Abstract
The novel human coronavirus was firstly emerged in December 2019 in Wuhan, China, and has spread rapidly around the world. There is no known specific effective treatment of COVID-19. The most commonly used agents against this disease both in Turkey and around the world include chloroquine, hydroxychloroquine, lopinavir/ritonavir, favipiravir, and remdesivir. In the study, we investigated the drug potential of molecules that the components of an important medicinal plant Hypericum perforatum by using molecular docking and drug possibility properties of these molecules. The molecular docking results showed that the most stable complex was obtained with COVID-19 main protease and hypericin/isohypericin ligands with − 11 kcal/mol binding energy. Furthermore, ADMET, drug-likeness features of compounds of H. perforatum were investigated using the rules of Lipinski, Veber, and Ghose. According to the results obtained, it has been shown that H. perforatum has the potential to be an effective drug in the COVID-19 pandemic. In the next stage, it is necessary to carry out the clinically necessary reliability studies of these components. It is thought that it can be used for the treatment of COVID-19 if our molecular docking results are found to be in high correlation with clinical studies.
Supplementary Information
The online version contains supplementary material available at 10.1007/s40495-021-00254-9.
Keywords: COVID-19, Hypericum perforatum, Molecular docking, ADMET, Drug-likeness drug
Introduction
Hypericum perforatum is commonly known as St. John’s wort around the world and in Turkey is known as yellow cantaron and blood grass. This species is highly important and remarkable because of its pharmacological effects like antidepressant, antiviral, and antibacterial properties. These features made it the most studied species of Hypericum [1].
Several studies of H. perforatum introduced that the chemical components of plant (naftodiantrone compounds (hypericin, psodohypericin), fluoroglusinols (hyperforin), flavonoids (hyperositis, quercetin), biflavones (biapigenin, amentoflavones) phenolic acids, (ferulic acid, caffeic acid), proanthocyanidins, and essential oils) have provided its pharmacological effects [2]. There are also studies showing that it has potent cytotoxic and proapoptotic effects against tumor cell lines and inhibits tumor-induced angiogenesis [3, 4].
These medicinal plant extracts include complex phytochemicals, and the complete molecular characteristics of these pharmacologically important chemicals are still unknown. However, napthodianthrones are a considerable compound of H. perforatum extracts, and hypericin is the best-characterized member of this class. Hypericin is one of the main components of H. perforatum. It can be extracted from the plant or chemically synthesized and has powerful cytotoxic and proapoptotic effects on cancer cells [5]. The molecular mechanism of this component is not known, but previous studies have reported the different cellular pathways related to survival, necrosis, or apoptosis of the cell [6]. Therewithal, further studies are needed to reveal the action of these compounds.
COVID-19 is a new strain of coronavirus that affects primarily aging humans with especially dyspnea. Alveolar-interstitial pneumonia develops in 20% of patients with acute respiratory distress syndrome. This virus spreads rapidly worldwide, and the World Health Organization (WHO) has announced it as a pandemic disease [7]. There is no specific treatment known so far. Thus, new approaches to drug design and discovery can be used as a promising tool for the discovery of some therapeutic drug candidates against COVID-19. In this sense, molecular docking has become a promising and useful tool for drug design and development and attracts researchers’ attention. With this useful tool, we tried to reveal the binding potential of the target protein and the ligands with drug potential [8, 9].
Many drugs, including chloroquine, remdesivir, and hydroxychloroquine, have been shown to be effective against COVID-19 [10, 11]. Most of these drugs are HIV protease inhibitors. Chloroquine and hydroxychloroquine are thought to cause changes in glycolysis transferases in the vesicles of the endoplasmic reticulum or trans-Golgi complex at low pH [12]. In the study, we have performed a docking procedure with COVID-19 (PDB ID: 6LU7) and main components of H. perforatum as a ligand.
