arabidopsis (Kinoshita et al. 2012), and to mediate defense responses of tomato and potato plants upon infection (D'Ippolito et al. 2016). GH3s are known to mediate different responses to biotic and abiotic stress (Park et al. 2007; Zhang et al. 2007, 2009; Ding et al. 2008; Du et al. 2012; Kirungu et al. 2019). A note of caution is due here since certain GH3 enzymes that function as IAA amido synthetases, as GH3.3, GH3.5, and GH3.6, present a promiscuous conjugating activity toward different substrates like salicylic acid, jasmonic acid, and benzoic acid (Zhang et al. 2007; Gutierrez et al. 2012; Westfall et al. 2016), at least in vitro. Hence, GH3-related phenotypes might be the result of the perturbation of the homeostasis of not just IAA, but also of other phytohormones.
The methyl ester of IAA, meIAA, also serves as an auxin storage form in plants. IAA methylation at the carboxyl group is mediated by IAA CARBOXYL METHYLTRANSFERASE1 (IAMT1) (Zubieta et al. 2003; Qin et al. 2005). As an inactive form of auxin, melAA does not interfere with the auxin signaling machinery (Li et al. 2008; Abbas et al. 2018). Nevertheless, the application of meIAA results in auxin-related phenotypes in plants (Qin et al. 2005), a dynamic that can be attributed to the hydrolysis of meIAA to IAA by METHYLESTERASE 17 (MES17) and related enzymes (Yang et al. 2008). Research using knockout iamt1 mutants revealed that auxin methylation has little impact on auxin levels, both locally or in whole seedlings (Abbas et al. 2018; Takubo et al. 2020). meIAA is a nonpolar compound that is transported in plants by both passive influx and PIN-mediated efflux and, thus, affects IAA gradients rather than IAA levels (Li et al. 2008; Abbas et al. 2018). Specific expression of IAMT1 in the hypocotyl endodermis was shown to be important for gravitropic growth (Abbas et al. 2018). The role of auxin methylation in regulating plant development is, however, still under debate (Takubo et al. 2020).
Irreversible IAA Catabolites
The amide-linked IAA-Asp and IAA-Glu conjugates, unlike other IAA-amino acid conjugates, are not hydrolyzed back to IAA in planta (Östin et al. 1998; Rampey et al. 2004) and are thus considered catabolites. These two irreversible conjugates are found in plants at much higher levels than the reversible IAA-aa conjugates (Kowalczyk and Sandberg 2001; Pěnčík et al. 2009). GH3 IAA-amido synthetases show different substrate preferences for Asp and Glu (Staswick et al. 2005; Brunoni et al. 2020). GH3.17, and to a lesser extent GH3.5, is known to preferentially use Glu as a cosubstrate (Staswick et al. 2005). Accordingly, gh3.17 plants show remarkably reduced levels of IAA-Glu in their hypocotyls (Zheng et al. 2016) and roots (Di Mambro et al. 2017), while the gh3.1,2,3,4,5,6 sextuple mutant does not produce any IAA-Asp, although IAA-Glu production is up-regulated, and is likely supported by the still functional GH3.17 (Porco et al. 2016).
The major catabolic pathway that regulates IAA levels in plants is the irreversible oxidation of IAA to oxIAA (2-oxindole-3-acetic acid), with further glycosylation to oxIAA-glc (Östin et al. 1998; Kai et al. 2007; Kubeš et al. 2012; Novák et al. 2012; Pěnčík et al. 2013). There is extensive evidence that the levels of these oxidative catabolites rapidly increase after IAA application (Östin et al. 1998; Kubeš et al. 2012). Moreover, they are prevalent at higher levels than amide-linked catabolites at physiological conditions in algae, as well as in vascular and nonvascular land plants, which suggests that oxidation is a major pathway for IAA catabolism across the plant kingdom (Novák et al. 2012; Pěnčík et al. 2013; Záveská Drábková et al. 2015; Porco et al. 2016; Žižková et al. 2017). Conifers were found to be an exception, as conjugation and not oxidation dominates IAA homeostasis (Brunoni et al. 2020). The conversion of IAA to oxIAA is catalyzed by DIOXYGENASE FOR AUXIN OXIDATION (DAO) proteins (Zhao et al. 2013), which belong to the 2-oxoglutarate-dependent Fe(II) dioxygenase superfamily (Kawai et al. 2014; Nadi et al. 2018), while the UGT74D1 enzyme participates in the glycosylation of oxIAA to oxIAA-glc (Tanaka et al. 2014). Arabidopsis DAO1 accounts for most de novo IAA oxidation and is widely expressed in plant tissues. However, DAO1 loss-of-function results in only mild developmental defects. Because amide-linked catabolites are greatly increased in dao1 mutants, while decreased in DAO1 overexpressors, DAO and GH3 enzymes are proposed to function redundantly in regulating IAA levels (Mellor et al. 2016; Porco et al. 2016; Zhang et al. 2016). Despite this redundant IAA catabolism, multicellular modeling of auxin gradients has shown that impaired oxidation perturbs IAA levels in specific root tissues (Mellor et al. 2016), which is supported by the localized, albeit subtle, phenotypes of dao1-1 mutant roots (Porco et al. 2016; Zhang et al. 2016).
LOCALIZED IAA METABOLISM COORDINATES PLANT DEVELOPMENT
The identification and characterization of several genes related to auxin metabolism has revealed an intricate spatiotemporal orchestration of their localized expression (exemplified in Fig. 2) that turned out to contribute to the regulation of local auxin concentrations. YUCCA genes represent an excellent example of specialization, as several rounds of gene duplication have resulted in multiple YUCs that have unique expression domains (Zhao 2018). For example, a group of shoot- and root-specific YUCCAs has been defined (Won et al. 2011; Chen et al. 2014). YUC1 and YUC4 are specifically expressed in the shoot apical meristem, along with flower and leaf primordia (Cheng et al. 2006, 2007). In the embryo, YUC1, YUC4, YUC10, and YUC11 were found to be expressed in the apical cells, with each showing certain temporal changes in expression domain as embryo development progressed (Cheng et al. 2007). The quadruple yuc1 yuc4 yuc10 yuc11 mutant was found to lack the hypophysis, a root meristem precursor cell, and thus germinated without a primary root (Cheng et al. 2007). Localized expression of TAA1 in the developing embryo was also found to be critical for root and apical embryonic meristem specification (Stepanova et al. 2008; Robert et al. 2013). In roots, TAA1 is specifically expressed at the quiescent center (QC), while TAR2 expression was reported in the root provasculature (Stepanova et al. 2008). Disruption of both of these genes resulted in the complete loss of the stem cell niche and root growth abortion early after germination (Stepanova et al. 2008). This dynamic and localized expression, together with the developmental abnormalities noted for plants with loss-of-function mutations in the TAA1/TAR and YUC genes, supports a role for IPyA-dependent local auxin biosynthesis in embryo patterning, root meristem maintenance, gynoecium formation, and leaf and floral development (Cheng et al. 2006, 2007; Stepanova et al. 2008). Localized expression of CYP79B2/B3 genes at the root meristem and lateral root primordia initiation sites additionally suggests that the IAOx pathway participates in local IAA synthesis during root development (Ljung et al. 2005).
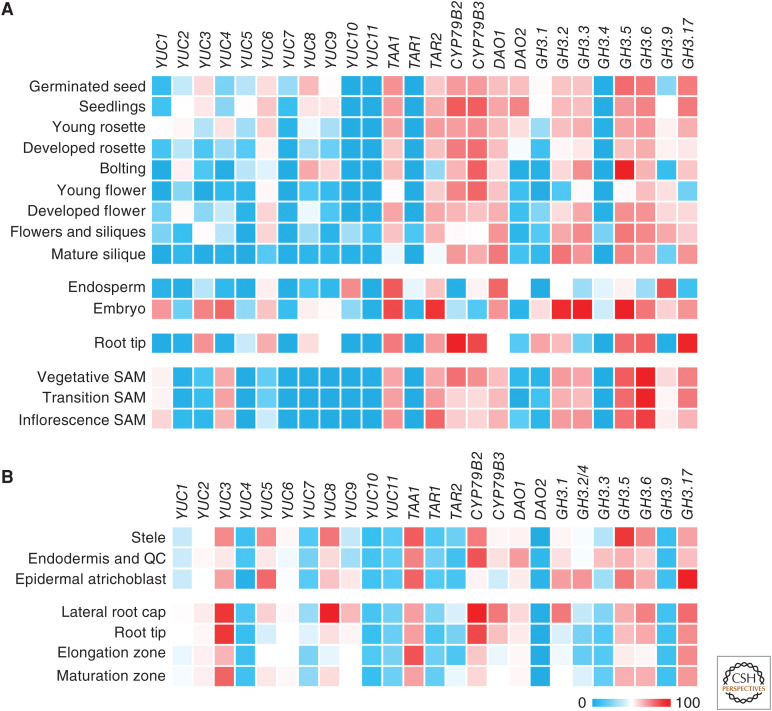
Figure 2.
Relative expression level of the main genes for auxin metabolism in different developmental stages, organs, and tissues from Arabidopsis. Data was retrieved from Genevestigator (genevestigator.com; Hruz et al. 2008) using datasets from (A) RNA-seq experiments, and (B) Affymetrix Arabidopsis ATH1 Genome Array. Results are expressed in percentage of expression potential (the maximum expression a gene reaches across all experiments). GH3.2 and GH3.4 share array probes in the results shown in B. AGI codes: YUC1 (At4G32540), YUC2 (At4G13260), YUC3 (At1G04610), YUC4 (At5G11320), YUC5 (At5G43890), YUC6 (At5G25620), YUC7 (At2G33230), YUC8 (At4G28720), YUC9 (At1G04180), YUC10 (At1G48910), YUC11 (At1G21430), TAA1 (At1G70560), TAR1 (At1G23320), TAR2 (At4G24670), CYP79B2 (At4G39950), CYP79B3 (At2G22330), DAO1 (At1G14130), DAO2 (At1G14120), GH3.1 (At2G14960), GH3.2 (At4G37390), GH3.3 (At2G23170), GH3.4 (At1G59500), GH3.5 (At4G27260), GH3.6 (At5G54510), GH3.9 (At2G47750), and GH3.17 (At1G28130).
Cooperation between local IAA biosynthesis and polar transport generates auxin concentration gradients that drive plant growth (Ikeda et al. 2009; Brumos et al. 2018). For example, the spatiotemporally coordinated expression of TAA1 and YUCs in the basal and apical embryo, together with the resulting auxin-triggered PIN polarization, was shown to define the apicobasal embryo axis (Robert et al. 2013; Wabnik et al. 2013). However, auxin transport cannot always compensate for deficiencies in local synthesis. Proper root development largely depends on auxin production in the root (Bhalerao et al. 2002; Chen et al. 2014), and localized auxin biosynthesis at the root QC was found to be sufficient for root meristem maintenance in the absence of functional polar transport (Brumos et al. 2018). Recently, the capacity of the roots to regenerate their tips after wounding was shown to be highly dependent on local auxin biosynthesis mediated by TAA1 and YUCs near the cut site and in the protoxylem and xylem-pole-pericycle, while PIN-mediated transport was found dispensable (Matosevich et al. 2020).
Localized auxin degradation was also found to play various roles in plant growth and development. The IAA-amido synthetase GH3.17 was shown to modulate hypocotyl elongation in response to shade and temperature independently of auxin transport and de novo biosynthesis (Zheng et al. 2016). GH3.17, GH3.5, and GH3.6 are specifically expressed in the lateral root cap (Di Mambro et al. 2019; Pierdonati et al. 2019), with GH3.17—at the very least—participating in the IAA degradation required to create an auxin minimum at the transition zone, which influences root meristem size (Di Mambro et al. 2019).
REGULATION OF AUXIN METABOLISM
Decades of intensive research have resulted in a deep understanding of the network of interconnected and redundant pathways involved in IAA synthesis and inactivation. To ensure plastic and coordinated plant development, these pathways need to be fine-tuned by an array of regulating mechanisms that control the activation/deactivation of various synthetic and catabolic routes. However, our understanding of the regulation of this complex homeostatic machinery is mainly based on research that has been conducted during the past decade. The regulatory mechanisms underlying IAA synthesis and inactivation include (1) metabolic regulation, (2) hormonal cross talk, (3) transcriptional regulation, (4) posttranslational modifications, and (5) subcellular compartmentalization.
