Abstract
Dysregulation of Wnt signaling is implicated in multiple ocular disorders. The roles of Wnt co-receptors LRP5 and LRP6 in Wnt signaling regulation remain elusive, as most retinal cells express both of the co-receptors. To address this question, LRP5 and LRP6 were individually knocked-out in a human retinal pigment epithelium cell line using the CRISPR-Cas9 technology. Wnt signaling activity induced by various Wnt ligands was measured using wild-type and the KO cell lines. The results identified three groups of Wnt ligands based on their co-receptor specificity: 1) activation of Wnt signaling only through LRP6, 2) through both LRP5 and LRP6 and 3) predominantly through LRP5. These results indicate that LRP5 and LRP6 have differential roles in Wnt singling regulation.
Keywords: Wnt signaling, Ocular disorder, LRP5, LRP6, CRISPR-Cas9 technology, Wnt ligand
1. Introduction
Wnt signaling plays a crucial role in embryogenesis and maintenance of homeostasis in adult tissues [1–3]. Dysregulation of Wnt signaling results in birth defects, and diseases such as cancer, age-related macular degeneration (AMD) and diabetic retinopathy (DR) [1–6]. Therefore, this ancient and highly conserved signaling pathway is pivotal for the embryonic development, physiological and pathological processes. Wnt signaling transduces through either a canonical or a non-canonical pathway [1–3]. In canonical Wnt signaling, binding of a Wnt ligand to a ternary complex of a Frizzled (FZD) receptor and a co-receptor low-density lipoprotein receptor-like protein 5 (LRP5) or LRP6 is required for the stabilization of cytosolic β-catenin [1–3,7]. In the absence of Wnt ligand binding, glycogen synthase kinase-3β (GSK-3β), a component of the destruction kinase complex, phosphorylates cytosolic β-catenin, leading to degradation [1–3]. When Wnt ligand binds to the receptor complex (a FZD receptor and LRP5 or LRP6), the co-receptors are phosphorylated, and the destruction complex is dissociated, leading to stabilization of cytosolic, non-phosphorylated β-catenin [1–3]. Accumulated non-phosphorylated β-catenin translocates into the nucleus and binds to a family of T cell factor/lymphoid enhancer factor (TCF/LEF-1) transcription factors, activating transcription of Wnt target genes including vascular endothelial growth factor (VEGF), cyclin D, c-Myc, interleukin-8 (IL-8), and matrix metalloproteinase-3 (MMP3) [1–3,7–12].
There are 19 Wnt ligands and 10 FZDs in mammals, while there are only two co-receptors, LRP5 and LRP6 [1–3]. Therefore, blocking LRP5 and/or LRP6 is considered a promising therapeutic approach to abrogate aberrant Wnt signal activation. LRP5/6 are both single-pass transmembrane receptors [1–3]. The extracellular domains of LRP5/6 contain the binding sites for Wnt ligands and Wnt antagonists [13–15]. Although LRP5 and LRP6 share 71% sequence homology, LRP5 knockout mice are viable, whereas LRP6 knockout mice die prenatally [1,16,17]. Thus, these co-receptors possess distinct functions at least during development (embryogenesis and vasculogenesis) and may mediate different physiological and pathological events in adult tissues [18]. Their functional differences in the complex Wnt biology remain uncertain, as most cell types in the retina express both LRP5 and LRP6 [19,20].
To define the distinct roles of the co-receptors in the regulation of Wnt signaling in the retinal degenerative diseases with aberrant activation of Wnt signaling, the present study generated LRP5 knockout (KO) and LRP6 KO in ARPE-19 [21], a cell line derived from the human RPE, using the CRISPR-Cas9 technology [22,23]. Using these KO cell lines, we examined specificities of canonical Wnt ligands toward LRP5 or LRP6 and regulatory roles of these co-receptors in Wnt signaling activation.
2. Materials and Methods
2.1. Generation of LRP5 KO and LRP6 KO RPE cell lines
The guide RNAs (gRNAs) to knock out the human LRP5 and LRP6 genes were designed using online applications (target finder and off-spotter). Prediction software showed that there is no potential off-target gene in the protein coding sequences for the designed LRP5 gRNA, while the LRP6 gRNA possesses 2 potential off-target genes with 4 mismatches in 20 bases of the target sequence. The synthesized gRNA oligos were hybridized and sub-cloned into the vector expressing both Cas9 nuclease and the designed gRNA (Plasmid ID, PX459) from Addgene (Cambridge, MA). Following sequence verification of gRNAs, the constructed vectors for knocking out the LRP5 and LRP6 genes were transfected into ARPE-19 cells, and transfected cells were selected with puromycin. Genomic DNA modification and protein expression of puromycinresistant (potential LRP5 KO and LRP6 KO) cells were verified by PCR product mismatch cleavage assay (using a Guide-it Mutation Detection kit (Clontech, Mountain View, CA) and Western blot analysis, respectively. Moreover, the genomic DNA sequences of the LRP5 and LRP6 genes containing respective gRNA target sites were analyzed by DNA sequencing. All DNA oligos and primers used in this study are shown in Supplementary Table 1.
2.2. Preparation of Wnt3A Conditioned Medium
Control L cell line (CRL-2648, ATCC, Manassas, VA) and Wnt3A cell line, L cells stably expressing Wnt3A (CRL-2647, ATCC), were cultured in low-glucose DMEM containing 10% FBS. Conditioned media from the control L cells (LCM) and Wnt3A cells (WCM) without G-418 were generated and collected according to ATCC’s recommendation followed by sterile filtration.
