Abstract
Ancient DNA sampling methods—although optimized for efficient DNA extraction—are destructive, relying on drilling or cutting and powdering (parts of) bones and teeth. As the field of ancient DNA has grown, so have concerns about the impact of destructive sampling of the skeletal remains from which ancient DNA is obtained. Due to a particularly high concentration of endogenous DNA, the cementum of tooth roots is often targeted for ancient DNA sampling, but destructive sampling methods of the cementum often result in the loss of at least one entire root. Here, we present a minimally destructive method for extracting ancient DNA from dental cementum present on the surface of tooth roots. This method does not require destructive drilling or grinding, and, following extraction, the tooth remains safe to handle and suitable for most morphological studies, as well as other biochemical studies, such as radiocarbon dating. We extracted and sequenced ancient DNA from 30 teeth (and nine corresponding petrous bones) using this minimally destructive extraction method in addition to a typical tooth sampling method. We find that the minimally destructive method can provide ancient DNA that is of comparable quality to extracts produced from teeth that have undergone destructive sampling processes. Further, we find that a rigorous cleaning of the tooth surface combining diluted bleach and UV light irradiation seems sufficient to minimize external contaminants usually removed through the physical removal of a superficial layer when sampling through regular powdering methods.
Over the past decade, the field of ancient DNA has experienced a rapid increase in the number of ancient genomes published each year (Slatkin and Racimo 2016) as a consequence of advances in ancient DNA sampling (Gamba et al. 2014; Damgaard et al. 2015), extraction (Dabney et al. 2013a; Rohland et al. 2018), and enrichment (Carpenter et al. 2013; Fu et al. 2013) techniques. As our ability to sequence large numbers of ancient individuals has increased, discussions about the destructive nature of ancient DNA sampling—which typically requires drilling or cutting and powdering ancient bones and teeth—have become more prominent (Makarewicz et al. 2017; Prendergast and Sawchuk 2018; Sirak and Sedig 2019). The identification of the osseous inner ear, and specifically the cochlea (located in the petrous portion of the temporal bone), as an optimal source of ancient DNA (Gamba et al. 2014; Pinhasi et al. 2015, 2019) is one of the driving factors in this revolution, making it possible to access ancient DNA from geographic regions with climatic conditions unfavorable to ancient DNA preservation. However, accessing this optimal source of ancient DNA results in the destruction of the inner ear morphology, which is a valuable source of morphological information (de León et al. 2018). Although there are protocols that reduce the destructive nature of sampling, by sampling from the ossicles of the inner ear (Sirak et al. 2020) or performing targeted drilling of the cochlea through the cranial base of complete or reconstructed crania (Sirak et al. 2017), some destruction (including that of morphologically informative inner ear components) is inevitable. As a consequence, this and other less invasive methods may be considered unsuitable in cases where samples are of particular anthropological value and are subject to stringent restrictions on permissible sampling practices.
Teeth are a valuable alternative to the sampling of the cochlea (Gamba et al. 2014; Damgaard et al. 2015), especially because they are particularly numerous in osteological collections, due to the fact that individuals have many more teeth than petrous bones and to their resistance to taphonomic decomposition. Despite this, little has been published outlining optimal practices for sampling from teeth. Traditionally, the standard practice has been to grind or drill large chunks of the tooth root to a powder (Rohland and Hofreiter 2007), as the crown enamel is largely inorganic and is therefore unlikely to contain a substantial amount of endogenous DNA (Higgins and Austin 2013). In an attempt to minimize potential external contaminants, the surface layer is often removed to access the “untouched” dentine and pulp. However, this practice removes some, if not all, of the thin layer of cementum that coats the inferior portion of dental roots.
The cellular cementum is rich in cementocytes, which are DNA-containing cells that remain encased in the mineral structure of the tooth after death (Bosshardt and Selvig 1997). Cementum also shares several histological properties with the cochlear region of the petrous that are thought to contribute to its high level of DNA preservation, including similarities between cementocytes (Zhao et al. 2016) and osteocytes, which are hypothesized to serve as repositories of ancient DNA in bones (Bell et al. 2008; Pinhasi et al. 2015). Like the cochlea, cementum also does not undergo remodeling (but, unlike the cochlea, it continues to accumulate throughout life) and the haphazard organization of collagen fibers in cementum resembles that of woven bone (Freeman 1994; Grzesik et al. 2000). Assessment of DNA preservation in ancient teeth shows that dental cementum contains a substantially higher proportion of endogenous DNA than dentine from the same tooth (Damgaard et al. 2015). Furthermore, in a direct comparison between cementum and petrous samples, Hansen et al. (2017) find that cementum and petrous yield a comparable amount of endogenous DNA in well-preserved samples, although in poorly preserved individuals, the petrous yields a higher proportion of endogenous molecules. The only published method for sampling DNA from the cementum specifically recommends a targeted method for extracting DNA from teeth using an “inside-out” approach that involves removing the crown and subsequently using a fine drill to remove as much pulp and dentine as possible from the tooth root to ultimately obtain a “case” of cementum (Damgaard et al. 2015). However, this valuable approach may still not be able to perfectly isolate the extremely thin and brittle layer of cementum, which ranges from 20–50 μm thick at the cementoenamel junction, to 150–200 μm thick at the apex of the root (Freeman 1994).
Here, we present an alternative, minimally destructive protocol for sampling ancient DNA from tooth cementum that does not require drilling or cutting, thereby maintaining the morphological integrity of the tooth (Fig. 1). The technique targets ancient DNA from the cementum of tooth roots by directly exposing the outermost layer of a portion of the tooth root to a lysis buffer for a short incubation period, following a nondestructive decontamination procedure. Similar less destructive methods have been reported in previous PCR-based mitochondrial ancient DNA studies (Rohland et al. 2004; Bolnick et al. 2012; Hofreiter 2012) and in forensic contexts (Correa et al. 2019). However, the ancient DNA obtained using these strategies was typically less well preserved and of a lesser quantity than DNA obtained using more destructive methods. Additionally, in some cases (Rohland et al. 2004; Hofreiter 2012), the hazardous chemicals used during sampling may have compromised safe handling and future chemical analyses of the remains. In this study, we conduct a systematic evaluation of the application of a minimally destructive sampling technique in a massively parallel sequencing context.
