Abstract
The wrapping of DNA around histone octamers challenges processes that use DNA as their template. In vitro, DNA replication through chromatin depends on histone modifiers, raising the possibility that cells modify histones to optimize fork progression. Rtt109 is an acetyl transferase that acetylates histone H3 before its DNA incorporation on the K56 and N-terminal residues. We previously reported that, in budding yeast, a wave of histone H3 K9 acetylation progresses ∼3–5 kb ahead of the replication fork. Whether this wave contributes to replication dynamics remained unknown. Here, we show that the replication fork velocity increases following deletion of RTT109, the gene encoding the enzyme required for the prereplication H3 acetylation wave. By using histone H3 mutants, we find that Rtt109-dependent N-terminal acetylation regulates fork velocity, whereas K56 acetylation contributes to replication dynamics only when N-terminal acetylation is compromised. We propose that acetylation of newly synthesized histones slows replication by promoting replacement of nucleosomes evicted by the incoming fork, thereby protecting genome integrity.
Eukaryotic cells protect their DNA by wrapping it within chromatin. The basic building blocks of chromatin are nucleosomes, which are histone octamers wrapped by ∼147 bp of DNA. During replication, DNA must dissociate from histones to allow progression of the replication fork. Once the fork has passed, nucleosomes rapidly reassemble, using both available and newly synthesized histones (Groth et al. 2007). This disassembly–reassembly process offers opportunities for regulating replication dynamics and coordinating replication with other processes that use DNA as their template (Hamperl and Cimprich 2016). Understanding the interplay between chromatin and DNA replication is therefore of great interest.
Nucleosome stability depends on the interactions between DNA and histones. Determinants of these interactions are the DNA sequence and the various modifications added to histones. Prominent among histone modifications is lysine acetylation, observed in different positions and, in particular, on the disordered histone N termini (Hong et al. 1993; Lee et al. 1993; Bauer et al. 1994). Histone modification regulates nucleosome stability by altering its electrostatic interactions with DNA and by recruiting regulatory proteins that assist in the assembly or disassembly of nucleosomes (Bannister and Kouzarides 2011; Owen-Hughes and Gkikopoulos 2012).
DNA replication is initiated from hundreds of sites that are distributed across the genome (Bell and Dutta 2002). These replication origins are bound by the origin recognition complex (ORC), which recruits the MCM complex and thus initiates assembly of the replication machinery (Bell and Stillman 1992). Upon firing, replication forks proceed bidirectionally until approaching forks that emanated from neighboring origins. The temporal replication dynamics therefore depend on the positioning of origins along the genome, the time in S phase when different origins fire, and the velocity at which replication forks proceed along the genome. Replication origins are relatively depleted of nucleosomes, and their surrounding chromatin is enriched with histone H3K4 methylation and H3K9/27 acetylation (MacAlpine et al. 2010; Eaton et al. 2011; Lubelsky et al. 2014; Miotto et al. 2016). Evidence from budding yeast suggests that histone modifications regulate origin firing. For example, tethering of Gcn5 (the catalytic subunit of the SAGA complex) to late origins advanced origin firing time (Vogelauer et al. 2002). Gcn5 acetylates H3 N-terminal lysines, implicating this acetylation in the regulation of origin firing.
Cells may regulate fork progression by modifying histones ahead of the progressing fork. In vitro reconstitution experiments have shown that replication through chromatin is significantly slower than replication of naked DNA but can be accelerated by the addition of chromatin chaperones and modifiers (Kurat et al. 2017). Among the factors facilitating fork progression was Gcn5, suggesting a role of H3 N-terminal acetylation in this process. Gcn5 was not sufficient for allowing fork progression through chromatin but promoted the function of the histone chaperone FACT (Orphanides et al. 1999; Belotserkovskaya et al. 2003). Of note, although FACT was essential for fork progression in this assay, CAF-1, which functions as the principal chromatin assembly factor on replicated DNA (Smith and Stillman 1989), was dispensable.
Chromatin modification may regulate fork velocity in vivo. We previously reported that in budding yeast, a wave of acetylated H3K9 precedes the replication fork by 3–5 kb (Bar-Ziv et al. 2016a). This wave coincides with the increased chromatin accessibility found ∼7 kb ahead of the fork (Rodriguez and Tsukiyama 2013). This prereplication was not dependent on Gcn5 but instead required Rtt109 (Bar-Ziv et al. 2016a). Rtt109 is an acetyltransferase that modifies H3 on its internal K56 residue and on several N-terminal lysine residues including nine, 14, 23, and 27 (Berndsen et al. 2008; Fillingham et al. 2008). Rtt109 is a replication-specific enzyme that modifies H3 before its DNA incorporation (Han et al. 2007a; Tsubota et al. 2007). This is in contrast to Gcn5, which acts primarily (although not exclusively (Burgess et al. 2010) on DNA-bound histones. Rtt109's roles during DNA replication include the protection of genomic stability, promotion of nucleosome assembly, and preservation of expression homeostasis (Driscoll et al. 2007; Li et al. 2008; Voichek et al. 2016b). To all these roles, Rtt109 contributes by acetylating H3K56. Acetylation of H3K56 promotes, for example, the interaction of H3 with CAF-1 and, in this way, increases the efficiency of nucleosome assembly at the wake of the fork (Li et al. 2008). H3K56ac is also important for buffering gene expression against the unbalanced gene dosage introduced by replication in S phase (Bar-Ziv et al. 2016b; Voichek et al. 2016a,b, 2018; Bar-Ziv et al. 2020). In contrast, the possible functional consequences of Rtt109 activity on the H3 N-terminal acetylation remain unknown. Our study was set to examine whether this modification contributes to DNA replication dynamics.
Results
Deletion of RTT109 increases replication fork velocity
Histone acetylation can reduce nucleosome stability and facilitate access to DNA (Hong et al. 1993; Lee et al. 1993; Bauer et al. 1994), raising the possibility that the H3K9ac wave progressing ahead of the replication fork contributes to replication dynamics. To begin testing this, we asked whether deleting RTT109, and thereby abolishing the H3K9ac wave, perturbs replication dynamics.
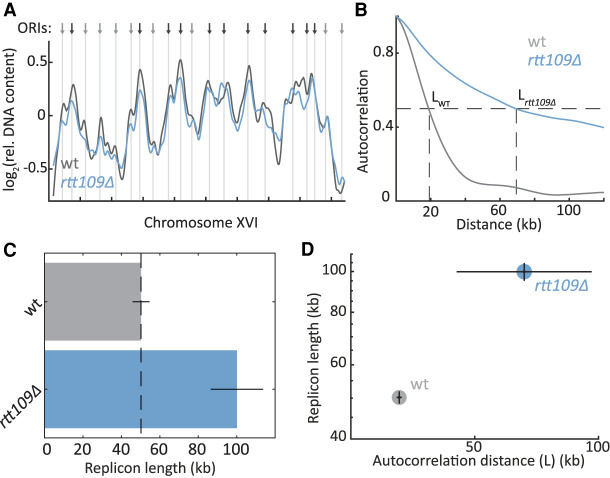
As a first approach for characterizing replication dynamics, we profiled the DNA content of an asynchronously growing cell population and applied an analysis we previously developed to characterize its replication dynamics (Müller et al. 2014; Gispan et al. 2016). In contrast to single-molecule techniques, this technique does not require the genetic and environmental perturbations necessary to enable efficient BrdU labelling that might affect DNA replication (Bianco et al. 2012). Briefly, a culture of asynchronous cells includes cells at different stages of S phase. Accordingly, the culture's DNA content contains a higher fraction of early replicating sequences than of late replicating ones, because these early replicating regions were duplicated in a larger fraction of cells. Plotting the relative DNA content along chromosomes identifies replication origins as peaks, as well as reports on origin firing time (or efficiency) by the height of these peaks (Fig. 1A). We previously showed that fork velocity and initiation rate shape these static replication profiles by modulating the frequency at which replication origins are passively replicated by forks emanating elsewhere, before having a chance to fire. Lowering initiation rate, for example, increases passive replication and therefore reduces the sharpness of the replication profile, whereas lowering fork velocity decreases passive replication and therefore increases profile sharpness. Through auto-correlation, or using the more robust principal component analysis, we can estimate the ratio of fork velocity to initiation rate. This ratio defines the typical length of DNA replicated from a highly efficient origin, and we therefore denote it by replicon length (Gispan et al. 2016).
