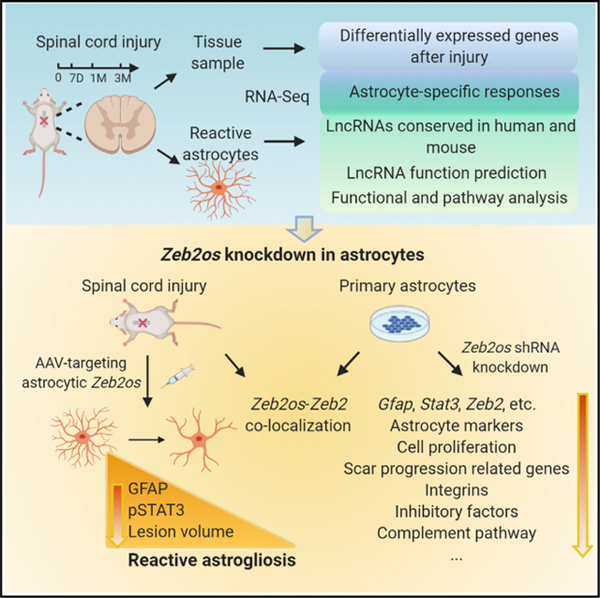
SUMMARY
Spinal cord injury (SCI) is one of the most devastating neural injuries without effective therapeutic solutions. Astrocytes are the predominant component of the scar. Understanding the complex contributions of reactive astrocytes to SCI pathophysiologies is fundamentally important for developing therapeutic strategies. We have studied the molecular changes in the injury environment and the astrocyte-specific responses by astrocyte purification from injured spinal cords from acute to chronic stages. In addition to protein-coding genes, we have systematically analyzed the expression profiles of long non-coding RNAs (lncRNAs) (>200 bp), which are regulatory RNAs that play important roles in the CNS. We have identified a highly conserved lncRNA, Zeb2os, and demonstrated using functional assays that it plays an important role in reactive astrogliosis through the Zeb2os/Zeb2/Stat3 axis. These studies provide valuable insights into the molecular basis of reactive astrogliosis and fill the knowledge gap regarding the function(s) of lncRNAs in astrogliosis and SCI.
Graphical Abstract
In Brief
Wei et al. comprehensively investigate the coding and long non-coding gene expression changes and astrocyte-specific responses in injured spinal cord tissue and purified astrocytes from acute to chronic stages. Bioinformatic and functional analysis identify a conserved lncRNA Zeb2os that plays an essential role in reactive astrogliosis through the Zeb2os/Zeb2/Stat3 axis.
INTRODUCTION
Spinal cord injury (SCI) is among the most devastating of neurological diseases (Petruska et al., 2013). Currently, there is no effective therapeutic solution for SCI. Astrocytes, the predominant component of the scar, can respond to the microenvironment of the injury, become reactive, and proliferate within a short time following SCI. Astrocytes play crucial roles in neural protection and repair, and can reduce the spread of inflammatory cells to undamaged tissue at the acute injury stage (Barres, 2008; Hackett and Lee, 2016; Khakh and Sofroniew, 2015; Mironets et al., 2016; Molofsky et al., 2012; Narang and Zheng, 2018; Sofroniew, 2014; Tran et al., 2018). Reactive astrocytes can also secret inhibitory molecules and block axonal regeneration (Filous and Silver, 2016; Hackett and Lee, 2016; Hara et al., 2017; Mironets et al., 2016; Su et al., 2014; Tran et al., 2018). Understanding the complex contributions of reactive astrocytes to SCI pathophysiologies is fundamentally important for developing therapeutic strategies.
To better understand the mechanisms of reactive astrogliosis in SCI, identify molecular targets in the injury environment, and improve axonal regeneration, we have investigated gene expression changes in SCI epicenter tissue and purified astrocytes from adult mouse spinal cords at both acute and chronic injury stages using RNA-sequencing (RNA-seq) (Cabili et al., 2011; Cuevas-Diaz Duran et al., 2019b; Mercer et al., 2008; Pastori and Wahlestedt, 2012; Sauvageau et al., 2013). The comparison of RNA-seq datasets of spinal cord tissue and purified astrocytes provided valuable information on the overall changes in gene expression in the injury environment also the astrocyte-specific responses and contributions. We also compared the RNA-seq datasets that we generated from tissue samples from mouse and rat SCI and identified the common bioprocesses between these widely used rodent SCI models.
We have previously systematically analyzed the expression profiles of long non-coding RNAs (lncRNAs; >200 nt in length and lacking an open reading frame) and protein-coding genes in SCI (Chen et al., 2013; Duran et al., 2017). LncRNAs regulate many biological processes including embryonic stem cell differentiation, neurogenesis, and cancer (Briggs et al., 2015; Cuevas-Diaz Duran et al., 2019a, 2019b; Dong et al., 2015; Sauvageau et al., 2013). Several studies including ours have revealed that lncRNAs are transcribed in tissue- and cell-type-specific manners to exert their functions (Cabili et al., 2011; Cuevas-Diaz Duran et al., 2019b). Gene expression profile analyses have revealed significant changes in lncRNA expression in rats or mice after SCI (Duran et al., 2017; Li et al., 2019). At the cellular level, lncRNAs regulate the expression of protein-coding RNAs and hence participate in neuronal death, demyelination, and glia activation (Li et al., 2019). Therefore, lncRNAs represent promising biomarkers for the diagnosis, treatment, and prognosis of SCI. However, there is still a significant knowledge gap regarding the functional role(s) and mechanisms of lncRNAs in reactive astrogliosis and SCI.
We have tested a number of candidate lncRNAs that are upregulated in reactive astrocytes after SCI in mice and identified a gene, Zeb2os, which is highly conserved with a human lncRNA. Zeb2os and its antisense protein-coding gene Zeb2 (zinc finger E-box-binding homeobox 2, which is also named Sip1, Smad-interacting protein 1) are both upregulated in astrocytes purified from injured spinal cords at 7 days (7D) post-injury and remain elevated at 1 and 3 months (1M and 3M) post-injury. Additionally, our RNAscope experiment showed that Zeb2os and Zeb2 co-localize in cultured primary astrocytes and SCI tissue sections. Zeb2 mutations have been reported in human Mowat-Wilson syndrome (Yamada et al., 2014), and important roles of Zeb2 in myelination in the CNS (Weng et al., 2012), Schwann cell differentiation (Wu et al., 2016b), and Bergmann glia development (He et al., 2018) have been described. When we submitted the present manuscript, Vivinetto et al. (2020) had just reported that Zeb2 plays a role in reactive astrogliosis at the acute injury stage, in agreement with our findings. Additionally, Zeb2os expression is highly correlated with that of an essential transcription factor (TF) in reactive astrogliosis, Stat3, at different stages of SCI. Signal transducers and activators of transcription 3 (STAT3) signaling can be triggered by upstream regulators such as ciliary neurotrophic factor (CNTF) and leukemia inhibitory factor (LIF), and STAT3 plays critical roles in reactive astrogliosis, upregulation of GFAP expression, and scar progression (Anderson et al., 2016; Herrmann et al., 2008; Liddelow and Barres, 2017; Liddelow et al., 2017). Here, we have demonstrated by short hairpin RNA (shRNA) gene knockdown (KD) that Zeb2os performs important functions during reactive astrogliosis in both primary astrocyte cultures and SCI animal models. Zeb2os KD in primary astrocytes led to significantly decreased expression of genes including Gfap, Zeb2, Stat3, and other genes involved in scar progression. Furthermore, our chromatin-immunoprecipitation sequencing (ChIP-seq) experiment revealed that STAT3 bound to Zeb2 promoter region. Thus, Zeb2os regulates Zeb2 and Stat3 directly or indirectly and STAT3 might regulate Zeb2 expression. Importantly, we verified that Zeb2os KD reduced reactive astrogliosis in mouse models of SCI. Finally, we have built a valuable resource (GEO: GSE153721) for the scientific community to investigate the functions of reactive astrocytes from acute to chronic stages after CNS injury.
RESULTS
Systematic analysis of coding and lncRNAs at the SCI epicenter at acute and chronic stages
In order to understand the molecular mechanisms involved in acute and chronic SCI pathologies, we carried out RNA-seq of SCI epicenter tissue from acute (2D and 7D) (Chen et al., 2013) and chronic (1M and 3M) stages after SCI (Figures S1A and S1B; Table S1). We used the moderate contusive SCI, which is a clinically relevant model with extensive scarring (Chen et al., 2013; Duran et al., 2017). A consensus dendrogram showed that all the samples were clustered well by stages, which suggests that the biological replicates were highly consistent (Figure S1C).
Recent studies have revealed that lncRNAs are abundant in the genome and may play critical roles in nervous system disorders, including SCI (Briggs et al., 2015). We have built a comprehensive lncRNA annotation database by combining GENCODE and NCBI annotations (Figures S1D–S1G; Table S2; STAR methods). To evaluate temporal transcriptional dynamics, we performed pairwise comparisons between samples from different time points following SCI and control and hierarchical clustering of 6,453 differentially expressed genes (DEGs) including 5,675 protein-coding genes and 778 lncRNAs that are DEGs in at least one comparison. Five clusters were sufficient to represent non-overlapping gene profiles with distinct enriched gene sets, as indicated in Figure 1A. For example, the expression of genes in cluster 2, which are enriched in gene sets associated with protein metabolism and synthesis, was elevated during acute stages and gradually decreased during chronic stages. The expression of genes in cluster 3, which are enriched in extracellular matrix-related gene sets, increased during chronic stages. Selected highly enriched gene sets from acute and chronic SCI stages are shown in Figures 1B and 1C. For example, genes encoding putative proteins with functions in cell proliferation and differentiation are enriched during acute stages, whereas those encoding putative cell-cell adhesion and extracellular matrix proteins are enriched in chronic stages.
Figure 1. Systematic analysis of coding and long non-coding RNAs in the SCI epicenter at acute and chronic stages in mouse.
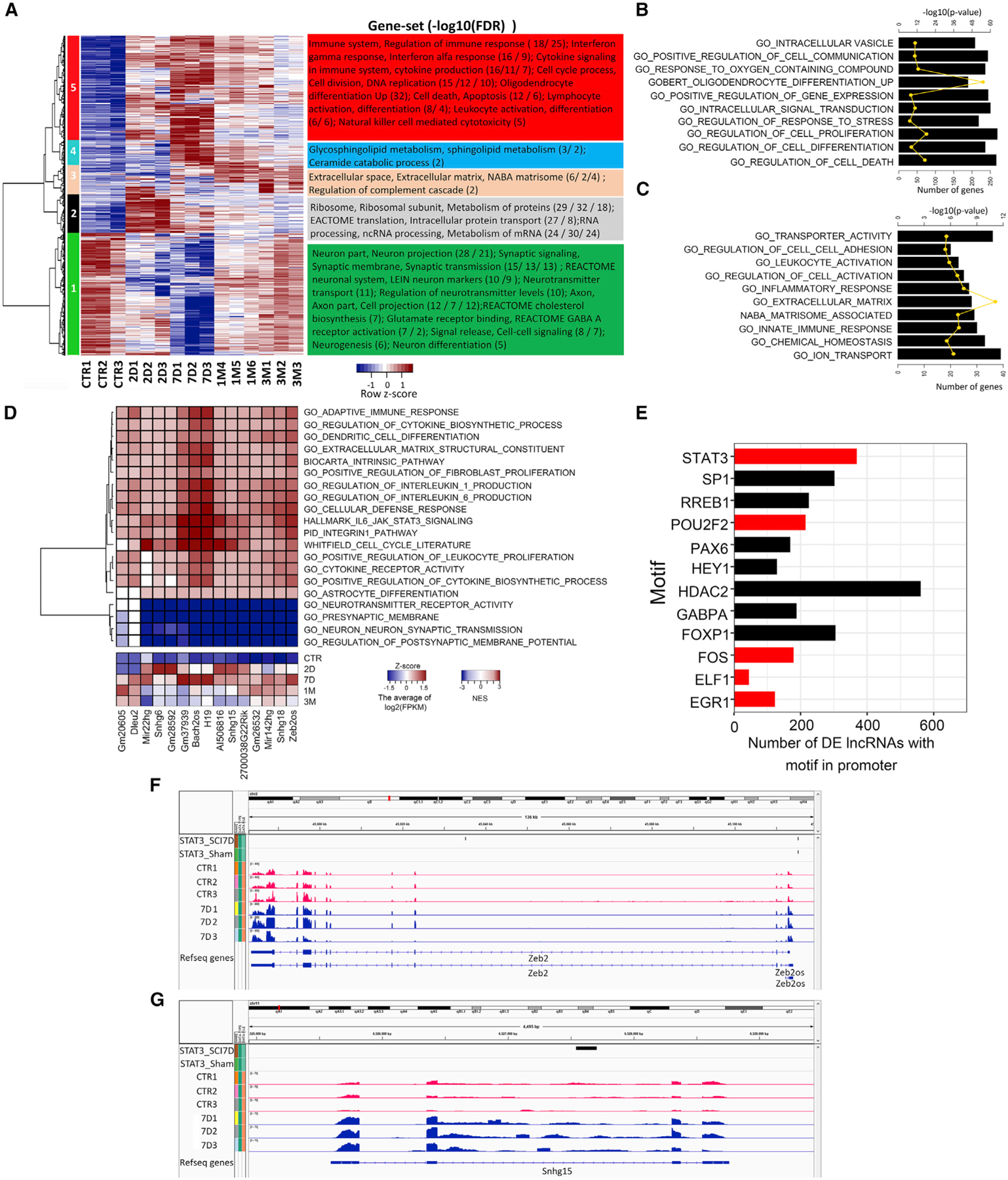
(A) Hierarchical cluster analysis showing expression patterns of DEGs over the SCI time course. The enriched gene sets in each cluster were identified using a hypergeometric test (FDR <0.05). CTR, sham control group.
(B and C) Gene set enrichment analysis. The most enriched gene sets for acute (B) and chronic (C) SCI stages (p value <0.05).
(D) Upper panel: heatmap representing an association matrix of upregulated conserved lncRNAs and enriched functional terms. Lower panel: the average of log2(FPKM) of selected lncRNAs.
(E) TFs with binding motifs found in the upstream regulatory regions (5 kb upstream and 1 kb downstream of transcription start site [TSS]) of more than 120 DE lncRNAs. The x axis indicates the number of DE lncRNAs containing binding motifs for particular TFs. DE TFs are shown in red and non-DE TFs in black. FDR <0.01.
(F and G) Genome browser views displaying STAT3 ChIP-seq binding signals in the top two tracks and gene expression in the remaining tracks for Zeb2 (F) and Sngh15 (G) within 5 kb upstream of the TSS of the genes and within the gene bodies.
Both mouse and rat are widely used animal models for SCI research, but due to their physiological and life-span differences, not all aspects of SCI injury pathology are comparable between the two. In order to identify the common bioprocesses during SCI between these species, we re-analyzed the DEGs from acute and chronic stages in moderate contusive SCI rat models using data published by our group and others with similar injury models and sample collection methods (Duran et al., 2017; Shi et al., 2017) (Figure S1H; Table S3). The enriched pathways of the common DEGs in mouse and rat after SCI were identified using gene set enrichment (MSigDB). DEGs related to STAT3 signaling, and the nuclear factor κB (NF-κB) and cell-cycle pathways were enriched mainly during acute stages, and those related to extracellular space and biological adhesion were enriched mainly during chronic stages (Figures S1I and S1J).
Taken together, our gene expression profile analyses have demonstrated significant changes in the expression of not only protein-coding genes but also lncRNAs in both mouse and rat models of SCI.
Differential expression of lncRNAs and TFs potentially regulating lncRNA expression changes after SCI
We found that although hundreds of lncRNAs are differentially expressed (DE) after SCI (Figure S1A), few of these have been previously studied (Li et al., 2019). To address the potential contribution of DE lncRNAs to the progression of SCI, we identified 297 DE lncRNAs that were upregulated at least in one time point after SCI. We analyzed the homology of upregulated lncRNAs in mouse and human using slncky software (Chen et al., 2016) and found 15 lncRNAs homologous between species. The results of ‘‘guilt-by-association’’ analysis (see STAR methods) showed that the expression of these lncRNAs is significantly correlated with that of protein-coding genes with functions such as immune response, cell differentiation, cell proliferation, and cytokine biosynthesis (Figure 1D). Some of these lncRNAs also exhibit a high degree of secondary structure conservation between species, which also suggests conserved functions (e.g., 2900097C17Rik, Figure S2A).