2.3. Western blot analysis
Following treatment or transfection, WT, LRP5 KO (KO5) and LRP6 KO (KO6) cells were harvested and total cellular proteins were extracted in lysis buffer. Western blot and densitometry analyses were carried out as described previously [24,25].
The following antibodies were used for Western blot analysis in this study: HRP- conjugated secondary antibodies (anti-mouse (Cat. No. PI-2000), anti-rabbit (Cat. No. PI-1000) were purchased from Vector Laboratory (Burlingame, CA). The anti-LRP5 (Cat. No. 5440), anti-phosphorylated LRP6 (Cat. No. 2568) antibodies were purchased from Cell Signaling Technology (Danvers, MA). A monoclonal anti-LRP6 antibody (2F1) was generated in our lab as described previously [26]. A monoclonal anti-β-actin antibody (Cat. No. A2228) was purchased from Sigma (St. Louis, MO). An anti-VEGF (Cat. No. sc-152) antibody was purchased from Santa Cruz Biotechnology (Dallas, TX).
2.4. Cell treatments, plasmid transfection and TCF/β-catenin activity (TOP-flash) assay
Equal numbers (7×104 cells/well) of cells were seeded in 24-well culture plates one day prior to transfection. The cells were treated with 50% of LCM or WCM for another 24 hrs, or co-transfected with an empty plasmid, plasmids expressing Wnt ligands (a gift from Dr. Xi He [27] through Addgene) together with Wnt reporter plasmids (a super TOP-flash vector [28] and a vector expressing Renilla luciferase (pRL-TK, Promega, Madison, WI) as an internal control) using Lipofectamine 2000 (ThermoFisher, Waltham, MA). For receptor rescue analysis, the plasmids expressing LRP5 [29] or LRP6 [15] were co-transfected with Wnt reporter plasmids. At 24 hrs post-transfection, TCF/β-catenin activity was measured with the dual luciferase reporter system (Promega) using a GloMax microplate reader (Promega) following the manufacturer’s instruction.
2.5. Cell growth curve
The cells were seeded in culture dishes at the same density (1×104 cells/dish). Then the cells were trypsinized, and the viable cells were counted using a hemocytometer after trypan blue staining for 5 consecutive days and the culture media were replaced once at day 3.
2.6. Statistical analysis
All the data are presented as mean ± SEM. The data were obtained from three independent biological experiments unless stated. Statistical analysis was performed using a Prism version 7 (GraphPad Software, San Diego, CA). Unpaired two-tailed Student’s t-test was used for comparison between two groups and one-way ANOVA with Tukey’s post hoc analysis was used for multiple group comparisons.
3. Results
3.1. LRP5 and LRP6 knockout in the ARPE-19 cell line using the CRISPR-Cas9 system
The LRP5 and an LRP6 genes were knocked-out in ARPE-19 cell line using the CRISPR-Cas9 system. PCR product mismatch cleavage assays demonstrated the expected genomic modifications in the LRP5 and LRP6 genes in these KO cell lines, without detectable off-target effects (Fig. 1A). Western blot analyses confirmed that LRP5 and LRP6 expression was abolished in the respective cell lines, suggesting a successful KO of the desired genes (Fig. 1B). In addition, the genomic DNA sequences of respective gRNA target sites in the LRP5 and LRP6 genes in respective KO cell lines (Suppl. Fig. 1), and also two putative off-target genes (SOGA1 and LMF1) for the LRP6 gRNA in the KO6 cells (Suppl. Fig. 2) were analyzed. We verified successful knockout of the LRP5 and LRP6 genes without off-target gene modifications. Furthermore, lithium chloride (LiCl), a GSK-3β inhibitor, intracellularly activated Wnt signaling in wild-type (WT), LRP5 KO (KO5) and LRP6 KO (KO6) cells to a similar extent (Fig. 1C), indicating that intracellular Wnt signaling components are intact in these KO cells.
Figure 1. LRP5 and LRP6 knockout (KO) in ARPE-19 cell line.
