Abstract
Lysosomal dysfunction caused by mutations in lysosomal genes results in lysosomal storage disorder (LSD), characterized by accumulation of damaged proteins and organelles in cells and functional abnormalities in major organs, including the heart, skeletal muscle, and liver. In LSD, autophagy is inhibited at the lysosomal degradation step and accumulation of autophagosomes is observed. Enlargement of the left ventricle (LV) and contractile dysfunction were observed in RagA/B cardiac-specific KO (cKO) mice, a mouse model of LSD in which lysosomal acidification is impaired irreversibly. YAP, a downstream effector of the Hippo pathway, was accumulated in RagA/B cKO mouse hearts. Inhibition of YAP ameliorated cardiac hypertrophy and contractile dysfunction and attenuated accumulation of autophagosomes without affecting lysosomal function, suggesting that YAP plays an important role in mediating cardiomyopathy in RagA/B cKO mice. Cardiomyopathy was also alleviated by downregulation of Atg7, an intervention to inhibit autophagy, whereas it was exacerbated by stimulation of autophagy. YAP physically interacted with transcription factor EB (TFEB), a master transcription factor that controls autophagic and lysosomal gene expression, thereby facilitating accumulation of autophagosomes without degradation. These results indicate that accumulation of YAP in the presence of LSD promotes cardiomyopathy by stimulating accumulation of autophagosomes through activation of TFEB.
Keywords: Cardiology
Keywords: Autophagy, Signal transduction
Introduction
Lysosomes are responsible for degrading cellular proteins, lipids, pathogens, and organelles sequestered through autophagy, thereby regulating a wide variety of physiological processes, including cellular quality control mechanisms, metabolism, and signaling (1, 2). Lysosomal dysfunction caused by genetic mutations in lysosomal genes results in lysosomal storage disorder (LSD), characterized by accumulation of damaged proteins and organelles in cells (3). Increasing lines of evidence demonstrate that aging and obesity also cause lysosomal dysfunction (4, 5). Lysosomal dysfunction leads to both morphological and functional abnormalities in major organs, including the heart, skeletal muscle, and liver. In the heart, lysosomal dysfunction causes severe hypertrophic cardiomyopathy and heart failure (6, 7). However, the underlying molecular mechanism mediating cardiomyopathy remains unclear and no effective treatment currently exists.
The Rag-Ragulator complex is a key regulator of lysosome function (8). Rag proteins are Ras-like small GTPases that are anchored on the surface of lysosomes by Ragulator, which also acts as a guanine nucleotide exchanger for Rag proteins A and B (RagA/B) (9, 10). Although Rag GTPases, consisting of RagA/B and RagC/D heterodimers, can bind directly to Raptor, a subunit of mechanistic target of rapamycin complex 1 (mTORC1), and are involved in the recruitment of mTORC1 to the lysosomal membrane (8, 11), Rag GTPases also indirectly bind to proton-pumping V-type ATPase through Ragulator. We demonstrated previously that loss of RagA/B induces lysosomal dysfunction with lysosomal pH elevation due to inactivation of proton-pumping V-type ATPase. Muscle-specific homozygous deletion of RagA/B driven by creatine kinase-Cre resulted in hypertrophic cardiomyopathy reminiscent of LSD (12). However, the underlying mechanism of cardiomyopathy observed in the RagA/B KO mice remains unknown. Thus, we here aimed to elucidate the molecular mechanism of cardiomyopathy in RagA/B KO mice.
The Hippo pathway is an evolutionarily conserved signaling pathway that serves as a key regulator of organ size (13). Yes-associated protein (YAP), a transcription cofactor and a major terminal effector of the Hippo pathway, promotes cell growth and survival by stimulating cell proliferation and inhibiting apoptosis (14). Activation of YAP induces tumorigenesis and tissue regeneration in many organs. Activation of YAP in the heart promotes survival of cardiomyocytes (CMs) (15) and induces cardiac repair and regeneration after myocardial infarction (16), thereby generally playing a salutary role. However, we recently discovered that persistent activation of YAP in the heart leads to cardiac dysfunction in response to stress, including pressure overload, by promoting dedifferentiation of CMs (17). Furthermore, nuclear accumulation of YAP in CMs is induced in the presence of high-fat diet consumption and superimposition of high blood pressure exacerbates the development of heart failure (18). Thus, activation of YAP in the heart can be either salutary or detrimental in a context-dependent manner. YAP associates with transcription factors, including the TEAD family transcription factors, thereby regulating a wide variety of downstream target genes, depending upon the availability of the transcription factors with which YAP partners. We discovered that another condition in which YAP is persistently activated in the heart is in cardiac-specific RagA/B KO (RagA/B cKO) mice, a mouse model of LSD, in which cardiomyopathy is induced by lysosomal dysfunction. Furthermore, we found that YAP physically interacts with transcription factor EB (TFEB), a transcription factor that controls lysosome biogenesis and autophagy.
Here, we hypothesize that activation of YAP caused by lysosomal dysfunction plays an important role in mediating hypertrophic cardiomyopathy. We asked (a) whether YAP is activated in the hearts of RagA/B KO mice, (b) whether YAP plays an important role in mediating the cardiomyopathy phenotype in RagA/B cKO mice, and (c) whether the YAP/TFEB pathway promotes autophagy and death of CMs in RagA/B cKO mice.
Results
RagA/B cKO mice exhibit severe hypertrophic cardiomyopathy reminiscent of LSD.
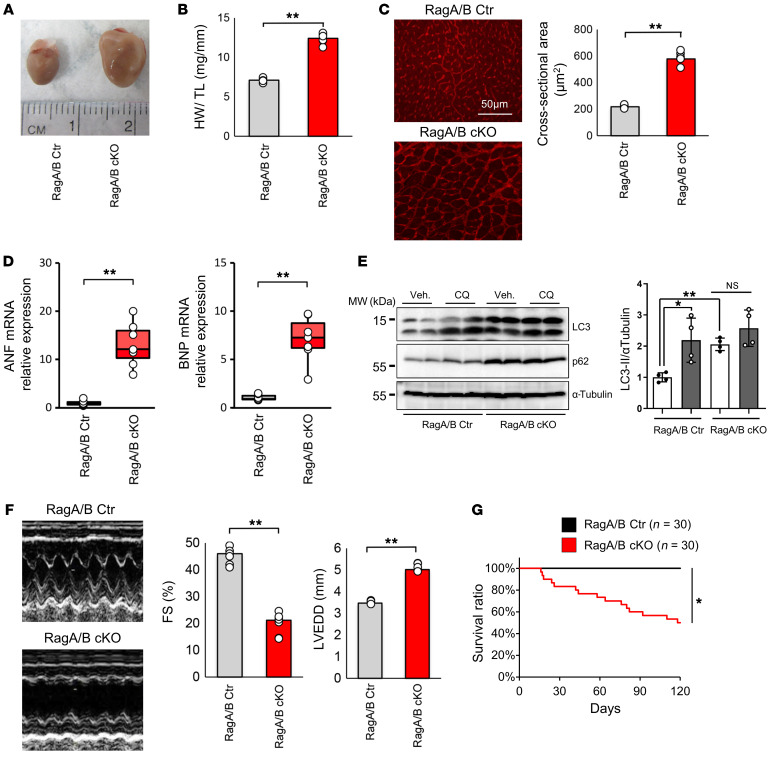
We have shown previously that mice with muscle-specific deletion of RagA/B (RagA/B mKO), generated using muscle creatine kinase promoter-driven Cre, develop marked left ventricular (LV) enlargement and cardiac dysfunction (12). Since RagA/B is downregulated not only in the heart but also in skeletal muscles in RagA/B mKO mice, in order to confirm that the cardiac defect is caused by downregulation of RagA/B in the heart alone, cardiac-specific homozygous RagA/B KO (RagA/B cKO) mice were generated, using Myh6-Cre–mediated deletion of floxed RagA/B alleles. Postmortem analyses indicated LV enlargement, increases in heart weight/tibial length (HW/TL), and increases in histologically evaluated CM cross-sectional area (CSA) at 12 weeks of age (Figure 1, A–C). RagA/B cKO mice also exhibited upregulation of hypertrophic markers, including atrial natriuretic factor (ANF) and brain natriuretic peptide (BNP) (Figure 1D). Similarly to RagA/B mKO mice, RagA/B cKO mice exhibited decreases in the mature form of cathepsin D and increases in LC3-II and p62/SQSTM1 in the heart (Supplemental Figure 2A; supplemental material available online with this article; https://doi.org/10.1172/JCI143173DS1), whereas chloroquine (CQ) treatment increased LC3-II in control mice, but not in RagA/B cKO mice (Figure 1E), consistent with lysosomal dysfunction and inhibition of autophagic flux. Echocardiographic examination at 12 weeks revealed that RagA/B cKO mice had a significantly lower LV fractional shortening (FS) and a significantly greater LV-end diastolic diameter (LVEDD) than control mice (Figure 1F). In order to evaluate whether cardiac hypertrophy develops secondary to LV dysfunction, the time courses of cardiac hypertrophy and LV dysfunction were evaluated in RagA/B cKO mice. RagA/B cKO mice exhibited LV dysfunction and dilation as early as 2 weeks, the earliest time point at which we could conduct echocardiography after weening (Supplemental Figure 1). Baseline cardiac hypertrophy, evidenced by increases in HW/TL, was also observed as early as 2 weeks, the earliest time point we examined (Supplemental Figure 1). Thus, both cardiac hypertrophy and LV dysfunction develop rapidly after birth in RagA/B cKO mice. Whether cardiac hypertrophy develops independently of LV dysfunction during early postnatal development remains to be clarified.
Figure 1. RagA/B cKO mice exhibit severe hypertrophy reminiscent of LSD.