Moreover, we have found TFs that are differentially expressed following SCI (Figure S1A). To identify TFs that regulate lncRNA expression during the progression of SCI pathology, we searched for TF-binding motifs in the regulatory regions of the DE lncRNAs. Occurrences of known and discovered ENCODE motifs were queried using FIMO, and a total of 50 significant TF motifs were found among the DE lncRNAs. These TF motifs were further filtered to include only those found in more than 120 promoter regions of the DE lncRNAs (Figure 1E). Interestingly, our results showed that 369 out of 778 (47%) DE lncRNA promoters contained STAT3 motifs (Figure 1E), which suggested that Stat3 may have important functions in the regulation of lncRNAs after SCI.
Previous studies have demonstrated that STAT3 is a critical TF in reactive astrogliosis after SCI (Herrmann et al., 2008). STAT3 can modulate astrocyte functions such as proliferation, migration, and cell-cycle processes (Herrmann et al., 2008). Astrocytes are the predominant component of the glial scar around the SCI epicenter. In order to comprehensively understand the molecular targets regulated by STAT3 after SCI, we performed a STAT3 ChIP-seq experiment using sham and injured spinal cord epicenter tissue encompassing the scar at 7D post SCI, and found 13,465 target genes for STAT3 after SCI. The specific genes that contained STAT3 binding peaks after SCI were enriched in the functions cell death and survival, cell morphology, and cellular growth and proliferation, among others (Figure S3A). To determine whether there was any correlation between the occurrence of STAT3 binding targets and the expression of any genes at 7D after SCI, we performed gene set enrichment analysis (GSEA) (Subramanian et al., 2005) (Figure S3B). GSEA revealed statistically significant enrichment of STAT3 (normalized enrichment score [NES] = 1.33, nominal p value = 0, and false discovery rate [FDR] = 0) targets among genes that were upregulated at 7D post-injury when compared to a random distribution. For example, our data revealed STAT3 binding peaks in the promoter regions of Zeb2 (Figure 1F). We also found 152 lncRNAs that are differentially expressed and have STAT3 binding peaks after SCI (Figure S3C). For example, lncRNA Snhg15 has STAT3 binding peaks and its expression increased after SCI (Figure 1G).
The dynamics of reactive astrocyte gene expression in a time course after SCI
To investigate the molecular basis of reactive astrogliosis, we purified astrocytes from the epicenter tissue of sham and moderately contused GFAP-Cre: R26-tdT adult mice at 7D, 1M, and 3M post-injury by fluorescence-activated cell sorting (FACS) based on GFAP-tdTomato expression using a published protocol (Figure S4A) (Cahoy et al., 2008; Zamanian et al., 2012; Zhang et al., 2014). Compared to tdTomato-negative cells or spinal cord tissue, qPCR of cell marker genes showed that tdTomato-positive cells expressed minimal levels of markers from contaminating oligodendrocyte lineage cells (Mbp), neurons (Tuj1), or microglia (Iba1) and high levels of the astrocyte marker Gfap, indicating that the isolated astrocytes were relatively pure (Figure 2A).
Figure 2. Purification of astrocytes over the SCI time course and RNA-seq.
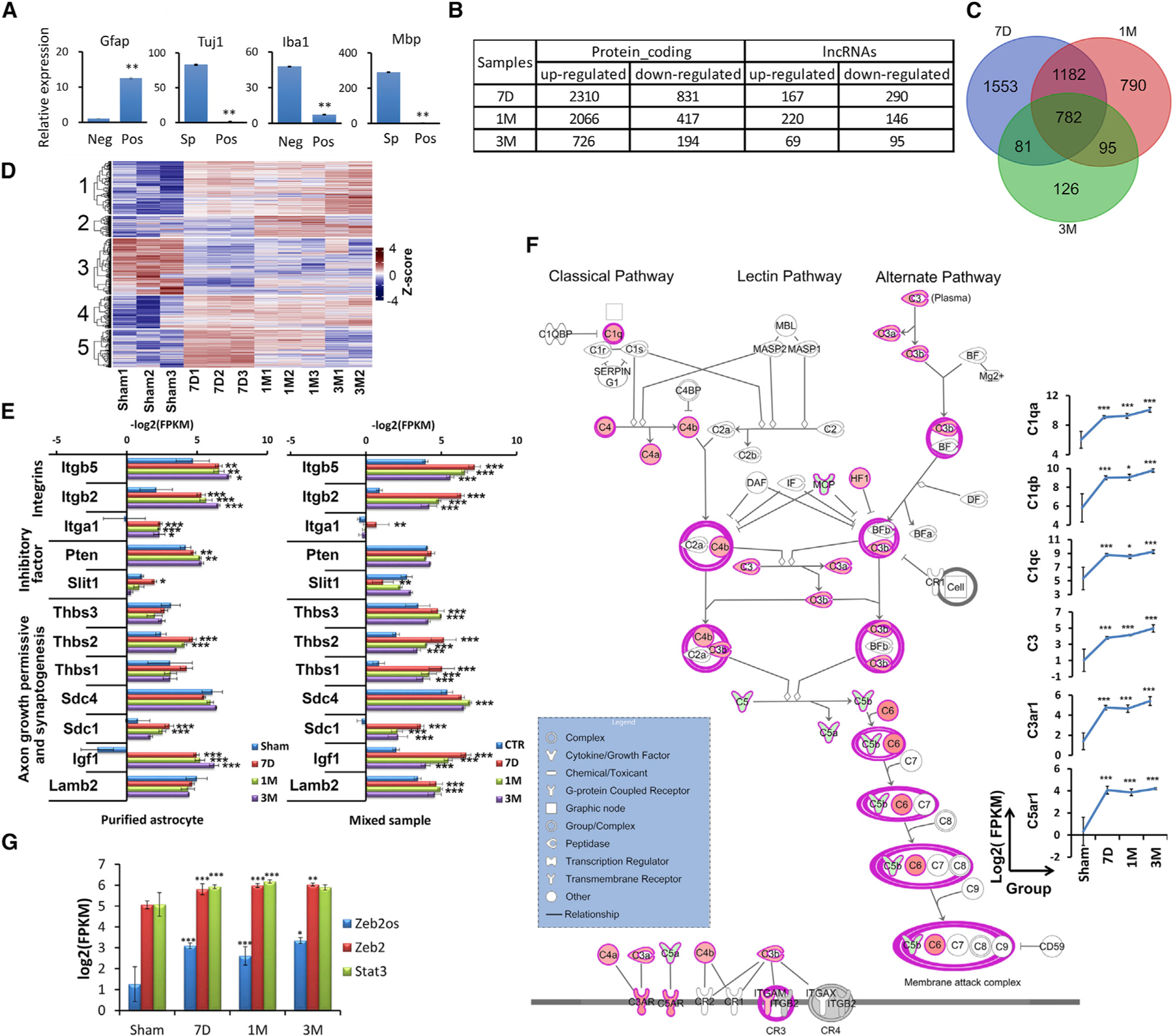
(A) qPCR result for the expression of different cell-type markers in FACS-sorted tdTomato-positive cells (pos) compared with sorted tdTomato-negative cells (neg) or uninjured spinal cord tissue (sp). Data are presented as means ± SEM; n = 3 independent experiments; **p < 0.01 compared with neg or sp (independent t test).
(B) Differentially expressed (DE) protein-coding genes and lncRNAs in purified astrocytes after SCI (log2|fold-change| >1, FDR <0.05, at least one sample fragments per kilobase of exon model per million reads mapped [FPKM] >1).
(C) Venn diagram depicting the extent of overlap between expression of DEGs at different time points in purified astrocytes.
(D) Hierarchical cluster analysis showing expression patterns of DEGs for each sample.
(E) Comparison of RNA-seq results for expression of integrins, inhibitory factors, and axon growth permissive and synaptogenesis factors in purified astrocytes and spinal cord tissue samples (*FDR <0.05, **FDR <0.01, ***FDR <0.001).
(F) Complement system pathway is enriched in purified astrocytes by RNA-seq at 7D after SCI (*FDR <0.05, **FDR <0.01, ***FDR <0.001).
(G) Transcript abundances of Zeb2os, Zeb2, and Stat3 in purified astrocytes from sham samples and SCI stages are correlated (*FDR <0.05, **FDR <0.01, ***FDR <0.001).
Our RNA-seq analysis provided valuable information on astrocyte gene expression dynamics after SCI, when the expression of thousands of protein-coding genes and hundreds of lncRNA genes changes (Figures 2B and 2C; Table S4). The enrichment analysis of the common DEGs from all stages showed that these genes were enriched not only in immune system and cytokine production but also in secretory vesicle and complement system functions (Figure S4B), which suggests that astrocytes might function during cell-cell communication through secreted vesicle and complement signaling. Additionally, we defined five expression patterns in astrocytes during the SCI time course (Figure 2D) and performed Ingenuity Pathway Analysis (IPA) of genes enriched in acute and chronic stages. Genes in cluster 5 that are highly expressed during acute stages (7D post-injury) are enriched in, for example, cell cycle (-log10(p value) = 6.273), Wnt/β-catenin signaling (-log10(p value) = 2.022), and RhoA signaling (-log10(p value) = 1.983). Genes in cluster 2 that are highly expressed in chronic stages (1M and 3M) are enriched in FGF (-log10(p value) = 3.025) and neuroinflammation signaling (-log10(p value) = 2.699). Our results also showed that some axon growth permissive molecules relatively decreased in chronic astrocytes (1M) compared to acute reactive astrocytes (7D), and some axon growth inhibitory factors and scar forming related genes increased in chronic astrocytes (1M) compared to acute reactive astrocytes (7D) (Table S5).
To reveal overall gene expression changes in the injury environment and the astrocyte-specific responses and contributions, we compared the expression of integrins, inhibitory factors, axon growth permissive and synaptogenesis factors between RNA-seq datasets of spinal cord tissue and purified astrocyte after SCI (Anderson et al., 2016; Chung et al., 2015; O’Shea et al., 2017). The levels of these factors are dynamically regulated across injury stages. Percentile ranking of gene expression in all samples showed 616 genes in cluster 2, including Lamb1, Ncam1, and Bmp7 (Figure S4C), with higher-ranking expression in purified astrocytes compared to bulk tissue samples, indicating that astrocytes is a critical cell type for expressing genes encoding these axon growth permissive factors after SCI. In contrast, the expression of genes encoding some other permissive and synaptogenesis-related factors such as thrombospondin 1 (Thbs1), Thbs3, syndecan-4 (Sdc4), and laminin subunit beta 2 (Lamb2) were significantly elevated in tissue samples at specific stages, but not in purified astrocytes (Figure 2E). Moreover, the expression of some inhibitory factors such as slit guidance ligand 1 (Slit1) at 7D and phosphatase and tensin homolog (Pten) at both 7D and 1M was also elevated in purified astrocytes but not in tissue samples (Figure 2E). Further, the expression of integrin-encoding genes including Itga1, Itgb2, and Itgb5 (Figure 2E) was significantly upregulated in both post-SCI tissue and purified astrocyte samples. Interestingly, the abundance of Itga1 transcripts increased in tissue samples only at 7D, but was elevated in purified astrocytes at all SCI stages (Figure 2E).
We analyzed enriched pathways among the DE genes expressed in purified astrocytes after SCI. Complement pathway (7D post-injury: -log10(p value) = 4.279) and STAT3 pathways (7D post-injury: -log10(p value) = 6.326) are remarkably upregulated among the enriched signaling pathways (Figures 2F and S4D). The complement cascade is an important component of the innate immune system that triggers early inflammatory responses, and many complement pathway genes have an effect on axon growth and neuronal functions (Peterson et al., 2015). The complement cascade is composed of classical, lectin, and alternative activation pathways which converge at the point of C3 activation. In this study, the expression of a large number of complement genes in classical and alternative pathways was significantly elevated at both acute (7D; shown in IPA schematic in Figure 2F) and chronic stages of SCI (e.g., C3 and C3ar1). In addition, the STAT3 pathway is known to regulate reactive astrogliosis after SCI (Ben Haim et al., 2015; Herrmann et al., 2008). Our data showed enrichment of STAT3 signaling pathways at all stages of SCI with increased expression of multiple components including STAT3 itself and downstream genes (Figure S4D).
Highly conserved lncRNA Zeb2os and its antisense protein-coding gene Zeb2 are upregulated in astrocytes after SCI and co-localize
We identified DE lncRNAs homologous to human genomic regions that contain single-nucleotide polymorphisms associated with diseases (Buniello et al., 2019; Landrum et al., 2014). The guilt-by-association analysis of these conserved lncRNAs in purified astrocytes revealed that Zeb2os has a highly significant positive correlation with astrocyte functions, STAT3 pathway, and integrin pathway (Figure S4E). Zeb2os is also a highly conserved in human (ZEB2-AS1) (Figure S2B). Our RNA-seq data showed that the expression Zeb2os and its antisense and overlapping protein-coding gene Zeb2 both increased in astrocytes after SCI at 7D and remain elevated at 1M and 3M (Figure 2G). Their expressions had high correlation at different SCI stages. Our observations are consistent with the increased expression of Zeb2 in the astrocyte gene expression dataset generated by Anderson et al. (2016) in crush-injured mice at 14 days post SCI compared to uninjured control mice. Zeb2os expression is also highly correlated with that of Stat3 over the injury time course (Figure 2G). RNAscope and immunostaining results verified that Zeb2os was expressed in primary astrocyte culture after scratch assays and in reactive astrocytes after SCI (Figures 3A and 3B).
Figure 3. Zeb2os is expressed and colocalized with Zeb2 in reactive astrocytes and injured spinal cord.
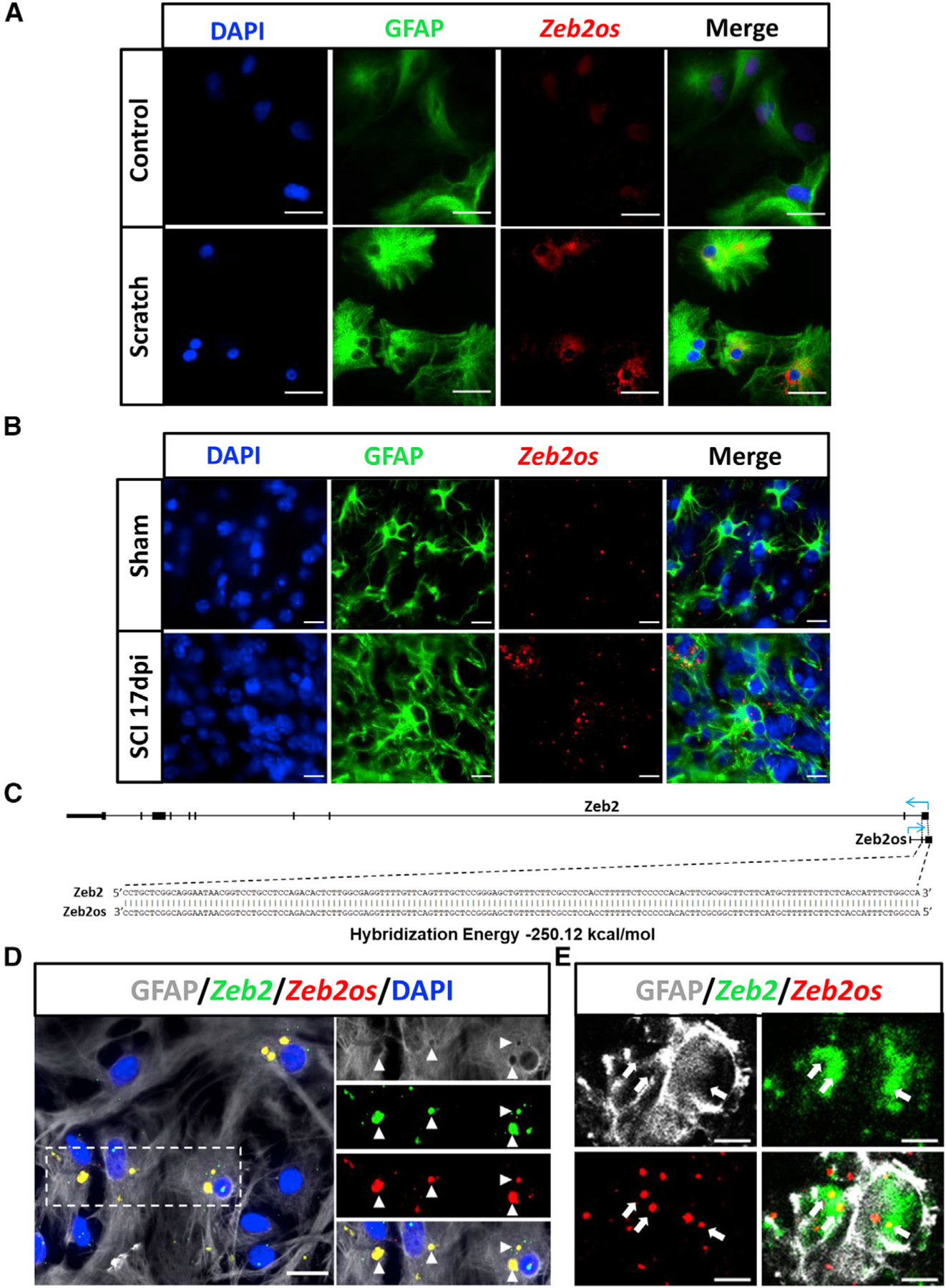
(A and B) Representative images showing the colocalization of Zeb2os mRNA (red) and GFAP protein (green) in vitro (scratched astrocytes after 48 h) (scale bars, 50 µm) (A), and in vivo (B) (sham and mice 17 days post-injury [dpi]) (scale bars, 20 µm). (C) Bioinformatic analysis of the probability of Zeb2os-Zeb2 interaction using IntaRNA 2.0.
