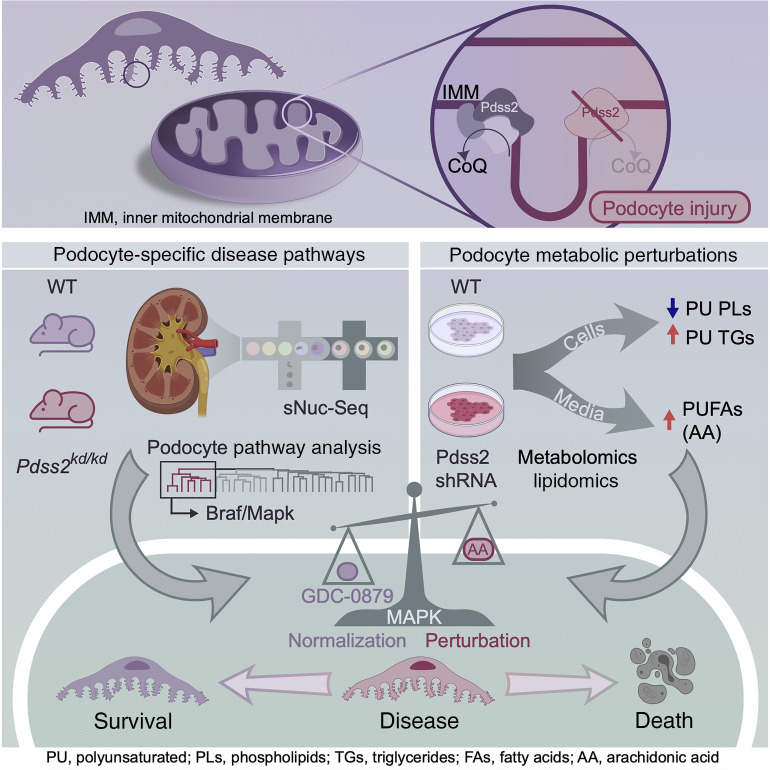
Abstract
Mutations affecting mitochondrial coenzyme Q (CoQ) biosynthesis lead to kidney failure due to selective loss of podocytes, essential cells of the kidney filter. Curiously, neighboring tubular epithelial cells are spared early in disease despite higher mitochondrial content. We sought to illuminate noncanonical, cell-specific roles for CoQ, independently of the electron transport chain (ETC). Here, we demonstrate that CoQ depletion caused by Pdss2 enzyme deficiency in podocytes results in perturbations in polyunsaturated fatty acid (PUFA) metabolism and the Braf/Mapk pathway rather than ETC dysfunction. Single-nucleus RNA-Seq from kidneys of Pdss2kd/kd mice with nephrotic syndrome and global CoQ deficiency identified a podocyte-specific perturbation of the Braf/Mapk pathway. Treatment with GDC-0879, a Braf/Mapk-targeting compound, ameliorated kidney disease in Pdss2kd/kd mice. Mechanistic studies in Pdss2-depleted podocytes revealed a previously unknown perturbation in PUFA metabolism that was confirmed in vivo. Gpx4, an enzyme that protects against PUFA-mediated lipid peroxidation, was elevated in disease and restored after GDC-0879 treatment. We demonstrate broader human disease relevance by uncovering patterns of GPX4 and Braf/Mapk pathway gene expression in tissue from patients with kidney diseases. Our studies reveal ETC-independent roles for CoQ in podocytes and point to Braf/Mapk as a candidate pathway for the treatment of kidney diseases.
Keywords: Cell Biology, Nephrology
Keywords: Chronic kidney disease, Genetic diseases, Mitochondria
Introduction
Deep mechanistic understanding of genetically defined diseases can provide insight into fundamental biology and potential therapeutic targets for the treatment of rare and prevalent diseases (1–7). Mitochondrial cytopathies are a group of heterogeneous multisystemic rare diseases with no satisfactory therapies (8). A commonly affected organ, the kidney, has the second highest mitochondrial content and oxygen consumption in the body (after the heart), a function concentrated in tubular epithelial cells (9, 10). Patients with mitochondrial cytopathies classically present with kidney dysfunction caused by tubular epithelial cell injury (11). However, a longstanding unsolved mystery has been that patients with mitochondrial mutations in the coenzyme Q (CoQ) biosynthesis pathway (PDSS1, PDSS2, COQ2, COQ6, COQ8B) do not have primary tubular epithelial injury. Instead, these patients present with podocyte dysfunction that causes nephrotic syndrome, a disease characterized by kidney filter damage and proteinuria, the spilling of essential proteins into the urine (11–22). Of particular interest, PDSS2 mediates the first committed step of CoQ biosynthesis (23–25).
CoQ (or ubiquinone [UQ]) has a well-established role in the electron transport chain (ETC) (23, 26) and ATP production. While mitochondrial dysfunction is a hallmark of both acute and chronic kidney diseases (27–30), recent work has demonstrated that mitochondria are dispensable for podocyte function and survival (31). Data from animal models of CoQ deficiency show rescue of kidney function without rescue of ETC activity following CoQ supplementation (25, 32). Together, these observations suggest that the mechanism of podocyte injury due to CoQ deficiency is independent of ETC function, and therefore, studies in nonpodocyte cells (i.e., patient fibroblasts) have failed to identify as-yet-uncharacterized podocyte-specific defects (22, 33, 34).
With mitochondrial dysfunction, lack of protection from lipid peroxidation sensitizes cells to death (35, 36). CoQ, a ubiquitous lipid present in all cellular membranes, protects against polyunsaturated fatty acid–mediated (PUFA-mediated) lipid peroxidation (37–39), and elevated markers of lipid peroxidation have been reported in the kidneys of CoQ-deficient mice (40). PUFAs, such as arachidonic acid (AA), and the enzyme that metabolizes PUFAs, COX2, are implicated in several kidney diseases (41–46). Harmful PUFAs are generated via retinoic acid signaling (47–49), but in some contexts, increased retinoic acid signaling has been shown to ameliorate podocyte injury (50–52). These conflicting data call for a deeper understanding of PUFAs, CoQ, and their regulatory pathways in podocytes.
Here we study homozygous kd/kd (kidney disease) mice, the result of a spontaneous missense mutation in Pdss2 (V117M, Pdss2kd/kd [KDKD]). These mice have nephrotic syndrome leading to kidney failure, which is similar to what occurs in humans with monogenic PDSS2 mutations (40, 53, 54). Unlike podocyte-specific knockout models (55–57), KDKD mice have global CoQ deficiency, which, together with the resolution afforded by single-nucleus RNA-Seq (sNuc-Seq), allowed us to identify a previously unknown podocyte-specific Braf/Mapk pathway driven by CoQ deficiency. Modulating this pathway in vivo with the Braf/Mapk-targeting compound GDC-0879 (58) rescued podocyte injury and kidney filter function. Integrated metabolomics and transcriptomics analyses identified a perturbation in PUFA metabolism. Treatment with GDC-0879 restored the abundance of glutathione peroxidase 4 (Gpx4), an enzyme that protects cells from lipid peroxidation. More generally, these pathways are broadly implicated in several human kidney diseases.
Results
Single-nuclei transcriptomics from kidneys of KDKD CoQ-deficient mice reveal several disease-associated cell states.
To identify the podocyte-specific pathways characterizing CoQ deficiency in vivo, we studied KDKD mice. Consistent with previous reports (25), 5-month-old mice had diffuse podocyte foot process effacement indicative of podocyte injury (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI141380DS1). To compare cell types at single-cell resolution, we performed sNuc-Seq of tissue from 5-month-old KDKD mice compared with age-matched controls (Supplemental Figure 1, B–D). We profiled 36,560 nuclei across 3 controls (16,119 nuclei) and 3 KDKD (20,441 nuclei) mice and performed unsupervised graph-based clustering followed by post hoc annotation based on canonical marker genes for kidney cell types (Supplemental Figure 2 and Supplemental Table 1). We identified all expected kidney cell types, including a podocyte cluster and 3 proximal tubule (PT) clusters (Figure 1A, Supplemental Figure 2, and refs. 59, 60).
Figure 1. SNuc-Seq from kidneys of Pdss2kd/kd mice reveals a podocyte-specific Mapk pathway, and treatment of mice with the Braf/Mapk-targeted compound GDC-0879 reverses kidney filter damage.
(A) UMAP visualization of single-nucleus transcriptomic profiles from three 5-month-old Pdss2kd/kd mice (KDKD) and 3 age-matched control mice (CTRL). Cell-type clusters identified by canonical marker genes. (B) Proportions of clusters across control and Pdss2kd/kd mice (left) with corresponding cell numbers (right). Pdss2kd/kd-specific clusters indicated with black arrowheads. CNT, connecting tubule; CD, collecting duct; PC, principal cell; IC, intercalated cell. (C) Analysis of leading edge genes from GSEA on differentially expressed genes between control and Pdss2kd/kd podocytes shows 3 pathway clusters: an actin cytoskeleton pathway (Actb, Actg1, Actn4, Actn1), a Mapk pathway (Raf1, Mapk1, Braf), and an ETC pathway (Uqcrq, Cox6a1, Uqcrh, Ndufs7). (D) Dot plot shows gene expression of Mapk pathway genes identified by GSEA in podocyte and PT clusters in control versus KDKD mice. (E) Urine albumin to creatinine ratio (UACR) of Pdss2kd/kd mice treated with 100 mg/kg/d or vehicle for 14 days. n = 5 animals per condition. Two-way ANOVA, Tukey’s multiple comparison test. ***P < 0.001.
Kidney cell types were equally represented across both KDKD and control mice, with the exception of 2 KDKD-enriched clusters: a Dock10/Vcam1+ cluster, named for 2 top enriched genes (Supplemental Table 1), and a mixed thick ascending limb (TAL) and distal convoluted tubule (DCT) (Mixed–TAL/DCT) cluster (Figure 1B and Supplemental Figure 3A). We hypothesized that these clusters represent disease-associated cell states. Further analysis of the Mixed–TAL/DCT cluster, which contained markers of both TAL (Slc12a1) and DCT (Slc12a3) (Supplemental Figure 2), revealed separate subpopulations of Slc12a1+ and Slc12a3+ cells that shared common expression of Dcdc2a, the top enriched gene (Supplemental Figure 3B and Supplemental Table 1). Dcdc2a, a ciliary gene mutated in hereditary nephronophthisis (61), was recently shown to be a marker of irreversible PT/acute kidney injury (62), suggesting it plays a role in both acute and chronic forms of kidney injury in proximal and distal tubular cells.
The disease-specific Dock10/Vcam1+ cluster was more difficult to classify because it lacked strong expression of canonical kidney markers (Supplemental Figure 2), but had high expression of genes involved in cytoskeleton regulation (Dock10), cell adhesion (Vcam1), and inflammation (Cxcl1) (Supplemental Figure 4A and Supplemental Table 1). We also noted that a fraction of this cluster expressed markers of parietal epithelial cells (PECs) of Bowman’s capsule, Cldn1 (63) and Pax8 (64), and of PEC activation, Cd44 (ref. 65 and Supplemental Figure 4B). To assess this cluster assignment in an unbiased manner, we determined cluster-cluster transcriptional similarity by calculating Spearman’s correlation coefficient between all clusters. This analysis showed that the Dock10/Vcam1+ cluster was most similar to PT cells (Supplemental Figure 4C). Since PT-like PECs extending into Bowman’s capsule have been previously described (66) and a PT-like, Vcam1+Cxcl1+ proliferative population has been reported in single-cell RNA-Seq analysis of another mouse model of kidney injury (59), we reasoned that the Dock10/Vcam1+ cluster likely represents an activated PEC state. PEC activation, an early step leading to kidney filter scarring (64, 65), is consistent with lesions observed in kidney biopsies of patients with PDSS2 mutations (12).
