Abstract
Brazilian red propolis (BRP) is a natural product widely known for its phenolic composition and strong antioxidant properties. In this study, we used the Box–Behnken Design (BBD) with Surface Response Methodology to optimize the extraction conditions for total phenolic content (TPC) and Trolox equivalent antioxidant capacity(TEAC) of bioactive phenolics from BRP. The extraction time, ethanol/water concentration and temperature, were tested. All variables had significant effects (p ≤ 0.05), with a desirability coefficient of 0.88. Under optimized conditions (90% ethanol at 80 °C for 30 min), the BRP extract showed a TPC of 129.00 ± 2.16 mg GAE/g and a TEAC of 3471.76 ± 53.86 µmol TE/g. Moreover, FRAP and ORAC assays revealed that the optimized BRP extract had 1472.86 ± 72.37 µmol Fe2+/g and 4339.61 ± 114.65 µmol TE/gof dry weight, respectively. Thirty-two phenolic compounds were tentatively identified by LC-QTOF-ESI-MS/MS, of which thirteen were found for the first time in BRP, including four flavones, one flavanol, two flavanones, two chalcones, and four isoflavonoids. Thus, our results highlight the importance of BRP as a source of a wide variety of phenolic compounds with significant antioxidant properties.
Keywords: isoflavonoids, bioactive compounds, Box–Behnkendesign, natural products, Apis mellifera
1. Introduction
Propolis or bee glue is a resinous balsamic substance collected by bees from plant exudates. It is naturally used to protect the hive and as an efficient antiseptic [1,2]. Historically, propolis has been used as a therapeutic substance in folk medicine, but recent advances in science and technology are increasing its commercial value in the food and pharmaceutical industries [3].
Among the different types of propolis occurring worldwide, Brazilian red propolis (BRP) stands out for its health benefits, which are attributed to a phenolic-rich composition, mainly isoflavonoids. The mechanisms of action of some BRP compounds, such as formononetin, vestitol, and neovestivol, were recently examined. These constituents were found to have strong antioxidant, anti-inflammatory, and antimicrobial properties [4,5].
To date, more than 200 compounds have been identified in BRP [4,6,7,8,9], most of which are polyphenols. Propolis composition is causally related to both its botanical source and environmental conditions. The main botanical sources of BRP are Dalbergia ecastaphyllum, a rich source of isoflavonoids, and Symphonia globulifera, a rich source of polyprenylated benzophenones (guttiferone E and oblongifolin B) and triterpenoids (β-amyrin and glutinol) [9].
Although several studies have correlated the presence of phenolics with the biological activity of BRP, mainly antimicrobial, antioxidant, anti-inflammatory, and anti-cancer properties [10,11,12], an optimization of extraction conditions has not been carried out thus far. The chemical extraction is the initial procedure for recovery of polyphenols from a natural product. Thus, choosing appropriate extraction conditions (e.g., sample-to-solvent ratio, solvent concentration, temperature, and extraction time) is utterly important as these may affect the final extract composition and bioactivity [13].
As stated by Riswanto et al. [14], the Response Surface Methodology (RSM) is a technique widely applied in the optimization of natural products due to its advantages compared to the traditional one-variable-at-a-time design. When combined with an experimental design like the Box–Behnken Design (BBD), it can be employed as a mathematical and statistical tool in natural product research. Due to its capacity of reducing the number of experiments required to find optimal conditions, BBD has been effectively used to optimize polyphenol extraction [13,15].
Several studies have reported the phenolic composition of BRP, but none of them were carried out under optimized extraction conditions. In addition, the few studies addressing atentative identification of phenolicsby High-Resolution Mass Spectrometrywere not carried out in negative mode.Here, the optimization procedureallowedus toidentifyseveral unknownphenolic compounds in the BRP extract.
Our study hypothesis was that the optimized BRP extract had a greater number of phenolic compounds—many of which yet unknown—and stronger antioxidant activity.
Therefore, the aim of this study was to establish the optimal extraction conditions for the recovery of antioxidant compounds from BRP and to evaluate the new phenolic composition of the optimized extracts by LC-ESI-QTOF-MS/MS in negative mode.
2. Materials and Methods
2.1. Chemicals
The following chemicals were used in this study: Folin–Ciocalteau reagent (DinamicaQuimicaContemporanea, Diadema, SP, Brazil); sodium carbonate, potassium chloride, ethanol (EtOH); monobasic and dibasic potassium phosphate. The standards ({±})-6-hydroxy-2,5,7,8-tetramethylchromane-2-carboxylic acid (Trolox), gallic acid, diammonium salt (ABTS) and potassium peroxydisulfate, fluorescein sodium salt and 2,20-azobis(2-methylpropionamidine) dihydrochloride (AAPH) were purchased from Sigma-Aldrich (St. Louis, MO, USA). All other reagents and solvents were of analytical grade.
2.2. Propolis Collection and Extraction
BRP samples were collected from the internal parts of Apis mellifera L. (Apidae) beehives located in the city of Maceió (9°40′ S, 35°41′ W), Alagoas State, Northeastern Brazil. Access to the Brazilian genetic heritage was previously obtained in accordance with the Brazilian legislation SECEX/CGEN Ordinance No. 1. Approval for sample collection was obtained via the SISGEN platform under accession number A5A0509.
Propolis samples were crushed with liquid nitrogen, weighted (0.5 g), mixed with 50 mL of solvent and remained in sealed tubes in a bath shaker (Gyromax 929, Amerex) for the time and temperature established in the experimental design (Table 1). After that, BRP ethanolic extracts were kept overnight at −20 °C until complete wax decantation. The supernatant solution was filtered, concentrated on a rotary evaporator at 110 mbar and 50 °C, lyophilized, and then used in the analysis of antioxidant activity and total phenolic content. All extraction procedures were carried out in triplicate.
Table 1.
Box–Behnken Design for the extraction of antioxidants compounds from Brazilian red propolis extracts.
| Run | Independent Variables | Dependent Variables | ||||||
|---|---|---|---|---|---|---|---|---|
| Coded Values | Real Values | TEAC (µmol TE/g) | TPC (mg GAE/g) | |||||
| Time (min) | Temp. °C | EtOH (%) | Time (min) | Temp. °C | EtOH (%) | |||
| 1 | −1 | −1 | 0 | 30 | 30 | 75 | 2560.36 | 125.76 |
| 2 | 1 | −1 | 0 | 90 | 30 | 75 | 2386.42 | 132.80 |
| 3 | −1 | 1 | 0 | 30 | 80 | 75 | 2804.74 | 136.17 |
| 4 | 1 | 1 | 0 | 90 | 80 | 75 | 2918.30 | 118.82 |
| 5 | −1 | 0 | −1 | 30 | 55 | 60 | 2370.61 | 120.30 |
| 6 | 1 | 0 | −1 | 90 | 55 | 60 | 2719.93 | 116.09 |
| 7 | −1 | 0 | 1 | 30 | 55 | 90 | 3106.62 | 127.64 |
| 8 | 1 | 0 | 1 | 90 | 55 | 90 | 3156.93 | 123.38 |
| 9 | 0 | −1 | −1 | 60 | 30 | 60 | 3169.87 | 118.82 |
| 10 | 0 | 1 | −1 | 60 | 80 | 60 | 2827.74 | 116.53 |
| 11 | 0 | −1 | 1 | 60 | 30 | 90 | 3200.06 | 109.49 |
| 12 | 0 | 1 | 1 | 60 | 80 | 90 | 3471.75 | 126.25 |
| 13 | 0 | 0 | 0 | 60 | 55 | 75 | 2544.55 | 133.00 |
| 14 | 0 | 0 | 0 | 60 | 55 | 75 | 2635.12 | 137.06 |
| 15 | 0 | 0 | 0 | 60 | 55 | 75 | 2622.18 | 134.98 |
Temp. = Temperature; EtOH = Ethanol; TEAC= Trolox equivalent antioxidant capacity; TPC= Total phenolic compounds.
