Abstract
An essential function of DNA topoisomerase IIα (TOP2α; 170 kDa, TOP2α/170) is to resolve DNA topologic entanglements during chromosome disjunction by introducing transient DNA double-stranded breaks. TOP2α/170 is an important target for DNA damage-stabilizing anticancer drugs, whose clinical efficacy is compromised by drug resistance often associated with decreased TOP2α/170 expression. We recently demonstrated that an etoposide-resistant K562 clonal subline, K/VP.5, with reduced levels of TOP2α/170, expresses high levels of a novel C-terminal truncated TOP2α isoform (90 kDa, TOP2α/90). TOP2α/90, the translation product of a TOP2α mRNA that retains a processed intron 19 (I19), heterodimerizes with TOP2α/170 and is a resistance determinant through a dominant-negative effect on drug activity. We hypothesized that genome editing to enhance I19 removal would provide a tractable strategy to circumvent acquired TOP2α-mediated drug resistance. To enhance I19 removal in K/VP.5 cells, CRISPR/Cas9 was used to make changes (GAG//GTAAAC→GAG//GTAAGT) in the TOP2α gene’s suboptimal exon 19/intron 19 5′ splice site (E19/I19 5′ SS). Gene-edited clones were identified by quantitative polymerase chain reaction and verified by sequencing. Characterization of a clone with all TOP2α alleles edited revealed improved I19 removal, decreased TOP2α/90 protein, and increased TOP2α/170 protein. Sensitivity to etoposide-induced DNA damage (γH2AX, Comet assays) and growth inhibition was restored to levels comparable to those in parental K562 cells. Together, the results indicate that our gene-editing strategy for optimizing the TOP2α E19/I19 5′ SS in K/VP.5 cells circumvents resistance to etoposide and other TOP2α-targeted drugs.
SIGNIFICANCE STATEMENT
Results presented here indicate that CRISPR/Cas9 gene editing of a suboptimal exon 19/intron 19 5′ splice site in the DNA topoisomerase IIα (TOP2α) gene results in circumvention of acquired drug resistance to etoposide and other TOP2α-targeted drugs in a clonal K562 cell line by enhancing removal of intron 19 and thereby decreasing formation of a truncated TOP2α 90 kDa isoform and increasing expression of full-length TOP2α 170 kDa in these resistant cells. Results demonstrate the importance of RNA processing in acquired drug resistance to TOP2α-targeted drugs.
Introduction
The human DNA topoisomerase II (TOP2α; 170 kDa, TOP2α/170) enzyme functions as a homodimer to resolve DNA topology by introducing transient DNA double-stranded breaks (DSBs) essential for chromosomal segregation during mitosis (Deweese and Osheroff, 2009; Nitiss, 2009). TOP2α/170 enzymatic activity is necessary for cell survival and is highly expressed in rapidly proliferating cells. This has made TOP2α/170 an important target in cancer therapy (Chen et al., 2013; Pommier et al., 2016). Type IIA topoisomerase interfacial inhibitors, such as etoposide, stabilize the enzyme-DNA complexes by insertion within the break sites generated by TOP2α/170, thereby inhibiting religation, resulting in DSBs and triggering cell death (Pommier and Marchand, 2011).
Resistance to TOP2α interfacial poisons is frequently associated with a reduction of TOP2α/170 expression levels or its altered subcellular localization (Ganapathi and Ganapathi, 2013; Capelôa et al., 2020). We previously demonstrated that acquired resistance to etoposide in a human K562 leukemia cell line, K/VP.5, is associated with decreased TOP2α/170 mRNA/protein expression levels and a dramatically increased expression of a novel TOP2α mRNA (University of California Santa Cruz Genome Browser accession number MH936673), which retains a processed intron 19 (I19) and encodes a 90-kDa TOP2α isoform now designated TOP2α/90 (Kanagasabai et al., 2017; Elton et al., 2020). Importantly, the TOP2α/90 isoform heterodimerizes with TOP2α/170, resulting in a dominant-negative effect with respect to etoposide-induced covalent TOP2α-DNA complexes, DNA damage, and cytotoxicity (Kanagasabai et al., 2018), thereby functioning as a resistance determinant.
Over 95% of genes undergo alternative pre-mRNA splicing, a process by which a single pre-mRNA is matured into multiple mRNA isoforms (Lee and Rio, 2015). This leads to the expression of different mRNA isoforms and is responsible for proteomic diversity. Several types of alternative splicing of a pre-mRNA have been described, including intron retention (Lee and Rio, 2015). Although most intron-retaining mRNA transcripts are susceptible to nuclear intron detention (Boutz et al., 2015) or nonsense-mediated decay (Kurosaki and Maquat 2016), some intron-retaining transcripts leave the nucleus and undergo translation to produce new protein isoforms with novel functions (Li et al., 2016; Uzor et al., 2018; Shoubridge et al., 2019; Wang et al., 2019b). This process seems to occur in a number of TOP2α intron-retaining mRNA variants that are translated into novel truncated TOP2α isoforms and play a role in chemoresistance (Harker et al., 1995; Yu et al., 1997; Mo and Beck, 1997; Kanagasabai et al., 2017, 2018; Elton et al., 2020).
Intron retention is regulated by a complex combination of cis- and trans-acting factors (Monteuuis et al., 2019). One cis-acting sequence feature is the presence of weak or suboptimal splice site (SS) at the 5′ and/or 3′ ends of the intron, which can impede the spliceosome’s ability to recognize introns that should be spliced out (Hicks et al., 2010; Huang et al., 2012; Eckert et al., 2016). Studies investigating intron retention events have demonstrated that removal of retained introns could be enhanced by strengthening the suboptimal 5′ SS by mutation in a minigene system (Wickramasinghe et al., 2015) or by CRISPR/CRISPR-associated protein 9 (Cas9) gene editing (Yue and Ogawa, 2018).
Since weak splice sites are inefficiently recognized by the spliceosome, which, in part, can lead to intron retention (Monteuuis et al., 2019), the human TOP2α gene was subjected to SS analyses (Splice Site Score Calculation; http://rulai.cshl.edu/new_alt_exon_db2/HTML/score.html). This analysis revealed that the TOP2α exon 19 (E19)/I19 5′ SS (GAG//GTAAAC) is suboptimal, with a score of 6.1 out of a maximum score of 12.4 for the optimal consensus 5′ SS (CAG//GTAAGT). Hence, we hypothesized that this weak SS influences I19 retention and that by mutating/gene editing the TOP2α E19/I19 5′ SS in etoposide-resistant K/VP.5 cells, sensitivity to etoposide would be restored.
Transfection experiments utilizing a TOP2α/Minigene (i.e., a plasmid that harbors a TOP2α gene segment encompassing E19 through exon 20 [E20]) demonstrated that mutating the suboptimal wild-type E19/I19 5′ SS (GAG//GTAAAC) to a consensus 5′ SS (CAG//GTAAGT) decreased I19 retention in K/VP.5 cells, providing “proof of concept” for CRISPR/Cas9 editing as a viable strategy to circumvent resistance. Therefore, the CRISPR/Cas9 system with homology-directed repair (HDR) (Jinek et al., 2012; Mali et al., 2013; Liang et al., 2017) was used to introduce specific gene edits (GAG//GTAAAC→GAG//GTAAGT) in the suboptimal TOP2α E19/I19 5′ SS in K/VP.5 cells. Notably, in K/VP.5 edited cells, intron 19 retention was attenuated, resulting in decreased formation of TOP2α/90, restoration of full-length TOP2α/170 levels, and increased etoposide-induced DNA damage and growth inhibitory effects comparable to those seen in parental K562 cells. Together, these results demonstrate that CRISPR/Cas9 editing of the TOP2α gene circumvents acquired drug resistance to etoposide and other TOP2α-targeted drugs.
Materials and Methods
Cell Culture and Acute Myeloid Leukemia Blasts.
Human K562 leukemia cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) (Corning, Manassas, VA) supplemented with 10% FBS. Etoposide-resistant K/VP.5 cells were selected and cloned subsequent to intermittent and eventually continuous exposure of K562 cells to 0.5 µM etoposide as previously described (Ritke and Yalowich, 1993). K/VP.5 and gene-edited clonal cells were maintained in DMEM/10% FBS with etoposide (0.5 µM) added every other week. CRISPR clones, generated from K/VP.5 cells, were maintained in DMEM/10% FBS. All experiments described below were performed utilizing cells growing in log phase. Deidentified blast cells from patients newly diagnosed with acute myeloid leukemia (AML) and from the same patients at relapse (who previously received TOP2α-targeted therapies) were obtained from The Ohio State University Comprehensive Cancer Center Leukemia Tissue Bank Shared Resource.
Human TOP2α/170 and TOP2α/90 Real-Time Polymerase Chain Reaction Assays.
Total RNA was isolated from K562, K/VP.5, CRISPR/Cas9-edited K/VP.5 cells, and blasts from patients with AML (matched pretreatment and relapse) using the RNA Easy Plus Mini Kit (cat. no. 74134; Qiagen, Germantown, MD). To ensure complete removal of contaminating DNA, an on-column digestion of DNA with RNase-free DNase (cat. no. 79254; Qiagen) was included during RNA purification. RNA (1 µg) was reverse transcribed using random hexamers and MultiScribe Reverse Transcriptase (High Capacity cDNA Reverse Transcription Kit, cat. no. 4368814; ThermoFisher Scientific, Waltham, MA) as previously described by our laboratory (Kanagasabai et al., 2017, 2018). Quantitative real-time polymerase chain reaction (PCR) experiments (total reaction volume 10 µl) were performed in duplicate using TaqMan Gene Expression hydrolysis probes (ThermoFisher Scientific) as previously described (Kanagasabai et al., 2017, 2018). TOP2α/170 mRNA expression levels were measured using a hydrolysis probe spanning the TOP2α E19/E20 boundary (5′-TCATGGTGAGATGTCACTAATGATG-3′) (TaqMan assay Hs01032135_m1), specific for TOP2α/170 cDNAs. TOP2α/90 mRNA expression levels were measured using a custom hydrolysis probe that spans the wild-type E19/I19 boundary (5′-TCATGGTGAGGTAAACACACAATCC-3′). A custom hydrolysis probe was also synthesized that harbored CRISPR/Cas9-mediated changes (bolded and underlined below) in the E19/I19 boundary (5′-TCATGCTGAGGTAAGTACACAATCC-3′) to specifically measure the expression levels of edited TOP2α/90 mRNAs transcribed in the K/VP.5/edit-3 cell line. Finally, a custom hydrolysis probe was synthesized that harbored the CRISPR/Cas9-mediated change (bolded and underlined) in the E19/E20 boundary (5′-TCATGCTGAGATGTCACTAATGATG-3′) to specifically measure the expression levels of edited TOP2α/170 mRNAs transcribed in the K/VP.5/edit-3 cell line. The relative mRNA expression levels of TOP2α/90, edited TOP2α/90, TOP2α/170, and edited TOP2α/170 in each cell line were normalized to TATA-binding protein (TaqMan assay Hs99999910_m1) expression using the 2−ΔΔCt method (Schmittgen and Livak, 2008).
Immunoassays.
K562, K/VP.5, and CRISPR/Cas9-edited K/VP.5 (with or without etoposide treatment) cellular extracts were subjected to Western blot analysis as previously described (Kanagasabai et al., 2017, 2018). Unless otherwise noted, 16 µg of protein was loaded into each well. Membranes were incubated overnight at 4°C with one of the following primary antibodies: a rabbit polyclonal antibody raised against the human TOP2α/90/170 N-terminal sequence (amino acids 14–27) (cat. no. ab74715; used at 1:1000 dilution; Abcam, Cambridge, MA), a mouse monoclonal glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibody (cat. no. sc-47724; used at 1:5000 dilution; Santa Cruz Biotechnology, Santa Cruz, CA), a mouse phosphorylated Ser-139 residue of the H2A histone family member X (γH2AX) monoclonal antibody (cat. no. sc-25330; used at 1:500 dilution; Santa Cruz Biotechnology). The membranes were subsequently incubated at room temperature for ∼3 hours with a donkey anti-rabbit or anti-mouse secondary antibody (1:5000 dilution) (Jackson Immuno Research, West Grove, PA). Finally, TOP2α isoforms, GAPDH, and γH2AX were detected using the Immun-Star or Clarity Max chemiluminescence kits (Bio-Rad Laboratories, Hercules, CA). All immunoassay images were acquired with the ChemiDoc XRS+ imaging system and analyzed with ImageLab software (Bio-Rad Laboratories).
TOP2α/Minigene Constructs.
