Abstract
Background
Coronavirus disease 2019 (COVID-19) is a highly variable condition. Validated tools to assist in the early detection of patients at high risk of mortality can help guide medical decisions.
Objective
We sought to validate externally, as well as in patients from the second pandemic wave in Europe, our previously developed mortality prediction model for hospitalized COVID-19 patients.
Methods
Three validation cohorts were generated: 2 external with 185 and 730 patients from the first wave and 1 internal with 119 patients from the second wave. The probability of death was calculated for all subjects using our prediction model, which includes peripheral blood oxygen saturation/fraction of inspired oxygen ratio, neutrophil-to-lymphocyte ratio, lactate dehydrogenase, IL-6, and age. Discrimination and calibration were evaluated in the validation cohorts. The prediction model was updated by reestimating individual risk factor effects in the overall cohort (N = 1477).
Results
The mortality prediction model showed good performance in the external validation cohorts 1 and 2, and in the second wave validation cohort 3 (area under the receiver-operating characteristic curve, 0.94, 0.86, and 0.86, respectively), with excellent calibration (calibration slope, 0.86, 0.94, and 0.79; intercept, 0.05, 0.03, and 0.10, respectively). The updated model accurately predicted mortality in the overall cohort (area under the receiver-operating characteristic curve, 0.91), which included patients from both the first and second COVID-19 waves. The updated model was also useful to predict fatal outcome in patients without respiratory distress at the time of evaluation.
Conclusions
This is the first COVID-19 mortality prediction model validated in patients from the first and second pandemic waves. The COR+12 online calculator is freely available to facilitate its implementation (https://utrero-rico.shinyapps.io/COR12_Score/).
Key words: COVID-19, IL-6, mortality risk, predictive model, second wave, external validation
Abbreviations used: ARDS, Acute respiratory distress syndrome; AUC, Area under the receiver-operating characteristic curve; COVID-19, Coronavirus disease 2019; ER, Emergency room; ICU, Intensive care unit; LDH, Lactate dehydrogenase; N/L, Neutrophil-to-lymphocyte; SpO2/FiO2, Peripheral blood oxygen saturation/fraction of inspired oxygen ratio
The coronavirus disease 2019 (COVID-19) outbreak started in December 2019 and since then has caused more than 77 million infections and more than a million and a half deaths.1 In Europe, infections by severe acute respiratory syndrome coronavirus 2 have occurred in 2 waves: a first wave from February to June 2020, and a second wave that started in August 2020 and peaked in November 2020.2 , 3
The overall infection-fatality rate among people diagnosed with COVID-19 is approximately 1%.4 Nonetheless, there is an extreme variation in clinical presentation, ranging from asymptomatic infection to severe pneumonia, multiorgan failure, and death. Multiple clinical and laboratory features have been associated with severity in patients with COVID-19,5 , 6 although most of them lack specificity. For example, age has strongly been associated with disease severity,4 , 5 yet there are elderly people with asymptomatic severe acute respiratory syndrome coronavirus 2 infection.7 , 8
In some patients, severe acute respiratory syndrome coronavirus 2 infection produces an immune dysregulation,9 , 10 triggering marked pulmonary and systemic inflammation, which can persist after viral clearance. This hyperinflammatory dysregulation, combined with other infection-derived complications such as thrombosis,11 is responsible for the disease severity. However, there are still many uncertainties about the pathophysiology of COVID-19. Until we have a better understanding of this disease, the availability of tools that allow risk stratification of infected patients can be useful for optimizing therapeutic management and improving patient prognosis.
During the first wave of COVID-19 cases in Europe, we developed a prediction model that estimates the probability of death in hospitalized patients with COVID-19, based on 5 parameters taken at, or soon after, hospital admission: peripheral blood oxygen saturation/fraction of inspired oxygen (SpO2/FiO2) ratio, neutrophil-to-lymphocyte (N/L) ratio, lactate dehydrogenase (LDH), IL-6, and age.12 This model was rigorously developed by logistic regression, showing high accuracy in the prediction of patient outcome (area under the receiver-operating characteristic curve [AUC], 0.94), and it was translated into the freely available COR+12 online calculator (https://utrero-rico.shinyapps.io/COR12_Score/). We have now externally validated this prediction model in 2 independent cohorts from 2 different tertiary hospitals (validation cohorts 1 and 2). In addition, we have internally validated the model in a prospective patient cohort recruited during the second wave (validation cohort 3). Finally, the model has been updated with the overall sum of patients from the development and validation cohorts.
Methods
Study design and population
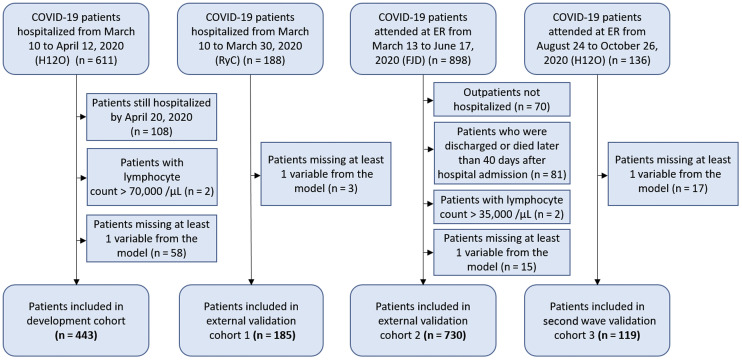
The validation of this prediction model12 was performed in 2 retrospective external cohorts from the first wave of COVID-19 cases, and in a prospective internal cohort from the second wave. Details on the development cohort from Hospital Universitario 12 de Octubre (Madrid) have already been published.12 The institutional Clinical Research Ethics Committee approved the study protocol (reference no. 20/167). The criteria for inclusion in the study were (a) being hospitalized with confirmed diagnosis of COVID-19 by real-time RT-PCR, (b) having data at admission, or within the first 4 days of hospitalization, on the 5 variables included in the model, and (c) having an outcome (discharge or death) within 40 days from hospital admission (40 days was the maximum time allowed for an outcome in the development cohort). Exclusion criteria were not meeting the inclusion criteria or having a hematological malignancy associated with increased lymphocyte count. A flowchart depicts the inclusion of patients in the different cohorts (Fig 1 ).
