Abstract
Non-communicable diseases (NCDs) kill more than 41 million people every year, accounting for 71% of all deaths globally. The prevalence of NCDs is estimated to be higher than that of infectious diseases in Africa by 2030. Precision medicine may help with early identification of cases, resulting in timely prevention and improvement in the efficacy of treatments. However, Africa has been lagging behind in genetic research, a key component of the precision medicine initiative. A number of genomic research initiatives which could lead to translational genomics are emerging on the African continent which includes the Non-communicable Diseases Genetic Heritage Study (NCDGHS) and the Men of African Descent and Carcinoma of the Prostate (MADCaP) Network. These offer a promise that precision medicine can be applied in African countries. This review evaluates the advances of genetic studies for cancer, hypertension, type 2 diabetes and body mass index (BMI) in Africa.
Keywords: Genetics, Africa, Cancer, Diabetes, Hypertension, Obesity
1. Introduction
Non-communicable diseases (NCDs) are the leading cause of global mortality, accounting for 71% of all deaths [1,2]. Notably, 80% of these NCD deaths are from four diseases, type 2 diabetes (T2D), cancer, respiratory diseases and cardiovascular disease, that annually have mortality rates of 1.6, 9.0, 3.9 and 17.9 million respectively [2]. Africa is experiencing a double burden of infectious diseases and NCDs. However, by 2030, deaths from NCDs in Africa are projected to exceed deaths due to communicable, maternal, perinatal and nutritional diseases combined [1]. Furthermore, it is estimated that more than 70% of the global mortality of cancers will occur in Africa within the next decade [3]. Urgent strategies are required to curb this NCD pandemic.
Strategies for preventing and controlling NCDs, such as hypertension and diabetes, are being implemented in Africa and integrated into HIV initiatives, given the double burden of infectious diseases and these NCDs [4]. These population-wide interventions include mass communication for promoting healthy lifestyles, through increased consumption of fruits and vegetables and physical activity [2,4]. However, the current treatment and preventative interventions are based on a “one size fits all” approach [5]. People respond differently to these interventions and some even experience unfavorable reactions. Precision medicine offers the hope that the right person will get the right treatment and help reduce adverse treatment reactions [5]. Through this approach high-risk individuals may be identified early and provided with requisite preventative and therapeutic options.
There are many ‘Omics’ technologies, including proteomics, genomics, and metabolomics, that are key to enhancing precision medicine [6]. However, for this review, we will focus on genomics. NCDs are heritable and extensive studies to unravel the genetic factors associated with these traits have been performed in European populations [7]. However, due to the genetic diversity within Africa and differences in linkage disequilibrium patterns between Europeans and Africans, the generalizability of these findings is limited [7]. The summation of genetic variants into polygenic risks scores seems promising to enable precision medicine efforts through stratification of individuals according to their genetic risk. However, the low representation of Africans in genome-wide association studies (GWAS) was noted to result in a 4.9-fold reduction in predictivity for 17 quantitative traits that include BMI in Africans compared to Europeans [8]. A number of initiatives such as the United Kingdom Biobank (UKBB) and Million Veteran programs are enhancing diversity to capture the genetic variation present in other populations and ethnic groups. Notably, the UKBB and MVP consist of 8000 and approximately 60,000 Africans respectively [9,10]. Nonetheless these are admixed Africans. Other ethnic groups, such as Europeans, are now reaching the point where millions of individual genomes are available for GWAS[11]. Such underrepresentation might lead to the exacerbation of health disparities[8]. Recently, a whole genome study of 426 continental Africans, representing 50 ethnolinguistic groups, reported great genetic diversity in continental Africans. From this, three million novel variants were discovered after comparing with thousands of other African genomes in public databases[12]. By relying on genetics of Africans in the diaspora, novel genetic variants associated with NCDs in continental Africans will be missed. Therefore, the representation of continental Africans needs to be prioritized.
This review will evaluate the advances that have been made on the African continent for genetic studies of NCDs, using cancer, diabetes, hypertension and obesity as examples. We will evaluate what has transpired in this field of study from 2009 to 2019, in Africa.