(D and E) Combination of RNAscope and immunohistochemistry showing Zeb2os mRNA (red) colocalized with Zeb2 mRNA (green) (indicated by triangles) in scratched astrocytes (D), Insets show a magnification of the boxed area (scale bar, 20 µm), and in injured spinal cord 17 days post-injury (E) (indicated by arrows) (scale bar, 3 µm).
LncRNAs are versatile molecules that can regulate gene transcription through RNA-RNA, RNA-DNA (e.g., binding to DNA promoter of TGs), or lncRNA-protein interactions. Our bioinformatics analysis using IntaRNA 2.0 (Mann et al., 2017) indicates a high probability of Zeb2os-Zeb2 interaction (Figure 3C). This result has been verified using a combination of RNAscope in situ hybridization and both in vitro and in vivo immunofluorescence with images captured using both an inverted fluorescence microscope and a confocal microscope (Figures 3D and 3E). We observed that co-localization of Zeb2os and Zeb2 mRNA occurred mostly at outer edge of astrocyte nuclei and cytoplasm.
Zeb2os plays an important role in reactive astrogliosis
To investigate the role of Zeb2os in astrocytes, we knocked down Zeb2os by shRNA using lentivirus in primary astrocyte cultures. Compared to the luciferase control, the Zeb2os KD group showed a significant decrease in the expression of Zeb2os, Zeb2, and Gfap, as shown by qPCR (Figure 4A). Thus, it is possible that Zeb2os plays a role in reactive astrogliosis by regulating Zeb2. Further, astrocyte proliferation significantly decreased in Zeb2os KD astrocytes, as shown by bromodeoxyuridine (BrdU) staining (Figure 4C). However, scratch wound assay showed astrocyte migration was not significantly affected by Zeb2os KD compared to control (Figures S5A–S5C). We also performed Zeb2 KD in primary astrocytes using shRNA. RNA-seq results showed that Zeb2 KD is successful (with ~6-fold reduction in Zeb2 expression). Zeb2 KD led to downregulation of Pten, Gsk3b, as well as the families of Adam, Integrin, Aqp, and Cdh genes (Figure S6A). We also found that knocking down Zeb2 decreased astrocyte proliferation shown by BrdU staining and Cspg4 expression verified by qPCR (Figure 4B). Note that CSPG4 (NG2) is a well-established oligodendrocyte precursor cell (OPC) marker, thereby we further verified its expression in primary cell culture by triple-staining of CSPG4, GFAP, and PDGFRα. Figure S5D showed CSPG4 is localized in GFAP-positive astrocytes, consistent with the previous study (Anderson et al., 2016). Additionally, there was a trend of reduced Gfap expression in KD group compared to control group but not statistically significant (Figure 4B). Therefore, Zeb2os KD caused a greater reduction in Gfap expression and astrocyte proliferation in cultured primary astrocytes than did Zeb2 KD.
Figure 4. Zeb2os and Zeb2 shRNA knockdown.
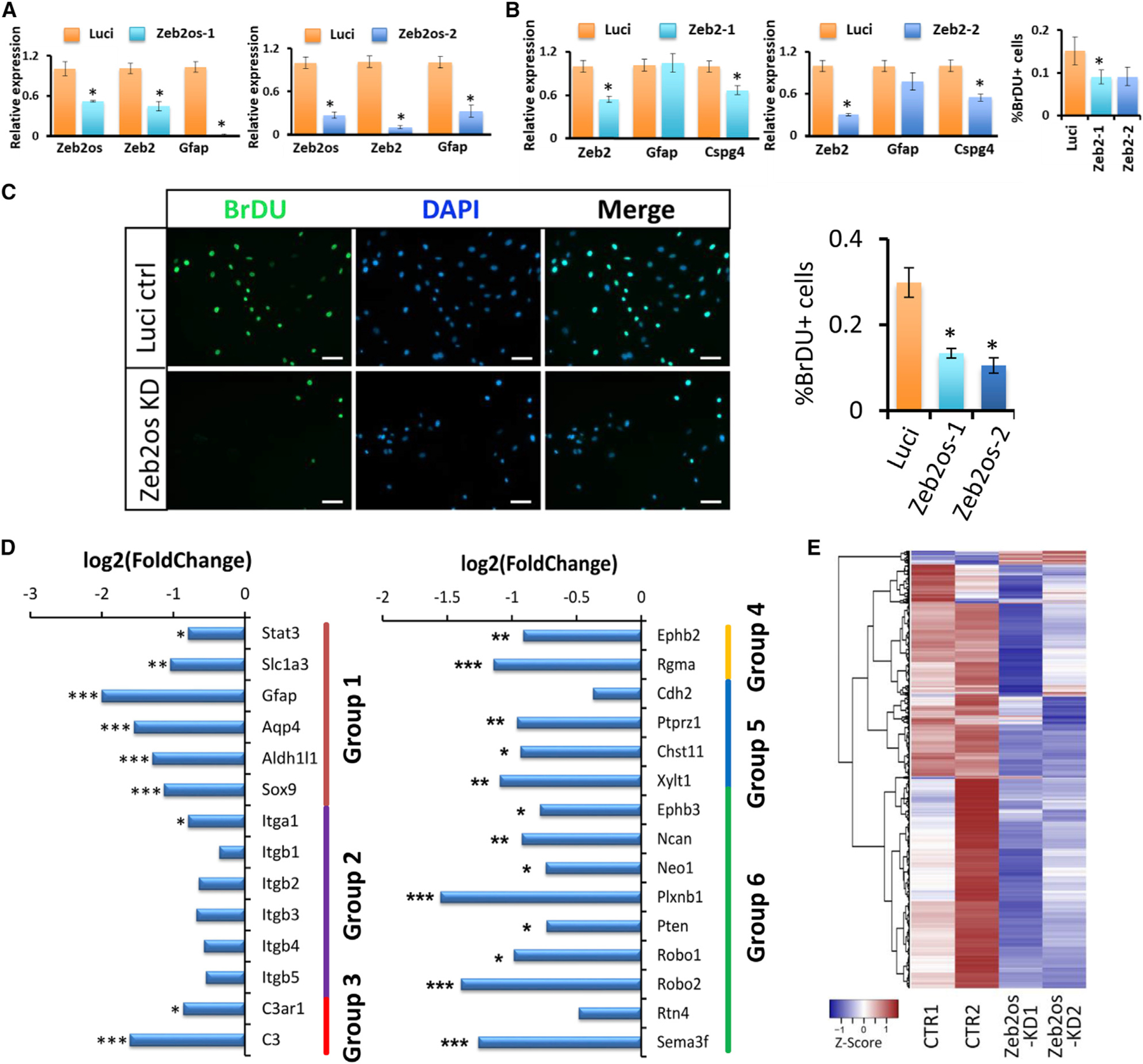
(A) qPCR analysis of Zeb2os KD in primary astrocytes using lentivirus. Data are presented as means ± SEM; n = 3 independent experiments; *p < 0.05 compared with Luci (independent t test); Luci, Luciferase shRNA control; Zeb2os-1 and Zeb2os-2, two different Zeb2os shRNA knockdown constructs.
(B) qPCR analysis of Zeb2 KD. Graph showing the comparison of BrdU-positive cells per unit field between the control and Zeb2 KD groups. The percentage of BrdU-expressing cells is significantly decreased in the Zeb2-1 KD group. Results are expressed as the mean ± SEM for each group (n = 3). *p < 0.05 compared with Luci (independent t test); Zeb2-1 and Zeb2-2, two Zeb2 shRNA knockdown constructs.
(C) DAPI (blue)/BrdU (green) double labeling in primary astrocytes from control (Luci ctrl) and Zeb2os KD groups. Comparison of BrdU-positive cells per unit field between the control and Zeb2os KD groups (scale bar, 100 µm). The percentage of BrdU-expressing cells is significantly decreased in the Zeb2os KD group. Results are expressed as the mean ± SEM for each group (n = 3). *p < 0.05 compared with Luci (independent t test).
(D) RNA-seq expression results for comparison of Zeb2os knockdown astrocytes to control. Group 1: Stat3 and astrocyte markers; group 2: integrins; group 3: complement genes; group 4: scar formation-related genes; group 5: SA markers; and group 6: inhibitory factors. *FDR <0.05, **FDR <0.01, ***FDR <0.001 (n = 2).
(E) Hierarchical cluster analysis of expression patterns of DEGs for Zeb2os KD (|fold-change| >1.5, FDR <0.05, at least one sample FPKM >1). CTR, control group.
We also performed RNA-seq using RNA samples from Zeb2os KD cells compared with those from negative control cells. Zeb2os KD in primary astrocytes affected the expression of a number of downstream genes (Figure 4D; Table S6). For example, the expression of RNAs encoding TF Stat3 and astrocyte markers such as Slc1a3, Gfap, Aqp4, and Aldh1l1 all significantly decreased in Zeb2os KD samples (group 1 in Figure 4D). Additionally, the expression of genes encoding integrin-family proteins also decreased (group 2 in Figure 4D). Moreover, we found that the expression of many genes related to scar development and inhibitory factors including Ephb2, Rgma (group 4 in Figure 4D), Ephb3, neurocan (Ncan), Neo1, Plxnb1, Pten, Robo1, Robo2, and Sema3f (group 6 in Figure 4D) (Anderson et al., 2016; Benson et al., 2005; Bundesen et al., 2003; Renault-Mihara et al., 2017; Wanner et al., 2008; Zhang et al., 2018) decreased in Zeb2os KD cells. Furthermore, in Zeb2os KD cells, the expression of previously reported scar-forming astrocyte (SA) marker genes, including Cdh2, phosphacan (Ptprz1), Chst11, and Xylt1 (group5 in Figure 4D) (Hara et al., 2017) also uniformly displayed either significant reduction or decreasing tendencies. These results suggest that knocking down Zeb2os might attenuate astrocyte scar progression. Therefore, we carried out functional and pathway analyses for downstream DEGs affected by Zeb2os KD (Figure 4E). Some of the signaling pathways affected by Zeb2os KD include the complement signaling pathway (-log10(p value) = 1.983) and the cell-cycle G1/S checkpoint regulation pathway (-log10(p value) = 2.23), which were significantly suppressed in Zeb2os KD astrocytes (Figures S6E and S6F), which is consistent with our BrdU cell proliferation result (Figure 4C). Most of the DEGs in the complement pathway such as C3 and C3ar1 (group 3 in Figures 4D and S6E) also showed significantly decreased expression in Zeb2os KD astrocytes.
To further verify the role of Zeb2os in reactive astrogliosis, we designed a Cre-dependent Zeb2os KD eGFP-adeno-associated virus (AAV) vector (Figure 5A) and validated its KD effect in primary astrocytes cultured from GFAP-Cre transgenic mice. qPCR of Zeb2os KD cells compared to controls showed significantly decreased expression of Zeb2os, Zeb2, and Gfap (Figure 5B), which was consistent with previous lentivirus results (Figure 4A). We then used AAV administration of control and Zeb2os KD constructs into GFAP-Cre SCI mice at 10 days post-injury (dpi) when the scar formation was well under way but not yet complete. Spinal cords were collected after another 7 days (17 dpi) following AAV injection (Figure 5A). Figure 5C showed that the Zeb2os KD AAV specifically infected astrocytes in the injured GFAP-Cre mouse spinal cord. To verify the AAV knockdown effect on Zeb2os in vivo, lncRNA was detected using RNAscope. Sections in one set of slides were used for co-detection of Zeb2os mRNA, egfp mRNA, and DRAQ5 Far-Red DNA dye. Zeb2os expression was then quantified in cells co-labeled with egfp and DRAQ5. We observed a significant reduction of Zeb2os expression after its knockdown compared to control (Figures 5D and 5E). Next, the impact of Zeb2os KD on reactive astrogliosis was assessed by measuring the expression of GFAP and phosphorylated STAT3 (pSTAT3) (Herrmann et al., 2008). The percentages of GFAP-immunoreactive (IR) areas in the Zeb2os KD AAV-transduced astrocytes (GFPIR) significantly decreased in the Zeb2os KD group compared to the control group (400, 800, and 1,200 µm rostral and 400 and 1,200 µm caudal to the injury epicenter) (Figures 6A and 6B). The percentages of pSTAT3-IR areas in the Zeb2os KD AAV-transduced astrocytes (GFP-IR) also significantly decreased in the Zeb2os KD group compared to the control group at the epicenter and surrounding injured segment (400 and 800 µm rostral and 800 µm caudal to the injury epicenter) (Figures 6C and 6D). Additionally, we evaluated whether Zeb2os KD affects lesion volume by measuring the lesion core using GFAP immunostaining. The lesion areas surrounded by astrocyte borders were regarded as lesion cores (Bellver-Landete et al., 2019; Butenschön et al., 2016; Renault-Mihara et al., 2011; Wanner et al., 2013). The result revealed a reduction of the lesion volume at 17 days post-SCI in Zeb2os KD-treated mice compared to the control group (Figure 6E). Furthermore, we assessed neuroinflammation by staining CD68, a common marker of microglia/macrophages (Cheng et al., 2018; Gallegos et al., 2020). Result showed the percentage of CD68-IR area in cross sections at various distance from epicenter showed no significant difference between Zeb2os KD and control groups, indicating the neuroinflammation was not exacerbated by Zeb2os KD (Figures S7A and S7B). We also used 5-HT (serotonin) immunostaining to evaluate whether knocking down Zeb2os affected serotonin axon sprouting after SCI. 5-HT-positive axons originated from the brainstem serotonergic neurons mainly innervate neurons at spinal ventral horns (Gallegos et al., 2020; Lee et al., 2010). We observed an increasing trend at 400, 800, and 1,600 µm rostral and 400, 1,600, and 2,000 µm caudal to the injury epicenter at 7D after Zeb2os KD although they were not statistically significant (Figures S7C and S7D). Taken together, the above results suggest Zeb2os KD can attenuate reactive astrogliosis by regulating GFAP and pSTAT3 expression and prevent the sequential damage by reducing the lesion size after SCI.
Figure 5. Zeb2os shRNA knockdown by AAV transduction in GFAP-Cre mice at 17 days after SCI.
