Abstract
Viperin is a radical S-adenosylmethionine (SAM) enzyme that inhibits viral replication by converting cytidine triphosphate (CTP) into 3′-deoxy-3′,4′-didehydro-CTP and by additional undefined mechanisms operating through its N- and C-terminal domains. Here, we describe crystal structures of viperin bound to a SAM analog and CTP or uridine triphosphate (UTP) and report kinetic parameters for viperin-catalyzed reactions with CTP or UTP as substrates. Viperin orients the C4′ hydrogen atom of CTP and UTP similarly for abstraction by a 5′-deoxyadenosyl radical, but the uracil moiety introduces unfavorable interactions that prevent tight binding of UTP. Consistently, kcat is similar for CTP and UTP whereas Km for UTP is much greater. The structures also show that nucleotide binding results in ordering of the C-terminal tail and reveal that this region contains a P-loop that binds the γ-phosphate of the bound nucleotide. Collectively, the results explain the selectivity for CTP and reveal a structural role for the C-terminal tail in binding CTP and UTP.
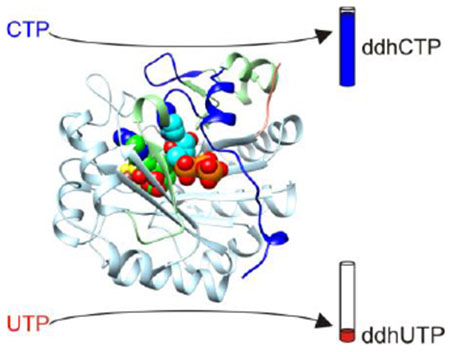
Graphical Abstract
Introduction
Antiviral defense mechanisms within the innate immune system include the production of interferons and the expression of hundreds of interferon-stimulated genes (ISGs).1–3 RSAD2 is an ISG that encodes viperin, an antiviral radical S-adenosylmethionine (SAM) enzyme.4–6 Early studies found that viperin associates with the endoplasmic reticulum (ER) and inhibits the replication of human cytomegalovirus, a double stranded DNA virus.5 Since then, viperin has been shown to inhibit the replication of several positive7–19 and negative20–23 sense single-stranded RNA viruses, and the retroviruses HIV-124 and equine infectious anemia virus.25 In addition, viperin has been shown to be a component of the Toll-like receptor 7 (TLR7) and TLR9 signaling pathways26–28 and a regulator of thermogenesis in adipose tissues29 and cellular levels of farnesyl pyrophosphate synthase.30
Recently, rat viperin was shown to convert cytidine triphosphate (CTP) into the RNA chain terminator 3′-deoxy-3′,4′-didehydro-CTP (ddhCTP), which inhibits the RNA-dependent RNA polymerases of dengue, West Nile, Zika, and hepatitis C viruses.31 Viperin activity, reported by the level of 5′-deoxyadenosine (5′-dA) formation, was also detected for other nucleotides, including cytidine diphosphate, deoxy-CTP, and uridine triphosphate (UTP), but was much lower than that obtained using CTP.31 The finding that the substrate of viperin is a triphosphate nucleotide is consistent with the active site structure of mouse viperin,32 which displays structural homology to the triphosphate binding site of the GTP-modifying radical SAM enzyme MoaA.33 However, the structural basis of substrate selectivity of viperin has not yet been demonstrated.
In addition to the radical-based mechanism, studies of various N- and C-terminal truncations suggest that additional undefined antiviral mechanisms operate through regions outside the radical SAM core domain. N-terminal deletions as short as typically 33-50 residues and short C-terminal truncations ranging from 1 to 17 residues lead to the loss of antiviral activity against several viruses.8–11, 13–15, 21, 24, 25 The N-terminal truncations remove an amphipathic helix that is believed to localize viperin to the cytosolic face of the ER34 suggesting that its subcellular localization can play an important role in viral inhibition. In contrast, the precise function of the C-terminal extension has remained elusive due in part to disorder observed in the final 25 residues of nucleotide-free structures of viperin,32 however, roles in binding an FeS cluster assembly protein14, 35 and in interfering with viral replication by binding viral or host proteins have been reported.9, 10, 16, 19
In the present study, we describe high resolution crystal structures of mouse viperin bound to S-adenosylhomocysteine (SAH; a structural analog of SAM) and CTP or UTP with an ordered C-terminal tail. In addition, we report kinetic parameters for ddhCTP and ddhUTP synthesis. The results provide a structural basis for the early steps of ddhCTP and ddhUTP formation, the high selectivity of viperin for CTP, and a role for the C-terminal tail in substrate binding.
Materials and Methods
Overexpression, purification, and crystallization.
Three mutations were made to mouse viperin to produce a crystallizable variant suitable for studying nucleotide binding. These include deletion of the first 44 residues, which removes the N-terminal membrane-associating helix,34 and mutation of Glu261 and Glu266 to alanine,36 which removes interactions between a surface loop and the positively charged barrel that occur in crystals of mouse viperin lacking nucleotides.32 The associated gene, MmviperinΔ44/E261A/E266A, was synthesized using codon optimization for expression in Escherichia coli and sub-cloned into pET28a for overexpression of the following product: NH2-MGSDKIHHHHHHSSGENLYFQG45…A261…A266…W362-COOH.
The viperin variant was prepared via quasi-anaerobic expression followed by anaerobic purification and crystallization.37, 38 E. coli Nico (DE3) cells were transformed with pET28a-MmviperinΔ44/E261A/E266A and pSuf39 for coexpression of the viperin fragment and iron-sulfur biosynthetic proteins. Large-scale cultures were grown at 37 °C in shaker flasks containing 1.85 L of selective minimal medium (1 X minimal medium salts, 40 mg/L kanamycin, 34 mg/L chloramphenicol, 6 g/L dextrose, 3 mM MgSO4, and 0.1 mM CaCl2) at a rotation rate of 180 rpm until the optical density reached 0.5-0.55. The cultures were then chilled at 4 °C for 2.5 h, and supplemented with L-Cys, Fe(NH4)2(SO4)2, and isopropyl β-D-1-thiogalactopyranoside to final concentrations of 0.28 mM, 0.086 mM, and 0.2 mM, respectively. Protein expression was induced for 20 h at 15 °C using a shaker rotation rate of 50 rpm. After induction, the cultures were chilled to 4 ºC, harvested via centrifugation at 6,000 x g and 4 ºC for 15 min, and flash frozen in liquid nitrogen.
Pellets were thawed in an anaerobic glovebox (Coy Laboratories), mixed with lysis buffer (50 mM HEPES, 400 mM NaCl, 25 mM imidazole, 5 mM dithiothreitol, 0.4 mg/mL lysozyme, and 1.9 kU benzonase, pH 7.3), and sonicated while maintained on ice. The lysate was centrifuged outside the glovebox at 60,000 x g for 20 min at 4 °C, then returned and subjected to nickel chelate chromatography using wash (50 mM HEPES, 400 mM NaCl, 25 mM imidazole, pH 7.3) and elution (50 mM HEPES, 400 mM NaCl, 250 mM imidazole, pH 7.6) buffers. The protein was transferred to (His)6 tag-cleavage buffer (25 mM HEPES, 225 mM NaCl, and 3 mM DTT, pH 7.4) using a BioRad Econo Pac 10DG desalting column and then incubated with tobacco etch virus protease for 7 h. The mixture was then subjected to subtractive nickel chelate chromatography, transferred to 5 mM HEPES, 27 mM NaCl, pH 7.1, and frozen in liquid nitrogen. Identical procedures were used to prepare MmviperinΔ4432 for the kinetic assays.
