Abstract
Purpose of Review
The hereditary spastic paraplegias (HSPs) are a group of disorders characterised by progressive lower limb weakness and spasticity. We address the challenges and controversies involved in the genetic diagnosis of HSP.
Recent Findings
There is a large and rapidly expanding list of genes implicated in HSP, making it difficult to keep gene testing panels updated. There is also a high degree of phenotypic overlap between HSP and other disorders, leading to problems in choosing the right panel to analyse. We discuss genetic testing strategies for overcoming these diagnostic hurdles, including the use of targeted sequencing gene panels, whole-exome sequencing and whole-genome sequencing. Personalised treatments for HSP are on the horizon, and a genetic diagnosis may hold the key to access these treatments.
Summary
Developing strategies to overcome the challenges and controversies in HSP may hold the key to a rapid and accurate genetic diagnosis.
Keywords: Hereditary spastic paraplegia, HSP, Diagnosis, Genetics, Whole-exome sequencing, Whole-genome sequencing
Introduction
The hereditary spastic paraplegias (HSPs) are a group of conditions characterised by progressive weakness and spasticity of the lower limbs [1, 2]. They can have autosomal dominant (AD), autosomal recessive (AR), X-linked and mitochondrial modes of inheritance [3]. The HSPs can be classified as either ‘pure’ (uncomplicated) or ‘complex’ (complicated). Pure forms involve lower limb spastic paraplegia and may include bladder involvement and subtle sensory signs such as impaired vibration sense. Complicated forms include additional neurological and non-neurological manifestations, such as cognitive impairment, dysarthria, optic atrophy and peripheral neuropathy [1]. There are also syndromic forms such as Silver syndrome (spastic paraparesis with distal amyotrophy predominantly of the hands). The different genetic forms are assigned spastic paraplegia loci (SPG), although the HSP genes may also be listed according to the new MDSGene nomenclature, e.g. SPAST-HSP for SPG4 [4]. The prevalence of AD HSP ranges from 0.5 to 5.5 per 100,000 and that of AR HSP from 0.3 to 5.3 per 100,000 [5]. Although the HSPs are rare, the progressive and disabling nature of these disorders means that they warrant greater attention from clinicians and researchers.
In this review, we discuss current challenges to reach a genetic diagnosis in HSP. These include (i) the large number of genes involved and the rapid rate of gene discovery, (ii) major phenotypic overlap between HSP and other disorders and (iii) disorders that mimic HSP. Further adding to the complexity is that a single HSP gene can have different patterns of inheritance, for example both autosomal dominant and recessive. Additionally, a single patient with HSP can have concurrent independent genetic diagnoses. Moreover, pseudodominant inheritance of autosomal recessive disease can occur when an individual with mutations on both copies of the gene has a partner carrying a heterozygous mutation, which may result in an affected offspring, a situation that typically occurs when there is a high carrier frequency in the population. In light of these challenges, we discuss the pros and cons of common genetic testing strategies in HSP such as multi-gene panels, whole-exome sequencing (WES) and whole-genome sequencing (WGS). An accurate, timely genetic diagnosis in HSP may become particularly relevant as new, targeted therapies are on the horizon.
Challenges to a Genetic Diagnosis
Multiple Genes and a Rapidly Increasing Gene List
There are many genes causative of HSP resulting in a high level of genetic heterogeneity. Different forms of HSP are assigned a genetic locus according to the order in which they are discovered (spastic paraplegia loci, SPG). Currently, the Online Mendelian Inheritance in Man (OMIM) lists 81 distinct genetic forms of HSP (Table 1, excluding SPG40 and for SPG65 see SPG45). Of these 81 genetic forms, 13 do not have a specific gene identified. Furthermore, while 55 had been identified in more than 1 family, twenty-six were reported in single families, warranting further confirmation.
Table 1.
Summary of genetic forms of hereditary spastic paraplegia
| Type | Gene | Location | Phenotype MIM number | Inheritance | Identified in more than 1 family with HSP (yes or no) | Allelic disorders/alternative gene-phenotype relationships, MIM number |
|---|---|---|---|---|---|---|
| SPG1 | L1CAM | Xq28 | MASA syndrome, CRASH syndrome, MIM303350 | XLR | Yes | Partial agenesis of the corpus callosum, MIM308840; hydrocephalus due to aqueductal stenosis, hydrocephalus with congenital idiopathic intestinal pseudoobstruction, hydrocephalus with Hirschsprung disease, MIM307000 |
| SPG2 | PLP1 | Xq22.2 | MIM312920 | XLR | Yes | Pelizaeus-Merzbacher disease, MIM312080 |
| SPG3A | ATL1 | 14q22.1 | MIM182600 | AD | Yes | Hereditary sensory neuropathy type ID, MIM613708 |
| SPG4 | SPAST | 2p22.3 | MIM182601 | AD | Yes | |
| SPG5A | CYP7B1 | 8q12.3 | MIM270800 | AR | Yes | Congenital bile acid synthesis defect type 3, MIM613812 |
| SPG6 | NIPA1 | 15q11.2 | MIM600363 | AD | Yes | |
| SPG7 | SPG7 | 16q24.3 | MIM607259 | AR | Yes | |
| SPG8 | KIAA0196 (WSHC5) | 8q24.3 | MIM603563 | AD | Yes | Ritscher-Schinzel syndrome 1, MIM220210 |
| SPG9A, B | ALDH18A1 | 10q24.1 | MIM601162, MIM616586 | AD | Yes | AD cutis laxa 3, MIM616603; AR cutis laxa type IIIA, MIM219150 |
| SPG10 | KIF5A | 12q13.3 | MIM604187 | AD | Yes | Neonatal intractable myoclonus, MIM617235 |
| SPG11 | SPG11 | 15q21.1 | MIM604360 | AR | Yes | Juvenile amyotrophic lateral sclerosis 5, MIM602099; axonal Charcot-Marie-Tooth disease type 2X, MIM616668 |
| SPG12 | RTN2 | 19q13.32 | MIM604805 | AD | Yes | |
| SPG13 | HSPD1 | 2q33.1 | MIM605280 | AD | Yes | Hypomyelinating leukodystrophy 4, MIM612233 |
| SPG14 | - | 3q27-q28 | MIM605229 | AR | No | |
| SPG15 | ZFYVE26 | 14q24.1 | MIM270700 | AR | Yes | |
| SPG16 | - | Xq11.2 | MIM300266 | XLR | No | |
| SPG17 | BSCL2 | 11q12.3 | Silver spastic paraplegia syndrome, MIM270685 | AD | Yes | Progressive encephalopathy with or without lipodystrophy, MIM615924; congenital generalised lipodystrophy type 2, MIM269700; distal hereditary motor neuropathy type VA, MIM600794 |
| SPG18 | ERLIN2 | 8p11.23 | MIM611225 | AR | Yes | |
| SPG19 | - | 9q | MIM607152 | AD | No | |
| SPG20 | SPG20 | 13q13.3 | Troyer syndrome, MIM2759002 | AR | Yes | |
| SPG21 | SPG21 | 15q22.31 | MAST syndrome, MIM248900 | AR | Yes | |
| SPG22 | SLC16A2 | Xq13.2 | Allan-Herndon-Dudley syndrome, MIM300523 | XL | Yes | |
| SPG23 | DSTYK | 1q32.1 | MIM270750 | AR | Yes | Congenital anomalies of kidney and urinary tract 1, MIM610805 |
| SPG24 | - | 13q14 | MIM607584 | AR | No | |
| SPG25 | - | 6q23-q24.1 | MIM608220 | AR | No | |
| SPG26 | B4GALNT1 | 12p11.1-q14 | MIM609195 | AR | Yes | |
| SPG27 | - | 10q22.1-q24.1 | MIM609041 | AR | No | |
| SPG28 | DDHD1 | 14q22.1 | MIM603940 | AR | Yes | |
| SPG29 | - | 1p31.1-p21.1 | MIM609727 | AD | No | |
| SPG30 | KIF1A | 2q37.3 | MIM610357 | AD, AR | Yes | AD mental retardation type 9, MIM 614255; hereditary sensory neuropathy type IIC, MIM614213 |
| SPG31 | REEP1 | 2p11.2 | MIM610250 | AD | Yes | Distal hereditary motor neuronopathy type VB, MIM614751 |
| SPG32 | - | 14q12-q21 | MIM611252 | AR | No | |
| SPG33 | ZFYVE27 | 10q24.2 | MIM610244 | AD | No | |
| SPG34 | - | Xq24-25 | MIM300750 | XLR | No | |
| SPG35 | FA2H | 16q23.1 | MIM612319 | AR | Yes | |
| SPG36 | - | 12q23-q24 | MIM613096 | AD | No | |
| SPG37 | - | 8p21.1-q13.3 | MIM611945 | AD | No | |
| SPG38 | - | 4p16-p15 | MIM612335 | AD | No | |
| SPG39 | PNPLA6 | 19p13.2 | MIM612020 | AR | Yes | Laurence-Moon syndrome, MIM245800; Boucher-Neuhauser syndrome MIM215470; Oliver-McFarlane syndrome MIM275400 |
| SPG40 | - | - | - | AD | No | |
| SPG41 | - | 11p14.1-p11.2 | MIM613364 | AD | No | |
| SPG42 | SLC33A1 | 3q25.31 | MIM612539 | AD | No | Congenital cataracts, hearing loss, and neurodegeneration, MIM614482 |
| SPG43 | C19orf12 | 19p13.11-q12 | MIM615043 | AR | No | Neurodegeneration with brain iron accumulation 4, MIM614298 |
| SPG44 | GJC2 | 1q42.13 | MIM613206 | AR | No | Hypomyelinating leukodystrophy 2, MIM608804 |
| SPG45 | NT5C2 | 10q24.3–q25.1 | MIM 613162 | AR | Yes | |
| SPG46 | GBA2 | 9p13.3 | MIM614409 | AR | Yes | |
| SPG47 | AP4B1 | 1p13.2 | MIM614066 | AR | Yes | |
| SPG48 | KIAA0415 | 7p22.1 | MIM613647 | AR | Yes | |
| SPG49 | TECPR2 | 14q32.31 | MIM615031 | AR | Yes | |
| SPG50 | AP4M1 | 7q22.1 | MIM612936 | AR | Yes | |
| SPG51 | AP4E1 | 15q21.2 | MIM613744 | AR | Yes | Familial persistent stuttering 1, MIM184450 |
| SPG52 | AP4S1 | 14q12 | MIM614067 | AR | Yes | |
| SPG53 | VPS37A | 8p22 | MIM614898 | AR | Yes | |
| SPG54 | DDHD2 | 8p11.23 | MIM615033 | AR | Yes | |
| SPG55 | C12orf65 | 12q24.31 | MIM615035 | AR | Yes | Combined oxidative phosphorylation deficiency 7, MIM613559 |
| SPG56 | CYP2U1 | 4q25 | MIM615030 | AR | Yes | Pseudoxanthoma elasticum |
| SPG57 | TFG | 3q12.2 | MIM604484 | AR | Yes | Hereditary motor and sensory neuropathy, Okinawa type, MIM604484 |
