Abstract
Background
Obstructive Sleep Apnea (OSA) is a highly prevalent and underdiagnosed sleep disorder. Recent studies suggest that OSA might disrupt the biological clock, potentially causing or worsening OSA-associated comorbidities. However, the effect of OSA treatment on clock disruption is not fully understood.
Methods
The impact of OSA and short- (four months) and long-term (two years) OSA treatment, with Continuous Positive Airway Pressure (CPAP), on the biological clock was investigated at four time points within 24 h, in OSA patients relative to controls subjects (no OSA) of the same sex and age group, in a case-control study. Plasma melatonin and cortisol, body temperature and the expression levels and rhythmicity of eleven clock genes in peripheral blood mononuclear cells (PBMCs) were assessed. Additional computational tools were used for a detailed data analysis.
Findings
OSA impacts on clock outputs and on the expression of several clock genes in PBMCs. Neither short- nor long-term treatment fully reverted OSA-induced alterations in the expression of clock genes. However, long-term treatment was able to re-establish levels of plasma melatonin and cortisol and body temperature. Machine learning methods could discriminate controls from untreated OSA patients. Following long-term treatment, the distinction between controls and patients disappeared, suggesting a closer similarity of the phenotypes.
Interpretation
OSA alters biological clock-related characteristics that differentially respond to short- and long-term CPAP treatment. Long-term CPAP was more efficient in counteracting OSA impact on the clock, but the obtained results suggest that it is not fully effective. A better understanding of the impact of OSA and OSA treatment on the clock may open new avenues to OSA diagnosis, monitoring and treatment.
Keywords: Obstructive sleep apnea, Continuous positive airway pressure (CPAP), Biological clock, Clock genes, Machine learning
Research in Context section.
Evidence before this study
Obstructive Sleep Apnea (OSA) is one of the most prevalent sleep disorders worldwide. Untreated, this disease has been associated with the development of a wide range of comorbidities, including hypertension, cardiovascular and metabolic diseases. Considerable efforts have been made to elucidate OSA-associated pathological mechanisms and impact on such a diverse range of comorbidities. Among these, the potential effect of OSA on the disruption of the biological clock has gained increasing attention. The biological clock regulates several biological processes of our body along the 24 h day, modulating physiology and behavior. Biological clock disruptions may exacerbate or play a causal role in the large-spectrum of adverse outcomes associated with OSA. However, only a few studies have explored the effects of OSA treatment on the clock.
Added value of this study
To the best of our knowledge, this study is the first comparing the effect of OSA and short (four months) and long-term (two years) Continuous Positive Airway Pressure (CPAP) treatment, the gold standard treatment for OSA, on biological clock-related characteristics. Our data reinforce that OSA impacts on the biological clock and show that short-term (four months) CPAP treatment does not counteract OSA effects on the clock. Long-term (two years) CPAP treatment does not fully re-establish the expression profile of clock genes, but shows evident ameliorations.
Implications of all the available evidence
Our data might partially explain the divergence of results regarding the impact of OSA treatment on the clock. Different treatment durations are frequent in different studies in the literature and may lead to different results and misleading interpretations regarding the effect of CPAP treatment on the clock of OSA patients. The fact that long-term (two years) treatment had more positive effects on the clock of OSA patients stresses the importance of patients’ compliance to treatment and is a call for further studies. On the other hand, the persistence of alterations even after long-term (two years) treatment suggests that CPAP might not fully re-establish clock-related characteristics, which reinforces the need for new approaches not only in OSA diagnosis but also OSA treatment. A deeper understanding of how OSA and OSA treatment affect the biological clock may lead to a better OSA clinical management.
Alt-text: Unlabelled box
1. Introduction
Obstructive Sleep Apnea (OSA) is one of the most common sleep disorders worldwide, estimated to affect nearly 1 billion people [1,2]. Its prevalence has substantially increased over the last decades and it is expected to increase continuously, mainly due to obesity and aging, two main OSA risk factors [1], [2], [3], [4]. This sleep-related breathing disorder is mainly characterized by recurrent breathing interruptions, caused by complete (apnea) or partial (hypopnea) obstructions in the upper airway during sleep, while respiratory efforts continue. This causes repetitive hypoxia/reoxygenation cycles that frequently promote arousals and micro arousals, leading to a fragmented and thus non-restorative sleep [5]. Thus, OSA patients commonly complain of fatigue and daytime sleepiness, which not only impacts their life quality, but also increases their risk for motor vehicle and workplace accidents [6]. Additionally, untreated OSA has been associated with the development of several conditions such as cardiovascular [7] and metabolic diseases [8], with increasing evidence of higher susceptibility to cancer [9], neurodegenerative disorders [10], depression [11] and musculoskeletal disorders [12]. Yet, 80 – 90 % of the OSA cases are estimated to be undiagnosed and thus untreated [1,2]. The gold standard for OSA treatment relies on continuous positive airway pressure (CPAP), which consists on the emission of pressurized air through a mask to avoid the collapse of the upper airways during sleep [5]. However, the low compliance of some patients to CPAP treatment, together with the controversial effects of CPAP treatment on OSA symptoms and comorbidities, questions the efficacy of this palliative strategy [13,14]. To counteract OSA burden, it is thus urgent to improve OSA diagnosis and treatment. In this context, several studies have explored OSA impact at the cellular and molecular levels to search for potential OSA biomarkers and to elucidate the mechanisms that may underlie OSA contribution to the development of such a large spectrum of associated comorbidities [15], [16], [17], [18], [19], [20]. Among these, OSA impact on biological rhythms have recently gained more attention [17,[21], [22], [23], [24], [25], [26]].
In mammals, several biological processes display circa 24 h rhythms that modulate physiology and behavior. Biological rhythms (also known as circadian rhythms) are generated by an endogenous timing system composed of a central and several peripheral clocks. The central clock, located in the suprachiasmatic nucleus, in the hypothalamus, is responsible for the synchronization of peripheral clocks within the body [27,28]. These exist in virtually all cells of our body and regulate the timing of various cellular and molecular processes, such as cell division, DNA-damage response and metabolism [27,29,30]. Consequently, multiple physiological and behavioural processes show marked circadian rhythms, among which body temperature, hormone levels (e.g. cortisol and melatonin), heart rate, blood pressure and sleep-wake cycles [28,30,31]. Core-clock genes and proteins are interconnected into a complex regulatory network of interlocked transcriptional/translational feedback loops that generate robust oscillations of approximately 24 h [29,[32], [33], [34]]. The core-clock network includes members of Brain and muscle ARNT‐like protein (BMAL1 and BMAL2), Circadian locomotor output cycles kaput (CLOCK) and RAR‐related orphan receptor (RORa, RORβ, and RORc) gene and protein families (positive limb); and members of Period (PER1, PER2, and PER3), Cryptochrome (CRY1 and CRY2) and the nuclear receptor, reverse strand of ERB (REV‐ERBα and REV‐ERBβ) gene and protein families (negative limb) [29,[32], [33], [34]]. Additional regulators, such as the transcription factors DEC1 and DEC2, and kinases (e.g. CSNK1ε) add further stability to this oscillatory network [32,35]. Core-clock proteins further control the expression of multiple target genes (clock-controlled genes), involved in numerous processes (e.g. cell cycle, metabolism, immune response, hormone regulation) [28,32,36]. As a consequence, disruptions in biological rhythms have been shown to promote several pathologies and lifespan decrease [28,31,[37], [38], [39]].
In the past years, impaired biological rhythms have been reported in OSA patients, in particular regarding hormone secretion [40], cytokine release [41], [42], [43], platelet activity [44], ocular [45] and blood pressure [40,[46], [47], [48], [49]]. Recent studies have also described alterations in the expression of several clock genes in peripheral blood mononuclear cells (PBMCs) of OSA patients [21], [22], [23], [24], [25]. Studies in OSA mouse models revealed that a short-term exposure (10 – 14 days) to intermittent hypoxia, during mice inactive phase, is sufficient to promote the dysregulation of clock genes expression in multiple cell types and lead to inter-tissue circadian misalignment. This suggests that OSA might impact on biological rhythms at an early stage of the disease and further contribute to/aggravate pathological mechanisms [50,51]. On the contrary, the effect of OSA treatment on the amelioration/ re-establishment of healthy rhythms is not so consensual [21,22,40] and has been less explored.
In this study, we investigated the impact of OSA and OSA short- (four months) and long-term (two years) CPAP treatment in the biological clock, by monitoring the levels and temporal profile of physiological clock-related outputs, namely, plasma melatonin, cortisol and body temperature, and the expression of core-clock genes in PBMCs, in 34 male OSA patients and seven control subjects (no OSA) of the same sex and age group (middle-age). We performed a detailed computational analysis using machine learning methods in the obtained data sets to better characterize the extent of OSA impact on the biological clock and the efficacy of OSA treatment on the re-establishment of biological clock-related characteristics.
Our results show that OSA promotes robust alterations in the levels and diurnal profiles of clock outputs and leads to the dysregulation of several clock genes. In addition, we show for the first time that short- (four months) and long-term (two years) CPAP treatment have a differential effect on the biological clock. Alterations in the expression of clock genes are not fully re-established with neither short- (four months) nor long-term (two years) OSA CPAP treatment. Nevertheless, long-term (two years) CPAP treatment leads to significant ameliorations in the levels and diurnal profiles of the assessed physiological clock outputs. Using a machine learning-based clustering analysis, we showed that OSA patients can be distinguished from control subjects based on the expression levels of clock genes and most strikingly, that the expression of clock genes in long-term (two years) treated OSA patients resembles the profiles of the control group. These observations suggest the existence of a direct effect of OSA and OSA CPAP treatment on biological rhythms and highlight a role for the biological clock as a promising tool in OSA diagnosis and monitoring of treatment response.
2. Materials and methods
2.1. Ethics
This study was approved by the Ethical Committees of the Faculty of Medicine of University of Coimbra (022-CE-2016; CE-011/2019) and of Coimbra Hospital and University center (CHUC, CHUC-013–19), Coimbra, Portugal. Experimental procedures were performed in accordance with ethical guidelines and regulations as laid down in the 1964 Declaration of Helsinki and its amendments; in 2016/679 provisions of the Regulation of the European Parliament and of the Council of April 27th 2016 (General Data Protection Regulation); and in the Portuguese Law n.° 12/2005 of January 26th and its regulation set forth in 131/2014 Decree-Law of August 29th 2014. Informed consent was obtained from all participants before the beginning of the study.
2.2. Study design
This case-control study only included male subjects given OSA-associated sex-specific differences [52] and sexual dimorphisms in body clocks [53]. Potential OSA patients and control subjects (age ≥ 18 years) observed in the Sleep Unit of Coimbra Hospital and University center, Coimbra, Portugal, between February 2016 and December 2019, were recruited to the current study, based on the presence or absence, respectively, of OSA-associated characteristics/symptoms, such as high cervical perimeter (> 40 cm), loud snoring, reported breathing interruptions or excessive daytime sleepiness. Selected subjects were consecutively admitted for full-night polysomnography (PSG) at the sleep unit (baseline visit, t0M). An ID was assigned to each subject to assure standardisable and blind data collection. Sleep studies were staged and respiratory events were scored according to the American Academy of Sleep Medicine (AASM) Scoring Manual v 2.4, 2012. Diagnosis and classification of OSA severity were based on the respiratory disturbance index (RDI) that considers the number of apneas, hypopneas and respiratory effort-related arousals (RERAs) per hour of sleep. According to AASM guidelines, subjects were classified as disease-free (control subjects, RDI < 5), mild OSA patients (5 ≤ RDI < 15), moderate OSA patients (15 ≤ RDI < 30) or severe OSA patients (RDI ≥ 30) [54].
For inclusion in the study, middle-aged male subjects who were not doing shift work were considered, given the known impact of age and shift work on the biological clock [55,56], and the impact of age on OSA progression [57]. Patients with OSA positive diagnosis, about to initiate CPAP treatment, not working nightshifts, were included in the study. In accordance, from the 47 patients that completed the baseline visit, 34 male OSA patients confirmed to be eligible and were included in the study. The criteria for inclusion of control subjects was based on OSA negative diagnosis, age (same age group of OSA patients - middle-age, 36 – 65 years) and absence of nightshifts. In accordance, seven control subjects were included.
All 34 OSA patients completed the baseline visit (t0M) between February 2016 and December 2019 and control subjects between March 2016 and November 2019. Control subjects underwent only the baseline visit whereas OSA patients underwent two more visits, after four months of CPAP treatment (t4M, short-term treatment) and after two years of CPAP treatment (t24M, long-term treatment). All OSA patients completed the t4M visit (between June 2016 and April 2020) and 16 patients completed the t24M visit (between June 2018 and November 2019). Treatment-associated visits (t4M and t24M) did not implied sleeping overnight in the Sleep Unit.
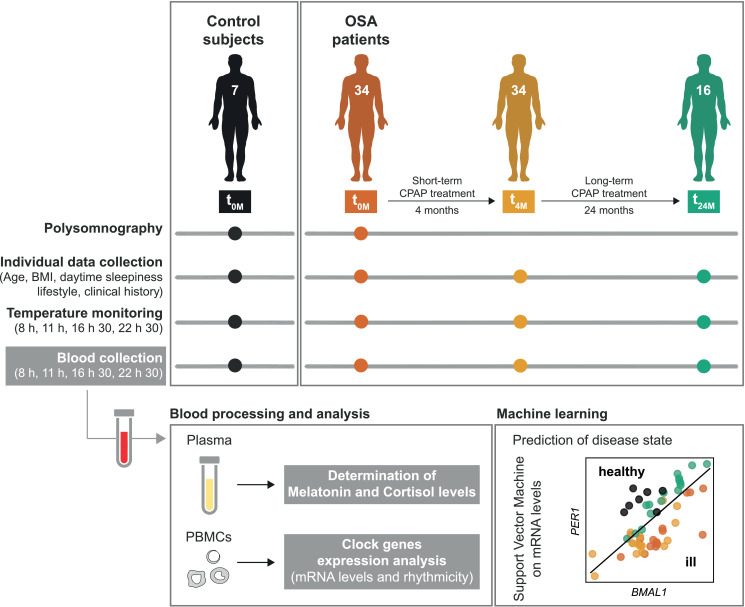
Individual data (age, BMI, daytime sleepiness, lifestyle and clinical history) were collected from controls at baseline (t0M), and from OSA patients at the three different visits of the study (t0M, t4M and t24M). In addition, at each visit (t0M, t4M and t24M), at four time points within the 24 h day (8, 11, 16:30 and 22:30 h, selected in accordance with the sleep unit routine), body temperature was monitored and blood was collected for analysis of melatonin and cortisol plasma levels and clock genes expression (biological clock-related characteristics). All participants were allowed to leave the Sleep Unit between the four time points, to maintain their daily routine. The study design is summarized in Fig. 1.