We localized the Dock10/Vcam1+ cells using immunofluorescence microscopy by Vcam1 expression and compared their localization to that of cells stained by PT (megalin, Lrp2) and podocyte markers (synaptopodin, Synpo). A portion of Vcam1+ cells were noted to have apical Lrp2 and basolateral Vcam1 expression, suggestive of an injured PT population (Supplemental Figure 4D). Additionally, Vcam1+ cells were present in cells surrounding Synpo-positive podocytes, consistent with their identity as an activated PEC cell state (Supplemental Figure 4E). We validated Vcam1 and Dock10 colocalization in cells surrounding podocytes (Nphs2) by in situ chain reaction hybridization (HCR) (67), an advanced version of FISH (see Methods). We noted coexpression of Vcam1 and Dock10 in cells adjacent to podocin-positive (Nphs2-positive) podocytes (Supplemental Figure 4F). Additionally, we validated the colocalization of Vcam1 with the PEC activation marker Cd44 (Supplemental Figure 4G) and colocalization of Cd44 with Cldn1, a general PEC marker (Supplemental Figure 4H). These Vcam1/Cd44/Cldn1+ cells surrounded Synpo+ glomeruli, suggesting that they are activated PECs based both on marker expression and localization in Bowman’s capsule. Finally, PEC activation in glomeruli of KDKD mice was histologically evident by periodic acid–Schiff (PAS) staining, as indicated by increased numbers of PECs with prominent nucleoli in Bowman’s capsule of KDKD mouse kidney tissue (Supplemental Figure 5A). In conclusion, our analysis revealed what we believe to be potentially novel markers of PEC activation (Dock10, Vcam1) and other disease-associated cell states, providing a snapshot of how different kidney epithelial cells respond to podocyte injury.
GDC-0879 reverses podocyte injury caused by CoQ deficiency.
Given the central role of podocytes in the pathogenesis of nephrotic syndrome, we focused our attention on transcriptional programs in podocytes. In particular, we sought to understand podocyte-specific effects in comparison with PT cells, which are unaffected by conditional Pdss2 knockout (25). Beyond clear histologic evidence (Supplemental Figure 1A), podocyte injury in KDKD mice was supported by transcriptional downregulation of genes critical to podocyte function: Synpo, Wt1, and Plce1 (Supplemental Figure 5B).
To look for disease-driven podocyte pathways, we applied gene set enrichment analysis (GSEA) on the podocyte cluster comparing upregulated genes in KDKD versus control cells, followed by hierarchical and k-means clustering of the leading-edge genes (see Methods) (Supplemental Table 2). This analysis revealed 2 pathway sets (Figure 1C): (a) genes associated with the actin cytoskeleton, a general feature of injured podocytes (68); and (b) genes associated with Braf/Mapk signaling, a complex signaling pathway that integrates cell stimuli to regulate many cell functions (69). A third set of genes was associated with the ETC, but was not specific to podocytes; KDKD PT also had upregulated expression of ETC genes (Supplemental Figure 5C).
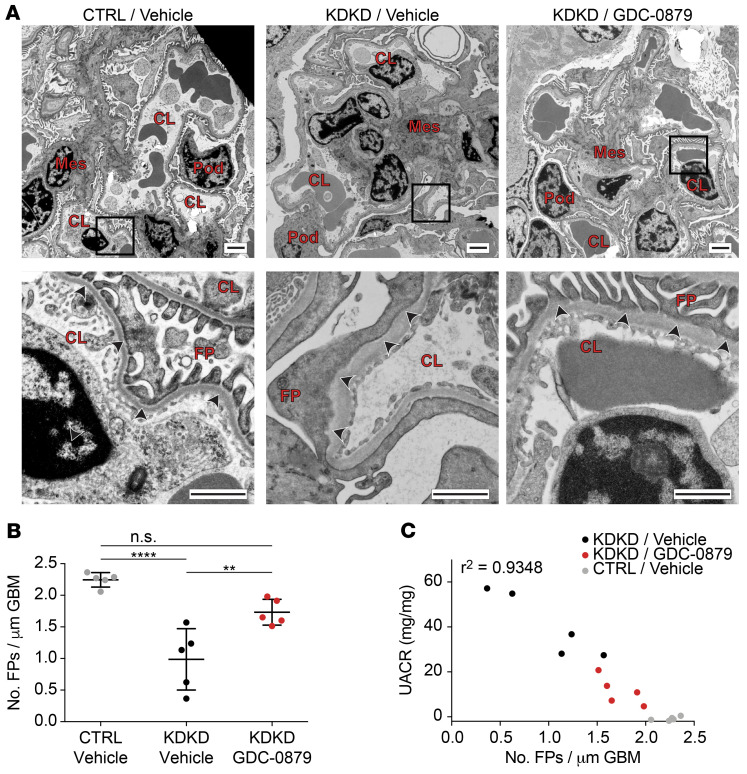
We were particularly intrigued by the discovery of a Braf/Mapk pathway perturbation in podocytes because the alteration in gene expression was podocyte specific (Figure 1D). We had previously identified this pathway in vitro as a putative signaling node for podocyte survival (70). In vitro, the Braf-targeted agent GDC-0879, a drug developed for melanoma treatment (58), protects podocytes from a wide array of cellular stressors including ER stress and lipid-mediated cytotoxicity (70). We asked whether in vivo treatment with GDC-0879 could rescue podocyte injury. Four-month-old KDKD mice with established proteinuria (specifically albuminuria, the hallmark of a damaged kidney filter) (Supplemental Figure 5D) were treated with either GDC-0879 or vehicle (71, 72). After a 14-day treatment, GDC-0879–treated animals had significantly reduced albuminuria (Figure 1E). Electron microscopy of glomeruli showed rescue of podocyte foot process effacement (Figure 2A) and a restoration in the number of foot processes (Figure 2B) in GDC-treated versus vehicle-treated animals. Further, loss of foot processes correlated with the magnitude of proteinuria (Figure 2C). To our knowledge, these data provide the first in vivo evidence of a Braf/Mapk-targeting compound as a therapeutic strategy for kidney disease.
Figure 2. Histologic analysis of mouse kidney tissue demonstrates podocyte filter barrier rescue by GDC-0879.
(A) Electron micrographs of kidneys of control, vehicle-treated KDKD, and GDC-0879–treated KDKD mice to assess integrity of the kidney filter, measured by the appearance and number of podocyte foot processes (arrowheads). Diffuse foot process simplification (called effacement) is seen in glomeruli of vehicle-treated KDKD mice. Rescue of foot process effacement is observed following treatment with GDC-0879. Pod, podocyte; Mes, mesangial cell; CL, capillary lumen; FP, podocyte foot processes. Scale bars: 2 μm (upper); 1 μm (lower). (B) Vehicle-treated KDKD mice have significantly reduced podocyte foot process numbers, which is reversed following treatment with GDC-0879. n = 5 animals per condition; 14–16 images quantified per animal.Shapiro-Wilk normality test, 1-way ANOVA, Tukey’s multiple comparison test. (C) Decreased number of foot processes correlates with escalating individual animal proteinuria levels. **P < 0.01; ****P < 0.0001.
Altered lipid profiles in Pdss2-depleted podocytes.
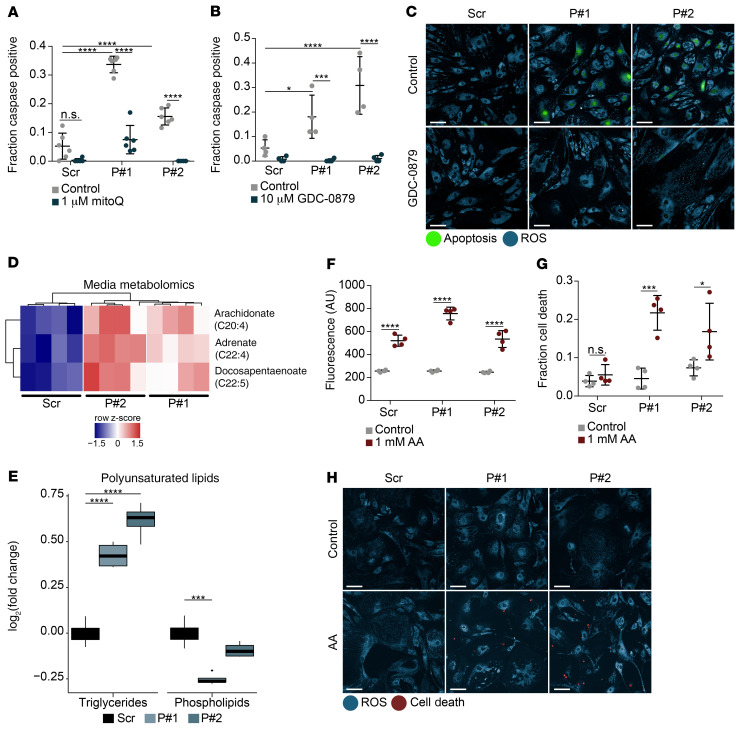
To investigate the specific molecular mechanisms leading to podocyte injury in CoQ deficiency, we generated an in vitro model of Pdss2 depletion in immortalized mouse podocytes. Given the podocyte is a rare cell type (60), an in vitro system allows for in-depth functional analysis that could not be accomplished by analysis of bulk kidney tissue. We validated protein depletion (Supplemental Figure 6A; see complete unedited blots in the supplemental material) and functional deficiency, as indicated by decreased abundance of the end pathway metabolites CoQ9 and CoQ10 (Supplemental Figure 6B). We also found increased apoptosis in Pdss2-depleted, CoQ-deficient podocytes that was rescued by mitoQ, a mitochondrial-targeted analog of CoQ (Figure 3A). In line with in vivo data, and serving as additional validation for the in vitro podocyte cell system, we found that GDC-0879 also rescued apoptosis (Figure 3, B and C).
Figure 3. Metabolomics reveal altered lipid/PUFA profiles in CoQ-deficient Pdss2-depleted podocytes.
(A) Caspase-positive quantification of podocytes expressing either scrambled or Pdss2-targeted shRNAs (P#1, P#2) and rescue of apoptosis with mitoQ. n = 6. (B) Apoptosis quantification after 1-week GDC-0879 treatment of Pdss2-depleted podocytes and scrambled controls. n = 4. (C) Live-cell fluorescence imaging of cellular ROS (blue) and apoptosis (green) in cells treated with GDC-0879 (10 μM). Scale bars: 100 μm. (D) Heatmap of statistically significant metabolites (FDR < 10%, Benjamini-Hochberg correction on Student’s t test) from conditioned media of Pdss2-depleted podocytes versus scrambled controls. n = 4. (E) Log2 fold change of polyunsaturated TAGs and polyunsaturated PLs in Pdss2-depleted podocytes versus scrambled controls. n = 4. Polyunsaturated lipids limited to lipids with 4–8 double bonds. (F) Cellular ROS quantification after AA treatment for 12 hours. n = 4. (G) Cell viability quantification after AA treatment for 84 hours, n = 4. (H) Live cell fluorescence imaging of cellular ROS (blue) and cell death (red) in cells treated with AA (1 mM) for 84 hours. Scale bars: 100 μm. Two-way ANOVA, Tukey’s multiple comparison test, unless otherwise noted. *P < 0.05; ***P < 0.001; ****P < 0.0001.
To better understand the mechanisms leading to podocyte dysfunction, we evaluated 4 known functions of CoQ. We assessed ETC function by a mitochondrial stress test using the Seahorse Flux Analyzer, but saw no decrease in oxygen consumption rates (OCR) in Pdss2-depleted podocytes either in standard or glucose-free media (Supplemental Figure 6, C and D). Glycolysis (as measured by extracellular acidification rate [ECAR]) was also unaffected in these cells and was appropriately diminished in the setting of glucose starvation (Supplemental Figure 6, C and D). Uridine levels, a readout of pyrimidine nucleotide biosynthesis, were unchanged (Supplemental Figure 6E). Mitochondrial permeability transition pore (MPTP) opening and cellular ROS generation (as measured by oxidation of the detector dye CellROX Orange), assessed by high-content live cell imaging, were not meaningfully altered in Pdss2-depleted podocytes compared with scrambled control (Supplemental Figure 6, F and G).
