Abstract
Over the last two decades, extensive studies have been performed at the molecular level to understand the evolution of carnivorous plants. As fruits, the repertoire of protein components in the digestive fluids of several carnivorous plants have gradually become clear. However, the quantitative aspects of these proteins and the expression mechanisms of the genes that encode them are still poorly understood. In this study, using the Australian sundew Drosera adelae, we identified and quantified the digestive fluid proteins. We examined the expression and methylation status of the genes corresponding to major hydrolytic enzymes in various organs; these included thaumatin-like protein, S-like RNase, cysteine protease, class I chitinase, β-1, 3-glucanase, and hevein-like protein. The genes encoding these proteins were exclusively expressed in the glandular tentacles. Furthermore, the promoters of the β-1, 3-glucanase and cysteine protease genes were demethylated only in the glandular tentacles, similar to the previously reported case of the S-like RNase gene da-I. This phenomenon correlated with high expression of the DNA demethylase DEMETER in the glandular tentacles, strongly suggesting that it performs glandular tentacle-specific demethylation of the genes. The current study strengthens and generalizes the relevance of epigenetics to trap organ-specific gene expression in D. adelae. We also suggest similarities between the trap organs of carnivorous plants and the roots of non-carnivorous plants.
Keywords: Carnivorous plants, defense-related proteins, DEMETER, DNA methylation, Drosera, epigenetic regulation, organ-specific DNA demethylation, sundew, tissue-specific DNA demethylation
Lance-leaf sundew provides an intriguing example of organ-specific DNA demethylation, which allows β-1, 3-glucanase, cysteine protease, and S-like RNase genes to be exclusively expressed in the glandular tentacles.
Introduction
Carnivory is not restricted to animals. Some plants also behave like ‘flesh eaters’ and are referred to as carnivorous plants (Charles Darwin originally called them ‘insectivorous plants’: Darwin, 1875; Lloyd, 1942; Juniper et al., 1989). Carnivorous plants have evolved the ability to trap and digest prey, such as arthropods and small animals, and absorb nutrients from the resulting digest. This ability has allowed them to grow in nutrient-deficient habitats. The carnivorous plants seem to have emerged between 1.9 and 95.1 million years ago (Sadowski et al., 2015; Fleischmann et al., 2018). They are thought to have evolved independently at least nine or ten times in five orders of flowering plants, namely Lamiales, Ericales, Caryophyllales, Oxalidales and Poales (Albert et al., 1992; Givnish, 2015; Fleischmann et al., 2018). Carnivorous plants presently account for at least ~600 species in ~20 genera, ~12 families and five orders of angiosperms (Ellison and Gotelli, 2009; Givnish, 2015).
Carnivorous plants have elaborate leaves or stolons that function as traps, which are grouped into five types (Darwin, 1875; Lloyd, 1942; Juniper et al., 1989; Król et al., 2012; Ellison and Adamec, 2018). These include adhesive traps [e.g. Drosera (sundews) and Pinguicula (butterworts)], snap traps [Dionaea (Venus flytrap) and Aldrovanda (waterwheel plant)], pitfall traps [e.g. Nepenthes (tropical pitcher plants), Sarracenia (American pitcher plants) and Cephalotus (Albany pitcher plant)], suction traps [Utricularia (bladderworts)], and lobster pot traps [Genlisea (corkscrew plants)]. There are two long-standing questions for these plants: how did their ancestors become equipped with the trap leaves and how did they acquire the mechanisms of prey digestion (Darwin, 1875; Lloyd, 1942; Juniper et al., 1989; Renner and Specht, 2013; Ellison and Adamec, 2018; Pavlovič and Mithöfer, 2019)? Recent studies have gradually begun to provide answers to these questions.
Trap leaf acquisition may have originated in hairy leaves and ‘foliar feeding’ in non-carnivorous plants (Fernández and Eichert, 2009; Fernández and Brown, 2013). Hairy leaves can hold raindrops that capture insects by the surface tension of water. Some insects could drown, rot, and finally release nutrients, and this phenomenon may have caused the evolution of the leaves of the ancestors of carnivorous plants. Phylogenetic data suggest that the adhesive trap is the origin of most carnivorous plants (Müller et al., 2004; Heubl et al., 2006). Various trap leaves are thought to have been formed by the adhesive trap evolving into clam shell-, pitcher- or bladder-like leaves. Furthermore, recent studies have gradually shed light on the molecular mechanisms of trap leaf formation. For Cephalotus follicularis or Utricularia gibba, the gene expression that controls the adaxial and abaxial domains is suggested to generate leaf specialization (Fukushima et al., 2017; Whitewoods et al., 2020). It has also been further demonstrated that these changes affect growth polarity, which generates cell division orientation and forms leaves with unusual shapes (Whitewoods et al., 2020). The cell division orientation may also be important for shaping the pitcher leaves of Sarracenia purpurea (Fukushima et al., 2015).
Elucidation of the structural and functional characteristics of the proteins in the digestive fluid and their genes have greatly advanced in the last two decades. Athauda et al. (2004) purified, characterized and sequenced two novel aspartic proteases (nepenthesins I and II) from the digestive fluid of Nepenthes distillatoria. Subsequently, a class I chitinase and an S-like ribonuclease (RNase) were found in the digestive fluids of D. rotundifolia and D. adelae, respectively (Matušíková et al., 2005; Okabe et al., 2005a, b). Class I chitinases break down the exoskeletons of arthropods (mainly insects), releasing the bound nitrogen, and allowing other enzymes to access and degrade internal tissues. Furthermore, these enzymes are also induced in response to pathogen attack or wounding in non-carnivorous plants (Liao et al., 1994; Wu et al., 1994; Hamel and Bellemare, 1995; Gijzen et al., 2001; Zhao et al., 2011). S-like RNases are structurally similar to S-RNases, but functionally different from the S-RNases that function in self-incompatibility (McClure et al., 1989; Huang et al., 1994; McClure et al., 2011). The S-like RNases in non-carnivorous plants respond to the senescence of leaves and flowers, phosphate starvation, pathogen attack and wounding (Nürnberger et al., 1990; Taylor et al., 1993; Bariola et al., 1994; Green, 1994; Ye and Droste, 1996; Galiana et al., 1997; Kariu et al., 1998; Irie, 1999; Hugot et al., 2002).
Okabe and colleagues discovered an S-like RNase in the digestive fluid of D. adelae, and hypothesized that carnivorous plants may utilize the defense-related proteins for carnivory (Okabe et al., 2005a). Since then, many such proteins, including proteases, nucleases, chitinases, glucanases, phosphatases, lipases, peroxidases, lipid transfer proteins (LTPs) and thaumatin-like proteins (TLPs), have been identified in the digestive fluids of carnivorous plants (Hatano and Hamada, 2008, 2012; Schulze et al., 2012; Nishimura et al., 2013; Bemm et al., 2016; Lee et al., 2016; Rottloff et al., 2016; Fukushima et al., 2017; Krausko et al., 2017; Matušíková et al., 2018; Kocáb et al., 2020). Amongst them, some had amino acid residues that are conserved only among carnivorous plants (Renner and Specht, 2012; Nishimura et al., 2014; Arai et al., 2015; Fukushima et al., 2017), substantiating the hypothesis proposed by Okabe et al. (2005a).
The expression mechanism of the proteins in the digestive fluid differs, depending on the prey-trapping system. In the snap traps, the protein expression is induced. In the pitcher traps and adhesive traps, some enzymes are constitutively expressed, some have induced expression, and others show basically constitutive expression which is occasionally enhanced by the stimulus of trapping prey (Eilenberg and Zilberstein, 2008; Matušíková et al., 2018; Pavlovič and Mithöfer, 2019). Regarding induced expression, mechanical or chemical stimuli induce the expression of the proteins in the digestive fluid (Scala et al., 1969; Matušíková et al., 2005; Eilenberg et al., 2006; Rottloff et al., 2011; Pavlovič et al., 2014), and in some cases jasmonates are involved in this expression (Escalante-Pérez et al., 2011; Libiaková et al., 2014; Bemm et al., 2016; Böhm et al., 2016; Yilamujiang et al., 2016; Krausko et al., 2017; Pavlovič et al., 2017; Pavlovič and Mithöfer, 2019). In non-carnivorous plants, jasmonates regulate the responses to necrotrophic pathogens and herbivore attacks (Zhang et al., 2017), suggesting that carnivorous plants employ the plant defense signaling pathway for inducible expression. On the other hand, the mechanism of constitutive expression has remained poorly understood. The only exception is the study by Nishimura et al. (2013) which suggested that the S-like RNase DA-I is constitutively and exclusively expressed in the glandular tentacles of D. adelae (Nishimura et al., 2013). Furthermore, the report also proposed that epigenetic regulation is involved in this organ-specific gene expression.
The current study was performed with two purposes: to identify the ‘actors’ that play in the hypothetical epigenetic regulation of da-I expression, and to clarify the repertoire, relative amounts and expression mechanisms of other proteins in the digestive fluid of D. adelae. We identified and quantitated the proteins in the digestive fluid by a proteomic approach, and examined the relationship between the expression of genes encoding major hydrolytic enzymes and DNA methylation. Our results suggest that D. adelae ensures that some of these genes are constitutively expressed via unmethylation, in a glandular tentacle-specific manner, for carnivory.
Materials and methods
Plant material
D. adelae was purchased from Y’s Exotics (http://ys-exotics.com). The growth conditions have been described previously (Nishimura et al., 2013).
Database construction for protein identification
The RNA-seq data of D. adelae shoots were downloaded from the DNA Data Bank of Japan (DDB) Sequence Read Archive (accession number: DRR051750; Fukushima et al., 2017). After cleaning and quality checks, 31 780 142 confident reads were assembled into 49 512 contigs with an average length of 628 bases, using the de Bruijn graph-based de novo assembly program in CLC Genomics Workbench version 8.0.2 (Qiagen, Hilden, Germany) with word size 23, bubble size 50, and minimum contig length 200 bp. Using TransDecoder (Haas et al., 2013), we then identified 20 741 contigs with putative protein-coding regions larger than or equal to 100 amino acids. Among the possible open reading frames (ORFs) in each contig, the most plausible ORF was selected based on the sequence length and two homology searches: a BLASTP search using the UniProt database and an HMMER search using the Pfam-A database for the protein-motifs search, which generated a protein database. Moreover, all protein sequences previously identified in the digestive fluid of D. adelae were collected from UniProt and added to our database. To obtain a non-redundant data set, sequences with ≥90% similarity to each other were removed using Cluster Database at High Identity with Tolerance (CD-HIT; Li and Godzik, 2006), which left 19 829 unique contigs in total. The protein sequences in the database were annotated by collating with the Arabidopsis thaliana sequences in UniProt/SwissProt, using BLASTP (E-value<10–4). The name of the highest scoring protein was used for the annotation.
Protein digestion
The sticky digestive fluid of D. adelae was collected according to the method described by Okabe et al. (2005a). The proteins in the digestive fluid were digested with two proteases: Asp-N (Roche Diagnostics, Mannheim, Germany) and chymotrypsin (Promega, Madison, WI, USA). In the Asp-N digestion, a 25 µl solution containing 5 µl digestive fluid, 10 mM DTT, 4.8 M urea and 30 mM Tris-HCl (pH 7.5) was first incubated at 37 °C for 90 min. Iodoacetamide was then added to the solution to a final concentration of 50 mM. After an incubation at ~25 °C for 30 min in the dark, the solution was diluted with 50 mM NH4CO3 to lower the urea concentration to less than 1 M. Subsequently, Asp-N was added to the solution at a final concentration of 1.2 ng µl-1. The solution was incubated at 37 °C overnight. Formic acid was then added to a final concentration of 0.1%, to stop the reaction. The resulting sample was desalted with a GL-Tip SDB (GL Sciences, Tokyo, Japan). After vacuum centrifugation, the peptides were finally dissolved in 50 µl of a solution containing 2% acetonitrile and 0.02% formic acid.
The chymotrypsin digestion was slightly different from the procedure described above. EDTA was added to the solution to a final concentration of 10 mM, and Tris-HCl (pH 8.0) was used in the reduction step with DTT. For dilution, 100 mM Tris-HCl (pH 8.0) was used after the alkylation step with iodoacetamide. In the digestion step, chymotrypsin and CaCl2 were added to final concentrations of 12.2 ng µl-1 and 10 mM, respectively.