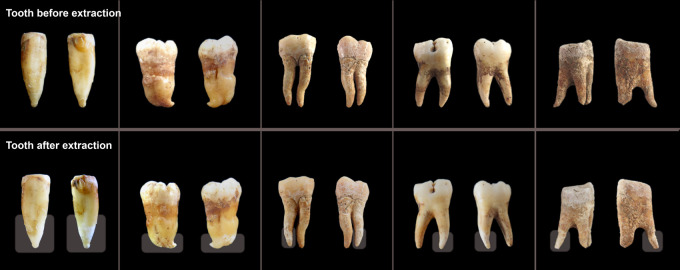
Figure 1.
Examples of teeth before and after minimally destructive extraction. Teeth which have been sampled using this minimally destructive extraction protocol were photographed prior to (top) and ∼24 h after (bottom) extraction. Through the use of parafilm to protect regions of the tooth that are not targeted during sampling, such as the crown, sample degradation is primarily restricted to the lower portion of the targeted tooth roots, and the overall morphology of the tooth remains intact. The region targeted for sampling (i.e., not covered by parafilm) is indicated by a transparent box in the after images. Note that these are representative examples of the typical impact of sampling using this method on ancient teeth of high quality (two left-most teeth) or moderate quality (three right-most teeth). Data from these teeth are not reported in this study. For before and after images of the tooth roots upon which sequencing was done during this study, see Supplemental Figure S1.
Results
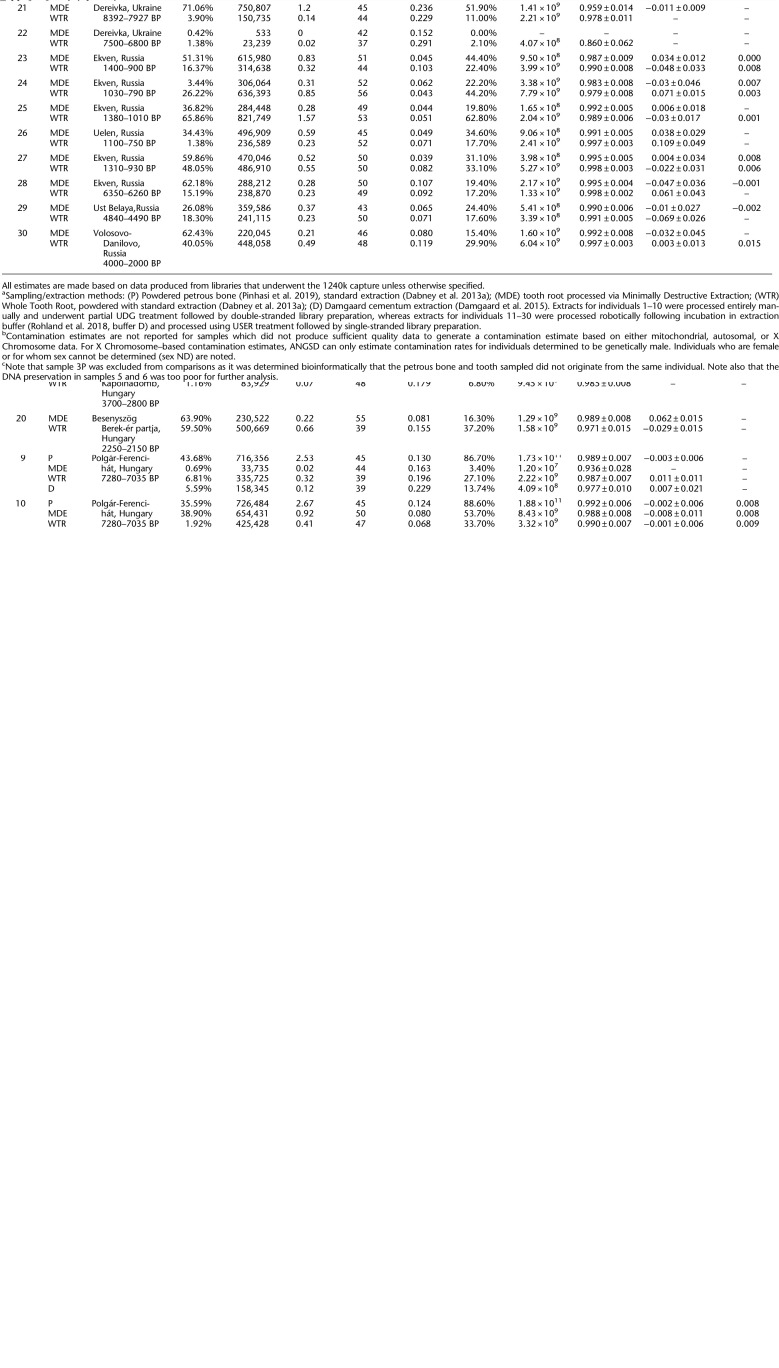
We selected thirty ancient individuals (Table 1; Supplemental Table S1) for a comparative analysis of the quality of ancient DNA—as measured through metrics such as the proportion of endogenous molecules of shotgun data, sample complexity, and contamination rate—that could be obtained from an individual using this minimally destructive extraction method versus standard sampling procedures that rely on cutting and powdering tooth samples. From each individual, we sampled a single multirooted tooth, from which the roots were removed via cutting (note that the tooth roots were cut in order to make it possible to process the samples using several independent methods, but cutting is not required by the minimally destructive sampling protocol) and each was randomly assigned to undergo one of the following extraction treatments. We extracted ancient DNA from a tooth root that was processed using the minimally destructive extraction protocol described in this paper (Method “MDE” for “Minimally Destructive Extraction”) and a second whole tooth root of the same tooth, that was completely powdered via milling (Method “WTR” for “Whole Tooth Root”). In four cases where a third tooth root was available, we extracted ancient DNA using the cementum-targeting approach described by Damgaard et al. (2015) (Method “D” for “Damgaard”). We also generated extracts from powder produced from petrous bones for 10 of the same individuals using the method described by Pinhasi et al. (2019) (Method “P” for “Petrous”). In one case (individual 3), we discovered through subsequent bioinformatic analyses that the petrous bone and tooth sampled did not originate from the same individual, and we therefore exclude the petrous bone results from further analyses. DNA preservation in two individuals (5 and 6) was uniformly poor, with no more than 10,000 sequences aligning to the 1.24 million sites captured through targeted enrichment (out of ∼5 million unique reads sequenced) from any of the libraries generated. Furthermore, all of these double-stranded libraries exhibited C-to-T damage rates at the terminal ends of molecules of less than 3%—the recommended minimum threshold for assessing ancient DNA authenticity in partially UDG-treated libraries (Rohland et al. 2015). These samples are considered to have “failed” screening for authentic ancient DNA and are not included in the statistical analyses. Additionally, individual 22 yielded relatively poor results for both treatments. Only 533 reads (out of ∼4 million unique reads sequenced) aligned to the 1.24 million sites targeted in the nuclear genome for the MDE treatment, making it impossible to calculate several of the reported metrics. Although we did obtain enough reads (23,239 reads out of ∼18 million unique reads sequenced) for some analyses to produce results for the tooth root that underwent Method WTR, the relatively low rate of mitochondrial match to the consensus (0.860) suggests that this sample is likely contaminated. Based on these results, we also chose to exclude individual 22 from statistical analyses. However, we note that there are no significant changes to the reported statistics when the excluded individuals are included in calculations for which metrics from both treatments are available (Supplemental Table S2). For all statistical calculations, we included data from all other samples, which were processed as either double-stranded (samples 1–10) or single-stranded (samples 11–30) libraries. Results where each of these methods was analyzed separately are reported in Supplemental Table S2.