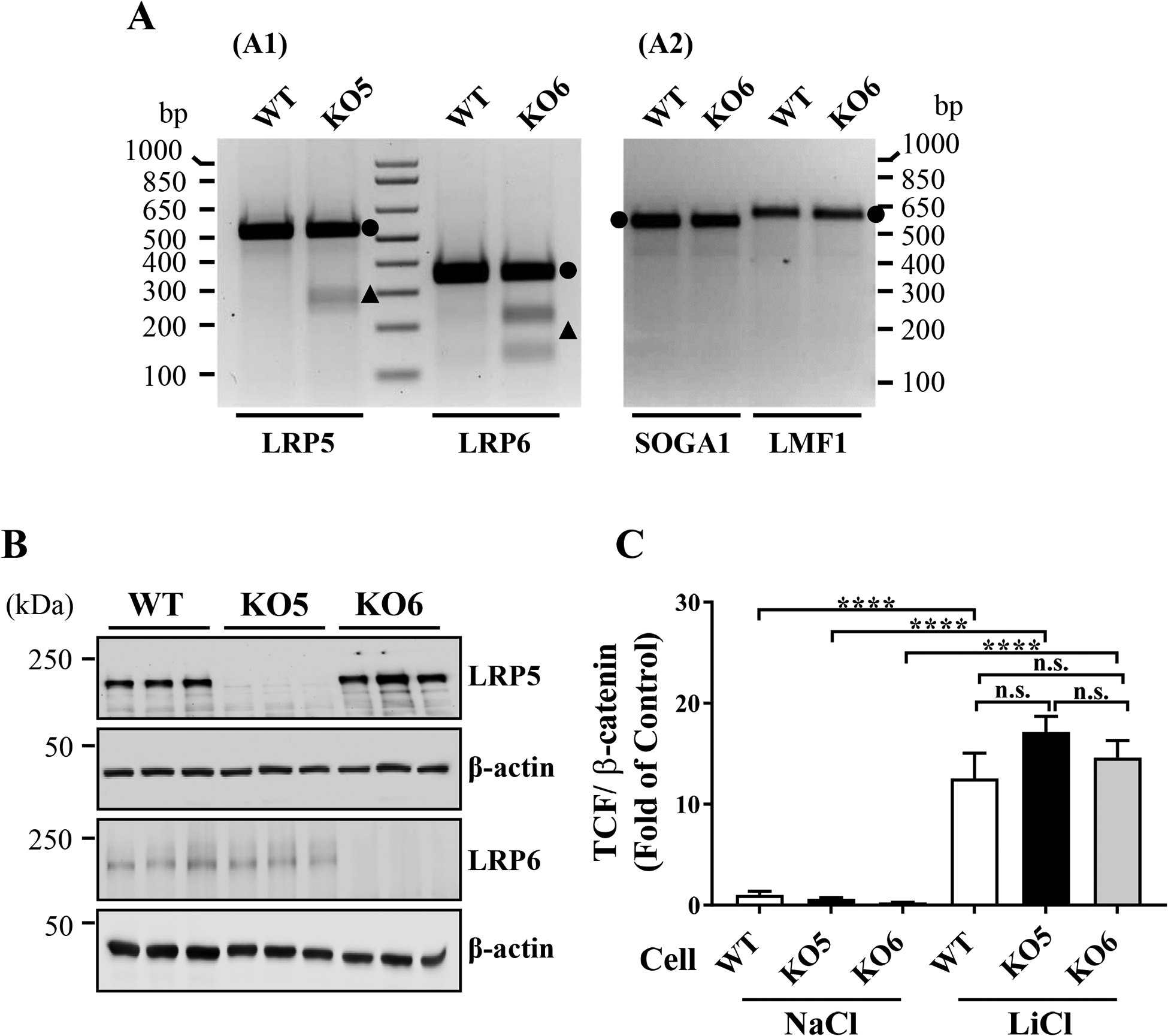
(A) Genomic DNA modifications in LRP5 KO (KO5) and LRP6 KO (KO6) ARPE-19 cell lines were verified using PCR product mismatch cleavage assay. Compared to wild-type (WT) cells, the assay demonstrated indel formation in the LRP5 gene in the KO5 cell line and the LRP6 gene in the KO6 cell line (A1), but no indel formation in the potential off-target genes for the LRP6 gRNA (SOGA1 and LMF1) genes (A2). Agarose gel electrophoresis showed full-length PCR products (circles) and cleaved fragments (triangles). (B) Equal amounts of total cellular proteins from three independent experimental samples of WT, KO5 and KO6 cell lines were immunoblotted for LRP5, LRP6 and β-actin, respectively. (C) TCF/β-catenin (Wnt signaling) activities in WT, KO5 and KO6 cell lines were measured after 24 hrs treatment of NaCl or LiCl. TCF/β-catenin activities are presented as fold of control in mean ± SEM, and one-way ANOVA with Tukey’s post hoc analysis was performed (n=9, n.s.: not significant, **** P<0.0001).
3.2. Impacts of LRP5 KO and LRP6 KO on cell morphology and growth rate
KO5 and KO6 cell lines showed morphology similar to that of WT cells (Suppl. Fig. 3A). However, the KO6 cell line grew at a significantly higher rate compared to the KO5 cell line and WT cells, while the KO5 cell line showed a growth rate lower than that of WT cells (Suppl. Fig. 3B).
3.3. Ligand specificities of the Wnt co-receptors
To examine differential specificities toward Wnt ligands, TCF/β-catenin activities in the WT, KO5 and KO6 cells were measured at 24 hrs after transfection of plasmids expressing Wnt3A, Wnt1 and Wnt2 ligands. Wnt3A increased TCF/β-catenin activity in WT and KO5 cells compared to the empty control plasmid, but not in KO6 cells (Fig. 2A), suggesting that Wnt3A activated canonical Wnt signaling only through LRP6. Next, Wnt1 increased TCF/β-catenin activity in all WT, KO5 and KO6 cells, but with lesser extent in KO6 cells (Fig. 2B), suggesting that Wnt1 ligand activates canonical Wnt signaling through both LRP5 and LRP6, and is more affected by LRP6. In contrast, Wnt2 increased TCF/β-catenin activity in WT and KO6 cells, while the Wnt2-induced Wnt activation was diminished in KO5 cells (Fig. 2C), suggesting that Wnt2 activated canonical Wnt signaling predominantly through LRP5. Further, the Western blot analysis of Wnt3A, Wnt1 and Wnt2 showed similar expression levels in these cell lines, suggesting that the observed differences in Wnt signaling activation were not due to different expression levels of Wnt ligands.
Figure 2. Ligand specificity for individual Wnt co-receptors.