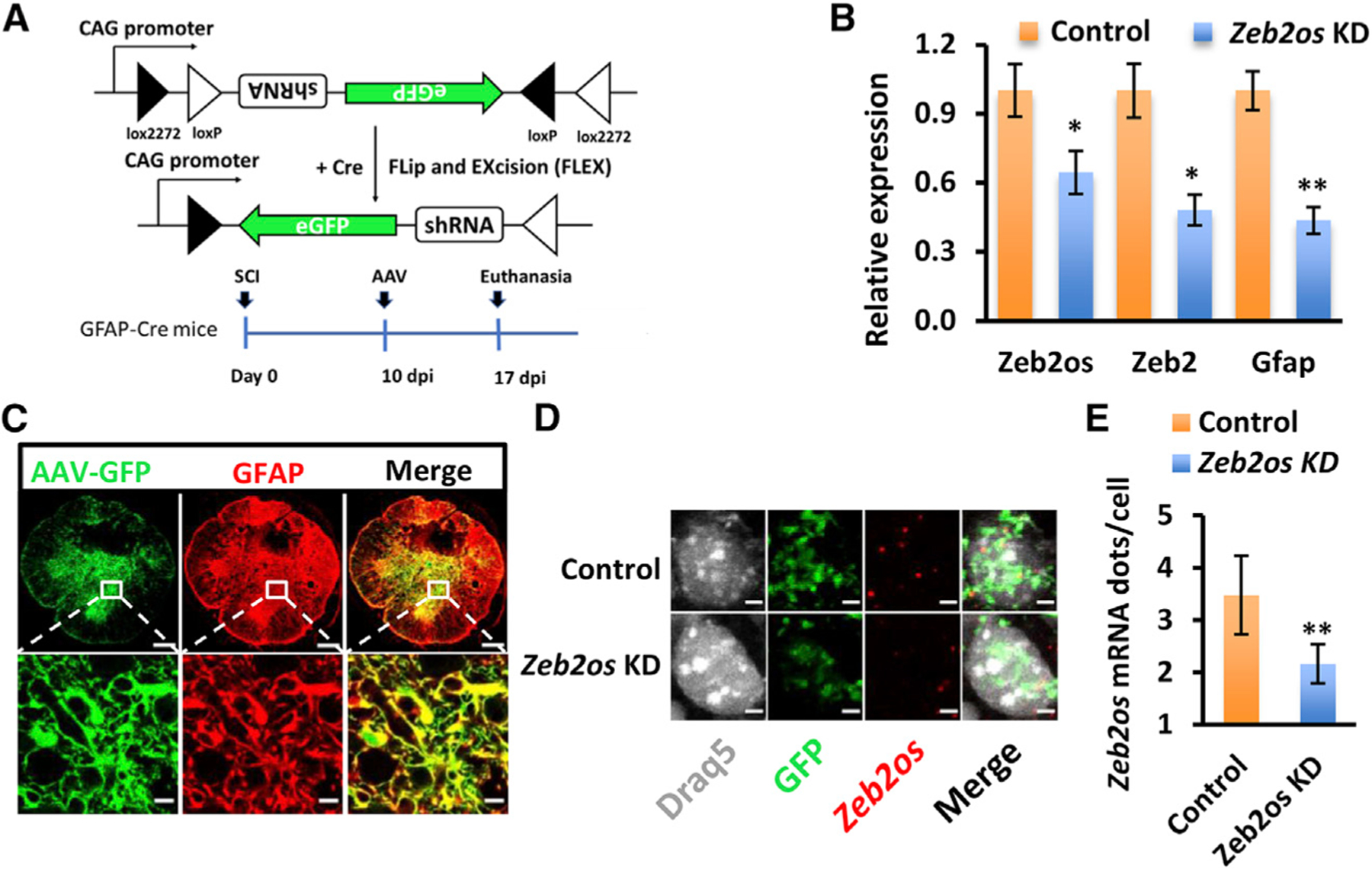
(A) Design of Cre-dependent Zeb2os shRNA KD AAV and experimental timeline of SCI followed by AAV injection in GFAP-Cre transgenic mice. Graph showing shRNA KD using AAV generated consistent results as lentivirus KD.
(B) qPCR analysis of gene expression in Zeb2os KD in primary astrocytes using AAV. Data are presented as means ± SEM; n = 3 independent experiments; *p < 0.05, **p < 0.01, Zeb2os KD versus control (independent t test).
(C) AAV transduction marker GFP (green) colocalization with GFAP (red) in spinal cord astrocytes. Top row: whole spinal cord section (Zeb2os KD rostral 800 µm distal to epicenter); bottom row: magnified region (box). (scale bar, 200 µm, top row; scale bar, 10 µm, bottom row).
(D) RNAscope and immunohistochemistry showing Zeb2os mRNA (red) expression in Zeb2os KD AAV-transduced astrocytes (green) compared to the eGFP AAV-transduced control. DRAQ5 was used for nuclear staining (white) (scale bar, 5 µm).
(E) Expression of Zeb2os mRNA dot number per cell in AAV transduced astrocytes using RNAscope (n = 3–6). Data are presented as mean ± SEM; **p < 0.01, compared with control group (independent t test).
Figure 6. Zeb2os shRNA knockdown by AAV transduction reduced GFAP and pSTAT3 expression as well as lesion volume in GFAP-Cre mice at 17 days after SCI.
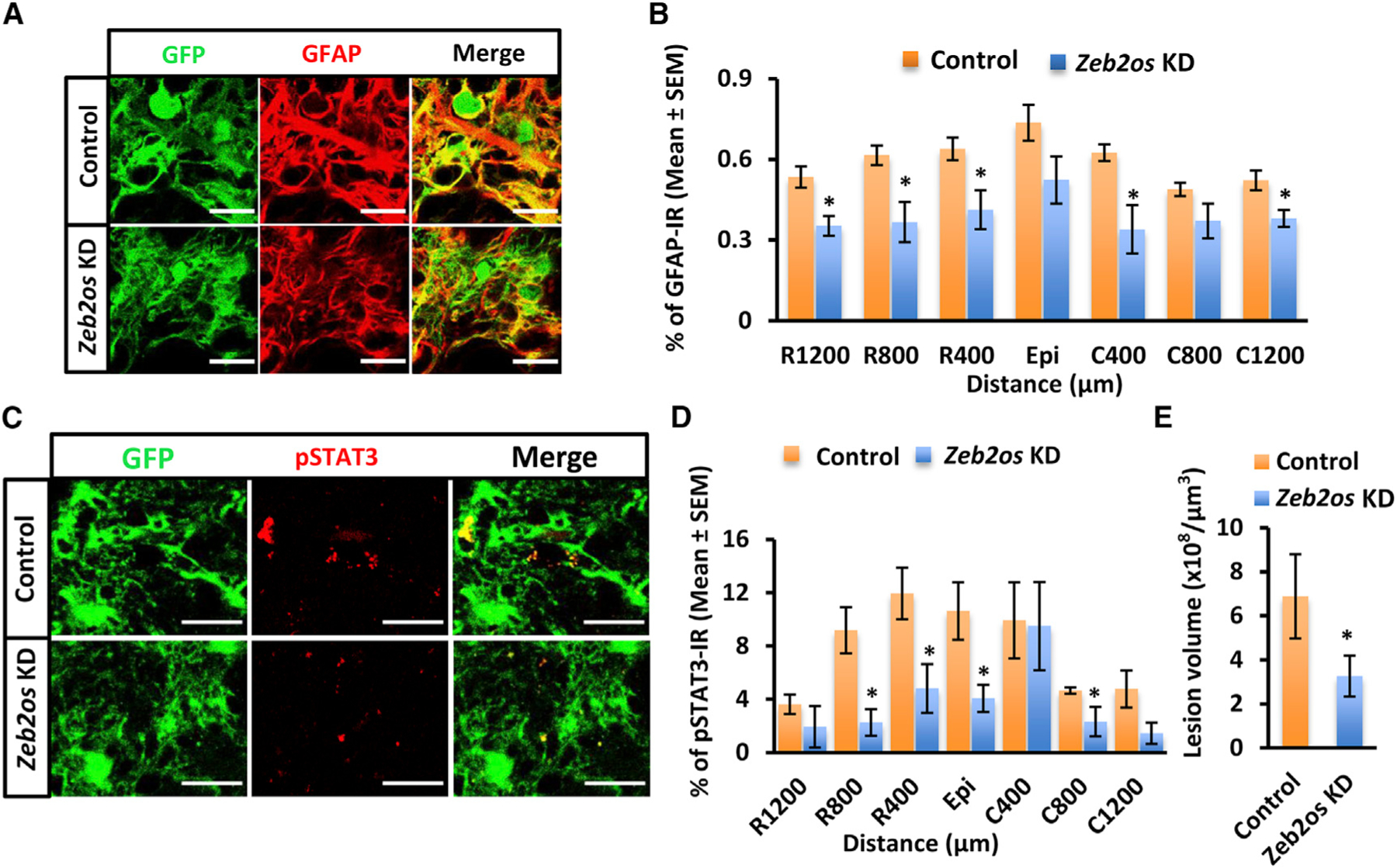
(A) Immunohistochemistry of GFAP (red) expression in Zeb2os KD in astrocytic scar compared with control (scale bar, 20 µm).
(B) Mean percentage of GFAP-immunoreactive area in the AAV transduced region of spinal cord sections at various distances from the SCI epicenter (Epi) (n= 5–7).
(C) Immunohistochemistry of pSTAT3 (red) expression in Zeb2os KD astrocytes compared with control (scale bar, 20 µm).
(D) Mean percentage of pSTAT3-immunoreactive area in the AAV transduced region of spinal cord sections at various distances from the SCI epicenter (Epi) (n = 4–6).
(E) Quantification of the lesion volume using GFAP immunostaining (n = 3).
Data are presented as mean ± SEM; *p < 0.05, **p < 0.01, compared with control group (independent t test) (B, D and E).
DISCUSSION
Astrocyte activation is an important biological response after CNS injuries and during neurodegenerative diseases. There is debate in the field as to whether astrocytes, by responding to the injury microenvironment, play inhibitory or beneficial/neuroprotective roles (Anderson et al., 2016, 2018; Filous and Silver, 2016; Khakh and Deneen, 2019; Tran et al., 2018). The comparison of RNA-seq datasets from spinal cord tissue and purified astrocytes reveals the overall gene expression changes in the injury environment and the astrocyte-specific responses and contributions. We found both axon growth permissive and inhibitory factors from purified astrocytes after SCI, including CSPGs, integrin-family genes, fibrosis-associated genes, and axon growth permissive factors such as Lamb1, Ncam1, and Bmp7 (Figure S4C), which indicates that astrocytes are a critical cell type for secreting both of these factors. Our current study paved the way for single cell RNA-seq of purified astrocytes, which will help to delineate the states or phenotypes of reactive astrocytes in the future.
There is a significant knowledge gap in the functional role of lncRNAs during reactive astrogliosis, the mechanisms of which are not well understood. Between 70% and 90% of the mammalian genome is transcribed into RNA at some point during development; however, less than 2% of the genome is associated with protein-coding genes (Cabili et al., 2011; Carninci et al., 2005; Birney et al., 2007; Guttman et al., 2009; Kapranov et al., 2007). By targeting RNA, we are significantly increasing the number of potential molecular targets and enabling new classes of mechanistic discoveries (Li et al., 2019). In our SCI RNA-seq results, we found the upregulated lncRNAs with homologous lncRNAs in the human genome that are significantly correlated with protein-coding genes functionally enriched in immune response, cell differentiation, and proliferation. This suggests that these lncRNAs may play roles in SCI pathophysiological processes. Thus, we have tested a number of candidates and found a gene of interest, Zeb2os, which is highly conserved with a human lncRNA ZEB2-AS1. We have shown that Zeb2os expression has high correlation over the injury time course with the expression of Zeb2 and Stat3. We have demonstrated by functional assays that Zeb2os plays an important functional role in reactive astrogliosis. Virus-mediated knockdown of Zeb2os in primary astrocytes leads to significantly reduction in astrocyte proliferation as shown by BrdU staining, and decreased expression of many genes including Gfap, Zeb2, Stat3, Ncan, phosphacan, and integrins, which may be involved in scar progression.
STAT3 is an essential TF in reactive astrogliosis and scar progression (Anderson et al., 2016; Herrmann et al., 2008; Liddelow and Barres, 2017; Liddelow et al., 2017). Our ChIP-seq experiment showed that STAT3 bound to the Zeb2 promoter region, thus Zeb2os could regulate Zeb2 and Stat3 either directly or indirectly and STAT3 might regulate Zeb2 expression. The functions of Stat3 in cell proliferation and cell-cycle processes (Wanner et al., 2013) were consistent with genes and pathways affected by Zeb2os KD. Therefore, lncRNA Zeb2os is a previously unknown regulator of Stat3, in addition to its well-known upstream regulatory pathways.
In order to better understand the effects of Stat3, Zeb2, or Zeb2os, in reactive astrogliosis, we have analyzed Stat3 KO astrocyte RNA-seq dataset (Anderson et al., 2016) and performed RNA-seq using Zeb2 knockdown primary astrocyte (Table S7). We compared the DEGs in astrocytes after Zeb2os KD or Stat3 KO. We found 471 DEGs are common between Stat3 KO and Zeb2os KD astrocytes (Figure S6B), and 311 DEGs are common between Zeb2os and Zeb2 KD astrocytes (Figure S6C). As mentioned above, Zeb2os KD caused a significant reduction in Stat3 expression in primary astrocyte culture (Figure 4D, group 1) and pSTAT3 expression in injured spinal cord (Figures 6C and 6D). Interestingly, IPA analysis for common DEGs between Zeb2os KD and Stat3 KO shows these genes are enriched in complement system, JAK/Stat signaling, PTEN signaling, and cyclins and cell-cycle regulation pathways, etc. (Figure S6D). The IPA analysis showed that Zeb2os KD unique genes are enriched in, for example, Wnt/β-catenin signaling (-log(p value) = 2.43) and cell-cycle pathways (-log(p value) = 2.68); while Stat3 KO unique genes are enriched in TREM1 signaling (-log(p value) = 12.4), Toll-like receptor signaling (-log(p value) = 4.57), and NF-κB signaling (-log(p value) = 3.43), etc. These results suggested Zeb2os regulates astrocyte functions through STAT3 pathway.
RNA-seq result shows that Zeb2 shRNA successfully knocks down its expression in astrocytes (with ~6-fold reduction in expression). Our results showed stronger effect on astrogliosis by Zeb2os KD than Zeb2 KD (Figure 4A–4C). GFAP and other astrocytic genes, as well as proliferation decreased more significantly in Zeb2os KD primary astrocytes than Zeb2 KD (Figure 4A–4C). The comparison of affected DEGs in Zeb2os KD and Zeb2 KD showed that Zeb2os KD unique genes are enriched in Wnt/β-catenin signaling (-log(p-value) = 2.47), complement system(-log(p-value) = 2.29) and cell cycle (-log(p-value) = 1.97) etc. Some of the Zeb2 KD unique DEGs enriched pathways include Ephrin receptor signaling (-log(p-value) = 2.94) and RhoA signaling (-log(p-value) = 2.97) (Murai and Pasquale, 2011; Renault-Mihara and Okano, 2017). Thereby, Zeb2 only mediates the effects of Zeb2os partially. Beltran et al. (2008) reported that Zeb2os regulates Zeb2 at translational levels during epithelial-mesenchymal transition in cancer cells. Our results show that in astrocytes Zeb2os knockdown significantly down-regulated Zeb2 at transcriptional level by qPCR. Unlike in cancer cells, Chd1 (E-cadherin) is almost absent in CNS cell types (http://jiaqianwulab.org/resource.htm). Astrocyte migration did not change significantly in Zeb2os KD in a scratch wound assay (Figures S5A–S5C). RNAscope experiments showed that Zeb2os RNA is expressed and colocalized with Zeb2 RNA in reactive astrocytes. LncRNAs are versatile molecules that can regulate target genes through RNA-RNA, RNA-DNA, or RNA-protein interactions.
Overall, our in vivo studies confirmed that Zeb2os KD reduced reactive astrogliosis and lesion volume without exacerbating neuroinflammation. Although not statistically significant, there is a trend of increased axon regeneration/sprouting at 7D after Zeb2os KD. Longer survival times after Zeb2os KD or/and combining with rehabilitation in the future might promote significant axonal regeneration (Hawthorne et al., 2011). Furthermore, we found Zeb2os KD suppressed expression of genes in the complement pathway. Stat3 KO RNA-seq data also showed complement pathway is affected (Figure S6D). Previous studies reported that complement genes are upregulated in reactive astrocytes, and that inactivating C3ar (C3aRKO) in Alzheimer’s disease mouse models can reduce disease pathology and gliosis (Lian et al., 2016; Liddelow et al., 2017). Additionally, our functional enrichment and pathway analysis showed that DEGs in purified astrocytes from the injured spinal cord were enriched in complement system. Whether complement genes are related to scar formation is a very interesting topic for future investigation. Based on previous studies and our current findings, we developed the working model that lncRNA Zeb2os functions in regulating reactive astrogliosis through Zeb2os/Zeb2/Stat3 axis and other target genes (TG) (Figure 7). The detailed mechanisms are our next phase of studies.