The pcDNA3.1(+) mammalian expression plasmid (ThermoFisher Scientific) was linearized by NheI/XbaI digestion in CutSmart buffer according to the manufacturer’s protocol (New England BioLabs, Ipswich, MA). Using human genomic DNA as a template, a 1355–base pair (bp) fragment encompassing the TOP2α gene from just inside the beginning of E19 through E20 was PCR amplified by utilizing sense (5′-CCCAAGCTGGCTAGCGTCAGAGAAAGGTTTTGTTTACT-3′) and antisense (5′-CCCTCTAGACTCGAGCTGAGCATTGTAAAGATGTATCG-3′) primers employing standard PCR procedures with a proofreading polymerase, CloneAmp HiFi (cat. no. 639298; Takara Bio Inc., Kusatsu, Shiga, Japan). The primer sequences homologous to the regions just downstream of the beginning of TOP2α E19 and immediately upstream of the end of TOP2α E20 are bolded. The sense and antisense primers also harbor 15-nucleotide extensions (not bolded) that are homologous to ends of the NheI/XbaI linearized pCR3.1 plasmid and are necessary for “In-Fusion” cloning methodology (In-Fusion HD Cloning Kit, cat. no. 638916; Takara Bio Inc). The TOP2α E19/I19/E20 amplicon was gel purified and subcloned into the NheI/XbaI linearized pCR3.1 plasmid according to the manufacturer’s protocol. The authenticity and orientation of the inserts relative to the cytomegalovirus promoter were confirmed by dideoxy sequencing. The resulting recombinant pCR/TOP2α/E19/I19/E20 “minigene” plasmid was designated TOP2α/Minigene1.
The suboptimal wild-type E19/I19 5′ SS (GAG//GTAAAC) harbored in the TOP2α/Minigene1 plasmid was mutated to a consensus 5′ SS (CAG//GTAAGT) using a Q5 Site-Directed Mutagenesis kit (cat. no. E0554S; New England Biolabs) following the manufacturer’s instructions. The primers used for mutagenesis were sense (5′-CAGGTAAGTACACAATCCATGAAACC-3′) and antisense (5′-ACCATGATGATAAGAAGACATTTCAGC-3′). Importantly, these primers were designed with their 5′ ends annealing back-to-back (inverse PCR). The nucleotides that were mutated are shown in bold print and were confirmed by dideoxy sequencing. The mutated consensus E19/I19 5′ SS plasmid was designated TOP2α/Minigene2.
Transfection and TOP2α/Minigene1/2 PCR Amplification.
K562 and K/VP.5 cells (2.25 × 106 cells in 100 µl per condition) were transfected with TOP2α/Minigene1 or TOP2α/Minigene2 (5.0 µg plasmid) by electroporation technology (Nucleofector Kit V; Lonza, Basel, Switzerland), according to the manufacturer’s instructions. Twenty-four hours after transfection, total RNA was isolated from transfected K562 and K/VP.5 cells and reverse transcribed as described above. The cDNA was subsequently PCR amplified using a T7 forward primer (5′-CGAAATTAATACGACTACTATAGG-3′) and a TOP2α I19 reverse primer (5′-GCAGACTTATGAATATCCCTGCAGG-3′) or a TOP2α E20 reverse primer (5′-GCAAGAGGTTTAGATTATTGCTACC-3′) using AmpliTaq DNA Polymerase (cat. no. N8080160; ThermoFisher). PCR products were fractionated by electrophoresis on a 1.5% agarose gel, and PCR amplicons were visualized under UV light after staining with 0.5 mg/ml ethidium bromide. Images were captured using the ChemiDoc XRS1 imaging system and analyzed using ImageLab software (Bio-Rad Laboratories).
Genomic Cleavage Detection.
TrueGuide hypoxanthine-guanine phosphoribosyltransferase (HPRT1) positive control CRISPR RNA (crRNA) (5′-GCAUUUCUCAGUCCUAAACA-3′) (cat. no. A35517), custom TrueGuide TOP2α crRNA #1 (5′-GTCTTCTTATCATCATGGTG-3′), TrueGuide TOP2α crRNA #2 (5′-GAAATGTCTTCTTATCATCA-3′), TrueGuide TOP2α crRNA #3 (5′-TATAATGCTTTCTGGAAACA-3′), and TrueGuide 72-bp trans-activating CRISPR RNA (tracrRNA) (cat. no. A35517) were obtained from ThermoFisher. Each TrueGuide RNA is chemically modified (2′O-methyl analogs and phosphorothioate linkages) to increase editing efficiency and protect against nuclease degradation. All TrueGuide crRNAs were individually annealed to a common tracrRNA scaffold according to the manufacturer’s instructions for a final crRNA:tracrRNA duplex (gRNA) concentration of 20 μM. The gRNAs (0.5 µg) were individually incubated with 2 µg TrueCut Cas9 Protein v2 (cat. no. A36498; ThermoFisher) for 15 minutes to form Cas9 protein/gRNA ribonucleoprotein complexes. These complexes were subsequently transfected into etoposide-resistant K/VP.5 cells by electroporation technology (Nucleofector Kit V; Lonza, Basel, Switzerland) according to the manufacturer’s instructions and as reported previously (Kanagasabai et al., 2017).
To determine if the gene-specific Cas9 protein/gRNA ribonucleoprotein complexes created on-target DSBs within the TOP2α E19/I19 boundary sequence, K/VP.5 cells (2 × 106) were lysed 48 hours after transfection using cell lysis buffer/Proteinase K (GeneArt Genomic Cleavage Detection (GCD) Kit (cat. no. A24372; ThermoFisher). Genomic DNA (1 µl of lysate) at the TOP2α locus between E18 and I19 was then PCR amplified (50 µl reaction volume) using the following primers: GCD TOP2α E18 forward (5′-GATCTATCCCTTCTATGGTGG-3′) and GCD TOP2α I19 reverse (5′-CAGAAATCAAAGGGCAAGCAG-3′). Positive control PCR experiments were also performed at the HPRT1 intron 2 locus using the following primers: GCD HPRT1 forward (5′-AGAGGAGGGCCTTACTAATTAC-3′) and GCD HPRT1 reverse 5′-CATGCATAGCCAGTGCTTGAG-3′). The PCR amplicons were subsequently denatured, reannealed, and incubated with T7 endonuclease I (i.e., structure-selective enzyme that recognizes and cleaves mismatched DNA) to detect insertions/deletions (indels) created by nonhomologous end joining (NHEJ). The digested and nondigested PCR products were fractionated by electrophoresis on a 2% agarose gel, and images were captured as described above. The following equation was used to the calculate the cleavage efficiency of HPRT1 and TOP2α gRNA/Cas9: cleavage efficiency = {1 – [(1 – fraction cleaved)1/2]} × 100, where fraction cleaved = (sum of cleaved band intensities)/(sum of the cleaved and parental band intensities).
Genome Editing of the TOP2α E19/I19 5′ SS.
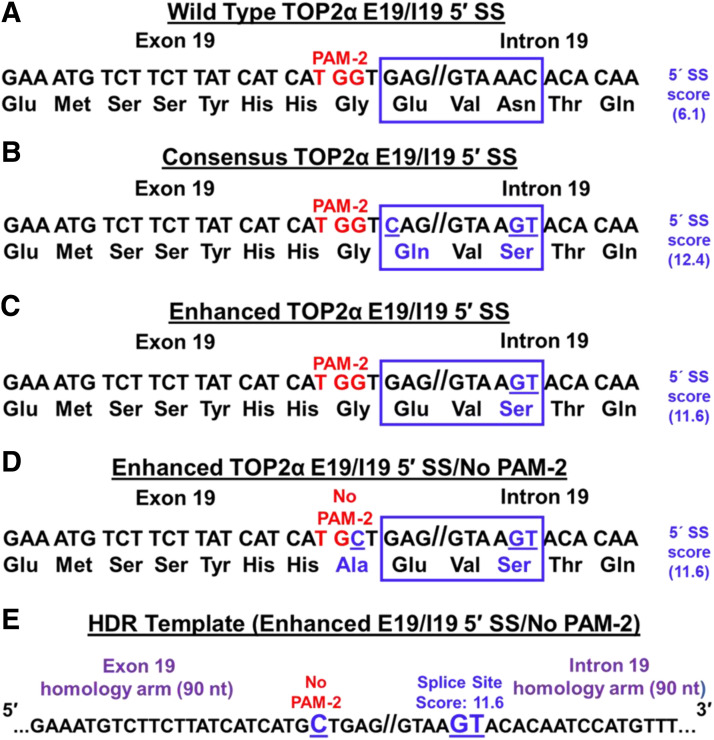
The suboptimal TOP2α E19/I19 5′ SS (GAG/GTAAAC; splicing score, 6.1) was gene-edited (GAG/GTAAGT; splicing score, 11.6) utilizing the TOP2α crRNA #2:tracrRNA duplex (gRNA-2) (see above), TrueCut Cas9 Protein, and a 180-nucleotide symmetric single-stranded oligonucleotide repair template (Ultramer Oligo; Integrated DNA Technologies, Coralville, Iowa; see Supplemental Table 1 for complete sequence) designated Enhanced E19/I19 5′ SS/No PAM-2. This repair template equally spans TOP2α E19/I19 and harbors the desired two 5′ SS nucleotide changes. Moreover, this repair template harbors an additional nucleotide change to eliminate the protospacer-adjacent motif (#2 (PAM-2; see Fig. 5) to avoid recutting of edited alleles upon the repeated transfections necessary to edit all three TOP2α alleles present in the clonal K562 cell line, K/VP.5 (Cioe et al., 1981; Zhou et al., 2019), at the E19/I19 5′ SS. Finally, the repair template was chemically modified (phosphorothioate linkages) at the first and last two nucleotides to protect against nuclease degradation.
Fig. 5.
Strategy to use CRISPR/Cas9 and HDR to edit the suboptimal TOP2α E19/I19 5′ SS and PAM-2. (A) The TOP2α E19/I19 gene boundary sequence is shown. The suboptimal E19/I19 5′ SS sequence to be edited by CRISPR/Cas9 is boxed in blue. The “weak” SS score is also denoted. The P AM-2 site is denoted in red. (B) The proposed sequence changes to generate a “consensus” E19/I19 5′ SS sequence is boxed, underlined, and denoted in blue as are the resultant amino acid changes. The “consensus” SS score is also denoted. (C) The proposed sequence changes to “enhance” the E19/I19 5′ SS sequence is boxed, underlined, and denoted in blue as is the resultant amino acid change. The “enhanced” SS score is also denoted. (D) The proposed sequence changes to “enhance” the E19/I19 5′ SS plus eliminate the PAM-2 site are shown in the blue box and by the change in the PAM-2 sequence (TGG→TGC). The resultant amino acid changes are shown in blue. The “enhanced” SS score is also denoted. (E) To introduce the proposed changes in the suboptimal TOP2α E19/I19 5′ SS and PAM-2, an ssODN HDR template (denoted “Enhanced E19/I19 5′ SS/No PAM-2”) was synthesized.
Genome editing of the TOP2α E19/I19 5′ SS was conducted as follows. Briefly, gRNA-2 (0.5 µg) was incubated with 2 µg TrueCut Cas9 Protein v2 and 5 µM TOP2α E19/I19 repair template (designated Enhanced E19/I19 5′ SS/No PAM-2; Fig. 5E and Supplemental Table 1) for 15 minutes according to the manufacturer’s instructions. This mixture was then transfected into K/VP.5 cells (2.25 × 106 cells in 100 µl) by electroporation as reported previously (Kanagasabai et al., 2017). Forty-eight hours later, K/VP.5 cells (50,000) were lysed for Cas9 targeting and repair efficiency using the GCD assay described above. After verification of successful on-target genome editing generated by NHEJ, the remaining transfected K/VP.5 cells were plated using limiting dilution cloning in ten 96-well plates (0.8 cells per well). Aliquots (∼25–50,000 cells) from single-cell clones were subsequently lysed with GCD buffer (see above) ∼2 weeks after plating. Supernatants were assayed for HDR by genomic quantitative PCR (qPCR) using a probe specific for the edited TOP2α E19/I19 5′ SS (5′-TCATGCTGAGGTAAGTACACAATCC-3′) to identify colonies with at least one TOP2α edited allele. After the first round of transfection, multiple colonies with one edited allele were identified by qPCR and confirmed by sequencing. A single TOP2α allele edited clone, designated K/VP.5/edit-1, was subjected to an additional round of transfection with gRNA-2, Cas9, and the TOP2α E19/I19 repair template, followed by limiting dilution cloning. Genomic qPCR using a probe specific for the edited TOP2α E19/I19 5′ SS and the suboptimal wild-type TOP2α E19/I19 5′ SS qPCR probe was used to identify gene-edited K/VP.5 clones with two and three TOP2α edited alleles, now designated K/VP.5/edit-2 and K/VP.5/edit-3 (see Fig. 6).
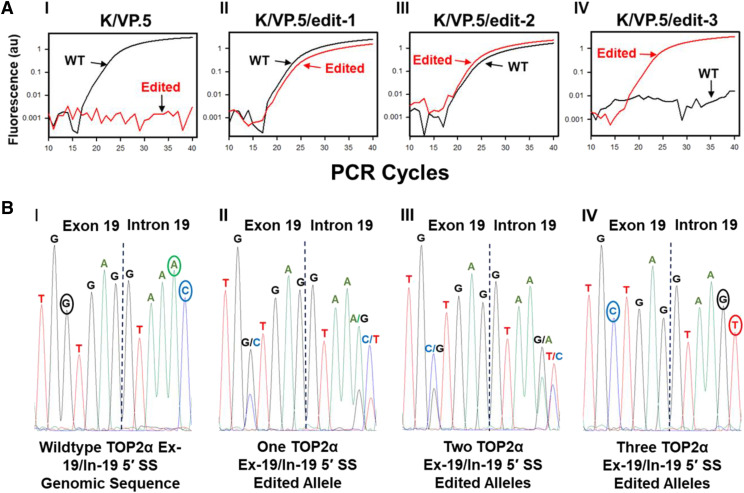
Fig. 6.
qPCR selection and sequence analysis of CRISPR/Cas9-edited TOP2α E19/I19 5′ SS clonal cell lines. (A) Amplification plots of qPCR reactions from K/VP.5 and K/VP.5/edit-1,2,3 cells (labeled I–IV) using wild-type and edited, specific E19/I19 boundary qPCR probes. (B) Electropherograms of the genomic sequence of the TOP2α E19/I19 gene boundary in K/VP.5 and K/VP.5/edit-1–3 cells (labeled I–IV).