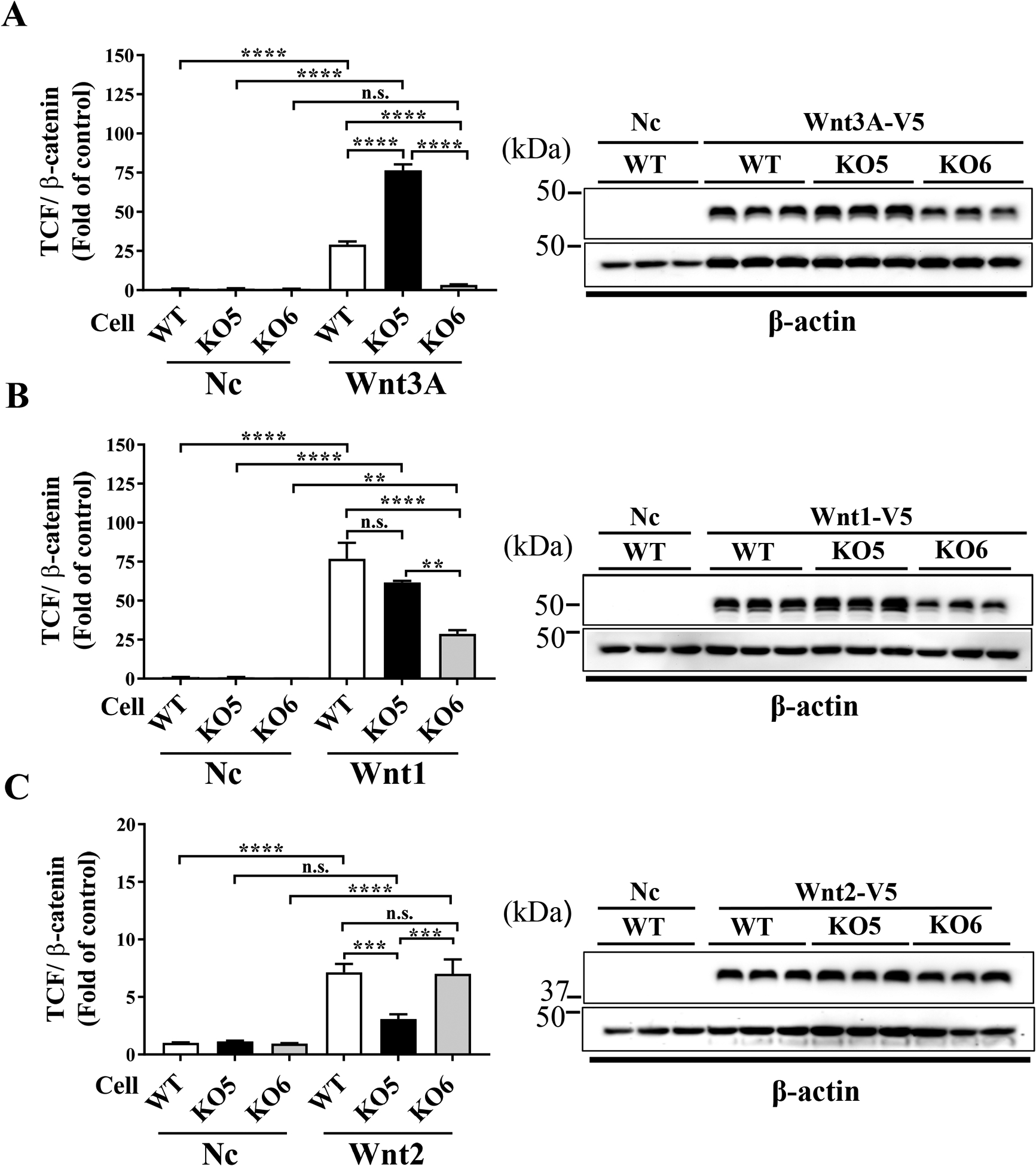
TCF/β-catenin activities in WT, KO5 and KO6 cells were measured at 24 hrs post-transfection of plasmids expressing Wnt3A (A), Wnt1 (B), Wnt2 (C) or an empty control plasmid (Nc), respectively. Representative Western blot images showed levels of each Wnt ligand (upper) and β–actin (lower) as a loading control. TCF/β-catenin activities are presented as fold of the empty plasmid control in WT cells, and one-way ANOVA with Tukey’s post hoc analysis was performed (n=3 (Wnt1), n=9 (Wnt2 and Wnt3A), n.s.: not significant, ** P<0.01, ***P<0.001, **** P<0.0001).
It should be noted that we have obtained more than one independent clones for each KO cell line, and these clones showed results similar to those in the original clone reported in the manuscript (data not shown), indicating that the different TCF/β-catenin activities in KO5 and KO6 cell lines were ascribed to the knock-out of the LRP5 and LRP6 genes.
Using the same strategy, we have examined co-receptor specificities of 6 more canonical Wnt ligands (Suppl. Fig. 4). The results showed that the examined Wnt ligands can be categorized into three groups based on their specificities toward LRP5 and LRP6 (Table 1): Group 1: Wnt ligands activate Wnt signaling only through LRP6 (Wnt3, Wnt3A, Wnt6, Wnt8A, and Wnt8B). Group 2: Wnt ligands activate Wnt signaling through both LRP5 and LRP6 (Wnt1, Wnt7B and Wnt9A). Group 3: Wnt ligands activate Wnt signaling predominantly through LRP5 (Wnt2).