Sequential window acquisition of all theoretical fragment ion spectra mass spectrometry (SWATH-MS) analysis
The SWATH-MS analysis was performed in two steps. In the first step, ion libraries were generated using information-dependent acquisition (IDA), and in the second, label-free quantification was performed based on the SWATH acquisition. For IDA, the digested peptides were analysed using a Prominence nano system (Shimadzu, Kyoto, Japan) coupled with a TripleTOF 4600 System (AB Sciex, Framingham, MA, USA). Each 4 µl sample was injected onto the trap column (Monolith Trap C18-50–150, 50 µm × 150 mm, Hitachi High-Tech Fielding Corporation, Tokyo, Japan) and desalted with solvent A (2% acetonitrile and 0.1% formic acid in water) at a flow rate of 4 µl min-1 for 5 min. Next, the valve position was switched, and the trapped peptides were eluted from a C18 analytical reversed-phased column (MonoCap C18, 50 µm × 150 mm, GL Sciences) at a flow rate of 300 nl min-1, with a linear gradient of 2–30% solvent B (98% acetonitrile and 0.1% formic acid in water) over 35 min. The gradient was subsequently increased from 30% to 95% solvent B in 5 min, held for 10 min, and then re-equilibrated with solvent A for 30 min. The valve position was switched back during re-equilibration. The eluate was directed into the nanospray ionization source of the mass spectrometer, with the following source conditions: ion source gas 1, 20 psi; curtain gas, 20 psi; interface heater temperature, 150 °C; ion spray voltage floating, 2300 V. In the MS analysis, we used the positive ion mode over the mass range of m/z 400–1250, with an accumulation time of 250 ms. The 10 most intense precursor ions exceeding 150 counts s-1 with charge states of 2–4 were selected for collision-induced dissociation fragmentation, with an ion tolerance of 50 mDa. The dynamic exclusion time was set to 12 s. Tandem mass spectrometry (MS/MS) spectra were acquired over the mass range of m/z 100–1500 with an accumulation time of 100 ms, using the rolling collision energy with a collision energy spread of 15 V.
For the SWATH acquisition, the liquid chromatography (LC) and source conditions were the same as above, and the MS/MS conditions were set using 100 variable windows (https://sciex.com/community/Asset/00001409/vw100_ces_5_10.txt) provided by AB Sciex (Framingham, MA, USA), across the precursor mass range of 400–1250 m z-1. A 50 ms survey scan (400–1250 m z-1) was acquired at the beginning of each cycle, and MS/MS spectra were collected from 100–1500 m z-1 for 25 ms, resulting in a cycle time of 2.7 s.
Ion library generation
We used the ProteinPilot 5.0 software (AB Sciex) with the Paragon Algorithm (Shilov et al., 2007) to identify the proteins. For each experiment, all IDA data were combined and searched against the putative proteolytic digests of the D. adelae proteins described above. The parameters were as follows: sample type, identification; Cys alkylation, iodoacetamide; digestion, Asp-N or chymotrypsin; instrument, TripleTOF 4600; ID focus, biological modifications; search effort, thorough ID. The detected protein threshold was set to 1.3, corresponding to a confidence level of 95%. The resulting files were used as the ion libraries for subsequent SWATH processing.
Protein quantification
The SWATH data were processed using MS/MS (ALL) with SWATH Acquisition MicroApp 2.0 in PeakView 2.2 (AB Sciex). The parameters were set as follows: 1000 peptides per protein, five transitions per peptide, peptide confidence threshold of 99%, false discovery rate (FDR) threshold of 1%, excluding modified peptides, excluding shared peptides, 5 min of extracted ion chromatogram (XIC) extraction window and 50 ppm of XIC mass tolerance. The retention time was calibrated with endogenous peptides. Based on the resulting data, we excluded reverse sequences and peptides with scores of infinity or FDR ≥1%, and then extracted the intensities of peptides common to two biological replicates. Protein intensities were subsequently calculated by summing all of the peptides for a given protein, and normalized based on the total ion intensity of each sample. Finally, the identified proteins were quantified by the intensity-based absolute quantification (iBAQ) algorithm (Schwanhäusser et al., 2011). The iBAQ values were obtained by the protein intensity divided by the number of peptides theoretically generated by a digestion with Asp-N or chymotrypsin (Supplementary Tables S1; S2).
Quantification of mRNA
Total RNA was isolated from the glandular tentacles, their heads and stalks, leaves with glandular tentacles removed (hereafter referred to as laminas), roots and inflorescences, using the cetyltrimethyl ammonium bromide-based method (Bekesiova, et al., 1999). The samples were treated with RNase-free DNase I (Promega). First-strand cDNAs were synthesized using ReverTra Ace reverse transcriptase (Toyobo, Osaka, Japan), oligo (dt)20 and random primers. Real-time PCR was performed with THUNDERBIRD SYBR qPCR Mix (Toyobo) and specific primers (Supplementary Table S3). Based on the geNorm (Vandesompele et al., 2002) analysis, actin gene, gene encoding eukaryotic initiation factor 4A (eIF4A), and gene encoding TIP41 (TAP42 interacting protein of 41 kDa)-like protein (TIP41) were selected as the most suitable reference genes. Gene expression was normalized to that of the geometric mean of the reference genes, using the modified Pfaffl method (Zifkin et al., 2012).
Determination of upstream sequences of genes encoding digestive enzymes
We named the genes encoding the cysteine protease, class I chitinase, β-1, 3-glucanase, TLP and HEL, as Cysp1, Chi1, Glu1, Tlp1, and Hel1, respectively. Genomic DNA was isolated from leaves, as described above (Bekesiova et al., 1999), and thermal asymmetric interlaced (TAIL)-PCR (Liu et al., 1995; Liu and Whittier, 1995) was performed. The resulting amplified fragments were cloned into the pUC19 vector and sequenced. The putative transcription start sites (TSSs) were determined based on the RNA-seq information (Fukushima et al., 2017), and the cis-DNA elements in the region upstream of the TSS were identified by PlantPAN3.0 (Chow et al., 2019).
Bisulfite sequencing
Genomic DNA samples purified from the glandular tentacles, laminas, roots and inflorescences were treated with sodium bisulfite, using an EpiTect Fast Bisulfite Conversion Kit (Qiagen). Each organ sample was then treated as follows. Using the KOD -Multi & Epi (Toyobo) enzymes and the primers with overhangs for the second PCR (Supplementary Table S4), the following sequences were amplified: Cysp1, positions from –336 to +167; Chi1, –432 to +36; Glu1, –358 to +90; Tlp1, –424 to +49; and Hel1, –440 to +52. The PCR reactions were repeated three times for each target, and generated 15 full-length replicates in total. After purification, they were combined and the mixture was diluted to 1 ng µl-1 with 10 mM Tris-HCl (pH 8.0). Then, using 1 µl of the solution, 10 rounds of PCR amplification were performed with KAPA HiFi HotStart Ready Mix (Roche) and barcode adapters (Fasmac, Kanagawa, Japan). The products were purified with Agencourt AMPure XP beads (Beckman Coulter, Brea, CA, USA) and diluted to 12 ng µl-1 with 10 mM Tris-HCl (pH 8.0). Equal amounts of the PCR products of all the organ samples thus obtained were finally combined, and the mixture was sequenced on an Illumina MiSeq (Illumina, San Diego, CA, USA) with 300 bp paired-end reads. The sequencing was performed by Fasmac (Kanagawa, Japan). Using Bisulfighter (Saito et al., 2014), the resulting reads were mapped to the reference sequences, and the percentages of methylated cytosines were determined as follows: the number of cytosines divided by the number of total reads mapped at the same position.
Identification of cytosine-5 DNA methyltransferase and DNA demethylase
The hidden Markov model profiles of the conserved key domains of cytosine-5 DNA methyltransferase (C5-MTase) (PF00145) and DNA demethylase (PF15628) were downloaded from the Pfam-A database. Using the profiles and HMMER, all possible C5-MTases and DNA demethylases among the D. adelae proteins were identified. InterPro was used to confirm and classify each putative C5-MTase or DNA demethylase. In cases where the protein sequences were inadequate for this analysis, we clarified the flanking sequences by inverse PCR or TAIL-PCR. Each protein was named by comparison with the A. thaliana sequences in UniProt/SwissProt, using BLASTP (Supplementary Table S5). The protein isoelectric points and molecular weights were determined using ProtParam (https://web.expasy.org/protparam/).
Results
Proteins identified in the digestive fluid
In the current study, we did not perform any treatment that could potentially induce gene expression, including prey attachment, and collected the sticky digestive fluid of D. adelae (Fig. 1). Using LC-MS/MS-based analysis, we identified 26 proteins in the sticky digestive fluid, of which 19 were novel (Table 1; Supplementary Tables S1; S2; Fig. S1). More than half of the 26 proteins were defense-related proteins, and 11 proteins among them were pathogenesis-related (PR) proteins, which are induced in response to infection by pathogens, including bacteria, fungi, viruses, and viroids (van Loon et al., 2006). Based on iBAQ, the major proteins in the digestive fluid were judged to be TLP, S-like RNase, LTP, cysteine protease, and class I chitinase (Table 1; Supplementary Tables S1; S2). Except for LTP, these proteins are hydrolytic enzymes. S-like RNase and cysteine protease generate ribonucleotides and peptides, respectively. Peptides and ribonucleotides are thought to be used as is, or as the source of amino acids or nitrogen and phosphates, respectively (Nürnberger et al., 1990; Bariola et al., 1994; Okabe et al., 2005a; Godlewski and Adamczyk, 2007; Adamczyk et al., 2009; Nishimura et al., 2014). Chitin is a major structural polysaccharide of arthropods, mollusks and fungi, and class I chitinase hydrolyses the β-1, 4-linkage of chitin, allowing other hydrolytic proteins to penetrate and break down internal tissues. TLP has antifungal activity, and its proposed mechanism of action involves β-1, 3-glucanase activity to disrupt the cell walls of pathogenic fungi (Grenier et al., 1999; Zareie et al., 2002). β-1, 3-glucanase and hevein-like protein (HEL; Brunner et al., 1998), which are considered as hydrolytic enzymes, were also present in the digestive fluid, although they were less abundant than the above-mentioned proteins.
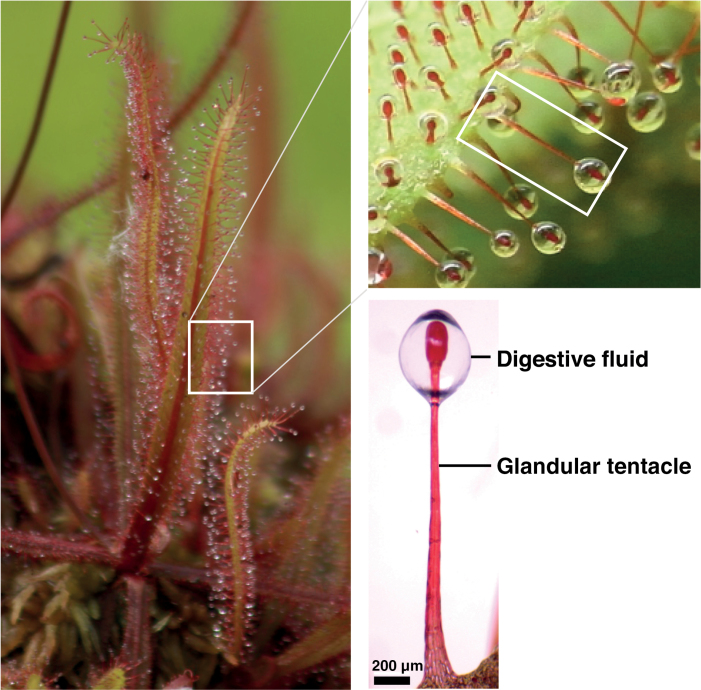
Fig. 1.
The Australian sundew D. adelae and its digestive fluid. The digestive fluid is secreted from the glandular cells forming the tips of the tentacles (scale bar, 200 μm).
Table 1.
