Abstract
CONTEXT:
Lung cancer is the leading cause of cancer-related deaths worldwide. The constitutive activation of multiple signaling pathways is the major cause of carcinogenesis.
AIMS:
The study evaluates the frequency of Kirsten rat sarcoma virus (KRAS) protein overexpression and correlates with clinicopathological and histomorphological features in non-small cell lung carcinoma (NSCLC)-adenocarcinoma.
SETTINGS AND DESIGN:
Tertiary hospital-based retrospective and prospective case series included 100 cases of NSCLC-adenocarcinoma.
MATERIALS AND METHODS:
The basic panel of Immunohistochemistry including Napsin-A, thyroid transcription factor-1 (TTF-1), and markers for squamous differentiation, p-40 was used in formalin-fixed paraffin-embedded tissue blocks. The KRAS monoclonal antibody (9.13, Thermo Fisher Scientific, USA) was used.
STATISTICAL ANALYSIS USED:
The IBM-Statistical Package for the Social Sciences (SPSS) (SPSS, International Business Machines Corporation, New York, NY, USA) analysis software, version 16 was used for all statistical calculations.
RESULTS:
KRAS protein expressed in 28.0% (28/100) cases. Cases were grouped as KRAS positive and negative. TTF-1 and Napsin-A were expressed in 89.25% (n = 25) and 92.86% (n = 26) cases, respectively. Stage IV clinical disease was identified in 55% of cases, and 36.84% of cases had a mean survival between 6 and 12 months. In KRAS positive group, the most common pattern of cellular arrangement was acinar/loose clusters pattern present in 64.29% (n = 21) and 75.0% (n = 18) cases followed by the solid pattern present in 42.86% of cases (n = 12), respectively. Necrosis was identified in 57.14% (n = 16) cases. Mucin pattern was present in 32.14% of cases (n = 9), which was significantly different when compared with the KRAS negative group (P = 0.036).
CONCLUSIONS:
This finding may imply that KRAS mutations may not be entirely triggered by alterations induced by carcinogens in smoke. KRAS gene is frequently mutated in pulmonary tumors. It should be tested in NSCLC owing to its predictive and prognostic effects.
Keywords: Histomorphology, Kirsten rat sarcoma virus, non-small cell lung carcinoma-adenocarcinoma, thyroid transcription factor-1
Introduction
Worldwide cancer-related mortality is the highest for lung cancer. In lung cancer, the constitutive activation of multiple signaling pathways with molecular alterations of the specific components of the pathway is the major cause of carcinogenesis.[1] The mitogen-activated protein (MAP) kinase pathway is a key pathway involved in the development and progression of pulmonary carcinogenesis.[2] The pathway is activated through several factors, including hormones, cytokines, chemokines, growth factors, and mitogens. Kirsten rat sarcoma virus (KRAS) is a member of the RAS gene family that encodes G proteins with intrinsic GTPase activity. KRAS is an oncogenic driver mutation in nonsmall cell lung carcinomas (NSCLCs) with prognostic and predictive implications. RAS proteins serve as molecular switches, regulating intracellular signal transduction pathways in response to stimulation of cell surface receptors. Epidermal growth factor receptors are receptor tyrosine kinases, which, when activated through epidermal growth factor leads to sequential activation of the RAS pathway. This is then followed by sequential activation of the MAP kinase pathway.[3,4,5] KRAS mutations result in the loss of intrinsic GTPase activity leading to the deregulation of cell proliferation signals.[6]
The frequency of KRAS protein expression in NSCLC, particularly adenocarcinoma phenotype, has been inconsistently documented particularly in the Indian population. Therefore, this study was undertaken with the objectives to document the frequency of KRAS protein overexpression and to co-relate the KRAS protein overexpression with clinicopathological and histomorphological features in NSCLC-adenocarcinoma phenotype.