Materials and Methods
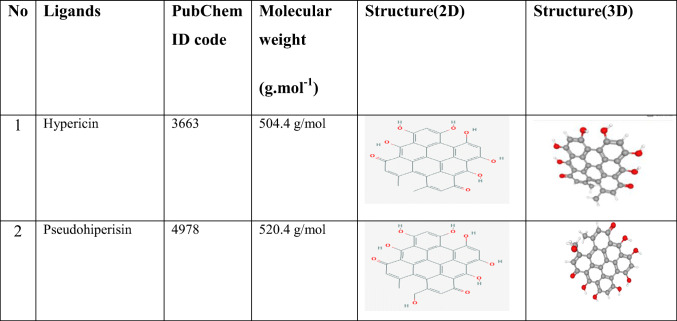
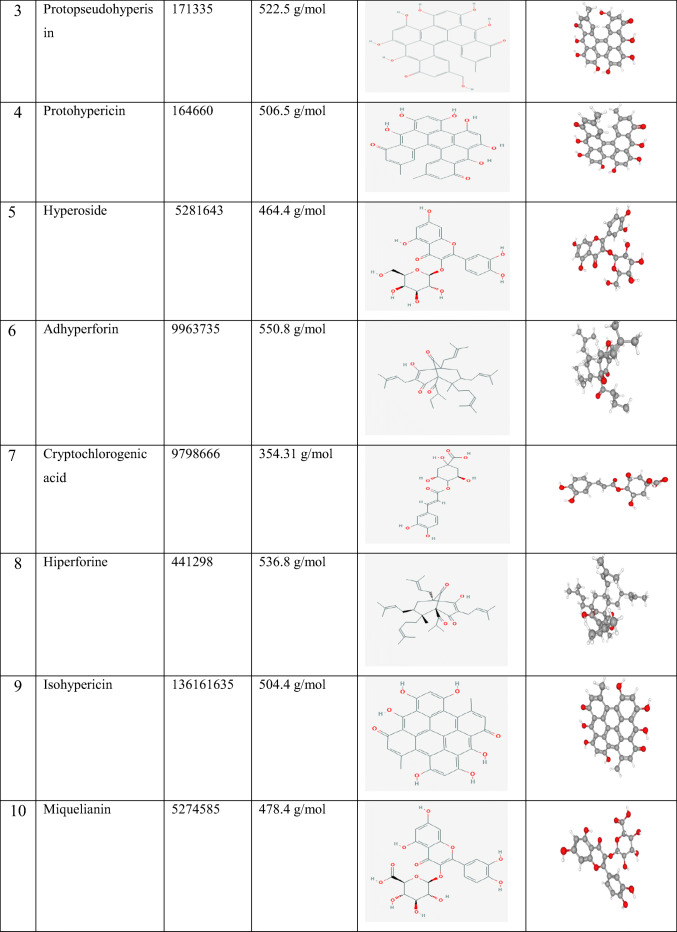
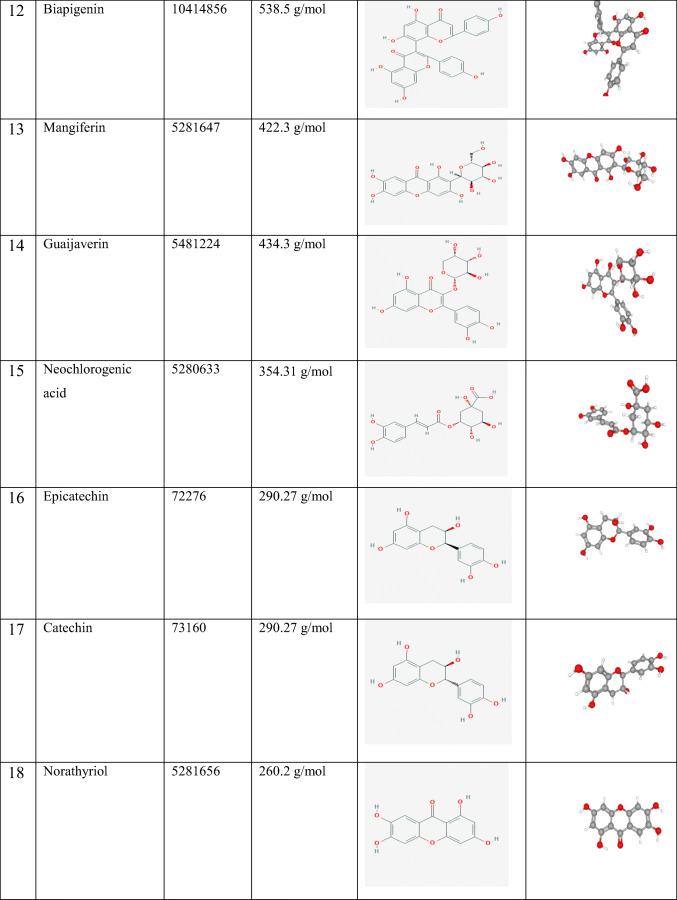
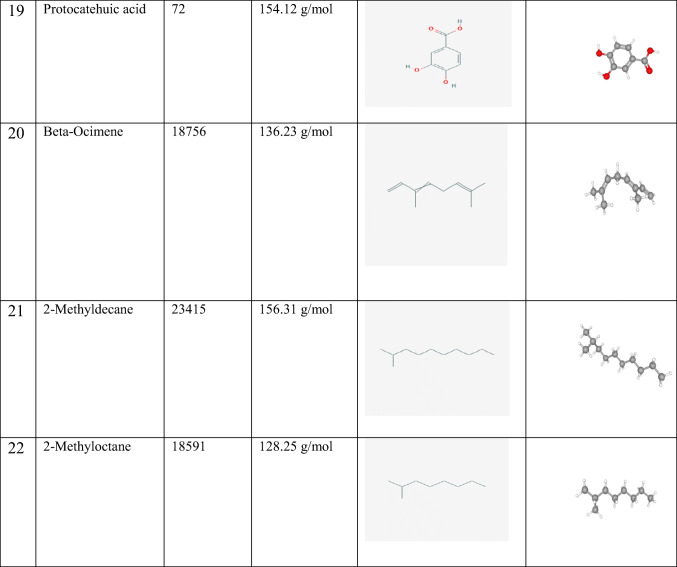
Molecular docking calculations were performed in Autodock Vina software [13]. The water molecules and cofactors were removed from the protein to clearly see the protein-ligand interaction. COVID-19 main protease [14] was used as a protein, and the structure of this protein was freely available from the RCSB Protein Data Bank as a 3D theoretical model (PDB ID: 6LU7) (https://www.rcsb.org/structure/6LU7). Ligands used in the study and their properties are given in Table 1. Ligands were obtained from PubChem (https://pubchem.ncbi.nlm.nih.gov/). Protein-ligand interactions were screened by Molegro Molecular Viewer 2.5 (Molegro Molecular viewer free software) (http://www.molegro.com) [15].
Table 1.
Ligands used in the study and their properties
The binding potential of chloroquine and hydroxychloroquine has been reviewed as a control ligand. 2D structure of the ligands was converted to energy-minimized 3D structure. The binding energy results calculated by Vina are presented in Table 2. All protein and ligands were validated before performing the in silico computations [16]. Protein and ligand interactions can be observed in Figs. 1–19 (Supplementary file 1).
Table 2.
Target protein and drug candidate molecules (ligands) molecular docking results
| Ligands | Binding energy (kcal/mol) | H bound and a.a. side |
|---|---|---|
| Chloroquine | − 5.6 kcal/mol | 1 (Trp 218) |
| Hydroxychloroquine | − 7.0 kcal/mol | 0 |
| Hypericin | − 11 kcal/mol | 6 (Asp 197x2; Lys 137; Leu 287x3) |
| Protopseudohypericin | − 9.9 kcal/mol | 5 (Tyr 239x2; Leu 271, Leu 272; Thr 199) |