Fig. 1.
Scheme of the study design. Seven control subjects (no OSA) and 34 OSA patients were included in this study, after the Polysomnography test (PSG, baseline visit, t0M). OSA patients undergoing Continuous Positive Airway Pressure (CPAP) treatment were followed after four months (short-term, t4M, 34 patients) and two years (long-term, t24M, 16 patients) of treatment. Individual data [age, body mass index (BMI), lifestyle and clinical history] were collected from controls at baseline and from OSA patients at the three different visits of the study (t0M, t4M and t24M). In addition, body temperature was monitored and blood samples were collected throughout the day (at 8, 11, 16:30 and 22:30 h), at each visit, in accordance with the sleep unit routine. Blood samples were processed for plasma and peripheral blood mononuclear cells (PBMCs) isolation. Melatonin and cortisol levels were determined in plasma samples. PBMCs were further processed for analysis of clock genes expression levels and rhythmicity. Based on these data, health status was predicted using machine learning (support vector machine) for a better comprehension of the obtained data.
2.3. Individual data collection
Data from the PSG test (PSG report) were obtained from each participant of the study at baseline (t0M). In addition, age, body mass index – BMI, daytime sleepiness (Epworth Sleepiness Scale - ESS [58]), lifestyle (sleep/wake routine, diet, meal schedules, physical exercise) and clinical history (comorbidities and medication) were obtained from controls at baseline (t0M), and from OSA patients at the three different visits of the study (t0M, t4M and t24M), through questionnaires and clinical information provided by the Sleep Unit. At t4M and t24M, information regarding CPAP treatment compliance (percentage of use ≥ 4 h per night) and the number of apneas and hypopneas per hour of sleep (Apnea-Hypopnea Index – AHI, follows the same guidelines as RDI [54]) were also collected from each OSA patient (CPAP report).
2.4. Body temperature
Axillary temperature was measured twice in OSA patients at t0M, t4M and t24M, and control subjects, at four time points within 24 h (8, 11, 16:30 and 22:30 h), in accordance with the sleep unit routine, through an electronic thermometer. The average between the two measurements was used.
2.5. Blood samples collection and processing
Blood samples were collected at four time points within 24 h (8, 11, 16:30 and 22:30 h), in accordance with the sleep unit routine, from OSA patients at t0M, t4M and t24M, and control subjects.
Blood (15–20 mL) was drawn from the antecubital vein, using a 21‐gage needle, into K2EDTA Tubes (#367,839, BD Vacutainer) and processed. Blood samples were diluted in 25 mL of Phosphate Buffered Saline (PBS) 1 x and were slowly added on the top of 10 mL of Histopaque, density 1.077 g/mL (#10,771, Sigma-Aldrich), in 50 mL falcon tubes (11,809,650, Fisher Scientific). Samples were centrifuged at 800 g, for 20 mins, at room temperature (RT), with the brake off. Plasma was immediately aliquoted and stored at - 80 ⁰C while interface bands, containing PBMCs, were aspirated into new 50 mL falcon tubes. PBMCs were washed with PBS 1 x up to the 50 mL mark, followed by centrifugation at 600 g, for 10 mins, at RT, with the brake on. Supernatants were aspirated and PBMCs’ pellets were further resuspended in 3 mL of PBS 1x. Cells were aliquoted into 1 mL tubes, and centrifuged at 300 g, for 15 mins, at RT, with the brake on. Supernatants were aspirated and PBMCs aliquots were stored at - 80 ⁰C until further analysis.
2.6. Melatonin and cortisol plasma levels
Levels of plasma melatonin and cortisol were determined by human-specific commercial ELISA assays (EH3344 and EH0641 respectively, Fine Test), according to manufacturer's instructions. Absorbance was read at 450 nm on a SpectraMax Plus 384 Microplate Reader (Molecular Devices). Results were interpolated from standard curves generated by plotting the concentration of the standards of each kit against their absorbance, using a sigmoidal 4PL curve where X is log (concentration).
2.7. Clock genes expression analysis
Total RNA was extracted from PBMCs using the miRCURY RNA Isolation kit – Cell and Plant (#300,110, Exiqon) according to manufacturer's instructions. DNAse digestion was performed during the process to exclude contamination with genomic DNA. Total RNA was quantified by optical density (OD) measurements using the ND-1000 Nanodrop Spectrophotometer (Thermo Scientific) and purity was assessed by the OD ratio at 260 and 280 nm.
To assure removal of genomic DNA, samples were further treated with DNAse (79,254, Qiagen) in solution. Final volume was adjusted to 10 μL, containing 1 μL of DNAse buffer, 0.5 μL of DNAse and 1000 ng of mRNA. After 30 mins incubation at 37 ⁰C, 1 μL of 20 mM EDTA, pH=8, was added to each sample and incubated at 65 ⁰C, for 10 mins, to stop DNAse reaction. RNA samples were converted into cDNA, using the iScript cDNA Synthesis Kit (Bio-Rad), according to manufacturer's instructions.
The mRNA levels of BMAL1, CLOCK, PER1–3, CRY1–2, CSNK1ε, REV-ERBα, REV-ERBβ and DEC1 of all the enrolled subjects were assessed by real-time quantitative reverse transcriptase polymerase chain reaction (qRT-PCR), using the iCycler iQ™ Real-Time PCR Detection System (Bio-Rad). Primer sequences and qPCR conditions are listed in Table S1. Relative gene expression was calculated according to the ΔCT method of Livak et al. [59]. Hypoxanthine-guanine phosphoribosyltransferase (HPRT), Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and β−2-microglobulin (β−2 M) were used as housekeeping genes, as validated by BestKeeper, Genorm and Normfinder algorithms. Gene expression data is shown as ΔΔCT values, relative to the average of the ΔCt of all time points of all control subjects (average of all time points of all controls is equal to 0), except for the machine learning analysis, where the ΔCT values were used without normalization by controls.
2.8. Statistical analysis
If not stated otherwise, statistical analyses were performed using Graph Pad Prism 7.00 (GraphPad Software, Inc). Based on data distribution, non-parametric t-tests (Mann-Whitney tests) were performed to detect statistically significant differences between control and each OSA patients’ group (t0M, t4M or t24M), at each time of the day. Statistically significant differences promoted by CPAP treatment were further evaluated in patients that completed the three visits of the study (t0M, t4M and t24M), through non-parametric pairwise one-way ANOVA (Friedman tests), followed by Dunn's correction of multiple comparisons (Type I error was adjusted). All statistical tests performed were two-sided, with statistical significance set at 0.05. No values were excluded.
2.9. Rhythmicity analysis
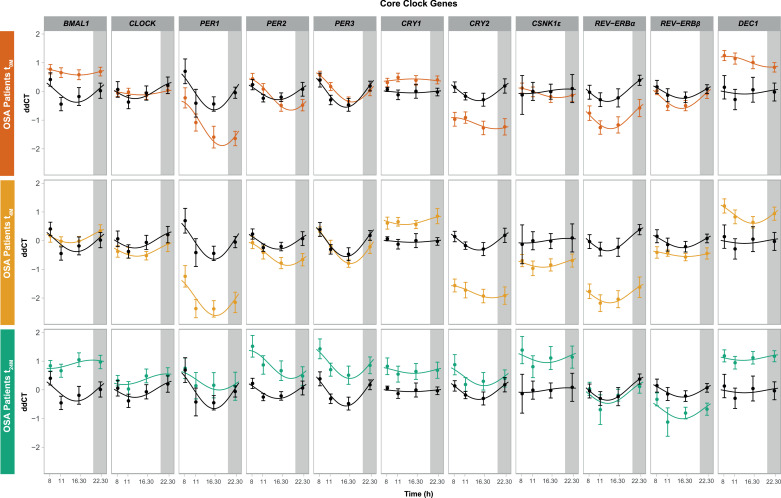
For the detection of circadian oscillating genes, harmonic regression fitting was used as implemented in the R package HarmonicRegression [60], setting the period to 24 h. Harmonic regression fits a linear model to time-series using the function y(t) = m + a × sin (2 × π × t/ω) + b × cos (2 × π × t/ω). Estimated amplitudes (formulated as A= √ (a2 + b2)) and acrophases (formulated as tan φ = b/a) along with p-values (according to F-test) and Benjamini-Hochberg adjusted p-values (q-values) can be found in Table S2 and S3 for mean gene expression values and for each individuals time course data, respectively. A representative harmonic regression curve was fitted to the mean of all participants of the study and results were visualized using ΔΔCT values normalized to the average of control donors. The summary plot using each data from each participant of the study is depicted using the standard error of mean and the figures plotted in R programming software using ggplot2 package (v.3.3.2). The original ∆∆CT values are depicted as points in the plots and the time points were connected using lines for the individual time course of each participant (Fig. S1) whereas a smooth harmonic regression curve was used for connecting the summary plot (Fig. 4).
Fig. 4.
The temporal expression of core-clock genes in peripheral blood mononuclear cells is affected by OSA and OSA Continuous Positive Airway Pressure (CPAP) treatment in patients, compared to control subjects. Clock genes expression oscillations (BMAL1,CLOCK, PER1–3, CRY1–2, CSNK1ε, REV-ERBα, REV-ERBβ and DEC1) along day (8, 11, 16:30 and 22:30 h) in PBMCs of OSA patients at the three visits of the study, relative to control subjects (black line). (a) OSA patients at diagnosis (t0M, orange line); (b) OSA patients after short-term (four months) CPAP treatment (t4M, yellow line); (c) OSA patients after long-term (two years) CPAP treatment (t24M, green line). Results are presented using harmonic regression for the data collected at 8, 11, 16:30 and 22:30 h, following the summarization of each participant data using the standard error of the mean ΔΔCT values ± SEM of 34 OSA patients at t0M, 34 patients at t4M and 16 patients at t24M.
2.10. Machine learning
For machine learning analysis, the data set was restricted to the 16 patients that completed all three visits of the study (t0M, t4M and t24M), plus the 7 controls.
For machine learning, the python package scikit-learn was used. For the set of n = 23 subjects (7 controls and 16 patients), let xi denote the data set consisting of the temporal means of the genetic profiles for the mRNAs measured in subject i, i from 1 to n. The subjects were separated into two or three groups (clusters) using the algorithm K-means clustering with k = 2 or 3 and the standard parametrization of sklearn.cluster.KMeans(). K-means clustering minimizes the within-cluster sum-of-squares criterion in the resulting clusters, which is given as
where C are the k clusters and for each cluster μj is the mean of the elements in the cluster. To predict the condition of a given subject (e.g. control versus patient as label), we trained a linear support vector machine (SVM, sklearn.svm.LinearSVC() with max_iter=90,000) using leave-one-out cross validation, i.e. for each element of the data set, the SVM is trained on the data excluding the chosen element, and then tested on this element. A linear support vector machine was chosen because of its simplicity and interpretability, which is advantageous when considering the size of the training set. To quantify classification success, the mean F1 score (F1 = 2 * (precision * recall) / (precision + recall)) was used, as implemented under sklearn.metrics.f1_score() with average='macro'. Individual F1 scores of both labels are also reported. The F1 score goes from zero (worst) to one (best), and a boot strap approach allows us to estimate a confidence interval for the F1 score, i.e. for each element of the data set, we train and test the SVM on the restricted data which lacks this element, and calculate the mean and standard deviation (std) of the resulting set of N F1 scores, where N is the number of elements in the data set. The 90 % confidence interval is then given as mean ± 1.645 * std/sqrt(N), where sqrt() is the square root function.
2.11. Role of funding
Funding sources had no role in study design, data collection, data analysis, interpretation, decision to publish or preparation of the manuscript.
3. Results
3.1. OSA impacts on some of the major clock outputs and only long-term (two years) CPAP treatment is effective on their re-establishment
The current study enrolled 34 male OSA patients and seven controls (no OSA) of the same sex and age-group. Participants were first characterized based on age, BMI, daytime sleepiness (ESS), lifestyle and clinical history (table 1), and sleep-related characteristics, assessed by PSG (table 2). OSA patients and control subjects did not show statistically significant differences in daytime sleepiness (p = 0.448, Mann-Whitney test) or BMI (p = 0.08, Mann-Whitney test) (table 1). OSA patients showed several comorbidities, with the most common being hypertension (73 %), metabolic conditions (71 %), among which diabetes and/or dyslipidaemia, and cardiovascular diseases (35 %). Usual medication is depicted in table 1. Significant differences were also found in sleep-related parameters assessed by PSG, namely, in the percentage of deep sleep, number of obstruction events, number of arousals and awakenings and blood oxygen saturation levels (SpO2) during sleep (see table 2). Among the group of OSA patients, four (12 %) patients were diagnosed with mild (5 ≤ RDI < 15), eight (23 %) with moderate (15 ≤ RDI < 30) and 22 (6 5 %) patients with severe (RDI ≥ 30) OSA.
Table 1.