2. The progress of genetics studies for NCDs on the African continent
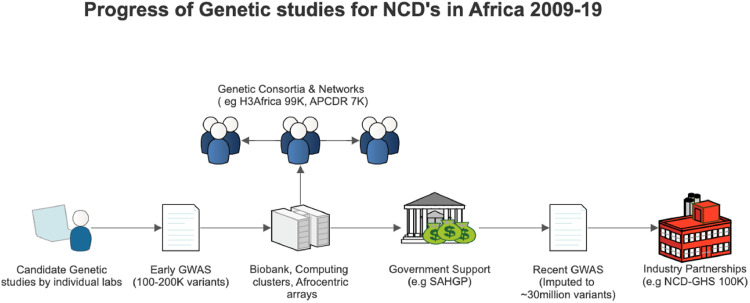
Early genetic studies were primarily performed in a few South African and Tunisian labs and were led by small research groups. Advances in technology subsequently led to genome-wide association studies (GWAS) studies which spurred population genetics approaches in larger GWAS consortia and led to the design of Afrocentric genotyping arrays. These above-mentioned aspects are described in detail, in the following sections and illustrated in Fig. 1.
Fig. 1.
Schematic outline of the progress, development and support of genetics research in Africa. Human Hereditary and Health in Africa (H3Africa) is ongoing and has recruited about 99,408 participants, African Partnerships of Chronic Disease Research (APCDR) has 7000 participants, and the Non-communicable Diseases Genetic Heritage Study (NCD-GHS) is planning to recruit 100,000 participants. The South African Human Genome Project (SAHGP) was funded by the government of South Africa.
2.1. Candidate Gene studies
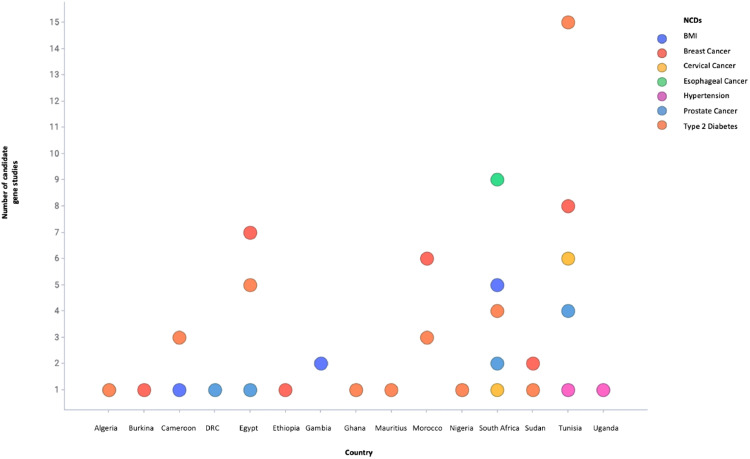
Due to limited capacity and resources, candidate gene studies were commonly used for evaluating the genetics of NCDs in South Africa and Tunisia (Fig. 2). These studies were targeted on particular genetic sites of interest and thus a limited number of SNPs, ranging from one to 250, in smaller studies of hundreds of participants were evaluated. Notably, some key genetic factors were associated with breast, cervical, oesophageal and prostate cancer which are the most common cancers in Africa [13], [14], [15]. Moreover, the rs12255372 variant of TCF7L2 was also associated with obesity in Cameroonians [16]. The majority of these early studies used genetic variants associated with NCDs in European and Asian populations that were limited by the differences in allele frequencies and LD pattern in Africans.
Fig. 2.
Number of candidate genetic studies of selected NCDs that have been conducted in African countries from 2009 to 2019.DRC, Democratic Republic of Congo.
As a consequence of the sparse genetic data in continental Africans, African American participants were used for replication efforts. For instance, the association of the rs1121980 and rs7204609 variants of the FTO gene with BMI in West Africans were replicated in African Americans [17]. The implication of this limitation was the inability to properly explore the genetic diversity of Africans. Although candidate studies are still being conducted for cancer, this approach has been criticized for poor replication even in Europeans and is thought to be biased by our incomplete understanding of gene functions[18]. These are some of the challenges that led to the quest for hypothesis free approaches such as GWAS for evaluating the genetic determinants of complex traits [19].
2.2. GWAS of NCDs in Africa
With the gradual advancement of GWAS in Africa, the utilization of genotyping arrays, such as the Metabochip, enabled the evaluation of the associations of about 200,000 genetic variants with NCDs [20]. Although, the Metabochip was a targeted candidate gene based array that lacked full genome-wide coverage, its use in evaluating genetic associations for BMI and hypertension led to notable results in Africans [21], [22], [23]. Some of these include the association of SEC16B variants with BMI in black South Africans which had not been identified in previous candidate gene studies (Table 1) [22]. The Affymetrix genotyping array, comprised of about two million SNPs, helped to provide some insights of the genetic determinants of NCDs in Africa. Through this genotyping array a novel gene, SEMA4D, was found to be associated with BMI in West Africans [24]. Other novel genes such as ZRANB3 were found to be uniquely associated with T2D in continental Africans [25]. However, most GWAS genotyping arrays were designed based on the genetic variation of European populations and therefore, might have missed the population-specific variants associated with NCDs in African populations [26].