| Hyperocide | − 9.5 kcal/mol | 5 (Asn274; Glu 270; Arg 279; Asn 277x2) |
| Cryptochlorogenic acid | − 8.1 kcal/mol | 6 (Gly 183; Phe 181; Arg 188; Glu 55; Arg 40x2) |
| Isohypericin | − 11.0 kcal/mol | 4 (Asp 197x2; Lys 137; Tyr 239) |
| Mangiferin | − 9.7 kcal/mol | 6 (Thr 199; Asp 289; Leu 287; Tyr 239x2; Leu 271) |
| Neochlorogenic acid | − 8.6 kcal/mol | 2 (Thr 292; Thr 111) |
| Catechin | − 7.4 kcal/mol | 4 (Leu 287; Thr 199; Asp 197; Tyr 237) |
| Protocatehuic acid | − 5.9 kcal/mol | 4 (Asn 277x2; Arg 279; Asn 221) |
| 2-Methyldecane | − 5.0 kcal/mol | 0 |
| Pseudohypericin | − 10.7 kcal/mol | 5(Thr 199, Tyr 237, Leu 272, Leu 287x2) |
| Protohypericin | − 9.7 kcal/mol | 5 (Asn 238; Thr 199; Tyr 239x2; Leu 271; Leu 272) |
| Adhyperforin | − 8.8 kcal/mol | 1 (Leu 287) |
| Hiperforine | − 7.9 kcal/mol | 2 (Arg 279; Asn 274) |
| Miquelianin | − 9.8 kcal/mol | 3 (Tyr 239; Leu 287; Thr 199) |
| Biapigenin | − 9.8 kcal/mol | 6 (Leu 271; Leu 287x2; Tyr 239; Thr 199; Asn 238) |
| Guaijaverin | − 9.3 kcal/mol | 5 (Asn 274; Glu 270; Arg 279; Asn 277x2) |
| Epicatechin | − 7.9 kcal/mol | 5 (Tyr 237; Thr 199; Asp 289; Lys 137;Tyr 239) |
| Norathyriol | − 7.1 kcal/mol | 3 (Arg 105; Gln 110; Thr 111) |
| Beta-Ocimene | − 5.2 kcal/mol | 0 |
| 2-Methyloctane | − 4.3 kcal/mol | 0 |
Drug Likeness and ADMET Prediction for the Components
Currently, computer-based ADMET analyses are gaining for drug discovery [17]. ADMET analyses are used to decide the pharmacological structure from the perspective of drug discovery (http://biosig.unimelb.edu.au/pkcsm/prediction). Pharmacokinetics and drug-likeness prediction of drug candidate molecule(s) were performed by online tools SwissADME (http://www.sib.swiss) (http://www.swissadme.ch/index.php) [18, 19] and admetSAR (http://lmmd.ecust.edu.cn/admetsar2/) [20]. In addition, these toxicological predictions have applied to Lipinski, Ghose, and Veber rules and bioavailability scores [21–23].