RsaI Restriction Enzyme Analysis of CRISPR/Cas9-Edited K/VP.5 Cells.
K/VP.5, K/VP.5/edit-1, K/VP.5/edit-2, and K/VP.5/edit-3 cells were lysed using cell GCD lysis buffer/Proteinase K as described above, and genomic DNA (1 µl of lysate) at the TOP2α locus between E18/intron 18/E19/I19 was PCR amplified (50 µl reaction volume) using the following primers; GCD TOP2α E18 forward (5′-GATCTATCCCTTCTATGGTGG-3′) and GCD TOP2α I19 reverse (5′-CAGAAATCAAAGGGCAAGCAG-3′). Ten microliters of the PCR reaction was then digested by RsaI (cat. no. R0167S; New England Biolabs) according to the manufacturer’s instructions. The digested and nondigested PCR products were fractionated by electrophoresis on a 2% agarose gel, and images were captured as described above (see Fig. 7).
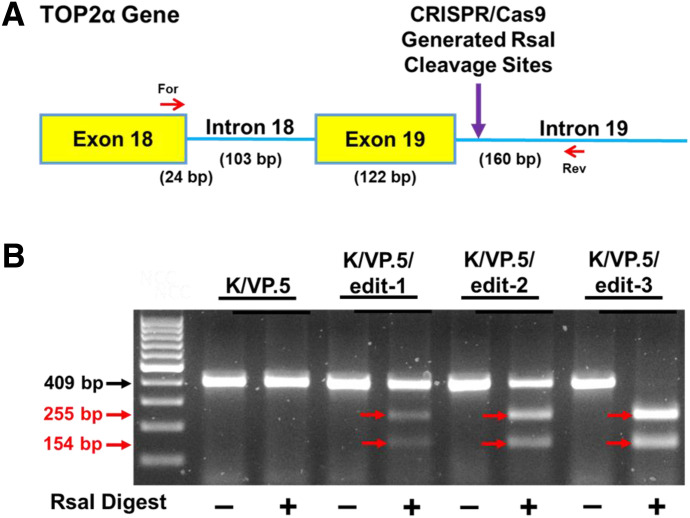
Fig. 7.
Validation of CRISPR/Cas9 editing of the TOP2α E19/I19 5′ SS in K/VP.5 cells by Rsa1 analysis. (A) Schematic representation of the E18 through I19 portion of the TOP2α gene. Red arrows denote the forward (For) and reverse (Rev) primers used for the identification of CRISPR editing of the TOP2α E19/I19 5′ SS using RsaI endonuclease. Purple arrow denotes site where Rsa1 creates double-stranded breaks of CRISPR-edited PCR amplicons (B) Ethidium bromide–stained agarose gel of fractionated Rsa1-treated PCR amplicons from K/VP.5 and K/VP.5/edit-1–3 DNA. The black arrow denotes the parental PCR amplicon. Expected sizes of parental and daughter PCR amplicons are indicated in red.
RNA Sequencing and Bioinformatics Analyses.
DNA-free RNA was isolated from K562, K/VP.5, and K/VP.5/edit-3 cells using the RNA Easy Plus Mini Kit (cat. no. 74134; Qiagen, Germantown, MD) with on-column digestion with RNase-Free DNase (cat. no. 79254). Two rounds of purification were used to assure high purity and DNase-free RNA isolation. RNA sequencing (RNA-seq) libraries were prepared at The Ohio State University Comprehensive Cancer Center Genomics Shared Resource and sequenced from quadruplicate samples from each cell line. Paired-end RNA-seq was performed on an Illumina HiSEq 4000 platform at the Genomics Services Laboratory of The Research Institute at Nationwide Children’s Hospital, Columbus, OH. Illumina 150-bp paired-end RNA-seq raw reads from K562 and K/VP.5 RNA samples were mapped to the human reference genome GRCh38 using Hierarchical Indexing for Spliced Alignment for Transcripts version 2.1.0 9 (Kim et al., 2015), converted to bigwig coverage tracks using deepTools (Ramírez et al., 2016), and visualized using the Integrative Genomics Viewer (Robinson et al., 2011). Gene counts were generated with featureCounts (Liao et al., 2014) as described in Gadepalli et al. (2019). Gene expression was quantified as log2 counts per million, and differential expression analysis was performed using R limma voom function (Ritchie et al., 2015). RNA-seq data are available in the Gene Expression Omnibus with accession number GSE163013.
Growth Inhibitory Assays.
Log-phase K562 cells, K/VP.5 cells, and gene-edited K/VP.5 clonal cells were adjusted to 1–1.5 × 105 cells per milliliter and incubated for 48 hours with DMSO as control solvent (final concentration 0.5%) and with various concentrations of a number of drugs, after which cells were counted on a model Z1 DUAL Coulter counter (Beckman Coulter, Indianapolis, IN). The extent of growth beyond the starting concentration in drug-treated versus control cells was expressed ultimately as percent inhibition of control growth. The 50% growth inhibitory values for each drug and each cell line were derived from replicate experiments performed on separate days fitting the concentration-response (inhibition) curves to a four-parameter logistic equation using Sigmaplot 14 (Systat Software, Inc., San Jose, CA).
DNA Damage (Comet) Assays.
Alkaline (pH 13, detects primarily single-stranded breaks) single-cell gel electrophoresis (Comet) assays were performed according to the manufacturer’s protocol (CometAssay Kit, cat. no. 4250-050-K; Trevigen, Gaithersburg, MD) and as previously described by our laboratory (Vlasova et al., 2011; Kanagasabai et al., 2017, 2018). Briefly, K562, K/VP.5, and K/VP.5/edit-3 cells were washed and resuspended in buffer (25 mM HEPES, 10 mM glucose, 1 mM MgCl2, 5 mM KCl, 130 mM NaCl, 5 mM NaH2PO4, pH 7.4). Cells were subsequently incubated with 2 or 10 μM etoposide or DMSO (solvent control) for 30 minutes at 37°C. The treated cells were washed with ice-cold buffer and resuspended to 0.28 × 106 cells per milliliter and then further diluted in low melt agarose. After alkaline electrophoresis (of ∼2000 cells) and subsequent staining with a fluorescent DNA intercalating dye, SYBR Gold, the migrating fragments (comet tail) from the nucleoid (comet head) were visualized and the images captured by fluorescence microscopy. The Olive tail moment (Olive, 2002) was quantified by the ImageJ processing program with the open-source software tool OpenComet (Gyori et al., 2014; www.cometbio.org). Olive tail moments from more than 100 cells per sample condition were determined.
Data Analysis.
Statistical analysis was performed using Sigma-Plot 14. All data were expressed as means ± S.D. Unless noted otherwise, groupwise differences were analyzed using a two-tailed paired Student’s t test with no adjustment for multiple comparisons. A P value of less than 0.05 was considered statistically significant with 95% confidence intervals (CIs) noted in figure legends.
Results
TOP2α Expression in Etoposide-Resistant K562 Cells.
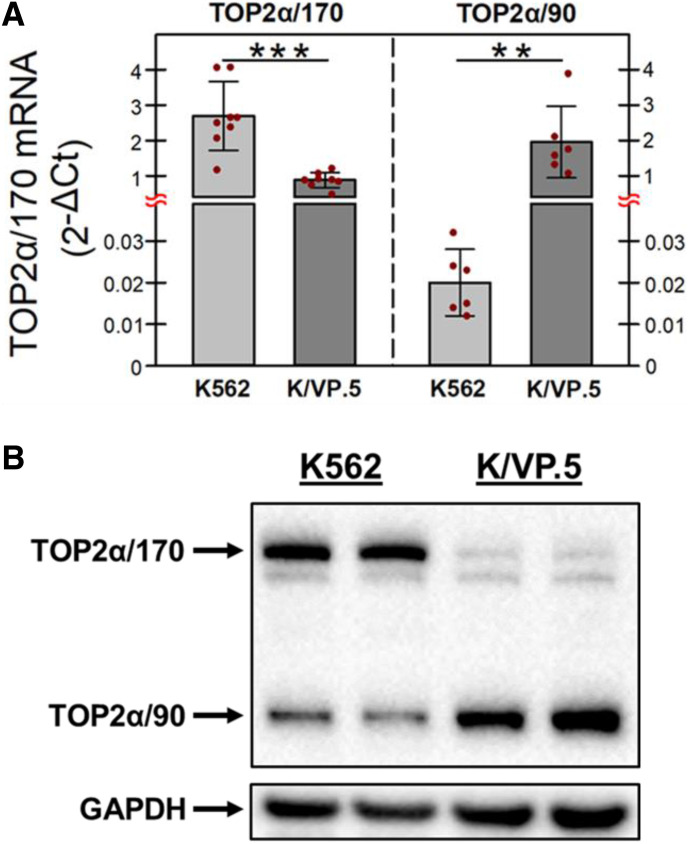
Our laboratory previously established an etoposide-resistant K562 cell line by intermittent/continuous treatment with 0.5 μM etoposide followed by limiting dilution to isolate and then characterize a clonal K/VP.5 cell line (Ritke and Yalowich, 1993; Ritke et al., 1994). Recent studies utilizing K/VP.5 cells established that TOP2α/170 mRNA/protein levels were attenuated and TOP2α/90 mRNA/protein levels were augmented compared with parental K562 cells (Kanagasabai et al., 2017, 2018; Elton et al., 2020). To recapitulate these findings, K562 and K/VP.5 cDNAs were subjected to qPCR utilizing TaqMan hydrolysis primers/probes. The results of these experiments demonstrated that TOP2α/170 mRNA expression in K/VP.5 cells was reduced to 33% of that found in parental K562 cells, whereas TOP2α/90 mRNA expression was increased 97-fold in K/VP.5 cells compared with parental K562 cells (Fig. 1A). In K562 cells, TOP2α/90 mRNA levels were extremely low compared with TOP2α/170 mRNA levels (Fig. 1A, light gray bars), whereas in K/VP.5 cells TOP2α/90 mRNA levels exceeded those of TOP2α/170 by ∼2-fold (Fig. 1A, dark gray bars), reflecting I19 retention. Figure 1B also recapitulates our previous reports of decreased TOP2α/170 protein in K/VP.5 cells, currently to a level 8% that of parental K562 cells, whereas TOP2α/90 protein expression was 2-fold higher in K/VP.5 cells compared with K562 cells.
Fig. 1.
TOP2α/170 and TOP2α/90 mRNA and protein levels in K562 and drug-resistant K/VP.5 cells. (A) qPCR experiments were performed utilizing K562 and K/VP.5 cDNAs and a TaqMan hydrolysis assay specific for TOP2α/170 and TOP2α/90 mRNAs. Results shown are means ± S.D. from six or eight paired RNA/cDNA isolations/determinations performed on separate days; P < 0.001, comparing the differences in mean calculated 2−ΔCt values for K/VP.5 vs. K562 TOP2α/170 mRNA, 95% CI (1.17, 2.46); P = 0.005, comparing the differences in mean calculated 2−ΔCt values for K/VP.5 vs. K562 TOP2α/90 mRNA, 95% CI (0.87, 2.99). (B) Representative immunoassay using K562 and K/VP.5 cellular lysates. Blots were probed with antibodies specific for the N-terminal portion of TOP2α/170/90 (i.e., amino acids 14–27) or for GAPDH. **P < 0.01; ***P < 0.001.
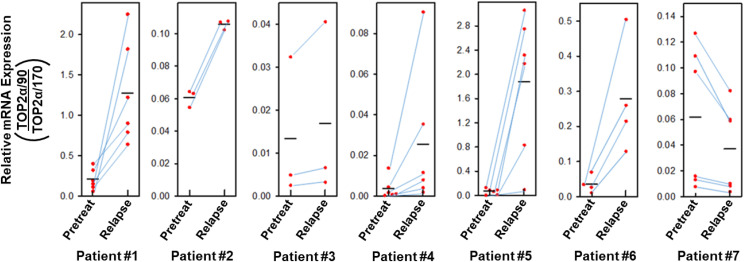
Since TOP2α/90 heterodimerizes with TOP2α/170 and is dominant-negative for etoposide-induced DNA damage and cytotoxicity compared with TOP2α/170 homodimers (Kanagasabai et al., 2018), it was of interest that the ratio of TOP2α/90 to TOP2α/170 mRNA/protein was greater in resistant K/VP.5 compared with parental K562 cells (Fig. 1, A and B). Therefore, we next investigated whether there would similarly be an increase in the TOP2α/90:TOP2α/170 mRNA ratio in AML blasts from patients who relapsed after initial standard-of-care treatment with TOP2α-targeted agents. Figure 2 demonstrated that, in six of seven patients, there was an increase in the TOP2α/90:TOP2α/170 mRNA ratio after relapse. Evaluation of the mean values for TOP2α/90:TOP2α/170 mRNA ratios pretreatment and after relapse for all seven patients indicated an average 4.01-fold increase that was statistically significant [P = 0.036; 95% CI (1.14, 14.16)]. These results suggested that the TOP2α/90 to TOP2α/170 ratio may be a correlate/determinant of acquired drug resistance and pointed to the potential importance of alternative RNA processing/intron retention as a biomarker and/or as a target for modulation/circumvention of drug resistance to TOP2α-targeted agents.