(A) Representative images of control (Ctr) and RagA/B cKO mouse hearts at 12 weeks old. (B) Quantitative analysis of total HW/TL at 12 weeks. n = 6. (C) Left: representative micrographs of WGA staining of the LV. Right: quantitative analysis of CSA at 12 weeks. n = 6. (D) Relative ANF and BNP mRNA expression at 12 weeks. n = 7. (E) Representative immunoblots of 1.5-month old control and RagA/B cKO mouse heart homogenates. Mice were treated with CQ or vehicle for 4 hours before euthanasia. Quantitative analyses of LC3-II/α-tubulin are shown. n = 4. (F) Representative echocardiographic tracings of control and RagA/B cKO mouse hearts at 12 weeks. Quantitative analyses of FS and LVEDD are shown. n = 6. (G) Kaplan-Meier survival curves. Results are expressed as mean ± SEM in B, C, E, and F. In D, results are shown as box plots, showing the median (center line) and IQR. Whiskers represent minima and maxima within 1.5 IQR as indicated. *P < 0.05; **P < 0.01, ANOVA.
RagA/B cKO mice exhibited elevated interstitial fibrosis compared with control mice (Supplemental Figure 2B). Furthermore, RagA/B cKO mice showed significantly more TUNEL-positive CMs than control mice (Supplemental Figure 2C). RagA/B cKO mice also exhibited less nuclear staining of HMGB1 (19) in CMs than control mice (Supplemental Figure 2D). These results suggest that cardiac-specific downregulation of RagA/B increases both apoptosis and necrosis of CMs in vivo. Kaplan-Meier survival analysis showed that RagA/B cKO mice had a significantly greater mortality rate than control mice (Figure 1G). Electron microscopic analyses indicated that the number of autophagosomes and autolysosomes was increased in myocardial sections obtained from RagA/B cKO mice compared with those from control mice. In addition, prominent accumulation of glycogen was observed in myocardial sections from RagA/B cKO mice (Supplemental Figure 2, E and F). Taken together, these data indicate that RagA/B cKO mice exhibit hypertrophic cardiomyopathy reminiscent of LSD.
YAP was upregulated in the hearts of RagA/B cKO mice.
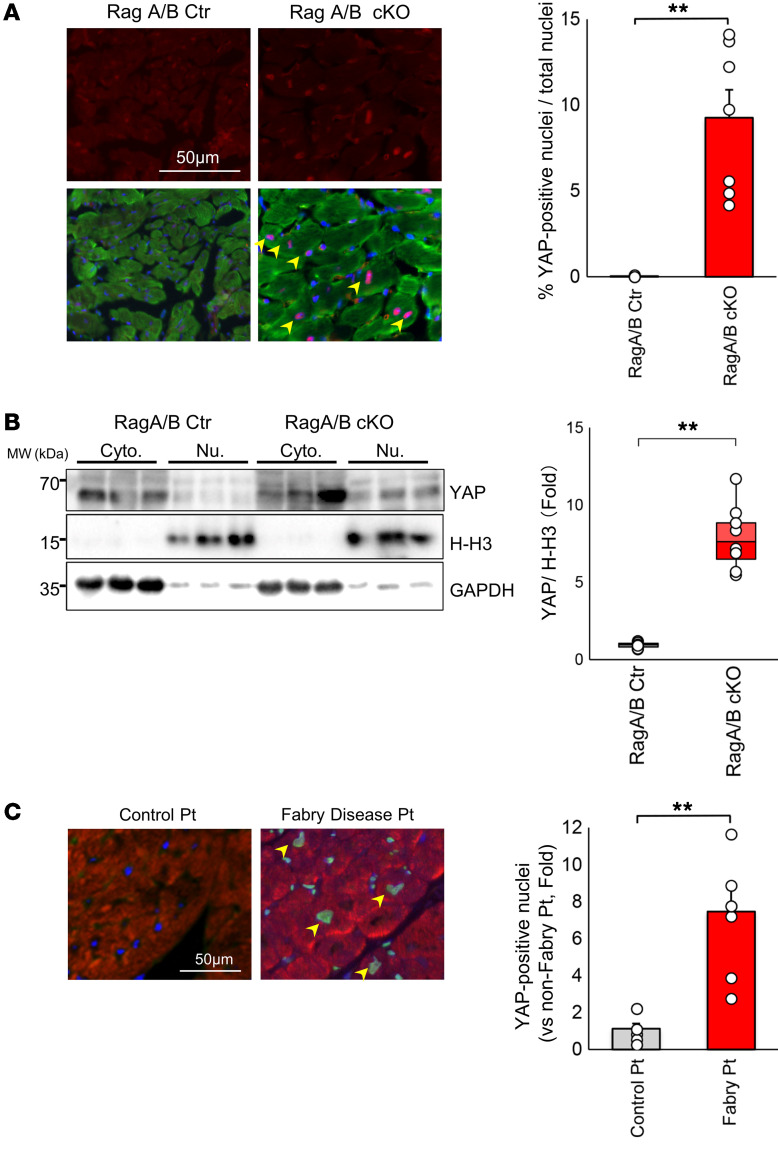
Inactivation of the Hippo pathway or activation of YAP, a major transcription factor cofactor of the Hippo pathway, induces enlargement of the fetal heart and the liver in mice through increased cell proliferation (13). We have shown recently that activation of YAP and its downstream mechanisms, including miR-206 and Akt, mediate cardiac hypertrophy in vitro and in vivo (15, 17, 20, 21). We therefore investigated whether YAP is activated in the hearts of RagA/B cKO mice. Immunostaining and Western blot analyses of the heart revealed that YAP was significantly upregulated in the cytosol and the nuclei of CMs in RagA/B cKO hearts (Figure 2, A and B). Quantitative analysis showed that the level of YAP in the nucleus was significantly higher in RagA/B cKO mice than in control mice (Figure 2, A and B). Quantitative PCR analyses indicated that target genes of YAP, including Ctgf and Runx2, were significantly upregulated in RagA/B cKO hearts (Supplemental Figure 3A). RagA/B cKO hearts also exhibited increases in cell-cycle reentry, as indicated by increases in phospho-histone H3 (Supplemental Figure 3B), and the markers of CM dedifferentiation, as indicated by upregulation of β-myosin heavy chain (β-MHC) and α-smooth muscle actin and downregulation of cardiac troponin T (Supplemental Figure 3, C and D), reminiscent of the cardiac phenotype observed in Hippo-deficient mice in the presence of pressure overload (17). Taken together, these results suggest that YAP is upregulated and that signs of CM dedifferentiation are observed in RagA/B cKO mouse hearts. We also evaluated protein expression of YAP in the heart in myocardial biopsy specimens obtained from patients with untreated Fabry disease, an LSD. The nuclear expression of YAP in CMs was significantly elevated in these patients compared with in those without Fabry disease (Figure 2C).
Figure 2. Nuclear accumulation of YAP in RagA/B cKO mouse hearts.
(A) Representative micrographs and quantitative analysis of nuclear YAP (red, YAP; green, sarcomeric actin; blue, DAPI). Symbols indicate mean percentage of YAP-positive nuclei/total nuclei calculated from more than 20 power fields per experiment. Arrowheads indicate YAP-positive nuclei in cardiomyocytes. n = 7. (B) Representative images of YAP immunoblots of cytosolic and nuclear fractions. n = 8. For A and B, analyses were carried out at 12 weeks of age. (C) Representative micrographs and quantitative analysis of nuclear YAP in the heart. Heart specimens obtained from control patients without Fabry disease (n = 8) and from Fabry disease patients (n = 7) were subjected to immunostaining with anti-YAP and anti-sarcomeric actin antibodies. Quantitative analysis is shown on the right. Symbols indicate relative mean YAP-positive nuclei/total nuclei calculated from more than 10 high-power fields per experiment. The mean YAP-positive nuclei/total nuclei in control patient hearts was define as 1. Results are expressed as mean ± SEM in A and C. In B, results are shown as box plots, showing the median (center line) and IQR. Whiskers represent minima and maxima within 1.5 IQR as indicated. **P < 0.01, ANOVA.
We next investigated the mechanism by which YAP is upregulated in RagA/B cKO mice. mRNA expression of YAP in the RagA/B cKO mouse heart did not differ significantly from that in the control mouse heart (Supplemental Figure 4A). Treatment with MG132, a proteasome inhibitor, or bafilomycin A1, a vacuolar H+ATPase inhibitor, increased the level of YAP in cultured CMs, suggesting that YAP is degraded through both proteasome- and lysosome-dependent mechanisms (Supplemental Figure 4B). Staining of cultured CMs with anti-YAP antibody showed that MG132 and bafilomycin A1 increased the level of YAP in both the cytosol and the nucleus (Supplemental Figure 4C). A previous study reported that YAP coimmunoprecipitates with the autophagy cargo receptor p62 in mouse embryonic fibroblasts (MEFs) in the presence of CQ (22). We confirmed that YAP coimmunoprecipitates with p62 in CMs in the presence of CQ, a condition of lysosome suppression similar to that observed in RagA/B mKO mice (ref. 12 and Supplemental Figure 4D). Since autophagy is suppressed in both RagA/B mKO and cKO mice, in order to test whether suppression of autophagy is sufficient to increase the level of YAP, we evaluated the levels of YAP in atg7 KO MEFs and in adult CMs freshly isolated from the hearts of cardiac-specific atg7 KO (atg7 cKO) mice (Supplemental Figure 4, E and F). The level of YAP was 1.8-fold higher in atg7 KO MEF cells than in control MEF cells and 5.8-fold higher in CMs isolated from atg7 cKO mouse hearts than in those from control mouse hearts. These results suggest that suppression of autophagic degradation leads to accumulation of YAP in RagA/B cKO hearts. YAP is degraded through a β-TcCP E3 ligase-dependent mechanism when the upstream Hippo kinases, namely the Mst1/2-Lats1/2 pathway, are activated (23). However, phosphorylation of Mst1 at Thr183, a measure of its activity, in RagA/B cKO mice did not differ significantly from that in WT mice (Supplemental Figure 4G). Thus, it is unlikely that the accumulation of YAP occurs due to suppression of ubiquitin proteasome degradation in RagA/B cKO mice.