| SPG58 | KIF1C | 17p13.2 | AR spastic ataxia 2, MIM611302 | AR | Yes | |
| SPG59 | USP8 | 15q21.2 | - | AR | No | |
| SPG60 | WDR48 | 3p22.2 | - | AR | No | |
| SPG61 | ARL6IP1 | 16p12.3 | MIM615685 | AR | Yes | |
| SPG62 | ERLIN1 | 10q24.31 | MIM615681 | AR | Yes | |
| SPG63 | AMPD2 | 1p13.3 | MIM615686 | AR | No | Pontocerebellar hypoplasia type 9, MIM615809 |
| SPG64 | ENTPD1 | 10q24.1 | MIM615683 | AR | Yes | |
| SPG66 | ARSI | 5q32 | - | AR | No | |
| SPG67 | PGAP1 | 2q33.1 | - | AR | No | AR mental retardation 42, MIM615802 |
| SPG68 | KLC2 | 11q13.1 | Spastic paraplegia, optic atrophy, and neuropathy, MIM609541 | AR | Yes | |
| SPG69 | RAB3GAP2 | 1q41 | - | AR | No | Martsolf syndrome, MIM212720; Warburg micro syndrome 2, MIM614225 |
| SPG70 | MARS1 | 12q13.3 | - | AR | No | AD Charcot-Marie-Tooth disease type 2, MIM616280; interstitial lung and liver disease, MIM615486 |
| SPG71 | ZFR | 5p13.3 | - | AR | No | |
| SPG72 | REEP2 | 5q31 | MIM615625 | AD, AR | Yes | |
| SPG73 | CPT1C | 19q13.33 | MIM616282 | AD | Yes | |
| SPG74 | IBA57 | 1q42.13 | MIM616451 | AR | No | Multiple mitochondrial dysfunctions syndrome 3, MIM615330 |
| SPG75 | MAG | 19q13.12 | MIM616680 | AR | Yes | |
| SPG76 | CAPN1 | 11q13.1 | MIM616907 | AR | Yes | |
| SPG77 | FARS2 | 6p25.1 | MIM617046 | AR | Yes | Combined oxidative phosphorylation deficiency 14, MIM614946 |
| SPG78 | ATP13A2 | 1p36.13 | MIM617225 | AR | Yes | Kufor-Rakeb syndrome, MIM606693 |
| SPG79 | UCHL1 | 4p13 | MIM615491 | AR | Yes | ?Parkinson disease 5, susceptibility to, MIM613643 |
| SPG80 | UBAP1 | 9p13.3 | MIM618418 | AD | Yes | |
| SPG81 | SELENOI | 2p23.3 | MIM618768 | AR | Yes | |
| SPG82 | PCYT2 | 17q25.3 | MIM618770 | AR | Yes | |
| SPG83 | HPDL | 1p34.1 | MIM619027 | AR | Yes | |
| Unassigned | RNF170 | 8p11.21 | AR | Yes | AD sensory ataxia 1, MIM608984 | |
| Unassigned | FAR1 | 11p15.3 | AR | Yes | Peroxisomal fatty acyl-CoA reductase 1 disorder, MIM616154 | |
| Unassigned | NEFL | 8p21.2 | AR | Yes | Charcot-Marie-Tooth disease, dominant intermediate G, MIM 617882; Charcot-Marie-Tooth disease type 1F, MIM607734; Charcot-Marie-Tooth disease type 2E, MIM607684 | |
| Unassigned | VPS13D | 1p36.22-p36.21 | AR | Yes | Spinocerebellar ataxia, autosomal recessive 4, MIM607317 | |
| Unassigned | TUBB4A | 19p13.3 | AD | Yes | Dystonia 4, MIM128101; hypomyelinating leukodystrophy 6, MIM612438 | |
| Unassigned | VCP | 9p13.3 | AD | Yes | Charcot-Marie-Tooth disease type 2Y, MIM 616687; frontotemporal dementia and/or amyotrophic lateral sclerosis 6, MIM613954; inclusion body myopathy with early-onset Paget disease and frontotemporal dementia 1, MIM167320 | |
| Unassigned | POLR3A | 10q22.3 | AR | Yes | Hypomyelinating leukodystrophy 7 with or without oligodontia and-or hypogonadotropic hypogonadism, MIM607694; Wiedemann-Rautenstrauch syndrome, MIM264090 |
Information extracted from OMIM [6]
AD, autosomal dominant; AR, autosomal recessive; XLR, X-linked recessive
Due to the rapid rate of progress of HSP research, new genes are being identified on a regular basis. Examples of recently identified HSP genes include UCHL1 (SPG79), UBAP1 (SPG80), SELENOI (SPG81), PCYT2 (SPG82), HPDL (SPG83), and those not yet assigned a locus (RNF170 and FAR1) [7–16]. Some genes are much rarer than others, and it cannot be excluded that certain mutations may be ‘private’ to individual families. For example, a SCL33A1 mutation was implicated as a cause of AD HSP (SPG42) in a large Chinese pedigree [17], but mutations in this gene were not identified in a large sample of European HSP cases [18]. In contrast, multiple groups have reported that UBAP1 causes AD HSP with a pure phenotype [8–11]. This suggests that UBAP1 mutations are a relatively frequent cause of HSP, and that UBAP1 warrants inclusion on current HSP gene testing panels.
Overlap with Other Inherited Disorders
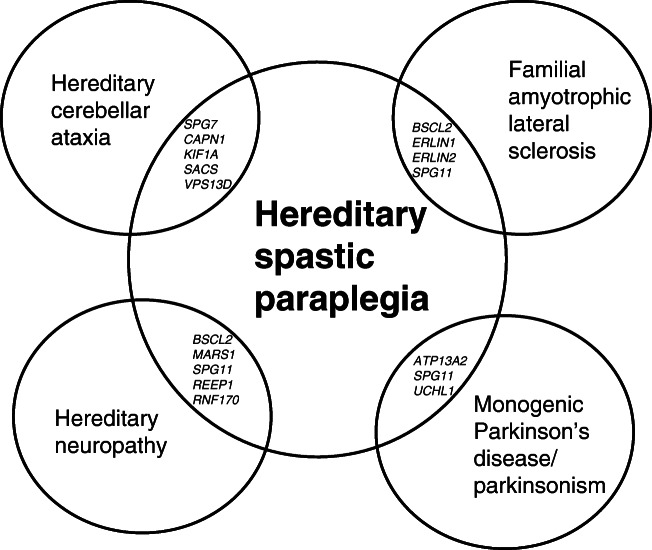
There is a large overlap between HSP and other disorders such as inherited forms of hereditary ataxia, peripheral neuropathy, amyotrophic lateral sclerosis (ALS) and Parkinson’s disease. Twenty-eight of 81 genetic forms of HSP are assigned alternative phenotypes on OMIM (Table 1) and this presents further diagnostic complexity (Fig. 1). Genetic testing is often performed with gene panels that are tailored to a specific disease category, and therefore an accurate clinical classification becomes a critical step able to significantly influence the diagnostic yield.
Fig. 1.
Examples of overlapping genes and shared phenotypes with hereditary spastic paraplegia
Overlap with the Hereditary Cerebellar Ataxias
Inherited ataxias commonly overlap with HSP [19], with a typical example being SPG7 [20, 21]. SPG7 mutations result in mitochondrial dysfunction [22] and may present with ataxia evolving to spastic ataxia phenotypes, as well as other features such as ophthalmoplegia and ptosis. SPG7 accounted for 2.3% of cerebellar ataxia cases in an Italian population [23]. Similarly, mutations in CAPN1 cause HSP with or without ataxia [24–27]. It has been suggested that ataxia and spasticity should not be considered separate phenotypes, but rather as existing on a ‘continuous ataxia-spasticity disease spectrum’ [19]. KIF1A mutations can cause both HSP and ataxia phenotypes (discussed below) [28•]. Mutations in SACS cause autosomal recessive spastic ataxia of Charlevoix-Saguenay (ARSACS), a disorder characterised by the triad of cerebellar ataxia, peripheral neuropathy, and spasticity; however not all features of the triad may be present and there is a phenotypic overlap with the AR HSP with a thin corpus callosum (AR-HSP-TCC) [29]. VPS13D mutations cause a recessive ataxia-spasticity spectrum movement disorder [30] but have also been reported to cause a pure or complicated form of HSP (Table 1) [31]. Additionally, HSP-like phenotypes can also be caused by expansions in triplet-repeat ataxia loci [32] and thus, may not be detected on a sequencing panel.
Overlap with the Inherited Neuropathies
Many forms of HSP overlap with the inherited neuropathies. Notable examples include mutations in BSCL2, which cause Silver syndrome, a complicated form of HSP in which affected individuals present with early-onset hand muscle wasting and leg spasticity [33]. BSCL2 mutations can also cause a range of phenotypes with lower motor neurone involvement including multifocal motor neuropathy with conduction block, Charcot-Marie-Tooth neuropathy type 2 and distal hereditary motor neuropathy type V [33, 34]. SPG11 mutations are a major cause of AR-HSP-TCC [35], but may also cause AR Charcot-Marie Tooth disease [36]. Mutations in MARS1 cause AR HSP complicated by cognitive impairment and nephrotic syndrome [37], as well as AD Charcot-Marie-Tooth Disease type 2 U [38]. Mutations in REEP1, the cause of SPG31, have been shown to cause distal hereditary motor neuropathy type V (Table 1). Recessive RNF170 mutations have recently been confirmed as a cause of HSP [14, 39]**, but a heterozygous mutation in RNF170 (p.Arg199Cys) was found to cause autosomal dominant late-onset progressive sensory ganglionopathy as a cerebellar ataxia, neuropathy, and vestibular areflexia syndrome (CANVAS) mimic [40].
Overlap with Hereditary Amyotrophic Lateral Sclerosis
There are many shared genes between HSP and ALS. For example, mutations in ERLIN1 have been implicated in SPG62, but may also be the cause of a slowly progressive early-onset ALS [41]. ERLIN2 mutations, causing SPG18, can evolve into rapidly progressive ALS [42] or cause juvenile primary lateral sclerosis [43]. Notably, Erlin1 and erlin2 are highly homologous endoplasmic reticulum membrane proteins that assemble into a ring-shaped complex [44]. Other examples of HSP genes implicated as causing ALS phenotypes include SPG11 [45] and BSCL2 [34].
Overlap with Monogenic Parkinson Disease
SPG11 has been linked with parkinsonism or dystonia-parkinsonism. This is highlighted by a recent study which showed that disruption of presynaptic dopaminergic pathways was a widespread phenomenon in individuals with SPG11 mutations, even without clinical manifestations of parkinsonism [46]. Of note, patients were unresponsive to levodopa, a finding which may relate to post-synaptic damage [46].
Recently, ATP13A2 mutations have been described as a cause of HSP complicated by cognitive impairment, cerebellar ataxia, and axonal motor and sensory polyneuropathy (SPG78) [47]. Mutations in this gene were first reported as a cause of an AR form of early-onset parkinsonism with pyramidal degeneration and dementia known as Kufor-Rakeb syndrome [48].
There has been a suggestion of a link between UCHL1 and Parkinson’s disease [49], although this association has not been confirmed. Mutations in UCHL1 have subsequently been implicated in an early-onset neurodegenerative syndrome, which may be considered HSP complicated by optic atrophy, cerebellar ataxia, seizures, myotonia, fasciculations, dorsal column signs, facial dysmorphism, myopathic facies, microcephaly and fasciculations [50, 51].
