Abstract
Background
Despite advances in our understanding of the genetic causes of hypertrophic cardiomyopathy (HCM), a large portion of this patient population do not carry sarcomere gene mutations when screened. It remains largely unknown why patients without sarcomere mutations develop asymmetric myocardial hypertrophy.
Methods
We performed a retrospective analysis of probands with HCM who underwent genetic testing to determine if clinical phenotypes were different depending on sarcomere mutation status. A medical history, three generation family history and clinical phenotyping were performed on 127 probands with HCM. Genetic screening was performed using clinically available HCM genetic testing panels.
Results
We found that probands with HCM with pathogenic sarcomere mutations were over three times more likely to have a family history of HCM (66% vs 17%, p<0.0001) and were diagnosed with HCM at a much younger age (32 vs 51 years old, p<0.0001). In contrast, probands with HCM without sarcomere mutations were significantly more obese (body surface area p=0.003, body mass index p=0.04 adjusted for age) and were more likely to present with left ventricular outflow tract obstruction (p=0.0483).
Conclusion
Patients with sarcomere mutation negative HCM present at an older age and are more obese compared with patients with sarcomere mutation positive HCM. The role of ageing and obesity in asymmetric myocardial hypertrophy warrants further investigation.
Keywords: cardiomyopathy, hypertrophic, cardiomyopathies, genetic diseases, inborn
Key questions.
What is already known about this subject?
Sarcomere gene mutations are the primary genetic cause of hypertrophic cardiomyopathy (HCM).
However, a large number of patients with HCM do not have sarcomere gene mutations and it remains poorly defined why these patients develop asymmetric myocardial hypertrophy.
What does this study add?
We found that patients with sarcomere gene mutation negative HCM presented at an older age, were significantly more obese, and were much less likely to have a family history of HCM.
How might this impact on clinical practice?
These findings suggest non-genetic risk factors such as obesity and ageing may contribute to asymmetric myocardial hypertrophy in some patients with HCM.
Further studies will need to be performed to determine how these risk factors impact asymmetric myocardial remodelling and if targeted risk factor modification can impact disease progression.
Introduction
Hypertrophic cardiomyopathy (HCM), a disease typically characterised by abnormal asymmetric growth of the ventricular myocardium, is a common genetic cause of heart failure and sudden death. The prevalence of this condition is ≈1:500 and over the last 30 years there has been tremendous advancement in understanding the genetic underpinnings of this disorder.1–4 The genes implicated in HCM encode for a diverse array of proteins involved in the proper function of the cardiomyocyte sarcomere. Despite advances in understanding the genetics of this disease, the role of genetic testing in diagnosis and disease management continues to evolve.5 Importantly, 30%–60% of individuals with HCM do not carry a germline mutation when screened for sarcomere gene mutations.6
The identification of sarcomere gene mutations provided a foundation from which to study the mechanisms controlling cardiomyocyte hypertrophy and pathological myocardial remodelling. Through research spanning two decades, sarcomere gene mutations have been shown to cause perturbations in cardiomyocyte calcium handling, energy utilisation and sarcomere function.7–12 Importantly, selective methods to correct these abnormalities are actively being pursued as therapeutic strategies in patients with HCM. It remains unclear why patients with sarcomere mutation negative HCM develop myocardial hypertrophy and if different treatment strategies are required for this patient population.
In order to explore these questions, we comprehensively evaluated a cohort of probands with HCM to identify similarities and differences between subjects with sarcomere mutation positive and negative HCM.
Methods
Study population
The diagnosis of HCM in proband subjects (n=127) was defined using the following criteria: left ventricular hypertrophy with a maximal wall thickness (LVWTmax) ≥15 mm in the absence of aortic stenosis or hypertension. In subjects with a previous diagnosis of hypertension, evidence of asymmetric myocardial hypertrophy as defined by an interventricular septal thickness to left ventricular posterior wall thickness ratio >1.5 and LVWTmax >15 mm were required. All patients met with a physician and genetic counsellor and a three generation family history was documented. All subjects had a comprehensive physical examination, seated non-invasive blood pressure measurement, 12-lead ECG and Doppler echocardiography performed. Subjects were excluded from our analysis if they were not screened for sarcomere gene mutations or if they did not have complete diagnostic imaging to verify phenotype status.
