Abstract
INTRODUCTION:
Recent studies suggest both sex-specific genetic risk factors and those shared between dementia and stroke are involved in dementia pathogenesis.
METHODS:
We performed both single-variant and gene-based genome-wide association studies of >11,000 whole genome sequences from the Women’s Health Initiative cohort to discover loci associated with dementia, adjusting for age, ethnicity, stroke, and venous thromboembolism status. Evidence for prior evidence of association and differential gene expression in dementia-related tissues and samples was gathered for each locus.
RESULTS:
Our multiethnic studies identified significant associations between variants within APOE, MYH11, FZD3, SORCS3, and GOLGA8B and risk of dementia. Ten genes implicated by these loci, including MYH11, FZD3, SORCS3, and GOLGA8B were differentially expressed in the context of Alzheimer’s disease.
DISCUSSION:
Our association of MYH11, FZD3, SORCS3, and GOLGA8B with dementia is supported by independent functional studies in human subjects, model systems, and associations with shared risk factors for stroke and dementia.
Keywords: Stroke, whole-genome sequence, sex-specific, women, dementia
1. Background
Over 20 years ago, the APOE ε4 allele was identified as a major genetic risk factor for late-onset Alzheimer’s disease (AD) and dementia [1, 2]. Genome-wide association studies (GWAS) have identified >30 additional loci contributing to AD risk, with several more identified by exome-based studies [3]. A substantial proportion of dementia risk remains unexplained. This may be because GWAS are typically underpowered for detecting associations with rare variants. Dementia also exhibits considerable phenotypic heterogeneity, and it is often difficult to distinguishing AD from vascular or other subtypes of dementia [4–6].
There is a complex relationship between vascular risk factors and dementia. Both AD and cerebrovascular disease increase in prevalence with age and share risk factors like hypertension, APOE genotype, smoking, and diabetes mellitus [6]. Stroke and dementia are each risk factors for the other, and there is accumulating evidence they share susceptibility genes and pathways [7]. There may also be sex-specific or hormonal differences that influence genetic susceptibility to AD or dementia (ex., [8]).
This analysis of >11,000 post-menopausal women from the Women’s Health Initiative study (WHI) enriched for vascular disease aims to discover new genetic loci associated with dementia. Our analysis of whole genome sequence (WGS) data assesses variation across the allele frequency spectrum, including both coding and non-coding variants. We find strong association between dementia and APOE, as well as novel loci involving MYH11, FZD3, SORCS3, and GOLGA8B. Independent transcriptomic studies reveal these loci are also associated with gene expression in the brain and that some of those genes are differentially expressed in AD and dementia.
2. Methods
2.1. The Women’s Health Initiative (WHI)
The WHI is a prospective study of postmenopausal women representing a socio-demographically diverse population, recruiting 161,808 women between 1993 and 1998, (Supplemental Methods) [9]. The WHI included an Observational Study (WHI-OS) and randomized Clinical Trials (WHI-CT), including a Hormone Therapy Trials (HT). Both the WHI-CT and WHI-OS cohorts have been actively followed for > 25 years. WGS data were collected for 11,085 WHI participants through the Trans-Omics for Precision Medicine (TOPMed) project sponsored by the National Heart, Lung and Blood Institute (NHLBI) using a centralized, rigorous approach (https://www.nhlbiwgs.org/topmed-whole-genome-sequencing-methods-freeze-8). Freeze 8 of the TOPMed data were aligned to the GRCh38 human reference and cleaned as previously described (Supplemental Methods [10]).
The WHI TOPMed participants included all eligible and consenting women with incident stroke (N=4,852) or venous thromboembolism (VTE; N=1,162), women with coronary heart disease (CHD; N=1,797), and 4,216 controls matched on age and ethnicity. APOE ε2/ε3/ε4 genotypes were defined using WGS genotypes at rs7412 (avg. read depth [DP] = 39) and rs429358 (DP = 40). 1,608 cases of dementia were identified by self-report, medical history updates, and/or death certificate information (Supplemental Methods). Comparisons with dementia status adjudicated by an expert panel suggests the classification of dementia in this sample has high specificity, but likely under-reports the true number of dementia cases. Although the majority of dementia cases in the United States are affected by AD (60–80%), cerebrovascular disease, lewy body disease, and frontotemporal dementia (FTD) each represent the cause of dementia in 5–10% of cases and up to 50% of dementia cases show mixed pathology [11].
2.2. Statistical analyses
Single-variant association testing between dementia and single nucleotide variants (SNVs) and short insertions/deletions was performed using SAIGE [12] implemented in ENCORE (https://encore.sph.umich.edu/), controlling type 1 error by adjusting for relatedness and sample size imbalances (Supplemental Methods). Tests were restricted to variants with minor allele frequency (MAF) >0.1% (N=877,506,482). Covariates included age at enrollment, self-reported ethnicity, stroke and VTE status, assignment in WHI-CT vs. WHI-OS, randomization arm for those in the HT trial, and principal components (PCs) 1–10 to control for population stratification. Genome-wide significance was defined as P < 5 × 10−8, and lead variants as those with the smallest P value at a locus significantly associated with dementia. For any loci with novel associations, we performed sensitivity analyses stratified by stroke status, APOE status, history of HT at baseline, and self-reported ancestry.
Aggregation tests improve the statistical power to identify associations driven by rare variants and can aid interpretation when variants are aggregated by biological features like genes. We applied the SKAT-O test [13], which allows for different variants within the same gene to have opposing effects. For variants with MAF ≤ 5%, we performed gene-based testing used the same covariates as the single-variant GWAS, where the “gene set” included all non-synonymous and splice junction variants within a gene. The genome-wide significance threshold was determined with a Bonferroni correction for the number of tested genes (N=18,750, P < 2.67 × 10−6).
2.3. Variant annotation
Variants were annotated Variant Effect Predictor (VEP; release 100 [14]) and Ensembl Regulatory Build (release 97 [15]) to identify their consequences in coding regions and regulatory features. Variants were intersected with published GWAS hits, expression quantitative trait loci (eQTL), transcription factor binding site (TFBS) motifs, and chromatin, histone, and DNAseI hypersensitivity sites (DHS) marks using HaploReg v4.1 (https://pubs.broadinstitute.org/mammals/haploreg/haploreg.php). The genomic context of variants associated with dementia was plotted using LocusZoom (http://my.locuszoom.org).
2.4. Evidence for validation from GWAS
Our study design, a secondary analysis of WGS and phenotype data collected for other purposes, prohibits true replication [16]: no data sets share many of the study’s attributes and no GWAS share the same covariates. As prior studies have shown a strong genetic correlation between clinically-defined AD and family history of AD or dementia [3], we assembled evidence of association between the genes containing our association signals and AD-related phenotypes in different populations and under different statistical models using GenomicsDB (v.40beta; https://beta.niagads.org/genomics/app), which provides summary statistics from AD-related studies from NIAGADS [17] and the NHGRI-EBI GWAS catalog [18].
2.5. Evidence for validation from expression studies
We hypothesize that the variants identified by our GWAS may influence dementia risk by altering gene expression; if so, we would expect to see evidence for differential gene expression between AD cases and controls in the appropriate tissues or cells. Lead variants from our GWAS were extracted from AD-specific eQTL studies to nominate genes whose expression they may influence. We then determined if these genes were differentially expressed between those with and without dementia or AD within several recent studies of gene expression.