Figure 7.
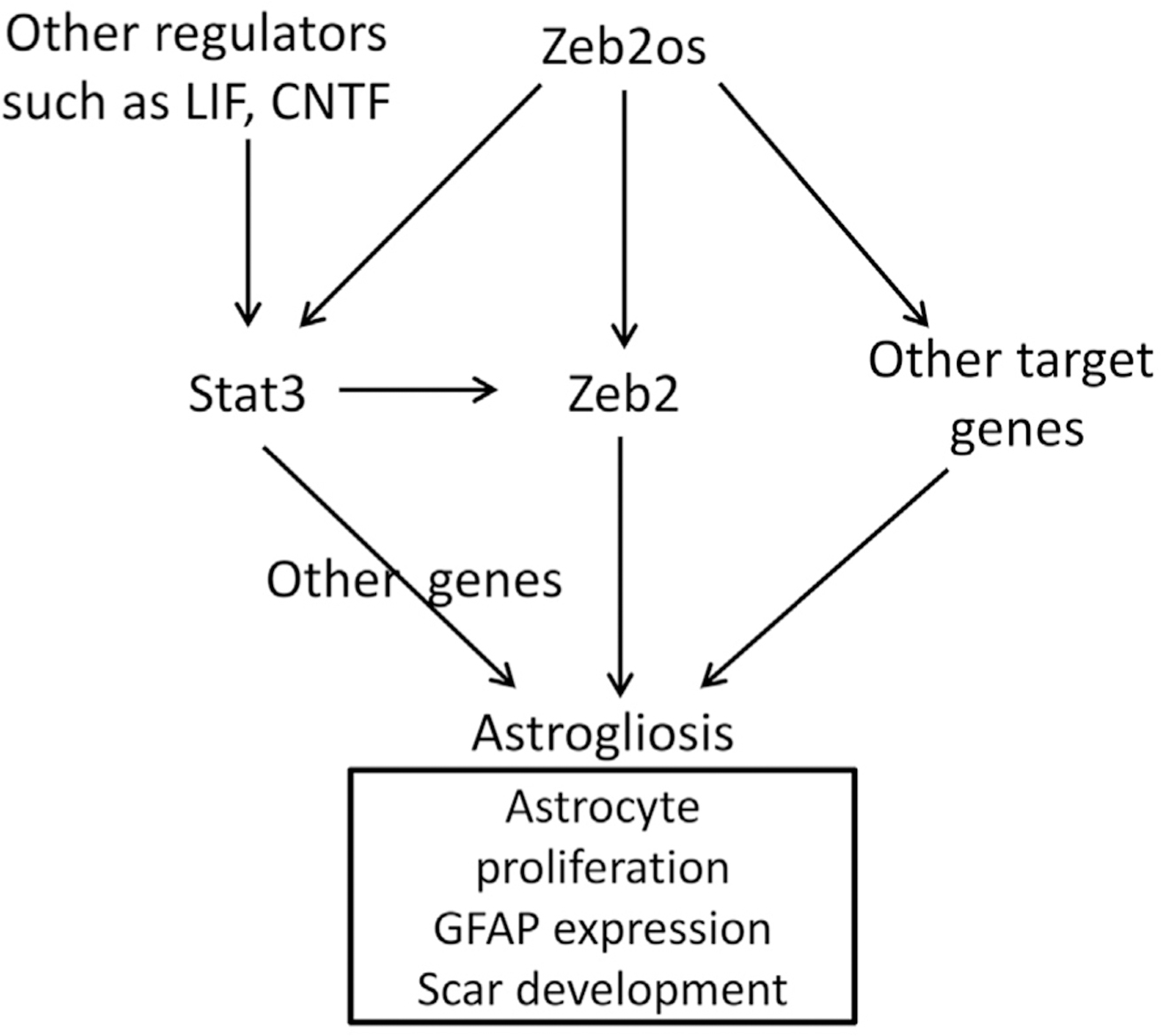
Working model of a Zeb2os/Zeb2/Stat3 axis in reactive astrogliosis
It is possible that Zeb2os functions differently at different stages of SCI. It might have neuroprotective effects at early stages but inhibitory effects at later stages. Previous studies have provided evidence that after injury astrocytes change over time from an initially beneficial state (reactive astrocytes [RA]) to a maladaptive state (scar-forming astrocytes [SA]) that inhibits neuroregeneration (Hara et al., 2017). RAs serve adaptive functions during the early stages of SCI by secluding inflammatory cells, limiting injury to a confined area, and promoting tissue repair (Filous and Silver, 2016; Mattucci et al., 2019; Tran et al., 2018), whereas SAs express significantly higher of scar-related CSPGs and SLIT proteins (Hara et al., 2017; Silver et al., 2014; Wu et al., 2016a). Attenuating astrocytic scar progression instead of removing the scar can promote functional improvement after SCI (Hara et al., 2017). On the other hand, preventing the formation of the astrocytic scar or the full astrocyte response to injury in the early stages after SCI can worsen SCI pathologies (Anderson et al., 2016; Vivinetto et al., 2020). Interestingly, Vivinetto et al. (2020) reported very recently that targeted knockout of Zeb2 in astrocytes (starting at 3 days before injury and for 7 continuous days after SCI) reduced reactive astrogliosis, increased injury size, and resulted in greater functional deficits. These results suggest that Zeb2 is important in regulating reactive astrogliosis and neuroprotection during the acute stage after SCI. Our current study is a comprehensive investigation of the expression of coding and long non-coding genes from acute to chronic SCI stages and demonstrates that Zeb2os has important roles in reactive astrogliosis and that regulation of Zeb2 expression could be a mechanism by which it regulates reactive astrogliosis during acute SCI. It is interesting to note that Zeb2os remains upregulated at the chronic SCI stages and regulates the expression of genes related to SA, suggesting that Zeb2os might also play an important role in scar progression at the later stages of SCI, including chronic SCI.
In summary, we have demonstrated by shRNA gene knockdown and functional assays that Zeb2os plays an important functional role in reactive astrogliosis both in vitro and in vivo. Modulating the astrocytic scar together with growth factors, growth-supportive substrates, and chemoattractive molecules (Anderson et al., 2018) could further enhance neuroregeneration after SCI. Continued investigation will be important for revealing the functions of Zeb2os during scar progression in chronic SCI and could lead to the development of RNA-based therapies for SCI and other neurological diseases involving reactive astrogliosis.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Jia Qian Wu (Jiaqian2009.wu@gmail.com).
Materials availability
We will make the materials developed available to the scientific community. This includes transgenic animals, reagents and cells. We may request reasonable payment for maintenance and transport of materials, and/or a completed Materials Transfer Agreement.
Data and code availability
The accession number for the raw and processed sequencing data reported in this paper is GEO : GSE153721.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mouse models
All animal care and surgical interventions were undertaken in strict accordance with the approval of the Animal Welfare Committee at the University of Texas Health Science Center at Houston (UTHealth). Animals were housed in standard laboratory conditions with 12 light/12 dark cycle and with free access to water and food. To obtain purified astrocytes for use in RNA-Seq, transgenic mouse model GFAP-Cre:R26-tdT (12–14 weeks old) was generated by mating heterozygous GFAP-Cre (B6.Cg-Tg(Gfap-cre)73.12Mvs/J) mice (JAX, Stock No. 012886) with R26-tdT (B6.Cg-Gt(ROSA)26Sortm9(CAG-tdTomato)Hze/J) mice (also known as: Ai9 or Ai9(RCL-tdT) (JAX, Stock No.007909)). Young mice were genotyped by PCR to detect the Cre sequence using DNA extracted from mouse tails at 18 days after birth. C57BL/6J (JAX, Stock No.000664) mice (12–14 weeks old) were used for collecting samples for RNA-Seq and ChIP-Seq. For AAV injection, heterozygous GFAP-Cre mice (16–20 weeks old) were generated by mating homozygous GFAP-Cre with C57BL/6J mice. All the mice were randomly assigned to experimental groups.
Primary astrocyte culture
Primary astrocytes were isolated from postnatal mouse brain at postnatal day 4 (P4)–P7 using a Neural Tissue Dissociation Kit (P) (Miltenyi Biotec). Suspensions of single live cells were cultured in DMEM media (Corning) supplemented with 10% FBS, 100 units/ml of penicillin and 100 µg/ml of streptomycin (Thermo Fisher Scientific). Microglia, oligodendrocyte progenitor cells, and neurons were removed by shaking at 220 rpm overnight at 7D after plating. After the shaking procedure, the majority of the cells that remain attached were astrocytes. The astrocytes were seeded into cell culture plates and ready for further applications.
METHOD DETAILS
Surgery procedures
A moderate contusive injury was performed using the Infinite Horizons Impactor (Precision Systems and Instrumentation) at the T9 level of the exposed spinal cord (60 kdyn) as described in our previous study (Chen et al., 2013; Duran et al., 2017). Animals in the sham control group received a dorsal laminectomy only. After surgery, bladder compression, hydration, and nutritional support were applied as needed to aid recovery. After transcardial perfusion with PBS, 5-mm segment of SCI epicenter tissue was harvested at 7D, 1M, or 3M post-injury for RNA-Seq, and tissue for ChIP-Seq was harvested at 7D post-injury. Tissues from three mice were pooled together for each biological replicate.
For AAV injection, GFAP-Cre mice were anesthetized with a mixed solution of ketamine (80 mg/kg ip) and xylazine (10 mg/kg ip) and the sutured incision was reopened to re-expose the dorsal surface of injured spinal cord at the T9 spinal cord segment to receive AAV injection at 10 days post-injury. One microliter of AAV (AAV Zeb2os KD-1 and AAV Zeb2os KD-2 mixed equally) was injected to a depth of 0.8 mm at the SCI epicenter, and then 0.5 µl/site was also injected to a depth of 1.0 mm at 0.5 mm both rostrally and caudally from the epicenter (a total of 2 µL injected). Spinal cords were then harvested at 17 days post-injury for further applications.
Tissue dissociation and astrocyte purification
Epicenter spinal cord tissue for astrocyte purification was harvested after transcardial perfusion with PBS at respective time points and dissociated using a mixed digestion solution of 0.01% papain (Worthington Biochemical), 0.025% trypsin-EDTA (Thermo Fisher Scientific), 0.01% DNase I (Worthington Biochemical), and 0.01% L-cysteine (Calbiochem) in DMEM (Corning) with 0.02% BSA (Thermo Fisher Scientific) for 15 min at 37°C with gentle shaking (70 rpm). The cells were separated from the undigested pellet by triturating with a wide-tipped, fire-polished glass Pasteur pipette eight times. After repeating digestion and separation twice, the cells were resuspended in DMEM and 4% BSA and the suspension was filtered using a 40-µm cell strainer by centrifugation. The cells were then resuspended in buffer containing Myelin Removal Beads II (Miltenyi Biotec) following the manufacturer’s protocol to remove myelin. Next, tdTomato-positive cells for RNA library construction were collected by FACS using a BD FACS Aria II flow cytometer (BD Biosciences).
RNA isolation and RNA-Seq
RNA libraries were prepared as described in previous studies (Chen et al., 2013; Duran et al., 2017). Briefly, RNA was isolated from either tissue samples or cells using TRIzol (Invitrogen) following the manufacturer’s protocol. About 300–500 ng total RNA was used to construct each RNA-Seq library. The paired-end sequencing libraries were prepared using NEBNext® Ultra Directional RNA Library Prep Kit for Illumina (New England Biolabs) following the manufacturer’s instructions. Sequencing was performed using the Illumina HiSeq2000, HiSeq2500 or Novaseq sequencing instrument.
STAT3 ChIP-Seq
ChIP was performed using STAT3 antibody (sc-482x, Santa Cruz Biotechnology) in dissociated spinal cord cells. ChIP DNA and input DNA were extracted using phenol:chloroform:isoamyl alcohol (25:24:1) as described in our previous studies (Dong et al., 2015; Dong et al., 2018). The ChIP-Seq libraries were constructed using the DNA SMART ChIP-Seq Kit (Clontech Laboratories) following the manufacturer’s instructions and were sequenced on the Illumina HiSeq 2000 Sequencer to obtain 75-bp paired-end sequencing reads.
Astrocyte scratch assay
The primary astrocytes were isolated as mentioned above. To perform scratch assay, primary astrocytes were cultured on poly-Lornithine-coated (Advanced BioMatrix) microscope slides (Thermo Fisher Scientific). After the cells reached about 80% confluence, in vitro “wounds” were created by two diagonal line scratches across the astrocyte monolayer using a P-200 pipette tip. The cells were then fixed with 4% paraformaldehyde (PFA) for 15 mins at 48 hours after scratch assay and used for immunohistochemistry or RNAscope assays.
Similar model was also applied to assess the effects of Zeb2os KD on the astrocyte migration associated with wound closing. Astrocytes were first transduced with control and Zeb2os KD AAVs for 5 days and seeded in 6-well plate at a density of 1 × 106 cells/ml. A single scratch wound was created across the cell monolayer using P-20 pipette tip in all wells. After washed with regular growth medium, cells were then plated into an environmental chamber for 2 days. Four wound fields within each well were randomly selected and repeatedly imaged at fixed time intervals (every 3 h) with 10x objective of Olympus IX81. Wound closure was measured using the wound confluence (% confluence) over a time course and the area under the curve (AUC) of the % confluence function plotted against time were also calculated to observe the wound closing efficacy (Parmentier-Batteur et al., 2011). All statistical analysis was performed using an independent t test (n = 4).
Plasmid construction
For lentivirus-mediated gene knockdown, Zeb2 KD shRNA containing plasmids were purchased from Sigma (SHCLNG-NM_015753). As there is no commercial available shRNA constructs for lncRNA Zeb2os, the shRNAs were designed and ligated into a pLKO.1-puro backbone as described in a previous study (Dong et al., 2015). The shRNAs were strand-specific (directional) (sequences are listed in Table S1). We used two plasmids for each Zeb2os shRNA KD (Zeb2os-1 and Zeb2os-2) and Zeb2 shRNA KD (Zeb2-1 and Zeb2-2), which exhibited greater than 50% knockdown efficiency. The luciferase lentiviral shRNA (Luci) construct expressing a shRNA sequence that targets firefly luciferase and has no significant homology to any mammalian transcript was used as a control.
To generate AAV-mediated Zeb2os knockdown plasmids (pAAV-CAG-FLEX-shRNA-eGFP), the enhanced green fluorescent protein (eGFP) fragment was combined with the Zeb2os shRNA fragments and inversely ligated into a floxed backbone (Figure 5A), pAAV-CAG-FLEX-SwaI-SV40pA, which was a gift from Dr. Haipeng Xue at UTHealth and sequenced verified. The recombined construct with inversely oriented eGFP only was used as a control (pAAV-CAG-FLEX-eGFP).
Viral packaging and transduction
Lentivirus were packaged for Zeb2 and Zeb2os KD as described in a previous study (Dong et al., 2015). To knock down the expression of target genes, primary astrocytes were cultured in 6-well plates to 70% confluence and then transduced with either sh-Luci or shRNA lentivirus for 24 hours. Puromycin (Sigma-Aldrich) at 1 µg/ml was used to select the transduced cells for 5 days, and then cells were collected for qPCR, BrDU assay, and RNA-Seq.