Evaluation for sarcomere gene mutations
After appropriate genetic counselling and following patient consent, blood samples were obtained and sent to either Harvard Partners Laboratory of Molecular Medicine (Boston, Massachusetts, USA) or GeneDx (Gaithersburg, Maryland, USA) for sequencing and variant classification following American College of Medical Genetics-accepted guidelines.13 The following genes were sequenced in all subjects: ACTC1, MYBPC3, MYH7, MYL2, MYL3, TNNC1, TNNI3, TNNT2, TPM1, PRKAG2, LAMP2, GLA and TTR.14 Subjects who tested positive for pathogenic DNA variants in PRKAG2, LAMP2, GLA or TTR genes were excluded from analysis (n=2). Sarcomere gene variants classified as pathogenic, or likely pathogenic, were considered sarcomere gene mutations. All DNA variants classified as sarcomere mutations had a minor allele frequency <1×10−4.15
Echocardiographic imaging
Doppler echocardiography was performed on all subjects to assess LVWTmax, posterior or minimum left ventricular wall thickness, left ventricular function and the measurement of the peak left ventricular outflow tract (LVOT) gradient. LVOT obstruction was defined as peak gradient ≥30 mm Hg at rest or with exercise.
Statistical analysis
Mann-Whitney U test was used to assess continuous variables and Fisher’s exact test was used to assess categorical data. To adjust for age, we performed multiple regression analyses for each outcome individually with sarcomere mutation as the main predictor. Age was adjusted for by entering into the model with non-linear effects using the restricted cubic spline.
Results
Family history of HCM is much less common in subjects with sarcomere mutation negative HCM
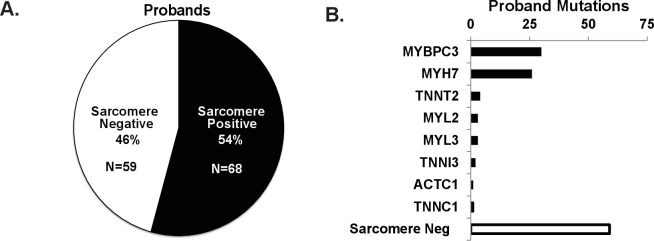
Of the 127 probands with HCM who underwent genetic screening for sarcomere gene mutations, 54% (n=68) tested positive for a pathogenic sarcomere mutation and 46% (n=59) tested negative for a sarcomere mutation (figure 1A and online supplemental table S1). The most common sarcomere genes with mutations were Myosin Binding Protein C3 (MYBPC3) and Myosin Heavy Chain 7 (MYH7) (figure 1B). Family history of HCM was over three times more common in subjects with sarcomere positive HCM compared with subjects with sarcomere negative HCM (p<0.0001, figure 2).
Figure 1.
Sarcomere mutation status and family history of HCM. (A) Sarcomere mutation prevalence in subjects with a clinical diagnosis of HCM (n=127). (B) Distribution of mutations across sarcomere genes screened. HCM, hypertrophic cardiomyopathy.
Figure 2.
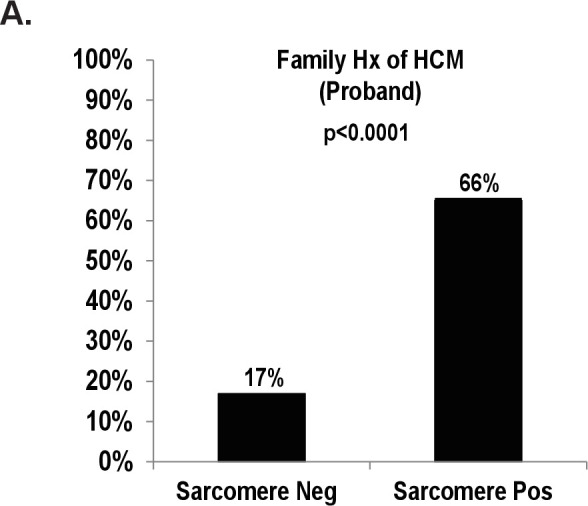
Family history of HCM in sarcomere mutation negative and positive probands. (A) Subjects with sarcomere mutation negative HCM and sarcomere mutation positive HCM with a family history of HCM (%). HCM, hypertrophic cardiomyopathy.
