Abstract
The ubiquitin-proteasome system (UPS) is a key player in the maintenance of cellular protein homeostasis (proteostasis). Since proteasome function declines upon aging leading to the acceleration of its progression and the manifestation of age-related pathologies, many attempts have been performed towards proteasome activation as a strategy to promote healthspan and longevity. The marine environment hosts a plethora of organisms that produce a vast array of primary and secondary metabolites, the majority of which are unique, exhibiting a wide spectrum of biological activities. The fact that these biologically important compounds are also present in edible marine organisms has sparked the interest for elucidating their potential health-related applications. In this review, we focus on the antioxidant, anti-aging, anti-aggregation and anti-photoaging properties of various marine constituents. We further discuss representatives of marine compounds classes with regard to their potential (direct or indirect) action on UPS components that could serve as UPS modulators and exert beneficial effects on conditions such as oxidative stress, aging and age-related diseases.
Keywords: Proteasome, Marine natural products, Anti-aging, Anti-aggregation, Antioxidant, Anti-photoaging
Graphical abstract
Highlights
-
•
Proteasome has been implicated in oxidative stress, aging and age-related diseases.
-
•
Various marine natural products exert strong antioxidant activities.
-
•
Various marine natural products possess anti-aging and anti-aggregation properties.
-
•
Various marine natural products can serve as potential proteasome modulators.
-
•
Various marine natural products represent promising anti-photoaging agents.
Abbreviations
- 6-OHDA
6-hydroxydopamine
- AAA ATPase
ATPase associated with different cellular activities
- AAPH
2,2′-azobis (2-amidinopropane) dihydrochloride
- AChE
acetylcholinesterase
- AD
Alzheimer's disease
- AKT-1/-2
protein kinase B
- AP-1
activator protein 1
- ARE
antioxidant response element
- BChE
butyrylcholinesterase
- BDNF
brain-derived neurotrophic factor
- bZip
basic region leucine zipper
- CAT
catalase
- ChAT
choline acetyltransferase
- C-L
caspase-like activity
- CNC
cap ‘n’ collar
- CP
core particle
- CT-L
chymotrypsin-like activity
- Cul-Rbx
Cullin-RING box
- D-Gal
d-galactose
- DHA-OOH
DHA hydroperoxide
- DPPH
1,1-diphenyl-2-picrylhydrazyl
- DUBs
deubiquitinating enzymes
- ECM
extracellular matrix
- ERK
extracellular-signal-regulated kinase
- FDA
Food and Drug Administration
- FOXO
forkhead box O
- Glb1
β-galactosidase 1
- GPx
glutathione peroxidase
- GSH
glutathione
- GSK-1
serum- and glucocorticoid-inducible kinase 1
- GSSG
glutathione disulfide
- GST
glutathione S-transferase
- HD
Huntington's disease
- HO-1
heme oxygenase-1
- HSF-1
heat shock factor 1
- IDRs
intrinsically disordered regions
- IGF-1
insulin-like growth factor-1
- IIS pathway
insulin/insulin-like growth factor-1 pathway
- INF-γ
interferon-γ
- INrf2
inhibitor of Nrf2
- Keap1
Kelch-like ECH-associated protein 1
- MAA
mycosporine-like amino acid
- MAPK
mitogen-activated protein kinase
- MDA
malondialdehyde
- MMP
matrix metalloproteinase
- MPP+
1-methyl-4-phenylpyridinum
- MPTP
1-methyl-4-phenyl-1, 2, 3, 6-tetrahydropyridine
- NF-kB
nuclear factor-kB
- NQO-1
NADPH quinone oxidoreductase-1
- Nrf2
nuclear factor erythroid 2-related factor 2
- PD
Parkinson's disease
- PDK-1
3-phosphoinositide-dependent protein kinase-1
- PGPH
peptidylglutamyl-peptide hydrolyzing activity
- PI3K
phosphatidylinositol-3-OH kinase
- PIP2
phosphatidylinositol 4,5-bisphosphate
- PIP3
phosphatidylinositol 3,4,5-trisphosphate
- PKC
protein kinase C; polyQ, polyglutamine
- RNS
reactive nitrogen species
- ROS
reactive oxygen species
- RP
regulatory particle
- Rpn
RP non-ATPase
- Rpt
Regulatory particle triple-A protein
- RPTP
receptor protein tyrosine phosphatase
- RTK
receptor tyrosine kinase
- SA-β-Gal
senescence-associated β-galactosidase
- SKN-1
skinhead 1; SOD, superoxide dismutase
- tBHP
tert-butyl hydroperoxide
- TGFRI
transforming growth factor β receptor I
- T-L
trypsin-like activity
- UPS
ubiquitin-proteasome system
- wt
wild type
- γ-GCS
γ-glutamylcysteine synthetase
1. The ubiquitin-proteasome system
The ubiquitin-proteasome system (UPS) is a highly conserved pathway among eukaryotic organisms [1]. Its role is to surveil protein homeostasis and to maintain the balance between continuous synthesis of new proteins and elimination of abnormal, damaged, short-lived or regulatory proteins by determining the time and the rate that a protein will be degraded. The proteasome is the core of this system. It is a multicatalytic enzyme complex located in the cytosol and nucleus [2,3]. Its functions are not limited to the maintenance of proteostasis; the proteasome is involved in a wide array of biological processes, including cell differentiation and proliferation, DNA repair and apoptosis, regulation of gene expression and response to stress [4,5], among others, highlighting its pivotal role for cell survival. The proteasome consists of a concave barrel-shaped proteolytic core particle (CP or 20S proteasome) and the regulatory particle (RP) known as the 19S RP. When one 19S RP caps the 20S core, it gives rise to the 26S proteasome conformation, while the attachment of the 19S RP in both sides of the CP leads to the formation of the 30S proteasome conformation [1,3]. Since both 30S and 26S conformations are frequently mentioned as 26S proteasome in the literature [6], we will refer to both conformations as 26S proteasome hereafter.
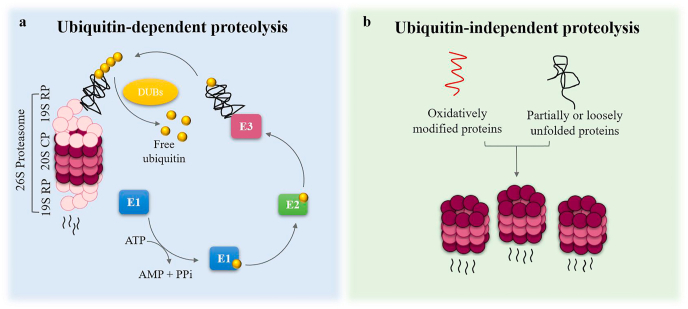
Ubiquitin is a crucial molecule for the UPS-mediated degradation of proteins. It is a 76 amino acid globular protein required for the ATP-dependent proteolysis [7]. The role of ubiquitin is to modify/tag the protein substrate that is destined for degradation with a process called ubiquitination. Three enzymes catalyze ubiquitination; E1 ubiquitin-activating enzyme, E2 ubiquitin-conjugating enzyme and E3 ubiquitin ligase enzyme [2,8]. E1 activates ubiquitin in an ATP-dependent manner. Activated ubiquitin is transferred to an E2 enzyme. The final step of ubiquitination is mediated by an E3 ubiquitin ligase, which is responsible for the ubiquitin transfer from the E2 enzyme to the substrate that will be degraded. This process is repeated to form a poly-ubiquitin chain that facilitates substrate recognition by the proteasome. Four ubiquitin moieties are the standard form of ubiquitination (tetra-ubiquitination); however, mono-ubiquitination and multiple mono-ubiquitinations have also been reported [9]. E3 ligases are essential regulators of the ubiquitination cascade as they control the efficiency and the specificity of this reaction. Besides, the several hundreds of E3 ligases that are present in human cells, in contrast with few dozens of E2 and the only two E1 enzymes, reflect their crucial role [2,10]. As the protein substrate attaches to the 19S RP, deubiquitinating enzymes (DUBs; Rpn11, USP14 and UCH37) remove the ubiquitin chain moieties to promote the substrate translocation into the core and to prevent the ubiquitin molecules from being degraded along with the substrate that they are bound to [2,11,12] (Fig. 1a). All proteins mentioned above are considered as UPS constituents.
Fig. 1.
The ubiquitin-dependent and ubiquitin-independent proteolysis. (a) The ubiquitin-dependent proteolysis is a three-step process that requires the presence of ubiquitin molecules and ATP consumption. E1 ubiquitin-activating enzyme, E2 ubiquitin-conjugating enzyme and E3 ubiquitin ligase enzyme are responsible for tagging the protein substrate with ubiquitin molecules. This process is repeated to generate a poly-ubiquitin chain, the standard form of which is tetra-ubiquitin. The ubiquitinated protein is recognized, unfolded and degraded by the 26S proteasome after the deubiquitinating enzymes (DUBs) remove the ubiquitin molecules from the protein substrate. The released free ubiquitin molecules will be used in another ubiquitination cycle. (b) Oxidatively modified proteins and partially or loosely unfolded proteins are degraded mainly by the 20S proteasome in a ubiquitin/ATP-independent manner.
Apart from the ubiquitin-dependent protein degradation, ubiquitin-independent proteolysis also occurs (Fig. 1b) [5]. This type of protein degradation is mainly conducted by the 20S proteasome. Studies have shown that the 20S proteasome plays a leading role in maintaining proteostasis under stress conditions, especially oxidative stress, where the cell has to cope with multiple damaged proteins [[13], [14], [15], [16]]. The narrow entrance of the 20S CP, as well as the lack of the 19S subunits (that facilitate substrate unfolding), have led to the assumption that partially or loosely unfolded proteins are targets of ubiquitin-independent proteolysis. More specifically, these are proteins that either have intrinsically disordered regions (IDRs) or proteins that have lost their tertiary structure [16,17].
1.1. 20S proteasome
Four heptameric rings are stacked to form the 700 kDa barrel-shaped complex of the 20S CP. This complex consists of two β-rings that are located in the center of the CP and two α-rings covering each side of the barrel. There are seven different, but structurally similar, α- and β-subunits that form the configuration α1-7β1-7β1-7α1-7. The inner β-subunits are forming a chamber in which the protein degradation takes place. Three β-subunits in each ring (β1, β2 and β5) have proteolytic activity with a different cleavage preference, which is attributed to the threonine residue of their catalytic centers [2,5,8]. The β1 subunit possesses a caspase-like activity (C-L) [also known as peptidylglutamyl-peptide hydrolyzing (PGPH) activity], the β2 subunit has a trypsin-like activity (T-L) and the β5 subunit confers a chymotrypsin-like activity (CT-L) [5,18]. The C-L, T-L and CT-L proteolytic activities lead to the cleavage of the peptide backbone after acidic, basic or hydrophobic residues, respectively [2]. The amino (N)-terminal tails of α-subunits form the gate of the proteasome with a diameter of approximately 13 Å [2,5,6,18]. This narrow aperture serves as the proteasome gate allowing the protein substrate to enter the proteolytic chamber. The N-terminal residues of α3 subunit have an important role as they control substrate entrance and gate opening [19,20].
1.2. 19S regulatory particle
Because of the small pore diameter of the 20S CP, folded proteins are not able to access the proteolytic chamber of the proteasome and therefore escape from degradation. Thus, the protein substrate must be unfolded [2,8]. In this step, 19S RP has a crucial role as it is responsible for the recognition, binding, deubiquitination, unfolding and subsequent translocation of the protein substrate to the 20S core after opening its α-gate [2,5,19]. RP is composed of 19 subunits, and it can be biochemically divided into the base and the lid [4,8]. The base consists of 10 subunits; six AAA ATPases arranged in a heterohexameric ring and four non-ATPase subunits. The AAA ATPase subunits are known as the Regulatory particle triple-A protein 1 (Rpt1)- Rpt6 [1,5], while the additional four are the RP non-ATPase 1 (Rpn1), Rpn2, Rpn10 and Rpn13. The lid is formed by nine non-ATPase subunits; Rpn3, Rpn5-Rpn9, Rpn11, Rpn12 and Sem 1. The lid functions as a bridge that connects the 20S core proteasome and the 19S RP. Rpn3, Rpn7, Rpn8 and Rpn11 (from the lid) and Rpt3–Rpt6 and Rpn2 subunits (from the base) are the subunits that facilitate this function [5]. Rpn1, Rpn10 and Rpn13 serve as ubiquitin receptors that recognize the ubiquitinated protein substrate [1,2,5,6,8,18,19], while Rpn11 subunit is a DUB with metalloprotease activity that cleaves the ubiquitin chains before substrate degradation [1,5,6]. Finally, the ring formed by the AAA ATPase subunits along with Rpn1, Rpn2 and Rpn13 (from the base) attaches to the α-ring of the CP [8], being responsible for the gate regulation and translocation of the substrate into the 20S CP [1].
1.3. Immunoproteasome
Immunoproteasomes are tissue-specific versions of proteasomes. They were initially identified to be continuously expressed in the cells of the immune system but they were then revealed to be expressed in almost all tissues upon stimulation by proinflammatory cytokines, interferon (INF)-γ and tumor necrosis factor (TNF)-α [21]. Immunoproteasomes are formed when β1, β2 and β5 catalytic subunits are substituted by β1i, β2i and β5i immunosubunits, respectively, following de novo synthesis and assembly [22]. The β2i subunit has a T-L activity, while both β1i (unlike the 20S β1 subunit) and β5i immunoproteasome subunits possess a CT-L activity thus explaining the enhanced CT-L activity exhibited by the immunoproteasomes [23]. Due to the difference in the cleavage sites, immunoproteasomes generate peptides that facilitate antigen presentation by the major histocompatibility complex (MHC) class I (peptides with hydrophobic C-termini that fit in the groove of MHC class I molecules) [[22], [23], [24]]. Apart from their implication in the immune response, immunoproteasomes participate in cellular adaptation to oxidative stress through the selective degradation of oxidized proteins [13,23,25]. In support, mice lacking one or two immunoproteasome subunits are more susceptible to oxidative stress [26].
The ring-shaped 11S regulator complex PA28 (also known as REG) is another important proteasome regulator that has been suggested to preferentially bind to the immunoproteasome and to increase its activity [13,23]. Three isoforms of PA28 have been described; the heteroheptamer PA28α and PA28β and the homoheptamer PA28γ [27]. It has been suggested that the 11S regulator enhances the selectivity of immunoproteasomes for oxidized proteins, thus playing a pivotal role in conditions where oxidative stress is excessive such as aging or various age-related diseases [25,28].
2. Proteasome implication in aging and disease
The integrity of the proteome is challenged upon aging progression [29]. The age-related decline of the proteostasis network, one of the hallmarks of aging [30], impedes the cells from maintaining a functional proteome [29,31]. As one of the main systems that surveil protein quality control, the proteasome has a pivotal role in this process. Indeed, it is well established that proteasome function and content are diminished with progressive aging [32,33]. Consequently, the accumulation of damaged, misfolded or aggregated proteins along with their escape from proteasome elimination confers to the onset and progression of aging and age-related diseases [31,34,35].
The proteasome has been shown to get impaired in various levels during aging; downregulated expression and/or post-translational modifications of subunits, disassembly, accumulation of damaged, aggregated or cross-linked proteins that clog the proteasome, resulting in decreased proteasome activities [29,32,34]. Many studies have revealed the correlation between increasing age and the decrease in proteasome activity in various mammalian tissues, including lymphocytes, lens, skeletal muscle, epidermis, adipose, retina, lung, muscle, heart, brain, spinal cord, hippocampus and cortex [29,32,[34], [35], [36]], while controversy has been observed for various tissues, including the liver [32,34,35,37]. Partial proteasome inhibition in young cells elicited an irreversible, premature senescence-like phenotype [38], supporting its causal role in the aging process. In addition, in vivo studies argued for the age-related proteasome impairment. Deletion of UMP1, a key regulator of the proteasome assembly, led to proteasome dysfunction, increased levels of oxidized proteins and reduction of cell survival in Saccharomyces cerevisiae [39]. In contrast, overexpression of UMP1 increased CT-L activity and extended the lifespan in yeast [40]. Aging-associated disturbance of the 26S proteasome assembly has also been found in adult and old brains of the killifish Nothobranchius furzeri [41] and old Drosophila melanogaster [42]. In contrast, overexpression of β5 proteasome subunit in adult flies increased CT-L activity, reduced ubiquitinated protein levels and extended their lifespan [43]. Likewise, overexpression of rpn-6 or pbs-5 (homolog of β5) proteasome subunits in Caenorhabditis elegans resulted in prolonged lifespan and healthspan improvement [44,45]. Tomaru et al. also illustrated that decreased CT-L activity afflicted lifespan in a transgenic mouse model [46]. On the other hand, the naked mole rat Heterocephalus glaber, the longest-lived rodent, has been shown to have higher CT-L and T-L activities compared to the control animals, which possibly attribute to their extended lifespan [47]. Similarly, fibroblast cultures from healthy centenarians revealed that proteasome function and activity remained unaltered compared to the ones in cultures from old (but not centenarians) donors [48], highlighting that “successful” aging seems to be correlated with functional proteasomes.
The proteasome also has a pivotal role in the progression of various age-related diseases [49], especially neurodegenerative diseases (specifically the ones that are categorized as proteinopathies), including Alzheimer's (AD), Parkinson's (PD), and Huntington's (HD) diseases. These pathologies are characterized by the accumulation of aggregate-prone neurotoxic proteins, such as Aβ peptide and tau in AD, α-synuclein in PD and mutant huntingtin containing a long polyglutamine (polyQ) tract at the N-terminus of the protein in HD [49]. Studies have revealed a correlation between UPS malfunction and the initiation and aggravation of the disease pathogenesis. Indeed, proteasome failure to clear aggregated, misfolded or damaged proteins subsequently led to their accumulation in neurons and, ultimately, to disease progression [50]. Dysregulation of the UPS has been observed in the brain of AD [51,52], PD [53] and HD patients [54]. Aβ peptide, α-synuclein and mutant huntingtin have been revealed to interact directly with the 26S proteasome and to negatively affect its activity [43,44]. These observations suggest a vicious cycle in which age-related decreased proteasome content and function feed the accumulation of damaged proteins, while this accumulation further hinders proteasome activity. However, interventions aiming at proteasome activation seem to be beneficial for alleviating the age-related phenotypes [34,35].
2.1. Insulin/insulin-like growth factor-1 (IGF-1) signaling (IIS) pathway
Aging pathways can directly modulate elements of the proteostasis network, including the proteasome, as a mechanism for lifespan extension. The IIS pathway is a very well conserved pathway among organisms that regulates the homeostasis of nutrients, growth, aging and longevity [30,55,56]. It was the first aging pathway discovered following the identification of the C. elegans mutants age-1 (encoding the sole phosphatidylinositol-3-OH kinase, PI3K) and daf-2 (encoding the IGF-1 receptor) that led to a dramatically extended lifespan [57]. Further studies revealed that these two genes belong to a single pathway, the IIS pathway [57]. This pathway is triggered by insulin-like peptides that bind to the IGF-1/DAF-2 receptor. This binding leads to the activation of the PI3K/AGE-1, which in turn converts phosphatidylinositol 4, 5-bisphosphate (PIP2) into phosphatidylinositol 3, 4, 5-trisphosphate (PIP3); the process is facilitated by the PTEN/DAF-18 phosphatase. This signal activates downstream kinases; 3-phosphoinositide-dependent protein kinase 1 (PDK-1), protein kinase B (AKT-1/-2) and serum- and glucocorticoid-inducible kinase 1 (GSK-1). These kinases mediate the phosphorylation of the forkhead box O FOXO/DAF-16 transcription factor. The phosphorylated form of FOXO/DAF-16 is inactive in the cytoplasm. In contrast, upon downregulation of the IIS pathway, activation of FOXO/DAF-16 is enhanced. It is translocated into the nucleus and initiates the transcription of genes involved in longevity. Besides, the IIS pathway has been strongly implicated in age-related diseases [58,59], in which the activation of the downstream transcription factors alleviates the disease phenotypes.
Heat shock factor 1 (HSF-1) and nuclear factor erythroid 2-related factor 2 (Nrf2)/skinhead 1 (SKN-1) are also key transcription factors that interact with the IIS pathway and modulate stress resistance, detoxification and longevity. HSF-1 and Nrf2/SKN-1 are activated under conditions that attenuate the function of the IIS pathway [55,56,[59], [60], [61]]. Sirtuins that represent another nutrient-sensing pathway related to longevity [30,55,60] have been revealed to regulate the FOXO/DAF-16 transcription factor [55,56]. Sirtuins are NAD+-dependent protein deacetylases; their overexpression has been found to prolong lifespan [55]. Overexpression of the sirtuin gene sir-2.1 in C. elegans extended the nematode's lifespan in a daf-16-dependent manner [62], while in mammals SIRT1 has been found to deacetylate FOXO and modulate its downstream targets that are related to the oxidative stress response [63].