Results and Discussion
Molecular docking results obtained from this study indicated a strong interaction between COVID-19 main protease and potential drug candidates. The binding strength was defined by use of scoring function based on the Lamarckian generic algorithm. The binding free energy may include electrostatic, hydrogen bonding, and van der Waals interactions [14]. The least binding energy refers to the most stable binding between protein and the ligand. According to these results, the best stable binding score was obtained between tested protein and hypericin/isohypericin ligands with same score (− 11 kcal/mol binding energy). All of the docked structures were visualized in Molegro Molecular viewer (free academic).
In this study, human intestinal absorption, aqueous solubility levels, BBB penetration levels, CYP inhibition (CYP2D6, CYP3A4, CYP1A2, CYP2C19, CYP2C6, CYP2D6), hepatotoxicity, AMES toxicity, max. tolerated, dose etc. of compounds of H. perforatum were analyzed by prediction models. ADMET (absorption, distribution, metabolism, excretion, toxicity) analysis shows that compounds predicted good human intestinal absorption and no hepatotoxicity (Supplementary file 2).
Drug-likeness can be characterized as a complex balance of different structural properties that determines whether a compound is a drug. These features are mainly lipophilicity, hydrogen bonding properties, molecule size, pharmacophoric features, and many others [24]. In addition, drug-likeness results of compounds are shown in Table 3.
Table 3.
