Summary
Advanced optical methods combined with various probes pave the way toward molecular imaging within living cells. However, major challenges are associated with the need to enhance the imaging resolution even further to the subcellular level for the imaging of larger tissues, as well as for in vivo studies. High scattering and absorption of opaque tissues limit the penetration of light into deep tissues and thus the optical imaging depth. Tissue optical clearing technique provides an innovative way to perform deep-tissue imaging. Recently, various optical clearing methods have been developed, which provide tissue clearing based on similar physical principles via different chemical approaches. Here, we introduce the mechanisms of the current clearing methods from fundamental physical and chemical perspectives, including the main physical principle, refractive index matching via various chemical approaches, such as dissociation of collagen, delipidation, decalcification, dehydration, and hyperhydration, to reduce scattering, as well as decolorization to reduce absorption.
Subject areas: Optical Imaging, Biological Sciences, Research Methodologies, Biological Sciences Tools
Graphical abstract
Subject areas: Optical Imaging; Biological Sciences; Research Methodologies; Biological Sciences Tools
Introduction
Biomedical photonics is currently one of the fastest-growing fields in the life sciences, connecting research in physics, optics, and electrical engineering coupled with medical or biological applications and allowing structural and functional analysis of tissues and cells with resolution and contrast unattainable by any other method. Advanced optical methods combined with the various probes pave the way toward real molecular imaging within living cells. However, major challenges are associated with the need to enhance the imaging resolution even further to the subcellular level for the imaging of larger tissue blocks or organs, as well as for in vivo studies. The high scattering of turbid tissues limits the penetration of visible and near-infrared light, and both the imaging resolution and the contrast decrease as the light propagates deeper into the tissue (Tuchin et al., 1997). Various optical imaging techniques have been developed for deeper tissue imaging (Gratton, 2011; Wang et al., 2018a; Ueda et al., 2020; Zhang et al., 2019). Alternatively, reducing tissue scattering and absorption could also significantly enhance the optical imaging of deep-tissue structures (Tuchin, 2006; Zhu et al., 2013; Yu et al., 2018). For example, White created a transparent adult zebrafish to study cancer pathology and cancer development in real time (White et al., 2008), but this method is unsuitable for studies in other animals or humans.
Tissue optical clearing technique provides an innovative way to perform deep-tissue imaging by making tissue more transparent and reducing the attenuation of light by using various optical clearing agents (OCAs) (Tuchin et al., 1997; Tuchin, 2006; Zhu et al., 2013) and has been applied in many biomedical fields (Kurihara et al., 2015; Silva Santisteban et al., 2017; Kubota et al., 2017; Lai et al., 2018; Yin et al., 2019). The mismatch in the refractive index (RI) between scattering particles and background medium causes the strong scattering of tissue (Tuchin, 2006). The introduction of high RI chemical agents into tissue and/or the efflux of water out of tissue by the hyperosmolarity of agents would increase the RI of the background medium. The removal of high RI scattering particles, such as lipids and proteins, can also enhance the RI matching in tissue to reduce the scattering of tissue. In addition, some endogenous pigments with strong absorption in biological tissues, such as heme, melanin, etc., are also essential factors limiting the optical imaging performance in deep tissue. Removal of these pigments reduces the attenuation of energy during light transport to facilitate deep penetration into thick tissues.
RI matching can reduce the scattering of tissue and make tissue transparent, which has been regarded as the essential foundation of tissue optical clearing. According to this principle, OCAs with higher refractive indices should induce better optical clearing efficacy to biological tissues. LaComb et al. supported this mechanism by examining the clearing capabilities in striated muscle and tendon (Lacomb et al., 2008). However, the optical clearing efficacy in skin did not directly correlate with the refractive indices of OCAs (Choi et al., 2005; Mao et al., 2008), but other reports demonstrated that the optical clearing effect of the skin induced by alcohols is positively correlated with the number of hydroxyl groups (Mao et al., 2008), which may be explained by the dissociation of collagen caused by OCAs (Hirshburg et al., 2007). Many studies have proved that dehydration is also one of the important mechanisms for skin optical clearing (Tuchin et al., 1997; Yu et al., 2011; Rylander et al., 2006; Xu and Wang, 2003). It is well known that tissue is composed of various components, including water, proteins, lipids, mineral compositions, and many other molecules, and the concentrations of these components vary in different types of tissues. Therefore, the variety of optical clearing mechanisms under discussion should result from complex interactions between agents and tissues (Tainaka et al., 2014, 2016; Richardson and Lichtman, 2015).
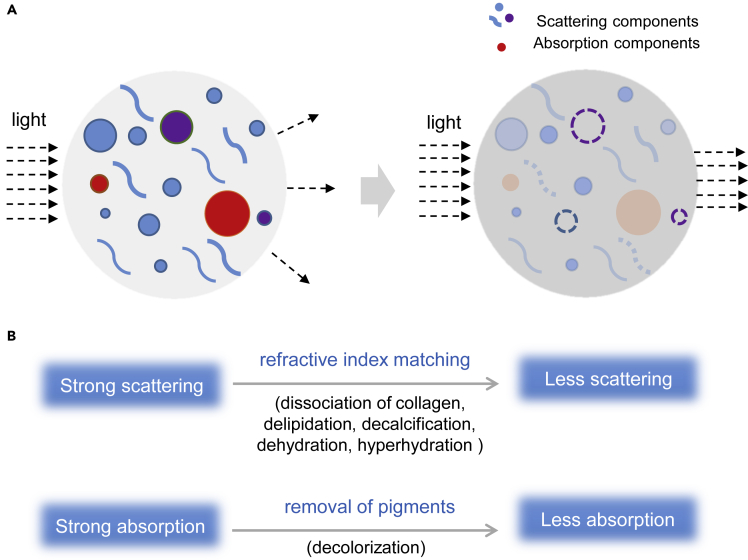
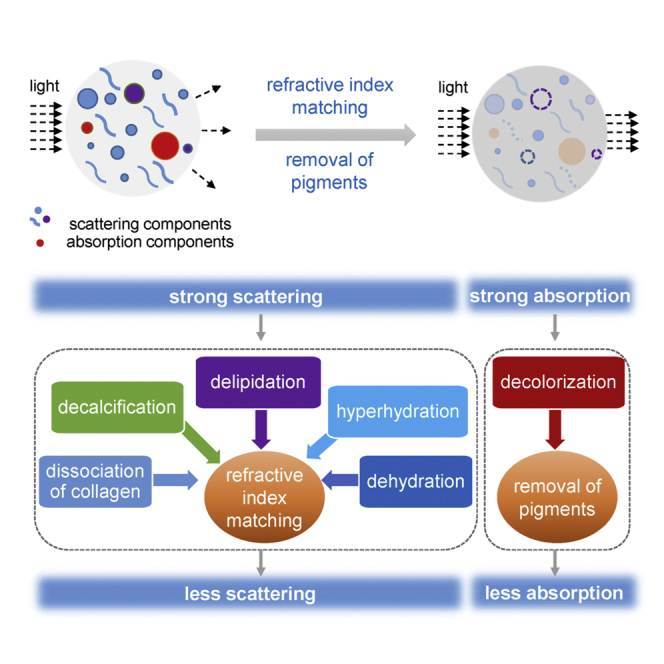
In this review, we focus on the mechanisms of tissue optical clearing from fundamental physical or chemical perspectives. The main physical principles include the reductions of light scattering via RI matching and light absorption via removal of pigments in tissues. Both reductions result from the interactions between chemicals and biomolecules, including collagen dissociation, delipidation, decalcification, dehydration, and hyperhydration for RI matching and decolorization for pigment removal (Figure 1).
Figure 1.
Principles of tissue optical clearing
Reducing light scattering and light absorption are the main physical principles of tissue optical clearing. RI matching can reduce scattering, and removal of pigments can reduce light absorption. Various chemical-biomolecule interactions contribute to these physical effects, resulting in tissue optical clearing.
Refractive index matching
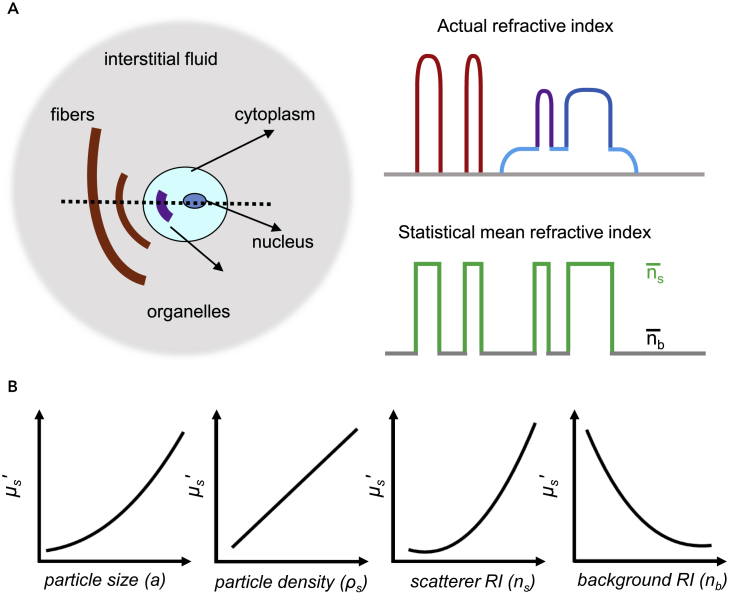
As mentioned above, RI matching is the main concept of tissue optical clearing. Tissues are densely packed with many types of substances, including scattering particles with higher RI (1.39–1.52), i.e., fibers, cell membrane, cell nucleus, and cell organelles, and surrounding media with lower RI (1.33–1.37), i.e., interstitial fluid and/or cytoplasm (Figure 2A). As shown in the right panel of Figure 2A, the actual refractive indices of these tissue structural elements vary, but the index variations can be represented by a statistically equivalent volume of discrete particles of the same index (ns) but different size (Tuchin, 2006). The difference in the refractive indices of these complex components and structures results in strong light scattering and prevents light traveling in tissues to impede deep imaging. RI matching by adjusting these RI differences was proposed to address this issue.
Figure 2.
Refractive index matching and tissue optical clearing
(A) Sketch map of biological tissue components and RI difference in different components (adapted from the study by [Tuchin, 2006]).
(B) Variation of the reduced scattering coefficient with different parameters at a specific wavelength, including particle diameter (a), particle density (ρs), scatterer RI (ns), and background RI (nb).
As biological tissues have complicated structures, the interactions between tissues and different OCAs are very complex, which prevents the understanding of the optical clearing mechanisms. Tissue-simulating phantoms with high homogeneity and stability, such as Intralipid, are an available and adequate model to block the biological effect of tissues and give a direct answer to the physical mechanism of optical clearing (Tuchin, 2002). To elucidate the physical mechanism of optical clearing, Wen et al. investigated the changes in the optical properties of Intralipid caused by different OCAs (Wen et al., 2009).
To acquire the reduced scattering coefficient (μs’) of the mixtures, Graaff et al. proposed a simplified Mie theory (Equation 1) (Graaff et al., 1992):
| (Equation 1) |
where a represents the mean diameter of the particles, ρs is the volume density of the particles, λ is the wavelength, and ns and nb are the RI of scatterers and background, respectively. The RI matching factor (i.e., m) is defined by ns divided by nb. According to this formula, the scattering property is largely determined by the RI matching factor, but changes of size and density of the scattering particles also contribute to the change of the scattering property. Figure 2B shows the variations of the reduced scattering coefficient with the individual parameters of particle size, particle density, scatterer RI, or background RI while all other parameters, except for the studied parameter, are kept constant.
Based on this simplified Mie theory, Wen et al. predicted the scattering property of Intralipid solution. The theoretical prediction matches well with the experimentally measured scattering coefficient. OCAs made the Intralipid solutions transparent, and the clearing effect of the OCAs increased in the order of 1,2-propanediol, 1,4-butanediol, glycerol, and dimethyl sulfoxide (DMSO), in parallel to the RI, which increased in the same order. Then, a further simplified formula was deduced at 589 nm (wavelength of commonly used light source of Abbe refractometers) where ns of the Intralipid solution was calculated to be 1.475:
| (Equation 2) |
When the size and density of scatterers remain unchanged, k is a constant. By fitting Equation 2 to the experimental data, k of 739.8 was obtained (Wen et al., 2009). If OCAs keep the solution uniform and do not change the particle size, the formula can be used to predict the OCA-induced change of the scattering property. A higher background RI was correlated with a stronger decrease in the reduced scattering coefficient and a better optical clearing effect. This reveals that increasing the background RI results in RI matching of the mixture. Regarding the physical principle, increasing RI matching to reduce light scattering of tissue is one essential mechanism of tissue optical clearing.