HSP Mimics: Other Mendelian Causes and Management Implications
HSP may be due to mutations in many other genes outside of the SPG loci, typically causing complicated phenotypes. For example, pathogenic variants in OPA3 can cause an optic atrophy plus syndrome, characterised by optic atrophy and lower limb spasticity [52]. Mutations in PEX16 have been shown to cause HSP complicated by cerebellar ataxia and dystonia [53, 54]. TUBB4A mutations have been initially described as a cause of whispering dysphonia (DYT4 dystonia) [55], but have subsequently been reported as a cause of HSP [56].
Several of the HSP mimics may be neurometabolic disorders with whose timely diagnosis has relevant implications for therapeutic strategies and management [57]. These disorders may have distinctive clinical features and biochemical findings (Table 2). Important examples include mutations in ABCD1, the gene associated with adrenoleukodystrophy and adrenomyeloneuropathy, which can cause spastic paraplegia in males and carrier females [24•]. Dopa-responsive dystonia may be misdiagnosed as HSP and is typically responsive to levodopa therapy [58]. Recently, combined homocysteinaemia with methylmalonic aciduria due to pathogenic recessive variants in the MMACHC gene has been highlighted as a treatable cause of HSP [59]. Testing urine methylmalonic acid and serum homocysteine levels and sequencing the MMACHC gene is critical when this rare condition is suspected [59]. Severe 5,10-methylenetetrahydrofolate reductase deficiency has also been reported as a cause of a complicated HSP phenotype, responsive to treatment with betaine and vitamins [60]. Additionally, cerebrotendinous xanthomatosis may mimic HSP and is treatable with chenodeoxycholic acid [61].
Table 2.
Examples of ‘treatable’ inherited mimics in HSP
| Disorder | Genetic basis | Mode of inheritance | Additional clinical features | Biochemical findings | Treatment | References |
|---|---|---|---|---|---|---|
| Adrenoleukodystrophy, MIM 300100; adult adrenomyeloneuropathy, MIM 300100 | ABCD1 | XLR | Sphincter disturbances, sexual dysfunction, adrenocortical dysfunction | Elevated very long chain fatty acids | Corticosteroid replacement therapy for adrenal insufficiency | Kim et al. [24•], Raymond et al. [62] |
| Argininemia, MIM 207800 | ARG1 | AR | Dystonia, dementia, peripheral neuropathy, epilepsy | Newborn screening, elevation of plasma arginine concentration | Measures to reduce ammonia, such as protein-restricted diet, branched-chained amino acids supplement and sodium benzoate. | Tsang et al. [63] |
| Biotinidase deficiency, MIM 253260 | BTD | AR | Seizures, hypotonia, limb weakness, ataxia, developmental delay, visual impairment, hearing loss, cutaneous abnormalities | Newborn screening or deficient biotinidase enzyme activity in serum/plasma | Treatment with biotin | Wolf [64], Wolf [65] |
| Primary coenzyme Q10 deficiency 8, MIM | COQ7 | AR | Primary coenzyme Q10 (CoQ10) deficiency is usually associated fatal neonatal encephalopathy with hypotonia, multiple-system atrophy-like phenotype, dystonia, spasticity, seizures, intellectual disability, sensorineural hearing loss, steroid-resistant nephrotic syndrome, hypertrophic cardiomyopathy | Reduced levels of CoQ10 in skeletal muscle or reduced activities of complex I+III and II+III of the mitochondrial respiratory chain on frozen muscle homogenates | 2,4-Dihydroxybenzoate bypass treatment, high-dose oral CoQ10 supplementation | Wang et al. [66], Salviati et al. [67] |
| Cerebrotendinous xanthomatosis, MIM 213700 | CYP27A1 | AR | Cerebellar signs, intellectual impairment, seizures, peripheral neuropathy, cataract, tendon xanthomas | Elevated levels of cholestanol and bile alcohols in serum and urine | Chenodeoxycholic acid | Nicholls et al. [61], Verrips et al. [68] |
| DOPA-responsive dystonia, MIM 128230 | GCH1 | AD, AR | Foot dystonia, later development of parkinsonism, diurnal variation in symptoms, dramatic and sustained response to levodopa | Reduced concentrations of total biopterin and total neopterin in the cerebrospinal fluid | Levodopa/decarboxylase inhibitor | Fan et al. [58] |
| Methylmalonic aciduria and homocystinuria cblC type MIM 277400 | MMACHC | AR | Cognitive impairment (5/8), spastic dysuria (3/8), personality change and depression (3/8), ataxia (2/8), seizures (2/8), limb numbness (2/8) and developmental delay (2/8). When patients were diagnosed, the mean serum homocysteine level, the methylmalonic acid level in urine, the serum propionylcarnitine (C3) level and the ratios of C3-to-acetylcarnitine (C2) and free carnitine (C0) were all dramatically elevated. Cranial MRIs showed nothing remarkable except mild brain atrophy. | Elevated urine methylmalonic acid and serum homocysteine levels | Intramuscular cobalamin, oral betaine and folate | Wei et al. [59] |
| Homocystinuria due to MTHFR deficiency, MIM 236250 | MTHFR | AR | Polyneuropathy, behavioural abnormalities, cognitive impairment, psychosis, seizures, leukoencephalopathy | Severe hyperhomocysteinemia associated with the characteristic amino acid profile | Betaine and vitamins | Lossos et al. [60] |
| Phenylketonuria, MIM 261600 | PAH | AR | Cognitive impairment | Serum phenylalanine concentrations | Classic phenylketonuria diet/protein restricted diet | Kasim et al. [69] |
| Dystonia 9, MIM 601042; GLUT1 deficiency syndrome 1, MIM 606777; GLUT1 deficiency syndrome 2, MIM 612126; Stomatin-deficient cryohydrocytosis with neurologic defects, MIM 608885 | SLC2A1 | AD | Seizures, delayed neurologic development, acquired microcephaly, intermittent ataxia, paroxysmal exercise-induced dyskinesia, choreo-athetosis, alternating hemiplegia | Cerebrospinal fluid analysis for hypoglycorrhachia | Ketogenic diet | Verrotti et al. [70] |
AD, autosomal dominant; AR, autosomal recessive; XLR, X-linked recessive
HSP Mimics: Overlap with Disorders Without Clear Mendelian Inheritance
Several disorders that do not have a readily recognisable monogenic cause may be difficult to differentiate from HSP, such as primary lateral sclerosis (PLS). PLS is a degenerative, mainly sporadic neuronopathy with primarily upper motor neurone features [71]. PLS frequently presents with spastic paraplegia, affects older, predominantly male patients and invariably progresses to involve cervical and bulbar regions [71]. However, the disease often remains as an isolated spastic paraplegia for many years and bulbar symptoms can appear after 10 years in up to 20% of patients [71]. Consequently, in the absence of family history, PLS and HSP may be clinically indistinguishable for longer than a decade [71]. However, cortical excitability studies may be used to differentiate these two conditions in a clinical setting [72], and genetic testing for HSP genes may also help [73].
It may also be challenging to differentiate between HSP and cerebral palsy. HSP may be distinguishable from spastic diplegic cerebral palsy by the absence of perinatal risk factors for brain injury and normal brain imaging, or specific findings indicative of an HSP syndrome, such as thinning of the corpus callosum [74]. Genetic testing may also be helpful, for example, a patient with childhood onset, non-progressive, spastic diplegia with no previous family history of HSP was long considered as affected by cerebral palsy, until his son also developed the same phenotype: genetic testing in these patients disclosed a heterozygous pathogenic variant in ATL1 (SPG3A) which had arisen de novo in the affected parent [75].
There may also be diagnostic uncertainty in differentiating HSP from multiple sclerosis. A personal observation is that patients may be referred to the neurogenetics clinic with HSP, only to find evidence of demyelinating lesions consistent with MS on upon repeating brain or spinal cord MRI. Conversely, mutations in HSP genes may be identified in individuals formerly diagnosed with MS. For example, SPG2 has been shown to mimic MS [76], and rare variants in genes including KIF5A and REEP1 were identified in patients with primary progressive MS [77].
When Should a Complex Disorder Be Diagnosed as HSP?
It may be difficult to decide when to categorise a disorder as HSP when the phenotype is complex. A chief consideration should be whether lower extremity weakness and spasticity are the predominant clinical manifestations [78]. For example, ATP13A2 mutations are known to cause Kufor-Rakeb syndrome [48], neuronal ceroid lipofuscinosis [79] and neurodegeneration with brain iron accumulation (NBIA) [80]. More recently, ATP13A2 mutations have been described as a cause of HSP complicated by cognitive impairment, cerebellar ataxia, and axonal motor and sensory polyneuropathy (SPG78) [47]. However, there is debate over whether an HSP predominant phenotype is a clinical outlier and if a new HSP locus was warranted [81]. Similarly, hypomorphic mutations in POLR3A were reported as a cause of HSP and ataxia [82], however, other authors considered that this condition should be defined as a ‘POLR3-related disorder’ instead [83].
HSP Genes with Different Modes of Inheritance
Variants in some HSP genes may be inherited with different modes of transmission, adding further complexity to the interpretation of genetic findings. As an example, biallelic mutations in KIF1A cause spastic paraplegia, distal wasting, peripheral neuropathy and mild cerebellar signs (AR SPG30) [84]. KIF1A mutations can also cause hereditary sensory and autonomic neuropathy type 2 with AR inheritance (Table 1). However, de novo dominant KIF1A mutations may result in a phenotypic spectrum overlapping with AR SPG30 including mental retardation, speech delay, epilepsy, optic nerve atrophy, thinning of the corpus callosum, periventricular white matter lesion and microcephaly [85–88]. A recent study showed that heterozygous mutations in KIF1A may result in two distinct phenotypes, a pure to complex HSP phenotype and a congenital or early-onset ataxia phenotype [28•]. Additionally, mutations in REEP2 have been identified in families with both AD and AR inheritance [37, 89]. A mutation in REEP2 has been found to cause AD HSP with a pure, early-onset phenotype [89], while the AR form is characterised by early-onset HSP with delayed motor milestones and normal cognition [37]. Similarly, ATL1 mutations are usually associated with dominant HSP (SPG3A), but recessive mutations in ATL1 have been shown to cause both pure and complex forms of HSP [90, 91].
Individuals with Concurrent Independent Genetic Diagnoses
Individuals presenting with HSP may have concurrent independent genetic diagnoses, further complicating genetic testing. As an example, a recent study showed two possible genetic diagnoses in a non-consanguineous family with 3 affected siblings: two brothers with intellectual impairment and spastic paraplegia, and a sister with behavioural disturbance and pes cavus. All affected siblings carried a maternally inherited interstitial 15q duplication and a paternally inherited REEP1 variant [92•]. In this case, it was thought that the 15q duplication was causing intellectual impairment and behavioural abnormalities, with supportive evidence from methylation and functional studies. On the other hand, the dominant HSP phenotype was attributed to the REEP1 variant. This in keeping with a large study of 7374 consecutive unrelated patients referred to a clinical diagnostic laboratory for WES, which demonstrated multiple molecular diagnoses in 4.9% of cases in whom WES was informative [93]. The results of these studies suggest that perhaps we too often claim a ‘phenotypic expansion’ to explain a phenotype that is different or more complicated than previously reported for a given gene, while in some of these cases the reason would be a ‘double hit’ and not a phenotypic expansion.
