Abstract
CRISPR-Cas9 deletion (CRISPR-del) is the leading approach for eliminating DNA from mammalian cells and underpins a variety of genome-editing applications. Target DNA, defined by a pair of double-strand breaks (DSBs), is removed during nonhomologous end-joining (NHEJ). However, the low efficiency of CRISPR-del results in laborious experiments and false-negative results. By using an endogenous reporter system, we show that repression of the DNA-dependent protein kinase catalytic subunit (DNA-PKcs)—an early step in NHEJ—yields substantial increases in DNA deletion. This is observed across diverse cell lines, gene delivery methods, commercial inhibitors, and guide RNAs, including those that otherwise display negligible activity. We further show that DNA-PKcs inhibition can be used to boost the sensitivity of pooled functional screens and detect true-positive hits that would otherwise be overlooked. Thus, delaying the kinetics of NHEJ relative to DSB formation is a simple and effective means of enhancing CRISPR-deletion.
CRISPR-Cas9 technology enables a variety of loss-of-function perturbations to study the functions of genomic elements in their natural context and engineer natural and unnatural mutations (Cong et al. 2013; Mali et al. 2013; Doench 2018). One such application, CRISPR deletion (CRISPR-del), is the primary means of permanently removing targeted genomic regions. CRISPR-del uses a pair of CRISPR-Cas9 complexes to introduce double-strand breaks (DSBs) at two sites flanking the target region. Paired DSBs are repaired by the endogenous nonhomologous end-joining (NHEJ) pathway, which frequently ejects the intervening fragment, resulting in a genomic deletion (Yang et al. 2013; Maddalo et al. 2014; Ho et al. 2015; Vidigal and Ventura 2015). Less frequently, the target region may be inverted without deletion (Canver et al. 2014; Antoniani et al. 2018; Watry et al. 2020). Cas9 complexes are recruited to the two edges of the target region via a user-defined pair of single guide RNAs (sgRNAs, or “pgRNAs” for the pair) (Pulido-Quetglas et al. 2017), delivered by transfection or viral transduction (Aparicio-Prat et al. 2015; Vidigal and Ventura 2015).
CRISPR-del has been used to successfully remove genomic regions from 101 to 106 bp (Canver et al. 2014). This range has enabled researchers to test the function of a variety of genomic elements, including gene regulatory sequences (Canver et al. 2015; Mochizuki et al. 2018; Gasperini et al. 2019), noncoding RNAs (Han et al. 2014; Ho et al. 2015; Holdt et al. 2016; Koirala et al. 2017; Xing et al. 2017), and structural elements (Huang et al. 2018). Engineered deletions have also been used to model human mutations (Lupiañez et al. 2015; Nelson et al. 2016).
CRISPR-del is readily scaled to high-throughput pooled screens, delivered via lentiviral libraries of thousands of pgRNAs (Aparicio-Prat et al. 2015; Vidigal and Ventura 2015). This has been used to discover long noncoding RNAs (lncRNAs) regulating cancer cell proliferation (Zhu et al. 2016; Liu et al. 2018) and to map cis-regulatory regions of key protein-coding genes (Diao et al. 2017; Gasperini et al. 2017). Screens are most commonly performed in transformed cell lines, for reasons both scientific (identification of novel cancer genes) and practical (requirement for large and homogeneous cell populations) (Zhu et al. 2016; Liu et al. 2018; Esposito et al. 2019). Lentiviral delivery of pooled libraries introduces practical constraints, requiring both sgRNAs of a pair to be encoded in a single vector (to ensure simultaneous delivery), and low multiplicity of infection (MOI; for each cell to receive a low number of perturbations) (Vidigal and Ventura 2015; Zhu et al. 2016; Gasperini et al. 2017; Doench 2018; Esposito et al. 2019).
The principal drawback of CRISPR-del is the low efficiency with which targeted alleles are deleted. Studies on cultured cells typically report efficiencies in the range 0%–50% of alleles, and often <20% (Mandal et al. 2014; Thomas et al. 2020), with similar estimates from individual clones (Canver et al. 2014; Aparicio-Prat et al. 2015; Ho et al. 2015; Vidigal and Ventura 2015; Pulido-Quetglas et al. 2017). Indeed, a recent publication reported high variation in the efficiencies of pgRNA targeting the same region, including many that yielded negligible deletion (Thomas et al. 2020). Transfection typically yields greater efficiency than viral transduction, possibly owing to higher sgRNA levels (Mangeot et al. 2019), but is incompatible with pooled screening. Low deletion efficiency introduces the need to isolate homozygous knockout clones by laborious expansion of hundreds of single cells (Aparicio-Prat et al. 2015). Resulting clones may not be representative of the general population (Stojic et al. 2018).
More important than these practical costs, is the potential impact of low deletion rates on the ability to discern bona fide functional effects arising from a given mutation (Thomas et al. 2020). Nonperforming pgRNAs are a particular problem for pooled CRISPR-del screens, where they reduce statistical power and lead to false-negative results. To overcome this, researchers are obliged to increase the number of deletion constructs per target and the number of cells screens, resulting in lower throughputs and increased costs (Doench et al. 2016; Sanson et al. 2018). Consequently, any method to improve CRISPR-del efficiency promises to streamline experiments and promote the discovery of presently overlooked functional elements.
CRISPR-del depends on NHEJ, the principal pathway for repairing Cas9-induced DSBs (Brinkman et al. 2018). In mammalian cells, the NHEJ pathway is divided in three steps: (1) DSB recognition and end binding, in which the exposed DNA ends are recognized by the Ku70/80 complex (Chang et al. 2017); (2) end processing (resection of mismatched nucleotides and DNA resynthesis), based on activation of the nuclease Artemis and Pol X family polymerases by the serine-threonine kinase DNA-PKcs (Chang et al. 2017); and (3) end ligation by DNA ligase complex (DNA ligase 4, XRCC4, and XLF) (Chang et al. 2017).
For some applications of CRISPR, notably precise genome editing using homologous recombination (HR), efficiency can be substantially improved through pharmaceutical strategies (Yeh et al. 2019). Here, editing events are rare, and HR is the rate-limiting step (Mao et al. 2008; Miyaoka et al. 2016). Two alternative strategies have been successful: (1) direct stimulation of homology-directed repair (HDR) (Lin et al. 2014a; Song et al. 2016; Riesenberg and Maricic 2018; Yeh et al. 2019); (2) suppression of the competing NHEJ pathway at early stages through inhibition of Ku70/80 complex (Fattah et al. 2008; Riesenberg and Maricic 2018; Yeh et al. 2019) or DNA-dependent protein kinase catalytic subunit (DNA-PKcs) (Robert et al. 2015; Riesenberg and Maricic 2018; Riesenberg et al. 2019; Yeh et al. 2019) or, at late phases, via DNA ligase 4 inhibition (Chu et al. 2015; Maruyama et al. 2015; Riesenberg and Maricic 2018; Yeh et al. 2019). To date, however, there are no reported methods for pharmacological enhancement of CRISPR-del.
Upon paired DSB, NHEJ can promote several distinct outcomes: (1) the intermediate fragment is removed, and the distal DNA edges ligated (leading to successful deletion); (2) each of the two DSB is repaired independently, leaving small indels at both target sites (no deletion); (3) the intermediate fragment is flipped, and the DSB are repaired (creating an inversion). A fourth possibility is that DSBs are repaired via HDR using the sister's chromatid as a template, which can be retargeted by the CRISPR complex. Systematic studies have concluded that genomic deletion is only the second most frequent outcome, after scenario 2 (up to 70% of the cases) (Canver et al. 2014; Antoniani et al. 2018). This implies that the greatest impediment to CRISPR-del is the process in which both DSBs occur at different times, creating genomic scars that prevent further deletion. Only when both CRISPR complexes act in a synchronized fashion can deletion occur (Fig. 1A). From this, we hypothesize that deletion will be favored when paired DSBs occur on a timescale shorter than that required for NHEJ.
Figure 1.
Poor deletion efficiency of paired sgRNAs and CRISPR-deletion (CRISPR-del) model. (A) Model for CRISPR-del and its improvement by inhibition of nonhomologous end-joining (NHEJ). (B) Reanalysis of significant growth-promoting gene “hits” identified in the CRISPR-del negative selection screen in Huh7 cells from Zhu et al. (2016). Each hit is targeted by several individual paired sgRNAs. Histogram displays the percentage of paired sgRNAs that effectively perturb their target gene. The dashed line represents the median (42.9%). (C) Scatterplot displays paired sgRNAs targeting gene hits from B. The x-axis indicates the mean of the two sgRNAs predicted on-target efficiency (calculated with the Rule Set II scoring algorithm from Doench et al. 2016); y-axis, log2-transformed fold-change in paired sgRNA detection after cell passaging (growth phenotype). R indicates Pearson's correlation; P, P-value.