Based on this principle, various strategies have been employed to achieve RI matching, such as the removal of low RI water, removal of high RI lipid, dissociation of protein followed by the import of RI matching solutions, etc. In most tissue optical clearing methods, high RI reagents or cocktails termed as RI matching solutions were employed for RI matching (Table 1). RI matching solutions mainly include hydrophilic and hydrophobic reagents. The former mainly include a high concentration of sugars, sugar alcohols, contrast agents, or amides with high RIs (1.38–1.52), such as glycerol (Chung et al., 2013; Hama et al., 2011, 2015; Zhu et al., 2019; Zhao et al., 2018), sucrose (Tsai et al., 2009; Susaki et al., 2014; Chen et al., 2017), fructose (Ke et al., 2013; Hou et al., 2015; Hildebrand et al., 2020), sorbitol (Yang et al., 2014; Economo et al., 2016; Zhu et al., 2020), PEG8000 (Kuwajima et al., 2013), iohexol (Yang et al., 2014; Treweek et al., 2015; Ke et al., 2016; Li et al., 2017), iodixanol (Murray et al., 2015), diatrizoic acid (Lin et al., 2007), and N-methylnicotinamide (Tainaka et al., 2018; Kubota et al., 2017). Some other molecules, such as sulfoxides (e.g., DMSO) (Economo et al., 2016; Hama et al., 2015), 2,2′-thiodiethanol (TDE) (Costantini et al., 2015; Di Giovanna et al., 2018; Mizutani et al., 2018), and anionic molecules (e.g., phosphoric acid) (Umezawa et al., 2019), were also employed to match the RI. Notably, the final RI matching solution is often comprised of cocktails of these reagents, such as the dPROTOS in ELAST (entangled link-augmented stretchable tissue-hydrogel) protocol, which uses a mixture of iohexol, TDE, and DMSO (Ku et al., 2020). As hydrophobic solution, mainly ether or ester reagents are used, such as methyl salicylate (Spalteholz, 1914), benzyl alcohol-benzyl benzoate (1:2, BABB) (Schwarz et al., 2015; Cai et al., 2019), dibenzyl ether (DBE) (Erturk et al., 2012a), diphenyl ether (DPE) (Yi et al., 2019), ethyl-3-phenylprop-2-enoate (ECi) (Klingberg et al., 2017; Masselink et al., 2019), PEGMMA500 (Jing et al., 2018), and others. These reagents are miscible with organic solvents and have a high RI (1.55–1.56), so they can effectively infiltrate into tissues and play a role of homogenization in optical clearing. Instead of the RI matching solution, Kellner et al. employed (liquid) curing with resin monomer mixtures to a polymer matched to the tissues (Kellner et al., 2016).
Table 1.
Common RI matching solutions
| Type | Chemicals | RI | Clearing methods or cocktails |
|---|---|---|---|
| Hydrophilic reagents | Glycerol | 1.48 | Scale, CLARITY, ScaleS, SOCW, FOCM |
| Fructose | 1.49 | SeeDB, FRUIT | |
| Sucrose | 1.44 | CUBIC, UbasM | |
| Sorbitol | 1.47 | SOCS, sRIMS, DMSO/Sorbitol, ScaleS, MACS, FOCM | |
| Xylitol | 1.39 | ClearSee, ePro-ClearSee | |
| Polyethylene glycol | 1.46–1.47 | ClearT2, PEGASOS | |
| Iohexol | – | RIMS, Ce3D, SeeDB2 | |
| Iodixanol | 1.41 | SWITCH | |
| Diatrizoic acid | – | FocusClear, Stochastic electrotransport | |
| Antipyrine | 1.59 | CUBIC-R, CUBIC-RA | |
| N-methylnicotinamide | 1.58 | CUBIC-R, CUBIC-RA | |
| Nicotinamide | 1.47 | CUBIC-R, CUBIC-R+ | |
| N-methyl-d-glucamine | – | Stochastic electrotransport, SWITCH | |
| DMSO | 1.48 | DMSO/Sorbitol, ScaleS, FOCM | |
| TDE | 1.52 | TDE/PBS, TOMEI, LUCID, CLARITY | |
| Phosphoric acid | 1.43 | Phosphoric acid (8.5–14.2 M) | |
| Hydrophobic reagents | Methyl salicylate | 1.54 | Spalteholz's technique |
| Benzyl alcohol | 1.54 | BABB, uDISCO, vDISCO | |
| Benzyl benzoate | 1.57 | BABB, uDISCO, vDISCO, PEGASOS | |
| DBE | 1.56 | 3DISCO, iDISCO, iDISCO+, FDISCO, Adipo-Clear, CalAdipoClear | |
| DPE | 1.58 | uDISCO | |
| ECi | 1.56 | Ethanol-ECi, 2ECi | |
| Resin | 1.56 | CRISTAL |
The other strategy for the reduction of the RI difference is to expand tissues for more homogeneous RI inside. The development of expansion microscopy provided a new perspective for super-resolution imaging based on sufficient tissue expansion with swellable hydrogels (Chen et al., 2015; Chang et al., 2017). Moreover, ePACT (PACT-based expansion clearing) (Treweek et al., 2015) and MAP (magnified analysis of the proteome) (Ku et al., 2016) are also based on hydrogel embedding to expand entire organs, showing high tissue transparency. The Ueda group developed CUBIC-X to obtain a single-cell-resolution whole-brain atlas (Murakami et al., 2018).
Dissociation of collagen
In investigations on tissue optical clearing, various OCAs, including alcohols, sugars, organic acids, and organic solvents, have been widely used. Choi et al. showed that the optical clearing efficacy of chemical agents does neither correlate with RI nor osmolality, and the hydroxy-terminated organic compounds demonstrated the highest optical clearing potential (OCP) (Choi et al., 2005). Furthermore, Mao et al. found that alcohol OCAs with more hydroxyl groups have better optical clearing efficacy with higher relative transmittance for porcine skin (e.g., 2.00 for glycerol versus 1.38 for 1-butanol after 60-min treatment) (Mao et al., 2008). The above experimental results demonstrated that skin optical clearing efficacy was correlated with the molecular structure of OCAs, which should be due to some interactions between OCAs and the molecular components of the skin, i.e., collagen fibers.
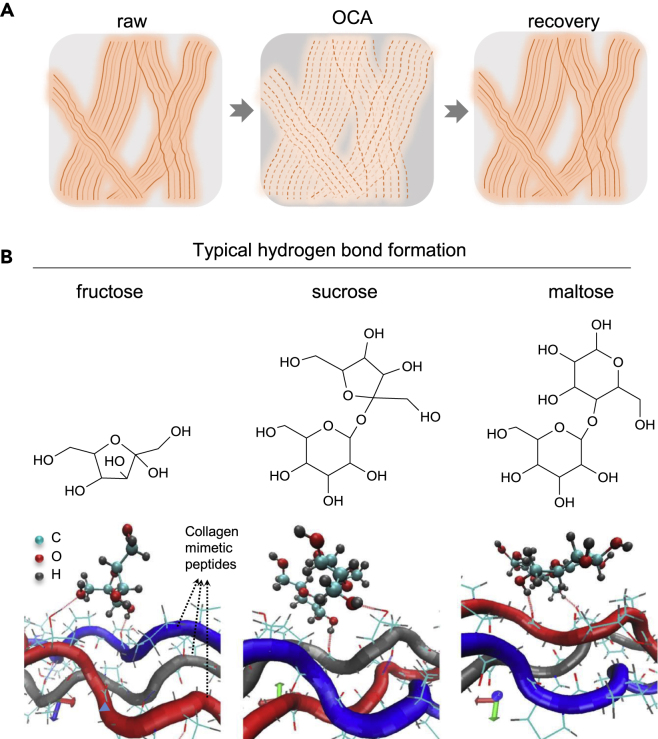
Collagen fibers are widely distributed in various biological tissues, such as skin dermis, corneal stroma, muscle, tendon, etc. Collagen fibers have complex self-assembled structures, which make them strong scattering sources in biological tissues (Yeh and Hirshburg, 2006). Synthetic collagen-like peptides have been used to demonstrate that self-assembly of secondary and tertiary structures of collagen I is driven by interactions of specific hydrophobic and electrostatic binding sites. The stability of these structures is enhanced by inductive (electronegativity) effects conferred by hydroxyproline residues. For higher-order structures, hydrogen bonding is the primary bonding force between collagen triple helices. OCAs with multiple hydroxyl groups can have strong electronegativity, which may screen the hydrogen bonds in collagen triple helices to trigger the dissociation of the higher-order structure of collagen. As the screening of the hydrogen bonds in collagen triple helices is a non-covalent effect, the effect of the OCAs on the dissociation of collagen can be reversible. Yeh et al. observed the dissociation of collagen fibers of in vitro skin tissues after immersion in OCA (e.g., glycerol) and a recovery of the collagen fibrous structure after removal of excess glycerol and subsequent application of PBS (phosphate buffer saline) by second-harmonic generation imaging (Yeh et al., 2003). In addition, the destabilization of high-order collagen structures resulted in primary light scatterers with smaller size, which may also contribute to skin optical clearing according to Equation 1 (Hirshburg et al., 2007). Based on these results, we illustrated the dissociation or looseness of collagen structures in Figure 3A.
Figure 3.
Dissociation of collagen fibers
(A) Sketch map of the reversible effect of OCAs (e.g., glycerol).
(B) Chemical structures of fructose, sucrose, and maltose, and schematic representation of the hydrogen bond formation of these sugars with representative collagen-mimetic peptides, respectively. The more hydrogen bonds form, the higher the amount of disrupted hydration shells and water-mediated hydrogen bonds. The average numbers of hydrogen bonds formed per ps for fructose, sucrose, and maltose are 1.254, 1.307, and 1.292, respectively, with fructose exhibiting the best OCP (adapted from the study by [Feng et al., 2016]).
Hirshburg et al. used molecular dynamics (MD) simulation to explore the hydrogen bond formation between alcohols and collagen molecules (Hirshburg et al., 2010). They divided the hydrogen bond bridges between alcohols and collagens into different types. The index of the type of hydrogen bond bridge is related to the number of carbon atoms of the alcohols in a hydrogen bond bridge. Higher bridge types are further expanded across the collagen surface and can disrupt collagen-collagen and collagen-water interactions potentially better than lower bridge types. Thus, the optical clearing efficacy of alcohols with hydroxyl group pairs with longer distance on the carbon chain will be better than that of alcohols only having hydroxyl groups adjacent to each other.
Considering that sugars also have hydroxyl groups, Zhu's group focused on monosaccharides and saccharides (Wang et al., 2014; Feng et al., 2016). MD simulation was applied to evaluate the OCPs of fructose, glucose, and ribose according to their propensities to form hydrogen bonds and bridges. Previous studies showed that the tertiary structures and triple helices of collagen were stabilized and organized for higher-order assembly by water bridges and a hydration shell (Hirshburg et al., 2006; Kuznetsova et al., 1998; Ravikumar and Hwang, 2008; Ravikumar et al., 2007). Furthermore, the formation of hydrogen bonds and hydrogen bond bridges between chemical agents and collagen disrupts the water bridges and hydration shell, resulting in the destabilization of the higher-order structures of collagen (Yeh and Hirshburg, 2006; Hirshburg et al., 2007). Thus, the propensity of sugars to form hydrogen bonds and bridges can be considered as a measure of their optical clearing efficacy. The more hydrogen bonds and bridges a sugar forms, the more effectively water bridges and hydration shell are disrupted. Wang et al. found that fructose forms more hydrogen bonds and bridges than the other sugars, while ribose forms the fewest hydrogen bonds and bridges, which is in agreement with the experimental results in skin tissues (Wang et al., 2014).
Apart from the number of hydroxyl groups, Hirshburg et al. found that an increase of the molecular size of the OCAs could also increase the efficacy of tissue optical clearing (Hirshburg, 2009). Considering that disaccharides have a larger molecular size and more hydroxyl groups than the above three monosaccharides, Zhu's group further investigated the OCPs of sucrose and maltose as representative disaccharides by theoretical simulation and compared the results with ex vivo and in vivo experiments of fructose (Feng et al., 2016). Figure 3B shows the chemical structures of the three agents and the hydrogen bonds they formed with collagen-mimetic peptide. It is expected that the more hydrogen bonds and hydrogen bond bridges an agent forms, the higher the amount of disrupted hydration shell and water-mediated hydrogen bonds. The average numbers of hydrogen bonds formed between an agent and two collagen-mimetic peptides per ps are 1.254, 1.307, and 1.292 for fructose, sucrose, and maltose, respectively. This indicates that sucrose has the strongest ability to disrupt the hydration shell and water-mediated hydrogen bonds. Therefore, the quality of skin OCP is predicted to decrease from sucrose over maltose to fructose. The results of the theoretical simulation were experimentally confirmed, which effectively prevents blindness and time-consuming experiments.
Among clearing reagents, urea is a serendipitously discovered reagent with remarkable clearing capability. In Scale, urea can induce obvious tissue expansion and transparency (Hama et al., 2011). It is hypothesized that urea relaxes protein scaffolds that are involved in solid tissue frameworks, such as collagen fibers, by forming hydrogen bonds and disturbing hydrogen-bonding networks of proteins (Tainaka et al., 2016). In ScaleS, N-acetyl-L-hydroxyproline (trans-1-acetyl-4-hydroxyl-proline) was also introduced to solvate collagen fibers to relax entangled protein networks (Hama et al., 2015).
Dissociation or relaxation of dense collagen fibers may render the tissue optically clear or accelerate molecular influx into tissues, playing an important role in tissue optical clearing.
Delipidation
Besides the proteins such as collagen described above, lipid is another cellular component with a high RI in most biological tissues. The lipid content reaches up to more than 70% in adipose tissue, and it is about 20% in white matter or tongue. Most soft tissues contain about 10% lipids. Lipid in tissues does not only cause light scattering within the tissue due to its high RI but also hinder the influx of the external medium as the main component of membranes. Thus, tissue delipidation is important for the full clearing of tissues and permeation of external molecules.
Removal of lipid is mainly achieved by specific solvents or water-soluble molecules (e.g., detergents and aminoalcohols) in current tissue optical clearing techniques (Table 2).
Table 2.