Drug-likeness results of compounds
| Ligand | Drug-likeness | Bioavailability score | ||
|---|---|---|---|---|
| Lipinski | Ghose | Veber | ||
| Hypericin | No; 2 violations: MW > 500, NHorOH > 5 | No; 3 violations: MW > 480, WLOGP > 5.6, MR > 130 | No; 1 violation: TPSA > 140 | 0,17 |
| Pseudohypericin | No; 2 violations: MW > 500, NHorOH > 5 | No; 2 violations: MW > 480, MR > 130 | No; 1 violation: TPSA > 140 | 0,17 |
| Protopseudohypericin | No; 2 violations: MW > 500, NHorOH > 5 | No; 2 violations: MW > 480, MR > 130 | No; 1 violation: TPSA > 140 | 0,17 |
| Protohypericin | No; 2 violations: MW > 500, NHorOH > 5 | No; 2 violations: MW > 480, MR > 130 | No; 1 violation: TPSA > 140 | 0,17 |
| Hyperocide | No; 2 violations: NorO > 10, NHorOH > 5 | No; 1 violation: WLOGP < − 0.4 | No; 1 violation: TPSA > 140 | 0,17 |
| Adhyperforin | No; 2 violations: MW > 500, MLOGP > 4.15 | No; 4 violations: MW > 480, WLOGP > 5.6, MR > 130, #atoms > 70 | No; 1 violation: rotors > 10 | 0,56 |
| Cryptochlorogenic acid | Yes; 1 violation: NHorOH > 5 | No; 1 violation: WLOGP < − 0.4 | No; 1 violation: TPSA > 140 | 0,11 |
| Hiperforine | No; 2 violations: MW > 500, MLOGP > 4.15 | No; 4 violations: MW > 480, WLOGP > 5.6, MR > 130, #atoms > 70 | No; 1 violation: rotors > 10 | 0,56 |
| Isohypericin | No; 2 violations: MW > 500, NHorOH > 5 | No; 3 violations: MW > 480, WLOGP > 5.6, MR > 130 | No; 1 violation: TPSA > 140 | 0,17 |
| Miquelianin | No; 2 violations: NorO>10, NHorOH>5 | No; 1 violation: WLOGP < − 0.4 | No; 1 violation: TPSA > 140 | 0,11 |
| Biapigenin | No; 2 violations: MW > 500, NHorOH > 5 | No; 2 violations: MW > 480, MR > 130 | No; 1 violation: TPSA > 140 | 0,17 |
| Mangiferin | No; 2 violations: NorO > 10, NHorOH > 5 | No; 1 violation: WLOGP < − 0.4 | No; 1 violation: TPSA > 140 | 0,17 |
| Guaijaverin | No; 2 violations: NorO > 10, NHorOH > 5 | Yes | No; 1 violation: TPSA > 140 | 0,17 |
| Neochlorogenic acid | Yes; 1 violation: NHorOH > 5 | No; 1 violation: WLOGP < − 0.4 | No; 1 violation: TPSA > 140 | 0,11 |
| Epicatechin | Yes | Yes | Yes | 0,55 |
| Catechin | Yes | Yes | Yes | 0,55 |
| Norathyriol | Yes | Yes | Yes | 0,55 |
| Protocatehuic acid | Yes | No; 3 violations: MW<160, MR<40, #atoms < 20 | Yes | 0,56 |
| Beta-Ocimene | Yes | No; 1 violation: MW < 160 | Yes | 0,55 |
| 2-Methyldecane | Yes; 1 violation: MLOGP > 4.15 | No; 1 violation: MW < 160 | Yes | 0,55 |
| 2-Methyloctane | Yes; 1 violation: MLOGP > 4.15 | No; 1 violation: MW < 160 | Yes | 0,55 |
According to Lipinski’s rule (Pfizer’s rule, Lipinski’s rule of five, RO5), the active drug has no more than one violation of the following properties including molecular weight (MW) ≤ 500, LogP ≤ 5, hydrogen bond acceptors ≤ 10, and hydrogen bond donors ≤ 5 [21]. According to Veber rules, the active drug has total hydrogen bonds ≤ 12, rotatable bonds ≤ 10, and polar surface area (PSA) ≤ 140 tend to have oral bioavailability ≥ 20% [22]. According to Ghose rules, active drug has Log P(− 0.4~5.6), MR (molar refractivity (40~150), MW (160~480), number of atoms (20~70), and PSA < 140 [23].
Based on the drug-likeness analysis, guaijaverin, epicatechin, catechin, norathyriol, protocatechuic acid, beta-ocimene, 2-methyldecane, and 2-methyloctane were found in accordance with the Lipinski’s, Veber’s, or Ghose’s rule. However, Lipinski’s rule of five may not apply to natural compounds. The only half of all FDA-approved small-molecule drugs are both used and compatible with the “rule of five” [25]. Therefore, it has the potential to be used as a medicine in other molecules.
Conclusion
Hypericum sp., used for many years as a medicinal plant for different treatments, has recently become popular with research for its different properties. This medicinal plant has long been used due to the beneficial effects it has on human health [26–29]. In our research, the possibility of this useful plant being used against COVID-19, which our country and other countries in the world have been fighting for a long time, has been investigated. Based on the results, it was concluded that H. perforatum could be effective against COVID-19 and it is hypothesized that compounds may be screened to in vitro and in vivo experimental analyses to indicate its inhibitory potency.
