Abstract
Leukemia cell-intrinsic somatic mutations and cytogenetic abnormalities have been used to define risk categories in acute myeloid leukemia (AML). In addition, since the immune microenvironment might influence prognosis and somatic mutations have been demonstrated to modulate the immune microenvironment in AML, there is need for developing and evaluating an immune prognostic model (IPM) derived from mutations associated with poor prognosis. Based on AML cases with intermediate and adverse-cytogenetic risk in the Cancer Genome Atlas (TCGA) database, 64 immune-related differentially expressed genes (DEGs) among patients with RUNX1, TP53, or ASXL1 mutations and patients without these mutations were identified. After Cox proportional hazards analysis, an IPM composed of PYCARD and PEAR1 genes was constructed. IPM defined high-risk (IPM-HR) independently predicted lower 2-year overall survival (OS) rates in both patients with intermediate and adverse-cytogenetic risks and non-M3 patients in the TCGA AML cohort. The poor prognostic impact of IPM-HR on OS was further validated by GSE71014, 37642, and 10358 downloaded from the Gene Expression Omnibus (GEO) database. Furthermore, IPM-HR was remarkably associated with higher proportions of CD8+ T cells and regulatory T cells (Tregs), lower proportions of eosinophils, and higher expression of the checkpoint molecules CTLA-4, PD-1, and LAG3 in the TCGA non-M3 AML cohort. In summary, we developed and validated an IPM derived from mutations related with poor prognosis in AML, which would provide new biomarkers for patient stratification and personalized immunotherapy.
Subject terms: Haematological cancer, Leukaemia, Acute myeloid leukaemia, Cancer, Immunology
Introduction
Cytogenetic abnormalities are the backbone of risk stratification in acute myeloid leukemia (AML), a malignant hematological disease, and more than 2/3 of patients with AML are classified as intermediate and adverse-cytogenetic risk groups1–3. Over the past two decades, several somatic mutations have been proven to be strongly prognostic in AML and have been incorporated into risk categories in both NCCN guidelines and ELN recommendations4–8. For example, AML patients with unfavorable cytogenetic risk harboring RUNX1, TP53, or ASXL1 mutations are defined as adverse risk categories.
In addition to the leukemia cell-intrinsic mechanism, the immune microenvironment plays an important role in the pathogenesis of AML and might influence the prognosis of patients with this disease9–11. Furthermore, some leukemia-related somatic mutations have been demonstrated to modulate the immune microenvironment in AML12–18. For instance, the RUNX1 mutation has been demonstrated to modulate nuclear factor (NF)-kB signaling and promote inflammatory signaling in the bone marrow microenvironment12. TP53 activates transcription of critical regulators of the innate immune response, and dysregulation of pathways downstream of mutated TP53 may mediate resistance to chemotherapy13–16. ASXL1 plays an important role in the microenvironment to support normal hematopoiesis, and mutant ASXL1 proteins gain functions that promote myeloid leukemogenesis17,18. Therefore, we speculate that the poor prognosis of patients with AML harboring RUNX1, TP53, or ASXL1 mutations may be partly caused by the specific influences of these mutations on the leukemia-associated immune system.
At present, the immune cell-specific gene expression profiles have been clarified. Based on multiple gene expression data and their association with treatment outcomes, the immune prognostic model (IPM) has been widely used to stratify patients with solid tumors, such as colorectal cancer19, hepatocellular carcinoma20,21, and lung cancer22. Nevertheless, no study has established an IPM in AML so far.
In the present study, we downloaded gene expression data of AML cohorts from The Cancer Genome Atlas (TCGA) database, identified the expression of RUNX1, TP53, and ASXL1 mutation-associated genes, and established an IPM. Furthermore, we demonstrated that the IPM-defined risk strongly predicted overall survival (OS) in the TCGA AML cohort and this was validated by three GEO datasets.
Results
RUNX1, TP53, and ASXL1 mutations associated with immune profile in AML with intermediate and adverse-cytogenetic risk in the TCGA cohort
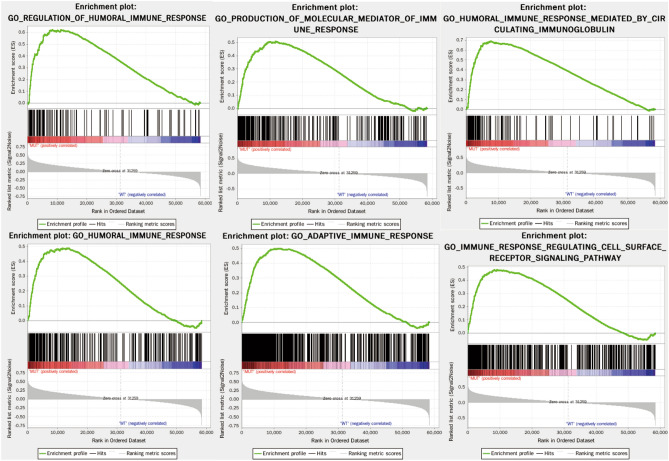
According to NCCN and ELN guidelines, non-favorable cytogenetic risk AML patients with RUNX1, TP53, or ASXL1 mutations were defined as adverse risk categories. Thus, we adopted AML patients with intermediate and adverse-cytogenetic risk to explore the RUNX1, TP53, and ASXL1 mutations associated with the immune signature. Of 173 AML patients in TCGA database, a total of 116 patients with available gene expression profile and mutation status had intermediate and adverse-cytogenetic risk and were analyzed. The RUNX1, TP53, and ASXL1 mutation frequencies were 12.1% (14/116), 9.5% (11/116), and 1.7% (2/116), respectively. A total of 25 patients had at least one somatic mutation (RUNX1, TP53, or ASXL1) and were categorized as the MUT group, and the remaining 91 patients had no mutations in RUNX1, TP53, and ASXL1 and were categorized as the WT group. GSEA analysis of the MUT and WT groups showed that the MUT group genetic profiles were significantly enriched in 445 biological processes, and six immune-related biological processes were included: REGULATION_OF_HUMORAL_IMMUNE_RESPONSE (NES = 1.80, size = 128, P = 0.031), PRODUCTION_OF_MOLECULAR_MEDIATOR_OF_IMMUNE_RESPONSE (normalized enrichment score, NES = 1.79, size = 272, P = 0.014), HUMORAL_IMMUNE_RESPONSE_MEDIATED_BY_CIRCULATING_IMMUNOGLOBULIN (NES = 1.77, size = 137, P = 0.036), HUMORAL_IMMUNE_RESPONSE (NES = 1.75, size = 337, P = 0.032), ADAPTIVE_IMMUNE_RESPONSE (NES = 1.67, size = 603, P = 0.033), and IMMUNE_RESPONSE_REGULATING_CELL_SURFACE_RECEPTOR_SIGNALING_PATHWAY (NES = 1.62, size = 482, P = 0.032) (Fig. 1). Thus, 1958 immune-related genes were obtained from these six immune-related processes.