AAV viruses were prepared by the Gene Vector Core at the Baylor College of Medicine. Briefly, AAV serotype 5 vectors including Zeb2os KD and control vectors, RC5 Rep-Cap plasmid, and pAdΔF6 helper plasmid were prepared using three-plasmid transfection using the iMFectin Poly DNA Transfection Reagent (GenDEPOT) in the HEK293T cell line. AAV purification was performed based on the method developed by Ayuso et al. (Ayuso et al., 2010). The titers (control: 1.58 × 1013g.c./ml (genome copies/ml), KD: AAV Zeb2os-1: 3.07 × 1013g.c./ml and AAV Zeb2os-2: 4.03 × 1013g.c./ml) were determined by real time PCR as previously published (Ljungberg et al., 2012). To verify the Zeb2os KD effect of the transduced AAV, Cre-expressing astrocytes were isolated from GFAP-Cre postnatal mice following the above method and were transduced with control and Zeb2os KD AAVs. Approximately 1.4 × 105 cells/well in 24-well plate were transduced with AAV. Cells were harvested at 7D post-transduction for assessment by qPCR.
Quantitative RT-PCR (qPCR) analysis
Total mRNA was isolated from primary astrocytes and tissue using TRIzol, then cDNA synthesis and qPCR were performed as described in our previous study (Dong et al., 2015). Relative mRNA expression was then calculated using comparative methods (2−ΔΔCT) using Gapdh or β-actin as expression references. Averages and SEM values were calculated from three independent experiments. The primers were designed as described in a previous study (Zhang et al., 2014). Zeb2os and Zeb2 primer sequences are listed in “KEY RESOURCES TABLE.”
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-Stat3 | Santa Cruz | Cat# sc-482x; RRID: AB_632440 |
| Mouse monoclonal anti-BrDU | Cell Signaling Technology | Cat# 50230; RRID: AB_2799369 |
| Rabbit polyclonal anti-GFAP | Agilent | Cat# Z0334; RRID: AB_10013382 |
| Rat monoclonal anti-GFAP | Thermofisher | Cat# 13–0300; RRID: AB_2532994 |
| Chicken polyclonal anti-GFAP | Millipore | Cat# AB5541; RRID: AB_177521 |
| Chicken polyclonal anti-GFP | Millipore | Cat# AB16901; RRID: AB_90890 |
| Rabbit monoclonal anti-GFP | Thermofisher | Cat# G10362; RRID: AB_2536526 |
| Goat polyclonal anti-GFP | Novus | Cat# NB 100–1770; RRID: AB_523903 |
| Rabbit polyclonal anti-CSPG4 (NG2) | Millipore | Cat# AB5320; RRID: AB_91789 |
| Rabbit anti-phosphorylated STAT3 (pSTAT3) (tyr705) | Cell Signaling Technology | Cat# 9145S; RRID: AB_2491009 |
| Rat monoclonal anti-CD68 | Bio-Rad | Cat# MCA1957GA; RRID: AB_324217 |
| Rabbit polyclonal anti-5-HT (Serotonin) | ImmunoStar | Cat# 20080; RRID: AB_572263 |
| Donkey anti-chicken FITC | Jackson ImmunoResearch Labs | Cat# 703–095-155; RRID: AB_2340356 |
| Donkey anti-goat FITC | Jackson ImmunoResearch Labs | Cat# 705–095-003; RRID: AB_2340400 |
| Donkey anti-chicken AMCA | Jackson ImmunoResearch Labs | Cat# 703–155-155; RRID: AB_2340361 |
| Donkey anti-rabbit Cy5 | Jackson ImmunoResearch Labs | Cat# 711–175-152; RRID: AB_2340607 |
| Donkey anti-rat TRITC | Jackson ImmunoResearch Labs | Cat# 712–025-150; RRID: AB_2340635 |
| Goat anti-mouse Alexa Fluor 488 | Thermofisher | Cat# A-11029; RRID: AB_2534088 |
| Bacterial and virus strains | ||
| NEB Stable Competent E. coli (High Efficiency) | New England Biolabs | C3040H |
| MISSION Zeb2 shRNA Bacterial Glycerol Stock | Sigma | SHCLNG-NM_015753 |
| pLKO.1-Luciferase-puro | This paper | N/A |
| pLKO.1-Zeb2os shRNA-puro | This paper | N/A |
| pLKO.1-Zeb2 shRNA-puro | This paper | N/A |
| pAAV-CAG-FLEX-eGFP | Gene Vector Core at the Baylor College of Medicine | N/A |
| pAAV-CAG-FLEX-Zeb2os shRNA-eGFP | Gene Vector Core at the Baylor College of Medicine | N/A |
| Biological samples | ||
| Postnatal C57BL/6J primary astrocytes | This paper | N/A |
| Postnatal GFAP-Cre primary astrocytes | This paper | N/A |
| Adult GFAP-Cre:R26-tdT purified astrocytes | This paper | N/A |
| Adult C57BL/6J spinal cords | This paper | N/A |
| Adult GFAP-Cre spinal cords | This paper | N/A |
| HEK293T cells | ATCC | CRL-3216 |
| Chemicals, peptides, and recombinant proteins | ||
| Phusion High-Fidelity PCR Master Mix with HF Buffer | New England Biolabs | M0531L |
| Papain | Worthington Biochemical | LS003126 |
| Trypsin-EDTA (0.25%) | Thermofisher | 25200056 |
| Deoxyribonuclease I (DNase I) | Worthington Biochemical | LS002138 |
| L-Cysteine, Hydrochloride | Calbiochem | 2430 |
| DMEM | Corning | 10-013-CM |
| Bovine Serum Albumin (BSA) | Thermofisher | BP9706100 |
| DPBS | Thermofisher | 14190144 |
| Myelin Removal Beads II | Miltenyi Biotec | 130-096-733 |
| TRIzol | Thermofisher | 15596026 |
| NEBNext Multiplex Oligos for Illumina | New England Biolabs | E7335S |
| Phenol:Chloroform:Isoamyl Alcohol | Thermofisher | 15-593-031 |
| FBS | Thermofisher | 16140-071 |
| Penicillin-Streptomycin | Thermofisher | 15140122 |
| Poly-L-Ornithine | Advanced BioMatrix | 5058 |
| Paraformaldehyde | Sigma | 158127–500G |
| Neural Tissue Dissociation Kit (P) | Miltenyi Biotec | 130–092-628 |
| Lipofectamine 3000 | Thermofisher | L3000015 |
| iMFectin Poly DNA Transfection Reagent | GenDEPOT | I7200 |
| Ethanol | Thermofisher | 241ACS200CSGL |
| RNase/DNase-Free Water | Thermofisher | 10977023 |
| DNase I | Thermofisher | 18068015 |
| DAPI | Santa Cruz | sc-3598 |
| Sucrose | Sigma | S1888 |
| O.C.T. Compound | Thermofisher | 23-730-571 |
| Triton X-100 | Sigma | T8787 |
| Normal Donkey Serum | Jackson ImmunoResearch Labs | 017-000-121 |
| ProLong Gold Antifade Mountant | Thermofisher | P10144 |
| DRAQ5 | Thermofisher | 62251 |
| PBS | Quality Biological | J3734L |
| Critical commercial assays | ||
| NEBNext Ultra Directional RNA Library Prep Kit for Illumina | New England Biolabs | E7420S |
| DNA SMART ChIP-Seq Kit | Takara Bio | 634865 |
| RNAscope 2.5 HD Reagent Kit-RED | Advanced Cell Diagnostics | 322350 |
| RNAscope Multiplex Fluorescent V2 Assay | Advanced Cell Diagnostics | 323100 |
| RNAscope Probe Mm-Zeb2os | Advanced Cell Diagnostics | 466611 |
| RNAscope Probe Mm-Zeb2-C2 | Advanced Cell Diagnostics | 436391-C2 |
| RNAscope Probe EGFP-C3 | Advanced Cell Diagnostics | 400281-C3 |
| RNAscope Negative Control Probe- DapB | Advanced Cell Diagnostics | 310043 |
| Deposited data | ||
| Raw sequencing data | GEO | GEO: GSE153721 |
| Experimental models: organisms/strains | ||
| Mouse: C57BL/6J | Jackson Laboratory | JAX: 000664 |
| Mouse: GFAP-Cre (B6.Cg-Tg(Gfap-cre) 73.12Mvs/J) | Jackson Laboratory | JAX: 012886 |
| Mouse: R26-tdT (B6.Cg-Gt(ROSA) 26Sortm9(CAG-tdTomato)Hze/J) | Jackson Laboratory | JAX: 007909 |
| Mouse: GFAP-Cre:R26-tdT | This paper | N/A |
| Oligonucleotides | ||
| Zeb2os-1 sense shRNA | This paper | AGCTTAAAAAGGAGAAAGGT CAGAGGAACTTCTCGAGAAG TTCCTCTGACCTTTCTCCGAT |
| Zeb2os-1 antisense shRNA | This paper | ATCGGAGAAAGGTCAGAGGA ACTTCTCGAGAAGTTCCTCTG ACCTTTCTCCTTTTTA |
| Zeb2os-2 sense shRNA | This paper | AGCTTAAAAAGAAGGGATTTCC TGGAGAATACTCGAGTATTCTC CAGGAAATCCCTTCGAT |
| Zeb2os-2 antisense shRNA | This paper | ATCGAAGGGATTTCCTGGAGAAT ACTCGAGTATTCTCCAGGAAATC CCTTCTTTTTA |
| Zeb2 forward | This paper | 5′-GTTCGGCATGAACCCATTTAG-3′ |
| Zeb2 reverse | This paper | 5′-TCCTCTGGTATTTCCTCCTTTG-3′ |
| Zeb2os forward | This paper | 5′-GATCTCAGTGTGCAGTGTATGT-3′ |
| Zeb2os reverse | This paper | 5′-GTCCTAAGTTCCTCTGACCTTTC-3′ |
| Recombinant DNA | ||
| pLKO.1-puro | Sigma | SHC001 |
| pLKO.1-Zeb2 shRNA-puro | Sigma | SHCLNG-NM_015753 |
| pLKO.1-Luci-puro | This paper | N/A |
| pLKO.1-Zeb2os shRNA-puro | This paper | N/A |
| pAAV-CAG-FLEX-SwaI-SV40pA | Dr. Haipeng Xue in UTH | N/A |
| pAAV-CAG-FLEX-eGFP | This paper | N/A |
| pAAV-CAG-FLEX-Zeb2os shRNA-eGFP | This paper | N/A |
| Software and algorithms | ||
| ImageJ/Fiji | NIH |
https://imagej.nih.gov/ij/index.html; RRID: SCR_003070 |
| Prism | GraphPad Software |
https://www.graphpad.com:443/; RRID:SCR_002798 |
| R Studio | R Studio | https://rstudio.com/ |
| Fastqc | Babraham Institute | https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ |
| RCircos | Zhang et al., 2013 | https://cran.r-project.org/web/packages/RCircos/index.html |
| Bowtie2 | Langmead and Salzberg, 2012 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| SPP | Kharchenko et al., 2008 | https://sites.google.com/a/brown.edu/bioinformatics-in-biomed/spp-r-from-chip-seq |
| TopHat v2.1.0 | Kim et al., 2013 | https://ccb.jhu.edu/software/tophat/downloads/ |
| Cufflinks v2.2.1 | Trapnell et al., 2012 | http://cole-trapnell-lab.github.io/cufflinks/releases/v2.2.1/ |
| htseq-count | Anders et al., 2015 | https://github.com/simon-anders/htseq |
| Ingenuity Pathway Analysis (IPA) | QIAGEN | https://www.qiagenbioinformatics.com |
| DESeq2 | Anders and Huber, 2010 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| Integrative Genomics Viewer (IGV) | Robinson et al., 2017 | http://software.broadinstitute.org/software/igv/ |
| GSEA | Subramanian et al., 2005 | https://www.gsea-msigdb.org/gsea/index.jsp |
| slncky.v1.0 | Chen et al., 2016 | https://slncky.github.io/index.html |
| IntaRNA 2.0 | Mann et al., 2017 | https://github.com/BackofenLab/IntaRNA |
| NUPACK | Zadeh et al., 2011 | http://www.nupack.org/partition/new |
| BEDTools | Quinlan and Hall, 2010 | https://bedtools.readthedocs.io/en/latest/ |
| MEME | MEME Suite | http://meme-suite.org/index.html |
| Other | ||
| BD FACS Aria II flow cytometer | BD Biosciences | https://www.bdbiosciences.com/en-us |
| Infinite Horizons Impactor | Precision Systems and Instrumentation | IH-0400 Impactor |
| Leica cryostat | Leica Microsystems | CM1950 |
| Illumina HiSeq2000 | Psomagen | https://psomagen.com |
| Illumina HiSeq2500 | Psomagen | https://psomagen.com |
| Illumina Novaseq6000 | Psomagen | https://psomagen.com |
| Zeiss Observer Z1 motorized Inverted Fluorescence Microscope | Zeiss | https://www.zeiss.com/microscopy/us/products/light-microscopes/axio-observer-for-biology.html |
| Leica TCS SP5 Confocal Laser Scanning Microscope | Leica | https://www.leica-microsystems.com/products/confocal-microscopes/p/leica-tcs-sp5 |
| Nikon Eclipse TE2000E Widefield Fluorescence Microscope | Nikon | https://www.microscope.healthcare.nikon.com |
| Olympus IX81 motorized inverted Fluorescence Microscope | Olympus | https://www.olympus-lifescience.com |
| Reactive astrocyte RNA-seq database | (Anderson et al., 2016) | https://astrocyte.rnaseq.sofroniewlab.neurobio.ucla.edu/homesci |
Cell proliferation assay by BrdU
Puromycin-selected lentivirus-transduced primary astrocytes from both KD and control groups were incubated in medium with the BrdU labeling reagent (Thermo Fisher Scientific) for 60 min and were subsequently fixed with 70% ethanol for 20 min at RT. After washing three times with PBS, cells were treated with 1.5M HCl (Sigma-Aldrich) for 30 min and were stained with mouse BrdU antibody (Cell Signaling), Alexa Fluor 488-conjugated goat anti-mouse IgG (H+L) antibody (Invitrogen), and DAPI. Three independent incubations were carried out and results were obtained by counting from 10 random fields for each experiment. Images were captured using a Zeiss Observer Z1 inverted fluorescence microscope at 20x magnification.
RNAscope in situ hybridization
RNAscope (Advanced Cell Diagnostics; ACD) in situ hybridization was performed on tissue sections and primary cultured cells. RNAscope 2.5 HD Assay-RED (ACD) and Multiplex Fluorescent V2 Assay (ACD) were used as required to label single and multiplex probes, respectively. The assays were performed according to the manufacturer’s instructions for fixed-frozen tissue and adherent cells. RNAscope probes Mm-Zeb2os (ACD), Mm-Zeb2-C2 (ACD), and egfp (ACD) were used in this study, and the RNAscope negative control probe DapB (ACD) was used as a negative control.
Immunohistochemistry and immunocytochemistry staining
Mice were transcardially perfused with PBS followed by 4% PFA, and then spinal cord segments from 1 cm rostral to 1 cm caudal of the injury epicenter (2 cm total length) were harvested and cryoprotected in serial sucrose (20% and 30%) and embedded into optimal cutting temperature compound (Thermo Fisher Scientific). Spinal cords were transversely sectioned at 20 µm thickness using a cryostat (Leica CM1950; Leica Microsystems, Wetzlar, Germany) and sections were mounted on serial Superfrost Plus Gold Microscope slides (Thermo Fisher Scientific). For unbiased assessment, 1 in 10 series were processed for immunohistochemistry in one of two sets. Two sets of slides were used for immunostaining or RNAscope assays with the following procedures. After washing with PBS-T (0.2% Triton X-100 in PBS; Sigma-Aldrich) three times, tissue sections or cultured cells on slides were blocked with PBS-T containing 10% donkey serum (Jackson ImmunoResearch) for 1 hour, and then incubated with primary antibodies diluted in blocking solution at 4°C overnight. The primary antibodies used in this study included rabbit anti-GFAP (1:500, Agilent Z0334), rat anti-GFAP (1:200, Invitrogen, 13-0300), chicken anti-GFAP (1:500, EMD Millipore, ab5541), chicken anti-GFP (1:400, EMD Millipore, ab16901), rabbit anti-GFP (1:200, Invitrogen, G10362), goat anti-GFP (1:700, Novus Biologicals, NB100-1770), rabbit anti-CSPG4 (NG2) (1:200, EMD Millipore, ab5320), rabbit anti-pSTAT3 (tyr705; 1:300, Cell Signaling Technology, 9145S), rat anti-CD68 (1:500, Bio-Rad, MCA1957GA) and rabbit anti-5-HT (serotonin) (1:250, ImmunoStar, 20080). After washing three times with PBS-T, the sections or cells were incubated with secondary antibodies for 1 hour at RT and coverslips were applied with ProLong Gold Antifade Mountant (Thermo Fisher Scientific). The secondary antibodies used in this study included FITC-conjugated donkey anti-chicken (1:200, Jackson ImmunoResearch, 703-095-155), FITC-conjugated donkey anti-goat (1:400, Jackson ImmunoResearch, 705-095-003), AMCA-conjugated donkey anti-chicken (1:300, Jackson ImmunoResearch, 703-155-155), Cy5-conjugated donkey anti-rabbit (1:200, Jackson ImmunoResearch, 711-175-152), TRITC-conjugated donkey anti-rat (1:200, Jackson ImmunoResearch, 712-025-150). DAPI (1:1000, Santa Cruz Biotechnology, SC3598) and DRAQ5 (1:1000, Thermo Fisher Scientific, 62251) were used for staining of DNA in nuclei.