Metabolic Regulation of IAA Metabolism
It has been well established that IAA biosynthesis is controlled by feedback inhibitory mechanisms dependent on IAA levels (Ljung et al. 2001a). Auxin perception and signaling was shown to cause the down-regulation of genes related to the IPyA biosynthetic pathway and, consequently, to decrease endogenous IAA levels in a feedback loop (Suzuki et al. 2015; Takato et al. 2017). Likewise, the impaired conversion of IAA-aa conjugates, IBA, and meIAA to free IAA has been shown to up-regulate the expression of genes associated with IPyA-dependent auxin synthesis (Spiess et al. 2014). Moreover, the disrupted oxidative degradation of auxin in Arabidopsis dao1 mutant plants up-regulates not only redundant GH3-mediated degradation, but also de novo auxin biosynthesis, which is a rather counterintuitive way of returning to homeostasis after an increase in auxin (Mellor et al. 2016; Porco et al. 2016).
Chorismate, the terminal product of the shikimate pathway, is a common precursor in the synthesis of the aromatic amino acids Trp, tyrosine (Tyr), and phenylalanine (Phe) (Maeda and Dudareva 2012). While the Tyr and Phe biosynthetic routes share common intermediates, Trp synthesis proceeds independently. However, a recent investigation revealed metabolic interplay between the Phe and IAA biosynthesis pathways. Phenylpyruvate, an intermediate in cytosolic Phe synthesis, can also serve as an amino acceptor in the Trp-to-IPyA conversion by Trp amino transferases and, thus, modulates IAA biosynthesis in response to Phe fluctuations (Lynch et al. 2020). Such metabolic interplay might also play a role under specific stresses, such as wounding (Lynch et al. 2020).
A screening for suppressors of the impaired shade-avoidance response observed in the TAA1 mutant sav3-1 identified the aminotransferase VAS1, which converts IPyA back to Trp and, thus, limits IAA synthesis (Zheng et al. 2013). VAS1 uses the ethylene intermediate methionine as a preferred amino donor, which revealed a link in the metabolic control of auxin and ethylene biosynthesis (Zheng et al. 2013). VAS1-mediated regulation is specifically, but most likely not exclusively, required for shade-induced elongation of hypocotyl and petioles (Zheng et al. 2013) and for the control of parthenocarpy in Solanaceae species (Matsuo et al. 2020).
Additional metabolic control of the IPyA pathway relies on the glycosylation of IPyA to IPyA-glc by UGT76F1, which was recently identified as a fine-tuning mechanism in the modulation of IPyA availability for IAA biosynthesis during light- and temperature-induced hypocotyl elongation (Chen et al. 2020).
Hormonal Cross Talk Modulates IAA Metabolism
The conversion of chorismate to anthranilate, which represents the first, rate-limiting step in the Trp biosynthetic pathway, is catalyzed by the anthranilate synthase complex (Niyogi and Fink 1992). Previous research has identified anthranilate production as a central hub in the modulation of IAA biosynthesis by other hormonal pathways. A screen for mutants with altered responses to ethylene identified two mutants, wei2 and wei7, which harbored mutations in genes encoding anthranilate synthase α1 (ASA1) and β1 (ASB1) subunits, respectively (Stepanova et al. 2005). The ASA1 promoter was later found to be a direct target of ETHYLENE RESPONSE FACTOR 1 (ERF1), which up-regulates ASA1 expression in response to ethylene, resulting in increased auxin biosynthesis and root growth inhibition (Mao et al. 2016). ASA1 was also found to be at the core of jasmonate-auxin metabolic cross talk (Sun et al. 2009), in a mechanism that relies on the direct up-regulation of ASA1 expression by the jasmonate-responsive ETHYLENE RESPONSE FACTOR 109 (ERF109) (Cai et al. 2014). ERF109 additionally binds to YUC2 promoter to stimulate auxin biosynthesis in response to an increase in jasmonate levels (Cai et al. 2014). YUC8 and YUC9 might also be involved in jasmonate-regulated auxin biosynthesis, as their expression is promoted by methyl jasmonate (Hentrich et al. 2013).
The interplay between auxin and cytokinin has long been recognized as crucial to plant growth and development (Moubayidin et al. 2009). In Arabidopsis, cytokinin signal transduction was shown to modulate the rate of IAA biosynthesis (Jones et al. 2010). This metabolic cross talk involves the cytokinin-mediated up-regulation of YUC1 and YUC4 at the gynoecium primordia (Müller et al. 2017) and of YUC8 in roots (Di et al. 2016). The cytokinin-response transcriptional effector ARABIDOPSIS RESPONSE REGULATOR 1 (ARR1) promotes auxin biosynthesis in the stem cell niche via the up-regulation of ASB1 (Moubayidin et al. 2013). While a direct molecular link for cytokinin-regulated auxin biosynthesis remains unknown, cytokinin was found to directly modulate IAA degradation. More specifically, ARR1 directly binds to GH3.17 and activates its transcription in response to cytokinin, thus promoting auxin degradation (Di Mambro et al. 2017). This regulatory module was found to be a prerequisite for root meristem size determination (Di Mambro et al. 2017, 2019).
Transcriptional Regulation of IAA Metabolism
The coordinated response of auxin biosynthesis and inactivation to specific physiological and environmental cues is mediated by the direct modulation that specific transcription factors and components of the epigenetic machinery exert on the expression of auxin metabolic genes. Although little is known about the transcriptional control of auxin metabolism, several transcription factors and epigenetic regulators have been identified and associated to the control of auxin metabolism during specific plant developmental responses (see Tables 1 and 2).
Table 1.
Transcription factors that have been shown to directly modulate the expression of genes related to IAA metabolism
| Target | Transcriptional regulator(s) | Structural type | Biological process(es) | References |
|---|---|---|---|---|
| YUC1 | ARR1, ARR10, ARR12 | B-ARR | Shoot stem cell niche maintenance | Meng et al. 2017 |
| SUP | C2H2-type zinc finger | Floral patterning | Xu et al. 2018 | |
| YUC4 | LEC2 | B3 domain | Embryogenesis | Stone et al. 2008 |
| SHI/STY1 | RING-like zinc finger | Leaf and flower development | Eklund et al. 2010 | |
| ARR1, ARR10, ARR12 | B-ARR | Shoot stem cell niche maintenance | Meng et al. 2017 | |
| LEC2, FUS3 | B3 domain | Lateral root formation | Tang et al. 2017 | |
| SUP | C2H2-type zinc finger | Floral patterning | Xu et al. 2018 | |
| CRC | C2C2 type zinc finger | Floral determinacy | Yamaguchi et al. 2018 | |
| AG | MADS | Floral determinacy | Yamaguchi et al. 2018 | |
| YUC5 | IDD14, IDD15, IDD16 | IDD | Organ morphogenesis, gravitropism | Cui et al. 2013 |
| YUC8 | PIF4 | bHLH | Hypocotyl elongation at high temperature | Franklin et al. 2011 |
| YUC9 | EIN3 | EIL | Aluminum-induced root growth inhibition | Liu et al. 2016b |
| TAA1 | PIF4 | bHLH | Hypocotyl elongation at high temperature | Franklin et al. 2011 |
| IDD14, IDD15, IDD16 | IDD | Organ morphogenesis, gravitropism | Cui et al. 2013 | |
| SPT | bHLH | Gynoecium development | Reyes-Olalde et al. 2017 | |
| ARR1s | B-ARR | Light-induced tissue-specific IAA synthesis | Yan et al. 2017 | |
| CYP79B2 | PIF4 | bHLH | Hypocotyl elongation at high temperature | Franklin et al. 2011 |
| UGT76F1 | PIF4 | bHLH | Hypocotyl elongation at high temperature | Chen et al. 2020 |
| OsYUC8 | OsEIL1 | EIL | Ethylene-mediated primary root elongation | Qin et al. 2017 |
| ZmGH3.2 | ZmDREB2A | AP2/ERF | Longevity of maize seed | Han et al. 2020 |
| GH3.2 GH3.6 | MYB30 | R2R3-MYB | Root growth | Zhao and Xue 2020 |
(Os) Oryza sativa, (Zm) Zea mays; otherwise Arabidopsis thaliana.
Table 2.
Epigenetic regulators that have been shown to directly modulate the expression of genes involved in IAA biosynthesis in a specific biological process
| Target | Epigenetic regulator(s) | Biological process(es) | References |
|---|---|---|---|
| YUC1 | LHP1 | Floral patterning (−) | Xu et al. 2018 |
| CLF-PRC2 | |||
| YUC2 | CMT3, DRM1, DRM2 | Leaf development/growth (−) | Forgione et al. 2019 |
| LOCUS_77297 | Ambient temperature (−) | Gyula et al. 2018 | |
| YUC4 | LHP1 | Floral patterning (−) | Xu et al. 2018 |
| CLF-PRC2 | |||
| CHR11 | Floral determinacy (+) | Yamaguchi et al. 2018 | |
| CHR17 | |||
| YUC8 | MRG2 | Hypocotyl elongation (shade) (+) | Peng et al. 2018 |
| HDA9 | Hypocotyl elongation (temperature) (+) | van der Woude et al. 2019 | |
| YUC9 | ARP4 | Hypocotyl elongation (shade) (−) | Lee and Seo 2017 |
| YUC10 | FIS2-PRC2 | Endosperm development (−) | Figueiredo et al. 2015 |
(ARP4) ACTIN-RELATED PROTEIN 4, (CHR11) CHROMATIN REMODELLING 11, (CLF) CURLY LEAF, (CMT3) CHROMOMETHYLASE 3, (DRM1) DOMAINS REARRANGED 1, (FIS2) FERTILIZATION INDEPENDENT SEED 2, (HDA9) HISTONE DEACETYLASE 9, (LHP1) LIKE HETEROCHROMATIN 1, (MRG2) MORF-RELATED GENE 2, (PRC2) POLYCOMB REPRESSIVE COMPLEX 2. The (-) and (+) reflect transcriptional repression and activation of the target, respectively.
Specific posttranslational histone modifications, also termed histone marks, are associated with an active or repressed transcriptional state. Whole-genome occupancy studies of the histone repressive mark H3K27me3 found that epigenetic mechanisms control auxin-related genes. In comparisons of dividing and differentiated cells, differential H3K27me3 modifications were observed at genes involved in auxin biosynthesis (YUCs, CYPs, TAA1/TARs, SUR1, NITs), inactivation (GH3s, IAMT), transport (PINs, AUX/LAXs), and signaling (TIR1/AFBs, IAAs, ARFs), thus revealing that this histone mark exerts a profound effect on auxin action (Lafos et al. 2011; He et al. 2012). YUC1 and YUC4, for example, were found to be specifically involved in early auxin-mediated de novo root regeneration, in direct correlation with an H3K27me3 drop along their promoter regions (Chen et al. 2016). The Polycomb Repressive Complex 2 (PRC2) accessory protein LIKE HETEROCHROMATIN 1 (LHP1) (Derkacheva et al. 2013) are recruited to YUC1, YUC2, YUC4, YUC5, YUC6, YUC8, YUC9, and YUC10 promoters to control their expression (Rizzardi et al. 2011). Beyond general correlations and whole-genome comparisons, specific mechanisms related not only to histone modifications—but also DNA methylation and small RNAs—have been shown to alter auxin homeostasis by regulating the transcription of YUCs (for review, see Mateo-Bonmatí et al. 2019).
During the first steps of flower determination—a process in which floral meristem cells stop proliferating and initiate a floral organ primordium—flower primordia formation requires auxin-driven rapid cell expansion and elongation. The required increase in auxin levels is, at least in part, achieved by the activation of YUC4 by the chromatin-remodeling factors CHROMATIN REMODELLING 11 (CHR11) and CHR17, both of which are specifically recruited to the YUC4 promoter during floral primordium formation (Yamaguchi et al. 2018). Later in floral organ development, the C2H2-type zinc-finger transcription factor SUPERMAN (SUP) actively represses YUC1 and YUC4 expression and, thereby, auxin biosynthesis at the boundaries between carpels and stamen primordia (Xu et al. 2018). SUP-mediated transcriptional silencing of YUC1 and YUC4 further involves the recruitment of members of the PRC2 machinery, such as CURLY LEAF (CLF) or LHP1, for the trimethylation of H3K27 (Xu et al. 2018).