Table 1.
Sample information
Physical impact of minimally destructive extraction protocol
We photographed each tooth root processed using the minimally destructive extraction protocol immediately prior to extraction and 24 h after extraction to allow for the complete drying of the roots (Supplemental Fig. S1). A slight degradation of the outer tooth root surface is visible for many of the samples, as the portion of the tooth root exposed to extraction buffer shows a visible change in color and/or diameter relative to the unexposed portion. In the case of two of the most poorly preserved samples (individuals 5 and 6), the tooth roots—one of which broke in two when cut from the tooth crown—crumbled during removal of the parafilm that covered the tops of the roots after the incubation in extraction buffer. These results suggest that users should exercise caution when applying this method to very friable teeth that are already susceptible to crumbling or being crushed.
Comparison of minimally destructive extraction protocol versus powder-based extraction protocols
Following bioinformatic processing, we generated summary statistics for each extract, including metrics of sample complexity and contamination rates (Table 1; Supplemental Table S1). In the following section, for each individual we compare the quality of ancient DNA yielded by the minimally destructive extraction method (Method MDE) to that produced by the destructive, traditional sampling methods (Methods WTR and P), using a Wilcoxon signed-rank test. The null hypothesis is that the difference between pairs of data generated using Method MDE and Method WTR or P follows a symmetric distribution around zero. The alternative hypothesis is that the difference between the paired data does not follow a symmetric distribution around zero. A threshold of P-value = 0.05 is used to denote significance which can only be achieved if there are a minimum of six samples represented in each test for which the data can be compared. As there were only four samples processed using Method D, we were unable to perform statistical comparisons involving these data.
Extraction efficiency
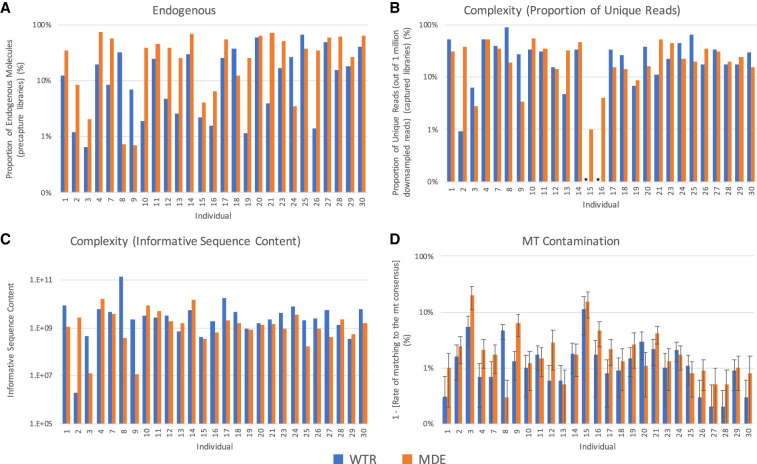
In order to assess the efficiency of the minimally destructive extraction method, we first compare the proportion of endogenous molecules (i.e., molecules that align to the human reference genome, hg19) in samples produced using each extraction method and sequenced via shotgun (i.e., precapture) sequencing. Although we observe a high degree of variability (Fig. 2A; Table 1) between treatment types for each individual, there is a statistically significant difference in the proportion of endogenous molecules sequenced using the MDE and WTR methods (P-value = 0.004), with an average of 35.8% and 18.8% endogenous molecules for each extraction method, respectively. These results support previous assertions that the outer cementum layer of the tooth root, which is targeted by the MDE method, contains a higher proportion of endogenous molecules than other portions of the tooth root (Damgaard et al. 2015). In contrast, we do not observe a significant difference in the proportion of endogenous molecules between Methods MDE and P (P-value = 1.000) (Supplemental Fig. S2A), with an average of 36.4% endogenous molecules observed when sampling from the petrous. These results are again consistent with claims that the petrous and tooth cementum both contain relatively high proportions of endogenous molecules (Damgaard et al. 2015; Hansen et al. 2017). Although the high proportion of endogenous molecules obtained using the MDE method is promising, measuring the fraction of endogenous molecules in a sample does not tell us about the total amount of DNA obtained using each method.
Figure 2.
Sample quality. A comparison of the quality of data produced by WTR (Whole Tooth Root) and MDE (Minimally Destructive Extraction) methods in samples that passed quality filtering. (A) The proportion of endogenous molecules in data obtained via shotgun sequencing. (B) The complexity of each sample, as measured by the proportion of unique reads out of 1,000,000 reads sequenced. Asterisks indicate that the total number of unique reads sequenced was below 1,000,000 for the specified sample, and therefore complexity estimates could not be generated. (C) The complexity of each sample, as measured by informative sequence content. (D) The rate of contamination is compared by considering the rate of matching to mitochondrial consensus sequence. Error bars indicate the 95% confidence interval. Only samples that passed quality screening are shown. Plots showing comparisons with samples generated using Method P are shown in Supplemental Figure S2.
We therefore consider the overall complexity—the number of unique molecules contained within a single library—using two metrics. In the first metric, we consider the proportion of unique molecules sequenced in each sample, after down-sampling to 1,000,000 sequences that align to the 1.24 million SNPs targeted during capture (Fig. 2B). This is a useful metric for comparison between samples as it is not biased by differences in sequencing depth across samples. However, as this metric is calculated using sequence data for samples that underwent targeted enrichment capture, a process that may introduce bias into the data, we therefore also consider a second complexity metric, the informative sequence content (Fig. 2C; Glocke and Meyer 2017). This metric quantifies the relative proportion of molecules that were successfully amplified from each sample using quantitative PCR (qPCR) analysis. The results are calibrated using the proportion of endogenous molecules and average length of molecules measured in the shotgun sequencing data, reflecting the number of sequences in the DNA extracts that can be aligned to the human genome.
