Abstract
Precision medicine is now pivotal to design patients' specific treatment strategies with the aim of prolonging progression and overall survival. In this regard, invasive tumor tissue testing has so far been the golden standard for making cancer diagnosis, but has limitations. Cell-free tumor DNA (ctDNA), a form of liquid biopsy, is a noninvasive biomarker that can be isolated from patients' blood and other biofluids. An increasing body of evidence has demonstrated clinical utility of plasma ctDNA profiling to select patients for genomic-driven therapies. Analyses of mutations in plasma ctDNA have shown high accuracy and more rapid identification of mutations, allowing matching patients for specific therapies with equivalent clinical efficacy to that of the tissue profiling. In the clinical setting, ctDNA has been recently implemented to select patients with specific genomic alterations to targeted treatments, and a few molecular tests have been approved for use in non-small-cell lung, prostate, ovarian, and breast cancers. However, standardization of ctDNA collection, storage, and analysis methods would be critical to facilitate the wide adoption of ctDNA technology in routine clinical practice. This review summarizes how we can exploit ctDNA analysis to treat cancer patients, and explains how the results should be interpreted. In addition, we focus on how ctDNA could be used in the future as a marker of minimal residual disease to guide adjuvant therapy, as an immuno-oncology biomarker in patients treated with immune checkpoint blockade drugs, and as an early cancer detection marker to screen the asymptomatic population.
Key words: liquid biopsy, circulating tumor DNA, cancer treatment, next generation sequencing, genomics, noninvasive, early detection
Highlights
-
•
Liquid biopsy-based cell-free tumor DNA (ctDNA) may detect and allow matching and monitoring patients’ genomic alterations to specific therapies.
-
•
In the clinics, ctDNA has been proven to guide therapy in non-small-cell lung, prostate, ovarian, and breast cancer patients.
-
•
The detection of minimal residual disease to guide adjuvant therapy represents a potential clinical use of ctDNA.
-
•
In ctDNA, tumor mutational burden and microsatellite instability may inform eligibility for an immune checkpoint inhibitor.
-
•
Standardization procedures for ctDNA detection and analysis are still needed.
Introduction
Timely and efficient identification of genomic alterations remain an obstacle to tailor cancer treatments and to achieve the goals of precision medicine.
Tumor tissue analysis is the golden standard for making cancer diagnosis and requires either formaldehyde-fixed and paraffin-embedded or frozen tumor biopsies. Methods to identify mutations or other genomic alterations require those invasive, difficult-to-obtain clinical biopsies, or alternatively the use of archival specimens, which may not reflect the current genomic heterogeneity of the disease, and that may preclude clinical interventions, including assessment for new precision medicines.1,2
Peripheral blood sampling is a minimally invasive approach that may substitute for tissue as a source of tumor-derived material. A significant proportion of cell-free tumor DNA (ctDNA) seems to be released by degraded or dying cancer cells into the blood of cancer patients. Plasma ctDNA analysis, a type of liquid biopsy, has opened previously unexpected perspectives for monitoring cancer genomics in the peripheral blood, and has shown to be useful for (i) revealing new targets for therapy selection, (ii) monitoring cancer evolution during the administration of targeted and immunotherapies, (iii) assessing minimal residual disease (MRD), (iv) uncovering intratumor heterogeneity, and potentially (v) helping making cancer diagnosis.1, 2, 3, 4
Clinical indications of plasma ctDNA
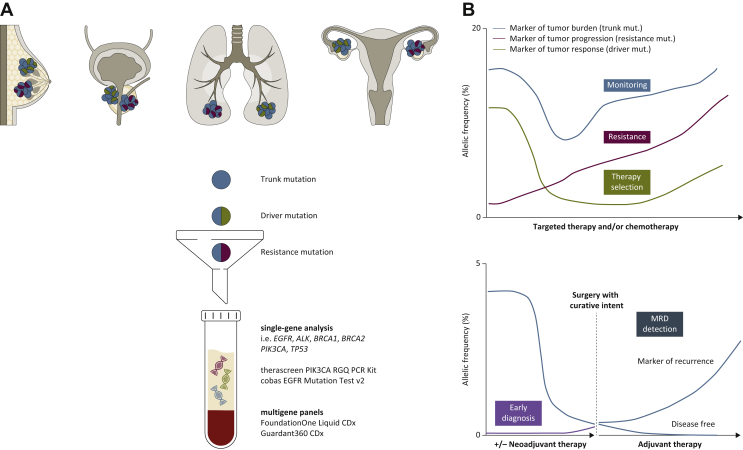
An increasing body of evidence is demonstrating clinical utility of plasma ctDNA profiling to select patients for genomic-driven therapies (Figure 1). Analyses of mutations in plasma ctDNA have shown high concordance and more rapid identification of mutations, allowing matching patients for specific therapies with equivalent clinical efficacy to that of the standard-of-care tissue profiling.5,6
Figure 1.
