Graphical abstract
Keywords: Head and neck squamous cell carcinoma, Cancer immunotherapy, Prognostic signature, Bioinformatics, Molecular docking
Abstract
Immunotherapy has become a promising therapeutic option for Head and neck squamous cell carcinoma (HNSC). However, only a small percentage of patients could benefit from it, and the overall prognosis was far from satisfactory. In this study, by comprehensively computational analyses of hundreds of HNSC samples, a prognostic signature composed of 13 immune-related genes (IRGs) was constructed. The results of the analyses in multiple datasets indicated that our signature had high predictive accuracy and could serve as an independent prognostic predictor. Based on this signature and multiple clinical variables, we also established a prognostic nomogram to quantitatively predict the survival risk of individual patients. Moreover, this signature could accurately predict survival, reflect the immune microenvironment, and predict immunotherapy efficacy among HNSC patients. Two potential drugs (doxorubicin and daunorubicin) were also identified via Connectivity Map and molecular docking, which could be used for HNSC combination therapy. Taken together, we developed and validated a robust IRG-based prognostic signature to monitor the prognosis of HNSC, which could provide a solid foundation for individualized cancer immunotherapy.
1. Introduction
Head and neck squamous cell carcinoma (HNSC) is one of the most prevalent malignant tumors globally, accounting for over 90% of all types of head and neck cancer. Approximately 600,000 people are diagnosed with new cases of HNSC annually worldwide [1]. Due to the special location and complicated types, HNSC not only damages the appearance, basic physiological functions, sensory functions and language functions, but also significantly influences the life quality of patients. However, the therapeutic approach of HNSC was single for a long period, without satisfactory outcomes. Until recent years, immunotherapy and targeted therapy have been incorporated into HNSC treatment, which undoubtedly provides strong support for cancer management [2], [3].
The immune system is vitally involved in controlling tumor growth and progression, while the tumor immune microenvironment is closely associated with the prognosis of malignant tumors [4], [5]. Some clinical studies have demonstrated that immune checkpoint inhibitors (ICIs), such as Nivolumab and Pembrolizumab, exert a good anti-tumor effect in HNSC [6], [7]. At present, the availability of cancer-related databases and the application of bioinformatics have enabled the development of prognostic biomarkers in multiple types of malignancies. A guideline concerning the administration of immunotherapy drugs in patients with recurrent or metastatic HNSC also has stated that PD-1 expression, tumor mutation burden (TMB), and immune gene markers could be used as biomarkers for HNSC [8]. Although extensive researches have been carried out on developing the signature of HNSC, existing studies have barely explored a robust prognostic signature based on immune genes. We believe that the construction of an immune-related prognostic signature could especially suitable to monitor the prognosis of HNSC patients treated with immunotherapy.
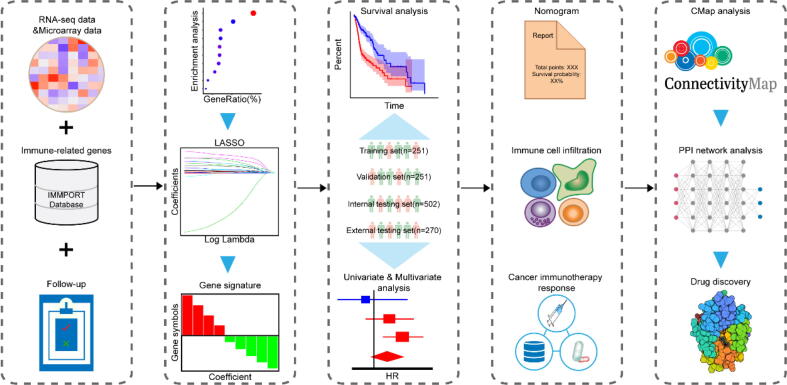
In this study, based on clinical and transcriptomic data of HNSC samples, an immune-related gene-based prognostic signature was constructed and validated in multiple datasets. We also thoroughly investigated the correlation with immune cell infiltration and immunotherapy response through comprehensive analyses. Moreover, we combined Connectivity Map analysis and molecular docking to identify potential small molecule drugs. The experimental technical roadmap was summarized in Fig. 1. Hopefully, this robust prognostic signature could provide a foundation for HNSC treatment.
Fig. 1.
Experimental technical roadmap.
2. Methods
2.1. Acquisition of patient specimens and immune-related genes
Clinical and transcriptomic data for HNSC samples were downloaded from The Cancer Genome Atlas (TCGA) data portal (https://portal.gdc.cancer.gov/) (TCGA cohort, n = 502) and Gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/gds/) (GSE65858 cohort, n = 270) [9], [10]. The TCGA cohort was randomly divided into two parts of equal size, including a training set and a validation set. Moreover, Entire TCGA cohort was used as the internal testing set, while GSE65858 cohort served as the external testing set. Patient demographics and clinical characteristics of the included datasets were summarized in Table S1. A list of IRGs was additionally collected from the Immunology Database and Analysis Portal (ImmPort) database (https://www.immport.org/home) [11], which is a powerful platform sharing the immunology data for cancer researches.
2.2. Identification of DEIRGs and functional enrichment analysis
We compared 502 HNSC samples and 44 normal samples to identify differentially expressed genes (DEGs) using R package “limma”, with the thresholds of |log2(Fold Change)| >1 and P-value < 0.05 [12]. Afterwards, differentially expressed IRGs (DEIRGs) were extracted from these DEGs, followed by plotting of volcano plot of DEIRGs using R package “ggplot2” [13], and Venn diagram by Venn diagram web tool (http://bioinformatics.psb.ugent.be/webtools/Venn/). Functional enrichment analyses were performed using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) 6.8 [14] based on Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) database [15], [16]. To be specific, GO terms, including molecular function (MF), cellular component (CC) as well as biological process (BP), and KEGG signaling pathways were enriched based on the standard of P-value < 0.05. Ultimately, the top 10 most significant terms or pathways were revealed and visualized by R package “ggplot2” [13].
2.3. Construction of IRGPI
Among these DEIRGs, the prognosis-related IRGs were identified and subjected to subsequent analysis. Firstly, the univariate Cox proportional hazard regression model was used to analyze the correlation between DEIRGs and overall survival (OS) in the training set, followed by the Least Absolute Shrinkage and Selection Operator (LASSO) penalized Cox proportional hazards regression for selecting the optimal subset of prognosis-related IRGs [17]. The risk score was then calculated using expression data multiplied by the regression coefficient from the LASSO regression model. The formula was as follows: risk score = [Expression level of Gene 1 * coefficient] + [Expression level of Gene 2 * coefficient] + … + [Expression level of Gene n * coefficient]. Patients were additionally separated into high- and low- risk groups according to the median value of risk score.
2.4. Validation and evaluation of IRGPI
In order to evaluate the reliability of the IRGPI in predicting prognosis, we plotted the Kaplan-Meier (K-M) survival curves in the training set, validating set and testing set by R package “survival” [18]. To further reflect the sensitivity and specificity of IRGPI, we performed a time-dependent receiver operating characteristic (ROC) curve analysis via R package “survivalROC” [19]. In addition, univariate and multivariate analyses of OS for IRGPI and clinicopathological factors were carried out in entire TCGA cohort and GSE65858 cohort using R package “survival” [18]. Moreover, the association of IRGPI with different clinicopathological factors was analyzed using independent t-tests.
2.5. Construction of prognostic nomogram
The C-index was calculated via R package “pROC” to evaluate the predictive accuracy between multiple clinical models and a model with clinical variables and IRGPI. Based on the above results, we constructed a prognostic nomogram to quantitatively estimate the survival risk of HNSC patients. Moreover, calibration curves were used to compare the predictive and observed survival possibility. The diagram of both nomogram and calibration curves were plotted via R package “rms” [20].
2.6. Assessment of immune cell infiltration in tumors
Based on the principle of linear support vector regression, CIBERSORT was utilized to estimate the relative percentage of immune cell infiltration in the RNA-seq data [21]. The correlation between these immune cells and risk score was analyzed, followed by survival analysis.
2.7. Tumor mutation burden (TMB) analysis
The mutation data containing somatic variants of HNSC patients was collected for mutation profile analysis by R package “maftools” [22], followed by calculation of TMB and correlation analysis between TMB and the risk score.
2.8. Immunophenoscore (IPS) analysis
To better predict the response of immune checkpoint inhibitors (ICIs), the expression of genes comprising major components of tumor immunity was integrated, followed by the acquisition of IPS from The Cancer Immunome Atlas (TCIA) (https://tcia.at/home) [23]. Besides, the expression of several prominent checkpoints was also explored.
2.9. Connectivity Map analysis
Connectivity Map (CMap, https://portals.broadinstitute.org/cmap/) is an online analysis platform based on genome-wide transcriptional expression data for the study on the mechanism of action and repurposing of small molecules [24], [25]. CMap contains more than 7000 expression profiles representing 1309 compounds. Based on its pattern-matching algorithms, a positive score indicates the induction effect of the small molecules on the query signature, and a negative score indicates the inhibition effect. Herein, CMap was used to screen for potential small-molecule drugs against IRGPI with the cutoff of mean connective score < −0.2 and P-Value < 0.05. The chemical structures of these drugs were collected from PubChem (https://pubchem.ncbi.nlm.nih.gov/) [26].
2.10. Construction of the protein–protein interaction (PPI) network and screening of the key target
To screen the IRGPI’s key target, all genes were mapped to the STRING database (https://string-db.org/), a database of known and predicted PPIs [27]. Afterwards, the PPI network with a combined score of >0.4 was constructed. Subsequently, a topological analysis was carried out using the Network Analyzer plug-in contained in Cytoscape, and the main topological parameters of the PPI network were obtained. In this research, three centrality algorithms, including degree centrality, closeness centrality and betweenness centrality, was adopted to identify the key target.
