Abstract
The Coronavirus Disease-2019 (COVID-19) imposed public health emergency and affected millions of people around the globe. As of January 2021, 100 million confirmed cases of COVID-19 along with more than 2 million deaths were reported worldwide. SARS-CoV-2 infection causes excessive production of pro-inflammatory cytokines thereby leading to the development of “Cytokine Storm Syndrome.” This condition results in uncontrollable inflammation that further imposes multiple-organ-failure eventually leading to death. SARS-CoV-2 induces unrestrained innate immune response and impairs adaptive immune responses thereby causing tissue damage. Thus, understanding the foremost features and evolution of innate and adaptive immunity to SARS-CoV-2 is crucial in anticipating COVID-19 outcomes and in developing effective strategies to control the viral spread. In the present review, we exhaustively discuss the sequential key immunological events that occur during SARS-CoV-2 infection and are involved in the immunopathogenesis of COVID-19. In addition to this, we also highlight various therapeutic options already in use such as immunosuppressive drugs, plasma therapy and intravenous immunoglobulins along with various novel potent therapeutic options that should be considered in managing COVID-19 infection such as traditional medicines and probiotics.
Keywords: COVID-19, cytokine storm, immunology, immunotherapy, SARS-CoV-2
1. Introduction
In December 2019, an outbreak of pneumonia occurred in Wuhan city of China that rapidly spread around the globe and posed serious public health emergency [1]. On 9 January 2020, it was officially announced that novel coronavirus 2019-nCoV is the reason behind the outbreak in Wuhan, China [2]. Later International Committee on Taxonomy named the novel coronavirus as Severe Respiratory Disease Syndrome-Coronavirus-2 (SARS-CoV-2) and World Health Organization (WHO) named the disease as Coronavirus Disease-2019 or COVID-19 [3]. Coronaviruses (CoVs) cause infection in humans and animals and are found to be responsible for various respiratory, renal, gastrointestinal and neurological disorders. CoVs are classified into four genera viz. alpha, beta, gamma and delta CoVs. Alpha and beta CoVs infect humans and usually cause upper respiratory tract infections but in some patients also cause lower respiratory tract infections [4]. SARS-CoV-2 belongs to the family Coronaviridae, order Nidovirales, genus Betacoronavirus and subgenus Sarbecovirus [5]. SARS-CoV-2 is the 7th CoV in the list that is reported to cause infections in humans. The other six CoVs are SARS-CoV, MERS-CoV, HKUI, NL63, OC43 and 229E. SARS-CoV and MERS-CoV cause various fatal and respiratory diseases like SERS-CoV-2 whereas HKU1, NL63, OC43 and 229E cause only minor symptoms [6]. SARS-CoV was found to be responsible for an epidemic in 2002–2003 which started from China and Asia Pacific regions and affected around 8000 people across 37 countries with fatality rate of 10% [7, 8]. Common symptoms observed in SARS-CoV infected patients were fever, dyspnea, dry cough and hypoxemia [9]. Middle East Respiratory Syndrome-Coronavirus (MERS-CoV) is a 2C beta CoV and was first reported in 2012 from Saudi Arabia [10]. MERS-CoV caused severe pneumonia and renal failure in infected patients [11]. The SARS-CoV-2 virus shares 79.6% sequence similarity with the SARS-CoV virus but SARS-CoV-2 is found to be more pathogenic [12]. Due to its pathogenicity and easy transmission from human to human WHO declared COVID-19 as pandemic disease on 11 March 2020. As of January 2021, there are 100 million confirmed cases of COVID-19 worldwide with over 2 million reported deaths. SARS-CoV-2 cause mild respiratory disorders to acute pneumonia and multiple organ failure and in severe cases can eventually lead to death [13]. Whole genome sequencing revealed that the SARS-CoV-2 is more closely related to bat CoV RaTGI3 which was isolated from Rhinolophus affinis with 96.2% sequence similarity [14].
Immune system plays a pivotal role in the pathogenesis of COVID-19. SARS-CoV-2 induces unrestrained innate immune response and impairs adaptive immune responses leading to widespread tissue damage. Till now, there is no effective treatment available for COVID-19. Knowledge of immunopathogenesis of COVID-19 will help in designing suitable immune therapy for the treatment of SARS-CoV-2 infection. In this review, we have discussed the pathogenesis and immunopathogenesis of COVID-19 along with the potential immunotherapeutic interventions that can be targeted toward the dysregulated immune system. We also discuss the plausible relevance of gut microbiota and probiotics in COVID-19.
2. Structure of SARS-CoV-2
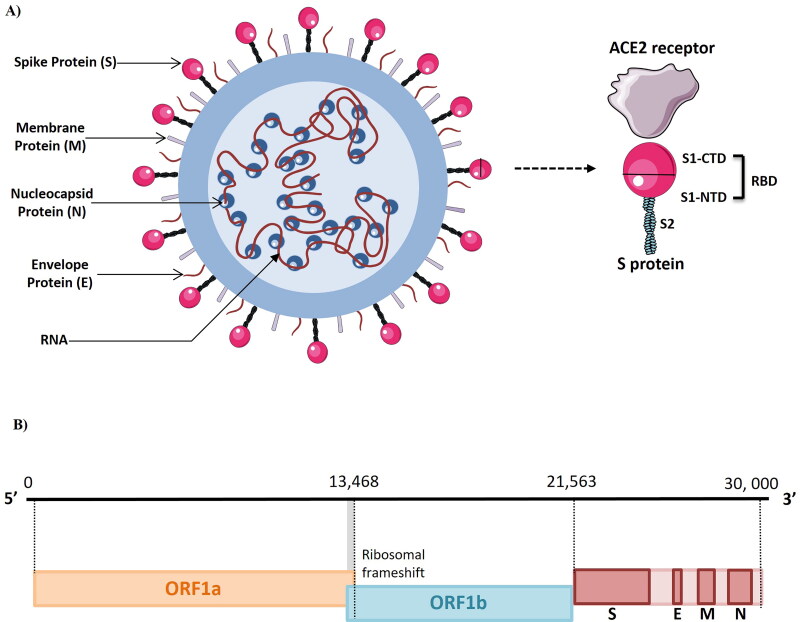
The novel CoV is an enveloped, positive sense, single stranded RNA virus with a genome size of 26 000 to 32 000 nucleotides encoding 14 open reading frames (ORFs). The first two large ORFs (orf1ab and orf1a) which are present at the 5′ end cover almost two third of the genome (20 kb; Figure 1). They constitute the replicase gene which contains 16 nonstructural proteins (nsps). Replicase gene is required for replication and transcription. Replicase gene codes for two polyproteins: pp1a (contains the 1–11 nsps) and pp1ab gene (contains the 12–16 nsps). The 3′ end of the genome which is around 10 kb encodes 4 structural and 8 accessory proteins. The structural proteins consist of spike (S) protein, membrane (M) protein, envelope (E) protein and the nucleocapsid (N) protein [15]. S protein allows the entry of virus into the host cell. M and E protein regulate virus assembly and N protein facilitates RNA synthesis. S protein is projected from the membrane surface and gives the crown like appearance to the virus [16]. The S protein is 1255 amino acids long and consists of three domains: large N terminal ectodomain (NTD), single transmembrane domain and small cytoplasmic endodomain (CTD). The NTD consists of single S subunit which is cleaved by the host proteases such as transmembrane protease serine 2 (TMPRSS2) into two subunits: S1 and S2. The M protein is the most abundant protein of the virus and is responsible for providing shape to the virion. M protein interacts with the spike protein and allows its incorporation into the viral envelope. E protein is required for pathogenesis, envelope formation, assembly and release of viruses from infected cells. N protein binds to the RNA in beads on a string conformation and forms the ribonucleic protein complex named as nucleocapsid. Both of these domains are required for binding of N protein to RNA [17]. Hemagglutinin esterase (HE) is present on the viral surface and is the fifth structural protein. It acts as hemagglutinin that binds to surface glycoproteins via sialic acid and enhance the S protein mediated viral entry into the host [18].
Figure 1.
(A, B) Structure and genome organization of SARS-CoV-2 (Figure illustrated with the help of https://smart.servier.com/. ).
3. Pathogenesis of COVID-19
SARS-CoV-2 is transmitted from infected patients to healthy individuals through direct contact or via spread of respiratory droplets from the infected patients [19]. The median incubation period of the SARS-CoV-2 infection is approximately 5.1 days with 97.5% of the population developing symptoms within 11.5 days [20]. The common symptoms associated with the infection are dry cough, fever, pain, weakness, chest tightness, loss of smell and taste, dyspnea, accompanied by acute respiratory distress syndrome (ARDS) [21, 22]. ARDS is a severe disease which leads to respiratory failure and is characterized by hypoxemia, difficulty in breathing and onset of pulmonary edema. It leads to damage in lung endothelium and alveolar epithelium [23]. RNAaemia (detectable viral load of SARS-CoV-2 in serum) and acute cardiac injury has also been observed in some of the COVID-19 patients [21]. Immunocompromised individuals like those who have diabetes, cardiovascular diseases and hypertension are estimated to be at higher risk of COVID-19 infection [24]. Mortality rate is reported to be higher in elderly population. Recently, a study reported that 73.9% of the infected individuals less than the age of 60 did not develop symptoms. Thus, indicating that elderly people are highly susceptible to developing symptoms [25]. Gender difference also affects the severity and mortality of the disease with men being more prone to infection and along with enhanced mortality with respect to women [26]. Hypoalbuminemia, liver dysfunction and renal abnormalities are also observed in severe cases of COVID-19 [27–29]. About 8–12% of COVID-19 patients also have acute cardiac injury due to systemic inflammation [30].
SARS-CoV-2 virus shares 79% similarity with the SARS-CoV virus. Like SARS-CoV it also binds to the human Angiotensin Converting Enzyme 2 (ACE2) that is expressed by various cells including lungs [31]. Apart from the lungs, surface expression of ACE2 is also observed on the epithelial cells of the small intestine, mucosa of the oral cavity, venous, arterial endothelial cells and arterial smooth muscles of all organs [32, 33]. This forms one of the major reason behind the multiorgan failure observed in case of SARS-CoV-2 [34]. ACE2 is homologue of ACE which produces angiotensin II from angiotensin I. Angiotensin II is the major protein of Renin Angiotensin System (RAS) and is required for vasoconstriction and for other biological functions. It has been reported that ACE2 negatively regulates RAS by suppressing the angiotensin II level, and thus, prevents from ARDS [35]. ACE2 receptor is expressed on the ciliated airway epithelial cells and on the type 2 alveolar cells of the lungs [36]. SARS-CoV-2 interacts with the ACE2 receptor with the help of S protein. S protein is cleaved into two subunits (S1 and S2) by the host protease TMPRSS2 at the boundary of the S1–S2 [37] (Figure 2). S1 subunit exhibits the receptor binding domain whereas S2 subunit facilitates the membrane fusion [38].
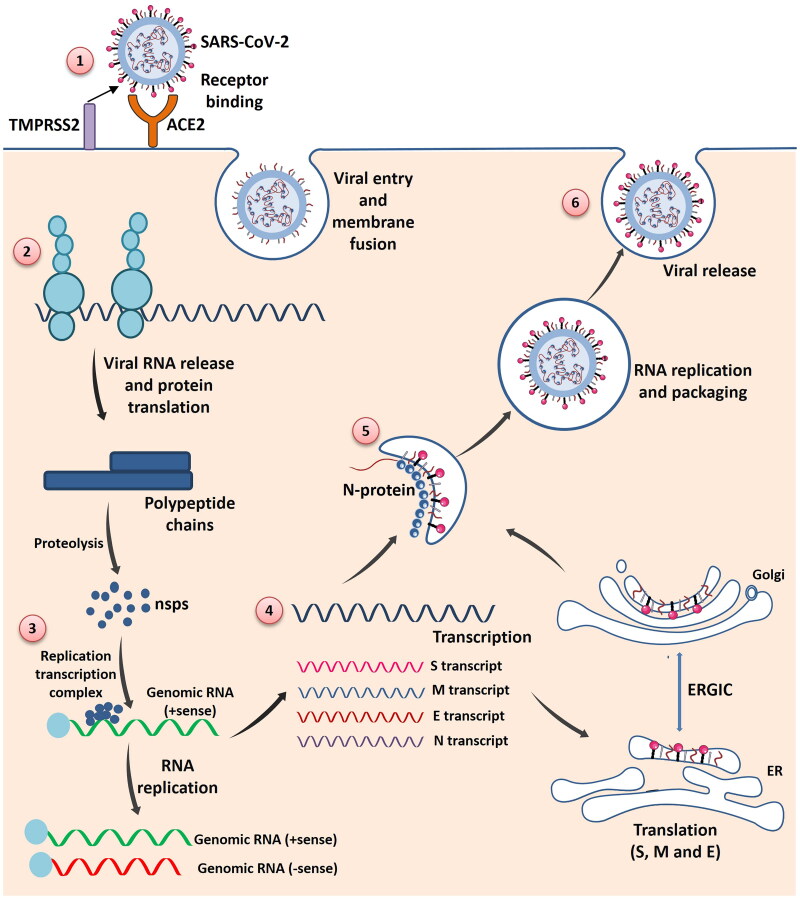
Figure 2.
The schematic diagram of SARS-CoV-2 invasion and replication into host cells in step wise manner: (1) SARS-CoV-2 enter into the cell by binding to the ACE2 receptor present on the host cell membrane via S protein which mediates viral-host membrane fusion and viral entry. (2) Entry of SARS-CoV-2 results in the uncoating of viral RNA into cytoplasm which then undergoes translation to produce polyproteins pp1a and pp1b, which are further processed by virus-encoded proteinases into individual nonstructural proteins (nsps). (3) Replication transcription complex is formed by some of the nsps. Replication transcription complex uses the (+) stand genomic RNA as template. Following replication (+) stand genomic RNA is produced which becomes the genome of the new viral particle. (4) Subgenomic RNAs synthesized through transcription are translated into viral structural proteins: S (Spike) protein, M (Membrane) protein, E (Envelope) protein, N (Nucleocapsid) protein. (5) N protein combined with the (+) strand genomic RNA to form the nucleoprotein complex. S, E and M proteins enter into endoplasmic reticulum (ER) and transported to Golgi apparatus. The nucleoprotein complex and S, E and M proteins are further assembled in the ER-Golgi intermediate compartment (ERGIC) to form mature virion. (6) Virions are released from the host cells by exocytosis (Figure illustrated with the help of https://smart.servier.com/. ).
Interestingly, it has been observed that the RBD domains of the SARS-CoV and SARS-CoV-2 shares 72% similarity in the amino acids sequence [39]. Residues required for attachment to ACE2 in RBD are also highly conserved in both the viruses [1]. Molecular modeling and biophysical techniques revealed that RBD of the SARS-CoV-2 binds more strongly and with higher affinity to the ACE2 receptor than the SARS-CoV [40]. ACE2 binding ridge in SARS-CoV-2 is more compact [12]. Cryo-electron microscopy structure of novel CoV S protein trimer at the resolution of 3.5 Ao revealed that RBD domain of the S trimer rotate in a conformation that is more accessible to the ACE2 [40]. SARS-CoV-2 also consists of different loop having flexible glycyl residues in place of rigid propyl residues that are present in SARS-CoV [39]. It has a very different furin like cleavage site in the S protein that is not present in other SARS like CoVs [41]. Thus, we can conclude that these features may be one of the reasons responsible for increased pathogenicity of SARS-CoV-2.
4. Immunopathogenesis of COVID-19
Viral infection stimulates the host immune system which is initiated by the activation of innate immune cells that recognizes the molecular patterns associated with the pathogens. Failure of the innate immune responses in eliminating the virus leads to the activation of adaptive immune system. Induction of innate and adaptive immune cells stimulate the secretion of cytokines or chemokines like interleukin-6 (IL-6), interferon-γ (IFN-γ), interferon-γ-inducible protein-10 (IP-10) and monocyte chemoattractant protein-1 (MCP)-1. These cytokines and chemokines promote the influx of monocytes/macrophages and neutrophils from the blood to the site of infection. These cells secrete the cytotoxic substances to clear the viral infection. Normally, this response is capable of eliminating the virus but sometimes immune system is dysregulated which leads to the disruption of the immune homeostasis [42]. In case of SARS-CoV-2 infection intense inflammatory response against the virus leads to excessive production of pro-inflammatory cytokines which thereby promote lung pathogenesis and respiratory failure (Figure 3). Pro-inflammatory cytokines induce uncontrolled accumulation of monocytes/macrophages and neutrophils at the site of infection. These cells then produce cytotoxic substances like reactive oxygen species (ROS) which leads to cell death and tissue damage [42]. In further sections, we have discussed the immunopathogenesis of SARS-CoV-2 infection in more details (Figure 4 and Table 1).
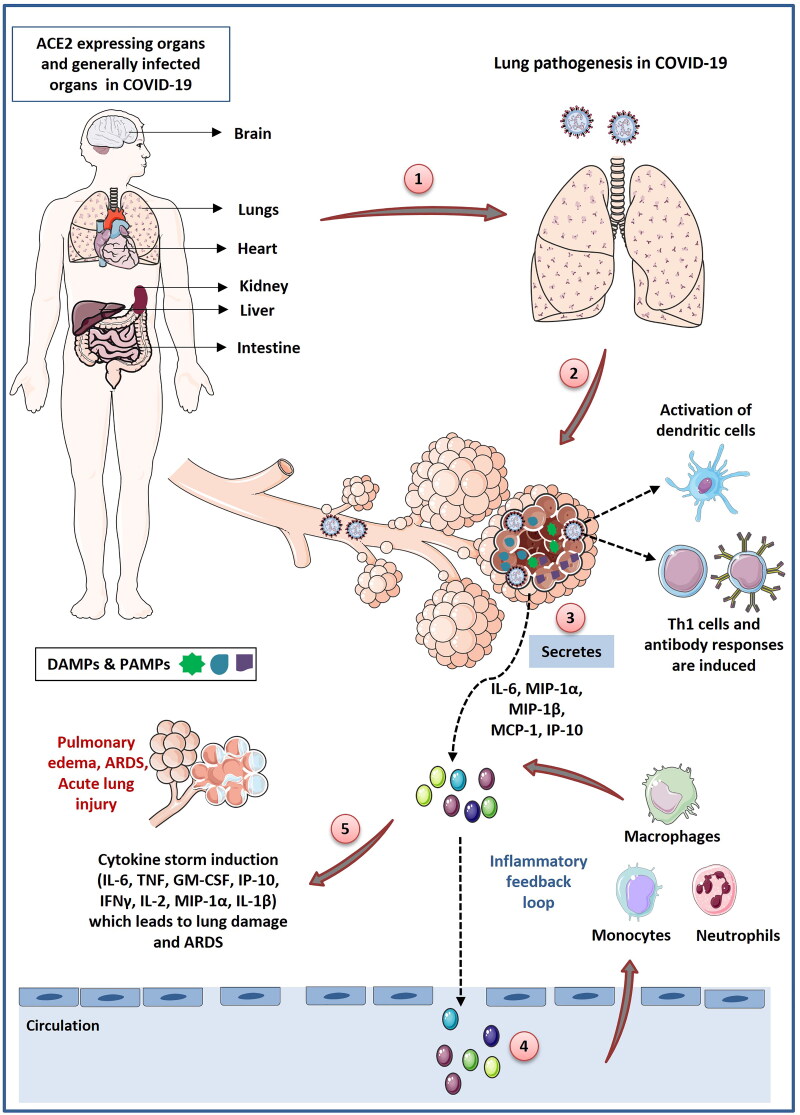
Figure 3.
Schematic representation of immunopathogenesis during COVID-19 in a step wise manner: (1) SARS-CoV-2 gain entry into the lung cells through ACE2 receptor. ACE2 receptor is expressed by different organs of the body like brain, lungs, kidney, liver and intestine. SARS-CoV-2 frequently infects these organs but the pathogenesis of SARS-CoV-2 infection initiates from lungs and cause major damage to the lungs. Lung infection starts when SARS-CoV-2 enter into type 2 alveolar cells through ACE2 receptor. (2) Virus replicates into the lung cells and induce cells to undergo pyroptosis and secrete damage associated molecular patterns (DAMPs). (3) DAMPs are recognized by adjoining epithelial cells, alveolar macrophages and endothelial cells which stimulate the secretion of various pro-inflammatory cytokines and chemokines like IL-6, IL-10, MIP1-α, MIP1-1β, MCP-1. (4) Release of theses cytokines and chemokines induce the recruitment of monocytes, macrophages and neutrophils into lungs which further secrete the proinflammatory cytokines and form the inflammatory feedback loop. (5) During SARS-CoV-2 infection immune response is dysregulated which leads to the persistent recruitment of immune cells which promote overproduction of inflammatory cytokines which cause various lung disorders like ARDS and acute lung injury. SARS-CoV-2 infection induce delayed activation of dendritic cells that cause defective T cell response. Most of the T lymphocytes in SARS-CoV-2 infection are induced to become Th1 cells. SARS-CoV-2 infection also induce delayed antibody response (Figure illustrated with the help of https://smart.servier.com/. ).