Fig. 2.
Effects on TOP2α/90:TOP2α/170 mRNA ratios in patients with AML upon post-therapy relapse. Total RNA samples were isolated from blast cells obtained from patients newly diagnosed with AML and the same patients at relapse, after receiving TOP2α-targeting therapy. qPCR experiments were performed utilizing TaqMan hydrolysis probes specific for TOP2α/170 and TOP2α/90 mRNAs as described in Materials and Methods and as used previously (Kanagasabai et al., 2017). From each patient’s samples (pretreatment and at relapse), cDNAs were generated in parallel from isolated RNAs by reverse transcription, and qPCR reactions were subsequently performed in parallel using a common master reaction mix. In addition, these evaluations were repeated on separate days by generation of cDNAs with subsequent qPCR reactions performed from each pair of patient’s samples. The ratio of TOP2α/90:TOP2α/170 mRNA pretreatment and after relapse was calculated and plotted as coordinate plots with point-line-point connections for each pair of evaluations for each patient. For patient #1, one of the sets of values connected by point-line-point for pretreatment (0.4) and relapse (0.9) were indicated in a previous publication (Kanagasabai et al., 2018). The black horizontal bars represent the mean values from repeated paired measures. Evaluation of the mean values for TOP2α/90:TOP2α/170 mRNA ratios pretreatment and after relapse for all seven patients indicated an average 4.01-fold increase that was statistically significant after performing a paired t test on log transformed values; P = 0.036 [95% CI (1.14, 14.16)].
TOP2α Minigene Constructs for Evaluation of I19 Retention.
Our laboratory previously demonstrated that the TOP2α/90 isoform is the translation product of an mRNA that shares the first 19 exons with the TOP2α/170 transcript (Kanagasabai et al., 2017). However, the TOP2α/90 mRNA retains a processed I19 and lacks the published TOP2α/170 transcript sequences from exon 20 to exon 35 (Kanagasabai et al., 2017, 2018; Elton et al., 2020). As a result of I19 retention, TOP2α/90 (i.e., 786 amino acids, 90,076 Da) is approximately one-half the size of the wild-type TOP2α/170 protein (i.e., 1531 amino acids, 174,385 Da) and does not harbor the active site tyrosine (Tyr-805) required for wild-type TOP2α/170 to generate DNA strand breaks (Deweese and Osheroff, 2009; Nitiss, 2009), which may account for its dominant-negative effects upon heterodimerization with TOP2α/170 (Kanagasabai et al., 2018).
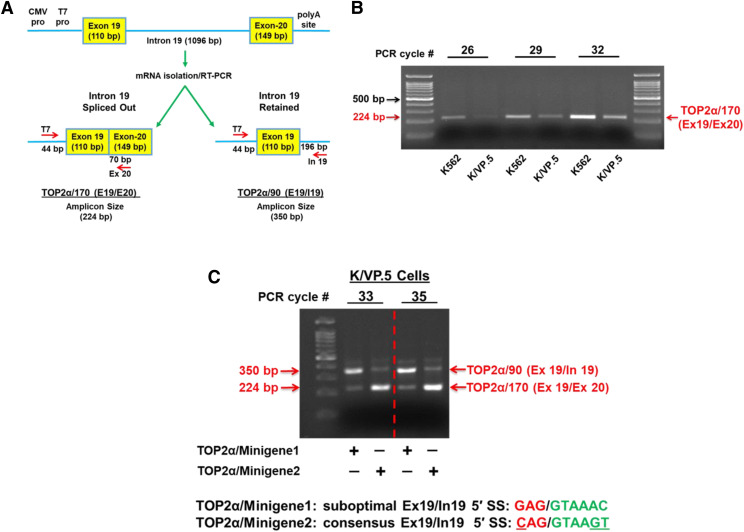
To examine the mechanism(s) responsible for TOP2α I19 retention and resultant increased TOP2α/90 protein levels, our laboratory generated a minigene expression construct (Cooper, 2005), designated TOP2α/Minigene1, which incorporated E19, I19, and E20 (Fig. 3A). To measure the relative expression levels of surrogate TOP2α/170 mRNAs (E19/E20; i.e., I19 spliced out) transcribed and processed from the plasmid, TOP2α/Minigene1 was transiently transfected into K562 and K/VP.5 cells, and total RNA was isolated, treated with DNAse I, and then reverse-transcribed for PCR. To ensure that only minigene-expressed mRNAs were amplified, a T7 primer (forward) was used in all PCR reactions (Fig. 3A). PCR analysis of the minigene-expressed TOP2α/170 mRNAs demonstrated that I19 was appropriately spliced out (224-bp amplicon) in both K562 and K/VP.5 cells and that K562 cells exhibited higher levels of properly processed TOP2α/170 mRNAs (Fig. 3B). Importantly, the minigene analysis correlated with the qPCR data shown in Fig. 1A, which demonstrated that TOP2α/170 mRNA expression levels were greater in K562 compared with K/VP.5 cells. Therefore, transfection of the TOP2α gene fragment (i.e., E19, I19, and E20) within TOP2α/Minigene1 recapitulated the TOP2α/170 mRNA expression levels observed in nontransfected K562 and K/VP.5 cells (Fig. 1A) indicating that RNA processing of the TOP2α/Minigene1-derived transcripts mimics endogenous TOP2α mRNA splicing at the E19/I19 boundary and is amenable for further mechanistic studies.
Fig. 3.
Effects of optimizing the weak TOP2α E19/I19 5′ SS harbored in a TOP2α minigene construct on TOP2α/170 mRNA expression in etoposide-resistant K/VP.5 cells. (A) Schematic representation of the TOP2α gene segment that was subcloned into the pcDNA3.1 expression plasmid. Minigene-expressed and processed TOP2α/170 (E19/E20; i.e., I19 spliced out) and TOP2α/90 (E19/I19; i.e., I19 retained) PCR amplicon sizes are shown. CMV, cytomegalovirus; polyA, XXXX; pro, promoter. (B) K562 and K/VP.5 cells were transfected with the TOP2α/Minigene1 construct (i.e., harboring the weak TOP2α E19/I19 5′ SS). RNA was isolated and reverse transcribed, and PCR experiments were performed. Ethidium bromide–stained agarose gel fractionated TOP2α/170 amplicons are denoted with the red arrow. The PCR cycle number is indicated to ensure the linear range of amplification and allows for the direct comparison of the intensity of the TOP2α/170 amplicon from K562 cells versus K/VP.5 cells. (C) K/VP.5 cells were cotransfected with TOP2α/Minigene1 and TOP2α/Minigene2 (i.e., harboring a consensus TOP2α E19/I19 5′ SS). RNA was isolated and reverse transcribed, and multiplex PCR experiments were performed. Ethidium bromide–stained agarose gel fractionated TOP2α/90 and TOP2α/170 amplicons are denoted. The PCR cycle number is indicated to ensure the linear range of amplification and allows for the direct comparison of the intensity of the TOP2α/90 and TOP2α/170 amplicon from K/VP.5 cells.
Enhanced TOP2α I19 retention observed in K/VP.5 compared with K562 cells is likely the result of a combinatorial effect of cis-element SS sequences and the dysregulation of trans-acting factors (Lee and Rio, 2015; Monteuuis et al., 2019). Cis-elements are recognition sequences used by splicing-initiating factors to recruit the splicing machinery, and the recognition efficiency of SS is governed by base pair homology with these splicing-initiating factors (Lee and Rio, 2015; Monteuuis et al., 2019). Interestingly, the TOP2α E19/I19 5′ SS is suboptimal (GAG//GTAAAC), differing from the consensus splice site (CAG//GTAAGT) by three nucleotides, and has a splicing “score” of 6.1 out of a maximum of 12.4 (Splice Site Score Calculation).
To investigate whether the weak TOP2α E19/I19 5′ SS contributes to I19 retention, the wild-type 5′ SS was mutated from GAG//GTAAAC to CAG//GTAAGT to generate the consensus TOP2α E19/I19 5′ SS in the TOP2α/Minigene1 construct (designated TOP2α/Minigene2) (Fig. 3C). K/VP.5 cells were transiently transfected with the suboptimal wild-type 5′ SS minigene (TOP2α/Minigene1) or with the consensus 5′ SS minigene (TOP2α/Minigene2) construct. The relative expression of the surrogate TOP2α/90 and TOP2α/170 mRNAs transcribed and processed from the minigenes was determined by multiplex PCR using a T7 forward primer and TOP2α/90 and TOP2α/170 specific reverse primers (Fig. 3A; Materials and Methods and Supplemental Table 1).
Multiplex PCR analysis of the minigene-expressed mRNAs in K/VP.5 cells transfected with the wild-type 5′ SS TOP2α/Minigene1 construct demonstrated that the TOP2α/90 mRNA (E19/I19; i.e., I19 retained) 350-bp amplicon levels were greater than the TOP2α/170 mRNA (E19/E20, I19 spliced out) 224-bp amplicon levels at PCR cycles 33 and 35 (Fig. 3C). In contrast, multiplex PCR analysis of the minigene-expressed mRNAs in K/VP.5 cells transfected with the consensus 5′ SS TOP2α/Minigene2 construct demonstrated that TOP2α/170 mRNA (E19/E20, I19 spliced out) 224-bp amplicon levels were now greater than the TOP2α/90 (E19/I19; I19 retained) 350-bp amplicon levels (Fig. 3C). Hence, optimizing the TOP2α E19/I19 5′ SS resulted in reduced intron retention, decreased surrogate TOP2α/90 mRNA expression, and increased surrogate TOP2α/170 mRNA (Fig. 3C). These promising preliminary results prompted our use of CRISPR/Cas9 gene editing to optimize the TOP2α E19/I19 5′ SS, diminish I19 retention in K/VP.5 cells, increase TOP2α/170 expression, and circumvent drug resistance.
gRNA-2 Directs Cas9 Cleavage in the TOP2α E19/I19 Boundary Sequence.
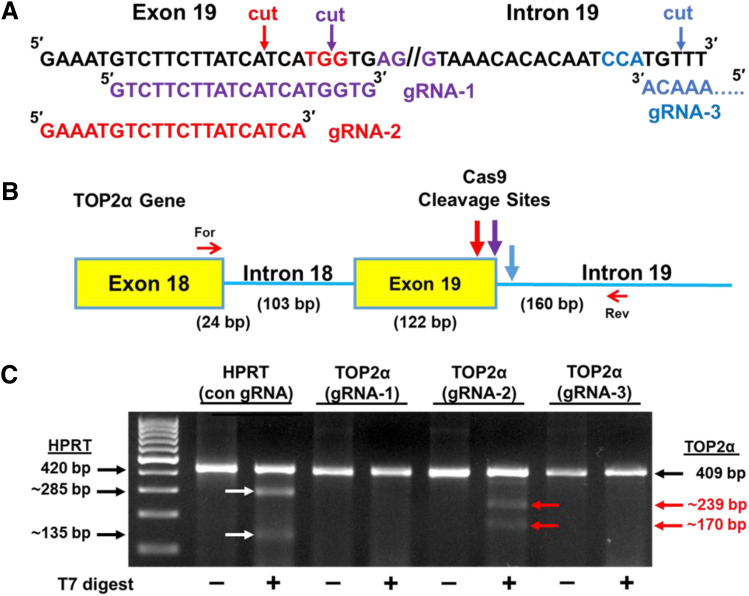
Our results above with the TOP2α minigene plasmids, together with previously published findings (Wickramasinghe et al., 2015; Yue and Ogawa, 2018), clearly demonstrated that optimizing a weak 5′ SS can improve the efficiency of intron removal. Therefore, we next used the CRISPR/Cas9 system (Jinek et al., 2012; Mali et al., 2013; Liand et al., 2017) to introduce specific gene edits in the suboptimal TOP2α E19/I19 5′ SS through HDR to enhance splicing out of I19. The gRNA/Cas9 complex binds to the target site, and the Cas9 nuclease introduces a blunt-end DSB three bases upstream of a three-nucleotide sequence motif (NGG), known as the protospacer-adjacent motif (PAM) (Jinek et al., 2012; Mali et al., 2013). Thus, the TOP2α E19/I19 boundary sequence (200 bp) was analyzed for PAM sequence motifs utilizing the CRISPR/Cas9 Target Online Predictor (CCTop) (https://cctop.cos.uni-heidelberg.de/) (Stemmer et al., 2015). Although 12 PAM sites were identified (not shown), only three candidate PAM/gRNAs (Fig. 4A, color-coded) were examined since they would target Cas9 in close proximity to the intended mutations (Fig. 5), which should maximize editing efficiency (Paquet et al., 2016; Liang et al., 2017). Of the three PAM/gRNAs studied, PAM/gRNA-2 had the highest CRISPRater efficacy score of 0.71 compared with 0.60 for PAM/gRNA-1 and 0.50 for PAM/gRNA-3 (Labuhn et al., 2018).
Fig. 4.
