Abstract
Chronic inflammatory diseases such as rheumatoid arthritis (RA), Juvenile Idiopathic Arthritis (JIA), psoriasis, and inflammatory bowel disease (IBD) are characterized by systemic as well as local tissue inflammation, often with a relapsing-remitting course. Tissue–resident memory T cells (TRM) enter non-lymphoid tissue (NLT) as part of the anamnestic immune response, especially in barrier tissues, and have been proposed to fuel chronic inflammation. TRM display a distinct gene expression profile, including upregulation of CD69 and downregulation of CD62L, CCR7, and S1PR1. However, not all TRM are consistent with this profile, and it is now more evident that the TRM compartment comprises a heterogeneous population, with differences in their function and activation state. Interestingly, the paradigm of TRM remaining resident in NLT has also been challenged. T cells with TRM characteristics were identified in both lymph and circulation in murine and human studies, displaying similarities with circulating memory T cells. This suggests that re-activated TRM are capable of retrograde migration from NLT via differential gene expression, mediating tissue egress and circulation. Circulating ‘ex-TRM’ retain a propensity for return to NLT, especially to their tissue of origin. Additionally, memory T cells with TRM characteristics have been identified in blood from patients with chronic inflammatory disease, leading to the hypothesis that TRM egress from inflamed tissue as well. The presence of TRM in both tissue and circulation has important implications for the development of novel therapies targeting chronic inflammation, and circulating ‘ex-TRM’ may provide a vital diagnostic tool in the form of biomarkers. This review elaborates on the recent developments in the field of TRM in the context of chronic inflammatory diseases.
Keywords: tissue–resident memory T cells, re(circulation), chronic inflammation
1. Introduction
1.1. TRM: Drivers of Chronic Inflammation
Characteristic of chronic inflammatory diseases is their self-perpetuating nature. Chronic inflammation can affect virtually any organ or tissue in the body including skin, gut, joint, muscles, and the central nervous system. Tissue–resident memory T cells (TRM) are known to be canonically present in tissues, playing a vital role in immune responses, but are also suspected of contributing adversely to chronic inflammatory conditions [1]. Specifically, TRM have been implicated in the hyper-response in inflamed environments due to their potential capability of cross-talk with other immune cells. Murine studies have demonstrated the ‘immune sentinel’ function of TRM by examining the functional response of these cells in mouse models of vitiligo and colitis, respectively [1,2]. The production of immune cell recruiting cytokines and expression of genes involved in the recruitment of innate immune cells supports the hypotheses that TRM have a crucial role in chronic inflammation by adding to the immune-inflammatory state [1,2]. This review aims to summarize the recent developments in the field of TRM with a focus on their role in chronic inflammation. Additionally, the concept of TRM recirculation and the impact this may have on exacerbation of chronic inflammatory diseases will be discussed.
1.2. TRM Phenotype
TRM are typically present in every tissue compartment and can be found residing in even higher numbers in barrier tissues such as the skin and gut [3]. The key markers of TRM are considered to be the C-type lectin CD69, and the αE integrin CD103 [1]. However not all TRM populations express CD103, and it is mainly expressed by CD8+ TRM and to a lesser extent CD4+ TRM. Possibly, CD103 expression is enriched in certain non-lymphoid tissue (NLT) compartments, such as the epithelium [1,4]. CD69 antagonizes S1PR1, which is required for migration of T cells out of tissues, hence S1PR1 inhibition facilitates tissue-residency [4]. In murine tissues, TRM populations that do not express CD69 have also been identified [4]. The variation of CD69 and CD103 expression on TRM between tissues indicates that TRM might comprise of subpopulations that differ between tissues.
Although transcriptional and functional TRM core signatures have been identified [3,5] there is considerable heterogeneity of TRM populations reflected in surface markers, cytokine/chemokine, and transcription factor expression. Therefore, it is important to use multiple markers to identify TRM and to include their functional profile. Furthermore, heterogeneity has not only been observed between TRM populations from different tissues, but also within TRM population in one tissue compartment. For example, a comparative study found that the majority of TRM in human skin were CD4+CD69+CD103– cells (dermis), with a mixed epidermal population (CD4+CD103+ and CD8+CD103+). Skin from naïve mice showed CD103+/−CD4+ TRM to be most prevalent, with a large proportion of dermal CD4+ memory T cells retaining their ability to circulate [5].
Studies in secondary lymphoid tissues (SLTs) of both humans and mice demonstrate that CD4+ and CD8+ TRM are present in lymph nodes (LNs) and the spleen, with up to 30–50% of these cells expressing phenotypes and transcriptional profiles compatible with TRM cells [3,6,7]. In the human LN, CD8+ TRM display phenotypic, functional, and epigenetic signatures associated with tissue residency, while exhibiting a higher proliferative capacity as well. However, TRM also exhibit an organ-specific signature compared with other sites, with increased expression of TCF-1, LEF-1, CXCR5, and CXCR4, and reduced expression of effector molecules [8]. This suggests that the LN niche is a site for extended TRM maintenance and quiescence.
These studies identify distinct ‘subsets’ of TRM, some displaying more effector capacity and others displaying more proliferative capacity Taken together, TRM are a highly heterogeneous pool of cells and presumably capable of plasticity as well as homing, allowing them to exist in a range of phenotypes. Other surface markers and transcription factors that are part of the characteristic TRM expression profile are summarized in Table 1.
Table 1.
Summary of the characteristic transcriptional profile of tissue–resident memory T cells (TRM). In this table, a selection of transcripts that are part of the key transcriptional profile displayed by TRM is shown. It must be noted that the markers displayed in this table do not occur on every population of TRM.
| Transcript | Gene | Up- (↑) or Down (↓) Regulated | Expression-Human vs. Mouse | References |
|---|---|---|---|---|
| Surface markers | ||||
| CD69 | Cd69 | ↑ | Both | [3,9,10] |
| CD103 | Itgae | ↑ | Both | [3,9,10] |
| CD49a | Itga1 | ↑ | Both | [3,9,10] |
| CD101 | Cd101 | ↑ | Both | [3] |
| CD62L | Sell | ↓ | Human | [3,9,10] |
| CXCR6 | Cxcr6 | ↑ | Both | [3,10] |
| CX3CR1 | Cx3cr1 | ↑ | Human | [3] |
| CCR7 | Ccr7 | ↓ | Human | [3,9] |
| PD-1 | Pdcd1 | ↑ | Human | [3,10] |
| S1PR1 | S1pr1 | ↓ | Both | [3,9,10] |
| Intracellular proteins | ||||
| DUSP6 | Dusp6 | ↑ | Both | [3,10] |
| KLF2 | Klf2 | ↓ | Both | [3,10] |
| KLF3 | Kl3 | ↓ | Human | [3,10] |
| Eomes | Eomes | ↓ | Human | [9,11] |
| T-bet | Tbx21 | ↓ * | Human | [11] |
| Blimp1 | Prdm1 | ↑ | Mouse | [12] |
| Hobit | Zfp683 | ↑ | Mouse | [12] |
| Runx3 | Runx3 | ↑ | Mouse & Human TIL ** | [13] |
| Id3 | Id3 | ↑ | Mouse | [14] |
| Nr4a1 | Nr4a1 | ↑ | Human | [9] |
* T-bet is downregulated, but some expression of T-bet is required for TRM survival [9]. ** TIL-tumor infiltrating lymphocytes.
1.3. TRM Development
A study from 2016 showed that in early life, memory T cells (mostly TEM) were found in higher frequencies in mucosal barrier tissues than in secondary lymphoid organs (SLOs) and circulation, compared to young adults where TEM cells were predominant in all mucosal sites (>90%). The majority of TEM cells in all pediatric tissue are CD69+, while circulating TEM were CD69−. A lower frequency of pediatric mucosal CD8+ TEM co-express CD69 and CD103 (compared to >90% in adult mucosa), implying that early life memory T cells in mucosa are not fully differentiated TRM cells. As such, the sequestering of early life memory T cells to mucosal barrier tissue and not to draining LN is suggestive of ‘local in-situ priming’ to inhaled and ingested antigens [15]. The inkling is that inhalation and ingestion of food and the microbiome itself may results in antigen triggers that prime memory T cells in early life and lead to TRM seeding of tissue compartments. There is also a consideration of tissue repletion. Once tissue compartments have been filled (early life seeding of TRM), a competitive dynamic is established, and strong triggers (infections) are required to direct further seeding. Memory precursor T cells and/or TEM cells both may play a role in the seeding dynamics. Thus, inflammation may not be the only driving force behind seeding of tissue compartments especially in early life, though it is one of the established mechanisms, especially in terms of TEM cells.
Memory T cell subsets consist of circulating and non-circulating subsets, the former of which comprises TCM and TEM, recirculating through lymphoid and non-lymphoid tissues respectively [16]. In the event of pathogen challenge, the non-circulating TRM in peripheral tissues are poised for an immune response [17]. Recently, TRM have been shown to potentially repopulate the lymphoid TCM and TEM compartments, alluding to the possibility that the development of these cells may be underscored by an epigenetic program, allowing developmental plasticity and an imprinted memory of prior localization [18,19].
TRM development is presumed to be driven by antigen exposure, as well as cytokines and chemokines in the local tissue environment. TGFβ has been shown to drive TRM generation (by downregulation of transcription factors Eomes and T-bet), as well as induce CD103 expression [20]. Recent studies suggest that TRM can sustain themselves in the tissue. For example, it was shown that TRM proliferated in situ upon re-stimulation with antigen in the female reproductive tract as well as the skin of mice, respectively [21,22]. IL-15 is thought to be essential for long-term survival of TRM [11]. This retention and survival of TRM dependent on the local availability of cytokines and factors (creating organ-specific dependencies) may influence the ability of egress of these cells during an immune challenge [11].