Proteins in the digestive fluid
| Protein name a | Corresponding proteins in A. thaliana | Putative physiological function c | Abundance rank d | |||
|---|---|---|---|---|---|---|
| Name a | E-value | Defense-related protein b | Asp-N | Chymotrypsin | ||
| Thaumatin-like protein (BCF79772) | Osmotin-like protein OSM34 (P50700) | 6.6E-97 | ● (PR-5) | Antifungal activity | 1 | 1 |
| Unknown (Contig_14 (Fragment)) | No Blast hit with E-value <10–4 | - | - | ? | 4 | 2 |
| S-like RNase (BAE16663) | Ribonuclease 1 (P42813) | 4.2E-96 | ● | Prey digestion | 3 | 3 |
| Lipid transfer protein (BAW35429) | No Blast hit with E-value <10–4 | - | - | ? | 2 | 5 |
| Cysteine protease (BAW35427) | Senescence-specific cysteine protease SAG12 (Q9FJ47) | 4.5E-108 | ● | Prey digestion | 5 | 4 |
| Lipid transfer protein (Contig_35636) | Non-specific lipid-transfer protein 11 (Q2V3C1) | 4.7E-13 | ● (PR-14) | Antifungal or antibacterial activity | 6 | - |
| Unknown (Contig_1360) | No Blast hit with E-value <10–4 | - | - | ? | 7 | 6 |
| Class I chitinase (BAR13254) | Basic endochitinase B (P19171) | 1.5E-151 | ● (PR-3) | Antifungal activity | 8 | 7 |
| Cysteine-rich repeat secretory protein (Contig_23860) | Cysteine-rich repeat secretory protein 38 (Q9LRJ9) | 6.5E-50 | ? | ? | 9 | 8 |
| Lipid transfer protein (Contig_25610 (Fragment)) | Non-specific lipid-transfer protein 11 (Q2V3C1) | 1.5E-05 | ● (PR-14) | Antifungal or antibacterial activity | 10 | - |
| β-1, 3-glucanase (BAR13253) | Glucan endo-1,3-beta-glucosidase, acidic isoform (P33157) | 1.2E-110 | ● (PR-2) | Antifungal activity | 12 | - |
| S1/P1 nuclease (Contig_14977 (Fragment)) | Endonuclease 2 (Q9C9G4) | 6.7E-51 | ? | Prey digestion | - | 10 |
| Lipid transfer protein (Contig_29120) | Non-specific lipid-transfer protein 4 (Q9LLR6) | 5.1E-06 | ● (PR-14) | Antifungal or antibacterial activity | 14 | 9 |
| Hevein-like protein (BAR13255) | Hevein-like preproprotein (P43082) | 1.0E-82 | ● (PR-4) | Antifungal activity | 11 | 12 |
| GDSL lipase (Contig_30899) | GDSL esterase/lipase APG (Q9LU14) | 2.3E-37 | ? | Prey digestion | 15 | 11 |
| Polyvinylalcohol dehydrogenase (FAA01288) | No Blast hit with E-value <10–4 | - | - | ? | 13 | 13 |
| Lipid transfer protein (Contig_30505) | Non-specific lipid-transfer protein 11 (Q2V3C1) | 3.5E-11 | ● (PR-14) | Antifungal or antibacterial activity | 16 | - |
| Polygalacturonase inhibitor (Contig_23225 (Fragment)) | Polygalacturonase inhibitor 1 (Q9M5J9) | 6.7E-49 | ● | Antifungal activity | 17 | - |
| Valine-tRNA ligase (Contig_1096 (Fragment)) | Valine--tRNA ligase, mitochondrial 1 (P93736) | 0 | ? | ? | - | 14 |
| Class IV chitinase (Contig_23195 (Fragment)) | Endochitinase EP3 (Q9M2U5) | 1.7E-31 | ● (PR-3) | Antifungal activity | 18 | - |
| S1/P1 nuclease (Contig_3798) | Endonuclease 4 (F4JJL0) | 5.2E-138 | ? | Prey digestion | - | 15 |
| Basic secretory protein (BAR13256) | No Blast hit with E-value <10–4 | - | - | ? | 19 | - |
| Serine/threonine protein kinase (Contig_9565 (Fragment)) | Serine/threonine-protein kinase Nek1 (Q9SLI2) | 0 | ? | ? | - | 16 |
| Class III peroxidase (Contig_31040 (Fragment)) | Peroxidase 51 (Q9SZE7) | 4.1E-29 | ● (PR-9) | Prey digestion | 20 | - |
| Calmodulin-binding protein (Contig_21026 (Fragment)) | Calmodulin-binding protein 60 B (Q9FKL6) | 4.4E-80 | ? | ? | - | 17 |
| Class III peroxidase (Contig_46408 (Fragment)) | Peroxidase 73 (Q43873) | 3.0E-33 | ● (PR-9) | Prey digestion | 21 | - |
a Names in the parentheses indicate the protein ID. The letters starting from ‘Contig’ indicate sequences obtained by de novo assembly based on the DRA data DRR051750 (Fukushima et al., 2017), and the others indicate sequences registered with NCBI or UniProt. b ‘PR’ indicates a group of pathogenesis-related proteins. ‘?’ indicates that we could not judge whether the protein was a defense-related protein. c ‘?’ indicates that function was unpresumable. dTable S1 and Table S2 are the bases of ranking. The proteins are listed in descending order based on the normalized average rank of each protein, which was obtained as follows: the abundance rank of each protein in the Asp-N digestion and that in the chymotrypsin digestion were divided by 21 and 17 respectively, and the resulting numbers were averaged (in case where only one digestion hit some protein, its rank in the relevant digestion was used as it was).
Expression of hydrolytic enzymes
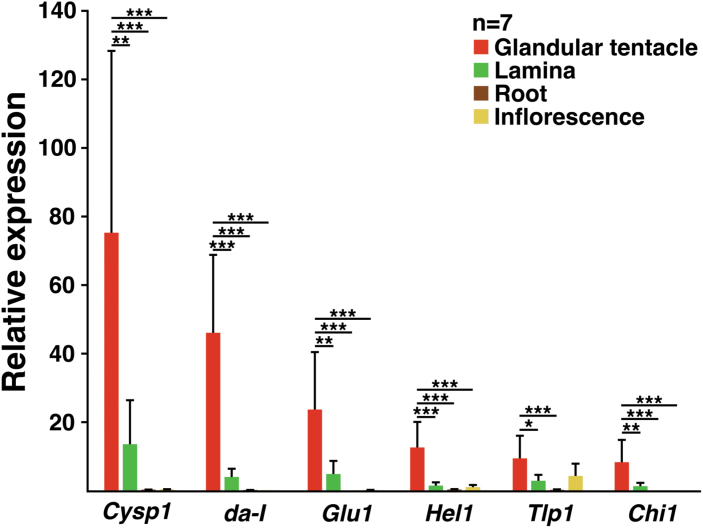
We examined the expression of genes encoding the six hydrolytic enzymes in the digestive fluid. For this experiment, D. adelae was dissected into glandular tentacles, laminas, roots and inflorescence, as described previously (Nishimura et al., 2013). All genes encoding hydrolytic enzyme were almost exclusively expressed in the trap organ (glandular tentacles), at considerably higher amounts than that of the geometric mean of the reference genes (actin, eIF4A, TIP41; Fig. 2). We previously reported the glandular tentacle-specific expression of da-I (Nishimura et al., 2013). However, the fold-expression of this gene was much higher in the present analysis. This may be related to the ‘seasonal vital difference’ of the plant: in the current study, glandular tentacles were dissected in June (rainy season in Japan), while in the previous study (Nishimura et al., 2013), they were dissected at the end of August. The former plants were more healthy than the latter, under our growth conditions (Nishimura et al., 2013). The high expression of Cysp1 and da-I in the glandular tentacles correlated with their corresponding protein contents, which were quite high (Table 1).
Fig. 2.
The genes encoding the major hydrolytic enzymes in the digestive fluid are expressed in a glandular tentacle-specific manner. The expression of Cysp1, da-I, Glu1, Hel1, Tlp1, and Chi1 in glandular tentacles, laminas, roots and inflorescences were examined and normalized to that of the geometric mean of the reference genes (actin, eIF4A, TIP41). The values are presented as the mean ±SD (n=7 biological replicates). P values were calculated by Tukey’s HSD test (*P<0.05; **P<0.01, ***P<0.001).
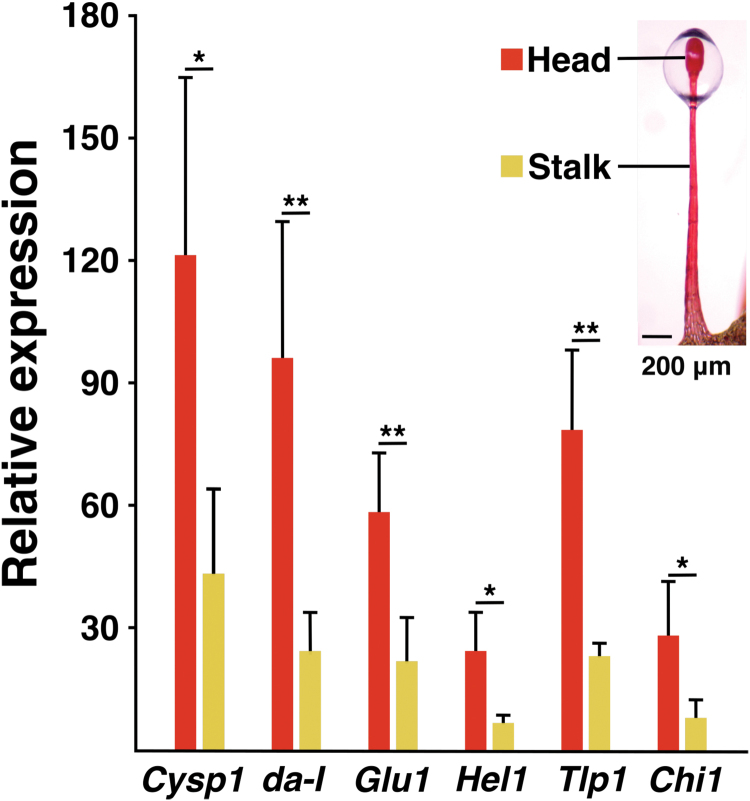
The glandular tentacles can be divided into two parts: head and stalk (Lloyd, 1942; Williams and Pickard, 1974; Outenreath and Dauwalder, 1982; Naidoo and Heneidak, 2013). Thus, we also examined the expression of Cysp1, da-I, Chi1, Glu1, Tlp1, and Hel1 in each part (Fig. 3), and found that most of the transcripts collected from the glandular tentacles were derived from the heads. In Drosera capensis, the head is mostly composed of the outer digestive gland cells (ODGC) and the inner digestive gland cells (IDGC; Williams and Pickard, 1974; Outenreath and Dauwalder, 1982). Given the similarity between D. capensis and D. adelae (Cameron et al., 2002), the six genes of interest were also considered to be transcribed in these cells in D. adelae. Lower amounts of these gene transcripts were also found in the stalks (Fig. 3). The stalk mostly consists of outer and inner stalk cells, and it also has sessile gland cells (SGC). Most of all of these cells may be implicated in this phenomenon.
Fig. 3.
Expression of Cysp1, da-I, Glu1, Hel1, Tlp1 and Chi1 in the head and stalk parts of glandular tentacles. Gene expression was normalized to that of the geometric mean of the reference genes (actin, eIF4A, TIP41). The values are presented as the mean ±SD (n=4 biological replicates). P values were calculated by Student’s t-test. (*P<0.05; **P<0.01).
DNA methylation profiles of genes encoding hydrolytic enzymes
In eukaryotes, DNA methylation is a chemical modification of genomes that affects gene expression and genome stability (Law and Jacobsen, 2010; Zhang et al., 2018). DNA methylation in plants commonly occurs in three cytosine sequence contexts: CG, CHG and CHH (where characters C and G represent nucleotides C and G, and H means non-G nucleotides; Zhang et al., 2006; Cokus et al., 2008; Lister et al., 2008). We previously reported that da-I, which is highly and specifically expressed in glandular tentacles, is only unmethylated in that organ, and highly methylated in other organs (Nishimura et al., 2013). This suggested that DNA methylation controls the expression of da-I. To determine whether the same phenomenon is observed in the hydrolytic enzyme genes described above, we analysed the DNA methylation profiles of the regions upstream of their TSSs.