Age, body mass index, daytime sleepiness, lifestyle and clinical history of the control subjects and OSA patients enrolled in this study (baseline, t0M).
| Control subjects n = 7 | OSA patients n = 34 | |
|---|---|---|
| Age | 49.7 ± 3.2 | 54.6 ± 1.8 |
| BMI | 28.1 ± 1.3 | 31.3 ± 0.8 |
| Daytime sleepiness – ESS | 8.6 ± 2.9 | 9.8 ± 0.7 |
| Sleep/wake cycles | ||
| Bedtime, h | 23.7 ± 0.4 | 23.6 ± 0.2 |
| Wake up time, h | 7.1 ± 0.2 | 7.3 ± 0.2 |
| Hours in bed, h | 6.9 ± 0.4 | 6.7 ± 0.2 |
| Diet | ||
| Healthy diet, y/n (%) | 6/7 (86%) | 30/33 (91%) |
| Regular diet, y/n (%) | 6/7 (86%) | 25/33 (76%) |
| Number of meals per day, average (n) | 4[7] | 4 [33] |
| Physical activity, y/n (%) | 3/7 (43%) | 8/33 (24%) |
| Frequency, average/week | 2 times | 3 times |
| At night | – | – |
| Diagnosed conditions | ||
| Hypertension, y/n (%) | – | 25/34 (73%) |
| Metabolic disorders, y/n (%) | 2/7 (29%) | 24/34 (71%) |
| Cardiovascular diseases, y/n (%) | – | 12/34 (35%) |
| Musculoskeletal disorders, y/n (%) | – | 9/34 (26%) |
| Anxiety, y/n (%) | 1/7 (14%) | 8/34 (23%) |
| Gastrointestinal disorders, y/n (%) | 1/7 (14%) | 6/34 (18%) |
| Benign prostatic hyperplasia, y/n (%) | 1/7 (14%) | 5/34 (15%) |
| Mood disorders, y/n (%) | 1/7 (14%) | 2/34 (6%) |
| Asthma/ COPD, y/n (%) | 1/7 (14%) | 2/34 (6%) |
| Kidney diseases, y/n (%) | – | 2/34 (6%) |
| Inflammatory diseases, y/n (%) | – | 2/34 (6%) |
| Endocrine disorders, y/n (%) | 1/7 (14%) | 1/34 (3%) |
| Medication | ||
| Antihypertensives, y/n (%) | – | 25/34 (73%) |
| Anticholesterolemic agents, y/n (%) | 2/7 (29%) | 23/34 (68%) |
| Hypoglicemic/Antidiabetic agents, y/n (%) | – | 11/34 (32%) |
| Heart drugs, y/n (%) | – | 10/34 (29%) |
| Anxiolytics, y/n (%) | 1/7 (14%) | 8/34 (23%) |
| Drugs for gastrointestinal system, y/n (%) | – | 10/34 (29%) |
| Antigout agents, y/n (%) | – | 6/34 (18%) |
| Anti-inflammatories, y/n (%) | – | 4/34 (12%) |
| Medication for BPH, y/n (%) | 1/7 (14%) | 4/34 (12%) |
| Antidepressants, y/n (%) | 2/7 (29%) | 4/34 (15%) |
| Asthma/COPD drugs, y/n (%) | 1/7 (14%) | 2/34 (6%) |
| Antihistamines, y/n (%) | 1/7 (14%) | 2/34 (6%) |
| Pain killers, y/n (%) | – | 2/34 (6%) |
| Thyroid drugs, y/n (%) | 1/7 (14%) | 1/34 (3%) |
| Drugs for kidney diseases, y/n (%) | – | 1/34 (3%) |
| Antipsychotics, y/n (%) | 1/7 (14%) | – |
| Anticonvulsants, y/n (%) | 1/7 (14%) | – |
BMI, Body mass index. ESS, Epworth sleepiness scale. COPD, Chronic Obstructive Pulmonary Disease. BPH, Benign prostatic hyperplasia. y/n, number of subjects included in the category per total number of subjects, in the group, from who data is available. Data is presented as mean ± SEM when applicable.
Table 2.
Sleep-related characteristics, assessed by Polysomnography, of control subjects and OSA patients.
| Control subjects n = 7 | OSA patients t0Mn = 34 | p-value | |
|---|---|---|---|
| Total sleep time, h | 6.5 ± 0.5 | 5.6 ± 0.2 | 0.122 |
| Sleep efficiency,% | 86.5 ± 2.9 | 74.5 ± 2.6 | 0.085 |
| Sleep staging | |||
| NREM,% of TST | 85.5 ± 0.9 | 86.6 ± 1.0 | 0.557 |
| Light sleep,% NREM | 60.2 ± 7.5 | 74.3 ± 2.2 | 0.090 |
| Deep sleep,% of NREM | 25.2 ± 8.1 | 12.3 ± 1•6 | * |
| REM,% of TST | 14.5 ± 0.9 | 13.4 ± 1.0 | 0.557 |
| Apnea índex | 0.1 ± 0.1 | 13.7 ± 3.6 | ** |
| Hypopnea índex | 4.1 ± 0.7 | 32.0 ± 3.6 | *** |
| AHI | 4.2 ± 0.7 | 45.7 ± 4.5 | **** |
| RERAs, count | 11.0 ± 6.3 | 18.2 ± 5.3 | 0.781 |
| RDI | 4.3 ± 0.7 | 46.0 ± 4.4 | **** |
| In NREM sleep | 3.4 ± 1.0 | 45.9 ± 4.7 | **** |
| In REM sleep | 9.4 ± 1.6 | 45.7 ± 3.7 | ** |
| Arousals and awakenings índex | 17 ± 2.0 | 40.6 ± 2.9 | ** |
| SpO2 mean,% | 94.5 ± 0.5 | 91.6 ± 0.5 | * |
| In NREM sleep | 94.5 ± 0.5 | 91.1 ± 0.5 | ** |
| In REM sleep | 94.3 ± 0.3 | 88.6 ± 0.9 | ** |
| SpO2 minimum,% | 89 ± 0.9 | 76.8 ± 1.9 | *** |
| SpO2 < 90%,% of TST | 0.1 ± 0.1 | 22.4 ± 4.3 | *** |
NREM, Non-rapid eye movement sleep. REM, Rapid eye movement sleep. TST, Total sleep time. Apnea index, number of apneas per hour of sleep. Hypopnea index, number of hypopneas per hour of sleep. AHI, Apnea-Hypopnea Index, number of apneas and hypopneas per hour of sleep. RERAs, Respiratory-effort related arousals. RDI, Respiratory disturbance index, number of apneas, hypopneas and respiratory effort-related arousals (RERAs) per hour of sleep. Arousals and awakenings index, number of arousals and awakenings per hour of sleep. SpO2, Peripheral capillary oxygen saturation. Data is presented as mean ± SEM. p-value, p-value for comparison between control subjects and OSA patients at t0M, calculated using Mann-Whitney tests: * p < 0.05; ** p < 0.01; *** p < 0.001; **** p < 0.0001.
OSA patients were followed from the moment of their diagnosis by PSG (t0M), up to four months (t4M, short-term treatment) and two years of CPAP treatment (t24M, long-term treatment). All 34 patients completed both t0M and t4M visits of the study and 16 patients completed t24M visit. CPAP treatment compliance and response are shown in table 3. The majority of OSA patients complied with CPAP treatment, using the mask for more than 4 h per night, both at t4M and t24M. Both short- and long-term CPAP were effective at reducing the number of obstruction episodes during sleep, to values below 5 [p < 0.0001 (****), Friedman test, 16 patients], and daytime sleepiness [p < 0.0001 (****), Friedman test, 13 patients] in OSA patients (table 3), suggesting that both treatment durations improved OSA condition. Patients’ BMI, comorbidities and medication remained constant throughout the study.
Table 3.
Follow-up of OSA patients before (t0M) and after short (four months, t4M) and long-term (two years, t24M) Continuous Positive Airway Pressure (CPAP) treatment.
| OSA patients t0M (n = 16) | OSA patients t4M (n = 16) | OSA patients t24M (n = 16) | p-value | |
|---|---|---|---|---|
| CPAP treatment compliance,% | 83.5 ± 5.8 | 83.2 ± 6.4 | 0.515 | |
| AHI | 41.9 ± 7.5 | 1.0 ± 0.2 #### | 1.1 ± 0.2 #### | **** |
| Daytime sleepiness – ESS | 10.7 ± 0.9 | 6.8 ± 1.4 ## | 4.7 ± 1.0 ### | **** |
| BMI | 30.9 ± 1.2 | 30.1 ± 0.8 | 0.649 |
CPAP treatment compliance, percentage of days with 4 h or more of continuous positive airway pressure, per night. AHI, Apnea-Hypopnea Index. ESS, Epworth sleepiness scale. BMI, Body mass index. Data is presented as mean ± SEM when applicable. p-value, p-value for comparison between two groups (t0M and t24M or t4M and t24M), calculated using Wilcoxon matched-pairs signed rank tests, or three groups (t0M, t4M and t24M), calculated using Friedman tests: **** p < 0.0001. Friedman tests were followed by Dunn's multiple comparison tests between t0M, t4M and t24M, where # in each group section represents statistical significant differences relative to t0M: ## p < 0.01, ### p < 0.001, #### p < 0.0001.
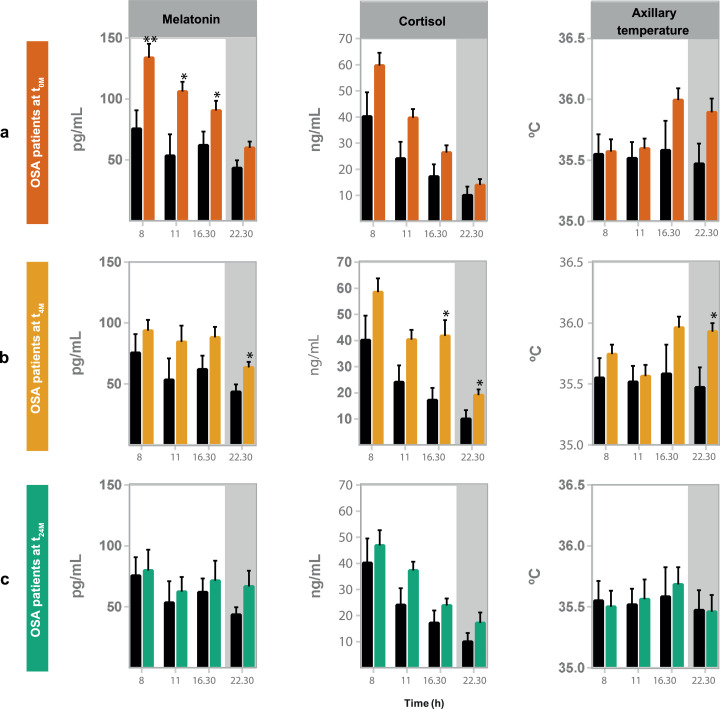
To evaluate OSA impact on the biological clock, we started by assessing clock outputs, namely, plasma levels of melatonin and cortisol and body temperature, surrogate markers of the biological clock, along the day (8, 11, 16:30 and 22:30 h), in OSA patients in comparison with control subjects (Fig. 2). OSA patients showed increased plasma melatonin levels during the day (at 8, 11 and 16:30 h), especially in the early morning (8 h) (Fig. 2a). Short-term CPAP treatment (four months, t4M) significantly reduced melatonin levels during the daytime (Fig. 2b, table 4). Yet, higher melatonin plasma and cortisol levels and body temperature became more pronounced at night (22:30 h) at t4M, relative to control subjects (Fig. 2b). Long-term (two years, t24M) treatment re-established the levels and diurnal profiles of plasma melatonin, cortisol and especially body temperature (Fig. 2c, table 4).
Fig. 2.
OSA impact in the levels and profiles of plasma melatonin, cortisol and body temperature is no longer observed upon long-term (two years) Continuous Positive Airway Pressure (CPAP) treatment. Levels of major clock outputs (plasma melatonin, cortisol and body temperature) measured along day (at 8, 11, 16:30 and 22:30 h) in OSA patients at the three visits of the study, relative to control subjects (black line). (a) OSA patients at diagnosis (t0M, orange line); (b) OSA patients after four months of CPAP (t4M, yellow line); (c) OSA patients after two years of CPAP treatment (t24M, green line). Plasma melatonin and cortisol are presented as mean ± SEM of 16 (melatonin) and 17 (cortisol) OSA patients at t0M and t4M and 9 patients at t24M. Body temperature is presented as mean ± SEM of 26 OSA patients at t0M, 34 patients at t4M and 16 patients at t24M. Non-parametric student's t-tests (Mann-Whitney tests) were used to assess differences between t0M, t4M or t24M and controls subjects, at each time point along the day: * p < 0.05; ** p < 0.01.
Table 4.
Short (four months, t4M) and long-term (two years, t24M) Continuous Positive Airway Pressure (CPAP) treatment promote differential alterations in plasma melatonin levels and body temperature of OSA patients (t0M).
| Clock output | Time of the day | Friedman test p-value | |||
|---|---|---|---|---|---|
| OSA patients t0M | OSA patients t4M | OSA patients t24M | |||
| Melatonin (pg/mL) | 8 h | 133.1 ± 14.0 | 91.9 ± 12.4 (*) | 79.8 ± 17.0 (*) | * |
| 11 h | 117.5 ± 9.8 | 91.1 ± 23.2 (*) | 62.5 ± 12.0 (*) | * | |
| 16:30 h | 91.1 ± 12.7 | 86.9 ± 11.3 | 71.5 ± 16.3 | ||
| 22:30 h | 58.5 ± 9.4 | 57.6 ± 4.5 | 64.0 ± 14.7 | ||
| Cortisol (ng/mL) | 8 h | 52.4 ± 5.5 | 46.5 ± 5.2 | 46.9 ± 5.8 | |
| 11 h | 42.1 ± 4.8 | 32.6 ± 3.2 | 37.2 ± 3.4 | ||
| 16:30 h | 27.4 ± 3.7 | 30.4 ± 5.1 | 23.8 ± 2.7 | ||
| 22:30 h | 16.8 ± 2.9 | 13.8 ± 3.0 | 14.9 ± 3.8 | ||
| Body temperature ( °C) | 8 h | 35.6 ± 0.1 | 35.9 ± 0.2 | 35.6 ± 0.1 | |
| 11 h | 35.5 ± 0.1 | 35.8 ± 0.1 | 35.4 ± 0.2 | ||
| 16:30 h | 36.2 ± 0.1 | 36.4 ± 0.1 | 35.8 ± 0.1 (*) (##) | ** | |
| 22:30 h | 36.2 ± 0.1 | 35.9 ± 0.1 | 35.5 ± 0.2 | ||
Data presented as mean ± SEM, at each time point of the day, from 7–10 OSA patients that completed long-term (two years) Continuous Positive Airway Pressure (CPAP) treatment (two years follow-up). Friedman tests were performed at each time point, throughout time (from t0M to t24M) to visualize how much variables have changed throughout treatment: * p < 0.05; ** p < 0.01. In addition, Friedman tests were followed by Dunn's multiple comparisons tests to visualize statistical differences relative to t0M (marked as *) and between short- and long-term treatment (marked as #), in each group column: (*) p < 0.05; (##) p < 0.01.