YAP deletion in RagA/B cKO mice alleviated cardiac hypertrophy and improved cardiac function.
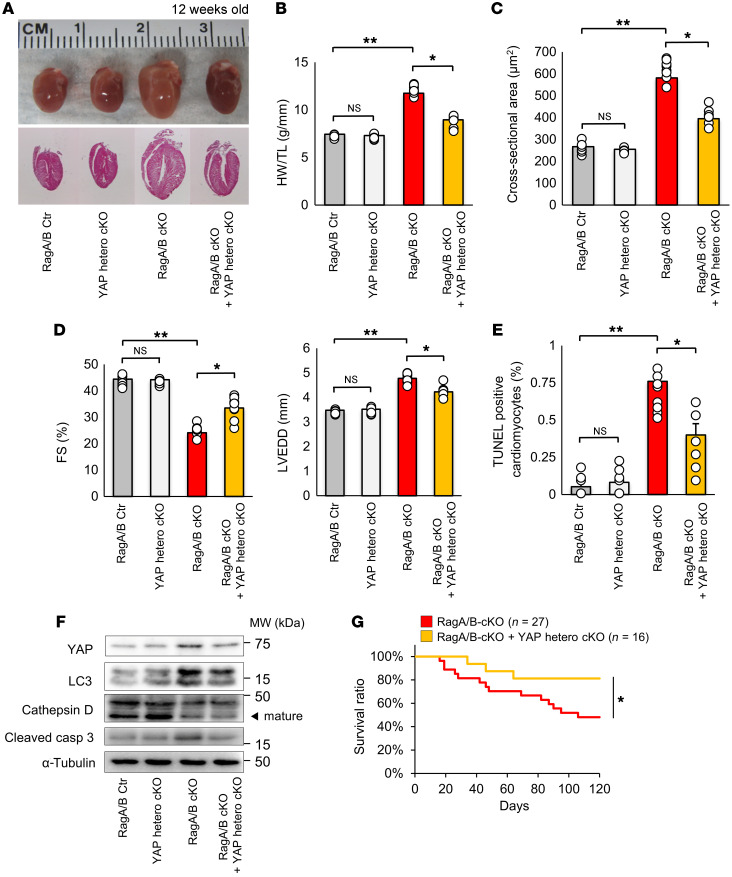
In order to investigate the role of endogenous YAP in mediating the development of cardiomyopathy in RagA/B cKO mice, we crossed RagA/B cKO mice with cardiac-specific heterozygous Yap KO (Yap hcKO) mice. Both heart size and HW/TL were significantly smaller in RagA/B cKO mice with cardiac-specific heterozygous deletion of YAP (RagA/B cKO+Yap hcKO) than in RagA/B cKO mice without YAP deletion at 3 months of age (Figure 3, A and B). CM CSA was significantly smaller in RagA/B cKO+Yap hcKO mice than in RagA/B cKO mice, indicating attenuation of cardiac hypertrophy (Figure 3C). RagA/B cKO+Yap hcKO mice exhibited a significantly higher FS and smaller LVEDD than RagA/B cKO mice, as evaluated by echocardiography (Figure 3D). RagA/B cKO+Yap hcKO mice also exhibited significantly fewer TUNEL-positive CMs than RagA/B cKO mice (Figure 3E). Furthermore, RagA/B cKO+Yap hcKO mice showed significantly lower levels of cleaved caspase-3 than RagA/B cKO mice (Figure 3F and Supplemental Figure 5). Kaplan-Meier survival analyses showed that RagA/B cKO+Yap hcKO mice had a significantly better survival rate than RagA/B cKO mice (Figure 3G).
Figure 3. Heterozygous deletion of YAP in RagA/B cKO mice reduces cardiac hypertrophy and alleviates cardiac dysfunction.
(A) Representative whole-mount images and sagittal sections of control, YAP hcKO, RagA/B cKO, and RagA/B-cKO+YAP hcKO mouse hearts at 12 weeks old. (B) Quantitative analysis of HW/TL at 12 weeks. n = 8. (C) Quantitative analysis of CM CSA. n = 8. (D) Quantitative analysis of FS and LVEDD, as evaluated with echocardiographic measurements. n = 8. (E) Quantitative analysis of TUNEL-positive CMs. n = 8. Symbols indicate percentage of TUNEL-positive cardiomyocytes calculated from more than 10 high-power fields per experiment. (F) Representative immunoblots of heart homogenates. Blots run in parallel, contemporaneously, using identical samples are shown. n = 8. Analyses were carried out at 12 weeks of age in A–F. (G) Kaplan-Meier survival curves. Results are expressed as mean ± SEM. *P < 0.05; **P < 0.01, ANOVA.
In order to further elucidate the role of endogenous YAP in the development of cardiomyopathy in adult RagA/B cKO mice, we treated 3-month-old RagA/B cKO mice with verteporfin, a small molecule that inhibits interaction between YAP and TEAD transcription factors (ref. 24 and Supplemental Figure 6A). Similarly to the effect of genetic downregulation of YAP, verteporfin treatment alleviated cardiac hypertrophy and dysfunction in RagA/B cKO mice (Supplemental Figure 6, B–E). Verteporfin treatment also decreased the number of TUNEL-positive CMs in RagA/B cKO mice (Supplemental Figure 6F), suggesting that endogenous YAP partially mediates cardiac hypertrophy and dysfunction in RagA/B cKO mice. Furthermore, verteporfin treatment improved cell viability in CMs transduced with shRNA-targeting RagA/B (sh-RagA/B) (Supplemental Figure 6G). These results suggest that YAP negatively affects the survival of CMs in the presence of RagA/B downregulation in a cell-autonomous manner. Although genetic downregulation of YAP with Myh6-Cre may affect myocyte proliferation during fetal development, verteporfin treatment inhibits the activity of YAP postnatally. Thus, these results demonstrate that postnatal suppression of YAP inhibits the development of cardiomyopathy in RagA/B cKO mice. Furthermore, verteporfin treatment reversed established cardiomyopathy in RagA/B cKO mice.
Normalization of YAP failed to rescue the lysosomal dysfunction in the presence of Rag A/B knockdown.
Lysosome function is markedly impaired in RagA/B mKO mice (12). We investigated to determine whether the salutary effect of YAP downregulation in Rag A/B cKO mice is mediated through improvement of lysosomal function. Downregulation of endogenous YAP in RagA/B cKO mice did not restore the level of mature cathepsin D in the heart in vivo (Figure 3F and Figure 4A). sh-RagA/B–mediated downregulation of RagA/B in cultured CMs decreased the level of mature cathepsin D in vitro (Figure 4B), suggesting that downregulation of RagA/B induces lysosomal dysfunction in a cell-autonomous manner. In this condition, neither downregulation of endogenous YAP nor overexpression of YAP affected the level of mature cathepsin D (Figure 4B). Downregulation of RagA/B induced neutralization of lysosomal pH in cultured CMs, consistent with what was observed in the RagA/B mKO mouse heart (12). Again, neither downregulation of endogenous YAP nor overexpression of YAP in this condition affected the pH in lysosomes (Figure 4C). Thus, YAP does not affect lysosomal function in CMs, and the rescue of the cardiomyopathy phenotype in RagA/B cKO mice via partial downregulation of YAP was likely not mediated through restoration of lysosomal function.
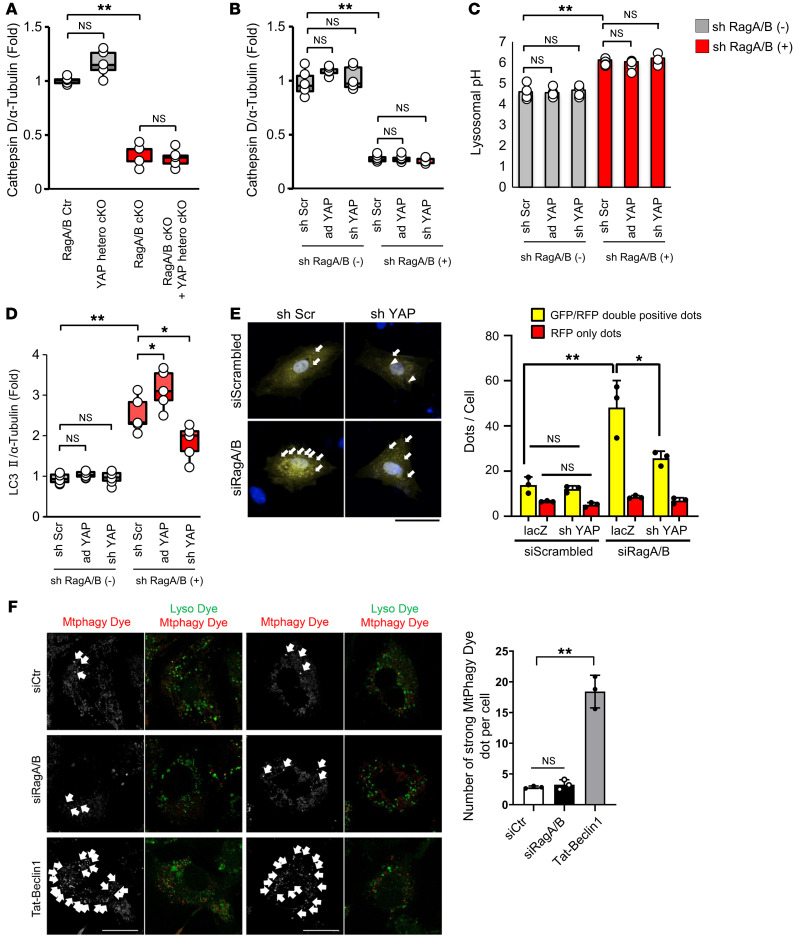
Figure 4. Downregulation of YAP attenuates autophagy without affecting lysosomal function in the presence of RagA/B downregulation.