Fig 1.
Flowchart of patients included in the study. FJD, Hospital Universitario Fundación Jiménez Díaz; H12O, Hospital Universitario 12 de Octubre; RyC, Hospital Universitario Ramon y Cajal.
The external validation cohort 1 was composed of 188 patients hospitalized between March 10 and March 30, 2020, at the Hospital Universitario Ramón y Cajal (Madrid) with IL-6 measurement. Three patients were not included because of missing LDH measurements.
The external validation cohort 2 included 898 patients who attended the emergency room (ER) at either Hospital Universitario Infanta Elena or Hospital Universitario Fundación Jiménez Díaz (Madrid) from March 13 to June 17, 2020, had confirmed COVID-19, and had an IL-6 measurement. These 2 hospitals share a centralized laboratory and electronic clinical records system and have therefore been considered as 1 cohort. One hundred sixty-eight patients who did not meet the inclusion criteria were dismissed, and the remaining 730 patients were included in the final analysis.
The model was further validated in a prospective internal cohort with second wave COVID-19 patients (validation cohort 3). One hundred thirty-six patients who attended the ER from August 24 to October 26 at the Hospital Universitario 12 de Octubre (Madrid), who had confirmed COVID-19 and an IL-6 measurement, were included in the second wave validation cohort 3. Seventeen patients had missing values, and 119 patients were included in the final analysis.
Data collection
Clinical electronic medical records were reviewed by researchers, and data were manually collected at the Hospital 12 de Octubre and Hospital Ramón y Cajal (validation cohorts 1 and 3). All data were revised by at least 2 independent researchers. At Hospital Infanta Elena and Hospital Fundación Jiménez Díaz (validation cohort 2), clinical data were extracted by means of big data/artificial intelligence processes from individual electronic medical records and, then, reviewed and refined by 4 independent researchers. Recorded data included demographic information, laboratory findings, length of hospital stay, intensive care unit (ICU) admission, and outcome. SpO2/FiO2 was used to assess respiratory function. SpO2/FiO2 shows a good correlation with the partial pressure of arterial oxygen (PaO2)/FiO2 ratio (SpO2/FiO2 = 64 + 0.84 × PaO2/FiO2),13 and was available for all the patients. Acute respiratory distress syndrome (ARDS) was classified according to Berlin criteria: SpO2/FiO2 more than 315, no ARDS; 315 to 235, mild ARDS; 148 to 235, moderate ARDS; less than 148, severe ARDS.14
Laboratory measurements
IL-6 was measured with the BD Cytometric Bead Array human IL-6 flex set (BD Biosciences, San Jose, Calif) using a BD Canto II flow cytometer at the Hospital 12 de Octubre and Hospital Ramón y Cajal. Results were analyzed with FCAP Array software v3.0 (BD Biosciences). At Hospital Fundación Jimenez Diaz, IL-6 was measured by Roche chemiluminescent immunoassay. Only IL-6 measurements before tocilizumab therapy were included in the study. Other laboratory parameters such as LDH, neutrophils, and lymphocytes were measured as part of standard of care. All laboratory measurements were taken on the same day.
The variables included in the prediction model (SpO2/FiO2, N/L ratio, LDH, IL-6, and age) were taken at hospital admission or in the first 4 days of the hospitalization. Most measurements were taken at the ER in the external validation cohort 2 and the second wave validation cohort 3, whereas measurements were taken with a median of 2 days from hospital admission in the external validation cohort 1 (Table I ).
Table I.
Demographic, clinical, and laboratory characteristics of patients in the development, external, and second wave internal validation cohorts
| Characteristic | Development cohort (H12O) (N = 443) | External validation cohort 1 (RyC) (N = 185) | External validation cohort 2 (FJD) (N = 730) | Second wave validation cohort 3 (H12O) (N = 119) | P |
|---|---|---|---|---|---|
| Age (y), median (IQR) | 53 (45-60) | 63 (53-72) | 69 (56-82) | 63 (51-76) | <.0001 |
| Sex: male, n (%) | 281 (63.4) | 131 (70.8) | 400 (54.8) | 79 (66.4) | <.0001 |
| ARDS classification on test day, n, (%) | <.0001 | ||||
| None | 255 (57.6) | 55 (29.7) | 450 (61.7) | 72 (60.5) | |
| Mild | 99 (22.3) | 53 (28.6) | 111 (15.2) | 29 (24.4) | |
| Moderate | 30 (6.8) | 21 (11.4) | 44 (6.0) | 8 (6.7) | |
| Severe | 59 (13.3) | 56 (30.3) | 125 (17.1) | 10 (8.4) | |
| SARS-CoV-2 RT-PCR result, positive, n (%) | 314 (70.9) | 185 (100) | 730 (100) | 119 (100) | .16 |
| Time from hospital admission to laboratory measurements (d), median (IQR) | 2 (1-4) | 2 (2-3) | 1 (1-2) | 0 (0-0) | <.0001 |
| Length of hospital stay (d), median, (IQR) | 8 (6-13) | 9 (6-13) | 8 (5-15) | 8 (6-12) | .19 |
| ICU admission, n (%) | 34 (7.7) | 35 (18.9) | 106 (14.5) | 15 (12.6) | <.0001 |
| Death, n (%) | 33 (7.4) | 44 (23.8) | 78 (10.7) | 18 (15.41) | <.0001 |
| Prediction model variables (except age), median (IQR) | |||||
| SpO2/FiO2 | 346 (263-452) | 258 (133-339) | 343 (251-448) | 343 (272-445) | <.0001 |
| N/L Ratio | 4.3 (2.4-8.6) | 6.1 (3.6-10.5) | 5.3 (2.8-9.5) | 7.0 (4.0-11.5) | <.0001 |
| LDH (U/L) | 350 (278-454) | 375 (308-478) | 284 (222-376) | 395 (323-463) | <.0001 |
| IL-6 (pg/mL) | 19 (5-48) | 33 (13-61) | 29 (9-65) | 31 (17-72) | <.0001 |
FJD, Hospital Universitario Fundación Jiménez Díaz; H12O, Hospital Universitario 12 de Octubre; IQR, interquartile range; RyC, Hospital Universitario Ramon y Cajal; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Statistical analysis
Continuous numerical data were represented as median and interquartile range and compared using the Mann-Whitney U test, or Kruskal-Wallis test when relevant. Categorical variables were represented as N and percentage, and compared using the chi-square test. The Fisher exact test was used when appropriate. The potential of each variable in the model to be used individually as a biomarker was evaluated using AUC analysis.
Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD) guidelines for validation of multivariate prediction models were followed.15 The logistic regression model in the development cohort was probability of death = 1 / (1 + EXP (− (−7.6991 – 0.0076 × (SpO2/FiO2) + 0.0547 × (N/L ratio) + 0.0046 × LDH + 0.0043 × IL-6 + 0.0682 × age))). This model was applied to the 3 validation cohorts. The validity of the prediction model was assessed by evaluating discrimination and calibration.16, 17, 18 Discrimination describes the ability of the model to distinguish a patient who will survive from a patient who will die. Discrimination of the model was evaluated by AUC analysis in the validation cohorts. The improved discrimination of the model, in comparison with each individual parameter, was evaluated using De Long test. Calibration of a model describes the agreement between the predicted and the observed mortality. Calibration was assessed (a) by the intercept alpha (A), which compares the mean of all predicted risks and the mean observed risk, and in a perfect prediction would be 0, and (b) by the slope beta (B), which in good predictions is close to 1. Calibration results were represented as calibration plots and a circular barplot. The C-statistic does not appear in the text because for binary outcomes it corresponds to the AUC of the model.
The prediction model was updated with the compiled data from all the cohorts, by reestimating the intercept and effect of each individual risk factor.19 The updated model was probability of death = 1 / (1 + EXP (8.3777 + 0.0071 × (SpO2/FiO2) − 0.0326 × (N/L ratio) − 0.0046 × LDH − 0.0027 × IL6 − 0.0852 × age)). K-fold cross-validation (k = 10) for a generalized linear model was used to validate the updated model. The updated model excluding IL-6 was probability of death = 1 / (1 + EXP (7.7260 + 0.0070 × (SpO2/FiO2) − 0.0364 × (N/L ratio) − 0.0044 × LDH − 0.0798 × age)).
Youden index was used for model cutoff selection, and with this cutoff sensitivity, specificity, negative predictive value, and positive predictive value were calculated. Time-to-event curves were plotted by the Kaplan-Meier method, and differences were compared with the log-rank test to analyze the ability of the score for stratification across risk categories. Hazard ratio was calculated using Cox regression method. Throughout the analysis, only patients with available data were compared and the cohorts’ size is specified in figures and tables. P less than .05 was considered statistically significant. Data sets can be made available upon formal request to the corresponding author. All the analysis was performed with R v4.0.3.
Results
Patient demographic and clinical characteristics
The demographic, clinical, and laboratory characteristics of the patients in the external validation cohorts 1 and 2, and in the second wave validation cohort 3, are presented in Table I. Most patients in the 3 validation cohorts were men, in their sixties, and were hospitalized for a median of 8 days. Median age was higher in the validation cohorts compared with the development cohort, and it was highest in the external validation cohort 2 (median age, 69 years). Patient severity was also greater in the validation cohorts compared with the development cohort, as measured by the rate of ICU admission, which ranged from 12.6% to 18.9%, and the mortality rate, which ranged from 10.7% to 23.8%. The external validation cohort 1 exhibited the highest degree of disease severity, with 18.9% of patients admitted to ICU and 23.8% patients dying during hospitalization.
Analysis of prediction model variables according to disease outcome
In the development cohort, the 5 variables with the highest association with mortality and the highest contribution to the predictive capacity of the model were included in the final prediction model. These variables (SpO2/FiO2, N/L ratio, LDH, IL-6, and age) differed statistically between the development and validation cohorts (Table I). Nonetheless, in the validation cohorts, all the variables were significantly increased in patients who died compared with those who survived, except for SpO2/FiO2 in the second wave validation cohort 3, which was relatively high in nonsurvivors (Table II ). Overall, patients who died were significantly older and had significantly lower SpO2/FiO2 and higher levels of N/L ratio, LDH, and IL-6 than patients who survived.
Table II.
