Abstract
Purpose:
Determination of genotypic/phenotypic features of GATAD2B-associated neurodevelopmental disorder (GAND).
Methods:
Fifty GAND subjects were evaluated to determine consistent genotypic/phenotypic features. Immunoprecipitation assays utilizing in vitro transcription-translation products were used to evaluate GATAD2B missense variants’ ability to interact with binding partners within the Nucleosome Remodeling and Deacetylase (NuRD) complex.
Results:
Subjects had clinical findings that included macrocephaly, hypotonia, intellectual disability, neonatal feeding issues, polyhydramnios, apraxia of speech, epilepsy, and bicuspid aortic valves. Forty-one novel GATAD2B variants were identified with multiple variant types (nonsense, truncating frameshift, splice-site variants, deletions and missense). Seven subjects were identified with missense variants that localized within two Conserved Region domains (CR1 or CR2) of the GATAD2B protein. Immunoprecipitation assays revealed several of these missense variants disrupted GATAD2B interactions with its NuRD complex binding partners.
Conclusions:
A consistent GAND phenotype was caused by a range of genetic variants in GATAD2B that include loss-of-function and missense subtypes. Missense variants were present in conserved region domains that disrupted assembly of NuRD complex proteins. GAND’s clinical phenotype had substantial clinical overlap with other disorders associated with the NuRD complex that involve CHD3 and CHD4, with clinical features of hypotonia, intellectual disability, cardiac defects, childhood apraxia of speech, and macrocephaly.
Keywords: GATAD2B, NuRD complex, Apraxia of speech, Chromatin remodeling, Macrocephaly
INTRODUCTION
The Nucleosome Remodeling and Deacetylase (NuRD) Complex is involved in the management of genomic integrity, stem cell differentiation, and neurodevelopment.1,2 This important multi-enzyme complex regulates transcription by linking two independent chromatin-regulating activities: histone deacetylase and ATP-dependent nucleosome remodeling activity.1,3 Several neurodevelopmental disorders have been associated with variants in NuRD subunit proteins that include CHD3, CHD4, and GATAD2B.4–8 Previously identified clinical features shared by each of these dominant disorders include intellectual disability, motor delays and distinct facies.4–8 De novo CHD4 variants were also associated with hearing loss, bony fusions, palatal abnormalities, hypogonadotrophic hypogonadism, and heart defects (MIM# 617159; CHD4-related syndrome (CHD4RS) or Sifrim-Hitz-Weiss syndrome (SIHIWES));5–7 while de novo CHD3 variants were associated with neonatal feeding issues, childhood apraxia of speech, joint laxity, undescended testes, and high forehead with frontal bossing (MIM#618205; CHD3-related syndrome (CHD3RS) or Snijders Blok-Campeau syndrome (SNIBCPS)).4 Both disorders were also associated with enlarged CSF spaces and macrocephaly.4–6 To date, previous reports of GATAD2B-associated neurodevelopmental disorder (GAND; MIM 615074) had identified a total of ten subjects that possessed deletion or truncating variants in GATAD2B and presented with intellectual disability, impaired language development, strabismus, and characteristic facies.8–12
In this report, we have characterized the phenotypic data of fifty subjects with multiple types of GATAD2B variants with the purpose of defining the common genetic and clinical features of GAND. Pathogenic variants included deletions (3), nonsense (17), truncating frameshift (16), splice-site (7), and missense (7) changes. Missense variants have not been previously associated with GAND and all of them were located within two highly conserved domains, Conserved Region-1 (CR1) and Conserved Region-2 (CR2) of the GATAD2B protein (also known as p66β) (Fig. 1, Table S1). Several of these missense variants were shown to disrupt interactions between the GATAD2B protein and its NuRD binding partners. All subjects had phenotypes that included intellectual disability, impaired language development, strabismus, and characteristic facies. Several additional phenotypic features were identified that included polyhydramnios, neonatal feeding difficulties, anisocoria, aortic valve defects, epilepsy, abnormal brain MRIs, and macrocephaly. This expanded GAND phenotype closely overlaps other NuRD-associated neurodevelopmental disorders indicating that these associated proteins (CHD3, CHD4, and GATAD2B) may have converging molecular functions and result in clinically-related syndromes.
Fig. 1. Genomic and Protein Schematic Diagram with GAND variants.
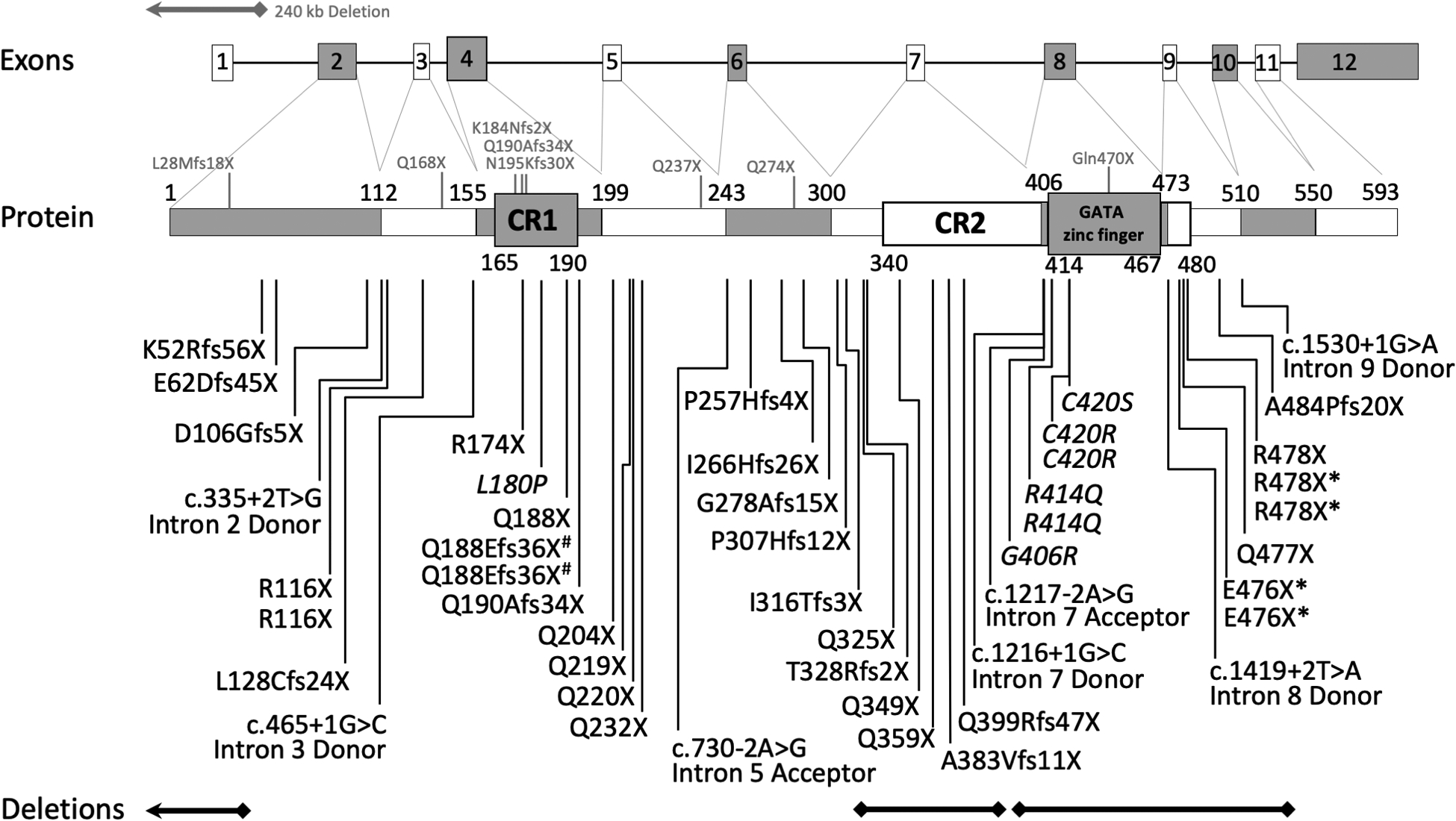
All reported pathogenic GATAD2B variants from our study are represented as deletions, splice-site or protein changes below the protein diagram. Genomic deletions are represented as bars below the figure. Previously reported variants and deletions are represented with grey letters and bars above the diagram. * = monozygotic twins; # = somatic mosaic family
MATERIALS AND METHODS
Standard Protocol Approvals, Registrations and Patient Consents
Research protocols were approved through an institutional review board (CSMC IRB protocol Pro00037131). The families of fifty subjects identified with GATAD2B variants gave informed consent and were enrolled in our retrospective study. Subjects GAND21, GAND24, GAND26, GAND27, GAND29, GAND30, GAND32–35 were identified and referred to our study through the Deciphering Developmental Disorders (DDD) study (UK Research Ethics Committee approval: 10/H0305/83, granted by the Cambridge South REC).13 Authorization for disclosure of recognizable subjects in photographs and information was obtained by parents.