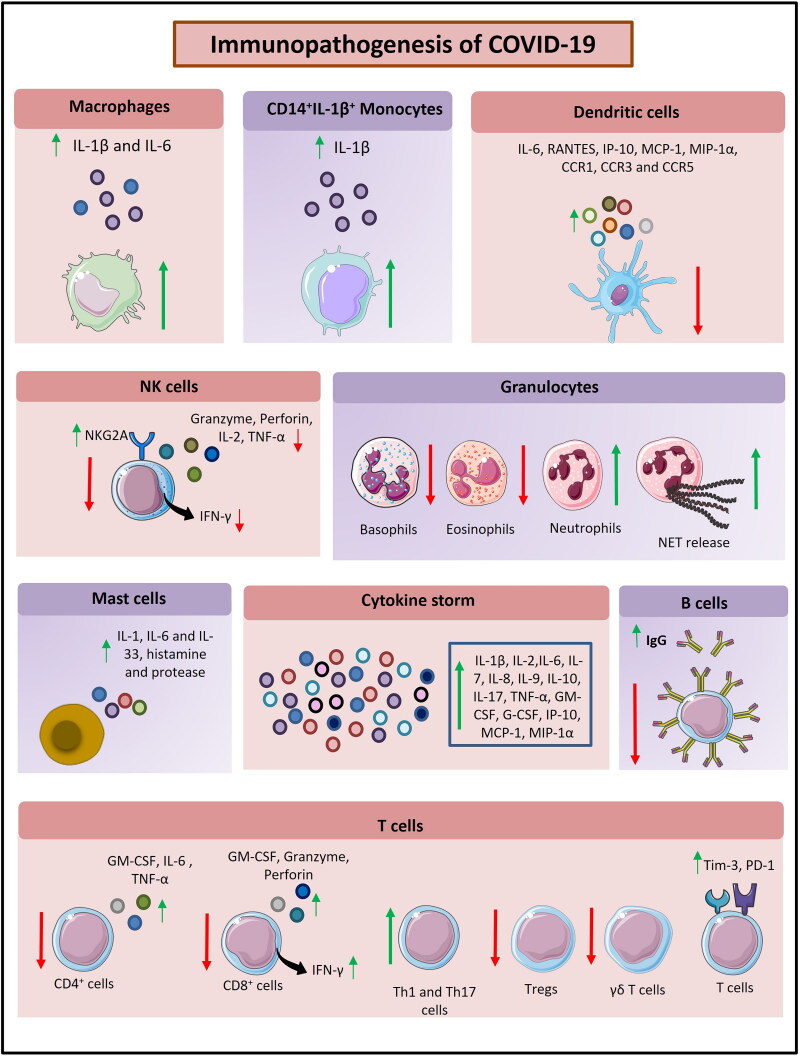
Figure 4.
The schematic diagram depicting immunopathogenesis of COVID-19 (Figure illustrated with the help of https://smart.servier.com/. ).
Table 1.
Response of various immune cells in COVID-19.
| S. No. | Immune cells | Response to SARS-CoV-2 infection | Treatment options | References |
|---|---|---|---|---|
| 1. | Monocytes and Macrophages |
|
|
[43] |
| 2. | Dendritic cells (DCs) |
|
|
[44] [45] |
| 3. | Natural Killer (NK) cells |
|
|
[46] [47] |
| 4. | Neutrophils |
|
|
[48] |
| 5. | Eosinophils |
|
|
[49] |
| 6. | Mast cells |
|
|
[50] [51] [52] [53] [54] |
| 7. | T cells |
|
|
[47] |
| 8. | B cells |
|
|
[55] |
4.1. Immune evasion by SARS-CoV-2
Innate immune cells are activated when they recognize pathogen associated molecular patterns (PAMPs) and damage associated molecular patterns (DAMPs) with the help of pattern recognition receptors present on them (PRRs). PRRs include the toll like receptors (TLRs), NOD like receptor (NLR), RIG-1 like receptor (RLR), C-type lectin-like receptors (CLRs) and melanoma differentiation associated protein 5 (MDA5). Recognition of viral genes activate the interferon regulatory (IRF)-3, IRF-7 and nuclear factor kappa-light-chain enhancer of activated B cells (NF-кB) thereby promoting their nuclear translocation. Generally, activation of TLR3/7 promotes nuclear translocation of IRF-3 and NF-кB and activation of RIG/MDA5 stimulate the nuclear translocation of IRF-3 [56]. Type 1 IFNs provides the first line of defence by preventing the spread of viral infection. It induces the expression of IFN stimulated genes (ISGs) by activating the JAK-STAT signaling pathway [4]. Successful activation of type 1 IFN response inhibits viral replication and prevents spread of viral infection. Innate immune response depends heavily on the type 1 IFN responses for eliminating the virus.
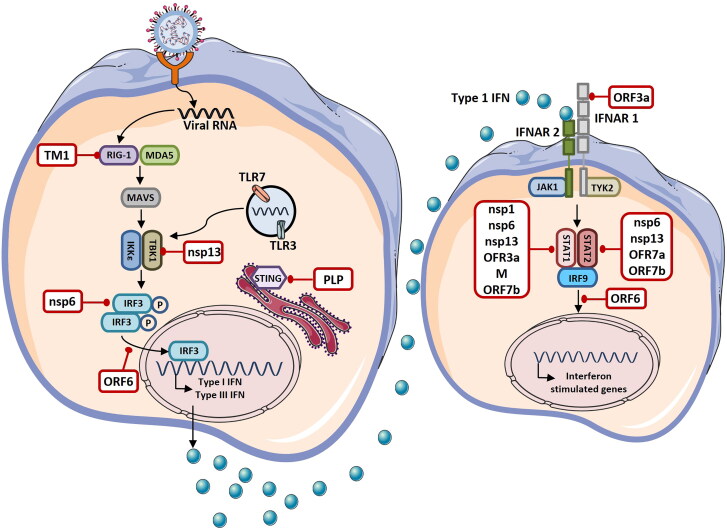
SARS-CoV-2 virus evades the innate immune response by suppressing the antiviral type 1 IFN responses. It is observed that SARS-CoV-2 infection stimulate low levels of antiviral cytokines IFN-α and IFN-β. Several studies on the SARS-CoV virus revealed that it antagonizes the type 1 IFN responses through various mechanisms (Figure 5). Versteeg et al. demonstrated that SARS-CoV prevented IFN production by shielding the viral RNA from the host cellular sensory molecules [57]. Furthermore, it was observed that SARS-CoV nsp1 inhibited host immune response by modulating the production of type 1 IFNs. nsp 1 mutant showed higher expressions of type I IFN [58]. Other viral proteins viz. nsp1, nsp7, nsp15 and ORFs like ORF3b, ORF6 and ORF9b also suppressed the activation of type 1 IFN [59]. Another study reported that SARS-CoV prevented IFN production by disrupting the stimulator of IFN genes (STING) signaling which is required for activation of IRF3 that induces the production of IFN [60]. IFN production is also inhibited via first transmembrane domain (TM1) of the M protein which is present in the N-terminal. TM1 can bind to the RIG-I, TNF receptor associated factor 3 (TRAF3), TANK-binding kinase1 (TBK1) and homolog IкB kinase epsilon (IKKє). By binding to them M protein inhibits the binding of these molecules with other downstream effectors, and thus, preventing induction of type 1 IFN [61]. Similarly, from in vitro studies it has been reported that SARS-CoV inhibits the expression of IFN-β in macrophages and stimulates the production of chemokines like IP-10 and MCP-1[62]. Channappanavar and Perlman showed that SARS-CoV infection delayed type 1 INFα/β responses inducing the accumulation of inflammatory monocytes and macrophages along with inhibiting T cells mediated immune response against the virus [63]. Delayed type 1 IFN response and monocytes/macrophages recruitment are found to be the major reasons for the lethal pneumonia and ARDS observed in SARS patients [63]. Melo et al. showed that SARS-CoV-2 like SARS-CoV stimulate lower levels of type 1 and type 3 IFN responses. This leads to restrained ISG expression and induction of high expression of cytokines like IL-6 and IL-1Rα and chemokines like C-X-C motif ligand 2 (CXCL2) and CXCL8. CXCL2 and CXCL8 promote immune cells infiltration [64]. Hadjadj et al. demonstrated that immune cells in peripheral blood of critically ill COVID-19 patients showed reduced type I IFN response. These patients also had higher levels of IL-6 and TNF-α [65]. Furthermore, Lucas et al. showed that patients with mild symptoms had decreased levels of type 1 and type 3 IFN responses [66]. Thus, it can be concluded that SARS-CoV-2 cause dysregulation of innate immunity by modulating the type 1 IFN responses.
Figure 5.
Immune evasion by Coronavirus. Coronavirus is sensed by various pathogen recognition receptors (PRRs) present in the host cell like toll like receptor (TLR)-3, TLR-7, retinoic acid inducible gene (RIG)-1 and melanoma differentiation associated protein 5 (MDA5). Recognition of Coronavirus by PRRs stimulate the production of type I and type 3 interferons (IFNs). IFNs are then secreted in autocrine and paracrine manner and induce the expression of interferon stimulated genes via JAK-STAT signaling pathway leading to the activation of antiviral response. Several viral proteins (shown in white boxes) encoded by the Coronavirus antagonize various steps of IFN signaling and escape the immune response. TM1: First Transmembrane Domain, MAVS: Mitochondrial Antiviral-Signaling, IKKε: IкB Kinase Epsilon, TBK1: TANK-Binding Kinase 1, IRF: Interferon Regulatory Factor, STING: Stimulator of IFN Genes, PLP: Papain Like Protease, NSPs: Nonstructural Proteins, ORFs: Open Reading Frames, IFNAR: Interferon- Alpha/Beta Receptor, JAK1: Janus Kinase 1, TYK2: Tyrosine Kinase 2, STAT: Signal Transducer and Activator of Transcription 2 (Figure illustrated with the help of https://smart.servier.com/. ).
4.2. Cytokine storm
One of the major reasons for the lung pathogenesis in COVID-19 patients is the over-production of proinflammatory cytokines. It was observed that early enhancement in levels of cytokines lead to worst outcomes [66]. Dysregulated immune system in COVID-19 patients results in development of cytokine storm which promotes lung inflammation. Severe COVID-19 patients have increased levels of cytokines such as IFN-γ, IL-1, IL-6, IL-8, IL-12 and transforming growth factor β (TGF-β) and chemokines C-C motif ligand 2 (CCL2), CXCL9 and CXCL10 [43]. COVID-19 patients also showed elevated levels of plasma IL-2, IL-7, IL-10, granulocyte-macrophage colony stimulating factor (GM-CSF), macrophage inflammatory protein 1-α (MIP1-α), tumor necrosis factor-α (TNF-α) and MCP-1 [21]. It was observed that patients who were in non-survivor group showed higher levels of IL-6 throughout the clinical course than patients in the survivor group [67]. In severe cases, cytokine storm violently attacks the body and causes ARDS, multiple organ failure eventually leading to death [4].
Cytokine storm induce unchecked influx of immune cells especially monocytes and neutrophils into the lungs. These cells secrete various inflammatory cytokines and chemokines such as IL-1β, IL-6, TNF-α, CCL2, CCL7 and CCL12 that further enhance disease severity. Earlier studies reported that IL-1β promoted pyroptosis defined as non-programmed cell death upon pathogenic infection [68]. Uncontrolled cell infiltrations promote lung damage due to excessive secretion of toxic substances like proteases and ROS. Furthermore, it causes alveolar damage, pulmonary edema, hyaline membrane formation and desquamation of pneumocytes which are defined as the earlier signs of ARDS [69, 70].
4.3. Complement activation
Complement proteins are the component of innate immunity and it has been reported that complement activation is also related with pathogenesis of ARDS. In case of SARS-CoV infection, activation of complement component C3 was found to be responsible for pathogenesis. Furthermore, studies reported that depletion of C3 in SARS-CoV infected mice reduced lung injury. Recently, it has been revealed that C3 deficient mice also have reduced levels of IL-6 and other chemokines and cytokines with decreased influx of neutrophils and monocytes [71]. Various studies have shown the role of complement activation in COVID-19. Significantly elevated levels of complement component C5b-9 and C5a were observed in the plasma of severe COVID-19 patients [72]. In study of Gao et al. it has been observed that N protein of SARS-CoV, MERS-CoV and SARS-CoV-2 binds to the mannan-binding lectin-associated serine protease 2 (MASP)-2 which is an enzymatic initiator of lectin pathway. N protein: MASP-2 interaction promotes abnormal complement activation and lung injury whereas blockade of this interaction suppressed the complement activation and prevented inflammation and lung injury [73]. Complement activation may also promote coagulopathy, thrombosis and microvascular injury that are widely reported in COVID-19 patients [74–76]. Moreover, in COVID-19 patients enhanced deposition of complement components C5b-9, C4d and MASP-2 are observed in the lung and are associated with microvascular injury and thrombosis [76]. Diao et al. observed that acute renal failure in COVID-19 patients is associated with increased depositions of C59-b in the kidney tubules [77]. Inhibition of C3 with anti-C3 agents like AMY-101 and with pathway inhibitors like lectin pathway can be beneficial in preventing induced lung injury in COVID-19 [71].
4.4. Dysregulation of immune cells in COVID-19
4.4.1. Monocytes and macrophages
Single stranded RNA viruses like CoVs activate the monocytes/macrophages through the TLR7/8 and stimulate type 1 IFN responses and cytokine production which leads to suppression of viral infection [78]. However, SARS-CoV-2 dysregulated the activity of monocytes and macrophages that further promotes lung injury and respiratory disorders like ARDS. Single cell RNA sequencing (ScRNA) of the bronchoalveolar lavage fluid (BALF) revealed that there was abundance of monocyte derived macrophages in the lungs of severe COVID-19 patients [79]. Similarly, single cell transcriptome of peripheral blood revealed that there was increased number of circulatory monocytes in COVID-19 patients [73]. Increased numbers of monocytes were also observed during the early recovery stage (ERS). ScRNA of peripheral blood during ERS revealed that there were increased counts of classical CD14+ monocytes and CD14+IL-1β+ monocytes in circulation of COVID-19 patients. These cells further enter into the pulmonary circulation and cause lung infection [14, 80]. Remarkably, increased numbers of monocyte derived FCN1+ macrophages have also been observed in BALF than the alveolar macrophages in severe patients. Alveolar macrophages are found to be the major type of macrophages in BALF [79].
In COVID-19 patients increased expression of inflammatory cytokines such as TNF-α, IL-1β, IL-2, IL-6, IL-7, GM-CSF, IP-10, MIP-1α and MCP-1 had been observed [43, 81]. Macrophages are known to play an important role in induction of these inflammatory cytokines. It has been observed that critically ill COVID-19 infected patients showed increased levels of CCL2, CCL3, CCL20, CXCL1, CXCL3, CXCL10, IL-8, IL-1β and TNF-α expressing inflammatory macrophages [82]. Sustained production of IL-1β and IL-6 from the circulating monocytes and macrophages resulted in macrophage activation syndrome (MAS) in COVID-19 patients that leads to severe respiratory failure [83]. MAS is characterized by the increased production of IL-6 along with higher levels of hemophagocytic macrophages [84]. In macrophages, NOD like receptor family, pyrin domain containing 3 (NLRP3) inflammasome regulates the release of proinflammatory cytokines IL-1β and IL-18 [85]. COVID-19 patients also have activated MAPK-interferon pathway as revealed by the single cell profiling and has very imperative role in immune response produced in case of COVID-19 [86]. Immunosuppression of inflammatory cytokines is considered as one of the treatment options for the control of COVID-19. Drugs like Ankinra (IL-1 antagonist) and Tocilizumab (IL-6 receptor antagonist) were suggested as treatment options for the inhibition of inflammatory response but results from a recent randomized double blind placebo controlled trial showed that Tocilizumab was not effective in saving lives of people infected with SARS-CoV-2 [43, 87]. Recently, Lee et al. performed ScRNA sequencing on the PMBCs of healthy donors, severe and mild COVID-19 patients and severe influenza patients. They observed that monocytes of severe COVID-19 patients showed higher type 1 IFN response along with TNF/IL-1β derived inflammation. Type 1 IFN-based inflammatory responses were also observed in severe influenza cases. This showed that macrophages derived type 1 IFN response plays an important role in inducing inflammation in severe COVID-19 cases [88]. Bao et al. showed that human ACE2 (hACE2) receptor also has role in pathogenesis caused by macrophages. In SARS-CoV-2 infected transgenic mice expressing hACE2, fatal pneumonia and infiltration and accumulation of macrophages in alveolar interstitium has been observed in consistent to the COVID-19 patients. Viral antigens have also been spotted in these macrophages [33, 69, 89]. Macrophage activation affects not only lungs but other organs also. In postmortem analysis of six deceased COVID-19 patients it has been observed that infection of tissue resident macrophages (CD169+) of spleen and lymph nodes by SARS-CoV-2 is found to be responsible for pathogenesis. Immunohistochemistry in the patients revealed that these tissue resident macrophages of spleen and lymph nodes express ACE2 receptor along with nucleocapsid protein which is also spotted in these cells. In situ TUNEL staining revealed that SARS-CoV-2 induce IL-6 production from macrophages that promote lymphocyte necrosis, and thus, might be the cause of pathogenesis in both spleen and lymph nodes [90].
4.4.2. Dendritic cells
DCs are the most potent professional antigen presenting cells that also play a vital role in preventing viral infections. Many viruses use the respiratory tract to get entry into the host and cause infection in the lungs which leads to the activation of innate and adaptive immune response. DCs are activated at the initial stages of the viral infection and provide antiviral immunity [91]. Activated DCs migrate from various tissues to the lymphoid organs and present antigens to the T cells that further activate specific adaptive immune response. Various studies showed that SARS virus evade the innate immune response by impairing the activity of DCs through different mechanisms. A study reported that infection of DCs with SARS-CoV resulted in suppression of MHC-I expression on DCs that further delayed the expression of IFN-α [92]. Another study reported that SARS-CoV induced the plasmacytoid DCs (pDCs) to produce prolonged production of type 1 IFNs. This leads to immune exhaustion and accumulation of monocytes, macrophages and neutrophils at the site of infection [93]. It actually suppresses the expression of antiviral genes like IFN-α, IFN-β and IFN-γ. It also induces the increased expression of proinflammatory cytokines and chemokines such as TNF-α, IL-6, CCL5, IP-10, MCP-1, MIP-1α, CCR1, CCR3 and CCR5. The SARS infection does not affect the expression of TLRs (TLR-1 to TLR-10). It stimulates the enhanced expression of TNF related apoptosis inducing ligand (TRAIL) with no effect on the expression of FAS ligand [94, 95]. The increased production of chemokines induces the migration of DCs to the lymph nodes but increased expression of TRAIL promotes the apoptosis of lymphocytes on their interaction with DCs. Thus, DCs induced apoptosis of T cells results in suppression of virus specific adaptive immune response. Zhao et al. showed that SARS infection also delayed the activation of DCs and their migration to the draining lymph nodes which leads to deficient adaptive T cell response [44]. Furthermore, a study reported by Tseng et al. showed that SARS infection leads to maturation of DCs and induced the expression of MHC-II and costimulatory molecules [96]. Impaired DC activity is also observed in MERS. MERS-CoV infects the monocyte derived DCs and induce markedly elevated expression of CCL5, IL-12, IP-10 and IFN-γ along with MHC-II molecules and co-stimulatory molecule CD86 [97].
SARS-CoV-2 like SARS-CoV and MERS-CoV affects the activity of DCs. Depletion of both pDCs and conventional DCs was observed in the severe COVID-19 patients [98]. It was observed that mammalian target of rapamycin (mTOR)-signaling and IFN-α production was diminished in the pDCs of SARS-CoV-2 infected patients [99]. In COVID-19 patients delayed activation and migration of DCs from the site of infection, i.e., from lungs to the draining lymph nodes was also observed. It can, thus, be concluded that delayed activation and migration of DCs might be responsible for the slow viral clearance observed in the COVID-19 patients [100]. Severe COVID-19 patients have lymphopenia as they have reduced counts of CD4+ and CD8+ T cells and have decreased expression of TCR. Reduced number of T cells and expression of TCR might be due to interaction of these cells with the DCs which promote their apoptosis as in case of SARS-CoV. Depletion of pDCs in COVID-19 patients is responsible for the impaired activity of IFN-α observed in these patients as these are the major producers of IFN-α [65]. From metatranscriptomic sequencing of BALF cells it was revealed that COVID-19 patients have more abundant DCs than the healthy people [101]. Chen et al. showed that cytokine storm caused by the SARS infection induced the influx of pDCs into the lungs [102]. Thus, cytokine storm induced in COVID-19 patients might promote influx of DCs into the lungs. However, ScRNA of BALF revealed that severe COVID-19 patients have reduced number of pDCs and myeloid derived DCs (mDCs) than the patients with mild symptoms.
CoVs interacts with DCs through several ways. Using mice models, it has been demonstrated that CoVs interacts with the DC in receptor dependent manner by using the murine DC marker CEACAM1a. A study reported that blockade of this receptor via anti-CEACAM1a monoclonal antibody (mAb) CC1 prevented infection of DCs [103]. DCs express DC-SIGN (CD209) which is C-type lectin present on their membrane. It is used by various viruses like human immunodeficiency virus (HIV) to interact with the DCs [103]. Jeffers et al. showed that CD209 expression induce the susceptibility of SARS-CoV infection [104]. It has been reported that single nucleotide polymorphism (SNP) in the promoter region of the CD209 enhanced the disease severity in SARS infection [100]. Thus, it might be possible that CoVs infect DCs via CD209. Through ScRNA it is revealed that DCs also express the ACE2 receptor [100], and thus, SARS-CoV-2 can directly infect the DCs through ACE2 receptor.
4.4.3. Natural killer cells
Natural killer (NK) cells are cytotoxic cells that play crucial role in providing immunity against the virus [105]. NK cells recognize the pathogens with the help of its receptors known as natural cytotoxicity receptors (NCRs) like NKp46 and NKp44. Activation of these markers leads to the killing of virus infected cells [106]. DCs can also activate the NK cells. During influenza virus infection it was observed that influenza infected DCs activate the NK cells by recognizing its receptors NKG2D and NKp46 [107]. The role of NK cells has been deciphered in various viral infections such as influenza, respiratory syncytial viruses and HIV.