Table 1.
List of GWAS studies that have been conducted in Africa for NCDs from 2009 to 2019.
| Study title | Year Published | Sample Size | Number of SNPs evaluated | Countries where studies were done | Traits/Disease evaluated | Number of Significant hits | Replication cohorts | Number of Replicated significant hits |
|---|---|---|---|---|---|---|---|---|
| Genome scan study of prostate cancer in Arabs: identification of three genomic regions with multiple prostate cancer susceptibility loci in Tunisians [43] | 2013 | 221 | 534,781 | Tunisia | Prostate cancer | 14 | 337 Qatari and Saudi Arabians | None |
| A genome-wide association study of prostate cancer in West African men [44] | 2014 | 932 | 2,837,019 | Ghana | Prostate cancer | 30 | 10 068 African Americans (African Ancestry Prostate Cancer GWAS Consortium) | 1 |
| Genome-wide analysis identifies an African-specific variant in SEMA4D associated with body mass index[24] | 2017 | 1411 | 2,217, 748 | West Africa | BMI | 6 | 1506 West Africans and 9020 African Americans (ARIC,CFS,HUFS,JHS and MESA) | 1 |
| Genetic variants in SEC16B are associated with body composition in black South Africans [22] | 2018 | 1926 | 125,878 | South Africa | BMI, Body composition | 12 | None | |
| Insights Into the Genetics of Blood Pressure in Black South African Individuals: The Birth to Twenty Cohort [21] | 2018 | 1947 | 125,878 | South Africa | Hypertension | 10 | None | |
| ZRANB3 is an African-specific type 2 diabetes locus associated with beta-cell mass and insulin response [25] | 2019 | 5231 | ~18 million | Nigeria, Ghana and Kenya | T2D | 14 | 2578 South Africans (AADM, DDS and DCC meta-analysis) | 2 |
| Genome-wide association study of type 2 diabetes in Africa [45] | 2019 | 4347 | 2,141,465 | South Africa, Nigeria, Ghana and Kenya | T2D | 5 | 4347 Continental Africans (AADM, DDS, DCC meta-analysis) | 2 |
| Uganda Genome Resource Enables Insights into Population History and Genomic Discovery in Africa [32] |
2019 | 6400 | 24, 419, 014 | Uganda | 34 cardiometabolic traits (hypertension and BMI) | 2 novel for BMI | 14124 Continental Africans AADM, DDS,DDC and UGR (meta-analysis) | 2 |
T2D = type 2 diabetes, BMI = body mass index, ARIC = Atherosclerosis Risk in Communities study, CFS = Cleveland Family Study, HUFS = Howard University Family Study, Jackson Heart Study = JHS, MESA = Multi-Ethnic Study of Atherosclerosis, DDS = Durban Diabetes Study, DCC = Durban Case Control Study, UGR = Uganda Genome Resource
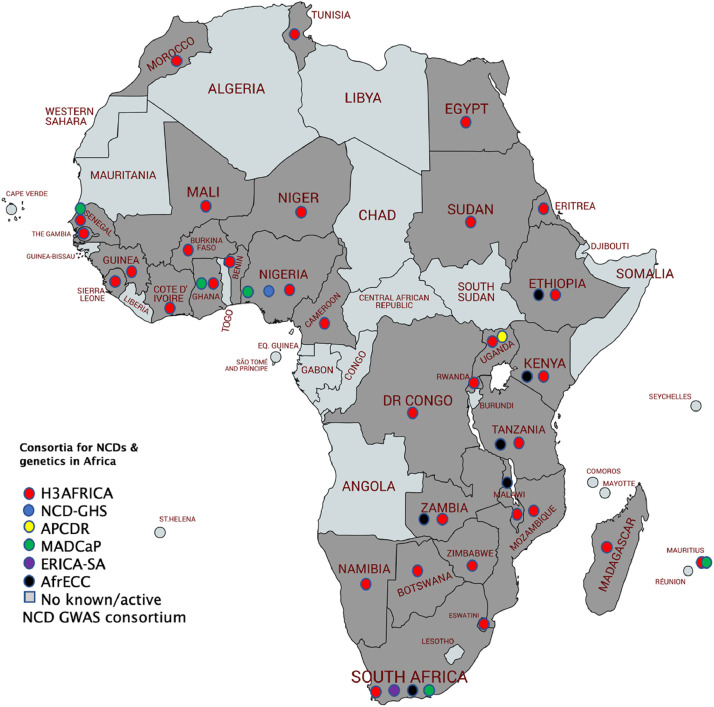
Despite the improved efficacy to discover more genetic variants associated with NCDs through GWAS in African populations, most studies were underpowered to adequately detect significant associations [17]. Notably, only two GWASs have investigated the genetic variants associated with cancer in Africa [27,28]. These GWASs were performed in Tunisian and Ghanaian men with prostate cancer. In Tunisia, 14 SNPs located in 9p24, 17q21 and 22q13 were identified as associated with prostate cancer [27]. This study included 90 prostate cancer cases and 131 controls in the discovery stage and 155 prostate cancer cases and 182 controls in the replication stage. However, this small scale GWAS failed to replicate 14 SNPs in Qatari and Saudi Arabian men with prostate cancer [27]. Another small scale GWAS of 474 prostate cancer cases and 458 healthy controls in Ghana identified rs34575154, rs985081 and rs2185710 to be associated