Plasma cell-free tumor DNA (ctDNA) clinical utility as a liquid biopsy.
(A) ctDNA can be shed by various tumor types, including breast, lung, prostate, and ovarian cancers. Each lesion can harbor different alterations (trunk mutation, blue, shared by all the clones as it occurs early in the tumorigenesis; driver mutation, green, and resistance mutation, red, which can emerge after treatment). Once in the blood, ctDNA can be collected by venipuncture, and the sample is then processed to obtain plasma. After isolation, ctDNA can be assessed to investigate for molecular alterations by two approaches: the single-gene analysis (PCR-based methods such as droplet digital PCR, upper panel), or the multigene panel analysis (next-generation sequencing, lower panel). Four tests have by now been approved by the US Food and Drug Administration (FDA) for clinical use: the therascreen PIK3CA RGQ PCR Kit and the cobas EGFR Mutation Test v2, and FoundationOne Liquid CDx and Guardant360 CDx, for the single-gene and multigene analysis, respectively. (B) ctDNA as a biomarker with different dynamics, where the percentage of mutational allelic frequency is quantified throughout all the collected ctDNA timepoints. Upper panel: ctDNA is assessed for three biomarkers: (i) for patients' monitoring during treatment, with a trunk mutation, as a marker of tumor burden, blue line; (ii) for therapy selection, with a driver mutation, as a marker of tumor response to therapy, green line; and (iii) with a resistance mutation, as a marker of tumor progression and resistance to therapy, red line. Lower panel: ctDNA used as a marker of early tumor detection and diagnosis in patients with early stage cancer or in asymptomatic population (purple line). In addition, ctDNA can be used to detect minimal residual disease (MRD, blue line) in cancer patients undergoing surgery with curative intent, with or without prior neoadjuvant treatment. If ctDNA after surgery is still detected, then the patient could be guided to receive adjuvant therapy and monitored for further disease recurrence; if ctDNA after surgery is not detected, then the patient could be considered disease free.
Currently, the metastatic setting is where liquid biopsies provide the best opportunity for their clinical utility, while guiding either the first-line or subsequent lines of systemic therapy. In non-small-cell lung cancer (NSCLC),7 ctDNA has enabled the identification and monitoring of several actionable genomic mutations in EGFR, ERBB2, MET, ALK, ROS1, and RET. Similarly, plasma ctDNA has been exploited in breast cancer patients to detect PIK3CA, AKT, HER2, ESR1 mutations and match patients' cancer having those actionable mutations to specific targeted therapies.8
ctDNA has proven useful to detect molecular alterations associated with primary and acquired resistance to targeted agents, with the ultimate goal of directing additional, more effective lines of therapies that may improve clinical outcome.3 ctDNA analyses were also fundamental in showing that a subset of colorectal cancer (CRC) patients acquire a plethora of EGFR, KRAS, NRAS, BRAF, ERBB2, and PIK3CA mutations as mechanisms of resistance to anti-EGFR drugs.3 In breast cancer, ESR1 mutations and PTEN loss mediate resistance to aromatase inhibitors and PI3K-alpha inhibition, respectively.9 Uncovering resistance mutations faster using a blood sample offers the possibility to provide action in highly metastatic patients, tailoring specific therapy and enrolling patients in clinical trials.
Currently, EGFR mutations and ALK rearrangements for NSCLC, PIK3CA mutations for hormone receptor-positive HER2-negative breast cancer, and BRCA1 and 2 mutations (germline and/or somatic) for ovarian cancer and castration-resistance prostate cancer were cleared as US Food and Drug Administration (FDA) companion diagnostic tests and have been used to select patients with advanced cancers for targeted therapy (Table 1).
Table 1.