MmviperinΔ44/E261A/E266A (0.2 mM) containing a [4Fe-4S] cluster was cocrystallized with SAH (6 mM) and CTP (10 mM) or UTP (10 mM) using the hanging drop vapor diffusion method. Solutions of viperin and the ligands were mixed with reservoir solutions in a 1:1 ratio at 23 °C. Typical reservoir solutions contained 100 mM Bis-tris propane, pH 6.2-6.8, 200 mM NaCl, and 19-22% (w/v) polyethylene glycol (PEG) 4000 or 6000; the PEG concentration was increased 18-21% in the cryoprotectant.
X-ray image collection and processing.
Crystals were irradiated with X-rays having wavelength λ = 0.9791 Å at NE-CAT beamline 24-ID-C of the Advanced Photon Source (Argonne National Laboratory). X-ray images were recorded at 1 °/s for a CTP or UTP-containing crystal positioned 255 or 340 mm, respectively, from a PILATUS 6M-F detector. The recorded intensities were indexed, integrated, and scaled using HKL2000.40
Structure determination and refinement.
The structures of the CTP- and UTP-bound complexes were determined by molecular replacement using PHASER41 with the nucleotide-free structure as the search model.32 Manual structure building was performed using COOT42 and automatic refinement was performed using PHENIX43 with a maximum likelihood target function and translation/libration/screw motion (TLS) restraints.44 Structural and electron density illustrations were prepared using Chimera45 and PyMOL.46
Kinetic assay for mouse viperin with CTP.
Mouse viperin kinetic assays were performed by varying the concentration of CTP from 10 μM to 100 μM in 100 μL reaction buffer containing 200 mM ammonium formate, pH 8.2, 100 μM SAM, 2 μM viperin and 3 mM dithionite. All the reaction components except dithionite were incubated at room temperature for 5 min. Reactions were initiated by adding dithionite and incubated for 5 min at 37 °C, then quenched by mixing with 100 μL acetonitrile and 50 μM UTP as an internal standard. After centrifuging at 17,000 g for 20 min, the supernatants were analyzed by LC-MS on a Shimazu LC-20AD coupled with a Thermo Scientific LCQ Fleet mass spectrometer. Assay mixtures were loaded on a Phenomenex Kinetex 5 μm EVO C18 column (50 mm × 2.1 mm) equilibrated with 100% buffer A (water with 0.1% acetic acid) and 0% buffer B (acetonitrile with 0.1% acetic acid). An isocratic wash of 100% A was applied from 0-3min, and 0-90% B from 3-6 min before returning to 0% A from 6-8 min. Detection of products and substrates was conducted using electrospray in negative mode with mass scan window from 400 to 500. The amounts of ddhCTP in the reactions were calculated by multiplying the initial CTP concentration with the area ratio (ddhCTP/CTP) in the extracted-ion chromatogram.
Kinetic assay for mouse viperin with UTP.
Mouse viperin kinetic assays were performed by varying the concentration of UTP ranging from 25 μM to 500 μM in 100 μL reaction buffer containing 200 mM ammonium formate, pH 8.2, 100 μM SAM, 5 μM viperin and 3 mM dithionite. All the reaction components except dithionite were mixed and incubated at room temperature for 5 min before introducing dithionite. Reactions were initiated by adding dithionite and incubated for 10 min at 37 °C, then quenched by mixing 100 μL acetonitrile and 100 μM CTP as an internal standard. The samples were treated and analyzed as previously described for the assay with CTP. The amounts of ddhUTP in the reactions were calculated by multiplying the initial UTP concentration with the area ratio (ddhUTP/UTP) in the extracted-ion chromatogram.
Results and Discussion
Crystal structure analysis
Crystal structures of a mouse viperin fragment (residues 45-362) with two surface loop point mutations (E261A/E266A)36 bound to a [4Fe-4S] cluster, SAH, and CTP or UTP were determined at 1.5 and 1.9 Å resolution, respectively (Figure 1A and Tables 1 and 2). The associated electron density maps show well defined electron density for the [4Fe-4S] clusters and ligands (Figure S1). The crystals of both complexes belong to space group P212121 and have similar unit cell dimensions. The asymmetric units contain two viperin molecules and display noncrystallographic twofold symmetry involving an interface formed by the mutated surface loop (Figure S2A,B). However, the structure of the mutated loop is similar to that of wild-type viperin (Figure S2C). Analysis of the protein interfaces and surfaces using PISA suggests that viperin is monomeric.47
Figure 1. Overall structure of mouse viperin.
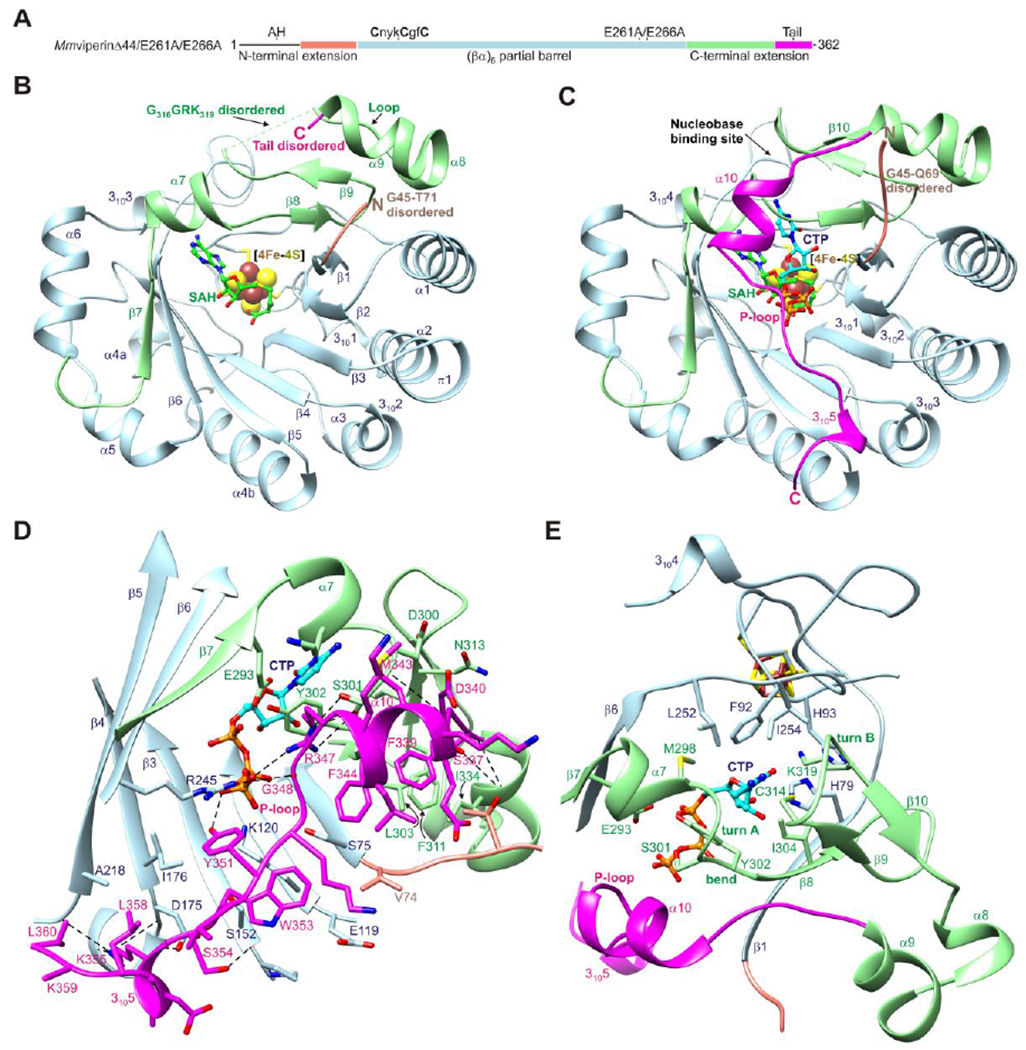
(A) One-dimensional schematic representation of viperin fragment cocrystallized with CTP or UTP showing structural organization and coloring scheme used in subsequent figures. AH denotes the amphipathic helix, which is not part of the viperin fragment. (B) Ribbon representation of MmviperinΔ44 bound to a [4Fe-4S] cluster and SAH (PDB code 5VSL).32 Secondary structure elements and disordered regions of the N- and C-terminal extensions are indicated. (C) Ribbon representation of MmviperinΔ44/E261A/E266A bound to a [4Fe-4S] cluster, SAH, and CTP. Changes in secondary structure assignments and new secondary structure elements observed in the CTP-bound complex are indicated. (D) Packing of C-terminal tail against the barrel opening and Pγ of CTP. (E) Nucleobase binding site. The residues surrounding the cytosine moiety from the core domain have a similar architecture in the nucleotide-free structure, whereas those from the C-terminal extension form turn A and a β-hairpin containing turn B. CTP and SAH are shown as balls and sticks and the [4Fe-4S] cluster as spheres. Potential hydrogen bonds and other electrostatic interactions are shown as dashed lines.