Figure 1.
DNA replication in the absence of the Rtt109 acetyl transferase. (A) RTT109 deletion affects the replication profile: The relative DNA abundance along Chromosome XVI is plotted for wild-type (wt; gray) and rtt109Δ (blue) strains. Arrows and vertical lines indicate ORIs, as defined by OriDB (Yabuki et al. 2002): black, early; gray, late. (B–D) RTT109 deletion increases replicon length: Shown is the autocorrelation (B), calculated over the full replication profiles, and replicon length (C), calculated through principal component analysis, for wt (gray) and rtt109Δ (blue) strains. The distance at which the autocorrelation reaches half maximum (L) is shown (dashed line) and used in D. Error bars, SE between biological repeats.
We applied this analysis to compare the replicon lengths of wild-type and RTT109-deleted cells (Fig. 1C). Following our hypothesis that the H3K9ac wave promotes fork velocity, we expected RTT109 deletion to reduce replication speed, leading to a shorter replicon length. However, contrasting our expectation, autocorrelation was in fact broader, and replicon length increased by ∼100% (100 kb vs. 50 kb) in RTT109-deleted cells (Fig. 1B–D).
The increased replicon length of RTT109-deletion mutants could reflect lower initiation frequency or increased fork velocity. To distinguish between these possibilities, we followed the replication dynamics in synchronized cultures (Fig. 2A–C). We arrested cells at the end of G1 using mating pheromone and released them into S phase, taking samples at 3-min intervals for profiling DNA content. This tight time course allowed measuring the temporal increase in DNA content at each locus and, from this, quantifying the fork velocity and origin initiation frequency (Methods).
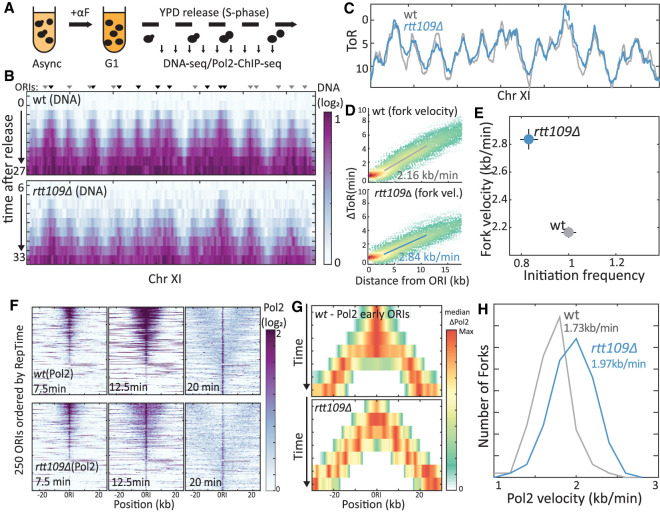
Figure 2.
RTT109 deletion increases fork velocity. (A) Experimental scheme. For details, see text. (B) Temporal replication profile measured by DNA content: Shown is the measured DNA content (color-coded) along Chromosome XI at the indicated times for wt (top) and RTT109-deleted (middle) cells. Triangles highlight ORIs (Yabuki et al. 2002), with the color differing between early (black) and late (gray) ORIs. (C) Time of replication (ToR), at each locus, defined as the time when DNA content increased by 40%. For details, see Methods. (D,E) RTT109 deletion accelerates replication: Each point in D represents a locus, replicated by a fork emanating from an early ORI (n = 107 or 91 ORIs for wt and rtt109Δ, respectively). Shown is the locus replication time relative to that of its ORI (ΔToR) as a function of its distance. Solid line is linear regression. Fork velocity calculated from slope is indicated and shown, together with the calculated initiation frequency (Supplemental Fig. S1B) in D (see Methods). (F) Temporal replication measured by Pol2 localization: Shown in F is the Pol2 abundance (color-coded) around 250 active ORIs at the indicated times. ORIs are ordered by their reported replication time (RepTime) (Yabuki et al. 2002): wt profiles on top, rtt109Δ profiles on the bottom. (G,H) RTT109 deletion increases fork velocity: Shown is the median Pol2 occupancy around early ORIs at the indicated times in wt (top) and RTT109-deleted (bottom) cells. The distribution of Pol2 velocity, as calculated from the temporal spread around each ORI is shown in G. n > 100 ORIs in two ChIP experiments that were analyzed per strain; median values are indicated (see Methods).
RTT109 deletion led to a moderate decrease in initiation rate (∼15%) (Fig. 2E; Supplemental Fig. S1B). More interesting, for us, was its effect on fork velocity, which increased by ∼30% (Fig. 2D,E). Thus, although the fork velocity found in wild-type cells (2.16 kb/min) was in good agreement with previous estimates, fork velocity in the RTT109-deleted strain was 2.84 kb/min (Fig. 2D,E). These results are in qualitative agreement with the increased replicon length found in asynchronous cells but predict a smaller increase than was in fact observed (60% vs. 100%). This difference may reflect the two experimental platforms (Batrakou et al. 2020) but more likely results from differences in the experimental conditions and, in particular, the need to synchronize cells in G1 before measuring their S-phase progression. Indeed, during this synchronization, cell mass increases, histone expression is repressed, and gene expression is reprogrammed, all of which could affect replication dynamics, likely at the initiation phase.
Our results suggest that Rtt109 activity results in a slower replication fork. To validate this result further, we followed more directly the temporal progression of the leading strand DNA polymerase (Pol2) itself. To this end, we released G1-synchronized cells into fresh media and followed them as they progressed synchronously through S phase, sampling cells at 2.5-min intervals for mapping Pol2's genomic localization using chromatin immunoprecipitation (ChIP)–seq. As expected, early time points showed distinct Pol2 peaks at early origins (Fig. 2F; Supplemental Fig. S1E,F). The starting occupancy of Pol2 at origins of replication initiation (ORIs) was tightly correlated between wild-type and RTT109-deleted cells, confirming the conserved hierarchy of origin firing time and showing no apparent activation of dormant origins (Supplemental Fig. S1C,D). As expected, with time, the Pol2 peaks gradually widened and moved away from the ORIs (Fig. 2F,G; Supplemental Fig. S1E,F). Fork velocities estimated from this spread were slower than previous estimates, perhaps owing to the epitope labeling of Pol2. Still, also here, deletion of RTT109 led to a clear ∼15% increase in fork velocity (Fig. 2H). Considering recent reports on transcription replication conflicts, we searched for locus-specific effects of RTT109 deletion on fork velocity. However, we could not detect such local effects. We thus concluded that Rtt109 activity uniformly slows down the progression of the replication fork.
