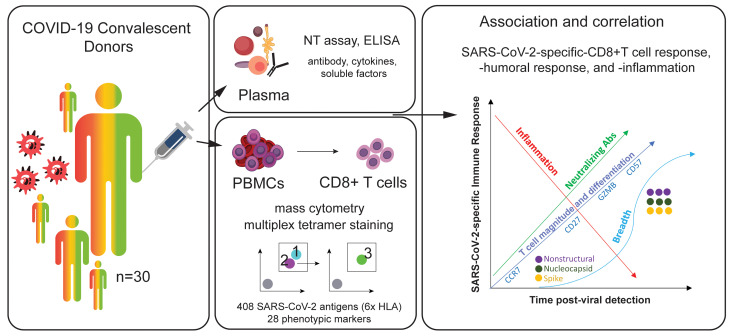
Abstract
Characterization of the T cell response in individuals who recover from severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection is critical to understanding its contribution to protective immunity. A multiplexed peptide-MHC tetramer approach was used to screen 408 SARS-CoV-2 candidate epitopes for CD8+ T cell recognition in a cross-sectional sample of 30 coronavirus disease 2019 convalescent individuals. T cells were evaluated using a 28-marker phenotypic panel, and findings were modelled against time from diagnosis and from humoral and inflammatory responses. There were 132 SARS-CoV-2–specific CD8+ T cell responses detected across 6 different HLAs, corresponding to 52 unique epitope reactivities. CD8+ T cell responses were detected in almost all convalescent individuals and were directed against several structural and nonstructural target epitopes from the entire SARS-CoV-2 proteome. A unique phenotype for SARS-CoV-2–specific T cells was observed that was distinct from other common virus-specific T cells detected in the same cross-sectional sample and characterized by early differentiation kinetics. Modelling demonstrated a coordinated and dynamic immune response characterized by a decrease in inflammation, increase in neutralizing antibody titer, and differentiation of a specific CD8+ T cell response. Overall, T cells exhibited distinct differentiation into stem cell and transitional memory states (subsets), which may be key to developing durable protection.
Keywords: COVID-19, Immunology
Keywords: Cellular immune response, MHC class 1, T cells
Introduction
The emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) rapidly evolved into a global pandemic. To date, over 75 million cases spanning 191 countries or territories have been reported, with more than 1.6 million deaths attributed to coronavirus disease 2019 (COVID-19). The clinical spectrum of SARS-CoV-2 infection is highly variable, spanning from asymptomatic or subclinical infection to severe or fatal disease (1, 2). Characterization of the immune response to SARS-CoV-2 is urgently needed in order to better inform more effective treatment strategies, including antivirals and rationally designed vaccines.
Antibody responses to SARS-CoV-2 have been shown to be heterogenous, whereby male sex, advanced age, and hospitalization status are associated with higher titers of antibodies (3). Low or even undetectable neutralizing antibodies in some individuals with rapid decline in circulating antibodies to SARS-CoV-2 after resolution of symptoms underscores the need to assess the role of the cellular immune response (4). Multiple studies suggest that T cells are important in the immune response against SARS-CoV-2, and may mediate long-term protection against the virus (5–9).
To date, studies that have evaluated SARS-CoV-2–specific T cells in convalescent individuals have focused on either characterization of responses to selected, well-defined SARS-CoV-2 epitopes, or broad assessment of T cell reactivity against overlapping peptide libraries (6–10). The assessment of the complete SARS-CoV-2 reactive T cell pool in the circulation remains challenging, and there is still much to be learned from capturing both the breadth (i.e., number of epitope-specific T cell responses recognized) and depth of T cell response (i.e., comprehensive phenotype) to natural SARS-CoV-2 infection. A study by Peng et al. indicated that the majority of those who recover from COVID-19 exhibit robust and broad SARS-CoV-2–specific T cell responses (8). Further, those who manifest mild symptoms displayed a greater proportion of polyfunctional CD8+ T cell responses compared with severely diseased cases, suggesting a role of CD8+ T cells in ameliorating disease severity.
Many current COVID-19 vaccine candidates primarily incorporate the SARS-CoV-2 spike protein to elicit humoral immunity (11–13). However, whether these approaches will induce long-term protection against SARS-CoV-2 infection or severe COVID-19 remain unknown. Gaining insight into the immune response that is induced by natural SARS-CoV-2 infection will be key to advancing vaccine design. Specifically, there is a need to identify what T cells are targeting in the viral proteome, their functional characteristics, and how these might correlate with disease outcomes. In this study, our analytical strategy progressed beyond these earlier findings by identifying dozens of epitopes recognized by CD8+ T cells that spanned different viral proteins in COVID-19 convalescent subjects, and simultaneously revealed the unmanipulated phenotypic profiles of these cells. These new findings can be exploited to further guide epitope selection for rationally designed vaccine candidates and vaccine assessment strategies.
Results
SARS-CoV-2–specific CD8+ T cell response in COVID-19 convalescent donors is broad and targets the whole-virus proteome.
To study the SARS-CoV-2–specific CD8+ T cell repertoire in COVID-19 convalescent donors, a mass cytometry–based multiplexed tetramer staining approach was employed to identify and characterize (i.e., phenotype) SARS-CoV-2–specific T cells ex vivo. A total of 30 convalescent plasma donors (confirmed by PCR at time of infection) with HLA-A*01:01, HLA-A*02:01, HLA-A03:01, HLA-A*11:01, HLA-A*24:02, and HLA-B*07:02 alleles were evaluated (3). The individuals included 18 males and 12 females ranging between 19 and 77 years old who were a median of 42.5 days (interquartile range 37.5–48.0 days) from initial diagnosis (Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/JCI145476DS1). The population was grouped into tertiles according to their overall anti–SARS-CoV-2 IgG titers, based on semiquantitative ELISA results against SARS-CoV-2 S protein (Supplemental Table 2). Additional plasma-derived parameters such as neutralizing antibody titers, inflammatory cytokines, and chemokines were used to associate the cellular SARS-CoV-2–specific T cell response with the humoral and inflammatory response. There was a strong correlation between the donors’ anti-S IgG levels and the neutralizing antibody activity (Supplemental Figure 1A). Levels of some inflammatory mediators were associated with age, sex, neutralizing antibody activity, and neutralizing antibody titers (Supplemental Figure 1, B–D).
Hundreds of candidate epitopes spanning the complete SARS-CoV-2 genome were recently identified as potential targets for a CD8+ T cell response to SARS-CoV-2 (14, 15). A triple-coded multiplexed peptide-MHC tetramer staining approach was used to screen 408 potential epitopes for recognition by T cell responses across 6 different HLA alleles: HLA-A*01:01, HLA-A*02:01, HLA-A03:01, HLA-A*11:01, HLA-A*24:02, and HLA-B*07:02 (16, 17). In addition, CD8+ T cells were probed for reactivity against up to 20 different SARS-CoV-2–unrelated control peptides per HLA for each sample (CMV-, EBV-, influenza-, adenovirus-, and MART-1–derived epitopes; Supplemental Table 3). The detection of bona fide antigen-specific T cells was based on the assessment of several objective criteria such as signal versus noise, consistency between 2 technical replicates, and detection threshold. In this study, an average limit of detection of 0.0024% (bootstrapping confidence interval of 0.0017 and 0.005 under a confidence level of 95%) was achieved for antigen-specific T cells. Depending on the individual’s HLA allele repertoire, between 48 and 220 peptides were simultaneously screened per participant.