Deficiency of NK cells lead to various viral infections and by inducing depletion of NK cells, viruses protected themselves by enhancing own survival [108]. Interestingly, it has also been observed that SARS-CoV-2 also evades the immune system by depleting the NK cell population. Yu et al. showed that the level of NK cells starts falling from day 16 of the disease onset [109]. ScRNA sequencing of BALF also revealed that COVID-19 patients have decreased levels of NK cells than their healthy counterparts [98]. Analysis of lung NK cells revealed that most of the NK cells that reside in human lungs are characterized by CD56dimCD16+ phenotype. Few NK cells also express tissue residency marker CD69 which consist of mostly CD16- NK cells (immature cells) and CD16+ cells (less differentiated cells). Maucourant et al. reported that highly activated NK cells were present in the peripheral blood of COVID-19 patients. They also observed that in severe cases peripheral NK cells showed higher expression of markers like perforin, NKG2C and Ksp37 [110]. NK cells in COVID-19 infected patients are found to be functionally exhausted as they highly express NKG2A. Higher expression of NKG2A is associated with the exhaustion of NK cells. NK cells of COVID-19 patients expressed low levels of TNF-α, IL-2, CD107a, granzyme B and IFN-γ which further suggests that they are functionally exhausted. Functional exhaustion of NK cells is one of the mechanisms through which SARS-CoV-2 evades immunity [47]. Another study reported that NK cells of COVID-19 patients express higher levels of exhaustion marker: T cell immunoglobulin and mucin-domain containing-3 (Tim-3). Expression of exhaustion marker Tim-3 is associated with the disease severity [65]. Viruses can spread the infection by decreasing the cytotoxicity of NK cells. Recently, a study reported that influenza virus decreased the cytotoxicity of NK cells by inducing their apoptosis [111]. SARS-CoV-2 is also reported to inhibit cytotoxicity of NK cells in an IL-6 dependent manner. COVID-19 patients possess higher levels of IL-6 cytokine which downregulated the cytotoxicity of NK cells by reducing the perforin and granzyme secretion [46]. Recently, it was also observed that Tocilizumab enhanced the activity of NK cells [73]. Single cell sequencing revealed that NK cells do not express the ACE2 receptor [112]. Thus, SARS-CoV-2 does not infect the NK cells directly but reduce the capability of NK cells by decreasing their cytotoxicity and inducing their exhaustion.
4.4.4. Neutrophils
Neutrophils are the most abundant polymorphonuclear leucocytes that provide first line of innate immune defence and kill microbes by phagocytosis and degranulation [113]. The role of neutrophils in providing antiviral defence has not been deciphered yet but it is observed that neutrophils can prevent viral infection by releasing various chemokines, cytokines and antimicrobial compounds [114]. One of the mechanisms by which neutrophils eradicate pathogens is through neutrophil extracellular traps (NETs) formation. NETs trap the bacteria, viruses, parasites, fungi and neutralize them to provide antimicrobial defence [115]. NET formation stimulates thrombosis by activating platelets and by inducing coagulation which promote various cardiovascular diseases like stroke and myocardial infraction [116]. NET formation is also found to be linked with various lung diseases like cystic fibrosis, obstructive pulmonary disease, acute lung injury and ARDS [116, 117]. In case of COVID-19, in vitro study revealed that sera of virus infected patients induced NET release [117]. This shows that NET formation in one of the mechanisms through which neutrophils increase the severity of COVID-19. Several studies prove the relevance of NET in COVID-19 pathogenesis [117].
It has been found that neutrophils have major role in the pathogenesis of COVID-19 that induces the proinflammatory response observed in COVID-19. In analysis of 82 patients it was observed that 74.3% of the patients showed neutrophilia whereas 94.5% of the patients showed higher neutrophil to lymphocyte ratio (NLR) [118]. ScRNA analysis of the BALF revealed that severe COVID-19 patients contain higher proportions of neutrophils than patients who had mild symptoms [79]. Interestingly, one of the study demonstrated that pregnant women infected with COVID-19 showed significant enhancement in neutrophils than in the non-pregnant counterparts [119]. Recently using machine learning algorithm Mathew et al. observed that neutrophil activation is the strongest predictor of COVID-19 severity. Activated neutrophils express resistin, lipocalin-2, IL-8, HGF (Hepatocyte Growth Factor) and G-CSF. Neutrophil activation was observed on the first day of the hospitalization only in the patients who later require ICU [120]. Altogether, from these studies it can be concluded that enhanced neutrophils population and activation and higher NLR can be utilized as biomarker to access disease severity.
In consistent to SARS-CoV infection neutrophil infiltrations have also been observed in COVID-19 patients. Cytokine storm promote the accumulation of neutrophils in lung [121]. Higher expression of neutrophil chemokine CXCR2 receptor along with delayed type 1 IFN response has been found to be responsible for accumulation of neutrophils in lungs of COVID-19 patients [63, 65]. It has been found that increased levels of IL-1β by monocytes in the circulation induce the accumulation of neutrophils and CD8+ T cells in the lung [21, 68]. Infiltrating neutrophils promote disease severity by inducing the production of chemokines and cytokines such as IL-1β, IL-6, IL-8, TNF-α, TGF-β and MCP-1 [122]. Neutrophils also produce cytotoxic substances like ROS, proteases and leukotrienes that further lead to various respiratory disorders like acute lung injury and ARDS [42, 65]. It has been observed that majority of COVID-19 patients who were diagnosed with thrombosis have higher levels of neutrophils and the respective markers of NET in the serum [117]. Furthermore, Shi et al. reported that COVID-19 patients who had higher levels of neutrophil activation marker calprotectin had higher need of mechanical ventilation than those who had lower levels of calprotectin [123]. Increased levels of IL-1β in COVID-19 patients stimulate NET release and promote lung damage [124] (Figure 6). It is reported that viable SARS-CoV-2 can directly promote the release of NET from neutrophils. NET release by SARS-CoV-2 is dependent on ACE2, serine protease, protein arginine deamination-4 (PAD-4) and virus replication [125]. It is observed that children suffering from Kawasaki disease (KD) which is characterized by inflammation in blood vessels and heart damage is common in children under the age 5. It is reported that that KD patients have increased number of neutrophils and have dysregulated NET formation. This indicates that children might be more susceptible to COVID-19 due to higher levels of neutrophils and NET release [126]. Altogether, these studies showed that increased number of neutrophils enhances the severity of COVID-19 infection through several ways, thus, it is necessary to control the dysregulated activity of neutrophils to lessen the severity.
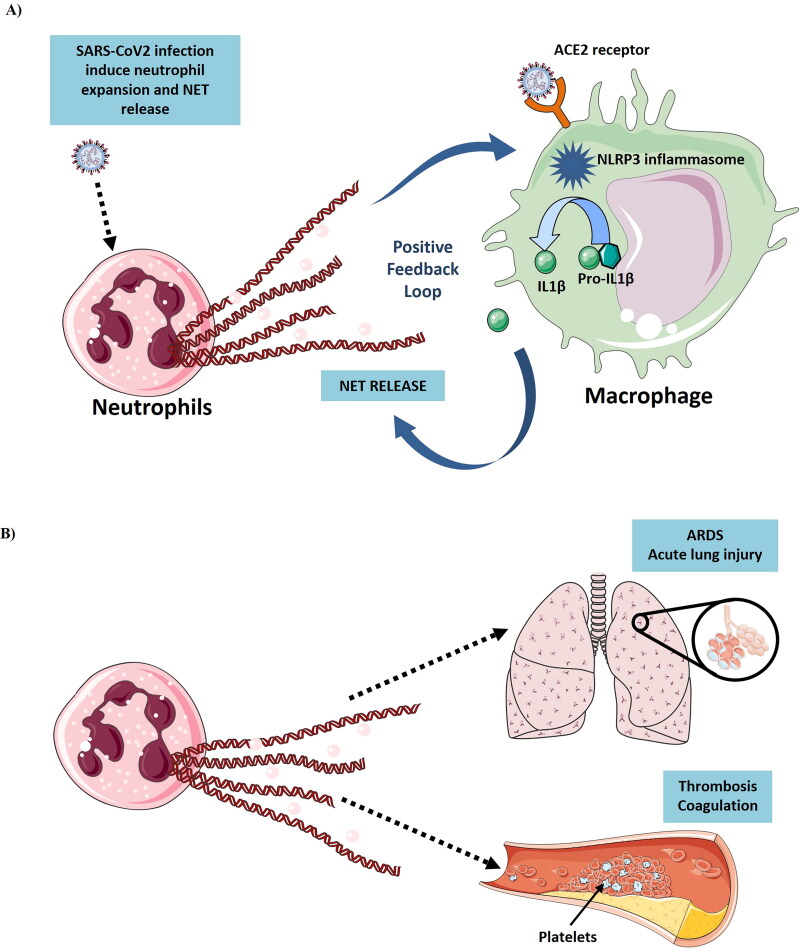
Figure 6.
(A) Induction of Neutrophil Extracellular Traps (NETs) from neutrophils by SARS-CoV-2 infection. SARS-CoV-2 induce the neutrophil expansion and NET release. NET release promote the IL-1β secretion from macrophages which further leads to inflammation. SARS-CoV-2 also induce the IL-1β secretion from macrophages by activating the NLRP3 inflammasome activation. IL-1β further stimulate NET release and forms a positive feedback loop. (B) NET release causes ARDS, thrombosis and coagulation in COVID-19 patients (Figure illustrated with the help of https://smart.servier.com/. ).
4.4.5. Basophils
Basophils are the least common circulating granulocytes. Generally, basophils secrete Th2 cell type cytokines like IL-4 and IL-13 and provide a link between the innate and adaptive immunity. Basophils are responsible for various Th2 cytokine mediated inflammatory diseases like allergy [127]. Degranulation of activated basophils release histamine, proteolytic enzymes, proteoglycans, leukotrienes, prostaglandins, etc. Several cytokines and chemokines such as IL-3, IL-4, IL-6, IL-9, IL-13, IL-25, CCL5, MIP-1A, MCP-1 and GM-CSF are also released during degranulation [128]. Their role in various viral diseases like influenza and HIV has been well established. Several studies revealed that influenza virus enhances the release of histamine from basophils [129, 130]. Recently, basophil dysregulation has also been observed in case of COVID-19 where it was observed that severe COVID-19 patients have reduced number of basophils as compared to non-severe patients [131]. Thus, it can be inferred that SARS-CoV-2 impairs the immune system by reducing the level of basophils and decreased counts of basophils can be correlated with the severity of COVID-19.
4.4.6. Eosinophils
Eosinophils are the circulatory granulocytes but apart from circulation granulocytes are also localized in the gastrointestinal tract and in the lungs. Single stranded RNA viruses induce the eosinophils through TLR7 and promote the cytokine production and degranulation of eosinophils. Furthermore, activation of eosinophils also induces the production of superoxide and nitric oxide in eosinophils and improves their survival. A study revealed that eosinophils provide the antiviral immunity by inducing the expression of MHC-II and costimulatory molecule CD86 as reported in case of influenza virus [132].
Various studies highlighted the relevance of eosinophils in SARS-CoV-2 infection. Severe COVID-19 patients have decreased eosinophil count and enhancement in the eosinophil levels can be used as a recovery marker [133]. Analysis of routine blood of hospitalized COVID-19 patient for 26 days revealed that eosinophil levels start decreasing within one week of the hospitalization. Lowest value was observed on the seventh day of the hospitalization. There were very low counts of eosinophils on tenth day and after approximately 12 days of the hospitalization levels of the eosinophils start recovering [134]. Liu et al. also reported that eosinophil levels were low at the time of hospitalization and recover before discharge [49]. These studies showed that enhanced eosinophil count can be used as markers of improvement in COVID-19. Chen et al. reported that non-survivors have reduced levels of eosinophil than the survivors and thus eosinophils can be employed as an indicator of disease severity [135]. It was earlier hypothesized that asthma patients must be at higher risk of getting infected with SARS-CoV-2 due to lack of antiviral immunity in them. Surprisingly, it has been observed that asthma patients showed lesser susceptibility to COVID-19 because of higher eosinophils count and type 2 cytokines IL-4 and IL-13. This shows the significance of eosinophils in preventing COVID-19. One of the mechanisms through which SARS-CoV-2 impaired the host immune response is via depleting eosinophils count. Altogether, corticosteroids, allergen immunotherapy and monoclonal antibodies against IgE used for the treatment of asthma might also decrease the risk of asthmatic patients to get infected with SARS-CoV-2 [136].
4.4.7. Mast cells
Mast cells are distributed throughout the body but predominantly they are localized at the body surfaces that are in contact with the external environment like skin, the nasal cavity, submucosa of the respiratory tract and intestine. In these body parts mast cells reside under the epithelium in connective tissue and form the barrier against the microbes [137]. Mast cells are known regulators of both innate and adaptive immune responses and various other physiological processes like vasodilation, vascular homeostasis, angiogenesis and venom detoxification. It has been reported that mast cells are also responsible for development of various diseases like asthma, allergy, anaphylaxis, gastrointestinal and cardiovascular disorders [137]. The role of mast cells in various bacterial, parasitic and bacterial infections like HIV, dengue virus has also been reported [138]. Several studies showed that viruses stimulate mast cells through TLRs that further induce inflammatory response by stimulating the secretion of inflammatory chemical compounds like TNF-α, tryptase, IL-1, IL-6 and CCL3 [53, 138]. Mast cells were also reported to increase lung injury in H5N1 influenza viral infection by secreting proinflammatory compounds like histamine, IFN-γ and tryptase. Inhibition of mast cell degranulation by Ketotifen prevented lung pathogenesis and viral infection [138]. Mast cells produce type 1 and 3 IFNs in response to viral infection but unsuitable inflammatory response of mast cells is responsible for vascular leakage and fibrosis observed in viral infections [139].
Few studies are reported till date on the role of mast cells in COVID-19 but it is observed that mast cells participate in the inflammatory response that is produced in COVID-19. SARS-CoV-2 infection induces mast cells to secrete early inflammatory compounds histamine and protease whereas late infection induces production of inflammatory cytokines IL-1, IL-3 and IL-6. It was observed that suppression of production of these cytokines by using anti-inflammatory cytokines of IL-1 family like IL-37 and IL-38 prevented SARS-CoV-2 infection [50, 51]. This shows that suppression of inflammation due to mast cells can be an effective treatment for the prevention of SARS-CoV-2 infection and various drugs are considered for this. It was observed that Famotidine can prevent the SARS-CoV-2 infection by targeting the activity of histamine receptor H2 [52]. Mast cells stabilizers were also considered which can control the serum TNF levels [140]. Gigante et al. hypothesized that Sodium chromo glycate and Palmitoylethanolamide can prevent the SARS-CoV-2 infection. Sodium chromo glycate is a well-known mast cell stabilizer that prevents inflammation by supressing the secretion of inflammatory compounds from the mast cells. It prevented lung inflammation in viral infected mice and improved their survival. Conversely, Palmitoylethanolamide is an endogenous fatty acid and nuclear factor agonist. It regulates mast cell homeostasis by downregulating the secretion of TNF-α and histamine and is reported to prevent respiratory tract infection during common cold and influenza [53]. Recently, a study suggested a novel mast cell stabilizer “chromones” which are cheap, anti-inflammatory and safest treatment option against COVID-19 [141]. Flavonoid Luteolin is also considered for preventing inflammation induced by mast cells. Luteolin exhibits antiviral and anti-inflammatory properties and it also suppresses the activity of serine proteases and prevents the entry of SARS-CoV-2 into host cells by binding to the S protein. An analogue of Luteolin called Tetramethoxyluteolin suppresses the release of proinflammatory cytokines and chemokines such as TNF, IL-1β, CCL2 and CCL5 from human mast cells [54].
Apart from producing inflammatory response mast cells can also induce lung injury through other mechanism. It is observed that mast cells produce and store enzymatically active renin which is homologous to the renin produced in human kidneys. Release of renin by the degranulation of bronchial mast cells lead to the formation of angiotensin II that further promotes bronchoconstriction and also upregulated the RAS activity in airways [142]. A study revealed that α and β chymases secreted by the mast cells also generate angiotensin II from angiotensin I [143]. Tryptases and chymases are serine proteinases and are chief constituents of mast cell granules that induce the influx of inflammatory cells [144]. Serine proteases are also required for priming of S protein. Mast cells thus have very important role in the pathogenesis of COVID-19 but their role need to be further explored.
4.4.8. T helper cells
CD4+ and CD8+ T cells play a vital role in virus clearance. Activated CD4+T cells produce cytokines that further induce the activation of other immune cells especially CD8+T cells and B cells for killing the pathogens. CD8+ T cells exhibit potential to directly kill the virus infected target cells. Among T helper cells, Th1 cells generally play major role in preventing viral infections. It has been observed that like SARS-CoV, SARS-CoV-2 also impairs the host adaptive immune response. In similarity to SARS-CoV infected patients, reduction in CD4+ and CD8+ Tlymphocytes (lymphopenia) has also been observed in COVID-19 patients [131, 145]. T cells of COVID-19 patients also showed significantly higher expression of exhaustion markers PD-1 and Tim-3 during symptomatic stages [146]. In COVID-19 patients, CD4+T cells predominantly express IFN-γ, TNF-α and IL-2 cytokines and it has been found that in severe patients, levels of these cytokines get reduced in comparison to mildly infected patients [47].
There are various reasons responsible for the depletion of T cells in the COVID-19 patients. It is observed that the number of T cells (CD4+ and CD8+) in peripheral blood is negatively interrelated with the serum levels of inflammatory cytokines IL-6, IL-10 and TNF-α. Severe SARS-CoV-2 infected patients have higher levels of these cytokines along with reduced T cells population. Recovered patients have decreased level of IL-6, IL-10 and TNF-α and have restored T cell population [90]. Type 1 IFN is also found to be induced in response to SARS-CoV-2 infection and it is observed that induction of type 1 IFN and TNF-α directly cause the observed lymphopenia. Previous reported studies demonstrated that type 1 IFN and TNF-α cytokines control the recirculation of lymphocytes by preventing the egress of lymphocytes from lymphoid organs [147, 148]. Postmortem analysis of six deceased COVID-19 patients revealed that SARS-CoV-2 induce the apoptosis of lymphocytes. It does this by upregulating the Fas expression and induces the macrophages to secrete IL-6 cytokine which further promote lymphocyte apoptosis [90]. MAS and reduced expression of Human Leukocyte Antigen-DR isotype (HLA-DR) is observed in critically ill COVID-19 infected patients and is found to be associated with the depletion of CD4+ T cells. MAS are recognized by higher production of IL-6 and thus blocking of IL-6 with Tocilizumab enhanced the counts of circulating lymphocytes. This shows that IL-6 secreted by macrophages might be responsible for lymphopenia observed in severe COVID-19 patients [83]. Lymphocytes infiltration can also be responsible for their reduce levels in peripheral blood. ScRNA of BALF revealed the infiltration of lymphocytes into lungs in COVID-19 patients [79]. Similarly, infiltration of lymphocytes into lungs is found in postmortem biopsy reports of deceased critically ill COVID-19 patients [69].
Various studies analyzed the adaptive immune response against the SARS-CoV-2 and it is observed that large number of SARS-CoV-2 epitopes are recognized by T cells [149]. Epitope mapping of SARS-CoV revealed that most of the adaptive immune response was found to be raised against the structural proteins viz. S, M, E and N proteins [150]. In SARS-CoV-2 infection majority of the T cells response observed are mounted against the S protein [151]. S protein primarily induces CD4+ cells which are mainly characterized by CD69+CD137+ markers. Majority of these CD4+ T cells are of central memory type as they express CD45RA and CCR7. S protein also activates the CD8+ T cells. Like CD4+ T cells most of these activated CD8+ T cells exhibits CD69+CD137+ phenotype and are of effector memory type (CCR7-) or of terminally differentiated effector T cell type and produce IFN-γ [151]. Huang et al. showed that all SARS-CoV-2 infected patients exhibit structural peptide specific CD4+ T cells response and 80% of the patients also harbored peptides specific CD8+ T cells [86]. Surprisingly, it has been observed that 20% of the healthy controls those were never exposed to SARS-CoV-2 also possessed virus specific T cells [86]. Furthermore, Maters et al. showed that some of these preexisting memory CD4+ T cells found to be reactive against SARS-CoV-2 also possess cross reactivity against common cold coronaviruses. This showed that these memory CD4+ T cells are raised in the healthy donors due to their previous exposure with the common cold coronaviruses and would also provide immunity against SARS-CoV-2 [152]. But recent studies have shown that protective immunity due to seasonal CoVs is short lived and provide little relief against SARS-CoV-2 infection [115, 153, 154]. It was observed that reinfection due to seasonal CoVs occurred frequently after a year of the infection [155].
It is reported that CD4+ T cells plays a vital role in preventing the SARS infection and deficiency of these cells hamper viral clearance that further promote lung inflammation. Majority of the CD4+ T cells in COVID-19 are induced to become Th1 that produce GM-CSF and other cytokines which stimulate the IL-6 producing CD14+CD16+ monocytes. These monocytes and Th1 cells accumulate in the pulmonary circulation and cause lung damage [151, 156]. GM-CSF producing CD4+ T cells are indicators of disease severity and are responsible for various inflammatory diseases like juvenile arthritis, multiple sclerosis and sepsis [157]. Within the CD4+ T cells, higher proportion of CCR6+ Th17 cells has also been observed in COVID-19 patients but the role of Th17 cells in COVID-19 is yet unknown [69, 151]. Th2 response was also observed in severe cases as there were higher levels of IL-2, IL-6 and IL-10 present in critically ill COVID-19 patients [151, 158]. CD4+ T cells of the severe COVID-19 patients produce low levels of IFN-γ than the non-severe patients (Chen et al., 2020). CD8+ T cells of COVID-19 patients are found to be functionally exhausted and showed enhanced expression of IFN-γ, TNF-α along with high levels of granzyme B and perforin (degranulated state). IFN-γ and TNF-α expressing CD8+ T cells were found to be present more in severe COVID-19 patients than the patients who have mild infection [159]. CD8+ T cells of COVID-19 patients admitted in hospitals express higher levels of GM-CSF than non-ICU patients [160]. Reduced number of T regulatory cells (Tregs) and naive T cells are also observed in COVID-19 patients [131]. Earlier a study reported that Tregs induced recovery in mice suffering from fatal pneumonia [161] and thus reduction in their numbers in COVID-19 patients might promote lethal pneumonia. Previous studies reported that γδ T cells exhibits strong antiviral activity and protected from pneumonia caused by influenza virus [159, 162]. In case of COVID-19, reduction in number of γδ T cells had been observed in the peripheral blood and showed higher expression of activation marker CD25 and CD4 [73, 163]. Bcl-6+ T follicular helper (TFH) cells are also found to be diminished in COVID-19 [164]. Phenotypic alterations were also observed in the circulating unconventional T cells (MAIT, γδ T cells and iNKT cells) of the COVID-19 patients. Highly activated unconventional T cells were detected in the airways of COVID-19 patients. It was also observed that the expression of CD69 activation marker on the blood MAIT and iNKT cells is the indicator of COVID-19 severity [165].