H3K56 acetylation is dispensable for the Rtt109-dependent replication slowdown
All previously reported functions of Rtt109 depend on H3K56 acetylation. This acetylation increases the affinity of H3 to CAF-1, the principal chaperone mediating replication-coupled nucleosome assembly, raising the possibility that Rtt109 slows replication by promoting CAF-1-dependent histone incorporation (Li et al. 2008). To examine this, we measured replicon length in mutants individually deleted of the three CAF-1 subunits (Fig. 3A, left). Deletion of either RLF2 or CAC2, the two CAF-1-specific subunits, led to a slight decrease in replicon length (∼5%). Deletion of MSI1 led to a stronger effect (∼50% increase in replicon length), but this subunit also functions outside of the complex (Johnston et al. 2001). Therefore, perturbing the CAF-1 complex has little effect on replicon length, making it unlikely that the Rtt109 replication phenotype depends on its role in CAF-1-dependent nucleosome deposition.
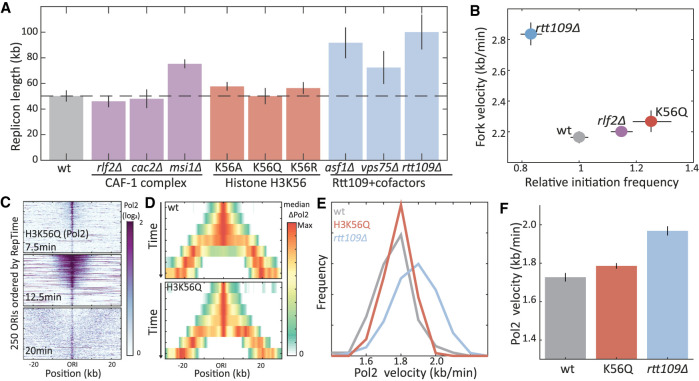
Figure 3.
Histone H3K56 acetylation does not account for Rtt109-dependent replication slowdown. (A) Replicon length is invariant to H3K56-related perturbations: Shown are the replicon lengths of the indicated substitution and deletion mutants. (B) H3K56 mutation does not increase fork velocity: Shown are the fork velocity and initiation frequency of the indicated mutants, as defined from the DNA content of cells progressing synchronously through S phase (for details, see Supplemental Fig. S2). (C–F) H3K56 mutation does not promote Pol2 progression: same as Figure 2, F through H, for the indicated mutants.
To examine more directly the possibility that Rtt109 slows replication by acetylating H3K56, we mutated the H3K56 residue into alanine, arginine, or glutamine (H3K56A, H3K56R, and H3K56Q) and examined the effect of these substitutions on replicon length (Fig. 3A, middle). Of note, although none of the mutants can be acetylated, they display different electrochemical properties: By electrostatic charge, glutamine is similar to acetylated lysine, whereas arginine imitates the unacetylated form (Hecht et al. 1995). By bulkiness, alanine is the most distinct, as it is smaller than glutamine, arginine, or lysine. No significant change in replicon length was observed for any of the three H3K56 mutants. As an additional test, we measured replicon length in mutants deleted of the two Rtt109 cofactors, Asf1 and Vps75 (Fig. 3A, right). Both histone chaperones are required for Rtt109-dependent H3 N-terminal acetylation in vivo, but only Asf1 is needed for H3K56 acetylation (Fillingham et al. 2008). Replicon length increased not only in ASF1-deleted (∼85%) but also in VPS75-deleted cells (∼45%), supporting the independence of this replication phenotype from H3K56 acetylation.
Mutations of H3K56, therefore, do not increase replicon length, contrasting RTT109 deletion. Replicon length, however, reports on the ratio between fork velocity and initiation rate, leaving open the possibility that H3K56's contribution to fork velocity is masked by a similar contribution to initiation rate. We therefore measured replication dynamics of these mutants more directly, following DNA content (Fig. 3B; Supplemental Fig. S2) and Pol2 occupancy (Fig. 3C–F) of cells progressing synchronously through S phase. In these assays, deleting RLF2, or mutating H3K56 to glutamine (H3K56Q), increased the initiation frequency (10%–25%). Fork velocity, however, remained invariant to both mutations. Therefore, H3K56ac does not account for the Rtt109-dependent slowdown of the replication fork.
H3 N-terminal acetylation can partially account for Rtt109-dependent replication slowdown
Following our finding that H3K56 is dispensable for Rtt109's role in slowing replication, we asked whether this phenotype depends on Rtt109's catalytic activity. To this end, we engineered two Rtt109 mutants, one deficient of all acetylase activities (DD287, 288AA) (Han et al. 2007a) and one that specifically perturbs histone N-terminal acetylation (K290Q) (Radovani et al. 2013). By measuring replicon lengths, we found that both mutations increased replicon length by approximately twofold, similar to a RTT109 deletion (Fig. 4A). These results pointed at H3 N-terminal acetylation as the likely process through which Rtt109 slows replication fork progression.
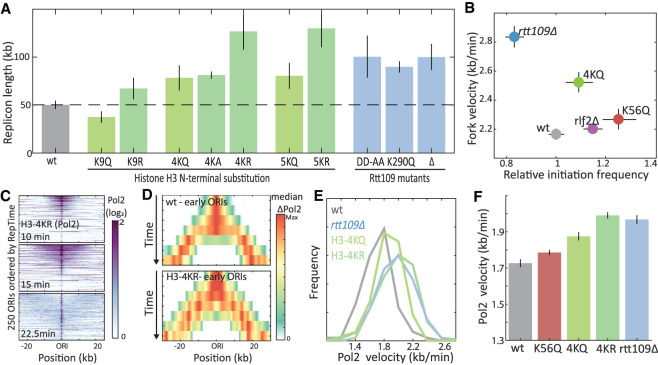
Figure 4.
Histone 3 N-terminal acetylation mutants increase replication fork velocity. (A) Replicon length increases in mutants deficient of histone H3 N-terminal acetylation: Shown are the replicon lengths of the indicated mutants. (B) H3 N-terminal mutations increase fork velocity: Shown are the fork velocity and initiation frequency of the indicated mutants, as defined from DNA content of cells progressing synchronously through S phase (for details, see Supplemental Fig. S3). (C–F) H3 N-terminal mutations accelerate Pol2 progression: same as Figure 2, F through H, for the indicated mutants.
To examine more directly the role of H3 N-terminal acetylation in slowing the replication fork, we mutated the respective lysine residues. We accounted for possible redundancies between these residues by simultaneously mutating one (K9), four (K4,9,14,18), or five (K9,14,18,23,27) residues. In each case, the lysine residues were mutated to alanine (A), glutamine (Q), or arginine (R). Measuring replicon lengths of these mutants, we found that, similar to a RTT109 deletion, most mutations increased replicon length (Fig. 4A). This increase was proportional to the number of mutated residues and was further dependent on the identity of the lysine replacements. Thus, glutamine and alanine were moderately disturbing (+60%–65%), whereas arginine, which best mimics unacetylated lysine and possibly a RTT109 deletion, led to the most pronounced increase in replicon length. The increase in replicon length in 4KR and 5KR mutants surpassed that of the RTT109 deletion strain, likely indicating an additional, Rtt109-independent effect. Thus, mutations in H3 N-terminal lysines change replicon length similarly to the RTT109 deletion.
To test whether these mutants change the replication fork velocity, we followed synchronized cultures. Quantifying DNA content in 4KQ mutants progressing synchronously through S phase revealed a 15% increase in fork velocity in these mutants (Fig. 4B; Supplemental Fig. S3). Profiling Pol2 progression of 4KQ and 4KR mutants showed an 8% and 15% increase in fork velocity, respectively, the latter being the same as observed in the RTT109-deleted strain using this same assay (Fig. 4C–F). Therefore, mutating N-terminal H3 lysines increases replication fork velocity, as observed in RTT109-deleted cells.