Materials and Methods
The current study was a tertiary care hospital-based, retrospective, and prospective case series conducted after obtaining approval from the Institutional Ethical Committee. The study included 100 cases of NSCLC-adenocarcinoma phenotype. The study included biopsies from both primary (n = 88) and metastatic (n = 12) sites. The biopsies were categorized as per the 2015 World Health Organization criteria for biopsy classification of lung tumors. A basic panel of immunohistochemistry (IHC) including Napsin-A, thyroid transcription factor-1 (TTF-1), and markers for squamous differentiation, namely P-40, were used. Cases categorized as NSCLC-adenocarcinoma or adenocarcinoma with squamous differentiation was included. Cases of pure NSCLC-squamous cell carcinoma phenotype were excluded from the study. Complete demographic, clinical, and radiological details were documented in all the cases. The tumors were staged clinically using the tumor size, lymph node status, and distant metastasis status. The tumor, node, and metastasis staging system was used as per the American Joint Committee on Cancer cancer staging guidelines of the eighth edition. KRAS protein overexpression testing: The testing for KRAS protein overexpression was done using IHC. The KRAS monoclonal antibody (9.13) from Thermo Fisher Scientific (USA) was used. IHC was performed on formalin-fixed paraffin-embedded (FFPE) tissue blocks. The FFPE tissue blocks were sectioned using a microtome (Leica, Germany) at a thickness of 3–4 μm and mounted on glass slides coated with tissue bond (Biocare, USA). These coated slides with the tissue sections were fixed overnight in an oven at 60°C. Xylene was used for de-paraffinization, followed by rehydration using a series of graded alcohol. Blocking was done to quench the endogenous peroxidase activity using 3% hydrogen peroxide in methanol for 30 min. This was followed by antigen retrieval using the sodium citrate buffer (pH-6.0) done in Pascal (DAKO Cytomation, California). The sections were incubated with the primary antibody. The KRAS antibody was incubated for1 h at a dilation of 1:50. Treatment with a polymer-based secondary antibody kit (Dakopatts, Envision kit, Denmark) was used. The visualization was done as per the manufacturer's instructions using diaminobenzidine. All the sections were adequately counterstained with hematoxylin and mounted for light microscopy visualization. All the slides were run in batches, including positive and negative controls.
The positive control used for KRAS IHC was KRAS mutant colorectal adenocarcinoma, while negative controls of the batch were cases where the primary antibody was omitted.
Assessment of Kirsten rat sarcoma virus protein overexpression
Cytoplasmic/membranous staining was assessed. The percentage of tumor cells and the intensity of staining were assessed. The cases were categorized as score 0: Absence of staining/barely perceptible staining in the tumor cells, score 1: Weak cytoplasmic/membranous staining in >10% tumor cells or strong cytoplasmic/membranous staining in <10% tumor cells, score 2: Strong cytoplasmic/membranous staining in >10% tumor cells. Cases with scores of 0 or 1 were considered negative, while cases with a score of 2 were considered as positive.
Histomorphological analysis
A comprehensive histomorphological analysis was performed in all cases. Various histomorphological parameters were analyzed, including the pattern of cellular arrangement, tumor differentiation, nuclear pleomorphism, and the presence of necrosis.
Statistical analysis
The IBM-Statistical Package for Social Sciences (SPSS) (SPSS, International Business Machines Corporation, New York, NY, USA) analysis software, version 16 was used for all statistical calculations. The categorical variables were compared using the Chi-square test. Two-sided tests were used for calculation of all P values, and P ≤ 0.05 was considered significant. The histomorphology of the cases with KRAS protein overexpression versus the KRAS negative cases was additionally analyzed.