Main delipidation chemicals in tissue clearing techniques
| Group | Chemicals | Clearing methods or cocktails |
|---|---|---|
| Solvents | THF | 3DISCO, FDISCO, vDISCO |
| DCM | 3DISCO, uDISCO, iDISCO(+), Adipo-Clear, CalAdipoClear, SHANEL | |
| Aminoalcohols | Quadrol | ScaleCUBIC-1, vDISCO, PEGASOS |
| Triethanolamine | ScaleCUBIC-2, RTF | |
| N-butyldiethanolamine | CUBIC-L | |
| Aliphatic amine | 1,3-BAC | CUBIC-HL |
| Detergents | Triton X-100 | Scale, UbasM, SUT, Ce3D, CUBIC, vDISCO |
| Saponin | SeeDB2 | |
| SDS | CLARITY and its derivatives | |
| SDC | SeeNet, ClearSee | |
| SDBS | USOCA | |
| CHAPS | SHANEL | |
| Oligosaccharides | methyl-cyclodextrin | ScaleS, vDISCO |
| γ-cyclodextrin | ScaleS |
In solvent-based clearing methods, delipidation is usually accompanied by dehydration (Richardson and Lichtman, 2015). Dodt's group first used ethanol as a dehydration reagent and BABB as a RI matching reagent to clear tissues (Dodt et al., 2007; Becker et al., 2008; Jahrling et al., 2009; Jährling et al., 2008). However, these reagents did not achieve sufficient transparency in adult tissues containing a high degree of lipids, such as the rodent spinal cord and brainstem. To overcome this limitation, they replaced ethanol by tetrahydrofuran (THF) and introduced dichloromethane (DCM) to improve the tissue transparency due to the good lipid-solvating capability of these solvents (Becker et al., 2012; Erturk et al., 2012b). Furthermore, the combination of THF with dibenzyl ether (DBE) resulted in the 3DISCO method, providing an improved tissue transparency and fluorescence signal (Erturk et al., 2012a). In general, for delipidation, DCM was also used only for short incubation times because of the trade-off between transparency and endogenous fluorescence preservation (Erturk et al., 2012a; Pan et al., 2016). Zhao et al. adopted DCM for delipidation in SHANEL (small-micelle-mediated human organ efficient clearing and labeling) clearing of human organs (Zhao et al., 2020). Other alcohols, such as 1-propanediol and tert-butanol (Schwarz et al., 2015; Pan et al., 2016; Jing et al., 2018), were also used for delipidation in solvent-based clearing methods such as FluoClearBABB (Schwarz et al., 2015), uDISCO (Pan et al., 2016), and PEGASOS (Jing et al., 2018) method.
Among water-soluble molecules, detergents and aminoalcohols play an important role in lipid removal.
The ScaleA2 clearing agent proposed by Miyawaki's group is composed of urea, glycerol, and Triton X-100 (Hama et al., 2011). Triton X-100 is a commonly used nonionic detergent in cell biology that can dissolve lipids to increase the cell permeability by incorporating the lipids into a micelle. Corresponding to the components of ScaleA2, Ueda's group screened a lot of chemicals including polyhydric alcohols, detergents, and hydrophilic small molecules (Susaki et al., 2014). To evaluate the ability to dissolve brain tissue, fixed brain homogenate was mixed with different solutions, and the OD600 (optical density at 600 nm) was measured, which was expected to correlate with the potential solubility of brain lipids (Susaki et al., 2014). All the detergents induced a decrease of the OD600 of the brain homogenate to a certain extent. Among the tested detergents, Triton X-100 and sodium deoxycholate (SDC) showed the highest levels of solubilizing activity. In addition, a series of aminoalcohols, including Quadrol and triethanolamine, also exhibited a considerable tissue solubilizing activity (Susaki et al., 2014), indicating potential clearing capability. It is speculated that the cationic amino group in aminoalcohols contributes to solvating anionic phospholipids. In other words, aminoalcohols presumably solubilize phospholipids by electrostatic interaction. Taking account of the clearing performance and fluorescence preservation, a cocktail of urea, Quadrol, and Triton X-100 was mixed as the first reagent in the CUBIC protocol, i.e., ScaleCUBIC-1 (Susaki and Ueda, 2016), and triethanolamine was also included in ScaleCUBIC-2 to further increase tissue transmittance (Susaki et al., 2014; Tainaka et al., 2014). In addition, for the whole-body profiling of cancer metastasis, the optimized cocktail of 10 w%/10w % N-butyldiethanolamine/Triton X-100 (termed CUBIC-L) was reported for highly effective delipidation (Kubota et al., 2017). Recently, the aliphatic amine 1,3-bis(aminomethyl)cyclohexane (1,3-BAC) also showed high lipid solubility and contributed to rapid delipidation in the CUBIC-HL cocktail (Tainaka et al., 2018).
Saponin is a widely used mild nonionic detergent that was also utilized for delipidation to permeabilize cellular membranes in tissue clearing methods. For instance, it was used to facilitate the permeation of iohexol in SeeDB2 clearing (Ke et al., 2016) and the diffusion of hydrogel monomer solution into human or zebrafish tissue in CLARITY clearing (Chung et al., 2013).
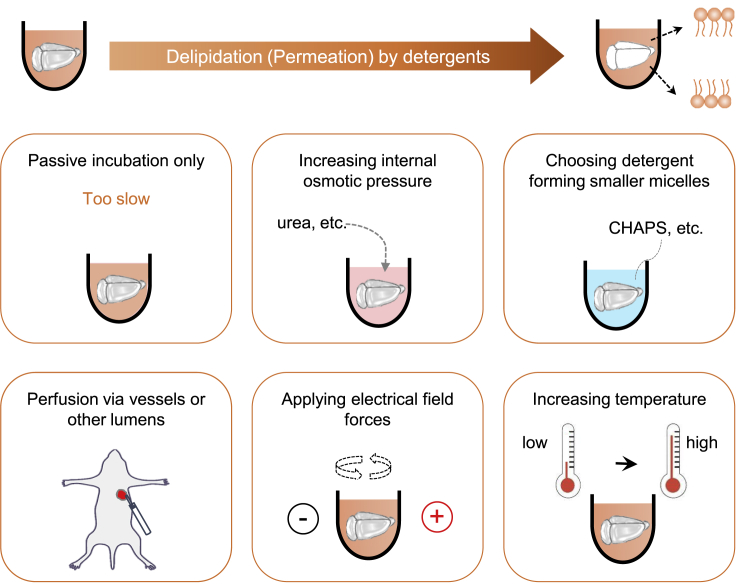
Sodium dodecyl sulfate (SDS) is an ionic detergent with strong lipid dissolving capability. All CLARITY-related clearing methods are based on delipidation by SDS. Passive diffusion of detergent micelles into tissues is very slow, presumably due to their large molecular size (Tainaka et al., 2016; Zhao et al., 2020). To achieve sufficient delipidation or permeation in large tissues, the diffusion of detergents within the tissues needs to be accelerated. The utilization of urea in Scale, CUBIC, and SUT methods can promote the passive influx of detergents by increasing the internal osmotic pressure (Hama et al., 2011; Susaki et al., 2014, 2015; Tainaka et al., 2014; Wang et al., 2018b). Some other strategies, including electric field potential, perfusion, and thermal energy, have also been utilized to promote the diffusion of detergents, such as SDS (Figure 4).
Figure 4.
Delipidation by detergents
Passive incubation for the diffusion of detergents is very slow. Permeation of detergents into tissues can be promoted by increasing the internal osmotic pressure via the introduction of hyperhydration chemicals such as urea; choosing a detergent that forms smaller micelles, such as CHAPS; delivering the detergents by perfusion via the vasculature or other lumens; applying electrical field forces for ionic detergent micelles via different patterns; and increasing the temperature to enhance molecular movement.
In 2013, Chung et al. proposed the distinct clearing method CLARITY, which is based on hydrogel embedding and electrophoresis (Chung et al., 2013). In this method, SDS was used to remove the lipid of hydrogel-embedded samples, and the removal was accelerated by an electric field potential applied in the electrophoresis approach. However, its application was limited to low electric fields, as using high electric fields can damage tissue structures (Kim et al., 2015; Lee et al., 2016). Chung's group introduced stochastic electrotransport by using a rotational electric field (Kim et al., 2015), which can selectively transport highly electromobile molecules to rapidly render tissue transparent without tissue damage. They further applied the stochastic electrotransport method to SHIELD (Park et al., 2018) and eFLASH (Yun et al., 2019) for rapid delipidation or acceleration of probe penetration. In the recently published ELAST protocol (Ku et al., 2020), SDS was also used for delipidation after gelation.
Perfusion is another strategy to reinforce detergent diffusion within tissues. Yang et al. first introduced the perfusion-assisted agent release method PARS (Yang et al., 2014). This method uses the blood circulatory system or the cerebrospinal fluid route to deliver detergents to the whole body or target organs for rapid delipidation, facilitating highly efficient whole-organ and whole-body clearing after RIMS (refractive index matching solution) treatment (Yang et al., 2014). Moreover, Tainaka et al. utilized transcardial perfusion of half-diluted ScaleCUBIC-1 combined with external immersion for enhancing whole-organ and whole-body clearing (Tainaka et al., 2014).
In several protocols, thermal activation of diffusion was also adopted to accelerate SDS permeation based on the temperature dependency of solute diffusion kinetics. Yu et al. reported the elevated temperature-induced acceleration of PACT and concluded that 42–47°C is an alternative temperature range for PACT, while higher temperatures would damage the sample by a strong surfactant solution (Yu et al., 2017). Chung's group introduced SWITCH-based glutaraldehyde fixation to secure tissue structures. The SWITCH-processed samples endured prolonged incubation in SDS at high temperatures up to 80°C (Murray et al., 2015). In the SWITCH method, the thermal energy considerably increased the passive clearing speed without noticeable tissue damage. Notably, with the increase of temperature, the SDS micelle volume decreases, and the number of micelles increases (Hammouda, 2013). This facilitates SDS micelle diffusion into tissue, which may be another reason for the thermal reinforcement of SDS permeation besides the increased molecule kinetics.
SDC is another ionic detergent, which has been used for delipidation in SeeNet (Miyawaki et al., 2020). Although the light transmittance of SDC-cleared tissues is slightly lower than that of SDS-cleared tissues, SDC-cleared tissues exhibited a spatially uniform RI across the samples and had no opaque areas. SDC has also been used in plant clearing protocols, such as ClearSee (Kurihara et al., 2015) and ePro-ClearSee (Nagaki et al., 2017). Sodium dodecylbenzenesulfonate (SDBS), as an ionic detergent, was used to extract lipids in the clearing of the skull by USOCA (urea-based skull optical clearing agent) (Zhang et al., 2018). Recently, Zhao et al. reported that the zwitterionic detergent CHAPS (3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate) forms smaller micelles than Triton X-100 and SDS, which could penetrate more rapidly and deeply into tissue, allowing full permeabilization of aged human organs (Zhao et al., 2020) (Figure 4). Based on the CHAPS tissue permeabilization approach, SHANEL was proposed to clear and label stiff human organs.
In addition, some oligosaccharides were employed in clearing protocols for delipidation (mainly for the extraction of cholesterol), such as methyl-cyclodextrin (Cai et al., 2019; Hama et al., 2015) and γ-cyclodextrin (Hama et al., 2015).
Decalcification
Notably, in hard bone or calcified tissue, the mineral components make the RI higher (1.55–1.65) than the other tissue components (Ohmi et al., 2000). Therefore, decalcification is indispensable for high transparency of bone clearing or whole-body clearing. To clear bone tissues or the whole body, decalcification was carried out prior to the other steps in most clearing methods. Some mineral acids, such as nitric acid, hydrochloric acid, and weak organic acids (e.g., formic acid, etc.), were used in histological decalcification (Susaki and Ueda, 2016; Callis and Sterchi, 1998; Gomes et al., 2008; Begum et al., 2010). Most of these acids (e.g., nitric acid) can achieve fast decalcification but would cause damage to the bone tissues (Sanjai et al., 2012; Sangeetha et al., 2013).
Chelating reagents, such as ethylenediaminetetraacetic acid and its disodium salt (both abbreviated as EDTA), can also be used for decalcification. EDTA causes little damage to tissue and can preserve the activity of antigens and certain enzymes in tissues, making it suitable for subsequent immunochemistry (Sanjai et al., 2012; Sangeetha et al., 2013). It has been utilized in many in vitro clearing methods, such as PACT-deCal (Woo et al., 2016), Bone-CLARITY (Greenbaum et al., 2017), PEGASOS (Jing et al., 2018), BoneClear (Wang et al., 2019), vDISCO (Cai et al., 2019), etc., for bone clearing or whole-body clearing. Konno et al. also used EDTA for the decalcification of crustaceans prior to whole-mount clearing (Konno and Okazaki, 2018). Zhu's group introduced EDTA in SOCS (skull optical clearing solution) and SOCW (skull optical clearing window) techniques to create a transparent cranial window for in vivo imaging of the cortex (Wang et al., 2012; Zhao et al., 2018).