A prediction of this, is that deletion can be promoted by reducing the rate of NHEJ, thereby increasing the likelihood that both DSBs co-occur. Here, we test this hypothesis, by examining the effect on CRISPR-del efficiency of pharmacologically slowing the rate of NHEJ during DSB formation, through the inhibition of DNA-PKcs (iDNA-PKcs).
Results
A quantitative endogenous reporter for CRISPR-del
CRISPR-del is an important loss-of-function perturbation for both low- and high-throughput studies, yet its relatively low efficiency remains a major impediment. We reanalyzed data from a published high-throughput pooled screen, in which growth promoting genes were each targeted by several pgRNAs (Zhu et al. 2016). Less than half of pgRNAs targeting “hit” genes yielded a detectable phenotype, indicating that the majority of screened pgRNAs do not efficiently delete their targets (Fig. 1B). Similar results were reported by Canver et al. (2014), where 65% of pgRNAs yielded deletion efficiency <20% (Canver et al. 2014). Furthermore, there was no correlation between phenotype and average on-target score of the two sgRNAs (Fig. 1C; Zhu et al. 2016), suggesting that deletion efficiency does not simply depend on the aggregate of sgRNA quality.
To identify factors capable of improving CRISPR-del efficiency, we designed a gene-based reporter system: CRISPR Deletion Endogenous Reporter (CiDER). Such a system should be quantitative, sensitive, and practical and be able to closely model the CRISPR-del process by targeting endogenous genes rather than plasmids. We focused on genes encoding cell-surface proteins, as they can be rapidly and sensitively detected by flow cytometry (Bausch-Fluck et al. 2015). A number of candidates were considered with criteria of (1) nonessentiality for cell viability and proliferation (Luo et al. 2008; Meyers et al. 2017; Tsherniak et al. 2017; https://depmap.org), (2) high expression in human cell lines (Thul et al. 2017; http://www.proteinatlas.org), (3) lack of overlap with other genomic elements that could lead to false-positive detection, and (4) availability of flow cytometry grade antibody. Consequently, we selected PLXND1 encoding the plexin D1 protein (Supplemental Fig. S1), present in three genomic copies in HeLa cells and HEK293T and two in HCT116 cells (Cancer Cell Line Encyclopedia [CCLE]) (Barretina et al. 2012; Lin et al. 2014b; https://portals.broadinstitute.org/ccle/data; www.hek293genome.org).
We conceived an experimental setup in which successful CRISPR-del leads to loss of PLXND1 expression but unsuccessful events do not. In this scheme, the gene's first exon is targeted for deletion by a series of pgRNAs recognizing the non-protein-coding regions upstream (promoter) and downstream (first intron) (Fig. 2A). Successful deletions of the first exon are expected to silence protein expression, but indels from individual sgRNAs do not affect the protein sequence directly and should not lead to silencing. It should also be noted that inversions will also silence plexin D1 expression and be called as mutations by this reporter, and therefore, results should be interpreted with care. Finally, we also designed a control pgRNA that directly targets the open reading frame (ORF), expected to yield maximal protein silencing independent of deletion (designated positive control, P+).
Figure 2.
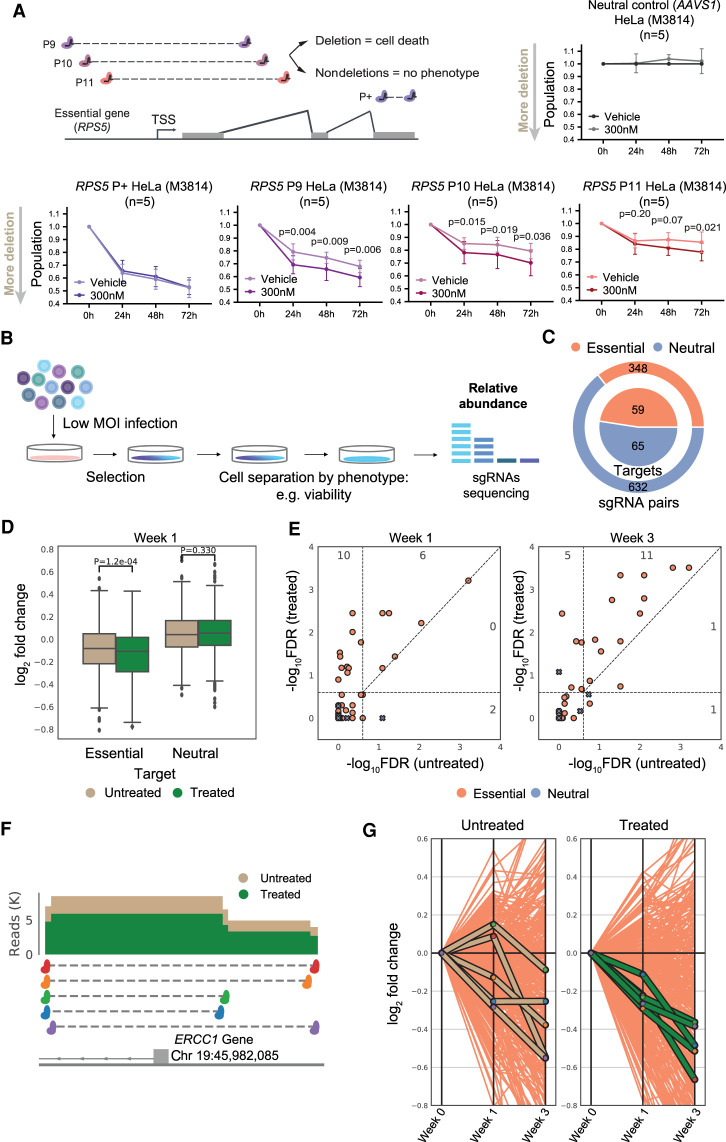
CiDER reporter system identifies DNA-PKcs inhibition as a means to increase CRISPR-del efficiency. (A) The CiDER endogenous reporter relies on a series of sgRNA pairs targeting exon 1 of PLXND1 gene, whose protein product is detected by flow cytometry. (B) CiDER measurements of CRISPR-del efficiency over a range of times between sgRNA delivery and cell harvesting (mean, standard deviation). (C) Independent evaluation of CRISPR-del efficiency in sorted plexin D1–negative and D1–positive cells. (Top) Representative fluorescent activated cell sorting (FACS) plot. Plexin D1–negative (APC-) and Plexin D1–positive cells (APC+) are gated. (Bottom) The fraction of nondeleted alleles in each population quantified by qPCR (mean, standard deviation). The black bar corresponds to a nontargeting control used for normalization. Green bars correspond to sorted HeLa cells with pgRNAs targeting PLXND1 (P3). (D) CRISPR-del efficiency measured by CiDER in HeLa upon DNA-PKcs inhibition (mean, standard deviation, one-tailed paired t-test). (E, top) Representative raw flow cytometry plots for HeLa upon DNA-PKcs inhibition. Plexin D1–positive cells are gated, and numbers correspond to percentage of cells. (Bottom) Representative images of individual sorted, stained HeLa cells. (F) CiDER measurements of CRISPR-del efficiency in HeLa upon DNA-PKcs inhibition with indicated small molecules (values: mean plexin D1–positive cells; *P < 0.05, one-tailed paired t-test).
We used flow cytometry to evaluate plexin D1 protein levels (Fig. 2B). Positive control sgRNAs (P+) yielded ∼90% knockout efficiency. We observed wide variability in the deletion efficiency of pgRNAs, from Pair1 (P1), displaying minimal efficacy, to the most efficient P4, yielding ∼40% deletion. Therefore, these pgRNAs achieve deletion efficiencies that are comparable to previous studies (Canver et al. 2014; Pulido-Quetglas et al. 2017). Measured deletion rates were consistent across biological replicates (Fig. 2B).
We next assessed whether flow cytometry was a good surrogate for genomic deletion and whether loss of plexin D1 expression required mutation of all alleles within a cell. To do so, we separated plexin D1–negative and plexin D1–positive cell populations via fluorescence-activated cell sorting (FACS) and detected remaining “wild-type” alleles (referring to nondeleted and noninverted alleles) with a genomic quantitative PCR (quantitative CRISPR PCR [QC-PCR]) (Pulido-Quetglas et al. 2017). We observed that plexin D1–negative cells show an almost complete loss of wild-type alleles (Fig. 2C; note the inverted scale used for QC-PCR). In summary, the flow cytometry–based CiDER assay is conservative: It classifies only homozygous knockout cells as “mutant,” and remaining wild-type and heterozygous cells as “wild type.” Furthermore, the greater a cell's ploidy for PLXND1, the more stringent this definition becomes.