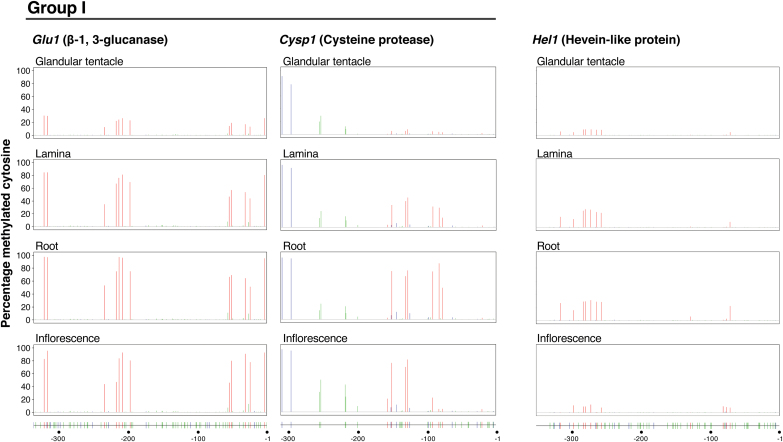
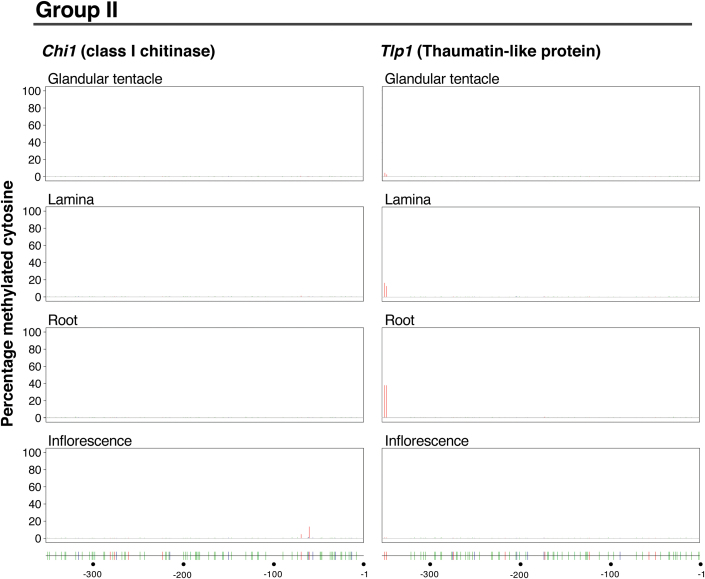
The DNA methylation profiles for Glu1 and Cysp1 (Fig.4) were somewhat similar to that of da-I (Nishimura et al., 2013). On the other hand, the profiles for Chi1 and Tlp1 considerably differed from the da-I profile: for these genes, methylation was generally not found or quite restricted. We named da-I, Glu1 and Cysp1 as the group I genes, and Chi1 and Tlp1 as the group II genes. The Hel1 profile appeared to be in between those of the two groups. The methylation profile of Glu1 was the closest to that of da-I (Nishimura et al., 2013), in the sense that the extent of methylation at the CG sites was generally high except for glandular tentacles and that the methylation at CHG and CHH sites was generally low in all organs examined (these sites were almost unmethylated in Glu1), although a small population of cells had methylated CG sites in the glandular tentacles in the case of Glu1 (Fig. 4). The differentially methylated regions (DMRs) for the CG sites of Glu1 were −322 to −317, −235 to −198 and −55 to −4, and the percentages of the methylation in this order were 30%, 21%, and 18% in glandular tentacles, 84%, 66%, and 56% in laminas, 97%, 79%, and 69% in roots, and 89%, 69%, and 77% in inflorescences, respectively. The CG sites in Cysp1 were almost unmethylated in the glandular tentacles, and were highly, regionally high, or moderately methylated in roots, inflorescences and laminas, respectively. For the methylations at the CHG and CHH sites in the gene, the profiles were almost the same amongst the organs, and the methylation extents were very low in the region from ~ −200 to −1. The DMRs for the CG sites of Cysp1 were from positions −157 to −128 and −92 to −78, and the percentages in this order were 5% and 4% in glandular tentacles, 31% and 26% in laminas, 55% and 70% in roots, and 62% and 10% in inflorescences, respectively. Regarding Hel1, the population of cells with methylated CG sites was small, and methylated CHG and CHH sites were not detected in all organs. For Hel1, the position of the DMR was from −317 to −258 and the percentages were 8% in glandular tentacles, 22% in laminas, 27% in roots, and 7% in inflorescences.
Organ-specific expression of cytosine-5 DNA methyltransferases and DNA demethylases
Plant C5-MTases can be divided into three families: methyltransferase (MET) family, chromomethylase (CMT) family, and domains rearranged methyltransferase (DRM) family (Finnegan et al., 1996; Lindroth et al., 2001; Cao and Jacobsen, 2002; Stroud et al., 2014). Plants also have a DNA demethylase family. In A. thaliana, this family comprises four members: REPRESSOR OF SILENCING 1 (ROS1), DEMETER (DME), DEMETER-LIKE PROTEIN 2 (DML2) and DML3 (Choi et al., 2002; Gong et al., 2002; Ortega-Galisteo et al., 2008), which can excise the methyl group of 5-meC from all cytosine sequence contexts (Agius et al., 2006; Gehring et al., 2006; Morales-Ruiz et al., 2006; Penterman et al., 2007; Zhu et al., 2007). We then analysed the expression of the C5-MTases and DNA demethylases in the relevant organs. To obtain the sequence information of these enzymes, the D. adelae proteins were screened using HMMER, which allowed us to identify the putative complete or partial coding sequences of three C5-MTase and two DNA demethylase genes (Supplementary Table S5).
The three C5-MTases were named DaMET1, DaCMT3, and DaDRM2 (Supplementary Table S5). DaMET1 was confirmed to have two replication foci domains, two bromo adjacent homology (BAH) domains and a C-terminal DNA MTase domain, usually found in the MET family. This family is used for maintenance methylation and methylates CG (Finnegan et al., 1996; Kankel et al., 2003). DaCMT3 was also confirmed to have a BAH domain and a chromodomain within the C-terminal DNA MTase domain, as commonly found among the CMT family enzymes. The CMT3 binds to H3K9me2 with its BAH and chromodomain, and maintains CHG methylation (Du et al., 2012). Regarding DaDRM2, we found two ubiquitin-associated domains in its N-terminal region and a DNA MTase domain in its C-terminal region, and thus confirmed that it belongs to the DRM family. This family is used for both the de novo methylation in all sequence contexts (CG, CHG and CHH) and the maintenance of CHH methylation, through the RNA-directed DNA methylation pathway (Matzke and Mosher, 2014). Although the sequence of DaCMT3 was partial, it was used in subsequent analysis because it contained all of the characteristic domains of CMT.
Each of the two putative DNA demethylases identified in the current study contains a helix-hairpin-helix Gly/Pro/Asp domain, a permuted single zinc finger-CXXC unit domain and an RNA recognition motif domain (Law and Jacobsen, 2010), which are also present in most plant DNA demethylases. Furthermore, the search against the A. thaliana protein database (UniProtKB/Swiss-Prot) revealed that one was most similar to DME, and the other was similar to ROS1. Thus, they were named DaDME and DaROS1, respectively (Supplementary Table S5).
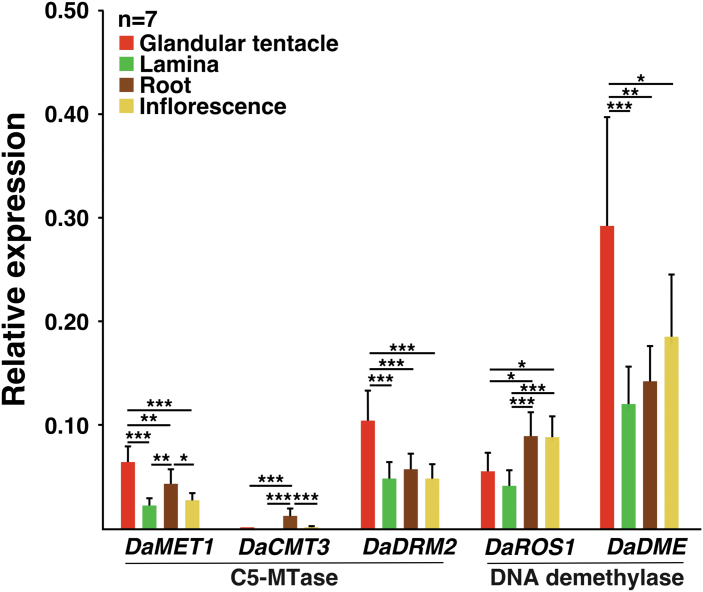
The relative expression of the identified genes (DaMET1, DaCMT3, DaDRM2, DaDME and DaROS1) in glandular tentacles, laminas, roots and inflorescences were analyzed by qPCR (Fig. 5). The expression of three genes DaMET1, DaDRM2 and DaDME were higher in the glandular tentacles than in the other organs. Notably, in the glandular tentacles, the expression of DaDME was considerably higher than those of the other four genes. DaCMT3 expression was almost undetectable in all organs except roots.
Fig. 5.
Expression of the genes encoding C5-MTase and DNA demethylase. Using glandular tentacles, laminas, roots and inflorescences, the expression of the relevant genes was examined and normalized to that of the geometric mean of the reference genes (actin, eIF4A, TIP41). The values are represented as the mean ±SD (n=7 biological replicates). P values were calculated by Tukey’s HSD test (*P<0.05; **P<0.01; ***P<0.001).
Discussion
Similarity of proteins in the digestive fluids of D. adelae and other carnivorous plants
Many of the major proteins in the digestive fluid of D. adelae (Table 1) are also present in other carnivorous plants; e.g. D. muscipula: an S-like RNase, cysteine proteases, a class I chitinase, a β-1, 3-glucanase(s), a TLP, LTPs and a HEL (Schulze et al., 2012; Nishimura et al., 2013; Bemm et al., 2016); Nepenthes alata: an S-like RNase, β-1, 3-glucanases, a TLP(s) and LTPs (Hatano and Hamada, 2008, 2012; Rottloff et al., 2016; Fukushima et al., 2017); C. follicularis: an S-like RNase, a β-1, 3-glucanase and TLPs (Nishimura et al., 2013; Fukushima et al., 2017); and S. purpurea: β-1, 3-glucanases and a TLP (Fukushima et al., 2017). The major proteins in the digestive fluid of D. muscipula are very similar to those of D. adelae (Schulze et al., 2012). A β-1, 3-glucanase and TLP are also predominant in the digestive fluid of N. alata (Hatano and Hamada, 2008). Importantly, many of these proteins also function in the roots of non-carnivorous plants (Nóbrega et al., 2005; Basu et al., 2006; De-la-Peña et al., 2008, 2010; Shinano et al., 2011, 2013).
The protease in the digestive fluid of D. adelae belongs to the cysteine protease family (Table 1). Similarly, D. muscipula predominantly secretes cysteine proteases in the digestive fluid (Schulze et al., 2012; Bemm et al., 2016). On the other hand, Nepenthes and C. follicularis mainly secrete aspartic proteases (Hatano and Hamada, 2008, 2012; Rottloff et al., 2016; Fukushima et al., 2017), similar to the D. capensis digestive fluid that has an aspartic protease (Takahashi et al., 2009; Krausko et al., 2017). At present, we do not understand the reason for these differences. There are two possibilities to consider whether D. adelae actually has an aspartic protease in its digestive fluid. Firstly, D. adelae could indeed lack aspartic proteases in its digestive fluid; secondly, the proteomic analysis was not exhaustive and the aspartic protease could remain undetected. We expect the former possibility to be more likely, for the following reason. Although the database constructed for protein identification might not be exhaustive, it contained the sequences of 13 putative aspartic proteases. If an aspartic protease was present in the digestive fluid of D. adelae, then there is a strong possibility that it would have been detected by the analysis. However, we only detected the cysteine protease CYSP1. It must also be noted that the constructed database was based on the transcriptome data of shoots (Fukushima et al., 2017). Thus, it seems that the expression of the aspartic proteases is either very low or does not occur in the glandular tentacles of D. adelae.