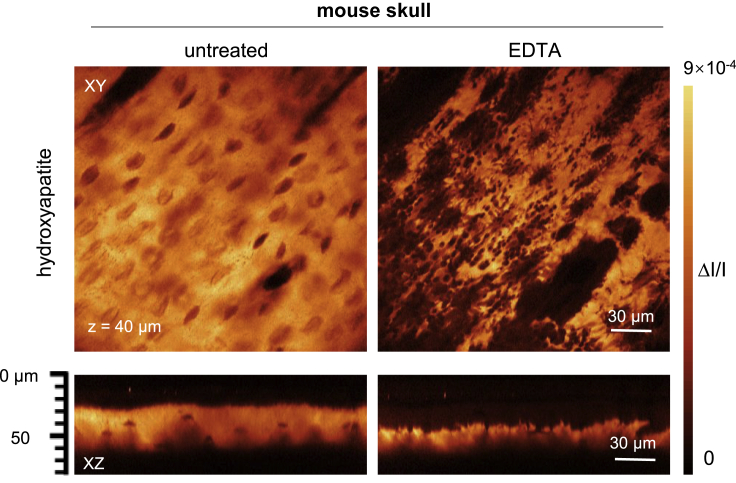
EDTA achieves very gentle decalcification mainly by capturing calcium ions from the surface of the apatite crystal and thus weakens the microstructure of the bone (Chen et al., 2019). By stimulated Raman scattering (SRS) imaging, Chen et al. directly visualized the microstructural variation of hydroxyapatite in the skull before and after treatment with EDTA (Chen et al., 2019). XY images at a depth of 40 μm in the same location of the skull showed that after 5–10 min of EDTA immersion, a large number of cavities formed around the bone lacunae, indicating hydroxyapatite corruption (Figure 5). XZ projection images showed that the thickness of hydroxyapatite in the skull was significantly reduced after EDTA treatment.
Figure 5.
Microstructural variation of hydroxyapatite
SRS images of the mouse skull before and after topical treatment with EDTA. A large number of cavities formed after EDTA immersion and the thickness of hydroxyapatite reduced, indicating corruption of hydroxyapatite (adapted from the study by [Chen et al., 2019]).
Tainaka et al. found that decalcification with EDTA would be substantially facilitated by neutralization with organic bases instead of inorganic bases. After screening different EDTA derivatives (e.g., PDTA, CyDTA, DOTA, EGTA, etc.) and organic bases, they concluded that a cocktail combining EDTA and imidazole (named CUBIC-B) is the optimal decalcification solution (Tainaka et al., 2018).
Recently, Muller et al. reported CalAdipoClear method (Muller et al., 2020), which used Morse solution consisted of one part 45% formic acid/one part 0.68 mM sodium citrate dihydrate for decalcification of spinal columns, followed by immunolabeling.
Dehydration
Water occupies 70–80% of the volume of tissues and exhibits a much lower RI (∼1.33) than proteins and lipids (>1.44). Regarding the physical mechanism, when water within tissues is substituted by chemicals with high RI, the scattering of tissue will be reduced (Tainaka et al., 2016). Therefore, OCA-induced tissue dehydration is regarded as an important mechanism of tissue optical clearing.
Rylander et al. found that air immersion induces a similar but slower increase in tissue transparency than OCA immersion (e.g., DMSO, glycerol) (Rylander et al., 2006). For air immersion, only water is volatile and can evaporate from the tissues, hence inducing substantial mass loss. After OCA immersion, the skin mass also decreases. This result indicates water loss via efflux (dehydration) during clearing, which should be at least more significant than the weight loss of tissues due to OCA influx. Rylander et al. also studied the tissues' ultrastructure and found that dehydration by DMSO or glycerol increases the packing density of individual scattering particles (e.g., collagen fibrils and organelles), while the size does not change significantly. The increase of the scattering particle volume fraction contributed substantially to optical clearing in collagenous and cellular tissue. Xu et al. used dual-wavelength analysis to estimate the water content of porcine muscles based on diffuse reflectance spectroscopy (Xu and Wang, 2003). Instead of dual-wavelength analysis, Yu et al. used a partial least-squares regression model based on reflectance spectroscopy to calculate the water content in absolute values (Yu et al., 2011). Both groups concluded that the optical clearing effect is strongly correlated with dehydration. These previous studies identified that dehydration has a major contribution to the reduction of light scattering. Dehydration can also change the particle size, reduce the background RI, and promote the influx of high RI reagents or mixtures for final RI matching.
During the development of tissue optical clearing techniques, many reagents were employed for tissue dehydration, mainly including hydrophilic agents or water-miscible polar solvents (Chung et al., 2013) (Table 3). As hydrophilic agents, some highly concentrated polyalcohols, such as glycerol (Hama et al., 2011; Chung et al., 2013), sucrose (Susaki et al., 2014; Tsai et al., 2009; Chen et al., 2017), fructose (Ke et al., 2013; Hou et al., 2015), sorbitol (Yang et al., 2014; Economo et al., 2016; Zhu et al., 2020), and xylitol (Kurihara et al., 2015), as well as contrast agents, such as iohexol (Yang et al., 2014; Treweek et al., 2015; Ke et al., 2016; Li et al., 2017) and iodixanol (Murray et al., 2015), were used to dehydrate tissue samples based on the osmotic difference. In general, these highly concentrated agents produce a higher osmolarity in external than internal tissues, thereby inducing efflux of water and influx of external molecules. At the same time, these agents have hydroxyl groups to form hydrogen bonds with proteins, showing a higher hydrogen-bonding ability than water (Hirshburg et al., 2010). Hence, samples that were dehydrated with these agents demonstrated a certain degree of transparency.
Table 3.
Main dehydration reagents for tissue dehydration
| Type | Chemicals | Features | Clearing methods or cocktails |
|---|---|---|---|
| Hydrophilic reagents | DMSO | Dehydration also induces a degree of transparency | DMSO/Sorbitol, FOCM |
| Glycerol | Scale, CLARITY | ||
| Sucrose | Single treatment, CUBIC, UbasM | ||
| Fructose | SeeDB, FRUIT | ||
| Sorbitol | DMSO/Sorbitol | ||
| Xylitol | ClearSee | ||
| Iohexol | RIMS, Ce3D, SeeDB2 | ||
| Iodixanol | SWITCH | ||
| Water-miscible polar solvents | Ethanol | Dehydration induces tissue shrinkage | BABB, Ethanol-ECi |
| Methanol | iDISCO+, Adipo-Clear, CalAdipoClear | ||
| 1-propanol | FluoClearBABB, 2ECi | ||
| Tert-butanol | FluoClearBABB, uDISCO, PEGASOS | ||
| THF | 3DISCO, FDISCO, vDISCO |
Water-miscible polar solvents play an essential role in mediating the substitution of water with high RI aromatic solvents within tissues, as aromatic solvents with high RI (n > 1.5) are generally immiscible with water. Classically, biological tissues were dehydrated with alcohols, such as ethanol (Dodt et al., 2007; Becker et al., 2008; Jahrling et al., 2009; Klingberg et al., 2017; Kellner et al., 2016). Initially, the tissues were dehydrated by successively increasing the ethanol concentration to 100%. As the hydrogen-bonding ability of ethanol is significantly lower than that of water, the tissues were shrunk and hardened after dehydration and showed no sign of transparency like hydrophilic agents. To achieve efficient clearing of lipid-rich tissues, such as the spinal cord and brainstem, Dodt's group selected THF as a highly effective and GFP (green fluorescent protein)-friendly dehydrating reagent among thousands of dehydration chemicals instead of ethanol (Becker et al., 2012; Erturk et al., 2012b). This polar ether-based protocol has achieved extremely rapid dehydration (Becker et al., 2012; Erturk et al., 2012a, 2012b) due to its highly permeable kinetics and has provided improved transparency of cleared large samples after RI matching due to its highly efficient lipid solubility.
In recent years, some other alcohols have been used as dehydration agents in solvent-based clearing methods for improved fluorescence or morphology preservation. For example, Schwarz et al. reported the FluoClearBABB protocol that uses 1-propanol or tert-butanol for dehydration (Schwarz et al., 2015), and the Ertürk group developed the uDISCO method (Pan et al., 2016) that achieves an improved fluorescence by using tert-butanol for dehydration. Renier et al. reported the iDISCO+ protocol wherein THF dehydration is replaced by methanol dehydration (Renier et al., 2016), resulting in less damage to the tissue morphology and enabling the automated registration to a reference brain atlas for automated analysis. Methanol has also been applied for dehydration of adipose tissues (Chi et al., 2018), as well as some clinical specimens (Scott et al., 2014; Van Royen et al., 2016).
Hyperhydration
Many tissue optical clearing methods benefit from the high hydration of hyperhydration agents, such as urea and urea-like chemicals (Table 4), which enhance the penetration of clearing reagents into the tissues to achieve better transparency.
Table 4.
Comparison of some hyperhydration agents
| Chemicals | Chemical structures | Group | RI | Water solubility | Clearing methods or cocktails |
|---|---|---|---|---|---|
| Urea | 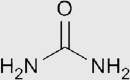 |
Amide | 1.40 | 108g/100mL | Scale, ScaleS, CUBIC and its derivatives |
| Formamide | 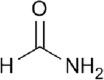 |
Amide | 1.45 | Miscible | ClearT/T2 |
| Thiourea | 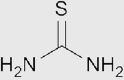 |
Thioamide | 1.53 | 13.6g/100mL | Not used in clearing due to its low solubility |
| 1,3-BAC | 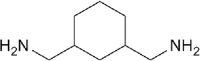 |
Aliphatic amine | 1.49 | Soluble | CUBIC-HL |
| MXDA | 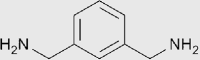 |
Aromatic amine | 1.57 | Miscible | MACS |
Among the hyperhydration agents, urea was serendipitously discovered to have good clearing capability (Hama et al., 2011). As a small, uncharged molecule with two NH2 groups, urea can be both a proton donor and acceptor in hydrogen bond formation, resulting in a strong hydration ability (Jedlovszky and Idrissi, 2008). Therefore, urea can disturb hydrogen-bonding networks of proteins and nucleic acids as a relatively weak denaturant (Hua et al., 2002; Priyakumar et al., 2009). Moreover, urea can increase the membrane fluidity to enhance the molecular flux (Barton et al., 1999; Feng et al., 2002), which also contributes to an enhanced permeation of reagents.
Urea is used in many hyperhydration-based clearing methods. The ScaleA2 cocktail contains 4 M urea, 10% glycerol, and 0.1% of the nonionic detergent Triton X-100 (Hama et al., 2011). This cocktail achieves good transparency in the mouse brain and embryos but causes obvious tissue swelling, presumably because urea relaxes protein scaffolds involved in solid tissue frameworks, such as collagen fibers, as described above. In addition, the internal osmotic pressure is increased, which induces an influx of small external molecules, including water and other ingredients. For instance, Hou et al. utilized a cocktail of fructose and urea in the FRUIT protocol (Hou et al., 2015), which achieved greater tissue transparency with a shorter clearing time than fructose alone in the SeeDB protocol. The Miyawaki group further proposed ScaleS, which combines urea, sorbitol, and DMSO and uses a gradient of concentrations to enhance the penetration (Hama et al., 2015). This protocol not only accelerated the clearing process compared to the sorbitol/DMSO protocol proposed by Economo et al. (Economo et al., 2016) but also showed excellent preservation of fluorescence and ultrastructure. Both FRUIT and ScaleS protocols demonstrated the importance of the gradient in solute concentration and the balance of ionic osmotic pressure. Notably, FRUIT and ScaleS were compatible with the lipophilic dye DiI, suggesting that urea would not disturb the lipid bilayer membrane.
Furthermore, Ueda's group developed CUBIC (Susaki et al., 2014) based on Scale. In this method, aminoalcohol, Triton X-100, and urea were mixed to produce ScaleCUBIC-1 for the removal of lipids and looseness of tissue structure. Then, ScaleCUBIC-2 with high RI was used to match the RI of the tissue. This group also described some other CUBIC-derived protocols that use other molecules, such as CUBIC-cancer analysis based on the immersion of N-butyldiethanolamine and antipyrine/nicotinamide for whole-body profiling of cancer metastasis (Kubota et al., 2017), CUBIC-X protocol based on imidazole and antipyrine reagents to expand and clear the samples (Murakami et al., 2018), as well as a series of additional CUBIC protocols (I-IV) (Tainaka et al., 2018).
Kuwajima et al. developed a formamide-based clearing protocol termed ClearT (Kuwajima et al., 2013). Formamide is a small, uncharged denaturant with both hydrogen donor and acceptor groups (Figure 6). The hydration energy of urea is expected to be much higher than that of formamide (Jedlovszky and Idrissi, 2008). However, ClearT demonstrated a more efficient clearing performance than ScaleA2, probably owing to the delipidation by highly concentrated formamide, which can also solubilize lipids as an aprotic polar solvent like N, N-dimethylformamide and DMSO (Dapper et al., 1987). Similar to urea, 1,3-BAC in the CUBIC-HL cocktail has two NH2 groups (Table 4), suggesting a high hydration ability. Together with the high lipid solubility of 1,3-BAC described above, CUBIC-HL provides a rapid clearing process. It should be noted that the urea analog thiourea (Figure 6), has a relatively low solubility and was rejected to be used in clearing reagents (Susaki et al., 2014), indicating that not all analogs demonstrate good clearing potential.