Finally, the observed loss of plexin D1 was not owing to large indels or disruption of gene regulatory elements at individual sgRNA target sites (Kosicki et al. 2018), because control experiments with single sgRNAs showed no loss of plexin D1 (Supplemental Fig. S2). In CiDER, we have a reproducible and practical reporter of CRISPR-del at a range of efficiencies.
Inhibition of DNA-PKcs during DSB formation increases CRISPR-del efficiency
We hypothesized that reducing the rate of NHEJ during DSB formation would favor CRISPR-del, by increasing the chance that both DSBs will co-occur (Fig. 1A). We tested DNA-PKcs, a DNA end-binding factor at the first step of NHEJ pathway, for which a number of small-molecule inhibitors are available (Harnor et al. 2017). We began by treating HeLa cells with the inhibitor M3814 (IC50 = 3 nM) (Fuchss et al. 2014; Zenke et al. 2016; Riesenberg et al. 2019) at two concentrations (300 and 900 nM). Importantly, cells constitutively expressing Cas9 were treated for an 18-h time window, 4 h after sgRNA plasmid delivery by transfection. Thus, DNA-PKcs was inhibited immediately before sgRNA expression. This resulted in improved deletion rates for all four pgRNAs, including a 17-fold increase for P1, which otherwise displays negligible deletion under normal conditions (Fig. 2D,E; Supplemental Fig. S3A,B).
The effect of inhibition of DNA-PKcs (iDNA-PKcs) was concentration dependent, and concentrations <300 nM were ineffective (Supplemental Fig. S4A). Inhibition of DNA-PKcs for <18 h did not improve CRISPR-del efficiency, whereas longer treatments showed slightly improved deletion rates (Supplemental Fig. S4B). Chronic iDNA-PKcs can be toxic for some cell types; therefore, we opted for 18-h treatments in this study. We hypothesized that observed improvements of CRISPR-del are a consequence of repression, but not complete abolition, of DNA-PKcs and NHEJ activity. Consistent with this, we evaluated levels of active (phospho) DNA-PKcs under conditions used here (M3814 300 nM, 18-h treatment), and observed substantial residual activity (Supplemental Fig. S4C). Similar observations were recently reported by Zenke et al. (2020).
We next investigated whether iDNA-PKcs alters the nature of editing events taking place at target sites. We performed Sanger sequencing at a number of deleted alleles and observed a mild reduction in the preciseness of editing (Supplemental Fig. S5A,B).
A more important issue is the potential effect of iDNA-PKcs on inversion rates: It is possible that CiDER miscalls increase in inversions as the observed increases in deletions. The literature indicates that inversions occur at nonnegligible rates, but less frequently than deletions. Canver et al. (2014) estimated an overall deletion rate, across 17 targets of varying size, to occur at approximately one-third the rate of deletions (5.8% vs. 14.2%, respectively). A more recent study found this ratio to be approximately one-half (Watry et al. 2020). We evaluated how this might affect our results. First, we cloned and sequenced nondeleted alleles from vehicle and M3814-treated HeLa and observed a similar rate of inversions (8/14 cases and 8/17 cases, respectively) (Supplemental Fig. S7A,C). Second, we used quantitative PCR with inverted primers to evaluate the effect of M3814 (Supplemental Fig. S3C), observing either no change in inversion rates (HeLa, HEK293T) or a moderate approximately 1.5-fold increase, less than the overall approximately 2.5-fold gain in loss of function (HCT116) (Fig. 3B). In summary, this leads us to conclude that observed change in loss of function we observe owing to iDNA-PKcs is mainly attributable to deletions.
Figure 3.
DNA-PKcs inhibition boosts CRISPR-del independent of cell, readout, or sgRNA delivery method. (A) CRISPR-del efficiency of CiDER in HeLa upon DNA ligase 4 inhibition (mean). (B) CiDER measurement of CRISPR-del efficiency in HCT116 and HEK293T cell lines upon DNA-PKcs inhibition (mean, standard deviation, one-tailed paired t-test). (C) CRISPR-del efficiency at MALAT1-enhancer upon DNA-PKcs inhibition. (Left) The fraction of WT allele quantified by qPCR (mean, standard deviation, one-tailed paired t-test). (Center) The pgRNAs, PCR primers, and a representative agarose gel from the genomic PCR of the region. (Right) Quantification of the KO band from the gel. (D) CRISPR-del efficiency of CiDER in HeLa as a result of initiating DNA-PKcs inhibition at 24 h after lentiviral infection (MOI = 0.3).
Our hypothesis also predicts that other inhibitors of DNA-PKcs yield a similar effect. We treated cells with four other commercially available inhibitors at a concentration of 10 µM: KU57788 (IC50 = 14 nM), NU7026 (IC50 = 230 nM), LTURM34 (IC50 = 34 nM), and DMNB (IC50 = 15 µM) (Fig. 2F). Each one yielded increases in CRISPR-del efficiency to varying degrees, correlating with published differences on the inhibition potency (Mohiuddin and Kang 2019). As expected based on previous literature, KU57788 gave the strongest effect (Mohiuddin and Kang 2019), and DMNB gave the weakest effect, likely owing to its high IC50.
We were curious whether improved deletion depends on inhibition specifically of DNA-PKcs or more generally on NHEJ. To answer this, we used SCR7 pyrazine to inhibit another step in NHEJ, the final ligation by DNA ligase 4. In contrast to iDNA-PKcs, this treatment did not improve deletion efficiency (Fig. 3A). This might be expected, because at this late stage, the NHEJ machinery (DNA end-binding and processing complex) has already brought together the free DNA ends. Thus, CRISPR-del efficiency improvements depend specifically on inhibition of DNA-PKcs activity. Altogether, we have shown that pharmacological inhibition of NHEJ at the DNA-PKcs step yields enhanced deletion of PLXND1 reporter in HeLa cells.
Generality of deletion enhancement by DNA-PKcs inhibition
We next assessed whether iDNA-PKcs is more generally effective across cell lines, genomic targets, and sgRNA delivery modalities.
We began by replicating CiDER experiments in two widely used cell lines, HCT116 and HEK293T (Fig. 3B; Hart et al. 2015; Li and Richard 2016; Liu et al. 2017; Thomas et al. 2020). Both have baseline CRISPR-del efficiency below HeLa, possibly owing to weaker NHEJ activity (Miyaoka et al. 2016). Nevertheless, iDNA-PKcs enhanced deletion in both cell backgrounds.
We next assessed whether these effects hold for other loci. We previously established QC-PCR to measure rates of deletion at the MALAT1 enhancer region (see primer setup in Fig. 3C, central panel; Pulido-Quetglas et al. 2017). For three out of four pgRNAs, we observed a significant enhancement of deletion with M3814 treatment of HeLa (Fig. 3C). It should be noted that the pgRNAs used here are more efficient (≥80%) than for PLXND1, reducing the scope for large improvements in deletion efficiency. Similar but weaker results were also observed for HCT116 (two out of four pairs) and HEK293T cells (one out of four pairs) (Supplemental Fig. S6A,B). We confirmed that M3814 treatment does not substantially alter the nature of editing at deleted junctions by Sanger sequencing (Supplemental Fig. S7A,B). We next assessed whether nondeleted alleles carry genomic scars at the sgRNA sites. Because we did not manage to amplify the full-length PCR product from PLXND1, we amplified nondeleted alleles from the MALAT1 enhancer region and performed Sanger sequencing of one of the junctions. We detected high rates of sgRNA-disrupting mutations at target sites (5/6 for untreated, 5/9 for treated samples), supporting the notion that mutations from uncoordinated DSBs prevent subsequent deletion (Supplemental Fig. S7A,C).
CRISPR-del is often used to edit nontransformed cells; therefore, we sought to test the effectiveness of iDNA-PKcs under such conditions. However, poor delivery of Cas9 was a substantial impediment in the various human primary fibroblast cell types we tested. In human induced pluripotent cells (hiPSCs), we managed to deliver Cas9 protein and sgRNAs and observed promising but not statistically significant increase in deletion with M3814 treatment in three biological replicates (Supplemental Fig. S8; Supplemental Methods). Future studies will be dedicated to optimize CRISPR-del in pluripotent and other nontransformed cells.