Liquid biopsy companion diagnostic indicationsa
| Test | Tumor type | Genomic alteration detected | Therapy |
|---|---|---|---|
| FoundationOne Liquid CDx | Metastatic NSCLC | EGFR Exon 19 deletions and EGFR Exon 21 L858R substitution | Osimertinib |
| Gefitinib, Erlotinib | |||
| ALK rearrangements | Alectinib | ||
| Metastatic castration-resistant prostate cancer | BRCA1, BRCA2, ATM mutations | Rucaparib, Olaparib | |
| Metastatic ovarian cancer | BRCA1, BRCA2 alterations | Rucaparib | |
| Metastatic HR+/HER2− breast cancer | PIK3CA mutations C420R, E542K, E545A, E545D [1635G>T only], E545G, E545K, Q546E, Q546R; and H1047L, H1047R, and H1047Y | Alpelisib | |
| Guardant360 CDx | Metastatic NSCLC | EGFR exon 19 deletions, L858R, and T790M∗ | Osimertinib |
| cobas EGFR Mutation Test v2 | Metastatic NSCLC | Plasma (Erlotinib): Exon 19 deletions and L858R exon 19 deletions and L858R. Osimertinib: Exon 19 deletions, L858R and T790M exon 19 deletions, L858R and T790M∗ Gefitinib: Exon 19 deletions and L858R exon 19 deletions and L858R |
Osimertinib Erlotinib, Osimertinib, Gefitinib |
| therascreen PIK3CA RGQ PCR Kit | Metastatic HR+/HER2− breast cancer | 11 Mutations in the PIK3CA gene [exon 7: C420R; exon 9: E542K, E545A, E545D (1635G>T only), E545G, E545K, Q546E, Q546R; and exon 20: H1047L, H1047R, H1047Y] | Alpelisib |
NSCLC, non-small-cell lung cancer.
At the time this article was prepared, FoundationOne Liquid CDx and Guardant360 are intended to provide tumor mutation profiling and are to be used by qualified health care professionals in oncology for patients with the solid tumors indicated above.
An unmet clinical need: MRD detection and adjuvant therapy
A current struggle is whether we should treat all patients after a therapy with curative intent, or whether one should wait and identify if MRD becomes apparent during subsequent follow up.
Evidence suggests that adjuvant therapy might improve overall survival in some cases,10 and that ctDNA analysis could identify patients for whom remains evidence of residual, radiographically occult cancer.11 Although it seems that pretreatment ctDNA levels alone do not hold prognostic power, serial ctDNA sampling may be useful to predict relapse in patients with MRD and to discriminate those cases where adjuvant treatment can be spared from those who need it.
Three research groups have so far tried to show how ctDNA analysis can be exploited to detect MRD and guide adjuvant treatment: (i) Tie et al.12,13 performed two prospective cohort studies (stage II and stage III colon cancer, respectively) where ctDNA resulted to have more prognostic power than standard clinicopathologic characteristics; (ii) Reinert et al.14 performed a prospective study on CRC patients in stages I-III where plasma ctDNA was assessed before and after surgery by a personalized tumor-informed next-generation sequencing analysis. Patients who displayed positive ctDNA after day 30 since surgery were seven times more likely to recur compared to those with negative ctDNA; (iii) Tarazona et al.15 recently reported similar data. They obtained and tested longitudinal ctDNA from patients with resected, localized colon cancer and found that ctDNA positivity after surgery was highly correlated with cancer recurrence.
Furthermore, the IDEA-France data16 from a retrospective analysis reported that a post-operative positive ctDNA is an independent negative prognostic marker for cancer relapse in stage III CRC patients receiving oxaliplatin-based adjuvant chemotherapy.
Overall, all these studies combined effectively show that ctDNA can be exploited as an accurate biomarker of MRD. Several clinical trials [i.e. DYNAMIC II (ACTRN12615000381583), COBRA (NCT0406810) and CIRCULATE (NCT04120701), C-TRAK TN (NCT03145961)], are currently ongoing to define whether ctDNA can be considered a surrogate biomarker of MRD in larger cohorts of patients, or whether the detection of cancer cells after standard treatment could trigger additional treatment. Of note, it remains unknown whether more patients could be cured with the optimization of a liquid biopsy to detect MRD or if this would simply delay by some months the development of metastasis.
More recently, Parikh et al.17,18 have shown that combining the analysis of epigenomic and genomic alterations in ctDNA has improved sensitivity for recurrence detection in CRC patients within 1 year after surgery, with a non-tumor-informed, plasma-only assay.
ctDNA for early cancer detection and screening of the asymptomatic population
Liquid biopsy approaches, combined with the development of more sensitive techniques, have the potentials to transform cancer screening or diagnosis in an asymptomatic population. Chabon et al.19 have optimized their method for the analysis of ctDNA, named cancer personalized profiling by deep sequencing (CAPP-Seq),20 to enable the screening of individuals at risk of NSCLC. The Lung-CLiP study results using ctDNA analyses for NSCLC screening achieved by applying a CAPP-Seq and machine-learning method highlights two important messages. First, ctDNA was present prior to therapy in most of the tested patients, and its presence was strongly prognostic.19 Second, the majority of the somatic mutations found in the ctDNA of NSCLC patients and of risk-matched controls were nonrecurrent, and revealed clonal hematopoiesis.