with high Gleason score prostate cancer [28]. However, these SNPs were not successfully replicated in 5,096 African Americans men with prostate cancer. There are currently ongoing GWASs across Africa aiming to identify genetic variants associated with cancer through various research groups and consortia including Evolving Risk Factors for Cancers in African Populations (ERICA), African Female Breast Cancer Epidemiology (AFBRECANE), and Africa Esophageal Cancer Consortium (AfrECC). For a summary of relevant consortia, see Table 2. A map showing the study sites of respective NCD and genetic consortia in Africa is presented in Fig. 3. Due to the efforts of these consortia there is great hope that the next frontier of genetic discoveries will be in Africa.
Table 2.
Consortia for NCDs and genetics in Africa.
| Name of Consortia | Aims and Objectives | Target Sample sizes | African countries covered |
|---|---|---|---|
| Human Hereditary and Health in Africa (H3Africa) https://h3africa.org/ |
To create and support a pan-continental network of laboratories that will be equipped to apply leading-edge research to the study of the complex interplay between environmental and genetic factors which determines disease susceptibility and drug responses in African populations. | 99,408 | 30 countries located in the North, South East, Central and West Africa |
| Non-communicable Diseases Genetic Heritage Study (NCD-GHS) https://actginitiative.com/ |
To assess the burden, spectrum and genetic determinants of various cardio-metabolic traits and diseases in Nigeria. | 100,000 | Nigeria |
| African Partnership of Chronic Disease Research (APCDR) https://www.apcdr.org/ |
It was an international network of research groups which worked together to facilitate and promote collaborative research of chronic diseases across Africa. | 7000 | Uganda |
| Men of African Descent and Carcinoma of the Prostate (MADCaP) https://www.madcapnetwork.org/ |
To assess genomic susceptibility to carcinoma of the prostate in African descent populations. | 576, recruitment ongoing | Ghana, Senegal, Nigeria, Mauritius, South Africa |
| Evolving Risk Factors for Cancers in African Populations (ERICA-SA) https://www.samrc.ac.za/intramural-research-units/evolving-risk-factors-cancers-african-populations-erica-sa |
To identify genetic variants associated with breast, cervical and oesophageal cancer. | 7000, recruitment ongoing | South Africa |
| Africa Esophageal Cancer Consortium (AfrECC) https://dceg.cancer.gov/ research/cancer-types/esophagus/afrecc |
To coordinate etiologic and molecular studies of esophageal cancer in high-risk populations within Africa. | 12,000 | Ethiopia, Kenya, Tanzania, Malawi, Zambia, South Africa |
Fig. 3.
NCDs and genetics consortia coverage in Africa. The Human Hereditary and Health in Africa (H3Africa), is the largest and widest consortium in Africa, to date (over 30 African countries). Non-communicable Diseases Genetic Heritage Study (NCD-GHS) is based in Nigeria. The African Partnerships of Chronic Disease Research (APCDR) is in Uganda, the Evolving Risk Factors for Cancers in African Populations (ERICA-SA) is underway in South Africa, the Africa Esophageal Cancer Consortium (AfrECC) is underway in six countries, and the Men of African Descent and Carcinoma of the Prostate (MADCaP) recruitment is underway in four countries. The consortia coverage above is based on the study sites of the respective consortia.