T-bet dependent IL-15 signaling has been shown to induce expression of the transcription factor Hobit in mice [12]. The dependence on these cytokines varies between tissues, as reviewed by an extensive comparative study [23]. The transcription factors Hobit, Blimp1, and Runx3 were shown to regulate differentiation of CD8+ TRM in mice, by stimulating expression of genes required for tissue residency and suppressing genes that mediate tissue egress [12,13]. A comparison between the gene signature of mouse TRM with human TRM showed that most genes followed a similar expression pattern [3]. However, there are also clear discrepancies between TRM in mice and humans. Hobit, for instance, is a key transcription factor that drives TRM function in mice [12,24] but expression was observed to be variable in human TRM, and may not be differentially expressed in TRM compared with other memory T cells [3,25,26]. Vieira Braga et al. observed increased Hobit expression in human effector-like human CD8+ T cells, but not in naive or memory CD8+ T cells [27]. This raises the possibility that TRM with an effector phenotype express increased Hobit, but other TRM subsets do not.
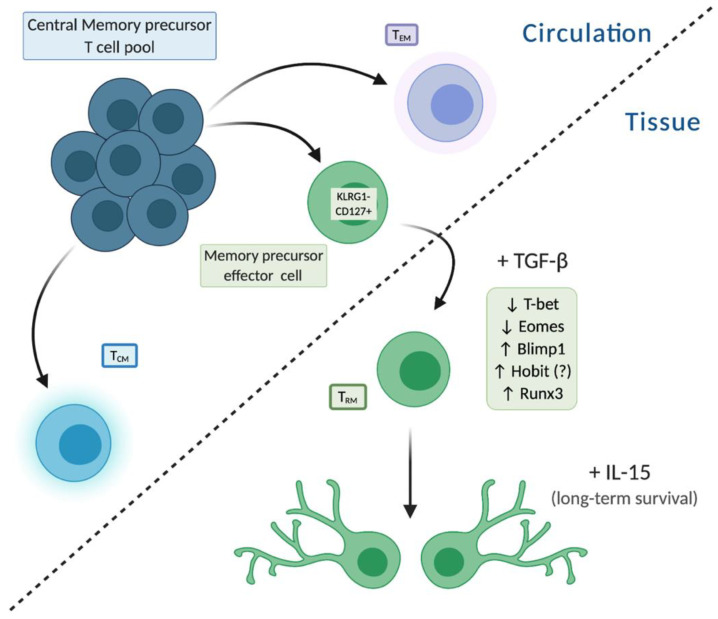
To truly differentiate between activated (CD69+CD62L−) effector T cells and CD69+ TRM cells, it may be crucial to perform full transcriptome analysis at a single cell level, as well as T cell receptor sequencing and analysis of other phenotypic markers. In inflamed tissues an expression gradient of CD69 (CD69hi vs. CD69lo) may discern TRM from TEM, alongside other markers specific to the two different pools of cells, but this has to be proven. The key cytokines and transcription factors that drive TRM differentiation are summarized in Figure 1. More recent studies have identified roles for other transcription factors and epigenetic changes in the differentiation of CD8+ TRM, which are reviewed by Chen et al. [28].
Figure 1.
Key cytokines and transcription factors for the development of tissue–resident memory T cells (TRM). TRM are derived from memory precursors effector cells that express low KLRG1 and high CD127. Analysis of development has shown that TRM displayed plasticity intermediate between tissue central-memory (TCM) and tissue effector-memory (TEM), and are not as terminally differentiated as previously believed. The central cytokine that induces TRM differentiation is transforming growth factor β (TGFβ). TGFβ induces downregulation of T-bet and Eomes, which is essential for TRM formation. Blimp1, Hobit, and Runx3 play a role in TRM differentiation as well. Upregulation of Hobit is observed in mice TRM, but only in a subset of human TRM.
2. TRM Are Enriched at Sites of Chronic Inflammation at Barrier and Non-Barrier Tissues
In humans, various CD4+ and CD8+ TRM (sub)populations were found to be enriched in inflammatory diseases that manifest at barrier tissues, such as inflammatory bowel disease (IBD) in the gut [24] and psoriasis in the skin [29], but also in disease manifested in non-barrier tissues, including diabetes type I [30], Sjogren’s syndrome [31], lupus nephritis [32], multiple sclerosis (MS) [33,34] and several arthritic diseases [35,36,37,38,39] such as Juvenile Idiopathic Arthritis (JIA) [35,36], psoriatic arthritis [37], ankylosing spondylitis [38], and rheumatoid arthritis (RA) [39]. The recent findings on TRM in inflammatory diseases in various tissues is summarized in Table 2 below.
Table 2.
Populations of tissue–resident memory T cells (TRM) that are enriched in barrier and non-barrier autoimmune diseases. This table provides an overview of the populations of TRM that were observed to be significantly enriched in several inflammatory diseases. The fourth column states the markers that were used to distinguish the enriched TRM population in that particular study.
| Chronic Inflammatory Diseases. | Tissue Enrichment | Specific Markers/Factors/Genes Associated | Characteristics of Enriched TRM Population | Ref |
|---|---|---|---|---|
| Psoriasis | Skin | IL-17 | CD49a+ CD103+ CD8+ | [40] |
| CD103+ CD8+ | [41] | |||
| Vitiligo | Skin | IFN-γ, CXCR3 | CCR7− CD69+ CD103+ CD8+ | [42] |
| CD69+ CD103+ CD8+ | [43] | |||
| Atopic dermatitis | Skin | CCL1, IL13, IL26, ANXA1, ANXA2 | CD69+ CD103+ CD8+ | [44] |
| Alopecia areata | Skin | ITGAE | CD103+ CD8+ | [45] |
| CD69+ or CD103+ (CD4 or CD8 not specified) | [46] | |||
| Inflammatory bowel disease | Gut | CD161, β7 | CCR7− CD69+ CD4+ | [47] |
| CXCR6, CD101, KLF2lo | CD69+ CD103+ (CD4 or CD8 not specified) | [24] | ||
| Type 1 diabetes | Pancreas | IFN-γ, IL-18, IL-22 | CD69+ CD103+ CD8+ | [30] |
| Sjögren’s syndrome | Nerve/Connective tissue | IFN-γ | CD69+ CD103+ CD8+ | [31] |
| Lupus nephritis | Connective tissue | JAK/STAT, TNF-α, IFN-γ | CD103+ CD8+ | [32] |
| Multiple sclerosis | Central nervous system | CD69− | CD103+ CD8+ | [34] |
| CXCR6, Ki67 | PD-1+ CD44+ CD49a+ CD69+ CD103+/– CD8+ | [33] | ||
| Juvenile idiopathic arthritis | Joint | ITGAE, ITGA, CXCR6 | PD-1+ CD69+ CD8+ | [35] |
| DUSP6, ITGAE, CXCR6, PD-1 | PD-1+ CD8+, PD-1+ CD39+/CD161+ CD4+ and PD-1+ CD39−/CD161− CD4+ |
[36] | ||
| Psoriatic arthritis | Joint/Skin | PD-1, ITGAE, ZNF683, CXCR6, β7, CLA, CD49a | IL-17A+ CD69+ and/or CD103+ CD8+ | [37] |
| Ankylosing spondylitis | Connective tissue/Joint | β7, CD29, IL-10, CXCR6 | CD49a+ CD103+ CD8+ T cells | [38] |
| Rheumatoid arthritis | Connective tissue/Joint | PD-1, Blimp, 1, CD44 | PD-1+ CXCR5− CD69+ CD4+ | [48] |
| CXCR6, CD49a, CD101, PD-1, Ki-67 | CD69+ CD103+/− CD8+ | [39] |
2.1. Skin
For chronic inflammatory diseases of the skin, such as vitiligo and psoriasis, a role for TRM in re-occurring lesions and disease flares has long been suspected [29,49]. The markers that are most widely used to identify TRM in the skin are CD69, CD103, and CD49a. A study looking into lesional and non-lesional skin samples from psoriasis patients found that the epidermis with active psoriasis was massively infiltrated by CD8+ TRM, both compared to non-lesional skin and healthy skin, with a 100-fold increase of TRM in active psoriatic lesions [40]. Moreover, significant enrichment of CD103+CD8+ TRM in lesional psoriatic skin compared to healthy controls was also observed [41]. Similarly, identification of TRM populations in vitiligo patients portrayed that CD8+ TRM were present in both dermis and epidermis of vitiligo skin and CD69+CD103+CD8+ TRM were observed to be significantly enriched in vitiligo patients (independent from disease status) compared to psoriatic skin and healthy controls [42]. Furthermore, a significant increase of melanocyte-reactive TRM in the lesional skin of vitiligo patients was noted [43].
Recent studies have also investigated TRM populations in other autoimmune skin disorders. A novel study set up to identify TRM in the skin of atopic dermatitis (AD) patients used single cell RNA sequencing to analyse both lesional and non-lesional skin, as well as skin from healthy controls. CD69+CD103+CD8+ T cells, but not CD69+CD103+CD4+ T cells, were significantly expanded in lesional skin from AD patients in comparison with non-lesional AD skin and healthy controls [44]. Additionally, in alopecia areata (AA), a disease caused by attack of the bulb region of the hair follicle by autoreactive CD8+ T cells, higher numbers of peribulbar CD103+ CD8+ T cells and increased CD69+ and CD103+ T cell numbers were found to be present in lesional skin from AA patients compared to non-lesional skin and healthy controls [45,46,50].
Consistent with the hypothesis of a role for TRM in the mediation of chronic inflammation, increased production of pro-inflammatory cytokines was found in TRM populations at lesional sites. A study observed that CD103+CD8+ T cells in psoriatic skin expressed IL-17 and IL-22 mRNA [40], noting that particularly CD49a−CD103+CD8+ TRM in the skin predominantly produced IL-17 upon stimulation, and that this subset was enriched in psoriasis [51]. Another study had similar findings, with T cells in explants of lesional psoriatic skin that were activated with pan-T cell activating antibody OKT-3 poised for IL-17A production [52]. It was also demonstrated that CD49a+CD103+CD8+ TRM in the skin of vitiligo patients had increased capability of IFN-γ production. Moreover, these cells recognized melanocyte-derived antigens. Increased expression of granzyme B and perforin was also found. Ex vivo, more co-expression of granzyme B and perforin was observed in CD49a+CD103+CD8+ TRM compared to CD49a− TRM, suggesting higher cytotoxic capacity in CD49a+ TRM [51]. An examination of TEM from perilesional skin of vitiligo patients found that these TEM included TRM populations. The TRM from skin of vitiligo patients with active disease displayed increased IFN-γ and TNF-α production compared to stable disease and healthy controls, suggesting that TRM in vitiligo are poised for production of these cytokines [42].