Neither complexity metric finds a statistically significant difference between complexity measured in samples prepared using Method MDE versus Method WTR (P-value = 0.792 and 0.107, for the first and second complexity metrics, respectively), suggesting that using a minimally destructive extraction method does not result in loss of genetic data when sampling from teeth (Fig. 2B; Table 1). Although we find no statistically significant difference between samples prepared using Method MDE versus Method P using the first complexity metric (P-value = 0.091), we do detect a significant difference using the second metric (P-value = 0.043) (Supplemental Fig. S2B,C). We note that the power of this analysis is limited due to the low number of comparisons we were able to make (N = 7); therefore, this comparison may warrant further study, particularly because previous studies have found that the rates of ancient DNA preservation in cementum versus petrous samples are dependent upon sample preservation (Hansen et al. 2017).
Contamination rate
We were concerned that extracting ancient DNA directly from the outer layer of the tooth root might result in a higher rate of contamination in the sample, especially due to the increased potential for exposure of this region to contaminants during handling. Standard sampling protocols typically involve the physical removal of the outermost layer of bone or tooth prior to sampling, using a sanding disc or a sandblaster, whereas the minimally destructive extraction method specifically targets this outer layer following a superficial chemical (bleach) and brief (5–10 min) ultraviolet decontamination. We therefore compare the relative contamination rates between sampling methods using a variety of metrics. First, we compare the rate of matching to the mitochondrial consensus sequence (Fu et al. 2013). A minimum threshold of 95% is typically applied during screening of ancient DNA for population genetic studies. We observe substantial variability in contamination rate between and within individuals for all treatment methods (Fig. 2D; Table 1). Although we detect a significant difference between mitochondrial match to consensus rates between the MDE and WTR methods (P-value = 0.004), the average difference between these two methods is small (97.0% and 98.2%, respectively). Notably, this signal appears to be driven by less well-preserved samples. If we restrict to only comparisons where both samples have a minimum of 10% endogenous DNA (n = 13), we do not detect a significant difference in mt match to the consensus rate between the two extraction methods (P = 0.09), and if we raise this minimum threshold to 20% endogenous (n = 7), the comparison becomes even less significant (P = 0.87). Conversely, if we examine only the most poorly preserved specimens by restricting to cases where both samples have a maximum percent endogenous of 30% (n = 9), we still observe a significant difference between the two treatments (P = 0.03), suggesting that the loss of significance observed in the most well-preserved samples is not the result of a loss of statistical power due to the smaller number of samples analyzed. Further, we observe no significant difference between Methods MDE and P (P = 0.310) (Supplemental Fig. S2D).
Next, we estimate the autosomal rate of contamination, using the tool ContamLD (Nakatsuka et al. 2020), which measures the breakdown of linkage disequilibrium in a sequenced individual, a process which is accelerated by increased contamination. We again estimate relatively low rates of contamination across all samples and find no significant difference in contamination rates between Methods MDE and WTR (P-value = 0.490) or between Methods MDE and P (P-value = 0.893).
We also estimate contamination rates in the individuals who are identified as genetically male using ANGSD (Korneliussen et al. 2014). We obtain low estimates of contamination (≤2.5%) across all male samples (Table 1). Comparing the X Chromosome contamination estimates for the six genetically male individuals for whom there was enough data to produce estimates for both treatment types, we do not detect a significant difference between the MDE and WTR methods (P-value = 0.293). Taken together, these three estimates of contamination suggest that, in practice, the UV and bleach decontamination protocol used for the MDE method performs similarly to the physical surface removal decontamination steps implemented in the destructive protocols and is sufficient to produce ancient DNA data of analyzable quality.
We considered the read length distribution and frequency of C-to-T damage in the terminal bases of reads that aligned to the human genome (hg19) that were obtained via shotgun sequencing (i.e., precapture). Authentic ancient DNA is thought to consist of characteristically short fragments, with very few reads longer than 100 base pairs (Sawyer et al. 2012; Dabney et al. 2013b; Glocke and Meyer 2017); therefore, the read length distribution is used as a general metric to assess ancient DNA authenticity. We find that all samples appear to have read length profiles characteristic for authentic ancient DNA (Supplemental Fig. S3), and we do not observe a significant difference in median length of reads obtained using Methods MDE and WTR (P-value = 0.375). A weakly significant difference is observed between reads obtained using Methods MDE and P (P-value = 0.034) (Table 1), consistent with previous observations of systematic differences between DNA preservation in petrous and cementum samples (Parker et al. 2020); however, we also note that the use of a bleach-based decontamination step in the MDE protocol may have contributed to this observed pattern.
Endogenous ancient DNA samples are also thought to exhibit a high rate of C-to-T damage, particularly in the terminal bases. Using a partial or USER UDG treatment for double-stranded and single-stranded libraries, respectively (Rohland et al. 2015; Gansauge et al. 2020), we removed this damage in the interior of each molecule, while retaining it in the terminal bases. Therefore, we are able to use the frequency of these errors to assess ancient DNA authenticity. For samples processed using Methods MDE and WTR (P-value = 0.249), we observe no significant difference in the frequencies of C-to-T damage in terminal bases at the 5′ end of molecules that aligned to the human genome (hg19), obtained via shotgun sequencing. However, the distribution of damage rates in samples processed using Method P is significantly different from Method MDE (P-value = 0.028), with higher rates of damage observed in libraries produced using Method P in most (8/9) cases, again suggesting that there may be systematic differences between DNA preservation in petrous and tooth samples (Table 1; Supplemental Figs. S4, S5).
Although we were unable to perform statistical comparisons between the two cementum targeting methods (MDE and D) due to the small sample size, we note that we observed substantial variability between the performance of these two methods across all metrics (Supplemental Fig. S1; Supplemental Table S1). For all of the comparisons that we highlight (endogenous proportion, mt contamination rate, and both measures of complexity), neither method outperformed the other for all individuals in any category. These results suggest that both of these approaches are capable of producing high quality ancient DNA data, although further study would be required to determine whether either method is optimal from a data quality perspective. However, we emphasize that our primary concern in describing Method MDE is to produce an alternative, minimally destructive ancient DNA sampling approach. Like Method WTR, Method D involves the complete morphological destruction of the sampled tooth root and is therefore not comparable to Method MDE from a preservation standpoint.