Similarity between trap organs and roots as deduced by the proteins in the digestive fluids
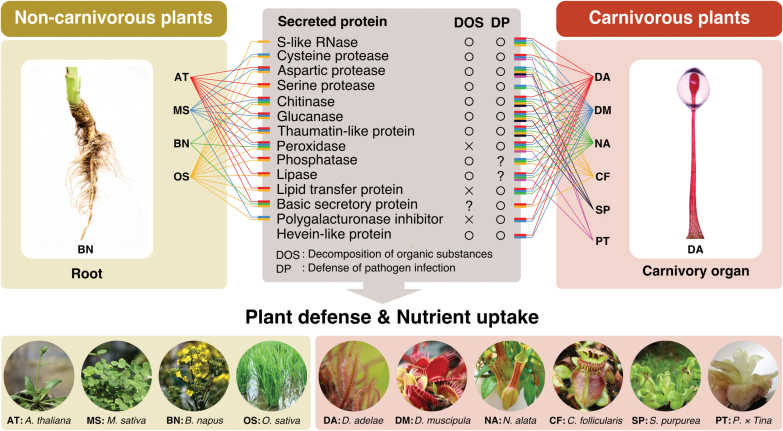
The carnivory organ (glandular tentacles) of D. adelae is similar to roots in terms of function, i.e. both organs can incorporate nutrients, protect themselves against pathogen infection, and establish symbiotic relationships with microorganisms. As for nutrient incorporation from prey, the S-like RNase DA-I and the cysteine protease CYSP1, which are highly abundant in the digestive fluid (Table 1), are thought to play the central role in D. adelae. In non-carnivorous plants, nucleases and proteases are considered to be secreted from roots upon phosphate or nitrogen deficiency, thereby allowing plants to efficiently absorb the nutrients present in soil (Chen et al., 2000; Godlewski and Adamczyk, 2007; Paungfoo-Lonhienne et al., 2008; Adamczyk et al., 2008, 2009; Adamczyk, 2014). Except for these enzymes, many of the proteins in the digestive fluid were ‘defense-related proteins’, of which most were PR proteins (Table 1). For example, chitinase can decompose the cell walls of fungi (Schlumbaum et al., 1986; Mauch et al., 1988; Sela-Buurlage et al., 1993), and TLP has antifungal activity (Grenier et al., 1999; Zareie et al., 2002). Thus, these proteins are thought to be used in a defense system against fungal attacks. Similarly, the major proteins in the digestive fluids of D. muscipula, N. alata, C. follicularis and S. purpurea were also defense-related proteins (Hatano and Hamada, 2008, 2012; Schulze et al., 2012; Nishimura et al., 2013; Bemm et al., 2016; Rottloff et al., 2016; Fukushima et al., 2017), although their expression in D. muscipula is inductive. In non-carnivorous plants, including A. thaliana, Medicago sativa, Brassica napus and Oryza sativa, many of the defense-related proteins shown in Fig. 6 are constitutively or inducibly secreted from the roots to the rhizosphere (Nóbrega et al., 2005; Basu et al., 2006; De-la-Peña et al., 2008, 2010; Shinano et al., 2011, 2013). Thus, there is close similarity between the digestive fluid proteins of carnivorous plants and those secreted from the roots of non-carnivorous plants (Fig. 6). The S-like RNases and proteases also play an important role in the self-defense system, which functions upon wounding and pathogen attack (Ye and Droste, 1996; Galiana et al., 1997; Kariu et al., 1998; Hugot et al., 2002; Hou et al., 2018). We did not examine the symbiotic relationship of carnivorous plants with microorganisms. However, carnivory organs generally harbor symbiotic bacteria and fungi that facilitate nutrient incorporation (Albino et al., 2006; Cao et al., 2015; Chan et al., 2016; Bittleston et al., 2018; Sirová et al., 2018). Similarly, the roots of non-carnivorous plants generally receive benefits from these microorganisms (Lugtenberg and Kamilova, 2009; Berendsen et al., 2012; Martin et al., 2017).
Fig. 6.
Similarity between the proteins in the digestive fluids of carnivorous plants and those secreted from the roots of non-carnivorous plants. The relevant proteins are aspartic protease, basic secretory protein, chitinase, cysteine protease, glucanase, hevein-like protein, lipase, lipid transfer protein, peroxidase, phosphatase, polygalacturonase inhibitor, S-like RNase, serine protease and thaumatin-like protein (shown in the center panel). Many or some of them were found in the digestive fluids of D. adelae (abbreviated as DA), D. muscipula (DM), N. alata (NA), C. follicularis (CF), S. purpurea (SP) and Pinguicula × Tina (PT) (right panel), and also found to be secreted from the roots of A. thaliana (AT), M. sativa (MS), B. napus (BN) and O. sativa (OS) (left panel). DOS and DP stand for ‘decomposition of organic substances’ and ‘defense of pathogen infection’, respectively. The data sources were as follows: A. thaliana, Basu et al., 2006, De-la-Peña et al., 2008, 2010, Tran et al., 2010; M. sativa, De-la-Peña et al., 2008; B. napus, Basu et al., 2006; O. sativa, Shinano et al., 2011, 2013; D. muscipula, Schulze et al., 2012, Bemm et al., 2016; N. alata, Hatano and Hamada, 2008, 2012, Rottloff et al., 2016, Fukushima et al., 2017; C. follicularis, Fukushima et al., 2017; S. purpurea, Fukushima et al., 2017; P. × Tina, Kocáb et al., 2020.
In addition to these comparable features, recent studies have revealed another similarity between carnivory organs and roots. Transporters are involved in nutrient absorption via roots. The same transporters are used in the carnivory organs of some carnivorous plants, presumably to absorb nutrients from prey: e.g. N. alata, D. muscipula and C. follicularis use ammonium transporter 1 (AMT1; Schulze et al., 1999; Scherzer et al., 2013; Fukushima et al., 2017), and D. muscipula uses phosphate transporter 1 (PHT1; Bemm et al., 2016). Moreover, transcriptome analyses of various D. muscipula tissues demonstrated closest similarity between its glandular tissues and roots, for the genes implicated in protein metabolism, transport and stress responses (Bemm et al., 2016; Palfalvi et al., 2020). Thus, carnivorous plants may have generally acquired the ability to absorb nutrients from their prey via leaves as an auxiliary mechanism for adaptation to nutrient- poor habitats.
Expression of genes encoding hydrolytic enzymes
The hydrolytic enzyme-encoding genes Cysp1, Chi1, Glu1, Tlp1 and Hel1 were almost exclusively expressed in glandular tentacles (Fig. 2). Furthermore, the heads generated the greatest proportion of the transcripts (Fig. 3). According to the studies using D. capensis, the head of the glandular tentacle contains ODGC, IDGC, endodermal cells forming a continuous layer that separate the digestive gland cells from the central conducting tissues, tracheids, and neck cells (Williams and Pickard, 1974; Outenreath and Dauwalder, 1982). Amongst these, ODGC and IDGC form the majority of the cell population. On the other hand, the stalk mostly consists of outer and inner stalk cells, and it also has SGC (Lloyd, 1942; Williams and Pickard, 1974; Naidoo and Heneidak, 2013). The cell composition in the glandular tentacles seems similar between D. adelae and D. capensis (Cameron et al., 2002). Accordingly, for heads, the major population of the transcripts of the six genes shown in Fig. 2 were most likely derived from ODGC and IDGC. For stalks, these genes may be expressed in most or all of the cells described above. For the inner stalk cells, a study using D. rotundifolia previously reported that induction-independent chitinase transcripts are confined to the cells (Matušíková et al., 2005). The study using D. capensis showed that SGC also has a secretory function (Naidoo and Heneidak, 2013). The secreted fluid from the SGC may be transported into the head area.
DNA methylation profiles of the group I genes (da-I, Glu1 and Cysp1) and those of the group II genes (Chi1 and Tlp1) were distinct (for da-I, Nishimura et al., 2013; for the other genes, Fig. 4). The CG sites of Glu1 and Cysp1 were almost unmethylated in the upstream region of each TSS in glandular tentacles, but highly methylated in laminas, roots and inflorescences (Fig. 4). These profiles were very similar to those of da-I (Nishimura et al., 2013). Therefore, the glandular tentacle-specific expression of these genes may also be explained in terms of epigenetic regulation, based on open and closed chromatin structures, as previously hypothesized for da-I expression by Nishimura et al. (2013). A transcription factor-implicated mechanism is also possible, as some transcription factors cannot bind to their target sequences when they are methylated, thus preventing transcription initiation (Mann et al., 2013; O’Malley et al., 2016; Yin et al., 2017). If the latter mechanism actually exists, then some DNA motifs that act in cis should be identified in the DMRs: da-I, –349 to –307, –257 to –205, –139 to –97 and –50 to –24 (Nishimura et al., 2013); Glu1, −322 to −317, −235 to −198 and −55 to −4; and Cysp1, −157 to −128 and −92 to −78. However, we could not detect any known cis DNA element or trans-acting factors that are likely to be responsible for this hypothetical mechanism (Okabe et al., 2005b; Supplementary Fig. S2).
Fig. 4.
Organ-dependent DNA methylation profiles of the genes encoding the major hydrolytic enzymes. Focusing on the genes Glu1, Cysp1, Hel1, Chi1 and Tlp1, the DNA methylation status of the upstream region of the TSS was examined in several organs, including glandular tentacles, laminas, roots and inflorescences. Histograms show the percentages of methylation at cytosine residues in CG, CHG and CHH contexts: CG, red; CHG, blue; CHH, green (averages of three individual plants). The lower diagrams show the positions of cytosines in the same contexts with the same colors. According to the methylation characteristics, the genes were grouped as group I, almost unmethylated only in glandular tentacles; group II, almost unmethylated in all organs examined.
On the other hand, the upstream regions of the TSSs of Chi1 and Tlp1 (group II) were almost completely unmethylated in all organs (Fig. 4). However, the expression of these genes was glandular tentacle-specific (Fig. 2). These observations strongly suggest that glandular cells contain some key, but unknown transcription factor(s) that can drive the transcription of these genes. Notably, the genes encoding the S-like RNases dm-I of D. muscipula and cf-I of C. follicularis have characteristics similar to those of the group II genes (Nishimura et al., 2013). Thus, the orthologs are not necessarily regulated in the same way. In the case of Hel1, its methylation profiles were in between those of groups I and II (Fig. 4). Considering its extent of methylation, however, DNA methylation may be irrelevant to its expression. We found a PHR1 (phosphate starvation response 1)-binding sequence (P1BS), a phosphate starvation responsive sequence (Rubio et al., 2001), in the region spanning from −317 to −258 (Fig. 4; Supplementary Fig. S2). Phosphate starvation may activate PHR1, which could then bind to P1BS and trigger Hel1 expression in glandular tentacles. The P1BS motif is also present in the da-I promoter (Okabe et al., 2005b) and the promoter of the RNase T2 gene that shows trap-specific expression in U. gibba (Oropeza-Aburto et al., 2020) which seems very intriguing.
Implications of the glandular tentacle-specific DNA demethylation
The methylation profiles of the group I genes (Fig. 4; Nishimura et al., 2013) suggested two possibilities: demethylation of these genes occurs in a glandular tentacle-specific manner, or their methylation specifically occurs in laminas, roots and inflorescences. The expression of DaMET1 and DaDRM2 was very low in laminas, roots and inflorescences, while in the glandular tentacles, DaDME expression was considerably higher than those of the other genes (Fig. 5). DaMET1 and DaDRM2 are MTases and DaDME is a demethylase. Thus, the data shown in Fig. 5 support the possibility that the demethylation of the group I genes (da-I, Glu1 and Cysp1) occurs in a glandular tentacle-specific manner. To our knowledge, tissue-specific DNA demethylation by DME confers distinct gene expression during gamete formation and nodule development in non-carnivorous plants (Gehring et al., 2009; Hsieh et al., 2009; Ibarra et al., 2012; Satgé et al., 2016). Thus, we may consider the glandular tentacle-specific demethylation of the group I genes as a similar type of gene regulation, in the sense that organ-specific DNA demethylation by DME presumably occurs in the differentiation process of glandular tentacles. Clearly, follow on work should examine the timing of promoter methylation or demethylation in the process of D. adelae organogenesis.
Conclusions
Our results substantiate the following hypothesis, in which a part was previously hypothesized regarding the abundant presence of DA-I in the digestive fluid (Okabe et al., 2005a). The ancestors of D. adelae must have engendered the development of carnivory in their leaves to adapt to nutrient-deficient habitats and, at the same time, establish self-defense mechanisms against pathogen attack and mechanical injury caused by insects. Furthermore, these features could be attained by slight modifications of the expression mechanisms of a set of genes that are generally used for specific functions of roots, including the epigenetic regulation shown in this study. A somewhat similar phenomenon occurs in the roots of some non-carnivorous plants: phosphate or nitrogen starvation causes the plants to express genes for nutrient absorption in the roots by altering DNA methylation patterns (Secco et al., 2015; Yong-Villalobos et al., 2015; Mager and Ludewig, 2018). To understand the evolution of carnivorous plants, it seems absolutely necessary to further explore similarities between roots of non-carnivorous plants and trap-organs of carnivorous plants, such as in gene usage and regulatory mechanisms of gene expression.
Supplementary data
The following supplementary data are available at JXB online.