During angiosperm fertilization, auxin biosynthesis is constitutively repressed in maternal-derived tissues by the action of the FERTILIZATION-INDEPENDENT SEED-PRC2 (FIS-PRC2) complex (Figueiredo et al. 2015). Mutations in genes encoding subunits of this complex lead to premature expression of YUC10 in the nonfertilized diploid central cell, which will result in the development of empty seeds. Recent reports indicate that additional mechanisms, driven by EMSY-like Tudor/Agenet H3K36me3 histone readers EMSY-Like protein 1 (EML1) and EML3, actively repress auxin biosynthesis, transport, and signaling during seed coat and endosperm development (Milutinovic et al. 2019).
Auxin integrates internal and external signals as sugar levels, shade, and temperature into regulated plant growth responses (Sairanen et al. 2012; for review, see Zhao 2018). A good model to exemplify such interaction is the auxin biosynthesis-driven hypocotyl elongation in response to shade or high temperature (Gray et al. 1998; Tao et al. 2008). In the hypocotyls of plants grown under normal light conditions, YUC9 is actively repressed by the action of two AT-HOOK-CONTAINING NUCLEAR-LOCALIZED (AHL) proteins, AHL27 and AHL29 (Lee and Seo 2017). AHL29 recruits ACTIN-RELATED PROTEIN 4 (ARP4), a member of the SWI2/SNF2-RELATED1 (SWR1) chromatin-remodeling complex, to the YUC9 regulatory region to promote the deposition of histone variant H2A.Z and, hence, block the access of RNApol II to the DNA (Lee and Seo 2017). Similar mechanisms for transcriptional regulation are driven by ARP6 under normal temperatures (Kumar and Wigge 2010). However, independent mechanisms trigger YUC8 expression in plant hypocotyls to facilitate their elongation under shade or high temperature. The transcription factor PHYTOCHROME-INTERACTING FACTOR 7 (PIF7) and the H3K4me3/H3K36me3-binding protein Morf-Related Gene 2 (MRG2) bind to the YUC8 promoter in response to shade, and facilitate its transcription by allowing the acetylation of H3 and H4 (Peng et al. 2018). At high temperature, HISTONE DEACETYLASE 9 (HDA9) accumulates, facilitating the H2A.Z removal from the YUC8 locus and providing a looser chromatin environment that allows PIF4-mediated activation of YUC8 transcription (van der Woude et al. 2019). High temperatures additionally promote auxin biosynthesis through the temperature-specific recruitment of PIF4 to the promoters of the IPyA glycosylase UGT76F1 and the IAOx-pathway-related CYP79B2 gene to repress and promote their transcription, respectively, by unknown epigenetic mechanisms (Chen et al. 2020).
DNA methylation of cytosines is another important epigenetic mark mostly associated with gene repression that has also been shown to participate in auxin homeostasis. In animals, DNA methylation is almost entirely restricted to CG dinucleotides while plant DNA can additionally be methylated at CHG and CHH sequence contexts (with H representing A, T, or C) (Zhang et al. 2018; Gallego-Bartolomé 2020). The cytosine methylation is mediated by RNA-directed DNA methylation (RdDM), in which 24-nt small interfering RNA (siRNA) guides the DNA methyltransferases DOMAINS REARREANGED METHYLTRANSFERASE 1 (DRM1) and DRM2 to the target region (Zhang et al. 2018). Following DNA replication, cell-specific methylation patterns are maintained by another subset of DNA methyltransferases (METHYLTRANSFERASE 1 [MET1], CHROMOMETHYLASE 2 [CMT2], and CMT3) which, in contrast to DRMs, function in a context-specific manner, targeting CG, CHH, and CHG sequences, respectively (Zhang et al. 2018). Interestingly, several phenotypes that have been linked with auxin deficiency, such as root agravitropism, aberrant embryogenesis, and vascular disorders, were also observed in a triple drm1 drm2 cmt3 mutant (Forgione et al. 2019). Tissue-specific expression analyses in drm1 drm2 cmt3 seedlings revealed that the auxin biosynthetic genes YUC2 and TAA1 were specifically up-regulated in leaves. Me-DIP (methylated DNA immunoprecipitation)-PCR experiments further confirmed a reduction of non-GC DNA methylation at the YUC2 promoter, thus linking RdDM with the regulation of auxin biosynthesis (Forgione et al. 2019). A search for thermoresponsive regulatory RNAs identified a novel temperature-regulated 24-nt siRNA, coined Locus_77297, in the vicinity of the YUC2 promoter (Gyula et al. 2018). The expression of this siRNA in leaves was positively correlated with CHH methylation at the YUC2 promoter. Plants grown at high temperatures showed severe reductions in both Locus_77297 expression and CHH methylation, which triggered the up-regulation of YUC2 (Gyula et al. 2018).
Posttranslational Regulation of IAA Metabolism
After transcriptional control, additional regulatory mechanisms also govern protein function posttranslationally. Posttranslational modifications (PTMs) refer to covalent modifications that generally modulate protein folding or activity. There is recent evidence that auxin homeostasis is also modulated by PTMs. Arabidopsis TAA1 phosphorylation at the Thr101 was shown to be triggered by auxin perception itself, and to serve as an on/off switch controlling the activity of the enzyme, and therefore IAA biosynthesis (Wang et al. 2020). DAO1 auxin oxidase is known to be barely induced by IAA (Porco et al. 2016). Instead, experiments in rice demonstrated that DAO activity is regulated posttranslationally by substrate-mediated multimerization, which involves the IAA-triggered formation of DAO dimers that show increased affinity for IAA (Takehara et al. 2020). Interestingly, IAA-triggered TAA1 phosphorylation also enables this enzyme to dimerize with homologous TAR enzymes, a dynamic that likely regulates IAA biosynthesis through the control of several isoenzymes (Wang et al. 2020). Whether DAO dimerization also depends on IAA-triggered phosphorylation remains unexplored, and future research might identify a common mechanism for the posttranslational control of auxin metabolism.
Subcellular Compartmentalization of the IAA Metabolism
The cellular compartmentalization of bioactive IAA, IAA metabolites, and IAA metabolic enzymes represents yet another mechanism through which intracellular levels of IAA are regulated (Skalický et al. 2018). The first compartmentalization of IAA regards its biosynthesis, as the central precursor Trp is produced in chloroplasts, and the IAOx-pathway-related CYP79B2 and CYP79B3 contain a chloroplast transit peptide (Hull et al. 2000). Nevertheless, the major routes for IAA biosynthesis and degradation are believed to take place in the cytosol, as the enzymes TAA1, YUC1, YUC2, YUC3, YUC6, YUC11, and DAO1 have been shown to share a cytoplasmic localization (Stepanova et al. 2008; Tao et al. 2008; Zhao et al. 2013; Kriechbaumer et al. 2016; Porco et al. 2016; Zhang et al. 2016). The IAA amido synthetase GH3.17 is also localized to the cytosol (Di Mambro et al. 2019). However, TAR2, YUC5, YUC7, YUC8, and YUC9 colocalize with ER-membrane markers (Kriechbaumer et al. 2016). YUC4 is expressed as two tissue-specific splice variants, with one localized to the cytosol and the other localized to the cytosolic face of the endoplasmic reticulum (ER) membrane (Kriechbaumer et al. 2012). This suggests that part of IAA biosynthesis is compartmentalized to the ER. In line with this hypothesis, isolated ER microsomes were shown to significantly contribute to IPyA-dependent IAA biosynthesis (Kriechbaumer et al. 2015, 2016). Additionally, the IAA amidohydrolases ILR1, IAR3, and ILL2 have been shown to localize to the ER (Sanchez Carranza et al. 2016).
Together with the apparent subcellular localization of the IAA metabolism, active transport between organelles and the cytosol is a major factor determining cellular IAA homeostasis. The atypical members of the PIN family of auxin transporters PIN5, PIN6, and PIN8, together with PIN-LIKES proteins (PILS), have been detected at the ER (Mravec et al. 2009; Barbez et al. 2012; Dal Bosco et al. 2012; Ding et al. 2012; Simon et al. 2016). Genetic analyses on these PINs and PILS mutants and overexpressors suggest that ER-compartmentalization of IAA regulates auxin signaling by limiting the available cytosolic IAA that can enter to the nucleus (Mravec et al. 2009; Béziat et al. 2017; Feraru et al. 2019). Genetic manipulation of the ER transporters leads to altered levels of IAA as well as the IAA-Asp and IAA-Glu conjugates (Mravec et al. 2009; Dal Bosco et al. 2012; Ding et al. 2012; Simon et al. 2016), which further suggests that the compartmentalization of certain IAA metabolites is important to maintaining auxin homeostasis. Vacuole-associated auxin transporters that move IAA out of the vacuole have also been found (Ranocha et al. 2013; Liu et al. 2016a), pointing to a role of the vacuole in intracellular IAA regulation (Fig. 1). The presence of IAA precursors, as well as oxIAA and IAA-glc, in Arabidopsis vacuoles suggests that this organelle is involved in at least part of IAA metabolism (Ranocha et al. 2013). Additionally, the IBA vacuolar transporter TOB1 was shown to move IBA from the cytoplasm to the vacuole, which provides further evidence that the vacuole participates in subcellular IAA metabolism (Michniewicz et al. 2019).
CONCLUDING REMARKS AND FUTURE PERSPECTIVES
Auxin is mainly produced in plants from Trp through the formation of IPyA by the action of plant-conserved members of the TAA and YUC enzyme families. The IPyA pathway is a major and essential pathway for auxin biosynthesis, while other parallels routes for auxin production turned out to have minor or environmentally restricted roles during plant development. Irreversible IAA degradation by the redundant action of DAO oxidases and GH3 amido synthetases additionally control auxin concentrations across plant tissues. Research on when and where these genes are expressed has revealed a strict requirement of localized auxin metabolism for regulated plant growth and development. Major breakthroughs during the past decade have shed light on the transcriptional control of auxin biosynthesis, as several transcription factors and epigenetic regulators have been described to control local auxin production during specific developmental processes in response to environmental and physiological cues (Tables 1 and 2). However, we are only starting to understand the molecular basis for such regulation. As such, the transcriptional control of genes associated with auxin inactivation remains largely unexplored. Whether the so-far-known regulatory mechanisms operate in a similar way across different tissues and/or developmental times needs further exploration. Moreover, when considering the number of transcription factors that have been reported to directly control auxin biosynthesis (Table 1), it seems probable that many additional epigenetic mechanisms for the regulation of auxin metabolism will be elucidated in the coming years. The recently described posttranslational modulation of the activity of the auxin biosynthetic enzyme TAA1 (Wang et al. 2020) and the auxin oxidase DAO (Takehara et al. 2020) represents a thrilling starting point to explore similar mechanisms controlling the activity of other enzymes from the pathway.
Despite extensive efforts to unravel intracellular IAA dynamics to understand the role of organelles such as the ER and the vacuole on cellular auxin homeostasis, many critical questions remain unanswered. For example, the subcellular localization of several enzymes involved in IAA metabolism, notably, most of the GH3s, remains unexplored. Moreover, our understanding of how various IAA metabolites are distributed among different organelles, and how this compartmentalization influences intracellular auxin levels, transport, and signaling, is still at a rudimentary level. Fluorescent activated cell sorting has been employed to characterize the hormone distribution in different Arabidopsis root cell types (Petersson et al. 2009; Antoniadi et al. 2015). Sorting pure fractions of organelles by fluorescent activated organelle sorting (FAOS), a technique successfully used in mammalian cells (Gauthier et al. 2008), followed by high-resolution IAA metabolite profiling is expected to advance our knowledge regarding how the intracellular compartmentalization of different enzymes and metabolites influences auxin homeostasis (Novák et al. 2017; Skalický et al. 2018). Novel techniques, however, will be required to study the dynamic changes of subcellular IAA distribution and metabolism in response to different stimuli in planta. As most of the IAA is sensed in the nucleus, determination of IAA levels in nuclei during different responses to auxin in living plants, and the specific role of other cell compartments in modulating the available IAA to be sensed, remains an exciting challenge.