Clinical Genetic Evaluations
Genomic DNA was extracted from saliva or blood and genetic testing was performed and analyzed per previous protocols for clinically-available trio-based exome sequencing14,15 (N=44; the DDD study; the Undiagnosed Diseases Network; GeneDx, Gaithersburg, MD; Centogene, Rostock, Germany; Baylor College of Medicine, Houston, TX), intellectual disability panels (N=4; GeneDx, Gaithersburg, MD; University of Chicago, Chicago, IL; Genome Diagnostics Nijmegen, Nijmegen, NL), and chromosomal SNP microarrays (N=1; GeneDx, Gaithersburg, MD).
Genome sequencing and analysis
Subject GAND50 was diagnosed via genome sequencing analysis. Genomic DNA was extracted from the subject and her unaffected parents. Genome sequencing was performed at Human Longevity Inc (San Diego, CA) on the Illumina Hiseq/novaseq platform to an average depth of 35–40X per sample. Approximately 450M 150bp-paired-end reads (132Gb of sequences) were generated across the genome for each sample using Illumina HiSeq X system with median insert size of ~340bp. Data analysis was performed on the DNAnexus platform using a UCLA custom built pipeline that incorporates BWA-mem v0.7.5 for alignment;16 Picard v2.0.1 (http://broadinstitute.github.io/picard/) for PCR and optical duplicate marking; GATK v3.417–19 for depth of coverage analysis, indel realignment, base quality score recalibration, haplotype calling, joint genotyping, and variant recalibration; and PLINK v1.0720 for determining regions of excess homozygosity. GoldenHelix VarSeq™ v1.4.5 (Golden Helix, Inc., Bozeman, MT, www.goldenhelix.com) was used for variant annotation, filtering and interpretation. Manta v1.4.0,20 ERDS v1.1,21 and CNVnator v0.3.322 were used for structural variant (SV) detection. Annotation of SV calls for genic content was performed using in-house scripts and validated by manual inspection and Sanger confirmation at a CAP/CLIA accredited laboratory (UCLA Orphan Disease Testing Center). No clinically significant de novo, homozygous, hemizygous or compound heterozygous SNVs or small indels were identified. However, a 1.5Kb de novo deletion, NC_000001.10: g.153788220_153789753del, was observed in the proband that removes a single exon in GATAD2B (exon 7 in transcript NM_020699.3), resulting in a predicted frameshift. No common SVs with similar breakpoints are observed in the Database of Genomic Variants23 or among 2,504 individuals called for SV by the 1000 Genomes Project.24
Clinical Data
Retrospective clinical, diagnostic, and neuro-diagnostic information was collected, analyzed and reported from medical records and family interviews. Physical exams were performed whenever possible. Please note that the average values of the reported parameters are based on available data with raw data supplied in parentheses wherever pertinent.
Plasmids
Full-length genes for mouse methyl-binding domain protein-3 (MBD3; UniProt ID:Q9Z2D8), and human methyl-binding domain 2-alpha (MBD2-alpha; UniProt ID:Q9UBB5), GATAD2B (UniProt ID:Q8WXI9), and partial gene sequences for the C-terminal domains (CTD) of Chromodomain-helicase DNA binding protein-3 (CHD3; UniProt ID:Q12873), Chromodomain-helicase DNA binding protein-4 (CHD4; UniProt ID:Q14839) and Chromodomain-helicase DNA binding protein-5 (CHD5; UniProt ID:Q8TDI0) were cloned into pcDNA3.1 expression vectors to generate HA-tagged GATAD2B, and FLAG-tagged MBD2, MBD3, and the CTD of CHD3 (residues 1246–1944), CHD4 (residues 1230–1912) and CHD5 (residues 1218–1954). Five missense variants (L180P, G406R, R414Q, C420R, and C420S) were introduced into the HA-GATAD2B plasmid using site-directed mutagenesis.
In vitro Rabbit Reticulocyte Lysate Protein Expression and Immunoprecipitation Assays
Protein expression and immunoprecipitation experiments were performed as previously described.25,26 In vitro transcription-translation in rabbit reticulocyte lysates (IVT) and pulldown studies were used to generate interaction data for: i) FLAG-tagged MBD2 or MBD3 co-expressed with versions of HA-GATAD2B (Wild Type (WT) or L180P) and ii) FLAG-tagged C1-C2 region26 of CHD3, CHD4, and CHD5 co-expressed with HA-GATAD2B (WT, G406R, R414Q, C420R, or C420S). In all experiments, FLAG-fusion proteins were immobilized on α-FLAG affinity beads and used as baits to pull down the co-expressed HA-GATAD2B proteins. Wild-type HA-GATAD2B was expressed individually as a negative control. Lysates to which no plasmids were added were also used as negative controls. In each case, 10% of inputs and 50% of elutions were loaded on an SDS-PAGE gel and PVDF membranes were probed with α-HA-HRP (#2999S, Cell Signalling Technology, Danver, MA, USA) in 1:20,000 dilution and α-FLAG-HRP (#A8592, Sigma Aldrich, USA) in 1:80,000 dilution.
RESULTS
GATAD2B subjects and demographics
Fifty subjects were enrolled in our study. Most subjects were unrelated, although two sets of identical twins and one set of non-twin siblings were identified. Subjects were located in the United States, Canada, United Kingdom, Australia, Portugal, Spain, United Arab Emirates, and Brazil. Fifty-eight percent of subjects were female (29/50). Parents were primarily of Caucasian background (95.5%); a minority of parents were of other backgrounds (Asian (1.5%), Arabic (2%), Hispanic (2%) and African descent (1%)). The average age at diagnosis was 6.8 years (SD 4.6). Average age at enrollment in our study was 7.0 years (SD 4.5; range: 20 months to 21 years).
GATAD2B pathogenic variants
Forty-three different GATAD2B pathogenic variants were identified in the fifty enrolled subjects, forty-one of these were novel. Almost all subjects (96%) had de novo pathogenic variants, with two subjects inheriting variants via parental mosaicism. Five of the novel variants were found in multiple subjects (p.R116X (2 unrelated subjects); p.Q188Efs36X (2 subjects, siblings via paternal mosaicism); p.R414Q (2 unrelated subjects); p.C420R (2 unrelated subjects); p.E476X (2 subjects, monozygotic twins); p.R478X (3 subjects, monozygotic twins and one unrelated child). Two pathogenic variants in our cohort had been previously reported (c.1217–2 A>G splice-site; p.Q190Afs34X)8,9 (Fig. 1 and Table S1).
Multiple types of variants were identified in our subjects that included: nonsense (17; 34%), truncating frameshift (16; 32%), splice-site (7; 14%), deletions (3; 6%), and missense (7; 14%) changes. One deletion (~1.5kb) involved exon 7 and another deletion (~3.5kb) involved exons 8–11. A third deletion (~175kb) extended from the non-coding exon 1 of GATAD2B upstream to include seven other genes of unknown function terminating within the NUP21OL gene. Interestingly, the missense variants were all located within the Conserved Region-1 (CR1; p.L180P) or Conserved Region-2 (CR2; p.G406R, p.R414Q, p.C420R, C420S) domains of the GATAD2B protein. These domains have important interactions with MDB and CHD proteins within the NuRD complex (Fig. 1).25–28
Facial features
In addition to macrocephaly, the majority of GAND subjects also had distinct facial features. Dysmorphisms were evaluated in thirty-seven children whose families provided adequate photos. Findings included a high wide forehead/frontal bossing (100%), prominent supraorbital ridges (62.2%), posteriorly angulated ears (59%), ocular hypertelorism (78.4%), down-slanting palpebral fissures (45.6%), epicanthal folds (29.7%), prominent or bulbous nasal tip (83.8%), wide nasal base (35.1%), elongated wide nose (35.1%), short philtrum (51.3%), small recessed jaw (24.3%), and a pointed chin (91.9%)(Fig. 2, Table S2). Face2Gene image analysis (FDNA Inc., USA) was used on photographs of 19 GAND subjects to produce a composite model of the facial features associated with GAND as compared to age-, ethnicity-, and gender-matched controls. The facial features of the GAND cohort was significantly different from healthy controls (N=19; area under the curve [AUC] value of 0.933 of receiver operating characteristic [ROC] curve, p-value: 0.004).