Fig. S1. FASTA file of amino acid sequences of the proteins subjected to LC-MS/MS analysis
Fig. S2. Upstream sequences and cis-DNA elements of Cysp1, Chi1, Glu1, Tlp1 and Hel1
Table S1. Summary of iBAQ values in the Asp-N digestion
Table S2. Summary of iBAQ values in the chymotrypsin digestion
Table S3. PCR primers used in the Real-Time PCR
Table S4. PCR primers used in the bisulfite sequencing
Table S5. C5-MTases and DNA demethylases identified in D. adelae
Data availability
The mass spectrometry proteomics data have been deposited to the Proteome Xchange Consortium via the jPOST partner repository (Okuda et al., 2017) with the dataset identifier PXD022147. The bisulfite sequence data have been deposited to DDBJ (accession number:DRA010963). Sequence data can be found in the DDBJ data libraries under the accession numbers LC037411 (Bsep1), LC037409 (Chi1), LC549487 (Cysp1), LC037408 (Glu1), LC037410 (Hel1), LC547507 (Tlp1), BR001645 (polyvinylalcohol dehydrogenase gene), YAAA01000001 (DaMET1), YAAA01000002 (DaCMT3), YAAA01000003 (DaDRM2), YAAA01000004 (DaROS1), YAAA01000005 (DaDME), LC589199 (eIF4A) and LC589200 (TIP41).
Acknowledgements
We thank Satoshi Miyanaga at the Center for Advanced Biomedical Sciences for the SWATH-MS analysis. This study was supported in part by Grants-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology-JAPAN (MEXT; to TO, 20K06777).
Glossary
Abbreviations
- AMT1
ammonium transporter 1
- BAH
bromo adjacent homology
- C5-MTase
cytosine-5 DNA methyltransferase
- CMT
chromomethylase
- DME
DEMETER
- DML
DEMETER-LIKE PROTEIN
- DMR
differentially methylated region
- DRM
domains rearranged methyltransferase
- FDR
false discovery rate
- HEL
hevein-like protein
- iBAQ
intensity-based absolute quantification
- IDA
information-dependent acquisition
- IDGC
inner digestive-gland cells
- LTP
lipid transfer protein
- MET
methyltransferase
- ODGC
outer digestive-gland cells
- P1BS, PHR1
-binding sequence
- PHR1
phosphate starvation response 1
- PR
pathogenesis-related
- RNase
ribonuclease
- ROS1
REPRESSOR OF SILENCING 1
- SGC
sessile gland cells
- SWATH-MS
sequential window acquisition of all theoretical fragment ion spectra mass spectrometry
- TAIL
thermal asymmetric interlaced
- TLP
thaumatin-like protein
- TSS
transcription start site
- XIC
extracted ion chromatogram.
Author contributions
NA and TO conceived and designed the study; NA, YO, SJ and YH performed the experiments; NA, YO, and TO analysed the data; NA and TO wrote the manuscript. All authors read and approved the final manuscript.
References
- Adamczyk B. 2014. Characterization of proteases secreted by leek roots. Russian Journal of Plant Physiology 61, 714–717. [Google Scholar]
- Adamczyk B, Godlewski M, Smolander A, Kitunen V. 2009. Degradation of proteins by enzymes exuded by Allium porrum roots-a potentially important strategy for acquiring organic nitrogen by plants. Plant Physiology and Biochemistry 47, 919–925. [DOI] [PubMed] [Google Scholar]
- Adamczyk B, Godlewski M, Zimny J, Zimny A. 2008. Wheat (Triticum aestivum) seedlings secrete proteases from the roots and, after protein addition, grow well on medium without inorganic nitrogen. Plant Biology 10, 718–724. [DOI] [PubMed] [Google Scholar]
- Agius F, Kapoor A, Zhu JK. 2006. Role of the Arabidopsis DNA glycosylase/lyase ROS1 in active DNA demethylation. Proceedings of the National Academy of Sciences, USA 103, 11796–11801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert VA, Williams SE, Chase MW. 1992. Carnivorous plants: phylogeny and structural evolution. Science 257, 1491–1495. [DOI] [PubMed] [Google Scholar]
- Albino U, Saridakis DP, Ferreira MC, Hungria M, Vinuesa P, Andrade G. 2006. High diversity of diazotrophic bacteria associated with the carnivorous plant Drosera villosa var. villosa growing in oligotrophic habitats in Brazil. Plant and Soil 287, 199–207. [Google Scholar]
- Arai N, Nishimura E, Kikuchi Y, Ohyama T. 2015. Functional analyses of carnivorous plant-specific amino acid residues in S-like ribonucleases. Biochemical and Biophysical Research Communications 465, 108–112. [DOI] [PubMed] [Google Scholar]
- Athauda SB, Matsumoto K, Rajapakshe S, et al. 2004. Enzymic and structural characterization of nepenthesin, a unique member of a novel subfamily of aspartic proteinases. The Biochemical Journal 381, 295–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bariola PA, Howard CJ, Taylor CB, Verburg MT, Jaglan VD, Green PJ. 1994. The Arabidopsis ribonuclease gene RNS1 is tightly controlled in response to phosphate limitation. The Plant Journal 6, 673–685. [DOI] [PubMed] [Google Scholar]
- Basu U, Francis JL, Whittal RM, Stephens JL, Wang Y, Zaiane OR, Goebel R, Muench DG, Good AG, Taylor GJ. 2006. Extracellular proteomes of Arabidopsis thaliana and Brassica napus roots: analysis and comparison by MudPIT and LC-MS/MS. Plant and Soil 286, 357–376. [Google Scholar]
- Bekesiova I, Nap JP, Mlynarova L. 1999. Isolation of high quality DNA and RNA from leaves of the carnivorous plant Drosera rotundifolia. Plant Molecular Biology Reporter 17, 269–277. [Google Scholar]
- Bemm F, Becker D, Larisch C, et al. 2016. Venus flytrap carnivorous lifestyle builds on herbivore defense strategies. Genome Research 26, 812–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berendsen RL, Pieterse CM, Bakker PA. 2012. The rhizosphere microbiome and plant health. Trends in Plant Science 17, 478–486. [DOI] [PubMed] [Google Scholar]
- Bittleston LS, Wolock CJ, Yahya BE, Chan XY, Chan KG, Pierce NE, Pringle A. 2018. Convergence between the microcosms of Southeast Asian and North American pitcher plants. eLife. Doi: 10.7554/eLife.36741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhm J, Scherzer S, Krol E, et al. 2016. The venus flytrap Dionaea muscipula counts prey-induced action potentials to induce sodium uptake. Current Biology 26, 286–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner F, Stintzi A, Fritig B, Legrand M. 1998. Substrate specificities of tobacco chitinases. The Plant Journal 14, 225–234. [DOI] [PubMed] [Google Scholar]
- Cameron K, Wurdack K, Jobson R. 2002. Molecular evidence for the common origin of snap-traps among carnivorous plants. American Journal of Botany 89, 1503–1509. [DOI] [PubMed] [Google Scholar]
- Cao X, Jacobsen SE. 2002. Role of the Arabidopsis DRM methyltransferases in de novo DNA methylation and gene silencing. Current Biology 12, 1138–1144. [DOI] [PubMed] [Google Scholar]
- Cao HX, Schmutzer T, Scholz U, Pecinka A, Schubert I, Vu GT. 2015. Metatranscriptome analysis reveals host-microbiome interactions in traps of carnivorous Genlisea species. Frontiers in Microbiology 6, 526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan XY, Hong KW, Yin WF, Chan KG. 2016. Microbiome and biocatalytic bacteria in monkey cup (Nepenthes Pitcher) digestive fluid. Scientific Reports 6, 20016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen DL, Delatorre CA, Bakker A, Abel S. 2000. Conditional identification of phosphate-starvation-response mutants in Arabidopsis thaliana. Planta 211, 13–22. [DOI] [PubMed] [Google Scholar]
- Choi Y, Gehring M, Johnson L, Hannon M, Harada JJ, Goldberg RB, Jacobsen SE, Fischer RL. 2002. DEMETER, a DNA glycosylase domain protein, is required for endosperm gene imprinting and seed viability in Arabidopsis. Cell 110, 33–42. [DOI] [PubMed] [Google Scholar]
- Chow CN, Lee TY, Hung YC, Li GZ, Tseng KC, Liu YH, Kuo PL, Zheng HQ, Chang WC. 2019. PlantPAN3.0: a new and updated resource for reconstructing transcriptional regulatory networks from ChIP-seq experiments in plants. Nucleic Acids Research 47, D1155–D1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, Haudenschild CD, Pradhan S, Nelson SF, Pellegrini M, Jacobsen SE. 2008. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature 452, 215–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin C. 1875. Insectivorous plants. London: John Murray. [Google Scholar]
- De-la-Peña C, Badri DV, Lei Z, Watson BS, Branda MM, Silva-Filho MC, Sumner LW, Vivanco JM. 2010. Root secretion of defense-related proteins is development-dependent and correlated with flowering time. Journal of Biological Chemistry 285, 30654–30665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De-la-Peña C, Lei Z, Watson BS, Sumner LW, Vivanco JM. 2008. Root-microbe communication through protein secretion. Journal of Biological Chemistry 283, 25247–25255. [DOI] [PubMed] [Google Scholar]
- Du J, Zhong X, Bernatavichute YV, et al. 2012. Dual binding of chromomethylase domains to H3K9me2-containing nucleosomes directs DNA methylation in plants. Cell 151, 167–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilenberg H, Pnini-Cohen S, Schuster S, Movtchan A, Zilberstein A. 2006. Isolation and characterization of chitinase genes from pitchers of the carnivorous plant Nepenthes khasiana. Journal of Experimental Botany 57, 2775–2784. [DOI] [PubMed] [Google Scholar]
- Eilenberg H, Zilberstein A. 2008. Carnivorous pitcher plants – Towards understanding the molecular basis of prey digestion. In Teixeira da Silva JA, ed. Floriculture, ornamental and plant biotechnology: advances and topical issues. Isleworth: Global Science Books, 287–294. [Google Scholar]
- Ellison AM, Adamec L. 2018. Carnivorous plants: physiology, ecology, and evolution. Oxford: Oxford University Press. [Google Scholar]
- Ellison AM, Gotelli NJ. 2009. Energetics and the evolution of carnivorous plants–Darwin’s ‘most wonderful plants in the world’. Journal of Experimental Botany 60, 19–42. [DOI] [PubMed] [Google Scholar]
- Escalante-Pérez M, Krol E, Stange A, Geiger D, Al-Rasheid KA, Hause B, Neher E, Hedrich R. 2011. A special pair of phytohormones controls excitability, slow closure, and external stomach formation in the Venus flytrap. Proceedings of the National Academy of Sciences, USA 108, 15492–15497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández V, Brown PH. 2013. From plant surface to plant metabolism: the uncertain fate of foliar-applied nutrients. Frontiers in Plant Science 4, 289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández V, Eichert T. 2009. Uptake of hydrophilic solutes through plant leaves: current state of knowledge and perspectives of foliar fertilization. Critical Reviews in Plant Science 28, 36–68. [Google Scholar]
- Finnegan EJ, Peacock WJ, Dennis ES. 1996. Reduced DNA methylation in Arabidopsis thaliana results in abnormal plant development. Proceedings of the National Academy of Sciences, USA 93, 8449–8454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischmann A, Schlauer J, Smith SA, Givnish TJ. 2018. Evolution of carnivory in angiosperms. In: Ellison A, Adamec L, eds. Carnivorous plants: physiology, ecology, and evolution. Oxford: Oxford University Press, 22–41. [Google Scholar]
- Fukushima K, Fang X, Alvarez-Ponce D, et al. 2017. Genome of the pitcher plant Cephalotus reveals genetic changes associated with carnivory. Nature Ecology and Evolution. Doi: 10.1038/s41559-016-0059 [DOI] [PubMed] [Google Scholar]
- Fukushima K, Fujita H, Yamaguchi T, Kawaguchi M, Tsukaya H, Hasebe M. 2015. Oriented cell division shapes carnivorous pitcher leaves of Sarracenia purpurea. Nature Communications. doi: 10.1038/ncomms7450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galiana E, Bonnet P, Conrod S, Keller H, Panabières F, Ponchet M, Poupet A, Ricci P. 1997. RNase activity prevents the growth of a fungal pathogen in tobacco leaves and increases upon induction of systemic acquired resistance with elicitin. Plant Physiology 115, 1557–1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring M, Bubb KL, Henikoff S. 2009. Extensive demethylation of repetitive elements during seed development underlies gene imprinting. Science 324, 1447–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring M, Huh JH, Hsieh TF, Penterman J, Choi Y, Harada JJ, Goldberg RB, Fischer RL. 2006. DEMETER DNA glycosylase establishes MEDEA polycomb gene self-imprinting by allele-specific demethylation. Cell 124, 495–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gijzen M, Kuflu K, Qutob D, Chernys JT. 2001. A class I chitinase from soybean seed coat. Journal of Experimental Botany 52, 2283–2289. [DOI] [PubMed] [Google Scholar]