ACKNOWLEDGMENTS
We apologize to the authors who have made valuable contributions to the field, but whose research we could not include because of space constraints. Research in the laboratory of Karin Ljung is supported by grants from the Swedish Foundation for Strategic Research (Vinnova), the Knut and Alice Wallenberg Foundation (KAW), the Swedish Research Councils VR and Formas, and Carl Tryggers Stiftelse för Vetenskaplig Forskning. R.C.-S. held a postdoctoral fellowship from Kempestiftelserna (JCK-1111). E.M.-B. holds a postdoctoral fellowship from Kempestiftelserna (JCK-1811).
Footnotes
Editors: Dolf Weijers, Karin Ljung, Mark Estelle, and Ottoline Leyser
Additional Perspectives on Auxin Signaling available at www.cshperspectives.org
REFERENCES
- Abbas M, Hernández-García J, Pollmann S, Samodelov SL, Kolb M, Friml J, Hammes UZ, Zurbriggen MD, Blázquez MA, Alabadí D. 2018. Auxin methylation is required for differential growth in Arabidopsis. Proc Natl Acad Sci 115: 6864–6869. 10.1073/pnas.1806565115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abel S, Theologis A. 2010. Odyssey of auxin. CSH Perspect Biol 2: a004572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoniadi I, Plačková L, Simonovik B, Doležal K, Turnbull C, Ljung K, Novák O. 2015. Cell-type-specific cytokinin distribution within the Arabidopsis primary root apex. Plant Cell 27: 1955–1967. 10.1105/tpc.15.00176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbez E, Kubeš M, Rolčík J, Béziat C, Pěnčík A, Wang B, Rosquete MR, Zhu J, Dobrev PI, Lee Y, et al. 2012. A novel putative auxin carrier family regulates intracellular auxin homeostasis in plants. Nature 485: 119–122. 10.1038/nature11001 [DOI] [PubMed] [Google Scholar]
- Barlier I, Kowalczyk M, Marchant A, Ljung K, Bhalerao R, Bennett M, Sandberg G, Bellini C. 2000. The SUR2 gene of Arabidopsis thaliana encodes the cytochrome P450 CYP83B1, a modulator of auxin homeostasis. Proc Natl Acad Sci 97: 14819–14824. 10.1073/pnas.260502697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel B, Fink GR. 1995. ILR1, an amidohydrolase that releases active indole-3-acetic acid from conjugates. Science 268: 1745–1748. 10.1126/science.7792599 [DOI] [PubMed] [Google Scholar]
- Bartel B, LeClere S, Magidin M, Zolman BK. 2001. Inputs to the active indole-3-acetic acid pool: de novo synthesis, conjugate hydrolysis, and indole-3-butyric acid β-oxidation. J Plant Growth Regul 20: 198–216. 10.1007/s003440010025 [DOI] [Google Scholar]
- Bernardi J, Lanubile A, Li QB, Kumar D, Kladnik A, Cook SD, Ross JJ, Marocco A, Chourey PS. 2012. Impaired auxin biosynthesis in the defective endosperm18 mutant is due to mutational loss of expression in the ZmYuc1 gene encoding endosperm-specific YUCCA1 protein in maize. Plant Physiol 160: 1318–1328. 10.1104/pp.112.204743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Béziat C, Barbez E, Feraru MI, Lucyshyn D, Kleine-Vehn J. 2017. Light triggers PILS-dependent reduction in nuclear auxin signalling for growth transition. Nat Plants 3: 17105. 10.1038/nplants.2017.105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalerao RP, Eklöf J, Ljung K, Marchant A, Bennett M, Sandberg G. 2002. Shoot-derived auxin is essential for early lateral root emergence in Arabidopsis seedlings. Plant J 29: 325–332. 10.1046/j.0960-7412.2001.01217.x [DOI] [PubMed] [Google Scholar]
- Boerjan W, Cervera MT, Delarue M, Beeckman T, Dewitte W, Bellini C, Caboche M, Van Onckelen H, Van Montagu M, Inzé D. 1995. Superroot, a recessive mutation in Arabidopsis, confers auxin overproduction. Plant Cell 7: 1405–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumos J, Robles LM, Yun J, Vu TC, Jackson S, Alonso JM, Stepanova AN. 2018. Local auxin biosynthesis is a key regulator of plant development. Dev Cell 47: 306–318.e5. 10.1016/j.devcel.2018.09.022 [DOI] [PubMed] [Google Scholar]
- Brunoni F, Collani S, Casanova-Sáez R, Šimura J, Karady M, Schmid M, Ljung K, Bellini C. 2020. Conifers exhibit a characteristic inactivation of auxin to maintain tissue homeostasis. New Phytol 226: 1753–1765. 10.1111/nph.16463 [DOI] [PubMed] [Google Scholar]
- Cai XT, Xu P, Zhao PX, Liu R, Yu LH, Xiang CB. 2014. Arabidopsis ERF109 mediates cross-talk between jasmonic acid and auxin biosynthesis during lateral root formation. Nat Commun 5: 5833. 10.1038/ncomms6833 [DOI] [PubMed] [Google Scholar]
- Chen Q, Dai X, De-Paoli H, Cheng Y, Takebayashi Y, Kasahara H, Kamiya Y, Zhao Y. 2014. Auxin overproduction in shoots cannot rescue auxin deficiencies in Arabidopsis roots. Plant Cell Physiol 55: 1072–1079. 10.1093/pcp/pcu039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Tong J, Xiao L, Ruan Y, Liu J, Zeng M, Huang H, Wang JW, Xu L. 2016. YUCCA-mediated auxin biogenesis is required for cell fate transition occurring during de novo root organogenesis in Arabidopsis. J Exp Bot 67: 4273–4284. 10.1093/jxb/erw213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Huang XX, Zhao SM, Xiao DW, Xiao LT, Tong JH, Wang WS, Li YJ, Ding Z, Hou BK. 2020. IPya glucosylation mediates light and temperature signaling to regulate auxin-dependent hypocotyl elongation in Arabidopsis. Proc Natl Acad Sci 117: 6910–6917. 10.1073/pnas.2000172117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Dai X, Zhao Y. 2006. Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev 20: 1790–1799. 10.1101/gad.1415106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Dai X, Zhao Y. 2007. Auxin synthesized by the YUCCA flavin monooxygenases is essential for embryogenesis and leaf formation in Arabidopsis. Plant Cell 19: 2430–2439. 10.1105/tpc.107.053009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen JD, Bandurski RS. 1982. Chemistry and physiology of the bound auxins. Annu Rev Plant Biol 33: 403–430. 10.1146/annurev.pp.33.060182.002155 [DOI] [Google Scholar]
- Cook SD. 2019. An historical review of phenylacetic acid. Plant Cell Physiol 60: 243–254. 10.1093/pcp/pcz004 [DOI] [PubMed] [Google Scholar]
- Cui D, Zhao J, Jing Y, Fan M, Liu J, Wang Z, Xin W, Hu Y. 2013. The Arabidopsis IDD14, IDD15, and IDD16 cooperatively regulate lateral organ morphogenesis and gravitropism by promoting auxin biosynthesis and transport. PLoS Genet 9: e1003759. 10.1371/journal.pgen.1003759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dal Bosco C, Dovzhenko A, Liu X, Woerner N, Rensch T, Eismann M, Eimer S, Hegermann J, Paponov IA, Ruperti B, et al. 2012. The endoplasmic reticulum localized PIN8 is a pollen-specific auxin carrier involved in intracellular auxin homeostasis. Plant J 71: 860–870. 10.1111/j.1365-313X.2012.05037.x [DOI] [PubMed] [Google Scholar]
- Davies PJ, Mitchell EK. 1972. Transport of indoleacetic acid in intact roots of Phaseolus coccineus. Planta 105: 139–154. 10.1007/BF00385573 [DOI] [PubMed] [Google Scholar]
- Davies RT, Goetz DH, Lasswell J, Anderson MN, Bartel B. 1999. IAR3 encodes an auxin conjugate hydrolase from Arabidopsis. Plant Cell 11: 365–376. 10.1105/tpc.11.3.365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delarue M, Prinsen E, Onckelen HV, Caboche M, Bellini C. 1998. Sur2 mutations of Arabidopsis thaliana define a new locus involved in the control of auxin homeostasis. Plant J 14: 603–611. 10.1046/j.1365-313X.1998.00163.x [DOI] [PubMed] [Google Scholar]
- Derkacheva M, Steinbach Y, Wildhaber T, Mozgová I, Mahrez W, Nanni P, Bischof S, Gruissem W, Hennig L. 2013. Arabidopsis MSI1 connects LHP1 to PRC2 complexes. EMBO J 32: 2073–2085. 10.1038/emboj.2013.145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di DW, Wu L, Zhang L, An CW, Zhang TZ, Luo P, Gao HH, Kriechbaumer V, Guo GQ. 2016. Functional roles of Arabidopsis CKRC2/YUCCA8 gene and the involvement of PIF4 in the regulation of auxin biosynthesis by cytokinin. Sci Rep 6: 36866. 10.1038/srep36866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Mambro R, De Ruvo M, Pacifici E, Salvi E, Sozzani R, Benfey PN, Busch W, Novák O, Ljung K, Di Paola L, et al. 2017. Auxin minimum triggers the developmental switch from cell division to cell differentiation in the Arabidopsis root. Proc Natl Acad Sci 114: E7641–E7649. 10.1073/pnas.1705833114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Mambro R, Svolacchia N, Dello Ioio R, Pierdonati E, Salvi E, Pedrazzini E, Vitale A, Perilli S, Sozzani R, Benfey PN, et al. 2019. The lateral root cap acts as an auxin sink that controls meristem size. Curr Biol 29: 1199–1205.e4. 10.1016/j.cub.2019.02.022 [DOI] [PubMed] [Google Scholar]
- Ding X, Cao Y, Huang L, Zhao J, Xu C, Li X, Wang S. 2008. Activation of the indole-3-acetic acid-amido synthetase GH3-8 suppresses expansin expression and promotes salicylate- and jasmonate-independent basal immunity in rice. Plant Cell 20: 228–240. 10.1105/tpc.107.055657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z, Wang B, Moreno I, Dupláková N, Simon S, Carraro N, Reemmer J, Pěnčík A, Chen X, Tejos R, et al. 2012. ER-localized auxin transporter PIN8 regulates auxin homeostasis and male gametophyte development in Arabidopsis. Nat Commun 3: 941. 10.1038/ncomms1941 [DOI] [PubMed] [Google Scholar]
- D'Ippolito S, Vankova R, Joosten MH, Casalongué CA, Fiol DF. 2016. Knocking down expression of the auxin-amidohydrolase IAR3 alters defense responses in Solanaceae family plants. Plant Sci 253: 31–39. 10.1016/j.plantsci.2016.09.008 [DOI] [PubMed] [Google Scholar]
- Du H, Wu N, Fu J, Wang S, Li X, Xiao J, Xiong L. 2012. A GH3 family member, OsGH3-2, modulates auxin and abscisic acid levels and differentially affects drought and cold tolerance in rice. J Exp Bot 63: 6467–6480. 10.1093/jxb/ers300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eklund DM, Ståldal V, Valsecchi I, Cierlik I, Eriksson C, Hiratsu K, Ohme-Takagi M, Sundström JF, Thelander M, Ezcurra I, et al. 2010. The Arabidopsis thaliana STYLISH1 protein acts as a transcriptional activator regulating auxin biosynthesis. Plant Cell 22: 349–363. 10.1105/tpc.108.064816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eklund DM, Ishizaki K, Flores-Sandoval E, Kikuchi S, Takebayashi Y, Tsukamoto S, Hirakawa Y, Nonomura M, Kato H, Kouno M, et al. 2015. Auxin produced by the indole-3-pyruvic acid pathway regulates development and gemmae dormancy in the liverwort Marchantia polymorpha. Plant Cell 27: 1650–1669. 10.1105/tpc.15.00065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feraru E, Feraru MI, Barbez E, Waidmann S, Sun L, Gaidora A, Kleine-Vehn J. 2019. PILS6 is a temperature-sensitive regulator of nuclear auxin input and organ growth in Arabidopsis thaliana. Proc Natl Acad Sci 116: 3893–3898. 10.1073/pnas.1814015116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueiredo DD, Batista RA, Roszak PJ, Köhler C. 2015. Auxin production couples endosperm development to fertilization. Nat Plants 1: 15184. 10.1038/nplants.2015.184 [DOI] [PubMed] [Google Scholar]