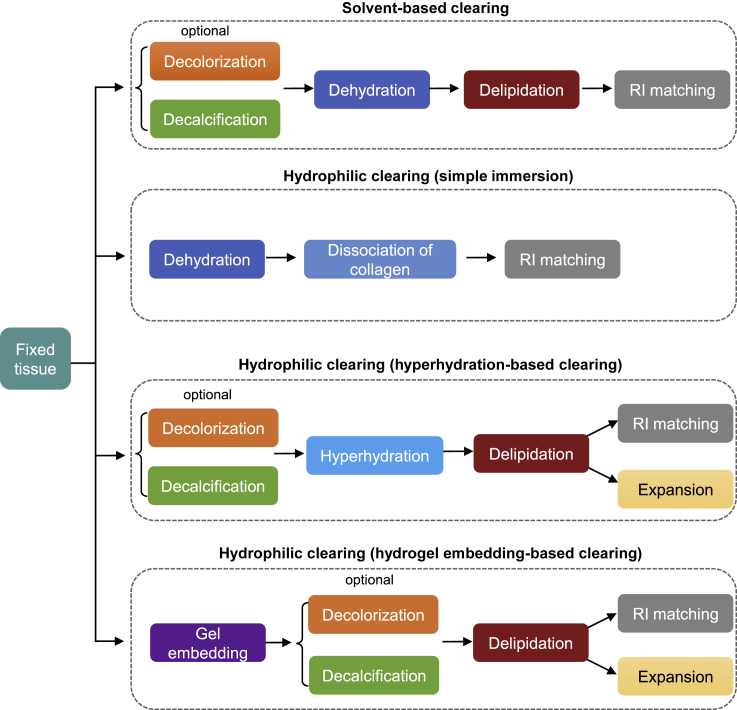
Figure 6.
Different processes involved in tissue clearing methods
Solvent-based clearing mainly includes an adequate dehydration followed by delipidation and RI matching. Hydrophilic clearing involves three types, simple immersion, hyperhydration-based clearing, and hydrogel embedding-based clearing. Simple immersion mainly includes dehydration and dissociation of collagen accompanying with RI matching. Hyperhydration-based clearing relies on the strong hydration capability of urea or other chemicals, followed by delipidation, RI matching, or expansion. Hydrogel embedding-based clearing starts with gel embedding, followed by lipid extraction, RI matching, or tissue expansion. The use of decolorization and decalcification in above processes depends on the tissue types.
Recently, the Zhu group first introduced m-xylylenediamine (MXDA) to tissue clearing and developed the MACS (MXDA-based aqueous clearing system) clearing method with ultrafast clearing speed, robust compatibility with lipophilic dyes, and fine morphology maintenance (Zhu et al., 2020). Similar to urea, MXDA has two NH2 groups (Table 4) and is expected to have great hyperhydration ability (Jedlovszky and Idrissi, 2008). Owing to its hyperhydration ability, as well as the high RI, MXDA contributed to the rapid clearing protocol MACS, showing much faster clearing than Scale and CUBIC series methods.
Some other clearing methods also utilize hyperhydration of urea or urea analogs. For example, Chen et al. proposed a urea-based amino-sugar mixture termed UbasM (Chen et al., 2017), Wang et al. developed SUT consisting of urea, Triton X-100, and SDS (Wang et al., 2018b), Lai et al. combined N-methylglucamine with TDE and iohexol for OPTIClear clearing (Lai et al., 2018), and Li et al. reported Ce3D (Li et al., 2017) utilizing N-methylacetamide.
Decolorization
Except for the scattering, the penetration of light in tissue also suffers from the absorption of some endogenous pigments, such as heme, lipofuscin, melanin, and other body pigments (Weissleder, 2001; Tuchin, 2016; Schnell et al., 1999; Shamim et al., 2014), which limits the imaging depth. Hence, decolorization is an important strategy besides RI matching for clearing large tissues to achieve deep detection. Table 5 summarizes some decolorization reagents for different kinds of pigments.
Table 5.
Decolorization reagents for different pigments
| Pigments | Chemicals/cocktails | Limitations | Clearing methods or cocktails |
|---|---|---|---|
| Heme | Hydrogen peroxide | Quenching of protein fluorescence | iDISCO, iDISCO+ |
| Sodium hydroxide (NaOH) | pH shifting at higher concentration of erythrocytes | – | |
| Ammonium solution | Potential pH shifting like NaOH | PEGASOS | |
| Acid acetone | Significant loss of GFP signal | – | |
| Quadrol | – | CUBIC, PEGASOS, vDISCO, Bone CLARITY | |
| THEED | Significant loss of GFP signal, but the signal is still detectable | Deep-Clear | |
| N-butyldiethanolamine | – | CUBIC-L | |
| N-methyldiethanolamine (in combination with CHAPS) | – | SHANEL | |
| 1,3-BAC | – | CUBIC-HL | |
| 1-methylimidazole | – | CUBIC-P | |
| MXDA | – | MACS | |
| SDS | Request for gel embedding | CLARITY and its derivatives | |
| Melanin | Hydrogen peroxide | Quenching of protein fluorescence | EyeCi, FlyClear, Deep-Clear |
| Lipofuscin | Copper(II) sulfate (CuSO4) | Quenching of protein fluorescence | – |
| Sudan Black | Turning black of tissues | PACT | |
| Ommochromes & pterins | THEED | Significant loss of GFP signal, but the signal is still detectable | FlyClear, Deep-Clear |
| Maillard reaction | Thioglycerol | – | SeeDB, Ce3D, hFRUIT, SWITCH |
| Sodium sulfite | – | SWITCH | |
| Chlorophyll | Chloral hydrate | Quenching of protein fluorescence | – |
| Triton X-100 | Insufficient chlorophyll removal | Scale-like | |
| SDC | – | ClearSee | |
| SDS | Request for gel embedding | PEA-CLARITY | |
| Ethanol (in combination with acetic acid) | Quenching of protein fluorescence | TOMEI-I |
Most of the pigments could be efficiently bleached by several oxidative treatments (Lyon et al., 2012). Hydrogen peroxide (H2O2) is one of the most widely used bleaching chemicals. It can directly degrade the pigment structure and also degrade the protein structure at the same time. However, such harsh bleaching is not conducive to preserving fluorescent proteins, and strategies to relabel these fluorophores are required. Renier et al. used H2O2 to bleach the samples prior to immunostaining to reduce the autofluorescence in the tissues in iDISCO protocols and its variants (Renier et al., 2014, 2016; Liu et al., 2020). Zhao et al. also utilized oxidative bleaching in SHANEL labeling of human tissues (Zhao et al., 2020).
Melanin is a prominent pigment of the vertebrate retinal pigment epithelium and very poorly soluble in both lipid- and water-based solvents. At present, bleaching of melanin can only be achieved by harsh oxidative treatment, e.g., with H2O2 (Henning et al., 2019; Kim and Assawachananont, 2016; Pende et al., 2020).
Removal of heme is important for clearing whole organs, especially heme-rich tissues. Simple buffer perfusion can effectively remove red blood cells in vessels. However, in specific tissues and organs, such as the spleen, there is still substantial residual blood after perfusion. Treatment of samples with strong acids (e.g., acid acetone) or strong alkalis (e.g., NaOH) can dissociate heme, thereby achieving efficient pigment bleaching (Kristinsson and Hultin, 2004). Several recently published clearing protocols can also efficiently decolorize heme. The Ueda group (Tainaka et al., 2014, 2018) discovered that aminoalcohols, such as Quadrol and N-butyldiethanolamine, could achieve unexpected tissue decolorization by efficiently eluting the heme chromophore in blood. Zhao et al. utilized N-methyldiethanolamine in combination with CHAPS for blood decolorization (Zhao et al., 2020). It was speculated that the structures of aminoalcohols resemble those of the globulin chain, resulting in a possible competition between aminoalcohols, oxygen, and histidine for the binding site of heme porphyrin, which may facilitate heme release and contribute to the decolorization effect (Tainaka et al., 2016). In addition, Ueda's group also screened out 1,3-BAC and 1-methylimidazole with high decolorization capability and chose them as the main components in CUBIC-HL and CUBIC-P, respectively (Tainaka et al., 2018). Recently, Zhu et al. (Zhu et al., 2020) found that MXDA showed excellent decolorizing capability, and its decolorization principle was different from that of Quadrol (releasing hemin) used in CUBIC-series methods but similar to that of NaOH (releasing Fe). In addition, SDS used in CLARITY (Chung et al., 2013) and its derivatives (Yang et al., 2014) can also decolorize heme-rich tissues such as the kidney and spleen. It is speculated that the denaturation of the holoenzyme (heme-hemoglobin) by SDS elutes the charged heme molecules (Tainaka et al., 2016), which could be supported by electrophoretic force in an electrophoretic tissue clearing setup. These protocols provide a milder decolorization effect for tissue clearing.
Except for heme, lipofuscin can be found in a wide range of cell types as the breakdown product of old red blood cells (Schnell et al., 1999). It may be quenched with either copper(II) sulfate (CuSO4) or Sudan black B (Schnell et al., 1999; Hu, 2014; Oliveira et al., 2010). B. Hu adopted Sudan black B to stain thick and even whole mouse brain to decrease background fluorescence while preserving most of the fluorescent protein signal (Hu, 2014). Treweek et al. also used Sudan black B for pre-PACT tissue staining to reduce high autofluorescent background (Treweek et al., 2015).
Ommochromes and pterins are other classes of abundant and poorly soluble pigments in invertebrate species. Pende et al. introduced THEED (2,2′,2″,2″′-(Ethylenedinitrilo)-tetraethanol) to replace Quadrol in ScaleCUBIC-1 for FlyClear clearing, resulting in a complete depigmentation of the eyes of D. melanogaster, which contain both ommochromes and pterins, while preserving endogenous GFP and mCherry signal (Pende et al., 2018). Recently, they further modified the FlyClear protocol and chemistry to achieve decolorization of different kinds of pigments, including pterins, ommochromes, heme, carotenoids, and melanin, and proposed DEEP-Clear (Pende et al., 2020). A permeabilization step with acetone at −20°C prior to solution-1 (a solution in FlyClear) treatment led to an increased depigmentation speed in the DEEP-Clear protocol (Pende et al., 2020). It should be noted that this protocol has to be combined with peroxide pretreatment for the removal of melanin in zebrafish and axolotl eyes, resulting in the damage of transgenic fluorophores. Hence, a relabeling strategy of endogenous fluorophores is still required.
The Maillard reaction would induce browning and autofluorescence accumulation. In some clearing protocols, such as SeeDB (Ke et al., 2013) and FRUIT (Hou et al., 2015), fructose is used as the clearing agent, and in SWITCH (Murray et al., 2015), based on glutaraldehyde fixation, discoloration would reduce light transmission. Thioglycerol and sodium sulfite (Li et al., 2017; Ke et al., 2013; Hou et al., 2015; Murray et al., 2015; Hildebrand et al., 2020) have been used to suppress the Maillard reaction to partially improve tissue transparency. Some other compounds such as aminoguanidine, epicatechin, vitamins, and amino acids are also useful Maillard reaction inhibitors (Lund and Ray, 2017), which may be used in clearing protocols in future studies.
In addition to animals, plant tissues also exhibit some pigments, such as chlorophyll, which has strong autofluorescence and is the main component that impedes fluorescence observation. Acidified chloral hydrate is the most commonly used reagent for clearing plant tissues, but it is not compatible with endogenous fluorescence (Kurihara et al., 2015; Warner et al., 2014). Warner et al. developed a Scale-like clearing solution containing urea, glycerol, and Triton X-100 to clear plant tissues with good compatibility with endogenous fluorescent proteins (Warner et al., 2014). Based on this, Kurihara et al. developed ClearSee method, which uses urea, xylitol, and SDC and renders leaves transparent with no green pigmentation, while leaves treated with Scale-like solution retained the green coloration and showed limited transparency (Kurihara et al., 2015). Some organic solvents, such as ethanol, have also been used to extract chlorophyll during clearing of plant tissues with TOMEI (Hasegawa et al., 2016). In addition, Palmer et al. developed PEA-CLARITY for 3D imaging of whole plant organs and successfully removed chlorophyll and other pigments using the ionic detergent SDS, as well as lipids (Palmer et al., 2015).
Figure 6 summarizes different processes involved in tissue clearing methods, showing main mechanisms potentially involved in each kind of method and how they are connected. In a broader sense, mechanical optical clearing by mechanical means (e.g., compression) can also achieve optical clearing, as it increases imaging resolution and contrast comparable with chemical tissue clearing. Izquierdo-Román et al. utilized localized compression to induce the tissue optical clearing effect, which was speculated to be related to reduced tissue thickness and local tissue dehydration due to lateral water expulsion (Izquierdo-Román et al., 2011). Compression was also utilized by Ku et al. in the ELAST protocol to accelerate clearing and molecular labeling (Ku et al., 2020).
Concluding remarks
In summary, the fundamental physical principles of tissue optical clearing are the reductions of light scattering via RI matching and light absorption via the removal of pigments. These physical mechanisms are based on different chemical-biomolecule interactions, such as dissociation of collagen, delipidation, decalcification, dehydration, and hyperhydration for RI matching and decolorization for the removal of absorption components. Current tissue optical clearing techniques are based on a single interaction or a combination of several interactions.
Based on these basic principles, plenty of tissue optical clearing methods have emerged in the past decades. The development of various clearing techniques provides new solutions to 3D mapping of various tissues, facilitating the understanding of many biological structures and events. Exploration and analysis of the physical mechanisms and chemical strategies are the foundation for the further development of clearing methods with applications in various fields.
In the future, clearing of larger samples, such as intact primate organs, including human organs, is expected. With future efforts on the labeling strategy, high-throughput imaging, and corresponding mass data processing, tissue optical clearing may provide more insights into the fundamental questions in biological events and many discoveries and also play an essential role in clinical diagnosis applications.
Limitations of the study
Many excellent works about tissue optical clearing methods and applications have not been discussed in this review due to the limited space. Additional articles may have been published on the topic since submission of the manuscript.