Prediction model variables measured in survivor and nonsurvivor patients
| Variables | Survivors |
Nonsurvivors |
P | ||
|---|---|---|---|---|---|
| Median | IQR | Median | IQR | ||
| Development cohort (H12O) | |||||
| SpO2/FiO2 | 352 | 269-457 | 107 | 101-209 | <.0001 |
| N/L Ratio | 4.0 | 2.3-7.8 | 17.3 | 8.8-30.3 | <.0001 |
| LDH (U/L) | 341 | 269-443 | 537 | 391-715 | <.0001 |
| IL-6 (pg/mL) | 17 | 5-44 | 86 | 20-225 | <.0001 |
| Age (y) | 52 | 44-59 | 65 | 56-71 | <.0001 |
| External validation cohort 1 (RyC) | |||||
| SpO2/FiO2 | 296 | 204-346 | 106 | 100-190 | <.0001 |
| N/L Ratio | 5.6 | 3.4-10.3 | 8.5 | 5.0-12.3 | .009 |
| LDH (U/L) | 354 | 296-425 | 512 | 395-622 | <.0001 |
| IL-6 (pg/mL) | 28 | 10-54 | 55 | 26-87 | .0004 |
| Age (y) | 59 | 50-69 | 73 | 69-79 | <.0001 |
| External validation cohort 2 (FJD) | |||||
| SpO2/FiO2 | 350 | 266-448 | 129 | 103-341 | <.0001 |
| N/L Ratio | 4.8 | 2.7-8.9 | 9.1 | 6.1-16.8 | <.0001 |
| LDH (U/L) | 277 | 220-361 | 371 | 249-440 | <.0001 |
| IL-6 (pg/mL) | 28 | 8-63 | 45 | 14-143 | .0004 |
| Age (y) | 67 | 55-80 | 84 | 77-90 | <.0001 |
| Second wave validation cohort 3 (H12O) | |||||
| SpO2/FiO2 | 343 | 291-443 | 299 | 98-429 | .12 |
| N/L Ratio | 6.3 | 3.9-10.0 | 14.8 | 6.7-22.7 | .007 |
| LDH (U/L) | 389 | 317-457 | 440 | 382-681 | .021 |
| IL-6 (pg/mL) | 30 | 16-57 | 78 | 33-195 | .005 |
| Age (y) | 58 | 49-71 | 78 | 69-85 | .016 |
FJD, Hospital Universitario Fundación Jiménez Díaz; H12O, Hospital Universitario 12 de Octubre; IQR, interquartile range; RyC, Hospital Universitario Ramon y Cajal.
Validation of the mortality prediction model
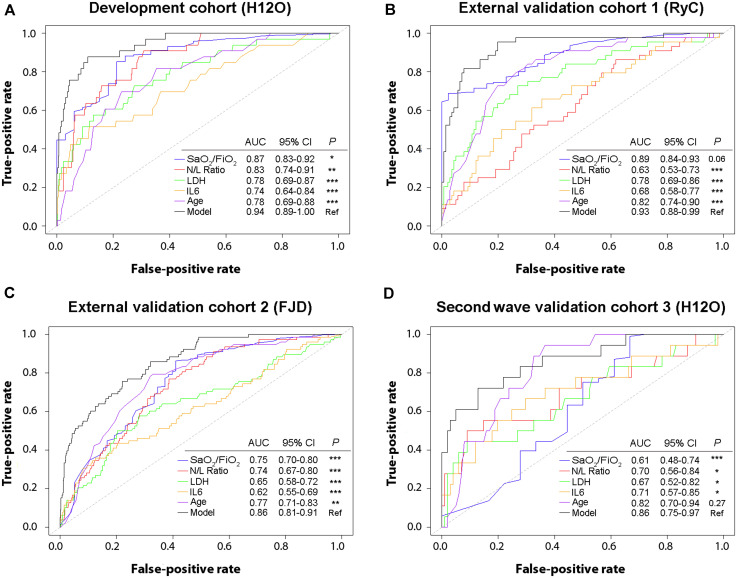
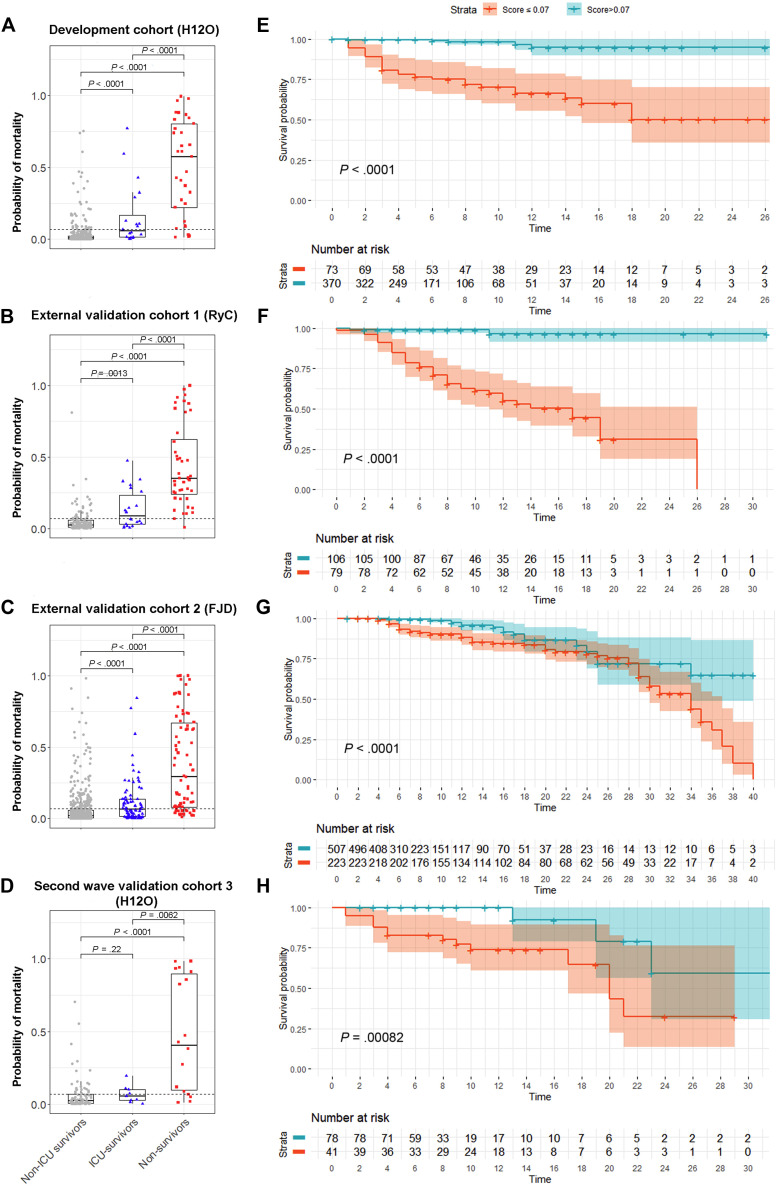
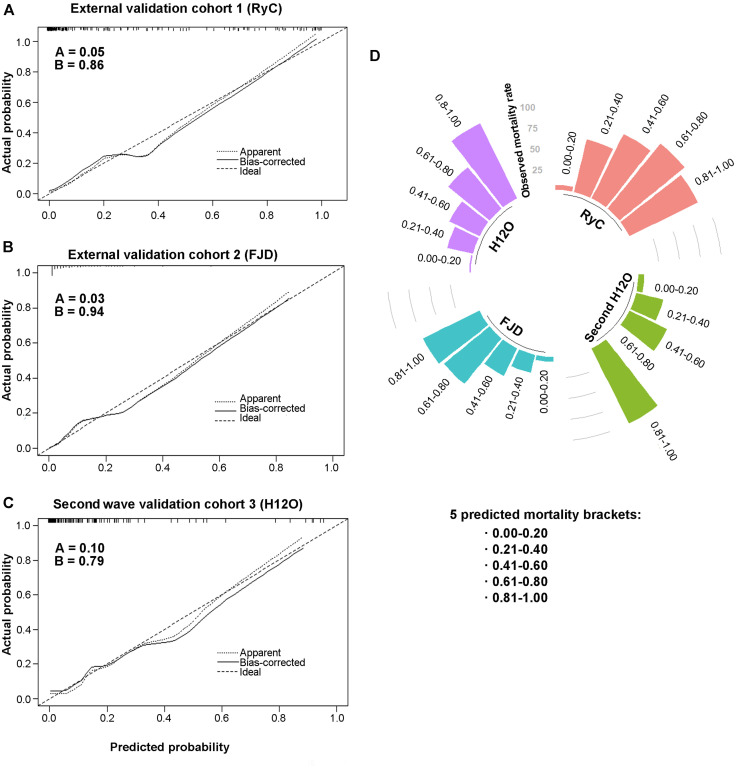
We first evaluated the generalizability of our prediction model in 2 external cohorts with a large number of patients from the first wave of COVID-19 in Spain. Validation of the model demonstrated a good predictive performance in both, the external validation cohort 1 (N = 185) (AUC = 0.93; 95% CI, 0.88-0.99) and the external validation cohort 2 (N = 730) (AUC = 0.86; 95% CI, 0.81-0.91) (Fig 2 ). Notably, the model showed a superior predictive capacity with the highest AUC, when compared with each individual risk factor, except for SpO2/FiO2 in the external validation cohort 1 (P = .06). We applied the model to the patients in both external cohorts and found, as expected, that the probability of dying was significantly higher in nonsurvivors, followed by patients who survived after intensive care, followed by patients who survived without the need for intensive care (Fig 3 ). The Youden index–based cutoff generated during the development of the prediction model was 0.07, with a 0.88 sensitivity, 0.89 specificity, 0.38 positive predictive value, and 0.99 negative predictive value. Using this cutoff, Kaplan-Meier analysis showed a very significant difference in survival for patients with low and high risk of death in both external cohorts (P < .0001; Fig 3). We next assessed the calibration of the model, that is, the agreement between the observed and the model-predicted mortality in the external validation