Table 1.
X-ray data collection statistics.
|
MmviperinΔ44/E261A/E266A SAH CTP |
MmviperinΔ44/E261A/E266A SAH UTP |
|
|---|---|---|
| Beamline | APS 24-ID-C | APS 24-ID-C |
| λ (Å) | 0.9791 Å | 0.9791 Å |
| Space group | P212121 | P212121 |
| a (Å) | 36.5 | 36.5 |
| b (Å) | 142.6 | 141.3 |
| c (Å) | 143.7 | 143.5 |
| Resolution range (Å) | 50.0-1.45 | 50.0-1.90 |
| Resolution of highest shell (Å) | 1.50-1.45 | 1.97-1.90 |
| No. of reflections | 474,193 | 180,923 |
| No. of unique reflections | 131,974 | 58,202 |
| Redundancy | 3.6 (3.6)a | 3.1 (3.0) |
| % Complete | 98.9 (99.1) | 96.6 (97.3) |
| <I/σI> | 8.6 (2.0) | 8.2 (2.1) |
| Rmerge (%) | 9.7 (56.3) | 14.6 (56.6) |
Values in parentheses are associated with the highest resolution shell.
Table 2.
Structure refinement statistics.
|
MmviperinΔ44/E261A/E266A SAH CTP |
MmviperinΔ44/E261A/E266A SAH UTP |
|
|---|---|---|
| No. of reflections | 131,887 | 58,170 |
| No. of reflections in working set | 125,188 | 55,266 |
| Resolution (Å) | 39.8-1.45 | 45.3-1.9 |
| No. of protein atoms | 4,994 | 4,903 |
| No. of waters | 959 | 756 |
| No. of ligand atoms | 148 | 149 |
| RMSD bonds (Å) | 0.013 | 0.008 |
| RMSD angles (°) | 1.8 | 0.953 |
| Rwork (%) | 14.1 | 14.6 |
| Rfree (%) | 16.0 | 18.2 |
| Ramachandran analysis | ||
| Most favored (%) | 89.0 | 88.7 |
| Additional allowed (%) | 11.0 | 11.1 |
| Generously allowed (%) | 0.0 | 0.2 |
Overall structure of the CTP-bound complex: ordering of the C-terminal extension
Previous studies showed that nucleotide-free viperin contains a (βα)6 partial barrel and has a disordered N-terminal extension and a partially ordered C-terminal extension (Figure 1B).32 Binding of CTP results in ordering of the C-terminal extension and the formation of a well-ordered nucleotide binding site in which a 25-residue tail folds over the barrel opening (Figure 1C). The N-terminal extension remains disordered, consistent with previous predictions of intrinsic disorder in this region.32
Ordering of the C-terminal tail introduces a six residue α-helix (α10) followed by an eight residue P-loop that binds the γ-phosphate (Pγ) of CTP and a 310-helix (3105) (Figure 1C). The C-terminal tail forms several hydrophobic and electrostatic interactions with residues near the N-terminal sides of the barrel β-strands, with the P-loop residues Arg347 and Tyr351 forming a salt bridge and a hydrogen bond with Pγ, respectively (Figure 1D). In one monomer, all but the final two residues (Asp361 and Trp362) become ordered, whereas in the second, these two residues become partially ordered through interactions between Trp362 and crystallographically related viperin molecules. The structure thus shows that the C-terminal truncations of 17 residues or the final residue of human viperin (Figure S3) that lead to decreased inhibition of several viruses are associated with removal of the P-loop and solvent-exposed C-terminal tryptophan.
Ordering of the nucleobase binding site involves the formation of two Type I β-turns involving residues M298KDS301 (turn A) and C314TGG317 (turn B) and a β-hairpin (β9-β10; residues 312-321) that is rotated out of alignment with the remaining strands of the barrel (Figure 1E). Turn A forms at the C-terminal end of helix α7, which is shortened from seven (295-301) to four residues (295-298) with Lys299, Asp300, and Ser301 forming hydrogen bonds with the N4 amino group of cytosine, the backbone of Asp340 of the C-terminal tail, and the side chain of Arg347 of the P-loop, respectively (Figure S4A). In addition, the side chain of Tyr302 undergoes a 100° change in its χ1 angle, packs against the cytosine and ribose moieties, and forms hydrogen bonds with a Pα oxygen atom of CTP and the side chain of Glu293 from β7. In the nucleotide-free structure, the β-hairpin region forms a loop with residues G316GRK319 disordered and with Cys314 located at the cytosine binding site.32 In the CTP-bound complex, the β-hairpin is stabilized by turn B, hydrogen bonds between the backbones of Asp320 and Asn313, and several additional hydrogen bonds with the cytosine moiety of CTP and other protein segments (Figure S4B). These include hydrogen bonds between the side chain of Cys314 and the N4 atom, the side chain of Lys319 and the O2 atom, the backbones of Gly317 and Glu255, and the side chain of Asn313 and the backbone of F336.
Active site architecture: structural basis of C4′ radical formation
The structure of viperin bound to CTP displays the canonical active site architecture of radical SAM enzymes with additional capping by the C-terminal tail. SAH adopts the canonical SAM conformation48 as observed in the nucleotide-free structure.32 CTP is surrounded by 25 residues from the core domain and C-terminal extension, 18 of which making direct contacts (d < 3.5 Å) (Figure 2A).
Figure 2. Active site architecture.