Finally, we were concerned that the use of parafilm to cover portions of the tooth roots that we did not want exposed to the extraction buffer could serve as a possible source of contamination. We therefore created a parafilm extraction control, in which a small strip of parafilm (comparable in size to that used for covering the tooth roots) was added to a tube of extraction buffer and underwent sample processing along with the MDE samples and regular extraction blanks. We observe very few reads associated with this parafilm blank (Supplemental Table S1), suggesting that the use of parafilm does not serve as a significant source of contamination in the MDE method.
Discussion
This minimally destructive sampling protocol enables extraction of ancient DNA from the cementum portion of tooth roots that is of similar quality to ancient DNA obtained from teeth using traditional, destructive sampling methods that rely on powder produced through drilling or cutting and powdering. This is true with regard to both the amount of DNA that it is possible to obtain and the levels of contamination detected in the samples. In contrast, our results suggest that DNA sampled from the petrous bone exhibits more complexity than DNA sampled from the tooth cementum, indicating that there is still justification for choosing to sample from petrous bones over teeth when trying to maximize the chances of successfully sequencing ancient DNA, particularly in cases where sample preservation is poor—a circumstance in which ancient DNA sampled from petrous has previously been found to be of higher quality than in cementum (Hansen et al. 2017). However, the physical damage to the sampled tooth is substantially reduced and the overall morphological integrity of the sampled tooth is retained (with minimal degradation of the outer layer of the tooth root that is exposed to the extraction buffer) when using this minimally destructive sampling protocol, making this an optimal sampling method of teeth in cases where sample preservation is of the highest priority. The decision to use this minimally destructive method therefore requires an assessment of curatorial needs to maximize preservation versus the likelihood of obtaining analyzable data from the specimen.
One of the major concerns surrounding an extraction protocol that targets the outer surface of an ancient sample is the potential for an increase in contamination, as this outer surface may come in direct contact with various contaminants, particularly during handling. Because the majority of samples selected for ancient DNA analysis have been excavated and manipulated without any consideration for potential future genetic studies, this is of particular concern. Although destructive methods physically remove the outermost layer of bones and teeth to reduce contamination, we instead applied a bleach and UV decontamination procedure to the tooth before processing. We detected little difference in contamination rates between samples processed using this minimally destructive decontamination and sampling method and those processed using standard destructive methods. Further, these results suggest that this protocol, which includes a bleach-based decontamination step, does not significantly reduce DNA yields as compared to the WTR protocol, which does not involve the use of bleach. These results contrast with previously described bleach-based decontamination methods that involve soaking the sample in bleach for an extended period of time (e.g., Higgins et al. 2013), in which a substantial reduction in DNA yield was observed. By targeting the outer cementum tooth surface directly, this method maximizes the proportion of cementum matrix which is being digested and minimizes the amount of dentine sampled when compared to other cementum-targeting methods (Damgaard et al. 2015) which sample a significant proportion of the inner dentine layer in addition to the cementum. Furthermore, we find that parafilm can be used to protect portions of the tooth that users do not wish to sample (i.e., the tooth crown) from significant exposure to the extraction buffer without increasing contamination rates.
Although these results show that this minimally destructive approach is a promising alternative to destructive sampling methods that are traditionally applied to ancient teeth, we stress that further research is needed to determine whether it is recommended to opt for this sampling method in all circumstances. Particularly, we note that the majority of teeth chosen for this analysis were of moderate to excellent preservation status. The two most poorly preserved individuals included in this study contained too little DNA to allow for comparisons to be made between Methods MDE and WTR, and the tooth roots processed via Method MDE sustained damage during processing. Further study of the utility of this method on less well-preserved teeth is therefore of great interest.
We also note that this method may not be optimal for researchers who are interested in co-analyzing ancient pathogen DNA. This minimally destructive protocol targets the outer cementum layer of the tooth root, whereas ancient pathogen DNA is often best preserved in the inner pulp layer of teeth, which is highly vascularized and therefore more likely to be exposed to pathogens that are carried in the blood during the individual's life (Margaryan et al. 2018). Therefore, sampling the entire tooth root (or the dental pulp specifically) may be a better option for researchers who hope to obtain pathogen DNA in their analyses. This is another important factor that custodians of skeletal remains should consider when making decisions about which ancient DNA sampling protocol best fits their needs.
As the impact on gross dental morphology is minimal, this approach enables the preservation of samples for future morphological analyses. Furthermore, previous studies have shown that exposure to the chemicals used for ancient DNA extraction (namely EDTA and Proteinase K) do not affect a specimen's suitability for subsequent biochemical analyses, such as radiocarbon (AMS C14) dating (Korlević et al. 2018). Therefore, teeth processed using this minimally destructive protocol would remain suitable for future biochemical analyses.
This minimally destructive extraction method significantly reduces the amount of physical destruction caused by ancient DNA extraction, creating no holes or cuts in the sampled tooth or bone, while also shortening the overall length of the extraction protocol, without meaningfully increasing the amount of contamination. This method therefore may make it possible to extract ancient DNA from individuals that would otherwise be unavailable for ancient DNA study due to the destructive nature of traditional sampling methods.
Methods
All ancient DNA analyses were performed in dedicated clean rooms at the University of Vienna and Harvard Medical School. For individuals 1–10, skeletal sampling, preparation, and DNA extraction were performed at the University of Vienna. Library preparation, targeted enrichment capture, and sequencing were performed at Harvard Medical School. For individuals 11–30, skeletal sampling was performed at the University of Vienna, while all other processing was performed at Harvard Medical School.
Sampling
We selected skeletal elements from 30 ancient individuals of varying archaeological age, geographic origin, and degree of preservation for analysis (Table 1). From each individual, we selected a single multirooted tooth for sampling. For the first 10 individuals, we also selected a temporal bone for sampling. We UV-irradiated each tooth in a cross-linker for 5–10 min on each side, in order to remove as much surface contamination as possible. We then cut off the roots of each tooth using a diamond cutting disc and a hand-held Dremel drill, treating each root separately in all subsequent analyses. From each individual, we randomly selected one tooth root (Method MDE) for minimally destructive extraction. These tooth roots were subject to additional surface cleaning by wiping the teeth clean with a 2% bleach solution and rinsing with 95% ethanol, followed by UV-irradiation for 5–10 min on each side. We prepared the second set of tooth roots (Method WTR) by removing the extreme outer surface of each tooth root using a sanding disc and drill, and milling the root in a Retsch MM400 mixer mill for a total of 60 sec with a 10-sec break after 30 sec to produce a powder. For the four triple-rooted teeth included in the first round of sampling (from individuals 1, 2, 7, and 9), we performed a drilling-based targeted cementum extraction as described in Damgaard et al. (2015), first removing the extreme outer surface of the tooth root using a sanding disc and dentistry drill, and then removing as much of the interior pulp and dentine portion of the tooth root as possible using a dental drill and burr. The remaining outer portion of the tooth root, which is enriched in cementum relative to the root as a whole, was milled to produce a powder. We note that, although there may be underlying differences in the DNA preservation in the individual roots of each tooth, due to the random assignment of each individual's tooth roots to each treatment, we do not expect this preservation variability to bias the results of subsequent statistical analyses. Additionally, we obtained ∼50 mg of bone powder from the petrous portion of each of the 10 selected temporal bones, using standard methods (Method P) (Pinhasi et al. 2019).