Importantly, various studies have deciphered how FOXO/DAF-16 modulates the UPS [[64], [65], [66]]. Under conditions of reduced IIS pathway, a DAF-16-dependent enhancement of the UPS activity was detected in the intestinal cells of the nematode C. elegans. More specifically, activation of DAF-16 led to repression of the proteasome-associated deubiquitinating enzyme UBH-4 that in turn promoted enhanced UPS activity. UBH-4 and the mammalian ortholog UCHL5 were revealed to interact with the RPN-13 proteasome subunit [67,68] and to negatively regulate UPS activity [64]. DAF-16 was shown to be essential for the increased proteasome activity detected in nematodes lacking their germline. More specifically, Vilchez and colleagues revealed that DAF-16 regulated the rpn-6.1 subunit of the 19S RP by elevating its expression in these animals that subsequently resulted in increased proteasome function [45]. In support, the increase in proteasome content and function that was observed following 20S proteasome activation through pbs-5 overexpression in C. elegans was found to be dependent on DAF-16/FOXO transcription factor, among others [44]. Finally, FOXO1 was recently shown to directly regulate the expression of β5 20S proteasome catalytic subunit and, hence, proteasome activity [69]. Consequently, it is possible that compounds that affect the IIS pathway may act at least partially through the FOXO/DAF-16-dependent UPS activation.
2.2. Nrf2
Nrf2 regulates the expression of many proteasome genes and, consequently, positively modulates its activity [[70], [71], [72], [73], [74]]. Nrf2 belongs to the cap ‘n' collar (CNC) subfamily of the basic region leucine zipper (bZip) transcription factors [75]. Under normal conditions, Nrf2 is retained in the cytoplasm via binding with a specific cytosolic inhibitor, namely inhibitor of Nrf2 (INrf2) or Kelch-like ECH-associated protein 1 (Keap1) [[76], [77], [78]]. The Keap1/Nrf2 complex binds the Cul3-RbX1 holoenzyme, a Cullin-RING box (Cul-Rbx) E3 ubiquitin ligase, which ubiquitinates Nrf2. Nrf2 is then driven for degradation by the 26S proteasome [71,75,76,78]. On the other hand, upon oxidative stress, Keap1 gets oxidized and loses its ability to bind Nrf2, which is then released into the cytosol [71,76]. Additionally, various kinases [such as protein kinase C (PKC), several mitogen-activated protein kinases (MAPKs) or PI3K] phosphorylate Nrf2, which is then translocated into the nucleus [71,76,77]. Nrf2 then heterodimerizes with small Maf (Musculoaponeurotic fibrosarcoma) or c-Jun proteins to bind to the antioxidant response elements (ARE) located in the promoter regions of various genes involved in antioxidant response [75,78]. Such genes include NQO-1 (NADPH quinone oxidoreductase-1), HO-1 (Heme oxygenase-1), GSTs (Glutathione S-transferases), γ-GCS (γ-Glutamylcysteine synthetase) and several proteasome subunits [70,71,78], among others.
Various natural terrestrial compounds have been identified to increase proteasome content and function through Nrf2 activation as reviewed by Chondrogianni et al., 2020 and references therein [79]. More specifically, the triterpenoids hederagenin and 18α-glycyrrhetinic acid (18α-GA) have been found to enhance proteasome activities in an Nrf2-dependent manner, to extend cellular lifespan and to confer protection against oxidative stress in HFL-1 primary human fibroblasts [72]. Similar results were obtained in C. elegans, where treatment with 18α-GA led to enhanced proteasome activity and prolonged the lifespan of the nematodes in a SKN-1-dependent manner while it also conferred resistance to proteotoxicity [80]. Quercetin, a flavonoid belonging to polyphenols and known for its ability to induce Nrf2 activation [81], was also found to stimulate proteasome function with subsequent anti-aging and antioxidant effects [82]. The Ginkgo biloba extract (EGb), which contains 24% ginkgo-flavone glycosides and 6% terpenoids, has been also identified as an Nrf2 activator that could enhance proteasome activity through the induction of proteasome subunits gene expression [83,84]. Concurrently, the beneficial properties of EGb in cells expressing polyQ proteins has been also reported [84].
2.3. HSF-1
HSF-1 is the main transcription factor that regulates the heat shock response. Under normal conditions, HSF-1 remains inactive, while upon stress, it accumulates to the nucleus and triggers the expression of heat shock response genes [85]. In nematodes, reduced HSF-1 activity led to decreased lifespan and promoted polyQ aggregate formation [86]. On the contrary, HSF-1 overexpression extended C. elegans lifespan in a DAF-16-dependent manner [62,87], thus strengthening its involvement in longevity.
Various studies have related HSF-1 signaling with the UPS function. Recently, Bianchi and coworkers deciphered that HSF-1 is required for the transcriptional induction of the poly-ubiquitin gene ubiquitin C (UBC) under oxidative stress and proteotoxic conditions. As UBC is one of the four genes encoding ubiquitin, its upregulation under stress conditions is related to the increased cellular demand for ubiquitin to eliminate toxic misfolded proteins [88]. We have also shown that HSF-1 is an essential transcription factor in inducing lifespan extension and resistance to oxidative stress following 20S proteasome activation through pbs-5 overexpression in C. elegans, as HSF-1 mutants that overexpress pbs-5 subunit did not exhibit the abovementioned beneficial properties [44]. In addition, HSF-1 has been revealed to promote the expression of UPS components (various E2 and E3 ligases) to facilitate linker-cell type-death, a morphologically conserved non-apoptotic cell death process occurring during C. elegans and vertebrate development [89]. Finally, carqueja hydroalcoholic extract (GHE), derived from the South Africa native plant Baccharis trimera was found to possess antioxidant and anti-aggregation properties. More specifically, GHE protected against Αβ1-42-induced paralysis in an AD nematode model through enhanced protein homeostasis represented by increased proteasome activity and upregulated hsp-4 and hsp-16.2 gene expression [90].
3. The proteasome as a regulator of oxidative stress
Aerobic organisms need oxygen (O2) to survive and inevitably produce reactive oxygen species (ROS). The term ROS refers to both free radicals and non-radicals. The free radicals are derived from oxygen, including superoxide anion (O2•‾), hydroxyl (•OH) and peroxyl (ROO•) radicals, while the non-radicals include hydrogen peroxide (H2O2) and singlet oxygen, among others, that can also generate free radicals [76,77,91]. Except for ROS, reactive nitrogen species (RNS) also exist, including nitric oxide (•NO) radicals [71,92]. Endogenous and exogenous sources can produce ROS/RNS; mitochondrial metabolism is the primary endogenous source that generates superoxide anion radicals, while smoke, air pollutants and radiation are representative exogenous sources [70,77,92]. Importantly, low levels of ROS/RNS are essential for cells since they serve as messengers for signaling cascades, including PI3K/Akt, MAPK, Wnt-β-catenin pathways [77]. However, ROS/RNS can target lipids, proteins and DNA and therefore cause cellular damage [77,91]. When the equilibrium between oxidants and antioxidants is disturbed in favor of the oxidants, oxidative stress occurs. To maintain redox balance, organisms have a network of antioxidant defense systems that consists of antioxidant enzymes and nonenzymatic antioxidants. Paradigms of nonenzymatic antioxidants are l-γ-glutamyl-L-cysteinyl glycine (glutathione, GSH), vitamin C and E, which scavenge ROS or prevent their formation [77,93]. Superoxide dismutases (SODs), glutathione peroxidases (GPxs) and catalases (CATs) are among the enzymatic antioxidants [77]. SODs, like Cu, ZnSOD (SOD1) or MnSOD (SOD2), can transform superoxide anion radicals into less reactive hydrogen peroxide and oxygen. Hydrogen peroxide is then converted to water (H2O) mainly through GPxs, which are present in the cytoplasm and mitochondria, CATs, which are heme-containing enzymes mainly located in the peroxisomes, and peroxiredoxins that are peroxidases consisted of six isoforms that localize in the cytosol, mitochondria and endoplasmic reticulum, among other cellular compartments [71,76,77,94]. GSH is an essential antioxidant and the ratio of GSH to its oxidized form glutathione disulfide (GSSG) is an indicator of the cellular redox status [71]. GSH protects protein thiol groups against oxidation via its reaction with reactive species or through the recruitment of GPxs [77].
Besides the primary antioxidant defense system, proteolysis and several transcription factors, such as the transcription factor Nrf2 that is activated upon oxidative stress, are also recruited to combat the consequences of oxidative stress in cells [70,76]. Ubiquitin-independent proteasome-mediated proteolysis serves in the degradation of oxidized proteins [71,91,93,95]. Since the (at least partially) unfolded state is a typical characteristic of oxidatively modified proteins, the 20S proteasome is recruited [70,96]. Studies have shown that the 20S proteasome is more resistant to oxidative stress than the 26S proteasome [13,70,91,96]. Indeed, the exposure of the 20S and 26S proteasomes to hydrogen peroxide revealed that the 20S proteasome activity was not affected by oxidative stress, while the 26S proteasome function was impaired [97,98]. Additionally, studies have deciphered that under oxidative stress conditions, the 26S proteasome is dissociated into the 20S CP and the 19S RP, leading to increased levels of the 20S proteasome [91,95,96]. Also, redox-dependent post-translational modifications, such as S-glutathionylation [99], have been shown to enhance the activity of the 20S proteasome and its further involvement in the redox regulation [70,71].
ROS play a crucial role in the aging process and age-related diseases. According to the Free Radical Theory of Aging, proposed by Harman in 1956 [100] that has evolved since then to integrate new evidence into the overall theory [101], aging is linked, among others, to the excessive accumulation of free radical damage and its deleterious effects in cells and tissues [100,102]. Indeed, when high ROS levels are present for an extended period in an organism, they can serve as mediators of cellular senescence progression [103]. Furthermore, heavily oxidatively modified and cross-linked proteins are resistant to degradation. During aging, reduced proteasome activity, decline of Nrf2 function and ineffective cellular response to oxidative stress lead to damaged protein accumulation that accelerates aging and triggers the pathogenesis of several diseases [13,70,77,93,96,104]. On the contrary, interventions that lead to increased expression of the antioxidant enzymes, controlled Nrf2 activation, and proteasome function enhancement are beneficial both for longevity and age-related diseases by alleviating the ROS-related effects, among others [35,74,75,105].
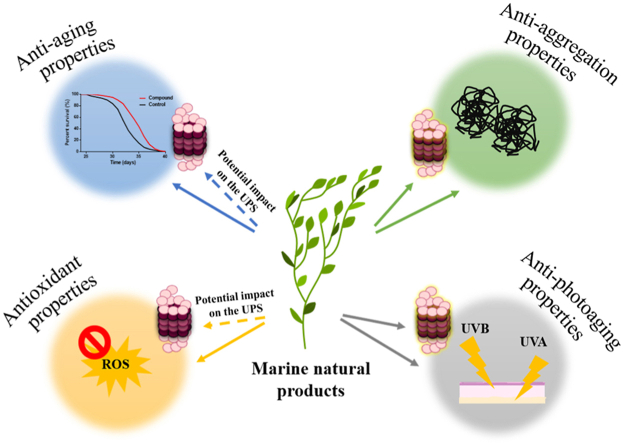
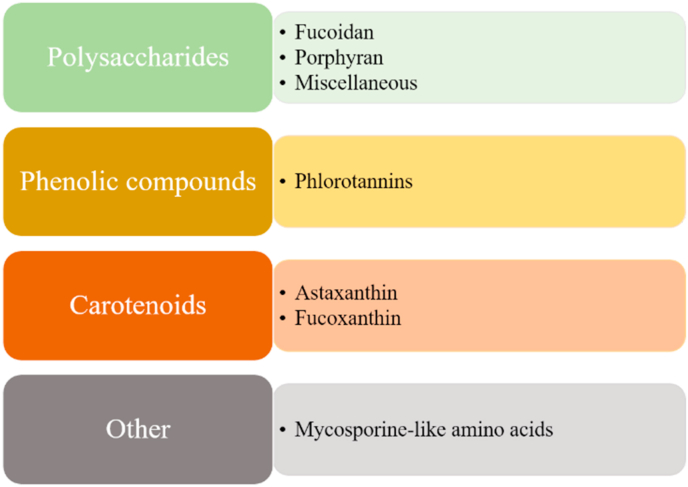
4. Marine-derived compounds with antioxidant, anti-aging and anti-aggregation properties as potential UPS modulators
Since the oceans cover the biggest part of Earth and represent the largest habitat, more than half of the recorded biodiversity consists of marine organisms. To adapt to a plethora of distinct, and in some cases, extreme environments, marine organisms have developed biochemical pathways that allow the production of a variety of compounds that may serve as an excellent reservoir of active biomolecules [106,107]. Indeed, to date, many marine natural products have been identified to display a wide range of beneficial properties, including antioxidation, anti-aging or anti-aggregation. Although these protective properties have been linked with proteasome modulation for several compounds derived from terrestrial organisms as reviewed by Panagiotidou and Chondrogianni, 2020 and references therein [108], very few marine compounds have been studied directly for their potential to modulate the UPS (in terms of 20S/26S proteasome complex modulation and/or modulation of the ubiquitination machinery including modulation of E1, E2, E3 ligases). Nevertheless, many of them have been shown to affect various pathways that have been associated with the UPS expression and function, such as the IIS pathway and the FOXO/DAF-16 transcription factor or the Nrf2 and the HSF-1 signaling pathways. In this context, these compounds may represent potential UPS modulators and it is possible that these properties will be revealed in the near future. Herein, we describe the few marine compounds that have been revealed to activate directly the proteasome and its function. Moreover, we present the current knowledge on marine-derived compounds that have been shown to serve as antioxidant, anti-aging or anti-aggregation agents and we suggest their potential to represent UPS modulators in an indirect manner through the modulation of UPS-associated pathways. In this context, polysaccharides, phenolic compounds and carotenoids are of great interest with regard to their properties to delay aging and age-related diseases (Fig. 2).
Fig. 2.
Marine classes of natural products and their major representatives. The illustrated marine compounds have been suggested to possess beneficial properties against aging, photoaging and age-related diseases.
Finally, since multiple marine organisms live in shallow waters, they are exposed to UV radiation during their lifetime. Consequently, they have developed photoprotective mechanisms to combat the subsequent adverse effects. Hence, the involved compounds may represent promising anti-photoaging agents. Therefore, photoaging will be also presented in the last section of this review along with its link with the UPS and the various marine compounds with known anti-photoaging properties. Moreover, those ones for which the link with the UPS is already known will be further discussed.
4.1. Polysaccharides
Carbohydrates, also known as saccharides, include mono-, di-, oligo- and poly-saccharides [109]. Among them, polysaccharides serve as structural components and have a role in energy storage in marine and terrestrial organisms [109,110]. However, polysaccharides derived from marine organisms, especially the sulfated ones, have attracted significant attention in recent years due to their bioactivity, with marine macroalgae (or seaweeds) being the most important source of non-animal sulfated polysaccharides [[111], [112], [113]].
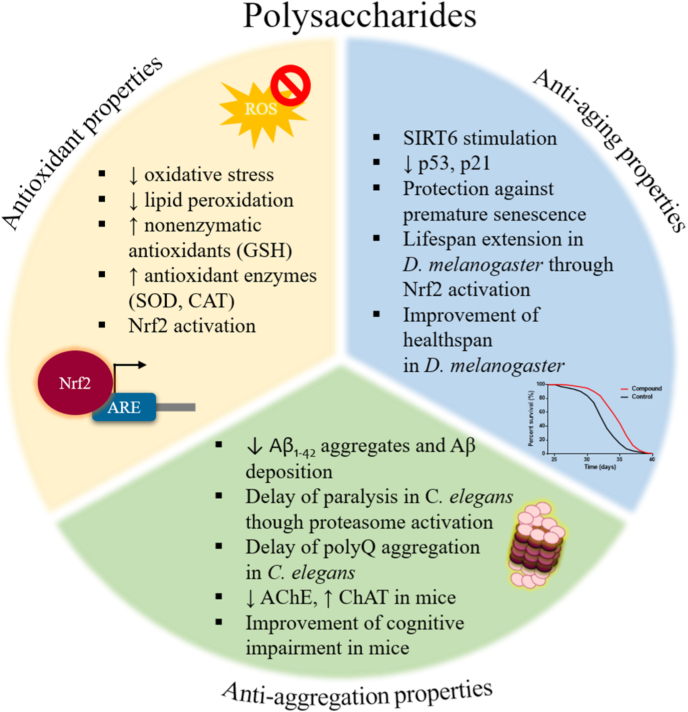
Three algal phyla are the most abundant source of sulfated polysaccharides, namely Rhodophyta (red macroalgae), Ochrophyta (brown macroalgae) and Chlorophyta (green macroalgae) [111,114]. The sulfated polysaccharides that are present in red algae are galactans that are further classified into the carrageenans and agarans [113,115,116]. Ulvans are the major polysaccharides derived from green macroalgae, while fucans represent the most common sulfated polysaccharides found in brown algae [115,116]. According to the macroalgal source, the obtained polysaccharides possess unique structures and, subsequently, different biological activities [113]. Their variety is not only related to the algal source, as different polysaccharides have been extracted even from the same species depending on the location and the time of algal harvest [112,116]. The relationship between the structure of polysaccharides and their biological properties is difficult to be established due to the complexity of their structures [113,115], yet they are very promising compounds. Their antioxidant, anti-aging and anti-aggregation properties are summarized in Fig. 3.
Fig. 3.
The antioxidant, anti-aging and anti-aggregation properties of marine-derived polysaccharides.
4.1.1. Fucoidan
The water-soluble polysaccharide fucoidan is one of the best-studied fucans derived from the cell wall of brown algae [114,115,117,118]. It was first isolated in 1913 and it is a complex sulfated polysaccharide with a wide variety of chemical structures depending on the species from which it is extracted [115,119]. Fucoidans are neither present in other marine algal phyla nor terrestrial plants. Since fucoidans derived from marine invertebrates do not possess such heterogeneous and complex structures, the algal-extracted fucoidans are considered more attractive for applications in the biomedical field [120].
The antioxidant capacity of fucoidans is the most significant property of these biopolymers, with fucoidans from brown seaweeds, such as Sargassum filipendula and Laminaria japonica, exerting in vitro antioxidant activity stronger than vitamin C [113,121,122]. In a recent study, Ryu and colleagues showed that fucoidan could effectively reduce oxidative stress. Treatment of human keratinocyte cells (HaCaT) with fucoidan increased the HO-1 and SOD1 mRNA and protein expression levels in a dose-dependent manner through the Nrf2/ERK signaling pathway [123]. Fucoidan isolated from Ecklonia cava was shown to possess antioxidant activity against peroxyl radicals in vitro. Its antioxidant activities were further evaluated against 2,2′-azobis (2-amidinopropane) dihydrochloride (AAPH)-induced oxidative stress in Vero cells (African green monkey kidney fibroblasts) and zebrafish (Danio rerio), where fucoidan also exerted protective effects against ROS production [124].
Zhang and colleagues revealed the anti-aging properties of a fucoidan fraction (with a molecular weight of 52.7 kDa and 15.3% sulfate content) isolated from the brown alga Sargassum fusiforme [125]. More specifically, the treatment of D. melanogaster with this fraction extended its lifespan and increased the SOD, CAT and GSH-Px activities, yet reduced the content of malondialdehyde (MDA) that, as the main index of lipid peroxidation, is a reflection of the ROS-induced cellular damage [96]. These observations point out that this fraction restored the antioxidant defense system that tends to decline upon aging. Finally, activation of the Keap1/Nrf2/ARE signaling pathway was found to mediate the observed lifespan extension and this activation subsequently contributed to the recorded heat stress resistance [125]. Finally, fucoidans extracted from Fucus distichus and Fucus vesiculosus were found to be potent stimulators of SIRT6, an NAD+-dependent histone deacetylase, in vitro [126].
