The Gram-negative bacterium Yersinia pestis is responsible for deadly plague, a zoonotic disease established in stable foci in the Americas, Africa, and Eurasia. Its persistence in the environment relies on the subtle balance between Y. pestis-contaminated soils, burrowing and nonburrowing mammals exhibiting variable degrees of plague susceptibility, and their associated fleas. Transmission from one host to another relies mainly on infected flea bites, inducing typical painful, enlarged lymph nodes referred to as buboes, followed by septicemic dissemination of the pathogen.
KEYWORDS: Yersinia pestis, epidemiology, lice, paleomicrobiology, plague
SUMMARY
The Gram-negative bacterium Yersinia pestis is responsible for deadly plague, a zoonotic disease established in stable foci in the Americas, Africa, and Eurasia. Its persistence in the environment relies on the subtle balance between Y. pestis-contaminated soils, burrowing and nonburrowing mammals exhibiting variable degrees of plague susceptibility, and their associated fleas. Transmission from one host to another relies mainly on infected flea bites, inducing typical painful, enlarged lymph nodes referred to as buboes, followed by septicemic dissemination of the pathogen. In contrast, droplet inhalation after close contact with infected mammals induces primary pneumonic plague. Finally, the rarely reported consumption of contaminated raw meat causes pharyngeal and gastrointestinal plague. Point-of-care diagnosis, early antibiotic treatment, and confinement measures contribute to outbreak control despite residual mortality. Mandatory primary prevention relies on the active surveillance of established plague foci and ectoparasite control. Plague is acknowledged to have infected human populations for at least 5,000 years in Eurasia. Y. pestis genomes recovered from affected archaeological sites have suggested clonal evolution from a common ancestor shared with the closely related enteric pathogen Yersinia pseudotuberculosis and have indicated that ymt gene acquisition during the Bronze Age conferred Y. pestis with ectoparasite transmissibility while maintaining its enteric transmissibility. Three historic pandemics, starting in 541 AD and continuing until today, have been described. At present, the third pandemic has become largely quiescent, with hundreds of human cases being reported mainly in a few impoverished African countries, where zoonotic plague is mostly transmitted to people by rodent-associated flea bites.
INTRODUCTION
Plague, caused by the bacterial pathogen Yersinia pestis, has been recognized by doctors and populations as a unique nosological entity for centuries because it is the sole disease characterized by swollen lymph nodes referred to as buboes to cause deadly epidemics. Historical sources in Europe have led to the delineation of three plague pandemics. The first pandemic, known as the Justinian Plague, devastated the Mediterranean Basin from 541 to 750/767 CE (1) and invaded northern Europe as far as Germany and England (2). The second pandemic, lasting from 1346 to the 18th century, including the so-called “Black Death” period of 1346 to 1353 (3), killed an estimated one-third of the European population (4). The third pandemic probably began in 1772 in the Chinese province of Yunnan (5, 6) and spread worldwide on the eve of the 20th century following human movement via steamship and railroad (7). Y. pestis infection may cause five principal forms of plague, including bubonic, septicemic, pneumonic, meningeal, and pharyngeal plague (8). In addition, Y. pestis may cause skin ulceration at its portal of entry, reported as carbuncles and ulcers, along with pustules, spots, petechiae, bruising, and gangrene (9). The plague etiology was resolved in 1894 in Hong Kong by the Swiss-French physician Alexandre Yersin, who was affiliated with the Pasteur Institute and who also contributed to resolving part of the cycle of transmission of Y. pestis involving the rat (Rattus rattus) and its ectoparasites (Xenopsylla cheopis) (10).
Obtaining a comprehensive overview of various sources of infection and various routes of transmission of Y. pestis in populations is critical for preventing and surveying plague. Cumulative field observations in plague foci combined with the critical review of data issuing from paleomicrobiological, anthropological, and historical studies continue to shed new light on questions related to the reservoirs, sources, transmission, and vectors of Y. pestis and to provide new avenues for addressing these questions.
Here, we critically review data reported in the English literature and some non-English publications to provide a comprehensive view of the various cycles of plague transmission as a basis for determining the appropriate medical management of plague in the countries in which it remains a zoonotic disease and a public health concern.
MODERN PLAGUE
Microbiology of Plague
Y. pestis is one of the three human-pathogenic Yersinia species, along with Yersinia pseudotuberculosis and Yersinia enterocolitica (8). Y. pestis is a nonmotile, nonsporulated, aerobic Gram-negative bacillus or coccobacillus exhibiting a hairpin morphology after Gram staining and growing within 24 to 72 h at a temperature range of 4 to 40°C (optimum, 28 to 30°C) at pH 7.4 (8). The sources and phenotypic characteristics of Y. pestis isolates allow their classification based upon the following: the region of isolation and circulation and the main hosts; the biochemical pattern, including the fermentation of rhamnose, melibiose, arabinose, glycerol, and melecytose, denitrification, fibrinolytic and coagulase activity, pesticin production and susceptibility, and dependence on amino acid sources; the frequency of mutation from a Pgm+ to a Pgm– phenotype in 10 generations; and virulence in guinea pigs (11). In particular, the biochemical pattern divides Y. pestis isolates into Y. pestis subsp. pestis, considered typically human (divided into the Intermedium, Antiqua, Medievalis, and Orientalis biovars), and Y. pestis subsp. microtus, considered typically zoonotic (11–13). Y. pestis bv. Orientalis isolates are unique in their capacity to stabilize the prophage YpfΦ (a filamentous phage) in their chromosome, which probably confers Y. pestis a selective advantage under natural conditions (14). Y. pestis is a gammaproteobacterium with a 4.60- to 4.65-Mb genome exhibiting numerous insertion sequences, intragenomic recombination and lateral gene transfer events, and remnants of an enteric life cycle (15, 16). The Y. pestis CO92 reference strain (biovar Orientalis) harbors three plasmids, a 70- to 75-kb plasmid common to the three human-pathogenic Yersinia species (designated pCD1, pCad, pVW, pYV, or pLcr) encoding the type III secretion system (T3SS) (including Yersinia outer proteins, or Yops) preventing the host’s immune response, the V antigen (LcrV), which is also implicated in immunosuppressive activity, and the yersiniabactin siderophore system gene (ybt), allowing Y. pestis to acquire iron from blood (8, 17). A 100- to 110-kb plasmid (designated pFra/Tox, pFra, pTox, pMT1, or pYT) encodes the capsular F1 glycoprotein antigen and the Yersinia murine toxin Ymt, allowing the survival of Y. pestis in the flea gut (18), and a 9.5-kb plasmid (designated pPst, pPla, pPCP1, or pYP) encodes the plasminogen activator Pla and pesticin, a bacteriocin (8). However, the plasmid contents can be altered by successive subculturing (19). Additional plasmids have also been characterized, illustrating the plasticity and ongoing evolution of Y. pestis (12, 20–22). The pPCP1 plasmid encodes the plasminogen activator Pla, regarded as a major virulence factor promoting the systemic spread of Y. pestis from peripheral sites (23). The pla gene is an outer membrane omptin member, and omptins are detected in several Gram-negative bacteria, including animal and plant pathogens (24, 25). Ectoparasite bites may provoke discrete local inflammation (26) at the skin portal of entry of Y. pestis, which then spreads via the lymphatic route toward regional lymph nodes, in which pathogen growth results in the development of a bubo (27). Y. pestis further spreads via the lymph (28, 29) and blood vessels to the spleen and liver and causes rapidly fatal septicemia, with dissemination in the lungs (resulting in secondary pneumonic plague) and the meninges and cerebrospinal fluid (causing meningitis). The hematogenous dissemination of the bacteria may cause intravascular coagulation and endotoxic shock (30). During this process, Y. pestis rapidly multiplies in tissues, being protected from the immune system by serum resistance (31) and the evasion of innate immune functions, including the neutralization of immune cells mediated by the T3SS (encoded by the virulence plasmid pYV/pCD1) (32), the absence of detectable pathogen-associated molecular patterns (30), and the modulation of host innate immune cell interactions (33). Thus, Y. pestis is a facultative intracellular bacterium that multiplies in macrophages (34). A role of the mediator of inflammation MyD88 has been identified in the pathology of primary pneumonic plague, with MyD88 exhibiting a biphasic inflammatory response that ultimately limits systemic infection in a mouse model (35).
Plague in the 21st Century
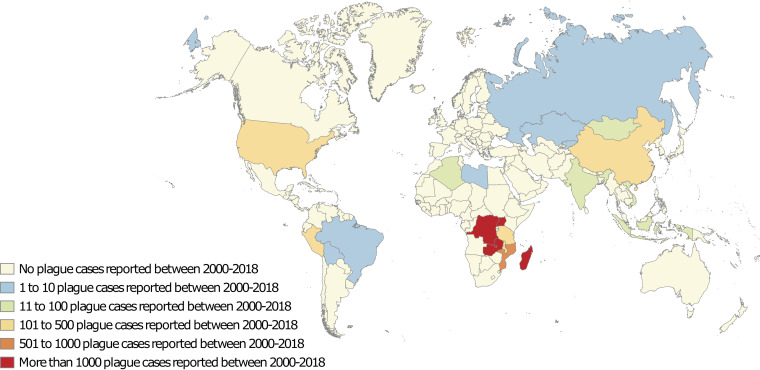
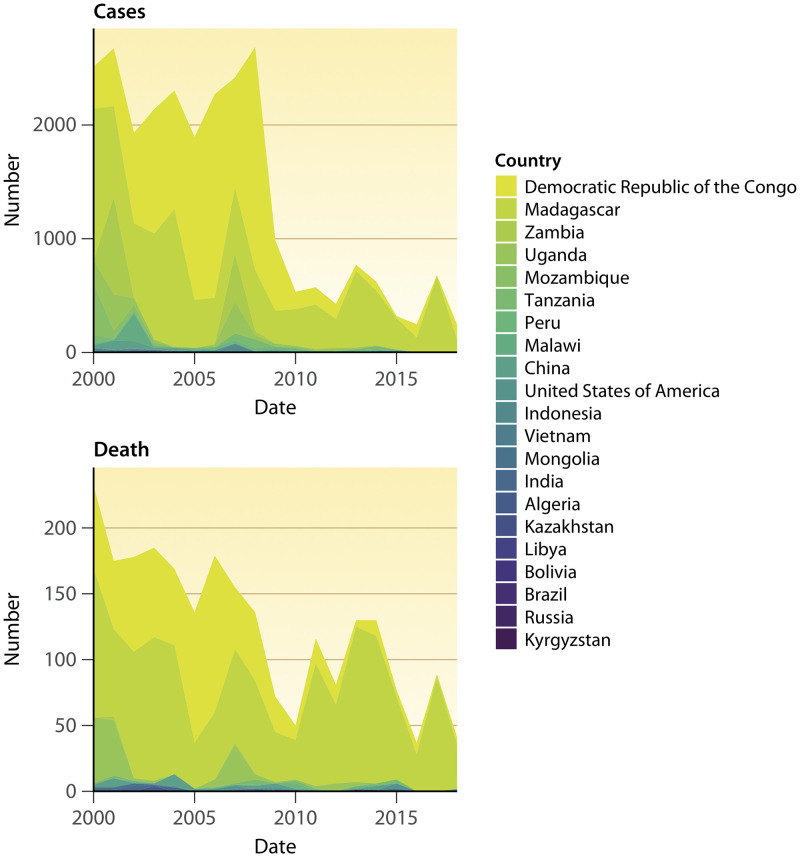
The natural history of plague is shaped by time and geography (Table 1). The geographical extension of plague can be retrieved from World Health Organization (WHO) notifications, but plague has not been a WHO-notifiable disease since 2005, with the exception of pulmonary plague cases and cases documented in countries where the disease is nonendemic (36) (Fig. 1 and 2; Table 1). In the last few decades, 21 countries scattered across the Americas, Africa, and Asia have reported a total of 26,237 plague cases to the WHO under different epidemiology regimens (data from 2000 to 2018) (37–39). In the Americas, 371 plague cases have been reported over the last 19 years, corresponding mainly to sporadic cases in the United States and clustered cases in South America (9, 40–45). Africa has experienced the largest number of plague cases worldwide (97%) (46–56), including 25,409 cases reported mainly by the Democratic Republic of Congo and Madagascar. In Madagascar, plague foci are located in the center and north of the island, especially in the Ambalavao and Tsiroanomandidy districts. In endemic highland foci, rats were found to be 1,000 times more resistant to plague than rats collected along the coast (57). Xenopsylla fleas feeding on these rats and spreading plague among susceptible rodents are considered essential key factors responsible for dissemination of plague in Madagascar (58). Accordingly, most cases are bubonic (transmitted by fleas), with possible secondary pneumonic forms (58–61). In 2017, a large outbreak accounted for approximately 2,400 cases, mostly of the pneumonic plague form (78%); however, only 32 cases were microbiologically confirmed (59). In northern Africa, Algeria reported 15 cases in 2003 (62), and 13 cases were reported in Libya near Tobruk in 2009 (63). Molecular analyses revealed two different strains, Y. pestis IP1860-64 bv. Orientalis in Algeria and Y. pestis IP1973-75 bv. Medievalis in Libya, suggesting that several independent plague foci exist in these two countries (63). In Mongolia in 2010, most recorded bubonic plague cases were traced to marmots (75.2%) (64, 65), similar to the situation in Kyrgyzstan and Russia, where two cases were contracted by marmot hunters (37, 66–68). These data indicate the diversity of current plague sources, including mammals such as dogs, cats, camels, and rodents and one species of bird (65). Some characteristics common to most plague foci include (i) locations in low-density rural zones (this characteristic may explain the low number of cases, because contacts between human populations and vectors are infrequent) (6), (ii) higher altitudes relative to the rest of the country (36, 40, 48, 65, 69–75), (iii) locations in arid or semiarid zones (low precipitation) (6), (iv) locations in or around saline soil (76, 77), (v) the presence of at least two hosts/vectors and one species of flea, and (vi) enzootic epidemiology seemingly regulated by subtle interplay between resistant hosts (arising from pressure selection) and susceptible hosts.
TABLE 1.
Currently available epidemiological information for the 19 countries that have reported plague cases over the last 19 yearsa
| Country | Localization of plague foci | Characteristic(s) of plague foci | Current main source | Reference(s) |
|---|---|---|---|---|
| Democratic Republic of the Congo | Ituri, Haut-Uele districts, North Kivu province | Dry tropical, tropical mountain zones, elevation > 1,000 m | Mastomys natalensis, Arvicanthis abysinnicus, Lemniscomys striatus, Mus minutoides, Rattus rattus, fleas | 48, 71, 192, 245 |
| Madagascar | Center, North, and East of the country | Higher elevation than in coastal region rural zones | Rattus rattus, shrew, fleas | 57–61, 183, 347 |
| Uganda | West Nile regions | Plague risk increases during the dry season and with an elevation of >1,300 m | Rats, fleas | 36, 49, 50, 51, 146, 184 |
| Tanzania | Lushoto, Mbulu, Monduli district | Brush-furred mice, Praomys spp., African dormice, zebra mice, commensal rats, fleas | 52–54, 56, 289 | |
| Zambia | Namwala, Sinda, Chama, Lundazi, Zambezi district | Semiarid zones | Gerbils, rats, fleas | 46, 47, 55 |
| Brazil | Pedra branca, Feira de Santana areas | Semiarid and xeric zones, plague risk increases with precipitation, elevation > 200 m | Grass mice, Oryzomys spp., vesper mice, Bolomy spp., Amazonian marsh rat, gray short-tailed opossum, Marsupialia, Oligoryzomys, Cerradomys, Spix's yellow-toothed cavy, Sao Lourenço punaré, fleas | 45–47, 290, 360 |
| Peru | Cajamarca, Otuzco, Ascope, Trujillo, Pacasmayo regions | Desert and arid zones, plague risk increases with an elevation of >1,300 m | Askodon, Oryzomys, cotton rat, Phyllotis, Rattus rattus, guinea pig, fleas | 42, 45 |
| United States | West of the country | Arid and saline zones, winter/spring rainfall, high elevation < 850 m | Prairie dog, squirrel, chipmunk, mountain lion, cat, dog, coyote, marmot, rabbit, hare, mice, vole, rat, fleas | 41, 69, 70, 76, 93, 138, 353 |
| China | North, Northeast, and South of the country; Junggar Basin | Plague risk increases with an elevation of >3,100 m; arid, desert, and subtropical zones; calcium- and iron-enriched environment | Marmot, Tibetan sheep, Spermophilus dauricus, dog, Meriones, rat, rabbit, mice, fleas | 74, 271–276 |
| Kazakhstan | Lake Balkhash areas, Aktyubinsk, Atyrau, Kyzylorda districts | Arid, saline, and desert zones | Camel, great gerbil, rodents, fleas | 102, 277, 279 |
| Mongolia | West of the country | High elevation > 650 m, desert and arid zones | Pallas's pika, Daurian pika, Siberian marmot, gray marmot, long-tailed suslik, Redcheeked suslik, Russian hamster, silver mountain vole, flat-headed vole, narrow-headed vole, Brandt's vole, Mongolian gerbil, midday gerbil, great gerbil, Siberian five-toed jerboa, corsac fox, Siberian polecat, mountain weasel, northern wheatear, fleas | 64, 65 |
| Vietnam | Central highlands region | High elevation zones > 850 m, plague risk increases during the dry season | Rattus rattus, Pacific rat, Rattus norvegicus, Asian house shrew, tree shrew, fleas | 75 |
| Algeria | Oran, Mascara, Laghouat areas | Arid and saline zones (Chott) | Camel, Rattus norvegicus, Rattus rattus, Meriones shawii, Psammomys obsesus, Mus spretus, Apodemus sylvaticus, Cricidura russula, fleas | 62, 77, 268, 269 |
| India | Andhra Pradesh, Karnataka, Tamil Nadu areas | Elevation > 300 m, tropical zones | Rattus rattus, Mus musculus, Indian gerbil, lesser bandicoot rat, fleas | 99, 105 |
| Libya | Nofilia area | Camel, rodents, fleas | 63 | |
| Indonesia | Central and West Java regions | High elevation > 1,000 m, humid zones | Rattus rattus, fleas | 436 |
| Bolivia | Franz Tomayo, Andres Ibanez provinces | Tropical and subtropical dry zones, plague risk increases with an elevation of >1,300 m | Akodon, Rattus rattus, fleas | 45 |
| Russia | Mountain-Altai, Tuva (Mongun-Taigin) areas | High elevation, arid and desert zones | Camel, marmot, rodents, fleas | 67, 68, 96, 154, 278, 280, 281, 283 |
| Kyrgyzstan | Tien Shan, Alai, Talas mountains | High elevation > 2,700 m, continental climate | Marmot, mountain vole, rodents, fleas | 66, 283, 284 |
Including the locations of plague foci, characteristics of plague foci, and plague sources based on data published in the peer-reviewed literature.
FIG 1.
World map of plague cases reported to the World Health Organization in the 21st century (39).
FIG 2.
All plague cases and plague deaths by year and country reported by the World Health Organization over the last 19 years (39).
Natural Sources of Y. pestis
Y. pestis in soil.
The hypothesis that soil might be a source of plague was proposed prior to the 1894 discovery of Y. pestis. In 1882, in a report to the British authorities, General Osbert Chadwik raised concerns regarding the wastewater disposal systems in the Taipingshan District of Hong Kong, leading to the idea that “the soil of Taipingshan was typically soaked with sewage discharged from dysfunctional drains and through the broken floors of the buildings above” (78). Equally widespread was the fear that the soil under inappropriately constructed houses in Taipingshan might become a receptacle of plague-infected bacteria, objects, or infected bodies falling through the porous nonintact floors of homes. An outbreak in Hong Kong in 1894 subsequently confirmed these fears, and the health commission proposed the burning of infected houses to “purify” the soil, which was believed to be the source of the infection (79). In an 1894 publication, Alexandre Yersin wrote that he had isolated one attenuated Y. pestis strain from the soil 4 to 5 cm below the surface of one house that was the home of plague victims in Hong Kong (80), and Kitasato reported the isolation of plague bacilli from “soil dust.” Alexandre Yersin verified his observations by collecting soil specimens from other houses where plague victims lived and from negative-control houses; he isolated the plague agent from 4 of 10 test houses and from none of the negative controls. Alexandre Yersin further confirmed his observations in Canton, where in contrast to Hong Kong, houses had not been disinfected. He found plague bacilli 20 to 30 cm below the surface but not at a depth of 1 m (79). At this time, the plague was considered by plague experts to be caused by a telluric bacterium from soil contaminated by feces from rats or other infected animals; even the direct inhalation of Y. pestis-contaminated soil was reported to cause primary pulmonary plague (81, 82). These preliminary data were not confirmed until 70 years later, when Karimi reported the isolation of Y. pestis from a burrow where plague-infected mammals had died approximately 11 months previously in Iran (83). More recently, Y. pestis was isolated under natural conditions in Arizona from soil under the corpse of a mountain lion 3 weeks after the death of the animal (84). These natural observations were completed by isolating the Algeria3 strain (biotype Orientalis) from saline soil (40 g/liter NaCl) collected at the edge of a saltwater lake in Algeria (77). It was recently found that such observations could be extended to the United States, where the location of significant plague foci is correlated with the aridity and salinity of the soil (76). These field observations were correlated with experimental observations recorded during the 16-month persistence of Y. pestis in soil by Mollaret (85). These experimental observations have been refined and have demonstrated the persistence of a virulent Y. pestis Orientalis biovar in soil for up to 280 days (86). The persistence of plague in soil contrasts with the limited survival of Y. pestis on steel or glass, where it survives for less than 72 h (87). Altogether, at least four Y. pestis strains have been isolated from soil (77, 80, 83, 84). Only the Y. pestis strain isolated in Hong Kong appears to be attenuated, as reported by Alexandre Yersin, which is potentially explained by the so-called dormancy phenomenon (80). Indeed, Y. pestis has the capacity to pass from a coccobacillus to an L-form in response to environmental stresses, such as extreme salinity, low temperatures, or the conditions of the flea host (77, 88, 89). The L-form might be a persistent form of Y. pestis outside mammalian hosts under extreme environmental conditions, as suggested by the conservation of the pst and pim genes despite their lack of function during mammalian infection (90). Moreover, under nonextreme conditions, Y. pestis remains alive and virulent in soil for long periods of time (85, 86) and in soil amoeba trophozoites, which may serve as temporary reservoirs (91, 92).
(i) Y. pestis and protozoa.
In the environment, Y. pestis may reside in protozoa, including amoebae (93). In 1999, Domaradsky hypothesized that the plague was a protonosis of the soil living in protozoan vegetative and cyst forms (94). Based on studies of soil ecology in Russia, it was suspected that Y. pestis could engage in intracellular interactions with protozoan cysts and blue-green algae (formerly cyanobacteria), enabling long-term preservation in the soil (95). Accordingly, several amoebae, belonging mainly to the Acanthamoeba genus (including A. castellani), were isolated from the soil in a natural plague focus in the Caspian region at a concentration of 300,000 cells/g of soil (96). Further experimental studies involving the coculture of soil free-living amoebae and Y. pestis demonstrated the survival and replication of Y. pestis for more than 48 h in Dictyostelium discoideum trophozoites (92) and for a minimum of 5 days after phagocytosis by A. castellani trophozoites (91). Similar to macrophages, amoeba infection is dependent on Y. pestis phoP, which encodes a transcriptional regulator (17, 97), and the Y. pestis type III secretion system (T3SS) (98). Similarly, protozoa present in rodent and lagomorph digestive tracts might host Y. pestis and act as temporary reservoirs (94).
Y. pestis in animals.
More than 200 species of mammals (rodents principally) can be infected with Y. pestis, and plague-resistant species are regarded as a source of Y. pestis (8, 9, 93, 99). However, not all mammalian species are plague susceptible, and species such as grasshopper mice (100), marmots (101), great gerbils (102), four-striped mice (103), deer mice (104), rats (105), California voles (106), kangaroo rats (93), and dogs (107–109) are suspected to be selectively plague resistant, resulting from selection pressure during relatively long periods of coexistence with Y. pestis (105). Although Gunnison’s and black-tailed prairie dogs are highly plague susceptible (showing almost 100% mortality) (93), some research hypotheses suggest that a few resistant animals might exist among these populations (69, 110). The subtle balance between plague-susceptible and plague-resistant species has been advocated for use in the modeling of plague epizootics (93). Nevertheless, resistant hosts appear to harbor very low quantities of bacteria in their blood, too low to infect fleas, for which the minimum required concentration is 10 million CFU/ml (111). Carnivores can be infected after the ingestion of contaminated rodents (100). Additionally, camels in the Maghreb, the Middle East, and Asia are highly susceptible to plague following contact with dead rodent-contaminated carcasses or excrement (6, 112). Camels were reported to develop plague after experimental subcutaneous inoculation with Y. pestis (2/4 died) and Y. pestis inhalation (6/6 died, with primary pneumonic symptoms), whereas fodder ingestion induced bubonic plague (3/3 recovered after exhibiting bubonic plague symptoms with submaxillary buboes) (113). A second set of experiments with experimental parameters closer to natural infection conditions was performed in 1954 to 1956. In total, 28 Bactrian camels were infected by natural camel ectoparasites, such as blocked Xenopsylla and Coptopsylla flea species and ticks (Hyalomma asiaticum and Ornithodoros tartakovskyi) that had previously fed on infected guinea pigs. The blocked fleas successfully transmitted plague to the eight camels they bit. The ticks transmitted plague to the camels only in rare cases; when they did so, it was within 1 to 2 days after infection, and the authors suggested mechanical transmission via the infection of their buccal parts (112). These experiments indicated that the susceptibility of camels to plague varied based on the route of contamination. Concerning domesticated animals, canids are relatively resistant to plague (9). Indeed, among 10 dogs experimentally infected with the Y. pestis 195/P strain via oral and parenteral routes, all 10 showed signs of infection; however, none of them died from plague, and after 7 days, the dogs recovered. All the dogs developed antiplague antibodies, which were present up to 300 days postinfection (107). Accordingly, it was recently demonstrated that the FRP1 receptor in mammals promotes the translocation of bacterial effectors; the absence of this receptor in canids confers enhanced resistance to plague (109). Nevertheless, dogs can present primary pneumonic plague and act as a source of contamination in humans (41). Additionally, domesticated cats can be infected following the hunting and consumption of infected wild prey, such as rodents, or by flea bites. Wild animals, livestock, and domesticated animals are therefore sources of plague for other animals and humans living in contact with them.
Plague Transmission
Transmission of Y. pestis to animal populations.
Y. pestis-contaminated soil can be a source of infection in mammals. This finding was demonstrated in a series of experiments performed by H. Mollaret et al., who exposed various species of Meriones to soil contaminated with Y. pestis (114). These experiments led to the conclusions that humid soil is a better contaminant than dry soil, that burrowing Meriones species are more likely to die from plague than nonburrowing Meriones species, and that some burrowing Meriones species are more susceptible to plague than others. However, it was not possible to determine the exact route of contamination (i.e., whether Meriones individuals inhaled or ingested contaminated soil or both) when a hemorrhagic digestive tract and hemorrhagic pneumonia were present (114). Further experiments performed 50 years later involve susceptible Swiss-Webster mice in which a scarified paw was left in contact with soil that had been contaminated for 10 days with blood containing Y. pestis (>108 CFU/ml). In this experiment, only 1 of 104 animals became infected, and none of the other mice seroconverted, suggesting that this route of contamination is unlikely to sustain epizootics (115). Beyond studies involving soil, the potential of plants to serve as a plague source in animals has been poorly investigated, except in the Russian literature (95, 116, 117). The colonization of Impatiens walleriana plants by the Y. pestis EV strain was observed after scarifying and immersing stems of this plant in an infected solution (117). This observation was met with skepticism and was poorly cited. Field observations incorporating a large number of plants, including negative-control plants, should clearly be performed to determine precisely whether plants play any role in the epidemiology of plague. Similarly, the ability of fleas to become infected through contact with soil remains questionable: one study showed greater vector efficiency in mice (50% infection against 23% in the control group) when Oropsylla montana was in contact with soil infected with wild flea feces (118). In parallel, it was shown that Y. pestis-contaminated flea feces could contaminate soil and burrows (89, 90, 118–120). The inhalation or ingestion of contaminated soil material associated with burrowing activities remains to be investigated in the field using appropriate experiments. Indeed, very few experiments have been aimed at observing the contamination of mammals via the oral route. Paul-Louis Simond failed to orally infect rats, monkeys, and squirrels by feeding them an infected culture or infected spleen, liver, blood, feces, and urine samples (27). Furthermore, tests in the rodents Mus musculus, Zygodontomys pixuna, and R. rattus in which doses of 5.46 to 9.62 log10 viable Y. pestis bacteria were administered intragastrically yielded 100% mortality, similar to the observations made following the addition of 108 Y. pestis bacteria/ml of drinking water to infect mice, which resulted in death within 3 days. Notably, Y. pestis was not found in any rodent fecal samples, which is related to the fact that the Y. pestis strain used for challenge does not survive at pH less than 3 (121). A confirmatory study showed that 3/20 Onychomys leucogaster individuals from a parental population exposed to plague and 7/20 O. leucogaster individuals from a plague-naive parental population died after they were fed plague-infected mice, whereas 4/20 and 13/20 individuals of these species, respectively, survived and developed positive serology (100). These laboratory experiments demonstrate the capacity of rodents to be infected via the oral route either by eating contaminated food or by burrowing into contaminated soil.
(i) Flea-borne transmission in animal populations.
