Abstract
The COVID-19 pandemic has wreaked havoc on the whole world, taking over half a million lives and capsizing the world economy in unprecedented magnitudes. With the world scampering for a possible vaccine, early detection and containment are the only redress. Existing diagnostic technologies with high accuracy like RT-PCRs are expensive and sophisticated, requiring skilled individuals for specimen collection and screening, resulting in lower outreach. So, methods excluding direct human intervention are much sought after, and artificial intelligence-driven automated diagnosis, especially with radiography images, captured the researchers’ interest. This survey marks a detailed inspection of the deep learning–based automated detection of COVID-19 works done to date, a comparison of the available datasets, methodical challenges like imbalanced datasets and others, along with probable solutions with different preprocessing methods, and scopes of future exploration in this arena. We also benchmarked the performance of 315 deep models in diagnosing COVID-19, normal, and pneumonia from X-ray images of a custom dataset created from four others. The dataset is publicly available at https://github.com/rgbnihal2/COVID-19-X-ray-Dataset. Our results show that DenseNet201 model with Quadratic SVM classifier performs the best (accuracy: 98.16%, sensitivity: 98.93%, specificity: 98.77%) and maintains high accuracies in other similar architectures as well. This proves that even though radiography images might not be conclusive for radiologists, but it is so for deep learning algorithms for detecting COVID-19. We hope this extensive review will provide a comprehensive guideline for researchers in this field.
Keywords: COVID-19, Deep learning, Radiography, Automated detection, Medical imaging, SARS-CoV-2
Introduction
COVID-19 has become a great challenge for humanity. Fast transmission, the ever-increasing number of deaths, and no specific treatment or vaccine made it one of the biggest problems on earth right now. It has already been characterized as a pandemic by the World Health Organization (WHO) and is being compared to the Spanish flu of 1920 that took millions of lives. Even though the fatality rate of the disease is only 2–3% [1], the more significant concern is its rapid spreading among humans. The reproductive number of the virus is between 1.5 and 3.5 [2], making it highly contagious. Therefore, early diagnosis is essential to contain the virus. This, however, has proved to be very difficult as the virus can stay inactive in humans approximately 5 days before showing any symptoms [3]. Even with symptoms, COVID-19 is hard to be distinguished from the common flu.
At present, one of the most accurate ways of diagnosing COVID-19 is by a test called Reverse Transcription Polymerase Chain Reaction (RT-PCR) [4]. Since the coronavirus is an RNA virus, its genetic material is reverse transcribed to get a complementary DNA or cDNA. This can then be amplified by polymerase chain reaction or PCR, making it easier to measure. However, it is a complicated and time-consuming process, taking almost 2–3 h and demands the involvement of an expert. Newer technology can produce results in 15 min, but it is costly. Even then, there have been studies showing that RT-PCR can yield false negatives [5]. There are newer machines that can autonomously carry on the tests, eliminating human errors and health risks associated with it. However, it is both costly and unavailable in many parts of the world. Moreover, RT-PCR only detects the presence of viral RNA. It cannot prove that the virus is alive and transmissible [6]. The testing material is also of scarcity due to the sheer number of cases in the pandemic, leading to increasing costs.
Another method of COVID-19 detection is antibody testing [7]. It aims to detect the antibody, generated as an immune response of the COVID-19-affected body. This testing method was designed for mass testing for the already affected. It is cheap and fast, producing results in 15 min and can be carried out in a modest laboratory. However, the problem is that the average incubation period of the coronavirus is 5.2 days [3], and antibodies are often not generated before a week from infection and sometimes even later than that. Thus, early containment is not possible. Also, this testing method is susceptible to both false positives and false negatives due to the cases of minor symptoms. Thus, in terms of early detection and containment, this method is not quite up to the task.
Since the outbreak of this disease, researchers have been trying to find a way to detect COVID-19 that is fast, cheap, and reliable. One of the prominent ideas is to diagnose COVID-19 from radiography images. Studies show that one of the first affected organs in coronavirus cases is the lungs [8]. Thus, radiography images of the lungs could give some insight on their condition. Radiologists, however, often fail to diagnose COVID-19 successfully solely from the images due to the similarity between COVID-19-affected lung images and pneumonia-affected lung images and sometimes even normal lung images. Besides, manual interpretation may suffer from inter and intra-radiologist variance and be influenced by different factors such as emotion and fatigue. Recent advances in deep learning regarding such diagnostic problems allow Computer-Aided Diagnosis (CAD) to reach new heights with its ability to learn from high-dimensional features automatically built from the data. Especially during this pandemic, when expert radiologists are conclusively experiencing difficulties diagnosing COVID-19, CAD seems to be the top candidate to assist the radiologists and doctors in the diagnosis. Works like [9–11] and many more are showing the potential of deep learning–driven CAD to face this pandemic.
Not just in the diagnosis of the virus, but there have been many works done with deep learning applied to almost all the sectors affected by the coronavirus. And the flood of such works have resulted in a number of surveys relating to the role of artificial intelligence in this pandemic situation covering not just diagnosis but also clinical management and treatment [12], image acquisition [13], infodemiology, infoveillance, drug and vaccine discovery [14], mathematical modeling [15], economic intervention [16] etc. including discussions of various datasets [12, 13, 15–18].
The overall observation is that most of the surveys done to date tried to cover a wider extent of the domain instead of depicting an exhaustive overview in one direction. However, we are motivated to focus on what we consider to be the most important aspect of fighting the dreadful disease, and that is detection and diagnosis. Throughout this work, we dispensed a comprehensive discussion on available datasets, existing approaches, research challenges with probable solutions, and future pathways for deep learning empowered automated detection of COVID-19. In addition to the qualitative assay, we provided a quantitative analysis that comprises of extensive experimentation using 315 deep learning models. We tried to investigate some of the key questions here: (1) What are the key challenges to diagnose the disease from radiography data? (2) Which CNN architecture performs the best to extract distinct features from the X-ray modality? (3) How transfer learning can be utilized and how well does it perform being pre-trained on the widely used ImageNet dataset [19]? To the best of our knowledge, this is the very first survey that includes a benchmark study of 315 deep learning models in diagnosis of COVID-19.
The rest of the paper is organized as follows. In “Related Work,” we present a study on related works. Subsequently, Section “Radiography-Based Automated Diagnosis: Hope or Hype” describes the radiography based diagnosis process. We shed light on some challenges in radiography-based diagnosis in “Challenges.” In “Description of Available Datasets,” we describe the publicly available radiography datasets. Detailed analysis of deep learning–based approaches of COVID-19 diagnosis is presented in “Deep Learning–Based Diagnosis Approaches”. Section “Image Prepossessing” reviews different data preprocessing techniques. Section “Comparative Analysis” provides a quantitative analysis of some state-of-the-art deep learning architectures on our compiled dataset. Finally, we conclude this paper with future research directions in “Discussion and Future Directions” and concluding remarks in “Conclusion.”
Related Work
This paper gives a comprehensive overview of the automated detection of COVID-19 through data-driven deep learning techniques. Previously, Ulhaq et al. [12] discussed the existing literature on deep learning–based computer vision algorithms relating to the diagnosis, prevention, and control of COVID-19. They also discussed the infection management and treatment-related deep learning algorithms along with a brief discussion about some of the existing datasets. However, they included a pre-print (non peer-reviewed) version of some papers which limit the acceptability of the work.
In [14], the authors discussed the role of AI and big data in fighting COVID-19. In addition to the existing deep learning architectures available for detection and diagnosis of COVID-19, they discussed the existing SIR (Susceptible, Infected, and Removed) models and other deep models for identification, tracking, and outbreak prediction. The survey also included different speech and text analysis methods, cloud-based algorithms for infodemiology and infoveillance, deep learning algorithms for drug repurposing, big data analysis–based outbreak prediction, virus tracking, vaccine, and drug discovery, etc. They did not, however, show any comparative quantitative analysis of the reviewed works.
Shi et al. [13] reviewed medical imaging techniques in battling COVID-19. Various contact-less image acquisition techniques, deep learning–based segmentation of lungs and lesion, X-ray and CT screenings, and severity analysis of COVID-19 along with some publicly open datasets are included in the work. Nevertheless, they did not provide any quantitative analysis of existing methods either. Also, their discussions on the existing datasets are somewhat inadequate.
In [15], the authors discussed mathematical modeling of the pandemic with SIR, SEIR (Susceptible, exposed, infected, and removed) and SIQR (Susceptible, infected, quarantined, and recovered) models.
In [17], the authors gave a comparative list of publicly available datasets consisting of the image data of COVID-19 cases. However, they did not shed any light on the works done so far and also did not provide any pathways for future research in this domain. In contrast to other studies, Latif et al. [16] discussed deep learning algorithms for risk assessment and prioritization of patients in addition to diagnosis.
In [18], Nguyen discussed deep learning–driven medical imaging, IoT-driven approaches for the pandemic management, and even Natural Language Processing (NLP)–based approaches for COVID-19 news analysis. However, like others, they did not give any comparative analysis either.
Most of the discussed papers have tried to cover a wide area of topics and, thus, lacked detailed discussions in a single domain. None of the papers provides a quantitative analysis of the works discussed, something that can be very helpful to the researchers. Our work aims to overcome these limitations by focusing on a single domain, COVID-19 detection. We also provide a quantitative and comparative analysis of 315 different deep learning algorithms, something that is yet to be done by any other survey papers. A summary of all the survey papers covered in this section is presented in Table 1.
Table 1.
Related surveys on AI techniques for detecting COVID-19 from radiography images
| Study | Key topics | No. of reviewed papers1 | No. of discussed datasets2 | Benchmarking deep models3 |
|---|---|---|---|---|
| Ulhaq et al. [12] | Vision-based diagnosis, control and treatment | 21 | 6 | – |
| Pham et al. [14] | AI and big data-based diagnosis, outbreak prediction, and biomedicine | 32 | 2 | – |
| Shi et al. [13] | AI-based image acquisition, segmentation, and diagnosis | 14 | 4 | – |
| Kalkreuth et al. [17] | COVID-19 dataset listing | 4 | 12 | – |
| Latif et al. [16] | AI-based COVID-19 diagnosis, pandemic modeling, dataset description, and bibliometric analysis | 25 | 5 | – |
| Nguyen [18] | AI-based COVID-19 diagnosis, modeling, text mining, and dataset description | 12 | 10 | - |
| Mohamadou et al. [15] | Mathematical modeling of pandemic and COVID-19 diagnosis | 20 | 6 | – |
| Our study | Deep learning–based COVID-19 diagnosis | 38 | 16 | Benchmarked 315 deep models that comprises the combinations of 15 CNNs and 21 classifiers |
1Diagnosis-related papers, 2Radiography-based datasets, 3“-” means not applicable for the paper
The Italic entries signify our contributions
Radiography-Based Automated Diagnosis: Hope or Hype
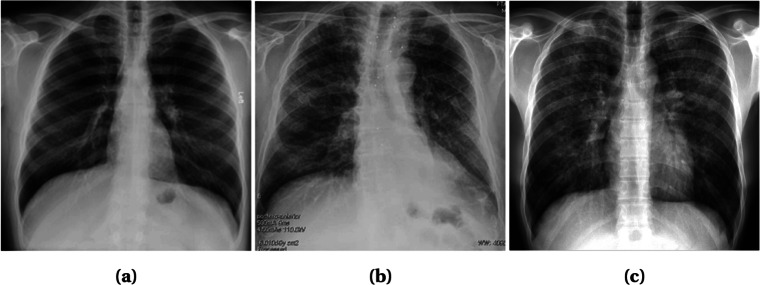
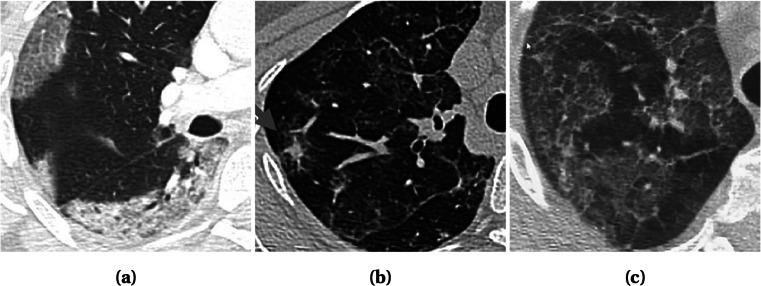
Radiography images, i.e., chest X-ray and computed tomography (CT), can be used to diagnose COVID-19 as the disease primarily affects the respiratory system of the human body [8]. The primary findings of chest X-rays are those of atypical pneumonia [20] and organizing pneumonia [21]. The most common finding in chest radiography images is Ground Glass Opacity (GGO), which refers to an area with increased attenuation in the lung. As shown in Fig. 1, a chest X-ray image shows some hazy grey shade instead of black with fine white blood vessels. In contrast, CT scans show GGO [8] (Fig. 2a), and in severe cases, consolidation (Fig. 2b). Chest images sometimes also show something called “crazy paving” (Fig. 2c), which refers to the appearance of GGO with a superimposed interlobular and intralobular septal thickening. These findings can be seen in isolation or combination. They may occur in multiple lobes and affect in the peripheral area of the lungs.
Fig. 1.
X-ray images with different infection types: a Patchy GGOs present at both lungs; b Nuanced parenchymal thickenings; and c GGOs with some interstitial prominence. Images obtained from [26]
Fig. 2.
CT scan showing different infection types: a Subpleural GGOs with consolidations in all lobes; b GGOs with probable partially resolved consolidations; and c Scattered GGOs with band consolidations. Images obtained from [26]
It is worth noting that chest CT is considered to be more sensitive [22] for early COVID-19 diagnosis than chest X-ray since chest X-ray may remain normal for 4–5 days after start of symptoms where CT scan shows a typical pattern of GGO and consolidation. Besides, CT scans can show the severity of the disease [23]. A recent study performed on 51 COVID-19 patients shows that the sensitivity of CT for COVID-19 infection was 98% compared to RT-PCR sensitivity of 71% [24].
However, CT scans are difficult to obtain in COVID-19 cases. It is mainly due to the difficulty of decontamination issues regarding patient transports to the CT suites. As a matter of fact, the American College of Radiology dictates that CT scan-related hassles can disrupt the availability of such radiology services [25]. Another problem is that CT is not available worldwide, and in most cases, expensive and, thus, has a low outreach. This is why, despite the lesser sensitivity and quality, chest X-rays are the most common method of diagnosis and prognosis of not only COVID-19 cases but most other lung-related abnormalities.
The main problem is that these findings are not only found in COVID-19 cases but also in pneumonia cases. In many mild COVID-19 cases, the symptoms are similar to that of the common cold and sometimes show no different than that of normal lungs. Even though Research in [27] has indicated that the radiography image of COVID-19-affected lungs differs from the image of bacterial pneumonia-affected lungs. In COVID-19 cases, the effect is more likely to be scattered diffusely across both lungs, unlike typical pneumonia. However, in the early stages, even expert radiologists are often unable to detect or distinguish between COVID-19 and pneumonia.