2.11. Molecular docking
Firstly, the crystal structure of the key target of IRGPI was obtained from the RCSB PDB (http://www.rcsb.org/) based on the optimal available resolution [28]. Secondly, the Schrödinger’s protein preparation wizard module was utilized to prepare the protein crystallographic structure [29]. Thirdly, the Schrödinger’s LigPrep module was adopted to obtain the 3D structures and energy minimization of the potential small-molecule drugs from CMap analysis. Finally, based on the protein's specific known active sites, the Schrödinger’s Glide module was adopted for molecular docking [30].
2.12. Statistical analysis
Univariate and multivariate Cox regressions analyses were performed via R package “survival” [18], shown as hazard ratios (HRs) and 95% confidence intervals (CIs). The difference among various clinical factors was analyzed by independent t-tests. A P-value < 0.05 indicated a statistical significance.
3. Results
3.1. Identification of DEIRGs and functional enrichment analysis
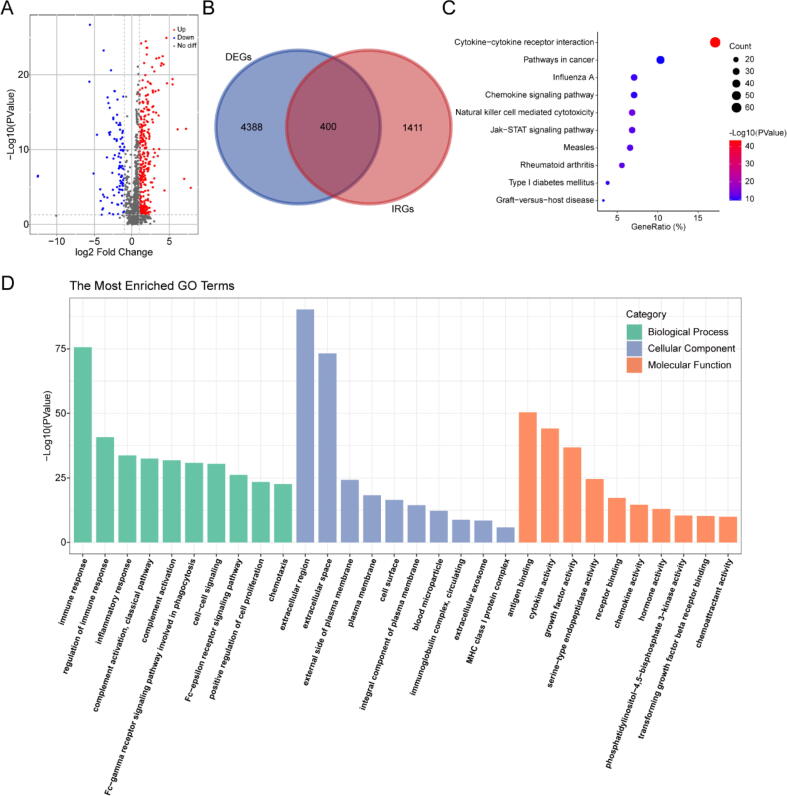
According to the filtering criteria, a total of 4788 DEGs were identified in 502 HNSC samples after comparison with 44 normal samples, including 3605 up-regulated genes and 1183 down-regulated genes. Besides, 1811 unique IRGs were collected from the ImmPort database. Therefore, 400 DEIRGs were extracted and displayed in an overlap region of the Venn diagram, including 305 up-regulated genes and 95 down-regulated genes (Fig. 2A-B). To explore the potential mechanisms, GO/KEGG functional enrichment analysis for these DEIRGs was performed. As shown in Fig. 2C, “cytokine-cytokine receptor interaction” was the most related signaling pathway to DEIRGs. “Immune response”, “extracellular region”, and “antigen binding” were the most frequent terms among BP, CC, and MF, respectively (Fig. 2D). Both biological processes and signaling pathways were closely related to immunity.
Fig. 2.
Differentially expressed immune-related genes (DEIRGs) and functional enrichment analysis. (A) Volcano plot showing DEIRGs between head and neck squamous cell carcinoma and normal tissues. (B) Venn diagram visualizing the intersections between DEGs and IRGs. (C) The top 10 most significantly enriched KEGG signaling pathways. (D) GO enrichment analysis. Green, blue, and orange parts indicated biological process, cellular component, and molecular function, respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.2. Construction of the IRGPI
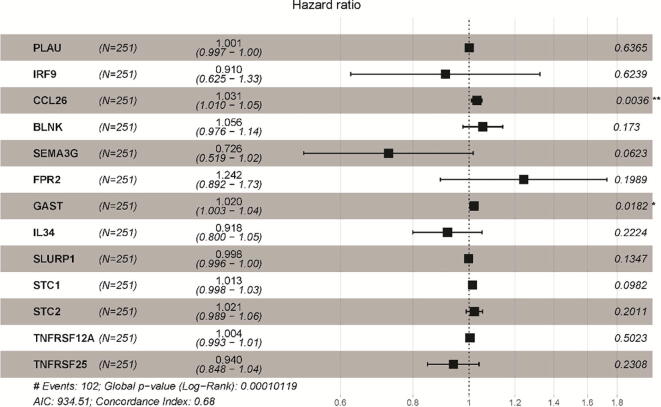
To explore DEIRGs closely associated with HNSC prognosis, a univariate Cox regression analysis was employed. As a result, 49 DEIRGs were ascertained as OS-related DEIRGs and selected for subsequent LASSO regression analysis (Fig. S1), which gave rise to an optimal subset consisting of 13 IRGs: PLAU, IRF9, CCL26, BLNK, SEMA3G, FPR2, GAST, IL34, SLURP1, STC1, STC2, TNFRSF12A and TNFRSF25 (Fig. 3). Further, the association of 13 IRGs with OS or progression free survival (PFS) was analyzed by univariate Cox regression analysis (Fig. S2). The results showed that 13 IRGs were significantly correlated with OS of HNSC patients (P < 0.05), 6 of 13 IRGs (PLAU, CCL26, IL34, STC1, STC2, TNFRSF12A) were significantly related to PFS of HNSC patients (P < 0.05). Besides, the expression levels of these 13 IRGs were significantly increased in a wide variety of tumor tissues in comparison with normal tissues (Fig. S3). Based on multivariate Cox regression analysis, the forest plot of hazard ratios revealed that the majority of these genes were hazardous factors. Ultimately, we established an IPGPI and calculated the risk score based on the relative expression level of these IRGs and their corresponding regression coefficients as follows: risk score = [Expression level of PLAU * 0.000511] + [Expression level of IRF9 * (-0.02022)] + [Expression level of CCL26 * 0.002514] + [Expression level of BLNK * (-0.00517) ] + [Expression level of SEMA3G * (-0.14066)] + [Expression level of FPR2 * 0.105068] + [Expression level of GAST * 0.018375] + [Expression level of IL34 * (-0.0564)] + [Expression level of SLURP1 * (-0.00147)] + [Expression level of STC1 * 0.011072] + [Expression level of STC2 * 0.011025] + [Expression level of TNFRSF12A * 0.003982] + [Expression level of TNFRSF25 * (-0.06533)].
Fig. 3.
The forest plot of multivariate Cox regression analysis of each gene in IRGPI.
3.3. Assessment of the predictive ability of IRGPI
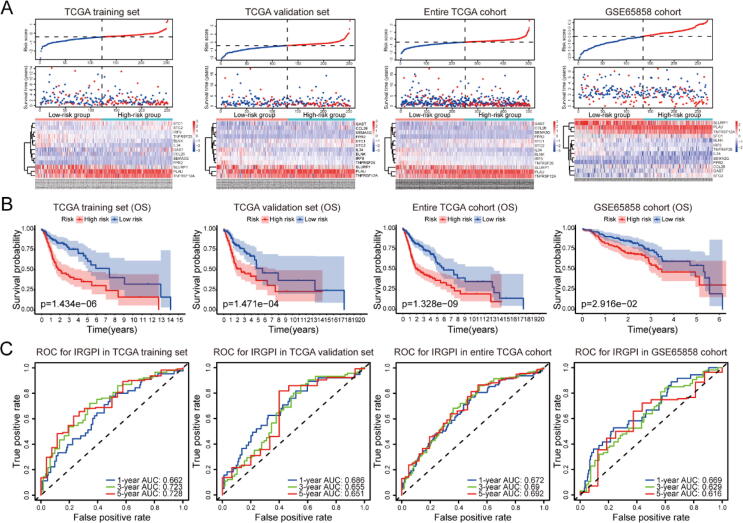
In both training set and testing sets, risk scores of all patients were calculated, followed by classification into high- and low-risk groups according to the median value (Fig. 4A). As expected, low-risk patients exhibited better survival probability than high-risk patients (Fig. 4B, P < 0.05), which was also observed in the analysis of progression free survival (Fig. S4). Besides, time-dependent ROC curves were adopted to explore the 1-year, 3-year, and 5-year mortality for IRGPI in multiple datasets (Fig. 4C). The results showed that most of the area under curves (AUCs) of 1-year, 3-year, and 5-year were greater than 0.65 in these datasets, indicating the good potential of our IRGPI in monitoring mortality.
Fig. 4.
Assessment of the predictive ability of IRGPI in TCGA training set, TCGA validation set, entire TCGA cohort, and GSE65858 cohort. (A) Risk score distribution, survival status, and 13 IRGs expression patterns of patients in both high- and low-risk groups. (B) The Kaplan-Meier analysis of OS for HNSC patients in multiple datasets. (C) Time-dependent ROC curve analysis of IRGPI in multiple datasets.