DNA extraction
We prepared selected tooth roots (Method MDE) for minimally destructive extraction by recording the initial weight of the tooth root, then isolating the targeted portion of the tooth root using parafilm (Supplemental Fig. S6; see Supplemental Note S1 for a step-by-step description of the minimally destructive extraction method). We targeted the lower portion of the tooth root, where cellular cementum is concentrated. All other surfaces were wrapped in UV-decontaminated parafilm in order to prevent significant contact with the extraction buffer. The tooth roots were placed in 750 µL–1 mL of extraction buffer (0.45 M EDTA, 0.25 mg/mL Proteinase K, pH 8.0) (defined in Rohland and Hofreiter 2007) with the exposed portion pointing down and incubated for 2.5 h at 37°C, shaking gently. Following incubation, the roots were removed from the extraction buffer, which was then processed according to standard ancient DNA extraction procedures. Samples from individuals 1–10 underwent manual ancient DNA extraction, as described in Dabney et al. (2013a), with modifications. The MinElute columns were replaced with a pre-assembled spin column device (Roche) (as described in Korlević et al. 2015). We washed with 650 µL of PE buffer (Qiagen) and spun at 6000 rpm for 1 min. Following dry spin, we isolated the DNA by placing the spin column in a fresh 1.5-mL collection tube, and 25 µL TET buffer was pipetted onto the column's silica membrane, which was incubated at room temperature for 10 min and then spun at maximum speed for 30 sec. We repeated this step, producing a total of 50 µL of DNA extract. Samples from individuals 11–30 underwent robotic extraction following incubation, using the robotic protocol described in Rohland et al. (2018), using binding buffer D.
For samples processed using Methods WTR, P, and D, sampled powders were incubated overnight (∼18 h) in extraction buffer at 37°C, with gentle shaking. For samples from individuals 1–10, up to 50 mg powder was incubated in 1 mL extraction buffer, which then underwent manual extraction, as described above. For samples from individuals 11–30, ∼37 mg of bone powder was incubated in 750 µL extraction buffer and then underwent robotic extraction, as described above.
Negative controls were prepared alongside ancient DNA extracts for all extraction batches. In each case, extraction buffer was added to an empty tube prior to incubation, and the negative control was treated identically to all other samples during subsequent processing. Additionally, we generated one parafilm extraction control, by incubating a piece of UV-decontaminated parafilm in extraction buffer overnight in order to determine whether the parafilm coverings used to protect the ends of the tooth roots might be a potential source of contamination.
Following incubation in the extraction buffer, the roots were rinsed with 95% ethanol in order to remove any remaining extraction buffer and air-dried at room temperature for 24 h. The samples were then reweighed to assess the total amount of dental material digested.
Library preparation, enrichment, and sequencing
We prepared double-stranded (samples 1–10) or single-stranded (samples 11–30) libraries from 10 µL of each extract using UDG-treatment methods, as described in Rohland et al. (2015) and Gansauge et al. (2020), respectively. These methods remove ancient DNA damage at the interior of each DNA sequence, while preserving characteristic ancient DNA damage at the terminal ends of the molecules, to be used for ancient DNA authentication during bioinformatic processing. We enriched libraries for human DNA via targeted enrichment of the mitochondrial genome and at 1.24 million SNP sites that are informative for population genetic analyses (Fu et al. 2015; Haak et al. 2015; Mathieson et al. 2015). Following enrichment, libraries were sequenced on an Illumina NextSeq 500 machine, with 2 × 76 or 2 × 101 cycles, with an additional 2 × 7 or 2 × 8 cycles used for identification of indices, for double-stranded and single-stranded libraries, respectively.
Bioinformatic processing
We trimmed molecular adapters and barcodes from sequenced reads, and the merged paired-end reads, requiring an overlap of 15 base pairs (allowing up to three mismatches of low base quality [<20] or one mismatch of high base quality [≥20]) using custom software (https://github.com/DReichLab/ADNA-Tools). We then aligned the merged sequences to both the mitochondrial RSRS genome (Behar et al. 2012) and the hg19 human reference sequence using samse in BWA (v0.6.1) (Li and Durbin 2009). We note that we aligned to the hg19 human reference sequence in order to be consistent with the majority of previously published ancient DNA and do not expect that aligning to GRCh38 would in any way alter the findings of this study, which focuses on assessing the overall quality of the ancient DNA that we generated rather than performing population genetic comparisons. We identified duplicate reads, defined as having the same start and end position and orientation and a shared DNA barcode (unique quadruple barcode combinations are inserted during library preparation), and retained only the copy with the highest quality sequence.
We assessed ancient DNA authenticity using several metrics. We used the tool ContamMix (Fu et al. 2014) to determine the rate of matching between mitochondrial reads and the consensus sequence. We used the tool ContamLD to estimate the rate of contamination in the autosomes, based on the degree of breakdown of linkage disequilibrium observed in each library relative to a panel of representative individuals from the 1000 Genomes Project (Nakatsuka et al. 2020). We determined the amount of contamination in the X Chromosome for male individuals using the tool ANGSD (Korneliussen et al. 2014). Finally, we estimated the rate of C-to-T substitution at the terminal ends of molecules for each sample (Jónsson et al. 2013), and we studied the lengths of sequenced molecules.
We assessed the quality of ancient DNA observed by measuring the percent of endogenous (unique reads that align to the human genome), coverage (average number of reads aligning to each of the 1.24 million targeted SNP sites), and overall complexity of the sample—assessed by determining the proportion of unique reads sequenced, after randomly down-sampling to 1,000,000 on-target reads, or by measuring the informative sequence content (Glocke and Meyer 2017), in order to minimize bias caused by differences in sequencing depth.
Data access
All sequencing data generated in this study have been submitted to the European Nucleotide Archive (ENA; https://www.ebi.ac.uk/ena) under accession number PRJEB32750.