2.3. Experimental Deisgn and Optimization
The following independent variables were considered: Time (X1) (30–90 min), temperature (X2) (30–80 °C), and percentage of ethanol/water (X3) (60–90%, v/v). Following the Box–Behnken design, 15 experiments with 3 central points were performed to determine the effects of these independent parameters on two dependent responses (TEAC and TPC) (Table 1). RSM was performed to investigate the relationship between the independent and dependent variables. The quadratic polynomial model is represented by the following equation:
| (1) |
where, Y is the dependent variable (antioxidant activity) for the independent responses (X1–X3); and β0, βi, βii, and βij are constant coefficients of intercept, linear, quadratic, and interaction terms.
2.4. Total Phenolic Content
The analysis of total phenolic content (TPC) was performed according to the Folin–Ciocalteau spectrophotometric method, with some modifications. Aliquots of 20 µL of the standard solution (gallic acid) or BRP extract and 100 µL of the Folin–Ciocalteau solution (10% in water) were pipetted into the wells of a microplate. After 5 min, 75 µL of a 7.5% sodium carbonate aqueous solution were added to each well. A control was prepared by replacing the sample with distilled water. The absorbance was measured at 740 nm in a microplate reader (Molecular Devices, LLC, Sunnyvale, CA, USA) after 40 min. The TPC was calculated by linear regression using gallic acid as a standard, and the results were expressed as mg of gallic acid equivalents (GAE) per g of dry extract [16]. All samples were analyzed in triplicate.
2.5. Antioxidant Activity
2.5.1. Ferric Reducing Antioxidant Power (FRAP) Assay
Briefly, 20 μL of BRP extract were mixed with 30 mL of water and 200 μL of FRAP reagent (prepared fresh daily) in a 96-well microplate. The FRAP reagent consisted of 10 volumes of 300 mmol/L acetate buffer (pH 3.6), one volume of 20 mmol/L FeCl3, and one volume of 10 mmol/L TPTZ in 40 mmol/L HCl [16]. The absorbance was measured at 595 nm in a microplate reader (Molecular Devices, LLC, Sunnyvale, CA, USA) after 8 min. Water was used as a blank; ferrous sulphate solutions (100 to 700 µM) were used for calibration; and the FRAP value was calculated by linear regression. The assay was performed in triplicate, and the results were estimated as µmol Fe2+ /g of dry extract.
2.5.2. Peroxyl Radical (ROO●)
Briefly, 30 μL of BRP extract plus 60 μL of fluorescein and 110 μL of an AAPH solution were transferred to a microplate. The reaction was performed at 37 °C and the absorbance was measured every minute for 2 h at 485 nm (excitation) and 528 nm (emission) in a microplate reader (Molecular Devices, LLC, Sunnyvale, CA, USA). Trolox standard was used at concentrations ranging from 12.5 to 400 μM. The results were expressed as μmol/Trolox equivalents (TE) per g of dry extract [17]. The assay was carried out in triplicate.
2.5.3. Trolox Equivalent Antioxidant Capacity (TEAC) Assay
The antioxidant capacity of the BRP extract was determined based on free radical ABTS, with modifications [16]. The ABTS radical was diluted in 75 mM potassium phosphate buffer (Ph 7.4) to an absorbance of 0.700 ± 0.01 at 734 nm. Aliquots of 20 µL of Trolox or BRP extract and 220 µL of ABTS radical solution were transferred to the wells and kept at room temperature protected from light. After 6 min of reaction, the absorbance was read at 734 nm using the potassium phosphate buffer as a blank. Trolox was used as a standard at concentrations ranging from 12.5 to 200 µM, and the results were expressed as µmol Trolox equivalents (TE) per g of dry extract.
2.6. High-Resolution Mass Spectrometry Analysis (LC-ESI-QTOF-MS/MS)
Liquid chromatography analysis was carried out in a chromatograph (Shimadzu Co., Tokyo) with a LC-30AD quaternary pump and SPD-20A photodiode array detector (PDA). Reversed phase chromatography was performed using a Phenomenex Luna C18 column (4.6 × 250 mm × 5 μM). A high-resolution mass spectrometer (MAXIS 3G—Bruker Daltonics, Bremen, Germany) was equipped with a Z-electrospray (ESI) interface operating in negative ion mode with a nominal resolution of 60,000 m/z. Twenty microliters of the BRP extract were injected into the liquid chromatography system. The analytical conditions were set as follows: Nebulizer at 2 Bar; dry gas at 8 L/min; temperature at 200 °C and HV at 4500 V. The mobile phase consisted of two solvents: (A) Water/acetic acid (99.5/0.5, v/v) and (B) methanol. The flow rate was 1 mL/min, and the gradient was initiated with 30% B, increasing to 40% B (15 min), 50% B (30 min), 60% B (45 min), 75% B (65 min), 75% B (85 min), 90% B (95 min), decreasing to 30% B (105 min). The run was complete after 114 min. An external calibration was carried out in MAXIS 3G—Bruker Daltonics 4.3 software to check for mass precision and data analysis. The tentative identification of the compounds was performed by comparing their exact mass (m/z) and MS2 spectra in negative mode to the database available in the literature and commercial standards (naringenin, pinocembrin, isoliquiritigenin, daidzein, formononetin andbiochanin A) from Sigma-Aldrich (St. Louis, MO, USA).
2.7. Data Analysis
STATISTICA 7.0 software was used to analyze the multivariate factorial design (ANOVA) and to determine the significance of the variables. The accuracy of the mathematical models was estimated by the coefficient of determination (R2) and the F-test (p < 0.05). All assays were carried out in triplicate, and the values were expressed as mean ± standard deviation.
3. Results and Discussion
3.1. Fitting the Models to Data
In this study, ANOVA was used to evaluate the accuracy of the RSM models (Table 2). The p-values (p < 0.05) observed for both response variables were considered significant, indicating that the developed models are appropriate to represent the relationship between the independent parameters and the response variables.
Table 2.
The effects of different parameters on Trolox equivalent antioxidant capacity (TEAC) and total phenolic content (TPC) outcomes during optimization of Brazilian red propolis ethanolic extracts.
| Term | SS | df | MS | F-Value | p-Value | Remarks |
|---|---|---|---|---|---|---|
| TEAC | ||||||
| Model | 1,247,423.00 | 9 | 138,602.55 | 603.14 | <0.0001 | significant |
| 8019.54 | 1 | 8019.54 | 34.89 | 0.0183 | ||
| 74,849.67 | 1 | 74,849.67 | 325.63 | 0.0020 | ||
| 425,485 | 1 | 425,484.7 | 1851.06 | 0.0003 | ||
| 83.58 | 1 | 83.58 | 0.3636 | 0.5347 | ||
| 144,174 | 1 | 144,173.8 | 627.22 | 0.0010 | ||
| 530,460 | 1 | 530,460.1 | 2307.80 | 0.0002 | ||
| 1153.35 | 1 | 1153.35 | 5.02 | 0.1092 | ||
| 1.87 | 1 | 1.87 | 0.0081 | 0.9254 | ||
| 94,179.02 | 1 | 94,179.02 | 409.73 | 0.0016 | ||
| Residual | 1149.29 | 5 | 229.86 | |||
| Lack of Fit | 846.99 | 3 | 282.33 | 1.87 | 0.3673 | non-significant |
| Pure error | 302.30 | 2 | 151.15 | |||
| Total | 1,248,572 | 14 | ||||
| R2 | 0.9991 | |||||
| Adj-R2 | 0.9974 | |||||
| TPC | ||||||
| Model | 627.04 | 9 | 69.67 | 34.75 | 0.0006 | significant |
| 134.88 | 1 | 134.88 | 67.27 | 0.0004 | ||
| 10.38 | 1 | 10.38 | 5.18 | 0.0719 | ||
| 9.45 | 1 | 9.45 | 4.71 | 0.0820 | ||
| 39.29 | 1 | 39.29 | 19.60 | 0.0068 | ||
| 250.65 | 1 | 250.65 | 125.01 | <0.0001 | ||
| 81.65 | 1 | 81.65 | 40.72 | 0.0014 | ||
| 5.97 | 1 | 5.97 | 2.98 | 0.1449 | ||
| 1.10 | 1 | 1.10 | 0.5473 | 0.4927 | ||
| 132.72 | 1 | 132.72 | 66.20 | 0.0005 | ||
| Residual | 10.02 | 5 | 2.00 | |||
| Lack of Fit | 1.78 | 3 | 0.5938 | 0.1441 | 0.9251 | non-significant |
| Pure error | 8.24 | 2 | 4.12 | |||
| Total | 637.07 | 14 | ||||
| R2 | 0.9843 | |||||
| Adj-R2 | 0.9559 |
X1: Time (min), X2: Temperature (°C), X3: EtOH (%), SS: Sum of squares, DF: Degree of freedom, MS: Mean square, R2: Quadratic correlation coefficient; Adj-R2: Adjusted quadratic correlation coefficient. TEAC: Trolox equivalent antioxidant capacity; TPC: Total phenolic content.