Interanimal transmission can be achieved by animal ectoparasites, and approximately 80 flea species are listed as common vectors of Y. pestis among rodents (12, 122, 123). In 1897, one of Kitasato’s mentors, Masanori Ogata, suspected fleas of playing a role in the transmission of plague (124) and successfully infected mice with a suspension of ground fleas that had fed on an infected rat (93). Simultaneously, Paul-Louis Simond et al. discovered plague bacilli in rat flea intestines in Cutch-Mandvi, India (125). Shortly thereafter, Simond caught an infected rat covered with fleas in the home of a plague victim in Karachi and harvested fleas from a stray cat lurking near his hotel. The experiment consisted of trapping the infected rat in a glass bottle, preventing it from moving, and placing the cat’s fleas on the dying rat. Finally, a naive plague rat was placed in a suspended cage close to the dying rat. Simond found that the healthy rat died 5 days after the death of the infected rat without experiencing any direct contact with the infected rat. Paul-Louis Simond concluded that plague transmission from rat to rat, rat to human, and human to human could be mediated by fleas of any kind and that flea feces are contagious when used to inoculate rats (27). Currently, two major routes of plague transmission by fleas have been identified. First, early-phase or mass transmission was discovered between 1904 and 1947 (119). It has been shown that from 3 h to 7 days after infection and before blocking, the flea has the ability to infect a healthy animal during its next blood meal (104, 126). In this model, the fleas immediately become infectious (104) (Y. pestis can survive for only 3 h on a flea’s mouthparts) (127) after their first plague blood meal because of the partial, early, ephemeral blockage of the proventriculus. Indeed, the ingested bacteria accumulate and form a small conglomerate in the proventriculus during a blood meal, and only a blood pulse clears this conglomerate and provokes blood reflux, carrying a few bacteria to the biting point (119). The efficiency of transmission varies greatly among flea species; for example, a transmission efficiency (TE) of 6.4% has been reported for X. cheopis (128), 17.88% for Oropsylla tuberculata cynomuris, 4.54% for Oropsylla hirsuta (129), and 7.7% to 10% for Oropsylla montana (130). Nosopsyllus fasciatus (0.213 ± 0.157 expected transmission per flea [ETF]) and Orchopeas sexdentus sexdentus (0.170 ± 0.138 ETF) (126) are quite effective vectors, while Aetheca wagneri (TE = 1.03%), Ctenocephalides felis (TE = 0.57%) (104, 131), Opisodasys nesiotus, Megabothris abantis, Malaraeus telchinum, and Diamanus montanus (126) appear to be very inefficient vectors. Two major limitations of early-phase/mass transmission implications were proposed by Hinnebusch et al. (119): first, an inoculum concentration higher than 108 bacteria/ml, which is required for effective transmission, is naturally achieved in mice but not in most other mammals except very shortly before death. Second, the number of bacteria transmitted individually by each flea is extremely low (less than 10 individual bacteria are sufficient to infect susceptible mammals) (132), and early-phase transmission (EPT) is effective only in a host that is highly sensitive to and has been bitten by a large number of fleas (133). EPT is ineffective for X. cheopis at low temperatures of ∼10°C (134). It is very difficult to draw conclusions from these studies, because the conditions under which the experiments were conducted differ greatly, leading to heterogeneous results. Indeed, the strains, the bacterial concentrations, the incubation times, and the hosts of the fleas differ. Sometimes, fleas can be blocked rapidly, and EPT can be confused with later-stage transmission. The results are also host dependent, as shown in a study conducted by Bland et al., who observed that 10% to 28% of fleas that fed on bacteremic rats or guinea pig blood showed the reflux of bacteria into their esophagus within the first 24 h postinfection and exhibited an increased vectorial capacity during EPT (135). Therefore, new experiments using standardized methods (as described above) to evaluate and compare transmission efficiencies are needed to understand the relative importance of EPT. The following second mechanism, referred to as “biofilm-dependent transmission,” was described in articles in 1914 and 1915 by Bacot and Martin demonstrating the vectorial capacity of blocking X. cheopis and Ceratophyllus fasciatus in rats (136, 137): fleas can develop a bacterial biofilm in the proventricular valve of the midgut from 1 to 3 days after ingestion (119, 126, 138). The biofilm partially or completely blocks the midgut such that the fleas can no longer feed and ultimately starve. The behavior of the fleas changes drastically; during the last days of their life, they relentlessly attempt to feed, considerably increasing the number of bites they inflict and the opportunities for Y. pestis transmission. Once blocked, aspirated blood comes into contact with the bacterial biofilm, mixing Y. pestis into noninfected blood. The fleas do not swallow the blood and thus release Y. pestis-infected blood at the biting point, sometimes infecting the host. The vectorial capacity of blocked fleas is much higher than that experienced during EPT. For example, blocked X. cheopis bites result in 25 to 50% transmission compared with the 0 to 10% probability of transmission during EPT. Flea proventriculus blockade allowing Y. pestis transmission is believed to involve approximately 25 genes (119). The factors that reportedly influence blocking and transmission efficiency include low temperature (139–142), seasonality (143, 144), the flea species involved and their proventricular morphology (93), and feeding frequency (131). These data indicate that the blocking mechanism is more efficient than EPT for the transmission of Y. pestis and probably contributes to the cycle of plague transmission (119). The interzoonotic persistence of Y. pestis may rely on the tissue sequestration of Y. pestis in plague-resistant hosts (depending on the plague focus, geography, and climate) (8, 145); the subtle balance among host susceptibility, bacteremia, and fleas (93, 104, 146); and the long-term carriage of Y. pestis by unblocked fleas during the hibernation or renewal of their hosts (depending on the flea stage [147], temperature [139–142], and sex [148] and the host population affecting the dynamics of the plague) (130). Interplay among all of these factors could occur in a particular space and season.
Transmission of Y. pestis to human populations.
Plague is a zoonotic infection that can be contracted through direct contact with animals, including contact with animal carcasses, animal bites, and the consumption of animal meat and its derivatives, and through indirect contact, mainly mediated by animal ectoparasite contact (9, 40, 41, 99). Plague transmission has been reported after handling the carcasses of mountain lions (84, 149), wild coyotes (150), camels (151–153), rats (27), goats (153), marmots (154), Tibetan sheep (155), guinea pigs (156), rabbits (157), and dogs (158). In some cases, carcass manipulation during necropsy may lead to primary pneumonic infection through the inhalation of infectious aerosol droplets (149, 155), to primary bubonic plague following presumed passage through any skin breach caused by butchering carcass (47, 150–153, 155–157), or to intestinal plague (155). However, the precise mechanisms and exact type of skin breach leading to Y. pestis transmission have not been clearly explored, and whether all Y. pestis strains are transmissible through any type of skin breach remains unknown. Animal carcass handling results primarily in bubonic plague, characterized by enlarged lymph nodes around the area of the lymphatic drainage of the portal of entry and local skin lesions, which have been poorly described under these conditions (47, 150–153, 155–157). In the Chinese plague focus on the Qinghai-Tibet Plateau, plague cases were traced to the slaughtering or skinning of diseased Tibetan sheep and exhibited a high mortality rate of 60%. Genome sequence data indicated an epidemiological chain from the local marmot Marmota himalayana to sheep and, then, to humans. Interanimal transmission from marmots to sheep was hypothesized to take place through marmot ectoparasites (155). In Tibetan sheep, seropositivity for the F1 antigen was detected in 5/7 provinces on the Qinghai-Tibet Plateau, with a prevalence ranging from 0.33% to 5.2% (159). Dogs and cats are occasional sources of transmission of pneumonic, bubonic, and septicemic plague to their owners after the ingestion of Y. pestis-infected marmots (160), the sniffing of a dead prairie dog (161), contact with dead chipmunks, squirrels, wood rats, and their fleas (41, 162–164), or contact with unknown environmental sources (158, 165, 166). Animal bite-transmitted plague has been reported in Gunnison’s prairie dog in New Mexico (167). The consumption of Y. pestis-contaminated animal meat and products is a reemerging form of zoonotic plague transmission. Y. pestis-contaminated products may include urine, which is recommended as a remedy in the Islamic world (168), dairy products, meat, and liver from goats, dromedaries, sheep, and guinea pigs (151–153, 155, 156). Liver and meat cooked at temperatures of ≤68.3°C might represent a risk for Y. pestis transmission, as it has been experimentally documented that Y. pestis is inactive above this temperature in meat (169). The consumption of uncooked or insufficiently cooked Y. pestis-contaminated food may result in a rare form of pharyngeal and meningeal plague, as reported in Maghreb and Asian areas (Table 1) (151–153). The classic pattern of transmission of Y. pestis from rodent populations, such as rat populations, to humans through ectoparasites, such as rat fleas, was established as dogma after the work of Paul-Louis Simond in 1898 in India (125) during the third pandemic wave. The zoonotic transmission of plague from wild animals and fleas leads to sporadic cases and limited outbreaks of plague; it may not by itself explain large epidemics. Furthermore, this transmission route has not resumed during the current epidemiology of the plague observed in the United States and Maghreb (Table 1). The current epidemiological cycle of plague most often involves an animal reservoir (rodents, mainly R. rattus) and a person infected by the inoculation of the bacterium via the bite of a flea that previously fed on an infected animal. This model, which excludes all human-to-human transmission (Fig. 3 and 4) (and thus implies a relatively slow spread of the disease), appears to be incompatible with the high rate of the territorial expansion of the Black Death recovered from historical sources (on the order of 1.5 to 6 km/day) (170). The spread of this epidemic to northern Europe, where the black rat was absent in the Middle Ages, also undermines this model (171–176). Finally, while it is agreed that the “eastern” rat flea (X. cheopis) has been the main vector of plague epidemics since the end of the 19th century, its role in spreading the Black Death is currently disputed. Because this species is of tropical origin, it would be difficult for it to acclimatize to the European climate, in accord with the absence of fossil discoveries of X. cheopis in Europe, despite its highly resistant exoskeleton, whereas remains of Pulex irritans have been discovered at these latitudes (177).
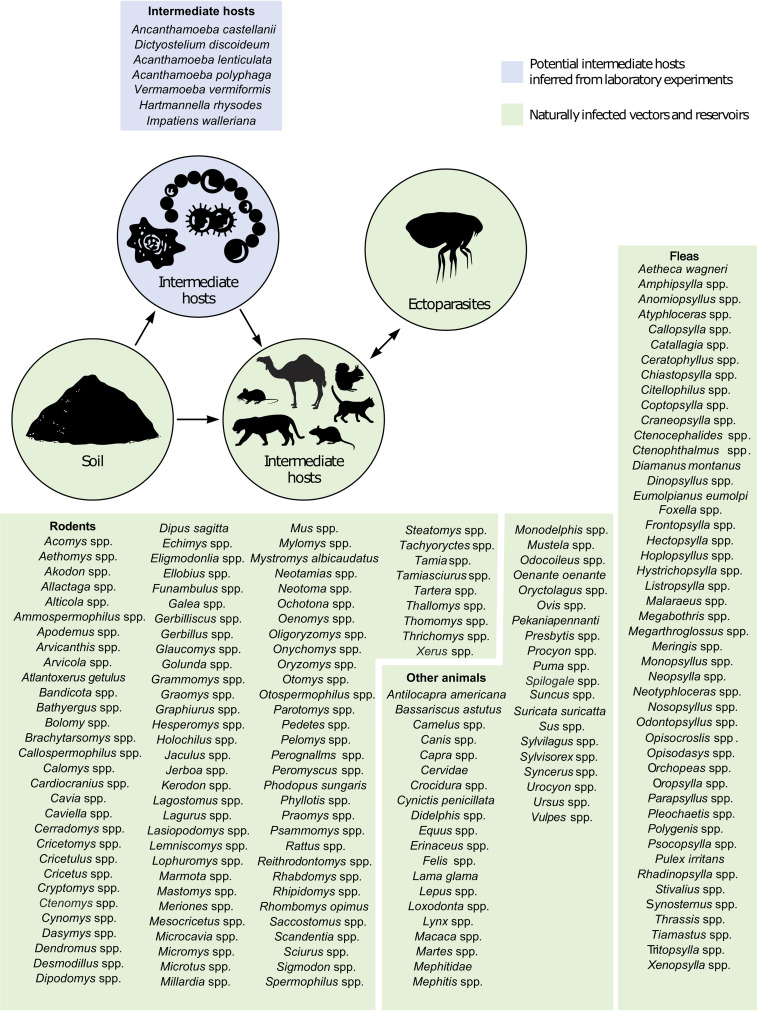
FIG 3.
Scheme of the natural epidemiology of plague representing all documented natural and animal reservoirs, intermediate reservoirs, sources of infection, and vectors for humans described in the literature. Green represents field isolation and laboratory-confirmed sources. Blue represents potential sources/reservoirs inferred from laboratory experiments.
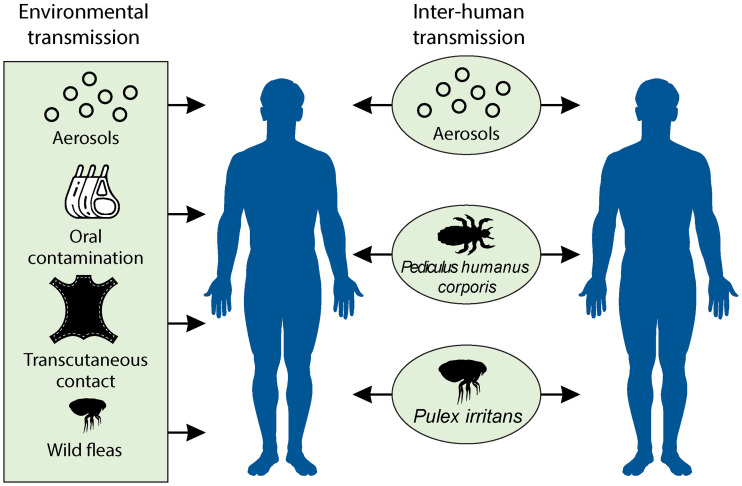
FIG 4.
Different routes of interhuman transmission and human infection from plague sources (as described in Fig. 3). Green represents field observations of confirmed plague sources, such as aerosol transmission, the consumption of raw or poorly cooked meat, transcutaneous contamination by carcass skinning, and nonhuman flea bites. Once humans are infected, effective interhuman transmission can occur through aerosols (in the case of pulmonary plague) and human ectoparasites, such as body and head lice and human fleas (P. irritans).
(i) Aerosol transmission.
Once Y. pestis has been introduced into a human population, it can be transmitted from one pneumonic plague patient to other individuals via droplet transmission. The Manchurian plague episode of 1910 to 1911 is the classic example cited to illustrate the interhuman droplet transmission of Y. pestis (178–180). The major limitation of the interpretation of more recent episodes attributed to pneumonic plague is the lack of the appropriate documentation of patients. For example, the 1924-1925 Los Angeles plague outbreak (8, 181) was thought to be pneumonic and highly contagious, but quarantine measures were ineffective because the majority of the patients diagnosed suffered from a secondary pulmonary form consecutive to a bubonic form caused by rodents and wild fleas (182). The recent investigation of a pneumonic plague outbreak in Madagascar led to the isolation of Y. pestis from two patients and the seroconversion of two additional patients, resulting in four microbiologically diagnosed cases from 14 suspected cases (28.7%) (183). Therefore, there has been an overestimation of the droplet transmissibility of pneumonic plague, which seems to be an effective route of transmission only in the final stage of the disease, when the patient can no longer move, and only through very close contact (≤1 m; based on experimental data) (182) for a prolonged period of time (182, 184). In Uganda, a precise investigation indicated that two index patients transmitted Y. pestis to only one caregiver each and not to 23 additional untreated close contacts (184). In China, an investigation indicated that three index patients exposed 214 contacts over a period of 3 to 13 days. All contacts were quarantined, and no secondary cases were reported (166). Furthermore, it was estimated from eight documented pneumonic outbreaks that a pneumonic plague patient can infect an average of 1.3 other persons (185). The careful analysis of documented pneumonic plague clusters indicates that the transmission of Y. pestis via respiratory droplets requires face-to-face exposure to a coughing patient, as can occur during funerals via close contact with coughing people who may have been exposed to the pathogen while visiting or attending plague victims before they died (58). The droplet transmission of plague by pneumonic patients remains very difficult to evaluate, but field observations and mathematical models indicate that the rate of transmission is very low and cannot sustain a large epidemic among the human population (186).
(ii) Human ectoparasites.
(a) Human lice. In 1909, during an outbreak of epidemic typhus in Tunis, Charles Nicolle and his team discovered a role of lice in the spread and transmission of the disease. Through his observations, he also demonstrated that the clothes of patients suffering from typhus could infect other people through aerosol transmissions (187, 188). This observation made 110 years ago remains crucial to this day for understanding the vector role of lice. The human transmission of Y. pestis by lice remains a controversial issue, even though several lines of evidence support this route of interhuman transmission of Y. pestis by the human louse Pediculus humanus corporis (186, 189). This hypothesis is supported by the paleomicrobiological observation in second-pandemic plague victims of coinfection with Y. pestis and Bartonella quintana, whose main vector is human body lice (190, 191), and the recent observation of Y. pestis in body lice collected from people living in areas of plague foci in the Congo (192); additionally, the codetection of B. quintana and Y. pestis in head and body lice (193) and experimental data that clearly demonstrate the capacity of body lice to transmit Y. pestis have been reported (194–198). Such transmission of Y. pestis was observed by G. Blanc and M. Baltazard in a cluster of bubonic plague in households in Morocco during World War II (195). These authors demonstrated that a body louse could be infected when feeding on a septicemic patient and then remain alive for 7 days while producing infectious feces and could, thus, transmit plague. Spontaneous infection was also demonstrated in Pediculus humanus capitis in 1903 by Herzog and in 1916 by de Raadt (194). When there is a plague outbreak involving an infested louse population, plague-infected lice have always been identified in epidemic situations when they are looked for. All reported evidence indicates that the louse is a unique vector that can be infected by deadly pathogens, including Rickettsia prowazekii, B. quintana, and Borrelia recurrentis. The only conditions required for lice to transmit any pathogen, including Y. pestis, are (i) that the pathogen ingested with blood remains viable in the digestive tract and the feces and (ii) that repeated louse bites provoke a local allergic response, inducing itching. The skin lesions that develop following self-scratching allow the penetration of pathogens present in the feces into the broken skin (Fig. 5A) (199). Indeed, the louse often defecates while feeding on the skin, and the distance between the bite point and the infected feces that are deposited is <4 mm (Fig. 5B). As seen in Fig. 5A, it is clear that such skin lesions in heavily louse-infected patients make it possible for them to be infected by pathogens. In fact, similar to all other louse-transmitted diseases, Y. pestis is introduced by autoinoculation by scratching the skin where the lice defecate. In contrast to the situation in fleas, mosquitoes, or ticks, the vectorial capacity of lice does not depend on a complex cycle shaped by genetic markers leading to the inoculation of the bacteria through saliva during blood meals. In lice, viable bacteria swallowed with the blood will remain intact and infectious in louse feces. Based on these data, human lice could be an effective vector of plague because of their continuous presence on the human body or clothes (200). This hypothesis is supported by an animal model demonstrating the transmissibility of the bacillus by body lice on rabbits (196, 198), the observation of small family plague outbreaks related to body lice in Morocco in the 1940s (195), and the current detection of Y. pestis in head and body lice in the Congo (192, 193). However, the presence of Y. pestis in ancient human ectoparasites has never been tested. Nevertheless, a recent model of second-pandemic mortality in nine European localities indicated that the human ectoparasite model (including P. humanus and P. irritans) fit the historical data better than the rat ectoparasite model and the pneumonic plague model at seven of the nine localities (186). Indeed, massive plague outbreaks (25 to 50% of the human population affected) can occur in the event of massive infestation of humans by body lice causing widespread pediculosis, increasing the chance of Y. pestis entry into the blood (Fig. 5).
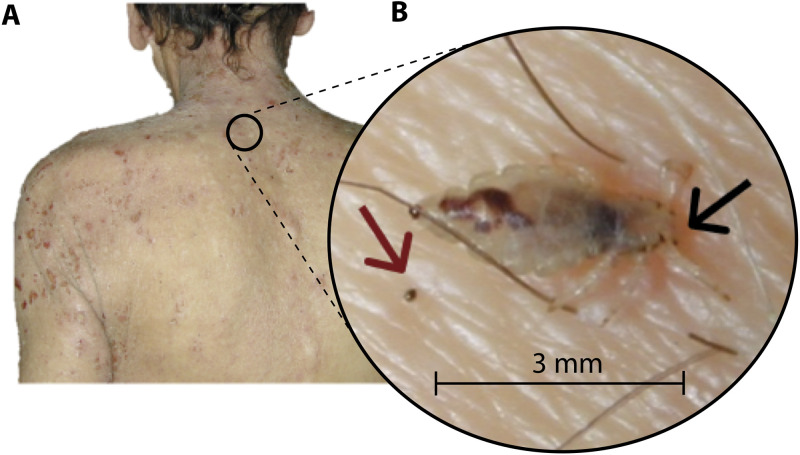
FIG 5.
Example of pediculosis, showing a body louse defecating on skin while feeding. (A) Photograph of a severe pediculosis affecting the back, arms, and neck found on a homeless person in Marseille. (B) Photograph of a Pediculus humanus corporis collected from a homeless person’s skin in Marseille. In this photograph, we see a body louse defecating (body louse feces are indicated by a red arrow) while taking a blood meal (the biting point is indicated by a black arrow). The body louse feces are deposited approximately 3 mm from the biting point. This proximity greatly increases the chance of the penetration of feces inside the broken skin (biting point) during scratching. (Both photos courtesy of Philippe Brouqui, reproduced with permission.)
(b) Human fleas. P. irritans, commonly known as the “human flea,” has been present in Eurasian human populations since the fourth millennium BC and has been recorded at more than 220 archeological sites dating from the Neolithic to the postmedieval period (177). Since the discovery of the rat flea (R. rattus-X. cheopis) model in 1898 by Paul-Louis Simond (27), scientists have hypothesized that plague is always transmitted from rodents to humans. Therefore, in the early 20th century, studies focused mainly on rodent ectoparasites, and research on the role of (nearly) strictly human fleas (9) in the spread of plague, have long been neglected (133). However, the Indian commission reported the collection of 85 P. irritans specimens in the houses of plague victims and the identification of only one infected flea (133). In 1904, Verjbitski succeeded in infecting P. irritans and showed that a batch of 10 fleas could transmit plague to a rat (194). During the Moroccan epidemic of Aït Imour in 1940, the French scientists Blanc and Baltazard (437) studied the role of P. irritans (after having demonstrated the transmission ability of body lice) to explain part of the family plague cluster that they observed in this area (in 1932, a French scientist noted that the number of human fleas in Moroccan dwellings was extremely high, suggesting that fleas might have played a role in the interhuman transmission of plague [201]). Their results showed the presence of P. irritans in the houses of plague victims during the epidemic episode. This observation was later confirmed by M. Baltazard in Turkey, Iraq, Syria, Iran, and Kurdistan, where rats were not found, while P. irritans was found in the houses and tents of nomads, and Pediculus humanus corporis was found in clothes (197). Experiments conducted in 1940 with the fleas harvested in Moroccan houses showed the ability of P. irritans to conserve the plague bacillus in its body for at least 21 days (after feeding on last-stage septicemic plague victims) and the contamination of flea feces with virulent Y. pestis for at least 5 days under natural conditions. Concerning the transmission of plague by flea bites, three guinea pigs bitten twice daily by 600 P. irritans fleas developed typical plague lesions, such as carbuncles and buboes, and died from plague (133). Nevertheless, the P. irritans blocking capacity was incredibly low (119), and its ability to transmit plague via EPT is almost nonexistent. Indeed, only 3 of 38 EPT experiments involving 20 fleas led to a host infection (119); however, as demonstrated by Blanc and Baltazard, the fleas can transmit plague via EPT under extreme conditions (600 fleas fed twice per day on a single guinea pig). However, P. irritans was found to be spontaneously infected during plague outbreaks (93, 126, 133, 202). Although the individual flea transmission rate remained low throughout EPT (0 to 10%), the percentage of rodents infected (rats, guinea pigs, and squirrels) by a group of 10 to 100 fleas ranged between 10 and 100% (135). While some authors argue that early-phase/mass transmission might explain the rapid spread and great mortality during epizootic episodes in populations of species such as prairie dogs (93, 130, 203), other scholars emphasize that given the mass transmission ability of P. irritans and the alleged absence of X. cheopis, P. irritans might have been among the most effective interhuman vectors during the Black Death (130, 204, 205). Accordingly, a debated epidemiological study suggested that P. irritans and P. humanus corporis may mediate interhuman transmission, explaining the ancient outbreak dynamics to some extent (194, 206). Although the vectorial capacity of P. irritans associated with biting seems to be extremely low, human infection may follow the introduction of infected feces at skin breaches, as reported for lice (196). Indeed, it has been shown that P. irritans digests its blood meal rapidly and defecates large amounts of feces containing virulent Y. pestis shortly after feeding to clear itself of infection (119). Human ectoparasite transmission via infected feces is currently the most parsimonious hypothesis for explaining ancient and modern interhuman transmission of plague.
Clinical Aspects
The clinical characteristics of plague partially depend on the route of contamination. People of all ages and both sexes are susceptible to plague, although plague cases have been reported in children, with a low predominance of males, over the last several decades (108). Genetic susceptibility or resistance to plague remains a controversial issue. Observations have been reported regarding whether the protective role of the CCR5-delta32 mutation, which confers resistance to HIV infection, also confers resistance to plague (207–209). The most recent studies identified the FPR1R190W allele (109) or pyrin variants (210) as possible candidates implicated in human plague resistance. As the flea-borne transmission of Y. pestis is its most common route of transmission worldwide, bubonic plague is the most frequent clinical form of plague, developing 2 to 10 days after inoculation with Y. pestis (108). Intriguingly for a vector-borne infection, skin lesions at the portal of entry are not well described and are seldom mentioned in recent reports, whereas skin lesions described as carbuncles, which were the cardinal sign of the infection during the second historical pandemic and were sporadically reported at the beginning of the third pandemic, are no longer being reported in the 21st century (27, 205, 211). According to Simond, the carbuncle is a result of necrosis following the development of a phlycten. A phlycten occurs at the inoculation point of Y. pestis via an ectoparasite bite, and the carbuncles reported to occur systematically during the second pandemic represent a bodily indicator of the point of entry of Y. pestis (27). Patients suffer from nonspecific signs and symptoms, including chills, fever, myalgias, arthralgias, and weakness (99). Much more evocative of the diagnosis in patients exposed to areas of plague endemicity are enlarged lymph nodes, which are painful, tender, and swollen and are referred to as “buboes,” draining the site of inoculation. Femoral (~31%) and inguinal (~24%) nodes are the most frequent, followed in frequency by axillary (~22%) and cervical (~9%) nodes (80, 99). While the development of any lesion at the site of inoculation is rarely reported, a careful examination of the site may reveal a local skin inflammation papule, pustule, scab, or ulcer (9). The buboes resulting from plague are distinguishable from enlarged lymph nodes due to other causes because of their association with systemic signs of toxemia and rapid onset (212). Moreover, plague is a cause of clustered cases of febrile, enlarged lymph nodes that can be confused with tularemia but lead to an unambiguous diagnosis in a deadly epidemic situation. Bubonic plague rapidly responds usually to appropriate antibiotic therapy (reducing mortality from 60% to 5%) (9), while the lymph nodes remain enlarged and tender for 1 week. If not treated with an effective antibiotic, the patient can become increasingly toxemic and develop a septicemic form of the plague. Septicemic plague can be either primary, in the absence of buboes, or secondary to a bubonic form and is characterized by rapidly progressive, overwhelming toxemia. The patient may present gastrointestinal symptoms, including nausea, vomiting, diarrhea, and abdominal pain, confusing the diagnosis. Septicemic plague is diagnosed from a positive blood culture (213) and may evolve to a pneumonic form. In the absence of rapid supportive therapy combined with effective antibiotic treatment, septicemic plague is fulminant and fatal (mortality range of 30 to 100% according to the WHO). Pneumonic plague is the most rapidly fatal form of plague and is characterized by two clinical phenomena: primary pneumonic plague with an incubation period of 2 to 4 days after contact with a coughing patient, and secondary pneumonic plague occurring after the dissemination of Y. pestis bacteria to the lungs during an episode of primary bubonic or septicemic plague. In primary pneumonic plague, the onset is sudden, including chills, fever, chest pain, cough, dyspnea, and hemoptysis. Without treatment, the case fatality rate approaches 100% but is between 25 and 50% when appropriate treatment is administered within 24 h after the onset of symptoms (59, 214).
Pharyngitis, gastrointestinal, or tonsillar plague is a rare form of plague characterized by anterior cervical lymphadenitis that is diagnosed in patients who consume raw or poorly cooked contaminated meat, such as camel meat (112, 151–153). It can also occur in persons who catch human ectoparasites such as fleas and lice with their teeth (a common practice among indigenous people in Ecuador) (99, 194) or who acquire infection from patients with pneumonia (206). Meningeal plague is an unusual form of plague that follows insufficiently treated bubonic plague, while pleuritis, endophthalmitis, and myocarditis are exceptional forms of plague (99, 215).
Diagnosis of plague.
The laboratory diagnosis of plague remains tedious. Early diagnosis is of major interest to start antibiotic treatment as quickly as possible and prevent severe complications leading to death. Its diagnosis relies on the isolation and culture of Y. pestis and the detection of Y. pestis-specific biomolecules from clinical samples. Many types of clinical samples can be used for Y. pestis diagnosis, including bubo aspirates, respiratory tract samples (i.e., sputum), blood, pharyngeal swabs, and urine. Sample quality is important, and the sampling procedure must be adapted to the suspected clinical form. For example, in the case of suspected pneumonic plague, deep respiratory secretions are required for diagnosis (33) due to tropism toward the lower respiratory tract, and viscous samples should be liquefied and homogenized (216). The gold standard is based on the isolation and detection of Y. pestis by culture from clinical samples. Clinical samples could be handled in a biosafety level 2 laboratory, but it is mandatory to perform the isolation, culture, and manipulation of Y. pestis in a biosafety level 3 laboratory, in line with national regulations. The easily culturable bacteria can grow under routine culture in solid or liquid media (brain heart infusion broth, sheep blood agar, or MacConkey agar) after 24 to 72 h of incubation at 28 to 37°C (28°C is the optimum temperature) under aerobic conditions. Selective solid agar medium supplemented with cefsulodin-irgasan-novobiocin (CIN medium) is recommended by the WHO to limit the growth of contaminant bacteria from respiratory tract, pharyngeal, and sputum samples. CIN medium can be improved with the addition of irgasan, cholate salts, crystal violet, and nystatin (BIN medium) for the isolation of Y. pestis from complex samples, such as respiratory tract, pharyngeal, or environmental samples (217). Previously commonly used automated identification systems may fail to identify Y. pestis colonies (218), and the first-line identification of colonies can be achieved by using matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) (219, 220), PCR, or phage lysis (221). To improve sensitivity, molecular assays have been developed as standard PCR assays, allowing Y. pestis DNA detection within 3 to 4 h or in less than 2 h for real-time PCR (33). Four main genes are targeted: the pla, caf1, inv, and yopM genes (108). The pla gene, encoding a plasminogen activator regarded as a major virulence factor of Y. pestis (222, 223), is present at 150 to 200 copies per bacterium (17), resulting in a high sensitivity of detection (100 CFU/ml in sputum) (224). However, the pla gene has also been detected in Citrobacter koseri (225), Escherichia coli (226), and rats (227), thus limiting the specificity of this detection method. The yopM gene, located on the Yersinia pYV/pCD1 virulence plasmid, which is present in approximately four copies per bacterium (33), shares 99.84% similarity with sequences from Y. pseudotuberculosis and 93.91% similarity with sequences from Y. enterocolitica strains. The chromosomal inv gene is also present in Y. pseudotuberculosis, but an insertion has increased the size of this gene in Y. pestis (228) (1,100 bp versus 400 bp for Y. pseudotuberculosis). The caf1 gene is considered to be specific to Y. pestis and is present in approximately one or two copies per bacterium (17). PCR can be multiplexed; for example, in the case of suspected bubonic plague, a multiplex quantitative PCR (qPCR) assay targeting the pla and caf1 genes has been validated only on bubo aspirates (229). In cases of positive results for pla and caf1, the presence of Y. pestis can be considered to be positively identified (33), but in the case of discordant or uncertain results, it is recommended to perform another multiplex qPCR assay, including the 1,100-bp inv gene (33, 230). Moreover, a portable real-time PCR instrument has been developed (231) for the triplex detection of Y. pestis, Bacillus anthracis, and Francisella tularensis. This instrument includes an embedded Y. pestis assay based solely on the detection of the pla gene but presents the limitations mentioned above. Furthermore, several commercially available assays incorporate Y. pestis as a target in a multiplex format in association with B. anthracis and F. tularensis (232) or target 17 pathogens, including Y. pestis (233). Both types of assays are reported to be usable at the point of care. During field trials, plague diagnosis can be achieved in approximately 90 min. Real-time PCR reagents with specific primers and probes targeting the nonspecific 3a sequence (234) and caf1 sequence that can be stored at room temperature have been developed to facilitate diagnosis in remote locations (235). Alternatively, the indirect detection of live Y. pestis cells can be achieved by the qPCR detection of Y. pestis-specific bacteriophages, such as ϕA1122 and L-413C (236). This method has been established with artificial clinical samples and presents a low sensitivity of 103 CFU/ml for ϕA1122 and 105 CFU/ml for L-413C and a high specificity for L-413C, while ϕA1122 can be detected in some Y. pseudotuberculosis strains. The detection of Y. pestis DNA can be achieved using the loop-mediated isothermal amplification method, but this method has been validated using only mock-infected animal samples and needs to be approved for sputum samples (237).