Various studies have also evaluated the neuroprotective effects of fucoidan. In primary cultures of rat basal forebrain cholinergic neurons, fucoidan improved both neuronal survival and cell morphology against Αβ-induced toxicity. Co-incubation of fucoidan with Aβ1-42 peptide did not show any ability of the polysaccharide to block Aβ aggregation in vitro, implying a different mechanism of action. Indeed, pre-treated cells with fucoidan showed that it impeded ROS generation induced by the Aβ peptide and maintained the phosphorylated form of PKC, which is otherwise blocked in the presence of Aβ [127]. Furthermore, treatment with fucoidan from Undaria pinnatifida secured cell viability and increased SOD activity and GSH content in PC12 cells (derived from a pheochromocytoma of rat adrenal medulla) challenged with a combination of Aβ25-35 and d-galactose (D-Gal); elevated levels of galactitol, a D-Gal metabolite, in the brain cannot be metabolized leading to neuronal loss and aging [128]. In the same study, fucoidan ameliorated spatial and memory impairment in ICR mice that were injected with D-Gal. Also, fucoidan restored SOD activity and GSH content in the animals blood serum, two proteins that are both impaired due to increased free radicals induced by Aβ25-35 and D-Gal [128]. Moreover, in a rat AD model, oral administration of fucoidan attenuated cognitive impairment and increased SOD and GSH-Px levels while it decreased MDA production [129]. Alghazwi et al. found that fucoidans from the brown seaweeds F. vesiculosus and U. pinnatifida protected PC12 cells against cytotoxicity induced by hydrogen peroxide and Aβ1-42. Moreover, fucoidan from both species reduced Aβ1-42 aggregates [130]. The positive effect of fucoidan has also been described in a PD model; fucoidan increased GSH, SOD, GSH-Px, CAT activities and total antioxidant capacity (a measure of the nonenzymatic antioxidant defense system [131]) in the substantia nigra of C57BL/6 mice injected with 1-methyl-4-phenyl-1, 2, 3, 6-tetrahydropyridine (MPTP) [132].
A direct relationship between fucoidan and proteasome function was also suggested by Wang et al. Administration of fucoidan to the GMC101 C. elegans strain (transgenic nematodes expressing human Aβ1-42 peptide in their body wall muscle cells) delayed the paralysis onset of the worms through reduced Aβ protein levels and deposition. In addition, while Aβ was found to impair proteasome activity in the nematodes, fucoidan inhibited the decrease of pbs-2 and pbs-5 gene expression and elevated proteasome activity. Fucoidan treatment also restored the expression of the heat shock protein hsp-16.2, which is involved in the protection against oxidative stress [133]. Two additional studies have implied that fucoidan can modulate proteasome function; fucoidan treatment of breast [134] or lung cancer cells [135] reduced the expression of transforming growth factor β receptor I (TGFRI) and TGFRII proteins, which participate in the epithelial to mesenchymal transition, via the ubiquitin-dependent proteasome degradation. The addition of MG132 proteasome inhibitor impeded fucoidan's ability to induce the degradation of these two proteins.
4.1.2. Porphyran
Porphyran is a sulfated polysaccharide and one of the best-studied agarans. It is produced by red algae from Porphyra species, which are edible seaweeds found in East and Southeast Asia. They are commonly known as nori, laver or gim and they have been used as drugs in traditional Chinese medicine for many decades. The cell walls of Porphyra algae are rich in porphyrans [114,115,136].
A sulfated polysaccharide fraction isolated from the red alga Porphyra haitanensis was identified to have antioxidant activity in vivo. Administration of this fraction increased the activity of SOD and GSH-Px and the total antioxidant capacity in aging Kunming mice, while MDA production was reduced. Importantly, the impact of this fraction was similar or stronger than that of vitamin C, which was used as a positive control [137]. The same research group revealed that the antioxidant activity of P. haitanensis was not limited in the particular fraction, as similar positive results were observed following administration of another fraction of the alga, differing in the sulfate and 3,6-anhydrogalactose content [138].
Additional studies have deciphered that porphyrans can alleviate cell damage and aging. Zhang and colleagues showed that porphyran from P. haitanensis secured cell viability following hydrogen peroxide-induced premature senescence in human lung WI-38 fibroblasts. More specifically, porphyran could restore the cellular morphological changes following exposure to hydrogen peroxide, while it diminished the proportion of cells positive for senescence-associated β-galactosidase activity (SA-β-Gal) [139], a widely used biomarker of senescent cells [140]. At the same time, porphyran downregulated the expression of p53 and p21 proteins, the activation of which is associated with DNA damage induced by oxidative stress that subsequently drives cellular senescence [139]. Porphyran has also exhibited anti-aging properties in vivo; two polysaccharide fractions isolated from P. haitanensis extended the lifespan of D. melanogaster and improved their resistance to thermal stress. These fractions improved the flies' vitality by encouraging the mating process. However, the involved mechanisms were not elucidated [141].
Finally, a recent study suggested the neuroprotective effects of a porphyran fraction extracted from P. haitanensis. In an AD-mouse model, porphyran ameliorated the adverse effects of Aβ1-40 peptide on learning and memory. Administration of porphyran increased the choline acetyltransferase (ChAT) activity in the hippocampus and decreased the acetylcholinesterase (AChE) activity in both the hippocampus and the cortex of the mice. These two enzymes have an essential role in proper brain function. Nevertheless, the exact mechanism of action was not revealed [142], highlighting the necessity to elucidate the way the porphyrans act on aging and age-related diseases progression.
4.1.3. Miscellaneous
The partially purified polysaccharides extracted from Chlorella pyrenoidosa were found to possess antioxidant activities in vitro through scavenging of the superoxide anion, hydroxyl and 1,1-diphenyl-2-picrylhydrazyl (DPPH) radicals. D. melanogaster treated with this polysaccharide-rich fraction exhibited lifespan extension, which was accompanied by increased activities of the antioxidant enzymes SOD, CAT and GPx. Nevertheless, the correlation between the enhancement of the antioxidant enzymes activities and the observed lifespan extension was not verified, implying another mechanism of action [143]. In a recent study, Wang and colleagues deciphered the anti-aging properties of polysaccharides derived from the marine microalga Gracilaria lemaneiformis. These polysaccharides extended the lifespan and improved the healthspan of the wild type (wt) C. elegans. Moreover, in a nematode model for HD (AM141 strain), they delayed the progression of the polyQ aggregation. These positive effects were mediated through the enhancement of the DAF-16 nuclear translocation and its subsequent activation [144].
4.2. Phenolic compounds
Phenolic compounds are classified as phenols or polyphenols based on the number of the incorporated phenol groups [145]. Their synthesis is mediated via the shikimic acid and acetate-malonate pathways [117,145]. Phenolic compounds are secondary metabolites of algae involved in a wide range of biological functions, such as signaling, defense and stress resistance [117,120]. The most common types of polyphenolic compounds found in algae are phenolic acids, flavonoids and phlorotannins, among others [145].
Phlorotannins are the most interesting polyphenolic compounds. They are very hydrophilic compounds and their molecular sizes range between 126 Da and 650 kDa [[145], [146], [147], [148]]. They are composed of oligomers or polymers of phloroglucinol (1,3,5-trihydroxynenzene) monomeric units biosynthesized through the acetate-malonate pathway [120,[146], [147], [148], [149]]. Fuhalols, phlorethols, fucols, fucophloroethols, eckols and carmalols are classes of phlorotannins, which are generated by several units of phloroglucinol linked to each other in different ways (Fig. 4) [145,147,148]. Phlorotannins are present mainly in brown algae [[145], [146], [147],149], they exhibit seasonal variations [147,149] and they are absent from terrestrial plants [120]. Among the brown algae, the edible seaweed Ecklonia cava has been reported as one of the most abundant sources of phlorotannins [145].
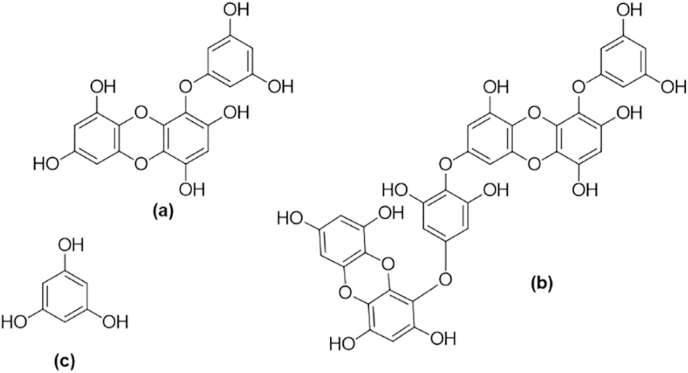
Fig. 4.
The chemical structures of the marine-derived phlorotannins (a) eckol, (b) dieckol, and (c) phloroglucinol.
Many studies have shown that phlorotannins derived from brown algae are strong antioxidants. Phlorotannins (eckol, phlorofucofuroeckol A, dieckol, 8,8′-bieckol) extracted from the brown algae Eisenia bicyclis, E. cava and Ecklonia kurome were found to exert both antioxidant and radical scavenging activities against superoxide anion radicals and DPPH. Indeed, these compounds are more effective than terrestrial polyphenols (e.g., catechin (−)-epigallocatechin-3-gallate) and vitamins (e.g., ascorbic acid, α-tocopherol), [150]. Kang and coworkers described the antioxidant properties of eckol. It enhanced the activity of HO-1 by upregulating its expression in Chinese hamster lung V79-4 fibroblasts. HO-1 activation resulted in protection against hydrogen peroxide-induced oxidative stress. This process was mediated through Nrf2 activation in an ERK/PI3K/Akt-dependent manner [151]. In another study, extracts of 17 algal species were evaluated for their antioxidant activity; extracts from Ulva pertusa, Symphyocladia latiuscula and Ecklonia stolonifera were shown to inhibit ROS production in Wistar rat kidney homogenates in vitro, with the extract of E. stolonifera exhibiting the strongest antioxidant activity [152]. Dieckol, eckol, eckstolonol, triphloroethol A and phloroglucinol isolated from E. cava inhibited intracellular ROS formation, reduced lipid peroxidation and prevented cell death against AAPH-induced oxidative stress in zebrafish embryos [153]. These observations support the intense antioxidant activity of these compounds in vivo. The extract from the brown alga Ascophyllum nodosum, a seaweed rich in phlorotannins, was found to protect human epithelial cells from tert-butyl hydroperoxide (tBHP)-induced oxidative stress by reducing ROS production. Moreover, A. nodosum extract increased SIRT1 activity [154]. Phloroglucinol possesses antioxidant activity, as in vitro studies showed that it scavenged nitric oxide, superoxide anion and hydroxyl radicals in a dose-dependent manner. These results were also confirmed in LLC-PK1 porcine kidney cells, where phloroglucinol alleviated oxidative stress, increased cell viability and decreased lipid peroxidation. At the same time, phloroglucinol exhibited anti-aging properties; treatment of WI-38 cells with phloroglucinol protected cell viability and reduced MDA production against premature senescence induced by hydrogen peroxide [155].
Research towards the discovery of phlorotannins as potential compounds against neurodegenerative diseases has been also performed. A phlorotannin-rich extract from E. cava, which was found to exert antioxidant activities in vitro, protected PC12 and human neuroblastoma SH-SY5Y cells against hydrogen peroxide and AAPH-induced oxidative stress. It was also found to inhibit the activity of AChE and butyrylcholinesterase (BChE) enzymes, thus pinpointing its potential neuroprotective effect [156]. Phloroglucinol offered protection in SH-SY5Y cells against hydrogen peroxide-induced cytotoxicity [157]. More specifically, cells pretreated with phloroglucinol exhibited decreased ROS levels and lipid peroxidation content, while lower levels of carbonylated proteins (a marker of protein oxidation) and DNA damage were also recorded [157]. The same results were observed following induction of cytotoxicity in SH-SY5Y cells with 6-hydroxydopamine (6-OHDA) [158]. In addition, phloroglucinol was found to increase both the protein expression and activities of CAT and GPx and to reverse Nrf2 reduction in the nucleus. Also, phloroglucinol ameliorated 6-OHDA-induced motor functional deficits in a rat PD model [158], highlighting its beneficial properties in vivo. In support, oral administration of phloroglucinol in an AD mouse model attenuated the cognitive impairment of the animals. Phloroglucinol reduced Αβ protein levels and the number of amyloid plaques in the brain, while it also decreased lipid peroxidation [159]. The advantages of phloroglucinol were also explained by Yang et al. [160]; Aβ1-42-induced ROS levels were reduced following treatment with phloroglucinol in immortalized mouse hippocampal HT22 cells. Moreover, in an AD-transgenic mouse model, phloroglucinol alleviated memory deficits, albeit no changes in Aβ levels were recorded between transgenic and control animals.
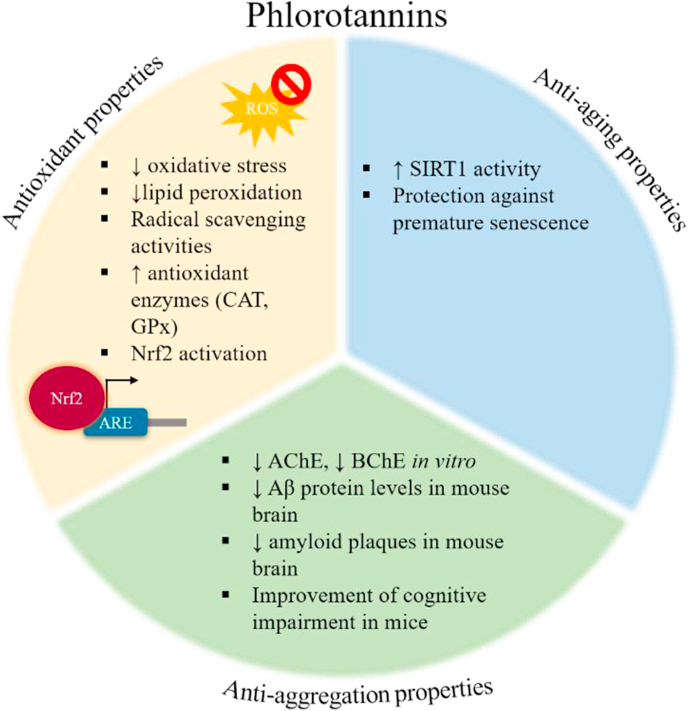
All the studies mentioned above could lead to the assumption that the beneficial effects of phlorotannins might be attributed to their scavenging and antioxidant activities. Especially for phloroglucinol, studies have indicated that it has an electron-rich component that might function as a stabilizer for free radicals [158,160]. Moreover, its ability to activate UPS-related pathways, such as the Nrf2/ARE pathway, strengthens the possibility of serving as a potent anti-aging and neuroprotective compound via the UPS modulation. The antioxidant, anti-aging and anti-aggregation properties of phlorotannins are summarized in Fig. 5.
Fig. 5.
The antioxidant, anti-aging and anti-aggregation properties of marine-derived phlorotannins.
4.3. Carotenoids
Carotenoids are natural pigments that are typically composed of 40 carbon atoms [[161], [162], [163]]. The end of their carbon chains may be completed by rings or oxygenated functional groups, giving rise to carotenes and xanthophylls, respectively [[161], [162], [163]]. However, there are carotenoids with more complex structures, such as violaxanthin and fucoxanthin [164]. Most carotenoids have the ability to absorb blue and violet light (400–500 nm), which attributes their colour ranging from red to yellow, depending on their structure and concentration, as well as their potential interactions with proteins [163]. They are lipophilic compounds found in membranes (usually thylakoid membranes) and oily droplets or in lipid vesicles in the cytosol [162,163,165].
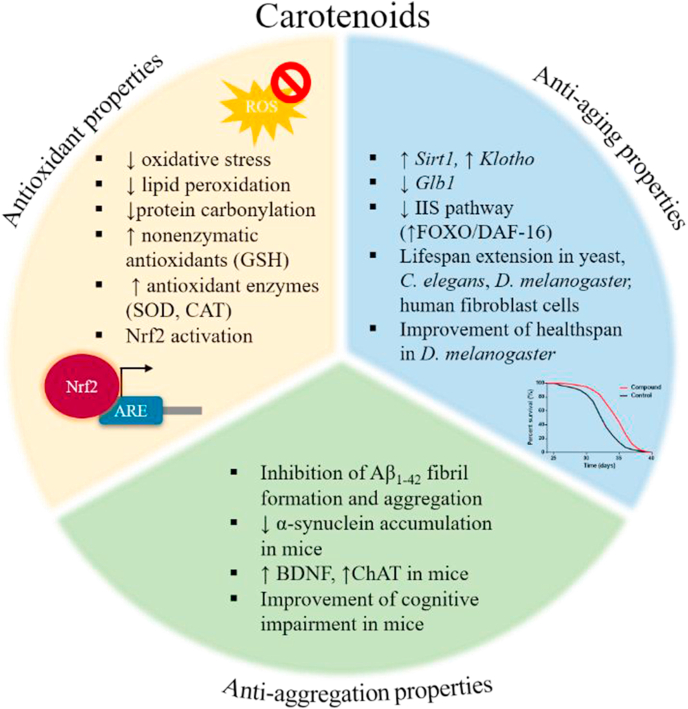
Carotenoids serve a variety of functions in marine organisms, such as cellular structure, photosynthesis, photoprotection and protection against stressful environmental conditions, during which organisms induce carotenogenesis to produce carotenoids [120,162,163,166]. One of the most significant benefits of carotenoids is related to their antioxidant properties [165]. They are quenchers of singlet oxygen, scavengers of free radicals or Nrf2/ARE activators [163,165]. Nevertheless, under certain conditions [163], they can switch to pro-oxidant compounds [163,165] being effective in states such as cancer, where they induce apoptosis to damaged cells by stimulating the cellular response to oxidative stress. The antioxidant, anti-aging and anti-aggregation properties of marine-derived carotenoids are summarized in Fig. 6.
Fig. 6.
The antioxidant, anti-aging and anti-aggregation properties of marine-derived carotenoids.
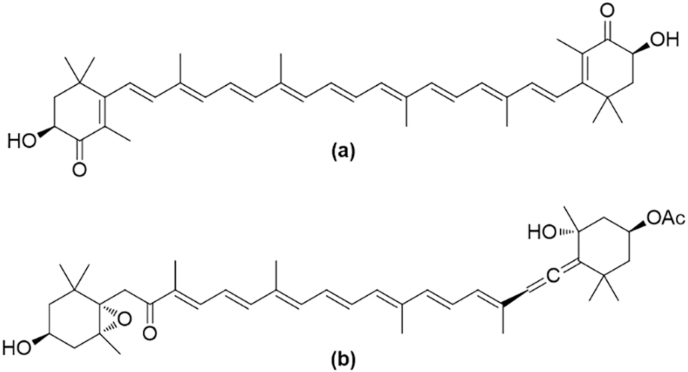
4.3.1. Astaxanthin
Astaxanthin (3,3′-dihydroxy-β, β′-carotene-4,4′-dione) is a red-orange keto-carotenoid pigment that belongs to xanthophylls (Fig. 7a). It is present in marine organisms, such as macroalgae, microalgae, bacteria and crustaceans (shrimps, crabs, krill). However, animals cannot biosynthesize astaxanthin; they obtain it through diet, which endows them the red colour they have in their flesh, skin or exoskeleton. The green microalga Haematococcus pluvialis is the most abundant source of astaxanthin [[167], [168], [169]]. The United States Food and Drug Administration (FDA) has approved astaxanthin as a colour additive in animals and fish foods since 1999 [165,168]. Its antioxidant activity is among its most significant characteristics, as it is found to surpass those of vitamin E and β-carotene [167,170,171]. In addition, astaxanthin can pass the blood-brain barrier. These observations, along with the so far lack of side effects from its use, have made astaxanthin more attractive, especially in the field of aging and neurodegenerative diseases [169,[172], [173], [174]].
Fig. 7.
The chemical structures of marine-derived carotenoids (a) astaxanthin and (b) fucoxanthin.
The antioxidant activity of astaxanthin has been established through in vitro and in vivo studies. Treatment of immortalized human hepatoma HepG2 cells with astaxanthin increased the intracellular levels of GSH and the total antioxidant activity. Astaxanthin induced Nrf2 expression and its downstream target genes, namely HO-1, NQO-1 and GSTM2 [175]. These results may suggest the dependency of its antioxidant character on the Nrf2/ARE pathway. Administration of astaxanthin was compared in Swiss albino young and old mice to elucidate its potential antioxidant activity between the different age groups. Astaxanthin alleviated oxidative stress in the tested brain areas via reducing lipid peroxidation, nitric oxide radical levels and advanced protein oxidation products. On the other hand, astaxanthin increased the activity of CAT and SOD and the GSH levels. Even though astaxanthin improved the oxidative markers in both age groups, the improvement in young mice was more considerable [176]. Furthermore, astaxanthin was found to exert antioxidant activity in the D-Gal-induced brain aging of rats via the decrease of MDA levels and protein carbonylation, while it alleviated the reduction of GSH-Px and SOD activities and protected against DNA damage. Importantly, astaxanthin administration restored the expression of the brain-derived neurotrophic factor (BDNF) in the brain of aged rats, which regulates neuronal development and survival and is also implicated in the regulation of cognition and memory storage [177].