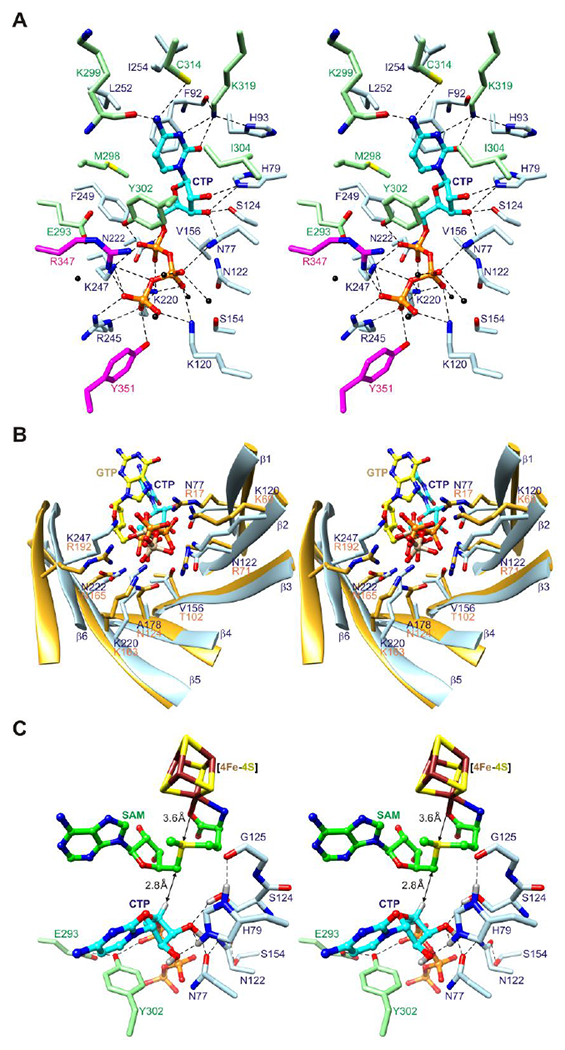
(A) Stereoview of CTP binding site. The side chains of twenty-five residues from the core domain and C-terminal extension, including two from the C-terminal tail (sticks), and five water molecules (black spheres) surround CTP (balls and sticks). Potential hydrogen bonds and other electrostatic interactions are shown as dashed lines. (B) Stereoview comparison of triphosphate binding residues of the (βα)6 partial barrels of viperin and MoaA. Eight residues in MoaA (PDB code 2FB333) bind the triphosphate moiety of GTP (balls and sticks with yellow colored carbon atoms and tan colored phosphorus atoms), five of which (Arg17, Lys69, Lys163, Asn165, and Arg192) have the same barrel locations as residues of viperin that bind the triphosphate moiety of CTP. (C) Stereoview model of substrate complex. SAM is modeled based on SAH coordinates.
The cytosine ring is situated between several hydrophobic residues and forms hydrogen bonds with residues of turns A and B. The N4 amino group forms a hydrogen bond with the backbone carbonyl of Lys299 and has an N4…S separation distance (3.6 Å) and N4-H…S angle (120°) with the sulfur atom of Cys314 consistent with values for amide N-H…S hydrogen bonds in proteins.49 The O2 atom forms a hydrogen bond with the ε-amino group of Lys319 located near turn B. The N3 atom is also within hydrogen bonding distance of the Lys319 Nζ atom, although the ε-amino group is better aligned to form additional hydrogen bonds with the backbone carbonyl oxygen atom of Phe92 and the Nδ1 atom of His93.
The triphosphate moiety of CTP makes sixteen potential hydrogen bonds or other electrostatic interactions (d < 3.5 Å) with inward pointing side chains of the barrel and P-loop. These occur between Pα oxygen atoms and the side chains of Lys220, Asn222, Lys247, and Tyr302, between Pβ oxygen atoms and the side chains of Asn77, Lys120, Lys220, and Lys247, and between Pγ oxygen atoms and the side chains of Lys120, Lys220, Arg245, Lys247, Arg347, and Tyr351. Four additional hydrogen bonds are formed with water molecules, including three with oxygen atoms of Pβ and one with an oxygen atom of Pγ. Notably, five of the phosphate binding residues occupy equivalent barrel sites used by MoaA to bind the triphosphate moiety of GTP, as predicted previously from structural alignments using the nucleotide-free structure (Figure 2B).32
The ribose moiety of CTP adopts a 2′-endo conformation and packs between SAH, Tyr302, and hydrophilic residues that make hydrogen bonds with its hydroxyl groups. The O2′-hydroxyl group forms a hydrogen bond with the Nε2 atom of His79 and the O3′-hydroxyl group forms hydrogen bonds with the hydroxyl group of Ser124 and the side chain carbonyl oxygen atom of Asn77. The O3′ and O5′ atoms are also within hydrogen bonding distance of the Nε2 atom of His79 and the Nδ2 atom of Asn222, respectively. The C4′ atom is positioned 3.8 Å from the C5′ atom of SAH with its hydrogen suitably oriented for abstraction by a 5′-deoxyadenosyl radical for generation of a C4′ radical (Figure 2C).
Reaction kinetics and structural basis of substrate selectivity
Recent biochemical studies have shown that rat viperin exhibits high catalytic activity for the conversion of CTP to ddhCTP31 [see also the additional biochemical studies of human viperin and fungal viperin like enzymes (VLEs)50–52 with other substrates]. Among other ribonucleoside triphosphates studied, conversion of UTP could also be detected but the efficiency is much lower than the reaction with CTP.31 To further characterize the substrate selectivity, we measured kinetic parameters for the mouse viperin fragment MmviperinΔ4432 with CTP or UTP as substrates (Figure 3A and B and Figure S5). For CTP, the kcat value is 0.6 min−1, but we could not obtain the exact Km value because even at the lowest concentration tried, 10 μM, the enzyme was already saturated. Thus, the Km value should be much smaller than 10 μM, and we estimate that it could be around 1 μM. For UTP, the kcat is only slightly smaller, 0.5 min−1, but the Km value, 40 μM, is estimated to be 40 times greater than that for CTP. These results show that mouse viperin strongly prefers CTP as the substrate and the selectivity mainly comes from the lower Km value for CTP. Since Km is an upper estimate of Kd, the data suggest that viperin binds CTP more tightly than UTP.
Figure 3. Kinetic studies and structural basis of substrate selectivity.