Competing interest statement
The authors declare no competing interests.
Supplementary Material
Acknowledgments
We thank Iñigo Olalde and Nathan Nakatsuka for contributions to the bioinformatic analyses. E.H. was supported by a graduate student fellowship from the Max Planck–Harvard Research Center for the Archaeoscience of the Ancient Mediterranean (MHAAM). D.R. is an Investigator of the Howard Hughes Medical Institute and this work was also supported by a John Templeton Foundation grant 61220. T.H., T.S., J.D., and K.K. were supported by a grant from the Hungarian Research, Development and Innovation Office, project number FK128013. A.A. was supported by a grant from the National Research, Development and Innovation Office (Grant K124326).
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.267534.120.
References
- Behar DM, van Oven M, Rosset S, Metspalu M, Loogväli E-L, Silva NM, Kivisild T, Torroni A, Villems R. 2012. A “Copernican” reassessment of the human mitochondrial DNA tree from its root. Am J Hum Genet 90: 675–684. 10.1016/j.ajhg.2012.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell LS, Kayser M, Jones C. 2008. The mineralized osteocyte: a living fossil. Am J Phys Anthropol 137: 449–456. 10.1002/ajpa.20886 [DOI] [PubMed] [Google Scholar]
- Bolnick DA, Bonine HM, Mata-Míguez J, Kemp BM, Snow MH, LeBlanc SA. 2012. Nondestructive sampling of human skeletal remains yields ancient nuclear and mitochondrial DNA. Am J Phys Anthropol 147: 293–300. 10.1002/ajpa.21647 [DOI] [PubMed] [Google Scholar]
- Bosshardt DD, Selvig KA. 1997. Dental cementum: the dynamic tissue covering of the root. Periodontol 2000 13: 41–75. 10.1111/j.1600-0757.1997.tb00095.x [DOI] [PubMed] [Google Scholar]
- Carpenter ML, Buenrostro JD, Valdiosera C, Schroeder H, Allentoft ME, Sikora M, Rasmussen M, Gravel S, Guillén S, Nekhrizov G, et al. 2013. Pulling out the 1%: whole-genome capture for the targeted enrichment of ancient DNA sequencing libraries. Am J Hum Genet 93: 852–864. 10.1016/j.ajhg.2013.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correa H, Carneiro L, Yoshitake N, Carneiro A, Bizo G. 2019. Powder -free DNA extraction from post-mortem teeth. J Forensic Res 10: 448. 10.13140/RG.2.2.21490.66242 [DOI] [Google Scholar]
- Dabney J, Knapp M, Glocke I, Gansauge M-T, Weihmann A, Nickel B, Valdiosera C, García N, Pääbo S, Arsuaga J-L, et al. 2013a. Complete mitochondrial genome sequence of a Middle Pleistocene cave bear reconstructed from ultrashort DNA fragments. Proc Natl Acad Sci 110: 15758–15763. 10.1073/pnas.1314445110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabney J, Meyer M, Pääbo S. 2013b. Ancient DNA damage. Cold Spring Harb Perspect Biol 5: a012567. 10.1101/cshperspect.a012567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damgaard PB, Margaryan A, Schroeder H, Orlando L, Willerslev E, Allentoft ME. 2015. Improving access to endogenous DNA in ancient bones and teeth. Sci Rep 5: 11184. 10.1038/srep11184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de León MSP, Koesbardiati T, Weissmann JD, Milella M, Reyna-Blanco CS, Suwa G, Kondo O, Malaspinas A-S, White TD, Zollikofer CP. 2018. Human bony labyrinth is an indicator of population history and dispersal from Africa. Proc Natl Acad Sci 115: 4128–4133. 10.1073/pnas.1717873115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman E. 1994. Periodontium. In Oral histology: development, structure, and function (ed. Ten Cate AR), pp. 276–312. Mosby, St. Louis. [Google Scholar]
- Fu Q, Meyer M, Gao X, Stenzel U, Burbano HA, Kelso J, Pääbo S. 2013. DNA analysis of an early modern human from Tianyuan Cave, China. Proc Natl Acad Sci 110: 2223–2227. 10.1073/pnas.1221359110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Q, Li H, Moorjani P, Jay F, Slepchenko SM, Bondarev AA, Johnson PL, Aximu-Petri A, Prüfer K, de Filippo C, et al. 2014. Genome sequence of a 45,000-year-old modern human from western Siberia. Nature 514: 445–449. 10.1038/nature13810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Q, Hajdinjak M, Moldovan OT, Constantin S, Mallick S, Skoglund P, Patterson N, Rohland N, Lazaridis I, Nickel B, et al. 2015. An early modern human from Romania with a recent Neanderthal ancestor. Nature 524: 216–219. 10.1038/nature14558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamba C, Jones ER, Teasdale MD, McLaughlin RL, Gonzalez-Fortes G, Mattiangeli V, Domboróczki L, Kővári I, Pap I, Anders A, et al. 2014. Genome flux and stasis in a five millennium transect of European prehistory. Nat Commun 5: 5257. 10.1038/ncomms6257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gansauge M-T, Aximu-Petri A, Nagel S, Meyer M. 2020. Manual and automated preparation of single-stranded DNA libraries for the sequencing of DNA from ancient biological remains and other sources of highly degraded DNA. Nat Protoc 15: 2279–2300. 10.1038/s41596-020-0338-0 [DOI] [PubMed] [Google Scholar]
- Glocke I, Meyer M. 2017. Extending the spectrum of DNA sequences retrieved from ancient bones and teeth. Genome Res 27: 1230–1237. 10.1101/gr.219675.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzesik WJ, Cheng H, Oh JS, Kuznetsov SA, Mankani MH, Uzawa K, Robey PG, Yamauchi M. 2000. Cementum-forming cells are phenotypically distinct from bone-forming cells. J Bone Miner Res 15: 52–59. 10.1359/jbmr.2000.15.1.52 [DOI] [PubMed] [Google Scholar]
- Haak W, Lazaridis I, Patterson N, Rohland N, Mallick S, Llamas B, Brandt G, Nordenfelt S, Harney E, Stewardson K, et al. 2015. Massive migration from the steppe was a source for Indo-European languages in Europe. Nature 522: 207–211. 10.1038/nature14317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen HB, Damgaard PB, Margaryan A, Stenderup J, Lynnerup N, Willerslev E, Allentoft ME. 2017. Comparing ancient DNA preservation in petrous bone and tooth cementum. PLoS One 12: e0170940. 10.1371/journal.pone.0170940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins D, Austin JJ. 2013. Teeth as a source of DNA for forensic identification of human remains: a review. Sci Justice 53: 433–441. 10.1016/j.scijus.2013.06.001 [DOI] [PubMed] [Google Scholar]