2.3. Consortium wide genetic studies
Through the funding of the Wellcome Trust and United States National Institutes of Health (NIH), a great boost to the study of the genetics in Africa was ignited in 2010 when the H3Africa consortium was developed [29]. These funds have led to the establishment of Biobanks, capacity development program initiatives, mentoring opportunities for early-career academics, and the establishment of multi-country collaborative studies. Combined, these efforts have significantly improved research output and scope on the African continent. H3Africa has networks and working groups such as the H3Africa Bioinformatics Network (H3ABioNet) and the Cardiovascular H3Africa Innovation Resource (CHAIR). Notably, through the efforts of the H3Africa Bioinformatics Network (H3ABioNet) a robust Afrocentric genotyping array was developed using 350 whole genome sequences from H3Africa, Wellcome Sanger and 1000Genomes reference panels to enhance the coverage of more population-specific variants [30].CHAIR is a key partnership of H3Africa projects which among its objectives seeks to explore the genetic determinants of NCDs in Africa. CHAIR is working towards overcoming the limitations of smaller sample sizes in GWAS, by harmonizing the genetic and phenotype datasets of H3Africa projects to enhance the discovery of novel associations for cardiometabolic traits [31].
Early cohorts from Uganda, West Africa and South Africa were also combined recently under the African Partnership of Chronic Disease Research (APCDR) Network resulting in a landmark paper exploring 34 cardiometabolic traits [32]. This laid a foundation that will be leveraged by other H3Africa studies in further exploring the genetics of NCDs [32]. In this paper, improvements in the imputation accuracy of the 1000 genomes version 3 reference panel of Africans were noted when it was combined with whole genome sequences from Uganda and the African Variation Genome Programme (AVGP). Notably the number of successfully imputed variants (info score > 0.3) increased by 1.5 million in East African populations and 8 million in Baganda [32]. This shows the importance of having a diverse reference panel of Africans for imputation efforts in Africa. Two novel signals for BMI were discovered through the meta-analysis of the APCDR, America Diabetes Mellitus (AADM) study, Durban Diabetes (DDS) study and Durban Case Control study (DCC)[32]. Moreover, a consortium of esophageal cancer researchers in Africa, along the eastern coast of Africa from Ethiopia to South Africa, known as the Africa's Oesophageal Cancer Corridor, is conducting the largest GWAS of esophageal cancer in the continent (Fig. 3) [33]. This project is funded by the NIH and aims to unravel key genetic factors and biological pathways involved in the pathogenesis of esophageal cancer in Africa, thereby providing a platform for risk prediction and precision medicine.
Attempts are now being made to evaluate genetic susceptibly using polygenic risk scores (PRS) for cancer in the Men of African Descent and Carcinoma of the Prostate (MADCaP) Network. Over and above the H3Africa array, this network has designed a genotyping array to aid the identification of Afrocentric variants for cancer and to explore polygenic risk scores [34]. An earlier study of a genetic risk score containing 100 European derived genetic variants found that the top 10% of Ugandan men had a 4.86-fold (95%CI:2.70, 8.76) increase in prostate cancer risk [35]. However, through the evidence from the MADCaP network, the heterogeneity of PRS prediction within continental Africans was depicted [34], implying findings from Uganda may not be applied to other African countries. Thus, more efforts are required to evaluate the possible heterogeneity of PRS prediction within the continent. The limited clinical utility of polygenic risk scores due to low discriminative ability has been reported even in Europeans [36,37]. Therefore, novel approaches to enhance PRS prediction are required.
2.4. Industry and government partnerships
Aside from the scientific community, some African governments are now seeing the value of genetic studies. Notably, the South African government supported a landmark initiative for sequencing the genomes of indigenous and local Africans through the South Africa Genome Project (SAHGP) [29]. These genetic resources, in addition to others in the African continent, were pivotal in the development of the African genotyping array and will be instrumental for future precision medicine efforts. The potential value of the genetics of Africans in leading to the development of novel therapies has also attracted the attention of African industry partners. Notably, the blockbuster PCSK9 inhibitors were developed to mimic the effects of the cholesterol lowering PCSK9 variants that are more prevalent in individuals of African ancestry [38], [39], [40]. The NCD-GHS study is a unique ground-breaking initiative, which will also help enhance precision medicine efforts in Africa by establishing a genetic resource for studying NCDs in 100,000 participants (Fig. 1). There is hope that more similar partnerships may increase as industries gradually become involved in collaborative efforts with academic institutions to translate findings for clinical application.