Amidst such a frightful situation, deep learning–driven CAD seems a logical solution. Deep learning can extract and learn from high-dimensional features humans are not even able to comprehend. So, it should be able to deliver in this dire situation as well. Moreover, there has already been a flood of such approaches recently with good results, showing hope in this crisis period. However, many such hopeful ideas have turned into false hopes in the past. This work investigates the different works relating to deep learning aided CAD to resolve whether this is our hope, or if it is only another hype.
Challenges
As discussed in the previous section, radiography image can pave an efficient way to the detection of the COVID-19 at an earlier stage. However, the unavailability and quality issues related to COVID-19 radiography images introduce challenges in the diagnosis process while effecting the accuracy of the detection model. Here we discuss some of the major challenges faced by the researchers in the detection of COVID-19 from radiography images.
Scarcity of COVID-19 Radiography Images
Significant numbers of radiography image datasets are made available by the researchers to facilitate collaborative efforts for combating COVID-19. However, they contain at most a few hundreds of radiography images of confirmed COVID-19 patients. As a result, poor predictions are made by the models being over-fitted by insufficient data which puts a cap on the potential of deep learning [28]. Data augmentation techniques can be used to increase the dataset volume. Common augmentation techniques like flipping and rotating can be applied to image data with satisfactory results [29]. Transfer learning is another alternative to deal with insufficient data size while reducing generalization errors and over-fitting. Significant numbers of works integrated data augmentation and transfer learning into deep learning algorithms to obtain pleasing performances [30–40]. Few shot learning [41] and zero shot learning [42] are also plausible solutions to the data insufficiency problem.
Class Imbalance
This is a common problem faced while detecting COVID-19 from radiography images. Radiography datasets consisting of a sufficient number of X-ray images of pneumonia-affected and normal lungs are available on a large scale. In contrast, for being a completely new disease, the number of images of COVID-19-affected lungs are significantly less than that of normal and pneumonia-affected lungs. As a result, the model becomes prone to giving poor predictions to the minority class. Re-sampling of the datasets is often performed as a solution to the class imbalance problem, which attempts to make the model unbiased from the majority classes. In random under sampling strategy, samples for the minor classes are duplicated randomly, whereas samples from the majority classes are removed randomly in random over sampling methods to mitigate the class imbalance of a dataset [43]. However, over sampling may result in over-fitting [44], which can be reduced adopting any of the improved oversampling techniques—Synthetic Minority Over Sampling (SMOTE)[45], borderline SMOTE [46], and safe level SMOTE [47]. Another technique to deal with the class imbalance problem is to assign weights to the cost function, which ensures that the deep learning model gives equal importance to every class. Some of the works applied data augmentation, e.g., LV et al. [48] employed a Module Extraction technique (MoEx) where standardized features of one image are mixed with normalized features of other images. In another work, Punn et al. [49] manifested class average and random oversampling as an alternative method to data augmentation.
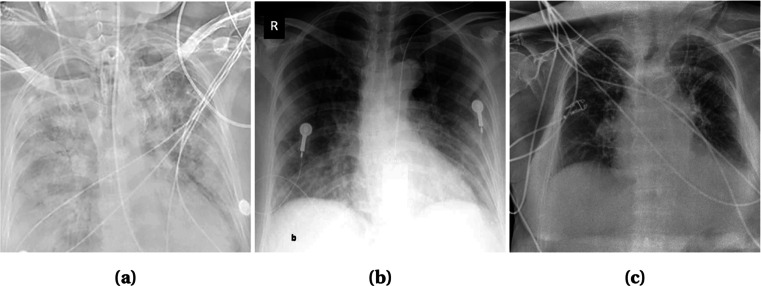
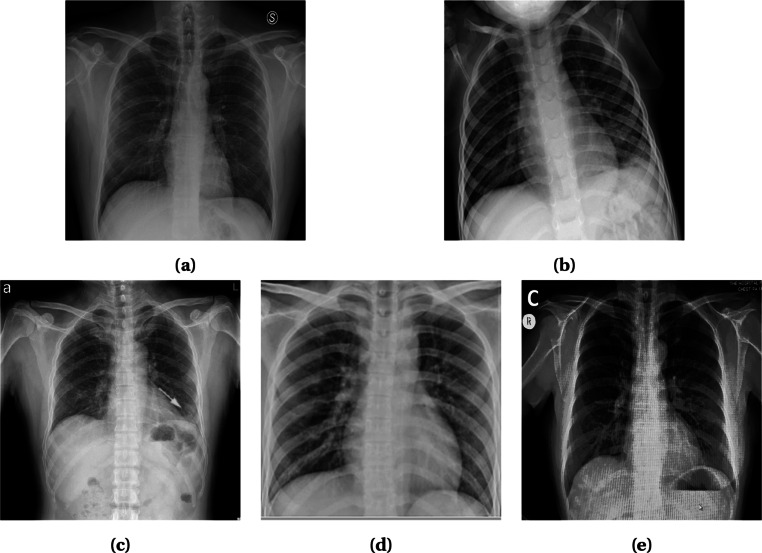
Artifacts/Textual Data and Low Contrast Images
In radiography images, artifacts like wires, probes, or necklaces are often present as depicted in Fig. 3. Even image annotation with textual data (e.g., annotation of the right and left side of an image with “R” and “L,” respectively) is a common practice. These artifacts hamper the learning of a model and lead to poor prediction results. Although textual data (Fig. 3b) can be erased manually by covering it with a black rectangle [32], it is time consuming. A more advanced efficient way is to use a mask of two lungs (for X-ray images) and concatenate with the original image [35, 48, 49]. Thus, the unnecessary areas are ignored and the focus is only given on the interested areas. Mask can be generated using U-Net[48] or binary thresholding [35, 49]. In CT images, the lungs are segmented to focus on the infectious regions [50]. Segmentation tools include U-Net, VB-Net, BCDU-Net, and V-Net which are used in [51–53] and [54], respectively. In some cases, image quality issues such as low contrast (Fig. 3a) introduces challenges in the detection process. To overcome this problem, histogram equalization and other similar methods can be applied [30, 35, 48]. Authors in [48] used Contrast Limited Adaptive Histogram Equalization (CLAHE) which is an advanced version of histogram equalization aiming to reduce over amplification of noise in near constant regions. Additionally, in [35], histogram equalization combined with Perona-Malik filter (PMF) and unsharp masking edge enhancement is applied to facilitate contrast enhancement, edge enhancement, and noise elimination on the entire chest X-ray image dataset. Some literature works are also observed to exclude the faulty images from their dataset [37, 52, 55, 56].
Fig. 3.
X-ray images of some faulty images. a Low Contrast with wire around Image. b Textual data on top left corner and probes on chest. c Wires over the chest. Images obtained from [26]
Similar Clinical Manifestations
In many cases, viral pneumonia shows similar symptoms as COVID-19 which makes it difficult to distinguish them. Additionally, mild COVID-19 cases often show no or mild symptoms, and thus, often indistinguishable from the normal lung images to the naked eye. In the worst-case scenario, these result in low detection probability, i.e., low true positive rate (TPR) and high false negative rate (FNR) for COVID-19 cases. The consequence is that the subjects who are screened as COVID-19 negative in false negative cases may end up contaminating others without attempting for a second test. This suggests that a trade-off should be made between sensitivity (true positive) and specificity (true negative) [57]. In [58], the authors argued that the trade-off should be kept as low as possible to make the model highly sensitive (in contrast to low specificity).
Description of Available Datasets
A large volume of well-labeled data can improve the network quality in deep learning while preventing over-fitting and poor predictions. It is a hard task to collect good-quality data then labeling those accordingly and for uncharted territory like the novel Coronavirus, the hurdles are even bigger. However, time demands to tackle this peril at hand. Therefore, many researchers around the world have been working on creating standard datasets. In this section, we discuss some of these datasets in detail.
COVID-19 Image Data Collection [26]:
This publicly available dataset consists of chest X-ray and CT images of individuals suspected with COVID 19 and pneumonia patients. They were collected through doctors, hospitals, and other public sources. 434 images were labeled from the gathered 542 COVID-19 images, and among them, the X-ray and CT images numbered 462 and 80, respectively. There are about 408 Anterior-to-Posterior (AP) or Posterior-to-Anterior (PA) images and 134 Anterior-to-Posterior Supine (AP Supine) images of the patients. The metadata of 380 subjects marked 276 COVID-19-positive cases where 150 male, 91 female patients, and the rest of them were unlabeled.
Actualmed COVID-19 Chest X-ray Dataset Initiative[59]:
A similar dataset of Anterior-to-Posterior (AP) and Posterior-to-Anterior (PA) views of chest X-rays including metadata have been published recently. This open sourced dataset consists of 238 images where cases with COVID-19, no findings and inconclusive images tallied 58, 127, and 53 respectively.
Figure1 COVID-19 Chest X-ray Dataset Initiative [60]:
Another dataset with 55 images of Anterior-to-Posterior (AP) and Posterior-to-Anterior (PA) view of chest X-ray was released with public accessibility. The X-ray images from 48 subjects labeled 10 males and 7 females and the rest remained unlabeled. The dataset enlisted 35 confirmed COVID-19, 3 no findings, and 15 inconclusive images. The age of the subjects ranges from 28 to 77.
COVID-19 Radiography Database [61]:
A substantial dataset is made based on the chest X-ray images. This dataset comprises such data of COVID-19-positive patients, normal individuals, and viral pneumonia-infected people. The latest release has Posterior-to-Anterior (PA) view images of 219 COVID-19 positives, 1341 normal, and 1345 viral pneumonia specimens. The Italian SIRM dataset[62], COVID Chest X-ray Dataset by Cohen J et al. [26], 43 different articles and chest X-ray (pneumonia) images database by Mooney P et al. [63] has led considerable facilitation for this dataset.
COVIDx [64]:
This dataset was developed by combining the following 5 publicly available dataset: Cohen J et al. [26], ActualMed COVID-19 Chest X-ray Dataset Initiative [59], Figure1 COVID-19 Chest X-ray Dataset Initiative [60], RSNA Pneumonia Detection Challenge dataset [65], and COVID-19 Radiography Database [61]. The augmented dataset consisted of 13989 images from 13870 individuals. The findings recorded 7966 normal images, 5459 images of pneumonia patients, and 473 images of COVID-19-positive patients.
Augmented COVID-19 X-ray Dataset [66]:
This dataset accumulated an equal number (310) of positive and negative COVID-19 images from 5 well-known datasets. They are as follows: COVID Chest X-ray Dataset [26], Italian SIRM [62] radiography dataset, Radiopaedia [67], RSNA Pneumonia Detection Challenge [65], and Chest X-ray Images (pneumonia) [63]. Later using data augmentation methods like flipping, rotating, translation, and scaling, the number was enhanced to 912 COVID-19-positive images and 912 COVID-19-negative images.
COVID-CT Dataset [68]:
This CT image-based radiography dataset has been well-acknowledged by radio experts. It consists of 349 and 463 CT images from 216 COVID-19-positive patients and 55 COVID-19-negative patients respectively. As the data collection from two categories implied, the labeling of the data has been done in two classes: COVID-19 positive and COVID-19 negative.
Extensive COVID-19 X-ray and CT Chest Images Dataset [69]:
Unlike the other datasets, this contains both X-ray images and CT scans, with a total number of augmented images reaching 17,099. Amongst them, 9544 of them are X-rays, and the rest are CT scans. The dataset has been created for binary classification between COVID and non-COVID. Among the X-ray images, there are 5500 non-COVID images and 4044 COVID images. In the case of the CT scans, there are 2628 non-COVID images and 5427 COVID images.
COVID-19 CT Segmentation Dataset [70]:
This dataset consists of 100 axial CT images of more than 40 COVID-positive patients. The images were segmented in three labels by radiologists. They are ground glass, consolidation, and pleural effusion. This dataset provides another nine volumetric CT scans that include 829 slices. Amongst them, 373 have been annotated positive and segmented by a radiologist, with lung masks provided for more than 700 slices.
COVID-19 X-ray Images [71]:
This is a dataset in Kaggle that contains both chest X-ray and CT images with the tally of the number of images up to 373. This dataset includes 3 classes, COVID-19, streptococcus pneumonia, and others, including SARS, MERS, and ARDS. The dataset comes with metadata that includes information such as sex, age, medical status of the patients, and other related medical status.
Researchers around the world have made their dataset open for everyone to facilitate the research scopes. The Italian SIRM [62] Covid-19 dataset, a Twitter thread reader [72] with chest X-ray images, Radiopaedia [67], COVID-19 BSTI Imaging Database [73] etc. are some notable dataset which have been made public recently. The Italian SIRM records 115 confirmed COVID-19 cases. The thread reader from Twitter assembled 134 chest X-ray images of 50 COVID-19 cases along with gender, age, and symptom information of each. The Radiopaedia provides chest radiography images of 153 normal, 135 COVID-19 positives, and 485 pneumonia-affected patients. BSTI Imaging Database consists of comprehensive reports and chest radiography images (both X-ray and CT scans) of 59 COVID-19-positive patients. Coronacases.org [74] is a website dedicated to COVID-19 cases, which includes 10 such cases with extensive patient reports. Eurorad.org [75] contains 39 radiography images of COVID-19-positive patients. Images include both X-ray and CT scans along with extensive clinical reports. All the datasets mentioned above are being used for model training either directly or after going through some data augmentation process.
A summary of the discussed datasets is presented in Table 2.
Table 2.