- Higgins D, Kaidonis J, Townsend G, Hughes T, Austin JJ. 2013. Targeted sampling of cementum for recovery of nuclear DNA from human teeth and the impact of common decontamination measures. Investig Genet 4: 18. 10.1186/2041-2223-4-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofreiter M. 2012. Nondestructive DNA extraction from museum specimens. In Ancient DNA: methods and protocols (ed. Shapiro B, Hofreiter M), pp. 93–100. Springer, New York. [DOI] [PubMed] [Google Scholar]
- Jónsson H, Ginolhac A, Schubert M, Johnson PL, Orlando L. 2013. mapDamage2.0: fast approximate Bayesian estimates of ancient DNA damage parameters. Bioinformatics 29: 1682–1684. 10.1093/bioinformatics/btt193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korlević P, Gerber T, Gansauge M-T, Hajdinjak M, Nagel S, Aximu-Petri A, Meyer M. 2015. Reducing microbial and human contamination in DNA extractions from ancient bones and teeth. BioTechniques 59: 87–93. 10.2144/000114320 [DOI] [PubMed] [Google Scholar]
- Korlević P, Talamo S, Meyer M. 2018. A combined method for DNA analysis and radiocarbon dating from a single sample. Sci Rep 8: 4127. 10.1038/s41598-018-22472-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korneliussen TS, Albrechtsen A, Nielsen R. 2014. ANGSD: analysis of next generation sequencing data. BMC Bioinformatics 15: 356. 10.1186/s12859-014-0356-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25: 1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarewicz C, Marom N, Bar-Oz G. 2017. Palaeobiology: Ensure equal access to ancient DNA. Nature 548: 158. 10.1038/548158a [DOI] [PubMed] [Google Scholar]
- Margaryan A, Hansen HB, Rasmussen S, Sikora M, Moiseyev V, Khoklov A, Epimakhov A, Yepiskoposyan L, Kriiska A, Varul L, et al. 2018. Ancient pathogen DNA in human teeth and petrous bones. Ecol Evol 8: 3534–3542. 10.1002/ece3.3924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieson I, Lazaridis I, Rohland N, Mallick S, Patterson N, Roodenberg SA, Harney E, Stewardson K, Fernandes D, Novak M, et al. 2015. Genome-wide patterns of selection in 230 ancient Eurasians. Nature 528: 499–503. 10.1038/nature16152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsuka NJ, Harney E, Mallick S, Mah M, Patterson N, Reich DE. 2020. ContamLD: estimation of ancient nuclear DNA contamination using breakdown of linkage disequilibrium. Genome Biol 21: 199. 10.1186/s13059-020-02111-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker C, Rohrlach AB, Friederich S, Nagel S, Meyer M, Krause J, Bos KI, Haak W. 2020. A systematic investigation of human DNA preservation in medieval skeletons. Sci Rep 20: 18225. 10.1038/s41598-020-75163-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinhasi R, Fernandes D, Sirak K, Novak M, Connell S, Alpaslan-Roodenberg S, Gerritsen F, Moiseyev V, Gromov A, Raczky P, et al. 2015. Optimal ancient DNA yields from the inner ear part of the human petrous bone. PLoS One 10: e0129102. 10.1371/journal.pone.0129102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinhasi R, Fernandes DM, Sirak K, Cheronet O. 2019. Isolating the human cochlea to generate bone powder for ancient DNA analysis. Nat Protoc 14: 1194–1205. 10.1038/s41596-019-0137-7 [DOI] [PubMed] [Google Scholar]
- Prendergast ME, Sawchuk E. 2018. Boots on the ground in Africa's ancient DNA ‘revolution’: archaeological perspectives on ethics and best practices. Antiquity 92: 803–815. 10.15184/aqy.2018.70 [DOI] [Google Scholar]
- Rohland N, Hofreiter M. 2007. Ancient DNA extraction from bones and teeth. Nat Protoc 2: 1756–1762. 10.1038/nprot.2007.247 [DOI] [PubMed] [Google Scholar]
- Rohland N, Siedel H, Hofreiter M. 2004. Nondestructive DNA extraction method for mitochondrial DNA analyses of museum specimens. BioTechniques 36: 814–821. 10.2144/04365ST05 [DOI] [PubMed] [Google Scholar]
- Rohland N, Harney E, Mallick S, Nordenfelt S, Reich D. 2015. Partial uracil–DNA–glycosylase treatment for screening of ancient DNA. Phil Trans R Soc B 370: 20130624. 10.1098/rstb.2013.0624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohland N, Glocke I, Aximu-Petri A, Meyer M. 2018. Extraction of highly degraded DNA from ancient bones, teeth and sediments for high-throughput sequencing. Nat Protoc 13: 2447–2461. 10.1038/s41596-018-0050-5 [DOI] [PubMed] [Google Scholar]
- Sawyer S, Krause J, Guschanski K, Savolainen V, Pääbo S. 2012. Temporal patterns of nucleotide misincorporations and DNA fragmentation in ancient DNA. PLoS One 7: e34131. 10.1371/journal.pone.0034131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirak KA, Sedig JW. 2019. Balancing analytical goals and anthropological stewardship in the midst of the paleogenomics revolution. World Archaeol 51: 560–573. 10.1080/00438243.2019.1617190 [DOI] [Google Scholar]
- Sirak KA, Fernandes DM, Cheronet O, Novak M, Gamarra B, Balassa T, Bernert Z, Cséki A, Dani J, Gallina JZ, et al. 2017. A minimally-invasive method for sampling human petrous bones from the cranial base for ancient DNA analysis. BioTechniques 62: 283–289. 10.2144/000114558 [DOI] [PubMed] [Google Scholar]
- Sirak K, Fernandes D, Cheronet O, Harney E, Mah M, Mallick S, Rohland N, Adamski N, Broomandkhoshbacht N, Callan K, et al. 2020. Human auditory ossicles as an alternative optimal source of ancient DNA. Genome Res 30: 427–436. 10.1101/gr.260141.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatkin M, Racimo F. 2016. Ancient DNA and human history. Proc Natl Acad Sci 113: 6380–6387. 10.1073/pnas.1524306113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao N, Foster B, Bonewald L. 2016. The cementocyte—an osteocyte relative? J Dent Res 95: 734–741. 10.1177/0022034516641898 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.