Figure 1.
Comparison between significantly enriched immune-related phenotypes in the patients of the MUT and WT groups based on GSEA analysis.
Differentially expressed immune-related genes between the MUT and WT groups
In order to identify more candidate immune-related DEGs, two different methods were used. Firstly, 897 DEGs (822 upregulated and 75 downregulated) were identified between the MUT and WT groups (|log2FC|≥ 1.0 and FDR < 0.05), and 25 of them were shown to be involved in immune-related biological processes (FDR < 0.05) by gene ontology (GO) enrichment analysis. Secondly, of the 1,958 immune-related genes obtained from GSEA analysis, 51 genes were found to be differentially expressed between the MUT and WT groups (|log2FC|≥ 1.0 and FDR < 0.05).Finally, excluding 12 repetitive DEGs, 64 DEGs in total between MUT and WT groups were shown to be immune-related and used for the design of the IPM.
Establishment of an IPM and evaluation of its predictive ability
Of 116 AML patients with intermediate and adverse-cytogenetic risk in TCGA database, 107 patients had survival information and survival analysis was performed. Their median follow-up period was 305 days (range: 0–2861 days) and the 2-year OS rate was 37.7% (95% confidence interval [CI]: 27.5–47.7%).
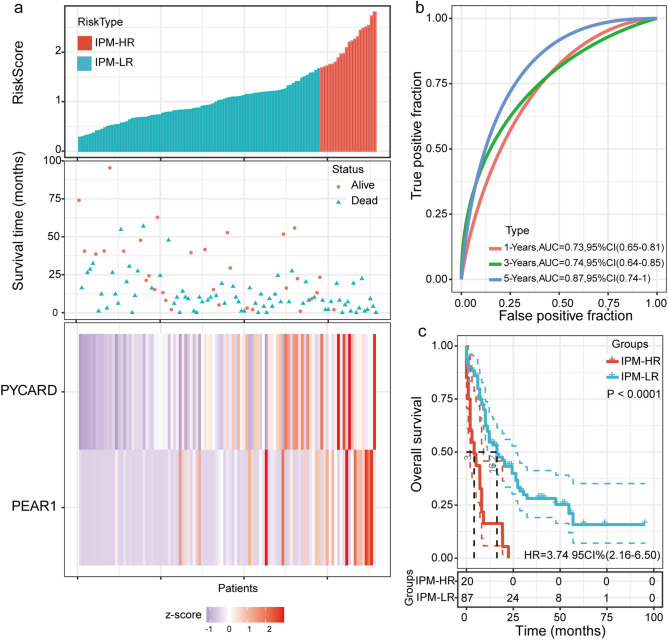
Univariate Cox regression analysis revealed that three of the 64 immune-related DEGs were significantly related to OS (Table S1). However, only two genes, PYD and CARD domain-containing (PYCARD) and platelet endothelial aggregation receptor 1 (PEAR1), were shown to be significantly prognostic (i.e., P < 0.05) in multivariate Cox regression analysis (both were poor prognostic factors). Then, an IPM based on these two genes was established by Cox proportional hazards analysis, and a risk score to predict prognostic value was calculated. The median IPM defined risk score for AML patients with intermediate and adverse-cytogenetic risk in the TCGA cohort was 1.07 (range: 0.28–2.81), and the risk score distribution and gene expression data are shown in Fig. 2a. As shown in Fig. 2b, the AUC of the IPM for OS was 0.73 at one year, 0.74 at three years, and 0.87 at five years.
Figure 2.
Establishment of an IPM and evaluation of its predictive ability in TCGA AML patients with intermediate and adverse-cytogenetic risk. (a) Risk score distribution and the corresponding survival and gene expression data; (b) Time-dependent ROC curve of the IPM; (c) Comparison between overall survival in the IPM-HR and IPM-LR groups.
The ROC curves showed that 1.67 had a maximum Youden index (0.22) among all values. Thus, 1.67 was identified as the optimal cutoff value of the IPM defined risk score, risk score ≥ 1.67 and < 1.67 were defined as the IPM-HR and IPM-LR groups, respectively. In AML patients with intermediate and adverse-cytogenetic risk, 20 (18.7%) and 87 (81.3%) patients were individually categorized into the IPM-HR and IPM-LR groups, respectively. As a result, patients in the IPM-HR group had a significantly lower 2-year OS rate than the patients in the IPM-LR group (0 vs. 45.9% [95% CI: 34.1–56.9%], P < 0.0001, Fig. 2c; Table S2).
Relationship between IPM-defined risk and patient characteristics in the TCGA cohort
As shown in Table S3, of 107 AML patients with intermediate and adverse-cytogenetic risk, IPM-HR was significantly related to RUNX1 mutation (P = 0.023) and advanced age (P < 0.0001). However, the IPM defined risk had no relationship with sex, white blood cell (WBC) counts, hemoglobin (HB) levels, platelet (PLT) counts, percentage of bone marrow (BM) blast, FAB subtype, ASXL1 mutation, TP53 mutation, FLT3-ITD mutation, NPM1 mutation, CEBPA biallelic mutation, WT1 mutation, and DNMT3A mutation (all P > 0.05).
IPM-defined risk independently predicted poor outcomes in the TCGA cohort
As shown in Table S2, in AML patients with intermediate and adverse-cytogenetic risk, older age, TP53 mutation, and DNMT3A mutation were all significantly related to a lower 2-year OS rate in addition to IPM-HR (all P < 0.05). Multivariable analysis showed that IPM-HR, older age, TP53 mutation, FLT3-ITD, and DNMT3A mutations were all independent adverse prognostic factors for OS in AML patients with intermediate and adverse-cytogenetic risk (Table 1).
Table 1.