- Forgione I, Wołoszynska M, Pacenza M, Chiappetta A, Greco M, Araniti F, Abenavoli MR, Van Lijsebettens M, Bitonti MB, Bruno L. 2019. Hypomethylated drm1 drm2 cmt3 mutant phenotype of Arabidopsis thaliana is related to auxin pathway impairment. Plant Sci 280: 383–396. 10.1016/j.plantsci.2018.12.029 [DOI] [PubMed] [Google Scholar]
- Franklin KA, Lee SH, Patel D, Kumar SV, Spartz AK, Gu C, Ye S, Yu P, Breen G, Cohen JD, et al. 2011. Phytochrome-interacting factor 4 (PIF4) regulates auxin biosynthesis at high temperature. Proc Natl Acad Sci 108: 20231–20235. 10.1073/pnas.1110682108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frick EM, Strader LC. 2018. Roles for IBA-derived auxin in plant development. J Exp Bot 69: 169–177. 10.1093/jxb/erx298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallavotti A, Barazesh S, Malcomber S, Hall D, Jackson D, Schmidt RJ, McSteen P. 2008. Sparse inflorescence1 encodes a monocot-specific YUCCA-like gene required for vegetative and reproductive development in maize. Proc Natl Acad Sci 105: 15196–15201. 10.1073/pnas.0805596105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego-Bartolomé J. 2020. DNA methylation in plants: mechanisms and tools for targeted manipulation. New Phytol 227: 38–44. 10.1111/nph.16529 [DOI] [PubMed] [Google Scholar]
- Gallei M, Luschnig C, Friml J. 2020. Auxin signalling in growth: Schrödinger's cat out of the bag. Curr Opin Plant Biol 53: 43–49. 10.1016/j.pbi.2019.10.003 [DOI] [PubMed] [Google Scholar]
- Gao Y, Dai X, Aoi Y, Takebayashi Y, Yang L, Guo X, Zeng Q, Yu H, Kasahara H, Zhao Y. 2020. Two homologous INDOLE-3-ACETAMIDE (IAM) HYDROLASE genes are required for the auxin effects of IAM in Arabidopsis. J Genet Genom 47: 157–165. 10.1016/j.jgg.2020.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier DJ, Sobota JA, Ferraro F, Mains RE, Lazure C. 2008. Flow cytometry-assisted purification and proteomic analysis of the corticotropes dense-core secretory granules. Proteomics 8: 3848–3861. 10.1002/pmic.200700969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glawischnig E, Hansen BG, Olsen CE, Halkier BA. 2004. Camalexin is synthesized from indole-3-acetaldoxime, a key branching point between primary and secondary metabolism in Arabidopsis. Proc Natl Acad Sci 101: 8245–8250. 10.1073/pnas.0305876101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WM, Östin A, Sandberg G, Romano CP, Estelle M. 1998. High temperature promotes auxin-mediated hypocotyl elongation in Arabidopsis. Proc Natl Acad Sci 95: 7197–7202. 10.1073/pnas.95.12.7197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubb CD, Zipp BJ, Ludwig-Müller J, Masuno MN, Molinski TF, Abel S. 2004. Arabidopsis glucosyltransferase UGT74B1 functions in glucosinolate biosynthesis and auxin homeostasis. Plant J 40: 893–908. 10.1111/j.1365-313X.2004.02261.x [DOI] [PubMed] [Google Scholar]
- Gutierrez L, Mongelard G, Floková K, Păcurar DI, Novák O, Staswick P, Kowalczyk M, Păcurar M, Demailly H, Geiss G, et al. 2012. Auxin controls Arabidopsis adventitious root initiation by regulating jasmonic acid homeostasis. Plant Cell 24: 2515–2527. 10.1105/tpc.112.099119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyula P, Baksa I, Tóth T, Mohorianu I, Dalmay T, Szittya G. 2018. Ambient temperature regulates the expression of a small set of sRNAs influencing plant development through NF-YA2 and YUC2. Plant Cell Environ 41: 2404–2417. 10.1111/pce.13355 [DOI] [PubMed] [Google Scholar]
- Hall PJ. 1980. Indole-3-acetyl-myo-inositol in kernels of Oryza sativa. Phytochemistry 19: 2121–2123. 10.1016/S0031-9422(00)82206-2 [DOI] [Google Scholar]
- Han Q, Chen K, Yan D, Hao G, Qi J, Wang C, Dirk LMA, Bruce Downie A, Gong J, Wang J, et al. 2020. ZmDREB2A regulates ZmGH3.2 and ZmRAFS, shifting metabolism towards seed aging tolerance over seedling growth. Plant J 104: 1268–1282. 10.1111/TPJ.14922 [DOI] [PubMed] [Google Scholar]
- He C, Chen X, Huang H, Xu L. 2012. Reprogramming of H3K27me3 is critical for acquisition of pluripotency from cultured Arabidopsis tissues. PLoS Genet 8: e1002911. 10.1371/journal.pgen.1002911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentrich M, Böttcher C, Düchting P, Cheng Y, Zhao Y, Berkowitz O, Masle J, Medina J, Pollmann S. 2013. The jasmonic acid signaling pathway is linked to auxin homeostasis through the modulation of YUCCA8 and YUCCA9 gene expression. Plant J 74: 626–637. 10.1111/tpj.12152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P. 2008. Genevestigator V3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinformatics 2008: 420747. 10.1155/2008/420747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull AK, Vij R, Celenza JL. 2000. Arabidopsis cytochrome P450s that catalyze the first step of tryptophan-dependent indole-3-acetic acid biosynthesis. Proc Natl Acad Sci 97: 2379–2384. 10.1073/pnas.040569997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda Y, Men S, Fischer U, Stepanova AN, Alonso JM, Ljung K, Grebe M. 2009. Local auxin biosynthesis modulates gradient-directed planar polarity in Arabidopsis. Nat Cell Biol 11: 731–738. 10.1038/ncb1879 [DOI] [PubMed] [Google Scholar]
- Ishimaru K, Hirotsu N, Madoka Y, Murakami N, Hara N, Onodera H, Kashiwagi T, Ujiie K, Shimizu B, Onishi A, et al. 2013. Loss of function of the IAA-glucose hydrolase gene TGW6 enhances rice grain weight and increases yield. Nat Genet 45: 707–711. 10.1038/ng.2612 [DOI] [PubMed] [Google Scholar]
- Jackson RG, Kowalczyk M, Li Y, Higgins G, Ross J, Sandberg G, Bowles DJ. 2002. Over-expression of an Arabidopsis gene encoding a glucosyltransferase of indole-3-acetic acid: phenotypic characterisation of transgenic lines. Plant J 32: 573–583. 10.1046/j.1365-313X.2002.01445.x [DOI] [PubMed] [Google Scholar]
- Jakubowska A, Kowalczyk S. 2005. A specific enzyme hydrolyzing 6-O(4-O)-indole-3-ylacetyl-β-d-glucose in immature kernels of Zea mays. J Plant Physiol 162: 207–213. 10.1016/j.jplph.2004.05.015 [DOI] [PubMed] [Google Scholar]
- Jayasinghege CPA, Ozga JA, Nadeau CD, Kaur H, Reinecke DM. 2019. TIR1 auxin receptors are implicated in the differential response to 4-Cl-IAA and IAA in developing pea fruit. J Exp Bot 70: 1239–1253. 10.1093/jxb/ery456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones B, Gunnerås SA, Petersson SV, Tarkowski P, Graham N, May S, Dolezal K, Sandberg G, Ljung K. 2010. Cytokinin regulation of auxin synthesis in Arabidopsis involves a homeostatic feedback loop regulated via auxin and cytokinin signal transduction. Plant Cell 22: 2956–2969. 10.1105/tpc.110.074856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kai K, Wakasa K, Miyagawa H. 2007. Metabolism of indole-3-acetic acid in rice: identification and characterization of N-β-d-glucopyranosyl indole-3-acetic acid and its conjugates. Phytochemistry 68: 2512–2522. 10.1016/j.phytochem.2007.05.040 [DOI] [PubMed] [Google Scholar]
- Kawai Y, Ono E, Mizutani M. 2014. Evolution and diversity of the 2-oxoglutarate-dependent dioxygenase superfamily in plants. Plant J 78: 328–343. 10.1111/tpj.12479 [DOI] [PubMed] [Google Scholar]
- King JJ, Stimart DP, Fisher RH, Bleecker AB. 1995. A mutation altering auxin homeostasis and plant morphology in Arabidopsis. Plant Cell 7: 2023–2037. 10.2307/3870148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita N, Wang H, Kasahara H, Liu J, Macpherson C, Machida Y, Kamiya Y, Hannah MA, Chua NH. 2012. IAA-Ala Resistant3, an evolutionarily conserved target of miR167, mediates Arabidopsis root architecture changes during high osmotic stress. Plant Cell 24: 3590–3602. 10.1105/tpc.112.097006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirungu JN, Magwanga RO, Lu P, Cai X, Zhou Z, Wang X, Peng R, Wang K, Liu F. 2019. Functional characterization of Gh_A08G1120 (GH3.5) gene reveal their significant role in enhancing drought and salt stress tolerance in cotton. BMC Genet 20: 62. 10.1186/s12863-019-0756-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong W, Li Y, Zhang M, Jin F, Li J. 2015. A novel Arabidopsis microRNA promotes IAA biosynthesis via the indole-3-acetaldoxime pathway by suppressing SUPERROOT1. Plant Cell Physiol 56: 715–726. 10.1093/pcp/pcu216 [DOI] [PubMed] [Google Scholar]
- Korasick DA, Enders TA, Strader LC. 2013. Auxin biosynthesis and storage forms. J Exp Bot 64: 2541–2555. 10.1093/jxb/ert080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczyk M, Sandberg G. 2001. Quantitative analysis of indole-3-acetic acid metabolites in Arabidopsis. Plant Physiol 127: 1845–1853. 10.1104/pp.010525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriechbaumer V, Wang P, Hawes C, Abell BM. 2012. Alternative splicing of the auxin biosynthesis gene YUCCA4 determines its subcellular compartmentation. Plant J 70: 292–302. 10.1111/j.1365-313X.2011.04866.x [DOI] [PubMed] [Google Scholar]
- Kriechbaumer V, Seo H, Park WJ, Hawes C. 2015. Endoplasmic reticulum localization and activity of maize auxin biosynthetic enzymes. J Exp Bot 66: 6009–6020. 10.1093/jxb/erv314 [DOI] [PubMed] [Google Scholar]
- Kriechbaumer V, Botchway SW, Hawes C. 2016. Localization and interactions between Arabidopsis auxin biosynthetic enzymes in the TAA/YUC-dependent pathway. J Exp Bot 67: 4195–4207. 10.1093/jxb/erw195 [DOI] [PubMed] [Google Scholar]
- Kubeš M, Yang H, Richter GL, Cheng Y, Młodzińska E, Wang X, Blakeslee JJ, Carraro N, Petrášek J, Zažímalová E, et al. 2012. The Arabidopsis concentration-dependent influx/efflux transporter ABCB4 regulates cellular auxin levels in the root epidermis. Plant J 69: 640–654. 10.1111/j.1365-313X.2011.04818.x [DOI] [PubMed] [Google Scholar]
- Kumar SV, Wigge PA. 2010. H2a.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell 140: 136–147. 10.1016/j.cell.2009.11.006 [DOI] [PubMed] [Google Scholar]