Given that expected CoQ functions were not perturbed in Pdss2-depleted podocytes, we hypothesized that podocyte injury may not only be due to depletion of CoQ, but also due to broader perturbations in metabolic pathways. To assess these alterations, we performed mass spectrometry–based metabolite profiling (metabolomic) studies on cell extracts and conditioned media of Pdss2-depleted podocytes. While bulk lipidomics have been done from kidneys of KDKD mice, these studies were not representative of podocytes, which are a rare kidney cell type (32). Therefore, we opted for the in vitro podocyte system to characterize podocyte-specific metabolic changes. Of all measured metabolites from conditioned media, only 3 were differentially abundant (DA) in Pdss2-depleted podocytes (FDR < 0.1): the PUFAs AA (C20:4), adrenic acid (C22:4), and docosapentaenoic acid (C22:5; Figure 3D and Supplemental Table 3). Building on previous work that has demonstrated PUFA susceptibility in CoQ-deficient yeast (73, 74), we identified an increase in endogenous PUFA abundance secondary to Pdss2 depletion. The majority of DA metabolites in the cell lysates were also lipid species (95/116 DA metabolites; Supplemental Figure 7A, and Supplemental Table 3). We further characterized these lipid metabolites by dividing them into 14 subclasses (Supplemental Figure 7B) and looking at overall abundance across cell conditions. Of the 14 classes, plasmalogens (a class of glycerophospholipids enriched in PUFAs) and sphingomyelins (SM) were significantly decreased, and triglycerides (TAG) were significantly increased in Pdss2-depleted podocytes compared with control cells (Supplemental Figure 7B). Additionally, when the TAGs were further subdivided by degree of saturation, polyunsaturated TAGs were most prominently abundant in Pdss2-depleted cells (Supplemental Figure 7C). Metabolomics revealed a perturbation in PUFA metabolism with increased polyunsaturated TAGs, decreased polyunsaturated phospholipids (PLs), and elevated endogenous PUFAs (Figure 3, D and E). To functionally assess the role of PUFAs, we treated Pdss2-depleted podocytes with AA, one of the 3 DA fatty acids. This treatment generally increased ROS (Figure 3F), but caused selective cell death only in Pdss2-depleted podocytes (Figure 3, G and H), demonstrating their susceptibility to PUFA-induced injury.
Next, we investigated the pathways responsible for the harmful perturbations in PUFA metabolism. GSEA on bulk RNA-Seq data from Pdss2-depleted podocytes compared with scrambled control (Supplemental Table 4) revealed retinol (retinoid acid [RA]) metabolism as the only significant positively enriched pathway in Pdss2-depleted podocytes (Figure 4A). We hypothesized that the positively (RA) and negatively enriched pathways (cell-cycle regulation; Figure 4A) from the RNA-Seq experiment were the result of increased RA receptor–mediated (Rar-mediated) transcription, given the known role of RA in podocyte differentiation (50–52). In other cell types, all-trans RA (atRA) has been associated with increased Pla2 enzymatic activity (47, 48) that releases PUFAs, such as AA, from PLs. atRA has also been associated with increased AA release (49) and increased incorporation of AA into TAGs (75, 76), reminiscent of the polyunsaturated TAGs measured in Pdss2-depleted podocytes. The agreement between these prior studies and the metabolomic changes we observed in Pdss2-depleted podocytes (Figure 3, D and E) suggested that the RA pathway might contribute to podocyte injury in the context of CoQ deficiency. Since cells deficient in CoQ are at risk for increased lipid peroxidation (37, 38) and PUFAs are known to promote cellular injury through lipid peroxidation (39), we hypothesized that RA signaling in podocytes is context dependent: it may be beneficial in settings such as viral-induced dedifferentiation, as previously shown (50, 51, 77), but injurious in the setting of CoQ deficiency, as demonstrated by Pdss2-deficient podocyte cell death.
Figure 4. Retinoic acid exacerbates podocyte injury and increases the abundance of PUFAs, whose effect on Mapk signaling is mitigated by GDC-0879.
(A) Significantly enriched KEGG pathways following GSEA on bulk RNA-Seq data from Pdss2-depleted podocytes versus scrambled controls. n = 3. (B) Luminescence readout from RARE -luciferase reporter assay following 24 hours of treatment with 100 nM atRA or 48 hours of treatment with 10 μM GGdP. Both increase Rar-mediated transcription. n = 3. (C) Luminescence readout from RARE-luciferase reporter assay, showing Rar-mediated transcription from Pdss2-depleted podocytes versus scrambled controls at baseline and after either 10 μM GGdP or 100 nM atRA. n = 3. (D) Cell death quantification after atRA treatment for 4 days. n = 4. Two-way ANOVA, Šidák’s multiple comparison test. (E) Live cell fluorescence imaging of cellular ROS (blue) and cell death (red) in cells treated with atRA (1 μM) for 84 hours. Scale bars: 100 μm. (F) Heatmap of statistically significant metabolites (FDR <10%, Benjamini-Hochberg correction on Student’s t test) from podocyte-conditioned media treated with 1 μM atRA versus DMSO control. n = 3. (G) Representative Western blot of P-ERK1/2 and total ERK1/2. Increased ERK1/2 phosphorylation following treatment with AA (500 μM) is reversed with GDC-0879 (10 μM). (H) Densitometric Western blot quantification of n = 4 biological replicates. Two-way ANOVA, Tukey’s multiple comparison test, unless otherwise noted. **P < 0.01; ***P < 0.001; ****P < 0.0001.
In support of elevated RA signaling, the carboxylic acid form of geranyl-geranyl pyrophosphate (GGdP), the substrate that likely accumulates in podocytes in the absence of Pdss2, can increase Rar-mediated transcription (78). We tested for changes in Rar-mediated transcription using a luciferase reporter assay (79). Treatment with the Pdss2 substrate GGdP phenocopied atRA treatment by increasing luciferase expression (Figure 4B and Supplemental Figure 8, A and B). Upon stimulation with either GGdP or atRA, Pdss2-depleted podocytes had significantly elevated luciferase expression over controls (Figure 4C). Differences between Pdss2-depleted podocytes and controls could not be detected prior to stimulation, likely because the low luciferase signal at baseline was below the sensitivity of the assay. Following atRA treatment, Pdss2-depleted podocytes had increased cell death over scrambled controls (Figure 4, D and E), demonstrating atRA-mediated injury in the context of CoQ deficiency. Lipidomic analysis of conditioned media from atRA-treated podocytes showed that PUFAs (linolenic acid, C18:2) were significantly more abundant and that several polyunsaturated PLs were significantly decreased in abundance after atRA treatment, which was similar to what occurred with Pdss2-depleted podocytes (Figure 4F and Supplemental Table 5). We concluded that Rar-mediated transcription contributes to the observed lipid perturbations in Pdss2-depleted podocytes and specifically that, in this context, (a) GGdP increases Rar-mediated transcription; (b) Pdss2-depleted podocytes have elevated levels of Rar-mediated transcription; (c) further stress through this pathway is harmful to podocytes; and (d) atRA treatment phenocopies the lipidomic changes observed in Pdss2-depleted podocytes.
Having elucidated the RA pathways upstream of PUFA generation, we shifted our focus to downstream Mapk signaling. ER stress has been previously shown to inhibit Mapk signaling, and GDC-0879 treatment paradoxically restores it by priming and stabilizing WT Braf-mediated phosphorylated ERK (P-ERK) activity in response to ER stress (70, 80). Accordingly, in podocytes treated with the ER stressor thapsigargin, GDC-0879 increased ERK1/2 phosphorylation, restoring Mapk signaling (Supplemental Figure 8C; see complete unedited blots in the supplemental material). Conversely, we found that AA treatment increased podocyte Mapk signaling (similar to cancer cells; ref.58), as demonstrated by increased ERK1/2 phosphorylation. Most interestingly, GDC-0879 reversed this effect back toward baseline, as evidenced by decreased ERK1/2 phosphorylation (Figure 4, G and H; see complete unedited blots in the supplemental material). Together, these findings suggest that GDC-0879 prevents both AA-induced increases and ER stress–induced decreases in Mapk signaling, thus maintaining podocyte homeostasis.
Podocyte-specific lipid metabolism pathways validated in vivo.
We next sought to validate transcriptomic readouts of PUFA perturbations in vivo. Markers of lipid peroxidation have been previously described in the kidneys of KDKD mice, but without cellular specificity (40). We reasoned that the resolution of sNuc-Seq might allow us to identify podocyte-specific transcriptomic changes related to PUFA metabolism. As an indirect readout of elevated PUFAs, we used transcriptomic signatures derived from an in vitro screen of structurally diverse free fatty acids (FFAs) (our unpublished observations). Using this data set, we generated signatures of both upregulated (PUFA upregulated genes) and downregulated (PUFA downregulated genes) genes specific for exposure to PUFAs relative to other FFA families, including saturated and monounsaturated fatty acids (see Methods, Supplemental Figure 8D). Scoring single cells for the PUFA-specific transcriptomic signature showed an increase in expression of the predicted upregulated signature (PUFA up) and a decrease of expression of the predicted downregulated signature (PUFA down) specifically in KDKD podocytes, but not in KDKD PT cells (Figure 5A and Supplemental 8E). In testing these signatures across all clusters (Wilcoxon’s rank sum test), we found that only the podocyte cluster showed a significant shift in both signatures; it also had the largest effect size in both directions compared with all kidney cell clusters (Figure 5B). Therefore, these sensitive transcriptional signatures provide support for increased exposure of podocytes to PUFAs in KDKD mice.
Figure 5. Podocyte-specific lipid metabolism pathways identified in vivo independently validate pathways derived in vitro.
(A) Violin plot of PUFA gene signatures (PUFA up: upregulated PUFA gene signature; PUFA down: downregulated PUFA gene signature) in podocytes versus all PT clusters combined. (B) Cohen’s d effect size in PUFA gene signatures across all clusters with significance determined by 1-tailed Wilcoxon’s rank sum test (alternative hypothesis: control left-shifted compared with KDKD for PUFA up; control right-shifted compared with KDKD for PUFA down). (C) Dot plot shows gene expression of Gpx4 in podocyte and PT clusters in control versus KDKD mice. (D) Immunofluorescence staining for Gpx4 and podocytes (Synpo) in control versus KDKD mice shows increased Gpx4 expression in KDKD glomeruli. Scale bars: 100 μm. (E) Violin plots of individual glomeruli Gpx4 immunofluorescence quantification in vehicle-treated control and KDKD mice and GDC-0879-treated KDKD mice. n = 4 animals per condition; 2 sections analyzed per animal. Wilcoxon’s rank sum test with Bonferroni’s correction (n = 3 comparisons). *P < 0.05; **P < 0.01; ****P < 0.0001.
Gpx4 encodes glutathione peroxidase, an enzyme that protects cells from PUFA-induced lipid peroxidation (39). Compared with PT cells, we found that Gpx4 expression was upregulated in podocytes (Figure 5C). We validated this finding at the protein level by immunofluorescence staining and similarly found increased Gpx4 protein expression in the filter units of KDKD mice (called glomeruli; Figure 5D). Using machine-learning analysis software to analyze immunofluorescence images, we quantified the expression of Gpx4 within individual glomeruli of controls and KDKD vehicle-treated and KDKD GDC-0879–treated mice (see Methods, Supplemental Figure 9A). Gpx4 expression was significantly increased in KDKD vehicle-treated animals compared with control animals, and most importantly, Gpx4 expression was significantly decreased following treatment with GDC-0879 (Figure 5E). Taken together, these experiments (a) provided in vivo validation for the podocyte-specific PUFA-generating pathways identified in vitro; and (b) suggested that treatment with GDC-0879 reverses lipid peroxidation-mediated podocyte injury.