Based on the statistical analysis, the F-values observed (603.14 and 34.75 for TEAC and TPC, respectively) were significant and the model fitted well, as the p-value was lower than 0.05. The p-value is used to estimate whether F is large enough to indicate statistical significance, and values lower than 0.05 for this parameter indicate that the developed model is statistically significant [18]. Hence, the independent variables affected the TEAC and TPC. The p-values for Lack of Fit in both models were greater than 0.05 (0.3673 and 0.9251, TEAC and TPC respectively). This function is performed by comparing the variability of the residuals in the current model with the variability in the observations under repeated conditions of the factors [19]. The coefficient of determination (R2) estimates the proportion of variation in the response that can be attributed to the model rather than to random error [20]. In our study, the R2 values were 0.9991 and 0.9843 for TEAC and TPC, respectively, whereas the Adj-R2 values were 0.9974 and 0.9559 for TEAC and TPC. High Adj-R2 values are indicative of a high correlation between the observed and the predicted values [18]. Based on the estimated results, it is evident that the developed models have high adequacy and accuracy.
The contribution of coefficients for the response variable is presented in Table 2. It can be observed that the linear (, and ) and quadratic terms ( and ) and the interactive effects () affected the TEAC, while in TPC the highly significant terms were the linear (), quadratic ( and ) and interactive (). The other coefficients were not significant (p > 0.05). Equations considering only the significant terms by RSM models were fitted to predict the responses, which are given below:
| (2) |
| (3) |
where, Y represents the predicting responses;and , and represent time, temperature and % ethanol, respectively.
3.2. Response Surface Analysis
3.2.1. The Effect of Solvent Concentration on TEAC and TPC
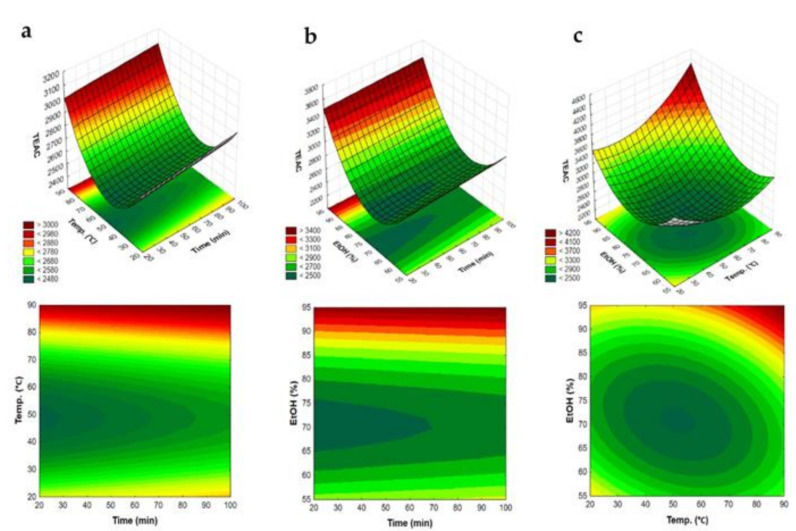
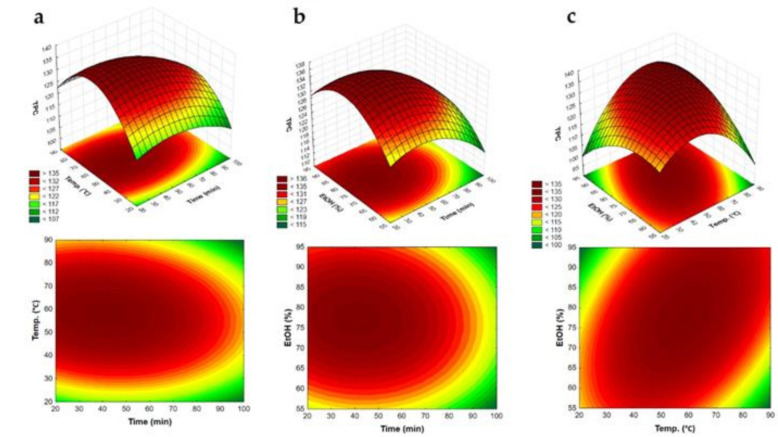
As shown in Figure 1 and Figure 2, solvent concentration significantly altered the content of bioactive compounds recovered from the BRP extract and its antioxidant activity. As the ethanol concentration increased, the efficiency of both phenolic extraction and antioxidant activity was greatly improved. TEAC values raised from 2700 to 3200 µmol TE/g as the ethanol concentration was increased from 75% to 90%. Similarly, TPC values raised from 123 to 135 mg GAE/g as the ethanol concentration was increased from 60% to 90%.
Figure 1.
Response surface plot showing the combined effect of temperature (°C) (a), time (min) (b), and EtOH (%) (c) on the TEAC of Brazilian red propolis extracts.
Figure 2.
Response surface plot showing the combined effect of temperature (°C) (a), time (min) (b), and EtOH (%) (c) on the TPC of Brazilian red propolis extracts.
Our data suggest that the interaction between temperature and ethanol concentration was highly significant. Hence, the phenolic content and TEAC values can be optimized if these parameters are increased. Yet, no improvement in phenolic extraction and antioxidant activity was observed at the highest ethanol concentration (90%) and temperatures below 30 °C. A similar outcome was reported by Roselló-Soto et al. [21] during the optimization of Tiger Nuts byproducts. The authors showed that an increase in ethanol concentration significantly increased the extraction yield at temperatures above 40 °C. Oldoni et et al. [22] also reported that the optimized conditions for extraction of phenolic compounds with antioxidant activity from propolis were 80 °C and 70% ethanol. As proposed by Yang et al. [23], after modifying and penetrating through the cell wall, ethanol affects cell components and improves the chemical extraction, particularly of polyphenols.
Although it has not been extensively discussed in optimization studies, the extraction equipment plays a determining role in the efficiency of the optimization process. When experiments are carried out in non-sealed tubes, volatile solvents or nonpolar compounds may evaporate as rocking and sonification produce heat and increase the solvent temperature. Therefore, the use of an appropriate flask to perform the analysis is as important as the experimental design itself.
3.2.2. The Effect of Temperature on TEAC and TPC
Temperature is one of the variables most frequently examined in natural product optimization studies due to its effects on the content and availability of bioactive compounds, mainly polyphenols. In our study, we found that as the temperature increased so did the TEAC of the extract. The best results were obtained at the highest tested temperature (80°C) 3471.75 µmol TE/g (Figure 1). This value is higher than those found by Andrade et al. [24] when testing the extraction of red, green, and brown propolis at 35 °C (2913.55 ± 95.26; 2214.96 ± 20.61 and 1868.45 ± 131.39 µmol TE/g, respectively).
Moreover, the results showed that when the temperature was escalated from 30 °C to 55 °C, the TPC increased from 122 to 128 mg GAE/g, but it decreased slightly when the temperature was further extended (Figure 2). Maran at al. [18] pointed out that higher temperatures enhance the efficiency of phenolic extraction by decreasing the viscosity and density of the extract. Thereby, higher temperatures enable the solvent to penetrate deeper into the sample matrix and have more contact with the surface area. However, if the temperature is excessively elevated, bioactive compounds may decompose or vaporize.