Independent prognostic factors for OS of AML patients in TCGA cohort.
| AML patients with intermediate and adverse-cytogenetic risk | Non-M3 AML patients | |||||
|---|---|---|---|---|---|---|
| No. of patients | HR(95%CI) | P value | No. of patients | HR(95%CI) | P value | |
| IPM | ||||||
| IPM-LR | 87 | 1.00 | < 0.0001 | 104 | 1.00 | < 0.0001 |
| IPM-HR | 20 | 3.22 (1.71–6.05) | 22 | 3.40 (1.84–6.27) | ||
| Age | ||||||
| Age < 60y | 52 | 1.00 | 0.040 | 60 | 1.00 | 0.030 |
| Age ≥ 60y | 55 | 1.71 (1.03–2.85) | 66 | 1.72 (1.06–2.81) | ||
| TP53 | ||||||
| Wild type | 97 | 1.00 | 0.001 | 114 | 1.00 | < 0.0001 |
| Mutation | 10 | 4.05 (1.82–9.0) | 10 | 4.26 (2.0–9.06) | ||
| FLT3-ITD | ||||||
| ( −) | 86 | 1.00 | 0.033 | 99 | 1.00 | 0.027 |
| ( +) | 21 | 1.94 (1.06–3.58) | 25 | 1.95 (1.08–3.52) | ||
| DNMT3A | ||||||
| Wild type | 74 | 1.00 | 0.006 | 90 | 1.00 | 0.006 |
| Mutation | 33 | 2.07 (1.23–3.48) | 34 | 2.08 (1.24–3.50) | ||
We further evaluated the prognostic impact of IPM risk score on OS in TCGA non-M3 AML patients. Of 126 patients with survival information, 22 (17.5%) and 104 (82.5%) patients were individually categorized into the IPM-HR and IPM-LR groups by a cutoff value of 1.67, and the IPM-HR group had a significantly lower 2-year OS rate than the IPM-LR group (0 vs. 49.6% [95% CI: 38.5–59.8%], P < 0.0001). Univariate analysis showed that IPM-HR, older age, TP53 mutation, DNMT3A mutation, and intermediate and adverse-cytogenetic risk were all significantly related to a lower 2-year OS rate (all P < 0.05, Table S4). Similar to the results of patients with intermediate and adverse-cytogenetic risk, IPM-HR, older age, TP53 mutation, FLT3-ITD, and DNMT3A mutation were all independent adverse prognostic factors for OS in non-M3 AML patients (Table 1).
Validation of the prognostic impact of IPM-defined risk in the GEO database
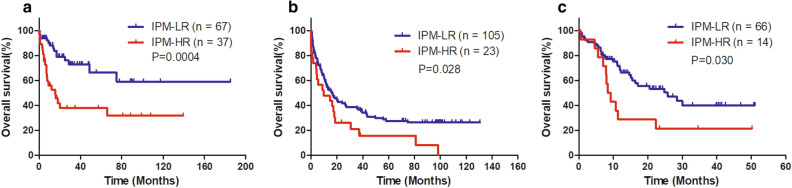
A total of 104 AML cases with normal karyotype from GSE71014, 128 non-M3 AML cases from GSE37642, and 80 non-M3 AML cases from GSE10358 were used to verify the prognostic significance of the poor prognostic mutation-derived IPM defined risk. Patients in each cohort had their IPM risk score calculated and were divided into the IPM-HR and IPM-LR groups based on the individual ROC curve determined cutoff values. Consistent with the results in TCGA cohort, patients in the IPM-HR group had significantly lower 2-year OS rates than those in the IPM-LR group (GSE71014: 37.9 [95% CI:21.1–54.7%] vs 79.1% [95% CI: 65.0–88.0%], P = 0.0004, Fig. 3a; GSE37642: 26.1% [95% CI: 10.6–44.7%] vs 42.5% [95% CI: 32.8–51.8%], P = 0.028, Fig. 3b; GSE10358: 21.4% [95% CI: 5.2–44.8%] vs 53.4% [95% CI: 39.3–65.6%], P = 0.030, Fig. 3c).
Figure 3.
Validation of the prognostic impact of IPM defined risk on OS in three GEO datasets (a) OS of AML cases with normal karyotype from GSE71014 (b) OS of non-M3 AML cases from GSE37642 (c) OS of non-M3 AML cases from GSE10358.
Immune cell infiltration landscapes and checkpoint molecule analysis of IPM-HR and IPM-LR patients
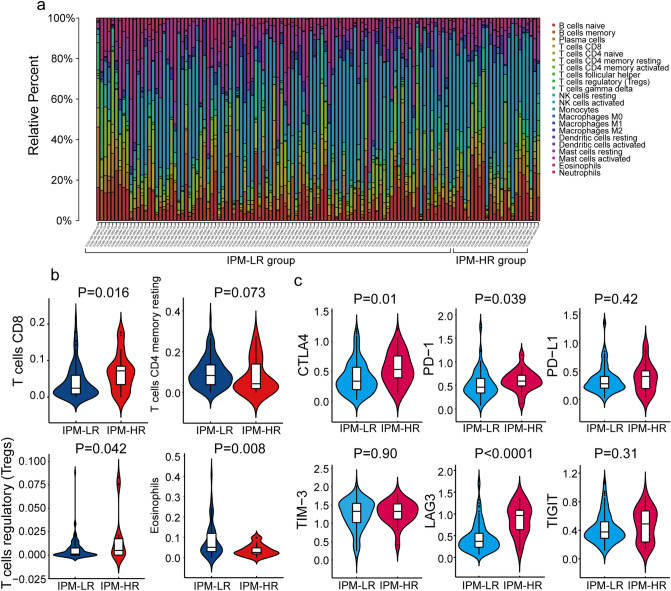
For 126 non-M3 AML patients in TCGA cohort, the proportions of 22 immune cell types were estimated using the CIBERSORT method and compared between the IPM-HR and IPM-LR groups (Fig. 4a). Patients in the IPM-HR group had significantly higher proportions of CD8 T cells and regulatory T cells (Tregs), lower proportions of eosinophils, and tended to have significantly lower proportions of resting CD4 memory T cells compared with patients in the IPM-LR group (P = 0.016, 0.042, 0.008, and 0.073, Fig. 4b).
Figure 4.