Acknowledgments
This work was supported by the National Key Research and Development Program of China (Grant No. 2017YFA0700501), the National Natural Science Foundation of China (Grant Nos. 61860206009, 91749209, 81961138015, 81870934), and the Innovation Fund of WNLO.
Author contributions
Conceptualization, D.Z.; Writing—Original Draft, T.T.Y., J.T.Z., D.Y.L., and D.Z.; Visualization, T.T.Y.; Supervision, D.Z.
Declaration of interests
The authors declare no competing interests.
References
- Barton K.N., Buhr M.M., Ballantyne J.S. Effects of urea and trimethylamineN-oxide on fluidity of liposomes and membranes of an elasmobranch. Am. J. Physiol. 1999;276:R397–R406. doi: 10.1152/ajpregu.1999.276.2.R397. [DOI] [PubMed] [Google Scholar]
- Becker K., Jahrling N., Kramer E.R., Schnorrer F., Dodt H.-U. Ultramicroscopy: 3D reconstruction of large microscopical specimens. J. Biophotonics. 2008;1:36–42. doi: 10.1002/jbio.200710011. [DOI] [PubMed] [Google Scholar]
- Becker K., Jahrling N., Saghafi S., Weiler R., Dodt H.-U. Chemical clearing and dehydration of GFP expressing mouse brains. PLoS One. 2012;7:e33916. doi: 10.1371/journal.pone.0033916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begum F., Zhu W., Namaka M.P., Frost E.E. A novel decalcification method for adult rodent bone for histological analysis of peripheral-central nervous system connections. J. Neurosci. Methods. 2010;187:59–66. doi: 10.1016/j.jneumeth.2009.12.013. [DOI] [PubMed] [Google Scholar]
- Cai R., Pan C., Ghasemigharagoz A., Todorov M.I., Forstera B., Zhao S., Bhatia H.S., Parra-Damas A., Mrowka L., Theodorou D. Panoptic imaging of transparent mice reveals whole-body neuronal projections and skull-meninges connections. Nat. Neurosci. 2019;22:317–327. doi: 10.1038/s41593-018-0301-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callis G., Sterchi D. Decalcification of bone literature review and practical study of various decalcifying agents, methods, and their effects on bone histology. J. Histotechnol. 1998;21:49–58. [Google Scholar]
- Chang J.B., Chen F., Yoon Y.G., Jung E.E., Babcock H., Kang J.S., Asano S., Suk H.J., Pak N., Tillberg P.W. Iterative expansion microscopy. Nat. Methods. 2017;14:593–599. doi: 10.1038/nmeth.4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F., Tillberg P.W., Boyden E.S. Optical imaging. Expansion microscopy. Science. 2015;347:543–548. doi: 10.1126/science.1260088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Li G., Li Y., Li Y., Zhu H., Tang L., French P., Mcginty J., Ruan S. UbasM: an effective balanced optical clearing method for intact biomedical imaging. Sci. Rep. 2017;7:12218. doi: 10.1038/s41598-017-12484-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Liu S., Liu H., Tong S., Tang H., Zhang C., Yan S., Li H., Yang G., Zhu D. Coherent Raman scattering unravelling mechanisms underlying skull optical clearing for through-skull brain imaging. Anal. Chem. 2019;91:9371–9375. doi: 10.1021/acs.analchem.9b02624. [DOI] [PubMed] [Google Scholar]
- Chi J., Wu Z., Choi C.H.J., Nguyen L., Tegegne S., Ackerman S.E., Crane A., Marchildon F., Tessier-Lavigne M., Cohen P. Three-dimensional adipose tissue imaging reveals regional variation in beige fat biogenesis and PRDM16-dependent sympathetic neurite density. Cell Metab. 2018;27:226–236.e3. doi: 10.1016/j.cmet.2017.12.011. [DOI] [PubMed] [Google Scholar]
- Choi B., Tsu L., Chen E., Ishak T.S., Iskandar S.M., Chess S., Nelson J.S. Determination of chemical agent optical clearing potential using in vitro human skin. Lasers Surg. Med. 2005;36:72–75. doi: 10.1002/lsm.20116. [DOI] [PubMed] [Google Scholar]
- Chung K., Wallace J., Kim S.Y., Kalyanasundaram S., Andalman A.S., Davidson T.J., Mirzabekov J.J., Zalocusky K.A., Mattis J., Denisin A.K. Structural and molecular interrogation of intact biological systems. Nature. 2013;497:332–337. doi: 10.1038/nature12107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantini I., Ghobril J.P., Di Giovanna A.P., Allegra Mascaro A.L., Silvestri L., Mullenbroich M.C., Onofri L., Conti V., Vanzi F., Sacconi L. A versatile clearing agent for multi-modal brain imaging. Sci. Rep. 2015;5:9808. doi: 10.1038/srep09808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dapper C.H., Valivullah H.M., Keenan T.W. Use of polar aprotic solvents to release membranes from milk lipid globules. J. Dairy Sci. 1987;70:760–765. doi: 10.3168/jds.S0022-0302(87)80071-1. [DOI] [PubMed] [Google Scholar]
- Di Giovanna A.P., Tibo A., Silvestri L., Mullenbroich M.C., Costantini I., Allegra Mascaro A.L., Sacconi L., Frasconi P., Pavone F.S. Whole-brain vasculature reconstruction at the single capillary level. Sci. Rep. 2018;8:12573. doi: 10.1038/s41598-018-30533-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodt H.-U., Leischner U., Schierloh A., Jahrling N., Mauch C.P., Deininger K., Deussing J.M., Eder M., Zieglgansberger W., Becker K. Ultramicroscopy: three-dimensional visualization of neuronal networks in the whole mouse brain. Nat. Methods. 2007;4:331–336. doi: 10.1038/nmeth1036. [DOI] [PubMed] [Google Scholar]
- Economo M.N., Clack N.G., Lavis L.D., Gerfen C.R., Svoboda K., Myers E.W., Chandrashekar J. A platform for brain-wide imaging and reconstruction of individual neurons. Elife. 2016;5:e10566. doi: 10.7554/eLife.10566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erturk A., Becker K., Jahrling N., Mauch C.P., Hojer C.D., Egen J.G., Hellal F., Bradke F., Sheng M., Dodt H.-U. Three-dimensional imaging of solvent-cleared organs using 3DISCO. Nat. Protoc. 2012;7:1983–1995. doi: 10.1038/nprot.2012.119. [DOI] [PubMed] [Google Scholar]
- Erturk A., Mauch C.P., Hellal F., Forstner F., Keck T., Becker K., Jahrling N., Steffens H., Richter M., Hubener M. Three-dimensional imaging of the unsectioned adult spinal cord to assess axon regeneration and glial responses after injury. Nat. Med. 2012;18:166–171. doi: 10.1038/nm.2600. [DOI] [PubMed] [Google Scholar]
- Feng W., Shi R., Ma N., Tuchina D.K., Tuchin V.V., Zhu D. Skin optical clearing potential of disaccharides. J. Biomed. Opt. 2016;21:081207. doi: 10.1117/1.JBO.21.8.081207. [DOI] [PubMed] [Google Scholar]
- Feng Y., Yu Z.-W., Quinn P.J. Effect of urea, dimethylurea, and tetramethylurea on the phase behavior of dioleoylphosphatidylethanolamine. Chem. Phys. Lipids. 2002;114:149–157. doi: 10.1016/s0009-3084(01)00198-0. [DOI] [PubMed] [Google Scholar]
- Gomes S.A., Dos Reis L.M., De Oliveira I.B., Noronha I.L., Jorgetti V., Heilberg I.P. Usefulness of a quick decalcification of bone sections embedded in methyl methacrylate[corrected]: an improved method for immunohistochemistry. J. Bone Miner. Metab. 2008;26:110–113. doi: 10.1007/s00774-007-0788-2. [DOI] [PubMed] [Google Scholar]
- Graaff R., Aarnoudse J.G., Zijp J.R. Reduced light scattering properties for mixtures of spherical particles: a simple approximation derived from Mie calculations. Appl. Opt. 1992;31:1370–1376. doi: 10.1364/AO.31.001370. [DOI] [PubMed] [Google Scholar]
- Gratton E. Applied physics. Deeper tissue imaging with total detection. Science. 2011;331:1016–1017. doi: 10.1126/science.1201542. [DOI] [PubMed] [Google Scholar]
- Greenbaum A., Chan K.Y., Dobreva T., Brown D., Balani D.H., Boyce R., Kronenberg H.M., Mcbride H.J., Gradinaru V. Bone CLARITY: clearing, imaging, and computational analysis of osteoprogenitors within intact bone marrow. Sci. Transl. Med. 2017;9:eaah6518. doi: 10.1126/scitranslmed.aah6518. [DOI] [PubMed] [Google Scholar]
- Hama H., Hioki H., Namiki K., Hoshida T., Kurokawa H., Ishidate F., Kaneko T., Akagi T., Saito T., Saido T. ScaleS: an optical clearing palette for biological imaging. Nat. Neurosci. 2015;18:1518–1529. doi: 10.1038/nn.4107. [DOI] [PubMed] [Google Scholar]
- Hama H., Kurokawa H., Kawano H., Ando R., Shimogori T., Noda H., Fukami K., Sakaue-Sawano A., Miyawaki A. Scale: a chemical approach for fluorescence imaging and reconstruction of transparent mouse brain. Nat. Neurosci. 2011;14:1481–1488. doi: 10.1038/nn.2928. [DOI] [PubMed] [Google Scholar]
- Hammouda B. Temperature effect on the nanostructure of SDS micelles in water. J. Res. Natl. Inst. Stand. Technol. 2013;118:151–167. doi: 10.6028/jres.118.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa J., Sakamoto Y., Nakagami S., Aida M., Sawa S., Matsunaga S. Three-dimensional imaging of plant organs using a simple and rapid transparency technique. Plant Cell Physiol. 2016;57:462–472. doi: 10.1093/pcp/pcw027. [DOI] [PubMed] [Google Scholar]
- Henning Y., Osadnik C., Malkemper E.P. EyeCi: optical clearing and imaging of immunolabeled mouse eyes using light-sheet fluorescence microscopy. Exp. Eye Res. 2019;180:137–145. doi: 10.1016/j.exer.2018.12.001. [DOI] [PubMed] [Google Scholar]
- Hildebrand S., Schueth A., Wangenheim K.V., Mattheyer C., Pampaloni F., Bratzke H., Roebroeck A.F., Galuske R.A.W. hFRUIT: an optimized agent for optical clearing of DiI-stained adult human brain tissue. Sci. Rep. 2020;10:9950. doi: 10.1038/s41598-020-66999-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirshburg J., Choi B., Nelson J.S., Yeh A.T. Collagen solubility correlates with skin optical clearing. J. Biomed. Opt. 2006;11:040501. doi: 10.1117/1.2220527. [DOI] [PubMed] [Google Scholar]
- Hirshburg J., Choi B., Nelson J.S., Yeh A.T. Correlation between collagen solubility and skin optical clearing using sugars. Lasers Surg. Med. 2007;39:140–144. doi: 10.1002/lsm.20417. [DOI] [PubMed] [Google Scholar]
- Hirshburg J.M. Texas A&M Univ.; 2009. Chemical Agent Induced Reduction of Skin Light Scattering Ph.D. diss. [Google Scholar]
- Hirshburg J.M., Ravikumar K.M., Hwang W., Yeh A.T. Molecular basis for optical clearing of collagenous tissues. J. Biomed. Opt. 2010;15:055002. doi: 10.1117/1.3484748. [DOI] [PubMed] [Google Scholar]
- Hou B., Zhang D., Zhao S., Wei M., Yang Z., Wang S., Wang J., Zhang X., Liu B., Fan L. Scalable and DiI-compatible optical clearance of the mammalian brain. Front. Neuroanat. 2015;9:19. doi: 10.3389/fnana.2015.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B. Huazhong Univ. Sci. & Technol.; 2014. Study on Reducing Background Fluorescence in Mouse Brain. Ph.D. diss. [Google Scholar]
- Hua L., Zhou R., Thirumalaic D., Berne B.J. Urea denaturation by stronger dispersion interactions with proteins than water implies a 2-stage unfolding. Proc. Natl. Acad. Sci. U S A. 2002;105:16928–16933. doi: 10.1073/pnas.0808427105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izquierdo-Román A., Vogt W.C., Hyacinth L., Rylander C.G. Mechanical tissue optical clearing technique increases imaging resolution and contrast through ex vivo porcine skin. Laser Surg. Med. 2011;43:814–823. doi: 10.1002/lsm.21105. [DOI] [PubMed] [Google Scholar]
- Jahrling N., Becker K., Dodt H.-U. 3D-reconstruction of blood vessels by ultramicroscopy. Organogenesis. 2009;5:227–230. doi: 10.4161/org.5.4.10403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jährling N., Becker K., Kramer E.R., Dodt H.-U. 3D-Visualization of nerve fiber bundles by ultramicroscopy. Med. Laser Appl. 2008;23:209–215. [Google Scholar]
- Jedlovszky P., Idrissi A. Hydration free energy difference of acetone, acetamide, and urea. J. Chem. Phys. 2008;129:164501. doi: 10.1063/1.2992584. [DOI] [PubMed] [Google Scholar]
- Jing D., Zhang S., Luo W., Gao X., Men Y., Ma C., Liu X., Yi Y., Bugde A., Zhou B.O. Tissue clearing of both hard and soft tissue organs with the PEGASOS method. Cell Res. 2018;28:803–818. doi: 10.1038/s41422-018-0049-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke M.T., Fujimoto S., Imai T. SeeDB: a simple and morphology-preserving optical clearing agent for neuronal circuit reconstruction. Nat. Neurosci. 2013;16:1154–1161. doi: 10.1038/nn.3447. [DOI] [PubMed] [Google Scholar]