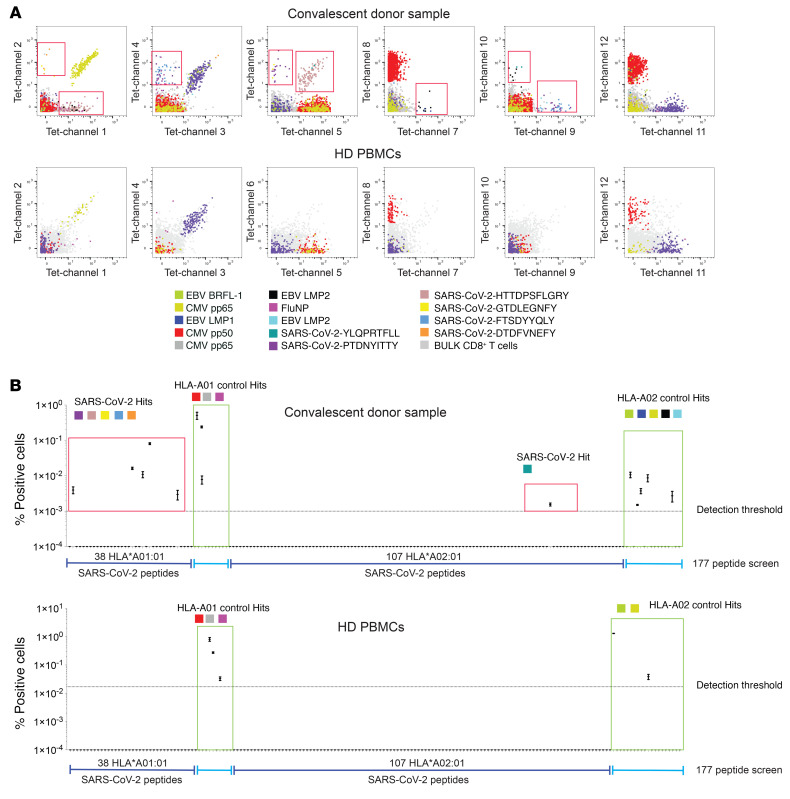
Figure 1 shows an example of the identification of antigen-specific CD8+ T cells in a COVID-19 convalescent donor screened for a total of 145 SARS-CoV-2 antigen candidates and 32 common (SARS-CoV-2 unrelated) control antigens across 2 HLA alleles. CD8+ T cells reactive to 6 different SARS-CoV-2 epitopes and 8 control antigens were detected, including peptides derived from influenza (FLU), Epstein Barr virus (EBV), and cytomegalovirus (CMV). In parallel, commercially obtained healthy donor PBMCs were run and similar common virus antigen specificities were identified. Notably, SARS-CoV-2–specific CD8+ T cells were not detected in any of the healthy donors recruited before the official SARS-CoV-2 pandemic (n = 4).
Figure 1. Identification and characterization of SARS-CoV-2–specific CD8+ T cells from SARS-CoV-2 convalescent donors.
(A) Representative staining for SARS-CoV-2–specific CD8+ T cells from a convalescent donor sample. Healthy donor PBMCs were run in parallel. Red boxes indicate SARS-CoV-2–specific T cell hits. (B) Screening example probing for 145 SARS-CoV-2 candidate antigens (HLA-A02 and HLA01) and 31 SARS-CoV-2–unrelated control antigens. Screening data show the values and means from the 2 technical replicates (2 staining configurations). Bona fide antigen-specific T cells were defined based on different objective criteria set (see Methods).
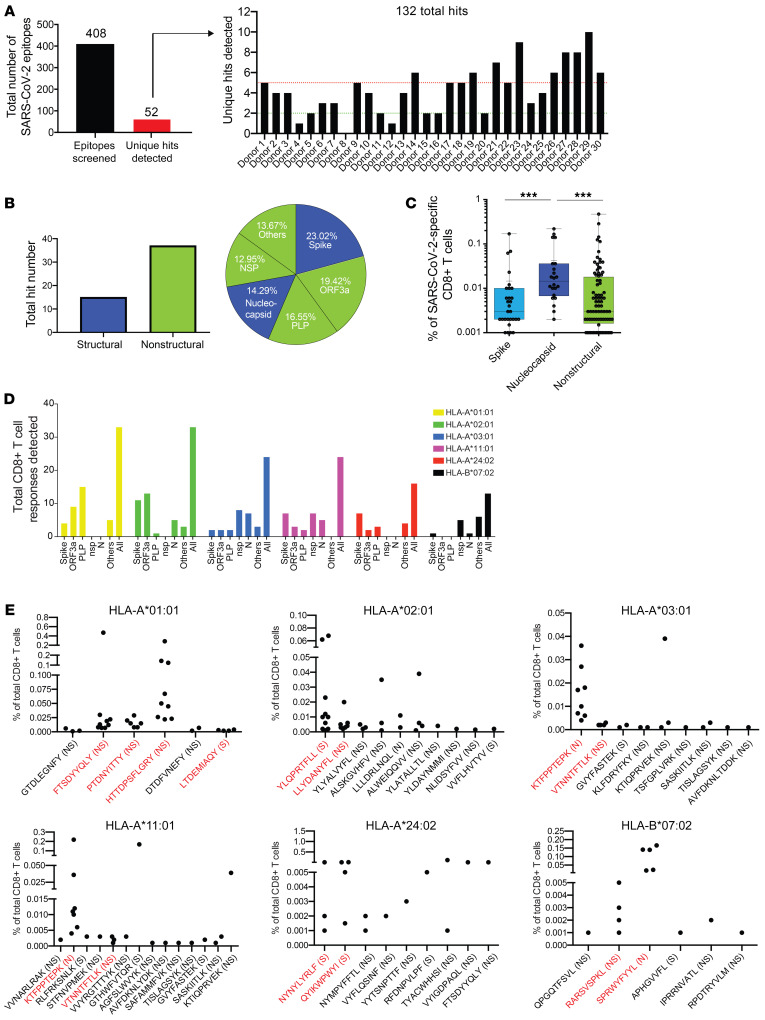
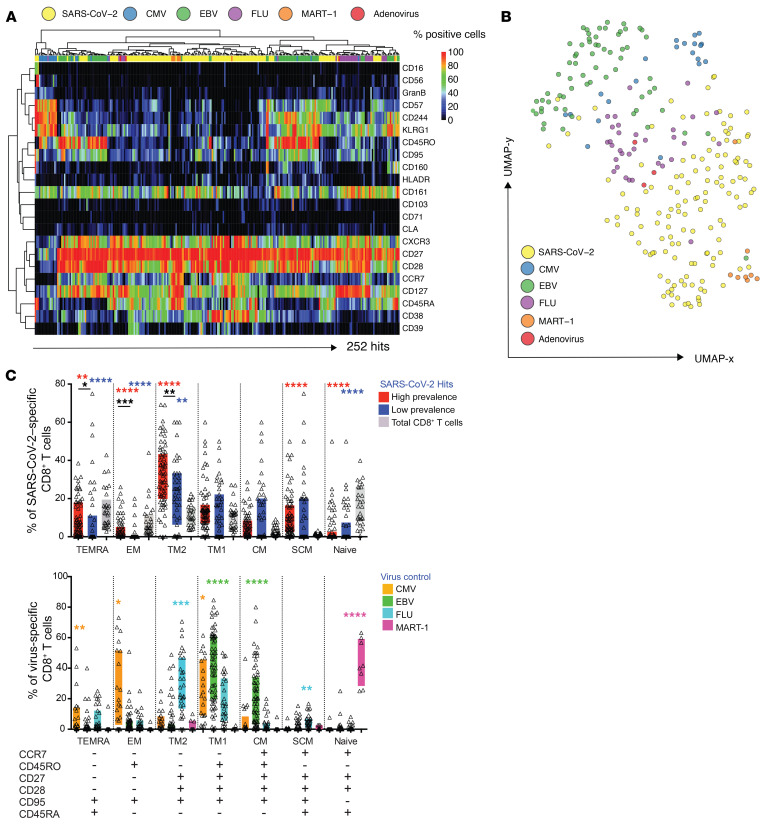
Among all 408 SARS-CoV-2 peptide candidates tested in the 30 convalescent donor samples, we detected 52 unique epitope reactivities (hits) out of a total of 132 SARS-CoV-2 T cell responses (Figure 2A). Almost all individuals screened demonstrated a CD8+ T cell response against SARS-CoV-2 (29/30), and individual hits ranged from 0 to 13 with greater than 40% of all individuals showing more than 5 different SARS-CoV-2 specificities. The frequency of these cells ranged from 0.001% to 0.471% of total CD8+ T cells (Supplemental Table 4). In addition, a total of 130 T cell hits against common control peptides were detected in these donor samples (0.001% to 1.074% of total CD8+ T cells, Supplemental Table 4). Interestingly, the majority of unique T cell hits were directed against epitopes associated with nonstructural proteins (NSP) such as papain-like protease (PLP) and open reading frame 3a protein (ORF3a) (Figure 2B). Of all the hits that were detected in the cross-sectional sample, the most common reactivities were against spike (structural, 23.02%) and ORF3a (nonstructural, 19.42%). By contrast, nucleocapsid-specific CD8+ T cells had significantly higher frequencies as compared with spike- or NSP-specific T cells, which was driven primarily through their detection in HLA-A*03:01-, HLA-A*11:01-, and HLA-B*07:02-positive donor samples (Figure 2C and Supplemental Figure 2A). The total number of epitopes targeted was distributed differently across the individual HLA alleles that were tested (Figure 2D and Supplemental Figure 2B), whereby T cell responses were identified against 6 to 14 different epitopes per allele (Figure 2E). For the purpose of the study, events detected in at least 3 donor samples or in more than 35% of donors for each allele group were defined as SARS-CoV-2 high-prevalence epitope hit responses.