Stimulation of T cell response is essential for controlling the spread of infection but proper induction of memory T cell response is also required for preventing further reinfections. SARS-CoV-2 infection is very recent and thus it is not possible to study the memory T cell response in COVID-19 patients. But there are studies on memory T cell response in SARS-CoV patients and can be referred to understand the memory T cell response in SARS-CoV-2 virus. Analysis of lymphocytes in two SARS-CoV patients revealed that memory CD4+ T cells were recovered from the patient even 12 months after the disease onset [166]. Another study reported that CD8+ memory T cells surpasses the CD4+ memory T cells in SARS-CoV recovered patients and were detected in serum even 9 and 11 years after the infection. Other studies also reported the presence of memory T cells against the SARS infection [167, 168]. All the memory T cells were observed to be induced against the structural proteins of SARS-CoV [169]. But the studies of memory response in SARS-CoV cannot be correlated with the studies on the SARS-CoV-2. Analysis of blood of SARS-CoV-2 infected patients 2 weeks after discharge revealed that there was decreased level of virus specific T cell response than the IgG specific antibody response [162]. Recently it was observed that memory CD4+ and CD8+ T cells induced against SARS-CoV-2 have half-life of 3–5 months [170]. Further studies are needed to understand the virus specific T cell memory response in the recovered patients.
4.4.9. B cells
Antibodies against SARS-CoV-2 are produced by highly varied TCR and BCR V (D) J recombination [86]. In SARS-CoV infection seroconversion take place after 10–11 days of the infection [171]. Like SARS-CoV delayed antibody response take place in SARS-CoV-2 infection. Neutralizing antibodies (NAbs) mostly comprise of IgG and IgM antibodies against the SARS-CoV-2 and start appearing after 7–14 days of the disease onset [172]. Most of the NAbs are produced against the RBD of the S protein and nucleocapsid protein [173]. RBD specific monoclonal antibodies are isolated from the COVID-19 patients. It has been observed that these antibodies have great neutralizing ability and possess specificity only for SARS-CoV-2 and does not cross react with the RBD of SARS-CoV and MERS-CoV [174].
B cells not only protect from the primary infection but also provide immunity against further challenges by producing memory B cell responses. Memory B cells produce secondary immune response and protect from reinfections by producing an immediate response against the antigen. Knowledge of the memory B cells response against the SARS-CoV-2 is of the great interest. It will be very helpful in designing vaccine strategies for combating COVID-19. Memory B cell response in SARS-CoV and MERS-CoV was studied by various groups. During SARS infection it was observed that recovered patients had higher and sustainable NAbs against the S and N proteins of SARS-CoV [175]. IgG antibody and NAbs against the SARS-CoV were recovered in the patients even after 2 years of the disease onset [176]. Liu et al. analyzed IgG and NAbs at regular interval for 2 years in SARS-CoV infected patients. They observed that levels of the IgG antibody and NAbs were higher at four months after the infection and then decreased thereafter. IgG and NAbs decreased markedly after the 16 months [177]. In another study however it was reported that IgG+ memory B cells were higher after 2 months of the infection and then decreased by 6th and 8th months [178]. Memory IgG antibodies specific for SARS-CoV were not detected 6 years after the infection which shows that memory B cell response does not protect after a longer period [168]. NAbs were detected against the MERS-CoV in the 86% of the patients even nearly 3 years after the outbreak [179]. But as SARS-CoV-2 infection is very recent there is no substantial data related to the memory B cell response in COVID-19. But there are some studies which may be helpful in understanding the memory B cell response against SARS-CoV-2. In study of Thevarajan et al. it had been observed that stimulation of CD19+CD27+CD38hi antibody secreting B cells is concomitant with the increase in TFH cells [180]. TFH cells help in the development of memory B cells response. ScRNA of peripheral blood mononuclear cells revealed that population of plasma B cells exists in recovered patients [73]. It had been observed that primary infection of SARS-CoV-2 can protect from the further reinfections [89]. RBD specific memory IgG antibodies were observed in the COVID-19 patients [174]. Study from Addetia et al. also had revealed that NAbs produced against SARS-CoV-2 can protect from reinfection. They showed that crew members of US fishing vessel who had NAbs against CoVs before departure were protected against SARS-CoV-2 infection caused during the departure. Conversely, crew members who did not have the NAbs against SARS-CoV-2 got infected [181]. In another study, Gudbjartsson et al. tested the levels of SARS-CoV-2 antibodies in around 30 000 people including more than 1200 people who had recovered from the COVID-19 infection. They observed that more than 90% of the recovered patients had antibodies against SARS-CoV-2. Antibodies levels peaked after 2 months of diagnosis and did not decline until 4 months [182]. These studies have shown that memory response exists against SARS-CoV-2 but different studies provide different perspective of memory response in COVID-19. Seow et al. analyzed the antibody responses in 65 infected people for 94 days. They observed that among them 19 patients had higher levels of antibodies than people with mild disease. They also observed that in most of the people antibody levels started falling about a month after the infection [183]. This study declined the possibility of designing a vaccine based on NAbs. Recently, a study by To et al. provided the first evidence of reinfection. They observed that a Hong Kong man who was first infected with SARS-CoV-2 in April was found infected again four months later (142 days) with different variant of SARS-CoV-2. This study also shows that immunity against SARS-CoV-2 can wane after sometime [184]. But again two recent studies have declined the possibility that memory response against SARS-CoV-2 diminish after sometime. In study of Wajnberg et al. which included more than 30 000 SARS-CoV-2 infected individuals it was observed that in 90% of the infected people antibodies against SARS-CoV-2 persisted for at least 5 months after the infection [185]. Similar results were reported by Dan et al. They analyzed the circulating memory response against SARS-CoV-2 and observed that spike IgG antibody remained for at least 6 months after the infections [170].
Antibodies usually protect from the infections but sometimes can also enhance the severity of the viral infection by mediating the entry of virus into the host cells, referred as antibody dependent enhancement of infection (ADE). ADE occurs when complex of virus and antibody comes in contact with cells having Fc receptors. Antibodies bind to the virus through the S protein on one end and to the Fc receptor on another end. Binding of antibody-virus complex to the Fc receptor bearing cells induces its internalization and entry of virus into the cell [186, 187]. ADE phenomenon hampers the activation of proper immune response against SARS-CoV-2. It is observed that IgG antibody against the S protein promote polarization of macrophages into proinflammatory phenotype which leads to acute lung injury. Blocking of Fcγ receptors inhibited the inflammatory response of macrophages and prevented lung injury. This shows that ADE might be responsible for dysregulation of macrophages activity [188]. SARS-CoV is also reported to infect macrophages in IgG dependent ADE by binding to the FcγRII receptors [189]. It was observed that monoclonal antibodies against the RBD of MERS also mediated the entry of MERS-CoV into host cell. Thus, ADE can create a huge problem in designing a vaccine against the virus. However, in contrast to the above discussion a recent study showed that NAbs against the SARS-CoV-2 RBD has not promoted ADE mediated viral entry [190].
5. Age and immune responses against COVID-19
It has been suggested that hyperinflammatory immune response is one of the reasons for higher mortality rate in aged population [191]. A study demonstrated that older people have higher expression of prostaglandin D2 which impairs the migration of respiratory DCs to draining lymph nodes and thus the induction of specific T cell response [192]. As age advances, lymph nodes undergo some changes that further effect the formation and maintenance of naive T and B cells. Recently, a study reported that with age activation, proliferation and differentiation of T cells is also impaired [193]. Thus, it is plausible to suggest that impairment in immune response during ageing enhance the severity of COVID-19 in aged people.
In contrast to aged population children rarely gets SARS-CoV-2 viral infection. Children infected with virus showed mild symptoms and rarely infection progresses in them into the severe stage [194]. It has been observed that only 6% of the children below the age of 18 have severe symptoms whereas 50% of them developed only mild symptoms [195]. Furthermore, a recent study has shown that people below the age of 20 are less susceptible to household transmission whereas people above the age 60 are highly susceptible [196]. Surprisingly, it has been observed that immune system plays crucial role in protection of children from COVID-19. It is observed that children have decreased levels of proinflammatory cytokines TNF-α and IL-6 that further reduces the neutrophil infiltration and lung injury. Along with the reduced proinflammatory cytokine levels, higher levels of immunomodulatory cytokines such as IL-10 and IL-13 has also been observed in children. Moreover, children have decreased expression of ACE2 and TMPRSS2 is lung epithelium and exhibits better capability of repairing the lung infection [194]. Thus, it can be concluded that multiple factors work in synergistic manner to protect children from developing severe COVID-19 disease. But the results of recent research have shown that viral load of SARS-CoV-2 infected children is significantly higher than the adults with severe disease [197]. This study thus contradicts with the previous results which claim that children are at low risk of COVID-19.
6. Immunotherapies for COVID-19
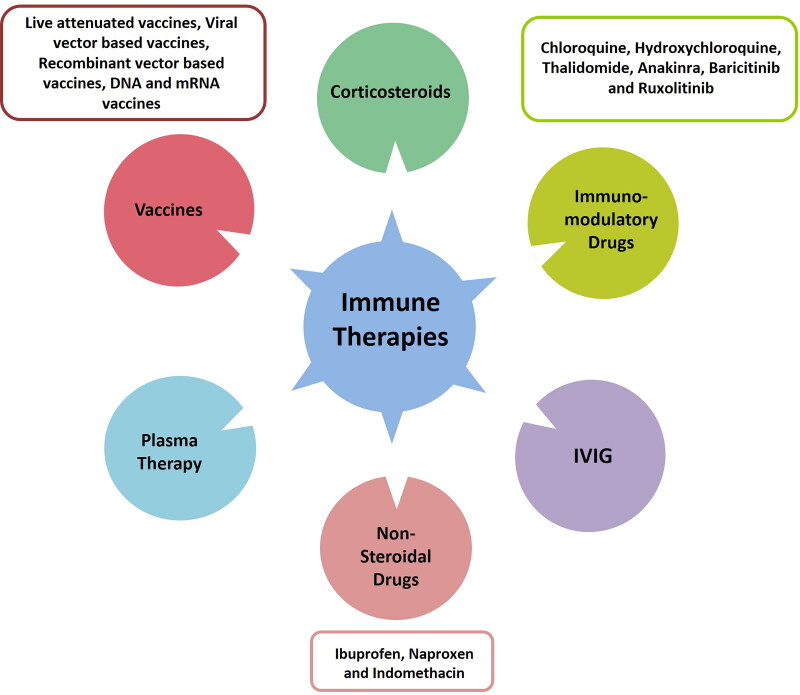
Currently, there is no effective therapy against SARS-CoV-2 infection. Inhibition of inflammatory response caused by dysregulated immune response can be an effective therapy in preventing COVID-19. Below we discussed some of the immunotherapeutic strategies that can be employed as potential treatments option for COVID-19 (Figures 7 and 8).
Figure 7.
Different types of immunotherapies for prevention of COVID 19.
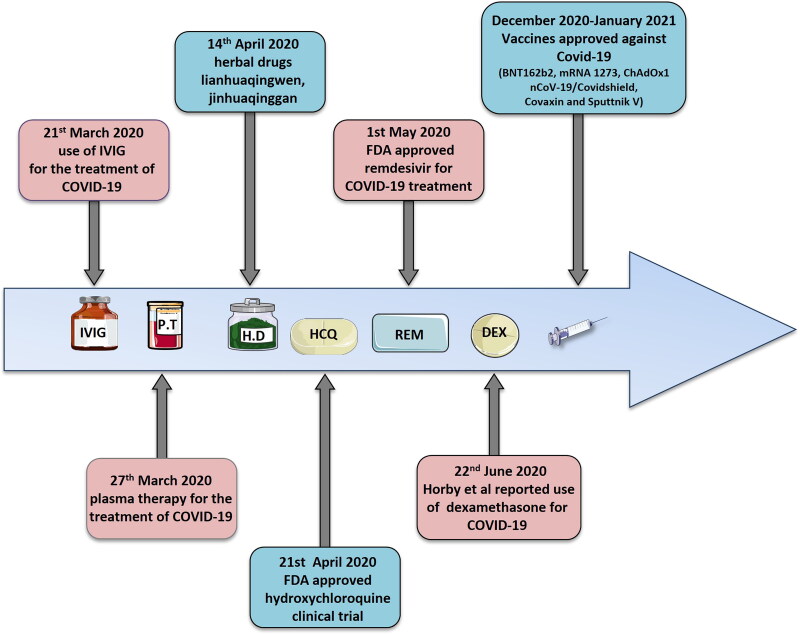
Figure 8.
Timeline of COVID-19-therapies: Advancements in the COVID-19 therapies. IVIG (Intravenous immunoglobulin), P.T (Plasma therapy), H.D (Herbal drugs), HCQ (Hydroxychloroquine), REM (Remdesivir), DEX (Dexamethasone).
6.1. Immunosuppressive drugs
Thalidomide is an anti-inflammatory, anti-angiogenic and anti-fibrotic immune modulator. Interestingly, it has been observed that thalidomide in combination with low dose glucocorticoid prevented pneumonia in SARS-CoV-2 infected patients [198]. Currently clinical trials are in progress for Thalidomide to further explore its role in prevention of COVID-19 (ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT04273529, 2020). Recently, a study demonstrated that Lianhuaqingwen, a traditional Chinese medicine prevented SARS-CoV-2 infection by significantly reducing the pro-inflammatory cytokines and chemokines such as TNF-α, IL-6, CXCL10 and MCP-1 levels [199]. Clinical trials on Tocilizumab and on anti-TNF-α antibody are also in progress [45]. Clinical trials for drugs against GM-CSF such as Lenzilumab, Gimsilumab and Namilumab are also under way [200–202] (Figure 9). Several studies reported the immunomodulatory properties of a common antimalarial drug Chloroquine. Clinical applications of chloroquine are increasing day by day. For years, Chloroquine is being used in chronic kidney disease, oncology, cardiovascular and rheumatological disorders. Currently, Chloroquine is suggested for the treatment of COVID-19. Antiviral therapy with Chloroquine recovers the levels of NK cells and CD8+ T cells during convalescent period in COVID-19 patients [47]. A systemic review on Chloroquine which includes six studies and various ongoing trials suggested that there is evidence of effectiveness of Chloroquine in preventing COVID-19. But for better understanding of anti-viral properties of Chloroquine some high-quality effective trials are recommended [203]. Hydroxychloroquine is also an anti-inflammatory drug that works by same mechanism as Chloroquine. Hydroxychloroquine has been well established as an immunomodulatory drug. Furthermore, Hydroxychloroquine prevents cellular autophagy, an important step for innate and adaptive immune system activation [204]. Hydroxychloroquine is preferred over Chloroquine for treatment of malaria as it is safer. It is reported to be three times more effective than Chloroquine in treatment of SARS-CoV-2 infection at a dose of 400 mg twice along with maintenance dose of 200 mg twice daily for 4 days [205]. Hydroxychloroquine reduced viral load in COVID-19 patients and Azithromycin in addition to Hydroxychloroquine reinforced its anti-viral potential and makes it more efficient in eliminating the virus [206]. Zheng et al. suggested that cross reactive antibodies are responsible for cytokine storm and their inhibition can prevent the SARS infection. They proposed that inhibition of activated memory B cells in early stage of COVID-19 patients can prevent the production of cross-reactive antibodies. mTOR inhibitors can potentially inhibit activation of memory B cells and can be used for the treatment of COVID-19 [207].
Figure 9.
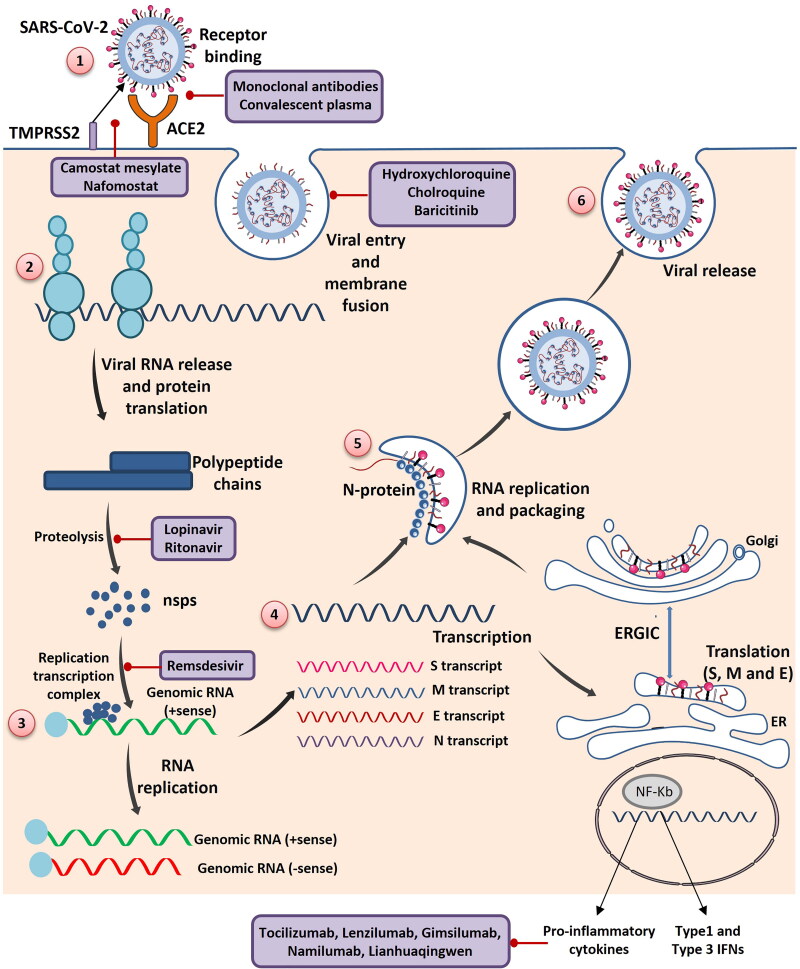
The schematic representation of SARS-CoV-2 life cycle in step wise manner depicting the potential therapeutic approaches against SARS-CoV-2: (1) SARS-CoV-2 enter into the cell by binding to the ACE2 receptor present on the host cell membrane via S protein which mediates viral-host membrane fusion and viral entry. (2) Entry of SARS-CoV-2 results in the uncoating of viral RNA into cytoplasm which then undergoes translation to produce polyproteins pp1a and pp1b, which are further processed by virus-encoded proteinases into individual nonstructural proteins (nsps). (3) Replication transcription complex is formed by some of the nsps. Replication transcription complex uses the (+) stand genomic RNA as template. Following replication (+) stand genomic RNA is produced which becomes the genome of the new viral particle. (4) Subgenomic RNAs synthesized through transcription are translated into viral structural proteins: S (Spike) protein, M (Membrane) protein, E (Envelope) protein, N (Nucleocapsid) protein. (5) N protein combined with the (+) strand genomic RNA to form the nucleoprotein complex. S, E and M proteins enter into endoplasmic reticulum (ER) and transported to Golgi apparatus. The nucleoprotein complex and S, E and M proteins are further assembled in the ER-Golgi intermediate compartment (ERGIC) to form mature virion. (6) Virions are released from the host cells by exocytosis. Many potential therapeutic approaches are considered for the management of COVID-19. These therapeutics act on various steps of the SARS-CoV-2 life cycle. Monoclonal antibodies and Convalescent plasma targets the S proteins of SARS-CoV-2 from interacting with the ACE2 receptor. Camostat mesylate and Nafomostat targets the serine protease (TMPRSS2) and prevents the S protein cleavage which is required for the viral fusion to the host cells. Lopinavir and Ritonavir targets the proteolysis of polypeptide chains. Remsdesivir prevents the replication of SARS-CoV-2 and drugs like Tocilizumab, Lenzilumab, Gimsilumab, Namilumab and Lianhuaqingwen inhibit the pro-inflammatory response.
6.2. Plasma therapy
In passive antibody therapy, antibodies against a particular agent are given to the infected person that provides immediate relief to infected person. In case of COVID-19, the source of antibodies against the SARS-CoV-2 is the serum of patient who recovered from COVID-19, i.e., convalescent sera [208]. Earlier, convalescent plasma therapy has also been employed for the treatment of SARS, MERS and influenza A virus pandemic in 2009 [209–211]. Recently, several studies evidenced that convalescent sera can be an effective therapy for the treatment of COVID-19. It is reported that treatment of 10 severe COVID-19 patients with single dose of 200 mL of convalescent sera markedly enhanced the NAbs against the SARS-CoV-2 virus [55]. Furthermore, Zhang et al. reported that all severely ill patients who received convalescent sera along with supportive care recovered from COVID-19 infection [212]. Convalescent sera administration improved health of the patients [213]. Food and drug administration (FDA) also approved the use of convalescent sera for treatment of critically ill COVID-19 patients [214].
6.3. Immunosuppression by corticosteroids and nonsteroidal drugs
Corticosteroids are steroid hormones and classified into glucocorticoids and mineralocorticoids [215]. Glucocorticoids are involved in various essential biological processes like metabolism, development, reproduction, cardiovascular functions, water and electrolyte balance, growth and development [216]. Glucocorticoids exhibits immunomodulatory properties and are being increasingly used for the treatment of various immune related disorders like inflammatory bowel disease (IBD), asthma, rheumatoid arthritis (RA), allergy, septic shock and multiple sclerosis [215]. Corticosteroids were previously considered for the treatment of MERS, SARS and influenza virus [217, 218].