- Givnish TJ. 2015. New evidence on the origin of carnivorous plants. Proceedings of the National Academy of Sciences, USA 112, 10–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godlewski M, Adamczyk B. 2007. The ability of plants to secrete proteases by roots. Plant Physiology and Biochemistry 45, 657–664. [DOI] [PubMed] [Google Scholar]
- Gong Z, Morales-Ruiz T, Ariza RR, Roldán-Arjona T, David L, Zhu JK. 2002. ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell 111, 803–814. [DOI] [PubMed] [Google Scholar]
- Green PJ. 1994. The ribonucleases of higher plants. Annual Review of Plant Physiology and Plant Molecular Biology 45, 421–445. [Google Scholar]
- Grenier J, Potvin C, Trudel J, Asselin A. 1999. Some thaumatin-like proteins hydrolyse polymeric beta-1,3-glucans. The Plant Journal 19, 473–480. [DOI] [PubMed] [Google Scholar]
- Haas BJ, Papanicolaou A, Yassour M, et al. 2013. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nature Protocols 8, 1494–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamel F, Bellemare G. 1995. Characterization of a class I chitinase gene and of wound-inducible, root and flower-specific chitinase expression in Brassica napus. Biochimica et Biophysica Acta 1263, 212–220. [DOI] [PubMed] [Google Scholar]
- Hatano N, Hamada T. 2008. Proteome analysis of pitcher fluid of the carnivorous plant Nepenthes alata. Journal of Proteome Research 7, 809–816. [DOI] [PubMed] [Google Scholar]
- Hatano N, Hamada T. 2012. Proteomic analysis of secreted protein induced by a component of prey in pitcher fluid of the carnivorous plant Nepenthes alata. Journal of Proteomics 75, 4844–4852. [DOI] [PubMed] [Google Scholar]
- Heubl G, Bringmann G, Meimberg H. 2006. Molecular phylogeny and character evolution of carnivorous plant families in Caryophyllales—revisited. Plant Biology 8, 821–830. [DOI] [PubMed] [Google Scholar]
- Hou S, Jamieson P, He P. 2018. The cloak, dagger, and shield: proteases in plant–pathogen interactions. Biochemical Journal 475, 2491–2509. [DOI] [PubMed] [Google Scholar]
- Hsieh TF, Ibarra CA, Silva P, Zemach A, Eshed-Williams L, Fischer RL, Zilberman D. 2009. Genome-wide demethylation of Arabidopsis endosperm. Science 324, 1451–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Lee HS, Karunanandaa B, Kao TH. 1994. Ribonuclease activity of Petunia inflata S proteins is essential for rejection of self-pollen. The Plant Cell 6, 1021–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugot K, Ponchet M, Marais A, Ricci P, Galiana E. 2002. A tobacco S-like RNase inhibits hyphal elongation of plant pathogens. Molecular Plant-Microbe Interactions 15, 243–250. [DOI] [PubMed] [Google Scholar]
- Ibarra CA, Feng X, Schoft VK, et al. 2012. Active DNA demethylation in plant companion cells reinforces transposon methylation in gametes. Science 337, 1360–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irie M. 1999. Structure-function relationships of acid ribonucleases: lysosomal, vacuolar, and periplasmic enzymes. Pharmacology & Therapeutics 81, 77–89. [DOI] [PubMed] [Google Scholar]
- Juniper BE, Robins RJ, Joel DM. 1989. The carnivorous plants. London: Academic Press. [Google Scholar]
- Kankel MW, Ramsey DE, Stokes TL, Flowers SK, Haag JR, Jeddeloh JA, Riddle NC, Verbsky ML, Richards EJ. 2003. ArabidopsisMET1 cytosine methyltransferase mutants. Genetics 163, 1109–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kariu T, Sano K, Shimokawa H, Itoh R, Yamasaki N, Kimura M. 1998. Isolation and characterization of a wound-inducible ribonuclease from Nicotiana glutinosa leaves. Bioscience, Biotechnology, and Biochemistry 62, 1144–1151. [DOI] [PubMed] [Google Scholar]
- Kocáb O, Jakšová J, Novák O, Petřík I, Lenobel R, Chamrád I, Pavlovič A. 2020. Jasmonate-independent regulation of digestive enzyme activity in the carnivorous butterwort Pinguicula × Tina. Journal of Experimental Botany 71, 3749–3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krausko M, Perutka Z, Šebela M, Šamajová O, Šamaj J, Novák O, Pavlovič A. 2017. The role of electrical and jasmonate signalling in the recognition of captured prey in the carnivorous sundew plant Drosera capensis. New Phytologist 213, 1818–1835. [DOI] [PubMed] [Google Scholar]
- Król E, Płachno BJ, Adamec L, Stolarz M, Dziubińska H, Trebacz K. 2012. Quite a few reasons for calling carnivores ‘the most wonderful plants in the world’. Annals of Botany 109, 47–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law JA, Jacobsen SE. 2010. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nature Reviews. Genetics 11, 204–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee L, Zhang Y, Ozar B, Sensen CW, Schriemer DC. 2016. Carnivorous nutrition in pitcher plants (Nepenthes spp.) via an unusual complement of endogenous enzymes. Journal of Proteome Research 15, 3108–3117. [DOI] [PubMed] [Google Scholar]
- Li W, Godzik A. 2006. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22, 1658–1659. [DOI] [PubMed] [Google Scholar]
- Liao YC, Kreuzaler F, Fischer R, Reisener H, Tiburzy R. 1994. Characterization of a wheat class Ib chitinase gene differentially induced in isogenic lines by infection with Puccinia graminis. Plant science 103, 177–187. [Google Scholar]
- Libiaková M, Floková K, Novák O, Slováková L, Pavlovič A. 2014. Abundance of cysteine endopeptidase dionain in digestive fluid of Venus flytrap (Dionaea muscipula Ellis) is regulated by different stimuli from prey through jasmonates. PLoS One. Doi: 10.1371/journal.pone.0104424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindroth AM, Cao X, Jackson JP, Zilberman D, McCallum CM, Henikoff S, Jacobsen SE. 2001. Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science 292, 2077–2080. [DOI] [PubMed] [Google Scholar]
- Lister R, O’Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, Ecker JR. 2008. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 133, 523–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YG, Mitsukawa N, Oosumi T, Whittier RF. 1995. Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. The Plant Journal 8, 457–463. [DOI] [PubMed] [Google Scholar]
- Liu YG, Whittier RF. 1995. Thermal asymmetric interlaced PCR: automatable amplification and sequencing of insert end fragments from P1 and YAC clones for chromosome walking. Genomics 25, 674–681. [DOI] [PubMed] [Google Scholar]
- Lloyd FE. 1942. The carnivorous plants. Waltham, MA: Chronica Botanica. [Google Scholar]
- Lugtenberg B, Kamilova F. 2009. Plant-growth-promoting rhizobacteria. Annual Review of Microbiology 63, 541–556. [DOI] [PubMed] [Google Scholar]
- Mager S, Ludewig U. 2018. Massive loss of DNA methylation in nitrogen-, but not in phosphorus-deficient zea mays roots is poorly correlated with gene expression differences. Frontiers in Plant Science 9, 497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann IK, Chatterjee R, Zhao J, He X, Weirauch MT, Hughes TR, Vinson C. 2013. CG methylated microarrays identify a novel methylated sequence bound by the CEBPB|ATF4 heterodimer that is active in vivo. Genome Research 23, 988–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin FM, Uroz S, Barker DG. 2017. Ancestral alliances: plant mutualistic symbioses with fungi and bacteria. Science 356, eaad4501. [DOI] [PubMed] [Google Scholar]
- Matušíková I, Pavlovič A, Renner T. 2018. Biochemistry of prey digestion and nutrient absorption. In Ellison A, Adamec L, eds. Carnivorous plants: physiology, ecology, and evolution. Oxford: Oxford University Press, 207–220. [Google Scholar]
- Matušíková I, Salaj J, Moravcíková J, Mlynárová L, Nap JP, Libantová J. 2005. Tentacles of in vitro-grown round-leaf sundew (Drosera rotundifolia L.) show induction of chitinase activity upon mimicking the presence of prey. Planta 222, 1020–1027. [DOI] [PubMed] [Google Scholar]
- Matzke MA, Mosher RA. 2014. RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nature Reviews. Genetics 15, 394–408. [DOI] [PubMed] [Google Scholar]
- Mauch F, Mauch-Mani B, Boller T. 1988. Antifungal hydrolases in pea tissue: II. Inhibition of fungal growth by combinations of chitinase and beta-1,3-glucanase. Plant Physiology 88, 936–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure B, Cruz-García F, Romero C. 2011. Compatibility and incompatibility in S-RNase-based systems. Annals of Botany 108, 647–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure BA, Haring V, Ebert PR, Anderson MA, Simpson RJ, Sakiyama F, Clarke AE. 1989. Style self-incompatibility gene products of Nicotiana alata are ribonucleases. Nature 342, 955–957. [DOI] [PubMed] [Google Scholar]
- Morales-Ruiz T, Ortega-Galisteo AP, Ponferrada-Marín MI, Martínez-Macías MI, Ariza RR, Roldán-Arjona T. 2006. DEMETER and REPRESSOR OF SILENCING 1 encode 5-methylcytosine DNA glycosylases. Proceedings of the National Academy of Sciences, USA 103, 6853–6858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller K, Borsch T, Legendre L, Porembski S, Theisen I, Barthlott W. 2004. Evolution of carnivory in Lentibulariaceae and the Lamiales. Plant Biology 6, 477–490. [DOI] [PubMed] [Google Scholar]
- Naidoo Y, Heneidak S. 2013. Morphological investigation of glandular hairs on Drosera capensis leaves with an ultrastructural study of the sessile glands. Botany 91, 234–241. [Google Scholar]
- Nishimura E, Jumyo S, Arai N, Kanna K, Kume M, Nishikawa J, Tanase J, Ohyama T. 2014. Structural and functional characteristics of S-like ribonucleases from carnivorous plants. Planta 240, 147–159. [DOI] [PubMed] [Google Scholar]
- Nishimura E, Kawahara M, Kodaira R, Kume M, Arai N, Nishikawa J, Ohyama T. 2013. S-like ribonuclease gene expression in carnivorous plants. Planta 238, 955–967. [DOI] [PubMed] [Google Scholar]
- Nóbrega FM, Santos IS, Cunha MD, Carvalho AO, Gomes VM. 2005. Antimicrobial proteins from cowpea root exudates: inhibitory activity against Fusarium oxysporum and purification of a chitinase-like protein. Plant and Soil 272, 223–232. [Google Scholar]
- Nürnberger T, Abel S, Jost W, Glund K. 1990. Induction of an extracellular ribonuclease in cultured tomato cells upon phosphate starvation. Plant Physiology 92, 970–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okabe T, Yoshimoto I, Hitoshi M, Ogawa T, Ohyama T. 2005a. An S-like ribonuclease gene is used to generate a trap-leaf enzyme in the carnivorous plant Drosera adelae. FEBS Letters 579, 5729–5733. [DOI] [PubMed] [Google Scholar]
- Okabe T, Futatsuya C, Tanaka O, Ohyama T. 2005b. Structural analysis of the gene encoding Drosera adelae S-like ribonuclease DA-I. Journal of Advanced Science 17, 218–224. [Google Scholar]
- Okuda S, Watanabe Y, Moriya Y, et al. 2017. jPOSTrepo: an international standard data repository for proteomes. Nucleic Acids Research 45, D1107–D1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Malley RC, Huang SC, Song L, Lewsey MG, Bartlett A, Nery JR, Galli M, Gallavotti A, Ecker JR. 2016. Cistrome and epicistrome features shape the regulatory DNA landscape. Cell 165, 1280–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oropeza-Aburto A, Cervantes-Pérez S, Albert V, Herrera-Estrella L. 2020. Agrobacterium tumefaciens mediated transformation of the aquatic carnivorous plant Utricularia gibba. Plant Methods. Doi: 10.1186/s13007-020-00592-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega-Galisteo AP, Morales-Ruiz T, Ariza RR, Roldán-Arjona T. 2008. Arabidopsis DEMETER-LIKE proteins DML2 and DML3 are required for appropriate distribution of DNA methylation marks. Plant Molecular Biology 67, 671–681. [DOI] [PubMed] [Google Scholar]