Analysis of the immune cells infiltration landscapes and checkpoint molecules between IPM-HR and IPM-LR groups in non-M3 AML patients from the TCGA cohort: (a) Relative proportion of immune cells infiltration in IPM-HR and IPM-LR patients; (b) Violin plots showing significantly different proportions of immune cells between IPM-HR and IPM-LR patients; (c) Violin plots showing the expression of different immune checkpoint molecules between IPM-HR and IPM-LR patients.
Then, we compared the expression of critical immune checkpoint molecules (CTLA-4, PD-1, PD-L1, TIM-3, LAG-3, and TIGIT) between the IPM-HR and IPM-LR groups. The expression of CTLA-4, PD-1, and LAG3 in the IPM-HR group was significantly higher than that in the IPM-LR group in non-M3 AML patients in TCGA cohort (P = 0.010, 0.039, and < 0.0001, Fig. 4c).
Discussion
In the present study, we developed an RUNX1, TP53, and ASXL1 mutation derived IPM based on TCGA AML cohort, and demonstrated that the IPM-defined risk independently predicted OS in AML, which was validated by GSE databases.
The first step in establishing an IPM is the identification of candidate genes. The somatic mutations associated with adverse risk included RUNX1, TP53, ASXL1, and FLT3-ITD in both ELN recommendations and NCCN guidelines. Because the prognostic role of FLT3-ITD is related to both the NPM1 mutation status and FLT3-ITD mutation load, and the mutation load information was unavailable in TCGA database, only RUNX1, TP53, and ASXL1 mutation status were considered for IPM development in the present study. For the comparison between poor-prognostic and non-poor prognostic groups, we identified DEGs in two ways in order to select as many candidates as possible, that is, immune-related genes in both GSEA and GO analysis. After univariate and multivariate Cox regression analyses, we established an IPM composed of two gene expression profiles with individual weights. Then, the cutoff value was determined by ROC curve analysis and used to group patients into the IPM-HR and -LR groups in each cohort, respectively. The survival analysis demonstrated that IPM defined high risk as an independent poor prognostic factor in both intermediate and adverse-cytogenetic risk AML patients and non-M3 AML patients in TCGA cohorts. Furthermore, this impact was individually validated in one cohort of AML with normal karyotype and two cohorts of non-M3 AML, which were downloaded from GEO databases. These results illustrate the usefulness of our IPM and also reflect that the immune microenvironment is involved in the prognosis of AML.
The IPM we developed was composed of PYCARD and PEAR1 genes. PYCARD is a 22-kD protein containing an N-terminal pyrin domain (PYD) and a C-terminal caspase activation and recruitment domain (CARD)26,27. PYCARD expression is found in tumor cells, tumor-associated macrophages, normal epithelial cells, and non-tumor adjacent tissues26–31. It is silenced by methylation in many tumors, preventing tumor cells from apoptosis, which supports its role as a tumor suppressor28–31. Subsequent studies found that PYCARD is a central adaptor molecule of the inflammasome complex, which mediates the secretion of inflammatory cytokines (i.e., IL-1β and IL-18)27,32. Inflammation contributes to tumor development and progression, and inflammatory cytokines contribute to tumor promotion33. Therefore, in the context of cancer development and progression, PYCARD may exert opposing functions, either tumor-suppressing by inducing apoptosis or tumor-promoting by secretion of inflammatory cytokines within the tumor microenvironment. Contradictory results exist in the clinical relevance of PYCARD expression in various solid tumors34–36. PEAR1 is a transmembrane protein that is mainly expressed on platelets, hematopoietic stem cells, and endothelial cells. It sustains αIIbβ3 activation in aggregating platelets and attenuates megakaryopoiesis by controlling the degree of Akt phosphorylation37,38. Moreover, methylation-controlled PEAR1 expression can activate platelets and have an impact on inflammation39–41. Both in vitro and in vivo studies in endothelial cells showed an inverse correlation between PEAR1 expression and vascular assembly, which implies that PEAR1 modifies neo-angiogenesis42. Angiogenesis is a hallmark of cancer, and acute leukemia has been demonstrated to have increased angiogenesis, as assessed by microvessel density, compared to normal controls43. In addition, PEAR1 regulates the early stages of hematopoietic differentiation44. Both expressions of PYCARD and PEAR1 were poor prognostic indicators in our developed and validated IPM, whereas none of them has ever been reported in AML to date. Considering the published studies, we suspected that these may represent distinct mechanisms of the effect of microenvironment on AML, and mass cytometry or single cell sequencing followed by functional studies would be the way to clarify this.
Some studies hypothesized that during tumor development in immune-competent hosts, less immunogenic cancer cells are selected and immunosuppressive networks are established to evade antitumor immune responses45,46. Decreasing the expression of cancer antigens and immunoreactive cells such as follicular helper T cells, and increasing immunosuppressive molecules and cells such as Treg cells and tumor-associated macrophages are immunosuppressive mechanisms of cancers47,48. In this study, we found that patients in the IPM-HR group had higher proportions of CD8+ T cells and Tregs but lower fractions of eosinophils and CD4+ memory resting T cells than the IPM-LR group in non-M3 AML patients in the TCGA cohort. Then, we investigated the expression of immune checkpoints between the IPM-HR and IPM-LR groups. The IPM-HR patients had significantly higher expression of CTLA-4, PD-1, and LAG3 than the IPM-LR patients. CTLA4 and PD-1 have been demonstrated to play important roles in hampering T cell immunity against hematological malignancies and immune evasion of AML49–52. Antibodies targeting both the PD-1 and CTLA-4 pathways have shown efficacy in a variety of solid tumors and in some hematologic malignancies in clinical trials33,53–55. CD4+ T helper (Th) cells expressing upregulated PD-1 and/or LAG3 were identified together with CD86+ and/or ICOS-LG + myeloid blasts in the bone marrow of patients with AML, which may induce Th cell exhaustion and limit antitumor immunity56. Cytotoxic T lymphocytes (CTLs) are CD8+ T cells that play an important role in antitumor immune responses, but CD8+ T cells in the bone marrow of AML patients had reduced killing capacity and higher levels of PD-1 expressed57. CD4+ T cells can differentiate into Tregs, which are important in immunosuppressive mechanisms that lead to the inhibition of proliferation and cytokine production of other T cells58. Increased numbers of Tregs in solid tumors have been associated with worse outcomes and lead to the suppression of antitumor immunity59. Our results suggest that IPM-HR patients have higher expression of some immune checkpoint molecules and more Tregs forming an immunosuppressive environment, which may lead to a poor prognosis. Thus, IPM-HR patients may benefit from immunotherapeutic strategies targeting Tregs and immune checkpoint inhibitors.