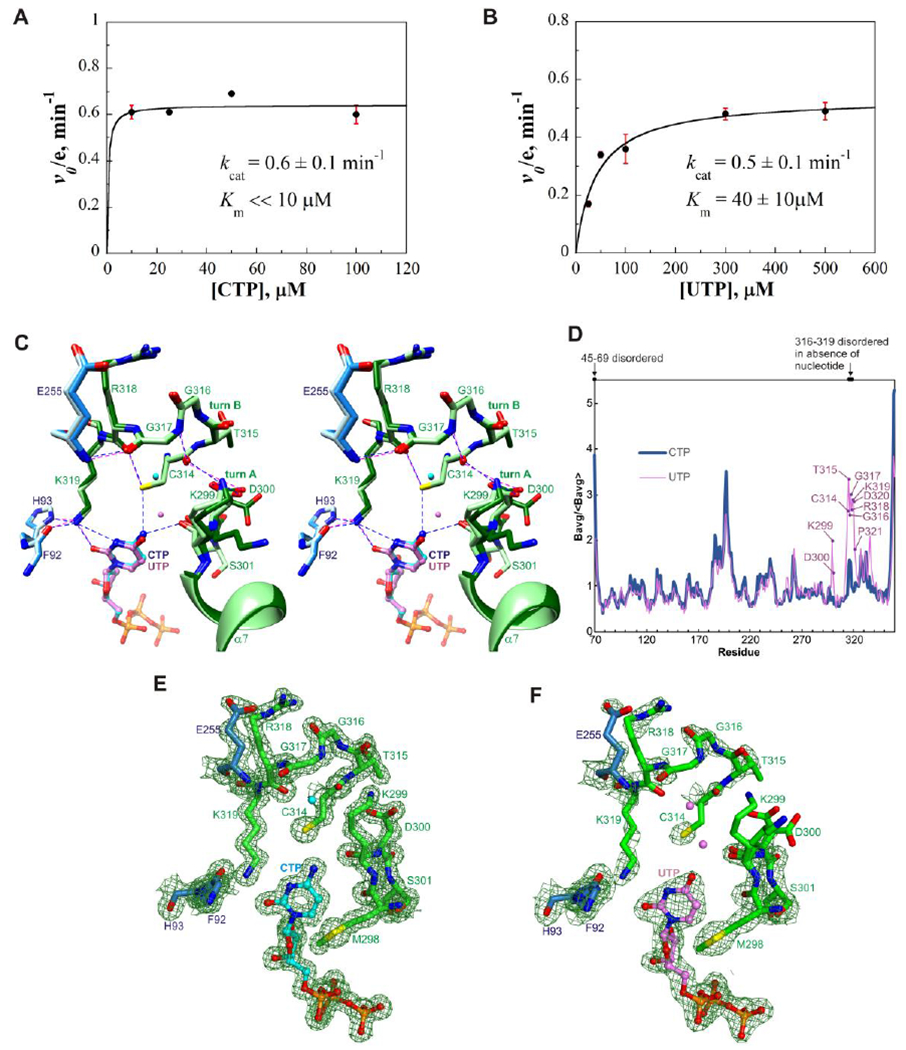
(A) Plot of initial rate of ddhCTP production versus CTP concentration (with 100 μM SAM). The enzyme was saturated even at the lowest substrate concentration, 10 μM. The kcat is 0.6±0.1 min−1. (B) Plot of initial rate of ddhUTP production versus UTP concentration (with 100 μM SAM). Km for UTP = 40±10 μM. kcat = 0.5±0.1 min−1. Each value is the mean of three replicates ± s.d. (C) Stereoview superimposition of nucleobase binding sites of the CTP and UTP-bound complexes. Core domain residues are colored light blue and dodger blue, and C-terminal extension residues light green and dark green, respectively. CTP and UTP are shown as balls-and-sticks with carbon atoms colored cyan and light purple, respectively; nearby water molecules are shown as spheres and colored similarly. The hydrophobic residues have similar structures in the two complexes and are omitted for clarity. Potential hydrogen bonds and other electrostatic interactions are shown as blue and magenta dashed lines in the CTP and UTP-bound complexes, respectively. (D) Average main chain B-factors for viperin bound to CTP or UTP. The averages are normalized by the average for all residues. Residues in the UTP-bound complex showing significantly higher B-factors relative to the CTP-bound complex are indicated. (E) Electron density of the cytosine binding site. (F) Electron density of the uracil binding site. Panels E and F show composite omit maps computed using PHENIX with default settings and drawn at a contour level of 1.2 times the root mean square value of the map using PyMOL.46
Consistent with the kinetic parameters, the overall structures of the UTP- and CTP-bound complexes are very similar (root mean square deviation of 0.13 Å after backbone superimposition) and the ribose and triphosphate moieties of UTP and CTP make the same interactions with viperin, but the uracil moiety is much less effective than the cytosine moiety at binding and ordering turns A and B. Substitution of an oxo group for an amino group at the ring 4-position -- resulting in a protonated rather than deprotonated nitrogen atom at the 3-position -- introduces three unfavorable interactions that are reflected in local structural differences (Figure 3C), increased B-factors (Figure 3D), and weaker electron density (Figure 3E and F). First, unlike N4 of CTP, O4 of UTP is unable to make a hydrogen bond with the backbone carbonyl of Lys299 and accordingly the O4⋯O separation distance (3.7 Å) is greater than the N4⋯O distance (3.0 Å). A second unfavorable contact, also related to O4, disrupts hydrogen bonding involving Cys314. In the CTP-bound complex, the sulfur atom of Cys314 is 3.5 Å from both N4 of CTP and the carbonyl oxygen atom of Arg318, suggesting roles of hydrogen bond acceptor and donor, respectively. In the UTP-bound complex, the sulfur atom is instead situated between two carbonyl oxygen atoms and thus an unfavorable interaction arises (notably, the S⋯O4 distance is 4.0 Å). Finally, the highly ordered conformation of Lys319 in the CTP-bound complex suggests that N3 of UTP introduces potential bad contacts by placing its proton less than 3.0 Å from the ε-amino nitrogen and Cε atoms, which direct protons towards N3 of the cytosine ring. Collectively, the results suggest that the higher selectivity for CTP compared to UTP is associated with the higher degree of chemical complementarity between the cytosine moiety and the nucleobase binding site.
Conserved and variable residues in viperins and viperin-like enzymes (VLEs)
Previous structure-based sequence analysis identified several VLEs having conserved [4Fe-4S] cluster and SAM binding residues, and several additional active site residues conserved over the six kingdoms.32 Beyond the CTP binding site, the VLEs conserve the Cys84, Cys88, and Cys91 sites, which bind the [4Fe-4S] cluster; the Asn85 and Asn158 sites, which stabilize the barrel base; the Phe/Tyr90, Phe/Tyr92, and Val224 sites, which form the SAM adenine binding site; the Ser180 and Arg194 sites, which bind the SAM carboxylate and hydroxyl groups; and a GGE motif,53, 54 which binds the SAM amino group. Additional sequence comparisons based on the CTP-bound complex show that the triphosphate binding site is highly conserved whereas the ribose binding site is less conserved and the nucleobase binding site is remarkably variable (Figure 4, Figure 5, and Figure S6).
Figure 4. Degree of conservation of residues of the CTP binding site of viperins or VLEs.
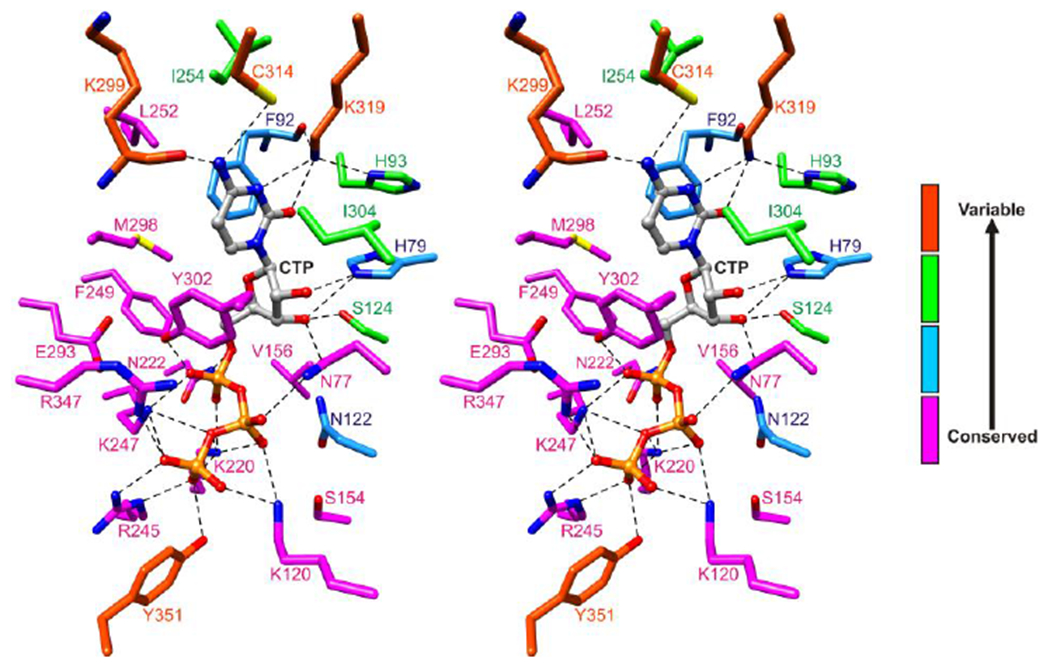
Twenty-five residues surrounding CTP are colored according to the number of different types of amino acids observed at the corresponding sites in forty-six viperins or VLEs from the six kingdoms (Figure S6). Sites showing absolute conservation or predicted conservative substitutions are colored magenta, high conservation with one predicted non-conservative substitution colored blue, moderate conservation with two to four predicted non-conservative substitutions colored green, and variable with five or more predicted non-conservative substitutions colored orange red.