Taking the example of psoriasis, skin with clinically resolved psoriasis was demonstrated to contain increased IL-17 mRNA expressing CD103+CD8+ TRM, and an enrichment of IL-23 responsive CD103+CCR6+IL23R+CD8+ T cells [40]. In addition, pathogenic IL17A-producing T cell clones were found to be present in both active psoriatic plaques as well as in resolved skin lesions, but not in healthy control skin [53]. Moreover, in disease-naïve non-lesional skin of psoriatic patients, an enrichment of IL-17A and IFN-γ producing CCR6+ T cells [54], and IL-17A producing CD103+CD8+ T cells [41], compared to healthy controls was observed. Together these studies demonstrate that IL-17 and IFN-γ producing resident T cells are present in non-affected skin as well as resolved psoriatic lesions, and that these TRM cells are poised for re-initiation of inflammation. Therefore, TRM may play a role in the initiation of psoriasis pathogenesis as well as disease flares [41,54].
2.2. Gut
CD69+CD103+ TRM are also numerous in the intestine, with mostly CD8+ TRM in the intestinal epithelium and both CD4+ TRM and CD8+ TRM in the lamina propria [4]. Inflammatory bowel disease (IBD) is a chronic inflammatory disease of the intestine, and includes Crohn’s Disease (CD) and ulcerative colitis (UC). IBD is thought to be caused by an overactive immune response against the gut microbiome, and is characterized by relapsing flares, which suggests that TRM may be involved in the pathogenesis [24].
Two studies extensively investigated the presence of TRM in IBD. Bishu et al. investigated colon tissue (both epithelium and lamina propria) from CD patients, discerning CD4+ TRM using CD69+ and CCR7− expression. CD4+ TRM constituted the majority of mucosal memory CD4+ T cells and the CD4+ TRM compartment was expanded, with absolute numbers increased in CD patients compared to controls. CD8+ TRM cells were not found to be enriched in CD patients compared to controls [53]. Colon samples from patients with active CD showed an increase in IL-17A producing CD4+ TRM, portraying a TH17 signature. CD4+ TRM were also found to be the major source of TNF-α production in CD patients ex vivo [47]. Zundler et al. investigated the presence of TRM in IBD patients using core characteristics of TRM (as summarized in Table 1) and observed enrichment of CD69+CD4+ T cells in the lamina propria of IBD patients with increased expression of CXCR6, CD103 and CD101, and decreased expression of KLF2 [24]. They also demonstrated that gut CD69+ T cells (from IBD patients) produce elevated amounts of the pro-inflammatory cytokines IFN-γ, IL-13, IL-17A, and TNF-α compared to CD69− T cells. In addition, a high number of CD4+ intestinal TRM was associated with shorter flare-free survival in these patients [24]. In line with this it was noted that gut-resident CD4+ T cells produced increased IL-17 compared to circulating CD4+ T cells, which was even more evident in patients with IBD [55]. CD161+CD4+ T cells in CD patients were found to display elevated production of IL-17 and IL-22, and higher expression of IL-23R, as is characteristic of a TH17 phenotype (stimulated by IL-1b and IL-23) [56]. Taken together, these studies show that CD4+ TRM in IBD patients have increased capabilities for the production of pro-inflammatory cytokines, including IL-17A and TNF-α, and may drive disease flares of IBD.
The role of CD8+ TRM in IBD has been studied less extensively. A study on CD8+ TRM in healthy gut samples demonstrated that the CD103+CD8+ TRM in the lamina propria expressed granzyme B and perforin and were potent producers of IFN-γ, IL-2, and TNF-α upon activation. This suggests that CD8+ TRM participate in the protective immune response of the gut [57]. CD8+ TRM numbers were also found to be significantly reduced in the intestinal epithelium of IBD patients with quiescent disease compared to healthy controls, postulating that the decreased barrier immunity in IBD pathogenesis may be due to TRM deficiency [58]. This touches upon the dual role TRM can have in inflammatory disease: on one hand they play a role in tissue homeostasis and integrity, protecting the host from overzealous inflammation, on the other hand they may sustain inflammation once a perpetual loop of inflammation has been instigated.
2.3. Joints
Arthritic inflammatory diseases are characterized by chronic, recurring inflammation in the joints. In JIA, a form of chronic arthritis that begins in children before the age of sixteen, there is accumulation of both CD4+ and CD8+ T cells in the synovium of inflamed joints. Petrelli et al. examined the CD8+ T cell population in the synovial fluid (SF) of JIA patients, and observed that a PD-1 expressing subset of CD8+ T cells was highly enriched in comparison with peripheral blood (PB) of JIA patients and healthy controls [35]. The PD-1+CD8+ T cell subset displayed a hallmark TRM transcriptional profile with upregulation of ITGAE, ITGA1, and CXCR6, and downregulation of S1PR1, KLF3, and SELL, compared to SF PD-1−CD8+ T cells. Gene set enrichment analysis (GSEA) showed that the PD-1+CD8+ T cells from JIA SF do not display an exhausted gene signature but instead are enriched for signature effector genes compared to their PD-1− counterparts. PD-1+CD8+ T cells showed increased expression of Ki-67, and increased expression of pro-inflammatory cytokines IFN-γ, TNF-α, and granzyme B. In addition, PD-1+CD8+ T cells were clonally expanded, and their TCRs barely overlapped with PD-1−CD8+ T cells, suggesting that these are local antigen-driven pathogenic cells [35].
Recently, subsets of CD4+ T cells in the SF of JIA patients were also identified displaying a TRM phenotype. Characteristic TRM gene patterns included low CD62L and CCR7 expression and increased DUSP6, ITGAE (encoding CD103), CXCR6, and PD-1 expression [36]. The PD-1+CD4+ SF TRM were found to secrete GM-CSF and IL-21, and these cells were shown to be enriched not only in the SF of JIA, but in the inflamed intestine of IBD patients as well. This suggests that PD-1+CD4+ TRM participate in chronic inflammation in several diseases. In line with the data of Petrelli et al., PD-1+CD8+ T cells present in the same SF samples were also demonstrated to produce GM-CSF and IL-21. Therefore, both PD-1+CD8+ and PD1+CD4+ TRM populations seem to have the potential to drive chronic inflammation in JIA [36], and joint or tissue inflammation in general. Steel et al. investigated the phenotype of IL-17A expressing CD8+ T cells, Tc17 cells, which were enriched in the SF of actively inflamed joints from patients with psoriatic arthritis compared to PB. Virtually all of the IL-17A+CD8+ T cells in the SF displayed a memory phenotype, and 65% expressed PD-1. Gene profiling showed that IL-17A+CD8+ T cells expressed genes characteristic of the TRM phenotype compared to IL-17A+CD4+ T cells. These hallmarks include increased expression of ITGAE (encoding CD103) and ZNF683 (encoding the transcription factor Hobit), CXCR6 and decreased expression of S1PR1 [37]. Using flow cytometry, they further confirmed high CD103 expression on IL-17A+CD8+ T cells and these cells often co-expressed β7 integrin (a gut homing marker), cutaneous lymphocyte antigen (CLA) and CD49a (skin homing markers). Finally, examination of the relationship between CD69 and CD103 expression and IL-17A production demonstrated that CD69−CD103−CD8+ T cells contained minimal IL-17A producing T cells, whereas CD69 and/or CD103 expressing cells contained higher fractions of IL-17A producing T cells [37]. A large fraction of the IL-17A-producing cells also expressed pro-inflammatory cytokines IFN-γ and/or TNF-α and high levels of granzyme B.
Analysis of the SF of patients with ankylosing spondylitis identified a distinct population of memory CD103+β7+CD49a+CD29+ CD8+ T cells that was enriched in the SF of ankylosing spondylitis patients compared to PB. The β7 integrin was often co-expressed with CD103, and CD29 was often co-expressed with CD49a, thus allowing for identification of these cells using only CD103 and CD49a expression. Enrichment of CD49a+CD103+CD8+ T cells was not observed in the SF compared to PB in RA patients. CD49a+CD103+CD8+ T cells displayed a TRM-like transcriptional profile, with elevated expression of IL-10 and CXCR6, and a lack of CD62L and S1PR1 [38]. It was demonstrated that the TRM produced IL-17A and granzyme B in both resting and stimulated conditions, and IL-10 and TNF-α when stimulated.
Very recently, a study identified a population of T cells that expressed TRM characteristics using flow cytometry, enriched in the SF of adult RA patients. The cells in this population were memory CD69+CD103+ or CD103−CD8+ T cells. Protein expression of these cells was consistent with the characteristic TRM phenotype, including increased CXCR6, CD49a, and CD101 expression, and decreased S1PR1 and KLF2 expression compared to CD69−CD8+ T cells. PD-1 and Ki-67 expression were also significantly elevated in these CD69+CD8+ T cells [39]. A significant increase of IFN-γ and TNF-α expression in TRM upon stimulation with anti-CD3 antibody was observed, although this increase was seen in CD69−CD8+ T cells as well. However, TRM displayed higher levels of granzyme B and perforin expression than CD69−CD8+ T cells, which further increased when cells were stimulated with IL-15, but not when stimulated with anti-CD3 antibody [39]. In summary, it has been demonstrated that SF from inflamed joints in arthritic inflammatory diseases contains highly activated CD4+ and CD8+ TRM with pro-inflammatory properties. It should be noted that the SF is an exudate of the inflamed synovium, not truly a tissue. Identifying TRM in SF is therefore not an entirely direct proof of the involvement of TRM in the tissue inflammation. However, SF can be accessed much less invasively than synovial tissue [35,36,37,38,48,59] and SF T cells have been shown to display overlapping phenotypic and functional profiles with synovial tissue derived TRM [35,36,37,60,61].
