Abstract
Structured RNA elements within the internal ribosome entry site (IRES) of hepatitis C virus (HCV) genome hijack host cell machinery for translation initiation through a cap-independent mechanism. Here, using a phage display selection, we obtained two antibody fragments (Fabs), HCV2 and HCV3, against HCV IRES that bind the RNA with dissociation constants of 32 ± 7 nM and 37 ± 8 nM respectively, specifically recognizing the so-called junction IIIabc (JIIIabc). We used these Fabs as crystallization chaperones and determined the high-resolution crystal structures of JIIIabc – HCV2 and – HCV3 complexes at 1.81-Å and 2.75-Å resolution respectively, revealing an anti-parallel four-way junction with the so-called IIIa and IIIc sub-domains brought together through tertiary interactions. The RNA conformation observed in the structures supports the structural model for this region derived from cryo-EM data for the HCV IRES – 40S ribosome complex, suggesting that the tertiary fold of the RNA pre-organizes the domain for interactions with the 40S ribosome. Strikingly, both Fabs and the ribosomal protein eS27 not only interact with a common subset of nucleotides within the JIIIabc but also use physio-chemically similar sets of protein residues to do so, suggesting that the RNA surface is well-suited for interactions with proteins, perhaps analogous to the ‘hot spot’ concept elaborated for protein-protein interactions. Using a rabbit reticulocyte lysate-based translation assay with a bicistronic reporter construct, we further demonstrated that Fabs HCV2 and HCV3 specifically inhibit the HCV IRES-directed translation, implicating disruption of the JIIIabc – ribosome interaction as a potential therapeutic strategy against HCV.
INTRODUCTION
Translation initiation in most eukaryotes involves the interaction of a 5ꞌ-cap structure of the mRNA with the translation initiation factors (eIFs).1,2 In the canonical mechanism, the cap-binding protein eIF4E recognizes the 5ꞌ-cap and recruits the scaffolding protein eIF4G, which then binds eIF4B, the helicase eIF4A and poly(A)-binding protein. Interactions between poly(A)-binding protein and the poly(A) tail circularize the mRNA. After recruitment of the 40S ribosome and eIF3, the complex scans the mRNA to find a suitable translation start site.1,2 Numerous viral genomes, and a subset of cellular mRNAs, bypass this canonical translation initiation mechanism however, initiating translation with cap-independent mechanisms that involve cis-acting, structured RNA elements, termed internal ribosome entry sites (IRESs).2–4 An IRES recruits the ribosome directly or via interactions with translation initiation factors in a cap-independent manner and positions the ribosome at, or upstream of, the translation start site.
In hepatitis C virus (HCV) – a member of Flaviviridae family and a major human pathogen – translation initiation is driven by the direct interaction of the IRES elements in the 5ꞌ-UTR of the positive sense ssRNA viral genome with the 40S ribosome and initiation factor 3 (eIF3).5–12 The 341-nt long 5ꞌ-UTR shows well-defined secondary structure domains (designated I to IV, Fig. 1a and Supplementary Fig. 1) that are highly conserved among isolates and strains.13–18 Previous mutation and deletion analyses using monocistronic and bicistronic reporter assays show that full translation activity requires 333 nucleotides (nts 40–372) of the viral genome, which include nucleotides 40–341 of the 5ꞌ-UTR, the AUG start codon (nts A342-G344) and nucleotides 345–372 of the coding region (see Supplementary Fig. 1 for the sequence).8,13,15,19,20 Nevertheless, several biochemical and structural studies have revealed that domains II and III (nts 40–341) contain all the structural features essential for the 40S ribosome binding and translation initiation.8,13,15,19,20
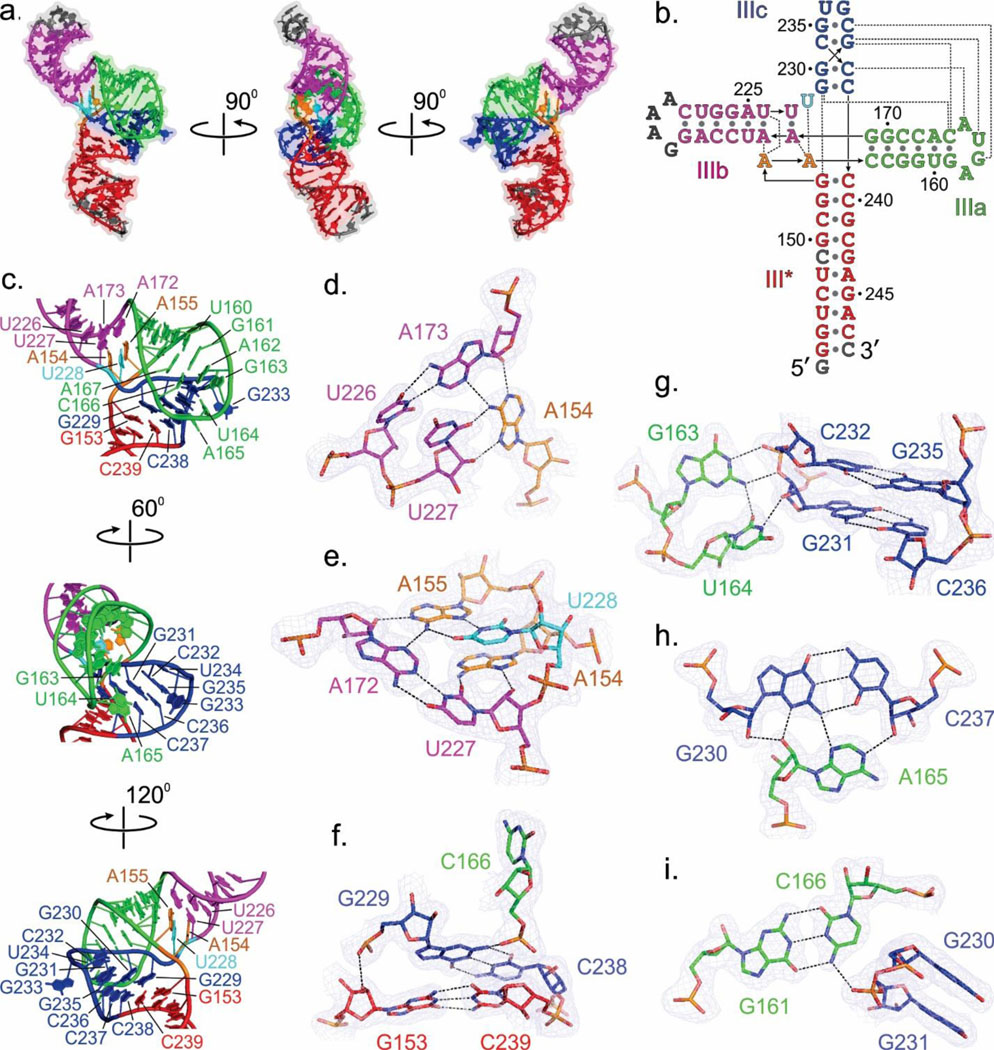
Fig. 1.
Overall structures of the HCV IRES JIIIabc in complex with Fabs HCV2 and HCV3. (a) Secondary structure of HCV IRES (genotype 1b) showing domains I – IV according to Brown et al.14 and Honda et al.16 Dotted box highlights the JIIIabc. Numbering depicts the approximate nucleotide position (b) The JIIIabc crystallization construct. Nucleotides in gray represent mutations or insertions compared to the wild-type (genotype 1b)13,15 sequence. (c) Crystal structure of the JIIIabc – HCV2 and (d) JIIIabc – HCV3 complexes solved at 1.81-Å and 2.75-Å resolution, respectively. (e) Superposition of the JIIIabc structures from JIIIabc – HCV2 and JIIIabc – HCV3 complexes. The JIIIabc structure is almost identical in both complexes; the HCV2 and HCV3 Fabs bind to the same region of the RNA with different orientations. Figures b-d and the corresponding labels are colored analogously for facile comparison.
