Abstract
Influenza A viruses (IAVs) are the causative agents of one of the most important viral respiratory diseases in pigs and humans. Human and swine IAV are prone to interspecies transmission, leading to regular incursions from human to pig and vice versa. This bidirectional transmission of IAV has heavily influenced the evolutionary history of IAV in both species. Transmission of distinct human seasonal lineages to pigs, followed by sustained within-host transmission and rapid adaptation and evolution, represent a considerable challenge for pig health and production. Consequently, although only subtypes of H1N1, H1N2, and H3N2 are endemic in swine around the world, extensive diversity can be found in the hemagglutinin (HA) and neuraminidase (NA) genes, as well as the remaining six genes. We review the complicated global epidemiology of IAV in swine and the inextricably entangled implications for public health and influenza pandemic planning.
Host-adapted influenza A viruses (IAVs) are the cause of one of the most important viral respiratory diseases in animals and humans. Repeated outbreaks and rapid spread of genetically and antigenically distinct IAVs represent a considerable challenge for swine production. Zoonotic IAV was recently ranked the number one priority during a One Health workshop for disease prioritization in the United States (https://www.cdc.gov/onehealth/pdfs/us-ohzdp-report-508.pdf), with swine being one of the animal hosts posing the greatest risk for zoonotic IAV. IAV is a primary example of an infectious disease challenge for both human and animal health, and swine IAV requires One Health strategies to appropriately respond to zoonotic infections of swine IAV in humans (“variant” IAV), to develop pandemic prevention plans, and to minimize reverse zoonoses of human seasonal IAV transmission to swine (Kasowski et al. 2011).
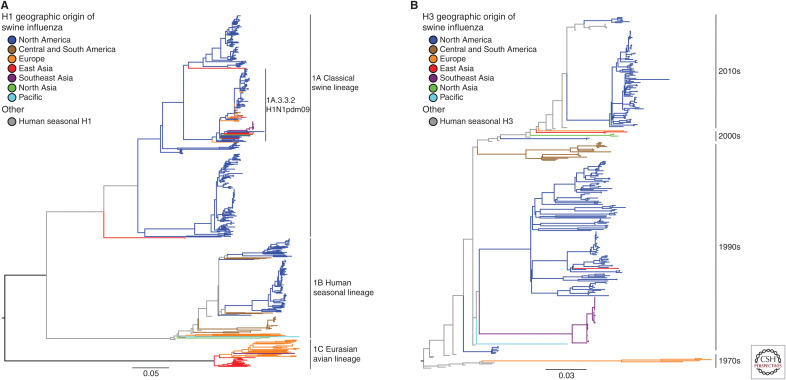
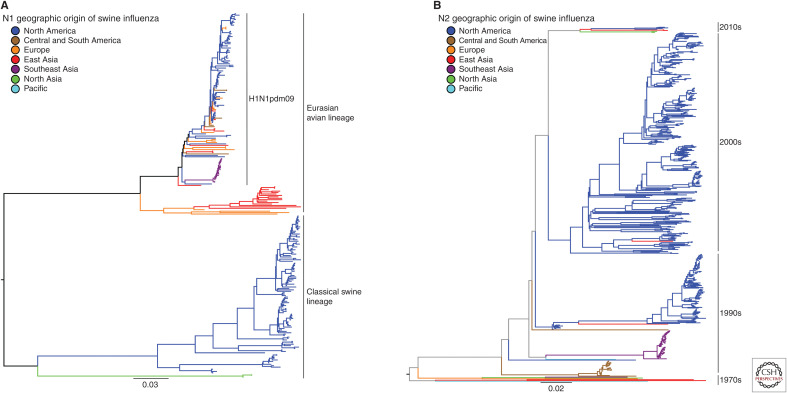
Viruses of the H1N1, H1N2, and H3N2 subtypes are endemic in swine around the world, and, despite only three circulating subtypes, the hemagglutinin (HA) and neuraminidase (NA) genes show tremendous diversity. Much of the observed IAV diversity is the result of two-way transmission between swine and humans (Ma et al. 2009; Nelson et al. 2012; Lewis et al. 2016; Rajao et al. 2018a), followed by antigenic drift and shift within swine host populations, and viral diffusion through live animal transport and trade. These complex evolutionary processes were highlighted by the emergence of the triple reassortant H3N2 viruses in the United States and subsequent maintenance of the triple reassortant internal gene (TRIG) constellation (Vincent et al. 2008). These dynamics contributed to the generation of numerous genetically and antigenically distinct lineages cocirculating in swine around the world. Then, the repercussions of these evolutionary events were starkly shown with the swine-origin pandemic in 2009 (Mena et al. 2016), the subsequent impact of the continued 2009 pandemic H1N1 (H1N1pdm09) introductions into pig populations (Nelson et al. 2015c), and ongoing detections of contemporaneous human seasonal H3N2 spillovers into swine (Ngo et al. 2012; Rajão et al. 2015; Krog et al. 2017; Wong et al. 2018; Zeller et al. 2018a). The genetic diversity of H1 and H3 viruses in swine is summarized in Figure 1A,B and the N1 and N2 neuraminidase genes in swine are summarized in Figure 2A,B.
Figure 1.
Phylogeny of H1 and H3 influenza A virus hemagglutinin genetic lineages of contemporary influenza A virus in global swine. The best-known tree was inferred using maximum likelihood methods for 637 H1 swine hemagglutinin gene sequences showing the three major lineages collected globally from 2016 to September 2019, and from the United States from March 2019 to September 2019: 1A classical swine lineage; 1B human seasonal lineage; and 1C Eurasian avian lineage (A). For the H3 HA, there were 335 swine HA genes collected globally from 2016 to September 2019 and from the United States from March 2019 to September 2019 (B). “Other” designations in H1 and H3 represent reference human or swine HA genes that have yet to be classified within the global nomenclature system, or lack current evidence of sustained transmission. Branch color represents geographic groups and include North America with data from Canada, Mexico, and United States; Central and South America with data from Argentina, Brazil, and Chile; Europe with data from Belgium, Czech Republic, Denmark, England, France, Germany, Italy, and Spain; East Asia with data from China, Japan, and South Korea; Southeast Asia with data from Thailand and Vietnam; North Asia with data from Russia; and the Pacific with data from Australia. Each tree is midpoint-rooted for clarity; all branch lengths are drawn to scale; and the scale bar indicates the number of nucleotide substitutions per site.
Figure 2.
The major N1 and N2 neuraminidase genetic lineages and geographic distribution of contemporary influenza A virus in global swine. (A) The best-known tree was inferred using maximum likelihood methods for 255 swine N1 NA sequences collected globally from 2016 to September 2019 and from the United States from March 2019 to September 2019; and (B) 552 swine N2 NA sequences collected globally from 2016 to September 2019, and from the United States from March 2019 to September 2019. Branch color represents geographic groups and include North America with data from Canada and United States; Central and South America with data from Argentina, Brazil, and Chile; Europe with data from Czech Republic, France, Germany, Italy, and Spain; East Asia with data from China, Japan, and South Korea; Southeast Asia with data from Thailand and Vietnam; North Asia with data from Russia; and the Pacific with data from Australia. The lineage of N1 NA is indicated by solid black lines on the right of the tree in A. The decade of introduction of human seasonal N2 NA gene into swine populations is indicated by solid black lines on the right of the tree in B (e.g., the 1970s includes the introduction in Europe, the 2000s includes the 2002 lineage in North American swine, etc.); the trees are midpoint-rooted for clarity; branch lengths are drawn to scale; and the scale bars indicate the number of nucleotide substitutions per site.
Human seasonal IAV adaptation to swine is likely situational to each event (Rajao et al. 2018b). Although species barriers exist between human and swine IAV, swine-adapted H1 and H3 IAV maintain receptor binding site (RBS) profiles for α2,6 sialic acid-binding preference, in contrast to IAV maintained in waterfowl and domestic poultry with α2,3 sialic acid-binding preference. However, human seasonal H3N2 and pre-2009 H1N1 viruses that spilled into swine underwent dramatic changes in the process of adaptation, including loss of one or several glycosylation sites of the HA, multiple amino acid substitutions across the genome, and frequent reassortment with endemic swine IAV, including the acquisition of different NA and internal gene segment lineages (Busch et al. 2008; Rajão et al. 2015). These adaptive changes quickly separate the emerging swine lineage from the seeding human seasonal virus. The exceptions are human H1N1pdm09 viruses that spill into swine, usually maintaining all eight gene segments of the H1N1pdm09 without acquiring swine lineage gene segments or adaptation mutations required for infection and transmission in swine (Nelson et al. 2015c; Gao et al. 2017; Chastagner et al. 2018). This is not the case with endemic swine strains that readily reassort with the H1N1pdm09, acquiring multiple gene segments, most notably the H1N1pdm09 M gene, with concomitant evolution of the swine HA and NA proteins (Liang et al. 2014; Watson et al. 2015; Rajão et al. 2017; Takemae et al. 2017).
The extraordinary genetic and antigenic diversity of H1 and H3 swine IAV is arguably the greatest challenge to controlling infection and to the development of broadly effective vaccines. Likewise, antigenically variable swine viruses pose a threat to humans if human population immunity no longer recognizes the resultant swine lineages. This diversity and the human–swine interface are incredibly important in the context of variant IAV infections in humans. Dramatic increases in the detection of variant IAV infections began with H3N2 viruses in the United States in 2012 (Epperson et al. 2013), and these viruses are regularly detected in the United States (Jhung et al. 2013; Choi et al. 2015; Greenbaum et al. 2015; Duwell et al. 2018; Pulit-Penaloza et al. 2018a) and to a lesser degree in other countries (Resende et al. 2017; Xie et al. 2018; Lu et al. 2019). The risk of variant infection is likely dependent on animal production systems, animal–human interfaces (e.g., live animal markets, exhibition practices), the ecology of the virus, and other less tangible factors (Karesh et al. 2012).
CLINICAL ASPECTS OF INFLUENZA IN PIGS
Swine influenza was historically characterized as a seasonal respiratory disease, primarily in weaned pigs with waning maternal immunity. Today, clinical disease in the United States still peaks during times of the year associated with fluctuations in temperature and decreased ventilation, similar to the human influenza season (Janke 2013). In U.S. pigs, a primary peak is typically observed November–December with a secondary spike March–April (Anderson et al. 2013; Walia et al. 2019), with a similar trend in Canada (Poljak et al. 2014). However, contemporary influenza illness and diagnosis can be found at any time of the year (https://influenza.cvm.iastate.edu; Zeller et al. 2018a), in nearly all age groups of pigs, even suckling pigs from sows with high titers of influenza-specific serum antibodies (Allerson et al. 2013; Corzo et al. 2013). The detection of virus in sow farms and suckling pigs is likely due to introduction of antigenically distinct strains to a herd, as well as variability in populations with mixed levels and specificity of immunity that promotes maintenance of endemic strains.
Influenza vaccines are primarily used in adult sows to protect the gestating sow and her suckling piglets or during the grow/finish phase of production to decrease IAV disease, lung lesions, and transmission (Beaudoin et al. 2012; Van Reeth and Ma 2013; Vincent et al. 2017). Vaccinating piglets may be desired in some clinical situations, but the presence of maternal antibodies interferes with vaccine efficacy of inactivated vaccines. Whole inactivated virus (WIV) with adjuvant, live-attenuated influenza virus (LAIV), and an alphavirus vectored RNA replicon particle (RP) vaccine (Vander Veen et al. 2012, 2013) are licensed in the United States and available commercially for use in swine. WIV and RP can also be formulated with autogenous or custom farm-based strains. The production of “off-the-shelf” commercially available efficacious WIV vaccines is difficult because of the number of strains in the vaccine required to immunize against all antigenically distinct circulating IAV strains in swine (Rajao et al. 2018a; Bolton et al. 2019). Formulating effective vaccines is further challenged by the difficulty in updating vaccine seed viruses given the emergence of novel lineages, antigenic drift, the time needed to approve and license veterinary vaccine products, maternal antibody interference, and the lack of adequate mucosal and cell-mediated immune responses (Kitikoon et al. 2006; Wesley and Lager 2006; Platt et al. 2011). Further contributing to the observed evolution and subsequent antigenic diversity, positive selection at antibody epitopes has been shown in swine HA genes and may be associated with partially effective vaccines (Kitikoon et al. 2013).