openhrt-2020-001560supp001.pdf (27.4KB, pdf)
Patients with sarcomere mutation negative HCM present at an older age
Proband subjects with sarcomere positive HCM (n=68) were diagnosed 18 years earlier than probands with sarcomere negative HCM (n=59) (median 32.5 vs 51 years old, p<0.0001, table 1). To control for an effect of family history on age of presentation, we performed a subgroup analysis of probands with only HCM with no family history of HCM (n=72). We found that probands with HCM with sarcomeric mutations but with no family history of HCM still presented 15 years earlier in life compared with probands with sarcomere negative HCM (median 37 vs 52 years old, p=0.0004). Therefore, probands with HCM with sarcomeric mutations presented at an earlier age regardless of family history. Both sarcomere positive and negative probands had a similar gender distribution. There was a slightly higher blood pressure in the probands with sarcomere negative HCM (~4 mm Hg), but when adjusted for age, this was not statistically different.
Table 1.
Baseline characteristics of proband subjects with HCM
| Sarcomere negative N=59 |
Sarcomere positive N=68 |
P value | Adjusted p value | |
| Age at diagnosis (years) | 51.0 (39.5,59.0) 48±14.5 |
32.5 (16.8,46.3) 32±16.8 |
<0.0001 | |
| Male (n) | 38 (64%) | 35 (51%) | 0.15 | |
| Systolic BP (mm Hg) | 123 (112,137) 126.5±18.9 |
120 (107,128) 119±16.3 |
0.0253 | 0.398 |
| Diastolic BP (mm Hg) | 74 (67,81) 74.3±9.5 |
70 (62,78) 69.5±10.1 |
0.0242 | 0.196 |
| Mean arterial pressure (mm Hg) | 91.3 (83.3,99.2) 91.7±11.4 |
86.7 (78.3,93.3) 85.9±10.9 |
0.0163 | 0.232 |
| BSA (m2) | 2.14 (1.95,2.22) 2.09±0.25 |
1.91 (1.73,2.11) 1.90±0.33 |
0.0008 | 0.003 |
| BMI (kg/m2) | 31.1 (27.7,34.9) 31.7±5 |
27.7 (23.3,33.7) 28.2±6.9 |
0.0026 | 0.037 |
All values expressed as median (25% percentile, 75% percentile) and mean±SD.
BMI, body mass index; BP, blood pressure; BSA, body surface area; HCM, hypertrophic cardiomyopathy.
Patients with sarcomere mutation negative HCM are more obese
In addition to the sarcomere negative probands presenting at an older age, these subjects were also more obese with larger body surface area (BSA) and body mass index (BMI) (table 1, age adjusted p=0.003 (BSA) and p=0.037 (BMI)).
Patients with sarcomere mutation negative HCM are more likely to have LVOT obstruction
The degree of asymmetric myocardial hypertrophy was less in the sarcomere mutation negative subjects and there was a trend to lower LVWTmax (table 2). When LVWTmax was indexed to BSA, there was a significant difference in the groups, reflective of the increased obesity in sarcomere mutation negative subjects (table 2). LVOT obstruction (>30 mm Hg) was more prevalent in subjects with sarcomere mutation negative HCM (table 2).
Table 2.
Myocardial hypertrophy and LVOT obstruction
| Sarcomere negative N=59 |
Sarcomere positive N=68 |
P value | |
| LVWTmax (mm)* | 20 (18,22) 20.3±4.1 |
21 (18,27) 22.6±6.5 |
0.0686 |
| Ratio LVWTmax:LVWTmin* | 1.8 (1.57,2.1) 1.84±0.4 |
2.1 (1.66,3.0) 2.48±1.1 |
0.0011 |
| LVWTmax/BSA (mm/mm2)* | 9.6 (8.1,10.7) 9.9±2.3 |
10.9 (8.9,13.8) 12.3±4.8 |
0.0006 |
| LVOT obstruction (%)† | 32 (54) | 24 (35) | 0.0483 |
| LVOT gradient (mm Hg)* | 105 (43,141) 98.5±56.3 |
71 (43,91) 75±51.3 |
0.1050 |
*Values expressed as median (25% percentile, 75% percentile) and mean±SD.
†LVOT obstruction defined as LVOT gradient >30 mm Hg.
BSA, body surface area; LVOT, left ventricular outflow tract; LVWTmax, maximum left ventricular wall thickness; LVWTmin, minimum left ventricular wall thickness.