3.2. OSA promotes alterations in expression levels and the temporal expression profiles of several clock genes that are not fully re-established by neither short- (four months) nor long-term (two years) CPAP treatment
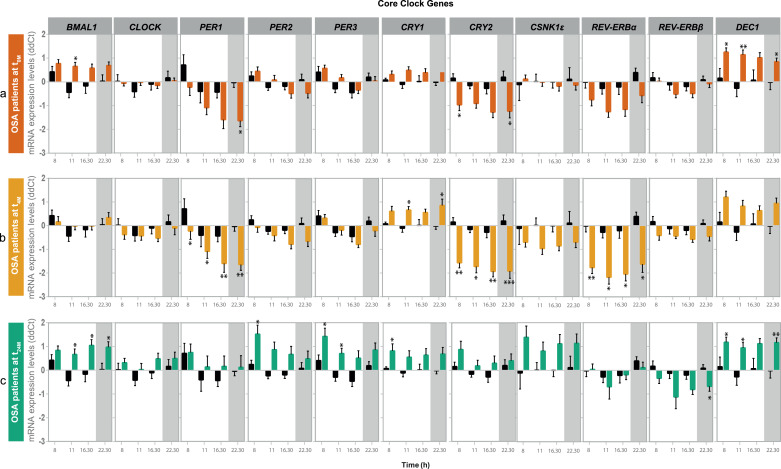
We further explored the impact of OSA on biological clock-related characteristics by assessing the expression levels of eleven clock genes (BMAL1, CLOCK, PER1-3, CRY1-2, CSNK1ε, REV-ERBα, REV-ERBβ and DEC1) along the four time points of the study, in PBMCs of OSA patients, compared to control subjects. OSA patients showed increased morning BMAL1 and DEC1 expression levels and decreased expression of PER1, CRY2 and REV-ERBα (the latter not statistically significant), at night, relative to control subjects. No alterations were observed in the expression levels of CLOCK, PER2, PER3, CRY1, REV-ERBβ and CSNK1ε in PBMCs of OSA patients (Fig. 3a).
Fig. 3.
OSA causes alterations in the expression of several core-clock genes in peripheral blood mononuclear cells (PBMCs) that are not fully re-established with neither short- (four months) or long-term (two years) Continuous Positive Airway Pressure (CPAP) treatment. Expression levels of 11 clock genes (BMAL1,CLOCK, PER1–3, CRY1–2, CSNK1ε, REV-ERBα, REV-ERBβ and DEC1) along day (8, 11, 16:30 and 22:30 h) in PBMCs of OSA patients at the three visits of the study, relative to control subjects (black line). (a) OSA patients at diagnosis (t0M, orange line); (b) OSA patients after four months of CPAP treatment (t4M, yellow line); (c) OSA patients after two years of CPAP treatment (t24M, green line). Data are presented as mean ΔΔCT values ± SEM of 34 OSA patients at t0M, 34 patients at t4M and 16 patients at t24M. Non-parametric student's t-tests (Mann-Whitney tests) were used to assess differences between t0M, t4M or t24M and controls subjects, at each time point along the day: * p < 0.05; ** p < 0.01; *** p < 0.001; **** p < 0.0001.
CPAP treatment impacted on the expression of all the evaluated clock genes in PBMCs, with less evident effects on CRY1, REV-ERBβ and DEC1. The clock genes whose expression changed the most along the CPAP treatment follow-up period include REV-ERBα, PER1, PER2 and CRY2 (table 5). OSA short-term (four months) treatment promoted an evident re-establishment of BMAL1 expression in OSA patients, relative to control subjects (Fig. 3b). On the contrary, PER1 and CRY2 decreased expression were more pronounced at all the evaluated time points of the day in PBMCs from OSA patients at t4M (Fig. 3b, table 5). In addition, short-term (four months) treatment promoted a decrease in REV-ERBα expression at all the evaluated time points and an increase in CRY1 gene expression at 11 and 22:30 h, relative to control subjects. Short-term treatment also promoted a decrease in CSNK1ε expression levels at t4M, in comparison to patients at t0M (Fig. 3b, table 5). After long-term (two years) OSA treatment, PER1, CRY2 and REV-ERBα expression were re-established in comparison to control subjects (Fig. 3c). Long-term (two years) CPAP treatment promoted a significant increase in the expression of these genes throughout the day (table 5). On the contrary, BMAL1 and DEC1 were overexpressed again in OSA patients at t24M relative to control subjects. In addition, PER2, PER3 and CRY1 showed to be overexpressed and REV-ERBβ less expressed, at specific time points, in PBMCs of OSA patients at t24M (Fig. 3c, table 5).
Table 5.
Short (four months, t4M) and long-term (two years, t24M) Continuous Positive Airway Pressure (CPAP) treatment promote differential alterations in clock genes expression in peripheral blood mononuclear cells of OSA patients (t0M).
| Gene | Time of the day | mRNA levels |
Friedman test p-value | ||
|---|---|---|---|---|---|
| OSA patients t0M | OSA patients t4M | OSA patients t24M | |||
| BMAL1 | 8 h | 0.739 ± 0.243 | 0.048 ± 0.303 | 0.839 ± 0.183 | * |
| 11 h | 0.754 ± 0.255 | −0.125 ± 0.238 (*) | 0.615 ± 0.237 | * | |
| 16:30 h | 0.543 ± 0.254 | 0.188 ± 0.233 | 1.048 ± 0.241 (#) | * | |
| 22:30 h | 0.721 ± 0.241 | 0.071 ± 0.256 | 0.961 ± 0.251 | ||
| CLOCK | 8 h | −0.102 ± 0.173 | −0.742 ± 0.205 | 0.321 ± 0.182 (#) | * |
| 11 h | −0.027 ± 0.180 | −0.702 ± 0.269 | −0.049 ± 0.262 | ||
| 16:30 h | −0.263 ± 0.179 | −0.707 ± 0.229 | 0.484 ± 0.240 (##) | ** | |
| 22:30 h | −0.031 ± 0.177 | −0.490 ± 0.235 | 0.483 ± 0.292 (#) | * | |
| PER1 | 8 h | −1.253 ± 0.394 | −1.751 ± 0.344 | 0.752 ± 0.356 (##) | ** |
| 11 h | −1.792 ± 0.323 | −2.802 ± 0.383 | 0.066 ± 0.522 (##) | ** | |
| 16:30 h | −2.623 ± 0.501 | −2.814 ± 0.340 | 0.160 ± 0.440 (**) (#) | ** | |
| 22:30 h | −2.181 ± 0.381 | −2.787 ± 0.379 | 0.134 ± 0.525 (##) | ** | |
| PER2 | 8 h | 0.131 ± 0.293 | −0.351 ± 0.307 | 1.518 ± 0.378 (##) | ** |
| 11 h | 0.086 ± 0.332 | −0.597 ± 0.351 | 0.805 ± 0.338 | * | |
| 16:30 h | −0.625 ± 0.307 | −0.862 ± 0.272 | 0.667 ± 0.342 (*) (#) | ** | |
| 22:30 h | −0.561 ± 0.306 | −0.996 ± 0.291 | 0.469 ± 0.347 (#) | * | |
| PER3 | 8 h | 0.362 ± 0.226 | 0.157 ± 0.199 | 1.428 ± 0.345 | |
| 11 h | 0.239 ± 0.257 | −0.637 ± 0.300 | 0..649 ± 0.236 | * | |
| 16:30 h | −0.483 ± 0.231 | −0.912 ± 0.191 | 0.509 ± 0.317 (##) | * | |
| 22:30 h | −0.231 ± 0.130 | −0.584 ± 0.236 | 0.853 ± 0.314 (#) | * | |
| CRY1 | 8 h | 0.127 ± 0.237 | 0.262 ± 0.149 | 0.821 ± 0.290 | |
| 11 h | 0.360 ± 0.262 | 0.508 ± 0.195 | 0.478 ± 0.287 | ||
| 16:30 h | 0.127 ± 0.253 | 0.390 ± 0.197 | 0.641 ± 0.276 | ||
| 22:30 h | 0.198 ± 0.231 | 0.557 ± 0.219 | 0.650 ± 0.300 | ||
| CRY2 | 8 h | −1.626 ± 0.384 | −1.819 ± 0.333 | 0.876 ± 0.352 (*) (#) | ** |
| 11 h | −1.070 ± 0.369 | −2.233 ± 0.405 (*) | 0.118 ± 0.247 (##) | ** | |
| 16:30 h | −1.799 ± 0.365 | −2.104 ± 0.361 | 0.298 ± 0.305 (##) | ** | |
| 22:30 h | −1.826 ± 0.390 | −2.310 ± 0.491 | 0.376 ± 0.320 (#) | * | |
| CSNK1ε | 8 h | −0.404 ± 0.223 | −1.03 ± 0.287 | 1.385 ± 0.475 (##) | * |
| 11 h | −0.251 ± 0.240 | −1.512 ± 0.299 (*) | 0.723 ± 0.399 (#) | ** | |
| 16:30 h | −0.746 ± 0.266 | −1.134 ± 0.262 | 1.109 ± 0.402 (#) | * | |
| 22:30 h | −0.641 ± 0.214 | −1.229 ± 0.277 | 1.096 ± 0.411 (#) | * | |
| REV-ERBα | 8 h | −1.723 ± 0.267 | −1.916 ± 0.318 | 0.036 ± 0.229 (**) (###) | **** |
| 11 h | −1.965 ± 0.255 | −2.574 ± 0.296 | −0.765 ± 0.546 (##) | ** | |
| 16:30 h | −2.077 ± 0.358 | −2.453 ± 0.361 | −0.186 ± 0.205 (#) | * | |
| 22:30 h | −1.432 ± 0.193 | −2.532 ± 0.275 | 0.147 ± 0.263 (*) (####) | **** | |
| REV-ERBβ | 8 h | −0.207 ± 0.243 | −0.270 ± 0.184 | −0.332 ± 0.225 | |
| 11 h | −0.733 ± 0.290 | −0.507 ± 0.138 | −1.112 ± 0.528 | ||
| 16:30 h | −0.684 ± 0.236 | −0.481 ± 0.191 | −0.809 ± 0.204 | ||
| 22:30 h | −0.319 ± 0.250 | −0.373 ± 0.202 | −0.612 ± 0.218 | ||
| DEC1 | 8 h | 1.415 ± 0.233 | 1.443 ± 0.236 | 1.183 ± 0.209 | |
| 11 h | 1.496 ± 0.202 | 1.097 ± 0.320 | 0.921 ± 0.234 | ||
| 16:30 h | 1.296 ± 0.304 | 1.056 ± 0.250 | 1.117 ± 0.218 | ||
| 22:30 h | 1.066 ± 0.227 | 1.051 ± 0.238 | 1.161 ± 0.206 | ||
Data presented as mean mRNA expression levels ± SEM, at each time point, from 13–16 OSA patients that completed long-term Continuous Positive Airway Pressure (CPAP) treatment (two years follow-up). Friedman tests were performed at each time point, throughout time (from t0M to t24M) to visualize how much variables have changed throughout treatment: * p < 0.05; ** p < 0.01; *** p < 0.001; **** p < 0.0001. In addition, Friedman tests were followed by Dunn's multiple comparisons tests to visualize statistical differences relative to t0M (marked as *) and between short- and long-term treatment (marked as #), in each group column: (* or #) p < 0.05; (** or ##) p < 0.01; (###) p < 0.001; (####) p < 0.0001.
We next explored the similarity of the data sets to a 24-hours rhythmic oscillation, by carrying out a harmonic regression analysis. We used a harmonic regression fitting with a period of 24 h to analyze the average data of all patients (n t0M = 34, n t4M = 34 and n t24M = 16, Fig. 4) as well as each individual patient time course data (Fig. S1). The results of the harmonic regression analysis for the mean gene expression for each time point across all subjects and for each individual time course data (for each individual subject) are provided in Table S2 and Table S3. Our results point to an impact of CPAP treatment in the rhythmic expression of core-clock genes in the follow-up data (four months and two years after treatment) as compared to the controls and patients at t0M (Fig. 4). We also observed an overall higher expression of PER1, PER2, PER3, CRY2, CSNK1ε and CLOCK following long-term CPAP treatment (two years, t24M) relative to t0M (Fig. 4). Interestingly, the temporal expression profiles of the core-clock genes in the group of patients analysed following long-term CPAP treatment (two years, t24M) resembled the ones of the control group. For some of the core-clock genes, such as CLOCK and CRY1, we observed alterations in their rhythmic profiles at earlier time points after the beginning of treatment (four months, t4M). This tendency became more evident at the later follow-up visit (two years, t24M).