Table 1:
Classification of Wnt ligands specificities toward LRP5/LRP6 receptors
| Only through LRP6 | Both LRP5 and LRP6 | Predominantly through LRP5 |
|---|---|---|
| Wnt3 | Wnt1 (more affected by LRP6) | Wnt2 |
| Wnt3A | Wnt7B | |
| Wnt6 | Wnt9A (more affected by LRP5) | |
| Wnt8A | ||
| Wnt8B |
3.4. Verification of Wnt signaling activation by Wnt3A conditioned media
To verify the results of luciferase-based TCF/β-catenin activity assay by the Wnt3A expression plasmid, we further examined Wnt signaling activation by Wnt3A-conditioned media (WCM) at: 1) the co-receptor level, by phosphorylation of LRP6 and 2) the expression level of Wnt target genes (e.g. VEGF). WCM significantly increased levels of phosphorylated LRP6 (pLRP6) in WT and KO5 cells, but not in KO6 cells, compared to that of control LCM (Fig. 3A). It should be noted that weak signals in pLRP6 Western blot analysis in KO6 cells were likely non-specific signals possibly due to cross-reaction of the anti-pLRP6 antibody with LRP5, as our KO6 cells did not show detectable total LRP6 (Fig. 1B). Further, WCM increased levels of VEGF, one of the Wnt target genes, in WT and KO5 cells, but not in KO6 cells, compared to LCM (Fig. 3B). These results indicated that Wnt3A activated canonical Wnt signaling through LRP6, as demonstrated by increased levels of pLRP6 and expression of VEGF in WT and KO5 cells, but not in KO6 cells, consistent with the result from the TCF/β-catenin activity assay (Fig. 2A).
Figure 3. Confirmation of Wnt signaling activation by Wnt3A conditioned media.
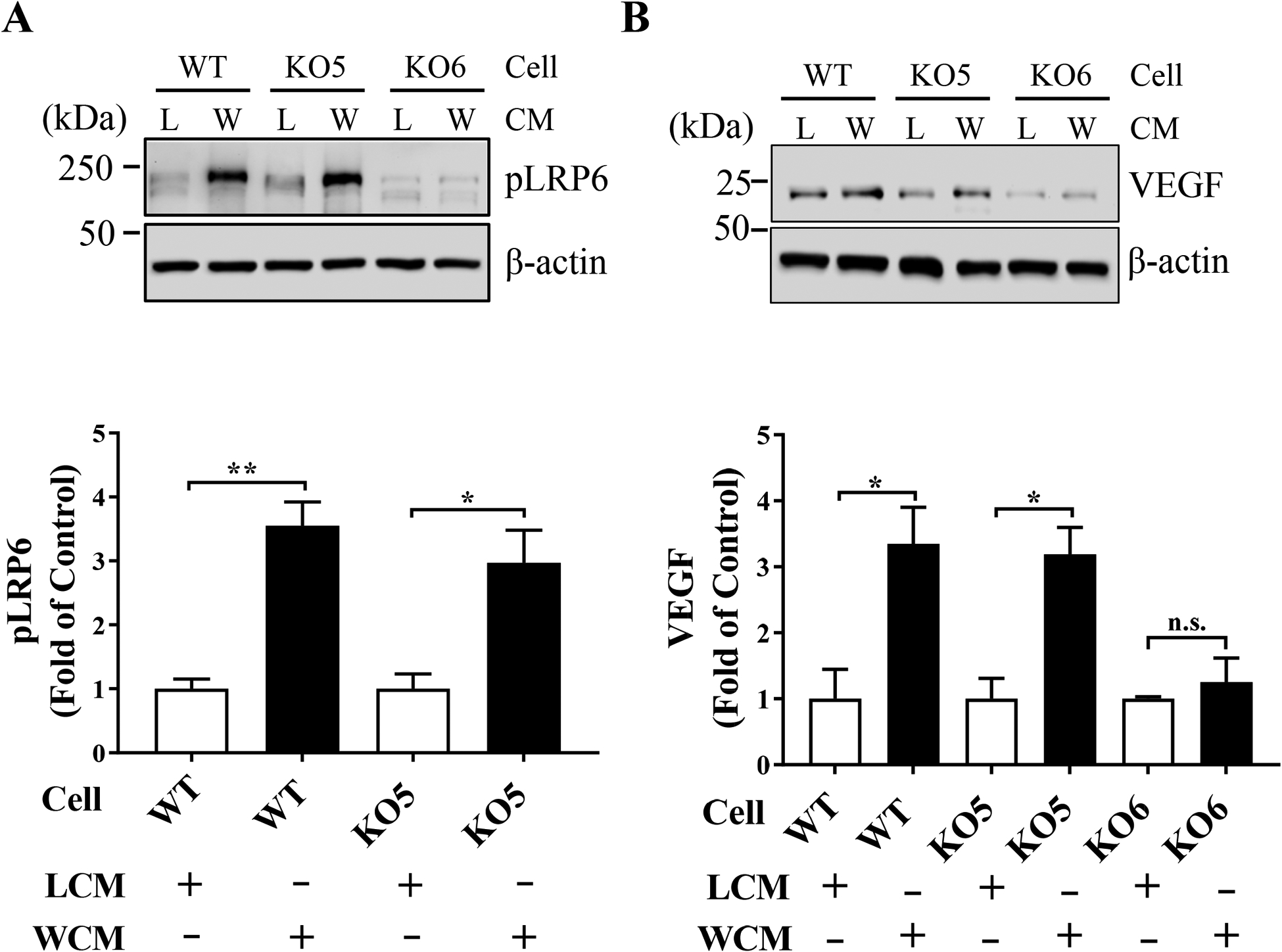
WT, KO5 and KO6 cells were treated with a control L-cell conditioned medium (L) or Wnt3A conditioned medium (W) for 24 hrs. Whole cell lysates of the treated cells were used for Western blot analyses. Representative images showed levels of phosphorylated LRP6 (pLRP6) (A) and VEGF (B) with β-actin as a loading control. Levels of pLRP6 and VEGF were semi-quantified by densitometry, normalized by β-actin and presented as fold of control in L-cell medium-treated cells (n=3, * P<0.05, ** P<0.01 by Student’s t-test).