Discussion
There have been significant advances in our understanding of the genetic and molecular mechanisms regulating the development of HCM caused by sarcomere gene mutations. However, approximately half of all patients with HCM who undergo genetic screening do not have a sarcomere mutation and the cause of HCM in these individuals remains poorly defined. We compared HCM with and without sarcomere mutations and discovered that family history, age of diagnosis and obesity were quite different between the cohorts.
We found that probands with sarcomere mutation positive HCM were much more likely to have a family history of HCM. This correlation between family history and sarcomere mutation status in HCM has also been reported in HCM cohorts from other centres.16–20 It would be logical that since sarcomere mutations are associated with an autosomal dominant inheritance pattern that patients who harbour sarcomere mutations would be likely to have a family history of disease. However, the much lower rate of clinically affected family members in pedigrees of patients with sarcomere negative HCM suggests that the cause of asymmetric myocardial hypertrophy may be related to multiple different factors. One possibility is that these patients harbour germline mutations with very low penetrance that are not routinely screened for. Alternatively, the more likely explanation is that there are non-heritable somatic mutations or non-genetic environmental factors (or a combination of these two factors) that contribute to the development of asymmetric myocardial hypertrophy in these patients. Our results suggest that ageing and obesity may serve as potential non-genetic factors that contribute to disease development in patients with sarcomere negative HCM.
We found that subjects with sarcomere negative HCM were more obese compared with subjects with sarcomere positive HCM. Supporting our results is earlier work which showed that obesity is independently associated with an increase in left ventricular mass and may affect progression of heart failure in a cohort of subjects with non-genotyped HCM.21 22 More recently, Fumagalli et al again demonstrated that obesity is associated with long-term adverse events in HCM overall.23 Our results suggest that obesity may have a greater impact on disease development in the population with sarcomere negative HCM and this finding was independent of age. Obesity has been associated with increased heart mass and myocardial wall thickness in multiple studies and remains an independent risk factor for left ventricular hypertrophy after controlling for other comorbidities.24–26 In addition, myocardial hypertrophy has been shown to be reversible in morbidly obese individuals after aggressive weight loss.27 These data further confirm that obesity contributes to myocardial growth. Multiple mechanisms have been proposed for how obesity increases myocardial growth. These include changes in systemic haemodynamics and alterations in circulating hormone levels, both of which can directly alter cardiomyocyte growth. Further investigation is required to determine how obesity influences asymmetric myocardial growth in HCM and if aggressive weight loss can reverse this remodelling process.
In addition to obesity, we also found that patients with sarcomere negative HCM were diagnosed much later in life. Importantly, we found that the later age of presentation in probands with sarcomere negative HCM persisted even when individuals with a family history of HCM were excluded from the comparison. Therefore, family history does not appear to be confounder of this association. These results suggest that another potential driver of asymmetric myocardial hypertrophy in HCM may be related to normal ageing. Indeed, ageing has been associated with increasing left ventricular hypertrophic remodelling.28–30 Similar to obesity, there are multiple mechanisms through which ageing could increase left ventricular hypertrophy. Some proposed mechanisms are ageing-related increased vascular stiffness, myocardial fibrosis and ageing-related cardiomyocyte dysfunction.30 It remains to be elucidated how ageing influences the asymmetric myocardial hypertrophy seen in HCM.
Limitations
This was a cohort that was collected from a single centre and therefore may not be representative of patient populations in other geographical areas. However, it should be noted that the distribution of sarcomere gene mutations in our cohort was similar to datasets from other cohorts in the USA and Europe, with MYBPC3 and MYH7 gene mutations accounting for ~80% of the identified mutations in our probands with HCM.6 In addition, we found a similar trend with other studies with regard to patients with sarcomere positive HCM being much more likely to have a family history of HCM.16–20
Conclusion
A large portion of patients with HCM do not test positive for sarcomere gene mutations and the cause of disease in these individuals remains elusive. In our study, we found that patients with sarcomere negative HCM are much less likely to have a family history of HCM, present later in life and are more obese than patients with HCM with sarcomere gene mutations. It will be important to investigate how obesity and ageing influence asymmetric myocardial remodelling and if therapies targeting these pathways can benefit this patient population.
Acknowledgments
The authors would like to acknowledge statistical support from Jianhui Zhu, UPMC Heart and Vascular Institute.
Footnotes
Twitter: @deFeriaMD, @kottmd
Contributors: AEdF, AEK and JRB designed the study, collected data, performed the analysis and wrote the manuscript.
Competing interests: None declared.