Figure 2. Breadth and magnitude of SARS-CoV-2–specific CD8+ T cells.
(A) Bar plots summarizing the absolute numbers of SARS-CoV-2 antigen specificities detected across donors within cross-sectional sample. Out of 408 SARS-CoV-2 peptide candidates, 52 unique peptide hits were detected. Between 0 and 13 unique hits were detected in each donor sample (5 or more hits in > 40% of all donors). In total, 132 SARS-CoV-2–specific T cell hits were detected. (B) Delineation of T cell reactivities against the SARS-CoV-2 proteome. The majority of epitope hits detected derived from nonstructural SARS-CoV-2 proteins. Pie chart displaying the percentages of epitopes detected derived from structural (nucleocapsid, spike) and nonstructural (NSP, PLP, ORF3a, others) proteins spanning the full proteome of SARS-CoV-2. (C) Frequencies of SARS-CoV-2–specific T cells reactive with epitopes derived from spike, nucleocapsid, and nonstructural proteins. Highest frequencies were detected for T cells targeting peptides from the nucleocapsid protein. Each dot represents 1 hit. ***P < 0.001. Kruskal-Wallis test. P values were adjusted for multiple testing using the Benjamini-Hochberg method to control the false discovery rate. (D) Numbers of epitope-specific T cell responses from the different protein categories detected across all 6 HLA alleles tested. (E) Definition of high- and low-prevalence hits per HLA allele. Plots showing individual peptide hits for each allele. Each dot represents 1 hit. High-prevalence epitope hits are indicated in red and were defined as events detected in at least 3 donor samples or in more than 35% of donors for each allele group. NS, nonstructural; N, nucleocapsid; S, spike.
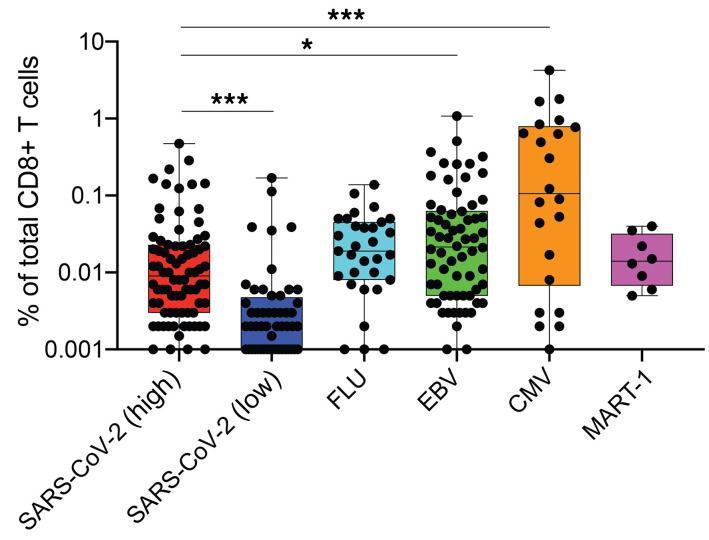
Based on these criteria, at least 2 peptides per HLA allele were defined as high-prevalence response hits (Figure 2E). Of note, the frequencies of high-prevalence SARS-CoV-2–specific T cells were significantly higher as compared with their low-prevalence counterparts (Figure 3). Frequencies of high-prevalence SARS-CoV-2–specific T cells were similar to those of FLU-specific T cells detected in the same cross-sectional sample, but significantly lower than frequencies of T cells reactive for EBV or CMV peptides (Figure 3). In summary, these data show a reliable detection of multiple SARS-CoV-2 T cell hits and indicate a broad recognition of epitopes by CD8+ T cell responses against the SARS-CoV-2 proteome during recovery from COVID-19.
Figure 3. Comparison of frequencies of SARS-CoV-2–specific T cell and T cells reactive with influenza, EBV, CMV, or endogenous MART-1 epitopes.
The percentage of SARS-CoV-2–specific T cells was higher for epitopes categorized as high-prevalence hits but lower than the frequencies of T cells reactive with EBV or CMV antigens detected. *P < 0.05, ***P < 0.001. Kruskal-Wallis test. P values were adjusted for multiple testing using the Benjamini-Hochberg method to control the false discovery rate.
SARS-CoV-2–specific CD8+ T cells exhibit a unique phenotype and can be classified into different memory subsets.
Our multiplexed tetramer staining approach enables deep phenotypic characterization of antigen-specific T cells. By using a panel comprising 28 markers that were dedicated to T cell identification and profiling, including several markers indicative of T cell differentiation (Supplemental Table 5), the phenotypic profiles of all SARS-CoV-2–specific T cells detected in this cross-sectional sample were further analyzed.
To compare the phenotypes of antigen-specific T cells targeting different SARS-CoV-2 proteins, the frequencies of T cells expressing all markers were determined (Figure 4A). Despite some phenotypic heterogeneity, the majority of SARS-CoV-2–specific T cells grouped together and were distinct from T cells that were specific for CMV-, EBV-, or FLU-derived epitopes detected in the same samples. The same outcome was reached when displaying the data as a 2D UMAP plot (Figure 4B). SARS-CoV-2–specific T cells showed an intermediate phenotype between MART-1–specific T cells, which are predominantly naive (CCR7hi and CD45RAhi), and memory FLU-specific T cells (18).
Figure 4. SARS-CoV-2–specific CD8+ T cells display a unique phenotype and can be categorized into different subsets.
(A) Heatmap summarizing the expression frequencies of all phenotypic markers analyzed among the total pool of SARS-CoV-2–specific and unrelated control antigen–specific CD8+ T cells detected in the same cross-sectional sample. The majority of SARS-CoV-2–specific T cells cluster differently from common virus-specific T cells. Antigen specificities and phenotypic markers were clustered using Pearson correlation coefficients as distance measure. (B) UMAP plot showing the clustering of all antigen-specific T cells by antigen category. SARS-CoV-2–specific CD8+ T cells occupy the lower region of the 2D map. Clustering is based on the expression of all phenotypic markers assessed. Each dot represents 1 hit. (C) Differentiation profiles of SARS-CoV-2–specific CD8+ T cells and common virus control antigen–specific T cells. Based on the expression of the markers below the bar diagrams, antigen-specific and total CD8+ T cells were categorized into distinct states of differentiation. SARS-CoV-2–specific T cells were enriched in SCM and TM2 cells. Control virus hits could be separated into distinct subsets dependent on the target epitope. *P < 0.1, **P < 0.01, ***P < 0.001, ****P < 0.0001. Wilcoxon rank sum test. SCM, stem cell memory cells; TM, transitional memory cells; TEMRA, terminal effector memory cells reexpressing CD45RA; EM, effector memory cells; CM, central memory cells.
An early differentiated memory phenotype was recently described for SARS-CoV-2–specific T cells (9). SARS-CoV-2–specific T cells were separated into subpopulations based on the stages of T cell differentiation, further split into high- and low-prevalence response hits as defined earlier, and their frequencies compared with one another, as well as with total CD8+ T cells. Likewise, these were compared with the differentiation profiles of T cells reactive against common virus antigens and MART-1. The classification into functionally different T cell subsets following antigen encounter is based on the expression of different marker combinations, which describe a progressive T cell differentiation and allow to delineate a dynamic transition between memory and effector cell function (Figure 4C and Supplemental Figure 3) (19). When compared with the total CD8+ T cell population, SARS-CoV-2–specific T cells were significantly enriched for cells with stem-cell memory (SCM) and transitional memory cells 2 (TM2) phenotypes. More specifically, high-prevalence SARS-CoV-2–specific T cells were skewed toward a phenotype that is typical of terminal effector memory cells reexpressing CD45RA (TEMRA), effector memory cells (EM cells), and TM2 cells, whereas their low-prevalence counterparts were enriched with SCM and central memory (CM) cells. In contrast, MART-1–specific T cells were naive, FLU-specific T cells were predominantly of a TM2 phenotype, EBV-specific T cells were largely characterized by TM1 and CM phenotypes, and CMV-specific T cells were more differentiated as reflected by a strong effector component.