3. Outstanding questions
Although the establishment of different consortia are a crucial development on the continent, there remain some key questions and opportunities to be further explored by these initiatives. Some of these include: Are the genetic determinants of the NCDs the same in continental Africans? How efficient are the current methods of polygenic risk scores predictions for African populations? The current databases such as FUMA, GTEX [41], etc. that are used to determine the functional role of the identified genetic variants are improving the representation of African Americans through initiatives such as TOPMed (Trans-Omics for Precision Medicine) . However since African Americans are admixed and greater genetic diversity exists in continental Africans, there is a need to include/create an Afrocentric database such as the African Functional Genomics Resource (AFGR)[42] of variants which might not be in these databases but are significantly associated with NCDs in continental Africans[12]. Furthermore, elucidation of causality and related studies such as Mendelian randomization and randomized clinical trials will be required in the future to give robust evidence that may help in the application of these findings in precision medicine. We also acknowledge that multi-omics approaches are required to enhance the realization of precision medicine in Africa. There is a growing number of microbiome studies for obesity and diabetes in the African continent which will also be informative in this regard
4. Conclusion
Africa is a continent with great genetic diversity that promises to be beneficial in the global elucidation of the pathogenesis of NCDs. There has, however, been a lag in the utilization of these unique genetic resources to facilitate the implementation of precision medicine on the continent. However, this is understandable as not many robust signals of association for NCDs have been documented. Nevertheless, a solid foundation to facilitate efforts towards the discovery of novel genetic advances has been set by funding bodies through platforms like the H3Africa as well as industry and academic partnerships such as the recently launched NCD-GHS study. There is great anticipation that Africa is well on its way to employ precision medicine in the efforts to curb the raging NCDs pandemic.
5. Search strategy
We used the keywords Africa, genetics, blood pressure/hypertension, cancer, diabetes, BMI, obesity to select the papers in the PubMed database from 2009 to 2019.
Author contributions
TC and SF conceptualized the paper. TC, ABK, OHO and TM conducted the literature search and wrote the first draft. All authors read, provided critical feedback and approved the final version of the paper.
Declaration of Interests
We have none to declare.
Acknowledgments/Funding
The funders stated therein had no role in the paper design, data collection, data analysis, interpretation and writing of the paper. TC is an international training fellow supported by the Wellcome Trust grant (214205/Z/18/Z). SF is an international Intermediate fellow funded by the Wellcome Trust grant (220740/Z/20/Z) at the MRC/UVRI and LSHTM. S.F. received support from NIH U01MH115485 and the Makerere University-Uganda Virus Research Institute Centre of Excellence for Infection and Immunity Research and Training (MUII). MUII is supported through the DELTAS Africa Initiative (grant 107743). The DELTAS Africa Initiative is an independent funding scheme of the African Academy of Sciences (AAS), Alliance for Accelerating Excellence in Science in Africa (AESA), and supported by the New Partnership for Africa\220s Development Planning and Coordinating Agency (NEPAD Agency) with funding from the Wellcome Trust (107743) and the U.K. government. TM received funding from the Department of Medicine, University of Cape Town (under the Mayosi research group's RHDGen Fellowship (Wellcome Trust, H3Africa project - grant number: 099313/B/12/A)), and the UCT Crasnow Travel Scholarship. T.M. also received funding from the Population Health Research Institute (PHRI) and McMaster University through the inaugural Bongani Mayosi UCT-PHRI Scholarship 2019/2020.
Contributor Information
Tinashe Chikowore, Email: tinashedoc@gmail.com.
Segun Fatumo, Email: Segun.Fatumo@lshtm.ac.uk.
References
- 1.Dicker D, Nguyen G, Abate D. Global, regional, and national age-sex-specific mortality and life expectancy, 1950–2017: a systematic analysis for the global burden of disease study 2017. Lancet. 2018;392(10159):1684–1735. doi: 10.1016/S0140-6736(18)31891-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bigna JJ, Noubiap JJ. The rising burden of non-communicable diseases in sub-Saharan Africa. Lancet Global Health. 2019;7(10):e1295–e12e6. doi: 10.1016/S2214-109X(19)30370-5. [DOI] [PubMed] [Google Scholar]
- 3.Parkin DM, Bray F, Ferlay J, Jemal A. Cancer in Africa 2012. Cancer Epidemiol Biomarkers Prev. 2014;23(6):953–966. doi: 10.1158/1055-9965.EPI-14-0281. [DOI] [PubMed] [Google Scholar]
- 4.Patel P, Rose CE, Collins PY. Noncommunicable diseases among HIV-infected persons in low-income and middle-income countries: a systematic review and meta-analysis. AIDS (London, England) 2018;32(Suppl 1):S5. doi: 10.1097/QAD.0000000000001888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arena R, Ozemek C, Laddu D. Applying precision medicine to healthy living for the prevention and treatment of cardiovascular disease. Curr Probl Cardiol. 2018;43(12):448–483. doi: 10.1016/j.cpcardiol.2018.06.001. [DOI] [PubMed] [Google Scholar]