Pseudodominant Inheritance and Intronic Variants
In a recent study, a patient with spastic paraplegia and ataxia was investigated with WES, revealing a novel missense variant in SPG7 (c.2195T>C; p.Leu732Pro) [94•]. To seek a second variant, WGS was performed, revealing an unreported, deep intronic variant (c.286 + 853A>G), shown to activate a cryptic splice site [94•]. The deep intronic variant would not have been identified with WES alone, highlighting the usefulness of WGS to increase diagnostic yield [94•]. Furthermore, it sheds light on the apparent dominant pattern of inheritance of SPG7 [95], which may be due to the mutation on the other allele being missed [94•]. Another report highlights the importance of an intronic variant in POLR3A, a gene previously associated with hypomyelinating leukodystrophy type 7 (Table 1), as a frequent cause of HSP and cerebellar ataxia [82]. Compound heterozygous mutations in POLR3A were found in approximately 3.1% of index cases of HSP and cerebellar ataxia, with over 80% carrying the same intronic mutation (c.1909+22G>A) which activates a cryptic splice site [82]. This suggests that non-coding DNA variants may account for a substantial number of unsolved cases of HSP.
Strategies for a Genetic Diagnosis
There are several options for reaching a genetic diagnosis in individuals with HSP and it can be challenging for the clinician to decide upon which approach to adopt. Different strategies include targeted sequencing gene panels, whole-exome sequencing (WES), or whole-genome sequencing (WGS) (Table 3). Targeted sequencing gene panels are commonly used but will overlook a diagnosis if the mutation is in a gene that is outside the panel. Furthermore, gene panels also are not reliable in detecting copy number variants (CNVs), structural variants (SVs) and intronic variants. WES can be a useful approach but again may not be reliable for CNVs, SVs, and will fail to detect deep intronic variants. WGS may be the most complete approach [24•, 54, 96], with uniformity of coverage that allows for the accurate detection of CNVs, SVs [97, 98], in addition to the detection of non-coding variants. However, this approach is limited by the expense and difficulty processing, storing, and interpreting the large amounts of genomic data. Both WES and WGS allow for testing of many genes and so may not be restricted to single panel, e.g. patients can be tested for both ‘ataxia’ and ‘HSP’ genes in a single test [24•]. The WES and WGS data can be used in several ways. For example, a panel of relevant HSP genes may be analysed, such as those listed in Table 1. A larger, less specific panel of genes can also be interrogated, such as the TruSight One ‘clinical exome’—a panel of 4813 genes that have been associated with human disease [99]. WES or WGS family studies may provide valuable additional information regarding segregation of genetic variants with the disease phenotype. For example, parent-child trios of healthy parents and an affected child may facilitate the detection of homozygous, compound heterozygous or de novo variants.
Table 3.
Comparison of different approaches for the genetic diagnosis of hereditary spastic paraplegia
| Technique | Pros | Cons |
|---|---|---|
| Targeted sequencing panels |
- Less expensive* - Reduce incidental findings |
- Gene list may be restrictive, missing unexpected findings or mutations in genes implicated in overlapping phenotypes - Inadequate coverage of CNVs, SVs; MLPA may be required - Inadequate coverage of deep non-coding variants |
| Whole-exome sequencing |
- Gene panel not restrictive - Less expensive compared to whole-genome sequencing* |
- Inadequate coverage of CNVs, SVs; MLPA may be required s- Inadequate coverage of deep non-coding variants - Challenge of incidental findings |
| Whole-genome sequencing |
- Gene panel not restrictive - Detection of CNVs, SVs (e.g. deletions in SPAST) - Detection of non-coding variants (see example of deep intronic variants reported in SPG7) |
- Expensive* - Challenge of processing, storing and analysing large amounts of data - Challenge of incidental findings |
*Note that to our knowledge, a cost-effectiveness study for genetic testing in hereditary spastic paraplegia comparing the different approaches has not yet been performed.
It is critical to remember that CNVs (e.g. exonic deletions in SPAST [100]) are important to consider and may require a separate test (e.g. multiplex ligation probe amplification or MLPA), unless using a method that provides reliable detection such as WGS. Furthermore, testing for repeat expansion disorders will often require a separate test such as a fluorescent repeat-primed PCR assay. However, a recent study suggests that long repeat expansions may be detectable from PCR-free WGS data using a software tool called ExpansionHunter [101]. Furthermore, a homoplasmic m.9176 T>C mutation in the mitochondrial ATP6 gene has been found to cause HSP. WES may allow for the detection of mitochondrial point mutations using ‘off-target reads’, providing additional diagnoses [102]. WGS provides exceptionally high coverage of the mitochondrial genome, allowing for accurate detection of mitochondrial point mutations even at low levels of heteroplasmy [103]. Hypothesis-free methods such as WES and WGS may also detect multiple concurrent genetic defects, as described above [92•].
Benefits of a Genetic Diagnosis in HSP
There are numerous benefits of a genetic diagnosis in HSP which may prompt the decision to undertake genetic testing. As an example, it may provide for prognostic information and facilitate genetic counselling and family planning. It may also allow for a prenatal diagnosis/preimplantation genetic diagnosis.
A genetic diagnosis rarely leads to findings with direct management implications (as discussed earlier, see Table 2). However, it may hold future value in that it could be used to enrol patients in clinical trials that target the disease mechanism. A targeted, disease-modifying treatment appears most likely for two forms of HSP—SPG4 and SPG5.
Microtubule-targeting drugs hold great promise for HSP due to SPAST mutations (SPG4). Supporting this concept, vinblastine has been shown to ameliorate the disease phenotype in a Drosophila model of SPG4 [104]. Additionally, microtubule-targeting drugs have been shown to rescue axonal swellings in cortical neurons in a mouse model of SPG4 [105]. In human patient-derived olfactory neurosphere-derived cells, SPAST mutations result in decreased levels of acetylated α-tubulin, a marker of stabilised microtubules, as well as reduced speed of peroxisome trafficking [106]. Tubulin binding drugs such as taxol, vinblastine, epothilone D and noscapine may increase acetylated alpha tubulin and thereby restore axonal transport, directly targeting the mechanism involved in SPG4 [106, 107].
Several genes associated with HSP phenotypes disturb lipid metabolic pathways as a potential therapeutic target, including CYP7B1, EPT1, PCYT2, DDHD1, DDHD2, PNPLA6, B4GALNT1, CYP2U1, FA2H, GBA2, PLA2G6, ATP13A2, BSCL2, C19orf12, ERLIN2, SPART, SPAST, SPG11, SPG15, ATL1 and REEP1 [108]. SPG5 is a recessive cause of HSP due to mutations in the CYP7B1 gene encoding a distinct microsomal oxysterol-7α-hydroxylase. This enzyme is involved in the degradation of cholesterol into primary bile acids. CYP7B1 deficiency results in accumulation of neurotoxic oxysterols, with elevation of 25-hydroxycholesterol (25-OHC) and 27-hydroxycholesterol (27-OHC) in the plasma and a much higher increase of 27-OHC in the CSF [109, 110]. Two recent studies have explored the use of drugs to lower cholesterol biomarkers in HSP. A study by Marelli and colleagues used atorvastatin, chenodeoxycholic acid and resveratrol in 21 patients with SPG5A and assessed 25-OHC and 27-OHC as diagnostic biomarkers [111••]. Treatment with atorvastatin decreased plasma 27-OHC but did not change the 27-OHC to total cholesterol ratio or 25-OHC levels. Marelli and colleagues also identified an abnormal bile acids profile in patients with SPG5, with a reduction in total serum bile acids and a decrease of ursodeoxycholic and lithocholic acids in comparison to deoxycholic acid. Treatment with chenodeoxycholic acid restored the bile acid profile. The authors concluded that atorvastatin and chenodeoxycholic acid may be worth considering for the treatment of SPG5A. A randomised placebo control trial by Schols and colleagues found that atorvastatin treatment reduced 27-OHC and 25-OHC in the serum, although 27-OHC was not significantly reduced in the cerebrospinal fluid [109••]. It is important to note that both these trials have demonstrated a reduction in cholesterol/bile acid biomarkers, but without benefit in terms of clinical, imaging, or electrophysiological outcome measures.
A more recent study explored the use of intravenous formulated mouse and human CYP7B1 mRNA in mice lacking the endogenous Cyp7b1 gene mutated in SPG5A. Results indicated that the treatment was safe and demonstrated a reduction in neurotoxic oxysterols in the liver, serum and to some degree in the brain, suggesting that this may be a valid strategy for the treatment of this condition [112].
Conclusion
The genetic diagnosis of HSP is complex and can represent a major challenge for clinicians. The complexity arises in part because of the high degree of genetic heterogeneity, with over 80 different genetic forms, and a growing number of genes being identified. Furthermore, there is a high level of phenotypic complexity, with HSP clinically and genetically overlapping with a variety of neurological phenotypes, including inherited forms of cerebellar ataxia, ALS, Parkinson’s disease, and peripheral neuropathy. There are many conditions that mimic HSP that the clinician should be alert for, and it may be particularly important to detect the rare HSP mimics that have management implications.
An understanding of the genetic and phenotypic complexity underlying HSP is essential to guide genetic testing strategies. Gene panels are commonly used, but the gene panel itself needs to be comprehensive to encompass the large number of genes involved. Panels must be regularly curated given that the rapid rate of gene discovery as they can quickly become obsolete. Furthermore, gene panels are typically based on a specific phenotypic category, and a genetic diagnosis may be missed if the responsible mutation is in a gene outside of that disease category. Thus, directed testing approaches such as gene panels may miss unanticipated findings [54].
Hypothesis-free approaches such as clinical WES, WES and WGS somewhat overcome the potential problems of gene panels by allowing for the potential interrogation of many relevant genes. However, clinical WES and WES may miss certain mutation types such as CNVs, SVs and repeat expansions, potentially detectable with WGS. In fact, WGS may be the most comprehensive method for coverage and detection of mutation types but is unfortunately limited by cost.
Next-generation sequencing has greatly improved our ability to detect a genetic diagnosis in HSP. Yet, large studies have shown that the diagnostic rate for HSP is still only about 45–50% of cases [113, 114]. The genetic diagnosis of HSP still represents a great challenge for clinicians, and there are no clear guidelines available about which approach to choose. However, it will become increasingly important to identify a genetic diagnosis in a rapid and accurate manner for enrolment in clinical trials and as targeted treatments become available.
Acknowledgements
We would like to thank Dr Smitha Sukumar for editing the manuscript.
Funding
KK receives funding to study dystonia from the Paul Ainsworth Family Foundation and receives a Working Group Co-Lead Award from the Michael J. Fox Foundation, Aligning Science Across Parkinson’s (ASAP) initiative.
Compliance with Ethical Standards
Conflict of Interest
Lydia Saputra and Kishore Raj Kumar have no conflict of interest.
Human and Animal Rights and Informed Consent
This article does not contain any original studies with human or animal subjects performed by any of the authors.
Footnotes
This article is part of the Topical Collection on Genetics
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Lydia Saputra, Email: lydia.saputra@health.nsw.gov.au.
