Abstract
Previous studies have shown that nitric oxide (NO) supplements may prevent bone loss and fractures in preclinical models of estrogen deficiency. However, the mechanisms by which NO modulates bone anabolism remain largely unclear. Argininosuccinate lyase (ASL) is the only mammalian enzyme capable of synthesizing arginine, the sole precursor for nitric oxide synthase–dependent (NOS-dependent) NO synthesis. Moreover, ASL is also required for channeling extracellular arginine to NOS for NO production. ASL deficiency (ASLD) is thus a model to study cell-autonomous, NOS-dependent NO deficiency. Here, we report that loss of ASL led to decreased NO production and impairment of osteoblast differentiation. Mechanistically, the bone phenotype was at least in part driven by the loss of NO-mediated activation of the glycolysis pathway in osteoblasts that led to decreased osteoblast differentiation and function. Heterozygous deletion of caveolin 1, a negative regulator of NO synthesis, restored NO production, osteoblast differentiation, glycolysis, and bone mass in a hypomorphic mouse model of ASLD. The translational significance of these preclinical studies was further reiterated by studies conducted in induced pluripotent stem cells from an individual with ASLD. Taken together, our findings suggest that ASLD is a unique genetic model for studying NO-dependent osteoblast function and that the NO/glycolysis pathway may be a new target to modulate bone anabolism.
Keywords: Bone Biology, Metabolism
Keywords: Mouse models, Nitric oxide, Osteoclast/osteoblast biology
Introduction
Nitric oxide (NO) is an important signaling molecule that influences a wide range of physiological processes, including bone metabolism. Bone is a highly dynamic tissue that is maintained by the balance between bone-degrading osteoclasts, bone-forming osteoblasts, and mechanosensing osteocytes. The study of NO metabolism in bone is clinically important because NO supplements have been reported to improve bone formation, increase bone strength, and prevent estrogen-deficiency osteoporosis in preclinical models (1–4). Furthermore, anecdotal studies have shown that this may be relevant in humans, as use of NO supplementation has been shown to be associated with decreased fracture risk (5, 6). However, the treatment effects of NO supplements on bone mass have been controversial, and much like the action of NO itself, context dependent. For example, one clinical trial failed to demonstrate a beneficial effect of NO supplements on bone loss (7). Collectively, while there is a substantial amount of data supporting an anabolic function of NO to promote bone formation, the mechanisms underlying NO-mediated action in vivo are unclear.
Metabolic programming in progenitor cells influences cell fate and differentiation capacity (8–12). Accumulating evidence suggests that osteoblasts utilize glucose metabolism via glycolysis as their main energy source (13, 14). Importantly, the anabolic response of bone to parathyroid hormone and Wnt proteins, such as Wnt7b and Wnt3a, has been shown to be due in part to the stimulation of aerobic glycolysis in osteoblast lineage cells (15–17). At the same time, physiological (i.e., low) levels of NO have been reported to promote glycolysis in a variety of cell types, such as skeletal muscle cells, neurons, and adipocytes (18–21). However, it is not known whether NO enhances glucose metabolism to facilitate osteoblast differentiation.
NO is generated from L-arginine by 3 nitric oxide synthase (NOS) enzymes. All 3 NOS isoforms (NOS1, -2, and -3) have been identified in bone cells (22–24). NOS-knockout (NOS-KO) mice have been used to examine the role of NO in bone homeostasis. Mice with KO of endothelial NOS (eNOS, or NOS3) demonstrate decreased bone mass due to reduced osteoblast activity and decreased mineralization (25, 26). Mice with KO of neuronal NOS (nNOS, or NOS1) exhibit increased bone mass with decreased osteoclast and osteoblast numbers in vivo; however, in vitro assays have shown that more osteoclasts are formed from these KO mice, suggesting a non–cell-autonomous mechanism (27). The studies evaluating the effects of loss of inducible NOS (iNOS, or NOS2) on bone have yielded inconsistent results. iNOS-KO mice have been reported to have no bone abnormalities (28), and to also have increased bone mass in association with reduced osteoclast-mediated bone resorption (29). Triple-NOS-KO mice with deletion of all 3 NOS isoforms have been shown to exhibit higher bone mass (30). In these mice, high bone mass was attributed to high bone turnover, with increased bone formation and resorption, which is different from each of the single-NOS-KO models (30). Together, murine models that globally lack NOS isoforms (nNOS, iNOS, and eNOS) exhibit contrasting bone phenotypes, in part due to the non–cell-autonomous effects as well as potential compensation by the other NOS isoforms.
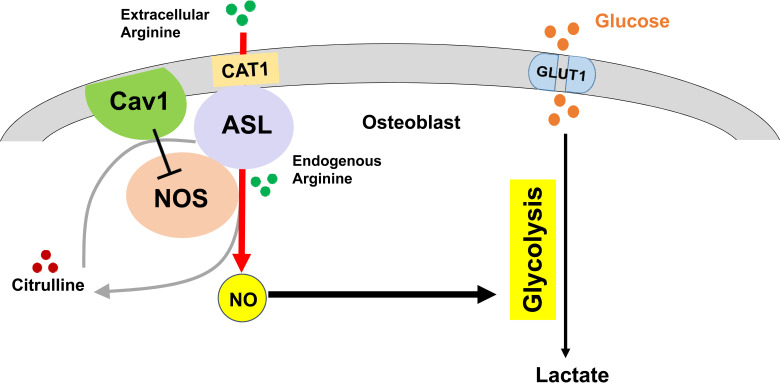
One of the key obstacles in understanding the role of NO in osteoblasts and bone anabolism is that the currently available models are limited by non–cell-autonomous sources of NO and redundancy of the NOS isoforms. Models that can bypass such redundancy and restrict NO deficiency to just the osteoblast lineage cells could have a significant impact in understanding the cell-specific roles of NO in bone homeostasis. We have previously shown that argininosuccinate lyase (ASL), a urea cycle enzyme that is also essential for the citrulline/NO cycle, is a key regulator of NO metabolism (31, 32). ASL is not only the sole enzyme required for generating arginine, the precursor for NOS-dependent NO production; it also facilitates the transport of extracellular arginine into an NO-synthesis complex that includes heat shock protein 90 (HSP90), the arginine transporter cationic amino acid transporter 1 (CAT-1), and argininosuccinate synthase 1 (ASS1) (31). ASL’s distinct enzymatic and structural functions integrate both intracellular and extracellular pathways for generating NO. Whereas ASL is critical for maintaining this “positive” NO synthesis complex, it has also been reported that binding of caveolin 1 (Cav-1) to eNOS inhibits NO production (33–35). Caveolins (Cav-1, -2, and-3) are a family of membrane proteins that constitute caveolae, which are flask-shaped organelle invaginations in the plasma membrane (36, 37). Cav-1 is the primary coat protein that maintains the structure of caveolae. Cav-1 and Cav-2 are expressed in most cells, while Cav-3 is specifically expressed in the muscle (38, 39). The main functions of caveolae include cholesterol transport, endocytosis, and signal transduction (40, 41). Most studies utilizing endothelial cells have shown that there is a direct interaction of Cav-1 with eNOS, and that the binding of Cav-1 to eNOS leads to the inhibition of eNOS activity (33, 35, 42–44). Hence, changing the balance of the positive versus negative regulators involved in NO synthesis may regulate dynamic NO production. Therefore, in the present study, we tested the hypothesis that inhibiting Cav-1 may be a potential therapeutic strategy to restore NO production in the context of ASL deficiency (ASLD), while also testing the NO-specific contribution to the ASLD phenotype.
Results
Deletion of Asl in osteoblast lineage cells leads to decreased NO production, osteoblast differentiation, and mineralization.
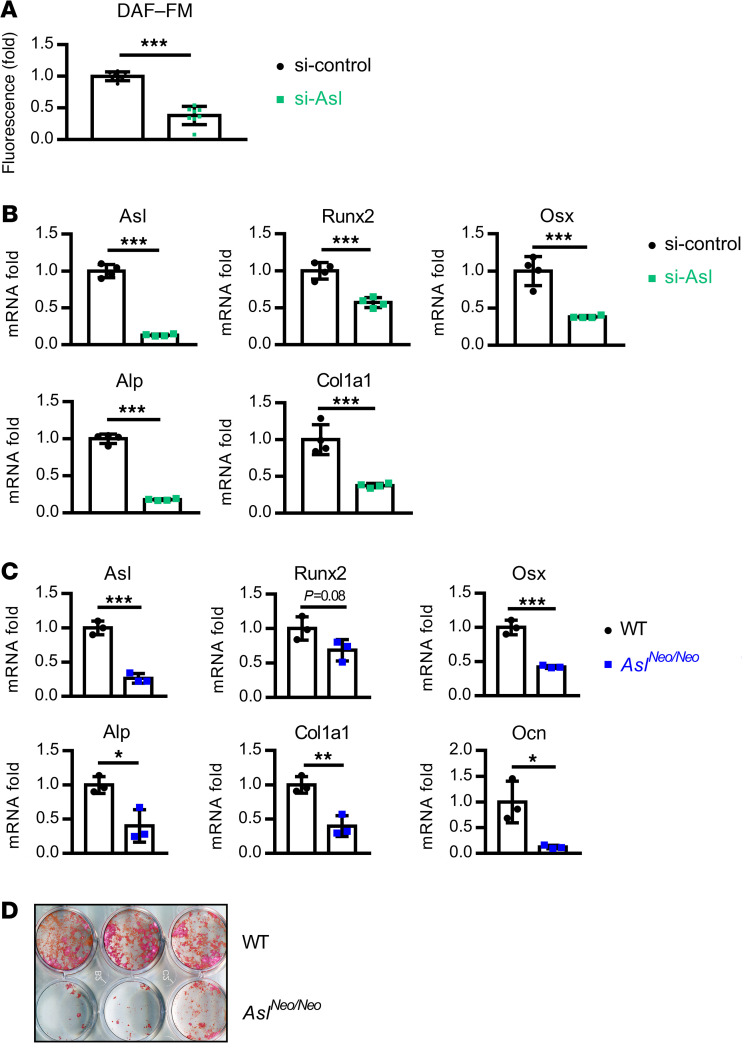
We and others have shown that deletion of Asl leads to decreased NO production in various cell types, including endothelial cells, neurons, and enterocytes (32, 45, 46). To test whether downregulation of Asl also results in decreased NO production in osteoblast lineage cells, we used small interfering RNA–mediated (siRNA-mediated) knockdown of Asl in ST2 cells, a mouse bone marrow–derived stromal cell line. NO production was measured by the DAF-FM diacetate (4-amino-5-methylamino-2′,7′-difluorofluorescein diacetate) assay, which is based on the fluorescence intensity of a fluorescent dye that reacts with NO autoxidation products. NO production was significantly decreased after knockdown of Asl in ST2 cells (Figure 1A). To determine the role of NO in osteoblast differentiation, we induced osteoblastic differentiation in ST2 cells. Knockdown of Asl resulted in decreased expression of osteoblast-marker genes, including Runx2 (Runt-related transcription factor 2), Osx (osterix), Alp (alkaline phosphatase), and Col1a1 (collagen type I α 1 chain) as shown by real-time quantitative RT-PCR (qPCR) (Figure 1B). The same effect was found to be reproduced in knockdown of Asl in MC3T3 cells, another osteoblast precursor cell line derived from mouse calvaria cells (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/JCI138935DS1).