3.4. Relationship between IRGPI and patient prognosis
For comparison of the predictive power of IRGPI with multiple clinicopathologic factors, univariate and multivariate analysis on OS was conducted in entire TCGA cohort and GSE65858 cohort. Consequently, tumor status, N stage, and IRGPI were significantly associated with OS in univariate and multivariate analysis in entire TCGA cohort (P < 0.05) (Table 1); while only age and IRGPI had a significant correlation with OS in univariate and multivariate analysis in GSE65858 cohort (P < 0.05) (Table 1). Overall, IRGPI was more sensitive than other clinicopathologic factors, which could be used as an independent prognostic predictor.
Table 1.
Univariate and multivariate Cox regression analyses of IRGPI and other clinicopathological factors for OS in entire TCGA cohort and GSE65858 cohort.
| Overall survival | Univariate analysis |
Multivariate analysis |
||||
|---|---|---|---|---|---|---|
| HR | 95%CI | P-value | HR | 95%CI | P-value | |
| Entire TCGA cohort | ||||||
| Age | 1.011 | 0.993–1.028 | 0.228 | 1.011 | 0.992–1.030 | 0.259 |
| Gender (male vs. female) | 0.765 | 0.514–1.137 | 0.185 | 0.705 | 0.463–1.074 | 0.103 |
| Tumor status (with tumor vs. tumor free) | 9.330 | 6.242–13.947 | <0.001 | 6.463 | 4.246–9.839 | <0.001 |
| Tumor grade | 1.184 | 0.884–1.587 | 0.257 | 0.950 | 0.695–1.299 | 0.749 |
| Pathological stage | 1.866 | 1.399–2.489 | <0.001 | 1.266 | 0.801–2.001 | 0.313 |
| T stage | 1.361 | 1.121–1.653 | 0.002 | 0.954 | 0.721–1.262 | 0.740 |
| N stage | 1.748 | 1.424–2.146 | <0.001 | 1.382 | 1.048–1.822 | 0.022 |
| IRGPI (high-risk vs. low-risk) | 3.178 | 2.265–4.458 | <0.001 | 2.068 | 1.416–3.022 | <0.001 |
| GSE65858 cohort | ||||||
| Age | 1.027 | 1.006–1.048 | 0.013 | 1.028 | 1.007–1.051 | 0.011 |
| Gender (male vs. female) | 1.046 | 0.617–1.771 | 0.868 | 1.086 | 0.637–1.852 | 0.762 |
| UICC stage | 1.615 | 1.204–2.168 | 0.001 | 1.177 | 0.735–1.887 | 0.497 |
| T stage | 1.512 | 1.222–1.872 | <0.001 | 1.234 | 0.937–1.625 | 0.135 |
| N stage | 1.415 | 1.129–1.774 | 0.003 | 1.195 | 0.867–1.648 | 0.275 |
| M stage | 3.705 | 1.634–8.400 | 0.002 | 2.254 | 0.907–5.599 | 0.080 |
| IRGPI (high-risk vs. low-risk) | 1.739 | 1.186–2.552 | 0.005 | 1.815 | 1.219–2.702 | 0.003 |
3.5. Relevance between the IRGPI and clinicopathologic factors
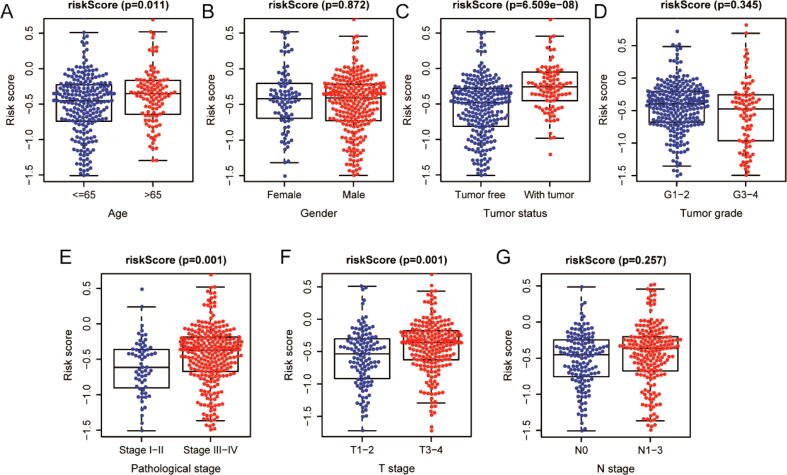
To further explore the correlation between IRGPI and multiple clinicopathologic factors, correlation analysis was carried out via independent t-tests. As shown in Fig. 5, age, tumor status, pathological stage, and T stage were significantly and positively correlated with IRGPI in entire TCGA cohort. Additionally, higher risk scores generally appeared on increased age, tumor-bearing status, advanced pathological stage, and advanced T stage (P < 0.05).
Fig. 5.
The correlations between the IRGPI and multiple clinicopathological factors in entire TCGA cohort. (A) age; (B) gender; (C) tumor status; (D) tumor grade; (E) pathological stage; (F) T stage; (G) N stage.
3.6. Construction and calibration of the nomogram
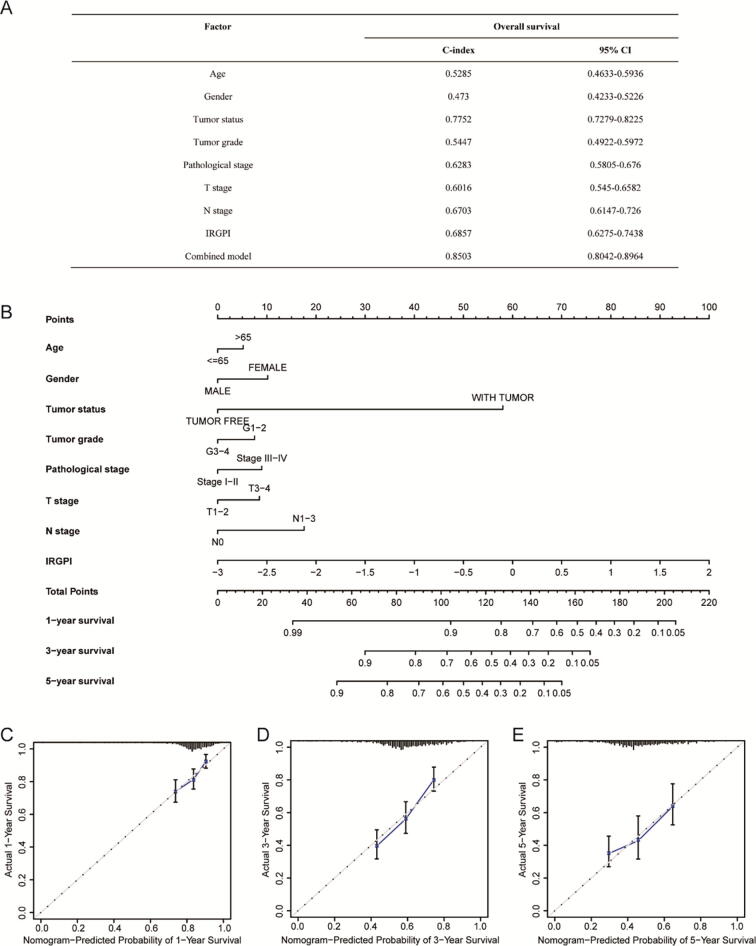
We compared the C-index among multiple clinical models and a combined model that included clinical variables and IRGPI. The combination of the IRGPI with clinical variables had a higher C-index (0.8503; 95% CI: 0.8042–0.8964) than the IRGPI or the clinical variables alone (Fig. 6A). The results indicated that the combined model could improve prognostic accuracy for HNSC patients. To quantitatively predict the survival probability of individual patients, we further established the prognostic nomogram integrating the IRGPI and multiple clinical variables (Fig. 6B). Furthermore, the calibration curves of the prognostic nomogram were applied, showing good consistency between predictive and observed 1-, 3-, and 5-year survival in entire TCGA cohort (Fig. 6C–E).
Fig. 6.
Construction and calibration of nomogram in HNSC patients. (A) Comparison of the predictive power of the prognostic models in entire TCGA cohort. C-index: Harrell’s concordance index; CI: confidence interval. Combined model: IRGPI + age + gender + tumor status + tumor grade + pathological stage + T stage + N stage. (B) Nomogram to predict 1-, 3-, and 5-year OS in entire TCGA cohort. (C-E) Calibration curves of nomogram on the consistency between predictive and observed 1-, 3-, and 5-year outcomes in entire TCGA cohort. Dashed line at 45° indicated perfect prediction. Actual performances of the nomogram were shown in blue lines. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.7. TIME changing associated with the IRGPI
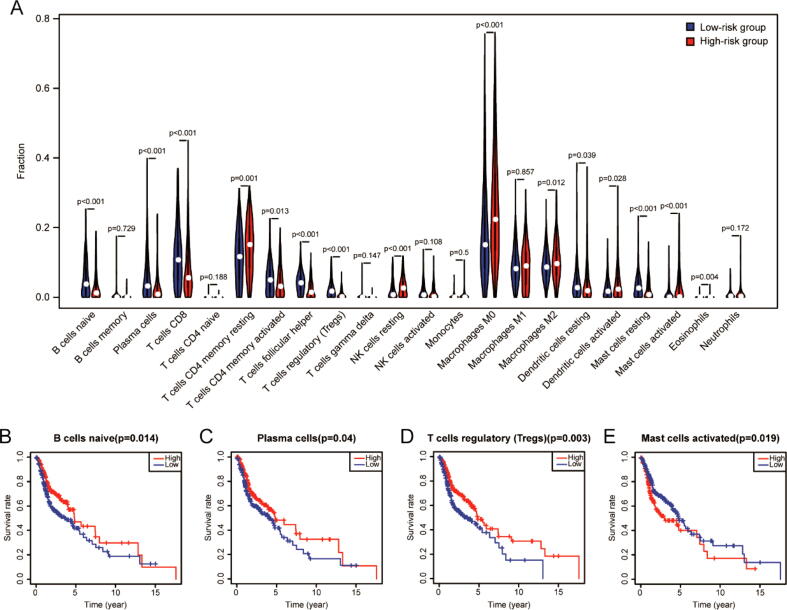
In order to explore the correlation between the specific type of immune cells and IRGPI, the CIBERSORT algorithm was applied to estimate the infiltration of 22 immune cells in each HNSC sample. The relative proportion and correlation matrix of these 22 immune cells were shown in Fig. S5. Among the 22 cell types, over half of the immune cells were significantly correlated with risk score. The relative proportion of some cells had a negative correlation with risk score, such as naive B cells and CD8 T cells, while others had a positive correlation with risk score, such as resting memory CD4 T cells and M2 macrophages (P < 0.05, Fig. 7A). Additionally, survival analysis revealed that the relative proportion of naive B cells (Fig. 7B), plasma cells (Fig. 7C), regulatory T cells (Fig. 7D) and activated mast cells (Fig. 7E) were significantly related to survival rate (P < 0.05). Among them, naive B cells, plasma cells, regulatory T cells with lower levels in high-risk groups were related to poorer OS, and activated mast cells with higher levels in high-risk groups were associated with poorer OS.