Kishore Raj Kumar, Email: kkum4618@uni.sydney.edu.au.
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
- 1.Kumar KR, Blair NF, Sue CM. An update on the hereditary spastic paraplegias: new genes and new disease models. Mov Disord Clin Pract. 2015;2(3):213–223. doi: 10.1002/mdc3.12184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fink JK, Heiman-Patterson T, Bird T, Cambi F, Dube MP, Figlewicz DA, Fink JK, Haines JL, Heiman-Patterson T, Hentati A, Pericak-Vance MA, Raskind W, Rouleau GA, Siddique T. Hereditary spastic paraplegia: advances in genetic research. Hereditary Spastic Paraplegia Working group. Neurology. 1996;46(6):1507–1514. doi: 10.1212/wnl.46.6.1507. [DOI] [PubMed] [Google Scholar]
- 3.Finsterer J, Loscher W, Quasthoff S, Wanschitz J, Auer-Grumbach M, Stevanin G. Hereditary spastic paraplegias with autosomal dominant, recessive, X-linked, or maternal trait of inheritance. J Neurol Sci. 2012;318(1-2):1–18. doi: 10.1016/j.jns.2012.03.025. [DOI] [PubMed] [Google Scholar]
- 4.Marras C, Lang A, van de Warrenburg BP, Sue CM, Tabrizi SJ, Bertram L, Mercimek-Mahmutoglu S, Ebrahimi-Fakhari D, Warner TT, Durr A, Assmann B, Lohmann K, Kostic V, Klein C. Nomenclature of genetic movement disorders: recommendations of the international Parkinson and movement disorder society task force. Mov Disord. 2016;31(4):436–457. doi: 10.1002/mds.26527. [DOI] [PubMed] [Google Scholar]
- 5.Ruano L, Melo C, Silva MC, Coutinho P. The global epidemiology of hereditary ataxia and spastic paraplegia: a systematic review of prevalence studies. Neuroepidemiology. 2014;42(3):174–183. doi: 10.1159/000358801. [DOI] [PubMed] [Google Scholar]
- 6.McKusick VA. Mendelian inheritance in man and its online version, OMIM. Am J Hum Genet. 2007;80(4):588–604. doi: 10.1086/514346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Das Bhowmik A, Patil SJ, Deshpande DV, Bhat V, Dalal A. Novel splice-site variant of UCHL1 in an Indian family with autosomal recessive spastic paraplegia-79. J Hum Genet. 2018;63(8):927–933. doi: 10.1038/s10038-018-0463-6. [DOI] [PubMed] [Google Scholar]
- 8.•.Farazi Fard MA, Rebelo AP, Buglo E, Nemati H, Dastsooz H, Gehweiler I, et al. Truncating mutations in UBAP1 cause hereditary spastic paraplegia. Am J Hum Genet. 2019;104(4):767–773. doi: 10.1016/j.ajhg.2019.03.001\. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gu S, Chen CA, Rosenfeld JA, Cope H, Launay N, Flanigan KM, Waldrop MA, Schrader R, Juusola J, Goker-Alpan O, Milunsky A, Schlüter A, Troncoso M, Pujol A, Tan QKG, Schaaf CP, Meng L. Truncating variants in UBAP1 associated with childhood-onset nonsyndromic hereditary spastic paraplegia. Hum Mutat. 2019;41:632–640. doi: 10.1002/humu.23950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin X, Su HZ, Dong EL, Lin XH, Zhao M, Yang C, Wang C, Wang J, Chen YJ, Yu H, Xu J, Ma LX, Xiong ZQ, Wang N, Chen WJ. Stop-gain mutations in UBAP1 cause pure autosomal-dominant spastic paraplegia. Brain. 2019;142(8):2238–2252. doi: 10.1093/brain/awz158. [DOI] [PubMed] [Google Scholar]
- 11.Nan H, Ichinose Y, Tanaka M, Koh K, Ishiura H, Mitsui J, Mizukami H, Morimoto M, Hamada S, Ohtsuka T, Tsuji S, Takiyama Y. UBAP1 mutations cause juvenile-onset hereditary spastic paraplegias (SPG80) and impair UBAP1 targeting to endosomes. J Hum Genet. 2019;64(11):1055–1065. doi: 10.1038/s10038-019-0670-9. [DOI] [PubMed] [Google Scholar]
- 12.Ahmed MY, Al-Khayat A, Al-Murshedi F, Al-Futaisi A, Chioza BA, Pedro Fernandez-Murray J, et al. A mutation of EPT1 (SELENOI) underlies a new disorder of Kennedy pathway phospholipid biosynthesis. Brain. 2017;140(3):547–554. doi: 10.1093/brain/aww318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vaz FM, McDermott JH, Alders M, Wortmann SB, Kolker S, Pras-Raves ML, et al. Mutations in PCYT2 disrupt etherlipid biosynthesis and cause a complex hereditary spastic paraplegia. Brain. 2019;142(11):3382–3397. doi: 10.1093/brain/awz291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.•.Wagner M, Osborn DPS, Gehweiler I, Nagel M, Ulmer U, Bakhtiari S, et al. Bi-allelic variants in RNF170 are associated with hereditary spastic paraplegia. Nat Commun. 2019;10(1):4790. doi: 10.1038/s41467-019-12620-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Husain RA, Grimmel M, Wagner M, Hennings JC, Marx C, Feichtinger RG, Saadi A, Rostásy K, Radelfahr F, Bevot A, Döbler-Neumann M, Hartmann H, Colleaux L, Cordts I, Kobeleva X, Darvish H, Bakhtiari S, Kruer MC, Besse A, Ng ACH, Chiang D, Bolduc F, Tafakhori A, Mane S, Ghasemi Firouzabadi S, Huebner AK, Buchert R, Beck-Woedl S, Müller AJ, Laugwitz L, Nägele T, Wang ZQ, Strom TM, Sturm M, Meitinger T, Klockgether T, Riess O, Klopstock T, Brandl U, Hübner CA, Deschauer M, Mayr JA, Bonnen PE, Krägeloh-Mann I, Wortmann SB, Haack TB. Bi-allelic HPDL variants cause a neurodegenerative disease ranging from neonatal encephalopathy to adolescent-onset spastic paraplegia. Am J Hum Genet. 2020;107(2):364–373. doi: 10.1016/j.ajhg.2020.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferdinandusse S, McWalter K, Te Brinke H, Lodewijk IJ, Mooijer PM, Ruiter JPN, et al. An autosomal dominant neurological disorder caused by de novo variants in FAR1 resulting in uncontrolled synthesis of ether lipids. Genet Med. 2020. 10.1038/s41436-020-01027-3. [DOI] [PMC free article] [PubMed]
- 17.Lin P, Li J, Liu Q, Mao F, Li J, Qiu R, Hu H, Song Y, Yang Y, Gao G, Yan C, Yang W, Shao C, Gong Y. A missense mutation in SLC33A1, which encodes the acetyl-CoA transporter, causes autosomal-dominant spastic paraplegia (SPG42) Am J Hum Genet. 2008;83(6):752–759. doi: 10.1016/j.ajhg.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schlipf NA, Beetz C, Schule R, Stevanin G, Erichsen AK, Forlani S, et al. A total of 220 patients with autosomal dominant spastic paraplegia do not display mutations in the SLC33A1 gene (SPG42) Eur J Hum Genet. 2010;18(9):1065–1067. doi: 10.1038/ejhg.2010.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Synofzik M, Schule R. Overcoming the divide between ataxias and spastic paraplegias: Shared phenotypes, genes, and pathways. Mov Disord. 2017;32(3):332–345. doi: 10.1002/mds.26944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kang C, Liang C, Ahmad KE, Gu Y, Siow SF, Colebatch JG, Whyte S, Ng K, Cremer PD, Corbett AJ, Davis RL, Roscioli T, Cowley MJ, Park JS, Sue CM, Kumar KR. High degree of genetic heterogeneity for hereditary cerebellar ataxias in Australia. Cerebellum. 2019;18(1):137–146. doi: 10.1007/s12311-018-0969-7. [DOI] [PubMed] [Google Scholar]
- 21.Kumar KR, Blair NF, Vandebona H, Liang C, Ng K, Sharpe DM, Grünewald A, Gölnitz U, Saviouk V, Rolfs A, Klein C, Sue CM. Targeted next generation sequencing in SPAST-negative hereditary spastic paraplegia. J Neurol. 2013;260(10):2516–2522. doi: 10.1007/s00415-013-7008-x. [DOI] [PubMed] [Google Scholar]
- 22.Wali G, Kumar KR, Liyanage E, Davis RL, Mackay-Sim A, Sue CM. Mitochondrial function in hereditary spastic paraplegia: deficits in SPG7 but not SPAST patient-derived stem cells. Front Neurosci. 2020;14:820. doi: 10.3389/fnins.2020.00820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mancini C, Giorgio E, Rubegni A, Pradotto L, Bagnoli S, Rubino E, Prontera P, Cavalieri S, di Gregorio E, Ferrero M, Pozzi E, Riberi E, Ferrero P, Nigro P, Mauro A, Zibetti M, Tessa A, Barghigiani M, Antenora A, Sirchia F, Piacentini S, Silvestri G, de Michele G, Filla A, Orsi L, Santorelli FM, Brusco A. Prevalence and phenotype of the c.1529C>T SPG7 variant in adult-onset cerebellar ataxia in Italy. Eur J Neurol. 2019;26(1):80–86. doi: 10.1111/ene.13768. [DOI] [PubMed] [Google Scholar]
- 24.•.Kim A, Kumar KR, Davis RL, Mallawaarachchi AC, Gayevskiy V, Minoche AE, et al. Increased diagnostic yield of spastic paraplegia with or without cerebellar ataxia through whole-genome sequencing. Cerebellum. 2019. 10.1007/s12311-019-01038-0This study suggests it may be helpful to investigate spastic paraplegia and ataxia as a single disease spectrum or common entity, using WGS to interrogate both HSP and ataxia genes. [DOI] [PubMed]
- 25.Wang Y, Hersheson J, Lopez D, Hammer M, Liu Y, Lee KH, Pinto V, Seinfeld J, Wiethoff S, Sun J, Amouri R, Hentati F, Baudry N, Tran J, Singleton AB, Coutelier M, Brice A, Stevanin G, Durr A, Bi X, Houlden H, Baudry M. Defects in the CAPN1 gene result in alterations in cerebellar development and cerebellar ataxia in mice and humans. Cell Rep. 2016;16(1):79–91. doi: 10.1016/j.celrep.2016.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shetty A, Gan-Or Z, Ashtiani S, Ruskey JA, van de Warrenburg B, Wassenberg T, Kamsteeg EJ, Rouleau GA, Suchowersky O. CAPN1 mutations: expanding the CAPN1-related phenotype: from hereditary spastic paraparesis to spastic ataxia. Eur J Med Genet. 2018;62:103605. doi: 10.1016/j.ejmg.2018.12.010. [DOI] [PubMed] [Google Scholar]
- 27.Gan-Or Z, Bouslam N, Birouk N, Lissouba A, Chambers DB, Veriepe J, et al. Mutations in CAPN1 cause autosomal-recessive hereditary spastic paraplegia. Am J Hum Genet. 2016;98(5):1038–1046. doi: 10.1016/j.ajhg.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.•.Nicita F, Ginevrino M, Travaglini L, D’Arrigo S, Zorzi G, Borgatti R, et al. Heterozygous KIF1A variants underlie a wide spectrum of neurodevelopmental and neurodegenerative disorders. J Med Genet. 2020. 10.1136/jmedgenet-2020-107007Recent study showing that heterozygousKIF1Avariants can cause both HSP and ataxia phenotypes. [DOI] [PubMed]
- 29.Synofzik M, Soehn AS, Gburek-Augustat J, Schicks J, Karle KN, Schule R, et al. Autosomal recessive spastic ataxia of Charlevoix Saguenay (ARSACS): expanding the genetic, clinical and imaging spectrum. Orphanet J Rare Dis. 2013;8:41. doi: 10.1186/1750-1172-8-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seong E, Insolera R, Dulovic M, Kamsteeg EJ, Trinh J, Bruggemann N, et al. Mutations in VPS13D lead to a new recessive ataxia with spasticity and mitochondrial defects. Ann Neurol. 2018;83(6):1075–1088. doi: 10.1002/ana.25220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koh K, Ishiura H, Shimazaki H, Tsutsumiuchi M, Ichinose Y, Nan H, Hamada S, Ohtsuka T, Tsuji S, Takiyama Y. VPS13D-related disorders presenting as a pure and complicated form of hereditary spastic paraplegia. Mol Genet Genomic Med. 2020;8(3):e1108. doi: 10.1002/mgg3.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bettencourt C, Quintans B, Ros R, Ampuero I, Yanez Z, Pascual SI, et al. Revisiting genotype-phenotype overlap in neurogenetics: triplet-repeat expansions mimicking spastic paraplegias. Hum Mutat. 2012;33(9):1315–1323. doi: 10.1002/humu.22148. [DOI] [PubMed] [Google Scholar]
- 33.Ito D BSCL2-related neurologic disorders/seipinopathy. In: Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJH, Stephens K et al., editors. GeneReviews((R)). Seattle (WA)1993.
