Significance Statement
Microscopic analysis of urinary sediment is one of the most fundamental tests in nephrology. Urinary cells, however, have not been characterized in a standardized, unbiased manner. Single-cell transcriptomics of urine, of subjects with diabetic kidney disease and controls, were used to characterize 23,082 urinary cells in an unbiased manner. Combined analysis of urinary, kidney, and bladder cells indicated the technique can detect almost all kidney cell types and a variety of bladder cell types in human urine. This pilot study provides a reference dataset for future urinary single-cell characterization.
Keywords: diabetic kidney disease, urine, single-cell transcriptomics, RNA sequencing
Abstract
Background
Microscopic analysis of urine sediment is probably the most commonly used diagnostic procedure in nephrology. The urinary cells, however, have not yet undergone careful unbiased characterization.
Methods
Single-cell transcriptomic analysis was performed on 17 urine samples obtained from five subjects at two different occasions, using both spot and 24-hour urine collection. A pooled urine sample from multiple healthy individuals served as a reference control. In total 23,082 cells were analyzed. Urinary cells were compared with human kidney and human bladder datasets to understand similarities and differences among the observed cell types.
Results
Almost all kidney cell types can be identified in urine, such as podocyte, proximal tubule, loop of Henle, and collecting duct, in addition to macrophages, lymphocytes, and bladder cells. The urinary cell–type composition was subject specific and reasonably stable using different collection methods and over time. Urinary cells clustered with kidney and bladder cells, such as urinary podocytes with kidney podocytes, and principal cells of the kidney and urine, indicating their similarities in gene expression.
Conclusions
A reference dataset for cells in human urine was generated. Single-cell transcriptomics enables detection and quantification of almost all types of cells in the kidney and urinary tract.
Microscopic analysis of the cells of the urinary sediment is one of the oldest diagnostic tools used in nephrology. The urine of almost every patient referred to the renal clinic is examined under a microscope.1 In the past, a variety of cells has been described in the urine of patients with CKD.2
Several early studies indicated that tubule cells are shed into the urine. Extensive studies by Racusen et al.3 in the early 1990s characterized these cells as proximal tubule (PT) cells, on the basis of their morphologic characteristics and expression of gamma glutamyl transferase,4–10 but in their study urinary PT cell numbers did not correlate with kidney disease severity.2 Some urinary cells stain positive for nephrin, indicating that viable podocytes are also shed into the urine. Recently it has also been proposed that undifferentiated kidney progenitor cells can also be found in the urine.10–13
Although these studies indicate the feasibility of capturing kidney cells from the human urine, the methods used have several limitations. Importantly, most studies only analyzed a limited number of markers, therefore it remains unclear how well urinary cells globally resemble kidney cells.2,14 Second, not all cells can be cultured using standard conditions, resulting in a potential detection bias. Finally, cells in culture transdifferentiate or dedifferentiate, and often fail to express their cell type of original markers.
Single-cell RNA sequencing (RNA-seq) is a revolutionary new method for an unbiased genome-wide characterization of individual cells at scale.15 Single-cell analysis of the mouse and human kidneys has generated an initial map of gene expression for most cells in the kidney, and allowed us to move from a morphotype-based cell characterization (such as shape, color, and location) to a more objective transcriptome-based cell-type definition.16,17 Single-cell sequencing of the kidney has helped define cell type–specific changes, cell fractions and cell-cell interactions in kidney disease conditions, thus allowing us to develop new diagnostics and therapeutics. In a recent study, Arazi et al.14 was able to capture 577 immune cells from the urine of patients with lupus nephritis. Similarly, in a recent paper by Menon et al.18 the authors were able to identify epithelial cells, immune cells, and kidney cells in the urine, but not specific cell types.
Here we used single-cell RNA-seq to generate a reference expression map for cells in the human urine. We show that most kidney cells, not just podocytes and PT cells, can be identified in the urine. The urinary cell–type composition was subject specific and reasonably stable in different collection methods and over time. Urinary cells clustered together with kidney and bladder cells, such as urinary podocytes clustered with kidney podocytes, and principal cells of the kidney and urine clustered together.
Methods
Study Design
Transformative Research in Diabetic Nephropathy (TRIDENT) is a multicenter observational cohort study that enrolls, follows, and performs multiomics characterization of subjects with diabetic kidney disease (DKD).19 This urinary single-cell study is an ancillary study of TRIDENT, which was approved by the University of Pennsylvania Institutional Review Board and by the TRIDENT Steering Committee. Informed consent was obtained from each participant.
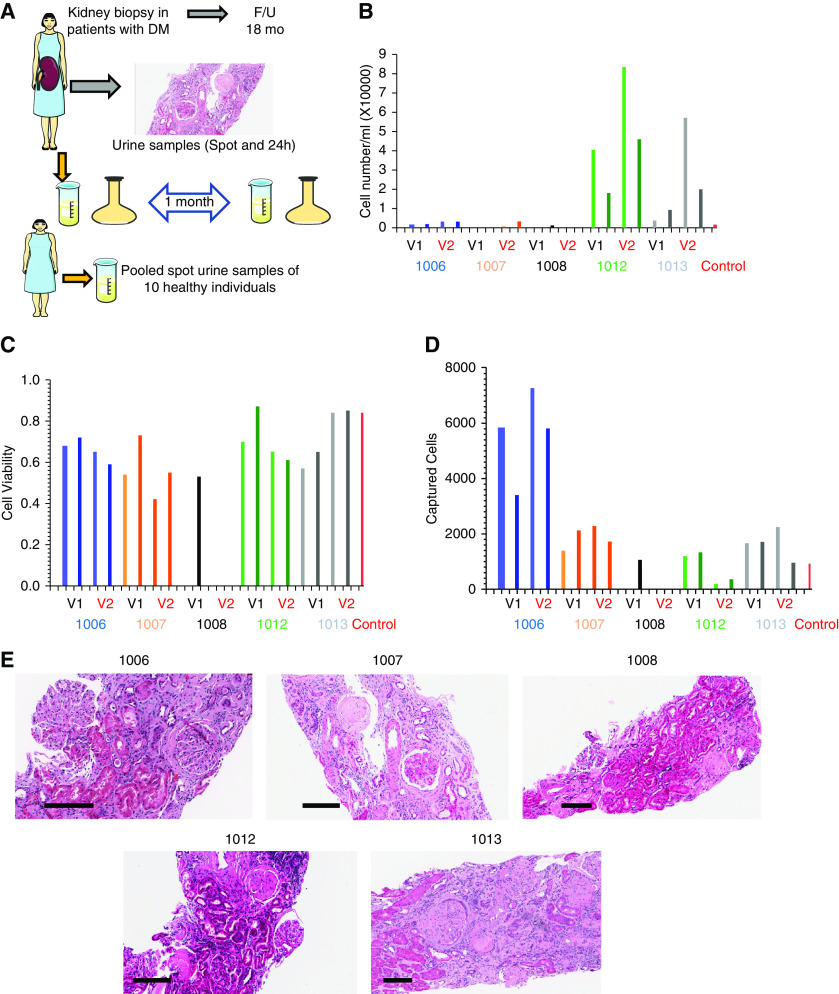
Due to the time sensitivity of the sample processing, five patients at the University of Pennsylvania were approached to participate in this ancillary study. As part of TRIDENT, all participants underwent a diagnostic kidney biopsy. Kidney biopsies were processed and changes in the glomerulus, tubules, interstitium, and vasculature were scored by light, electron, and immunofluorescence microscopic evaluations (Figure 1A). We collected clinical and demographic information on patients at enrollment. All patients were followed for 18 months after their kidney biopsy and their kidney function was monitored. They provided spot and 24-hour urine samples, then returned 1 month later and provided a second set of spots and 24-hour urine samples. Once the samples were obtained, they were immediately transported on ice to the laboratory for processing. Freshly voided control samples were pooled from ten anonymous healthy individuals.
Figure 1.
Study design and urine characteristics. (A) Subjects with diabetes underwent diagnostic kidney biopsy. Tissue samples analyzed and scored by light and electron microscopy and immunofluorescence. Subjects were followed for 18 months. Spot and 24-hour urine samples were collected 1 month apart and analyzed. Ten spot urine samples from ten healthy individuals as control were obtained and pooled for the single-cell RNA-seq study. (B) Sample level, urinary cell number per ml urine (y-axis), x-axis is each patient, collection method, and time. (C) Sample level, urinary cell viability (y-axis), x-axis is each patient, collection method, and time. (D) Total captured cell number per sample, each column represents one sample, lighter and darker shades of the same color represent spot urine sample and the darker color shows the 24-hour urine collection. V1 and V2 represent visit 1 and visit 2, respectively, for each subject. (E) Images of periodic acid–Schiff-stained kidney section of each kidney biopsy. Scale bars are shown at the left bottom of each image represent 200 µm. DM, diabetes mellitus; F/U, follow up.