HISTORICAL EVOLUTION OF SUSTAINED LINEAGES OF IAV IN SWINE GLOBALLY
1A
A phylogenetically informed swine H1 nomenclature system was developed to define the three major cocirculating lineages, 1A, 1B, and 1C (Fig. 1A; Anderson et al. 2016). The 1A swine lineage was derived from the human Spanish influenza pandemic in 1918 (Koen 1919; Shope 1931). These viruses formed the classical swine H1N1 (cH1N1) lineage that spread globally, and the HA genes formerly named α-, β-, or γ-clades commonly found in North America (Vincent et al. 2009). 1A viruses were enzootic in swine populations throughout Asia, including Thailand and Japan (Takemae et al. 2008; Choi et al. 2013; Zhu et al. 2013). The 1A lineage includes the H1 HA of the H1N1pdm09, a virus with NA and M genes from the Eurasian avian H1N1 swine lineage in addition to TRIG and classical swine lineage genes. Although an immediate precursor with all eight gene segments of the human H1N1pdm09 was not found in pigs before 2009, characterization of IAV isolated from pigs in Mexico between 2010 and 2014 revealed the presence of multiple genotypes containing segments that shared a common ancestor with the H1N1pdm09 virus (Mena et al. 2016). Following the rapid spread of H1N1pdm09 in humans, this virus was repeatedly re-introduced into swine populations globally, with a dominant contribution being the reassortment of its internal genes with existing endemic H1 and H3 swine viruses. The 1A HA clades are found paired with an N1 NA gene from the classical swine lineage or H1N1pdm09 lineage (Fig. 2A; Anderson et al. 2015; Walia et al. 2019) or N2 NA genes derived from the 1998 or 2002 human seasonal lineages (Fig. 2B; Nelson et al. 2011).
1B
In the United Kingdom, an H1N2 virus was described in the 1990s that contained the HA gene of a human seasonal H1N1 virus, now classified as 1B.1, with the remainder of the genes from the predominant circulating H3N2 swine IAV. Human seasonal H1 IAV also spilled into U.S. swine in the early 2000s and acquired TRIG genes similar to those found in contemporary triple-reassortant viruses (Vincent et al. 2009). These H1 HAs were genetically and antigenically distinct from those of classical swine lineage H1 viruses and were classified as 1B.2.2 and 1B.2.1 (δ-1 and δ-2, respectively) (Lorusso et al. 2011). Argentina also reported distinct human seasonal 1B.2 viruses (Pereda et al. 2011). In Brazil, H1N2 viruses with H1 HA and N2 NA genes of human seasonal origin and internal genes from H1N1pdm09 were similarly detected (Biondo et al. 2014), followed by detection of an additional lineage of H1N2 reassortants with the H1N1pdm09 internal genes and human seasonal surface genes (Nelson et al. 2015b). Chile reported two human seasonal lineage H1 viruses paired with N1 or N2 NAs that are each distinct from those found in other South American countries (Nelson et al. 2015a). A more recent introduction of a human seasonal H1N1 from the 2000s was detected in Vietnam (Baudon et al. 2018). In Australia, H1 viruses likely derived from human seasonal H1 viruses from the 1970s and 1990s and distinct from other global swine IAV went undetected for many years in swine (Wong et al. 2018). The 1B.2 HA clades in the United States are found paired with N2 NA genes derived from the 1998 or 2002 human seasonal origin (Nelson et al. 2011; Anderson et al. 2015; Walia et al. 2019), whereas the European 1B.1 H1 HAs are primarily paired with a different human seasonal N2 NA introduced in the 2000s (Fig. 2B; Watson et al. 2015).
1C
In the 1970s, a lineage derived from an avian H1N1 virus was detected in Europe, spread to Asia, and became known as the Eurasian avian (EA) 1C lineage. The 1C H1N1 lineage (in Europe, colloquially referred to as H1avN1) remained a dominant lineage in Eurasia, and components of its internal gene constellation played an important role in the modern ecology of IAV. IAV with the HA and NA of the 1C H1N1 lineage were also reported in Mexico (Mena et al. 2016). The NA paired with 1C.1 and 1C.2 HA is most often the N1 of avian origin maintained from the initial introduction (Fig. 2A) or N2 acquired from 1980s and 2000s human seasonal introductions (Fig. 2B; Watson et al. 2015).
H3
Swine H3N2 are best defined by the time period of circulation of the putative ancestral human seasonal virus and can thus be classified by decade of introduction and contemporary strains given a numerical clade definition (Fig. 1B). The most recent common ancestor of contemporary H3N2 viruses in European swine populations was a 1970s human lineage H3N2 virus (Van Reeth et al. 2012). These H3N2 viruses reassorted with the 1C H1avN1 swine IAV in the 1980s, acquiring its internal gene cassette (Castrucci et al. 1993). In the 1990s, a triple-reassortant H3N2 virus was identified containing HA, NA, and PB1 gene segments derived from human seasonal H3N2, the PB2 and PA gene segments derived from avian IAV, and the NP, M, and NS gene segments from the cH1N1 swine IAV (Zhou et al. 1999). These H3N2 viruses persisted and evolved over time into defined phylogenetic clades, with “Cluster-IV” H3 (C-IV, 1990.4) sustaining and expanding in diversity to the present day (Walia et al. 2019), and with the TRIG constellation persisting in both H3N2 and reassorted H1 viruses (Vincent et al. 2008). H3N2 viruses of two lineages were also reported in Mexico, one a likely importation of C-IV H3 from North America, and the second a unique human seasonal introduction in the 1990s, 1990.2 (Nelson et al. 2015a). A further 1990s introduction was reported in Brazil, 1990.5 (Nelson et al. 2015b).
A reassorted H3N2 with an H3 HA from the early 2000s human seasonal H3N2 viruses with internal genes from the H1N1pdm09 virus was reported in Argentina (Dibárbora et al. 2013). An introduction of a human seasonal H3N2 virus in the late 2000s occurred in Vietnam (Ngo et al. 2012), with the lineage subsequently detected in China (He et al. 2018). Also, in the 2000s, a human seasonal H3 HA emerged in Danish pigs, reassorted with a swine N2 NA and internal genes from the H1N1pdm09 virus, 2000.3 (Krog et al. 2017). Then, during the 2010–2011 human influenza season, a distinct human seasonal H3N2 virus transmitted to U.S. swine and sustained onward transmission (Rajão et al. 2015). This more contemporary H3N2 lineage, 2010.1, was genetically and antigenically distinct from the 1998 H3N2 lineage C-IV viruses in the United States and quickly became dominant, but did not replace the C-IV H3 lineage in U.S. swine (Zeller et al. 2018a). During the same decade, a second distinct human seasonal H3N2 virus transmitted to U.S. swine, 2010.2, and continues to be detected at low levels (Zeller et al. 2018b).
Diversity of swine IAV in Chinese, South Korean, Vietnamese, and other Asian swine herds was shaped by the intercontinental movement of swine (Vijaykrishna et al. 2011; Nelson et al. 2015d). European H3N2 viruses, along with North American TRIG viruses, were introduced in the late 1990s to early 2000s and continue to be detected. In Thailand, swine H3N2 viruses contained human seasonal HA and NA genes similar to those of European H3N2 viruses, as well as a unique human seasonal H3N2 virus introduced in the 1990s, 1990.3. In Australia, an H3 lineage derived from spillover of human seasonal viruses from the 1990s, but distinct from other global swine IAV, was detected (Wong et al. 2018). Swine H3 HAs are almost exclusively paired with N2 NA lineages of human seasonal virus origin, frequently from the same initial human seasonal spillover event (Fig. 2B).
CURRENT STATUS OF IAV IN SWINE
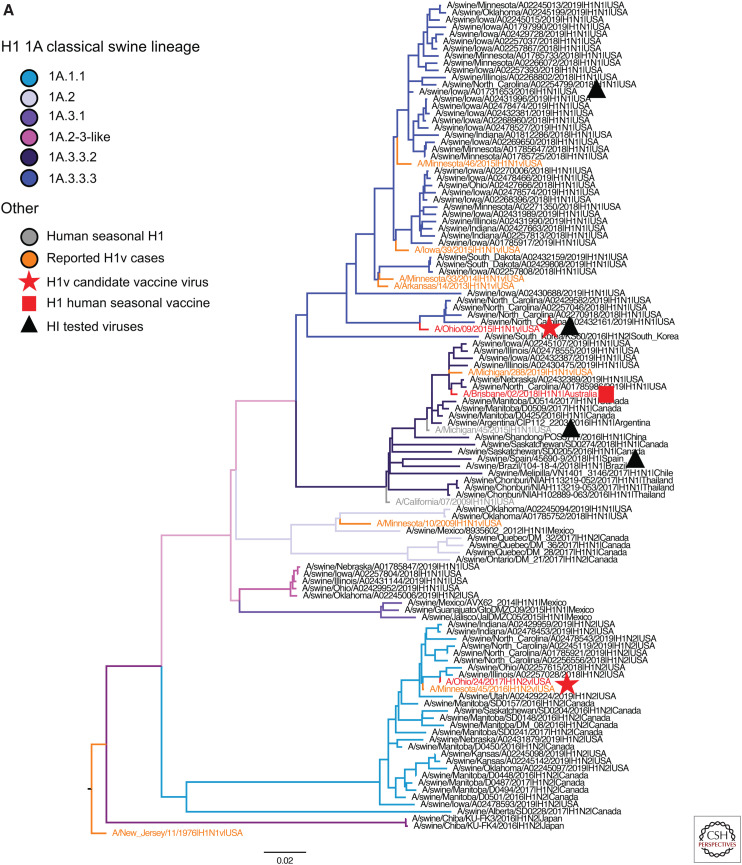
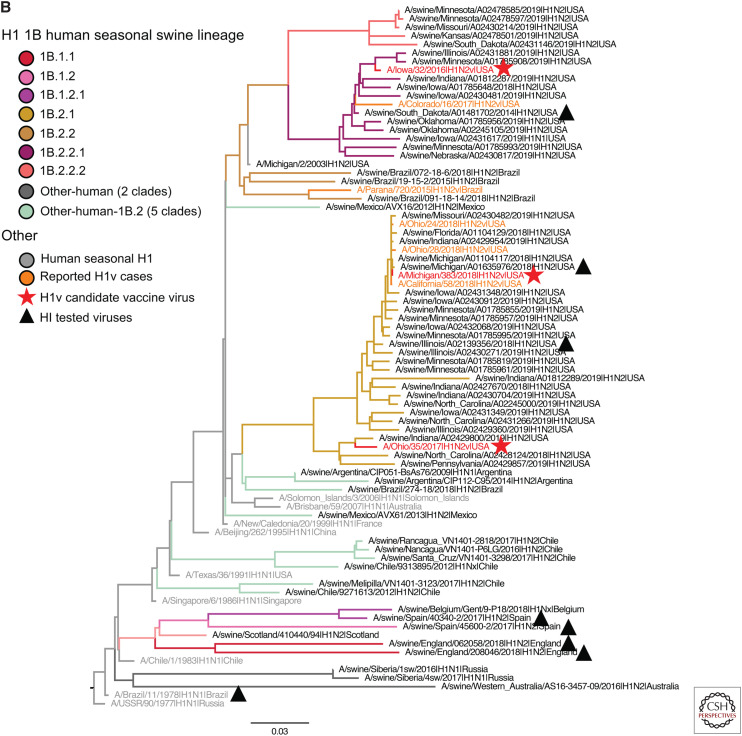
To quantify the currently circulating diversity of swine IAV, we collated H1N1, H1N2, and H3N2 influenza A virus HA sequences. Analyses were restricted to IAV genes detected from March 2019 to September 2019 in the United States because systematic surveillance adequately captured genetic diversity within this time frame. In contrast, surveillance for IAV in swine outside of the United States is limited; consequently, detections from 2016 to 2019 were considered contemporary as these data better captured the diversity of swine IAV circulating in these populations. The swine IAV data and World Health Organization (WHO)-recommended human seasonal H1 and H3 HA vaccine sequences and reported variant sequences were downloaded from GISAID (Global Initiative on Sharing All Influenza Data) (Shu and McCauley 2017) and the Influenza Research Database (Zhang et al. 2017). Unpublished strains from undersurveilled regions were made available through the joint World Organization for Animal Health–Food and Agriculture Organization (OIE-FAO) influenza (OFFLU) swine network and were included in analyses when that data represented unique genetic clades. The sequences for each subtype were aligned, and the best-known maximum-likelihood phylogeny for each alignment was inferred using IQ-TREE v1.6.10 (Nguyen et al. 2015). To better visualize the evolutionary relatedness of the swine genetic data to human vaccine strains and candidate vaccine viruses, we proportionally subsampled the genetic clades from Figure 1 by subtype and lineage, ensuring that all unique genetic clades were maintained, and present these data in Figure 3.