Quantitative analyses of images
To verify the Zeb2os KD with RNAscope, one set of slides was simultaneously labeled with Zeb2os mRNA, egfp mRNA, and DRAQ5. A z stack of 1-µm-thick images was captured through the depth of the section using a 63x oil objective on a Leica TCS SP5 Confocal Laser Scanning Microscope. As AAV injections (negative control and KD construct) were used to selectively target Cre-expressing astrocytes, the egfp-in situ hybridized (egfp-ISH) area was defined as AAV-transduced astrocytes (Figure 5D). Zeb2os expression in single cells defined as colabeling with egfp-ISH and DRAQ5 in the region of interest was measured following the guidelines recommended by ACD using ImageJ/Fiji (v1.52n, NIH, https://imagej.nih.gov/). A total of at least 200 cells in nine different sections (spaced 200 µm apart) from each group were measured. Zeb2os expression was presented as the average number of dots representing labeling with Zeb2os mRNA in each cell (dots/cell) in the region of interest. Mean dots/cell values were calculated and statistically analyzed using independent t tests (n = 3–6).
To assess the extent of reactive astrogliosis and lesion volume impacted by Zeb2os KD, one set of the slides was counterstained with GFP, GFAP and DAPI, and the other set with GFP and pSTAT3. Images of the injury site were captured starting from the epicenter and every 400 µm traversing rostrally and caudally at 20x magnification on a Zeiss Observer Z1 inverted microscope. GFAP-IR and pSTAT3-IR area were quantified based on previously studies with modifications (Cao et al., 2001; Gallegos et al., 2020; Mattugini et al., 2019) using ImageJ/Fiji (v1.52n). Briefly, a set threshold was used to automatically calculate the areas of GFP-IR and the GFAP-IR within GFP-IR were analyzed using ImageJ/Fiji. The ratios of GFAP-IR to GFP-IR area were calculated on the sections every 400 µm apart over a total distance of 1,200 µm rostral and caudal to the injury epicenter and were compared between KD and control groups (n = 5–7 mice). The percentage of pSTAT3 was quantified following the same method (n = 4–6 mice). Statistical analysis was performed as using independent t tests. The person who analyzed the images was blinded to the experimental groups.
Additionally, for lesion volume quantification, we manually outlined and measured the lesion areas surrounded by GFAP-positive astrocytic processes from rostral 1,200 µm to the caudal 1,200 µm (spaced by 400 µm) in the cross sections of each cord using ImageJ/Fiji (Bellver-Landete et al., 2019; Butenschön et al., 2016; Renault-Mihara et al., 2011; Wanner et al., 2013). The total lesion volume was measured by summing the individual subvolumes which was calculated by multiplying the cross-sectional lesion area (A) x distance between sections (400 µm) (n = 3 mice). Independent t test was used to statistically analyze the differences of lesion volume between groups.
Furthermore, another set of slides was counterstained with CD68, 5-HT and GFP to evaluate neuroinflammation and serotonin axon sprouting after SCI. Images were captured on the sections every 400 µm over a total distance of 2,000 mm rostral and caudal to the epicenter at 20x magnification. To assess neuroinflammation, total area of cross section was manually traced and cross-sectional CD68-IR area was measured using ImageJ following the same method as previously described (Cheng et al., 2018; Gallegos et al., 2020). The percentage of CD68-IR to the total area was calculated at all measured sections (n = 5–6). To assess the sprouting of serotonin axons, an area of approximately 229 µm diameter circle in the ventral horns was applied as the region of interest (ROI) and 5-HT-IR area within the ROI was measured by setting a threshold (Gallegos et al., 2020; Lee et al., 2010). The percentage of 5-HT-IR area was calculated by dividing 5-HT-IR area by ROI area (n = 5). The difference of mean values between groups were compared using independent t tests.
lncRNA annotation and categorization
We surveyed public lncRNA databases for sequences with known and predicted annotations. We combined lncRNA annotations from GENCODE M14 and NCBI (GCF_000001635.25_GRCm38.p5). Annotations were filtered by removing lncRNA transcripts smaller than 200 nt and those with overlapping exons using BEDTools (Quinlan and Hall, 2010). The resulting lncRNA set consists of 35,368 transcripts representing 21,811 genes. Annotated lncRNA transcripts were classified based on their mapped genomic location relative to the closest protein-coding gene transcript, according to a previously published classification (Derrien et al., 2012; Duran et al., 2017). We also found another category (“others”) in which the genic region of genes encoding lncRNAs and their neighboring protein-coding genes overlapped without any overlaps between their exons.
RNA-Seq analysis
The quality of all sequenced samples (mouse and rat) was analyzed using FastQC. Read mapping, transcript assembly, and expression estimation were performed using an in-house pipeline (Duran et al., 2017). High-quality reads were mapped to the mouse mm10 or rat Rnor6 reference genomes. Mapping and transcript assembly were performed using TopHat v2.1.0 (Kim et al., 2013) and Cufflinks v2.2.1 (Trapnell et al., 2012), respectively, using default parameters (Duran et al., 2017). FPKM (Fragments Per Kilobase of transcript per Million mapped reads) values were obtained for genes and transcripts included in the combined annotation file. To avoid FPKM inflation, FPKM values of less than 0.1 were set to 0.1 (Quackenbush, 2002). A Euclidean distance matrix of log2-transformed quantile normalized FPKM was used to perform hierarchical cluster analysis following Ward’s method (Duran et al., 2017). Read counts corresponding to genes and transcripts were also obtained using htseq-count (Anders et al., 2015). The gene read counts of samples were normalized and compared to control samples using DESeq2 (Anders and Huber, 2010). Only DEGs were used in downstream analysis. Enriched pathways were identified by IPA (QIAGEN, https://www.qiagenbioinformatics.com). A total of 1,747 TFs in mouse collected from AnimalTFDB 2.0 (Zhang et al., 2015) and TcoF-DB v2 (Schmeier et al., 2017) were used for further analysis.
ChIP-Seq analysis
All reads were mapped to the mouse mm10 reference genome using Bowtie2 (Langmead and Salzberg, 2012), and only unique mapped reads were used for subsequent analysis. STAT3 ChIP-Seq data were analyzed using the TF pipeline from ENCODE (Landt et al., 2012). Peaks for STAT3 were called using SPP (Kharchenko et al., 2008) and filtered to exclude regions blacklisted by ENCODE, and peaks with an FDR > 0.05 or fold-enrichment < 5. The resulting peaks were annotated within 5 kb upstream of the TSS of a gene and within its gene body. A Circos plot of lncRNAs with STAT3-bound peaks was drawn using the RCircos R library (Zhang et al., 2013). The STAT3-bound peaks and gene expression were visualized using the Integrative Genomics Viewer (IGV) (Robinson et al., 2017).
Predicting potential functions of lncRNAs
We performed “guilt-by-association” analysis to predict the potential functions of DE lncRNAs, as described in our previous studies (Duran et al., 2017). First, we calculated the Pearson correlation between the expression of lncRNAs and protein-coding genes. Then, the ranked list of protein-coding genes for each lncRNA was used in GSEA (Subramanian et al., 2005) to identify significantly enriched gene sets from MSiGdb (Liberzon et al., 2011). Gene sets with an FDR < 0.25 were used to generate an association matrix. Normalized enrichment scores (NES) were used to designate significantly enriched gene sets.
Gene set enrichment analysis
The MSigDB gene sets for mouse were downloaded from http://bioinf.wehi.edu.au/software/MSigDB/. Analysis of the enrichment of gene sets was performed using a hypergeometric statistical test (phyper R function). Gene sets with an FDR < 0.05 were considered significant.
Comparison of transcriptomes of purified astrocyte and tissue samples
The percentile rankings of FPKM for each gene identified in each purified astrocyte and tissue samples were calculated for further analysis. We removed genes with FPKM < 1 across all samples, as these genes are not expressed or are expressed at very low levels in all samples. Percentile rankings were subjected to hierarchical cluster analysis using Ward’s method (Duran et al., 2017).
lncRNA genes homologous between mouse and human
A set of 27,908 lncRNA transcripts encoded by the human genome were downloaded from GENCODE (GRCH38, version 27) and lncRNA-encoding genes homologous between mouse and human were identified using slncky.v1.0 (Chen et al., 2016). To reduce the number of possible spurious homologies between pairs of mouse and human lncRNAs, we filtered the homologous transcripts by requiring either transcript-transcript identity greater than 60% with the FDR set at 0.01) or at least one conserved splice site (Chen et al., 2016). Finally, 671 highly conserved lncRNA transcript pairs representing 362 lncRNAs in the mouse genome were identified between mouse and human.
Mouse lncRNAs with human homologs harboring disease-associated SNPs
GWAS (Genome-wide association study) data (v1.0.1) was downloaded from GWAS Catalog (Buniello et al., 2019) and Clinically Relevant Sequence Variations (clinvar_20180128) data was downloaded from NCBI (Landrum et al., 2014). The diseases or traits associated with the homologous lncRNA-encoding genes in human and mouse genome were collected.
Analysis of RNA secondary structure
RNA structure plays key roles in the functions of lncRNAs (Novikova et al., 2012). First, we used Meme tools to detect consensus regions in the primary transcript sequences of lncRNAs that are homologous between mice and humans (Bailey and Elkan, 1994). We then predicted the secondary structures of these conserved regions using NUPACK software with minimum free energy (MFE) (Zadeh et al., 2011).
QUANTIFICATION AND STATISTICAL ANALYSIS
All statistical analyses were performed with GraphPad Prism 5.0 software. Further statistical details are indicated in the figure legends. The number of animals per group was based on the previous experience and calculated with StatMate software (GraphPad) to provide at least 80% calculable power with the following parameters: probability of type I error (0.05), conservative effect size (0.25). The values of n refer to the number of mice used in the animal experiments. In other sections, n refers to the number of experimental repeats. Data are presented as the means ± standard error of the mean (SEM) or mean ± standard deviation (SD). Differences between experimental groups were analyzed using independent t test (paired, two-sided). A p value less than 0.05 was considered statistically significant. For all imaging and quantifications, personnel measuring the experimental results were blinded to the experimental groups.
Supplementary Material
Highlights.
Transcriptomes reveal expression dynamics of both coding and long non-coding RNAs
Zeb2os and Zeb2 are upregulated in astrocytes in acute and chronic stages and co-localize
Knockdown of Zeb2os leads to reduced astrogliosis, lesion size, and Zeb2 and Stat3 expression
ACKNOWLEDGMENTS
We thank Dr. Haipeng Xue, Zhengmei Mao and Chrystine Gallegos for their invaluable technical assistance and discussion. We also thank Dr. Haipeng Xue for providing us with pAAV vector. We are grateful to Dr. Richard Qing Lu for providing the anti-Zeb2 antibody. We thank Ms. Mary Ann Cushman for editing this manuscript. This work was supported by grants from the NIH, United States (R01 NS088353, R21 NS113068–01, and R21 EY028647–01), The Staman Ogilvie Fund-Memorial Hermann Foundation, and Mission Connect, a program of The Institute for Rehabilitation and Research (TIRR) Foundation.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2021.108721.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Anders S, and Huber W (2010). Differential expression analysis for sequence count data. Genome Biol 11, R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, and Huber W (2015). HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson MA, Burda JE, Ren Y, Ao Y, O’Shea TM, Kawaguchi R, Coppola G, Khakh BS, Deming TJ, and Sofroniew MV (2016). Astrocyte scar formation aids central nervous system axon regeneration. Nature 532, 195–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson MA, O’Shea TM, Burda JE, Ao Y, Barlatey SL, Bernstein AM, Kim JH, James ND, Rogers A, Kato B, et al. (2018). Required growth facilitators propel axon regeneration across complete spinal cord injury. Nature 561, 396–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayuso E, Mingozzi F, Montane J, Leon X, Anguela XM, Haurigot V, Edmonson SA, Africa L, Zhou S, High KA, et al. (2010). High AAV vector purity results in serotype- and tissue-independent enhancement of transduction efficiency. Gene Ther 17, 503–510. [DOI] [PubMed] [Google Scholar]
- Bailey TL, and Elkan C (1994). Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol 2, 28–36. [PubMed] [Google Scholar]
- Barres BA (2008). The mystery and magic of glia: a perspective on their roles in health and disease. Neuron 60, 430–440. [DOI] [PubMed] [Google Scholar]
- Bellver-Landete V, Bretheau F, Mailhot B, Vallières N, Lessard M, Janelle ME, Vernoux N, Tremblay ME, Fuehrmann T, Shoichet MS, and Lacroix S (2019). Microglia are an essential component of the neuroprotective scar that forms after spinal cord injury. Nat. Commun 10, 518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltran M, Puig I, Peña C, García JM, Alvarez AB, Peña R, Bonilla F, and de Herreros AG (2008). A natural antisense transcript regulates Zeb2/Sip1 gene expression during Snail1-induced epithelial-mesenchymal transition. Genes Dev 22, 756–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben Haim L, Ceyzériat K, Carrillo-de Sauvage MA, Aubry F, Auregan G, Guillermier M, Ruiz M, Petit F, Houitte D, Faivre E, et al. (2015). The JAK/STAT3 pathway is a common inducer of astrocyte reactivity in Alzheimer’s and Huntington’s diseases. J. Neurosci 35, 2817–2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson MD, Romero MI, Lush ME, Lu QR, Henkemeyer M, and Parada LF (2005). Ephrin-B3 is a myelin-based inhibitor of neurite outgrowth. Proc. Natl. Acad. Sci. USA 102, 10694–10699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E, Stamatoyannopoulos JA, Dutta A, Guigó R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, et al. ; ENCODE Project Consortium; NISC Comparative Sequencing Program; Baylor College of Medicine Human Genome Sequencing Center; Washington University Genome Sequencing Center; Broad Institute; Children’s Hospital Oakland Research Institute (2007). Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447, 799–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs JA, Wolvetang EJ, Mattick JS, Rinn JL, and Barry G (2015). Mechanisms of Long Non-coding RNAs in Mammalian Nervous System Development, Plasticity, Disease, and Evolution. Neuron 88, 861–877. [DOI] [PubMed] [Google Scholar]
- Bundesen LQ, Scheel TA, Bregman BS, and Kromer LF (2003). Ephrin-B2 and EphB2 regulation of astrocyte-meningeal fibroblast interactions in response to spinal cord lesions in adult rats. J. Neurosci 23, 7789–7800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buniello A, MacArthur JAL, Cerezo M, Harris LW, Hayhurst J, Malangone C, McMahon A, Morales J, Mountjoy E, Sollis E, et al. (2019). The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res 47 (D1), D1005–D1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butenschön J, Zimmermann T, Schmarowski N, Nitsch R, Fackelmeier B, Friedemann K, Radyushkin K, Baumgart J, Lutz B, and Leschik J (2016). PSA-NCAM positive neural progenitors stably expressing BDNF promote functional recovery in a mouse model of spinal cord injury. Stem Cell Res. Ther 7, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, and Rinn JL (2011). Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev 25, 1915–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS, Xing Y, Lubischer JL, Krieg PA, Krupenko SA, et al. (2008). A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J. Neurosci 28, 264–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao QL, Zhang YP, Howard RM, Walters WM, Tsoulfas P, and Whit-temore SR (2001). Pluripotent stem cells engrafted into the normal or lesioned adult rat spinal cord are restricted to a glial lineage. Exp. Neurol 167, 48–58. [DOI] [PubMed] [Google Scholar]