Urine Collection
All steps were performed on ice or at 4°C. Once the urine volume was measured, the sample was transferred into a 50 ml conical tube and centrifuged at 1000 rpm for 5 minutes. The supernatant was carefully removed, leaving about 1 ml volume in the tube. Cells then were suspended in 1× Dulbecco’s PBS (DPBS) and centrifuged at 1000 rpm for 5 minutes. This process was repeated twice. The pelleted cells were suspended in 1 ml of 1× DPBS and passed through a 40 µm cell strainer. After centrifugation at 1000 RPM for 5 minutes, the cell pellet was resuspended in the appropriate volume of 1× DPBS (resuspension volume was approximated on the basis of the pellet size). Cell number and viability were analyzed using Countess Auto Counter (C10227; Invitrogen). Due to the low cell number in the control urine samples, the control samples were combined, and the subsequent steps were performed on a single pooled control urine sample.
Single Cell Barcoding, Library Preparation, and Sequencing
cDNA libraries were prepared according to the manufacturer’s protocol (Single Cell 3′ Reagent Kits v3; 10× Genomics).16,17 In brief, cell suspensions, reverse transcription master mix, and partitioning oil were loaded on a single-cell “B” chip, then run on the Chromium Controller. mRNA was reverse transcribed within the droplets at 53°C for 45 minutes. cDNA was amplified for 12 cycles total on a BioRad C1000 Touch thermocycler. The cDNA was size selected using SpriSelect beads (Beckman Coulter) with a ratio of SpriSelect reagent volume to sample volume of 0.6.
cDNA quality was examined on an Agilent Bioanalyzer High Sensitivity DNA chip. cDNA was fragmented using the proprietary fragmentation enzyme blend for 5 minutes at 32°C, followed by end repair and A-tailing at 65°C for 30 minutes. cDNA was double-sided size selected using SpriSelect beads. Sequencing adaptors were ligated to the cDNA at 20°C for 15 minutes. cDNA was amplified using a sample-specific index oligo as primer, followed by another round of double-sided size selection using SpriSelect beads. Final library quality was analyzed on an Agilent Bioanalyzer High Sensitivity DNA chip. cDNA libraries were sequenced on a HiSeq4000 platform.
Primary Single-Cell Data Processing
FASTQ files from each 10× single-cell run were processed by Cell Ranger v3.0.2 (10× Genomics). Human genome reference Ensembl 93 (GRCh38.p12) and a custom gene model on the basis of UCSC genes were used to generate gene expression matrix for each cell.
Data Processing and Computational Analyses
Seurat objects from the aligned outputs (from multiple samples) were created, where genes expressed in more than three cells and cells with at least 300 genes were retained. A merged Seurat object was obtained using “MergeSeurat” function of Seurat v (3.1.4).20 Filtering of cells on the basis of quality control metrics was implemented. Cells with >15% mitochondrial counts were filtered out.
Data Normalization and Cell Population Identification
Raw unique molecular identifier (UMI) counts were normalized with a scale factor of 10,000 UMIs per cell and subsequently natural log transformed with a pseudocount of 1. The top 3000 highly variable genes were identified using the method “vst,” then data were scaled and the total number of UMI and the percentage of UMI arising from mitochondrial genes were regressed out. The scaled values were then subjected to principle component (PC) analysis and dimension reduction. Then, we used the Harmony package and “RunHarmony” function to integrate the samples.21 A shared nearest-neighbor network was created on the basis of the Euclidean distances between cells in a multidimensional PC space (the first 30 PC were used) and a fixed number of neighbors per cell, which was used to generate a two-dimensional Uniform Manifold Approximation and Projection for visualization. To identify cell-type markers, we used Seurat’s “FindAllMarkers” function, which calculates log-fold changes, percentages of expression within and outside a group, and P values of Wilcoxon-Rank sum test, comparing a group to all cells outside that specific group including adjustments for multiple testing.
Ambient RNA Contamination Removal and Doublet Finder
To determine the fraction of ambient RNA in each single cell, the SoupX package was used.22 The RNA expression in each cell was then corrected using the ambient mRNA expression profile and estimated contamination. After this correction, we reran the Seurat clustering pipeline as previously described. In addition, we identified probable doublets in our cells using the DoubletFinder function.23 To understand the contribution of cell doublets, we repeated our original clustering.
Integration of Urine Single-Cell Data with Kidney Single Nucleus and Bladder Single-Cell RNA-Seq
To understand similarities between urine, kidney, and bladder cells, our urine single-cell dataset was integrated with a previously published kidney single-nucleus dataset and a bladder single-cell dataset.24,25 Two approaches were used for the integration: (1) the Harmony method,21 (2) and the anchor strategy.26 The Harmony method, as previously described, first places cells into multiple subset clusters then projects cells to a shared embedding, in which cells are grouped by cell type rather than origins of the datasets. The anchor strategy first identifies shared genes, called anchors, between datasets and then datasets are combined using these anchors.26 In the anchor method, after splitting the datasets on the basis of their origin, steps of normalization and highly variable gene identification was performed. Using the “FindIntegrationAnchors” function, anchors that could connect two datasets were determined. The two datasets were integrated using the “IntegrateData” function. This was followed by scaling dimension reduction, clustering, and Uniform Manifold Approximation and Projection visualization.
Data Access
The raw data and the count matrix for all genes and cells after filtering the analyzed urine single-cell samples can be accessed on the National Center for the Biotechnology Information database (GSE157640).
Results
Study Subject Characteristics
The clinical and demographics of the participants are shown in Table 1. We enrolled three male and two female subjects, with a relatively wide age range of 47–72 years. The eGFR range among patients was 21–39 ml/min per 1.73 m2. One patient (1013) had nephrotic range proteinuria on enrollment (urine protein to creatinine ratio >3500 mg/g). Subjects donated spot and 24-hour urine samples on two occasions, separated by 1 month. Most subjects (except 1013) had a stable eGFR during the 18 months of follow-up. Three patients had Renal Pathology Society classification III DKD lesions. We used a single control urine sample pooled from ten anonymous volunteers.
Table 1.
The clinical and pathologic characteristics of the enrolled subjects
| Characteristics | Patient | ||||
|---|---|---|---|---|---|
| 1006 | 1007 | 1008 | 1012 | 1013 | |
| Age, yr | 72 | 48 | 50 | 68 | 47 |
| Gender | M | M | M | F | F |
| BMI, kg/m2 | 28 | 33 | 30 | 37 | 50 |
| BP, mm Hg | 188/70 | 144/77 | 129/61 | 101/60 | 176/81 |
| eGFR, ml/min per 1.73 m2 | 22 | 39 | 22 | 23 | 21 |
| ΔeGFR, ml/min per 1.73 m2 | 0.59 | −2.3 | 12.01 | 4.76 | −12.7 |
| UPCR, mg/g | 1400 | 1100 | 800 | 1500 | 5000 |
| ΔUPCR, mg/g | −700 | 130 | 1140 | 320 | 900 |
| UA | |||||
| Epi | 0–2 | 0–10 | 0 | 0–5 | 10–20 |
| WBC | 0–1 | 0–5 | 0–5 | 0–5 | 5–10 |
| RBC | 0–2 | 0–2 | 0–2 | 0 | 2–5 |
| RPS class | 2 | 3 | 2 | 3 | 3 |
| Global sclerosis (%) | 26–50 | 1–25 | 26–50 | 26–50 | 26–50 |
| Interstitial fibrosis (%) | 50 | 80 | 50 | 40 | 60 |
| Mesangial matrix increase | Mod | Severe | Mod | Mod | Severe |
| Interstitial lymphocytes | Mod | Mod | Mild | Mild | Mod |
| Arterial sclerosis | Severe | Mild | Mild | Mod | Severe |
| Foot process effacement (%) | 30 | 70 | 30 | 30 | 50 |
| Avg GBM thickness (nm) | 431 | 491 | 722 | 785 | 1120 |
BMI, body mass index; UPCR, urine protein to creatinine ratio; UA, urinalysis; epi, epithelial cells; WBC, white blood cells; RBC, red blood cells; RPS class, Renal Pathology Society classification; mod: moderate; GBM, glomerular basement membrane.