Evidence for eQTLs was collected from an Accelerating Medical Partnerships for Alzheimer’s Disease (AMP-AD) consortium meta-analysis of RNA-sequencing (RNA-seq) data of brain tissues (N = 2,051) collected from the Mayo study [19], the Religious Orders Study (ROS) and Rush Memory and Aging (MAP) study [20], and the Mount Sinai Brain Bank study (MSBB)[21][22] (https://www.synapse.org/#!Synapse:syn17015233). Differentially expressed genes (DEGs) were identified in the AMP-AD RNA-seq data by a sex-stratified meta-analysis comparing AD cases and controls (N = 2,114) [23] (https://www.synapse.org/#!Synapse:syn11914606). AMP-AD eQTLs and DEGs were defined using the reported FDR < 0.05 thresholds. DEGs from a study of single-cell RNA-seq data collected from ROS samples with little to no AD pathology, early-stage, or late-stage AD pathology (N=48) [24] were defined by a FDR-adjusted P < 0.01 and absolute log2 fold change (log2FC) >0.25. Expression microarray data from the frontal cortex were used to identify DEGs for AD, vascular dementia (VaD), and FTD versus controls (N=140) [5], with DEGs defined as those with a log2FC ≥ 1.2 and P ≤ 0.05. Additional details for each of these studies are provided in Supplemental Methods.
3. Results
3.1. Sample summary
Table 1 describes the characteristics of the WHI sample, stratified by dementia status. These 11,085 women have a mean age at baseline of 62 years, self-identifying as non-Hispanic white (81%), African American (13%), Hispanic (3%), Asian/Pacific Islander (2%), Native American/Alaskan (0.5%), or other (0.7%). Compared to women without dementia, those with dementia were older, less likely to be current smokers, and more likely to have had an incident stroke during follow-up. As expected, the APOE ε4 (rs429358) carrier frequency was higher among dementia cases than controls (33% vs. 22%) who more closely resembled reference populations (26%; non-Finnish Europeans [25]). Similarly, the frequency of APOE ε2 (rs7412) was higher among controls and these reference samples (14% and 15%) than dementia cases (11%).
Table 1.
Summary statistics and sample description for the WHI data set.
| Characteristic | No dementia | Dementia |
|---|---|---|
| N | 9,477 | 1,608 |
| Age in years (SD) | 66.2 (6.9) | 68.4 (5.2) |
| Race/ethnicity (%) | ||
| White | 81 | 84 |
| Black | 13 | 11 |
| Hispanic | 3 | 2 |
| Asian | 2 | 1 |
| Other | 1 | 1 |
| Current smoker (%) | 8.1 | 5.8 |
| Body mass index (kg/m2) | 28.9 | 28.0 |
| Incident stroke (%) | 43 | 50 |
| Incident venous thromboembolism (%) | 11 | 10 |
| Diabetes (%) | 6.8 | 6.2 |
| Systolic blood pressure (mmHg) | 132 | 132 |
| Diastolic blood pressure (mmHg) | 76 | 75 |
| Treated Hypertension (%) | 45.3 | 44.3 |
| APOE ε2 carrier (%) | 14 | 11 |
| APOE ε4 carrier (%) | 22 | 33 |
3.2. Single variant association testing
The missense variant defining the APOE ε4 allele and dementia was strongly associated with dementia (rs429358: OR = 1.89, 95%CI 1.68–2.12, P = 7.69 × 10−27), along with 30 additional variants with P < 5 × 10−8 (Supplemental Figure 1; Table 2). We also observe significant evidence of association between three additional loci and dementia: an SNV at the MYH11 locus, nine SNVs at the FZD3 locus, and an SNV at the SORCS3 locus. Within intron 8 of MYH11, rs10852375 (16:15778040, A>T, DP = 34, MAF=0.41) is significantly associated with an increased risk of dementia (OR = 1.27, 95%CI: 1.17– 1.37, P = 1.70 × 10−9). The association at FZD3 is led by a downstream variant, rs352214 (8:28577124, C>G, DP = 35, MAF = 0.41), significantly associated with reduced risk of dementia (OR = 0.81, 95%CI: 0.75– 0.87, P = 4.15 × 10−8, MAF = 0.41). Within intron 14 of SORCS3, rs76590698 (10:105189362, G>C, DP = 36, MAF = 0.0062) is associated with a sharply increased risk of dementia (OR = 4.36, 95%CI: 2.57–7.40, P = 4.91 × 10−8).
Table 2.
Variants significantly associated with dementia in single-variant association tests.
| Locus | CHR | POS | A1 | A2 | A2 freq | β | SE(β) | OR | P |
|---|---|---|---|---|---|---|---|---|---|
| FZD3 | 8 | 28498654 | C | T | 0.5848 | 0.2130 | 0.0388 | 0.8082 | 4.17E-08 |
| 8 | 28524903 | G | T | 0.5917 | 0.2132 | 0.0390 | 0.8080 | 4.77E-08 | |
| 8 | 28540845 | T | C | 0.5917 | 0.2129 | 0.0390 | 0.8083 | 4.96E-08 | |
| 8 | 28560229 | T | TC | 0.5940 | 0.2135 | 0.0391 | 0.8078 | 4.84E-08 | |
| 8 | 28567614 | A | G | 0.5937 | 0.2140 | 0.0391 | 0.8073 | 4.45E-08 | |
| 8 | 28567992 | A | C | 0.5938 | 0.2138 | 0.0391 | 0.8075 | 4.62E-08 | |
| 8 | 28568746 | G | A | 0.5937 | 0.2140 | 0.0391 | 0.8073 | 4.45E-08 | |
| 8 | 28570481 | T | C | 0.5941 | 0.2139 | 0.0391 | 0.8074 | 4.53E-08 | |
| 8 | 28577124 | C | G | 0.5911 | 0.2141 | 0.0390 | 0.8073 | 4.15E-08 | |
| SORCS3 | 10 | 105189362 | G | C | 0.0062 | 1.4726 | 0.2700 | 4.3605 | 4.91E-08 |
| MYH11 | 16 | 15778040 | A | T | 0.4127 | 0.2363 | 0.0393 | 1.2666 | 1.75E-09 |
| APOE | 19 | 44883210 | G | GTAA | 0.1161 | 0.4800 | 0.0620 | 1.6160 | 9.94E-15 |
| 19 | 44884202 | C | G | 0.1163 | 0.4750 | 0.0617 | 1.6080 | 1.37E-14 | |
| 19 | 44884339 | G | A | 0.1163 | 0.4760 | 0.0617 | 1.6096 | 1.23E-14 | |
| 19 | 44884873 | G | A | 0.1213 | 0.4597 | 0.0606 | 1.5835 | 3.24E-14 | |
| 19 | 44885243 | A | G | 0.2298 | 0.3365 | 0.0474 | 1.4000 | 1.28E-12 | |
| 19 | 44887076 | A | G | 0.2365 | 0.3476 | 0.0475 | 1.4156 | 2.62E-13 | |