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. ; FANTOM Consortium; RIKEN Genome Exploration Research Group and Genome Science Group (Genome Network Project Core Group) (2005). The transcriptional landscape of the mammalian genome. Science 309, 1559–1563. [DOI] [PubMed] [Google Scholar]
- Chen K, Deng S, Lu H, Zheng Y, Yang G, Kim D, Cao Q, and Wu JQ (2013). RNA-seq characterization of spinal cord injury transcriptome in acute/subacute phases: a resource for understanding the pathology at the systems level. PLoS ONE 8, e72567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Shishkin AA, Zhu X, Kadri S, Maza I, Guttman M, Hanna JH, Regev A, and Garber M (2016). Evolutionary analysis across mammals reveals distinct classes of long non-coding RNAs. Genome Biol 17, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Zheng Y, Bu P, Qi X, Fan C, Li F, Kim DH, and Cao Q (2018). Apolipoprotein E as a novel therapeutic neuroprotection target after traumatic spinal cord injury. Exp. Neurol 299 (Pt A), 97–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung WS, Allen NJ, and Eroglu C (2015). Astrocytes Control Synapse Formation, Function, and Elimination. Cold Spring Harb. Perspect. Biol 7, a020370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuevas-Diaz Duran R, Wang CY, Zheng H, Deneen B, and Wu JQ (2019a). Brain Region-Specific Gene Signatures Revealed by Distinct Astrocyte Subpopulations Unveil Links to Glioma and Neurodegenerative Diseases. eNeuro 6, ENEURO.0288-18.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuevas-Diaz Duran R, Wei H, Kim DH, and Wu JQ (2019b). Invited Review: Long non-coding RNAs: important regulators in the development, function and disorders of the central nervous system. Neuropathol. Appl. Neurobiol 45, 538–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, et al. (2012). The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res 22, 1775–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Chen K, Cuevas-Diaz Duran R, You Y, Sloan SA, Zhang Y, Zong S, Cao Q, Barres BA, and Wu JQ (2015). Comprehensive Identification of Long Non-coding RNAs in Purified Cell Types from the Brain Reveals Functional LncRNA in OPC Fate Determination. PLoS Genet 11, e1005669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Cuevas-Diaz Duran R, You Y, and Wu JQ (2018). Identifying Transcription Factor Olig2 Genomic Binding Sites in Acutely Purified PDGFRalpha+ Cells by Low-cell Chromatin Immunoprecipitation Sequencing Analysis. J. Vis. Exp (134), 57547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duran RC, Yan H, Zheng Y, Huang X, Grill R, Kim DH, Cao Q, and Wu JQ (2017). The systematic analysis of coding and long non-coding RNAs in the sub-chronic and chronic stages of spinal cord injury. Sci. Rep 7, 41008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filous AR, and Silver J (2016). “Targeting astrocytes in CNS injury and disease: A translational research approach”. Prog. Neurobiol 144, 173–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallegos C, Carey M, Zheng Y, He X, and Cao QL (2020). Reaching and Grasping Training Improves Functional Recovery After Chronic Cervical Spinal Cord Injury. Front. Cell. Neurosci 14, 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. (2009). Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458, 223–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett AR, and Lee JK (2016). Understanding the NG2 Glial Scar after Spinal Cord Injury. Front. Neurol 7, 199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara M, Kobayakawa K, Ohkawa Y, Kumamaru H, Yokota K, Saito T, Kijima K, Yoshizaki S, Harimaya K, Nakashima Y, and Okada S (2017). Interaction of reactive astrocytes with type I collagen induces astrocytic scar formation through the integrin-N-cadherin pathway after spinal cord injury. Nat. Med 23, 818–828. [DOI] [PubMed] [Google Scholar]
- Hawthorne AL, Hu H, Kundu B, Steinmetz MP, Wylie CJ, Deneris ES, and Silver J (2011). The unusual response of serotonergic neurons after CNS Injury: lack of axonal dieback and enhanced sprouting within the inhibitory environment of the glial scar. J. Neurosci 31, 5605–5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Yu K, Lu F, Wang J, Wu LN, Zhao C, Li Q, Zhou X, Liu H, Mu D, et al. (2018). Transcriptional Regulator ZEB2 Is Essential for Bergmann Glia Development. J. Neurosci 38, 1575–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann JE, Imura T, Song B, Qi J, Ao Y, Nguyen TK, Korsak RA, Takeda K, Akira S, and Sofroniew MV (2008). STAT3 is a critical regulator of astrogliosis and scar formation after spinal cord injury. J. Neurosci 28, 7231–7243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermüller J, Hofacker IL, et al. (2007). RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 316, 1484–1488. [DOI] [PubMed] [Google Scholar]
- Khakh BS, and Deneen B (2019). The Emerging Nature of Astrocyte Diversity. Annu. Rev. Neurosci 42, 187–207. [DOI] [PubMed] [Google Scholar]
- Khakh BS, and Sofroniew MV (2015). Diversity of astrocyte functions and phenotypes in neural circuits. Nat. Neurosci 18, 942–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharchenko PV, Tolstorukov MY, and Park PJ (2008). Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat. Biotechnol 26, 1351–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, and Salzberg SL (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14, R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landrum MJ, Lee JM, Riley GR, Jang W, Rubinstein WS, Church DM, and Maglott DR (2014). ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res 42, D980–D985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landt SG, Marinov GK, Kundaje A, Kheradpour P, Pauli F, Batzoglou S, Bernstein BE, Bickel P, Brown JB, Cayting P, et al. (2012). ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res 22, 1813–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JK, Geoffroy CG, Chan AF, Tolentino KE, Crawford MJ, Leal MA, Kang B, and Zheng B (2010). Assessing spinal axon regeneration and sprouting in Nogo-, MAG-, and OMgp-deficient mice. Neuron 66, 663–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Ho IHT, Li X, Xu D, Wu WKK, Chan MTV, Li S, and Liu X (2019). Long non-coding RNAs in the spinal cord injury: Novel spotlight. J. Cell. Mol. Med 23, 4883–4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian H, Litvinchuk A, Chiang AC, Aithmitti N, Jankowsky JL, and Zheng H (2016). Astrocyte-Microglia Cross Talk through Complement Activation Modulates Amyloid Pathology in Mouse Models of Alzheimer’s Disease. J. Neurosci 36, 577–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberzon A, Subramanian A, Pinchback R, Thorvaldsdóttir H, Tamayo P, and Mesirov JP (2011). Molecular signatures database (MSigDB) 3.0. Bioinformatics 27, 1739–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liddelow SA, and Barres BA (2017). Reactive Astrocytes: Production, Function, and Therapeutic Potential. Immunity 46, 957–967. [DOI] [PubMed] [Google Scholar]
- Liddelow SA, Guttenplan KA, Clarke LE, Bennett FC, Bohlen CJ, Schirmer L, Bennett ML, Münch AE, Chung WS, Peterson TC, et al. (2017). Neurotoxic reactive astrocytes are induced by activated microglia. Nature 541, 481–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljungberg MC, Ali YO, Zhu J, Wu CS, Oka K, Zhai RG, and Lu HC (2012). CREB-activity and nmnat2 transcription are down-regulated prior to neurodegeneration, while NMNAT2 over-expression is neuroprotective, in a mouse model of human tauopathy. Hum. Mol. Genet 21, 251–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann M, Wright PR, and Backofen R (2017). IntaRNA 2.0: enhanced and customizable prediction of RNA-RNA interactions. Nucleic Acids Res 45 (W1), W435–W439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattucci S, Speidel J, Liu J, Kwon BK, Tetzlaff W, and Oxland TR (2019). Basic biomechanics of spinal cord injury - How injuries happen in people and how animal models have informed our understanding. Clin. Biomech. (Bristol, Avon) 64, 58–68. [DOI] [PubMed] [Google Scholar]
- Mattugini N, Bocchi R, Scheuss V, Russo GL, Torper O, Lao CL, and Götz M (2019). Inducing Different Neuronal Subtypes from Astrocytes in the Injured Mouse Cerebral Cortex. Neuron 103, 1086–1095.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Sunkin SM, Mehler MF, and Mattick JS (2008). Specific expression of long noncoding RNAs in the mouse brain. Proc. Natl. Acad. Sci. USA 105, 716–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mironets E, Wu D, and Tom VJ (2016). Manipulating extrinsic and intrinsic obstacles to axonal regeneration after spinal cord injury. Neural Regen. Res 11, 224–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molofsky AV, Krencik R, Ullian EM, Tsai HH, Deneen B, Richardson WD, Barres BA, and Rowitch DH (2012). Astrocytes and disease: a neurodevelopmental perspective. Genes Dev 26, 891–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murai KK, and Pasquale EB (2011). Eph receptors and ephrins in neuronastrocyte communication at synapses. Glia 59, 1567–1578. [DOI] [PubMed] [Google Scholar]
- Narang A, and Zheng B (2018). To Scar or Not to Scar. Trends Mol. Med 24, 522–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novikova IV, Hennelly SP, and Sanbonmatsu KY (2012). Structural architecture of the human long non-coding RNA, steroid receptor RNA activator. Nucleic Acids Res 40, 5034–5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Shea TM, Burda JE, and Sofroniew MV (2017). Cell biology of spinal cord injury and repair. J. Clin. Invest 127, 3259–3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmentier-Batteur S, Finger EN, Krishnan R, Rajapakse HA, Sanders JM, Kandpal G, Zhu H, Moore KP, Regan CP, Sharma S, et al. (2011). Attenuation of scratch-induced reactive astrogliosis by novel EphA4 kinase inhibitors. J. Neurochem 118, 1016–1031. [DOI] [PubMed] [Google Scholar]
- Pastori C, and Wahlestedt C (2012). Involvement of long noncoding RNAs in diseases affecting the central nervous system. RNA Biol 9, 860–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson SL, Nguyen HX, Mendez OA, and Anderson AJ (2015). Complement protein C1q modulates neurite outgrowth in vitro and spinal cord axon regeneration in vivo. J. Neurosci 35, 4332–4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petruska JC, Hubscher CH, and Rabchevsky AG (2013). Challenges and opportunities of sensory plasticity after SCI. Front. Physiol 4, 231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quackenbush J (2002). Microarray data normalization and transformation. Nat. Genet 32 (Suppl), 496–501. [DOI] [PubMed] [Google Scholar]
- Quinlan AR, and Hall IM (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renault-Mihara F, and Okano H (2017). STAT3-regulated RhoA drives reactive astrocyte dynamics. Cell Cycle 16, 1995–1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renault-Mihara F, Katoh H, Ikegami T, Iwanami A, Mukaino M, Yasuda A, Nori S, Mabuchi Y, Tada H, Shibata S, et al. (2011). Beneficial compaction of spinal cord lesion by migrating astrocytes through glycogen synthase kinase-3 inhibition. EMBO Mol. Med 3, 682–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renault-Mihara F, Mukaino M, Shinozaki M, Kumamaru H, Kawase S, Baudoux M, Ishibashi T, Kawabata S, Nishiyama Y, Sugai K, et al. (2017). Regulation of RhoA by STAT3 coordinates glial scar formation. J. Cell Biol 216, 2533–2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Wenger AM, Zehir A, and Mesirov JP (2017). Variant Review with the Integrative Genomics Viewer. Cancer Res 77, e31–e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauvageau M, Goff LA, Lodato S, Bonev B, Groff AF, Gerhardinger C, Sanchez-Gomez DB, Hacisuleyman E, Li E, Spence M, et al. (2013). Multiple knockout mouse models reveal lincRNAs are required for life and brain development. eLife 2, e01749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmeier S, Alam T, Essack M, and Bajic VB (2017). TcoF-DB v2: update of the database of human and mouse transcription co-factors and transcription factor interactions. Nucleic Acids Res 45 (D1), D145–D150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi LL, Zhang N, Xie XM, Chen YJ, Wang R, Shen L, Zhou JS, Hu JG, and Lü HZ (2017). Transcriptome profile of rat genes in injured spinal cord at different stages by RNA-sequencing. BMC Genomics 18, 173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver J, Schwab ME, and Popovich PG (2014). Central nervous system regenerative failure: role of oligodendrocytes, astrocytes, and microglia. Cold Spring Harb. Perspect. Biol 7, a020602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofroniew MV (2014). Astrogliosis. Cold Spring Harb. Perspect. Biol 7, a020420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Z, Niu W, Liu ML, Zou Y, and Zhang CL (2014). In vivo conversion of astrocytes to neurons in the injured adult spinal cord. Nat. Commun 5, 3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, and Mesirov JP (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran AP, Warren PM, and Silver J (2018). The Biology of Regeneration Failure and Success After Spinal Cord Injury. Physiol. Rev 98, 881–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, and Pachter L (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc 7, 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivinetto AL, Kim ID, Goldberg DC, Fones L, Brown E, Tarabykin VS, Hill CE, Cho S, and Cave JW (2020). Zeb2 Is a Regulator of Astrogliosis and Functional Recovery after CNS Injury. Cell Rep 31, 107834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanner IB, Deik A, Torres M, Rosendahl A, Neary JT, Lemmon VP, and Bixby JL (2008). A new in vitro model of the glial scar inhibits axon growth. Glia 56, 1691–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanner IB, Anderson MA, Song B, Levine J, Fernandez A, Gray-Thompson Z, Ao Y, and Sofroniew MV (2013). Glial scar borders are formed by newly proliferated, elongated astrocytes that interact to corral inflammatory and fibrotic cells via STAT3-dependent mechanisms after spinal cord injury. J. Neurosci 33, 12870–12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng Q, Chen Y, Wang H, Xu X, Yang B, He Q, Shou W, Chen Y, Higashi Y, van den Berghe V, et al. (2012). Dual-mode modulation of Smad signaling by Smad-interacting protein Sip1 is required for myelination in the central nervous system. Neuron 73, 713–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Klaw MC, Kholodilov N, Burke RE, Detloff MR, Côté MP, and Tom VJ (2016a). Expressing Constitutively Active Rheb in Adult Dorsal Root Ganglion Neurons Enhances the Integration of Sensory Axons that Regenerate Across a Chondroitinase-Treated Dorsal Root Entry Zone Following Dorsal Root Crush. Front. Mol. Neurosci 9, 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu LM, Wang J, Conidi A, Zhao C, Wang H, Ford Z, Zhang L, Zweier C, Ayee BG, Maurel P, et al. (2016b). Zeb2 recruits HDAC-NuRD to inhibit Notch and controls Schwann cell differentiation and remyelination. Nat. Neurosci 19, 1060–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada Y, Nomura N, Yamada K, Matsuo M, Suzuki Y, Sameshima K, Kimura R, Yamamoto Y, Fukushi D, Fukuhara Y, et al. (2014). The spectrum of ZEB2 mutations causing the Mowat-Wilson syndrome in Japanese populations. Am. J. Med. Genet. A 164A, 1899–1908. [DOI] [PubMed] [Google Scholar]
- Zadeh JN, Steenberg CD, Bois JS, Wolfe BR, Pierce MB, Khan AR, Dirks RM, and Pierce NA (2011). NUPACK: Analysis and design of nucleic acid systems. J. Comput. Chem 32, 170–173. [DOI] [PubMed] [Google Scholar]
- Zamanian JL, Xu L, Foo LC, Nouri N, Zhou L, Giffard RG, and Barres BA (2012). Genomic analysis of reactive astrogliosis. J. Neurosci 32, 6391–6410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Meltzer P, and Davis S (2013). RCircos: an R package for Circos 2D track plots. BMC Bioinformatics 14, 244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O’Keeffe S, Phatnani HP, Guarnieri P, Caneda C, Ruderisch N, et al. (2014). An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci 34, 11929–11947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HM, Liu T, Liu CJ, Song S, Zhang X, Liu W, Jia H, Xue Y, and Guo AY (2015). AnimalTFDB 2.0: a resource for expression, prediction and functional study of animal transcription factors. Nucleic Acids Res 43, D76–D81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Wu Y, Xie F, Zhong Y, Wang Y, Xu M, Feng J, Charish J, Monnier PP, and Qin X (2018). RGMa mediates reactive astrogliosis and glial scar formation through TGFb1/Smad2/3 signaling after stroke. Cell Death Differ 25, 1503–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the raw and processed sequencing data reported in this paper is GEO : GSE153721.