Fig. 7.
Immune cell infiltration analysis. (A) The correlation of IRGPI with immune cell infiltration. The blue and red violin represented the low- and high-risk group, respectively, and the white points in the violin indicated median values. The relationships between OS and naive B cells (B), plasma cells (C), regulatory T cells (D), and activated mast cells (E). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.8. Relationship between IRGPI and TMB
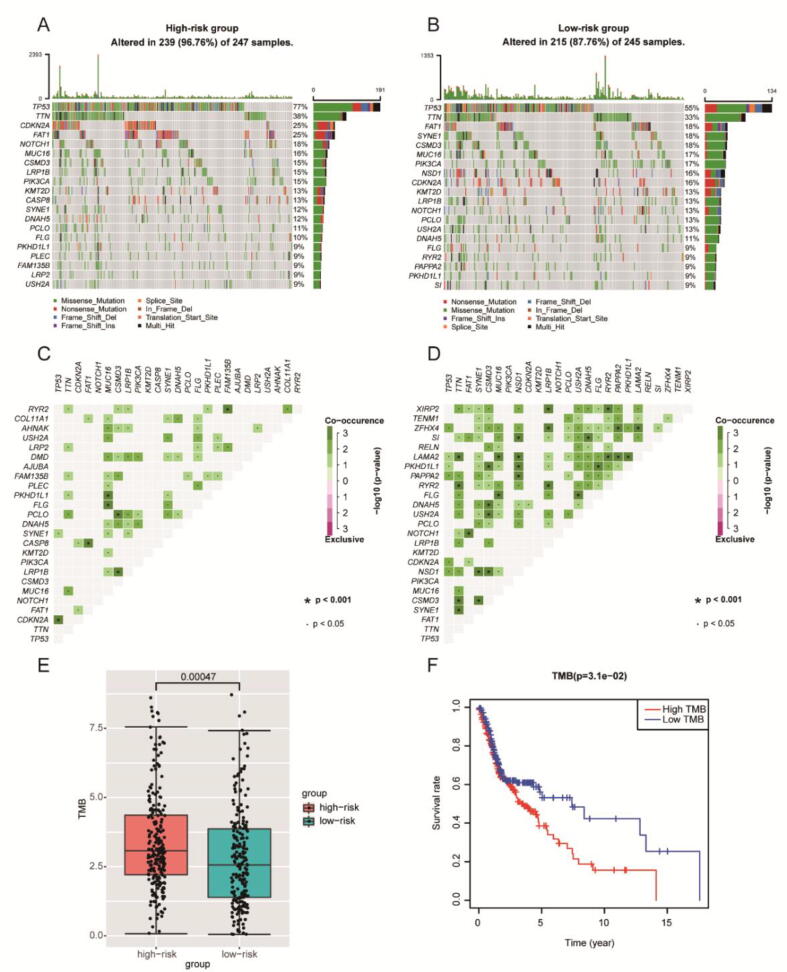
To explore the correlation between IRGPI and TMB, available somatic mutation data were analyzed in entire TCGA cohort. The overall mutation profile with statistical analysis was revealed in Fig. S6. The frequently mutated genes in low- and high-risk groups were summarized in Fig. 8A-B, with TP53 being the most frequently mutant one. And more coincident associations across mutated genes existed in the low-risk group (Fig. 8C-D). Besides, the high-risk group had a higher TMB, which was associated with worse OS (P < 0.05) (Fig. 8E-F).
Fig. 8.
The mutation profile and TMB of low- and high-risk groups. Mutation profile of the high-risk group (A) and low-risk group (B). The coincident and exclusive correlation across mutated genes in the high-risk group (C) and low-risk group (D). (E) The association of IRGPI with TMB. (F) The relationship between TMB and OS.
3.9. Prediction of ICIs response
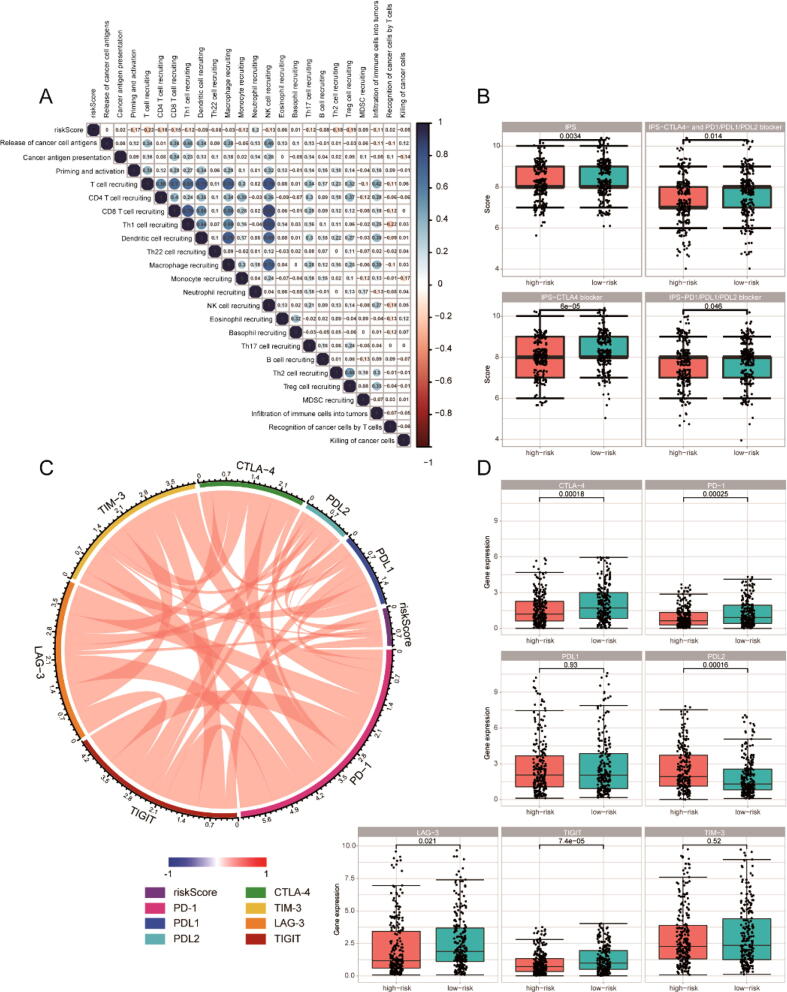
Due to the close association of immunity with tumors, the correlation between IRGPI and anti-cancer immune responses was investigated. As a result, IRGPI was negatively correlated with most of anti-cancer immune responses (Fig. 9A). In consideration of the importance of ICIs in immunotherapy, the response of patients on ICIs should be explored. Notably, IPS was conducive to predict this response, and the correlation between the IRGPI and IPS in HNSC patients was established. The scores of IPS, IPS-CTLA4 blocker, IPS-PD1/PD-L1/PD-L2 blocker, and IPS-CTLA4 + PD1/PD-L1/PD-L2 blocker were used for estimation of the potential of ICIs' application. As shown in Fig. 9B, there was a significant increase of all IPS scores in the low-risk group, suggesting more immunogenicity on ICIs. Besides, the correlation between IRGPI and some critical immune checkpoints was also explored, revealing that IRGPI was related to all these immune checkpoints (Fig. 9C). Moreover, the expression of these critical immune checkpoints was investigated, showing that the expression of CTLA-4, PD-1, LAG-3, and TIGIT were significantly higher in the low-risk group than the high-risk group (Fig. 9D). In order to validate the performance of IRGPI predicting ICIs response, we also analyzed the correlation between IRGPI and the response of cancer patients treated with ICIs in multiple cohorts [31], [32], [33]. The results showed that patients with low risk score were more likely to be responders, which indicated that IRGPI had good potential in predicting ICIs response (Fig. S7).
Fig. 9.
The correlation between the IRGPI and immune checkpoints. (A) Correlation matrix of IRGPI and anti-cancer immune responses. (B) The relationship between IRGPI and IPS. (C) Correlation of IRGPI with several prominent checkpoints. (D) Gene expression of prominent checkpoints.