Overall, we established and validated an IPM based on two immune microenvironment-related genes associated with RUNX1, TP53, or ASXL1 mutation status, which independently predicted the overall survival of AML patients. IPM-HR was related to a higher proportion of Tregs and higher expression of checkpoint molecules CTLA-4, PD-1, and LAG3. The current IPM may provide a new biomarker for stratification and immunotherapeutic strategies for AML.
Materials and methods
Database
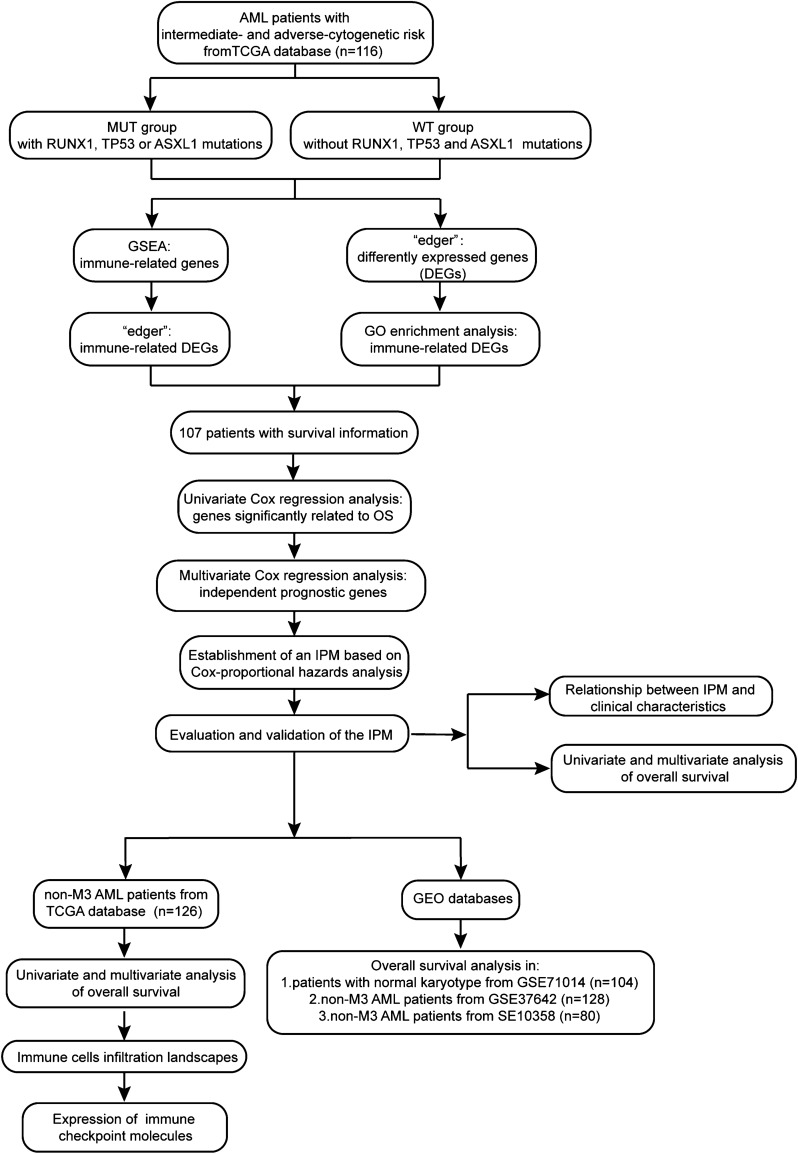
The workflow is shown in Fig. 5. The somatic mutation status (workflow type: VarScan2 Variant Aggregation and Masking), transcriptional profiles, and the corresponding clinical and overall survival (OS) data of 173 AML patients were downloaded from The Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/). The gene expression profile was measured experimentally using the Illumina HiSeq 2000 RNA Sequencing platform. The gene symbols were annotated based on Homo sapiens GRCh38.91.chr.gtf file (http://asia.ensembl.org/index.html). Log2 transformations were performed for all gene expression data. The study reported herein fully satisfies the TCGA publication requirements (http://cancergenome.nih.gov/publications/publicationguidelines). The definition of cytogenetic risk and risk related somatic mutations were based on NCCN guidelines7. Of 173 AML patients in TCGA database, 116 patients with intermediate and adverse-cytogenetic risk had available gene expression profile and mutation status while 107 patients had survival information..
Figure 5.
Workflow chart of data generation and analysis.
To validate the predictive ability of the IPM based on TCGA data, 3 GEO cohorts with survival information were used, they are GSE71014 including 104 AML patients with normal karyotype (based on GPL10558 Illumina HumanHT-12 V4.0 expression beadchip), GSE37642 including 128 non-M3 AML patients and GSE10358 including 80 non-M3 AML patients (based on GPL570 Affymetrix Human Genome U133 Plus 2.0 Array), and the individual clinical and OS information were downloaded from the GEO database (https://www.ncbi.nlm.nih.gov/geo/).
Identification of differentially expressed genes
First, the raw counts of gene expression data from TCGA were normalized using a weighted trimmed mean of log ratios-based method24. To obtain differentially expressed genes (DEGs) between patients with RUNX1, TP53, or ASXL1 mutations (MUT group) and without RUNX1, TP53, and ASXL1 mutations (WT group), the “edger” package in R software (Version 3.6.2; https://www.r-project.org/) was used. |log2FC|≥ 1.0 and FDR < 0.05 were selected as DEGs.
Functional enrichment analysis of the DEGs
Functional enrichment analysis of DEGs between the MUT and WT groups was performed based on clusterProfiler, enrichplot, and org.Hs.eg.db packages to identify the immune-related DEGs involved in the immune-related biological processes (BP) of Gene Ontology (GO) categories. FDR < 0.05 was considered statistically significant.
Gene set enrichment analysis (GSEA)
We used TCGA genomewide expression profiles and selected an annotated gene set file (c5.bp.v6.2.symbols.gm) as the reference gene set to perform GSEA (Version 3.0; http://software.broadinstitute.org/gsea/index.jsp) analysis for identifying immunological pathways and corresponding immune-related genes differ between the MUT and WT groups23. The GSEA threshold for significantly enriched immune-related functional annotations was set at P < 0.05, false discovery rate (FDR) < 0.25, and a normalized enrichment score > 1.5. Likely, the “edger” package in R software with criteria of |log2FC|≥ 1.0 and FDR < 0.05 was also used to identify the immune-related DEGs in immune-related genes obtained from GSEA.
