Abstract
Acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors (EGFR-TKIs), such as erlotinib, remains a major challenge in the targeted therapy of non-small cell lung cancer (NSCLC). HKB99 is a novel allosteric inhibitor of phosphoglycerate mutase 1 (PGAM1) that preferentially suppresses cell proliferation and induces more apoptosis in acquired erlotinib-resistant HCC827ER cells compared with its parental HCC827 cells. In this study we identified the molecular biomarkers for HKB99 response in erlotinib-resistant HCC827ER cells. We showed that HCC827ER cells displayed enhanced invasive pseudopodia structures as well as downregulated plasminogen activator inhibitor-2 (PAI-2). Meanwhile, PAI-2 knockdown by siPAI-2 candidates decreased the sensitivity of HCC827 parental cells to erlotinib. Moreover, HKB99 (5 μM) preferentially inhibited the invasive pseudopodia formation and increased the level of PAI-2 in HCC827ER cells. Collectively, this study provides new insight into the role of PAI-2 in regulating the sensitivity of erlotinib resistant NSCLC cells to PGAM1 inhibitor. Furthermore, PAI-2 level might be considered as a potential biomarker for predicting the efficacy of the PGAM1 allosteric inhibitor on the erlotinib resistant NSCLC cells.
Keywords: non-small cell lung cancer, erlotinib resistance, phosphoglycerate mutase 1, allosteric inhibitor, HKB99, plasminogen activator inhibitor-2
Introduction
Lung cancer is a malignant tumor with high global morbidity and mortality, and the incidence of lung cancer in China is quickly increasing [1]. Lung cancer is pathologically divided into small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC), which can be further divided into lung adenocarcinoma, squamous cell carcinoma, and large cell carcinoma [2]. Epidermal growth factor receptor tyrosine kinase inhibitors (EGFR-TKIs), such as erlotinib, gefitinib and afatinib, are the first-line treatment for patients with EGFR mutation-positive lung adenocarcinoma [3–5]. However, the development of drug resistance is inevitable and presents a great challenge to the long-term success of TKI treatment [6].
Plasminogen activator inhibitor-2 (PAI-2), also known as the second member of clade B serine protease inhibitors (SERPINB2), has been thoroughly shown to be an inhibitor of extracellular urokinase plasminogen activator (uPA) [7]. Low levels of PAI-2 are correlated with high metastasis and poor prognosis in lung cancer [8–10]. Another study reported that in comparison to parental cells, the expression of PAI-2 is decreased in gefitinib-resistant NSCLC cells, which increased invasiveness and metastasis by lengthening invadopodia-like structures around the cells [11].
Metabolic reprogramming is well documented as one of the hallmarks of cancer [12]. Phosphoglycerate mutase 1 (PGAM1) plays a key role in cancer metabolism by regulating aerobic glycolysis and anabolic biosynthesis [13]. In addition, PGAM1 is reported to promote homologous recombination repair by regulating the deoxyribonucleotide triphosphate (dNTP) pool [14]. In addition to its metabolic functions, PGAM1 also directly interacts with α-smooth muscle actin (ACTA2) to promote cancer cell migration [15]. The expression of PGAM1 is upregulated in multiple cancers, such as lung cancer, urothelial bladder cancer, and pancreatic cancer, and it is closely related to the prognosis of patients [16–18], which further indicates that PGAM1 is a potential target for tumor therapy.
We recently discovered a novel PGAM1 allosteric inhibitor, HKB99, which blocks the structure of PGAM1 by affecting the metabolic activity and nonmetabolic function of PGAM1, and it inhibits tumor growth and metastasis, and overcomes the resistance of NSCLC to erlotinib [19]. In addition, while it does not change the growth in parent HCC827 cells, HKB99 preferentially inhibits the growth of erlotinib-resistant HCC827ER cells; however, it remains unclear what molecular biomarkers might be predictive of HKB99 sensitivity.
Here, we established erlotinib-resistant HCC827ER cells and found that the expression of PAI-2 protein was downregulated and the formation of invasive pseudopodia was enhanced in HCC827ER cells. After knockdown of PAI-2, HCC827 parental cells became resistant to erlotinib. In addition, the PGAM1 allosteric inhibitor HKB99 can effectively inhibit the formation of invasive pseudopodia and increase the level of PAI-2 in HCC827ER cells.