Urinary Cell Number and Viability
Total urinary cell numbers showed relatively large variability. The urinary cell number was higher in female urine samples compared with males. Cell viability in the control urine sample was 84% and ranged from 42% to 87% in the DKD samples. We did not observe important differences between spot or 24-hour urine samples or between different subjects (Figure 1C). We aimed for a cell capture rate of 10,000 cells per sample; however, the cell capture rate showed significant heterogeneity. It is possible the microfluidics device did not capture the large squamous epithelial cells that are often present in the female urine, as capture rate was lower in urine samples obtained from women. It is also important to note that one subject (1008) had a low urinary cell count, and we were unable to capture cells in three out of the four samples (Figure 1D). This patient had the lowest proteinuria (800 mg/g) and the most preserved eGFR during follow-up. The urinary cell number was also very low in our control samples, even after pooling urine from multiple donors. The final analysis included 18 samples, 17 DKD, and one pooled control urine sample, indicating an overall 86% success rate to generate single-cell libraries.
Urine Contains a Large Number of Epithelial Cells
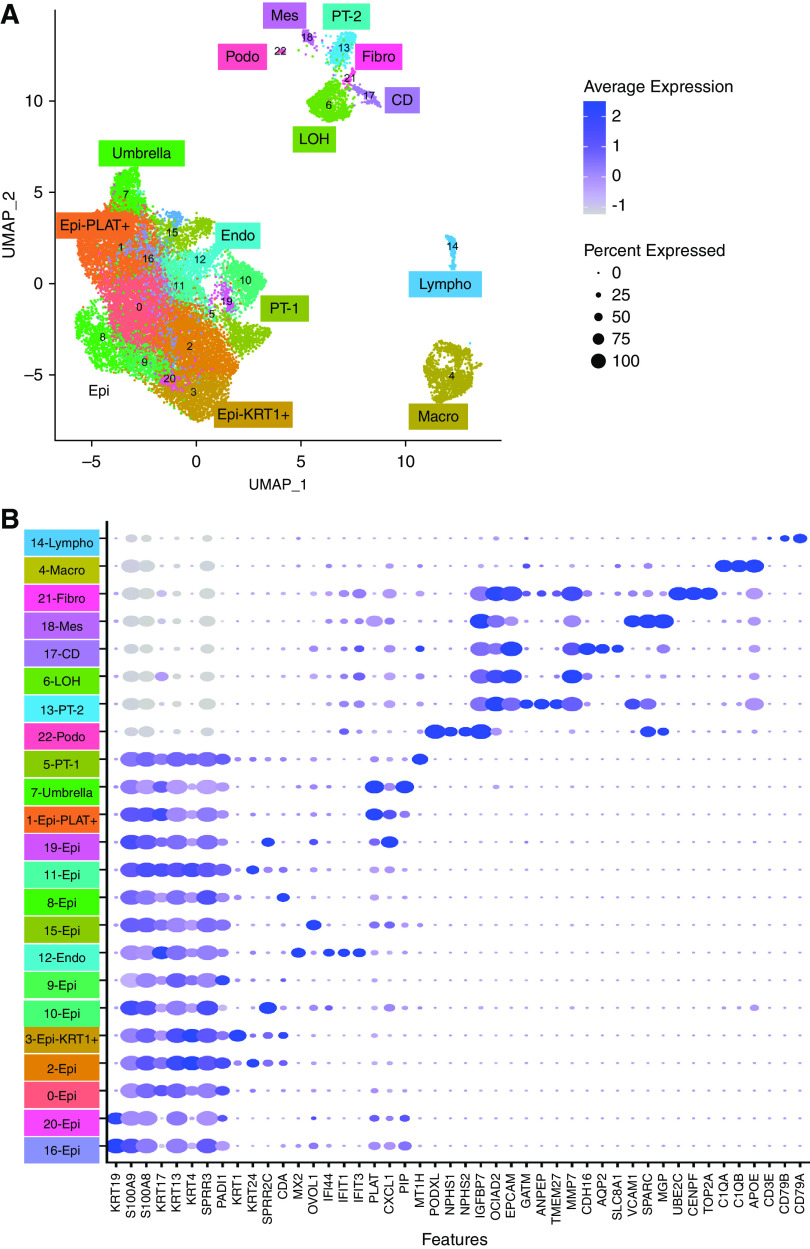
In total, we sequenced 41,620 cells and after quality control, such as setting the minimal number of detected genes and mitochondrial read percentage, we only retained 23,082 for further analysis (Supplemental Figure 1, Supplemental Table 1). This was critical as we found that inclusion of poor-quality cells (mitochondrial percentage of up to 50%) interfered with clear clustering and cell-type identification (Supplemental Figure 1F). The total number of reads did not correlate with the captured versus filtered cell number (Supplemental Figure 1G). Overall, the fraction of ambient RNA in urine samples ranged from 1% to 33%, with the mean of 7.77% as estimated by SoupX22 (Supplemental Figure 2A). The fraction of ambient RNA was much higher in 24-hour urine samples than spot urine specimens, which might be an indicator of an overall higher number of dead cells and RNA release in the 24-hour collection. We found no major differences when we clustered the data without batch correction (Supplemental Figure 2B) or Harmony-based integration (Supplemental Figure 2C) (Figure 2A).
Figure 2.
Single-cell survey of the human urine. (A) Uniform Manifold Approximation and Projection (UMAP) dimension reduction of 23,089 urinary cells identified in 17 DKD and one pooled control urine samples. Clusters 0, 2, 8, 9, 10, 11, 15, 16, 19, and 20 were called Epi. (B) Bubble dot plots of the top cluster specific genes. The size of the dot indicates expression percentage and the darkness of the color indicates average expression. Epi, variety epithelial cells; Epi-PLAT+, PLAT-positive cells; Epi-KRT1+, KRT1-positive cells; umbrella, umbrella cells; endo, endothelial cells; podo, podocytes; LOH, loop of Henle; fibro, fibroblast; CD, collecting duct principal cell; macro, macrophages; lympho, lymphocytes; mes, mesenchymal cells.
Our clustering indicated 23 discrete clusters separated into three relatively distinct large groups (Figure 2A). The second largest group contained cells positive for kidney epithelial markers, such as EPCAM, OCIAD2, and CRYAB (Figure 2B). We identified a distinct cluster that was highly positive for the collecting duct marker AQP2 (cluster 17). There was a cluster (cluster 6) highly positive for MMP7, which is mostly expressed by loop of Henle cells in the kidney. Another group (cluster 13) of cells expressed GATM and EPCAM, markers of PT cells. Another cluster (cluster 22) was positive for podocyte markers, such as NPHS1 and NPHS216,17,27(Figure 2B). In addition, we also found a group of cells that was positive for fibroblast markers, such as PTTG1, CENPF, UBE2C, and TOP2A (cluster 21).
The smallest group contained immune cells. One of the clusters (cluster 14) was positive for classic lymphocyte markers, such as CD79, and another (cluster 4) for classic macrophage markers, such as C1Q (Figure 2B). The complete list of cell-type specific differentially expressed genes is available in Supplemental Table 2.
Group 1, the largest group, separated into several subclusters (Figure 2A). Multiple clusters were positive for KRT4, KRT13, and KRT17; these are known markers of renal pelvic and bladder epithelial cells.25 Interestingly, KRT4-positive cells were also positive for myeloid markers S100A8 and S100A928 (Supplemental Figure 2D). In our analysis it did not seem that the expression of epithelial (KRT) and myeloid (S100) markers by the same cells was related to cell doublets (Supplemental Figure 1H) or to ambient RNA contamination (Figure 2A, Supplemental Figure 2, B and C). Cluster 3 was positive for KRT1, raising the possibility that these might be skin or genital-related cells. We identified two clusters positive for the bladder cell marker KRT19.25 This large group also included cells from cluster, expressing MT1, which has been previously identified as a marker of PT.29 Cluster 7 was positive for PLAT, FXYD3, and TSPAN1 potential markers of umbrella cells of the bladder.25 It is interesting to note this cluster also included cluster 12, cells expressing IFI44, IFIT1, IFIT3 markers of kidney endothelial cells.30
Cell Fractions are Subject Specific and Reasonably Stable over Time and Collection Method
As a first step, we have compared the distribution of cells in the control and DKD urine samples. Although single-cell sequencing could be biased by batch effect, we found that all DKD urine samples (Figure 3, A and C) contributed to the kidney-specific and immune cell clusters, indicating a lesser role of a technical batch effect (Supplemental Figure 1I). There were very few kidney and immune cells in the urine of control subjects (Figure 3B). The differences of distribution of urinary cells were significant between DKD and control samples measured by chi-squared test (X2=167.11, P<0.0001) (Figure 3B).
Figure 3.
