Abstract
EGLN1 encodes the hypoxia-inducible factor (HIF) pathway prolyl hydroxylase 2 (PHD2) that serves as an oxygen-sensitive regulator of HIF activity. The EGLN1 locus exhibits a signature of positive selection in Tibetan and Andean populations and is associated with hemoglobin concentration in Tibetans. Recent reports provide evidence for functional roles of protein-coding variants within the first exon of EGLN1 (rs186996510, rs12097901) that are linked to an adaptive signal in Tibetans, yet whether these same variants are present and contribute to adaptation in Andean highlanders is unknown. We determined the frequencies of these adaptive Tibetan alleles in Quechua Andeans resident at high altitude (4,350 m) in addition to individuals of Nepali ancestry resident at sea level. The rs186996510 C (minor) allele previously found at high frequency in Tibetans is absent in Andean (G: 100%) and rare among Nepali (C: 11.8%, G: 88.2%) cohorts. The minor G allele of rs12097901 is found at similarly low frequencies in Andeans (G: 12.7%, C: 87.3%) and Nepalis (G: 23.5%, C: 76.5%) compared to Tibetans. These results suggest that adaptation involving EGLN1 in Andeans involves different mechanisms than those described in Tibetans. The precise Andean adaptive variants remain to be determined.
Keywords: Andean, EGLN1, high altitude, rs12097901, rs186996510, Tibetan
1. INTRODUCTION
High-altitude environments impose strong selective pressures that have resulted in rapid genetic adaptations. Andean, Tibetan, and Ethiopian highlanders living at elevations greater than 3,500 m above sea level show signatures of evolutionary selection at various genetic loci that contain hypoxia inducible factor (HIF) pathway and non-HIF-related genes (Bigham et al., 2010; Moore, 2017; Simonson, 2015). These candidate genes likely underlie distinct physiological traits observed in these populations.
The HIF pathway gene EGLN1, which encodes the oxygen-sensitive enzyme prolyl hydroxylase 2 (PHD2), is contained within a region exhibiting one of the strongest signals of selection in Tibetans and shows evidence of convergent evolution in Tibetans and Andeans (Bigham et al., 2010, 2013; Foll, Gaggiotti, Daub, Vatsiou, & Excoffier, 2014). Variants in linkage disequilibrium at this locus are associated with hemoglobin (Hb) in Tibetans at high altitudes (Simonson et al., 2010). In the presence of oxygen, PHD2 hydroxylates and targets HIF 1α subunits for degradation, thereby regulating HIF activity (Jaakkola et al., 2001; Ke & Costa, 2006) and influencing physiological responses to hypoxia. Recent studies demonstrate that the missense variant rs186996510 (c.12C > G), in combination with the linked missense variant c.380G > C, results in a gain-of-function (lower Km) phenotype in vitro that dampens the hypoxia response and could alter erythroid progenitor proliferation (Lorenzo et al., 2014). These variants also result in loss of PHD2 function and increased HIF activation via defective binding of cochaperone p23 (Song et al., 2014). Furthermore, noncoding variants within the first intron of EGLN1 are associated with increased gene expression and high-altitude pulmonary edema in a population studied in India (Aggarwal et al., 2010), suggesting non-protein coding variants at this locus might also be involved in regulation of EGLN1 and the HIF pathway.
In addition to reports of adaptive significance in Tibetans (Bigham et al., 2010; Hu et al., 2017; Peng et al., 2011; Simonson et al., 2010, 2012; Wuren et al., 2014; Yi et al., 2010), single-nucleotide polymorphism (SNP)-based analyses indicate EGLN1 is under selection in Andean populations as well (Bigham et al., 2009, 2010). Another study used whole-genome sequences and identified EGLN1 as a top hypoxia-response gene showing evidence of selection in Andeans, although they did not identify EGLN1 in the top 1% of all genes under selection (Crawford et al., 2017). Further examination of tagging SNPs at this locus did not identify associations with Hb in Bolivians (3600 m) and Peruvians (4300 m) (Bigham et al., 2009, 2013). Because of the high GC content of the first exon of EGLN1, limited sequence information has been available and, to our knowledge, the frequency of c.12C > G, c.380G > C variants have not been reported in Andeans (Figure 1). We therefore sequenced this region in DNA from Andean highlanders (Cerro de Pasco, Peru, 4350 m), to determine whether variants associated with functional adaptation in Tibetans (Lorenzo et al., 2014) are also present in this population.