The development of rapid diagnostic tests (RDTs) allows F1 antigen detection in 15 min at concentrations as low as 0.5 ng/ml, which is of major interest in a deadly epidemic situation (238). This assay has been validated for the diagnosis of bubonic plague with both a specificity and a sensitivity of 100% in clinical samples and Y. pestis strains but needs to be evaluated for pneumonic plague because of false-positive and false-negative results observed in sticky sputum or saliva (33, 238). RDTs are easy to use, provide rapid results at remote locations, and are 5 to 50 times less expensive than other molecular tests (238). RDTs are available at the Pasteur Institute of Madagascar but have not been commercialized. Hsu et al. developed another specific strip assay that has been favorably evaluated in clinical and mouse samples, with a minimum concentration of 4 ng/ml for the F1 protein and 103 CFU/ml for Y. pestis; to the best of our knowledge, this assay is not commercially available (239). Upconverting phosphor technology-based immunochromatographic assay (UPT-ICA) has been developed for the quantitative detection of Y. pestis in 15 min with a high specificity and an effective minimum concentration of 104 CFU/ml (100 CFU/test) (240, 241). This test can be applied in the field to various sample types, including blood, fresh or decomposed viscera (useful for zoonotic surveillance), and powdered material (useful for bioterrorism investigation), and can tolerate a wide pH range (pH 2 to 12) and high viscosity (unlike RDTs). Serological diagnoses are also commercially available, such as F1 capsular antigen capture ELISA, requiring a minimum concentration of 4 ng/ml and exhibiting a specificity of approximately 98% in sera and a sensitivity of 90.1% in serum and 100% in bubo aspirates (242). The results of these assays are similar for bubo aspirates but are more sensitive for serum than those reported by Chanteau el al. (sensitivity of 100% in bubo aspirates, 52% in serum, and 58% in urine specimens) for the F1 antigen test developed by the Naval Medical Research Institute (243). The gold standard diagnosis of plague remains difficult in countries of endemicity because infrastructure, resources, and logistics are often limited in remote areas. Furthermore, its diagnosis is time-consuming and is impossible outside biosafety level 3 laboratories. RDTs have become an effective (showing the same sensitivity and specificity as other molecular tools), time-saving, economical alternative to molecular diagnostic tests (PCR) adapted for field diagnosis at the point of care and can be performed by nontechnically trained personnel (213). Nevertheless, the diagnosis of plague is often uncertain, depending on the sample quality, invasion stage, and/or available technical support (244); for example, only 23% of patients with suspected pneumonic plague and 35% of patients with suspected bubonic plague received a laboratory-confirmed diagnosis (by rapid antigen detection, PCR, or culture) during the 2017 plague epidemic in Madagascar (59). This situation of underdocumentation is clearly detrimental to patients and to people in contact with them, as the epidemiological and clinical aspects of epidemics may not achieve a reasonable positive predictive value for the plague and may obscure other infectious diseases, such as leptospirosis (245).
Treatment of plague.
At the beginning of the microbiological era, Alexandre Yersin built upon his discovery of Y. pestis and became the first to develop an antiserum for the treatment of plague patients. He developed the antiserum in collaboration with Calmette, Roux, and Borrel at the Pasteur Institute in Paris in 1895. The antiserum was first used to treat 23 Chinese plague patients in 1896, and only two of these patients died, lowering the mortality rate to 9% (246). These encouraging results led other physicians to use this treatment, but the lack of standardization among the methods they applied, such as the great diversity of the animals used for antiserum production and the variation in the doses given, led to unconvincing results. However, according to a report by Meyer et al. (247) based on the use of antiserum over a decade in Asia, Africa, and South America, this treatment is predicted to have reduced mortality from 82% without treatment to 32% following the injection of the serum. Antiserum can be coupled with sulfapyridine to achieve better efficiency; for example, in Egypt in 1940, only 12 of 69 plague patients treated in this manner died (248). Treatment with antisera, which confers only short-term protection and causes severe side effects, was progressively replaced by the development of new molecules, such as sulfonamides (preceded a year earlier by the intramuscular injection of sulfonamide prontosil with encouraging results [249] and used for the first time in East Africa and India in 1938 [250, 251]). In Madagascar in 1940, Girard reported the curing of 28 of 37 bubonic plague patients (mortality rate, 24%) by the injection of sulfapyridine, whereas all treated pneumonic plague patients died (252). In fact, this treatment proved effective only when administered within 3 days of the onset of symptoms, even in the case of pneumonic plague, as demonstrated by the recovery of three such patients in Madagascar in 1947 (253). At the beginning of the 1940s, Wagle et al. demonstrated the superiority of sulfathiazole over classic antiserum in patients suffering from all forms of plague in India (254). In 1947, the first case of pulmonary plague was cured with streptomycin (255), which later cured five infected patients in Argentina (249). This new antibiotic was very effective and was able to cure patients in whom sulfonamide failed (256). Chloramphenicol and oxytetracycline were used in the early 1950s with the same efficiency as streptomycin (249). Cumulative evidence led to an updating of the recommendations regarding the antibiotic treatment of suspected and confirmed cases of plague. Streptomycin, which is the historical reference antibiotic and is still used in Madagascar, is no longer available in most countries, and it has been shown that gentamicin alone or in combination with tetracycline is an acceptable substitute (257). Gentamicin and doxycycline were shown to be equivalent in curing patients, except during the terminal stage (258). According to WHO and Centers for Disease Control and Prevention (CDC) recommendations, antibiotics that are commonly used against Enterobacteriaceae, such as streptomycin, gentamicin, levofloxacin, ciprofloxacin, doxycycline, moxifloxacin, and chloramphenicol, are proven to be effective against plague if given promptly (257–261). Antibiotic treatments should be continued for 10 to 14 days, and improvement is clinically evident 2 to 3 days after the initiation of antibiotic treatment, although fever may persist for several more days. Supportive therapy should be undertaken in the case of septic shock and septicemic plague. Very few antibiotic-resistant Y. pestis isolates have been described. In Madagascar, one isolate was reported to be resistant to eight antimicrobial agents, including those recommended for the treatment (streptomycin, chloramphenicol, and tetracycline) and prophylaxis (sulfonamide and tetracycline) of plague, as well as ampicillin, kanamycin, and spectinomycin (262). Streptomycin resistance may be plasmid transferable (263). A second isolate was reported to be resistant to streptomycin (264). In these strains, all resistance genes were carried by a conjugative plasmid consisting of approximately 150-kb and 40-kb sequences. Horizontal gene transfer in fleas may be the source of the antibiotic-resistant Y. pestis strains isolated from plague patients in Madagascar (263). Further, ampicillin- or tetracycline-resistant isolates have been generated in fleas and rats in Madagascar (264), whereas the 150-kb plasmid backbone has been shown to be broadly disseminated among multidrug-resistant zoonotic pathogens associated with agriculture (265). However, none of the 50 Y. pestis isolates generated during the 2017 epidemic in Madagascar were antibiotic resistant (59). Experimental data indicate that Y. pestis strains that are resistant to fluoroquinolones and rifampin can be easily selected in the laboratory, with no medical relevance thus far (266).
Prevention of plague.
The measures for primary prevention prior to potential exposure to Y. pestis may include the avoidance of areas with known epizootic plague (42, 51, 63, 65, 66, 71, 96, 99, 267–290). In areas where plague is endemic, it is good practice to avoid contact with obviously sick or dead animals and to report such animals to the health department. Regarding potentially contaminated ectoparasites, people should avoid exposure to fleas from diseased rodents, dress in protective clothing, use repellents to avoid exposure to ectoparasites when outdoors, and apply insect repellent containing diethyltoluamide to the legs and ankles. Additionally, it is good practice to apply repellents and insecticides to clothes and outer bedding, to wear gloves and masks when handling dead animals and carcasses, and to cook meat on an open-flame grill or a clamshell-type electric grill (169). In 1946, killed whole-cell vaccines were developed and preventively administered to soldiers, but these vaccines conferred only short-term protection and did not protect against primary pulmonary forms of the disease (291). Furthermore, a live vaccine developed from an attenuated Y. pestis EV76 strain was demonstrated to protect against bubonic and pulmonary plague but potentially caused major side effects, such as general malaise, severe headaches, and pyrexia (292). Several vaccines, including a live attenuated Y. pestis EV76 strain, formalin-inactivated whole-cell plague, and heat-killed whole-cell plague (F1 fraction), can be administered via aerosol (EV76), subcutaneously (EV76, heat-killed CSL vaccine; CSL Ltd., Victoria Australia) or intramuscularly (formalin-inactivated Greer vaccine; Greer Laboratories Inc., North Carolina). F1 fraction vaccines appear to show low efficacy (<60%) and cause side effects in 35% of vaccinees (293). There is not enough evidence to evaluate the effectiveness of any plague vaccine or the relative effectiveness of the vaccines and their tolerability. Circumstantial data from observational studies suggest that killed types of vaccines may be more effective against bubonic plague and have fewer adverse effects than attenuated types of vaccines. There is no evidence regarding the potential of vaccines to help control plague outbreaks (293). Furthermore, there is currently no vaccine recommended by the WHO or the CDC for preventing plague. Some vaccines are under development but will not be available in the immediate future (294). Secondary prevention in the case of potential exposure to Y. pestis relies on the administration of tetracycline or trimethoprim-sulfamethoxazole to people who are bitten by fleas during a local outbreak, are exposed to tissues or fluids from a plague-infected animal, live in households with a bubonic plague patient (since they may also be exposed to infected fleas), or are in close contact with a person or pet with suspected plague pneumonia (257). In a mass casualty situation, oral therapy with doxycycline, tetracycline, or ciprofloxacin is recommended (261, 295), and the use of the last option has been supported by an animal model, although a recent evaluation indicated that doxycycline should be considered a first-line antibiotic in the management of bioterrorism agents, including Y. pestis (296). The prevention of human-to-human transmission from patients with pneumonic plague must be achieved by maintaining confirmed and suspected cases under droplet precaution and negative-pressure isolation when available (297) for at least 48 days after the initiation of antibiotic treatment (298). Additionally, wearing gowns, gloves, surgical masks, and eye protection is strongly recommended to stop the spread of the disease (299).
Plague control is partly based on the active surveillance of sentinel animals such as wild carnivores, which generally produce antibodies against Y. pestis without suffering mortality (with the exception of some felids) (9). Wild carnivores become infected by eating infected prey (rodents) and are therefore a good indicator of infection among rodent populations; thus, testing one carnivore is equal to the testing of hundreds of rodents (300). In areas of epizootics, it is mandatory to eliminate food and shelter for rodents around homes, workplaces, and certain recreation areas, such as picnic sites or camping grounds, where people congregate. Bushes, rock piles, junk, and food sources, including pet food, should be removed. Pets (cats and dogs) should be treated for fleas regularly (9). Similarly, the control of human plague outbreaks relies mainly on the rapid confirmation of the diagnosis and treatment of confirmed and suspected cases. Killing rodent fleas and rodents using appropriate licensed insecticides and rodenticides or rodent traps is mandatory during plague outbreaks according to WHO recommendations. Nevertheless, during an epizootic situation involving endangered species, flea control must be prioritized (9). New control strategies are essentially reliant on monitoring (301, 302) or on the modeling of susceptible rodent populations combined with climate variation and environmental factors to predictively evaluate epizootic risk and human-related cases (9, 303, 304). The active long-term surveillance of plague foci coupled with the rapid response of health care professionals during epizootics helps to successfully reduce human cases.
ANCIENT PLAGUE
History of Plague
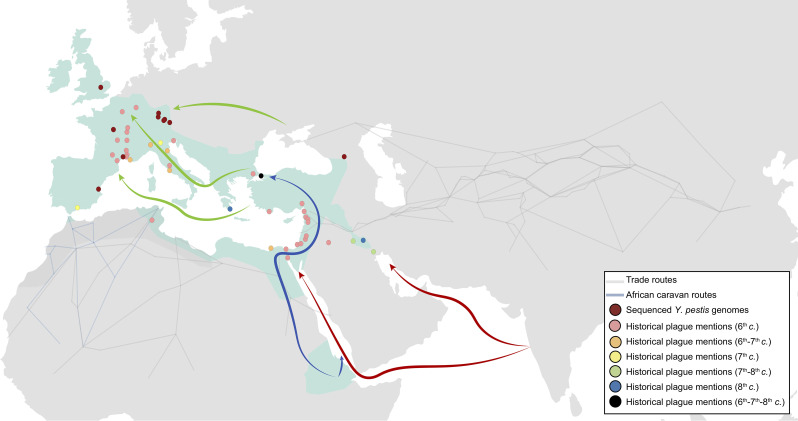
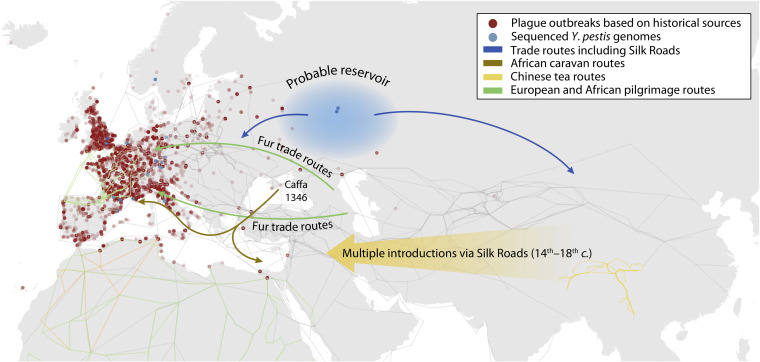
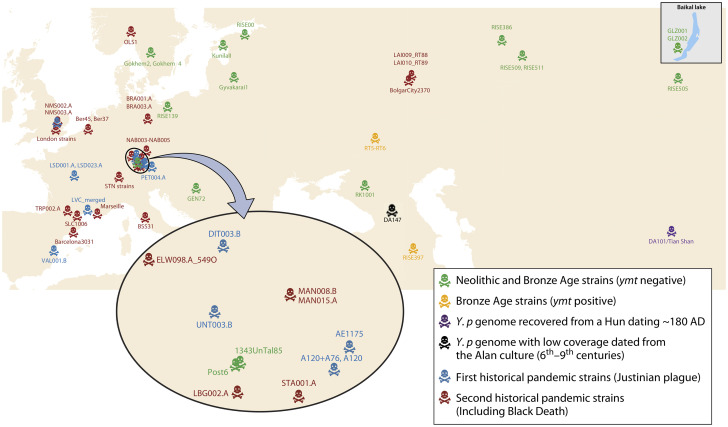
Historical sources.
Several historical epidemics referred to as “plague” by ancient authors have been related in historical sources, based largely on the accounts of direct witnesses of epidemic episodes. The information from these accounts is of great value to scientists, who can extract valuable data from these documents to broaden the scope of the knowledge of plague, including its epidemiology. Sources of modern knowledge about plague include preserved historical archives, diaries of witnesses and contemporaries, preserved historical paintings, related artifacts, and biological archives, including human remains (most often the skeletons of the victims of these past epidemics). Furthermore, plague is the subject of a wealth of artistic output in Europe. Plague epidemics were the archetype disaster, threatening all of humankind and leading to a profusion of representations, attesting to their impact on people’s mentality (305–311). This historical material constitutes a source of knowledge that can be interpreted in a scientific manner after distinguishing between representations of reality (e.g., regarding the localization of buboes and the management of plague-infected corpses) and symbolic and pictorial material (e.g., a living baby feeding from the breast of its dead mother). All of these representations have made deep impressions on the collective imagination of the ancient and contemporary European population.
(i) First pandemic: the Plague of Justinian (541 to 750/767).
This pandemic, which was named after the emperor of the Roman Empire of the Orient, Justinian I (emperor from 527 to 565), started in 541 in the Egyptian port of Pelusium (312, 313) and was initially recorded around the Mediterranean Basin between 541 and 544 during the first wave called the “plague of Justinian,” after which it returned from 558 to 750/767 in 14 to 21 additional waves called the “first pandemic” (1, 8, 314). According to Evagrius Scholasticus, the plague originated in Ethiopia (315), although this assumption remains controversial (316, 317), and an alternative hypothesis has been proposed (see below). The disease exhibited an endemic character, spreading through recurrent epidemic outbreaks over a long period (from the mid-6th to mid-8th centuries) across a wide geographical area (Europe and Mediterranean Basin). Indeed, the Justinian Plague primarily affected banks and ports and then penetrated deep inland via human movements and trade routes and along the Loire and Rhone valleys to Germany (2, 318, 319), England, and Ireland (2) (Fig. 6). Based on contemporary sources, the symptoms that were described (headache, fever, buboes, and rapid death) provide a clinical picture clearly indicating that plague was the etiological agent of this pandemic (314). The historian Procopius described these epidemics as follows: “It generally happened that those whose buboes grew large and suppurated, recovered from the disease, which seemed to spend its violence upon these tumors; while in those whose buboes remained without suppuration, it had an unfavorable termination” (320). According to Gregory of Tours, “Death was sudden. A wound the shape of a serpent would appear on groin or armpit and the man would be so overcome by the poison as to die on the second or third day” (321). The origins of the plague and causes of its decline and temporary eradication over 3 centuries in affected European regions remain unknown (2). The human and social consequences of this pandemic remain controversial. Some authors put forward estimates ranging from 15 to 100 million victims, equivalent to 25% to 60% of the estimated human population (322, 323), and hypothesize severe social and economic consequences that initiated the collapse of classical antiquity (322, 324). However, in a recent study based on historical and archaeological data, it was suggested that the death toll has been greatly overestimated and was probably 0.1% of the estimated human population, suggesting that the Justinian Plague was not significantly influential of demographic, political, and social change (1).
FIG 6.
World map of the first plague pandemic (541 to 750/767). Regions historically affected by plague are represented in green, and regions potentially affected by plague (western part of North Africa) are in dark gray. The biological hypothesis of plague diffusion is represented by green (Eastern European origin) and red (Indian origin) arrows, while the debated historical hypothesis of plague diffusion is represented by a blue arrow (Ethiopian origin). The map was generated in QGIS 3.4. The mapped regions and roads are based on the Digital Atlas of Roman and Medieval Civilizations (DARMC; https://darmc.harvard.edu).
(ii) Second pandemic (1346 to 18th century).
“Consult historians, they are silent; ask physicians, they are stupefied; seek the answer from philosophers, they shrug their shoulders, furrow their brows, and with fingers pressed against their lips, bid you be silent. Will posterity believe these things, when we who have seen it can scarcely believe it, thinking it a dream except that we are awake and see these things with our open eyes, and when we know that what we bemoan is absolutely true, as in a city fully lit by the torches of its funeral we head for home, finding our longed-for security in its emptiness? O happy people of the next generation, who will not know these miseries and most probably will reckon our testimony as a fable!” These few lines from the Florentine poet Petrarch clearly set the scene for the return of plague in the middle of the 14th century (325). The second pandemic likely began at the end of the 1330s in central Asia, probably in present-day Kazakhstan, Russia, or China (3, 326–329). After the alleged first episode of bacteriological warfare in 1346 (330, 331), plague spread via sea routes from the port of Caffa to Constantinople and then to the whole of North Africa and western Europe (Fig. 7). During the 14th century, people testified to this first epidemic assault, and Father Franciscan Michel Platensis of Piazza described it as follows: “In the month of October 1347, arrived from Genoa in the port of Messina, 12 ships having fled the plague that the Lord had sent them as punishment for their sins. They brought a disease so contagious that it was enough to talk to those who were afflicted to be mortally wounded without hope of healing” (332). During the first few months, it was easy to follow the route of the disease and the chronology of the infected cities. The route of dispersion then became more erratic because the plague followed travelers and commodities and stopped at their staging posts. The first attack of the plague, which occurred during the first 7 years of the second pandemic (1346 to 1353), killed between one-quarter and one-third of the human population and is historically referred to as the Black Death (333). The name Black Death does not refer to the color of the corpses of the plague victims. The expression refers to the figurative meaning of the adjective (e.g., lugubrious and appalling). Accordingly, the term Black Death does not date from the 14th century and is completely anachronistic (334). The people of the Middle Ages used the terms “great mortality,” “bumps disease,” and “epidemic” to describe the plague. Furthermore, some places were affected several times by recurrent epidemics, as reported by a chronicler of the city of Orvieto (335): “The first general plague occurred in 1348 and was the strongest.” Then, this author adds, “Second plague, 1363; Third plague, 1374; Fourth plague, 1383; Fifth plague, 1389…Sixth Plague, 1410.” Biraben summarized the endemicity of the disease perfectly as follows: “From that point onward, and until 1670, the plague raged every year in Europe, sometimes in vast territories, sometimes only in certain localities, but without skipping a single annual link in this long and painful chain” (336). From 1347 until at least the middle of the 17th century, the plague became a common fact of life in western European societies. After 1670, the plague became rarer, although some outbreaks still occurred during the 18th century, as recorded in Marseille and Provence between 1720 and 1722 (337), in Messina and its surrounding area in 1743 (338), and in Moscow and the surrounding area in 1771 (339, 340). Plague had considerable consequences for social life. It contributed to ending the Hundred Years’ War between the French and English kingdoms, which disrupted the economic expansion of the continent and affected the workforce (341). It is difficult to pinpoint the terms of the second pandemic. Indeed, Antoine-Jean Gros painted plague-affected soldiers of Bonaparte engaged in the Campaign of Egypt in Jaffa in 1799 (342). The plague was still present in Malta in 1813 (343), in Tunisia between 1818 and 1820 (344), and in Egypt between 1834 and 1835 (345). Finally, the epidemic that struck Constantinople in 1839 (346) is considered the last manifestation of the second pandemic in Europe (332). As reported above, although a period of several centuries separates the first from the second pandemic, the chronological break between the second and third pandemics is much less obvious. Indeed, the first manifestation of the third pandemic occurred in 1772 in Yunnan Province in southwestern China (3, 6, 347), while the plague that was still raging in Europe at that time was attributed to the second pandemic.
FIG 7.
Map of the second pandemic (1346 to 18th century), including the so-called Black Death (1346 to 1353) and pestis secunda (1357 to 1366) episodes. Brown arrows indicate the well-known starting point of the Black Death in 1346 (city of Caffa) and the probable spread of the plague to Europe and Africa via land and maritime routes. Blue arrows indicate the first hypothesis regarding the dynamics of the second pandemic, in which plague was introduced from Eastern Europe to Western Europe before settling in one or several reservoirs and disappearing, followed by reintroduction in Asia giving rise to the third pandemic. The yellow arrow indicates the second hypothesis, indicating that plague was also introduced to Europe from Central Asia by successive waves from the 14th to the 18th centuries. This hypothesis excludes the existence of temporary plague reservoirs in Europe. The plague would have been introduced by successive waves over 4 centuries in Europe, mainly via silk roads and fur roads, by establishing several permanent foci (still existing) along these roads and spreading to Europe along maritime and terrestrial routes. The map was generated in QGIS 3.4. The mapped regions and roads are based on the Digital Atlas of Roman and Medieval Civilizations (DARMC; https://darmc.harvard.edu).
(iii) Third historical pandemic, 1772 to 1945.
The third plague pandemic is likely to have originated from the province of Yunnan in southwestern China, where it was registered as early as 1772 in the city of Dali (348) before becoming endemic in southwestern China in the 1850s and 1860s (8). Thriving because of troop and refugee movements during the rebellion of the Mohammedans (99), the plague reached the city of Canton in March 1894, causing more than 60,000 deaths and rapidly spreading to Hong Kong in May of the same year (10). Between 1899 and 1900, the plague invaded all continents, including Asia (India in 1896 and Japan in 1899), the Middle East (Saudi Arabia and Turkey in 1897) (332, 349), Africa (Madagascar in 1898), Oceania (Brisbane in 1899 and Sydney in 1900), Europe (Lisbon in 1899 and Glasgow in 1900), and North and South America (Brazil, Paraguay, and Honolulu in 1899 and San Francisco in 1900) (8, 332, 350). Plague was thus recorded in more than 100 countries during the ongoing third pandemic (350). The rapid spread of plague along steamboat and rapid train routes worried health authorities in Europe. Because of the memory of previous, devastating epidemics, an emergency meeting of European health authorities took place in Venice in 1897 to attempt to stem and control the possible return of the plague to Europe (332). Despite the reinforced quarantine system, bubonic plague returned to Lisbon in 1899 and Glasgow in 1900, claiming 37 lives (351, 352). Furthermore, the plague reached territories where it had never previously been documented, including South America, the United States, South Africa, and Australia. Following its introduction to these supposedly plague-free regions, Y. pestis established permanent plague foci in all of these regions, with the exception of Australia. It is interesting to compare the outcome of third plague pandemic in the United States and Australia. The plague entered both countries during approximately the same period (San Francisco, California, United States, in 1896 and 1900 [99, 353]; Brisbane and Sydney, Australia, in 1899 and 1900 [354, 355]) via the maritime route. In the United States, the plague remained a disease of harbor towns until the 1930s, and it emerged as an inland endemic infection in the 1950s, affecting populations of local rodents, including prairie dogs, squirrels, and chipmunks, and establishing endemicity in the semiarid southern states (41, 76, 353, 356). The plague entered Australia through Brisbane harbor in 1899 and Sydney 1 year later, where the affected districts were isolated, the inhabitants were evacuated, and more than 100,000 rats were killed to prevent the spread of the disease (354). The reasons why the plague did not become permanently established in Australia after its introduction, in contrast to the situation in the United States, remain unknown and are possibly linked to the inability of Y. pestis to establish permanent plague foci in the Australian soil (6, 76, 356). From 1910 to 1911, a massive presumed pneumonic plague outbreak occurred in Manchuria, causing approximately 60,000 deaths (180). Following this epidemic, the “ten thousand nations” international plague conference in Mukden (now Shenyang) was held from 3 to 28 April 1911 (357) and marked a turning point in the international fight against the plague. At this meeting, international plague scientists (including S. Kitasato) and government delegates discussed disease reservoirs, vectors, transmission, and etiology and preventive measures to be applied internationally in the event of new plague epidemics. The Mukden conference established a system of international cooperation to fight the spread of the plague during a time when steamboats and railroads were emerging (357). In France, the final cases were recorded in Marseille in 1919 to 1920 (358), and in Paris, the final cases were recorded in June 1920 (“Ragman’s plague”) (359). During this episode, a total of 90 cases of bubonic and pulmonary plague resulted in 30 deaths. The first cases were recorded in children after a boat transporting coal from London arrived in northern Paris. Notably, this pulmonary plague episode began at the same time as Spanish influenza, and confusion existed between the two diseases. Genomic analyses indicated that, in most cases, a single introduction was followed by regional dispersion, as recently illustrated in Brazil (360). Following World War II, the last cases of plague in Europe occurred in 1945 in Corsica and Italy and in 1947 in Kaliningrad (332, 351). The third pandemic had already accounted for more than 26 million cases and more than 12 million deaths in India and China alone (8, 99).
(iv) Undocumented historical epidemics.
Regarding ancient periods, it is difficult to recognize the clinical symptoms of plague by reading the very rare and imprecise available sources. Thus, the descriptions of Greek and Chinese authors are very uncertain (361, 362). Other sources may report epidemics originating from an entirely different pathogen under the name “plague.” Sometimes, we can only infer the possible occurrence of an epidemic on the basis of metaphorical texts, which are mostly theological in nature, making it impossible to recognize objective clinical symptoms in the episodes described in the Bible and Koran and in nonreligious texts, which may report plague episodes lacking any current microbiological confirmation. The most ancient record of a putative plague outbreak can be found in the Old Testament in the Book of Samuel, where the plague was interpreted as a divine punishment. God allegedly sent the plague to the Philistine city of Ashdod in retaliation for the theft of the Ark of Covenant (363). In antiquity, the first epidemic is reported by Homer’s Aoidos at the beginning of The Iliad, where he describes an outbreak of plague driven by Apollo against the Greek army of Agamemnon (364). “The Plague of Thebes” described by Sophocles in the tragedy Oedipus Rex was recently reported as being attributable to Brucella abortus (365). The same applies to the “Plague of Athens” reported by Thucydides in 430 to 426 BC (366), which was controversially documented as typhoid fever (367, 368). The final outbreak reported by a Greek historian was the “Plague of Syracuse” in 396 BC among the Carthaginian army during the siege of the city. In this chronicle, Diodorus Siculus, who was not a direct witness of the epidemic, reported that “now the plague first attacked the Libyans, and, as many of them perished, at first they buried the dead, but later, both because of the multitude of corpses and because those who tended the sick were seized by the plague, no one dared approach the suffering … For by reason of the stench of the unburied and the miasma from the marshes, the plague began with a catarrh; then came a swelling in the throat; gradually burning sensations ensued, pains in the sinews of the back, and a heavy feeling in the limbs; then dysentery supervened and pustules upon the whole surface of the body.” According to Diodorus, these symptoms were similar to those endured by the Greeks 30 years earlier during the Plague of Athens (369). The occidental Roman Empire also experienced several large-scale epidemics that were allegedly attributable to plague, but this hypothesis was never formally tested. According to Saez, the tragic event of the “Antonine Plague” during the 2nd century killed approximately 10% of the Roman population and played a substantial role in initiating the decline of the Roman Empire (370). The Greek surgeon Galen witnessed this outbreak, and a critical reading of his writings suggests that smallpox might have been the etiological agent (371, 372). Subsequently, the plague of Cyprian, named after the bishop of Carthage, occurred on the periphery of the Mediterranean Basin from 249 to 270 (373). Although Cyprian left some testimonials about this epidemic, documentation related to this episode is extremely sparse. Nevertheless, coins have been discovered from this period that are inscribed with the words “Apoll Salutari,” indicating the seeking of protection from Apollo, who governs illness, and archaeologists uncovered a Roman mass grave in the ancient city of Thebes dating from the same period (374). Considered together, these observations support the veracity of an epidemic episode, but questions remain as to whether the etiological agent was Y. pestis. In addition to historical documentation, archaeological documentation is currently essential to determine the history of plague and its epidemiology. Recently, several research teams have verified the presence of Y. pestis in the remains of individuals dating from the Neolithic era. These molecular biological studies, which were performed using skeletons discovered in Russia, Sweden, central Europe, and Asia, showed the presence of Y. pestis DNA in individuals aged approximately 4,000 to 5,000 years (375–379). Despite the extreme importance of these recent studies that have identified plague bacilli, there are no data other than genetic data that might help to understand the impacts of these epidemics.