Results
The age range of the patients varied from 26 to 80 years with a mean age of 55.75 years and 34% of cases were between the age ranges of 51–60 years. The male-to-female ratio was 1.8:1. The site of the biopsy was from the respiratory tree in 88% cases, while biopsies were obtained from metastatic sites in 12% (n = 12/100) cases, which were following Bone and vertebral biopsies 5, lymph node 2, liver 1, brain 1, soft tissue deposits in chest wall, epigastric region and scapular region in 1 case each. At all metastatic sites, the primary was established in the lung with the aid of IHC. Bone and vertebrae were the most common metastatic sites (n = 5/12). Most patients had combination of clinical symptoms that included cough (n = 89), breathlessness (n = 76), hemoptysis (n = 66), weight loss (n = 63), and chest pain (n = 27). A history of smoking was obtained in all cases and 37% of cases were categorized as current/ex-smokers, while 63% of cases were nonsmokers.
The clinical staging, including the tumor (T stage), lymph node status (N stage) and metastatic (M stage) status, was documented in 65 cases. The clinical-stage was IV in 46.23% (n = 32) cases. The one time follow-up/survival was documented in 72 patients. The survival varied from 1 month to 20 months with a mean survival of 7.6 months.
The immunostaining for P-40 in >50% tumor cells mixed with cells expressing TTF-1 was observed in 4 cases, and these tumors were categorized as NSCLC-adenocarcinoma with squamous differentiation, while 96 cases were categorized as NSCLC-adenocarcinoma phenotype based on both or either Napsin or TTF1 expression. TTF-1 and Napsin-A were positive in 86% and 88% cases, respectively.
KRAS protein expression was identified in 28% of cases [Figure 1]. The characteristics of KRAS positive cases as compared to the study group are shown in Table 1. KRAS was expressed in one case with squamous differentiation. TTF-1 and Napsin-A were expressed in 89.25% and 92.86% cases, respectively. Stage IV clinical disease was identified in 55% of cases and 36.84% cases had a mean survival between 6 and 12 months. A comparison was made between the KRAS positive versus the KRAS negative groups; however, none of the categories were statistically significant [Table 1].
Figure 1.
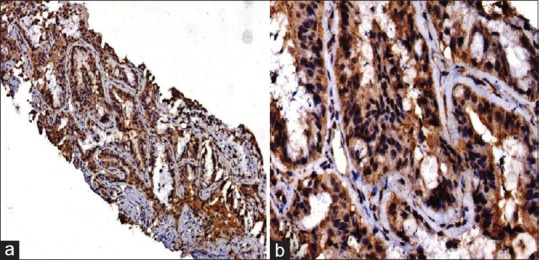
(a and b) Kirsten rat sarcoma virus protein over-expression using immunohistochemistry with strong cytoplasmic and membranous staining (a = diaminobenzidine × 100, b = Diaminobenzidine × 200)
Table 1.
Sociodemographic, clinical and histological features of KRAS+versus KRAS-cases
| Feature | KRAS+(n=28), n(%) | KRAS− (n=72), n(%) | P |
|---|---|---|---|
| Age(years) | |||
| <40 | 4(14.3) | 6(8.33) | 0.372 |
| >40 | 24(85.7) | 66(91.67) | |
| Gender | |||
| Male | 16(57.14) | 49(68.06) | 0.304 |
| Female | 12(42.86) | 23(31.94) | |
| Site of biopsy | |||
| Respiratory tree | 25(89.29) | 63(87.5) | 0.805 |
| Metastatic site | 3(10.71) | 9(12.5) | |
| Smoking history | |||
| Smoker/ex-smoker | 9(32.14) | 28(38.89) | 0.530 |
| Nonsmoker | 19(67.86) | 44(61.11) | |
| Educational status | |||
| Primary | 10(35.71) | 18(25) | 0.599 |
| Metric | 5(17.85) | 11(15.2) | |
| Higher/graduate | 2(7.14) | 10(13.88) | |
| Illiterate | 11(39.28) | 33(45.83) | |
| Economical class | |||
| Poor | 19(67.85) | 53(73.61) | 0.841 |
| Middle/upper middle | 4(14.28) | 8(11.11) | |