In a recent study, aging was induced in mice through D-Gal treatment and jet lag. Administration of astaxanthin ameliorated the impaired locomotor activity. Besides, astaxanthin induced SOD, CAT and GSH-Px activities and decreased lipid peroxidation in the aged mice. The expression of the antioxidant genes Cat, Gpx 1, Sod1 and Sod2, the anti-aging gene Sirt1 and the aging-suppressor gene Klotho were induced, while the expression of β-galactosidase 1 (Glb1) gene was decreased [178]. Interestingly, the anti-aging mechanism of Klotho may be related to FOXO activation and the inhibition of oxidative stress via the induction of the MnSOD expression [179]. In support, Sudharshan and coworkers showed that astaxanthin protected S. cerevisiae antioxidant deficient strains against hydrogen peroxide-induced oxidative stress by scavenging ROS. Also, astaxanthin exerted anti-aging properties since it improved the chronological lifespan in yeast via ROS reduction and offered protection against apoptosis [180]. In C. elegans, astaxanthin was found to extend the lifespan of the wt nematodes and the age-1 long-lived mutants via the modulation of the IIS pathway. Astaxanthin promoted the DAF-16/FOXO nuclear translocation and decreased the mitochondrial ROS production. Moreover, in astaxanthin-exposed animals, the expression of the DAF-16 target genes, namely sod-3, sod-5, ctl-1 and ctl-2 was induced [181]. Finally, the anti-aging potential of astaxanthin was also revealed in D. melanogaster. Wt flies treated with astaxanthin had a longer survival time than the control-treated flies. Similarly, in SODn108 mutant flies, astaxanthin improved their lifespan, as well as their locomotor activity. In addition, astaxanthin protected flies against mortality induced by oxidative stress [182]. Given the effects of astaxanthin on IIS and Nrf2 pathways (both shown to affect the UPS), one cannot rule out the possibility that this compound may act as a UPS activator.
As for the neuroprotective properties of astaxanthin, it protected SH-SY5Y cells against Aβ25-35-induced cytotoxicity via the enhancement of HO-1 expression in a concentration-dependent manner through the activation of the ERK signaling pathway [183]. In PC12 cells treated with 1-methyl-4-phenylpyridinum (MPP+), astaxanthin also attenuated ROS production and induced HO-1 mRNA expression [184], which supports the involvement of the Nrf2/ARE pathway in astaxanthin's mechanism of action. Furthermore, astaxanthin treatment protected SH-SY5Y cells against cytotoxicity induced by DHA hydroperoxide (DHA-OOH), 6-OHDA [185] and MPP+ [186] through reduction of ROS production. Besides, astaxanthin inhibited the MPTP-induced α-synuclein levels in the substantia nigra of a PD mouse model, pointing out its neuroprotective role in vivo [186]. Finally, similar results were revealed in PC12 cells challenged with increased Aβ1-42 levels [187].
4.3.2. Fucoxanthin
Fucoxanthin is an orange xanthophyll found in brown seaweeds, microalgae or diatoms, such as Phaeodactylum tricornutum and the edible algae U. pinnatifida, Hijika fusiformis, Sargassum fulvellum and L. japonica [165,168,188,189]. Its structure contains an allene moiety, an epoxide group and a conjugated carbonyl group in a polyene chain (Fig. 7b) [165,188,189].
Many studies have shown that fucoxanthin exerts a broad range of biological activities [120,168,189]. Research on the metabolism and toxicity of fucoxanthin revealed fuxoxanthinol and amarouciaxanthin A as the main metabolic products. More specifically, fucoxanthin is hydrolyzed in the gastrointestinal tract by digestive enzymes to fucoxanthinol, which is then converted to amarouciaxanthin A in the liver. Concerning the toxicity, no adverse effects have been observed in animal studies, whereas those that include humans are limited [165,188,189]. Nevertheless, two clinical trials have been launched to elucidate the beneficial effects of daily consumption of fucoxanthin [[189], [190], [191]]. It is worth noting that the FDA has allowed fucoxanthin extracted from P. tricornutum as a dietary supplement [189].
Liu and colleagues suggested that fucoxanthin may act as a pro-oxidant. Six hours incubation of murine embryonic liver BNL CL.2 cells with fucoxanthin induced intracellular ROS production. At the same time, fucoxanthin induced Nrf2 activation via its nuclear translocation, which subsequently upregulated both the mRNA and protein expression levels of HO-1, NQO-1 genes in a concentration-dependent manner. The researchers attributed these effects to the activation of the Nrf2/ARE pathway via the ERK/p38 pathway [192]. Furthermore, fucoxanthin protected against hydrogen peroxide-induced oxidative stress in vitro [193,194] by blocking ROS generation, rescuing cell viability and inducing antioxidant enzymes. The antioxidant activity of fucoxanthin was mediated through the Nrf2 activation in a PI3K-dependent manner [194].
The beneficial effects of fucoxanthin have also been investigated in aging and age-related diseases. Fucoxanthin increased the lifespan of wt D. melanogaster and improved healthspan as shown by the enhancement of fecundity, fertility, food consumption and locomotor activity [195,196]. Fucoxanthin conferred resistance to oxidative stress in the presence of paraquat (a generator of superoxide anion radicals) in flies and induced the expression of genes related to stress response, DNA repair, detoxification of ROS, heat shock response and apoptosis; CncC (the homolog of the mammalian Nrf2 gene) and dSir2/SIRT1 were involved in these responses. These results were dependent on the administered dose of fucoxanthin and the sex of the flies [195]. Nevertheless, contradictory results were also shown by another group [196]. Transcriptome analysis revealed that fucoxanthin downregulated the expression of Hsp70Ab gene, yet it induced the mRNA expression of the antioxidant enzyme SOD3. Importantly, in old flies, fucoxanthin administration induced the expression of 49 genes involved in the MAPK and Wnt signaling pathways and autophagy [196]. Moreover, fucoxanthin prolonged the lifespan of C. elegans [195], yet the mechanism was not revealed. In a recent study, late passage human embryonic lung fibroblasts treated with fucoxanthin exhibited decreased SA-β-Gal activity, while fucoxanthin protected cells against oxidative stress induced by the electron chain complex I inhibitor rotenone. Finally, transcriptome analysis in young and late passage cells treated with fucoxanthin revealed that it promoted differential expression of genes involved in aging-related pathways, such as Wnt and FOXO signaling pathways [197]. Since various proteasome subunits have been shown to be regulated by FOXO/DAF-16 transcription factor [44,45,64,69], the potential fucoxanthin-mediated proteasome modulation cannot be ruled out.
In addition, fucoxanthin inhibited Aβ1-42 fibril formation and aggregation in vitro [187,198] and rescued cells from apoptosis [187]. Xiang and colleagues suggested that this effect was mediated via hydrophobic interactions of fucoxanthin with the Aβ peptide. Also, in Αβ1-42 oligomer-treated mice, fucoxanthin administration alleviated cognitive impairment and improved spatial learning and memory. Moreover, it enhanced the activity of SOD, CAT and GSH in the hippocampus of mice, while it restored the reduced expression of BDNF and ChAT [198]. Data also showed that fucoxanthin possesses neuroprotective properties since it reduced apoptosis in mice with traumatic brain injury. Fucoxanthin administration reduced oxidative stress and decreased lipid peroxidation, while it activated the Nrf2/ARE pathway and Nrf2-dependent autophagy [199]. These results could also imply a potential implication of the UPS. If we take into account that Nrf2 activation induces the transcription of proteasome subunits and the fact that there is a cross-talk between the proteasome and autophagy under stress conditions [200], fucoxanthin could indeed modulate the UPS. Finally, fucoxanthin reduced oxidative stress and protected SH-SY5Y cells and primary cerebellar granule neurons against hydrogen peroxide-induced toxicity. These effects were mediated through the activation of the PI3K/Akt pathway and the inhibition of the pro-apoptotic ERK signaling pathway [201,202].
4.4. Other marine-derived compounds
Besides the compounds belonging to the chemical classes that have been discussed above, other marine-derived compounds have also been characterized as anti-aging or anti-aggregation agents. The extracts from the red algae Acanthophora spicifera, Jania acutilobum and Peyssonnelia sp., which are rich in lipids, prolonged the lifespan of a small multicellular invertebrate animal belonging to the phylum Rotifera named Brachionus manjavacas. Although the extracts possessed antioxidant activity, the observed lifespan extension was not linked to this property [203]. Furthermore, a benzo-1, 3-oxazine xyloketal derivative extracted from the mangrove-derived fungus Xylaria sp. prolonged the lifespan of wt C. elegans and improved its healthspan. It also protected the nematodes from heat stress and delayed the paralysis progression in an AD nematode model (CL4176 strain). Data from docking studies and an hsf-1 mutant of C. elegans revealed that these effects were dependent on HSF-1 [204]. Although proteasome activities were not examined in the animals treated with the extract, given the interconnection of HSF-1 signaling pathway with the proteasome, it is possible that the extract affected the UPS status, among others. The extract of the ubiquitous Mediterranean brown alga Padina pavonica alleviated the adverse effects in viability induced by Aβ42 and α-synuclein aggregates in yeast and D. melanogaster. The extract inhibited Aβ42 and α-synuclein fibril formation in vitro, although the exact mechanism was not revealed [205]. Moreover, zonarol, a sesquiterpene hydroquinone from the brown alga Dictyopteris undulata, ensured cell viability under oxidative stress conditions in vitro by activating the Nrf2/ARE signaling pathway [206]. Finally, gracilins, norditerpenes isolated from the sponge Spongionella sp., have exerted neuroprotective activity. More specifically, gracilins reduced tau phosphorylation in vitro through the inhibition of ERK activation. Likewise, gracilins improved the cognitive impairment in an AD mouse model and inhibited ERK and tau phosphorylation, while Aβ42 levels were found reduced in the mouse brain [207]. Taking into account that gracilins have been suggested to have antioxidant activity and to promote Nrf2 nuclear translocation in vitro [208], we can hypothesize that the neuroprotective properties of these sponge-derived secondary metabolites may be attributed to the Nrf2 activation that may, in turn, modulate the proteasome, among others.
5. Marine-derived compounds against photoaging
Skin is the biggest organ in vertebrates that executes multiple functions, with the maintenance of organismal homeostasis and the protection against harmful microorganisms and substances being among the most significant ones [209,210]. Skin is composed of three layers; epidermis, dermis and subcutaneous tissue [211]. Just like all our body organs, skin also undergoes aging. Apart from intrinsic (chronological) aging, which is an inevitable process occurring in all living organisms, extrinsic aging also affects the skin. Extrinsic aging is caused by environmental factors, such as air pollution, smoking, poor nutrition and exposure to sunlight [209,210,[212], [213], [214], [215]]. Sun exposure (especially the solar UVA and UVB radiation) is the main factor affecting skin photoaging [209,210,213,215], initially suggested in 1969 [210,216]. Several visible signs characterize skin photoaging; coarse wrinkles, laxity, loss of elasticity and hyperpigmentation differentiate photoaging from the intrinsically aged skin [209,210,213], while both types of skin aging share almost the same molecular pathways [213,214].
Many molecular mechanisms have been involved in photoaging [210,211,217]. DNA is directly or indirectly affected by UV radiation. UVB absorption can directly damage the DNA by stimulating pyrimidine dimer generation that results in mutations and chromosomal instability [210,213,214,217]. Skin cells (fibroblasts, keratinocytes) exposed to UVB radiation exhibit DNA damage, cell cycle arrest and various hallmarks of senescence (i.e., SA-β-Gal positivity) [140,210]. The same results were also observed in vivo [210]. On the contrary, UVA radiation indirectly affects DNA since DNA cannot absorb this range of solar radiation. When skin is exposed to UVA, the latter mediates a reaction that produces singlet oxygen radicals, leading to purine base modifications and subsequent DNA mutations [214,217]. Moreover, in contrast with the UVB radiation that mainly affects the epidermis, UVA can penetrate both the epidermis and dermis [213,215], resulting in detrimental effects in the extracellular matrix (ECM). The dermis is composed of the ECM that is structured from collagen, elastin and glycosaminoglycans, which are responsible for the maintenance of the skin strength, elasticity and hydration [211,213].
ROS participate in the process of photoaging with deleterious effects on ECM. During UV exposure, ROS inhibit the receptor protein tyrosine phosphatases (RPTPs) activity and induce the phosphorylation of receptor tyrosine kinases (RTKs), activating downstream signaling pathways [210,211]. Activation of MAPKs results in the induction of the downstream transcription factors, namely activator protein 1 (AP-1) and nuclear factor-kB (NF-kB). AP-1 and NF-kB regulate the transcription of several matrix metalloproteinases (MMPs). The activation of these two transcription factors leads to the increased expression of MMPs, which are secreted in the skin by keratinocytes and dermal fibroblasts, resulting in increased fragmentation of the ECM by triggering the degradation of collagen fibrils [210,211,214,217,218].
Data have shown that the proteasome is also implicated in photoaging. In fact, the decline of proteasome activity was observed in epidermal cells and dermal fibroblasts from middle-aged and aged healthy donors [219,220], as well as in vitro in aged keratinocytes [219] and UV-radiated cells [221,222]. Decreased proteasome activity was accompanied by elevated levels of oxidatively modified proteins [219,221,222] and ROS [220,221], while increased expression of MMP-1, the main matrix metalloproteinase responsible for collagen degradation [217], was also detected [221]. Transcriptomic analysis comparing samples from young and aged skin has not revealed any significant differences in the protein expression of almost all proteasome subunits with the exception of PSMD8 subunit, which encodes the Rpn12 lid subunit. PSMD8 was found reduced by almost 30% in aged skin [223,224]. In contrast, Imbert and colleagues stated that proteasome activation by a dimerized tripeptide led to the reduction of the UVB-induced protein carbonylation and improvement of the senescent morphology of aged keratinocytes in vitro and in skin biopsies [222]. Furthermore, WI-38/T cells that were stably transfected with β5 proteasome subunit exhibited lower mRNA levels of MMP-1 compared to cells transfected with an empty vector [221]. These results suggest that proteasome activation may be a promising strategy against photoaging.
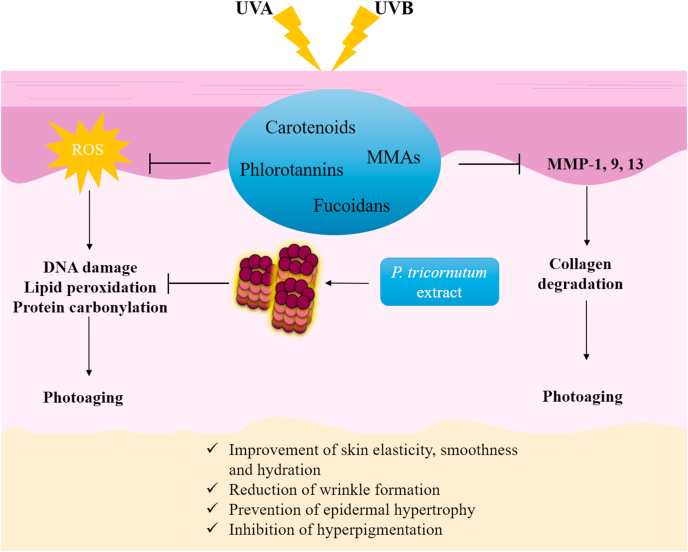
Marine organisms, and in particular those living in the euphotic zone, have received considerable attention in this field. Due to their habitat, they are exposed to extreme environmental conditions, including continuous sun exposure. Thus, they produce photoprotective substances to minimize the harmful damage they are subjected to [107,225]. These characteristics have driven to the exploitation of marine-derived extracts or compounds in the pharmaceutical and cosmetic industries as anti-photoaging and photoprotective products. Their anti-photoaging properties are summarized in Fig. 8.
Fig. 8.
The photoprotective mechanisms of marine-derived compounds.
Exposure to the sun, especially the solar UVA and UVB radiation, promotes reactive oxygen species (ROS) production that enhances DNA damage, lipid peroxidation and protein carbonylation, major causative factors of photoaging. Moreover, sun exposure stimulates the expression of several matrix metalloproteinases (MMPs) that result in increased collagen fibrils degradation contributing to photoaging. Marine-derived compounds (MAAs; Mycosporine-like amino acids, fucoidans, phlorotannins, and carotenoids) prevent photoaging through reduction of ROS generation and inhibition of MMPs expression. The extract from P. tricornutum stimulates proteasome activity resulting in decreased cellular levels of carbonylated and oxidatively damaged proteins. Consequently, marine-derived compounds alleviate photoaging markers leading to improvement of skin elasticity, smoothness and hydration, reduction of wrinkle formation, prevention of epidermal hypertrophy and inhibition of hyperpigmentation.
Mycosporine-like amino acids (MAAs) are among the fascinating substances that can be found in a plethora of marine organisms, including cyanobacteria, phytoplankton, sponges and marine algae, among others [107]. Red algae produce most of the MAAs that have been identified so far [107]. MAAs are typically small (<400 Da), water-soluble and colourless molecules composed of a cyclohexanone or cyclohexenimine chromophore conjugated to the nitrogen substituent of an amino acid or imino alcohol [217,226,227]. The type and concentration of MAAs isolated from marine organisms vary among species and depend on harvesting sites, season, climate, the depth that the organism lives in, and various other environmental factors [107,227]. MAAs' absorption ranges between 268 and 362 nm, covering the UVA and UVB spectrum [227]. They are considered the most potent compounds in nature that can absorb UVA radiation [217].
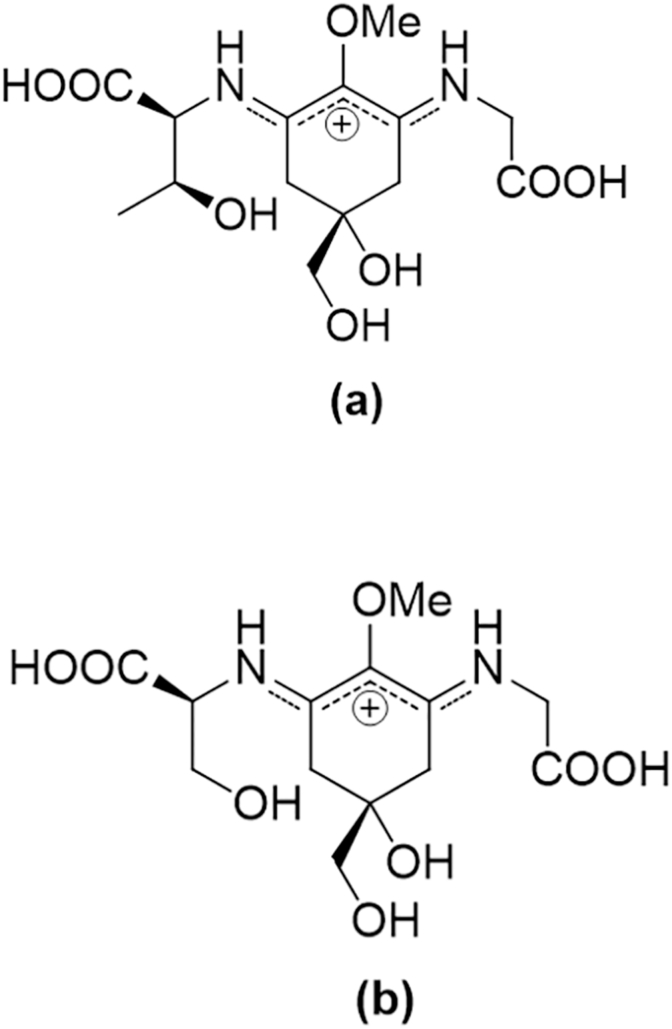
Antioxidation, anti-photoaging, photoprotection and protection against DNA damage are characteristic biological properties of MAAs [107,217,226,227]. Moreover, the fact that the MAAs exert antioxidant activity and suppress the stress induced by UV radiation via the energy dissipation as heat in the absence of ROS generation [217,226,227] is a desirable advantage, as both UV and ROS are closely related to photoaging. Porphyra-334 (Fig. 9a), one of the most studied MAA isolated from Porphyra umbilicalis, was identified as a photoprotectant. Treatment of human skin fibroblasts with porphyra-334 inhibited the UVA-induced ROS intracellular accumulation and exerted anti-aging activity. Moreover, it protected cells against collagen degradation and decreased elastase activity, which are related to wrinkle formation. At the same time, incubation of the UVA-exposed cells with porphyra-334 inhibited the mRNA expression of MMP-1 [228]. In another study, porphyra-334 was applied twice daily on the inner side of the forearm in 20 women. These areas were irradiated with UVA twice a week in a range that mimics the daily UVA radiation that the skin is exposed to. Porphyra-334 offered protection against UVA by reducing lipid peroxidation, improving skin elasticity, smoothness and hydration [229]. The combination of the MAAs, porphyra-334 and shinorine (Fig. 9b), extracted from the red alga Porphyra rosengurttii was applied topically to the dorsal skin of albino hairless mice (Skh: hr-1) followed by UV radiation. The two MAAs acted photoprotectively, rescued the decreased activity of SOD and CAT antioxidant enzymes and elevated the Hsp 70 expression levels [230]. Finally, bioinformatic analysis revealed that mycosporine-glycine, mycosporine-glycine-valine and porphyra-334 were potential activators of the Nrf2 transcription factor, while this observation was confirmed in vitro for porphyra-334 [227,231,232].