cohorts. As shown in the calibration plots, the model performed remarkably well at predicting patient mortality (Fig 4 ). The slopes in the external validation cohorts 1 and 2 were close to 1 (0.86 and 0.94, respectively), and the intercepts were close to 0 (0.05 and 0.03, respectively). We have depicted in a circular barplot how the observed mortality increased steadily as the predicted mortality increased (Fig 4). Altogether, these results indicate a good performance of the model and a minimal difference between predicted and observed values, and, therefore, they validate the model externally.
Fig 2.
Comparison of the capacity to predict mortality between the model and the individual risk factors (SpO2/FiO2, N/L ratio, LDH, IL-6, and age) in each patient cohort. A, The classification performance of the model was better than the individual risk factors in the development cohort. B, The classification performance of the model was better than the individual risk factors in the external validation cohort 1, except for SpO2/FiO2. C, The classification performance of the model was better than the individual risk factors in the external validation cohort 2. D, The classification performance of the model was better than the individual risk factors in the second wave validation cohort 3, except for age. Classification performance was compared with De Long test. FJD, Hospital Universitario Fundación Jiménez Díaz; H12O, Hospital Universitario 12 de Octubre; RyC, Hospital Universitario Ramon y Cajal.
Fig 3.
The prediction model accurately identified patients at high risk of dying in the validation cohorts. A-D, The probability of dying predicted by the model was significantly higher in nonsurvivors (red), than in survivors who required intensive care (blue) and than in survivors who did not require intensive care (gray), in all cohorts. Dashed lines indicate the model’s optimal cutoff for mortality (0.07). E-H, Using the model’s optimal cutoff, Kaplan-Meier analysis showed a very different survival between the groups with low and high risk of death (P < .001, for all cohorts). FJD, Hospital Universitario Fundación Jiménez Díaz; H12O, Hospital Universitario 12 de Octubre; RyC, Hospital Universitario Ramon y Cajal. Color shades represent the 95% CI. Time is indicated in days.
Fig 4.
Calibration analysis depicting the predicted vs observed mortality. A-C, Calibration curves were close to the diagonal dotted line, which represents ideal calibration in which predicted and observed risks are identical. Intercept (Fig 4, A) and slope (Fig 4, B) are shown for each validation cohort. D, Patients were grouped in 5 brackets of increasing probability of death (0%-20%, 21%-40%, 41%-60%, 61%-80%, and 81%-100%). The circular barplot shows how the observed mortality increased steadily as the predicted mortality increased. Of note, in the 2 external validation cohorts, mortality in the highest risk bracket was lower than predicted. Observed mortality rate is represented in concentric circles. FJD, Hospital Universitario Fundación Jiménez Díaz; H12O, Hospital Universitario 12 de Octubre; RyC, Hospital Universitario Ramon y Cajal.