Fig. 2. Photographs of affected individuals with associated with variant types. a-e:

Nonsense or truncating frameshift variants: a. GAND19 b. GAND18 c. GAND33 & GAND34 d. GAND53 e. GAND20 f-k: Missense variants: f. GAND28 g. GAND15 h. GAND52 i. GAND32 j. GAND55 k. GAND49 l-q: Splice-site variants: l. GAND17 m. GAND27 n. GAND42 o. GAND21 p. GAND6 q-v: Nonsense or truncating frameshift variants: q. GAND40 r. GAND12 s. GAND3 t. GAND13 u. GAND8 v. GAND36 w. GAND2 x. composite of 19 photographs of GAND subjects and y. 19 healthy controls (FDNA Inc. USA).
Birth and Development
Average parental age at birth was in the early thirties. The average gestational age at birth was 38.0 weeks (SD 1.8). Polyhydramnios (45%) was the most common complication during pregnancy. The average birthweight and birth length were within normal limits. Macrocephaly was seen in 60.4% of subjects at birth, with an average birth head circumference of 36.7 cm (SD 1.7). The average percentile rank of the birth head circumferenece was ~87 (SD 23.5). Macrocephaly at older timepoints was present in 91.8% of subjects, with average head circumference percentile score of 96.0 (SD 6.4) (Tables 1 and S3).
Table 1.
Phenotypic Findings
| Phenotypic Findings | Affected/Total (%) |
|---|---|
| Neonatal | |
| Polyhydramnios | 19/45 (42.2) |
| Neonatal Feeding Issues | 41/50 (82.0) |
| Infantile Hypotonia | 50/50 (100) |
| Neurology | |
| Intellectual Disability/DD | 50/50 (100) |
| Macrocephaly-Birth | 26/43 (60.4) |
| Macrocephaly-Current | 45/49 (91.8) |
| Seizures/Epilepsy | 12/50 (24) |
| Abnormal MRI | 27/45 (60) |
| Non-Verbal (<4 years) | 5/15 (33.3)* |
| Non-Verbal (>4 years) | 7/35 (20)* |
| Non-Ambulatory (<3 years) | 3/9 (33.3)* |
| Non-Ambulatory (>3 years) | 2/41 (4.9)* |
| Wide-based Gait | 41/45 (91.1) |
| Orthotics | 25/41 (61.0) |
| Deafness | 1/50 (2) |
| Behavioral | |
| Good Eye Contact | 49/50 (98) |
| Social Reciprocity | 49/50 (98) |
| Not Toilet Trained (<4 years) | 15/15 (100) |
| Not Toilet Trained (>4 years) | 22/35 (62.9) |
| Cardiac | |
| Bicuspid Aortic Valve | 5/50 (10) |
| Cardiac intervention | 2/50 (4) |
| Ophthalmologic | |
| Strabismus | 44/50 (88) |
| Anisocoria | 7/46 (13.6) |
| Astigmatism | 19/46 (41.3) |
| Hyperopia | 14/46 (30.4) |
| Myopia | 10/46 (21.7) |
| Optic Nerve Hypoplasia | 4/44 (9.1) |
| Deafness | 1/50 (2.0) |
| Dysmorphology | |
| Prominent Forehead | 37/37 (100) |
| Hypertelorism | 29/37 (78.4) |
| Prominent/Bulbous Nasal Tip | 31/37 (83.8) |
| Pointed Chin | 34/37 (91.9) |
specific ages of non-verbal and non-ambulatory subjects are noted in Table S4.
All developmental milestones were abnormal from early infancy, including feeding, language, motor ability, and intellectual ability. Infantile hypotonia was present in all subjects (100%), with most being described as “floppy” in infancy. Children sat up on average at 14.7 months (SD 5.2) and most children were ambulatory by an average age of 33.1 months (SD 12.6). Five subjects were non-ambulatory (ages: 1.3, 2, 2.5, 5, and 6 years) (Table S4). Most ambulatory children had a persistent unsteady gait (91%). Infantile feeding issues and gastro-esophageal reflux disease were reported in 82% of subjects. Two subjects required gastrostomy-tube placement (4%). Reflux disease often resolved with age, but most children had persistent oro-mechanical issues and excessive drooling. (Tables 1, S3, S4, and S5)
All subjects had intellectual disability associated with expressive and receptive language issues. The average age of first spoken words was 3.9 years (SD 2.0), although none of the subjects attained an extensive vocabulary. Eleven subjects were non-verbal (ages: 1.3, 2, 3, 3, 3, 5, 5, 6, 6, 11, 14, and 21 years) (Tables S4, S5). All children over 7 years could follow a one-step command (Table S5), although 20% of subjects 0–3 years and 10.5% in the 4–7 years age group could not. A substantial number of subjects never attained complete toilet-training (Table S5). Almost all subjects exhibited normal eye contact and social reciprocity (98% each).
Neurological, neuroimaging, and ophthalmological findings
Epilepsy was present in 24% of subjects. Interestingly, 58% of all subjects underwent electroencephalograms (EEGs) for suspected seizure events, with most EEGs being normal (79.3%). Primary generalized and focal features were noted in several subjects (13.7% and 6.9%; respectively). Most epileptic subjects were well-controlled with anti-seizure medication (75%; 8/12). MRI of the brain was performed on 88% of subjects, with abnormalities being present in 60% of all neuroimaging studies (Tables 1, S6). Predominant findings were enlarged extra-axial spaces/ventriculomegaly (35.6%), white matter signal abnormalities (22.2%), thin corpus callosum (8.9%), and hypomyelination (13.3%) (Fig. 3). Ophthalmological findings included strabismus (88%), astigmatism (41.3%), hypermetropia (30.4%), myopia (21.7%), anisocoria (13.6%) and optic nerve hypoplasia (9.1%) (Table 1).
Fig. 3. Neuroimaging of GAND8 and GAND53.
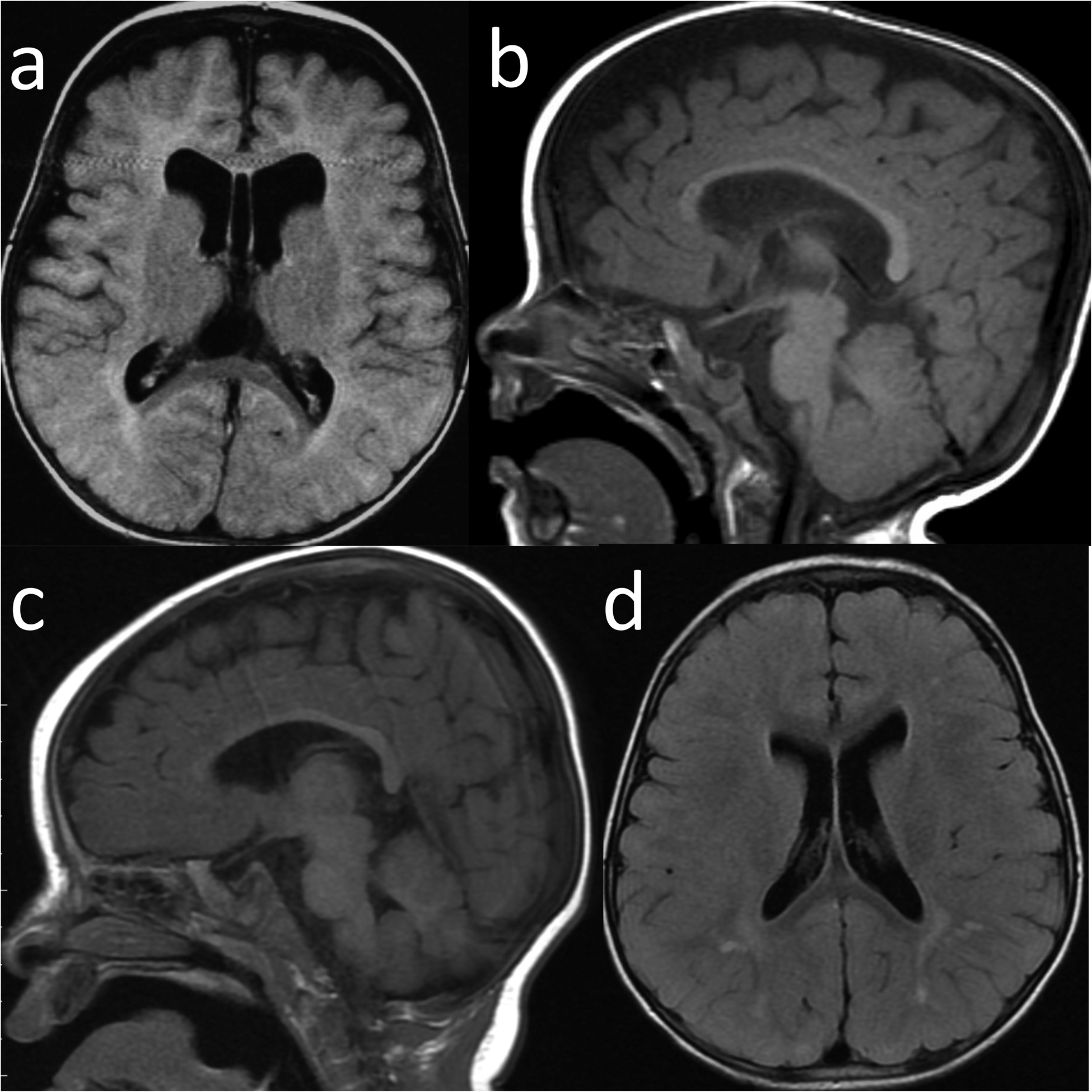
a-b: GAND8 a. Axial FLAIR image showed enlarged subarachnoid spaces and nonobstructive ventriculomegaly of the lateral ventricles (cavum vergae was also present) b. Sagittal T1 showed macrocephaly, enlarged extra-axial fluid, and a thin corpus callosum c-d: GAND53 c. Sagittal T1 with normal corpus callosum and structures (prominence of the extraaxial CSF spaces previously seen on earlier imaging had resolved) d. Axial FLAIR image showing multiple punctate and patchy foci of nonspecific signal hyperintensity scattered in the bilateral cerebral white matter, particularly involving the parietal lobes.