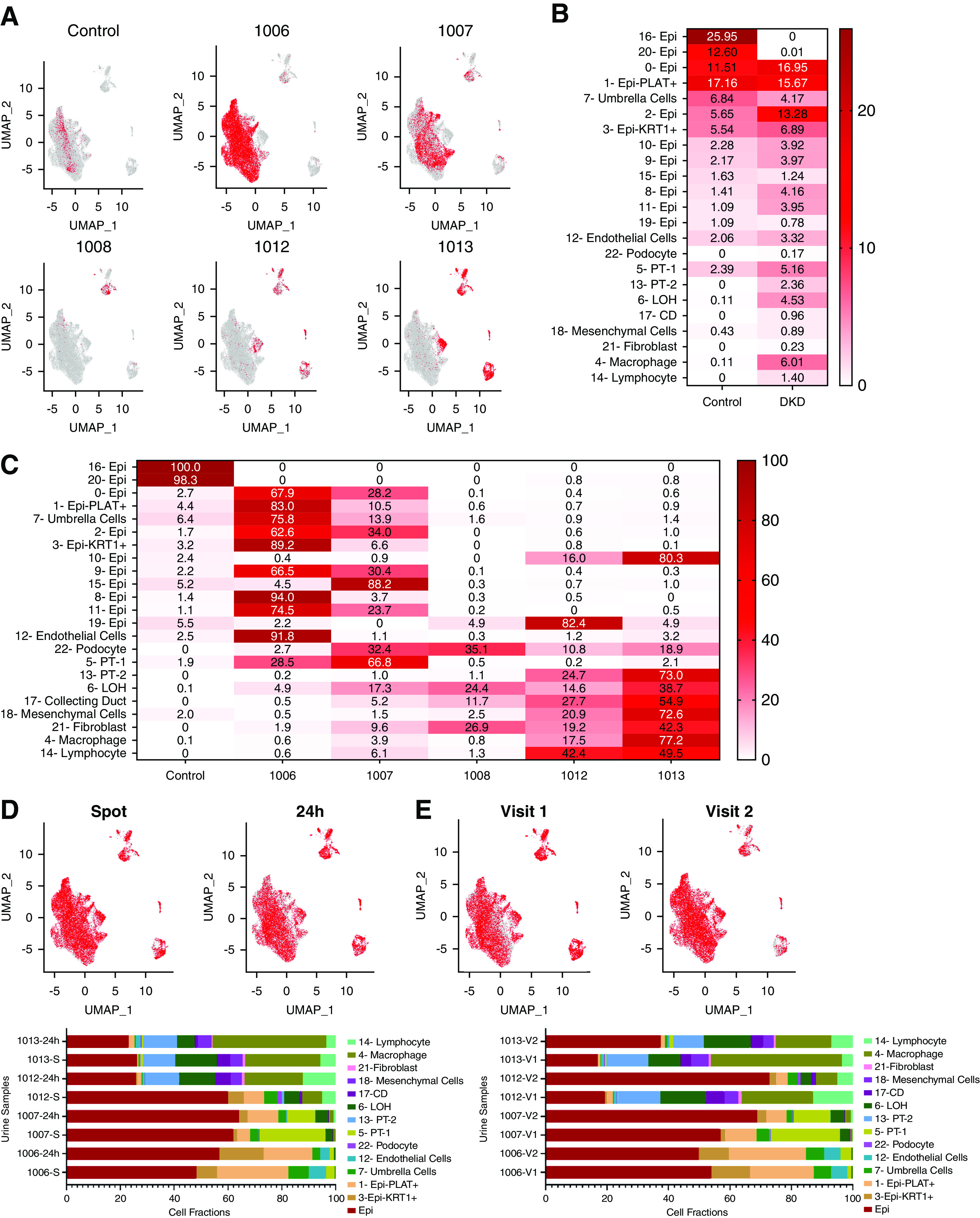
Distribution of cells in control and DKD samples. (A) Subject-level feature plot of cells over the Uniform Manifold Approximation and Projection (UMAP) of all cells (gray). All cells analyzed for a single subject (visit 1 [V1] and V2, spot and 24-hour) were combined and shown in red. (B) The fractions of cells in each cell cluster in control and DKD urine samples. Darker color indicates higher percentage of cells. (C) The fractions of cells in each cell cluster in each sample. Darker color indicates a higher percentage of cells. (D) The stability of urinary cell number on the basis of the collection method. The collection method (spot versus 24-hour) level feature models (colored red) over all cells in the UMAP (gray) (upper panel). Cumulative urine cell fractions of spot and 24-hour urine samples (the two visits were combined) (lower panel). (E) The stability of urinary cell number on the basis of time. V1 or V2 level feature models (colored red) over all cells in the UMAP (gray) (upper panel). Cumulative urine cell fractions of visits 1 and 2 samples (combined spot and 24-hour collection) (lower panel). Epi, variety epithelial cells; Epi-PLAT+, PLAT-positive cells; Epi-KRT1+, KRT1-positive cells; LOH, loop of Henle; CD, collecting duct principal cell.
Next, we examined the distribution of cell types in each urine specimen to understand whether we can observe a specific pattern. We observed that cell fractions varied substantially between patients (Figure 3C). For example, subjects 1006 and 1007 mostly had epithelial cells in their urine, whereas 1012 and 1013 had more kidney cells. Next, we analyzed the stability of the cell fractions between collection method and visits (Figure 3, D and E). There was no observable difference in cell identification when spot and 24-hour urine samples were compared, indicating cells remained viable in the 24-hour collection (Figure 3D). Furthermore, we also found the cell fractions were reasonably stable and remained subject specific when comparing cell fractions between the two different collection time points (Figure 3E).
Cells in the Urine Cocluster with Kidney and Bladder Cells
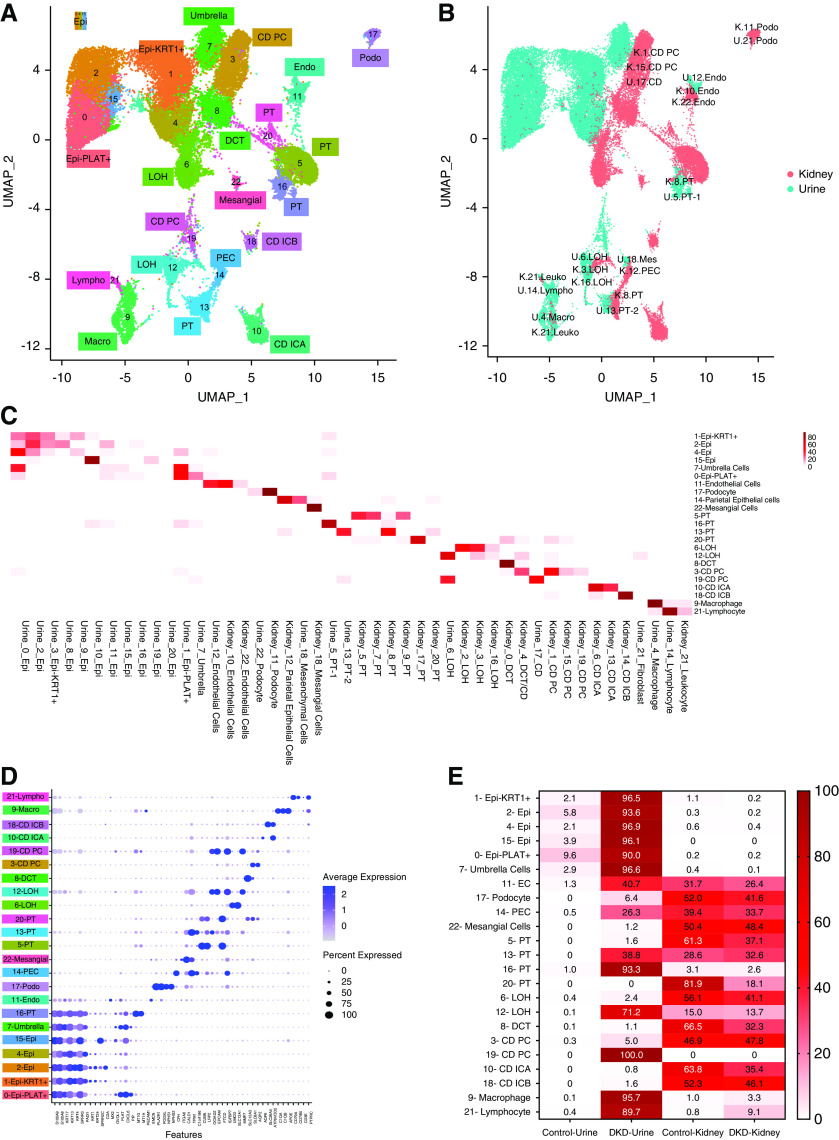
To understand the origin of the urinary cells and their relatedness to cells in the kidney and the urinary tract, we integrated our urine single-cell dataset with published human kidney single nucleus and bladder single-cell RNA-sequencing datasets. Human kidney single-nucleus RNA-sequencing datasets obtained from three control and three patients with DKD were downloaded.24 We used two different strategies to integrate the urine and kidney cells as implemented in Harmony21 and anchor packages26 (detailed in Methods).