Now, their use is also being suggested for the treatment of severe COVID-19 cases to suppress lung inflammation [21]. Recently, a study revealed that no association exists between corticosteroids treatment and virus clearance in case of COVID-19 [219]. Clinical outcomes from other studies also do not support the theory that corticosteroids can prevent lung inflammation in COVID-19 patients [220]. But recently in a randomized clinical trial (2100 participants) in United Kingdom (UK) it has been observed that synthetic glucocorticoid dexamethasone suppressed the deaths by one third in the COVID-19 patients who were on ventilators. But dexamethasone showed no impact on the non-severe patients but it was very impactful in case of severe patients that require ventilators and reduced their chance of dying by 20% [221]. Moreover, another study reported that short term treatment with dexamethasone significantly reduced the C- reactive protein levels and the length of stay of COVID-19 patients in hospitals [222].
Nonsteroidal drugs (NSDs) are the common prescribed drugs indorsed for reducing pain and inflammation. NSDs inhibit cyclooxygenase-2 (COX-2) enzyme which are required for prostaglandins induced pain and inflammation [223]. It has been observed that NSDs like ibuprofen and aspirin suppressed antibody production and prevented MHC restricted antigen presentation in DCs [224]. Because of their immunosuppressive properties NSDs are recommended for the inhibition of SARS-CoV-2 infection. Naproxen and Indomethacin are the two NSDs that are mainly suggested for COVID-19 infection. Naproxen is an antiviral drug that prevented infection due to influenza A and B virus [225] and can be an effective therapy for COVID-19 [226]. Indomethacin has anti-inflammatory and antiviral activities and reported to inhibit SARS-CoV infection by suppressing RNA synthesis in vitro in monkey VERO cells [227]. In vivo studies showed reduced level of canine CoVs in dogs, and thus, can be considered for suppressing SARS-CoV-2 infection [228].
6.4. Intravenous immunoglobulins
Intravenous immunoglobulins (IVIG) is the mixture of immunoglobulins pooled from thousands of normal individuals [229]. IVIG has been employing for the treatment of various neurological, hematogical, nephrological, dermatological, rheumatological and immune related disorders [230]. Several mechanisms are being involved in preventive activities of IVIG. A study found that IVIG promoted cytokine balance, inhibited auto-reactive T cells, reduced antibody production from B cells and also reduced macrophage activity [231]. Recently, the relevance of IVIG has also been explored in case of COVID-19. It has been observed that administration of IVIG at a dose of 0.3–0.5g/kg for 5 days to critically ill COVID-19 patients those were not responding to other treatment options reduced the progression of pulmonary lesions and improved their clinical and respiratory conditions [232]. Furthermore, another study reported that usage of IVIG for treatment of severely ill COVID-19 patients decreased the need of mechanical ventilation, reduced the length of hospital stay and decreased the mortality rate [233]. It was observed that all IVIG treated patients had satisfactory recovery with no adverse effects [234]. These studies raise the possibility that IVIG may provide a better way in the treatment of COVID-19.
6.5. Vaccines
There is an urgent requirement of vaccine for the prevention of SARS-CoV-2 infection because of its highly transmissible nature and pathogenesis. Before SARS-CoV-2 there were epidemics due to SARS-CoV and MERS-CoV. Several vaccines like inactivated virus vaccines, live attenuated virus vaccines, viral vector vaccines, subunit vaccines and DNA vaccines have been designed against SARS-CoV and MERS-CoV. But so far, they have been tested only on animals and no data on their safety and efficacy in case of human is available [235, 236]. Thus, these vaccines cannot be recommended for the treatment of SARS-CoV-2 infection. But the knowledge that has been gained in making these vaccines is now being used for designing a better SARS-CoV-2 vaccine. All over the world within few weeks of SARS-CoV-2 outbreak many research groups started making vaccines. After 2 months of the outbreak it was observed that over 37 pharmaceutical companies or research institutes were in the race of making COVID-19 vaccine [237]. Right now, various biopharmaceutical companies in collaboration with research institutes are designing several types of vaccines like DNA vaccines, RNA vaccines, inactivated vaccines, viral vector-based vaccines and recombinant vaccines for COVID-19 (Table 2). Some of these vaccines have shown positive results in clinical trials. Pfizer and BioNtech vaccine candidate, BNT162b2 has found to be more than 90% effective in preventing COVID-19. These companies have received emergency use authorization (EUA) in various countries like UK, America, Europe, Australia and have already started vaccinating people [238–241]. Moderna vaccine candidate, mRNA-1273 was found to be 94.5% effective in phase 3 clinical trials and the vaccine is already approved by America [241, 242]. Phase 3 clinical trials of Oxford-AstraZeneca COVID-19 vaccine, ChAdOx1 nCoV-19 or Covidshield (in India) has shown that the vaccine is 70% effective in preventing COVID-19 and have already started vaccinating people in UK and India [241, 243]. The Russian Direct Investment Fund (RDIF) and Gameleya center have also announced their vaccine candidate, SPUTNIK-V with 92% efficacy in phase 3 clinical trials and is already approved by countries like Argentina and Belarus [244]. Bharat biotech vaccine candidate Covaxin is also approved by Indian government and the Indian government has already started vaccinating its citizens [245]. Although the process of vaccination has started in various countries it is still early to estimate whether these vaccines will be able to provide effective immunity against COVID-19 as postvaccination data is still awaited.
Table 2.
Status of different types of COVID-19 vaccines.
| S.No | Type of vaccine | Name | Developer | Trial phase | Registration number/Clinical trial number |
|---|---|---|---|---|---|
| 1. | RNA-based vaccine | BNT162 | Pfizer/BioNTech | Phase 3 | NCT04368728 |
| 2. | mRNA-based vaccine | mRNA 1273 | Moderna | Phase 3 | NCT04283461 |
| 3. | Viral vectored vaccine | Inactivated vaccine | Wuhan Institute of Biological Products, China National Pharmaceutical group (Sinopharm) | Phase 3 | ChiCTR2000031809 |
| 4. | Adenovirus-based vaccine | Ad5-nCoV | CanSino Biologics | Phase 3 | NCT04313127 |
| 5. | Inactivated vaccine | CoronaVac | Sinovac | Phase 3 | NCT04456595 |
| 6. | Adenovirus vectored vaccine | ChAdOx1 nCoV-19 or Covidshield |
The University of Oxford | Phase 3 | NCT04324606 |
| 7. | Adenovirus vectored vaccine | Sputnik V | Gamaleya Research Institute of epidiomology and Microbiology, Health Ministry of the Russian Federation | Phase 3 | NCT04530396 |
| 8. | Recombinant vaccine | NVX-CoV2373 | Novavax | Phase 3 | NCT04533399 |
| 9. | Whole virion inactivated vaccine | Covaxin | Bharat Biotech | Phase 3 | NCT04471519 |
| 10. | Inactivated vaccine | BBIBP-CorV | Beijing Institute of Biological Products, China National Pharmaceutical group (Sinopharm) | Phase1/2 | – |
| 11. | DNA vaccine | GX-19 | Genexine | Phase 1/2 | NCT04445389 |
| 12. | mRNA-based vaccine | LUNAR-COV19 | Arcturus Therapeutics and Duke-NUS Medical School | Phase 1/2 | – |
| 13. | DNA vaccine | INO-4800 | Inovio Pharmaceuticals | Phase 1/2 | NCT04447781 |
| 14. | mRNA-based vaccine | CVnCoV | CureVac | Phase 1 | NCT04515147 |
| 15. | Recombinant subunit vaccine | SCB-2019 | Clover Biopharmaceuticals | Phase 1 | NCT04405908 |
| 16. | Recombinant vaccine | COVAX-19 | Vaxine Pty Ltd. | Phase 1 | NCT04453852 |
| 17. | Recombinant vaccine | Plant-based adjutant COVID-19 vaccine | Medicago | Phase 1 | NCT04450004 |
| 18. | Lentivirus vector vaccine | COVID-19 aAPC | Shenzhen Geno-Immune Medical Institute | Phase 1 | NCT04299724 |
6.6. Monoclonal antibodies
Designing antibodies against the RBD of the S protein is one of the strategies used to combat COVID-19. RBD of S protein binds to the ACE2 receptor and mediate viral entry. Blockade of RBD binding to the S protein by antibodies against it can prevent the viral entry into host cells and thus prevents SARS-CoV-2 infection [246]. In 2010, Berry et al. successfully designed mAbs against the SARS-CoV RBD which cross neutralized and prevented its binding to the ACE2 receptor [247]. Study of Berry et al. showed the potential and feasibility of mAbs as vaccine for combating COVID-19. SARS-CoV RBD specific human mAbs can also be explored against the SARS-CoV-2. After screening of various mAbs, it has been observed that one SARS-CoV specific mAbCR3022 mAb bound effectively with the SARS-CoV-2 RBD thus; provide a treatment option for COVID-19 [248]. Moreover, 47D11 mAb designed against the conserved epitope in the SARS2-S-S1b domain was also found to neutralize SARS-CoV-2 [5]. Lei et al. designed fusion protein comprised of extracellular domain of the ACE2 receptor and fc region of the human IgG1 antibody. The fusion protein showed high binding affinity for both SARS-CoV and SARS-CoV-2. They observed that fusion protein was capable of neutralizing pseudotyped SARS-CoV and SARS-CoV-2 viruses [13]. Not only the RBD but other parts of the S protein can also be targeted for vaccine preparation. Chi et al. isolated the mAbs from the convalescent SARS-CoV-2 infected patients and observed that a mAb named as 4A8 showed high potency of neutralizing the SARS-CoV-2. 4A8 showed no binding with RBD but to the N terminal domain of the S protein. This showed that mAbs targeting N terminal domain can also acts as promising therapeutic against COVID-19 [249]. Recently Liu et al. isolated 61 mABs against SARS-CoV-2 from 5 severe hospitalized patients. They observed that 19 antibodies among these potently neutralized SARS-CoV-2 in vitro. Epitope mapping revealed that these antibodies were directed against the RBD and NTD of the S protein [250]. Mercado et al. also showed the efficacy of adenovirus serotype 26 (AD26) vector-based vaccine in 52 rhesus macaques. They showed that AD26 induced NAbs and protected against SARS-CoV-2 challenge [251]. Furthermore, Baum et al. made a cocktail of two potent mAbs (REGN10987 + REGN10933) against the SARS-CoV-2 S protein. They demonstrated that the antibody cocktail reduced the viral load in rhesus macaques and golden hamsters [252].
6.7. Herbal drugs
A number of traditional Chinese medicines (TCM) and herbal drugs like Gypsum, Semen armeniacaeamarae, Liquorice, Scutellariabaicalenishas been reported for the treatment of COVID-19 [253]. On 14 April 2020 China approved the usage of herbal drugs Xuebijing for severe SARS-CoV-2 infection and Lianhuaqingwen and Jinhuaqinggan for the treatment of mild conditions. TCM and herbal drugs are observed beneficial in preventing SARS-CoV-2 infection as they have antiviral and anti-inflammatory properties. It has been reported that approximately 60 107 COVID-19 patients were treated with TCM or herbal drugs [254]. But some of these herbal drugs have been found to shown adverse effects which limit their use in COVID-19 treatment [255]. TCMs are sometimes adulterated and are not processed properly [256].
Indian Ayurveda also has great potential of treating deadly diseases and can replace the TCM in treatment of COVID-19. Ayurveda which means “The science of life” is the oldest medicinal science was originated in India dating back 5000 years. Ayurveda recommends the use of traditional Indian medicines, medicinal plants and spices for the treatment of diseases. Medicinal herbs promote immune system development and exhibits anti-inflammatory, anti-cancer and anti-oxidant properties. Medicinal herbs modulate the immune system and maintain immune homeostasis [257]. Several ayurvedic plants exhibits immunomodulators like: Curcuma longa (Cucumin), Tinosporacordifolia (Guduchi), Andographispaniculate, Dioscorea japonica, Boerhaaviadiffusa (Punarnavine), Withaniasomnifera (Ashwagandha), Allium sativum (Garlic), Terminaliaarjuna and Mangiferaindica [258]. Ministry of AYUSH, Government of India also proposed guidelines for use of ayurvedic herbs during COVID-19 pandemic. Recently, a group published an intervention plan based on ayurvedic plants for the treatment of COVID-19 [254]. A recent study which collected data from 163 countries also demonstrated the role of spices in COVID-19. The study had shown that correlation exits between the total cases of COVID-19/million and the gram of spices consumed/capita/day. They observed that nations which consumed less amount of spices/capita had more cases of COVID-19/million than high spice consuming nations [259]. But till now there is no study which directly proves the role of Indian Ayurvedic herbs or medicines in the treatment of COVID-19. Nevertheless, Ayurvedic herbs have the potential of curing various diseases and thus there should be clinical trials on further investigating their role in preventing COVID-19. Ayurvedic herbs can also provide a safe and better option in place of TCM.
7. Inhibitors of viral entry and replication
Apart from immunosuppression, blocking of viral entry into cells can be an effective way of preventing the SARS-CoV-2 infection. SARS-CoV-2 enters into the cells with the help of S protein and required host protease TMPRSS2 and cathepsin L for S protein priming. Hoffman et al. showed that viral infection can be prevented by blocking the viral entry using the inhibitors for TMPSS2 protease. They used camostat mesylate to block TMPSSS2 and observed that it had prevented viral entry and thus viral infection. Hoffman et al. also showed that NAbs produced in the convalescent patients against SARS-CoV-2 S proteins also blocked S protein mediated viral entry [37]. Earlier Yamamoto et al. showed that Nafomostat, an inhibitor of TMPSS2 had inhibited S protein mediated entry of MERS virus [260]. Two antiviral drugs remdesivir and chloroquine has also been reported to prevent SARS-CoV-2 infection. Remdesivir acts on post virus entry. It is an adenosine analogue and gets incorporated into the viral RNA chains which cause premature termination. Beigel et al. had shown that Remdesivir decreased the recovery time in hospitalized COVID-19 patients to 11 days from 15 days [261]. Remdesivir is widely used for the treatment of COVID-19 and on May 1st FDA EUA to the Remdesivir for COVID-19 treatment. On August 28 FDA expanded the EUA to treat hospitalized COVID-19 patients [262]. Chloroquine which is common antimalarial drug inhibits viral entry into the host cell. Chloroquine interferes with the glycosylation of S protein [263]. SARS virus enter into the host cell by endocytosis also and thus SARS infection can also be prevented by inhibiting the regulators of endocytosis like AP-2 associated protein kinase 1 (AAK1). Richardson et al. suggested through computational studies that Baricitinib which is one of the best AAKI inhibitors can also be used for the treatment of COVID-19. It also binds to G-associated kinase which also regulates endocytosis [264]. Some studies target the SARS-CoV-2 main protease (CoV Mpro). CoV Mpro cleaves polyproteins required for the replication and transcription of COVID-19 and inhibition of CoV Mpro will prevent viral multiplication. Computer aided drug designing and bioinformatics tools are used for designing the inhibitors for CoV Mpro. Jin et al. designed inhibitor M3 which inhibits the activity CoV Mpro by fitting into the substrate binding pocket [265]. Carmofur which is an antineoplastic agent can also inhibit CoV Mpro and is suggested for treatment of COVID-19 [266].
8. Gut microbiota and COVID-19
Microbiota is the collection of microbes like bacteria, protozoa, archaea, fungi, viruses and eukaryotes that colonize in or on the host [267]. The aggregate of microbes that reside in the gastrointestinal tract is termed as gut microbiota (GM). Human GM consists of over 100 trillion microbes encoding more than three million genes. GM is considered as an essential organ of our body [268]. GM regulates various biological processes in our body like they increase nutrient absorption, enhance extraction of energy from food and maintain integrity of gut. One of the most important functions of the GM is that it promotes immune system development. A study reported that germ free mice have reduced number of CD4+ T cells in the lamina propria and IgA producing B cells [269]. They have smaller number of payer’s patches with reduced size, poorly developed B and T cells zones in spleen [270]. Germ free mice also possessed imbalanced Th1/Th2 response [271]. GM is mainly dominated by Firmicutes and Bacteroidetes phyla and constitutes over 90% of the total bacterial species localized in gut. Other microbial phyla like Proteobacteria, Actino-bacteria, Fusobacteria and Verrucomicrobiaare present in smaller proportions [272]. GM regulates various body functions. But alterations in GM composition can cause various gastrointestinal, metabolic, respiratory, autoimmune, psychological and liver diseases [273]. Alternation is GM composition is known as dysbiosis. A study reported that excessive intake of medications; antibiotics and irradiation altered the GM [274]. As altered GM leads to several diseases it might also be possible that GM gets altered during SARS-CoV-2 infection. Several studies showed that elderly people and immunocompromised people having cardiovascular diseases, hypertension and diabetes are at much higher risk of COVID-19. Dysbiosis is also one of the reasons for cardiovascular diseases, diabetes and hypertension and elderly people also have altered GM [275]. This further suggests the role of dysbiosis in COVID-19. Dysbiosis leads to dysregulated immune response and promote inflammatory response. Dysbiosis induced enhanced production of Th17 cells and proinflammatory cytokines like TNF-α, IL-1β and IL-6. Several studies revealed that dysbiosis lead to development of various inflammatory diseases like IBD [276], colorectal cancer [277] and RA [278]. Thus, it might be possible that altered GM is behind the inflammatory response reported in COVID-19.
A recent study published has shown that COVID-19 patients have altered GM during hospitalization. In one of the studied COVID-19 patient, GM was dominated by pathogens and was depleted of beneficial microbes. It was observed that there was positive correlation between the abundance of Clostridium ramnosum, Clostridium hathewayi and Coprobacillus and COVID-19 severity. Abundance of Faecalibacterium prausnitziiwas negatively correlated with COVID-19 severity [279]. This study thus has shown the role of GM in COVID-19, but further research is required to establish the relation between dysbiosis and SARS-CoV-2 infection.
It is also reported that like gut, lung also has its own microbiota denoted as lung microbiota. Lung microbe is dominated by Firmicutes, Bacteroidetes and Proteobacteria. Surprisingly, it has been revealed that GM regulates pulmonary health and there is effect of lung inflammation on GM also. Thus, GM and lung interconnect with each other and affect each other’s development. This bidirectional interconnection between the GM and lung is referred as gut-lung axis. Gut-lung axis increases the likelihood that SARS-CoV-2 infection might affect the GM. It was also observed that GM also has role in ARDS which further increase the possibility that GM has role in COVID-19 [280]. Interestingly, a study has reported that GM colonization in germ free mice reduced the expression of colonic ACE2 [281]. This again confirms the role of GM in SARS-CoV-2 pathogenesis.
To further test the role of GM in COVID-19, a clinical trial (NCT04355741) is under progress. The aim of the trial is to test if there is any difference of GM composition between the patients those having mild disease and those having severe disease. The study will assess is there any relation between the GM composition and the rate of mortality, length of stay in hospitals and the duration of mechanical ventilation. The outcome of this study will explain further the role of GM in COVID-19. There should be more studies performed to find out if there is any correlation of dysbiosis found in patients having cardiovascular diseases, hypertension and diabetes and their increased chances of getting infected with SARS-CoV-2.
9. Probiotics for treatment and management of COVID-19
Probiotics which are defined as “live organisms that when administered in adequate amounts confer health benefits on the host.” Probiotics are used as therapeutics for the treatment of various gastrointestinal and metabolic disorders like IBD, irritable bowel syndrome, antibiotic associated diarrhoea, obesity, osteoporosis and diabetes [282, 283]. Probiotics modulate the GM, regulate immune system and maintain the gut integrity [284–286]. Probiotics modify GM and suppress inflammation by modulating the immune system. We propose here that probiotics can also be an effective therapy for the treatment and management of COVID-19. China’s National Administration of Traditional Chinese Medicine and China’s National Health Commission also proposed the use of probiotics for the treatment of SARS-CoV-2 infection. Administration of probiotics- Enterococcus faecalis, Lactobacillus rhamnosus GG and Bacillus subtilis to the severely ill pneumonia patients (on ventilator support) significantly reduced the need of ventilators [287]. Thus, probiotics can be used to reduce the demand of mechanical ventilation in COVID-19 patients. In China it was observed that 2–36% of the COVID-19 patients who were given antibiotics had diarrhoea [287]. Probiotics replace the harmful bacteria in gut and prevent gut dysbiosis in several diseases like IBD, colorectal cancer, diabetes and osteoporosis. Thus, probiotics can also be used for preventing dysbiosis in COVID-19 [279].
Another mechanism through which probiotics can prevent SARS-CoV-2 infection is by modulating the immune system. Probiotics showed anti-inflammatory properties [284, 285]. Probiotic combination of L. rhamnosus, B. lactis and B. longum decreased the production of IL-β and IL-6 and unregulated the expression of IL-10 [288]. L. fermentum and L. salivarius downregulated the expression of inflammatory cytokine IL-Iβ in DSS induced colitis mouse model [289]. Probiotics mixture VSL#3 decreased the level of TNF-α in peripheral blood and prevented sickness behavior [290]. A study reported that L. rhamnosus suppressed the production of TNF-α from macrophages [291]. Many other studies proved the role of probiotics in preventing inflammation [292, 293]. Thus, the anti-inflammatory properties of probiotics and their role in preventing dysbiosis should be exploited for preventing SARS-CoV-2 infection. More future research should be done on the pathogenesis and efficacy of probiotics in humans. Probiotics role in COVID-19 should be explored because probiotics can provide a cheap and effective way of treating and managing novel CoV 2019.
10. Conclusion
SARS-CoV-2 enforced public health crisis with an unprecedented challenge for the development of successful therapeutics. SARS-CoV-2 infection impairs the immune system leading to various inflammatory responses and in severe cases death, but till now there is no specific therapy available for COVID-19. Thus, there is an exigent need to identify a potential therapeutic compound that can be employed against COVID-19. This review summarizes various mechanisms of SARS-CoV-2 immunopathogenesis and available therapeutic interventions for the cure of COVID-19. Control of inflammatory response is vital for targeting the viral infection, and therefore, it is imperative that the mechanisms behind the hyperinflammation are further elucidated to design better therapeutic strategy to restrict the viral spread. In the last section, we also summarized the hidden potential of probiotics that can provide a safe and inexpensive option in the treatment and management of COVID-19.
Acknowledgements
AB, LS, CS, ZA, BV, PKM, GCM and RKS acknowledge the Department of Biotechnology, AIIMS, New Delhi, India for providing infrastructural facilities. ZA thanks Dr. Harisingh Gour Central University, Sagar (MP). All figures (except figure 7) are created with the help of https://smart.servier.com/.