- Lafos M, Kroll P, Hohenstatt ML, Thorpe FL, Clarenz O, Schubert D. 2011. Dynamic regulation of H3K27 trimethylation during Arabidopsis differentiation. PLoS Genet 7: e1002040. 10.1371/journal.pgen.1002040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam HK, McAdam SA, McAdam EL, Ross JJ. 2015. Evidence that chlorinated auxin is restricted to the Fabaceae but not to the Fabeae. Plant Physiol 168: 798–803. 10.1104/pp.15.00410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeClere S, Tellez R, Rampey RA, Matsuda SP, Bartel B. 2002. Characterization of a family of IAA-amino acid conjugate hydrolases from Arabidopsis. J Biol Chem 277: 20446–20452. 10.1074/jbc.M111955200 [DOI] [PubMed] [Google Scholar]
- Lee K, Seo PJ. 2017. Coordination of matrix attachment and ATP-dependent chromatin remodeling regulate auxin biosynthesis and Arabidopsis hypocotyl elongation. PLoS ONE 12: e0181804. 10.1371/journal.pone.0181804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann T, Janowitz T, Sánchez-Parra B, Alonso MP, Trompetter I, Piotrowski M, Pollmann S. 2017. Arabidopsis NITRILASE 1 contributes to the regulation of root growth and development through modulation of auxin biosynthesis in seedlings. Front Plant Sci 8: 36. 10.3389/fpls.2017.00036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Hou X, Tsuge T, Ding M, Aoyama T, Oka A, Gu H, Zhao Y, Qu LJ. 2008. The possible action mechanisms of indole-3-acetic acid methyl ester in Arabidopsis. Plant Cell Rep 27: 575–584. 10.1007/s00299-007-0458-9 [DOI] [PubMed] [Google Scholar]
- Liu X, Barkawi L, Gardner G, Cohen JD. 2012. Transport of indole-3-butyric acid and indole-3-acetic acid in Arabidopsis hypocotyls using stable isotope labeling. Plant Physiol 158: 1988–2000. 10.1104/pp.111.191288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Zhang L, Luo Y, Xu M, Fan Y, Wang L. 2016a. Interactions of Oryza sativa OsCONTINUOUS VASCULAR RING-LIKE 1 (OsCOLE1) and OsCOLE1-INTERACTING PROTEIN reveal a novel intracellular auxin transport mechanism. New Phytol 212: 96–107. 10.1111/nph.14021 [DOI] [PubMed] [Google Scholar]
- Liu G, Gao S, Tian H, Wu W, Robert HS, Ding Z. 2016b. Local transcriptional control of YUCCA regulates auxin promoted root-growth inhibition in response to aluminium stress in Arabidopsis. PLoS Genet 12: e1006360. 10.1371/journal.pgen.1006360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Chen TT, Xiao DW, Zhao SM, Lin JS, Wang T, Li YJ, Hou BK. 2019. OsIAGT1 is a glucosyltransferase gene involved in the glucose conjugation of auxins in rice. Rice 12: 92. 10.1186/s12284-019-0357-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljung K, Bhalerao RP, Sandberg G. 2001a. Sites and homeostatic control of auxin biosynthesis in Arabidopsis during vegetative growth. Plant J 28: 465–474. 10.1046/j.1365-313X.2001.01173.x [DOI] [PubMed] [Google Scholar]
- Ljung K, Östin A, Lioussanne L, Sandberg G. 2001b. Developmental regulation of indole-3-acetic acid turnover in Scots pine seedlings. Plant Physiol 125: 464–475. 10.1104/pp.125.1.464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljung K, Hull AK, Celenza J, Yamada M, Estelle M, Normanly J, Sandberg G. 2005. Sites and regulation of auxin biosynthesis in Arabidopsis roots. Plant Cell 17: 1090–1104. 10.1105/tpc.104.029272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig-Müller J, Walz A, Slovin JP, Epstein E, Cohen JD, Dong W, Town CD. 2005. Overexpression of maize IAGLU in Arabidopsis thaliana alters plant growth and sensitivity to IAA but not IBA and 2,4-D. J Plant Growth Regul 24: 127–141. 10.1007/s00344-004-0006-6 [DOI] [Google Scholar]
- Ludwig-Müller J, Jülke S, Bierfreund NM, Decker EL, Reski R. 2009. Moss (Physcomitrella patens) GH3 proteins act in auxin homeostasis. New Phytol 181: 323–338. 10.1111/j.1469-8137.2008.02677.x [DOI] [PubMed] [Google Scholar]
- Lynch JH, Qian Y, Guo L, Maoz I, Huang XQ, Garcia AS, Louie G, Bowman ME, Noel JP, Morgan JA, et al. 2020. Modulation of auxin formation by the cytosolic phenylalanine biosynthetic pathway. Nat Chem Biol 16: 850–856. 10.1038/s41589-020-0519-8 [DOI] [PubMed] [Google Scholar]
- Maeda H, Dudareva N. 2012. The shikimate pathway and aromatic amino acid biosynthesis in plants. Annu Rev Plant Biol 63: 73–105. 10.1146/annurev-arplant-042811-105439 [DOI] [PubMed] [Google Scholar]
- Mao JL, Miao ZQ, Wang Z, Yu LH, Cai XT, Xiang CB. 2016. Arabidopsis ERF1 mediates cross-talk between ethylene and auxin biosynthesis during primary root elongation by regulating ASA1 expression. PLoS Genet 12: e1005760. 10.1371/journal.pgen.1005760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashiguchi K, Tanaka K, Sakai T, Sugawara S, Kawaide H, Natsume M, Hanada A, Yaeno T, Shirasu K, Yao H, et al. 2011. The main auxin biosynthesis pathway in Arabidopsis. Proc Natl Acad Sci 108: 18512–18517. 10.1073/pnas.1108434108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateo-Bonmatí E, Casanova-Sáez R, Ljung K. 2019. Epigenetic regulation of auxin homeostasis. Biomolecules 9: 623. 10.3390/biom9100623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matosevich R, Cohen I, Gil-Yarom N, Modrego A, Friedlander-Shani L, Verna C, Scarpella E, Efroni I. 2020. Local auxin biosynthesis is required for root regeneration after wounding. Nat Plants 6: 1020–1030. 10.1038/s41477-020-0737-9 [DOI] [PubMed] [Google Scholar]
- Matsuo S, Miyatake K, Endo M, Urashimo S, Kawanishi T, Negoro S, Shimakoshi S, Fukuoka H. 2020. Loss of function of the Pad-1 aminotransferase gene, which is involved in auxin homeostasis, induces parthenocarpy in Solanaceae plants. Proc Natl Acad Sci 117: 12784–12790. 10.1073/pnas.2001211117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthes MS, Best NB, Robil JM, Malcomber S, Gallavotti A, McSteen P. 2019. Auxin EvoDevo: conservation and diversification of genes regulating auxin biosynthesis, transport, and signaling. Mol Plant 12: 298–320. 10.1016/j.molp.2018.12.012 [DOI] [PubMed] [Google Scholar]
- Mellor N, Band LR, Pěnčík A, Novák O, Rashed A, Holman T, Wilson MH, Voß U, Bishopp A, King JR, et al. 2016. Dynamic regulation of auxin oxidase and conjugating enzymes AtDAO1 and GH3 modulates auxin homeostasis. Proc Natl Acad Sci 113: 11022–11027. 10.1073/pnas.1604458113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng WJ, Cheng ZJ, Sang YL, Zhang MM, Rong XF, Wang ZW, Tang YY, Zhang XS. 2017. Type-B ARABIDOPSIS RESPONSE REGULATORs specify the shoot stem cell niche by dual regulation of WUSCHEL. Plant Cell 29: 1357–1372. 10.1105/tpc.16.00640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michniewicz M, Ho CH, Enders TA, Floro E, Damodaran S, Gunther LK, Powers SK, Frick EM, Topp CN, Frommer WB, et al. 2019. TRANSPORTER OF IBA1 links auxin and cytokinin to influence root architecture. Dev Cell 50: 599–609.e4. 10.1016/j.devcel.2019.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen MD, Hansen CH, Wittstock U, Halkier BA. 2000. Cytochrome P450 CYP79B2 from Arabidopsis catalyzes the conversion of tryptophan to indole-3-acetaldoxime, a precursor of indole glucosinolates and indole-3-acetic acid. J Biol Chem 275: 33712–33717. 10.1074/jbc.M001667200 [DOI] [PubMed] [Google Scholar]
- Milutinovic M, Lindsey BE III, Wijeratne A, Hernandez JM, Grotewold N, Fernández V, Grotewold E, Brkljacic J. 2019. Arabidopsis EMSY-like (EML) histone readers are necessary for post-fertilization seed development, but prevent fertilization-independent seed formation. Plant Sci 285: 99–109. 10.1016/j.plantsci.2019.04.007 [DOI] [PubMed] [Google Scholar]
- Moubayidin L, Di Mambro R, Sabatini S. 2009. Cytokinin-auxin crosstalk. Trends Plant Sci 14: 557–562. 10.1016/j.tplants.2009.06.010 [DOI] [PubMed] [Google Scholar]
- Moubayidin L, Di Mambro R, Sozzani R, Pacifici E, Salvi E, Terpstra I, Bao D, van Dijken A, Dello Ioio R, Perilli S, et al. 2013. Spatial coordination between stem cell activity and cell differentiation in the root meristem. Dev Cell 26: 405–415. 10.1016/j.devcel.2013.06.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mravec J, Skůpa P, Bailly A, Hoyerová K, Křeček P, Bielach A, Petrášek J, Zhang J, Gaykova V, Stierhof YD, et al. 2009. Subcellular homeostasis of phytohormone auxin is mediated by the ER-localized PIN5 transporter. Nature 459: 1136–1140. 10.1038/nature08066 [DOI] [PubMed] [Google Scholar]
- Müller CJ, Larsson E, Spíchal L, Sundberg E. 2017. Cytokinin-auxin crosstalk in the gynoecial primordium ensures correct domain patterning. Plant Physiol 175: 1144–1157. 10.1104/pp.17.00805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadi R, Mateo-Bonmatí E, Juan-Vicente L, Micol JL. 2018. The 2OGD superfamily: emerging functions in plant epigenetics and hormone metabolism. Mol Plant 11: 1222–1224. 10.1016/j.molp.2018.09.002 [DOI] [PubMed] [Google Scholar]
- Nafisi M, Goregaoker S, Botanga CJ, Glawischnig E, Olsen CE, Halkier BA, Glazebrook J. 2007. Arabidopsis cytochrome P450 monooxygenase 71A13 catalyzes the conversion of indole-3-acetaldoxime in camalexin synthesis. Plant Cell 19: 2039–2052. 10.1105/tpc.107.051383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niehaus TD, Patterson JA, Alexander DC, Folz JS, Pyc M, MacTavish BS, Bruner SD, Mullen RT, Fiehn O, Hanson AD. 2019. The metabolite repair enzyme Nit1 is a dual-targeted amidase that disposes of damaged glutathione in Arabidopsis. Biochem J 476: 683–697. 10.1042/BCJ20180931 [DOI] [PubMed] [Google Scholar]
- Niyogi KK, Fink GR. 1992. Two anthranilate synthase genes in Arabidopsis: defense-related regulation of the tryptophan pathway. Plant Cell 4: 721–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonhebel HM. 2015. Tryptophan-independent indole-3-acetic acid synthesis: critical evaluation of the evidence. Plant Physiol 169: 1001–1005. 10.1104/pp.15.01091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normanly J, Cohen JD, Fink GR. 1993. Arabidopsis thaliana auxotrophs reveal a tryptophan-independent biosynthetic pathway for indole-3-acetic acid. Proc Natl Acad Sci 90: 10355–10359. 10.1073/pnas.90.21.10355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novák O, Hényková E, Sairanen I, Kowalczyk M, Pospíšil T, Ljung K. 2012. Tissue-specific profiling of the Arabidopsis thaliana auxin metabolome. Plant J 72: 523–536. 10.1111/j.1365-313X.2012.05085.x [DOI] [PubMed] [Google Scholar]
- Novák O, Napier R, Ljung K. 2017. Zooming in on plant hormone analysis: tissue- and cell-specific approaches. Annu Rev Plant Biol 68: 323–348. 10.1146/annurev-arplant-042916-040812 [DOI] [PubMed] [Google Scholar]
- Okrent RA, Wildermuth MC. 2011. Evolutionary history of the GH3 family of acyl adenylases in rosids. Plant Mol Biol 76: 489–505. 10.1007/s11103-011-9776-y [DOI] [PubMed] [Google Scholar]