Figure 1. Deletion of Asl in osteoblast lineage cells leads to decreased NO production, osteoblast differentiation, and mineralization.
(A) NO level was measured by DAF-FM fluorescence intensity in ST2 cells after siRNA-mediated Asl knockdown. n = 8. (B) mRNA levels of Asl, Runx2, Osx, Alp, and Col1a1 by qPCR in ST2 cells. Asl-knockdown ST2 cells were differentiated in osteogenic medium for 3 days. n = 4. (C) mRNA levels of Asl, Runx2, Osx, Alp, Col1a1, and osteocalcin (Ocn) by qPCR in BMSC-derived osteoblasts from WT and AslNeo/Neo mice. BMSCs were cultured in osteogenic medium for 14 days. n = 3. (D) Alizarin red staining of BMSC-derived osteoblasts from WT and AslNeo/Neo mice. BMSCs were cultured in osteogenic medium for 14 days. n = 3. Data are presented as mean ± SD. *P < 0.05; **P < 0.01; ***P < 0.005 by Student’s t test.
To determine whether Asl-deficient osteoblasts have an intrinsic defect in differentiation and mineralization, primary bone marrow stromal cells (BMSCs) were generated from the bone marrow of WT and AslNeo/Neo mice, a hypomorphic mouse model of ASLD. AslNeo/Neo mice have approximately 25% residual Asl expression (31). BMSCs were cultured in osteogenic medium for 14 days and expression of osteoblast-marker genes and staining for mineralization were evaluated. Consistent with the results from in vitro differentiation of ST2 cells and MC3T3 cells, the ability of BMSCs to differentiate into osteoblasts and mineralization capacity of cells derived from AslNeo/Neo mice were significantly reduced as compared with WT mice (Figure 1, C and D).
Deletion of Asl in osteoblast lineage cells leads to downregulation of glycolysis pathway.
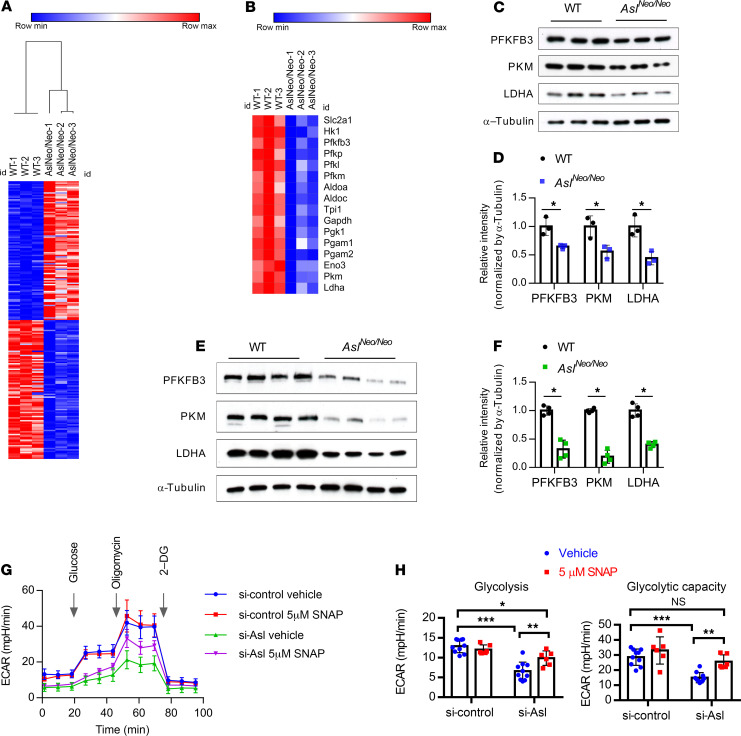
To further investigate the possible molecular mechanisms underlying osteoblast differentiation, we performed RNA sequencing (RNA-seq) in BMSC-derived osteoblasts from WT and AslNeo/Neo mice. A heatmap depicting the top 100 upregulated and downregulated genes is shown in Figure 2A. Several canonical pathways were enriched in a core analysis in Ingenuity Pathway Analysis (IPA) (Supplemental Figure 2). Notably, the glycolysis pathway was indicated to be inhibited in AslNeo/Neo mouse–derived osteoblasts (z score of –2.1). Most of the genes in the glycolysis pathway were significantly downregulated (Figure 2B and Supplemental Excel file). These expression data were confirmed by immunoblotting of several enzymes of the glycolysis pathway including PFKFB3 (6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3), PKM (M2 isoform of pyruvate kinase), and LDHA (lactate dehydrogenase A) in BMSC-derived osteoblasts from WT and AslNeo/Neo mice (Figure 2, C and D). Furthermore, protein levels of PFKFB3, PKM, and LDHA were also reduced in long-bone protein extracts from AslNeo/Neo mice as compared with WT mice (Figure 2, E and F).
Figure 2. Deletion of Asl in osteoblast lineage cells leads to downregulation of glycolysis pathway.
(A) Heatmap of the top 200 differentially expressed transcripts (100 upregulated and 100 downregulated) detected using RNA-seq in BMSC-derived osteoblasts from WT and AslNeo/Neo mice. BMSCs were cultured in osteogenic medium for 14 days. n = 3. (B) Heatmap of downregulated glycolysis pathway–related genes in BMSC-derived osteoblasts from WT and AslNeo/Neo mice. (C) Protein levels of PFKFB3, PKM, and LDHA by Western blot and (D) quantification of Western blot from WT and AslNeo/Neo BMSC–derived osteoblasts. n = 3. (E) Protein levels of PFKFB3, PKM, and LDHA by Western blot and (F) quantification of Western blot from WT and AslNeo/Neo mouse long-bone protein extracts. n = 4. (G and H) Seahorse assay of glycolytic activity (ECAR) in Asl-knockdown ST2 cells (siRNA mediated) with 5 μM NO donor S-nitroso-N-acetylpenicillamine (SNAP) or 5 μM DMSO (vehicle) 18-hour treatment (n = 6–10; data are representative of 3 independent experiments). 2-DG, 2-deoxyglucose. Data are presented as mean ± SD. Student’s t test (D and F) and 2-way ANOVA followed by Tukey’s multiple-comparison test (H). *P < 0.05; **P < 0.01; ***P < 0.005. NS, not significant (P > 0.05).
Many studies have suggested that glucose metabolism through aerobic glycolysis is the predominant source of energy that promotes osteoblast differentiation (13, 14). Thus, we examined whether changes in the level of glycolytic gene expression actually affected glycolytic function. We quantified the extracellular acidification rate (ECAR, which measures glycolysis) in Asl-deficient cells using the Seahorse XFe96 Analyzer. Both the basal and capacity for glycolysis, as indicated by ECAR, were significantly reduced with Asl knockdown in ST2 cells (Figure 2, G and H). Importantly, the basal level of glycolysis was partially restored and the capacity for glycolysis was fully restored with the addition of the NO donor S-nitroso-N-acetylpenicillamine (SNAP) (Figure 2, G and H). In contrast, N-acetylpenicillamine (NAP) (negative control compound of SNAP) did not increase ECAR levels in Asl-knockdown ST2 cells (Supplemental Figure 3). Collectively, these data suggest that NO is important for maintaining a normal glycolytic state during osteoblast differentiation.
NO also plays important roles in regulating mitochondrial biogenesis and function. We did not observe alterations in mitochondrial DNA copy number after knockdown of Asl in ST2 cells (Supplemental Figure 4A). We further evaluated mitochondrial function by measuring oxygen consumption rate (OCR, an indicator of oxidative phosphorylation) via Seahorse assay. We observed decreased OCR levels in BMSCs and BMSC-derived osteoblasts from AslNeo/Neo mice as compared with WT mice (Supplemental Figure 4, B–E). Because glycolysis contributes to the majority of ATP production in osteoblast lineage cells (47), we focused on the potential role of NO in enhancing glycolysis.
Heterozygous deletion of Cav-1 restores NO production and glycolytic ability.
The overall production of NO is determined by a balance between positive and negative regulators. ASL is a positive regulator of NO; in contrast, Cav-1 has been reported to be a negative regulator of NO production (refs. 33, 35; and Figure 3A). We thus hypothesized that deletion of Cav-1 may restore NO production and rescue the Asl deficiency–induced phenotype, thereby also supporting the NO specificity of the ASLD phenotype. To investigate the role of ASL and Cav-1 in the regulation of NO production and bone mass in vivo, we crossed Asl+/Neo;Cav-1+/– mice with Asl+/Neo mice to generate AslNeo/Neo mice and AslNeo/Neo;Cav-1+/– mice. As complete absence of Cav-1 in mice results in systemic manifestations including abnormal lipid metabolism, pulmonary hypertension, and cardiomyopathy (48–50), we compared AslNeo/Neo mice to AslNeo/Neo Cav-1+/– mice.