Using Harmony integration, we identified 23 discrete clusters (Figure 4A). We plotted the source of cells in the integrated clusters (kidney and urine) on the basis of the primary clusters (Figure 4C). We observed a strong, almost one-on-one, correlation between urinary and kidney cell clusters. Urinary cells showed strong similarities to kidney cells, such as podocytes from the urine and kidney had formed a single, discrete cluster. The principal and intercalated cells of the collecting duct of the urine formed discrete clusters with corresponding cell types from the kidney. We have identified four distinct PT cell clusters. First, we note that control and diseased PT cells already showed important diversity in the published kidney dataset, such as cluster 20, which almost exclusively contained cells from the control kidney, and cluster 5, which contained cells both from diabetic and control kidneys. The PT cluster 13 contained urinary, control, and DKD kidney cells (Figure 4C). This cluster was positive with TPM1 and OCIAD2, and expressed only low levels of traditional PT markers, such as CUBN or LRP2, indicating these cells might be dedifferentiated PT cells. Cluster 16, which was located close to the kidney PT cells, contained PT cells with high levels of MT1H and MT1G.29
Figure 4.
Integration of urine single cell with human kidney single nuclei datasets. (A) Uniform Manifold Approximation and Projection (UMAP) of Harmony-based integration of urinary and kidney cells. (B) UMAP of Harmony-based integration of urinary and kidney cells colored by the sample of origin. Blue indicates the urine origin and pink shows the cells originated from kidney. Each cluster is labelled with the cells of origin. (C) The percent of cells in each integrated (urine and kidney) cluster (y-axis) that came from each original cluster (x-axis). (D) Bubble dot plots of the top cell-type–specific differentially expressed genes in the integrated clusters of urine and kidney samples. The size of the dot indicates the expression percentage and the darkness of the color indicates average expression. (E) The fraction of cells in each integrated cluster and their sample of origin. The data are colored by the percentage of cell. Epi, variety epithelial cells; Epi-PLAT+, PLAT-positive cells; Epi-KRT1+, KRT1-positive cells; umbrella, umbrella cells; endo, endothelial cells; EC, endothelial cells; LOH, loop of Henle; CD PC, collecting duct principal cells; CD ICA, collecting duct intercalated cells A; CD ICB, collecting duct intercalated cells B; DCT, distal convoluted tubule; mesangial, mesangial cells; podo, podocytes; PEC, parietal epithelial cells; macro, macrophages; lympho, lymphocytes; U, urine; K, kidney; leuko, leukocyte; mes, mesenchymal cells.
The loop of Henle cells separated into two clusters. Cluster 12 contained cells from DKD urine, and control and DKD kidney samples. These cells were positive for MMP7, IGFBP7, and EPCAM (Figure 4E). We identified both principal and intercalated cells of the collecting duct in the urine sample. Urinary lymphocytes clustered closely with kidney lymphocytes. It is interesting to note that cluster 12 of urine, which was positive for the endothelial markers, IFI44, IFIT1, IFIT3,30 clustered closely with kidney endothelial cells, suggesting even endothelial cells are shed into the urine. We verified our integration analysis using the anchor method, and found consistent results (Supplemental Figure 3).
Next, we wanted to understand whether we could identify bladder cells in the urine by integrating our urine single-cell RNA-seq data with single-cell RNA-seq dataset from the human bladder25(Supplemental Figure 4). We found that urinary cells expressing PLAT and TSPAN1 clustered closely with bladder umbrella cells (cluster 20). It is interesting to note that bladder and kidney fibroblasts clustered together (cluster 21). In addition, the cluster we initially labeled mesenchymal cells in the urine clustered with smooth muscle cells, indicating shared gene markers expressed by those two clusters (cluster 12). It should be noted that cells labeled as loop of Henle in the urine clustered with intermediate cells of the bladder, mostly due to a shared gene marker of MMP7 (cluster 11). We believe this cluster likely contains multiple types of cells. Urinary endothelial and bladder endothelial cells did not cocluster. We observed an important overlap between urine lymphocytes and bladder lymphocytes (clusters 16 and 19) and some between bladder macrophages and urinary monocytes (cluster 8). We again validated our integration results using the anchor model (Supplemental Figure 5) with minor differences. An important difference was the coclustering of urinary clusters 0 and 8 with basal cells and TNNT1-positive cells of the bladder, respectively (clusters 1 and 13).
Combined Analysis of Human Urine, Bladder, and Kidney Cells
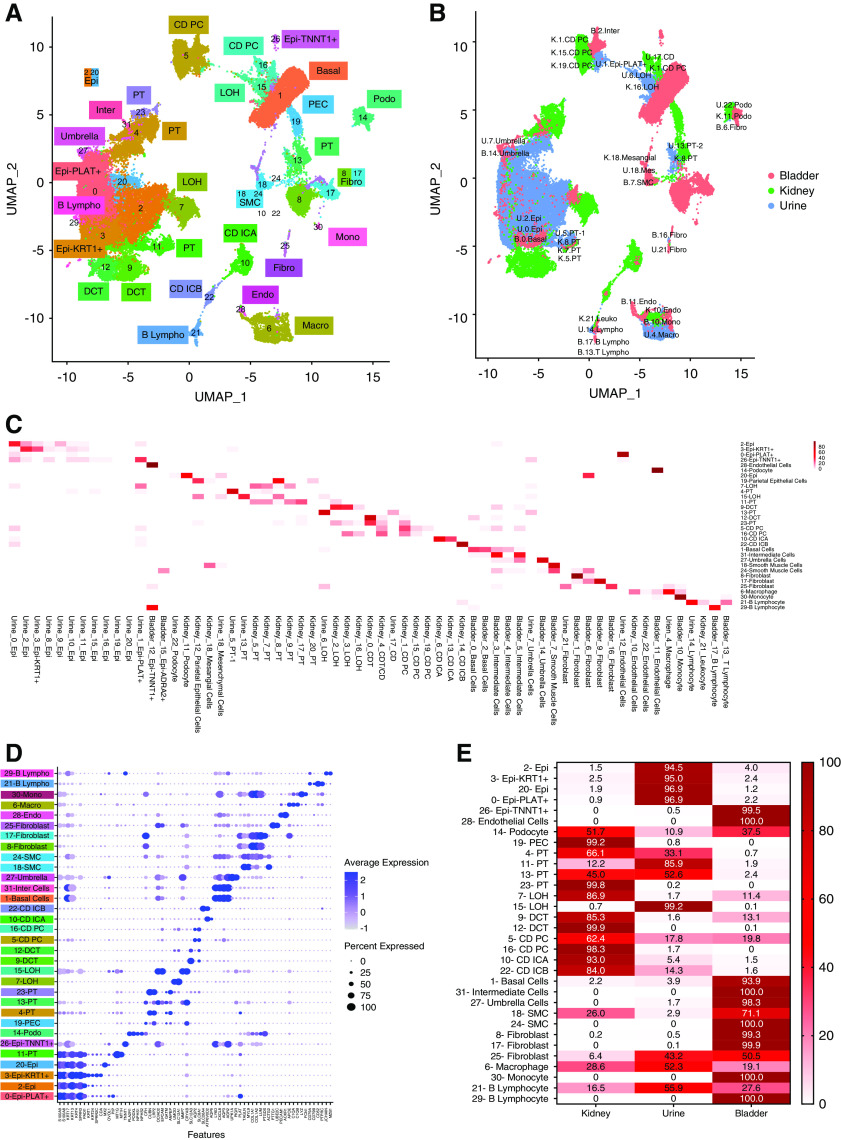
Finally, we integrated the urine, bladder single-cell, and human kidney nucleus datasets to understand relatedness of the analyzed cells (Figure 5, Supplemental Figure 6). We plotted the source of cells in the integrated clusters (kidney, bladder, and urine) on the basis of the primary clusters, and found a strong almost one-to-one relationship (Figure 5, B and C).
Figure 5.