3.10. Screening of small-molecule drugs
The CMap analysis was conducted to screen for small-molecule drugs against IRGPI. Totally, 27 drugs were identified: Arachidonic acid, Doxorubicin, Xamoterol, Levodopa, Camptothecin, Ranitidine, Piromidic acid, Minaprine, Ethaverine, Daunorubicin, Lomefloxacin, Captopril, Clonidine, Dihydroergocristine, Vorinostat, Ribavirin, Levonorgestrel, Fluorometholone, Apigenin, Doxycycline, Norfloxacin, Tretinoin, Wortmannin, Trichostatin A, Etilefrine, Cyanocobalamin and Pyrithyldione (Table 2).
Table 2.
Potential targeted therapeutic drugs for IRGPI based on CMap analysis.
| CMap name | Mean connective score | n | Enrichment | P-Value | Specificity | Percent non-null |
|---|---|---|---|---|---|---|
| Arachidonic acid | −0.592 | 3 | −0.812 | 0.0133 | 0.0128 | 100 |
| Doxorubicin | −0.57 | 3 | −0.798 | 0.01695 | 0.2181 | 100 |
| Xamoterol | −0.536 | 3 | −0.806 | 0.01462 | 0.0343 | 100 |
| Levodopa | −0.51 | 5 | −0.703 | 0.00505 | 0.0152 | 80 |
| Camptothecin | −0.499 | 3 | −0.773 | 0.024 | 0.2614 | 100 |
| Ranitidine | −0.498 | 5 | −0.661 | 0.01059 | 0 | 80 |
| Piromidic acid | −0.489 | 4 | −0.772 | 0.00547 | 0.0214 | 100 |
| Minaprine | −0.483 | 5 | −0.617 | 0.02233 | 0.0236 | 80 |
| Ethaverine | −0.47 | 4 | −0.721 | 0.01239 | 0.0191 | 75 |
| Daunorubicin | −0.47 | 4 | −0.66 | 0.03032 | 0.1754 | 75 |
| Lomefloxacin | −0.465 | 6 | −0.639 | 0.00638 | 0.0051 | 83 |
| Captopril | −0.463 | 5 | −0.607 | 0.02637 | 0.0423 | 80 |
| Clonidine | −0.462 | 4 | −0.631 | 0.04518 | 0.0148 | 75 |
| Dihydroergocristine | −0.449 | 4 | −0.74 | 0.00897 | 0.0308 | 75 |
| Vorinostat | −0.412 | 12 | −0.552 | 0.00068 | 0.3097 | 66 |
| Ribavirin | −0.407 | 4 | −0.688 | 0.02039 | 0.0921 | 75 |
| Levonorgestrel | −0.402 | 6 | −0.525 | 0.04676 | 0.1913 | 66 |
| Fluorometholone | −0.4 | 4 | −0.651 | 0.03503 | 0.057 | 75 |
| Apigenin | −0.379 | 4 | −0.686 | 0.02103 | 0.1304 | 75 |
| Doxycycline | −0.308 | 5 | −0.587 | 0.03507 | 0.0622 | 80 |
| Norfloxacin | −0.289 | 5 | −0.614 | 0.02357 | 0.0263 | 60 |
| Tretinoin | −0.272 | 22 | −0.342 | 0.00826 | 0.1397 | 50 |
| Wortmannin | −0.269 | 18 | −0.321 | 0.04013 | 0.3758 | 50 |
| Trichostatin A | −0.264 | 182 | −0.441 | 0 | 0.2772 | 51 |
| Etilefrine | −0.254 | 4 | −0.653 | 0.03428 | 0.0426 | 50 |
| Cyanocobalamin | −0.243 | 4 | −0.642 | 0.03945 | 0.0432 | 50 |
| Pyrithyldione | −0.207 | 4 | −0.657 | 0.03191 | 0.0986 | 50 |
3.11. Screening of key target
To screen the key target of IRGPI, we constructed a PPI network that contains 19 interactive proteins. As shown in Fig. 10, PLAU was located at the hub of the network. Furthermore, three algorithms were adopted to calculate the whole network, and all these 19 targets were ranked according to the results of three centrality algorithms (Table S2). Notably, PLAU ranked first in all three centrality algorithms. Therefore, PLAU was considered the key target.
Fig. 10.
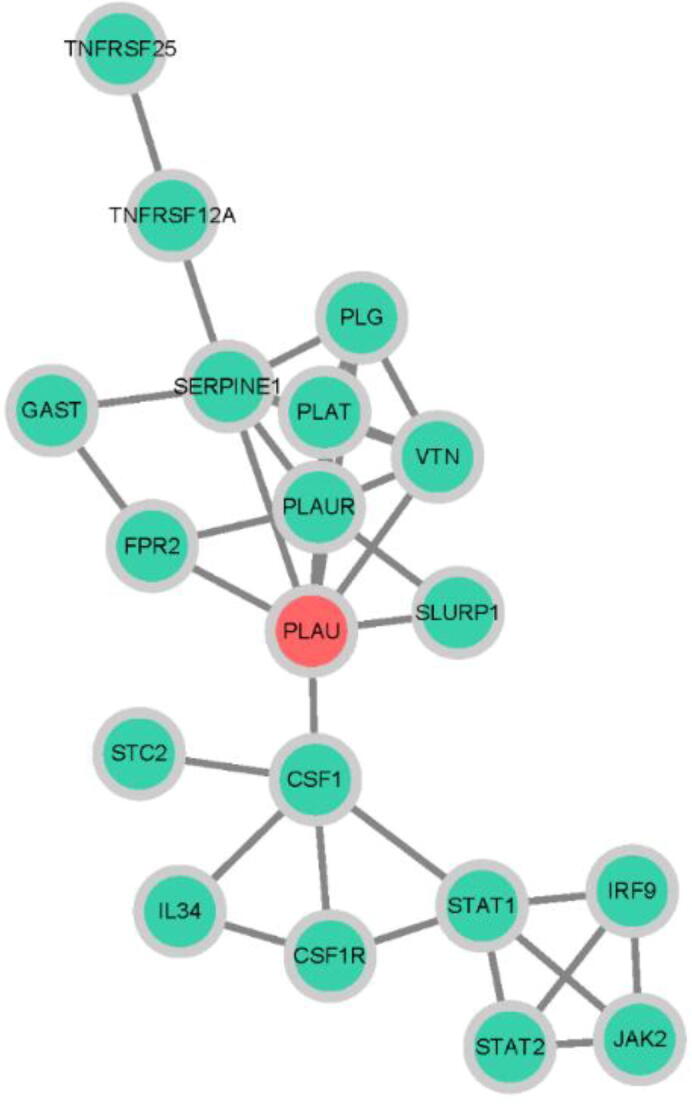
Protein-protein interaction network. The red node represents the key target, which was calculated by three centrality algorithms. Green nodes represent the other 18 targets that make up the network. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.12. Molecular docking
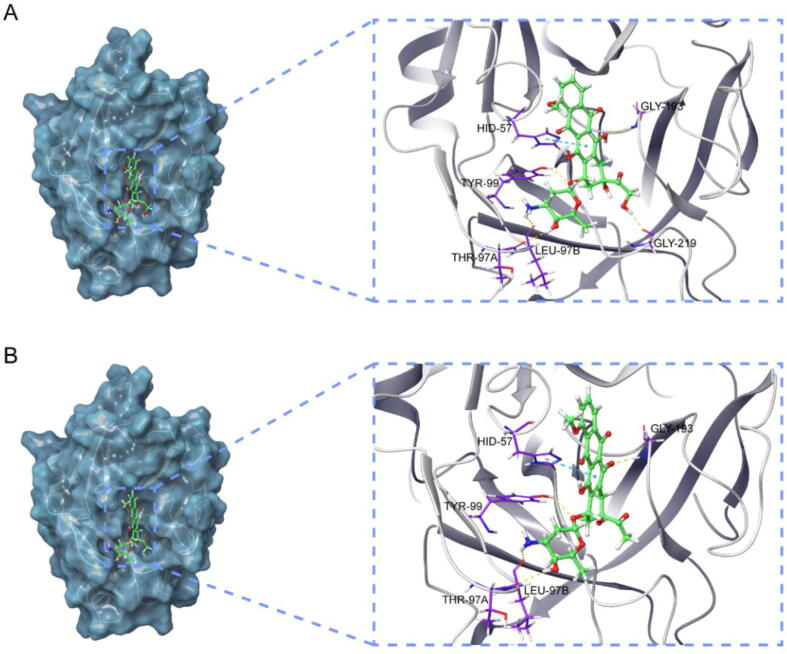
Molecular docking is an efficient and fast method for compound screening, which uses free binding energy to infer binding stability. Table 3 showed the top 10 compounds that bind well to the key target. Compared with the native ligand, doxorubicin and daunorubicin possessed the lower binding energy towards PLAU, which suggested that these two compounds displayed a high affinity towards PLAU. The 3D interaction diagrams of these two compounds with PLAU were displayed to visualize the docking results. The interaction diagram of doxorubicin at the active site of PLAU (Fig. 11A) revealed hydrogen bonds' formation with the key residues TYP-99, GLY-219, THR-97A, and LEU-97B. Meanwhile, a pi-pi stacking interaction with HID-57 also contributed to stabilizing the ligand at the active site. Similarly, daunorubicin also relied on four hydrogen bonds and one pi-pi stacking interaction to maintain its favorable binding with PLAU (Fig. 11B). The difference was that one of the hydrogen bonds of daunorubicin is formed with GLY-193 rather than GLY-219.
Table 3.
The results of molecular docking.
| Rank | Pubchem CID | Compound name | Glide gscore |
|---|---|---|---|
| 1 | 31703 | Doxorubicin | −8.062 |
| 2 | 30323 | Daunorubicin | −8.018 |
| 3 | 3306 | Etilefrine | −6.63 |
| 4 | 107715 | Dihydroergocristine | −6.512 |
| 5 | 6047 | Levodopa | −6.132 |
| 6 | 4539 | Norfloxacin | −5.891 |
| 7 | 54671203 | Doxycycline | −5.578 |
| 8 | 24360 | Camptothecin | −5.507 |
| 9 | 4855 | Piromidic Acid | −5.357 |
| 10 | 5311498 | Cyanocobalamin | −4.993 |
| Native ligand | 137349240 | 3-Azanyl-5-(Azepan-1-Yl)-N-Carbamimidoyl-6-(Furan-2-Yl)Pyrazine-2-Carboxamide | −7.457 |
Fig. 11.