3.2.3. The Effect of Extraction Time on TEAC and TPC
Figure 1 and Figure 2 show the results of different extraction times on TEAC and TPC outcomes. Although this variable was not strongly associated with TEAC values, a shorter extraction time (30 min) yielded better results. As for TPC, the optimal extraction time was also 30 min. When the time was extended from 60 to 90 min, the number of phenolic compounds recovered was drastically reduced. As stated in the literature, an extended extraction time increases the probability of oxidation, epimerization, and degradation of bioactive compounds [25]. Thus, a prolonged extraction procedure may not be appropriate for all types of natural products [21]. On the other hand, Oldoni et al. [22] reported an increase in the TPC of a propolis type produced in Southern Brazil when using 80% ethanol at 70 °C for 45 min. Yusof et al. [26] observed that the optimal conditions for extraction of phenolics from a Malaysian propolis were 80% ethanol at 60 °C for 25 min. Importantly, we note that the optimal extraction time reported by these authors was shorter than that found in our study, and that it was not possible to predict the extraction efficiency after 60 min.
3.2.4. Optimization and Validation of RSM Models
The optimization of the independent parameters–time (min), ethanol (%) and temperature (°C)—was carried out based on the desirability coefficient (0.8780) to obtain the highest TEAC and TPC (See Supplementary Materials Figure S1). The TEAC and TPC values predicted by the model under optimal conditions (90% ethanol, 80°C, 30 min) were 3550.8 ± 70 µmol TE/g and 132 ± 6.48 mg GAE/g, respectively. The RSM model was validated by comparing the experimental data (n = 3) with the predicted values. The actual TEAC and TPC values obtained under optimal conditions were 3471.76 ± 53.86 µmol TE/g and 129.00 ± 2.16 mg GAE/g, respectively. These findings are similar to the predicted data, indicating that the method is suitable to determine the optimal conditions for extraction of phenolic compounds with antioxidant activity from BRP samples.
3.3. Antioxidant Activity of the Optimized BRP Extract
The antioxidant activity of the optimized BRP extract was evaluated in vitro by single electron transfer and hydrogen atom transfer assays. The FRAP method is based on the reduction of Fe3+ into Fe2+ by antioxidant compounds in the presence of 2,4,6-tris-(2-pyridyl)-s-triazine (TPTZ), forming a colored complex with Fe2+ at 593 nm [27]. The FRAP value obtained for the optimized BRP extract was 1472.86 ± 72.37 µmol Fe2+/g of dry extract. These values were higher than those reported by Calegari et al. [27], who determined the antioxidant activity of 30 propolis samples collected in the states of Paraná and Santa Catarina, Brazil. The authors used Fourier transform near-infrared (FTNIR) spectroscopy and obtained FRAP values ranging from 61.9 to 1770 µmol Fe2+/g of dry weight. Andrade et al. [24] reported a FRAP value of 633.18 ± 40.20 µmol TE/g of dry weight for the BRP extract, while Oldoni et al. [22] found 259.30 ± 9.50 µmol Fe2+/g of dry weight for optimized propolis samples from the state of Paraná, Brazil. When evaluating Croatian propolis from five locations in Adriatic Sea islands, Sveĉnjak et al. [28] observed reducing FRAP activity from 0.1 to 0.8 mmol Fe2+/g of dry weight.
The oxygen radical absorbance capacity (ORAC) assay measures antioxidant inhibition of the peroxyl radical via hydrogen atom transfer reactions. This method is suitable to detect both hydrophilic and hydrophobic antioxidants [29]. For that reason, it is commonly used to determine the antioxidant capacity of different types of natural products, including propolis. In our study, the ORAC value of the BRP extract was 4339.61 ± 114.65 µmol TE/gof dry extract. El-Guendouz et al. [30] examined 24 different samples of Moroccan propolis and found ORAC values ranging from 630.39 ± 33.79 to 1723.28 ± 33.79 µmol TE/g of dry weight. Using 95% ethanol for extraction, Sun et al. [31] reported that Beijing propolis extract had an ORAC value of 1433.72 ± 120 µmol TE/g of dry weight. Finally, a high correlation between the phenolic composition and antioxidant activity of the optimized extract was observed in our study (ABTS, r2 = 0.9925; FRAP = −0.7321 and ORAC = 0.8152).
3.4. Characterization of Phenolic Compounds in the Optimized BRPExtract by LC-ESI-QTOF-MS/MSAnalysis
The qualitative analysis of the BRP extract composition was carried by LC-ESI-QTOF-MS/MS. The compounds were tentatively identified by comparing their m/z values and MS2 spectra in negative mode to the literature findings and corresponding standards.
As shown in Table 3, LC-MS/MS analysis revealed the presence of 32 phenolic compounds in the optimized BRP extract, including flavones, flavanones, flavanonols, chalcones, isoflavonoids, quinone, coumarin, and their derivatives.
Table 3.
LC-ESI-QTOF-MS/MS analysis of phenolic compounds present in Brazilian red propolis.
| Compound | Putative Compound Name | RT (min) | ProposedFormula | [M–H]−(m/z) | MS/MS Fragments (m/z) |
|---|---|---|---|---|---|
| Flavones | |||||
| 1 | Chrysin | 28.6 | C15H10O4 | 253.0510 | 253.0507; 119.0483; 195.0438; 224.0481; 209.0614 |
| 2 | Tricin | 34.2 | C17H14O7 | 329.0677 | 329.0667; 299.0218; 271.0263; 243.0289 |
| 3 | Genkwanin | 36.2 | C16H12O5 | 283.0624 | 268.0360; 283.0583; 269.0397 |
| 4 | Hispidulin | 37.5 | C16H12O6 | 299.0565 | 284.0331; 227.0354; 255.0301; 212.0483 |
| 5 | 8-Hydroxy-5-methoxyflavanone | 44.2 | C16H14O4 | 269.0831 | 254.0589; 252.0437; 195.0451; 210.0685 |
| 6 | Acacetin | 54.2 | C16H12O5 | 283.0617 | 268.0382; 211.0408; 269.042 |
| Flavanones | |||||
| 7 | Liquiritigenin | 25.1 | C15H12O4 | 255.0667 | 119.0495; 135.0083; 255.0656; 120.0526 |
| 8 | Naringenin * | 32.6 | C15H12O5 | 271.0619 | 119.0487; 151.0029; 254.0596; 271.0609; 165.0207 |
| 9 | Pinocembrin * | 48.6 | C15H12O4 | 255.0668 | 255.0678; 240.0426; 151.0034; 133.0285; 213.0540; |
| 10 | 5,6-Dihydroxy-3′,4′-dimethoxyflavanone | 48.7 | C17H16O6 | 315.0882 | 315.0881;151.0037; 235.0636; 255.1042; 121.0292; |
| 11 | 6-Hydroxyflavanone | 57.3 | C15H12O3 | 239.0722 | 239.0732; 135.0091; 197.0643 |
| Chalcones | |||||
| 12 | Isoliquiritigenin * | 41.1 | C15H12O4 | 255.0676 | 119.0496; 135.0082; 120.0531; 151.0384; 255.0665 |