Available radiography datasets to detect COVID-19. Here, NC means COVID-19 negative
| Sl. No. | Dataset | Date of publication | Modality | Class | Description | Works on the dataset | Highest accuracy |
|---|---|---|---|---|---|---|---|
| 1 | COVID-19 Image Data Collection [26] | February 15, 2020a | Chest X-ray CT scan | COVID-19 | 462 X-ray 80 CT scan | Apostolopoulos et al. [55] Narin et al. [76] Minaee et al. [36] LV et al. [48] Punn et al. [49] | 99.18% [55] |
| 2 | Actualmed COVID-19 Chest X-ray Dataset initiative [59] | April 20, 2020a | Chest X-ray | COVID-19 | 238 X-ray | – | – |
| 3 | Figure1 COVID-19 Chest X-ray Dataset Initiative [60] | April 3, 2020a | Chest X-ray | COVID-19 | 55 X-ray | – | – |
| 4 | COVID-19 Radiography Dataset [61] | March 28, 2020 | Chest X-ray | COVID-19 Normal Pneumoinia | 219 COVID-19 1341 Normal 1345 Pneumonia | Chowdhury et al. [31] Bassi et al. [32] Zhang et al. [77] | 98.3% [31] |
| 5 | COVIDx [39] | March 19, 2020a | Chest X-ray | COVID-19 Pneumonia Normal | 473 COVID-19 5459 Pneumonia 7966 Normal | Ucar et al. [38] Farooq et al. [34] Wang et al. [39] Karim et al. [35] | 98.26% [38] |
| 6 | Augmented COVID-19 X-ray-Images Dataset [66] | March 26, 2020 | Chest X-ray | COVID-19 NC (COVID Negative) | 912 COVID-19 912 NC | Alqudah et al. [78] | 95.2% |
| 7 | Italian SIRM COVID-19 Database[62] | March 3, 2020 | Chest CT scan | COVID-19 | 115 COVID-19 cases | Apostolopoulos et al. [55] Bassi et al. [32] Yamac et al. [40] Apostolopoulos et al. [79] Hall et al. [33] | 99.18% [55] |
| 8 | Radiopaedia[67] | September 8, 2020b | Chest X-ray CT scan | COVID-19 | 135 COVID-19 485 Pneumonia 153 Normal | Apostolopoulos et al. [55] Yamac et al. [40] Hall et al. [33] | 99.18% [55] |
| 9 | COVID-CT Dataset [68] | March 28, 2020a | Chest CT scan | COVID-19 Normal | 349 COVID-19 463 Normal | He at al. [80] | 86% [80] |
| 10 | Thread reader [72] | March 28, 2020 | Chest X-ray | COVID-19 | 50 COVID-19 cases | Yamac et al. [40] | 95.90% [40] |
| 11 | Extensive COVID-19 X-ray and CT Chest Images Dataset [69] | June 13, 2020 | Chest X-ray CT scan | COVID-19 Normal | 9471 COVID 8128 NC | - | - |
| 12 | COVID-19 CT segmentation dataset [70] | April 13, 2020 | Chest CT scan | Positive (COVID-19) Negative | 373 Positive 456 Negative | Chen et al. [81] | 89% [81] |
| 13 | COVID-19 X-ray Images [71] | May 15, 2020 | Chest X-ray CT scan | COVID-19 Streptococcus Other | 309 COVID 17 Streptococcus 47 Other | - | - |
| 14 | COVID-19 BSTI Imaging Database [73] | September 8, 2020b | Chest X-ray CT scan | COVID-19 Positive | 59 COVID-19 cases | Maghdid et al. [82] | 94.1% [82] |
| 15 | Coronacases.org [74] | September 8, 2020b | Chest CT scan | COVID-19 cases | 10 COVID-19 Positive cases | - | - |
| 16 | Eurorad.org [75] | September 8, 2020b | Chest X-ray CT scan | COVID-19 Positive | 39 COVID-19 cases | - | - |
a First commit
b Last accessed
Deep Learning–Based Diagnosis Approaches
In this section, we explore the recent literature on radiography-based COVID-19 diagnosis using deep learning–based methods by arranging it into two groups. The first group includes studies that detect COVID-19 from chest X-rays whereas the latter discuses CT scan-based works.
X-ray-Based Diagnosis
Recently, a lot of work have been published on COVID-19 detection from chest X-ray images. The detection problem is mostly modeled as a classification problem of 3 classes: COVID-19-affected lungs, normal lungs, and pneumonia-affected lungs. Here we discuss some recent works on X-ray based COVID-19 diagnosis grouping them according to the used deep learning approaches.
Transfer Learning
There remains a scarcity of standard, large volume dataset to train deep learning models for COVID-19 detection. The existing deep convolutional neural networks like ResNet, DenseNet, and VGGNet have the setbacks of having a deep structure with excessively large parameter sets and lengthy training time. Whereas Transfer Learning (TL) surmounts most of these offsets. In transfer learning, knowledge acquired from the training on one dataset is reused in another task with a related dataset, yielding improved performance and faster convergence. In the case of COVID-19 detection, the models are often pre-trained on ImageNet [19] dataset and then fine-tuned with the X-ray images. Many researchers have used TL after pre-training their network on an existing standard dataset [30–40, 48, 49, 55, 76, 77, 79, 83, 84]. Some works even pre-trained their model twice. For example, ChexNet [85] is one such network with twofold pre-train on ImageNet [86] and ChestX-ray-14 [87].
Ensemble Learning
Ensemble learning uses an augmentative learning technique that combines predictions from multiple models to generate more accurate results. It improves the model prediction results by reducing the generalization error and variance. In [88], ensemble learning technique is used by combining 12 models (Resnet-18,50,101,152, WideResnet-50,101, ResNeXt-50,101, MobileNet-v1, Densenet-121,169,201) to get better results. Karim et al. [35] have also ensembled 3 models (ResNet18, VGG19, DenseNet161). However, authors in [37], used this a bit differently by using only one model (i.e., ResNet18), then fine-tuning it with three different datasets and finally ensembling the three networks to get the final result. Ensemble learning has contributed significantly towards achieving an accurate result for COVID-19 detection.
Domain Adaptation
Domain adaptation supports the modeling of a relatively new target domain by adapting learning patterns from a source domain, which has similar characteristics as the target domain. Chest X-ray image of a COVID- 19 patient has a different distribution but similar characteristics as that of pneumonia; thus, Domain Adaptation technique can be used. This technique was used by Zhang et al. [77] for creating COVID-DA where the discrepancy of data distribution and task differences was handled by using feature adversarial adaptation and a novel classifier scheme, respectively. Employing this learning method marked a noticeably improved result in detecting COVID-19.
Cascaded Network
Radiography-based COVID-19 detection suffers from the data scarcity. Introducing cascaded network architecture in a small dataset facilitates dense neurons in the network while avoiding the overfitting problem [89]. LV et al. [48] cascaded two networks (ResNet-50 and DenseNet-169) to classify COVID-19 samples. After a subject got classified as viral pneumonia from the 3 classes (normal, bacterial, and viral pneumonia) using ResNet, it was fed into DenseNet169 for the final classification as COVID-19. The infectious regions were concentrated on with an attention mechanism technique Squeeze-Excitation (SE) [90]. Contrast Limited Adaptive Histogram Equalization improved their image quality, and an additional Module Excitation (MoEx) [91] with the two networks enhanced the imaging features. Both the cascaded networks, ResNet-50 and DenseNet-50 gained high accuracy of 85.6% and 97.1%, respectively.
Other Approaches
Wang et al. [39] designed the COVID-Net architecture optimizing the human-machine collaborative design strategy. They also fabricated a customized dataset for the network training. Lightweight design pattern, architectural diversity, and selective long-range connectivity supported its reliability with an accuracy of 93.3% for detecting COVID-19. Optimizing the COVIDx dataset with Data Augmentation and pretraining the model with ImageNet also contributed to the high accuracy. Ozturk et al. [92] proposed Dark-COVIDNet evolved from DarkNet-19. It boasted 17 convolutional layers optimizing different filtering on each layer. Punn et al. [49] used NASNetLarge to detect COVID-19. Both proposed models performed well.
Computed Tomography-Based Diagnosis
CT scan images are scarce as it is expensive and unavailable in many parts of the world. CT scan images still have shown better performance in COVID-19 detection as it provides more information and features than X-ray. Many works have used deep learning for segmentation of CT images [81, 93, 94]. Authors in [52] even determined the severity of COVID-19 case followed by the conversion time from mild to severe case. In [51–54, 56, 93–95], authors used 3D images as input to detect the infectious regions from COVID-19 patients. Most works applied machine learning based methods. Here we discuss some of these works clustered according to the algorithms used.
Joint Regression and Classification, Support Vector Machine
Machine learning (ML) algorithms, such as Logistic Regression (LR) and Support Vector Machine (SVM), hold superiority over CNN if we probe the complicacy of learning millions of parameters and ease of use. Even though LR and SVM are not that efficient in learning very high-dimensional features, the high-definition CT scan images redress the need for it in this application. So, it can be stated that LR and SVM provide effective results in COVID-19 detection with some attributive advantages. For example, in [50], authors formed four datasets by taking various patch sizes from 150 CT scans. For extracting features, they applied Grey Level Co-occurrence Matrix (GLCM), Local Directional Pattern (LDP), Grey Level Run Length Matrix (GLRLM), Grey-Level Size Zone Matrix (GLSZM), and Discrete Wavelet Transform (DWT) algorithms to the different patches of the images before applying SVM for COVID-19 classification with GLSZM, which resulted in a better performance. In [52], authors proposed a joint regression and classification algorithm to detect the severe COVID-19 cases and estimate conversion time from mild to severe manifestation of the disease. They used logistic regression for classification and linear regression for estimating the conversion time. They also attempted to reduce the influence of outliers and class imbalance, by giving more weight to the important class. Both of the proposed models showed satisfactory performance.
Random Forest and Deep Forest
Random forest algorithm relies on building decision trees and ensembling those to make predictions. Its simplicity and diversity made it applicable to a wide range of classification applications while reducing computational complexity. In [96], an infection Size-Adaptive Random Forest (iSARF) method is proposed where the location-specific features are extracted using VB-Net toolkit [97] and the optimal feature set is obtained using the Least Absolute Shrinkage and Selection Operator (LASSO). The selected features are then fed into the random forest algorithm for the classification. In [98], the authors proposed an Adaptive Feature Selection guided Deep Forest (AFS-DF) for the classification task. They also extracted the location-specific features using VB-Net and utilized these features to construct an N random forest. The adaptive feature selection was opted for minimizing redundancy before cascading multiple layers to facilitate the learning of a deep, distinguishable feature representation for COVID-19 classification.
Multi-view Representation Learning
In classification and regression tasks single-view learning is commonly used as a standard for its straightforward nature. However, multi-view representation learning is getting attention in recent times because of its ability to acquire multiple heterogeneous features to describe a given problem with improved generalization. In [54], authors employed multi-view representation learning for COVID-19 classification among community-acquired pneumonia (CAP). V-Net[99] is used to extract different radiomics and handcrafted features from CT images before applying a latent representation based classifier to detect COVID-19 cases.
Hypergraph Learning
In [100], a hypergraph was constructed where each vertex represents COVID-19 or community-acquired pneumonia (CAP) case. They extracted both radiomics and regional features; thus, two groups of hyperedges were employed. For each group of hyperedges, k-nearest neighbor was applied by keeping each vertex as centroid and found related vertices depending on features. Then the related vertices connected together by making edges. It is not impossible to have noisy, unstable or abnormal image in a dataset. To keep away these images, two types of uncertainty score were calculated: aleatoric uncertainty for noisy or abnormal data and epistemic uncertainty for the model’s inablity to distinguish two different cases. These uncertainty score referred to the quality of image and used as the weight of vertices. Label propagation algorithm [101] was run on the hypergraph generating a label propagation matrix, which was then used to test new cases.
CNN Architecture
The quality of CT images coupled with the expansive computational power of recent technologies make deep learning the most potent candidate for COVID-19 detection. Many state-of-the-art deep CNN architectures like ResNet, DenseNet, and Alexnet have already been used for COVID-19 diagnosis [80–82, 94, 95]. These models are generally pre-trained on the ImageNet dataset before being fine-tuned using CT image datasets to avoid learning millions of parameters from scratch. However, the insufficiency of chest CT image data of COVID-19-affected patients results in the augmentation of training data in many cases [51, 80, 81, 95]. In [53], authors proposed COVIDCT-Net where they used BCDU-Net [102] to segment infectious areas before feeding it into a CNN for classification. DeCovNet was proposed in [51] where a pre-trained U-Net[103] is used to segment the 3D volume of the lung image before being fed into a deep CNN architecture.
Attention Mechanism
To find the infectious regions better, attention mechanism was applied in [56, 81, 93, 94] and others. Authors in [81] used Locality Sensitive Hashing Attention [104] in their residual attention U-Net model. In [93], an online 3D class activation mapping (CAM)[105] was used with ResNet-34 architecture. Authors in [94] applied Feature Pyramid Network (FPN)[106] with ResNet-50 to extract top k-features of an image before feeding them to an attention module (Recurrent Attention CNN [107]) to detect the infections regions from the images. In [56], authors used location attention with the model learning the location of infectious patches in the image by calculating relative distance-from-edge with extra weight.
The details of the works that used deep learning for diagnosing COVID-19 is given in Table 3.
Table 3.