| Lower | 5(17.85) | 11(15.27) | |
| Histological phenotype-(presence of squamous differentiation) | |||
| Adenocarcinoma with squamous differentiation(primary and metastatic) | 1(3.57) | 3(4.17) | 0.891 |
| Adenocarcinoma with the absence of squamous differentiation(primary and metastatic) | 27(96.43) | 69(95.83) | |
| TTF-1 | |||
| Positive | 25(89.29) | 61(84.72) | 0.554 |
| Negative | 3(10.71) | 11(15.28) | |
| Napsin-A | |||
| Positive | 26(92.86) | 62(86.11) | 0.351 |
| Negative | 2(7.14) | 10(13.89) | |
| T stage(n=65) | |||
| T1 | 0(0) | 3(6.67) | 0.402 |
| T2 | 5(25) | 6(13.33) | |
| T3 | 4(20) | 17(37.78) | |
| T4 | 11(55) | 19(42.22) | |
| N stage(n=65) | |||
| N0 | 8(40) | 13(28.89) | 0.478 |
| N1 | 2(10) | 11(24.44) | |
| N2 | 7(35) | 17(37.78) | |
| N3 | 3(15) | 4(8.89) | |
| M stage(n=65) | |||
| M0 | 9(45) | 24(53.33) | 0.535 |
| M1 | 11(55) | 21(46.67) | |
| Clinical stage(n=65) | |||
| I | 0(0) | 1(2.22) | 0.682 |
| II | 3(15) | 4(8.89) | |
| III | 6(30) | 19(42.22) | |
| IV | 11(55) | 21(46.67) | |
| Survival(n=72) | |||
| Up to 3 months | 3(15.79) | 14(26.42) | 0.636 |
| >3 months–6 months | 6(31.58) | 10(18.86) | |
| >6 months–12 months | 7(36.84) | 20(37.74) | |
| >12 months | 3(15.79) | 9(16.98) |
KRAS: Kirsten rat sarcoma virus, TTF: Thyroid transcription factor
Histomorphological analysis
The histomorphological features of the cases that harbored KRAS protein expression versus the KRAS negative cases were analyzed. The majority of cases had mixed histomorphological patterns. In the KRAS positive group, the most common pattern of cellular arrangement was acinar pattern/loose clusters present in 64.29%–75% of cases followed by the solid pattern present in 42.86% of cases. Micropapillary/papillary pattern was least common and was documented in only 10.71% of cases. Nuclear pleomorphism was moderate/severe in 71.43% cases. Necrosis was identified in 57.14% of cases. Mucin (extra-cellular or intra-cellular) was present in 32.14% cases, which was statistically significant (P = 0.036) when compared with the KRAS negative group [Table 2 and Figure 2].
Table 2.
Histomorphological characteristics of KRAS+versus KRAS−cases
| Histomorphological characteristics | KRAS positive(n=28), n(%) | KRAS negative(n=72), n(%) | P |
|---|---|---|---|
| Cellular arrangement | |||
| Solid | |||
| Present | 12(42.86) | 28(38.89) | 0.716 |
| Absent | 16(57.14) | 44(61.11) | |
| Acinar | |||
| Present | 18(64.29) | 36(50) | 0.198 |
| Absent | 10(35.71) | 36(50) | |
| Papillary/micropapillary | |||
| Present | 3(10.71) | 12(16.67) | 0.454 |
| Absent | 25(89.29) | 60(83.33) | |
| Lepidic | |||
| Present | 5(17.86) | 7(9.72) | 0.265 |
| Absent | 23(82.14) | 65(90.28) | |
| Singly scattered/loose clusters | |||
| Present | 21(75) | 61(84.72) | 0.255 |
| Absent | 7(25) | 11(15.28) | |
| Nuclear pleomorphism | |||
| Mild | 8(28.57) | 11(15.28) | 0.128 |
| Moderate orsevere | 20(71.43) | 61(84.72) | |
| Tumor differentiation | |||
| Well | 8(28.57) | 8(11.11) | 0.065 |
| Moderate | 11(39.29) | 43(59.72) | |
| Poor | 9(32.14) | 21(29.16) | |
| Mucin(extracellular or intracytoplasmic) | |||
| Present | 9(32.14) | 10(13.89) | 0.036* |
| Absent | 19(67.86) | 62(86.11) | |
| Necrosis | |||
| Present | 16(57.14) | 29(40.28) | 0.127 |
| Absent | 12(42.86) | 43(59.72) | |
KRAS: Kirsten rat sarcoma virus
Figure 2.