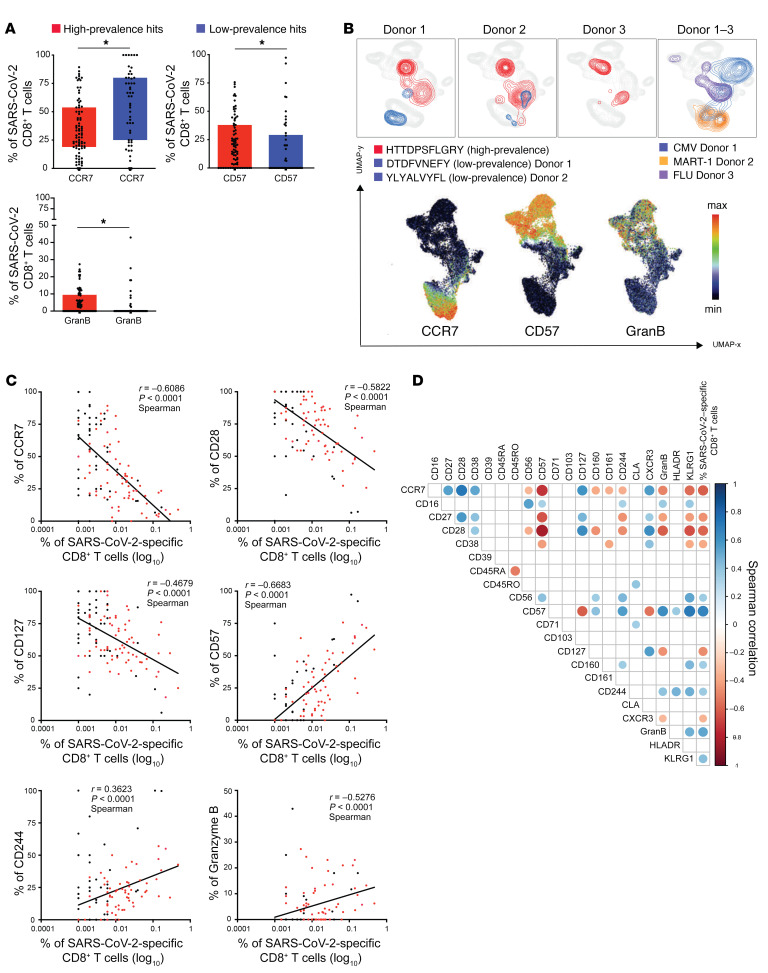
Expansion of highly differentiated SARS-CoV-2–specific CD8+ T cells in convalescent donors.
To gain further insight into the phenotypes of SARS-CoV-2–specific CD8+ T cells, the expression of all the phenotypic markers were compared between T cells exhibiting high-prevalence and those exhibiting low-prevalence epitope responses. Similar to our findings in the total pool of SARS-CoV-2–specific CD8+ T cells, a heterogenous marker expression was detected across these cells, but no specific clustering with respect to the epitope response prevalence (Supplemental Figure 4A). To further compare the phenotypes of T cells from high- versus low-prevalence epitope response categories, the high dimensionality of the data set was reduced and the phenotypic information plotted from Supplemental Figure 4A using principal component analysis (PCA) (Supplemental Figure 4B). The PCA displayed a skewing of high-prevalence SARS-CoV-2–specific T cells toward late T cell differentiation (CD57 and CD45RA), in contrast to the low-prevalence response hits characterized by early differentiation markers (CD27, CD28, CCR7). In order to quantify this spatial distribution, the individual expression of all markers was evaluated and the frequencies for each marker compared between the high- and low-prevalence response hits. Significantly higher frequencies of T cells expressing CD57 and Granzyme B were detected among high-prevalence SARS-CoV-2–specific T cells, whereas the frequencies of CCR7-expressing cells were substantially higher among the low-prevalence hit responses (Figure 5A). These findings were further confirmed when overlaying the SARS-CoV-2–specific T cells on a 2D UMAP plot created based on the full phenotypic panel (Figure 5B). The majority of T cells that had been categorized as high-prevalence response hits were associated with the expression of CD57 and Granzyme B, while their low-frequency counterparts detected in the same donors were characterized by high CCR7 expression.
Figure 5. Expansion of highly differentiated SARS-CoV2–specific CD8+ T cells in convalescent donors.
(A) Box plots showing differences in the expression of markers between high- and low-prevalence response hits. High-prevalent response hits showed a higher expression of markers associated with differentiation. Each dot represents 1 donor. *P < 0.05. Kruskal-Wallis test. P values were adjusted for multiple testing using the Benjamini-Hochberg method to control the false discovery rate. (B) UMAP plot showing the relative position of high- and low-prevalence response hits in the high-dimensional space. Data from 3 donors are shown. (C) Scatterplots showing the correlations between SARS-CoV-2–specific T cell frequencies and differentiation marker expression. The magnitude of antigen-specific T cells correlated with the expression of markers associated with T cell differentiation. The correlations were calculated with the Spearman’s rank-order test. Red dots are high prevalence response hits. (D) Correlogram showing the correlation between all phenotypic markers and frequencies of SARS-CoV-2–specific T cells. Later stage differentiation markers positively correlated with higher frequency SARS-CoV-2–specific T cells. Spearman’s correlation coefficients were indicated by a heat scale whereby blue color shows positive linear correlation, and red color shows negative linear correlation. Only significant correlations are shown (*P < 0.05, P values were adjusted for multiple testing using the Bonferroni method).
High-prevalence SARS-CoV-2–specific T cells were detected at a higher frequency (Figure 3) as compared with their low-prevalence counterparts. Therefore, assessment of the magnitude of the SARS-CoV-2–specific T cell response was also correlated with their phenotypes. Interestingly, a negative correlation between the frequency of SARS-CoV-2–specific CD8+ T cells and the expression of markers associated with early T cell differentiation was observed (CD28, CCR7, CD127, CD27, CD38, and CXCR3; Figure 5, C and D). In contrast, the level of expression of markers that are associated with late-stage T cell differentiation (CD244, CD57, Granzyme B, and KLRG1) correlated positively with increasing frequencies of SARS-CoV-2–specific CD8+ T cells.
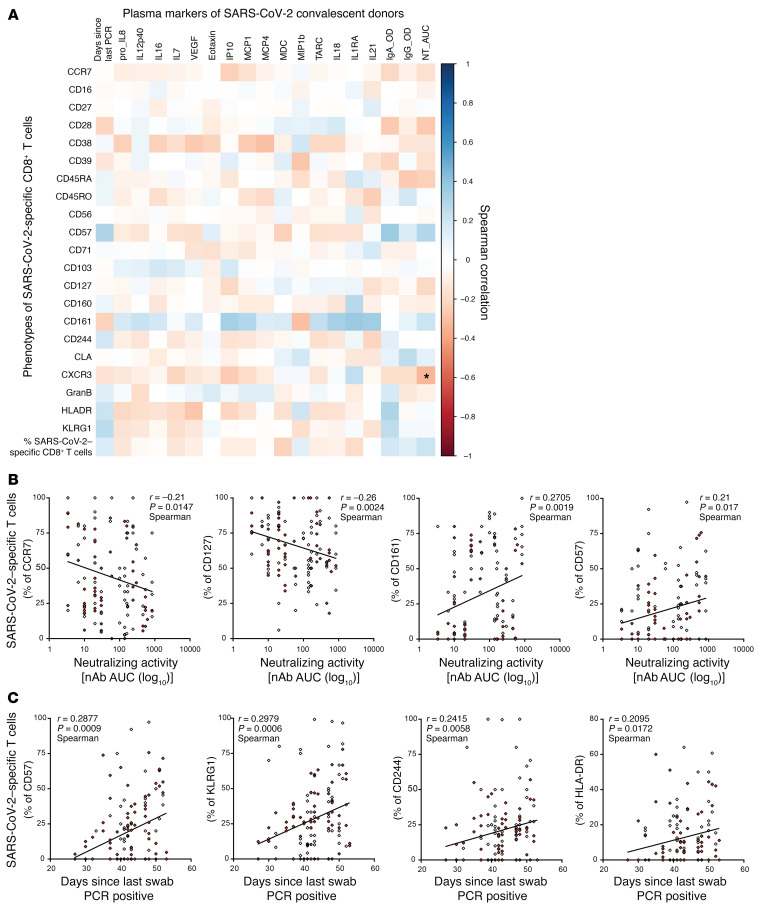
Time-dependent evolution of SARS-CoV-2–specific CD8+ T cell response, inflammation, and humoral immune response.