Effects of gRNA-2 on Cas9 cleavage at the TOP2α E19/I19 5′ boundary sequence. (A) Sequence of the TOP2α E19/I19 5′ boundary sequence is shown. Candidate PAM sites and corresponding gRNAs are color coded. Arrows denote where Cas9 will generate a DSB. (B) Schematic representation of the E18 through I19 region of the TOP2α gene. The large color-coded arrows denote sites where gRNA-directed Cas9 cleavage and NHEJ generation of indels can occur. The small red arrows denote the forward (For) and reverse (Rev) primers used for the GCD assay. (C) Ethidium bromide–stained agarose gel fractionated GCD PCR amplicons before and after treatment with T7 endonuclease I. The parental and T7 endonuclease I cleaved daughter PCR amplicons are indicated, and their respective sizes are denoted. A positive HPRT1 control (con) GCD assay is also shown.
To determine the most efficient PAM/gRNA site, each corresponding gRNA was complexed with Cas9 and transiently transfected into K/VP.5 cells. Forty-eight hours post-transfection, cells were assayed for indels created by NHEJ (Qi et al., 2013) due to Cas9 cleavage in the TOP2α E19/I19 boundary sequence. Briefly, cells were lysed using the GCD lysis buffer, followed by PCR using a forward primer on E18 and a reverse primer on I19 to amplify the CRISPR/Cas9 target region (Fig. 4B). PCR amplicons were then denatured and reannealed to evaluate Cas9-induced breaks and NHEJ-mediated mismatches (heteroduplex DNA) targeted/cleaved by T7 endonuclease I. GCD analysis of PAM/gRNAs 1–3 revealed that only PAM-2/gRNA-2 (color-coded red) effectively guided Cas9 to the target site (Fig. 4C). The calculated Cas9 cleavage efficiency (Liang et al., 2017) in the TOP2α E19/I19 boundary by the TOP2α gRNA-2 was 7.3% compared with the optimized control HPRT1 gRNA of 14.0%. All subsequent experiments were performed with TOP2α gRNA-2.
CRISPR/Cas9: Strategy to Edit the TOP2α E19/I19 5′ SS.
Since the wild-type TOP2α E19/I19 5′ SS is suboptimal (GAG//GTAAAC) (Fig. 5A, blue box), analyses were performed to determine the impact of specific gene edits on the 5′ SS scores. These analyses demonstrated that if the TOP2α E19/I19 5′ SS (splicing “score” of 6.1) was edited to a consensus 5′ SS (splicing “score” of 12.4) (Splice Site Score Calculation), as was done for the TOP2α/Minigene2 construct (Fig. 3C), then a nonconservative amino acid change would result (Glu→Gln) in the TOP2α/170 protein (Fig. 5B, blue box). In contrast, by editing only the last two nucleotides of the TOP2α E19/I19 5′ SS (AC→GT) (Fig. 5C), the 5′ SS score would be enhanced (from 6.1→11.6) with no amino acid change in the TOP2α/170 protein sequence (compare Fig. 5, A and C). Therefore, we initially planned to include only the AC→GT nucleotide change (Fig. 5C) in the mutagenic single-stranded oligo DNA nucleotide (ssODN) repair template (not shown). However, K562 and drug-resistant K/VP.5 cells are triploid with three copies of the TOP2α gene (Cioe et al., 1981; Zhou et al., 2019), presenting challenges for successful isolation of a clone with gene editing of the E19/I19 5′ SS in all three alleles after a single Cas9, gRNA, repair template cotransfection.
Since only PAM-2/gRNA-2 effectively guided Cas9 to the target site (Fig. 4C), a repair template was designed based on the TOP2α E19/I19 gene boundary sequences changes shown in Fig. 5D to include the AC→GT alterations and to eliminate PAM-2. This strategy was implemented to avoid recutting of already edited alleles upon subsequent rounds of transfection, to assure editing of all three TOP2α alleles, and to result in an optimized TOP2α E19/I19 5′ SS. The G→C modification in the PAM-2 site (Fig. 5, D and E) resulted in a very conservative amino acid change, Gly→Ala (compare Fig. 5, A and D), in the TOP2α protein. The symmetric 180-nucleotide ssODN HDR template (Fig. 5E, denoted as Enhanced E19/I19 5′ SS/No PAM-2) with homology to both TOP2α E19 and I19 was synthesized and used in all CRISPR/Cas9 transfection experiments.
CRISPR/Cas9: qPCR Selection and Sequence Analysis of Edited TOP2α E19/I19 Clonal Cell Lines.
K/VP.5 cells were transfected with gRNA-2 (Fig. 4A; Supplemental Table 1), Cas9 protein, and the HDR template (Fig. 5E). Forty-eight hours post-transfection, cells were seeded at 0.8 cells per well in 96-well plates and allowed to grow for 2 weeks. Cell aliquots harvested from single-colony wells were lysed and screened by genomic DNA qPCR.
We have previously validated the specificity of a qPCR hybridization probe (5′-TCATGGTGAGGTAAACACACAATCC-3′) for the wild-type TOP2α E19/I19 boundary to demonstrate that the TOP2α/90 truncated isoform was encoded by an mRNA that harbors a retained and processed I19 (Kanagasabai et al., 2017, 2018; Elton et al., 2020). To discriminate between the wild-type TOP2α E19/I19 and the CRISPR/Cas9-edited TOP2α E19/I19 boundary, an additional custom qPCR hybridization probe containing the edited sequence (5′-TCATGCTGAGGTAAGTACACAATCC-3′) was synthesized. Nontransfected K/VP.5 cell control experiments demonstrated no signal from this custom E19/I19 edited qPCR probe (Fig. 6AI, red line). Only the wild-type TOP2α E19/I19 boundary probe (black line) yielded a positive genomic qPCR signal (Fig. 6AI). Sanger sequencing verified the wild-type TOP2α genomic sequence (Fig. 6BI). In contrast, when CRISPR/Cas9-edited K/VP.5 cell lysates were screened (∼60 clonal cell colonies), several clones were identified where the qPCR signal for the edited TOP2α E19/I19 boundary probe signal (red line) appeared a few PCR cycles after the wild-type (black line) TOP2α E19/I19 boundary probe (Fig. 6AII). Sanger sequence analysis demonstrated that both wild-type and edited genomic sequence were present in the TOP2α E19/I19 boundary, with the wild-type sequence predominant at all three of the edited sites (Fig. 6BII). Together, these results suggested that one of the three TOP2α E19/I19 boundary alleles was edited in this clonal cell line (now designated K/VP.5/edit-1).
The K/VP.5/edit-1 clonal cell line was subjected to an additional transfection with gRNA-2, Cas9 protein, and the TOP2α HDR template. Forty-eight hours post-transfection, K/VP.5/edit-1 cells were seeded at 0.8 cells per well, grown 2 weeks followed by screening of cell lysates from single clonal colonies by genomic DNA qPCR utilizing the wild-type and CRISPR/Cas9-edited TOP2α E19/I19 boundary probes. From the second transfection (∼60 clonal colonies screened), a clonal cell population was identified where the edited TOP2α E19/I19 boundary probe signal (red line) appeared earlier than the wild-type (black line) TOP2α E19/I19 boundary probe (Fig. 6AIII). Sanger sequence analysis again demonstrated that both wild-type and edited genomic sequences were present in the TOP2α E19/I19 boundary (Fig. 6BIII). However, the CRISPR/Cas9-edited sequence was now predominant in the electropherogram at all three mutated sites (Fig. 6BIII). Together, these results suggested that two of the three TOP2α E19/I19 boundary alleles were edited in this clonal cell line (now designated K/VP.5/edit-2).
Finally, from this same second transfection, another clonal cell line was identified where the genomic qPCR results of a cellular lysate yielded a positive PCR signal (red line) only with the edited TOP2α E19/I19 boundary probe (Fig. 6AIV), suggesting that all three TOP2α E19/I19 boundary alleles were edited. Sanger sequence analysis verified that only the edited sequence was present all three mutated sites (Fig. 6BIV) in this clonal cell line (now designated K/VP.5/edit-3).
RsaI Analysis Validation of CRISPR/Cas9 Editing of the TOP2α E19/I19 5′ SS in K/VP.5 Cells.
Fortuitously, editing the last two nucleotides of the TOP2α E19/I19 5′ SS (AC→GT) (Fig. 5C) introduced a restriction site for RsaI endonuclease (GT↓AC). Successful TOP2α gene editing would allow for RsaI digestion at the E19/I19 boundary. Thus, an independent assay with RsaI endonuclease was carried out to validate the CRISPR-edited TOP2α gene clones. Cell lysates from the CRISPR-generated cell lines (K/VP.5/edit-1→K/VP.5/edit-3) and parental K/VP.5 cells were used as templates for PCR reactions performed with a forward primer that anneals to TOP2α E18 and a reverse primer that anneals to I19 (Fig. 7A). Based on the location of the forward and reverse primers, the expected size of the parental PCR amplicon is 409 bp. After RsaI digestion of putative gene-edited clones, the expected sizes of gene-edited clones are 265 and 154 bp (Fig. 7A). As expected, RsaI did not result in cleavage in nonedited K/VP.5 cells (Fig. 7B). In contrast, partial RsaI digestion of the 409-bp band was evident in K/VP.5/edit-1 lysates with reduction in this parental amplicon. There was greater intensity of cleaved bands in K/VP.5/edit-2 lysates along with greater reduction in the parental PCR 409-bp amplicon. These results are consistent with successful CRISPR/Cas9 editing of one and two TOP2α E19/I19 5′SS alleles in K/VP.5/edit-1 and K/VP.5/edit-2 clonal cell lines, respectively. Importantly, the 409-bp amplicon from K/VP.5/edit-3 lysates was completely digested by RsaI (Fig. 7B) indicating, together with qPCR and sequencing data (Fig. 6), that all the three TOP2α E19/I19 5′ SS alleles were edited by CSRISP/Cas9 and HDR.
Modulation of TOP2α/170 and TOP2α/90 Expression in K/VP.5 Cells with CRISPR/Cas9-Edited TOP2α E19/I19 5′ SS.
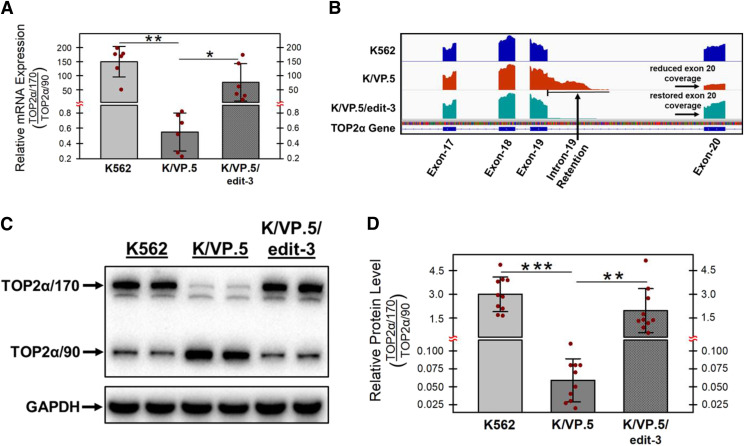
Figure 1A’s qPCR evaluations demonstrate a very high ratio of TOP2α/170:TOP2α/90 mRNA in K562 cells (gray bars) and a much lower ratio in drug-resistant K/VP.5 cells (dark gray bars), reflecting both a decrease in TOP2α/170 mRNA and an increase in TOP2α/90 mRNA due to I19 retention in these drug-resistant cells. We have posited that gene editing of the suboptimal TOP2α E19/I19 5′ SS in K/VP.5 cells will decrease I19 retention and restore levels of TOP2α/170 mRNA and protein relative to those of TOP2α/90. Separate qPCR experiments (Fig. 8A) evaluating TOP2α/170 and TOP2α/90 mRNA in K562 and KVP.5 cells recapitulated Fig. 1A’s results, indicating that TOP2α/170:TOP2α/90 mRNA ratios were statistically significantly decreased in K/VP.5 cells compared with K562 cells. In K/VP.5/edit-3 cells compared with K/VP.5 cells, the TOP2α/170:TOP2α/90 mRNA ratio was statistically significantly increased (Fig. 8A), consistent with our central hypothesis and a putative indication for circumvention of drug resistance.
Fig. 8.