Previous structural and functional studies, including NMR and cryo-EM structural analyses, have revealed important features of the IRES – ribosome interactions.10,21–27 Consistent with the outcomes of those studies, a recent 3.9-Å resolution cryo-EM structure of the HCV IRES-40S subunit complex (PDB code: 5A2Q25) determined by the focused refinement of the human 80S ribosome-HCV IRES complex demonstrates that domain II of the HCV IRES assumes a L-shaped structure that reaches across the 40S subunit and into the 40S-60S intersubunit space, interacting with the ribosomal proteins uS7, uS11 and uS25.25 Domain III – the largest domain within the IRES (nts 119–330) – consists of several stem-loops (designated sub-domains IIIa to IIIf) involved in three-way and four-way junctions and pseudoknot structures (Supplementary Fig. 1).22,25,28–30 The base of the domain III, including sub-domains IIIe and IIIf, forms a four-way junction and a pseudoknot involving the IIIf loop and the nucleotides 325–330.25,28 This region interacts with the proteins (eS1 and eS28) and 18S rRNA components of the 40S ribosome.25 In the middle of domain III, sub-domain IIId forms part of a three-way junction and interacts with 18S rRNA.25 The apical region of domain III contains a four-way junction (designated JIIIabc) involving the central domain III stem and sub-domains IIIa, IIIb and IIIc, and IIIa and IIIc interact with ribosomal protein eS27 (Supplementary Fig. 1).25 Although the highest resolution (3.9-Å) cryo-EM structure thus far lacks stem-loop IIIb, earlier chemical and enzymatic foot-printing, UV-cross-linking and quantitative binding assays have shown that IIIb interacts with eIF3, playing a crucial role for initiating the HCV translation.1,7,8,31,32 More recently, a modest resolution (9.3-Å) cryo-EM reconstruction of a similar IRES from classical swine flu virus (CSFV) in complex with the 40S ribosome and eIF3 revealed that sub-domain IIIb binds to eIF3, displacing it from the canonical 40S-eIF3 complex.11
Structural analysis of IRES elements both as isolated RNA domains and as higher order complex with ribosomes contributes to our understanding IRES function. High-resolution X-ray and NMR structures of many fragments of the HCV IRES have been reported previously, including domains II, IIIb, IIIc, IIId, IIIef-IV, and the IIIabc junction (JIIIabc).22,23,30,32–34 Interestingly, the JIIIabc crystal structure (2.8-Å resolution, PDB code: 1KH6)33 captured the four-way junction in a conformation that is distinct from the conformation observed in 3.9-Å resolution cryo-EM structure of the IRES – ribosome complex (Supplementary Fig. 2).25,33 The four-way junction in the former structure assumes a parallel conformation with two coplanar sets of stacked helices – IIIa stacks on IIIb and IIIc on the remaining helix (designated III*).33 The overall architecture separates IIIa and IIIc and positions domain IIIb close to the III* helix.33 In contrast, the same junction in the context of the full IRES bound to the 40S ribosomal subunit (3.9-Å resolution cryo-EM structure) adopts an antiparallel conformation, placing IIIb and III* in an opposite direction and juxtaposing IIIa and IIIc to form a binding platform for the ribosomal protein eS27.25 It is noteworthy that the crystal structure corresponds to a dimeric form of the JIIIabc construct, which contains the same junction nucleotides and flanking base pairs in the helices IIIa, IIIb and III* as the monomeric form (cryo-EM structure) but loop IIIc is engaged with the same loop of the neighboring molecule to form a helical stem (Supplementary Fig. 2).25,33 The dimerization of the JIIIabc in the crystal might have arisen due to crystallization conditions. It is possible that both parallel and antiparallel monomeric conformations may exist in solution,35 but ribosome binding stabilizes the antiparallel conformation.
We have been developing chaperone-assisted RNA crystallography using antibody-derived antigen binding fragments (Fabs) as chaperones.36–42 Fabs that bind RNA targets specifically are obtained through screening of phage libraries displaying Fabs bearing limited, prescribed sequence diversity in their complementarity determining regions (CDRs).36–42 Here, we target the HCV IRES RNA and obtain two Fabs, termed HCV2 and HCV3, that specifically bind the JIIIabc domain within the IRES. Using these Fabs as crystallization chaperones, we solved high-resolution crystal-structures of JIIIabc – HCV2 and – HCV3 complexes at 1.81-Å and 2.75-Å resolution, respectively. The overall architecture of JIIIabc crystal structure closely resembles that modeled from lower resolution (3.9-Å) cryo-EM data of the HCV IRES in complex with the human 40S ribosome,25 establishing that the JIIIabc domain folds into a structure pre-organized for the ribosome and eIF3 interactions. Despite using the entire HCV IRES (333 nts) as the target for phage display Fab selection, both Fabs bind to JIIIabc in the same region, which serves as a docking site for ribosomal protein eS27, implicating a potential ‘hot spot’ within the IRES for protein interactions – a concept well-elaborated for protein-protein interactions.43–45 We also demonstrate that both Fabs HCV2 and HCV3 effectively inhibit HCV IRES-mediated translation in a concentration dependent manner while having no effect on canonical translation. These findings demonstrate proof-of-principle that antibodies to RNA domains have potential as crystallization chaperones, as tools to identify vulnerable regions within pathogenic RNAs for targeting small molecules, and as a potential class of translation inhibitors for developing antiviral therapeutics.
RESULTS AND DISCUSSION
Selection of Fabs against HCV IRES using phage display.
The RNA construct used for phage display selection contained 333-nucleotides from the HCV IRES (nts 40 to 372, genotype 1b)16,18 with an additional 3ꞌ-overhang sequence that hybridizes to a biotinylated DNA oligonucleotide (Supplementary Fig. 3), allowing immobilization of the RNA-DNA hybrid construct on streptavidin-coated magnetic beads. We have previously selected Fabs that specifically bind to RNA using phage libraries displaying Fabs derived from a humanized Fab4D5 framework bearing reduced codon diversity in the CDRs, the hypervariable loops that endow antibody repertoires with the capacity to bind a wide variety of antigens.36,37,39,41,42 To isolate Fabs that specifically bind to HCV IRES, we performed in vitro phage display selection using a phage library termed YSGR that contains constant CDRs-L1 and -L2 from the parent Fab4D5, encodes equal proportions of Y and S at variable positions in CDR-L3, -H1, and -H2, and encodes 38% Y, 25% S, 25% G, and 12% R in each CDR-H3 position. Additionally, this library was designed to have variable length between 6 – 17 residues in CDR-H3. The prescribed restriction in amino acid types reduces the gap between the practical size of the library (~1011) and the theoretical diversity (>1050 if all CDR positions were randomized across all amino acid types). We carried out three rounds of phage display selection against the HCV IRES RNA. We obtained several clones that showed a positive RNA binding response in a phage ELISA assay. We expressed these Fabs individually as soluble proteins in E. coli using a phagemid expression vector, purified them by affinity and ion-exchange chromatography to obtain RNase free Fabs, and tested their binding affinity for the 333-nt HCV IRES using a filter binding assay. We found that two Fabs, termed HCV2 and HCV3 hereafter, bound to the HCV IRES with dissociation constants (Kds) of 32 ± 7 nM and 37 ± 8 nM, respectively, as determined by a filter binding assay in 10 mM Tris-HCl (pH 7.4), 10 mM MgCl2 and 200 mM NaCl buffer at 23°C (Supplementary Fig. 3). These two Fabs were advanced to further analysis and crystallization trials.
Crystallization and structure determination of JIIIabc in complex with HCV2 and HCV3.
To test the ability of Fabs HCV2 and HCV3 to serve as chaperones for the HCV IRES crystallization, we set up crystallization trials for the intact, 333-nt HCV IRES in complex with either Fab HCV2 or HCV3 using the hanging drop vapor diffusion method. However, we did not obtain any useful crystals. Next, we performed truncation analysis to approximate the epitope regions recognized by Fabs HCV2 and HCV3 within the HCV IRES. Binding assays for Fabs HCV2 and HCV3 with several truncated RNA constructs derived from the wild-type HCV IRES revealed that both Fabs recognized a region located around the four-way junction structure – JIIIabc. Based on truncation analysis, we prepared an isolated JIIIabc construct (Supplementary Fig. 4) similar to that crystallized previously by Kieft et al.33 in a dimeric form (Supplementary Fig. 2). This construct also bound to Fabs HCV2 and HCV3 with affinities (Kd = 28 ± 8 nM and 35 ± 11 nM, respectively, see Supplementary Fig. 4) similar to those of the full-length HCV IRES (Kd = 32 ± 7 nM and 37 ± 8 nM with HCV2 and HCV3, respectively). Additionally, isolated sub-domain constructs IIIa, IIIc and IIIb/IIIc did not bind either Fab, indicating a requirement for the intact JIIIabc tertiary structure in Fab recognition (Supplementary Fig. 5). Following these tests, we set up crystallization trials of the JIIIabc – Fab complexes and obtained crystals that diffracted to ~ 5-Å resolution with poor data quality. Given the tendency of this construct to dimerize under the crystallization conditions, we prepared a second JIIIabc RNA construct for crystallization (Fig. 1b), which was similar to the previous construct but contained extended IIIb and III* helical stems (see supplementary note 1 and Supplementary Fig. 4 for details).
After preparing the Fabs HCV2 and HCV3 complexes with the RNA, we set up the crystallization trials. We observed robust crystals for both JIIIabc – HCV2 and JIIIabc – HCV3 complexes, but more co2nditions yielded crystals for former complex compared to the latter (44 and 8 different conditions, respectively, out of 480 conditions screened for each complex). We observed no crystals in analogous trials using only the RNA, implicating a role for the Fabs in facilitating the JIIIabc crystallization. The crystals for JIIIabc – HCV2 and JIIIabc – HCV3 diffracted to 1.81-Å and 2.75-Å resolution, respectively. To solve the crystal structure of JIIIabc – HCV2 complex, we readily obtained the initial phases by molecular replacement using the Fab portion of the previous crystal structure of Fab HAVx in complex with domain V from the hepatitis A virus IRES (PDB code: 6MWN).42 The high-resolution electron density map obtained from the molecular replacement allowed unambiguous modeling of the RNA nucleotides. After iterative rounds of model building and refinement at 1.81-Å resolution, the final values of Rfree and Rwork were 20.89% and 17.35%, respectively. For the JIIIabc – HCV3 complex, the structure of the RNA and Fab HCV2 without CDR loops were used as the molecular replacement models. After several rounds of model building and refinement process at 2.75-Å resolution, the final values of Rfree and Rwork were 22.22% and 18.60%, respectively. The structural models of the JIIIabc – HCV2 and JIIIabc – HCV3 complexes from the final refinements along with the 2|Fo|− |Fc| electron density maps are shown in Supplementary Fig. 6. Details of data collection and refinement statistics for both complexes are provided in Table 1.
