Abstract
Objective
This study assesses the correlation between MDR1 gene polymorphism and clopidogrel resistance (CR) in Hui patients with coronary heart disease (CHD) who were treated with percutaneous coronary intervention (PCI).
Methods
The study includes 204 Ningxia Hui patients with CHD who were treated with PCI. These patients were divided into two groups: those who with CR and others were non-clopidogrel resistant (NCR), according to the results of the patients’ platelet aggregation rate, which was tested by adenosine diphosphate-induced turbidimetry on the second postoperative day. C3435T and C1236T genotypes and alleles were tested by polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP).
Results
The CR rate was 24.0%, and there were 3 genotypes of C3435T and C1236T. For C3435T, the distribution frequency of the 3435TT genotype and T allele was significantly higher in the CR group than in the NCR group. For C1236T, no significant difference was found between the two groups.
Conclusion
Hui patients who had CHD were treated with PCI. CR was most likely to occur in those who had the T allele of MDR1 in gene C3435T.
Keywords: coronary heart disease, MDR1 gene, clopidogrel resistance, gene polymorphism, platelet aggregation rate
Background
Coronary heart disease (CHD) is a major cardiovascular condition that threatens human health and requires percutaneous coronary intervention (PCI) treatment. Postoperative aspirin combined with clopidogrel antiplatelet therapy can effectively reduce adverse events such as stent thrombosis.1–4 However, some patients have clopidogrel resistance (CR), which can lead to thromboembolisms.5 Genetic factors are a major mechanism for the occurrence of CR and are related to region, race and population.
The MDR1 gene encodes the protein P-glycoprotein, which has a molecular weight of approximately 170,000. P-glycoprotein is a membrane transporter depended on adenosine triphosphate (ATP)6 and is widely distributed in many excretory organs and protective tissues in the human bodies. P-glycoprotein is closely related to the concentration of drugs in body. Clopidogrel can inhibit platelet aggregation, but its intestinal absorption is regulated by P-glycoprotein. Thus, P-glycoprotein has an important effect on the in vivo concentration of clopidogrel.
Currently, the MDR1 genetic markers C1236T and C3435T are the research hotspots.7,8 However, studies on the correlation between the MDR1 gene and clopidogrel are mostly limited to the Han population, and no such studies have been conducted on Hui population. Therefore, the present studies selected the genotype and allele related to CR in Hui patients who had CHD and received PCI and evaluated the correlation between the MDR1 gene polymorphism and CR.
Research Object and Methods
Patients
The studies include 204 patients (130 males and 74 females, 63.7% vs 36.3%) who range from 18 to 80 years old. These patients have been diagnosed with CHD and had accepted PCI treatment for the first time in Yinchuan First People’s Hospital between May 2015 and January 2017.
The inclusion group patients were required to have neither ethnic intermarriage within 3 generations nor related by blood. During their PCI treatment, the patients would receive 1 to 2 stent implantations, long-term oral administration of 100 mg of aspirin (Bayer), and one 300 mg dose of clopidogrel (Sanofi), followed by a maintenance dose of 75 mg daily. Patients also received 40 mg atorvastatin calcium tablets (Pfizer) and a subcutaneous injection of low-molecular-weight heparin according to their weight. Patients were excluded if they were averse to platelet therapy or had pre-existing medical conditions: a previous coronary artery bypass graft or PCI, severe liver or kidney disease, digestive system diseases, acute and chronic hematological diseases, tumors or other end-stage diseases, or severe cardiac insufficiency. Patients would also be excluded if they recently took platelet glycoprotein IIb/IIIa inhibitors, oral rifampin Li Fuping, erythromycin, or proton pump inhibitors. Participation in the study was voluntary. Patients have signed informed consent, and their treatments have been overseeing by trained medical personnel.
Diagnostic Criteria
Diagnostic criteria for CHD: The World Health Organization and the Chinese Medical Association (2007) issued the guidelines for diagnosis and treatment of CHD, including involvement of the left main stem, circumflex branch, or anterior descending branch confirmed by coronary angiography and diagnosis of stenosis of more than 50% in the main coronary artery or its branches.
Clopidogrel resistance: CR patients had a platelet aggregation rate of more than 50%; Non-clopidogrel resistant (NCR) patients had a rate below 50%.9,10
Research Methods
Collect General Information
With admission, the general clinical data of patients were collected by trained medical personnel (Table 1).
Table 1.
Comparison of General Clinical Data Between Group CR and Group NCR
| Subjects | CR Group (n=49) | NCR Group (n=155) | T/X2 value | P value |
|---|---|---|---|---|
| Man/cases (%) | 28 (57.1) | 102 (65.8) | 1.209 | 0.272 |
| Age (years) | 60.73±9.94 | 61.52±9.01 | −0.516 | 0.606 |
| Hypertension/cases (%) | 27 (55.1) | 94 (60.6) | 0.474 | 0.491 |
| Diabetes/cases (%) | 16 (32.7) | 50 (32.3) | 0.003 | 0.959 |
| Smoking/cases (%) | 15 (30.6) | 45 (29.0) | 0.045 | 0.832 |
| BMI (Kg·m−2) | 25.22±3.57 | 25.05±3.39 | 0.303 | 0.762 |
| Systolic pressure (mmHg) | 129.47±22.06 | 134.02±22.39 | −1.244 | 0.215 |
| Diastolic pressure (mmHg) | 76.61±14.10 | 80.22±12.67 | −1.690 | 0.093 |
| Triglyceride (mmol·L−1) | 2.13±1.42 | 1.87±1.23 | 1.236 | 0.218 |
| Cholesterol (mg·dL−1) | 3.93±1.24 | 3.98±2.55 | −0.131 | 0.896 |
| High density lipoprotein (mmol·L−1) | 0.99±0.21 | 0.93±0.21 | 1.771 | 0.078 |
| Fasting blood glucose (mmol·L−1) | 6.95±2.89 | 6.15±2.19 | 2.041 | 0.043* |
| Uric acid (mmol·L−1) | 321.27±86.32 | 327.20±92.06 | −0.248 | 0.804 |
| Number of stents (Pieces) | 1.30±0.46 | 1.33±0.47 | −0.306 | 0.760 |
Note: *P < 0.05 was statistically significant.
Collect Peripheral Blood Samples
EDTA anticoagulant tubes and sodium citrate anticoagulant tubes were used to collect 2 mL specimens of fasting venous blood in the morning of the first postoperative day. The blood samples in the EDTA anticoagulant tube were centrifuged at 3500 rpm, and DNA was extracted from the middle and lower portions of the tubes after −80°C refrigeration. The blood in the sodium citrate tube was used to determine the platelet aggregation rate 2 hours after collection.
Experimental Methods
Extraction of DNA from peripheral blood: Blood samples were extracted and preserved, and a TIANGEN kit was used to extract genomic DNA from the peripheral blood.
Polymerase chain reaction (PCR) amplification: Primer sequences for C3435T and for C1236T were designed according to the literature:11
C3435T primer sequence: P1: 5-TGCTGGTCCTGAAGTTGATCTGTGAAC-3′, P2: 5′-ACATTAGGCAGTGACTCGATGAAGGCA-3′
C1236T primer sequence: P1: 5′-TCTTTGTCACTTTATCCAGC-3′, P2: 5′-TCTCACCATCCCCTCTGT-3′.
PCR reaction conditions: DNA materials were pre-denatured at 94°C for 5 minutes, denatured for 30 seconds at 94°C, annealed at 60°C, extended at 72°C for one minute. This thermal cycling was repeated 30 times. The procedure was then extended at 72°C for 10 minutes. The primer sequence was compounded by Shanghai Biotechnology Co., Ltd.
Restriction enzyme digestion: The restriction endonuclease of the C3435T locus was DpnII, and the restriction endonuclease of the C1236T locus was Eco0109I; the reaction system was incubated for 15 minutes at 37°C.
The platelet aggregation test: The blood samples in the sodium citrate anticoagulant tube were centrifuged at a speed of 300–500 RPM for 5 mins, then drew the upper plasma to make the Platelet-rich plasma (PRP) and counted the number of platelet. The rest blood were centrifuged at a speed of 3000 RPM for 10 mins to make the Platelet-poor plasma (PPP). And 300 ul PRP and PPP were put into the platelet aggregation instrument (LBY-NJ4) to determine the relative platelet concentration. Finally, 3 ul ADP was added to determine the maximum platelet aggregation rate.
Statistical Methods
SPSS 19.0 software was used to analyze the data. The measurement data were expressed as mean ± standard deviation, while counting data were expressed in frequencies. A X2-test was used to measure the data of the 2 groups, and a t-test was used to compare the groups’ measurement data. Logistic regression analysis showed the correlation between the CR, genotype, blood sugar, and other factors. A value of P < 0.05 was considered statistically significant.
Results
Comparison of General Clinical Data
Table 1 shows the patients’ general clinical data.
Electrophoretic Results
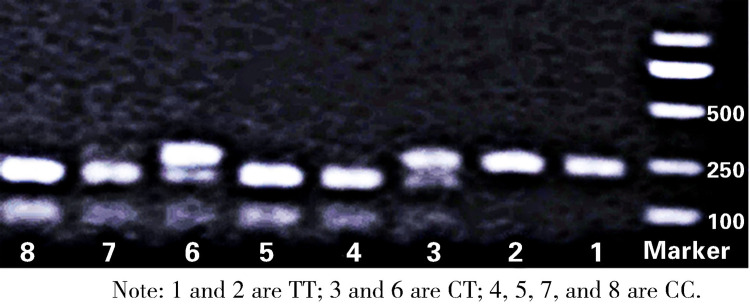
After the MDR1 gene C1236T site was cleaved, the CC type was cut into 379 bp and 123 bp, and 2 fluorescent bands were visible, while the CT type was cut into 502 bp, 379 bp, and 123 bp, and 3 fluorescent bands were visible. However, the TT type could not be cleaved; hence, only one fluorescent band (502 bp) was observed. After the C3435T site was cleaved, the CT type was cut into 248 bp, 190 bp, and 58 bp, and 2 fluorescent bands were visible, while the CC type was cut into 190 bp and 58 bp, and one fluorescent band was visible. However, the TT type could not be cut by endonuclease identification; hence, only one fluorescent band (248 bp) was visible (Figure 1).
Figure 1.
Electrophoresis of the restriction enzyme cleaved product of C3435T.
Genotype and Allele Frequency Distribution
Significant differences were seen in the distribution of the C3435T genotype and allele, respectively, between the CR and NCR groups (P < 0.05,Table 2). On the other hand, the C1236T genotype and allele frequencies showed no crucial differences when the CR and NCR groups were compared (P > 0.05, Table 3).
Table 2.
Comparison of Genotype Frequencies and Allelic Frequencies of C3435T Between CR and NCR Groups
| C3435T | Genotype, n (%) | Allele, n (%) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| CC | CT | TT | X2 value | P value | C | T | X2 value | P value | |
| CR group | 14 (28.6) | 18 (36.7) | 17 (34.7) | 6.998 | 0.030* | 46 (46.9) | 52 (53.1) | 8.185 | 0.004* |
| NRC group | 69 (44.5) | 58 (37.4) | 28 (18.1) | 196 (63.2) | 114 (36.8) | ||||
Note: *P < 0.05 was statistically significant.
Table 3.
Comparison of Genotype Frequencies and Allelic Frequencies of C1236T Between CR and NCR Groups
| C1236T | Genotype, n (%) | Allele, n (%) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| CC | CT | TT | X2 value | P value | C | T | X2 value | P value | |
| CR group | 12 (24.5) | 17 (34.7) | 20 (40.8) | 1.306 | 0.521 | 41 (41.8) | 57 (58.2) | 0.732 | 0.392 |
| NRC group | 40 (25.8) | 65 (41.9) | 50 (32.3) | 145 (46.8) | 165 (53.2) | ||||
The Hardy–Weinberg Balance Test
The Hardy–Weinberg equilibrium test was performed on the genotype distribution of the subjects’ MDR1 gene C1236T and C3435T loci. For the C1236T locus, the CR group scored X2 = 1.857, P = 0.395, and the NCR group had X2 = 1.884, P = 0.390. For the C3435T locus: The CR group had X2 = 1.507, P = 0.471, and the NCR group scored X2 = 2.882, P = 0.237. The P-values of both groups were greater than 0.05, indicating these subjects were from the same large group with good representation.
The Correlation Between MDR1 Gene Polymorphism and Clopidogrel Resistance
A two-element logistic regression analysis was performed with CR as the dependent variable. The independent variables included diabetes, fasting blood glucose levels, hypertension history, total cholesterol, body mass index, and C3435T and C1236T alleles (that were compared with the CC type, which acted as a dummy variable). The 3435TT locus (odds ratio [OR] =2.992, P = 0.010) and fasting blood glucose levels (OR = 2.479, P = 0.001) were independent risk factors for CR, while other factors were not correlated with CR.
Discussion
Since the specific criteria for CR are not consistent, different studies have been using different methods to measure platelet function, which makes the incidence of CR vary. The overall incidence of CR has been reported to be within 4%–31%.12,13 In the present studies, we used the method defined by Barragan14 to test the platelet aggregation rate of 204 patients. According to Barragan, a value of more than 50% was defined as CR, and the patients received 600 mg of clopidogrel 6 hours before the operation. The incidence of CR was 24%, which was consistent with the above findings. Other studies have shown that taking 600 mg of clopidogrel after 4 hours can be effective; taking 300 mg can induce the strongest effect after 24 hours; and a dose of 75 mg/d can inhibit platelet aggregation for 3–7 days after the operation.15 The subjects in our study underwent elective PCI surgery, and a 300 mg dose of clopidogrel was given after 4 hours, followed by a dose of 75 mg/d for maintenance. Although this was not strictly in accordance with the treatment protocol of 600 mg, this dose could also reach a steady state and inhibit platelet action.
Previous studies showed that the allele distribution and C3435T locus were significantly different when Asian and African16 were compared. The allele frequencies of locus T in African population in Ghana, Kenya, Sudan, and other African countries were 17%, 17%, and 27%, respectively, which were significantly lower than those in the Caucasus region (42%), India (62%), and among other Asian population.16 However, the TT genotype frequency in the African population was nearly zero (0% in Ghana, 4% in Kenya, and 6% in Sudan), which was observably lower than that in other population.17
This shows that the distribution of polymorphism is different among population depending on their races and regions. In the studies of healthy Han subjects conducted by Kim,18 the frequencies of the CC, CT, and TT types in C3435T were 35.1%, 49%, and 15.9%, respectively. In our present studies, among the 204 Hui patients, the CC, CT, and TT genotypes accounted for 40.7%, 37.3%, and 22.1%, respectively, and the C and T alleles accounted for 59.3% and 40.7%, respectively. Compared to the results reported by Kim, there was no significant difference in genotype distributions (P > 0.05), suggesting there is no significant difference in C3435T polymorphism between the Hui and Han peoples.
In the present studies on Hui patients, the CC, CT, and TT genotype frequencies of the C1236T loci were 25.5%, 40.2%, and 34.3%, respectively, which were all significantly different compared to the Han population19 whose CC, CT and TT genotype frequencies were 14.4%, 38%, and 47.6%, respectively; P < 0.05. This suggests that C1236T polymorphism exists not only in different regions but divergent ethnic groups.
Different genetic polymorphisms of the MDR1 C3435T locus have different impacts on CR. Studies have shown that patients with MDR1 CT or TT mutations have a higher rate of clinical endpoint events compared to those who carry the CC wild-type genes. Furthermore, several large clinical trials have shown that subjects with the C3435T locus TT mutant gene have lower rates of clopidogrel metabolism and inhibited platelet activities and, thereby, have an increasing risk of thromboembolic events.9,20 In addition, the present studies found that the TT genotypes and T alleles of the C3435T locus were dominant in the CR group, while the CC genotypes and C alleles were dominant in the NCR group.
The differences between these 2 groups are statistically significant (P = 0.004), which suggest the mutation of the T locus may lead to CR, which is consistent with the results of the above studies. It is also reported that subjects who carries the wild-type C3435T locus have the maximum risk of a thromboembolism event,21 which may due to the different functions of the alleles and the subsequent activity of encoding functional proteins. Due to the different frequencies of alleles in different races and regions, there are differences in the activities in coding functional proteins. For the C1236T site, Sibbing22 found that the CC wild type had a lower platelet inhibition rate when compared to the CT+TT mutant. Furthermore, other domestic and foreign studies did not reveal a significant correlation between C1236T and antiplatelet drug resistance.23 The polymorphism of this locus was not associated with CR in the present studies.
A study has shown that patients with myocardial infarction whose blood glucose level is higher than 8.5 mmol/l are usually low in response to clopidogrel and prone to clopidogrel resistance,24 which is consistent with the results of the present study. The different levels of fasting blood glucose between the 2 groups may interfere with the relationship between genetic polymorphism and CR. Hence, this variable should be controlled in future large-sample studies in order to reduce the impact of these interference factors on the results.
In order to clarify the correlation between MDR1 gene polymorphism and CR, a multivariate logistic regression equation was used for the analysis. Taking CR as the dependent variable, the C3435T and C1236T genotypes (compared with the CC types, which acted as a dummy variable) were the model’s independent variables. The results revealed that the TT genotype of the C3435T locus was an independent risk factor for CR (OR = 2.992, P = 0.010, 95% CI: 1.301–6.880). In addition, fasting blood glucose was also an independent risk factor for CR (P = 0.001, OR = 2.479, 95% CI: 1.438–4.274), which is consistent with the report of Hochholzer.25
In the present studies, we found that the MDR1 C3435T locus T allele mutation was associated with CR in Hui patients who had got PCI-treated CHD, but there was no significant relationship between the C1236T genetic polymorphism and CR in this patient group. However, due to the restriction of conditions, the platelet levels were not measured before drugs were taken. Meanwhile, the sample size was too small to be workable. Hence, these results need to be further confirmed by multicenter studies with larger sample sizes and higher experimental standards. Nevertheless, CR may be identified through genetic studies, which provide a theoretical basis for the individualization, safety, and rationale of clinical medication protocols for patients suffering from CHD after PCI.
Acknowledgments
We are particularly grateful to all the people who have offered us help on our articles.
Funding Statement
Source of supports: Ningxia Science and Technology Support Project (Grant No. 2012ZYS212). The funding body plays no role in designing the studies and collections, analysis, and interpretation of data and writing the manuscript as well.
Data Sharing Statement
We declared that materials described in the manuscript, including all relevant raw data, will be freely available to any scientists who wish to use them for non-commercial purposes without breaching participants’ confidentiality.
Ethics Approval and Consent to Participation
This study was conducted with approval of the Ethics Committee of Yinchuan First People’s Hospital. This study was conducted in accordance with the declaration of Helsinki. Written informed consent has been obtained from all participants.
Consent for Publication
All participants have signed documents with informed consent.
Disclosure
The authors declare that they have no competing interests.
References
- 1.Liu YM, Liu NF. Advances in clinical and genetic studies of clopidogrel response variability. Chin J Cardiovasc Dis. 2010;38:759–762. [PubMed] [Google Scholar]
- 2.Juliao RL, Ling V. A surface glycoprotein modulating drug permeability in Chinese hamster ovary cell mutants. Pharmacogenomics. 2011;12:114–144. [DOI] [PubMed] [Google Scholar]
- 3.Barragan P, Bouvier JL, Roqu Ebert PO. Resistance to thienopyridines: clinical detection of coronary stent thrombosis by monitoring of vasodilator stimulated phosphoprotein phosphorylation. Catheter Cardiovasc Interv. 2003;59(3):295–302. doi: 10.1002/ccd.10497 [DOI] [PubMed] [Google Scholar]
- 4.Bliden KP, Di CJ, Tantry US. Increased risk in patients with high platelet aggregation receiving chronic clopidogrel therapy undergoing percutaneous coronary intervention: is the current anti-platelet therapy adequate? J Am Coll Cardio. 2007;49(6):657–666. doi: 10.1016/j.jacc.2006.10.050 [DOI] [PubMed] [Google Scholar]
- 5.Simon T, Verstuyft C, Mary-Krause M. Genetic determinants of response to clopidogrel and cardiovascular events. N Engl J Med. 2009;360(4):363–375. doi: 10.1056/NEJMoa0808227 [DOI] [PubMed] [Google Scholar]
- 6.Zhou TT. Current status of clopidogrel and gene polymorphism. Chin J Clin Pharm. 2017;13:110–113. [Google Scholar]
- 7.Gutierrez-Rubio SA, Quintero-Ramos A, Durán-Cárdenas A. 1236 C/T and 3435 C/T polymorphisms of the ABCB1 gene in Mexican breast cancer patients. Genet Mol Res. 2015;14(1):1250–1259. doi: 10.4238/2015.February.13.3 [DOI] [PubMed] [Google Scholar]
- 8.Bai XC, Liu M. To investigate the effect of clopidogrel resistance on the prognosis of elderly patients with acute coronary syndrome undergoing coronary intervention. Chin J Mod Drug Appl. 2018;23:89–90. [Google Scholar]
- 9.Karaźniewicz-Łada M, Danielak D, Rubiś B, et al. Impact of common ABCB1 polymorphism on pharmacokinetics and pharmacodynamics of clopidogrel and its metabolites. J Clin Pharm Ther. 2015;40(2):226–231. doi: 10.1111/jcpt.12236 [DOI] [PubMed] [Google Scholar]
- 10.Chen F. Clinical study on cytochrome P4502C19*2 gene polymorphism and clopidogrel resistance and recurrent cardiovascular events. Taiyuan: Second Clinical Medical College of Shanxi Medical University; 2014. [Google Scholar]
- 11.Su J Association between ABCB1 gene polymorphism and clopidogrel resistance; 2014.