Archaeology of Plague
The interest of archaeologists and biological anthropologists in plague burials is relatively recent. It was only in the mid-1980s to 1990s that the first excavations of the mass graves or funeral complexes of plague victims were carried out (380–384). Since that time, researchers have understood the interest of these sites and these human remains, which constitute true “biological archives.” These “archives” provide information on both the process of death (practices implemented for the care of the deceased during a health crisis) and the dead themselves, including certain parameters that make it possible to characterize the victims (sex, age, health status, etc.). In the years that followed, archaeologists and anthropologists continued to explore burial sites with large numbers of multiple and simultaneous burials. The first sites that attracted attention were associated with the end of the second pandemic (17th and 18th centuries). These efforts notably enabled research projects to connect these mass graves with conserved historical archives and to place the archaeological human populations studied in their social and chrono-cultural context. It is therefore at this type of site (major complexes and epidemics of the modern era) that the first collaborations with paleomicrobiologists were established (385), allowing the formal identification of the pathogenic agent responsible for the Marseille and Provençal epidemic of 1720 to 1722. Since the mid-1990s, research on the burial sites of plague victims has continued to increase in many European countries and in chronological contexts much broader than those initially considered. Today, nearly 100 sites with variable numbers of burials of plague victims have been the subject of cross-discipline studies, bringing together various complementary academic fields (archeology, anthropology, microbiology, and genetics). At the first sites, archeologists thought that the funeral areas related to plague epidemics were composed only of mass graves, within long trenches or large pits. This type of structure is found, for example, in the long parallel trenches of the Abbey of San Salvatore (3, 386), the long trenches and large pits, respectively, at the London sites of East Smithfield (387, 388) and the New Churchyard (326, 389), the great pit of Saints Màrtirs Just i Pastor in Barcelona (390, 391), the three large pits, including over 300 individuals, from the Rue des Trente Six Ponts site in Toulouse (326), and the Observance pit in Marseille (392, 393). All of these sites range from the 14th to the beginning of the 18th centuries and therefore cover the entire second plague pandemic. However, archaeoanthropologists have recently questioned the presence of burials presenting slight anomalies from conventional burial practices, such as the existence of small multiple burials (grouping from 2 to 10 individuals) in regular cemeteries. The observation of certain archaeological characteristics does not mark a profound change in funeral practices, as imposed by a major demographic crisis, but provides signs hinting at the simultaneous death of some members of the community. This type of anomaly might be related to the burial of the first or last victims of an epidemic or to the funeral management practices inherent to an epidemic that is successfully controlled by administrative and health measures. In all cases, these small multiple burials deserve special attention because some of them may have been associated with the burial of plague victims according to paleomicrobiological studies. This is particularly likely in several contexts from the 14th century, such as the cemetery of the churches of St. Nicolay and St. Clement of Oslo in Norway (3, 394), the Saint-Laurent-de-la-Cabrerisse cemetery in France (3, 395), or the Laishevo III cemetery in Russia (326). It is also the case for archeological contexts related to the Justinian Plague, such as the cemetery of Dittenheim in Germany (2, 396) or the cemetery of the Horts in Lunel-Viel in France (2, 397). Thus, relevant paleoepidemiological studies are no longer based solely on the large funeral complexes known as “disaster mass graves” but are also based on small multiple burials and even individual burials in “regular” cemeteries. Based on these discoveries, researchers have therefore reconsidered the data from ancient excavations. These advances have made it possible to associate other small multiple burials initially discovered in traditional funeral spaces with plague episodes. Thus, the presence of Y. pestis has been confirmed in skeletons excavated in 1984/85 in Germany at the site of St. Leonhardi in Manching-Pichl and in 1979 at the Russian site of Laishevo III (326). In some cases, this has allowed a reexamination of old excavation data, such as data from the Estonian site of Kunila II studied in 1948, which revealed that two adult men died from plague (375, 398). The recovery of these ancient data combined with renewed scientific attention toward these small original funeral structures has allowed us to “excavate” the plague bacillus from burials prior to our own era and in totally unexpected chronological contexts (thousands of years before the “first known historical pandemic” of antiquity), such as at the Kunila II site in Estonia, Beli Manastir in Croatia, Gyvakarai in Lithuania, Rasshevatskiy in Russia, and even Haunstetten graves in Germany (375, 398–401). These data have demonstrated the importance of close scientific collaboration between historians, archaeologists, anthropologists, microbiologists, and geneticists, enabling the identification of Y. pestis from a funerary context and within a well-established chronological framework.
Paleomicrobiology of Plague
Detection of ancient biomolecules.
In 1998, one of the first ancient bacterial DNA studies was performed on ancient dental pulp, which is a complex tissue embedded in the teeth and a source of DNA and proteins, from which a 133-bp fragment of the Y. pestis rpoB gene was amplified (385). While the use of dental pulp was fortuitous, we now understand that ancient dental pulp still contains morphologically intact blood cells (402), which may include blood-borne pathogens such as Y. pestis (7). Furthermore, dental pulp is sufficiently protected from the external environment by dentine and enamel to be characterized by low levels of contamination (403). Regarding Y. pestis DNA, initial studies aimed to detect specific sequences of Y. pestis, including the chromosomal sequences of the rpoB gene, encoding the beta-subunit of RNA polymerase (385, 404), the glpD gene (190, 191, 404, 405), the F1 antigen gene (caf1M), located on the pMT1 plasmid (3, 404, 406, 407), and the pPCP1 plasmid-borne pla gene sequence (2, 3, 318, 319, 326, 385, 387, 390, 392, 404, 408–410). The detection of ancient DNA sequences can be performed by sequencing PCR products resulting from multiplex real-time PCR (190) and suicide PCR (409). Suicide PCR avoids the need for positive controls and uses PCR primer pairs only once to avoid any risk of in-laboratory contamination (409). The detection of ancient Y. pestis can also be achieved by immuno-PCR or rapid diagnostic testing based on the detection of the F1 antigen specific to Y. pestis (404, 407, 411–413) or by paleoproteomics relying on the detection of host and blood-borne pathogen proteins (414).
(i) Ancient genome sequencing.
Studies have aimed to recover large genomic fragments, starting with pPCP1 plasmid sequences from the London Black Death site of East Smithfield and dated from 1348 to 1350 (406) and entire genomes of ancient Y. pestis strains (7). These studies have indicated a higher yield of ancient DNA recovered from teeth (37%) than from bones (5.7%) (406). The first draft genome dating from 1348 to 1350 was reported in 2011 (East Smithfield strain) and was reconstructed using ancient DNA from teeth and bones from the plague-confirmed East Smithfield collection (387). The obtained reads were mapped to the CO92 Y. pestis bv. Orientalis reference genome, and the unmapped reads were assembled separately. The size of this first ancient Y. pestis genome reached approximately 4 million base pairs. The coverage of pPCP1 plasmid DNA reached 98.68%, C and T nucleotide damage was revealed, and an average length of 55 bp was observed for chromosomal and plasmid DNA reads, which is characteristic of ancient DNA (average length < 100 bp) (387, 415). With regard to the applied experimental approach, the total DNA extracted from ancient specimens is enriched for Y. pestis DNA using microarrays (318, 319, 387, 390, 392, 406) or via in-solution capture (2, 326, 375, 378, 408), followed by high-throughput next-generation sequencing (NGS) (7, 416). Alternatively, specific sequences of Y. pestis can be extracted in silico from the ultradeep sequencing of human genomes from anthropological material to reconstruct complete ancient genomes (376, 377, 379, 417). By combining the technical approaches reported above, a total of 88 ancient Y. pestis genomes have been reported to date (available from the European Nucleotide Archive or NCBI GenBank databases) (Fig. 8). These ancient genomes were all recovered from individuals from Eurasia, covering a long period from 5000 BP (before the present) to 1722 (376, 392). The analysis of ancient Y. pestis genome sequences offers a unique opportunity to explore the genetic evolution of the pathogen through observation rather than deduction from phylogenetic reconstructions based on the whole-genome sequencing of modern isolates (17). Indeed, subsequent deductions have indicated that Y. pestis may have diverged from Y. pseudotuberculosis approximately 13,000 to 79,000 years ago (7), based on genes obtained from the acquisition of two associated virulence plasmids, pFra/pMT1 and pPLA/pPCP1 (418), and that a genome reduction of approximately 10% occurred, including the loss of genes associated with virulence and metabolism in Y. pseudotuberculosis (genome size, 4.72 Mb) (33, 419, 420).
FIG 8.
Map of ancient Yersinia pestis (Y. p) genomes.
(ii) Paleogenomic study of prehistoric plague, 5000 BP to 800 BC.
Sixteen genomes predating historical pandemics have been sequenced, proving the endemicity of plague infection among human populations in the Late Neolithic-Bronze Age period (LNBA) (∼5000 BP to ∼3500 BP). The most ancient strain of Gok2 (which diverged from all other Y. pestis strains 5,700 years BP) is basal to all known modern and ancient Y. pestis genomes, while the other Bronze Age genomes were found to belong to an independent lineage emerging between 6000 and 5000 BP in Eurasia and probably spreading in the context of Yamnaya-related steppe expansion during the fifth millennium BP (376, 379). This first divergence yielded the basal lineages 0.PE7 and 0.PE2, which still exist today, and two extinct lineages from the Neolithic (Gok2) and Bronze Ages (376). The second divergence occurred 4,000 years ago on the Eurasian steppes and yielded the extinct RT5 lineage, the extant 0.PE4 lineage (Microtus), and other lineages that may be ancestral to those responsible for the historical pandemics (378). These genomes specifically lack a 20-kb plasmid region comprising the virulence-associated Yersinia murine toxin gene (ymt) (375–377, 379). The ymt gene encodes a phospholipase D implicated in the survival and replication of Y. pestis inside the flea gut. The presence of several virulence-associated genes, including pla, caf1, ureD, rcsA, flhD, pde2, and DFR4, indicates that these genomes belong to ancestral strains of Y. pestis associated with reduced invasiveness and, possibly, host adaptation (375, 378). The two easternmost Bronze Age genomes (GLZ001 and GLZ002) exhibit a lack of ympt1.66c, which is involved in the initial within-macrophage survival of the bacterium and, thus, bubonic plague progression (379). Furthermore, the lack of the ymt gene, which plays a key role in the flea-borne transmission of plague through late-stage biofilm-dependent transmission (18), also suggests that alternative transmission scenarios might have occurred during the Late Neolithic-Early Bronze Age (LN-EBA) period. However, fleas do not require the ymt gene for the early-phase transmission (EPT) of plague (203, 421), but this mechanism is considered to be poorly efficient, and it has been extensively debated whether it has the capacity to drive massive epidemics (119, 130, 186, 204, 422). All of these elements may explain the absence of mass graves during this period. The appearance of the ymt gene combined with altered sequence-suggested inactivation or loss of the ureD, rcsA, flhD, pde2, and pde3 genes contributed to effective flea-borne transmission and an increase in bubonic plague forms. This event was dated to approximately 3,800 years ago based on two individuals found in Russia but possibly began as early as 5,000 years ago (33, 378). These data highlight the coexistence of ymt-negative and ymt-positive strains in Eurasia between 5,000 and 3,000 years ago, suggesting different transmission cycles and different disease symptoms, although the exact mechanisms of transmission and the vectors and sources involved remain unclear (7). Concerning the spread of plague, researchers have hypothesized the occurrence of dispersion through the early trade networks, rather than through human migration (376), and have suggested that plague might have contributed to the decline of Neolithic societies by causing large deadly outbreaks, although there is currently no genomic (absence of the ymt gene) or archaeological (absence of mass graves) evidence that plague could have been epidemic in nature. Nevertheless, Neolithic plague may have migrated from the Eurasian steppes toward western Europe, which potentially continued during the Bronze Ages through lineages that eventually became extinct (376).
(iii) Paleogenomic study of the first historical pandemic, 541 to 750/767.
The pandemic of 541 to 750/767 was confirmed as being caused by Y. pestis by three paleomicrobiological studies that recovered genomes from 10 individuals from Germany, France, England, and Spain (2, 318, 319). Genomic analyses suggested the existence of great diversity among the Y. pestis strains that were circulating during the sixth to eighth centuries in Europe (2), classifying all of these strains into the same lineage, with no contemporary representatives, diverging between the clades 0.ANT1, 0.ANT2, and 0.ANT5. The ED1001 strain recovered from Edix-Hill (Great Britain) is ancestral to all the other Justinian strains. Interestingly, four strains, including the Lunel-Viel and Saint-Doulchard strains, diverged from the Altenerding cluster through a polytomy; these strains belonged to a subsequent wave that was different from the original wave of 541 to 544. In Saint-Doulchard, one of the two studied genomes was ancestral to the second one, illustrating for the first time the coexistence of two independent Y. pestis genomes at the same site (2). These data led to a reinterpretation of the historical sources that had once suggested that the Justinian Plague was restricted to the shores of the Mediterranean Basin (8). Paleomicrobiological data indicated that the Justinian Plague was a continental plague that reached what is currently Germany and the United Kingdom (2, 318, 319). Three unique deletions were observed in pheA, the YPO2283 region, and the celB gene. Notably, a 45-kb and a 49-kb deletion including the mgtB and mgtC virulence-associated genes have been observed in some tardive strains of both the first and second pandemics (2, 326). It has been hypothesized that this genomic decay found only within end-of-epidemic strains might have been significant for the disappearance of these two pandemics (2). Concerning the origin of the Justinian Plague, some researchers have hypothesized a central Asian localization (417). Indeed, the framework for these genomes comes from the 0.ANT1, 0.ANT2, and 0.ANT5 lineages, which were isolated in China or Kyrgyzstan (66, 327). The basal strain of all the first-pandemic genomes was isolated from the Hun people in the Tian Shan Mountains, China (417). The Tian Shan mountain lineage may be the origin of the Justinian Plague, although 3 centuries separate this genome and the beginning of the pandemic in the Mediterranean Basin. Alternatively, the Justinian Plague may have spread along the Red Sea and Indian Ocean maritime routes connecting the Byzantine Empire and India (322) (Fig. 6). However, these claims based on genomic and phylogenetic analyses contradict certain historical sources. Procopius, who experienced this pandemic, described the plague as originating from Egypt and then spreading to Palestine and, finally, Constantinople (423). Evagrius described an “Ethiopian” origin of this outbreak. The possible African origin of the first pandemic is among the most scientifically debated issues (316, 317). In fact, in the absence of an ancient African Y. pestis genome, the current data do not allow the determination of the origin and spread of this pandemic (2).
(iv) Paleogenomic study of the second historical pandemic, 1346 to 18th century.
In total, 56 Y. pestis genomes dating from the second pandemic in Eurasia have been sequenced (3, 326, 387, 390, 392, 408) (Table 2). These genomes have been identified through phylogenetic analyses as belonging to the second-pandemic branch, indicating that this pandemic was caused by a single introduction of one Y. pestis lineage (326, 390). In this lineage, four genomes have been shown to be identical to that of the Black Death strains (Barcelona3031, NAB003, NMS003, and OSL1), supporting the hypothesis of a single wave entering Europe and further spreading via maritime and terrestrial routes (326, 390). The Laishevo strain (LAI009), dating from 1300 to 1400 in Russia, was identified as the most ancestral form of the Y. pestis strain that entered Europe and caused the second pandemic, confirming the hypothesis of plague foci present approximately 2,000 km away in northeastern Crimea before the first plague was introduced to southern Europe in 1347 (326). Furthermore, the analysis of these 56 Y. pestis genomes (Table 2) provided considerable evidence of microevolution into the same lineage at the end of the second pandemic (post-Black Death lineage), as follows: the Ellwangen strain gave rise to (at least) two distinct post-Black Death clades, where the first clade includes the German and Swiss strains from the 15th to 17th centuries and the second clade includes the Observance strains and the London strains from the 17th to 18th centuries. All the lineages were derived from the Black Death isolates and are likely to have evolved separately, leading to their divergence from the Black Death strains from approximately the 16th and 18th centuries. The post-Black Death lineage no longer exists, which could partially explain the disappearance of plague in the 18th century (326, 390). A study analyzing 34 ancient second-pandemic genomes identified a deleted region associated with virulence-related genes (mgtB, mgtC, and inv genes) that are vital for macrophage colonization (associated with decreased virulence in mice) (424) in genomes dated to the end of the second pandemic. The same deletion has been observed in late first-pandemic genomes. The functions of these genes in mammalian or arthropod plague conservation remain to be established (2, 326). The second-pandemic genomes are closely related to the genome sequence of current isolates, suggesting very few differences in virulence factors between current and ancient strains. Nevertheless, the Black Death strains and released Justinian Plague genomes (318, 319, 387) exhibit a 15-kb genomic island corresponding to DFR4 (difference region 4), which is deleted in all contemporary Y. pestis bv. Orientalis strain isolates but is present in some Y. pestis bv. Antiqua isolates, such as the Antiqua and Nairobi strains, some Y. pestis bv. Medievalis isolates, Pestoides A to D isolates, and Y. pseudotuberculosis (425). This region potentially includes virulence factor genes such as ccm2A, whose role in human infection is poorly understood. Analyses of genomes from the second historical pandemic suggest a single introduction of plague (based on the absence of genetic diversity among the sequenced strains) from Asia/eastern Europe into Europe in 1347 via terrestrial and maritime routes, followed by the persistence of plague in several temporary but undetermined foci (now extinct) to give rise to the western European phenomenon of endemic plague until the 18th century. Shortly after its introduction to Europe, the Black Death strain returned to Asia to give rise to the third pandemic (318, 326, 387, 390, 392). However, this hypothetical scenario seems very unlikely according to historical and epidemiological studies (3, 426, 427). An alternative second hypothesis argues that the plague originated in eastern Europe/central Asia and spread along trade routes (particularly the Silk Road), thus creating relatively permanent plague foci along these routes. The plague may have spread from such secondary foci through successive waves in western Europe along terrestrial and maritime trade routes (such as fur roads) (427) or through human migration (428) from the 14th to the 18th centuries (3, 429). Climate data showed that events such as episodes of aridity or significant fluctuations were decisive in the second-pandemic plague introduction and reintroductions (428, 429). Furthermore, the Asian origin of the second pandemic is debatable (3, 326–328, 430). A recent study indicates the lack of historical and molecular evidence of plague in 14th century eastern China. The whole-genome sequencing of Y. pestis animal isolates from modern plague foci in China showed genomic diversity greater than that observed in medieval European strains. Based on this observation, the most recent study led to the inference that the northeastern European plague reservoir acted as a second-pandemic source (326).
TABLE 2.
Summary of all sequenced genomes of Y. pestis belonging to the second pandemica
| Archeological site | City | Country | No. of genome(s) | Dating (yrs) | Strain name(s) | Sample accession no. | Study accession no. | Reference(s) |
|---|---|---|---|---|---|---|---|---|
| East Smithfield burial ground | London | England | 5 | 1348–1350 | Individual 8124 | SRX096049 | SRP008060 | 387 |
| Individual 11972 | SRX096048 | |||||||
| Individual 8291 | SRX096047 | |||||||
| Individual 6330 | SRX096046 | |||||||
| East Smithfield | SRX096021 | |||||||
| Observance | Marseille | France | 5 | 1722 | OBS107 | ERS1020860 | PRJEB12163 | 392 |
| OBS110 | ERS1020861 | |||||||
| OBS116 | ERS1020862 | |||||||
| OBS124 | ERS1020863 | |||||||
| OBS137 | ERS1020864 | |||||||
| Saints Màrtirs Just i Pastor | Barcelona | Spain | 1 | 1300–1420 | Barcelona3031 | ERS1124787 | PRJEB13664, PRJEB29990 | 326, 390 |
| Ust'-Jerusalem necropolis and Bolgar City mausoleum | Bolgar | Russia | 1 | 1298–1388 | BolgarCity2370 | ERS1124788 | ||
| Marktplatz | Ellwangen | Germany | 1 | 1485–1627 | Ellwangen549_O | ERS1124789 | ||
| ERS1124790 | ||||||||
| ERS3607398 | ||||||||
| Saint-Laurent-de-la-Cabrerisse | Saint-Laurent-de-la-Cabrerisse | France | 1 | 1348 | SLC1006 | ERS2865271 | PRJEB24499 | 3 |
| Bergen op Zoom | Bergen op Zoom | The Netherlands | 2 | 1358–1360 | Ber37 | ERS2865267 | ||
| Ber45 | ERS2865268 | |||||||
| Churches of St. Nicolay and St. Clement | Oslo | Norway | 1 | 1349–1350 | OSL1 | ERS2865270 | ||
| Abbadia San Salvatore | Abbadia San Salvatore | Italy | 1 | 1348 | BSS31 | ERS2865269 | ||
| Laishevo III cemetery | Laishevo | Russia | 2 | 1300–1400 | LAI009 | ERS3607399 | PRJEB29990 | 326 |
| LAI010 | ERS3607400 | |||||||
| 16 Rue des Trente Six Ponts | Toulouse | France | 1 | 1347–1350 | TRP002 | ERS3607426 | ||
| The New Churchyard | London | England | 5 | 1560–1635 | BED024 | ERS3607391 | ||
| BED028 | ERS3607392 | |||||||
| BED030 | ERS3607393 | |||||||
| BED034 | ERS3607394 | |||||||
| BED038 | ERS3607395 | |||||||
| Augustinian Friary | Cambridge | England | 2 | 1475–1536 | NMS002 | ERS3607408 | ||
| NMS003 | ERS3607409 | |||||||
| Sankt Johans Freidhof | Nabburg | Germany | 3 | 1292–1392 | NAB003 | ERS3607404 | ||
| NAB004 | ERS3607405 | |||||||
| NAB005 | ERS3607406 | |||||||
| ERS3607407 | ||||||||
| St. Leonhardi | Manching-Pichl | Germany | 2 | 1283–1390 | MAN008 | ERS3607402 | ||
| MAN015 | ERS3607403 | |||||||
| Possenhofener Str. 3 | Starnberg | Germany | 1 | 1433–1523 | STA001 | ERS3607410 | ||
| Kirchhof St. Johannis | Landsberg am Lech | Germany | 1 | 1455–1632 | LBG002 | ERS3607401 | ||
| Nägeligasse | Stans | Switzerland | 15 | 1485–1635 | STN002 | ERS3607411 | ||
| STN004 | ERS3607412 | |||||||
| STN005 | ERS3607413 | |||||||
| STN007 | ERS3607414 | |||||||
| STN008 | ERS3607415 | |||||||
| STN011 | ERS3607416 | |||||||
| STN012 | ERS3607417 | |||||||
| STN013 | ERS3607418 | |||||||
| STN014 | ERS3607419 | |||||||
| STN018 | ERS3607420 | |||||||
| STN019 | ERS3607421 | |||||||
| STN020 | ERS3607422 | |||||||
| STN021 | ERS3607423 | |||||||
| STN031 | ERS3607424 | |||||||
| STN032 | ERS3607425 | |||||||
| Domlinden 12 | Brandenburg an der Havel | Germany | 2 | 1618–1648 | BRA001 | ERS3607396 | ||
| BRA003 | ERS3607397 | |||||||
| Aguonu g. 10 | Vilnius | Lithuania | 4 | 1440–1621 | AGU007 | ERS4398260 | PRJEB37508 | 408 |
| AGU010 | ERS4410876 | |||||||
| AGU020 | ERS4398261 | |||||||
| AGU025 | ERS4398262 | |||||||
| ERS4398263 |
Based on the phylogenetic tree or dating and including project numbers for read accession.
AREAS OF UNCERTAINTY: PROSPECTIVE STUDIES
This review highlights areas of uncertainty regarding plague dynamics. Clearly, the lack of any ancient genomes from outside Eurasia biases our understanding of historical plague origins and spreading, which may be partially related to cultural bias, as most ancient plague studies have been performed by European researchers, including the discovery of the plague bacillus by Alexandre Yersin. The recovery of ancient Y. pestis genomes in Africa might be of value for resolving the ongoing controversies regarding the sources of plague and their persistence over several centuries (2, 326). Concerning plague vectors, the role of bedbugs (Cimex lectularius), which have long been suspected to be an effective interhuman vector (27, 194), should be investigated on the basis of current concepts and techniques and proper methodology. In 1897, Imagiva reported a human case of plague possibly caused by bedbugs (431). In 1910, Walker found Y. pestis-infected bedbugs in a camp where plague was raging and successfully transmitted the plague to rats via these bedbugs. Interestingly, Walker found no fleas during this outbreak (432). Subsequently, bedbugs were commonly found to be infected (sometimes in plague patients’ beds) during plague outbreaks, as reported by Pollitzer (99). Unfortunately, the previous experiments were all carried out at the beginning of the 20th century using methods that did not allow researchers to indisputably prove the infection of bedbugs by Y. pestis. Nevertheless, these experiments indicated that virulent Y. pestis colonizes bedbug stomachs and feces for >100 days, thereby a host infection; however, the underlying mechanisms and efficiency remain to be investigated (99, 194, 433, 434). Outbreaks of the alleged pneumonic form have remained a debated issue since the confusing observation of Streptococcus pneumoniae in blood from plague patients by Kitasato in 1894. Kitasato’s patients were clearly coinfected with Y. pestis and S. pneumoniae, and the culture conditions used by Kitasato favored the growth of S. pneumoniae (435). The propensity for these coinfections during plague outbreaks remains unquantified, as a syndromic approach is not routinely used in cases of suspected pneumonic plague. For example, during the last major outbreak in Madagascar in 2017, only 32 of 1,846 probable or suspected pneumonic plague cases were laboratory confirmed, leaving the possibility that other pathogens were responsible for pneumonia (59). This possibility is not only speculative, as a leptospirosis outbreak was retrospectively found to be nested within a suspected pneumonic plague outbreak (245). The relative efficiency of numerous plague vectors, such as fleas or body lice, remains extremely difficult to evaluate because of the lack of uniformity of the methods used in the laboratory (depending on the susceptibility of the hosts, the infectious doses used, the strains involved, and the choice of the temporal dynamics of infection and postinfection); sometimes, these methods are contrived and do not approach the actual reality that can be observed in the field.
CONCLUSION
Y. pestis is among the oldest reported and deadliest opportunistic human pathogens. During the Neolithic period at least, plague raged across Eurasia, causing outbreaks and possibly leading to a temporary decline in centenarian cultures. Approximately 3,800 years ago, Y. pestis acquired the ymt gene, enabling it to become a more efficient flea-borne bacterium. The Justinian Plague (541 to 750/767) remains controversial as to its demographic, political, and social consequences, unlike the second pandemic (1346 to 18th century), which killed at least 30% of the European population, made a profound impression on the collective imagination, and led to a deep restructuring of medieval society. The mortality rates resulting from the current third pandemic (1772 to present) have been incredibly low (except at its beginning) due to the first treatments and the use of antibiotics, which made it possible to reduce the mortality rate from 50 to 100% in the absence of treatment to approximately 10 to 25%. The most recent study indicates that Y. pestis is a telluric bacterium that exists in a free state, an L-form, or inside amoeba, and soil could serve as a reservoir. More than 200 mammals (mainly rodents) have been found to be infected by plague, probably orally, resulting from digging in soil. Human contamination from wild sources may occur from flea and tick bites, transcutaneous contact, aerosol inhalation, or oral ingestion (raw meat consumption). The most recent evidence indicates that interhuman transmission may be due mainly to human ectoparasites, such as body lice and fleas (aerosol transmission appears to be very low). The current data indicate that the epidemiology of plague is extremely complex because it is intrinsically linked to a given environment and time. For example, rats and fleas were found to be effective vectors in 1898 in India, but this scheme cannot be applied to effective reservoirs and vectors in ancient and modern plague outbreaks. The plague in the United States seems to be linked mainly to squirrels or chipmunks. In North Africa and the Middle East, outbreaks are linked to the consumption of raw or poorly cooked meat from camels or goats, and Tibetan cases are linked to sheep and marmot skinning. In regions where plague is endemic, efforts have been made to prevent human cases by performing “sentinel” animal surveys and vaccination. In addition, to support the appropriate management of patients and provide a rapid and accurate microbiological diagnosis, we recommend evaluation in point-of-care laboratories, some of which are currently operating in a few remote regions in Africa. In addition to the direct diagnosis of disease in humans, the direct detection of Y. pestis at the point of care among potential sources and vectors could facilitate our understanding of how plague epidemics are sustained.
ACKNOWLEDGMENT
We thank Oleg Mediannikov for his kind help with the translation of scientific articles from Russian to French.
Biographies

R. Barbieri has been a Ph.D. student in microbiology and biological anthropology at the IHU Méditerranée Infection, developing paleomicrobiology, with a particular interest in past plague, source, and dynamics.

M. Signoli is Senior Researcher at the CNRS and Director of the ADES research unit (UMR 7268) at the Marseille Faculty of Medicine. An anthropologist specializing in the study of past epidemics, he works on these contexts using two types of sources: biological archives, i.e., the victims of these epidemics exhumed during archaeological operations, and historical archives, i.e., documents written by contemporaries of these epidemic crises. He has thus conducted and participated in numerous interdisciplinary studies on the plague, but also on other pathogens (tuberculosis, typhus, pox virus, etc.).

D. Chevé is an anthropologist, a Ph.D. in biocultural anthropology, and an associate researcher at UMR ADES (AMU-CNRS-EFS, Marseille, France), where she is coresponsible for a theme on the anthropology of life. Her work focuses on bodies (states, practices, and representations) at the real and symbolic levels and their relationships to health, the environment, and society, in particular to epidemics. She is codirector of the journal “Corps” (CNRS-Editions, Paris, France).

C. Costedoat is an Associate Professor in population genetics and HDR of Aix-Marseille University at the UMR 7268 Bio-Cultural Anthropology, Ethics and Health Law (ADES-AMU, CNRS, French Blood Establishment PACA Corse) research unit. Her research activities on genome-environment relations focus on the human model and in particular on the impact of the pathogenic environment on the genome and of the cultural footprint on the diversity of populations. Her dual affiliation in environmental science and anthropology allows her to address public health issues through the prism of interdisciplinarity, from field studies to data analysis.

S. Tzortzis is a Ph.D. in anthropology, an engineer of the French Ministry of Culture (Direction Régionale des Affaires Culturelles, Service Régional de l’Archéologie de Provence Alpes-Côte d’Azur, Aix-en-Provence), and a member of the Unité Mixte de Recherche 6578 ADES (Anthropologie Bio-Culturelle, Droit, Ethique, Santé, Marseille). As an archeoanthropologist, he has led excavations of multiple burial sites linked to plague epidemics of the Modern Times (17th to 18th centuries).

G. Aboudharam, a dentist by training, is a cofounder of the field of paleomicrobiology, inventing all the techniques for the manipulation of ancient dental pulp, leading to the discovery of Yersinia pestis in ancient human specimens, with further interest in ancient blood-borne infections at large.

D. Raoult M.D., Ph.D., who specializes in infectious diseases, is a professor of microbiology at the Faculty of Medicine of Marseille, Aix-Marseille University. In 1984, he created ex nihilo his research laboratory, the Rickettsia Unit, in the frame of which he serendipitously created the field of paleomicrobiology. He accordingly started working on plague more than 20 years ago, and his unit has now become the Research Unit in Infectious and Tropical Emergent Diseases (URMITE), collaborating with the CNRS (National Center for Scientific Research), the IRD (Institute of Research for Development), and INSERM (National Institute of Health and Medical Research). In 2011, he became the Director of the University Hospital Institute Mediterranée Infection, which is a 600-person medical institute focused on infectious diseases. This facility includes the largest diagnostic and research microbiology laboratory in France. As of 2020, Prof. Raoult has published over 2,800 indexed publications.

M. Drancourt M.D., Ph.D., is a professor of medical microbiology in IHU Méditerranée Infection, Marseille Medical School, and head of the Microbiology Laboratory at Marseille’s public hospital. His research topics include paleomicrobiology and past plague epidemics.