Overview of some COVID-19 detection approaches using deep learning. Trn train, Val validation, Tst test, TV train and validation, CV cross-validation, ACC accuracy, PRE precision, REC recall, SEN sensitivity, SPE specificity, F1-SCR F1 score, CM calculated from Confusion Matrix, AUC area under curve
| Sl. No. | Study | Method | Modality | Class | Dataset | Train-test split | Performance |
|---|---|---|---|---|---|---|---|
| 1 | LV et al. [48] | Casecade SEMENet | Chest X-ray | COVID-19, Pneumonia, Normal | [26, 108] | Trn: 6386 images Val: 456 images Tst: 456 images | ACC: 97.14% F1-SCR: 97% |
| 2 | Bassi et al. [32] | CheXNet [85] | Chest X-ray | COVID-19, Pneumonia, Normal | [26, 61, 62, 108] | Trn: 80% Val: 20% Tst: 180 images | ACC: 97.8% PRE: 98.3% REC: 98.3% |
| 3 | Yamac et al. [40] | CSEN (CheXNet [85]) | Chest X-ray | COVID-19, Viral, and Bacterial Pneumonia | [62, 63, 67, 72] | Stratified 5-fold CV | ACC: 95.9% SEN: 98.5% SPE: 95.7% |
| 4 | Zhang et al. [77] | COVID-DA (Domain Adaptation) | Chest X-ray | COVID-19, Pneumonia, Normal | [61, 65] | Trn: 10,718 images Tst: 945 images | AUC: 0.985 PRE: 98.15% REC: 88.33% F1-SCR: 92.98% |
| 5 | Goodwin et al. [88] | 12 Models Ensembled | Chest X-ray | COVID-19, Normal, Pneumonia | [26] | Trn: 80% Val: 10% Tst: 10% | ACC: 89.4% PRE: 53.3% REC: 80% F1-SCR: 64% |
| 6 | Misra et al. [37] | ResNet-18 | Chest X-ray | COVID-19, Normal, Pneumonia | [26, 65] | Trn: 90% Tst: 10% | ACC: 93.3% PRE: 94.4% REC: 100% |
| 7 | Abbas et al. [30] | DeTraC- ResNet18 | Chest X-ray | COVID-19, Normal, SARS | [26] | Trn: 70% Tst: 30% | ACC: 95.12% SEN: 97.91% SPE: 91.87% |
| 8 | Chowdhury et al. [31] | SqueezNet (Best Model) | Chest X-ray | COVID-19, Viral Pneumonia, Normal | [61] | 5-fold CV | ACC: 98.3% PRE: 100% REC: 96.7% F1-SCR: 100% |
| 9 | Farooq et al. [34]. | COVID-ResNet | Chest X-ray | COVID-19, Normal. Viral, and Bacterial Pneumonia | [39] | Trn: 13,675 images Tst: 300 images | ACC: 96.23% PRE: 100% REC: 100% F1-SCR: 100% |
| 10 | Alqudah et al. [78] | AOCT-Net | Chest X-ray | COVID-19, NonCovid-19 | [66] | 10-fold CV | ACC: 95.2% SEN: 93.3% SPE: 100% PRE: 100% |
| 11 | Hall et al. [33] | 3 Models Ensembled | Chest X-ray | COVID-19, Pneumonia | [26, 62, 67] | 10-fold CV | ACC: 91.24% SPE: 93.12% SEN: 78.79% |
| 12 | Hemdan et al. [109] | COVIDX-Net | Chest X-ray | COVID-19, Normal | [26] | Trn: 80% Tst: 20% | ACC: 90% PRE: 83% REC: 100% F1-SCR: 91% |
| 13 | Apostolo- poulos et al. [79] | VGG19 (Best Model) | Chest X-ray | COVID-19, Normal, Pneumonia | [26, 62, 65] | 10-fold CV | ACC: 93.48% SEN: 92.85% SPE: 98.75% |
| 14 | Apostolo- poulos et al. [55] | Mobile- Netv2.0 (from scratch) | Chest X-ray | COVID-19, nonCOVID-19 | [26, 62, 65, 67] | 10-fold CV | ACC: 99.18% SEN: 97.36% SPE: 99.42% |
| 15 | Karim et al. [35] | Deep COVID- Explainer | Chest X-ray | COVID-19, Normal. Viral, and Bacterial Pneumonia | [62, 65, 110] | 5-fold CV | ACC: 96.77% (CM) PRE: 90% REC: 83% |
| 16 | Majeed et al. [83] | CNNx | Chest X-ray | COVID-19, Normal. Viral, and Bacterial Pneumonia | [26, 62, 65, 108] | Trn: 5327 images Tst: 697 images | SEN: 93.15% SPE: 97.86% |
| 17 | Minaee et al. [36] | SqueezNet (Best Model) | Chest X-ray | COVID-19, nonCOVID-19 | [26, 111] | Trn: 2496 images Tst: 3040 images | ACC: 97.73% (CM) SEN: 97.50% SPE: 97.80% |
| 18 | Narin et al. [76] | ResNet50 (Best Model) | Chest X-ray | COVID-19, Normal | [26, 63] | 5-fold CV | ACC: 98% PRE: 100% REC: 96% SPE: 100% |
| 19 | Punn et al. [49] | NASNetLarge (Best Model) | Chest X-ray | COVID-19, Normal, Pneumonia | [26, 65] | Trn: 1266 images Val: 87 images Tst: 108 images | ACC: 96% PRE: 88% REC: 91% SPE: 94% |
| 20 | Ozturk et al. [92] | DarkCovidNet | Chest X-ray | COVID-19, Normal, Pneumonia | [26, 112] | 5-fold CV | ACC: 87.20% PRE: 89.96% REC: 92.18% |
| 21 | Sethy et al. [84] | ResNet50 +SVM | Chest X-ray | COVID-19, Normal | [26, 63] | Trn: 60% Val: 20% Tst: 20% | ACC: 95.38% F1-SCR: 95.52% MCC: 90.76% |
| 22 | Wang et al. [64] | COVIDNet | Chest X-ray | COVID-19, Normal, Pneumonia | [39] | Trn: 13,675 images Tst: 300 images | ACC: 93.30% PRE: 98.90% REC: 91% |
| 23 | Ucar et al. [38] | COVIDiag-nosisNet | Chest X-ray | COVID-19, Normal, Pneumonia | [26, 39, 108] | Trn: 80% Val: 10% Tst: 10% | ACC: 98.26% PRE: 98.26% REC: 98.26% SPE: 99.13% |
| 24 | Sun et al. [98] | AFS-DF (Deep-Forest) | Chest CT scan | COVID-19, Pneumonia | Not available | 5-fold CV | ACC: 91.79% SPE: 89.95% SEN: 93.05% AUC: 96.35% |
| 25 | Javaheri et al. [53] | COVID-CTNet | Chest CT scan | COVID-19, Pneumonia, Normal | Not available | Trn: 90% Val: 10% Tst: 20 cases | ACC: 90.00% SEN: 83.00% SPE: 92.85% |
| 26 | Kang et al. [54] | Multiview Representaion Learning | Chest CT scan | COVID-19, Pneumonia | Not available | Trn: 70% Tst: 30% | ACC: 95.5% SEN: 96.6% SPE: 93.2% |
| 27 | Donglin et al. [100] | UVHL (Hypergraph Learning) | Chest CT scan | COVID-19, Pneumonia | Not available | 10-fold CV | ACC: 89.79% SEN: 93.26% SPE: 84% PPV: 90.06% |
| 28 | Zhu et al. [52] | Joint regression and Classification | Chest CT scan | COVID-19, Severity estimation | Not available | 5-fold CV | ACC: 85.91% |
| 29 | Ouyang et al. [93] | Attention ResNet34 +Dual Sampling | Chest CT scan | COVID-19, Pneumonia | [65] | TV set: 2186 images Tst: 2796 images | AUC: 0.944 ACC: 87.5% SEN: 86.9% SPE: 90.1% F1-SCR: 82.0% |
| 30 | Chen et al. [81] | Residual Attention U-Net | Chest CT scan | COVID-19 (Segmentation) | [62] | 10-fold CV | ACC: 89% PRE: 95% DSC: 94% |
| 31 | He et al. [80] | DenseNet169 (Self-supervised Transfer Learning) | Chest CT scan | COVID-19, nonCOVID-19 | [68] | Trn: 60% Val: 15% Tst: 25% | ACC: 86% F1-SCR: 85% AUC: 94% |
| 32 | Maghdid et al. [82] | Modified AlexNet | Chest CT scan | COVID-19, Normal | [26, 73] | Trn: 50% Val: 50% Tst: 17 images | ACC: 94.1% SPE: 100% SEN: 90% |
| 33 | Maghdid et al. [82] | Modified AlexNet | Chest X-ray | COVID-19, Normal | [26, 73] | Trn: 50% Val: 50% Tst: 50 images | ACC: 94% SPE: 88% SEN: 100% |
| 34 | Butt et al. [56] | ResNet18 +Location Attention | Chest CT scan | COVID-19, Normal, Viral Pneumonia | Not available | TV set: 85.4% Tst: 14.6% | ACC: 86.7% PRE: 86.7% REC: 81.3% F1-SCR: 83.90% |
| 35 | Song et al. [94] | DRE-Net | Chest CT scan | COVID-19, Normal, Pneumonia | Not available | Trn: 60% Val: 10% Tst: 30% | ACC: 86% PRE: 79% REC: 96% F1-SCR: 97% |
| 36 | Zheng et al. [51] | DeCovNet | Chest CT scan | COVID-19 | Not available | Trn: 499 images Tst: 131 images | ACC: 90.10% SEN: 90.70% SPE: 91.10% |
| 37 | Barstugan et al. [50] | GLSZM+SVM (Best Model) | Chest CT scan | COVID-19, nonCOVID-19 | [62] | 10-fold CV | ACC: 98.71% SEN: 97.56% SPE: 99.68% PRE: 99.62% |
| 38 | Shi et al. [96] | iSARF (Random Forest) | Chest CT scan | COVID-19 Pneumonia | Not available | 5-fold CV | ACC: 87.9% SEN: 90.7% SPE: 83.3% |
| 39 | Gozes et al. [95] | 2D and 3D CNN (ResNet-50) | Chest CT scan | COVID-19 Normal | Not available | Trn: 50 patients Tst: 157 patients | SEN: 98.2% SPE: 92.2% AUC: 0.996 |
Image Prepossessing
Image prepossessing is a crucial step in radiography image classification with deep learning, simply because the smallest details in the images can veer the model to learn completely different features and make disparate predictions. Especially when it comes to COVID-19 images, where it is already difficult for models to learn to classify from two very similar classes (pneumonia and COVID-19), preprocessing becomes an important step. This is not only credited to the similarity between the classes but also the imbalance present in the datasets currently available. In this section, we discuss the various preprocessing methods used by different works in COVID-19 detection. A summary of the discussed techniques is also given in Table 4.
Table 4.
A summary of different prepossessing approaches employed in deep learning-based automated detection of COVID-19
| Preprocessing | Approach | Method | Reference |
|---|---|---|---|
| Addressing data imbalance | Data augmentation | Rotation, Translation, Cropping, Flipping etc. | [30–40, 51, 55, 66, 79, 81, 88, 95] |
| Class resampling | Undersampling | [113, 114] | |
| Oversampling | [37, 49, 93, 114] | ||
| Loss function | Focal Loss[115] | [51] | |
| Weighted Loss[116] | [49, 52, 88] | ||
| Image segmentation | Segmentation model | VB-Net[97] | [52, 93, 96, 98, 100] |
| U-Net[103] | [48, 51, 95] | ||
| V-Net[99] | [54] | ||
| BCDU-Net[102] | [53] | ||
| Image quality enhancement | Contrast enhancement | Histogram Equalizetion | [30, 35, 48, 49, 51, 80, 93] |
| Brightness changing | Adding or subtracting every pixel by a constant value | [38, 51, 80] | |
| Noise removal | Perona-Malik filter [117] | [35] | |
| Adaptive Total Variation [118] | [49] | ||
| Edge sharpening | Unsharp Masking | [35] |
Data Augmentation
Imbalance dataset is a prevalent issue in COVID-19 classification due to the scarcity of COVID-19 images. As discussed before in Section “Challenges,” it can cause wrongful predictions. One way to solve this is data augmentation, which is a strategy to increase the number of images by using various image transformation methods such as rotation, scaling, translation, flipping, and changing the pixels’ brightness. It creates diversity in images without actually collecting new ones. It is one of the simplest ways to deal with imbalanced datasets, and thus, is used in most of the works regarding COVID-19 classification, the list of which, is given in Table 4.
Class Resampling
This is a popular method and has seen much success in overcoming data imbalance. In [113], authors randomly omitted images from the majority class (viral pneumonia, bacterial pneumonia, and normal) to balance the dataset, which is called random undersampling or RUS. The opposite is done in [37, 49, 93] where the authors randomly oversampled the minority class (COVID-19), which is called random oversampling or ROS. Both methods are widely used for their simplicity and effectiveness. In [114], authors experimented several undersampling and oversampling methods with multiclass and hierarchical classification models.
Focal Loss and Weighted Loss
Focal loss [115] is another method to solve the issue of class imbalance. It puts more weight on the hard to classify objects and decreases the weight on easy and correct predictions. A scaling factor is added to the cross-entropy loss function, and it decreases as the confidence in a prediction goes up. This was used in [77] for addressing the class imbalance issue. On the other hand, the weighted loss function is a more common technique to balance the data among different classes, which puts more weight on the minority class and less on the majority. In [49, 88], and [52], authors have used weighted loss function and obtained good results.
Image Quality Enhancement
In medical image analysis, every pixel of the image is important for the proper diagnosis of the disease. This is even more applicable when it comes to diagnosing COVID-19 as it bears so much similarity with pneumonia. Thus, image quality enhancement is applied in many works regarding COVID-19 detection. For this, the most prominent technique is increasing the contrast of the image that allows the features to stand out more. Histogram equalization used in [30, 35, 48, 49, 51, 80, 93], is an example of that. In [38, 51, 80], authors applied brightness adjustment for enhancing image quality. In [35, 49], authors applied Perona-Malik Filter [117] and Adaptive total variation [118], respectively for noise removal. Unsharp masking is used in [35] for edge sharpening.
Image Segmentation
Image segmentation involves dividing an image into segments to simplify image analysis and focus only on the important part. Thus, in the case of COVID-19 detection, image segmentation is often a critical preprocessing step that can allow the model to learn from only the affected organ, namely, the lungs, to provide more accurate predictions. One of the most well-known algorithms for medical image segmentation is U-net [103], which has a superior design of skip connections between different stages of the network (used in [81] and [95]). Probably, the most famous derivations of U-Net is VB-Net [97], which is used in [52, 96, 98], and [100]. There are other architectures available for segmentations such as V-Net [99] used in [54], BCDU-Net [102] used in [53] and other works like [56, 81] and [94] that have incorporated image segmentation into their deep models rather than going through segmentation as a preprocessing step.
Comparative Analysis
As we have seen so far, researchers’ continuous efforts have already elicited deep learning–based automated diagnosis of COVID-19 as a potential research avenue. However, despite having several publicly accessible datasets (See “Description of Available Datasets”), no “benchmark” dataset has been released yet that can be used to assess the performance of different methods’ ability to detect COVID-19 using the same standard. Therefore, different authors reported the performance of their method based on different datasets and evaluation protocols. Being motivated by the urge to compare the models on the same scale, here we present a comparative quantitative analysis of 315 deep models that consists of the combinations of 15 convolutional neural network (CNN) models and 21 classifiers using our customized dataset. Note that, we release our dataset along with train-test split and models to be publicly available. 1
Dataset Description
To conduct the analysis, we compile a dataset of our own that includes X-ray images from 4 different data sources, [26, 59, 60], and [61]. The dataset contains 7879 distinct images in total for 3 different classes: COVID-19, normal, and pneumonia, where the pneumonia class has both viral and bacterial infections. However, we have not taken all the available images of the 4 datasets, but only the frontal X-ray (Posterior-to-Anterior) images, leaving out the side views. We selected datasets from the above sources as they are fully accessible to the research community and the public. To the best of our knowledge, the images are annotated by radiologists, and therefore, the reliability of ground truth labels is ensured.
The dataset is split into training (60%) and test (40%) sets using Scikit-learn’s train_test_split module, which, given the split percentage, randomly splits the data between the two sets due to a random shuffle. The shuffling algorithm uses D.E. Knuth’s shuffle algorithm (also called Fisher-Yates) [119] and the random number generator in [120]. The split is also stratified to ensure the presence of the percentage of class samples in each set. The distributions of samples across the classes and train-test set are shown in Table 5.
Table 5.
Data distribution of chest X-ray images among 3 different classes: COVID-19, normal, and pneumonia
| Class | Total no. of samples | Training data | Test data |
|---|---|---|---|
| COVID-19 | 683 | 410 (5.2%) | 273 (3.5%) |
| Normal | 2924 | 1754 (22.3%) | 1170 (14.8%) |
| Pneumonia | 4272 | 2563 (32.5%) | 1709 (21.7%) |
| Total | 7879 | 4727 (60%) | 3152 (40%) |
Method
The overall detection problem is posed as a multi-class classification problem that consists of two major components: feature extraction and learning a classifier.
Feature Extraction
For the feature extraction step, we utilize transfer learning and select a CNN model which has been trained on ImageNet [86], a dataset containing 1.2 million images of 1000 categories. The last fully connected layer, classification layer, and softmax layer are removed from the model and the rest is considered as a feature extractor that computes a feature vector for each image.
Learning a Classifier
We forward the extracted features to a learning model, which is then trained using 5-fold cross-validation on the training set. From Table 1, we notice an imbalance of sample distribution across the classes (COVID-19: 8.7%, normal: 37.1%, and pneumonia: 52.2%). Therefore, to analyze the deep models’ performance at the presence of class imbalance problem, we have done experimentation with three different approaches: a weighted cost function, upsampling the training dataset, and downsampling the training dataset.