We considered it relevant to further validate the utility of this prediction model with patients from the second wave of COVID-19 cases in Spain. We built a prospective internal validation cohort with patients admitted to the same hospital as the development cohort, but approximately 6 months apart (validation cohort 3, N = 119). The mortality prediction model also maintained a good performance during the second wave (AUC = 0.86; 95% CI, 0.75-0.97) (Fig 2, D). The probability of dying was calculated for all patients on admission, and it was significantly higher in nonsurvivors than in patients who survived, either with intensive care requirement (P < .01) or without it (P < .0001) (Fig 3, D). Kaplan-Meier analysis also showed a very significant difference in survival for patients below or above the model threshold during the second wave (P < .001) (Fig 3, H). The calibration curve of the model performance on the second wave validation cohort 3 also did reasonably well, with a slope of 0.79 and an intercept of 0.10. These results validate the accuracy of our mortality prediction model in patients during the second wave.
Update of the mortality prediction model
Because we observed significant differences in the variables included in the model between the cohorts, we revised the model with the patients from the entire 4 cohorts (N = 1477) with the aim of increasing its generalizability. In the updated model, the 5 variables had little collinearity (demonstrated by variation inflation factor < 1.4 for each of the 5 parameters included in the model) and significantly contributed to the model’s prediction capacity (data not shown). Age increased its weight in the updated model, whereas SpO2/FiO2, N/L ratio, and IL-6 reduced it (see the 2 model equations in the Methods section). The classifying accuracy of the updated model was very robust (AUC = 0.91; 95% CI, 0.87-0.94) (Fig 5 , A), and it was not affected by patients’ sex (see Fig E1 in this article’s Online Repository at www.jacionline.org). Cross-validation of the updated model showed a substantial agreement between the predicted and observed mortality (accuracy of 0.91 [0.89-0.93] and kappa coefficient 0.51 [0.42-0.60]). An updated model including only SpO2/FiO2, N/L ratio, LDH, and age was also developed, to be used in settings without IL-6 availability (see model equation in the Methods section). The classifying accuracy of the 4-variable updated model was also robust (AUC = 0.89; 95% CI, 0.86-0.92); however, it was significantly lower than that of the updated model including IL-6 (P < .05).
Fig 5.
The updated model accurately classified patients at risk of dying. A, The prediction model was revised with the sum of patients from development and validation cohorts (N = 1477). AUC of the updated model was 0.91 (95% CI, 0.87-0.94), with optimal cutoff in 0.107. B, Kaplan-Meier analysis based on Youden index optimal cutoff showed a very different survival between the groups with low and high risk of death (P < .0001). Color shades represent the 95% CI. Time is indicated in days. C, The predicted probability of death in nonsurvivors (red) was significantly higher than in survivors who required intensive care (blue) (P < .0001), and than in survivors who did not require intensive care (gray) (P < .0001). Dashed line indicates optimal cutoff for mortality (0.107).
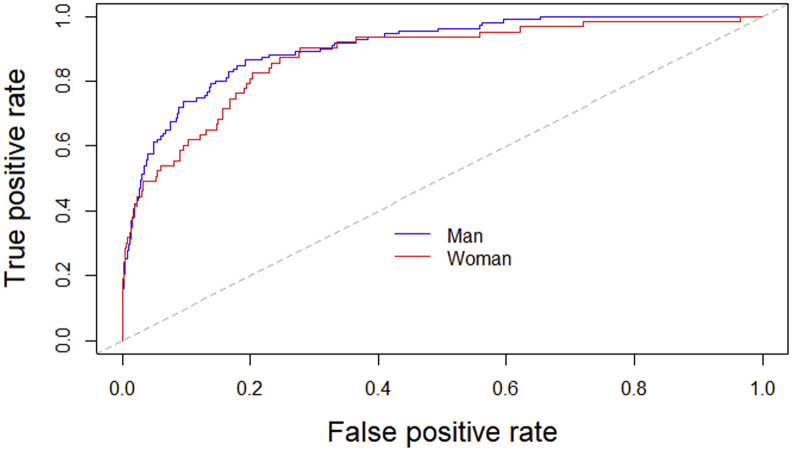
Fig E1.
Classification performance of the updated model by sex. The capacity of the updated model to predict mortality was similar in men (AUC, 0.91; 95% CI, 0.87-0.94) than in women (AUC, 0.87; 95% CI, 0.82-0.93), with no statistical difference in accuracy between sex (De Long test, P = .27).
The Youden index–based cutoff generated for the updated model containing 5 variables was 0.107, with a 0.85 sensitivity, 0.81 specificity, 0.37 positive predictive value, and 0.98 negative predictive value. Kaplan-Meier analysis showed very clear differences in survival for patients below or above this model threshold (P < .0001; Fig 5, B). Of note, the probability of survival in the low-risk group started to decrease after the third week of hospitalization, suggesting that the model decreased its accuracy in patients with long hospital stays.
Clinical significance of the updated prediction model
Respiratory function is a key factor evaluated in patients with COVID-19 to assess their disease severity. As an example of the clinical utility of this prediction model, we analyzed its utility at predicting fatal outcome in patients without ARDS at the time of evaluation. In the overall cohort, 832 patients had no ARDS on admission, of whom 37 (4.4%) finally died. The cutoff value established in the prediction model classified 755 patients as low risk and 77 patients as high risk. In the low-risk group, 17 (2.3%) patients died, whereas in the high-risk group, 20 (26%) patients passed away. Kaplan-Meier analysis showed that patients classified as high risk by the model survived significantly less than patients classified as low risk (P < .0001) (Fig 6 ). Moreover, patients without ARDS classified as high risk had 8.76 times more risk of dying than low-risk patients (hazard ratio, 8.76; 95% CI, 5.74-13.36; P < .0001). These results indicate that the model can be helpful at identifying patients at high risk of dying, who a priori may not be considered as potentially severe.
Fig 6.