REFERENCES
- 1.Mordechai L, Eisenberg M, Newfield TP, Izdebski A, Kay JE, Poinar H. 2019. The Justinianic Plague: an inconsequential pandemic? Proc Natl Acad Sci U S A 116:25546–25554. doi: 10.1073/pnas.1903797116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Keller M, Spyrou MA, Scheib CL, Neumann GU, Kröpelin A, Haas-Gebhard B, Päffgen B, Haberstroh J, Ribera I Lacomba A, Raynaud C, Cessford C, Durand R, Stadler P, Nägele K, Bates JS, Trautmann B, Inskip SA, Peters J, Robb JE, Kivisild T, Castex D, McCormick M, Bos KI, Harbeck M, Herbig A, Krause J. 2019. Ancient Yersinia pestis genomes from across Western Europe reveal early diversification during the First Pandemic (541–750). Proc Natl Acad Sci U S A 116:12363–12372. doi: 10.1073/pnas.1820447116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Namouchi A, Guellil M, Kersten O, Hänsch S, Ottoni C, Schmid BV, Pacciani E, Quaglia L, Vermunt M, Bauer EL, Derrick M, Jensen AØ, Kacki S, Cohn SK, Stenseth NC, Bramanti B. 2018. Integrative approach using Yersinia pestis genomes to revisit the historical landscape of plague during the Medieval Period. Proc Natl Acad Sci U S A 115:E11790–E11797. doi: 10.1073/pnas.1812865115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Slack P. 1989. The black death past and present. 2. Some historical problems. Trans R Soc Trop Med Hyg 83:461–463. doi: 10.1016/0035-9203(89)90247-2. [DOI] [PubMed] [Google Scholar]
- 5.Tan J, Liu Y, Shen E, Zhu W, Wang W, Li R, and, Yang L. 2002. Towards the Atlas of Plague and Its Environment in the People’s Republic of China: idea, principle and methodology of design and research results. Huan Jing Ke Xue 23:1–8. [PubMed] [Google Scholar]
- 6.Bramanti B, Stenseth NC, Walløe L, Lei X. 2016. Plague: a disease which changed the path of human civilization, p 1–26. In Yang R, Anisimov A (ed), Yersinia pestis: retrospective and perspective. Springer, Heidelberg, Germany. doi: 10.1007/978-94-024-0890-4_1. [DOI] [PubMed] [Google Scholar]
- 7.Spyrou MA, Bos KI, Herbig A, Krause J. 2019. Ancient pathogen genomics as an emerging tool for infectious disease research. Nat Rev Genet 20:323–340. doi: 10.1038/s41576-019-0119-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perry RD, Fetherston JD. 1997. Yersinia pestis—etiologic agent of plague. Clin Microbiol Rev 10:35–66. doi: 10.1128/CMR.10.1.35-66.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abbott RC, Rocke TE. 2012. Plague. U.S. Geological Survey Circular 1372. https://pubs.usgs.gov/circ/1372.
- 10.Yersin A. 1894. Sur la peste de HongKong (Lundi 30 juillet 1894). C R Hebd Séances Acad Sci 119:356. [Google Scholar]
- 11.Platonov ME, Evseeva VV, Dentovskaya SV, Anisimov AP. 2013. Molecular typing of Yersinia pestis. Mol Genet Microbiol Virol 28:41–51. doi: 10.3103/S0891416813020067. [DOI] [PubMed] [Google Scholar]
- 12.Anisimov AP, Lindler LE, Pier GB. 2004. Intraspecific diversity of Yersinia pestis. Clin Microbiol Rev 17:434–464. doi: 10.1128/CMR.17.2.434-464.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vogler AJ, Keim P, Wagner DM. 2016. A review of methods for subtyping Yersinia pestis: from phenotypes to whole genome sequencing. Infect Genet Evol 37:21–36. doi: 10.1016/j.meegid.2015.10.024. [DOI] [PubMed] [Google Scholar]
- 14.Derbise A, Chenal‐Francisque V, Pouillot F, Fayolle C, Prévost M-C, Médigue C, Hinnebusch BJ, Carniel E. 2007. A horizontally acquired filamentous phage contributes to the pathogenicity of the plague bacillus. Mol Microbiol 63:1145–1157. doi: 10.1111/j.1365-2958.2006.05570.x. [DOI] [PubMed] [Google Scholar]
- 15.Carniel E. 2014. Subtle genetic modifications transformed an enteropathogen into a flea-borne pathogen. Proc Natl Acad Sci U S A 111:18409–18410. doi: 10.1073/pnas.1421887112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Achtman M, Zurth K, Morelli G, Torrea G, Guiyoule A, Carniel E. 1999. Yersinia pestis, the cause of plague, is a recently emerged clone of Yersinia pseudotuberculosis. Proc Natl Acad Sci U S A 96:14043–14048. doi: 10.1073/pnas.96.24.14043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parkhill J, Wren BW, Thomson NR, Titball RW, Holden MT, Prentice MB, Sebaihia M, James KD, Churcher C, Mungall KL, Baker S, Basham D, Bentley SD, Brooks K, Cerdeño-Tárraga AM, Chillingworth T, Cronin A, Davies RM, Davis P, Dougan G, Feltwell T, Hamlin N, Holroyd S, Jagels K, Karlyshev AV, Leather S, Moule S, Oyston PC, Quail M, Rutherford K, Simmonds M, Skelton J, Stevens K, Whitehead S, Barrell BG. 2001. Genome sequence of Yersinia pestis, the causative agent of plague. Nature 413:523–527. doi: 10.1038/35097083. [DOI] [PubMed] [Google Scholar]
- 18.Hinnebusch BJ, Rudolph AE, Cherepanov P, Dixon JE, Schwan TG, Forsberg A. 2002. Role of Yersinia murine toxin in survival of Yersinia pestis in the midgut of the flea vector. Science 296:733–735. doi: 10.1126/science.1069972. [DOI] [PubMed] [Google Scholar]
- 19.Leal-Balbino TC, Leal NC, Lopes CV, de Almeida AMP. 2004. Differences in the stability of the plasmids of Yersinia pestis cultures in vitro: impact on virulence. Mem Inst Oswaldo Cruz 99:727–732. doi: 10.1590/S0074-02762004000700011. [DOI] [PubMed] [Google Scholar]
- 20.Protsenko OA, Filippov AA, Kutyrev VV. 1991. Integration of the plasmid encoding the synthesis of capsular antigen and murine toxin into Yersinia pestis chromosome. Microb Pathog 11:123–128. doi: 10.1016/0882-4010(91)90005-U. [DOI] [PubMed] [Google Scholar]
- 21.Zsigray RM, Hopper JB, Zukowski K, Chesbro WR. 1985. Integration of the Vwa plasmid into the chromosome of Yersinia pestis strains harboring F′ plasmids of Escherichia coli. Infect Immun 47:670–673. doi: 10.1128/IAI.47.3.670-673.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chu MC, Dong XQ, Zhou X, Garon CF. 1998. A cryptic 19-kilobase plasmid associated with US isolates of Yersinia pestis: a dimer of the 9.5-kilobase plasmid. Am J Trop Med Hyg 59:679–686. doi: 10.4269/ajtmh.1998.59.679. [DOI] [PubMed] [Google Scholar]
- 23.Sebbane F, Jarrett CO, Gardner D, Long D, Hinnebusch BJ. 2006. Role of the Yersinia pestis plasminogen activator in the incidence of distinct septicemic and bubonic forms of flea-borne plague. Proc Natl Acad Sci U S A 103:5526–5530. doi: 10.1073/pnas.0509544103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lane MC, Lenz JD, Miller VL. 2013. Proteolytic processing of the Yersinia pestis YapG autotransporter by the omptin protease Pla and the contribution of YapG to murine plague pathogenesis. J Med Microbiol 62:1124–1134. doi: 10.1099/jmm.0.056275-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haiko J, Laakkonen L, Westerlund-Wikström B, Korhonen TK. 2011. Molecular adaptation of a plant-bacterium outer membrane protease towards plague virulence factor Pla. BMC Evol Biol 11:43. doi: 10.1186/1471-2148-11-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Akhoundi M, Sereno D, Marteau A, Bruel C, Izri A. 2020. Who bites me? A tentative discriminative key to diagnose hematophagous ectoparasites biting using clinical manifestations. Diagnostics 10:308. doi: 10.3390/diagnostics10050308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Simond P-L. 1898. La propagation de la peste. Ann Inst Pasteur 10:626–687. [Google Scholar]
- 28.Gonzalez RJ, Miller VL. 2016. A deadly path: bacterial spread during bubonic plague. Trends Microbiol 24:239–241. doi: 10.1016/j.tim.2016.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.John ALS, Ang WG, Huang M-N, Kunder CA, Chan EW, Gunn MD, Abraham SN. 2014. S1P-dependent trafficking of intracellular Yersinia pestis through lymph nodes establishes buboes and systemic infection. Immunity 41:440–450. doi: 10.1016/j.immuni.2014.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Montminy SW, Khan N, McGrath S, Walkowicz MJ, Sharp F, Conlon JE, Fukase K, Kusumoto S, Sweet C, Miyake K, Akira S, Cotter RJ, Goguen JD, Lien E. 2006. Virulence factors of Yersinia pestis are overcome by a strong lipopolysaccharide response. Nat Immunol 7:1066–1073. doi: 10.1038/ni1386. [DOI] [PubMed] [Google Scholar]
- 31.Thomson JJ, Plecha SC, Krukonis ES. 2019. Ail provides multiple mechanisms of serum resistance to Yersinia pestis. Mol Microbiol 111:82–95. doi: 10.1111/mmi.14140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bi Y. 2016. Immunology of Yersinia pestis infection, p 273–292. In Yang R, Anisimov A (ed), Yersinia pestis: retrospective and perspective. Springer, Heidelberg, Germany. [DOI] [PubMed] [Google Scholar]
- 33.Demeure CE, Dussurget O, Fiol GM, Le Guern A-S, Savin C, Pizarro-Cerdá J. 2019. Yersinia pestis and plague: an updated view on evolution, virulence determinants, immune subversion, vaccination, and diagnostics. Genes Immun 20:357–370. doi: 10.1038/s41435-019-0065-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li B, Yang R. 2008. Interaction between Yersinia pestis and the host immune system. Infect Immun 76:1804–1811. doi: 10.1128/IAI.01517-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Olson RM, Dhariwala MO, Mitchell WJ, Anderson DM. 2019. Yersinia pestis exploits early activation of MyD88 for growth in the lungs during pneumonic plague. Infect Immun 87:e00757-18. doi: 10.1128/IAI.00757-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eisen RJ, Griffith KS, Borchert JN, MacMillan K, Apangu T, Owor N, Acayo S, Acidri R, Zielinski-Gutierrez E, Winters AM, Enscore RE, Schriefer ME, Beard CB, Gage KL, Mead PS. 2010. Assessing human risk of exposure to plague bacteria in northwestern Uganda based on remotely sensed predictors. Am J Trop Med Hyg 82:904–911. doi: 10.4269/ajtmh.2010.09-0737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bertherat E. 2016. Plague around the world, 2010–2015. Wkly Epidemiol Rec 91:89–94.26922822 [Google Scholar]
- 38.Bertherat E. 2019. Plague around the world in 2019. Wkly Epidemiol Rec 94:289–292. [Google Scholar]
- 39.WHO. 2010. –2017. Wkly Epidemiol Rec 85: 509–512 to 93: 693–708. https://www.who.int/wer/en/. [Google Scholar]
- 40.Danforth M, Novak M, Petersen J, Mead P, Kingry L, Weinburke M, Buttke D, Hacker G, Tucker J, Niemela M. 2016. Investigation of and response to 2 plague cases, Yosemite National Park, California, USA, 2015. Emerg Infect Dis 22:2045. doi: 10.3201/eid2212.160560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Campbell SB, Nelson CA, Hinckley AF, Kugeler KJ. 2019. Animal exposure and human plague, United States, 1970–2017. Emerg Infect Dis 25:2270–2273. doi: 10.3201/eid2512.191081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ruiz A. 2001. Plague in the Americas. Emerg Infect Dis 7:539–540. doi: 10.3201/eid0707.017718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Giles TA, Greenwood AD, Tsangaras K, Giles TC, Barrow PA, Hannant D, Abu-Median A-B, Yon L. 2016. Detection of a Yersinia pestis gene homologue in rodent samples. PeerJ 4:e2216. doi: 10.7717/peerj.2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Giles J, Peterson AT, Almeida A. 2011. Ecology and geography of plague transmission areas in northeastern Brazil. PLoS Negl Trop Dis 5:e925. doi: 10.1371/journal.pntd.0000925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schneider MC, Najera P, Aldighieri S, Galan DI, Bertherat E, Ruiz A, Dumit E, Gabastou JM, Espinal MA. 2014. Where does human plague still persist in Latin America? PLoS Negl Trop Dis 8:e2680. doi: 10.1371/journal.pntd.0002680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nyirenda SS, Hang’ombe BM, Machang’u R, Mwanza J, Kilonzo BS. 2017. Identification of risk factors associated with transmission of plague disease in eastern Zambia. Am J Trop Med Hyg 97:826–830. doi: 10.4269/ajtmh.16-0990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nyirenda SS, Hang’ombe BM, Kilonzo BS. 2016. Factors that precipitated human plague in Zambia from 1914 to 2014—an overview for a century (100 years). J Zoonotic Dis 1:1–14. [Google Scholar]
- 48.Abedi AA, Shako J-C, Gaudart J, Sudre B, Ilunga BK, Shamamba SKB, Diatta G, Davoust B, Tamfum J-JM, Piarroux R, Piarroux M. 2018. Ecologic features of plague outbreak areas, Democratic Republic of the Congo, 2004–2014. Emerg Infect Dis 24:210–220. doi: 10.3201/eid2402.160122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Amatre G, Babi N, Enscore RE, Ogen-Odoi A, Atiku LA, Akol A, Gage KL, Eisen RJ. 2009. Flea diversity and infestation prevalence on rodents in a plague-endemic region of Uganda. Am J Trop Med Hyg 81:718–724. doi: 10.4269/ajtmh.2009.09-0104. [DOI] [PubMed] [Google Scholar]
- 50.MacMillan K, Enscore RE, Ogen-Odoi A, Borchert JN, Babi N, Amatre G, Atiku LA, Mead PS, Gage KL, Eisen RJ. 2011. Landscape and residential variables associated with plague-endemic villages in the West Nile region of Uganda. Am J Trop Med Hyg 84:435–442. doi: 10.4269/ajtmh.2011.10-0571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moore SM, Monaghan A, Borchert JN, Mpanga JT, Atiku LA, Boegler KA, Montenieri J, MacMillan K, Gage KL, Eisen RJ. 2015. Seasonal fluctuations of small mammal and flea communities in a Ugandan plague focus: evidence to implicate Arvicanthis niloticus and Crocidura spp. as key hosts in Yersinia pestis transmission. Parasit Vectors 8:1–15. doi: 10.1186/s13071-014-0616-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Laudisoit A, Leirs H, Makundi RH, Van Dongen S, Davis S, Neerinckx S, Deckers J, Libois R. 2007. Plague and the human flea. Emerg Infect Dis 13:687–693. doi: 10.3201/eid1305.061084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Makundi RH, Massawe AW, Mulungu LS, Katakweba A, Mbise TJ, Mgode G. 2008. Potential mammalian reservoirs in a bubonic plague outbreak focus in Mbulu District, northern Tanzania, in 2007. Mammalia 72:253–257. doi: 10.1515/MAMM.2008.038. [DOI] [Google Scholar]
- 54.Ziwa MH, Matee MI, Hang’ombe BM, Lyamuya EF, Kilonzo BS. 2014. Plague in Tanzania: an overview. Tanzan J Health Res 15:252–258. doi: 10.4314/thrb.v15i4.7. [DOI] [PubMed] [Google Scholar]
- 55.Hang'ombe BM, Nakamura I, Samui KL, Kaile D, Mweene AS, Kilonzo BS, Sawa H, Sugimoto C, Wren BW. 2012. Evidence of Yersinia pestis DNA from fleas in an endemic plague area of Zambia. BMC Res Notes 5:72. doi: 10.1186/1756-0500-5-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Njunwa KJ, Mwaiko GL, Kilonzo BS, Mhina JIK. 1989. Seasonal patterns of rodents, fleas and plague status in the Western Usambara Mountains, Tanzania. Med Vet Entomol 3:17–22. doi: 10.1111/j.1365-2915.1989.tb00469.x. [DOI] [PubMed] [Google Scholar]
- 57.Andrianaivoarimanana V, Kreppel K, Elissa N, Duplantier J-M, Carniel E, Rajerison M, Jambou R. 2013. Understanding the persistence of plague foci in Madagascar. PLoS Negl Trop Dis 7:e2382. doi: 10.1371/journal.pntd.0002382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Duplantier J-M, Duchemin J-B, Chanteau S, Carniel E. 2005. From the recent lessons of the Malagasy foci towards a global understanding of the factors involved in plague reemergence. Vet Res 36:437–453. doi: 10.1051/vetres:2005007. [DOI] [PubMed] [Google Scholar]
- 59.Randremanana R, Andrianaivoarimanana V, Nikolay B, Ramasindrazana B, Paireau J, ten Bosch QA, Rakotondramanga JM, Rahajandraibe S, Rahelinirina S, Rakotomanana F, Rakotoarimanana FM, Randriamampionona LB, Razafimbia V, De Dieu Randria MJ, Raberahona M, Mikaty G, Le Guern A-S, Rakotonjanabelo LA, Ndiaye CF, Rasolofo V, Bertherat E, Ratsitorahina M, Cauchemez S, Baril L, Spiegel A, Rajerison M. 2019. Epidemiological characteristics of an urban plague epidemic in Madagascar, August–November, 2017: an outbreak report. Lancet Infect Dis 19:537–545. doi: 10.1016/S1473-3099(18)30730-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tsuzuki S, Lee H, Miura F, Chan YH, Jung S-M, Akhmetzhanov AR, Nishiura H. 2017. Dynamics of the pneumonic plague epidemic in Madagascar, August to October 2017. Euro Surveillance 22:17-00710. doi: 10.2807/1560-7917.ES.2017.22.46.17-00710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Andrianaivoarimanana V, Piola P, Wagner DM, Rakotomanana F, Maheriniaina V, Andrianalimanana S, Chanteau S, Rahalison L, Ratsitorahina M, Rajerison M. 2019. Trends of human plague, Madagascar, 1998–2016. Emerg Infect Dis 25:220–228. doi: 10.3201/eid2502.171974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bertherat E, Bekhoucha S, Chougrani S, Razik F, Duchemin JB, Houti L, Deharib L, Fayolle C, Makrerougrass B, Dali-Yahia R, Bellal R, Belhabri L, Chaieb A, Tikhomirov E, Carniel E. 2007. Plague reappearance in Algeria after 50 years, 2003. Emerg Infect Dis 13:1459–1462. doi: 10.3201/eid1310.070284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cabanel N, Leclercq A, Chenal-Francisque V, Annajar B, Rajerison M, Bekkhoucha S, Bertherat E, Carniel E. 2013. Plague outbreak in Libya, 2009, unrelated to plague in Algeria. Emerg Infect Dis 19:230–236. doi: 10.3201/eid1902.121031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Riehm JM, Vergnaud G, Kiefer D, Damdindorj T, Dashdavaa O, Khurelsukh T, Zöller L, Wölfel R, Le Flèche P, Scholz HC. 2012. Yersinia pestis lineages in Mongolia. PLoS One 7:e30624. doi: 10.1371/journal.pone.0030624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Galdan B, Baatar U, Molotov B, Dashdavaa O. 2010. Plague in Mongolia. Vector Borne Zoonotic Dis 10:69–75. doi: 10.1089/vbz.2009.0047. [DOI] [PubMed] [Google Scholar]
- 66.Eroshenko GA, Nosov NY, Krasnov YM, Oglodin YG, Kukleva LM, Guseva NP, Kuznetsov AA, Abdikarimov ST, Dzhaparova AK, Kutyrev VV. 2017. Yersinia pestis strains of ancient phylogenetic branch 0.ANT are widely spread in the high-mountain plague foci of Kyrgyzstan. PLoS One 12:e0187230. doi: 10.1371/journal.pone.0187230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Anisimov AP. 2002. Yersinia pestis factors, assuring circulation and maintenance of the plague pathogen in natural foci ecosystems. Report 1. Mol Gen Mikrobiol Virusol 2002:3–23. (In Russian.) [PubMed] [Google Scholar]
- 68.Anisimov AP. 2002. Factors of Yersinia pestis providing circulation and persistence of plague pathogen in ecosystems of natural foci. Communication 2. Mol Gen Mikrobiol Virusol 2002:3–11. (In Russian.) [PubMed] [Google Scholar]
- 69.Cully JF, Barnes AM, Quan TJ, Maupln G. 1997. Dynamics of plague in a Gunnison’s prairie dog colony complex from New Mexico. J Wildl Dis 33:706–719. doi: 10.7589/0090-3558-33.4.706. [DOI] [PubMed] [Google Scholar]
- 70.Salkeld DJ, Eisen RJ, Stapp P, Wilder AP, Lowell J, Tripp DW, Albertson D, Antolin MF. 2007. The potential role of swift foxes (Vulpes velox) and their fleas in plague outbreaks in prairie dogs. J Wildl Dis 43:425–431. doi: 10.7589/0090-3558-43.3.425. [DOI] [PubMed] [Google Scholar]
- 71.Neerinckx SB, Peterson AT, Gulinck H, Deckers J, Leirs H. 2008. Geographic distribution and ecological niche of plague in sub-Saharan Africa. Int J Health Geogr 7:54. doi: 10.1186/1476-072X-7-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Parmenter RR, Yadav EP, Parmenter CA, Ettestad P, Gage KL. 1999. Incidence of plague associated with increased winter-spring precipitation in New Mexico. Am J Trop Med Hyg 61:814–821. doi: 10.4269/ajtmh.1999.61.814. [DOI] [PubMed] [Google Scholar]
- 73.Ettestad P, Targhetta J, Brown T, Cheek J, Gage KL, Bueno R, Enscore RE, Eisen RJ, Montenieri JA, Biggerstaff BJ, Reynolds PJ. 2007. Residence-linked human plague in New Mexico: a habitat-suitability model. Am J Trop Med Hyg 77:121–125. doi: 10.4269/ajtmh.2007.77.121. [DOI] [PubMed] [Google Scholar]
- 74.Ben-Ari T, Neerinckx S, Agier L, Cazelles B, Xu L, Zhang Z, Fang X, Wang S, Liu Q, Stenseth NC. 2012. Identification of Chinese plague foci from long-term epidemiological data. Proc Natl Acad Sci U S A 109:8196–8201. doi: 10.1073/pnas.1110585109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pham HV, Dang DT, Tran Minh NN, Nguyen ND, Nguyen TV. 2009. Correlates of environmental factors and human plague: an ecological study in Vietnam. Int J Epidemiol 38:1634–1641. doi: 10.1093/ije/dyp244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Barbieri R, Texier G, Keller C, Drancourt M. 2020. Soil salinity and aridity specify plague foci in the United States of America. Sci Rep 10:6186. doi: 10.1038/s41598-020-63211-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Malek MA, Bitam I, Levasseur A, Terras J, Gaudart J, Azza S, Flaudrops C, Robert C, Raoult D, Drancourt M. 2017. Yersinia pestis halotolerance illuminates plague reservoirs. Sci Rep 7:40022. doi: 10.1038/srep40022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chu C. 2013. Combating nuisance: sanitation, regulation, and the politics of property in colonial Hong Kong, p 17–36. In Peckham R, Pomfret DM (ed), Imperial contagions: medicine, hygiene, and cultures of planning in Asia. Hong Kong University Press, Hong Kong. [Google Scholar]
- 79.Lynteris C. 2017. A ‘suitable soil’: plague’s urban breeding grounds at the dawn of the third pandemic. Med Hist 61:343–357. doi: 10.1017/mdh.2017.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yersin A. 1894. La peste bubonique à Hong Kong. Ann Inst Pasteur 8:662–667. [Google Scholar]
- 81.Farrar R. 1902. Plague as a soil infection. BMJ 2:454–456. [Google Scholar]
- 82.Gordon-Tucker EF. 1904. Plague: a soil disease. Ind Med Gaz 39:316. [PMC free article] [PubMed] [Google Scholar]
- 83.Karimi Y. 1963. Natural preservation of plague in soil. Bull Soc Pathol Exot Filiales 56:1183–1186. (In French.) [PubMed] [Google Scholar]
- 84.Eisen RJ, Petersen JM, Higgins CL, Wong D, Levy CE, Mead PS, Schriefer ME, Griffith KS, Gage KL, Beard CB. 2008. Persistence of Yersinia pestis in soil under natural conditions. Emerg Infect Dis 14:941–943. doi: 10.3201/eid1406.080029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mollaret HH. 1963. Conservation expérimentale de la peste dans le sol. Bull Soc Pathol Exot Filiales 56:1168–1182. [PubMed] [Google Scholar]
- 86.Ayyadurai S, Houhamdi L, Lepidi H, Nappez C, Raoult D, Drancourt M. 2008. Long-term persistence of virulent Yersinia pestis in soil. Microbiology (Reading) 154:2865–2871. doi: 10.1099/mic.0.2007/016154-0. [DOI] [PubMed] [Google Scholar]
- 87.Rose LJ, Donlan R, Banerjee SN, Arduino MJ. 2003. Survival of Yersinia pestis on environmental surfaces. Appl Environ Microbiol 69:2166–2171. doi: 10.1128/AEM.69.4.2166-2171.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pawlowski DR, Metzger DJ, Raslawsky A, Howlett A, Siebert G, Karalus RJ, Garrett S, Whitehouse CA. 2011. Entry of Yersinia pestis into the viable but nonculturable state in a low-temperature tap water microcosm. PLoS One 6:e17585. doi: 10.1371/journal.pone.0017585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bazanova LP, Maevskiĭ MP, Khabarov AV. 1997. An experimental study of the possibility for the preservation of the causative agent of plague in the nest substrate of the long-tailed suslik. Med Parazitol (Mosk) Oct-Dec 1997:37–39. (In Russian.) [PubMed] [Google Scholar]
- 90.Easterday WR, Kausrud KL, Star B, Heier L, Haley BJ, Ageyev V, Colwell RR, Stenseth NC. 2012. An additional step in the transmission of Yersinia pestis? ISME J 6:231–236. doi: 10.1038/ismej.2011.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Benavides-Montaño JA, Vadyvaloo V. 2017. Yersinia pestis resists predation by Acanthamoeba castellanii and exhibits prolonged intracellular survival. Appl Environ Microbiol 83:e00593-17. doi: 10.1128/AEM.00593-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Markman DW, Antolin MF, Bowen RA, Wheat WH, Woods M, Gonzalez-Juarrero M, Jackson M. 2018. Yersinia pestis survival and replication in potential ameba reservoir. Emerg Infect Dis 24:294–302. doi: 10.3201/eid2402.171065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gage KL, Kosoy MY. 2005. Natural history of plague: perspectives from more than a century of research. Annu Rev Entomol 50:505–528. doi: 10.1146/annurev.ento.50.071803.130337. [DOI] [PubMed] [Google Scholar]
- 94.Domaradsky IV. 1999. Is not plague a “protonosis”? (the role of Protozoa in the epizootiology of plague). Med Parazitol (Mosk) Apr-Jun 1999:10–13. (In Russian.) [PubMed] [Google Scholar]
- 95.Litvin V, Pushkareva VI, Emel’ianenko EN. 2004. Biocenosis of the natural foci of sapronotic infections (the results of 15-year observations). Zh Mikrobiol Epidemiol Immunobiol Jul-Aug 2004:102–108. (In Russian.) [PubMed] [Google Scholar]
- 96.Koshel’ EI, Anisimova LV, Novichkova LA, Vidyaeva NA, Guseva NP, Eroshenko GA, Kutyrev VV. 2015. A study on the taxonomy of soil amoebas from Caspian plague foci based on an analysis of ribosomal operon sequences. Genetika 51:39–45. (In Russian.) doi: 10.1134/S1022795415010056. [DOI] [PubMed] [Google Scholar]
- 97.Rebeil R, Jarrett CO, Driver JD, Ernst RK, Oyston PC, Hinnebusch BJ. 2013. Induction of the Yersinia pestis PhoP-PhoQ regulatory system in the flea and its role in producing a transmissible infection. J Bacteriol 195:1920–1930. doi: 10.1128/JB.02000-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Viboud GI, Bliska JB. 2005. Yersinia outer proteins: role in modulation of host cell signaling responses and pathogenesis. Annu Rev Microbiol 59:69–89. doi: 10.1146/annurev.micro.59.030804.121320. [DOI] [PubMed] [Google Scholar]
- 99.Pollitzer R, World Health Organization. 1954. Plague. World Health Organization, Geneva, Switzerland. https://apps.who.int/iris/handle/10665/41628. [Google Scholar]
- 100.Thomas RE, Beard ML, Quan TJ, Carter LG, Barnes AM, Hopla CE. 1989. Experimentally induced plague infection in the northern grasshopper mouse (Onychomys leucogaster) acquired by consumption of infected prey. J Wildl Dis 25:477–480. doi: 10.7589/0090-3558-25.4.477. [DOI] [PubMed] [Google Scholar]
- 101.Biggins DE, Kosoy MY. 2001. Influences of introduced plague on North American mammals: implications from ecology of plague in Asia. J Mammal 82:906–916. doi:. [DOI] [Google Scholar]
- 102.Schmid BV, Jesse M, Wilschut LI, Viljugrein H, Heesterbeek JAP. 2012. Local persistence and extinction of plague in a metapopulation of great gerbil burrows, Kazakhstan. Epidemics 4:211–218. doi: 10.1016/j.epidem.2012.12.003. [DOI] [PubMed] [Google Scholar]
- 103.Shepherd AJ, Leman PA, Hummitzsch DE. 1986. Experimental plague infection in South African wild rodents. J Hyg (Lond) 96:171–183. doi: 10.1017/S0022172400065943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Eisen RJ, Holmes JL, Schotthoefer AM, Vetter SM, Montenieri JA, Gage KL. 2008. Demonstration of early-phase transmission of Yersinia pestis by the mouse flea, Aetheca wagneri (Siphonaptera: Ceratophylidae), and implications for the role of deer mice as enzootic reservoirs. J Med Entomol 45:1160–1164. doi: 10.1093/jmedent/45.6.1160. [DOI] [PubMed] [Google Scholar]
- 105.Lewnard JA, Townsend JP. 2016. Climatic and evolutionary drivers of phase shifts in the plague epidemics of colonial India. Proc Natl Acad Sci U S A 113:14601–14608. doi: 10.1073/pnas.1604985113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hubbert WT, Goldenberg MI. 1970. Natural resistance to plague: genetic basis in the vole (Microtus californicus). Am J Trop Med Hyg 19:1015–1019. doi: 10.4269/ajtmh.1970.19.1015. [DOI] [PubMed] [Google Scholar]
- 107.Rust JH, Cavanaugh DC, O'Shita R, Marshall JD. 1971. The role of domestic animals in the epidemiology of plague. I. Experimental infection of dogs and cats. J Infect Dis 124:522–526. doi: 10.1093/infdis/124.5.522. [DOI] [PubMed] [Google Scholar]
- 108.Butler T. 2009. Plague into the 21st century. Clin Infect Dis 49:736–742. doi: 10.1086/604718. [DOI] [PubMed] [Google Scholar]
- 109.Osei-Owusu P, Charlton TM, Kim HK, Missiakas D, Schneewind O. 2019. FPR1 is the plague receptor on host immune cells. Nature 574:57–62. doi: 10.1038/s41586-019-1570-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Pauli JN, Buskirk SW, Williams ES, Edwards WH. 2006. A plague epizootic in the black-tailed prairie dog (Cynomys ludovicianus). J Wildl Dis 42:74–80. doi: 10.7589/0090-3558-42.1.74. [DOI] [PubMed] [Google Scholar]
- 111.Engelthaler DM, Gage KL. 2000. Quantities of Yersinia pestis in fleas (Siphonaptera: Pulicidae, Ceratophyllidae, and Hystrichopsyllidae) collected from areas of known or suspected plague activity. J Med Entomol 37:422–426. doi: 10.1093/jmedent/37.3.422. [DOI] [PubMed] [Google Scholar]
- 112.Fedorov VN. 1960. Plague in camels and its prevention in the USSR. Bull World Health Organ 23:275–281. [PMC free article] [PubMed] [Google Scholar]
- 113.Nikanorov SM. 1922. On the question of the rôle of camels in the epidemiology of plague in Astrachan. Herald Microbiol Epidemiol, vol 1, part 2. [Google Scholar]
- 114.Mollaret HH, Karimi Y, Eftekhari M, Baltazard M. 1963. La peste de fouissement. Bull Soc Pathol Exot 56:1186–1193. [PubMed] [Google Scholar]
- 115.Boegler KA, Graham CB, Montenieri JA, MacMillan K, Holmes JL, Petersen JM, Gage KL, Eisen RJ. 2012. Evaluation of the infectiousness to mice of soil contaminated with Yersinia pestis-infected blood. Vector Borne Zoonotic Dis 12:948–952. doi: 10.1089/vbz.2012.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Rivkus YZ, Mitropolsky OV, Bochkaroff VM, Blummer AG, Bochkaroff SV. 1993. Desert vegetation as a possible component in plague parasitocoenosis. Bull Mosk Obschestva Ispyt Prirody 98:3–13. [Google Scholar]
- 117.Rivkus I, Bochkarev VM. 2000. The colonization of plants by Yersinia pestis EV in an experiment. Zh Mikrobiol Epidemiol Immunobiol May–Jun 2000:40–41. (In Russian.) [PubMed] [Google Scholar]
- 118.Jones RT, Vetter SM, Gage KL. 2013. Exposing laboratory-reared fleas to soil and wild flea feces increases transmission of Yersinia pestis. Am J Trop Med Hyg 89:784–787. doi: 10.4269/ajtmh.13-0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hinnebusch BJ, Jarrett CO, Bland DM. 2017. “Fleaing” the plague: adaptations of Yersinia pestis to its insect vector that lead to transmission. Annu Rev Microbiol 71:215–232. doi: 10.1146/annurev-micro-090816-093521. [DOI] [PubMed] [Google Scholar]
- 120.Konnonov NP, Popov NV, Velichko LN, Kniazeva TV. 2009. Phenomenon of Yersinia pestis biofilm formation in flea organism. Parazitologiia 43:330–337. [PubMed] [Google Scholar]
- 121.Butler T, Fu YS, Furman L, Almeida C, Almeida A. 1982. Experimental Yersinia pestis infection in rodents after intragastric inoculation and ingestion of bacteria. Infect Immun 36:1160–1167. doi: 10.1128/IAI.36.3.1160-1167.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hinnebusch BJ. 2005. The evolution of flea-borne transmission in Yersinia pestis. Curr Issues Mol Biol 7:197–212. [PubMed] [Google Scholar]
- 123.Kislichkina AA, Platonov ME, Vagaiskaya AS, Bogun AG, Dentovskaya SV, Anisimov AP. 2019. Rational taxonomy of Yersinia pestis. Mol Genet Microbiol Virol 34:110–117. doi: 10.3103/S0891416819020058. [DOI] [Google Scholar]
- 124.Ogata M. 1897. Ueber die Pestepidemie in Formosa. Centralbl Bakteriol Parasitenkd 21:774. [Google Scholar]
- 125.Simond M, Godley ML, Mouriquand PD. 1998. Paul-Louis Simond and his discovery of plague transmission by rat fleas: a centenary. J R Soc Med 91:101–104. doi: 10.1177/014107689809100219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Burroughs AL. 1947. Sylvatic plague studies. The vector efficiency of nine species of fleas compared with Xenopsylla cheopis. J Hyg (Lond) 45:371–396. doi: 10.1017/S0022172400014042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Bibikova VA. 1977. Contemporary views on the interrelationships between fleas and the pathogens of human and animal diseases. Annu Rev Entomol 22:23–32. doi: 10.1146/annurev.en.22.010177.000323. [DOI] [PubMed] [Google Scholar]
- 128.Eisen RJ, Wilder AP, Bearden SW, Montenieri JA, Gage KL. 2007. Early-phase transmission of Yersinia pestis by unblocked Xenopsylla cheopis (Siphonaptera: Pulicidae) is as efficient as transmission by blocked fleas. J Med Entomol 44:678–682. doi: 10.1603/0022-2585(2007)44[678:ETOYPB]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 129.Wilder AP, Eisen RJ, Bearden SW, Montenieri JA, Tripp DW, Brinkerhoff RJ, Gage KL, Antolin MF. 2007. Transmission efficiency of two flea species (Oropsylla tuberculata cynomuris and Oropsylla hirsuta) involved in plague epizootics among prairie dogs. EcoHealth 5:205. doi: 10.1007/s10393-008-0165-1. [DOI] [PubMed] [Google Scholar]
- 130.Eisen RJ, Bearden SW, Wilder AP, Montenieri JA, Antolin MF, Gage KL. 2006. Early-phase transmission of Yersinia pestis by unblocked fleas as a mechanism explaining rapidly spreading plague epizootics. Proc Natl Acad Sci U S A 103:15380–15385. doi: 10.1073/pnas.0606831103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Bland DM, Hinnebusch BJ. 2016. Feeding behavior modulates biofilm-mediated transmission of Yersinia pestis by the cat flea, Ctenocephalides felis. PLoS Negl Trop Dis 10:e0004413. doi: 10.1371/journal.pntd.0004413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lorange EA, Race BL, Sebbane F, Joseph Hinnebusch B. 2005. Poor vector competence of fleas and the evolution of hypervirulence in Yersinia pestis. J Infect Dis 191:1907–1912. doi: 10.1086/429931. [DOI] [PubMed] [Google Scholar]
- 133.Blanc G, Baltazard M. 1941. Plague infection of P. irritans. C R Acad Sci 213:813–816. [Google Scholar]
- 134.Schotthoefer AM, Bearden SW, Vetter SM, Holmes J, Montenieri JA, Graham CB, Woods ME, Eisen RJ, Gage KL. 2011. Effects of temperature on early-phase transmission of Yersina pestis by the flea, Xenopsylla cheopis. J Med Entomol 48:411–417. doi: 10.1603/ME10155. [DOI] [PubMed] [Google Scholar]
- 135.Bland DM, Jarrett CO, Bosio CF, Hinnebusch BJ. 2018. Infectious blood source alters early foregut infection and regurgitative transmission of Yersinia pestis by rodent fleas. PLoS Pathog 14:e1006859. doi: 10.1371/journal.ppat.1006859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Bacot AW, Martin CJ. 1914. LXVII. Observations on the mechanism of the transmission of plague by fleas. J Hyg (Lond) 13:423–439. [PMC free article] [PubMed] [Google Scholar]
- 137.Bacot AW. 1915. LXXXI. Further notes on the mechanism of the transmission of plague by fleas. J Hyg (Lond) 14:774–776.3. [PMC free article] [PubMed] [Google Scholar]
- 138.Eskey CR, Haas VH. 1939. Plague in the western part of the United States. Public Health Rep (1896–1970) 54:1467–1481. doi: 10.2307/4582984. [DOI] [Google Scholar]
- 139.Cavanaugh DC. 1971. Specific effect of temperature upon transmission of the plague bacillus by the oriental rat flea, Xenopsylla cheopis. Am J Trop Med Hyg 20:264–273. doi: 10.4269/ajtmh.1971.20.264. [DOI] [PubMed] [Google Scholar]
- 140.Kreppel KS, Telfer S, Rajerison M, Morse A, Baylis M. 2016. Effect of temperature and relative humidity on the development times and survival of Synopsyllus fonquerniei and Xenopsylla cheopis, the flea vectors of plague in Madagascar. Parasit Vectors 9:82. doi: 10.1186/s13071-016-1366-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Hinnebusch BJ, Fischer ER, Schwan TG. 1998. Evaluation of the role of the Yersinia pestis plasminogen activator and other plasmid-encoded factors in temperature-dependent blockage of the flea. J Infect Dis 178:1406–1415. doi: 10.1086/314456. [DOI] [PubMed] [Google Scholar]
- 142.Kartman L. 1969. Effect of differences in ambient temperature upon the fate of Pasteurella pestis in Xenopsylla cheopis. Trans R Soc Trop Med Hyg 63:71–75. doi: 10.1016/0035-9203(69)90068-6. [DOI] [PubMed] [Google Scholar]
- 143.Krasnov BR, Khokhlova IS, Fielden LJ, Burdelova NV. 2001. Development rates of two Xenopsylla flea species in relation to air temperature and humidity. Med Vet Entomol 15:249–258. doi: 10.1046/j.0269-283x.2001.00295.x. [DOI] [PubMed] [Google Scholar]
- 144.Eads DA, Hoogland JL. 2017. Precipitation, climate change, and parasitism of prairie dogs by fleas that transmit plague. J Parasitol 103:309–320. doi: 10.1645/16-195. [DOI] [PubMed] [Google Scholar]
- 145.Wimsatt J, Biggins DE. 2009. A review of plague persistence with special emphasis on fleas. J Vector Borne Dis 46:85–99. [PubMed] [Google Scholar]
- 146.Vetter SM, Gage KL, Borchert JN, Enscore RE, Wilder AP, Amatre G, Atiku LA, Montenieri JA, Eisen RJ, Van Wyk K, Holmes JL, Babi N, Bearden SW. 2008. Early-phase transmission of Yersinia pestis by cat fleas (Ctenocephalides felis) and their potential role as vectors in a plague-endemic region of Uganda. Am J Trop Med Hyg 78:949–956. doi: 10.4269/ajtmh.2008.78.949. [DOI] [PubMed] [Google Scholar]
- 147.Bazanova LP, Zhovtyĭ IF, Maevskiĭ MP, Klimov VT, Popkov AF. 1991. The seasonal dynamics of blocking in the flea Citellophorus tesquorum altaicus from the Tuva natural plague focus. Med Parazitol (Mosk) Jan-Feb 1991:24–26. (In Russian.) [PubMed] [Google Scholar]
- 148.Bazanova LP, Voronova GA, Tokmakova EG. 2000. Differences in the blocking of the proventriculus in male and female Xenopsylla cheopis (Siphonaptera: Pulicidae). Parazitologiia 34:56–59. (In Russian.) [PubMed] [Google Scholar]
- 149.Wong D, Wild MA, Walburger MA, Higgins CL, Callahan M, Czarnecki LA, Lawaczeck EW, Levy CE, Patterson JG, Sunenshine R, Adem P, Paddock CD, Zaki SR, Petersen JM, Schriefer ME, Eisen RJ, Gage KL, Griffith KS, Weber IB, Spraker TR, Mead PS. 2009. Primary pneumonic plague contracted from a mountain lion carcass. Clin Infect Dis 49:e33–e38. doi: 10.1086/600818. [DOI] [PubMed] [Google Scholar]
- 150.Von Reyn CF, Barnes AM, Weber NS, Quan T, Dean WJ. 1976. Bubonic plague from direct exposure to a naturally infected wild coyote. Am J Trop Med Hyg 25:626–629. doi: 10.4269/ajtmh.1976.25.626. [DOI] [PubMed] [Google Scholar]
- 151.Arbaji A, Kharabsheh S, Al-Azab S, Al-Kayed M, Amr ZS, Abu Baker M, Chu MC. 2005. A 12-case outbreak of pharyngeal plague following the consumption of camel meat. Ann Trop Med Parasitol 99:789–793. doi: 10.1179/136485905X65161. [DOI] [PubMed] [Google Scholar]
- 152.Saeed AAB, Al-Hamdan NA, Fontaine RE. 2005. Plague from eating raw camel liver. Emerg Infect Dis 11:1456–1457. doi: 10.3201/eid1109.050081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Christie AB, Chen TH, Elberg SS. 1980. Plague in camels and goats: their role in human epidemics. J Infect Dis 141:724–726. doi: 10.1093/infdis/141.6.724. [DOI] [PubMed] [Google Scholar]
- 154.Jones SD, Amramina AA. 2018. Entangled histories of plague ecology in Russia and the USSR. Hist Philos Life Sci 40:49. doi: 10.1007/s40656-018-0220-3. [DOI] [PubMed] [Google Scholar]
- 155.Dai R, Wei B, Xiong H, Yang X, Peng Y, He J, Jin J, Wang Y, Zha X, Zhang Z, Liang Y, Zhang Q, Xu J, Wang Z, Li W. 2018. Human plague associated with Tibetan sheep originates in marmots. PLoS Negl Trop Dis 12:e0006635. doi: 10.1371/journal.pntd.0006635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Gabastou JM, Proaño J, Vimos A, Jaramillo G, Hayes E, Gage K, Chu M, Guarner J, Zaki S, Bowers J, Guillemard C, Tamayo H, Ruiz A. 2000. An outbreak of plague including cases with probable pneumonic infection, Ecuador, 1998. Trans R Soc Trop Med Hyg 94:387–391. doi: 10.1016/S0035-9203(00)90114-7. [DOI] [PubMed] [Google Scholar]
- 157.Von Reyn CF, Barnes AM, Weber NS, Hodgin UG. 1976. Bubonic plague from exposure to a rabbit: a documented case, and a review of rabbit-associated plague cases in the United States. Am J Epidemiol 104:81–87. doi: 10.1093/oxfordjournals.aje.a112276. [DOI] [PubMed] [Google Scholar]
- 158.Runfola JK, House J, Miller L, Colton L, Hite D, Hawley A, Mead P, Schriefer M, Petersen J, Casaceli C, Erlandson KM, Foster C, Pabilonia KL, Mason G, Douglas JM, Centers for Disease Control and Prevention (CDC). 2015. Outbreak of human pneumonic plague with dog-to-human and possible human-to-human transmission—Colorado, June–July 2014. MMWR Morb Mortal Wkly Rep 64:429–434. [PMC free article] [PubMed] [Google Scholar]
- 159.Dai R, Qi M, Xiong H, Yang X, He J, Zhang Z, Yang H, Jin J, Li X, Xin Y, Yang Y, Li C, Li Z, Xu J, Wang Z, Li W, Wei B. 2019. Serological epidemiological investigation of Tibetan sheep (Ovis aries) plague in Qinghai, China. Vector-Borne Zoonotic Dis 19:3–7. doi: 10.1089/vbz.2017.2257. [DOI] [PubMed] [Google Scholar]
- 160.Ge P, Xi J, Ding J, Jin F, Zhang H, Guo L, Zhang J, Li J, Gan Z, Wu B, Liang J, Wang X, Wang X. 2015. Primary case of human pneumonic plague occurring in a Himalayan marmot natural focus area Gansu Province, China. Int J Infect Dis 33:67–70. doi: 10.1016/j.ijid.2014.12.044. [DOI] [PubMed] [Google Scholar]
- 161.Schaffer PA, Brault SA, Hershkowitz C, Harris L, Dowers K, House J, Aboellail TA, Morley PS, Daniels JB. 2019. Pneumonic plague in a dog and widespread potential human exposure in a veterinary hospital, United States. Emerg Infect Dis 25:800–803. doi: 10.3201/eid2504.181195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Werner SB, Weidmer CE, Nelson BC, Nygaard GS, Goethals RM, Poland JD. 1984. Primary plague pneumonia contracted from a domestic cat at South Lake Tahoe, Calif. JAMA 251:929–931. doi: 10.1001/jama.1984.03340310043018. [DOI] [PubMed] [Google Scholar]
- 163.Gage KL, Dennis DT, Orloski KA, Ettestad P, Brown TL, Reynolds PJ, Pape WJ, Fritz CL, Carter LG, Stein JD. 2000. Cases of cat-associated human plague in the Western US, 1977–1998. Clin Infect Dis 30:893–900. doi: 10.1086/313804. [DOI] [PubMed] [Google Scholar]
- 164.Doll JM, Zeitz PS, Ettestad P, Bucholtz AL, Davis T, Gage K. 1994. Cat-transmitted fatal pneumonic plague in a person who traveled from Colorado to Arizona. Am J Trop Med Hyg 51:109–114. doi: 10.4269/ajtmh.1994.51.109. [DOI] [PubMed] [Google Scholar]
- 165.Wang H, Cui Y, Wang Z, Wang X, Guo Z, Yan Y, Li C, Cui B, Xiao X, Yang Y, Qi Z, Wang G, Wei B, Yu S, He D, Chen H, Chen G, Song Y, Yang R. 2011. A dog-associated primary pneumonic plague in Qinghai Province, China. Clin Infect Dis 52:185–190. doi: 10.1093/cid/ciq107. [DOI] [PubMed] [Google Scholar]
- 166.Li Y, Li D, Shao H, Li H, Han Y. 2016. Plague in China 2014—all sporadic case report of pneumonic plague. BMC Infect Dis 16:85. doi: 10.1186/s12879-016-1403-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Melman SD, Ettestad PE, VinHatton ES, Ragsdale JM, Takacs N, Onischuk LM, Leonard PM, Master SS, Lucero VS, Kingry LC, Petersen JM. 2018. Human case of bubonic plague resulting from the bite of a wild Gunnison’s prairie dog during translocation from a plague‐endemic area. Zoonoses Public Health 65:e254–e258. doi: 10.1111/zph.12419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Lammens H. 2013. Islam: beliefs and institutions. Routledge, Abingdon, United Kingdom. [Google Scholar]
- 169.Porto-Fett AC, Juneja VK, Tamplin ML, Luchansky JB. 2009. Validation of cooking times and temperatures for thermal inactivation of Yersinia pestis strains KIM5 and CDC-A1122 in irradiated ground beef. J Food Prot 72:564–571. doi: 10.4315/0362-028X-72.3.564. [DOI] [PubMed] [Google Scholar]
- 170.Christakos G, Olea RA, Yu H-L. 2007. Recent results on the spatiotemporal modelling and comparative analysis of Black Death and bubonic plague epidemics. Public Health 121:700–720. doi: 10.1016/j.puhe.2006.12.011. [DOI] [PubMed] [Google Scholar]
- 171.Walløe L. 2008. Medieval and modern bubonic plague: some clinical continuities. Med Hist Suppl 2008:59–73. doi: 10.1017/S0025727300072094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Hufthammer AK, Walløe L. 2013. Rats cannot have been intermediate hosts for Yersinia pestis during medieval plague epidemics in Northern Europe. J Archaeol Sci 40:1752–1759. doi: 10.1016/j.jas.2012.12.007. [DOI] [Google Scholar]
- 173.Benedictow OJ. 2016. The Black Death and later plague epidemics in the Scandinavian countries, p 307–343. De Gruyter, Berlin, Germany. [Google Scholar]
- 174.Karlsson G. 1996. Plague without rats: the case of fifteenth-century Iceland. J Mediev Hist 22:263–284. doi: 10.1016/S0304-4181(96)00017-6. [DOI] [Google Scholar]
- 175.Audoin-Rouzeau F, Vigne J-D. 1994. La colonisation de l’Europe par le rat noir (Rattus rattus). Rev Paléobiol 13:125–145. [Google Scholar]
- 176.Brothen JA. 1996. Population decline and plague in late medieval Norway. Ann Demogr Hist (Paris) 1996:137–149. doi: 10.3406/adh.1996.1915. [DOI] [PubMed] [Google Scholar]
- 177.Panagiotakopulu E, Buckland PC. 2017. A thousand bites—insect introductions and late Holocene environments. Quat Sci Rev 156:23–35. doi: 10.1016/j.quascirev.2016.11.014. [DOI] [Google Scholar]
- 178.Gamsa M. 2006. The epidemic of pneumonic plague in Manchuria 1910–1911. Past Present 190:147–183. doi: 10.1093/pastj/gtj001. [DOI] [Google Scholar]
- 179.Nishiura H. 2006. Epidemiology of a primary pneumonic plague in Kantoshu, Manchuria, from 1910 to 1911: statistical analysis of individual records collected by the Japanese Empire. Int J Epidemiol 35:1059–1065. doi: 10.1093/ije/dyl091. [DOI] [PubMed] [Google Scholar]
- 180.Chernin E. 1989. Richard Pearson Strong and the Manchurian epidemic of pneumonic plague, 1910–1911. J Hist Med Allied Sci 44:296–319. doi: 10.1093/jhmas/44.3.296. [DOI] [PubMed] [Google Scholar]
- 181.Viseltear AJ. 1974. The pneumonic plague epidemic of 1924 in Los Angeles. Yale J Biol Med 47:40–54. [PMC free article] [PubMed] [Google Scholar]
- 182.Kool JL, Weinstein RA. 2005. Risk of person-to-person transmission of pneumonic plague. Clin Infect Dis 40:1166–1172. doi: 10.1086/428617. [DOI] [PubMed] [Google Scholar]
- 183.Ramasindrazana B, Andrianaivoarimanana V, Rakotondramanga JM, Birdsell DN, Ratsitorahina M, Rajerison M. 2017. Pneumonic plague transmission, Moramanga, Madagascar, 2015. Emerg Infect Dis 23:521–524. doi: 10.3201/eid2303.161406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Begier EM, Asiki G, Anywaine Z, Yockey B, Schriefer ME, Aleti P, Ogden-Odoi A, Staples JE, Sexton C, Bearden SW, Kool JL. 2006. Pneumonic plague cluster, Uganda, 2004. Emerg Infect Dis 12:460–467. doi: 10.3201/eid1203.051051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Gani R, Leach S. 2004. Epidemiologic determinants for modeling pneumonic plague outbreaks. Emerg Infect Dis 10:608–614. doi: 10.3201/eid1004.030509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Dean KR, Krauer F, Walløe L, Lingjærde OC, Bramanti B, Stenseth NC, Schmid BV. 2018. Human ectoparasites and the spread of plague in Europe during the Second Pandemic. Proc Natl Acad Sci U S A 115:1304–1309. doi: 10.1073/pnas.1715640115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Nicolle C. 1910. Recherches expérimentales sur le typhus exanthématique entreprises à l’Institut Pasteur de Tunis pendant l’année 1909. Ann Inst Pasteur, p 243–275. [Google Scholar]
- 188.Gross L. 1996. How Charles Nicolle of the Pasteur Institute discovered that epidemic typhus is transmitted by lice: reminiscences from my years at the Pasteur Institute in Paris. Proc Natl Acad Sci U S A 93:10539–10540. doi: 10.1073/pnas.93.20.10539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Raoult D. 2016. A personal view of how paleomicrobiology aids our understanding of the role of lice in plague pandemics. Microbiol Spectr 4:PoH-0001-2014. doi: 10.1128/microbiolspec.PoH-0001-2014. [DOI] [PubMed] [Google Scholar]
- 190.Tran T-N-N, Signoli M, Fozzati L, Aboudharam G, Raoult D, Drancourt M. 2011. High throughput, multiplexed pathogen detection authenticates plague waves in medieval Venice, Italy. PLoS One 6:e16735. doi: 10.1371/journal.pone.0016735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Tran T-N-N, Forestier CL, Drancourt M, Raoult D, Aboudharam G. 2011. Brief communication: co-detection of Bartonella quintana and Yersinia pestis in an 11th-15th burial site in Bondy, France. Am J Phys Anthropol 145:489–494. doi: 10.1002/ajpa.21510. [DOI] [PubMed] [Google Scholar]
- 192.Piarroux R, Abedi AA, Shako J-C, Kebela B, Karhemere S, Diatta G, Davoust B, Raoult D, Drancourt M. 2013. Plague epidemics and lice, Democratic Republic of the Congo. Emerg Infect Dis 19:505. doi: 10.3201/eid1903.121542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Drali R, Shako J-C, Davoust B, Diatta G, Raoult D. 2015. A new clade of African body and head lice infected by Bartonella quintana and Yersinia pestis—Democratic Republic of the Congo. Am J Trop Med Hyg 93:990–993. doi: 10.4269/ajtmh.14-0686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Chabaud A-G. 1947. Les arthropodes vecteurs de la peste bubonique (suite et fin). Ann Parasitol Hum Comp 22:357–379. doi: 10.1051/parasite/1947225357. [DOI] [Google Scholar]
- 195.Blanc G, Baltazard M. 1941. Recherches expérimentales sur la peste. L’infection du pou de l’homme. Pediculus corporis de Geer. C R Hebd Séances Acad Sci 213:e51. [Google Scholar]
- 196.Houhamdi L, Lepidi H, Drancourt M, Raoult D. 2006. Experimental model to evaluate the human body louse as a vector of plague. J Infect Dis 194:1589–1596. doi: 10.1086/508995. [DOI] [PubMed] [Google Scholar]
- 197.Baltazard M. 1960. Déclin et destin d’une maladie infectieuse: la peste. Bull World Health Organ 23:247–262. [PMC free article] [PubMed] [Google Scholar]
- 198.Ayyadurai S, Sebbane F, Raoult D, Drancourt M. 2010. Body lice, Yersinia pestis orientalis, and black death. Emerg Infect Dis 16:892–893. doi: 10.3201/eid1605.091280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Raoult D, Roux V. 1999. The body louse as a vector of reemerging human diseases. Clin Infect Dis 29:888–911. doi: 10.1086/520454. [DOI] [PubMed] [Google Scholar]
- 200.Boutellis A, Abi-Rached L, Raoult D. 2014. The origin and distribution of human lice in the world. Infect Genet Evol 23:209–217. doi: 10.1016/j.meegid.2014.01.017. [DOI] [PubMed] [Google Scholar]
- 201.Delanoë P. 1932. L’importance de la puce de l’homme, Pulex irritans L., dans les épidémies de peste au Maroc. Bull Soc Pathol Exot 25:958–960. [Google Scholar]
- 202.Ratovonjato J, Rajerison M, Rahelinirina S, Boyer S. 2014. Yersinia pestis in Pulex irritans fleas during plague outbreak. Emerg Infect Dis 20:1414–1415. doi: 10.3201/eid2008.130629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Johnson TL, Hinnebusch BJ, Boegler KA, Graham CB, MacMillan K, Montenieri JA, Bearden SW, Gage KL, Eisen RJ. 2014. Yersinia murine toxin is not required for early-phase transmission of Yersinia pestis by Oropsylla montana (Siphonaptera: Ceratophyllidae) or Xenopsylla cheopis (Siphonaptera: Pulicidae). Microbiology (Reading) 160:2517–2525. doi: 10.1099/mic.0.082123-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Blanc G. 1956. Une opinion non conformiste sur le mode de transmission de la peste. Rev Hyg Med Soc 4:535–562. [PubMed] [Google Scholar]
- 205.Cohn SK. 2008. Epidemiology of the Black Death and successive waves of plague. Med Hist 52:74–100. doi: 10.1017/S0025727300072100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Laforce FM, Acharya IL, Stott G, Brachman PS, Kaufman AF, Clapp RF, Shah NK. 1971. Clinical and epidemiological observations on an outbreak of plague in Nepal. Bull World Health Organ 45:693. [PMC free article] [PubMed] [Google Scholar]
- 207.Sabeti PC, Walsh E, Schaffner SF, Varilly P, Fry B, Hutcheson HB, Cullen M, Mikkelsen TS, Roy J, Patterson N, Cooper R, Reich D, Altshuler D, O'Brien S, Lander ES. 2005. The case for selection at CCR5-Δ32. PLoS Biol 3:e378. doi: 10.1371/journal.pbio.0030378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Elvin SJ, Williamson ED, Scott JC, Smith JN, De Lema GP, Chilla S, Clapham P, Pfeffer K, Schlöndorff D, Luckow B. 2004. Evolutionary genetics: ambiguous role of CCR5 in Y. pestis infection. Nature 430:418–418. doi: 10.1038/nature02822. [DOI] [PubMed] [Google Scholar]
- 209.Mecsas J, Franklin G, Kuziel WA, Brubaker RR, Falkow S, Mosier DE. 2004. Evolutionary genetics: CCR5 mutation and plague protection. Nature 427:606–606. doi: 10.1038/427606a. [DOI] [PubMed] [Google Scholar]
- 210.Park YH, Remmers EF, Lee W, Ombrello AK, Chung LK, Shilei Z, Stone DL, Ivanov MI, Loeven NA, Barron KS, Hoffmann P, Nehrebecky M, Akkaya-Ulum YZ, Sag E, Balci-Peynircioglu B, Aksentijevich I, Gül A, Rotimi CN, Chen H, Bliska JB, Ozen S, Kastner DL, Shriner D, Chae JJ. 2020. Ancient familial Mediterranean fever mutations in human pyrin and resistance to Yersinia pestis. Nat Immunol 21:857–811. doi: 10.1038/s41590-020-0705-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Chicoyneau F. 1721. Relation de la peste de Marseille contenant ses syntomes, son prognostic, sa curation, & celle des bubons & des charbons. Donnée par messieurs Chicoyneau, Verny & Soullier. par ordre de messrs les comandant, gouverneur & echevins de la ville. https://books.google.fr/books?hl=fr&lr=&id=qE1FRF666bgC.
- 212.Friedewald VE. 2008. Clinical guide to bioweapons and chemical agents, p 167–171. Springer-Verlag, London, United Kingdom. [Google Scholar]
- 213.Nikiforov VV, Gao H, Zhou L, Anisimov A. 2016. Plague: clinics, diagnosis and treatment, p 293–312. In Yang R, Anisimov A (ed), Yersinia pestis: retrospective and perspective. Springer, Heidelberg, Germany. [DOI] [PubMed] [Google Scholar]
- 214.Pechous RD, Sivaraman V, Stasulli NM, Goldman WE. 2016. Pneumonic plague: the darker side of Yersinia pestis. Trends Microbiol 24:190–197. doi: 10.1016/j.tim.2015.11.008. [DOI] [PubMed] [Google Scholar]
- 215.Becker TM, Poland JD, Quan TJ, White ME, Mann JM, Barnes AM. 1987. Plague meningitis—a retrospective analysis of cases reported in the United States, 1970–1979. West J Med 147:554. [PMC free article] [PubMed] [Google Scholar]
- 216.Stokell JR, Khan A, Steck TR. 2014. Mechanical homogenization increases bacterial homogeneity in sputum. J Clin Microbiol 52:2340–2345. doi: 10.1128/JCM.00487-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Ber R, Mamroud E, Aftalion M, Tidhar A, Gur D, Flashner Y, Cohen S. 2003. Development of an improved selective agar medium for isolation of Yersinia pestis. Appl Environ Microbiol 69:5787–5792. doi: 10.1128/AEM.69.10.5787-5792.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Tourdjman M, Ibraheem M, Brett M, DeBess E, Progulske B, Ettestad P, McGivern T, Petersen J, Mead P. 2012. Misidentification of Yersinia pestis by automated systems, resulting in delayed diagnoses of human plague infections—Oregon and New Mexico, 2010–2011. Clin Infect Dis 55:e58–e60. doi: 10.1093/cid/cis578. [DOI] [PubMed] [Google Scholar]
- 219.Ayyadurai S, Flaudrops C, Raoult D, Drancourt M. 2010. Rapid identification and typing of Yersinia pestis and other Yersinia species by matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry. BMC Microbiol 10:285. doi: 10.1186/1471-2180-10-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Lasch P, Wahab T, Weil S, Pályi B, Tomaso H, Zange S, Kiland Granerud B, Drevinek M, Kokotovic B, Wittwer M, Pflüger V, Di Caro A, Stämmler M, Grunow R, Jacob D. 2015. Identification of highly pathogenic microorganisms by matrix-assisted laser desorption ionization–time of flight mass spectrometry: results of an interlaboratory ring trial. J Clin Microbiol 53:2632–2640. doi: 10.1128/JCM.00813-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Schofield DA, Molineux IJ, Westwater C. 2009. Diagnostic bioluminescent phage for detection of Yersinia pestis. J Clin Microbiol 47:3887–3894. doi: 10.1128/JCM.01533-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Sodeinde OA, Subrahmanyam YV, Stark K, Quan T, Bao Y, Goguen JD. 1992. A surface protease and the invasive character of plague. Science 258:1004–1007. doi: 10.1126/science.1439793. [DOI] [PubMed] [Google Scholar]
- 223.Welkos SL, Friedlander AM, Davis KJ. 1997. Studies on the role of plasminogen activator in systemic infection by virulent Yersinia pestis strain C092. Microb Pathog 23:211–223. doi: 10.1006/mpat.1997.0154. [DOI] [PubMed] [Google Scholar]
- 224.Loïez C, Herwegh S, Wallet F, Armand S, Guinet F, Courcol RJ. 2003. Detection of Yersinia pestis in sputum by real-time PCR. J Clin Microbiol 41:4873–4875. doi: 10.1128/JCM.41.10.4873-4875.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Armougom F, Bitam I, Croce O, Merhej V, Barassi L, Nguyen T-T, La Scola B, Raoult D. 2016. Genomic insights into a new Citrobacter koseri strain revealed gene exchanges with the virulence-associated Yersinia pestis pPCP1 plasmid. Front Microbiol 7:340. doi: 10.3389/fmicb.2016.00340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Hänsch S, Cilli E, Catalano G, Gruppioni G, Bianucci R, Stenseth NC, Bramanti B, Pallen MJ. 2015. The pla gene, encoding plasminogen activator, is not specific to Yersinia pestis. BMC Res Notes 8:535. doi: 10.1186/s13104-015-1525-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Janse I, Hamidjaja RA, Reusken C. 2013. Yersinia pestis plasminogen activator gene homolog in rat tissues. Emerg Infect Dis 19:342–344. doi: 10.3201/eid1902.120659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Simonet M, Riot B, Fortineau N, Berche P. 1996. Invasin production by Yersinia pestis is abolished by insertion of an IS200-like element within the inv gene. Infect Immun 64:375–379. doi: 10.1128/IAI.64.1.375-379.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Riehm JM, Rahalison L, Scholz HC, Thoma B, Pfeffer M, Razanakoto LM, Al Dahouk S, Neubauer H, Tomaso H. 2011. Detection of Yersinia pestis using real-time PCR in patients with suspected bubonic plague. Mol Cell Probes 25:8–12. doi: 10.1016/j.mcp.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 230.Tsukano H, Itoh K, Suzuki S, Watanabe H. 1996. Detection and identification of Yersinia pestis by polymerase chain reaction (PCR) using multiplex primers. Microbiol Immunol 40:773–775. doi: 10.1111/j.1348-0421.1996.tb01140.x. [DOI] [PubMed] [Google Scholar]
- 231.Mölsä M, Hemmilä H, Katz A, Niemimaa J, Forbes KM, Huitu O, Stuart P, Henttonen H, Nikkari S. 2015. Monitoring biothreat agents (Francisella tularensis, Bacillus anthracis and Yersinia pestis) with a portable real-time PCR instrument. J Microbiol Methods 115:89–93. doi: 10.1016/j.mimet.2015.05.026. [DOI] [PubMed] [Google Scholar]
- 232.Banada PP, Deshpande S, Banik S, Shah D, Koshy R, Patel B, Kwiatkowski R, Persing D, Alland D. 2019. Multiplex detection of three select agents directly from blood by use of the GeneXpert system. J Clin Microbiol 57:e00036-19. doi: 10.1128/JCM.00036-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Gay-Andrieu F, Magassouba N, Picot V, Phillips CL, Peyrefitte CN, Dacosta B, Doré A, Kourouma F, Ligeon-Ligeonnet V, Gauby C, Longuet C, Scullion M, Faye O, Machuron JL, Miller M. 2017. Clinical evaluation of the BioFire FilmArray bioThreat-E test for the diagnosis of Ebola virus disease in Guinea. J Clin Virol 92:20–24. doi: 10.1016/j.jcv.2017.04.015. [DOI] [PubMed] [Google Scholar]
- 234.Qi Z, Wu Y, Li Y, Li C, Yang X, Zhang Q, Xin Y, Jin Y, Wei R, Cui Y. 2015. 3a-negative Yersinia pestis, China. Infect Dis Transl Med 1:61–62. [Google Scholar]
- 235.Qu S, Shi Q, Zhou L, Guo Z, Zhou D, Zhai J, Yang R. 2010. Ambient stable quantitative PCR reagents for the detection of Yersinia pestis. PLoS Negl Trop Dis 4:e629. doi: 10.1371/journal.pntd.0000629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Sergueev KV, He Y, Borschel RH, Nikolich MP, Filippov AA. 2010. Rapid and sensitive detection of Yersinia pestis using amplification of plague diagnostic bacteriophages monitored by real-time PCR. PLoS One 5:e11337. doi: 10.1371/journal.pone.0011337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Feng N, Zhou Y, Fan Y, Bi Y, Yang R, Zhou Y, Wang X. 2018. Yersinia pestis detection by loop-mediated isothermal amplification combined with magnetic bead capture of DNA. Braz J Microbiol 49:128–137. doi: 10.1016/j.bjm.2017.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Chanteau S, Rahalison L, Ralafiarisoa L, Foulon J, Ratsitorahina M, Ratsifasoamanana L, Carniel E, Nato F. 2003. Development and testing of a rapid diagnostic test for bubonic and pneumonic plague. Lancet 361:211–216. doi: 10.1016/S0140-6736(03)12270-2. [DOI] [PubMed] [Google Scholar]
- 239.Hsu H-L, Chuang C-C, Liang C-C, Chiao D-J, Wu H-L, Wu Y-P, Lin F-P, Shyu R-H. 2018. Rapid and sensitive detection of Yersinia pestis by lateral-flow assay in simulated clinical samples. BMC Infect Dis 18:402. doi: 10.1186/s12879-018-3315-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Yan Z, Zhou L, Zhao Y, Wang J, Huang L, Hu K, Liu H, Wang H, Guo Z, Song Y, Huang H, Yang R. 2006. Rapid quantitative detection of Yersinia pestis by lateral-flow immunoassay and up-converting phosphor technology-based biosensor. Sens Actuators B Chem 119:656–663. doi: 10.1016/j.snb.2006.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Zhang P, Liu X, Wang C, Zhao Y, Hua F, Li C, Yang R, Zhou L. 2014. Evaluation of up-converting phosphor technology-based lateral flow strips for rapid detection of Bacillus anthracis Spore, Brucella spp., and Yersinia pestis. PLoS One 9:e105305. doi: 10.1371/journal.pone.0105305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Splettstoesser WD, Rahalison L, Grunow R, Neubauer H, Chanteau S. 2004. Evaluation of a standardized F1 capsular antigen capture ELISA test kit for the rapid diagnosis of plague. FEMS Immunol Med Microbiol 41:149–155. doi: 10.1016/j.femsim.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 243.Chanteau S, Rahalison L, Ratsitorahina M, Rasolomaharo M, Boisier P, O’Brien T, Aldrich J, Keleher A, Morgan C, Burans J. 2000. Early diagnosis of bubonic plague using F1 antigen capture ELISA assay and rapid immunogold dipstick. Int J Med Microbiol 290:279–283. doi: 10.1016/S1438-4221(00)80126-5. [DOI] [PubMed] [Google Scholar]
- 244.Drancourt M, Michel-Lepage A, Boyer S, Raoult D. 2016. The point-of-care laboratory in clinical microbiology. Clin Microbiol Rev 29:429–447. doi: 10.1128/CMR.00090-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Bertherat E, Mueller M, Shako J-C, Picardeau M. 2014. Discovery of a leptospirosis cluster amidst a pneumonic plague outbreak in a miners’ camp in the Democratic Republic of the Congo. Int J Environ Res Public Health 11:1824–1833. doi: 10.3390/ijerph110201824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Yersin A. 1897. Sur la peste bubonique (sérothérapie). Ann Inst Pasteur 11:81–93. [Google Scholar]
- 247.Meyer KF, Quan SF, Mccrumb F, Larson A. 1952. Effective treatment of plague. Ann N Y Acad Sci 55:1228–1274. doi: 10.1111/j.1749-6632.1952.tb22687.x. [DOI] [PubMed] [Google Scholar]
- 248.Kamal AM, Gayed I, Anwar M. 1941. On the epidemiology and treatment of plague in Egypt. “The 1940 epidemic”. J Egypt Public Health Assoc 16:31–103. [Google Scholar]
- 249.Butler T. 2014. Plague history: Yersin’s discovery of the causative bacterium in 1894 enabled, in the subsequent century, scientific progress in understanding the disease and the development of treatments and vaccines. Clin Microbiol Infect 20:202–209. doi: 10.1111/1469-0691.12540. [DOI] [PubMed] [Google Scholar]
- 250.Carman JA. 1938. Prontosil in the treatment of Oriental plague. East Afr Med J 14:362–366. [Google Scholar]
- 251.Vine RS. 1938. Plague in the Nilgiris. J R Army Med Corps 71:382–395. [Google Scholar]
- 252.Girard G. 1941. Treatment of plague by sulphonamides, experimental and human. Bull Soc Pathol Exot 34:43–54. [Google Scholar]
- 253.Favarel R, Carriere M, Chartres A. 1948. Guérison de trois cas de peste pulmonaire primitive par le traitement sulfamide. Bull Soc Pathol Exot 41:515–523. [Google Scholar]
- 254.Wagle PM, Sokhey SS, Dikshit BB, Ganapathy K. 1941. Chemotherapy in plague. Ind Med Gaz 76:29–32. [PMC free article] [PubMed] [Google Scholar]
- 255.Girard G. 1949. La streptomycine, médication héroïque de la peste. Rev Coloniale Med Chir 21:2–4. [PubMed] [Google Scholar]
- 256.Haddad C, Valero A. 1948. Streptomycin in bubonic plague. BMJ 1:1026–1027. doi: 10.1136/bmj.1.4560.1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Boulanger LL, Ettestad P, Fogarty JD, Dennis DT, Romig D, Mertz G. 2004. Gentamicin and tetracyclines for the treatment of human plague: review of 75 cases in New Mexico, 1985–1999. Clin Infect Dis 38:663–669. doi: 10.1086/381545. [DOI] [PubMed] [Google Scholar]
- 258.Mwengee W, Butler T, Mgema S, Mhina G, Almasi Y, Bradley C, Formanik JB, Rochester CG. 2006. Treatment of plague with gentamicin or doxycycline in a randomized clinical trial in Tanzania. Clin Infect Dis 42:614–621. doi: 10.1086/500137. [DOI] [PubMed] [Google Scholar]
- 259.Kwit N, Nelson C, Kugeler K, Petersen J, Plante L, Yaglom H, Kramer V, Schwartz B, House J, Colton L, Feldpausch A, Drenzek C, Baumbach J, DiMenna M, Fisher E, Debess E, Buttke D, Weinburke M, Percy C, Schriefer M, Gage K, Mead P. 2015. Human plague—United States, 2015. MMWR Morb Mortal Wkly Rep 64:918–919. doi: 10.15585/mmwr.mm6433a6. [DOI] [PubMed] [Google Scholar]
- 260.Apangu T, Griffith K, Abaru J, Candini G, Apio H, Okoth F, Okello R, Kaggwa J, Acayo S, Ezama G, Yockey B, Sexton C, Schriefer M, Mbidde EK, Mead P. 2017. Successful treatment of human plague with oral ciprofloxacin. Emerg Infect Dis 23:553–555. doi: 10.3201/eid2303.161212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.CDC. 2020. Plague: resources for clinicians. Centers for Disease Control and Prevention, Atlanta, GA. https://www.cdc.gov/plague/healthcare/clinicians.html. [Google Scholar]
- 262.Galimand M, Guiyoule A, Gerbaud G, Rasoamanana B, Chanteau S, Carniel E, Courvalin P. 1997. Multidrug resistance in Yersinia pestis mediated by a transferable plasmid. N Engl J Med 337:677–681. doi: 10.1056/NEJM199709043371004. [DOI] [PubMed] [Google Scholar]
- 263.Guiyoule A, Gerbaud G, Buchrieser C, Galimand M, Rahalison L, Chanteau S, Courvalin P, Carniel E. 2001. Transferable plasmid-mediated resistance to streptomycin in a clinical isolate of Yersinia pestis. Emerg Infect Dis 7:43–48. doi: 10.3201/eid0701.010106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Galimand M, Carniel E, Courvalin P. 2006. Resistance of Yersinia pestis to antimicrobial agents. Antimicrob Agents Chemother 50:3233–3236. doi: 10.1128/AAC.00306-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Welch TJ, Fricke WF, McDermott PF, White DG, Rosso M-L, Rasko DA, Mammel MK, Eppinger M, Rosovitz MJ, Wagner D, Rahalison L, Leclerc JE, Hinshaw JM, Lindler LE, Cebula TA, Carniel E, Ravel J. 2007. Multiple antimicrobial resistance in plague: an emerging public health risk. PLoS One 2:e309. doi: 10.1371/journal.pone.0000309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Ryzhko IV, Shcherbaniuk AI, Samokhodkina ED, Tsuraeva RI, Mishn’kin Bn Kasatkina IV, Zhigalova TA. 1994. Virulence of rifampicin and quinolone resistant mutants of strains of plague microbe with Fra+ and Fra− phenotypes. Antibiot Khimioter 39:32–36. [PubMed] [Google Scholar]
- 267.Munyenyiwa A, Zimba M, Nhiwatiwa T, Barson M. 2019. Plague in Zimbabwe from 1974 to 2018: a review article. PLoS Negl Trop Dis 13:e0007761. doi: 10.1371/journal.pntd.0007761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Malek MA, Bitam I, Drancourt M. 2016. Plague in Arab Maghreb, 1940–2015: a review. Front Public Health 4:112. doi: 10.3389/fpubh.2016.00112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Bitam I, Ayyadurai S, Kernif T, Chetta M, Boulaghman N, Raoult D, Drancourt M. 2010. New rural focus of plague, Algeria. Emerg Infect Dis 16:1639–1640. doi: 10.3201/eid1610.091854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Klein J-M, Alonso JM, Baranton G, Poulet AR, Mollaret HH. 1975. La peste en Mauritanie. Méd Mal Infect 5:198–207. doi: 10.1016/S0399-077X(75)80141-7. [DOI] [Google Scholar]
- 271.Nikitin AI, Maramovich AS, Bazanova LP, Okunev LP, Kosilko SA, Innokent’eva TI, Voronova GA. 2009. Epizootological characteristics of the natural foci of plague in China: a review of literature. Med Parazitol (Mosk) Jan-Mar 2009:51–58. (In Russian.) [PubMed] [Google Scholar]
- 272.Fang X, Yang R, Xu L, Liu Q, Dong X, Zhang R, Yu X, Qin C, Gong Z, Zhou D, Cui Y, Li Y, Ye R-Y, Lu L, Zhang J, Li G. 2012. Ecological-geographic landscapes of natural plague foci in China VII. Typing of natural plague foci. Zhonghua Liu Xing Bing Xue Za Zhi 33:1144–1150. (In Chinese.) [PubMed] [Google Scholar]
- 273.Shen X, Wang Q, Xia L, Zhu X, Zhang Z, Liang Y, Cai H, Zhang E, Wei J, Chen C, Song Z, Zhang H, Yu D, Hai R. 2010. Complete genome sequences of Yersinia pestis from natural foci in China. J Bacteriol 192:3551–3552. doi: 10.1128/JB.00340-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Wang P, Shi L, Zhang F, Guo Y, Zhang Z, Tan H, Cui Z, Ding Y, Liang Y, Liang Y, Yu D, Xu J, Li W, Song Z. 2018. Ten years of surveillance of the Yulong plague focus in China and the molecular typing and source tracing of the isolates. PLoS Negl Trop Dis 12:e0006352. doi: 10.1371/journal.pntd.0006352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Zhang Y, Luo T, Yang C, Yue X, Guo R, Wang X, Buren M, Song Y, Yang R, Cao H, Cui Y, Dai X. 2018. Phenotypic and molecular genetic characteristics of Yersinia pestis at an emerging natural plague focus, Junggar Basin, China. Am J Trop Med Hyg 98:231–237. doi: 10.4269/ajtmh.17-0195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Du H-W, Wang Y, Zhuang D-F, Jiang X-S. 2017. Temporal and spatial distribution characteristics in the natural plague foci of Chinese Mongolian gerbils based on spatial autocorrelation. Infect Dis Poverty 6:124. doi: 10.1186/s40249-017-0338-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Aikimbajev A, Meka-Mechenko T, Temiralieva G, Bekenov J, Sagiyev Z, Kaljan K, Mukhambetova AK. 2003. Plague in Kazakhstan at the present time. Przegl Epidemiol 57:593–598. [PubMed] [Google Scholar]
- 278.Bykov LT, Tsoi DC, Rakhimov KR. 1985. Results of using the serological method of epidemiological investigation of plague foci in the Muyunkum and Eastern Kyzylkum deserts in 1978–1982. J Hyg Epidemiol Microbiol Immunol 29:369–376. [PubMed] [Google Scholar]
- 279.Lowell JL, Zhansarina A, Yockey B, Meka-Mechenko T, Stybayeva G, Atshabar B, Nekrassova L, Tashmetov R, Kenghebaeva K, Chu MC, Kosoy M, Antolin MF, Gage KL. 2007. Phenotypic and molecular characterizations of Yersinia pestis isolates from Kazakhstan and adjacent regions. Microbiology (Reading) 153:169–177. doi: 10.1099/mic.0.29059-0. [DOI] [PubMed] [Google Scholar]
- 280.Korzun VM, Chipanin EV, Balakhonov SV, Denisov AV, Rozhdestvenskiĭ EN, Mihaĭlov EP, Iarygina MB, Kosilko SA. 2014. Change in the habitat of Yersinia pestis in the Gorno-Altaisk natural focus of plague. Med Parazitol (Mosk) Oct-Dec 2014:11–19. (In Russian.) [PubMed] [Google Scholar]
- 281.Kotti BK. 2011. Value of fleas in the natural foci of plague in the Caucasus. Med Parazitol (Mosk) Oct-Dec 2011:28–30. (In Russian.) [PubMed] [Google Scholar]
- 282.Cherchenko II, Dyatlov AI. 1976. Broader investigation into the external environment of the specific antigen of the infectious agent in epizootiological observation and study of the structure of natural foci of plague. J Hyg Epidemiol Microbiol Immunol 20:221–228. [PubMed] [Google Scholar]
- 283.Asvarov BM, -M Gaziev SG, Khasaev SM, Gruba VP, Grizhebovskiĭ GM. 2001. The epizootic situation on the territories with natural foci of plague in Chechen Republic and in the Republic of Ingushetia. Zh Mikrobiol Epidemiol Immunobiol Nov-Dec 2001:66–68. (In Russian.) [PubMed] [Google Scholar]
- 284.Sariyeva G, Abdel Z, Shabunin A, Sagiyev Z, Abdikarimov S, Bazarkanova G, Kendirbaev D, Maimulov R, Dzhaparova A, Sofeikov V, Abdirassilova A, Mussagaliyeva R, Kurmanov B, Aitbaeva Z, Almazbek D. 2018. Current status of the Sari-Dzhas natural focus of plague, Kyrgyzstan: epizootic activity and marmot population. Vector Borne Zoonotic Dis 18:524–532. doi: 10.1089/vbz.2017.2200. [DOI] [PubMed] [Google Scholar]
- 285.Mostafavi E, Shahraki AH, Japoni-Nejad A, Esmaeili S, Darvish J, Sedaghat MM, Mohammadi A, Mohammadi Z, Mahmoudi A, Pourhossein B, Ghasemi A, Gyuranecz M, Carniel E. 2017. A field study of plague and tularemia in rodents, Western Iran. Vector Borne Zoonotic Dis 17:247–253. doi: 10.1089/vbz.2016.2053. [DOI] [PubMed] [Google Scholar]
- 286.Gascuel F, Choisy M, Duplantier J-M, Débarre F, Brouat C. 2013. Host resistance, population structure and the long-term persistence of bubonic plague: contributions of a modelling approach in the Malagasy focus. PLoS Comput Biol 9:e1003039. doi: 10.1371/journal.pcbi.1003039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Eisen RJ, Borchert JN, Mpanga JT, Atiku LA, MacMillan K, Boegler KA, Montenieri JA, Monaghan A, Gage KL. 2012. Flea diversity as an element for persistence of plague bacteria in an East African plague focus. PLoS One 7:e35598. doi: 10.1371/journal.pone.0035598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Janssens PG, Pattyn SR. 1994. Plague in Zaire. Verh K Acad Geneeskd Belg 56:281–360. Discussion 360–361. (In Dutch.) [PubMed] [Google Scholar]
- 289.Neerinckx S, Peterson AT, Gulinck H, Deckers J, Kimaro D, Leirs H. 2010. Predicting potential risk areas of human plague for the Western Usambara Mountains, Lushoto District, Tanzania. Am J Trop Med Hyg 82:492–500. doi: 10.4269/ajtmh.2010.09-0426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Da Costa E, de CV, Sobreira M, Leal NC, De Almeida AMP. 2017. Rodents and other small mammal reservoirs in plague foci in northeastern Brazil. J Infect Dev Ctries 11:426–430. doi: 10.3855/jidc.8271. [DOI] [PubMed] [Google Scholar]
- 291.Williamson ED, Oyston PCF. 2012. The natural history and incidence of Yersinia pestis and prospects for vaccination. J Med Microbiol 61:911–918. doi: 10.1099/jmm.0.037960-0. [DOI] [PubMed] [Google Scholar]
- 292.Meyer KF. 1970. Effectiveness of live or killed plague vaccines in man. Bull World Health Organ 42:653. [PMC free article] [PubMed] [Google Scholar]
- 293.Jefferson T, Demicheli V, Pratt M. 1998. Vaccines for preventing plague. Cochrane Database Syst Rev 1:CD000976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.CDC. 2018. Plague: prevention. Centers for Disease Control and Prevention, Atlanta, GA. https://www.cdc.gov/plague/prevention/index.html. [Google Scholar]
- 295.Inglesby TV, Dennis DT, Henderson DA, Bartlett JG, Ascher MS, Eitzen E, Fine AD, Friedlander AM, Hauer J, Koerner JF, Layton M, McDade J, Osterholm MT, O'Toole T, Parker G, Perl TM, Russell PK, Schoch-Spana M, Tonat K. 2000. Plague as a biological weapon: medical and public health management. JAMA 283:2281–2290. doi: 10.1001/jama.283.17.2281. [DOI] [PubMed] [Google Scholar]
- 296.Brouillard JE, Terriff CM, Tofan A, Garrison MW. 2006. Antibiotic selection and resistance issues with fluoroquinolones and doxycycline against bioterrorism agents. Pharmacotherapy 26:3–14. doi: 10.1592/phco.2006.26.1.3. [DOI] [PubMed] [Google Scholar]
- 297.Kman NE, Nelson RN. 2008. Infectious agents of bioterrorism: a review for emergency physicians. Emerg Med Clin North Am 26:517–547. doi: 10.1016/j.emc.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 298.Darling RG, Catlett CL, Huebner KD, Jarrett DG. 2002. Threats in bioterrorism I: CDC category A agents. Emerg Med Clin North Am 20:273–309. doi: 10.1016/S0733-8627(02)00005-6. [DOI] [PubMed] [Google Scholar]
- 299.Weant KA, Bailey AM, Fleishaker EL, Justice SB. 2014. Being prepared: bioterrorism and mass prophylaxis: part I. Adv Emerg Nurs J 36:226–238. doi: 10.1097/TME.0000000000000029. [DOI] [PubMed] [Google Scholar]
- 300.Gage KL, Montenieri JA, Thomas RE. 1994. The role of predators in the ecology, epidemiology, and surveillance of plague in the United States, p 200–206. In Halverson WS, Crabb AC (ed), Proceedings of the Sixteenth Vertebrate Pest Conference. University of California Press, Davis, CA. [Google Scholar]
- 301.Shi G, Ju C, Zhang R, Zhang Z, Sun J, Wang M, Zhang X, Ye X, Zhu Z, Xing J. 2015. Risk assessments and control strategies of plague in five key surveillance counties, Zhejiang province. Zhonghua Yu Fang Yi Xue Za Zhi 49:896–900. (In Chinese.) [PubMed] [Google Scholar]
- 302.Bevins SN, Baroch JA, Nolte DL, Zhang M, He H. 2012. Yersinia pestis: examining wildlife plague surveillance in China and the USA. Integr Zool 7:99–109. doi: 10.1111/j.1749-4877.2011.00277.x. [DOI] [PubMed] [Google Scholar]
- 303.Stenseth NC, Samia NI, Viljugrein H, Kausrud KL, Begon M, Davis S, Leirs H, Dubyanskiy VM, Esper J, Ageyev VS, Klassovskiy NL, Pole SB, Chan K-S. 2006. Plague dynamics are driven by climate variation. Proc Natl Acad Sci U S A 103:13110–13115. doi: 10.1073/pnas.0602447103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Davis RM, Cleugh E, Smith RT, Fritz CL. 2008. Use of a chitin synthesis inhibitor to control fleas on wild rodents important in the maintenance of plague, Yersinia pestis, in California. J Vector Ecol 33:278–284. doi: 10.3376/1081-1710-33.2.278. [DOI] [PubMed] [Google Scholar]
- 305.Boeckl CM. 2001. Giorgio Vasari’s “San Rocco Altarpiece”: tradition and innovation in plague iconography. Artibus et Historiae 22:29–40. doi: 10.2307/1483649. [DOI] [Google Scholar]
- 306.Boeckl CM. 2000. Images of plague and pestilence: iconography and iconology. Truman State University Press, Kirksville, MO. [Google Scholar]
- 307.Barker S. 2005. Plague art in early modern Rome: divine directives and temporal remedies, p 45–63. In Bailey GA, Jones PM, Mormando F, Worcester TW (ed), Hope and healing: painting in Italy in a time of plague, 1500–1800. University of Chicago Press, Chicago, IL. [Google Scholar]
- 308.Jordan L. 1981. The iconography of death in western medieval art to 1350, p 62–66. PhD dissertation. University of Notre Dame, Notre Dame, IN. [Google Scholar]
- 309.Polzer J. 1982. Aspects of the fourteenth-century iconography of death and the plague, p 107–130. In Williman D (ed), The Black Death: the impact of the fourteenth century plague. Center for Medieval and Early Renaissance Studies, Binghamton, NY. [Google Scholar]
- 310.Chevé D. 2005. Les corps et les lieux du fléau: un regard de philosophe naturaliste. Gassendi et l’épidémie de peste de Digne en 1629. Le corps de l'Alpin. Perceptions, représentations, modifications. Editions Des Hautes Alpes, p 157–166. Anthropologie des Populations Alpines. [Google Scholar]
- 311.Chevé-Aicardi D. 2003. Les corps de la Contagion. Etude anthropologique des représentations iconographiques de la peste (XVIème–Xxème siècles en Europe). PhD dissertation. Aix-Marseille Université, Marseille, France. [Google Scholar]
- 312.Little LK (ed). 2007. Plague and the end of antiquity: the pandemic of 541–750. Cambridge University Press, New York, NY. [Google Scholar]
- 313.Tsiannis C, Poulakou-Rebelakou E, Petridou E. 2009. The Red Sea and the port of Clysma. A possible gate of Justinian’s plague. Gesnerus 662:209–217. [PubMed] [Google Scholar]
- 314.Stathakopoulos DC. 2017. Famine and pestilence in the late Roman and early Byzantine empire: a systematic survey of subsistence crises and epidemics. Routledge, Abingdon, United Kingdom. [Google Scholar]
- 315.Dols MW. 1974. Plague in early Islamic history. J Am Orient Soc 94:371–383. doi: 10.2307/600071. [DOI] [Google Scholar]
- 316.Sarris P. 2002. The Justinianic plague: origins and effects. Cont Change 17:169–182. doi: 10.1017/S0268416002004137. [DOI] [Google Scholar]
- 317.Allen P. 1979. The” Justinianic” plague. Byzantion 49:5–20. [PubMed] [Google Scholar]
- 318.Wagner DM, Klunk J, Harbeck M, Devault A, Waglechner N, Sahl JW, Enk J, Birdsell DN, Kuch M, Lumibao C, Poinar D, Pearson T, Fourment M, Golding B, Riehm JM, Earn DJD, Dewitte S, Rouillard J-M, Grupe G, Wiechmann I, Bliska JB, Keim PS, Scholz HC, Holmes EC, Poinar H. 2014. Yersinia pestis and the plague of Justinian 541–543 AD: a genomic analysis. Lancet Infect Dis 14:319–326. doi: 10.1016/S1473-3099(13)70323-2. [DOI] [PubMed] [Google Scholar]
- 319.Feldman M, Harbeck M, Keller M, Spyrou MA, Rott A, Trautmann B, Scholz HC, Päffgen B, Peters J, McCormick M, Bos K, Herbig A, Krause J. 2016. A high-coverage Yersinia pestis genome from a sixth-century Justinianic Plague victim. Mol Biol Evol 33:2911–2923. doi: 10.1093/molbev/msw170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Dewing HB. 1914. Procopius. W. Heinemann, London, United Kingdom. [Google Scholar]
- 321.Grégoire de Tours. 1823. Histoire des Francs. J.-L.-J. Brière, Paris, France. [Google Scholar]
- 322.Harper K. 2017. The fate of Rome: climate, disease, and the end of an empire. Princeton University Press, Princeton, NJ. [Google Scholar]
- 323.Mitchell S. 2014. A history of the later Roman Empire, AD 284–641, 2nd ed. John Wiley & Sons, Hoboken, NJ. [Google Scholar]
- 324.Meier M. 2016. The ‘Justinianic Plague’: the economic consequences of the pandemic in the eastern Roman empire and its cultural and religious effects. Early Mediev Europe 24:267–292. doi: 10.1111/emed.12152. [DOI] [Google Scholar]
- 325.Watkins RN. 1972. Petrarch and the Black Death: from fear to monuments. Stud Renaiss 19:196–223. doi: 10.2307/2857093. [DOI] [PubMed] [Google Scholar]
- 326.Spyrou MA, Keller M, Tukhbatova RI, Scheib CL, Nelson EA, Andrades Valtueña A, Neumann GU, Walker D, Alterauge A, Carty N, Cessford C, Fetz H, Gourvennec M, Hartle R, Henderson M, von Heyking K, Inskip SA, Kacki S, Key FM, Knox EL, Later C, Maheshwari-Aplin P, Peters J, Robb JE, Schreiber J, Kivisild T, Castex D, Lösch S, Harbeck M, Herbig A, Bos KI, Krause J. 2019. Phylogeography of the second plague pandemic revealed through analysis of historical Yersinia pestis genomes. Nat Commun 10:1–13. doi: 10.1038/s41467-019-12154-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Cui Y, Yu C, Yan Y, Li D, Li Y, Jombart T, Weinert LA, Wang Z, Guo Z, Xu L, Zhang Y, Zheng H, Qin N, Xiao X, Wu M, Wang X, Zhou D, Qi Z, Du Z, Wu H, Yang X, Cao H, Wang H, Wang J, Yao S, Rakin A, Li Y, Falush D, Balloux F, Achtman M, Song Y, Wang J, Yang R. 2013. Historical variations in mutation rate in an epidemic pathogen, Yersinia pestis. Proc Natl Acad Sci U S A 110:577–582. doi: 10.1073/pnas.1205750110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Morelli G, Song Y, Mazzoni CJ, Eppinger M, Roumagnac P, Wagner DM, Feldkamp M, Kusecek B, Vogler AJ, Li Y, Cui Y, Thomson NR, Jombart T, Leblois R, Lichtner P, Rahalison L, Petersen JM, Balloux F, Keim P, Wirth T, Ravel J, Yang R, Carniel E, Achtman M. 2010. Yersinia pestis genome sequencing identifies patterns of global phylogenetic diversity. Nat Genet 42:1140–1143. doi: 10.1038/ng.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Norris J. 1977. East or West? The geographic origin of the Black Death. Bull Hist Med 51:1–24. [PubMed] [Google Scholar]
- 330.Wheelis M. 2002. Biological warfare at the 1346 siege of Caffa. Emerg Infect Dis 8:971–975. doi: 10.3201/eid0809.010536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Wheelis M. 2004. A short history of biological warfare and weapons, p 15–31. In The implementation of legally binding measures to strengthen the biological and toxin weapons convention. Springer, New York, NY. [Google Scholar]
- 332.Signoli M. 2012. Reflections on crisis burials related to past plague epidemics. Clin Microbiol Infect 18:218–223. doi: 10.1111/j.1469-0691.2012.03787.x. [DOI] [PubMed] [Google Scholar]
- 333.Benedictow OJ, Benedictow OL. 2004. The Black Death, 1346–1353: the complete history. Boydell & Brewer, Woodbridge, United Kingdom. [Google Scholar]
- 334.Brossolet J, Mollaret HH. 1994. Pourquoi la peste?: le rat, la puce et le bubon. Gallimard, Paris, France. [Google Scholar]
- 335.Carpentier E. 1962. Une ville devant la peste: oevieto et la peste noire de 1348. Imprimerie Nationale, Paris, France. [Google Scholar]
- 336.Biraben JN. 1975. Les hommes et la peste en France et dans les pays européens et méditerranéens. Mouton, Paris, France. [Google Scholar]
- 337.Bertrand J-B. 1723. Relation historique de tout ce qui s’ est passé à Marseille pendant la dernière peste. https://books.google.fr/books?id=0gAkfiqKFaAC.