| 13 | 2′,4′-Dihydroxychalcone | 41.9 | C15H12O3 | 239.0723 | 239.0709; 197.0609; 135.0085; 198.0667 |
| 14 | 7-hydroxyflavanone | 42.2 | C15H12O3 | 239.0719 | 197.0610; 135.0085; 239.0732; 198.0643 |
| 15 | 2′,6′-dihydroxy-4′-methoxydihydrochalcone | 45.2 | C16H16O4 | 271.0990 | 254.0590; 135.0444; 109.0287; |
| 16 | 2′-Hydroxy-4′-methoxychalcone | 49.9 | C16H14O3 | 253.0879 | 237.0552; 255.0665; 253.0872; 136.0169; 161.0239 |
| Isoflavonoids | |||||
| 17 | Daidzein * | 28.7 | C15H10O4 | 253.0511 | 253.0513; 208.0523; 119.0488; 135.0089 |
| 18 | Calycosin | 31.9 | C16H12O5 | 283.0617 | 268.0353; 211.0422; 224.0506; 239.0313; 267.0665 |
| 19 | Dihydrobiochanin A | 34.1 | C16H14O5 | 285.0776 | 270.0541; 109.0289; 161.0242; 285.0767 |
| 20 | Vestitone | 34.5 | C16H14O5 | 285.0776 | 270.0535; 161.0240; 109.0286; 271.0607 |
| 21 | Vestitol | 41.4 | C16H16O4 | 271.0987 | 135.0450; 109.0282; 149.0604; 147.0452; 271.0986; 256.0747 |
| 22 | Neovestitol | 41.8 | C16H16O4 | 271.0990 | 135.0360; 109.0217;256.0555; 197.0482; 212.0707 |
| 23 | Formononetin * | 43.9 | C16H12O4 | 267.0666 | 252.0431; 254.0594; 223.0404; 195.0456; 253.0483 |
| 24 | Demethyl medicarpin | 45.2 | C15H12O4 | 255.0673 | 255.0668; 105.0189; 151.0032; 107.0118; 213.0532 |
| 25 | Medicarpin | 48.6 | C16H14O4 | 269.0827 | 254.0594; 225.0540; 105.0191; 121.0300; 133.0287 |
| 26 | Biochanin A * | 52.0 | C16H12O5 | 283.0619 | 268.0389; 239.0354; 211.0393; 132.0202;195.4450 |
| 27 | 5,4′-Dihydroxy-7-methoxyisoflavone | 53.1 | C16H12O5 | 283.0619 | 268.0383; 211.0422; 223.0402; 224.0506; |
| 28 | 3,9-Dimethoxypterocarpan | 63.0 | C17H16O4 | 283.0988 | 253.0515; 225.0564; 268.0754; 183.0456; 254.0554 |
| Flavonols | |||||
| 29 | 7-Hydroxy-6-methoxydihydroflavonol | 30.9 | C16H14O5 | 285.0743 | 270.0534; 268.0383; 78.9984; 123.0078 |
| Neoflavonoids | |||||
| 30 | Dalbergin | 38.3 | C16H12O4 | 267.0667 | 252.0465; 224.0503; 195.0451; 267.0650; 204.9615 |
| Polyprenylated benzophenones | |||||
| 31 | Guttiferone E/Xanthochymol | 92.2 | C38H50O6 | 601.3567 | 109.0291; 108.0214; 202.9997; 177.0198; 335.1285 |
| 32 | Oblongifolin B | 93.8 | C38H50O6 | 601.3569 | 109.0292; 108.0216; 176.0146; 307.1362 |
Bold values indicate the main fragments; RT = retention time; [M−H]− (negative ionization mode) experimental mass of compound. * As compared to an authentic standard.
3.4.1. Flavonoids
Flavonoids are the main class of phenolic compounds in several natural products, including fruits, vegetables, roots, stems and flowers [32]. Multiple studies have revealed the beneficial effects of flavonoids extracted from propolis against human diseases [33]. In our study, a total of 28 flavonoids were identified in the BRP extract, which corresponded to the main chemical group present in the sample.
3.4.2. Flavones
Among the flavonoids detected in the BRP extract, six were flavones. Chrysin (compound 1 with [M–H]− at m/z 253.0510) yielded a fragment at m/z 253.0507 [34]. Tricin (compound 2 with [M–H]− at m/z 329.0667) was tentatively identified based on product ions at m/z 329.0667, 299.0218 [M–H–2CH3]−, 271.0263 [M–H–C2H2O2]−, and 243.0289 [M–H–C4H6O2]−. Genkwanin (compound 3 with [M–H]− at m/z 283.0624) yielded the predominant m/z 268 fragment due to the loss of CH2 from the m/z 283 fragment, resulting in a stable fragment structure [35]. Hispidulin (compound 4) was tentatively identified based on the [M–H]₋ ion at m/z 299.0565, with fragment ions at m/z 284.0331 [M–CH3], 227.0354 [M–CO2–CO]; 255.0301 [M–H–CO2] and 212.0483 [M–H–CO2–CO–CH3] [34]. Compound 5 was tentatively characterized as 8-Hydroxy-5-methoxyflavanon (m/z 269.0831) based on the m/z 254.0589 fragment. Finally, the characteristic [M–H]− ion at m/z 283.0617 and a major fragmentation at m/z 268.0382 were suggestive of acacetin (compound 6) [36].
3.4.3. Flavanones
Retro–Diels–Alder (RDA) is the pathway fragmentation commonly used by flavanones. Fragment ions resulting from RDA fragments are more abundant than the loss of other radical ions, such as CH3, CO, OH, or H2O [37]. Liquiritigenin (compound 7) was detected with [M–H]− at m/z 255.0667. Its identity was confirmed by comparing with data from a previous study, in which Dalbergia odorifera was characterized by LC-MS/MS and based on the spectrum of product ions at m/z 119.0495 ([M–H–C8H8O2]−) and m/z 135.0083, corresponding to breaks of [1,3A–H]− and [1,3B–H]− fragmen [38]. Four flavanones and derivates (compounds 8, 9, 10 and 11) were tentatively identified in the optimized BRP extract as naringenin, pinocembrin, 5,6-Dihydroxy-3′,4′-dimethoxyflavanon and 6-Hydroxyflavanone, according to the precursor ions [M–H]− at m/z 271.0619, 255.0668, 315.0882 and 239.0722, respectively. The identification of naringenin was confirmed by a product ion at m/z 119.0487 [39]. Pinocembrin was identified by comparing our findings with those of a previous report, where this compound was found in leaf extracts of Alpinia zerumbe, yielding the m/z 255.0678 fragment [40]. Pinocembrin is an important marker in BRP, because it is also found in D. ecastaphyllum [9]. The compound 5,6-Dihydroxy-3′,4′-dimethoxyflavanon (compound 10 with [M–H]− at m/z 315.0882), which was found for the first time in BRP, was tentatively identified based on a product ion at m/z 315.0881. Lastly, 6-Hydroxyflavanone displayed a product ion at m/z 239.0732 in the MS2 spectra [37].
3.4.4. Chalcones
Isoliquiritigenin (compound 12 with [M ₋ H]¯ at m/z 255.0676) was previously described in the literature and tentatively identified herein based on product ions at m/z 119.0496 and 135.0082 [41]. The compound 2′,4′-Dihydroxychalcone (compound 13), detected with [M–H]− at m/z 239.0723, was identified based on fragment ions at m/z 239.0709, 197.0609 and 135.0085 [42] This compound was previously reported as an efficient antiviral targeting HlyU in Vibrio vulnificus [43]. Compound 14 was assigned as 7-hydroxyflavanone (m/z 239.0719), yielding the m/z 197.0610 fragment [37]. Compound 15 was detected with [M–H]− at m/z 271.0990 and was tentatively identified as 2′,6′-dihydroxy-4′-methoxydihydrochalcone based on fragment ions at m/z 254.0590, 135.0444 and 109.0287. Compound 16, with [M–H]− at m/z 271.0990, was identified as 2′-Hydroxy-4′-methoxychalcone (C16H13O3−), yielding fragment ions at m/z 237.0552, 255.0665 and 253.0872. This compound was previously found in orange-yellow resin from Zuccagnia punctate [19].