- 6.Seyhan AA, Carini C. Are innovation and new technologies in precision medicine paving a new era in patients centric care? J Transl Med. 2019;17(1):114. doi: 10.1186/s12967-019-1864-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duncan L, Shen H, Gelaye B. Analysis of polygenic risk score usage and performance in diverse human populations. Nat Commun. 2019;10(1):3328. doi: 10.1038/s41467-019-11112-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martin AR, Kanai M, Kamatani Y, Okada Y, Neale BM, Daly MJ. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat Genet. 2019;51(4):584–591. doi: 10.1038/s41588-019-0379-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gaziano JM, Concato J, Brophy M. Million Veteran Program: a mega-biobank to study genetic influences on health and disease. J Clin Epidemiol. 2016;70:214–223. doi: 10.1016/j.jclinepi.2015.09.016. [DOI] [PubMed] [Google Scholar]
- 10.Bahcall OG. UK Biobank — a new era in genomic medicine. Nat Rev Genet. 2018;19(12):737. doi: 10.1038/s41576-018-0065-3. [DOI] [PubMed] [Google Scholar]
- 11.Evangelou E, Warren HR, Mosen-Ansorena D. Genetic analysis of over 1 million people identifies 535 new loci associated with blood pressure traits. Nat Genet. 2018;50(10):1412–1425. doi: 10.1038/s41588-018-0205-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choudhury A, Aron S, Botigué LR. High-depth African genomes inform human migration and health. Nature. 2020;586(7831):741–748. doi: 10.1038/s41586-020-2859-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kamiza AB, Kamiza S, Mathew CG. HLA-DRB1 alleles and cervical cancer: a meta-analysis of 36 case-control studies. Cancer Epidemiol. 2020;67 doi: 10.1016/j.canep.2020.101748. [DOI] [PubMed] [Google Scholar]
- 14.Kamiza AB, Kamiza S, Singini MG, Mathew CG. Association of TP53 rs1042522 with cervical cancer in the sub-Saharan African population: a meta-analysis. Trop Med Int Health. 2020;25(6):666–672. doi: 10.1111/tmi.13397. [DOI] [PubMed] [Google Scholar]
- 15.Shan J, Mahfoudh W, Dsouza SP. Genome-Wide Association Studies (GWAS) breast cancer susceptibility loci in Arabs: susceptibility and prognostic implications in Tunisians. Breast Cancer Res Treat. 2012;135(3):715–724. doi: 10.1007/s10549-012-2202-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ngwa EN, Sobngwi E, Atogho-Tiedeu B. Association between the rs12255372 variant of the TCF7L2 gene and obesity in a Cameroonian population obesity. BMC Res Notes. 2015;8(1) doi: 10.1186/s13104-015-1661-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adeyemo A, Chen G, Zhou J. FTO genetic variation and association with obesity in West Africans and African Americans. Diabetes. 2010;59(6):1549–1554. doi: 10.2337/db09-1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jorgensen TJ, Ruczinski I, Kessing B, Smith MW, Shugart YY, Alberg AJ. Hypothesis-driven candidate gene association studies: practical design and analytical considerations. Am J Epidemiol. 2009;170(8):986–993. doi: 10.1093/aje/kwp242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bush WS, Moore JH. Chapter 11: genome-wide association studies. PLoS Comput Biol. 2012;8(12) doi: 10.1371/journal.pcbi.1002822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lombard Z, Crowther NJ, van der Merwe L, Pitamber P, Norris SA, Ramsay M. Appetite regulation genes are associated with body mass index in black South African adolescents: a genetic association study. BMJ Open. 2012;2(3) doi: 10.1136/bmjopen-2012-000873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hendry LM, Sahibdeen V, Choudhury A, Norris SA, Ramsay M, Lombard Z. Insights into the genetics of blood pressure in black South African individuals: the birth to twenty cohort. BMC Med Genom. 2018;11(1):2. doi: 10.1186/s12920-018-0321-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sahibdeen V, Crowther NJ, Soodyall H. Genetic variants in SEC16B are associated with body composition in black South Africans. Nutr Diabetes. 2018;8(1):43. doi: 10.1038/s41387-018-0050-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Munthali RJ, Sahibdeen V, Kagura J. Genetic risk score for adult body mass index associations with childhood and adolescent weight gain in an African population. Genes Nutr. 2018;13:24. doi: 10.1186/s12263-018-0613-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen G, Doumatey AP, Zhou J. Genome-wide analysis identifies an African-specific variant in SEMA4D associated with body mass index. Obesity (Silver Spring) 2017;25(4):794–800. doi: 10.1002/oby.21804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adeyemo AA, Zaghloul NA, Chen G. ZRANB3 is an African-specific type 2 diabetes locus associated with beta-cell mass and insulin response. Nat Commun. 2019;10(1):3195. doi: 10.1038/s41467-019-10967-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rosenberg NA, Huang L, Jewett EM, Szpiech ZA, Jankovic I, Boehnke M. Genome-wide association studies in diverse populations. Nat Rev Genet. 2010;11(5):356–366. doi: 10.1038/nrg2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shan J, Al-Rumaihi K, Rabah D. Genome scan study of prostate cancer in Arabs: identification of three genomic regions with multiple prostate cancer susceptibility loci in Tunisians. J Transl Med. 2013;11:121. doi: 10.1186/1479-5876-11-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cook MB, Wang Z, Yeboah ED. A genome-wide association study of prostate cancer in West African men. Hum Genet. 2014;133(5):509–521. doi: 10.1007/s00439-013-1387-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Choudhury A, Aron S, Sengupta D, Hazelhurst S, Ramsay M. African genetic diversity provides novel insights into evolutionary history and local adaptations. Hum Mol Genet. 2018;27(R2):R209–RR18. doi: 10.1093/hmg/ddy161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.H3ABioNet. H3Africa genotyping chip. 2015. https://www.h3abionet.org/resources/h3africa-chip (accessed January 12, 2021).