To examine the relationship between inflammation, humoral immunity, and the T cell response, the frequencies of SARS-CoV-2–specific CD8+ T cells were evaluated against their IgG to Spike titer and neutralizing antibody activity (measured by NT AUC, Figure 6A). Interestingly, although the phenotypic clustering of SARS-CoV-2–specific CD8+ T cells was not associated with IgG titer tertiles (Supplemental Figure 4A), NT AUC correlated negatively with expression of markers associated with an immature or early differentiated phenotype (CCR7, CD28, CD45RA, CD127, CXCR3), while correlating positively with CD57 and CD161 (Figure 6, A and B). Next, assessment of the association between inflammatory molecules and SARS-CoV-2–specific T cells was conducted. Inflammation can indirectly regulate the persistence of antigen-specific T cells in the absence of TCR stimulation or during chronic infection by modulating the homeostatic cytokine profile (20, 21). Overall, the correlation between inflammatory mediators and the expression of individual markers on SARS-CoV-2–specific T cells, or the T cell frequency, remained weak (Figure 6A). Finally, the evolution of the SARS-CoV-2–directed T cell response against time based on the last detection of SARS-CoV-2–specific mRNA was modeled in each donor (Supplemental Table 1). Interestingly, an increase in the breadth of the specific CD8+ T cell response was observed during the resolution phase of the disease, peaking at approximately 6 weeks (Supplemental Figure 5). Longer recovery time was associated with higher frequencies of cells expressing markers of terminal T cell differentiation (CD57, CD244, and KLRG-1) and activation (HLA-DR), indicating a positive correlation between recovery time and T cell maturation (Figure 6, A and C). Plasma levels of several cytokines (IL-18, TARC, MCP-1, VEGF) also decreased over time, suggesting a negative correlation between recovery time and inflammation (Supplemental Figure 1A).
Figure 6. Time-dependent evolution of SARS-CoV-2–specific CD8+ T cell response, inflammation, and humoral immune response.
(A) Correlation matrix showing the associations between frequencies and phenotypic markers of SARS-CoV-2–specific T cells and plasma markers and recovery time (days since PCR). Spearman correlation (blue: positive correlation, red: negative correlation). *P < 0.05, P values were adjusted for multiple testing using the Bonferroni method. (B) Scatterplots showing the correlations between marker expression on SARS-CoV-2–specific T cells and neutralizing antibody activity. Higher expression of markers associated with T cell differentiation was associated with a stronger neutralizing antibody activity. (C) Scatterplots showing the correlations between marker expression on SARS-CoV-2–specific T cells and recovery time. The expression of markers associated with late-stage differentiation correlated with the donors’ recovery time (days since last swab was PCR positive). Correlations were calculated with the Spearman’s rank order test. Red dots indicate high-prevalence response hits.
These data suggest that during early recovery from COVID-19, an overall, time-dependent decrease in inflammation is associated with sustained and effective antibody neutralizing activity with progressive differentiation of a broad and functional SARS-CoV-2–specific CD8+ T cell response (Supplemental Figure 6).
Discussion
An improved understanding of natural immunity to SARS-CoV-2 is needed to advance development of prevention strategies and/or treatment options for COVID-19. Recent findings suggest that T cells confer protection, whereby virus-specific memory T cell responses have been demonstrated in the majority of those who recover from COVID-19 even in the absence of detectable circulating antibodies (9). Moreover, the detection of T cells that are specific for the original SARS-CoV nucleoprotein in patients years after infection highlights the potential role of T cells in generating long-lasting immunity against the virus (7). A mass cytometry–based peptide-MHC-tetramer staining strategy (17) was applied, whereby 408 SARS-CoV-2 candidate epitopes were screened spanning 6 different HLA alleles. This enabled an ex vivo identification and true phenotypic characterization of virus-specific T cells in COVID-19 convalescent individuals without an in vitro culture or stimulation bias, which could affect the cellular phenotype, in contrast to prior studies using overlapping peptide pools (6–8).
The high detection rate of SARS-CoV-2–specific CD8+ T cells across these COVID-19 convalescent donors is consistent with previous reports (6–8, 22). In addition to the detection of T cells reactive with epitopes previously described by others, over a third (i.e., 35%) of the antigen-specific T cells identified here have not been previously reported (Supplemental Table 6), thereby highlighting the sensitivity of the adopted screening approach (8, 9, 22–27). However, given the low frequencies of many of these CD8+ T cells, it is possible that these were below the detection threshold since T cell counts are very low in acutely infected patient samples (28). The T cell response in our study was directed against the full SARS-CoV-2 proteome with the majority of CD8+ T cells targeting epitopes derived from internal and/or nonstructural virus proteins, which is in agreement with the recent findings by others (8, 22). Moreover, half of the high-prevalence response hits identified for each HLA comprised antigens derived from NSPs. In total, 12 highly prevalent SARS-CoV-2–specific CD8+ T cell responses were identified, several of which overlapped with the immunodominant peptides detected by others (8), while some differed by the HLA type or the viral proteins that were assessed. The overall breadth and magnitude of the SARS-CoV-2–specific CD8+ T cell response may depend on the viral load, the severity of the disease, and the priming of the T cell response, which may be affected by the inflammatory environment or the site of the initial priming (i.e., the lungs), resulting in a delayed onset of adaptive immunity and T cell repertoire diversity. Therefore, the collective findings support inclusion of a broad repertoire of SARS-CoV-2 epitopes in future vaccine designs (8).
A unique phenotype for SARS-CoV-2–specific T cells was observed that was distinct from other common virus-specific T cells detected in the same cross-sectional sample. In particular, an enrichment in cells with a stem cell and transitional memory phenotype was observed as compared with total and other virus-specific T cells. A similar early differentiated memory phenotype was recently described for SARS-CoV-2–specific T cells, and was further characterized by polyfunctionality and proliferative capacity (9, 22, 29). The potential of stem cell memory T cells (TSCMs) to differentiate into various T cell memory subsets might contribute to durable protection against SARS-CoV-2 in COVID-19 convalescent donors. The potential role of TSCM in SARS-CoV-2 immune protection remains to be assessed in larger cohorts with longitudinal follow-up studies.
Higher T cell frequencies were observed in high-prevalence epitope responses, and an increased expression of late differentiation markers (CD57, Granzyme B) versus early differentiation markers (CCR7) was observed in high- versus low-prevalence epitope responses, respectively. Overall, the increased expression of markers associated with T cell differentiation correlated with the frequencies of SARS-CoV-2–specific T cells detected in this cross-sectional sample. The evolving profiles of antigen-specific T cell responses during the resolution phase of the disease (i.e., viral clearance and resolution of the inflammation) suggest a continuous proliferation and dynamic differentiation of TSCM into effector memory CD8+ T cells. Our findings bring new insights into the viral targets and dynamics of the SARS-COV-2–specific CD8+ T cell response. Nevertheless, it remains to be investigated whether a T cell response to a broad diversity of epitopes is relevant at the early and acute stages of the disease, and whether they have a protective role at the primary site of infection, as observed in influenza virus–induced respiratory disease (30). Likewise, it will be important to better understand the phenotypic kinetics of SARS-CoV-2–specific T cells and their contribution to long-term protection. Future studies should also examine these longitudinal CD8+ T cell responses and characteristics in patients with severe or life-threatening COVID-19 infection.