3. Recirculation of Tissue–Resident Memory T Cells
3.1. TRM Migrate Out of Non-Lymphoid Tissue into Circulation
In the context of chronic inflammation, the occurrence of disease flares or relapses is evident in multiple diseases such as JIA (multiple joints affected in the same patient) and CD (multiple inflammatory patches). The tendency of recurrent inflammatory flares and expansion of the disease to, sometimes anatomically separated, locations, has prompted the hypothesis of potential TRM recirculation. Are recirculating TRM cells indeed capable of triggering recurrent inflammatory bursts as well as potentially spreading disease to ‘non-affected’ areas of the body? Most of our knowledge about TRM recirculation comes from animal studies. Early indications for the ability of TRM to egress from non-lymphoid tissue (NLT) came from the study of Beura et al., in which they investigated the origin and transcriptional profile of secondary lymphoid organ (SLO) TRM [62]. They used a C57BL/6J mouse model, into which naive lymphocytic choriomeningitis virus (LCMV)-specific transgenic CD8+ T cells were transferred. Subsequently, these mice were infected with lymphocytic choriomeningitis virus (LCMV) of the Armstrong strain, inducing an acute infection. After clearance of the LCMV infection, the LCMV-specific P14 CD8+ memory T cells were examined. A CD69+CD62Llo tissue resident population of P14 CD8+ memory T cells was observed in all examined SLOs. Tissue residency of this CD8+ T cell population was confirmed with a parabiosis experiment.
Further observation of these TRM in lymph nodes (LNs) showed that local re-stimulation of NLT resulted in accumulation of these cells in draining LN, but not in distant LN. These data indicate that NLT CD8+ TRM migrate from NLT into LN upon reactivation, and are a source of SLO TRM. Administration of a molecule that inhibits S1PR1 resulted in a reduction of TRM in draining LN after reactivation, suggesting a role for S1PR1 in tissue egress by TRM [18]. Finally, it was demonstrated that TRM could differentiate into TCM, by upregulation of CD62L. Downregulation of CD69 and CD103 was observed on these ‘ex-TRM’ as well. Of note, traces of the TRM phenotype did remain. For instance, CCR9 expression was only slowly downregulated and still higher on ex-TRM than in other circulating cells 100 days after infection [18]. Finally, in another study the same group also identified a population of CD4+ TRM in SLO of Armstrong LMCV infected mice, which shared transcriptional characteristics with CD4+ TRM from NLT [63].
Fonseca et al. further examined the egress of CD8+ TRM from NLT in mice. They generated C57BL/6J mice with OT-I TRM in their skin, i.e., ovalbumin specific CD8+ T cells. After reactivation of these TRM, skin-derived cells could be found in both draining and distant LN. Furthermore, circulating T cells, but not TRM, were depleted in this mouse model. Ten days after re-stimulation of TRM, OT-I T cells were observed in the blood. These cells displayed transient retention of a TRM phenotype (CD103 expression and lack of CD62L expression), thus these cells were referred to as ex-TRM. Similar results were obtained when reactivating TRM in the female reproductive tract instead of the skin [58]. Additionally, it was demonstrated that TRM intravenously transferred into recipient mice were observed in the same tissue compartment in the recipient as in the donor. This suggests that TRM have a predilection for returning to their parental tissue. Bias for return to the tissue compartment of origin was still observed in ex-TRM in which a tertiary immune response was induced, suggesting maintenance of an epigenetic TRM profile. This indicates that ex-TRM have a propensity to re-acquire a TRM phenotype [18].
Another study examined TRM in human skin, defined as CD69+CD103+CLA+ T cells. Tissue explant cultures with human skin were performed, and a CD69- population of T cells were detected. Tissue exit was associated with downregulation of CD69 though less pronounced than in the CD8+ T cell compartment. Egress of CD103+ CLA+CD4+ T cells was also studied in vivo using human skin transplants on immune-deficient mice. After 50 days, the T cell population of the spleen was analyzed and the same population was observed [64]. This suggest that skin TRM had egressed from the human skin explant in this mouse model as well. CD103+CLA+CD4+ T cells were abundant in the skin in both epidermis and dermis, but constituted less than 2% of CLA+CD4+ memory T cells in circulation. The very same population was also observed in lymph of the human thoracic duct, indicating a CD4+ T cell population in the circulation of humans that mirrors a population of skin TRM [64]. Based on these data, it can be postulated that CD4+ TRM egress from NLT via LN into circulation, similar to CD8+ TRM. Together, these studies indicate that tissue egress by TRM occurs in both mice and humans, in the CD8+ as well as CD4+ TRM compartment.
3.2. Ex-TRM, Present in the Circulation, Share Characteristics with TRM in Tissue, But Also Display Molecules Characteristic of Circulating T Cells
After identification of TRM-like cells in SLOs and circulation, one study used mass cytometry to analyze the phenotype of blood CLA+ ex-TRM. A specific cluster, comprised of CD103+CLA+CD4+ T cells, was observed within the CLA+CD4+ T cell population. Cells of this cluster expressed CCR4, CCR6 and CCR10, which are indicative for skin tropism, and CD101 and β7 integrin, while lacking CD27, CCR7 and CXCR3 expression [64]. This phenotype was shared by CD103+CLA+CD4+ T cells in the skin. Additional transcriptional profiling demonstrated that blood CD103+CLA+CD4+ T cells displayed a distinct transcriptional signature (compared to memory CD4+CLA+CD103−CCR− and CD4+CLA+CD103−CCR7+ T cells in circulation), which was also significantly enriched in skin CD103+CLA+CD4+ T cells. The shared transcriptional profile confirmed the phenotype observed with mass cytometry, and further included increased expression of ITGAE, CD101, CXCR6 and TWIST1, and decreased expression of Eomes. Moreover, TCRβ sequencing of CLA+CD4+ memory T cells showed that CD103+CLA+CD4+ T cells from skin and blood are strongly clonally related, whereas little overlap was found with other CLA+CD4+ memory T cell populations. Finally, a lack of CD69 expression was observed in CD4+ ex-TRM in the blood, while this molecule was highly expressed on CD4+ TRM in the skin. This implicates that skin TRM were able to egress from tissue by downregulating CD69 [64].
A murine study examined the phenotype of SLO CD8+ TRM using the markers CD103, CD122, Ly6C, CD27, CD62L, CXCR3, CCR9, KLRG1, CX3CR1, CD69, and CD44. Based on these markers, SLO TRM were shown to be more similar to NLT TRM (from either skin, small intestine or female reproductive tract) than to TCM and TEM, although SLO TRM did comprise a population distinct from NLT TRM. SLO CD8+ TRM shared part of the characteristic TRM gene expression profile with NLT TRM, whereas expression of other genes was more closely related to TCM. Thus, SLO CD8+ TRM were demonstrated to be a memory T cell population that is similar to but distinct from NLT TRM and shares characteristics with TCM [62].
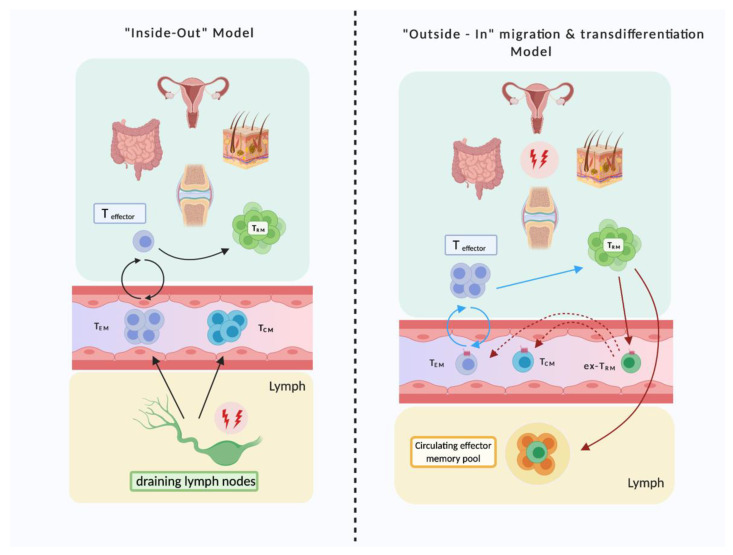
These findings further support the notion that both human and mouse TRM are capable of tissue egress and that ex-TRM have undergone changes in gene expression that mediate egress and circulation, for example downregulation of CD69 and transient upregulation of CD62L. As such, Fonseca et al. have proposed a new model for anamnestic immune responses, an ‘outside-in’ immune response [18]. Historically, the memory T cell population was divided into TCM and TEM. TCM reside predominantly in SLOs and differentiate into various T cell subtypes upon reactivation with antigen. TEM patrol in blood and TRM surveil in NLT [65]. The ‘inside-out’ model (left panel, Figure 2) describes primary immune responses. TRM were proposed to reside permanently in NLT, where they would proliferate locally upon antigen encounter and thus provide protection [21,22]. This model can be seen as ‘inside-out’, because TCM are activated in SLOs, i.e., ‘deeper’ tissues, after which TCM and TEM would shape the systemic recall immune response, proliferating and migrating towards affected tissue sites. The proposed ‘outside-in’ model (right panel, Figure 2) suggests that TRM (which are activated in barrier tissues) participate in the systemic immune response by potentially contributing to the circulating memory T cell pool, egressing and differentiating into TCM and TEM (epigenetic plasticity), and even populating other tissues [18,66]. Moreover, there has been further speculation that this systemic distribution of TRM might contribute to a broad protection against pathogens, if pathogens were to escape local defense [66]. Circulating ex-TRM populations could also serve to re-populate NLT in the event of depletion of the local TRM population [18].
Figure 2.
Models of the recall immune response by memory T cells. The left panel shows the ‘inside-out’ model, where TRM are local tissue mediators of the anamnestic immune response, proliferating inside their tissue upon reactivation. TCM and TEM constitute the systemic recall response in this model. TCM are activated in deeper tissues i.e., draining lymph nodes at site of infection, proliferating and migrating towards affected tissue sites. The right panel shows the recently proposed ‘outside-in’ model. In this mode anamnestic immune response is generated at site of infection. TRM are activated at barrier tissues and migrate inwards, to lymph and circulation, with the potential to differentiate into TCM and TEM (highlighted by the arrows in red). The blue arrows represent the T cell traffic under chronic inflammatory (permanently activated) states [18,21,22,65,66].
3.3. Indications for Recirculation of TRM in Chronic Inflammatory Disease
As previously mentioned, studies have highlighted the presence and role of TRM in several tissues permeated with chronic inflammation. The outside-in model described above, proposes that circulating ex-TRM could be contributing to the expansion of inflammatory sites in affected tissue, as has been suggested in the several disease settings such as psoriasis [41,53,54], IBD [36], and JIA [35,36]. Flow cytometry was implemented to examine the phenotype of CD4+ T cells in the synovium of patients with active JIA. These cells displayed distinct characteristics compared to T cells in circulation, including increased expression of CD69, PD1, CTLA-4, Ki-67, CCR5, and CCR6, and decreased expression of CCR7 [35,36,67]. It was hypothesized that if a small fraction of the pro-inflammatory synovial CD4+ T cells would leave the inflamed synovium, a population of blood T cells with similarities to synovial T cells should exist. Indeed, a small subset of PB T cells that displayed the same characteristics as synovial T cells was identified, albeit with lower CD69 expression. These T cells were labelled ‘circulating pathogenic lymphocytes’ (CPLs). To further investigate these CPLs, next-generation sequencing of the TCRβ chain was performed. Both synovial T cells and CPLs displayed decreased clonal diversity compared to total blood CD4+ T cells. Moreover, clustering of TCRβ sequences showed that CPLs shared more clonotypes with their synovial counterparts than other blood T cells. It was also demonstrated that CPLs had increased capacity for the production of IL-17, IFN-γ, and TNF-α compared to other non-CPL CD4+ T cells in the blood. This suggests that these cells retain their active, pro-inflammatory profile in circulation. In addition, CPLs correlated with disease activity in JIA and RA and also with resistance to therapy in JIA (Figure 3) [67].
Figure 3.
T cells resembling tissue–resident memory T cells (TRM) from various tissues have been identified. Subsets of memory T cells with characteristics of TRM were identified in the circulation of Juvenile Idiopathic Arthritis (JIA), RA, and AS (with inflammatory bowel disease (IBD)) patients. These cells displayed an activated, effector pathogenic phenotype, and were clonally related to their tissue counterparts. This suggests that these cells may be ex-TRM, having egressed from the tissue of origin. Cells resembling synovial fluid (SF) Tregs were also identified in circulation.
Tregs are a crucial part of suppressive immunity and have been shown to differ phenotypically and functionally in non-diseased and diseased states [60,68,69,70]. It has been shown that Tregs are part of the TRM compartment i.e., CD69-expressing Treg in synovial, uterus, and gut tissue [60,71]. Tregs from chronic inflammatory settings are phenotypically/functionally different from healthy tissue Treg [36,60,70,72] and it has also been postulated that Tregs can re-circulate [72]. By studying the PB of JIA patients, it was observed that a specific subset of Tregs, which was positive for HLA-DR, was significantly increased in patients who did not respond to therapy compared with patients who reached inactive disease in response to therapy. These Tregs were as labelled ‘inflammation associated Tregs’ (iaTreg). iaTregs showed increased CCR5, Ki-67, and CTLA-4, compared to other Tregs, indicating that they can home to tissue and have been recently activated. Moreover, next-generation sequencing of the TCRβ chain demonstrated that iaTregs were clonally related to synovial Tregs, more than to Tregs in the blood. Finally, expansion of iaTregs was observed in the PB of adults with active RA. Together, these data show that a specific subset of Tregs that is clonally more similar to synovial Tregs compared to other blood Treg, is expanded in the PB of both JIA and RA patients (Figure 3) [72]. When examining TRM in conjunction with Treg populations, though CD69 and CD103 are adequate to construe it, there is some selectivity within different compartments in a tissue (i.e., increased CD103 expression in the epithelial compartment compared to the lamina propria in the gut). Tissue migration capacities by upregulation of integrins is induced in the lymph nodes, and can be driven by inflammatory stimuli (such as infections) but also less harmful antigens derived from food or commensals (as seen in early life seeding of naïve peripheral tissues such as the gut). Once in the tissue, cells may differentiate/adapt through tissue-specific, antigen, and inflammatory cues both for Treg and non-Treg cells [60,69,70,71,73]. It is currently unknown how selective maintenance in different tissues is regulated.
Other T cell populations in SF and PB of JIA patients were also investigated. Several clusters of CD4+ T cells in the SF were identified, two of which expressed TRM associated genes. These TRM-like cell clusters were enriched in the SF of JIA patients compared to healthy controls. One of the two clusters did not only display gene expression associated with TRM, but also with circulating T cells. This subset expressed IL-10, Eomes, PRF1, TNFRSF9, and genes encoding granzymes, which indicate resemblance to type 1 regulatory T (Tr1)-like cells. Furthermore, analysis of the PB of JIA patients led to the identification of a small blood CD4+ T cell cluster that mirrored gene expression of the Tr1-like cell resembling cluster in the SF and was clonally related to the Tr1-like cluster [36].
Taken together, findings indicate that there is recirculation of both Treg and pathogenic CD4+ T cells between blood and SF in JIA patients. Recirculation may contribute to the evolution of the disease over time, i.e., the involvement of multiple joints over the disease course. It may even be responsible for spreading of the disease to different organs, such as the gut (or the skin). Qaiyum et al. identified a population of CD103+β7+CD49a+CD29+CD8+ T cells that was enriched in the SF of ankylosing spondylitis patients compared to PB. Because the β7 integrin is a gut homing molecule, and ankylosing spondylitis is associated with IBD, the authors speculated that the population of T cells they identified might originate from the gut [38]. This notion was further investigated by Guggino et al., where paired samples from gut, synovial tissue, SF and PB from ankylosing spondylitis patients and controls were examined. Using flow cytometry, they observed that CD8+CD69+CD103+ TRM were expanded in not only SF, but also in gut and PB of ankylosing spondylitis patients compared to healthy controls. The majority of these cells in SF and PB expressed β7 integrin. These data support the hypothesis that TRM from the gut recirculate to PB and inflamed joints in patients with ankylosing spondylitis (Figure 3) [74].
As such, disease-associated TRM entering other tissues via circulation might contribute to off-site or systemic disease-associated symptoms that often occur in autoimmune diseases. However, current data only provide indications for this, and further elucidation of these circulating pathogenic TRM populations and their association with disease symptoms is warranted.
4. Summary and Outlook
Post-pathogen exposure, TRM become established in peripheral tissue. In the case of re-exposure to antigen TRM, as is well known, mount an immune response at the site of infection, or as we suggest, in settings of chronic inflammation, are capable of not only taking on an aggressive effector function but egressing and through developmental plasticity, contributing to the effector memory and effector cell pools. In this instance, they may be comparable to TIL-like cells in cancer settings, functionally overlapping with TEM. Interestingly overlap in phenotype and transcriptome has been demonstrated for inflamed joint- and tumor-derived Treg [69]. The transient expression of CD69 upon activation may be a gradient along which we can discern TRM from TEM, though TCR sequencing and single cell transcriptomics may provide a robust additional layer of characterization. Though the exact trigger of ex-TRM in autoimmunity remains yet unknown, further examination of the ex-TRM/egressed TRM-like subsets may provide insight into the mechanism(s) by and the motive for which these cells have entered circulation.
Novel concepts of TRM recirculation and pro-inflammatory, effector-like functions of TRM may add to the paradigm of diagnosing, treating, and understanding the etiology of autoimmune diseases and chronic inflammation. TRM cells are crucial in maintaining homeostasis, providing a stringent immune front in tissues. However, their involvement in chronic inflammation and the degree of damage induced by relapsing episodes abrogate efforts made to manage diseases successfully. The heterogeneity of the TRM population alludes to the difficulty of aptly identifying and potentially therapeutically targeting these mostly tissue embedded cells. Moreover, the enrichment of TRM found in non-lesional tissue of patients suggests that TRM may contribute to predisposition for chronic inflammation, resulting in a higher likelihood of relapsing episodes that require further medical and lifestyle interventions. The burden of disease in conditions with a relapsing-remitting course is understandably difficult for patients and also challenging for clinicians to manage and/or treat. The concept of TRM recirculation further denotes the contribution of these cells in spreading of the disease and may also add to off-site or systemic disease-associated symptoms that often occur in autoimmune diseases. For instance, in the case of JIA-associated uveitis, further investigation into the critical role of immune cells and response in this setting would vastly benefit patients from debilitating long-term ocular damage [75].
As such, early targeted therapy could be an interesting approach to halt disease progression. Targeting TRM would not be a clear-cut approach since there are several variables that prove difficult to estimate in terms of their identification, placement, function, and numbers. For example, what would be an ideal number of TRM to target in autoimmune diseases? Additionally, the potential side effects of targeting TRM are unknown and may be detrimental as these cells are immune sentinels and vital for homeostasis. Targeting T cells in tissue may also prove to be technically difficult, but this may be circumvented by targeting/blocking (re)migration of these immune cells. In CD, the biological therapeutic Vedolizumab effectively targets immune cells via α4β7 (a marker for homing to gut tissue). The standard of care for patients with CD is a treat-to-target approach, with an early, aggressive attempt at inflammatory control. However, the majority of patients are intolerant to conventional therapy and gradually intolerant to TNF-α therapy [76]. In the GEMINI-LTS trial, patients treated with Vedolizumab at four-week intervals resulted in 83% [n = 100/120] and 89% [n = 62/70] of patients in remission after 104 and 152 weeks, respectively, showing remarkable improvement and positive long-term response [77]. Similarly, MS patients can be effectively treated with the α4-integrin inhibitor Natalizumab, blocking T cell migration over the blood-brain barrier, as well as to the intestines. However, this treatment is also linked to progressive multifocal leukoencephalopathy (an opportunistic viral infection), stressing the delicate balance in such settings [78].