Table 1:
X-ray crystallography data collection and structure refinement statistics.
| Data collection | JIIIabc – HCV2 complex | JIIIabc – HCV3 complex |
|---|---|---|
| Space group | C 1 2 1 | P 43 |
| Resolution (Å) | 61.36 – 1.807 (1.872 – 1.807) | 173.29 – 2.75 (2.848 – 2.75) |
| Cell dimensions | ||
| a, b, c (Å) α, β, γ (°) |
49.705, 161.223, 96.978 90, 102.658, 90 |
173.289, 173.289, 140.49 90, 90, 90 |
| Rmerge (%) | 4.042 (113.4) | 14.75 (107.2) |
| I/σI | 15.66 (0.92) | 9.31 (1.11) |
| CC1/2 | 0.999 (0.481) | 0.965 (0.399) |
| Completeness (%) | 98.73 (94.58) | 98.54 (99.03) |
| Redundancy | 3.9 (3.7) | 4.7 (4.8) |
| Refinement | ||
| No. reflections | 67010 (6420) | 106172 (10622) |
| Rwork/Rfree (%) | 17.35/20.89 | 18.60/22.22 |
| R.M.S deviations | ||
| Bond angles (°) | 0.970 | 1.06 |
| Bond length (Å) | 0.006 | 0.008 |
| Average B-factor, all atoms (Å2) | 55.0 | 76.00 |
| Ramachandran plot of protein residues | ||
| Preferred regions (%) | 96.79 | 96.00 |
| Allowed regions (%) | 3.21 | 4.00 |
| Number of residues | ||
| RNA | 68 | 68 |
| Protein | 440 | 437 |
| Values in the parentheses are for the highest resolution shell | ||
Overall structure of the JIIIabc in complex with HCV2 and HCV3.
We observed crystals of JIIIabc – HCV2 and JIIIabc – HCV3 complexes in several overlapping crystallization conditions and most of the crystals were morphologically similar. However, the crystals that yielded the highest resolution diffraction datasets for solving the structures originated from non-overlapping crystallization conditions (see above) and had different crystal morphologies (rectangular prism and diamond shaped crystals for JIIIabc – HCV2 and JIIIabc – HCV3, respectively), underscoring the value of having multiple crystallization chaperones. Consistent with the morphological differences in the crystals, JIIIabc – HCV2 and JIIIabc – HCV3 complexes crystallized in the C 1 2 1 and P 43 space group lattices, respectively. The former contained one and the latter three RNA – Fab complexes per asymmetric unit (Supplementary Fig. 7). Nevertheless, the final structures of each Fab and RNA from the JIIIabc – HCV2 and JIIIabc – HCV3 complexes (Figs. 1c and d, respectively) were almost identical except for Fab CDRs and two RNA nucleotides (Figs. 1c–e and Supplementary Fig. 7). In the JIIIabc – HCV2 structure, the first G nucleotide remains stacked within the helical stem but in JIIIabc – HCV3, it flips out of the helix to make crystal contacts with the neighboring Fab molecule (Supplementary Fig. 7). Additionally, G233 flips out of the IIIc helical stack in both complexes but in opposite directions (Supplementary Fig. 7) to interact extensively with Fab residues form CDRs L3 and H2 in HCV2 and L3, H2 and H3 in HCV3.
In the crystal structures of both JIIIabc – HCV2 and – HCV3 complexes, the majority of the macromolecular interactions involve either Fab-Fab or Fab-RNA interaction. Analysis of buried surface area of JIIIabc – HCV2 and JIIIabc – HCV3 structures using PDBePISA46 revealed that including the Fab-RNA binding interface, Fab-mediated contacts account for 72% and 89%, respectively, of the buried surface area in the crystal packing, suggesting a prominent role of the Fabs as RNA crystallization chaperones in both cases. Nevertheless, RNA – RNA contacts occur in these crystals, accounting for 28% and 11% of the buried surface area in the crystal lattice of JIIIabc – HCV2 and JIIIabc – HCV3, respectively. In the JIIIabc – HCV2 complex, two symmetry mate RNA molecules make two sets of tetra-loop – receptor interactions47,48 involving helical stem IIIa and the IIIb GAAA tetra-loop (Supplementary Fig. 8), whereas in JIIIabc – HCV3, the RNA-RNA contacts occur between two RNA copies within the asymmetric unit via GAAA tetra-loop – tetra-loop interactions47,48 (Supplementary Fig. 8).
In the JIIIabc crystal structure (Fig. 2a), a four-way junction architecture organizes the four helices designated III*, IIIa, IIIb and IIIc into two sets of co-linearly stacked helices, in which IIIa and IIIc stack on IIIb and III*, respectively (Figs. 2a, b). The IIIc and III* helices form a nearly continuous A-form helix with no unpaired nucleotides in between, whereas nucleotides A154, A155 and U228 interrupt the continuity of the IIIa and IIIb helical stack and make tertiary interactions with the IIIa – IIIb interface (Figs. 2a, b). This arrangement of the helices within the four-way junction juxtaposes the sub-domains IIIa and IIIc to enable loop (IIIa) – helix receptor (IIIc) type tertiary interactions (Figs. 2b, c), forming a platform for Fab HCV2 or HCV3 binding (Figs. 1c–e). Recognition of this tertiary structure by the Fabs accounts for the observation that isolated IIIa and IIIc hairpins fail to bind either Fab. The secondary structures within each helix match previous predictions for the full-length IRES (see Figs. 1b, 2b and Supplementary Fig. 1). Our crystal structure of JIIIabc supports the model of the HCV IRES – ribosome complex derived from 3.9-Å resolution cryo-EM data (Supplementary Fig. 9). The similarity of the RNA structures in all three complexes, the 40S ribosome, Fab HCV2, or Fab HCV3, strongly suggests that the JIIIabc conformation is pre-organized for binding the ribosome and eIF3. If the RNA lacked pre-organization and sampled multiple conformations dynamically, then eS27, HCV2, and HCV3 would each have to bind and stabilize the same RNA conformation, which seems unlikely considering that the three proteins have overlapping but not identical binding epitopes.
Fig. 2.
Structural features of the JIIIabc four-way junction. (a) Overall architecture of the JIIIabc based on the crystal structure of JIIIabc – HCV2 complex. (b) Secondary structure of the JIIIabc derived from the crystal structure. The dotted lines represent the tertiary interactions. (c) Structural features of the JIIIabc four-way junction. (d-i) Specific interactions between the nucleotides organizing the four-way junction. The dashed lines reflect heteroatoms within hydrogen bonding distance. Blue mesh represents the 2|Fo| - |Fc| electron density map at 1σ contour level and carve radius 1.8 Å. All figures and the labels are colored analogously for facile comparison.
Structural features of the four-way junction in JIIIabc.
Here, we elaborate on the detailed structural features of the four-way junction and provide a comprehensive description of the JIIIabc architecture based on our two high-resolution crystal structures (Figs. 2b–i). In the III* – IIIb joining region, unpaired nucleotides A154 and A155 engage in tertiary interactions with the A172-U227 and A173-U226 base pairs of IIIb as well as with unpaired U228 at the IIIb – IIIc junction (Figs. 2b–e). A155 makes Watson-Crick/Sugar Edge and Hoogsteen/Watson-Crick interactions with A172 and U228, respectively, forming a U228•A155•A172-U227 tetrad (Fig. 2e). Similarly, A154 makes Watson-Crick/Sugar Edge and Hoogsteen/Sugar Edge interactions with A173 and U227, respectively, thereby organizing a second tetrad U227•A154•A173-U226 (Fig. 2d). Consistent with the important structural role of these interactions for the integrity of JIIIabc and IRES-mediated translation, mutation of any nucleotide associated with these networks of interactions (A154G, A155G, U248C, A172U, A173U) significantly lowered IRES-mediated translation of a downstream reporter gene by disrupting the ribosome and eIF3 interactions with JIIIabc.8,33 Compensatory base-pair mutations A172-U227 to U172-A227 or A173-U226 to U173-A226 retained nearly wild-type IRES activity.33
The junction architecture intertwines the IIIa and IIIc stem loops (Figs. 2a, c). The 3’ strand of helix IIIc traverses the major groove of helix IIIa but makes no direct contact, and the terminal loop of helix IIIa docks into the minor groove of helix IIIc. This docking involves tertiary interactions between four nucleotides (G163, U164, A165 and C166) from helix IIIa and four base pairs (G229-C238, G230-C237, G231-C236 and C232-G235) from the IIIc helical stem (Figs. 2b, c). The phosphate oxygen of G229 interacts with the 2ꞌ-OH of G153 of III* (Fig. 2f). The G163’s N1H and exocyclic amine make hydrogen bonding interactions with C232’s phosphate oxygen and G231’s O3ꞌ, respectively (Fig. 2g). Additionally, U164 N3 and O2 make hydrogen bonds to G231’s 2ꞌ-OH and G163’s exocyclic amine (Fig. 2g). A165 engages in A-minor interactions with the G230-C237 base pair (Fig. 2h). Finally, C166 engages in a canonical base pair with G161 and uses its exocyclic amine and phosphate oxygen to form hydrogen bonds to the phosphate backbone at G231 and the exocyclic amine of G229, respectively (Figs. 2f, i). These interactions stabilize the juxtaposition of IIIa and IIIc to make a well-organized platform for Fabs HCV2 and HCV3 binding.
Structural features of the Fab – RNA binding interface.
As expected, Fabs HCV2 and HCV3 interact with JIIIabc through their CDRs. In the JIIIabc – HCV2 complex, CDRs L3 and H2 interact with sub-domain IIIc and CDR H3 forms a topologically smooth surface that mediates interactions with sub-domain IIIa (Fig. 3a). In the JIIIabc – HCV3 complex, CDRs L3, H1, H2 and H3 form a deep pocket that accommodates the loop of sub-domain IIIc. In addition, the loop of the sub-domain IIIa wraps around CDR H3 and makes contact with the surfaces of CDRs L2 and H1 located on opposite sides of CDR H3 (Fig. 3b).