Fig. 9.
The chemical structures of the marine-derived mycosporine-like amino acids (a) porphyra-334 and (b) shinorine.
Fucoidans have also been extensively investigated for their anti-photoaging properties. Fucoidan was found to inhibit the UVB-induced mRNA and protein expression of MMP-1 in vitro in a UVB-dependent manner since fucoidan-treated cells without prior exposure to UVB radiation did not exhibit any changes. Also, fucoidan induced the mRNA and protein expression of the type I procollagen [233,234]. In addition, topical application of low molecular weight fucoidan in the skin of HR-1 hairless mice exposed to UVB radiation alleviated photoaging markers (wrinkle formation, inflammation) and decreased the mRNA expression of MMP-1, -9 and -13. Moreover, it increased GSH activity and decreased MDA content and superoxide anion radicals in mice after UVB exposure [235]. Oral administration of fucoidan isolated from the sporophylls of U. pinnatifida in UVB-irradiated mice also exerted protective activity [236]. Furthermore, carrageenans (sulfated polysaccharides extracted from red algae) were found to exert photoprotective activities [107]. Incubation of UVB-treated HaCaT cells with carrageenan rescued cell viability, decreased ROS production and acted as a free radical scavenger in a way similar to vitamin E [237].
Studies revealed that phlorotannins (phloroglucinol, eckol, dieckol) isolated from E. cava and E. stolonifera decreased the intracellular ROS accumulation and prevented DNA damage in cells exposed to UVB radiation [238,239]. Besides, eckol and dieckol from E. stolonifera inhibited MMP-1 expression in human dermal fibroblasts after UVB radiation [239]. Treatment of UVB-exposed HaCaT cells with phloroglucinol inhibited lipid peroxidation, DNA damage and protein carbonylation while scavenging ROS and restoring SOD, CAT and GSH activities [240]. The anti-photoaging properties of phlorotannins were further supported by in vivo experiments. In UVB-radiated zebrafish, dieckol reduced ROS and nitric oxide radical production, rescued cell death and inhibited hyperpigmentation [241,242], while phloroglucinol alleviated lipid peroxidation, protein carbonylation and DNA damage in Balb/c mice after UVB radiation. At the same time, it decreased the number of mast cells, which are involved in the initiation of UVB-induced oxidative stress and inflammation, and improved the epidermal and dermal thickness in the dorsal skin of the radiated mice [243].
Carotenoids have also been proposed as suitable agents for photoaging prevention. Their absorption range, their ability to act photoprotectively in the photosynthetic organisms, along with the observation that they accumulate in both terrestrial and marine plants following enhanced UVB radiation, make them attractive targets [107]. In astaxanthin-treated human dermal fibroblasts, the MMP-1 mRNA and protein expression levels were significantly reduced after UVA radiation [244]. Moreover, astaxanthin administration to UV-radiated hairless mice prevented wrinkle formation, ameliorated skin thickness and reduced the loss of collagen fibers and ROS generation [245,246]. The orally administered astaxanthin was found accumulated in the dermis and epidermis of these mice [245]. Furthermore, fucoxanthin protected human fibroblasts against UVB radiation by maintaining cell viability and reducing ROS levels and DNA damage [247]. In vivo, the topical application of fucoxanthin in hairless mice prevented wrinkle formation and epidermal hypertrophy and reduced the expression of MMP-13 after UVB exposure [248].
In another study, an extract of the diatom P. tricornutum exerted potent photoprotectant activity via proteasome activity stimulation. More specifically, its isopropanol extract, which contained 99% fatty acids and 1% xanthophylls, increased the three proteasome activities both in human purified proteasome and in treated human keratinocyte cells. Proteasome activation was also maintained after UVA and UVB radiation of the treated cells, which was accompanied by a decrease of the oxidatively modified proteins [249]. The P. tricornutum lipid-rich extract was also found to reduce the carbonylated proteins in the human stratum corneum, the outermost layer of the epidermis, after applying the extract twice a day for three weeks [250].
6. Conclusions
The proteasome is a multicatalytic enzymatic complex that orchestrates many cellular functions and assures the maintenance of proteostasis. However, its function declines upon aging, leaving the cell unprotected at the mercy of damaged, misfolded or aggregated proteins, a condition that accelerates the aging process and constitutes a high-risk factor for age-related diseases. Due to the significant role of the proteasome in the surveillance of protein quality control systems, multiple studies have been performed to find means to “rescue” its function as the organism ages, with many of them directed to natural compounds. From ancient times, human beings use natural products for cosmetic purposes, improvement of health or therapeutic reasons (drugs). Thus, significant efforts have been devoted to deciphering the beneficial biological activities of natural compounds and correlate them with conditions that affect their well-being; aging and diseases are among these conditions. Even though various terrestrial compounds have been revealed to enhance proteasome activity and act protectively against aging, marine natural products have been largely unexplored. Despite the fact that only a few studies have directly linked a marine metabolite with the proteasome, many others have illustrated marine-derived compounds to modulate pathways/molecules known to be potentially involved in proteasome modulation, such as FOXO/DAF-16 and Nrf2 transcription factors [34,35,64,65,72,80]. Still, the precise mechanism through which marine natural products act has to be elucidated to strengthen their potential link with proteasome activation. Moreover, efforts have been made towards the improvement of bioaccessibility and bioavailability of marine products. Enhancement of those properties along with their ability to cross the blood-brain barrier could convert marine products to “natural weapons” to prevent or even treat age-related diseases. Taking into account that significant progress has also been achieved towards more efficient and cost-effective methods for the production of marine natural products, the oceans and their “hidden treasures” represent a very promising source of compounds to be used towards longevity and healthspan improvement.
Funding
We acknowledge funding by the European Union and Greek national funds through the Operational Program Competitiveness, Entrepreneurship and Innovation under the Program “Special Actions Aquaculture – Industrial Materials – Open Innovation in Culture” (project ALGOSMETIC, Τ6ΥΒΠ-00474). We also acknowledge funding by Greece and the European Union (European Social Fund- ESF) through the Operational Program “Human Resources Development, Education and Lifelong Learning 2014–2020″ in the context of the project “Proteostasis” (MIS 5050341).
Declaration of competing interest
None.
Acknowledgments
We apologize to all the authors whose work we could not cite due to space limitations.
Contributor Information
Mary Α. Vasilopoulou, Email: mvasilopoulou@eie.gr.
Efstathia Ioannou, Email: eioannou@pharm.uoa.gr.
Vassilios Roussis, Email: roussis@pharm.uoa.gr.
Niki Chondrogianni, Email: nikichon@eie.gr.
References
- 1.Gu Z.C., Enenkel C. Proteasome assembly, cell. Mol. Life Sci. 2014;71:4729–4745. doi: 10.1007/s00018-014-1699-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yu H., Matouschek A. Recognition of client proteins by the proteasome. Annu. Rev. Biophys. 2017;46:149–173. doi: 10.1146/annurev-biophys-070816-033719. [DOI] [PubMed] [Google Scholar]
- 3.Enenkel C. Proteasome dynamics. Biochim. Biophys. Acta Mol. Cell Res. 2014;1843:39–46. doi: 10.1016/j.bbamcr.2013.03.023. [DOI] [PubMed] [Google Scholar]
- 4.Bard J.A.M., Goodall E.A., Greene E.R., Jonsson E., Dong K.C., Martin A. Structure and function of the 26S proteasome. Annu. Rev. Biochem. 2018;87:27.1–27.28. doi: 10.1146/annurev-biochem. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rousseau A., Bertolotti A. Regulation of proteasome assembly and activity in health and disease. Nat. Rev. Mol. Cell Biol. 2018;19:697–712. doi: 10.1038/s41580-018-0040-z. [DOI] [PubMed] [Google Scholar]
- 6.Collins G.A., Goldberg A.L. The logic of the 26S proteasome. Cell. 2017;169:792–806. doi: 10.1016/j.cell.2017.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zheng N., Shabek N. Ubiquitin ligases: structure, function, and regulation. Annu. Rev. Biochem. 2017;86:129–157. doi: 10.1146/annurev-biochem-060815-014922. [DOI] [PubMed] [Google Scholar]
- 8.Saeki Y. Ubiquitin recognition by the proteasome. J. Biochem. 2017;161:113–124. doi: 10.1093/jb/mvw091. [DOI] [PubMed] [Google Scholar]
- 9.Livneh I., Kravtsova-Ivantsiv Y., Braten O., Kwon Y.T., Ciechanover A. Monoubiquitination joins polyubiquitination as an esteemed proteasomal targeting signal. Bioessays. 2017;39 doi: 10.1002/bies.201700027. [DOI] [PubMed] [Google Scholar]
- 10.Deshaies R.J., Joazeiro C.A.P. RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 11.Finley D., Prado M.A. The proteasome and its network: engineering for adaptability. Cold Spring Harb. Perspect. Biol. 2020;12 doi: 10.1101/cshperspect.a033985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grice G.L., Nathan J.A. The recognition of ubiquitinated proteins by the proteasome. Cell. Mol. Life Sci. 2016;73:3497–3506. doi: 10.1007/s00018-016-2255-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pickering A.M., Davies K.J.A. Prog. Mol. Biol. Transl. Sci. Elsevier B.V.; 2012. Degradation of damaged proteins: the main function of the 20S proteasome; pp. 227–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raynes R., Pomatto L.C.D., Davies K.J.A. Degradation of oxidized proteins by the proteasome: distinguishing between the 20S, 26S, and immunoproteasome proteolytic pathways. Mol. Aspect. Med. 2016;50:41–55. doi: 10.1016/j.mam.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davies K.J.A. Degradation of oxidized proteins by the 20S proteasome. Biochimie. 2001;83:301–310. doi: 10.1016/S0300-9084(01)01250-0. [DOI] [PubMed] [Google Scholar]
- 16.Ben-Nissan G., Sharon M. Regulating the 20S proteasome ubiquitin-independent degradation pathway. Biomolecules. 2014;4:862–884. doi: 10.3390/biom4030862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Njomen E., Tepe J.J. Proteasome activation as a new therapeutic approach to target proteotoxic disorders. J. Med. Chem. 2019;62:6469–6481. doi: 10.1021/acs.jmedchem.9b00101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Budenholzer L., Cheng C.L., Li Y., Hochstrasser M. Proteasome structure and assembly. J. Mol. Biol. 2017;429:3500–3524. doi: 10.1016/j.jmb.2017.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Livneh I., Cohen-Kaplan V., Cohen-Rosenzweig C., Avni N., Ciechanover A. The life cycle of the 26S proteasome: from birth, through regulation and function, and onto its death. Cell Res. 2016;26:869–885. doi: 10.1038/cr.2016.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Groll M., Bajorek M., Köhler A., Moroder L., Rubin D.M., Huber R., Glickman M.H., Finley D. A gated channel into the proteasome core particle. Nat. Struct. Biol. 2000;7:1062–1067. doi: 10.1038/80992. [DOI] [PubMed] [Google Scholar]
- 21.Ebstein F., Kloetzel P.M., Krüger E., Seifert U. Emerging roles of immunoproteasomes beyond MHC class i antigen processing. Cell. Mol. Life Sci. 2012;69:2543–2558. doi: 10.1007/s00018-012-0938-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kniepert A., Groettrup M. The unique functions of tissue-specific proteasomes. Trends Biochem. Sci. 2014;39:17–24. doi: 10.1016/j.tibs.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 23.Johnston-Carey H.K., Pomatto L.C.D., Davies K.J.A. The Immunoproteasome in oxidative stress, aging, and disease. Crit. Rev. Biochem. Mol. Biol. 2016;51:268–281. doi: 10.3109/10409238.2016.1172554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kloetzel P.M. Generation of major histocompatibility complex class I antigens: functional interplay between proteasomes and TPPII. Nat. Immunol. 2004;5:661–669. doi: 10.1038/ni1090. [DOI] [PubMed] [Google Scholar]
- 25.Pickering A.M., Koop A.L., Teoh C.Y., Ermak G., Grune T., Davies K.J.A. The immunoproteasome, the 20S proteasome and the PA28αβ proteasome regulator are oxidative-stress-adaptive proteolytic complexes. Biochem. J. 2010;432:585–594. doi: 10.1042/BJ20100878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hussong S.A., Kapphahn R.J., Phillips S.L., Maldonado M., Ferrington D.A. Immunoproteasome deficiency alters retinal proteasome's response to stress. J. Neurochem. 2010;113:1481–1490. doi: 10.1111/j.1471-4159.2010.06688.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stadtmueller B.M., Hill C.P. Proteasome activators. Mol. Cell. 2011;41:8–19. doi: 10.1016/j.molcel.2010.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pickering A.M., Linder R.A., Zhang H., Forman H.J., Davies K.J.A. Nrf2-dependent induction of proteasome and Pa28αβ regulator are required for adaptation to oxidative stress. J. Biol. Chem. 2012;287:10021–10031. doi: 10.1074/jbc.M111.277145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vilchez D., Saez I., Dillin A. The role of protein clearance mechanisms in organismal ageing and age-related diseases. Nat. Commun. 2014;5:1–13. doi: 10.1038/ncomms6659. [DOI] [PubMed] [Google Scholar]
- 30.López-Otín C., Blasco M.A., Partridge L., Serrano M., Kroemer G. The hallmarks of aging. Cell. 2013;153:1194. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hipp M.S., Kasturi P., Hartl F.U. The proteostasis network and its decline in ageing. Nat. Rev. Mol. Cell Biol. 2019;20:421–435. doi: 10.1038/s41580-019-0101-y. [DOI] [PubMed] [Google Scholar]
- 32.Carrard G., Bulteau A.L., Petropoulos I., Friguet B. Impairment of proteasome structure and function in aging. Int. J. Biochem. Cell Biol. 2002;34:1461–1474. doi: 10.1016/S1357-2725(02)00085-7. [DOI] [PubMed] [Google Scholar]
- 33.Kevei É., Hoppe T. Ubiquitin sets the timer: impacts on aging and longevity. Nat. Struct. Mol. Biol. 2014;21:290–292. doi: 10.1038/nsmb.2806. [DOI] [PubMed] [Google Scholar]
- 34.Chondrogianni N., Sakellari M., Lefaki M., Papaevgeniou N., Gonos E.S. Proteasome activation delays aging in vitro and in vivo. Free Radic. Biol. Med. 2014;71:303–320. doi: 10.1016/j.freeradbiomed.2014.03.031. [DOI] [PubMed] [Google Scholar]
- 35.Papaevgeniou N., Chondrogianni N. Methods Mol. Biol. Humana Press Inc.; 2016. UPS activation in the battle against aging and aggregation-related diseases: an extended review. [DOI] [PubMed] [Google Scholar]
- 36.Baraibar M.A., Friguet B. Prog. Mol. Biol. Transl. Sci. first ed. Elsevier B.V.; 2012. Changes of the proteasomal system during the aging process; pp. 249–275. [DOI] [PubMed] [Google Scholar]
- 37.Bellavista E., Martucci M., Vasuri F., Santoro A., Mishto M., Kloss A., Capizzi E., Degiovanni A., Lanzarini C., Remondini D., Dazzi A., Pellegrini S., Cescon M., Capri M., Salvioli S., D'Errico-Grigioni A., Dahlmann B., Grazi G.L., Franceschi C. Lifelong maintenance of composition, function and cellular/subcellular distribution of proteasomes in human liver. Mech. Ageing Dev. 2014;141–142:26–34. doi: 10.1016/j.mad.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 38.Chondrogianni N., Stratford F.L.L., Trougakos I.P., Friguet B., Rivett A.J., Gonos E.S. Central role of the proteasome in senescence and survival of human fibroblasts. Induction of a senescence-like phenotype upon its inhibition and resistance to stress upon its activation. J. Biol. Chem. 2003;278:28026–28037. doi: 10.1074/jbc.M301048200. [DOI] [PubMed] [Google Scholar]
- 39.Chen Q., Ding Q., Thorpe J., Dohmen R.J., Keller J.N. RNA interference toward UMP1 induces proteasome inhibition in Saccharomyces cerevisiae: evidence for protein oxidation and autophagic cell death. Free Radic. Biol. Med. 2005;38:226–234. doi: 10.1016/j.freeradbiomed.2004.10.019. [DOI] [PubMed] [Google Scholar]
- 40.Chen Q., Thorpe J., Dohmen J.R., Li F., Keller J.N. Ump1 extends yeast lifespan and enhances viability during oxidative stress: central role for the proteasome? Free Radic. Biol. Med. 2006;40:120–126. doi: 10.1016/j.freeradbiomed.2005.08.048. [DOI] [PubMed] [Google Scholar]
- 41.Kelmer Sacramento E., Kirkpatrick J.M., Mazzetto M., Baumgart M., Bartolome A., Di Sanzo S., Caterino C., Sanguanini M., Papaevgeniou N., Lefaki M., Childs D., Bagnoli S., Terzibasi Tozzini E., Di Fraia D., Romanov N., Sudmant P.H., Huber W., Chondrogianni N., Vendruscolo M., Cellerino A., Ori A. Reduced proteasome activity in the aging brain results in ribosome stoichiometry loss and aggregation. Mol. Syst. Biol. 2020 doi: 10.15252/msb.20209596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vernace V.A., Arnaud L., Schmidt‐Glenewinkel T., Figueiredo‐Pereira M.E. Aging perturbs 26S proteasome assembly in Drosophila melanogaster. Faseb. J. 2007;21:2672–2682. doi: 10.1096/fj.06-6751com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nguyen N.N., Rana A., Goldman C., Moore R., Tai J., Hong Y., Shen J., Walker D.W., Hur J.H. Proteasome β5 subunit overexpression improves proteostasis during aging and extends lifespan in Drosophila melanogaster. Sci. Rep. 2019;9:3170. doi: 10.1038/s41598-019-39508-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chondrogianni N., Georgila K., Kourtis N., Tavernarakis N., Gonos E.S. 20S proteasome activation promotes life span extension and resistance to proteotoxicity in Caenorhabditis elegans. Faseb. J. 2015;29:611–622. doi: 10.1096/fj.14-252189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vilchez D., Morantte I., Liu Z., Douglas P.M., Merkwirth C., Rodrigues A.P.C., Manning G., Dillin A. RPN-6 determines C. elegans longevity under proteotoxic stress conditions. Nature. 2012;489:263–268. doi: 10.1038/nature11315. [DOI] [PubMed] [Google Scholar]
- 46.Tomaru U., Takahashi S., Ishizu A., Miyatake Y., Gohda A., Suzuki S., Ono A., Ohara J., Baba T., Murata S., Tanaka K., Kasahara M. Decreased proteasomal activity causes age-related phenotypes and promotes the development of metabolic abnormalities. Am. J. Pathol. 2012;180:963–972. doi: 10.1016/j.ajpath.2011.11.012. [DOI] [PubMed] [Google Scholar]