Identification of disease-exacerbating TRM cells in circulation will not only open avenues for novel therapeutics but may also allow crucial monitoring (biomarkers) of disease progression. For instance, the disease course of JIA is unpredictable. Currently, JIA patients are only ‘allowed’ to be treated with biologicals (e.g., TNFα blockers or IL-6 blockade) when they are unresponsive to the first-line treatment methotrexate. However, unresponsiveness to methotrexate occurs in 30–50% of patients. If resistance to methotrexate therapy could be reliably predicted, patients could be treated with biologicals instantly. The window of opportunity, a period shortly after disease onset during which an optimal effect of treatment can be achieved, would then be utilized. Recirculating ex-TRM may serve as prognostic biomarkers of disease severity in chronic inflammation and autoimmune disease, and may even predict resistance to therapy.
Acknowledgments
Figures are created with BioRender.com.
Author Contributions
A.A.K.S. and J.v.d.G. wrote the manuscript with support and supervision from S.J.V., J.v.L. and F.v.W. All authors approved the final version of the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
Anoushka Samat is funded by ReumaNederland, grant number 19-1-403. Femke van Wijk is supported by a VIDI grant, number 91714332 from the Netherlands Organization for Health Research and Development (ZonMw). The Department of Pediatric Rheumatology and Immunology is supported by ReumaNederland, grant number LLP10.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Schenkel J.M., Masopust D. Tissue-Resident Memory T Cells. Immunity. 2014;41:886–897. doi: 10.1016/j.immuni.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Richmond J.M., Strassner J.P., Rashighi M., Agarwal P., Garg M., Essien K.I., Pell L.S., Harris J.E. Resident Memory and Recirculating Memory T Cells Cooperate to Maintain Disease in a Mouse Model of Vitiligo. J. Investig. Dermatol. 2019;139:769–778. doi: 10.1016/j.jid.2018.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kumar B., Ma W., Miron M., Granot T., Guyer R., Carpenter D., Senda T., Sun X., Ho S., Lerner H., et al. Human Tissue-Resident Memory T Cells Are Defined by Core Transcriptional and Functional Signatures in Lymphoid and Mucosal Sites. Cell Rep. 2017;20:2921–2934. doi: 10.1016/j.celrep.2017.08.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mueller S.N., Mackay L.K. Tissue-resident memory T cells: Local specialists in immune defence. Nat. Rev. Immunol. 2016;16:79–89. doi: 10.1038/nri.2015.3. [DOI] [PubMed] [Google Scholar]
- 5.Szabo P.A., Miron M., Farber D.L. Location, location, location: Tissue resident memory T cells in mice and humans. Sci. Immunol. 2019;4:eaas9673. doi: 10.1126/sciimmunol.aas9673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thome J.J., Yudanin N., Ohmura Y., Kubota M., Grinshpun B., Sathaliyawala T., Kato T., Lerner H., Shen Y., Farber D.L. Spatial Map of Human T Cell Compartmentalization and Maintenance over Decades of Life. Cell. 2014;159:814–828. doi: 10.1016/j.cell.2014.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schenkel J.M., Fraser K.A., Masopust D. Cutting Edge: Resident Memory CD8 T Cells Occupy Frontline Niches in Secondary Lymphoid Organs. J. Immunol. 2014;192:2961–2964. doi: 10.4049/jimmunol.1400003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miron M., Kumar B.V., Meng W., Granot T., Carpenter D.J., Senda T., Chen D., Rosenfeld A.M., Zhang B., Lerner H., et al. Human Lymph Nodes Maintain TCF-1hi Memory T Cells with High Functional Potential and Clonal Diversity throughout Life. J. Immunol. 2018;201:2132–2140. doi: 10.4049/jimmunol.1800716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mackay L., Rahimpour A., Ma J., Collins N., Stock A., Hafon M., Vega-Ramos J., Lauzurica P., Mueller S., Stefanovic T., et al. The developmental pathway for CD103+CD8+ tissue-resident memory T cells of skin. Nat. Immunol. 2013;14:1294–1301. doi: 10.1038/ni.2744. [DOI] [PubMed] [Google Scholar]
- 10.Siracusa F., Durek P., McGrath M., Sercan-Alp Ö., Rao A., Du W., Cendón C., Chang H., Heinz G., Mashreghi M., et al. CD69 + memory T lymphocytes of the bone marrow and spleen express the signature transcripts of tissue-resident memory T lymphocytes. Eur. J. Immunol. 2019;49:966–968. doi: 10.1002/eji.201847982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mackay L.K., Wynne-Jones E., Freestone D., Pellicci D.G., Mielke L.A., Newman D.M., Braun A., Masson F., Kallies A., Belz G.T., et al. T-box Transcription Factors Combine with the Cytokines TGF-β and IL-15 to Control Tissue-Resident Memory T Cell Fate. Immunity. 2015;43:1101–1111. doi: 10.1016/j.immuni.2015.11.008. [DOI] [PubMed] [Google Scholar]
- 12.Mackay L., Minnich M., Kragten N., Liao Y., Nota B., Seillet C., Zaid A., Man K., Preston S., Freestone D., et al. Hobit and Blimp1 instruct a universal transcriptional pro-gram of tissue residency in lymphocytes. Science. 2016;352:459–463. doi: 10.1126/science.aad2035. [DOI] [PubMed] [Google Scholar]
- 13.Milner J.J., Toma C., Yu B., Zhang K., Omilusik K., Phan A.T., Wang D., Getzler A.J., Nguyen T., Crotty S., et al. Runx3 programs CD8+ T cell residency in non-lymphoid tissues and tumours. Nat. Cell Biol. 2017;552:253–257. doi: 10.1038/nature24993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Milner J.J., Toma C., He Z., Kurd N.S., Nguyen Q.P., McDonald B., Quezada L., Widjaja C.E., Witherden D.A., Crowl J.T., et al. Heterogenous Populations of Tissue-Resident CD8+ T Cells Are Generated in Response to Infection and Malignancy. Immunity. 2020;52:808–824.e7. doi: 10.1016/j.immuni.2020.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thome J.J., Bickham K.L., Ohmura Y., Kubota M., Matsuoka N., Gordon C., Granot T., Griesemer A., Lerner H., Kato T., et al. Early-life compartmentalization of human T cell differentiation and regulatory function in mucosal and lymphoid tissues. Nat. Med. 2016;22:72–77. doi: 10.1038/nm.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sallusto F., Geginat J., Lanzavecchia A. Central Memory and Effector Memory T Cell Subsets: Function, Generation, and Maintenance. Annu. Rev. Immunol. 2004;22:745–763. doi: 10.1146/annurev.immunol.22.012703.104702. [DOI] [PubMed] [Google Scholar]
- 17.Masopust D., Soerens A.G. Tissue-Resident T Cells and Other Resident Leukocytes. Annu. Rev. Immunol. 2019;37:521–546. doi: 10.1146/annurev-immunol-042617-053214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fonseca R., Beura L., Quarnstrom C., Ghoneim H., Fan Y., Zebley C., Scott M., Fares-Frederickson N., Wijeyesinghe S., Thompson E., et al. Developmental plasticity allows out-side-in immune responses by resident memory T cells. Nat. Immunol. 2020;21:412–421. doi: 10.1038/s41590-020-0607-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Künzli M., King C.G. Resident Memory T Cells Escape ‘Home Quarantine’. Trends Immunol. 2020;41:454–456. doi: 10.1016/j.it.2020.04.010. [DOI] [PubMed] [Google Scholar]
- 20.Watanabe R., Gehad A., Yang C., Scott L.L., Teague J.E., Schlapbach C., Elco C.P., Huang V., Matos T.R., Kupper T.S., et al. Human skin is protected by four functionally and phenotypically discrete populations of resident and recirculating memory T cells. Sci. Transl. Med. 2015;7:279ra39. doi: 10.1126/scitranslmed.3010302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park S.L., Zaid A., Hor J.L., Christo S.N., Prier J.E., Davies B., Alexandre Y.O., Gregory J.L., Russell T.A., Gebhardt T., et al. Local proliferation maintains a stable pool of tissue-resident memory T cells after antiviral recall responses. Nat. Immunol. 2018;19:183–191. doi: 10.1038/s41590-017-0027-5. [DOI] [PubMed] [Google Scholar]
- 22.Beura L.K., Mitchell J.S., Thompson E.A., Schenkel J.M., Mohammed J., Wijeyesinghe S., Fonseca R., Burbach B.J., Hickman H.D., Vezys V., et al. Intravital mucosal imaging of CD8+ resident memory T cells shows tissue-autonomous recall responses that amplify secondary memory. Nat. Immunol. 2018;19:173–182. doi: 10.1038/s41590-017-0029-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mackay L.K., Kallies A. Transcriptional Regulation of Tissue-Resident Lymphocytes. Trends Immunol. 2017;38:94–103. doi: 10.1016/j.it.2016.11.004. [DOI] [PubMed] [Google Scholar]