Figure 5. Amino acid sequence alignment of viperins or VLEs from the six kingdoms.
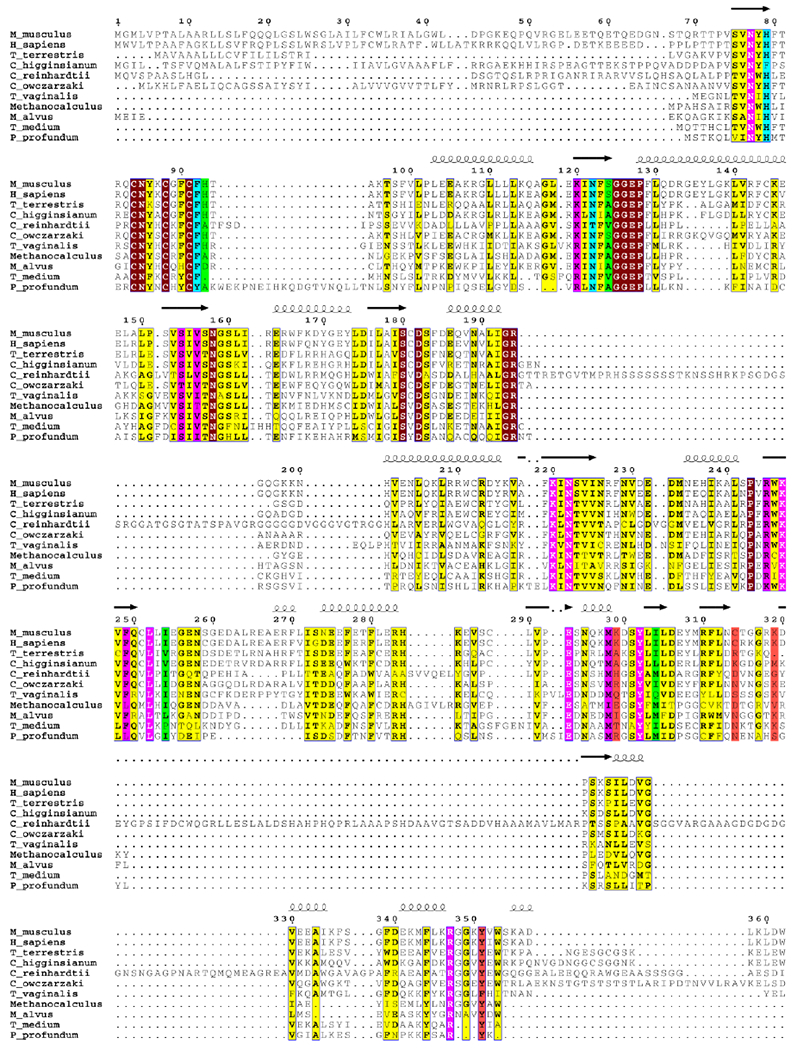
25 residues surrounding CTP are highlighted using the coloring system used in Figure 4. Other absolutely conserved sites are highlighted in maroon and other mostly conserved sites are highlighted in yellow. The viperin or VLE sequences aligned are from Homo sapiens (NP_542388.2), Mus musculus (AAH57868.1), Trichomonas vaginalis G3 (XP_001324419.1), Capsaspora owczarzaki ATCC 30864 (XP_004363934.1), Chlamydomonas reinhardtii (XP_001693132.1), Colletotrichum higginsianum IMI 349063 (XP_018157122.1), Thielavia terrestris NRRL 8126 (XP_003653480.1), Methanocalculus sp. 52_23 (KUK70247.1), Candidatus Methanomethylophilus alvus (WP_048097766.1), Photobacterium profundum (WP_011218337), and Treponema medium ATCC 700293 (EPF28955.1). Sequences were aligned using MUSCLE55 and illustrated using ESPript 3.0 (http://espript.ibcp.fr56).
Many animal VLEs conserve all of the 25 amino acids that closely surround CTP (Figure 2A), although minor variations occur at the Lys299 and Ser124 sites (Figure S6A). The protist Capsaspora owczarzaki VLE contains an asparagine at the Cys314 site whereas the Trichomonas vaginalis VLE contains a serine at this site and shows variation at the Ile304 and Tyr351 sites (Figure 5 and Figure S6B). The green alga Chlamydomonas reinhardtii VLE has an alanine at the His93 site and an insertion of 95 amino acids near turn B (Figure 5 and Figure S6C). Fungal, archaebacterial, and bacterial VLEs show variation at the Lys299, Cys314, and Lys319 sites, with fungal VLEs also showing variation at the His79 and Tyr351 sites, archaebacterial VLEs at the His93 site, and bacterial VLEs at the His93 and Tyr351 sites (noting however that most of the bacterial VLEs lacking a tyrosine at the Tyr351 site have a tyrosine at the G348 site) (Figure 5 and Figure S6D–F). Interestingly, the genes encoding the VLEs of Vibrio vulnificus CMCP6 and various Photobacteria (P. profundum ATCC BAA-1252, P. swingsii CAIM 1393, and P. phosphoreum ANT220) reside next to a putative thymidylate kinase gene suggesting a possible function of DNA modification.
The structure-based sequence analysis suggests that the VLEs are members of a family of radical SAM enzymes having a conserved architecture well-suited for performing radical reactions on nucleotides or structurally related molecules. Nearly all of the triphosphate binding residues are conserved, including all protruding inward from the barrel, Tyr302, and Arg347 from the P-loop. The high degree of conservation of the Arg347 site implies that P-loops are utilized throughout this family, possibly for selection of triphosphate nucleotides. The VLEs showing variations in the ribose and/or nucleobase binding sites are interesting systems for future studies.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by a National Institutes of Health grants DK-067081 (S.E.E) and by the Howard Hughes Medical Institute (H.L.). The work is based upon research conducted at the Advanced Photon Source on the Northeastern Collaborative Access Team beamlines, which are supported by award P30 GM124165 from the NIH. Use of the Advanced Photon Source is supported by the U.S. Department of Energy, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357.
Footnotes
Supporting Information
The Supporting Information containing Figures S1–S6 is available free of charge on the ACS Publications website at DOI: https://doi.org/10.1021/acs.biochem.9b00741
Electron density of CTP, UTP, and [4Fe-4S] clusters (Figure S1), noncrystallographic symmetry and mutated loop structure (Figure S2), amino acid sequence alignment of human and mouse viperins (Figure S3), structures of turns A and B in the CTP-bound complex (Figure S4), mass spectrometry results for kinetic studies (Figure S5), and amino acid sequence alignment of mouse viperin and other viperins or VLEs from the six kingdoms (Figure S6).