| 19 | 44888997 | C | T | 0.1384 | 0.4982 | 0.0579 | 1.6457 | 7.32E-18 | |
| 19 | 44891079 | T | C | 0.1181 | 0.4743 | 0.0615 | 1.6069 | 1.21E-14 | |
| 19 | 44891712 | T | G | 0.2322 | 0.3632 | 0.0477 | 1.4380 | 2.60E-14 | |
| 19 | 44892362 | A | G | 0.1274 | 0.4677 | 0.0593 | 1.5964 | 2.99E-15 | |
| 19 | 44892457 | T | C | 0.2363 | 0.3428 | 0.0475 | 1.4090 | 5.42E-13 | |
| 19 | 44892587 | G | A | 0.0890 | 0.4428 | 0.0690 | 1.5571 | 1.43E-10 | |
| 19 | 44892652 | C | G | 0.1230 | 0.4859 | 0.0603 | 1.6257 | 7.71E-16 | |
| 19 | 44892887 | C | T | 0.1248 | 0.4767 | 0.0599 | 1.6108 | 1.66E-15 | |
| 19 | 44892962 | C | T | 0.2338 | 0.3485 | 0.0478 | 1.4169 | 3.18E-13 | |
| 19 | 44893408 | G | T | 0.2046 | 0.3383 | 0.0489 | 1.4025 | 4.64E-12 | |
| 19 | 44903416 | G | A | 0.2793 | 0.2717 | 0.0433 | 1.3123 | 3.34E-10 | |
| 19 | 44906745 | G | A | 0.0915 | 0.6185 | 0.0698 | 1.8562 | 7.53E-19 | |
| 19 | 44908684 | T | C | 0.1365 | 0.6367 | 0.0594 | 1.8903 | 7.69E-27 | |
| 19 | 44912456 | G | A | 0.1142 | 0.4912 | 0.0632 | 1.6342 | 7.43E-15 | |
| 19 | 44912678 | G | T | 0.1143 | 0.4896 | 0.0631 | 1.6317 | 8.71E-15 | |
| 19 | 44912921 | G | T | 0.2397 | 0.2972 | 0.0465 | 1.3460 | 1.70E-10 | |
| 19 | 44913484 | C | T | 0.2435 | 0.3108 | 0.0465 | 1.3645 | 2.39E-11 | |
| 19 | 44915533 | T | C | 0.2197 | 0.2892 | 0.0471 | 1.3353 | 8.62E-10 | |
| 19 | 44916825 | A | C | 0.1034 | 0.5508 | 0.0660 | 1.7347 | 7.31E-17 | |
| 19 | 44917997 | G | A | 0.1210 | 0.4450 | 0.0605 | 1.5605 | 1.92E-13 | |
| 19 | 44918903 | C | G | 0.1514 | 0.5068 | 0.0557 | 1.6600 | 9.60E-20 | |
| 19 | 44919589 | G | A | 0.1662 | 0.4569 | 0.0535 | 1.5791 | 1.43E-17 | |
| 19 | 44919689 | A | G | 0.1670 | 0.4615 | 0.0535 | 1.5865 | 5.95E-18 | |
| 19 | 44923868 | T | A | 0.1222 | 0.4409 | 0.0603 | 1.5541 | 2.60E-13 | |
| 19 | 44924977 | G | A | 0.1347 | 0.3856 | 0.0576 | 1.4706 | 2.21E-11 |
CHR: chromosome, POS: hg38 sequence position, A1: allele 1, A2: allele 2, A2 freq: frequency of the A2 allele, β: beta coefficient from analysis model, SE: standard error, OR: odds ratio for minor allele. Lead SNVs are defined by the smallest P value within an associated locus and are highlighted in bold font.
For the two common SNVs associated with dementia outside the APOE region, rs10852375 and rs352214, we performed additional analyses stratified by APOE ε4 carrier status, stroke status, prior use of hormone therapy (past or current vs. never), and self-reported ancestry (Supplemental Tables 1–4). We observed no evidence of an interaction with either APOE ε4 carrier or stroke status (P > 0.05, Supplemental Tables 1 and 3), though there was significant evidence of an interaction between rs352214 and hormone therapy status (P = 0.041, Supplemental Table 2), where the minor allele was more strongly associated with reduced risk of dementia among hormone therapy users (OR=0.82, 95%CI: 0.75– 0.89; P <0.0001) than non-users (OR=0.92, 95%CI: 0.84–1.00; P =0.04). For both rs10852375 and rs352214, the direction of effect was consistent across ancestry groups. rs1085375 was significantly associated with dementia in the European- (OR=1.27, 95%CI: 1.16–1.39, P <1E-05), African- (OR=1.29, 95%CI: 1.02–1.63, P =0.0360) and Hispanic-Americans (OR=2.16, 95%CI: 1.22–3.83, P =0.0090), while rs352214 was associated with dementia in both the European-American (OR=0.79, 95%CI: 0.72–0.87, P <1E-05) and Other (OR=0.47, 95%CI: 0.23–0.99, P =0.0460) subsets (Supplemental Table 4).
3.3. Gene-based association testing
An association between GOLGA8B and dementia (P =1.22 × 10−6) reached the genome-wide significance threshold and was driven by eight rare coding changes. These rare variants have a maximum alternate allele count of two, five of which were unique to cases (Supplemental Table 5). Most variants observed in cases were either frameshift or inframe deletions, while most variants observed in controls were base-pair substitutions. None of the genes at the single-variant GWAS loci exhibited association with dementia in the gene-based analyses: P-values for APOE, MYH11, FZD3, and SORCS3 were 0.36, 0.60, 0.23, and 0.08, respectively.
3.4. Variant annotation.
rs10852375 falls within intron 8 of MYH11 (Figure 1A) and both a CTCF binding site and a TFBS active in bipolar neurons and ≥ 12 cell/tissue types. rs10852375 occurs at a high information position across seven TFBS motifs and is a known eQTL for NDE1 and MYH11 in lymphoblastic cell lines, for MYH11 in whole blood, NPIPA5 in the thyroid, and AF001548.5 in the brain and other cell types. rs352214 falls within a large LD block 2.9kb 3’ of FZD3 (Figure 1B) and enhancer histone marks in five cell/tissue types including derived neurospheres. rs352214 is predicted to alter five TFBS motifs and is a known eQTL for FZD3 in testis. Finally, the rare variant rs76590698 falls within intron 14 of SORCS3 (Figure 1C) and a promoter flanking region active in seven cell/tissue types including astrocytes, has enhancer-like histone marks in 10 tissue types including brain, and is at a high information position for four TFBS motifs. These features suggest that the functional consequences of our lead SNVs are likely to be regulatory and influence gene expression rather than protein structure.
Figure 1. LocusZoom plots for rs10852375 (A) and rs352214 (B).