Cardiac findings
Many subjects underwent cardiology consultations and echocardiograms (34%). One subject had left pulmonary artery stenosis, while four other subjects had a bicuspid aortic valve (8%). Two of these subjects underwent balloon valvulotomy. One of these two children had neonatal critical aortic stenosis and subsequently developed aortic regurgitation that required a Ross cardiac procedure for correction.
Evaluation of cohorts with specific variant types
All of the above evaluations were subdivided into cohorts consisting of nonsense, truncating frameshift, deletions, splice-site, and missense variants. There were no obvious or dramatic differences between these variant groups with regards to the above-mentioned parameters, with the exception of the frequency of epilepsy in subjects with missense variants (57%), compared to the total (24%) (Tables S2, S4, and S6).
Disruption of NuRD interactions by GATAD2B missense variants
GAND-associated missense variants were all located in conserved regions of GATAD2B: namely CR1 (L180P) and CR2 (G406R, R414Q, C420R, C420S). The CR1 motif is known to bind MBD2/3 NuRD subunits through the formation of a coiled coil.37 The CR2 region has been shown to interact with the C-terminal region of CHD4.26,36 In order to understand the effect of the GATAD2B missense variants in the context of NuRD assembly, we performed pairwise interaction experiments to assess the binding of GATAD2B to MBD or CHD proteins. Full-length HA-tagged GATAD2B (wildtype or missense variants) was co-expressed with either FLAG-tagged MBD2/3 or the FLAG-tagged C-terminal region of CHD proteins (the C1-C2 region that follows the helicase domain) and their interaction examined in pulldown experiments.26 The L180P variant abrogated the interaction of GATAD2B with both MBD2 and MBD3 (Fig. 4a). The L180 residue lies at the interface of the GATAD2-MBD coiled-coil, and the leucine to proline substitution is likely to disrupt both the interaction interface and the intrinsic helical propensity of the CR1 region. As compared to wild type GATAD2B, the CR2-region variants variable effects on the interactions between GATAD2B and the CHD paralogues (Fig. 4b). The C420R and C420S variants substantially inhibited these interactions with the CHD paralogues as compared to wild-type controls, whereas R414Q variant caused a mild reduction in these interactions. The G406R variant showed no clear effect. The C420 residue is one of the zinc-binding ligands in the GATAD2B zinc-finger domain found in CR2 and substitution of this cysteine is likely to severely disrupt the structure of the CR2 domain. In contrast, G406 and R414 are upstream of the zinc finger domain and are predicted to be in a region that is unstructured – consistent with their smaller (R414Q) or negligible (G406R) effect on the GATAD2B-CHD4 interaction. As compared to controls, the effect of these variants on interactions between GATAD2B and the CHD paralogues was consistent with the close sequence similarity between the CHD paralogues in the C2 region of these proteins (Fig. 4b).26
Fig. 4. GAND-associated missense variants L180P, R414Q, C420S and C420R disrupt binding to NuRD components CHD3/4/5 or MBD2/3.
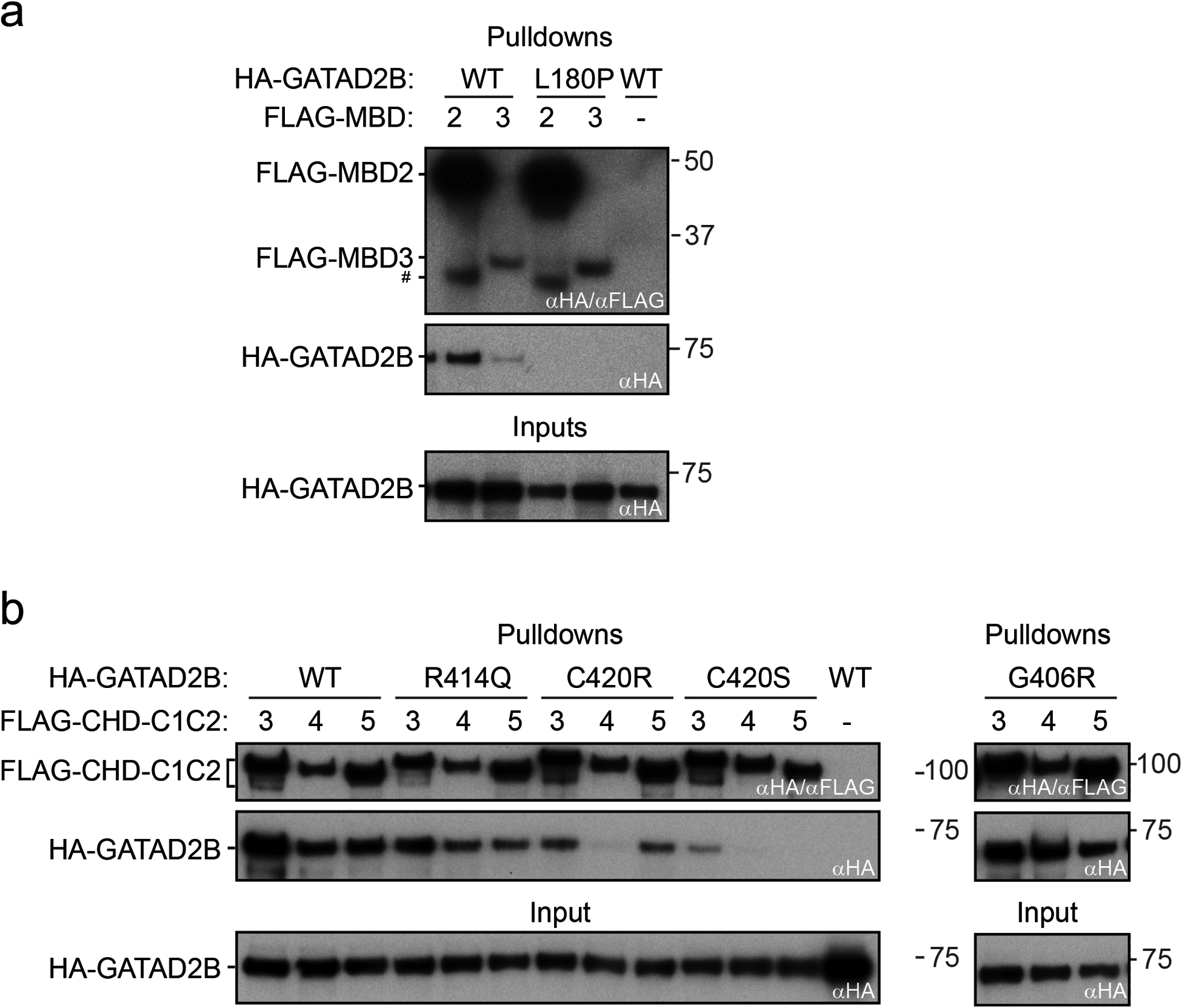
a) FLAG-tagged MBD2 or MBD3 were co-expressed with HA-GATAD2B (wild type (WT) or L180P) by in vitro transcription-translation (IVT) in a rabbit reticulocyte lysate. b) The FLAG-tagged C1-C2 domains of CHD3, CHD4 or CHD5 was co-expressed with HA-GATAD2B (WT, R414Q, C420R, C420S, and G406R) in IVTs. In all experiments, FLAG-fusion proteins were immobilized on αFLAG affinity beads and used as baits to pull down the co-expressed HA-GATAD2B. As a negative control, wild-type (WT) HA-GATAD2B was added to beads to which no FLAG fusion protein had been immobilized. In each case, 10% of inputs and 50% of elutions were loaded on an SDS-PAGE and proteins were detected by Western blot, using αHA and/or αFLAG antibodies.