- Östin A, Kowalyczk M, Bhalerao RP, Sandberg G. 1998. Metabolism of indole-3-acetic acid in Arabidopsis. Plant Physiol 118: 285–296. 10.1104/pp.118.1.285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JE, Park JY, Kim YS, Staswick PE, Jeon J, Yun J, Kim SY, Kim J, Lee YH, Park CM. 2007. GH3-mediated auxin homeostasis links growth regulation with stress adaptation response in Arabidopsis. J Biol Chem 282: 10036–10046. 10.1074/jbc.M610524200 [DOI] [PubMed] [Google Scholar]
- Patten CL, Blakney AJ, Coulson TJ. 2013. Activity, distribution and function of indole-3-acetic acid biosynthetic pathways in bacteria. Crit Rev Microbiol 39: 395–415. 10.3109/1040841X.2012.716819 [DOI] [PubMed] [Google Scholar]
- Pěnčík A, Rolčík J, Novák O, Magnus V, Barták P, Buchtík R, Salopek-Sondi B, Strnad M. 2009. Isolation of novel indole-3-acetic acid conjugates by immunoaffinity extraction. Talanta 80: 651–655. 10.1016/j.talanta.2009.07.043 [DOI] [PubMed] [Google Scholar]
- Pěnčík A, Simonovik B, Petersson SV, Henyková E, Simon S, Greenham K, Zhang Y, Kowalczyk M, Estelle M, Zažímalová E, et al. 2013. Regulation of auxin homeostasis and gradients in Arabidopsis roots through the formation of the indole-3-acetic acid catabolite 2-oxindole-3-acetic acid. Plant Cell 25: 3858–3870. 10.1105/tpc.113.114421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pěnčík A, Casanova-Sáez R, Pilařová V, Žukauskaitė A, Pinto R, Micol JL, Ljung K, Novák O. 2018. Ultra-rapid auxin metabolite profiling for high-throughput mutant screening in Arabidopsis. J Exp Bot 69: 2569–2579. 10.1093/jxb/ery084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng M, Li Z, Zhou N, Ma M, Jiang Y, Dong A, Shen WH, Li L. 2018. Linking PHYTOCHROME-INTERACTING FACTOR to histone modification in plant shade avoidance. Plant Physiol 176: 1341–1351. 10.1104/pp.17.01189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersson SV, Johansson AI, Kowalczyk M, Makoveychuk A, Wang JY, Moritz T, Grebe M, Benfey PN, Sandberg G, Ljung K. 2009. An auxin gradient and maximum in the Arabidopsis root apex shown by high-resolution cell-specific analysis of IAA distribution and synthesis. Plant Cell 21: 1659–1668. 10.1105/tpc.109.066480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips KA, Skirpan AL, Liu X, Christensen A, Slewinski TL, Hudson C, Barazesh S, Cohen JD, Malcomber S, McSteen P. 2011. Vanishing tassel2 encodes a grass-specific tryptophan aminotransferase required for vegetative and reproductive development in maize. Plant Cell 23: 550–566. 10.1105/tpc.110.075267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierdonati E, Unterholzner SJ, Salvi E, Svolacchia N, Bertolotti G, Dello Ioio R, Sabatini S, Di Mambro R. 2019. Cytokinin-dependent control of GH3 group II family genes in the Arabidopsis root. Plants 8: 94. 10.3390/plants8040094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piotrowski M. 2008. Primary or secondary? Versatile nitrilases in plant metabolism. Phytochemistry 69: 2655–2667. 10.1016/j.phytochem.2008.08.020 [DOI] [PubMed] [Google Scholar]
- Pollmann S, Müller A, Piotrowski M, Weiler EW. 2002. Occurrence and formation of indole-3-acetamide in Arabidopsis thaliana. Planta 216: 155–161. 10.1007/s00425-002-0868-4 [DOI] [PubMed] [Google Scholar]
- Porco S, Pěnčík A, Rashed A, Voß U, Casanova-Sáez R, Bishopp A, Golebiowska A, Bhosale R, Swarup R, Swarup K, et al. 2016. Dioxygenase-encoding AtDAO1 gene controls IAA oxidation and homeostasis in Arabidopsis. Proc Natl Acad Sci 113: 11016–11021. 10.1073/pnas.1604375113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulet A, Kriechbaumer V. 2017. Bioinformatics analysis of phylogeny and transcription of TAA/YUC auxin biosynthetic genes. Int J Mol Sci 18: 1791. 10.3390/ijms18081791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin G, Gu H, Zhao Y, Ma Z, Shi G, Yang Y, Pichersky E, Chen H, Liu M, Chen Z, et al. 2005. An indole-3-acetic acid carboxyl methyltransferase regulates Arabidopsis leaf development. Plant Cell 17: 2693–2704. 10.1105/tpc.105.034959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin H, Zhang Z, Wang J, Chen X, Wei P, Huang R. 2017. The activation of OsEIL1 on YUC8 transcription and auxin biosynthesis is required for ethylene-inhibited root elongation in rice early seedling development. PLoS Genet 13: e1006955. 10.1371/journal.pgen.1006955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampey RA, LeClere S, Kowalczyk M, Ljung K, Sandberg G, Bartel B. 2004. A family of auxin-conjugate hydrolases that contributes to free indole-3-acetic acid levels during Arabidopsis germination. Plant Physiol 135: 978–988. 10.1104/pp.104.039677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranocha P, Dima O, Nagy R, Felten J, Corratgé-Faillie C, Novák O, Morreel K, Lacombe B, Martinez Y, Pfrunder S, et al. 2013. Arabidopsis WAT1 is a vacuolar auxin transport facilitator required for auxin homoeostasis. Nat Commun 4: 2625. 10.1038/ncomms3625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Olalde JI, Zúñiga-Mayo VM, Serwatowska J, Chavez Montes RA, Lozano-Sotomayor P, Herrera-Ubaldo H, Gonzalez-Aguilera KL, Ballester P, Ripoll JJ, Ezquer I, et al. 2017. The bHLH transcription factor SPATULA enables cytokinin signaling, and both activate auxin biosynthesis and transport genes at the medial domain of the gynoecium. PLoS Genet 13: e1006726. 10.1371/journal.pgen.1006726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzardi K, Landberg K, Nilsson L, Ljung K, Sundås-Larsson A. 2011. TFL2/LHP1 is involved in auxin biosynthesis through positive regulation of YUCCA genes. Plant J 65: 897–906. 10.1111/j.1365-313X.2010.04470.x [DOI] [PubMed] [Google Scholar]
- Robert HS, Grones P, Stepanova AN, Robles LM, Lokerse AS, Alonso JM, Weijers D, Friml J. 2013. Local auxin sources orient the apical-basal axis in Arabidopsis embryos. Curr Biol 23: 2506–2512. 10.1016/j.cub.2013.09.039 [DOI] [PubMed] [Google Scholar]
- Robert HS, Park C, Gutièrrez CL, Wojcikowska B, Pěnčík A, Novák O, Chen J, Grunewald W, Dresselhaus T, Friml J, et al. 2018. Maternal auxin supply contributes to early embryo patterning in Arabidopsis. Nat Plants 4: 548–553. 10.1038/s41477-018-0204-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sairanen I, Novák O, Pěnčík A, Ikeda Y, Jones B, Sandberg G, Ljung K. 2012. Soluble carbohydrates regulate auxin biosynthesis via PIF proteins in Arabidopsis. Plant Cell 24: 4907–4916. 10.1105/tpc.112.104794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez Carranza AP, Singh A, Steinberger K, Panigrahi K, Palme K, Dovzhenko A, Dal Bosco C. 2016. Hydrolases of the ILR1-like family of Arabidopsis thaliana modulate auxin response by regulating auxin homeostasis in the endoplasmic reticulum. Sci Rep 6: 24212. 10.1038/srep24212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu-Mitao Y, Kakimoto T. 2014. Auxin sensitivities of all Arabidopsis Aux/IAAs for degradation in the presence of every TIR1/AFB. Plant Cell Physiol 55: 1450–1459. 10.1093/pcp/pcu077 [DOI] [PubMed] [Google Scholar]
- Simon S, Skůpa P, Viaene T, Zwiewka M, Tejos R, Klima P, Čarná M, Rolčík J, De Rycke R, Moreno I, et al. 2016. PIN6 auxin transporter at endoplasmic reticulum and plasma membrane mediates auxin homeostasis and organogenesis in Arabidopsis. New Phytol 211: 65–74. 10.1111/nph.14019 [DOI] [PubMed] [Google Scholar]
- Skalický V, Kubeš M, Napier R, Novák O. 2018. Auxins and cytokinins—the role of subcellular organization on homeostasis. Int J Mol Sci 19: 3115. 10.3390/ijms19103115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiess GM, Hausman A, Yu P, Cohen JD, Rampey RA, Zolman BK. 2014. Auxin input pathway disruptions are mitigated by changes in auxin biosynthetic gene expression in Arabidopsis. Plant Physiol 165: 1092–1104. 10.1104/pp.114.236026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick PE, Serban B, Rowe M, Tiryaki I, Maldonado MT, Maldonado MC, Suza W. 2005. Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell 17: 616–627. 10.1105/tpc.104.026690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova AN, Hoyt JM, Hamilton AA, Alonso JM. 2005. A link between ethylene and auxin uncovered by the characterization of two root-specific ethylene-insensitive mutants in Arabidopsis. Plant Cell 17: 2230–2242. 10.1105/tpc.105.033365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova AN, Robertson-Hoyt J, Yun J, Benavente LM, Xie DY, Doležal K, Schlereth A, Jürgens G, Alonso JM. 2008. TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 133: 177–191. 10.1016/j.cell.2008.01.047 [DOI] [PubMed] [Google Scholar]
- Stepanova AN, Yun J, Robles LM, Novák O, He W, Guo H, Ljung K, Alonso JM. 2011. The Arabidopsis YUCCA1 flavin monooxygenase functions in the indole-3-pyruvic acid branch of auxin biosynthesis. Plant Cell 23: 3961–3973. 10.1105/tpc.111.088047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone SL, Braybrook SA, Paula SL, Kwong LW, Meuser J, Pelletier J, Hsieh TF, Fischer RL, Goldberg RB, Harada JJ. 2008. Arabidopsis LEAFY COTYLEDON2 induces maturation traits and auxin activity: implications for somatic embryogenesis. Proc Natl Acad Sci 105: 3151-3156. 10.1073/pnas.0712364105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strader LC, Bartel B. 2011. Transport and metabolism of the endogenous auxin precursor indole-3-butyric acid. Mol Plant 4: 477–486. 10.1093/mp/ssr006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara S, Hishiyama S, Jikumaru Y, Hanada A, Nishimura T, Koshiba T, Zhao Y, Kamiya Y, Kasahara H. 2009. Biochemical analyses of indole-3-acetaldoxime-dependent auxin biosynthesis in Arabidopsis. Proc Natl Acad Sci 106: 5430–5435. 10.1073/pnas.0811226106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Xu Y, Ye S, Jiang H, Chen Q, Liu F, Zhou W, Chen R, Li X, Tietz O, et al. 2009. Arabidopsis ASA1 is important for jasmonate-mediated regulation of auxin biosynthesis and transport during lateral root formation. Plant Cell 21: 1495–1511. 10.1105/tpc.108.064303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Yamazaki C, Mitsui M, Kakei Y, Mitani Y, Nakamura A, Ishii T, Soeno K, Shimada Y. 2015. Transcriptional feedback regulation of YUCCA genes in response to auxin levels in Arabidopsis. Plant Cell Rep 34: 1343–1352. 10.1007/s00299-015-1791-z [DOI] [PubMed] [Google Scholar]
- Szerszen JB, Szczyglowski K, Bandurski RS. 1994. iaglu, a gene from Zea mays involved in conjugation of growth hormone indole-3-acetic acid. Science 265: 1699–1701. 10.1126/science.8085154 [DOI] [PubMed] [Google Scholar]