- 31.Owolabi MO, Akpa OM, Made F. Data resource profile: cardiovascular H3africa innovation resource (CHAIR) Int J Epidemiol. 2019;48(2):366–367g. doi: 10.1093/ije/dyy261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gurdasani D, Carstensen T, Fatumo S. Uganda genome resource enables insights into population history and genomic discovery in Africa. Cell. 2019;179(4):984–1002.e36. doi: 10.1016/j.cell.2019.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van Loon K, Mwachiro MM, Abnet CC. The African esophageal cancer consortium: a call to action. J Glob Oncol. 2018;4 doi: 10.1200/JGO.17.00163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harlemon M, Ajayi O, Kachambwa P. A custom genotyping array reveals population-level heterogeneity for the genetic risks of prostate cancer and other cancers in Africa. Cancer Res. 2020 doi: 10.1158/0008-5472.CAN-19-2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Du Z, Lubmawa A, Gundell S. Genetic risk of prostate cancer in Ugandan men. Prostate. 2018;78(5):370–376. doi: 10.1002/pros.23481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chikowore T, van Zyl T, Feskens EJ, Conradie KR. Predictive utility of a genetic risk score of common variants associated with type 2 diabetes in a black South African population. Diabetes Res Clin Pract. 2016;122:1–8. doi: 10.1016/j.diabres.2016.09.019. [DOI] [PubMed] [Google Scholar]
- 37.Lewis CM, Vassos E. Polygenic risk scores: from research tools to clinical instruments. Genome Med. 2020;12(1):44. doi: 10.1186/s13073-020-00742-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cohen JC. Emerging LDL therapies: using human genetics to discover new therapeutic targets for plasma lipids. J Clin Lipidol. 2013;7:S1–S5. doi: 10.1016/j.jacl.2013.03.005. 3 0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hooper AJ, Marais AD, Tanyanyiwa DM, Burnett JR. The C679X mutation in PCSK9 is present and lowers blood cholesterol in a Southern African population. Atherosclerosis. 2007;193(2):445–448. doi: 10.1016/j.atherosclerosis.2006.08.039. [DOI] [PubMed] [Google Scholar]
- 40.Chikowore T, Sahibdeen V, Hendry LM. C679X loss-of-function PCSK9 variant is associated with lower fasting glucose in black South African adolescents: birth to twenty plus cohort. J Clin Transl Endocrinol. 2019;16 doi: 10.1016/j.jcte.2019.100186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science. 2020;369(6509):1318. doi: 10.1126/science.aaz1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Montgomery SB. African functional genomics resource (AFGR). 2020. https://github.com/smontgomlab/AFGR (accessed January 12,2021 2021).
- 43.Shan J, Al-Rumaihi K, Rabah D. Genome scan study of prostate cancer in Arabs: identification of three genomic regions with multiple prostate cancer susceptibility loci in Tunisians. J Transl Med. 2013;11:121. doi: 10.1186/1479-5876-11-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cook MB, Wang Z, Yeboah ED. A genome-wide association study of prostate cancer in West African men. Hum Genet. 2014;133(5):509–521. doi: 10.1007/s00439-013-1387-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen J, Sun M, Adeyemo A. Genome-wide association study of type 2 diabetes in Africa. Diabetologia. 2019;62(7):1204–1211. doi: 10.1007/s00125-019-4880-7. [DOI] [PMC free article] [PubMed] [Google Scholar]