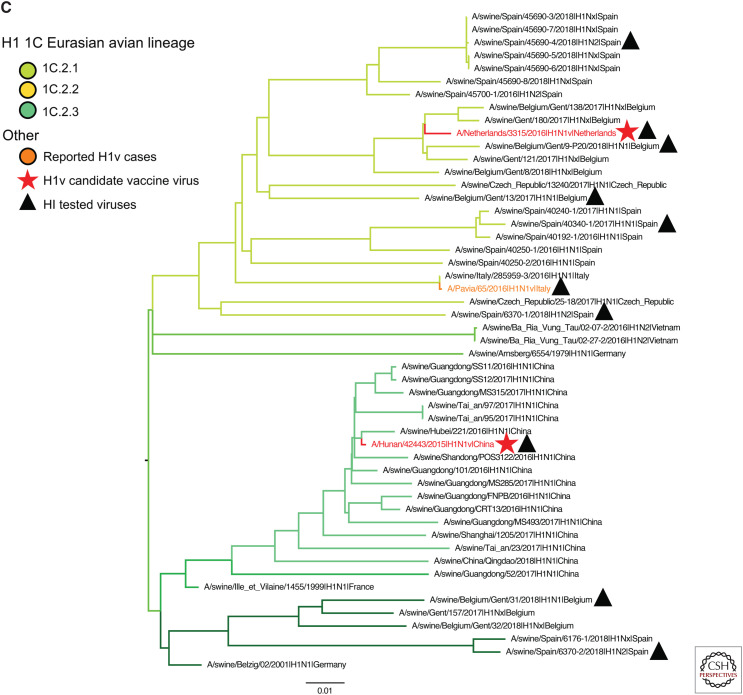
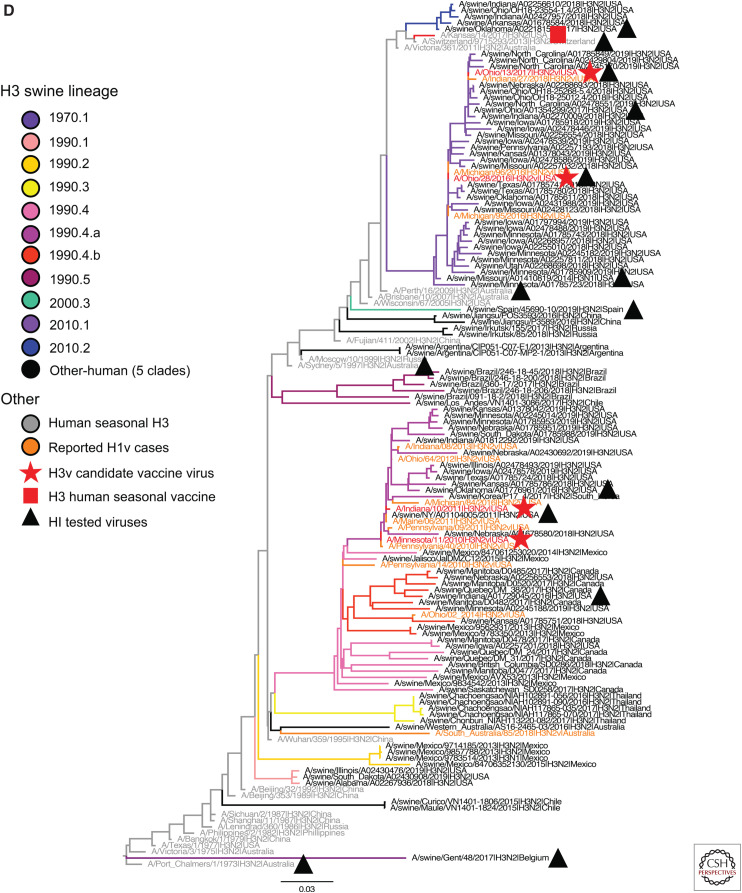
Figure 3.
H1 and H3 influenza A virus hemagglutinin phylogenetic lineages of contemporary influenza A virus in global swine. The trees were inferred using maximum likelihood methods for randomly subsampled HA gene sequences, maintaining proportional representation of swine clades detected in the United States from March 2019 to September 2019 and from remaining global countries from 2016 to September 2019. (A) The 1A classical swine lineage; (B) 1B human seasonal lineage; (C) 1C Eurasian avian lineage; and (D) H3 swine lineages. Branch color represents swine genetic clade; reported human variant strain names are colored in orange; candidate vaccine virus strains are indicated by a red star; H1 and H3 human seasonal vaccine strains by a red square; and strains tested by hemagglutination inhibition assays as antigen or antisera by a black triangle. The trees are midpoint-rooted for clarity; branch lengths are drawn to scale; and the scale bars indicate the number of nucleotide substitutions per site.
The 1A lineage contained 637 viruses from six genetic clades detected in 11 countries (Fig. 3A). 1A.1.1 viruses, similar to α-H1 (n = 153, United States, Canada, and Japan); 1A.2 viruses, similar to β-H1, (n = 13, United States and Canada); 1A.3 viruses, similar to γ-H1, (n = 230, 10 countries); and 1A.3.4 (n = 8, United States). The only clade within the 1A lineage with wide global distribution was the H1N1pdm09 clade (1A.3.3.2) with 134 viruses in 12 countries (Argentina, Canada, Chile, China, France, Japan, Mexico, Russia, Spain, South Korea, Thailand, and the United States). The remaining clades were more limited in geographic spread, potentially reflecting the dissemination of viruses with specific agricultural trade routes (Nelson et al. 2015d). For example, the 1A.1.1 clade was detected in Canada and the United States (n = 82); the 1A.3.2 clade was detected in Mexico and the United States (γ-2 H1); and the 1A.3.3.3 viruses were isolated in South Korea and the United States (γ-H1). Two clades were restricted to single countries, 1A.3.1 in Mexico (n = 3), and 1A.2-3-like (n = 8) in the United States.
The 1B lineage contained 201 viruses from 13 genetic clades collected in 11 countries (Fig. 3B). The 1B.1 viruses derived from the H1N2 viruses first detected in Great Britain in the 1990s were sparsely represented (n = 5). The three clades, detected in England, Spain, and Belgium, showed long branch lengths, suggesting additional diversity not captured by current surveillance efforts. The 1B.2 viruses (δ-1a, δ-1b, and δ-2; n = 126, United States) were the majority of detected 1B strains. In addition to the named 1B clades, the 1B.2 clade since 2016 contains viruses from additional human-to-swine transmission episodes (n = 7) in Argentina, Brazil, Chile, Mexico, Australia, and Russia. Although infrequently detected, they are likely circulating in these regional swine populations because of periodic evidence of now-extinct human seasonal HA genes being maintained in swine (classified as “Other-human” or “Other-human-1B.2”).
The 1C lineage consisted of 49 viruses from three genetic clades collected in seven countries in Europe and Asia. Notably, these viruses were not detected in the United States or Canada (Fig. 3C). During this time period, we only identified clade 1C.2 viruses in continental Europe and Asia. Within the 1C.2 clade, 3 third-order divisions were detected: 1C.2.1 (n = 26) in Belgium, Czech Republic, Italy, Spain, and Vietnam; 1C.2.2 (n = 6) in Belgium, Germany, and Spain; and 1C.2.3 (n = 16) in China.
The H3 lineage contained 309 viruses from eight genetic clades collected in 11 countries (Fig. 3D). The major contemporary H3 lineages circulating in swine truly reflect the tangled nature of IAV in swine and humans; with the exception of the 1980s, there were swine lineages from each decade from 1970 to present continually circulating in global swine populations. In contrast to the H1 1A lineage, the majority of these H3 lineages are geographically restricted—that is, the 2010.1 (n = 92) and 2010.2 (n = 6) clades only circulate in the United States, the 1970.1 (n = 4, related to A/Port Chalmers/1/1973) and 2000.3 (n = 1) clades were only detected in Europe (France, Spain, and Denmark), the 1990.5 clade was only detected in Brazil (n = 16) and Russia (n = 1), and the 1990.3 clade was only detected in Thailand (n = 45). The exception to this is the 1990s “Cluster IV” H3N2 (1990.4, n = 129) that was detected in the United States, Canada, Mexico, and Korea. A unique clade H3.1990.1 (n = 8) was only found in the United States, and there were separate detections of Other-human-2000s in Canada (n = 2), China (n = 2), and Russia (n = 3).
The level of surveillance in swine increased globally in the years following the emergence of the H1N1pdm09 and revealed the presence of swine IAV in regions not previously known to have endemic IAV or the presence of novel gene lineages not previously recognized in swine (Anderson et al. 2013; Simon et al. 2014; Vincent et al. 2014). However, funding for surveillance in pigs has fallen victim to short-term memory, budget cuts, and changes in disease priorities for swine health. Although surveillance activity in swine remains high in the United States, data from 2019 was extremely sparse from the rest of the world. Despite these limitations, 30 genetic clades of H1 and H3 viruses were detected in the past 3 years.
RISK OF CONTEMPORARY SWINE IAV FOR HUMANS
Zoonotic transmission of swine IAV has been documented for several decades and generally results in an influenza-like illness similar to that of human seasonal IAV with limited onward human-to-human transmission. The most dramatic exception to this was the 2009 H1N1 pandemic. Novel IAVs that are capable of infecting humans are concerning, but those that are capable of human-to-human transmission and those for which human population immunity is lacking are of particular concern. Since 2010, 465 people in the United States were infected with variant IAVs of swine origin, with 430 of these detections being H3N2v, 25 H1N2v, and 10 H1N1v (https://www.cdc.gov/flu/weekly/fluviewinteractive.htm). The H3N2v cases were from two lineages of H3 that circulate in the United States. Before 2016, H3N2v infections were caused by viruses containing HA genes from the 1990.4 C-IV H3 lineage (Jhung et al. 2013; Greenbaum et al. 2015). Although originally derived from human seasonal H3 viruses circulating in the 1990s, these viruses are antigenically distinct from contemporary seasonal H3N2 in humans, prompting the development of candidate vaccine viruses (CVVs) targeting the 1990.4 C-IV viruses (https://www.who.int/influenza/resources/documents/2011_09_h5_h9_vaccinevirusupdate.pdf?ua=1). A substantial proportion of adolescents and young adults were shown to have cross-reactive antibodies against 2011–2012 1990.4 (C-IV) H3N2v viruses. However, children and older adults lacked such protective antibodies (https://www.ncbi.nlm.nih.gov/pubmed/22495226; Skowronski et al. 2012; Liu et al. 2017), and H3N2 seasonal vaccines did not appear to protect against the 1990.4 (C-IV) H3N2v virus (Skowronski et al. 2012; Houser et al. 2013). Consequently, people that never received a 1990s H3 vaccine and/or natural infection may lack cross-reactive antibodies and adult immunity to this swine lineage may have waned. Since 2016, the majority of H3N2v cases resulted from infection with H3.2010.1 HA lineage viruses (Bowman et al. 2017; Duwell et al. 2018). These viruses were poorly recognized by ferret antisera raised to circulating seasonal H3N2 vaccine viruses and the 1990.4 C-IV lineage CVVs, leading to development of H3.2010.1 lineage-specific CVVs (https://www.who.int/influenza/vaccines/virus/201609_zoonotic_vaccinevirusupdate.pdf?ua=1). Because the vast majority of cases of H3N2v of both lineages were in children with close contact to pigs and long exposure time at agricultural fairs, all of these factors point to a unique set of circumstances that collectively might have increased the odds for H3N2v infection in these spillover events. In addition to H3N2v virus infections reported in the United States, several other countries have reported sporadic human infection with H3N2v viruses, including Australia, Canada, China, and Vietnam (Gregory et al. 2001; Olsen et al. 2006; Robinson et al. 2007; Bastien et al. 2010; Freidl et al. 2014; Lu et al. 2019; https://www.who.int/influenza/vaccines/virus/201902_zoonotic_vaccinevirusupdate.pdf?ua=1). Whereas C-IV viruses from Canada were similar to strains detected in the United States, viruses from Asia and Australia, although genetically related to viruses detected in swine in these regions, were genetically and antigenically distinct from contemporary human strains.