Epistatic analysis links the H3 N-terminal replication phenotype to its acetylation by Rtt109
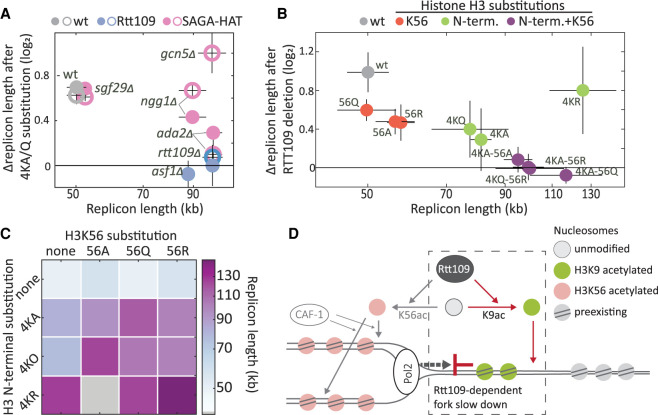
Our results above, showing that fork velocity increases upon mutating either Rtt109 or its H3 N-terminal lysine substrates, suggest that Rtt109 suppresses fork velocity by acetylating these same residues. These H3 N-terminal lysines, however, are acetylated not only by Rtt109 but also by Gcn5, leaving the question of whether the H3 N-terminal replication phenotype is Gcn5 dependent. We addressed this using a genetic analysis, reasoning that if the phenotype of H3 N-terminus mutants is Gcn5 dependent, expression of this phenotype would require a functional Gcn5 complex. To examine this, we determined the replication profile of GCN5-deleted cells and the consequences of H3 N-terminal substitutions in this background. Mutating H3 N-terminal lysine residues to glutamine in GCN5-deleted cells increased replicon length to a similar extent as it did in the wild-type background. Repeating this experiment in several other deletion mutants important for Gcn5 function led to a similar, although weaker effect (Fig. 5A). Therefore, the replication phenotype of H3 N-terminal mutants is not Gcn5 dependent.
Figure 5.
Rtt109 acts via H3 N-terminal acetylation to reduce fork velocity. (A) Genetic interaction between histone N-terminal acetylation and Rtt109 or SAGA/SLIK complexes. Relative replicon length change (log2) after an N-terminal substitution (4KQ, open circle; 4KA, closed circle) in wt (gray), RTT109-deleted (blue), or SAGA-HAT-core subunit-deleted (pink) cells is plotted against their replicon length (x-axis in log scale). (B) Genetic interactions between RTT109 and histone acetylation during DNA replication. For each lysine H3 N-terminal and K56 substitution (naming as in Figs. 3A, 4A), and their combinations, the relative replicon length (log2) after RTT109 deletion is plotted against its replicon length without RTT109 deletion. Gray indicates wt; red, H3K56 substitutions; green, N-terminal substitutions; purple, combined H3K56 and N-terminal substitutions; x-axis in log scale. (C) Synergistic effect of H3K56 and H3 N-terminal substitutions on DNA replication dynamics. The replicon lengths of mutants with combinations of H3K56 (columns) and H3 N-terminal 4K (rows) substitutions are shown. Colored squares indicate the replicon length after combining N-terminal mutation; gray squares indicate missing data for 4KR + K56A. (D) A working model: Histone H3 N-terminal acetylation by Rtt109 promotes nucleosome deposition in front of the fork and thus slows DNA replication.
To further confirm that the H3 N-terminal replication phenotype is Rtt109 dependent, we performed the same genetic analysis with Rtt109. In this case, no additional replication effect was observed when mutating the H3 N-terminal lysines to alanine or to glutamine in a RTT109-deleted background. This suggests that the H3 N-terminal replication phenotype is indeed Rtt109 dependent (Fig. 5A).
Finally, we wished to verify that Rtt109 affects replication dynamics exclusively through its known acetylation targets. We therefore combined the H3K56 and H3 N-terminal replacements and examined the consequences of deleting RTT109 in these backgrounds (Fig. 5B). Replicon length of mutants incapable of both H3 N-terminal and H3K56 acetylation remained invariant to RTT109 deletion, consistent with Rtt109's replication phenotype being fully dependent on its known targets. This analysis further revealed that mutating H3K56 in the background of H3 N-terminal mutants does increase replicon length (>35%), contrasting its limited effect in the wild-type background (<15%) (Fig. 5C), raising the possibility that H3K56 acetylation can serve as a back-up for H3 N-terminal acetylation (Fig. 5C).
Discussion
The doubling of DNA content during S phase requires the production of new histones to allow nucleosome assembly on replicated DNA. A specific set of marks is added to these newly synthesized histones before DNA incorporation. By acetylating newly synthesized histone H3 on K56 and N-terminal residues, Rtt109 contributes to the reshaping of the chromatin landscape during replication. Previous studies ascribe functional roles to H3K56ac, showing that it promotes CAF-1-dependent chromatin assembly at the wake of the fork, reduces genomic instabilities, and ensures expression homeostasis by suppressing transcription from replicated DNA (Voichek et al. 2016b). Here, we reveal, for the first time, a role of Rtt109-dependent H3 N-terminal acetylation, showing that this acetylation contributes to replication dynamics by slowing the replication fork. Tight control of fork velocity appears to be important for optimal genomic stability, as perturbations that either reduced this velocity, like nucleotide depletion (Bester et al. 2011), or increased it, for example, through PARP inhibition (Maya-Mendoza et al. 2018), were found to reduce genome stability.
Acetylated H3K56 accumulates on replicated DNA and thereby marks replicated regions. In contrast, Rtt109-dependent H3K9ac forms a wave progressing ∼3–5 kb ahead of the fork. Although the lack of highly specific antibodies prevents an analogous study of other N-terminal lysine residues, it is likely that considering their known redundancies, they show the same wave-like pattern as does H3K9ac. We therefore attribute the Rtt109-dependent replication slowdown to this prereplication acetylation wave. This, however, raises the question of how Rtt109, which only functions on free nucleosomes (Han et al. 2007b), specifically affects nucleosomes ahead of the fork.
Based on our data and the existing literature, we suggest the following working model (Fig. 5D). Overwinding of DNA ahead of the replication fork is known to evict nucleosomes, similar to the eviction shown during transcription (Corless and Gilbert 2016). We assume that H3 N-terminal acetylation by Rtt109 promotes the reassembly of nucleosomes in these regions. This reassembly protects the DNA until the fork approaches but will also slow it down, leading to the effect we observe. One possible candidate that might preferentially incorporate these acetylated histones ahead of the fork is FACT, as this chaperone is known to be associated with the DNA helicase in front of the fork (Gambus et al. 2006; Formosa 2012; Yang et al. 2016) and its function, at least in vitro, is facilitated by H3 N-terminal acetylation (Kurat et al. 2017; Pathak et al. 2018). Note, however, that to allow histone incorporation at distances ∼3–5 kb ahead of the fork would require the dissociation of FACT from the helicase. Another candidate is Vps75, which stimulates Rtt109-dependent acetylation of residues on the H3 N-terminus and promotes histone deposition on DNA in a manner that is dependent on Rtt109 (Berndsen et al. 2008).
Our working model therefore suggests that Rtt109 regulates two parallel pathways for nucleosome assembly: First, through H3K56 acetylation, Rtt109 promotes the CAF-1-pathway, which assembles nucleosomes on newly replicated DNA behind the fork. Second, through H3 N-terminal acetylation, it promotes deposition of nucleosomes ahead of the fork. The lack of H3K56ac ahead of the fork confirms that a distinct population of H3 exists, in which the H3 N-terminus, but not H3K56, is acetylated. The existence of such a population is further supported by recent in vitro experiments that mapped the different pathways by which Rtt109 acetylates H3 on its N-terminal residues and K56 (Cote et al. 2019), as well as the observation that the FACT complex does not associate with K56 acetylated histones (Foltman et al. 2013). This working model therefore puts our results in a framework of existing literature and points at future experiments required to establish its validity.