3.5. Rescuing effect of co-receptor expression on Wnt signaling in their respective knockout cell lines
We further examined whether restored expression of the Wnt co-receptors (LRP5 and LRP6) can rescue the abrogated TCF/β-catenin activity in KO5 and KO6 cells. TCF/β-catenin activity assay showed that transfection of the LRP5 expression plasmid restored Wnt2-mediated Wnt signaling in KO5 cells (Suppl. Fig. 5A). Similarly, transfection of an LRP6 expression plasmid restored Wnt3A-mediated Wnt signaling activation in KO6 cells (Suppl. Fig. 5B). These results further support that intracellular Wnt signaling components in KO5 and KO6 cells remain intact.
3.6. Both LRP5 and LRP6 mediate A2E-induced Wnt signal activation
Finally, we investigated whether an AMD stressor, A2E, activated Wnt signaling through either LRP5 or LRP6. TCF/β-catenin activity assay showed that A2E substantially induced Wnt signal activation in WT, KO5 and KO6 cells. However, KO5 and KO6 cells showed significant reductions in A2E-induced Wnt signaling compared to that of WT cells, and there was no significant difference between KO5 and KO6 cells, suggesting A2E activates Wnt signaling through both LRP5 and LRP6 (Suppl. Fig. 6).
4. Discussion
Extensive studies have established that dysregulation of Wnt signaling plays key pathogenic roles in ocular diseases including diabetic retinopathy and AMD [1–6]. However, most cell types in the retina express both LRP5 and LRP6 [19,20], and thus, their individual roles in mediating Wnt signaling are not completely understood. Therefore, it is imperative to fill this critical knowledge gap in the regulation of Wnt signaling. The present study for the first time knocked out LRP5 and LRP6 individually in RPE cells using the CRISPR system [21–23] and identified differential roles of LRP5 and LRP6 in mediating Wnt signaling. Our studies for the first time demonstrated that most Wnt ligands activate Wnt signaling through LRP6, suggesting a predominant role of LRP6 in mediating Wnt signaling, and that LRP6 may have a higher functional significance at least in the retina relative to LRP5. These findings may explain why LRP6 KO is embryonically lethal [1,16], whereas LRP5 KO does not affect viability of mice, although it causes retinal vascular pathologies [1,17].
An earlier study reported that LRP5 knock-down in mammary epithelial cells resulted in a lower growth rate relative to WT and LRP6 knock-down cells [30]. Consistent with this study, our LRP5 KO ARPE-19 cells grew at a slower rate relative to WT and LRP6 KO cells, although the KO5 and KO6 cells showed morphology similar to that of WT cells (Suppl. Fig. 3). We also demonstrated that LRP6 KO cells showed a higher growth rate than that of WT and LRP5 KO cell lines (Suppl. Fig. 3). The difference in the growth rate between LRP6 KO RPE cells and LRP6 knock-down mammary epithelial cells can be attributed to the different origins of the cells (mammary epithelial vs RPE cells) or methods of gene ablation (shRNA-mediated knock-down vs complete knockout using the CRISPR-Cas9 system).
In the present study, 9 canonical Wnt ligands were examined for their differential specificities toward co-receptors using the LRP5 KO and LRP6 KO cells (Table 1 and Suppl. Fig. 4). A previous study using mouse embryonic fibroblasts (MEFs) reported that Wnt3A activates Wnt signaling only through LRP6, and Wnt1 activates canonical Wnt signaling through both LRP5 and LRP6 [31]. This study was consistent with our current findings. Also earlier studies using AMD models showed that Wnt3A was upregulated in the eye cup in the laser-induced choroidal neovascularization model [6], and Wnt3A, Wnt7A and Wnt10A mRNAs were significantly upregulated in the retinas of oxygen-induced retinopathy mice model [32], suggesting that, at least, Wnt3A and its receptor LRP6 play roles in pathological neovascularization in AMD. Our study allowed us to analyze co-receptor specificities of Wnt ligands expressed in the retina and filled a knowledge gap regarding activation of Wnt signaling through the co-receptors.
VEGF is one of the Wnt target genes and a key pathogenic factor in wet AMD [10,33]. Currently, anti-VEGF drugs are widely used clinically to treat pathological angiogenesis and vascular dysfunction in cancer, wet AMD and DR [33–35]. However, these therapies are not always effective in all patients. Therefore, blocking Wnt signaling has potential to become an effective approach for those patients who do not respond to the anti-VEGF therapy, since Wnt signaling is an up-stream signaling pathway regulating multiple pro-angiogenic and pro-inflammatory factors in addition to VEGF. Combination of a Wnt signaling blocker and an anti-VEGF drug may generate synergistic effects. Blocking LRP6 may be a promising therapeutic approach, as LRP6 is a dominant receptor and ocular injection of an LRP6-blocking antibody showed partial alleviation of pathogenic features in AMD and DR [6,26].