The updated model-predicted mortality in patients with no respiratory distress at the time of evaluation. A, In the overall cohort, 832 patients had no ARDS in the beginning of their hospitalization. Within these patients, the predicted probability of death in nonsurvivors (red) was significantly higher than in survivors who required intensive care (blue) (P < .0001), and than in survivors who did not require intensive care (gray) (P < .0001). Dashed line indicates optimal cutoff for mortality (0.107). B, Within the patients without ARDS initially, patients classified as high risk by the cutoff survived significantly less than the low-risk patients (P < .0001).
Discussion
Here, we validate our previously published mortality prediction model for hospitalized COVID-19 patients12 in 2 independent external validation cohorts and in a second wave validation cohort (Fig 1). From a methodological point of view, this work reinforces the utility of BigData-oriented frameworks to structure and extract patients’ information from electronic clinical records. Artificial intelligence–based data mining has already been successfully used to obtain results regarding COVID-19 diagnosis20 and outcome prediction.21
The present prediction model, which includes 5 easily collected clinical and laboratory biomarkers (namely SpO2/FiO2, N/L ratio, LDH, IL-6, and age), performed with high accuracy in the 2 external validation cohorts, with AUC of 0.93 and 0.86 and well-fitted calibration curves (Fig 2, Fig 3, Fig 4). The data for the external validation were collected retrospectively, from patients hospitalized with COVID-19 in 2 unrelated hospitals during the first wave of the pandemic in Spain.
This external validation is important because it showed that, despite significant differences between the development and the validation cohorts, the prediction model worked with high accuracy in the latter. There were differences in the biomarkers included in the model between the cohorts (Table I). The model was developed in a younger and less severe cohort than the validation cohorts, as shown by higher SpO2/FiO2 and lower ICU admission and death rates. In addition, levels of IL-6 were measured by cytometric bead array in the development cohort and the external validation cohort 1, whereas they were measured by chemiluminescence in the external validation cohort 2. It was relevant to prove that different laboratory techniques could be used without altering the model performance, especially regarding IL-6, whose measurement is not as standardized as that of other inflammation markers. Finally, the measurement of the biomarkers included in the predictive model took place in the first days of hospitalization in the development cohort and the external validation cohort 1, whereas it occurred at ER in the external validation cohort 2. The model performed similarly for inpatients and ER patients who were subsequently hospitalized. These considerations demonstrate that this prediction model can work in different clinical settings.
To our knowledge, this is the first mortality prediction model validated among patients from the second wave of COVID-19. There are a number of epidemiological and clinical practice differences between the first wave and the second wave of the pandemic. For example, during the second wave, patients have sought medical help sooner, within less days from symptom onset, and severe patients have received corticoids and heparin more precociously. Despite this changing practice over time, the model accuracy in the internal validation cohort was good, yielding an AUC of 0.86 and a well-fitted calibration curve. These results validate the use of this prediction model in patients during the second and possibly successive waves.
To improve the predictive performance in any clinical setting, we have updated the model by reestimating the weights of the 5 biomarkers in an overall cohort including all patients from the 4 cohorts (N = 1477). The accuracy of the updated model was excellent (AUC, 0.91), and the cutoff allowed for the correct separation of low- and high-risk patients (Fig 5). Of note, the survival curves of these 2 patient groups were remarkably distinct since the beginning of hospitalization. In addition, this updated model can be useful to discriminate between patients with likely fatal outcome and those who are likely to survive, among patients without respiratory distress at the time of evaluation (Fig 6). SpO2/FiO2 was a worse individual biomarker in the external validation cohort 2 and in the second wave cohort, both being cohorts in which evaluation was made mostly at the ER (Fig 2). In this ER setting, the use of the updated model can greatly improve the detection of high-risk patients.
A limitation of this study is, however, related to the accuracy of the updated model in the low mortality risk group in the long-term (Fig 5, B). Of the initially 1083 patients classified as low risk, 26 (2.4%) died. Three weeks after evaluation, only 51 low-risk patients were still hospitalized, of whom 8 (15.7%) died. This shows that the probability of survival decreases in patients classified as low risk if they have not been discharged after 3 weeks. This should be taken into account for patients who have a prolonged hospital stay and suggests that it could be helpful to periodically reassess their mortality risk. Additional limitations to this study are the need for further external validation of the updated model and the need for impact studies in different populations and health care systems to be conducted.
Conclusions
Our mortality prediction model has been validated in 3 large validation cohorts, 2 external and 1 from the second wave. Moreover, the model has been updated, showing good discrimination and excellent calibration. This suggests that the updated model is likely to be generalizable to other populations and clinical settings, and that its predictive performance should be accurate when applied to its target population, patients with COVID-19 who are attended at ER and require hospitalization, and patients with COVID-19 who have recently been hospitalized. This model, and the COR+12 online calculator, could potentially assist in efficient classification of patients with COVID-19, and contribute to guide medical decisions.
Clinical implications.
This updated model has been developed with patients from first and second COVID-19 waves and provides the probability of dying, even in patients without respiratory distress at the time of evaluation.
Acknowledgments
We thank all patients, nurses, and medical colleagues who contributed to this study.
Footnotes
This study was supported by the Instituto de Salud Carlos III, Spanish Ministry of Science and Innovation (COVID-19 research call COV20/00181)—cofinanced by the European Development Regional Fund “A way to achieve Europe.” R.L.-G. holds a research contract “Rio Hortega” (CM19/00120), B.A. a research contract “Juan Rodes” (JR17/00020), and M.F.-R., P.M., and M.C. a research contract “Miguel Servet” (CP18/00073, CP16/00116, and CP17/00006, respectively), all from the Instituto de Salud Carlos III, Spanish Ministry of Science and Innovation. R.L.-R. is sponsored by the IIS-Fundación Jiménez Díaz-UAM Genomic Medicine Chair.
Disclosure of potential conflict of interest: The authors declare that they have no relevant conflicts of interest.