In conclusion, we suggest that by acetylating residues on the H3 N-terminus, Rtt109 promotes the reassembly of nucleosomes evicted by superhelical stresses emanating from the incoming fork. By doing so, Rtt109 reduces replication speed and potentially protects genome stability.
Methods
Saccharomyces cerevisiae strains
Deletion strains, except bar1Δ, were generated via transformation and homologous recombination of the desired gene with a G418 resistance cassette. Point mutations in both histone H3 genes (HHT1 and HHT2) and BAR1 deletion were generated via CRISPR editing (Mans et al. 2015; Anand et al. 2017). Strains for ChIP (Pol2 tagged with HA and Mcm7 tagged with Myc) were generated via homologs recombination with the tagging plasmids (pYM17+ and pYM18+). All strains and plasmids used in the study are listed in Supplemental Tables S1 and S2.
Growth conditions of cells harvested for DNA extraction
Cells were grown in yeast extract–peptone–dextrose (YPD) medium for at least 24 h until stationary phase and then diluted in fresh YPD to a very low OD600 and grown overnight until reaching an OD600 of 0.15–0.25. For asynchronous cultures, 1.5 mL of cells was taken at this point, media discarded, and frozen in liquid nitrogen. Additionally, cells were taken for DNA staining and fixated with 70% ethanol at 4°C. For synchronized experiments of strains without BAR1 deletion, cells were grown similarly and washed once with fresh YPD before adding alpha factor to a final concentration of 5 µg/mL. Cells were incubated for 3 h, washed once with fresh YPD, resuspended in fresh YPD, and incubated for periods of 1–3 h. Samples for DNA extraction and DNA staining were collected every 3–5 min and treated as described above. For time-resolved experiments with bar1Δ strains, the initial growth was as described above, but no additional wash was performed before alpha factor addition. Alpha factor was added to a final concentration of 7.5 ng/mL. After 3 h of incubation, cells were washed twice with fresh YPD supplemented with 50 µg/mL Pronase (Sigma-Aldrich) and then released into fresh YPD containing 50 µg/mL Pronase for the time-resolved sample collection described above.
Sample collection for ChIP of Pol2-HA (Pol2 subunit) during S phase
Cells lacking BAR1 and tagged with HA on Pol2 and Myc on Mcm7 were grown for a time-resolved experiment as described above. After release from alpha factor into fresh YPD supplemented with Pronase, cells were collected every 2.5 min and immediately cross-linked with final 1% formaldehyde for ChIP. In parallel, additional cells were collected for FACS staining. After 20 min, cells were quenched with glycine (125 mM final concentration) for 5 min and then washed twice with ice-cold, double-distilled water; pellets were flash-frozen in liquid nitrogen and stored at −80°C.
Library preparation and DNA sequencing
DNA was extracted either by blending the cells in 300 µL lysis buffer (50 mM HEPES at pH 7.5, 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate) with 0.5-mm zirconium oxide beads in a Bullet Blender 24 (Next Advance) for 1 min at level 8, or by resuspension in sorbitol 1 M + 0.1 M EDTA, addition of 0.016 kilounits lyticase in 1 M sorbitol and incubation for 30 min at 30°C. Cleared lysate was sonicated for 20 min (30 sec on, 30 sec off; low intensity) in a Bioruptor plus (Diagenode) cooled water bath sonicator, resulting in an average DNA fragment size of ∼200 bp. Lysates were RNase A treated (10 µg per sample in 100 µL lysis buffer) for 1 h at 37°C and then Proteinase K treated (20 µg per sample) for an additional 2 h at 37°C. Thirty to 50 µL of the lysate was taken from each sample, and a multiplexed library for sequencing was prepared as previously described (Blecher-Gonen et al. 2013). Libraries were sequenced in an Illumina HiSeq 2500 with 50-bp paired-end sequencing or in an Illumina NextSeq 500 with 50-bp paired-end sequencing.
Library preparation using Tn5 tagmentation and DNA sequencing
DNA was extracted by resuspension in sorbitol 1 M + 0.1 M EDTA, addition of 0.016 kilounits lyticase in 1 M sorbitol and incubation for 30 min at 30°C. Lysates were RNase A treated (10 µg per sample in 100 µL lysis buffer as described above) for 1 h at 37°C and then Proteinase K treated (20 µg per sample) for an additional 2 h at 37°C. Twenty-five microliters of the lysate was taken from each sample and cleaned with a X2.3 solid-phase reversible immobilization (SPRI) bead cleanup. Tagmentation reaction was then performed using homemade 2× tagmentation (TD) buffer (as described by Wang et al. 2013)—1.25 µL 2× TD buffer and 0.25 µL Tn5 enzyme per sample (1 µL cleaned DNA)—final reaction volume was 2.5 µL. The reaction was performed for 8 min at 55°C. Tagmented DNA was then amplified with KAPA HiFi hotstart ready mix PCR (13 cycles) with barcoded Tn5 primers, resulting in multiplexed libraries. PCR products were then cleaned with an additional X0.8 SPRI beads cleanup, and final libraries were sequenced on an Illumina NextSeq 500 with 50-bp sequencing from R1, 15 bp from R2, and both 8-bp indices.
ChIP of Pol2-HA and library preparation with tagmentation
ChIP was performed as described by Gutin et al. (2018) but with the on-bead library preparation substituted with on-bead tagmentation as described by Schmidl et al. (2015). In detail, cell pellets were thawed on ice, washed with 1 M ice-cold sorbitol, and resuspended in Buffer Z supplemented with 10 mM β-mercaptoethanol (5 µL buffer per 1 OD600 of cells). Cells were then treated with Zymolase 100T (0.5 units per 1 OD600 of cells) for 30 min at 30°C, and spheroplasts were then pelleted for 5 min at 6500g and 4°C and resuspended in lysis buffer with NP-40 (NP) buffer (supplemented with Protease inhibitors [PIs], 500 µM spermidine, and 1mM β-mercaptoethanol). After cell lysis, nuclei were pelleted at 16,000g for 10 min at 4°C and resuspended in NP buffer again. Sonication followed for 25 min (30 sec on, 30 sec off) at high intensity in a Bioruptor plus (Diagenode) cooled water bath sonicator. Resulting lysates were vortexed for 30 sec, kept on ice for at least 30 min, vortexed again, and centrifuged for 10 min at 16,000g at 4°C. Around 3 µg of HA antibody (produced in-house by the Weizmann Antibody Unit using a hybridoma cell line of the 12CA5 clone) was added to each supernatant, and samples were incubated for 2.5 h or overnight at 4°C with gentle tumbling. Twenty microliters of Protein G beads suspended in radioimmunoprecipitation assay (RIPA) buffer (supplemented with PI) was then added to each sample, and an additional 1-h incubation in 4°C tumbling followed. Samples were then magnetized and washed six times with RIPA buffer, three times with RIPA 500 buffer, three times with lithium chloride (LiCl) wash buffer, three times with 10 mM Tris (pH 7.5), and once with 10 mM Tris (pH 8) off the magnet (all wash buffers were supplemented with PI). Tagmentation reaction was performed using commercial Tn5 (Illumina) with 7.5 µL 2× TD buffer and 0.25 µL Tn5 enzyme per sample (final reaction volume was 15 µL). Beads were resuspended in the tagmentation mix, and the reaction was incubated for 10 min at 37°C. The reaction was stopped on ice with RIPA buffer, and beads were washed three times with RIPA buffer. Samples were resuspended in chromatin elution buffer, treated with 0.5 µg RNase A for 30 min at 37°C and 50 µg Proteinase K for 2 h at 37°C, and then de-cross-linked for 12–16 h at 65°C. DNA was isolated with 2.2X SPRI bead purification and then amplified with KAPA HiFi hotstart ready mix PCR (after preactivation at 98 for 3 min, 14 cycles) with barcoded Tn5 primers, resulting in multiplexed libraries. Libraries were sequenced by an Illumina NextSeq 500 or NovaSeq kits with 50-bp paired-end sequencing.