In summary, to study the distinct role of LRP5 and LRP6 in the retina and RPE, we successfully generated the co-receptor KO cells using the CRISPR-Cas9 technology without disturbing the intracellular Wnt signaling cascade or detectable off-target effect. As ARPE-19 cells have a high transfection efficiency, our KO cells can be used as models to examine other genes interacting with LRP5 and LRP6 and can be used to examine the roles of LRP5 and LRP6 in disease–induced aberrant Wnt signaling activation.
Supplementary Material
Highlights.
LRP5 and LRP6, Wnt co-receptors are both co-expressed in RPE cells.
LRP5 and LRP6 individually knockout in a RPE cells line using CRISPR-Cas9 technology.
LRP5 and LRP6 showed differential ligand specificities for canonical Wnt ligands.
More Wnt ligand activate Wnt signaling through LRP6.
Acknowledgements:
We thank Dr. Xi He from the Harvard University for kindly providing the LRP6 expression plasmid for this study. We also thank Dr. Lori Garman from the Quantitative Analysis Core at Oklahoma Medical Research Foundation for assistance with biostatistical analyses. This study was supported by National Institutes of Health (NIH; EY018659, EY012231, EY019309, EY028949, GM122744), Juvenile Diabetes Research Foundation (JDRF; 2-SRA-2019-711-S-B), Presbyterian Health Foundation (PHF; Research Support Grant Program and Bridge Grant Program).
Abbreviations:
- AMD
Age-related macular degeneration
- CRISPR
Clustered Regularly Interspaced Short Palindromic Repeats
- DR
Diabetic retinopathy
- FGF
Fibroblast growth factor
- FZD
Frizzled
- GSK3β
Glycogen Synthase Kinase 3β
- gRNAs
guide RNAs
- KO
Knock out
- KO5
Knock out of LRP5
- KO6
Knock out of LRP6
- LRP
Low-density lipoprotein receptor-related protein
- MEF
mouse embryonic fibroblast
- MMP
Matrix metallopeptidase
- RPE
Retinal pigment epithelial
- shRNA
Short hairpin RNA
- TOP flash
TCF Reporter Plasmid
- TCF/LEF-1
T cell factor/lymphoid enhancer factor
- VEGF
Vascular endothelial growth factor
- WT
Wild type
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of competing interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- [1].He X, Semenov M, Tamai K, et al. , LDL receptor-related proteins 5 and 6 in Wnt/beta-catenin signaling: arrows point the way, Development (Cambridge, England) 131 (2004) 1663–1677, 10.1242/dev.01117 [DOI] [PubMed] [Google Scholar]
- [2].MacDonald BT, Tamai K, He X, Wnt/beta-catenin signaling: components, mechanisms, and diseases, Developmental cell 17 (2009) 9–26, 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Kikuchi A, Yamamoto H, Sato A, et al. , New insights into the mechanism of Wnt signaling pathway activation, International review of cell and molecular biology 291 (2011) 21–71, 10.1016/b978-0-12-386035-4.00002-1. [DOI] [PubMed] [Google Scholar]
- [4].Chen Q, Ma JX, Canonical Wnt signaling in diabetic retinopathy, Vision Res 139 (2017) 47–58, 10.1016/j.visres.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Tuo J, Wang Y, Cheng R, et al. , Wnt signaling in age-related macular degeneration: human macular tissue and mouse model, J Transl Med 13 (2015) 330, 10.1186/s12967-015-0683-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Hu Y, Chen Y, Lin M, et al. , Pathogenic role of the Wnt signaling pathway activation in laser-induced choroidal neovascularization, Invest Ophthalmol Vis Sci 54 (2013) 141–154, 10.1167/iovs.12-10281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Cong F, Schweizer L, Varmus H, Wnt signals across the plasma membrane to activate the beta-catenin pathway by forming oligomers containing its receptors, Frizzled and LRP, Development (Cambridge, England) 131 (2004) 5103–5115, 10.1242/dev.01318. [DOI] [PubMed] [Google Scholar]
- [8].Hoppler S, Kavanagh CL, Wnt signalling: variety at the core, Journal of cell science 120 (2007) 385–393, 10.1242/jcs.03363. [DOI] [PubMed] [Google Scholar]
- [9].Masckauchan TN, Shawber CJ, Funahashi Y, et al. , Wnt/beta-catenin signaling induces proliferation, survival and interleukin-8 in human endothelial cells, Angiogenesis 8 (2005) 43–51, 10.1007/s10456-005-5612-9. [DOI] [PubMed] [Google Scholar]
- [10].Zhang X, Gaspard JP, Chung DC, Regulation of vascular endothelial growth factor by the Wnt and K-ras pathways in colonic neoplasia, Cancer Res 61 (2001) 6050–6054. [PubMed] [Google Scholar]
- [11].Tetsu O, McCormick F, Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells, Nature 398 (1999) 422–426, 10.1038/18884. [DOI] [PubMed] [Google Scholar]
- [12].He TC, Sparks AB, Rago C, et al. , Identification of c-MYC as a target of the APC pathway, Science 281 (1998) 1509–1512, 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- [13].Mao B, Wu W, Li Y, et al. , LDL-receptor-related protein 6 is a receptor for Dickkopf proteins, Nature 411 (2001) 321–325, 10.1038/35077108 [DOI] [PubMed] [Google Scholar]