Appendix
References
- 1.Coronavirus Resource Center https://coronavirus.jhu.edu/ Available at:
- 2.Cacciapaglia G., Cot C., Sannino F. Second wave COVID-19 pandemics in Europe: a temporal playbook. Sci Rep. 2020;10:15514. doi: 10.1038/s41598-020-72611-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.World Health Organization https://who.maps.arcgis.com/apps/opsdashboard/index.html#/ead3c6475654481ca51c248d52ab9c61 Available at:
- 4.Pastor-Barriuso R., Perez-Gomez B., Hernan M.A., Perez-Olmeda M., Yotti R., Oteo-Iglesias J. Infection fatality risk for SARS-CoV-2 in community dwelling population of Spain: nationwide seroepidemiological study. BMJ. 2020;371:m4509. doi: 10.1136/bmj.m4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Williamson E.J., Walker A.J., Bhaskaran K., Bacon S., Bates C., Morton C.E. Factors associated with COVID-19-related death using OpenSAFELY. Nature. 2020;584:430–436. doi: 10.1038/s41586-020-2521-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Del Valle D.M., Kim-Schulze S., Huang H.H., Beckmann N.D., Nirenberg S., Wang B. An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat Med. 2020;26:1636–1643. doi: 10.1038/s41591-020-1051-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Patel M.C., Chaisson L.H., Borgetti S., Burdsall D., Chugh R.K., Hoff C.R. Asymptomatic SARS-CoV-2 infection and COVID-19 mortality during an outbreak investigation in a skilled nursing facility. Clin Infect Dis. 2020;71:2920–2926. doi: 10.1093/cid/ciaa763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borras-Bermejo B., Martinez-Gomez X., San Miguel M.G., Esperalba J., Anton A., Martin E. Asymptomatic SARS-CoV-2 infection in nursing homes, Barcelona, Spain, April 2020. Emerg Infect Dis. 2020;26:2281–2283. doi: 10.3201/eid2609.202603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lucas C., Wong P., Klein J., Castro T.B.R., Silva J., Sundaram M. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature. 2020;584:463–469. doi: 10.1038/s41586-020-2588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuri-Cervantes L., Pampena M.B., Meng W., Rosenfeld A.M., Ittner C.A.G., Weisman A.R. Comprehensive mapping of immune perturbations associated with severe COVID-19. Sci Immunol. 2020;5:eabd7114. doi: 10.1126/sciimmunol.abd7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ackermann M., Verleden S.E., Kuehnel M., Haverich A., Welte T., Laenger F. Pulmonary vascular endothelialitis, thrombosis, and angiogenesis in Covid-19. N Engl J Med. 2020;383:120–128. doi: 10.1056/NEJMoa2015432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laguna-Goya R., Utrero-Rico A., Talayero P., Lasa-Lazaro M., Ramirez-Fernandez A., Naranjo L. IL-6-based mortality risk model for hospitalized patients with COVID-19. J Allergy Clin Immunol. 2020;146:799–807.e9. doi: 10.1016/j.jaci.2020.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rice T.W., Wheeler A.P., Bernard G.R., Hayden D.L., Schoenfeld D.A., Ware L.B. Comparison of the SpO2/FIO2 ratio and the PaO2/FIO2 ratio in patients with acute lung injury or ARDS. Chest. 2007;132:410–417. doi: 10.1378/chest.07-0617. [DOI] [PubMed] [Google Scholar]
- 14.Ranieri V.M., Rubenfeld G.D., Thompson B.T., Ferguson N.D., Caldwell E., Fan E. Acute respiratory distress syndrome: the Berlin Definition. JAMA. 2012;307:2526–2533. doi: 10.1001/jama.2012.5669. [DOI] [PubMed] [Google Scholar]
- 15.Moons K.G., Altman D.G., Reitsma J.B., Collins G.S. New guideline for the reporting of studies developing, validating, or updating a multivariable clinical prediction model: the TRIPOD statement. Adv Anat Pathol. 2015;22:303–305. doi: 10.1097/PAP.0000000000000072. [DOI] [PubMed] [Google Scholar]
- 16.Steyerberg E.W., Vergouwe Y. Towards better clinical prediction models: seven steps for development and an ABCD for validation. Eur Heart J. 2014;35:1925–1931. doi: 10.1093/eurheartj/ehu207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moons K.G., Kengne A.P., Grobbee D.E., Royston P., Vergouwe Y., Altman D.G. Risk prediction models: II. External validation, model updating, and impact assessment. Heart. 2012;98:691–698. doi: 10.1136/heartjnl-2011-301247. [DOI] [PubMed] [Google Scholar]
- 18.Alba A.C., Agoritsas T., Walsh M., Hanna S., Iorio A., Devereaux P.J. Discrimination and calibration of clinical prediction models: users’ guides to the medical literature. JAMA. 2017;318:1377–1384. doi: 10.1001/jama.2017.12126. [DOI] [PubMed] [Google Scholar]
- 19.Siregar S., Nieboer D., Versteegh M.I.M., Steyerberg E.W., Takkenberg J.J.M. Methods for updating a risk prediction model for cardiac surgery: a statistical primer. Interact Cardiovasc Thorac Surg. 2019;28:333–338. doi: 10.1093/icvts/ivy338. [DOI] [PubMed] [Google Scholar]
- 20.Jin C., Chen W., Cao Y., Xu Z., Tan Z., Zhang X. Development and evaluation of an artificial intelligence system for COVID-19 diagnosis. Nat Commun. 2020;11:5088. doi: 10.1038/s41467-020-18685-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clift A.K., Coupland C.A.C., Keogh R.H., Diaz-Ordaz K., Williamson E., Harrison E.M. Living risk prediction algorithm (QCOVID) for risk of hospital admission and mortality from coronavirus 19 in adults: national derivation and validation cohort study. BMJ. 2020;371:m3731. doi: 10.1136/bmj.m3731. [DOI] [PMC free article] [PubMed] [Google Scholar]