Establishment and validation of an immune prognostic model
Univariate Cox regression analysis was performed using the R package “survival” to evaluate correlations between the DEG expression levels and OS of TCGA AML patients with intermediate and adverse-cytogenetic risk. DEGs with P < 0.05 by univariate Cox regression analysis were identified as alternative prognostic genes. Among the immune genes that were significant in the univariate Cox regression analysis, a sub-selection of immune genes involved in prognosis was determined by multivariate Cox regression analysis, and in this analysis, genes were regarded as significant at P < 0.05. Finally, an IPM was constructed based on Cox-proportional hazards analysis, and the risk score derived from the IPM was calculated by utilizing the “predict” function in R software to assess each patient’s risk. Patients with available survival data were separated into IPM-defined high-risk (IPM-HR) and IPM-defined low-risk (IPM-LR) groups using the optimal cutoff obtained from a receiver operating characteristic (ROC) curve. The predictability of the IPM was evaluated by area under ROC curve (AUC); the higher the value of the AUC, the better the predictability of the model.
Estimation of immune cell-type fractions
Cell type identification by estimating relative subsets of RNA transcripts (CIBERSORT) is an approach to characterize the cell composition of complex tissues based on gene expression profiles, and has been demonstrated to be highly consistent with ground truth estimations in many cancers25. We uploaded normalized gene expression data with standard annotation files to the CIBERSORT web portal. A leukocyte gene signature matrix termed LM22 was used to distinguish 22 immune cell types, including myeloid subsets, natural killer (NK) cells, plasma cells, naïve and memory B cells, and seven T cell types. We used CIBERSORT in combination with the LM22 signature matrix to estimate the fractions of 22 immune cell types between the IPM-HR and IPM-LR AML groups. The sum of all estimated immune cell type fractions was 100% for each sample.
Statistical analysis
Pairwise comparisons of the variables between groups were performed using the Mann–Whitney U test for continuous variables and Fisher's exact test for categorical variables. Survival functions were estimated using the Kaplan–Meier method and compared using the log-rank test. Variables associated with P ≤ 0.25 in the univariate analysis were entered into a multivariate analysis. Comparisons of immune cell type fractions and checkpoint molecules between the IPM-HR and IPM-LR groups were performed using the Mann–Whitney U test. The level for a statistically significant difference was set at P < 0.05. The SPSS 22.0 software package (SPSS Inc., Chicago, IL) and GraphPad Prism 5 (GraphPad Software Inc., La Jolla, CA) were used for data analysis.
Supplementary Information
Author contributions
Y.-Z.Q. designed the study. Material preparation, data collection and analysis were performed by F.-T.D., J.W. and L.Y. F.-T.D. and J.W. wrote the manuscript. Y.-Z.Q. revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (82070153).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Feng-Ting Dao and Jun-Wang.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-84190-0.
References
- 1.Slovak ML, Kopecky KJ, Cassileth PA, Harrington DH, Theil KS, Mohamed A, et al. Karyotypic analysis predicts outcome of preremission and postremission therapy in adult acute myeloid leukemia: A Southwest Oncology Group/Eastern Cooperative Oncology Group Study. Blood. 2000;96:4075–4083. doi: 10.1182/blood.V96.13.4075. [DOI] [PubMed] [Google Scholar]
- 2.Byrd JC, Mrózek K, Dodge RK, Carroll AJ, Edwards CG, Arthur DC, et al. Pretreatment cytogenetic abnormalities are predictive of induction success, cumulative incidence of relapse, and overall survival in adult patients with de novo acute myeloid leukemia: Results from Cancer and Leukemia Group B (CALGB 8461) Blood. 2002;100:4325–4336. doi: 10.1182/blood-2002-03-0772. [DOI] [PubMed] [Google Scholar]
- 3.Ferrara F, Schiffer CA. Acute myeloid leukaemia in adults. Lancet. 2013;381:484–495. doi: 10.1016/S0140-6736(12)61727-9. [DOI] [PubMed] [Google Scholar]
- 4.Papaemmanuil E, Gerstung M, Bullinger L, Gaidzik VI, Paschka P, Roberts ND, et al. Genomic classification and prognosis in acute myeloid leukemia. N. Engl. J. Med. 2016;374:2209–2221. doi: 10.1056/NEJMoa1516192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Metzeler KH, Herold T, Rothenberg-Thurley M, Amler S, Sauerland MC, Görlich D, et al. Spectrum and prognostic relevance of driver gene mutations in acute myeloid leukemia. Blood. 2016;128:686–698. doi: 10.1182/blood-2016-01-693879. [DOI] [PubMed] [Google Scholar]
- 6.Medinger M, Passweg JR. Acute myeloid leukaemia genomics. Br. J. Haematol. 2017;179:530–542. doi: 10.1111/bjh.14823. [DOI] [PubMed] [Google Scholar]
- 7.National Comprehensive Cancer Network. NCCN clinical practice guidelines in oncology: acute myeloid leukemia (version 3.2019). 2019. http://www.nccn.org. [DOI] [PubMed]
- 8.Döhner H, Estey E, Grimwade D, Amadori S, Appelbaum FR, Büchner T, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 2017;129:424–447. doi: 10.1182/blood-2016-08-733196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nahas MR, Stroopinsky D, Rosenblatt J, Cole L, Pyzer AR, Anastasiadou E, et al. Hypomethylating agent alters the immune microenvironment in acute myeloid leukaemia (AML) and enhances the immunogenicity of a dendritic cell/AML vaccine. Br. J. Haematol. 2019;185:679–690. doi: 10.1111/bjh.15818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Isidori A, Salvestrini V, Ciciarello M, Loscocco F, Visani G, Parisi S, et al. The role of the immunosuppressive microenvironment in acute myeloid leukemia development and treatment. Expert Rev. Hematol. 2014;7:807–818. doi: 10.1586/17474086.2014.958464. [DOI] [PubMed] [Google Scholar]