Materials and methods
Cell lines and reagents
HCC827 and HCC827ER cells were cultured in RPMI-1640 (Gibco) supplemented with 10% fetal bovine serum at 37 °C in 5% CO2 in a humidified atmosphere, which was carried out according to ATCC protocols. Erlotinib-resistant HCC827ER cells were kindly donated by Dr. Zhuang (Ren Ji Hospital, School of Medicine, Shanghai Jiao Tong University, Shanghai, China). Erlotinib was purchased from APExBIO (A3397) and reconstituted in DMSO (Sigma-Aldrich).
Cell viability assays
NSCLC cells were seeded in 96-well plates at a density of 3000 cells per well and were allowed to adhere overnight. HKB99 was added, in a dilution series, to the cells, and they were incubated for 72 h. Cell viability was determined by Cell Counting Kit-8 (CCK8) colorimetric assay (Dojindo).
Western blotting analysis
NSCLC cells were lysed with RIPA lysis buffer (Beyotime) containing PMSF protease inhibitor (Beyotime). The concentration of protein in the lysate was determined by BCA (Thermo Scientific), and then the samples were adjusted until they all had the same concentration. The protein samples were incubated with preheated 5% SDS and were exposed to strong vortexing for 2–3 s, boiled for 10 min, separated by SDS-PAGE and transferred to PVDF membranes (Immobilon-P, Millipore). Membranes were blocked for 1 h at room temperature with 5% milk in 1× Tris-buffered saline Tween-20 (TBST), and then they were blotted with primary antibodies at 4 °C overnight. The following antibodies were used: anti-PGAM 1 (Santa Cruz), anti-PAI2 (Santa Cruz), and anti-β-actin (Genetex). After washing with 1× TBST three times, membranes were incubated with horseradish peroxidase-conjugated anti-rabbit IgG (dilution, 1:5000) or anti-mouse IgG (dilution, 1:5000) antibodies at room temperature for 1 h, and the immunoblots were visualized with an ECL detection kit (Thermo Fisher Scientific).
Immunoblotting fluorescence
Cells in the logarithmic growth phase were plated into dishes. When at approximately 80% confluence, the cells were fixed by treatment with 4% paraformaldehyde at room temperature for 15 min. The fixed cells were blocked with 1% BSA containing 0.1% Triton X-100 for 30 min at room temperature. Cells were incubated with FICT-phalloidin (YESEN, 40735ES75) or anti-PAI2 (Santa Cruz) at 4 °C overnight. DAPI (0.5 μg/mL, Vector Laboratories, Burlingame, CA, USA) was used to counterstain the nuclei. The images were obtained using a Leica TCP-SP8 confocal microscope (Leica, Wetzlar, Germany) and a Zeiss ApoTome microscope (Carl Zeiss, Jena, Germany); the scale bar is equal to 50 μm.
siRNA transfection
RNA interference of PAI2 was performed using 25-bp siRNA duplexes purchased from GenePharma. Cells were plated at 30%–40% confluence in OPTI-MEM serum-free medium, and then they were transfected with a specific siRNA duplex using Lipofectamine 3000 (Invitrogen) according to the manufacturer’s instructions. Target sequences for siRNAs were as follows: siPGAM1 #1: 5′-CCA-GTG-GAA-GAC-GAA-TGT-A-3′, siPGAM1 #2: 5′-AGG-TTC-TCA-GTC-TAA-GCT-A-3′, siPAI2 #1: 5′-AGG-CAC-AAG-CTG-CAG-ATA-A-3′, siPAI2 #3: 5′-AGG-CAG-TAG-ACT-TCC-TAG-A-3′, and siPAI2 #4: 5′-GTA-GTT-ATC-CTG-ATG-CGA-T-3′. Experiments were performed 24 h or 72 h after transfection. Cells transfected with a nonspecific siRNA duplex were used as controls for direct comparison.
Cell proliferation assays
NSCLC cells were seeded in 96-well plates at a density of 3000 cells per well. Then, the cells were incubated for 72 h and scanned every 4 h by an IncuCyte Live Cell Analysis system (Sartorius).
Statistical analysis
All experiments were repeated more than three times. The following statistical parameters are described in the figure legends: the exact value of n, dispersion and precision measures and statistical significance. Differences between two groups were analyzed using two-tailed unpaired Student’s t-tests. Kaplan–Meier survival analysis of PGAM1 and PAI2 expression was performed with a log-rank test based on a database from TCGA. A P value of < 0.05 was considered significant (*P < 0.05; **P < 0.01; ***P < 0.001; and ****P < 0.0001). Statistical analyses were performed using Graph Pad Prism 7.0 (GraphPad Software, https://www.graphpad.com/scientificsoftware/prism).