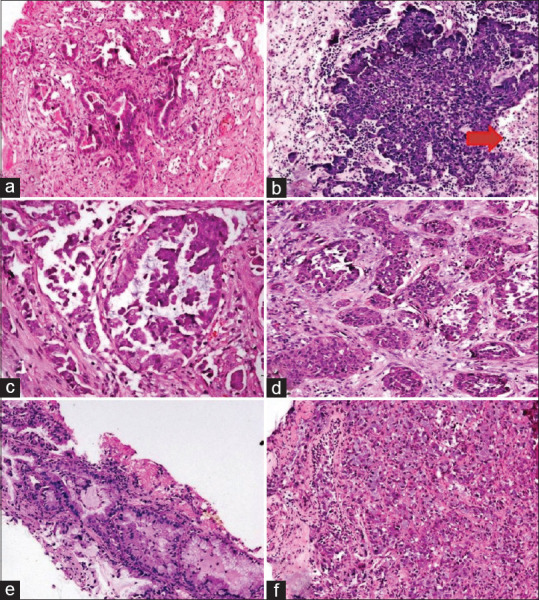
Histomorphological patterns of Kirsten rat sarcoma virus mutant cases: (a) Acinar pattern, (b) Loose clusters with necrosis (red arrow), (c) Micropapillary pattern, (d) Solid nests, (e and f) Presence of intra cytoplasmic mucin (a-f = H and E, ×200)
Survival analysis
The one time follow-up/survival was documented in 72 patients, including 19 KRAS positive and 53 KRAS-negative patients. The estimation of survival functions was performed using the Kaplan-Meier analysis [Figure 3]. A comparison of overall survival between the KRAS positive versus the KRAS negative cases were not significant (P = 0.636). In KRAS positive group, survival was lower as compared to KRAS negative group [Table 3].
Figure 3.
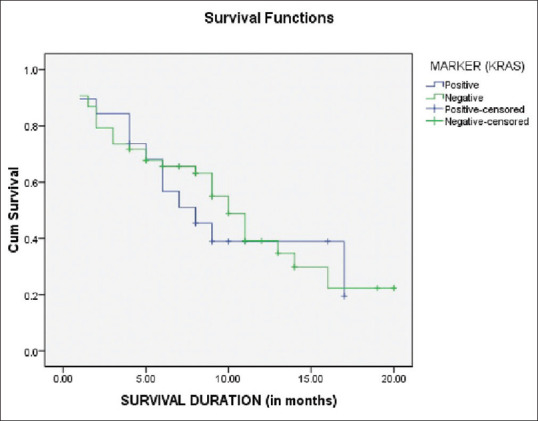
Kaplan–Meier curve for survival time of kirsten rat sarcoma virus positive versus kirsten rat sarcoma virus negative cases
Table 3.