Structures and orthogonal views of the pocket of binding between the well-matched chemicals and key target. (A) Structures and orthogonal views of the pocket of binding between doxorubicin and PLAU. (B) Structures and orthogonal views of the pocket of binding between daunorubicin and PLAU.
4. Discussion
In recent years, tumor immunotherapy has set off a wave, which has also been widely recognized as an effective strategy for managing solid tumors [34]. Therefore, immunotherapy has become a promising treatment option for HNSC patients [35], [36]. Numerous clinical studies have evaluated the efficacy of ICIs alone or combined with radiotherapy, chemotherapy, and other immune therapies in HNSC [37], [38], [39]. As the first approved ICI in treating platinum-refractory recurrent or metastatic HNSC, pembrolizumab and nivolumab increased the OS of patients as well as disease-free survival, which declares that HNSC has entered the era of immunotherapy [40], [41]. However, the therapeutic response varies from person to person. Moreover, only a small proportion of patients could experience objective clinical benefit. Therefore, it is urgent to develop a robust indicator to monitor prognosis and predict immunotherapy response.
A strong relationship between immune and tumor has been reported in the literature [42], [43], [44]. The tremendous diversity and plasticity have promise immune exerting multifaceted functions on tumorigenicity and progression. Although some studies have established prognostic indicators based on lncRNA or miRNA [45], [46], we still believe that the signature based on immune genes is more suitable to predict the prognosis of immunotherapy. In this study, multiple genomic data enabled the identification of 400 DEIRGs in HNSC tissues, of which biological process and signaling pathway were most relevant to immunity. The optimal indicator IRGPI was further identified, which include 13 IRGs: PLAU, IRF9, CCL26, BLNK, SEMA3G, FPR2, GAST, IL34, SLURP1, STC1, STC2, TNFRSF12A and TNFRSF25.
According to the reports, among these 13 IRGs, some genes may be involved in immunomodulatory activities, such as IL34 [47], FPR2 [48], SLURP1 [49], STC1 [50], and STC2 [51], while other genes are closely correlated with immune cells. For instance, IRF9 [52] and BLNK [53] are separately required for the survival of CD8+ T cell or B lymphocytes, while PLAU [54] and TNFRSF25 [55] play decisive roles in the suppressive function of Tregs. Besides, CCL26 [56] is involved in the hypoxia-mediated monocyte migration, and TNFRSF25 [57] functions as a costimulatory receptor for memory CD8+ T cells. Previous studies have also suggested the prognostic value of these genes. For example, PLAU [58], [59] is related to OS of HNSC patients, while STC2 [60] and FPR2 [61] are involved in HNSC metastasis. BLNK was also reported to have significant potential as candidate therapeutics for the clinical management of HNSC [62]. Moreover, SEMA3G [63] is a potential target for anti-tumor migration and invasion, typically associates with survival advantage. In addition, the interactions between these 13 genes were explored by GAIL (Gene-Gene Association Inference based on biomedical literature) [64]. As shown in Fig. S8, the result showed that PLAU interacts with FPR2, SEMA3G, and TNFRSF12A, while TNFRSF25 has an interaction with TNFRSF12A and BLNK. The gene STC1 and STC2 influence each other. The interactions between the other genes remain unknown and deserve further exploration.
Stability is crucial for a model, so K-M survival analysis and time-dependent ROC curves were applied to assess the predictive ability of IRGPI. The results in multiple datasets suggested that our prognostic signature had good potential in monitoring prognosis. Since there were some clinicopathologic factors related to OS, we further compare the predictive capabilities of IRGPI with them, and found that IRGPI could be used as an independent prognostic predictor.
To further explore the potential of IRGPI in clinical applications, we comprehensively examined the correlation of IRGPI with immune cell infiltration, TMB and IPS. Firstly, the relative proportion of 22 types immune cells in each HNSC sample was assessed, most of which were found to be related to IRGPI. Naive B cells, plasma cells, regulatory T cells which have lower levels in the high-risk group, and activated mast cells which have higher levels in the high-risk group were related to worse OS, partially explaining the survival status of the high-risk group. A previous study has shown that regulatory T cells could further subdivide HNSC and highly expressed in “inflamed” tumors for improving OS [65]. Secondly, TMB that can reflect the total number of mutations in tumor cells was analyzed in entire TCGA cohort. Consistent with literature reports [66], the high-risk group had a higher TMB and a poorer OS. Finally, we evaluated the response to ICIs. Since immune checkpoint therapy is selective, it is critical to identify patients who may benefit from it. As a reliable tool, IPS was established on the grounds of the gene expression that determines the immunogenicity of the tumors, including immunomodulators, MHC molecules, effector cells as well as immunosuppressive cells [23]. Our study revealed that higher scores of IPS, IPS-CTLA4, IPS-PD1/PD-L1/PD-L2, and IPS-CTLA4 + PD1/PD-L1/PD-L2 appeared in the low-risk group. Intriguingly, all seven prominent checkpoints were associated with IRGPI, among which CTLA-4, PD-1, LAG-3 as well as TIGIT were highly expressed in the low-risk group. All these findings implied that the low-risk group had more immunogenicity on ICIs. Nevertheless, the prognostic value of our signature should be further estimated in prospective cohort studies.
In order to find some potential small molecule drugs for the clinic, we identified 27 small-molecule drugs that have an inhibitory effect on the signature by CMap analysis. Thereinto, camptothecin, a potent topoisomerase inhibitor, was reported to reduce the risk of developing HNSC [67], and its analogs exhibited a good anti-tumor efficacy toward esophageal squamous cell carcinoma [68]. Ribavirin is not only effective in inhibiting nasopharyngeal carcinoma but also reduces the risk of HCV-related oral cancer [69], [70]. These identified small molecules may be applicable in the clinical treatment of HNSC in the future, or used in conjunction with existing drugs to enhance the therapeutic effect in treating HNSC.
Meanwhile, we constructed a PPI network for these IRGs that constitute the prognostic signature. Among 19 interactive targets, PLAU was considered as the key target. Previous studies have reported that PLAU could regulate cell proliferation, migration and invasion, closely related to the initiation and prognosis of HNSC [71], [72]. We found doxorubicin and daunorubicin, two well-known chemotherapy drugs, had a higher affinity towards PLAU than the native ligand by molecular docking. Although the connection with PLAU has not been studied yet, the anti-cancer effects of these two compounds have been widely recognized [73], [74], [75]. Considering the potential to suppress the expression level of PLAU, doxorubicin and daunorubicin may be used in conjunction with cancer immunotherapy to improve the HNSC high-risk group’s prognosis. Even so, these potential compounds and their specific molecular mechanism need more in-depth exploration and experimental verification.
In conclusion, we developed an IRG-based prognostic signature of HNSC and proved its predictive capability in multiple datasets, as well as explored potential small molecule drugs for clinical combination therapy. Hopefully, this signature could provide a solid foundation for individualized cancer immunotherapy.
5. Author statement
Weijie Qiang, Yifei Dai, Xiaoyan Xing and Xiaobo Sun were involved in the conception and design of study. Weijie Qiang and Yifei Dai were involved in the acquisition, analysis, and interpretation of all data. Weijie Qiang was involved in the draft of the manuscript. Yifei Dai, Xiaoyan Xing and Xiaobo Sun edited the manuscript; and all authors read and gave final approval to submit the manuscript.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgments
This study was supported by Major Program of the National Natural Science Foundation of China (No. 81891012), CAMS Innovation Fund for Medical Sciences (CIFMS) (No. 2020-I2M-2-011), Major Scientific and Technological Special Project for Significant New Drugs Formulation (No. 2018ZX09711001-009), and National natural science foundation of China (No. U1812403-5-3).
Data availability
The datasets obtained in this study or materials are available from the corresponding authors on reasonable request.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2021.01.046.
Contributor Information
Xiaoyan Xing, Email: xyxing@implad.ac.cn.