- [14].Cheng Z, Biechele T, Wei Z, et al. , Crystal structures of the extracellular domain of LRP6 and its complex with DKK1, Nat Struct Mol Biol 18 (2011) 1204–1210, 10.1038/nsmb.2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Zhang B, Abreu JG, Zhou K, et al. , Blocking the Wnt pathway, a unifying mechanism for an angiogenic inhibitor in the serine proteinase inhibitor family, Proceedings of the National Academy of Sciences of the United States of America 107 (2010) 6900–6905, 10.1073/pnas.0906764107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Pinson KI, Brennan J, Monkley S, et al. , An LDL-receptor-related protein mediates Wnt signalling in mice, Nature 407 (2000) 535–538, 10.1038/35035124 [DOI] [PubMed] [Google Scholar]
- [17].Kato M, Patel MS, Levasseur R, et al. , Cbfa1-independent decrease in osteoblast proliferation, osteopenia, and persistent embryonic eye vascularization in mice deficient in Lrp5, a Wnt coreceptor, The Journal of cell biology 157 (2002) 303–314, 10.1083/jcb.200201089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Joiner DM, Ke J, Zhong Z, et al. , LRP5 and LRP6 in development and disease, Trends Endocrinol Metab 24 (2013) 31–39, 10.1016/j.tem.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Xia CH, Lu E, Zeng J, et al. , Deletion of LRP5 in VLDLR knockout mice inhibits retinal neovascularization, PLoS One 8 (2013) e75186, 10.1371/journal.pone.0075186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Chen Y, Hu Y, Lu K, et al. , Very low density lipoprotein receptor, a negative regulator of the wnt signaling pathway and choroidal neovascularization, The Journal of biological chemistry 282 (2007) 34420–34428, 10.1074/jbc.M611289200 [DOI] [PubMed] [Google Scholar]
- [21].Dunn KC, Aotaki-Keen AE, Putkey FR, et al. , ARPE-19, a human retinal pigment epithelial cell line with differentiated properties, Experimental eye research 62 (1996) 155–169, 10.1006/exer.1996.0020. [DOI] [PubMed] [Google Scholar]
- [22].Ran FA, Hsu PD, Wright J, et al. , Genome engineering using the CRISPR-Cas9 system, Nature Protocols 8 (2013) 2281–2308, 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Mali P, Yang L, Esvelt KM, et al. , RNA-guided human genome engineering via Cas9, Science 339 (2013) 823–826, 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Murray AR, Chen Q, Takahashi Y, et al. , MicroRNA-200b downregulates oxidation resistance 1 (Oxr1) expression in the retina of type 1 diabetes model, Invest Ophthalmol Vis Sci 54 (2013) 1689–1697, 10.1167/iovs.12-10921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Schneider CA, Rasband WS, Eliceiri KW, NIH Image to ImageJ: 25 years of image analysis, Nature methods 9 (2012) 671–675, 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Lee K, Hu Y, Ding L, et al. , Therapeutic potential of a monoclonal antibody blocking the Wnt pathway in diabetic retinopathy, Diabetes 61 (2012) 2948–2957, 10.2337/db11-0300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].MacDonald BT, Hien A, Zhang X, et al. , Disulfide bond requirements for active Wnt ligands, The Journal of biological chemistry 289 (2014) 18122–18136, 10.1074/jbc.M114.575027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Veeman MT, Slusarski DC, Kaykas A, et al. , Zebrafish prickle, a modulator of noncanonical Wnt/Fz signaling, regulates gastrulation movements, Current biology : CB 13 (2003) 680–685, 10.1016/s0960-9822(03)00240-9 [DOI] [PubMed] [Google Scholar]
- [29].He X, Cheng R, Huang C, et al. , A novel role of LRP5 in tubulointerstitial fibrosis through activating TGF-β/Smad signaling, Signal transduction and targeted therapy 5 (2020) 45, 10.1038/s41392-020-0142-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Chin EN, Martin JA, Kim S, et al. , Lrp5 Has a Wnt-Independent Role in Glucose Uptake and Growth for Mammary Epithelial Cells, Mol Cell Biol 36 (2015) 871–885, 10.1128/MCB.00800-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Goel S, Chin EN, Fakhraldeen SA, et al. , Both LRP5 and LRP6 receptors are required to respond to physiological Wnt ligands in mammary epithelial cells and fibroblasts, The Journal of biological chemistry 287 (2012) 16454–16466, 10.1074/jbc.M112.362137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Wang Z, Liu CH, Huang S, et al. , Wnt Signaling in vascular eye diseases, Progress in retinal and eye research 70 (2019) 110–133, 10.1016/j.preteyeres.2018.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Lynch SS, Cheng CM, Bevacizumab for neovascular ocular diseases, The Annals of pharmacotherapy 41 (2007) 614–625, 10.1345/aph.1H316. [DOI] [PubMed] [Google Scholar]
- [34].Bagri A, Kouros-Mehr H, Leong KG, et al. , Use of anti-VEGF adjuvant therapy in cancer: challenges and rationale, Trends in molecular medicine 16 (2010) 122–132, 10.1016/j.molmed.2010.01.004. [DOI] [PubMed] [Google Scholar]
- [35].Cheung N, Wong IY, Wong TY, Ocular anti-VEGF therapy for diabetic retinopathy: overview of clinical efficacy and evolving applications, Diabetes care 37 (2014) 900–905, 10.2337/dc13-1990. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