- Weighted Cost Function: Being inspired by the work in [116], we have applied a weighted cost function which ensures each class to be given same relative importance by assigning more weight on the minority class and less on the majority.
where L is total categorical cross-entropy loss. Lnormal, Lpneumonia, and LCOVID denote the cross-entropy losses for normal, pneumonia, and COVID, respectively. Weight of each class is calculated using the following formula:1
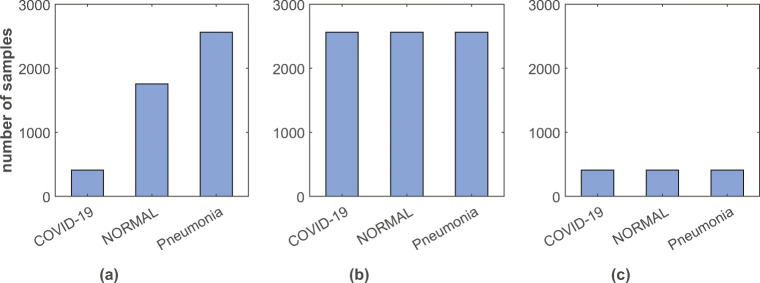
where wi is the weight of class i, n is the total number of observations in the dataset, ni is the total number of observations in class i, and k is the total number of classes in the dataset.2 Upsampling: Upsampling is the method of randomly replicating samples from the minority class. First, we separate observations from each class. Next, we resample the minority classes (normal and COVID-19), setting the number of samples to match the majority class (pneumonia). Thus, the ratio of the three classes becomes 1:1:1 as shown in Fig. 4b.
Downsampling: Downsampling involves randomly eliminating samples from the majority class to prevent from dominating the learning algorithm. It has a similar process, like upsampling. After separating samples from each class, we resample the majority classes (pneumonia and normal), setting the number of samples to match that of the minority class (COVID-19). After that, the ratio of the three classes becomes 1:1:1 (see Fig. 4c).
Fig. 4.
Distribution of samples among 3 different classes in a original, b upsampled, and c downsampled training dataset
Evaluation Process
15 CNN architectures are used to extract feature vectors, and these are fed to 21 classifiers. Therefore, 15 × 21 = 315 models are experimented with a general cross-entropy and the weighted cross-entropy given in (1). Thus, a total of 630 models are deployed with the dataset and ranked according to the performance metrics. In addition, the performance of the top-5 models is evaluated with the upsampled and downsampled training dataset. This study thus includes a total of 640 models for performance benchmarking.
Following is the list of CNN models that have been used to assess the feature extraction step (number of features extracted from each image is given in the parenthesis):
Alexnet (4096) [121],
Xception (2048) [122],
InceptionV3 (2048) [123],
InceptionResNetV2 (1536) [124],
VGG16 (4096) [125],
ResNet50 (2048) [126],
MobileNetV2 (1280) [127],
DarkNet53 (1024) [128],
DarkNet19 (1000) [129],
GoogleNet (1024) [130],
DenseNet-201 (1920) [131],
ShuffleNet (544) [132],
NasNetMobile (1054) [133],
ResNet18 (512) [126], and
VGG19 (4096) [125].
The architectures of these models are shown in Table 6.
Table 6.
Architectural details of the CNNs employed for performance benchmarking in COVID-19 detection (zoom in for better visualization)
| Layer | Patch size/stride | Depth | Output size |
|---|---|---|---|
| (a) ResNet-50 | |||
| Convolution | 7×7×64/2 | 1 | 112×112 |
| Max pool | 3×3/2 | 1 | 56×56 |
| Convolution | 1×1×64 | 3 | 56×56 |
| 3×3×64 | |||
| 1×1×256 | |||
| Convolution | 1×1×128 | 4 | 28×28 |
| 3×3×128 | |||
| 1×1×512 | |||
| Convolution | 1×1×256 | 6 | 14×14 |
| 3×3×256 | |||
| 1×1×1024 | |||
| Convolution | 1×1×512 | 3 | 7×7 |
| 3×3×512 | |||
| 1×1×2048 | |||
| Average pool | − | 1 | 1×1000 |
| Fully connected | |||
| Softmax | |||
| (b) GoogleNet | |||
| Convolution | 7×7/2 | 1 | 112×112×64 |
| Max pool | 3×3/2 | 0 | 56×56×64 |
| Convolution | 3×3/1 | 2 | 56×56×192 |
| Max pool | 3×3/2 | 0 | 28×28×192 |
| Inception(3a) | - | 2 | 28×28×256 |
| Inception(3b) | - | 2 | 28×28×480 |
| Max pool | 3×3/2 | 0 | 14×14×480 |
| Inception(4a) | - | 2 | 14×14×512 |
| Inception(4b) | - | 2 | 14×14×512 |
| Inception(4c) | - | 2 | 14×14×512 |
| Inception(4d) | - | 2 | 14×14×528 |
| Inception(4e) | - | 2 | 14×14×832 |
| Max pool | 3×3/2 | 0 | 7×7×832 |
| Inception(5a) | - | 2 | 7×7×832 |
| Inception(5b) | - | 2 | 7×7×1024 |
| Average pool | 7×7/1 | 0 | 1×1×1024 |
| Dropout(40%) | - | 0 | 1×1×1024 |
| Linear | - | 1 | 1×1×1024 |
| Softmax | - | 0 | 1×1×1024 |
| (c) VGG-16 | |||
| Convolution | 3×3×64/1 | 2 | 224×224×64 |
| Max pool | 3×3/2 | 1 | 112×112×64 |
| Convolution | 3×3×128/1 | 2 | 112×112×128 |
| Max pool | 3×3/2 | 1 | 56x56x128 |
| Convolution | 3×3×256/1 | 2 | 56×56×256 |
| 1×1×256/1 | 1 | ||
| Max pool | 3×3/2 | 1 | 28×28×256 |
| Convolution | 3×3×512/1 | 2 | 28×28×512 |
| 1×1×512/1 | 1 | ||
| Max pool | 3×3/2 | 1 | 14×14×512 |
| Convolution | 3×3×512/1 | 2 | 14×14×512 |
| 1×1×512/1 | 1 | ||
| Max pool | 3×3/2 | 1 | 7×7×512 |
| Fully connected | - | 2 | 1×4096 |
| Softmax | - | 1 | 1×1000 |
| (d) AlexNet | |||
| Convolution | 11×11/4 | 1 | 55×55×96 |
| Max pool | 3×3/2 | 1 | 27×27×96 |
| Convolution | 5×5/1 | 1 | 27×27×256 |
| Max pool | 3×3/2 | 1 | 13×13×256 |
| Convolution | 3×3/1 | 1 | 13×13×384 |
| Convolution | 3×3 | 1 | 13×13×384 |
| Convolution | 3×3 | 1 | 13×13×256 |
| Max pool | 3×3/2 | 1 | 6×6×256 |
| Fully connected | - | 2 | 1×4096 |
| Softmax | - | 1 | 1×1000 |
| (e) DarkNet-53 | |||
| Convolution | 3×3×32/1 | 1 | 256×256×32 |
| Convolution | 3×3×64/2 | 1 | 128×128×64 |
| Convolution | 1×1×32/1 | 1 | 128×128 |
| Convolution | 3×3×64/1 | ||
| Residual | - | ||
| Convolution | 3×3128/2 | 1 | 64×64 |
| Convolution | 1×1×64/1 | 2 | 64×64 |
| Convolution | 3×3×128/1 | ||
| Residual | - | ||
| Convolution | 3×3×256/2 | 1 | 32×32 |
| Convolution | 1×1×128/1 | 8 | 32×32 |
| Convolution | 3×3×256/1 | ||
| Residual | - | ||
| Convolution | 3×3×512/2 | 1 | 16×16 |
| Convolution | 1×1×256/1 | 8 | 16×16 |
| Convolution | 3×3×512/1 | ||
| Residual | - | ||
| Convolution | 3×3×1024/2 | 1 | 8×8 |
| Convolution | 1×1×512/1 | 4 | 8×8 |
| Convolution | 3×3×1024/1 | ||
| Residual | - | ||
| Average pool | - | 1 | 1×1000 |
| FC | |||
| Softmax | |||
| (f ) ShuffleNet | |||
| Convolution | 3×3/2 | 2 | 112×112 |
| Max pool | 3×3/2 | 2 | 56×56 |
| Stage 2 | -/2 | 1 | 28×28 |
| -/1 | 3 | 28×28 | |
| Stage 3 | -/2 | 1 | 14×14 |
| -/1 | 7 | 14×14 | |
| Stage 4 | -/2 | 1 | 28×28 |
| -/1 | 3 | 7×7 | |
| Global pool | 7×7 | - | 1×1 |
| FC | - | - | 1×1000 |
| (g)MobileNetV2 | |||
| Convolution | 3×3/2 | 1 | 112×112×32 |
| Bottleneck | -/1 | 1 | 112×112×16 |
| Bottleneck | -/2 | 2 | 56×56×24 |
| Bottleneck | -/2 | 3 | 28×28×32 |
| Bottleneck | -/2 | 4 | 14×14×64 |
| Bottleneck | -/1 | 3 | 14×14×96 |
| Bottleneck | -/2 | 3 | 7×7×160 |
| Bottleneck | -/1 | 1 | 7×7×320 |
| Convolution | 1×1/1 | 1 | 7×7×1280 |
| Average pool | 7×7/- | 1 | 1×1×1280 |
| Convolution | 1×1/1 | - | k |
| (h) DenseNet-201 | |||
| Convolution | 7×7/2 | 1 | 112×112 |
| Max pool | 3×3/2 | 1 | 56×56 |
| Dense block (1) | 1×1 | 6 | 56×56 |
| 3×3 | |||
| Transition layer (1) | 1×1 | 1 | 56×56 |
| 3×3/2, avg pool | 28×28 | ||
| Dense block (2) | 1×1 | 12 | 28×28 |
| 3×3 | |||
| Transition layer (2) | 1×1 | 1 | 28×28 |
| 3×3/2 ,avg pool | 14×14 | ||
| Dense block (3) | 1×1 | 48 | 14×14 |
| 3×3 | |||
| Transition layer (3) | 1×1 | 1 | 14×14 |
| 3×3/2, avg pool | 7×7 | ||
| Dense block (4) | 1×1 | 32 | 7×7 |
| 3×3 | |||
| Average pool | 7×7 | 1 | 1×1 |
| FC | 1000 | ||
| Softmax | - | ||
| (i) Xception | |||
| Convolution | 3×3×32/2 | 1 | 149×149×32 |
| 3×3×64 | 147×147×64 | ||
| Separable convolution | 3×3×128/1 | 2 | 147×147×128 |
| Max pool | 3×3/2 | 1 | 74×74×128 |
| Separable convolution | 3×3×256/1 | 2 | 74×74×256 |
| Max pool | 3×3/2 | 1 | 37×37×256 |
| Separable convolution | 3×3×728/1 | 2 | 37×37×728 |
| Max pool | 3×3/2 | 1 | 19×19×728 |
| Separable convolution | 3×3×728/1 | 24 | 19×19×728 |
| Separable convolution | 3×3×728/1 | 1 | 19×19×728 |
| 3×3×1024/1 | 19×19×1024 | ||
| Max pool | 3×3/2 | 1 | 10×10×1024 |
| Separable convolution | 3×3×1536/1 | 1 | 10×10×1536 |
| 3×3×2048/1 | 10×10×2048 | ||
| Average pool | - | - | 1×1×2048 |
| Optional FC | - | - | 1×1×1000 |
| Logistic regression | - | - | 1×1×1000 |
(a) In ResNet-18, the convolution layers 2 to 5 contain 2 successive convolutions instead of 3 and each is repeated only 2 times
(b) InceptionV3 incorporates factorized 7×7 convolutions and takes in different sized inputs. InceptionResNetV2 incorporates residual networks with factorized convolutions, as well as several reduction blocks
(c) In VGG-19, the 3rd, 4th, and 5th convolution layer has all 3×3 convolutions and is repeated 4 times each instead of 3 successive convolutions
(e) DarkNet-19 has no residual networks, with only max pooling layers reducing the image
List of classifiers we tried for the comparative analysis:
Tree-Based[134]: Fine, Medium, Coarse
Discriminant Analysis[135]: Linear, Quadratic
Support vector machine[136]: Linear , Quadratic , Cubic, Coarse Gaussian, Medium Gaussian
K-Nearest Neighbor[137]: Fine, Medium, Cubic, Cosine, Coarse, Weighted
Ensemble: BoostedTree[138], BaggedTree[139], SubspaceDiscriminant, Subspace KNN[140], RusBoostedTree
The overall performance is evaluated using accuracy, COVID-19 accuracy, weighted precision, weighted recall, weighted specificity, and weighted F1-score. Here we have used weighted average of performance metrics instead of average as the dataset is imbalanced. This gives same weight to each class.
Analysis of Result
We present a comparative analysis in Table 7 among 5 top performing models in four categories, (a) models that employ a general cross-entropy cost function, (b) models that employ a weighted cost function, (c) models that are trained on the upsampled dataset, and (d) models that are trained on the downsampled dataset. The models are evaluated using different evaluation metrics on the test dataset.
Table 7.