FIGURE 1.
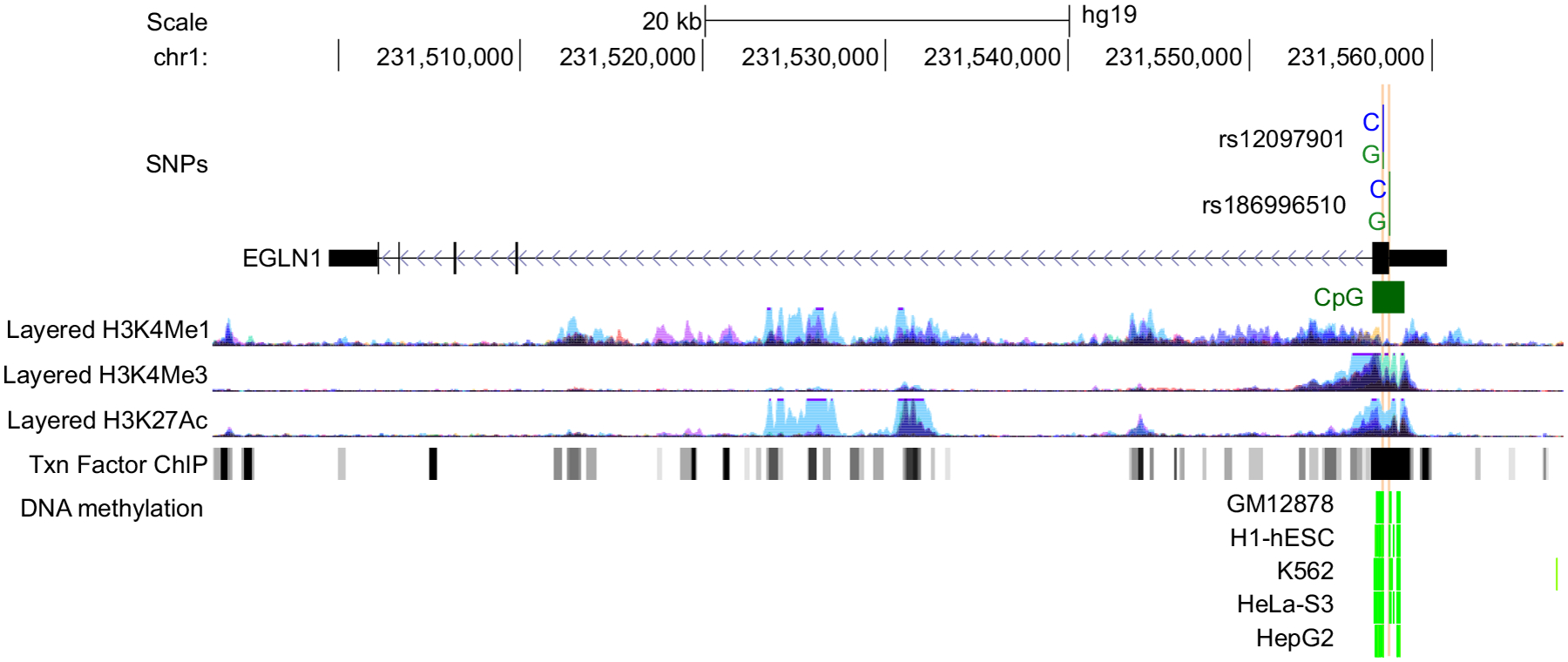
The EGLN1 region and location of rs186996510 and rs12097901 in the first exon. The first exon contains a CpG island (highlighted in green) and histone modifications (primarily H3K27Ac and H3K4Me3 marks). H3K4Me1 and H3K4Me1 marks are often found near regulatory elements and H3K4Me3 marks are often found near promoters. Peak color in the histone modification tracks indicate cell type (light blue = human umbilical vein endothelial cells (HUVEC), dark blue = K562 cells, light purple = normal human epidermal keratinocytes (NHEK) cells. Regions of transcription factor binding are shown in the Txn Factor ChIP track. DNA methylation by reduced representation bisulfite sequencing from ENCODE/HudsonAlpha is displayed for multiple cell types in light green. SNPs, single-nucleotide polymorphisms
2. MATERIALS AND METHODS
2.1. Subjects
This study was approved by the University of California, San Diego Human Research Protections Program and was conducted in accordance with Declaration of Helsinki principles. Men and non-pregnant women ages 19 to 65 years old were included as part of a larger study. Nepali (N = 17) subjects were recruited in San Francisco, California, and Andean subjects were recruited in Cerro de Pasco, Peru (N = 106). Subjects were excluded during a preliminary screening session if they demonstrated a history of cardiovascular/pulmonary disease. Andean subjects with chronic mountain sickness or excessive erythrocytosis (Hb ≥ 21 g/dL in men and Hb ≥ 19 g/dL in women) were included. Subjects were also excluded if they did not have at least three generations of ancestry in Cerro de Pasco, Peru (>2,500 m) or Nepal. Andean subjects must have been born in Cerro de Pasco and could not have undergone blood transfusions or phlebotomies in the previous 6 months, or journeys to lower altitude for more than 7 days during the previous 6 months.
2.2. Genotyping
Genomic DNA was obtained from peripheral blood samples. Blood samples were taken from the antecubital vein and stored immediately at 4°C. DNA was isolated from buffy coat within 24 hours using a Gentra Puregene blood kit (Qiagen, Venlo, Netherlands). Genomic DNA quality and purity was confirmed via NanoDrop 2000 (Thermo Fisher Scientific, Waltham, MA) and gel electrophoresis.
A 1025 bp sequence of the first exon of EGLN1 was amplified with polymerase chain reaction (PCR) using a Lucigen FailSafe 2X PreMix H (Middleton, WI) and Integrated DNA Technologies ([IDT], Skokie, IL) forward (CCC-CTATCTCTCTCCCCG) and reverse (CCTGTCCAGCA-CAAACCC) primers. PCR was performed on a QuantStudio 3 Real-Time PCR System (Thermo Fisher Scientific) with the following settings: an initial denaturation step at 95°C for 4 minutes followed by 30 cycles of denaturation at 95°C for 30 seconds, annealing at 51°C for 1 minute, and elongation at 68°C for 70 seconds. There was a final elongation at 68°C for 5 minutes.
The PCR amplicon was enzymatically purified (GeneWiz PCR cleanup) and sequenced via Sanger sequencing with the GeneWiz difficult template protocol (GeneWiz, South Plain-field, NJ). Four sequencing primers were designed with the PrimerQuest Tool from IDT and optimal sequence results were obtained with the following reverse primer (ACA-GATGCCGTGCTTGTT). All sequences were aligned with Clustal Omega multiple sequence alignment (EMBL-EBI, Cambridgeshire, UK) and c.12C > G and c.380G > C SNP genotypes were confirmed manually and recorded for each individual.
2.3. Statistical tests
To determine whether putatively adaptive alleles previously identified in Tibetans were found at different frequencies compared to our Andean and Nepali samples, we used Pearson chisquare tests in Excel (Microsoft, Redmond, WA). Andean and Nepali allele frequencies were compared to Tibetan frequencies separately at each of two SNPs, resulting in four tests, and therefore a Bonferroni corrected p value of 0.0125 was required to reject the null hypothesis that allele frequencies were the same across both groups in each comparison.
3. RESULTS
3.1. Genotype frequencies
The first SNP rs186996510 (hg38; chr1:231421877, + strand), has a reported allele frequency of C: 0.4% (145/39074), G: 99.6% (38929/39074) in the general human population (UCSC Genome Browser, Santa Cruz, CA). Lorenzo et al. (2014) reported increased minor “C” allele frequencies in Tibetans (C: 80.8%, G: 19.2%; genotypes: G/G: 15.4% (4/26); G/C: 7.7% (2/26); C/C: 76.9% [20/26]) (Figure 2). We found that the minor “C” allele is absent in Andeans (genotypes: G/G: 100% [106/106]) and less common in Nepalis (C: 11.8%, G: 88.2%; genotypes: G/G: 82.4% [14/17]; G/C: 11.8% [2/17]; C/C: 5.9% [1/17]) compared to Tibetans. Andean and Nepali minor allele C frequencies were significantly different than those previously reported in Tibetans (both p < 0.01).
FIGURE 2.
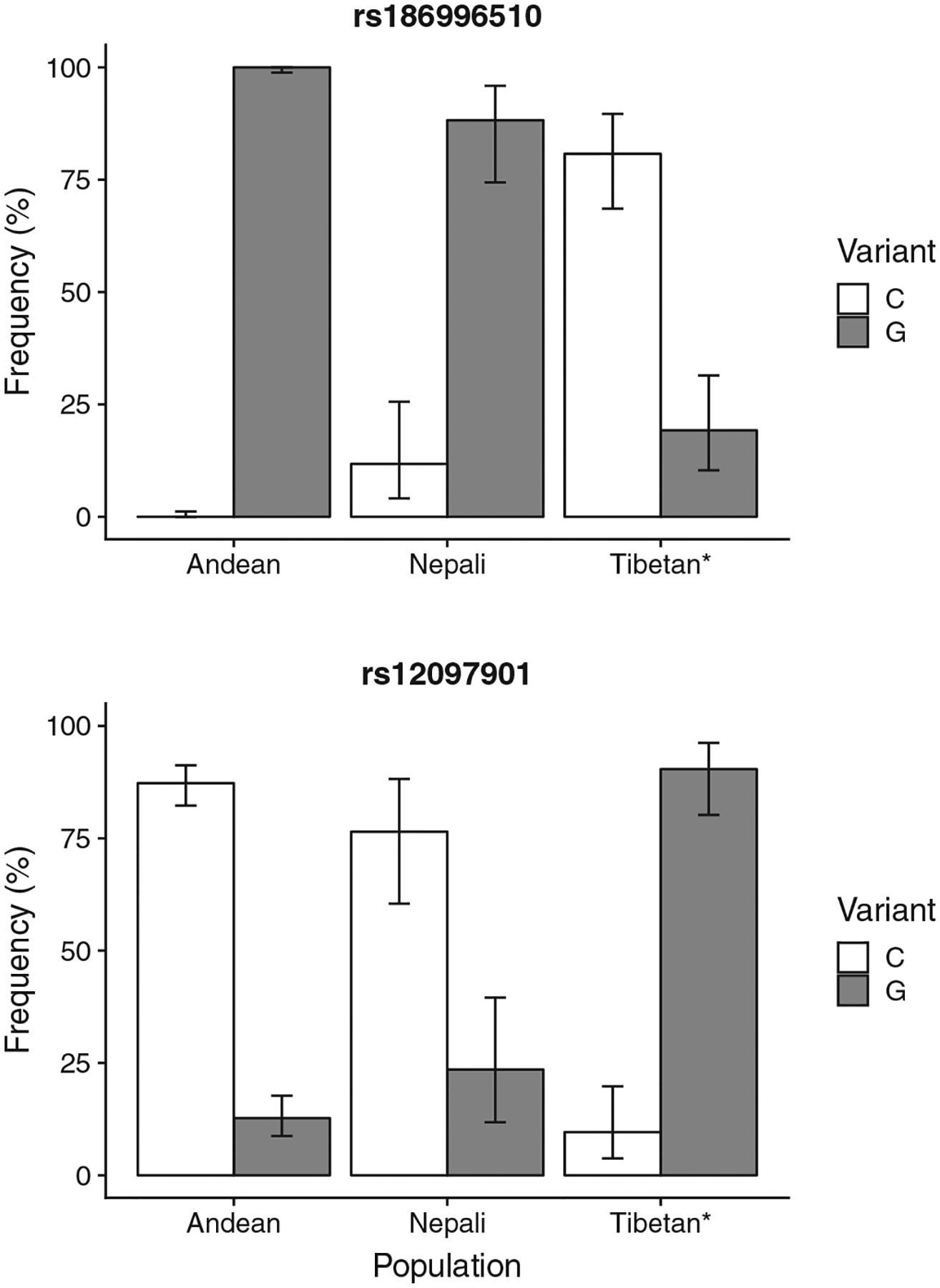
Allele frequencies at two EGLN1 single-nucleotide polymorphisms (SNPs) (rs186996510, rs12097901) in our Andean (N = 106) and Nepali (N = 17) samples compared to previously reported Tibetan (N = 26) allele frequencies. *Data reported in Lorenzo et al. (2014). Error bars indicate 95% confidence intervals for binomial data using the Jeffrey’s prior
The second SNP rs12097901 (hg38; chr1:231421509, + strand) has a reported allele frequency of C: 73.9% (3780/5116), G: 26.1% (1336/5116) in the general human population (UCSC Genome Browser). In our data set, the minor G allele was found at lower frequency in Andeans (G: 12.7%, C: 87.3%; genotypes: C/C: 76.4% [81/106]; C/G: 21.7% [23/106]; G/G: 1.9% [2/106]) and Nepalis (C: 76.5, G: 23.5%; genotypes: C/C: 64.7% [11/17]; C/G: 23.5% [4/17]; G/G: 11.8% [2/17]) compared to previous reports in Tibetans (C: 9.6%, G: 90.4%; genotypes: C/C: 0.0% [0/26]; C/G: 19.2% [5/26]; G/G: 80.8% [21/26]) (Figure 2) (Lorenzo et al., 2014). Andean and Nepali minor allele G frequencies were significantly different than those previously reported in Tibetans (both p < 0.01).
4. DISCUSSION
The EGLN1 adaptive Tibetan alleles c.12C > G (rs186996510) and 380G > C (rs12097901) that are in linkage disequilibrium with adaptive tagging SNPs in Tibetans are absent and at lower frequencies, respectively, in Andeans. Given the association between Hb and this locus in Tibetans, it remains to be determined whether specific variants at this locus are associated with this phenotype in Andeans, who generally exhibit higher, but variable, Hb at high altitudes (Beall, 2007). The two variants increase PHD2 activity by lowering its Km value for oxygen, and erythroid progenitor cells from Tibetan individuals with c.[12C > G; 380G > C] variants show impaired proliferation in hypoxia compared to wild-type controls (Lorenzo et al., 2014; Song et al., 2014). Therefore, these adaptive alleles may be protective against high-altitude polycythemia, and the absence of these variants in Andeans may underlie the prevalence of maladaptive phenotypes less commonly observed among Tibetans.
Considering previous reports of adaptive signals at this locus in Andeans (Bigham et al., 2009, 2010), we suspect other variants, potentially with regulatory function, are targeted by selection in this population. Epigenetic modifications (e.g., via DNA methylation and histone modifications) that influence gene activity and expression likely play an important role in high-altitude adaptation because many of the key players in the hypoxic-response pathway are epigenetically regulated. Universal histone modification may provide a mechanism for a short-term, plastic response to hypoxia, likely by influencing Jumonji C domain-containing (JmjC) histone demethylases (Shmakova, Batie, Druker, & Rocha, 2014). The first exon of EGLN1 contains a CpG island, which is prone to methylation, and is further susceptible to histone modifications (Figure 1). Additionally, Crawford et al. (2017) demonstrate unique distributions of long undifferentiated EGLN1 haplotypes in Andeans, which may suggest that a particular haplotype was selected for upon entering the high-altitude environment.
The two adaptive Tibetan variants examined here are located within the CpG island of EGLN1, and genetic differences can impact the extent to which epigenetic modifications occur (Bell et al., 2011; Moen et al., 2013). Recent work suggests that over 80% of genetic variants that disrupt CpG motifs influence DNA methylation status locally and up to 10 kb from the site of the mutation (Zhi et al., 2013). There is also evidence for differential EGLN1 gene expression and high-altitude pulmonary edema susceptibility in individuals from India (Agarwal et al. 2010) and methylation status at select sites of EGLN1 in Andeans with and without excessive erythrocytosis (Julian 2017). EGLN1 also contains an upstream hypoxia response element (GGTGTACGTGCAGAGCGC), which serves as the binding site for HIF-1 (Brown & Rupert, 2014), and the availability of this site likely depends on the methylation status of the EGLN1 CpG region. Andeans may have different epigenetic regulation of EGLN1 compared to Tibetan highlanders as a result of genetic variance in this noncoding region, making it more or less prone to modification. The extent to which these c.[12C > G; 380G > C] Tibetan variants, and/or others in noncoding regions, influence epigenetic regulation of EGLN1 remain to be determined.
Both the Andean and Nepali populations demonstrate absent or lower allele frequencies of c.12C > G and 380G > C, similar to those reported for the general population (UCSC Genome Browser). However, these alleles have been generally underreported because this region of the EGLN1 exon 1 has high CG content and typically demonstrates poor sequencing coverage in next-generation sequencing data (Benjamini & Speed, 2012; Dohm, Lottaz, Borodina, & Himmelbauer, 2008) (e.g., this region was not successfully sequenced in the 54 genomes from the high-coverage Complete Genomics discovery panel) (Drmanac et al., 2010). Additional sequence analyses of this underreported region are needed.
The lack of c.12C > G and low frequency of 380G > C in Nepali individuals likely reflects distinct population histories in these groups. The Nepali individuals (11 of the 17) were originally from Kathmandu, two from Bhojpur, and the remaining four from Bhaktapur, Lalipur, Gorkha, and Sailung, Nepal, with at lease three previous generations in the same area. Given the large amount of genetic admixture in the Nepali population from north and south of the Himalaya, it is likely that these individuals, primarily from the southern Himalayan region, may have more Indian admixture and therefore fewer high-altitude adaptive Tibetan alleles (Cole et al., 2017).
Based on the lack of Tibetan variants found in Andeans using our targeted sequencing protocol, these variants together are likely not the direct targets of selection in Andeans. Indeed, rs186996510 is found at low frequency in East Asian populations and was likely a poor candidate for selection in early population migrations into the Americas. There is evidence that other genes involved in cardiac responses to hypoxia (VEGFB and ELTD1 in Collas) as well as other major HIF-pathway regulators (EPAS1) are the primary targets of selection in Andean populations (Eichstaedt et al., 2014, 2017). However, given previous reports of selection signals at the EGLN1 locus in Andeans, it will be important to determine the putatively adaptive variants in this population, if there is physiological relevance of 380G > C and other genetic/epigenetic changes, and how these factors contribute to individual responses to hypoxia such as the differences observed in Tibetan versus Andean populations.
ACKNOWLEDGMENTS
The authors thank the research team at the Instituto de Investigaciones de Altura in Cerro de Pasco, Peru, for assistance with sample collection in Peru. We would also like to thank Pavan Shrestha for his assistance with recruitment of Nepali participants, Dr. Philip Bickler for the kind use of his laboratory space at the University of California, San Francisco, and Elizabeth Blue for R code. This research was further supported by resources from the Center for Physiological Genomics of Low Oxygen (CPGLO) at the University of California, San Diego, by an Investigators Programme grant from Science Foundation Ireland (12/IP/1727), and by a Wellcome Trust grant (107544/Z/15/Z) to support the UPCH team.
Funding information
University of California, San Diego, Grant/Award Number: Office of Research Affairs Frontiers of Innovation; Science Foundation Ireland, Grant/Award Number: 12/IP/1727; National Institutes of Health, Grant/Award Number: 5F32HL131218
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicts of interest to disclose.
REFERENCES
- Aggarwal S, Negi S, Jha P, Singh PK, Stobdan T, Pasha MAQ, … Mukerji M (2010). EGLN1 involvement in high-altitude adaptation revealed through genetic analysis of extreme constitution types defined in Ayurveda. PNAS, 107(44), 18961–18966. 10.1073/pnas.1006108107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beall C (2007). Two routes to functional adaptation: Tibetan and Andean high-altitude natives. PNAS, 104(Suppl 1), 8655–8660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell JT, Pai AA, Pickrell JK, Gaffney DJ, Degner J, Gilad Y, & Pritchard JK (2011). DNA methylation patterns associatewith genetic and gene expression variation in HapMap cell lines. Genome Biology, 12, R10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, & Speed TP (2012). Summarizing and correcting the GC content bias in high-throughput sequencing. Nucleic Acids Research, 40(10), 1–14. 10.1093/nar/gks001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigham A, Bauchet M, Pinto D, Mao X, Akey JM, Mei R, … Shriver MD (2010). Identifying signatures of natural selection in Tibetan and Andean populations using dense genome scan data. PLoS Genetics, 6(9), e1001116. 10.1371/journal.pgen.1001116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigham AW, Mao X, Mei R, Brutsaert T, Wilson MJ, Julian CG, … Shriver MD (2009). Identifying positive selection candidate loci for high-altitude adaptation in Andean populations. Human Genomics, 4(2), 79–90. 10.1186/1479-7364-4-2-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigham AW, Wilson MJ, Julian CG, Kiyamu M, Vargas E, Leon-Velarde F, … Shriver MD (2013). Andean and Tibetan patterns of adaptation to high altitude. American Journal of Human Biology, 25, 190–197. 10.1002/ajhb.22358. [DOI] [PubMed] [Google Scholar]
- Brown CJ, & Rupert JL (2014). Hypoxia and environmental epigenetics. High Altitude Medicine & Biology, 15 (3), 323–330. 10.1089/ham.2014.1016. [DOI] [PubMed] [Google Scholar]
- Cole AM, Cox S, Jeong C, Petousi N, Aryal DR, Droma Y, … Cavalleri GL (2017). Genetic structure in the Sherpa and neighboring Nepalese populations. BMC Genomics, 18(1), 1–10. 10.1186/s12864-016-3469-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford JE, Amaru R, Song J, Julian CG, Racimo F, Cheng JY, … Nielsen R (2017). Natural selection on genes related to cardiovascular health in high-altitude adapted Andeans. American Journal of Human Genetics, 101(5), 752–767. 10.1016/j.ajhg.2017.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohm JC, Lottaz C, Borodina T, & Himmelbauer H (2008). Substantial biases in ultra-short read data sets from high-throughput DNA sequencing. Nucleic Acids Research, 36(16), e105. 10.1093/nar/gkn425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drmanac R, Sparks AB, Callow MJ, Halpern AL, Burns NL, Kermani BG, … Reid CA (2010). Human genome sequencine using unchained base reads on self-assembling DNA nanoarrays. Science, 327, 78–82. 10.1126/science.1181498. [DOI] [PubMed] [Google Scholar]
- Eichstaedt CA, Antão T, Pagani L, Cardona A, Kivisild T, & Mormina M (2014). The Andean adaptive toolkit to counteract high altitude maladaptation: Genome-wide and phenotypic analysis of the Collas. PLoS One, 9(3), e93314. 10.1371/journal.pone.0093314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichstaedt CA, Pagani L, Antao T, Inchley CE, Cardona A, Mörseburg A, … Kivisild T (2017). Evidence of early-stage selection on EPAS1 and GPR126 genes in Andean high altitude populations. Scientific Reports, 7(1), 1–9. 10.1038/s41598-017-13382-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foll M, Gaggiotti OE, Daub JT, Vatsiou A, & Excoffier L (2014). Widespread signals of convergent adaptation to high altitude in Asia and America. American Journal of Human Genetics, 95 (4), 394–407. 10.1016/j.ajhg.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Petousi N, Glusman G, Yu Y, Bohlender R, Tashi T, … Huff CD (2017). Evolutionary history of Tibetans inferred from whole-genome sequencing. PLOS Genetics, 13(4), e1006675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaakkola P, Mole DR, Tian Y-M, Wilson MI, Gielbert J, Gaskell SJ, … Ratcliffe PJ (2001). Targeting of HIF-α to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science, 292, 468–472. 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- Julian CG (2017). Epigenomics and human adaptation to high altitude. J Appl Physiol, 123, 1362–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke Q, & Costa M (2006). Hypoxia-inducible factor-1 (HIF-1). Molecular Pharmacology, 70(5), 1469–1480. 10.1124/mol.106.027029.ABBREVIATIONS. [DOI] [PubMed] [Google Scholar]
- Lorenzo FR, Huff C, Myllymäki M, Olenchock B, Swierczek S, Tashi T, … Prchal JT (2014). A genetic mechanism for Tibetan high-altitude adaptation. Nature Genetics, 46, 951–956. 10.1038/ng.3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moen EL, Zhang X, Mu W, Delaney SM, Wing C, Mcquade J, … Zhang W (2013). Genome-wide variation of cytosine modifications between European and African populations and the implications for complex traits. Genetics, 194(4), 987–996. 10.1534/genetics.113.151381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore LG (2017). Measuring high-altitude adaptation. Journal of Applied Physiology, 123, 1371–1385. 10.1152/japplphysiol.00321.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Yang Z, Zhang H, Cui C, Qi X, Luo X, … Su B (2011). Genetic variations in Tibetan populations and high-altitude adaptation at the Himalayas. Molecular Biology and Evolution, 28(2), 1075–1081. 10.1093/molbev/msq290. [DOI] [PubMed] [Google Scholar]
- Shmakova A, Batie M, Druker J, & Rocha S (2014). Chromatin and oxygen sensing in the context of JmjC histone demethylases. Biochemical Journal, 462(3), 385–395. 10.1042/BJ20140754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonson TS (2015). Altitude adaptation: A glimpse through various lenses. High Altitude Medicine & Biology, 16(2), 125–137. 10.1089/ham.2015.0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonson TS, McClain DA, Jorde LB, & Prchal JT (2012). Genetic determinants of Tibetan high-altitude adaptation. Human Genetics, 131, 527–533. 10.1007/s00439011-1109-3. [DOI] [PubMed] [Google Scholar]
- Simonson TS, Yang Y, Huff CD, Yun H, Qin G, Witherspoon DJ, … Ge R (2010). Genetic evidence for high-altitude adaptation in Tibet. Science, 329, 72–75. [DOI] [PubMed] [Google Scholar]
- Song D, Li LS, Arsenault PR, Tan Q, Bigham AW, Heaton-Johnson KJ, … Lee FS (2014). Defective Tibetan PHD2 binding to p23 links high altitude adaption to altered oxygen sensing. Journal of Biological Chemistry, 289(21), 14656–14665. 10.1074/jbc.M113.541227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson JA, Watson CJ, Mccann A, & Baugh J (2010). Epigenetics, the epicenter of the hypoxic response. Epigenetics, 5 (4), 293–296. 10.4161/epi.5.4.11684. [DOI] [PubMed] [Google Scholar]
- Wuren T, Simonson TS, Qin G, Xing J, Huff CD, Witherspoon DJ, … Ge R-L (2014). Shared and unique signals of high-altitude adaptation in geographically distinct Tibetan populations. PloS One, 9(3), e88252. 10.1371/journal.pone.0088252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi X, Liang Y, Huerta-Sanchez E, Jin X, Cuo ZXP, Pool JE, … Wang J (2010). Sequencing of 50 human exomes reveals adaptation to high altitude. Science, 329, 75–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhi D, Aslibekyan S, Irvin MR, Claas SA, Borecki IB, Ordovas JM, … Arnett DK (2013). SNPs located at CpG sites modulate genome-epigenome interaction. Epigenetics, 8(8), 802–806. 10.4161/epi.25501. [DOI] [PMC free article] [PubMed] [Google Scholar]