Effects of optimizing the TOP2α intron 19 5′ SS in K/VP.5 on TOP2α/170 mRNA and protein expression in K/VP.5 cells. (A) qPCR analyses were performed from RNA samples isolated from K562, K/VP.5, and KVP.5/edit-3 cells utilizing TaqMan hydrolysis probes specific for TOP2α/170 and TOP2α/90 mRNAs as previously described (Kanagasabai et al., 2017). Results shown are means ± S.D. from six cellular RNA isolation experiments performed on separate days; P = 0.001 comparing the difference in mean values for K562 and K/VP.5 mRNAs, 95% CI (93.1, 206.1); P = 0.038 comparing the difference in mean values for K/VP.5/edit-3 vs. K/VP.5 mRNAs, 95% CI (6.4, 145.5). (B) Visualization of retained intron 19 of TOP2α RNA-seq genome coverage tracks showing the intron 19 retention event in K/VP.5 cells and the restoration of intron removal in K/VP.5/edit-3 cells. Reduced coverage denoted for exon 20 indicates fewer full-length TOP2α/170 reads in K/VP.5 cells. Increased exon 20 coverage in K/VP.5/edit-3 cells indicates restoration of intron 19 removal and more full-length TOP2α/170 reads consistent with expression of greater full-length mature TOP2α/170 mRNA. (C) Representative immunoassay using K562, K/VP.5, and KVP.5/edit-3 cellular lysates. Blots were probed with antibodies specific for the N-terminal portion of TOP2α/170/90 (i.e., amino acids 14–27) or for GAPDH. (D) The ratio of TOP2α/170 to TOP2α/90 protein expression levels was calculated from multiple immunoassays from K562, K/VP.5, and KVP.5/edit-3 cellular lysates. Results shown are means ± S.D. from ten experiments performed on separate days; P < 0.001 comparing the difference in mean values for K562 and K/VP.5 cell TOP2α/170 to TOP2α/90 ratios, 95% CI (2.17, 3.71); P = 0.002 comparing the difference in mean values for K/VP.5/edit-3 and K/VP.5 cell TOP2α/170 to TOP2α/90 ratios, 95% CI (0.83, 2.89). *P < 0.05; **P < 0.01; ***P < 0.001.
To further confirm that our CRISPR/Cas9 strategy effectively diminished I19 retention, total transcriptome analysis was performed by RNA-seq in K562, K/VP.5, and K/VP.5/edit-3 cells. Results (Fig. 8B) clearly visualize I19 retention events in K/VP.5 cell TOP2α RNA-seq genome coverage tracks. In addition, the reduced genome coverage tracks for E20 in K/VP.5 cells indicated, as expected, that there were fewer full-length TOP2α/170 reads in these resistant cells. In K/VP.5/edit-3 cells, there was restoration of I19 removal as well as recovery of full-length TOP2α/170 reads, consistent with increased full-length mature TOP2α/170 mRNA (Fig. 8B). Finally, in K/VP.5/edit-3 cells, there was decreased TOP2α/90 and restoration of TOP2α/170 protein levels comparable to those in parental K562 cells (Fig. 8C) along with a corresponding statistically significant increase in the TOP2α/170:TOP2α/90 protein ratios (Fig. 8D).
Circumvention of Etoposide Resistance in K/VP.5 Cells with CRISPR/Cas9-Edited TOP2α E19/I19 5′ SS.
We previously demonstrated that, in K562 and K/VP.5 cells, etoposide activity was directly related to TOP2α/170 mRNA/protein and inversely correlated with the expression of TOP2α/90 mRNA/protein (Kanagasabai et al., 2017; 2018). Since, in K/VP.5/edit-3 cells, TOP2α/90 mRNA/protein levels were decreased and TOP2α/170 mRNA/protein levels were increased (Fig. 8A, C and D), we next evaluated whether etoposide activity in this gene-edited clonal cell line would be enhanced consistent with circumvention of resistance.
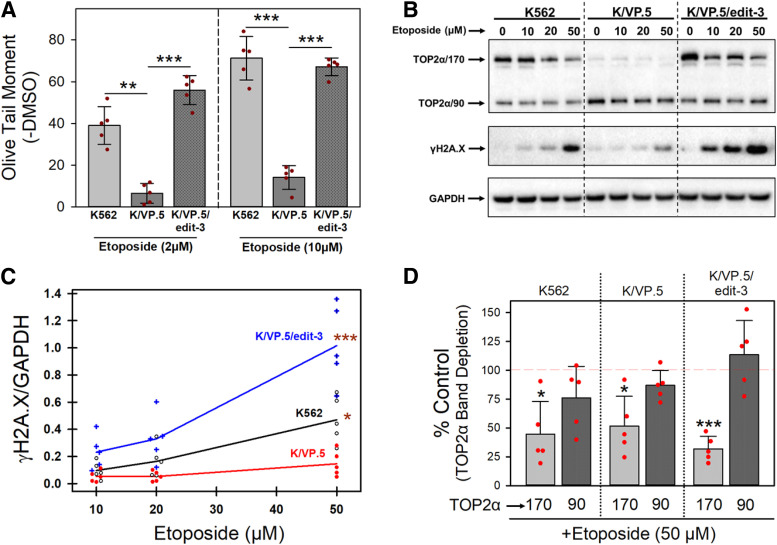
Alkaline single-cell gel electrophoresis (Comet) assays (Olive, 2002) were performed to assess DNA strand breaks (Olive Tail Moment) in K562, K/VP.5, and K/VP.5/edit 3 cells incubated for 30 minutes with 2 and 10 µM etoposide (Fig. 9A). Etoposide induced concentration-dependent DNA strand breaks in K562 cells, which were attenuated in resistant K/VP.5 cells and restored in K/VP.5/edit-3 cells to levels comparable to those in the parental K562 cells. As performed (at pH 13), Comet assays detected primarily single-stranded breaks (Luke et al., 2010). Therefore, etoposide-induced DNA DSBs were also evaluated using expression of phosphorylated H2AX (γH2AX) as an endpoint (Rogakou et al., 1998). Cells were incubated for 30 minutes with DMSO solvent control or etoposide (10–50 µM) followed by lysis and immunoblotting with anti-γH2AX antibody. Etoposide induced a concentration-dependent expression of γH2AX in all three cells lines with evident attenuation of DSBs in resistant K/VP.5 cells and restoration of etoposide-induced DSBs in K/VP.5/edit-3 cells (Fig. 9B). These results were consistent with the profile observed with Comet assays assessing single-stranded breaks (Fig. 9A). Quantitation of γH2AX results from five experiments performed on separate days demonstrated a concentration-dependent increase in etoposide-induced γH2AX in all three cells lines, with a statistically significant increase in DSBs induced in K562 cells (P = 0.023) and K/VP.5/edit-3 cells (P < 0.001) compared with K/VP.5 cells at 50 µM etoposide (Fig. 9C).
Fig. 9.
Effects of optimizing the TOP2α E19/I19 5′ SS on sensitivity to etoposide-induced DNA damage. (A) K562, K/VP.5, and KVP.5/edit-3 cells were incubated with etoposide (2 or 10 μM) or DMSO (control) for 30 minutes followed by alkaline (pH 13) Comet assays. Results shown are means ± S.D. for five cellular experiments run on separate days. For all experimental conditions in each experiment, more than 100 cells were evaluated by OpenComet software. At 2 µM etoposide, P = 0.005 comparing the difference in mean values for K562 and K/VP.5 cells, 95% CI (16.8, 48.3); P < 0.001 comparing the difference in mean values for K/VP.5/edit-3 and K/VP.5 cells, 95% CI (39.7, 59.2). At 10 µM etoposide, P < 0.001 comparing the difference in mean values for K562 and K/VP.5 cells, 95% CI (41.5, 72.7); P < 0.001 comparing the difference in mean values for K/VP.5/edit-3 and K/VP.5 cells, 95% CI (44.9, 61.1). (B) Representative immunoblot from whole cell lysates of K562, K/VP.5, and K/VP.5/edit-3 cells treated with DMSO or etoposide (10, 20, 50 µM) for 30 minutes. The blot was probed with an antibody specific for the N-terminal portion of TOP2α/170/90 (i.e., amino acids 14–27) and with γH2AX and GAPDH antibodies. (C) Scattergram of etoposide-induced DNA double-stranded breaks assessed by γH2AX formation. Lines connect the mean values at each etoposide concentration and are derived from five separate cellular experiments performed on separate days under the same conditions as shown in Fig. 9B. P = 0.023 comparing K562 and K/VP.5 cells at 50 µM etoposide; P < 0.001 comparing K/VP.5/edit-3 and K/VP.5 cells at 50 µM etoposide. Statistical analysis was performed by a repeated measured one-way ANOVA using multiple comparisons vs. the K/VP.5 cell control group (Holm-Sidak method). (D) Etoposide (50 µM) induced TOP2α/170 and TOP2α/90 band depletion. Results shown are means ± S.D. for five separate cellular/immunoblot experiments identical to the representative experiment shown in Fig. 9B. For etoposide-induced TOP2α/170 band depletion compared with 100% DMSO controls: P = 0.012 K562 cells, 95% CI (20.1, 90.4); P = 0.014 K/VP.5 cells, 95% CI (16.3, 80.3); P < 0.001 K/VP.5/edit-3 cells, 95% CI (54.5, 81.7). For etoposide-induced TOP2α/90 band depletion compared with 100% controls: P = 0.122 K562 cells; P = 0.088 K/VP.5 cells; P = 0.365 K/VP.5/edit cells. *P < 0.05; **P < 0.01; ***P < 0.001.
In parallel with increased etoposide-induced DSBs (γH2AX), there was concentration-dependent “band depletion” of TOP2α/170 in K562, K/VP.5, and K/VP.5/edit-3 cells (Fig. 9B), consistent with etoposide-induced formation of high molecular weight TOP2α/DNA covalent complexes prevented from entering gels (Kaufmann and Svingen, 1999). At 50 µM etoposide there was a statistically significant depletion of TOP2α/170 compared with DMSO controls in all three cell lines (Fig. 9D). In contrast, there was no statistically significant decrease in TOP2α/90 consistent with the lack of active site Tyr-805 in this truncated isoform (Kanagasabai et al., 2017, 2018) and the presumed loss of ability to form covalent complexes with DNA. Together, drug-induced DNA damage studies (Fig. 9, A and B) support the hypothesis that optimizing the TOP2α E19/I19 5′ SS sensitizes previously resistant K/VP.5 cells as a consequence of 1) enhanced splicing out of I19, 2) reduced production of TOP2α/90, and 3) restoration of TOP2α/170 levels.
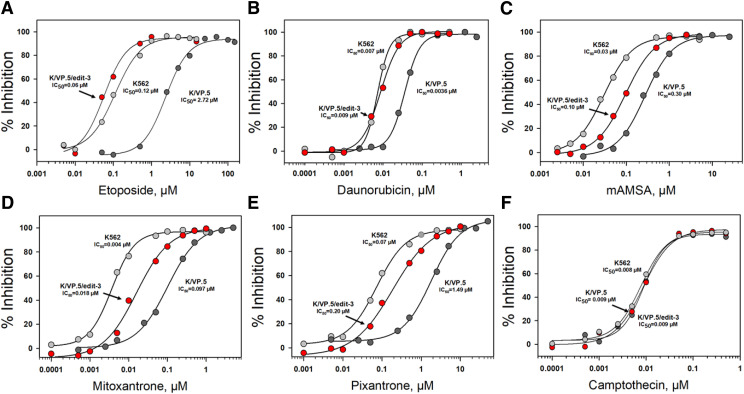
Next, 48-hour growth inhibition assays were performed in K562, K/VP.5, and K/VP.5/edit cells treated continuously with etoposide, other TOP2α-targeted agents, the topoisomerase I inhibitor camptothecin, and the microtubule inhibitor podophyllotoxin (Fig. 10; Table 1). Compared with K562 cells, K/VP.5 cells were 21-fold resistant to etoposide, with complete circumvention of resistance observed in K/VP.5/edit-3 cells (Fig. 10A; Table 1). Separate experiments in K562, K/VP.5, K/VP.5/edit-1, KVP.5/edit-2, and K/VP.5/edit-3 cells revealed a progressive restoration of TOP2α/170 protein levels compared with K562 cells (Supplemental Fig. 1). Moreover, based on IC50 values from replicate etoposide-induced growth inhibition assays (Supplemental Fig. 2; Supplemental Table 2), there was partial but progressive reversal of resistance in the one TOP2α allele- and two TOP2α allele-edited K/VP.5/edit-1 and K/VP.5/edit-2 cells, respectively, with complete circumvention of resistance in K/VP.5/edit-3 cells, consistent with results shown in Fig. 10 and Table 1. These results validate the import of the E19/I19 5′ SS for intron retention, expression of TOP2α/170 mRNA/protein, and drug sensitivity. Of note, doubling times for all five cell lines were similar (Supplemental Table 2), indicating that gene editing did not alter growth characteristics.
Fig. 10.
Growth inhibitory effects of etoposide and other topoisomerase-targeted drugs in K562, K/VP.5, and K/VP.5/edit-3 cells. Log-phase cells were incubated for 48 hours with various concentrations of etoposide (A), daunorubicin (B), mAMSA, (C), mitoxantrone (D), pixantrone (E), and camptothecin (F) after which cells were counted on an electronic particle counter (Z1 Coulter counter). The extent of growth beyond the starting concentration in drug-treated vs. DMSO controls was expressed ultimately as percent inhibition. Shown are representative concentration-response (inhibitory) curves for each of the tested drugs with 50% inhibitory concentrations (IC50 values) indicated. Compilation of replicate experiments performed on different days is shown in Table 1.
TABLE 1.