Evaluation of top-5 models in terms of performance metrics
| Cost function unchanged | Weighted cost function | Up-sampling | Downsampling | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Architecture | Dense- | Dense- | Res- | Dark- | Dense- | Dense- | Dense- | Res- | Dark- | Dense- | Dense- | Dense- | Res- | Dark- | Dense- | Dense- | Dense- | Res- | Dark- | Dense- |
| net201 | net201 | net50 | net53 | net201 | net201 | net201 | net50 | net53 | net201 | net201 | net201 | net50 | net53 | net201 | net201 | net201 | net50 | net53 | net201 | |
| Classifier | QSVM | ESD | QSVM | QSVM | LSVM | QSVM | ESD | QSVM | QSVM | LSVM | QSVM | ESD | QSVM | QSVM | LSVM | QSVM | ESD | QSVM | QSVM | LSVM |
| Validation accuracy | 98.18 | 97.91 | 97.60 | 97.75 | 97.52 | 98.16 | 97.82 | 97.93 | 97.61 | 97.54 | 99.39 | 98.47 | 99.36 | 99.23 | 98.56 | 97.31 | 97.39 | 97.61 | 97.97 | 96.50 |
| Test accuracy | 98.73 | 98.45 | 98.41 | 98.19 | 98.22 | 98.76 | 98.35 | 98.41 | 98.19 | 98.22 | 98.60 | 97.79 | 98.16 | 97.53 | 97.93 | 97.14 | 96.92 | 96.68 | 96.29 | 96.44 |
| Weighted precision | 98.73 | 98.45 | 98.41 | 98.19 | 98.23 | 98.76 | 98.36 | 98.41 | 98.19 | 98.23 | 98.60 | 97.78 | 98.16 | 97.55 | 97.95 | 97.20 | 96.98 | 96.72 | 96.32 | 96.56 |
| Weighted recall | 98.89 | 98.58 | 98.64 | 98.32 | 98.31 | 98.93 | 98.47 | 98.62 | 98.32 | 98.31 | 98.76 | 97.86 | 98.35 | 97.60 | 98.00 | 97.19 | 97.02 | 96.88 | 96.57 | 96.58 |
| Weighted F1-score | 98.81 | 98.51 | 98.53 | 98.25 | 98.27 | 98.84 | 98.41 | 98.51 | 98.25 | 98.26 | 98.68 | 97.81 | 98.24 | 97.59 | 97.96 | 97.17 | 97.97 | 96.77 | 96.42 | 96.51 |
| Weighted specificity | 98.73 | 98.44 | 98.34 | 98.35 | 98.23 | 98.77 | 98.43 | 98.42 | 98.23 | 98.31 | 98.60 | 97.83 | 98.17 | 97.58 | 97.98 | 97.28 | 97.03 | 96.78 | 96.36 | 96.68 |
QSVM quadratic SVM, ESD ensemble subspace discriminant, LSVM linear SVM
The Italic entries signify the highest performing architecture for that metric
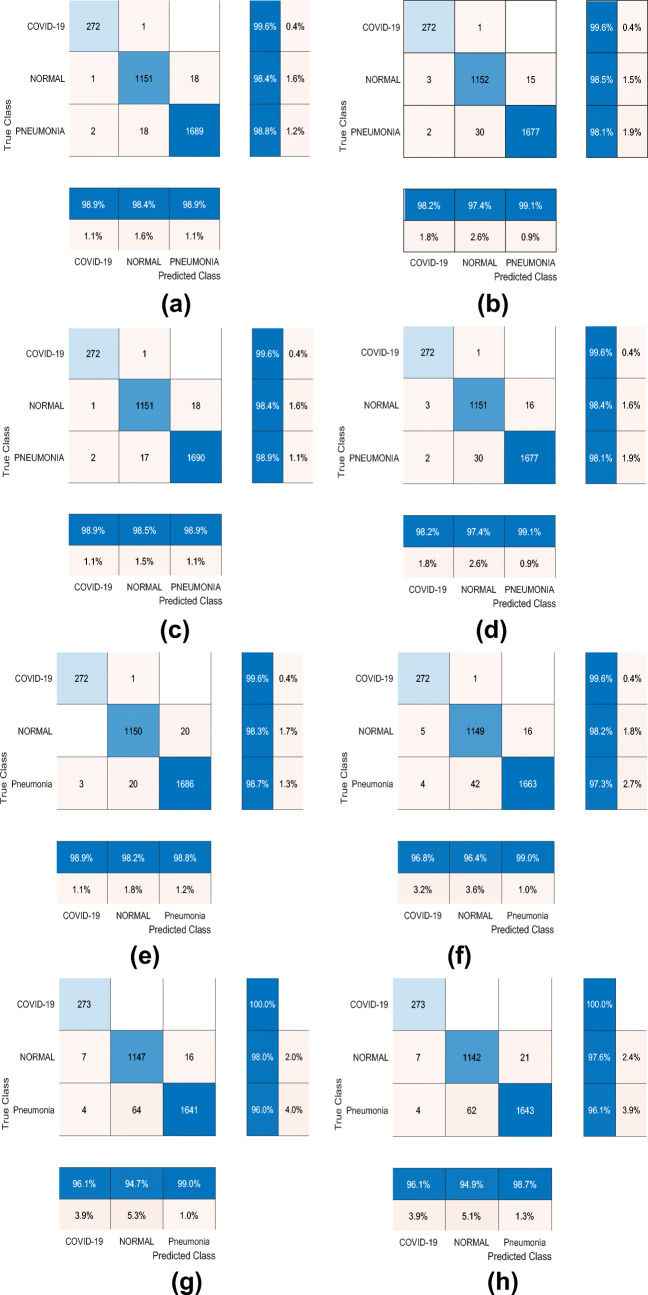
From Table 7, we can see that the Densenet201-Quadratic SVM model outperforms other model in terms of performance metrics. By adopting the weighted cost function as given in (1), we observe slight improvement in the result for the best model. However, the change varies differently across the models. Some models’ performance get worse (e.g., Alexnet-CoarseTree, Shufflenet-MediumTree, Alexnet-QuadraticSVM), some obtain better results (e.g., GoogleNet-BaggedTrensemble, Resnet50-CoarseKNN, NasNetMobile-Bagged Tree Ensemble etc.) and others do not change significantly. Note that, we consider at least 0.6% change (minimum 20 samples) in test accuracy as a significant change. We also show confusion matrix of some of the top performing models in Fig. 5.
Fig. 5.
a–h Confusion matrix of two top performing models generated using four different settings: general cost function, weighted cost function, upsampling training-set, and downsampling training-set. In the figure, GC, WC, QSVM, and ESD denote general cost function, weighted cost function, Quadratic SVM and Ensemble Subspace Discriminant respectively
Here we note few observations that are based on the performance analysis of 640 models:
As we extracted features from the last pooling layer of each CNN architecture, the number of features is in mid range (between 1000 and 5000). For this dimension of the feature vector, SVM (Linear,Quadratic,cubic) and Ensemble classifiers (Subdiscriminant, Bagged, Boosted) are found to outperform other classifiers.
Using Densenet201 as CNN architecture gives better result than other CNN architectures.
Weighted cost function performs better than the two alternative approaches, i.e., upsampling and downsampling.
With Darknet53-Quadratic SVM model, the accuracy of COVID-19 class is 100%. The model however detects three samples of pneumonia class to be COVID-19.
The validation accuracy and the test accuracy of the models are almost the same.
Misclassified COVID Samples
We present some of the COVID samples in Fig. 6 that have been misclassified by 78% of the models when trained with the general cost function and the weighted cost function. For COVID-positive cases, generally a chest X-ray shows high opacity in the peripheral part of the image. For patients with serious condition, multiple consolidations present across the image. In contrast, discontinuity of diaphragmatic shadow and subtle consolidation in chest X-ray differentiates pneumonia from other diseases. On the other hand, normal samples do not have any signs of consolidation or high opacity or any discontinuity of the diaphragmatic shadow.
Fig. 6.
Some miss-classified COVID samples. a COVID-19 miss-classified as normal; b–e COVID-19 miss-classified as pneumonia
According to an expert radiologist, the X-ray sample shown in Fig. 6a has low opacity in the left and right upper lobes and absence of consolidation even in the peripheral part of the lung, which is similar to normal lung condition. In contrast, the COVID samples shown in Fig. 6b–e are misclassified as pneumonia. These samples, according to the radiologist, are hard to diagnose even by an expert in this domain. More tests like CT scan and RT-PCR are required to confirm the status.
Feature Visualization
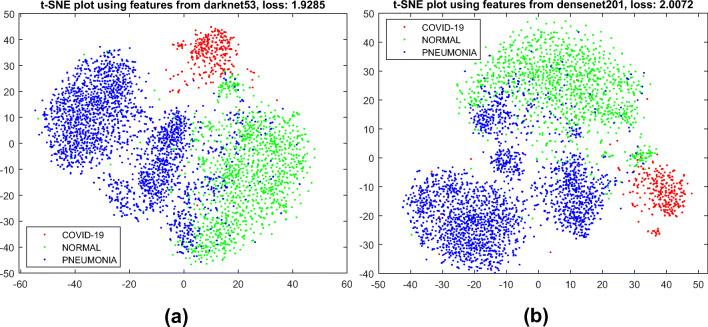
As we mentioned earlier in “Method,” the CNN models we utilize here as feature extractor are pre-trained on the ImageNet dataset. Even though none of the classes we consider here is included in Imagenet, the CNN architectures produce very good features that are distinctive across the classes.
We visualize the features we obtain via transfer learning in Fig. 7 using 2D t-SNE (t-distributed stochastic neighbor embedding) [141] plot. One can notice that the features of three different classes are quite well separated and easy for a simple machine learning model to do the classification.
Fig. 7.
2D t-SNE [141] visualization of the extracted features obtained by a DarkNet53 and b DenseNet201 from our X-ray image dataset
Performance with Lower Dimensional Features
Here we re-investigate the performance of the models using lower dimensional features. We apply t-SNE to the originally extracted higher dimensional features and fed the two-dimensional feature generated by t-SNE into the classifiers we have mentioned in “Method.” Table 8 shows the performance measures of top-5 models.
Table 8.
Performance analysis of the models using t-SNE output as feature (top-5 models)
| Architecture | Res-net50 | Res-net50 | Dense-net201 | Dark-net53 | Dense-net201 |
|---|---|---|---|---|---|
| Classifier | WKNN | EBT | WKNN | WKNN | EBT |
| Validation accuracy | 95.37 | 95.20 | 95.43 | 94.39 | 95.37 |
| Test accuracy | 96.51 | 96.26 | 95.94 | 95.88 | 95.84 |
| COVID-19 accuracy | 97.80 | 96.70 | 98.53 | 99.63 | 98.53 |
| Weighted precision | 96.52 | 96.29 | 95.97 | 95.91 | 95.89 |
| Weighted sensitivity | 96.88 | 96.69 | 96.28 | 96.37 | 96.16 |
| Weighted F1-score | 96.70 | 96.47 | 96.10 | 96.12 | 95.99 |
| Weighted specificity | 96.56 | 96.34 | 96.23 | 95.97 | 95.93 |
WKNN weighted KNN, EBT ensemble bagged tree
The Italic entries signify the highest performing architecture for that metric
From Table 8, one can see that the Resnet50-Weighted KNN model gives the best result. It is to be noticed that, with the originally obtained higher dimensional features, Denenet201-Quadratic SVM performs the best. Here are our observations:
Using only 2D features, the best accuracy we obtained is 96.5%. This model gives about 2% more error than the previously obtained one. However, this method is quicker and simpler than the previous one.
We found KNN classifiers (Weighted, Fine, Medium) and Ensemble Bagged Tree to perform better than other classifiers. We think ese classifiers’ ability to learn from lower dimensional space make these perform better than the alternatives.
Discussion and Future Directions
One of the major issues in the deep learning–based automated diagnosis of COVID-19 from the radiography images is the requirement of providing a large annotated dataset, prepared by an expert physician or radiologist to train the deep model. Although a major number of recent works made their dataset of radiography images publicly available, they contain at most a few hundreds of images in the COVID-19 class. Compiling a dataset with sufficient images in the COVID-19 class requires collecting radiography images of confirmed COVID-19 patients from reliable and authentic sources which is a challenging task. The challenge is further intensified due to the requirement of the proper annotation of the collected data. To deal with the aforementioned challenges, Few-shot, Zero-shot, and Deep Reinforcement Learning (DRL) can be adopted in this domain for ensuring minimal human intervention in the diagnosis.
Few-shot learning enables a deep model to learn information from only a handful of labeled examples per category. Recently, Few-shot learning has been applied on chest X-ray images to classify some lung diseases where it performed promisingly well [142]. However, Few-shot learning has great potential to detect COVID-19 as well, which has not been explored yet. On the other hand, Zero-shot learning has the amazing capacity of recognizing a test sample which it has not seen during training. Therefore, Zero-shot learning can also be a resort to minimizing the issue regarding the scarcity of training data of COVID-19 class. In addition, DRL can also alleviate the need for good-quality images and the hassle of proper annotations. If the problem can be modeled as a Markov’s Decision Process, DRL algorithms can perform remarkably well. As far as we know, the application of DRL in the detection of COVID-19 is not done yet. However, the success of DRL in varied fields, e.g., image processing tasks [143, 144]; image captioning [145]; video captioning [146]; automated diagnosis [147, 148], shows the potential of DRL to be effectively utilized in the diagnosis of COVID-19.
Conclusion
Due to scientists’ and researchers’ constant conscientious effort to unbolt new methods for the effectuation of COVID-19 detection with the power of deep learning, the research in this domain has made significant progress in a short time. However, due to the absence of a benchmark dataset, researchers have reported performances based on different datasets and evaluation protocols, rendering the idea of comparative analysis far-fetched. This work is inclined towards addressing this issue. Here we discussed some of the existing challenges and methods of COVID-19 detection from radiography images along with a description of the existing datasets. We have also proposed our customized dataset comprising four others. Our finding is that the primary challenge in any COVID-19 detection is the data imbalance issue due to the scarcity of COVID-19 image data. Therefore, to analyze the deep models’ performance at the presence of skewness in the data, we have done extensive experimentation on 315 deep models that comprise the combinations of 15 existing deep CNN architectures and 21 classifiers. In the experimentation, we utilized three approaches: a weighted cost function, upsampling the training dataset, and downsampling the training dataset to find the most competent solution in addressing the data imbalance issue. Our results show that deep CNNs boast high-performance metrics all-around in COVID-19 classification, with DenseNet-201 with quadratic SVM classifier achieving the best results among all, demonstrating its potential as a viable solution for early diagnosis of COVID-19. We also observed that the weighted cost function outperforms the other two alternative approaches. We believe that the comprehensive analysis provided in this work can facilitate future enthusiasts working in this domain. Nevertheless, this review may be considered an early work as many more deep learning approaches will probably be applied and tested on detecting COVID-19 to search for the best possible fit. We believe that our efforts will have a positive impact on the fight against COVID-19 during this pandemic situation.
Acknowledgments
The authors would like to thank Syeda Faiza Ahmed from the Department of Robotics and Mechatronics Engineering, University of Dhaka for her contribution to proofreading.
Compliance with Ethical Standards
Conflict of Interest
The authors declare that they have no conflict of interest.
Ethical Approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Footnotes
The dataset and models are available at https://github.com/rgbnihal2/COVID-19-X-ray-Dataset
This article belongs to the Topical Collection: Data-Driven Artificial Intelligence approaches to Combat COVID-19
Guest Editors: Mufti Mahmud, M. Shamim Kaiser, Nilanjan Dey, Newton Howard, Aziz Sheikh
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Sejuti Rahman, Email: sejuti@gmail.com.
Sujan Sarker, Email: sujan@du.ac.bd.
Md Abdullah Al Miraj, Email: miraj2465@gmail.com.
Ragib Amin Nihal, Email: rgbnihal2@gmail.com.
A. K. M. Nadimul Haque, Email: a.k.m.n.haque13@gmail.com
Abdullah Al Noman, Email: a.a.noman.rmedu@gmail.com.
References
- 1.Guan WJ, Ni ZY, Hu Y, Liang WH, Ou CQ, He JX, et al. Clinical characteristics of coronavirus disease 2019 in China. New England J Med. 2020;382(18):1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tavakoli M, Carriere J, Torabi A. 2020. Robotics, smart wearable technologies, and autonomous intelligent systems for healthcare during the COVID-19 pandemic: an analysis of the state of the art and future vision. Advanced intelligent systems;n/a(n/a):2000071. Available from: https://onlinelibrary.wiley.com/doi/abs/10.1002/aisy.202000071.