Results
PAI-2 is downregulated in erlotinib-resistant NSCLC cells
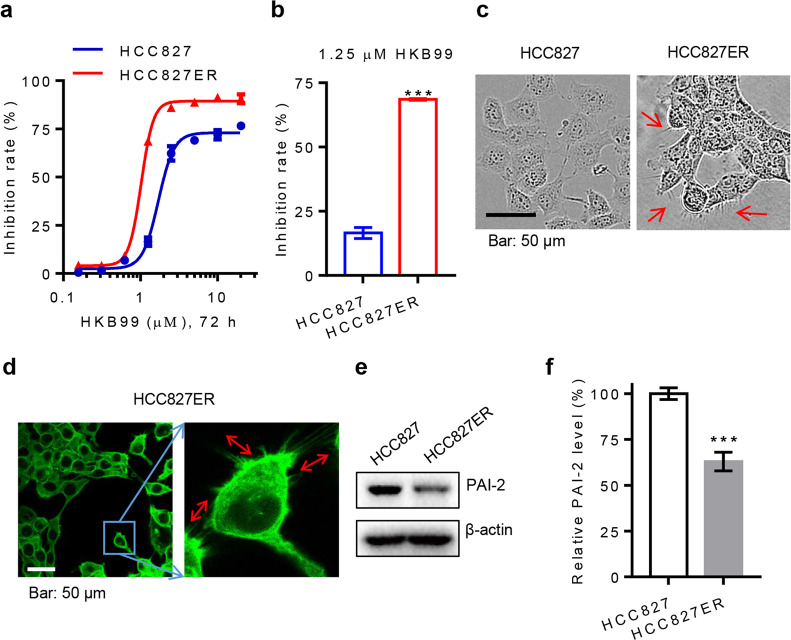
HKB99 has been identified as a brand new allosteric inhibitor of PGAM1 that exhibits multiple antitumor properties, including inhibiting tumor growth and metastasis in erlotinib-resistant NSCLC cells [19]. In accordance with our previous study, we confirmed that HKB99 preferentially suppresses cellular proliferation in acquired erlotinib-resistant HCC827ER cells, and it has higher potency (IC50 value of 1.020 μM) and efficacy than is achieved in the parental HCC827 cells (IC50 value of 1.705 μM) (Fig. 1a, b). Therefore, we wanted to explore whether there is some potential biomarker for predicting the efficacy of HKB99 on erlotinib-resistant NSCLC cells.
Fig. 1.
Increased invasive pseudopodia structures and downregulation of PAI-2 in HCC827ER cells. a Cells were treated with HKB99 at various concentrations for 72 h. b The indicated cells were treated with 1.25 μM HKB99 for 72 h. The inhibition of the cell growth rate was then analyzed by CCK8 assay. c Cellular morphology was observed by phase-contrast microscopy. d F-actin was stained with FITC phalloidin. Representative images were detected by laser confocal fluorescence microscopy. e Protein levels of PAI-2 were measured by immunoblotting of HCC827 and HCC827ER cells. f Grayscale analysis of PAI-2 described in e was from three independent experiments. Error bars represent the mean ± SD of triplicates. ***P < 0.001. Scale bar, 50 μm
First, enhanced invadopodia-like structural protrusions were observed under phase-contrast microscopy in HCC827ER cells that were not observed in parental HCC827 cells (Fig. 1c). The result of F-actin staining also showed many protrusions with actin filaments around the HCC827ER cells (Fig. 1d). As a previous study demonstrated that such alterations in cell morphology are closely associated with the downregulation of PAI-2 in gefitinib-resistant NSCLC cells, we then explored whether the level of PAI-2 was changed in erlotinib-resistant NSCLC cells [11]. Accordingly, the PAI-2 protein level was significantly lower in HCC827ER cells (approximately 60% relative to parental HCC827 cells, Fig. 1e, f). Collectively, our results suggest that PAI-2 is downregulated and that there is an acquisition of invasive pseudopodia in acquired erlotinib-resistant NSCLC cells.