- 24.Zundler S., Becker E., Spocinska M., Slawik M., Parga-Vidal L., Stark R., Wiendl M., Atreya R., Rath T., Leppkes M., et al. Author Correction: Hobit- and Blimp-1-driven CD4+ tissue-resident memory T cells control chronic intestinal inflammation. Nat. Immunol. 2019;20:514. doi: 10.1038/s41590-019-0360-y. [DOI] [PubMed] [Google Scholar]
- 25.Oja A.E., Piet B., Helbig C., Stark R., Van Der Zwan D., Blaauwgeers H., Remmerswaal E.B.M., Amsen D., Jonkers R.E., Moerland P.D., et al. Trigger-happy resident memory CD4+ T cells inhabit the human lungs. Mucosal Immunol. 2018;11:654–667. doi: 10.1038/mi.2017.94. [DOI] [PubMed] [Google Scholar]
- 26.Snyder M.E., Finlayson M.O., Connors T.J., Dogra P., Senda T., Bush E., Carpenter D., Marboe C., Benvenuto L., Shah L., et al. Generation and persistence of human tissue-resident memory T cells in lung transplantation. Sci. Immunol. 2019;4:eaav5581. doi: 10.1126/sciimmunol.aav5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Braga F.A.V., Hertoghs K.M.L., Kragten N.A.M., Doody G.M., Barnes N.A., Remmerswaal E.B.M., Hsiao C.-C., Moerland P.D., Wouters D., Derks I.A.M., et al. Blimp-1 homolog Hobit identifies effector-type lymphocytes in humans. Eur. J. Immunol. 2015;45:2945–2958. doi: 10.1002/eji.201545650. [DOI] [PubMed] [Google Scholar]
- 28.Chen Y., Zander R., Khatun A., Schauder D.M., Cui W. Transcriptional and Epigenetic Regulation of Effector and Memory CD8 T Cell Differentiation. Front. Immunol. 2018;9:2826. doi: 10.3389/fimmu.2018.02826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen L., Shen Z. Tissue-resident memory T cells and their biological characteristics in the recurrence of inflam-matory skin disorders. Cell. Mol. Immunol. 2020;17:64–75. doi: 10.1038/s41423-019-0291-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuric E., Seiron P., Krogvold L., Edwin B., Buanes T., Hanssen K., Skog O., Dahl-Jørgensen K., Korsgren O. Demonstration of Tissue Resident Memory CD8 T Cells in Insulitic Lesions in Adult Patients with Recent-Onset Type 1 Di-abetes. Am. J. Pathol. 2017;187:581–588. doi: 10.1016/j.ajpath.2016.11.002. [DOI] [PubMed] [Google Scholar]
- 31.Gao C., Yao Y., Li L., Yang S., Chu H., Tsuneyama K., Li X., Gershwin M., Lian Z. Tissue-Resident Memory CD 8+ T Cells Acting as Mediators of Salivary Gland Damage in a Murine Model of Sjögren’s Syndrome. Arthritis Rheumatol. 2019;71:121–132. doi: 10.1002/art.40676. [DOI] [PubMed] [Google Scholar]
- 32.Zhou M., Guo C., Li X., Huang Y., Li M., Zhang T., Zhao S., Wang S., Zhang H., Yang N. JAK/STAT signaling controls the fate of CD8+CD103+ tissue-resident memory T cell in lupus nephritis. J. Autoimmun. 2020;109:102424. doi: 10.1016/j.jaut.2020.102424. [DOI] [PubMed] [Google Scholar]
- 33.Fransen N.L., Hsiao C.-C., Van Der Poel M., Engelenburg H.J., Verdaasdonk K., Vincenten M.C.J., Remmerswaal E.B.M., Kuhlmann T., Mason M.R.J., Hamann J., et al. Tissue-resident memory T cells invade the brain parenchyma in multiple sclerosis white matter lesions. Brain. 2020;143:1714–1730. doi: 10.1093/brain/awaa117. [DOI] [PubMed] [Google Scholar]
- 34.Machado-Santos J., Saji E., Tröscher A.R., Paunovic M., Liblau R., Gabriely G., Bien C.G., Bauer J., Lassmann H. The compartmentalized inflammatory response in the multiple sclerosis brain is composed of tissue-resident CD8+ T lymphocytes and B cells. Brain. 2018;141:2066–2082. doi: 10.1093/brain/awy151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Petrelli A., Mijnheer G., Van Konijnenburg D.P.H., Van Der Wal M.M., Giovannone B., Mocholi E., Vazirpanah N., Broen J.C., Hijnen D., Oldenburg B., et al. PD-1+CD8+ T cells are clonally expanding effectors in human chronic inflammation. J. Clin. Investig. 2018;128:4669–4681. doi: 10.1172/JCI96107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maschmeyer P., Heinz G., Skopnik C., Lutter L., Mazzoni A., Heinrich F., Stuckrad S., Wirth L., Tran C., Riedel R., et al. Antigen-driven PD-1 + TOX + BHLHE40 + and PD-1 + TOX + EOMES + T lymphocytes regulate juvenile idiopathic arthritis in situ. Eur. J. Immunol. 2020 doi: 10.1002/eji.202048797. [DOI] [PubMed] [Google Scholar]
- 37.Steel K.J.A., Srenathan U., Ridley M., Durham L.E., Wu S., Ryan S.E., Hughes C.D., Chan E., Kirkham B.W., Taams L.S. Polyfunctional, Proinflammatory, Tissue-Resident Memory Phenotype and Function of Synovial Interleukin-17A+ CD 8+ T Cells in Psoriatic Arthritis. Arthritis Rheumatol. 2019;72:435–447. doi: 10.1002/art.41156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qaiyum Z., Gracey E., Yao Y., Inman R.D. Integrin and transcriptomic profiles identify a distinctive synovial CD8+ T cell subpopulation in spondyloarthritis. Ann. Rheum. Dis. 2019;78:1566–1575. doi: 10.1136/annrheumdis-2019-215349. [DOI] [PubMed] [Google Scholar]
- 39.Jung J.H., Lee J.S., Kim Y., Lee C., Yoo B., Shin E., Hong S. Synovial fluid CD69 + CD8 + T cells with tissue-resident phenotype mediate perforin-dependent citrullination in rheumatoid arthritis. Clin. Transl. Immunol. 2020;9:e1140. doi: 10.1002/cti2.1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cheuk S., Wikén M., Blomqvist L., Nylén S., Talme T., Ståhle M., Eidsmo L. Epidermal Th22 and Tc17 Cells Form a Localized Disease Memory in Clinically Healed Psoriasis. J. Immunol. 2014;192:3111–3120. doi: 10.4049/jimmunol.1302313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vo S., Watanabe R., Koguchi-Yoshioka H., Matsumura Y., Ishitsuka Y., Nakamura Y., Okiyama N., Fujisawa Y., Fujimoto M. CD 8 resident memory T cells with interleukin 17A-producing potential are accumulated in disease-naïve nonlesional sites of psoriasis possibly in correlation with disease duration. Br. J. Dermatol. 2019;181:410–412. doi: 10.1111/bjd.17748. [DOI] [PubMed] [Google Scholar]
- 42.Boniface K., Jacquemin C., Darrigade A.-S., Dessarthe B., Martins C., Boukhedouni N., Vernisse C., Grasseau A., Thiolat D., Rambert J., et al. Vitiligo Skin Is Imprinted with Resident Memory CD8 T Cells Expressing CXCR3. J. Investig. Dermatol. 2018;138:355–364. doi: 10.1016/j.jid.2017.08.038. [DOI] [PubMed] [Google Scholar]
- 43.Richmond J.M., Strassner J.P., Zapata L., Garg M., Riding R.L., Refat M.A., Fan X., Azzolino V., Tovar-Garza A., Tsurushita N., et al. Antibody blockade of IL-15 signaling has the potential to durably reverse vitiligo. Sci. Transl. Med. 2018;10:eaam7710. doi: 10.1126/scitranslmed.aam7710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.He H., Suryawanshi H., Morozov P., Gay-Mimbrera J., Del Duca E., Kim H.J., Kameyama N., Estrada Y., Der E., Krueger J.G., et al. Single-cell transcriptome analysis of human skin identifies novel fibroblast subpopulation and enrichment of immune subsets in atopic dermatitis. J. Allergy Clin. Immunol. 2020;145:1615–1628. doi: 10.1016/j.jaci.2020.01.042. [DOI] [PubMed] [Google Scholar]
- 45.Li J., van Vliet C., Rufaut N., Jones L., Sinclair R., Carbone F. Laser Capture Microdissection Reveals Transcriptional Abnormalities in Alopecia Areata before, during, and after Active Hair Loss. J. Investig. Dermatol. 2016;136:715–718. doi: 10.1016/j.jid.2015.12.003. [DOI] [PubMed] [Google Scholar]
- 46.Del Duca E., Ruano Ruiz J., Pavel A., Sanyal R., Song T., Gay-Mimbrera J., Zhang N., Estrada Y., Peng X., Renert-Yuval Y., et al. Frontal fibrosing alopecia shows robust T helper 1 and Janus kinase 3 skewing. Br. J. Dermatol. 2020;183:1083–1093. doi: 10.1111/bjd.19040. [DOI] [PubMed] [Google Scholar]
- 47.Bishu S., El Zaatari M., Hayashi A., Hou G., Bowers N., Kinnucan J., Manoogian B., Muza-Moons M., Zhang M., Grasberger H., et al. CD4+ Tissue-resident Memory T Cells Expand and Are a Major Source of Mucosal Tumour Necrosis Factor α in Active Crohn’s Disease. J. Crohn’s Coliti. 2019;13:905–915. doi: 10.1093/ecco-jcc/jjz010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rao D.A., Gurish M.F., Marshall J.L., Slowikowski K., Fonseka K.S.C.Y., Liu Y., Donlin L.T., Henderson L.A., Wei K., Mizoguchi F., et al. Pathologically expanded peripheral T helper cell subset drives B cells in rheumatoid arthritis. Nat. Cell Biol. 2017;542:110–114. doi: 10.1038/nature20810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Watanabe R. Protective and pathogenic roles of resident memory T cells in human skin disorders. J. Dermatol. Sci. 2019;95:2–7. doi: 10.1016/j.jdermsci.2019.06.001. [DOI] [PubMed] [Google Scholar]
- 50.Strazzulla L., Wang E., Avila L., Lo Sicco K., Brinster N., Christiano A., Shapiro J. Alopecia areata: Disease characteristics, clinical evaluation, and new perspectives on pathogenesis. J. Am. Acad. Dermatol. 2018;78:1–12. doi: 10.1016/j.jaad.2017.04.1141. [DOI] [PubMed] [Google Scholar]