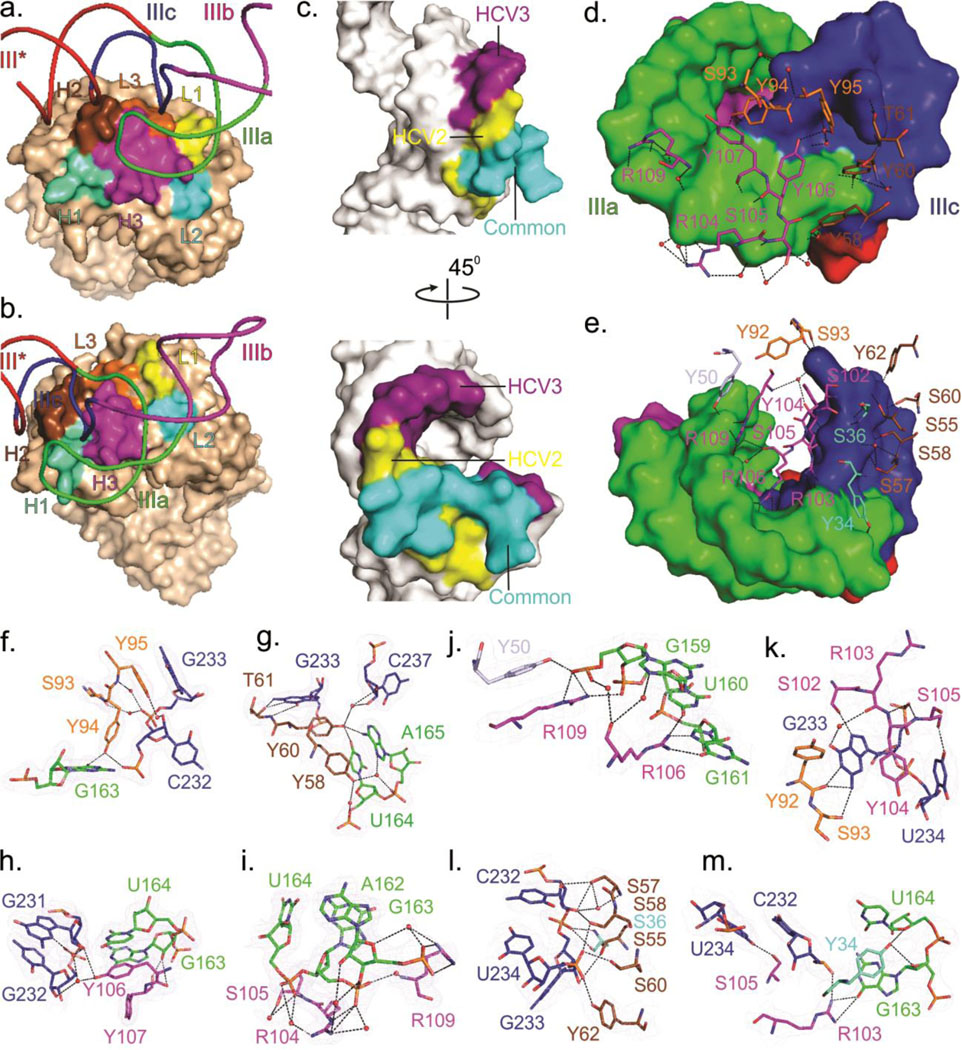
Fig. 3.
Structural features of the Fab-RNA binding interfaces. (a) Overall view of the binding interface of JIIIabc bound to Fab HCV2 or (b) Fab HCV3. (c) RNA surface within the binding interface, highlighting the common and specific regions recognized by Fabs HCV2 and HCV3. (d) Surface of the RNA interface and the Fab residues involved in binding interactions for JIIIabc – HCV2 and (e) for JIIIabc – HCV3 complexes. (f-m) Interactions between the RNA and the Fab CDR residues for JIIIabc – HCV2 (f-i) and JIIIabc – HCV3 complexes (j-m). Orange-red spheres represent water molecules and the dashed lines reflect heteroatoms within hydrogen bonding distance. Blue mesh represents the 2|Fo| - |Fc| electron density map at 1σ contour level and carve radius 1.8 Å. All figures and the labels are colored analogously for facile comparison.
Although they bind in different global orientations (Fig. 1e), Fabs HCV2 and HCV3 recognize a significantly overlapping surface of the RNA (Fig. 3c). Recognition involves stacking, electrostatic, and hydrogen bonding (direct and water-mediated) interactions (Figs. 3f–m, see Supplementary Note 2 for detailed description of the interactions). Fab HCV2 interacts with the four unpaired nucleotides (A162, G163, U164 and A165) from the IIIa loop and an unpaired nucleotide (G233) and three base-paired nucleotides (G231, C232 and C237) from IIIc (Supplementary Figs. 10, 11). Fab HCV3-RNA interactions involve two unpaired nucleotides (G163, U164) and three base-paired nucleotides (G159, U160 and G161) from IIIa, and two unpaired nucleotides (G233 and U234) and a base-paired nucleotide (C232) from IIIc (Supplementary Figs. 10, 11). Fab HCV2 utilizes six tyrosines, two serines, two arginines and a threonine from CDRs L3, H2 and H3 only (Fig. 3d), whereas Fab HCV3 utilizes five tyrosines, eight serines and three arginines from all CDRs except L1 (Fig. 3e).
Fabs HCV2 and HCV3 mimic ribosomal protein eS27 in binding JIIIabc.
In the 3.9-Å resolution cryo-EM structure,25 the ribosomal protein eS27 interacts directly with IIIa and IIIc sub-domains within JIIIabc via contacts to many of the same nucleotides that the Fabs HCV2 and HCV3 interact with (Figs. 4a–j). Specifically, two lysines (K42 and K82), an arginine (R80), a serine (S78) and an aspartic acid (D34) of eS27 interact with the sub-domain IIIc nucleotides G233, U234, C236 and C237 (Figs. 4e–h), whereas a tyrosine (Y41) and a lysine (K36) residue of the protein interact with the IIIa sub-domain nucleotides G163 and U164 (Figs. 4e, h). Thus, not only do all three proteins interact with the same five nucleotides (IIIa G163 and U164, and IIIc G233, U234 and C237) but use physiochemically congruous sets of protein residues to do so (see Figs. 3d–m, 4e–j, and Supplementary Figs. 10, 11). The emergence of two Fabs that bind to the same region within the JIIIabc as eS27 binds, despite phage display screening against the entire HCV IRES (333 nts), may suggest a tendency of Fabs to target functional RNA surfaces involved in protein interactions. On the other hand, this outcome may reflect the restricted amino acid types (YSGR) used for generating CDR diversity.
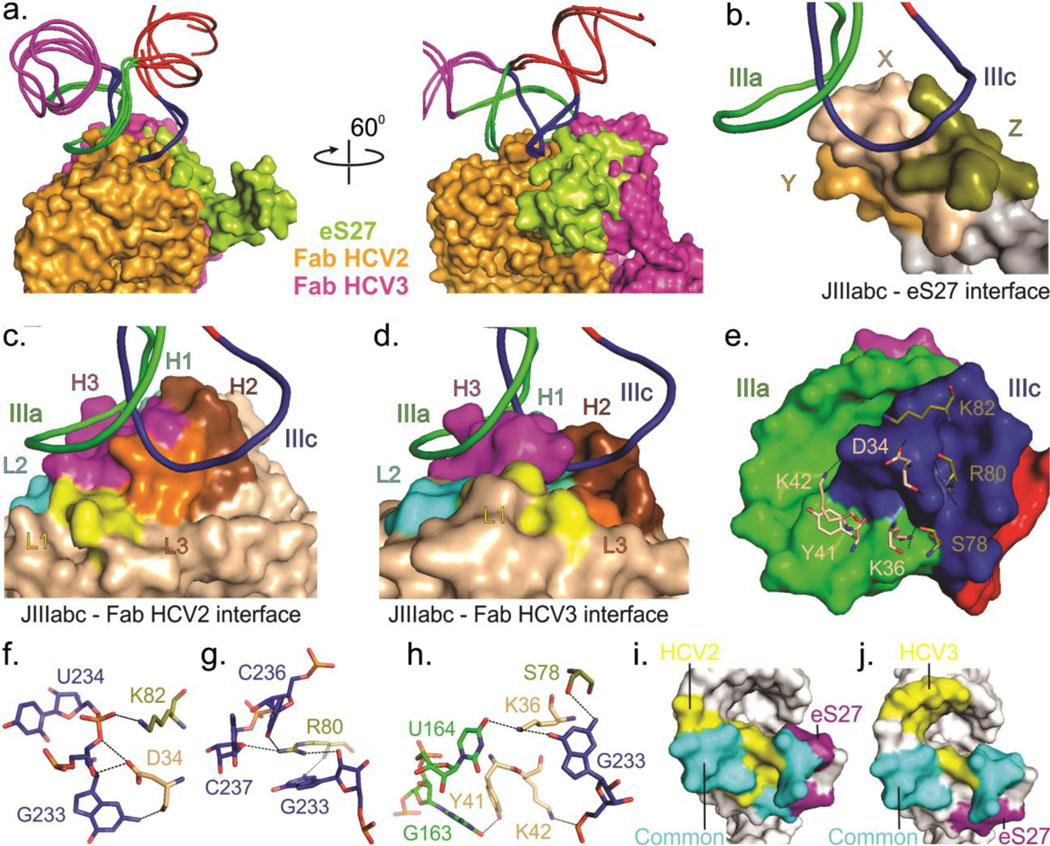
Fig. 4.
Comparison of binding interfaces of JIIIabc – HCV2, JIIIabc – HCV3 and JIIIabc – eS27 complexes. (a) Superposition of the JIIIabc portion of the JIIIabc – HCV2, JIIIabc – HCV3 (crystal structures) and JIIIabc – eS27 (3.9-Å cryo-EM model, PDB code: 5A2Q)25 complexes. Molecular surfaces of the Fabs HCV2, HCV3 and protein eS27 are shown in orange, purple and lime-green colors. (b-d) Overall view of the binding interfaces for JIIIabc – eS27 (b), JIIIabc – HCV2 (c) and JIIIabc – HCV3 (d) complexes, highlighting that these proteins recognize a common epitope (IIIa and IIIc sub-domains) within the JIIIabc RNA. X, Y and Z in eS27 molecular surface are arbitrary assignments. (e) Molecular surface of the JIIIabc interface (3.9-Å cryo-EM model) and the eS27 residues that are involved in binding interactions with the RNA epitope. (f-h) Specific interactions between the RNA epitope and the eS27 residues. The dashed lines reflect hetero-atoms within hydrogen bonding distance. (i-j) Molecular surfaces of the JIIIabc showing a common and different recognition surfaces within the RNA epitopes by HCV2 and eS27 (i) and by eS27 and HCV3 (j). Unless specified, all figures and the labels are colored analogously for facile comparison.
Similar to recognition site within the JIIIabc for eS27, HCV2 and HCV3, RNA recognition within signal recognition particle (SRP) – 14 kDa – 16 kDa protein complexes (PDB code: 1E8O) also involves protein docking to an RNA surface created by the juxtaposition of two interacting RNA loops.49 This mode of molecular recognition renders protein binding dependent on RNA tertiary structure formation and therefore provides a strategy for enhancing binding specificity relative to recognition of isolated stem-loops, which occur in high frequency throughout the transcriptome.
Inhibition of HCV IRES-mediated translation by Fabs HCV2 and HCV3.
Previous studies including the cryo-EM structures of HCV IRES in complex with the human ribosome revealed that JIIIabc is one of the essential domains for the IRES interactions with the 40S ribosome,7–12,21,24–27,29,32 specifically, through the interactions of a ribosomal protein eS27 with the IIIa and IIIc sub-domains – the same region that Fabs HCV2 and HCV3 bind to. Consequently, we investigated whether these Fabs could hinder the IRES interactions with the ribosome to inhibit the translation mediated by the HCV IRES. We performed in vitro translation assays with rabbit reticulocyte lysate using a bicistronic reporter construct encoding both Firefly and Renilla luciferase (Fig. 5a).
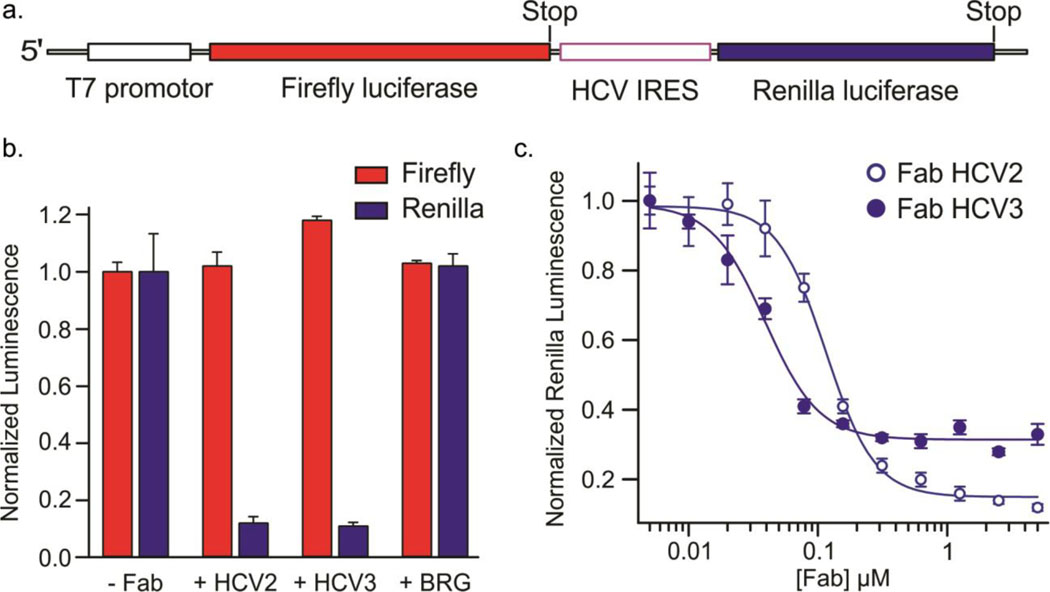
Fig. 5.
Inhibition of HCV IRES-mediated in vitro translation by Fabs HCV2 and HCV3 in a rabbit reticulocyte lysate-based assay. (a) Schematic of the dsDNA template used for generating the corresponding bicistronic mRNA for translation via a T7 promotor-controlled transcription. (b) Normalized luminescence corresponding to the expression of Firefly (red bars) and Renilla (blue bars) luciferases in the absence and presence of 5 μM of Fab HCV2, HCV3 or BRG. Fab BRG possesses the same scaffold domains as Fabs HCV2 and HCV3 but does not bind to the HCV IRES. (c) Normalized luminescence corresponding to the expression of Renilla luciferase with varying concentration of Fab HCV2 (solid circles) and HCV3 (hollow circles). Error bars in b and c represent the standard deviations from three independent experiments.
The canonical translation mechanism directs the Firefly luciferase expression, whereas the HCV IRES directs Renilla luciferase expression. We prepared the dsDNA template for the construct via PCR and generated the corresponding mRNA for translation via T7 promotor-controlled transcription in situ. Firefly and Renilla luciferase expression levels within each reaction were quantified using a dual luciferase reporter assay that measures the luminescence activity of each luciferase. Specific binding of the Fab HCV2 or HCV3 to the JIIIabc RNA of HCV IRES would be expected to inhibit the Renilla luciferase expression without interfering with the Firefly luciferase expression.
In the presence of Fab HCV2 or HCV3 (5 μM), expression of Renilla luciferase decreased to the background level, whereas corresponding Firefly expression remained unaffected (Fig. 5b). As a control, we tested the effect of a different RNA-binding Fab on translation. Fab BRG binds to a short sequence of RNA that adopts a stem loop conformation.39 Although it possesses the same scaffold domains as HCV2 and HCV3, it has distinct CDR sequences and does not bind to the HCV IRES. Inclusion of 5 μM Fab BRG in the translation assay had no effect on the expression of either luciferase (Fig. 5b), suggesting that Fab HCV2 or HCV3 specifically inhibit Renilla luciferase translation by binding to the HCV IRES, presumably hindering its interaction with the 40S ribosome. Similar translation assays with varying concentration of added Fabs revealed IC50 values of 115 ± 10 nM and 40 ± 10 nM for HCV2 and HCV3, respectively (Fig. 5c), which are similar to the dissociation constants of these Fabs with the HCV IRES (32 ± 7 nM and 37 ± 8 nM, respectively).
Implications for IRES domains as therapeutic targets.
Due to its central role in translation of the viral genome and strong conservation among isolates and strains, the HCV IRES has been recognized as an attractive target for the development of the therapeutics that inhibit viral translation. Our cell lysate-based in vitro translation assays demonstrate that binding of Fabs HCV2 and HCV3 to junction IIIabc of HCV IRES specifically inhibits IRES-mediated translation, presumably by blocking the interactions between the IRES and the 40S ribosome or the eIF3 complex, which represent essential interactions for initiating translation driven by HCV and HCV-like IRESs. Given that HCV infection causes widespread hepatitis C and hepatocellular carcinoma, affecting over 150 million humans worldwide50 and current therapeutic strategies, though effective, are expensive and can cause serious side effects,51,52 antibodies against HCV IRES could provide potential alternative therapeutic molecules. Of course, antibodies serve as highly effective drugs in their own right, but their targets generally reside outside of the cell. Future advances in protein delivery technologies may render intracellular targets, including viral RNA domains, vulnerable to the therapeutic potential of antibodies.
Numerous approaches have been developed to inhibit HCV translation by targeting highly conserved IRES elements, yielding a set of oligonucleotide, peptide and small molecule ligands.53–56 So far, domain II stands as the only HCV IRES element for which a small-molecule translation inhibitor has been developed.53,57 Our finding that Fabs HCV3 and HCV2 can inhibit translation in a concentration-dependent manner suggests that disruption of the interaction between JIIIabc and the ribosome could provide a therapeutic strategy against HCV, implicating JIIIabc as a possible target also for small molecule inhibitors. In this regard, the ability of Fabs HCV2 and HCV3 to facilitate crystallization of the RNA domain provides an avenue toward structure-based inhibitor design and optimization. The ability to engineer Fabs that bind RNA motifs with high affinity and specificity enables a targeted perturbation of RNA-RNA, RNA-protein, or RNA-ligand interactions. Analysis of the functional consequences of Fab binding in vitro and in cellulo (through reformatting as genetically encoded intrabodies) can facilitate dissection of RNA signaling networks and illuminate both the functional roles and druggability of the target motifs.58
CONCLUSIONS
The HCV IRES JIIIabc binds 40S ribosome and eIF3, playing an essential role in viral genome translation. Using phage display selection against the HCV IRES, we obtained two Fabs, HCV2 and HCV3 that bind the RNA with high affinity, specifically recognizing the JIIIabc region. Using these Fabs as crystallization chaperones, we determined the high-resolution crystal-structures of JIIIabc – HCV2 and – HCV3 complexes at 1.81-Å and 2.75-Å resolution, respectively, revealing an anti-parallel four-way junction with the IIIa and IIIc domains juxtaposed through tertiary interactions. The structure of the junction strongly agrees with the RNA model derived from 3.9-Å resolution cryo-EM structure of HCV IRES – 40S ribosome complex, suggesting that the domain adopts a pre-organized architecture for ribosome interactions. In these structures, the IIIa – IIIc juxtaposition creates a platform for protein recognition by Fabs HCV2 and HCV3 and ribosomal protein eS27. Strikingly, both Fabs and eS27 interact with a common subset of nucleotides, suggesting that this surface could be a “hot spot” for protein docking. In rabbit reticulocyte lysate translation assays using a bicistronic reporter assay, Fabs HCV2 and HCV3 specifically inhibited HCV IRES-directed translation, implicating disruption of the JIIIabc – eS27 interaction as a potential therapeutic strategy against HCV. Consistent with our previously reported Fab-RNA complexes, the RNA recognition strategies adopted by Fabs HCV2 and HCV3 are mostly similar to those observed for natural RNA binding proteins, suggesting that Fabs could serve as a unique and versatile scaffold with the potential to bind a wide variety of RNAs for applications beyond RNA crystallography.
METHODS
RNA synthesis and purification.
DNA templates for transcription reactions of full-length HCV IRES constructs used for phage display selection and crystallization were produced by PCR amplification of a plasmid (generously provided by Evgeny V. Pilipenko, University of Chicago) template containing the HCV IRES sequence following a T7-promotor, whereas the DNA templates for transcription reactions of isolated JIIIabc constructs were prepared by PCR amplification of ssDNA oligomer with T7-promotor sequence purchased from Integrated DNA Technologies (IDT, www.idtdna.com). The first two nucleotides of the reverse primer contained 2′-OMe modifications to reduce transcriptional heterogeneity at the 3′ end.59 RNA was prepared by in-vitro transcription for 3 hours at 37 °C in buffer containing 40 mM Tris-HCl pH 8.0, 2 mM spermidine, 10 mM NaCl, 25 mM MgCl2, 10 mM DTT, 40 U/ml RNase Inhibitor, 5 U/ml TIPPase, 5 mM of each NTPs, 50 pmol/ml DNA template, 50 μg/ml homemade T7 RNA polymerase.60 Transcription reactions were quenched by adding 5 U/ml RNase free DNase I (Promega, www.promega.com) and incubating at 37 °C for 30 minutes. Following DNase treatment and phenol-chloroform extraction, the RNA was purified by denaturing polyacrylamide gel electrophoresis. The corresponding RNA band was visualized by UV shadowing and excised from the gel. RNA was eluted overnight at 4°C in 10 mM Tris, pH 8.0, 2 mM EDTA, 300 mM NaCl buffer. The buffer for eluted RNA was exchanged 3 times for pure water using 10 kDa-cutoff size-exclusion column (Amicon, www.emdmillipore.com). RNA was collected, aliquoted and stored at −80 °C until further use.
Phage display selection.
Phage display to select a Fab that binds the RNA of interest was performed by following a similar strategy as described elsewhere.36,37 The HCV IRES RNA construct that contained an additional 3ꞌ overhang sequence, 5ꞌ-AGG UCG ACU CUA GAG GAU CCC CGG (x-module) was hybridized with the biotinylated DNA oligonucleotide, 5ꞌ-Biotin-ACC GGG GAT CCT CTA GAG TC (see Supplementary Fig. 3) and this RNA-DNA hybrid was immobilized on the streptavidin-coated magnetic beads via biotin-streptavidin interaction. For the first round, 500 nM of RNA was immobilized by using a pre-determined amount of beads required for complete immobilization and then incubated with 1013 cfu (colony forming units) of phage for 15 min in 1 ml of selection buffer, PBS (8 mM Na2HPO4, 1.5 mM KH2PO4, 137 mM NaCl, and 3 mM KCl, pH 7.4), 0.05% Tween 20, 10 mM Tris-HCl (pH 7.4), 10 mM MgCl2 and 200 mM NaCl, supplemented with 0.1 mg/ml BSA, 0.1 mg/ml streptavidin, and 1 unit/μL RNase inhibitor (NEB, www.neb.com). The solution was then removed, and the beads were washed twice with the selection buffer. In subsequent rounds, purified phage pools were first incubated with streptavidin beads in the selection buffer for 30 min, and the supernatant was used for the subsequent selection on a King Fisher magnetic particle processor (Thermo Electron Corporation, www.gogenlab.com). The 1011 cfu of Phages were incubated for 15 min with (50 nM in 2nd and 5 nM in 3rd round) of the RNA in 100 μl of the selection buffer, supplemented with 0.1 mg/ml BSA, 1 unit/μl RNase inhibitor, and 1.5 μM x-module DNA-RNA hybrid as a competitor. Streptavidin magnetic beads were then added to the solution for 15 min to allow the capture of the biotinylated RNA construct together with the bound phage. The beads were then blocked with 50 μM biotin, washed five times with the buffer, and eluted in 50 μL of elution buffer (PBS, and 1 μg/ml biotinylated RNaseA). The biotinylated RNase A was removed from the resulting phage library by incubation with streptavidin beads. After each round of selection, recovered phages were amplified as described previously.36,37 After 3rd round of selection, the phages were sequenced.
Fab expression and purification.
Enriched output clones from the 3rd round of selection was tested for binding to target RNA using phage ELISA. For the phage ELISA assay, the RNA construct was immobilized through the x-module via biotin-neutravidin interactions, and ELISA assay was performed according to published protocols.61 Clones that showed a positive binding response in a phage ELISA assay were reformatted for soluble protein expression with the introduction of a stop codon on phagemids using Q5 site-directed mutagenesis kit (NEB, www.neb.com). Fabs were expressed and purified first on a small scale (100–250 mL culture) as described elsewhere.36,37,61 No affinity tag was employed in the purification, which is described briefly below. Binding affinity of each clone with the RNA lacking the x-module was determined by filter binding assay. Fabs that bound the RNA with desirable affinity were then expressed on a larger scale (4 liter culture) according to published protocols.29,30,55 Both small and large scale Fab production methods essentially followed similar steps and yielded pure and RNase free Fabs. Briefly, phagemids with stop codon were transformed into 55244 chemical competent cells (www.atcc.org) and directly inoculated a starter culture with 100 μg/ml ampicillin. This overnight culture was then used to inoculate 2xYT media and grown for 24 hours at 30°C. Culture was centrifuged at room temperature, cell pellet was resuspended in the same volume of CRAP-Pi media61 with 100 μg/ml ampicillin and grown for 24 hours at 30°C. Collected cell pellets were lysed in PBS buffer using 0.4 mg/ml of lysozyme (Millipore Sigma, www.sigmaaldrich.com), and Fab proteins were purified using the AKTAxpress fast protein liquid chromatography (FPLC) purification system (Amersham, www.gelifesciences.com) as described previously.36,37 The lysate in PBS buffer (pH 7.4) was loaded into a protein A column, the eluted Fab in 0.1 M acetic acid was dialyzed back into the buffer PBS (pH 7.4) and loaded into a protein G column. The eluted Fab from protein G column in 0.1 M glycine (pH 2.7) dialyzed into 50 mM NaOAc, 50 mM NaCl buffer (pH 5.5) and loaded into a heparin column. Finally, the eluted Fab in 50 mM NaOAc, 2 M NaCl (pH 5.5) was dialyzed back into 1x PBS (pH 7.4), concentrated, and analyzed by 12% SDS-PAGE using Coomassie Blue R-250 staining for visualization. Aliquots of Fab samples were tested for RNase activity using the RNaseAlert kit (Ambion, www.thermofisher.com). The aliquots of Fab samples were flash frozen in liquid nitrogen and stored at −80°C until further use.
Fab-RNA binding affinity measurements.
The binding constants of the HCV IRES and JIIIabc constructs to our identified binders were determined by nitrocellulose filter binding assay as reported previously.38 Briefly, ~30 pmol of RNA was 5’−32P radiolabeled and purified using size exclusion spin columns Micro Bio-Spin™ P-30 (Bio-Rad, www.bio-rad.com). A constant amount of radiolabeled RNA was incubated at 90°C for 1 minute and then at 50°C for 10 minutes in a binding buffer containing either 10 mM Tris-HCl (pH 7.4), 10 mM MgCl2 and 200 mM NaCl. The sample was cooled to room temperature for 10 minutes and incubated for 30 minutes with Fab HCV2 or HCV3 ranging from 2 nM to 2 μM in a final volume of 40 μL. The Bio-Dot apparatus from Bio-Rad was assembled by placing a BA85 nitrocellulose filter (Whatman, www.gelifesciences.com) at the top and Hybond filter at the bottom (Amersham, www.gelifesciences.com) and wells were pre-equilibrated with 100 μL of the binding buffer. The Fab-RNA complex was applied and washed 2 times with 100 μl of the same buffer at a time. Both filters were air dried, exposed to Phosphor-Imager screens, scanned with a Typhoon Trio imager (GE Healthcare) and the amount of RNA retained in each of the filters was quantified by using Image Quant software (Molecular Dynamics). The dissociation constants were calculated by fitting the data of fraction of RNA retained in the nitrocellulose membrane versus the concentration of the Fab to the equation: , where F represent the fraction of bound RNA at a given concentration of the Fab, Kd is the dissociation constant and F0 and Fmax are the minimum and maximum fractions of the bound RNA, respectively.
Crystallization.
An aliquot of RNA sample was refolded in a folding buffer containing 10 mM Tris-HCl (pH 7.4), 10 mM MgCl2 and 200 mM NaCl. For refolding, RNA was heated at 90°C for 1 minute in water, added appropriate volume of 10x folding buffer and then incubated at 50°C for 10 minutes in 1x folding buffer followed by incubation at room temperature for 10 minutes and in ice for 5 minutes. The refolded RNA was then incubated with 1.1 equivalents of the Fab and concentrated to 5 mg/ml using 10 kDa-cutoff, Amicon Ultra-15 column (www.emdmillipore.com). The formation of Fab–RNA complex was confirmed by native polyacrylamide gel electrophoresis (nPAGE). To decrease the number of nucleation events, Fab-RNA complexes were then passed through 0.2 μm cutoff, Millipore centrifugal filter units (www.emdmillipore.com). A Mosquito liquid handling robot (TTP Labtech, ttplabtech.com) was used to set up high-throughput hanging-drop vapor-diffusion crystallization screens at room temperature using commercially available screening kits from Hampton Research and Jena Bioscience. Crystals appeared and grew to full size within a week at 22 °C. Select conditions were further optimized for pH, precipitant and salt concentration to grow larger crystals using the hanging drop vapor diffusion method. For cryoprotection, drops containing suitable crystals were brought to 10% glycerol, keeping all other compositions same. Crystals were immediately flash-frozen in liquid nitrogen after being fished out from the drops and taken to Argonne National Laboratory for collecting the X-ray diffraction data. The crystals that were grown in 0.2 M ammonium acetate, 0.1 M bis-tris pH 5.5, 45% 2-methyl-2,4-pentanediaol at 22°C for JIIIabc – HCV2, and 0.1 M HEPES pH 7.5, 1.4 M sodium citrate tribasic dihydrate at 22°C for JIIIabc – HCV3 diffracted to 1.81-Å and 2.75-Å resolution, respectively.
Structural data collection, processing and analysis.
The X-ray diffraction data sets were collected at the Advanced Photon Source NE-CAT section beamline 24-ID-C and 24-ID-E. All the datasets were then integrated and scaled using its on-site RAPD automated programs (https://rapd.nec.aps.anl.gov/rapd/). Initial phases were obtained by molecular replacement with the Fab portion of the previously reported structure of Fab HAVx in complex with domain V from the hepatitis A virus IRES (PDB code: 6MWN)42 as the search model using Phaser on Phenix.62 Except for the CDRs, sequences of Fab HCV2, HCV3 and HAVx are identical. Iterative model building and refinement was performed by using COOT,63 and Phenix package.62 RNA was built unambiguously by modeling the individual nucleotides into the electron density map obtained from the molecular replacement. During the refinement, default NCS option in Phenix was selected. Most of the water molecules were automatically determined by Phenix during refinement. However, some water molecules were added manually for the positive electron density in the map based on their possibility to form hydrogen bonds with protein or RNA residues. Solvent-accessible surface area and area of interaction were calculated using PDBePISA (http://www.ebi.ac.uk/pdbe/pisa/).46 All structure related figures were made in PyMOL (Schrodinger, www.pymol.org) and figure labels were edited in CorelDraw (Corel Corporation, http://www.corel.com).
Translation inhibition assay.
The DNA template for the in vitro translation assay (see Fig. 4a) was generated via PCR amplification of pFR_HCV_xb plasmid, which was a gift from Phillip A. Sharp (Addgene plasmid # 11510, http://www.addgene.org/11510/).64 The rabbit reticulocyte lysate-based translation assay was performed by using a coupled transcription-translation kit (TnT® Coupled Reticulocyte Lysate Systems, www.promega.com) according to the manufacturer’s protocol. About 10 μg of the DNA template was used and the reaction was carried out with or without the Fab in a final volume of 50 μL for 2 hours at 30°C. After translation, the expression levels of the luciferases were detected by measuring the luminescence signals (Synergy Neo2 plate reader, www.biotek.com), which were obtained by using a dual-luciferase reporter assay kit (Dual-Luciferase® Reporter Assay System, www.promega.com) according to the manufacturer’s protocol. The luciferase activity measurements were performed for a 10 μL of the reaction aliquot and each reaction sample was measured in triplicate.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by grants from the National Institutes of Health (R01AI081987, R01GM102489) and the Chicago Biomedical Consortium with support from the Searle Funds at The Chicago Community Trust to J. Piccirilli. The crystallographic work is based on research conducted at the Advanced Photon Source on the Northeastern Collaborative Access Team beamline, 24-ID-C and 24-ID-E, which is supported by a grant from the National Institute of General Medical Sciences (P41 GM103403) from the National Institutes of Health. This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02–06CH11357. We would like to thank staff of the Advanced Photon Source at Argonne National Laboratory for providing technical advice during data collection. We are thankful to P. Rice (University of Chicago) for guiding X-ray data interpretation and structure solving, and Y. Chan (University of Chicago) for assisting with the translation construct design. We also thank Piccirilli laboratory members, especially M. Disare and H. Rees, for critical comments on the manuscript.
Footnotes
ACCESSION NUMBERS
Atomic coordinates and structure factors for the reported crystal structures have been deposited with the Protein Data Bank under accession numbers 6U8D and 6U8K for JIIIabc – HCV2 and JIIIabc – HCV3 complexes, respectively.
SUPPLEMENTARY INFORMATION
Supplementary information includes supplementary notes and supplementary figures. This material is available free of charge via the internet at http://pubs.acs.org.
CONFLICT OF INTEREST
Authors declare no competing interests.
REFERENCES
- 1.Jackson RJ, Hellen CU & Pestova TV The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 11, 113 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hinnebusch AG, Ivanov IP & Sonenberg N Translational control by 5′-untranslated regions of eukaryotic mRNAs. Science 352, 1413–1416 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jan E, Mohr I & Walsh D A cap-to-tail guide to mRNA translation strategies in virus-infected cells. Annu. Rev. Virol. 3, 283–307 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Jaafar ZA & Kieft JS Viral RNA structure-based strategies to manipulate translation. Nat. Rev. Microbiol, 1 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang C, Sarnow P & Siddiqui A Translation of human hepatitis C virus RNA in cultured cells is mediated by an internal ribosome-binding mechanism. J. Virol. 67, 3338–3344 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pestova TV, Shatsky IN, Fletcher SP, Jackson RJ & Hellen CU A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 12, 67–83 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sizova DV, Kolupaeva VG, Pestova TV, Shatsky IN & Hellen CU Specific interaction of eukaryotic translation initiation factor 3 with the 5′ nontranslated regions of hepatitis C virus and classical swine fever virus RNAs. J. Virol. 72, 4775–4782 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.KIEFT JS, ZHOU K, JUBIN R & DOUDNA JA Mechanism of ribosome recruitment by hepatitis C IRES RNA. RNA 7, 194–206 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.. Otto GA & Puglisi JD The pathway of HCV IRES-mediated translation initiation. Cell 119, 369–380 (2004). [DOI] [PubMed] [Google Scholar]
- 10.Fraser CS & Doudna JA Structural and mechanistic insights into hepatitis C viral translation initiation. Nat. Rev. Microbiol. 5, 29–38 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Hashem Y, Des Georges A, Dhote V, Langlois R, Liao HY, Grassucci RA, Pestova TV, Hellen CU & Frank J Hepatitis-C-virus-like internal ribosome entry sites displace eIF3 to gain access to the 40S subunit. Nature 503, 539 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Niepmann M Hepatitis C virus RNA translation. in Hepatitis C Virus: From Molecular Virology to Antiviral Therapy 143–166 (Springer, 2013). [Google Scholar]
- 13.Bukh J, Purcell RH & Miller RH Sequence analysis of the 5’noncoding region of hepatitis C virus. Proc. Natl. Acad. Sci. U.S.A. 89, 4942–4946 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown EA, Zhang H, Ping L-H & Lemon SM Secondary structure of the 5′ nontranslated regions of hepatitis C virus and pestivirus genomic RNAs. Nucleic Acids Res. 20, 5041–5045 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Honda M, Ping L-H, Rijnbrand RC, Amphlett E, Clarke B, Rowlands D & Lemon SM Structural requirements for initiation of translation by internal ribosome entry within genome-length hepatitis C virus RNA. Virology 222, 31–42 (1996). [DOI] [PubMed] [Google Scholar]
- 16.Honda M, Rijnbrand R, Abell G, Kim D & Lemon SM Natural variation in translational activities of the 5′ nontranslated RNAs of hepatitis C virus genotypes 1a and 1b: evidence for a long-range RNA-RNA interaction outside of the internal ribosomal entry site. J. Virol. 73, 4941–4951 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kato N Genome of human hepatitis C virus (HCV): gene organization, sequence diversity, and variation. Microb. Comp. Genomics 5, 129–151 (2000). [DOI] [PubMed] [Google Scholar]
- 18.Forton DM, Karayiannis P, Mahmud N, Taylor-Robinson SD & Thomas HC Identification of unique hepatitis C virus quasispecies in the central nervous system and comparative analysis of internal translational efficiency of brain, liver, and serum variants. J. Virol. 78, 5170–5183 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han J, Shyamala V, Richman K, Brauer M, Irvine B, Urdea M, Tekamp-Olson P, Kuo G, Choo Q & Houghton M Characterization of the terminal regions of hepatitis C viral RNA: identification of conserved sequences in the 5’untranslated region and poly (A) tails at the 3’end. Proc. Natl. Acad. Sci. U.S.A. 88, 1711–1715 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rijnbrand R, Bredenbeek P, van der Straaten T, Whetter L, Inchauspé G, Lemon S & Spaan W Almost the entire 5′ non-translated region of hepatitis C virus is required for cap-independent translation. FEBS Lett. 365, 115–119 (1995). [DOI] [PubMed] [Google Scholar]