Growth inhibitory effects of anticancer drugs in K562, K/VP.5, and K/VP.5/edit-3 cells
| Anticancer agent | K562 cells (IC50)a | K/VP.5 cells (IC50) | K/VP.5/edit-3 cells (IC50) | Relative resistanceb (K/VP.5/K562) | Relative resistanceb (K/VP.5/edit-3/K562) |
|---|---|---|---|---|---|
| nM | nM | nM | |||
| Etoposide | 146.8 ± 27.7 (5)c | 3131 ± 1162 (5) | 66.0 ± 6.3 (5) | 21.6 | 0.5 |
| Teniposide | 15.5 ± 1.9 (5) | 291.2 ± 26.0 (5) | 10.1 ± 0.9 (5) | 18.8 | 0.7 |
| Daunorubicin | 7.4 ± 0.5 (6) | 34.5 ± 3.8 (6) | 7.5 ± 1.3 (5) | 4.7 | 1.0 |
| mAMSA | 28.8 ± 4.8 (5) | 274.0 ± 33.4 (5) | 94.0 ± 15.9 (5) | 9.6 | 3.3 |
| Mitoxantrone | 3.6 ± 0.6 (5) | 104.0 ± 22.1 (5) | 14.0 ± 4.0 (5) | 29.7 | 4.0 |
| Pixantrone | 81.4 ± 12.1 (5) | 1457 ± 227 (5) | 233.4 ± 47.7 (5) | 18.1 | 2.9 |
| Camptothecin | 11.2 ± 5.0 (5) | 10.6 ± 3.5 (5) | 10.1 ± 2.7 (5) | 0.9 | 0.9 |
| Podophyllotoxin | 10.9 ± 0.4 (3) | 10.9 ± 1.3 (3) | 10.5 ± 1.1 (3) | 1.0 | 1.0 |
Fifty percent inhibitory concentration (IC50) in a 48-h growth inhibition assay.
IC50 of K/VP.5 or K/VP.5/edit-3 cells divided by that of the parental K562 cell line.
Mean ± S.D.; numbers in parentheses, number of independent experiments performed on different days.
Figure 10 and Table 1 also indicated 5–30-fold crossresistance to teniposide, daunorubicin, mAMSA, pixantrone, and mitoxantrone, respectively (Table 1). As with etoposide, complete circumvention of resistance was observed with teniposide (Table 1) and daunorubicin (Fig. 10B; Table 1) in K/VP.5/edit-3 cells, whereas resistance was extensively but not completely reversed with mAMSA (Fig. 10C), mitoxantrone (Fig. 10D), and pixantrone (Fig. 10E; Table 1). Finally, there was no resistance to camptothecin (Fig. 10F; Table 1) or podophyllotoxin (Table 1) in K/VP.5 or K/VP.5/edit-3 cells compared with K562 cells, thus validating the specificity of the resistance phenotype in K/VP.5 cells for TOP2α-targeted agents and indicating that gene editing did not impact the sensitivity to two agents with intracellular targets differing from TOP2α. Together, the results were again consistent with optimization of the E19/I19 5′ SS playing an important role in diminishing intron retention leading to restored TOP2α/170 mRNA/protein and greater sensitivity to TOP2α-targeting agents.
Discussion
Chemoresistance to TOP2α-targeted agents can result from a number of molecular mechanisms (Ganapathi and Ganapathi, 2013; Capelôa et al., 2020). In the case of K/VP.5 cells, acquired resistance to etoposide and similar TOP2α poisons is due, in part, to decreased TOP2α/170 expression associated with the increased expression of a C-terminal truncated isoform, TOP2α/90 (Fig. 1, A and B), as a result of intron 19 retention and processing (Kanagasabai et al., 2017, 2018). Importantly, the truncated TOP2α/90 isoform lacks the active site Tyr-805 required to generate DNA strand breaks and is a determinant of chemoresistance through a dominant-negative effect related to heterodimerization with TOP2α/170, leading to reduced drug-induced TOP2α/170-DNA covalent cleavage complexes, attenuated DNA damage, and decreased cytotoxic action (Kanagasabai et al., 2017, 2018).
Intron retention affects ∼80% of protein coding genes in humans and is characterized by the inclusion of one or more introns in mature mRNA transcripts (Middleton et al., 2017). The SS sequences that define the 5′ and/or 3′ SS retained introns are often weak (Monteuuis et al., 2019). Importantly, several studies have demonstrated that the strengthening of a weak 5′ SS by mutagenesis could enhance the splicing out of retained introns (Huang et al., 2012; Wickramasinghe et al., 2015; Eckert et al., 2016; Yue et al., 2017). Here, we used CRISPR/Cas9 gene editing to optimize the weak TOP2α E19/I19 5′ SS in K/VP.5 cells to improve splicing and circumvent drug resistance.
The CRISPR/Cas9 gene editing system requires a gRNA, which comprises the crRNA (required for DNA targeting) and the tracrRNA (necessary for nuclease activity) (Jinek et al., 2012; Cong et al., 2013; Mali et al., 2013). To direct the Cas9 nuclease, the 20-nucleotide crRNA sequence must be complementary to the target DNA, and a three-nucleotide sequence motif (NGG), known as the PAM, must also be present in the targeted locus (Fig. 4A). Once bound to the target DNA, the Cas9 nuclease introduces a blunt-end DSB three bases upstream of the PAM (Fig. 4A) (Jinek et al., 2012; Cong et al., 2013; Mali et al., 2013). Cas9-induced DSBs are predominantly repaired by the error-prone NHEJ pathway, which results in nonspecific indels (Hsu et al., 2014).
Importantly, however, DSBs can also be repaired by HDR by utilizing exogenous custom repair templates, thus allowing knock-in of specific mutations (Cong et al., 2013). Nevertheless, high editing efficiency requires gRNA targeting close to the intended mutation (Paquet et al., 2016; Liang et al., 2017). Therefore, we focused on the three candidate PAMs identified by CCTop (Stemmer et al., 2015) near the suboptimal/weak TOP2α E19/I19 5′ SS (Fig. 4A). Interestingly, GCD assays demonstrated that only gRNA-2 effectively targeted Cas9 to introduce DSBs in the TOP2α E19/I19 boundary sequence (Fig. 4, A–C). Given that all three gRNAs anneal with genomic DNA sequences that are in close proximity to each other, differences in accessibility to their TOP2α E19/I19 boundary targets cannot account for the lack of targeting by gRNA-1 and -3 (Uusi-Mäkelä et al., 2018). It is possible that the lack of gRNA-1 and -3 activity results from either the failure to form a functional Cas9-gRNA complex or internal interactions between the crRNA and tracrRNA that may interfere with Cas9-mediated cleavage (Thyme et al., 2016).
To expedite the screening of the edited TOP2α E19/I19 5′ SS in K/VP.5 cells, single-cell clones isolated by limiting dilution were subjected to genomic DNA qPCR utilizing hybridization probes that discriminated between the wild-type TOP2α E19/I19 and the CRISPR/Cas9-edited TOP2α E19/I19 boundary (compare Fig. 6, AI and AIV). Of the ∼60 clonal cell lines screened, nine contained the desired three genomic edits in one TOP2α allele based on qPCR (Fig. 6AII). Sanger sequencing (Fig. 6BII) from one of the clones was consistent with one TOP2α allele edited based on the electropherogram peaks with no indication of indels in the two nonedited TOP2α alleles. Therefore, this clonal cell line (designated K/VP.5/edit-1) was then subjected to an additional round of transfection, and ∼60 new clonal cell lines were screened by genomic DNA qPCR. Five clones were identified that had the desired edits in two TOP2α alleles (Fig. 6, AIII and BIII; i.e., K/VP.5/edit-2). One clonal cell line was characterized that had all three TOP2α alleles properly edited across the TOP2α E19/I19 5′ boundary (Fig. 6, AIV and BIV; i.e., K/VP.5/edit-3). Finally, RsaI restriction enzyme analysis validated the interpretation of the CRISPR/Cas9 editing of TOP2α alleles in K/VP.5/edit-1–3 clonal cell lines (Fig. 7B).
Characterization of K/VP.5/edit-3 clonal cells revealed that TOP2α/90 protein levels were reduced, whereas TOP2α/170 levels were increased compared with K/VP.5 cells and were at similar levels to those in parental K562 cells (Fig. 8C). In addition, TOP2α/170:TOP2α/90 mRNA and protein ratios were statistically significantly increased compared with etoposide-resistant K/VP.5 cells and were comparable to the parental K562 cells (Fig. 8, A and D). Together, these results strongly suggested that CRISPR/Cas9 editing of the TOP2α E19/I19 5′ SS improved the removal of I19. RNA-seq genome coverage track reads demonstrated that I19 retention events were decreased in K/VP.5/edit-3 clonal cells compared with K/VP.5 cells (Fig. 8B), thus independently validating our qPCR and Western blot data.
Importantly, functional studies demonstrated that sensitivity to etoposide-induced DNA damage (Fig. 9, A–C) and etoposide-induced growth inhibition (Fig. 10A; Supplemental Table 2; Table 1) was restored in K/VP.5/edit-3 cells to levels comparable to those found in parental K562 cells. The lack of etoposide-induced band depletion for TOP2α/90 in all cell lines (Fig. 9D) is consistent with the absence of the active site Tyr-805 in this truncated isoform, preventing formation of covalent complexes with DNA. Sensitivity to additional TOP2α poisons was also completely (e.g., daunorubicin) (Fig. 10B; Table 1) or partially (e.g., mAMSA, mitoxantrone, pixantrone) restored (Fig. 10, C–E; Table 1) in K/VP.5/edit-3 cells.
CRISPR/Cas9 off-target effects might have played a role in the circumvention of the drug resistance phenotype observed in K/VP.5/edit-3 cells (Figs. 9 and 10). To limit mutations (indels) and unwanted chromosomal translocations, we transiently transfected K/VP.5 cells with Cas9/gRNA-2 ribonucleoprotein complexes to restrict the temporal activity of these complexes, improve precision, and reduce off-target effects (Kim et al., 2014). Given that the doubling times for all our cell lines (gene-edited or not) were similar (Supplemental Table 2), CRISPR/Cas9 gene editing did not significantly alter the growth characteristics of K/VP.5 edited cells. In addition, evaluating our RNA-seq data, essential genes (i.e., required for cell survival) (Wang et al., 2019a) were not differentially expressed (fold change cutoff of 2 at 10% false discovery rate) in gene-edited K/VP.5/edit-3 compared with K/VP.5 cells (Supplemental Fig. 3, denoted in red), consistent with a lack of important off-target effects. Additionally, the CCTop algorithm (Stemmer et al., 2015) was used to predict potential Cas9/gRNA-2 off-target genes where induced DSBs could result in unwanted indels and chromosomal translocations. Given that more than four mismatches between the gRNA and target DNA would prevent Cas9-mediated DSB induction (Cong et al., 2013; Hsu et al., 2013; Cho et al., 2014), potential gRNA-2 off-target sites were predicted, allowing four mismatches and querying the human genomic sequence (Homo sapiens GRCh38/hg38). A total of 137 off-target sites were identified. Of these, only 55 were expressed in K/VP.5 and K/VP.5/edit-3 cells. Of the top 20 putative Cas9/gRNA-2 off-target sites/genes, 18 were not differentially expressed (fold change cutoff of 2 at 10% false discovery rate) comparing gene-edited K/VP.5/edit-3 cells and K/VP.5 cells, whereas the two lowest expressing genes in this group exhibited slightly greater than a 2-fold change (Supplemental Fig. 3, denoted in blue), suggesting that off-target effects did not contribute to the overall phenotype observed in K/VP.5/edit-3 cells. Of note, the DNA topoisomerase IIβ gene E19/I19 5′ SS was one of the top 20 putative Cas9/gRNA-2 off-target sites/genes. To demonstrate that this gene was not targeted by Cas9/gRNA-2, genomic DNA was isolated from K/VP.5/edit-3 cells, and the DNA topoisomerase IIβ E19/I19 boundary was analyzed by a GCD assay; no indels were present (data not shown), suggesting that this gene was not targeted by Cas9/gRNA-2. Finally, circumvention of resistance in K/VP.5/edit-3 cells was observed only to those drugs targeting TOP2α with no changes in sensitivity to camptothecin, a TOP1 inhibitor, or to podophyllotoxin, a microtubule inhibitor (Table 1). Although CRISPR/Cas9 off-target effects may occur in K/VP.5/edit-3 cells, results suggest that the predominant phenotype associated with circumvention of resistance appears to be driven by the modifications in the TOP2α E19/I19 5′ SS.
Importantly, the same suboptimal/weak TOP2α E19/I19 5′ SS is present in both parental K562 and resistant K/VP.5 cells (Kanagasabai et al., 2017, 2018). Hence, increased TOP2α I19 retention in K/VP.5 cells is likely due to aberrant spliceosome function/effectors in this acquired resistant cell line. Our CRISPR/Cas9 editing experiments demonstrated circumvention of resistance by optimizing the TOP2α E19/I19 5′ SS, thereby improving splicing out of I19, decreasing production of TOP2α/90, and increasing levels of TOP2α/170 in spite of putative/presumed alterations in spliceosome function. Future studies will focus on the identification and characterization of splicing factors and cis-elements regulating TOP2α I19 retention in acquired resistance to TOP2α-targeting drugs.
Acknowledgments
We are grateful to the patients (and healthy volunteers) who provided tissue samples for these studies to The Ohio State University Comprehensive Cancer Center Leukemia Tissue Bank Shared Resource (supported by National Institutes of Health National Cancer Institute P30 CA016058) and to Christopher Manring, Clinical Laboratory Manager, who facilitated our obtaining these samples.