Predictive value of K-RAS expression on mean and median overall survival in patients of non-small cell lung carcinoma-adenocarcinoma
| KRAS over-expression | Mean | Median | ||||||
|---|---|---|---|---|---|---|---|---|
| Estimate | SE | 95% CI | Estimate | SE | 95% CI | |||
| Lower bound | Upper bound | Lower bound | Upper bound | |||||
| Positive | 9.636 | 1.528 | 6.640 | 12.631 | 8.000 | 1.975 | 4.129 | 11.871 |
| Negative | 10.407 | 1.047 | 8.355 | 12.459 | 10.000 | 0.960 | 8.119 | 11.881 |
| Overall | 10.349 | 0.894 | 8.597 | 12.101 | 10.000 | 1.004 | 8.032 | 11.968 |
Estimation is limited to the largest survival time if it is censored. CI: Confidence interval; SE: Standard deviation, KRAS: Kirsten rat sarcoma virus
Discussion
KRAS is an oncogene with predictive and prognostic implications. KRAS is one of the most frequently mutated genes in various solid tumors. The frequency of KRAS mutations in NSCLC in the Western population varies from 11% to 38%.[7,8,9] In the south East Asian region, the prevalence of KRAS mutations has been documented between 3% and 30%. In the current study KRAS protein overexpression was identified in 28% of cases. The frequency documented in this study is higher than those that have been documented in previous studies published from the Indian sub-continent[10,11,12] [Table 4].
Table 4.
The frequency of KRAS mutations in the studies from the Indian sub-continent
| Authors/year of publication | Frequency of KRAS mutations(%) |
|---|---|
| Das etal./2015[10] | 4.5 |
| Bhaumik etal./2016[11] | 6.4 |
| Singh etal./201912 | 19.2 |
| Present study | 28 |
KRAS: Kirsten rat sarcoma virus
In the current study, KRAS mutations were frequent in males with an age >40 years. This finding is in concordance with the results of the study conducted by Singh et al. and contrary to the findings of Das et al., where KRAS mutations were frequent in females.[10,11,12] In the current study, 67.86% of cases were nonsmokers. This finding is contrary to the findings published previously.[10,11,12] This finding may imply that the development of KRAS mutations may not be entirely triggered by alterations induced by carcinogens in smoke. The possible implication of alternative triggers in the KRAS oncogenic pathways may be the plausible cause of the development of tumors in the absence of smoking exposure, although larger studies are warranted to derive conclusive results. Among the studies published from the western population majority of the KRAS mutant cases were males with a history of smoking.[13]
The majority of cases in the current study, in which KRAS protein overexpression was identified, were NSCLC with the adenocarcinoma phenotype. In the study conducted by Das et al., the authors failed to document any specific histological subtype in the KRAS positive sub-group.[10]
TTF-1 was expressed in 89.29% of cases with KRAS protein overexpression. TTF-1 is a major regulatory factor and plays an important role in lung development. TTF-1 expressions are associated with differentiation and in the maintenance of functions of the terminal respiratory unit. The expression of TTF-1 in the majority of cases with KRAS protein overexpression may suggest that tumors with KRAS phenotype are derived from a similar histogenetic pathway.[14,15]
In the current study, the clinical stage was IV in 55% cases amongst the group with KRAS protein overexpression. KRAS is a highly aggressive oncogene. The expression of KRAS in NSCLC causes resistance to the conventional chemotherapy and radiotherapy treatment regimens that are administered in cases of advanced and metastatic tumors. In addition, KRAS mutations also confer resistance to the tyrosine kinase inhibitor therapy that is administered in cases that harbor epidermal growth factor receptor mutations.[16,17,18] In India, the majority of cases are generally diagnosed in advanced stages, where the mainstay of treatment is either targeted therapy of standard chemotherapy or radiotherapy. The presence of KRAS mutations may be a possible justification for nonresponse or poor response to therapy in advanced cases of NSCLC.[10,12]
The histomorphology of the cases with KRAS protein overexpression versus the KRAS negative cases has been inconsistently documented in the literature with rare publications. In the current study, detailed histomorphological analysis has been done. The KRAS mutant cases had acinar/loose clusters pattern arrangement with the presence of necrosis in the majority of the cases. The presence of mucin was statistically significant (P = 0.036) when compared with the KRAS negative group. Among the studies published from the Indian sub-continent, the histomorphology was only analyzed by Singh et al. The authors, however, failed to ascertain any specific pattern or morphology to the KRAS mutant cases.[12,19,20] It appears that KRAS protein is expressed in a fair number of lung adenocarcinoma and needs to be confirmed by molecular studies. The study is limited by being confined to protein expression.