3.3. The effect of OSA and short- (four months) and long-term (two years) CPAP treatment on biological clocks persists among different subgroups of OSA patients
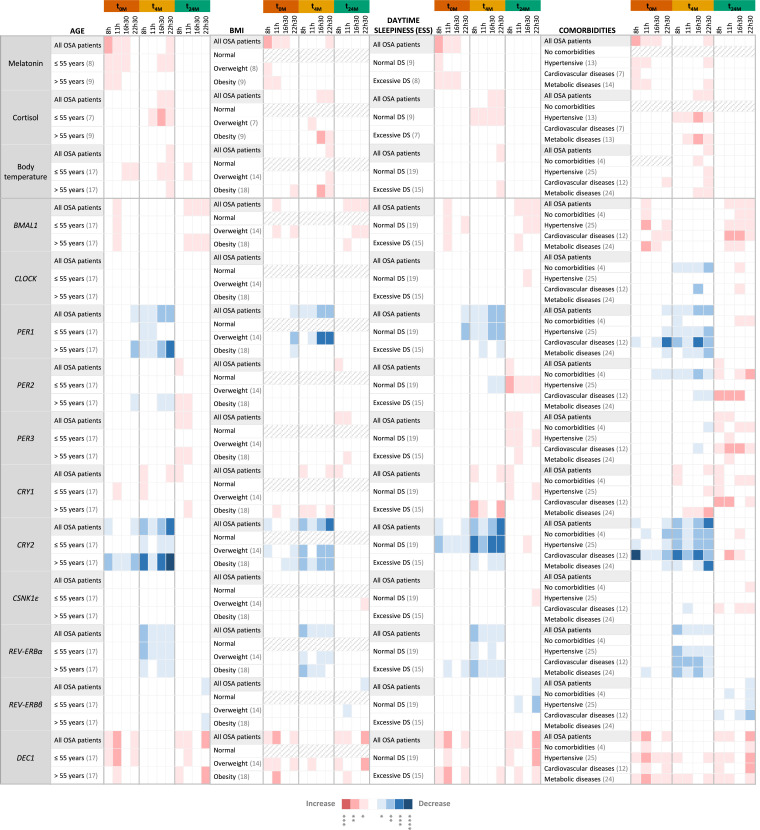
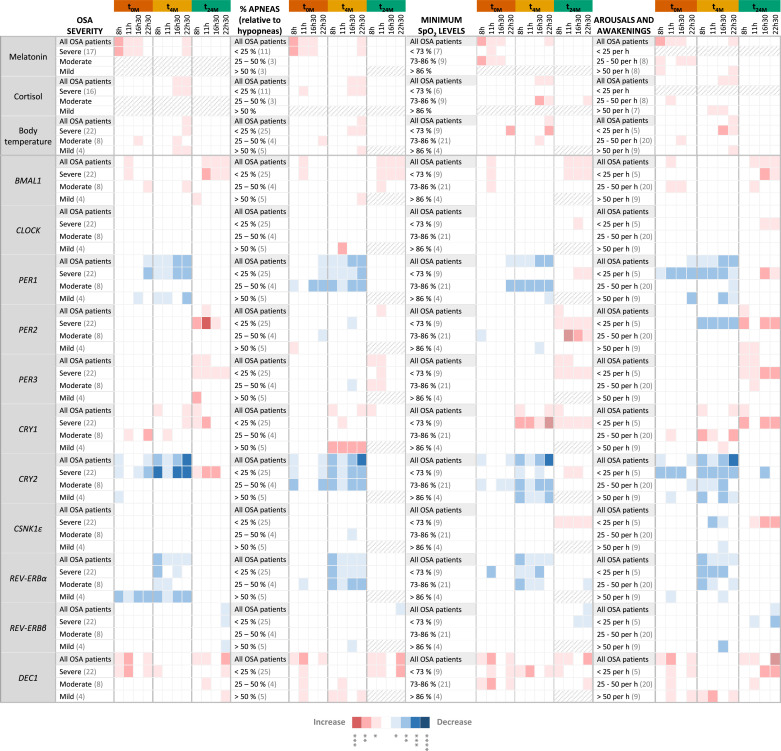
In an attempt to discriminate the specific effect of OSA and CPAP treatment on clock-related characteristics, patients were divided into subgroups according to age, BMI, daytime sleepiness or comorbidities (Fig. 5) and PSG results (OSA severity; percentage of apneas over hypopneas; minimum SpO2 levels; or the number of arousals and awakenings; Fig. 6), registered at baseline (t0M), and compared to control subjects. The previously observed alterations in clock outputs and expression of clock genes were very consistent among the different subgroups. Still, some differences were observed in specific patient subgroups.
Fig. 5.
The effect of OSA and OSA short- (four months) and long-term (two years) Continuous Positive Airway Pressure (CPAP) treatment on clock outputs and core-clock genes expression persists in different OSA patient subgroups despite their differences in age, body mass index (BMI), daytime sleepiness and clinical history. Heatmap showing significant alterations in the levels of major clock outputs (plasma melatonin, cortisol and body temperature) and clock genes expression (BMAL1,CLOCK, PER1–3, CRY1–2, CSNK1ε, REV-ERBα, REV-ERBβ and DEC1), in peripheral blood mononuclear cells, along day (8, 11, 16:30 and 22:30 h), in OSA patient subgroups, before (t0M) and after four months (t4M) and two years (t24M) of CPAP treatment, relative to control subjects (seven). OSA patients were stratified according to age (≤ 55 years; > 55 years), BMI (normal; overweight; obese), daytime sleepiness (DS), assessed through the Epworth Sleepiness Scale - ESS (normal DS; excessive DS) or comorbidities (no comorbidities; hypertension; cardiovascular diseases; metabolic diseases) at baseline (t0M). The number of patients in each subgroup is shown in brackets in each subgroup. Gray slashes represent subgroups with less than three subjects, not evaluated. Results of all OSA patients at t0M, t4M, and t24M are used as a reference. A color rule (red: increase, blue: decrease) is used to highlight statistically significant alterations, evaluated by non-parametric Student t-tests (Mann-Whitney tests) between each OSA group (t0M, t4M or t24M) and the control group, at each time point of the day. The association between color and p-value is shown below the table.
Fig. 6.
The effect of OSA and OSA short- (four months) and long-term (two years) Continuous Positive Airway Pressure (CPAP) treatment on clock outputs and core-clock genes expression persists in different OSA patient subgroups despite their differences in sleep variables observed by PSG. Heatmap showing significant alterations in the levels of major clock outputs (plasma melatonin, cortisol and body temperature) and clock genes expression (BMAL1,CLOCK, PER1–3, CRY1–2, CSNK1ε, REV-ERBα, REV-ERBβ and DEC1), in peripheral blood mononuclear cells, along day (8, 11, 16:30 and 22:30 h), in OSA patients subgroups, before (t0M) and after four months (t4M) and two years (t24M) of CPAP treatment, relative to control subjects (seven). OSA patients were stratified according to sleep parameters monitored at baseline (t0M), namely, OSA severity (mild; moderate; severe), percentage of apneas relative to hypopneas (< 25%,; 25 – 50%; > 50%), minimum SpO2 levels (< 73%; 73 – 86%; > 86%) and arousals and awakenings index (< 25 per hour; 25 – 50 per hour; > 50 per hour). The number of patients in each subgroup is shown in brackets in each subgroup. Gray slashes represent subgroups with less than 3 subjects, not evaluated. Results of all OSA patients at t0M, t4M, and t24M are used as a reference. A color rule (red: increase, blue: decrease) is used to highlight statistically significant alterations, evaluated by non-parametric Student t-tests (Mann-Whitney tests) between each OSA group (t0M, t4M or t24M) and the control group, at each time point of the day. The association between color and p-value is shown below the table.
Whereas few subgroups of OSA patients showed increased plasma cortisol levels at t0M relative to control subjects, several subgroups showed increased body temperature in the late afternoon (16:30) and/or night (22:30), in addition to higher melatonin plasma levels (Fig. 5, Fig. 6). At t4M, the increased plasma cortisol levels observed relative to control subjects were visible in more subgroups, especially in hypertensive OSA patients, who showed increased cortisol levels at all measured time points (Fig. 5). At t24M, alterations in clock outputs were re-established in all OSA subgroups with few exceptions (Fig. 5, Fig. 6).
Regarding the expression of clock genes, at t0M, particular subgroups showed additional alterations, namely in CLOCK, PER2, CRY1 and/or REV-ERBα expression, at specific time points of the day. OSA patients with no comorbidities showed alterations in BMAL1, PER2, CRY2 and DEC1 expression relative to controls (Fig. 5). No alterations were found in PER3, CSNK1ε and REV-ERBβ gene expression levels in PBMCs of any of the evaluated OSA subgroups in comparison to control subjects. At t4M, overweight patients, patients with hypertension or with metabolic disorders, patients with excessive daytime sleepiness, patients with more than 25 % of apneas over hypopneas, patients with lower minimum SpO2 levels and patients that woke up more than 50 times per hour of sleep continued to show alterations in BMAL1 and/or DEC1 gene expression (Fig. 5, Fig. 6). In addition, we also observed other gene expression alterations in specific OSA patients’ subgroups, namely in the expression of CLOCK, PER2, CSNK1ε and REV-ERBβ (Fig. 5, Fig. 6). At t24M, some OSA subgroups still showed alterations in PER1 and/or CRY2 gene expression, namely, patients with no comorbidities and patients with cardiovascular diseases, patients with normal daytime sleepiness, severe OSA patients, patients with lower minimum SpO2 levels and patients with less arousals and awakenings (Fig. 5, Fig. 6). Particular subgroups also showed increased expression of CSNK1ε and CLOCK (Fig. 5, Fig. 6). The subgroups of older OSA patients (more than 55 years old), patients with comorbidities, severe OSA patients, patients with lower SpO2 levels and patients with lower awakening and arousal indexes show more gene expression alterations after short- (four months) and long-term (two years) CPAP treatment than the other subgroups, relative to control subjects.
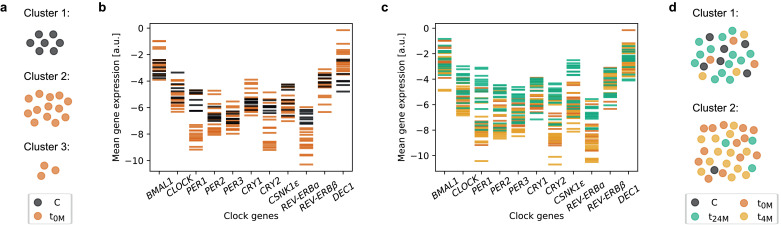
3.4. Gene expression in the group of OSA patients after long-term (two years) treatment resembles the gene expression of controls
Given the observed difference in terms of physiological and molecular data, we further investigated whether a separation between control and patient groups was possible using the obtained datasets. We thus used an unsupervised clustering tool to test whether the algorithm automatically separates controls from the patient group. Clustering based on the mean gene expression levels of controls and patients at t0M, in two clusters (k-means clustering with k = 2) resulted in one cluster containing only patients (12 patients) and a second cluster containing mixed elements of four patients and seven controls. A subsequent clustering in three clusters (k-means clustering with k = 3) based on the same data resulted in one cluster containing only controls, and two clusters with patients (Fig. 7a). These results suggested that there is indeed a salient difference between patients and controls, which the clustering algorithm detected solely based on the mean expression levels of the core-clock genes analysed. The underlying difference, not evident by eye (Fig. 7b), was indeed confirmed by supervised classification. We trained a linear support vector machine using a leave-one-out approach, providing the mean genetic expression levels as input and using “control” versus “patient” as labels for the required output. The prediction of the health status, i.e. whether the subject is part of the control or patient group, was then tested on the subject that was not used for training. Repeating this for all subjects, we found that all but one subject of the data set can be correctly classified, which is reflected in a good F1 score of 0.95 [individual F1 scores of 0.97 and 0.93, 90 % confidence interval (CI) of 0.93 ± 0.01].
Fig. 7.
OSA patients show a differential mean expression of core-clock genes in peripheral blood mononuclear cells, compared to control subjects, that is less evident upon two years of Continuous Positive Airway Pressure (CPAP) treatment. Subjects were clustered based on the mean expression of each clock-gene at the four time points of the day. (a) Clustering of controls and patients before treatment (t0M) results in three cluster, one with all controls (black), and two with patients (orange). (b) Mean mRNA levels of the evaluated clock genes color-coded by patients at t0M (orange) and controls (black). (c) Mean mRNA levels of the evaluated clock genes color-coded by study visit, before treatment (t0M, orange), after four months of treatment (t4M, yellow) and after two years of treatment (t24M, green). (d) Clustering of all subjects in two clusters results in one cluster with predominantly controls (black) and patients at t24M (green), and the other cluster with predominantly patients at t0M (orange) and at t4M (yellow).
Next, we tested whether it is possible to differentiate between the data of patients retrieved at t0M, t4M (four months of CPAP treatment) or t24M (two years of CPAP treatment). For this we labelled the patient data by visit of the study and applied the same computational machine learning algorithm as above, i.e. supervised classification based on a linear support vector machine trained as before. Our classification showed a limited prediction success in predicting the visit of the study, i.e. t0M, t4M or t24M [F1 score of 0.73, individual scores (0.625, 0.625, 0.94), 90 % CI of 0.73 ± 0.004]. Yet, a closer look at the prediction results showed that the F1 score calculated solely for predictions labelled as t24M was higher, with a value of 0.94 for t24M, which suggested that the latter can be distinguished from the data recorded at t0M and t4M. Indeed, training the classifier to distinguish t24M from the other two visits (t0M and t4M) resulted in an improved classification with a mean F1 score of 0.90 [individual scores (0.94, 0.87), 90 % CI of 0.91 ± 0.004]. The underlying difference between t24M, and the other two visits seems to be even visible by eye (see Fig. 7c). The observed difference is particularly salient for PER1.
We have so far observed that the data of patients at t0M differ from controls and differ as well from patients at t24M. In the following, we asked whether the patient data at t24M is actually closer to the controls than to the data from the same patients at the beginning of the study (t0M). To test this, we pooled the patient data of t0M, t4M and t24M with the data from the controls. An unsupervised clustering in two clusters (k-means clustering with k = 2) resulted for these data in two clusters with predominantly controls and patient data from t24M in one cluster, and patient data from t0M and t4M in the other cluster (Fig. 7d). This suggested that the patient data after long-term (two years) treatment is in most cases more similar to the data of the controls than to the data of the same patients before or after short-term (four months) treatment. Training a machine learning classifier to distinguish controls and patient data at t24M from patient data at t0M or t4M results in a good classification success with an F1 score of 0.92 [individual scores (0.94, 0.91), 90 % CI of 0.93 ± 0.002]. Patients before and after short-term (four months) treatment thus show a different gene expression profile compared to controls and patients after long-term (two years) treatment, which are more similar to each other.
We finally evaluated if our results change when using the available physiological data (plasma melatonin, cortisol and body temperature, 28 subjects) for the machine learning algorithm, instead of the mean-gene expression levels used so far, regarding the impact of the treatment. Restricting the data set to the 28 subjects with additional data on plasma melatonin, cortisol and body temperature allows to compare the additional information provided by non-genetic data. While the non-genetic data alone did not lead to good predictions, neither as time-course data, nor using the temporal means, adding the temporal means of the non-genetic data to the temporal means of the genetic data increases the prediction success from an F1 score of 0.96 to 1.0 [individual scores (0.96, 0.97) and (1.0, 1.0), 90 % CI of 0.96 ± 0.005 and 1.0 ± 0.0, all evaluated to 1.0].
Next, we evaluated whether using the temporal mean of the genetic data for predictions is indeed advantageous, or whether the additional information contained in the full time-course of the genetic data might improve results. Using the non-averaged mRNA time-course data instead of the temporal mean results in a reduced classification performance with an F1 score of 0.75 [individual scores (0.79, 0.71), 90 % CI of 0.76 ± 0.006], and using mean-normalized time-course data reduced the prediction success even more to an F1 score of 0.62 [individual scores (0.73, 0.51), 90 % CI of 0.66 ± 0.009]. Replacing the temporal mean with the data measured at one of the four time points results in reduced prediction performance with an F1 score between 0.84 and 0.90 [individual scores (0.87, 0.81) and (0.92, 0.88), 90 % CIs of 0.85 ± 0.005 and 0.90 ± 0.002]. This suggested that the information distinguishing controls and treated patients from patients before or after short-term (four months) treatment is found in the temporal mean of clock genes expression levels.