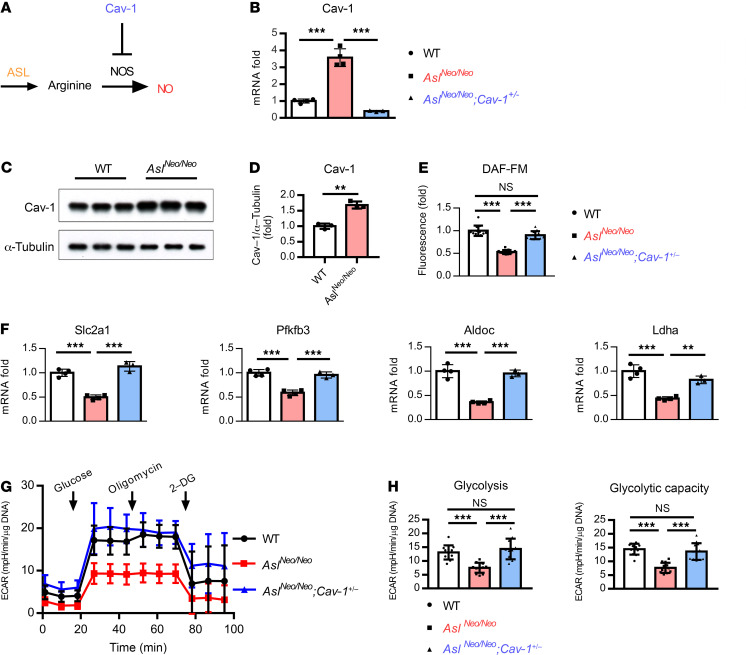
Figure 3. Heterozygous deletion of Cav-1 restores NO production and glycolytic ability.
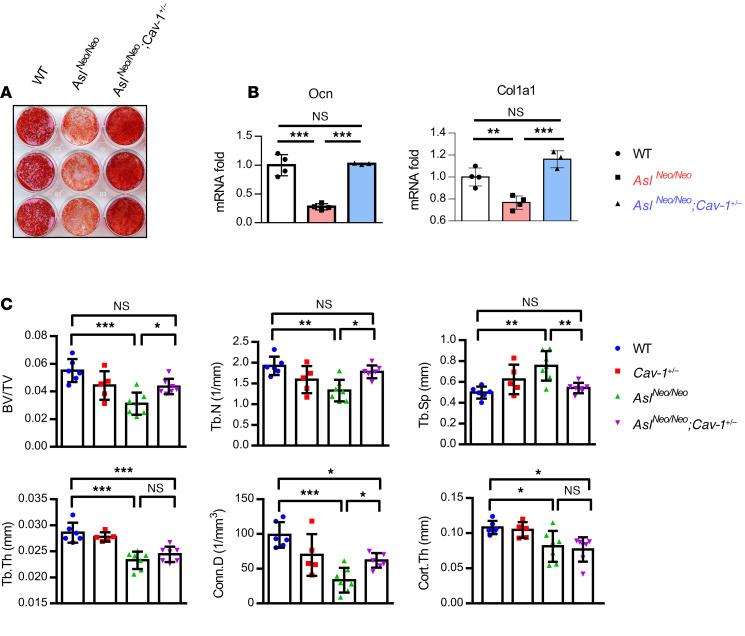
(A) Schematic figure of Asl and Cav-1 in the regulation of NO synthesis. (B) mRNA and (C and D) protein levels of Cav-1 in BMSC-derived osteoblasts from WT and AslNeo/Neo mice. BMSCs were cultured in osteogenic medium for 14 days. n = 3–4. (E) DAF-FM fluorescence intensity of BMSCs derived from WT, AslNeo/Neo, and AslNeo/Neo;Cav-1+/– mice. n = 12 wells/mouse. Data are representative of 2 independent experiments. (F) mRNA levels of glycolytic genes Slc2a1, Pfkfb3, Aldoc (aldolase, fructose-bisphosphate C), and Ldha from BMSC-derived osteoblasts from WT, AslNeo/Neo, and AslNeo/Neo;Cav-1+/– mice. BMSCs were cultured in osteogenic medium for 14 days. n = 3–4. (G and H) Seahorse assay of ECAR from WT, AslNeo/Neo, and AslNeo/Neo;Cav-1+/– mouse BMSC–derived osteoblasts. n = 12 wells/mouse. 2-DG, 2-deoxyglucose. Data are representative of 3 independent and biological repeats and are presented as mean ± SD. Student’s t test for 2 groups or 1-way ANOVA followed by Tukey’s multiple-comparison test for multiple groups. **P < 0.01; ***P < 0.005. NS, not significant (P > 0.05).
First, we measured Cav-1 expression in BMSC-derived osteoblasts. Cav-1 mRNA and protein levels were upregulated in BMSC-derived osteoblasts from AslNeo/Neo mice compared with WT mice (Figure 3, B–D), indicating an accumulation of the negative regulator of NO synthesis in the absence of ASL. NO production was decreased in BMSCs from AslNeo/Neo mice compared with WT mice; however, with heterozygous loss of Cav-1, NO synthesis was restored (Figure 3E). Consistently, compared with WT mouse–derived osteoblasts, AslNeo/Neo mouse–derived osteoblasts had decreased expression of glycolytic genes, as well as decreased glycolytic ability as shown by ECAR (Figure 3, F–H). In addition, we found that genes in the glycolysis pathway were expressed at higher levels in osteoblasts derived from AslNeo/Neo;Cav-1+/– mice as compared with AslNeo/Neo mice (Figure 3F). Furthermore, glycolytic rate and glycolytic capacity in BMSC-derived osteoblasts from AslNeo/Neo;Cav-1+/– mice were higher as compared with AslNeo/Neo mice (Figure 3, G and H).
Heterozygous deletion of Cav-1 partially rescues low bone mass in AslNeo/Neo mice.
Because heterozygous Cav-1 deletion restored NO production and glycolytic capacity in cells derived from AslNeo/Neo mice, we next tested whether osteoblast differentiation ability and mineralization were also rescued in AslNeo/Neo;Cav-1+/– mice. Alizarin red staining showed that the mineralization capacity was restored in AslNeo/Neo;Cav-1+/– as compared with AslNeo/Neo BMSC–derived osteoblasts (Figure 4A). mRNA levels of osteoblastic marker genes Col1a1 and Ocn (osteocalcin) were higher in BMSC-derived osteoblasts from AslNeo/Neo;Cav-1+/– mice as compared with AslNeo/Neo mice (Figure 4B). Micro–computed tomography (μCT) analysis of femurs revealed that AslNeo/Neo mice exhibited a low-bone-mass phenotype compared with WT mice (Figure 4C). Heterozygous Cav-1 deletion in AslNeo/Neo;Cav-1+/– mice partially rescued the low bone mass in AslNeo/Neo mice (Figure 4C). Specifically, the parameters bone volume/total volume (BV/TV), trabecular number (Tb.N), and trabecular separation (Tb.Sp) were completely rescued, and connectivity density (Conn.D) was partially rescued, while trabecular thickness (Tb.Th) and cortical thickness (Cort.Th) were not rescued (Figure 4C). The fact that Cav-1+/– mice did not have any obviously altered bone phenotype reiterates the notion that partially inhibiting Cav-1 in the context of ASLD could be of therapeutic benefit (Figure 4C).
Figure 4. Heterozygous deletion of Cav-1 partially rescues low bone mass in AslNeo/Neo mice.
(A) Alizarin red staining and (B) mRNA levels of Ocn and Col1a1 by qPCR of BMSC-derived osteoblasts from WT, AslNeo/Neo, and AslNeo/Neo;Cav-1+/– mice. BMSCs were differentiated in osteogenic medium for 14 days. n = 3–4. (C) μCT analysis of distal femur from WT, Cav-1+/-, AslNeo/Neo, and AslNeo/Neo;Cav-1+/– mice (n = 5–7). Male, postnatal days 21–23. BV/TV, bone volume/tissue volume ratio; Tb.N, trabecular number; Tb.Th, trabecular thickness; Tb.Sp, trabecular separation; Conn.D, connectivity density; Cort.Th, cortical thickness. Data are presented as mean ± SD. One-way ANOVA followed by Tukey’s multiple-comparison test. *P < 0.05; **P < 0.01; ***P < 0.005. NS, not significant (P > 0.05).
Asl deletion in the osteoblast lineage leads to decreased bone mass due to impaired osteoblast differentiation and function.
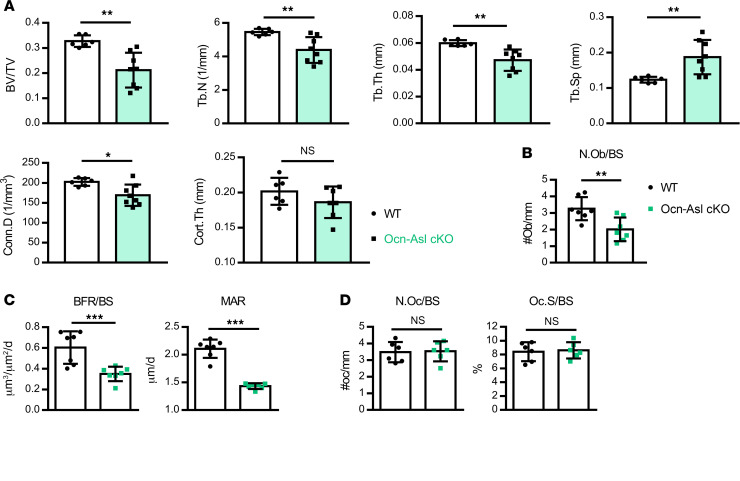
To investigate the osteoblast-specific function of ASL-dependent NO in vivo, Aslfl/fl mice were crossed with Ocn-Cre–transgenic mice to obtain Aslfl/fl;Ocn-Cre+ conditional KO (Ocn-Asl–cKO) mice. Ocn-Cre deletes the floxed alleles in differentiated and mature osteoblasts. μCT analysis in 3-month-old mice showed that Ocn-Asl–cKO mice exhibited lower bone mass as compared with the control mice (Aslfl/fl) (Figure 5A). BV/TV, Tb.N, Tb.Th, and Conn.D were all reduced, while Tb.Sp was increased, and Cort.Th was unaltered in Ocn-Asl–cKO mice compared with control mice (Figure 5A).