Financial support and sponsorship
The study was funded by Intramural Research Grant (IEC 12/17), Dr. Ram Manohar Lohia Institute of Medical Sciences, Lucknow, India.
Conflicts of interest
There are no conflicts of interest.
Acknowledgment
The authors wish to acknowledge the Department of Biosciences, Integral University, Lucknow for providing MCN NO (IU/R&D/2020-MCN000799), and Ph.D. registration to Mr. Rahul Kumar Pandey.
References
- 1.Travis WD, Brambilla E, Nicholson AG, Yatabe Y, Austin JH, Beasley MB, et al. The 2015 World Health Organization classification of lung tumors: Impact of genetic, clinical and radiologic advances since the 2004 classification. J Thorac Oncol. 2015;10:1243–60. doi: 10.1097/JTO.0000000000000630. [DOI] [PubMed] [Google Scholar]
- 2.Dhillon AS, Hagan S, Rath O, Kolch W. MAP kinase signalling pathways in cancer. Oncogene. 2007;26:3279–90. doi: 10.1038/sj.onc.1210421. [DOI] [PubMed] [Google Scholar]
- 3.Roberts PJ, Der CJ. Targeting the Raf-MEK-ERK mitogen-activated protein kinase cascade for the treatment of cancer. Oncogene. 2007;26:3291–310. doi: 10.1038/sj.onc.1210422. [DOI] [PubMed] [Google Scholar]
- 4.Santarpia L, Lippman SM, El-Naggar AK. Targeting the MAPK-RAS-RAF signaling pathway in cancer therapy. Expert Opin Ther Targets. 2012;16:103–19. doi: 10.1517/14728222.2011.645805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Luca A, Maiello MR, D'Alessio A, Pergameno M, Normanno N. The RAS/RAF/MEK/ERK and the PI3K/AKT signaling pathways: Role in cancer pathogenesis and implications for therapeutic approaches. Expert Opin Ther Target. 2012;16(Suppl 2):S17–27. doi: 10.1517/14728222.2011.639361. [DOI] [PubMed] [Google Scholar]
- 6.Román M, Baraibar I, López I, Nadal E, Rolfo C, Vicent S, et al. KRAS oncogene in non-small cell lung cancer: Clinical perspectives on the treatment of an old target. Mol Cancer. 2018;17:33. doi: 10.1186/s12943-018-0789-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schmid K, Oehl N, Wrba F, Pirker R, Pirker C, Filipits M. EGFR/KRAS/BRAF mutations in primary lung adenocarcinomas and corresponding locoregional lymph node metastases. Clin Cancer Res. 2009;15:4554–60. doi: 10.1158/1078-0432.CCR-09-0089. [DOI] [PubMed] [Google Scholar]
- 8.Serizawa M, Koh Y, Kenmotsu H, Isaka M, Murakami H, Akamatsu H, et al. Assessment of mutational profile of Japanese lung adenocarcinoma patients by multitarget assays: A prospective, single-institute study. Cancer. 2014;120:1471–81. doi: 10.1002/cncr.28604. [DOI] [PubMed] [Google Scholar]
- 9.Arrieta O, Cardona AF, Federico BG, Gallo A, Campos-Parra AD, Serrano S, et al. Genotyping non-small cell lung cancer (NSCLC) in Latin America. J Thorac Oncol. 2011;6:1955–9. doi: 10.1097/JTO.0b013e31822f655f. [DOI] [PubMed] [Google Scholar]
- 10.Das BR, Bhaumik S, Ahmad F, Mandsaurwala A, Satam H. Molecular spectrum of somatic EGFR and KRAS gene mutations in non small cell lung carcinoma: Determination of frequency, distribution pattern and identification of novel variations in Indian patients. Pathol Oncol Res. 2015;21:675–87. doi: 10.1007/s12253-014-9874-7. [DOI] [PubMed] [Google Scholar]