- 11.Hanahan D, Coussens LM. Accessories to the crime: Functions of cells recruited to the tumor microenvironment. Cancer Cell. 2012;21:309–322. doi: 10.1016/j.ccr.2012.02.022. [DOI] [PubMed] [Google Scholar]
- 12.Nakagawa M, Shimabe M, Watanabe-Okochi N, Arai S, Yoshimi A, Shinohara A, et al. AML1/RUNX1 functions as a cytoplasmic attenuator of NF-κB signaling in the repression of myeloid tumors. Blood. 2011;118:6626–6637. doi: 10.1182/blood-2010-12-326710. [DOI] [PubMed] [Google Scholar]
- 13.Shatz M, Menendez D, Resnick MA. The human TLR innate immune gene family is differentially influenced by DNA stress and p53 status in cancer cells. Can. Res. 2012;72:3948–3957. doi: 10.1158/0008-5472.CAN-11-4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cui Y, Guo G. Immunomodulatory function of the tumor suppressor p53 in host immune response and the tumor microenvironment. Int. J. Mol. Sci. 2016;17:1942. doi: 10.3390/ijms17111942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H, Lakshmikanth T, Garofalo C, Enge M, Spinnler C, Anichini A, et al. Pharmacological activation of p53 triggers anticancer innate immune response through induction of ULBP2. Cell Cycle. 2011;10:3346–3358. doi: 10.4161/cc.10.19.17630. [DOI] [PubMed] [Google Scholar]
- 16.Mendez LM, Posey RR, Pandolfi PP. The interplay between the genetic and immune landscapes of AML: Mechanisms and implications for risk stratification and therapy. Front. Oncol. 2019;9:1162. doi: 10.3389/fonc.2019.01162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang P, Chen Z, Li R, Guo Y, Shi H, Bai J, et al. Loss of ASXL1 in the bone marrow niche dysregulates hematopoietic stem and progenitor cell fates. Cell Discov. 2018;4:4. doi: 10.1038/s41421-017-0004-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Asada S, Fujino T, Goyama S, Kitamura T. The role of ASXL1 in hematopoiesis and myeloid malignancies. Cell Mol. Life Sci. 2019;76:2511–2523. doi: 10.1007/s00018-019-03084-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao X, Liu J, Liu S, Yang F, Chen E. Construction and Validation Of An Immune-Related Prognostic Model Based on TP53 status in colorectal cancer. Cancers. 2019;11:1722. doi: 10.3390/cancers11111722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Long J, Wang A, Bai Y, Lin J, Yang X, Wang D, et al. Development and validation of a TP53-associated immune prognostic model for hepatocellular carcinoma. EBioMedicine. 2019;42:363–374. doi: 10.1016/j.ebiom.2019.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Z, Zhu J, Liu Y, Liu C, Wang W, Chen F, et al. Development and validation of a novel immune-related prognostic model in hepatocellular carcinoma. J. Transl. Med. 2020;18:67. doi: 10.1186/s12967-020-02255-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luo C, Lei M, Zhang Y, Zhang Q, Li L, Lian J, et al. Systematic construction and validation of an immune prognostic model for lung adenocarcinoma. J. Cell Mol. Med. 2020;24:1233–1244. doi: 10.1111/jcmm.14719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods. 2015;12:453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Masumoto J, Taniguchi S, Ayukawa K, Sarvotham H, Kishino T, Niikawa N, et al. ASC, a novel 22-kDa protein, aggregates during apoptosis of human promyelocytic leukemia HL-60 cells. J. Biol. Chem. 1999;274:33835–33838. doi: 10.1074/jbc.274.48.33835. [DOI] [PubMed] [Google Scholar]
- 27.Martinon F, Hofmann K, Tschopp J. The pyrin domain: a possible member of the death domain-fold family implicated in apoptosis and inflammation. Curr. Biol. 2001;11:R118–R120. doi: 10.1016/s0960-9822(01)00056-2. [DOI] [PubMed] [Google Scholar]
- 28.Ohtsuka T, Liu XF, Koga Y, Kitajima Y, Nakafusa Y, Ha CW, et al. Methylation-induced silencing of ASC and the effect of expressed ASC on p53-mediated chemosensitivity in colorectal cancer. Oncogene. 2006;25:1807–1811. doi: 10.1038/sj.onc.1209204. [DOI] [PubMed] [Google Scholar]
- 29.Ohtsuka T, Ryu H, Minamishima YA, Macip S, Sagara J, Nakayama KI, et al. ASC is a Bax adaptor and regulates the p53-Bax mitochondrial apoptosis pathway. Nat. Cell Biol. 2004;6:121–128. doi: 10.1038/ncb1087. [DOI] [PubMed] [Google Scholar]
- 30.Parsons MJ, Patel P, Brat DJ, Colbert L, Vertino PM. Silencing of TMS1/ASC promotes resistance to anoikis in breast epithelial cells. Can. Res. 2009;69:1706–1711. doi: 10.1158/0008-5472.CAN-08-2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hong S, Hwang I, Lee YS, Park S, Lee WK, Fernandes-Alnemri T, et al. Restoration of ASC expression sensitizes colorectal cancer cells to genotoxic stress-induced caspase-independent cell death. Cancer Lett. 2013;331:183–191. doi: 10.1016/j.canlet.2012.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Srinivasula SM, Poyet JL, Razmara M, Datta P, Zhang Z, Alnemri ES. The PYRIN-CARD protein ASC is an activating adaptor for caspase-1. J. Biol. Chem. 2002;277:21119–21122. doi: 10.1074/jbc.C200179200. [DOI] [PubMed] [Google Scholar]
- 33.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 34.Machida EO, Brock MV, Hooker CM, Nakayama J, Ishida A, Amano J, et al. Hypermethylation of ASC/TMS1 is a sputum marker for late-stage lung cancer. Can. Res. 2006;66:6210–6218. doi: 10.1158/0008-5472.CAN-05-4447. [DOI] [PubMed] [Google Scholar]
- 35.Zhang Z, Tan S, Zhang L. Prognostic value of apoptosis-associated speck-like protein containing a CARD gene promoter methylation in resectable non-small-cell lung cancer. Clin. Lung Cancer. 2006;8:62–65. doi: 10.3816/CLC.2006.n.035. [DOI] [PubMed] [Google Scholar]