- Ke M.T., Nakai Y., Fujimoto S., Takayama R., Yoshida S., Kitajima T.S., Sato M., Imai T. Super-resolution mapping of neuronal circuitry with an index-optimized clearing agent. Cell Rep. 2016;14:2718–2732. doi: 10.1016/j.celrep.2016.02.057. [DOI] [PubMed] [Google Scholar]
- Kellner M., Heidrich M., Lorbeer R.A., Antonopoulos G.C., Knudsen L., Wrede C., Izykowski N., Grothausmann R., Jonigk D., Ochs M. A combined method for correlative 3D imaging of biological samples from macro to nano scale. Sci. Rep. 2016;6:35606. doi: 10.1038/srep35606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.Y., Assawachananont J. A new method to visualize the intact subretina from retinal pigment epithelium to retinal tissue in whole mount of pigmented mouse eyes. Transl. Vis. Sci. Technol. 2016;5:6. doi: 10.1167/tvst.5.1.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.Y., Cho J.H., Murray E., Bakh N., Choi H., Ohn K., Ruelas L., Hubbert A., Mccue M., Vassallo S.L. Stochastic electrotransport selectively enhances the transport of highly electromobile molecules. Proc. Natl. Acad. Sci. U S A. 2015;112:E6274–E6283. doi: 10.1073/pnas.1510133112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingberg A., Hasenberg A., Ludwig-Portugall I., Medyukhina A., Mann L., Brenzel A., Engel D.R., Figge M.T., Kurts C., Gunzer M. Fully automated evaluation of total glomerular number and capillary tuft size in nephritic kidneys using lightsheet microscopy. J. Am. Soc. Nephrol. 2017;28:452–459. doi: 10.1681/ASN.2016020232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konno A., Okazaki S. Aqueous-based tissue clearing in crustaceans. Zool. Lett. 2018;4:13. doi: 10.1186/s40851-018-0099-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristinsson H.G., Hultin H.O. Changes in trout hemoglobin conformations and solubility after exposure to acid and alkali pH. J. Agric. Food Chem. 2004;52:3633–3643. doi: 10.1021/jf034563g. [DOI] [PubMed] [Google Scholar]
- Ku T., Guan W., Evans N.B., Sohn C.H., Albanese A., Kim J.G., Frosch M.P., Chung K. Elasticizing tissues for reversible shape transformation and accelerated molecular labeling. Nat. Methods. 2020;17:609–613. doi: 10.1038/s41592-020-0823-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku T., Swaney J., Park J.Y., Albanese A., Murray E., Cho J.H., Park Y.G., Mangena V., Chen J., Chung K. Multiplexed and scalable super-resolution imaging of three-dimensional protein localization in size-adjustable tissues. Nat. Biotechnol. 2016;34:973–981. doi: 10.1038/nbt.3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota S.I., Takahashi K., Nishida J., Morishita Y., Ehata S., Tainaka K., Miyazono K., Ueda H.R. Whole-body profiling of cancer metastasis with single-cell resolution. Cell Rep. 2017;20:236–250. doi: 10.1016/j.celrep.2017.06.010. [DOI] [PubMed] [Google Scholar]
- Kurihara D., Mizuta Y., Sato Y., Higashiyama T. ClearSee: a rapid optical clearing reagent for whole-plant fluorescence imaging. Development. 2015;142:4168–4179. doi: 10.1242/dev.127613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwajima T., Sitko A.A., Bhansali P., Jurgens C., Guido W., Mason C. ClearT: a detergent- and solvent-free clearing method for neuronal and non-neuronal tissue. Development. 2013;140:1364–1368. doi: 10.1242/dev.091844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsova N., Chi S.L., Leikin S. Sugars and polyols inhibit fibrillogenesis of type I collagen by disrupting hydrogen-bonded water bridges between the helices. Biochemistry. 1998;37:11888–11895. doi: 10.1021/bi980089+. [DOI] [PubMed] [Google Scholar]
- Lacomb R., Nadiarnykh O., Carey S., Campagnola P.J. Quantitative second harmonic generation imaging and modeling of the optical clearing mechanism in striated muscle and tendon. J. Biomed. Opt. 2008;13:021109. doi: 10.1117/1.2907207. [DOI] [PubMed] [Google Scholar]
- Lai H.M., Liu A.K.L., Ng H.H.M., Goldfinger M.H., Chau T.W., Defelice J., Tilley B.S., Wong W.M., Wu W., Gentleman S.M. Next generation histology methods for three-dimensional imaging of fresh and archival human brain tissues. Nat. Commun. 2018;9:1066. doi: 10.1038/s41467-018-03359-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E., Choi J., Jo Y., Kim J.Y., Jang Y.J., Lee H.M., Kim S.Y., Lee H.J., Cho K., Jung N. ACT-PRESTO: rapid and consistent tissue clearing and labeling method for 3-dimensional (3D) imaging. Sci. Rep. 2016;6:18631. doi: 10.1038/srep18631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Germain R.N., Gerner M.Y. Multiplex, quantitative cellular analysis in large tissue volumes with clearing-enhanced 3D microscopy (Ce3D) Proc. Natl. Acad. Sci. U S A. 2017;114:E7321–E7330. doi: 10.1073/pnas.1708981114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H.H., Lai J.S., Chin A.L., Chen Y.C., Chiang A.S. A map of olfactory representation in the Drosophila mushroom body. Cell. 2007;128:1205–1217. doi: 10.1016/j.cell.2007.03.006. [DOI] [PubMed] [Google Scholar]
- Liu T., Yang L., Han X., Ding X., Li J., Yang J. Local sympathetic innervations modulate the lung innate immune responses. Sci. Adv. 2020;6:eaay1497. doi: 10.1126/sciadv.aay1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund M.N., Ray C.A. Control of maillard reactions in foods: strategies and chemical mechanisms. J. Agric. Food Chem. 2017;65:4537–4552. doi: 10.1021/acs.jafc.7b00882. [DOI] [PubMed] [Google Scholar]
- Lyon H., Andersen A., Hasselager E., Hoyer P.-E., Moller M., Prento P., Deurs B.V. Springer Science & Business Media; 2012. Theory and Strategy in Histochemistry: A Guide to the Selection and Understanding of Techniques. [Google Scholar]
- Mao Z., Zhu D., Hu Y., Wen X., Han Z. Influence of alcohols on the optical clearing effect of skin in vitro. J. Biomed. Opt. 2008;13:021104. doi: 10.1117/1.2892684. [DOI] [PubMed] [Google Scholar]
- Masselink W., Reumann D., Murawala P., Pasierbek P., Taniguchi Y., Bonnay F., Meixner K., Knoblich J.A., Tanaka E.M. Broad applicability of a streamlined ethyl cinnamate-based clearing procedure. Development. 2019;146:dev166884. doi: 10.1242/dev.166884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyawaki T., Morikawa S., Susaki E.A., Nakashima A., Takeuchi H., Yamaguchi S., Ueda H.R., Ikegaya Y. Visualization and molecular characterization of whole-brain vascular networks with capillary resolution. Nat. Commun. 2020;11:1104. doi: 10.1038/s41467-020-14786-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani H., Ono S., Ushiku T., Kudo Y., Ikemura M., Kageyama N., Yamamichi N., Fujishiro M., Someya T., Fukayama M. Transparency-enhancing technology allows three-dimensional assessment of gastrointestinal mucosa: a porcine model. Pathol. Int. 2018;68:102–108. doi: 10.1111/pin.12627. [DOI] [PubMed] [Google Scholar]
- Muller P.A., Schneeberger M., Matheis F., Wang P., Kerner Z., Ilanges A., Pellegrino K., Del Marmol J., Castro T.B.R., Furuichi M. Microbiota modulate sympathetic neurons via a gut-brain circuit. Nature. 2020;583:441–446. doi: 10.1038/s41586-020-2474-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami T.C., Mano T., Saikawa S., Horiguchi S.A., Shigeta D., Baba K., Sekiya H., Shimizu Y., Tanaka K.F., Kiyonari H. A three-dimensional single-cell-resolution whole-brain atlas using CUBIC-X expansion microscopy and tissue clearing. Nat. Neurosci. 2018;21:625–637. doi: 10.1038/s41593-018-0109-1. [DOI] [PubMed] [Google Scholar]
- Murray E., Cho J.H., Goodwin D., Ku T., Swaney J., Kim S.Y., Choi H., Park Y.G., Park J.Y., Hubbert A. Simple, scalable proteomic imaging for high-dimensional profiling of intact systems. Cell. 2015;163:1500–1514. doi: 10.1016/j.cell.2015.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaki K., Yamaji N., Murata M. ePro-ClearSee: a simple immunohistochemical method that does not require sectioning of plant samples. Sci. Rep. 2017;7:42203. doi: 10.1038/srep42203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmi M., Ohnishi Y., Yoden K., Haruna M. In vitro simultaneous measurement of refractive index and thickness of biological tissue by the low coherence interferometry. IEEE Trans. Biomed. Eng. 2000;47:1266–1270. doi: 10.1109/10.867961. [DOI] [PubMed] [Google Scholar]
- Oliveira V.C., Carrara R.C., Simoes D.L., Saggioro F.P., Carlotti C.G., Jr., Covas D.T., Neder L. Sudan Black B treatment reduces autofluorescence and improves resolution of in situ hybridization specific fluorescent signals of brain sections. Histol. Histopathol. 2010;25:1017–1024. doi: 10.14670/HH-25.1017. [DOI] [PubMed] [Google Scholar]
- Palmer W.M., Martin A.P., Flynn J.R., Reed S.L., White R.G., Furbank R.T., Grof C.P. PEA-CLARITY: 3D molecular imaging of whole plant organs. Sci. Rep. 2015;5:13492. doi: 10.1038/srep13492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan C., Cai R., Quacquarelli F.P., Ghasemigharagoz A., Lourbopoulos A., Matryba P., Plesnila N., Dichgans M., Hellal F., Erturk A. Shrinkage-mediated imaging of entire organs and organisms using uDISCO. Nat. Methods. 2016;13:859–867. doi: 10.1038/nmeth.3964. [DOI] [PubMed] [Google Scholar]
- Park Y.G., Sohn C.H., Chen R., Mccue M., Yun D.H., Drummond G.T., Ku T., Evans N.B., Oak H.C., Trieu W. Protection of tissue physicochemical properties using polyfunctional crosslinkers. Nat. Biotechnol. 2018;37:73–83. doi: 10.1038/nbt.4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pende M., Becker K., Wanis M., Saghafi S., Kaur R., Hahn C., Pende N., Foroughipour M., Hummel T., Dodt H.U. High-resolution ultramicroscopy of the developing and adult nervous system in optically cleared Drosophila melanogaster. Nat. Commun. 2018;9:4731. doi: 10.1038/s41467-018-07192-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pende M., Vadiwala K., Schmidbaur H., Stockinger A.W., Murawala P., Saghafi S., Dekens M.P.S., Becker K., Revilla I.D.R., Papadopoulos S.C. A versatile depigmentation, clearing, and labeling method for exploring nervous system diversity. Sci. Adv. 2020;6:eaba0365. doi: 10.1126/sciadv.aba0365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priyakumar U.D., Hyeon C., Thirumalai D., Mackerell A.D., Jr. Urea destabilizes RNA by forming stacking interactions and multiple hydrogen bonds with nucleic acid bases. J. Am. Chem. Soc. 2009;131:17759–17761. doi: 10.1021/ja905795v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravikumar K.M., Humphrey J.D., Hwang W. Spontaneous unwinding of a labile domain in a collagen triple helix. J. Mech. Mater. Struct. 2007;2:999–1010. [Google Scholar]
- Ravikumar K.M., Hwang W. Region-specific role of water in collagen unwinding and assembly. Proteins: Struct. Funct. Bioinform. 2008;72:1320–1332. doi: 10.1002/prot.22026. [DOI] [PubMed] [Google Scholar]
- Renier N., Adams E.L., Kirst C., Wu Z., Azevedo R., Kohl J., Autry A.E., Kadiri L., Umadevi Venkataraju K., Zhou Y. Mapping of brain activity by automated volume analysis of immediate early genes. Cell. 2016;165:1789–1802. doi: 10.1016/j.cell.2016.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renier N., Wu Z., Simon D.J., Yang J., Ariel P., Tessier-Lavigne M. iDISCO: a simple, rapid method to immunolabel large tissue samples for volume imaging. Cell. 2014;159:896–910. doi: 10.1016/j.cell.2014.10.010. [DOI] [PubMed] [Google Scholar]
- Richardson D.S., Lichtman J.W. Clarifying tissue clearing. Cell. 2015;162:246–257. doi: 10.1016/j.cell.2015.06.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rylander C.G., Stumpp O.F., Milner T.E., Kemp N.J., Mendenhall J.M., Diller K.R., Welch A.J. Dehydration mechanism of optical clearing in tissue. J. Biomed. Opt. 2006;11:041117. doi: 10.1117/1.2343208. [DOI] [PubMed] [Google Scholar]