- 3.Lauer SA, Grantz KH, Bi Q, Jones FK, Zheng Q, Meredith HR, et al. The incubation period of coronavirus disease 2019 (COVID-19) from publicly reported confirmed cases: estimation and application. Annals Int Med. 2020;172(9):577–582. doi: 10.7326/M20-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tang YW, Schmitz JE, Persing DH, Stratton CW. Laboratory diagnosis of COVID-19: current issues and challenges. J Clinical Microbiol. 2020;58(6). [DOI] [PMC free article] [PubMed]
- 5.Zhang JJ, Cao YY, Dong X, Wang BC, Liao MY, Lin J, et al. Distinct characteristics of COVID-19 patients with initial rRT-PCR-positive and rRT-PCR-negative results for SARS-CoV-2. Allergy. 2020;75(7):1806—1812. doi: 10.1111/all.14316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Joynt GM, Wu WK. Understanding COVID-19: What does viral RNA load really mean? The Lancet Infectious Diseases. 2020;20(6):635–636. doi: 10.1016/S1473-3099(20)30237-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo L, Ren L, Yang S, Xiao M, Chang D, Yang F, et al. Profiling early humoral response to diagnose novel coronavirus disease (COVID-19) Clin Infect Dis. 2020;71(15):778–785. doi: 10.1093/cid/ciaa310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li M. Chest CT features and their role in COVID-19. Radiol Infect Dis. 2020;7(2):51–54. doi: 10.1016/j.jrid.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li L, Qin L, Xu Z, Yin Y, Wang X, Kong B, et al. Artificial intelligence distinguishes COVID-19 from community acquired pneumonia on chest CT. Radiology. 2020;296(2):200905. doi: 10.1148/radiol.2020200905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ardakani AA, Kanafi AR, Acharya UR, Khadem N, Mohammadi A. Application of deep learning technique to manage COVID-19 in routine clinical practice using CT images: results of 10 convolutional neural networks. Comput Biol Med. 2020;121:103795. doi: 10.1016/j.compbiomed.2020.103795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang L, Han R, Ai T, Yu P, Kang H, Tao Q, et al. Serial quantitative chest ct assessment of covid-19: deep-learning approach. Radiology: Cardiothoracic Imaging. 2020;2(2):e200075. doi: 10.1148/ryct.2020200075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ulhaq A, Khan A, Gomes D, Pau M. 2020. Computer vision for COVID-19 control: a survey. arXiv:200409420. [DOI] [PMC free article] [PubMed]
- 13.Shi F, Wang J, Shi J, Wu Z, Wang Q, Tang Z, et al. 2020. Review of artificial intelligence techniques in imaging data acquisition, segmentation and diagnosis for COVID-19. IEEE Rev Biomed Eng. 10.1109/RBME.2020.2987975. [DOI] [PubMed]
- 14.Pham QV, Nguyen DC, Hwang WJ, Pathirana PN, et al. Artificial intelligence (AI) and big data for coronavirus (COVID-19) pandemic: a survey on the state-of-the-arts. Preprints (preprint). 2020; Available from: 10.20944/preprints202004.0383.v1. [DOI] [PMC free article] [PubMed]
- 15.Mohamadou Y, Halidou A, Kapen PT. A review of mathematical modeling, artificial intelligence and datasets used in the study, prediction and management of COVID-19. Appl Intell. 2020;50(11):1–13. doi: 10.1007/s10489-020-01770-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Latif S, Usman M, Manzoor S, Iqbal W, Qadir J, Tyson G, et al. 2020. Leveraging data science to combat COVID-19: a comprehensive review. TechRxiv preprint. [DOI] [PMC free article] [PubMed]
- 17.Kalkreuth R, Kaufmann P. 2020. COVID-19: A survey on public medical imaging data resources. arXiv:200404569.
- 18.Nguyen TT. Artificial intelligence in the battle against coronavirus (COVID-19): a survey and future research directions. Preprint. 2020;10. Available from: 10.13140/RG.2.2.36491.23846.
- 19.Deng J, Dong W, Socher R, Li LJ, Li K, Fei-Fei L. Imagenet: a large-scale hierarchical image database. 2009 IEEE conference on computer vision and pattern recognition. IEEE; 2009. p. 248–255.
- 20.Kooraki S, Hosseiny M, Myers L, Gholamrezanezhad A. Coronavirus (COVID-19) outbreak: what the department of radiology should know. J Am Coll Radiol. 2020;17(4):447–451. doi: 10.1016/j.jacr.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kanne JP, Little BP, Chung JH, Elicker BM, Ketai LH. Essentials for radiologists on COVID-19: an update—radiology scientific expert panel. Radiological Society of North America. 2020;296(2):E113–E114. doi: 10.1148/radiol.2020200527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu B, Xing Y, Peng J, Zheng Z, Tang W, Sun Y, et al. Chest CT for detecting COVID-19: a systematic review and meta-analysis of diagnostic accuracy. Eur Radiol. 2020;30(10):1. doi: 10.1007/s00330-020-06934-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu J, Wu X, Zeng W, Guo D, Fang Z, Chen L, et al. Chest CT findings in patients with coronavirus disease 2019 and its relationship with clinical features. Investigative Radiology. 2020;55(5):257. doi: 10.1097/RLI.0000000000000670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fang Y, Zhang H, Xie J, Lin M, Ying L, Pang P, et al. Sensitivity of chest CT for COVID-19: comparison to RT-PCR. Radiology. 2020;296(2):200432. doi: 10.1148/radiol.2020200432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jacobi A, Chung M, Bernheim A, Eber C. Portable chest x-ray in coronavirus disease-19 (COVID-19): a pictorial review. Clin Imag. 2020;64:35–42. doi: 10.1016/j.clinimag.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cohen JP. COVID-19 image data collection. Github; 2020. online; last accessed July-13,2020. https://github.com/ieee8023/covid-chestxray-dataset.
- 27.Weinstock MB, Echenique A, DABR JWR, Leib A, ILLUZZI FA. Chest x-ray findings in 636 ambulatory patients with COVID-19 presenting to an urgent care center: a normal chest x-ray is no guarantee. J Urgent Care Med. 2020;14(7):13–8. [Google Scholar]
- 28.Guo Y, Liu Y, Oerlemans A, Lao S, Wu S, Lew MS. Deep learning for visual understanding: a review. Neurocomputing. 2016;187:27–48. doi: 10.1016/j.neucom.2015.09.116. [DOI] [Google Scholar]
- 29.Perez L, Wang J. 2017. The effectiveness of data augmentation in image classification using deep learning. arXiv:171204621.
- 30.Abbas A, Abdelsamea MM, Gaber MM. 2020. Classification of COVID-19 in chest x-ray images using DeTraC deep convolutional neural network. arXiv:200313815. [DOI] [PMC free article] [PubMed]
- 31.Chowdhury ME, Rahman T, Khandakar A, Mazhar R, Kadir MA, Mahbub ZB, et al. 2020. Can AI help in screening viral and COVID-19 pneumonia? arXiv:200313145.
- 32.Bassi PR, Attux R. 2020. A deep convolutional neural network for COVID-19 detection using chest x-rays. arXiv:200501578.
- 33.Hall LO, Paul R, Goldgof DB, Goldgof GM. 2020. Finding covid-19 from chest x-rays using deep learning on a small dataset. arXiv:200402060.
- 34.Farooq M, Hafeez A. 2020. Covid-resnet: a deep learning framework for screening of covid19 from radiographs. arXiv:200314395.
- 35.Karim M, Döhmen T, Rebholz-Schuhmann D, Decker S, Cochez M, Beyan O, et al. 2020. Deepcovidexplainer: explainable COVID-19 predictions based on chest x-ray images. arXiv:200404582.
- 36.Minaee S, Kafieh R, Sonka M, Yazdani S, Soufi GJ. 2020. Deep-covid: predicting covid-19 from chest x-ray images using deep transfer learning. arXiv:200409363. [DOI] [PMC free article] [PubMed]
- 37.Misra S, Jeon S, Lee S, Managuli R, Kim C. 2020. Multi-channel transfer learning of chest x-ray images for screening of COVID-19. arXiv:200505576.
- 38.Ucar F, Korkmaz D. COVIDiagnosis-Net: Deep Bayes-SqueezeNet based diagnostic of the coronavirus disease 2019 (COVID-19) from x-ray images. Med Hypotheses. 2020;140:109761. doi: 10.1016/j.mehy.2020.109761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang L, Wong A. COVID-Net: a tailored deep convolutional neural network design for detection of COVID-19 cases from chest radiography images. arXiv; 2020. Available from: https://github.com/lindawangg/COVID-Net. [DOI] [PMC free article] [PubMed]
- 40.Yamac M, Ahishali M, Degerli A, Kiranyaz S, Chowdhury ME, Gabbouj M. 2020. Convolutional sparse support estimator based Covid-19 recognition from x-ray images. arXiv:200504014. [DOI] [PMC free article] [PubMed]
- 41.Wang Y, Yao Q, Kwok JT, Ni LM. Generalizing from a few examples: a survey on few-shot learning. ACM Computing Surveys (CSUR) 2020;53(3):1–34. doi: 10.1145/3386252. [DOI] [Google Scholar]
- 42.Wang W, Zheng VW, Yu H, Miao CA. survey of zero-shot learning: settings, methods, and applications. ACM Transactions on Intelligent Systems and Technology (TIST) 2019;10(2):1–37. [Google Scholar]
- 43.Van Hulse J, Khoshgoftaar TM, Napolitano A. Experimental perspectives on learning from imbalanced data. Proceedings of the 24th international conference on Machine learning; 2007. p. 935–942.
- 44.Chawla NV, Japkowicz N, Kotcz A. Special issue on learning from imbalanced data sets. ACM SIGKDD Explorations Newsletter. 2004;6(1):1–6. doi: 10.1145/1007730.1007733. [DOI] [Google Scholar]
- 45.Chawla NV, Bowyer KW, Hall LO, Kegelmeyer WP. SMOTE: synthetic minority over-sampling technique. J Artif Intell Res. 2002;16:321–357. doi: 10.1613/jair.953. [DOI] [Google Scholar]
- 46.Han H, Wang WY, Mao BH. Borderline-SMOTE: a new over-sampling method in imbalanced data sets learning. International conference on intelligent computing. Springer; 2005. p. 878–887.
- 47.Bunkhumpornpat C, Sinapiromsaran K, Lursinsap C. Safe-level-smote: safe-level-synthetic minority over-sampling technique for handling the class imbalanced problem. Pacific-Asia conference on knowledge discovery and data mining. Springer; 2009. p. 475–482.
- 48.Lv D, Qi W, Li Y, Sun L, Wang Y. 2020. A cascade network for detecting COVID-19 using chest x-rays. arXiv:200501468.
- 49.Punn NS, Agarwal S. 2020. Automated diagnosis of COVID-19 with limited posteroanterior chest X-ray images using fine-tuned deep neural networks. arXiv:200411676. [DOI] [PMC free article] [PubMed]
- 50.Barstugan M, Ozkaya U, Ozturk S. 2020. Coronavirus (covid-19) classification using ct images by machine learning methods. arXiv:200309424.
- 51.Zheng C, Deng X, Fu Q, Zhou Q, Feng J, Ma H, et al. 2020. Deep learning-based detection for COVID-19 from chest CT using weak label. medRxiv. 10.1101/2020.03.12.20027185.
- 52.Zhu X, Song B, Shi F, Chen Y, Hu R, Gan J, et al. 2020. Joint prediction and time estimation of COVID-19 developing severe symptoms using chest CT scan. arXiv:200503405. [DOI] [PMC free article] [PubMed]
- 53.Javaheri T, Homayounfar M, Amoozgar Z, Reiazi R, Homayounieh F, Abbas E, et al. 2020. CovidCTNet: an open-source deep learning approach to identify covid-19 using CT image. arXiv:200503059. [DOI] [PMC free article] [PubMed]
- 54.Kang H, Xia L, Yan F, Wan Z, Shi F, Yuan H, et al. Diagnosis of coronavirus disease 2019 (covid-19) with structured latent multi-view representation learning. IEEE Trans Med Imaging. 2020;39(8):2606–2614. doi: 10.1109/TMI.2020.2992546. [DOI] [PubMed] [Google Scholar]
- 55.Apostolopoulos ID, Aznaouridis SI, Tzani MA. Extracting possibly representative COVID-19 biomarkers from X-ray images with deep learning approach and image data related to pulmonary diseases. J Med Biol Eng. 2020;40(3):1. doi: 10.1007/s40846-020-00529-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Butt C, Gill J, Chun D, Babu BA. Deep learning system to screen coronavirus disease 2019 pneumonia. Appl Intell. 2020:1. 10.1007/s10489-020-01714-3. [DOI] [PMC free article] [PubMed]
- 57.Long SS, Prober CG, Fischer M, (eds). 2018. Index, 5th ed.. Elsevier. Available from: http://www.sciencedirect.com/science/article/pii/B9780323401814003030.
- 58.Zhang J, Xie Y, Li Y, Shen C, Xia Y. 2020. Covid-19 screening on chest x-ray images using deep learning based anomaly detection. arXiv:200312338.
- 59.Chung A. Actualmed COVID-19 chest x-ray dataset. Github; 2020. online; last accessed July-13,2020. https://github.com/agchung/Actualmed-COVID-chestxray-dataset.
- 60.Chung A. Figure-1 COVID chest x-ray datset. Github; 2020. online; last accessed July-13,2020. Available from: https://github.com/agchung/Figure1-COVID-chestxray-dataset.
- 61.Rahman T. COVID-19 radiography dataset. Kaggle; 2020. Accessed 2 June 2020. Available from: https://www.kaggle.com/tawsifurrahman/covid19-radiography-database.
- 62.SIRM. COVID-19 database. Society of Medical and Interventional Radiology; 2020. Accessed 3 June 2020. Available from: https://www.sirm.org/category/senza-categoria/covid-19/.
- 63.Mooney P. Chest x-ray images (pneumonia). Kaggle; 2018. Accessed 2 June 2020. Available from: https://www.kaggle.com/paultimothymooney/chest-xray-pneumonia.
- 64.Wang L, Wong A. 2020. COVID-Net: a tailored deep convolutional neural network design for detection of COVID-19 cases from chest X-ray images. arXiv:200309871. [DOI] [PMC free article] [PubMed]
- 65.RSNA. RSNA pneumonia detection challenge. Radiological Society of North America; 2018. Accessed 7 June 2020. Available from: https://www.kaggle.com/c/rsna-pneumonia-detection-challenge/.
- 66.Ali Mohammad Alqudah SQ. Augmented COVID-19 x-ray images dataset. Mendeley Data; 2020. Accessed 3 June 2020. Available from: 10.17632/2fxz4px6d8.4.
- 67.Radiopaedia. Radiopaedia; 2020. Accessed 7 June 2020. Available from: https://radiopaedia.org/.
- 68.Zhao J, Zhang Y, He X, Xie P. COVID-CT-Dataset: a CT scan dataset about COVID-19. arXiv:200313865. 2020;Available from: https://github.com/UCSD-AI4H/COVID-CT.
- 69.Walid El-Shafai FAES. Extensive COVID-19 X-ray and CT chest images dataset. Mendeley Data; 2020. Accessed 8 September, 2020. Available from: 10.17632/8h65ywd2jr.3.
- 70.COVID-19 CT segmentation dataset; 2020. Accessed 8 September 2020. Available from: http://medicalsegmentation.com/covid19/.
- 71.COVID-19 X-ray images. Kaggle;. Accessed 8 September 2020. Available from: https://www.kaggle.com/bachrr/covid-chest-xray.