- 34.Musacchio T, Zaum AK, Uceyler N, Sommer C, Pfeifroth N, Reiners K, et al. ALS and MMN mimics in patients with BSCL2 mutations: the expanding clinical spectrum of SPG17 hereditary spastic paraplegia. J Neurol. 2017;264(1):11–20. doi: 10.1007/s00415-016-8301-2. [DOI] [PubMed] [Google Scholar]
- 35.Stevanin G, Santorelli FM, Azzedine H, Coutinho P, Chomilier J, Denora PS, Martin E, Ouvrard-Hernandez AM, Tessa A, Bouslam N, Lossos A, Charles P, Loureiro JL, Elleuch N, Confavreux C, Cruz VT, Ruberg M, Leguern E, Grid D, Tazir M, Fontaine B, Filla A, Bertini E, Durr A, Brice A. Mutations in SPG11, encoding spatacsin, are a major cause of spastic paraplegia with thin corpus callosum. Nat Genet. 2007;39(3):366–372. doi: 10.1038/ng1980. [DOI] [PubMed] [Google Scholar]
- 36.Montecchiani C, Pedace L, Lo Giudice T, Casella A, Mearini M, Gaudiello F, Pedroso JL, Terracciano C, Caltagirone C, Massa R, St George-Hyslop PH, Barsottini OGP, Kawarai T, Orlacchio A. ALS5/SPG11/KIAA1840 mutations cause autosomal recessive axonal Charcot-Marie-Tooth disease. Brain. 2016;139(Pt 1):73–85. doi: 10.1093/brain/awv320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Novarino G, Fenstermaker AG, Zaki MS, Hofree M, Silhavy JL, Heiberg AD, Abdellateef M, Rosti B, Scott E, Mansour L, Masri A, Kayserili H, al-Aama JY, Abdel-Salam GMH, Karminejad A, Kara M, Kara B, Bozorgmehri B, Ben-Omran T, Mojahedi F, Mahmoud IGED, Bouslam N, Bouhouche A, Benomar A, Hanein S, Raymond L, Forlani S, Mascaro M, Selim L, Shehata N, al-Allawi N, Bindu PS, Azam M, Gunel M, Caglayan A, Bilguvar K, Tolun A, Issa MY, Schroth J, Spencer EG, Rosti RO, Akizu N, Vaux KK, Johansen A, Koh AA, Megahed H, Durr A, Brice A, Stevanin G, Gabriel SB, Ideker T, Gleeson JG. Exome sequencing links corticospinal motor neuron disease to common neurodegenerative disorders. Science. 2014;343(6170):506–511. doi: 10.1126/science.1247363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gonzalez M, McLaughlin H, Houlden H, Guo M, Yo-Tsen L, Hadjivassilious M, Speziani F, Yang XL, Antonellis A, Reilly MM, Züchner S, Inherited Neuropathy Consortium (INC) Exome sequencing identifies a significant variant in methionyl-tRNA synthetase (MARS) in a family with late-onset CMT2. J Neurol Neurosurg Psychiatry. 2013;84(11):1247–1249. doi: 10.1136/jnnp-2013-305049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.de Sainte Agathe JM, Mercier S, Mahe JY, Pereon Y, Buratti J, Tissier L, et al. RNF170-related hereditary spastic paraplegia: confirmation by a novel mutation. Mov Disord. 2020. 10.1002/mds.28371. [DOI] [PubMed]
- 40.Cortese A, Callegari I, Curro R, Vegezzi E, Colnaghi S, Versino M, et al. Mutation in RNF170 causes sensory ataxic neuropathy with vestibular areflexia: a CANVAS mimic. J Neurol Neurosurg Psychiatry. 2020;91(11):1237–1238. doi: 10.1136/jnnp-2020-323719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tunca C, Akcimen F, Coskun C, Gundogdu-Eken A, Kocoglu C, Cevik B, et al. ERLIN1 mutations cause teenage-onset slowly progressive ALS in a large Turkish pedigree. Eur J Hum Genet. 2018;26(5):745–748. doi: 10.1038/s41431-018-0107-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Amador MD, Muratet F, Teyssou E, Banneau G, Danel-Brunaud V, Allart E, et al. Spastic paraplegia due to recessive or dominant mutations in ERLIN2 can convert to ALS. Neurol Genet. 2019;5(6):e374. doi: 10.1212/NXG.0000000000000374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Al-Saif A, Bohlega S, Al-Mohanna F. Loss of ERLIN2 function leads to juvenile primary lateral sclerosis. Ann Neurol. 2012;72(4):510–516. doi: 10.1002/ana.23641. [DOI] [PubMed] [Google Scholar]
- 44.Pednekar D, Wang Y, Fedotova TV, Wojcikiewicz RJ. Clustered hydrophobic amino acids in amphipathic helices mediate erlin1/2 complex assembly. Biochem Biophys Res Commun. 2011;415(1):135–140. doi: 10.1016/j.bbrc.2011.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Daoud H, Zhou S, Noreau A, Sabbagh M, Belzil V, Dionne-Laporte A, et al. Exome sequencing reveals SPG11 mutations causing juvenile ALS. Neurobiol Aging. 2012;33(4):839 e5–839 e9. doi: 10.1016/j.neurobiolaging.2011.11.012. [DOI] [PubMed] [Google Scholar]
- 46.Faber I, Martinez ARM, Martins CR, Jr, Maia ML, Souza JP, Lourenco CM, et al. SPG11-related parkinsonism: clinical profile, molecular imaging and l-dopa response. Mov Disord. 2018;33(10):1650–1656. doi: 10.1002/mds.27491. [DOI] [PubMed] [Google Scholar]
- 47.Estrada-Cuzcano A, Martin S, Chamova T, Synofzik M, Timmann D, Holemans T, Andreeva A, Reichbauer J, de Rycke R, Chang DI, van Veen S, Samuel J, Schöls L, Pöppel T, Mollerup Sørensen D, Asselbergh B, Klein C, Zuchner S, Jordanova A, Vangheluwe P, Tournev I, Schüle R. Loss-of-function mutations in the ATP13A2/PARK9 gene cause complicated hereditary spastic paraplegia (SPG78) Brain. 2017;140(2):287–305. doi: 10.1093/brain/aww307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ramirez A, Heimbach A, Grundemann J, Stiller B, Hampshire D, Cid LP, et al. Hereditary parkinsonism with dementia is caused by mutations in ATP13A2, encoding a lysosomal type 5 P-type ATPase. Nat Genet. 2006;38(10):1184–1191. doi: 10.1038/ng1884. [DOI] [PubMed] [Google Scholar]
- 49.Leroy E, Boyer R, Auburger G, Leube B, Ulm G, Mezey E, Harta G, Brownstein MJ, Jonnalagada S, Chernova T, Dehejia A, Lavedan C, Gasser T, Steinbach PJ, Wilkinson KD, Polymeropoulos MH. The ubiquitin pathway in Parkinson’s disease. Nature. 1998;395(6701):451–452. doi: 10.1038/26652. [DOI] [PubMed] [Google Scholar]
- 50.Bilguvar K, Tyagi NK, Ozkara C, Tuysuz B, Bakircioglu M, Choi M, Delil S, Caglayan AO, Baranoski JF, Erturk O, Yalcinkaya C, Karacorlu M, Dincer A, Johnson MH, Mane S, Chandra SS, Louvi A, Boggon TJ, Lifton RP, Horwich AL, Gunel M. Recessive loss of function of the neuronal ubiquitin hydrolase UCHL1 leads to early-onset progressive neurodegeneration. Proc Natl Acad Sci U S A. 2013;110(9):3489–3494. doi: 10.1073/pnas.1222732110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Osaka H, Wang YL, Takada K, Takizawa S, Setsuie R, Li H, Sato Y, Nishikawa K, Sun YJ, Sakurai M, Harada T, Hara Y, Kimura I, Chiba S, Namikawa K, Kiyama H, Noda M, Aoki S, Wada K. Ubiquitin carboxy-terminal hydrolase L1 binds to and stabilizes monoubiquitin in neuron. Hum Mol Genet. 2003;12(16):1945–1958. doi: 10.1093/hmg/ddg211. [DOI] [PubMed] [Google Scholar]
- 52.Arif B, Kumar KR, Seibler P, Vulinovic F, Fatima A, Winkler S, Nürnberg G, Thiele H, Nürnberg P, Jamil AZ, Brüggemann A, Abbas G, Klein C, Naz S, Lohmann K. A novel OPA3 mutation revealed by exome sequencing: an example of reverse phenotyping. JAMA Neurol. 2013;70(6):783–787. doi: 10.1001/jamaneurol.2013.1174. [DOI] [PubMed] [Google Scholar]
- 53.Kumar KR, Wali G, Davis RL, Mallawaarachchi AC, Palmer EE, Gayevskiy V, Minoche AE, Veivers D, Dinger ME, Mackay-Sim A, Cowley MJ, Sue CM. Expanding the spectrum of PEX16 mutations and novel insights into disease mechanisms. Mol Genet Metab Rep. 2018;16:46–51. doi: 10.1016/j.ymgmr.2018.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kumar KR, Wali GM, Kamate M, Wali G, Minoche AE, Puttick C, Pinese M, Gayevskiy V, Dinger ME, Roscioli T, Sue CM, Cowley MJ. Defining the genetic basis of early onset hereditary spastic paraplegia using whole genome sequencing. Neurogenetics. 2016;17(4):265–270. doi: 10.1007/s10048-016-0495-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lohmann K, Wilcox RA, Winkler S, Ramirez A, Rakovic A, Park JS, Arns B, Lohnau T, Groen J, Kasten M, Brüggemann N, Hagenah J, Schmidt A, Kaiser FJ, Kumar KR, Zschiedrich K, Alvarez-Fischer D, Altenmüller E, Ferbert A, Lang AE, Münchau A, Kostic V, Simonyan K, Agzarian M, Ozelius LJ, Langeveld APM, Sue CM, Tijssen MAJ, Klein C. Whispering dysphonia (DYT4 dystonia) is caused by a mutation in the TUBB4 gene. Ann Neurol. 2013;73(4):537–545. doi: 10.1002/ana.23829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kancheva D, Chamova T, Guergueltcheva V, Mitev V, Azmanov DN, Kalaydjieva L, Tournev I, Jordanova A. Mosaic dominant TUBB4A mutation in an inbred family with complicated hereditary spastic paraplegia. Mov Disord. 2015;30(6):854–858. doi: 10.1002/mds.26196. [DOI] [PubMed] [Google Scholar]
- 57.Shribman S, Reid E, Crosby AH, Houlden H, Warner TT. Hereditary spastic paraplegia: from diagnosis to emerging therapeutic approaches. Lancet Neurol. 2019;18(12):1136–1146. doi: 10.1016/S1474-4422(19)30235-2. [DOI] [PubMed] [Google Scholar]