- Sangeetha R., Uma K., Chandavarkar V. Comparison of routine decalcification methods with microwave decalcification of bone and teeth. J. Oral Maxillofac. Pathol. 2013;17:386–391. doi: 10.4103/0973-029X.125204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjai K., Kumarswamy J., Patil A., Papaiah L., Jayaram S., Krishnan L. Evaluation and comparison of decalcification agents on the human teeth. J. Oral Maxillofac. Pathol. 2012;16:222–227. doi: 10.4103/0973-029X.99070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell S.A., Staines W.A., Wessendorf M.W. Reduction of lipofuscin-like autofluorescence in fluorescently labeled tissue. J. Histochem. Cytochem. 1999;47:719–730. doi: 10.1177/002215549904700601. [DOI] [PubMed] [Google Scholar]
- Schwarz M.K., Scherbarth A., Sprengel R., Engelhardt J., Theer P., Giese G. Fluorescent-protein stabilization and high-resolution imaging of cleared, intact mouse brains. PLoS One. 2015;10:e0124650. doi: 10.1371/journal.pone.0124650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott G.D., Blum E.D., Fryer A.D., Jacoby D.B. Tissue optical clearing, three-dimensional imaging, and computer morphometry in whole mouse lungs and human airways. Am. J. Respir. Cell Mol. Biol. 2014;51:43–55. doi: 10.1165/rcmb.2013-0284OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamim G., Ranjan S.K., Pandey D.M., Ramani R. Biochemistry and biosynthesis of insect pigments. Eur. J. Entomol. 2014;111:149–164. [Google Scholar]
- Silva Santisteban T., Rabajania O., Kalinina I., Robinson S., Meier M. Rapid spheroid clearing on a microfluidic chip. Lab Chip. 2017;18:153–161. doi: 10.1039/c7lc01114h. [DOI] [PubMed] [Google Scholar]
- Spalteholz W. Leipzig, S. Hirzel; 1914. Über das Durchsichtigmachen von menschlichen und tierischen Präparaten und seine theoretischen Bedingungen. [Google Scholar]
- Susaki E.A., Tainaka K., Perrin D., Kishino F., Tawara T., Watanabe T.M., Yokoyama C., Onoe H., Eguchi M., Yamaguchi S. Whole-brain imaging with single-cell resolution using chemical cocktails and computational analysis. Cell. 2014;157:726–739. doi: 10.1016/j.cell.2014.03.042. [DOI] [PubMed] [Google Scholar]
- Susaki E.A., Tainaka K., Perrin D., Yukinaga H., Kuno A., Ueda H.R. Advanced CUBIC protocols for whole-brain and whole-body clearing and imaging. Nat. Protoc. 2015;10:1709–1727. doi: 10.1038/nprot.2015.085. [DOI] [PubMed] [Google Scholar]
- Susaki E.A., Ueda H.R. Whole-body and whole-organ clearing and imaging techniques with single-cell resolution: toward organism-level systems biology in mammals. Cell Chem. Biol. 2016;23:137–157. doi: 10.1016/j.chembiol.2015.11.009. [DOI] [PubMed] [Google Scholar]
- Tainaka K., Kubota S.I., Suyama T.Q., Susaki E.A., Perrin D., Ukai-Tadenuma M., Ukai H., Ueda H.R. Whole-body imaging with single-cell resolution by tissue decolorization. Cell. 2014;159:911–924. doi: 10.1016/j.cell.2014.10.034. [DOI] [PubMed] [Google Scholar]
- Tainaka K., Kuno A., Kubota S.I., Murakami T., Ueda H.R. Chemical principles in tissue clearing and staining protocols for whole-body cell profiling. Annu. Rev. Cell Dev. Biol. 2016;32:713–741. doi: 10.1146/annurev-cellbio-111315-125001. [DOI] [PubMed] [Google Scholar]
- Tainaka K., Murakami T.C., Susaki E.A., Shimizu C., Saito R., Takahashi K., Hayashi-Takagi A., Sekiya H., Arima Y., Nojima S. Chemical landscape for tissue clearing based on hydrophilic reagents. Cell Rep. 2018;24:2196–2210.e9. doi: 10.1016/j.celrep.2018.07.056. [DOI] [PubMed] [Google Scholar]
- Treweek J.B., Chan K.Y., Flytzanis N.C., Yang B., Deverman B.E., Greenbaum A., Lignell A., Xiao C., Cai L., Ladinsky M.S. Whole-body tissue stabilization and selective extractions via tissue-hydrogel hybrids for high-resolution intact circuit mapping and phenotyping. Nat. Protoc. 2015;10:1860–1896. doi: 10.1038/nprot.2015.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai P.S., Kaufhold J.P., Blinder P., Friedman B., Drew P.J., Karten H.J., Lyden P.D., Kleinfeld D. Correlations of neuronal and microvascular densities in murine cortex revealed by direct counting and colocalization of nuclei and vessels. J. Neurosci. 2009;29:14553–14570. doi: 10.1523/JNEUROSCI.3287-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuchin V.V. Tissue optics and photonics: light-tissue interaction II. J. Biomed. Photon. Eng. 2016;2:030201. [Google Scholar]
- Tuchin V.V. SPIE Press; 2002. Handbook of Optical Biomedical Diagnostics. [Google Scholar]
- Tuchin V.V. SPIE Press; 2006. Optical Clearing of Tissues and Blood. [Google Scholar]
- Tuchin V.V., Maksimova I.L., Zimnyakov D.A., Kon I.L., Mavlyutov A.H., Mishin A.A. Light propagation in tissues with controlled optical properties. J. Biomed. Opt. 1997;2:401–417. doi: 10.1117/12.281502. [DOI] [PubMed] [Google Scholar]
- Ueda H.R., Dodt H.U., Osten P., Economo M.N., Chandrashekar J., Keller P.J. Whole-brain profiling of cells and circuits in mammals by tissue clearing and light-sheet microscopy. Neuron. 2020;106:369–387. doi: 10.1016/j.neuron.2020.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezawa M., Haruguchi S., Fukushima R., Sekiyama S., Kamimura M., Soga K. Rapid increase in transparency of biological organs by matching refractive index of medium to cell membrane using phosphoric acid. RSC Adv. 2019;9:15269–15276. doi: 10.1039/c9ra01445d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Royen M.E., Verhoef E.I., Kweldam C.F., Van Cappellen W.A., Kremers G.J., Houtsmuller A.B., Van Leenders G.J. Three-dimensional microscopic analysis of clinical prostate specimens. Histopathology. 2016;69:985–992. doi: 10.1111/his.13022. [DOI] [PubMed] [Google Scholar]
- Wang J., Ma N., Shi R., Zhang Y., Yu T.T., Zhu D. Sugar-induced skin optical clearing: from molecular dynamics simulation to experimental demonstration. IEEE J. Sel. Top. Quant. 2014;20:7101007. [Google Scholar]
- Wang J., Zhang Y., Xu T.H., Luo Q.M., Zhu D. An innovative transparent cranial window based on skull optical clearing. Laser Phys. Lett. 2012;9:469–473. [Google Scholar]
- Wang Q., Liu K., Yang L., Wang H., Yang J. BoneClear: whole-tissue immunolabeling of the intact mouse bones for 3D imaging of neural anatomy and pathology. Cell Res. 2019;29:870–872. doi: 10.1038/s41422-019-0217-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Ouzounov D.G., Wu C., Horton N.G., Zhang B., Wu C.H., Zhang Y., Schnitzer M.J., Xu C. Three-photon imaging of mouse brain structure and function through the intact skull. Nat. Methods. 2018;15:789–792. doi: 10.1038/s41592-018-0115-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Zhang J., Fan G., Zhao H., Wang X., Zhang J., Zhang P., Wang W. Imaging transparent intact cardiac tissue with single-cell resolution. Biomed. Opt. Express. 2018;9:423–436. doi: 10.1364/BOE.9.000423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner C.A., Biedrzycki M.L., Jacobs S.S., Wisser R.J., Caplan J.L., Sherrier D.J. An optical clearing technique for plant tissues allowing deep imaging and compatible with fluorescence microscopy. Plant Physiol. 2014;166:1684–1687. doi: 10.1104/pp.114.244673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissleder R. A clearer vision for in vivo imaging. Nat. Biotechnol. 2001;19:316–317. doi: 10.1038/86684. [DOI] [PubMed] [Google Scholar]
- Wen X., Tuchin V.V., Luo Q., Zhu D. Controling the scattering of intralipid by using optical clearing agents. Phys. Med. Biol. 2009;54:6917–6930. doi: 10.1088/0031-9155/54/22/011. [DOI] [PubMed] [Google Scholar]
- White R.M., Sessa A., Burke C., Bowman T., Leblanc J., Ceol C., Bourque C., Dovey M., Goessling W., Burns C.E. Transparent adult zebrafish as a tool for in vivo transplantation analysis. Cell Stem Cell. 2008;2:183–189. doi: 10.1016/j.stem.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo J., Lee M., Seo J.M., Park H.S., Cho Y.E. Optimization of the optical transparency of rodent tissues by modified PACT-based passive clearing. Exp. Mol. Med. 2016;48:e274. doi: 10.1038/emm.2016.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X.Q., Wang R.K.K. The role of water desorption on optical clearing of biotissue: studied with near infrared reflectance spectroscopy. Med. Phys. 2003;30:1246–1253. doi: 10.1118/1.1576228. [DOI] [PubMed] [Google Scholar]
- Yang B., Treweek J.B., Kulkarni R.P., Deverman B.E., Chen C.K., Lubeck E., Shah S., Cai L., Gradinaru V. Single-cell phenotyping within transparent intact tissue through whole-body clearing. Cell. 2014;158:945–958. doi: 10.1016/j.cell.2014.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh A.T., Choi B., Nelson J.S., Tromberg B.J. Reversible dissociation of collagen in tissues. J. Invest. Dermatol. 2003;121:1332–1335. doi: 10.1046/j.1523-1747.2003.12634.x. [DOI] [PubMed] [Google Scholar]
- Yeh A.T., Hirshburg J. Molecular interactions of exogenous chemical agents with collagen - implications for tissue optical clearing. J. Biomed. Opt. 2006;11:014003. doi: 10.1117/1.2166381. [DOI] [PubMed] [Google Scholar]
- Yi Y., Men Y., Jing D., Luo W., Zhang S., Feng J.Q., Liu J., Ge W.P., Wang J., Zhao H. 3-dimensional visualization of implant-tissue interface with the polyethylene glycol associated solvent system tissue clearing method. Cell Prolif. 2019;52:e12578. doi: 10.1111/cpr.12578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X., Yu T., Chen B., Xu J., Chen W., Qi Y., Zhang P., Li Y., Kou Y., Ma Y. Spatial distribution of motor endplates and its adaptive change in skeletal muscle. Theranostics. 2019;9:734–746. doi: 10.7150/thno.28729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu T., Qi Y., Gong H., Luo Q., Zhu D. Optical clearing for multiscale biological tissues. J. Biophotonics. 2018;11:e201700187. doi: 10.1002/jbio.201700187. [DOI] [PubMed] [Google Scholar]
- Yu T., Qi Y., Zhu J., Xu J., Gong H., Luo Q., Zhu D. Elevated-temperature-induced acceleration of PACT clearing process of mouse brain tissue. Sci. Rep. 2017;7:38848. doi: 10.1038/srep38848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu T., Wen X., Tuchin V.V., Luo Q., Zhu D. Quantitative analysis of dehydration in porcine skin for assessing mechanism of optical clearing. J. Biomed. Opt. 2011;16:095002. doi: 10.1117/1.3621515. [DOI] [PubMed] [Google Scholar]
- Yun D.H., Park Y.-G., Cho J.H., Kamentsky L., Evans N.B., Albanese A., Xie K., Swaney J., Sohn C.H., Tian Y. Ultrafast immunostaining of organ-scale tissues for scalable proteomic phenotyping. bioRxiv. 2019 doi: 10.1101/660373. [DOI] [Google Scholar]
- Zhang C., Feng W., Zhao Y., Yu T., Li P., Xu T., Luo Q., Zhu D. A large, switchable optical clearing skull window for cerebrovascular imaging. Theranostics. 2018;8:2696–2708. doi: 10.7150/thno.23686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B.W., Gong W., Wu C.X., Hu L.J., Zhu X.P., Si K. Multidither coherent optical adaptive technique for deep tissue two-photon microscopy. J. Innov. Opt. Health Sci. 2019;12:1942003. [Google Scholar]
- Zhao S., Todorov M.I., Cai R., Maskari R.A., Steinke H., Kemter E., Mai H., Rong Z., Warmer M., Stanic K. Cellular and molecular probing of intact human organs. Cell. 2020;180:796–812 e719. doi: 10.1016/j.cell.2020.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Yu T., Zhang C., Li Z., Luo Q., Xu T., Zhu D. Skull optical clearing window for in vivo imaging of the mouse cortex at synaptic resolution. Light Sci. Appl. 2018;7:17153. doi: 10.1038/lsa.2017.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu D., Larin K.V., Luo Q., Tuchin V.V. Recent progress in tissue optical clearing. Laser Photon. Rev. 2013;7:732–757. doi: 10.1002/lpor.201200056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Yu T., Li Y., Xu J., Qi Y., Yao Y., Ma Y., Wan P., Chen Z., Li X. MACS: rapid aqueous clearing system for 3D mapping of intact organs. Adv. Sci. 2020;7:1903185. doi: 10.1002/advs.201903185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., Huang L., Zheng Y., Song Y., Xu Q., Wang J., Si K., Duan S., Gong W. Ultrafast optical clearing method for three-dimensional imaging with cellular resolution. Proc. Natl. Acad. Sci. U S A. 2019;116:11480–11489. doi: 10.1073/pnas.1819583116. [DOI] [PMC free article] [PubMed] [Google Scholar]