Xiaobo Sun, Email: xbsun@implad.ac.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Jou A., Hess J. Epidemiology and molecular biology of head and neck cancer. Oncol Res Treat. 2017;40:328–332. doi: 10.1159/000477127. [DOI] [PubMed] [Google Scholar]
- 2.Ferris R.L., Blumenschein G., Fayette J., Guigay J., Colevas A.D., Licitra L. Nivolumab vs investigator’s choice in recurrent or metastatic squamous cell carcinoma of the head and neck: 2-year long-term survival update of CheckMate 141 with analyses by tumor PD-L1 expression. Oral Oncol. 2018;81:45–51. doi: 10.1016/j.oraloncology.2018.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cohen E.E.W., Soulières D., Le Tourneau C., Dinis J., Licitra L., Ahn M.-J. Pembrolizumab versus methotrexate, docetaxel, or cetuximab for recurrent or metastatic head-and-neck squamous cell carcinoma (KEYNOTE-040): a randomised, open-label, phase 3 study. Lancet. 2019;393(10167):156–167. doi: 10.1016/S0140-6736(18)31999-8. [DOI] [PubMed] [Google Scholar]
- 4.Candeias S.M., Gaipl U.S. The immune system in cancer prevention, development and therapy. Anticancer Agents Med Chem. 2016;16:101–107. doi: 10.2174/1871520615666150824153523. [DOI] [PubMed] [Google Scholar]
- 5.Fridman W.H., Zitvogel L., Sautès–Fridman C., Kroemer G. The immune contexture in cancer prognosis and treatment. Nat Rev Clin Oncol. 2017;14(12):717–734. doi: 10.1038/nrclinonc.2017.101. [DOI] [PubMed] [Google Scholar]
- 6.Specenier P. Nivolumab in squamous cell carcinoma of the head and neck. Expert Rev Anticancer Ther. 2018;18(5):409–420. doi: 10.1080/14737140.2018.1456337. [DOI] [PubMed] [Google Scholar]
- 7.Larkins E., Blumenthal G.M., Yuan W., He K., Sridhara R., Subramaniam S. FDA approval summary: pembrolizumab for the treatment of recurrent or metastatic head and neck squamous cell carcinoma with disease progression on or after platinum-containing chemotherapy. Oncologist. 2017;22(7):873–878. doi: 10.1634/theoncologist.2016-0496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cohen E.E.W., Bell R.B., Bifulco C.B., Burtness B., Gillison M.L., Harrington K.J. The Society for Immunotherapy of Cancer consensus statement on immunotherapy for the treatment of squamous cell carcinoma of the head and neck (HNSCC) J Immunother Cancer. 2019;7(1):184. doi: 10.1186/s40425-019-0662-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weinstein J.N., Collisson E.A., Mills G.B., Shaw K.R.M., Ozenberger B.A., Ellrott K. The cancer genome atlas pan-cancer analysis project. Nat Genet. 2013;45(10):1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Y.u., Gao B., Tan P.Y., Handoko Y.A., Sekar K., Deivasigamani A. Genome-wide CRISPR knockout screens identify NCAPG as an essential oncogene for hepatocellular carcinoma tumor growth. FASEB J. 2019;33(8):8759–8770. doi: 10.1096/fsb2.v33.810.1096/fj.201802213RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bhattacharya S., Andorf S., Gomes L., Dunn P., Schaefer H., Pontius J. ImmPort: Disseminating data to the public for the future of immunology. Immunol Res. 2014;58(2-3):234–239. doi: 10.1007/s12026-014-8516-1. [DOI] [PubMed] [Google Scholar]
- 12.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. https://doi.org/10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed]
- 13.Ginestet C. ggplot2: Elegant Graphics for Data Analysis. J R Stat Soc Ser A (Statistics Soc 2011;174:245–6. https://doi.org/10.1111/j.1467-985x.2010.00676_9.x.
- 14.Huang D.W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 15.Pang J.S., Li Z.K., Lin P., Wang X.D., Chen G., Yan H.B. The underlying molecular mechanism and potential drugs for treatment in papillary renal cell carcinoma: a study based on TCGA and Cmap datasets. Oncol Rep. 2019;41:2089–2102. doi: 10.3892/or.2019.7014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gene Ontology Consortium The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32(90001):258D–261D. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friedman J., Hastie T., Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw. 2010;33:1–22. doi: 10.18637/jss.v033.i01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Therneau TM. A Package for Survival Analysis in S. Version 2.38. CRAN Website - Http//Cran.r-ProjectOrg/Package=survival 2015.
- 19.Heagerty P.J., Lumley T., Pepe M.S. Time-dependent ROC curves for censored survival data and a diagnostic marker. Biometrics. 2000;56:337–344. doi: 10.1111/j.0006-341X.2000.00337.x. [DOI] [PubMed] [Google Scholar]
- 20.Harrell Jr FE. rms: Regression Modeling Strategies. R package version 5.0-0. CRAN; 2016.
- 21.Newman A.M., Liu C.L., Green M.R., Gentles A.J., Feng W., Xu Y. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12(5):453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mayakonda A., Lin D.-C., Assenov Y., Plass C., Koeffler H.P. Maftools: Efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018;28(11):1747–1756. doi: 10.1101/gr.239244.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Charoentong P., Finotello F., Angelova M., Mayer C., Efremova M., Rieder D. Pan-cancer immunogenomic analyses reveal genotype-immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep. 2017;18(1):248–262. doi: 10.1016/j.celrep.2016.12.019. [DOI] [PubMed] [Google Scholar]
- 24.Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, et al. The connectivity map: Using gene-expression signatures to connect small molecules, genes, and disease. Science (80-) 2006;313:1929–35. https://doi.org/10.1126/science.1132939. [DOI] [PubMed]
- 25.Lamb J. The Connectivity Map: A new tool for biomedical research. Nat Rev Cancer. 2007;7(1):54–60. doi: 10.1038/nrc2044. [DOI] [PubMed] [Google Scholar]
- 26.Kim S, Chen J, Cheng T, Gindulyte A, He J, He S, et al. PubChem 2019 update: Improved access to chemical data. Nucleic Acids Res 2019;47:D1102–9. https://doi.org/10.1093/nar/gky1033. [DOI] [PMC free article] [PubMed]
- 27.Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 2019;47:D607–13. https://doi.org/10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed]
- 28.Berman H.M., Battistuz T., Bhat T.N., Bluhm W.F., Bourne P.E., Burkhardt K. The protein data bank. Acta Crystallogr Sect D Biol Crystallogr. 2002;58(6):899–907. doi: 10.1107/S0907444902003451. [DOI] [PubMed] [Google Scholar]
- 29.Schrödinger. Maestro | Schrödinger. Schrödinger Release 2018-1 2018.
- 30.Friesner R.A., Murphy R.B., Repasky M.P., Frye L.L., Greenwood J.R., Halgren T.A. Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J Med Chem. 2006;49:6177–6196. doi: 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- 31.Riaz N., Havel J.J., Makarov V., Desrichard A., Urba W.J., Sims J.S. Tumor and microenvironment evolution during immunotherapy with nivolumab. Cell. 2017;171(4):934–949.e16. doi: 10.1016/j.cell.2017.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Auslander N., Zhang G., Lee J.S., Frederick D.T., Miao B., Moll T. Robust prediction of response to immune checkpoint blockade therapy in metastatic melanoma. Nat Med. 2018;24(10):1545–1549. doi: 10.1038/s41591-018-0157-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao J., Chen A.X., Gartrell R.D., Silverman A.M., Aparicio L., Chu T. Immune and genomic correlates of response to anti-PD-1 immunotherapy in glioblastoma. Nat Med. 2019;25(3):462–469. doi: 10.1038/s41591-019-0349-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pham T., Roth S., Kong J., Guerra G., Narasimhan V., Pereira L. An update on immunotherapy for solid tumors: a review. Ann Surg Oncol. 2018;25(11):3404–3412. doi: 10.1245/s10434-018-6658-4. [DOI] [PubMed] [Google Scholar]
- 35.Przybylski K., Majchrzak E., Weselik L., Golusiński W. Immunotherapy of head and neck squamouscell carcinoma (HNSCC). Immune checkpoint blockade. Otolaryngol Pol. 2018;72(4):1–5. doi: 10.5604/0030665710.5604/01.3001.0012.4367. [DOI] [PubMed] [Google Scholar]
- 36.Bauman J.E., Cohen E., Ferris R.L., Adelstein D.J., Brizel D.M., Ridge J.A. Immunotherapy of head and neck cancer: Emerging clinical trials from a National Cancer Institute Head and Neck Cancer Steering Committee Planning Meeting. Cancer. 2017;123(7):1259–1271. doi: 10.1002/cncr.30449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Migden M.R., Rischin D., Schmults C.D., Guminski A., Hauschild A., Lewis K.D. PD-1 blockade with cemiplimab in advanced cutaneous squamous-cell carcinoma. N Engl J Med. 2018;379(4):341–351. doi: 10.1056/NEJMoa1805131. [DOI] [PubMed] [Google Scholar]
- 38.Saâda-Bouzid E., Defaucheux C., Karabajakian A., Coloma V.P., Servois V., Paoletti X. Hyperprogression during anti-PD-1/PD-L1 therapy in patients with recurrent and/or metastatic head and neck squamous cell carcinoma. Ann Oncol Off J Eur Soc Med Oncol. 2017;28(7):1605–1611. doi: 10.1093/annonc/mdx178. [DOI] [PubMed] [Google Scholar]
- 39.Yu Y., Lee N.Y. JAVELIN Head and Neck 100: A Phase III trial of avelumab and chemoradiation for locally advanced head and neck cancer. Futur Oncol. 2019;15(7):687–694. doi: 10.2217/fon-2018-0405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ferris R.L., Blumenschein G., Fayette J., Guigay J., Colevas A.D., Licitra L. Nivolumab for recurrent squamous-cell carcinoma of the head and neck. N Engl J Med. 2016;375(19):1856–1867. doi: 10.1056/NEJMoa1602252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seiwert T.Y., Burtness B., Mehra R., Weiss J., Berger R., Eder J.P. Safety and clinical activity of pembrolizumab for treatment of recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-012): an open-label, multicentre, phase 1b trial. Lancet Oncol. 2016;17(7):956–965. doi: 10.1016/S1470-2045(16)30066-3. [DOI] [PubMed] [Google Scholar]