Figure 5. Asl deletion in osteoblast lineage cells leads to decreased bone mass due to decreased bone formation.
(A) μCT analysis of distal femur of WT (Aslfl/fl) and Ocn-Asl–cKO mice. Bone parameters from distal femur of 3-month-old male mice. (B–D) Histomorphometric analysis of the distal femur of 3-month-old male WT (Aslfl/fl) and Ocn-Asl–cKO mice. (B) Osteoblast number/bone surface (N.Ob/BS). (C) Dynamic bone formation by double calcein labeling. BFR/BS, bone formation rate; MAR, mineral apposition rate. (D) Parameters osteoclast surface/bone surface (Oc.S/BS) and osteoclast number/bone surface (N.Oc/BS). n = 6–8/group. Data are presented as mean ± SD. Student’s t test. *P < 0.05; **P < 0.01; ***P < 0.005. NS, not significant (P > 0.05).
To determine the in vivo cellular phenotype caused by loss of Asl in the osteoblast lineage, we performed bone histomorphometric analyses for both osteoblasts and osteoclasts in WT and Ocn-Asl–cKO mice. Static bone histomorphometry showed that osteoblast number (N.Ob/BS) was lower in Ocn-Asl–cKO mice compared with WT mice (Figure 5B). Dynamic histomorphometry using double calcein labeling showed that Ocn-Asl–cKO mice exhibited lower bone formation rate (BFR/BS) and mineral apposition rate (MAR) (Figure 5C). In addition, histomorphometry demonstrated that osteoclast surface (Oc.S/BS) and number (N.Oc/BS) were unaltered in Ocn-Asl–cKO mice compared with WT mice (Figure 5D). Taken together, these results show that the low-bone-mass phenotype in the Ocn-Asl–cKO mice is primarily due to decreased osteoblast-mediated bone formation. These results also support a critical role of cell-autonomous NO in the maintenance of in vivo bone formation.
Effect of ASL deficiency on osteoblast function and bone mineral density in humans with ASLD.
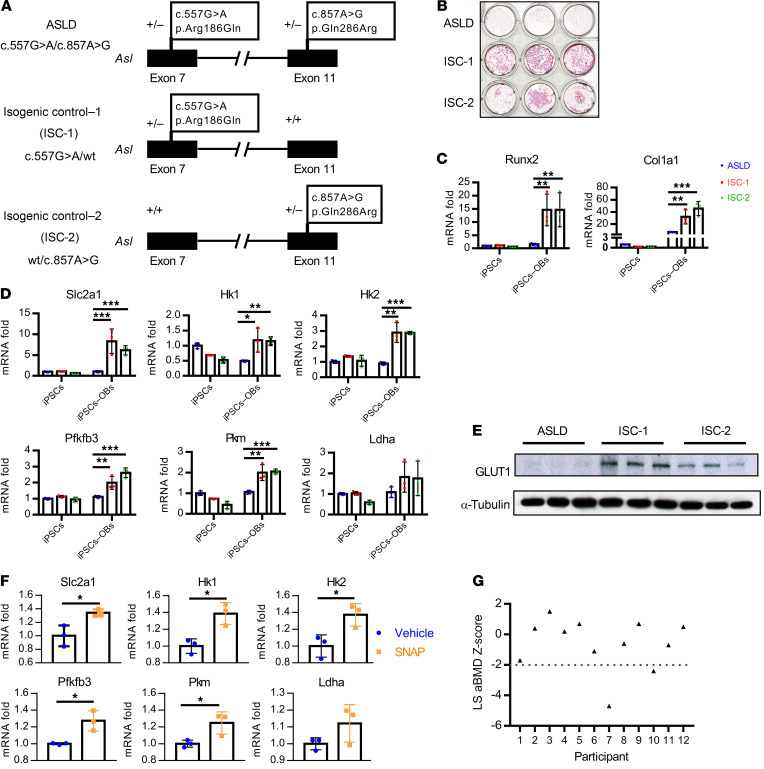
To evaluate the effect of loss of ASL on human osteoblasts, we generated induced pluripotent stem cells (iPSCs) from an individual with ASLD (pathogenic variants in ASL c.557G>A or p.Arg186Gln and c.857A>G or p.Gln286Arg, NCBI reference sequence NM_001024943.1; ref. 32). This individual was diagnosed with ASLD at 3 years of age, with hypertension at 5 years of age, and has not had any documented episodes of hyperammonemia (i.e., plasma ammonia levels >100 μmol/L; ref. 51). We also established 2 isogenic control iPSC lines from this parental line, each with a heterozygous variant in ASL (c.557G>A or c.857A>G) by correcting 1 of the 2 variants (Figure 6A), using a helper-dependent adenovirus system to mediate homologous recombination (ref. 52; Supplemental Methods, and Supplemental Figures 5 and 6).
Figure 6. Effect of ASL deficiency on osteoblast function and bone mineral density in humans with ASLD.
(A) Schematic figure of correction of each of the variants of the compound heterozygous mutations of an individual with ASLD. (B) Alizarin red staining of ASLD and 2 isogenic control iPSC lines, differentiated in osteogenic medium for 21 days. n = 3. (C) mRNA levels of osteoblast marker genes Runx2, Col1a1, and (D) glycolytic genes Slc2a1, Hk1, Hk2, Pfkfb3, Pkm, and Ldha by qPCR, before (iPSCs) and after iPSCs were differentiated in osteogenic medium to osteoblasts (iPSCs→OBs) for 14 days. n = 3. (E) GLUT1 protein levels by Western blot. n = 3. iPSCs were differentiated in osteogenic medium for 14 days. n = 3. (F) mRNA levels of glycolytic genes Slc2a1, Hk1, Hk2, Pfkfb3, Pkm, and Ldha by qPCR. iPSCs from an individual with ASLD were differentiated in osteogenic medium and treated with 5 μM DMSO or 5 μM SNAP for 12 days. n = 3. Data are presented as mean ± SD. Student’s t test. *P < 0.05; **P < 0.01; ***P < 0.005. (G) Dot plot of aBMD z scores of lumbar spine (LS) regions L1–L4 in 12 individuals with ASLD. Two individuals had low bone density (L1–L4 z scores less than –2.0).
We evaluated osteoblast differentiation and mineralization capacity using iPSCs from the individual with ASLD as compared to the 2 corrected, isogenic cell lines. The mineralization capacity was markedly increased in both corrected, isogenic cell lines compared with the ASLD parental line, when iPSCs were cultured in osteogenic medium for 21 days followed by alizarin red staining (Figure 6B). The osteoblast differentiation capacity was also increased in the 2 isogenic, corrected iPSC lines as compared with the parental ASLD iPSCs, as assessed by expression of the osteoblast markers Runx2 and Col1a1 after culturing in osteogenic medium for 14 days (Figure 6C).
We also assessed mRNA levels of genes in the glycolysis pathway, before and after iPSCs were differentiated in osteogenic medium for 14 days. We found that mRNA levels of the glycolytic genes Slc2a1 (solute carrier family 2 member 1), HK1 (hexokinase 1), HK2, Pfkfb3, Pkm, and Ldha were significantly higher in the 2 isogenic, corrected cells compared with ASLD iPSCs after differentiation in osteogenic medium, indicating that correcting 1 of the 2 Asl variants was sufficient to restore glycolytic ability (Figure 6D). This is consistent with the recessive inheritance of ASLD and absence of an apparent clinical phenotype in carriers of ASLD. In addition, the protein level of glucose transporter 1 (GLUT1) was dramatically upregulated in the 2 isogenic, corrected iPSCs compared with parental ASLD iPSC–derived osteoblasts (Figure 6E). Importantly, when the iPSCs from the ASLD subject were treated with SNAP during osteoblast differentiation, the mRNA levels of glycolytic genes were upregulated (Figure 6F). These results suggest that iPSCs derived from the individual with ASLD also have decreased osteoblast differentiation ability, which is associated with impaired glycolysis.
ASLD is a rare Mendelian disorder; thus, to assess bone turnover markers and areal bone mineral density (aBMD) in humans with ASLD, we leveraged an ongoing clinical study (NCT03064048) that was being conducted at Texas Children’s Hospital and Baylor College of Medicine. In this sample, 12 individuals (9 males, 3 females) with ASLD were evaluated (Supplemental Table 2). The median age of participants was 9 years (range, 6–21). Seven participants had history of hyperammonemia and 3 had undergone liver transplantation. In these 12 individuals, plasma markers that are representative of bone formation (i.e., OCN and bone-specific alkaline phosphatase [BsALP]), bone resorption (C-terminal telopeptide [CTX]), and aBMD using dual-energy X-ray absorptiometry were assessed. The bone turnover markers were within the age-specific normative values (Supplemental Table 3). The proportion of individuals with ASLD with low bone density defined by a z score of less than –2.0 at the lumbar spine (0.166; n = 2) was statistically higher when compared with the expected proportion (0.0228) from the age- and gender-matched general population data (Figure 6G). The 2 individuals with low bone density did not have any history of fractures. Three individuals with lumbar spine aBMD above a z score of –2.0 had history of fractures after trauma.