This study has limitations. Foremost is the relatively small sample size. The need to generate a well-characterized sample set limited the number of subjects that could be included. Second, the study was confined to a sampling of COVID-19 convalescent individuals from the greater Baltimore/Washington DC area. As such, this is a geographically restricted population and may not be broadly representative. Third, a low proportion of those who were evaluated had been hospitalized. While this limited our ability to investigate T cell responses in those who were severely ill, it afforded insight into those with milder disease, which is a more commonly encountered form of COVID-19 and could alternatively be considered a strength of our study. Fourth, while the HLA types that were included account for approximately 73% of the continental US population, the technology was restricted to only 6 HLA types. Last, the study was cross-sectional and restricted to a relatively narrow time period. Specifically, individuals were evaluated 27 to 62 days after symptom resolution. At a minimum, they needed to be at least 28 days postresolution to donate convalescent plasma without additional testing (31). This limits the conclusions with respect to earlier and/or later in the convalescent period. Of note, even within the period that was evaluated, changes in the T cell and cytokine responses were observed over time. For example, those later in the convalescent period exhibited T cell maturation with effector cells remaining, possibly to clear residual infection. This is consistent with the cytokine data, demonstrating a time effect since diagnosis (32).
To our knowledge, this is one the most comprehensive and precise characterizations of SARS-CoV-2–specific CD8+ T cell epitope recognition and corresponding ex vivo T cell phenotypes in COVID-19 convalescent subjects to date. The discovery of hitherto undescribed SARS-CoV-2 T cell specificities, their unbiased phenotypic evaluation, and their correlation with overall inflammation greatly extends the current understanding about natural immunity to SARS-CoV-2. Knowing the combination of epitope targets and T cell profiles capable of differentiating into long-term mediators of protection may be pivotal for triggering a durable immune response. Based on these findings, it seems prudent to include several internal and nonstructural viral proteins in the rational design of a second-generation multivalent vaccine.
Methods
Sample selection, antibody titers, HLA typing, and cytokine testing.
The study samples were collected from individuals who were at least 18 years old, who had recovered from COVID-19, and who expressed a willingness to donate COVID-19 convalescent plasma (CCP). In order to qualify for CCP donation, individuals had to have a history of COVID-19 as confirmed by a molecular test (e.g., nasopharyngeal swab) for SARS-CoV-2 and meet all eligibility criteria for community blood donation (e.g., not having been pregnant within the 6 weeks prior to donation, and no history or socio-behavioral risk factors for the major transfusion transmissible infections such as HIV, hepatitis B and hepatitis C) (3). Eligible individuals were enrolled in the study under full, written informed consent, after which whole blood (25 mL) samples were collected. The samples were separated into plasma and PBMCs within 12 hours of blood collection, as previously described (3, 33, 34). Aliquots of plasma and PBMCs were stored at –80°C until further processing.
A subset of convalescent individuals was selected for evaluating SARS-CoV-2–specific CD8+ T cells using highly multiplexed mass cytometry. Among the first 118 eligible CCP donors, there were 87 individuals with at least 4 vials of PBMCs collected (each vial contains at least 5 million PBMCs). These individuals were grouped into tertiles (high, medium, and low IgG titers) according to overall anti–SARS-CoV-2 IgG titers based on EuroImmun ELISA results against SARS-CoV-2 (Supplemental Table 2) (3). Fifteen individuals were randomly selected from each tertile for HLA typing using the donor PMBC samples. HLA-A and HLA-B loci were tested from genomic DNA by next-generation sequencing using the TruSight HLA v1 Sequencing Panel (CareDx). Individuals matched for at least 2 HLA-A or HLA-B alleles (HLA-A*01:01, HLA-A*02:01, HLA-A*03:01, HLA-A*11:01, HLA-A*24:02, and HLA-B*07:02) were included in the subsequent analyses. The remainder of individuals matched for one HLA-A or HLA-B allele were randomly selected so that each tertile group comprised 10 different donors (total n = 30).
The 30 donor samples were transferred to ImmunoScape from Johns Hopkins University in the form of cryopreserved PBMCs. Each sample consisted of either one or 2 aliquots with an average cell number of 12.15 million cells and a viability above 95% per donor. Samples were thawed at 37°C and immediately transferred into complete RPMI medium (10% hiFCS, 1% penicillin/streptomycin/glutamine, 10 mM HEPES, 55 μM 2-mercaptoethanol (2-ME) supplemented with 50 U/mL Benzonase (MilliporeSigma). Aliquots derived from the same donors were combined and all samples were enriched for T cells by removing CD14- and CD19-expressing cells using a column-based magnetic depletion approach according to the manufacturer’s recommendations (Miltenyi). Healthy donor PBMCs (STEMCELL) matched for at least one of the donor HLA alleles were included in each experiment as control for specific T cell identification.
SARS-CoV-2 neutralizing antibody (nAb) titers against 100 50% tissue culture infectious doses (TCID50) per 100 μL were determined using a microneutralization (NT) assay, as previously described (3). The nAb titer was calculated as the highest plasma dilution that prevented cytopathic effect (CPE) in 50% of the wells tested. nAb AUC values were estimated using the exact number of wells protected from infection at every plasma dilution.
Highly sensitive, multiplexed sandwich immunoassays using MULTI-ARRAY electrochemiluminescence detection technology (MesoScale Discovery) were used for the quantitative evaluation of 35 different human cytokines and chemokines in plasma samples from eligible CCP donors (IFN-γ, IL-1β, IL-2, IL-4, IL-6, IL-8, IL-10, IL-12p70, IL-13, TNF-α, GM-CSF, IL-1α, IL-5, IL-7, IL-12/IL23p40, IL-15, IL-16, IL-17A, TNF-β, VEGF-A, Eotaxin, MIP-1β, Eotaxin-3, TARC, IP-10, MIP-1α, MCP-1, MDC, MCP-4, IL-18, IL-1RA, G-CSF [CSF3], IFN-2a, IL-33, and IL-21), as previously described (32). Cytokine and chemokine concentrations were calculated per manufacturer protocol (MSD Discovery Workbench analysis software) and were considered detectable if both runs of each sample had a signal greater than the analyte- and plate-specific lower limit of detection (LLOD) (i.e., 2.5 SDs of the plate-specific blank). Cytokine and chemokine concentrations (pg/mL) from both runs of each analyte were averaged.
Peptides.
A total of 408 unique SARS-CoV-2 candidate peptide epitopes spanning 6 HLAs (HLA-A*01:01, HLA-A*02:01, HLA-A*03:01, HLA-A*11:01, HLA-A*24:02, and HLA-B*07:02) were selected based on recent predictions (Supplemental Table 3) (14, 15). For each of the HLA alleles tested, up to 20 different control peptides (SARS-CoV-2–unrelated epitopes) were also included in the screenings (Supplemental Table 3). All peptides were ordered from Genscript or Mimotopes, with a purity above 85% by HPLC purification and mass spectrometry. Lyophilized peptides were reconstituted at a stock concentration of 10 mM in DMSO.
Antibody staining panel setup.
Purified antibodies lacking carrier proteins (100 μg/antibody) were conjugated to DN3 MAXPAR chelating polymers loaded with heavy metal isotopes following the recommended labelling procedure (Fluidigm). A specific staining panel was set up consisting of 28 antibodies addressing lineage, phenotypic, and functional markers (Supplemental Table 5). All labelled antibodies were titrated and tested by assessing relative marker expression intensities on relevant immune cell subsets in PBMCs from healthy donors (STEMCELL). Antibody mixtures were prepared freshly and filtered using a 0.1 mM filter (Millipore) before staining.
Tetramer multiplexing setup.