(A) Quantitative analysis of immunoblotting for cathepsin D/α-tubulin in mouse hearts. n = 6. (B) Quantitative analysis of immunoblotting for cathepsin D/α-tubulin in cultured CMs. n = 5. (C) Quantitative analysis of lysosomal pH in cultured CMs. Relative intensity was obtained from more than 10 cells per each experiment. n = 4. (D) Quantitative analysis of immunoblotting for LC3II/α-tubulin in cultured CMs. n = 5. (E) Representative images of mRFP-GFP-LC3 puncta in cultured CMs in vitro. Quantitative analyses of LC3 puncta are shown on the right. Yellow puncta (left, indicated by arrows) and columns (right) indicate GFP-RFP double-positive puncta and represent autophagosomes, whereas red puncta (left, indicated by arrowheads) and columns (right) indicate GFP negative-RFP positive puncta and represent autolysosomes. Puncta were measured from more than 100 cells per experiment. n = 3. (F) Neonatal CMs were treated with siControl or siRagA/B for 60 hours or 5 μM TAT–beclin 1 for 24 hours. Cells were treated with 100 nM MtPhagy Dye and 1 μM Lyso dye. Strong MtPhagy Dye puncta colocalized with Lysodye (indicated by white arrows) were counted from more than 20 cells per experiment. n = 3. Scale bars: 20 μm. In A, B, and D, results are shown as box plots, showing the median (center line) and IQR. Whiskers represent minima and maxima within 1.5 IQR as indicated. In C, results are expressed as mean ± SEM. In E and F, results are expressed as mean ± SD. *P < 0.05; **P < 0.01, ANOVA.
Downregulation of endogenous YAP decreases autophagosome formation without affecting lysosomal function in CMs with RagA/B downregulation.
Autophagy is markedly impaired in RagA/B mKO mice (12). We next evaluated how downregulation of YAP affects the level of autophagy in CMs. Downregulation of RagA/B significantly increased the level of LC3II and the number of autophagosomes, evaluated by counting GFP-LC3/RFP-LC3 double-positive (yellow) puncta in cultured CMs transduced with tandem fluorescent LC3 (Figure 4, D and E). Since lysosomes do not function properly in the presence of RagA/B downregulation, the increases in LC3II and in yellow dots without a concurrent increase in red dots indicate accumulation of autophagosomes without an increase in autophagic flux. Mitophagic activity, as evaluated with MtPhagy Dye (25), was low and at background levels in CMs in which RagA/B was downregulated with siRNAs (Figure 4F). Treatment of control CMs with a low dose of TAT–Beclin 1, an inducer of mitophagy (26), prominently increased strong fluorescence of MtPhagy Dye, serving as a positive control in this experiment. Thus, mitophagy is not affected by downregulation of RagA/B.
Although shRNA-mediated downregulation of YAP (sh-YAP) did not affect the level of LC3II or the number of yellow dots in control CMs, it significantly decreased LC3II and yellow dots in CMs treated with sh-RagA/B (Figure 3F and Figure 4, D and E). On the other hand, overexpression of YAP further enhanced the increase in the level of LC3II in the presence of RagA/B downregulation (Figure 4D). Taken together, these results suggest that downregulation of endogenous YAP decreases autophagosome formation without affecting lysosomal function in CMs in the presence of RagA/B downregulation.
Cardiac dysfunction in RagA/B cKO mice is alleviated by suppression of autophagy and exacerbated by stimulation of autophagy.
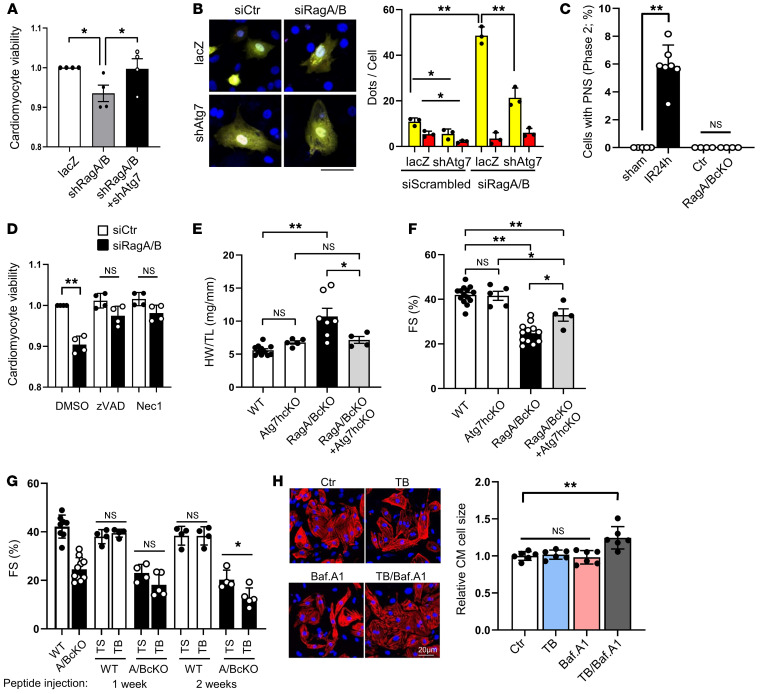
Previous studies demonstrated that accumulation of autophagosomes facilitates cell death in the presence of stable inactivation of lysosomes by CQ treatment (27–29). We therefore determined whether attenuation of autophagosome accumulation alleviates CM death in the presence of RagA/B downregulation. To this end, we evaluated the effect of interventions to inhibit autophagosome formation upon cell death in RagA/B knockdown CMs. Knockdown of Atg7 in conjunction with sh-RagA/B treatment alleviated CM cell death; this was accompanied by decreases in the accumulation of autophagosomes in the CMs (Figure 5, A and B). One mechanism mediating cell death in response to excessive activation of autophagy is autosis (30). Electron microscopic analyses indicated that cells with perinuclear space, an important feature of autosis (30), are identified more frequently in the heart after ischemia/reperfusion, but not in the RagA/B cKO mouse heart or in the control heart (Figure 5C). Decreased CM viability in response to RagA/B downregulation was alleviated in the presence of zVAD, an inhibitor of apoptosis, or Nec1, an inhibitor of necroptosis (Figure 5D). Thus, downregulation of RagA/B induces cell death in CMs through either apoptosis or necroptosis, but not autosis. Furthermore, cardiac hypertrophy and LV dysfunction observed in RagA/B cKO mice were partially alleviated when RagA/B cKO mice were crossed with Atg7 heterozygous cKO mice (Figure 5, E and F). We further investigated whether the cardiac dysfunction phenotype in RagA/B cKO mice is exacerbated when autophagy is stimulated. To this end, we injected either TAT–beclin 1, a cell-permeable and potent inducer of autophagy (31), or TAT-scrambled into control and RagA/B cKO mice. We have shown previously that injection with a low dose of TAT–beclin 1 upregulates autophagy in the heart (26). Injection of TAT–beclin 1, but not TAT-scrambled, exacerbated cardiac dysfunction, as indicated by decreases in FS, in RagA/B cKO mice, but not in WT mice (Figure 5G). These results suggest that the accumulation of autophagosomes without a concomitant increase in autophagic flux may be toxic in the heart. Cotreatment of cultured CMs with TAT–beclin 1 and bafilomycin A1 significantly increased the size of CMs, suggesting that accumulation of autophagosomes in the presence of lysosomal dysfunction directly stimulates cardiac hypertrophy in a cell-autonomous manner (Figure 5H). Known inducers of cardiac hypertrophy, including mTORC1 and Akt, were not significantly activated in RagA/B cKO mice (Supplemental Figure 7), consistent with our previous results. We next investigated the mechanism by which YAP affects autophagosome formation.
Figure 5. Suppression of autophagy alleviates cardiac dysfunction in RagA/B cKO mice.
(A and B) CMs were transduced with Ad-lacZ, Ad-sh-RagA+Ad-sh-RagB, or Ad-sh-RagA+Ad-sh-RagB+Ad-sh-Atg7. (A) Cell viability quantified by CellTiter-Blue assay. n = 4. Values were measured from more than 8 different wells per experiment. (B) CMs were transduced with Ad-tandem fluorescent LC3, and accumulation of autophagosomes (yellow puncta) and autolysosomes (red puncta) was evaluated. n = 3. Values were measured for more than 50 cells per experiment. Scale bar: 20 μm. (C) Percentage of cells with PNS. n = 4–7. In each mouse, cells with PNS were counted from more than 100 CMs. (D) Cells were treated with DMOS, zVAD, or Nec1 with siRagA/B or siCont. Cell viability quantified by CellTiter-Blue assay. n = 4. In each experiment, cell viability was evaluated from more than 8 different wells. (E) HW/TL in WT, Atg7 hcKO, RagA/B cKO, and RagA/B cKO+Atg7 hcKO mice at 2.5 months old. n = 4–11. (F) Percentage of FS in WT, Atg7 hcKO, RagA/B cKO, and RagA/B cKO+Atg7 hcKO mice at 2.5 months old. n = 4–14. (G) WT and RagA/B cKO mice were injected with TAT-scrambled (TS) or TAT–beclin 1 (TB) for 1 to 2 weeks. Quantitative analyses of echocardiographically evaluated percentage of FS are shown. n = 4–12. (H) Neonatal CMs were treated with 5 μM TAT–beclin 1, 20 nM bafilomycin A1 or TAT–beclin 1+Bafilomycin A1 for 24 hours. Cells were stained with cardiac troponin T, and cell size was quantified. n = 6. In each experiment, cell size was measured from more than 50 cells. Results are expressed as mean ± SD. *P < 0.05; **P < 0.01, ANOVA.
YAP regulates TFEB activity during lysosomal dysfunction.