DNA staining
To assess cell cycle synchronization efficiency and position along the cell cycle, we followed DNA staining of samples for every time point using flow cytometry. Briefly, EtOH-fixed cells (see above) were washed twice with 50 mM Tris-HCl (pH 8), resuspended in 1 mg/mL RNase A for 40 min at 37°C, washed twice with 50 mM Tris-HCl (pH 8), and resuspended in 2 mg/mL Proteinase K for 1-h incubation at 37°C. Then, cells were washed twice with 50 mM Tris-HCl (pH 8) again, resuspended in SYBR Green (Sigma-Aldrich S9430; 1:1000) and incubated in the dark for 1 h at room temperature. Cells were washed again, resuspended in 50 mM Tris-HCl (pH 8), and sonicated in the Bioruptor plus (Diagenode) for three cycles of 10 sec on and 20 sec off at low intensity. SYBR Green fluorescence was measured using BD LSRII system (BD Biosciences).
Data processing for dynamic DNA-seq samples
DNA libraries were demultiplexed using custom Python code and read one of each sample aligned to the S288C genome R64-1-1 using Bowtie with the parameters “-X 4000 -m 1 –best –strata” (Langmead et al. 2009). All subsequent processing steps were performed with MATLAB. First, the resulting alignments were extended to between 100 and 200 bp (based on the average fragment length) in the read direction. The whole genome was then divided into 500-bp bins, that is, loci, and the coverage of all bins normalized so the mean coverage is one. To adjust for possible loci-specific biases in coverage, the normalized occupancy of a bin i in a sample X was compared to the median occupancy of this bin in all G1-synchronized samples of the same sequencing run:
where Ci,x is the adjusted coverage of bin i in sample x; ci,x is the normalized coverage of bin i in sample x, and cn,x is the coverage of all bins in sample x; adjusted coverage was smoothened using the moving average of 11 bins along the chromosome.
These adjusted coverages only report on the relative DNA amount of a certain locus compared with the other loci, with decreases in relative DNA amount indicating a relatively slower replication of this locus and not DNA degradation. We therefore calculated the changes in total DNA analogous to that of Bar-Ziv et al. (2016a). To this end, the bins were clustered according to the dynamics of their adjusted coverage (or similarly their published replication times) and the median coverage in each cluster and each time point calculated. Next, the increase of total DNA at each time point was initially chosen so that the absolute DNA stays constant for the slowest replicating cluster and subsequently smoothened with a sigmoidal curve. If, owing to a possible underestimation of DNA replication in the slowest replicating clusters, the total DNA amount apparently did not double during S phase, the increase at each time point was multiplied by a constant factor to reach full DNA replication. The resulting DNA dynamics were subsequently verified with the DNA staining profiles. The resulting total DNA at each time point was subsequently used to calculate the copy number in each bin: log2(Di,x) = log2(Ci,x) + log2(Tx), with Tx being the total DNA content at time x and Di,x being the absolute DNA content of locus i at time x.
Replication time calculation
To calculate the replication time of each locus, that is, 500-bp bin, we used a linear fit over the four closest time points to estimate the exact time at which the copy number reached 1.4, that is, log2(1.4) = 0.5, meaning 40% of cells replicated the corresponding locus.
Initiation frequency calculation
By using the replication time of each locus, we first filtered the 410 possible ORIs (OriDB) according to three criteria to only select those for which initiation could be detected in the respective time course: (1) The latest replication time of the slowest locus between two active ORIs needs to be at least 1 min after the slower ORI; if two ORIs are not separated by such a locus, only the faster one can be an active ORI. (2) In the direct vicinity of the ORI, the replication time of the locus must be strongly correlated with its distance from the ORI (c ≥ 0.75). To adjust for differences in the exit from alpha factor between the time courses, we next defined the adjusted initiation time of the active ORIs as the difference between their replication time and the replication time of the five first active ORIs. The initiation time of an ORI is determined by the initiation frequency of a cell and an ORI-specific parameter. To distinguish between these two parameters, we used robust linear regression to compare the adjusted initiation time of the active ORIs against their replication time as determined by (Yabuki et al. 2002). The slope of the fit then indicates the relative delay between early and late ORIs in certain cells, which can be defined as the inverse initiation frequency of a strain: that is, high initiation frequency = short delay between early and late ORIs, and low initiation frequency = long delay between early and late ORIs.
Fork velocity calculation
For each ORI, we first defined the boundaries between two ORIs as the locus with the latest replication time, as well as the environment of each ORI as those points between the closest left and the right boundary; those boundaries do not need to be symmetric and might even be next to the ORI for passively replicating ORIs. Next, we selected good early ORIs (replication time <25 min according to the method of Yabuki et al. 2002) for which the correlation between replication time and distance from the ORI in each locus in its environment is >0.85. For each locus in the environment of these good ORIs, we next calculated the delay in replication time between it and the corresponding ORI (Fig. 2D). Next, we used robust linear regression to determine the relation between replication time delay and distance from the ORI for the close locus (i.e., distance >3 kb and <11 kb). The slope then corresponds to the inverse of the fork velocity in a specific strain.
Data processing and analysis of Pol2 ChIP-seq data
ChIP-Seq DNA libraries were demultiplexed using bcl2fastq (Illumina), and the paired-end data were subsequently aligned to the Saccharomyces cerevisiae genome R64-1-1 using Bowtie 2 with the options “‐‐end-to-end ‐‐trim-to 40 ‐‐very-sensitive”. Duplicates were subsequently determined with customized Picard and genome-wide coverage for concordantly aligned, unique read- pairs calculated with genomeCoverage from BEDTools (Quinlan and Hall 2010) using the parameters “-d -pc”. All further processing was performed using MATLAB. First, the total coverage was normalized so that the mean coverage in the unique regions of the genome (e.g., not telomeres, rRNA genes or transposons) was one and subdivided into 200-bp bins. To determine the Pol2 movement around the earliest 100 ORIs, we first smoothened the signal around each ORI (±25 kb) using a moving Gaussian filter with the width of 21 bins and σ of five bins and next subtracted the median, normalized coverage at each bin over the entire time course to account for bin-specific coverage differences (for processing, see Supplemental Fig. S1). Then, two different “blinded” persons manually fitted a line to the moving peaks along the time course of randomly selected ORIs from different genotypes and time courses. After fitting around 100 forks per time course (two time courses for rtt109Δ, H3-4KR H3-K56Q, and wild-type cells; one time course for H3-4KQ), the slope of the fits was used to calculate the Pol2 velocity for the individual forks.
For visualization of median Pol2 dynamics, the same calculation was performed but on the median occupancy across all early ORIs (RepTime < 21).
Data access
The DNA sequencing data generated in this study have been submitted to the NCBI BioProject database (https://www.ncbi.nlm.nih.gov/bioproject/) under accession number PRJNA631463. Newly generated scripts and custom code required to reproduce the work were uploaded to the Supplemental Material as Supplemental Code and to GitHub (https://github.com/barkailab/Frenkel2020).
Competing interest statement
The authors declare no competing interests.
Supplementary Material
Acknowledgments
We thank the Barkai laboratory members for helpful discussions. We thank Ariel Gispan for implementing the Replicon length analysis. We thank Gat Krieger, Michal Chapal, Sagie Brodsky, Tamar Gera, Assaf Biran, and Inbal Zigdon for critical reading and commenting on the manuscript. This project was supported by the U.S. National Science Foundation–U.S. Israel Binational Science Foundation–Molecular and Cellular Biosciences (NSF-BSF-MCB; 2019625), the Israel Science Foundation (ISF; 1738/15), and the Minerva Center (AZ 57 469407 65).