Taken together, these findings support the general applicability of iDNA-PKcs for boosting CRISPR-del, although optimization will likely be required on a cell-by-cell basis.
DNA-PKcs inhibition boosts sensitivity of high-throughput pooled screens
One particularly powerful application of CRISPR-del is in pooled functional screens (Esposito et al. 2019). Here, libraries of pgRNAs are delivered by lentivirus to cultured cells, and phenotypic consequences of deleting hundreds to thousands of genomic elements can be assayed in parallel.
We first asked whether iDNA-PKcs is practical under these conditions, by targeting the PLXND1 reporter with sgRNAs delivered by standard low-MOI lentiviral infection. In initial experiments, M3814 was added to cell media at the same time of lentiviral transduction, but almost no improvement in deletion efficiency was observed (Supplemental Fig. S9A). This is explained by the fact that lentiviruses require NHEJ for genomic integration (Li et al. 2001; Daniel et al. 2004). Therefore, we modified our protocol so as to leave sufficient time for viral integration before NHEJ inhibition (24 h was optimal) (Supplemental Fig. S9B). This resulted in a 2.7-fold increase in CRISPR-del efficiency (Fig. 3D).
Cell population is a widely used phenotypic readout of pooled screens, not only for its practicality but also because it is a proxy for fitness and can identify genes required for enhanced growth or survival of cancer cells (Doench 2018; Esposito et al. 2019). To test whether improved CRISPR-del translates into stronger phenotypes, we developed a simple assay for the effect of CRISPR-del on cell survival. Analogous to PLXND1 (Fig. 2A), we designed three pgRNAs targeting the first exon (but not ORF) of the essential gene, RPS5 (coding for the 40S ribosomal protein S5, P46782, UniProt): RPS5-P9, P10, P11. As expected, sgRNAs targeting the AAVS1 locus had no effect, whereas positive control sgRNAs targeting the RPS5 ORF (RPS5-P+) resulted in ∼47% mortality after 72 h (Fig. 4A). pgRNAs targeting the first exon (P9, P10, P11) resulted in a substantial mortality (32%, 21%, and 15%, respectively), which was significantly enhanced by addition of M3814 (41%, 30%, and 22%, respectively). Therefore, pharmacologically improved CRISPR-del yields enhanced cellular phenotypes.
Figure 4.
Applying DNA-PKcs inhibition to pooled CRISPR-del screens. (A) Testing the functional benefit of DNA-PKcs inhibition. (Left) Experimental setup, with pgRNAs targeting the transcription start site (TSS) of the essential gene RPS5. A positive control sgRNA pair (P+) target the gene open reading frame and thus cause loss of function independent of deletion. (Right) Viability assays with pgRNAs targeting the nonessential AAVS1 locus. (Bottom) Viability assays after RPS5 TSS deletion (mean, standard deviation, one-tailed paired t-test). (B) Design of a typical pooled high-throughput screen to identify essential genes. (C) Composition of pgRNA library targeting both essential genes’ TSS and nonessential neutral loci. (D) Log2 fold-change (LFC) in abundance of indicated pgRNAs compared with day 0. Significance calculated by two-tailed t-test. (E) Hits reported by MAGeCK at two timepoints. Negative log10 false-discovery rate (−log10FDR) for treated and untreated samples are indicated in the y- and x-axis, respectively. Each point represents a target. A hit is called a true positive (TP) if it is targeting an essential gene and has an FDR <0.25. Points above the diagonal indicate hits with a lower FDR (higher −log10FDR) in the treated sample. Numbers in plot reflect TPs in each combination of treated/untreated cells. (F) pgRNAs targeting ERCC1 TSS: read coverage for untreated and treated samples at 1 wk. (G) Fold-change variation for individual ERCC1 pgRNAs across timepoints: x-axis, timepoint (0, 1, and 3 wk); y-axis, LFC in abundance. Orange lines represent all pgRNAs targeting essential genes.
We sought to test the benefit of iDNA-PKcs in a realistic fitness screen in cancer cells (Fig. 4B; Zhu et al. 2016; Esposito et al. 2019). To do so, we created a library of pgRNAs targeting positive-control essential genes (348 pgRNAs, 59 target genes) and neutral-control intergenic regions (632 pgRNAs, 65 target regions) (Fig. 4C). Similar to above, pgRNAs for positive control regions flanked the gene TSS without targeting the ORF, so that loss of fitness should only be observed in the case of a deletion (Fig. 4A). We transduced Cas9-expressing HeLa (cervical carcinoma) with this library at low MOI (0.35) and then divided them into two populations, one of which was treated with M3814 (18 h, 300 nM). Cells were passaged for periods of 1 and 3 wk, to allow loss of fitness to manifest in decreases in pgRNA populations (“drop out”), and then pgRNA populations were counted by deep massively parallel sequencing.
We observed that M3814 treatment significantly enhanced drop-out of pgRNAs targeting essential genes, but not neutral controls, after 1 wk (Fig. 4D,F,G) and 3 wk (Supplemental Fig. S10B). Although we observed the expected inverse relationship between deletion size and efficiency (Canver et al. 2014), there was no preference for M3814 to increase the rate of shorter or longer deletions (Supplemental Fig. S10C). Next we combined the results from individual pgRNAs to identify significant gene hits, and we could correctly identify essential genes with greater sensitivity in M3814-treated versus untreated cells after 1 wk (16/59 27.1%% vs. 8/59 13.5%, respectively) and 3 wk (17/59 28.8% vs. 13/59 22.0%, respectively) (Fig. 4E). More generally, gene-level false-discovery rates (FDRs) were consistently lower in M3814-treated cells (Fig. 4E). On the contrary, M3814 led to no increase in false-positive predictions, as measured by neutral targets called as hits (Fig. 4E). A replicated screen at higher MOI (0.6) yielded similar findings (Supplemental Fig. S10A,B). In conclusion, pharmacological inhibition of DNA-PKcs can be used to improve the sensitivity of pooled functional screens based on CRISPR-del.
Discussion
The intrinsic DNA damage response underpins CRISPR-Cas9 genome editing and may be manipulated to favor desired editing outcomes. In the case of precise genome editing, efficiency has been substantially improved through pharmacological promotion of HDR and inhibition of the competing NHEJ pathway (Yeh et al. 2019). No such solutions have been developed for CRISPR-del, despite its being one of the most common CRISPR-Cas9 modalities with diverse scientific and technological applications (Han et al. 2014; Canver et al. 2015; Ho et al. 2015; Lupiañez et al. 2015; Holdt et al. 2016; Nelson et al. 2016; Koirala et al. 2017; Xing et al. 2017; Huang et al. 2018; Mochizuki et al. 2018; Gasperini et al. 2019).
We hypothesized that successful CRISPR-del requires paired DSBs to co-occur before NHEJ has time to act and thus may be enhanced by pharmacological inhibition of DNA-PKcs. This is initially counter-intuitive, as DNA-PKcs is a necessary step in the NHEJ pathway upon which CRISPR-del relies, and its inhibition is widely used to promote the competing HDR pathway (Robert et al. 2015; Riesenberg and Maricic 2018; Riesenberg et al. 2019; Yeh et al. 2019). However, rather than permanently blocking NHEJ, our protocol inhibits NHEJ to reduce the kinetics of DSB formation, presumably increasing the chance of two events co-occurring. This results in a significant enhancement of DNA deletion efficiency.
DNA-PKcs inhibition represents a practical option for a variety of CRISPR-del applications, from basic research to gene therapy. DNA-PKcs inhibitors are cheap and widely available. Deletion efficiency improved regardless of the inhibitor molecule, target region, sgRNA sequence, cell background, and delivery method. The exception was for nontransformed cells, where promising preliminary data were obtained in iPSCs, but extensive optimization of Cas9 delivery and treatment conditions will be necessary before concluding whether iDNA-PKcs is effective here. Particularly striking was the observation that a pgRNA that is ineffective under normal conditions achieved respectable rates of deletion with iDNA-PKcs (P1 in Fig. 2D). This suggests that the failure of many pgRNAs to efficiently delete DNA may arise not from their inability to promote DSBs but rather as a result of poor kinetic properties (e.g., a mismatch in kinetics between the two individual sgRNAs). Finally, this method (with minor modifications) is compatible with low-MOI lentiviral delivery and led to impressive gains in sensitivity of a pooled functional screen, an approach with rapidly growing adoption (Zhu et al. 2016; Diao et al. 2017; Gasperini et al. 2017; Liu et al. 2018).