Integration of urine single-cell, kidney single-nucleus, and bladder single-cell datasets. (A) Uniform Manifold Approximation and Projection (UMAP) of Harmony-based integration of urinary, bladder, and kidney cells. (B) UMAP of Harmony-based integration of urinary bladder and kidney cells colored by the sample of origin. Blue, green, and pink indicates the urine, kidney, and bladder origins, respectively. The origins of the cells are written in each plot. (C) The percent of cells in each integrated (kidney, urine, and bladder) cluster (y-axis) that originated from the original clusters (x-axis). (D) Bubble dot plots of the top cell-type–specific differentially expressed genes in the integrated clusters of urine, kidney, and bladder samples. The size of the dot indicates expression percentage and the darkness of the color indicates average expression. (E) The fraction of cells in each integrated cluster and their sample of origin. Epi, variety epithelial cells; Epi-PLAT+, PLAT-positive cells; Epi-KRT1+, KRT1-positive cells; Epi-TNNT1+, TNNT1-positive cells; PEC, parietal epithelial cells; LOH, loop of Henle; DCT, distal convoluted tubule; CD PC, collecting duct principal cells; CD ICA, collecting duct intercalated cells A; CD ICB, collecting duct intercalated cells B; SMC, smooth muscle cell; basal, basal cells; inter, intermediate cells; umbrella, umbrella cells; endo, endothelial cells; podo, podocyte; fibro, fibroblast; SMC, smooth muscle cell; macro, macrophages; mono, monocyte; lympho, lymphocytes; U, urine; K, kidney; B, bladder; leuko, leukocyte.
As we observed earlier urine cells coclustered both with kidney and bladder cells, indicating that we could potentially ascertain their origin. Again, urinary and kidney podocytes clustered well together (cluster 14). We observed multiple PT clusters (4, 11, 13, and 23) (Figure 5D, Supplemental Figure 6D). Again, the urine seemed to contain few DCT cells, or these cells were so severely degenerated in the urine that we could no longer identify them. Principal (cluster 5) and intercalated cells (cluster 22) of the collecting duct were readily identifiable in the urine. We were able to identify a variety of bladder cells in the urine such as basal cell, umbrella cells, fibroblasts, and smooth muscle cells. Lymphocytes (cluster 21) and macrophages (cluster 6) from the bladder, urine, and kidney clustered together. In this analysis we found that smooth muscle cells of the bladder coclustered with mesangial cells of the kidney and mesenchymal cells in the urine, mostly due to shared gene markers, especially MYL9 and TAGLN (cluster 18). As mesangial cells and vascular smooth muscle cells share multiple markers, it is possible the cell cluster originally labeled as mesangial cells in the kidney was a vascular smooth muscle cell cluster.
Urinary Single-Cell Profiling Provides a Read-Out for Kidney Disease Genes and Drug Targets
Next, we wanted to understand whether urinary single cell profiling could potentially be used to make diagnostic or therapeutic decisions.
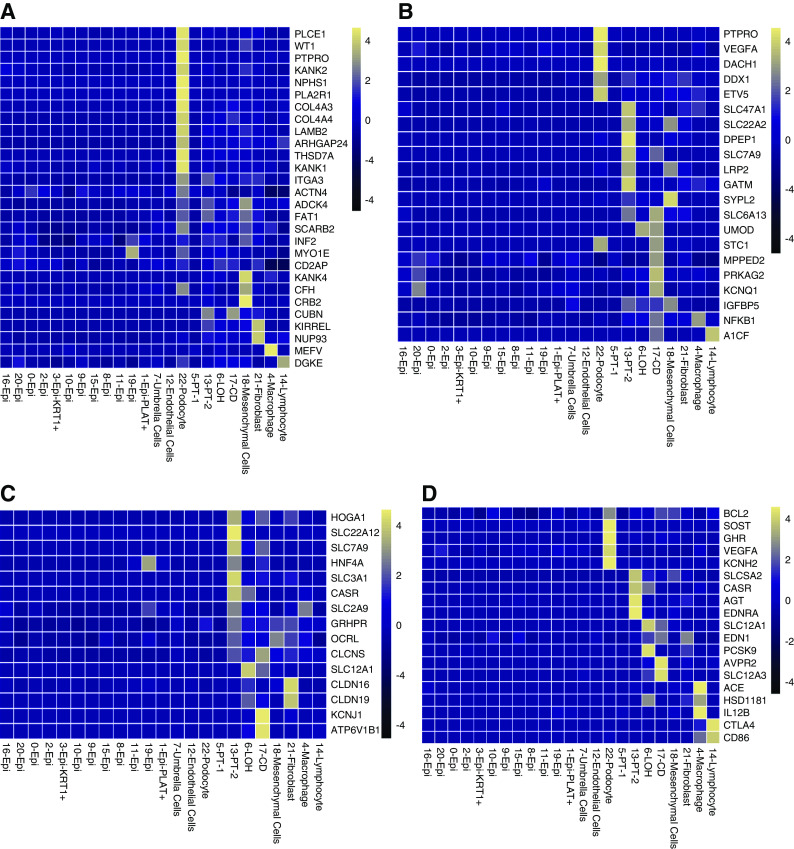
First, we have analyzed the expression of genes known to cause monogenic nephrotic syndrome. Figure 6A shows strong enrichment of nephrotic syndrome genes in urinary podocytes. Next, we examined the expression of genes nominated to mediate the effect of the polygenic eGFR GWAS.31,32 Similar to our previous findings, these genes showed strong enrichment for urinary PT expression (Figure 6B). Finally, we analyzed the expression of genes that showed association with kidney stones. We found enrichment for PT and collecting-duct cell expression of kidney stone–associated genes (Figure 6C).
Figure 6.
Urinary single-cell profiling provides a read-out for kidney disease genes and drug targets. (A) Urinary single cell-type specific expression enrichment of monogenic nephrotic syndrome genes. (B) Urinary single cell-type–specific expression enrichment of CKD-GWAS–nominated genes. (C) Urinary single cell-type–specific expression enrichment genes related to nephrolithiasis. (D) Urinary single cell-type–specific expression enrichment of Food and Drug Administration–approved drug target genes. Mean gene expression values were calculated in each cell type cluster. The color scheme of the heatmap is on the basis of z score distribution. In each heatmap, x-axis represents the urinary cell clusters and y-axis shows the genes. Epi, variety epithelial cells; Epi-PLAT+, PLAT-positive cells; Epi-KRT1+, KRT1-positive cells; PT, proximal tubule; LOH, loop of Henle; CD, collecting duct principal cells.
Finally, we analyzed the urine single cell-type expression of genes targeted by commonly used Food and Drug Administration–approved drugs (Figure 6D). We show that the urinary loop of Henle cells are positive for the loop diuretic target SLC12A1. Urinary collecting tubule cells expressed AVPR2 as a target gene for tolvaptan. Several Food and Drug Administration–approved drugs showed podocyte-enriched expression, such as BCL2, SOST, GHR, and VEGFA. PT cells were enriched for SLC5A2, the target gene of SGLT2 inhibitors and angiotensinogen, a key substrate in the renin angiotensin pathway.
Overall, our pilot study indicates that urine cells can be used to read-out for monogenic and complex kidney disease genes and targets of the most commonly used drugs, suggesting the method might be of use for precision medicine approaches.
Discussion
Examination of the urine sediment is considered one of the most basic diagnostic and prognostic procedures performed by nephrologists.1 Our study, is an important attempt to perform single-cell resolution profiling of all urinary cells of patients with diabetic kidney disease in a high throughput manner. Single-cell RNA-seq has been explored for its potential in kidney disease investigations and earlier studies relied on the use of kidney biopsy samples for sequencing.24,33 Our study set to establish the use of fresh urine as a biospecimen for single-cell RNA-seq.
Here, we show a new method of analyzing urinary cells using single-cell sequencing. We show that the method is technically feasible. Overall, both spot and 24-hour urine specimens appeared suitable for the urinary single-cell RNA-seq. The cellular diversity of the spot and 24-hour collecting was similar. We found that the 24-hour urine samples might suffer from greater ambient RNA contamination, which might be related to increased cell death during the extended collection time. We observed that healthy subjects and the patient with DKD with the most preserved kidney function had very few cells in their urine, potentially indicating that total urinary cell numbers might also be an indicator of kidney function.
It is also important to note that our study used stringent filtering and we were able to identify a variety of specific kidney epithelial cells. Urinary podocytes and collecting duct cells expressed the same marker genes observed in kidney single-cell analysis.24 We observed multiple different PT subclusters, with many showing signs of severe dedifferentiation. Indeed, kidney PT cells clustered better with PT cells profiled from patients with DKD and more distant from healthy PT cells.24 Although prior studies have analyzed a small number of immune,14 kidney, and epithelial cells,18 these studies were not able to characterize specific kidney tubule cells, whereas our work provides new gene expression marker data for urinary podocytes, PT cells, loop of Henle, and collecting duct cells.