4. Discussion
In the current study, we investigated the impact of OSA and OSA CPAP treatment (short and long-term) on clock-related characteristics in a cohort of 34 male OSA patients and seven control subjects (no OSA) of the same sex and age group. Major clock outputs (melatonin and cortisol, and body temperature) and expression levels and diurnal variations of 11 clock genes were evaluated at four time points along the day. The measured diurnal profiles of melatonin and cortisol plasma levels, body temperature and clock genes expression in control subjects are in accordance with what is described in the literature [61], [62], [63], [64], [65], [66], [67], [68], [69].
Only a few studies have explored the impact of OSA and even less of OSA treatment on the biological clock, and the data obtained is not consensual. Lemmer and colleagues inferred that OSA disturbs plasma melatonin levels (17 OSA patients), causing a loss of the well-known melatonin nocturnal increase, and that 8 weeks of CPAP treatment cannot correct this output [40]. Regarding the expression of clock genes, several alterations are reported in the literature, in a time-dependent manner, including on BMAL1, CLOCK, CSNK1ε, CRY1, CRY2, PER2 and PER3 expression, in PBMCs of OSA patients relative to control subjects [[22], [23], [24],70]. Yang et al. showed that OSA impairs the daily expression patterns of several clock genes in PBMCs of 133 patients, compared to 11 controls [24]. Whereas at 12 h, OSA reduces the expression of BMAL1, CLOCK, CRY1, CRY2, PER2, PER3 and CSNK1ε, at 18 h it increases BMAL1, CLOCK, CRY1 and PER3 expression levels. The most striking alteration was observed at midnight, when CRY1 and PER3 were significantly downregulated, particularly in severe OSA patients. No alterations were observed in PER1 expression [24]. However, Yang et al. did not evaluated how such alterations respond to OSA treatment. Moreira et al. showed that 1 month of OSA treatment was not able to revert the decrease observed in CLOCK expression at 8 h in OSA PBMCs (13 OSA patients, seven controls) [22]. On the contrary, Burioka and co-workers showed that 3 months of CPAP treatment re-established the impaired diurnal oscillation of PER1 mRNA expression in PBMCs of OSA patients (8 OSA patients, 8 control subjects) [21].
Our data showed that OSA impacts on clock outputs and on the expression of several clock genes in a time dependent manner. The most prominent changes are an increase in melatonin plasma levels and in BMAL1 and DEC1 expression in PBMCs during the morning, and a decrease in the night of PER1 and CRY2 expression, relative to control subjects. REV-ERBα expression also showed to be lower at all time points throughout the day, although not statistically significant with this group size. We did not found more marked alterations in the clock of severe OSA patients (based on RDI/AHI). However, the validity of RDI/AHI as an isolated indicator of OSA severity has been questioned [71]. These results are not in line with the prior published results, showing a different set of affected clock genes and different expression changes [24]. This variability might be attributed to differences in study cohorts and assessed time points of the day. In addition, OSA has been increasingly recognized as a very heterogeneous disorder, with high variability of symptoms, clinical manifestations, pathophysiological mechanisms and treatment response, which might also contribute to the observed discrepancies [72,73].
It is also difficult to discern whether the observed alterations in the clock of OSA patients are specific to OSA or promoted/affected by excessive weight, lifestyle (e.g. sleep and wake cycles, nightshifts, diet, feeding schedules, physical exercise) or comorbidities (e.g. hypertension, metabolic and cardiovascular diseases) and/or associated medication, in such a heterogeneous cohort. Indeed, when patients were sub-grouped based on age, BMI, daytime sleepiness, comorbidities or PSG data, we found different alterations in clock outputs (melatonin, cortisol and body temperature) and in gene expression or at different time points of the day in different OSA subgroups. Several OSA subgroups also showed increased CRY1 expression during the morning, lower PER2 expression and higher body temperature at night and lower REV-ERBα expression throughout the day. Such differences may result from the different contributions of the different variables. Yet, the core alterations observed in all groups of patients was very consistent among the subgroups, which might be indicative of an OSA-specific effect. Among OSA-promoted alterations, the fact that OSA patients with no comorbidities showed alterations in BMAL1, PER2, CRY2 and DEC1 expression points towards a more specific OSA effect on the expression of these genes. Because of the unknown contribution of each of the mentioned variables, we also opted for comparing the same patients before and after OSA treatment, as under these circumstances, the major variable changing is OSA (BMI, comorbidities and medication remained constant throughout the follow-up). The same OSA patients at t4M and t24M were also compared to t0M and both short and long-term CPAP treatment promoted significant alterations in clock-related characteristics, most of these in line with the alterations observed relative to control subjects, supporting the observed effect of OSA on the clock. Yet, other external variant that must be considered is the putative seasonal effect on the expression of clock genes, as this study was performed throughout several seasons. Dopico et al. showed that more than 4000 protein-coding mRNAs in PBMCs have seasonal expression profiles, among which BMAL1, CLOCK, CRY1, CSNK1ε and REV-ERBβ [74]. In accordance, we also explored the seasonal influence on the obtained data and we did not find any evident contribution (Fig. S2).
By using advanced computational tools such as unsupervised machine learning, we were able to better understand the extent of the differences observed between OSA patients and controls. Unsupervised clustering based on the temporal mean expression levels of clock genes was able to discriminate untreated OSA patients from control subjects, which reinforces OSA impact on the biological clock. Machine learning classification algorithm mostly relied on the expression of BMAL1, CLOCK and PER1 to distinguish OSA patients from controls. Interestingly, these and other clock genes whose expression we found to be more impaired in OSA patients (CRY2, DEC1) have been shown to be hypoxia sensitive [75], [76], [77], [78], [79]. BMAL1, PER1, CRY1–2 and DEC1, have hypoxia response elements, pertinent for binding of Hypoxia-inducible factors (HIF), key mediators of adaptation to changes in oxygen levels [75], [76], [77], [78], [79]. Gabryelska and colleagues have also recently showed that increased levels of subunit α of HIF-1, in serum of OSA patients (n = 10), were associated with an overexpression of several circadian clock proteins (PER1, CRY1 and CLOCK), relative to control subjects (n = 10) [25]. Variations in oxygen/carbon monoxide levels could be at the basis of the clock dysregulation in OSA [76,77,80]. However, we cannot discard the potential contribution of sleep fragmentation and autonomic dysregulation as well.
As several biological processes are under clock control, and clock genes per se also control the expression of multiple other genes, alterations in clock functioning and clock genes expression could be at the basis of OSA pleiotropic effects and the diverse multi-organ chronic morbidities associated with the disease. However, further research is needed to elucidate the causative mechanisms of the observed alterations. One should ask whether these alterations are a consequence of OSA or whether these can be at the basis of OSA development. Butler et al. showed that the circadian system has a strong contribution in determining the duration of apneas and hypopneas of OSA patients across the night, and that it potentially accounts for over half of the lengthening observed during normal overnight sleeps [81]. Other types of studies must be performed to explore the link between OSA and circadian dysfunction. Inducing OSA or clock dysfunction in an animal model could allow a better understanding of the reciprocal impact of each condition. The alterations observed in the current study also suggest a potential delay in the peak time of expression of some core-clock genes in OSA patients. Such alteration could potentially be associated to phase shifts in internal time, which are reported to occur in response to changes in environmental/internal time cues and can lead to desynchrony within the body, favouring the development of several diseases [82], [83], [84]. In this regard, it would be interesting to analyze other peripheral clocks, in addition to PBMCs, to evaluate OSA clocks misalignment.
Our results also show that neither short- (four months) or long-term (two years) CPAP treatment can fully revert OSA impact on the expression profile of core-clock genes. More strikingly, our results highlighted a differential response of the OSA clock to short- (four months) and long-term (two years) CPAP treatment. Short-term (four months) CPAP treatment was effective on re-establishing BMAL1 and DEC1 expression levels relative to controls, but the decreased expression of PER1 and CRY2 was accentuated in the time course dataset. In addition, the differential expression of other clock genes (increase of CRY1 and decrease of REV-ERBα expression, and in some subgroups also increase of CLOCK and decrease of PER2, CSNK1ε and REV-ERBβ expression) was also altered throughout the day. The additional increase of cortisol and body temperature at night further points to the potential phase delay in the biological rhythms of OSA patients that seems to be accentuated upon short-term (four months) treatment. On the other hand, long-term (two years) CPAP treatment was able to re-establish the evaluated clock outputs (melatonin, cortisol and body temperature) and PER1 and CRY2 expression levels, but promoted an overall increase in the expression of multiple clock genes (BMAL1, PER2, PER3, CRY1, REV-ERBβ, DEC1, and in some subgroups also CLOCK and CSNK1ε). Interestingly, we found that CPAP treatment led to more alterations in gene expression in subgroups of OSA patients that may have more severe OSA conditions or higher dysregulations of O2/CO2 levels (e.g. older patients, patients with comorbidities, patients with more obstruction events, patients that reach lower SpO2 levels during sleep, and patients with lower awakening and arousal indexes that might not re-establish oxygen levels so easily). One may speculate that O2/CO2 modulation by CPAP might have a higher impact in these patients and thus may lead to more gene expression alterations.
The reasons behind the observed differential response of the biological clock to short- (four months) and long-term (two years) CPAP treatment remains to be elucidated. We may speculate that there is a differential adaptation to CPAP treatment along time; though AHI is significantly reduced in short-term treated patients (t4M), OSA patients usually take time to get used to sleep with CPAP, and we did see better sleep indicators, evaluated through the Epworth Sleepiness Scale, in patients treated long-term (t24M); in the period of adaptation, CPAP may be initially perceived as a disturbance to the clock system and with time the clock may re-adapt to the new circumstances; or CPAP impact on the clock might not be a direct effect. In fact, Corda et al. showed that CPAP treatment for one week induces upper airway dilation, but anatomical and functional improvements are only observed later on (6 months of CPAP treatment) [85]. More studies are needed to better understand the long-term effect of CPAP treatment. Our study emphasizes its importance not only to better understand the effect and efficacy of CPAP treatment on the clock, but also on OSA conditions.
CPAP capacity to counteract OSA consequences and/or prevent OSA-comorbidities has been recently questioned [14]. Not only OSA diagnosis, but also treatment has been mainly guided by RDI/AHI indexes and sleepiness scales (e.g. ESS). Both short- (four months) and long-term (two years) CPAP were very effective on reducing AHI and daytime sleepiness of the enrolled patients, but might not be able to fully revert all OSA-associated outcomes, including alterations in the biological clock. On the other hand, we cannot rule out the possibility that CPAP treatment, as a modulator of airway pressure and thus gas levels, might affect biological clocks per se. Our results showed alterations in the expression profiles of PER2, PER3, CRY1, REV-ERBα and REV-ERBβ mainly after CPAP treatment, relative to control subjects, with REV-ERBα being more profoundly affected by CPAP treatment. Adamovich and co-workers have shown that cultured cells subjected to cycles of 12 h of 5 % oxygen followed by 12 h of 8 % oxygen show increased CRY1 expression levels only when oxygen levels are restored to 8 % [76]. Further research is needed to understand the effect of different gas levels on the modulation of the biological clock. This knowledge is likely to have a direct impact in clinical management and treatment of several breathing disorders as OSA.
Still, clustering based on data from controls and patients that completed the three visits of the study (t0M, t4M and t24M) indeed resulted in one cluster containing predominantly controls and long-term treated patients (two years, t24M), and another cluster containing patients without treatment and with short-term treatment (four months, t4M). While the clustering using controls and untreated patients suggests a fundamental difference in gene expression levels between controls and untreated patients, the latter suggests that long-term treatment (two years, t24M) recovers an expression phenotype state closer to controls. These results were supported by supervised classification. It remains to be shown which features distinguish controls and long-term treated patients from patients in the beginning of treatment. As a first hint, the observation that unsupervised clustering results in clusters which separate both groups suggests that the distinguishing features are rather strong.
To our knowledge, our study was carried out using the largest OSA cohort in which clock-related alterations were evaluated before and after treatment. Yet, the number of control subjects included was limited. As OSA is one of the most prevalent sleep disorders, strict criteria must be used for controls selection, based on clinical history, sleep quality and OSA screening. It is very challenging for sleep units to include sleep studies of potentially healthy subjects in their clinic routine, as PSG is costly, time-consuming, labor intensive and associated with long waiting lists [14]. This is a limitation that our study shares with other studies [24]. The small sample size may hide larger statistical significant differences due to the lack of statistical power or overweight others based on the selected subjects. Further studies, in a larger case-control study, with comparable numbers of controls and patients, are thus relevant to validate the obtained results in order to translate our finding to the clinics. We also observed considerable inter individual variability which limits statistical analysis and data interpretation. Assessing four time points along one day also constitutes a limitation that is difficult to overcome in human studies. The assessment of rhythmic parameters is especially affected by the number of time points. Most subjects did not show significant rhythmic oscillations likely due to the low number of time points. In accordance, the assessed rhythmic parameters are only an approximation. Ideally, for the evaluation of circadian rhythms, a larger number of time points, equally spread across 24 h, should be used for sampling, and preferably during two consecutive days [86]. However, this was not possible due to the clinic routine. The night phase was the most compromised phase of the 24 h day, as night time points would imply interfering with the participants' sleep. A later time point would be particularly relevant to visualize the melatonin peak, described to occur between 2 and 7 h [61,63,67,69], and the lower levels of core-body temperature reported after the mid-point of sleep [67,87], in healthy subjects. Even so, the obtained profiles in this study are in accordance with what has been described in the literature for similar time windows [61,63,67,69].
Altogether, this study provides evidences that OSA promotes alterations in biological clock-related characteristics that are ameliorated upon long-term (two years) CPAP treatment. The fact that long-term (two years) treatment had more positive effects on OSA clock-related alterations stresses the importance of patients’ compliance to treatment. Yet, the persistence of clock gene expression alterations even after long-term (two years) treatment questions the efficacy of CPAP treatment in fully re-establishing OSA-promoted alterations. This data reinforces the need of further studies and of new/complementary strategies more effective on OSA diagnosis and treatment. While machine learning is commonly used to assess the biological clock [88], our study in particular highlights the potential of machine learning approaches based on clock genes expression for OSA diagnosis and treatment response monitoring. A deeper understanding of how OSA and OSA treatment affects the biological clock may thus lead to a better OSA clinical management and might help to counteract the progression of OSA associated comorbidities.