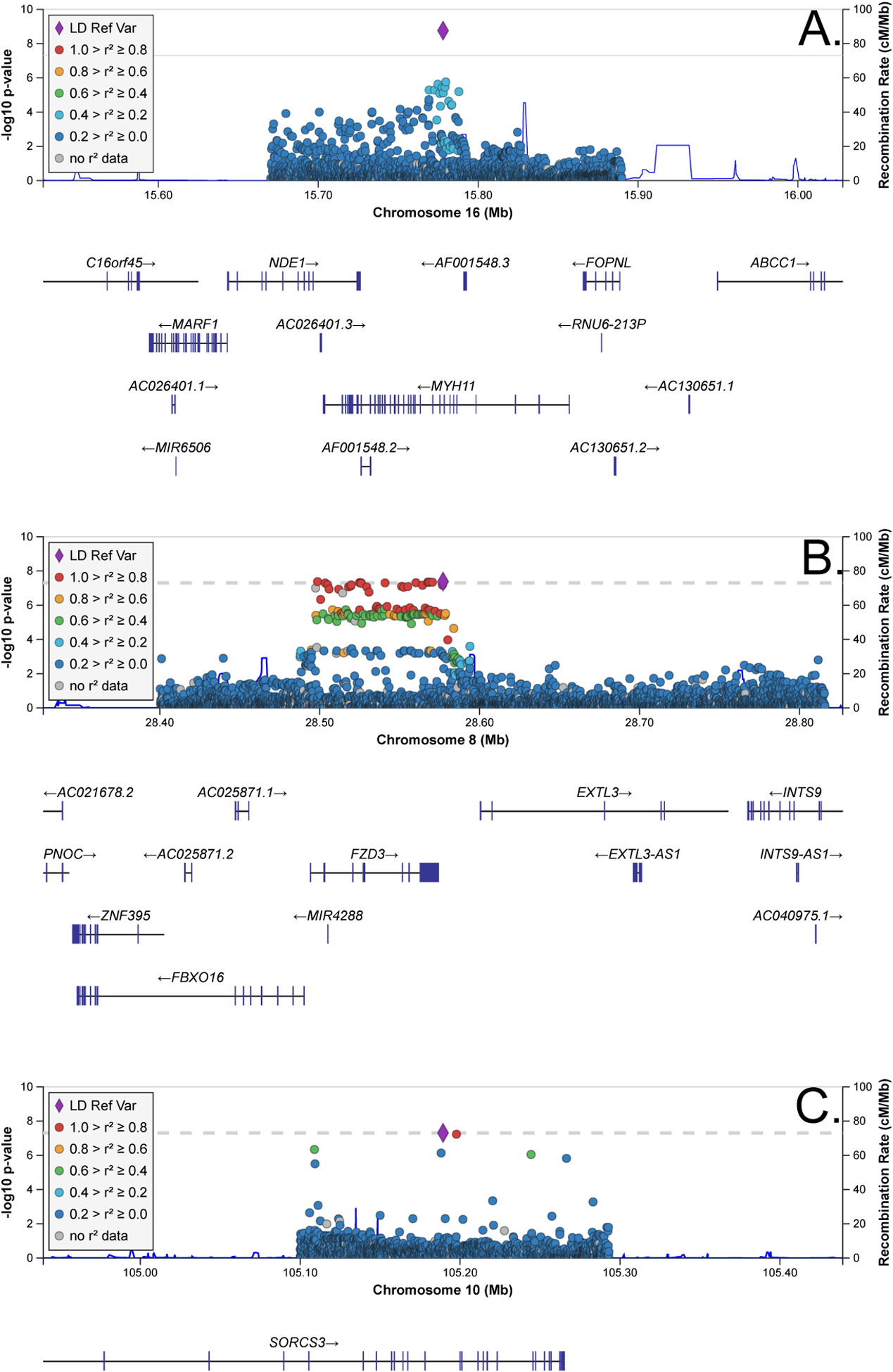
Chromosomal positions are given with respect to the hg38 genome reference.
3.5. Evidence for validation from GWAS
Variants within FZD3, MYH11, SORCS3, and GOLGA8B have prior evidence for association with AD-related traits. Genetic variants in SORCS3 are significantly associated with AD [26, 27], although large GWAS have not identified significant evidence of association between MYH11, FZD3, or GOLGA8B and AD (Supplemental Table 6). We do observe nominally significant evidence (P < 0.001) for association between FZD3 variants and AD in Europeans [28] and African-Americans [29], between MYH11 and AD in studies representing samples with European [30] or trans-ethnic ancestry [31], and between GOLGA8B variants and the presence of neuritic plaques characteristic of AD [32].
3.6. Evidence for validation in expression studies
Both rs10852375 and rs352214 are significant eQTLs while the gene implicated by the rare variant rs76590698 is differentially expressed in the AMP-AD study (Tables 3 and 4). Although rs76590698 was not observed in the AMP-AD eQTL data, both rs10852375 and rs352214 were significant eQTLs in the larger ROSMAP dorsolateral prefrontal cortex (DLPFC) sample and were nominally significant eQTLs in the smaller Mayo cerebellum and temporal cortex cohorts (CER, TCX; P < 0.05, Table 3). rs10852375 was a significant eQTL for NPIPA1 (β = −0.1833, FDR = 0.0487), NPIPA5 (β = 0.2798, FDR = 1.60 × 10−4), and an unprocessed pseudogene AC138969.2 (β = 0.1915, FDR = 0.0350) in the DLPFC, while rs352214 was a significant eQTL for FZD3 (β = −0.7323, FDR = 2.10E-35), CCDC25 (β = −0.2001, FDR = 0.0151), and DUSP4 (β = −0.1877, FDR = 0.0344). These brain-specific results are consistent with those in other tissue types (above) and identify new genes whose expression is associated with either common SNV.
Table 3.
Evidence rs10852375 and rs352214 are eQTLs in brain tissues.
| Tissue | rsID | GENE | SYMBOL | A1 | A2 | B | P | FDR |
|---|---|---|---|---|---|---|---|---|
| TCX | rs352214 | ENSG00000259366 | . | C | G | −0.2201 | 1.02E-02 | 3.23E-01 |
| TCX | rs352214 | ENSG00000279302 | MIR3622A | C | G | −0.1821 | 4.32E-02 | 5.78E-01 |
| DLPFC | rs352214 | ENSG00000147419 | CCDC25 | C | G | −0.2001 | 6.15E-04 | 1.51E-02 |
| DLPFC | rs352214 | ENSG00000120875 | DUSP4 | C | G | −0.1877 | 1.65E-03 | 3.44E-02 |
| DLPFC | rs352214 | ENSG00000104290 | FZD3 | C | G | −0.7323 | 1.65E-38 | 2.10E-35 |
| DLPFC | rs352214 | ENSG00000279302 | MIR3622A | C | G | −0.1268 | 3.85E-02 | 3.24E-01 |
| CER | rs10852375 | ENSG00000261819 | . | A | T | −0.2354 | 6.41E-03 | 2.51E-01 |
| CER | rs10852375 | ENSG00000091262 | ABCC6 | A | T | 0.1835 | 3.33E-02 | 5.33E-01 |
| CER | rs10852375 | ENSG00000133393 | FOPNL | A | T | −0.1877 | 2.92E-02 | 5.10E-01 |
| CER | rs10852375 | ENSG00000179889 | PDXDC1 | A | T | −0.1998 | 2.05E-02 | 4.44E-01 |
| TCX | rs10852375 | ENSG00000085721 | RRN3 | A | T | −0.1831 | 3.36E-02 | 5.83E-01 |
| DLPFC | rs10852375 | ENSG00000270580 | . | A | T | −0.1260 | 3.42E-02 | 3.61E-01 |
| DLPFC | rs10852375 | ENSG00000227827 | AC138969.2 | A | T | 0.1915 | 1.20E-03 | 3.50E-02 |
| DLPFC | rs10852375 | ENSG00000183426 | NPIPA1 | A | T | −0.1833 | 1.79E-03 | 4.87E-02 |
| DLPFC | rs10852375 | ENSG00000183793 | NPIPA5 | A | T | 0.2798 | 2.54E-06 | 1.60E-04 |
| DLPFC | rs10852375 | ENSG00000179889 | PDXDC1 | A | T | −0.1736 | 3.63E-03 | 8.42E-02 |
All results from the AMP-AD studies study where the evidence for an eQTL was nominally significant (P < 0.05; https://www.synapse.org/#!Synapse:syn17015233). Results with FDR < 0.05 are highlighted in bold font. A1: allele 1, A2: allele 2, β: beta coefficient from regression model, FDR: false discovery rate, CER: cerebellum from Mayo study, DLPFC: dorsolateral prefrontal cortex from ROSMAP study, TCX: temporal cortex from Mayo study.