Accession codes
The coordinates of viperin-SAH-CTP and viperin-SAH-UTP have been deposited in the Protein Data Bank under accession codes 6Q2P and 6Q2Q, respectively. The NCBI accession number of Mmviperin is AAH57868.1.
References
- (1).de Veer MJ, Holko M, Frevel M, Walker E, Der S, Paranjape JM, Silverman RH, and Williams BR (2001) Functional classification of interferon-stimulated genes identified using microarrays, J Leukoc Biol 69, 912–920. [PubMed] [Google Scholar]
- (2).Samuel CE (2001) Antiviral actions of interferons, Clin Microbiol Rev 14, 778–809, table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Schoggins JW, and Rice CM (2011) Interferon-stimulated genes and their antiviral effector functions, Curr Opin Virol 1, 519–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Boudinot P, Massin P, Blanco M, Riffault S, and Benmansour A (1999) vig-1, a new fish gene induced by the rhabdovirus glycoprotein, has a virus-induced homologue in humans and shares conserved motifs with the MoaA family, J Virol 73, 1846–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Chin KC, and Cresswell P (2001) Viperin (cig5), an IFN-inducible antiviral protein directly induced by human cytomegalovirus, Proc Natl Acad Sci U S A 98, 15125–15130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Zhu H, Cong JP, and Shenk T (1997) Use of differential display analysis to assess the effect of human cytomegalovirus infection on the accumulation of cellular RNAs: induction of interferon-responsive RNAs, Proc Natl Acad Sci U S A 94, 13985–13990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Chan YL, Chang TH, Liao CL, and Lin YL (2008) The cellular antiviral protein viperin is attenuated by proteasome-mediated protein degradation in Japanese encephalitis virus-infected cells, J Virol 82, 10455–10464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Fang J, Wang H, Bai J, Zhang Q, Li Y, Liu F, and Jiang P (2016) Monkey Viperin Restricts Porcine Reproductive and Respiratory Syndrome Virus Replication, PLoS One 11, e0156513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Helbig KJ, Carr JM, Calvert JK, Wati S, Clarke JN, Eyre NS, Narayana SK, Fiches GN, McCartney EM, and Beard MR (2013) Viperin is induced following dengue virus type-2 (DENV-2) infection and has anti-viral actions requiring the C-terminal end of viperin, PLoS Negl Trop Dis 7, e2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Helbig KJ, Eyre NS, Yip E, Narayana S, Li K, Fiches G, McCartney EM, Jangra RK, Lemon SM, and Beard MR (2011) The antiviral protein viperin inhibits hepatitis C virus replication via interaction with nonstructural protein 5A, Hepatology 54, 1506–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Jiang D, Guo H, Xu C, Chang J, Gu B, Wang L, Block TM, and Guo JT (2008) Identification of three interferon-inducible cellular enzymes that inhibit the replication of hepatitis C virus, J Virol 82, 1665–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Szretter KJ, Brien JD, Thackray LB, Virgin HW, Cresswell P, and Diamond MS (2011) The interferon-inducible gene viperin restricts West Nile virus pathogenesis, J Virol 85, 11557–11566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Teng TS, Foo SS, Simamarta D, Lum FM, Teo TH, Lulla A, Yeo NK, Koh EG, Chow A, Leo YS, Merits A, Chin KC, and Ng LF (2012) Viperin restricts chikungunya virus replication and pathology, J Clin Invest 122, 4447–4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Upadhyay AS, Vonderstein K, Pichlmair A, Stehling O, Bennett KL, Dobler G, Guo JT, Superti-Furga G, Lill R, Overby AK, and Weber F (2014) Viperin is an iron-sulfur protein that inhibits genome synthesis of tick-borne encephalitis virus via radical SAM domain activity, Cell Microbiol 16, 834–848. [DOI] [PubMed] [Google Scholar]
- (15).Van der Hoek KH, Eyre NS, Shue B, Khantisitthiporn O, Glab-Ampi K, Carr JM, Gartner MJ, Jolly LA, Thomas PQ, Adikusuma F, Jankovic-Karasoulos T, Roberts CT, Helbig KJ, and Beard MR (2017) Viperin is an important host restriction factor in control of Zika virus infection, Sci Rep 7, 4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Wang S, Wu X, Pan T, Song W, Wang Y, Zhang F, and Yuan Z (2012) Viperin inhibits hepatitis C virus replication by interfering with binding of NS5A to host protein hVAP-33, J Gen Virol 93, 83–92. [DOI] [PubMed] [Google Scholar]
- (17).Li W, Mao L, Cao Y, Zhou B, Yang L, Han L, Hao F, Lin T, Zhang W, and Jiang J (2017) Porcine Viperin protein inhibits the replication of classical swine fever virus (CSFV) in vitro, Virol J 14, 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Jiang D, Weidner JM, Qing M, Pan XB, Guo H, Xu C, Zhang X, Birk A, Chang J, Shi PY, Block TM, and Guo JT (2010) Identification of five interferon-induced cellular proteins that inhibit west nile virus and dengue virus infections, J Virol 84, 8332–8341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Panayiotou C, Lindqvist R, Kurhade C, Vonderstein K, Pasto J, Edlund K, Upadhyay AS, and Overby AK (2018) Viperin Restricts Zika Virus and Tick-Borne Encephalitis Virus Replication by Targeting NS3 for Proteasomal Degradation, J Virol 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Carlton-Smith C, and Elliott RM (2012) Viperin, MTAP44, and protein kinase R contribute to the interferon-induced inhibition of Bunyamwera Orthobunyavirus replication, J Virol 86, 11548–11557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Tang HB, Lu ZL, Wei XK, Zhong TZ, Zhong YZ, Ouyang LX, Luo Y, Xing XW, Liao F, Peng KK, Deng CQ, Minamoto N, and Luo TR (2016) Viperin inhibits rabies virus replication via reduced cholesterol and sphingomyelin and is regulated upstream by TLR4, Sci Rep 6, 30529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Wang X, Hinson ER, and Cresswell P (2007) The interferon-inducible protein viperin inhibits influenza virus release by perturbing lipid rafts, Cell Host Microbe 2, 96–105. [DOI] [PubMed] [Google Scholar]
- (23).Jumat MR, Huong TN, Ravi LI, Stanford R, Tan BH, and Sugrue RJ (2015) Viperin protein expression inhibits the late stage of respiratory syncytial virus morphogenesis, Antiviral Res 114, 11–20. [DOI] [PubMed] [Google Scholar]
- (24).Nasr N, Maddocks S, Turville SG, Harman AN, Woolger N, Helbig KJ, Wilkinson J, Bye CR, Wright TK, Rambukwelle D, Donaghy H, Beard MR, and Cunningham AL (2012) HIV-1 infection of human macrophages directly induces viperin which inhibits viral production, Blood 120, 778–788. [DOI] [PubMed] [Google Scholar]
- (25).Tang YD, Na L, Zhu CH, Shen N, Yang F, Fu XQ, Wang YH, Fu LH, Wang JY, Lin YZ, Wang XF, Wang X, Zhou JH, and Li CY (2014) Equine viperin restricts equine infectious anemia virus replication by inhibiting the production and/or release of viral Gag, Env, and receptor via distortion of the endoplasmic reticulum, J Virol 88, 12296–12310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Dumbrepatil AB, Ghosh S, Zegalia KA, Malec PA, Hoff JD, Kennedy RT, and Marsh ENG (2019) Viperin interacts with the kinase IRAK1 and the E3 ubiquitin ligase TRAF6, coupling innate immune signaling to antiviral ribonucleotide synthesis, J Biol Chem 294, 6888–6898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Jiang X, and Chen ZJ (2011) Viperin links lipid bodies to immune defense, Immunity 34, 285–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Saitoh T, Satoh T, Yamamoto N, Uematsu S, Takeuchi O, Kawai T, and Akira S (2011) Antiviral protein Viperin promotes Toll-like receptor 7- and Toll-like receptor 9-mediated type I interferon production in plasmacytoid dendritic cells, Immunity 34, 352–363. [DOI] [PubMed] [Google Scholar]