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.266510.120.
References
- Anand R, Beach A, Li K, Haber J. 2017. Rad51-mediated double-strand break repair and mismatch correction of divergent substrates. Nature 544: 377–380. 10.1038/nature22046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister AJ, Kouzarides T. 2011. Regulation of chromatin by histone modifications. Cell Res 21: 381–395. 10.1038/cr.2011.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Ziv R, Voichek Y, Barkai N. 2016a. Chromatin dynamics during DNA replication. Genome Res 26: 1245–1256. 10.1101/gr.201244.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Ziv R, Voichek Y, Barkai N. 2016b. Dealing with gene-dosage imbalance during S phase. Trends Genet 32: 717–723. 10.1016/j.tig.2016.08.006 [DOI] [PubMed] [Google Scholar]
- Bar-Ziv R, Brodsky S, Chapal M, Barkai N. 2020. Transcription factor binding to replicated DNA. Cell Rep 30: 3989–3995.e4. 10.1016/j.celrep.2020.02.114 [DOI] [PubMed] [Google Scholar]
- Batrakou DG, Müller CA, Wilson RHC, Nieduszynski CA. 2020. DNA copy-number measurement of genome replication dynamics by high-throughput sequencing: the sort-seq, sync-seq and MFA-seq family. Nat Protoc 15: 1255–1284. 10.1038/s41596-019-0287-7 [DOI] [PubMed] [Google Scholar]
- Bauer WR, Hayes JJ, White JH, Wolffe AP. 1994. Nucleosome structural changes due to acetylation. J Mol Biol 236: 685–690. 10.1006/jmbi.1994.1180 [DOI] [PubMed] [Google Scholar]
- Bell SP, Dutta A. 2002. DNA replication in eukaryotic cells. Annu Rev Biochem 71: 333–374. 10.1146/annurev.biochem.71.110601.135425 [DOI] [PubMed] [Google Scholar]
- Bell SP, Stillman B. 1992. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature 357: 128–134. 10.1038/357128a0 [DOI] [PubMed] [Google Scholar]
- Belotserkovskaya R, Oh S, Bondarenko VA, Orphanides G, Studitsky VM, Reinberg D. 2003. FACT facilitates transcription-dependent nucleosome alteration. Science 301: 1090–1093. 10.1126/science.1085703 [DOI] [PubMed] [Google Scholar]
- Berndsen CE, Tsubota T, Lindner SE, Lee S, Holton JM, Kaufman PD, Keck JL, Denu JM. 2008. Molecular functions of the histone acetyltransferase chaperone complex Rtt109–Vps75. Nat Struct Mol Biol 15: 948–956. 10.1038/nsmb.1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bester AC, Roniger M, Oren YS, Im MM, Sarni D, Chaoat M, Bensimon A, Zamir G, Shewach DS, Kerem B. 2011. Nucleotide deficiency promotes genomic instability in early stages of cancer development. Cell 145: 435–446. 10.1016/j.cell.2011.03.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco JN, Poli J, Saksouk J, Bacal J, Silva MJ, Yoshida K, Lin YL, Tourrière H, Lengronne A, Pasero P. 2012. Analysis of DNA replication profiles in budding yeast and mammalian cells using DNA combing. Methods 57: 149–157. 10.1016/j.ymeth.2012.04.007 [DOI] [PubMed] [Google Scholar]
- Blecher-Gonen R, Barnett-Itzhaki Z, Jaitin D, Amann-Zalcenstein D, Lara-Astiaso D, Amit I. 2013. High-throughput chromatin immunoprecipitation for genome-wide mapping of in vivo protein-DNA interactions and epigenomic states. Nat Protoc 8: 539–554. 10.1038/nprot.2013.023 [DOI] [PubMed] [Google Scholar]
- Burgess RJ, Zhou H, Han J, Zhang Z. 2010. A role for Gcn5 in replication-coupled nucleosome assembly. Mol Cell 37: 469–480. 10.1016/j.molcel.2010.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corless S, Gilbert N. 2016. Effects of DNA supercoiling on chromatin architecture. Biophys Rev 8: 245–258. 10.1007/s12551-016-0210-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cote JM, Kuo Y-M, Henry RA, Scherman H, Krzizike DD, Andrews AJ. 2019. Two factor authentication: Asf1 mediates crosstalk between H3 K14 and K56 acetylation. Nucleic Acids Res 47: 7380–7391. 10.1093/nar/gkz508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll R, Hudson A, Jackson SP. 2007. Yeast Rtt109 promotes genome stability by acetylating histone H3 on lysine 56. Science 315: 649–652. 10.1126/science.1135862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton ML, Prinz JA, MacAlpine HK, Tretyakov G, Kharchenko PV, MacAlpine DM. 2011. Chromatin signatures of the Drosophila replication program. Genome Res 21: 164–174. 10.1101/gr.116038.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillingham J, Recht J, Silva AC, Suter B, Emili A, Stagljar I, Krogan NJ, Allis CD, Keogh M-C, Greenblatt JF. 2008. Chaperone control of the activity and specificity of the histone H3 acetyltransferase Rtt109. Mol Cell Biol 28: 4342–4353. 10.1128/MCB.00182-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltman M, Evrin C, De Piccoli G, Jones Richard C, Edmondson Rick D, Katou Y, Nakato R, Shirahige K, Labib K. 2013. Eukaryotic replisome components cooperate to process histones during chromosome replication. Cell Rep 3: 892–904. 10.1016/j.celrep.2013.02.028 [DOI] [PubMed] [Google Scholar]
- Formosa T. 2012. The role of FACT in making and breaking nucleosomes. Biochim Biophys Acta 1819: 247–255. 10.1016/j.bbagrm.2011.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambus A, Jones RC, Sanchez-Diaz A, Kanemaki M, van Deursen F, Edmondson RD, Labib K. 2006. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat Cell Biol 8: 358–366. 10.1038/ncb1382 [DOI] [PubMed] [Google Scholar]
- Gispan A, Carmi M, Barkai N. 2016. Model-based analysis of DNA replication profiles: predicting replication fork velocity and initiation rate by profiling free-cycling cells. Genome Res 27: 310–319. 10.1101/gr.205849.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth A, Rocha W, Verreault A, Almouzni G. 2007. Chromatin challenges during DNA replication and repair. Cell 128: 721–733. 10.1016/j.cell.2007.01.030 [DOI] [PubMed] [Google Scholar]
- Gutin J, Sadeh R, Bodenheimer N, Joseph-Strauss D, Klein-Brill A, Alajem A, Ram O, Friedman N. 2018. Fine-resolution mapping of TF binding and chromatin interactions. Cell Rep 22: 2797–2807. 10.1016/j.celrep.2018.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamperl S, Cimprich KA. 2016. Conflict resolution in the genome: how transcription and replication make it work. Cell 167: 1455–1467. 10.1016/j.cell.2016.09.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Zhou H, Horazdovsky B, Zhang K, Xu R-M, Zhang Z. 2007a. Rtt109 acetylates histone H3 lysine 56 and functions in DNA replication. Science 315: 653–655. 10.1126/science.1133234 [DOI] [PubMed] [Google Scholar]
- Han J, Zhou H, Li Z, Xu RM, Zhang Z. 2007b. The Rtt109-Vps75 histone acetyltransferase complex acetylates non-nucleosomal histone H3. J Biol Chem 282: 14158–14164. 10.1074/jbc.M700611200 [DOI] [PubMed] [Google Scholar]
- Hecht A, Laroche T, Strahl-Bolsinger S, Gasser SM, Grunstein M. 1995. Histone H3 and H4 N-termini interact with SIR3 and SIR4 proteins: a molecular model for the formation of heterochromatin in yeast. Cell 80: 583–592. 10.1016/0092-8674(95)90512-X [DOI] [PubMed] [Google Scholar]