Discussion
The role of NO in bone metabolism has been studied in NOS-KO mice as well as a few clinical trials. Murine models that globally lack each isoform of NOS (nNOS, iNOS, and eNOS) and the triple-NOS-KO mice exhibit contrasting bone phenotypes, likely due to the redundancy among the NOS isoforms in these single-KO mice as well as the non–cell-autonomous effects of NO in all the models. Given that ASL is essential for all cell-autonomous, NOS-dependent NO production, we utilized models with the loss-of-function of ASL as an approach to study NO signaling in osteoblasts. We used multiple model systems including ASL knockdown in osteoblastic cell lines, a hypomorphic mouse model of ASLD, an osteoblast-specific KO model of ASLD, and human osteoblasts generated from iPSCs from an individual with ASLD, as well as clinical bone data from a convenient sample of individuals with ASLD. We show that loss of ASL in osteoblasts leads to a decrease in NO production, impairment of osteoblast differentiation and function, and a decrease in bone mass.
The results from the clinical study of bone turnover markers and aBMD in individuals with ASLD have to be interpreted within the context of limitations of the data set. The study sample size was small and the aBMD scores as presented represent the results compared with age- and gender-matched individuals from the general population, although they do serve to correlate with the significant findings of preclinical data generated by a variety of approaches. Multiple factors can influence the overall bone health and aBMD in individuals with ASLD including protein restriction, insufficient intake of micronutrients, repeated hospitalizations, neurocognitive and motor deficits, and concomitant medications, to name a few. With a small sample size, we were unable to control for all of these important clinical covariates. Given the complexity in the clinical presentations, we tested the clinical significance of our preclinical findings utilizing iPSCs from an individual with ASLD as a model of congenital human NO deficiency (32). To support the results from the mouse studies, iPSCs from the individual with ASLD also showed decreased osteoblast differentiation ability, decreased mineralization capacity, and decreased glycolytic gene expression as compared with the 2 isogenic control iPSC lines.
ASL is responsible for de novo synthesis of arginine. It is possible that arginine-derived metabolites such as urea, polyamine, and glutamate may contribute to potential pathomechanisms. However, we believe that NO is the major downstream mediator. ASL forms a complex with the arginine transporter CAT1 and NOS, and directs arginine to the NOS-NO synthetic complex. In the absence of ASL, this leads to selective loss of arginine substrate for NOS-dependent NO production. Moreover, ASLD cells are not arginine deficient in the presence of adequate extracellular arginine, which continues to be transported into other compartments within the cell by multiple mammalian plasma membrane transporters. Loss of ASL disrupts channeling of extracellular arginine to NOS via the CAT1 transporter, but not arginine transport into the cytoplasm in general (31). In support of this, patients, mice, and cells with ASLD still have NO deficiency when arginine is supplemented (31, 53, 54).
Given the diverse effects of NO on bone metabolism, it is important to dissect distinct biological effects and identify the precise roles of downstream mediators. In this study, we explored the possible pathways by comparing WT and Asl-deficient mouse–derived osteoblasts. RNA-seq revealed that many genes in the glycolysis pathway were downregulated in Asl-deficient osteoblasts. Accumulating evidence supports the notion that upregulation of glucose metabolism through aerobic glycolysis is a major metabolic pathway to meet energy demand and promote osteoblast-mediated bone formation (13). Moreover, some of the anabolic agents used to increase bone mass have been shown to drive glycolysis during osteoblast differentiation, thus underscoring an essential role of glycolysis in bone anabolism (14). It has been reported that NO can enhance glycolysis in a variety of cells, such as muscle cells and adipocytes (18–21). To our knowledge, this is among the first reports that NO enhances glycolysis in osteoblasts. It is likely that NO modulates glycolysis by multiple mechanisms. For example, we have shown that the mechanism of metabolic regulation by NO involves the transcriptional upregulation of Slc2a1 as well as multiple glycolytic genes in osteoblasts. GLUT1 has been reported to be the main glucose transporter to mediate glucose uptake in osteoblasts (55). In muscle cells and adipocytes, NO has been reported to increase glucose transporter 4 (GLUT4) expression and enhance glucose uptake (56–58). The specific mechanisms by which NO activates glycolysis are not well understood. Some studies have shown that the canonical soluble guanylyl cyclase (sGC)/cyclic guanosine monophosphate (cGMP) pathway is involved in glucose uptake in skeletal muscle cells; however, others have shown that sGC/cGMP-independent regulation is also involved (59).
In addition to activating the cGMP-dependent pathway, NO can directly modify protein thiols through the posttranslational modification S-nitrosylation. S-nitrosylation leads to changes in protein activity, protein-protein interactions, or subcellular location of the target proteins (60–62). Although a substantial number of enzymes in glycolysis pathways that can be S-nitrosylated have been identified in S-nitrosoproteomic analysis, for example in both plants and mammalian tissues (63–65), little is known about the consequences of this modification in physiological or pathological conditions. In the Seahorse assay, we observed that SNAP treatment partially rescued the glycolytic rate in Asl-knockdown ST2 cells. We also tested a different class of NO donor: NONOate (which releases NO with minimal S-nitrosylation effects), and we did not observe a rescue of glycolysis (data not shown). These data support the idea that S-nitrosylation may play an important role in enhancing glycolysis. Therefore, the identification and functional validation of S-nitrosylation in glycolytic proteins in osteoblasts may provide new insights into NO signaling in bone anabolism. In addition to the defective glycolytic phenotype, we observed that ASLD led to impaired oxygen consumption in BMSCs and BMSC-derived osteoblasts from AslNeo/Neo mice. It has been demonstrated that NO exerts diverse effects on mitochondrial functions, depending on the cellular context (66–69). Our data suggest that cell-autonomous and physiological levels of NO may promote mitochondrial oxygen consumption in osteoblast lineage cells.
In the present study, we confirmed the hypothesis that genetic alteration of positive and negative regulators in the NOS-NO synthetic complex could modulate NO production and subsequently influence in vivo bone mass. We showed that heterozygous deletion of Cav-1 resulted in increased NO production, overcoming the NO deficiency in the context of ASLD. We demonstrated partial restoration of bone mass in AslNeo/Neo;Cav-1+/– mice, accompanied with reversal of glycolytic gene expression. This strongly suggests that manipulating the balance of negative versus positive regulators of NO production could be a therapeutic way to modulate NO-mediated bone metabolism. However, Cav-1 downstream NO-independent pathways may also contribute to the rescue of the low bone mass in AslNeo/Neo mice. In addition, beyond supporting NO specificity of the ASLD phenotype here, targeting Cav-1 function may be a viable approach for treating ASLD and NO production in vivo. Together, our results address the contribution of ASLD as a nonredundant regulator of cell-autonomous NO production in bone metabolism.
Methods
Human studies.
To evaluate bone turnover and bone mineral density in individuals with ASLD, we leveraged an ongoing clinical trial that was being conducted at Texas Children’s Hospital (TCH) and Baylor College of Medicine (BCM) (NCT03064048). When participants were enrolled in the parent clinical study, ancillary study procedures to measure aBMD and plasma markers of bone turnover were implemented.
The inclusion criteria for enrollment were the following: (a) age between 6 and 50 years, (b) diagnosis of ASLD based on presence of argininosuccinic acid in plasma or urine or reduced ASL enzymatic activity from fibroblasts or red blood cells or 2 pathogenic or likely pathogenic variants in ASL, and (c) history of compliance with diet and treatment regimens. Individuals with the following characteristics were excluded: (a) clinical or laboratory abnormality of grade 3 or greater according to the Common Terminology Criteria for Adverse Events (CTCAE, version 4.0; https://evs.nci.nih.gov/ftp1/CTCAE/CTCAE_4.03/Archive/CTCAE_4.0_2009-05-29_QuickReference_8.5x11.pdf); (b) clinical or laboratory abnormalities not covered by the CTCAE, but representing a severe or severe or life-threatening condition; (c) known hypersensitivity to nitrite; and (d) inability to lie still for measurement of bone density. All females of child-bearing potential had to have a negative pregnancy test and were required to use an acceptable birth control method for the duration of the study. All males who had the potential to father children were required to use an acceptable form of birth control for the duration of the study. Information regarding the diagnosis of ASLD, history of hyperammonemia, prescribed diet, and concomitant medications including the use of nitrogen-scavenging medications was collected by review of medical records and interview of the participant and/or their parent(s). The presence of a history of hyperammonemia in a participant was defined by a plasma ammonia level greater than 100 μmol/L at least once during their lifetime. The prescribed protein intake from diet and supplemental formulae in each participant was collected based on review of the medical records. The percentage of the total protein intake per day as compared with the recommended daily intake (RDI) was calculated using the age- and stature-specific RDI data available from the United States Department of Agriculture (https://www.nal.usda.gov/fnic/dri-calculator/). Height and weight were measured by trained research staff at the Clinical Research Center of TCH. The z scores for height and weight were calculated using growth data from the Centers for Disease Control. Markers of bone turnover in plasma (i.e., BsALP, OCN, and CTX) were measured by the CLIA- and CAP-certified clinical diagnostic laboratory of TCH. aBMD was measured by dual-energy X-ray absorptiometry using a Hologic Discovery A densitometer that has been validated for clinical use. The aBMD measurements were performed by qualified technologists and interpreted by radiologists who were not involved in the design or implementation of the study. The lumbar spine aBMD z scores were generated using a software from Hologic as per the manufacturer’s protocol. Here, only cross-sectional results from the baseline study visit are presented. The comparison between the proportion of individuals with ASLD who had low bone density (z score less than –2.0 at the lumbar spine) with expected proportions from the general population was assessed using a 1-sample test of proportions. A P value of less than 0.05 was considered statistically significant.
Mice.
The generation of AslNeo/Neo mice and Aslfl/fl mice was previously described (31). To generate osteoblast-specific Asl-KO mice, Ocn-Cre transgenic mice (provided by Thomas Clemens, Johns Hopkins University, Baltimore, Maryland, USA; ref. 70) were bred with Aslfl/fl mice. Cav-1 gene–deleted mice were purchased from The Jackson Laboratory (B6-Cg-Cav1tm1Mis/J). To generate AslNeo/Neo;Cav-1+/– mice, Asl+/Neo mice were bred with Cav-1–/– mice to generate Asl+/Neo;Cav-1+/– mice. Asl+/Neo;Cav-1+/– mice were bred with Asl+/Neo mice to generate AslNeo/Neo mice and AslNeo/Neo;Cav-1+/– mice for experiments. All mice were maintained on the C57BL/6J background.