- (29).Eom J, Kim JJ, Yoon SG, Jeong H, Son S, Lee JB, Yoo J, Seo HJ, Cho Y, Kim KS, Choi KM, Kim IY, Lee HY, Nam KT, Cresswell P, Seong JK, and Seo JY (2019) Intrinsic expression of viperin regulates thermogenesis in adipose tissues, Proc Natl Acad Sci U S A 116, 17419–17428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Makins C, Ghosh S, Roman-Melendez GD, Malec PA, Kennedy RT, and Marsh EN (2016) Does Viperin Function as a Radical S-Adenosyl-l-methionine-dependent Enzyme in Regulating Farnesylpyrophosphate Synthase Expression and Activity?, J Biol Chem 291, 26806–26815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Gizzi AS, Grove TL, Arnold JJ, Jose J, Jangra RK, Garforth SJ, Du Q, Cahill SM, Dulyaninova NG, Love JD, Chandran K, Bresnick AR, Cameron CE, and Almo SC (2018) A naturally occurring antiviral ribonucleotide encoded by the human genome, Nature 558, 610–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Fenwick MK, Li Y, Cresswell P, Modis Y, and Ealick SE (2017) Structural studies of viperin, an antiviral radical SAM enzyme, Proc Natl Acad Sci U S A 114, 6806–6811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Hanzelmann P, and Schindelin H (2006) Binding of 5’-GTP to the C-terminal FeS cluster of the radical S-adenosylmethionine enzyme MoaA provides insights into its mechanism, Proc Natl Acad Sci U S A 103, 6829–6834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Hinson ER, and Cresswell P (2009) The N-terminal amphipathic alpha-helix of viperin mediates localization to the cytosolic face of the endoplasmic reticulum and inhibits protein secretion, J Biol Chem 284, 4705–4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Upadhyay AS, Stehling O, Panayiotou C, Rosser R, Lill R, and Overby AK (2017) Cellular requirements for iron-sulfur cluster insertion into the antiviral radical SAM protein viperin, J Biol Chem 292, 13879–13889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Mateja A, Devedjiev Y, Krowarsch D, Longenecker K, Dauter Z, Otlewski J, and Derewenda ZS (2002) The impact of Glu->Ala and Glu-->Asp mutations on the crystallization properties of RhoGDI: the structure of RhoGDI at 1.3 A resolution, Acta Crystallogr D Biol Crystallogr 58, 1983–1991. [DOI] [PubMed] [Google Scholar]
- (37).Fenwick MK, Mehta AP, Zhang Y, Abdelwahed SH, Begley TP, and Ealick SE (2015) Non-canonical active site architecture of the radical SAM thiamin pyrimidine synthase, Nat Commun 6, 6480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Mehta AP, Abdelwahed SH, Fenwick MK, Hazra AB, Taga ME, Zhang Y, Ealick SE, and Begley TP (2015) Anaerobic 5-Hydroxybenzimidazole Formation from Aminoimidazole Ribotide: An Unanticipated Intersection of Thiamin and Vitamin B12 Biosynthesis, J Am Chem Soc 137, 10444–10447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Hanzelmann P, Hernandez HL, Menzel C, Garcia-Serres R, Huynh BH, Johnson MK, Mendel RR, and Schindelin H (2004) Characterization of MOCS1A, an oxygen-sensitive iron-sulfur protein involved in human molybdenum cofactor biosynthesis, J Biol Chem 279, 34721–34732. [DOI] [PubMed] [Google Scholar]
- (40).Otwinoski Z, and Minor W (1997) Processing of X-ray diffraction data collected in oscillation mode, Method Enzymol 276, 307–326. [DOI] [PubMed] [Google Scholar]
- (41).McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, and Read RJ (2007) Phaser crystallographic software, J Appl Crystallogr 40, 658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Emsley P, Lohkamp B, Scott WG, and Cowtan K (2010) Features and development of Coot, Acta Crystallogr D 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Adams PD, Afonine PV, Bunkoczi G, Chen VB, Echols N, Headd JJ, Hung LW, Jain S, Kapral GJ, Grosse Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner RD, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, and Zwart PH (2011) The Phenix software for automated determination of macromolecular structures, Methods 55, 94–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Painter J, and Merritt EA (2006) Optimal description of a protein structure in terms of multiple groups undergoing TLS motion, Acta Crystallogr. D 62, 439–450. [DOI] [PubMed] [Google Scholar]
- (45).Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, and Ferrin TE (2004) UCSF Chimera--a visualization system for exploratory research and analysis, J Comput Chem 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- (46).DeLano WL (2002) The PyMOL Molecular Graphics System, DeLano Scientific, San Carlos, CA. [Google Scholar]
- (47).Krissinel E, and Henrick K (2007) Inference of macromolecular assemblies from crystalline state, J Mol Biol 372, 774–797. [DOI] [PubMed] [Google Scholar]
- (48).Layer G, Moser J, Heinz DW, Jahn D, and Schubert WD (2003) Crystal structure of coproporphyrinogen III oxidase reveals cofactor geometry of Radical SAM enzymes, EMBO J 22, 6214–6224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Zhou P, Tian F, Lv F, and Shang Z (2009) Geometric characteristics of hydrogen bonds involving sulfur atoms in proteins, Proteins 76, 151–163. [DOI] [PubMed] [Google Scholar]
- (50).Chakravarti A, Selvadurai K, Shahoei R, Lee H, Fatma S, Tajkhorshid E, and Huang RH (2018) Reconstitution and substrate specificity for isopentenyl pyrophosphate of the antiviral radical SAM enzyme viperin, J Biol Chem 293, 14122–14133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Ebrahimi KH, Carr SB, McCullagh J, Wickens J, Rees NH, Cantley J, and Armstrong FA (2017) The radical-SAM enzyme Viperin catalyzes reductive addition of a 5’-deoxyadenosyl radical to UDP-glucose in vitro, FEBS Lett 591, 2394–2405. [DOI] [PubMed] [Google Scholar]
- (52).Mikulecky P, Andreeva E, Amara P, Weissenhorn W, Nicolet Y, and Macheboeuf P (2018) Human viperin catalyzes the modification of GPP and FPP potentially affecting cholesterol synthesis, FEBS Lett 592, 199–208. [DOI] [PubMed] [Google Scholar]
- (53).Nicolet Y, and Drennan CL (2004) AdoMet radical proteins--from structure to evolution--alignment of divergent protein sequences reveals strong secondary structure element conservation, Nucleic Acids Res 32, 4015–4025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Sofia HJ, Chen G, Hetzler BG, Reyes-Spindola JF, and Miller NE (2001) Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: functional characterization using new analysis and information visualization methods, Nucleic Acids Res 29, 1097–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput, Nucleic Acids Res 32, 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Robert X, and Gouet P (2014) Deciphering key features in protein structures with the new ENDscript server, Nucleic Acids Res 42, W320–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