Human infections with H1v were also documented in Asia, Europe, and the Americas. Perhaps the most notable was the 1976 swine-origin H1N1 (1A.1) outbreak on a U.S. Army base in New Jersey that led to a country-wide immunization campaign (Gaydos et al. 2006). Subsequently, sporadic H1v infections were reported in the United States and elsewhere that were predominantly caused by 1A.2 and 1A.3 H1 clade viruses (Freidl et al. 2014), as well as 1B.2 viruses. Still other H1v infections in Europe and Asia were caused by endemic swine 1C.2 viruses. Since reporting of novel IAV became nationally notifiable to public health authorities in the United States in 2007 (https://wwwn.cdc.gov/nndss/downloads.html), 48 human infections with H1v viruses have been reported in the United States. Whereas most were caused by 1A.3 viruses, 13 cases in 2018 resulted from 1B.2.1 viruses genetically related to IAVs detected in swine at agricultural fairs (Rambo-Martin et al. 2019). Reverse zoonoses of H1N1pdm09 viruses (1A.3.3.2) into swine also resulted in zoonotic transmission of swine-origin H1N1pdm09 back into humans (https://www.who.int/influenza/vaccines/virus/201909_zoonotic_vaccinevirusupdate.pdf?ua=1), further confounding the transmission directionality of these viruses as well as the diagnostic capacity to distinguish their source. Antigenically, the H1v represent a diverse subset of viruses given their ancestry to 1918 precursors (1A), pre-2009 seasonal H1N1 (1B), Eurasian avian lineage (1C), and most recently, H1N1pdm09 (1A.3.3.2). Whereas each of these H1v lineages share antigenic similarity with circulating swine viruses, all but the H1N1pdm09-like viruses have drifted antigenically from either their human seasonal or avian ancestor. Serological testing of adult and child populations vaccinated with seasonal influenza vaccine showed that, although many 1A.3 viruses remain neutralized by human antisera, the 1B and 1C HA lineage H1v viruses have very little to no cross-reactivity, indicating a lack of immunity even in the vaccinated adult population (https://www.who.int/influenza/vaccines/virus/201809_zoonotic_vaccinevirusupdate.pdf?ua=1).
The question remains as to why some swine IAV clades cause variant infections. Although there were increased detections of 1990.4 (C-IV) H3N2 viruses in swine similar to the H3N2v viruses in humans detected in 2011–2012, and increased 2010.1 H3N2 virus detections in swine similar to the H3N2v virus detections in humans in 2016–2017, this cannot be the sole explanation for the increase in H3N2v cases, because there were also increases in other genetically dissimilar H3, as well as in subsets of H1 viruses in the U.S. swine population in the same time periods (Anderson et al. 2013, 2015; Walia et al. 2019). There may be specific virus properties that conferred a better “fit” for the H3N2v to infect humans; in fact, H3N2v viruses from 2009–2011 and 2011–2016 were capable of airborne transmission in ferrets (Pearce et al. 2012; Sun et al. 2018), the standard animal model used to assess mammalian transmissibility of influenza viruses. However, human H1v viruses were also shown to successfully infect and transmit between ferrets (Pulit-Penaloza et al. 2018a,b, 2019a,b). Moreover, some swine IAV have been shown to infect and transmit among ferrets without evidence of human zoonosis (Yen et al. 2011; Zhu et al. 2011; Pascua et al. 2012, 2013). The ferret transmission model appears to be a rather liberal litmus test for indicating potential for human infection by swine IAV or an indication that many of the globally circulating swine virus lineages are capable of such infection. Clearly, other viral factors that permit swine to human zoonotic infection remain largely undefined. These complex factors do not diminish the epidemic or pandemic risk of swine-adapted IAV if the viruses gained the ability to transmit from human-to-human, allowing further opportunity to evolve and adapt to the human host. However, except for the H1N1pdm09 virus, sustained human-to-human transmission has not been observed for swine influenza viruses, despite the apparent propensity to establish an infection. A better understanding of how the process of adaptation to a new host shapes the subsequent establishment and transmission of viruses among and between humans and pigs is critical for breaking the cycle.
PANDEMIC PREPAREDNESS: CANDIDATE VACCINE VIRUS DEVELOPMENT AND ASSESSMENT
Without the ability to accurately predict which of the 30 current clades of swine IAV may cause human infections or even pandemics, human vaccine preparedness efforts for swine strains are difficult. These efforts occur biannually at the WHO Vaccine Composition Meeting, where animal influenza activity data are presented concurrently with human seasonal influenza activity data. If an animal IAV clade is detected at high levels relative to other clades, variant cases are identified, and/or genetic and antigenic diversity of that clade is significantly drifted from previously recommended pre-pandemic CVVs and current human seasonal vaccine strains, a representative animal strain may be considered for development of a new CVV. The CVVs are shared among the WHO Global Influenza Surveillance and Response Network (GISRS) and with academic, governmental, and industry partners for research or commercial development (Robertson et al. 2011). Before the 2009 pandemic, the HA genes of CVVs were exclusively of avian origin, but more recently, several swine variant viruses have been selected based on confirmed variant infections and antigenic divergence from other CVVs and seasonal vaccines. The cross-reactivities of newly detected viruses are tested against monovalent ferret antisera raised against CVVs and/or seasonal vaccine strains and, when available, pooled sera obtained from vaccinated humans.
To determine whether the current CVVs and human seasonal vaccine components adequately captured the currently circulating diversity of swine IAV, we compared the HA1 amino acid sequences against existing CVVs and/or nearest human seasonal vaccines when a clade-specific CVV was absent, and summarized the results in Table 1. For each of the 30 contemporary swine clades, a majority consensus HA1 sequence was generated. For clades without a CVV, the comparison was made against the 2019–2020 northern hemisphere influenza season vaccine components, H1 (A/Brisbane/02/2018) or H3 (A/Kansas/14/2017), or the comparison strain was the most genetically similar historical human vaccine component. A total number of amino acid substitutions and substitutions in putative epitopes were calculated using custom Python scripts (Gerloff et al. 2014).
Table 1.
Percentage identity of contemporary swine HA1 consensus to clade candidate vaccine virus or human seasonal vaccine strain
| Contemporary swine consensusa | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CVV or vaccine | 1A.1.1 | 1A.3.3.3 | 1A.3.3.2 | Misc. 1A | 1B.2.1 | 1B.2.2.1 | 1B.2.2.2 | Brazil 1B.2.2 | Brazil 1B.2 | Misc. 1B.1 | 1C.2.1 | 1C.2.2 | 1C.2.3 | H3.90.4a | H3.90.4b | Misc. H3.90s | H3.90.1 | H3.10.1 | H3.10.2 | Epitope changes | |
| 1A.1.1 | A/Ohio/24/2017 | 97% | Sa, Sb | ||||||||||||||||||
| 1A.3.3.3 | A/Ohio/09/2015 | 92% | Sa, Sb, Ca1, Ca2 | ||||||||||||||||||
| Hu-Vac (1A.3.3.2) | A/Brisbane/02/2018 | 98% | Sa | ||||||||||||||||||
| Hu-Vac (1A.3.3.2) | A/Brisbane/02/2018 | 86%–88% | Sa, Sb, Ca1, Ca2, Cb, RBS | ||||||||||||||||||
| 1B.2.1 | A/Michigan/383/2018 | 99% | None | ||||||||||||||||||
| 1B.2.2.1 | A/Iowa/32/2016 | 99% | Ca2 | ||||||||||||||||||
| 1B.2.2.1 | A/Iowa/32/2016 | 91% | Ca2, Sb, RBS | ||||||||||||||||||
| 1B.2.2.1 | A/Iowa/32/2016 | 94% | Ca2, Sb, RBS | ||||||||||||||||||
| Other-hu-1B | A/New Caledonia/20/1999 | 95% | Cb, Sa, Sb, RBS | ||||||||||||||||||
| Other-hu-1B | A/Brazil/11/1978 | 93% | Cb, Sa, Sb, Ca2, RBS | ||||||||||||||||||
| 1C.2.1 | A/Netherlands/3315/2016 | 95% | Sa, Ca2, Cb, RBS | ||||||||||||||||||
| 1C.2.1 | A/Netherlands/3315/2016 | 92% | Sa, Sb, Ca2, Cb, RBS | ||||||||||||||||||
| 1C.2.3 | A/Hunan/42443/205 | 99% | None | ||||||||||||||||||
| H3.1990.4a | A/Indiana/10/2011 | 98% | D | ||||||||||||||||||
| H3.1990.4a | A/Indiana/10/2011 | 93% | A, B, C, D | ||||||||||||||||||
| Hu-Vac | A/Wuhan/359/1995 | 93% | A, B, C | ||||||||||||||||||
| Hu-Vac | A/Wuhan/359/1995 | 95% | C, D | ||||||||||||||||||
| H3.2010.1 | A/Ohio/28/2016 | 99% | A | ||||||||||||||||||
| H3.2010.1 | A/Ohio/13/2017 | 99% | None | ||||||||||||||||||
| Hu-Vac | A/Hong Kong/4801/2014 | 97% | A, C | ||||||||||||||||||
aHA1 swine clade consensus for 2016–2019 globally and March 2019–September 2019 United States.
Of the six swine 1A clades we detected in the contemporary data, three included a CVV or the human seasonal vaccine (Fig. 3A): 1A.1.1 CVV (A/Ohio/24/2017); 1A.3.3.3 CVV (A/Ohio/09/2015); and 1A.3.3.2 human seasonal vaccine (A/Brisbane/02/2018). Amino acid comparisons of swine HA1 clade consensus sequences and nearest CVV or vaccine strain are summarized in Table 1. The 1A.1.1 swine consensus sequence was 97% identical to the CVV (A/Ohio/24/2017), with two amino acid mutations occurring in known antigenic sites (Sa and Sb). The 1A.3.3.3 HA1 consensus sequence shared 92% identity with the CVV (A/Ohio/09/2015), with seven mutations in antigenic epitopes (Sa, Sb, Ca1, and Ca2) or the RBS. The 1A.3.3.2 (H1N1pdm09) swine consensus sequence shared 98% identity with a human vaccine strain (A/Brisbane/02/2018), but possessed one mutation in an antigenic site (Sa) and another in the RBS: notably, there was a 2019 1A.3.3.2 variant case. The remaining three clades (1A.2, 1A.3.1, and 1A.3.4) were detected at low levels, and the swine consensus sequences from each clade had 86%–88% identity to the human vaccine strain A/Brisbane/02/2018, with seven to 10 mutations in Sa, Sb, Ca1, Ca2, Cb, or in the RBS.