Patient consent for publication: Not required.
Ethics approval: The institutional review board at Vanderbilt University School of Medicine approved this study.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data availability statement: Data are available upon reasonable request.
References
- 1.Maron BJ, Gardin JM, Flack JM, et al. Prevalence of hypertrophic cardiomyopathy in a general population of young adults. echocardiographic analysis of 4111 subjects in the cardia study. coronary artery risk development in (young) adults. Circulation 1995;92:785–9. 10.1161/01.cir.92.4.785 [DOI] [PubMed] [Google Scholar]
- 2.Seidman CE, Seidman JG. Identifying sarcomere gene mutations in hypertrophic cardiomyopathy: a personal history. Circ Res 2011;108:743–50. 10.1161/CIRCRESAHA.110.223834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maron BJ, Maron MS, Semsarian C. Genetics of hypertrophic cardiomyopathy after 20 years: clinical perspectives. J Am Coll Cardiol 2012;60:705–15. 10.1016/j.jacc.2012.02.068 [DOI] [PubMed] [Google Scholar]
- 4.Sabater-Molina M, Pérez-Sánchez I, Hernández Del Rincón JP, et al. Genetics of hypertrophic cardiomyopathy: a review of current state. Clin Genet 2018;93:3–14. 10.1111/cge.13027 [DOI] [PubMed] [Google Scholar]
- 5.Gersh BJ, Maron BJ, Bonow RO, et al. 2011 ACCF/AHA guideline for the diagnosis and treatment of hypertrophic cardiomyopathy: Executive summary: a report of the American College of cardiology Foundation/American heart association Task force on practice guidelines. Circulation 2011;124:2761–96. 10.1161/CIR.0b013e318223e230 [DOI] [PubMed] [Google Scholar]
- 6.Alfares AA, Kelly MA, McDermott G, et al. Results of clinical genetic testing of 2,912 probands with hypertrophic cardiomyopathy: expanded panels offer limited additional sensitivity. Genet Med 2015;17:880–8. 10.1038/gim.2014.205 [DOI] [PubMed] [Google Scholar]
- 7.Fatkin D, McConnell BK, Mudd JO, et al. An abnormal Ca(2+) response in mutant sarcomere protein-mediated familial hypertrophic cardiomyopathy. J Clin Invest 2000;106:1351–9. 10.1172/JCI11093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lan F, Lee AS, Liang P, et al. Abnormal calcium handling properties underlie familial hypertrophic cardiomyopathy pathology in patient-specific induced pluripotent stem cells. Cell Stem Cell 2013;12:101–13. 10.1016/j.stem.2012.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crilley JG, Boehm EA, Blair E, et al. Hypertrophic cardiomyopathy due to sarcomeric gene mutations is characterized by impaired energy metabolism irrespective of the degree of hypertrophy. J Am Coll Cardiol 2003;41:1776–82. 10.1016/S0735-1097(02)03009-7 [DOI] [PubMed] [Google Scholar]
- 10.Sequeira V, Wijnker PJM, Nijenkamp LLAM, et al. Perturbed length-dependent activation in human hypertrophic cardiomyopathy with missense sarcomeric gene mutations. Circ Res 2013;112:1491–505. 10.1161/CIRCRESAHA.111.300436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tyska MJ, Hayes E, Giewat M, et al. Single-molecule mechanics of R403Q cardiac myosin isolated from the mouse model of familial hypertrophic cardiomyopathy. Circ Res 2000;86:737–44. 10.1161/01.RES.86.7.737 [DOI] [PubMed] [Google Scholar]
- 12.Spudich JA, Aksel T, Bartholomew SR, et al. Effects of hypertrophic and dilated cardiomyopathy mutations on power output by human β-cardiac myosin. J Exp Biol 2016;219:161–7. 10.1242/jeb.125930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of medical genetics and genomics and the association for molecular pathology. Genet Med 2015;17:405–23. 10.1038/gim.2015.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walsh R, Buchan R, Wilk A, et al. Defining the genetic architecture of hypertrophic cardiomyopathy: re-evaluating the role of non-sarcomeric genes. Eur Heart J 2017;38:ehw603. ehw603 [pii]. 10.1093/eurheartj/ehw603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walsh R, Thomson KL, Ware JS. Reassessment of Mendelian gene pathogenicity using 7,855 cardiomyopathy cases and 60,706 reference samples. Genet Med 2016. gim201690 [pii] (published Online First: 2016/08/18). [DOI] [PMC free article] [PubMed]