The CiDER method presented here has wide applications in identifying intrinsic and extrinsic factors that modulate CRISPR-del efficiency. Its advantages include simplicity, practicality, and faithful representation of the deletion process in cellular chromatin. However, it is incapable of distinguishing deletion from inversion, the latter being a nonnegligible editing outcome. In the future, it will be important to use methods capable of distinguishing deletion and inversion events, such as digital droplet PCR (ddPCR) (Antoniani et al. 2018; Watry et al. 2020), although it might not be practical for testing large numbers of samples and conditions. Although the CiDER and qPCR assays used here could not explicitly distinguish the ratios of deletion and insertion arising from pgRNA targeting, we found that most of the measured iDNA-PKcs-dependent changes arise from increases in deletion.
Methods
Reanalysis of CRISPR-del screen data
Raw and processed data were obtained from Zhu et al. (2016). sgRNA pairs were defined to contribute to a gene-level phenotype (Fig. 1B) based on the “good sgRNAs” column in Supplementary Table 5 of Zhu et al. (2016). To generate the results in Figure 1C, massively parallel sequencing data from the negative selection screen in Huh7 cells was reanalyzed using the MAGeCK algorithm (Li et al. 2014; https://sourceforge.net/p/mageck/wiki/Home/) to obtain the log2 fold-change (LFC) for each pgRNA. As the purpose of our analysis was to examine deletion efficiency, and sgRNAs targeting an ORF can inactivate a gene independent of deletion, we excluded pgRNAs having at least one sgRNA targeting a gene's ORF. Thus, from the 45 significant gene hits described in the paper (cutoff FDR < 0.25), the subset of 41 that were targeted for a deletion of their transcription start site (TSS) were selected and are displayed in Figure 1, B and C. The predicted on-target efficiency of each sgRNA was calculated using the Rule Set II system (Doench et al. 2016).
Cell culture
HeLa, HCT116, and HEK293T were cultured on Dulbecco's modified Eagles medium (DMEM; Sigma-Aldrich D5671) supplemented with 10% fetal bovine serum (FBS; Thermo Fisher Scientific 10500064), 1% L-glutamine (Thermo Fisher Scientific 25030024), 1% penicillin-streptomycin (Thermo Fisher Scientific 15140122). Cells were grown at 37°C and 5% CO2 and passaged every 2 d at 1:5 dilution.
pgRNA design and cloning
pgRNAs were designed using CRISPETa (http://crispeta.crg.eu/) and cloned into the pDECKO backbone as described previously (Pulido-Quetglas et al. 2017). Off-target filters did not allow fewer than three mismatches for each sgRNA sequence. No positive or negative masks were applied in the search. The minimum individual score was set at 0.2; the minimum paired score, at 0.4. The pgRNAs were then manually selected from the output list. All sgRNA sequences may be found in Supplemental Figure S11.
Inhibitors
All molecules used in this study are commercially available: M3814 (MedChemExpress HY-101570), KU57788 (MedChemExpress HY-11006), NU7026 (MedChemExpress HY-15719), LTURM34 (MedChemExpress HY-101667), DMNB (ToChris 2088), and SCR7 Pyrazine (Sigma-Aldrich SML1546). Ten millimolar stocks (and 5 mM for NU7026, owing to solubility limitations) were prepared by resuspension in dimethylsulfoxide (DMSO; Sigma-Aldrich D4540).
Transfection and lentiviral transduction
For transfection experiments, 70% confluent 12-well plates were transfected using Lipofectamine 2000 (Thermo Fisher Scientific 11668019) with 1250 ng of pDECKO plasmid following the provider's guidelines. After 6 h, transfection media was replaced for fresh complete DMEM (10% FBS, 1% L-glutamine, and 1% penicillin-streptomycin), and the corresponding small molecule was added to media for 18 h. The treatment was finished by replacing the media with complete DMEM. After 1 d, cells were selected with puromycin (2 µg/mL).
For lentiviral infection experiments, cells were spin-infected at a 0.3 MOI in the presence of DMEM (10% FBS, 1% L-glutamine) and hexadimethrine bromide (8 µg/mL; Sigma-Aldrich 107689) at 2000 rpm, during 1.5 h at 37°C. After 0, 5, 10, 24, 48, and 72 h, infection media was replaced for fresh complete DMEM (10% FBS, 1% L-glutamine, and 1% penicillin-streptomycin), and the corresponding small molecule was added to media for 18 h. The treatment was finished by replacing the media with complete DMEM and puromycin (2 µg/mL) to start the selection.
Flow cytometry
After 5 d of puromycin selection, cells were trypsinized, resuspended in PBS, and incubated for 30 min at room temperature (RT) with the human α-plexin D1 mouse monoclonal antibody (1:150 dilution; R&D systems MAB4160). Cells were washed twice with PBS and incubated for 30 min at RT with an α-mouse IgG secondary goat antibody conjugated to the APC fluorochrome (1:200 dilution; eBioscience 17-4010-82). Cells were washed and resuspended in PBS, processed with the LSRII SORP flow cytometer, and analyzed with FlowJo v10 software. A total of 10,000 cells per sample are sorted. Cell population is selected in the SSC-A/FSC-A plot. Single cells are gated in the FSC-H/FSC-A plot. Finally, the APC-positive population is set in the mCherry/APC plot in the control sample and expanded to all the other samples without modification. The fraction of plexin D1–negative singlet cells is calculated by gating plexin D1–positive singlet cells, normalizing to a nontargeting control and subtracting the value to one (negative cells = 1 − positive cells). An example of the gating strategy may be found in Supplemental Figure S12. Single-cell imaging was performed using ImageStream (Luminex) and analyzed with IDEAS software.
Genomic PCR
After 5 d of puromycin selection, cells were collected and genomic DNA (qDNA) was extracted using a GeneJET genomic DNA purification kit (Thermo Fisher Scientific K0722). Genomic PCR was performed using GoTaq G2 DNA polymerase (Promega M7841) from 10 ng gDNA. The PCR primers are the following ones: PLXND1 (forward, 5′-AGTCTAGGTTCTGAATCTGTCGC-3′; reverse, 5′-CAAACACTGTAGCTCTGCTCC-3′) and MALAT1 enhancer (forward, 5′-CCTGCTATGAACTGACCCATG-3′; reverse, 5′-CCTGAACAGTCAGTCCATGCT-3′).
TOPO cloning and Sanger sequencing of deleted alleles
The PLXND1 KO band and MALAT1 enhancer full-length and KO bands obtained with the genomic PCRs was gel-purified using the GeneJET gel extraction and DNA cleanup micro kit (Thermo Fisher Scientific K0831) and ligated into a backbone vector using the TOPO TA cloning kit (Thermo Fisher Scientific 45-0030). Stbl3-competent cells were transformed with the ligation product, colonies were expanded, and Sanger sequencing was performed (MicroSynth GmBH) using the same reverse primers as used for the PCR.
Genomic Quantitative PCR
After 5 d of puromycin selection, cells were collected and genomic DNA (qDNA) was extracted using a GeneJET genomic DNA purification kit (Thermo Fisher Scientific K0722). Quantitative real-time PCR (qPCR) from 10 ng of gDNA was performed using GoTaq qPCR master mix (Promega A6001) on a TaqMan Viia 7 real-time PCR system. Target sequence primers for PLXND1 “wild-type” amplification (forward, 5′-GGCAACTGTGTTTGGCACTG-3′; reverse, 5′-GGAACCAGAATGGTTCTTGCC-3′), PLXND1 inversion amplification (forward, 5′-GGCAACTGTGTTTGGCACTG-3′; reverse, 5′-CAAACACTGTAGCTCTGCTCC-3′), MALAT1 enhancer “wild-type” amplification (forward, 5′-GCTGGGGAATCCACAGAGAC-3′; reverse, 5′-CATCTCAGCCCTTGTTATCCTG-3′), and LDHA (forward, 5′-TGGGCAGTAGAAAGTGCAG-3′; reverse, 5′-TACCAGCTCCCACTCACAG-3′). Target sequence primers were normalized to primers targeting the distal, nontargeted gene LDHA. Data were normalized using the ΔΔCt method (Schmittgen and Livak 2008).