Cell culture.
The bone marrow–derived stromal cell line ST2 was provided by the laboratory of Fanxin Long (University of Pennsylvania, Philadelphia, Pennsylvania, USA). ST2 cells were maintained in α-MEM supplemented with 10% FBS and 1% penicillin/streptomycin (1% P/S).
To generate BMSCs, bone marrow cells were isolated from mouse long bones. Briefly, femurs and tibiae were dissected and both ends as epiphyses were cut off. Bone marrow was flushed from the long bones and passed through a 70-μm cell strainer. Cells were cultured in α-MEM containing 10% FBS and 1% P/S, with medium change every other day to remove nonadherent cells. On day 7, cells were dissociated with trypsin and plated for subsequent experiments.
For osteogenic induction, 100 μg/mL ascorbic acid (Sigma-Aldrich) and 5 mM β-glycerophosphate (Sigma-Aldrich) were added to α-MEM containing 10% FBS and 1% P/S with a change of medium every other day. The total differentiation days are stated in the Results section.
Osteogenic differentiation of human iPSCs.
The generation of iPSCs from an individual with ASLD was previously reported (compound hemizygous; ASLD 1-1; ref. 32). In this study, we generated 2 isogenic control iPSC lines by correcting each of the mutant variants using a helper-dependent adenovirus system (Supplemental Figures 5 and 6).
iPSCs were differentiated into osteoblast-like cells as previously described with some modifications (71). Briefly, Accutase-dissociated single iPSCs were first plated in gelatin-coated 12-well plates at a density of 4 × 105 cells/well. Osteoblast differentiation medium was supplemented with ROCK inhibitor (Y27632) for the first 2 days. The osteoblast differentiation medium consisted of 20% FBS, 2 mM glutamine, 1% nonessential amino acids, 0.1 mM β-mercaptoethanol, 10 mM β-glycerophosphate, 1 nM dexamethasone, and 50 μg/mL ascorbic acid (made fresh), with Knockout DMEM (Gibco) as the basal medium. The medium was changed every other day. The total differentiation days are stated in the Results section.
Skeletal analyses and bone histomorphometry.
Femurs were collected from 3-month-old male WT (Aslfl/fl) mice and Ocn-Asl–cKO mice. Femurs were collected on postnatal days 21–23 from male AslNeo/Neo and AslNeo/Neo;Cav-1+/– mice. μCT was performed using the Scanco μCT-40 system (Scanco Medical) to quantify bone parameters. Undecalcified femurs were embedded in plastic for sectioning. Trichrome and tartrate-resistant acid phosphatase (TRAP) staining were performed for osteoblast and osteoclast quantification (Bioquant Osteo Image Analysis System). For dynamic histomorphometry, calcein (20 mg/kg) was injected 2 times with an interval of 7 days before bone collection.
Gene expression analysis by qPCR.
RNA was extracted with TRIzol reagent, and cDNA was synthesized using a Superscript III First-Strand RT-PCR kit (Invitrogen). RNA expression was analyzed by qPCR with SYBR Green I reagent (Roche). Rpl19 was used as reference gene for normalization of mouse and cell line experiments. Hprt1 was used as reference gene for iPSC line experiments.
siRNA-mediated knockdown of Asl.
ST2 or MC3T3 cells were plated at 1 × 105 cells/well in 12-well plates overnight, and transfected with 10 pmol/well si-control (siGENOME Non-Targeting siRNA Pool 1; Dharmacon) or si-Asl (SMARTpool siRNAs; Dharmacon) using Lipofectamine RNAiMAX (Thermo Fisher Scientific) according to the manufacturer’s instructions.
RNA-seq and analysis.
BMSCs generated from primary bone marrow cells were differentiated in osteogenic medium for 14 days. Total RNA was extracted from cells using an AllPrep DNA/RNA/Protein Mini Kit (QIAGEN). Strand-specific libraries were generated using the TruSeq Stranded mRNA sample preparation kit according to the manufacturer’s instructions (Illumina, document 1000000040498 v00). Briefly, polyadenylated RNA from 1000 ng intact total RNA was purified using oligo-dT beads, fragmented, random primed, and reverse transcribed. Amplified cDNA was validated and quantified on an Agilent Bioanalyzer with the High-Sensitivity DNA chip. The purified libraries were normalized, pooled together, denatured, and diluted according to the manual instructions and followed by 1 × 75 bp sequencing on a NextSeq 500 (Illumina). Raw reads were aligned to a references genome (mm10, UCSC assembly) using TopHat v2.0.14 and Bowtie v2.10 with the following parameters: -no-coverage-search-library-type fr-firststrand –G were GFF file was obtained from Genome Reference Consortium Mouse Build 38, patch release 4. The distribution of alignments was analyzed using Cufflinks v2.2.1; FPKM values were quantile normalized. Differential expression was evaluated using Cuffdiff v2.2.1. Differentially expressed genes with a threshold of 1.5-fold change were used for IPA core analysis (Ingenuity Systems). The raw RNA-seq data were deposited in NCBI’s sequence read archive (SRA) database, with the BioProject accession number PRJNA681756. The heatmap was generated using the online Morpheus software (https://software.broadinstitute.org/morpheus/).
ECAR and OCR measurements with Seahorse assay.
For Seahorse (Agilent) tests using BMSC-derived osteoblasts, BMSCs generated from primary bone marrow cells were plated in XF96 cell culture microplates (Agilent) at a density of 3 × 104 cells/well in α-MEM supplemented with 10% FBS and 1% P/S, and differentiated in osteogenic medium for 7 days with medium change every other day. On the day of assay, for glycolysis tests, medium was switched to Seahorse XF assay medium (without glucose), and cells were incubated in a 37°C CO2-free incubator for 1 hour. The basal level of ECAR, and ECAR following sequential additions of glucose (final concentration was 10 mM), oligomycin (final concentration was 1 μM), and 2-deoxyglucose (2-DG; final concentration was 50 mM) were measured using the Seahorse XFe96 Analyzer. On the day of assay, for mitochondrial stress test (Agilent, Seahorse XF Cell Mito Stress Test Kit, 103015-100), medium was switched to Seahorse XF assay medium supplemented with 1 mM pyruvate, 2 mM glutamine, and 10 mM glucose, and cells were incubated in a 37°C CO2-free incubator for 1 hour. The basal level of OCR, and OCR following sequential additions of oligomycin (final concentration was 1.5 μM), FCCP (final concentration was 0.5 μM), and rotenone and antimycin A (final concentration was 0.5 μM) were measured using the Seahorse XFe96 Analyzer. After the assay, total DNA amount was measured for normalization (when BMSCs were differentiated in XF96 microplates for 7 days). For ST2 cell Seahorse assay, 48 hours after transfection of si-control or si-Asl, ST2 cells were differentiated in osteogenic medium for 3 days, dissociated with trypsin, plated in XF96 cell culture microplates (Agilent) at 3 × 104cells/well the day before the assay, and treated with vehicle control (DMSO), NAP (Cayman Chemical), or SNAP (Sigma-Aldrich) 18 hours before the assay. For Seahorse assay using BMSCs, BMSCs were plated in XF96 cell culture microplates at 3 × 104cells/well, 1 day before the assay. This cell density ensured similar numbers and high confluence of plated cells in each condition.
Western blot.
BMSCs were differentiated in osteogenic medium for 14 days, washed with cold PBS, and directly lysed in Laemmli buffer (Bio-Rad). To extract protein from the long bone, femurs and tibiae were dissected and soft tissue was removed, and both ends as epiphyses were removed. Bone marrow was removed from the long bones and the remainder of the long bones was chopped into small pieces and homogenized (IKA T25 Basic Ultra-Turrax Homogenizer) in RIPA buffer (Thermo Fisher Scientific [89900] and supplemented with Protease Inhibitor from Roche [11836170001]) at maximal speed. A standard protocol was used for Western blotting. In brief, samples were resolved in an SDS polyacrylamide gel, transferred onto a 0.45 μm PVDF membrane (Thermo Fisher Scientific), and incubated with the primary antibodies as follows: Cav-1 antibody (Cell Signaling Technology, 3267s), GLUT1 antibody (Cell Signaling Technology, 12939s), PFKFB3 antibody (Cell Signaling Technology, 13123s), PKM antibody (Abcam, ab137791), LDHA antibody (Abcam, ab47010), α-tubulin antibody (Invitrogen, T5168), and HRP-conjugated secondary antibodies. Western blot images were quantified with ImageJ (NIH).
Mitochondrial DNA copy number by real-time PCR.
Total DNA was extracted from cell samples using InstaGene Matrix (Bio-Rad, 7326030) according to the manufacturer’s instructions. The relative mitochondrial DNA copy number was determined by real-time PCR with specific primers for the mitochondrial gene MT-ND5 (forward: 5′-TTATTGAAGCAATTAATACCTGCAACAC-3′; reverse: 5′-CGGTTTTGTTATTGTTACGAAGTAAATG-3′), and was normalized to the nuclear gene GAPDH (forward: 5′-GCAAGAGAGGCCCTATCCCAA-3′; reverse: 5′-CTCCCTAGGCCCCTCCTGTTATT-3′).
DAF-FM test of NO.
DAF-FM diacetate (Thermo Fisher Scientific) was used to detect and quantify relative NO concentrations. Equal numbers of cells were labeled with 10 μM DAF-FM diacetate at room temperature for 45 minutes, washed twice in PBS, incubated in PBS for an additional 30 minutes, and NO signal was measured by fluorescence microplate reader.
Statistics.