- 11.Bhaumik S, Ahmad F, Das BR. Somatic mutation analysis of KRAS, BRAF, HER2 and PTEN in EGFR mutation-negative non-small cell lung carcinoma: Determination of frequency, distribution pattern and identification of novel deletion in HER2 gene from Indian patients. Med Oncol. 2016;33:117. doi: 10.1007/s12032-016-0828-7. [DOI] [PubMed] [Google Scholar]
- 12.Singh V, Guleria P, Malik PS, Mohan A, Thulkar S, Pandey RM, et al. Epidermal growth factor receptor (EGFR), KRAS, and BRAF mutations in lung adenocarcinomas: A study from India. Curr Probl Cancer. 2019;43:391–401. doi: 10.1016/j.currproblcancer.2018.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tam IY, Chung LP, Suen WS, Wang E, Wong MC, Ho KK, et al. Distinct epidermal growth factor receptor and KRAS mutation patterns in non-small cell lung cancer patients with different tobacco exposure and clinicopathologic features. Clin Cancer Res. 2006;12:1647–53. doi: 10.1158/1078-0432.CCR-05-1981. [DOI] [PubMed] [Google Scholar]
- 14.Stenhouse G, Fyfe N, King G, Chapman A, Kerr KM. Thyroid transcription factor 1 in pulmonary adenocarcinoma. J Clin Pathol. 2004;57:383–7. doi: 10.1136/jcp.2003.007138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Y, Wang R, Li Y, Pan Y, Hu H, Zang Y, et al. Negative thyroid transcription factor 1 expression defines an unfavorable subgroup of lung adenocarcinomas. J Thorac Oncol. 2015;10:1444–50. doi: 10.1097/JTO.0000000000000626. [DOI] [PubMed] [Google Scholar]
- 16.Kakegawa S, Shimizu K, Sugano M, Miyamae Y, Kaira K, Araki T, et al. Clinicopathological features of lung adenocarcinoma with KRAS mutations. Cancer. 2011;117:4257–66. doi: 10.1002/cncr.26010. [DOI] [PubMed] [Google Scholar]
- 17.Jia Y, Jiang T, Li X, Zhao C, Zhang L, Zhao S, et al. Characterization of distinct types of KRAS mutation and its impact on first-line platinum-based chemotherapy in Chinese patients with advanced non-small cell lung cancer. Oncol Lett. 2017;14:6525–32. doi: 10.3892/ol.2017.7016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lim SM, Kim EY, Kim HR, Ali SM, Greenbowe JR, Shim HS, et al. Genomic profiling of lung adenocarcinoma patients reveals therapeutic targets and confers clinical benefit when standard molecular testing is negative. Oncotarget. 2016;7:24172–8. doi: 10.18632/oncotarget.8138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rekhtman N, Ang DC, Riely GJ, Ladanyi M, Moreira AL. KRAS mutations are associated with solid growth pattern and tumor-infiltrating leukocytes in lung adenocarcinoma. Mod Pathol. 2013;26:1307–19. doi: 10.1038/modpathol.2013.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dong YJ, Cai YR, Zhou LJ, Su D, Mu J, Chen XJ, et al. Association between the histological subtype of lung adenocarcinoma, EGFR/KRAS mutation status and the ALK rearrangement according to the novel IASLC/ATS/ERS classification. Oncol Lett. 2016;11:2552–8. doi: 10.3892/ol.2016.4233. [DOI] [PMC free article] [PubMed] [Google Scholar]