- 58.Fan Z, Greenwood R, Felix AC, Shiloh-Malawsky Y, Tennison M, Roche M, et al. GCH1 heterozygous mutation identified by whole-exome sequencing as a treatable condition in a patient presenting with progressive spastic paraplegia. J Neurol. 2014;261(3):622–624. doi: 10.1007/s00415-014-7265-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wei Y, Zhou Y, Yuan J, Ni J, Qian M, Cui L, Peng B. Treatable cause of hereditary spastic paraplegia: eight cases of combined homocysteinaemia with methylmalonic aciduria. J Neurol. 2019;266(10):2434–2439. doi: 10.1007/s00415-019-09432-8. [DOI] [PubMed] [Google Scholar]
- 60.Lossos A, Teltsh O, Milman T, Meiner V, Rozen R, Leclerc D, Schwahn BC, Karp N, Rosenblatt DS, Watkins D, Shaag A, Korman SH, Heyman SN, Gal A, Newman JP, Steiner-Birmanns B, Abramsky O, Kohn Y. Severe methylenetetrahydrofolate reductase deficiency: clinical clues to a potentially treatable cause of adult-onset hereditary spastic paraplegia. JAMA Neurol. 2014;71(7):901–904. doi: 10.1001/jamaneurol.2014.116. [DOI] [PubMed] [Google Scholar]
- 61.Nicholls Z, Hobson E, Martindale J, Shaw PJ. Diagnosis of spinal xanthomatosis by next-generation sequencing: identifying a rare, treatable mimic of hereditary spastic paraparesis. Pract Neurol. 2015;15(4):280–283. doi: 10.1136/practneurol-2015-001117. [DOI] [PubMed] [Google Scholar]
- 62.Raymond GV, Moser AB, Fatemi A. X-linked adrenoleukodystrophy. In: Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJH, Stephens K et al., editors. GeneReviews((R)). Seattle (WA)1993. [PubMed]
- 63.Tsang JP, Poon WL, Luk HM, Fung CW, Ching CK, Mak CM, et al. Arginase deficiency with new phenotype and a novel mutation: contemporary summary. Pediatr Neurol. 2012;47(4):263–269. doi: 10.1016/j.pediatrneurol.2012.06.012. [DOI] [PubMed] [Google Scholar]
- 64.Wolf B. Biotinidase deficiency should be considered in individuals exhibiting myelopathy with or without and vision loss. Mol Genet Metab. 2015;116(3):113–118. doi: 10.1016/j.ymgme.2015.08.012. [DOI] [PubMed] [Google Scholar]
- 65.Wolf B. Biotinidase deficiency. In: Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJH, Stephens K et al., editors. GeneReviews((R)). Seattle (WA)1993.
- 66.Wang Y, Smith C, Parboosingh JS, Khan A, Innes M, Hekimi S. Pathogenicity of two COQ7 mutations and responses to 2,4-dihydroxybenzoate bypass treatment. J Cell Mol Med. 2017;21(10):2329–2343. doi: 10.1111/jcmm.13154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Salviati L, Trevisson E, Doimo M, Navas P. Primary coenzyme Q10 deficiency. In: Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJH, Stephens K et al., editors. GeneReviews((R)). Seattle (WA)1993. [PubMed]
- 68.Verrips A, Hoefsloot LH, Steenbergen GC, Theelen JP, Wevers RA, Gabreels FJ, et al. Clinical and molecular genetic characteristics of patients with cerebrotendinous xanthomatosis. Brain. 2000;123(Pt 5):908–919. doi: 10.1093/brain/123.5.908. [DOI] [PubMed] [Google Scholar]
- 69.Kasim S, Moo LR, Zschocke J, Jinnah HA. Phenylketonuria presenting in adulthood as progressive spastic paraparesis with dementia. J Neurol Neurosurg Psychiatry. 2001;71(6):795–797. doi: 10.1136/jnnp.71.6.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Verrotti A, Di Francesco L, Striano P. GLUT1 deficiency and pediatric-onset hereditary spastic paraplegia: a new association. Eur J Paediatr Neurol. 2019;23(2):233–234. doi: 10.1016/j.ejpn.2019.02.010. [DOI] [PubMed] [Google Scholar]
- 71.Vazquez-Costa JF, Bataller L, Vilchez JJ. Primary lateral sclerosis and hereditary spastic paraplegia in sporadic patients. An important distinction in descriptive studies. Ann Neurol. 2016;80(1):169–170. doi: 10.1002/ana.24671. [DOI] [PubMed] [Google Scholar]
- 72.Geevasinga N, Menon P, Sue CM, Kumar KR, Ng K, Yiannikas C, Kiernan MC, Vucic S. Cortical excitability changes distinguish the motor neuron disease phenotypes from hereditary spastic paraplegia. Eur J Neurol. 2015;22(5):826–831. doi: 10.1111/ene.12669. [DOI] [PubMed] [Google Scholar]
- 73.Brugman F, Veldink JH, Franssen H, de Visser M, de Jong JM, Faber CG, et al. Differentiation of hereditary spastic paraparesis from primary lateral sclerosis in sporadic adult-onset upper motor neuron syndromes. Arch Neurol. 2009;66(4):509–514. doi: 10.1001/archneurol.2009.19. [DOI] [PubMed] [Google Scholar]
- 74.Pearson TS, Pons R, Ghaoui R, Sue CM. Genetic mimics of cerebral palsy. Mov Disord. 2019;34(5):625–636. doi: 10.1002/mds.27655. [DOI] [PubMed] [Google Scholar]
- 75.Rainier S, Sher C, Reish O, Thomas D, Fink JK. De novo occurrence of novel SPG3A/atlastin mutation presenting as cerebral palsy. Arch Neurol. 2006;63(3):445–447. doi: 10.1001/archneur.63.3.445. [DOI] [PubMed] [Google Scholar]
- 76.Rubegni A, Battisti C, Tessa A, Cerase A, Doccini S, Malandrini A, Santorelli FM, Federico A. SPG2 mimicking multiple sclerosis in a family identified using next generation sequencing. J Neurol Sci. 2017;375:198–202. doi: 10.1016/j.jns.2017.01.069. [DOI] [PubMed] [Google Scholar]
- 77.Jia X, Madireddy L, Caillier S, Santaniello A, Esposito F, Comi G, Stuve O, Zhou Y, Taylor B, Kilpatrick T, Martinelli-Boneschi F, Cree BAC, Oksenberg JR, Hauser SL, Baranzini SE. Genome sequencing uncovers phenocopies in primary progressive multiple sclerosis. Ann Neurol. 2018;84(1):51–63. doi: 10.1002/ana.25263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fink JK. Hereditary spastic paraplegia: clinico-pathologic features and emerging molecular mechanisms. Acta Neuropathol. 2013;126(3):307–328. doi: 10.1007/s00401-013-1115-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bras J, Verloes A, Schneider SA, Mole SE, Guerreiro RJ. Mutation of the parkinsonism gene ATP13A2 causes neuronal ceroid-lipofuscinosis. Hum Mol Genet. 2012;21(12):2646–2650. doi: 10.1093/hmg/dds089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schneider SA, Paisan-Ruiz C, Quinn NP, Lees AJ, Houlden H, Hardy J, Bhatia KP. ATP13A2 mutations (PARK9) cause neurodegeneration with brain iron accumulation. Mov Disord. 2010;25(8):979–984. doi: 10.1002/mds.22947. [DOI] [PubMed] [Google Scholar]
- 81.de Bot S, Kamsteeg EJ, Van De Warrenburg BPC. Complicated hereditary spastic paraplegia due to ATP13A2 mutations: what’s in a name? Brain. 2017;140(12):e73. doi: 10.1093/brain/awx280. [DOI] [PubMed] [Google Scholar]
- 82.Minnerop M, Kurzwelly D, Wagner H, Soehn AS, Reichbauer J, Tao F, Rattay TW, Peitz M, Rehbach K, Giorgetti A, Pyle A, Thiele H, Altmüller J, Timmann D, Karaca I, Lennarz M, Baets J, Hengel H, Synofzik M, Atasu B, Feely S, Kennerson M, Stendel C, Lindig T, Gonzalez MA, Stirnberg R, Sturm M, Roeske S, Jung J, Bauer P, Lohmann E, Herms S, Heilmann-Heimbach S, Nicholson G, Mahanjah M, Sharkia R, Carloni P, Brüstle O, Klopstock T, Mathews KD, Shy ME, de Jonghe P, Chinnery PF, Horvath R, Kohlhase J, Schmitt I, Wolf M, Greschus S, Amunts K, Maier W, Schöls L, Nürnberg P, Zuchner S, Klockgether T, Ramirez A, Schüle R. Hypomorphic mutations in POLR3A are a frequent cause of sporadic and recessive spastic ataxia. Brain. 2017;140(6):1561–1578. doi: 10.1093/brain/awx095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gauquelin L, Tetreault M, Thiffault I, Farrow E, Miller N, Yoo B, et al. POLR3A variants in hereditary spastic paraplegia and ataxia. Brain. 2018;141(1):e1. doi: 10.1093/brain/awx290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Klebe S, Lossos A, Azzedine H, Mundwiller E, Sheffer R, Gaussen M, Marelli C, Nawara M, Carpentier W, Meyer V, Rastetter A, Martin E, Bouteiller D, Orlando L, Gyapay G, el-Hachimi KH, Zimmerman B, Gamliel M, Misk A, Lerer I, Brice A, Durr A, Stevanin G. KIF1A missense mutations in SPG30, an autosomal recessive spastic paraplegia: distinct phenotypes according to the nature of the mutations. Eur J Hum Genet. 2012;20(6):645–649. doi: 10.1038/ejhg.2011.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cheon CK, Lim SH, Kim YM, Kim D, Lee NY, Yoon TS, Kim NS, Kim E, Lee JR. Autosomal dominant transmission of complicated hereditary spastic paraplegia due to a dominant negative mutation of KIF1A, SPG30 gene. Sci Rep. 2017;7(1):12527. doi: 10.1038/s41598-017-12999-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Citterio A, Arnoldi A, Panzeri E, Merlini L, D’Angelo MG, Musumeci O, Toscano A, Bondi A, Martinuzzi A, Bresolin N, Bassi MT. Variants in KIF1A gene in dominant and sporadic forms of hereditary spastic paraparesis. J Neurol. 2015;262(12):2684–2690. doi: 10.1007/s00415-015-7899-9. [DOI] [PubMed] [Google Scholar]