All results are presented as mean ± SD. Statistically significant differences between 2 groups were determined by 2-tailed Student’s t test. Comparisons between multiple groups were performed by 1-way or 2-way ANOVA followed by Tukey’s multiple-comparison test. *P < 0.05; **P < 0.01; ***P < 0.005; NS, not significant (P > 0.05).
Study approval.
The study procedures were reviewed and approved by the Institutional Review Board (IRB) of Baylor College of Medicine. Informed written consent was obtained from all participants or their parents. All animal care and experimental procedures were approved by the IACUC of Baylor College of Medicine.
Author contributions
ZJ conceptualized the project, designed the experiments, performed most of the experiments, and wrote the manuscript. JK assisted with iPSC culture and differentiation. BD assisted with μCT analyses. MMJ assisted with real-time qPCR. YC assisted with histomorphometry. MG assisted with initial mouse breeding. DJP, DLT, and PN generated iPSC isogenic lines. SA, LCB, and SCSN performed human studies. BL supervised the project and wrote the manuscript.
Supplementary Material
Acknowledgments
We would like to thank Megan Bagos and Carrie Jiang for assistance with μCT analyses. This work was supported by the BCM Intellectual and Developmental Disabilities Research Center (P30 HD024064, U54 HD083092, and P50 HD103555) from the National Institute of Child Health & Human Development, the BCM Advanced Technology Cores with funding from the NIH (R01 DK114356 and UM1 HG006348), the Rolanette and Berdon Lawrence Bone Disease Program of Texas, and the BCM Center for Skeletal Medicine and Biology. This project was also supported by the BCM Human Stem Cell Core, which is funded in part by the institution and the NIH (P30 CA125123 and S10 OD028591) and NIH grants R01 DK102641 and R01 AR071741. LCB holds a Career Award for Medical Scientists from the Burroughs Wellcome Fund and was supported by NIH K08 DK106453.
Version 1. 12/29/2020
In-Press Preview
Version 2. 03/01/2021
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2021, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2021;131(5):e138935.https://doi.org/10.1172/JCI138935.
See the related Commentary at Nitric oxide and bone: The phoenix rises again.
Contributor Information
Zixue Jin, Email: zixue.jin@bcm.edu.
Jordan Kho, Email: kho@bcm.edu.
Brian Dawson, Email: bcd@bcm.tmc.edu.
Ming-Ming Jiang, Email: mjiang@bcm.edu.
Saima Ali, Email: saima.ali@bcm.edu.
Lindsay C. Burrage, Email: burrage@bcm.edu.
Monica Grover, Email: mgrover@stanford.edu.
Donna J. Palmer, Email: palmer@bcm.edu.
Dustin L. Turner, Email: Dustin.turner81@yahoo.com.
Philip Ng, Email: png@bcm.edu.
Sandesh C.S. Nagamani, Email: nagamani@bcm.edu.
Brendan Lee, Email: blee@bcm.edu.
References
- 1.Wimalawansa SJ. Nitroglycerin therapy is as efficacious as standard estrogen replacement therapy (Premarin) in prevention of oophorectomy-induced bone loss: a human pilot clinical study. J Bone Miner Res. 2000;15(11):2240–2244. doi: 10.1359/jbmr.2000.15.11.2240. [DOI] [PubMed] [Google Scholar]
- 2.Wimalawansa SJ, et al. Nitric oxide donor alleviates ovariectomy-induced bone loss. Bone. 1996;18(4):301–304. doi: 10.1016/8756-3282(96)00005-1. [DOI] [PubMed] [Google Scholar]
- 3.Hukkanen M, et al. Effect of nitric oxide donor nitroglycerin on bone mineral density in a rat model of estrogen deficiency-induced osteopenia. Bone. 2003;32(2):142–149. doi: 10.1016/S8756-3282(02)00955-9. [DOI] [PubMed] [Google Scholar]
- 4.Kalyanaraman H, et al. A novel, direct NO donor regulates osteoblast and osteoclast functions and increases bone mass in ovariectomized mice. J Bone Miner Res. 2017;32(1):46–59. doi: 10.1002/jbmr.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pouwels S, et al. Use of organic nitrates and the risk of hip fracture: a population-based case-control study. J Clin Endocrinol Metab. 2010;95(4):1924–1931. doi: 10.1210/jc.2009-2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rejnmark L, et al. Decreased fracture risk in users of organic nitrates: a nationwide case-control study. J Bone Miner Res. 2006;21(11):1811–1817. doi: 10.1359/jbmr.060804. [DOI] [PubMed] [Google Scholar]
- 7.Wimalawansa SJ, et al. Transdermal nitroglycerin therapy may not prevent early postmenopausal bone loss. J Clin Endocrinol Metab. 2009;94(9):3356–3364. doi: 10.1210/jc.2008-2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shum LC, et al. Energy metabolism in mesenchymal stem cells during osteogenic differentiation. Stem Cells Dev. 2016;25(2):114–122. doi: 10.1089/scd.2015.0193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shyh-Chang N, et al. Stem cell metabolism in tissue development and aging. Development. 2013;140(12):2535–2547. doi: 10.1242/dev.091777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guntur AR, et al. Osteoblast-like MC3T3-E1 cells prefer Glycolysis for ATP production but adipocyte-like 3T3-L1 cells prefer oxidative phosphorylation. J Bone Miner Res. 2018;33(6):1052–1065. doi: 10.1002/jbmr.3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jin Z, et al. Mitochondrial complex I activity suppresses inflammation and enhances bone resorption by shifting macrophage-osteoclast polarization. Cell Metab. 2014;20(3):483–498. doi: 10.1016/j.cmet.2014.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dirckx N, et al. Vhl deletion in osteoblasts boosts cellular glycolysis and improves global glucose metabolism. J Clin Invest. 2018;128(3):1087–1105. doi: 10.1172/JCI97794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karner CM, Long F. Glucose metabolism in bone. Bone. 2018;115:2–7. doi: 10.1016/j.bone.2017.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee WC, et al. Energy metabolism of the osteoblast: implications for osteoporosis. Endocr Rev. 2017;38(3):255–266. doi: 10.1210/er.2017-00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Esen E, et al. PTH promotes bone anabolism by stimulating aerobic glycolysis via IGF signaling. J Bone Miner Res. 2015;30(11):1959–1968. doi: 10.1002/jbmr.2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen H, et al. Increased glycolysis mediates Wnt7b-induced bone formation. FASEB J. 2019;33(7):7810–7821. doi: 10.1096/fj.201900201RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Esen E, et al. WNT-LRP5 signaling induces Warburg effect through mTORC2 activation during osteoblast differentiation. Cell Metab. 2013;17(5):745–755. doi: 10.1016/j.cmet.2013.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Etgen GJ, Jr, et al. Nitric oxide stimulates skeletal muscle glucose transport through a calcium/contraction- and phosphatidylinositol-3-kinase-independent pathway. Diabetes. 1997;46(11):1915–1919. doi: 10.2337/diab.46.11.1915. [DOI] [PubMed] [Google Scholar]
- 19.Stamler JS, Meissner G. Physiology of nitric oxide in skeletal muscle. Physiol Rev. 2001;81(1):209–237. doi: 10.1152/physrev.2001.81.1.209. [DOI] [PubMed] [Google Scholar]
- 20.Bolanos JP, et al. Regulation of glycolysis and pentose-phosphate pathway by nitric oxide: impact on neuronal survival. Biochim Biophys Acta. 2008;1777(7–8):789–793. doi: 10.1016/j.bbabio.2008.04.011. [DOI] [PubMed] [Google Scholar]
- 21.Roy D, et al. Insulin stimulation of glucose uptake in skeletal muscles and adipose tissues in vivo is NO dependent. Am J Physiol. 1998;274(4):E692–E699. doi: 10.1152/ajpendo.1998.274.4.E692. [DOI] [PubMed] [Google Scholar]
- 22.Basso N, Heersche JN. Effects of hind limb unloading and reloading on nitric oxide synthase expression and apoptosis of osteocytes and chondrocytes. Bone. 2006;39(4):807–814. doi: 10.1016/j.bone.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 23.Hukkanen MV, et al. Developmental regulation of nitric oxide synthase expression in rat skeletal bone. J Bone Miner Res. 1999;14(6):868–877. doi: 10.1359/jbmr.1999.14.6.868. [DOI] [PubMed] [Google Scholar]
- 24.Helfrich MH, et al. Expression of nitric oxide synthase isoforms in bone and bone cell cultures. J Bone Miner Res. 1997;12(7):1108–1115. doi: 10.1359/jbmr.1997.12.7.1108. [DOI] [PubMed] [Google Scholar]
- 25.Aguirre J, et al. Endothelial nitric oxide synthase gene-deficient mice demonstrate marked retardation in postnatal bone formation, reduced bone volume, and defects in osteoblast maturation and activity. Am J Pathol. 2001;158(1):247–257. doi: 10.1016/S0002-9440(10)63963-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Armour KE, et al. Defective bone formation and anabolic response to exogenous estrogen in mice with targeted disruption of endothelial nitric oxide synthase. Endocrinology. 2001;142(2):760–766. doi: 10.1210/endo.142.2.7977. [DOI] [PubMed] [Google Scholar]
- 27.van’t Hof RJ, et al. Regulation of bone mass and bone turnover by neuronal nitric oxide synthase. Endocrinology. 2004;145(11):5068–5074. doi: 10.1210/en.2004-0205. [DOI] [PubMed] [Google Scholar]
- 28.Watanuki M, et al. Role of inducible nitric oxide synthase in skeletal adaptation to acute increases in mechanical loading. J Bone Miner Res. 2002;17(6):1015–1025. doi: 10.1359/jbmr.2002.17.6.1015. [DOI] [PubMed] [Google Scholar]