Contributors
CC, ARA, JM and LSG were responsible for the study plan and design; CC, ARA and AR supervised the work; LSG, ARA, BS and CCA conducted the study and performed the experiments for clock outputs evaluation and gene expression analysis; JM evaluated patients and all the necessary clinical data; MF gathered all clinical data from all patients. AR, JH and MY were responsible for the design of the computational analysis. MY carried out the bioinformatics rhythmicity analysis in gene expression datasets; JH performed machine learning-based analysis with the obtained study datasets; LSG, MY and JH wrote the manuscript with input from all authors. All co-authors reviewed the manuscript and approved the final version to be submitted.
Declaration of Competing Interest
All the authors declare no conflicts of interests.
Acknowledgments
Acknowledgments
Work in Neuroendocrinology and Aging's laboratory was supported by the European Regional Development Fund (ERDF) through the Operational Programme for Competitiveness and internationalisation - COMPETE 2020 - and Portuguese national funds via FCT – Fundação para a Ciência e a Tecnologia, under the projects noOSAnoAGEING (POCI-01-0145-FEDER-029002, PTDC/MEC-MCI/29002/2017), HealthyAging 2020 (CENTRO-01-0145-FEDER-000012) and UIDB/04539/2020; and by the European Social Fund through POCH - Human Capital Operational Programme and Portuguese national funds via FCT under PD/BD/135497/2018. The work in AR group was supported by the German Federal Ministry of Education and Research (BMBF)—eBio-CIRSPLICE – [FKZ031A316] and by the Dr. Rolf M. Schwiete Stiftung. MY was additionally funded by the Einstein Foundation and the graduate school Berlin School of Integrative Oncology (BSIO), Charité Berlin. Funding sources had no role in study design, data collection, data analysis, interpretation, or writing of the report.
We would also like to thank the Sleep Medicine center team, of Coimbra Hospital and University Center, Coimbra, Portugal, for their collaboration and effort in patients’ recruitment, scheduling and clinical data collection and analysis, especially to Dr. Fátima Teixeira, Clara Santos and Judite Pereira. We would also like to thank the Nursery team of the Sleep Medicine center and the Nephrology team of Coimbra Hospital and University center, Coimbra, Portugal, and to Denny Marques, Ricardo Simões, Sara Carmo-Silva, Silvia Pinto and Rui Pinto, for their help on blood samples’ collection. We also thank all members of the Neuroendocrinology and Aging Group, from the Centre for Neuroscience and Cell Biology, University of Coimbra, Coimbra, Portugal, for all the input given to this work, particularly, Ana Santos-Carvalho and Ana Teresa Barros-Viegas, that further contributed to the protocol implementation and blood samples processing. Finally, we thank to Paulo Rodrigues Santos and Jani Sofia Almeida, from the Immunology and Oncology Laboratory, from the center for Neuroscience and Cell Biology, Coimbra, Portugal, for their contribution in cortisol assays analysis, and to Prof. Adérito Araújo and Prof. Carlos Tenreiro, from the Mathematics Department of the University of Coimbra, Coimbra, Portugal, for their help in the statistical analysis of this study.
Data Sharing Statement
The data that support the findings of this study are available from the corresponding authors, AR, CC and ARA, upon reasonable request.
Footnotes
Funding: Full list of funding sources at the end of the manuscript.
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2021.103248.
Contributor Information
Angela Relógio, Email: angela.relogio@charite.de, angela.relogio@medicalschool-hamburg.de.
Cláudia Cavadas, Email: ccavadas@ci.uc.pt.
Ana Rita Álvaro, Email: ritaa80@cnc.uc.pt.
Appendix. Supplementary materials
References
- 1.WHO . Global surveillance, prevention and control of chronic respiratory diseases: a comprehensive approach. In: Nikolai J.B., K, editors. Chronic respiratory disease. Geneva; 2007. pp. 1–155. [Google Scholar]
- 2.Benjafield A.V., Ayas N.T., Eastwood P.R., Heinzer R., Ip M.S.M., Morrell M.J. Estimation of the global prevalence and burden of obstructive sleep apnoea: a literature-based analysis. Lancet Respir Med. 2019;7(8):687–698. doi: 10.1016/S2213-2600(19)30198-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peppard P.E., Young T., Barnet J.H., Palta M., Hagen E.W., Hla K.M. Increased prevalence of sleep-disordered breathing in adults. Am J Epidemiol. 2013;177(9):1006–1014. doi: 10.1093/aje/kws342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lim D.C., Pack A.I. Obstructive sleep apnea: update and future. Annu Rev Med. 2017;68:99–112. doi: 10.1146/annurev-med-042915-102623. [DOI] [PubMed] [Google Scholar]
- 5.American Academy of Sleep Medicine . 3rd ed. American Academy of Sleep Medicine; Darien, IL: 2014. International classification of sleep disorders: diagnostic and coding manual. [Google Scholar]
- 6.Léger D., Stepnowsky C. The economic and societal burden of excessive daytime sleepiness in patients with obstructive sleep apnea. Sleep Med Rev. 2020;51 doi: 10.1016/j.smrv.2020.101275. [DOI] [PubMed] [Google Scholar]
- 7.Tietjens J.R., Claman D., Kezirian E.J., de Marco T., Mirzayan A., Sadroonri B. Obstructive sleep apnea in cardiovascular disease: a review of the literature and proposed multidisciplinary clinical management strategy. J Am Heart Assoc. 2019;8(1) doi: 10.1161/JAHA.118.010440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gaines J., Vgontzas A.N., Fernandez-Mendoza J., Bixler E.O. Obstructive sleep apnea and the metabolic syndrome: the road to clinically-meaningful phenotyping, improved prognosis, and personalized treatment. Sleep Med Rev. 2018;42:211–219. doi: 10.1016/j.smrv.2018.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pataka A., Bonsignore M.R., Ryan S., Riha R.L., Pepin J.L., Schiza S. Cancer prevalence is increased in females with sleep apnoea: data from the ESADA study. Eur Respir J. 2019;53(6) doi: 10.1183/13993003.00091-2019. [DOI] [PubMed] [Google Scholar]
- 10.Lajoie A.C., Lafontaine A.-.L., Kimoff R.J., Kaminska M. Obstructive sleep apnea in neurodegenerative disorders: current evidence in support of benefit from sleep apnea treatment. J Clin Med. 2020;9(2):297. doi: 10.3390/jcm9020297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jackson M.L., Tolson J., Bartlett D., Berlowitz D.J., Varma P., Barnes M. Clinical depression in untreated obstructive sleep apnea: examining predictors and a meta-analysis of prevalence rates. Sleep Med. 2019;62:22–28. doi: 10.1016/j.sleep.2019.03.011. [DOI] [PubMed] [Google Scholar]
- 12.Gaspar L., Sousa C., Álvaro A., Cavadas C., Mendes A. Common risk factors and therapeutic targets in obstructive sleep apnea and osteoarthritis: an unexpectable link? Pharmacol Res. 2020 doi: 10.1016/j.phrs.2020.105369. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 13.Gottlieb D.J., Punjabi N.M. Diagnosis and management of obstructive sleep apnea: a review. JAMA. 2020;323(14):1389–1400. doi: 10.1001/jama.2020.3514. [DOI] [PubMed] [Google Scholar]
- 14.Randerath W., Bassetti C.L., Bonsignore M.R., Farre R., Ferini-Strambi L., Grote L. Challenges and perspectives in obstructive sleep apnoea. Eur Respir J. 2018;52(3) doi: 10.1183/13993003.02616-2017. [DOI] [PubMed] [Google Scholar]
- 15.Gaspar L.S., Álvaro A.R., Moita J., Cavadas C. Obstructive sleep apnea and hallmarks of aging. Trends Mol Med. 2017;23(8):675–692. doi: 10.1016/j.molmed.2017.06.006. [DOI] [PubMed] [Google Scholar]
- 16.Ryan S. Adipose tissue inflammation by intermittent hypoxia: mechanistic link between obstructive sleep apnoea and metabolic dysfunction. J Physiol. 2017;595(8):2423–2430. doi: 10.1113/JP273312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.von Allmen D.C., Francey L.J., Rogers G.M., Ruben M.D., Cohen A.P., Wu G. Circadian dysregulation: the next frontier in obstructive sleep apnea research. Otolaryngol - Head Neck Surg (United States) 2018;159(6):948–955. doi: 10.1177/0194599818797311. [DOI] [PubMed] [Google Scholar]
- 18.Chen Y.C., Hsu P.Y., Hsiao C.C., Lin M.C. Epigenetics: a potential mechanism involved in the pathogenesis of various adverse consequences of obstructive sleep apnea. Int J Mol Sci. 2019;20(12):2937. doi: 10.3390/ijms20122937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Freitas L.S., Silveira A.C., Martins F.C., Costa-Hong V., Lebkuchen A., Cardozo K.H.M. Severe obstructive sleep apnea is associated with circulating microRNAs related to heart failure, myocardial ischemia, and cancer proliferation. Sleep Breath. 2020;24(4):1463–1472. doi: 10.1007/s11325-019-02003-1. [DOI] [PubMed] [Google Scholar]
- 20.Ryan S., Cummins E.P., Farre R., Gileles-Hillel A., Jun J.C., Oster H. Understanding the pathophysiological mechanisms of cardiometabolic complications in obstructive sleep apnoea – towards personalised treatment approaches. Eur Respir J. 2020;56(2) doi: 10.1183/13993003.02295-2019. [DOI] [PubMed] [Google Scholar]
- 21.Burioka N., Koyanagi S., Endo M., Takata M., Fukuoka Y., Miyata M. Clock gene dysfunction in patients with obstructive sleep apnoea syndrome. Eur Respir J. 2008;32(1):105–112. doi: 10.1183/09031936.00138207. [DOI] [PubMed] [Google Scholar]
- 22.Moreira S., Rodrigues R., Barros A.B., Pejanovic N., Neves-Costa A., Pedroso D. Changes in expression of the CLOCK gene in obstructive sleep apnea syndrome patients are not reverted by continuous positive airway pressure treatment. Front Med. 2017;4(187):1–5. doi: 10.3389/fmed.2017.00187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Canales M.T., Holzworth M., Bozorgmehri S., Ishani A., Weiner I.D., Berry R.B. Clock gene expression is altered in veterans with sleep apnea. Physiol Genomics. 2019;51(3):77–82. doi: 10.1152/physiolgenomics.00091.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang Lin, Lin Lin, Chen Friedman. Alternations of circadian clock genes expression and oscillation in obstructive sleep apnea. J Clin Med. 2019;8(10):1634. doi: 10.3390/jcm8101634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gabryelska A., Sochal M., Turkiewicz S., Białasiewicz P. Relationship between HIF-1 and circadian clock proteins in obstructive sleep apnea patients— preliminary study. J Clin Med. 2020;9(5):E1599. doi: 10.3390/jcm9051599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meira e Cruz M., Gozal D. Sleepiness and cardiometabolic impact of short sleep duration and OSA: what about the clock? Chest. 2019;156(6):1273–1274. doi: 10.1016/j.chest.2019.07.029. [DOI] [PubMed] [Google Scholar]
- 27.Albrecht U. Timing to perfection: the biology of central and peripheral circadian clocks. Neuron. 2012;74(2):246–260. doi: 10.1016/j.neuron.2012.04.006. [DOI] [PubMed] [Google Scholar]
- 28.Bass J., Lazar M.A. Circadian time signatures of fitness and disease. Science. 2016;354(6315):994–999. doi: 10.1126/science.aah4965. [DOI] [PubMed] [Google Scholar]
- 29.Fuhr L., Abreu M., Pett P., Relógio A. Circadian systems biology: when time matters. Comput Struct Biotechnol J. 2015;13:417–426. doi: 10.1016/j.csbj.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gaspar L., Álvaro A.R., Carmo-Silva S., Mendes A.F., Relógio A., Cavadas C. The importance of determining circadian parameters in pharmacological studies. Br J Pharmacol. 2019;176(16):2827–2847. doi: 10.1111/bph.14712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roenneberg T., Merrow M. The circadian clock and human health. Curr Biol. 2016;26(10):R432–R443. doi: 10.1016/j.cub.2016.04.011. [DOI] [PubMed] [Google Scholar]
- 32.Takahashi J.S. Transcriptional architecture of the mammalian circadian clock. Nat Rev Genet. 2017;18(3):164–179. doi: 10.1038/nrg.2016.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lehmann R., Childs L., Thomas P., Abreu M., Fuhr L., Herzel H. Assembly of a comprehensive regulatory network for the mammalian circadian clock: a bioinformatics approach. PLoS ONE. 2015;10(5) doi: 10.1371/journal.pone.0126283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Genov N., Castellana S., Scholkmann F., Capocefalo D., Truglio M., Rosati J. A multi-layered study on harmonic oscillations in mammalian genomics and proteomics. Int J Mol Sci. 2019;20(18):4585. doi: 10.3390/ijms20184585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Honma S., Kawamoto T., Takagi Y., Fujimoto K., Sato F., Noshiro M. Dec1 and Dec2 are regulators of the mammalian molecular clock. Nature. 2002;419(6909):841–844. doi: 10.1038/nature01123. [DOI] [PubMed] [Google Scholar]
- 36.Relógio A., Westermark P.O., Wallach T., Schellenberg K., Kramer A., Herzel H. Tuning the mammalian circadian clock: robust synergy of two loops. PLoS Comput Biol. 2011;7(12) doi: 10.1371/journal.pcbi.1002309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.El-Athman R., Relógio A. Escaping circadian regulation: an emerging hallmark of cancer? Cell Syst. 2018;6(3):266–267. doi: 10.1016/j.cels.2018.03.006. [DOI] [PubMed] [Google Scholar]