Supplementary Information
(DOCX 16762 kb)
(DOCX 100 kb)
Footnotes
This article is part of the Topical Collection on Natural Products: From Chemistry to Pharmacology
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Barnes J, Anderson LA, Phillipson JD. St John’s wort (Hypericum perforatum L.): a review of its chemistry, pharmacology and clinical properties. J Pharm Pharmacol. 2001;53(5):583–600. doi: 10.1211/0022357011775910. [DOI] [PubMed] [Google Scholar]
- 2.Hostettmann K, Wolfender JL. St. John's Wort and its Active Principles in Depression and Anxiety. Phytochemistry. 2005:5–20. 10.1007/3-7643-7338-5_2.
- 3.Momekov G, Ferdinandov D, Zheleva-Dimitrova D, Nedialkov P, Girreser U, Kitanov G. Cytotoxic effects of hyperatomarin, a prenylated phloroglucinol from Hypericum annulatum Moris subsp. annulatum, in a panel of malignant cell lines. Phytomedicine. 2008;15(11):1010–1015. doi: 10.1016/j.phymed.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 4.Schempp CM, Kirkin V, Simon-Haarhaus B, Kersten A, Kiss J, Termeer CC, et al. Inhibition of tumour cell growth by hyperforin, a novel anticancer drug from St. John’s wort that acts by induction of apoptosis. Oncogene. 2002;21(8):1242–50. 10.1038/sj.onc.1205190. [DOI] [PubMed]
- 5.Ercan FS, Azarkan SY, Ercan N, Koc M. Sequence variants of CYP345a1 and CYP6a14 gene regions in Tribolium castaneum (Coleoptera: Tenebrionidae) adults treated with the novel characterızed Bolanthus turcicus (Caryophyllaceae) extract. Mol Biol Res Commun. 2020;9(3):105–110. doi: 10.22099/mbrc.2020.35861.1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Theodossiou TA, Hothersall JS, De Witte PA, Pantos A, Agostinis P. The multifaceted photocytotoxic profile of hypericin. Mol Pharm. 2009;6(6):1775–1789. doi: 10.1021/mp900166q. [DOI] [PubMed] [Google Scholar]
- 7.Wang C, Horby PW, Hayden FG, Gao GF. A novel coronavirus outbreak of global health concern. Lancet. 2020;395(10223):470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McConkey BJ, Sobolev V, Edelman M. The performance of current methods in ligand–protein docking. Curr Sci. 2002:845–56. 10.2307/24107087.
- 9.Abdelli I, Hassani F, Bekkel Brikci S, Ghalem S. In silico study the inhibition of angiotensin converting enzyme 2 receptor of COVID-19 by Ammoides verticillata components harvested from Western Algeria. J Biomol Struct Dyn. 2020;14:1–14. doi: 10.1080/07391102.2020.1763199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yao X, Ye F, Zhang M, Cui C, Huang B, Niu P, Liu X, Zhao L, Dong E, Song C, Zhan S, Lu R, Li H, Tan W, Liu D. In vitro antiviral activity and projection of optimized dosing design of hydroxychloroquine for the treatment of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Clin Infect Dis. 2020;71(15):732–739. doi: 10.1093/cid/ciaa237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lim J, Jeon S, Shin HY, Kim MJ, Seong YM, Lee WJ, Choe KW, Kang YM, Lee B, Park SJ. Case of the index patient who caused tertiary transmission of COVID-19 infection in Korea: the application of lopinavir/ritonavir for the treatment of COVID-19 infected pneumonia monitored by quantitative RT-PCR. J Korean Med Sci. 2020;35(6):e79. doi: 10.3346/jkms.2020.35.e79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Devaux CA, Rolain JM, Colson P, Raoult D. New insights on the antiviral effects of chloroquine against coronavirus: what to expect for COVID-19? Int J Antimicrob Agents. 2020;55(5):105938. doi: 10.1016/j.ijantimicag.2020.105938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jin Z, Du X, Xu Y, Deng Y, Liu M, Zhao Y, et al. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582(7811):289–293. 10.1038/s41586-020-2223-y. [DOI] [PubMed]