3.4.5. Isoflavonoids
The isoflavones aglycones daidzein (compound 17, m/z 253.0511), formononetin (compound 22, m/z 267.0666) and biochanin A (compound 25, m/z 283.0619) were detected in the BRP extract. Daidzein yielded a product ion at m/z 253.0513 [M–C6H10O5]− as result of a loss of glucoside and another product ion at m/z 135.0089 [M–H–C8H6O]− [44]. The main fragment ions in the MS2 spectra of formononetin corresponded to successive losses of CH3, CHO, and CO [39]. Its MS2 spectra showed fragment ions at m/z 252.0431 [M–H–CH3]−, 223.0404 (C14H7O3) [M–H–CH3–CHO]− and 195.0456 [M–H–CH3–CHO–CO]−. A fragment ion at m/z 268.0389 [M–H–CH3]−, which was produced due to the loss of a CH3 group, and ions at m/z 239.0354 [M–CO2] and 211.0393 [M–CO2–CO], were suggestive of Biochanin A fragmentation [34]. Compound 18, with [M−H]− at m/z 283.0617 (C16H11O5), showed typical product ions at m/z 268.0353 (C15H8O5), 211.0422, 224.0506 and 239.0313. Therefore, it was tentatively classified as calycosin [45]. Dihydrobiochanin A (compound 19 with [M–H]− at m/z 285.0776) and vestitone (compound 20 with [M–H]− at m/z 285.0776) were characterized based on fragment ions at m/z 270.0541 and 270.0535, respectively [46,47]. Vestitol (compound 21), with [M–H]− at m/z 285.0776 (C16H15O4), displayed product ions at m/z 135.0450 (C8H7O2), 109.0282 (C6H5O2) and 149.0604 [48]. Even though compound 22 ([M–H]− at m/z 271.0990) showed a similar fragment to vestitol, it was tentatively identified as neovestitol based on the main fragment ions at m/z 135.0360,197.0482 and 212.0707 [5]. Vestitol and neovestitol have been previously isolated from BRP and were reported to have strong biological properties [5,9]. Compound 24 ([M–H]− at m/z 255.0673) was tentatively identified as dimethyl medicarpin based on fragment ions at m/z 255.0668, 151.0032, 107.0118 and 213.0532. Compound 25 was characterized as medicarpin according to the precursor ion at m/z 269.0827. In its MS2 spectra, the following typical product ions were detected: 254.0594([M–H–CH3•]−), 225.0540, 105.0191, 121.0300 (C7H5O2, 3,5A−)and 133.0287 [49]. Compound 27 (with [M–H]− at m/z 283.0619) and compound 28 (with [M–H]− at m/z 283.0988) were tentatively identified as 5,1′-Dihydroxy-7-methoxyisoflavone and 3,9-Dimethoxypterocarpan, respectively. In their MS2 spectra, 5,1′-Dihydroxy-7-methoxyisoflavone displayed fragment ions at m/z 268.0383 [M–H–CH3], 211.0422 [M–CO2–CO] and 223.0402 [M–H–CO–H2O] [34].
3.4.6. Flavonols, Neoflavonoids, Coumarins, and Polyprenylated Benzophenone Derivates
Dalbergin (compound 30 with [M–H]− at m/z 267.0667) yielded product ions at m/z 252.0465 and 224.0503, corresponding to the loss of a CO2 and further loss of H2O from the precursor ion [47]. Compound 29 (with [M–H]− at m/z 285.0743) was tentatively identified as 7-Hydroxy-6-methoxydihydroflavonol based on fragment ions at m/z 270.0534. Guttiferone E (Compound 31) displayed deprotonated molecular ion at m/z 601.3571. Its MS2 spectra showed fragment ions at m/z 109.0291 [C6O2H5]−, 108.0214, 202.9997, 177.0198 [M–H–C10H16O]−, 335.1285 [50]. Lastly, compound 32 (with [M–H]− at m/z 601.3670) was tentatively identified as oblongifolin B, yielding a fragment ion at m/z 109.0292.
In addition to comparing the tentative compounds with the literature, we further compared the data against the electronic database available from metadata-centric approaches, such as the Mass Bank of North America (MoNA) and the Mass Bank. The LC-QTOF-ESI-MS/MS analysis in negative mode enabled the identification for the first time in BRP of the following phenolic compounds: Flavones (tricin, genkwanin, hispidulin and 8-Hydroxy-5-methoxyflavanone), flavanones (5,6-Dihydroxy-3′,4′-dimethoxyflavanone and 6-Hydroxyflavanone), chalcones (2′,4′-Dihydroxychalcone and 2′,6′-dihydroxy-4′-methoxydihydrochalcone), isoflavonoids (dihydrobiochanin A, demethyl medicarpin, 5,4′-Dihydroxy-7-methoxyisoflavone and 3,9-Dimethoxypterocarpan) and flavanols (7-Hydroxy-6-methoxydihydroflavonol).
4. Conclusions
The optimization of conditions for extraction of antioxidant compounds from BRP extract was successfully performed using the Response Surface Methodology. The optimal extraction conditions for stronger antioxidant activity and higher phenolic content were 90% ethanol at 80 °C for 30 min. Under optimized conditions, BRP extract showed a TPC of 129.00 mg GAE/g. When examined for its antioxidant activity, the TEAC, FRAP and ORAC assays revealed that the optimized extract had 3471.76 µmol TE/g, 1472.86 µmol Fe2+/g and 4339.61 µmol TE/gof dry extract, respectively. Thirty-two phenolic compounds were tentatively identified by LC-QTOF-ESI-MS/MS, of which thirteen were found for the first time in BRP. Our study may guide further research in the field, since this is the first study that reported the optimization of the extraction of phenolic compounds from BRP samples. Collectively, our results highlight the importance of BRP as a source of a wide variety of phenolic compounds with significant antioxidant properties.
Acknowledgments
The authors thank beekeepers from Maceió, AL, Brazil, for providing the propolis samples.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-3921/10/2/297/s1, Figure S1 Coefficient of desirability for optimization of TEAC and TPC in Brazilian red propolis extracts as a function of time (min), temperature (°C) and EtOH concentration (%).
Author Contributions
Conceptualization, D.V.d.M. and S.M.d.A.; methodology, D.V.d.M., A.P.d.S.S., A.P.M., and S.M.d.A.; software, D.V.d.M.; validation, D.V.d.M. and S.M.d.A.; formal analysis, D.V.d.M.; investigation, D.V.d.M.; resources, S.M.d.A. and A.P.M.; data curation, D.V.d.M. and S.M.d.A.; writing—original draft preparation, D.V.d.M. and S.M.d.A.; writing—review and editing, S.M.d.A., M.I., A.P.d.S.S., and P.L.R.; visualization, D.V.d.M. and S.M.d.A.; supervision, S.M.d.A.; project administration, S.M.d.A., M.I., and P.L.R.; funding acquisition, D.V.d.M. and S.M.d.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by theCoordenação de Aperfeiçoamento de Pessoal de Nível Superior—CAPES (Finance Code 001), National Council for Scientific and Technological Development—CNPq (grant number 870493/1997-3) and The São Paulo Research Foundation—FAPESP (grant number 19/11248-0).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data is contained within the article and Supplementary Materials.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ghisalberti E.L. Propolis: A Review. Bee World. 1979;60:59–84. doi: 10.1080/0005772X.1979.11097738. [DOI] [Google Scholar]
- 2.Silva B.B., Rosalen P.L., Cury J.A., Ikegaki M., Souza V.C., Esteves A., Alencar S.M. Chemical composition and botanical origin of red propolis, a new type of Brazilian propolis. Evid. BasedComplement. Altern. Med. 2008;5:313–316. doi: 10.1093/ecam/nem059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Freires I.A., De Alencar S.M., Rosalen P.L. A pharmacological perspective on the use of Brazilian Red Propolis and its isolated compounds against human diseases. Eur. J. Med. Chem. 2016;110:267–279. doi: 10.1016/j.ejmech.2016.01.033. [DOI] [PubMed] [Google Scholar]