- 51.Cheuk S., Schlums H., Sérézal I.G., Martini E., Chiang S.C., Marquardt N., Gibbs A., Detlofsson E., Introini A., Forkel M., et al. CD49a Expression Defines Tissue-Resident CD8 + T Cells Poised for Cytotoxic Function in Human Skin. Immunity. 2017;46:287–300. doi: 10.1016/j.immuni.2017.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gallais Sérézal I., Classon C., Cheuk S., Barrientos-Somarribas M., Wadman E., Martini E., Chang D., Xu Landén N., Ehrström M., Nylén S., et al. Resident T Cells in Resolved Psoriasis Steer Tissue Responses that Stratify Clinical Outcome. J. Investig. Dermatol. 2018;138:1754–1763. doi: 10.1016/j.jid.2018.02.030. [DOI] [PubMed] [Google Scholar]
- 53.Matos T., O’Malley J., Lowry E., Hamm D., Kirsch I., Robins H., Kupper T., Krueger J., Clark R. Clinically resolved psoriatic lesions contain psoriasis-specific IL-17–producing αβ T cell clones. J. Clin. Investig. 2017;127:4031–4041. doi: 10.1172/JCI93396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sérézal I.G., Hoffer E., Ignatov B., Martini E., Zitti B., Ehrström M., Eidsmo L. A skewed pool of resident T cells triggers psoriasis-associated tissue responses in never-lesional skin from patients with psoriasis. J. Allergy Clin. Immunol. 2019;143:1444–1454. doi: 10.1016/j.jaci.2018.08.048. [DOI] [PubMed] [Google Scholar]
- 55.Hegazy A., West N., Stubbington M., Wendt E., Suijker K., Datsi A., This S., Danne C., Campion S., Duncan S., et al. Circulating and Tissue-Resident CD4 + T Cells With Reactivity to Intestinal Microbiota Are Abundant in Healthy Individuals and Function Is Altered During Inflammation. Gastroenterology. 2017;153:1320–1337. doi: 10.1053/j.gastro.2017.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kleinschek M.A., Boniface K., Sadekova S., Grein J., Murphy E.E., Turner S.P., Raskin L., Desai B., Faubion W.A., Malefyt R.D.W., et al. Circulating and gut-resident human Th17 cells express CD161 and promote intestinal inflammation. J. Exp. Med. 2009;206:525–534. doi: 10.1084/jem.20081712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bartolomé-Casado R., Landsverk O.J., Chauhan S.K., Richter L., Phung D., Greiff V., Risnes L.F., Yao Y., Neumann R.S., Yaqub S., et al. Resident memory CD8 T cells persist for years in human small intestine. J. Exp. Med. 2019;216:2412–2426. doi: 10.1084/jem.20190414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Noble A., Durant L., Hoyles L., McCartney A.L., Man R., Segal J., Costello S.P., Hendy P., Reddi D., Bouri S., et al. Deficient Resident Memory T Cell and CD8 T Cell Response to Commensals in Inflammatory Bowel Disease. J. Crohn’s Coliti. 2019;14:525–537. doi: 10.1093/ecco-jcc/jjz175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Smolen J., Aletaha D., McInnes I. Rheumatoid arthritis. Lancet. 2016;388:2023–2038. doi: 10.1016/S0140-6736(16)30173-8. [DOI] [PubMed] [Google Scholar]
- 60.Mijnheer G., van Wijk F. T-Cell Compartmentalization and Functional Adaptation in Autoimmune Inflammation: Lessons from Pediatric Rheumatic Diseases. Front. Immunol. 2019;10:940. doi: 10.3389/fimmu.2019.00940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wehrens E.J., Prakken B.J., Van Wijk F. T cells out of control—Impaired immune regulation in the inflamed joint. Nat. Rev. Rheumatol. 2012;9:34–42. doi: 10.1038/nrrheum.2012.149. [DOI] [PubMed] [Google Scholar]
- 62.Beura L.K., Wijeyesinghe S., Thompson E.A., Macchietto M.G., Rosato P.C., Pierson M.J., Schenkel J.M., Mitchell J.S., Vezys V., Fife B.T., et al. T Cells in Nonlymphoid Tissues Give Rise to Lymph-Node-Resident Memory T Cells. Immunity. 2018;48:327–338.e5. doi: 10.1016/j.immuni.2018.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Beura L.K., Fares-Frederickson N.J., Steinert E.M., Scott M.C., Thompson E.A., Fraser K.A., Schenkel J.M., Vezys V., Masopust D. CD4+ resident memory T cells dominate immunosurveillance and orchestrate local recall responses. J. Exp. Med. 2019;216:1214–1229. doi: 10.1084/jem.20181365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Klicznik M.M., Morawski P.A., Höllbacher B., Varkhande S.R., Motley S.J., Kuri-Cervantes L., Goodwin E., Rosenblum M.D., Long S.A., Brachtl G., et al. Human CD4+CD103+ cutaneous resident memory T cells are found in the circulation of healthy individuals. Sci. Immunol. 2019;4:eaav8995. doi: 10.1126/sciimmunol.aav8995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sallusto F., Lenig D., Förster R., Lipp M., Lanzavecchia A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nat. Cell Biol. 1999;401:708–712. doi: 10.1038/44385. [DOI] [PubMed] [Google Scholar]
- 66.Gratz I.K., Campbell D.J. Resident memory T cells show that it is never too late to change your ways. Nat. Immunol. 2020;21:359–360. doi: 10.1038/s41590-020-0637-1. [DOI] [PubMed] [Google Scholar]
- 67.Spreafico R., Rossetti M., Van Loosdregt J., A Wallace C., Massa M., Magni-Manzoni S., Gattorno M., Martini A., Lovell D.J., Albani S. A circulating reservoir of pathogenic-like CD4+T cells shares a genetic and phenotypic signature with the inflamed synovial micro-environment. Ann. Rheum. Dis. 2014;75:459–465. doi: 10.1136/annrheumdis-2014-206226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sakaguchi S., Mikami N., Wing J.B., Tanaka A., Ichiyama K., Ohkura N. Regulatory T Cells and Human Disease. Annu. Rev. Immunol. 2020;38:541–566. doi: 10.1146/annurev-immunol-042718-041717. [DOI] [PubMed] [Google Scholar]
- 69.Mijnheer G., Lutter L., Mokry M., van der Wal M., Fleskens V., Scholman R., Pandit A., Tao W., Wekking M., Vervoort S., et al. Conserved human effector Treg signature is reflected in transcriptomic and epigenetic landscape. bioRxiv. 2020 doi: 10.1101/2020.09.30.319962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Szabo P.A., Levitin H.M., Miron M., Snyder M.E., Senda T., Yuan J., Cheng Y.L., Bush E.C., Dogra P., Thapa P., et al. Single-cell transcriptomics of human T cells reveals tissue and activation signatures in health and disease. Nat. Commun. 2019;10:1–16. doi: 10.1038/s41467-019-12464-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wienke J., Brouwers L., Van Der Burg L.M., Mokry M., Scholman R.C., Nikkels P.G., Van Rijn B.B., Van Wijk F. Human Tregs at the materno-fetal interface show site-specific adaptation reminiscent of tumor Tregs. JCI Insight. 2020;5 doi: 10.1172/jci.insight.137926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rossetti M., Spreafico R., Consolaro A., Leong J.Y., Chua C., Massa M., Saidin S., Magni-Manzoni S., Arkachaisri T., Wallace C.A., et al. TCR repertoire sequencing identifies synovial Treg cell clonotypes in the bloodstream during active inflammation in human arthritis. Ann. Rheum. Dis. 2016;76:435–441. doi: 10.1136/annrheumdis-2015-208992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Poon M.M., Farber D.L. The Whole Body as the System in Systems Immunology. iScience. 2020;23:101509. doi: 10.1016/j.isci.2020.101509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Guggino G., Rizzo A., Mauro D., Macaluso F., Ciccia F. Gut-derived CD8+ tissue-resident memory T cells are expanded in the peripheral blood and synovia of SpA patients. Ann. Rheum. Dis. 2019:–2019–216456. doi: 10.1136/annrheumdis-2019-216456. [DOI] [PubMed] [Google Scholar]
- 75.Vastert S.J., Bhat P., Goldstein D.A. Pathophysiology of JIA-associated Uveitis. Ocul. Immunol. Inflamm. 2014;22:414–423. doi: 10.3109/09273948.2014.926937. [DOI] [PubMed] [Google Scholar]
- 76.Argollo M., Fiorino G., Peyrin-Biroulet L., Danese S. Vedolizumab for the treatment of Crohn’s disease. Expert Rev. Clin. Immunol. 2018;14:179–189. doi: 10.1080/1744666X.2018.1438189. [DOI] [PubMed] [Google Scholar]
- 77.Vermeire S., Loftus E.V., Colombel J.-F., Feagan B.G., Sandborn W.J., Sands B.E., Danese S., D’Haens G.R., Kaser A., Panaccione R., et al. Long-term Efficacy of Vedolizumab for Crohn’s Disease. J. Crohn’s Coliti. 2016;11:412–424. doi: 10.1093/ecco-jcc/jjw176. [DOI] [PubMed] [Google Scholar]
- 78.Dalla Costa G., Martinelli V., Moiola L., Sangalli F., Colombo B., Finardi A., Cinque P., Kolb E., Haghikia A., Gold R., et al. Serum neurofilaments increase at progressive multifocal leukoencephalopathy onset in natalizumab-treated multiple sclerosis patients. Ann. Neurol. 2019;85:606–610. doi: 10.1002/ana.25437. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.