- 15.Thomsen R, Christensen MH. MolDock: a new technique for high-accuracy molecular docking. J Med Chem. 2006;49(11):3315–3321. doi: 10.1021/jm051197e. [DOI] [PubMed] [Google Scholar]
- 16.Yalcin S, Sas EB, Cankaya N, Ercan F, Kurt M. The physical studies and interaction with anti-apoptotic proteins of 2-(bis (cyanomethyl) amino)-2-oxoethyl methacrylate molecule. arXiv preprint arXiv:1910.00900. 2019, DOI: 10.5488/CMP.22.33301
- 17.Ntie-Kang F, Lifongo LL, Mbah JA, Owono Owono LC, Megnassan E, Mbaze LM, Judson PN, Sippl W, Efange SM. In silico drug metabolism and pharmacokinetic profiles of natural products from medicinal plants in the Congo basin. In Silico Pharmacol 2013;1:12. doi: 10.1186/2193-9616-1-12. [DOI] [PMC free article] [PubMed]
- 18.Zoete V, Daina A, Bovigny C, Michielin O. SwissSimilarity: a web tool for low to ultra high throughput ligand-based virtual screening. J Chem Inf Model. 2016;56(8):1399–1404. doi: 10.1021/acs.jcim.6b00174. [DOI] [PubMed] [Google Scholar]
- 19.Daina A, Michielin O, Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep. 2017;7:42717. doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheng F, Li W, Zhou Y, Shen J, Wu Z, Liu G, Lee PW, Tang Y. admetSAR: a comprehensive source and free tool for assessment of chemical ADMET properties. J Chem Inf Model. 2012;52(11):3099–3105. doi: 10.1021/ci300367a. [DOI] [PubMed] [Google Scholar]
- 21.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev. 2001;46(1-3):3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 22.Veber DF, Johnson SR, Cheng HY, Smith BR, Ward KW, Kopple KD. Molecular properties that influence the oral bioavailability of drug candidates. J Med Chem. 2002;45(12):2615–2623. doi: 10.1021/jm020017n. [DOI] [PubMed] [Google Scholar]
- 23.Ghose AK, Viswanadhan VN, Wendoloski JJ. A knowledge-based approach in designing combinatorial or medicinal chemistry libraries for drug discovery. 1. A qualitative and quantitative characterization of known drug databases. J Comb Chem. 1999;1(1):55–68. doi: 10.1021/cc9800071. [DOI] [PubMed] [Google Scholar]
- 24.Turner JV, Agatonovic-Kustrin S. In book: Comprehensive Medicinal Chemistry II. 2007; DOI: 10.1016/B0-08-045044-X/00147-4
- 25.Zhang MQ, Wilkinson B. Drug discovery beyond the ‘rule-of-five’. Curr Opin Biotechnol. 2007;18(6):478–488. doi: 10.1016/j.copbio.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 26.Yalcin S. Molecular Docking, Drug Likeness, and ADMET Analyses of Passiflora compounds as P-glycoprotein (P-gp) inhibitor for the treatment of cancer. Curr Pharmacol Rep. 2020;6:1–12. doi: 10.1007/s40495-020-00241-6. [DOI] [Google Scholar]
- 27.Xiao CY, Mu Q, Gibbons S. The phytochemistry and pharmacology of Hypericum. Prog Chem Org Nat Prod. 2020;112:85–182. doi: 10.1007/978-3-030-52966-6_2. [DOI] [PubMed] [Google Scholar]
- 28.Zhang R, Ji Y, Zhang X, Kennelly EJ, Long C. Ethnopharmacology of Hypericum species in China: a comprehensive review on ethnobotany, phytochemistry and pharmacology. J Ethnopharmacol. 2020;254:112686. doi: 10.1016/j.jep.2020.112686. [DOI] [PubMed] [Google Scholar]
- 29.Allegra A, Tonacci A, Spagnolo EV, Musolino C, Gangemi S. Antiproliferative effects of St. John’s wort, its derivatives, and other Hypericum species in hematologic malignancies. Int J Mol Sci. 2020;22(1):146. doi: 10.3390/ijms22010146. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 16762 kb)
(DOCX 100 kb)