Recently, Liu et al.21 used an innovative targeted methylation-based approach and classifier to analyze, detect, and localize multiple cancer types in ctDNA among 6689 participants in a case–control substudy of The Circulating Cell-free Genome Atlas (CCGA; NCT02889978). This substudy involved >50 cancer types across stages. Not surprisingly, cancer detection improved at higher stages and cancer localization was accurate in 93% of cases.21 Results support the viability of using targeted methylation analysis of cell-free DNA in ongoing clinical trials in the intended use population for early cancer detection.
Other ongoing clinical studies [STRIVE (NCT03085888), SUMMIT (NCT03934866), PATHFINDER (NCT04241796)] assessing cancer patients, healthy individuals without cancer, and those at high risk for cancer will demonstrate the role of ctDNA (e.g. methylation analyses) for making cancer diagnosis.
Lastly, future approaches should also incorporate multi-omics approaches, combining genomic and standard serum markers: in this regard, CancerSEEK22 has been reported to be potentially successful in screening the asymptomatic population.
Liquid biopsy as an immuno-oncology biomarker
Tumor PD-L1 expression is the only approved biomarker, albeit imperfect, used in clinical practice to select a subset of patients who would benefit from checkpoint blockers drugs (e.g. NSCLC, head and neck, bladder, stomach, esophageal, and cervical cancer are selected for pembrolizumab based on PD-L1 expression).23,24 Such limitation has stimulated the investigation of other markers, both in tissue and in blood.
ctDNA, proteins, and cytotoxic T cells are suitable targets in liquid biopsy for cancer immunotherapy. The quantification of ctDNA levels as predictive and prognostic biomarkers has shown encouraging results in cohorts of patients treated with immune checkpoint inhibitors.25,26 Furthermore, it has been shown that ctDNA analysis can identify MRD in patients with NSCLC displaying long-term responses to anti-PD-L1 and predict the risk of eventual relapse.27
In ctDNA, tumor mutational burden28 and microsatellite instability29 (e.g. for metastatic cancer patients with microsatellite instability-High or mismatch-repair-deficient solid tumors) may inform eligibility for treatment with an immune checkpoint inhibitor. Blood samples are also a source of circulating T cells and T-cell receptors and may allow the analysis of the collection of immune cells with either cytotoxic or immunosuppressive roles.30
Data retrieved from a proper quantification of immuno-genomic biomarkers could guide immunotherapy approaches such as neoantigen vaccine or adoptive cell therapies.31
Another scenario where ctDNA may be useful is to rule out true progression from pseudoprogression, which has 5%-10% incidence with immuno-oncology agents in solid tumors.32 A recent study including 29 melanoma patients showed the ability of ctDNA to differentiate between true progression and pseudoprogression to anti-PD-1 antibodies (90% sensitivity and 100% specificity).33
These data suggest that the use of liquid biopsies could select patients for immunotherapy or reduce prolonged, unnecessary treatment in patients who would not be benefitting further from treatment with immune checkpoint blockade.
Other liquid biopsy rather than plasma
Several studies have reported the presence of tumor-derived nucleic acids in other body fluids, such as cerebrospinal fluid (CSF),34, 35, 36, 37, 38 saliva,39 and urine,40, 41, 42 providing additional if not complementary information about a patient's tumor, that might be missed by plasma or tissue tests alone.
CSF-derived ctDNA is particularly easy to investigate as it is not diluted by the normal DNA that usually can be found in the blood. A few studies have investigated CSF and paired plasma, and tumor tissue from patients with tumor confined to the central nervous system (glioblastoma and medulloblastoma) as well as brain metastases from lung or breast cancer.34 Somehow expectedly, plasma ctDNA was found to be lower as compared with CFS for tumors localized in the central nervous system, suggesting that the latter could be exploited to longitudinally monitor tumor burden.34 Although additional studies are needed, CSF-derived ctDNA will probably complement the use of other already established biomarkers, clinical parameters, and radiological imaging.
The presence of saliva-derived ctDNA has been exploited to detect HPV and genomic point mutations in patients with head and neck squamous-cell carcinoma.39 Saliva ctDNA was shown to be enriched for ctDNA originating from the oral cavity, while plasma ctDNA is enriched for tumor DNA from other sites.