Our study shows that almost all kidney cells can be identified in the urine. Urinary podocytes and collecting duct cells seemed to show the strongest and most consistent correlation with kidney cells. We found very few distal convoluted tubule cells in the urine, or these cells might have severely dedifferentiated limiting our ability to identify them. Consistent with the observed plasticity of PT cells,34 we found that PT cells were severely dedifferentiated in the urine samples. Urinary PT cells coclustered better with PT cells identified in DKD kidneys than those found in control healthy samples. We also observed important differences in PT subtypes, which will be important to follow.
Our study had several limitations; first, as a pilot study we included few patients and controls. We collected urine at different time points and with different methods to confirm the reproducibility and feasibility of this approach. Our study indicates significant differences in capturing efficiency between male and female urine samples. We have also observed that ambient RNA contamination was significantly higher in 24-hour urine collections, likely indicating increased cellular degradation during extended storage. Overall, our study optimistically indicates the cell composition of urine is stable over time and between different collection methods. It is encouraging that our study and a previous publication found correlation between urinary immune cells and immune cell fractions in the kidney, although the sample size in the prior publication was modest (n=8 and 13 subjects).14,18 It seems that a large prospective cohort study will be needed to establish the diagnostic and prognostic utility of urinary single cell sequencing.
In summary, in this study we present the clinical feasibility of performing single-cell RNA-seq on human urine samples. We generated a reference dataset and were able to identify most kidney and many bladder cells in human urine. Our analysis indicates that urinary cell profiles show patient specificity and modest variability between collection method and timing. Our study indicates the method should be further pursued for diagnoses of diseases of the kidney and genitourinary tract.
Disclosures
C. Boustany-Kari reports having an ownership interest in Pfizer and being a board member of BioCT, and is an employee of Boehringer Ingelheim. G. Halasz reports having an ownership interest in Regeneron Pharmaceuticals. J. Hogan reports consultancy agreements with Retrophin; receiving research funding from Achillion-site principal investigator (PI) for a clinical trial in C3G, Boeringer Ingelheim, Calliditassite PI for a clinical trial in IgA nephropathy, Complexa-site PI for a clinical trial in FSGS, Gilead, GSK, the National Institutes of Health for a study in membranous nephropathy, Omeros-site PI for a clinical trial in IgA nephropathy, Regeneron, Retrophin-site PI for DUET and DUPLEX Studies in FSGS, and the TRIDENT study for diabetic nephropathy; reports receiving honoraria from Alexion, Aurinia, Calliditas, Dimerix, Goldfinch Bio, GSK, Retrophin, Zyversa; receiving royalties for patent (pending) as a coinventor of WO2019213434A1; being a scientific advisor or member of Zyversa (Advisory board); and other interests/relationships as an UpToDate.com author with royalties for topics on calcium/phosphorous balance and monoclonal gammopathy of renal significance. K. Devalaraja-Narashimha reports having an ownership interest in Regeneron Pharmaceuticals. K. Devalaraja-Narashimha, G. Halasz, L. Morton, Y. Wei, Y. O. Zhu, and Y.Z. are employees of Regeneron Pharmaceuticals. K. Susztak reports consultancy agreements with Boehringer Ingelheim, Jnana, Kyowa Kirin, and Maze; receiving research funding from Bayer, Boehringer Ingelheim, Gilead, GSK, Lilly, Merck, Regeneron; receiving honoraria from Jnana and Maze; being a scientific advisor or member of the editorial board for Kidney International, Journal of Clinical Investigation. L. Morton reports having an ownership interest in Regeneron Pharmaceuticals; reports receiving research funding from Regeneron Pharmaceuticals; has patents and inventions with Regeneron Pharmaceuticals; and Speakers Bureau from Regeneron Pharmaceuticals. M. Balzer reports receiving research funding from the Else Kröner-Fresenius Foundation and the German Research Foundation (Deutsche Forschungsgemeinschaft). M. Palmer reports being an Editorial Board Member of the American Journal of Kidney Disease. R. Townsend reports consultancy agreements with Axio, IONIS, Medtronic, and Regeneron; reports receiving research funding from the National Institutes of Health; being a scientific advisor or member of the editorial boards of Pulse and the Journal of Clinical Hypertension; and reports receiving royalties as an UpToDate contributor. S. Almaani reports consultancy agreements with Aurinia Pharmaceuticals. S. Blady reports receiving research funding from BIPI, Gilead Sciences, GSK, and Regeneron. U. Patel is an employee of Gilead and reports current employment as Adjunct Professor at Duke University; reports past consultancy agreements with Ablative Solutions Inc (DSMB 2014–2016), Amgen (CEC 2008–2014; TSC 2013–2016), Angion Biomedica (DSMB 2011–2013), CSL Limited (DSMB 2011–2013), Eli Lilly & Co (CEC 2012–2016), Gilead Sciences, Inc (Ad Board 2013, DSMB 2014–2016), GSK (ESC 2014–2016), Keryx Pharmaceuticals (Ad Board 2013), Hospira Inc (Ad Board 2014); reports an ownership interest in Gilead Sciences, Inc.; reports receiving past research funding from clinical event classification committees for trials: Amgen, Eli Lilly & Co., GSK; reports receiving honoraria from Gilead Sciences, Inc. (Ad Board 2013), Hospira Inc (Ad Board 2014), and Keryx Pharmaceuticals (Ad Board 2013); being a current scientific advisor or membership of Goldfinch Bio, Board Observer and Kidney Health Initiative, Board of Directors; being a past scientific advisor or member of Advances in Chronic Kidney Disease (Editorial Board), American Heart Journal (Associate Editor), American Society of Nephrology (Public Policy Board), American Society of Pediatric Nephrology (Public Policy Committee), CJASN (Editorial Board), JASN (Editorial Board), and National Kidney Foundation (North Carolina Medical Advisory Board); other interests/relationships: National Kidney Disease Education Program Health Information Technology Working Group (member). Y. Wei reports ownership interest in Regeneron. Y. Zhu reports having an ownership interest in Regeneron Pharmaceuticals. All remaining authors have nothing to disclose.
Funding
This work is supported by the National Institutes of Health grants DK076077, DK087635, and DK105821. The TRIDENT study is supported by Boehringer Ingelheim, Gilead Sciences, Regeneron Pharmaceuticals, and GlaxoSmithKline Foundation. The funders have no influence on the reported results. We thank the University of Pennsylvania Diabetes Research Center for the use of the Core (P30-DK19525).
Supplementary Material
Acknowledgments
Kishor Devalaraja-Narashimha and Prof. Katalin Susztak designed and conceived the experiment, Dr. Matthew Palmer performed the pathological analysis of tissue samples, Dr. Jonathan Hogan, Dr. Raymond Townsend, and Shira Blady recruited the subjects, Rojesh Shrestha conducted the single-cell experiments. Shatakshee Chatterjee, Yi Wei, Yuan O. Zhu, Jihwan Park, Prof. Katalin Susztak, Katie Marie Sullivan, Gabor Halasz, Tong Zhou, Hailong Hu, Xin Sheng, and Amin Abedini conducted bioinformatics analysis with advice from Prof. Katalin Susztak, Michael S. Balzer, Ziyuan Ma, Kishor Devalaraja-Narashimha, and Lori Morton. Amin Abedini and Prof. Katalin Susztak wrote the manuscript and all authors edited and approved of the final manuscript.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
Contributor Information
Collaborators: The TRIDENT Study Investigators, Katalin Susztak, Raymond Townsend, Shira Blady, Matthew Palmer, Carine Boustany, Richard Urquhart, Paolo Guarnieri, Lea Sarov-Blat, Erding Hu, Lori Morton, Kishor Devalaraja, Uptal Patel, Shawn Badal, John Liles, Jonathan Rosen, Anil Karihaloo, Randy Luciano, Jonathan Hogan, Amy Mottl, Shweta Bansal, Salem Almaani, Christos Argyropoulos, Kirk Campbell, Tamara Isakova, Oliver Lenz, Harold Szerlip, Matthias Kretzler, Pietro Canetta, Jeffery Schelling, Rupali Avasare, Frank Brosius, Michael Ross, Nelson Kopyt, James Tumlin, Julia Scialla, Richard Lafayette, Manisha Singh, and Yan Zhong
Supplemental Material
This article contains the following supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2020050757/-/DCSupplemental.
Supplemental Table 1. Quality control metrics of the urine single cell RNA-seq data.
Supplemental Table 2. The list of cell-type specific genes of the human urinary cells. Epi, variety epithelial cells; Epi-PLAT+, PLAT positive cells; Epi-KRT1+, KRT1 positive cells; PT, proximal tubule; CD, collecting duct principal cells; mes, mesenchymal cells; fibro, fibroblast; podo, podocyte; macro, macrophages; lympho, lymphocytes.