Downregulation of PAI-2 is associated with erlotinib resistance in NSCLC cells
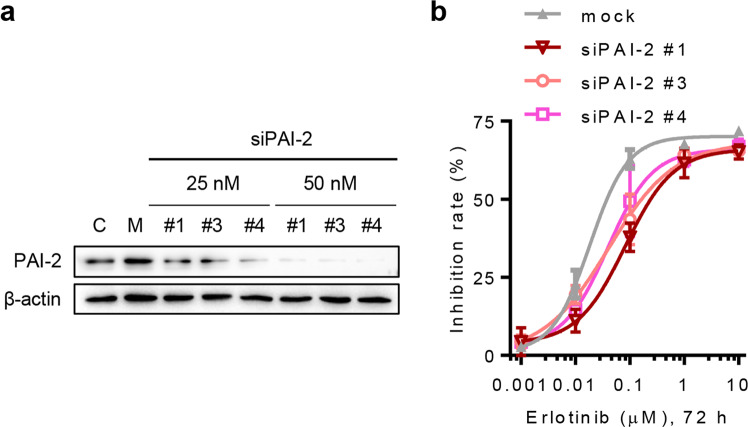
As the PAI-2 protein level is downregulated in HCC827ER cells, we wondered whether PAI-2 contributes to erlotinib resistance in NSCLC cells. To minimize the potential off-target effects, three siPAI-2 candidates were used to efficiently knock down the expression of PAI-2 in HCC827 cells (Fig. 2a). The results of CCK8 assays showed that the sensitivity of erlotinib treatment was reduced after HCC827 cells were transfected with the different siPAI-2 candidates (50 nM each) (Fig. 2b). In addition, knockdown of PAI-2 alone had no significant influence on the proliferation and cell morphology of HCC827 cells (Fig. S1). Together, these data suggest that PAI-2 deletion promotes erlotinib resistance in NSCLC cells.
Fig. 2.
Downregulation of PAI-2 induces resistance to erlotinib treatment in HCC827 cells. a siPAI-2 candidates were transfected into HCC827 cells at 25 and 50 nM and were incubated for 72 h. The knockdown efficiency of siPAI-2 candidates was then determined by immunoblotting. C, untreated control; M, 5 nM negative siRNA transfection. b After HCC827 cells were treated with various concentrations of erlotinib, 50 nM siPAI-2 candidates or 5 nM negative siRNA (mock) for 72 h, as indicated, and a CCK8 assay was used to measure cell viability. Error bars represent the mean ± SD of triplicates
HKB99 preferentially inhibits invasive pseudopodia formation and increases the level of PAI-2 in HCC827ER cells
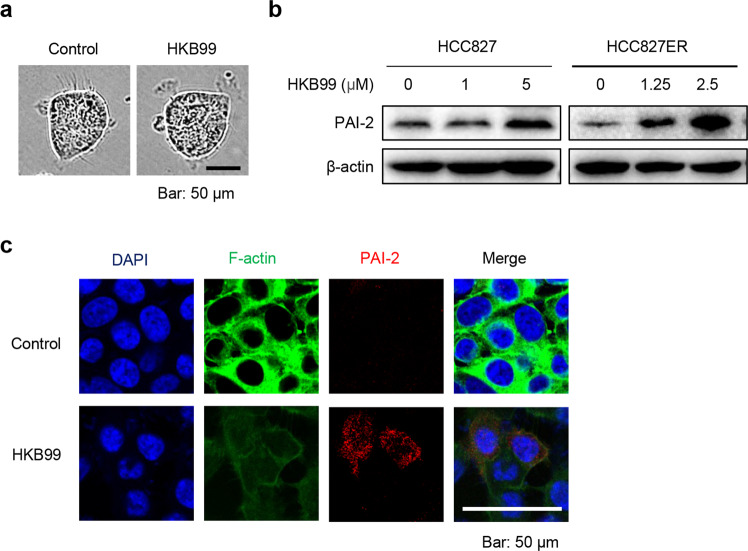
In accordance with our previous work, we observed that HKB99 quickly disrupted the invadopodia-like protrusions in HCC827ER cells (Fig. 3a). Combined with the result that HKB99 preferentially suppresses the proliferation of HCC827ER cells, we wondered whether HKB99 would also affect the level of PAI-2. As expected, HKB99 treatment of HCC827ER cells notably increased the level of PAI-2 after 6 h of short-term treatment; this was observed even after treatment with a low concentration of 1.25 μM, while no such upregulation of PAI-2 was observed until the HCC827 cells were treated with 5 μM HKB99 (Fig. 3b). We also treated HCC827 and HCC827ER cells with siPGAM1 or siPAI-2 individually. The data consistently showed that PGAM1 inhibition by siPGAM1 also increased the protein level of PAI-2, while siPAI2 had no effect on the level of PGAM1 (Fig. S2). The immunofluorescence results also showed that the level of F-actin was significantly decreased and that PAI-2 was increased in HCC827ER cells after 3 h of treatment with 5 μM HKB99 (Fig. 3c). Consistent with our hypothesis, HKB99 was shown to suppress invasive pseudopodia formation and upregulate PAI-2 in erlotinib-resistant NSCLC cells.