Urine-derived ctDNA has been shown to display tumor-specific genomic and epigenomic alterations in patients with urological, prostate, NSCLC, CRC, pancreatic cancer, and others.40, 41, 42, 43, 44, 45 Although assessing urine-derived ctDNA is more challenging due to the massive amount of normal DNA constantly released by the epithelial cells in the urinary tract, it represents, together with the saliva DNA, one of the completely noninvasive fluids, and therefore worth pursuing.
Overall, these evidence showed the importance of obtaining and analyzing bodily fluids in close proximity to the tumor anatomical localization in order to achieve higher sensitivity of ctDNA detection. If confirmed by future, larger studies, all the aforementioned fluids could be implemented into the clinical practice, to complement current routinely examinations, in both diagnosis and disease monitoring settings.
How to interpret liquid biopsy in the clinics
Currently, liquid biopsy tests do not have standardized procedures across workflows, and therefore lack reproducibility. In particular, the preanalytical phase is the most critical one, as it includes specimen collection and processing, transport and storage, ctDNA isolation, and quality controls. Errors within these steps can heavily affect the data generated in the following analytical steps, resulting in unreliable results which ultimately can lead to incorrect clinical decisions.
However, progress has been made, and there is now a general consensus among scientists on which tube is best to collect the fluid and on storage conditions. For instance, Cell-Free DNA BCT® RUO tubes are the best tubes for blood collection as they contain the preservative to stabilize the white blood cells and prevent them from lysis46; as for centrifugations, one should not use the brakes, but may need to discard the bottom of the sample to avoid taking cell debris. In addition, plasma storage at −80°C laboratory freezers is the accepted freezing condition. A consensus has not been reached yet on best practices overall, and other variables are also unknown.
Standardization of ctDNA collection, storage, and analysis methods would be critical to facilitate the wide adoption of ctDNA technology in routine clinical practice, and also to better interpret the results.
A negative result from a plasma specimen does not assure that the patient's tumor is negative for genomic findings. This could be attributed to the scarce amount of tumor DNA in blood, low test sensitivity, or the presence of ctDNA restricted to sanctuary sites.3,36 Patients who are negative for these mutations by plasma ctDNA specimens should be reflexed to routine biopsy and testing for mutations with the formaldehyde-fixed and paraffin-embedded sample type. By contrast, a positive liquid biopsy does not necessarily mean that a given mutation comes from tumor cells.
The role of clonal hematopoiesis of indeterminate potential, a condition in which there are age-dependent acquired mutations in hematopoietic progenitor cells in individuals without a clear diagnosis of malignancy, should be taken into consideration, so as its causes (e.g. aging, smoking) and consequences (e.g. myelodysplastic syndrome, acute myeloid leukemia, as well as cardiovascular disease, heart attacks, and strokes).47,48
The clinical significance of clonal hematopoiesis of indeterminate potential should be accounted for when interpreting ctDNA variants for both tumor genotyping and early cancer detection in the asymptomatic population.47,48 It is suggested that targeted sequencing would require concurrent sequencing of white blood cells/normal DNA to provide accurate results.47,48 Recently, it was observed that clonal hematopoiesis mutations, when compared with tumor-derived mutations of NSCLC patients, occur on longer cell-free DNA fragments and lack mutational signatures that are associated with tobacco smoking.19
In the setting of MRD, the improvement of technical aspects may contribute to the way ctDNA is used in the clinical practice. The collection of higher blood volume and the performance of novel analyses including assessing a higher number of mutations, assessing fragment size, adding epigenomics evaluation, exploiting multiple unique molecular identifiers to minimize PCR errors, and performing background polishing might increase assay sensitivity.49
A few international initiatives such as CANCER-ID,50 BloodPAC,51 and SPIDIA4P52 are making efforts to hopefully determine the standardization of liquid biopsy procedures, so that they can be more easily and rapidly implemented into clinical laboratories.
Conclusions
The ability of liquid biopsy to identify a real-time therapeutic target will make up-to-the-minute immuno-genomic analysis of the patient possible and will allow patients without available tissue to undergo novel personalized clinical trials. The presence of tumor-derived nucleic acids in other body fluids, such as CSF, saliva, and urine, may provide additional if not complementary information about a patient's tumor that might be missed by plasma or tissue tests alone.3,34,36
The choice of ctDNA assays in future prospective studies will need to balance considerations in sensitivity, specificity, availability, cost, and practicalities for implementation in the clinical practice. In addition, multigene panels with adequate positive predictive value, cost, and accessibility may allow the detection of immuno-genomic biomarkers and create more possibilities for matching patients for standard of care therapy or clinical trials.