- 87.Ylikallio E, Kim D, Isohanni P, Auranen M, Kim E, Lonnqvist T, et al. Dominant transmission of de novo KIF1A motor domain variant underlying pure spastic paraplegia. Eur J Hum Genet. 2015;23(10):1427–1430. doi: 10.1038/ejhg.2014.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ohba C, Haginoya K, Osaka H, Kubota K, Ishiyama A, Hiraide T, Komaki H, Sasaki M, Miyatake S, Nakashima M, Tsurusaki Y, Miyake N, Tanaka F, Saitsu H, Matsumoto N. De novo KIF1A mutations cause intellectual deficit, cerebellar atrophy, lower limb spasticity and visual disturbance. J Hum Genet. 2015;60(12):739–742. doi: 10.1038/jhg.2015.108. [DOI] [PubMed] [Google Scholar]
- 89.Esteves T, Durr A, Mundwiller E, Loureiro JL, Boutry M, Gonzalez MA, Gauthier J, el-Hachimi KH, Depienne C, Muriel MP, Acosta Lebrigio RF, Gaussen M, Noreau A, Speziani F, Dionne-Laporte A, Deleuze JF, Dion P, Coutinho P, Rouleau GA, Zuchner S, Brice A, Stevanin G, Darios F. Loss of association of REEP2 with membranes leads to hereditary spastic paraplegia. Am J Hum Genet. 2014;94(2):268–277. doi: 10.1016/j.ajhg.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Khan TN, Klar J, Tariq M, Anjum Baig S, Malik NA, Yousaf R, Baig SM, Dahl N. Evidence for autosomal recessive inheritance in SPG3A caused by homozygosity for a novel ATL1 missense mutation. Eur J Hum Genet. 2014;22(10):1180–1184. doi: 10.1038/ejhg.2014.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Willkomm L, Heredia R, Hoffmann K, Wang H, Voit T, Hoffman EP, Cirak S. Homozygous mutation in Atlastin GTPase 1 causes recessive hereditary spastic paraplegia. J Hum Genet. 2016;61(6):571–573. doi: 10.1038/jhg.2016.6. [DOI] [PubMed] [Google Scholar]
- 92.•.Aguilera-Albesa S, de la Hoz AB, Ibarluzea N, Ordonez-Castillo AR, Busto-Crespo O, Villate O, et al. Hereditary spastic paraplegia and intellectual disability: clinicogenetic lessons from a family suggesting a dual genetics diagnosis. Front Neurol. 2020;11:–41. 10.3389/fneur.2020.00041A recent study showing concurrent genetic diagnoses in a family with HSP. [DOI] [PMC free article] [PubMed]
- 93.Posey JE, Harel T, Liu P, Rosenfeld JA, James RA, Coban Akdemir ZH, Walkiewicz M, Bi W, Xiao R, Ding Y, Xia F, Beaudet AL, Muzny DM, Gibbs RA, Boerwinkle E, Eng CM, Sutton VR, Shaw CA, Plon SE, Yang Y, Lupski JR. Resolution of disease phenotypes resulting from multilocus genomic variation. N Engl J Med. 2017;376(1):21–31. doi: 10.1056/NEJMoa1516767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.•.Verdura E, Schluter A, Fernandez-Eulate G, Ramos-Martin R, Zulaica M, Planas-Serra L, et al. A deep intronic splice variant advises reexamination of presumably dominant SPG7 Cases. Ann Clin Transl Neurol. 2020;7(1):105–111. doi: 10.1002/acn3.50967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sanchez-Ferrero E, Coto E, Beetz C, Gamez J, Corao AI, Diaz M, et al. SPG7 mutational screening in spastic paraplegia patients supports a dominant effect for some mutations and a pathogenic role for p.A510V. Clin Genet. 2013;83(3):257–262. doi: 10.1111/j.1399-0004.2012.01896.x. [DOI] [PubMed] [Google Scholar]
- 96.Kumar KR, Cowley MJ, Davis RL. Next-generation sequencing and emerging technologies. Semin Thromb Hemost. 2019;45:661–673. doi: 10.1055/s-0039-1688446. [DOI] [PubMed] [Google Scholar]
- 97.Meienberg J, Bruggmann R, Oexle K, Matyas G. Clinical sequencing: is WGS the better WES? Hum Genet. 2016;135(3):359–362. doi: 10.1007/s00439-015-1631-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gross AM, Ajay SS, Rajan V, Brown C, Bluske K, Burns NJ, Chawla A, Coffey AJ, Malhotra A, Scocchia A, Thorpe E, Dzidic N, Hovanes K, Sahoo T, Dolzhenko E, Lajoie B, Khouzam A, Chowdhury S, Belmont J, Roller E, Ivakhno S, Tanner S, McEachern J, Hambuch T, Eberle M, Hagelstrom RT, Bentley DR, Perry DL, Taft RJ. Copy-number variants in clinical genome sequencing: deployment and interpretation for rare and undiagnosed disease. Genet Med. 2019;21(5):1121–1130. doi: 10.1038/s41436-018-0295-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kirk EP, Barlow-Stewart K, Selvanathan A, Josephi-Taylor S, Worgan L, Rajagopalan S, Cowley MJ, Gayevskiy V, Bittles A, Burnett L, Elakis G, Lo W, Buckley M, Colley A, Roscioli T. Beyond the panel: preconception screening in consanguineous couples using the TruSight One “clinical exome”. Genet Med. 2018;21:608–612. doi: 10.1038/s41436-018-0082-9. [DOI] [PubMed] [Google Scholar]
- 100.Beetz C, Nygren AO, Schickel J, Auer-Grumbach M, Burk K, Heide G, et al. High frequency of partial SPAST deletions in autosomal dominant hereditary spastic paraplegia. Neurology. 2006;67(11):1926–1930. doi: 10.1212/01.wnl.0000244413.49258.f5. [DOI] [PubMed] [Google Scholar]
- 101.Dolzhenko E, van Vugt J, Shaw RJ, Bekritsky MA, van Blitterswijk M, Narzisi G, et al. Detection of long repeat expansions from PCR-free whole-genome sequence data. Genome Res. 2017;27(11):1895–1903. doi: 10.1101/gr.225672.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wagner M, Berutti R, Lorenz-Depiereux B, Graf E, Eckstein G, Mayr JA, Meitinger T, Ahting U, Prokisch H, Strom TM, Wortmann SB. Mitochondrial DNA mutation analysis from exome sequencing-a more holistic approach in diagnostics of suspected mitochondrial disease. J Inherit Metab Dis. 2019;42(5):909–917. doi: 10.1002/jimd.12109. [DOI] [PubMed] [Google Scholar]
- 103.Puttick C, Kumar KR, Davis RL, Pinese M, Thomas DM, Dinger ME et al. <em>mity</em>: a highly sensitive mitochondrial variant analysis pipeline for whole genome sequencing data. bioRxiv. 2019:852210. 10.1101/852210.
- 104.Orso G, Martinuzzi A, Rossetto MG, Sartori E, Feany M, Daga A. Disease-related phenotypes in a Drosophila model of hereditary spastic paraplegia are ameliorated by treatment with vinblastine. J Clin Invest. 2005;115(11):3026–3034. doi: 10.1172/JCI24694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Fassier C, Tarrade A, Peris L, Courageot S, Mailly P, Dalard C, Delga S, Roblot N, Lefevre J, Job D, Hazan J, Curmi PA, Melki J. Microtubule-targeting drugs rescue axonal swellings in cortical neurons from spastin knockout mice. Dis Model Mech. 2013;6(1):72–83. doi: 10.1242/dmm.008946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Fan Y, Wali G, Sutharsan R, Bellette B, Crane DI, Sue CM, Mackay-Sim A. Low dose tubulin-binding drugs rescue peroxisome trafficking deficit in patient-derived stem cells in hereditary spastic paraplegia. Biol Open. 2014;3(6):494–502. doi: 10.1242/bio.20147641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wali G, Sue CM, Mackay-Sim A. Patient-derived stem cell models in SPAST HSP: disease modelling and drug discovery. Brain Sci. 2018;8(8):142. doi: 10.3390/brainsci8080142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rickman OJ, Baple EL, Crosby AH. Lipid metabolic pathways converge in motor neuron degenerative diseases. Brain. 2020;143(4):1073–1087. doi: 10.1093/brain/awz382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.••.Schols L, Rattay TW, Martus P, Meisner C, Baets J, Fischer I, et al. Hereditary spastic paraplegia type 5: natural history, biomarkers and a randomized controlled trial. Brain. 2017;140(12):3112–3127. doi: 10.1093/brain/awx273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Schule R, Siddique T, Deng HX, Yang Y, Donkervoort S, Hansson M, et al. Marked accumulation of 27-hydroxycholesterol in SPG5 patients with hereditary spastic paresis. J Lipid Res. 2010;51(4):819–823. doi: 10.1194/jlr.M002543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.••.Marelli C, Lamari F, Rainteau D, Lafourcade A, Banneau G, Humbert L, et al. Plasma oxysterols: biomarkers for diagnosis and treatment in spastic paraplegia type 5. Brain. 2018;141(1):72–84. doi: 10.1093/brain/awx297. [DOI] [PubMed] [Google Scholar]
- 112.Hauser S, Poenisch M, Schelling Y, Hoflinger P, Schuster S, Teegler A, et al. mRNA as a novel treatment strategy for hereditary spastic paraplegia type 5. Mol Ther Methods Clin Dev. 2019;15:359–370. doi: 10.1016/j.omtm.2019.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kara E, Tucci A, Manzoni C, Lynch DS, Elpidorou M, Bettencourt C, Chelban V, Manole A, Hamed SA, Haridy NA, Federoff M, Preza E, Hughes D, Pittman A, Jaunmuktane Z, Brandner S, Xiromerisiou G, Wiethoff S, Schottlaender L, Proukakis C, Morris H, Warner T, Bhatia KP, Korlipara LVP, Singleton AB, Hardy J, Wood NW, Lewis PA, Houlden H. Genetic and phenotypic characterization of complex hereditary spastic paraplegia. Brain. 2016;139(Pt 7):1904–1918. doi: 10.1093/brain/aww111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Schule R, Wiethoff S, Martus P, Karle KN, Otto S, Klebe S, et al. Hereditary spastic paraplegia: clinicogenetic lessons from 608 patients. Ann Neurol. 2016;79(4):646–658. doi: 10.1002/ana.24611. [DOI] [PubMed] [Google Scholar]