- 36.Wu L, Zhang C, Wang X, Ding X, Deng J, Liang H. Methylation of ASC/TMS1 promoter is associated with poor prognosis of patients with gastric cancer. Clin. Transl. Oncol. 2016;18:296–303. doi: 10.1007/s12094-015-1367-y. [DOI] [PubMed] [Google Scholar]
- 37.Kauskot A, Di Michele M, Loyen S, Freson K, Verhamme P, Hoylaerts MF. A novel mechanism of sustained platelet αIIbβ3 activation via PEAR1. Blood. 2012;119:4056–4065. doi: 10.1182/blood-2011-11-392787. [DOI] [PubMed] [Google Scholar]
- 38.Nanda N, Bao M, Lin H, Clauser K, Komuves L, Quertermous T, et al. Platelet endothelial aggregation receptor 1 (PEAR1), a novel epidermal growth factor repeat-containing transmembrane receptor, participates in platelet contact-induced activation. J. Biol. Chem. 2005;280:24680–24689. doi: 10.1074/jbc.M413411200. [DOI] [PubMed] [Google Scholar]
- 39.Izzi B, Pistoni M, Cludts K, Akkor P, Lambrechts D, Verfaillie C, et al. Allele-specific DNA methylation reinforces PEAR1 enhancer activity. Blood. 2016;128:1003–1012. doi: 10.1182/blood-2015-11-682153. [DOI] [PubMed] [Google Scholar]
- 40.Johnson AD. Pairing megakaryopoiesis methylation with PEAR1. Blood. 2016;128:890–892. doi: 10.1182/blood-2016-06-723940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Izzi B, Gianfagna F, Yang WY, Cludts K, De Curtis A, Verhamme P, et al. Variation of PEAR1 DNA methylation influences platelet and leukocyte function. Clin. Epigenet. 2019;11:151. doi: 10.1186/s13148-019-0744-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vandenbriele C, Kauskot A, Vandersmissen I, Criel M, Geenens R, Craps S, et al. Platelet endothelial aggregation receptor-1: A novel modifier of neoangiogenesis. Cardiovasc. Res. 2015;108:124–138. doi: 10.1093/cvr/cvv193. [DOI] [PubMed] [Google Scholar]
- 43.AbdElAal Asmaa A, Afify RA, Zaher AE, ElGammal MM, Atef AM. Study of prognostic significance of marrow angiogenesis assessment in patients with de novo acute leukemia. Hematology. 2015;20:504–510. doi: 10.1179/1607845415Y.0000000012. [DOI] [PubMed] [Google Scholar]
- 44.Krivtsov AV, Rozov FN, Zinovyeva MV, Hendrikx PJ, Jiang Y, Visser JW, et al. Jedi–A novel transmembrane protein expressed in early hematopoietic cells. J. Cell Biochem. 2007;101:767–784. doi: 10.1002/jcb.21232. [DOI] [PubMed] [Google Scholar]
- 45.Dunn GP, Bruce AT, Ikeda H, Old LJ, Schreiber RD. Cancer immunoediting: from immunosurveillance to tumor escape. Nat. Immunol. 2002;3:991–998. doi: 10.1038/ni1102-991. [DOI] [PubMed] [Google Scholar]
- 46.Schreiber RD, Old LJ, Smyth MJ. Cancer immunoediting: Integrating immunity's roles in cancer suppression and promotion. Science. 2011;331:1565–1570. doi: 10.1126/science.1203486. [DOI] [PubMed] [Google Scholar]
- 47.Zou W, Chen L. Inhibitory B7-family molecules in the tumour microenvironment. Nat. Rev. Immunol. 2008;8:467–477. doi: 10.1038/nri2326. [DOI] [PubMed] [Google Scholar]
- 48.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer. 2012;12:252–264. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang L, Gajewski TF, Kline J. PD-1/PD-L1 interactions inhibit antitumor immune responses in a murine acute myeloid leukemia model. Blood. 2009;114:1545–1552. doi: 10.1182/blood-2009-03-206672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou Q, Munger ME, Highfill SL, Tolar J, Weigel BJ, Riddle M, et al. Program death-1 signaling and regulatory T cells collaborate to resist the function of adoptively transferred cytotoxic T lymphocytes in advanced acute myeloid leukemia. Blood. 2010;116:2484–2493. doi: 10.1182/blood-2010-03-275446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pérez-García A, Brunet S, Berlanga JJ, Tormo M, Nomdedeu J, Guardia R, et al. CTLA-4 genotype and relapse incidence in patients with acute myeloid leukemia in first complete remission after induction chemotherapy. Leukemia. 2009;23:486–491. doi: 10.1038/leu.2008.339. [DOI] [PubMed] [Google Scholar]
- 52.Hobo W, Hutten T, Schaap N, Dolstra H. Immune checkpoint molecules in acute myeloid leukaemia: Managing the double-edged sword. Br. J. Haematol. 2018;181:38–53. doi: 10.1111/bjh.15078. [DOI] [PubMed] [Google Scholar]
- 53.Stamm H, Klingler F, Grossjohann EM, Muschhammer J, Vettorazzi E, Heuser M, et al. Immune checkpoints PVR and PVRL2 are prognostic markers in AML and their blockade represents a new therapeutic option. Oncogene. 2018;37:5269–5280. doi: 10.1038/s41388-018-0288-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hodi FS, O'Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N. Engl. J. Med. 2010;363:711–723. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Page DB, Postow MA, Callahan MK, Allison JP, Wolchok JD. Immune modulation in cancer with antibodies. Annu. Rev. Med. 2014;65:185–202. doi: 10.1146/annurev-med-092012-112807. [DOI] [PubMed] [Google Scholar]
- 56.Ozkazanc D, Yoyen-Ermis D, Tavukcuoglu E, Buyukasik Y, Esendagli G. Functional exhaustion of CD4+ T cells induced by co-stimulatory signals from myeloid leukaemia cells. Immunology. 2016;149:460–471. doi: 10.1111/imm.12665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jia B, Wang L, Claxton DF, Ehmann WC, Rybka WB, Mineishi S, et al. Bone marrow CD8 T cells express high frequency of PD-1 and exhibit reduced anti-leukemia response in newly diagnosed AML patients. Blood Cancer J. 2018;8:34. doi: 10.1038/s41408-018-0069-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Plitas G, Rudensky AY. Regulatory T cells: Differentiation and function. Cancer Immunol. Res. 2016;4:721–725. doi: 10.1158/2326-6066.CIR-16-0193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu C, Workman CJ, Vignali DA. Targeting regulatory T cells in tumors. FEBS J. 2016;283:2731–2748. doi: 10.1111/febs.13656. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.