Table 4.
Evidence our candidate genes are differentially expressed in brain between AD cases and controls AMP-AD data.
| Fixed effects model | ||||
|---|---|---|---|---|
| Symbol | Sex stratum | Z | P | FDR |
| AC138969.2 | FEMALE | −2.4354 | 1.49E-02 | 4.98E-02 |
| CCDC25 | FEMALE | −2.3297 | 1.98E-02 | 6.18E-02 |
| DUSP4 | FEMALE | −5.7706 | 7.90E-09 | 8.26E-07 |
| DUSP4 | MALE | −3.5100 | 4.00E-04 | 3.50E-03 |
| GOLGA8B | FEMALE | −5.5125 | 3.54E-08 | 2.58E-06 |
| GOLGA8B | MALE | −3.0663 | 2.17E-03 | 1.16E-02 |
| NDE1 | FEMALE | 3.8782 | 1.00E-04 | 1.10E-03 |
| NPIPA1 | MALE | −2.9887 | 2.80E-03 | 1.41E-02 |
| SORCS3 | FEMALE | −5.9265 | 3.10E-09 | 4.31E-07 |
Results from the AMP-AD meta-analysis where p < 0.05 (https://www.synapse.org/#!Synapse:syn11914606). Results with FDR < 0.05 are highlighted in bold. Z: Z statistic value, P: P value, FDR: false discovery rate. Results were comparable in the random effects model, with slightly weaker P values (data not shown).
We investigated the evidence for differential gene expression within AD-related analyses for 11 genes implicated by our genome scans either directly (MYH11, FZD3, GOLGA8B, SORCS3) or by eQTLs (AC138969.2, AF001548.5, CCDC25, DUSP4, FZD3, MYH11, NDE1, NPIPA1, NPIPA5). Within the microarray study of frontal cortex, MYH11 was differentially expressed in both AD (avg. log2FC = 1.31, min P = 0.0005) and FTD (log2FC = 1.51, P =0.0434), while DUSP4 was differentially expressed in VaD (log2FC = −1.86, P =0.0049). Seven of the 11 genes were significant DEGs in the AMP-AD meta-analysis (AC138969.2, CCDC25, DUSP4, GOLGA8B, NDE1, NPIPA1, SORCS3; Table 4). Five of these significant results were in the female-specific analyses. In each case, the association pattern was similar but slightly weaker in the random effects model (not shown). Nine of the 11 genes were significant DEGs in neurons (FDR < 0.05; Table 5). Among the excitatory neurons, DUSP4, MYH11, and NPIPA5 were significant DEGs when those with and without AD pathology were compared, CCDC25, DUSP4, and MYH11 were significant DEGs when those with early-stage pathology were compared with those without AD pathology, and CCDC25, DUSP4, and NPIPA5 were significant DEGs when those with early- and late-stage pathology were compared. In the inhibitory neurons, DUSP4 and MYH11 were significant DEGs in comparisons of those with and without AD pathology, and FZD3 was a significant DEG when those with early- vs. late-stage pathology were compared. Across the expression studies, validation support was observed for all but AF001548.5, with DUSP4, MYH11, CCDC25, GOLGA8B, and NPIPA5 having support from multiple sources.
Table 5.
Evidence our candidate genes are differentially expressed between samples with varying levels of AD pathology in neurons [24].
| Excitatory Neurons | Inhibitory Neurons | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| absent vs. present | absent vs. early | early vs. late | absent vs. present | absent vs. early | early vs. late | |||||||
| FDR | log2FC | FDR | log2FC | FDR | log2FC | FDR | log2FC | FDR | log2FC | FDR | log2FC | |
| CCDC25 | 6.18E-56 | −0.23 | 2.20E-72 | −0.37 | 1.22E-20 | 0.38 | 6.88E-05 | −0.11 | 2.39E-08 | −0.06 | 2.62E-05 | −0.15 |
| DUSP4 | 8.70E-14 | −0.58 | 6.19E-16 | −0.84 | 2.66E-04 | 0.66 | 2.18E-04 | −0.36 | 2.96E-02 | −0.02 | 1.13E-02 | −1.27 |
| FZD3 | 9.58E-26 | 0.08 | 5.34E-48 | 0.02 | 7.71E-28 | 0.18 | 5.43E-03 | 0.11 | 1.33E-09 | 0.01 | 7.27E-13 | 0.27 |
| GOLGA8B | 1.25E-51 | −0.08 | 1.12E-38 | 0.03 | 8.36E-01 | −0.37 | 3.76E-03 | 0.01 | 2.03E-03 | 0.12 | 3.33E-01 | −0.35 |
| MYH11 | 5.26E-09 | −0.52 | 2.28E-10 | −0.47 | 7.19E-03 | −0.17 | 3.09E-04 | −0.31 | 2.51E-03 | −0.19 | 7.26E-01 | −0.36 |
| NDE1 | 1.09E-02 | 0.24 | 1.04E-06 | 0.19 | 1.90E-08 | 0.16 | 1.87E-01 | −0.12 | 1.27E-02 | −0.11 | 1.10E-02 | −0.04 |
| NPIPA1 | 5.88E-14 | 0.14 | 1.31E-26 | 0.07 | 2.53E-18 | 0.18 | 1.47E-01 | 0.08 | 7.44E-03 | 0.14 | 7.89E-03 | −0.17 |
| NPIPA5 | 2.15E-03 | 0.33 | 4.06E-01 | 0.58 | 1.72E-05 | −0.91 | 4.57E-02 | 0.19 | 3.87E-01 | 0.61 | 3.96E-02 | −1.78 |
| SORCS3 | 1.60E-87 | −0.17 | 1.43E-74 | −0.11 | 4.27E-02 | −0.18 | 4.74E-08 | −0.04 | 4.40E-06 | 0.12 | 7.09E-01 | −0.50 |
FDR: false discovery rate-adjusted P-value from two-sided Wilcoxon-rank-sum test, log2FC: log2 fold change between comparison groups, bold: indicates when the FDR-adjusted P-value is < 0.01 and the absolute value of log2FC > 0.25, pathology: refers to evidence of Alzheimer’s disease related pathology in postmortem brain.