Protein extraction and western blot
After pgRNA transfection and 18 h of vehicle/M3814 treatment, HeLa cells were collected in urea lysis buffer containing 20 mM HEPES (pH 8.0), 9.0 M urea, 2.5 mM sodium pyrophosphate, 1 mM β-glycerol-phosphate, and 1 mM sodium orthovanadate. Lysates were sonicated and clarified by centrifugation. Protein concentration was measured using the Bio-Rad protein quantification reagent (Bio-Rad Laboratories 5000006), and total protein extracts (50 µg) were resolved by SDS-PAGE on a 6% acrylamide gel. Proteins were blotted onto PVDF membranes (Novex, Thermo Fisher Scientific IPFL00010). The membranes were blocked in 5% BSA in TBS- 0.1% tween and incubated within the indicated primary antibodies: total DNA-PKcs Y393 (Abcam ab32566), pS2056 DNA-PKcs (Abcam ab124918), α-tubulin (Sigma-Aldrich T6199). The signal was detected using the corresponding fluorescence secondary antibodies: IRDye 680RD goat anti-mouse (Li-Cor Biosciences 925-68070) and IRDye 800CW goat anti-rabbit (Li-Cor Biosciences 925-32211).
Cell viability assay
CellTiter-Glo 2.0 cell viability assay (Promega G9241) was performed upon puromycin selection (2 d post transfection). Three thousand cells per well were seeded in 96-well white polystyrene plates (Corning, Sigma-Aldrich CLS3610-48EA), and cell viability was measured in technical duplicates during four consecutive days (0, 24, 48, 72 h) according to the manufacturer's protocol. Luminescence was measured using a Tecan reader Infinite 200.
Generation of Cas9 stable cell lines
HeLa cells were infected with lentivirus carrying the Cas9-BFP (blue fluorescent protein) vector (Addgene 52962). HCT116 and HEK293T were transfected with the same vector using Lipofectamine 2000 (Thermo Fisher Scientific 11668019). All cell types were selected with blasticidin (4 µg/mL) for at least 5 d and selected for BFP-positive cells twice by FACS. HeLa cells were infected with lentivirus carrying the DDCas9-mVenus vector (Addgene 90085). HeLa cells with DDCas9-mVenus were selected with zeocin (70 µg/mL) for at least 5 d and selected for mVenus-positive cells by FACS.
Design of CRISPR-del library
pgRNAs were designed using CRISPETa (http://crispeta.crg.eu/) with the same parameters as in section “pgRNA Design and Cloning.” Three to 10 pgRNAs were designed to target essential protein-coding genes and intergenic regions as neutral controls. Essential genes were collected by literature search and encode essential ribosomal proteins and oncogenes. Neutral control regions comprise randomly sampled intergenic regions >10 kb from the nearest annotated gene. The library was synthesized and packaged into lentivirus by VectorBuilder (Germany). The library sequence is available in Supplemental File S1.
Pooled CRISPR-del screen for gene essentiality
HeLa DDCas9-mVenus cells were spin-infected with lentiviruses carrying the library of mixed pgRNAs at 0.35 and 0.6 MOI with coverage of 500-fold (pgRNA sequence to cells) in the presence of DMEM (10% FBS, 1% L-glutamine) and hexadimethrine bromide (8 µg/mL; Sigma-Aldrich 107689) at 2000 rpm for 1.5 h at 37°C. After the spin down, the infection media was replaced for fresh complete DMEM (10% FBS, 1% L-glutamine, and 1% penicillin-streptomycin). Sixteen hours post infection, media was replaced with complete DMEM and puromycin (2 µg/mL) to select for successfully infected cells. Cells were kept under selection for 6 d. After selection (7 d after infection) cells were split in three populations: One was collected (time point 0 [T0]) and stored at −20°C; the second was kept in culture, and stabilization of DD-Cas9 was effected by adding 400 nM Shield1 for 3 d; and the third was kept in culture, 400 nM Shield1 was added for 3 d, and 300 nM of M3814 was added to media for 18 h. Cells were kept in culture and passaged every 2 d. One week after T0 (time point 1 [T1]) and 3 wk after T0 (time point 3 [T3]), cells were collected and stored at −20°C. Total gDNA from all collected cells was extracted following the manufacturer's instructions with the blood and cell culture DNA midi kit (Qiagen). pgRNAs were amplified from genomic DNA using NEBNext Ultra II Q5 master mix (NEB), custom primers with the sequencing adaptors, and sequencing indexes. In total, 40 µg of each sample was amplified with 25 cycles in 50-µL reaction steps (2 µg of DNA at each step) at an annealing temperature of 52°C. All PCR products were pooled and purified with SPRIselect beads (Beckman Coulter) following the manufacturer's instructions. The quality and quantity of the sample preparation were checked with Agilent high sensitivity DNA chips with a Bioanalyzer (Agilent) and Qubit dsDNA HS assay kit (Invitrogen). Samples were sequenced at 3000-fold coverage (reads to pgRNA sequence) by paired-end (PE) 150-bp reads.
Screen data processing
FASTQ files were processed using CASPR (Bergadà-Pijuan et al. 2020) using adapters ACCG and AAAC for forward and reverse reads, respectively. The resulting count table was used for downstream analysis. LFCs for each pgRNA were computed from the count table normalized as the fraction of total reads per sample. To identify gene hits, the count table was used as input for MAGeCK (Li et al. 2014). For each timepoint, hits were detected using T0 as control, selecting an FDR gene test threshold of 0.25 (gene-test-fdr-threshold 0.25). MAGeCK output files were processed with an in-house Python script (Supplemental Code). Read counts and processed data are available in Supplemental Files S2 and S3.
Competing interest statement
The authors declare no competing interests.
Supplementary Material
Acknowledgments
We acknowledge administrative support from Basak Ginsbourger and Silvia Roesselet (DBMR, University of Bern). We thank Bill Keyes (IGBMC) and Norbert Polacek (DCB, University of Bern) for insightful feedback and discussions. We also thank Stefan Müller (DBMR, University of Bern) for his expertise with ImageStream and the other members of the FACS laboratory from the University of Bern for their advice. We acknowledge Taisia Polidori, Paulina Schaerer (DBMR, University of Bern), Sofia Nasif, and Oliver Mühlemann (DCB, University of Bern) for experimental support; Álvaro Andrades (Universidad de Granada) for the CCLE data; and the rest of the members of Johnson's laboratory for their valuable input. Andrea Maddalena (Department of Physiology, University of Bern) provided valuable advice regarding lentiviral transduction inhibition. Finally, we thank the anonymous reviewers whose insightful comments and suggestions prompted numerous improvements to the experimental data and their interpretation. This work was funded by the Swiss National Science Foundation through the National Center of Competence in Research (NCCR) “RNA & Disease,” by the Medical Faculty of the University and University Hospital of Bern, by the Helmut Horten Stiftung, Krebsliga Schweiz (4534-08-2018), and Science Foundation Ireland through Future Research Leaders award 18/FRL/6194.
Author contributions: N.B-G. and R.J. conceived and designed the experiments. N.B-G. and T.U. performed all the experiments. T.U. and H.A.G.-R. performed and analyzed the gene essentiality high-throughput screen. N.B-G. performed the analysis of public data. R.R. performed the western blot experiments. A.G. assisted and contributed to the results with different cell lines. M.M. and Y.Z. suggested DNA-PKcs inhibition to modulate NHEJ. C.P-Q. contributed on the design of the pgRNAs and analysis of public data. R.E. provided the solution to circumvent lentiviral infection problems. N.B-G. and R.J. wrote the whole manuscript with feedback from M.M. and Y.Z. Finally, R.J. directed the research.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.265736.120.
Freely available online through the Genome Research Open Access option.