Abbreviations
- AML
acute myeloid leukemia
- bp
base pair
- Cas9
CRISPR-associated protein 9
- CCTop
CRISPR/Cas9 Target Online Predictor
- CI
confidence interval
- CMV pro
cytomegalovirus promoter
- crRNA
CRISPR RNA
- DMEM
Dulbecco’s modified Eagle’s medium
- DSB
double-stranded break
- E19
exon 19
- E20
exon 20
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- GCD
genomic cleavage detection
- gRNA
crRNA:tracrRNA duplex
- γH2AX
phosphorylated Ser-139 residue of the H2A histone family member X
- HDR
homology-directed repair
- HPRT1
hypoxanthine-guanine phosphoribosyltransferase
- I19
intron 19
- indel
insertions/deletion
- NHEJ
nonhomologous end joining
- PAM
protospacer-adjacent motif
- PCR
polymerase chain reaction
- polyA site
polyadenylation site
- qPCR
quantitative PCR
- RNA-seq
RNA sequencing
- SS
splice site
- ssODN
single-stranded oligo DNA nucleotide
- TOP2α
DNA topoisomerase IIα protein
- TOP2α/170
TOP2α 170 kDa
- TOP2α/90
TOP2α 90 kDa
- T7 pro
T7 polymerase promoter
- tracrRNA
trans-activating CRISPR RNA
Authorship Contributions
Participated in research design: Hernandez, Carvajal-Moreno, Papa, Shkolnikov, Ozer, Yalowich, Elton.
Conducted experiments: Hernandez, Carvajal-Moreno, Papa, Shkolnikov.
Contributed new reagents or analytic tools: Li, Ozer.
Performed data analysis: Hernandez, Carvajal-Moreno, Papa, Shkolnikov, Li, Ozer, Yalowich, Elton.
Wrote or contributed to the writing of the manuscript: Hernandez, Elton, Yalowich, Li, Ozer.
Footnotes
This work was supported by National Institutes of Health National Cancer Institute [Grant R01 CA226906-01A1] (to J.C.Y. and T.S.E.)
 This article has supplemental material available at molpharm.aspetjournals.org.
This article has supplemental material available at molpharm.aspetjournals.org.
References
- Boutz PL, Bhutkar A, Sharp PA (2015) Detained introns are a novel, widespread class of post-transcriptionally spliced introns. Genes Dev 29:63–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capelôa T, Benyahia Z, Zampieri LX, Blackman MCNM, Sonveaux P (2020) Metabolic and non-metabolic pathways that control cancer resistance to anthracyclines. Semin Cell Dev Biol 98:181–191. [DOI] [PubMed] [Google Scholar]
- Chen SH, Chan NL, Hsieh TS (2013) New mechanistic and functional insights into DNA topoisomerases. Annu Rev Biochem 82:139–170. [DOI] [PubMed] [Google Scholar]
- Cho SW, Kim S, Kim Y, Kweon J, Kim HS, Bae S, Kim JS (2014) Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res 24:132–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cioe L, McNab A, Hubbell HR, Meo P, Curtis P, Rovera G (1981) Differential expression of the globin genes in human leukemia K562(S) cells induced to differentiate by hemin or butyric acid. Cancer Res 41:237–243. [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339:819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper TA (2005) Use of minigene systems to dissect alternative splicing elements. Methods 37:331–340. [DOI] [PubMed] [Google Scholar]
- Deweese JE, Osheroff N (2009) The DNA cleavage reaction of topoisomerase II: wolf in sheep’s clothing. Nucleic Acids Res 37:738–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert D, Andrée N, Razanau A, Zock-Emmenthal S, Lützelberger M, Plath S, Schmidt H, Guerra-Moreno A, Cozzuto L, Ayté J, et al. (2016) Prp4 kinase grants the license to splice: control of weak splice sites during spliceosome activation. PLoS Genet 12:e1005768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elton TS, Ozer HG, Yalowich JC (2020) Effects of DNA topoisomerase IIα splice variants on acquired drug resistance. Cancer Drug Resist 3:161–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadepalli VS, Ozer HG, Yilmaz AS, Pietrzak M, Webb A (2019) BISR-RNAseq: an efficient and scalable RNAseq analysis workflow with interactive report generation. BMC Bioinformatics 20 (Suppl 24):670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganapathi RN, Ganapathi MK (2013) Mechanisms regulating resistance to inhibitors of topoisomerase II. Front Pharmacol 4:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyori BM, Venkatachalam G, Thiagarajan PS, Hsu D, Clement MV (2014) OpenComet: an automated tool for comet assay image analysis. Redox Biol 2:457–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harker WG, Slade DL, Parr RL, Holguin MH (1995) Selective use of an alternative stop codon and polyadenylation signal within intron sequences leads to a truncated topoisomerase II alpha messenger RNA and protein in human HL-60 leukemia cells selected for resistance to mitoxantrone. Cancer Res 55:4962–4971. [PubMed] [Google Scholar]
- Hicks MJ, Mueller WF, Shepard PJ, Hertel KJ (2010) Competing upstream 5′ splice sites enhance the rate of proximal splicing. Mol Cell Biol 30:1878–1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, et al. (2013) DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol 31:827–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PD, Lander ES, Zhang F (2014) Development and applications of CRISPR-Cas9 for genome engineering. Cell 157:1262–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang SC, Ou AC, Park J, Yu F, Yu B, Lee A, Yang G, Zhou A, Benz EJ Jr (2012) RBFOX2 promotes protein 4.1R exon 16 selection via U1 snRNP recruitment. Mol Cell Biol 32:513–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E (2012) A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337:816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanagasabai R, Karmahapatra S, Kientz CA, Yu Y, Hernandez VA, Kania EE, Yalowich JC, Elton TS (2018) The novel C-terminal truncated 90-kDa isoform of topoisomerase IIα (TOP2α/90) is a determinant of etoposide resistance in K562 leukemia cells via heterodimerization with the TOP2α/170 isoform. Mol Pharmacol 93:515–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanagasabai R, Serdar L, Karmahapatra S, Kientz CA, Ellis J, Ritke MK, Elton TS, Yalowich JC (2017) Alternative RNA processing of topoisomerase IIα in etoposide-resistant human leukemia K562 cells: intron retention results in a novel C-terminal truncated 90-kDa isoform. J Pharmacol Exp Ther 360:152–163. [DOI] [PubMed] [Google Scholar]
- Kaufmann SH, Svingen PA (1999) Immunoblot analysis and band depletion assays. Methods Mol Biol 94:253–268. [DOI] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL (2015) HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12:357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Kim D, Cho SW, Kim J, Kim JS (2014) Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res 24:1012–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosaki T, Maquat LE (2016) Nonsense-mediated mRNA decay in humans at a glance. J Cell Sci 129:461–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labuhn M, Adams FF, Ng M, Knoess S, Schambach A, Charpentier EM, Schwarzer A, Mateo JL, Klusmann JH, Heckl D (2018) Refined sgRNA efficacy prediction improves large- and small-scale CRISPR-Cas9 applications. Nucleic Acids Res 46:1375–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Rio DC (2015) Mechanisms and regulation of alternative pre-mRNA splicing. Annu Rev Biochem 84:291–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Bor YC, Fitzgerald MP, Lee KS, Rekosh D, Hammarskjold ML (2016) An NXF1 mRNA with a retained intron is expressed in hippocampal and neocortical neurons and is translated into a protein that functions as an Nxf1 cofactor. Mol Biol Cell 27:3903–3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Potter J, Kumar S, Ravinder N, Chesnut JD (2017) Enhanced CRISPR/Cas9-mediated precise genome editing by improved design and delivery of gRNA, Cas9 nuclease, and donor DNA. J Biotechnol 241:136–146. [DOI] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W (2014) featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30:923–930. [DOI] [PubMed] [Google Scholar]
- Luke AM, Chastain PD, Pachkowski BF, Afonin V, Takeda S, Kaufman DG, Swenberg JA, Nakamura J (2010) Accumulation of true single strand breaks and AP sites in base excision repair deficient cells. Mutat Res 694:65–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM (2013) RNA-guided human genome engineering via Cas9. Science 339:823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton R, Gao D, Thomas A, Singh B, Au A, Wong JJ, Bomane A, Cosson B, Eyras E, Rasko JEJ, et al. (2017) IRFinder: assessing the impact of intron retention on mammalian gene expression. Genome Biol 18:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo YY, Beck WT (1997) Heterogeneous expression of DNA topoisomerase II alpha isoforms in tumor cell lines. Oncol Res 9:193–204. [PubMed] [Google Scholar]
- Monteuuis G, Wong JJL, Bailey CG, Schmitz U, Rasko JEJ (2019) The changing paradigm of intron retention: regulation, ramifications and recipes. Nucleic Acids Res 47:11497–11513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitiss JL (2009) Targeting DNA topoisomerase II in cancer chemotherapy. Nat Rev Cancer 9:338–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olive PL (2002) The comet assay. An overview of techniques. Methods Mol Biol 203:179–194. [DOI] [PubMed] [Google Scholar]
- Paquet D, Kwart D, Chen A, Sproul A, Jacob S, Teo S, Olsen KM, Gregg A, Noggle S, Tessier-Lavigne M (2016) Efficient introduction of specific homozygous and heterozygous mutations using CRISPR/Cas9. Nature 533:125–129. [DOI] [PubMed] [Google Scholar]
- Pommier Y, Marchand C (2011) Interfacial inhibitors: targeting macromolecular complexes. Nat Rev Drug Discov 11:25–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pommier Y, Sun Y, Huang SN, Nitiss JL (2016) Roles of eukaryotic topoisomerases in transcription, replication and genomic stability. Nat Rev Mol Cell Biol 17:703–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA (2013) Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152:1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F, Ryan DP, Grüning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dündar F, Manke T (2016) deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res 44:W160–W165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK (2015) Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritke MK, Yalowich JC (1993) Altered gene expression in human leukemia K562 cells selected for resistance to etoposide. Biochem Pharmacol 46:2007–2020. [DOI] [PubMed] [Google Scholar]
- Ritke MK, Roberts D, Allan WP, Raymond J, Bergoltz VV, Yalowich JC (1994) Altered stability of etoposide-induced topoisomerase II-DNA complexes in resistant human leukaemia K562 cells. Br J Cancer 69:687–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP (2011) Integrative genomics viewer. Nat Biotechnol 29:24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM (1998) DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem 273:5858–5868. [DOI] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3:1101–1108. [DOI] [PubMed] [Google Scholar]
- Shoubridge C, Jackson M, Grinton B, Berkovic SF, Scheffer IE, Huskins S, Thomas A, Ware T (2019) Splice variant in ARX leading to loss of C-terminal region in a boy with intellectual disability and infantile onset developmental and epileptic encephalopathy. Am J Med Genet A 179:1483–1490. [DOI] [PubMed] [Google Scholar]
- Stemmer M, Thumberger T, Del Sol Keyer M, Wittbrodt J, Mateo JL (2015) CCTop: an intuitive, flexible and reliable CRISPR/Cas9 target prediction tool. PLoS One 10:e0124633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thyme SB, Akhmetova L, Montague TG, Valen E, Schier AF (2016) Internal guide RNA interactions interfere with Cas9-mediated cleavage. Nat Commun 7:11750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uusi-Mäkelä MIE, Barker HR, Bäuerlein CA, Häkkinen T, Nykter M, Rämet M (2018) Chromatin accessibility is associated with CRISPR-Cas9 efficiency in the zebrafish (Danio rerio). PLoS One 13:e0196238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzor S, Zorzou P, Bowler E, Porazinski S, Wilson I, Ladomery M (2018) Autoregulation of the human splice factor kinase CLK1 through exon skipping and intron retention. Gene 670:46–54. [DOI] [PubMed] [Google Scholar]
- Vlasova II, Feng WH, Goff JP, Giorgianni A, Do D, Gollin SM, Lewis DW, Kagan VE, Yalowich JC (2011) Myeloperoxidase-dependent oxidation of etoposide in human myeloid progenitor CD34+ cells. Mol Pharmacol 79:479–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Wang M, Zhang W, Xiao T, Chen CH, Wu A, Wu F, Traugh N, Wang X, Li Z, et al. (2019) Integrative analysis of pooled CRISPR genetic screens using MAGeCKFlute. Nat Protoc 14:756–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Buolamwini JK (2019) A novel RNA variant of human concentrative nucleoside transporter 1 (hCNT1) that is a potential cancer biomarker. Exp Hematol Oncol 8:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickramasinghe VO, Gonzàlez-Porta M, Perera D, Bartolozzi AR, Sibley CR, Hallegger M, Ule J, Marioni JC, Venkitaraman AR (2015) Regulation of constitutive and alternative mRNA splicing across the human transcriptome by PRPF8 is determined by 5′ splice site strength. Genome Biol 16:201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q, Mirski SE, Sparks KE, Cole SP (1997) Two COOH-terminal truncated cytoplasmic forms of topoisomerase II α in a VP-16-selected lung cancer cell line result from partial gene deletion and alternative splicing. Biochemistry 36:5868–5877. [DOI] [PubMed] [Google Scholar]
- Yue M, Ogawa Y (2018) CRISPR/Cas9-mediated modulation of splicing efficiency reveals short splicing isoform of Xist RNA is sufficient to induce X-chromosome inactivation. Nucleic Acids Res 46:e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B, Ho SS, Greer SU, Zhu X, Bell JM, Arthur JG, Spies N, Zhang X, Byeon S, Pattni R, et al. (2019) Comprehensive, integrated, and phased whole-genome analysis of the primary ENCODE cell line K562. Genome Res 29:472–484. [DOI] [PMC free article] [PubMed] [Google Scholar]