- 338.Spiteri SC. 2017. Guarding against contagion: vigilance and the role of fortifications in Malta during the outbreak of Plague in Messina 1743. J Baroque Stud 2:177–200. [Google Scholar]
- 339.de Mertens C. 2019. An account of the plague which raged at Moscow, in 1771. Good Press, Glasgow, Scotland. [Google Scholar]
- 340.Samojlovič DS. 1787. Opuscules sur la peste qui en 1771 ravage a Moscou. https://books.google.fr/books?hl=fr&lr=&id=KbM_AAAAcAAJ.
- 341.Aberth J. 2013. From the brink of the apocalypse: confronting famine, war, plague and death in the later Middle Ages. Routledge, Abingdon, United Kingdom. [Google Scholar]
- 342.Mollaret HH, Brossollet J. 1968. À propos des “Pestiférés de Jaffa” de AJ Gros, p 263–308. Koninklijk Museum Voor Schone Kunsten, Antwerp, Belgium. [Google Scholar]
- 343.Calvert R. 1815. An account of the origin and progress of the Plague of Malta, in the year 1813. Med Chir Trans 6:1–64. [PMC free article] [PubMed] [Google Scholar]
- 344.Valensi L. 1969. Calamités démographiques en Tunisie et en Méditerranée orientale aux xviii e et xix e siècles. Ann Hist Sci Soc 1969:1540–1561. doi: 10.3406/ahess.1969.422187. [DOI] [Google Scholar]
- 345.La Rue GM. 2007. African slave women in Egypt, from ca. 1820 to the plague epidemic of 1834–1835. Women Slavery 1:168–189. [Google Scholar]
- 346.Pezzoni MA. 1842. Notice of cases of plague contracted in the Lazeretto of Constantinople, in a letter addressed to Dr. Davy. Med Chir Trans 25:167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Vogler AJ, Chan F, Nottingham R, Andersen G, Drees K, Beckstrom-Sternberg SM, Wagner DM, Chanteau S, Keim P. 2013. A decade of plague in Mahajanga, Madagascar: insights into the global maritime spread of pandemic plague. mBio 4:e00623-12. doi: 10.1128/mBio.00623-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Xu L, Liu Q, Stige LC, Ari TB, Fang X, Chan K-S, Wang S, Stenseth NC, Zhang Z. 2011. Nonlinear effect of climate on plague during the third pandemic in China. Proc Natl Acad Sci U S A 108:10214–10219. doi: 10.1073/pnas.1019486108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Hussein AG. 1955. Changes in the epidemiology of plague in Egypt, 1899–1951. Bull World Health Organ 13:27. [PMC free article] [PubMed] [Google Scholar]
- 350.Xu L, Stige LC, Leirs H, Neerinckx S, Gage KL, Yang R, Liu Q, Bramanti B, Dean KR, Tang H. 2019. Historical and genomic data reveal the influencing factors on global transmission velocity of plague during the Third Pandemic. Proc Natl Acad Sci U S A 116:11833–11838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Bramanti B, Dean KR, Walløe L, Stenseth NC. 2019. The Third Plague Pandemic in Europe. Proc Biol Sci 286:20182429. doi: 10.1098/rspb.2018.2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Dean KR, Krauer F, Schmid BV. 2019. Epidemiology of a bubonic plague outbreak in Glasgow, Scotland in 1900. R Soc Open Sci 6:181695. doi: 10.1098/rsos.181695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Kugeler KJ, Staples JE, Hinckley AF, Gage KL, Mead PS. 2015. Epidemiology of human plague in the United States, 1900–2012. Emerg Infect Dis 21:16–22. doi: 10.3201/eid2101.140564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Turner AJ. 2011. Disease control during the colonial period in Australia. Aust Vet J 89:239–242. doi: 10.1111/j.1751-0813.2011.00787.x. [DOI] [PubMed] [Google Scholar]
- 355.Robinson G. 1992. A sop to the “oi polloi”: capital, labour and the reform of New South Wales maritime administration 1867 to 1914. Aust J Pub Admin 51:104–116. doi: 10.1111/j.1467-8500.1992.tb01460.x. [DOI] [Google Scholar]
- 356.Ari TB, Neerinckx S, Gage KL, Kreppel K, Laudisoit A, Leirs H, Stenseth NC. 2011. Plague and climate: scales matter. PLoS Pathog 7:e1002160. doi: 10.1371/journal.ppat.1002160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Knab C. 2011. Plague times: scientific internationalism and the Manchurian Plague of 1910/1911. Itinerario 35:87–105. doi: 10.1017/S0165115312000083. [DOI] [Google Scholar]
- 358.Placidi T. 1921. La Peste à Marseille en 1920. Epidémiologie et Prophylaxie. Thèse pour obtenir le grade de docteur en médecine, par Thomas Placidi, élève de l’École de santé militaire. Imprimerie Ducros et Lombard, Valence-sur-Rhône, France. [Google Scholar]
- 359.Gueniot-Le Minor G. 1980. La peste des chiffonniers à Paris en 1920. Université de Paris VI, Faculte de Medecine Broussais-Hôtel Dieu, Paris, France. [Google Scholar]
- 360.Vogler AJ, Sahl JW, Leal NC, Sobreira M, Williamson CHD, Bollig MC, Birdsell DN, Rivera A, Thompson B, Nottingham R, Rezende AM, Keim P, Almeida AMP, Wagner DM. 2019. A single introduction of Yersinia pestis to Brazil during the 3rd plague pandemic. PLoS One 14:e0209478. doi: 10.1371/journal.pone.0209478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Sussman GD. 2011. Was the black death in India and China? Bull Hist Med 85:319–355. doi: 10.1353/bhm.2011.0054. [DOI] [PubMed] [Google Scholar]
- 362.Soupios MA. 2004. Impact of the plague in Ancient Greece. Infect Dis Clin North Am 18:45–51. doi: 10.1016/S0891-5520(03)00101-6. [DOI] [PubMed] [Google Scholar]
- 363.Khan IA. 2004. Plague: the dreadful visitation occupying the human mind for centuries. Trans R Soc Trop Med Hyg 98:270–277. doi: 10.1016/S0035-9203(03)00059-2. [DOI] [PubMed] [Google Scholar]
- 364.Macleod CW (ed). 1982. Homer: Iliad Book XXIV. Cambridge University Press, Cambridge, United Kingdom. [Google Scholar]
- 365.Kousoulis AA, Economopoulos KP, Poulakou-Rebelakou E, Androutsos G, Tsiodras S. 2012. The plague of Thebes, a historical epidemic in Sophocles’ Oedipus Rex. Emerg Infect Dis 18:153–157. doi: 10.3201/eid1801.AD1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Page DL. 1953. Thucydides’ description of the great plague at Athens. Class Q 3:97–119. doi: 10.1017/S0009838800003050. [DOI] [Google Scholar]
- 367.Papagrigorakis MJ, Yapijakis C, Synodinos PN, Baziotopoulou-Valavani E. 2006. DNA examination of ancient dental pulp incriminates typhoid fever as a probable cause of the Plague of Athens. Int J Infect Dis 10:206–214. doi: 10.1016/j.ijid.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 368.Shapiro B, Rambaut A, Gilbert MTP. 2006. No proof that typhoid caused the Plague of Athens (a reply to Papagrigorakis et al.). Int J Infect Dis 10:334–335. doi: 10.1016/j.ijid.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 369.Terrasson J. 1743. Histoire universelle de Diodore de Sicile. Chez François Changuion, Amsterdam, Netherlands. [Google Scholar]
- 370.Sáez A. 2016. The Antonine plague: a global pestilence in the II century dC. Rev Chilena Infectol 33:218–221. (In Spanish). doi: 10.4067/S0716-10182016000200011. [DOI] [PubMed] [Google Scholar]
- 371.Sabbatani S, Fiorino S. 2009. The Antonine Plague and the decline of the Roman Empire. Infez Med 17:261–275. (In Italian.) [PubMed] [Google Scholar]
- 372.Haas C. 2006. The Antonine plague. Bull Acad Natl Med 190:1093–1098. (In French.) . doi: 10.1016/S0001-4079(19)33258-3. [DOI] [PubMed] [Google Scholar]
- 373.Harper K. 2015. Pandemics and passages to late antiquity: rethinking the plague of c. 249–270 described by Cyprian. J Roman Archaeol 28:223–260. doi: 10.1017/S1047759415002470. [DOI] [Google Scholar]
- 374.Tiradritti F. 2014. Of kilns and corpses: Theban plague victims. Egypt Archaeol 44:15–18. [Google Scholar]
- 375.Andrades Valtueña A, Mittnik A, Key FM, Haak W, Allmäe R, Belinskij A, Daubaras M, Feldman M, Jankauskas R, Janković I, Massy K, Novak M, Pfrengle S, Reinhold S, Šlaus M, Spyrou MA, Szécsényi-Nagy A, Tõrv M, Hansen S, Bos KI, Stockhammer PW, Herbig A, Krause J. 2017. The Stone Age Plague and its persistence in Eurasia. Curr Biol 27:3683–3691.e8. doi: 10.1016/j.cub.2017.10.025. [DOI] [PubMed] [Google Scholar]
- 376.Rascovan N, Sjögren K-G, Kristiansen K, Nielsen R, Willerslev E, Desnues C, Rasmussen S. 2019. Emergence and spread of basal lineages of Yersinia pestis during the Neolithic decline. Cell 176:295–305.e10. doi: 10.1016/j.cell.2018.11.005. [DOI] [PubMed] [Google Scholar]
- 377.Rasmussen S, Allentoft ME, Nielsen K, Orlando L, Sikora M, Sjögren K-G, Pedersen AG, Schubert M, Van Dam A, Kapel CMO, Nielsen HB, Brunak S, Avetisyan P, Epimakhov A, Khalyapin MV, Gnuni A, Kriiska A, Lasak I, Metspalu M, Moiseyev V, Gromov A, Pokutta D, Saag L, Varul L, Yepiskoposyan L, Sicheritz-Pontén T, Foley RA, Lahr MM, Nielsen R, Kristiansen K, Willerslev E. 2015. Early divergent strains of Yersinia pestis in Eurasia 5,000 years ago. Cell 163:571–582. doi: 10.1016/j.cell.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378.Spyrou MA, Tukhbatova RI, Wang C-C, Valtueña AA, Lankapalli AK, Kondrashin VV, Tsybin VA, Khokhlov A, Kühnert D, Herbig A, Bos KI, Krause J. 2018. Analysis of 3800-year-old Yersinia pestis genomes suggests Bronze Age origin for bubonic plague. Nat Commun 9:2234. doi: 10.1038/s41467-018-04550-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Yu H, Spyrou MA, Karapetian M, Shnaider S, Radzevičiūtė R, Nägele K, Neumann GU, Penske S, Zech J, Lucas M, LeRoux P, Roberts P, Pavlenok G, Buzhilova A, Posth C, Jeong C, Krause J. 2020. Paleolithic to Bronze Age Siberians reveal connections with first Americans and across Eurasia. Cell 181:1232–1245.e20. doi: 10.1016/j.cell.2020.04.037. [DOI] [PubMed] [Google Scholar]
- 380.Antoine D. 2008. The archaeology of “plague.” Med Hist 52:101–114. doi: 10.1017/S0025727300072112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Cabezuelo U, Castex D. 1994. Le cimetière Saint-Pierre à Dreux (Eure-et-Loire). Dolmens, sarcophages et pierres tombales Les pratiques funéraires en Eure-et-Loir de la préhistoire à nos jours, p 68–69. Maison de l’Archéologie, Chartres, France. [Google Scholar]
- 382.Castex D. 1994. Mortalité, morbidité et gestion de l’espace funéraire au cours du Haut Moyen-Age: contribution spécifique de l’anthropologie biologique. PhD dissertation. University of Bordeaux 1, Bordeaux, France. [Google Scholar]
- 383.Signoli M, Chausserie-Laprée J, Dutour O. 1995. Étude anthropologique d’un charnier de la peste de 1720–1721 à Martigues. Préhist Anthropol Médit 4:173–189. [Google Scholar]
- 384.Dutour O, Signoli M, Georgeon E, DA Silva J. 1994. Le charnier de la Grande Peste de Marseille (rue Leca): données de la fouille de la partie centrale et premiers résultats anthropologiques. Préhist Anthropol Médit 3:191–203. [Google Scholar]
- 385.Drancourt M, Aboudharam G, Signoli M, Dutour O, Raoult D. 1998. Detection of 400-year-old Yersinia pestis DNA in human dental pulp: an approach to the diagnosis of ancient septicemia. Proc Natl Acad Sci U S A 95:12637–12640. doi: 10.1073/pnas.95.21.12637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Cambi F, Dallai L. 2000. Archeologia di un monastero: gli scavi a San Salvatore al monte Amiata. Archeol Mediev 27:193. [Google Scholar]
- 387.Bos KI, Schuenemann VJ, Golding GB, Burbano HA, Waglechner N, Coombes BK, McPhee JB, DeWitte SN, Meyer M, Schmedes S, Wood J, Earn DJD, Herring DA, Bauer P, Poinar HN, Krause J. 2011. A draft genome of Yersinia pestis from victims of the Black Death. Nature 478:506–510. doi: 10.1038/nature10549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 388.Hawkins D. 1990. The Black Death and the new London cemeteries of 1348. Antiquity 64:637–642. doi: 10.1017/S0003598X0007856X. [DOI] [Google Scholar]
- 389.Hartle R, Carty N, Henderson M, Knox EL, Walker D. 2017. The new churchyard: from Moorfields marsh to Bethlem burial ground, Brokers Row and Liverpool Street. Museum of London Archaeology, London, United Kingdom. [Google Scholar]
- 390.Spyrou MA, Tukhbatova RI, Feldman M, Drath J, Kacki S, Beltrán de Heredia J, Arnold S, Sitdikov AG, Castex D, Wahl J, Gazimzyanov IR, Nurgaliev DK, Herbig A, Bos KI, Krause J. 2016. Historical Y. pestis genomes reveal the European Black Death as the source of ancient and modern plague pandemics. Cell Host Microbe 19:874–881. doi: 10.1016/j.chom.2016.05.012. [DOI] [PubMed] [Google Scholar]
- 391.Kacki S, Castex D. 2014. La sépulture multiple de la Basillque des Saints Martyrs Just et Pastor: bio-archéologie des restes humains. QUARHIS 10:180–199. [Google Scholar]
- 392.Bos KI, Herbig A, Sahl J, Waglechner N, Fourment M, Forrest SA, Klunk J, Schuenemann VJ, Poinar D, Kuch M, Golding GB, Dutour O, Keim P, Wagner DM, Holmes EC, Krause J, Poinar HN. 2016. Eighteenth century Yersinia pestis genomes reveal the long-term persistence of an historical plague focus. Elife 5:e12994. doi: 10.7554/eLife.12994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393.Signoli M, Bello S, Dutour O. 1998. La rechute épidémique de la grande peste de Marseille [mai-juillet 1722]: le charnier de l’observance. Méd Trop 58:7–13. [PubMed] [Google Scholar]
- 394.Derrick M. 2018. Follobaneprosjektet F04 Klypen Øst og Saxegaardsgata 15. Arkeologisk utgravning mellom Bispegata og Loenga. Middelalderparken og Saxegaardsgata 15 & 17, Oslo, Norway: Norsk institutt for kulturminneforskning, Oslo, Norway. [Google Scholar]
- 395.Gaillard H, Kacki S, Puig C, Bénézet J, Corrochano A. 2010. Premiers résultats concernant le site des Jardins de Saint-Benoît (Saint-Laurent-de-la-Cabrerisse, Aude), pôle religieux et funéraire des Corbières. Archeol Midi Mediev 28:209–218. doi: 10.3406/amime.2010.1926. [DOI] [Google Scholar]
- 396.Haas-Gebhard B. 1998. Ein frühmittelalterliches Gräberfeld bei Dittenheim (D). M. Mergoil. Europe medievale 1. [Google Scholar]
- 397.Raynaud C. 2011. Les nécropoles de Lunel-Viel (Hérault) de l’Antiquité tardive au Moyen Âge. Editions de l'Association de la Revue Archéologique de Narbonnaise. Revue Archéologique de Narbonnaise, supplément 41. [Google Scholar]
- 398.Kriiska A, Lõugas L, Lõhmus M, Mannermaa K, Johanson K. 2007. New AMS dates from Estonian Stone Age burial sites. Estonian J Archaeol 11:83–121. [Google Scholar]
- 399.Rostunov VL. 2006. Opyt prekonstrukcii sakral’nogo postranstva rannykh kurganov Evropy I Severogo Kavkaza. Severo-Oseinskiy institute gumannitarnykh I sozial’nykh issledovanny, Vladikavkaz, Russia. [Google Scholar]
- 400.Stockhammer PW, Massy K, Knipper C, Friedrich R, Kromer B, Lindauer S, Radosavljević J, Wittenborn F, Krause J. 2015. Rewriting the central European Early Bronze Age chronology: evidence from large-scale radiocarbon dating. PLoS One 10:e0139705. doi: 10.1371/journal.pone.0139705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.Tebelškis P, Jankauskas R. 2006. The Late Neolithic grave at Gyvakarai in Lithuania in the context of current archaeological and anthropological knowledge. Archaeol Balt 6:8–21. [Google Scholar]
- 402.Barbieri R, Mai BHA, Chenal T, Bassi M-L, Gandia D, Camoin-Jau L, Lepidi H, Aboudharam G, Drancourt M. 2020. A 2,000-year-old specimen with intraerythrocytic Bartonella quintana. Sci Rep 10:1–8. doi: 10.1038/s41598-020-66917-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403.Drancourt M, Aboudharam G, Croce O, Armougom F, Robert C, Raoult D. 2017. Dental pulp as a source of low-contaminated DNA. Microb Pathog 105:63–67. doi: 10.1016/j.micpath.2017.02.007. [DOI] [PubMed] [Google Scholar]
- 404.Haensch S, Bianucci R, Signoli M, Rajerison M, Schultz M, Kacki S, Vermunt M, Weston DA, Hurst D, Achtman M, Carniel E, Bramanti B. 2010. Distinct clones of Yersinia pestis caused the black death. PLoS Pathog 6:e1001134. doi: 10.1371/journal.ppat.1001134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 405.Drancourt M, Signoli M, Dang LV, Bizot B, Roux V, Tzortzis S, Raoult D. 2007. Yersinia pestis Orientalis in remains of ancient plague patients. Emerg Infect Dis 13:332–333. doi: 10.3201/eid1302.060197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 406.Schuenemann VJ, Bos K, DeWitte S, Schmedes S, Jamieson J, Mittnik A, Forrest S, Coombes BK, Wood JW, Earn DJD, White W, Krause J, Poinar HN. 2011. Targeted enrichment of ancient pathogens yielding the pPCP1 plasmid of Yersinia pestis from victims of the Black Death. Proc Natl Acad Sci U S A 108:E746–E752. doi: 10.1073/pnas.1105107108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 407.Pusch CM, Rahalison L, Blin N, Nicholson GJ, Czarnetzki A. 2004. Yersinial F1 antigen and the cause of Black Death. Lancet Infect Dis 4:484–485. doi: 10.1016/S1473-3099(04)01099-0. [DOI] [PubMed] [Google Scholar]
- 408.Giffin K, Lankapalli AK, Sabin S, Spyrou MA, Posth C, Kozakaitė J, Friedrich R, Miliauskienė Ž, Jankauskas R, Herbig A, Bos KI. 2020. A treponemal genome from an historic plague victim supports a recent emergence of yaws and its presence in 15th century Europe. Sci Rep 10:1–13. doi: 10.1038/s41598-020-66012-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409.Raoult D, Aboudharam G, Crubézy E, Larrouy G, Ludes B, Drancourt M. 2000. Molecular identification by “suicide PCR” of Yersinia pestis as the agent of medieval black death. Proc Natl Acad Sci U S A 97:12800–12803. doi: 10.1073/pnas.220225197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 410.Wiechmann I, Grupe G. 2005. Detection of Yersinia pestis DNA in two early medieval skeletal finds from Aschheim (Upper Bavaria, 6th century A.D.). Am J Phys Anthropol 126:48–55. doi: 10.1002/ajpa.10276. [DOI] [PubMed] [Google Scholar]
- 411.Bianucci R, Rahalison L, Massa ER, Peluso A, Ferroglio E, Signoli M. 2008. Technical note: a rapid diagnostic test detects plague in ancient human remains: an example of the interaction between archeological and biological approaches (southeastern France, 16th–18th centuries). Am J Phys Anthropol 136:361–367. doi: 10.1002/ajpa.20818. [DOI] [PubMed] [Google Scholar]
- 412.Bianucci R, Rahalison L, Peluso A, Rabino Massa E, Ferroglio E, Signoli M, Langlois J-Y, Gallien V. 2009. Plague immunodetection in remains of religious exhumed from burial sites in central France. J Archaeol Sci 36:616–621. doi: 10.1016/j.jas.2008.10.007. [DOI] [Google Scholar]
- 413.Malou N, Tran T-N-N, Nappez C, Signoli M, Le Forestier C, Castex D, Drancourt M, Raoult D. 2012. Immuno-PCR—a new tool for paleomicrobiology: the plague paradigm. PLoS One 7:e31744. doi: 10.1371/journal.pone.0031744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 414.Barbieri R, Mekni R, Levasseur A, Chabrière E, Signoli M, Tzortzis S, Aboudharam G, Drancourt M. 2017. Paleoproteomics of the dental pulp: the plague paradigm. PLoS One 12:e0180552. doi: 10.1371/journal.pone.0180552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 415.Arning N, Wilson DJ. 2020. The past, present and future of ancient bacterial DNA. Microb Genom 6:mgen000384. doi: 10.1099/mgen.0.000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 416.Bos KI, Kühnert D, Herbig A, Esquivel-Gomez LR, Andrades Valtueña A, Barquera R, Giffin K, Kumar Lankapalli A, Nelson EA, Sabin S, Spyrou MA, Krause J. 2019. Paleomicrobiology: diagnosis and evolution of ancient pathogens. Annu Rev Microbiol 73:639–666. doi: 10.1146/annurev-micro-090817-062436. [DOI] [PubMed] [Google Scholar]
- 417.Damgaard P, de B, Marchi N, Rasmussen S, Peyrot M, Renaud G, Korneliussen T, Moreno-Mayar JV, Pedersen MW, Goldberg A, Usmanova E, Baimukhanov N, Loman V, Hedeager L, Pedersen AG, Nielsen K, Afanasiev G, Akmatov K, Aldashev A, Alpaslan A, Baimbetov G, Bazaliiskii VI, Beisenov A, Boldbaatar B, Boldgiv B, Dorzhu C, Ellingvag S, Erdenebaatar D, Dajani R, Dmitriev E, Evdokimov V, Frei KM, Gromov A, Goryachev A, Hakonarson H, Hegay T, Khachatryan Z, Khaskhanov R, Kitov E, Kolbina A, Kubatbek T, Kukushkin A, Kukushkin I, Lau N, Margaryan A, Merkyte I, Mertz IV, Mertz VK, Mijiddorj E, Moiyesev V, Mukhtarova G, et al. 2018. 137 ancient human genomes from across the Eurasian steppes. Nature 557:369–374. doi: 10.1038/s41586-018-0094-2. [DOI] [PubMed] [Google Scholar]
- 418.Chain PSG, Carniel E, Larimer FW, Lamerdin J, Stoutland PO, Regala WM, Georgescu AM, Vergez LM, Land ML, Motin VL, Brubaker RR, Fowler J, Hinnebusch J, Marceau M, Medigue C, Simonet M, Chenal-Francisque V, Souza B, Dacheux D, Elliott JM, Derbise A, Hauser LJ, Garcia E. 2004. Insights into the evolution of Yersinia pestis through whole-genome comparison with Yersinia pseudotuberculosis. Proc Natl Acad Sci U S A 101:13826–13831. doi: 10.1073/pnas.0404012101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 419.Achtman M, Morelli G, Zhu P, Wirth T, Diehl I, Kusecek B, Vogler AJ, Wagner DM, Allender CJ, Easterday WR, Chenal-Francisque V, Worsham P, Thomson NR, Parkhill J, Lindler LE, Carniel E, Keim P. 2004. Microevolution and history of the plague bacillus, Yersinia pestis. Proc Natl Acad Sci U S A 101:17837–17842. doi: 10.1073/pnas.0408026101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 420.McNally A, Thomson NR, Reuter S, Wren BW. 2016. ‘Add, stir and reduce’: Yersinia spp. as model bacteria for pathogen evolution. Nat Rev Microbiol 14:177–190. doi: 10.1038/nrmicro.2015.29. [DOI] [PubMed] [Google Scholar]
- 421.Vetter SM, Eisen RJ, Schotthoefer AM, Montenieri JA, Holmes JL, Bobrov AG, Bearden SW, Perry RD, Gage KL. 2010. Biofilm formation is not required for early-phase transmission of Yersinia pestis. Microbiology (Reading) 156:2216–2225. doi: 10.1099/mic.0.037952-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 422.Benedictow OJ. 2019. Epidemiology of plague: problems with the use of mathematical epidemiological models in plague research and the question of transmission by human fleas and lice. Can J Infect Dis Med Microbiol 2019:1542024. doi: 10.1155/2019/1542024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 423.Green MH, Jones L, Little LK, Schamiloglu U, Sussman GD. 2014. Yersinia pestis and the three plague pandemics. Lancet Infect Dis 14:918. doi: 10.1016/S1473-3099(14)70878-3. [DOI] [PubMed] [Google Scholar]
- 424.Ford DC, Joshua GW, Wren BW, Oyston PC. 2014. The importance of the magnesium transporter MgtB for virulence of Yersinia pseudotuberculosis and Yersinia pestis. Microbiology (Reading) 160:2710–2717. doi: 10.1099/mic.0.080556-0. [DOI] [PubMed] [Google Scholar]
- 425.Radnedge L, Agron PG, Worsham PL, Andersen GL. 2002. Genome plasticity in Yersinia pestis. Microbiology (Reading) 148:1687–1698. doi: 10.1099/00221287-148-6-1687. [DOI] [PubMed] [Google Scholar]
- 426.Barbieri R, Drancourt M, Raoult D. 2019. Plague, camels, and lice. Proc Natl Acad Sci U S A 116:7620–7621. doi: 10.1073/pnas.1901145116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 427.Bramanti B, Namouchi A, Schmid BV, Dean KR, Stenseth NC. 2019. Reply to Barbieri et al.: Out of the land of darkness: plague on the fur trade routes. Proc Natl Acad Sci U S A 116:7622–7623. doi: 10.1073/pnas.1902274116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 428.Yue RP, Lee HF, Wu CY. 2017. Trade routes and plague transmission in pre-industrial Europe. Sci Rep 7:1–10. doi: 10.1038/s41598-017-13481-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 429.Schmid BV, Büntgen U, Easterday WR, Ginzler C, Walløe L, Bramanti B, Stenseth NC. 2015. Climate-driven introduction of the Black Death and successive plague reintroductions into Europe. Proc Natl Acad Sci U S A 112:3020–3025. doi: 10.1073/pnas.1412887112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 430.Seifert L, Wiechmann I, Harbeck M, Thomas A, Grupe G, Projahn M, Scholz HC, Riehm JM. 2016. Genotyping Yersinia pestis in historical plague: evidence for long-term persistence of Y. pestis in Europe from the 14th to the 17th century. PLoS One 11:e0145194. doi: 10.1371/journal.pone.0145194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 431.Novikova E, Lalazarov G. 1931. The rôle of bedbugs in the epidemiology of plague. I. The duration of viability of plague virus in the body of the infected bedbug. Rev Microbiol Saratov 10:323–324. [Google Scholar]
- 432.Wu L, Chun JWH, Pollitzer R, Wu CY. 1936. Plague: a manual for medical and public health workers. Weishengshu National Quarantine Service, Shanghai Station, Shanghai, China. [Google Scholar]
- 433.Manning JVV. 1912. Bedbugs and bubonic plague. Med Rec (1866–1922) 82:148. [Google Scholar]
- 434.Walker EA. 1910. Transmission of plague in the absence of rats and rat fleas. Ind Med Gaz 45:93–94. [PMC free article] [PubMed] [Google Scholar]
- 435.Bibel DJ, Chen TH. 1976. Diagnosis of plague: an analysis of the Yersin-Kitasato controversy. Bacteriol Rev 40:633–651. doi: 10.1128/MMBR.40.3.633-651.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 436.Williams JE, Hudson BW, Turner RW, Saroso JS, Cavanaugh DC. 1980. Plague in Central Java, Indonesia. Bull World Health Organ 58:459. [PMC free article] [PubMed] [Google Scholar]
- 437.Blanc G, Baltazard M. 1941. Recherches expérimentales sur la peste. L’infection du pou de l’homme. Pediculus corporis de Geer. Maroc Med 216:39. [Google Scholar]