To screen for SARS-CoV-2–specific CD8+ T cells we set up a 3-metal combinatorial tetramer staining approach as described previously (17, 35). Briefly, specific peptide–MHC class I complexes were generated by incubating biotinylated UV-cleavable peptide HLA monomers in the presence of individual antigen candidates. For the generation of a triple-coded tetramer staining mixture, recombinant streptavidin was conjugated to heavy metal–loaded DN3 polymers (17) and 3 of 12 differently labelled streptavidin molecules were randomly combined by using an automated pipetting device (TECAN), resulting in a total of 220 unique possible combinations to encode single peptide candidates. Peptide exchange was performed at 100 μg/mL of HLA monomer in PBS with 50 μM peptides of interest in a 96-well plate. Peptides with similar sequences were assigned the same triple code to avoid multiple signals through potential T cell cross-reactivity. According to the donors’ HLA genotypes, total epitope screenings ranged from 49 to 220 peptides for individual samples, including SARS-CoV-2–unrelated control peptides. For tetramerization, each triple-coded streptavidin mixture was added in 3 steps to their corresponding exchanged peptide–MHC complexes to reach a final molar ratio of 1:4 (total streptavidin/peptide–MHC). The tetramerized peptide–MHC complexes were incubated with 10 μM of free biotin (MilliporeSigma) to saturate remaining unbound streptavidins. All tetramers were combined and concentrated (10 kDa cutoff filter) in cytometry buffer (PBS, 2% FCS, 2 mM EDTA, 0.05% sodium azide) before staining the cells. As internal control and to facilitate the detection of bona fide antigen-specific T cells we generated a second tetramer-staining configuration for each experiment using a completely different coding scheme for each peptide (17).
Sample staining and acquisition.
T cell–enriched donor samples and healthy donor PBMCs were split into 2 fractions and seeded at equal numbers in 2 wells of a 96-well plate. Cells were washed and each well was then stained with 100 μL of either 1 of the 2 tetramer configurations for 1 hour at room temperature. After 30 minutes, a unique metal–labelled (Cd-111 and Cd-113) anti-CD45 antibody was added into each of the wells to further barcode the cells that were stained with the different tetramer configurations. Cells were then washed twice and the 2 wells per sample were combined and stained with the heavy metal–labelled antibody mixtures for 30 minutes on ice and 200 μM cisplatin during the last 5 minutes for the discrimination of live and dead cells. Cells were washed and fixed in 2% paraformaldehyde in PBS overnight at 4°C. For intracellular staining, cells were incubated in 1× permeabilization buffer (Biolegend) for 5 minutes on ice and incubated with metal conjugated anti-GranzymeB antibodies for 30 minutes on ice. Samples from different donors were barcoded with a unique dual combination of bromoacetamidobenzyl-EDTA–linked (Dojindo) metal barcodes (Pd-102, Pd-104, PD106 and PD108, and Pd-110) for 30 minutes on ice. Cells were then washed and resuspended in 250 nM iridium DNA intercalator (Fluidigm) in 2% paraformaldehyde/PBS at room temperature. Cells were washed, pooled together, and adjusted to 0.5 million cells per milliliter H2O together with 1% equilibration beads (EQ Four element calibration beads, Fluidigm) for acquisition on a HELIOS mass cytometer (CyTOF, Fluidigm).
Data analysis.
After mass cytometry acquisition, signals for each parameter were normalized based on EQ beads (Fluidigm) added to each sample (36) and any zero values were randomized using a custom Rscript that uniformly distributes values between –1 and 0. Each sample was manually debarcoded followed by gating on live CD8+ and CD4+ T cells (CD45+ DNA+ cisplatin– CD3+ cells) from either staining configuration after gating out residual monocytes (CD14) and B cells (CD19) using FlowJo (Tree Star) software. Antigen-specific triple tetramer–positive cells (hits) were identified by an automated peptide-MHC gating method (17) and each hit was confirmed and refined using manual gating. The designation of bona fide antigen-specific T cells was further dependent on (a) the detection cut-off threshold (≥ 2 events to be detected in each staining configuration), (b) the frequency correspondence between the 2 tetramer staining configurations (ratio between the frequencies of a hit in either staining configuration to be ≤ 2), and (c) the background noise (frequencies of specific CD8+ T cell events must be greater than events from the corresponding CD4+ T cell population), as unbiased objective criteria for antigen-specificity assessment (16). Bulk T cells and true hits from both staining configurations were combined for assessing frequencies and phenotypic and statistical analysis.
Frequency values were calculated based on the percentage of the parent immune cell population. Phenotypic markers were gated individually for each sample and calculated as the percentage of positive cells. High-dimensional phenotypic profiles and sample distributions were shown using uniform manifold approximation and projection (37). Data analysis was performed using CYTOGRAPHER, ImmunoScape cloud-based analytical software, custom R-scripts, GraphPad Prism, and Flowjo software.
Statistics.
Comparative analyses of frequencies of cell subsets and marker expression between samples were performed using Wilcoxon rank sum tests, extended to Kruskal-Wallis tests by ranks for more than 2 levels in a grouping variable; resulting P values were adjusted for multiple testing using the Benjamini-Hochberg method to control the false discovery rate. Data are displayed as boxes and whiskers showing all data points (minimum to maximum). Correlations were calculated with the Spearman’s rank order test. A correlation matrix was calculated comparing phenotypic and serological marker variables in a pairwise fashion, using the corr.test function from the psych CRAN package; the corrplot package was subsequently used to graphically display the correlation matrix. Resulting P values were adjusted for multiple testing using the Bonferroni method. Spearman’s correlation coefficients were indicated by a heat scale whereby blue color shows positive linear correlation, and red color shows negative linear correlation. All statistical analyses were performed using GraphPad Prism and R. Statistical significance was set at a threshold of *P < 0.05, **P < 0.01, and ***P < 0.001.
Study approval.
The Johns Hopkins University School of Medicine IRB reviewed and approved the sample collection and overall study. Written informed consent was received from participants prior to inclusion in the study.
Author contributions
EMB, AART, DS, and SS contributed samples. TSB, KL, AP, HS, FK, MPB, SEB, OL, and MF collected experimental data. HK, DC, EUP, and MF analyzed data and performed statistical analysis. HK, BA, EWN, AN, and MF drafted the manuscript. BA, EWN, AN, MF, AART, ADR, EMB, and TCQ conceived and designed the study. All authors contributed intellectually and approved the manuscript.
Supplementary Material
Acknowledgments
The authors would like to thank the COVID-19 convalescent plasma study and laboratory teams, and all the donors for their participation in the study. We thank the National Institute of Infectious Diseases, Japan, for providing VeroE6TMPRSS2 cells and acknowledge the Centers for Disease Control and Prevention, BEI Resources, National Institute of Allergy and Infectious Diseases (NIAID), NIH, for SARS-Related Coronavirus 2, Isolate USA-WA1/2020, NR-5228. This work was supported in part by NIAID grants R01AI120938, R01AI120938S1, and R01AI128779 (to AART); NIAID grants AI052733, N272201400007C, and AI15207 (to AC); NIAID grant T32AI102623 (to EUP); funding from the Division of Intramural Research, NIAID, NIH (to OL, AR, and TQ); National Heart Lung and Blood Institute grants 1K23HL151826-01 (to EMB) and R01HL059842 (to AC); funding from Bloomberg Philanthropies (to AC); and the Department of Defence grant W911QY2090012 (to DS).
Version 1. 01/11/2021
In-Press Preview
Version 2. 03/01/2021
Electronic publication
Funding Statement
This work was supported in part by National Institute of Allergy and Infectious Diseases (NIAID) R01AI120938, R01AI120938S1 and R01AI128779 (A.A.R.T); NIAID AI052733, N272201400007C) and AI15207 (A.C.); NIAID T32AI102623 (E.U.P.); the Division of Intramural Research, NIAID, NIH (O.L., A.R., T.Q.); National Heart Lung and Blood Institute 1K23HL151826-01 (E.M.B) and R01HL059842 (A.C.). Bloomberg Philanthropies (A.C.); Department of Defence W911QY2090012 (D.S.).
Footnotes
Conflict of interest: HK, HS, FK, DC, BA, AN, EWN, and MF are shareholders or employees of ImmunoScape Pte Ltd. AN is on the Board of Directors of ImmunoScape Pte Ltd.
Copyright: © 2021, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2021;131(5):e145476.https://doi.org/10.1172/JCI145476.
Contributor Information
Hassen Kared, Email: hassen.kared@immunoscape.com.
Andrew D. Redd, Email: aredd2@jhmi.edu.
Evan M. Bloch, Email: ebloch2@jhmi.edu.
Tania S. Bonny, Email: tbonny1@jhu.edu.
Faris Kairi, Email: faris.kairi@immunoscape.com.
Daniel Carbajo, Email: daniel.carbajo@immunoscape.com.
Brian Abel, Email: brian.abel@immunoscape.com.
Evan W. Newell, Email: enewell@fredhutch.org.
Sarah E. Benner, Email: sbenner7@jhmi.edu.