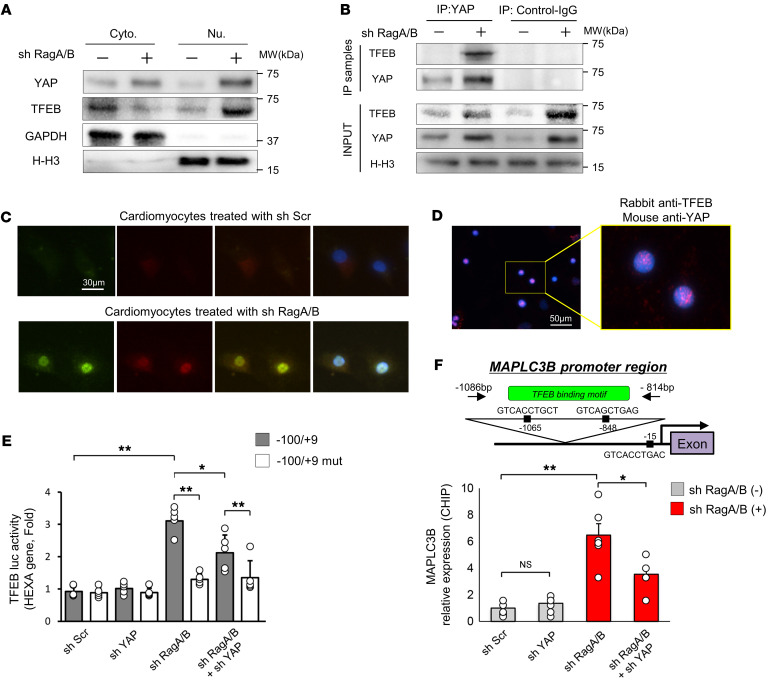
TFEB, a master regulator of autophagic and lysosomal gene expression, is translocated into the nucleus and activated in hearts with LSD (32). We have shown that RagA/B mKO hearts exhibit nuclear localization of TFEB (12). On the other hand, the level of TEAD, a well-established target of YAP, did not differ significantly between RagA/B cKO and control mouse hearts (Supplemental Figure 8, A and B). Both YAP and TFEB were translocated into the nuclei of CMs in the presence of RagA/B downregulation (Figure 6A). Coimmunoprecipitation assays showed that YAP interacts with TFEB in the CM nucleus when RagA/B is downregulated (Figure 6B). Likewise, coimmunostaining assays (Figure 6C) and proximity ligation assays (PLAs) (Figure 6D and Supplemental Figure 9A) indicated that YAP interacts with TFEB in the CM nucleus in the presence of RagA/B knockdown.
Figure 6. Endogenous YAP interacts with TFEB in CMs transduced with Ad-sh-RagA/B and promotes TFEB-mediated transcription.
(A and B) CMs were transduced with or without Ad-sh-RagA/B. (A) Representative immunoblots of cytosolic and nuclear fractions. Blots run in parallel, contemporaneously, using identical samples are shown. n = 5. (B) CM lysates were subjected to immunoprecipitation with anti-YAP or control antibody. Original lysates (input) and the immunoprecipitation samples were then subjected to immunoblot analyses. Representative images are shown. For input, a blot run in parallel, contemporaneously, using identical samples is shown. n = 5. (C) Triple immunostaining with anti-YAP and anti-TFEB antibodies and DAPI of CMs transduced with Ad-sh-RagA/B. n = 6. (D) PLAs using anti-YAP and anti-TFEB antibodies in CMs transduced with Ad-sh-RagA/B. Inset shows a 3.2-fold magnification of area indicated by yellow rectangle. n = 3. (E) TFEB reporter gene assays were conducted with neonatal CMs transduced with Ad-sh-Scr or Ad-sh-YAP in the presence or absence of Ad-sh-RagA/B. n = 6. (F) ChIP assays with anti-YAP antibody in CMs transduced with Ad-sh-Scr or Ad-sh-YAP in the presence or absence of Ad-sh-RagA/B. The precipitated chromatin was subjected to PCR to detect the presence of the TFEB-binding element in the rat MAPLC3B promoter, as indicated in the inset. n = 6. Results are expressed as mean ± SEM. *P < 0.05; **P < 0.01, ANOVA.
In order to evaluate the effect of YAP upon the transcriptional activity of TFEB, reporter gene assays were conducted using a luciferase reporter harboring the TFEB-binding site (–100/+9) (33). The activity of the reporter gene in CMs was increased in the presence of RagA/B knockdown, but this increase was attenuated in the presence of YAP downregulation (Figure 6E). RagA/B knockdown did not activate the reporter when there was a mutation in the TFEB-binding site (–100/+9 mut). ChIP assays showed that TFEB interacts with the MAPLC3B promoter, which contains a TFEB-binding site, in CMs in the presence of RagA/B downregulation. This interaction was attenuated in the presence of YAP knockdown (Figure 6F). Consistently, downregulation of RagA/B significantly upregulated mRNA expression of TFEB target genes in cultured CMs, an effect that was significantly attenuated in the presence of YAP downregulation (Supplemental Figure 9B). These results are consistent with the notion that TFEB upregulates genes involved in autophagosome formation (32) and that YAP positively affects the function of TFEB in CMs by acting as a transcription cofactor. Finally, we evaluated the role of TFEB in mediating CM death in the presence of RagA/B downregulation in vitro. Decreases in the viability of cultured CMs in response to RagA/B knockdown were significantly alleviated when TFEB was downregulated (Supplemental Figure 10), suggesting that endogenous TFEB plays a critical role in mediating the cell death–inducing effect of RagA/B downregulation. In contrast, adenovirus-mediated upregulation of TFEB failed to enhance RagA/B knockdown–induced cell death (Supplemental Figure 10), consistent with the notion that RagA/B downregulation and TFEB act on the same pathway for the induction of cell death and, therefore, do not have additive effects.
Discussion
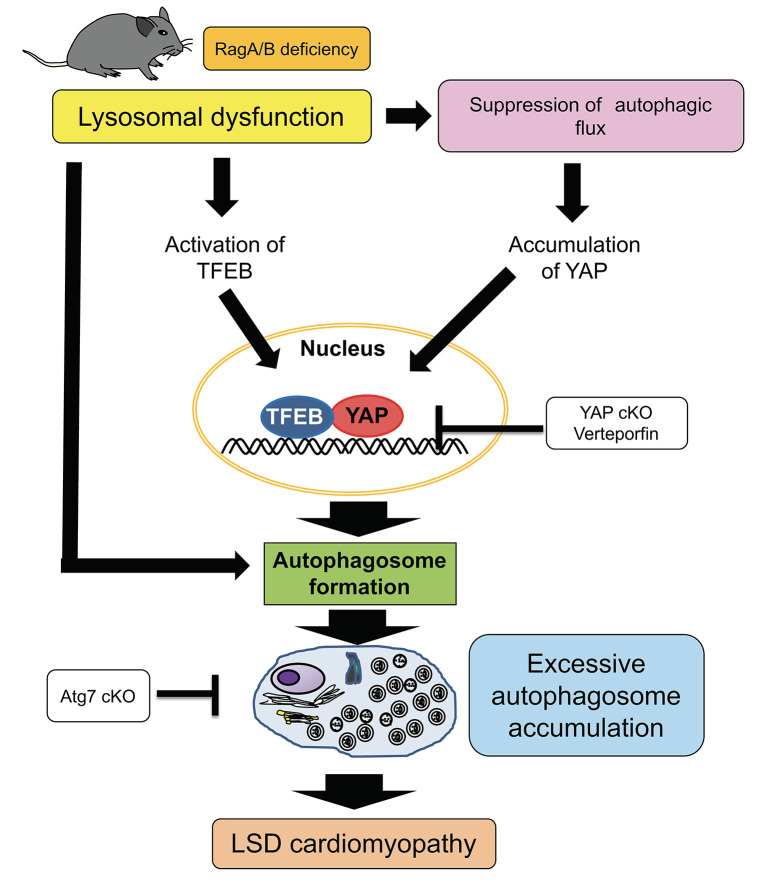
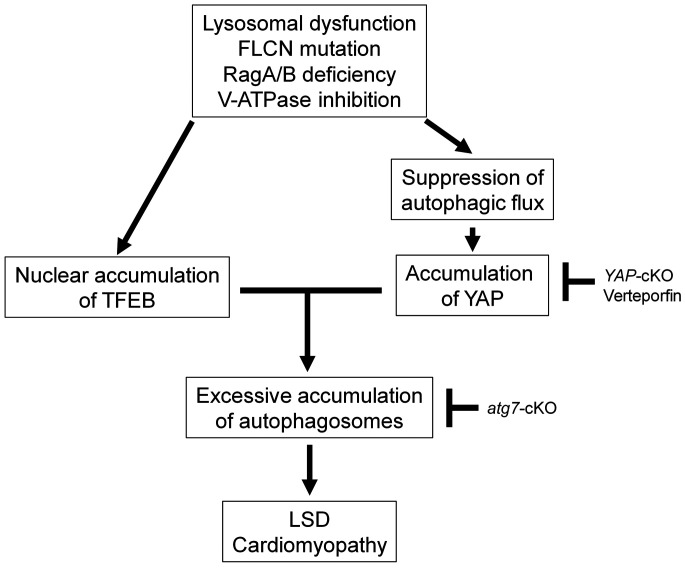
We here show that activation of autophagy through the YAP/TFEB pathway in the presence of an intractable block of autophagic flux at the lysosome level, such as permanent suppression of lysosomal acidification due to downregulation of RagA/B (12), promotes accumulation of autophagosomes and death of CMs, thereby promoting the development of cardiomyopathy in RagA/B cKO mice, a mouse model of LSD (Figure 7). Although previous studies have suggested stimulation of autophagy as a therapeutic option for LSD (34), caution should be exercised in evaluating whether lysosomal dysfunction is reversible; if not, as in the presence of a genetic mutation in lysosomal genes, interventions to alleviate accumulation of autophagosomes through suppression of autophagy should be developed.
Figure 7. Schematic representation of the current hypothesis.