- 12.Qiu XY, Jiao Z, Zhong LJ. Effects of MDR1 C1236T, G2677T/A and C3435T gene polymorphisms on cyclosporine pharmacokinetics in Chinese Han renal transplant patients. Chin J Clin Pharm. 2008;17:141–146. [Google Scholar]
- 13.Mega JL, Close SL, Wiviott SD. Genetic variants in ABCB1 and CYP2C19 and cardiovascular outcomes after treatment with clopidogrel and prasugrel in the TRITON-TIMI 38 trial: a pharmacogenetic analysis. Lancet. 2010;376(9749):1312–1319. doi: 10.1016/S0140-6736(10)61273-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wei HJ, Shi JT, Li XX, et al. Antiplatelet drug efficacy in clopidogrel-resistant patients after coronary stenting. J Clin Exp Med. 2017;16:79–82. [Google Scholar]
- 15.Bonello L, De Labriolle A, Scheinowitz M, et al. Emergence of the concept of platelet reactivity monitoring of response to thienopyridine. Heart. 2009;95(15):1214–1219. doi: 10.1136/hrt.2008.152660 [DOI] [PubMed] [Google Scholar]
- 16.Sun W, Li Y, Li J. Variant recurrent risk among stroke patients with different CYP2C19 phenotypes and treated with clopidogrel. Platelets. 2015;26(6):558–562. doi: 10.3109/09537104.2014.953044 [DOI] [PubMed] [Google Scholar]
- 17.Li HP. Association between CYP2C19 gene polymorphism and clopidogrel resistance. Tianjin: Graduate School of Tianjin Medical University; 2013. [Google Scholar]
- 18.Kim RB, Leake BF. Identification of functionally variant MDR1 alleles among European Americans and African Americans. Clin Pharmacol Ther. 2001;70(2):189–199. doi: 10.1067/mcp.2001.117412 [DOI] [PubMed] [Google Scholar]
- 19.Li YH, Wang YH. MDR1 gene polymorphisms and clinical relevance. Acta Genet Sin. 2006;33(2):93–104. doi: 10.1016/S0379-4172(06)60027-9 [DOI] [PubMed] [Google Scholar]
- 20.Pedersen RS, Nielsen F, Stage TB. CYP2C19*17 increases clopidogrel-mediated platelet inhibition but does not alter the pharmacokinetics of the active metabolite of clopidogrel. Clin Exp Pharmacol Physiol. 2014;41(11):870–878. doi: 10.1111/1440-1681.12297 [DOI] [PubMed] [Google Scholar]
- 21.Li Y, Tang HL, Hu YF. The gain-of-function variant allele CYP2C19*17: a double-edged sword between thrombosis and bleeding in clopidogrel-treated patients. J Thromb Haemost. 2012;10(2):199–206. doi: 10.1111/j.1538-7836.2011.04570.x [DOI] [PubMed] [Google Scholar]
- 22.Sibbing D, Koch W, Gebhard D. Cytochrome 2C19*17 allelic variant, platelet aggregation, bleeding events, and stent thrombosis in clopidogrel-treated patients with coronary stent placement. Circulation. 2010;121(4):512–518. doi: 10.1161/CIRCULATIONAHA.109.885194 [DOI] [PubMed] [Google Scholar]
- 23.Tiroch KA, Sibbing D, Koch W. Protective effect of the CYP2C19 *17 polymorphism with increased activation of clopidogrel on cardiovascular events. Am Heart J. 2010;160(3):506–512. doi: 10.1016/j.ahj.2010.06.039 [DOI] [PubMed] [Google Scholar]
- 24.Carlquist JF, Knight S, Horne BD. Cardiovascular risk among patients on clopidogrel anti-platelet therapy after placement of drug-eluting stents is modifi ed by genetic variants in both the CYP2C19 and ABCB1 genes. Thromb Haemost. 2013;109(04):744–754. doi: 10.1160/TH12-05-0336 [DOI] [PubMed] [Google Scholar]
- 25.Pare G, Mehta SR, Yusuf S. Effects of CYP2C19 genotype on outcomes of clopidogrel treatment. N Engl J Med. 2010;363(18):1704–1714. doi: 10.1056/NEJMoa1008410 [DOI] [PubMed] [Google Scholar]