Eshan U. Patel, Email: epatel6@jhmi.edu.
Kirsten Littlefield, Email: klittl20@jhu.edu.
Oliver Laeyendecker, Email: olaeyen@jhmi.edu.
Shmuel Shoham, Email: sshoham1@jhmi.edu.
David Sullivan, Email: dsulliv7@jhmi.edu.
Arturo Casadevall, Email: acasade1@jhu.edu.
Andrew Pekosz, Email: apekosz@jhsph.edu.
Alessandra Nardin, Email: alessandra.nardin@immunoscape.com.
Michael Fehlings, Email: michael.fehlings@immunoscape.com.
Thomas C. Quinn, Email: tquinn@jhmi.edu.
References
- 1.Guan WJ, et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382(18):1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang C, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klein SL, et al. Sex, age, and hospitalization drive antibody responses in a COVID-19 convalescent plasma donor population. J Clin Invest. 2020;130(11):6141–6150. doi: 10.1172/JCI142004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Long QX, et al. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat Med. 2020;26(8):1200–1204. doi: 10.1038/s41591-020-0965-6. [DOI] [PubMed] [Google Scholar]
- 5.Braun J, et al. SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature. 2020;587(7833):270–274. doi: 10.1038/s41586-020-2598-9. [DOI] [PubMed] [Google Scholar]
- 6.Grifoni A, et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181(7):1489–1501. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Le Bert N, et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. 2020;584(7821):457–462. doi: 10.1038/s41586-020-2550-z. [DOI] [PubMed] [Google Scholar]
- 8.Peng Y, et al. Broad and strong memory CD4+ and CD8+ T cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19. Nat Immunol. 2020;21(11):1336–1345. doi: 10.1038/s41590-020-0782-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sekine T, et al. Robust T cell immunity in convalescent individuals with asymptomatic or mild COVID-19. Cell. 2020;183(1):158–168. doi: 10.1016/j.cell.2020.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ni L, et al. Detection of SARS-CoV-2-specific humoral and cellular immunity in COVID-19 convalescent individuals. Immunity. 2020;52(6):971–977. doi: 10.1016/j.immuni.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Folegatti PM, et al. Safety and immunogenicity of the ChAdOx1 nCoV-19 vaccine against SARS-CoV-2: a preliminary report of a phase 1/2, single-blind, randomised controlled trial. Lancet. 2020;396(10249):467–478. doi: 10.1016/S0140-6736(20)31604-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu J, et al. DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science. 2020;369(6505):806–811. doi: 10.1126/science.abc6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu FC, et al. Safety, tolerability, and immunogenicity of a recombinant adenovirus type-5 vectored COVID-19 vaccine: a dose-escalation, open-label, non-randomised, first-in-human trial. Lancet. 2020;395(10240):1845–1854. doi: 10.1016/S0140-6736(20)31208-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grifoni A, et al. A sequence homology and bioinformatic approach can predict candidate targets for immune responses to SARS-CoV-2. Cell Host Microbe. 2020;27(4):671–680. doi: 10.1016/j.chom.2020.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prachar M, et al. Identification and validation of 174 COVID-19 vaccine candidate epitopes reveals low performance of common epitope prediction tools. Scientific Reports. 2020;10(1):20465. doi: 10.1038/s41598-020-77466-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fehlings M, et al. Late-differentiated effector neoantigen-specific CD8+ T cells are enriched in peripheral blood of non-small cell lung carcinoma patients responding to atezolizumab treatment. J Immunother Cancer. 2019;7(1):249. doi: 10.1186/s40425-019-0695-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Newell EW, et al. Combinatorial tetramer staining and mass cytometry analysis facilitate T-cell epitope mapping and characterization. Nat Biotechnol. 2013;31(7):623–629. doi: 10.1038/nbt.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pittet MJ, et al. High frequencies of naive Melan-A/MART-1-specific CD8(+) T cells in a large proportion of human histocompatibility leukocyte antigen (HLA)-A2 individuals. J Exp Med. 1999;190(5):705–715. doi: 10.1084/jem.190.5.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mahnke YD, et al. The who’s who of T-cell differentiation: Human memory T-cell subsets. Eur J Immunol. 2013;43(11):2797–2809. doi: 10.1002/eji.201343751. [DOI] [PubMed] [Google Scholar]
- 20.Elsaesser H, et al. IL-21 is required to control chronic viral infection. Science. 2009;324(5934):1569–1572. doi: 10.1126/science.1174182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kared H, et al. Galectin-9 and IL-21 mediate cross-regulation between Th17 and Treg cells during acute hepatitis C. PLoS Pathog. 2013;9(6):e1003422. doi: 10.1371/journal.ppat.1003422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schulien I, et al. Characterization of pre-existing and induced SARS-CoV-2-specific CD8+ T cells. Nat Med. 2021;27(1):78–85. doi: 10.1038/s41591-020-01143-2. [DOI] [PubMed] [Google Scholar]
- 23.Ferretti AP, et al. Unbiased screens show CD8+ T cells of COVID-19 patients recognize shared epitopes in SARS-CoV-2 that largely reside outside the spike protein. Immunity. 2020;53(5):1095–1107.E3. doi: 10.1016/j.immuni.2020.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. doi: 10.21203/rs.3.rs-33197/v1. Gangaev A, et al. Profound CD8 T cell responses towards the SARS-CoV-2 ORF1ab in COVID-19 patients [preprint]. Posted on Research Square June 4, 2020. [DOI]
- 25. doi: 10.1101/2020.08.26.267724. Quadeer AA, et al. Epitopes targeted by T cells in convalescent COVID-19 patients [preprint]. Posted on bioRxiv August 26, 2020. [DOI]
- 26.Shomuradova AS, et al. SARS-CoV-2 epitopes are recognized by a public and diverse repertoire of human T cell receptors. Immunity. 2020;53(6):S1074–S1257. doi: 10.1016/j.immuni.2020.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. doi: 10.1101/2020.07.31.20165647. Snyder TM, et al. Magnitude and dynamics of the T-cell response to SARS-CoV-2 infection at both individual and population levels [preprint]. Posted on medRxiv September 17, 2020. [DOI]
- 28.Diao B, et al. Reduction and functional exhaustion of T cells in patients with coronavirus disease 2019 (COVID-19) Front Immunol. 2020;11:827. doi: 10.3389/fimmu.2020.00827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neidleman J, et al. SARS-CoV-2-specific T cells exhibit phenotypic features reflecting robust helper function, lack of terminal differentiation, and high proliferative potential. Cell Reports Med. 2020;1(6):100081. doi: 10.1016/j.xcrm.2020.100081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pizzolla A, et al. Resident memory CD8+ T cells in the upper respiratory tract prevent pulmonary influenza virus infection. Sci Immunol. 2017;2(12):eaam6970. doi: 10.1126/sciimmunol.aam6970. [DOI] [PubMed] [Google Scholar]
- 31.Bloch EM, et al. Deployment of convalescent plasma for the prevention and treatment of COVID-19. J Clin Invest. 2020;130(6):2757–2765. doi: 10.1172/JCI138745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bonny TS, et al. Cytokine and chemokine levels in COVID-19 convalescent plasma. Open Forum Infect Dis. 2021;8(2):ofaa574. doi: 10.1093/ofid/ofaa574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Benner SE, et al. SARS-CoV-2 antibody avidity responses in COVID-19 patients and convalescent plasma donors. J Infect Dis. 2020;222(12):1974–1984. doi: 10.1093/infdis/jiaa581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Patel EU, et al. Comparative performance of five commercially available serologic assays to detect antibodies to SARS-CoV-2 and identify individuals with high neutralizing titers. J Clin Microbiol. doi: 10.1128/JCM.02257-20. [published online November 2, 2020]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fehlings M, et al. Author correction: checkpoint blockade immunotherapy reshapes the high-dimensional phenotypic heterogeneity of murine intratumoural neoantigen-specific CD8+ T cells. Nat Commun. 2018;9(1):3000. doi: 10.1038/s41467-018-05468-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Finck R, et al. Normalization of mass cytometry data with bead standards. Cytom Part J Int Soc Anal Cytol. 2013;83(5):483–494. doi: 10.1002/cyto.a.22271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Becht E, et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat Biotechnol. doi: 10.1038/nbt.4314. [published online December 3, 2018]. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.