- 47.Rodriguez K.A., Edrey Y.H., Osmulski P., Gaczynska M., Buffenstein R. Altered composition of liver proteasome assemblies contributes to enhanced proteasome activity in the exceptionally long-lived naked mole-rat. PloS One. 2012;7 doi: 10.1371/journal.pone.0035890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chondrogianni N., Petropoulos I., Franceschi C., Friguet B., Gonos E.S. Fibroblast cultures from healthy centenarians have an active proteasome. Exp. Gerontol. 2000;35:721–728. doi: 10.1016/s0531-5565(00)00137-6. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=11053662 [DOI] [PubMed] [Google Scholar]
- 49.Zheng Q., Huang T., Zhang L., Zhou Y., Luo H., Xu H., Wang X. Dysregulation of ubiquitin-proteasome system in neurodegenerative diseases. Front. Aging Neurosci. 2016;8 doi: 10.3389/fnagi.2016.00303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zheng C., Geetha T., Babu J.R. Failure of ubiquitin proteasome system: risk for neurodegenerative diseases. Neurodegener. Dis. 2014;14:161–175. doi: 10.1159/000367694. [DOI] [PubMed] [Google Scholar]
- 51.Keller J.N., Hanni K.B., Markesbery W.R. Impaired proteasome function in Alzheimer's disease. J. Neurochem. 2000;75:436–439. doi: 10.1046/j.1471-4159.2000.0750436.x. [DOI] [PubMed] [Google Scholar]
- 52.López Salon M., M L., C E.M., S E.F., P J.M. Defective ubiquitination of cerebral proteins in alzheimer's disease. J. Neurosci. Res. 2000;62 doi: 10.1002/1097-4547(20001015)62:2<302::AID-JNR15>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 53.McNaught K.S.P., Belizaire R., Isacson O., Jenner P., Olanow C.W. Altered proteasomal function in sporadic Parkinson's disease. Exp. Neurol. 2003;179:38–46. doi: 10.1006/exnr.2002.8050. [DOI] [PubMed] [Google Scholar]
- 54.Seo H., Sonntag K.C., Isacson O. Generalized brain and skin proteasome inhibition in Huntington's disease. Ann. Neurol. 2004;56:319–328. doi: 10.1002/ana.20207. [DOI] [PubMed] [Google Scholar]
- 55.Kenyon C.J. The genetics of ageing. Nature. 2010;464:504–512. doi: 10.1038/nature08980. [DOI] [PubMed] [Google Scholar]
- 56.Mathew R., Pal Bhadra M., Bhadra U. Insulin/insulin-like growth factor-1 signalling (IIS) based regulation of lifespan across species. Biogerontology. 2017;18:35–53. doi: 10.1007/s10522-016-9670-8. [DOI] [PubMed] [Google Scholar]
- 57.Kenyon C. The first long-lived mutants: discovery of the insulin/IGF-1 pathway for ageing. Philos. Trans. R. Soc. B Biol. Sci. 2011;366:9–16. doi: 10.1098/rstb.2010.0276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hesp K., Smant G., Kammenga J.E. Caenorhabditis elegans DAF-16/FOXO transcription factor and its mammalian homologs associate with age-related disease. Exp. Gerontol. 2015;72:1–7. doi: 10.1016/j.exger.2015.09.006. [DOI] [PubMed] [Google Scholar]
- 59.Altintas O., Park S., Lee S.J.V. The role of insulin/IGF-1 signaling in the longevity of model invertebrates, C. elegans and D. melanogaster, BMB Rep. 2016;49:81–92. doi: 10.5483/BMBRep.2016.49.2.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pan H., Finkel T. Key proteins and pathways that regulate lifespan. J. Biol. Chem. 2017;292:6452–6460. doi: 10.1074/jbc.R116.771915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Murphy C.T., Hu P.J. Insulin/insulin-like growth factor signaling in C. elegans. Worm. 2013:1–43. doi: 10.1895/wormbook.1.164.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mukhopadhyay A., Oh S.W., Tissenbaum H.A. Worming pathways to and from DAF-16/FOXO. Exp. Gerontol. 2006;41:928–934. doi: 10.1016/j.exger.2006.05.020. [DOI] [PubMed] [Google Scholar]
- 63.Brunet A., Sweeney L.B., Sturgill J.F., Chua K.F., Greer P.L., Lin Y., Tran H., Ross S.E., Mostoslavsy R., Cohen H.Y., Hu L.S., Cheng H.L., Jedrychowski M.P., Gygi S.P., Sinclair D.A., Alt F.W., Greenberg M.E. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. 80. [DOI] [PubMed] [Google Scholar]
- 64.Matilainen O., Arpalahti L., Rantanen V., Hautaniemi S., Holmberg C.I. Insulin/IGF-1 signaling regulates proteasome activity through the deubiquitinating enzyme UBH-4. Cell Rep. 2013;3:1980–1995. doi: 10.1016/j.celrep.2013.05.012. [DOI] [PubMed] [Google Scholar]
- 65.Webb A.E., Brunet A. FOXO transcription factors: key regulators of cellular quality control. Trends Biochem. Sci. 2014;39:159–169. doi: 10.1016/j.tibs.2014.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Motosugi R., Murata S. Dynamic regulation of proteasome expression. Front. Mol. Biosci. 2019;6:30. doi: 10.3389/fmolb.2019.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li S., Armstrong C.M., Bertin N., Ge H., Milstein S., Boxem M., Vidalain P.O., Han J.D.J., Chesneau A., Hao T., Goldberg D.S., Li N., Martinez M., Rual J.F., Lamesch P., Xu L., Tewari M., Wong S.L., Zhang L.V., Berriz G.F., Jacotot L., Vaglio P., Reboul J., Hirozane-Kishikawa T., Li Q., Gabel H.W., Elewa A., Baumgartner B., Rose D.J., Yu H., Bosak S., Sequerra R., Fraser A., Mango S.E., Saxton W.M., Strome S., Van Den Heuvel S.v., Piano F., Vandenhaute J., Sardet C., Gerstein M., Doucette-Stamm L., Gunsalus K.C., Harper J.W., Cusick M.E., Roth F.P., Hill D.E., Vidal M. A map of the interactome network of the metazoan C. elegans. Science 84. 2004;303:540–543. doi: 10.1126/science.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yao T., Song L., Xu W., DeMartino G.N., Florens L., Swanson S.K., Washburn M.P., Conaway R.C., Conaway J.W., Cohen R.E. Proteasome recruitment and activation of the Uch37 deubiquitinating enzyme by Adrm1. Nat. Cell Biol. 2006;8:994–1002. doi: 10.1038/ncb1460. [DOI] [PubMed] [Google Scholar]
- 69.Kapetanou M., Tain L.S., Nespital T., Partridge L., Gonos E.S. FoxO1 is a novel regulator of 20S proteasome subunits expression and activity. Front. Cell Dev. Biol. 2021;9:169. doi: 10.3389/FCELL.2021.625715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lefaki M., Papaevgeniou N., Chondrogianni N. Redox regulation of proteasome function. Redox Biol. 2017;13:452–458. doi: 10.1016/j.redox.2017.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Korovila I., Hugo M., Castro J.P., Weber D., Höhn A., Grune T., Jung T. Proteostasis, oxidative stress and aging. Redox Biol. 2017;13:550–567. doi: 10.1016/j.redox.2017.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kapeta S., Chondrogianni N., Gonos E.S. Nuclear erythroid factor 2-mediated proteasome activation delays senescence in human fibroblasts. J. Biol. Chem. 2010;285:8171–8184. doi: 10.1074/jbc.M109.031575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kwak M.-K., Wakabayashi N., Greenlaw J.L., Yamamoto M., Kensler T.W. Antioxidants enhance mammalian proteasome expression through the keap1-Nrf2 signaling pathway. Mol. Cell Biol. 2003;23:8786–8794. doi: 10.1128/mcb.23.23.8786-8794.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pajares M., Cuadrado A., Rojo A.I. Modulation of proteostasis by transcription factor NRF2 and impact in neurodegenerative diseases. Redox Biol. 2017;11:543–553. doi: 10.1016/j.redox.2017.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ma Q. Role of Nrf2 in oxidative stress and toxicity. Annu. Rev. Pharmacol. Toxicol. 2013;53:401–426. doi: 10.1146/annurev-pharmtox-011112-140320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang J., Wang X., Vikash V., Ye Q., Wu D., Liu Y., Dong W. ROS and ROS-mediated cellular signaling. Oxid. Med. Cell. Longev. 2016 doi: 10.1155/2016/4350965. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang Y., Du Y., Le W., Wang K., Kieffer N., Zhang J. Redox control of the survival of healthy and diseased cells. Antioxidants Redox Signal. 2011;15:2867–2902. doi: 10.1089/ars.2010.3685. [DOI] [PubMed] [Google Scholar]
- 78.Niture S.K., Khatri R., Jaiswal A.K. Regulation of Nrf2 - an update. Free Radic. Biol. Med. 2014;66:36–44. doi: 10.1016/j.freeradbiomed.2013.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chondrogianni N., Vasilopoulou M.A., Kapetanou M., Gonos E.S. Proteasome modulation: a way to delay aging? In: Rattan S.I.S., editor. Encycl. Biomed. Gerontol. Academic Press; Oxford: 2020. pp. 92–104. [DOI] [Google Scholar]
- 80.Papaevgeniou N., Sakellari M., Jha S., Tavernarakis N., Holmberg C.I., Gonos E.S., Chondrogianni N. 18 alpha-Glycyrrhetinic acid proteasome activator decelerates aging and alzheimer's disease progression in Caenorhabditis elegans and neuronal cultures. Antioxidants Redox Signal. 2016;25:855–869. doi: 10.1089/ars.2015.6494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tanigawa S., Fujii M., Hou D.X. Action of Nrf2 and Keap1 in ARE-mediated NQO1 expression by quercetin. Free Radic. Biol. Med. 2007;42:1690–1703. doi: 10.1016/j.freeradbiomed.2007.02.017. [DOI] [PubMed] [Google Scholar]
- 82.Chondrogianni N., Kapeta S., Chinou I., Vassilatou K., Papassideri I., Gonos E.S. Anti-ageing and rejuvenating effects of quercetin. Exp. Gerontol. 2010;45:763–771. doi: 10.1016/j.exger.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 83.Liu X.P., Goldring C.E.P., Copple I.M., Wang H.Y., Wei W., Kitteringham N.R., Park B.K. Extract of Ginkgo biloba induces phase 2 genes through Keap1-Nrf2-ARE signaling pathway. Life Sci. 2007;80:1586–1591. doi: 10.1016/j.lfs.2007.01.034. [DOI] [PubMed] [Google Scholar]
- 84.Stark M., Behl C. The Ginkgo biloba extract EGb 761 modulates proteasome activity and polyglutamine protein aggregation, Evidence-Based Complement. Alternative Med. 2014 doi: 10.1155/2014/940186. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Anckar J., Sistonen L. Regulation of HSF1 function in the heat stress response: implications in aging and disease. Annu. Rev. Biochem. 2011;80:1089–1115. doi: 10.1146/annurev-biochem-060809-095203. [DOI] [PubMed] [Google Scholar]
- 86.Hsu A.L., Murphy C.T., Kenyon C. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science. 2003;300:1142–1145. doi: 10.1126/science.1083701. 80. [DOI] [PubMed] [Google Scholar]
- 87.Morley J.F., Morimoto R.I. Regulation of longevity in Caenorhabditis elegans by heat shock factor and molecular chaperones. Mol. Biol. Cell. 2004;15:657–664. doi: 10.1091/mbc.E03-07-0532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bianchi M., Crinelli R., Arbore V., Magnani M. Induction of ubiquitin C (UBC) gene transcription is mediated by HSF1: role of proteotoxic and oxidative stress. FEBS Open Bio. 2018;8:1471–1485. doi: 10.1002/2211-5463.12484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kinet M.J., Malin J.A., Abraham M.C., Blum E.S., Silverman M.R., Lu Y., Shaham S. HSF-1 activates the ubiquitin proteasome system to promote non-apoptotic developmental cell death in C. elegans. Elife. 2016;5 doi: 10.7554/eLife.12821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Paiva F.A., Bonomo L. de F., Boasquivis P.F., de Paula I.T.B.R., Guerra J.F. da C., Leal W.M., Silva M.E., Pedrosa M.L., Oliveira R. de P. Carqueja (Baccharis trimera) protects against oxidative stress and β-amyloid-induced toxicity in Caenorhabditis elegans. Oxid. Med. Cell. Longev. 2015:740162. doi: 10.1155/2015/740162. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Reeg S., Grune T. Protein oxidation in aging: does it play a role in aging progression? Antioxidants Redox Signal. 2015;23:239–255. doi: 10.1089/ars.2014.6062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ye Z.W., Zhang J., Townsend D.M., Tew K.D. Oxidative stress, redox regulation and diseases of cellular differentiation. Biochim. Biophys. Acta Gen. Subj. 2015;1850:1607–1621. doi: 10.1016/j.bbagen.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bader N., Grune T. Biol. Chem. 2006. Protein oxidation and proteolysis; pp. 1351–1355. [DOI] [PubMed] [Google Scholar]
- 94.Rhee S.G., Kang S.W., Jeong W., Chang T.S., Yang K.S., Woo H.A. Intracellular messenger function of hydrogen peroxide and its regulation by peroxiredoxins. Curr. Opin. Cell Biol. 2005;17:183–189. doi: 10.1016/j.ceb.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 95.Kriegenburg F., Poulsen E.G., Koch A., Krüger E., Hartmann-Petersen R. Redox control of the ubiquitin-proteasome system: from molecular mechanisms to functional significance. Antioxidants Redox Signal. 2011;15:2265–2299. doi: 10.1089/ars.2010.3590. [DOI] [PubMed] [Google Scholar]
- 96.Höhn A., König J., Grune T. Protein oxidation in aging and the removal of oxidized proteins. J. Proteomics. 2013;92:132–159. doi: 10.1016/j.jprot.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 97.Reinheckel T., Sitte N., Ullrich O., Kuckelkorn U., Davies K.J.A., Grune T. Comparative resistance of the 20 S and 26 S proteasome to oxidative stress. Biochem. J. 1998;335:637–642. doi: 10.1042/bj3350637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Reinheckel T., Ullrich O., Sitte N., Grune T. Differential impairment of 20S and 26S proteasome activities in human hematopoietic K562 cells during oxidative stress. Arch. Biochem. Biophys. 2000;377:65–68. doi: 10.1006/abbi.2000.1717. [DOI] [PubMed] [Google Scholar]
- 99.Silva G.M., Netto L.E.S., Simões V., Santos L.F.A., Gozzo F.C., Demasi M.A.A., Oliveira C.L.P., Bicev R.N., Klitzke C.F., Sogayar M.C., Demasi M. Redox control of 20S proteasome gating. Antioxidants Redox Signal. 2012;16:1183–1194. doi: 10.1089/ars.2011.4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Harman D. Aging: a theory based on free radical and radiation chemistry. J. Gerontol. 1956;11:298–300. doi: 10.1093/geronj/11.3.298. [DOI] [PubMed] [Google Scholar]
- 101.Viña J., Borras C., Abdelaziz K.M., Garcia-Valles R., Gomez-Cabrera M.C. The free radical theory of aging revisited: the cell signaling disruption theory of aging. Antioxidants Redox Signal. 2013;19:779–787. doi: 10.1089/ars.2012.5111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wickens A.P. Respir. Physiol. 2001. Ageing and the free radical theory; pp. 379–391. [DOI] [PubMed] [Google Scholar]
- 103.Höhn A., Weber D., Jung T., Ott C., Hugo M., Kochlik B., Kehm R., König J., Grune T., Castro J.P. Happily (n)ever after: aging in the context of oxidative stress, proteostasis loss and cellular senescence. Redox Biol. 2017;11:482–501. doi: 10.1016/j.redox.2016.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhang H., Davies K.J.A., Forman H.J. Oxidative stress response and Nrf2 signaling in aging. Free Radic. Biol. Med. 2015;88:314–336. doi: 10.1016/j.freeradbiomed.2015.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chondrogianni N., Voutetakis K., Kapetanou M., Delitsikou V., Papaevgeniou N., Sakellari M., Lefaki M., Filippopoulou K., Gonos E.S. Proteasome activation: an innovative promising approach for delaying aging and retarding age-related diseases. Ageing Res. Rev. 2015;23:37–55. doi: 10.1016/j.arr.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 106.Pangestuti R., Kim S.K. Neuroprotective effects of marine algae. Mar. Drugs. 2011;9:803–818. doi: 10.3390/md9050803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Pangestuti R., Siahaan E.A., Kim S.K. Photoprotective substances derived from marine algae. Mar. Drugs. 2018;16 doi: 10.3390/md16110399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Panagiotidou E., Chondrogianni N. We are what we eat: ubiquitin-proteasome system (UPS) modulation through dietary products. Adv. Exp. Med. Biol. 2020;1233:329–348. doi: 10.1007/978-3-030-38266-7_15. [DOI] [PubMed] [Google Scholar]
- 109.Ahmed A.B.A., Adel M., Karimi P., Peidayesh M. Adv. Food Nutr. Res. 2014. Pharmaceutical, cosmeceutical, and traditional applications of marine carbohydrates; pp. 197–220. [DOI] [PubMed] [Google Scholar]
- 110.Ruocco N., Costantini S., Guariniello S., Costantini M. Polysaccharides from the marine environment with pharmacological, cosmeceutical and nutraceutical potential. Molecules. 2016;21 doi: 10.3390/molecules21050551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ngo D.H., Kim S.K. Sulfated polysaccharides as bioactive agents from marine algae. Int. J. Biol. Macromol. 2013;62:70–75. doi: 10.1016/j.ijbiomac.2013.08.036. [DOI] [PubMed] [Google Scholar]
- 112.Pangestuti R., Kim S.K. Marine-derived bioactive materials for neuroprotection. Food Sci. Biotechnol. 2013;22:1175–1186. doi: 10.1007/s10068-013-0200-z. [DOI] [Google Scholar]
- 113.De Jesus Raposo M.F., De Morais A.M.B., De Morais R.M.S.C. Marine polysaccharides from algae with potential biomedical applications. Mar. Drugs. 2015;13:2967–3028. doi: 10.3390/md13052967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kim J.H., Lee J.E., Kim K.H., Kang N.J. Beneficial effects of marine algae-derived carbohydrates for skin health. Mar. Drugs. 2018;16 doi: 10.3390/md16110459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Jiao G., Yu G., Zhang J., Ewart H.S. Chemical structures and bioactivities of sulfated polysaccharides from marine algae. Mar. Drugs. 2011;9:196–223. doi: 10.3390/md9020196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Muhamad I.I., Zulkifli N., a/p Selvakumaran S., Lazim N.A.M. Bioactive algal-derived polysaccharides: multi-functionalization, therapeutic potential and biomedical applications. Curr. Pharmaceut. Des. 2019;25:1147–1162. doi: 10.2174/1381612825666190618152133. [DOI] [PubMed] [Google Scholar]
- 117.Wang H.M.D., Li X.C., Lee D.J., Chang J.S. Potential biomedical applications of marine algae. Bioresour. Technol. 2017;244:1407–1415. doi: 10.1016/j.biortech.2017.05.198. [DOI] [PubMed] [Google Scholar]
- 118.Kang H.K., Seo C.H., Park Y. The effects of marine carbohydrates and glycosylated compounds on human health. Int. J. Mol. Sci. 2015;16:6018–6056. doi: 10.3390/ijms16036018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kylin H. Biochemistry of sea algae. Phys. Chem. 1913;83:171–197. [Google Scholar]
- 120.Balboa E.M., Conde E., Moure A., Falqué E., Domínguez H. In vitro antioxidant properties of crude extracts and compounds from brown algae. Food Chem. 2013;138:1764–1785. doi: 10.1016/j.foodchem.2012.11.026. [DOI] [PubMed] [Google Scholar]
- 121.Costa L.S., Fidelis G.P., Cordeiro S.L., Oliveira R.M., Sabry D.A., Câmara R.B.G., Nobre L.T.D.B., Costa M.S.S.P., Almeida-Lima J., Farias E.H.C., Leite E.L., Rocha H.A.O. Biological activities of sulfated polysaccharides from tropical seaweeds. Biomed. Pharmacother. 2010;64:21–28. doi: 10.1016/j.biopha.2009.03.005. [DOI] [PubMed] [Google Scholar]
- 122.Wang J., Zhang Q., Zhang Z., Li Z. Antioxidant activity of sulfated polysaccharide fractions extracted from Laminaria japonica. Int. J. Biol. Macromol. 2008;42:127–132. doi: 10.1016/j.ijbiomac.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 123.Ryu M.J., Chung H.S. Fucoidan reduces oxidative stress by regulating the gene expression of HO-1 and SOD-1 through the Nrf2/ERK signaling pathway in HaCaT cells. Mol. Med. Rep. 2016;14:3255–3260. doi: 10.3892/mmr.2016.5623. [DOI] [PubMed] [Google Scholar]
- 124.Kim E.A., Lee S.H., Ko C.I., Cha S.H., Kang M.C., Kang S.M., Ko S.C., Lee W.W., Ko J.Y., Lee J.H., Kang N., Oh J.Y., Ahn G., Jee Y.H., Jeon Y.J. Protective effect of fucoidan against AAPH-induced oxidative stress in zebrafish model. Carbohydr. Polym. 2014;102:185–191. doi: 10.1016/j.carbpol.2013.11.022. [DOI] [PubMed] [Google Scholar]
- 125.Z Y., X M., H C., L A., C J., G C., Z X., Y C., Tong H., W M., C P. Sargassum fusiforme fucoidan SP2 extends the lifespan of Drosophila melanogaster by upregulating the Nrf2-mediated antioxidant signaling pathway. Oxid. Med. Cell. Longev. 2019 doi: 10.1155/2019/8918914. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Rahnasto-Rilla M.K., McLoughlin P., Kulikowicz T., Doyle M., Bohr V.A., Lahtela-Kakkonen M., Ferrucci L., Hayes M., Moaddel R. The identification of a SIRT6 activator from brown algae Fucus distichus. Mar. Drugs. 2017;15 doi: 10.3390/md15060190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Jhamandas J.H., Wie M.B., Harris K., MacTavish D., Kar S. Fucoidan inhibits cellular and neurotoxic effects of β-amyloid (Aβ) in rat cholinergic basal forebrain neurons. Eur. J. Neurosci. 2005;21:2649–2659. doi: 10.1111/j.1460-9568.2005.04111.x. [DOI] [PubMed] [Google Scholar]
- 128.Wei H., Gao Z., Zheng L., Zhang C., Liu Z., Yang Y., Teng H., Hou L., Yin Y., Zou X. Protective effects of fucoidan on Aβ25-35 and D-gal-induced neurotoxicity in PC12 cells and D-gal-induced cognitive dysfunction in mice. Mar. Drugs. 2017;15 doi: 10.3390/md15030077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Gao Y., Li C., Yin J., Shen J., Wang H., Wu Y., Jin H. Fucoidan, a sulfated polysaccharide from brown algae, improves cognitive impairment induced by infusion of Aβ peptide in rats. Environ. Toxicol. Pharmacol. 2012;33:304–311. doi: 10.1016/j.etap.2011.12.022. [DOI] [PubMed] [Google Scholar]
- 130.Alghazwi M., Smid S., Karpiniec S., Zhang W. Comparative study on neuroprotective activities of fucoidans from Fucus vesiculosus and Undaria pinnatifida. Int. J. Biol. Macromol. 2019;122:255–264. doi: 10.1016/j.ijbiomac.2018.10.168. [DOI] [PubMed] [Google Scholar]
- 131.Bartosz G. Non-enzymatic antioxidant capacity assays: limitations of use in biomedicine. Free Radic. Res. 2010;44:711–720. doi: 10.3109/10715761003758114. [DOI] [PubMed] [Google Scholar]
- 132.Luo D., Zhang Q., Wang H., Cui Y., Sun Z., Yang J., Zheng Y., Jia J., Yu F., Wang X., Wang X. Fucoidan protects against dopaminergic neuron death in vivo and in vitro. Eur. J. Pharmacol. 2009;617:33–40. doi: 10.1016/j.ejphar.2009.06.015. [DOI] [PubMed] [Google Scholar]
- 133.Wang X., Yi K., Zhao Y. Food Funct., Royal Society of Chemistry. 2018. Fucoidan inhibits amyloid-β-induced toxicity in transgenic: Caenorhabditis elegans by reducing the accumulation of amyloid-β and decreasing the production of reactive oxygen species; pp. 552–560. [DOI] [PubMed] [Google Scholar]
- 134.Hsu H.Y., Lin T.Y., Hwang P.A., Tseng L.M., Chen R.H., Tsao S.M., Hsu J. Fucoidan induces changes in the epithelial to mesenchymal transition and decreases metastasis by enhancing ubiquitin-dependent tgfβ receptor degradation in breast cancer. Carcinogenesis. 2013;34:874–884. doi: 10.1093/carcin/bgs396. [DOI] [PubMed] [Google Scholar]
- 135.Hsu H.Y., Lin T.Y., Wu Y.C., Tsao S.M., Hwang P.A., Shih Y.W., Hsu J. Fucoidan inhibition of lung cancer in vivo and in vitro: role of the Smurf2-dependent ubiquitin proteasome pathway in TGFβ receptor degradation. Oncotarget. 2014;5:7870–7885. doi: 10.18632/oncotarget.2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Priyan Shanura Fernando I., Kim K.N., Kim D., Jeon Y.J. Algal polysaccharides: potential bioactive substances for cosmeceutical applications. Crit. Rev. Biotechnol. 2019;39:99–113. doi: 10.1080/07388551.2018.1503995. [DOI] [PubMed] [Google Scholar]
- 137.Zhang Q., Li N., Zhou G., Lu X., Xu Z., Li Z. In vivo antioxidant activity of polysaccharide fraction from Porphyra haitanesis (Rhodephyta) in aging mice. Pharmacol. Res. 2003;48:151–155. doi: 10.1016/s1043-6618(03)00103-8. [DOI] [PubMed] [Google Scholar]
- 138.Zhang Q., Li N., Liu X., Zhao Z., Li Z., Xu Z. The structure of a sulfated galactan from Porphyra haitanensis and its in vivo antioxidant activity. Carbohydr. Res. 2004;339:105–111. doi: 10.1016/j.carres.2003.09.015. [DOI] [PubMed] [Google Scholar]
- 139.Zhang Z., Wang X., Su H., Pan Y., Han J., Zhang T., Mao G. Effect of sulfated galactan from Porphyra haitanensis on H2O2-induced premature senescence in WI-38 cells. Int. J. Biol. Macromol. 2018;106:1235–1239. doi: 10.1016/j.ijbiomac.2017.08.123. [DOI] [PubMed] [Google Scholar]
- 140.Hernandez-Segura A., Nehme J., Demaria M. Hallmarks of cellular senescence. Trends Cell Biol. 2018;28:436–453. doi: 10.1016/j.tcb.2018.02.001. [DOI] [PubMed] [Google Scholar]
- 141.Zhao T., Zhang Q., Qi H., Liu X., Li Z. Extension of life span and improvement of vitality of Drosophila melanogaster by long-term supplementation with different molecular weight polysaccharides from Porphyra haitanensis. Pharmacol. Res. 2008;57:67–72. doi: 10.1016/j.phrs.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 142.Zhang Z., Wang X., Pan Y., Wang G., Mao G. The degraded polysaccharide from Pyropia haitanensis represses amyloid beta peptide-induced neurotoxicity and memory in vivo. Int. J. Biol. Macromol. 2020;146:725–729. doi: 10.1016/j.ijbiomac.2019.09.243. [DOI] [PubMed] [Google Scholar]
- 143.Chen Y., Liu X., Wu L., Tong A., Zhao L., Liu B., Zhao C. Physicochemical characterization of polysaccharides from Chlorella pyrenoidosa and its anti-ageing effects in Drosophila melanogaster. Carbohydr. Polym. 2018;185:120–126. doi: 10.1016/j.carbpol.2017.12.077. [DOI] [PubMed] [Google Scholar]
- 144.Wang X., Zhang Z., Zhou H., Sun X., Chen X., Xu N. The anti-aging effects of Gracilaria lemaneiformis polysaccharide in Caenorhabditis elegans. Int. J. Biol. Macromol. 2019;140:600–604. doi: 10.1016/j.ijbiomac.2019.08.186. [DOI] [PubMed] [Google Scholar]
- 145.Shanura Fernando I.P., Kim M., Son K.T., Jeong Y., Jeon Y.J. Antioxidant activity of marine algal polyphenolic compounds: a mechanistic approach. J. Med. Food. 2016;19:615–628. doi: 10.1089/jmf.2016.3706. [DOI] [PubMed] [Google Scholar]
- 146.Thomas N.V., Kim S.K. Potential pharmacological applications of polyphenolic derivatives from marine brown algae. Environ. Toxicol. Pharmacol. 2011;32:325–335. doi: 10.1016/j.etap.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 147.Catarino M.D., Silva A.M.S., Cardoso S.M., Fucaceae A source of bioactive phlorotannins. Int. J. Mol. Sci. 2017;18 doi: 10.3390/ijms18061327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Wijesekara I., Kim S.K., Li Y., Li Y.X. Phlorotannins as bioactive agents from brown algae. Process Biochem. 2011;46:2219–2224. doi: 10.1016/j.procbio.2011.09.015. [DOI] [Google Scholar]
- 149.Kim S.K., Himaya S.W.A. Adv. Food Nutr. Res. 2011. Medicinal effects of phlorotannins from marine brown algae; pp. 98–109. [DOI] [PubMed] [Google Scholar]
- 150.Shibata T., Ishimaru K., Kawaguchi S., Yoshikawa H., Hama Y. Ninet. Int. Seaweed Symp. Springer Netherlands; 2009. Antioxidant activities of phlorotannins isolated from Japanese Laminariaceae; pp. 255–261. [DOI] [Google Scholar]
- 151.Kim K.C., Kang K.A., Zhang R., Piao M.J., Kim G.Y., Kang M.Y., Lee S.J., Lee N.H., Surh Y.J., Hyun J.W. Up-regulation of Nrf2-mediated heme oxygenase-1 expression by eckol, a phlorotannin compound, through activation of Erk and PI3K/Akt. Int. J. Biochem. Cell Biol. 2010;42:297–305. doi: 10.1016/j.biocel.2009.11.009. [DOI] [PubMed] [Google Scholar]
- 152.Hye S.K., Hae Y.C., Ji Y.K., Byeng W.S., Hyun A.J., Jae S.C. Inhibitory phlorotannins from the edible brown alga Ecklonia stolonifera on total reactive oxygen species (ROS) generation. Arch Pharm. Res. (Seoul) 2004;27:194–198. doi: 10.1007/BF02980106. [DOI] [PubMed] [Google Scholar]
- 153.Kang M.C., Cha S.H., Wijesinghe W.A.J.P., Kang S.M., Lee S.H., Kim E.A., Song C.B., Jeon Y.J. Protective effect of marine algae phlorotannins against AAPH-induced oxidative stress in zebrafish embryo. Food Chem. 2013;138:950–955. doi: 10.1016/j.foodchem.2012.11.005. [DOI] [PubMed] [Google Scholar]
- 154.Dutot M., Fagon R., Hemon M., Rat P. Antioxidant, anti-inflammatory, and anti-senescence activities of a phlorotannin-rich natural extract from Brown Seaweed Ascophyllum nodosum. Appl. Biochem. Biotechnol. 2012;167:2234–2240. doi: 10.1007/s12010-012-9761-1. [DOI] [PubMed] [Google Scholar]
- 155.So M.J., Cho E.J. Phloroglucinol attenuates free radical-induced oxidative stress. Prev. Nutr. Food Sci. 2014;19:129–135. doi: 10.3746/pnf.2014.19.3.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Nho J.A., Shin Y.S., Jeong H.-R., Cho S., Heo H.J., Kim G.H., Kim D.-O. Neuroprotective effects of phlorotannin-rich extract from Brown seaweed Ecklonia cava on neuronal PC-12 and SH-SY5Y cells with oxidative stress. J. Microbiol. Biotechnol. 2020;30:359–367. doi: 10.4014/jmb.1910.10068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Kim H.S., Lee K., Kang K.A., Lee N.H., Hyun J.W., Kim H.S. Phloroglucinol exerts protective effects against oxidative stress-induced cell damage in SH-SY5Y cells. J. Pharmacol. Sci. 2012;119:186–192. doi: 10.1254/jphs.12056FP. [DOI] [PubMed] [Google Scholar]
- 158.Yang E.-J.E.-J., Ahn S., Ryu J., Choi M.M.-S., Choi S., Chong Y.H., Hyun J.-W.W., Chang M.-J., Kim H.S.H.-S., Zhang R., Hong B.-H., Yang E.-J.E.-J., Kang K.A., Choi M.M.-S., Kim K.C., Noh S.-J., Kim H.S.H.-S., Lee N.-H., Hyun J.-W.W., Kim H.S.H.-S. Phloroglucinol attenuates motor functional deficits in an animal model of Parkinson's disease by enhancing Nrf2 activity. PloS One. 2013;8 doi: 10.1371/journal.pone.0135686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Yang E.J., Mahmood U., Kim H., Choi M., Choi Y., Lee J.P., Cho J.Y., Hyun J.W., Kim Y.S., Chang M.J., Kim H.S. Phloroglucinol ameliorates cognitive impairments by reducing the amyloid β peptide burden and pro-inflammatory cytokines in the hippocampus of 5XFAD mice. Free Radic. Biol. Med. 2018;126:221–234. doi: 10.1016/j.freeradbiomed.2018.08.016. [DOI] [PubMed] [Google Scholar]
- 160.Yang E.J., Ahn S., Ryu J., Choi M.S., Choi S., Chong Y.H., Hyun J.W., Chang M.J., Kim H.S. Phloroglucinol attenuates the cognitive deficits of the 5XFAD mouse model of Alzheimer's disease. PloS One. 2015;10 doi: 10.1371/journal.pone.0135686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.De Jesus Raposo M.F., De Morais R.M.S.C., De Morais A.M.M.B. Health applications of bioactive compounds from marine microalgae. Life Sci. 2013;93:479–486. doi: 10.1016/j.lfs.2013.08.002. [DOI] [PubMed] [Google Scholar]
- 162.Guedes A.C., Amaro H.M., Malcata F.X. Microalgae as sources of carotenoids. Mar. Drugs. 2011;9:625–644. doi: 10.3390/md9040625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Rodriguez-Concepcion M., Avalos J., Bonet M.L., Boronat A., Gomez-Gomez L., Hornero-Mendez D., Limon M.C., Meléndez-Martínez A.J., Olmedilla-Alonso B., Palou A., Ribot J., Rodrigo M.J., Zacarias L., Zhu C. A global perspective on carotenoids: metabolism, biotechnology, and benefits for nutrition and health. Prog. Lipid Res. 2018;70:62–93. doi: 10.1016/j.plipres.2018.04.004. [DOI] [PubMed] [Google Scholar]
- 164.De Jesus Raposo M.F., De Morais A.M.M.B., De Morais R.M.S.C. Carotenoids from marine microalgae: a valuable natural source for the prevention of chronic diseases. Mar. Drugs. 2015;13:5128–5155. doi: 10.3390/md13085128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Gammone M.A., Riccioni G., D'Orazio N. Marine carotenoids against oxidative stress: effects on human health. Mar. Drugs. 2015;13:6226–6246. doi: 10.3390/md13106226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Varela J.C., Pereira H., Vila M., León R. Production of carotenoids by microalgae: achievements and challenges. Photosynth. Res. 2015;125:423–436. doi: 10.1007/s11120-015-0149-2. [DOI] [PubMed] [Google Scholar]
- 167.Higuera-Ciapara I., Félix-Valenzuela L., Goycoolea F.M. Astaxanthin: a review of its chemistry and applications. Crit. Rev. Food Sci. Nutr. 2006;46:185–196. doi: 10.1080/10408690590957188. [DOI] [PubMed] [Google Scholar]
- 168.Manivasagan P., Bharathiraja S., Santha Moorthy M., Mondal S., Seo H., Dae Lee K., Oh J. Marine natural pigments as potential sources for therapeutic applications. Crit. Rev. Biotechnol. 2018;38:745–761. doi: 10.1080/07388551.2017.1398713. [DOI] [PubMed] [Google Scholar]
- 169.Kidd P. Astaxanthin, cell membrane nutrient with diverse clinical benefits and anti-aging potential. Alternative Med. Rev. 2011;16:355–364. [PubMed] [Google Scholar]
- 170.Miki W. Biological functions and activities of animal carotenoids. Pure Appl. Chem. 1991;63:141–146. doi: 10.1351/pac199163010141. [DOI] [Google Scholar]
- 171.Shimidzu N., Goto M., Miki W. 1996. Carotenoids as Singlet Oxygen Quenchers in Marine Organisms. [Google Scholar]
- 172.Fakhri S., Aneva I.Y., Farzaei M.H., Sobarzo-Sánchez E. The neuroprotective effects of astaxanthin: therapeutic targets and clinical perspective. Molecules. 2019;24 doi: 10.3390/molecules24142640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Grimmig B., Kim S.H., Nash K., Bickford P.C., Douglas Shytle R. Neuroprotective mechanisms of astaxanthin: a potential therapeutic role in preserving cognitive function in age and neurodegeneration. GeroScience. 2017;39:19–32. doi: 10.1007/s11357-017-9958-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Galasso C., Orefice I., Pellone P., Cirino P., Miele R., Ianora A., Brunet C., Sansone C. On the neuroprotective role of astaxanthin: new perspectives? Mar. Drugs. 2018;16 doi: 10.3390/md16080247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Saw C.L.L., Yang A.Y., Guo Y., Kong A.N.T. Astaxanthin and omega-3 fatty acids individually and in combination protect against oxidative stress via the Nrf2-ARE pathway. Food Chem. Toxicol. 2013;62:869–875. doi: 10.1016/j.fct.2013.10.023. [DOI] [PubMed] [Google Scholar]
- 176.Al-Amin M.M., Akhter S., Hasan A.T., Alam T., Nageeb Hasan S.M., Saifullah A.R.M., Shohel M. The antioxidant effect of astaxanthin is higher in young mice than aged: a region specific study on brain. Metab. Brain Dis. 2015;30:1237–1246. doi: 10.1007/s11011-015-9699-4. [DOI] [PubMed] [Google Scholar]
- 177.Wu W., Wang X., Xiang Q., Meng X., Peng Y., Du N., Liu Z., Sun Q., Wang C., Liu X. Astaxanthin alleviates brain aging in rats by attenuating oxidative stress and increasing BDNF levels. Food Funct. 2014;5:158–166. doi: 10.1039/c3fo60400d. [DOI] [PubMed] [Google Scholar]
- 178.Ni Y., Wu T., Yang L., Xu Y., Ota T., Fu Z. Protective effects of astaxanthin on a combination of D-galactose and jet lag-induced aging model in mice. Endocr. J. 2018;65:569–578. doi: 10.1507/endocrj.EJ17-0500. [DOI] [PubMed] [Google Scholar]
- 179.Yamamoto M., Clark J.D., Pastor J.V., Gurnani P., Nandi A., Kurosu H., Miyoshi M., Ogawa Y., Castrillon D.H., Rosenblatt K.P., Kuro-O M. Regulation of oxidative stress by the anti-aging hormone klotho. J. Biol. Chem. 2005;280:38029–38034. doi: 10.1074/jbc.M509039200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Sj S., V B., S S., D M. Astaxanthin enhances the longevity of Saccharomyces cerevisiae by decreasing oxidative stress and apoptosis. FEMS Yeast Res. 2019;19 doi: 10.1093/FEMSYR/FOY113. [DOI] [PubMed] [Google Scholar]
- 181.Yazaki K., Yoshikoshi C., Oshiro S., Yanase S. Supplemental cellular protection by a carotenoid extends lifespan via Ins/IGF-1 signaling in caenorhabditis elegans. Oxid. Med. Cell. Longev. 2011 doi: 10.1155/2011/596240. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Huangfu J., Liu J., Sun Z., Wang M., Jiang Y., Chen Z.Y., Chen F. Antiaging effects of astaxanthin-rich alga haematococcus pluvialis on fruit flies under oxidative stress. J. Agric. Food Chem. 2013;61:7800–7804. doi: 10.1021/jf402224w. [DOI] [PubMed] [Google Scholar]
- 183.Wang H.Q., Sun X.B., Xu Y.X., Zhao H., Zhu Q.Y., Zhu C.Q. Astaxanthin upregulates heme oxygenase-1 expression through ERK1/2 pathway and its protective effect against beta-amyloid-induced cytotoxicity in SH-SY5Y cells. Brain Res. 2010;1360:159–167. doi: 10.1016/j.brainres.2010.08.100. [DOI] [PubMed] [Google Scholar]
- 184.Ye Q., Huang B., Zhang X., Zhu Y., Chen X. Astaxanthin protects against MPP+-induced oxidative stress in PC12 cells via the HO-1/NOX2 axis. BMC Neurosci. 2012;13 doi: 10.1186/1471-2202-13-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Liu X., Shibata T., Hisaka S., Osawa T. Astaxanthin inhibits reactive oxygen species-mediated cellular toxicity in dopaminergic SH-SY5Y cells via mitochondria-targeted protective mechanism. Brain Res. 2009;1254:18–27. doi: 10.1016/j.brainres.2008.11.076. [DOI] [PubMed] [Google Scholar]
- 186.Lee D.H., Kim C.S., Lee Y.J. Astaxanthin protects against MPTP/MPP+-induced mitochondrial dysfunction and ROS production in vivo and in vitro. Food Chem. Toxicol. 2011;49:271–280. doi: 10.1016/j.fct.2010.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Alghazwi M., Smid S., Musgrave I., Zhang W. In vitro studies of the neuroprotective activities of astaxanthin and fucoxanthin against amyloid beta (Aβ 1-42 ) toxicity and aggregation. Neurochem. Int. 2019;124:215–224. doi: 10.1016/j.neuint.2019.01.010. [DOI] [PubMed] [Google Scholar]
- 188.D'Orazio N., Gemello E., Gammone M.A., De Girolamo M., Ficoneri C., Riccioni G. Fucoxantin: a treasure from the sea. Mar. Drugs. 2012;10:604–616. doi: 10.3390/md10030604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Bae M., Kim M.B., Park Y.K., Lee J.Y. Health benefits of fucoxanthin in the prevention of chronic diseases. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2020 doi: 10.1016/j.bbalip.2020.158618. [DOI] [PubMed] [Google Scholar]
- 190.Fucoidan improves the metabolic profiles of patients with non-alcoholic fatty liver disease (NAFLD) - full text view - ClinicalTrials.gov. https://clinicaltrials.gov/ct2/show/NCT02875392 n.d.
- 191.Effect of fucoxanthin on the components of the metabolic syndrome, insulin sensitivity and insulin secretion - full text view - ClinicalTrials.gov. https://clinicaltrials.gov/ct2/show/NCT03613740 n.d.
- 192.Liu C.L., Chiu Y.T., Hu M.L. Fucoxanthin enhances HO-1 and NQO1 expression in murine hepatic BNL CL.2 Cells through activation of the Nrf2/ARE system partially by its pro-oxidant activity. J. Agric. Food Chem. 2011;59:11344–11351. doi: 10.1021/jf2029785. [DOI] [PubMed] [Google Scholar]
- 193.Zheng J., Piao M.J., Keum Y.S., Kim H.S., Hyun J.W. Fucoxanthin protects cultured human keratinocytes against oxidative stress by blocking free radicals and inhibiting apoptosis. Biomol. Ther. 2013;21:270–276. doi: 10.4062/biomolther.2013.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.W X., C Y.J., Q J., Z M.M., Z T.L., C M., L S.M., W G.C. Fucoxanthin exerts cytoprotective effects against hydrogen peroxide-induced oxidative damage in L02 cells. BioMed Res. Int. 2018;2018 doi: 10.1155/2018/1085073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Lashmanova E., Proshkina E., Zhikrivetskaya S., Shevchenko O., Marusich E., Leonov S., Melerzanov A., Zhavoronkov A., Moskalev A. Fucoxanthin increases lifespan of Drosophila melanogaster and Caenorhabditis elegans. Pharmacol. Res. 2015;100:228–241. doi: 10.1016/j.phrs.2015.08.009. [DOI] [PubMed] [Google Scholar]
- 196.Moskalev A., Shaposhnikov M., Zemskaya N., Belyi A., Dobrovolskaya E., Patova A., Guvatova Z., Lukyanova E., Snezhkina A., Kudryavtseva A. Transcriptome analysis reveals mechanisms of geroprotective effects of fucoxanthin in Drosophila. BMC Genom. 2018;19 doi: 10.1186/s12864-018-4471-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Guvatova Z., Dalina A., Marusich E., Pudova E., Snezhkina A., Krasnov G., Kudryavtseva A., Leonov S., Moskalev A. Protective effects of carotenoid fucoxanthin in fibroblasts cellular senescence. Mech. Ageing Dev. 2020 doi: 10.1016/j.mad.2020.111260. [DOI] [PubMed] [Google Scholar]
- 198.Xiang S., Liu F., Lin J., Chen H., Huang C., Chen L., Zhou Y., Ye L., Zhang K., Jin J., Zhen J., Wang C., He S., Wang Q., Cui W., Zhang J. Fucoxanthin inhibits β-amyloid assembly and attenuates β-amyloid oligomer-induced cognitive impairments. J. Agric. Food Chem. 2017;65:4092–4102. doi: 10.1021/acs.jafc.7b00805. [DOI] [PubMed] [Google Scholar]
- 199.Zhang L., Wang H., Fan Y., Gao Y., Li X., Hu Z., Ding K., Wang Y., Wang X. Fucoxanthin provides neuroprotection in models of traumatic brain injury via the Nrf2-ARE and Nrf2-autophagy pathways. Sci. Rep. 2017;7 doi: 10.1038/srep46763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Dikic I. Proteasomal and autophagic degradation systems. Annu. Rev. Biochem. 2017;86:193–224. doi: 10.1146/annurev-biochem-061516-044908. [DOI] [PubMed] [Google Scholar]
- 201.Yu J., Lin J.J., Yu R., He S., Wang Q.W., Cui W., Zhang J.R. Fucoxanthin prevents H2O2-induced neuronal apoptosis via concurrently activating the PI3-K/Akt cascade and inhibiting the ERK pathway. Food Nutr. Res. 2017;61 doi: 10.1080/16546628.2017.1304678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Lin J., Yu J., Zhao J., Zhang K., Zheng J., Wang J., Huang C., Zhang J., Yan X., Gerwick W.H., Wang Q., Cui W., He S. Fucoxanthin, a marine carotenoid, attenuates β -amyloid oligomer-induced neurotoxicity possibly via regulating the PI3K/akt and the ERK pathways in SH-SY5Y cells. Oxid. Med. Cell. Longev. 2017 doi: 10.1155/2017/6792543. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Snare D.J., Fields A.M., Snell T.W., Kubanek J. Lifespan extension of rotifers by treatment with red algal extracts. Exp. Gerontol. 2013;48:1420–1427. doi: 10.1016/j.exger.2013.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Zhou J.-B., Zheng Y.-L., Zeng Y.-X., Wang J.-W., Pei Z., Pang J.-Y. Marine derived xyloketal derivatives exhibit anti-stress and anti-ageing effects through HSF pathway in Caenorhabditis elegans. Eur. J. Med. Chem. 2018;148:63–72. doi: 10.1016/j.ejmech.2018.02.028. [DOI] [PubMed] [Google Scholar]
- 205.Briffa M., Ghio S., Neuner J., Gauci A.J., Cacciottolo R., Marchal C., Caruana M., Cullin C., Vassallo N., Cauchi R.J. Extracts from two ubiquitous Mediterranean plants ameliorate cellular and animal models of neurodegenerative proteinopathies. Neurosci. Lett. 2017;638:12–20. doi: 10.1016/j.neulet.2016.11.058. [DOI] [PubMed] [Google Scholar]
- 206.Shimizu H., Koyama T., Yamada S., Lipton S.A., Satoh T. Zonarol, a sesquiterpene from the brown algae Dictyopteris undulata, provides neuroprotection by activating the Nrf2/ARE pathway. Biochem. Biophys. Res. Commun. 2015;457:718–722. doi: 10.1016/j.bbrc.2015.01.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Leirós M., Alonso E., Rateb M.E., Houssen W.E., Ebel R., Jaspars M., Alfonso A., Botana L.M. Gracilins: Spongionella-derived promising compounds for Alzheimer disease. Neuropharmacology. 2015;93:285–293. doi: 10.1016/j.neuropharm.2015.02.015. [DOI] [PubMed] [Google Scholar]
- 208.Leirós M., Sánchez J., Alonso E., Rateb M., Houssen W., Ebel R., Jaspars M., Alfonso A., Botana L. Spongionella secondary metabolites protect mitochondrial function in cortical neurons against oxidative stress. Mar. Drugs. 2014;12:700–718. doi: 10.3390/md12020700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Wang A.S., Dreesen O. Biomarkers of cellular senescence and skin aging. Front. Genet. 2018;9 doi: 10.3389/fgene.2018.00247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Zhang S., Duan E. Fighting against skin aging: the way from bench to bedside. Cell Transplant. 2018;27:729–738. doi: 10.1177/0963689717725755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Shin J.W., Kwon S.H., Choi J.Y., Na J.I., Huh C.H., Choi H.R., Park K.C. Molecular mechanisms of dermal aging and antiaging approaches. Int. J. Mol. Sci. 2019;20 doi: 10.3390/ijms20092126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Cao C., Xiao Z., Wu Y., Ge C. Diet and skin aging-from the perspective of food nutrition. Nutrients. 2020;12 doi: 10.3390/nu12030870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Kammeyer A., Luiten R.M. Oxidation events and skin aging. Ageing Res. Rev. 2015;21:16–29. doi: 10.1016/j.arr.2015.01.001. [DOI] [PubMed] [Google Scholar]
- 214.Gonzaga E.R. Role of UV light in photodamage, skin aging, and skin cancer: importance of photoprotection. Am. J. Clin. Dermatol. 2009;10:19–24. doi: 10.2165/0128071-200910001-00004. [DOI] [PubMed] [Google Scholar]
- 215.Lephart E.D. Skin aging and oxidative stress: equol's anti-aging effects via biochemical and molecular mechanisms. Ageing Res. Rev. 2016;31:36–54. doi: 10.1016/j.arr.2016.08.001. [DOI] [PubMed] [Google Scholar]
- 216.Kligman A.M. Early destructive effect of sunlight on human skin. JAMA, J. Am. Med. Assoc. 1969;210:2377–2380. doi: 10.1001/jama.1969.03160390039008. [DOI] [PubMed] [Google Scholar]
- 217.Kageyama H., Waditee-Sirisattha R. Antioxidative, anti-inflammatory, and anti-aging properties of mycosporine-like amino acids: molecular and cellular mechanisms in the protection of skin-aging. Mar. Drugs. 2019;17 doi: 10.3390/md17040222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Cole M.A., Quan T., Voorhees J.J., Fisher G.J. Extracellular matrix regulation of fibroblast function: redefining our perspective on skin aging. J. Cell Commun. Signal. 2018;12:35–43. doi: 10.1007/s12079-018-0459-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Petropoulos I., Conconi M., Wang X., Hoenel B., Brégégère F., Milner Y., Friguet B. Increase of oxidatively modified protein is associated with a decrease of proteasome activity and content in aging epidermal cells. J. Gerontol. A. Biol. Sci. Med. Sci. 2000;55:B220–B227. doi: 10.1093/gerona/55.5.b220. [DOI] [PubMed] [Google Scholar]
- 220.Kozie R., Greussing R., Maier A.B., Declercq L., Jansen-Dürr P. Functional interplay between mitochondrial and proteasome activity in skin aging. J. Invest. Dermatol. 2011;131:594–603. doi: 10.1038/jid.2010.383. [DOI] [PubMed] [Google Scholar]
- 221.Catalgol B., Ziaja I., Breusing N., Jung T., Höhn A., Alpertunga B., Schroeder P., Chondrogianni N., Gonos E.S., Petropoulos I., Friguet B., Klotz L.O., Krutman J., Grune T. The proteasome is an integral part of solar ultraviolet a radiation-induced gene expression. J. Biol. Chem. 2009;284:30076–30086. doi: 10.1074/jbc.M109.044503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Imbert I., Gondran C., Oberto G., Cucumel K., Dal Farra C., Domloge N. Maintenance of the ubiquitin-proteasome system activity correlates with visible skin benefits. Int. J. Cosmet. Sci. 2010;32:446–457. doi: 10.1111/j.1468-2494.2010.00575.x. [DOI] [PubMed] [Google Scholar]
- 223.Chang A.L.S., Bitter P.H., Qu K., Lin M., Rapicavoli N.A., Chang H.Y. Rejuvenation of gene expression pattern of aged human skin by broadband light treatment: a pilot study. J. Invest. Dermatol. 2013;133:394–402. doi: 10.1038/jid.2012.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Ishii M.A., Miyachi K.J., Cheng B., Sun B.K. Aging-associated decline of epidermal PSMD8 contributes to impaired skin function. J. Invest. Dermatol. 2018;138:976–978. doi: 10.1016/j.jid.2017.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Pallela R., Na-Young Y., Kim S.K. Anti-photoaging and photoprotective compounds derived from marine organisms. Mar. Drugs. 2010;8:1189–1202. doi: 10.3390/md8041189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Chrapusta E., Kaminski A., Duchnik K., Bober B., Adamski M., Bialczyk J. Mycosporine-like amino acids: potential health and beauty ingredients. Mar. Drugs. 2017;15 doi: 10.3390/md15100326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Lawrence K.P., Long P.F., Young A.R. Mycosporine-like amino acids for skin photoprotection. Curr. Med. Chem. 2017;25:5512–5527. doi: 10.2174/0929867324666170529124237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Ryu J., Park S.J., Kim I.H., Choi Y.H., Nam T.J. Protective effect of porphyra-334 on UVA-induced photoaging in human skin fibroblasts. Int. J. Mol. Med. 2014;34:796–803. doi: 10.3892/ijmm.2014.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Schmid D., Cornelia S., Fred Z. UV-A sunscreen from red algae for protection against premature skin aging. Cosmetics. 2004:139–143. [Google Scholar]
- 230.de la Coba F., Aguilera J., de Gálvez M.V., Álvarez M., Gallego E., Figueroa F.L., Herrera E. Prevention of the ultraviolet effects on clinical and histopathological changes, as well as the heat shock protein-70 expression in mouse skin by topical application of algal UV-absorbing compounds. J. Dermatol. Sci. 2009;55:161–169. doi: 10.1016/j.jdermsci.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 231.Gacesa R., Dunlap W.C., Long P.F. Bioinformatics analyses provide insight into distant homology of the Keap1-Nrf2 pathway. Free Radic. Biol. Med. 2015;88:373–380. doi: 10.1016/j.freeradbiomed.2015.06.015. [DOI] [PubMed] [Google Scholar]
- 232.Ryu J., Kwon M.J., Nam T.J. Nrf2 and NF-κB signaling pathways contribute to porphyra-334-mediated inhibition of UVA-induced inflammation in skin fibroblasts. Mar. Drugs. 2015;13:4721–4732. doi: 10.3390/md13084721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Moon H.J., Kyong S.P., Mi J.K., Myeong S.L., Seok H.J., Imbs T.I., Zvyagintseva T.N., Ermakova S.P., Yong H.L. Effect of Costaria costata fucoidan on expression of matrix metalloproteinase-1 promoter, mRNA, and protein. J. Nat. Prod. 2009;72:1731–1734. doi: 10.1021/np800797v. [DOI] [PubMed] [Google Scholar]
- 234.Moon H.J., Lee S.H., Ku M.J., Yu B.C., Jeon M.J., Jeong S.H., Stonil V.A., Zvyagintseva T.N., Ermakova S.P., Lee Y.H. Fucoidan inhibits UVB-induced MMP-I promoter expression and down regulation of type I procollagen synthesis in human skin fibroblasts. Eur. J. Dermatol. 2009;19:129–134. doi: 10.1684/ejd.2008.0611. [DOI] [PubMed] [Google Scholar]
- 235.Kim Y.I., Oh W.S., Song P.H., Yun S., Kwon Y.S., Lee Y.J., Ku S.K., Song C.H., Oh T.H. Anti-photoaging effects of low molecular-weight fucoidan on ultraviolet B-irradiated mice. Mar. Drugs. 2018;16 doi: 10.3390/md16080286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Maruyama H., Tamauchi H., Kawakami F., Yoshinaga K., Nakano T. Suppressive effect of dietary fucoidan on proinflammatory immune response and MMP-1 expression in UVB-irradiated mouse skin. Planta Med. 2015;81:1370–1374. doi: 10.1055/s-0035-1557821. [DOI] [PubMed] [Google Scholar]
- 237.Thevanayagam H., Mohamed S.M., Chu W.L. Assessment of UVB-photoprotective and antioxidative activities of carrageenan in keratinocytes. J. Appl. Phycol. 2014;26:1813–1821. doi: 10.1007/s10811-013-0207-0. [DOI] [Google Scholar]
- 238.Heo S.J., Ko S.C., Cha S.H., Kang D.H., Park H.S., Choi Y.U., Kim D., Jung W.K., Jeon Y.J. Effect of phlorotannins isolated from Ecklonia cava on melanogenesis and their protective effect against photo-oxidative stress induced by UV-B radiation. Toxicol. Vitro. 2009;23:1123–1130. doi: 10.1016/j.tiv.2009.05.013. [DOI] [PubMed] [Google Scholar]
- 239.Joe M.J., Kim S.N., Choi H.Y., Shin W.S., Park G.M., Kang D.W., Yong K.K. The inhibitory effects of eckol and dieckol from Ecklonia stolonifera on the expression of matrix metalloproteinase-1 in human dermal fibroblasts. Biol. Pharm. Bull. 2006;29:1735–1739. doi: 10.1248/bpb.29.1735. [DOI] [PubMed] [Google Scholar]
- 240.Kim K.C., Piao M.J., Cho S.J., Lee N.H., Hyun J.W. Phloroglucinol protects human keratinocytes from ultraviolet B radiation by attenuating oxidative stress. Photodermatol. Photoimmunol. Photomed. 2012;28:322–331. doi: 10.1111/phpp.12010. [DOI] [PubMed] [Google Scholar]
- 241.Ko S.C., Cha S.H., Heo S.J., Lee S.H., Kang S.M., Jeon Y.J. Protective effect of Ecklonia cava on UVB-induced oxidative stress: in vitro and in vivo zebrafish model. J. Appl. Phycol. 2011;23:697–708. doi: 10.1007/s10811-010-9565-z. [DOI] [Google Scholar]
- 242.Cha S.H., Ko C.I., Kim D., Jeon Y.J. Protective effects of phlorotannins against ultraviolet B radiation in zebrafish (Danio rerio) Vet. Dermatol. 2012;23 doi: 10.1111/j.1365-3164.2011.01009.x. [DOI] [PubMed] [Google Scholar]
- 243.Piao M.J., Ahn M.J., Kang K.A., Kim K.C., Zheng J., Yao C.W., Cha J.W., Hyun C.L., Kang H.K., Lee N.H., Hyun J.W. Phloroglucinol inhibits ultraviolet B radiation-induced oxidative stress in the mouse skin. Int. J. Radiat. Biol. 2014;90:928–935. doi: 10.3109/09553002.2014.911990. [DOI] [PubMed] [Google Scholar]
- 244.Suganuma K., Nakajima H., Ohtsuki M., Imokawa G. Astaxanthin attenuates the UVA-induced up-regulation of matrix-metalloproteinase-1 and skin fibroblast elastase in human dermal fibroblasts. J. Dermatol. Sci. 2010;58:136–142. doi: 10.1016/j.jdermsci.2010.02.009. [DOI] [PubMed] [Google Scholar]
- 245.Komatsu T., Sasaki S., Manabe Y., Hirata T., Sugawara T. Preventive effect of dietary astaxanthin on UVA-induced skin photoaging in hairless mice. PloS One. 2017;12 doi: 10.1371/journal.pone.0171178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Li X., Matsumoto T., Takuwa M., Ali M.S.E.S., Hirabashi T., Kondo H., Fujino H. Protective Effects of astaxanthin supplementation against ultraviolet-induced photoaging in hairless mice. Biomedicines. 2020;8 doi: 10.3390/biomedicines8020018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Heo S.J., Jeon Y.J. Protective effect of fucoxanthin isolated from Sargassum siliquastrum on UV-B induced cell damage. J. Photochem. Photobiol. B Biol. 2009;95:101–107. doi: 10.1016/j.jphotobiol.2008.11.011. [DOI] [PubMed] [Google Scholar]
- 248.Urikura I., Sugawara T., Hirata T. Protective effect of fucoxanthin against UVB-induced skin photoaging in hairless mice. Biosci. Biotechnol. Biochem. 2011;75:757–760. doi: 10.1271/bbb.110040. [DOI] [PubMed] [Google Scholar]
- 249.Bulteau A.L., Moreau M., Saunois A., Nizard C., Friguet B. Algae extract-mediated stimulation and protection of proteasome activity within human keratinocytes exposed to UVA and UVB irradiation. Antioxidants Redox Signal. 2006;8:136–143. doi: 10.1089/ars.2006.8.136. [DOI] [PubMed] [Google Scholar]
- 250.Nizard C., Poggioli S., Heusèle C., Bulteau A.L., Moreau M., Saunois A., Schnebert S., Mahé C., Friguet B. Ann. N. Y. Acad. Sci. New York Academy of Sciences; 2004. Algae extract protection effect on oxidized protein level in human Stratum corneum; pp. 219–222. [DOI] [PubMed] [Google Scholar]