References
- Antoniani C, Meneghini V, Lattanzi A, Felix T, Romano O, Magrin E, Weber L, Pavani G, El Hoss S, Kurita R, et al. 2018. Induction of fetal hemoglobin synthesis by CRISPR/Cas9-mediated editing of the human β-globin locus. Blood 131: 1960–1973. 10.1182/blood-2017-10-811505 [DOI] [PubMed] [Google Scholar]
- Aparicio-Prat E, Arnan C, Sala I, Bosch N, Guigó R, Johnson R. 2015. DECKO: single-oligo, dual-CRISPR deletion of genomic elements including long non-coding RNAs. BMC Genomics 16: 846. 10.1186/s12864-015-2086-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehár J, Kryukov GV, Sonkin D, et al. 2012. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483: 603–607. 10.1038/nature11003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bausch-Fluck D, Hofmann A, Bock T, Frei AP, Mirkowska P, Härtlová A, Van EJ, Bourquin J. 2015. A mass spectrometric-derived cell surface protein atlas. PLoS One 10: e0121314. 10.1371/journal.pone.0121314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergadà-Pijuan J, Pulido-Quetglas C, Vancura A, Johnson R. 2020. CASPR, an analysis pipeline for single and paired guide RNA CRISPR screens, reveals optimal target selection for long non-coding RNAs. Bioinformatics 36: 1673–1680. 10.1093/bioinformatics/btz811 [DOI] [PubMed] [Google Scholar]
- Brinkman EK, Chen T, de Haas M, Holland HA, Akhtar W, van Steensel B. 2018. Kinetics and fidelity of the repair of Cas9-induced double-strand DNA breaks. Mol Cell 70: 801–813.e6. 10.1016/j.molcel.2018.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canver MC, Bauer DE, Dass A, Yien YY, Chung J, Masuda T, Maeda T, Paw BH, Orkin SH. 2014. Characterization of genomic deletion efficiency mediated by clustered regularly interspaced palindromic repeats (CRISPR)/Cas9 nuclease system in mammalian cells. J Biol Chem 289: 21312–21324. 10.1074/jbc.M114.564625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canver MC, Smith EC, Sher F, Pinello L, Sanjana NE, Shalem O, Chen DD, Schupp PG, Vinjamur DS, Garcia SP, et al. 2015. BCL11A enhancer dissection by Cas9-mediated in situ saturating mutagenesis. Nature 527: 192–197. 10.1038/nature15521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HHY, Pannunzio NR, Adachi N, Lieber MR. 2017. Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat Rev Mol Cell Biol 18: 495–506. 10.1038/nrm.2017.48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu VT, Weber T, Wefers B, Wurst W, Sander S, Rajewsky K, Kühn R. 2015. Increasing the efficiency of homology-directed repair for CrIsPr-Cas9-induced precise gene editing in mammalian cells. Nat Biotechnol 33: 543–548. 10.1038/nbt.3198 [DOI] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. 2013. Multiplex genome engineering using CRISPR/Cas systems. Science 339: 819–823. 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel R, Greger JG, Katz RA, Taganov KD, Wu X, Kappes JC, Skalka AM. 2004. Evidence that stable retroviral transduction and cell survival following DNA integration depend on components of the nonhomologous end joining repair pathway. J Virol 78: 8573–8581. 10.1128/JVI.78.16.8573-8581.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao Y, Fang R, Li B, Meng Z, Yu J, Qiu Y, Lin KC, Huang H, Liu T, Marina RJ, et al. 2017. A tiling-deletion-based genetic screen for cis-regulatory element identification in mammalian cells. Nat Methods 14: 629–635. 10.1038/nmeth.4264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench JG. 2018. Am I ready for CRISPR? A user's guide to genetic screens. Nat Rev Genet 19: 67–80. 10.1038/nrg.2017.97 [DOI] [PubMed] [Google Scholar]
- Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, Smith I, Tothova Z, Wilen C, Orchard R, et al. 2016. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol 34: 184–191. 10.1038/nbt.3437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito R, Bosch N, Lanzós A, Polidori T, Pulido-Quetglas C, Johnson R. 2019. Hacking the cancer genome: profiling therapeutically actionable long non-coding RNAs using CRISPR-Cas9 screening. Cancer Cell 35: 545–557. 10.1016/j.ccell.2019.01.019 [DOI] [PubMed] [Google Scholar]
- Fattah FJ, Lichter NF, Fattah KR, Oh S, Hendrickson EA. 2008. Ku70, an essential gene, modulates the frequency of rAAV-mediated gene targeting in human somatic cells. Proc Natl Acad Sci 105: 8703–8708. 10.1073/pnas.0712060105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchss T, Emde U, Buchstaller H-P, Mederski WKR. 2014. Arylquinazolines 2014; U.S. patent no. WO2014183850A1.
- Gasperini M, Findlay GM, McKenna A, Milbank JH, Lee C, Zhang MD, Cusanovich DA, Shendure J. 2017. CRISPR/Cas9-mediated scanning for regulatory elements required for HPRT1 expression via thousands of large, programmed genomic deletions. Am J Hum Genet 101: 192–205. 10.1016/j.ajhg.2017.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasperini M, Hill AJ, McFaline-Figueroa JL, Martin B, Kim S, Zhang MD, Jackson D, Leith A, Schreiber J, Noble WS, et al. 2019. A genome-wide framework for mapping gene regulation via cellular genetic screens. Cell 176: 377–390.e19. 10.1016/j.cell.2018.11.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Zhang J, Chen L, Shen B, Zhou J, Hu B, Du Y, Tate PH, Huang X, Zhang W. 2014. Efficient in vivo deletion of a large imprinted lncRNA by CRISPR/Cas9. RNA Biol 11: 829–835. 10.4161/rna.29624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harnor SJ, Brennan A, Cano C. 2017. Targeting DNA-dependent protein kinase for cancer therapy. ChemMedChem 12: 895–900. 10.1002/cmdc.201700143 [DOI] [PubMed] [Google Scholar]
- Hart T, Chandrashekhar M, Aregger M, Durocher D, Steinhart Z, Brown KR, Macleod G, Mis M, Zimmermann M, Fradet-Turcotte A, et al. 2015. High-resolution CRISPR screens reveal fitness genes and genotype-specific cancer liabilities. Cell 163: 1515–1526. 10.1016/j.cell.2015.11.015 [DOI] [PubMed] [Google Scholar]
- Ho T-T, Zhou N, Huang J, Koirala P, Xu M, Fung R, Wu F, Mo Y-Y. 2015. Targeting non-coding RNAs with the CRISPR/Cas9 system in human cell lines. Nucleic Acids Res 43: e17. 10.1093/nar/gku1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdt LM, Stahringer A, Sass K, Pichler G, Kulak NA, Wilfert W, Kohlmaier A, Herbst A, Northoff BH, Nicolaou A, et al. 2016. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat Commun 7: 12429. 10.1038/ncomms12429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Li K, Cai W, Liu X, Zhang Y, Orkin SH, Xu J, Yuan G. 2018. Dissecting super-enhancer hierarchy based on chromatin interactions. Nat Commun 9: 943. 10.1038/s41467-018-03279-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koirala P, Huang J, Ho T-T, Wu F, Ding X, Mo Y-Y. 2017. LncRNA AK023948 is a positive regulator of AKT. Nat Commun 8: 14422. 10.1038/ncomms14422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosicki M, Tomberg K, Bradley A. 2018. Repair of double-strand breaks induced by CRISPR–Cas9 leads to large deletions and complex rearrangements. Nat Biotechnol 36: 765–771. 10.1038/nbt.4192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Richard S. 2016. Sam68 functions as a transcriptional coactivator of the p53 tumor suppressor. Nucleic Acids Res 44: 8726–8741. 10.1093/nar/gkw582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Olvera JM, Yoder KE, Mitchell RS, Butler SL, Lieber M, Martin SL, Bushman FD. 2001. Role of the non-homologous DNA end joining pathway in the early steps of retroviral infection. EMBO J 20: 3272–3281. 10.1093/emboj/20.12.3272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Xu H, Xiao T, Cong L, Love MI, Zhang F, Irizarry RA, Liu JS, Brown M, Liu XS. 2014. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol 15: 554. 10.1186/s13059-014-0554-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Staahl BT, Alla RK, Doudna JA. 2014a. Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. eLife 3: e04766. 10.7554/eLife.04766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YC, Boone M, Meuris L, Lemmens I, Van Roy N, Soete A, Reumers J, Moisse M, Plaisance S, Drmanac R, et al. 2014b. Genome dynamics of the human embryonic kidney 293 lineage in response to cell biology manipulations. Nat Commun 5: 4767. 10.1038/ncomms5767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu SJ, Horlbeck MA, Cho SW, Birk HS, Malatesta M, He D, Attenello FJ, Villalta JE, Cho MY, Chen Y, et al. 2017. CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells. Science 355: eaah7111. 10.1126/science.aah7111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Cao Z, Wang Y, Guo Y, Xu P, Yuan P, Liu Z. 2018. Genome-wide screening for functional long noncoding RNAs in human cells by Cas9 targeting of splice sites. Nat Biotechnol 36: 1203–1210. 10.1038/nbt.4283 [DOI] [PubMed] [Google Scholar]