Funding Statement
This work was financially supported by projects: DST-SERB (EMR/2016/007158), Govt. of India, Intramural project from All India Institute of Medical Sciences (AIIMS), New Delhi-India and AIIMS-IITD collaborative project sanctioned to RKS. AB thank DST SERB project for research fellowship, LS and ZA thank UGC for research fellowship.
Authors’ contributions
RKS contributed in conceptualization and writing of the manuscript. AB, LS, CS, ZA, PKM BV, GCM participated in writing and editing of the review. RKS suggested and AB and LS created the illustrations.
Conflicts of interest
The authors declare no conflicts of interest.
References
- 1.Lan J, Ge J, Yu J, et al. . Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581(7807):215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 2.Wu JT, Leung K, Bushman M, et al. . Estimating clinical severity of COVID-19 from the transmission dynamics in Wuhan, China. Nat Med. 2020;26(4):506–510. doi: 10.1038/s41591-020-0822-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hassan SA, Sheikh FN, Jamal S, et al. . Coronavirus (COVID-19): a review of clinical features, diagnosis, and treatment. Cureus. 2020;12(3):e7355. doi: 10.7759/cureus.7355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li G, Fan Y, Lai Y, et al. . Coronavirus infections and immune responses. J Med Virol. 2020;92(4):424–432. doi: 10.1002/jmv.25685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang C, Li W, Drabek D, et al. . A human monoclonal antibody blocking SARS-CoV-2 infection. Nat Commun. 2020;11(1):2251. doi: 10.1038/s41467-020-16256-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andersen KG, Rambaut A, Lipkin WI, et al. . The proximal origin of SARS-CoV-2. Nat Med. 2020;26(4):450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drosten C, Gunther S, Preiser W, et al. . Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med. 2003;348(20):1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 8.Cheng VC, Lau SK, Woo PC, et al. . Severe acute respiratory syndrome coronavirus as an agent of emerging and reemerging infection. Clin Microbiol Rev. 2007;20(4):660–694. doi: 10.1128/CMR.00023-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ksiazek TG, Erdman D, Goldsmith CS, et al. . A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med. 2003;348(20):1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 10.Ramadan N, Shaib H.. Middle East respiratory syndrome coronavirus (MERS-CoV): a review. Germs. 2019;9(1):35–42. doi: 10.18683/germs.2019.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Groot RJ, Baker SC, Baric RS, et al. . Middle East respiratory syndrome coronavirus (MERS-CoV): announcement of the Coronavirus Study Group. J Virol. 2013;87(14):7790–7792. doi: 10.1128/JVI.01244-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shang J, Ye G, Shi K, et al. . Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581(7807):221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lei C, Qian K, Li T, et al. . Neutralization of SARS-CoV-2 spike pseudotyped virus by recombinant ACE2-Ig. Nat Commun. 2020;11(1):2070. doi: 10.1038/s41467-020-16048-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou P, Yang XL, Wang XG, et al. . A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu A, Peng Y, Huang B, et al. . Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020;27(3):325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li F. Structure, function, and evolution of coronavirus spike proteins. Annu Rev Virol. 2016;3(1):237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fehr AR, Perlman S.. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol Biol. 2015;1282:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boopathi S, Poma AB, Kolandaivel P.. Novel 2019 coronavirus structure, mechanism of action, antiviral drug promises and rule out against its treatment. J Biomol Struct Dyn. 2020;38:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rothan HA, Byrareddy SN.. The epidemiology and pathogenesis of Coronavirus Disease (COVID-19) outbreak. J Autoimmun. 2020;109:102433. doi: 10.1016/j.jaut.2020.102433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lauer SA, Grantz KH, Bi Q, et al. . The incubation period of Coronavirus Disease 2019 (COVID-19) From publicly reported confirmed cases: estimation and application. Ann Intern Med. 2020;172(9):577–582. doi: 10.7326/M20-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang C, Wang Y, Li X, et al. . Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vaira LA, Salzano G, Deiana G, et al. . Anosmia and ageusia: common findings in COVID-19 patients. Laryngoscope. 2020;130(7):1787. doi: 10.1002/lary28692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matthay MA, Zemans RL, Zimmerman GA, et al. . Acute respiratory distress syndrome. Nat Rev Dis Primers. 2019;5(1):18. doi: 10.1038/s41572-019-0069-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Almeida-Pititto B, Dualib PM, Zajdenverg L, et al. . Brazilian diabetes society study G. Severity and mortality of COVID 19 in patients with diabetes, hypertension and cardiovascular disease: a meta-analysis. Diabetol Metab Syndr. 2020;12(1):75. doi: 10.1186/s13098-020-00586-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poletti P, Tirani M, Cereda D, et al. . Age-specific SARS-CoV-2 infection fatality ratio and associated risk factors, Italy, February to April 2020. Eurosurveillance. 2020;25(31):2001383. doi: 10.2807/1560-7917.ES.2020.25.31.2001383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jin J-M, Bai P, He W, et al. . Gender differences in patients with COVID-19: focus on severity and mortality. Front Public Health. 2020;8:152. doi: 10.3389/fpubh.2020.00152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang J, Cheng A, Kumar R, et al. . Hypoalbuminemia predicts the outcome of COVID-19 independent of age and co-morbidity. J Med Virol. 2020;92(10):2152–2158. doi: 10.1002/jmv.26003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martinez-Rojas MA, Vega-Vega O, Bobadilla NA.. Is the kidney a target of SARS-CoV-2? Am J Physiol Renal Physiol. 2020;318(6):F1454–F1462. doi: 10.1152/ajprenal001602020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang C, Shi L, Wang FS.. Liver injury in COVID-19: management and challenges. Lancet Gastroenterol Hepatol. 2020;5(5):428–430. doi: 10.1016/S2468-1253(20)30057-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bansal M. Cardiovascular disease and COVID-19. Diabetes Metab Syndr. 2020;14(3):247–250. doi: 10.1016/jdsx202003013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang H, Penninger JM, Li Y, et al. . Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: molecular mechanisms and potential therapeutic target. Intensive Care Med. 2020;46(4):586–590. doi: 10.1007/s00134-020-05985-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hamming I, Timens W, Bulthuis ML, et al. . Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol. 2004;203(2):631–637. doi: 10.1002/path15702004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu H, Zhong L, Deng J, et al. . High expression of ACE2 receptor of 2019-nCoV on the epithelial cells of oral mucosa. Int J Oral Sci. 2020;12(1):8. doi: 10.1038/s41368-020-0074-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zaim S, Chong JH, Sankaranarayanan V, et al. . COVID-19 and multiorgan response. Curr Probl Cardiol. 2020;45(8):100618. doi: 10.1016/jcpcardiol2020100618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuba K, Imai Y, Penninger JM.. Angiotensin-converting enzyme 2 in lung diseases. Curr Opin Pharmacol. 2006;6(3):271–276. doi: 10.1016/jcoph200603001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sims AC, Baric RS, Yount B, et al. . Severe acute respiratory syndrome coronavirus infection of human ciliated airway epithelia: role of ciliated cells in viral spread in the conducting airways of the lungs. J Virol. 2005;79(24):15511. doi: 10.1128/JVI792415511-1515524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hoffmann M, Kleine-Weber H, Schroeder S, et al. . SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280. doi: 10.1016/jcell202002052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Babcock GJ, Esshaki DJ, Thomas WD, Jr., et al. . Amino acids 270 to 510 of the severe acute respiratory syndrome coronavirus spike protein are required for interaction with receptor. J Virol. 2004;78(9):4552–4560. doi: 10.1128/jvi7894552-456020042020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen Y, Guo Y, Pan Y, et al. . Structure analysis of the receptor binding of 2019-nCoV. Biochem Biophys Res Commun. 2020;525(1):135–140. 2020. doi: 10.1016/jbbrc202002071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wrapp D, Wang N, Corbett KS, et al. . Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–1263. doi: 10.1126/scienceabb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Coutard B, Valle C, de Lamballerie X, et al. . The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antiviral Res. 2020;176:104742. doi: 10.1016/jantiviral2020104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perlman S, Dandekar AA.. Immunopathogenesis of coronavirus infections: implications for SARS. Nat Rev Immunol. 2005;5(12):917–927. doi: 10.1038/nri1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mehta P, McAuley DF, Brown M, et al. . COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395(10229):1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao J, Van Rooijen N, et al. . Evasion by stealth: inefficient immune activation underlies poor T cell response and severe disease in SARS-CoV-infected mice. PLoS Pathog 2009;5(10):e1000636. doi: 10.1371/journalppat1000636 [DOI] [PMC free article] [PubMed]
- 45.Feldmann M, Maini RN, Woody JN, et al. . Trials of anti-tumour necrosis factor therapy for COVID-19 are urgently needed. Lancet. 2020;395(10234):1407–1409. doi: 10.1016/S0140-6736(20)30858-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cifaldi L, Prencipe G, Caiello I, et al. . Inhibition of natural killer cell cytotoxicity by interleukin-6: implications for the pathogenesis of macrophage activation syndrome. Arthritis Rheumatol 2015;67(11):3037–3046. doi: 10.1002/art39295 [DOI] [PubMed]
- 47.Zheng M, Gao Y, Wang G, et al. . Functional exhaustion of antiviral lymphocytes in COVID-19 patients. Cell Mol Immunol. 2020;17(5):533–535. doi: 10.1038/s41423-020-0402-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weber AG, Chau AS, Egeblad M, et al. . Nebulized in-line endotracheal dornase alfa and albuterol administered to mechanically ventilated COVID-19 patients: a case series. medRxiv. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu F, Xu A, Zhang Y, et al. . Patients of COVID-19 may benefit from sustained Lopinavir-combined regimen and the increase of Eosinophil may predict the outcome of COVID-19 progression. Int J Infect Dis. 2020;95:183–191. doi: 10.1016/j.ijid.2020.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kritas SK, Ronconi G, Caraffa A, et al. . Mast cells contribute to coronavirus-induced inflammation: new anti-inflammatory strategy. J Biol Regul Homeost Agents 2020;34(1):9–14. doi: 10.23812/20-Editorial-Kritas [DOI] [PubMed]
- 51.Ronconi G, Teté G, Kritas SK, et al. . SARS-CoV-2, which induces COVID-19, causes kawasaki-like disease in children: role of pro-inflammatory and anti-inflammatory cytokines. J Biol Regul Homeost Agents 2020;34(3):767–773. doi: 10.23812/EDITORIAL-RONCONI-E-59 [DOI] [PubMed]
- 52.Malone RW, Tisdall P, Fremont-Smith P, et al. . COVID-19: Famotidine, Histamine, Mast Cells, and Mechanisms. Research Square. 2020:rs.3.rs-30934. doi: 10.21203/rs.3.rs-30934/v2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gigante A, Aquili A, Farinelli L, et al. . Sodium chromo-glycate and palmitoylethanolamide: a possible strategy to treat mast cell-induced lung inflammation in COVID-19. Med Hypotheses. 2020;143:109856. doi: 10.1016/j.mehy.2020.109856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Theoharides TC. COVID-19, pulmonary mast cells, cytokine storms, and beneficial actions of luteolin. Biofactors. 2020;46(3):306–308. doi: 10.1002/biof.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Duan K, Liu B, Li C, et al. . Effectiveness of convalescent plasma therapy in severe COVID-19 patients. Proc Natl Acad Sci USA. 2020;117(17):9490–9496. doi: 10.1073/pnas2004168117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Felsenstein S, Herbert JA, McNamara PS, et al. . COVID-19: immunology and treatment options. Clin Immunol. 2020;215:108448. doi: 10.1016/j.clim.2020.108448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Versteeg GA, Bredenbeek PJ, van den Worm SHE, et al. . Group 2 coronaviruses prevent immediate early interferon induction by protection of viral RNA from host cell recognition. Virology. 2007;361(1):18–26. doi: 10.1016/j.virol.2007.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Narayanan K, Huang C, Lokugamage K, et al. . Severe acute respiratory syndrome coronavirus nsp1 suppresses host gene expression, including that of type I interferon, in infected cells. J Virol. 2008;82(9):4471–4479. doi: 10.1128/JVI.02472-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kindler E, Thiel V, Weber F.. Interaction of SARS and MERS coronaviruses with the antiviral interferon response. Adv Virus Res. 2016;96:219–243. doi: 10.1016/bs.aivir.2016.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sun L, Xing Y, Chen X, et al. . Coronavirus papain-like proteases negatively regulate antiviral innate immune response through disruption of STING-mediated signaling. PLoS One. 2012;7(2):e30802. doi: 10.1371/journal.pone.0030802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Siu KL, Chan CP, Kok KH, et al. . Suppression of innate antiviral response by severe acute respiratory syndrome coronavirus M protein is mediated through the first transmembrane domain. Cell Mol Immunol. 2014;11(2):141–149. doi: 10.1038/cmi.2013.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cheung CY, Poon LL, Ng IH, et al. . Cytokine responses in severe acute respiratory syndrome coronavirus-infected macrophages in vitro: possible relevance to pathogenesis. J Virol. 2005;79(12):7819–7826. doi: 10.1128/JVI.79.12.7819-7826.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Channappanavar R, Perlman S.. Pathogenic human coronavirus infections: causes and consequences of cytokine storm and immunopathology. Semin Immunopathol. 2017;39(5):529–539. doi: 10.1007/s00281-017-0629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Blanco-Melo D, Nilsson-Payant BE, Liu W-C, et al. . Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181(5):1036–1045.e9. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hadjadj J, Yatim N, Barnabei L, et al. Impaired type I interferon activity and exacerbated inflammatory responses in severe COVID-19 patients. Science 2020;369(6504):718–724. doi: 10.1126/scienceabc6027 [DOI] [PMC free article] [PubMed]
- 66.Lucas C, Wong P, Klein J, et al. . Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature. 2020;584(7821):463–469. doi: 10.1038/s41586-020-2588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhou F, Yu T, Du R, et al. . Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395(10229):1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rayamajhi M, Zhang Y, Miao EA.. Detection of pyroptosis by measuring released lactate dehydrogenase activity. Methods Mol Biol. 2013;1040:85–90. doi: 10.1007/978-1-62703-523-1_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xu Z, Shi L, Wang Y, et al. . Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med. 2020;8(4):420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tian S, Hu W, Niu L, et al. . Pulmonary pathology of early-phase 2019 novel coronavirus (COVID-19) pneumonia in two patients with lung cancer. J Thorac Oncol. 2020;15(5):700–704. doi: 10.1016/j.jtho.2020.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Risitano AM, Mastellos DC, Huber-Lang M, et al. . Complement as a target in COVID-19? Nat Rev Immunol. 2020;20(6):343–344. doi: 10.1038/s41577-020-0320-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cugno M, Meroni PL, Gualtierotti R, et al. . Complement activation in patients with COVID-19: a novel therapeutic target. J Allergy Clin Immunol. 2020;146(1):215–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Guo C, Li B, Ma H, et al. . Tocilizumab treatment in severe COVID-19 patients attenuates the inflammatory storm incited by monocyte centric immune interactions revealed by single-cell analysis. bioRxiv. 2020. doi: 10.1101/2020.04.08.029769 [DOI] [Google Scholar]
- 74.Llitjos J-F, Leclerc M, Chochois C, et al. . High incidence of venous thromboembolic events in anticoagulated severe COVID-19 patients. J Thromb Haemost. 2020;18(7):1743–1746. doi: 10.1111/jth.14869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Helms J, Kremer S, Merdji H, et al. . Neurologic features in severe SARS-CoV-2 infection. N Engl J Med. 2020;382(23):2268–2270. doi: 10.1056/NEJMc2008597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Magro C, Mulvey JJ, Berlin D, et al. . Complement associated microvascular injury and thrombosis in the pathogenesis of severe COVID-19 infection: a report of five cases. Transl Res. 2020;220:1–13. doi: 10.1016/j.trsl.2020.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Diao B, Wang C, Wang R, et al. . Human kidney is a target for novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection. medRxiv. 2020. doi:10.1101/2020.03.04.20031120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.de Marcken M, Dhaliwal K, Danielsen AC, et al. . TLR7 and TLR8 activate distinct pathways in monocytes during RNA virus infection. Sci Signal. 2019;12(605):eaaw1347. doi: 10.1126/scisignal.aaw1347. [DOI] [PubMed] [Google Scholar]
- 79.Liao M, Liu Y, Yuan J, et al. . The landscape of lung bronchoalveolar immune cells in COVID-19 revealed by single-cell RNA sequencing. Nature Medicine 2020;26:842–844. [DOI] [PubMed] [Google Scholar]
- 80.Wen W, Su W, Tang H, et al. . Immune cell profiling of COVID-19 patients in the recovery stageby single-cell sequencing. Cell Discov. 2020;6(1):31. doi: 10.1038/s41421-020-0168-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ong EZ, Chan YFZ, Leong WY, et al. . A dynamic immune response shapes COVID-19 progression. Cell Host Microbe. 2020;27(6):879–882.e2. doi: 10.1016/j.chom.2020.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chua RL, Lukassen S, Trump S, et al. COVID-19 severity correlates with airway epithelium-immune cell interactions identified by single-cell analysis. Nat Biotechnol. 2020;38(8):970–979. doi: 10.1038/s41587-020-0602-4. [DOI] [PubMed] [Google Scholar]
- 83.Giamarellos-Bourboulis EJ, Netea MG, Rovina N, et al. . Complex immune dysregulation in COVID-19 patients with severe respiratory failure. Cell Host Microbe. 2020;27(6):992–1000.e3. doi: 10.1016/j.chom.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tanaka T, Narazaki M, Kishimoto T.. Immunotherapeutic implications of IL-6 blockade for cytokine storm. Immunotherapy 2016;8(8):959–970. doi: 10.2217/imt-2016-0020 [DOI] [PubMed]
- 85.Chen IY, Moriyama M, Chang MF, et al. . Severe acute respiratory syndrome coronavirus viroporin 3a activates the NLRP3 inflammasome. Front Microbiol 2019;10:50 doi: 10.3389/fmicb201900050 [DOI] [PMC free article] [PubMed]
- 86.Huang L, Shi Y, Gong B, et al. . Blood single cell immune profiling reveals the interferon-MAPK pathway mediated adaptive immune response for COVID-19 . medRxiv. 2020. doi: 10.1101/2020.03.15.20033472. [DOI] [Google Scholar]
- 87.Stone JH, Frigault MJ, Serling-Boyd NJ, et al. . Efficacy of tocilizumab in patients hospitalized with COVID-19. N Engl J Med. 2020;383(24):2333–2344. doi: 10.1056/NEJMoa2028836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lee JS, Park S, Jeong HW, et al. . Immunophenotyping of COVID-19 and influenza highlights the role of type I interferons in development of severe COVID-19. Sci Immunol. 2020;5(49):eabd1554. doi: 10.1126/sciimmunol.abd1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bao L, Deng W, Gao H, et al. . Reinfection could not occur in SARS-CoV-2 infected rhesus macaques . bioRxiv. 2020. doi: 10.1101/2020.03.13.990226. [DOI] [Google Scholar]
- 90.Chen y, Feng Z, Diao B, et al. The novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) directly decimates human spleens and lymph nodes. medRxiv 2020. doi: 10.1101/2020.03.27.20045427. [DOI]
- 91.Peebles RS, Jr., Graham BS.. Viruses, dendritic cells and the lung. Respir Res. 2001;2(4):245–249. doi: 10.1186/rr63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Spiegel M, Schneider K, Weber F, et al. . Interaction of severe acute respiratory syndrome-associated coronavirus with dendritic cells. J Gen Virol 2006;87(Pt 7):1953–1960. doi: 10.1099/vir081624-0. [DOI] [PubMed]
- 93.Taghizadeh-Hesary F, Akbari H.. The powerful immune system against powerful COVID-19: a hypothesis. Med Hypotheses. 2020;140:109762. doi: 10.1016/j.mehy.2020.109762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Law HKW, Cheung CY, Sia SF, et al. . Toll-like receptors, chemokine receptors and death receptor ligands responses in SARS coronavirus infected human monocyte derived dendritic cells. BMC Immunol. 2009;10:35. doi: 10.1186/1471-2172-10-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lau YL, Peiris J, Law HKW, et al. . Role of dendritic cells in SARS coronavirus infection. Hong Kong Med J 2012;18(Suppl 3):28–30. [PubMed]
- 96.Tseng CT, Perrone LF-ZH, Zhu H, et al. . Severe acute respiratory syndrome and the innate immune responses: modulation of effector cell function without productive infection. J Immunol 2005;174(12):7977–7985. doi: 10.4049/jimmunol174127977. [DOI] [PubMed]
- 97.Chu H, Zhou J, Wong BH, et al. . Productive replication of Middle East respiratory syndrome coronavirus in monocyte-derived dendritic cells modulates innate immune response. Virology 2014;454-455:197–205. doi: 10.1016/jvirol201402018. [DOI] [PMC free article] [PubMed]
- 98.Wilk A, Rustagi A, Zhao N, et al. A single-cell atlas of the peripheral immune response to severe COVID-19. Nature Medicine 2020;26(7):1070–1076. [DOI] [PMC free article] [PubMed]
- 99.Arunachalam PS, Wimmers F, Mok CKP, et al. . Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans. Science. 2020;369(6508):1210–1220. doi: 10.1126/science.abc6261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Brufsky A, Marti JLG, Nasrazadani A, et al. . Boning up: amino-bisphophonates as immunostimulants and endosomal disruptors of dendritic cell in SARS-CoV-2 infection. J Transl Med. 2020;18(1):261. doi: 10.1186/s12967-020-02433-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhou Z, Ren L, Zhang L, et al. Heightened Innate Immune Responses in the Respiratory Tract of COVID-19 Patients. Cell Host Microbe. 2020;27(6):883–890e2. doi: 10.1016/jchom202004017. [DOI] [PMC free article] [PubMed]