- 4.Franchin M., Freires I.A., Lazarini J.G., Nani B.D., da Cunha M.G., Colón D.F., de Alencar S.M., Rosalen P.L. The use of Brazilian propolis for discovery and development of novel anti-inflammatory drugs. Eur. J. Med. Chem. 2018;153:49–55. doi: 10.1016/j.ejmech.2017.06.050. [DOI] [PubMed] [Google Scholar]
- 5.Franchin M., Colón D.F., Da Cunha M.G., Castanheira F.V.S., Saraiva A.L.L., Bueno-Silva B., Alencar S.M., Cunha T.M., Rosalen P.L. Neovestitol, an isoflavonoid isolated from Brazilian red propolis, reduces acute and chronic inflammation: Involvement of nitric oxide and IL-6. Sci. Rep. 2016;6:1–12. doi: 10.1038/srep36401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alencar S.M., Cadorin T.L., Castro M.L., Ribeiro I.S., Costa Neto C.M., Cury J.A., Rosalen P.L., Ikegaki M. Chemical composition and biological activity of a new type of Brazilian propolis: Red propolis. J. Ethnopharmacol. 2007;113:278–283. doi: 10.1016/j.jep.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 7.Piccinelli A.L., Lotti C., Campone L., Cuesta-Rubio O., Campo Fernandez M., Rastrelli L. Cuban and Brazilian red propolis: Botanical origin and comparative analysis by high-performance liquid chromatography-photodiode array detection/electrospray ionization tandem mass spectrometry. J. Agric. Food Chem. 2011;59:6484–6491. doi: 10.1021/jf201280z. [DOI] [PubMed] [Google Scholar]
- 8.Bueno-Silva B., Marsola A., Ikegaki M., Alencar S.M., Rosalen P.L. The effect of seasons on Brazilian red propolis and its botanical source: Chemical composition and antibacterial activity. Nat. Prod. Res. 2017;31:1318–1324. doi: 10.1080/14786419.2016.1239088. [DOI] [PubMed] [Google Scholar]
- 9.Ccana-Ccapatinta G.V., Mejía J.A.A., Tanimoto M.H., Groppo M., de Carvalho J.C.A.S., Bastos J.K. Dalbergia ecastaphyllum (L.) Taub. and Symphonia globulifera L.f.: The botanical sources of isoflavonoids and benzophenones in Brazilian red propolis. Molecules. 2020;25:2060. doi: 10.3390/molecules25092060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Franchin M., Cólon D.F., Castanheira F.V.S., da Cunha M.G., Bueno-Silva B., Alencar S.M., Cunha T.M., Rosalen P.L. Vestitol isolated from Brazilian red propolis inhibits neutrophils migration in the inflammatory process: Elucidation of the mechanism of action. J. Nat. Prod. 2016;79:954–960. doi: 10.1021/acs.jnatprod.5b00938. [DOI] [PubMed] [Google Scholar]
- 11.Oldoni T.L.C., Cabral I.S.R., d’Arce M.A.B.R., Rosalen P.L., Ikegaki M., Nascimento A.M., Alencar S.M. Isolation and analysis of bioactive isoflavonoids and chalcone from a new type of Brazilian propolis. Sep. Purif. Technol. 2011;77:208–213. doi: 10.1016/j.seppur.2010.12.007. [DOI] [Google Scholar]
- 12.Novak E.M., Silva M.S.E.C., Marcucci M.C., Sawaya A.C.H.F., Giménez-Cassina López B., Fortes M.A.H.Z., Giorgi R.R., Marumo K.T., Rodrigues R.F., Maria D.A. Antitumoural activity of Brazilian red propolis fraction enriched with xanthochymol and formononetin: An in vitro and in vivo study. J. Funct. Foods. 2014;11:91–102. doi: 10.1016/j.jff.2014.09.008. [DOI] [Google Scholar]
- 13.Abd El-Salam E.A., Morsy N.F.S. Optimization of the extraction of polyphenols and antioxidant activity from Malva parviflora L. leaves using Box–Behnken design. Prep. Biochem. Biotechnol. 2019;49:876–883. doi: 10.1080/10826068.2019.1633667. [DOI] [PubMed] [Google Scholar]
- 14.Riswanto F.D.O., Rohman A., Pramono S., Martono S. Application of response surface methodology as mathematical and statistical tools in natural product research. J. Appl. Pharm. Sci. 2019;9:125–133. doi: 10.7324/JAPS.2019.91018. [DOI] [Google Scholar]
- 15.Jovanović A.A., Đorđević V.B., Zdunić G.M., Pljevljakušić D.S., Šavikin K.P., Gođevac D.M., Bugarski B.M. Optimization of the extraction process of polyphenols from Thymus serpyllum L. herb using maceration, heat- and ultrasound-assisted techniques. Sep. Purif. Technol. 2017;179:369–380. doi: 10.1016/j.seppur.2017.01.055. [DOI] [Google Scholar]
- 16.Al-Duais M., Müller L., Böhm V., Jetschke G. Antioxidant capacity and total phenolics of Cyphostemmadigitatum before and after processing: Use of different assays. Eur. Food Res. Technol. 2009;228:813–821. doi: 10.1007/s00217-008-0994-8. [DOI] [Google Scholar]
- 17.Melo P.S., Massarioli A.P., Denny C., Dos Santos L.F., Franchin M., Pereira G.E., Vieira T.M.F.D.S., Rosalen P.L., De Alencar S.M. Winery by-products: Extraction optimization, phenolic composition and cytotoxic evaluation to act as a new source of scavenging of reactive oxygen species. Food Chem. 2015;181:160–169. doi: 10.1016/j.foodchem.2015.02.087. [DOI] [PubMed] [Google Scholar]
- 18.Prakash Maran J., Manikandan S., Vigna Nivetha C., Dinesh R. Ultrasound assisted extraction of bioactive compounds from Nephelium lappaceum L. fruit peel using central composite face centered response surface design. Arab. J. Chem. 2017;10:S1145–S1157. doi: 10.1016/j.arabjc.2013.02.007. [DOI] [Google Scholar]
- 19.Martín-García B., Pimentel-Moral S., Gómez-Caravaca A.M., Arráez-Román D., Segura-Carretero A. A Box-Behnken design for optimal green extraction of compounds from olive leaves that potentially activate the AMPK pathway. Appl. Sci. 2020;10:4620. doi: 10.3390/app10134620. [DOI] [Google Scholar]
- 20.Gaur P.K., Shanmugam S.K. Box-Behnken design–directed optimization of Wickerhamomycesanomalus–mediated biotransformation process to enhance the flavonoid profile of polyherbal extract. J. Pharm. Innov. 2020:1–12. doi: 10.1007/s12247-020-09467-9. [DOI] [Google Scholar]
- 21.Roselló-Soto E., Martí-Quijal F.J., Cilla A., Munekata P.E.S., Lorenzo J.M., Remize F., Barba F.J. Influence of temperature, solvent and pH on the selective extraction of phenolic compounds from tiger nuts by-products: Triple-TOF-LC-MS-MS characterization. Molecules. 2019;24:797. doi: 10.3390/molecules24040797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oldoni T.L.C., Oliveira S.C., Andolfatto S., Karling M., Calegari M.A., Sado R.Y., Maia F.M.C., Alencar S.M., Lima V.A. Chemical characterization and optimization of the extraction process of bioactive compounds from propolis produced by selected bees Apis mellifera. J. Braz. Chem. Soc. 2015;26:2054–2062. doi: 10.5935/0103-5053.20150186. [DOI] [Google Scholar]
- 23.Yang L., Jiang J.G., Li W.F., Chen J., Wang D.Y., Zhu L. Optimum extraction process of polyphenols from the bark of Phyllanthus emblica L. based on the response surface methodology. J. Sep. Sci. 2009;32:1437–1444. doi: 10.1002/jssc.200800744. [DOI] [PubMed] [Google Scholar]
- 24.Andrade J.K.S., Denadai M., de Oliveira C.S., Nunes M.L., Narain N. Evaluation of bioactive compounds potential and antioxidant activity of brown, green and red propolis from Brazilian northeast region. Food Res. Int. 2017;101:129–138. doi: 10.1016/j.foodres.2017.08.066. [DOI] [PubMed] [Google Scholar]
- 25.Vuong Q.V., Golding J.B., Stathopoulos C.E., Nguyen M.H., Roach P.D. Optimizing conditions for the extraction of catechins from green tea using hot water. J. Sep. Sci. 2011;34:3099–3106. doi: 10.1002/jssc.201000863. [DOI] [PubMed] [Google Scholar]
- 26.Yusof N., Munaim M.S.A., VelooKutty R. Optimization of total phenolic compounds extracted from propolis by ultrasound-assisted extraction. Chem. Eng. Commun. 2020:1–9. doi: 10.1080/00986445.2020.1761799. [DOI] [Google Scholar]