- Luo B, Cheung HW, Subramanian A, Sharifnia T, Okamoto M, Yang X, Hinkle G, Boehm JS, Beroukhim R, Weir BA, et al. 2008. Highly parallel identification of essential genes in cancer cells. Proc Natl Acad Sci 105: 20380–20385. 10.1073/pnas.0810485105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupiañez DG, Kraft K, Heinrich V, Krawitz P, Brancati F, Klopocki E, Horn D, Kayserili H, Opitz JM, Laxova R, et al. 2015. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell 161: 1012–1025. 10.1016/j.cell.2015.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddalo D, Manchado E, Concepcion CP, Bonetti C, Vidigal JA, Han Y, Ogrodowski P, Crippa A, Rekhtman N, De Stanchina E, et al. 2014. In vivo engineering of oncogenic chromosomal rearrangements with the CRISPR/Cas9 system. Nature 516: 423–427. 10.1038/nature13902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. 2013. RNA-guided human genome engineering via Cas9. Science 339: 823–826. 10.1126/science.1232033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal PK, Ferreira MR, Collins R, Meissner TB, Boutwell CL, Friesen M, Vrbanac V, Garrison BS, Stortchevoi A, Bryder D, et al. 2014. Efficient ablation of genes in human hematopoietic stem and effector cells using CRISPR/Cas9. Cell Stem Cell 15: 643–652. 10.1016/j.stem.2014.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangeot PE, Risson V, Fusil F, Marnef A, Laurent E, Blin J, Mournetas V, Massouridès E, Sohier TJM, Corbin A, et al. 2019. Genome editing in primary cells and in vivo using viral-derived Nanoblades loaded with Cas9-sgRNA ribonucleoproteins. Nat Commun 10: 45. 10.1038/s41467-018-07845-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Z, Bozzella M, Seluanov A, Gorbunova V. 2008. Comparison of nonhomologous end joining and homologous recombination in human cells. DNA Repair 7: 1765–1771. 10.1016/j.dnarep.2008.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama T, Dougan SK, Truttmann MC, Bilate AM, Ingram JR, Ploegh HL. 2015. Increasing the efficiency of precise genome editing with CrIsPr-Cas9 by inhibition of nonhomologous end joining. Nat Biotechnol 33: 538–542. 10.1038/nbt.3190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers RM, Bryan JG, McFarland JM, Weir BA, Sizemore AE, Xu H, Dharia NV, Montgomery PG, Cowley GS, Pantel S, et al. 2017. Computational correction of copy number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nat Genet 49: 1779–1784. 10.1038/ng.3984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyaoka Y, Berman JR, Cooper SB, Mayerl SJ, Chan AH, Zhang B, Karlin-neumann GA, Conklin BR. 2016. Systematic quantification of HDR and NHEJ reveals effects of locus, nuclease, and cell type on genome-editing. Sci Rep 6: 23549. 10.1038/srep23549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki Y, Chiba T, Kataoka K, Yamashita S, Sato T, Kato T, Takahashi K, Miyamoto T, Kitazawa M, Hatta T, et al. 2018. Combinatorial CRISPR/Cas9 approach to elucidate a far-upstream enhancer complex for tissue-specific Sox9 expression. Dev Cell 46: 794–806.e6. 10.1016/j.devcel.2018.07.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohiuddin IS, Kang MH. 2019. DNA-PK as an emerging therapeutic target in cancer. Front Oncol 9: 635. 10.3389/fonc.2019.00635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson CE, Hakim CH, Ousterout DG, Thakore PI, Moreb EA, Rivera RMC, Madhavan S, Pan X, Ran FA, Yan WX, et al. 2016. In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science 351: 403–408. 10.1126/science.aad5143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulido-Quetglas C, Aparicio-Prat E, Arnan C, Polidori T, Hermoso T, Palumbo E, Ponomarenko J, Guigo R, Johnson R. 2017. Scalable design of paired CRISPR guide RNAs for genomic deletion. PLoS Comput Biol 13: e1005341. 10.1371/journal.pcbi.1005341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riesenberg S, Maricic T. 2018. Targeting repair pathways with small molecules increases precise genome editing in pluripotent stem cells. Nat Commun 9: 2164. 10.1038/s41467-018-04609-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riesenberg S, Chintalapati M, Macak D, Kanis P, Maricic T, Pääbo S. 2019. Simultaneous precise editing of multiple genes in human cells. Nucleic Acids Res 47: e116. 10.1093/nar/gkz669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert F, Barbeau M, Éthier S, Dostie J, Pelletier J. 2015. Pharmacological inhibition of DNA-PK stimulates Cas9-mediated genome editing. Genome Med 7: 93. 10.1186/s13073-015-0215-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanson KR, Hanna RE, Hegde M, Donovan KF, Strand C, Sullender ME, Vaimberg EW, Goodale A, Root DE, Piccioni F, et al. 2018. Optimized libraries for CRISPR-Cas9 genetic screens with multiple modalities. Nat Commun 9: 5416. 10.1038/s41467-018-07901-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. 2008. Analyzing real-time PCR data by the comparative CT method. Nat Protoc 3: 1101–1108. 10.1038/nprot.2008.73 [DOI] [PubMed] [Google Scholar]
- Song J, Yang D, Xu J, Zhu T, Chen YE, Zhang J. 2016. RS-1 enhances CRISPR/Cas9- and TALEN-mediated knock-in efficiency. Nat Commun 7: 10548. 10.1038/ncomms10548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojic L, Lun ATLL, Mangei J, Mascalchi P, Quarantotti V, Barr AR, Bakal C, Marioni JC, Gergely F, Odom DT. 2018. Specificity of RNAi, LNA and CRISPRi as loss-of-function methods in transcriptional analysis. Nucleic Acids Res 46: 5950–5966. 10.1093/nar/gky437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JD, Polaski JT, Feng Q, De Neef EJ, Hoppe ER, McSharry MV, Pangallo J, Gabel AM, Belleville AE, Watson J, et al. 2020. RNA isoform screens uncover the essentiality and tumor-suppressor activity of ultraconserved poison exons. Nat Genet 52: 84–94. 10.1038/s41588-019-0555-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thul PJ, Åkesson L, Wiking M, Mahdessian D, Geladaki A, Ait Blal H, Alm T, Asplund A, Björk L, Breckels LM, et al. 2017. A subcellular map of the human proteome. Science 356: eaal3321. 10.1126/science.aal3321 [DOI] [PubMed] [Google Scholar]
- Tsherniak A, Vazquez F, Montgomery PG, Weir BA, Kryukov G, Cowley GS, Gill S, Harrington WF, Pantel S, Krill-Burger JM, et al. 2017. Defining a cancer dependency map. Cell 170: 564–576.e16. 10.1016/j.cell.2017.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidigal JA, Ventura A. 2015. Rapid and efficient one-step generation of paired gRNA CRISPR-Cas9 libraries. Nat Commun 6: 8083. 10.1038/ncomms9083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watry HL, Feliciano CM, Gjoni K, Takahashi G, Miyaoka Y, Conklin BR, Judge LM. 2020. Rapid, precise quantification of large DNA excisions and inversions by ddPCR. Sci Rep 10: 14896. 10.1038/s41598-020-71742-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y-H, Yao R-W, Zhang Y, Guo C-J, Jiang S, Xu G, Dong R, Yang L, Chen L-L. 2017. SLERT regulates DDX21 rings associated with Pol I transcription. Cell 169: 664–678.e16. 10.1016/j.cell.2017.04.011 [DOI] [PubMed] [Google Scholar]
- Yang H, Wang H, Shivalila CS, Cheng AW, Shi L, Jaenisch R. 2013. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell 154: 1370–1379. 10.1016/j.cell.2013.08.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh CD, Richardson CD, Corn JE. 2019. Advances in genome editing through control of DNA repair pathways. Nat Cell Biol 21: 1468–1478. 10.1038/s41556-019-0425-z [DOI] [PubMed] [Google Scholar]
- Zenke FT, Zimmermann A, Sirrenberg C, Dahmen H, Vassilev L, Pehl U, Fuchss T, Blaukat A. 2016. M3814, a novel investigational DNA-PK inhibitor: enhancing the effect of fractionated radiotherapy leading to complete regression of tumors in mice. In Proceedings of the 107th Annual Meeting of the American Association for Cancer Research, Abstract no. 1658. New Orleans, LA. AACR, Philadelphia. 10.1158/1538-7445.AM2016-1658 [DOI] [Google Scholar]
- Zenke FT, Zimmermann A, Sirrenberg C, Dahmen H, Kirkin V, Pehl U, Grombacher T, Wilm C, Fuchss T, Amendt C, et al. 2020. Pharmacologic inhibitor of DNA-PK, M3814, potentiates radiotherapy and regresses human tumors in mouse models. Mol Cancer Ther 19: 1091–1101. 10.1158/1535-7163.MCT-19-0734 [DOI] [PubMed] [Google Scholar]
- Zhu S, Li W, Liu J, Chen C-H, Liao Q, Xu P, Xu H, Xiao T, Cao Z, Peng J, et al. 2016. Genome-scale deletion screening of human long non-coding RNAs using a paired-guide RNA CRISPR–Cas9 library. Nat Biotechnol 34: 1279–1286. 10.1038/nbt.3715 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.