Fig. 3.
HKB99 preferentially inhibits invasive the formation of pseudopodia protrusions and increases the level of PAI-2 in HCC827ER cells. a HCC827ER cells were treated with 5 μM HKB99 for 3 h, and cell morphology was observed under a phase contrast microscope. b The protein level of PAI-2 was determined after HKB99 treatment of HCC827 and HCC827ER cells at the indicated concentration for 6 h by immunoblotting. c Detection of F-actin and PAI-2 proteins by immunofluorescence after HCC827ER cells were treated with 5 μM HKB99 for 3 h. Representative images were captured by laser scanning confocal microscopy. Scale bar, 50 μm
Discussion
Increasing evidence indicates that PAI-2 acts as a tumor suppressor. The PAI-2/uPA axis regulates stromal remodeling and local invasion in pancreatic cancer [20]. PAI-2 that translocates to the nucleus in normal cells exerts a DNA repair function that is blocked in tumor cells [7]. Another recent study showed that the kindlin-2/p53/SerpinB2 signaling axis regulates cellular senescence in breast cancer [21]. Furthermore, PAI-2 downregulation contributes to acquired resistance to cisplatin in head and neck carcinomas [22]. Additionally, low PAI-2 levels are associated with gefitinib resistance and the formation of invadopodia-like structure protrusions in NSCLC [11]. In accordance with recent studies, our results showed that PAI-2 levels were decreased and that more invasive pseudopodia structures were observed in erlotinib-resistant HCC827ER cells than were seen in the parental HCC827 cells. siPAI-2s-mediated knockdown of PAI-2 in HCC827 cells resulted in decreased sensitivity to erlotinib treatment. Taken together, these findings imply a potential role for PAI-2 in acquired resistance to erlotinib in NSCLC.
Metabolic enzymes are involved in acquired resistance to EGFR-TKIs in NSCLC treatment. Therefore, targeting key metabolic enzymes may represent a promising strategy for overcoming EGFR-TKI resistance in NSCLC targeted therapy [23, 24]. Our previous work identified a novel PGAM1 allosteric inhibitor, HKB99, which blocks the function of PGAM1, suppresses tumor growth and metastasis and overcomes erlotinib resistance in NSCLC by impacting both PGAM1’s metabolic activity and its non-metabolic function. Furthermore, HKB99 preferentially inhibits cell growth and induces more apoptosis in erlotinib-resistant HCC827ER cells than is observed in their parental HCC827 cells [19]. Here, we found that HKB99 preferentially inhibited invasive pseudopodia formation and increased the level of PAI-2 to a greater extent in HCC827ER cells than it did in parental HCC827 cells (Fig. 3b), which reveals a role of PAI-2 in regulating the sensitivity of erlotinib-resistant NSCLC cells to PGAM1 inhibitors. However, the specific signaling pathway contributing to the upregulation of PAI-2 by treatment with allosteric PGAM1 inhibitors still needs further investigation.
Previous work has reported that aberrant PGAM1 levels correlate with poor prognosis in NSCLC patients [16, 19, 25], whereas low expression of PAI-2 correlates with reduced survival in patients with lung adenocarcinomas [10]. Here, we observed that NSCLC patients with PGAM1high vs. PAI-2low had poorer prognosis than those with PGAM1low vs. PAI-2high expression (P = 0.0641; Fig. S3), which we determined using a combined survival analysis of an NSCLC cohort composed of 500 individuals containing PGAM1 and PAI-2 mRNA data, in which PGAM1 and PAI-2 expression was divided based on quartiles. Combined with the results from our current study, we conclude that it is worth exploring the clinical significance of PGAM1 and PAI-2 expression in EGFR-mutated lung cancer patients.