# = degradation product of FLAG-MBD2.
DISCUSSION
We report the genetic and clinical features of 50 individuals with GATAD2B-associated neurodevelopmental disorder (GAND). Prior to our work, a handful of GAND patients had been reported with distinct facies, infantile hypotonia, intellectual disability, limited language ability, and strabismus.8–12 Our large cohort has allowed us to identify an expanded range of new phenotypic and genotypic features associated with GAND. These new features included macrocephaly, frontal bossing, polyhydramnios, infantile feeding difficulties, cardiac defects, anisocoria, astigmatism, epilepsy (focal and/or primary generalized), and abnormal neuroimaging, as well as a variety of new variant types including missense variants.
The clinical phenotype of GAND subjects was relatively consistent across our cohort. Pregnancies were mostly without complications, with the exception of polyhydramnios (45%). Several children were noted to have macrocephaly on prenatal ultrasound and macrocephaly was frequently present at birth (69%), and became more common with age (91.8%). All subjects had global developmental delay and intellectual disability. All subjects had infantile hypotonia and delayed motor milestones. Most children learned to ambulate. A majority of subjects were noted to have infantile feeding difficulties and GERD (82%). These oromechanical issues were also associated with delayed and limited expressive language development in all subjects that was reminiscent of childhood apraxia of speech.4 Receptive language was not as severely affected, with most children being able to follow multi-step commands at older ages. Epilepsy was present in a minority of subjects (24%), and could be of focal or primary generalized onset. Most children responded well to anti-epileptic treatment. Neuroimaging was abnormal in the majority of subjects (60%), with common features including ventriculomegaly/enlarged CSF spaces, white matter signal abnormalities, hypomyelination, and thin corpora callosi. Almost all subjects made good eye contact and exhibited social reciprocity. Toilet training was not attained in the majority of subjects. Most subjects had ophthalmological issues that included strabismus (88%), which was sometimes associated with astigmatism (41.3%), anisocoria (13.6%) and/or hypoplastic optic nerves (9%). Lastly, a minority of subjects were born with bicuspid aortic valves (10%, as compared to 2% in the general population),29 with two subjects requiring surgical intervention. Of note, the two pairs of monozygotic twins in our cohort (GAND33/GAND34; GAND40/GAND41) had similar overall phenotypes with some differences. GAND33 developed epilepsy in her teens and continues to require seizure medication, while GAND34 does not have epilepsy. GAND33 has also plateauted developmentally, while GAND34 continues to progress with school. The other set of monozygotic twins shared very similar phenotypes, although GAND40 has had an easier time with school performance and GAND41 has been more adept at potty training. Of course, with only two sets of twins it is difficult to make any definitive comments on phenotypic trends of monozygotic siblings. As the NuRD complex plays an active epigenetic role in corticogenesis,30 the identification of more monozygotic twin sets will be valuable to determine variable aspects of the GAND phenotype, especially with regards to cortical function.
Almost all GAND variants were de novo, with a diverse range in variant types that included nonsense (34%), truncating frameshift (32%), splice-site variants (14%), and deletions (6%). Seven subjects had missense variants (14%), which had not been previously reported in GAND. All of the missense variants were located within the two conserved region domains (CR1 and CR2), of the GATAD2B protein. Aside from an increased incidence of epilepsy in subjects with missense variants as compared to subjects with other variants (57 to 19%, respectively), there were no dramatic phenotypic differences between subjects with missense versus loss-of-function (deletion, splice-site, nonsense, and truncating frameshift) variants (Table S4). Of course, in the context of these relatively low numbers of subjects (N = 7 vs. N = 43, respectively) that were across a range of ages, it is difficult to make any firm conclusions about specific variant types. If, with larger numbers, subjects with missense variants continue to be similar to subjects with loss-of-function variants, this similarity may indicate that haploinsufficiency of the GATAD2B protein may likely be the primary pathological mechanism in most GAND subjects.8
The GATAD2B protein functions as a subunit within the NuRD complex, which is an important regulator of gene expression during neurodevelopment. NuRD is a multi-protein holoenzyme possessing both histone deacetylase and chromatin remodeling activity. The complex can consist of an assortment of paralogous subunits, which in addition to GATAD2B (and its paralogue, GATAD2A) includes several scaffold proteins: i) Retinoblastoma binding proteins (RBBP4/RBBP7), ii) metastasis associated proteins (MTA1/MTA2/MTA3), and iii) methyl-domain binding proteins (MBD2/MBD3), along with enzymatically active subunits that include: iv) histone deacetylases (HDAC1/HDAC2), and v) chromatin-helicase DNA-binding proteins (CHD3/CHD4/CHD5).1,2 CDK2AP1 is also present in specific cell types.28 The MBD, GATAD2, and CHD subunits exist as interacting monomers;,28,30–33 therefore, in principle, many configurations can exist with unique combinations of these paralogues that may have specific regulatory functions.20 For example, recent work suggested that MBD2-NuRD converts open chromatin into compacted chromatin, while MBD3-NuRD was associated with more active promoter regions.31 Other reports indicate that the CHD proteins are developmentally regulated during corticogenesis, which influences the generation of neural progenitors and neurons, as well as their subsequent migration and laminar identity.30 Of note, the GATA zinc-finger proteins (GATAD2B and its paralogue, GATAD2A) may have distinct effects on NuRD activity. For example, GATAD2A plays a specific role in DNA repair,33 as well as having a role in repression of pluripotency factors during IPSC reprogramming;34 however, when GATAD2A expression was reduced, GATAD2B was incapable of rescuing the lost GATAD2A activity. To date, no specific roles for GATAD2B have been identified, although it has been shown to be the predominant GATAD2 gene expressed in the brain.35
One hypothesis regarding the general function of GATAD2 proteins is that they provide a “bridge” that links MBD proteins (via CR1) and the CHD chromatin remodeling proteins (via CR2) in the NuRD complex.2,25,26 GATAD2B-CR1’s interaction with MBD proteins occurs through coiled-coil domains within each protein that links GATAD2B with the core proteins of NuRD through MBD.26 In turn, the GATAD2-CR2 domain extends this bridge by its direct interactions with the carboxy-terminal C2 region of CHD proteins, thereby linking CHD proteins to the NuRD core.36 The validity of this bridge model is corroborated by the fact that all of our cohort’s pathogenic missense variants were located within these two regions. Furthermore, our immunoprecipitation assays indicated that several of these missense variants disrupted GATAD2B interactions as predicted by the model. For example, our L180P variant (analogous to L159 of GATAD2A) is located within the coiled-coil domain of CR1 (CR1-CC) that is a key contact point for MBD binding (Fig. S1).37 Our immunoprecipitation assays showed this variant prevented GATAD2B from interacting with MBD2 or MBD3 (Fig. 4a). This result is in line with the prediction that the substituted proline is likely to destabilize the CR1-CC α-helix that forms a coiled-coil with MBD. It had been shown previously that a GATAD2A CR1-CC missense change (K149R) abolished the interaction of GATAD2A with MBD proteins, as well as MBD-mediated transcriptional repression.27 Our other missense variants were clustered within the GATAD2B-CR2 domain. The C420S and C420R variants affect the first of four zinc-binding cysteines of the GATA zinc-finger domain and so would be predicted to disrupt the proper folding and the known interaction of this domain with the CHD carboxy-terminal C2 region.36 Our immunoprecipitation assays confirmed this to be the case. It is also important to note that several subjects with CHD3-related syndrome and CHD4-related syndrome have been reported with carboxy-terminal variants (CHD3: p.R1881L, p.F1935Efs108X; CHD4: p.R1870X) that lie within the CHD carboxy-terminal C2 region.4,7 These variants could also disrupt the GATAD2B-CHD bridge and lead to the exclusion of CHD activity from NuRD complexes causing their associated neurodevelopmental disorders.4,7,8 The R414Q variant was present in two subjects and resulted in a milder disruption of in vitro GATAD2B-CHD interactions. It may be that, in the context of expression within a complete NuRD complex, this substitution may be more disruptive or alter interactions with other proteins associated with NuRD activity. In contrast to the effects of these other missense variants, the G406R substitution did not alter the in vitro interactions between GATAD2B and CHD proteins. This variant involve highly-conserved GATAD2B-CR2 residue (Fig. S1) that is located in a region that has not been characterized structurally or functionally and that lies just N-terminal to the GATA zinc finger (Fig. S1). Further work to determine the mechanism by which this variant affects GATAD2B function is required; however, the nucleotide substitution (c.1216 G>C) associated with G406R involves the terminal nucleotide of exon 7 and so might interrupt proper splicing of intron 8. Whether these missense variants produce proteins capable of antagonizing CHD-MBD-NuRD interactions in a dominant negative manner or work via a dosage effect similar to haploinsufficiency with a similar resulting phenotype remains to be determined; nonetheless, their observed phenotypes are not dramatically altered from subjects with loss-of-function variants. Identification of more missense subjects and additional work are required to answer these questions.