Braf/Mapk pathway and lipid peroxidation gene expression correlate with kidney diseases in human tissue samples.
Given the role of lipid peroxidation and the Braf/Mapk pathway in different cellular contexts, from cancer to diabetes (35–38), we asked whether these pathways might also be relevant to common kidney diseases. Nephroseq (81) is a publicly available database that has aggregated a wide array of gene expression data sets. We investigated the expression of GPX4 and of genes identified by GSEA pathway analysis in KDKD mice (MAPK1, RAF1, and BRAF; Figure 1C) and NRAS (the most podocyte-specific Ras gene in the mouse sNuc-Seq data). We looked for either significant changes in expression between disease and control samples (P < 0.05 and fold change > 1.5), or significant correlations between gene expression and proteinuria (P < 0.05 and Pearson’s r > 0.5), the hallmark of podocyte-mediated kidney diseases (68).
The expression of Braf/Mapk pathway genes was increased across a wide spectrum of diseases, from focal segmental glomerulosclerosis (FSGS) (a disease of primary podocyte injury and loss) to lupus nephritis (caused by secondary, immune-mediated injury to podocytes) to chronic kidney disease (CKD) (driven by a variety of factors, both genetic and other, such as diabetes, exposure to toxins, etc.) (Figure 6A). As a control, we used SYNPO, a podocyte-specific gene whose decreased expression is associated with podocyte injury (82). Expression of RAF1, a member of the MAP3K RAF family (along with BRAF), was correlated with disease severity in human FSGS samples, as measured by proteinuria (ref. 83 and Figure 6, A and B). To explore the generalizability of lipid peroxidation as a driver for kidney diseases, we investigated the gene expression pattern for GPX4. Interestingly, GPX4 was also correlated significantly with proteinuria in human FSGS samples (Figure 6C), and RAF1 and GPX4 expression were correlated to one another (Figure 6D). Taken together, these results support the notion that the Braf/Mapk pathway may represent a common injury pathway for several kidney diseases.
Figure 6. Mapk and lipid peroxidation gene-expression profiles in human kidney diseases associated with podocyte injury.
(A) Summary of NephroSeq, version 5, data filtered for significant results (P < 0.05), fold change > 1.5 (disease versus control), and Pearson’s correlation coefficient, r > 0.5 (Proteinuria) show increased GPX4 and Mapk pathway gene expression in human kidney disease samples (left) and correlation with disease severity, as measured by proteinuria (right). SYNPO expression, used as a control, is expected to be decreased in diseases involving podocyte injury. (B) Scatter plot showing correlation between Mapk pathway gene expression, RAF1, and proteinuria in human FSGS. (C) Scatter plot showing correlation among lipid peroxidation gene expression, GPX4, and proteinuria in human kidney disease samples (FSGS). (D) Scatter plot showing correlation between GPX4 and RAF1 gene expression in human FSGS.
Discussion
In this study, we investigated mechanisms downstream of CoQ depletion due to Pdss2 enzyme deficiency in an effort to understand both basic podocyte biology and disease-specific pathways for targeted therapeutic intervention. In this context, our approach of combining hypothesis-driven mechanistic work with the targeted deployment of transcriptomics and metabolomics technologies led us to several insights.
First, sNuc-Seq allowed us to identify a disease-specific perturbation in the Mapk pathway. The reversal of podocyte injury with the Braf/Mapk-targeted agent GDC-0879 bolsters the idea that focusing on this pathway may be of therapeutic value. Beyond CoQ deficiency, we showed that the expression of relevant Mapk pathway genes correlates with human FSGS and disease severity. These data suggest the opportunity to repurpose GDC-0879 with a dosing regimen appropriate for kidney diseases and to learn from this strategy to develop targeted, podocyte-specific compounds.
Second, our systematic understanding of CoQ deficiency in vitro and in vivo supports a key conclusion that kidney injury in CoQ deficiency is not driven by a lack of ATP production in podocytes. Consistently, we observed no changes in mitochondrial respiration in podocytes lacking 60% of CoQ (Supplemental Figure 6B), suggesting that 40% CoQ is sufficient for podocytes to maintain functional respiration. More broadly, the fact that podocytes can effectively survive ETC inhibition is in agreement with a recent study showing that aerobic respiration in podocytes is dispensable (31). Our study thus contributes to an expanded understanding of mitochondrial energetics in podocytes.
Third, our data show that atRA-mediated transcriptional changes drive PUFA generation, in support of context-dependent signaling in the setting of CoQ deficiency. Our work provides a mechanistic framework: (a) increased PUFAs, the result of upregulated Rar transcriptional programs and (b) the absence of CoQ protection from PUFA-mediated lipid peroxidation combine to promote podocyte death. This concept of context-dependent Rar podocyte injury is supported by recent work demonstrating expression of retinoic acid receptor responder protein 1 (RARRES1) is correlated with kidney injury in FSGS and diabetic kidney disease (84). While our work was focused on mechanisms of kidney injury (Pdss2kdk/kd mice have isolated kidney disease; ref. 25), patients with CoQ deficiency frequently present with extrarenal symptoms, including encephalopathy (12, 18, 20–22, 85–87). Future research may explore the role of atRA and PUFA-mediated injury pathways in neuronal injury, and thus interventions targeting these pathways, such as GDC-0879, may confer benefit for the brain as well as the kidney.
While metabolomics had been previously performed from lysates of whole kidneys of KDKD mice (32), podocyte-specific changes had not been measured because podocytes represent a small fraction of all kidney cells. Here, we revealed a podocyte-specific metabolomic profile characterized by increased endogenous PUFAs. Furthermore, our studies now demonstrate that endogenous PUFAs promote podocyte injury, thus building on prior work showing that treatment with exogenous PUFAs promotes ROS generation in cancer cells (88) and cell death in CoQ-deficient yeast (73, 74). Further, we have linked this perturbation in PUFA metabolism to Mapk activation that was reversed by GDC-0879. We thus revealed a nuanced understanding of GDC-0879 as neither a strict inhibitor nor activator, but rather as a normalizer of BRAF signaling, ensuring homeostatic, adaptive Mapk signaling in podocytes.
In support of the generalizability of our findings, harmful levels of angiotensin II increase Gpx4 in rat glomeruli and Mapk signaling in cultured podocytes (89). GPX4 is also a genetic modifier of cell viability in the setting of mitochondrial dysfunction in cancer cells (35). In addition to cancer, lipid peroxidation is implicated in a wide range of chronic diseases, including atherosclerosis and neurodegeneration (90). Our study highlights a previously unrecognized role for lipid peroxidation in diseases driven by podocyte injury and kidney filter damage (FSGS, CKD), which represent a large and growing fraction of the global burden of chronic metabolic diseases (91).
One limitation of our study is that the mechanistic work was done using mouse model systems. Therefore, despite evidence of applicability to human kidney diseases in available patient cohorts, future studies in larger, independent patient cohorts will be needed. Furthermore, podocytes are a rare cell type in the kidney, so the small numbers of podocytes recovered by sNuc-Seq may have limited the statistical power to detect subtle changes between disease and control mice. Nevertheless, taken together, our data combining in vivo and in vitro multi-omic approaches have provided insights into kidney biology and broadly relevant mechanisms of mitochondrial dysfunction and lipid peroxidation. Most importantly, we reveal GDC-0879 as a readily testable strategy with broad therapeutic potential for kidney diseases.
Methods
See Supplemental Table 6 for reagents, antibodies, and primer sequences.
Cell culture
Conditionally immortalized mouse podocytes (RRID:CVCL_AS86) (92) were cultured under permissive conditions (33°C) in RPMI-1640 with 10% FBS, 100 U/ml penicillin, and 100 mg/ml streptomycin and IFN-γ on type I collagen. Induction of differentiation is mediated by thermoshift to 37°C without IFN-γ. Lentivirus transduction was performed between days 5 and 10 of differentiation, and assays were performed between days 10 and 17 of differentiation. HEK293 cells were cultured in DMEM supplemented with 10% FBS and penicillin/streptomycin.
Lentivirus production
HEK293 cells (60%–80% confluency in a 10 cm tissue culture dish) were triple transfected with the 2 helper plasmids pCMV-dR8.91 (2 μg) and VSV-G (1 μg) and the shRNA containing pLKO.1 (Sigma-Aldrich, SHC002 [Scr], TRCN0000253052 [no. 1], TRCN0000253054 [no. 2]) or RARE-luciferase–containing (where RARE indicates retinoic acid receptor element) vvPW plasmid (see below) (3 μg) using Lipofectamine 3000. Fresh antibiotic-containing media was added 20 hours after transfection. Virus was harvested 48 hours later, sterile filtered, aliquoted, and stored at –80°C. Podocytes were transduced with 20% to 30% virus, by volume, in the presence of 4 μg/ml polybrene for 20 hours and experiments were performed as described below.
Cloning of RARE-luciferase into lentivirus delivery plasmid
pGL3-RARE-luciferase was a gift from T. Michael Underhill (Department of Physiology, Faculty of Medicine and Dentistry, University of Western Ontario, London, Ontario, Canada) (Addgene plasmid 13458; http://n2t.net/addgene:13458; RRID:Addgene_13458). The RARE-luciferase promoter and open reading frame was subcloned into the vvPW lentivirus delivery plasmid by PCR amplification (Q5 Hot Start High-Fidelity 2× Master Mix) and digestion of the vvPW backbone (SbfI-HF; XhoI), followed by assembly and transformation into NEB 5-α competent cells (NEBuilder HiFi DNA Assembly Cloning Kit). Transformants were validated by Sanger sequencing followed by whole-plasmid sequencing.
Western blotting
shRNA knockdown.
Cells were collected for Western blotting 5 days following transduction as described above.
Mapk pathway experiments.
Mouse podocytes were differentiated as described above. For AA experiments, podocytes were serum starved for 24 hours, followed by treatment with GDC-0879 (10 μM or DMSO) for 24 hours, followed by treatment with AA (500 μM or BSA) for 5 minutes. For thapsigargin experiments, podocytes were serum starved for 24 hours, followed by treatment with GDC-0879 (10 μM or DMSO) and thapsigargin (2.5 μM or DMSO) for 24 hours. Densitometric analysis was performed with ImageJ (NIH). P-ERK and total ERK were normalized to respective loading controls (vinculin). Then, P-ERK was normalized to total ERK.
Proteins in lysis buffer with protease inhibitor cocktail and PhosSTOP Phosphatase Inhibitor (Sigma-Aldrich) in LDS sample buffer and reducing agent were boiled at 95°C for 5 minutes, separated in 4% to 12% SDS-PAGE gel, and transferred to nitrocellulose membranes. All washes were carried out with 0.1% Tween in PBS. Membranes were blocked for 1 hour at room temperature in 5% nonfat dry milk in PBS containing 0.1% Tween. Primary antibody incubation in blocking solution was performed for 1 hour at room temperature or overnight at 4°C. Secondary antibodies were applied for 1 hour at room temperature (in blocking solution). Immunoblots were detected using SuperSignal West Pico or Femto (Thermo Fisher Scientific) using the gel-imaging system G:BOX Chemi XT4 (Syngene) or ChemiDoc Imaging System (Bio-Rad).
RNA isolation
Podocytes were plated into 10 cm dishes (90 K/plate) for differentiation and transduced with shRNA lentivirus on day 7 of differentiation; RNA was collected on day 14 of differentiation. Total RNA extraction was performed using RNeasy Mini Kit. RNA was eluted using nuclease-free water, and total yield and purity of RNA were assessed using NanoDrop 2000 (Thermo Fisher).
cDNA library construction
RNA from cells were quantified using the Quant-iTTM RiboGreen RNA Assay Kit and normalized to 5 ng/μL. An automated variant of the Illumina TruSeq Stranded mRNA Sample Preparation Kit was used for library preparation from a 200 ng aliquot of RNA. This method preserves strand orientation of the RNA transcript and uses oligo dT beads to select mRNA from the total RNA sample. Following cDNA synthesis and enrichment, cDNA libraries were quantified with quantitative PCR (qPCR) using KAPA Library Quantification Kit for Illumina Sequencing Platforms and then pooled equimolarly.
Illumina sequencing
Pooled libraries were normalized to 2 nM and denatured using 0.1 N NaOH prior to sequencing. Flowcell cluster amplification and sequencing were performed according to the manufacturer’s protocols using either the HiSeq 2000 or HiSeq 2500. Each run was a 101 bp paired end with an 8-base index barcode read. Data were analyzed using the Broad Institute Picard Pipeline, which includes demultiplexing and data aggregation.
Seahorse flux analyzer
Podocytes were plated onto 15 cm dishes (220 K/plate) for differentiation. Podocytes were transferred to XF96 cell culture plates (Seahorse Bioscience) at a density of 50 K/well on differentiation day 6. Lentivirus transduction with shRNA was performed on differentiation day 7. Treatment with RPMI 1640 with or without glucose was added on differentiation day 15, and the assay was performed on differentiation day 17. OCR and ECAR measurements were performed using the XF96 Extracellular Flux Analyzer (Seahorse Bioscience). Prior to performing the assay, media was replaced with unbuffered RPMI 1640 and plates were placed in a 37°C, non-CO2 incubator for 1 hour. Baseline OCR and ECAR were measured repeatedly with 8 minutes of mixing, followed by 4 minutes of measuring, with 4 measurements before injection and 4 minutes following each of the following injections: oligomycin (1 μM); CCCP (5 μM); rotenone (1 μM). OCR and ECAR were automatically recorded and calculated by Seahorse XF-96 software.
High content imaging assays and image analysis
Podocytes were plated into CellCarrier-384 Ultra Microplates (125–150 cells/well) for differentiation. Lentivirus transduction with shRNA virus was performed on differentiation day 6. This was followed by drug treatments and imaging as follows.
Baseline apoptosis, rescue assays (mitoQ and GDC0879), and atRA treatment.
Initial treatment was given immediately after lentivirus removal on differentiation day 7. On differentiation day 10, media was replaced and supplemented with rescue treatment and live cell imaging dyes. Live cell imaging was carried out daily from differentiation day 10 through differentiation day 14.
Cell viability susceptibility assays.
Podocytes were maintained after lentiviral transduction until differentiation day 17, at which point podocytes were treated with AA and live cell imaging was performed daily for 5 days. All fluorescence imaging was performed using the Opera Phenix High-Content Screening System (PerkinElmer). For fluorescence imaging of cells (live cell or fixed cell imaging), CellCarrier Ultra microplates were used and a minimum of 9 fields were acquired per well. Image analysis for all imaging experiments was performed using Harmony software (PerkinElmer).
Caspase activation and DRAQ7 staining were used to calculate fractions of cells going through apoptosis or cell death, respectively. Single nuclei were first identified using a combination of digital phase contrast, caspase, and DRAQ7. The entire cell body was identified using an additional dye, either TMRM or CellROX Orange. Threshold fluorescence was then determined, and the fraction of positive cells was calculated. Further, large, podocyte-like cells were identified as having a cell area greater than 3500 μm2, based on higher expression of Synaptopodin (Supplemental Figure 9B). Mitochondrial membrane potential or ROS per cell was calculated from TMRM or CellROX Orange, respectively (Supplemental Figure 9C)
Gpx4 immunofluorescence quantification.
Whole glomeruli were identified based on synaptopodin expression. The mean expression of Gpx4 within the glomerulus was normalized to the mean expression in the immediate surrounding to normalize for technical differences in background staining (Supplemental Figure 9A).
Luciferase assay
Podocytes were plated into 96-well dishes (400–500 cells/well) for differentiation. Lentivirus transduction with RARE-luciferase virus was performed at differentiation day 5. For shRNA experiments, lentivirus transduction with shRNA virus and drug treatments were performed at differentiation day 9. The luciferase assay (ONE-Glo EX Luciferase Assay System) was performed after 28 hours. For drug treatments, luciferase assay was performed 5 to 8 days after delivery of RARE-luciferase virus and luminescence was measured 24 to 48 hours after treatment.
Metabolite extraction
For metabolomics performed on the shRNA lentivirus–transduced podocytes, podocytes were plated into 10 cm dishes (90 K/plate). Lentivirus transduction with shRNA virus was performed at differentiation day 7, and metabolite extraction was performed at differentiation day 14. For the atRA treatment, podocytes were plated into 6-well dishes (15 K/well). Treatment was added on differentiation day 8, and metabolite extraction was performed at differentiation day 13. Conditioned media was snap-frozen (500 μL). For nonpolar metabolite extraction, cells were washed with cold PBS (no Mg2+/no Ca2+), followed by extraction using HPLC grade 2 propanol. Cell extracts were incubated at 4°C for 1 hour and spun down (2846 g, 4°C, 10 minutes); supernatant was transferred to new tubes. Polar metabolites were extracted in 80% HPLC-grade methanol, scraped from plates after incubation at –80°C for 15 minutes, and spun down (2846 g, 4°C, 10 minutes). Supernatant was transferred to new tubes.
Metabolomics data acquisition:
Metabolites were profiled using 4 complimentary liquid chromatography tandem mass spectrometry (LC-MS/MS) methods designed to measure polar metabolites, FFAs, and lipids as described previously (93, 94). Chromatography was performed using Shimadzu Nexera X2 U-HPLC (Shimadzu Corp.) and mass spectrometry using Q Exactive/Exactive Plus Instruments (Thermo Fisher Scientific).
For the measurement of water-soluble metabolites, methanol cell extracts were dried down under N2 gas and resuspended in 10 μl of water. Metabolites from media and resuspended cell extracts (10 μl) were extracted with the addition of 90 μL of acetonitrile/methanol/formic acid (74.9/24.9/0.2; v/v/v) containing stable isotope-labeled internal standards (valine-d8, Sigma-Aldrich; and phenylalanine-d8, Cambridge Isotope Laboratories). The samples were centrifuged (10 minutes, 9,000 g, 4°C), and the supernatants (10 μL) were injected directly onto a 150 × 2 mm, 3 μm Atlantis HILIC column (Waters). MS analyses were carried out using positive ion mode ionization using full scan analysis over 70–800 m/z.
For the measurement of polar metabolites that ionize in negative mode, methanol extracts were centrifuged (10 minutes, 9,000 g, 4°C) and 10 μl of the supernatants was injected directly onto a 150 × 2.0 mm Luna NH2 column (Phenomenex). MS analyses were carried out using electrospray ionization in the negative ion mode using full-scan analysis over m/z 60–750 at 70,000 resolution and 3 Hz data acquisition rate
For lipid profiling and CoQ measurements, 200 μl of isopropanol cell extracts was dried under N2. Both cell and media lipids were extracted with isopropanol containing 1,2-didodecanoyl-sn-glycero-3-phosphocholine (Avanti Polar Lipids) either by directly resuspending dried cell extracts with 100 μl of solvent or 1:20 (vol media/vol solvent) extractions for media. After centrifugation, supernatants (2 μL) were injected directly onto a 100 × 2.1 mm, 1.7 μm ACQUITY BEH C8 column (Waters). Eluting compounds were analyzed with MS ionization in the positive ion mode using full-scan analysis over 220–1100 m/z. Lipid identities were denoted by total acyl carbon number and total number of double-bond number; the identity of CoQs was determined by matching the retention time and m/z of the authentic standards for CoQ9 and CoQ10. For the measurement of FFAs and metabolites of intermediate polarity, media (30 μL) were extracted using 90 μL of methanol containing PGE2-d4 as an internal standard (Cayman Chemical Co.) and centrifuged (10 minutes, 9,000 g, 4°C). The supernatants of media and cell methanol extracts (10 μL) were injected onto a 150 × 2.1 mm ACQUITY BEH C18 column (Waters). MS analyses were carried out using electrospray ionization in the negative ion mode using full-scan analysis over m/z 70–850 at 70,000 resolution and a 3 Hz data acquisition rate.
Raw data were processed using TraceFinder 3.3 or 4.0 software (Thermo Fisher Scientific) and Progenesis QI (Nonlinear Dynamics). For each method, metabolite identities were confirmed using authentic reference standards from an in-house collection of metabolites. HMDB identifiers were assigned to annotated compounds when available, using version 3.0 of the HMDB library. For lipid families, based on the inability of the methods used in assigning the position of double bonds in the acyl chains, representative HMDB IDs were chosen instead.
Mice kidney immunofluorescence
Kidneys were removed and rapidly frozen in Tissue-Tek OCT Compound (Sakura Finetek) using dry ice and methylbutane. Sagittal sections 5 μm thick were obtained using a Leica CM1950 cryostat, thaw mounted on microscope slides (Fisherbrand Superfrost Plus, Fisher Scientific), and air dried for 15 minutes. Sections were fixed by immersion in 4% PFA at 4°C for 10 minutes. All washes were carried out with PBS. Prior to staining, sections were incubated for 20 minutes at room temperature in PBS blocking solution containing 5% normal goat serum, 0.2% Triton X-100, and 2% BSA. Sections were incubated at 4°C overnight with primary antibodies diluted in blocking solution. Sections were incubated with secondary antibodies diluted in PBS containing 0.1% Triton X-100 for 2 hours at room temperature. Following 3 washes of 10 minutes in PBS, sections were incubated with 1:10,000 DAPI solution in PBS for 5 minutes. Sections were air dried and mounted using ProLong Gold Antifade Mountant.
Kidney histology
Kidney histology of 3 control and 3 KDKD mice was assessed. Light microscopy images of PAS, toluidine blue–stained sections as well as transmission electron micrographs were analyzed in a blinded fashion.
HCR
HCR was performed as previously described (67).All HCR v3 reagents (probes, hairpins, and buffers) were purchased from Molecular Technologies. Thin sections of tissue (10 μm) were mounted in 24-well glass-bottom plates coated with APTES (1:50). The following solutions were added to the tissue: 10% formalin for 15 minutes, 2 washes of 1× PBS, ice-cold 70% EtOH at –20 for 2 hours to overnight, 3 washes 5× SSCT with 0.2% Tween-20, hybridization buffer (Molecular Technologies) for 10 minutes, probes in hybridization buffer overnight, four 15-minute washes in wash buffer (Molecular Technologies), 3 washes in 5× SSCT, amplification buffer (Molecular Technologies) for 10 minutes, heat-denatured hairpins in amplification buffer overnight, three 15-minute washes in 5× SSCT DAPI (1:10,000), and storage/imaging in 5× SSCT. Imaging was performed on a spinning disk confocal (Yokogawa W1 on Nikon Eclipse Ti) operating NIS elements AR software. Image analysis and processing were performed on ImageJ Fiji.
Single-nuclei isolation and sequencing
Nuclei were isolated as previously described with modification (59). Nuclei were isolated with Nuclei EZ Lysis buffer supplemented with protease inhibitor and RNase inhibitor. Samples were cut into pieces smaller than 2 mm and homogenized with a Dounce homogenizer in 2 mL of ice-cold Nuclei EZ Lysis Buffer, then filtered through a 70 μm cell strainer. The samples were homogenized again, an additional 2 mL of lysis buffer was added, and samples were incubated on ice for 5 minutes. The homogenate was filtered through a 40 m cell strainer and centrifuged at 500 g for 5 minutes at 4°C. The pellet was resuspended and washed with 4 mL of the buffer, incubated on ice for 5 minutes, and centrifuged again. The pellet was resuspended in Nuclei Suspension Buffer (1XDPBS, 0.1% RNase inhibitor), filtered through a 30 μm cell strainer, and counted. To minimize batch effects, single-nuclei isolation of 1 control and 1 diseased kidney were performed in parallel until all 6 samples were completed. Library preparation and sequencing were performed on all 6 samples in parallel with one 10× sample loaded per round of sequencing.
Single nuclei were processed through the 10X Chromium 3′ Single Cell Platform using the Chromium Single Cell 3′ Library, Gel Bead, and Chip Kits (10x Genomics), following the manufacturer’s protocol. Briefly, 10,000 cells were added to each channel of a chip to be partitioned into gel beads in emulsion (GEMs) in the chromium instrument, followed by cell lysis and barcoded reverse transcription of RNA in the droplets. Breaking of the emulsion was followed by amplification, fragmentation, and addition of adapter and sample indices. Libraries were sequenced at a concentration of 1.8 pM on a Nextseq with a 150 cycle, version 2, kit with a read structure of Read 1 26bp, Read 2 98bp, Index 1 8bp, and Index 2 0bp. One 10× lane was loaded per round of sequencing, resulting in 400M reads per sample.
Animal experiments
B6/KDKD mice were purchased from Jackson Laboratory. KDKD mice harbor a spontaneous mutation in the gene encoding the subunit 2 of polyprenyl-diphosphate synthase (Pdss2), and their phenotype was previously described (40, 54). Mice were housed under a 12-hour light/12-hour dark cycle and given free access to food and water.
Single nucleus sequencing.
Animals were euthanized by rapid carbon dioxide narcosis, followed by cervical dislocation after disease onset (5 months). Kidneys were quickly removed. One kidney was frozen in the liquid phase of isopentane and precooled toward its freezing point (–80°C) with dry ice. The second kidney was fixed in neutral buffered formalin. Experiments were performed in 3 mice for each group.
GDC-0879 treatment.
Mutant mice were tested for proteinuria starting at 100 days of age. Following onset of proteinuria, animals were treated daily with 100 mg/kg or GDC–0857 or 0.5% methylcellulose/0.2% Tween 80 (MCT) by oral gavage for 14 days. Dose was based on the IC50 of the rescue of apoptosis in vitro of Pdss2 knockdown podocytes (500–700 nM, Supplemental Figure 9D) and prior work that determined that the in vivo IC50 for inhibiting tumor growth was 50-fold higher (i.e., desired in vivo concentration of ~30 μM), which was achieved with oral dosing of 100 mg/kg (71). Spot urine samples were collected, spun down, and stored at –80°C. Experiments were performed in 5 mice for each group. Animal euthanasia and tissue collection were performed as described above.
Urine assay.
For albuminuria quantification, 15 μl of urine at 1:50 dilution was analyzed by SDS-PAGE for day 1 and day 14 spot urine samples. BSA standards (0.12, 0.24, 0.47, 0.94, 1.9, 3.75, and 7.5 μg) were run to identify and quantify urinary albumin bands. Coomassie signals were quantified using ImageJ. BSA values were used for construction of a standard curve using linear regression. Sample band values were translated into albumin concentrations. For creatinine quantification, 5–10 μl of urine was sent for measurement by isotope dilution LC-MS/MS at the UAB/UCSD O’Brien Center Core.
Quantification and statistical analysis
Statistics.
Statistical analysis was performed and results plotted using Graphpad Prism, version 7.0. Data are presented as mean ± SD unless otherwise specified. Box plots are presented as median, with lower and upper hinges corresponding to the first and third quartiles. The upper/lower whiskers extend from the hinge to the largest/smallest value at most 1.5 IQR from the hinge. Data beyond the end of the whiskers are plotted individually. Statistical comparisons are as noted in figure legends. All other analysis was done in R/Bioconductor. Graphs were visualized using ggplot2.
Metabolomics analysis.
To preprocess the data, metabolites that had more than 30% missing values were filtered out. The remaining missing values were imputed with half of the minimum of the metabolite’s intensity. All metabolite intensities were log2 transformed, and samples were mean-centered. For differential abundance (DA) analysis, only annotated peaks were selected, and 2-tailed Student’s t test was performed (function: t.test) with either equal or unequal variances based on the results of an F test comparing variances (function: var.test). P values were adjusted for multiple comparisons using the Benjamini and Hochberg correction to calculate an FDR per metabolite. Metabolites were DA for FDR of less than 0.1 for at least 1 comparison (Scr vs. shRNA no. 1 or Scr vs. shRNA no. 2) and the log2 fold change in the same direction for both comparisons. Abundances of different lipid groups were calculated by summing mean-normalized abundances of all lipids of a class followed by calculating the log2 fold change relative to Scr samples.
Bulk RNA-Seq analysis.
Reads were aligned to the mm10 mouse genome using STAR aligner. RSEM, version 1.3.0, was used to estimate gene expression. FastQC was used to evaluate the quality of raw reads. Differential expression (DE) of individual genes was carried out using DESeq2 (95). For GSEA, DE using DESeq2 was using to compare Scr to all other samples. GSEA was carried out with the fgsea R package using KEGG pathways (http://software.broadinstitute.org/gsea/msigdb/collections.jsp). Rank list was generated by calculating as follows: –log10(P value) × SIGN(log2fc) per gene.
NephroSeq database data.
Data were obtained from the NephroSeq database, version 5 (81). For selected genes (BRAF, MAPK1, NRAS, RAF1, GPX4, SYNPO), all kidney expression data were downloaded for “Proteinuria Analysis” and “Disease vs. Control Analysis.” The data were filtered for P < 0.05, fold change > 1.5 (disease vs. control) and Pearson’s correlation coefficient, r (Proteinuria Analysis).
Single-nuclei sequencing analysis
Preprocessing of 10× droplet-based sequencing output.
Cellranger toolkit, version 2.1.1, was used to perform demultiplexing using the “cellranger mkfastq” command, and the “cellranger count” command for alignment to the mouse pre-mRNA transcriptome, cell-barcode partitioning, collapsing unique molecular identifier (UMI) to transcripts, and gene-level quantification. Genes were filtered to only include genes with expression in at least 5 cells. Cells were filtered to only include cells expressing a minimum of 200 genes and a maximum of 4000 genes. The percentage of reads mapping to mitochondrial genes was capped at 5%.
Unsupervised clustering and dimensionality reduction.
Downstream analysis was performed using Seurat R package (96), version 3.0. Data normalization (log1p[counts]), finding variable genes, and scaling (regularized negative binomial regression) were performed, and unwanted variation due to variation in nFeature_RNA and percent.mito were subsequently regressed out (SCTransform, vars.to.regress = c[“nFeature_RNA”,”percent.mito”]). Samples were then integrated using default settings (FindIntegrationAnchors, IntegrateData). Following data integration, the combined data set was rescaled (ScaleData, vars.to.regress = c[“nFeature_RNA”,”percent.mito”]). Dimensionality reduction was performed using principal component analyses (RunPCA) on the highly variable genes. ElbowPlot function was used to distinguish principal components (PCs) for further analysis. Fifteen components were found sufficient to capture variance. We identified molecularly distinct clusters using default parameters (FindNeighbors, FindClusters) and a resolution of 0.2. We computed an embedding of the data in 2-D space using uniform manifold approximation and projection (UMAP) in the PC space for visualization (RunUMAP, 15 PCs).
Assignment of cell-identity.
Cluster-enriched or marker genes were computed using Wilcoxon’s rank sum test (FindAllMarkers) for DE of genes in one cluster cell versus all other cells, filtering for genes with a logFC greater than 0.25. Cluster identities were assigned by comparing data-driven genes with a list of literature-curated genes for mature kidney cell types. For GSEA, enrichment of all genes was calculated with default parameters, except that logfc.threshold was set to 0 (FindMarkers). A small podocyte cluster (102 cells) was identified by strong coexpression of canonical markers (Synpo, Nphs1, Wt1).
Differential gene expression analysis.
For DE between control and KDKD samples, we performed pair-wise DE analysis (FindMarkers) with default parameters.
GSEA.
GSEA was carried out as described above on positive- and negative-enriched genes separately. Leading-edge genes were extracted for each pathway, and hierarchical clustering (hclust) was performed on the pathways based on overlap of leading-edge genes. K-means clustering (k = 3) was performed on the resulting dendrogram (cutree) to generate pathway clusters, and a list of the most highly represented leading-edge genes per cluster was generated.
Gene signatures.
For PUFA gene signatures, we performed RNA-Seq on cells treated with a range of different FFAs, including PUFAs. The complete gene-expression matrix was mapped to a pathway matrix by performing GSEA (97) on each sample independently, as described previously (98). Hierarchical clustering of the resulting matrix revealed a cluster of PUFAs (including AA) with a unique pathway signature. To generate the PUFA-specific gene list, we used the union of all gene sets of this unique PUFA signature and performed DE analysis against the untreated control. Only significantly differentially expressed genes (after correction for multiple testing, Bonferroni’s test) were considered to be PUFA-specific genes of interest.
A gene-signature score for each cell was calculated for the up- and downregulated genes separately (AddModuleScore). To determine statistically significant shifts in gene-signature score, Wilcoxon’s rank aum test was run with a 1-tailed alternative hypothesis: an increased/decreased mean gene-signature score in KDKD for PUFA upregulated/downregulated gene signature.
Study approval
All experiments in mice were performed according to a protocol approved by the Institutional Animal Care and Use Committee of the Columbia University Medical Center and were consistent with the NIH’s Guide for the Care and Use of Laboratory Animals (National Academies Press, 2011).
Data and code availability
All original microarray data were deposited in the NCBI’s Gene Expression Omnibus database (GEO GSE164274). Processed single nuclei RNA-Seq data are available at the Single Cell Portal (https://singlecell.broadinstitute.org/single_cell/study/SCP1245). Processed metabolomics data areavailable in Supplemental Tables 3 and 5. Associated GitHub code is available (Pdss2_2020; https://github.com/esidhom/Pdss2_2020
Author contributions
EHS and AG designed the study and interpreted the results. EHS performed experiments and data analysis. CK, MKA, KK, JAP, KAV, JLM, EJG, ERB, FC, and CBC assisted with experiments. NW assisted with analysis. MTT, AJBW, GK, MR, and CMQ performed mouse experiments. AW performed histologic analysis. EHS, JLS, and AG wrote the manuscript. AG supervised the project. All authors read and approved the manuscript.
Supplementary Material
Acknowledgments
We thank Marcia Haigis and Alison Ringel for their valuable advice on mitochondrial biology and metabolomic analysis. Also, we thank Lan Nguyen, Orit Rosenblatt-Rosen, and Aviv Regev, who provided advice on single-cell library preparation, and Vamsi Mootha for advice on Seahorse Flux Analyzer studies. This work was supported by a National Defense Science and Engineering Graduate fellowship (to EHS) and by NIH/National Institute of Diabetes and Digestive and Kidney Diseases grants F30DK112477 (to EHS) and DK103658, DK099465, and DK095045 (to AG).
Version 1. 01/14/2021
In-Press Preview
Version 2. 03/01/2021
Electronic publication
Footnotes
Conflict of interest: AG has a financial interest in Goldfinch Biopharma in the form of equity.
Copyright: © 2021, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2021;131(5):e141380.https://doi.org/10.1172/JCI141380.
Contributor Information
Eriene-Heidi Sidhom, Email: esidhom@broadinstitute.org.
Choah Kim, Email: ckim@broadinstitute.org.
Maria Kost-Alimova, Email: malimova@broadinstitute.org.
May Theng Ting, Email: mtt2134@cumc.columbia.edu.
Keith Keller, Email: khkeller@bwh.harvard.edu.
Julian Avila-Pacheco, Email: jravilap@broadinstitute.org.
Andrew J.B. Watts, Email: awatts1@bwh.harvard.edu.
Katherine A. Vernon, Email: kvernon@broadinstitute.org.
Jamie L. Marshall, Email: jmarshal@broadinstitute.org.
Estefanía Reyes-Bricio, Email: ebricio@broadinstitute.org.
Matthew Racette, Email: mracette@broadinstitute.org.
Nicolas Wieder, Email: nwieder@broadinstitute.org.
Giulio Kleiner, Email: giulio.kleiner@gmail.com.
Elizabeth J. Grinkevich, Email: egrinkev@broadinstitute.org.
Fei Chen, Email: chenf@broadinstitute.org.
Astrid Weins, Email: AWEINS@PARTNERS.ORG.
Clary B. Clish, Email: clary@broadinstitute.org.
Jillian L. Shaw, Email: jshaw@broadinstitute.org.
Catarina M. Quinzii, Email: cmq2101@cumc.columbia.edu.
Anna Greka, Email: agreka@bwh.harvard.edu.
References
- 1.Zhou Y, et al. A small-molecule inhibitor of TRPC5 ion channels suppresses progressive kidney disease in animal models. Science. 2017;358(6368):1332–1336. doi: 10.1126/science.aal4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yu H, et al. A role for genetic susceptibility in sporadic focal segmental glomerulosclerosis. J Clin Invest. 2016;126(3):1067–1078. doi: 10.1172/JCI82592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dvela-Levitt M, et al. Small molecule targets TMED9 and promotes lysosomal degradation to reverse proteinopathy. Cell. 2019;178(3):521–535. doi: 10.1016/j.cell.2019.07.002. [DOI] [PubMed] [Google Scholar]
- 4.Endo A. A historical perspective on the discovery of statins. Proc Jpn Acad Ser B Phys Biol Sci. 2010;86(5):484–493. doi: 10.2183/pjab.86.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Priori SG. Inherited arrhythmogenic diseases. Circ Res. 2004;94(2):140–145. doi: 10.1161/01.RES.0000115750.12807.7E. [DOI] [PubMed] [Google Scholar]
- 6.Imbrici P, et al. Therapeutic approaches to genetic ion channelopathies and perspectives in drug discovery. Front Pharmacol. 2016;7:1–28. doi: 10.3389/fphar.2016.00121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rossari F, et al. Past, present, and future of Bcr-Abl inhibitors: from chemical development to clinical efficacy. J Hematol Oncol. 2018;11(1):84. doi: 10.1186/s13045-018-0624-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.El-Hattab AW, Scaglia F. Mitochondrial cytopathies. Cell Calcium. 2016;60(3):199–206. doi: 10.1016/j.ceca.2016.03.003. [DOI] [PubMed] [Google Scholar]
- 9.O’Connor PM. Renal oxygen delivery: matching delivery to metabolic demand. Clin Exp Pharmacol Physiol. 2006;33(10):961–967. doi: 10.1111/j.1440-1681.2006.04475.x. [DOI] [PubMed] [Google Scholar]
- 10.Bhargava P, Schnellmann RG. Mitochondrial energetics in the kidney. Nat Rev Nephrol. 2017;13(10):629–646. doi: 10.1038/nrneph.2017.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Emma F, Salviati L. Mitochondrial cytopathies and the kidney. Nephrol Ther. 2017;13:S23–S28. doi: 10.1016/j.nephro.2017.01.014. [DOI] [PubMed] [Google Scholar]
- 12.Ozaltin F. Primary coenzyme Q10 (CoQ10) deficiencies and related nephropathies. Pediatr Nephrol. 2014;29(6):961–969. doi: 10.1007/s00467-013-2482-z. [DOI] [PubMed] [Google Scholar]
- 13.Vasta V, et al. Next-generation sequencing for mitochondrial diseases: A wide diagnostic spectrum. Pediatr Int. 2012;54(5):585–601. doi: 10.1111/j.1442-200X.2012.03644.x. [DOI] [PubMed] [Google Scholar]
- 14.Ashraf S, et al. ADCK4 mutations promote steroid-resistant nephrotic syndrome through CoQ10 biosynthesis disruption. J Clin Invest. 2013;123(12):5179–5189. doi: 10.1172/JCI69000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Emma F, et al. Renal involvement in mitochondrial cytopathies. Pediatr Nephrol. 2012;27(4):539–550. doi: 10.1007/s00467-011-1926-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Akchurin O, Reidy KJ. Genetic causes of proteinuria and nephrotic syndrome: Impact on podocyte pathobiology. Pediatr Nephrol. 2015;30(2):221–233. doi: 10.1007/s00467-014-2753-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Quinzii C, et al. A mutation in para-hydroxybenzoate-polyprenyl transferase (COQ2) causes primary coenzyme Q10 deficiency. Am J Hum Genet. 2006;78(2):345–349. doi: 10.1086/500092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Diomedi-Camassei F, et al. COQ2 nephropathy: a newly described inherited mitochondriopathy with primary renal involvement. J Am Soc Nephrol. 2007;18(10):2773–2780. doi: 10.1681/ASN.2006080833. [DOI] [PubMed] [Google Scholar]
- 19.Mollet J, et al. Prenyldiphosphate synthase, subunit 1 (PDSS1) and OH-benzoate polyprenyltransferase (COQ2) mutations in ubiquinone deficiency and oxidative phosphorylation disorders. J Clin Invest. 2007;117(3):765–772. doi: 10.1172/JCI29089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scalais E, et al. Early myoclonic epilepsy, hypertrophic cardiomyopathy and subsequently a nephrotic syndrome in a patient with CoQ10 deficiency caused by mutations in para-hydroxybenzoate-polyprenyl transferase (COQ2) Eur J Paediatr Neurol. 2013;17(6):625–630. doi: 10.1016/j.ejpn.2013.05.013. [DOI] [PubMed] [Google Scholar]
- 21.López LC, et al. Leigh syndrome with nephropathy and CoQ10 deficiency due to decaprenyl diphosphate synthase subunit 2 (PDSS2) mutations. Am J Hum Genet. 2006;79(6):1125–1129. doi: 10.1086/510023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heeringa SF, et al. COQ6 mutations in human patients produce nephrotic syndrome with sensorineural deafness. J Clin Invest. 2011;121(5):2013–2024. doi: 10.1172/JCI45693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Turunen M, et al. Metabolism and function of coenzyme Q. Biochim Biophys Acta. 2004;1660(1–2):171–199. doi: 10.1016/j.bbamem.2003.11.012. [DOI] [PubMed] [Google Scholar]
- 24.Kawamukai M. Biosynthesis of coenzyme Q in eukaryotes. Biosci Biotechnol Biochem. 2016;80(1):23–33. doi: 10.1080/09168451.2015.1065172. [DOI] [PubMed] [Google Scholar]
- 25.Peng M, et al. Primary coenzyme Q deficiency in Pdss2 mutant mice causes isolated renal disease. PLoS Genet. 2008;4(4):e1000061. doi: 10.1371/journal.pgen.1000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Acosta MJ, et al. Coenzyme Q biosynthesis in health and disease. Biochim Biophys Acta. 2016;1857(8):1079–1085. doi: 10.1016/j.bbabio.2016.03.036. [DOI] [PubMed] [Google Scholar]
- 27.Galvan DL, et al. The hallmarks of mitochondrial dysfunction in chronic kidney disease. Kidney Int. 2017;92(5):1051–1057. doi: 10.1016/j.kint.2017.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tran MT, et al. PGC1α drives NAD biosynthesis linking oxidative metabolism to renal protection. Nature. 2016;531(7595):528–532. doi: 10.1038/nature17184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li SY, et al. Increasing the level of peroxisome proliferator-activated receptor γ coactivator-1α in podocytes results in collapsing glomerulopathy. JCI Insight. 2017;2(14):e92930. doi: 10.1172/jci.insight.92930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Long J, et al. Long noncoding RNA Tug1 regulates mitochondrial bioenergetics in diabetic nephropathy. J Clin Invest. 2016;126(11):4205–4218. doi: 10.1172/JCI87927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brinkkoetter PT, et al. Anaerobic glycolysis maintains the glomerular filtration barrier independent of mitochondrial metabolism and dynamics. Cell Rep. 2019;27(5):1551–1566. doi: 10.1016/j.celrep.2019.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kleiner G, et al. CoQ10 supplementation rescues nephrotic syndrome through normalization of H2S oxidation pathway. Biochim Biophys Acta Mol Basis Dis. 2018;1864(11):3708–3722. doi: 10.1016/j.bbadis.2018.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quinzii CM, et al. Respiratory chain dysfunction and oxidative stress correlate with severity of primary CoQ 10 deficiency. FASEB J. 2008;22(6):1874–1885. doi: 10.1096/fj.07-100149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Quinzii CM, et al. Reactive oxygen species, oxidative stress, and cell death correlate with level of CoQ10 deficiency. FASEB J. 2010;24(10):3733–3743. doi: 10.1096/fj.09-152728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.To TL, et al. A compendium of genetic modifiers of mitochondrial dysfunction reveals intra-organelle buffering. Cell. 2019;179(5):1222–1238. doi: 10.1016/j.cell.2019.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim W, et al. Polyunsaturated fatty acid desaturation is a mechanism for glycolytic NAD+ recycling. Cell Metab. 2019;29(4):856–870. doi: 10.1016/j.cmet.2018.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bersuker K, et al. The CoQ oxidoreductase FSP1 acts parallel to GPX4 to inhibit ferroptosis. Nature. 2019;575(7784):688–692. doi: 10.1038/s41586-019-1705-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Doll S, et al. FSP1 is a glutathione-independent ferroptosis suppressor. Nature. 2019;575(7784):693–698. doi: 10.1038/s41586-019-1707-0. [DOI] [PubMed] [Google Scholar]
- 39.Yang WS, et al. Peroxidation of polyunsaturated fatty acids by lipoxygenases drives ferroptosis. Proc Natl Acad Sci. 2016;113(34):E4966–E4975. doi: 10.1073/pnas.1603244113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Quinzii CM, et al. Tissue-specific oxidative stress and loss of mitochondria in CoQ-deficient Pdss2 mutant mice. FASEB J. 2013;27(2):612–621. doi: 10.1096/fj.12-209361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hörl WH. Nonsteroidal anti-inflammatory drugs and the kidney. Pharmaceuticals (Basel) 2010;3(7):2291–2321. doi: 10.3390/ph3072291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hocherl K. Cyclosporine a suppresses cyclooxygenase-2 expression in the rat kidney. J Am Soc Nephrol. 2002;13(10):2427–2436. doi: 10.1097/01.ASN.0000031702.86799.B9. [DOI] [PubMed] [Google Scholar]
- 43.Shehadeh IH, et al. Indomethacin and the nephrotic syndrome. JAMA. 1979;241(12):1264–1266. [PubMed] [Google Scholar]
- 44.Cheng H, et al. Podocyte COX-2 exacerbates diabetic nephropathy by increasing podocyte (pro)renin receptor expression. J Am Soc Nephrol. 2011;22(7):1240–1251. doi: 10.1681/ASN.2010111149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cheng H, et al. Overexpression of cyclooxygenase-2 predisposes to podocyte injury. J Am Soc Nephrol. 2007;18(2):551–559. doi: 10.1681/ASN.2006090990. [DOI] [PubMed] [Google Scholar]
- 46.Cheng HF, et al. Cyclooxygenase-2 inhibitor blocks expression of mediators of renal injury in a model of diabetes and hypertension. Kidney Int. 2002;62(3):929–939. doi: 10.1046/j.1523-1755.2002.00520.x. [DOI] [PubMed] [Google Scholar]
- 47.Farooqui AA, et al. Retinoic acid-mediated phospholipase A2 signaling in the nucleus. Brain Res Rev. 2004;45(3):179–195. doi: 10.1016/j.brainresrev.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 48.Antony P, et al. Ca2+-independent phospholipases A2 and production of arachidonic acid in nuclei of LA-N-1 cell cultures: a specific receptor activation mediated with retinoic acid. Mol Brain Res. 2003;115(2):187–195. doi: 10.1016/S0169-328X(03)00207-9. [DOI] [PubMed] [Google Scholar]
- 49.Levine L. Stimulated release of arachidonic acid by agonists of the peroxisome proliferator-activated receptor and retinoic acid receptors. Prostaglandins Leukot Essent Fat Acids. 2001;65(4):229–232. doi: 10.1054/plef.2001.0316. [DOI] [PubMed] [Google Scholar]