- 102.Chen J, Lau YF, Lamirande EW, et al. . Cellular immune responses to severe acute respiratory syndrome coronavirus (SARS-CoV) infection in senescent BALB/c mice: CD4+ T cells are important in control of SARS-CoV infection. J Virol. 2010;84(3):1289–1301. doi: 10.1128/JVI.01281-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Turner BC, Hemmila E, Beauchemin N, et al. . Receptor-dependent coronavirus infection of dendritic cells. J Virol. 2004;78(10):5486–5490. doi: 10.1128/jvi78105486-54902004. [DOI] [PMC free article] [PubMed]
- 104.Jeffers SA, Tusell SF-G-RL, Gillim-Ross L F-HE, et al. . CD209L (L-SIGN) is a receptor for severe acute respiratory syndrome coronavirus. Proc Natl Acad Sci USA. 2004;101(44):15748–15753. doi: 10.1073/pnas0403812101. [DOI] [PMC free article] [PubMed]
- 105.Duev-Cohen A, Bar-On Y, Glasner A, et al. . The human 2B4 and NTB-A receptors bind the influenza viral hemagglutinin and co-stimulate NK cell cytotoxicity. Oncotarget. 2016;7(11):13093–13105. doi: 10.18632/oncotarget.7597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Glasner A, Zurunic A, Meningher T, et al. . Elucidating the mechanisms of influenza virus recognition by Ncr1. PLoS One. 2012;7(5):e36837. doi: 10.1371/journal.pone.0036837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Draghi M, Pashine A, Sanjanwala B, et al. . NKp46 and NKG2D recognition of infected dendritic cells is necessary for NK cell activation in the human response to influenza infection. J Immunol. 2007;178(5):2688–2698. doi: 10.4049/jimmunol.178.5.2688. [DOI] [PubMed] [Google Scholar]
- 108.van Erp EA, van Kampen MR, van Kasteren PB, et al. . Viral infection of human natural killer cells. Viruses. 2019;11(3):243. doi: 10.3390/v11030243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yu L, Tong Y, Shen G, et al. Immunodepletion with hypoxemia: a potential high risk subtype of Coronavirus Disease 2019. medRxiv, 2020. doi: 10.1101/2020.03.03.20030650. [DOI]
- 110.Maucourant C, Filipovic I, Ponzetta A, et al. . Natural killer cell immunotypes related to COVID-19 disease severity. Sci Immunol. 2020;5(50):eabd6832. doi: 10.1126/sciimmunol.abd6832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mao H, Tu W, Gin Q, et al. . Influenza virus directly infects human natural killer cells and induces cell apoptosis. J Virol. 2009;83(18):9215–9222. doi: 10.1128/JVI00805-09. [DOI] [PMC free article] [PubMed]
- 112.Travaglini KJ, Nabhan AN, Penland L, et al. . A molecular cell atlas of the human lung from single cell RNA sequencing. Nature. 2020;587:619–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Rosales C. Neutrophil: a cell with many roles in inflammation or several cell types? Front Physiol. 2018;9:113. doi: 10.3389/fphys.2018.00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Galani IE, Andreakos E.. Neutrophils in Viral Infections: Current Concepts and Caveats. J Leukoc Biol. 2015;98(4):557–564. doi: 10.1189/jlb4VMR1114-555R. [DOI] [PubMed] [Google Scholar]
- 115.Papayannopoulos V. Neutrophil extracellular traps in immunity and disease. Nat Rev Immunol. 2018;18(2):134–147. doi: 10.1038/nri2017105. [DOI] [PubMed] [Google Scholar]
- 116.Laridan E, Martinod K, De Meyer SF.. Neutrophil extracellular traps in arterial and venous thrombosis. Semin Thromb Hemost. 2019;45(1):86–93. doi: 10.1055/s-0038-1677040. [DOI] [PubMed] [Google Scholar]
- 117.Zuo Y, Zuo M, Yalavarthi S, et al. . Neutrophil Extracellular Traps and Thrombosis in COVID-19. medRxiv. 2020. doi: 10.1101/2020.04.30.20086736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zhang B, Zhou X, Qiu Y, et al. . Clinical characteristics of 82 cases of death from COVID-19. PLoS One. 2020;15(7):e0235458. doi: 10.1371/journalpone0235458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wang Z, Wang Z, Xiong G.. Clinical characteristics and laboratory results of pregnant women with COVID-19 in Wuhan. Int J Gynecol Obstet. 2020;150(3):312–317. doi: 10.1002/ijgo.13265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Mathew D, Giles JR, Baxter AE, et al. . Deep immune profiling of COVID-19 patients reveals distinct immunotypes with therapeutic implications. Science. 2020;369(6508):eabc8511. doi: 10.1126/science.abc8511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Tomar B, Anders H-J, Desai J, et al. . Neutrophils and neutrophil extracellular traps drive necroinflammation in COVID-19. Cells. 2020;9(6):1383. doi: 10.3390/cells9061383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Mahallawi WH, Khabour OF, Zhang Q, et al. . MERS-CoV infection in humans is associated with a pro-inflammatory Th1 and Th17 cytokine profile. Cytokine. 2018;104:8–13. doi: 10.1016/j.cyto.2018.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Shi H, Zuo Y, Yalavarthi S, et al. . Neutrophil Calprotectin Identifies Severe Pulmonary Disease in COVID-19. medRxiv. 2020. doi: 10.1002/JLB.3COVCRA0720-359R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Yaqinuddin A, Kashir J. Novel therapeutic targets for SARS-CoV-2-induced acute lung injury: targeting a potential IL-1н╡/neutrophil extracellular traps feedback loop. Med Hypotheses 2020;143:109906. doi: 10.1016/jmehy2020109906. [DOI] [PMC free article] [PubMed]
- 125.Veras FP, Pontelli MC, Silva CM, et al. . SARS-CoV-2-triggered neutrophil extracellular traps mediate COVID-19 pathology. J Exp Med. 2020;217(12). doi: 10.1084/jem.20201129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Thierry AR. Does the newly observed inflammatory syndrome in children demonstrate a link between uncontrolled neutrophil extracellular traps formation and COVID-19? Pediatr Res. 2020. doi: 10.1038/s41390-020-0996-1. [DOI] [PubMed]
- 127.Siracusa MC, Kim BS, Spergel JM, et al. . Basophils and allergic inflammation. J Allergy Clin Immunol. 2013;132(4):789–788. doi: 10.1016/j.jaci.2013.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Chirumbolo S. State-of-the-art review about basophil research in immunology and allergy: is the time right to treat these cells with the respect they deserve? Blood Transfus. 2012;10(2):148–164. doi: 10.2450/2011.0020-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Clementsen P, Jensen CB ,Jarløv , JO, et al. . Virus enhances histamine release from human basophils. Agents Actions 1988;23(3–4):165–167. doi: 10.1007/BF02142529. [DOI] [PubMed]
- 130.Jiang A-P, Jiang J-F, Guo M-G, et al. . Human blood-circulating basophils capture HIV-1 and Mediate Viral trans-infection of CD4+ T cells. J Virol. 2015;89(15):8050–8062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Qin C, Zhou L, Hu Z, et al. . Dysregulation of immune response in patients with coronavirus 2019 (COVID-19) in Wuhan, China. Clin Infect Dis. 2020;71(15):762–768. doi: 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lindsley AW, Schwartz JT, Rothenberg ME.. Eosinophil responses during COVID-19 infections and coronavirus vaccination. J Allergy Clin Immunol. 2020;146(1):1–7. doi: 10.1016/j.jaci.2020.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Bao J, Li C, Zhang K, et al. . Comparative analysis of laboratory indexes of severe and non-severe patients infected with COVID-19. Clin Chim Acta. 2020;509:180–194. doi: 10.1016/j.cca.2020.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lu G, Wang J.. Dynamic changes in routine blood parameters of a severe COVID-19 case. Clinica Chimica Acta 2020;508. doi: 10.1016/j.cca.2020.04.034. [DOI] [PMC free article] [PubMed]
- 135.Chen R, Sang L, Jiang M, et al. . Longitudinal hematologic and immunologic variations associated with the progression of COVID-19 patients in China. J Allergy Clin Immunol. 2020;146(1):89–100. doi: 10.1016/j.jaci.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Liu S, Zhi YA-O, Ying S.. COVID-19 and asthma: reflection during the pandemic. Clinical Reviews in Allergy & Immunology 2020;59(1):78–88. [DOI] [PMC free article] [PubMed]
- 137.Krystel-Whittemore M, Dileepan KN, Wood JG.. Mast cell: a multi-functional master cell. Frontiers in immunology 2016;6(620). [DOI] [PMC free article] [PubMed]
- 138.Hu Y, Jin Y, Han D, et al. . Mast cell-induced lung injury in mice infected with H5N1 influenza virus. J Virol. 2012;86:3347–3356. [DOI] [PMC free article] [PubMed]
- 139.Marshall JS, Portales-Cervantes L, Leong E.. Mast cell responses to viruses and pathogen products. IJMS. 2019;20(17):4241. doi: 10.3390/ijms20174241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ramos L, Peña G, Cai B, et al. . Mast cell stabilization improves survival by preventing apoptosis in sepsis. J Immunol. 2010;185(1):709–716. doi: 10.4049/jimmunol.1000273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Sestili P, Stocchi V.. Repositioning chromones for early anti-inflammatory treatment of COVID-19. Front Pharmacol. 2020;11:854. doi: 10.3389/fphar.2020.00854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Veerappan A, Reid AC, Estephan R, et al. . Mast cell renin and a local renin-angiotensin system in the airway: role in bronchoconstriction. Proceedings of the National Academy of Sciences 2008;105(4):1315–1320. [DOI] [PMC free article] [PubMed]
- 143.Caughey GH, Raymond W, Wolters PJ, et al. . Angiotensin II generation by mast cell alpha- and beta-chymases. Biochim Biophys Acta 2000;1480(1–2):245–257. doi: 10.1016/s0167-4838(00)00076-5. [DOI] [PubMed]
- 144.Miller HR, Pemberton AD.. Tissue-specific expression of mast cell granule serine proteinases and their role in inflammation in the lung and gut. Immunology 2002;105(4):375–390. doi: 10.1046/j1365-2567200201375x. [DOI] [PMC free article] [PubMed]
- 145.Cui W, Fan Y, Wu W, et al. . Expression of lymphocytes and lymphocyte subsets in patients with severe acute respiratory syndrome. Clin Infect Dis. 2003;37(6):857–859. doi: 10.1086/378587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Diao B, Wang C, Tan Y.. Reduction and functional exhaustion of T cells in patients with Coronavirus Disease 2019 (COVID-19). Front Immunol. 2020;11(827). doi: 10.3389/fimmu202000827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Satija N, Lal SK.. The molecular biology of SARS coronavirus. Ann N Y Acad Sci. 2007;1102:26–38. doi: 10.1196/annals.1408.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Shiow LR, Rosen DB, Brdickova N, et al. . CD69 acts downstream of interferon-alpha/beta to inhibit S1P1 and lymphocyte egress from lymphoid organs. Nature. 2006;440(7083):540–544. doi: 10.1038/nature04606. [DOI] [PubMed] [Google Scholar]
- 149.Campbell KM, Steiner G, Wells DK, et al. . Prediction of SARS-CoV-2 Epitopes across 9360 HLA Class I Alleles. bioRxiv. 2020. doi: 10.3389/fimmu202000827. [DOI] [Google Scholar]
- 150.Li CK-f, Wu H, Yan H, et al. . T cell responses to whole SARS coronavirus in humans. J Immunol. 2008;181(8):5490–5500. doi: 10.4049/jimmunol.181.8.5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Weiskopf D, Schmitz KS, Raadsen MP, et al. . Phenotype of SARS-CoV-2-specific T-cells in COVID-19 patients with acute respiratory distress syndrome. Sci Immunol. 2020;5(48):eabd2071. doi: 10.1126/sciimmunol.abd2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Mateus J, Grifoni A, Tarke A, et al. . Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans. Science. 2020;370(6512):89–94. doi: 10.1126/science.abd3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Poston D, Weisblum Y, Wise H, et al. . Absence of SARS-CoV-2 neutralizing activity in pre-pandemic sera from individuals with recent seasonal coronavirus infection. medRxiv. 2020. doi: 10.1101/2020.10.08.20209650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Anderson EM, Goodwin EC, Verma A, et al. . Seasonal human coronavirus antibodies are boosted upon SARS-CoV-2 infection but not associated with protection : medRxiv. 2020. doi: 10.1101/2020.11.06.20227215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Edridge AWD, Kaczorowska JM, Hoste ACR, et al. . Coronavirus protective immunity is short-lasting Nature Medicine 2020;26(11):1691–1693. [DOI] [PubMed] [Google Scholar]
- 156.Zhou Y, Fu B, Zheng X, et al. . Pathogenic T cells and inflammatory monocytes incite inflammatory storm in severe COVID-19 patients. Natl Sci Rev. 2020;7(6):998–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Huang H, Wang S, Jiang T, et al. High levels of circulating GM-CSF + CD4+ T cells are predictive of poor outcomes in sepsis patients: a prospective cohort study. Cell Mol Immunol. 2019;16(6):602–610. doi: 10.1038/s41423-018-0164-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Chen G, Wu D, Guo W, et al. . Clinical and immunological features of severe and moderate Coronavirus Disease 2019. J Clin Invest. 2020;130(5):2620–2629. doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Zheng J, Liu Y, Lau Y-L, et al. . нЁн╢-T cells: an Unpolished sword in human anti-infection immunity. Cell Mol Immunol 2013;10(1):50–57. doi: 10.1038/cmi201243. [DOI] [PMC free article] [PubMed]
- 160.Zhou Y, Fu B, Zheng X, et al. . Aberrant pathogenic GM-CSF + T cells and inflammatory CD14 + CD16+. Monocytes in severe pulmonary syndrome patients of a new coronavirus. National Science Review 2020. doi: 10.1093/nsr/nwaa041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Walter JM, Helmin KA, Abdala-Valencia H, et al. . Multidimensional assessment of alveolar T cells in critically ill patients. JCI Insight 2018;3(17):e123287. doi: 10.1172/jciinsight123287. [DOI] [PMC free article] [PubMed]
- 162.Dong C, Ni L, Ye F, et al. . Characterization of anti-viral immunity in recovered individuals infected by SARS-CoV-2. medRxiv. 2020. doi: 10.1101/2020.03.17.20036640. [DOI] [Google Scholar]
- 163.Lei L, Qian H, Yang X.et al. The phenotypic changes of γδ T cells in COVID-19 patients. J Cel Mol Med. 2020;24(19):11603–11606. [DOI] [PMC free article] [PubMed]
- 164.Kaneko N, Kuo HH, Boucau J, et al. . Loss of Bcl-6-expressing T follicular helper cells and germinal centers in COVID-19. Cell. 2020;183(1):143–157. doi: 10.1016/jcell202008025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Jouan Y, Guillon A, Gonzalez L, et al. . Functional alteration of innate T cells in critically ill COVID-19 patients . medRxiv. 2020. doi: 10.1101/2020.05.03.20089300. [DOI] [Google Scholar]
- 166.Libraty DH, O’Neil KM, Baker LM, et al. . Human CD4(+) memory T-lymphocyte responses to SARS coronavirus infection. Virol. 2007;368(2):317–321. doi: 10.1016/jvirol200707015. [DOI] [PMC free article] [PubMed]
- 167.Fan Y-Y, Huang Z-T, Li L, et al. . Characterization of SARS-CoV-specific memory T cells from recovered individuals 4 years after infection. Arch Virol. 2009;154(7):1093–1099. doi: 10.1007/s00705-009-0409-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Tang F, Quan Y, Xin Z-T, et al. . Lack of peripheral memory B cell responses in recovered patients with severe acute respiratory syndrome: a six-year follow-up study. J Immunol. 2011;186(12):7264–7268. doi: 10.4049/jimmunol.0903490. [DOI] [PubMed] [Google Scholar]
- 169.Ng OW, Chia A, Tan AT, et al. . Memory T cell responses targeting the SARS coronavirus persist up to 11 years post-infection.Vaccine 2016;34(17):2008–2014. doi: 10.1016/jvaccine201602063. [DOI] [PMC free article] [PubMed]
- 170.Dan JM, Mateus J, Kato Y, et al. . Immunological memory to SARS-CoV-2 assessed for greater than six months after infection. Science 2020;371(6529):eabf4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Hsueh PR, Huang L, Chen PJ, et al. . Chronological evolution of IgM, IgA, IgG and neutralisation antibodies after infection with SARS-associated coronavirus. Clin Microbiol Infect. 2004;10(12):1062–1066. doi: 10.1111/j1469-0691200401009x. [DOI] [PMC free article] [PubMed]
- 172.Haveri A, Smura T, Kuivanen S, et al. . Serological and molecular findings during SARS-CoV-2 infection: the first case study in Finland, January to February 2020. Euro Surveillance 2020;25(11):2000266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.To KK-W, Tsang OT-Y, Leung W-S, et al. . Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis. 2020;20(5):565–574. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Ju B, Zhang Q, Ge X, et al. Potent human neutralizing antibodies elicited by SARS-CoV-2 infection. Nat. 2020;584(7819):115–119. [DOI] [PubMed]
- 175.Zhang L, Zhang F, Yu W, et al. . Antibody responses against SARS coronavirus are correlated with disease outcome of infected individuals. J Med Virol. 2006;78(1):1–8. doi: 10.1002/jmv20499. [DOI] [PMC free article] [PubMed]
- 176.Cao W, Liu W, Zhang P-H, et al. . Disappearance of antibodies to SARS-associated coronavirus after recovery. N Engl J Med. 2007;357(11):1162–1163. doi: 10.1056/NEJMc070348. [DOI] [PubMed]
- 177.Liu W, Fontanet A, Zhang P-H, et al. . Two-year prospective study of the humoral immune response of patients with severe acute respiratory syndrome. J Infect Dis. 2006;193(6):792–795. doi: 10.1086/500469. [DOI] [PMC free article] [PubMed]
- 178.Traggiai E, Becker S, Subbarao K, et al. . An efficient method to make human monoclonal antibodies from memory B cells: potent neutralization of SARS coronavirus. Nature Medicine 2004;10(8):871–875. [DOI] [PMC free article] [PubMed]
- 179.Payne DC, Iblan I, Rha B, et al. . Persistence of antibodies against middle east respiratory syndrome coronavirus. Emerg Infect Dis. 2016;22(10):1824–1826. doi: 10.3201/eid2210.160706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Thevarajan I, Nguyen THO, Koutsakos M, et al. Breadth of concomitant immune responses prior to patient recovery: a case report of non-severe COVID-19. Nat Med. 2020;26(4):453–455. [DOI] [PMC free article] [PubMed]
- 181.Addetia A, Crawford KH, Dingens A, et al. . Neutralizing Antibodies Correlate with Protection from SARS-CoV-2 in Humans during a Fishery Vessel Outbreak with High Attack Rate. Virol. 2020. doi: 10.1128/JCM.02107-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Gudbjartsson DF, Norddahl GL, Melsted P, et al. Humoral Immune Response to SARS-CoV-2 in Iceland. N Engl J Med. 2020;383(18):1724–1734. doi: 10.1056/NEJMoa2026116. [DOI] [PMC free article] [PubMed]
- 183.Seow J, Graham C, Merrick B, et al. Longitudinal evaluation and decline of antibody responses in SARS-CoV-2 infection. Nat Microbiol. 2020. doi: 10.1038/s41564-020-00813-8. [DOI] [PMC free article] [PubMed]
- 184.To KK, Hung IF, Ip JD, et al. COVID-19 re-infection by a phylogenetically distinct SARS-coronavirus-2 strain confirmed by whole genome sequencing. Clin Infect Dis. 2020:ciaa1275. doi: 10.1093/cid/ciaa1275. [DOI] [PMC free article] [PubMed]
- 185.Wajnberg A, Amanat F, Firpo A, et al. . Robust neutralizing antibodies to SARS-CoV-2 infection persist for months. Science. 2020;370(6521):1227–1230. doi: 10.1126/science.abd7728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Taylor A, Foo S-S, Bruzzone R, et al. . Fc receptors in antibody-dependent enhancement of viral infections. Immunol Rev. 2015;268(1):340–364. doi: 10.1111/imr.12367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Wan Y, Shang J, Sun S, et al. Molecular mechanism for antibody-dependent enhancement of coronavirus entry. Virol. 2020;94(5):e02015–19. [DOI] [PMC free article] [PubMed]
- 188.Liu L, Wei Q, Lin Q, et al. Anti-spike IgG causes severe acute lung injury by skewing macrophage responses during acute SARS-CoV infection. JCI Insight. 2019;4(4):e123158. doi: 10.1172/jciinsight123158. [DOI] [PMC free article] [PubMed]
- 189.Yip M, Leung NHL, Leung N, et al. Antibody-dependent infection of human macrophages by severe acute respiratory syndrome coronavirus. 2014. [DOI] [PMC free article] [PubMed]
- 190.Quinlan BD, Mou H, Zhang L, et al. . The SARS-CoV-2 receptor-binding domain elicits a potent neutralizing response without antibody-dependent enhancement. bioRxiv. 2020. doi: 10.1101/2020.04.10.036418. [DOI] [Google Scholar]
- 191.Chen J, Kelley WJ, Goldstein DR.. Role of aging and the immune response to respiratory viral infections: potential implications for COVID-19. JI. 2020;205(2):313–320. doi: 10.4049/jimmunol.2000380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Zhao J, Zhao J, Legge K, et al. . Age-related increases in PGD(2) expression impair respiratory DC migration, resulting in diminished T cell responses upon respiratory virus infection in mice. J Clin Invest. 2011;121(12):4921–4930. doi: 10.1172/JCI59777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Nikolich-Zugich J, Knox KS, Rios CT, et al. . SARS-CoV-2 and COVID-19 in older adults: what we may expect regarding pathogenesis, immune responses, and outcomes. Geroscience. 2020;42(2):505–514. doi: 10.1007/s11357-020-00186-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Lingappan K, Karmouty-Quintana H, Davies J, et al. . Understanding the age divide in COVID-19: why are children overwhelmingly spared? Am J Physiol Lung Cell Mol Physiol. 2020;319(1):L39–L44. doi: 10.1152/ajplung.00183.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Dong Y, Mo X, Hu Y, et al. . Epidemiological characteristics of 2143 pediatric patients with 2019 Coronavirus Disease in China. Pediatrics. 2020;145(6):e20200702. doi: 10.1542/peds.2020-0702. [DOI] [PubMed] [Google Scholar]
- 196.Jing Q-L, Liu M-J, Yuan J, et al. . Household secondary attack rate of COVID-19 and associated determinants . medRxiv. 2020. doi: 10.1016/S1473-3099(20)30471-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Yonker LM, Neilan AM, Bartsch Y, et al. . Pediatric SARS-CoV-2: clinical presentation, infectivity, and immune responses. J Pediatr. 2020;227:45–52.e5. doi: 10.1016/j.jpeds.2020.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Chen C, Qi F, Shi K, et al. . Thalidomide combined with low-dose short-term glucocorticoid in the treatment of critical Coronavirus Disease 2019. Clin Transl Med. 2020;10(2):e35. doi: 10.1002/ctm2352020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Runfeng L, Yunlong H, Jicheng H, et al. . Lianhuaqingwen exerts anti-viral and anti-inflammatory activity against novel coronavirus (SARS-CoV-2). Pharmacol Res. 2020;156:104761. doi: 10.1016/jphrs2020104761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Humanigen. Humanigen Partners with CTI, a Leading Contract Research Organization, for Planned Phase III Study for Lenzilumab for Coronavirus Treatment. Burlingame, CA and Covington, KY: Humanigen; 2020. [Google Scholar]
- 201.Sciences R. Roivant announces development of anti-GM- CSF monoclonal antibody to prevent and treat acute respiratory distress syndrome (ARDS) in patients with COVID-19. Roivant Sciences 2020.