Currently, no other germline disorders have been associated with GATAD2A or most of the other NuRD proteins. Nevertheless, numerous mouse models have indicated the importance of each of these subunits in development.2 Besides GATAD2B, the only other NuRD complex genes that are associated with human disease are CHD3 and CHD4, which have a substantial phenotypic overlap with GAND (although an HDAC1 missense variant was reported in a large exomic screen associated with epilepsy without a detailed report of the phenotype).4–8,38 CHD4RS (or SIHIWES)) has been recently reported in 32 subjects, while CHD3RS (or SNIBCPS) has been reported in thirty-five subjects.4–7 Our expanded GAND phenotype indicates these three neurodevelopmental disorders share features that include macrocephaly, developmental delay, intellectual disability, infantile hypotonia, and ventriculomegaly. Interestingly, each of these disorders share some facial features that are associated with macrocephaly that include a high broad forehead, frontal bossing, ocular hypertelorism, and a wide nasal bridge (Fig. 2).4–8 Many GAND and CHD3RS subjects also had a prominent nose/nasal tip and pointed chins, while CHD4RS subjects had a small nose and ears with a square chin.4–8 Besides these facial characteristics, other variable features across these disorders include male genital abnormalities in both CHD3RS and CHD4RS, inguinal hernia in CHD3RS, and deafness in CHD4RS. Alternatively, GAND and CHD4RS are associated with congenital heart defects;5–7,29 whereas GAND and CHD3RS subjects presented with oromechanical dysfunction that was absent in CHD4RS that included neonatal feeding issues (and perhaps polyhydramnios) and language deficits reminiscent of childhood apraxia of speech.4,38
Childhood Apraxia of Speech (CAS) is a rare neurodevelopmental disorder associated with oromotor incoordination that limits fluent speech.39 Pathogenic variants in several members of the forkhead transcription factor family (FOXP1, MIM 613670; FOXP2, MIM 602081) have been associated with oromotor incoordination and language issues classified as CAS.39–41 Similar to CHD3,4 GATAD2B is one of a few documented proteins that interact with FOXP1/FOXP2 (via its CR2 domain and the zinc-finger/leucine-zipper repressor domain of FOXP),4,42 with GAND, CHD3RS, and FOXP1 subjects sharing several phenotypic features that include high forehead, frontal bossing, intellectual disability, and delayed language milestones. These data may indicate that these proteins may undergo important interactions during neurodevelopment and corticogenesis that have essential roles in language development. Further work on the specific interactions of CHD3, GATAD2B, and the FOXP proteins is warranted and may provide further clarification of the molecular events required for corticogenesis and language development.
In summary, our GAND cohort has provided a more established picture of the genotypic and phenotypic features of this disorder. At this time, this diagnosis has been associated with several developmental, cardiac, ophthalmological, and neurological issues. Therefore, we recommend any child who is genetically diagnosed with GAND undergo consultations with specialists in these fields. Additional evaluations should include speech, physical, and occupational therapy consultations. Notably, the identification of macrocephaly as a feature of GAND has aligned this phenotype with other NuRD-associated disorders that include CHD4RS and CHD3RS. One hypothesis for this could be that GATAD2B dosage effects cause associated deficiencies in CHD3 and/or CHD4 chromatin remodeling activity in GAND children by the decreased availability of haploinsufficient GATAD2B levels to bring CHD-associated chromatin remodeling activity to NuRD complexes. Therefore, it may be prudent to evaluate GAND subjects for additional clinical features seen in CHD3RS and CHD4RS (e.g. bony lesions, hearing loss, hernias, or genital abnormalities), as these findings may also be present in GAND, albeit at lower frequency (e.g. one male subject had an undescended atrophic testis, which is more frequent in CHD3RS).4
GAND-associated missense variants were another important finding within our cohort. These variants were only present within GATAD2B’s CR1 or CR2 domains, which are known to interact with other NuRD proteins. Whether these variants act via a dominant negative mechanism by sequestering NuRD components or by blocking their interactions with other effector proteins (e.g. CHD4, CHD3, FOXP) and/or altering its DNA binding will require further evaluations with affected samples. It is interesting to note that subjects possessing missense variants and loss-of-function variants had similar phenotypes for the most part, which may indicate that they act through similar dosage-dependent mechanisms. More work will be required to unravel these interesting questions with regards to GATAD2B, the NuRD complex, and their phenotypic overlap with other neurodevelopmental disorders.
Supplementary Material
ACKNOWLEDGEMENTS:
We are grateful and thank the subjects and their families for their help, for their providing medical information, and providing consent to post photographs in this article. We also would like to extend special thanks to the Helping Hands for GAND Foundation for their assistance and continued cooperation. We are grateful to Dr. Charles Simmons and Clive Svendsen for mentorship and Kelli Dejohn, Roxana Ramirez, Joanne Baez, and Carmela Brito for clinical and technical assistance. We would like to acknowledge Jaemin Kim and Yogesh Kushwaha for their contribution to the work. We would like to thank Barrington Burnett for their critical review of the manuscript. Research reported in this manuscript was supported by the NIH Common Fund, through the Office of Strategic Coordination/Office of the NIH Director under Award Number(s) [U01HG007672]. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. GAND50’s seq+analysis was supported by NIH National Center for Advancing Translational Science (NCATS) UCLA CTSI Grant Number UL1TR001881. JPM received funding from the National Health and Medical Research Council (APP1012161, APP1063301, APP1126357, APP1058916). TMP and this research was supported by the Cedars-Sinai institutional funding program and the Cedars-Sinai Diana and Steve Marienhoff Fashion Industries Guild Endowed Fellowship in Pediatric Neuromuscular and the Fashion Industries Guild Endowed Fellowship for the Undiagnosed Diseases Program. TMP is especially grateful for the wonderful and continued support from the Cedars-Sinai Fashion Industries Guild.
GLOSSARY
- GAND
GATAD2B-associated neurodevelopmental disorder
- GATAD2A/B
GATA zinc finger domain containing 2A or 2B
- CR1
Conserved Region 1
- CR2
Conserved Region 2
- NuRD
nucleosome remodeling and deactylase complex
- CHD
Chromodomain-helicase-DNA-binding protein
- MBD
methyl-CpG binding domain protein
- MTA
metastasis-associated protein
- RBBP
retinoblastoma associated protein
- HDAC
histone deacetyltransferase
Footnotes
SUPPLEMENTARY INFORMATION
Please see Supplementary Information Section
Disclosures: The authors declare no conflict of interest.
Contributor Information
Christine Shieh, David Geffen School of Medicine at UCLA, Los Angeles, CA, USA.
Natasha Jones, School of Life and Environmental Sciences, University of Sydney, NSW Australia.
Brigitte Vanle, Department of Psychiatry & Behavioral Neurosciences, Cedars-Sinai Medical Center, Los Angeles, CA 90048; Medical College of Wisconsin-Central Wisconsin, Wausau, WI, USA.