- 21.Spahn CM, Kieft JS, Grassucci RA, Penczek PA, Zhou K, Doudna JA & Frank J Hepatitis C virus IRES RNA-induced changes in the conformation of the 40s ribosomal subunit. Science 291, 1959–1962 (2001). [DOI] [PubMed] [Google Scholar]
- 22.Lukavsky PJ, Otto GA, Lancaster AM, Sarnow P & Puglisi JD Structures of two RNA domains essential for hepatitis C virus internal ribosome entry site function. Nat. Struc. Mol. Biol. 7, 1105 (2000). [DOI] [PubMed] [Google Scholar]
- 23.Lukavsky PJ, Kim I, Otto GA & Puglisi JD Structure of HCV IRES domain II determined by NMR. Nat. Struc. Mol. Biol. 10, 1033 (2003). [DOI] [PubMed] [Google Scholar]
- 24.Yamamoto H, Unbehaun A, Loerke J, Behrmann E, Collier M, Bürger J, Mielke T & Spahn CM Structure of the mammalian 80S initiation complex with initiation factor 5B on HCV-IRES RNA. Nat. Struc. Mol. Biol. 21, 721 (2014). [DOI] [PubMed] [Google Scholar]
- 25.Quade N, Boehringer D, Leibundgut M, Van Den Heuvel J & Ban N Cryo-EM structure of Hepatitis C virus IRES bound to the human ribosome at 3.9-A resolution. Nat. Commun. 6(2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson AG, Grosely R, Petrov AN & Puglisi JD Dynamics of IRES-mediated translation. Phil. Trans. R. Soc. B 372, 20160177 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yokoyama T, Machida K, Iwasaki W, Shigeta T, Nishimoto M, Takahashi M, Sakamoto A, Yonemochi M, Harada Y & Shigematsu H HCV IRES Captures an Actively Translating 80S Ribosome. Mol. Cell (2019). [DOI] [PubMed] [Google Scholar]
- 28.Wang C, Le S, Ali N & Siddiqui A An RNA pseudoknot is an essential structural element of the internal ribosome entry site located within the hepatitis C virus 5’noncoding region. RNA 1, 526–537 (1995). [PMC free article] [PubMed] [Google Scholar]
- 29.Lukavsky PJ Structure and function of HCV IRES domains. Virus Res. 139, 166–171 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berry KE, Waghray S, Mortimer SA, Bai Y & Doudna JA Crystal structure of the HCV IRES central domain reveals strategy for start-codon positioning. Structure 19, 1456–1466 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buratti E, Tisminetzky S, Zotti M & Baralle FE Functional analysis of the interaction between HCV 5′ UTR and putative subunits of eukaryotic translation initiation factor eIF3. Nucleic Acids Res. 26, 3179–3187 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Collier AJ, Gallego J, Klinck R, Cole PT, Harris SJ, Harrison GP, Aboul-ela F, Varani G & Walker S A conserved RNA structure within the HCV IRES eIF3-binding site. Nat. Struc. Mol. Biol. 9, 375 (2002). [DOI] [PubMed] [Google Scholar]
- 33.Kieft JS, Zhou K, Grech A, Jubin R & Doudna JA Crystal structure of an RNA tertiary domain essential to HCV IRES-mediated translation initiation. Nat. Struc. Mol. Biol. 9, 370 (2002). [DOI] [PubMed] [Google Scholar]
- 34.Rijnbrand R, Thiviyanathan V, Kaluarachchi K, Lemon SM & Gorenstein DG Mutational and structural analysis of stem-loop IIIC of the hepatitis C virus and GB virus B internal ribosome entry sites. J. Mol. Biol. 343, 805–817 (2004). [DOI] [PubMed] [Google Scholar]
- 35.MELCHER SE, WILSON TJ & LILLEY DM The dynamic nature of the four-way junction of the hepatitis C virus IRES. RNA 9, 809–820 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ye J-D, Tereshko V, Frederiksen JK, Koide A, Fellouse FA, Sidhu SS, Koide S, Kossiakoff AA & Piccirilli JA Synthetic antibodies for specific recognition and crystallization of structured RNA. Proc. Natl. Acad. Sci. U.S.A. 105, 82–87 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koldobskaya Y, Duguid EM, Shechner DM, Suslov NB, Ye J, Sidhu SS, Bartel DP, Koide S, Kossiakoff AA & Piccirilli JA A portable RNA sequence whose recognition by a synthetic antibody facilitates structural determination. Nat. Struct. Mol. Biol. 18, 100–106 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang H, Suslov NB, Li N-S, Shelke SA, Evans ME, Koldobskaya Y, Rice PA & Piccirilli JA A G-quadruplex–containing RNA activates fluorescence in a GFP-like fluorophore. Nat. Chem. Biol. 10, 686–691 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shao Y, Huang H, Qin D, Li N-S, Koide A, Staley JP, Koide S, Kossiakoff AA & Piccirilli JA Specific Recognition of a Single-Stranded RNA Sequence by a Synthetic Antibody Fragment. J. Mol. Biol. 428, 4100–4114 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shelke SA, Shao Y, Laski A, Koirala D, Weissman BP, Fuller JR, Tan X, Constantin TP, Waggoner AS & Bruchez MP Structural basis for activation of fluorogenic dyes by an RNA aptamer lacking a G-quadruplex motif. Nat. Commun. 9, 4542 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koirala D, Shelke SA, Dupont M, Ruiz S, DasGupta S, Bailey LJ, Benner SA & Piccirilli JA Affinity maturation of a portable Fab–RNA module for chaperone-assisted RNA crystallography. Nucleic Acids Res. 46, 2624–2635 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koirala D, Shao Y, Koldobskaya Y, Fuller JR, Watkins AM, Shelke SA, Pilipenko EV, Das R, Rice PA & Piccirilli JA A conserved RNA structural motif for organizing topology within picornaviral internal ribosome entry sites. Nat. Commun. 10, 3629 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bogan AA & Thorn KS Anatomy of hot spots in protein interfaces. J. Mol. Biol. 280, 1–9 (1998). [DOI] [PubMed] [Google Scholar]
- 44.Kortemme T & Baker D A simple physical model for binding energy hot spots in protein–protein complexes. Proc. Natl. Acad. Sci. U.S.A. 99, 14116–14121 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moreira IS, Fernandes PA & Ramos MJ Hot spots—A review of the protein–protein interface determinant amino-acid residues. Proteins: Struct., Funct., Bioinf. 68, 803–812 (2007). [DOI] [PubMed] [Google Scholar]
- 46.Krissinel E & Henrick K Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007). [DOI] [PubMed] [Google Scholar]
- 47.Jaeger L, Michel F & Westhof E Involvement of a GNRA tetraloop in long-range RNA tertiary interactions. J. Mol. Biol. 236, 1271–1276 (1994). [DOI] [PubMed] [Google Scholar]
- 48.Fiore JL & Nesbitt DJ An RNA folding motif: GNRA tetraloop–receptor interactions. Q. Rev. Biophys. 46, 223–264 (2013). [DOI] [PubMed] [Google Scholar]
- 49.Weichenrieder O, Wild K, Strub K & Cusack S Structure and assembly of the Alu domain of the mammalian signal recognition particle. Nature 408, 167 (2000). [DOI] [PubMed] [Google Scholar]
- 50.Thrift AP, El-Serag HB & Kanwal F Global epidemiology and burden of HCV infection and HCV-related disease. Nat. Rev. Gastroenterol. 14, 122 (2017). [DOI] [PubMed] [Google Scholar]
- 51.Pawlotsky J-M, Feld JJ, Zeuzem S & Hoofnagle JH From non-A, non-B hepatitis to hepatitis C virus cure. J. Hepatol. 62, S87–S99 (2015). [DOI] [PubMed] [Google Scholar]
- 52.Chhatwal J, Kanwal F, Roberts MS & Dunn MA Cost-effectiveness and budget impact of hepatitis C virus treatment with sofosbuvir and ledipasvir in the United States. Ann. Intern. Med. 162, 397 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Parsons J, Castaldi MP, Dutta S, Dibrov SM, Wyles DL & Hermann T Conformational inhibition of the hepatitis C virus internal ribosome entry site RNA. Nat. Chem. Biol. 5, 823 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Davis DR & Seth PP Therapeutic targeting of HCV internal ribosomal entry site RNA. Antivir. Chem. Chemother. 21, 117–128 (2011). [DOI] [PubMed] [Google Scholar]
- 55.Dibrov SM, Parsons J, Carnevali M, Zhou S, Rynearson KD, Ding K, Garcia Sega E, Brunn ND, Boerneke MA & Castaldi MP Hepatitis C Virus Translation Inhibitors Targeting the Internal Ribosomal Entry Site: Miniperspective. J. Med. Chem. 57, 1694–1707 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Moon J-S, Lee S-H, Kim E-J, Cho H, Lee W, Kim G-W, Park H-J, Cho S-W, Lee C & Oh J-W Inhibition of hepatitis C virus in mice by a small interfering RNA targeting a highly conserved sequence in viral IRES pseudoknot. PLoS One 11, e0146710 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dibrov SM, Ding K, Brunn ND, Parker MA, Bergdahl BM, Wyles DL & Hermann T Structure of a hepatitis C virus RNA domain in complex with a translation inhibitor reveals a binding mode reminiscent of riboswitches. Proc. Natl. Acad. Sci. U.S.A. 109, 5223–5228 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sha F, Gencer EB, Georgeon S, Koide A, Yasui N, Koide S & Hantschel O Dissection of the BCR-ABL signaling network using highly specific monobody inhibitors to the SHP2 SH2 domains. Proc. Natl. Acad. Sci. U.S.A. 110, 14924–14929 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kao C, Rüdisser S & Zheng M A simple and efficient method to transcribe RNAs with reduced 3′ heterogeneity. Methods 23, 201–205 (2001). [DOI] [PubMed] [Google Scholar]
- 60.Rio DC Expression and Purification of Active Recombinant T7 RNA Polymerase from E. coli. Cold Spring Harb. Protoc. 2013, pdb.prot078527 (2013). [DOI] [PubMed] [Google Scholar]
- 61.Paduch M, Koide A, Uysal S, Rizk SS, Koide S & Kossiakoff AA Generating conformation-specific synthetic antibodies to trap proteins in selected functional states. Methods 60, 3–14 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ & Grosse-Kunstleve RW PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Emsley P, Lohkamp B, Scott WG & Cowtan K Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Petersen CP, Bordeleau M-E, Pelletier J & Sharp PA Short RNAs repress translation after initiation in mammalian cells. Mol. Cell 21, 533–542 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.