- 27.Calegari M.A., Ayres B.B., dos Santos Tonial L.M., de Alencar S.M., Oldoni T.L.C. Fourier transform near infrared spectroscopy as a tool for predicting antioxidant activity of propolis. J. King Saud Univ. Sci. 2020;32:784–790. doi: 10.1016/j.jksus.2019.02.006. [DOI] [Google Scholar]
- 28.Svečnjak L., Marijanović Z., Okińczyc P., Kuś P.M., Jerković I. Mediterranean propolis from the adriatic sea islands as a source of natural antioxidants: Comprehensive chemical biodiversity determined by GC-MS, ftiratr, UHPLC-DAD-QQTOF-MS, DPPH and FRAP assay. Antioxidants. 2020;9:337. doi: 10.3390/antiox9040337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lewoyehu M., Amare M. Comparative evaluation of analytical methods for determining the antioxidant activities of honey: A review. Cogent Food Agric. 2019;5:5. doi: 10.1080/23311932.2019.1685059. [DOI] [Google Scholar]
- 30.El-Guendouz S., Aazza S., Lyoussi B., Bankova V., Popova M., Neto L., Faleiro M.L., Da Graça Miguel M. Moroccan Propolis: A Natural Antioxidant, Antibacterial, and Antibiofilm against Staphylococcus aureus with no induction of resistance after continuous exposure. Evid. Based Complement. Altern. Med. 2018;2018:1–19. doi: 10.1155/2018/9759240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun C., Wu Z., Wang Z., Zhang H. Effect of ethanol/water solvents on phenolic profiles and antioxidant properties of Beijing propolis extracts. Evid. Based Complement. Altern. Med. 2015;2015:1–9. doi: 10.1155/2015/595393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Panche A.N., Diwan A.D., Chandra S.R. Flavonoids: An overview. J. Nutr. Sci. 2016;5:e47. doi: 10.1017/jns.2016.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Watanabe M.A.E., Amarante M.K., Conti B.J., Sforcin J.M. Cytotoxic constituents of propolis inducing anticancer effects: A review. J. Pharm. Pharmacol. 2011;63:1378–1386. doi: 10.1111/j.2042-7158.2011.01331.x. [DOI] [PubMed] [Google Scholar]
- 34.Peng Q., Shang X., Zhu C., Qin S., Zhou Y., Liao Q., Zhang R., Zhao Z., Zhang L. Qualitative and quantitative evaluation of Oroxylum indicum (L.) Kurz by HPLC and LC-qTOF-MS/MS. Biomed. Chromatogr. 2019;33:e4657. doi: 10.1002/bmc.4657. [DOI] [PubMed] [Google Scholar]
- 35.Yuan L., Liang C., Diao X., Cheng X., Liao M., Zhang L. Metabolism studies on hydroxygenkwanin and genkwanin in human liver microsomes by UHPLC-Q-TOF-MS. Xenobiotica. 2018;48:332–341. doi: 10.1080/00498254.2017.1319991. [DOI] [PubMed] [Google Scholar]
- 36.Hossain M.B., Rai D.K., Brunton N.P., Martin-Diana A.B., Barry-Ryan A.C. Characterization of phenolic composition in lamiaceae spices by LC-ESI-MS/MS. J. Agric. Food Chem. 2010;58:10576–10581. doi: 10.1021/jf102042g. [DOI] [PubMed] [Google Scholar]
- 37.Chen Q., He L., Mo C., Zhang Z., Long H., Gu X., Wei Y. Rapid evaluation of chemical consistency of artificially induced and natural resinadraconis using Ultra-Performance Liquid Chromatography Quadrupole-Time-Of-Flight mass spectrometry-based chemical profiling. Molecules. 2018;23:1850. doi: 10.3390/molecules23081850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao X., Zhang S., Liu D., Yang M., Wei J. Analysis of flavonoids in Dalbergia odorifera by ultra-performance liquid chromatography with tandem mass spectrometry. Molecules. 2020;25:389. doi: 10.3390/molecules25020389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rini Vijayan K.P., Raghu A.V. Tentative characterization of phenolic compounds in three species of the genus Embelia by liquid chromatography coupled with mass spectrometry analysis. Spectrosc. Lett. 2019;52:653–670. doi: 10.1080/00387010.2019.1682013. [DOI] [Google Scholar]
- 40.Ghareeb M.A., Sobeh M., Rezq S., El-Shazly A.M., Mahmoud M.F., Wink M. HPLC-ESI-MS/MS profiling of polyphenolics of a leaf extract from Alpinia zerumbet (Zingiberaceae) and its anti-inflammatory, anti-nociceptive, and antipyretic activities in vivo. Molecules. 2018;23:3238. doi: 10.3390/molecules23123238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu Y., Wang P., Yang H., Sui F. UPLC–Q-TOF–MS and UPLC–MS/MS methods for metabolism profiles and pharmacokinetics of major compounds in XuanmaiGanjie Granules. Biomed. Chromatogr. 2019;33:1–13. doi: 10.1002/bmc.4449. [DOI] [PubMed] [Google Scholar]
- 42.Agüero M.B., Gonzalez M., Lima B., Svetaz L., Sánchez M., Zacchino S., Feresin G.E., Schmeda-Hirschmann G., Palermo J., Daniel Wunderlin A.N.D., et al. Argentinean propolis from Zuccagnia punctata cav. (Caesalpinieae) exudates: Phytochemical characterization and antifungal activity. J. Agric. Food Chem. 2010;58:194–201. doi: 10.1021/jf902991t. [DOI] [PubMed] [Google Scholar]
- 43.Imdad S., Batool N., Pradhan S., Chaurasia A.K., Kim K.K. Identification of 2,4-dihydroxychalcone as an antivirulence agent targeting hlyu, a master virulence regulator in vibrio vulnificus. Molecules. 2018;23:1492. doi: 10.3390/molecules23061492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peng H., Li W., Li H., Deng Z., Zhang B. Extractable and non-extractable bound phenolic compositions and their antioxidant properties in seed coat and cotyledon of black soybean (Glycine max (L.) merr) J. Funct. Foods. 2017;32:296–312. doi: 10.1016/j.jff.2017.03.003. [DOI] [Google Scholar]
- 45.Qi Y., Li S., Pi Z., Song F., Lin N., Liu S., Liu Z. Chemical profiling of Wu-tou decoction by UPLC-Q-TOF-MS. Talanta. 2014;118:21–29. doi: 10.1016/j.talanta.2013.09.054. [DOI] [PubMed] [Google Scholar]
- 46.Zhao C., Liu Y., Cong D., Zhang H., Yu J., Jiang Y., Cui X., Sun J. Screening and determination for potential α-glucosidase inhibitory constituents from Dalbergia odorifera T. Chen using ultrafiltration-LC/ESI-MSn. Biomed. Chromatogr. 2013;27:1621–1629. doi: 10.1002/bmc.2970. [DOI] [PubMed] [Google Scholar]
- 47.Zhong B., Robinson N.A., Warner R.D., Barrow C.J., Dunshea F.R., Suleria H.A.R. LC-ESI-QTOF-MS/MS characterization of seaweed phenolics and their antioxidant potential. Mar. Drugs. 2020;18:331. doi: 10.3390/md18060331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Omar R.M.K., Igoli J., Gray A.I., Ebiloma G.U., Clements C., Fearnley J., EdradaEbel R.A., Zhang T., De Koning H.P., Watson D.G. Chemical characterisation of Nigerian red propolis and its biological activity against Trypanosoma brucei. Phytochem. Anal. 2016;27:107–115. doi: 10.1002/pca.2605. [DOI] [PubMed] [Google Scholar]
- 49.Wang H.Y., Li T., Ji R., Xu F., Liu G.X., Li Y.L., Shang M.Y., Cai S.Q. Metabolites of medicarpin and their distributions in rats. Molecules. 2019;24:1966. doi: 10.3390/molecules24101966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.de Souza E.C.A., da Silva E.J.G., Cordeirob H.K.C., Filhob N.M.L., da Felipe Silvac M.A., dos Reisd D.L.S., Portod, Carla Pilaud E.J., da Costaa L.A.M.A., De A.D.L.S., Menezese C.F.A. Chemical compositions and antioxidant and antimicrobial activities of propolis Produced by Frieseomelittalongipes and Apis mellifera bees. Quim. Nova. 2018;41:485–491. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data is contained within the article and Supplementary Materials.