Nine variant viruses of the 1B lineage were sequenced and deposited in databases within the last 3 years. Of the 13 contemporary 1B clades detected in global swine, two contained a CVV: 1B.2.1 (A/Ohio/35/2017 and A/Michigan/383/2018) and 1B.2.2.1 (A/Iowa/32/2016). The swine 1B.2.1 HA1 consensus sequence was 99% identical to the A/Michigan/383/2018 CVV, with no mutations in known antigenic sites. Similarly, the swine 1B.2.2.1 HA1 consensus sequence had high identity (99%) to the CVV, with two mutations located in Ca2. Similarity of consensus HA1 sequences for the other swine 1B clades was dramatically lower. The swine 1B.2.2.2 HA1 consensus sequence had low identity to the A/Iowa/32/2016 CVV (91%), with mutations in Ca2, Sb, and the RBS. A unique human-to-swine spillover in Brazil, 1B.2.2, was most similar to the A/Iowa/32/2016 CVV (91%) with mutations in Ca2, Sb, and the RBS. Viruses from a second unique human-to-swine spillover circulating in Brazil (Other-human-1B.2 in Fig. 3B, and Brazil 1B.2 in Table 1) were most similar (94%) to a WHO human seasonal vaccine recommended in 2000 (A/New Caledonia/20/1999), with mutations in Cb, Sa, Sb, and the RBS. The 1B.1 viruses in Europe were most similar (93%) to a vaccine strain that was last recommended for use in 1984 (A/Brazil/11/1978), with mutations in Cb, Sa, Sb, Ca2, and the RBS. Genetic diversity for viruses in the 1B.1 clade was high; when individual swine HA1 genes were compared with A/Brazil/11/1978, the lowest similarity dropped to 88%.
Three variant viruses of the swine IAV 1C lineage were reported within the last 5 years from clades 1C.2.1 (A/Netherlands/3315/2016 and A/Pavia/65/2016) and 1C.2.3 (A/Hunan/42443/205). Two of these variant viruses were recommended as CVV strains; a 1C.2.1 (A/Netherlands/3315/2016) and a 1C.2.3 (A/Hunan/42443/2015). Swine IAV related to the 1C.2.1 strains continue to circulate in swine with these data representing 53% of the detected 1C viruses over the past 3 years (Fig. 3C). The HA1 consensus sequence for clade 1C.2.1 shared 95% identity with A/Netherlands/3314/2016, with differences at amino acid sites within epitopes Sa, Ca2, Cb, and the RBS. The HA1 consensus sequence for 1C.2.2 was most similar to the 1C.2.1 CVV (92%), with amino acid differences in epitopes Sa, Sb, Ca2, Cb, and the RBS. The 1C.2.3 (n = 16) viruses circulating in China were 99% similar to A/Hunan/42443/2015 CVV, with only two amino acid differences that were not located in known antigenic sites.
The swine 1990s H3 lineages contain two H3.1990.4a genetic clade CVVs (A/Indiana/10/2011 and A/Minnesota/11/2010). The H3.1990.4a CVV (Fig. 3D) had relatively high identity to the consensus sequence of contemporary swine H3.1990.4a strains (97.5%), although it had a single mutation in antigenic epitope D. The H3.1990.4b swine strains were significantly different from the most similar CVV (93% similarity to A/Indiana/10/2011) with mutations in antigenic epitopes A, B, C, and D. For the remaining data in the H3.1990.4 clade, the swine consensus sequence was different from the most similar human vaccine strain (93% identity to A/Wuhan/359/1995) with amino acid differences in antigenic epitopes A, B, and C. The H3.1990.1 consensus sequence was most similar to the human vaccine A/Wuhan/359/1995 (95%), with three differences in antigenic epitopes C and D. The H3.2010.1 clade had two current CVVs (A/Ohio/28/2016 and A/Ohio/13/2017), and the HA1 clade consensus sequence shared high similarity (99.1%) to these CVVs, but with changes in antigenic epitope A compared with A/Ohio/28/2016. Another sustained human-to-swine lineage of the 2010s (H3.2010.2, n = 6) was most similar to the 2016 H3 human vaccine component (97% identity to A/Hong Kong/4801/2014), and differed at amino acid sites within antigenic epitopes A and C.
To quantify whether this genetic variation had an impact on the antigenic diversity and potential coverage of swine-origin/variant CVVs, swine strains similar to the clade consensus were tested against reference ferret antisera to identify swine strains with eightfold or greater reduction in titer compared with the homologous titer. Hemagglutination inhibition (HI) assays were performed following Lin et al. (2010), but guinea pig red blood cells were used for measuring hemagglutination, and oseltamivir was omitted from a subset of the assays for H1 viruses. Although availability of HI test antigens was limited for representative viruses from several clades of swine IAV, the data illustrated gaps in coverage of swine IAV by current CVVs (Tables 2–5) and serve as a baseline for future assessment of antigenic diversity in swine IAV evolution in the context of human pandemic preparedness.
Table 2.
Swine 1A test antigens in hemagglutination inhibition assays with monovalent ferret antisera
| Serastrain | A/Ohio/9/2015a | A/Michigan/45/2015 H1N1 | A/Michigan/45/2015 H1N1 | A/Michigan/45/2015 H1N1 | |||
|---|---|---|---|---|---|---|---|
| Clade | Global | Antigen | Clade | 1A.3.3.3 | Hu-Vac | Hu-Vac | Hu-Vac |
| gamma.1b | 1A.3.3.3 | A/Ohio/9/2015 H1N1va | 2560 | 20 | 20 | ||
| gamma.3b | 1A.3.3.3 | A/swine/Iowa/A01731653/2016 H1N2 | 40 | 40 | |||
| pdmb | 1A.3.3.2 | A/swine/Spain/45690-9/2018 H1N1 | 80 | 1280 | |||
| Hu-Vac | 1A.3.3.2 | A/Michigan/45/2015 H1N1 | 2560 | 320 | 640 |
Blank cells are untested antigens and repeated sera reflect different antisera or testing that occurred at different laboratories.
aCVV or equivalent.
bRepresents current swine strains.
The swine 1A.3.3.3 viruses showed limited cross-reactivity in HI assays against a ferret antiserum raised to the human seasonal vaccine strain from the H1N1pdm09 (1A.3.3.2) clade, whereas the 1A.3.3.2 virus retained good cross-reactivity (Table 2). The 1B.2.1 and 1B.2.2.2 swine strains showed reduced heterologous titers to the 1B.2.1 CVV-like ferret antisera (Table 3). European 1B.1.1, 1B.1.2, and 1B.1.2.1 retained some reactivity with an ancestral human seasonal vaccine strain (Table 3), but with evidence of antigenic heterogeneity within the 1B strains, particularly in the 1B.1.2 clade. Representative swine 1C viruses were tested against ferret antisera raised against the CVV of 1C.2.3 (A/Hunan/42443/2015) and the variant 1C.2.1 (A/Pavia/65/2016) (Table 4). The A/Pavia/65/2016 antiserum showed low cross-reactivity to the contemporary 1C strains tested despite being in the same genetic clade as the A/Netherlands/3316/2016 CVV, showing within-clade antigenic diversity that may impact CVV coverage of this clade. The 1C.2.3 A/Hunan/42443/2015 serum was cross-reactive to the 1C.2.1 viruses.
Table 3.
Swine 1B test antigens in hemagglutination inhibition assays with monovalent ferret antisera
| Serastrain | A/Michigan/383/2018 H1N2a,b | A/Brazil/11/1978 | |||
|---|---|---|---|---|---|
| Clade | Global | Antigen | Clade | 1B.2.1 | Hu-Vac |
| delta.2c | 1B.2.1 | A/swine/Michigan/A01104117/2018 H1N2a,b | 640 | ||
| delta.2c | 1B.2.1 | A/swine/Illinois/A02139356/2018 H1N2 | 160 | ||
| delta.1Ac | 1B.2.2.1 | A/swine/South Dakota/A01481702/2014 H1N2 | 80 | ||
| Hu-Vac | Human | A/Brazil/11/1978 H1N1 | 10,240 | ||
| hu-likec | 1B.1.1 | A/swine/England/208046/2018 H1N2 | 5120 | ||
| hu-likec | 1B.1.1 | A/swine/England/062058/2018 H1N2 | 2560 | ||
| hu-likec | 1B.1.2 | A/swine/Spain/45600-2/2017 H1N2 | 1280 | ||
| hu-likec | 1B.1.2.1 | A/swine/Spain/40340-2/2017 H1N2 | 5120 |
Blank cells are untested antigens and repeated sera reflect different antisera or testing that occurred at different laboratories.
aA/swine/Michigan/A01104117/2018 HA represents A/Michigan/383/2018 H1N2v (100% AA identity HA1).
bCVV or equivalent.
cRepresents current swine strains.
Table 4.
Swine 1C test antigens in hemagglutination inhibition assays with monovalent ferret antisera
| Serastrain | A/Pavia/65/2016 | A/Hunan/42443/2015a | A/Hunan/42443/2015a | |||
|---|---|---|---|---|---|---|
| Clade | Global | Antigen | Clade | 1C.2.1 | 1C.2.3 | 1C.2.3 |
| EAb | 1C.2.1 | A/Netherlands/3315/2016a H1N1v | 160 | 80 | 80 | |
| EAb | 1C.2.1 | A/Pavia/65/2016 H1N1v | 2560 | |||
| EAb | 1C.2.1 | A/swine/Spain/40340-1/2017 H1N1 | 80 | 160 | 160 | |
| EAb | 1C.2.1 | A/swine/Belgium/Gent/13/2017 H1N1 | 160 | 640 | 640 | |
| EAb | 1C.2.1 | A/swine/Belgium/Gent/9-P20/2018 H1N1 | 160 | 40 | 160 | |
| EAb | 1C.2.1 | A/swine/Spain/45690-4/2018 H1N2 | 160 | 320 | 320 | |
| EAb | 1C.2.1 | A/swine/Spain/6370-1/2018 H1N2 | 80 | |||
| EAb | 1C.2.2 | A/swine/Belgium/Gent/31/2018 H1N1 | 320 | |||
| EAb | 1C.2.2 | A/swine/Spain/6370-2/2018 H1N2 | 160 | |||
| EAb | 1C.2.3 | A/Hunan/42443/2015a H1N1v | 80 | |||
Blank cells are untested antigens and repeated sera reflect different antisera or testing that occurred at different laboratories.
aCVV or equivalent.
bRepresents current swine strains; (EA) Eurasian avian lineage.
The H3.1970.1 and H3.2000.3 lineage viruses showed limited cross reactivity with the H3.2010.1 CVV strain (A/Ohio/28/2016) antisera (Table 5). The H3.1970.1 lineage virus also had limited cross reactivity with the closest related human vaccine strains A/Port Chalmers/1/1973 and A/Perth/16/2009. In contrast, the H3.2000.3 lineage strain retained cross reactivity with A/Perth/16/2009 and A/Sydney/5/1997 human vaccine strains. The contemporary H3.1990.4a and H3.1990.4b lineage viruses showed low-to-moderate cross reactivity to the CVV-like strain (A/Minnesota/11/2010).
Table 5.