- 16.Gruner C, Ivanov J, Care M, et al. Toronto hypertrophic cardiomyopathy genotype score for prediction of a positive genotype in hypertrophic cardiomyopathy. Circ Cardiovasc Genet 2013;6:19–26. 10.1161/CIRCGENETICS.112.963363 [DOI] [PubMed] [Google Scholar]
- 17.Bos JM, Will ML, Gersh BJ, et al. Characterization of a phenotype-based genetic test prediction score for unrelated patients with hypertrophic cardiomyopathy. Mayo Clin Proc 2014;89:727–37. 10.1016/j.mayocp.2014.01.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ingles J, Sarina T, Yeates L, et al. Clinical predictors of genetic testing outcomes in hypertrophic cardiomyopathy. Genet Med 2013;15:972–7. 10.1038/gim.2013.44 [DOI] [PubMed] [Google Scholar]
- 19.Lopes LR, Syrris P, Guttmann OP, et al. Novel genotype-phenotype associations demonstrated by high-throughput sequencing in patients with hypertrophic cardiomyopathy. Heart 2015;101:294–301. 10.1136/heartjnl-2014-306387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ko C, Arscott P, Concannon M, et al. Genetic testing impacts the utility of prospective familial screening in hypertrophic cardiomyopathy through identification of a nonfamilial subgroup. Genet Med 2018;20:69–75. 10.1038/gim.2017.79 [DOI] [PubMed] [Google Scholar]
- 21.Olivotto I, Maron BJ, Tomberli B, et al. Obesity and its association to phenotype and clinical course in hypertrophic cardiomyopathy. J Am Coll Cardiol 2013;62:449–57. 10.1016/j.jacc.2013.03.062 [DOI] [PubMed] [Google Scholar]
- 22.Lauer MS, Anderson KM, Kannel WB, et al. The impact of obesity on left ventricular mass and geometry. The Framingham heart study. JAMA 1991;266:231–6. [PubMed] [Google Scholar]
- 23.Fumagalli C, Maurizi N, Day SM, et al. Association of obesity with adverse long-term outcomes in hypertrophic cardiomyopathy. JAMA Cardiol 2020;5:65–72. 10.1001/jamacardio.2019.4268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wong CY, O'Moore-Sullivan T, Leano R, et al. Alterations of left ventricular myocardial characteristics associated with obesity. Circulation 2004;110:3081–7. 10.1161/01.CIR.0000147184.13872.0F [DOI] [PubMed] [Google Scholar]
- 25.Woodiwiss AJ, Libhaber CD, Majane OHI, et al. Obesity promotes left ventricular concentric rather than eccentric geometric remodeling and hypertrophy independent of blood pressure. Am J Hypertens 2008;21:1144–51. 10.1038/ajh.2008.252 [DOI] [PubMed] [Google Scholar]
- 26.Aurigemma GP, de Simone G, Fitzgibbons TP. Cardiac remodeling in obesity. Circ Cardiovasc Imaging 2013;6:142–52. 10.1161/CIRCIMAGING.111.964627 [DOI] [PubMed] [Google Scholar]
- 27.Ippisch HM, Inge TH, Daniels SR, et al. Reversibility of cardiac abnormalities in morbidly obese adolescents. J Am Coll Cardiol 2008;51:1342–8. 10.1016/j.jacc.2007.12.029 [DOI] [PubMed] [Google Scholar]
- 28.Ganau A, Saba PS, Roman MJ, et al. Ageing induces left ventricular concentric remodelling in normotensive subjects. J Hypertens 1995;13:1818–22. 10.1097/00004872-199512010-00058 [DOI] [PubMed] [Google Scholar]
- 29.Cheng S, Fernandes VRS, Bluemke DA, et al. Age-related left ventricular remodeling and associated risk for cardiovascular outcomes: the multi-ethnic study of atherosclerosis. Circ Cardiovasc Imaging 2009;2:191–8. 10.1161/CIRCIMAGING.108.819938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lakatta EG, Levy D. Arterial and cardiac aging: major shareholders in cardiovascular disease enterprises: Part II: the aging heart in health: links to heart disease. Circulation 2003;107:346–54. 10.1161/01.cir.0000048893.62841.f7 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
openhrt-2020-001560supp001.pdf (27.4KB, pdf)