- 38.El-Athman R., Fuhr L., Relógio A. A systems-level analysis reveals circadian regulation of splicing in colorectal cancer. EBioMedicine. 2018;33:68–81. doi: 10.1016/j.ebiom.2018.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mazzoccoli G., Vinciguerra M., Carbone A., Relógio A. The circadian clock, the immune system, and viral infections: the intricate relationship between biological time and host-virus interaction. Pathogens. 2020;9(2):83. doi: 10.3390/pathogens9020083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lemmer B., Scholtze J., Schmitt J. Circadian rhythms in blood pressure, heart rate, hormones, and on polysomnographic parameters in severe obstructive sleep apnea syndrome patients: effect of continuous positive airway pressure. Blood Press Monit. 2016;21(3):136–143. doi: 10.1097/MBP.0000000000000173. [DOI] [PubMed] [Google Scholar]
- 41.Entzian P., Linnemann K., Schlaak M., Zabel P. Obstructive sleep apnea syndrome and circadian rhythms of hormones and cytokines. Am J Respir Crit Care Med. 1996;153(3):1080–1086. doi: 10.1164/ajrccm.153.3.8630548. [DOI] [PubMed] [Google Scholar]
- 42.Burioka N., Miyata M., Fukuoka Y., Endo M., Shimizu E. Day-night variations of serum interleukin-6 in patients with severe obstructive sleep apnea syndrome before and after continuous positive airway pressure (CPAP) Chronobiol Int. 2008;25(5):827–834. doi: 10.1080/07420520802384101. [DOI] [PubMed] [Google Scholar]
- 43.Smith D.F., Hossain M.M., Hura A., Huang G., McConnell K., Ishman S.L. Inflammatory milieu and cardiovascular homeostasis in children with obstructive sleep apnea. Sleep. 2017;40(4):zsx022. doi: 10.1093/sleep/zsx022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barceló A., Piérola J., De La Peña M., Frontera G., Yañez A., Alonso-Fernández A. Impaired circadian variation of platelet activity in patients with sleep apnea. Sleep Breath. 2012;16(2):355–360. doi: 10.1007/s11325-011-0501-5. [DOI] [PubMed] [Google Scholar]
- 45.Pépin J.L., Chiquet C., Tamisier R., Lévy P., Almanjoumi A., Romanet J.P. Frequent loss of nyctohemeral rhythm of intraocular pressure restored by nCPAP treatment in patients with severe apnea. Arch Ophthalmol. 2010;128(10):1257–1263. doi: 10.1001/archophthalmol.2010.220. [DOI] [PubMed] [Google Scholar]
- 46.Nieto F.J., Young T.B., Lind B.K., Shahar E., Samet J.M., Redline S. Association of sleep-disordered breathing, sleep apnea, and hypertension in a large community-based study. Sleep heart health study. JAMA. 2000;283(14):1829–1836. doi: 10.1001/jama.283.14.1829. [DOI] [PubMed] [Google Scholar]
- 47.Lavie P., Yoffe N., Berger I., Peled R. The relationship between the severity of sleep apnea syndrome and 24-h blood pressure values in patients with obstructive sleep apnea. Chest. 1993;103(3):717–721. doi: 10.1378/chest.103.3.717. [DOI] [PubMed] [Google Scholar]
- 48.Smolensky M.H., Hermida R.C., Castriotta R.J., Portaluppi F. Role of sleep-wake cycle on blood pressure circadian rhythms and hypertension. Sleep Med. 2007;8(6):668–680. doi: 10.1016/j.sleep.2006.11.011. [DOI] [PubMed] [Google Scholar]
- 49.Butler M.P., Thosar S.S., Smales C., DeYoung P.N., Wu H., Hussain M.V. Effects of obstructive sleep apnea on endogenous circadian rhythms assessed during relaxed wakefulness; an exploratory analysis. Chronobiol Int. 2020;37(6):856–866. doi: 10.1080/07420528.2020.1740723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Durgan D.J., Crossland R.F., Bryan R.M. The rat cerebral vasculature exhibits time-of-day-dependent oscillations in circadian clock genes and vascular function that are attenuated following obstructive sleep apnea. J Cereb Blood Flow Metab. 2017;37(8):2806–2819. doi: 10.1177/0271678X16675879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Manella G., Aviram R., Bolshette N., Muvkadi S., Golik M., Smith D. Hypoxia induces a time- and tissue-specific response that elicits intertissue circadian clock misalignment. Proc Natl Acad Sci U S A. 2019;117(1):779–786. doi: 10.1073/pnas.1914112117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bonsignore M.R., Saaresranta T., Riha R.L., Riha R., Bonsignore M. Sex differences in obstructive sleep apnoea. Eur Respir Rev. 2019;28(154) doi: 10.1183/16000617.0030-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Anderson S.T., FitzGerald G.A. Sexual dimorphism in body clocks. Science. 2020;369(6508):1164–1165. doi: 10.1126/science.abd4964. [DOI] [PubMed] [Google Scholar]
- 54.American Academy of Sleep Medicine Task Force Sleep-related breathing disorders in adults: recommendations for syndrome definition and measurement techniques in clinical research. The report of an american academy of sleep medicine task force. Sleep. 1999;22:667–689. [PubMed] [Google Scholar]
- 55.James S.M., Honn K.A., Gaddameedhi S., Van Dongen H.P.A. Shift work: disrupted circadian rhythms and sleep—implications for health and well-being. Curr Sleep Med Rep. 2017;3(2):104–112. doi: 10.1007/s40675-017-0071-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hood S., Amir S. The aging clock: circadian rhythms and later life. J Clin Invest. 2017;127(2):437–446. doi: 10.1172/JCI90328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Edwards B.A., Wellman A., Sands S.A., Owens R.L., Eckert D.J., White D.P. Obstructive sleep apnea in older adults is a distinctly different physiological phenotype. Sleep. 2014;37(7):1227–1236. doi: 10.5665/sleep.3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sargento P., Perea V., Ladera V., Lopes P., Oliveira J. The Epworth Sleepiness Scale in Portuguese adults: from classical measurement theory to Rasch model analysis. Sleep Breath. 2015;19:693–701. doi: 10.1007/s11325-014-1078-6. [DOI] [PubMed] [Google Scholar]
- 59.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 60.Lück S., Thurley K., Thaben P.F., Westermark P.O. Rhythmic degradation explains and unifies circadian transcriptome and proteome data. Cell Rep. 2014;9(2):741–751. doi: 10.1016/j.celrep.2014.09.021. [DOI] [PubMed] [Google Scholar]
- 61.Selmaoui B., Lambrozo J., Touitou Y. Magnetic fields and pineal function in humans: evaluation of nocturnal acute exposure to extremely low frequency magnetic fields on serum melatonin and urinary 6-sulfatoxymelatonin circadian rhythms. Life Sci. 1996;58(18):1539–1549. doi: 10.1016/0024-3205(96)00128-2. [DOI] [PubMed] [Google Scholar]
- 62.Takata M., Burioka N., Ohdo S., Takane H., Terazono H., Miyata M. Daily expression of mRNAs for the mammalian Clock genes Per2 and clock in mouse suprachiasmatic nuclei and liver and human peripheral blood mononuclear cells. Jpn J Pharmacol. 2002;90(3):263–269. doi: 10.1254/jjp.90.263. [DOI] [PubMed] [Google Scholar]
- 63.Selmaoui B., Touitou Y. Reproducibility of the circadian rhythms of serum cortisol and melatonin in healthy subjects: a study of three different 24-h cycles over six weeks. Life Sci. 2003;73(26):3339–3349. doi: 10.1016/j.lfs.2003.05.007. [DOI] [PubMed] [Google Scholar]
- 64.Fukuya H., Emoto N., Nonaka H., Yagita K., Okamura H., Yokoyama M. Circadian expression of clock genes in human peripheral leukocytes. Biochem Biophys Res Commun. 2007;354(4):924–928. doi: 10.1016/j.bbrc.2007.01.063. [DOI] [PubMed] [Google Scholar]
- 65.Kusanagi H., Hida A., Satoh K., Echizenya M., Shimizu T., Pendergast J.S. Expression profiles of 10 circadian clock genes in human peripheral blood mononuclear cells. Neurosci Res. 2008;61(2):136–142. doi: 10.1016/j.neures.2008.01.012. [DOI] [PubMed] [Google Scholar]
- 66.Hastings M., O'Neill J.S., Maywood E.S. Circadian clocks: regulators of endocrine and metabolic rhythms. J Endocrinol. 2007;195(2):187–198. doi: 10.1677/JOE-07-0378. [DOI] [PubMed] [Google Scholar]
- 67.Bollinger T., Leutz A., Leliavski A., Skrum L., Kovac J., Bonacina L. Circadian clocks in mouse and human CD4+ T cells. PLoS ONE. 2011;6(12):e29801. doi: 10.1371/journal.pone.0029801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Boivin D.B., James F.O., Wu A., Cho-Park P.F., Xiong H., Sun Z.S. Circadian clock genes oscillate in human peripheral blood mononuclear cells. Blood. 2003;102(12):4143–4145. doi: 10.1182/blood-2003-03-0779. [DOI] [PubMed] [Google Scholar]
- 69.Cuesta M., Boudreau P., Cermakian N., Boivin D.B. Rapid resetting of human peripheral clocks by phototherapy during simulated night shift work. Sci Rep. 2017;7(1):16310. doi: 10.1038/s41598-017-16429-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Khurana S., Waidha K., Guleria R., Sharda S., Bose S. In-silico investigations of selective miRNA-gene targets and their validation studies in obstructive sleep apnea (OSA) patient cohorts. Comput Biol Chem. 2020;87(107264):1–11. doi: 10.1016/j.compbiolchem.2020.107264. [DOI] [PubMed] [Google Scholar]
- 71.Punjabi N.M. COUNTERPOINT: is the apnea-hypopnea index the best way to quantify the severity of sleep-disordered breathing? No. Chest. 2016;149(1):16–19. doi: 10.1378/chest.14-2261. [DOI] [PubMed] [Google Scholar]
- 72.Keenan B.T., Kim J., Singh B., Bittencourt L., Chen N.H., Cistulli P.A. Recognizable clinical subtypes of obstructive sleep apnea across international sleep centers: a cluster analysis. Sleep. 2018;41(3):zsx214. doi: 10.1093/sleep/zsx214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.McNicholas W., Bassetti C., Ferini-Strambi L., Pépin J., Pevernagie D., Verbraecken J. Challenges in obstructive sleep apnoea. Lancet Respir Med. 2018;6(3):170–172. doi: 10.1016/S2213-2600(18)30059-6. [DOI] [PubMed] [Google Scholar]
- 74.Dopico X.C., Evangelou M., Ferreira R.C., Guo H., Pekalski M.L., Smyth D.J. Widespread seasonal gene expression reveals annual differences in human immunity and physiology. Nat Commun. 2015;6(7000):1–13. doi: 10.1038/ncomms8000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Miyazaki K., Kawamoto T., Tanimoto K., Nishiyama M., Honda H., Kato Y. Identification of functional hypoxia response elements in the promoter region of the DEC1 and DEC2 genes. J Biol Chem. 2002;277(49):47014–47021. doi: 10.1074/jbc.M204938200. [DOI] [PubMed] [Google Scholar]
- 76.Adamovich Y., Ladeuix B., Golik M., Koeners M.P., Asher G. Rhythmic oxygen levels reset circadian clocks through HIF1α. Cell Metab. 2017;25(1):93–101. doi: 10.1016/j.cmet.2016.09.014. [DOI] [PubMed] [Google Scholar]
- 77.Wu Y., Tang D., Liu N., Xiong W., Huang H., Li Y. Reciprocal regulation between the circadian clock and hypoxia signaling at the genome level in mammals. Cell Metab. 2017;25(1):73–85. doi: 10.1016/j.cmet.2016.09.009. [DOI] [PubMed] [Google Scholar]
- 78.Sato F., Bhawal U.K., Yoshimura T., Muragaki Y. DEC1 and DEC2 crosstalk between circadian rhythm and tumor progression. J Cancer. 2016;7(2):153–159. doi: 10.7150/jca.13748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dimova E.Y., Jakupovic M., Kubaichuk K., Mennerich D., Chi T.F., Tamanini F. The circadian clock protein CRY1 is a negative regulator of HIF-1α. iScience. 2019;13:284–304. doi: 10.1016/j.isci.2019.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dioum E.M., Rutter J., Tuckerman J.R., Gonzalez G., Gilles-Gonzalez M.A., McKnight S.L. NPAS2: a gas-responsive transcription factor. Science (80-) 2002;298(5602):2385–2387. doi: 10.1126/science.1078456. [DOI] [PubMed] [Google Scholar]
- 81.Butler M.P., Smales C., Wu H., Hussain M.V., Mohamed Y.A., Morimoto M. The circadian system contributes to apnea lengthening across the night in obstructive sleep apnea. Sleep. 2015;38(11):1793–1801. doi: 10.5665/sleep.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Albrecht U. Timing to perfection: the biology of central and peripheral circadian clocks. Neuron. 2012;74(2):246–260. doi: 10.1016/j.neuron.2012.04.006. [DOI] [PubMed] [Google Scholar]
- 83.Moon J.H., Cho C.H., Son G.H., Geum D., Chung S., Kim H. Advanced circadian phase in mania and delayed circadian phase in mixed mania and depression returned to normal after treatment of bipolar disorder. EBioMedicine. 2016;11:285–295. doi: 10.1016/j.ebiom.2016.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Murray J.M., Sletten T.L., Magee M., Gordon C., Lovato N., Bartlett D.J. Prevalence of circadian misalignment and its association with depressive symptoms in delayed sleep phase disorder. Sleep. 2017;40(1):1–10. doi: 10.1093/sleep/zsw002. [DOI] [PubMed] [Google Scholar]
- 85.Corda L., Redolfi S., Taranto Montemurro L., Piana G.E., Bertella E., Tantucci C. Short- and long-term effects of CPAP on upper airway anatomy and collapsibility in OSAH. Sleep Breath. 2009;13(2):187–193. doi: 10.1007/s11325-008-0219-1. [DOI] [PubMed] [Google Scholar]
- 86.Hughes M.E., Abruzzi K.C., Allada R., Anafi R., Arpat A.B., Asher G. Guidelines for genome-scale analysis of biological rhythms. J Biol Rhythms. 2017;32(5):380–393. doi: 10.1177/0748730417728663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Krauchi K., Wirz-Justice A. Circadian rhythm of heat production, heart rate, and skin and core temperature under unmasking conditions in men. Am J Physiol - Regul Integr Comp Physiol. 1994;267(3 Pt 2):R819–R829. doi: 10.1152/ajpregu.1994.267.3.R819. [DOI] [PubMed] [Google Scholar]
- 88.Hesse J., Malhan D., Yalҫin M., Aboumanify O., Basti A., Relógio A. An optimal time for treatment—predicting circadian time by machine learning and mathematical modelling. Cancers (Basel) 2020;12(11):3103. doi: 10.3390/cancers12113103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.