Margaret Au, Department of Pediatrics Cedars-Sinai Medical Center, Los Angeles, CA, USA.
Alden Y. Huang, Institute for Precision Health, David Geffen School of Medicine, University of California, Los Angeles, Los Angeles, CA, USA.
Ana P.G. Silva, School of Life and Environmental Sciences, University of Sydney, NSW Australia.
Hane Lee, Department of Human Genetics and Pathology and Laboratory Medicine, David Geffen School of Medicine, University of California, Los Angeles, Los Angeles, CA, USA.
Emilie D. Douine, Department of Human Genetics, David Geffen School of Medicine, University of California, Los Angeles, Los Angeles, CA, USA.
Katheryn Grand, Department of Pediatrics, Cedars-Sinai Medical Center, Los Angeles, CA, USA.
Ingrid P. Taff, Department of Neurology, Hofstra School of Medicine, Great Neck NY, USA.
Mauricio R. Delgado, Department of Neurology and Neurotherapeutics, University of Texas Southwestern Medical Center and Texas Scottish Rite Hospital for Children, Dallas, TX, USA.
MJ Hajianpour, Department of Pediatrics, Division of Medical Genetics, East Tennessee State University, Quillen College of Medicine, TN, USA.
Andrea Seeley, Geisinger Medical Center, Danville, PA, USA.
Luis Rohena, Division of Genetics, Department of Pediatrics, Brooke Army Medical Center, Fort Sam Houston, TX 78234; Department of Pediatrics, UT Health San Antonio, Long School of Medicine, San Antonio, TX, USA.
Hilary Vernon, McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University, Balitmore MD USA.
Karen W. Gripp, Division of Medical Genetics, Al DuPont Hospital for Children, Wilmington, DE, USA.
Samantha A. Vergano, Division of Medical Genetics and Metabolism, Children’s Hospital of The King’s Daughters, Norfolk, VA, USA.
Sonal Mahida, Department of Neurogenetics, Kennedy Krieger Institute, Baltimore, MD, USA.
Sakkubai Naidu, Department of Neurology and Pediatrics, Johns Hopkins School of Medicine, Baltimore, Maryland. Hugo Moser Research Institute, Kennedy Krieger Institute, Baltimore, MD, USA.
Ana Berta Sousa, Serviço de Genética Médica, Hospital Santa Maria, CHULN, Lisboa, Portugal and Faculdade de Medicina de Lisboa, Universidade de Lisboa, Lisboa, Portugal.
Geoffrey Beek, Children’s Hospitals and Clinics of Minnesota Department of Genetics, Minneapolis, MN USA.
Donald Basel, Department of Pediatrics; Division of Genetics; Children’s Hospital of Wisconsin, Milwaukee, WI USA.
Judith Ranells, Division of Genetics and Metabolism, Department of Pediatrics, University of South Florida, Tampa, FL, USA.
Rosemarie Smith, Department of Pediatrics, Division of Genetics, Maine Medical Center, Portland, ME, USA.
Roman Yusupov, Division of Clinical Genetics, Joe DiMaggio Children’s Hospital, Hollywood, Florida, USA.
Mary-Louise Freckmann, Royal North Shore Hospital, St Leonards NSW 2065, Australia.
Lisa Ohden, Department of Genetic Counseling, Sanford Children’s Specialty Clinic, Sioux Falls, SD, USA.
Laura Davis-Keppen, Department of Pediatrics, Sanford School of Medicine of the University of South Dakota, Sioux Falls, SD, USA.
David Chitayat, The Prenatal Diagnosis and Medical Genetics Program, Department of Obstetrics and Gynecology, Mount Sinai Hospital, University of Toronto; Division of Clinical and Metabolic Genetics, Department of Pediatrics, The Hospital for Sick Children, University of Toronto, Toronto, Ontario, Canada..
James J. Dowling, Division of Neurology, Department of Pediatrics, The Hospital for Sick Children, Toronto, Ontario, Canada..
Richard Finkel, Division of Pediatric Neurology, Department of Pediatrics, Nemours Children’s Hospital, Orlando, FL, USA.
Andrew Dauber, Division of Endocrinology, Children’s National Health System, Washington, DC, USA.
Rebecca Spillmann, Department of Pediatrics, Division of Medical Genetics, Duke University Medical Center, Durham, NC, USA.
Loren D.M. Pena, Division of Human Genetics, Cincinnati Children’s Hospital Medical Center, Cincinnati, Ohio; Department of Pediatrics, University of Cincinnati College of Medicine, Cincinnati, Ohio, USA.
Kay Metcalfe, Manchester Centre for Genomic Medicine, Manchester University NHS FT, Manchester, UK.
Miranda Splitt, Institute of Genetic Medicine, Northern Genetics Service, Newcastle upon Tyne Hospitals Trust, Newcastle, UK.
Katherine Lachlan, Faculty of Medicine, University of Southampton, Southampton, UK & Human Development and Health Division, Wessex Clinical Genetics Service, University Hospitals of Southampton NHS Trust, Southampton, UK.
Shane A. McKee, Northern Ireland Regional Genetics Service, Belfast City Hospital, Belfast, UK.
Jane Hurst, Department of Clinical Genetics, NE Thames Genetics Service, Great Ormond Street Hospital, London, UK.
David R. Fitzpatrick, Medical Research Council Human Genetics Unit, University of Edinburgh, Edinburgh, UK.
Jenny E.V. Morton, West Midlands Regional Clinical Genetics Service and Birmingham Health Partners; Birmingham Women’s and Children’s Hospitals NHS Foundation Trust; Birmingham Women’s Hospital, Edgbaston, Birmingham, UK.
Helen Cox, West Midlands Regional Clinical Genetics Service and Birmingham Health Partners; Birmingham Women’s and Children’s Hospitals NHS Foundation Trust; Birmingham Women’s Hospital, Edgbaston, Birmingham, UK.
Sunita Venkateswaran, Division of Neurology, Department of Pediatrics, Children’s Hospital of Eastern Ontario, University of Ottawa, Ottawa, Ontario, Canada.
Juan I. Young, John P Hussman Institute for Human Genomics, University of Miami Miller School of Medicine, Miami, FL, USA.
Eric D. Marsh, Division of Neurology, Children’s Hospital of Philadelphia and Department of Neurology and Pediatrics, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, PA, USA.
Stanley F. Nelson, Department of Human Genetics, David Geffen School of Medicine, University of California, Los Angeles, Los Angeles, CA USA.
Julian A. Martinez, Department of Human Genetics; Division of Medical Genetics, Department of Pediatrics; David Geffen School of Medicine, University of California, Los Angeles, Los Angeles, CA USA.
John M. Graham, Jr, Department of Pediatrics, Medical Genetics, Cedars-Sinai Medical Center, Los Angeles, CA, USA
Usha Kini, Oxford Centre for Genomic Medicine, Oxford University Hospital NHS Foundation Trust, Oxford, UK.
Joel P. Mackay, School of Life and Environmental Sciences, University of Sydney, NSW Australia.
Tyler Mark Pierson, Department of Pediatrics, Cedars-Sinai Medical Center, Los Angeles, CA, USA; Department of Neurology, Cedars-Sinai Medical Center, Los Angeles, CA, USA; Board of Governors Regenerative Medicine Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA.