- Outenreath R, Dauwalder M. 1982. Ultrastructural and radioautographic studies of the digestive gland cells of Drosera capensis. I. Development and mucilage secretion. Journal of Ultrastructure Research 80, 71–88. [DOI] [PubMed] [Google Scholar]
- Palfalvi G, Hackl T, Terhoeven N, et al. 2020. Genomes of the venus flytrap and close relatives unveil the roots of plant carnivory. Current Biology 30, 2312–2320.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paungfoo-Lonhienne C, Lonhienne TG, Rentsch D, Robinson N, Christie M, Webb RI, Gamage HK, Carroll BJ, Schenk PM, Schmidt S. 2008. Plants can use protein as a nitrogen source without assistance from other organisms. Proceedings of the National Academy of Sciences, USA 105, 4524–4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlovič A, Jakšová J, Novák O. 2017. Triggering a false alarm: wounding mimics prey capture in the carnivorous Venus flytrap (Dionaea muscipula). New Phytologist 216, 927–938. [DOI] [PubMed] [Google Scholar]
- Pavlovič A, Krausko M, Libiaková M, Adamec L. 2014. Feeding on prey increases photosynthetic efficiency in the carnivorous sundew Drosera capensis. Annals of Botany 113, 69–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlovič A, Mithöfer A. 2019. Jasmonate signalling in carnivorous plants: copycat of plant defence mechanisms. Journal of Experimental Botany 70, 3379–3389. [DOI] [PubMed] [Google Scholar]
- Penterman J, Zilberman D, Huh JH, Ballinger T, Henikoff S, Fischer RL. 2007. DNA demethylation in the Arabidopsis genome. Proceedings of the National Academy of Sciences, USA 104, 6752–6757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renner T, Specht CD. 2012. Molecular and functional evolution of class I chitinases for plant carnivory in the caryophyllales. Molecular Biology and Evolution 29, 2971–2985. [DOI] [PubMed] [Google Scholar]
- Renner T, Specht CD. 2013. Inside the trap: gland morphologies, digestive enzymes, and the evolution of plant carnivory in the Caryophyllales. Current Opinion in Plant Biology 16, 436–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottloff S, Miguel S, Biteau F, et al. 2016. Proteome analysis of digestive fluids in Nepenthes pitchers. Annals of Botany 117, 479–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottloff S, Stieber R, Maischak H, Turini FG, Heubl G, Mithöfer A. 2011. Functional characterization of a class III acid endochitinase from the traps of the carnivorous pitcher plant genus, Nepenthes. Journal of Experimental Botany 62, 4639–4647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio V, Linhares F, Solano R, Martín AC, Iglesias J, Leyva A, Paz-Ares J. 2001. A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes & Development 15, 2122–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadowski EM, Seyfullah LJ, Sadowski F, Fleischmann A, Behling H, Schmidt AR. 2015. Carnivorous leaves from Baltic amber. Proceedings of the National Academy of Sciences, USA 112, 190–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito Y, Tsuji J, Mituyama T. 2014. Bisulfighter: accurate detection of methylated cytosines and differentially methylated regions. Nucleic Acids Research 42, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satgé C, Moreau S, Sallet E, et al. 2016. Reprogramming of DNA methylation is critical for nodule development in Medicago truncatula. Nature Plants 2, 16166. [DOI] [PubMed] [Google Scholar]
- Scala J, Iott K, Schwab DW, Semersky FE. 1969. Digestive secretion of Dionaea muscipula (Venus’s Flytrap). Plant Physiology 44, 367–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherzer S, Krol E, Kreuzer I, et al. 2013. The Dionaea muscipula ammonium channel DmAMT1 provides NH4+ uptake associated with Venus flytrap’s prey digestion. Current Biology 23, 1649–1657. [DOI] [PubMed] [Google Scholar]
- Schlumbaum A, Mauch F, Vögeli U, Boller T. 1986. Plant chitinases are potent inhibitors of fungal growth. Nature 324, 365–367. [Google Scholar]
- Schulze W, Frommer WB, Ward JM. 1999. Transporters for ammonium, amino acids and peptides are expressed in pitchers of the carnivorous plant Nepenthes. The Plant Journal 17, 637–646. [DOI] [PubMed] [Google Scholar]
- Schulze WX, Sanggaard KW, Kreuzer I, et al. 2012. The protein composition of the digestive fluid from the Venus flytrap sheds light on prey digestion mechanisms. Molecular & Cellular Proteomics 11, 1306–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. 2011. Global quantification of mammalian gene expression control. Nature 473, 337–342. [DOI] [PubMed] [Google Scholar]
- Secco D, Wang C, Shou H, Schultz MD, Chiarenza S, Nussaume L, Ecker JR, Whelan J, Lister R. 2015. Stress induced gene expression drives transient DNA methylation changes at adjacent repetitive elements. eLife. Doi: 10.7554/elife.09343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sela-Buurlage MB, Ponstein AS, Bres-Vloemans SA, Melchers LS, Van Den Elzen P, Cornelissen B. 1993. Only specific tobacco (Nicotiana tabacum) chitinases and [beta]-1,3-glucanases exhibit antifungal activity. Plant Physiology 101, 857–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilov IV, Seymourt SL, Patel AA, Loboda A, Tang WH, Keating SP, Hunter CL, Nuwaysir LM, Schaeffer DA. 2007. The paragon algorithm, a next generation search engine that uses sequence temperature values sequence temperature values and feature probabilities to identify peptides from tandem mass spectra. Molecular and Cellular Proteomics 6, 1638–1655. [DOI] [PubMed] [Google Scholar]
- Shinano T, Komatsu S, Yoshimura T, Tokutake S, Kong FJ, Watanabe T, Wasaki J, Osaki M. 2011. Proteomic analysis of secreted proteins from aseptically grown rice. Phytochemistry 72, 312–320. [DOI] [PubMed] [Google Scholar]
- Shinano T, Yoshimura T, Watanabe T, Unno Y, Osaki M, Nanjo Y, Komatsu S. 2013. Effect of phosphorus levels on the protein profiles of secreted protein and root surface protein of rice. Journal of Proteome Research 12, 4748–4756. [DOI] [PubMed] [Google Scholar]
- Sirová D, Bárta J, Šimek K, Posch T, Pech J, Stone J, Borovec J, Adamec L, Vrba J. 2018. Hunters or farmers? Microbiome characteristics help elucidate the diet composition in an aquatic carnivorous plant. Microbiome 6, 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud H, Do T, Du J, Zhong X, Feng S, Johnson L, Patel DJ, Jacobsen SE. 2014. Non-CG methylation patterns shape the epigenetic landscape in Arabidopsis. Nature Structural & Molecular Biology 21, 64–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Matsumoto K, Nishii W, Muramatsu M, Kubota K, Shibata C, Athauda SBP. 2009. Comparative studies on the acid proteinase activities in the digestive fluids of Nepenthes, Cephalotus, Dionaea, and Drosera. Carnivorous Plants Newsletter 38, 75–82. [Google Scholar]
- Taylor CB, Bariola PA, delCardayré SB, Raines RT, Green PJ. 1993. RNS2: a senescence-associated RNase of Arabidopsis that diverged from the S-RNases before speciation. Proceedings of the National Academy of Sciences, USA 90, 5118–5122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran HT, Qian W, Hurley BA, She YM, Wang D, Plaxton WC. 2010. Biochemical and molecular characterization of AtPAP12 and AtPAP26: the predominant purple acid phosphatase isozymes secreted by phosphate-starved Arabidopsis thaliana. Plant, Cell and Environment 33, 1789–1803. [DOI] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biology 3, RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Loon LC, Rep M, Pieterse CM. 2006. Significance of inducible defense-related proteins in infected plants. Annual Review of Phytopathology 44, 135–162. [DOI] [PubMed] [Google Scholar]
- Whitewoods CD, Gonçalves B, Cheng J, Cui M, Kennaway R, Lee K, Bushell C, Yu M, Piao C, Coen E. 2020. Evolution of carnivorous traps from planar leaves through simple shifts in gene expression. Science 367, 91–96. [DOI] [PubMed] [Google Scholar]
- Williams SE, Pickard BG. 1974. Connections and barriers between cells of Drosera tentacles in relation to their electrophysiology. Planta 116, 1–16. [DOI] [PubMed] [Google Scholar]
- Wu S, Kriz AL, Widholm JM. 1994. Molecular analysis of two cDNA clones encoding acidic class I chitinase in maize. Plant Physiology 105, 1097–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye ZH, Droste DL. 1996. Isolation and characterization of cDNAs encoding xylogenesis-associated and wounding-induced ribonucleases in Zinnia elegans. Plant Molecular Biology 30, 697–709. [DOI] [PubMed] [Google Scholar]
- Yilamujiang A, Reichelt M, Mithöfer A. 2016. Slow food: insect prey and chitin induce phytohormone accumulation and gene expression in carnivorous Nepenthes plants. Annals of Botany 118, 369–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Morgunova E, Jolma A, et al. 2017. Impact of cytosine methylation on DNA binding specificities of human transcription factors. Science 356, eaaj2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong-Villalobos L, González-Morales SI, Wrobel K, Gutiérrez-Alanis D, Cervantes-Peréz SA, Hayano-Kanashiro C, Oropeza-Aburto A, Cruz-Ramírez A, Martínez O, Herrera-Estrella L. 2015. Methylome analysis reveals an important role for epigenetic changes in the regulation of the Arabidopsis response to phosphate starvation. Proceedings of the National Academy of Sciences, USA 112, E7293–E7302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zareie R, Melanson DL, Murphy PJ. 2002. Isolation of fungal cell wall degrading proteins from barley (Hordeum vulgare L.) leaves infected with Rhynchosporium secalis. Molecular Plant-Microbe Interactions 15, 1031–1039. [DOI] [PubMed] [Google Scholar]
- Zhang H, Lang Z, Zhu JK. 2018. Dynamics and function of DNA methylation in plants. Nature Reviews. Molecular Cell Biology 19, 489–506. [DOI] [PubMed] [Google Scholar]
- Zhang X, Yazaki J, Sundaresan A, et al. 2006. Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell 126, 1189–1201. [DOI] [PubMed] [Google Scholar]
- Zhang L, Zhang F, Melotto M, Yao J, He SY. 2017. Jasmonate signaling and manipulation by pathogens and insects. Journal of Experimental Botany 68, 1371–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, Ma Y, Pan YH, Zhang CH, Yuan WX. 2011. A hevein-like protein and a class I chitinase with antifungal activity from leaves of the paper mulberry. Biomedical Chromatography 25, 908–912. [DOI] [PubMed] [Google Scholar]
- Zhu J, Kapoor A, Sridhar VV, Agius F, Zhu JK. 2007. The DNA glycosylase/lyase ROS1 functions in pruning DNA methylation patterns in Arabidopsis. Current Biology 17, 54–59. [DOI] [PubMed] [Google Scholar]
- Zifkin M, Jin A, Ozga JA, Zaharia LI, Schernthaner JP, Gesell A, Abrams SR, Kennedy JA, Constabel CP. 2012. Gene expression and metabolite profiling of developing highbush blueberry fruit indicates transcriptional regulation of flavonoid metabolism and activation of abscisic acid metabolism. Plant Physiology 158, 200–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the Proteome Xchange Consortium via the jPOST partner repository (Okuda et al., 2017) with the dataset identifier PXD022147. The bisulfite sequence data have been deposited to DDBJ (accession number:DRA010963). Sequence data can be found in the DDBJ data libraries under the accession numbers LC037411 (Bsep1), LC037409 (Chi1), LC549487 (Cysp1), LC037408 (Glu1), LC037410 (Hel1), LC547507 (Tlp1), BR001645 (polyvinylalcohol dehydrogenase gene), YAAA01000001 (DaMET1), YAAA01000002 (DaCMT3), YAAA01000003 (DaDRM2), YAAA01000004 (DaROS1), YAAA01000005 (DaDME), LC589199 (eIF4A) and LC589200 (TIP41).