4. Discussion
Variants within APOE, MYH11, FZD3, and SORCS3 were associated with dementia in the WHI. Two of these non-coding SNVs are common and associated with allele-specific differences in gene expression and appear to fall within regulatory elements. Evidence for differential gene expression between AD cases and controls supported the potential role for MYH11, FZD3, GOLGA8B, and SORCS3 in AD pathogenesis, along with AC138969.2, CCDC25, DUSP4, NDE1, and NPIPA1, and NPIPA5. Expression array studies of brain also supported a role for DUSP4 in VaD, and for MYH11 in FTD. Several of these genes are differentially expressed between those with varying levels of AD pathology in neurons. Gene-based tests detected significant association between dementia and rare coding changes in GOLGA8B, a gene whose expression differed significantly in the brain between AD cases and controls.
Our association of FZD3, MYH11, SORCS3, and GOLGA8B with dementia is supported by functional studies. Both MYH11 and FZD3 are differentially expressed in AD brain (ex. [33, 34]), and upregulation of FZD3 relieves the phenotype in mouse models of AD [35]. SORCS3 is differentially expressed in AD brain and is involved in APP processing [26]. Mouse models of SORCS3 have shown it is downregulated after Aβ plaque formation [36] and plays important roles in memory formation and synaptic plasticity [37]. Finally, GOLGA8B is significantly downregulated in the TCX in AD [38]. The genes identified by brain-based eQTL analyses of our lead variants also have biological ties to AD. DUSP4 is differentially expressed in AD hippocampus [39], and knock-out mice have impaired working memory and hippocampal function [40]. NDE1 expression levels in blood is a potential biomarker for AD [33], and CCDC25 is differentially expressed in the entorhinal cortex of mice with APOE ε3/ε4 versus ε3/ε3 genotypes [41].
Prior GWAS of AD have not implicated FZD3, MYH11, or GOLGA8B, possibly for reasons explained by our study design. The ascertainment for vascular disease in our sample has enriched for variants associated with both dementia and vascular disease. We observe variants within MYH11, FZD3, and SORCS3 are associated with shared risk factors for stroke and dementia, including smoking, hypertension, and diabetes-related traits [6, 7, 42–46], though there was no evidence that the lead SNVs at MYH11 or FZD3 were significantly associated with smoking, BMI, blood pressure, diabetes, LDL cholesterol, stroke, VTE, or coronary heart disease in WHI (data not shown). We note a recent AD GWAS identified significant associations unique to those with or without hypertension [47], suggesting GWAS stratified by different AD risk factors may find novel results. Similarly, recent studies highlight shared genetic architecture and pathways between AD and other causes of dementia [4, 5]; GWAS for dementia as defined in our study may enrich for those features shared across causes of dementia rather than those specific to AD. The WHI cohort is also exclusively female. Although the evidence for a significant sex effect on risk of AD is inconsistent [8], there is evidence for sex differences in the effect of APOE ε4, hypertension, and diabetes on risk of AD. In older WHI participants, hormone therapy was previously associated with cognitive impairment, persisting for years after medications were terminated (ex., [48]). We observed a significant interaction between rs352214 and hormone therapy, where the protective effect on dementia risk was strongest among hormone therapy users. FZD3 is a receptor within the WNT/beta-catenin signaling pathway, playing a role in both neurodegeneration and estrogen biosynthesis [49, 50]. Together, these results suggest alternative study designs may identify additional loci associated with dementia not captured by AD-specific GWAS.
Our study has limitations which likely reduced its power. Our phenotype definition is based upon self-report data and medical records rather than a systematic evaluation by neurologists or neuropathology data. Our dementia phenotype may underreport cases and our cases likely represent several forms of dementia. Phenotypic heterogeneity, as well as difficulty sequencing APOE may explain the weaker estimated effect size at rs429358 in our study compared to typical AD GWAS [2]. Our study was restricted to postmenopausal women and therefore results may not be generalizable to men. Similarly, the scarcity of comparable data sets or GWAS with similar diagnoses, exposures, and covariates for replication analyses makes it more challenging to generalize our results to other populations. Functional studies are needed to determine whether the variants associated with dementia in our study directly influence gene expression, alone or in combination with other variation. Molecular or cellular studies are also needed to assess the consequences of our GOLGA8B variants on the protein’s function.
We have demonstrated the importance of large and diverse WGS data sets to identify genetic risk factors for dementia. We identified a strong association between rs76590698 within SORCS3 and dementia, with large odds ratios for AD comparable to APOE ε4 and TREM2 R47H. Although rs76590698 and the TREM2 variant share both similar effect sizes (OR ~ 4) and allele frequencies in non-Finnish Europeans (0.5% vs. 0.2% [25]). However, rs76590698 is an intronic variant with annotations suggesting a potential effect on gene regulation, while TREM2 R47H is a loss-of-function variant. Furthermore, rs76590698 is much more common in Latino and East Asian populations (4.2% - 5% [25]), highlighting the importance of studying diverse populations when searching for genetic variants influencing disease risk.
In conclusion, this study has shown that genetic variation significantly associated with risk of dementia among post-menopausal women selected for a study of stroke implicate genes which are differentially expressed between AD cases and controls. These loci have previously been associated with shared risk factors for dementia and stroke. Future studies are needed to further investigate the associations between these loci and dementia, the roles the implicated genes may play in AD pathogenesis, and the potential influence of sex, stroke, hormone therapy, and ancestry.
Supplementary Material
Research in Context:
Systematic review:
The authors performed a literature review encompassing pre-prints and published articles and abstracts investigating the relationship between stroke, sex, and dementia. Previous studies provided evidence for shared risk factors between stroke and dementia, and inconsistent evidence for sex-specific effects on dementia risk.
Interpretation:
Genome scans revealed significant associations between genetic variation in APOE, FZD3, GOLGA8B, MYH11, and SORCS3 and dementia risk. Excluding the well-established APOE locus, these loci have not been widely associated with Alzheimer’s disease risk, but they have been associated with cognitive traits, shared risk factors for dementia and stroke, and/or differential gene expression in the brain between Alzheimer’s disease cases and controls. They also find evidence for an interaction between hormone therapy, FZD3 genotype, and dementia risk.
Future directions:
Further studies across diverse populations, stratified by sex, and enriched for known risk factors for dementia may reveal new genetic pathways influencing dementia.
Acknowledgements/Conflicts/Funding Sources
We thank the study participants and their families for their contributions to this work. The authors declare no conflicts of interest. Research funding was provided by National Institute on Aging (NIA) [grant number R01AG059737].
WGS for the TOPMed program was supported by the NHLBI. WGS for “NHLBI TOPMed: Women’s Health Initiative” (phs001237.v1.p1) was performed at the Broad Institute (HHSN268201500014C). Centralized read mapping and genotype calling, along with variant quality metrics and filtering were provided by the TOPMed Informatics Research Center (3R01HL-117626-02S1; contract HHSN268201800002I). Phenotype harmonization, data management, sample-identity QC, and general study coordination were provided by the TOPMed Data Coordinating Center (3R01HL-120393-02S1; contract HHSN268201800001I). We gratefully acknowledge the studies and participants who provided biological samples and data for TOPMed.