- Takato S, Kakei Y, Mitsui M, Ishida Y, Suzuki M, Yamazaki C, Hayashi KI, Ishii T, Nakamura A, Soeno K, et al. 2017. Auxin signaling through SCFTIR1/AFBs mediates feedback regulation of IAA biosynthesis. Biosci Biotechnol Biochem 81: 1320–1326. 10.1080/09168451.2017.1313694 [DOI] [PubMed] [Google Scholar]
- Takehara S, Sakuraba S, Mikami B, Yoshida H, Yoshimura H, Itoh A, Endo M, Watanabe N, Nagae T, Matsuoka M, et al. 2020. A common allosteric mechanism regulates homeostatic inactivation of auxin and gibberellin. Nat Commun 11: 2143. 10.1038/s41467-020-16068-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takubo E, Kobayashi M, Hirai S, Aoi Y, Ge C, Dai X, Fukui K, Hayashi KI, Zhao Y, Kasahara H. 2020. Role of Arabidopsis INDOLE-3-ACETIC ACID CARBOXYL METHYLTRANSFERASE 1 in auxin metabolism. Biochem Biophys Res Commun 527: 1033–1038. 10.1016/j.bbrc.2020.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Hayashi K, Natsume M, Kamiya Y, Sakakibara H, Kawaide H, Kasahara H. 2014. UGT74D1 catalyzes the glucosylation of 2-oxindole-3-acetic acid in the auxin metabolic pathway in Arabidopsis. Plant Cell Physiol 55: 218–228. 10.1093/pcp/pct173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang LP, Zhou C, Wang SS, Yuan J, Zhang XS, Su YH. 2017. FUSCA3 interacting with LEAFY COTYLEDON2 controls lateral root formation through regulating YUCCA4 gene expression in Arabidopsis thaliana. New Phytol 213: 1740–1754. 10.1111/nph.14313 [DOI] [PubMed] [Google Scholar]
- Tao Y, Ferrer JL, Ljung K, Pojer F, Hong F, Long JA, Li L, Moreno JE, Bowman ME, Ivans LJ, et al. 2008. Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133: 164–176. 10.1016/j.cell.2008.01.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terol J, Domingo C, Talón M. 2006. The GH3 family in plants: genome wide analysis in rice and evolutionary history based on EST analysis. Gene 371: 279–290. 10.1016/j.gene.2005.12.014 [DOI] [PubMed] [Google Scholar]
- Uzunova VV, Quareshy M, Del Genio CI, Napier RM. 2016. Tomographic docking suggests the mechanism of auxin receptor TIR1 selectivity. Open Biol 6: 160139. 10.1098/rsob.160139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Woude LC, Perrella G, Snoek BL, van Hoogdalem M, Novák O, van Verk MC, van Kooten HN, Zorn LE, Tonckens R, Dongus JA, et al. 2019. HISTONE DEACETYLASE 9 stimulates auxin-dependent thermomorphogenesis in Arabidopsis thaliana by mediating H2A.Z depletion. Proc Natl Acad Sci 116: 25343–25354. 10.1073/pnas.1911694116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Overbeek J. 1939. Is auxin produced in roots? Proc Natl Acad Sci 25: 245–248. 10.1073/pnas.25.5.245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Raalte MH. 1936. On the influence of glucose on auxin production by the root tip of Vicia faba. Proc K Akad Wetensch 39: 261–265. [Google Scholar]
- Wabnik K, Robert HS, Smith RS, Friml J. 2013. Modeling framework for the establishment of the apical-basal embryonic axis in plants. Curr Biol 23: 2513–2518. 10.1016/j.cub.2013.10.038 [DOI] [PubMed] [Google Scholar]
- Wang B, Chu J, Yu T, Xu Q, Sun X, Yuan J, Xiong G, Wang G, Wang Y, Li J. 2015. Tryptophan-independent auxin biosynthesis contributes to early embryogenesis in Arabidopsis. Proc Natl Acad Sci 112: 4821–4826. 10.1073/pnas.1503998112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Qin G, Cao M, Chen R, He Y, Yang L, Zeng Z, Yu Y, Gu Y, Xing W, et al. 2020. A phosphorylation-based switch controls TAA1-mediated auxin biosynthesis in plants. Nat Commun 11: 679. 10.1038/s41467-020-14395-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westfall CS, Sherp AM, Zubieta C, Alvarez S, Schraft E, Marcellin R, Ramirez L, Jez JM. 2016. Arabidopsis thaliana GH3.5 acyl acid amido synthetase mediates metabolic crosstalk in auxin and salicylic acid homeostasis. Proc Natl Acad Sci 113: 13917–13922. 10.1073/pnas.1612635113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won C, Shen X, Mashiguchi K, Zheng Z, Dai X, Cheng Y, Kasahara H, Kamiya Y, Chory J, Zhao Y. 2011. Conversion of tryptophan to indole-3-acetic acid by TRYPTOPHAN AMINOTRANSFERASES OF ARABIDOPSIS and YUCCAs in Arabidopsis. Proc Natl Acad Sci 108: 18518–18523. 10.1073/pnas.1108436108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright AD, Sampson MB, Neuffer MG, Michalczuk L, Slovin JP, Cohen JD. 1991. Indole-3-acetic acid biosynthesis in the mutant maize orange pericarp, a tryptophan auxotroph. Science 254: 998–1000. 10.1126/science.254.5034.998 [DOI] [PubMed] [Google Scholar]
- Xu Y, Prunet N, Gan ES, Wang Y, Stewart D, Wellmer F, Huang J, Yamaguchi N, Tatsumi Y, Kojima M, et al. 2018. SUPERMAN regulates floral whorl boundaries through control of auxin biosynthesis. EMBO J 37: e97499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada M, Greenham K, Prigge MJ, Jensen PJ, Estelle M. 2009. The TRANSPORT INHIBITOR RESPONSE2 gene is required for auxin synthesis and diverse aspects of plant development. Plant Physiol 151: 168–179. 10.1104/pp.109.138859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi N, Huang J, Tatsumi Y, Abe M, Sugano SS, Kojima M, Takebayashi Y, Kiba T, Yokoyama R, Nishitani K, et al. 2018. Chromatin-mediated feed-forward auxin biosynthesis in floral meristem determinacy. Nat Commun 9: 5290. 10.1038/s41467-018-07763-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y, Kamiya N, Morinaka Y, Matsuoka M, Sazuka T. 2007. Auxin biosynthesis by the YUCCA genes in rice. Plant Physiol 143: 1362–1371. 10.1104/pp.106.091561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z, Liu X, Ljung K, Li S, Zhao W, Yang F, Wang M, Tao Y. 2017. Type B response regulators act as central integrators in transcriptional control of the auxin biosynthesis enzyme TAA1. Plant Physiol 175: 1438–1454. 10.1104/pp.17.00878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Xu R, Ma CJ, Vlot AC, Klessig DF, Pichersky E. 2008. Inactive methyl indole-3-acetic acid ester can be hydrolyzed and activated by several esterases belonging to the AtMES esterase family of Arabidopsis. Plant Physiol 147: 1034–1045. 10.1104/pp.108.118224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa T, Ito M, Sumikura T, Nakayama A, Nishimura T, Kitano H, Yamaguchi I, Koshiba T, Hibara K, Nagato Y, et al. 2014. The rice FISH BONE gene encodes a tryptophan aminotransferase, which affects pleiotropic auxin-related processes. Plant J 78: 927–936. 10.1111/tpj.12517 [DOI] [PubMed] [Google Scholar]
- Yue J, Hu X, Huang J. 2014. Origin of plant auxin biosynthesis. Trends Plant Sci 19: 764–770. 10.1016/j.tplants.2014.07.004 [DOI] [PubMed] [Google Scholar]
- Záveská Drábková L, Dobrev PI, Motyka V. 2015. Phytohormone profiling across the Bryophytes. PLoS ONE 10: e0125411. 10.1371/journal.pone.0125411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Li Q, Li Z, Staswick PE, Wang M, Zhu Y, He Z. 2007. Dual regulation role of GH3.5 in salicylic acid and auxin signaling during Arabidopsis–Pseudomonas syringae interaction. Plant Physiol 145: 450–464. 10.1104/pp.107.106021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Wang B, Ouyang J, Li J, Wang Y. 2008. Arabidopsis indole synthase, a homolog of tryptophan synthase α, is an enzyme involved in the Trp-independent indole-containing metabolite biosynthesis. J Integra Plant Biol 50: 1070–1077. 10.1111/j.1744-7909.2008.00729.x [DOI] [PubMed] [Google Scholar]
- Zhang SW, Li CH, Cao J, Zhang YC, Zhang SQ, Xia YF, Sun DY, Sun Y. 2009. Altered architecture and enhanced drought tolerance in rice via the down-regulation of indole-3-acetic acid by TLD1/OsGH3.13 activation. Plant Physiol 151: 1889–1901. 10.1104/pp.109.146803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Lin JE, Harris C, Campos Mastrotti Pereira F, Wu F, Blakeslee JJ, Peer WA. 2016. DAO1 catalyzes temporal and tissue-specific oxidative inactivation of auxin in Arabidopsis thaliana. Proc Natl Acad Sci 113: 11010–11015. 10.1073/pnas.1604769113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Lang Z, Zhu JK. 2018. Dynamics and function of DNA methylation in plants. Nat Rev Mol Cell Biol 19: 489–506. 10.1038/s41580-018-0016-z [DOI] [PubMed] [Google Scholar]
- Zhao Y. 2018. Essential roles of local auxin biosynthesis in plant development and in adaptation to environmental changes. Annu Rev Plant Biol 69: 417–435. 10.1146/annurev-arplant-042817-040226 [DOI] [PubMed] [Google Scholar]
- Zhao CY, Xue HW. 2020. PI4Kγ2 interacts with E3 ligase MIEL1 to regulate auxin metabolism and root development. Plant Physiol 184: 933–944. 10.1104/pp.20.00799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Hull AK, Gupta NR, Goss KA, Alonso J, Ecker JR, Normanly J, Chory J, Celenza JL. 2002. Trp-dependent auxin biosynthesis in Arabidopsis: involvement of cytochrome P450s CYP79B2 and CYP79B3. Genes Dev 16: 3100–3112. 10.1101/gad.1035402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Zhang Y, Liu X, Zhang X, Liu S, Yu X, Ren Y, Zheng X, Zhou K, Jiang L, et al. 2013. A role for a dioxygenase in auxin metabolism and reproductive development in rice. Dev Cell 27: 113–122. 10.1016/j.devcel.2013.09.005 [DOI] [PubMed] [Google Scholar]
- Zheng Z, Guo Y, Novák O, Dai X, Zhao Y, Ljung K, Noel JP, Chory J. 2013. Coordination of auxin and ethylene biosynthesis by the aminotransferase VAS1. Nat Chem Biol 9: 244–246. 10.1038/nchembio.1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z, Guo Y, Novák O, Chen W, Ljung K, Noel JP, Chory J. 2016. Local auxin metabolism regulates environment-induced hypocotyl elongation. Nat Plants 2: 16025. 10.1038/nplants.2016.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Žižková E, Kubeš M, Dobrev PI, Přibyl P, Šimura J, Zahajská L, Záveská Drábková L, Novák O, Motyka V. 2017. Control of cytokinin and auxin homeostasis in cyanobacteria and algae. Ann Bot 119: 151–166. 10.1093/aob/mcw194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolman BK, Martinez N, Millius A, Adham AR, Bartel B. 2008. Identification and characterization of Arabidopsis indole-3-butyric acid response mutants defective in novel peroxisomal enzymes. Genetics 180: 237–251. 10.1534/genetics.108.090399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubieta C, Ross JR, Koscheski P, Yang Y, Pichersky E, Noel JP. 2003. Structural basis for substrate recognition in the salicylic acid carboxyl methyltransferase family. Plant Cell 15: 1704–1716. 10.1105/tpc.014548 [DOI] [PMC free article] [PubMed] [Google Scholar]