Acknowledgments
Funding
LDM-A acknowledges funding support from Merck Salud Foundation (Spain) and Sorigué; GS is supported by the ECOR fund for Medical Discoveries 2020 (no grant numbers).
Disclosure
LDM-A has received honoraria for participation in a speaker's bureau/consultancy from Roche; has performed/performs research collaboration with NanoString Technologies; has received education grant from BMS; and reports research to institution (Grifols, Gilead, MSD, Jansen, and AbbVie). GS has no disclosure to declare.
References
- 1.De Mattos-Arruda L., Cortes J., Santarpia L. Circulating tumour cells and cell-free DNA as tools for managing breast cancer. Nat Rev Clin Oncol. 2013;10(7):377–389. doi: 10.1038/nrclinonc.2013.80. [DOI] [PubMed] [Google Scholar]
- 2.Siravegna G., Mussolin B., Venesio T. How liquid biopsies can change clinical practice in oncology. Ann Oncol. 2019;30(10):1580–1590. doi: 10.1093/annonc/mdz227. [DOI] [PubMed] [Google Scholar]
- 3.Siravegna G., Marsoni S., Siena S., Bardelli A. Integrating liquid biopsies into the management of cancer. Nat Rev Clin Oncol. 2017;14(9):531–548. doi: 10.1038/nrclinonc.2017.14. [DOI] [PubMed] [Google Scholar]
- 4.De Mattos-Arruda L., Weigelt B., Cortes J. Capturing intra-tumor genetic heterogeneity by de novo mutation profiling of circulating cell-free tumor DNA: a proof-of-principle. Ann Oncol. 2014;25(9):1729–1735. doi: 10.1093/annonc/mdu239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leighl N.B., Page R.D., Raymond V.M. Clinical utility of comprehensive cell-free DNA analysis to identify genomic biomarkers in patients with newly diagnosed metastatic non-small cell lung cancer. Clin Cancer Res. 2019;25:4691–4700. doi: 10.1158/1078-0432.CCR-19-0624. [DOI] [PubMed] [Google Scholar]
- 6.Nakamura Y., Taniguchi H., Ikeda M. Clinical utility of circulating tumor DNA sequencing in advanced gastrointestinal cancer: SCRUM-Japan GI-SCREEN and GOZILA studies. Nat Med. 2020;26:1859–1864. doi: 10.1038/s41591-020-1063-5. [DOI] [PubMed] [Google Scholar]
- 7.Thress K.S., Brant R., Carr T.H. EGFR mutation detection in ctDNA from NSCLC patient plasma: a cross-platform comparison of leading technologies to support the clinical development of AZD9291. Lung Cancer. 2015;90(3):509–515. doi: 10.1016/j.lungcan.2015.10.004. [DOI] [PubMed] [Google Scholar]
- 8.Turner N.C., Kingston B., Kilburn L.S. Circulating tumour DNA analysis to direct therapy in advanced breast cancer (plasmaMATCH): a multicentre, multicohort, phase 2a, platform trial. Lancet Oncol. 2020;21(10):1296–1308. doi: 10.1016/S1470-2045(20)30444-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Razavi P., Dickler M.N., Shah P.D. Alterations in PTEN and ESR1 promote clinical resistance to alpelisib plus aromatase inhibitors. Nat Cancer. 2020;1(4):382–393. doi: 10.1038/s43018-020-0047-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 11.Garcia-Murillas I., Schiavon G., Weigelt B. Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Sci Transl Med. 2015;7(302):302ra133. doi: 10.1126/scitranslmed.aab0021. [DOI] [PubMed] [Google Scholar]
- 12.Tie J., Wang Y., Tomasetti C. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci Transl Med. 2016;8(346):346ra92. doi: 10.1126/scitranslmed.aaf6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tie J., Cohen J.D., Wang Y. Circulating tumor DNA analyses as markers of recurrence risk and benefit of adjuvant therapy for stage III colon cancer. JAMA Oncol. 2019;5(12):1710–1717. doi: 10.1001/jamaoncol.2019.3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reinert T., Henriksen T.V., Christensen E. Analysis of plasma cell-free DNA by ultradeep sequencing in patients with stages I to III colorectal cancer. JAMA Oncol. 2019;5(8):1124–1131. doi: 10.1001/jamaoncol.2019.0528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tarazona N., Gimeno-Valiente F., Gambardella V. Targeted next-generation sequencing of circulating-tumor DNA for tracking minimal residual disease in localized colon cancer. Ann Oncol. 2019;30(11):1804–1812. doi: 10.1093/annonc/mdz390. [DOI] [PubMed] [Google Scholar]
- 16.Taieb J., Taly V., Vernerey D. Analysis of circulating tumour DNA (ctDNA) from patients enrolled in the IDEA-FRANCE phase III trial: prognostic and predictive value for adjuvant treatment duration. Ann Oncol. 2019;30:v867. [Google Scholar]