Swine H3 test antigens in hemagglutination inhibition assays with monovalent ferret antisera
| Serastrain | A/swine/New York/A01104005/2011a,b | A/swine/New York/A01104005/2011a,b | A/swine/Missouri/A01410819/2014b,c | A/swine/Missouri/A01410819/2014b,c | A/Ohio/28/2016b | A/Indiana/27/2018 | A/swine/Oklahoma/A02218157/2017 | A/Port Chalmers/1/1973 | A/Sydney/5/1997 | A/Perth/16/2009 | A/Switzerland/9715293/2013 | A/Switzerland/9715293/2013 | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Clade | Antigen | Clade | 3.1990.4a | 3.1990.4a | 3.2010.1 | 3.2010.1 | 3.2010.1 | 3.2010.1 | 3.2010.2 | Hu-Vac | Hu-Vac | Hu-Vac | Hu-Vac | Hu-Vac |
| 3.1970.1d | A/swine/Gent/48/2017 H3N2 | 80 | 80 | 80 | ||||||||||
| 3.1990.4ad | A/swine/New York/A01104005/2011 H3N2a,b | 80 | 320 | <10 | 10 | 10 | 10 | 20 | 20 | |||||
| 3.1990.4ad | A/swine/Oklahoma/A01776961/2016 H3N2 | 40 | 160 | 10 | 10 | 20 | 20 | 20 | 20 | |||||
| 3.1990.4bd | A/swine/Indiana/A01729045/2016 H3N2 | <10 | 20 | 10 | 20 | 20 | 20 | 40 | 20 | |||||
| 3.2000.3d | A/swine/Spain/45690-10/2019 H3N2 | 80 | 80 | 320 | 160 | |||||||||
| 3.2010.1 | A/swine/Missouri/A01410819/2014 H3N1c,b | <10 | <10 | 640 | 640 | 80 | 40 | 80 | 40 | |||||
| 3.2010.1 | A/Ohio/28/2016 H3N2b | 10,240 | ||||||||||||
| 3.2010.1d | A/Indiana/27/2018 H3N2 | <10 | <10 | 20 | 20 | 1280 | 10 | 40 | 40 | |||||
| 3.2010.1d | A/swine/Ohio/A01354299/2017 H3N2 | <10 | <10 | 40 | 80 | 160 | 80 | 40 | 40 | |||||
| 3.2010.2d | A/swine/Oklahoma/A02218157/2017 H3N2 | 10 | <10 | 40 | 40 | 40 | 640 | 160 | 160 | |||||
| Hu-Vac | A/Port Chalmers/1/1973 H3N2 | 5120 | ||||||||||||
| Hu-Vac | A/Sydney/5/1997 H3N2 | 2560 | ||||||||||||
| Hu-Vac | A/Perth/16/2009 H3N2 | 2560 | ||||||||||||
| Hu-Vac | A/Switzerland/9715293/2013 H3N2 | <10 | 10 | 40 | 40 | 40 | 80 | 2560 | 2560 | |||||
Blank cells are untested antigens and repeated sera reflect different antisera or testing that occurred at different laboratories.
aA/swine/New York/A01104005/2011 represents A/Minnesota/11/2010 (99.7% AA identity HA1).
bCVV or equivalent.
cA/swine/Missouri/A01410819/2014 represents A/Ohio/28/2016 (97.6% AA identity HA1).
dRepresents current swine strains.
CONCLUDING REMARKS
IAV in swine is highly diverse, with sustained transmission in global pig populations of all three main lineages of H1 (1A, 1B, and 1C) and multiple lineages of H3 from human seasonal IAV established across several decades following the 1968 Hong Kong pandemic. The NA and other six gene segments also show a high degree of diversity. Following the spread of the H1N1pdm09 virus in humans, annual introduction of this human seasonal H1N1 virus into pigs has potentiated a decade of reassortment and diversification of HA and NA in endemic swine lineages. A consequence of this is a montage of swine IAV that have human-adapted H1N1pdm09 lineage genes and endemic swine surface glycoproteins. This diversity has important implications for swine health and control of IAV using vaccines. Additionally, the diversity of swine IAV significantly increases the challenge of pandemic preparedness for the global public health community. The global genetic diversity of swine IAV circulating from 2016 to present and of swine IAV in the United States over the past 6 months showed that most swine IAV were significantly different from the current H1 and H3 components of human IAV vaccines. Additionally, only nine of the 30 distinct genetic clades detected in swine globally currently contained a CVV or human seasonal vaccine, and the degree to which those CVVs provide protection is uncertain given observed genetic and antigenic differences identified in recently circulating swine viruses. Because human and swine IAV evolution are inherently tangled, a system to regularly and rapidly prioritize and evaluate evolving swine IAV in the context of human risk should be part of a comprehensive pandemic preparedness plan. Surveillance in swine must continue to be a priority for animal and public health, with priority given to geographic areas with high levels of swine IAV diversity, rapid evolution, production practices that support viral transmission and migration, as well as specific animal–human interfaces that promote greater contact between pigs and people.
ACKNOWLEDGMENTS
Antigenic data were generated by the Animal and Plant Health Agency (APHA) (United Kingdom) and by the National Animal Disease Center (NADC), U.S. Department of Agriculture Agricultural Research Service (USDA-ARS) (United States). Ferret sera was kindly provided by WHO-CC (Centers for Disease Control and Prevention [CDC]) and the National Institutes of Health (NIH)-Centers of Excellence in Influenza Research and Surveillance (CEIRS). We acknowledge contributions from Ian Brown, Steve Essen, and Susan Collins and the OFFLU Swine Group for expertise and sharing data. We thank the Influenza Division at the CDC for sharing custom Python scripts used to identify amino acid substitutions. Research reported in this review and the efforts of the authors, were supported by USDA-ARS, USDA-Animal and Plant Health Inspection Service (APHIS), and by an NIH-National Institute of Allergy and Infectious Diseases (NIAID) interagency agreement associated with CRIP (Center of Research in Influenza Pathogenesis), an NIAID-funded Center of Excellence in Influenza Research and Surveillance (CEIRS, HHSN272201400008C) to A.L.V. and N.S.L. J.C., Z.W.A., C.K.S., and J.B.K. were supported by an appointment to the USDA-ARS Research Participation Program administered by the Oak Ridge Institute for Science and Education (ORISE) through an interagency agreement between the U.S. Department of Energy (DOE) and USDA under contract number DE-AC05-06OR23100. This research used resources provided by the SCINet project of the USDA Agricultural Research Service, ARS project number 0500-00093-001-00-D. Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture, CDC, DOE, or ORISE. USDA and CDC are equal opportunity providers and employers. The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention or the Agency for Toxic Substances and Disease Registry.
This article has been made freely available online courtesy of TAUNS Laboratories.
Footnotes
Editors: Gabriele Neumann and Yoshihiro Kawaoka
Additional Perspectives on Influenza: The Cutting Edge available at www.perspectivesinmedicine.org
REFERENCES
- Allerson M, Deen J, Detmer SE, Gramer MR, Joo HS, Romagosa A, Torremorell M. 2013. The impact of maternally derived immunity on influenza A virus transmission in neonatal pig populations. Vaccine 31: 500–505. 10.1016/j.vaccine.2012.11.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson TK, Nelson MI, Kitikoon P, Swenson SL, Korslund JA, Vincent AL. 2013. Population dynamics of cocirculating swine influenza A viruses in the United States from 2009 to 2012. Influenza Other Respir Viruses 7: 42–51. 10.1111/irv.12193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson TK, Campbell BA, Nelson MI, Lewis NS, Janas-Martindale A, Killian ML, Vincent AL. 2015. Characterization of co-circulating swine influenza A viruses in North America and the identification of a novel H1 genetic clade with antigenic significance. Virus Res 201: 24–31. 10.1016/j.virusres.2015.02.009 [DOI] [PubMed] [Google Scholar]
- Anderson TK, Macken CA, Lewis NS, Scheuermann RH, Van Reeth K, Brown IH, Swenson SL, Simon G, Saito T, Berhane Y, et al. 2016. A phylogeny-based global nomenclature system and automated annotation tool for H1 hemagglutinin genes from swine influenza A viruses. mSphere 1: e00275. 10.1128/mSphere.00275-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastien N, Antonishyn NA, Brandt K, Wong CE, Chokani K, Vegh N, Horsman GB, Tyler S, Graham MR, Plummer FA, et al. 2010. Human infection with a triple-reassortant swine influenza A(H1N1) virus containing the hemagglutinin and neuraminidase genes of seasonal influenza virus. J Infect Dis 201: 1178–1182. 10.1086/651507 [DOI] [PubMed] [Google Scholar]
- Baudon E, Chu DKW, Tung DD, Thi Nga P, Vu Mai Phuong H, Le Khanh Hang N, Thanh LT, Thuy NT, Khanh NC, Mai LQ, et al. 2018. Swine influenza viruses in Northern Vietnam in 2013–2014. Emerg Microbes Infect 7: 123. 10.1038/s41426-018-0109-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudoin A, Johnson S, Davies P, Bender J, Gramer M. 2012. Characterization of influenza A outbreaks in Minnesota swine herds and measures taken to reduce the risk of zoonotic transmission. Zoonoses Public Health 59: 96–106. 10.1111/j.1863-2378.2011.01423.x [DOI] [PubMed] [Google Scholar]
- Biondo N, Schaefer R, Gava D, Cantão ME, Silveira S, Mores MA, Ciacci-Zanella JR, Barcellos DE. 2014. Genomic analysis of influenza A virus from captive wild boars in Brazil reveals a human-like H1N2 influenza virus. Vet Microbiol 168: 34–40. 10.1016/j.vetmic.2013.10.010 [DOI] [PubMed] [Google Scholar]
- Bolton MJ, Abente EJ, Venkatesh D, Stratton JA, Zeller M, Anderson TK, Lewis NS, Vincent AL. 2019. Antigenic evolution of H3N2 influenza A viruses in swine in the United States from 2012 to 2016. Influenza Other Respir Viruses 13: 83–90. 10.1111/irv.12610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman AS, Walia RR, Nolting JM, Vincent AL, Killian ML, Zentkovich MM, Lorbach JN, Lauterbach SE, Anderson TK, Davis CT, et al. 2017. Influenza A(H3N2) virus in swine at agricultural fairs and transmission to humans, Michigan and Ohio, USA, 2016. Emerg Infect Dis 23: 1551–1555. 10.3201/eid2309.170847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch MG, Bateman AC, Landolt GA, Karasin AI, Brockman-Schneider RA, Gern JE, Suresh M, Olsen CW. 2008. Identification of amino acids in the HA of H3 influenza viruses that determine infectivity levels in primary swine respiratory epithelial cells. Virus Res 133: 269–279. 10.1016/j.virusres.2008.01.014 [DOI] [PubMed] [Google Scholar]
- Castrucci MR, Donatelli I, Sidoli L, Barigazzi G, Kawaoka Y, Webster RG. 1993. Genetic reassortment between avian and human influenza A viruses in Italian pigs. Virology 193: 503–506. 10.1006/viro.1993.1155 [DOI] [PubMed] [Google Scholar]
- Chastagner A, Hervé S, Bonin E, Quéguiner S, Hirchaud E, Henritzi D, Béven V, Gorin S, Barbier N, Blanchard Y, et al. 2018. Spatiotemporal distribution and evolution of the A/H1N1 2009 pandemic influenza virus in pigs in France from 2009 to 2017: identification of a potential swine-specific lineage. J Virol 92: e00988. 10.1128/JVI.00988-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YK, Pascua PN, Song MS. 2013. Swine influenza viruses: an Asian perspective. Curr Top Microbiol Immunol 370: 147–172. [DOI] [PubMed] [Google Scholar]
- Choi MJ, Morin CA, Scheftel J, Vetter SM, Smith K, Lynfield R, The Variant Influenza Investigation Team. 2015. Variant influenza associated with live animal markets, Minnesota. Zoonoses Public Health 62: 326–330. 10.1111/zph.12139 [DOI] [PubMed] [Google Scholar]