- 29.Gyurko R, et al. Inducible nitric oxide synthase mediates bone development and P. gingivalis-induced alveolar bone loss. Bone. 2005;36(3):472–479. doi: 10.1016/j.bone.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 30.Sabanai K, et al. Genetic disruption of all NO synthase isoforms enhances BMD and bone turnover in mice in vivo: involvement of the renin-angiotensin system. J Bone Miner Res. 2008;23(5):633–643. doi: 10.1359/jbmr.080107. [DOI] [PubMed] [Google Scholar]
- 31.Erez A, et al. Requirement of argininosuccinate lyase for systemic nitric oxide production. Nat Med. 2011;17(12):1619–1626. doi: 10.1038/nm.2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kho J, et al. Argininosuccinate lyase deficiency causes an endothelial-dependent form of hypertension. Am J Hum Genet. 2018;103(2):276–287. doi: 10.1016/j.ajhg.2018.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bucci M, et al. In vivo delivery of the caveolin-1 scaffolding domain inhibits nitric oxide synthesis and reduces inflammation. Nat Med. 2000;6(12):1362–1367. doi: 10.1038/82176. [DOI] [PubMed] [Google Scholar]
- 34.Mineo C, Shaul PW. Circulating cardiovascular disease risk factors and signaling in endothelial cell caveolae. Cardiovasc Res. 2006;70(1):31–41. doi: 10.1016/j.cardiores.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 35.Trane AE, et al. Deciphering the binding of caveolin-1 to client protein endothelial nitric-oxide synthase (eNOS): scaffolding subdomain identification, interaction modeling, and biological significance. J Biol Chem. 2014;289(19):13273–13283. doi: 10.1074/jbc.M113.528695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Williams TM, Lisanti MP. The caveolin proteins. Genome Biol. 2004;5(3):214. doi: 10.1186/gb-2004-5-3-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rothberg KG, et al. Caveolin, a protein component of caveolae membrane coats. Cell. 1992;68(4):673–682. doi: 10.1016/0092-8674(92)90143-Z. [DOI] [PubMed] [Google Scholar]
- 38.Stan RV. Structure of caveolae. Biochim Biophys Acta. 2005;1746(3):334–348. doi: 10.1016/j.bbamcr.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 39.Scherer PE, et al. Identification, sequence, and expression of caveolin-2 defines a caveolin gene family. Proc Natl Acad Sci U S A. 1996;93(1):131–135. doi: 10.1073/pnas.93.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu P, et al. Multiple functions of caveolin-1. J Biol Chem. 2002;277(44):41295–41298. doi: 10.1074/jbc.R200020200. [DOI] [PubMed] [Google Scholar]
- 41.Gratton JP, et al. Caveolae and caveolins in the cardiovascular system. Circ Res. 2004;94(11):1408–1417. doi: 10.1161/01.RES.0000129178.56294.17. [DOI] [PubMed] [Google Scholar]
- 42.Garcia-Cardena G, et al. Endothelial nitric oxide synthase is regulated by tyrosine phosphorylation and interacts with caveolin-1. J Biol Chem. 1996;271(44):27237–27240. doi: 10.1074/jbc.271.44.27237. [DOI] [PubMed] [Google Scholar]
- 43.Feron O, et al. Endothelial nitric oxide synthase targeting to caveolae. Specific interactions with caveolin isoforms in cardiac myocytes and endothelial cells. J Biol Chem. 1996;271(37):22810–22814. doi: 10.1074/jbc.271.37.22810. [DOI] [PubMed] [Google Scholar]
- 44.Michel JB, et al. Reciprocal regulation of endothelial nitric-oxide synthase by Ca2+-calmodulin and caveolin. J Biol Chem. 1997;272(25):15583–15586. doi: 10.1074/jbc.272.25.15583. [DOI] [PubMed] [Google Scholar]
- 45.Lerner S, et al. ASL metabolically regulates tyrosine hydroxylase in the nucleus locus coeruleus. Cell Rep. 2019;29(8):2144–2153. doi: 10.1016/j.celrep.2019.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stettner N, et al. Induction of nitric-oxide metabolism in enterocytes alleviates colitis and inflammation-associated colon cancer. Cell Rep. 2018;23(7):1962–1976. doi: 10.1016/j.celrep.2018.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee WC, et al. Malic enzyme couples mitochondria with aerobic glycolysis in osteoblasts. Cell Rep. 2020;32(10):108108. doi: 10.1016/j.celrep.2020.108108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Drab M, et al. Loss of caveolae, vascular dysfunction, and pulmonary defects in caveolin-1 gene-disrupted mice. Science. 2001;293(5539):2449–2452. doi: 10.1126/science.1062688. [DOI] [PubMed] [Google Scholar]
- 49.Razani B, et al. Caveolin-1 null mice are viable but show evidence of hyperproliferative and vascular abnormalities. J Biol Chem. 2001;276(41):38121–38138. doi: 10.1074/jbc.M105408200. [DOI] [PubMed] [Google Scholar]
- 50.Zhao YY, et al. Persistent eNOS activation secondary to caveolin-1 deficiency induces pulmonary hypertension in mice and humans through PKG nitration. J Clin Invest. 2009;119(7):2009–2018. doi: 10.1172/JCI33338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nagamani SC, et al. Nitric-oxide supplementation for treatment of long-term complications in argininosuccinic aciduria. Am J Hum Genet. 2012;90(5):836–846. doi: 10.1016/j.ajhg.2012.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Palmer DJ, et al. Homology requirements for efficient, footprintless gene editing at the CFTR locus in human iPSCs with helper-dependent adenoviral vectors. Mol Ther Nucleic Acids. 2016;5(10):e372. doi: 10.1038/mtna.2016.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Erez A, et al. Argininosuccinate lyase deficiency-argininosuccinic aciduria and beyond. Am J Med Genet C Semin Med Genet. 2011;157C(1):45–53. doi: 10.1002/ajmg.c.30289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gladwin MT, Tejero J. Nitrite-NO bailout for a NOS complex too big to fail. Nat Med. 2011;17(12):1556–1557. doi: 10.1038/nm.2591. [DOI] [PubMed] [Google Scholar]
- 55.Wei J, et al. Glucose uptake and runx2 synergize to orchestrate osteoblast differentiation and bone formation. Cell. 2015;161(7):1576–1591. doi: 10.1016/j.cell.2015.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lira VA, et al. Nitric oxide increases GLUT4 expression and regulates AMPK signaling in skeletal muscle. Am J Physiol Endocrinol Metab. 2007;293(4):E1062–E1068. doi: 10.1152/ajpendo.00045.2007. [DOI] [PubMed] [Google Scholar]
- 57.Kellogg DL, et al. Neuronal nitric oxide synthase mediates insulin- and oxidative stress-induced glucose uptake in skeletal muscle myotubes. Free Radic Biol Med. 2017;110:261–269. doi: 10.1016/j.freeradbiomed.2017.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tanaka T, et al. Nitric oxide stimulates glucose transport through insulin-independent GLUT4 translocation in 3T3-L1 adipocytes. Eur J Endocrinol. 2003;149(1):61–67. doi: 10.1530/eje.0.1490061. [DOI] [PubMed] [Google Scholar]
- 59.McConell GK, et al. Skeletal muscle nitric oxide signaling and exercise: a focus on glucose metabolism. Am J Physiol Endocrinol Metab. 2012;303(3):E301–E307. doi: 10.1152/ajpendo.00667.2011. [DOI] [PubMed] [Google Scholar]
- 60.Hess DT, et al. Protein S-nitrosylation: purview and parameters. Nat Rev Mol Cell Biol. 2005;6(2):150–166. doi: 10.1038/nrm1569. [DOI] [PubMed] [Google Scholar]
- 61.Stamler JS, et al. Nitrosylation. the prototypic redox-based signaling mechanism. Cell. 2001;106(6):675–683. doi: 10.1016/S0092-8674(01)00495-0. [DOI] [PubMed] [Google Scholar]
- 62.Jaffrey SR, et al. Protein S-nitrosylation: a physiological signal for neuronal nitric oxide. Nat Cell Biol. 2001;3(2):193–197. doi: 10.1038/35055104. [DOI] [PubMed] [Google Scholar]
- 63.Doulias PT, et al. Nitric oxide regulates mitochondrial fatty acid metabolism through reversible protein S-nitrosylation. Sci Signal. 2013;6(256):rs1. doi: 10.1126/scisignal.2003252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Qiu C, et al. First nitrosoproteomic profiling deciphers the cysteine S-nitrosylation involved in multiple metabolic pathways of tea leaves. Sci Rep. 2019;9(1):17525. doi: 10.1038/s41598-019-54077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tan C, et al. Extensive protein S-nitrosylation associated with human pancreatic ductal adenocarcinoma pathogenesis. Cell Death Dis. 2019;10(12):914. doi: 10.1038/s41419-019-2144-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Brown GC. Regulation of mitochondrial respiration by nitric oxide inhibition of cytochrome c oxidase. Biochim Biophys Acta. 2001;1504(1):46–57. doi: 10.1016/S0005-2728(00)00238-3. [DOI] [PubMed] [Google Scholar]
- 67.Nisoli E, et al. Mitochondrial biogenesis in mammals: the role of endogenous nitric oxide. Science. 2003;299(5608):896–899. doi: 10.1126/science.1079368. [DOI] [PubMed] [Google Scholar]
- 68.Nisoli E, et al. Mitochondrial biogenesis by NO yields functionally active mitochondria in mammals. Proc Natl Acad Sci U S A. 2004;101(47):16507–16512. doi: 10.1073/pnas.0405432101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Litvinova L, et al. Nitric oxide and mitochondria in metabolic syndrome. Front Physiol. 2015;6:20. doi: 10.3389/fphys.2015.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang M, et al. Osteoblast-specific knockout of the insulin-like growth factor (IGF) receptor gene reveals an essential role of IGF signaling in bone matrix mineralization. J Biol Chem. 2002;277(46):44005–44012. doi: 10.1074/jbc.M208265200. [DOI] [PubMed] [Google Scholar]
- 71.Barruet E, Hsiao EC. Using human induced pluripotent stem cells to model skeletal diseases. Methods Mol Biol. 2016;1353:101–118. doi: 10.1007/7651_2014_171. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.