- 17.Parikh A.R., Van Seventer E.E., Boland G.M. A plasma-only integrated genomic and epigenomic circulating tumor DNA (ctDNA) assay to inform recurrence risk in colorectal cancer (CRC) J Clin Oncol. 2019;37(suppl 15):3602. [Google Scholar]
- 18.Parikh A., Van Seventer E., Boland G. Serial assessment of cell-free circulating tumor DNA (ctDNA) to assess treatment effect and minimal residual disease during neoadjuvant and adjuvant therapy in colorectal cancer. Ann Oncol. 2019;30:iv131. [Google Scholar]
- 19.Chabon J.J., Hamilton E.G., Kurtz D.M. Integrating genomic features for non-invasive early lung cancer detection. Nature. 2020;580:245–251. doi: 10.1038/s41586-020-2140-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Newman A.M., Lovejoy A.F., Klass D.M. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat Biotechnol. 2016;34(5):547–555. doi: 10.1038/nbt.3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu M.C., Oxnard G.R., Klein E.A. Sensitive and specific multi-cancer detection and localization using methylation signatures in cell-free DNA. Ann Oncol. 2020;31(6):745–759. doi: 10.1016/j.annonc.2020.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cohen J.D., Li L., Wang Y. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science. 2018;359(6378):926–930. doi: 10.1126/science.aar3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maleki Vareki S., Garrigós C., Duran I. Biomarkers of response to PD-1/PD-L1 inhibition. Crit Rev Oncol Hematol. 2017;116:116–124. doi: 10.1016/j.critrevonc.2017.06.001. [DOI] [PubMed] [Google Scholar]
- 24.Darvin P., Toor S.M., Sasidharan Nair V., Elkord E. Immune checkpoint inhibitors: recent progress and potential biomarkers. Exp Mol Med. 2018;50(12):165. doi: 10.1038/s12276-018-0191-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cabel L., Riva F., Servois V. Circulating tumor DNA changes for early monitoring of anti-PD1 immunotherapy: a proof-of-concept study. Ann Oncol. 2017;28:1996–2001. doi: 10.1093/annonc/mdx212. [DOI] [PubMed] [Google Scholar]
- 26.Bratman S.V., Yang S.Y.C., Iafolla M.A.J. Personalized circulating tumor DNA analysis as a predictive biomarker in solid tumor patients treated with pembrolizumab. Nat Cancer. 2020;1(9):873–881. doi: 10.1038/s43018-020-0096-5. [DOI] [PubMed] [Google Scholar]
- 27.Hellmann M.D., Nabet B.Y., Rizvi H. Circulating tumor DNA analysis to assess risk of progression after long-term response to PD-(L)1 blockade in NSCLC. Clin Cancer Res. 2020;26:2849–2858. doi: 10.1158/1078-0432.CCR-19-3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Samstein R.M., Lee C.-H., Shoushtari A.N. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet. 2019;51(2):202–206. doi: 10.1038/s41588-018-0312-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rothwell D.G., Ayub M., Cook N. Utility of ctDNA to support patient selection for early phase clinical trials: the TARGET study. Nat Med. 2019;25(5):738–743. doi: 10.1038/s41591-019-0380-z. [DOI] [PubMed] [Google Scholar]
- 30.Gros A., Parkhurst M.R., Tran E. Prospective identification of neoantigen-specific lymphocytes in the peripheral blood of melanoma patients. Nat Med. 2016;22(4):433–438. doi: 10.1038/nm.4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.De Mattos-Arruda L., Vazquez M., Finotello F. Neoantigen prediction and computational perspectives towards clinical benefit: recommendations from the ESMO Precision Medicine Working Group. Ann Oncol. 2020;31(8):978–990. doi: 10.1016/j.annonc.2020.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park H.J., Kim K.W., Pyo J. Incidence of pseudoprogression during immune checkpoint inhibitor therapy for solid tumors: a systematic review and meta-analysis. Radiology. 2020;297:87–96. doi: 10.1148/radiol.2020200443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee J.H., Long G.V., Menzies A.M. Association between circulating tumor DNA and pseudoprogression in patients with metastatic melanoma treated with anti–programmed cell death 1 antibodies. JAMA Oncol. 