- Corzo CA, Culhane M, Juleen K, Stigger-Rosser E, Ducatez MF, Webby RJ, Lowe JF. 2013. Active surveillance for influenza A virus among swine, midwestern United States, 2009–2011. Emerg Infect Dis 19: 954–960. 10.3201/eid1906.121637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibárbora M, Cappuccio J, Olivera V, Quiroga M, Machuca M, Perfumo C, Pérez D, Pereda A. 2013. Swine influenza: clinical, serological, pathological, and virological cross-sectional studies in nine farms in Argentina. Influenza Other Respir Viruses 7: 10–15. 10.1111/irv.12200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duwell MM, Blythe D, Radebaugh MW, Kough EM, Bachaus B, Crum DA, Perkins KA, Blanton L, Davis CT, Jang Y, et al. 2018. Influenza A(H3N2) variant virus outbreak at three fairs—Maryland, 2017. MMWR Morb Mortal Wkly Rep 67: 1169–1173. 10.15585/mmwr.mm6742a1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epperson S, Jhung M, Richards S, Quinlisk P, Ball L, Moll M, Boulton R, Haddy L, Biggerstaff M, Brammer L, et al. 2013. Human infections with influenza A(H3N2) variant virus in the United States, 2011–2012. Clin Infect Dis 57: S4–S11. 10.1093/cid/cit272 [DOI] [PubMed] [Google Scholar]
- Freidl GS, Meijer A, de Bruin E, de Nardi M, Munoz O, Capua I, Breed AC, Harris K, Hill A, Kosmider R, et al. 2014. Influenza at the animal-human interface: a review of the literature for virological evidence of human infection with swine or avian influenza viruses other than A(H5N1). Euro Surveill 19: 20793. 10.2807/1560-7917.ES2014.19.18.20793 [DOI] [PubMed] [Google Scholar]
- Gao S, Anderson TK, Walia RR, Dorman KS, Janas-Martindale A, Vincent AL. 2017. The genomic evolution of H1 influenza A viruses from swine detected in the United States between 2009 and 2016. J Gen Virol 98: 2001–2010. 10.1099/jgv.0.000885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaydos JC, Top FH Jr, Hodder RA, Russell PK. 2006. Swine influenza A outbreak, Fort Dix, New Jersey, 1976. Emerg Infect Dis 12: 23–28. 10.3201/eid1201.050965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerloff NA, Khan SU, Balish A, Shanta IS, Simpson N, Berman L, Haider N, Poh MK, Islam A, Gurley E, et al. 2014. Multiple reassortment events among highly pathogenic avian influenza A(H5N1) viruses detected in Bangladesh. Virology 450–451: 297–307. 10.1016/j.virol.2013.12.023 [DOI] [PubMed] [Google Scholar]
- Greenbaum A, Quinn C, Bailer J, Su S, Havers F, Durand LO, Jiang V, Page S, Budd J, Shaw M, et al. 2015. Investigation of an outbreak of variant influenza A(H3N2) virus infection associated with an agricultural fair—Ohio, August 2012. J Infect Dis 212: 1592–1599. 10.1093/infdis/jiv269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory V, Lim W, Cameron K, Bennett M, Marozin S, Klimov A, Hall H, Cox N, Hay A, Lin YP. 2001. Infection of a child in Hong Kong by an influenza A H3N2 virus closely related to viruses circulating in European pigs. J Gen Virol 82: 1397–1406. 10.1099/0022-1317-82-6-1397 [DOI] [PubMed] [Google Scholar]
- He P, Wang G, Mo Y, Yu Q, Xiao X, Yang W, Zhao W, Guo X, Chen Q, He J, et al. 2018. Novel triple-reassortant influenza viruses in pigs, Guangxi, China. Emerg Microbes Infect 7: 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houser KV, Katz JM, Tumpey TM. 2013. Seasonal trivalent inactivated influenza vaccine does not protect against newly emerging variants of influenza A (H3N2v) virus in ferrets. J Virol 87: 1261–1263. 10.1128/JVI.02625-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke BH. 2013. Clinicopathological features of Swine influenza. Curr Top Microbiol Immunol 370: 69–83. [DOI] [PubMed] [Google Scholar]
- Jhung MA, Epperson S, Biggerstaff M, Allen D, Balish A, Barnes N, Beaudoin A, Berman L, Bidol S, Blanton L, et al. 2013. Outbreak of variant influenza A(H3N2) virus in the United States. Clin Infect Dis 57: 1703–1712. 10.1093/cid/cit649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karesh WB, Dobson A, Lloyd-Smith JO, Lubroth J, Dixon MA, Bennett M, Aldrich S, Harrington T, Formenty P, Loh EH, et al. 2012. Ecology of zoonoses: natural and unnatural histories. Lancet 380: 1936–1945. 10.1016/S0140-6736(12)61678-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasowski EJ, Garten RJ, Bridges CB. 2011. Influenza pandemic epidemiologic and virologic diversity: reminding ourselves of the possibilities. Clin Infect Dis 52: S44–S49. 10.1093/cid/ciq010 [DOI] [PubMed] [Google Scholar]
- Kitikoon P, Nilubol D, Erickson BJ, Janke BH, Hoover TC, Sornsen SA, Thacker EL. 2006. The immune response and maternal antibody interference to a heterologous H1N1 swine influenza virus infection following vaccination. Vet Immunol Immunopathol 112: 117–128. 10.1016/j.vetimm.2006.02.008 [DOI] [PubMed] [Google Scholar]
- Kitikoon P, Nelson MI, Killian ML, Anderson TK, Koster L, Culhane MR, Vincent AL. 2013. Genotype patterns of contemporary reassorted H3N2 virus in US swine. J Gen Virol 94: 1236–1241. 10.1099/vir.0.051839-0 [DOI] [PubMed] [Google Scholar]
- Koen JS. 1919. A practical method for field diagnosis of swine diseases. Am J Vet Med 14: 468–470. [Google Scholar]
- Krog JS, Hjulsager CK, Larsen MA, Larsen LE. 2017. Triple-reassortant influenza A virus with H3 of human seasonal origin, NA of swine origin, and internal A(H1N1) pandemic 2009 genes is established in Danish pigs. Influenza Other Respir Viruses 11: 298–303. 10.1111/irv.12451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis NS, Russell CA, Langat P, Anderson TK, Berger K, Bielejec F, Burke DF, Dudas G, Fonville JM, Fouchier RA, et al. 2016. The global antigenic diversity of swine influenza A viruses. Elife 5: e12217. 10.7554/eLife.12217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H, Lam TT, Fan X, Chen X, Zeng Y, Zhou J, Duan L, Tse M, Chan CH, Li L, et al. 2014. Expansion of genotypic diversity and establishment of 2009 H1N1 pandemic-origin internal genes in pigs in China. J Virol 88: 10864–10874. 10.1128/JVI.01327-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YP, Gregory V, Collins P, Kloess J, Wharton S, Cattle N, Lackenby A, Daniels R, Hay A. 2010. Neuraminidase receptor binding variants of human influenza A(H3N2) viruses resulting from substitution of aspartic acid 151 in the catalytic site: a role in virus attachment? J Virol 84: 6769–6781. 10.1128/JVI.00458-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Veguilla V, Gross FL, Gillis E, Rowe T, Xu X, Tumpey TM, Katz JM, Levine MZ, Lu X. 2017. Effect of priming with seasonal influenza A(H3N2) virus on the prevalence of cross-reactive hemagglutination-inhibition antibodies to swine-origin A(H3N2) variants. J Infect Dis 216: S539–S547. 10.1093/infdis/jix093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorusso A, Vincent AL, Harland ML, Alt D, Bayles DO, Swenson SL, Gramer MR, Russell CA, Smith DJ, Lager KM, et al. 2011. Genetic and antigenic characterization of H1 influenza viruses from United States swine from 2008. J Gen Virol 92: 919–930. 10.1099/vir.0.027557-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Yi L, Jing Y, Tan H, Mai W, Song Y, Zou L, Liang L, Xiao H, Kang M, et al. 2019. A human infection with a novel reassortant H3N2 swine virus in China. J Infect 79: 174–187. 10.1016/j.jinf.2019.04.015 [DOI] [PubMed] [Google Scholar]
- Ma W, Lager KM, Vincent AL, Janke BH, Gramer MR, Richt JA. 2009. The role of swine in the generation of novel influenza viruses. Zoonoses Public Health 56: 326–337. 10.1111/j.1863-2378.2008.01217.x [DOI] [PubMed] [Google Scholar]
- Mena I, Nelson MI, Quezada-Monroy F, Dutta J, Cortes-Fernandez R, Lara-Puente JH, Castro-Peralta F, Cunha LF, Trovao NS, Lozano-Dubernard B, et al. 2016. Origins of the 2009 H1N1 influenza pandemic in swine in Mexico. Elife 5: e16777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson MI, Lemey P, Tan Y, Vincent A, Lam TT, Detmer S, Viboud C, Suchard MA, Rambaut A, Holmes EC, et al. 2011. Spatial dynamics of human-origin H1 influenza A virus in North American swine. PLoS Pathog 7: e1002077. 10.1371/journal.ppat.1002077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson MI, Gramer MR, Vincent AL, Holmes EC. 2012. Global transmission of influenza viruses from humans to swine. J Gen Virol 93: 2195–2203. 10.1099/vir.0.044974-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson M, Culhane MR, Rovira A, Torremorell M, Guerrero P, Norambuena J. 2015a. Novel human-like influenza A viruses circulate in swine in Mexico and Chile. PLoS Curr 7: ii. 10.1371/currents.outbreaks.c8b3207c9bad98474eca3013fa933ca6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson MI, Schaefer R, Gava D, Cantão ME, Ciacci-Zanella JR. 2015b. Influenza A viruses of human origin in swine, Brazil. Emerg Infect Dis 21: 1339–1347. 10.3201/eid2108.141891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson MI, Stratton J, Killian ML, Janas-Martindale A, Vincent AL. 2015c. Continual reintroduction of human pandemic H1N1 influenza A viruses into swine in the United States, 2009 to 2014. J Virol 89: 6218–6226. 10.1128/JVI.00459-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson MI, Viboud C, Vincent AL, Culhane MR, Detmer SE, Wentworth DE, Rambaut A, Suchard MA, Holmes EC, Lemey P. 2015d. Global migration of influenza A viruses in swine. Nat Commun 6: 6696. 10.1038/ncomms7696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo LT, Hiromoto Y, Pham VP, Le HT, Nguyen HT, Le VT, Takemae N, Saito T. 2012. Isolation of novel triple-reassortant swine H3N2 influenza viruses possessing the hemagglutinin and neuraminidase genes of a seasonal influenza virus in Vietnam in 2010. Influenza Other Respir Viruses 6: 6–10. 10.1111/j.1750-2659.2011.00267.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32: 268–274. 10.1093/molbev/msu300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen CW, Karasin AI, Carman S, Li Y, Bastien N, Ojkic D, Alves D, Charbonneau G, Henning BM, Low DE, et al. 2006. Triple reassortant H3N2 influenza A viruses, Canada, 2005. Emerg Infect Dis 12: 1132–1135. 10.3201/eid1207.060268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascua PN, Song MS, Lee JH, Baek YH, Kwon HI, Park SJ, Choi EH, Lim GJ, Lee OJ, Kim SW, et al. 2012. Virulence and transmissibility of H1N2 influenza virus in ferrets imply the continuing threat of triple-reassortant swine viruses. Proc Natl Acad Sci 109: 15900–15905. 10.1073/pnas.1205576109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascua PN, Song MS, Kwon HI, Lim GJ, Kim EH, Park SJ, Lee OJ, Kim CJ, Webby RJ, Webster RG, et al. 2013. The homologous tripartite viral RNA polymerase of A/swine/Korea/CT1204/2009(H1N2) influenza virus synergistically drives efficient replication and promotes respiratory droplet transmission in ferrets. J Virol 87: 10552–10562. 10.1128/JVI.01333-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce MB, Jayaraman A, Pappas C, Belser JA, Zeng H, Gustin KM, Maines TR, Sun X, Raman R, Cox NJ, et al. 2012. Pathogenesis and transmission of swine origin A(H3N2)v influenza viruses in ferrets. Proc Natl Acad Sci 109: 3944–3949. 10.1073/pnas.1119945109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereda A, Rimondi A, Cappuccio J, Sanguinetti R, Angel M, Ye J, Sutton T, Dibárbora M, Olivera V, Craig MI, et al. 2011. Evidence of reassortment of pandemic H1N1 influenza virus in swine in Argentina: are we facing the expansion of potential epicenters of influenza emergence? Influenza Other Respir Viruses 5: 409–412. 10.1111/j.1750-2659.2011.00246.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt R, Vincent AL, Gauger PC, Loving CL, Zanella EL, Lager KM, Jr KM, Kimura K, Roth JA. 2011. Comparison of humoral and cellular immune responses to inactivated swine influenza virus vaccine in weaned pigs. Vet Immunol Immunopathol 142: 252–257. 10.1016/j.vetimm.2011.05.005 [DOI] [PubMed] [Google Scholar]