- Hong L, Schroth GP, Matthews HR, Yau P, Bradbury EM. 1993. Studies of the DNA binding properties of histone H4 amino terminus. Thermal denaturation studies reveal that acetylation markedly reduces the binding constant of the H4 “tail” to DNA. J Biol Chem 268: 305–314. 10.1016/S0021-9258(18)54150-8 [DOI] [PubMed] [Google Scholar]
- Johnston SD, Enomoto S, Schneper L, McClellan MC, Twu F, Montgomery ND, Haney SA, Broach JR, Berman J. 2001. CAC3(MSI1) suppression of RAS2G19V is independent of chromatin assembly factor I and mediated by NPR1. Mol Cell Biol 21: 1784–1794. 10.1128/MCB.21.5.1784-1794.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurat CF, Yeeles JTP, Patel H, Early A, Diffley JFX. 2017. Chromatin controls DNA replication origin selection, lagging-strand synthesis, and replication fork rates. Mol Cell 65: 117–130. 10.1016/j.molcel.2016.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DY, Hayes JJ, Pruss D, Wolffe AP. 1993. A positive role for histone acetylation in transcription factor access to nucleosomal DNA. Cell 72: 73–84. 10.1016/0092-8674(93)90051-Q [DOI] [PubMed] [Google Scholar]
- Li Q, Zhou H, Wurtele H, Davies B, Horazdovsky B, Verreault A, Zhang Z. 2008. Acetylation of histone H3 lysine 56 regulates replication-coupled nucleosome assembly. Cell 134: 244–255. 10.1016/j.cell.2008.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubelsky Y, Prinz JA, DeNapoli L, Li Y, Belsky JA, MacAlpine DM. 2014. DNA replication and transcription programs respond to the same chromatin cues. Genome Res 24: 1102–1114. 10.1101/gr.160010.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAlpine HK, Gordân R, Powell SK, Hartemink AJ, MacAlpine DM. 2010. Drosophila ORC localizes to open chromatin and marks sites of cohesin complex loading. Genome Res 20: 201–211. 10.1101/gr.097873.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mans R, van Rossum HM, Wijsman M, Backx A, Kuijpers NG, van den Broek M, Daran-Lapujade P, Pronk JT, van Maris AJ, Daran JM. 2015. CRISPR/Cas9: a molecular Swiss army knife for simultaneous introduction of multiple genetic modifications in Saccharomyces cerevisiae. FEMS Yeast Res 15. 10.1093/femsyr/fov004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maya-Mendoza A, Moudry P, Merchut-Maya JM, Lee M, Strauss R, Bartek J. 2018. High speed of fork progression induces DNA replication stress and genomic instability. Nature 559: 279–284. 10.1038/s41586-018-0261-5 [DOI] [PubMed] [Google Scholar]
- Miotto B, Ji Z, Struhl K. 2016. Selectivity of ORC binding sites and the relation to replication timing, fragile sites, and deletions in cancers. Proc Natl Acad Sci U S A 113: E4810–E4819. 10.1073/pnas.1609060113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller CA, Hawkins M, Retkute R, Malla S, Wilson R, Blythe MJ, Nakato R, Komata M, Shirahige K, de Moura APS, et al. 2014. The dynamics of genome replication using deep sequencing. Nucleic Acids Res 42: e3. 10.1093/nar/gkt878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orphanides G, Wu WH, Lane WS, Hampsey M, Reinberg D. 1999. The chromatin-specific transcription elongation factor FACT comprises human SPT16 and SSRP1 proteins. Nature 400: 284–288. 10.1038/22350 [DOI] [PubMed] [Google Scholar]
- Owen-Hughes T, Gkikopoulos T. 2012. Making sense of transcribing chromatin. Curr Opin Cell Biol 24: 296–304. 10.1016/j.ceb.2012.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak R, Singh P, Ananthakrishnan S, Adamczyk S, Schimmel O, Govind CK. 2018. Acetylation-dependent recruitment of the FACT complex and its role in regulating Pol II occupancy genome-wide in Saccharomyces cerevisiae. Genetics 209: 743–756. 10.1534/genetics.118.300943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radovani E, Cadorin M, Shams T, El-Rass S, Karsou AR, Kim H-S, Kurat CF, Keogh M-C, Greenblatt JF, Fillingham JS. 2013. The carboxyl terminus of Rtt109 functions in chaperone control of histone acetylation. Eukaryotic Cell 12: 654–664. 10.1128/EC.00291-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez J, Tsukiyama T. 2013. ATR-like kinase Mec1 facilitates both chromatin accessibility at DNA replication forks and replication fork progression during replication stress. Genes Dev 27: 74–86. 10.1101/gad.202978.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidl C, Rendeiro AF, Sheffield NC, Bock C. 2015. ChIPmentation: fast, robust, low-input ChIP-seq for histones and transcription factors. Nat Methods 12: 963–965. 10.1038/nmeth.3542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S, Stillman B. 1989. Purification and characterization of CAF-I, a human cell factor required for chromatin assembly during DNA replication in vitro. Cell 58: 15–25. 10.1016/0092-8674(89)90398-X [DOI] [PubMed] [Google Scholar]
- Tsubota T, Berndsen CE, Erkmann JA, Smith CL, Yang L, Freitas MA, Denu JM, Kaufman PD. 2007. Histone H3-K56 acetylation is catalyzed by histone chaperone-dependent complexes. Mol Cell 25: 703–712. 10.1016/j.molcel.2007.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelauer M, Rubbi L, Lucas I, Brewer BJ, Grunstein M. 2002. Histone acetylation regulates the time of replication origin firing. Mol Cell 10: 1223–1233. 10.1016/S1097-2765(02)00702-5 [DOI] [PubMed] [Google Scholar]
- Voichek Y, Bar-Ziv R, Barkai N. 2016a. A role for Rtt109 in buffering gene-dosage imbalance during DNA replication. Nucleus 7: 375–381. 10.1080/19491034.2016.1216743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voichek Y, Bar-Ziv R, Barkai N. 2016b. Expression homeostasis during DNA replication. Science 351: 1087–1090. 10.1126/science.aad1162 [DOI] [PubMed] [Google Scholar]
- Voichek Y, Mittelman K, Gordon Y, Bar-Ziv R, Lifshitz Smit D, Shenhav R, Barkai N. 2018. Epigenetic control of expression homeostasis during replication is stabilized by the replication checkpoint. Mol Cell 70: 1121–1133.e9. 10.1016/j.molcel.2018.05.015 [DOI] [PubMed] [Google Scholar]
- Wang Q, Gu L, Adey A, Radlwimmer B, Wang W, Hovestadt V, Bähr M, Wolf S, Shendure J, Eils R, et al. 2013. Tagmentation-based whole-genome bisulfite sequencing. Nat Protoc 8: 2022–2032. 10.1038/nprot.2013.118 [DOI] [PubMed] [Google Scholar]
- Yabuki N, Terashima H, Kitada K. 2002. Mapping of early firing origins on a replication profile of budding yeast. Genes Cells 7: 781–789. 10.1046/j.1365-2443.2002.00559.x [DOI] [PubMed] [Google Scholar]
- Yang J, Zhang X, Feng J, Leng H, Li S, Xiao J, Liu S, Xu Z, Xu J, Li D, et al. 2016. The histone chaperone FACT contributes to DNA replication-coupled nucleosome assembly. Cell Rep 14: 1128–1141. 10.1016/j.celrep.2015.12.096 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.