- 202.Bioscience I. Initiation of two- centre compassionate use study involving namilumab in the treatment of individual patients with rapidly worsening COVID-19 infection in Italy. Izana Bioscience 2020.
- 203.Cortegiani A, Ingoglia G, Ippolito M, et al. . A systematic review on the efficacy and safety of chloroquine for the treatment of COVID-19. J Crit Care. 2020;57:279–283. doi: 10.1016/jjcrc202003005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Ben-Zvi I, Kivity S, Langevitz P, et al. . Hydroxychloroquine: from malaria to autoimmunity. Clin Rev Allergy Immunol. 2012;42(2):145–153. doi: 10.1007/s12016-010-8243-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Yao X, Ye F, Zhang M, et al. . In vitro antiviral activity and projection of optimized dosing design of hydroxychloroquine for the treatment of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Clin Infect Dis. 2020;71(15):732–739. doi: 10.1093/cid/ciaa237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Gautret P, Lagier JC, Parola P, et al. . Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial. Int J Antimicrob Agents. 2020; 56(1):105949. doi: 10.1016/jijantimicag2020105949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Zheng Y, Li R, Liu S.. Immunoregulation with mTOR inhibitors to prevent COVID-19 severity: a novel intervention strategy beyond vaccines and specific antiviral medicines. J Med Virol. 2020;92(9):1495–1500. doi: 10.1002/jmv26009.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Casadevall A, Pirofski L-A.. The convalescent sera option for containing COVID-19. J Clin Invest. 2020;130(4):1545–1548. doi: 10.1172/JCI138003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Cheng Y, Wong R, Soo YO, et al. . Use of convalescent plasma therapy in SARS patients in Hong Kong. Eur J Clin Microbiol Infect Dis. 2005;24(1):44–46. 2005. doi: 10.1007/s10096-004-1271-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Hung IF, To KK, Lee CK, et al. . Convalescent plasma treatment reduced mortality in patients with severe pandemic influenza A (H1N1) 2009 virus infection. Clin Infect Dis. 2011;52(4):447–456. doi: 10.1093/cid/ciq106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Ko JH, Seok H, Cho SY, et al. . Challenges of convalescent plasma infusion therapy in Middle East respiratory coronavirus infection: a single centre experience. Antivir Ther. 2018;23(7):617–622. doi: 10.3851/IMP3243. [DOI] [PubMed] [Google Scholar]
- 212.Zhang B, Liu S, Tan T, et al. . Treatment with convalescent plasma for critically ill patients with severe acute respiratory syndrome coronavirus 2 infection. Chest. 2020;158(1):e9–e13. doi: 10.1016/j.chest.2020.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Shen C, Wang Z, Zhao F, et al. . Treatment of 5 critically ill patients with COVID-19 with convalescent plasma. JAMA. 2020;323(16):1582–1589. 2020. doi: 10.1001/jama20204783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Tanne JH. COVID-19: FDA approves use of convalescent plasma to treat critically ill patients. BMJ. 2020;368:m1256. doi: 10.1136/bmjm1256. [DOI] [PubMed] [Google Scholar]
- 215.Ramamoorthy S, Cidlowski JA.. Corticosteroids: mechanisms of action in health and disease. Rheum Dis Clin North Am. 2016;42(1):15–31. doi: 10.1016/jrdc2015080022016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Timmermans S, Souffriau J, Libert C.. A general introduction to glucocorticoid biology. Front Immunol. 2019;10:1545. doi: 10.3389/fimmu.2019.01545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Arabi YM, Mandourah Y, Al-Hameed F, et al. . Corticosteroid therapy for critically ill patients with middle east respiratory syndrome. Am J Respir Crit Care Med. 2018;197(6):757–767. 2018. doi: 10.1164/rccm201706-1172OC. [DOI] [PubMed] [Google Scholar]
- 218.Lansbury LE, Rodrigo C, Leonardi-Bee J, et al. . Corticosteroids as adjunctive therapy in the treatment of influenza: an updated cochrane systematic review and meta-analysis. Crit Care Med. 2020;48(2):e98–e106. doi: 10.1097/CCM00000000000040932020. [DOI] [PubMed] [Google Scholar]
- 219.Zha L, Li S, Pan L, et al. . Corticosteroid treatment of patients with Coronavirus Disease 2019 (COVID-19). Med J Aust. 2020;212(9):416–420. doi: 10.5694/mja250577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Russell CD, Millar JE, Baillie JK.. Clinical evidence does not support corticosteroid treatment for 2019-nCoV lung injury. Lancet. 2020;395(10223):473–475. doi: 10.1016/S0140-6736(20)30317-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Ledford H. Coronavirus breakthrough: dexamethasone is first drug shown to save lives. Nature. 2020;582(7813):469–469. doi: 10.1038/d41586-020-01824-5. [DOI] [PubMed] [Google Scholar]
- 222.Selvaraj J, Sardar H, Vishnupriya V, et al. . Molecular docking analysis of amyloid precursor protein with compounds from the Australian cowplant. Bioinformation. 2020;16(7):561–566. doi: 10.6026/97320630016561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Wongrakpanich S, Wongrakpanich A, Melhado K, Rangaswami J.. A comprehensive review of non-steroidal anti-inflammatory drug use in the elderly. Aging Dis. 2018;9(1):143–150. doi: 10.14336/AD20170306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Manjili RH, Zarei M, Habibi M, et al. . COVID-19 as an acute inflammatory disease. J Immunol. 2020;205(1):12–19. doi: 10.4049/jimmunol2000413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Zheng W, Fan W, Zhang S, et al. . Naproxen exhibits broad anti-influenza virus activity in mice by impeding viral nucleoprotein nuclear export. Cell Rep. 2019;27(6):1875–1885. doi: 10.1016/jcelrep2019040532019. [DOI] [PubMed] [Google Scholar]
- 226.Kickbusch I, Leung G.. Response to the emerging novel coronavirus outbreak. BMJ. 2020;368:m406. doi: 10.1136/bmj.m406. [DOI] [PubMed] [Google Scholar]
- 227.Amici C, Di CA, Ciucci A, et al. . Indomethacin has a potent antiviral activity against SARS coronavirus. Antivir Ther. 2006;11(8):1021–1030. [PubMed] [Google Scholar]
- 228.Marinella MA. Indomethacin and resveratrol as potential treatment adjuncts for SARS-CoV-2/COVID-19. Int J Clin Pract. 2020;74(9):e13535. doi: 10.1111/ijcp13535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Galeotti C, Kaveri SV, Bayry J.. IVIG-mediated effector functions in autoimmune and inflammatory diseases. Int Immunol. 2017;29(11):491–498. doi: 10.1093/intimm/dxx039. [DOI] [PubMed] [Google Scholar]
- 230.Jolles S, Sewell WA, Misbah SA.. Clinical uses of intravenous immunoglobulin. Clin Exp Immunol. 2005;142(1):1–11. doi: 10.1111/j1365-2249200502834x2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Hartung HP. Advances in the understanding of the mechanism of action of IVIg. J Neurol. 2008;255 (Suppl 3):3–6. doi: 10.1007/s00415-008-3002-02008. [DOI] [PubMed] [Google Scholar]
- 232.Mohtadi N, Ghaysouri A, Shirazi S, et al. . Recovery of severely ill COVID-19 patients by intravenous immunoglobulin (IVIG) treatment: a case series. Virology. 2020;548:1–5. doi: 10.1016/jvirol202005006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Xie Y, Cao S, Dong H, et al. . Effect of regular intravenous immunoglobulin therapy on prognosis of severe pneumonia in patients with COVID-19. J Infect. 2020;81(2):318–356. doi: 10.1016/j.jinf.2020.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Cao W,Liu X,Bai T, et al. . High-dose intravenous immunoglobulin as a therapeutic option for deteriorating patients with Coronavirus Disease 2019. Open Forum Infectious Dis. 2020;7(3). doi: 10.1093/ofid/ofaa102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Graham RL, Donaldson EF, Baric RS.. A decade after SARS: strategies for controlling emerging coronaviruses. Nat Rev Microbiol. 2013;11(12):836–848. doi: 10.1038/nrmicro3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.de Wit E, van Doremalen N, Falzarano D, et al. . SARS and MERS: recent insights into emerging coronaviruses. Nat Rev Microbiol. 2016;14(8):523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Prompetchara E, Ketloy C, Palaga T.. Immune responses in COVID-19 and potential vaccines: lessons learned from SARS and MERS epidemic. Asian Pac J Allergy Immunol. 2020;38(1):1–9. doi: 10.12932/AP-200220-0772. [DOI] [PubMed] [Google Scholar]
- 238.Pfizer . Pfizer and Biontech announce vaccine candidate against COVID-19 achieved success in first interim analysis from phase 3 study. 2020.
- 239.Hindu T. Coronavirus | Pfizer seeks emergency use authorisation for its COVID-19 vaccine in India. 2020.
- 240.Focus R. COVID-19 vaccine tracker. 2020.
- 241.Agencies AJAN. Which countries have rolled out COVID vaccine? 2021.
- 242.BMJ T. COVID-19: Moderna vaccine is nearly 95% effective, trial involving high risk and elderly people shows. BMJ. 2020;371. doi: 10.1136/bmj.m4471. [DOI]
- 243.BBC . COVID-19: Oxford University vaccine is highly effective. 2020.
- 244.Trials C. Russia’s Sputnik V vaccine shows 92% efficacy in COVID-19 trial. 2020.
- 245.Poonawalla A. Coronavirus: India approves vaccines from Bharat Biotech and Oxford/AstraZeneca. BBC News India 2021.
- 246.Tai W, He L, Zhang X, et al. . Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell Mol Immunol. 2020;17(6):613–620. doi: 10.1038/s41423-020-0400-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Berry JD, Hay K, Rini JM, et al. . Neutralizing epitopes of the SARS-CoV S-protein cluster independent of repertoire, antigen structure or mAb technology. MAbs. 2010;2(1):53–66. doi: 10.4161/mabs.2.1.10788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Tian X, Li C, Huang A, et al. . Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg Microbes Infect. 2020;9(1):382–385. doi: 10.1080/22221751.2020.1729069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Chi X, Yan R, Zhang J, et al. . A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science. 2020;369(6504):650–655. doi: 10.1126/scienceabc6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Liu L, Wang P, Nair MS, et al. . Potent neutralizing monoclonal antibodies directed to multiple epitopes on the SARS-CoV-2 spike. Nature. 2020;584:450–456. [DOI] [PubMed] [Google Scholar]
- 251.Mercado NB, Zahn R, Wegmann F, et al. . Single-shot Ad26 vaccine protects against SARS-CoV-2 in rhesus macaques. Nature. 2020;586(7830):583–588. doi: 10.1038/s41586-020-2607-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Baum A, Fulton BO, Wloga E, et al. . Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science. 2020;369(6506):1014–1018. doi: 10.1126/science.abd0831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Fan T, Chen Y, Bai Y, et al. Analysis of medication characteristics of traditional Chinese medicine in treating COVID-19 based on data mining. Zhejiang Da Xue Xue Bao Yi Xue Ban. 2020;49(2):260–269. doi: 10.3785/j.issn.1008-9292.2020.03.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Rastogi S, Pandey DN, Singh RH.. COVID-19 pandemic: a pragmatic plan for Ayurveda Intervention. J Ayurveda Integr Med. 2020. doi:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Yang Y, Islam MS, Wang J, et al. . Traditional Chinese medicine in the treatment of patients infected with 2019-new coronavirus (SARS-CoV-2): a review and perspective. Int J Biol Sci. 2020;16(10):1708–1717. doi: 10.7150/ijbs.45538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Chan K, Zhang H, Lin ZX.. An overview on adverse drug reactions to traditional Chinese medicines. Br J Clin Pharmacol. 2015;80(4):834–843. doi: 10.1111/bcp.12598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Khodadadi S. Role of herbal medicine in boosting immune system. Immunopathol Persa. 2015;1(1):e01. [Google Scholar]
- 258.Singh N, Tma MS.. A review on herbal plants as immunomodulators. Int J Pharm Sci Res. 2016;7(9):3602–3610. [Google Scholar]
- 259.Elsayed Y, Khan NA.. Immunity-boosting spices and the novel coronavirus. ACS Chem Neurosci. 2020;11(12):1696–1698. doi: 10.1021/acschemneuro.0c00239. [DOI] [PubMed] [Google Scholar]
- 260.Yamamoto M, Matsuyama S, Li X, et al. . Identification of nafamostat as a potent inhibitor of middle east respiratory syndrome coronavirus S protein-mediated membrane fusion using the split-protein-based cell-cell fusion assay. Antimicrob Agents Chemother. 2016;60(11):6532–6539. doi: 10.1128/AAC.01043-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Beigel JH, Tomashek KM, Dodd LE, et al. . Remdesivir for the treatment of COVID-19 - preliminary report. N Engl J Med. 2020;383(19):1813–1826. doi: 10.1056/NEJMoa2007764. [DOI] [PubMed] [Google Scholar]
- 262.Hunt A . COVID-19 Update: FDA Broadens Emergency Use Authorization for Veklury (remdesivir) to Include All Hospitalized Patients for Treatment of COVID-19. 2020.
- 263.Wang M, Cao R, Zhang L, et al. . Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30(3):269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Richardson P, Griffin I, Tucker C, et al. . Baricitinib as potential treatment for 2019-nCoV acute respiratory disease. Lancet. 2020;395(10223):e30–e31. doi: 10.1016/S0140-6736(20)30304-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Jin Z, Du X, Xu Y, et al. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582(7811):289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 266.Jin Z, Zhao Y, Sun Y, et al. . Structural basis for the inhibition of SARS-CoV-2 main protease by antineoplastic drug carmofur. Nat Struct Mol Biol. 2020;27(6):529–532. doi: 10.1038/s41594-020-0440-6. [DOI] [PubMed] [Google Scholar]
- 267.Sommer F, Backhed F. The gut microbiota-masters of host development and physiology. Nat Rev Microbiol. 2013;11(4):227–238. doi: 10.1038/nrmicro2974. [DOI] [PubMed] [Google Scholar]
- 268.Rinninella E, Raoul P, Cintoni M, et al. . What is the healthy gut microbiota composition? A changing ecosystem across age, environment, diet, and diseases. Microorganisms. 2019;7(1):14. doi: 10.3390/microorganisms7010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Smith K, McCoy KD, Macpherson AJ.. Use of axenic animals in studying the adaptation of mammals to their commensal intestinal microbiota. Semin Immunol. 2007;19(2):59–69. doi: 10.1016/j.smim.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 270.Macpherson AJ, Harris NL.. Interactions between commensal intestinal bacteria and the immune system. Nat Rev Immunol. 2004;4(6):478–485. doi: 10.1038/nri1373. [DOI] [PubMed] [Google Scholar]
- 271.Mazmanian SK, Liu CH, Tzianabos AO, et al. . An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122(1):107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 272.Eckburg PB, Bik EM, Bernstein CN, et al. . Diversity of the human intestinal microbial flora. Science. 2005;308(5728):1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Wang B, Yao M, Lv L, et al. . The human microbiota in health and disease. Engineering. 2017;3(1):71–82. doi: 10.1016/J.ENG.2017.01.008. [DOI] [Google Scholar]
- 274.Williams NT. Probiotics. Am J Health Syst Pharm. 2010;67(6):449–458. doi: 10.2146/ajhp090168. [DOI] [PubMed] [Google Scholar]
- 275.Tang WH, Kitai T, Hazen SL.. Gut microbiota in cardiovascular health and disease. Circ Res. 2017;120(7):1183–1196. doi: 10.1161/CIRCRESAHA.117.309715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Takeuchi K, Maiden L, Bjarnason I.. Genetic aspects of intestinal permeability in inflammatory bowel disease. Novartis Found Symp. 2004;263:151–158. [PubMed] [Google Scholar]
- 277.Soler AP, Miller RD, Laughlin KV, et al. . Increased tight junctional permeability is associated with the development of colon cancer. Carcinogenesis. 1999;20(8):1425–1431. doi: 10.1093/carcin/20.8.1425. [DOI] [PubMed] [Google Scholar]
- 278.Bjarnason I, Williams P, So A, et al. . Intestinal permeability and inflammation in rheumatoid arthritis: effects of non-steroidal anti-inflammatory drugs. Lancet. 1984;2(8413):1171–1174. doi: 10.1016/S0140-6736(84)92739-9. [DOI] [PubMed] [Google Scholar]
- 279.Zuo T, Zhang F, Lui GCY, et al. . Alterations in gut microbiota of patients with COVID-19 during time of hospitalization. Gastroenterol. 2020;159(3):944–955.e8. doi: 10.1053/j.gastro.2020.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Dhar D, Mohanty A.. Gut microbiota and COVID-19- possible link and implications. Virus Res. 2020;285:198018. doi: 10.1016/j.virusres.2020.198018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Yang T, Chakraborty S, Saha P, et al. . Gnotobiotic rats reveal that gut microbiota regulates colonic mRNA of Ace2, the receptor for SARS-CoV-2 infectivity. Hypertension. 2020;76(1):e1–e3. doi: 10.1161/HYPERTENSIONAHA.120.15360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Verna EC, Lucak S.. Use of probiotics in gastrointestinal disorders: what to recommend? Therap Adv Gastroenterol. 2010;3(5):307–319. doi: 10.1177/1756283X10373814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Diaz de Barboza G, Guizzardi S, Tolosa de Talamoni N.. Molecular aspects of intestinal calcium absorption. World J Gastroenterol. 2015;21(23):7142–7154. doi: 10.3748/wjg.v21.i23.7142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Dar HY, Pal S, Shukla P, et al. . Bacillus clausii inhibits bone loss by skewing Treg-Th17 cell equilibrium in postmenopausal osteoporotic mice model. Nutrition. 2018;54:118–128. doi: 10.1016/j.nut.2018.02.013. [DOI] [PubMed] [Google Scholar]
- 285.Dar HY, Shukla P, Mishra PK, et al. . Lactobacillus acidophilus inhibits bone loss and increases bone heterogeneity in osteoporotic mice via modulating Treg-Th17 cell balance. Bone Rep. 2018;8:46–56. doi: 10.1016/j.bonr.2018.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Suez J, Zmora N, Segal E, et al. . The pros, cons, and many unknowns of probiotics. Nat Med. 2019;25(5):716–729. doi: 10.1038/s41591-019-0439-x. [DOI] [PubMed] [Google Scholar]
- 287.Mak JWY, Chan FKL, Ng SC.. Probiotics and COVID-19: one size does not fit all. Lancet Gastroenterol Hepatol. 2020;5(7):644–645. doi: 10.1016/S2468-1253(20)30122-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Sichetti M, De Marco S, Pagiotti R, et al. Anti-inflammatory effect of multistrain probiotic formulation (L. rhamnosus, B. lactis, and B. longum). Nutrition. 2018;53:95–102. doi: 10.1016/j.nut.2018.02.005. [DOI] [PubMed] [Google Scholar]
- 289.Rodriguez-Nogales A, Algieri F, Garrido-Mesa J, et al. . Differential intestinal anti-inflammatory effects of Lactobacillus fermentum and Lactobacillus salivarius in DSS mouse colitis: impact on microRNAs expression and microbiota composition. Mol Nutr Food Res. 2017;61(11):1700144. [DOI] [PubMed] [Google Scholar]
- 290.D’Mello C, Ronaghan N, Zaheer R, et al. . Probiotics improve inflammation-associated sickness behavior by altering communication between the peripheral immune system and the brain. J Neurosci. 2015;35(30):10821–10830. doi: 10.1523/JNEUROSCI.0575-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Pena JA, Versalovic J.. Lactobacillus rhamnosus GG decreases TNF-alpha production in lipopolysaccharide-activated murine macrophages by a contact-independent mechanism. Cell Microbiol. 2003;5(4):277–285. doi: 10.1046/j.1462-5822.2003.t01-1-00275.x. [DOI] [PubMed] [Google Scholar]
- 292.Plaza-Diaz J, Ruiz-Ojeda FJ, Vilchez-Padial LM, et al. . Evidence of the anti-inflammatory effects of probiotics and synbiotics in intestinal chronic diseases. Nutrients. 2017;9(6):555. doi: 10.3390/nu9060555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Liu Y, Alookaran JJ, Rhoads JM.. Probiotics in autoimmune and inflammatory disorders. Nutrients. 2018;10(10):1537. doi: 10.3390/nu10101537. [DOI] [PMC free article] [PubMed] [Google Scholar]