REFERENCES
- 1.Torchy MP, Hamiche A, Klaholz BP. Structure and function insights into the NuRD chromatin remodeling complex. Cell Mol Life Sci. 2015. doi: 10.1007/s00018-015-1880-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Basta J, Rauchman M. The Nucleosome Remodeling and Deacetylase Complex in Development and Disease. In: Translating Epigenetics to the Clinic.; 2017. doi: 10.1016/B978-0-12-800802-7.00003-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vignali M Distribution of acetylated histones resulting from Gal4-VP16 recruitment of SAGA and NuA4 complexes. EMBO J. 2000. doi: 10.1093/emboj/19.11.2629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Snijders Blok L, Rousseau J, Twist J, et al. CHD3 helicase domain mutations cause a neurodevelopmental syndrome with macrocephaly and impaired speech and language. Nat Commun. 2018. doi: 10.1038/s41467-018-06014-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sifrim A, Hitz MP, Wilsdon A, et al. Distinct genetic architectures for syndromic and nonsyndromic congenital heart defects identified by exome sequencing. Nat Genet. 2016. doi: 10.1038/ng.3627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weiss K, Terhal PA, Cohen L, et al. De Novo Mutations in CHD4, an ATP-Dependent Chromatin Remodeler Gene, Cause an Intellectual Disability Syndrome with Distinctive Dysmorphisms. Am J Hum Genet. 2016. doi: 10.1016/j.ajhg.2016.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weiss K, Lazar HP, Kurolap A, et al. The CHD4-related syndrome: a comprehensive investigation of the clinical spectrum, genotype–phenotype correlations, and molecular basis. Genet Med. 2019. doi: 10.1038/s41436-019-0612-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Willemsen MH, Nijhof B, Fenckova M, et al. GATAD2B loss-of-function mutations cause a recognisable syndrome with intellectual disability and are associated with learning deficits and synaptic undergrowth in Drosophila. J Med Genet. 2013. doi: 10.1136/jmedgenet-2012-101490 [DOI] [PubMed] [Google Scholar]
- 9.Hamdan FF, Srour M, Capo-Chichi JM, et al. De Novo Mutations in Moderate or Severe Intellectual Disability. PLoS Genet. 2014. doi: 10.1371/journal.pgen.1004772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luo X, Zou Y, Tan B, et al. Novel GATAD2B loss-of-function mutations cause intellectual disability in two unrelated cases. J Hum Genet. 2017. doi: 10.1038/jhg.2016.164 [DOI] [PubMed] [Google Scholar]
- 11.Ueda K, Yanagi K, Kaname T, Okamoto N. A novel mutation in the GATAD2B gene associated with severe intellectual disability. Brain Dev. 2019. doi: 10.1016/j.braindev.2018.10.003 [DOI] [PubMed] [Google Scholar]
- 12.Rabin R, Millan F, Cabrera-Luque J, Pappas J. Intellectual disability due to monoallelic variant in GATAD2B and mosaicism in unaffected parent. Am J Med Genet Part A. 2018. doi: 10.1002/ajmg.a.40667 [DOI] [PubMed] [Google Scholar]
- 13.Wright CF, Fitzgerald TW, Jones WD, et al. Genetic diagnosis of developmental disorders in the DDD study: A scalable analysis of genome-wide research data. Lancet. 2015. doi: 10.1016/S0140-6736(14)61705-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tanaka AJ, Cho MT, Millan F, et al. Mutations in SPATA5 Are Associated with Microcephaly, Intellectual Disability, Seizures, and Hearing Loss. Am J Hum Genet. 2015. doi: 10.1016/j.ajhg.2015.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010. doi: 10.1093/bioinformatics/btp698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010. doi: 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Depristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011. doi: 10.1038/ng.806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van der Auwera GA, Carneiro MO, Hartl C, et al. From fastQ data to high-confidence variant calls: The genome analysis toolkit best practices pipeline. Curr Protoc Bioinforma. 2013. doi: 10.1002/0471250953.bi1110s43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007. doi: 10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen X, Schulz-Trieglaff O, Shaw R, et al. Manta: Rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics. 2016. doi: 10.1093/bioinformatics/btv710 [DOI] [PubMed] [Google Scholar]
- 21.Zhu M, Need AC, Han Y, et al. Using ERDS to infer copy-number variants in high-coverage genomes. Am J Hum Genet. 2012. doi: 10.1016/j.ajhg.2012.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abyzov A, Urban AE, Snyder M, Gerstein M. CNVnator: an approach to discover, genotype, and characterize typical and atypical CNVs from family and population genome sequencing. Genome Res. 2011. doi: 10.1101/gr.114876.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.MacDonald JR, Ziman R, Yuen RKC, Feuk L, Scherer SW. The Database of Genomic Variants: A curated collection of structural variation in the human genome. Nucleic Acids Res. 2014. doi: 10.1093/nar/gkt958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sudmant PH, Rausch T, Gardner EJ, et al. An integrated map of structural variation in 2,504 human genomes. Nature. 2015. doi: 10.1038/nature15394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Low JKK, Webb SR, Silva APG, et al. CHD4 is a peripheral component of the nucleosome remodeling and deacetylase complex. J Biol Chem. 2016. doi: 10.1074/jbc.M115.707018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Torrado M, Low JKK, Silva APG, et al. Refinement of the subunit interaction network within the nucleosome remodelling and deacetylase (NuRD) complex. FEBS J. 2017. doi: 10.1111/febs.14301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brackertz M, Gong Z, Leers J, Renkawitz R. p66alpha and p66beta of the Mi-2/NuRD complex mediate MBD2 and histone interaction. Nucleic Acids Res. 2006. doi: 10.1093/nar/gkj437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sharifi Tabar M, Mackay JP, Low JKK. The stoichiometry and interactome of the Nucleosome Remodeling and Deacetylase (NuRD) complex are conserved across multiple cell lines. FEBS J. 2019. doi: 10.1111/febs.14800 [DOI] [PubMed] [Google Scholar]
- 29.Michelena HI, Prakash SK, Della Corte A, et al. Bicuspid aortic valve identifying knowledge gaps and rising to the challenge from the international bicuspid aortic valve consortium (BAVCON). Circulation. 2014. doi: 10.1161/CIRCULATIONAHA.113.007851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nitarska J, Smith JG, Sherlock WT, et al. A Functional Switch of NuRD Chromatin Remodeling Complex Subunits Regulates Mouse Cortical Development. Cell Rep. 2016. doi: 10.1016/j.celrep.2016.10.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Le Guezennec X, Vermeulen M, Brinkman AB, et al. MBD2/NuRD and MBD3/NuRD, Two Distinct Complexes with Different Biochemical and Functional Properties. Mol Cell Biol. 2006. doi: 10.1128/mcb.26.3.843-851.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoffmeister H, Fuchs A, Erdel F, et al. CHD3 and CHD4 form distinct NuRD complexes with different yet overlapping functionality. Nucleic Acids Res. 2017. doi: 10.1093/nar/gkx711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spruijt CG, Luijsterburg MS, Menafra R, et al. ZMYND8 Co-localizes with NuRD on Target Genes and Regulates Poly(ADP-Ribose)-Dependent Recruitment of GATAD2A/NuRD to Sites of DNA Damage. Cell Rep. 2016. doi: 10.1016/j.celrep.2016.09.037 [DOI] [PubMed] [Google Scholar]
- 34.Mor N, Rais Y, Sheban D, et al. Neutralizing Gatad2a-Chd4-Mbd3/NuRD Complex Facilitates Deterministic Induction of Naive Pluripotency. Cell Stem Cell. 2018. doi: 10.1016/j.stem.2018.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fagerberg L, Hallström BM, Oksvold P, et al. Analysis of the Human Tissue-specific Expression by Genome-wide Integration of Transcriptomics and Antibody-based Proteomics. Mol Cell Proteomics. 2014. doi: 10.1074/mcp.m113.035600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sher F, Hossain M, Seruggia D, et al. Rational targeting of a NuRD subcomplex guided by comprehensive in situ mutagenesis. Nat Genet. 2019. doi: 10.1038/s41588-019-0453-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gnanapragasam MN, Scarsdale JN, Amaya ML, et al. p66 -MBD2 coiled-coil interaction and recruitment of Mi-2 are critical for globin gene silencing by the MBD2-NuRD complex. Proc Natl Acad Sci. 2011. doi: 10.1073/pnas.1015341108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Helbig KL, Farwell Hagman KD, Shinde DN, et al. Diagnostic exome sequencing provides a molecular diagnosis for a significan proportion of patients with epilepsy. Genet. Med 2016. doi: 10.1038/gim.2015.186. [DOI] [PubMed] [Google Scholar]
- 39.Morgan AT, Murray E, Liégeois FJ. Interventions for childhood apraxia of speech. Cochrane Database Syst Rev. 2018. doi: 10.1002/14651858.CD006278.pub3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Estruch SB, Graham SA, Deriziotis P, Fisher SE. The language-related transcription factor FOXP2 is post-translationally modified with small ubiquitin-like modifiers. Sci Rep. 2016. doi: 10.1038/srep20911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eising E, Carrion-Castillo A, Vino A, et al. A set of regulatory genes co-expressed in embryonic human brain is implicated in disrupted speech development. Mol Psychiatry. 2018. doi: 10.1038/s41380-018-0020-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chokas AL, Trivedi CM, Lu MM, et al. Foxp1/2/4-NuRD interactions regulate gene expression and epithelial injury response in the lung via regulation of interleukin. J Biol Chem. 2010. doi: 10.1074/jbc.M109.088468 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