The WHI program is funded by NHLBI, the National Institutes of Health, and the U.S. Department of Health and Human Services through contracts, HHSN268201600018C, HHSN268201600001C, HHSN268201600002C, HHSN268201600003C, and HHSN268201600004C. This manuscript was prepared in collaboration with investigators of the WHI, and has been reviewed and/or approved by the WHI. WHI investigators include: Program Office: (National Heart, Lung, and Blood Institute, Bethesda, Maryland) Jacques Rossouw, Shari Ludlam, Joan McGowan, Leslie Ford, and Nancy Geller; Clinical Coordinating Center: (Fred Hutchinson Cancer Research Center, Seattle, WA) Garnet Anderson, Ross Prentice, Andrea LaCroix, and Charles Kooperberg; Investigators and Academic Centers: (Brigham and Women’s Hospital, Harvard Medical School, Boston, MA) JoAnn E. Manson; (MedStar Health Research Institute/Howard University, Washington, DC) Barbara V. Howard; (Stanford Prevention Research Center, Stanford, CA) Marcia L. Stefanick; (The Ohio State University, Columbus, OH) Rebecca Jackson; (University of Arizona, Tucson/Phoenix, AZ) Cynthia A. Thomson; (University at Buffalo, Buffalo, NY) Jean Wactawski-Wende; (University of Florida, Gainesville/Jacksonville, FL) Marian Limacher; (University of Iowa, Iowa City/Davenport, IA) Jennifer Robinson; (University of Pittsburgh, Pittsburgh, PA) Lewis Kuller; (Wake Forest University School of Medicine, Winston-Salem, NC) Sally Shumaker; (University of Nevada, Reno, NV) Robert Brunner; Women’s Health Initiative Memory Study: (Wake Forest University School of Medicine, Winston-Salem, NC) Mark Espeland.
The results published here are in whole or in part based on data obtained from the AMP-AD Knowledge Portal (doi:10.7303/syn2580853). Study data were provided by the following sources: The Mayo Clinic Alzheimers Disease Genetic Studies, led by Dr. Nilufer Taner and Dr. Steven G. Younkin, Mayo Clinic, Jacksonville, FL using samples from the Mayo Clinic Study of Aging, the Mayo Clinic Alzheimers Disease Research Center, and the Mayo Clinic Brain Bank. Data collection was supported through funding by NIA grants P50 AG016574, R01 AG032990, U01 AG046139, R01 AG018023, U01 AG006576, U01 AG006786, R01 AG025711, R01 AG017216, R01 AG003949, NINDS grant R01 NS080820, CurePSP Foundation, and support from Mayo Foundation. Study data includes samples collected through the Sun Health Research Institute Brain and Body Donation Program of Sun City, Arizona. The Brain and Body Donation Program is supported by the National Institute of Neurological Disorders and Stroke (U24 NS072026 National Brain and Tissue Resource for Parkinsons Disease and Related Disorders), the NIA (P30 AG19610 Arizona Alzheimers Disease Core Center), the Arizona Department of Health Services (contract 211002, Arizona Alzheimers Research Center), the Arizona Biomedical Research Commission (contracts 4001, 0011, 05-901 and 1001 to the Arizona Parkinson’s Disease Consortium) and the Michael J. Fox Foundation for Parkinsons Research. Study data were also provided by and the Rush Alzheimer’s Disease Center, Rush University Medical Center, Chicago. Data collection was supported through funding by NIA grants P30AG10161, R01AG15819, R01AG17917, R01AG30146, R01AG36836, U01AG32984, U01AG46152, the Illinois Department of Public Health, and the Translational Genomics Research Institute. Data were also generated from postmortem brain tissue collected through the Mount Sinai VA Medical Center Brain Bank and were provided by Dr. Eric Schadt from Mount Sinai School of Medicine.
Footnotes
Competing Interests: The authors declare no competing interests.
References
- [1].Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science. 1993;261:921–3. [DOI] [PubMed] [Google Scholar]
- [2].Belloy ME, Napolioni V, Greicius MD. A Quarter Century of APOE and Alzheimer’s Disease: Progress to Date and the Path Forward. Neuron. 2019;101:820–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Andrews SJ, Fulton-Howard B, Goate A. Interpretation of risk loci from genome-wide association studies of Alzheimer’s disease. Lancet Neurol. 2020;19:326–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Guerreiro R, Gibbons E, Tabuas-Pereira M, Kun-Rodrigues C, Santo GC, Bras J. Genetic architecture of common non-Alzheimer’s disease dementias. Neurobiol Dis. 2020;142:104946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Santiago JA, Bottero V, Potashkin JA. Transcriptomic and Network Analysis Identifies Shared and Unique Pathways across Dementia Spectrum Disorders. Int J Mol Sci. 2020;21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Attems J, Jellinger KA. The overlap between vascular disease and Alzheimer’s disease--lessons from pathology. BMC Med. 2014;12:206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Hachinski V, Einhaupl K, Ganten D, Alladi S, Brayne C, Stephan BCM, et al. Preventing dementia by preventing stroke: The Berlin Manifesto. Alzheimers Dement. 2019;15:961–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Pike CJ. Sex and the development of Alzheimer’s disease. J Neurosci Res. 2017;95:671–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Design of the Women’s Health Initiative clinical trial and observational study. The Women’s Health Initiative Study Group. Control Clin Trials. 1998;19:61–109. [DOI] [PubMed] [Google Scholar]
- [10].Taliun D, Harris DN, Kessler MD, Carlson J, Szpiech ZA, Torres R, et al. Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program. 2019. [DOI] [PMC free article] [PubMed]
- [11].2020 Alzheimer’s disease facts and figures. Alzheimers Dement. 2020. [DOI] [PubMed] [Google Scholar]
- [12].Zhou W, Nielsen JB, Fritsche LG, Dey R, Gabrielsen ME, Wolford BN, et al. Efficiently controlling for case-control imbalance and sample relatedness in large-scale genetic association studies. Nat Genet. 2018;50:1335–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Lee S, Emond MJ, Bamshad MJ, Barnes KC, Rieder MJ, Nickerson DA, et al. Optimal unified approach for rare-variant association testing with application to small-sample case-control whole-exome sequencing studies. Am J Hum Genet. 2012;91:224–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GR, Thormann A, et al. The Ensembl Variant Effect Predictor. Genome Biol. 2016;17:122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Zerbino DR, Wilder SP, Johnson N, Juettemann T, Flicek PR. The ensembl regulatory build. Genome Biol. 2015;16:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Konig IR. Validation in genetic association studies. Brief Bioinform. 2011;12:253–8. [DOI] [PubMed] [Google Scholar]
- [17].Kuzma A, Valladares O, Cweibel R, Greenfest-Allen E, Childress DM, Malamon J, et al. NIAGADS: The NIA Genetics of Alzheimer’s Disease Data Storage Site. Alzheimers Dement. 2016;12:1200–3. [Google Scholar]
- [18].Buniello A, MacArthur JAL, Cerezo M, Harris LW, Hayhurst J, Malangone C, et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019;47:D1005–D12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Allen M, Wang X, Burgess JD, Watzlawik J, Serie DJ, Younkin CS, et al. Conserved brain myelination networks are altered in Alzheimer’s and other neurodegenerative diseases. Alzheimers Dement. 2018;14:352–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].De Jager PL, Ma Y, McCabe C, Xu J, Vardarajan BN, Felsky D, et al. A multi-omic atlas of the human frontal cortex for aging and Alzheimer’s disease research. Sci Data. 2018;5:180142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Wang M, Beckmann ND, Roussos P, Wang E, Zhou X, Wang Q, et al. The Mount Sinai cohort of large-scale genomic, transcriptomic and proteomic data in Alzheimer’s disease. Sci Data. 2018;5:180185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Sieberts SK, Perumal T, Carrasquillo MM, Allen M, Reddy JS, Hoffman GE, et al. Large eQTL meta-analysis reveals differing patterns between cerebral cortical and cerebellar brain regions. 2019. [DOI] [PMC free article] [PubMed]
- [23].Logsdon BA, Perumal TM, Swarup V, Wang M, Funk C, Gaiteri C, et al. Meta-analysis of the human brain transcriptome identifies heterogeneity across human AD coexpression modules robust to sample collection and methodological approach. 2019.