- 72.Threadreader. Chest xray. Threadreader; 2020. Accessed 7 June, 2020. Available from: https://threadreaderapp.com/thread/1243928581983670272.html.
- 73.BSTI. COVID-19 british society of thoracic imaging database — the BRITISH SOCIETY OF THORACIC IMAGING. BSTI; 2020. Accessed 28 june 2020. Available from: https://www.bsti.org.uk/training-and-education/covid-19-bsti-imaging-database/.
- 74.Coronavirus Cases; Accessed 8 September 2020. Available from: https://coronacases.org/.
- 75.Europian Society of Radiology. Eurorad; Accessed 8 September 2020. Available from: https://eurorad.org/.
- 76.Narin A, Kaya C, Pamuk Z. 2020. Automatic detection of coronavirus disease (covid-19) using x-ray images and deep convolutional neural networks. arXiv:200310849. [DOI] [PMC free article] [PubMed]
- 77.Zhang Y, Niu S, Qiu Z, Wei Y, Zhao P, Yao J, et al. 2020. COVID-DA: Deep domain adaptation from typical pneumonia to COVID-19. arXiv:200501577.
- 78.Alqudah A, Qazan S, Alquran H, Qasmieh I, Alqudah A. COVID-19 detection from x-ray images using different artificial intelligence hybrid models. Jordan J Electr Eng. 2020;6:168. doi: 10.5455/jjee.204-1585312246. [DOI] [Google Scholar]
- 79.Apostolopoulos ID, Mpesiana TA. Covid-19: automatic detection from x-ray images utilizing transfer learning with convolutional neural networks. Phys Eng Sci Med. 2020;43(2):1. doi: 10.1007/s13246-020-00865-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.He X, Yang X, Zhang S, Zhao J, Zhang Y, Xing E, et al. 2020. Sample-efficient deep learning for COVID-19 diagnosis based on CT scans. medRxiv. 10.1101/2020.04.13.20063941.
- 81.Chen X, Yao L, Zhang Y. 2020. Residual attention U-Net for automated multi-class segmentation of COVID-19 chest CT images. arXiv:200405645.
- 82.Maghdid HS, Asaad AT, Ghafoor KZ, Sadiq AS, Khan MK. 2020. Diagnosing COVID-19 pneumonia from x-ray and CT images using deep learning and transfer learning algorithms. arXiv:2004.00038.
- 83.Majeed T, Rashid R, Ali D, Asaad A. 2020. Covid-19 detection using CNN transfer learning from X-ray Images. medRxiv. 10.1101/2020.05.12.20098954.
- 84.Sethy PK, Behera SK. Detection of coronavirus disease (covid-19) based on deep features. Preprints. 2020;2020:2020030300. [Google Scholar]
- 85.Rajpurkar P, Irvin J, Zhu K, Yang B, Mehta H, Duan T, et al. 2017. Chexnet: radiologist-level pneumonia detection on chest x-rays with deep learning. arXiv:171105225.
- 86.Deng J, Dong W, Socher R, Li LJ, Li K, Fei-Fei L. ImageNet: a large-scale hierarchical image database. 2009 IEEE conference on computer vision and pattern recognition; 2009. p. 248–255.
- 87.Wang X, Peng Y, Lu L, Lu Z, Bagheri M, Summers RM. Chestx-ray8: hospital-scale chest x-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases. Proceedings of the IEEE conference on computer vision and pattern recognition; 2017. p. 2097–2106.
- 88.Goodwin BD, Jaskolski C, Zhong C, Asmani H. 2020. Intra-model variability in COVID-19 classification using chest x-ray images. arXiv:200502167.
- 89.Littmann E, Ritter H. Generalization abilities of cascade network architecture. Advances in neural information processing systems; 1993. p. 188–195.
- 90.Hu J, Shen L, Sun G. Squeeze-and-excitation networks. Proceedings of the IEEE conference on computer vision and pattern recognition; 2018. p. 7132–7141.
- 91.Li B, Wu F, Lim SN, Belongie S, Weinberger KQ. 2020. On feature normalization and data augmentation. arXiv:200211102.
- 92.Ozturk T, Talo M, Yildirim EA, Baloglu UB, Yildirim O, Acharya UR. Automated detection of COVID-19 cases using deep neural networks with x-ray images. Comput Biol Med. 2020;03792. [DOI] [PMC free article] [PubMed]
- 93.Ouyang X, Huo J, Xia L, Shan F, Liu J, Mo Z, et al. 2020. Dual-sampling attention network for diagnosis of COVID-19 from community acquired pneumonia. IEEE Transactions on Medical Imaging. [DOI] [PubMed]
- 94.Song Y, Zheng S, Li L, Zhang X, Zhang X, Huang Z, et al. 2020. Deep learning enables accurate diagnosis of novel coronavirus (COVID-19) with CT images. medRxiv. 10.1101/2020.02.23.20026930. [DOI] [PMC free article] [PubMed]
- 95.Gozes O, Frid-Adar M, Greenspan H, Browning PD, Zhang H, Ji W, et al. 2020. Rapid AI development cycle for the coronavirus (covid-19) pandemic: initial results for automated detection & patient monitoring using deep learning CT image analysis. arXiv:200305037.
- 96.Shi F, Xia L, Shan F, Wu D, Wei Y, Yuan H, et al. 2020. Large-scale screening of covid-19 from community acquired pneumonia using infection size-aware classification. arXiv:200309860. [DOI] [PubMed]
- 97.Shan F, Gao Y, Wang J, Shi W, Shi N, Han M, et al. 2020. Lung infection quantification of covid-19 in CT images with deep learning. arXiv:200304655.
- 98.Sun L, Mo Z, Yan F, Xia L, Shan F, Ding Z, et al. 2020. Adaptive feature selection guided deep forest for COVID-19 classification with chest CT. arXiv:200503264. [DOI] [PMC free article] [PubMed]
- 99.Milletari F, Navab N, Ahmadi SA. V-net: fully convolutional neural networks for volumetric medical image segmentation. 2016 fourth international conference on 3D vision (3DV). IEEE; 2016. p. 565–571.
- 100.Di D, Shi F, Yan F, Xia L, Mo Z, Ding Z, et al. 2020. Hypergraph learning for identification of COVID-19 with CT imaging. arXiv:200504043. [DOI] [PMC free article] [PubMed]
- 101.Liu M, Zhang J, Yap PT, Shen D. View-aligned hypergraph learning for Alzheimer’s disease diagnosis with incomplete multi-modality data. Med Image Anal. 2017;36:123–134. doi: 10.1016/j.media.2016.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Azad R, Asadi-Aghbolaghi M, Fathy M, Escalera S. Bi-directional convLSTM U-net with densley connected convolutions. Proceedings of the IEEE International conference on computer vision workshops; 2019. p. 0–0.
- 103.Ronneberger O, Fischer P, Brox T. U-net: Convolutional networks for biomedical image segmentation. International conference on medical image computing and computer-assisted intervention. Springer; 2015. p. 234–241.
- 104.Kitaev N, Kaiser Ł, Levskaya A. 2020. Reformer: the efficient transformer. arXiv:200104451.
- 105.Zhou B, Khosla A, Lapedriza A, Oliva A, Torralba A. Learning deep features for discriminative localization. Proceedings of the IEEE conference on computer vision and pattern recognition; 2016. p. 2921–2929.
- 106.Lin TY, Dollár P, Girshick R, He K, Hariharan B, Belongie S. Feature pyramid networks for object detection. Proceedings of the IEEE conference on computer vision and pattern recognition; 2017. p. 2117–2125.
- 107.Fu J, Zheng H, Mei T. Look closer to see better: recurrent attention convolutional neural network for fine-grained image recognition. Proceedings of the IEEE conference on computer vision and pattern recognition; 2017. p. 4438–4446.
- 108.Kermany D, Zhang K, Goldbaum M. Labeled optical coherence tomography (OCT) and chest x-ray images for classification. Mendeley data. 2018:2. 10.17632/rscbjbr9sj.2.
- 109.Hemdan EED, Shouman MA, Karar ME. 2020. Covidx-net: A framework of deep learning classifiers to diagnose covid-19 in x-ray images. arXiv:200311055.
- 110.Wang L. COVIDx dataset. Github; 2020. online; last accessed July-13,2020. https://github.com/lindawangg/COVID-Net/blob/master/docs/COVIDx.md.
- 111.Irvin J, Rajpurkar P, Ko M, Yu Y, Ciurea-Ilcus S, Chute C, et al. Chexpert: a large chest radiograph dataset with uncertainty labels and expert comparison. Proceedings of the AAAI conference on artificial intelligence; 2019. p. 590–597.
- 112.Summers R. Chest xray 8 database. National In; 2017. EaltAccehssed 16 stituteJune,20 of H20. Available from: https://nihcc.app.box.com/v/ChestXray-NIHCC.
- 113.Khan AI, Shah JL, Bhat MM. Coronet: a deep neural network for detection and diagnosis of COVID-19 from chest x-ray images. Comput Meth Prog Bio. 2020;196:105581. doi: 10.1016/j.cmpb.2020.105581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Pereira RM, Bertolini D, Teixeira LO, Silla Jr CN, Costa YM. COVID-19 identification in chest X-ray images on flat and hierarchical classification scenarios. Comput Meth Prog Bio. 2020;194:105532. doi: 10.1016/j.cmpb.2020.105532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lin TY, Goyal P, Girshick R, He K, Dollár P. Focal loss for dense object detection. Proceedings of the IEEE international conference on computer vision; 2017. p. 2980–2988.
- 116.King G, Zeng L. Logistic regression in rare events data. Political Analysis. 2001;9(2):137–163. doi: 10.1093/oxfordjournals.pan.a004868. [DOI] [Google Scholar]
- 117.Kamalaveni V, Rajalakshmi RA, Narayanankutty K. Image denoising using variations of Perona-Malik model with different edge stopping functions. Procedia Comput Sci. 2015;58:673–682. doi: 10.1016/j.procs.2015.08.087. [DOI] [Google Scholar]
- 118.Szegedy C, Liu W, Jia Y, Sermanet P, Reed S, Anguelov D, et al. Going deeper with convolutions. Proceedings of the IEEE conference on computer vision and pattern recognition; 2015 . p. 1–9.
- 119.Knuth DE. The art of computer programming, Vol. 3. London: Pearson Education; 1997. [Google Scholar]
- 120.Marsaglia G. Random number generators. J Modern Appl Stat Methods. 2003;2(1):2. doi: 10.22237/jmasm/1051747320. [DOI] [Google Scholar]
- 121.Krizhevsky A, Sutskever I, Hinton G. ImageNet classification with deep convolutional neural networks. Neural Info Process Sys. 2012;01:25. [Google Scholar]
- 122.Chollet F. Xception: deep learning with depthwise separable convolutions. 2016;arXiv:1610.02357.
- 123.Szegedy C, Vanhoucke V, Ioffe S, Shlens J, Wojna Z. Rethinking the inception architecture for computer vision. 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2016.
- 124.Szegedy C, Ioffe S, Vanhoucke V. Inception-v4, Inception-ResNet and the impact of residual connections on learning. 2016; arXiv:1602.07261.
- 125.Simonyan K, Zisserman A. 2014. Very deep convolutional networks for large-scale image recognition. arXiv:14091556.
- 126.He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2016.
- 127.Sandler M, Howard AG, Zhu M, Zhmoginov A, Chen L. 2018. Inverted residuals and linear bottlenecks: mobile networks for classification, detection and segmentation. arXiv:1801.04381.
- 128.Redmon J, Farhadi A. 2018. Yolov3: an incremental improvement. arXiv:180402767.
- 129.Redmon J, Farhadi A. YOLO9000: better, faster, stronger. Proceedings of the IEEE conference on computer vision and pattern recognition; 2017. p. 7263–7271.
- 130.Szegedy C, Wei L, Yangqing J, Sermanet P, Reed S, Anguelov D, et al. Going deeper with convolutions. 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2015. p. 1–9.
- 131.Huang G, Liu Z, van der Maaten L, Weinberger KQ. Densely connected convolutional networks. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2017.
- 132.Zhang X, Zhou X, Lin M, Sun J. ShuffleNet: an extremely efficient convolutional neural network for mobile devices. 2018 IEEE/CVF Conference on computer vision and pattern recognition; 2018. p. 6848–6856.
- 133.Zoph B, Vasudevan V, Shlens J, Le QV. 2017. Learning transferable architectures for scalable image recognition. Available from: arXiv:1707.07012.
- 134.Safavian SR, Landgrebe D. A survey of decision tree classifier methodology. IEEE Trans Sys Man Cybern. 1991;21(3):660–674. doi: 10.1109/21.97458. [DOI] [Google Scholar]
- 135.McLachlan GJ. Discriminant analysis and statistical pattern recognition, Vol. 544. Hoboken: Wiley; 2004. [Google Scholar]
- 136.Kecman V. Support vector machines–an introduction. Support vector machines: theory and applications. Springer; 2005. p. 1–47.
- 137.Kramer O. K-nearest neighbors. Dimensionality reduction with unsupervised nearest neighbors. Springer; 2013. p. 13–23.
- 138.Drucker H, Cortes C. Boosting decision trees. 1995;8:479–485.
- 139.Breiman L. Bagging predictors. Machine Learning. 1996;24(2):123–140. [Google Scholar]
- 140.Ho TK. Nearest neighbors in random subspaces. Joint IAPR International Workshops on Statistical Techniques in Pattern Recognition (SPR) and Structural and Syntactic Pattern Recognition (SSPR). Springer; 1998. p. 640–648.
- 141.Van Der Maaten L. Accelerating t-SNE using tree-based algorithms. J Mach Learn Res. 2014;15(1):3221–3245. [Google Scholar]
- 142.Rajan D, Thiagarajan JJ, Karargyris A, Kashyap S. 2020. Self-training with improved regularization for few-shot chest x-ray classification. arXiv:200502231.
- 143.Cao Q, Lin L, Shi Y, Liang X, Li G. Attention-aware face hallucination via deep reinforcement learning. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition; 2017. p. 690–698.
- 144.Park J, Lee JY, Yoo D, So Kweon I. Distort-and-recover: Color enhancement using deep reinforcement learning. Proceedings of the IEEE Conference on computer vision and pattern recognition; 2018. p. 5928–5936.
- 145.Ren Z, Wang X, Zhang N, Lv X, Li LJ. Deep reinforcement learning-based image captioning with embedding reward. Proceedings of the IEEE conference on computer vision and pattern recognition; 2017. p. 290–298.
- 146.Li L, Gong B. End-to-end video captioning with multitask reinforcement learning. 2019 IEEE Winter Conference on Applications of Computer Vision (WACV). IEEE; 2019. p. 339–348.
- 147.Yu C, Liu J, Nemati S. 2019. Reinforcement learning in healthcare: a survey. arXiv:190808796.
- 148.Steimle LN, Denton BT. Markov decision processes for screening and treatment of chronic diseases. Markov decision processes in practice. Springer; 2017. p. 189–222.