Supplemental Figure 1. Quality control metrics of the urine single cell RNA-seq data.
Supplemental Figure 2. Ambient RNA contamination of urine single-cell samples.
Supplemental Figure 3. Integration of urine single cells with human DKD and control single-nucleus kidney datasets using anchor method.
Supplemental Figure 4. Integration of urine and bladder single-cell datasets by the Harmony method.
Supplemental Figure 5. Integration of urine and bladder single-cell datasets by anchor method.
Supplemental Figure 6. Integration of urine single-cell, kidney single nucleus, and bladder single-cell datasets using the anchor method.
References
- 1.Cavanaugh C, Perazella MA: Urine sediment examination in the diagnosis and management of kidney disease: Core curriculum 2019. Am J Kidney Dis 73: 258–272, 2019 [DOI] [PubMed] [Google Scholar]
- 2.Oliveira Arcolino F, Tort Piella A, Papadimitriou E, Bussolati B, Antonie DJ, Murray P, et al.: Human urine as a noninvasive source of kidney cells. Stem Cells Int 2015: 362562, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Racusen LC, Fivush BA, Andersson H, Gahl WA: Culture of renal tubular cells from the urine of patients with nephropathic cystinosis. J Am Soc Nephrol 1: 1028–1033, 1991 [DOI] [PubMed] [Google Scholar]
- 4.Dörrenhaus A, Müller JI, Golka K, Jedrusik P, Schulze H, Föllmann W: Cultures of exfoliated epithelial cells from different locations of the human urinary tract and the renal tubular system. Arch Toxicol 74: 618–626, 2000 [DOI] [PubMed] [Google Scholar]
- 5.Inoue CN, Sunagawa N, Morimoto T, Ohnuma S, Katsushima F, Nishio T, et al.: Reconstruction of tubular structures in three-dimensional collagen gel culture using proximal tubular epithelial cells voided in human urine. In Vitro Cell Dev Biol Anim 39: 364–367, 2003 [DOI] [PubMed] [Google Scholar]
- 6.Al-Malki AL: Assessment of urinary osteopontin in association with podocyte for early predication of nephropathy in diabetic patients. Dis Markers 2014: 493736, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakamura T, Ushiyama C, Suzuki S, Hara M, Shimada N, Ebihara I, et al.: Urinary excretion of podocytes in patients with diabetic nephropathy. Nephrol Dial Transplant 15: 1379–1383, 2000 [DOI] [PubMed] [Google Scholar]
- 8.Detrisac CJ, Mayfield RK, Colwell JA, Garvin AJ, Sens DA: In vitro culture of cells exfoliated in the urine by patients with diabetes mellitus. J Clin Invest 71: 170–173, 1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bharadwaj S, Liu G, Shi Y, Wu R, Yang B, He T, et al.: Multipotential differentiation of human urine-derived stem cells: Potential for therapeutic applications in urology. Stem Cells 31: 1840–1856, 2013 [DOI] [PubMed] [Google Scholar]
- 10.Lang R, Liu G, Shi Y, Bharadwaj S, Leng X, Zhou X, et al.: Self-renewal and differentiation capacity of urine-derived stem cells after urine preservation for 24 hours. PLoS One 8: e53980, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Da Sacco S, Sedrakyan S, Boldrin F, Giuliani S, Parnigotto P, Habibian R, et al.: Human amniotic fluid as a potential new source of organ specific precursor cells for future regenerative medicine applications. J Urol 183: 1193–1200, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rahman MS, Wruck W, Spitzhorn L-S, Nguyen L, Bohndorf M, Martins S, et al.: The fGf, tGfβ and Wnt axis modulate self-renewal of human SIX2+ urine derived renal progenitor cells. Sci Rep 10: 1–16, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Y, McNeill E, Tian H, Soker S, Andersson K-E, Yoo JJ, et al.: Urine derived cells are a potential source for urological tissue reconstruction. J Urol 180: 2226–2233, 2008 [DOI] [PubMed] [Google Scholar]
- 14.Arazi A, Rao DA, Berthier CC, Davidson A, Liu Y, Hoover PJ, et al.; Accelerating Medicines Partnership in SLE network: The immune cell landscape in kidneys of patients with lupus nephritis [published correction appears in Nat Immunol 20: 1404, 2019 10.1038/s41590-019-0473-3]. Nat Immunol 20: 902–914, 2019. 31209404 [Google Scholar]
- 15.Wang YJ, Kaestner KH: Single-cell RNA-seq of the pancreatic islets––a promise not yet fulfilled? Cell Metab 29: 539–544, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park J, Liu CL, Kim J, Susztak K: Understanding the kidney one cell at a time. Kidney Int 96: 862–870, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park J, Shrestha R, Qiu C, Kondo A, Huang S, Werth M, et al.: Single-cell transcriptomics of the mouse kidney reveals potential cellular targets of kidney disease. Science 360: 758–763, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Menon R, Otto EA, Sealfon R, Nair V, Wong AK, Theesfeld CL, et al.: SARS-CoV-2 receptor networks in diabetic and COVID-19-associated kidney disease. Kidney Int 98: 1502–1518, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Townsend RR, Guarnieri P, Argyropoulos C, Blady S, Boustany-Kari CM, Devalaraja-Narashimha K, et al.; TRIDENT Study Investigators: Rationale and design of the transformative research in diabetic nephropathy (TRIDENT) study [published correction appears in Kidney Int 97: 809, 2020 10.1016/j.kint.2020.02.005]. Kidney Int 97: 10–13, 2020. 31901339 [Google Scholar]
- 20.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R: Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36: 411–420, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Korsunsky I, Millard N, Fan J, Slowikowski K, Zhang F, Wei K, et al.: Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Methods 16: 1289–1296, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Young MD, Behjati S: SoupX removes ambient RNA contamination from droplet based single cell RNA sequencing data. BioRxiv 303727, 2020. 10.1101/303727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McGinnis CS, Murrow LM, Gartner ZJ: DoubletFinder: Doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Systems 8: 329–337.e4, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilson PC, Wu H, Kirita Y, Uchimura K, Ledru N, Rennke HG, et al.: The single-cell transcriptomic landscape of early human diabetic nephropathy. Proc Natl Acad Sci U S A 116: 19619–19625, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu Z, Liao J, Chen Y, Zou C, Zhang H, Cheng J, et al.: Single-cell transcriptomic map of the human and mouse bladders. J Am Soc Nephrol 30: 2159–2176, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, et al.: Comprehensive integration of single-cell data. Cell 177: 1888–1902.e21, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu H, Uchimura K, Donnelly EL, Kirita Y, Morris SA, Humphreys BD: Comparative analysis and refinement of human PSC-derived kidney organoid differentiation with single-cell transcriptomics. Cell Stem Cell 23: 869–881.e8, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nacken W, Roth J, Sorg C, Kerkhoff C: S100A9/S100A8: Myeloid representatives of the S100 protein family as prominent players in innate immunity. Microsc Res Tech 60: 569–580, 2003 [DOI] [PubMed] [Google Scholar]
- 29.Menon R, Otto EA, Hoover P, Eddy S, Mariani L, Godfrey B, et al.; Nephrotic Syndrome Study Network (NEPTUNE): Single cell transcriptomics identifies focal segmental glomerulosclerosis remission endothelial biomarker. JCI Insight 5: e133267, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dumas SJ, Meta E, Borri M, Goveia J, Rohlenova K, Conchinha NV, et al.: Single-cell RNA sequencing reveals renal endothelium heterogeneity and metabolic adaptation to water deprivation. J Am Soc Nephrol 31: 118–138, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pattaro C, Teumer A, Gorski M, Chu AY, Li M, Mijatovic V, et al.; ICBP Consortium; AGEN Consortium; CARDIOGRAM; CHARGe-Heart Failure Group; ECHOGen Consortium: Genetic associations at 53 loci highlight cell types and biological pathways relevant for kidney function. Nat Commun 7: 10023, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qiu C, Huang S, Park J, Park Y, Ko Y-A, Seasock MJ, et al.: Renal compartment–specific genetic variation analyses identify new pathways in chronic kidney disease. Nature Medicine 24: 1721–1731, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu H, Malone AF, Donnelly EL, Kirita Y, Uchimura K, Ramakrishnan SM, et al.: Single-cell transcriptomics of a human kidney allograft biopsy specimen defines a diverse inflammatory response. J Am Soc Nephrol 29: 2069–2080, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Edeling M, Ragi S, Huang S, Pavenstädt H, Susztak K: Developmental signalling pathways in renal fibrosis: the roles of Notch, Wnt and Hedgehog. Nature Reviews Nephrology 12: 426, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.