- [24].Mathys H, Davila-Velderrain J, Peng Z, Gao F, Mohammadi S, Young JZ, et al. Single-cell transcriptomic analysis of Alzheimer’s disease. Nature. 2019;570:332–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alfoldi J, Wang Q, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Reitz C, Tosto G, Vardarajan B, Rogaeva E, Ghani M, Rogers RS, et al. Independent and epistatic effects of variants in VPS10-d receptors on Alzheimer disease risk and processing of the amyloid precursor protein (APP). Transl Psychiatry. 2013;3:e256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Herold C, Hooli BV, Mullin K, Liu T, Roehr JT, Mattheisen M, et al. Family-based association analyses of imputed genotypes reveal genome-wide significant association of Alzheimer’s disease with OSBPL6, PTPRG, and PDCL3. Mol Psychiatry. 2016;21:1608–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet. 2013;45:1452–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Reitz C, Jun G, Naj A, Rajbhandary R, Vardarajan BN, Wang LS, et al. Variants in the ATP-binding cassette transporter (ABCA7), apolipoprotein E 4,and the risk of late-onset Alzheimer disease in African Americans. JAMA. 2013;309:1483–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Jun G, Ibrahim-Verbaas CA, Vronskaya M, Lambert JC, Chung J, Naj AC, et al. A novel Alzheimer disease locus located near the gene encoding tau protein. Mol Psychiatry. 2016;21:108–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Jun GR, Chung J, Mez J, Barber R, Beecham GW, Bennett DA, et al. Transethnic genome-wide scan identifies novel Alzheimer’s disease loci. Alzheimers Dement. 2017;13:727–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Beecham GW, Hamilton K, Naj AC, Martin ER, Huentelman M, Myers AJ, et al. Genome-wide association meta-analysis of neuropathologic features of Alzheimer’s disease and related dementias. PLoS Genet. 2014;10:e1004606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Bai Z, Stamova B, Xu H, Ander BP, Wang J, Jickling GC, et al. Distinctive RNA expression profiles in blood associated with Alzheimer disease after accounting for white matter hyperintensities. Alzheimer Dis Assoc Disord. 2014;28:226–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Folke J, Pakkenberg B, Brudek T. Impaired Wnt Signaling in the Prefrontal Cortex of Alzheimer’s Disease. Mol Neurobiol. 2019;56:873–91. [DOI] [PubMed] [Google Scholar]
- [35].Zhang L, Fang Y, Cheng X, Lian YJ, Xu HL. Silencing of Long Noncoding RNA SOX21-AS1 Relieves Neuronal Oxidative Stress Injury in Mice with Alzheimer’s Disease by Upregulating FZD3/5 via the Wnt Signaling Pathway. Mol Neurobiol. 2019;56:3522–37. [DOI] [PubMed] [Google Scholar]
- [36].Hermey G, Hoffmeister-Ullerich SA, Merz B, Gross D, Kuhl D, Kins S. Amyloidosis causes downregulation of SorLA, SorCS1 and SorCS3 expression in mice. Biol Chem. 2019;400:1181–9. [DOI] [PubMed] [Google Scholar]
- [37].Breiderhoff T, Christiansen GB, Pallesen LT, Vaegter C, Nykjaer A, Holm MM, et al. Sortilin-related receptor SORCS3 is a postsynaptic modulator of synaptic depression and fear extinction. PLoS One. 2013;8:e75006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Tan MG, Chua WT, Esiri MM, Smith AD, Vinters HV, Lai MK. Genome wide profiling of altered gene expression in the neocortex of Alzheimer’s disease. J Neurosci Res. 2010;88:1157–69. [DOI] [PubMed] [Google Scholar]
- [39].Annese A, Manzari C, Lionetti C, Picardi E, Horner DS, Chiara M, et al. Whole transcriptome profiling of Late-Onset Alzheimer’s Disease patients provides insights into the molecular changes involved in the disease. Sci Rep. 2018;8:4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Abdul Rahman NZ, Greenwood SM, Brett RR, Tossell K, Ungless MA, Plevin R, et al. Mitogen-Activated Protein Kinase Phosphatase-2 Deletion Impairs Synaptic Plasticity and Hippocampal-Dependent Memory. J Neurosci. 2016;36:2348–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Nuriel T, Larrea D, Guilfoyle DN, Pirhaji L, Shannon K, Arain H, et al. APOE4 is Associated with Differential Regional Vulnerability to Bioenergetic Deficits in Aged APOE Mice. 2019. [DOI] [PMC free article] [PubMed]
- [42].Jin YP, Di Legge S, Ostbye T, Feightner JW, Hachinski V. The reciprocal risks of stroke and cognitive impairment in an elderly population. Alzheimers Dement. 2006;2:171–8. [DOI] [PubMed] [Google Scholar]
- [43].Giri A, Hellwege JN, Keaton JM, Park J, Qiu C, Warren HR, et al. Trans-ethnic association study of blood pressure determinants in over 750,000 individuals. Nat Genet. 2019;51:51–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Kichaev G, Bhatia G, Loh PR, Gazal S, Burch K, Freund MK, et al. Leveraging Polygenic Functional Enrichment to Improve GWAS Power. Am J Hum Genet. 2019;104:65–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Liu M, Jiang Y, Wedow R, Li Y, Brazel DM, Chen F, et al. Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use. Nat Genet. 2019;51:237–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Gerlini R, Berti L, Darr J, Lassi M, Brandmaier S, Fritsche L, et al. Glucose tolerance and insulin sensitivity define adipocyte transcriptional programs in human obesity. Mol Metab. 2018;18:42–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Nazarian A, Arbeev KG, Yashkin AP, Kulminski AM. Genetic heterogeneity of Alzheimer’s disease in subjects with and without hypertension. Geroscience. 2019;41:137–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Espeland MA, Rapp SR, Manson JE, Goveas JS, Shumaker SA, Hayden KM, et al. Long-term Effects on Cognitive Trajectories of Postmenopausal Hormone Therapy in Two Age Groups. J Gerontol A Biol Sci Med Sci. 2017;72:838–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Inestrosa NC, Varela-Nallar L, Grabowski CP, Colombres M. Synaptotoxicity in Alzheimer’s disease: the Wnt signaling pathway as a molecular target. IUBMB Life. 2007;59:316–21. [DOI] [PubMed] [Google Scholar]
- [50].Qiao GY, Dong BW, Zhu CJ, Yan CY, Chen BL. Deregulation of WNT2/FZD3/beta-catenin pathway compromises the estrogen synthesis in cumulus cells from patients with polycystic ovary syndrome. Biochem Biophys Res Commun. 2017;493:847–54. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