- Poljak Z, Carman S, McEwen B. 2014. Assessment of seasonality of influenza in swine using field submissions to a diagnostic laboratory in Ontario between 2007 and 2012. Influenza Other Respir Viruses 8: 482–492. 10.1111/irv.12248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulit-Penaloza JA, Jones J, Sun X, Jang Y, Thor S, Belser JA, Zanders N, Creager HM, Ridenour C, Wang L, et al. 2018a. Antigenically diverse swine origin H1N1 variant influenza viruses exhibit differential ferret pathogenesis and transmission phenotypes. J Virol 92: e00095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulit-Penaloza JA, Pappas C, Belser JA, Sun X, Brock N, Zeng H, Tumpey TM, Maines TR. 2018b. Comparative in vitro and in vivo analysis of H1N1 and H1N2 variant influenza viruses isolated from humans between 2011 and 2016. J Virol 92: e01444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulit-Penaloza JA, Belser JA, Tumpey TM, Maines TR. 2019a. Mammalian pathogenicity and transmissibility of a reassortant Eurasian avian-like A(H1N1v) influenza virus associated with human infection in China (2015). Virology 537: 31–35. 10.1016/j.virol.2019.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulit-Penaloza JA, Belser JA, Tumpey TM, Maines TR. 2019b. Sowing the seeds of a pandemic? Mammalian pathogenicity and transmissibility of H1 variant influenza viruses from the swine reservoir. Trop Med Infect Dis 4: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajão DS, Gauger PC, Anderson TK, Lewis NS, Abente EJ, Killian ML, Perez DR, Sutton TC, Zhang J, Vincent AL. 2015. Novel reassortant human-like H3N2 and H3N1 influenza A viruses detected in pigs are virulent and antigenically distinct from swine viruses endemic to the United States. J Virol 89: 11213–11222. 10.1128/JVI.01675-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajão DS, Walia RR, Campbell B, Gauger PC, Janas-Martindale A, Killian ML, Vincent AL. 2017. Reassortment between swine H3N2 and 2009 pandemic H1N1 in the United States resulted in influenza A viruses with diverse genetic constellations with variable virulence in pigs. J Virol 91: e01763. 10.1128/JVI.01763-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajao DS, Anderson TK, Kitikoon P, Stratton J, Lewis NS, Vincent AL. 2018a. Antigenic and genetic evolution of contemporary swine H1 influenza viruses in the United States. Virology 518: 45–54. 10.1016/j.virol.2018.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajao DS, Vincent AL, Perez DR. 2018b. Adaptation of human influenza viruses to swine. Front Vet Sci 5: 347. 10.3389/fvets.2018.00347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambo-Martin BL, Keller MW, Wilson MM, Nolting JM, Anderson TK, Vincent AL, Bagal U, Jang Y, Neuhaus EB, Davis CT, et al. 2019. Mitigating pandemic risk with influenza A virus field surveillance at a swine-human interface. bioRxiv doi:10.1101/585588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resende PC, Born PS, Matos AR, Motta FC, Caetano BC, Debur MD, Riediger IN, Brown D, Siqueira MM. 2017. Whole-genome characterization of a novel human influenza A(H1N2) virus variant, Brazil. Emerg Infect Dis 23: 152–154. 10.3201/eid2301.161122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson JS, Nicolson C, Harvey R, Johnson R, Major D, Guilfoyle K, Roseby S, Newman R, Collin R, Wallis C, et al. 2011. The development of vaccine viruses against pandemic A(H1N1) influenza. Vaccine 29: 1836–1843. 10.1016/j.vaccine.2010.12.044 [DOI] [PubMed] [Google Scholar]
- Robinson JL, Lee BE, Patel J, Bastien N, Grimsrud K, Seal RF, King R, Marshall F, Li Y. 2007. Swine influenza (H3N2) infection in a child and possible community transmission, Canada. Emerg Infect Dis 13: 1865–1870. 10.3201/eid1312.070615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shope RE. 1931. Swine influenza: III. Filtration experiments and etiology. J Exp Med 54: 373–385. 10.1084/jem.54.3.373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu Y, McCauley J. 2017. GISAID: global initiative on sharing all influenza data—from vision to reality. Euro Surveill 22: 30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon G, Larsen LE, Dürrwald R, Foni E, Harder T, Van Reeth K, Markowska-Daniel I, Reid SM, Dan A, Maldonado J, et al. 2014. European surveillance network for influenza in pigs: surveillance programs, diagnostic tools and swine influenza virus subtypes identified in 14 European countries from 2010 to 2013. PLoS ONE 9: e115815. 10.1371/journal.pone.0115815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowronski DM, Janjua NZ, De Serres G, Purych D, Gilca V, Scheifele DW, Dionne M, Sabaiduc S, Gardy JL, Li G, et al. 2012. Cross-reactive and vaccine-induced antibody to an emerging swine-origin variant of influenza A virus subtype H3N2 (H3N2v). J Infect Dis 206: 1852–1861. 10.1093/infdis/jis500 [DOI] [PubMed] [Google Scholar]
- Sun X, Pulit-Penaloza JA, Belser JA, Pappas C, Pearce MB, Brock N, Zeng H, Creager HM, Zanders N, Jang Y, et al. 2018. Pathogenesis and transmission of genetically diverse swine-origin H3N2 variant influenza A viruses from multiple lineages isolated in the United States, 2011–2016. J Virol 92: e00665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemae N, Parchariyanon S, Damrongwatanapokin S, Uchida Y, Ruttanapumma R, Watanabe C, Yamaguchi S, Saito T. 2008. Genetic diversity of swine influenza viruses isolated from pigs during 2000 to 2005 in Thailand. Influenza Other Respir Viruses 2: 181–189. 10.1111/j.1750-2659.2008.00062.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemae N, Harada M, Nguyen PT, Nguyen T, Nguyen TN, To TL, Nguyen TD, Pham VP, Le VT, Do HT, et al. 2017. Influenza A viruses of swine (IAV-S) in Vietnam from 2010 to 2015: multiple introductions of A(H1N1)pdm09 viruses into the pig population and diversifying genetic constellations of Enzootic IAV-S. J Virol 91: e01490. 10.1128/JVI.01490-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Veen RL, Loynachan AT, Mogler MA, Russell BJ, Harris DL, Kamrud KI. 2012. Safety, immunogenicity, and efficacy of an alphavirus replicon-based swine influenza virus hemagglutinin vaccine. Vaccine 30: 1944–1950. 10.1016/j.vaccine.2012.01.030 [DOI] [PubMed] [Google Scholar]
- Vander Veen RL, Mogler MA, Russell BJ, Loynachan AT, Harris DL, Kamrud KI. 2013. Haemagglutinin and nucleoprotein replicon particle vaccination of swine protects against the pandemic H1N1 2009 virus. Vet Rec 173: 344. 10.1136/vr.101741 [DOI] [PubMed] [Google Scholar]
- Van Reeth K, Ma W. 2013. Swine influenza virus vaccines: to change or not to change—that's the question. Curr Top Microbiol Immunol 370: 173–200. [DOI] [PubMed] [Google Scholar]
- Van Reeth K, Brown IH, Olsen CW. 2012. Swine influenza. In Diseases of swine (ed. Zimmerman JJ). Wiley-Blackwell, Chichester, United Kingdom. [Google Scholar]
- Vijaykrishna D, Smith GJ, Pybus OG, Zhu H, Bhatt S, Poon LL, Riley S, Bahl J, Ma SK, Cheung CL, et al. 2011. Long-term evolution and transmission dynamics of swine influenza A virus. Nature 473: 519–522. 10.1038/nature10004 [DOI] [PubMed] [Google Scholar]
- Vincent AL, Ma W, Lager KM, Janke BH, Richt JA. 2008. Swine influenza viruses: a North American perspective. Adv Virus Res 72: 127–154. 10.1016/S0065-3527(08)00403-X [DOI] [PubMed] [Google Scholar]
- Vincent AL, Ma W, Lager KM, Gramer MR, Richt JA, Janke BH. 2009. Characterization of a newly emerged genetic cluster of H1N1 and H1N2 swine influenza virus in the United States. Virus Genes 39: 176–185. 10.1007/s11262-009-0386-6 [DOI] [PubMed] [Google Scholar]
- Vincent A, Awada L, Brown I, Chen H, Claes F, Dauphin G, Donis R, Culhane M, Hamilton K, Lewis N, et al. 2014. Review of influenza A virus in swine worldwide: a call for increased surveillance and research. Zoonoses Public Health 61: 4–17. 10.1111/zph.12049 [DOI] [PubMed] [Google Scholar]
- Vincent AL, Perez DR, Rajao D, Anderson TK, Abente EJ, Walia RR, Lewis NS. 2017. Influenza A virus vaccines for swine. Vet Microbiol 206: 35–44. 10.1016/j.vetmic.2016.11.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walia RR, Anderson TK, Vincent AL. 2019. Regional patterns of genetic diversity in swine influenza A viruses in the United States from 2010 to 2016. Influenza Other Respir Viruses 13: 262–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson SJ, Langat P, Reid SM, Lam TT, Cotten M, Kelly M, Van Reeth K, Qiu Y, Simon G, Bonin E, et al. 2015. Molecular epidemiology and evolution of influenza viruses circulating within European swine between 2009 and 2013. J Virol 89: 9920–9931. 10.1128/JVI.00840-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesley RD, Lager KM. 2006. Overcoming maternal antibody interference by vaccination with human adenovirus 5 recombinant viruses expressing the hemagglutinin and the nucleoprotein of swine influenza virus. Vet Microbiol 118: 67–75. 10.1016/j.vetmic.2006.07.014 [DOI] [PubMed] [Google Scholar]
- Wong FYK, Donato C, Deng YM, Teng D, Komadina N, Baas C, Modak J, O'Dea M, Smith DW, Effler PV, et al. 2018. Divergent human-origin influenza viruses detected in Australian swine populations. J Virol 92: e00316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie JF, Zhang YH, Zhao L, Xiu WQ, Chen HB, Lin Q, Weng YW, Zheng KC. 2018. Emergence of Eurasian avian-like swine influenza A (H1N1) virus from an adult case in Fujian Province, China. Virol Sin 33: 282–286. 10.1007/s12250-018-0034-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen HL, Forrest H, Cheung P, Wong D, Li O, Krauss S, Ferguson A, Crumpton JC, Jones J, Choy T, et al. 2011. Transmissibility of pandemic H1N1 and genetically related swine influenza viruses in ferrets. Influenza Other Respir Viruses 5: 85–87. [PubMed] [Google Scholar]
- Zeller MA, Anderson TK, Walia RW, Vincent AL, Gauger PC. 2018a. ISU FLUture: a veterinary diagnostic laboratory web-based platform to monitor the temporal genetic patterns of influenza A virus in swine. BMC Bioinformatics 19: 397. 10.1186/s12859-018-2408-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeller MA, Li G, Harmon KM, Zhang J, Vincent AL, Anderson TK, Gauger PC. 2018b. Complete genome sequences of two novel human-like H3N2 influenza A viruses, A/swine/Oklahoma/65980/2017 (H3N2) and A/Swine/Oklahoma/65260/2017 (H3N2), detected in swine in the United States. Microbiol Resour Announc 7: e01203. 10.1128/MRA.01203-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Aevermann BD, Anderson TK, Burke DF, Dauphin G, Gu Z, He S, Kumar S, Larsen CN, Lee AJ, et al. 2017. Influenza research database: an integrated bioinformatics resource for influenza virus research. Nucleic Acids Res 45: D466–D474. 10.1093/nar/gkw857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou NN, Senne DA, Landgraf JS, Swenson SL, Erickson G, Rossow K, Liu L, Yoon K, Krauss S, Webster RG. 1999. Genetic reassortment of avian, swine, and human influenza A viruses in American pigs. J Virol 73: 8851–8856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Zhou B, Fan X, Lam TT, Wang J, Chen A, Chen X, Chen H, Webster RG, Webby R, et al. 2011. Novel reassortment of Eurasian avian-like and pandemic/2009 influenza viruses in swine: infectious potential for humans. J Virol 85: 10432–10439. 10.1128/JVI.05352-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Webby R, Lam TT, Smith DK, Peiris JS, Guan Y. 2013. History of swine influenza viruses in Asia. Curr Top Microbiol Immunol 370: 57–68. [DOI] [PubMed] [Google Scholar]