- 42.Dumauthioz N., Labiano S., Romero P. Tumor resident memory T cells: new players in immune surveillance and therapy. Front Immunol. 2018;9:2076. doi: 10.3389/fimmu.2018.02076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wellenstein M.D., de Visser K.E. Cancer-cell-intrinsic mechanisms shaping the tumor immune landscape. Immunity. 2018;48(3):399–416. doi: 10.1016/j.immuni.2018.03.004. [DOI] [PubMed] [Google Scholar]
- 44.Bedognetti D., Hendrickx W., Ceccarelli M., Miller L.D., Seliger B. Disentangling the relationship between tumor genetic programs and immune responsiveness. Curr Opin Immunol. 2016;39:150–158. doi: 10.1016/j.coi.2016.02.001. [DOI] [PubMed] [Google Scholar]
- 45.Li J., Wang W., Xia P., Wan L., Zhang L., Yu L. Identification of a five-lncRNA signature for predicting the risk of tumor recurrence in patients with breast cancer. Int J Cancer. 2018;143(9):2150–2160. doi: 10.1002/ijc.v143.910.1002/ijc.31573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Siriwardhana C., Khadka V.S., Chen J.J., Deng Y. Development of a miRNA-seq based prognostic signature in lung adenocarcinoma. BMC Cancer. 2019;19:34. doi: 10.1186/s12885-018-5206-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin W., Xu D., Austin C.D., Caplazi P., Senger K., Sun Y. Function of CSF1 and IL34 in macrophage homeostasis, inflammation, and cancer. Front Immunol. 2019;10:1–18. doi: 10.3389/fimmu.2019.02019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stama M.L., Lacivita E., Kirpotina L.N., Niso M., Perrone R., Schepetkin I.A. Functional N-formyl peptide receptor 2 (FPR2) antagonists based on the ureidopropanamide scaffold have potential to protect against inflammation-associated oxidative stress. ChemMedChem. 2017;12:1839–1847. doi: 10.1002/cmdc.201700429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Swamynathan S., Buela K.-A., Kinchington P., Lathrop K.L., Misawa H., Hendricks R.L. Klf4 regulates the expression of slurp1, which functions as an immunomodulatory peptide in the mouse cornea. Investig Ophthalmol Vis Sci. 2012;53(13):8433–8446. doi: 10.1167/iovs.12-10759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu Y., Li Z., Jia W., Li M., Tang M. Upregulation of stanniocalcin-1 inhibits the development of osteoarthritis by inhibiting survival and inflammation of fibroblast-like synovial cells. J Cell Biochem. 2019;120(6):9768–9780. doi: 10.1002/jcb.v120.610.1002/jcb.28257. [DOI] [PubMed] [Google Scholar]
- 51.Lv H, Liu Q, Sun Y, Yi X, Wei X, Liu W, et al. Mesenchymal stromal cells ameliorate acute lung injury induced by LPS mainly through stanniocalcin-2 mediating macrophage polarization. Ann Transl Med 2020;8:334–334. https://doi.org/10.21037/atm.2020.02.105. [DOI] [PMC free article] [PubMed]
- 52.Huber M., Suprunenko T., Ashhurst T., Marbach F., Raifer H., Wolff S. IRF9 Prevents CD8+ T cell exhaustion in an extrinsic manner during acute lymphocytic choriomeningitis virus infection. J Virol. 2017;91(22) doi: 10.1128/JVI.01219-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tan J.-L., Wong S.-C., Gan S.-E., Xu S., Lam K.-P. The adaptor protein BLNK is required for b cell antigen receptor-induced activation of nuclear factor-κB and cell cycle entry and survival of B lymphocytes. J Biol Chem. 2001;276(23):20055–20063. doi: 10.1074/jbc.M010800200. [DOI] [PubMed] [Google Scholar]
- 54.He F., Chen H., Probst‐Kepper M., Geffers R., Eifes S., del Sol A. PLAU inferred from a correlation network is critical for suppressor function of regulatory T cells. Mol Syst Biol. 2012;8(1):624. doi: 10.1038/msb:2012.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Valatas V., Kolios G., Bamias G. TL1A (TNFSF15) and DR3 (TNFRSF25): A co-stimulatory system of cytokines with diverse functions in gut mucosal immunity. Front Immunol. 2019;10:583. doi: 10.3389/fimmu.2019.00583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hummitzsch L., Berndt R., Kott M., Rusch R., Faendrich F., Gruenewald M. Hypoxia directed migration of human naïve monocytes is associated with an attenuation of cytokine release: indications for a key role of CCL26. J Transl Med. 2020;18(1):404. doi: 10.1186/s12967-020-02567-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Slebioda T.J., Rowley T.F., Ferdinand J.R., Willoughby J.E., Buchan S.L., Taraban V.Y. Triggering of TNFRSF25 promotes CD8 + T-cell responses and anti-tumor immunity. Eur J Immunol. 2011;41(9):2606–2611. doi: 10.1002/eji.201141477. [DOI] [PubMed] [Google Scholar]
- 58.Yang K., Zhang S., Zhang D., Tao Q., Zhang T., Liu G. Identification of SERPINE1, PLAU and ACTA1 as biomarkers of head and neck squamous cell carcinoma based on integrated bioinformatics analysis. Int J Clin Oncol. 2019;24(9):1030–1041. doi: 10.1007/s10147-019-01435-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sepiashvili L., Hui A., Ignatchenko V., Shi W., Su S., Xu W. Potentially novel candidate biomarkers for head and neck squamous cell carcinoma identified using an integrated cell line-based discovery strategy. Mol Cell Proteomics. 2012;11(11):1404–1415. doi: 10.1074/mcp.M112.020933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang S., Ji Q., Chang B., Wang Y., Zhu Y., Li D. STC2 promotes head and neck squamous cell carcinoma metastasis through modulating the PI3K/AKT/Snail signaling. Oncotarget. 2017;8(4):5976–5991. doi: 10.18632/oncotarget.13355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hou X.-L., Ji C.-D., Tang J., Wang Y.-X., Xiang D.-F., Li H.-Q. FPR2 promotes invasion and metastasis of gastric cancer cells and predicts the prognosis of patients. Sci Rep. 2017;7(1) doi: 10.1038/s41598-017-03368-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Islam T., Rahman R., Gov E., Turanli B., Gulfidan G., Haque A. Drug targeting and biomarkers in head and neck cancers: insights from systems biology analyses. Omi A J Integr Biol. 2018;22(6):422–436. doi: 10.1089/omi.2018.0048. [DOI] [PubMed] [Google Scholar]
- 63.Zhou X., Ma L., Li J., Gu J., Shi Q., Yu R. Effects of SEMA3G on migration and invasion of glioma cells. Oncol Rep. 2012;28:269–275. doi: 10.3892/or.2012.1796. [DOI] [PubMed] [Google Scholar]
- 64.Couch D., Yu Z., Nam J.H., Allen C., Ramos P.S., da Silveira W.A. Gail: An interactive webserver for inference and dynamic visualization of gene-gene associations based on gene ontology guided mining of biomedical literature. PLoS ONE. 2019;14(7):e0219195. doi: 10.1371/journal.pone.0219195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Echarti A., Hecht M., Büttner-Herold M., Haderlein M., Hartmann A., Fietkau R. Cd8+ and regulatory T cells differentiate tumor immune phenotypes and predict survival in locally advanced head and neck cancer. Cancers (Basel) 2019;11(9):1398. doi: 10.3390/cancers11091398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang L., Liu Z., Li J., Huang T., Wang Y., Chang L. Genomic analysis of primary and recurrent gliomas reveals clinical outcome related molecular features. Sci Rep. 2019;9(1):16058. doi: 10.1038/s41598-019-52515-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu Z., Liu H., Han P., Gao F., Dahlstrom K.R., Li G. Apoptotic capacity and risk of squamous cell carcinoma of the head and neck. Eur J Cancer. 2017;72:166–176. doi: 10.1016/j.ejca.2016.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zou J., Li S., Chen Z., Lu Z., Gao J., Zou J. A novel oral camptothecin analog, gimatecan, exhibits superior antitumor efficacy than irinotecan toward esophageal squamous cell carcinoma in vitro and in vivo. Cell Death Dis. 2018;9(6):661. doi: 10.1038/s41419-018-0700-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xu M., Tao Z., Wang S., Jiang Y., Qu M. Suppression of oncogenic protein translation via targeting eukaryotic translation initiation factor 4E overcomes chemo-resistance in nasopharyngeal carcinoma. Biochem Biophys Res Commun. 2019;512(4):902–907. doi: 10.1016/j.bbrc.2019.03.118. [DOI] [PubMed] [Google Scholar]
- 70.Su T., Yang H., Tseng T., Chou S., Lin C., Liu C. Antiviral therapy in patients with chronic hepatitis C is associated with a reduced risk of parkinsonism. Mov Disord. 2019;34(12):1882–1890. doi: 10.1002/mds.v34.1210.1002/mds.27848. [DOI] [PubMed] [Google Scholar]
- 71.Zou B., Li J., Xu K., Liu J., Yuan D., Meng Z. Identification of key candidate genes and pathways in oral squamous cell carcinoma by integrated Bioinformatics analysis. Exp Ther Med. 2019;17:4089–4099. doi: 10.3892/etm.2019.7442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ge Y., Li W., Ni Q., He Y., Chu J., Wei P. Weighted gene co-expression network analysis identifies hub genes associated with occurrence and prognosis of oral squamous cell carcinoma. Med Sci Monit. 2019;25:7272–7288. doi: 10.12659/MSM.916025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Meredith A.-M., Dass C.R. Increasing role of the cancer chemotherapeutic doxorubicin in cellular metabolism. J Pharm Pharmacol. 2016;68(6):729–741. doi: 10.1111/jphp.12539. [DOI] [PubMed] [Google Scholar]
- 74.Rivankar S. An overview of doxorubicin formulations in cancer therapy. J Cancer Res Ther. 2014;10:853–858. doi: 10.4103/0973-1482.139267. [DOI] [PubMed] [Google Scholar]
- 75.Wang X., Wang Y., Fu M., Liu J., Yang Y., Yu Y. Inhibition of tumor metastasis by targeted daunorubicin and dioscin codelivery liposomes modified with pfv for the treatment of non-smallcell lung cancer. Int J Nanomed. 2019;14:4071–4090. doi: 10.2147/IJN.S194304. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets obtained in this study or materials are available from the corresponding authors on reasonable request.