2018;4(5):717–721. doi: 10.1001/jamaoncol.2017.5332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.De Mattos-Arruda L., Mayor R., Ng C.K.Y. Cerebrospinal fluid-derived circulating tumour DNA better represents the genomic alterations of brain tumours than plasma. Nat Commun. 2015;6(1):8839. doi: 10.1038/ncomms9839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pan W., Gu W., Nagpal S. Brain tumor mutations detected in cerebral spinal fluid. Clin Chem. 2015;61(3):514–522. doi: 10.1373/clinchem.2014.235457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seoane J., De Mattos-Arruda L., Le Rhun E., Bardelli A., Weller M. Cerebrospinal fluid cell-free tumour DNA as a liquid biopsy for primary brain tumours and central nervous system metastases. Ann Oncol. 2019;30(2):211–218. doi: 10.1093/annonc/mdy544. [DOI] [PubMed] [Google Scholar]
- 37.Escudero L., Llort A., Arias A. Circulating tumour DNA from the cerebrospinal fluid allows the characterisation and monitoring of medulloblastoma. Nat Commun. 2020;11(1):5376. doi: 10.1038/s41467-020-19175-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller A.M., Shah R.H., Pentsova E.I. Tracking tumour evolution in glioma through liquid biopsies of cerebrospinal fluid. Nature. 2019;565:654–658. doi: 10.1038/s41586-019-0882-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Y., Springer S., Mulvey C.L. Detection of somatic mutations and HPV in the saliva and plasma of patients with head and neck squamous cell carcinomas. Sci Transl Med. 2015;7(293):293ra104. doi: 10.1126/scitranslmed.aaa8507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Siravegna G., Sartore-Bianchi A., Mussolin B. Tracking aCAD-ALK gene rearrangement in urine and blood of a colorectal cancer patient treated with an ALK inhibitor. Ann Oncol. 2017;28(6):1302–1308. doi: 10.1093/annonc/mdx095. [DOI] [PubMed] [Google Scholar]
- 41.Dudley J.C., Schroers-Martin J., Lazzareschi D.V. Detection and surveillance of bladder cancer using urine tumor DNA. Cancer Discov. 2019;9(4):500–509. doi: 10.1158/2159-8290.CD-18-0825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith C.G., Moser T., Mouliere F. Comprehensive characterization of cell-free tumor DNA in plasma and urine of patients with renal tumors. Genome Med. 2020;12(1):23. doi: 10.1186/s13073-020-00723-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Crisafulli G., Mussolin B., Cassingena A. Whole exome sequencing analysis of urine trans-renal tumour DNA in metastatic colorectal cancer patients. ESMO Open. 2019;4(6):e000572. doi: 10.1136/esmoopen-2019-000572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Casadio V., Calistri D., Salvi S. Urine cell-free DNA integrity as a marker for early prostate cancer diagnosis: a pilot study. Biomed Res Int. 2013;2013:1–5. doi: 10.1155/2013/270457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fujii T., Barzi A., Sartore-Bianchi A. Mutation-enrichment next-generation sequencing for quantitative detection of KRAS mutations in urine cell-free DNA from patients with advanced cancers. Clin Cancer Res. 2017;23(14):3657–3666. doi: 10.1158/1078-0432.CCR-16-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zill O.A., Greene C., Sebisanovic D. Cell-free DNA next-generation sequencing in pancreatobiliary carcinomas. Cancer Discov. 2015;5(10):1040–1048. doi: 10.1158/2159-8290.CD-15-0274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Steensma D.P., Bejar R., Jaiswal S. Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood. 2015;126(1):9–16. doi: 10.1182/blood-2015-03-631747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Swanton C., Venn O., Aravanis A. Prevalence of clonal hematopoiesis of indeterminate potential (CHIP) measured by an ultra-sensitive sequencing assay: exploratory analysis of the Circulating Cancer Genome Atlas (CCGA) study. J Clin Oncol. 2018;36:12003. [Google Scholar]
- 49.Chakrabarti S., Xie H., Urrutia R., Mahipal A. The promise of circulating tumor DNA (ctDNA) in the management of early-stage colon cancer: a critical review. Cancers (Basel) 2020;12(10):2808. doi: 10.3390/cancers12102808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cancer-ID https://www.cancer-id.eu/ Available at:
- 51.BloodPAC https://www.bloodpac.org/ Available at:
- 52.SPIDIA https://www.spidia.eu/ Available at: