Abstract
As the COVID-19 pandemic continues, there is an imminent need for rapid diagnostic tools and effective antivirals targeting SARS-CoV-2. We have developed a novel bioluminescence-based biosensor to probe a key host-virus interaction during viral entry: the binding of SARS-CoV-2 viral spike (S) protein to its receptor, angiotensin-converting enzyme 2 (ACE2). Derived from Nanoluciferase binary technology (NanoBiT), the biosensor is composed of Nanoluciferase split into two complementary subunits, Large BiT and Small BiT, fused to the Spike S1 domain of the SARS-CoV-2 S protein and ACE2 ectodomain, respectively. The ACE2-S1 interaction results in reassembly of functional Nanoluciferase, which catalyzes a bioluminescent reaction that can be assayed in a highly sensitive and specific manner. We demonstrate the biosensor's large dynamic range, enhanced thermostability and pH tolerance. In addition, we show the biosensor's versatility towards the high-throughput screening of drugs which disrupt the ACE2-S1 interaction, as well as its ability to act as a surrogate virus neutralization assay. Results obtained with our biosensor correlate well with those obtained with a Spike-pseudotyped lentivirus assay. This rapid in vitro tool does not require infectious virus and should enable the timely development of antiviral modalities targeting SARS-CoV-2 entry.
Keywords: SARS-CoV2, COVID-19, Spike, ACE2, Receptor interaction, Luminescent biosensor
1. Introduction
With over a million new COVID-19 cases being reported weekly by WHO and cumulative death totals surpassing one million worldwide, there is an urgent need for novel antivirals and rapid diagnostic tools for SARS-CoV-2 (WHO 2020). The 30 kb SARS-CoV-2 genome encodes four structural genes: the envelope protein, the membrane protein, the nucleocapsid protein, and the transmembrane spike (S) glycoprotein. The S protein mediates viral attachment to ACE2, the SARS-CoV-2 cell receptor, and subsequent fusion. All coronaviruses utilize a homotrimeric S in their envelope to facilitate viral entry. The S protein comprises of S1 and S2 subunits. The conserved receptor binding domain (RBD) at the C-terminus of the S1 subunit is sufficient for binding to ACE2(Azad et al., 2020; Sun et al., 2020; Tai et al., 2020). Binding of S1 to ACE2 triggers a conformational change in the S protein from the pre-fusion state to a highly stable conformation mediating fusion. Given the importance of cell attachment and fusion in the SARS-CoV-2 life cycle, there is a major focus on inhibiting the Spike-ACE2 interaction for antiviral and vaccine development.
The establishment of high-throughput screening approaches to identify inhibitors of the ACE2-S1 interaction will facilitate accelerated SARS-CoV-2 antiviral drug discovery. To this end, reporter fragment complementation represents a powerful tool to investigate protein-protein interactions(Azad et al., 2014). Traditional schemes employ the use of split-luciferase schemes engineered by dissecting the luciferase gene from different species (e.g. Photinus pyralis, Gaussia princeps or Renilla reniformis) into two fragments(Azad et al., 2018; Paulmurugan and Gambhir 2003; Remy and Michnick 2006) (Ataei et al., 2013). These fragments weakly reassemble independently, but complementation is rescued when the fragments are fused to interacting protein partners – enabling catalysis of bioluminescence. This approach has been utilized to generate biosensors capable of directly analyzing protein-protein interactions(Azad et al., 2018) in the context of different pathways, including cellular apoptosis(Torkzadeh-Mahani et al., 2012), phosphoinositide signaling(Ataei et al., 2013), and viral infection(Deng et al., 2011; Wei et al., 2018). The application of the aforementioned split-luciferases schemes is limited due to several inherent weaknesses of the bioreporters, including weak stability, short half-lives of their catalyzed luminescent reactions, and large sizes.
To overcome these limitations, we applied the split reporter strategy with the recently developed Nanoluciferase (NanoLuc), engineered from deep sea luminous shrimp (Oplophorus gracilirostris)(Hall et al., 2012) to probe Spike S1-ACE2 interactions. Split NanoLuc schemes do not possess the limitations associated with traditional split luciferase reporters(Dixon et al., 2016; Kazem Nouri et al., 2019; Nouri et al., 2019). This system, termed NanoLuc Binary Technology (NanoBiT), dissects NanoLuc into two components, Small BiT (SmBiT) and Large BiT (LgBiT). These two components of the split reporter system display poor intrinsic affinity and strong conformational stability, creating an ideal split-reporter for investigating protein-protein interactions(Dixon et al., 2016). In addition, more robust luminescence is produced, relative to their traditional split-luciferase counterparts, when the fragments reassemble due to interaction of their protein partners. We designed a biosensor consisting of SARS-CoV-2 Spike S1-LgBiT and SmBiT-ACE2. When expressed in mammalian cells this pair of recombinant proteins provides a biosensor to sensitively detect S1-ACE2 interactions. The biosensor provides a simple and rapid assay to detect interactions in both cell lysates and supernatants of mammalian cells transfected with S1-LgBiT and SmBiT-ACE2 constructs.
2. Materials and methods
2.1. Cell culture
The HEK293T (CRL-3216) cell line was obtained from the American Type Culture Collections (Manassus, VA, USA). Cells were maintained in Dulbecco's modified Eagle's medium (DMEM) (Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS) (Thermo Fisher Scientific, Cat.#SH30396.03).
2.2. Plasmids
Codon optimized coding sequences for S1 and ACE2 ectomain were ordered from GenScript (Piscataway, NJ, USA). Sequences are shown in Table S1. Bacterial expression plasmid (pSb_init) encoding synthetic nanobody #45 targeting SARS-CoV-2 RBD was a kind gift from Dr. Markus Seeger (Addgene plasmid # 153526; http://n2t.net/addgene:153526; RRID:Addgene_153526)(Walter et al., 2020).
2.3. SDS-PAGE and immunoblotting
Whole cell lysates were obtained by lysing the HEK293T cells in RIPA buffer (Thermo Scientific), and 1X protease inhibitor cocktail (Roche, Basel, Switzerland) on ice. Protein concentration was determined by Pierce bicinchoninic acid (BCA) assay (Thermo Scientific, Cat.# 23225). 10 μg of cell extract were mixed into DTT-Laemmli buffer and boiled for 5 min. Samples were resolved using the NuPAGE SDS-PAGE system (Invitrogen, Carlsbad, CA, USA, Cat. # NP0322) for 1.5 h at constant voltage (150 V). Following gel electrophoresis, proteins were transferred to Immobilon-P polyvinylidene fluoride (PVDF) membrane (MilliporeSigma, Burlington, MA, USA). The PVDF membrane was blocked for 1 h in 5% milk in Tris-buffered saline with 0.1% Tween 20 (TBS-T), washed in TBS-T, then probed for 1 h at room temperature with mouse anti-FLAG (1:1000, MilliporeSigma, Cat.#F3165), anti-B-actin (1:5000, Thermofisher, Cat.#MA1-140) or with mouse anti-HA (1:5000, Thermofisher, Cat.#26183). Blots were then washed and incubated with anti-mouse (1:5000, MilliporeSigma, Cat.#A9044) for 1 h at room temperature. SuperSignalWest Pico PLUS Chemiluminescent Substrate (Thermo Fisher Scientific, Cat. #34577) was used to visualize the protein bands. Blots were imaged using the ChemiDoc MP imaging system (Bio-Rad Laboratories, Mississauga, ON, USA).
2.4. Transfection
HEK293T cells at 70% confluency were transfected with SmBiT-ACE2, LgBiT-S1, or co-transfected with both constructs using PolyJet transfection reagent (Signagen, MD, USA) following manufacturer's protocols. Cells were lysed 48 h post-transfection using 1X passive lysis buffer (Promega, Cat.# E1910) on ice and centrifuged to clear the lysate. If not used immediately, lysates are stored at -20 °C.
2.5. General biosensor assay method and recipes
Lyophilized native coelenterazine (Nanolight Technology, Cat # 303-500) is resuspended in 100% EtOH or 3N HCl to generate a 2 mM stock. Substrate salt buffer is composed of 45 mM EDTA (sodium salt), 30 mM sodium pyrophosphate, and 1.42M NaCl in ddH2O. Working reagent for lysed cell assays is made by adding 5 μL of 2 mM stock for each mL of salt buffer. For live cell assays 20 μL of 2 mM stock was added for each mL of salt buffer.
In an opaque white 96 well plate, 50 μL of working coelenterazine substrate is added to cell lysate(s) containing biosensor components immediately before reading luminescence. Synergy Microplate Reader (BioTek, Winooski, VT, USA) was used to measure luminescence.
2.6. Competition assays (with purified RBD or ACE2)
Recombinant RBD (230-30162- 100) and ACE2 (00707-01-05B) were purchased from RayBiotech respectively. For competition assays, RBD or ACE2 was incubated with cell lysates including SmBiT-ACE2 and S1-LgBiT, respectively. After 30 min incubation S1-LgBiT or SmBiT-ACE2 was added and luciferase assay was performed.
2.7. Temperature stability assays
HEK293T cells were co-transfected with SmBiT-ACE2 and LgBiT-S1 using PolyJet transfection reagent (Signagen, MD, USA) following manufacturer's protocols. 48 h post-transfection, cells were lysed using 1X passive lysis buffer then centrifuged to clear the lysate. For one set of experiments, cell lysate was incubated at 0, 4, 25, 30, 37, 42, 55, 60, 64, 72, or 80 °C for 30 min. For one set of experiments, cell lysate was incubated at 25, 30, or 37C for 0.5, 2, 4, 6, 8, 16, 20, or 24 h. Immediately after incubation 50 μL of coelenterazine substrate was added to 50 μL cell lysate and luminescence was read using Synergy Microplate Reader (BioTek, Winooski, VT, USA).
2.8. pH stability assay
HEK293T cells were co-transfected with SmBiT-ACE2 and LgBiT-S1 using PolyJet transfection reagent (Signagen, MD, USA) following manufacturer's protocols. 48 h post-transfection, cells were lysed using NP40 lysis buffer set to pH 2, 3, 4, 5, 6, 7, 7.6, 8, 9, 10, 11, or 12 using concentrated HCl or NaOH. The lysis reaction was incubated on ice for 30 min then centrifuged to clear the lysate. 50 μL of coelenterazine substrate was added to 50 μL cell lysate and luminescence was read using Synergy Microplate Reader (BioTek, Winooski, VT, USA).
2.9. Live & lysed cell assays
HEK293T cells were co-transfected with SmBiT-ACE2 and LgBiT-S1 using PolyJet transfection reagent (Signagen, MD, USA) following manufacturer's protocols. 48 h post-transfection, cells were harvested and counted. Cells were diluted to have 100-50000 cell/50 μL and assayed with or without lysis using passive lysis buffer. 50 μL of coelenterazine working substrate was added to 50 μL cell lysate and luminescence was read using Synergy Microplate Reader (BioTek, Winooski, VT, USA).
2.10. Co-transfection DNA gradient assay
HEK293T cells were seeded in a 6-well plate 24hrs prior to transfection. Cells were transfected with varying but equal quantities of SmBiT-ACE2 and LgBiT-S1 with the addition of empty pcDNA3.1 vector (to have an equal amount of DNA in each transfection) using PolyJet transfection reagent (Signagen, MD, USA) following manufacturer's protocols. Each well was lysed with 500 μL 1X PLB, centrifuged to clear lysate, then 50 μL of coelenterazine substrate was added to 50 μL cell lysate and luminescence was read using Synergy Microplate Reader (BioTek, Winooski, VT, USA).
2.11. Monoclonal antibody screening
For neutralization assay with monoclonal antibodies, 5 μg RBD-LgBiT containing cell lysate was incubated at 37 °C for 30 min with candidate antibodies or serum. Then 5 μg of SmBiT-ACE2-transfected cell lysate was added, and incubated for an additional 5 min at room temperature. Subsequently, luciferase assay was performed. The following monoclonal RBD antibodies were tested in the biosensor-based neutralization assay: 1A9 (GeneTex; GTX632604); 2414 (ActiveMotif 91349), 1414 (ActiveMotif 91361), 273074 (Abcam ab273074); 40592 (Sino Biological 40592-MM57); 9ACA (GenScript 5B7D7), 11D11F2 (GenScript), 10G6H5 (Genscript), HC2001 (GenScript), and L00847 (Genscript Biotech).
2.12. Synthetic nanobody purification
Sybody 45 was purified according to the procedure as previously described (Walter et al., 2020), except the gel filtration step was omitted. Sybody purity was determined by SDS-PAGE and the protein's concentration was determined by A280 using an extinction coefficient derived from ExPasy Protparam (E. Gasteiger 2005).
2.13. Pseudovirus assay
SARS-CoV-2 spike pseudotyped lentivirus were produced as previously described using plasmids kindly provided by Dr. Jesse Bloom (Fred Hutchinson Cancer Research Center, Seattle, WA) (Crawford et al., 2020). Briefly, HEK293T cells were co-transfected with HDM-IDTSpike-fixK, pHAGE-CMV-Luc2-IRES-ZsGreen-W, and pSPAX2. 48 h post-transfection, cell supernatants containing virus were harvested. For neutralization assay, viruses were incubated with RBD antibody at the indicated concentrations for 1 h prior to infection of HEK293T stably overexpressing ACE2/TMPRSS2 cells (Abe et al., 2020). 48 h post-transduction, cells were lysed in 1X PLB, and infection efficiency was assessed by luciferase assay using the Bright-Glo Luciferase Assay system (Promega).
2.14. Analytical performance evaluation
The analytical performance of the biosensor was evaluated by several metrics: detection limit Incubation Time (I.T.), Detection Limit (D.L.); Linear Range (L.R.); Signal Stability (SS) (refer to Supplementary Table 2). In addition, RSD for all figures is shown in Supplementary Table 3.
3. Results and discussion
3.1. Generation of biosensor for detecting S1-ACE2 interaction
We sought to develop a biosensor to measure interactions between the SARS-CoV-2 S protein and the host entry receptor ACE2 (Fig. 1 A). Previous work has demonstrated that RBD is sufficient to mediate Spike's interaction with ACE2, and several previously reported surrogate SARS-CoV-2 neutralization assays are based on interrogating the interaction between RBD and ACE2 (Abe et al., 2020; Tan et al., 2020). However, previous work has demonstrated that neutralizing antibodies targeting S1 epitopes outside of the RBD are capable of disrupting the Spike-ACE2 interaction. In addition, there may be differences in the ACE2 affinity of S1 compared to RBD (Wang et al., 2020) (Wong et al., 2004). Therefore, we selected the SARS-CoV-2 S1 domain and the ACE2 ectodomain as the protein interaction partners for our NanoBiT-based biosensor order to create a biosensor sensitive to a larger breadth of neutralizing antibodies and antivirals, and more representative of SARS-CoV-2 virus-receptor interaction. S1 is the smaller interacting partner; thus it was linked to LgBiT; whereas the larger ACE2 ectodomain was linked to SmBiT. Both sequences were codon optimized to maximize expression. To enable both cell lysate and supernatant based assays, the SmBiT-ACE2 coding sequence was preceded by an interleukin-12 secretion signal, while an IgK secretory leader sequence was added to the S1-LgBiT. Lastly, to ensure proper folding and minimize the NanoBiT fusion's effect on the S1/ACE2 interaction, a flexible glycine-serine linker was inserted between ACE2 or S1 and its respective linked NanoBiT component(Kazem Nouri et al., 2019).
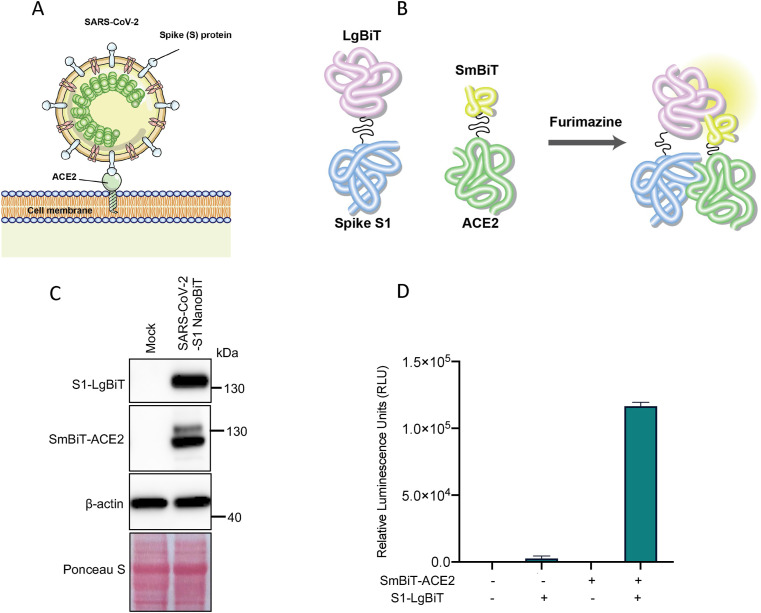
Fig. 1.
Establishment of SARS-CoV-2 S1-NanoBiT biosensor. (A) Schematic of SARS-CoV-2 viral entry with Spike (S) protein interaction with cellular receptor, ACE2. (B) Overall design of Nanoluciferase complementation-based biosensor of SARS-CoV-2 Spike S1 and ACE2 ectodomain interaction. (C) Immunoblot analysis of S1-LgBiT and SmBiT-ACE2 expression in SARS-CoV-2 S1 NanoBiT construct transfected HEK293T cells. β-actin and total protein loading are shown as controls. (D) Demonstration of the complementation-based nature of the designed S1-LgBiT and SmBiT-ACE2 interaction nano-luciferase complementation-based biosensor.
For the initial validation of our system, we transfected HEK293T cells with the expression constructs of S1-LgBiT and SmBiT-ACE2, either independently or in combination. As expected, luciferase assays performed on cells transfect with either NanoBiT fusion construct independently produce negligible amounts of background signal. When the constructs were co-transfected, robust signal was produced (greater than 1e5 RLU) with a signal-to-noise ratio (SNR) greater than 200-fold (Fig. 1D). These data establish the functionality of the SmBiT-ACE2/S1-LgBiT based biosensor, which we call SARS-CoV-2 S1 NanoBiT.
3.2. SARS-CoV-2 S1-NanoBiT possesses large dynamic range, high stability and sensitivity
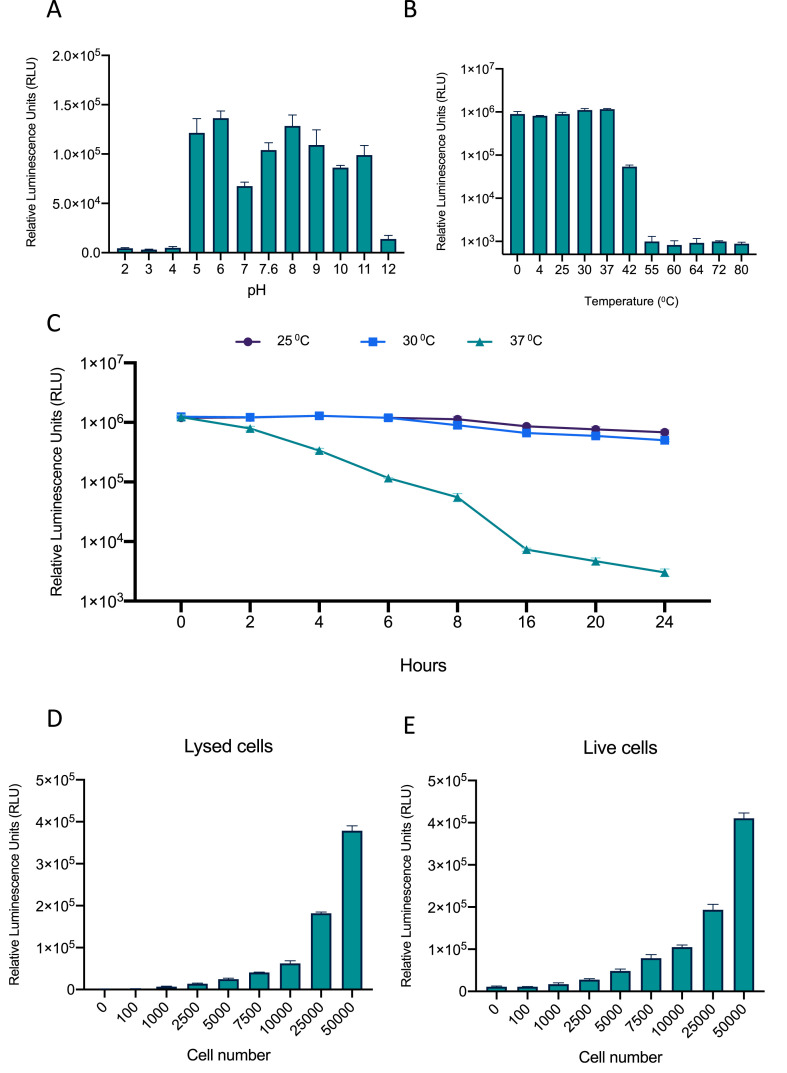
Traditional split luciferase reporters possess poor thermostability and pH tolerance(Law et al., 2006); however, the use of a NanoLuc complementation-based biosensor should overcome these limitations. In order to examine this, we tested the biosensor's efficacy over a range of pH's (2-12) and temperatures (0 °C–80 °C). The assay displayed a high signal-to-noise ratio over a pH range of 5–11, demonstrating a wide pH tolerance (Fig. 2 A). The assay also displayed thermostability between 0 °C and 42 °C (Fig. 2B). Outside of these optimal pH and temperature ranges, the biosensor produces increased variabililty. These are impressive properties of our NanoBiT-based biosensor, when compared to previously described firefly luciferase-based biosensors (Law et al., 2006; Said Alipour et al., 2009). Furthermore, we demonstrated the biosensor was stable even after incubation at 25 °C or 30 °C for 24 h (Fig. 2C). In fact, at 37 °C, the biosensor still produced detectable signal over background after 24 h. This impressive stability profile has important implications for biosensor shipping, transport and storage, which are critical to its application as a diagnostic tool.
Fig. 2.
Determination of the relative luciferase activity of the S1-ACE2 NanoBiT system under a variety of conditions. (A) Examination of pH stability of the S1-ACE2 biosensor looking at relative luminescence of cell lysates after lysis with NP40 lysis buffers ranging from pH 2-12. (B) Temperature stability of cell lysates containing the S1-ACE2 biosensor after a 30 min incubation at 0-80 °C. (C) Temperature stability of cell lysates containing the S1-ACE2 biosensor after 0-24 h incubations at 25, 30, or 37 °C. (D) S1-ACE2 biosensor activity measured in lysed cells (0-50000 Hek293T cells) 48 h post-transfection. (E) S1-ACE2 biosensor activity measured in live cells (0-50000 Hek293T cells) 48 h post-transfection.
We also wished to analyze the sensitivity of our biosensor, so we investigated the minimum cell numbers required for both live and lysate-based assay in order to produce detectable signal over noise (Fig. 2D-E). Our NanoBiT-based biosensor generated significant signal with as little as 2500 lysed or live cells. The requirement for low cells numbers to generate significant luminescent signal in combination with the enhanced thermostability and pH tolerance of our biosensor suggest the assay can be easily translated to high-throughput screening and in vivo applications.
3.3. SARS-CoV-2 S1 NanoBiT facilitates screening of drugs disrupting S1-ACE2 interaction
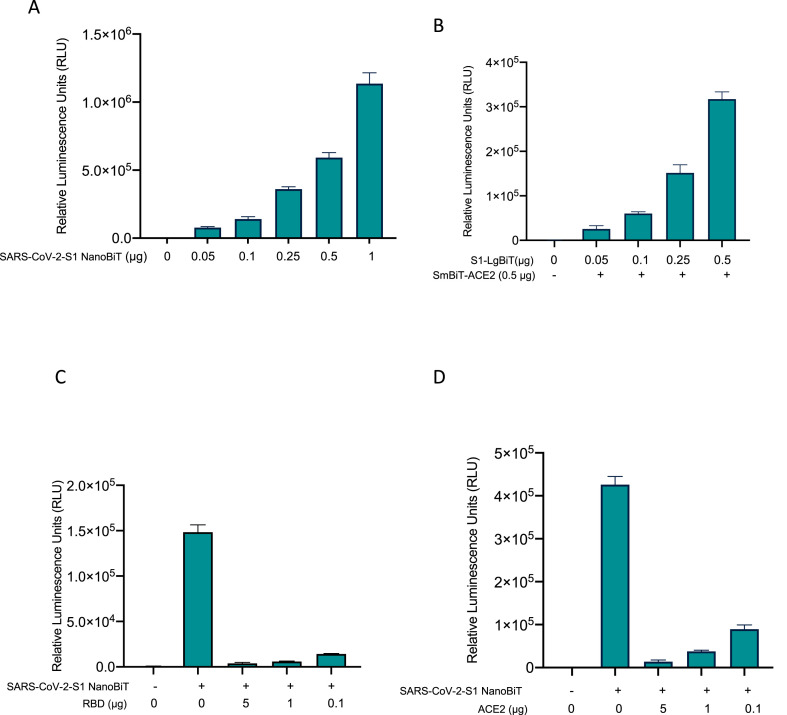
In order to examine the sensitivity of the assay, we transfected HEK293T cells in 6-well format with a dose range of the S1-LgBiT and SmBiT-ACE2 expression constructs (Fig. 3 A). As little as 50 ng of each construct produced discernible signal over background – demonstrating the high sensitivity of the assay. Furthermore, a dose-dependent effect (50 ng–1 μg) was observed suggesting that the biosensor produces luminescence in an S1-ACE2 interaction-specific manner. Also, we measured the luciferase activity using increasing amount of S1, while holding the ACE levels constant (Fig. 3B), and, in an independent experiment, using increasing amount of ACE2 while holding S1 levels constant (Supplementary Fig. 1). As expected, we observe a dose-dependent effect in line with S1-ACE2 interaction-specific signal. Collectively, these results suggest the SARS-CoV-2 S1 NanoBiT is a highly sensitive biosensor of the S1-ACE2 interaction.
Fig. 3.
SARS-CoV-2 S1-NanoBiT biosensor enables detection of molecule inhibitors of viral entry. (A) HEK293T cells were transfected with a dose range of LgBiT-S1 and SmBiT-ACE2 biosensor components, and 24 h post-transfection, luciferase assay was performed. (B) HEK293T cells were transfected with a dose range of LgBiT-S1 and 500 ng of the SmBiT-ACE2 biosensor component. 24 h post-transfection, luciferase assay was performed. (C–D) Biosensor assay was performed in the presence of dose range of soluble RBD (C) or ACE2 (D).
Next, we sought to demonstrate the utility of the biosensor to identify small molecule inhibitors of the S1-ACE2 interaction. Hence, we transfected HEK293T cells independently with either the S1-LgBIT or SmBiT-ACE2 constructs, and then combined the two cell lysates in the presence of recombinant RBD (Fig. 3C) or ACE2 (Fig. D). Both recombinant RBD and ACE2, through their actions as competitive inhibitors, are being considered as potential SARS-CoV-2 attachment inhibitors(Monteil et al., 2020; Tai et al., 2020). We pre-incubated SmBiT-ACE2 transfected cell lysate with mammalian recombinant RBD, and, subsequently added S1-LgBiT transfected cell lysate. As expected, recombinant RBD effectively outcompeted S1-LgBiT for ACE2 interaction, resulting in a dose-dependent loss of reporter signal (Fig. 3B). Analogously, we incubated S1-LgBiT containing lysate with soluble ACE2 prior to combination with the SmBiT-ACE2 lysate. Once again, the ACE2 ectodomain acted as an effective inhibitor of reporter complementation (Fig. 3C). This disruption occurred in a soluble ACE2 dose-dependent manner, consistent with competitive inhibition. These data serve to validate the biosensor's capacity to identify small molecule inhibitors of the S1-ACE2 interaction.
3.4. SARS-CoV-2 S1-NanoBiT serves as a surrogate virus neutralization assay
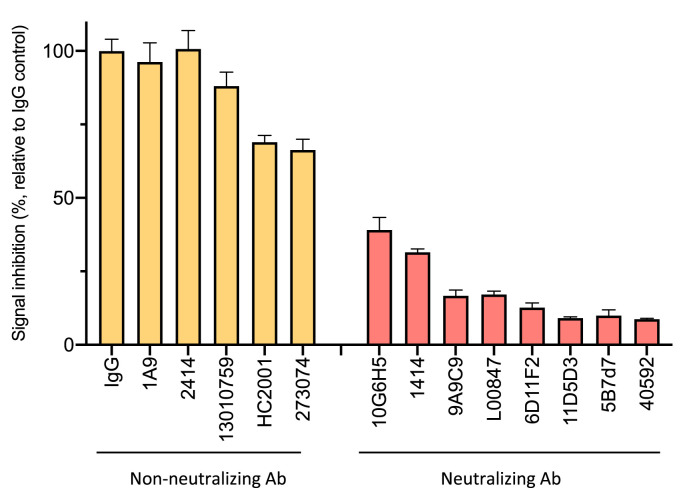
There is an immediate need for rapid diagnostics to screen SARS-CoV-2 seropositive patients. In order to investigate the capacity of our biosensor to measure neutralizing antibody levels, we screened a panel of 13 different monoclonal antibodies targeting RBD (Fig. 4 ). A subset of these antibodies have been reported to neutralize S pseudotyped lentivirus infection, and the results of our biosensor assay were consistent with this observation. In particular, antibody clones 40592, 9A9C9, 5B7d7m 11D5D3, 6D11F2, and L00847 showed high neutralization potency – causing a large reduction in S1-ACE2 interaction. On the other hand, other RBD-targeted antibodies appeared to target non-neutralizing epitopes (1A9, 2414, and 13010759). These data suggest our biosensor should be able to measure neutralizing antibody levels in sera, and identify SARS-CoV-2 seroconverters.
Fig. 4.
SARS-CoV-2 S1-NanoBiT biosensor is capable of detecting neutralizing antibodies. SARS-CoV-2 S1 biosensor assays were performed with an array of anti-RBD monoclonal antibodies or control IgG.
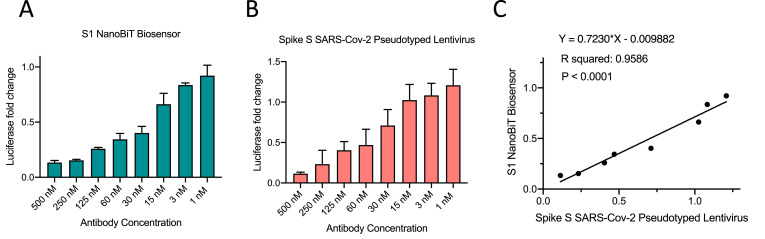
To further substantiate our biosensor's utility in detecting neutralizing antibodies, we performed a direct correlative comparison between the SARS-CoV-2 S1-NanoBiT biosensor and a widely accepted Spike pseudotyped lentivirus assay for SARS-CoV-2 neutralization antibody testing (Fig. 5 ). We tested the ability of both assays to detect the levels of a synthetic nanobody targeting RBD (Walter et al., 2020). Both assays showed a dose-dependent reduction in signal, and the two assays' results correlated quite well (Fig. 5C). This correlation analysis validates the application of the biosensor as a surrogate for virus neutralization assays.
Fig. 5.
Correlation analysis of pseudovirus neutralization assay and SARS-CoV-2 S1-NanoBiT biosensor. SARS-CoV-2 S1-NanoBiT assay (A) and spike pseudotyped lentivirus assay (B) were performed in the presence of a dose curve of synthetic nanobody targeting RBD. (C) Correlation and linear regression analyses were performed in GraphPad Prsim using Pearson's correlation coefficients. Statistical significance was calculated using two-tailed Student's t-test.
4. Conclusions
In summary, our study establishes SARS-CoV-2 S1-NanoBiT as a versatile and rapid tool for interrogating SARS-CoV-2 host-virus interaction at the viral entry stage. We characterized an optimal set of protein partners (SmBiT-ACE2 and S1-LgBiT) for this NanoBiT-mediated biosensor and demonstrated that this screening compatible assay's output may be measured with a standard luminometer. This virus-free assay is simple, accessible, rapid (<30 min), and sensitive. When using lysates, there is no requirement for live biological materials or biosafety containment. SARS-CoV-2 S1-NanoBiT produces robust signal, possesses an impressive safety profile and is accessible to the majority of the research community. While previous studies using split-luciferase reporters have examined viral protein interactions (Deng et al., 2011; Wei et al., 2018), to the best of our knowledge, this work is the first report using a Nanoluc complementation reporter-based assay to investigate host-virus interactions. We have validated the system's ability to probe monoclonal antibodies and recombinant proteins blocking the Spike/ACE2 interaction. Lastly, we applied the biosensor to demonstrate its ability to detect neutralizing antibody, and the results correlated well with an established pseudovirus assay. Our work highlights the untapped potential of the NanoLuc complementation-based reporter strategy in identifying antiviral drugs targeting other host-virus interactions.
CRediT authorship contribution statement
Taha Azad: Conceptualization, Methodology, Investigation, Formal analysis, Visualization, Writing – original draft, Reviewing & Editing. Ragunath Singaravelu: Conceptualization, Methodology, Investigation, Writing – original draft, Reviewing & Editing. Emily E.F. Fekete: Conceptualization, Methodology, Investigation, Writing – original draft, Reviewing & Editing. Zaid Taha: Writing – review & editing. Reza Rezaei: Investigation. Rozanne Arulanandam: Writing – review & editing. Stephen Boulton: Investigation, Writing – review & editing. Jean-Simon Diallo: Writing – review & editing. Carolina S. Ilkow: Writing – review & editing, All authors have read and agreed to the published version of the manuscript. John C. Bell: Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was funded by the generous support from the Ottawa Hospital Foundation and a grant from the Canadian Institutes of Health Research (#448323) to J.C.B. This work was also supported by a Fast Grant for COVID-19 Science to J.C.B. T.A. is funded by a CIHR Banting Fellowship. R.S. would like to thank the CIHR and MITACS Accelerate for funding support in the form of post-doctoral fellowships. E.E.F.F. is funded by a CIHR Frederick Banting and Charles Best Canada Graduate Scholarship and an OICR Lebovic Fellowship. Z.T. is funded by a NSERC post-graduate scholarship.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bios.2021.113122.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
ACE2 concentration-dependent increase in biosensor activity. HEK293T cells were transfected with a dose range of SmBiT-ACE2 construct and 500 ng of the S1-LgBiT biosensor component. 24 h post-transfection, luciferase assay was performed.
References
- Abe K.T., Li Z., Samson R., Samavarchi-Tehrani P., Valcourt E.J., Wood H., Budylowski P., Dupuis Ii A.P., Girardin R.C., Rathod B., Wang J., Barrios-Rodiles M., Colwill K., McGeer A., Mubareka S., Gommerman J.L., Durocher Y., Ostrowski M., McDonough K.A., Drebot M.A., Drews S.J., Rini J.M., Gingras A.-C. JCI Insight; 2020. A Simple Protein-Based Surrogate Neutralization Assay for SARS-CoV-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ataei F., Torkzadeh-Mahani M., Hosseinkhani S. A novel luminescent biosensor for rapid monitoring of IP3 by split-luciferase complementary assay. Biosens. Bioelectron. 2013;41:642–648. doi: 10.1016/j.bios.2012.09.037. [DOI] [PubMed] [Google Scholar]
- Azad T., Janse van Rensburg H.J., Lightbody E.D., Neveu B., Champagne A., Ghaffari A., Kay V.R., Hao Y., Shen H., Yeung B., Croy B.A., Guan K.L., Pouliot F., Zhang J., Nicol C.J.B., Yang X. A LATS biosensor screen identifies VEGFR as a regulator of the Hippo pathway in angiogenesis. Nat. Commun. 2018;9(1):1061. doi: 10.1038/s41467-018-03278-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azad T., Singaravelu R., Crupi M.J., Jamieson T., Dave J., Fekete E.E.F., Rezaei R., Taha Z., Boulton S., Martin N.T. Implications for SARS-CoV-2 vaccine design: fusion of spike glycoprotein transmembrane domain to receptor-binding domain induces trimerization. Membranes. 2020;10(9):215. doi: 10.3390/membranes10090215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azad T., Tashakor A., Hosseinkhani S. Split-luciferase complementary assay: applications, recent developments, and future perspectives. Anal. Bioanal. Chem. 2014;406(23):5541–5560. doi: 10.1007/s00216-014-7980-8. [DOI] [PubMed] [Google Scholar]
- Crawford K.H.D., Eguia R., Dingens A.S., Loes A.N., Malone K.D., Wolf C.R., Chu H.Y., Tortorici M.A., Veesler D., Murphy M., Pettie D., King N.P., Balazs A.B., Bloom J.D. Protocol and reagents for pseudotyping lentiviral particles with SARS-CoV-2 spike protein for neutralization assays. Viruses. 2020;12(5) doi: 10.3390/v12050513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Q., Wang D., Xiang X., Gao X., Hardwidge P.R., Kaushik R.S., Wolff T., Chakravarty S., Li F. Application of a split luciferase complementation assay for the detection of viral protein–protein interactions. J. Virol Methods. 2011;176(1):108–111. doi: 10.1016/j.jviromet.2011.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon A.S., Schwinn M.K., Hall M.P., Zimmerman K., Otto P., Lubben T.H., Butler B.L., Binkowski B.F., Machleidt T., Kirkland T.A., Wood M.G., Eggers C.T., Encell L.P., Wood K.V. NanoLuc complementation reporter optimized for accurate measurement of protein interactions in cells. ACS Chem. Biol. 2016;11(2):400–408. doi: 10.1021/acschembio.5b00753. [DOI] [PubMed] [Google Scholar]
- Gasteiger E., Hoogland C., Gattiker A., vDuvaud S., Wilkins M.R., Appel R.D., Bairoch A A. In: The Proteomics Protocols Handbook. Walker J.M., editor. Humana Press; 2005. Protein identification and analysis tools on the Expasy Server; pp. 571–607. [Google Scholar]
- Hall M.P., Unch J., Binkowski B.F., Valley M.P., Butler B.L., Wood M.G., Otto P., Zimmerman K., Vidugiris G., Machleidt T., Robers M.B., Benink H.A., Eggers C.T., Slater M.R., Meisenheimer P.L., Klaubert D.H., Fan F., Encell L.P., Wood K.V. Engineered luciferase reporter from a deep sea shrimp utilizing a novel imidazopyrazinone substrate. ACS Chem. Biol. 2012;7(11):1848–1857. doi: 10.1021/cb3002478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazem Nouri, Taha Azad, Lightbody E., Khanal P., Nicol C.J., Yang X. A kinome-wide screen using a NanoLuc LATS luminescent biosensor identifies ALK as a novel regulator of the Hippo pathway in tumorigenesis and immune evasion. Faseb. J. 2019;33(11):12487–12499. doi: 10.1096/fj.201901343R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law G.H.E., Gandelman O.A., Tisi L.C., Lowe C.R., Murray J.A.H. Mutagenesis of solvent-exposed amino acids in Photinus pyralis luciferase improves thermostability and pH-tolerance. Biochem. J. 2006;397(2):305–312. doi: 10.1042/BJ20051847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteil V., Kwon H., Prado P., Hagelkrüys A., Wimmer R.A., Stahl M., Leopoldi A., Garreta E., Hurtado del Pozo C., Prosper F., Romero J.P., Wirnsberger G., Zhang H., Slutsky A.S., Conder R., Montserrat N., Mirazimi A., Penninger J.M. Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2. Cell. 2020;181(4):905–913. doi: 10.1016/j.cell.2020.04.004. e907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nouri K., Azad T., Ling M., van Rensburg H.J.J., Pipchuk A., Shen H., Hao Y., Zhang J., Yang X. Identification of celastrol as a novel YAP-TEAD inhibitor for cancer therapy by high throughput screening with ultrasensitive YAP/TAZ-TEAD biosensors. Cancers. 2019;11(10) doi: 10.3390/cancers11101596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulmurugan R., Gambhir S.S. Monitoring Protein−Protein interactions using split synthetic Renilla luciferase protein-fragment-assisted complementation. Anal. Chem. 2003;75(7):1584–1589. doi: 10.1021/ac020731c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remy I., Michnick S.W. A highly sensitive protein-protein interaction assay based on Gaussia luciferase. Nat. Methods. 2006;3(12):977–979. doi: 10.1038/nmeth979. [DOI] [PubMed] [Google Scholar]
- Said Alipour B., Hosseinkhani S., Ardestani S.K., Moradi A. The effective role of positive charge saturation in bioluminescence color and thermostability of firefly luciferase. Photochem. Photobiol. Sci. 2009;8(6):847–855. doi: 10.1039/b901938c. [DOI] [PubMed] [Google Scholar]
- Sun C., Chen L., Yang J., Luo C., Zhang Y., Li J., Yang J., Zhang J., Xie L. 2020. SARS-CoV-2 and SARS-CoV Spike-RBD Structure and Receptor Binding Comparison and Potential Implications on Neutralizing Antibody and Vaccine Development. bioRxiv, 2020.2002.2016.951723. [Google Scholar]
- Tai W., He L., Zhang X., Pu J., Voronin D., Jiang S., Zhou Y., Du L. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell. Mol. Immunol. 2020;17:613–620. doi: 10.1038/s41423-020-0400-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan C.W., Chia W.N., Qin X., Liu P., Chen M.I.C., Tiu C., Hu Z., Chen V.C.-W., Young B.E., Sia W.R., Tan Y.-J., Foo R., Yi Y., Lye D.C., Anderson D.E., Wang L.-F. A SARS-CoV-2 surrogate virus neutralization test based on antibody-mediated blockage of ACE2–spike protein–protein interaction. Nat. Biotechnol. 2020;38(9):1073–1078. doi: 10.1038/s41587-020-0631-z. [DOI] [PubMed] [Google Scholar]
- Torkzadeh-Mahani M., Ataei F., Nikkhah M., Hosseinkhani S. Design and development of a whole-cell luminescent biosensor for detection of early-stage of apoptosis. Biosens. Bioelectron. 2012;38(1):362–368. doi: 10.1016/j.bios.2012.06.034. [DOI] [PubMed] [Google Scholar]
- Walter J.D., Hutter C.A.J., Zimmermann I., Wyss M., Egloff P., Sorgenfrei M., Hürlimann L.M., Gonda I., Meier G., Remm S., Thavarasah S., Plattet P., Seeger M.A. 2020. Sybodies Targeting the SARS-CoV-2 Receptor-Binding Domain. bioRxiv, 2020.2004.2016.045419. [Google Scholar]
- Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., Lu G., Qiao C., Hu Y., Yuen K.-Y., Wang Q., Zhou H., Yan J., Qi J. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181(4):894–904. doi: 10.1016/j.cell.2020.03.045. e899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X.-F., Gan C.-Y., Cui J., Luo Y.-Y., Cai X.-F., Yuan Y., Shen J., Li Z.-Y., Zhang W.-L., Long Q.-X., Hu Y., Chen J., Tang N., Guo H., Huang A.-L., Hu J.-L. Identification of compounds targeting hepatitis B virus core protein dimerization through a split luciferase complementation assay. Antimicrob. Agents Chemother. 2018;62(12) doi: 10.1128/AAC.01302-18. e01302-01318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . 2020. Coronavirus Disease 2019 (COVID-19) Weekly Epidemiology Update. [Google Scholar]
- Wong S.K., Li W., Moore M.J., Choe H., Farzan M. A 193-amino acid fragment of the SARS coronavirus S protein efficiently binds angiotensin-converting enzyme 2. J. Biol. Chem. 2004;279(5):3197–3201. doi: 10.1074/jbc.C300520200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
ACE2 concentration-dependent increase in biosensor activity. HEK293T cells were transfected with a dose range of SmBiT-ACE2 construct and 500 ng of the S1-LgBiT biosensor component. 24 h post-transfection, luciferase assay was performed.