In the hearts of RagA/B cKO mice, a mouse model of LSD, YAP is stabilized in the CM nucleus, interacts with TFEB, and stimulates autophagy. Since the function of lysosomes is severely and irreversibly impaired in this model of LSD, stimulation of autophagy is detrimental. In CMs with RagA/B downregulation, suppression of autophagy is protective, whereas activation of autophagy is detrimental. Together with our recent results showing that hyperactivation of YAP promotes dedifferentiation of CMs (17), we propose that YAP promotes heart failure in LSD.
Critical involvement of YAP in LSD.
We here show that YAP plays an essential role in mediating cardiac hypertrophy and LV dysfunction in RagA/B cKO mice. We have shown previously that persistent activation of YAP due to genetic deletion of WW45 or high-fat diet consumption promotes dedifferentiation of CMs, thereby exacerbating cardiac dysfunction, in the presence of pressure overload (17, 18). Although we observed that activation of YAP in RagA/B cKO mice is accompanied by CM dedifferentiation, including upregulation of phospho-histone H3–, β-MHC–, and α-SM actin–positive myocytes, given that systolic LV pressure was not elevated, it is unlikely that myocyte dedifferentiation alone is responsible for the drastic cardiac phenotype, including systolic dysfunction, observed in RagA/B cKO mice. TEAD, upregulation of which promotes YAP-induced CM dedifferentiation during pressure overload, is not significantly upregulated in RagA/B cKO mice. On the other hand, accumulation of autophagosomes was observed in RagA/B cKO mouse hearts, and interventions to increase or decrease autophagosomes exacerbated or alleviated, respectively, the cardiac phenotype in RagA/B cKO mice. We found that YAP promotes autophagy, particularly in the presence of lysosome inhibition, through binding to and stimulation of TFEB. Thus, we propose that YAP promotes cardiac hypertrophy and LV dysfunction by stimulating accumulation of autophagosomes in RagA/B cKO mice. Many forms of LSD are accompanied by cardiac hypertrophy. Since YAP is degraded through autophagy and autophagy is commonly inhibited in LSD, YAP may be a common driver of cardiac hypertrophy in LSD.
Autophagy promotes cell death in RagA/B cKO mice.
Increasing lines of evidence suggest that autophagy induces multiple forms of cell death (35). Although the molecular mechanism through which excessive autophagy induces cell death is not fully understood, autophagic cell death is often not mediated through massive degradation in lysosomes. Excessive autophagy can kill cells even in the complete absence of lysosomal degradation, and inhibition of autophagosome accumulation plays a cytoprotective role through inhibition of apoptosis (27, 28). Since cellular materials used for the generation of autophagosomes cannot be recycled when lysosomal degradation does not take place, cellular sources of autophagosomes, including both ER and mitochondrial membranes, are depleted, thereby inducing cellular dysfunction. We have shown recently that myocardial ischemia/reperfusion induces massive accumulation of autophagosomes and autosis, a unique form of autophagic cell death characterized by the presence of perinuclear space, in CMs (30). Although we have conducted an extensive search for the morphological features of autosis in CMs using electron microscopy (36), thus far we have only been able to identify accumulation of autophagosomes, rather than a complete signature of autotic cell death, in RagA/B cKO mice. We speculate that more severe indexes of autosis, including perinuclear space, may be less obvious in RagA/B cKO mice than in the ischemia/reperfusion model due to the chronic development of cardiomyopathy (30). We here show that CM death induced by downregulation of RagA/B is autophagy dependent, but occurs through apoptosis, necrosis, or necroptosis. Further investigation is needed to clarify how accumulation of autophagosomes leads to CM death in RagA/B cKO mice.
YAP promotes autophagosome formation through TFEB without restoring lysosomal pH in RagA/B cKO mice.
YAP is found within the autophagic cargo, and it accumulates when autophagy is inhibited in MEF cells, such as in the presence of CQ (22). Consistently, we found that YAP is accumulated when autophagy is inhibited in CMs. Since YAP interacts with p62/SQSTM1, a protein that interacts with LC3 through a short hydrophobic LC3-interacting region (37), it is likely that YAP is targeted for degradation through selective autophagy. Since YAP promotes autophagosome formation under our experimental conditions, accumulation of YAP may be part of a feedback mechanism intended to alleviate the block of autophagy. We show that YAP physically interacts with TFEB in the nucleus in RagA/B cKO hearts and positively regulates the transcriptional activity of TFEB, which would stimulate transcription of genes involved in autophagy and lysosome biogenesis (38). We have shown previously that TFEB is activated in RagA/B mKO mice (12). Here, we show that knockdown of RagA/B in cultured CMs induces nuclear translocation of TFEB in a cell-autonomous manner. Since TFEB is anchored at lysosomes through RagA/B under nutrient-rich conditions, it can be released and translocated to the nucleus in the absence of RagA/B (39). This allows YAP/TFEB interaction to stimulate the activity of TFEB. Importantly, however, YAP failed to normalize the elevated lysosomal pH caused by downregulation of RagA/B. This, then, creates a condition in which autophagosomes accumulate in the absence of degradation by lysosomes. In fact, TAT–beclin 1, a peptide that efficiently promotes autophagosome formation by mobilizing endogenous beclin 1 (31), exacerbated cardiomyopathy in RagA/B cKO mice, mimicking the effect of YAP. Since the cell death–promoting effect of RagA/B downregulation in cultured CMs was alleviated in the presence of TFEB knock-down, YAP-mediated stimulation of TFEB appears to make a substantial contribution to the overall effect of RagA/B downregulation in CMs.
A therapeutic approach for hypertrophic cardiomyopathy caused by LSD.
Recent studies have suggested that activation of TFEB is a viable therapeutic option for LSDs, increasing autophagosome processing and consequently improving protein and organelle quality-control mechanisms (32, 40). The fact that TFEB is a master regulator of lysosomal biogenesis provides this approach with a strong rationale. However, although stimulation of TFEB may improve lysosomal biogenesis, lysosomal dysfunction caused by genetic mutations, such as those found in LSD, may not necessarily be reversible, and attempts to promote autophagosome formation without concomitant upregulation of lysosomal function may induce excessive accumulation of autophagosomes. In fact, a loss of RagA/B function caused by downregulation of the tumor suppressor folliculin in mice induces dysregulated activation of TFE3, thereby leading to excessive glucogenesis and LSD-like phagocyte activation (41). Thus, caution should be exercised when autophagy is targeted, and attempts to normalize the balance between autophagosome formation and degradation should be considered. We here propose that interrupting YAP function as a cofactor of TFEB is an effective modality for preventing excessive accumulation of autophagosomes in a subset of LSDs.
Methods
Mouse models.
RagA/B floxed mice were previously described (12). To generate RagA/B cKO mice, we crossed transgenic mice harboring the floxed RagA/B allele with Myh6-Cre recombinase transgenic mice. Yap cKO and atg7 cKO mice have been described previously (15, 42). All genetically altered mice were generated on a C57BL/6 background.
Human heart samples.
Myocardial biopsy specimens were obtained from cardiomyopathy patients at Tohoku University Hospital. Diagnosis of Fabry disease was determined by α-galactosidase A mutation analysis or abnormally low activity of α-galactosidase A. Biopsy samples were fixed with 10% paraformaldehyde in phosphate-buffered saline (pH 7.4), paraffin embedded, and sectioned. Immunostaining was conducted using biopsy specimens from patients with and without Fabry disease. Control specimens were obtained from patients without Fabry disease, but with cardiomyopathy, such as dilated cardiomyopathy or ischemic cardiomyopathy, who were matched by age, sex, and body mass index (Supplemental Table 1).
Primary culture of neonatal rat CMs.
Primary cultures of CMs were prepared from 1-day-old Charles River Laboratories (Crl)/Wistar Institute (WI) BR-Wistar rats (Envigo as described previously) (43). A CM-rich fraction was obtained by centrifugation through a discontinuous Percoll gradient.
Echocardiography.
Echocardiography was performed as described previously with a 13 MHz linear ultrasound transducer (17). Two-dimensional guided M-mode measurements of LV internal diameter were obtained from at least 3 beats and then averaged. LVEDD was measured at the time of the apparent maximal LV diastolic dimension, and LV end-systolic dimension (LVESD) was measured at the time of the most anterior systolic excursion of the posterior wall.
Real-time quantitative PCR.
Total RNA was extracted from mouse hearts using the RNeasy Plus Universal Kit (QIAGEN). Total RNA was converted to cDNA using PrimeScript RT Master Mix (Takara). Quantitative real-time PCR was performed using the CFX 96 Real-Time PCR Detection System (Bio-Rad). The Ct value determined by CFX Manager Software (version 2.0, Bio-Rad) for all samples was normalized to housekeeping gene Gapdh, and the relative fold change was computed by the comparative Ct (ΔΔCt) method.
Histological analysis.
The heart was isolated, fixed with 4% phosphate-buffered paraformaldehyde, embedded in paraffin, and cut into 10 μm thick sections. Serial sections of the heart were stained with wheat germ agglutinin (WGA) for analysis of CM CSA and Picrosirius red for analysis of the myocardial fibrosis area. CSA was obtained by tracing the outlines of 100–200 CMs with a clear image of the nucleus from the LV per experiment. These analyses were performed by fluorescence microscopy (Eclipse E800, Nikon) and ImageJ Software (NIH).
Immunohistochemistry.
Mouse hearts were immersed in 4% phosphate-buffered paraformaldehyde for 24 hours at 4°C and then immersed in sucrose solution. Also, heart samples were embedded in OCT (Tissue-Tek; Miles Inc). Finally, snap-frozen cross sections (10 μm) were prepared. Neonatal CMs were cultured on coverslips and fixed in 4% phosphate-buffered paraformaldehyde. For immunofluorescence staining, an overnight incubation with specific antibodies was followed by 2 hours of incubation with secondary antibody conjugated with Alexa Fluor 488, 555, 594, or 647 dye (Life Technologies). Samples were washed and mounted on glass slides with a reagent containing DAPI (VECTASHIELD; Vector Laboratories). Cells were observed under a fluorescence microscope (Eclipse E800, Nikon).
PLAs.
Neonatal CMs treated with shRNA were fixed, permeabilized, and blocked as previously described (44). Incubation with primary antibodies (rabbit TFEB [Proteintech, catalog 13372-1-AP] and mouse YAP [Cell Signaling Technology, catalog 12395] in blocking solution) was performed at room temperature for 2 hours. Cells were washed 3 times for 5 minutes in PBS plus 0.1% Tween 20. The protein-protein interaction between TFEB and YAP was detected with secondary proximity probes (Rabbit-PLUS and Mouse-MINUS) (Olink Biosciences AB). The secondary proximity probes were incubated for 1 hour at 37°C. Cells were washed once for 5 minutes in 10 mM Tris-HCl (pH 7.5) plus 0.1% Tween 20 at 37°C, then twice for 5 minutes in PBS plus 0.1% Tween 20. All subsequent steps were performed according to the Duolink Proximity Ligation Assay Detection Kit protocol (Olink Biosciences AB).
Evaluation of apoptosis.
DNA fragmentation was detected in cardiac myocytes using TUNEL as described previously (45). Nuclear density was determined by counting DAPI-stained nuclei in 20 different fields for each sample.
Evaluation of autophagy.
Autophagy activity was evaluated by measuring LC3 conversion using Western blotting or by counting LC3 puncta using an mRFP-GFP-LC3 (Tf-LC3, where Tf indicates tandem fluorescence) assay as described previously (46). In the mRFP-GFP-LC3 assay, the probe expresses both GFP and mRFP-LC3. Low pH, such as inside the lysosome, quenches the fluorescent signal of GFP, but mRFP exhibits more stable fluorescence in the acidic lysosome. Therefore, autophagosomes and autolysosomes are labeled with mRFP/GFP double-positive (yellow) and mRFP single-positive (red) signals, respectively. CMs cultured on coverslips were transduced with Ad-Tf-LC3 for 48 hours, washed with PBS, and fixed with 4% paraformaldehyde in PBS. Fixed cells were mounted with DAPI-Containing Mounting Solution (VECTASHIELD, Vector Laboratories).
Detection of mitophagy.
Mitophagy in neonatal CMs was detected with the Mitophagy Detection Kit (MD01-10, Dojindo Molecular Technologies) according to the manufacturer’s instructions. Briefly, neonatal CMs were treated with 100 nM MtPhagy Dye in serum-free DMEM for 30 minutes at 37°C. After washing, CMs were incubated with CM culture media or treated with 5 μM of TAT–beclin 1 for 2 hours. Then, CMs were washed with serum-free DMEM and incubated with 1 μM Lyso Dye for 30 minutes at 37°C. Live CMs were observed by Nikon A1RSI confocal microscopy.
Immunoblots, antibodies, and reagents.
The following commercial primary antibodies were used: YAP (Cell Signaling Technology, catalog 14074 and catalog 12395), cathepsin D (Santa Cruz Biotechnology Inc., catalog sc377299), LC3 (MBL, catalog M186-3), p62/SQSTM1 (ORIGENE, catalog TA307334), Atg5 (Cell Signaling Technology, catalog 12994), Atg7 (Cell Signaling Technology, catalog 8558), RagA (Cell Signaling Technology, catalog 4357), RagB (Proteintech, catalog 13023-1-AP), GAPDH (Cell Signaling Technology, catalog 2118S), Histone H3 (Cell Signaling Technology, catalog 9717), cleaved caspase-3 (Cell Signaling Technology, catalog 9664), MYH7 (MilliporeSigma, catalog M8421), ACTA2 (MilliporeSigma, catalog A5228), TFEB (Proteintech, catalog 13372-1-AP), HMGB1 (abcam, ab18256), Mst1 (BD Transduction Laboratories, catalog 611052), phospho-MST1 (Thr183)/MST2 (Thr180) (Cell Signaling, catalog 3681), and α-tubulin (MilliporeSigma, catalog T6199). See complete unedited blots in the supplemental material.
Nuclear complex coimmunoprecipitation.
Nuclear complex coimmunoprecipitation was performed using the Nuclear Complex Co-IP Kit (Active motif, catalog 54001) according to the manufacturer’s instructions.
Transfection and luciferase assays.
Luciferase assays were performed as described previously (21). Briefly, cultured CMs were transfected using FuGENE 6 transfection reagent (Roche) according to the manufacturer’s instructions. HEXA luciferase reporter genes were gifts from Lorena Urbanelli (University of Perugia, Perugia, Italy) (33). Forty-eight hours after transfection, cells were lysed with Passive Lysis Buffer (Promega), and the transcriptional activity was measured using the luciferase assay system (Promega) with an OPTOCOMP I luminometer (MGM instruments).
RNA interference.
For siRNA interference, cells were transfected with Lipofectamine RNAi MAX (Thermo Fisher Scientific) and cultured for 60 hours before analysis. siRNA oligos were purchased from the Dharmacon ON-TARGETplus SMARTpool.
ChIP assays.
ChIP assay was performed using the SimpleChIP Plus Enzymatic Chromatin IP Kit (Cell Signaling Technology, catalog 9005) according to the manufacturer’s instructions. Briefly, cultured cardiac myocytes were fixed using 1% formaldehyde for 10 minutes. Cells were then washed with 1× PBS, and glycine was added to stop the crosslinking reaction. Then, cells were scraped, nuclei were isolated and lysed, and sheared chromatin was isolated after sonication. Immunoprecipitation reactions were carried out with chromatin extracts overnight at 4°C. ChIP samples were analyzed by PCR.
Cell viability assays.
The cell viability assay was performed as described previously (45, 47). Briefly, CM viability was measured by CellTiter-Blue assay (Promega) according to the manufacturer’s protocol.
Lysosomal pH measurement.
Cells grown on 6-well plates were incubated with 30 μg/ml Oregon Green 514–conjugated dextran overnight. The following day, cells were incubated in serum-free medium for 1 hour and then collected by trypsinization. Cells were resuspended in 200 μl of HBSS. The fluorescence of samples was measured at 530 nm by excitation at 440 nm and 490 nm at 30-second intervals for 10 minutes using a 96-well plate reader (Infinite M200, TECAN). The average of collected values was interpolated to a pH calibration curve that was plotted using control cell aliquots in nigericin-containing (10 μg/ml) buffered isotonic solutions (pH 3–7).
Verteporfin treatment.
Verteporfin was purchased from MilliporeSigma (SML0534) and dissolved in DMSO (100 mg/ml). A working solution was prepared at 10 mg/ml in PBS before use. Mice at 8 weeks of age were administered verteporfin or vehicle solution i.p. at a dose of 100 mg/kg every other day for 10 days (22). All mice were sacrificed and analyzed at 12 weeks old.
Statistics.
Data are presented either as bar graphs showing mean ± SEM or as box plots, showing the median (center line) and interquartile range (IQR). Whiskers represent minima and maxima within 1.5 IQR, as indicated. Sample sizes were determined on the basis of our previous studies (17) or using standard power analysis (statistical power ≥ 0.8 and α < 0.05). Indicated sample size (in figure legends) always refers to biological replicates (independent animals). No data were excluded from statistical analyses. Unless otherwise stated, statistical testing was performed using statistical analysis software (Excel Tokei 2015, Social Survey Research Information Co.). Student’s t test (paired or unpaired, as appropriate, 2 tailed) and 1-way ANOVA followed by Tukey’s honest significant difference tests were used for comparisons between 2 or multiple groups, respectively. Survival curves were analyzed by the Kaplan-Meier log-rank (Mantel-Cox) test. P < 0.05 was considered to be statistically significant.
Study approval.
All experiments involving animals were approved by the Rutgers New Jersey Medical School’s Institutional Animal Care and Use Committee. The study with human heart samples was approved by the Ethics Committee of Tohoku University Graduate School of Medicine. All patients provided written consent for the use of their heart tissues for research.
Author contributions
SI and JS conceived the project. SI and PZ designed the experiments. SI, JN, AS, SO, and SS conducted experiments. KLG and HS provided resources. SI and JS wrote the original draft. JN and JS reviewed and edited the manuscript. HS and JS supervised the project. JS acquired funding. The order of the co–first authors was determined alphabetically.
Supplementary Material
Acknowledgments
We thank Lorena Urbanelli (University of Perugia, Perugia Italy) for gifts of HEXA luciferase reporter genes. We thank Daniela Zablocki (Rutgers New Jersey Medical School) for assistance with the manuscript. This work was supported in part by US Public Health Service grants HL67724, HL91469, HL102738, HL112330, HL138720, HL144626, HL150881, and AG23039 (to JS), and by the Fondation Leducq Transatlantic Network of Excellence 15CBD04 (to JS). SI has been supported by a postdoctoral fellowship from the Japan Heart Foundation/Bayer Yakuhin Research Grant Abroad and Grants-in-Aid for Scientific Research (18K15876). JN has been supported by a postdoctoral fellowship from the American Heart Association (18POST34050036) and a merit award from the American Heart Association (20 Merit 35120374).
Version 1. 12/29/2020
In-Press Preview
Version 2. 03/01/2021
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2021, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2021;131(5):e143173.https://doi.org/10.1172/JCI143173.
See the related Commentary at YAP/TFEB pathway promotes autophagic cell death and hypertrophic cardiomyopathy in lysosomal storage diseases.
Contributor Information
Shohei Ikeda, Email: shouhei-ikeda@cardio.med.tohoku.ac.jp.
Jihoon Nah, Email: gn111@njms.rutgers.edu.
Akihiro Shirakabe, Email: s6042@nms.ac.jp.
Peiyong Zhai, Email: zhaipe@njms.rutgers.edu.
Shin-ichi Oka, Email: okash@njms.rutgers.edu.
Sebastiano Sciarretta, Email: sebastiano.sciarretta@uniroma1.it.
Kun-Liang Guan, Email: kuguan@ucsd.edu.
Hiroaki Shimokawa, Email: shimo@cardio.med.tohoku.ac.jp.
Junichi Sadoshima, Email: sadoshju@njms.rutgers.edu.
References
- 1.Ciechanover A. Proteolysis: from the lysosome to ubiquitin and the proteasome. Nat Rev Mol Cell Biol. 2005;6(1):79–87. doi: 10.1038/nrm1552. [DOI] [PubMed] [Google Scholar]
- 2.Mindell JA. Lysosomal acidification mechanisms. Annu Rev Physiol. 2012;74(1):69–86. doi: 10.1146/annurev-physiol-012110-142317. [DOI] [PubMed] [Google Scholar]
- 3.Futerman AH, van Meer G. The cell biology of lysosomal storage disorders. Nat Rev Mol Cell Biol. 2004;5(7):554–565. doi: 10.1038/nrm1423. [DOI] [PubMed] [Google Scholar]
- 4.Shirakabe A, et al. Aging and autophagy in the heart. Circ Res. 2016;118(10):1563–1576. doi: 10.1161/CIRCRESAHA.116.307474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Trivedi PC, et al. Glucolipotoxicity diminishes cardiomyocyte TFEB and inhibits lysosomal autophagy during obesity and diabetes. Biochim Biophys Acta. 2016;1861(12 Pt A):1893–1910. doi: 10.1016/j.bbalip.2016.09.004. [DOI] [PubMed] [Google Scholar]
- 6.Kushwaha SS, et al. Restrictive cardiomyopathy. N Engl J Med. 1997;336(4):267–276. doi: 10.1056/NEJM199701233360407. [DOI] [PubMed] [Google Scholar]
- 7.Arad M, et al. Glycogen storage diseases presenting as hypertrophic cardiomyopathy. N Engl J Med. 2005;352(4):362–372. doi: 10.1056/NEJMoa033349. [DOI] [PubMed] [Google Scholar]
- 8.Shen K, et al. The rag-ragulator complex regulates lysosome function and phagocytic flux in microglia. Cell Rep. 2016;14(3):547–559. doi: 10.1016/j.celrep.2015.12.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sancak Y, et al. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141(2):290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sengupta S, et al. Regulation of the mTOR complex 1 pathway by nutrients, growth factors, and stress. Mol Cell. 2010;40(2):310–322. doi: 10.1016/j.molcel.2010.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sancak Y, et al. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320(5882):1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim YC, et al. Rag GTPases are cardioprotective by regulating lysosomal function. Nat Commun. 2014;5:4241. doi: 10.1038/ncomms5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma S, et al. The Hippo pathway: biology and pathophysiology. Annu Rev Biochem. 2019;88:577–604. doi: 10.1146/annurev-biochem-013118-111829. [DOI] [PubMed] [Google Scholar]
- 14.Wackerhage H, et al. The Hippo signal transduction network in skeletal and cardiac muscle. Sci Signal. 2014;7(337):re4. doi: 10.1126/scisignal.2005096. [DOI] [PubMed] [Google Scholar]
- 15.Del Re DP, et al. Yes-associated protein isoform 1 (Yap1) promotes cardiomyocyte survival and growth to protect against myocardial ischemic injury. J Biol Chem. 2013;288(6):3977–3988. doi: 10.1074/jbc.M112.436311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang J, et al. The Hippo pathway in the heart: pivotal roles in development, disease, and regeneration. Nat Rev Cardiol. 2018;15(11):672–684. doi: 10.1038/s41569-018-0063-3. [DOI] [PubMed] [Google Scholar]
- 17.Ikeda S, et al. Hippo deficiency leads to cardiac dysfunction accompanied by cardiomyocyte dedifferentiation during pressure overload. Circ Res. 2019;124(2):292–305. doi: 10.1161/CIRCRESAHA.118.314048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ikeda S, et al. Yes-associated protein (YAP) facilitates pressure overload-induced dysfunction in the diabetic heart. JACC Basic Transl Sci. 2019;4(5):611–622. doi: 10.1016/j.jacbts.2019.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raucci A, et al. HMGB1: a signal of necrosis. Autoimmunity. 2007;40(4):285–289. doi: 10.1080/08916930701356978. [DOI] [PubMed] [Google Scholar]
- 20.Byun J, et al. Yes-associated protein (YAP) mediates adaptive cardiac hypertrophy in response to pressure overload. J Biol Chem. 2019;294(10):3603–3617. doi: 10.1074/jbc.RA118.006123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang Y, et al. miR-206 Mediates YAP-induced cardiac hypertrophy and survival. Circ Res. 2015;117(10):891–904. doi: 10.1161/CIRCRESAHA.115.306624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liang N, et al. Regulation of YAP by mTOR and autophagy reveals a therapeutic target of tuberous sclerosis complex. J Exp Med. 2014;211(11):2249–2263. doi: 10.1084/jem.20140341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao B, et al. The Hippo-YAP pathway in organ size control and tumorigenesis: an updated version. Genes Dev. 2010;24(9):862–874. doi: 10.1101/gad.1909210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu-Chittenden Y, et al. Genetic and pharmacological disruption of the TEAD-YAP complex suppresses the oncogenic activity of YAP. Genes Dev. 2012;26(12):1300–5. doi: 10.1101/gad.192856.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iwashita H, et al. Live cell imaging of mitochondrial autophagy with a novel fluorescent small molecule. ACS Chem Biol. 2017;12(10):2546–2551. doi: 10.1021/acschembio.7b00647. [DOI] [PubMed] [Google Scholar]
- 26.Shirakabe A, et al. Drp1-dependent mitochondrial autophagy plays a protective role against pressure overload-induced mitochondrial dysfunction and heart failure. Circulation. 2016;133(13):1249–1263. doi: 10.1161/CIRCULATIONAHA.115.020502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walls KC, et al. Lysosome dysfunction triggers Atg7-dependent neural apoptosis. J Biol Chem. 2010;285(14):10497–10507. doi: 10.1074/jbc.M110.103747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kessel DH, et al. ATG7 deficiency suppresses apoptosis and cell death induced by lysosomal photodamage. Autophagy. 2012;8(9):1333–1341. doi: 10.4161/auto.20792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boya P, et al. Inhibition of macroautophagy triggers apoptosis. Mol Cell Biol. 2005;25(3):1025–1040. doi: 10.1128/MCB.25.3.1025-1040.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nah J, et al. Upregulation of Rubicon promotes autosis during myocardial ischemia/reperfusion injury. J Clin Invest. 2020;130(6):2978–2991. doi: 10.1172/JCI132366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shoji-Kawata S, et al. Identification of a candidate therapeutic autophagy-inducing peptide. Nature. 2013;494(7436):201–206. doi: 10.1038/nature11866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Settembre C, et al. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332(6036):1429–1433. doi: 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Urbanelli L, et al. Oncogenic H-Ras up-regulates acid β-hexosaminidase by a mechanism dependent on the autophagy regulator TFEB. PLoS One. 2014;9(2):e89485. doi: 10.1371/journal.pone.0089485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gatto F, et al. AAV-mediated transcription factor EB (TFEB) gene delivery ameliorates muscle pathology and function in the murine model of Pompe Disease. Sci Rep. 2017;7(1):15089. doi: 10.1038/s41598-017-15352-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bialik S, et al. Autophagy-dependent cell death — where, how and why a cell eats itself to death. J Cell Sci. 2018;131(18):jcs215152. doi: 10.1242/jcs.215152. [DOI] [PubMed] [Google Scholar]
- 36.Liu Y, et al. Autosis is a Na+,K+-ATPase-regulated form of cell death triggered by autophagy-inducing peptides, starvation, and hypoxia-ischemia. Proc Natl Acad Sci U S A. 2013;110(51):20364–20371. doi: 10.1073/pnas.1319661110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kirkin V, et al. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol Cell. 2009;33(4):505–516. doi: 10.1016/j.molcel.2009.01.020. [DOI] [PubMed] [Google Scholar]
- 38.Raben N, Puertollano R. TFEB and TFE3: linking lysosomes to cellular adaptation to stress. Annu Rev Cell Dev Biol. 2016;32:255–278. doi: 10.1146/annurev-cellbio-111315-125407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martina JA, Puertollano R. Rag GTPases mediate amino acid-dependent recruitment of TFEB and MITF to lysosomes. J Cell Biol. 2013;200(4):475–491. doi: 10.1083/jcb.201209135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spampanato C, et al. Transcription factor EB (TFEB) is a new therapeutic target for Pompe disease. EMBO Mol Med. 2013;5(5):691–706. doi: 10.1002/emmm.201202176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Endoh M, et al. A FLCN-TFE3 Feedback loop prevents excessive glycogenesis and phagocyte activation by regulating lysosome activity. Cell Rep. 2020;30(6):1823–1834. doi: 10.1016/j.celrep.2020.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saito T, et al. An alternative mitophagy pathway mediated by Rab9 protects the heart against ischemia. J Clin Invest. 2019;129(2):802–819. doi: 10.1172/JCI122035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maejima Y, et al. Mst1 inhibits autophagy by promoting Beclin1-Bcl-2 interaction Nature Med. 2013;19(11):1478–1488. doi: 10.1038/nm.3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shao D, et al. A functional interaction between Hippo-YAP signalling and FoxO1 mediates the oxidative stress response. Nat Commun. 2014;5:3315. doi: 10.1038/ncomms4315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Del Re DP, et al. Mst1 promotes cardiac myocyte apoptosis through phosphorylation and inhibition of Bcl-xL. Mol Cell. 2014;54(4):639–350. doi: 10.1016/j.molcel.2014.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hariharan N, et al. Oxidative stress stimulates autophagic flux during ischemia/reperfusion. Antioxid Redox Signal. 2011;14(11):2179–2190. doi: 10.1089/ars.2010.3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ikeda Y, et al. Endogenous Drp1 mediates mitochondrial autophagy and protects the heart against energy stress. Circ Res. 2015;116(2):264–278. doi: 10.1161/CIRCRESAHA.116.303356. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.