- 50.Li X, et al. Induction of retinol dehydrogenase 9 expression in podocytes attenuates kidney injury. J Am Soc Nephrol. 2014;25(9):1933–1941. doi: 10.1681/ASN.2013111150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.He JC, et al. Retinoic acid inhibits HIV-1-induced podocyte proliferation through the cAMP pathway. J Am Soc Nephrol. 2007;18(1):93–102. doi: 10.1681/ASN.2006070727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vaughan MR, et al. ATRA induces podocyte differentiation and alters nephrin and podocin expression in vitro and in vivo. Kidney Int. 2005;68(1):133–144. doi: 10.1111/j.1523-1755.2005.00387.x. [DOI] [PubMed] [Google Scholar]
- 53.Lyon MF, Hulse EV. An inherited kidney disease of mice resembling human nephronophthisis. J Med Genet. 1971;8(1):41–48. doi: 10.1136/jmg.8.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Peng M, et al. Mutant prenyltransferase-like mitochondrial protein (PLMP) and mitochondrial abnormalities in kd/kd mice. Kidney Int. 2004;66(1):20–28. doi: 10.1111/j.1523-1755.2004.00702.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Licitra F, Puccio H. An overview of current mouse models recapitulating coenzyme Q10 deficiency syndrome. Mol Syndromol. 2014;5(3–4):180–186. doi: 10.1159/000362942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Widmeier, et al. Treatment with 2,4-dihydroxybenzoic acid prevents FSGS progression and renal fibrosis in podocyte-specific Coq6 knockout mice. J Am Soc Nephrol. 2019;30(3):393–405. doi: 10.1681/ASN.2018060625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Widmeier E, et al. ADCK4 deficiency destabilizes the coenzyme Q complex, which is rescued by 2,4-dihydroxybenzoic acid treatment. J Am Soc Nephrol. 2020;31(6):1191–1211. doi: 10.1681/ASN.2019070756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hoeflich KP, et al. Antitumor efficacy of the novel RAF inhibitor GDC-0879 is predicted by BRAFV600E mutational status and sustained extracellular signal-regulated kinase/mitogen-activated protein kinase pathway suppression. Cancer Res. 2009;69(7):3042–3051. doi: 10.1158/0008-5472.CAN-08-3563. [DOI] [PubMed] [Google Scholar]
- 59.Wu H, et al. Advantages of single-nucleus over single-cell RNA sequencing of adult kidney: rare cell types and novel cell states revealed in fibrosis. J Am Soc Nephrol. 2019;30(1):23–32. doi: 10.1681/ASN.2018090912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Park J, et al. Single-cell transcriptomics of the mouse kidney reveals potential cellular targets of kidney disease. Science. 2018;360(6390):758–763. doi: 10.1126/science.aar2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schueler M, et al. DCDC2 mutations cause a renal-hepatic ciliopathy by disrupting Wnt signaling. Am J Hum Genet. 2015;96(1):81–92. doi: 10.1016/j.ajhg.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kirita Y, et al. Cell profiling of mouse acute kidney injury reveals conserved cellular responses to injury. Proc Natl Acad Sci. 2020;117(27):15874–15883. doi: 10.1073/pnas.2005477117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gharib SA, et al. Transcriptional landscape of glomerular parietal epithelial cells. PLoS One. 2014;9(8):e0105289. doi: 10.1371/journal.pone.0105289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kuppe C, et al. Novel parietal epithelial cell subpopulations contribute to focal segmental glomerulosclerosis and glomerular tip lesions. Kidney Int. 2019;96(1):80–93. doi: 10.1016/j.kint.2019.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Smeets B, et al. Tracing the origin of glomerular extracapillary lesions from parietal epithelial cells. J Am Soc Nephrol. 2009;20(12):2604–2615. doi: 10.1681/ASN.2009010122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Haensly WE, et al. Proximal-tubule-like epithelium in Bowman’s capsule in spontaneously hypertensive rats. Changes with age. Am J Pathol. 1982;107(1):92–7. [PMC free article] [PubMed] [Google Scholar]
- 67.Choi HMT, et al. Third-generation in situ hybridization chain reaction: multiplexed, quantitative, sensitive, versatile, robust. Development. 2018;145(12):dev165753. doi: 10.1242/dev.165753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Greka A, Mundel P. Cell biology and pathology of podocytes. Annu Rev Physiol. 2012;74(1):299–323. doi: 10.1146/annurev-physiol-020911-153238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kurtzeborn K, et al. MAPK/ERK signaling in regulation of renal differentiation. Int J Mol Sci. 2019;20(7):1779. doi: 10.3390/ijms20071779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sieber J, et al. GDC-0879, a BRAFV600E inhibitor, protects kidney podocytes from death. Cell Chem Biol. 2018;25(2):175–184. doi: 10.1016/j.chembiol.2017.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wong H, et al. Pharmacodynamics of 2-{4-[(1 E )-1-(Hydroxyimino)-2,3-dihydro-1 H -inden-5-yl]-3-(pyridine-4-yl)-1 H -pyrazol-1-yl}ethan-1-ol (GDC-0879), a potent and selective B-Raf kinase inhibitor: understanding relationships between systemic concentrations, phosphory. J Pharmacol Exp Ther. 2009;329(1):360–367. doi: 10.1124/jpet.108.148189. [DOI] [PubMed] [Google Scholar]
- 72.Choo EF, et al. Disposition of GDC-0879, a B-RAF kinase inhibitor in preclinical species. Xenobiotica. 2009;39(9):700–709. doi: 10.1080/00498250902991827. [DOI] [PubMed] [Google Scholar]
- 73.Poon WW, et al. Sensitivity to treatment with polyunsaturated fatty acids is a general characteristic of the ubiquinone-deficient yeast coq mutants. Mol Aspects Med. 1997;18(SUPPL.):121–127. doi: 10.1016/s0098-2997(97)00004-6. [DOI] [PubMed] [Google Scholar]
- 74.Do TQ, et al. Enhanced sensitivity of ubiquinone-deficient mutants of Saccharomyces cerevisiae to products of autoxidized polyunsaturated fatty acids. Proc Natl Acad Sci. 1996;93(15):7534–7539. doi: 10.1073/pnas.93.15.7534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Petroni A, et al. Changes in arachidonic acid levels and formation and in lipid synthesis in the human neuroblastoma SK-N-BE during retinoic acid-induced differentiation. J Neurochem. 2002;67(2):549–556. doi: 10.1046/j.1471-4159.1996.67020549.x. [DOI] [PubMed] [Google Scholar]
- 76.Moon PO, et al. Effects of retinoic acid treatment on release of arachidonic acid by chondrogenic cells in response to ionophore A23187. Life Sci. 1986;38(16):1445–1450. doi: 10.1016/0024-3205(86)90556-4. [DOI] [PubMed] [Google Scholar]
- 77.Ratnam KK, et al. Role of the retinoic acid receptor-α in HIV-associated nephropathy. Kidney Int. 2011;79(6):624–634. doi: 10.1038/ki.2010.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Araki H, et al. Retinoid agonist activities of synthetic geranyl geranoic acid derivatives. Biochem Biophys Res Commun. 1995;209(1):66–72. doi: 10.1006/bbrc.1995.1471. [DOI] [PubMed] [Google Scholar]
- 79.Hoffman LM, et al. BMP action in skeletogenesis involves attenuation of retinoid signaling. J Cell Biol. 2006;174(1):101–113. doi: 10.1083/jcb.200604150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hatzivassiliou G, et al. RAF inhibitors prime wild-type RAF to activate the MAPK pathway and enhance growth. Nature. 2010;464(7287):431–435. doi: 10.1038/nature08833. [DOI] [PubMed] [Google Scholar]
- 81. University of Michigan. NephroSeq. https://www.nephroseq.org/resource/login.html Updated January, 2021. Accessed November 17, 2020.
- 82.Srivastava T, et al. Synaptopodin expression in idiopathic nephrotic syndrome of childhood. Kidney Int. 2001;59(1):118–125. doi: 10.1046/j.1523-1755.2001.00472.x. [DOI] [PubMed] [Google Scholar]
- 83.Wenderfer SE, Gaut JP. Glomerular diseases in children. Adv Chronic Kidney Dis. 2017;24(6):364–371. doi: 10.1053/j.ackd.2017.09.005. [DOI] [PubMed] [Google Scholar]
- 84.Chen A, et al. Soluble RARRES1 induces podocyte apoptosis to promote glomerular disease progression. J Clin Invest. 2020;130(10):5523–5535. doi: 10.1172/JCI140155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jakobs BS, et al. A novel mutation in COQ2 leading to fatal infantile multisystem disease. J Neurol Sci. 2013;326(1–2):24–28. doi: 10.1016/j.jns.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 86.Emmanuele V, et al. Heterogeneity of coenzyme Q10 deficiency: patient study and literature review. Arch Neurol. 2012;69(8):978–983. doi: 10.1001/archneurol.2012.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Salviati L, et al. Infantile encephalomyopathy and nephropathy with CoQ10 deficiency: a CoQ10-responsive condition. Neurology. 2005;65(4):606–608. doi: 10.1212/01.wnl.0000172859.55579.a7. [DOI] [PubMed] [Google Scholar]
- 88.Tanaka A, et al. Polyunsaturated fatty acids induce ovarian cancer cell death through ROS-dependent MAP kinase activation. Biochem Biophys Res Commun. 2017;493(1):468–473. doi: 10.1016/j.bbrc.2017.08.168. [DOI] [PubMed] [Google Scholar]
- 89.Hsu HH, et al. Downregulation of the antioxidant protein peroxiredoxin 2 contributes to angiotensin II-mediated podocyte apoptosis. Kidney Int. 2011;80(9):959–969. doi: 10.1038/ki.2011.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Stockwell BR, et al. Ferroptosis: a regulated cell death nexus linking metabolism, redox biology, and disease. Cell. 2017;171(2):273–285. doi: 10.1016/j.cell.2017.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jha V, et al. Chronic kidney disease: global dimension and perspectives. Lancet. 2013;382(9888):260–272. doi: 10.1016/S0140-6736(13)60687-X. [DOI] [PubMed] [Google Scholar]
- 92.Mundel P, et al. Induction of differentiation in cultured rat and human podocytes. J Am Soc Nephrol. 1997;8(5):697–705. doi: 10.1681/ASN.V85697. [DOI] [PubMed] [Google Scholar]
- 93.Paynter NP, et al. Metabolic predictors of incident coronary heart disease in women. Circulation. 2018;137(8):841–853. doi: 10.1161/CIRCULATIONAHA.117.029468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.O’Sullivan JF, et al. Dimethylguanidino valeric acid is a marker of liver fat and predicts diabetes. J Clin Invest. 2017;127(12):4394–4402. doi: 10.1172/JCI95995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Love MI, et al. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):1–21. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Butler A, et al. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36(5):411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mertins P, et al. Proteogenomics connects somatic mutations to signalling in breast cancer. Nature. 2016;534(7605):55–62. doi: 10.1038/nature18003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All original microarray data were deposited in the NCBI’s Gene Expression Omnibus database (GEO GSE164274). Processed single nuclei RNA-Seq data are available at the Single Cell Portal (https://singlecell.broadinstitute.org/single_cell/study/SCP1245). Processed metabolomics data areavailable in Supplemental Tables 3 and 5. Associated GitHub code is available (Pdss2_2020; https://github.com/esidhom/Pdss2_2020