In conclusion, our study suggests that the downregulation of PAI-2 is associated with acquired resistance to erlotinib in NSCLC cells. Moreover, the induction of PAI-2 by HKB99 might be considered a potential biomarker for predicting the efficacy of the PGAM1 allosteric inhibitor in NSCLC therapy.
Supplementary information
Acknowledgements
We are grateful for grants from the National Natural Science Foundation of China (81773748, 81473232, 21472026, 21877014, 81573018 and 81872841), the Construction Project of High Level Local Universities in Shanghai and Pharmacy (XD18011) and the Interdisciplinary Program of Shanghai Jiaotong University (YG2017MS82).
Author contributions
YS, LZ, and LZ conceived the project. QL, WMG, KH, MYL, JHZ, and HML designed and conducted the experiments. The entire project was supervised by HZC, and GLZ provided some essential materials, such as NSCLC cells. YS, LZ, and LZ wrote the manuscript with input from QL, WMG, and KH. All the authors contributed to the manuscript.
Competing interests
YS, LZ, HZC, QL, WMG, KH, and MYL are named inventors of pending patent applications (201810273938.9 and 201910284333.4, to the Chinese Patent Office) related to the work. The other authors declare no competing interests.
Footnotes
These authors contributed equally: Qian Liang, Wei-ming Gu, Ke Huang
Contributor Information
Liang Zhu, Email: zhuliang17@126.com.
Lu Zhou, Email: zhoulu@fudan.edu.cn.
Ying Shen, Email: yshen0510@sjtu.edu.cn.
Supplementary information
The online version of this article (10.1038/s41401-020-0399-1) contains supplementary material, which is available to authorized users.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. Cancer J Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Rodriguez-Canales J, Parra-Cuentas E, Wistuba II. Diagnosis and molecular classification of lung cancer. Cancer Treat Res. 2016;170:25–46. doi: 10.1007/978-3-319-40389-2_2. [DOI] [PubMed] [Google Scholar]
- 3.Maemondo M, Inoue A, Kobayashi K, Sugawara S, Oizumi S, Isobe H, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med. 2010;362:2380–8. doi: 10.1056/NEJMoa0909530. [DOI] [PubMed] [Google Scholar]
- 4.Wu YL, Zhou C, Liam CK, Wu G, Liu X, Zhong Z, et al. First-line erlotinib versus gemcitabine/cisplatin in patients with advanced EGFR mutation-positive non-small-cell lung cancer: analyses from the phase III, randomized, open-label, ENSURE study. Ann Oncol. 2015;26:1883–9. doi: 10.1093/annonc/mdv270. [DOI] [PubMed] [Google Scholar]
- 5.Wu YL, Zhou C, Hu CP, Feng J, Lu S, Huang Y, et al. Afatinib versus cisplatin plus gemcitabine for first-line treatment of Asian patients with advanced non-small-cell lung cancer harbouring EGFR mutations (LUX-Lung 6): an open-label, randomised phase 3 trial. Lancet Oncol. 2014;15:213–22. doi: 10.1016/S1470-2045(13)70604-1. [DOI] [PubMed] [Google Scholar]
- 6.Chong CR, Janne PA. The quest to overcome resistance to EGFR-targeted therapies in cancer. Nat Med. 2013;19:1389–400. doi: 10.1038/nm.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Majoros H, Ujfaludi Z, Borsos BN, Hudacsek VV, Nagy Z, Coin F, et al. SerpinB2 is involved in cellular response upon UV irradiation. Sci Rep. 2019;9:2753. doi: 10.1038/s41598-019-39073-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robert C, Bolon I, Gazzeri S, Veyrenc S, Brambilla C, Brambilla E. Expression of plasminogen activator inhibitors 1 and 2 in lung cancer and their role in tumor progression. Clin Cancer Res. 1999;5:2094–102. [PubMed] [Google Scholar]
- 9.He C, He P, Liu LP, Zhu YS. Analysis of expressions of components in the plasminogen activator system in high- and low-metastatic human lung cancer cells. J Cancer Res Clin Oncol. 2001;127:180–6. doi: 10.1007/s004320000192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ramnefjell M, Aamelfot C, Helgeland L, Akslen LA. Low expression of SerpinB2 is associated with reduced survival in lung adenocarcinomas. Oncotarget. 2017;8:90706–18. doi: 10.18632/oncotarget.21456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bae SY, Park HJ, Hong JY, Lee HJ, Lee SK. Down-regulation of SerpinB2 is associated with gefitinib resistance in non-small cell lung cancer and enhances invadopodia-like structure protrusions. Sci Rep. 2016;6:32258. doi: 10.1038/srep32258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 13.Hitosugi T, Zhou L, Elf S, Fan J, Kang HB, Seo JH, et al. Phosphoglycerate mutase 1 coordinates glycolysis and biosynthesis to promote tumor growth. Cancer Cell. 2012;22:585–600. doi: 10.1016/j.ccr.2012.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qu J, Sun W, Zhong J, Lv H, Zhu M, Xu J, et al. Phosphoglycerate mutase 1 regulates dNTP pool and promotes homologous recombination repair in cancer cells. J Cell Biol. 2017;216:409–24. doi: 10.1083/jcb.201607008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang D, Jin N, Sun W, Li X, Liu B, Xie Z, et al. Phosphoglycerate mutase 1 promotes cancer cell migration independent of its metabolic activity. Oncogene. 2017;36:2900–9. doi: 10.1038/onc.2016.446. [DOI] [PubMed] [Google Scholar]
- 16.Chen G, Gharib TG, Wang H, Huang CC, Kuick R, Thomas DG, et al. Protein profiles associated with survival in lung adenocarcinoma. Proc Natl Acad Sci U S A. 2003;100:13537–42. doi: 10.1073/pnas.2233850100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peng XC, Gong FM, Chen Y, Qiu M, Cheng K, Tang J, et al. Proteomics identification of PGAM1 as a potential therapeutic target for urothelial bladder cancer. J Proteom. 2016;132:85–92. doi: 10.1016/j.jprot.2015.11.027. [DOI] [PubMed] [Google Scholar]
- 18.Wen CL, Huang K, Jiang LL, Lu XX, Dai YT, Shi MM, et al. An allosteric PGAM1 inhibitor effectively suppresses pancreatic ductal adenocarcinoma. Proc Natl Acad Sci U S A. 2019;116:23264–73. doi: 10.1073/pnas.1914557116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang K, Liang Q, Zhou Y, Jiang LL, Gu WM, Luo MY, et al. A novel allosteric inhibitor of phosphoglycerate mutase 1 suppresses growth and metastasis of non-small-cell lung cancer. Cell Metab. 2019;30:1107–19. doi: 10.1016/j.cmet.2019.09.014. [DOI] [PubMed] [Google Scholar]
- 20.Harris NLE, Vennin C, Conway JRW, Vine KL, Pinese M, Cowley MJ, et al. SerpinB2 regulates stromal remodelling and local invasion in pancreatic cancer. Oncogene. 2017;36:4288–98. doi: 10.1038/onc.2017.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sossey-Alaoui K, Pluskota E, Szpak D, Plow EF. The Kindlin2-p53-SerpinB2 signaling axis is required for cellular senescence in breast cancer. Cell Death Dis. 2019;10:539. doi: 10.1038/s41419-019-1774-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang Z, Li H, Huang Q, Chen D, Han J, Wang L, et al. SERPINB2 down-regulation contributes to chemoresistance in head and neck cancer. Mol Carcinog. 2014;53:777–86. doi: 10.1002/mc.22033. [DOI] [PubMed] [Google Scholar]
- 23.Lei HM, Zhang KR, Wang CH, Wang Y, Zhuang GL, Lu LM, et al. Aldehyde dehydrogenase 1A1 confers erlotinib resistance via facilitating the reactive oxygen species-reactive carbonyl species metabolic pathway in lung adenocarcinomas. Theranostics. 2019;9:7122–39. doi: 10.7150/thno.35729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dong JK, Lei HM, Liang Q, Tang YB, Zhou Y, Wang Y, et al. Overcoming erlotinib resistance in EGFR mutation-positive lung adenocarcinomas through repression of phosphoglycerate dehydrogenase. Theranostics. 2018;8:1808–23. doi: 10.7150/thno.23177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun Q, Li S, Wang Y, Peng H, Zhang X, Zheng Y, et al. Phosphoglyceric acid mutase-1 contributes to oncogenic mTOR-mediated tumor growth and confers non-small cell lung cancer patients with poor prognosis. Cell Death Differ. 2018;25:1160–73. doi: 10.1038/s41418-017-0034-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.